Abstract
COVID-19, a new respiratory infectious disease, was first reported at the end of 2019, in Wuhan, China. Now, COVID-19 is still causing major loss of human life and economic productivity in almost all countries around the world. Early detection, early isolation, and early diagnosis of COVID-19 patients and asymptomatic carriers are essential to blocking the spread of the pandemic. This paper briefly reviewed COVID-19 diagnostic assays for clinical application, including nucleic acid tests, immunological methods, and Computed Tomography (CT) imaging. Nucleic acid tests (NAT) target the virus genome and indicates the existence of the SARS-CoV-2 virus. Currently, real-time quantitative PCR (qPCR) is the most widely used NAT and, basically, is the most used diagnostic assay for COVID-19. Besides qPCR, many novel rapid and sensitive NAT assays were also developed. Serological testing (detection of serum antibodies specific to SARS-CoV-2), which belongs to the immunological methods, is also used in the diagnosis of COVID-19. The positive results of serological testing indicate the presence of antibodies specific to SARS-CoV-2 resulting from being infected with the virus. Viral antigen detection assays are also important immunological methods used mainly for rapid virus detection. However, only a few of these assays had been reported. CT imaging is still an important auxiliary diagnosis tool for COVID-19 patients, especially for symptomatic patients in the early stage, whose viral load is low and different to be identified by NAT. These diagnostic techniques are all good in some way and applying a combination of them will greatly improve the accuracy of COVID-19 diagnostics.
Keywords: COVID-19, SARS-CoV-2, Nucleic acid testing, Immunological methods, CT imaging
1. Introduction
In late December 2019, a viral pneumonia respiratory infectious disease caused by a novel coronavirus was firstly reported in Wuhan, Hubei Province, China (Liu et al., 2020a). It resulted in infecting more than 80,000 people in China. The world health organization (WHO) named the disease COVID-19, short for Coronavirus Disease 2019 (WHO, 2020). Subsequently, scientific experts in the Coronavirus Study Group of the ICTV named the virus SARS-CoV-2 (Gorbalenya et al., 2020). On March 11, 2020, the WHO announced that COVID-19 constituted a global pandemic. According to the statistics released by Johns Hopkins University, as of December 14, 2021, the cumulative confirmed COVID-19 cases worldwide had exceeded 270,822,728 (over 22,000,000 cases in the US, India, and Brazil), and the cumulative number of deaths had exceeded 5,312,913 (over 470,000 deaths in the US, Brazil, and India).
SARS-CoV-2 is a positive-sense single-stranded RNA virus of the species Severe acute respiratory syndrome-related coronavirus belonging to the genus Betacoronavirus in the family Coronaviridae (Zhu et al., 2020a), some members of which are highly pathogenic to humans (Weiss and Navas-Martin, 2005; Cui et al., 2019). SARS-CoV-2 is the seventh coronavirus (HCoV-229E, HCoV-OC43, HCoV-NL63, HCoV-HKU1, SARS-CoV, MERS-CoV and SARS-CoV-2) found to be infectious in humans (Ludwig and Zarbock, 2020).
COVID-19 patients often suffer from fever, fatigue, cough, and dyspnea (Huang et al., 2020; Wang et al., 2020c). Severe cases rapidly evolve into acute respiratory distress syndrome (ARDS) and multiple organ failure (MOF), or even death (Yang et al., 2020). SARS-CoV-2 became a huge threat to the safety of human life (Wang et al., 2020a) and the world has changed greatly because of it. Over 2020, the value of the basic reproduction number (R0) of SARS-CoV-2 ranged from 1.8 to 3.6, indicating that the epidemic was spreading (Petersen et al., 2020). The infectivity and fatality rates of SARS-CoV-2 are both higher than those of influenza, which is also transmitted mainly through airborne routes (Chen, 2020; Wu and McGoogan, 2020). However, as the COVID-19 pandemic progressing, it was reported that the new SARS-CoV-2 delta variant had a shocking R0 of nearly 7 (Burki, 2021) and its incubation period was shorter than the original one (Zhang et al., 2021a). It is an even greater challenge for global governments and now we are in the early stages of a third wave according to the WHO (Tedros, 2021). To stop the spread of COVID-19, the identification of COVID-19 patients and contact tracing are especially important. Compared to previous infectious diseases, more advanced contact tracing technologies are being applied in the pandemic, such as big data and contact tracing smartphone apps (Maccari and Cagno, 2021). These technologies greatly improve the speed by which the relevant government departments respond to the outbreaks, but they also bring a lot of controversies such as privacy and ethics (Scassa, 2021). Additional measures, such as isolation and lockdown, to limit the movement of people were also applied which caused a lot of economic and social hardship as well (World Trade Organization, 2020; Zhang et al., 2021b). Nevertheless, the availability of assays for the widespread, rapid, and early detection of COVID-19 infections will reduce the social demands of contact tracing and prevent extensive and extended lockdowns, as well as reducing the risk of being infected by close contacts.
Early detection, early isolation, and early diagnostic of COVID-19 patients are vital for winning the fight against the pandemic. Like all virological diagnostics, nucleic acid tests (NAT) and immunological assays are the most widely used methods for SARS-CoV-2 detection and the diagnosis of COVID-19 patients and asymptomatic carriers (Yu et al., 2021; Eftekhari et al., 2021). CT imaging is still important for COVID-19 diagnosis, especially for symptomatic patients in the early stage, whose viral load is low and different to be identified by NAT (De et al., 2021; Brogna et al., 2021). The 8th guideline for diagnosis and treatment of COVID-19 released by the National Health Commission (NHC) of China still added CT imaging in the diagnostic criteria to avoid false-negative results (National Health Commission of the People's Republic of China, 2020). Over the past 18 months, after the initial panic along with shortage of diagnosis and treatment resources, effective detection systems (reagents and equipment production, testing capacity expand, and skilled personnel training.) have been developed and applied in the spread control. Besides, research and development of new assays are emerging with understanding viral pathogenesis. Therefore, this mini-review focused on the developed diagnostic assays of COVID-19. We hoped it would provide more thoughts for developing novel diagnostic assays.
2. Nucleic acid tests (NAT)
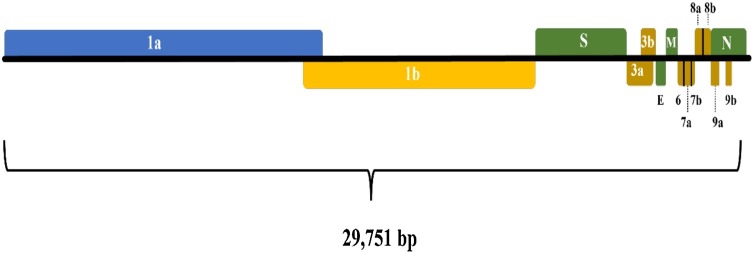
With the development of sequencing technology, the whole genome sequence of SARS-CoV-2 was rapidly obtained and published on NCBI website on January 5, 2020 (GenBank: MN908947.3, Wu et al., 2020). The SARS-CoV-2 genome consists of ORF1a and ORF1b (ORF1a and ORF1b coding 16 nonstructural proteins such as RNA-dependent RNA polymerase (Udugama et al., 2020), S (spike protein), E (envelop protein), M (membrane protein), and N (nucleocapsid protein) (Touma, 2020) (Fig. 1 ). Based on the genome sequence of SARS-CoV-2, the specific target sequences are selected and used in NAT assays development.
Fig. 1.
Genome structure of SARS-CoV-2.
2.1. Real-time PCR assays for detection of SARS-CoV-2
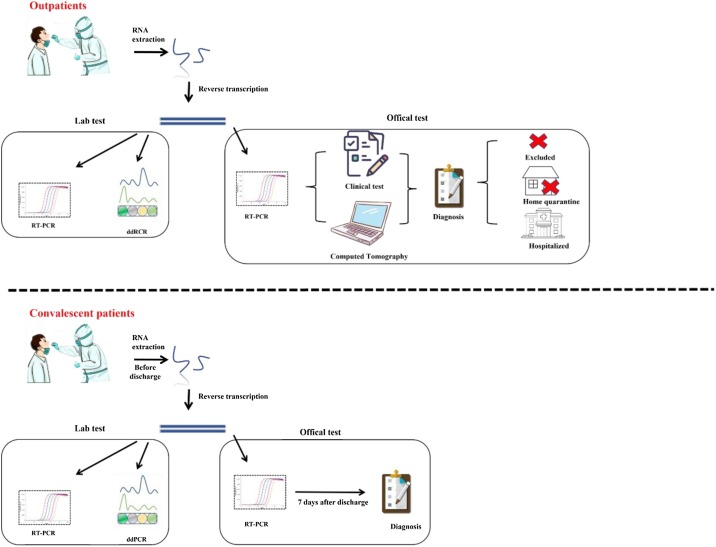
Real-time quantitative PCR (qPCR) is regarded as a gold standard for the detection of SARS-CoV-2 and is recommended by WHO and global CDCs. It is widely applied in departure and entry of people and goods, community screening for SARS-CoV-2 detection because of its rapidity and low cost. Chu et al. (2020) developed two one-step real-time reverse transcription PCR (rRT-PCR) methods to detect specific genes ORF1b and N, with the limit of detection (LOD) lower than 10 copies/reaction. Corman et al. (2020) reported the qPCR assay targeting E (envelope protein) gene and RdRp (RNA-dependent RNA polymerase) gene, and the LOD was 5.2 and 3.8 copies/reaction, respectively, while the sensitivity of N gene detection was slightly lower. Suo et al. (2020) reported a droplet digital PCR (ddPCR) assay has a limit significantly lower than RT-PCR (Fig. 2 ). Generally, it is recommended that COVID-19 patients can be identified only when two or more SARS-CoV-2 target gene sequences are detected as positive results. If only one target is positive, a retest is required. NAT with multiple target genes can effectively avoid false-positive results.
Fig. 2.
Research design for suspected outpatients and supposed convalescents by RT-PCR and ddPCR.
Because of the spread of infection, the demand for tests increases quickly, and the test results are time-sensitive. Researchers have tried many ways to improve the efficiency of qPCR. Ben-Ami et al. (2020) tested 184 samples in eight pools for RNA extraction and detection. Compared with testing individually, the results were not significantly affected. This strategy could greatly improve the efficiency of the test. Wuhan has tested 9,899,828 samples with this strategy in 16 days and the results were satisfactory. RNA extraction kits are the bottleneck of detection (Fomsgaard and Rosenstierne, 2020), and the lack of RNA extraction kits affect the response to the pandemic. Fukumoto et al. (2020), Eckel et al. (2020) and Hasan et al. (2020), and other researchers tried different methods to avoid the RNA extraction step. Unfortunately, the specificity and sensitivity of assays without the RNA extraction step were still inferior to those with the RNA extraction step. The false-negative rate of clinical samples was up to 83.0 % compared with standard PCR assays (Eckel et al., 2020). This might be contributed to the lower sensitivity of detection without the RNA extraction step (Pearson et al., 2021). An additional heat step might help improve the sensitivity (Hasan et al., 2020). Nevertheless, the virus RNA extraction and purification should be paid more attention to. Besides, there were also some studies tried other ways to increase detection efficiency such as comparing different samples treatment methods and thermocyclers (Ransom et al., 2020), designing double-quencher probes (Hirotsu et al., 2020), developing RT-PCR automated analyzer tool for SARS-CoV-2 detection (Dharavath et al., 2020).
2.2. Isothermal amplification assays for the detection of SARS-CoV-2
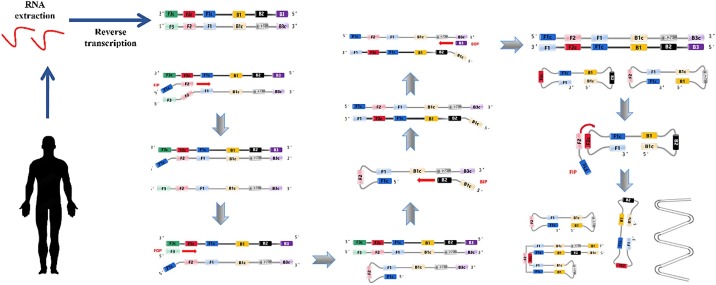
qPCR assay plays an important role in the clinical diagnosis of COVID-19 and there are numerous commercial products now (Iglói et al., 2020; Mostafa et al., 2020). In addition, various isothermal amplification assays such as loop-mediated isothermal amplification (LAMP) (Fig. 3 ) (Yoo et al., 2021), transcription-mediated amplification (TMA), reverse transcription recombinase–aided amplification (RT-RAA), multiple cross displacement amplification (MCDA), and recombinase-aided amplification (RPA), CHAnicking and extension amplification reaction (NEAR) have been developed for diagnosing COVID-19 (Table 1 ). These isothermal amplification assays are more sensitive and independent of a thermal cycler, which makes them more suitable in rapid, high-throughput, and low-cost assays development. Among them, LAMP is the most mature isothermal amplification method and the LAMP assays for SARS-CoV-2 detection are the most used isothermal amplification method.
Fig. 3.
Schematic procedure of reverse transcription loop-mediated isothermal amplification for SARS-CoV-2.
Table 1.
Isothermal amplification assays for the detection of SARS-CoV-2.
| Assay | Targeted gene | Detection limit | References |
|---|---|---|---|
| LAMP | N gene | 60 copies/μL | (Deng et al., 2021) |
| TMA | N gene | 5.5 copies/μL | (Gorzalski et al., 2020). |
| RT-RAA | ORF1ab gene | 0.04 copies/μL | (Wang et al., 2020d) |
| MCDA | N gene | 100 copies/μL | (Luu et al., 2021) |
| RPA | N gene and S gene | 0.05 copies/μL | (Xia and Chen, 2020) |
| CHA | ORF1ab and N genes | 2 copies/μL | (Zou et al., 2021) |
| NEAR | RdRp gene | 0.125 copies/μL | (Khan et al., 2020) |
2.3. CRISPR–CasN-based assays and other novel NAT assays for detection of SARS-CoV-2
Cas proteins including Cas 9, Cas 12, Cas 13, and Cas 14 can identify specific DNA/RNA sequences under a given condition and activate the activity of cleaving DNA or RNA fragments (Wang et al., 2020f). This makes Cas proteins particularly suitable in the detection of nucleic acid sequences. The detection assays based on Cas proteins have been developed. However, Cas protein could not detect target DNA when its concentration is lower than 1–10 nM (Guo et al., 2020). An isothermal amplification step was usually added before the detection of CRISPR–CasN-based assays. There have been several Cas-protein-based assays. For instance, based on RPA and Cas13a (namely SHERLOCK system), Zhang et al. (2020b) established an isothermal amplification method for detecting RNA of SARS-CoV-2, which could detect 20–200 aM SARS-CoV-2 within 1 h. Broughton et al. (2020) and Ding et al. (2020) developed CRISPR–Cas12-based assays in the detection of SARS-CoV-2. To avoid the additional nucleic acid amplification, Fozouni et al. (2020) used CRISPR-Cas13a and mobile phone microscopy to develop an amplification-free detection assay of SARS-CoV-2 and this assay had a sensitivity of 100 copies/uL in 30 min. Although CRISPR–CasN-based assays are far from mature, they have tremendous application potential in the diagnostic of COVID-19 and other diseases caused by viruses.
Other novel assays such as DNA nano-scaffold based on SARS-CoV-2 RNA triggered isothermal amplification (The reaction time was about 10 min and the reaction temperature is between 15–35 °C) (Jiao et al., 2020) and visual “naked-eye” detection assay based on thiol-modified ASO-capped AuNPs (this assay has a detection limit of 0.18 ng/μL of RNA) (Moitra et al., 2020) has also been developed. All these provide an alternative for SARS-CoV-2 detection.
2.4. Limitation of NAT methods
It is noteworthy that the false-negative results of RT-PCR and other NAT assays appeared frequently (Woloshin et al., 2020), which brings challenges to epidemic control. The reasons for false-negative results may be poor specificity of target genes or inadequate sensitivity of detection methods (Tahamtan and Ardebili, 2020). Wang et al. (2020b) identified 13 mutation sites in ORF (open reading frame) of the virus genome by comparing 95 strains of SARS-CoV-2, of which the mutation rate in ORF1a region was 29.47 %. It suggested that SARS-CoV-2 might have selective mutations, and certain regions must be avoided when designing primers and probes (Tian et al., 2020). These results suggest that each 10-fold increase in LoD is expected to increase the false-negative rate by 13 %, missing an additional one in eight infected patients (Arnaout et al., 2020). Chan et al. (2020) compared the three qPCR assays targeting RdRp gene, S (spike glycoprotein) gene, and N gene established by themselves with the NAT methods adopted by 30 laboratories in the European Union. The results showed that the detection methods currently applied in clinical practice need to strengthen the sensitivity to reduce false-negative results.
The sample collection methods also have an important impact on the accuracy of NAT results. Among them, nasopharyngeal swab sampling is the recommended method in most countries for detection of the nucleic acid of SARS-CoV-2 according to the characteristic of respiratory infectious disease. However, the nasopharyngeal swab sampling needs technical skills and puts the health care workers at risk of exposure. Saliva seems to be a good alternative. Saliva can be self-collected at a lower cost and few health care workers are needed. The virus RNA in saliva specimens is stable in 48 h (Matic et al., 2020) and SARS-CoV-2 in saliva can be detected even over one week (Williams et al., 2021). Saliva shows good concordance with a nasopharyngeal swab (or even better than) for detection of SARS-CoV-2 in most recent reports (Fan et al., 2020; Medeiros da Silva et al., 2020; Sakanashi et al., 2021; Sui et al., 2020). Notably, the collection way of saliva will affect the detection result. For example, hock-a-loogie saliva is better than saliva without cough (Fan et al., 2020; Yee et al., 2020). The U.S. Food and Drug Administration (FDA) had authorized the first in-home saliva collection for the detection of SARS-CoV-2 on May 08, 2020 (FDA, 2020). Besides, sputum is also another potential tool for detection (Lin et al., 2020).
Anyway, correct sampling operation and clinical application of new methods with high specificity and high sensitivity can reduce the risk of infection while sampling (Fabbris et al., 2021), false-negative results, and improve the accuracy of NAT methods in COVID-19 diagnosis.
3. Immunological methods
Immunological methods for COVID-19 diagnostics could be divided into two types: antibodies detection methods and antigens detection. For the former, we usually used serological tests to detect serum antibodies specific to SARS-CoV-2, which indicated if people were ever exposed to the virus or not. For the latter, we usually used prepared specific antibodies to develop detection methods, that was direct detection of the presence of SARS-CoV-2 antigens. They were valuable techniques for detecting COVID-19 patients with high viral load (that was super spreaders) at low cost (Pujadas et al., 2020).
3.1. Serological tests: detection of serum antibody specific to SARS-CoV-2
NAT assay usually indicates the current infection situation. Serological tests usually detect the antibody level in the serum, which indicates the situation of ever infection. IgM indicates an early viral infection and subsequent IgG indicates the later stages of infection (Zheng et al., 2020). Serological tests can be used in epidemiological investigation such as the investigation of pediatric medical workers exposed to varying levels of SARS-CoV-2 (Tu et al., 2020). In Hubei Province, China, COVID-19 patients, asymptomatic infection, and healthy people were tested by serological methods, with an accuracy of about 80 %, showing good specificity and sensitivity (Li et al., 2020; Xiang et al., 2020).
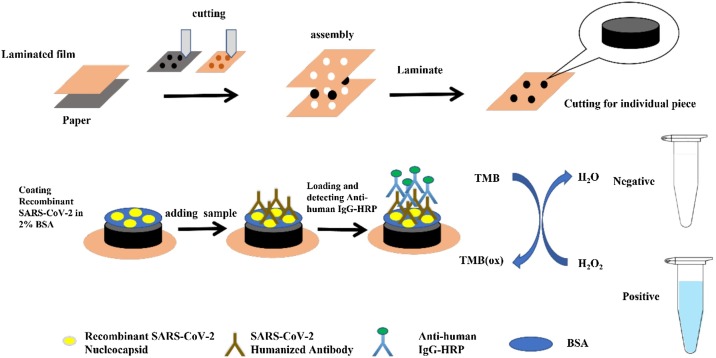
Enzyme-Linked Immunosorbent Assay (ELISA), Chemiluminescence Immunoassay (CLIA), and Lateral Flow Immunoassay (LFIA) are widely used in the detection of anti-SARS-CoV-2 antibodies (Nguyen et al., 2020). Researchers also tried to put forward other alternatives. Kasetsirikul et al. (2020) developed a paper-based ELISA assay which could be finished in 30 min with a cost of around 1.45–1.65 USD (Fig. 4 ). Dzimianski et al. (2020) developed a biolayer interferometry immunosorbent assay. This assay could obtain a semi-quantitative result in less than 20 min. It is not even the fastest detection assay. An electrochemical impedance-based detector developed by Rashed et al. could detect the serum antibody in less than 5 min (Rashed et al., 2021). Besides, some researchers also introduced a machine learning approach scoring patient samples for prior COVID-19 infection based on the antibody detection data to further improve the diagnostic efficiency (Cady et al., 2021). Most of the serological tests examine the IgG, IgM or both that are specific to SARS-CoV-2. Roda et al. (2021) developed dual lateral flow optical/chemiluminescence immunosensors for IgA which could be used to assess the efficacy of SARS-CoV-2 vaccines.
Fig. 4.
Schematic diagram of the fabrication and preparation process of the paper-based device for SARS-CoV-2 humanized antibody detection assay.
Some researchers believed that 30%–60% of infection is asymptomatic (Qiu, 2020). However, the asymptomatic carriers are also contagious, which may lead to a new outbreak. Nowadays, some countries and regions are in the late stage of the pandemic. Extensive serological tests can help to detect the previous infection, assess the progress of a regional epidemic situation, and predict the likely outcome of the epidemic.
3.2. Antigen detection: detection of specific antigens of SARS-CoV-2 based on specific antibody
The detection of serum antibodies is not suitable for rapid SARS-CoV-2 diagnostic. The developed rapid detection assays based on antibody tends to use a specific antibody to detect SARS-CoV-2 in samples directly or capture SARS-CoV-2 for further detection. Now, there have been some studies and commercial antigen detection kits for SARS-CoV-2 (Verma et al., 2020; Yuce et al., 2021; Azzi et al., 2021). Seo et al. (2020) immobilized antibody targeting SARS-CoV-2 spike protein on the graphene surface and prepared a field-effect transistor-based biosensor. This biosensor could detect SARS-CoV-2 with a concentration of 2.42 × 102 copies/mL in clinical samples. Karakuş et al. (2021) developed a colorimetric and electrochemical biosensor for SARS-CoV-2. The detection limit of it was 48 ng/mL SARS-CoV-2 spike antigen. Wang et al. (2021) developed a fluorescent immunochromatographic assay for SARS-CoV-2 based on multilayer quantum dot nanobead with a detection limit of 5 pg/mL SARS-CoV-2 NP antigen.
To avoid the long preparation period, Kim et al. (2020) used phage display technology to obtain single-chain variable fragment-crystallizable fragment (scFv-Fc) fusion antibodies specific to SARS-CoV-2 and developed a lateral flow immunoassay (LFIA)-based biosensor (Fig. 5 ). The reports of antigen detection kits were less than other detection methods although they were low cost and easy to use. This may be contributed to the long preparation period of specific antibodies and their sensitivity was generally lower than NAT methods (Liu and Rusling, 2021; Mak et al., 2020; Scohy et al., 2020).
Fig. 5.
Schematic showing the scFv-Fc-based LFIA.
Some researchers also tried other ways to detect antigens of SARS-Cov-2 without specific antibodies. Antibody analog such as aptamer is also applied in the detection of SARS-Cov-2. Chen et al. (2020) modified a SARS-CoV-2 specific DNA aptamer and selected three variants targeting SARS-CoV-2 N protein. These aptamers had potentials in diagnostic and had a short preparation period compared with a traditional antibody. Human angiotensin-converting enzyme 2 (ACE2) is the functional receptor of SARS-Cov-2 to bind with the SARS-Cov-2. The ACE2 can be detected by a commercial antibody. Based on it, Lee et al. (2021) developed a lateral flow immunoassay. It was interesting that the SARS-CoV S1 and MERS-CoV S1 proteins could not cause false-positive results although ACE2 was also the receptor of SARS-CoV and MERS-CoV.
These antigen detection assays based on specific antibodies or other macromolecules that can bind with SARS-CoV-2 are rapid and low-cost detection assays although the detection sensitivity is lower than NAT (Table 2 ). They may be suitable tools for rapid detection of COVID-19 to stop the spread. More attention should be paid to antigen detection.
Table 2.
Detection assays of specific antigens of SARS-CoV-2.
| Assay | Targeted antigen | Detection limit | References |
|---|---|---|---|
| ACE2-LFIA | Spike 1 antigen | 1.86 × 105 copies/mL | (Lee et al., 2021) |
| Fluorescent immunochromatographic assay | NP antigen | 5 pg/mL | (Wang et al., 2021) |
| Gold nanoparticle-based biosensor | Spike antigen | 48 ng/mL | (Karakuş et al., 2021) |
| Field-effect transistor-based biosensor | Spike protein | 2.42 copies/mL | (Seo et al., 2020) |
| ScFv-Fc biosensor | NP antigen | 2.5 × 104 pfu/reaction | (Kim et al., 2020) |
| Aptamer | NP antigen | 10 ng/mL | (Chen et al., 2020) |
3.3. Limitations of immunological methods
For the serological tests, all the detection assays are based on the detection of the antibodies specific to SARS-CoV-2 that are produced by the human immune system. The increase of the titer of the antibodies specific to SARS-CoV-2 is later than the infection (window period), so serological tests are not suitable for early diagnosis of COVID-19 (Wang, 2020; Ma et al., 2020). Serological tests are more suitable in the epidemiological survey and help to improve the accuracy of COVID-19 diagnosis. For example, the combination of serological and RT-PCR test has better sensitivity and specificity (Wang, 2020). Another important limitation of serological tests is the cross-reaction. The IgM and IgG specific to other viruses might also have the ability of binding SARS-CoV-2. For example, Zhu et al. (2020b) found the serum antibodies from SARS patients and immunized animals had a cross-reaction with SARS-CoV-2. This might bring a false positive result during traceability research. Researchers had noticed this and try to avoid it (La Rosa et al., 2021). For the same reason, the cross-reaction and long-period preparation of antibodies are also the limitations of antigen detection. This should be an important reason why there are only a few antigen detection assays.
4. CT imaging
Computed tomography imaging (CT imaging) is a kind of non-invasive medical imaging technique for diagnostics based on radiology. COVID-19 patients often have pulmonary inflammation. Chest CT examination can observe the imaging features of COVID-19 patients with multiple ground-glass opacity (GGO) in both lungs, which have the advantages of short time-consuming and high resolution (Wang et al., 2020e). Thus, CT imaging also became an important tool for the diagnosis of COVID-19.
4.1. Advantages and application of CT imaging in the diagnosis of COVID-19
The guideline for diagnosis and treatment of COVID-19 released by China NHC has included CT imaging in the diagnostic criteria to improve the accuracy of COVID-19 diagnosis. CT imaging can assist in the diagnosis of COVID-19. It was an important technique for the early detection of COVID-19 patients (Liu et al., 2020b).
Due to the low accuracy of NAT methods in COVID-19 diagnosis, CT imaging is recommended as an important diagnostic method for COVID-19 by researchers and clinicians. Fang et al. reported that the sensitivity of chest CT examination was higher than that of qPCR (98 % and 71 %, respectively) in the diagnosis of early COVID-19 cases in Hubei Province, China, suggesting the application of CT imaging to screen suspected patients with clinical symptoms and epidemiological history of COVID-19, especially when the test results of qPCR were negative (Fang et al., 2020; Gao et al., 2020). Ai et al. (2020) analyzed 1014 cases of COVID-19 patients, of which 601 (59 %) patients were positive for qPCR assays, while 888 (88 %) patients were positive for chest CT examinations. It suggested that CT imaging was extremely sensitive to the diagnosis of COVID-19 to be widely used for suspected cases, even as a preliminary examination for admission. In epidemic areas, CT imaging may be the primary diagnostic technique for COVID-19. The fact that chest CT might be beneficial in the early detection of cases of COVID-19 was also proved by a meta-analysis study (Mahmoud et al., 2020).
Researchers also developed tools based on deep learning to provides automated detection and quantification of pneumonia by automated CT imaging analysis (Nayak et al., 2021; Zhang et al., 2020a). This tool greatly reduces the workload although it cannot replace the role of the doctor now.
4.2. Limitations of CT imaging
Although CT imaging is an important tool for the diagnosis of COVID-19, there are some limitations of its application. Hope et al. proposed that the imaging features of pneumonia in COVID-19 patients were not specific to the disease. It might appear in a series of infectious and non-communicable diseases. Positive chest CT results were only credible when COVID-19 was highly suspected (Hope et al., 2020). The results of a chest CT examination for a definite diagnosis of COVID-19 should combine with NAT and immunological methods. In addition, the environmental safety of CT examinations should be considered. Given that COVID-19 is a highly contagious disease that can be spread through the air, the staff must take strict protective measures and the CT room should be thoroughly disinfected (Escudero et al., 2020; Kenarkoohi et al., 2020).
5. Conclusion and future trend/development/improvements
With the acceleration of globalization, emerging infectious diseases are likely to cause large-scale spread around the world. The high infectivity and high pathogenicity of COVID-19 pose a serious threat to the safety of human life. Early detection, early isolation, and early diagnosis of COVID-19 patients are especially important, which could effectively block the spread of the pandemic and return people to normal life orders as soon as possible. It challenges the ability of clinical diagnosis. In this review, we briefly reviewed diagnostic assays of COVID-19 for clinical application. Nucleic acid testing (NAT) can detect SARS-CoV-2 ribonucleic acid fragments and it (mainly qPCR) is widely applied in the departure and entry of people and goods, community screening. Other novel NAT assays such as isothermal amplification assays and CRISPR–CasN-based assays are also developed. The limitations of NAT are that the accuracy of its detection varied with the sensitivity, sampling methods, and so on. The researcher had also done a lot of work on them such as saliva sampling and pooled RNA extraction. These improved the detection efficiency and made large-scale testing in a short time became possible, which prevent the spread. However, different variants are emerging continuously, which makes diagnosis and treatment methods development more challenged.
Serological testing (namely detection of serum antibody specific to SARS-CoV-2), belongs to immunological methods, and also used in the diagnosis of COVID-19. However, the positive results of serological testing indicate the situation of infection at an undetermined time in the past, rather than the present situation. Compared with serological testing, there are fewer reports about the SARS-CoV-2 antigen detection based on the specific antibody. To improve the accuracy, a combination of NAT and serological testing is recommended. Besides, CT imaging is also another important auxiliary diagnosis tool for improving the diagnostic accuracy of COVID-19.
In total, Researchers are still working on better detection assay development to further improve the sensitivity and detection speed, increase throughput and reduce cost. They will make a great contribution to terminate the epidemic.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
The study was supported by the Shaanxi Provincial Natural Science Basic Research Program (No. 2020JZ-59).
References
- Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020 doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaout R., Lee R.A., Lee G.R., Callahan C., Yen C.F., Smith K.P., Arora R., Kirby J.E. 2020. SARS-CoV2 Testing: The Limit of Detection Matters. bioRxiv: the preprint server for biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzi L., Maurino V., Baj A., Dani M., d’Aiuto A., Fasano M., Lualdi M., Sessa F., Alberio T. Diagnostic salivary tests for SARS-CoV-2. J. Dent. Res. 2021;100(2):115–123. doi: 10.1177/0022034520969670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ami R., Klochendler A., Seidel M., Sido T., Gurel-Gurevich O., Yassour M., Meshorer E., Benedek G., Fogel I., Oiknine-Djian E., Gertler A., Rotstein Z., Lavi B., Dor Y., Wolf D.G., Salton M., Drier Y., Hebrew University-Hadassah C.-D.T. Large-scale implementation of pooled RNA extraction and RT-PCR for SARS-CoV-2 detection. Clin. Microbiol. Infect. 2020;26(9):1248–1253. doi: 10.1016/j.cmi.2020.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogna B., Brogna C., Petrillo M., Conte A.M., Benincasa G., Montano L., Piscopo M. Sars-cov-2 detection in fecal sample from a patient with typical findings of covid-19 pneumonia on CT but negative to multiple sars-cov-2 rt-pcr tests on oropharyngeal and nasopharyngeal swab samples. Medicina (Lithuania) 2021;57 doi: 10.3390/medicina57030290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton J.P., Deng X.D., Yu G.X., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C.Y., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020;38:870–U54. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burki T.K. Lifting of COVID-19 restrictions in the UK and the Delta variant. Lancet Respir. Med. 2021;9:e85. doi: 10.1016/s2213-2600(21)00328-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cady N.C., Tokranova N., Minor A., Nikvand N., Strle K., Lee W.T., Page W., Guignon E., Pilar A., Gibson G.N. Multiplexed detection and quantification of human antibody response to COVID-19 infection using a plasmon enhanced biosensor platform. Biosens. Bioelectron. 2021;171 doi: 10.1016/j.bios.2020.112679. 112679-112679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F., Yip C.C., To K.K., Tang T.H., Wong S.C., Leung K.H., Fung A.Y., Ng A.C., Zou Z., Tsoi H.W., Choi G.K., Tam A.R., Cheng V.C., Chan K.H., Tsang O.T., Yuen K.Y. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse transcription-pcr assay validated in vitro and with clinical specimens. J. Clin. Microbiol. 2020;58(5):e00310–00320. doi: 10.1128/JCM.00310-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. Pathogenicity and transmissibility of 2019-nCoV-A quick overview and comparison with other emerging viruses. Microbes Infect. 2020;22:69–71. doi: 10.1016/j.micinf.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.Q., Wu Q.H., Chen J., Ni X.H., Dai J.F. A DNA aptamer-based method for detection of SARS-CoV-2 nucleocapsid protein. Virol. Sin. 2020;35:351–354. doi: 10.1007/s12250-020-00236-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D.K.W., Pan Y., Cheng S.M.S., Hui K.P.Y., Krishnan P., Liu Y., Ng D.Y.M., Wan C.K.C., Yang P., Wang Q., Peiris M., Poon L.L.M. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin. Chem. 2020;66:549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brunink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet K., De Smet D., Ryckaert T., Laridon E., Heremans B., Vandenbulcke R., Demedts I., Bouckaert B., Gryspeerdt S., Martens G.A. Diagnostic Performance of Chest CT for SARS-CoV-2 infection in individuals with or without COVID-19 Symptoms. Radiology. 2021;298:E30–e37. doi: 10.1148/radiol.2020202708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H., Jayawardena A., Chan J., Tan S.M., Alan T., Kwan P. An ultra-portable, self-contained point-of-care nucleic acid amplification test for diagnosis of active COVID-19 infection. Sci. Rep. 2021;11:15176. doi: 10.1038/s41598-021-94652-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharavath B., Yadav N., Desai S., Sunder R., Mishra R., Ketkar M., Bhanshe P., Gupta A., Redhu A.K., Patkar N., Dutt S., Gupta S., Dutt A. A one-step, one-tube real-time RT-PCR based assay with an automated analysis for detection of SARS-CoV-2. Heliyon. 2020;6 doi: 10.1016/j.heliyon.2020.e04405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X., Yin K., Li Z., Liu C. 2020. All-in-One Dual CRISPR-Cas12a (AIOD-CRISPR) Assay: A Case for Rapid, Ultrasensitive and Visual Detection of Novel Coronavirus SARS-CoV-2 and HIV Virus. bioRxiv: the preprint server for biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzimianski J.V., Lorig-Roach N., O’Rourke S.M., Alexander D.L., Kimmey J.M., DuBois R.M. Rapid and sensitive detection of SARS-CoV-2 antibodies by biolayer interferometry. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-78895-x. 21738-21738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel F., Kusters F., Drossel B., Konert M., Mattes H., Schopf S. Variplex (TM) test system fails to reliably detect SARS-CoV-2 directly from respiratory samples without RNA extraction. Eur. J. Clin. Microbiol. Infect. Dis. 2020;39:2373–2377. doi: 10.1007/s10096-020-03983-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eftekhari A., Alipour M., Chodari L., Maleki Dizaj S., Ardalan M., Samiei M., Sharifi S., Zununi Vahed S., Huseynova I., Khalilov R., Ahmadian E., Cucchiarini M. A comprehensive review of detection methods for SARS-CoV-2. Microorganisms. 2021;9(2):232. doi: 10.3390/microorganisms9020232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escudero D., Antonio Boga J., Fernandez J., Forcelledo L., Balboa S., Albillos R., Astola I., Garcia-Prieto E., Elena Alvarez-Arguelles M., Martin G., Jimenez J., Vazquez F. SARS-CoV-2 analysis on environmental surfaces collected in an intensive care unit: keeping Ernest Shackleton’s spirit. ICMx. 2020:8. doi: 10.1186/s40635-020-00349-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbris C., Cestaro W., Menegaldo A., Spinato G., Frezza D., Vijendren A., Borsetto D., Boscolo-Rizzo P. Is oro/nasopharyngeal swab for SARS-CoV-2 detection a safe procedure? Complications observed among a case series of 4876 consecutive swabs. Am. J. Otolaryng. 2021;42 doi: 10.1016/j.amjoto.2020.102758. 102758-102758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J., Yu F., Wang X., Zou Q., Lou B., Xie G., Yang X., Chen W., Wang Q., Zhang D., Wang R., Feng B., Dong Y., Huang L., Teng Y., Deng Z., Yu L., Xu K., Sheng J., Zheng S., Chen Y. Hock-a-loogie saliva as a diagnostic specimen for SARS-CoV-2 by a PCR-based assay: a diagnostic validity study. Clin. Chim. Acta. 2020;511:177–180. doi: 10.1016/j.cca.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y., Zhang H., Xie J., Lin M., Ying L., Pang P., Ji W. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020;296(2) doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fomsgaard A.S., Rosenstierne M.W. An alternative workflow for molecular detection of SARS-CoV-2-escape from the NA extraction kit-shortage, Copenhagen, Denmark, March 2020. Eurosurveillance. 2020;25:6–9. doi: 10.2807/1560-7917.Es.2020.25.14.2000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fozouni P., Son S., Diaz de Leon Derby M., Knott G.J., Gray C.N., D’Ambrosio M.V., Zhao C., Switz N.A., Kumar G.R., Stephens S.I., Boehm D., Tsou C.-L., Shu J., Bhuiya A., Armstrong M., Harris A.R., Chen P.-Y., Osterloh J.M., Meyer-Franke A., Joehnk B., Walcott K., Sil A., Langelier C., Pollard K.S., Crawford E.D., Puschnik A.S., Phelps M., Kistler A., DeRisi J.L., Doudna J.A., Fletcher D.A., Ott M. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell. 2020;184(2):323–333. doi: 10.1016/j.cell.2020.12.001. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumoto T., Iwasaki S., Fujisawa S., Hayasaka K., Sato K., Oguri S., Taki K., Nakakubo S., Kamada K., Yamashita Y., Konno S., Nishida M., Sugita J., Teshima T. Efficacy of a novel SARS-CoV-2 detection kit without RNA extraction and purification. Int. J. Infect. Dis. 2020;98:16–17. doi: 10.1016/j.ijid.2020.06.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Liu J.Q., Wen H.J., Liu H., Hu W.D., Han X., Li C.X., Wang X.J. The unsynchronized changes of CT image and nucleic acid detection in COVID-19: reports the two cases from Gansu, China. Respir. Res. 2020;21:4. doi: 10.1186/s12931-020-01363-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., Penzar D., Perlman S., Poon L.L.M., Samborskiy D.V., Sidorov I.A., Sola I., Ziebuhr J., Grp C.S. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorzalski A.J., Tian H.L., Laverdure C., Morzunov S., Verma S.C., VanHooser S., Pandori M.W. High-Throughput Transcription-mediated amplification on the Hologic Panther is a highly sensitive method of detection for SARS-CoV-2. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Sun X.H., Wang X.G., Liang C., Jiang H.P., Gao Q.Q., Dai M.Y., Qu B., Fang S., Mao Y.H., Chen Y.C., Feng G.H., Gu Q., Wang R.R., Zhou Q., Li W. SARS-CoV-2 detection with CRISPR diagnostics. Cell Discov. 2020;6:4. doi: 10.1038/s41421-020-0174-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan M.R., Mirza F., Al-Hail H., Sundararaju S., Xaba T., Iqbal M., Alhussain H., Yassine H.M., Perez-Lopez A., Tang P. Detection of SARS-CoV-2 RNA by direct RT-qPCR on nasopharyngeal specimens without extraction of viral RNA. PLoS One. 2020;15 doi: 10.1371/journal.pone.0236564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsu Y., Mochizuki H., Omata M. Double-quencher probes improve detection sensitivity toward Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in a reverse-transcription polymerase chain reaction (RT-PCR) assay. J. Virol. Methods. 2020;284 doi: 10.1016/j.jviromet.2020.113926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope M.D., Raptis C.A., Henry T.S. Chest computed tomography for detection of coronavirus disease 2019 (COVID-19): don’t rush the science. Ann. Intern. Med. 2020;173(2):147–148. doi: 10.7326/M20-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglói Z., leven M., Abdel-Karem Abou-Nouar Z., Weller B., Matheeussen V., Coppens J., Koopmans M., Molenkamp R. Comparison of commercial realtime reverse transcription PCR assays for the detection of SARS-CoV-2. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao J., Duan C., Xue L., Liu Y., Sun W., Xiang Y. DNA nanoscaffold-based SARS-CoV-2 detection for COVID-19 diagnosis. Biosens. Bioelectron. 2020;167 doi: 10.1016/j.bios.2020.112479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakuş E., Erdemir E., Demirbilek N., Liv L. Colorimetric and electrochemical detection of SARS-CoV-2 spike antigen with a gold nanoparticle-based biosensor. Anal. Chim. Acta. 2021:1182. doi: 10.1016/j.aca.2021.338939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasetsirikul S., Umer M., Soda N., Sreejith K.R., Shiddiky M.J.A., Nam-Trung N. Detection of the SARS-CoV-2 humanized antibody with paper-based ELISA. Analyst. 2020;145(9):7680–7686. doi: 10.1039/d0an01609h. [DOI] [PubMed] [Google Scholar]
- Kenarkoohi A., Noorimotlagh Z., Falahi S., Amarloei A., Mirzaee S.A., Pakzad I., Bastani E. Hospital indoor air quality monitoring for the detection of SARS-CoV-2 (COVID-19) virus. Sci. Total Environ. 2020;748 doi: 10.1016/j.scitotenv.2020.141324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan P., Aufdembrink L.M., Engelhart A.E. Isothermal SARS-CoV-2 diagnostics: tools for enabling distributed pandemic testing as a means of supporting safe reopenings. ACS Synth. Biol. 2020;9(11):2861–2880. doi: 10.1021/acssynbio.0c00359. [DOI] [PubMed] [Google Scholar]
- Kim H.-Y., Lee J.-H., Kim M.J., Park S.C., Choi M., Lee W., Ku K.B., Kim B.T., Changkyun Park E., Kim H.G., Kim S.I. Development of a SARS-CoV-2-specific biosensor for antigen detection using scFv-Fc fusion proteins. Biosens. Bioelectron. 2020 doi: 10.1016/j.bios.2020.112868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Mancini P., Bonanno Ferraro G., Veneri C., Iaconelli M., Bonadonna L., Lucentini L., Suffredini E. SARS-CoV-2 has been circulating in northern Italy since December 2019: evidence from environmental monitoring. Sci. Total Environ. 2021;750 doi: 10.1016/j.scitotenv.2020.141711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-H., Choi M., Jung Y., Lee S.K., Lee C.-S., Kim J., Kim J., Kim N.H., Kim B.-T., Kim H.G. A novel rapid detection for SARS-CoV-2 spike 1 antigens using human angiotensin converting enzyme 2 (ACE2) Biosens. Bioelectron. 2021;171 doi: 10.1016/j.bios.2020.112715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Yi Y., Luo X., Xiong N., Liu Y., Li S., Sun R., Wang Y., Hu B., Chen W., Zhang Y., Wang J., Huang B., Lin Y., Yang J., Cai W., Wang X., Cheng J., Chen Z., Sun K., Pan W., Zhan Z., Chen L., Ye F. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. 2020;92(9):1518–1524. doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C., Xiang J., Yan M., Li H., Huang S., Shen C. Comparison of throat swabs and sputum specimens for viral nucleic acid detection in 52 cases of novel coronavirus (SARS-Cov-2)-infected pneumonia (COVID-19) Clin. Chem. Lab. Med. 2020 doi: 10.1515/cclm-2020-0187. [DOI] [PubMed] [Google Scholar]
- Liu G., Rusling J.F. COVID-19 antibody tests and their limitations. ACS Sens. 2021:0c02621. doi: 10.1021/acssensors.0c02621. [DOI] [PubMed] [Google Scholar]
- Liu R., Han H., Liu F., Lv Z., Wu K., Liu Y., Feng Y., Zhu C. Positive rate of RT-PCR detection of SARS-CoV-2 infection in 4880 cases from one hospital in Wuhan, China, from Jan to Feb 2020. Clin. Chim. Acta. 2020;505:172–175. doi: 10.1016/j.cca.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R.R., Zhu Y., Wu M.Y., Liu J., Ren R., Cao Q.L., Shen X.H., Chen G.Q., Li M. CT imaging analysis of 33 cases with the 2019 novel coronavirus infection. Zhonghua Yi Xue Za Zhi. 2020;100(13):1007–1011. doi: 10.3760/cma.j.cn112137-20200203-00182. [DOI] [PubMed] [Google Scholar]
- Ludwig S., Zarbock A. Coronaviruses and SARS-CoV-2: a brief overview. Anesth. Analg. 2020;131:93–96. doi: 10.1213/ane.0000000000004845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu L.D.W., Payne M., Zhang X., Luo L., Lan R. Development and comparison of novel multiple cross displacement amplification (MCDA) assays with other nucleic acid amplification methods for SARS-CoV-2 detection. Sci. Rep.-UK. 2021;11:1. doi: 10.1038/s41598-021-81518-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Zeng W., He H., Zhao D., Jiang D., Zhou P., Cheng L., Li Y., Ma X., Jin T. Serum IgA, IgM, and IgG responses in COVID-19. Cell. Mol. Immunol. 2020;17:773–775. doi: 10.1038/s41423-020-0474-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccari L., Cagno V. Do we need a contact tracing app? Comput. Commun. 2021;166:9–18. doi: 10.1016/j.comcom.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoud H., Taha M.S., Askoura A., Aleem M., Omran A., Aboelela S. Can chest CT improve sensitivity of COVID-19 diagnosis in comparison to PCR? A meta-analysis study. Egypt. J. Otolaryng. 2020;36:49. doi: 10.1186/s43163-020-00039-9. [DOI] [Google Scholar]
- Mak G.C., Cheng P.K., Lau S.S., Wong K.K., Lau C.S., Lam E.T., Chan R.C., Tsang D.N. Evaluation of rapid antigen test for detection of SARS-CoV-2 virus. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matic N., Stefanovic A., Leung V., Lawson T., Ritchie G., Li L., Champagne S., Romney M.G., Lowe C.F. Practical challenges to the clinical implementation of saliva for SARS-CoV-2 detection. Eur. J. Clin. Microbiol. Infect. Dis. 2020;40(2):447–450. doi: 10.1007/s10096-020-04090-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros da Silva R.C., Nogueira Marinho L.C., de Araujo Silva D.N., Costa de Lima K., Pirih F.Q., Luz de Aquino Martins A.R. Saliva as a possible tool for the SARS-CoV-2 detection: a review. Travel. Med. Infect. Dis. 2020;38 doi: 10.1016/j.tmaid.2020.101920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moitra P., Alafeef M., Dighe K., Frieman M.B., Pan D. Selective naked-eye Detection of SARS-CoV-2 mediated by n gene targeted antisense oligonucleotide capped plasmonic nanoparticles. ACS Nano. 2020;14(6):7617–7627. doi: 10.1021/acsnano.0c03822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostafa H.H., Hardick J., Morehead E., Miller J.-A., Gaydos C.A., Manabe Y.C. Comparison of the analytical sensitivity of seven commonly used commercial SARS-CoV-2 automated molecular assays. J. Clin. Virol. 2020;130 doi: 10.1016/j.jcv.2020.104578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak S.R., Nayak D.R., Sinha U., Arora V., Pachori R.B. Application of deep learning techniques for detection of COVID-19 cases using chest X-ray images: a comprehensive study. Biomed. Signal. Proc. 2021;64 doi: 10.1016/j.bspc.2020.102365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen N.N.T., McCarthy C., Lantigua D., Camci-Unal G. Development of diagnostic tests for detection of SARS-CoV-2. Diagnostics. 2020;10(11):905. doi: 10.3390/diagnostics10110905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson J.D., Trcka D., Lu S., Hyduk S.J., Jen M., Aynaud M.-M., Hernández J.J., Peidis P., Barrios-Rodiles M., Chan K., Woodgett J., Mazzulli T., Attisano L., Pelletier L., Cybulsky M.I., Wrana J.L., Bremner R. Comparison of SARS-CoV-2 indirect and direct RT-qPCR detection methods. Virol. J. 2021;18:99. doi: 10.1186/s12985-021-01574-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen E., Koopmans M., Go U., Hamer D.H., Petrosillo N., Castelli F., Storgaard M., Al Khalili S., Simonsen L. Comparing SARS-CoV-2 with SARS-CoV and influenza pandemics. Lancet. 2020;20(9):E238–E244. doi: 10.1016/s1473-3099(20)30484-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujadas E., Chaudhry F., McBride R., Richter F., Zhao S., Wajnberg A., Nadkarni G., Glicksberg B.S., Houldsworth J., Cordon-Cardo C. SARS-CoV-2 viral load predicts COVID-19 mortality. Lancet. Resp. Med. 2020;8:e70. doi: 10.1016/S2213-2600(20)30354-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J. Covert coronavirus infections could be seeding new outbreaks. Nature. 2020 doi: 10.1038/d41586-020-00822-x. [DOI] [PubMed] [Google Scholar]
- Ransom E.M., Potter R.F., Wallace M.A., Mitchell K.F., Yarbrough M.L., Burnham C.A., Anderson N.W., Parikh B.A. Comparison of extraction methods and thermocyclers for SARS-CoV-2 molecular detection using clinical specimens. J. Clin. Microbiol. 2020 doi: 10.1128/jcm.01622-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashed M.Z., Kopechek J.A., Priddy M.C., Hamorsky K.T., Palmer K.E., Mittal N., Valdez J., Flynn J., Williams S.J. Rapid detection of SARS-CoV-2 antibodies using electrochemical impedance-based detector. Biosens. Bioelectron. 2021;171 doi: 10.1016/j.bios.2020.112709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roda A., Cavalera S., Di Nardo F., Calabria D., Rosati S., Simoni P., Colitti B., Baggiani C., Roda M., Anfossi L. Dual lateral flow optical/chemiluminescence immunosensors for the rapid detection of salivary and serum IgA in patients with COVID-19 disease. Biosens. Bioelectron. 2021;172 doi: 10.1016/j.bios.2020.112765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakanashi D., Asai N., Nakamura A., Miyazaki N., Kawamoto Y., Ohno T., Yamada A., Koita I., Suematsu H., Hagihara M., Shiota A., Kurumiya A., Sakata M., Kato S., Muramatsu Y., Koizumi Y., Kishino T., Ohashi W., Yamagishi Y., Mikamo H. Comparative evaluation of nasopharyngeal swab and saliva specimens for the molecular detection of SARS-CoV-2 RNA in Japanese patients with COVID-19. J. Infect. Chemother. 2021;27(1):126–129. doi: 10.1016/j.jiac.2020.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scassa T. COVID-19 contact tracing: from local to global and back again. Int. J. E-Planning Res. 2021;10(2):45–58. doi: 10.4018/IJEPR.20210401.oa4. [DOI] [Google Scholar]
- Scohy A., Anantharajah A., Bodéus M., Kabamba-Mukadi B., Verroken A., Rodriguez-Villalobos H. Low performance of rapid antigen detection test as frontline testing for COVID-19 diagnosis. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo G., Lee G., Kim M.J., Baek S.H., Choi M., Ku K.B., Lee C.S., Jun S., Park D., Kim H.G., Kim S.J., Lee J.O., Kim B.T., Park E.C., Kim S.I. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in Human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- Sui Z., Zhang Y., Tu S., Xie J., Huang W., Peng T., Dong L., Yang J., Ouyang Y., Liu S., Li L., Wang Z., Peng K., Fang X., Dai X. Evaluation of saliva as an alternative diagnostic specimen source for SARS-CoV-2 detection by RT-dPCR. J. Infect. 2020 doi: 10.1016/j.jinf.2020.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo T., Liu X., Feng J., Guo M., Hu W., Guo D., Ullah H., Yang Y., Zhang Q., Wang X., Sajid M., Huang Z., Deng L., Chen T., Liu F., Xu K., Liu Y., Zhang Q., Liu Y., Xiong Y., Chen G., Lan K., Chen Y. ddPCR: a more accurate tool for SARS-CoV-2 detection in low viral load specimens. Emerg. Microbes Infect. 2020;9(1):1259–1268. doi: 10.1080/22221751.2020.1772678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahamtan A., Ardebili A. Real-time RT-PCR in COVID-19 detection: issues affecting the results. Expert Rev. Mol. Diagn. 2020;20(5):453–454. doi: 10.1080/14737159.2020.1757437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B., Gao F., Fock J., Dufva M., Hansen M. Homogeneous circle-to-circle amplification for real-time optomagnetic detection of SARS-CoV-2 RdRp coding sequence. Biosens. Bioelectron. 2020;165:112356. doi: 10.1016/j.bios.2020.112356. [DOI] [PubMed] [Google Scholar]
- Touma M. COVID-19: molecular diagnostics overview. J. Mol. Med. 2020;98:947–954. doi: 10.1007/s00109-020-01931-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu D., Shu J., Wu X., Li H., Xia Z., Zhang Y., Fang Y., Shen S., Guan W., Wang H., Huang Z., Wang G., Zhou X., Deng F. Immunological detection of serum antibodies in pediatric medical workers exposed to varying levels of SARS-CoV-2. J. Infect. 2020 doi: 10.1016/j.jinf.2020.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udugama B., Kadhiresan P., Kozlowski H.N., Malekjahani A., Osborne M., Li V.Y.C., Chen H., Mubareka S., Gubbay J.B., Chan W.C.W. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020;14:3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- Verma N., Patel D., Pandya A. Emerging diagnostic tools for detection of COVID-19 and perspective. Biomed. Microdevices. 2020;22(4):83. doi: 10.1007/s10544-020-00534-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P. Combination of serological total antibody and RT-PCR test for detection of SARS-COV-2 infections. J. Virol. Methods. 2020;283 doi: 10.1016/j.jviromet.2020.113919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Liu Z., Chen Z., Huang X., Xu M., He T., Zhang Z. The establishment of reference sequence for SARS-CoV-2 and variation analysis. J. Med. Virol. 2020;92:667–674. doi: 10.1002/jmv.25762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., Zhao Y., Li Y., Wang X., Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Cai K., He X., Shen X., Wang J., Liu J., Xu J., Qiu F., Lei W., Cui L., Ge Y., Wu T., Zhang Y., Yan H., Chen Y., Yu J., Ma X., Shi H., Zhang R., Li X., Gao Y., Niu P., Tan W., Wu G., Jiang Y., Xu W. Multiple-centre clinical evaluation of an ultrafast single-tube assay for SARS-CoV-2 RNA. Clin. Microbiol. Infect. 2020;26:1076–1081. doi: 10.1016/j.cmi.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Kang S., Tian R., Zhang X., Wang Y. Imaging manifestations and diagnostic value of chest CT of coronavirus disease 2019 (COVID-19) in the Xiaogan area. Clin. Radiol. 2020;75(5):341–347. doi: 10.1016/j.crad.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Zhang R., Li J. CRISPR/cas systems redefine nucleic acid detection: principles and methods. Biosens. Bioelectron. 2020;165 doi: 10.1016/j.bios.2020.112430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Yang X., Zheng S., Cheng X., Xiao R., Li Q., Wang W., Liu X., Wang S. Development of an ultrasensitive fluorescent immunochromatographic assay based on multilayer quantum dot nanobead for simultaneous detection of SARS-CoV-2 antigen and influenza A virus. Sens. Actuators B Chem. 2021;345 doi: 10.1016/j.snb.2021.130372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S.R., Navas-Martin S. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 2005;69:635–664. doi: 10.1128/mmbr.69.4.635-664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2020. Novel Coronavirus (2019-nCoV) Situation Report – 22.https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200211-sitrep-22-ncov.pdf?sfvrsn=fb6d49b1_2 [Google Scholar]
- Williams E., Isles N., Chong B., Bond K., Yoga Y., Druce J., Catton M., Ballard S.A., Howden B.P., Williamson D.A. Detection of SARS-CoV-2 in saliva: implications for specimen transport and storage. J. Med. Microbiol. 2021;70(2) doi: 10.1099/jmm.0.001285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woloshin S., Patel N., Kesselheim A.S. False negative tests for SARS-CoV-2 infection - challenges and implications. New Engl. J. Med. 2020;383:e38. doi: 10.1056/NEJMp2015897. [DOI] [PubMed] [Google Scholar]
- Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323(13):1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S.M., Chen X. Single-copy sensitive, field-deployable, and simultaneous dual-gene detection of SARS-CoV-2 RNA via modified RT-RPA. Cell Discov. 2020;6:4. doi: 10.1038/s41421-020-0175-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang F., Wang X., He X., Peng Z., Yang B., Zhang J., Zhou Q., Ye H., Ma Y., Li H., Wei X., Cai P., Ma W.L. Antibody detection and dynamic characteristics in patients with COVID-19. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Yu Y., Xu J., Shu H., Xia J., Liu H., Wu Y., Zhang L., Yu Z., Fang M., Yu T., Wang Y., Pan S., Zou X., Yuan S., Shang Y. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir. Med. 2020;8(5):475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee R., Truong T., Pannaraj P.S., Eubanks N., Gai E., Jumarang J., Turner L., Peralta A., Lee Y., Dien Bard J. Saliva is a promising alternative specimen for the detection of SARS-CoV-2 in children and adults. J. Clin. Microbiol. 2020;59(2) doi: 10.1128/jcm.02686-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H.M., Kim I.H., Kim S. Nucleic acid testing of sars‐cov‐2. Int. J. Mol. Sci. 2021;22:6150. doi: 10.3390/ijms22116150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.Y., Chan K.G., Yean C.Y., Ang G.Y. Nucleic acid-based diagnostic tests for the detection SARS-CoV-2: an update. Diagnostics. 2021:11. doi: 10.3390/diagnostics11010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuce M., Filiztekin E., Ozkaya K.G. COVID-19 diagnosis -a review of current methods. Biosens. Bioelectron. 2021;172 doi: 10.1016/j.bios.2020.112752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.T., Zhang J.S., Zhang H.H., Nan Y.D., Zhao Y., Fu E.Q., Xie Y.H., Liu W., Li W.P., Zhang H.J., Jiang H., Li C.M., Li Y.Y., Ma R.N., Dang S.K., Gao B.B., Zhang X.J., Zhang T. Automated detection and quantification of COVID-19 pneumonia: CT imaging analysis by a deep learning-based software. Eur. J. Nucl. Med. Mol. Imaging. 2020;47:2525–2532. doi: 10.1007/s00259-020-04953-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Abudayyeh O.O., Gootenberg J.S. 2020. A Protocol for Detection of COVID-19 Using CRISPR Diagnostics.https://www.broadinstitute.org/files/publications/special/COVID-19%20detection%20(updated).pdf [Google Scholar]
- Zhang M., Xiao J., Deng A., Zhang Y., Zhuang Y., Hu T., Li J., Tu H., Li B., Zhou Y., Yuan J., Luo L., Liang Z., Huang Y., Ye G., Cai M., Li G., Yang B., Xu B., Huang X., Cui Y., Ren D., Zhang Y., Kang M., Li Y., Guangdong Provincial Center for Disease, C, Prevention, G.G.C, Guangdong Provincial Institute of Public Health, G.G.C, School of Public Health, S.M.U.G.G.C, Guangzhou Center for Disease, C, Prevention, G.G.C, Foshan Center for Disease, C, Prevention, G.G.C, Maoming Center for Disease, C, Prevention, G.G.C, Zhanjiang Center for Disease, C, Prevention, G.G.C, Guangzhou Liwan District Center for Disease, C, Prevention, G.G.C, Guangzhou Panyu District Center for Disease, C, Prevention, G.G.C, Guangzhou Haizhu District Center for Disease, C, Prevention, G.G.C, Guangzhou Yuexiu District Center for Disease, C, Prevention, G.G.C, Foshan Nanhai District Center for Disease, C, Prevention, G.G.C, Maoming Dianbai District Center for Disease, C, Prevention, G.G.C, National Institute for Communicable Disease, C, Prevention, C.C.f.D.C, Prevention, B.C, Chinese Center for Disease, C, Prevention, B.C Transmission dynamics of an outbreak of the COVID-19 delta variant B.1.617.2 — Guangdong Province, China, May–June 2021a. China CDC Wkly. 2021;3:584–586. doi: 10.46234/ccdcw2021.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Litvinova M., Liang Y., Zheng W., Shi H., Vespignani A., Viboud C., Ajelli M., Yu H. The impact of relaxing interventions on human contact patterns and SARS-CoV-2 transmission in China. Sci. Adv. 2021;7(19):eabe2584. doi: 10.1126/sciadv.abe2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Yan M., Wang L., Luan L., Liu J., Tian X., Wan N. Analysis of the application value of serum antibody detection for staging of COVID-19 infection. J. Med. Virol. 2020;93(2):899–906. doi: 10.1002/jmv.26330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Yu D., Han Y., Yan H., Chong H., Ren L., Wang J., Li T., He Y. Cross-reactive neutralization of SARS-CoV-2 by serum antibodies from recovered SARS patients and immunized animals. Sci. Adv. 2020;6(45):eabc9999. doi: 10.1126/sciadv.abc9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou M., Su F., Zhang R., Jiang X., Xiao H., Yan X., Yang C., Fan X., Wu G. Rapid point-of-care testing for SARS-CoV-2 virus nucleic acid detection by an isothermal and nonenzymatic Signal amplification system coupled with a lateral flow immunoassay strip. Sens. Actuators B-Chem. 2021;342 doi: 10.1016/j.snb.2021.129899. [DOI] [PMC free article] [PubMed] [Google Scholar]
Web references
Tedros. WHO Director-General's opening remarks at the 8th meeting of the IHR Emergency Committee on COVID-19 – 14 July 2021 https://www.who.int/director-general/speeches/detail/who-director-general-s-opening-remarks-at-the-8th-meeting-of-the-ihr-emergency-committee-on-covid-19-14-july-2021.
World Trade Organization. World merchandise trade fell 14 % in volume, 21 % in value in Q2 amid global lockdown https://www.wto.org/english/news_e/news20_e/stat_23sep20_e.htm.
National Health Commission of the People's Republic of China. http://www.nhc.gov.cn/yzygj/s7653p/202008/0a7bdf12bd4b46e5bd28ca7f9a7f5e5a.shtml, accessed on August 19, 2020.
FDA. Coronavirus (COVID-19) Update: FDA Authorizes First Diagnostic Test Using At-Home Collection of Saliva Specimens. https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-first-diagnostic-test-using-home-collection-saliva. accessed May 08, 2020.