Abstract
The frequent loss of both INK4a and ARF in melanoma raises the question of which INK4a-ARF gene product functions to suppress melanoma genesis in vivo. Moreover, the high incidence of INK4a-ARF inactivation in transformed melanocytes, along with the lack of p53 mutation, implies a cell type-specific role for INK4a-ARF that may not be complemented by other lesions of the RB and p53 pathways. A mouse model of cutaneous melanoma has been generated previously through the combined effects of INK4aΔ2/3 deficiency (null for INK4a and ARF) and melanocyte-specific expression of activated RAS (tyrosinase-driven H-RASV12G, Tyr-RAS). In this study, we made use of this Tyr-RAS allele to determine whether activated RAS can cooperate with p53 loss in melanoma genesis, whether such melanomas are biologically comparable to those arising in INK4aΔ2/3−/− mice, and whether tumor-associated mutations emerge in the p16INK4a-RB pathway in such melanomas. Here, we report that p53 inactivation can cooperate with activated RAS to promote the development of cutaneous melanomas that are clinically indistinguishable from those arisen on the INK4aΔ2/3 null background. Genomewide analysis of RAS-induced p53 mutant melanomas by comparative genomic hybridization and candidate gene surveys revealed alterations of key components governing RB-regulated G1/S transition, including c-Myc, cyclin D1, cdc25a, and p21CIP1. Consistent with the profile of c-Myc dysregulation, the reintroduction of p16INK4a profoundly reduced the growth of Tyr-RAS INK4aΔ2/3−/− tumor cells but had no effect on tumor cells derived from Tyr-RAS p53−/− melanomas. Together, these data validate a role for p53 inactivation in melanomagenesis and suggest that both the RB and p53 pathways function to suppress melanocyte transformation in vivo in the mouse.
Melanocyte-specific H-RASV12G (Tyr-RAS) transgene expression in mice homozygous for the INK4aΔ2/3 mutant allele (null for both INK4a and ARF) generates a melanoma-prone condition (8). Tyr-RAS-induced melanomas arising in INK4aΔ2/3 heterozygotes invariably sustain deletions in the wild-type INK4a allele, and all such deletions cripple both p16INK4a and p19ARF coding sequences (8). Importantly, despite the high incidence of p53 mutations associated with the development of many different cancers, the p53 gene remains intact in these murine melanomas, a genetic profile that appears to hold true for human melanomas as well (see below). Indeed, it was the lack of p53 mutations in these INK4a-ARF-deficient melanomas and in spontaneously immortalized INK4a-ARF-deficient fibroblasts, coupled with high levels of p19ARF in p53 null cells (40), that suggested a genetic link between INK4a-ARF (specifically, p19ARF) and p53 (8, 27). In line with this genetic relationship, a clear biochemical link has been forged between p19ARF (p14ARF in humans) and p53 through the ability of p19ARF to block MDM2-induced degradation of p53 (26, 39, 54, 61). Correspondingly, tumors arising in p53 mutant mice maintain an intact INK4a-ARF locus (27), thus fortifying the view that p19ARF-MDM2-p53 constitutes a tumor suppressor pathway. This concept follows from the paradigm first proposed to explain the reciprocal pattern of INK4a and RB mutations in human cancers (reviewed in reference 44).
Evidence supporting a tumor suppression role for p19ARF is exceedingly clear in the mouse and derives from the cancer-prone phenotype of an ARF-specific knockout (11, 27) and from the antioncogenic effect of p19ARF on Myc and Ela transformation (39). In addition, p19ARF promotes p53-dependent apoptosis in the setting of aberrant cell proliferation brought about by loss of RB function in vivo (39) or by activation of numerous oncoproteins in primary cultured cells (10, 41, 62) and in vivo (11, 48). Evidence for a tumor suppressor role of p14ARF in humans has been mounting but will remain indirect in the absence of germ line ARF-specific mutations in cancer-prone kindreds (50). While the role of p14ARF in human cancer susceptibility remains unclear, the role of p16INK4a as a human tumor suppressor is irrefutable. Most compelling is the presence of germline mutations that compromise p16INK4a but preserve p14ARF function and confer hereditary susceptibility to melanoma and pancreatic adenocarcinoma (44). Curiously, a role of p16INK4a in tumor suppression may not be as prominent in the mouse, since the ARF-specific and INK4aΔ2/3 (null for INK4a and ARF) knockouts show similar phenotypes with respect to cellular immortalization and cancer susceptibility (24, 27, 49). However, a separate line of evidence has raised the possibility that p16INK4a is relevant in some murine cancer types, such as plasmacytoma (60). Together, these species differences imply that while p16INK4a has a critical tumor suppressor function in humans, its role may be less prominent in the mouse, where tumor suppression appears to be dominated by the p19ARF-p53 axis.
Mutation or deletion of p53 has been linked to >55% of all human cancers (17); however, the role of p53 in melanoma remains controversial. Mutational analyses by many groups have shown a very low incidence of point mutation or allelic loss of p53 in surgical specimens of primary and metastatic melanomas (2, 7, 32, 38), while others have estimated the incidence of p53 mutation to be 15 to 25% of primary and metastatic samples (1, 51, 59). Moreover, expression analysis by immunohistochemistry has revealed a significantly higher incidence of p53 overexpression, implying stabilizing point mutations, in metastatic melanomas than in primary melanomas (15), suggesting that loss of p53 function promotes disease progression. In contrast, other groups have reported a lack of correlation between p53 overexpression and stages of melanoma development (12, 43). Indeed, Zerp and colleagues had reported a lower frequency of p53 mutation in metastasis compared with primary melanoma lesions, implying that p53 mutation, although associated with human cutaneous melanoma arising in sun-exposed sites, does not contribute to melanoma pathogenesis and progression (59).
In this report, we sought to validate a role for functional p53 pathway inactivation in the pathogenesis of melanomas. We demonstrated that RAS activation and p53 loss cooperate to generate melanomas that are clinically indistinguishable from those arising on an INK4a-ARF null background. Furthermore, identification of alterations in key components of the RB pathway by comparative genomic hybridization (CGH) and candidate gene surveys supports a role for both the RB and p53 pathways in melanoma suppression in vivo.
MATERIALS AND METHODS
Mouse strains.
Tyrosinase enhancer-promoter-driven H-RASV12G transgenic mice (8) were crossed onto the p53 mutant background (Jackson Laboratory) and the INK4aΔ2/3−/− background (49) and observed for tumor development or apparent ill health. The Tyr-RAS p53 mutant mice analyzed in this study were of mixed genetic background (∼80% C57BL/6, 20% 129Sv) or N1 generation FVB backcross (50% FVB, 40% C57BL/6). The Tyr-RAS INK4aΔ2/3 null mice analyzed were of either mixed genetic background (65% C57BL/6, 25% CBA, 10% 129Sv) or N3 generation FVB backcross (83% FVB). No consistent difference was noted with respect to tumor latency and CGH profiles in either the mixed genetic background or the FVB backcross generation.
Mouse tumor surveillance and characterization.
Mice were observed biweekly for development of tumors or appearance of ill health. Premorbid animals or animals with significant tumor burdens were sacrificed, and detailed autopsies were performed. Tumor specimens were fixed in 10% formalin and embedded in paraffin for histological and immunological analysis as previously described (8). In cases in which sufficient specimens were available, primary tumors were adapted to culture to establish derivative cell lines.
Comparative genomic hybridization.
DNA was extracted from microdissected tumor tissue from paraffin-embedded tumor blocks by standard methods (4). Reference and test DNAs labeled with Alexa 594 dUTP (Molecular Bioprobes) and fluorescein-12-dUTP (NEN), respectively, were hybridized to normal metaphase chromosome spreads; chromosomes were identified by 4′,6′-diamidino-2-phenylindole (DAPI) counterstaining, and green-red fluorescence intensity profiles were obtained as previously described (4). Regions were called amplified if the tumor/reference ratio of a chromosomal arm exceeded 1.5 or if the ratio elevation involved a sharply demarcated segment of a chromosomal segment. For the amplification involving chromosome 15, both criteria were met, but in tumors that had focused amplifications involving chromosome 2 or 12, the amplified chromosomal segment was too small to yield a ratio of >1.5.
DNA and RNA analysis.
DNA was isolated from snap-frozen tumor specimens or from cell lines derived from primary tumors using the Puregene DNA isolation system (Gentra) according to manufacturer's protocol. Loss of heterozygosity (LOH) analysis of the p53 locus was done by allele-specific PCR using oligonucleotide primers directed against the wild-type and knockout p53 alleles (22). The wild-type p53 allele was amplified using primers 5′P53 (5′-ACAGCGTGGTGGTACCTTAT-3′) and 3′P53WT (5′-TATACTCAGAGCCGGCCT-3′), whereas the mutant allele was amplified by using primers 5′P53 and 3′P53KO (5′-CTATCAGGACATAGCGTTGG-3′). PCRs were performed in a 50-μl volume in 1× PCR buffer (Perkin-Elmer) in the presence of 4 μM MgCl2, 0.8 μM deoxynucleoside triphosphate mix, 1.25 U of AmpliTaq DNA polymerase (Perkin-Elmer), 200 ng of 5′P53, 150 ng of 3′P53KO, 75 ng of 3′P53WT, and 250 ng of genomic DNA. Samples were incubated at 94°C for 2 min, followed by 40 cycles of 94°C for 1 min, 62°C for 2 min, and 72°C for 2 min. PCR products were visualized by agarose gel electrophoresis and ethidium bromide staining.
For sequence analysis of the ARF coding sequence, total RNA was isolated from cultured melanoma cell lines using the Trizol reagent (Gibco BRL) according to manufacturer's protocol. A 2-μg RNA sample was used as a template in a reverse transcription reaction using Superscript II polymerase (Gibco BRL) primed with oligo(dT). The coding region of the ARF cDNA was amplified by PCR using oligonucleotide primers p19-1 (5′-GTCACAGTGAGGCCGCCGCTGAGGGA-3′) and p19-2 (5′-CTCTTGGGATTGGCCGCGAAGTTCCA-3′). The PCR product was subjected to direct DNA sequencing in both directions using the same primers as above.
To measure changes in gene copy number, genomic DNA was isolated from both primary tumor samples and derivative cell lines by the Puregene DNA isolation system (Gentra) according to manufacturer's protocol and analyzed by slot blot analysis. Blots were hybridized with random primed cDNA probes, and signals were quantitated by PhosphorImager analysis (Fuji BAS). DNA quantities were normalized to hybridization signals of at least two control probes in genomic regions without CGH-detected alteration. The ratio of normalized hybridization intensities on tumor DNA relative to diploid control DNA allowed copy number designations. The control probes used included a 400-bp BamHI/EcoRI fragment of mTERT (16), a 750-bp fragment of c-Myc exon 2, a 270-bp SalI/SphI fragment of cyclin D1, full-length ID2 from pID2k (55), and a 560-bp fragment of N-Myc exon 3.
Protein analysis.
Cell extracts were prepared from early-passage melanoma cell lines by lysis in radioimmunoprecipitation assay (RIPA) buffer in the presence of protease and phosphatase inhibitors. Lysates were sonicated briefly and clarified by centrifugation. All manipulations of the protein extracts were performed at 4°C. Proteins were quantitated by Bradford assay (Bio-Rad). For immunoprecipitation of p16INK4a complexes, 1 mg of cell extract was precleared by incubation with protein A-Sepharose (Sigma) and preimmune serum and then incubated for 1 h in the presence of anti-p16INK4a antibody M-16 (Santa Cruz). Following addition of protein A-Sepharose, extracts were incubated for an additional 3 h. Precipitated complexes were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred to polyvinylidene difluoride filters. The blots were probed with either the p16INK4a antibody used for immunoprecipitation or anti-CDK4 antibody C22 (Santa Cruz). For other Western blot analyses, 50 μg of cell lysates was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis or by the NuPAGE Bis Tris gel system (Novex) and transferred to polyvinylidene difluoride filters. The antibodies used in this study included the following: for c-Myc, 06-340 (UBI); for p19ARF, AEC40 (39); for RB, 14001A (Pharmingen). The following antibodies were from Santa Cruz Biotechnology: for p16INK4a, M-156; for p15INK4b, M-20; for p27KIP1, C-19; for p21CIP1, M-19; for Cdc25a, F-6; for cyclin D1, HD-11 and 72-13G; for cyclin D2, M-20; for cyclin D3, C-16; for β-catenin, C-18; for CDK6, C-21.
Production of retroviruses and infection of melanoma cell lines.
cDNAs for human p16INK4a and p27KIP1 were cloned into the pBABE puro retrovirus vector. The retrovirus vectors were transfected into the 293GPG packaging cell line (37) using the Lipofectamine 2000 reagent (Gibco BRL). Supernatants containing the retrovirus were filtered and applied to the melanoma cell lines in the presence of 4 μg of Polybrene per ml. Transduced cells were selected 24 h postinfection using 2.5 μg of puromycin per ml. Efficiency of selection was monitored by puromycin treatment of nontransduced cells. Following 48 h of selection, the selected cells were assayed for growth rates and colony formation after low-density seeding. Expression of the exogenous proteins was confirmed by Western blot analyses. Proliferation was assessed by seeding 8,000 cells/well in 12-well plates, followed by counting of viable cells in duplicate on consecutive days. For colony formation, 2,000 transduced cells were seeded on 10-cm-diameter dishes. After approximately 2 weeks, colonies were counted following trypan blue staining.
RESULTS
Loss of p53 can cooperate with activated H-RASV12G to promote melanoma development.
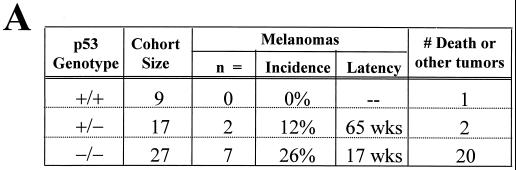
In our previous study, over a period of greater than 1 year of observation, only 1 of 49 Tyr-RAS INK4a+/+ p53+/+ mice developed melanoma that sustained a homozygous deletion of the INK4a locus (8). In contrast, Tyr-RAS mice of similar genetic background and harboring mutant p53 alleles readily developed melanomas, i.e., 2 of 17 Tyr-RAS p53+/− mice and 7 of 27 Tyr-RAS p53−/− mice, with average latencies of 65 and 17 weeks, respectively (Fig. 1A). As expected, additional tumor types (sarcoma and lymphoma) known to be associated with germline p53 mutation were observed and their early onset (average latency, 17 weeks) likely masked the development of additional melanomas in the p53-null cohort.
FIG. 1.
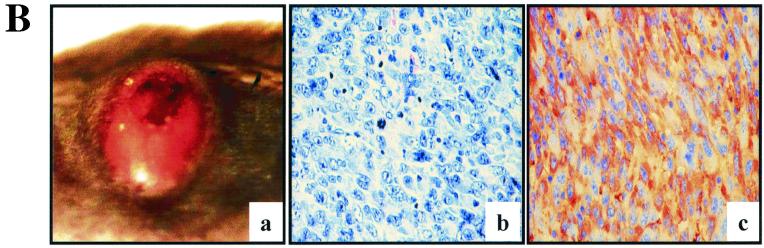
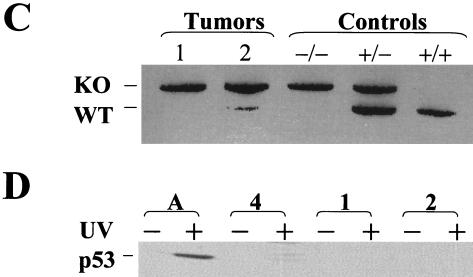
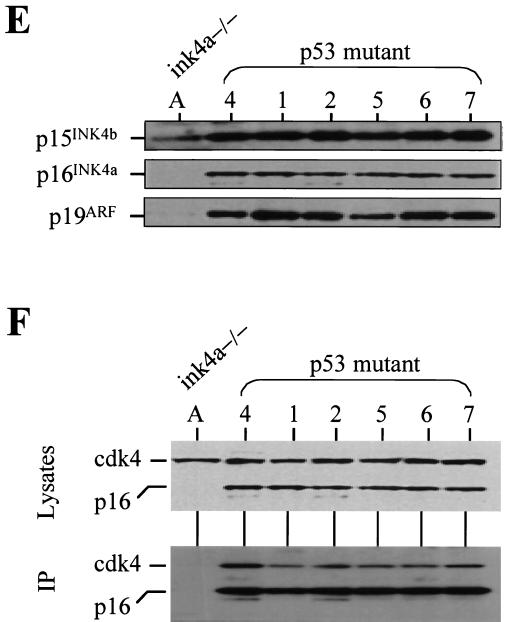
p53 deficiency and oncogenic RAS expression cooperate to induce melanoma. (A) Summary of tumor incidence in Tyr-RAS mice in relation to p53 status. (B) Part a, photograph of a nonpigmented cutaneous melanoma arising on the flank of animal 3. Part b, hematoxylin-and-eosin-stained tumor from animal 3 displaying nuclear pleomorphism and hyperchromasia. Part c, TRP1 immunopositivity demonstrating the melanocytic origin of the tumor. Histology and immunohistochemistry were performed as previously described (8). (C) LOH of p53 in primary melanoma specimens arising in Tyr-RAS p53+/− mice. DNA was isolated from melanomas arising in Tyr-RAS p53+/− mice (mice 1 and 2) was analyzed by multiplex PCR using primers specific for the wild-type and mutant p53 alleles. Allelotyping of normal DNA from mice of all three genotypes (p53+/+, p53+/−, and p53−/−) are presented as controls. The bands corresponding to the wild-type (WT) and knockout (KO) p53 alleles are indicated. (D) Immunoblot analysis of p53 in lysates from tumor cell lines that were either untreated (minus sign) or exposed to UV radiation at 100 J/m2 (plus sign). Cells were harvested 6 h following irradiation. Tumor A, a melanoma cell line arising in a Tyr-RAS INK4aΔ2/3−/− mouse, retains p53 function, while no p53 is induced in melanoma cell lines from Tyr-RAS p53+/− mice (mice 1 and 2). Tumor 4 is derived from a Tyr-RAS p53−/− mouse. (E) Immunoblot analyses of cell lysates from early-passage melanoma cell lines probed with specific antisera show that Tyr-RAS p53−/− melanomas retain expression of p15INK4b, p16INK4a, and p19ARF. (F) Coimmunoprecipitation analysis of melanoma cell lysates using an anti-p16INK4a antibody demonstrates that the p16INK4a expressed in the Tyr-RAS p53−/− melanomas is capable of binding to CDK4. At the top is an immunoblot analysis of melanoma cell lysates probed with antibodies to CDK4 and p16INK4A. At the bottom is an immunoblot of complexes immunoprecipitated (IP) with an anti-p16INK4a antibody and probed with antibodies to CDK4 and p16INK4a.
On the p53 mutant background, Tyr-RAS transgenic animals developed melanomas that were primarily cutaneous (Fig. 1B, a), with one exception that was ocular in origin (tumor 9). Compared to those arising in Tyr-RAS INK4aΔ2/3−/− mice, the RAS-induced p53 mutant melanomas were similarly amelanotic, highly vascular, and locally invasive but not metastatic (8). Microscopically, these dermal tumors were composed of highly pleomorphic, anaplastic cells with characteristic vacuolated nuclei (Fig. 1B, b). The melanocytic origin was confirmed by strong immunoreactivity to the melanocyte-specific marker tyrosinase-related protein 1 (TRP1) (Fig. 1B, c). Together, these data suggest that p53 mutation can cooperate with activated H-RASV12G to promote development of nonmetastatic melanomas that are clinically and histologically similar to those observed in Tyr-RAS INK4aΔ2/3−/− mice.
In the two cutaneous melanomas derived from Tyr-RAS p53+/− mice, allele-specific PCR revealed reduction to homozygosity for p53 (Fig. 1C). That the wild-type p53 allele was indeed lost was confirmed further by the lack of p53 stabilization following UVB irradiation of early-passage cell lines derived from these primary tumors, in contrast to the intact p53 response to UVB in Tyr-RAS INK4aΔ2/3−/− tumor cell lines (Fig. 1D). These findings, coupled with the decrease in melanoma latency observed on the p53−/− background (Fig. 1A), support a causal role for p53 loss in the genesis of melanoma in this Tyr-RAS model.
Status of INK4a-ARF in melanomas arising in Tyr-RAS p53 mutant mice.
The development of melanoma in p53 mutant mice provides an opportunity to assess whether loss of INK4a-ARF, particularly p16INK4a, is essential for melanomagenesis in mice. In human melanoma, p16INK4a function can be compromised on one of several levels, including deletion or point mutations of the ankyrin repeats of p16INK4a (44), a domain required for interaction with CDK4 and -6 (45), germ line CDK4 mutations that disrupt binding to p16INK4a (57), or cyclin D1 amplifications (20). Here, Western and Southern blot analyses of these mouse melanomas failed to detect gross deletion-rearrangement of INK4a-ARF sequences or a decrease in the levels of all three gene products encoded by this locus: p15INK4b, p16INK4a, and p19ARF (Fig. 1E; Southern blot not shown). The robust p19ARF expression displayed by the p53 mutant melanomas is consistent with the loss of a p53-mediated feedback loop in these tumors (27, 54). The functional integrity of the p16INK4a and CDK4 proteins was also substantiated by their mutual coimmunoprecipitation (Fig. 1F). Finally, the p19ARF transcripts from these melanomas were reverse transcription-PCR amplified for direct sequence analysis and found to be free of point mutations (data not shown). Thus, the products of the INK4a-ARF locus are expressed and remain structurally intact in melanomas arising in p53 mutant mice.
Comparative genomic hybridization of RAS-induced melanomas on INK4aΔ2/3 and p53 mutant backgrounds.
The lack of INK4a mutations in Tyr-RAS p53 mutant melanomas does not exclude the presence of mutations that target other components governing exit from the G1 phase of the cell cycle. CGH, a technique that permits analysis of the entire tumor genome for alterations in DNA copy number of chromosomal regions (23), was employed as a broad genomic screen to search for tumor-associated changes, particularly those involving loci encoding components that govern the G1/S transition. A total of 19 Tyr-RAS INK4aΔ2/3−/− and 9 Tyr-RAS p53 mutant melanomas were analyzed (Fig. 2 and Table 1).
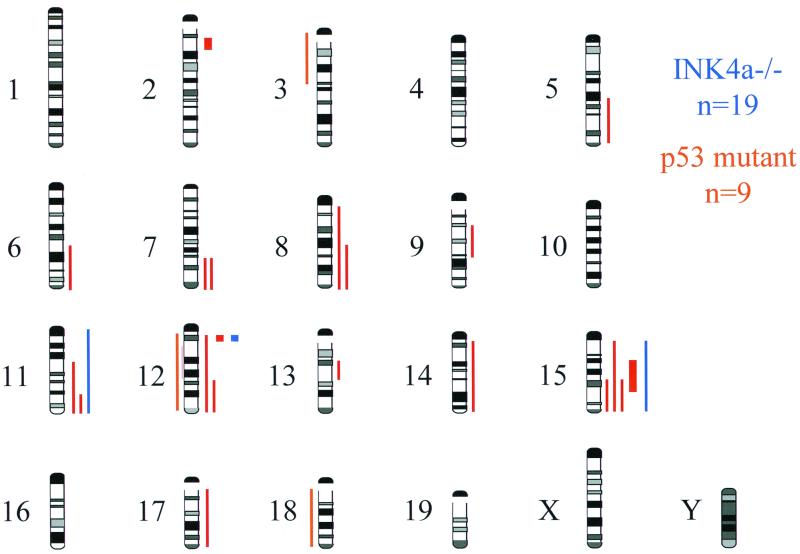
FIG. 2.
Chromosomal locations of DNA sequence copy number alterations detected by CGH in RAS-induced melanomas from 9 p53 mutant mice (red) and 19 INK4a-ARF mutant mice (blue). Gains are indicated by lines to the right of the chromosome ideograms, and losses are indicated by lines to the left. Amplifications are indicated by thick lines. A highly amplified focal region at chromosome 12A3 was detected in one Tyr-RAS p53−/− melanoma and one Tyr-RAS INK4a−/− melanoma. The N-myc gene is located in the proximity of these amplicons but was present in normal copy number (data not shown).
TABLE 1.
Summary of chromosomal changes detected by CGH in melanomas in relation to p53 and INK4a-ARF genotypea
| Tumor no. | p53 status | INK4a status | CGH-detected genomic event(s) |
|---|---|---|---|
| 1 | +/− | enh (6D-G, 7F, 9B-D, 11B-E, 15D3-F), dim (3A-E), amp (2B) | |
| 2 | +/− | enh (5E-G, 13A5-B), amp (15B3-E) | |
| 3 | −/− | None | |
| 4 | −/− | enh (7F, 8C-E, 11C-E, 13A5-B, 14, 15D3-F, 17) dim (12, 18) | |
| 5 | −/− | None | |
| 6 | −/− | amp (12A3) | |
| 7 | −/− | None | |
| 8 | −/− | enh (12D-F, 15) | |
| 9 | −/− | enh (8, 12) | |
| A | −/− | None | |
| A2 | −/− | None | |
| A3 | −/− | None | |
| A4 | −/− | enh (15) | |
| A5 | −/− | None | |
| A6 | −/− | None | |
| A7 | −/− | None | |
| A8 | −/− | None | |
| A9 | −/− | None | |
| A10 | −/− | None | |
| A11 | −/− | None | |
| A12 | −/− | None | |
| A13 | −/− | None | |
| A14 | −/− | None | |
| A15 | −/− | None | |
| A16 | −/− | amp (12A3) | |
| A17 | −/− | None | |
| A18 | −/− | None | |
| A19 | −/− | enh (11) |
Losses (dim), gains (enh), and amplifications (amp) are indicated (see Materials and Methods).
The p53 mutant melanomas possessed a greater degree of chromosomal gains and losses compared with the more euploid profile of the INK4aΔ2/3−/− melanomas (2.7 CGH-detected genomic events per tumor versus 0.16 event per tumor, respectively; P < 0.001; Fig. 2 and Table 1). The less aneuploid profile of INK4aΔ2/3−/−, relative to p53 mutant, melanomas is reminiscent of the benign cytogenetic profiles observed in a murine ARF−/− lymphoma model (48) and in ARF−/− fibroblasts (27). These findings are consistent with the concept that p19ARF is dispensable for p53-dependent control of genomic stability (25).
Overexpression of c-Myc in p53 mutant melanomas.
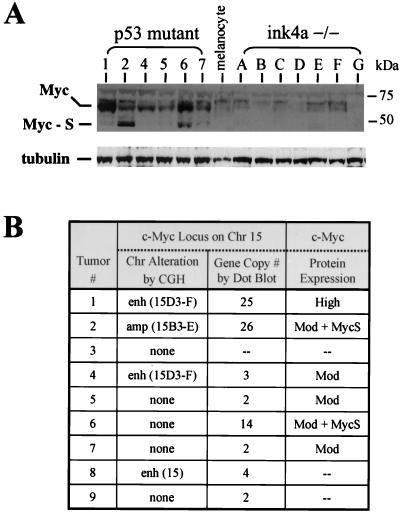
The most common chromosomal abnormality detected by CGH analysis in the p53 mutant melanomas was gain of chromosome 15 (four of nine mice; Table 1). Collectively, these alterations overlap in the central portion of chromosome 15, a region that encodes the c-Myc oncoprotein, a critical regulator of cellular proliferation (reviewed in reference 9). In dot blot analyses of genomic DNA isolated from primary tumors or their derivative cell lines, hybridization to c-Myc confirmed an increase in c-Myc gene copy number in these p53 mutant melanomas (Fig. 3B), relative to the signal of two internal control probes (see Materials and Methods). These conventional dot blot assays also revealed one additional tumor (no. 6) whose c-Myc gene dosage increase eluded detection by CGH (Fig. 3B), suggesting the presence of more focal gain. These five tumors possessed 1 to 24 extra copies of the c-Myc gene, as determined by quantitation of hybridization intensity (Fig. 3B; data not shown). Furthermore, by Western blot analysis, robust c-Myc protein expression can be detected in all six derivative cell lines of Tyr-RAS p53 mutant melanomas (Fig. 3A and B), at levels that greatly exceed that observed in Tyr-RAS INK4aΔ2/3−/− samples. The presence of Myc-S, measuring approximately 46 kDa, in tumors 2 and 6 is notable given that this N-terminally truncated c-Myc isoform is oncogenically -competent and often detected in transformed cells (52, 58).
FIG. 3.
Tyr-RAS p53−/− melanomas show elevated c-Myc expression and frequent amplification of the c-Myc locus. (A) Top, immunoblot showing that c-Myc expression is strongly elevated in p53 mutant melanoma cells relative to melanocytes (M) and INK4-ARF mutant melanomas (A to G). Note the expression of the ∼46-kDa short c-Myc isoform (Myc-S) in tumors 2 and 6. The migration of the molecular size markers is indicated to the right. Bottom, immunoblot showing expression of tubulin as a loading control. (B) Summary of genomic alterations at the c-Myc locus in p53 mutant melanomas. Hybridization analysis of melanoma DNA using c-Myc and control probes was used to determine the c-Myc gene copy number (see Materials and Methods). Note that tumor 6 harbored an amplification of c-Myc which was not detected by CGH. c-Myc expression levels (see panel A) are summarized in the rightmost column (Mod, moderate increase in expression). The expression of Myc-S is also indicated. Chr, chromosome.
That c-Myc protein expression was found to be elevated in all Tyr-RAS p53 mutant melanomas, regardless of c-Myc gene amplification status, indicates that mechanisms other than increased gene dosage also contribute to deregulated c-Myc expression. Previous cell culture-based studies have demonstrated that high levels of p53 can negatively regulate c-Myc expression (30, 42), raising the possibility that loss of p53-dependent repression accounts for elevated c-Myc expression in Tyr-RAS p53 mutant tumors. On the other hand, Tyr-RAS INK4aΔ2/3−/− melanomas display low levels of c-Myc (Fig. 3A) despite having undetectable levels of p53 (Fig. 1D, sample A, and data not shown), arguing against the existence of p53-mediated repression of c-Myc in transformed melanocytes. The up-regulation of c-Myc expression in all of the tumors tested, together with amplification of the c-Myc locus in a majority of tumors, is consistent with c-Myc dysregulation being an acquired oncogenic event in p53 mutant melanomas. These observations gain added significance in light of studies showing that c-Myc can bypass the G1 block conferred by p16INK4a due, in part, to c-Myc's ability to regulate G1 molecules operating parallel to and downstream from p16INK4a (reviewed in reference 9). The overexpression of Myc may also contribute to the karyotypic abnormalities detected in the p53 mutant tumors (13).
Alterations in expression of G1 regulators in p53 mutant melanomas.
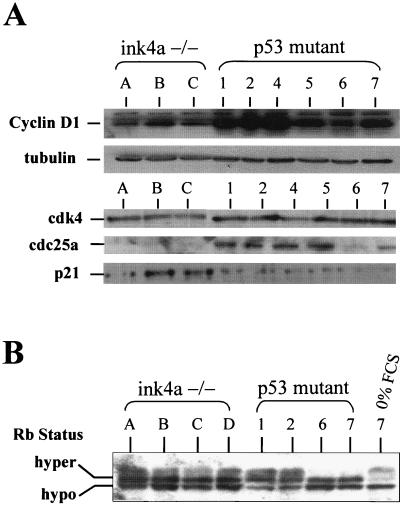
Gains in the distal region of chromosome 7 were detected by CGH in two of nine Tyr-RAS p53−/− melanomas examined (Fig. 2). This region encodes cyclin D1, a G1 cyclin that activates CDK4 and -6 kinase activity that, in turn, phosphorylates and inactivates RB. All p53 mutant melanomas were shown to express elevated cyclin D1 relative to those present in all of the Tyr-RAS INK4aΔ2/3−/− melanomas examined (Fig. 4A). Cyclins D2 and D3 were expressed at modest levels in both INK4aΔ2/3−/− and p53 mutant samples (data not shown). Given the equivalent levels of CDK4 and CDK6 kinase activity in all of the samples tested (data not shown), it is tempting to speculate that the increased cyclin D1 expression associated with loss of p53 is a functional equivalent to p16INK4a loss in the Tyr-RAS INK4aΔ2/3−/− melanomas.
FIG. 4.
Expression analysis of G1/S regulators in RAS-induced melanomas. (A) Top, immunoblot analysis of cyclin D1 levels in melanoma cell lysates from INK4a-ARF−/− and p53−/− tumors. Below is an immunoblot probed with α-tubulin as a loading control. Bottom, immunoblot of p21CIP1, Cdc25a, and CDK4 levels. (B) Western blot analysis of RB phosphorylation status in INK4aΔ2/3-and p53-null melanomas. Tumor 7 was grown in 0% fetal calf serum (FCS; rightmost lane) to show that RB is responsive by shifting to a hypophosphorylated state.
These CGH data prompted expression analysis of factors playing prominent roles in regulating G1 exit, including p21CIP1, p27KIP1, and Cdc25a. Levels of the general CDK inhibitor p21CIP1 were reduced in the p53 mutant tumors relative to those in INK4aΔ2/3−/− melanomas (Fig. 4A). This low level of p21CIP1 expression may act to enhance the assembly of active CDK4-D1 complexes (28). The Cdc25a phosphatase, a rate-limiting activator of the CDK2-cyclin E complex, showed higher levels of expression in all p53 mutant melanomas (Fig. 4A). Since Cdc25a can collaborate with activated RAS to effect cellular transformation and has been reported to be transcriptionally regulated by c-Myc (14, 47), it is possible that enhanced Cdc25a expression reflects elevated c-Myc activity in these p53 mutant tumors. Alternatively, since the levels of c-Myc and Cdc25a are not tightly correlated in the p53 mutant tumors, the increased Cdc25a expression may represent an independently acquired event that further facilitates G1 exit in melanomas with intact p16INK4a function.
RB-mediated G1/S transition is dysregulated in p53 mutant melanomas.
The finding of c-Myc and cyclin D1 overexpression, coupled with genetic and functional data linking c-Myc to the G1 cell cycle machinery, suggests that c-Myc and cyclin D1 dysregulation provides an alternative route to RB inactivation, a route that is functionally equivalent to p16INK4a loss in melanoma. Along these lines, the INK4aΔ2/3 -null and p53 mutant tumors have similar overall RB and E2F activity profiles. Specifically, Western blot analysis showed no differences in RB phosphorylation status between p53 mutant and INK4aΔ2/3 null melanomas during exponential growth under high- and low-serum conditions or during of confluence (Fig. 4B and data not shown). Correspondingly, E2F transcriptional activity levels assessed by transfection of an E2F reporter construct revealed highly variable, yet overlapping, trends between these p53 mutant and INK4aΔ2/3 null tumors (data not shown). Together, these data imply that dysregulation of the G1/S transition—either by p16INK4a loss in an INK4aΔ2/3-null background or by upregulation of c-Myc, cyclin D1, and/or Cdc25a in a p53 mutant background—is required for melanomagenesis.
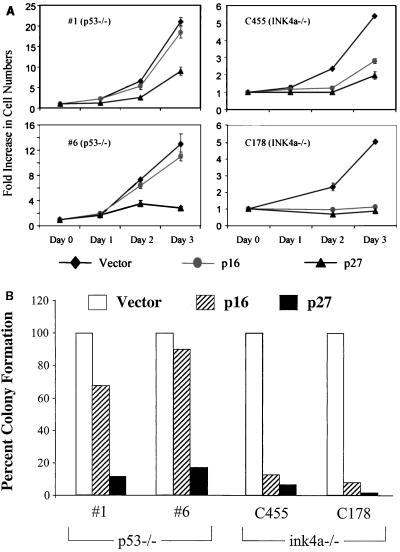
To further substantiate that overcoming RB-mediated G1 arrest is important in the growth of mouse melanomas and to provide evidence that the RAS-induced p53 mutant melanomas have acquired lesions that bypass this cell cycle block, we assessed the effect of p16INK4a and p27KIP1 expression on growth and low-density colony formation in four independently derived INK4aΔ2/3 null and three independently derived p53 mutant melanoma cell lines. These cell lines were transduced with retroviruses encoding the empty vector, p16INK4a, or p27KIP1. Levels of protein expression were comparable among the cell lines, as determined by Western blot analysis (data not shown). In RAS-induced INK4aΔ2/3 null melanoma cells, the expression of p16INK4a or p27KIP1 strongly inhibited growth and colony formation, relative to those transduced with the empty vector (Fig. 5A and B and data not shown). In contrast, p16INK4a did not substantially affect the growth and colony formation of the RAS-induced p53 mutant melanoma cell lines, although these cells were was strongly growth inhibited by p27KIP1 expression (Fig. 5A and B and data not shown). Given the high levels of c-Myc in the Tyr-RAS p53 mutant melanoma cell lines, these observations are in accord with several lines of evidence positioning the actions of c-Myc downstream of p16INK4a and at the level of the cyclin E-CDK2 complex (9). It is also possible that these differential responses are dictated, in part, by differences in the levels of Cdc25a and/or p21CIP1.
FIG. 5.
Effects of exogenous p16INK4a and p27KIP1 on the growth of Tyr-RAS melanoma cells on an INK4aΔ2/3-null or p53 mutant background. (A) Melanoma cell populations transduced with the indicated retroviruses were selected with puromycin for 2 days and then assayed for proliferation rates (see Materials and Methods). Data from two representative cell lines from each genotype are shown. Error bars indicate ranges of variability in duplicate samples assayed. (B) The relative colony-forming ability of the transduced cell lines was determined following low-density seeding (see Materials and Methods). The graph plots the number of colonies as a ratio compared to the number of colonies seen for transduction with the empty vector.
DISCUSSION
In summary, while p53 deficiency cooperates with oncogenic RAS to confer susceptibility to melanoma development, collateral somatic alterations in components known to impinge upon the RB-regulated G1/S transition are acquired to facilitate G1 exit. Oncogenic RAS contributes to malignant growth on a number of levels, including alteration of cell motility, cell survival, cell growth, and direct signaling to the G1 cell cycle machinery (reviewed in reference in 33). With respect to the cell cycle, RAS has been shown to increase assembly of active CDK4-cyclin D complexes. Sustained RAS expression can also induce antiproliferative signals, including up-regulation of p16INK4a and p21CIP1 (31). These consequences of RAS activation must be abrogated if a cell is to progress to malignancy. Since the p53 mutant melanomas sustain alterations in components regulating G1 exit, it follows that the proliferative signals induced by RAS are insufficient to drive malignant cell proliferation. Indeed, the frequent concurrence of RAS mutations and RB pathway defects in human cancers (34, 46) suggests that the oncogenic actions of RAS extend beyond the regulation of the RB restriction point.
The integral role of c-Myc in driving cellular proliferation is demonstrated by its ability to stimulate S-phase entry and shorten the cell cycle. c-Myc triggers G1 exit by both promoting an increase in cell mass (21) and modulating expression of genes that control the cell cycle (9). Most of these c-Myc gene targets regulate the activity of G1 CDKs. c-Myc expression in quiescent cells leads to a rapid induction of cyclin E-CDK2 kinase activity (53), whereas the expression of dominant-negative c-Myc or somatic deletion of c-Myc suppresses cyclin E-CDK2 activity (5, 35). c-Myc also activates the expression of cyclins D1 and D2 (6, 19) and CDK4 (18), leading to type D cyclin sequestration of p27KIP1 from CDK2 complexes. CDK2 activity is also promoted by the ability of c-Myc to repress the expression of p27KIP1 (56) and to induce expression of the CDK2 activator Cdc25a (14). Collectively, these genetic and functional data support a model in which c-Myc stimulates transition through G1/S (Fig. 6). Expression of either c-Myc or cyclin E allows cells to bypass p16INK4a-induced growth arrest, and it is likely that cyclin E-CDK2 is the key functional target of Myc. The resistance of the p53-null melanomas to growth inhibition by 16INK4a is consistent with a role for c-Myc in inactivating the RB-mediated restriction point. These results are also analogous to previous studies showing that p16INK4a functions upstream of Myc (3). However, definitive proof that c-Myc amplification or overexpression indeed plays a causal role in the genesis of RAS-induced p53 mutant melanomas requires further genetic studies of the type reported here for p53 and previously for RAS and INK4a (8). The studies described here establish that inactivation of the p53 pathway can play a causal role in the pathogenesis of melanoma. This, coupled with the functional link between p19ARF and p53 and the fact that INK4a-ARF loss typically occurs early in the development of human melanomas provides a rational explanation for the rare involvement of direct p53 mutations in this cancer type—presumably reflecting a diminished need for p53 inactivation in the face of ARF loss. Furthermore, the consistent finding of dysregulation of components governing G1 exit in these p53 mutant melanomas supports the dual importance of both the RB and p53 pathways in melanoma suppression in vivo. Finally, these results should motivate detailed analyses of c-Myc, cyclin D1, and cdc25a expression in human melanomas harboring p53 mutations, thereby providing additional targets for rational therapeutic intervention.
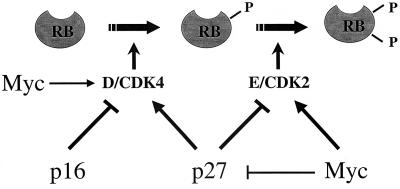
FIG. 6.
c-Myc and G1/S transition. The schematics show sequential phosphorylation of RB during the G1/S transition. RB phosphorylation is initiated by the activity of CDK4-cyclin D complexes and maintained by that of CDK2-cyclin E complexes. p16INK4a negatively regulates the activity of CDK4, and p27KIP1 inhibits the activity of cyclin E-CDK2, although it is also required for assembly of the active CDK4-cyclin D complex. c-Myc can activate the expression of cyclins D1 and D2 (6, 19) and CDK4 (18), leading to type cyclin sequestration of p27KIP1 from CDK2 complexes. CDK2 activity is also promoted by the abilities of c-Myc to repress the expression of p27KIP1 (29, 56), to downregulate p27KIP1 indirectly via upregulation of Cul1 (the SCF complex responsible for its degradation by ubiquitination [36]), and to induce expression of the CDK2 activator Cdc25a (14).
ACKNOWLEDGMENTS
We are grateful to Charles Sherr, Martine Roussel, Steven Artandi, Norman Sharpless, Matthew Meyerson, and Kornelia Polyak for critical reading of the manuscript and to Gregory David and Jim DeCaprio for helpful suggestions during the course of this work. We also thank Susan Charzan for excellent technical assistance.
This work was supported by grants from the NIH (K08AR02104-01) and the NCI (U01CA84313-01). N.B. is supported by the American Cancer Society-John Peter Hoffman Postdoctoral Fellowship. B.C.B. was supported by the Marvin and Roma Auerback Melanoma Research Fund. A.H. is an HHMI Medical Student Research Fellow. R.A.D. is an American Cancer Society Research Professor and a Steven and Michele Kirsch Foundation Investigator. L.C. is a V Foundation Scholar. Support from the DFCI Cancer Core grant to R.A.D. and L.C. is acknowledged. D.P., R.A.D., and L.C. are members of the NCI Mouse Models of Human Cancer Consortium.
REFERENCES
- 1.Akslen L A, Monstad S E, Larsen B, Straume O, Ogreid D. Frequent mutations of the p53 gene in cutaneous melanoma of the nodular type. Int J Cancer. 1998;79:91–95. doi: 10.1002/(sici)1097-0215(19980220)79:1<91::aid-ijc17>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 2.Albino A P, Vidal M J, McNutt N S, Shea C R, Prieto V G, Nanus D M, Palmer J M, Hayward N K. Mutation and expression of the p53 gene in human malignant melanoma. Melanoma Res. 1994;4:35–45. doi: 10.1097/00008390-199402000-00006. [DOI] [PubMed] [Google Scholar]
- 3.Alevizopoulos K, Vlach J, Hennecke S, Amati B. Cyclin E and c-Myc promote cell proliferation in the presence of p16INK4a and of hypophosphorylated retinoblastoma family proteins. EMBO J. 1997;16:5322–5333. doi: 10.1093/emboj/16.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bastian B C, LeBoit P E, Hamm H, Brocker E B, Pinkel D. Chromosomal gains and losses in primary cutaneous melanomas detected by comparative genomic hybridization. Cancer Res. 1998;58:2170–2175. [PubMed] [Google Scholar]
- 5.Berns K, Hijmans E M, Bernards R. Repression of c-Myc responsive genes in cycling cells causes G1 arrest through reduction of cyclin E/CDK2 kinase activity. Oncogene. 1997;15:1347–1356. doi: 10.1038/sj.onc.1201280. [DOI] [PubMed] [Google Scholar]
- 6.Bouchard C, Thieke K, Maier A, Saffrich R, Hanley-Hyde J, Ansorge W, Reed S, Sicinski P, Bartek J, Eilers M. Direct induction of cyclin D2 by Myc contributes to cell cycle progression and sequestration of p27. EMBO J. 1999;18:5321–5333. doi: 10.1093/emboj/18.19.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castresana J S, Rubio M P, Vazquez J J, Idoate M, Sober A J, Seizinger B R, Barnhill R L. Lack of allelic deletion and point mutation as mechanisms of p53 activation in human malignant melanoma. Int J Cancer. 1993;55:562–565. doi: 10.1002/ijc.2910550407. [DOI] [PubMed] [Google Scholar]
- 8.Chin L, Pomerantz J, Polsky D, Jacobson M, Cohen C, Cordon-Cardo C, Horner J W, 2nd, DePinho R A. Cooperative effects of INK4a and ras in melanoma susceptibility in vivo. Genes Dev. 1997;11:2822–2834. doi: 10.1101/gad.11.21.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dang C V. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Bio. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Stanchina E, McCurrach M E, Zindy F, Shieh S Y, Ferbeyre G, Samuelson A V, Prives C, Roussel M F, Sherr C J, Lowe S W. E1A signaling to p53 involves the p19(ARF) tumor suppressor. Genes Dev. 1998;12:2434–2442. doi: 10.1101/gad.12.15.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eischen C M, Weber J D, Roussel M F, Sherr C J, Cleveland J L. Disruption of the ARF-Mdm2–p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 1999;13:2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Essner R, Kuo C T, Wang H, Wen D R, Turner R R, Nguyen T, Hoon D S. Prognostic implications of p53 overexpression in cutaneous melanoma from sun-exposed and nonexposed sites. Cancer. 1998;82:309–316. doi: 10.1002/(sici)1097-0142(19980115)82:2<317::aid-cncr10>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 13.Felsher D W, Bishop J M. Transient excess of MYC activity can elicit genomic instability and tumorigenesis. Proc Natl Acad Sci USA. 1999;96:3940–4. doi: 10.1073/pnas.96.7.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galaktionov K, Chen X, Beach D. Cdc25 cell-cycle phosphatase as a target of c-myc. Nature. 1996;382:511–517. doi: 10.1038/382511a0. [DOI] [PubMed] [Google Scholar]
- 15.Grant S W, Kyshtoobayeva A S, Kurosaki T, Jakowatz J, Fruehauf J P. Mutant p53 correlates with reduced expression of thrombospondin-1, increased angiogenesis, and metastatic progression in melanoma. Cancer Detect Prev. 1998;22:185–194. doi: 10.1046/j.1525-1500.1998.0oa18.x. [DOI] [PubMed] [Google Scholar]
- 16.Greenberg R A, Allsopp R C, Chin L, Morin G B, DePinho R A. Expression of mouse telomerase reverse transcriptase during development, differentiation and proliferation. Oncogene. 1998;16:1723–1730. doi: 10.1038/sj.onc.1201933. [DOI] [PubMed] [Google Scholar]
- 17.Greenblatt M S, Bennett W P, Hollstein M, Harris C C. Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res. 1994;54:4855–4878. [PubMed] [Google Scholar]
- 18.Hermeking H, Rago C, Schuhmacher M, Li Q, Barrett J F, Obaya A J, O'Connell B C, Mateyak M K, Tam W, Kohlhuber F, Dang C V, Sedivy J M, Eick D, Vogelstein B, Kinzler K W. Identification of CDK4 as a target of c-MYC. Proc Natl Acad Sci USA. 2000;97:2229–2234. doi: 10.1073/pnas.050586197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoang A T, Cohen K J, Barrett J F, Bergstrom D A, Dang C V. Participation of cyclin A in Myc-induced apoptosis. Proc Natl Acad Sci USA. 1994;91:6875–6879. doi: 10.1073/pnas.91.15.6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hosokawa Y, Arnold A. Cyclin D1/PRAD1 as a central target in oncogenesis. J Lab Clin Med. 1996;127:246–252. doi: 10.1016/s0022-2143(96)90092-x. [DOI] [PubMed] [Google Scholar]
- 21.Iritani B M, Eisenman R N. c-Myc enhances protein synthesis and cell size during B lymphocyte development. Proc Natl Acad Sci USA. 1999;96:13180–13185. doi: 10.1073/pnas.96.23.13180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacks T, Remington L, Williams B O, Schmitt E M, Halachmi S, Bronson R T, Weinberg R A. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 23.Kallioniemi A, Kallioniemi O P, Sudar D, Rutovitz D, Gray J W, Waldman F, Pinkel D. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- 24.Kamijo T, Bodner S, van de Kamp E, Randle D H, Sherr C J. Tumor spectrum in ARF-deficient mice. Cancer Res. 1999;59:2217–2222. [PubMed] [Google Scholar]
- 25.Kamijo T, van de Kamp E, Chong M J, Zindy F, Diehl J A, Sherr C J, McKinnon P J. Loss of the ARF tumor suppressor reverses premature replicative arrest but not radiation hypersensitivity arising from disabled atm function. Cancer Res. 1999;59:2464–2469. [PubMed] [Google Scholar]
- 26.Kamijo T, Weber J D, Zambetti G, Zindy F, Roussel M F, Sherr C J. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc Natl Acad Sci USA. 1998;95:8292–8297. doi: 10.1073/pnas.95.14.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamijo T, Zindy F, Roussel M F, Quelle D E, Downing J R, Ashmun R A, Grosveld G, Sherr C J. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 28.LaBaer J, Garrett M D, Stevenson L F, Slingerland J M, Sandhu C, Chou H S, Fattaey A, Harlow E. New functional activities for the p21 family of CDK inhibitors. Genes Dev. 1997;11:847–862. doi: 10.1101/gad.11.7.847. [DOI] [PubMed] [Google Scholar]
- 29.Leone G, DeGregori J, Sears R, Jakoi L, Nevins J R. Myc and Ras collaborate in inducing accumulation of active cyclin E/Cdk2 and E2F. Nature. 1997;387:422–426. doi: 10.1038/387422a0. [DOI] [PubMed] [Google Scholar]
- 30.Levy N, Yonish-Rouach E, Oren M, Kimchi A. Complementation by wild-type p53 of interleukin-6 effects on M1 cells: induction of cell cycle exit and cooperativity with c-myc suppression. Mol cell Biol. 1993;13:7942–7952. doi: 10.1128/mcb.13.12.7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lloyd A C. Ras versus cyclin-dependent kinase inhibitors. Curr Opin Genet Dev. 1998;8:43–48. doi: 10.1016/s0959-437x(98)80060-9. [DOI] [PubMed] [Google Scholar]
- 32.Lubbe J, Reichel M, Burg G, Kleihues P. Absence of p53 gene mutations in cutaneous melanoma. J Investig Dermatol. 1994;102:819–821. doi: 10.1111/1523-1747.ep12381544. [DOI] [PubMed] [Google Scholar]
- 33.Malumbres M, Pellicer A. RAS pathways to cell cycle control and cell transformation. Front Biosci. 1998;3:887–912. doi: 10.2741/a331. [DOI] [PubMed] [Google Scholar]
- 34.Mangray S, King T C. Molecular pathobiology of pancreatic adenocarcinoma. Front Biosci. 1998;3:1148–1160. doi: 10.2741/a351. [DOI] [PubMed] [Google Scholar]
- 35.Mateyak M K, Obaya A J, Adachi S, Sedivy J M. Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 1997;8:1039–1048. [PubMed] [Google Scholar]
- 36.O'Hagan R C, Ohh M, David G, de Alboran I M, Alt F W, Kaelin W G, Jr, DePinho R A. Myc-enhanced expression of Cul1 promotes ubiquitin-dependent proteolysis and cell cycle progression. Genes Dev. 2000;14:2185–2191. doi: 10.1101/gad.827200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ory D S, Neugeboren B A, Mulligan R C. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc Natl Acad Sci USA. 1996;93:11400–11406. doi: 10.1073/pnas.93.21.11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Papp T, Jafari M, Schiffmann D. Lack of p53 mutations and loss of heterozygosity in non-cultured human melanocytic lesions. J Cancer Res Clin Oncol. 1996;122:541–548. doi: 10.1007/BF01213550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pomerantz J, Schreiber-Agus N, Liegeois N J, Silverman A, Alland L, Chin L, Potes J, Orlow I, Lee H W, Cordon-Cardo C, DePinho R A. The INK4a tumor suppressor gene product, p19ARF, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 40.Quelle D E, Zindy F, Ashmun R A, Sherr C J. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell. 1995;83:993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 41.Radfar A, Unnikrishnan I, Lee H W, DePinho R A. p19(Arf) induces p53-dependent apoptosis during Abelson virus-mediated pre-B cell transformation. Proc Natl Acad Sci USA. 1998;95:13194–13199. doi: 10.1073/pnas.95.22.13194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ragimov N, Krauskopf A, Navot N, Rotter V, Oren M, Aloni Y. Wild-type but not mutant p53 can repress transcription initiation in vitro by interfering with the binding of basal transcription factors to the TATA motif. Oncogene. 1993;8:1183–1193. [PubMed] [Google Scholar]
- 43.Rhim K J, Hong S I, Hong W S, Lee S Y, Lee D S, Jang J J. Aberrant expression of p53 gene product in malignant melanoma. J Korean Med Sci. 1994;9:376–381. doi: 10.3346/jkms.1994.9.5.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruas M, Peters G. The p16INK4a/CDKN2A tumor suppressor and its relatives. Biochim Biophy Acta. 1998;1378:115–77. doi: 10.1016/s0304-419x(98)00017-1. [DOI] [PubMed] [Google Scholar]
- 45.Russo A A, Tong L, Lee J O, Jeffrey P D, Pavletich N P. Structural basis for inhibition of the cyclin-dependent kinase Cdk6 by the tumor suppressor p16INK4a. Nature. 1998;395:237–243. doi: 10.1038/26155. [DOI] [PubMed] [Google Scholar]
- 46.Salgia R, Skarin A T. Molecular abnormalities in lung cancer. J Clin Oncol. 1998;16:1207–1217. doi: 10.1200/JCO.1998.16.3.1207. [DOI] [PubMed] [Google Scholar]
- 47.Santoni-Rugiu E, Falck J, Mailand N, Bartek J, Lukas J. Involvement of Myc activity in a G1/S-promoting mechanism parallel to the pRb/E2F pathway. Mol Cell Biol. 2000;20:3497–3509. doi: 10.1128/mcb.20.10.3497-3509.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmitt C A, McCurrach M E, de Stanchina E, Wallace-Brodeur R R, Lowe S W. INK4a/ARF mutations accelerate lymphomagenesis and promote chemoresistance by disabling p53. Genes Dev. 1999;13:2670–2677. doi: 10.1101/gad.13.20.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho R A. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 50.Sharpless N E, DePinho R A. The INK4A/ARF locus and its two gene products. Curr Opin Genet Dev. 1999;9:22–30. doi: 10.1016/s0959-437x(99)80004-5. [DOI] [PubMed] [Google Scholar]
- 51.Sparrow L E, Soong R, Dawkins H J, Iacopetta B J, Heenan P J. p53 gene mutation and expression in naevi and melanomas. Melanoma Res. 1995;5:93–100. doi: 10.1097/00008390-199504000-00004. [DOI] [PubMed] [Google Scholar]
- 52.Spotts G D, Patel S V, Xiao Q, Hann S R. Identification of downstream-initiated c-Myc proteins which are dominant-negative inhibitors of transactivation by full-length c-Myc proteins. Mol Cell Biol. 1997;17:1459–1468. doi: 10.1128/mcb.17.3.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Steiner P, Philipp A, Lukas J, Godden-Kent D, Pagano M, Mittnacht S, Bartek J, Eilers M. Identification of a Myc-dependent step during the formation of active G1 cyclin-cdk complexes. EMBO J. 1995;14:4814–4826. doi: 10.1002/j.1460-2075.1995.tb00163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stott F J, Bates S, James M C, McConnell B B, Starborg M, Brookes S, Palmero I, Ryan K, Hara E, Vousden K H, Peters G. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 1998;17:5001–5014. doi: 10.1093/emboj/17.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun X H, Copeland N G, Jenkins N A, Baltimore D. Id proteins Id1 and Id2 selectively inhibit DNA binding by one class of helix-loop-helix proteins. Mol Cell Biol. 1991;11:5603–5611. doi: 10.1128/mcb.11.11.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vlach J, Hennecke S, Alevizopoulos K, Conti D, Amati B. Growth arrest by the cyclin-dependent kinase inhibitor p27Kipl is abrogated by c-Myc. EMBO J. 1996;15:6595–6604. [PMC free article] [PubMed] [Google Scholar]
- 57.Wolfel T, Hauer M, Schneider J, Serrano M, Wolfel C, Klehmann-Hieb E, De P E, Hankeln T, Meyer z B K H, Beach D. A p16INK4a-insensitive CDK4 mutant targeted by cytolytic T lymphocytes in a human melanoma. Science. 1995;269:1281–1284. doi: 10.1126/science.7652577. [DOI] [PubMed] [Google Scholar]
- 58.Xiao Q, Claassen G, Shi J, Adachi S, Sedivy J, Hann S R. Transactivation-defective c-MycS retains the ability to regulate proliferation and apoptosis. Genes Dev. 1998;12:3803–3808. doi: 10.1101/gad.12.24.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zerp S F, van Elsas A, Peltenburg L T, Schrier P I. p53 mutations in human cutaneous melanoma correlate with sun exposure but are not always involved in melanomagenesis. Br J Cancer. 1999;79:921–926. doi: 10.1038/sj.bjc.6690147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang S, Ramsay E S, Mock B A. Cdkn2a, the cyclin-dependent kinase inhibitor encoding p16INK4a and p19ARF, is a candidate for the plasmacytoma susceptibility locus, Pctrl. Proc Natl Acad Sci USA. 1998;95:2429–2434. doi: 10.1073/pnas.95.5.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Y, Xiong Y, Yarbrough W G. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 62.Zindy F, Eischen C M, Randle D H, Kamijo T, Cleveland J L, Sherr C J, Roussel M F. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]