Abstract
The coronavirus disease 2019 (COVID-19) pandemic has highlighted the major shortcoming of healthcare systems globally in their inability to diagnose the disease rapidly and accurately. At present, the molecular approaches for diagnosing COVID-19 primarily use reverse transcriptase polymerase chain reaction (RT-PCR) to create and amplify cDNA from severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA. Although molecular tests are reported to be specific, false negatives are quite common. Furthermore, literally all these tests require a step involving RNA isolation which does not make them point-of-care (POC) in the true sense. Here, we report a lateral flow strip-based RNA extraction and amplification-free nucleic acid test (NAT) for rapid diagnosis of positive COVID-19 cases at POC. The assay uses highly specific 6-carboxyfluorescein (6-FAM) and biotin labeled antisense oligonucleotides (ASOs) as probes those are designed to target N-gene sequence of SARS-CoV-2. Additionally, we utilized cysteamine capped gold-nanoparticles (Cyst-AuNPs) to augment the signal further for enhanced sensitivity. Without any large-stationary equipment and highly trained staffers, the entire sample-to-answer approach in our case would take less than 30 min from a patient swab sample collection to final diagnostic result. Moreover, when evaluated with 60 clinical samples and verified with an FDA-approved TaqPath RT-PCR kit for COVID-19 diagnosis, the assay obtained almost 99.99% accuracy and specificity. We anticipate that the newly established low-cost amplification-free detection of SARS-CoV-2 RNA will aid in the development of a platform technology for rapid and POC diagnosis of COVID-19 and other pathogens.
Keywords: COVID-19, SARS-CoV-2, Antisense oligonucleotide, Lateral flow assay, Point-of-care, Gold-nanoparticles
1. Introduction
Coronavirus disease 2019 (COVID-19) is a respiratory disease with a wide variety of clinical outcomes, ranging from asymptomatic and mild illness to serious complications and death, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Grifoni et al., 2020; Hu et al., 2020). Rapid and accurate COVID-19 diagnosis with kits that are emergency use authorization (EUA) approved or with eventual FDA clearance remains crucial for monitoring and slowing the spread of the virus and avoiding future outbreaks (Carter et al., 2020; Huang et al., 2020; Ravi et al., 2020). However, the high volume of samples and complicated scenarios encountered in hospitals present significant challenges in terms of early detection and intensive surveillance (Alafeef et al., 2020b; Moitra et al., 2021; Vandenberg et al., 2021). Additionally, because of the limited sensitivity and specificity of existing technologies, false negative instances among infected individuals lead to widespread community transmission of COVID-19. Moreover, continuous mutation of the original SARS-CoV-2 genetic sequence makes its detection more difficult with the prevailing technologies (Wang et al., 2020).
Currently, three approaches, i.e., reverse-transcription polymerase chain reaction (RT-PCR), serological/immunological antigen-based tests and chest computed tomography (CT), are generally used for COVID-19 diagnosis (Lukas et al., 2020; Udugama et al., 2020; Valera et al., 2021). Of the above, chest CT is a key screening tool for patients with COVID-19 symptoms (Cai et al., 2020; Rong et al., 2021). Despite being widely available in cities, CT facilities typically do not reliably detect COVID-19 infection in its early stages, making them unsuitable for intensive patient surveillance. Separately, several immunodiagnostic and serological assays (Tables S1–S2) have also been developed to respectively detect either the presence of antibodies against COVID-19 or antigens of SARS-CoV-2 (Alafeef et al., 2019; Pokhrel et al., 2020). But the utility of most of these techniques are limited. While immunological tests suffer from a detectable antibody response at the early stages of infection, serological ones have the drawbacks of cross-reactivity with other pathogens, including other human coronaviruses (Liu and Rusling, 2021). Thus, both these approaches may contribute to high incidence of either false positive or false negative results, respectively.
At present, nucleic acid-based test is most popular towards the diagnosis of COVID-19 (Cherkaoui et al., 2021; Yüce et al., 2021). Here the detection principle majorly encounters the complex requirement of amplification of cDNA extracted from the viral SARS-CoV-2 RNA collected from bio-fluids of patients under observation (W. H. Feng et al., 2020). Based on this principle, numerous RT-PCR kits have been developed so far for diagnosis of COVID-19. While these RT-PCR kits are highly sensitive, there is still a significant prevalence of false negative results (20–40%) in instances where clinical symptoms and CT results prompted substantial concerns about the illness (Binnicker, 2020; H. W. Feng et al., 2020; He, J., Luo, L., Luo, Z., Lyu, J., Ng, M., Shen, X., Wen, 2020). Numerous factors have been identified as potentially contributing to the high prevalence of false negative results, with genetic mutation being the most concerning of them (“SARS-CoV-2 Viral Mutations: Impact on COVID-19 Tests,” 2021). Most of the RT-PCR primers target a particular segment of the SARS-CoV-2 genome, and the possibility of mutation and recombination increases the chance of false negative results. Additionally, current RT-PCR tests for diagnosing COVID-19 are complex and labor-intensive. All these limit the universal applicability of currently available commercial COVID-19 detection kits. Therefore, to control the COVID-19 pandemic, an easy and rapid diagnostic test (RDT) is needed to identify SARS-CoV-2 viral RNA with high sensitivity, accuracy and specificity for early detection of COVID-19 infection and successful monitoring of patients during and after treatment.
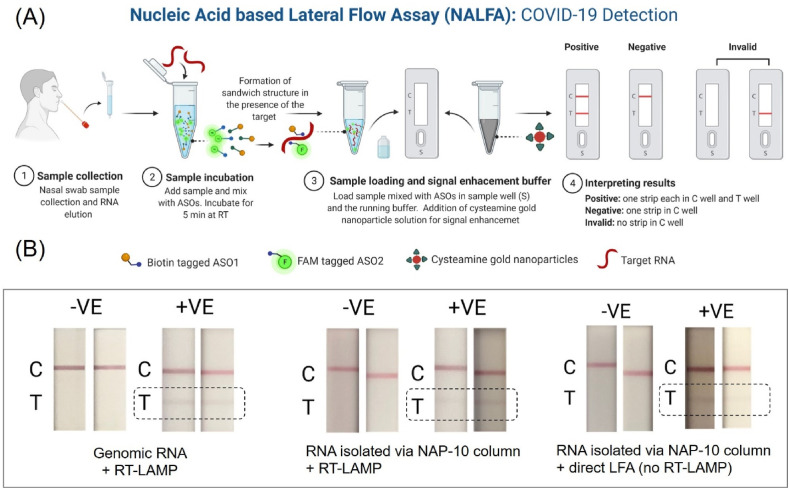
Herein, we have developed a nucleic acid based lateral flow assay (NALFA) which address all the above limitations of the existing techniques, as it can be used in resource scarce environment as well as at POC level without compromising the sensitivity and accuracy. Furthermore, our platform also addresses the current concerns associated with antigen or antibody based lateral flow assays because of its all-inclusive targeting approach covering two closely spaced but separate regions of a viral genome sequence, i.e., N-gene (nucleocapsid phosphoprotein). This ensures minimal false-negative outcome from the currently developed assay even if a single segment of SARS-CoV-2 genome undergoes mutation. Briefly, the developed platform can provide rapid detection of SARS-CoV-2 viral RNA using a lateral flow assay (LFA) platform when combined with suitably designed antisense oligonucleotides (ASO) that target the N-gene of SARS-CoV-2 (Fig. 1 A). The ASOs selectively recognize its target RNA sequence and demonstrate a change in color in the test line of the lateral flow strip, chemically augmented using gold nanoparticles (AuNPs) (Fig. 1A). The capability of the developed LFA has been confirmed using 30 COVID positive and 30 COVID negative patient samples validated by TaqPath RT–PCR test. Our findings showed that the assay could differentiate among positive and negative COVID-19 samples within less than 10 min after sample incubation, with an analytical limit of detection (LOD) of 0.02 copies/μL. Additionally, the sample-to-answer approach offers several advantages of being user-friendly, low-cost, and portable for POC diagnostics. Thus, the proposed approach has a high potential for intensive management of hospitalized patients, which is critical for avoiding relapse and controlling the current outbreak of COVID-19. Furthermore, the same platform can be adapted quickly using minimal modification to the antisense oligonucleotides for the detection of other emerging infectious diseases.
Fig. 1.
(A) Workflow of the direct lateral flow assay (LFA) platform; Optimization of LFA. (B) Results from SARS-CoV-2 genomic RNA (10 copies/μL) and RT-LAMP mediated LFA (left); results from RNA isolated via NAP-10 column and RT-LAMP mediated LFA (middle) and results from RNA isolated via NAP-10 column and direct LFA (right).
2. Results and discussion
We have recently developed four of the ASOs targeted towards SARS-CoV-2 N-gene. These ASOs are used to cap gold nanoparticles (AuNPs) which were then successfully used to selectively detect SARS-CoV-2 RNA by both photophysical (Moitra et al., 2020) and electrochemical (Alafeef et al., 2020a) means. However, it was realized that only two of the ASOs among the four have optimum RNA binding capability and hence AuNPs conjugated with those two ASOs were used to develop a nano-amplified colorimetric test for COVID-19 diagnosis (Alafeef et al., 2021). A comparative analysis of previously published COVID-19 detection technologies from our group is shown in Table S3. However, there is an immediate unfulfilled requirement for a point-of-care diagnostic test which can be implemented immediately without the use of any equipment but simultaneously provide rapid and accurate result with reasonable sensitivity and specificity. Herein we intend to develop a lateral flow assay which can alleviate these problems associated with rapid diagnosis of COVID-19.
2.1. Design of ASOs as lateral flow assay integratable probes for targeting SARS-CoV-2 viral RNA
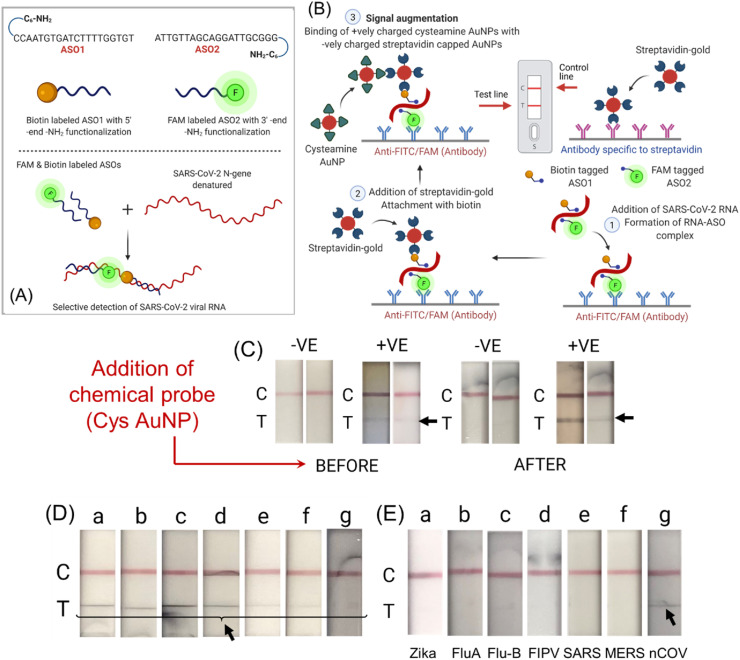
Two ASOs were designed to target two closely spaced but separate regions of N-gene sequence of SARS-CoV-2 isolate 2019-nCoV/USA-WA1-A12/2020. The ASOs were designed to target the N-gene sequence of SARS-CoV-2 as the analytical sensitivity for N-gene (8.3 copies per reaction) was found to be relatively low compared to RdRP and E genes (3.6 and 3.9 copies per reaction). Furthermore, we were also interested in improving the viability of the assay, which we realized, can be improved if a mostly conserved region of N-gene sequence can be targeted by the ASO probes. Recently, the genetic surveillance of SARS-CoV-2 strains circulating around the world has revealed a number of variants with one or more mutations that may affect detection by nucleic acid-based testing methods. We have listed down the currently known major SARS-CoV-2 variants of concern (including delta and omicron) and their associated N protein mutations (Table S4). It was observed that the currently known mutations associated with SARS-CoV-2 variants mostly occur at 39–41, 93–101, 606–617, 702–704 and 1131–1133 of N-gene sequence (Internet Resource, 2021). However, our designed ASOs uniquely target regions of the N gene (ASO1 starting 421 and ending with 440; ASO2 starting 443 and ending with 462) which is unaffected by the N-gene mutations among the current circulating variants. Thus, our assay is capable of detecting major variants of concerns or variants of interesting (including delta and omicron) without any deleterious effect on the assay performance. Furthermore, the mutations in other regions of the genome, such as in the spike protein or envelope protein, should have no impact on the performance of our assay. It was expected that 5′- end of the first ASO strand will come closer to the 3′- end of the second ASO strand only in presence of the target SARS-CoV-2 RNA sequence. These two ASOs, ASO1 and ASO2, are therefore labeled with either of biotin and 6-FAM at their 5′- and 3′- end to be integrated with the lateral flow approach (Fig. 2 A). Briefly, amine terminated six carbon spaced ASOs (25 μM) were reacted with 6-FAM (30 μM) by EDC coupling reaction overnight at room temperature in dark to get FAM labeled ASOs. Simultaneously, amine terminated six carbon spaced ASOs (25 μM) were mixed EDC/NHS/DMAP (37.5/37.5/0.25 μM respectively) along with biotin (30 μM) overnight at room temperature to get biotin labeled ASOs. All the ASOs after conjugation were purified by sephadex G-25 column (Fig. S1) using RNase free water as eluent to remove unbounded FAM and biotin (Ostadhossein et al., 2020; Srivastava et al., 2020). It was observed that under identical condition, the purified fraction of ASO2 conjugated with 6-FAM (ASO2-F) showed increased absorbance at 480 nm (Fig. S2) when compared to the fraction where ASO1 was conjugated with 6-FAM. This might be due to the presence of increased amount of ASO2-F in the purified fraction. We therefore chose ASO2-F as the preferred FAM conjugated ASO and ASO1 conjugated with biotin (ASO1-B) as the preferred biotinylated ASO to counter ASO2-F during the detection mechanism. Thus, ASO1-B and ASO2-F were subsequently used for sensing purposes using a lateral flow strip as shown in Fig. 2A.
Fig. 2.
(A) ASOs functionalized differentially with their sequences along with a schematic representation of labeling them with FAM and biotin. The figure demonstrates the proposed concept behind the targeting ability of the labeled ASOs to target gene sequence to enable visual detection of SARS-CoV-2. (B) Working principle of the designed nucleic acid based lateral flow assay. (C) Results before and after addition of cysteamine AuNPs. (D) Changes in the test line at different concentrations of the SARS- CoV-2 genomic RNA (a: 67250 copies/μL; b: 3362 copies/μL; c: 168 copies/μL; d: 8 copies/μL; e: 0.42 copies/μL; f: 0.02 copies/μL; g: 0.001 copies/μL). (E) Cross-reactivity of the designed LFA platform using different synthetic RNAs from various microorganisms (a: Zika; b: Influenza A; c: Influenza B; d: feline infectious peritonitis virus (FIPV); e: SARS-CoV; f: MERS-CoV and g: SARS-CoV-2).
2.2. Lateral flow assay (LFA) design and implementation
As a proof-of-concept experiment, genomic SARS-CoV-2 viral RNA was first reversed transcribed to cDNA and then amplified using suitably designed LAMP primers from a previous work (Alafeef et al., 2021). The amplified viral DNA was then mixed with FAM and biotin labeled ASOs and allowed to incubate at 65 °C for 5 min. The incubation ensures unfolding of DNA and optimum hybridization with the ASO strands. The resulting mixture (20 μL) along with running buffer (85 μL) was then added to a lateral flow strip. The result is available after 10 min and can be read with naked eyes. As the sample flows through the test strips FAM/biotin labeled ASOs bind to specific target SARS-CoV-2 viral gene. The test line (T) immobilized with anti-FAM antibody, captures this assembly using the FAM-labeled ASO 2. The streptavidin coated gold nanoparticles are then attracted by the biotin labeled ASO1 leading to the formation of a faint red T line (Fig. 2B). The biotin present at the control line (C) captures the remaining unreacted streptavidin-coated gold nanoparticles, which confirms that the lateral flow system is working properly (Fig. 2B).
2.3. RNA extraction-free LFA approach
Traditional nucleic acid detection approaches include the extraction and purification of nucleic acids from clinical samples. This step is generally time-consuming and expensive and requires advanced infrastructural facilities. Furthermore, other noncommercial nucleic acid separation methods, such as using a silica gel matrix or magnetic beads, need extensive skill that make it challenging to employ for localized POC screening. The complex nature of the current nucleic acid isolation techniques makes them unsuitable for use outside complex lab environments. A more direct approach, however, is required to improve the widespread availability of COVID-19 diagnosis kits, particularly in non-laboratory contexts such as port of entry and transit points (airports, railway stations and bus terminals), supermarkets, clinics, hospitals, urgent care facilities, and remote research organizations. To realize such an approach, herein, we used an RNA extraction-free technique that utilizes a Sephadex G25 size exclusion column (NAP-10) (Fig. S3) for rapid SARS-CoV-2 viral RNA isolation from de-identified clinical nasal swab samples. Briefly, NAP-10 column was equilibrated as per the manufacturer's protocol. The nasal/nasopharyngeal swab samples were stored in 1 mL VTM to prevent the virus from denaturation. For our assay, we used 40 μL of the VTM containing SARS-CoV-2 virus, mixed it with 20 μL of guanidine isothiocyanate containing lysis buffer and added to the NAP-10 column. RNase free water (1 mL) was added to the column and the eluted liquid containing RNA was collected in a low binding RNase free centrifuge tube. To confirm the isolation of target SARS-CoV-2 viral RNA through this process, we tested the presence of RNA through two separate RT-LAMP based techniques. Both of these techniques, fluorescence-based RT-LAMP assay (Figs. S4a and b) and nano-amplified colorimetric test (Figs. S4c and d), verified the presence of SARS-CoV-2 RNA in the eluent of NAP-10 column.
Next, the isolated RNA is reversed transcribed to cDNA and then amplified using suitably designed LAMP primers. The amplified DNA is then added to FAM and biotin labeled ASOs and allowed to incubate for 5 min at room temperature. The resulting mixture (20 μL) along with running buffer (85 μL) is then added to a lateral flow strip and result is read with naked eyes after 10 min (Fig. 1B). The mechanism of interaction among ASO1-B, ASO2-F, SARS-CoV-2 RNA, anti-FAM antibody and streptavidin coated gold particles over the LFA strip are the same as discussed above. We also compared the efficiency of currently developed LFA approach towards its selective detection of SARS-CoV-2 when (i) genomic RNA was amplified by aforementioned RT-LAMP method and (ii) RNA isolated from patient samples directly using NAP-10 column and then amplified with RT-LAMP approach (Fig. 1B). It was observed that the RNA isolated from NAP-10 column showed a comparable response as the genomic RNA, indicating the effective use of NAP-10 column towards the detection of RNA using the developed LFA approach.
2.4. Visual detection with a lateral flow strip
Our ultimate intention is to develop a LFA approach which is devoid of any amplification step. Hence, having realized the NAP-10 column approach would provide us similar sensitivity like the commercial kits, we have used the NAP-10 column isolated RNA directly for the LFA mediated SARS-CoV-2 detection. Briefly, isolated RNA is directly added to FAM and biotin labeled ASOs and allowed to incubate for 5 min at room temperature. The remaining steps and the principle of operation of the LFA is same as discussed above. It was observed that the direct LFA (Fig. 1B) i.e., RNA isolated using NAP-10 column without any amplification step showed comparable result (faint test line) when compared to RT-LAMP mediated LFA. Therefore, our approach can efficiently be used for diagnosing COVID-19 without any need for RNA extraction as well as amplification.
2.5. Signal augmentation
It was realized that the test line band is quite faint in intensity and hence there is a necessity to amplify its signal by other means. We hypothesized that the addition of positively charged molecules might increase the test line band intensity because of their inherent interaction with the negatively charged streptavidin coated gold particles (Kumar et al., 2020; Schwartz-Duval et al., 2020). Accordingly, we introduced several cationic nanoparticles (e.g. cysteine capped AuNPs, cysteamine capped AuNPs, Tris (2-aminoethyl)amine) capped AuNPs) as well as cationic small molecules (e.g. methylene blue) to bind with the streptavidin coated gold nanoparticles accumulated on the test line as shown in (Fig. S5). The complete working principle of the developed LFA platform has been schematically represented in Fig. 2B. It is evident that the visibility and contrast of the test line (T) has been increased significantly when cysteamine capped gold nanoparticles (Cyst-AuNPs) were used as signal enhancer molecule as shown in Fig. 2C. This is probably due to the strong electrostatic interaction between the positively charged Cyst-AuNPs with negatively charged streptavidin coated AuNPs. It only takes 10 min to complete the signal augmentation process with the lateral flow strip, from the loading of Cyst-AuNPs to signal readout.
2.6. Raman spectroscopy
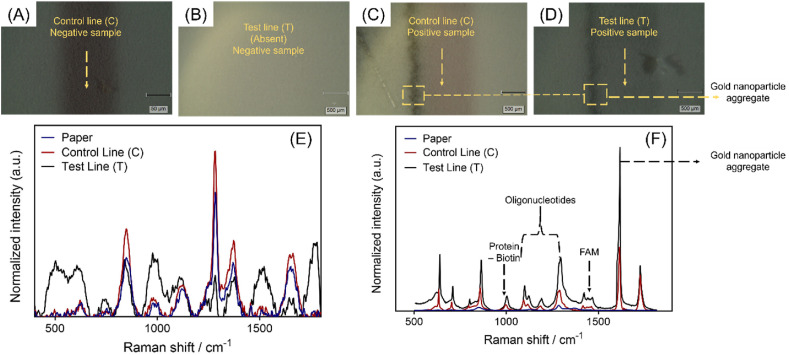
Raman spectroscopy was employed to characterize the lateral flow strips. Fig. 3 A and Fig. 3B depicts a Raman microscope image of the control line (C) and test line (T) respectively from a LFA strip ran with negative sample. It can be observed from the microscopic image that the test line (T) is absent from the lateral flow strip and only the control line (C) is visible. On the other hand, Raman microscopic image of control line (C) and test line (T) from a lateral flow strip tested with a SARS–CoV–2 positive sample are represented in Fig. 3C–D. As we can see from the Raman spectral analysis of the LFA strip tested with a negative clinical sample, there is no clear differentiation between normalized Raman intensity values obtained from the paper, control line (C), and the test line (T) (Fig. 3E). However, the lateral flow strip tested with a positive sample showed much higher as well as clear differentiation for the Raman intensities between paper, control line (C), and the test line (T) (Fig. 3F). In this case, the characteristic peaks were also observed at 1000 cm−1, 1099 cm−1, 1217 cm−1 1421 cm −1, and 1617 cm−1 which can be assigned to the presence of biotin, oligonucleotides and gold nanoparticle aggregates (Gearheart et al., 2001; Huang et al., 2019; Torreggiani and Fini, 1998). Further, the intensity of the Raman peak increased sharply at 1617 cm−1 which could be due to the enhanced surface-enhanced Raman scattering (SERS) caused by the gold nanoparticle aggregates formed due to the electrostatic interaction between negatively charged streptavidin capped AuNPs and positively charged cysteamine coated AuNPs.
Fig. 3.
Raman microscopic images of lateral flow strip tested with (A-B) negative and (C-D) positive clinical sample respectively. Raman spectra of a lateral flow strip tested with a (E) negative and (F) positive clinical sample.
2.7. Sensitivity and specificity
Once we established that RNA isolated through NAP-10 column could be directly utilized (without RT-LAMP) for detecting SARS-CoV-2, we further validated the LFA platform and tested it for COVID-19 diagnosis. To establish the sensitivity, we utilized SARS-CoV-2 genomic RNA to investigate the LFA system's analytical limit of detection for COVID-19 diagnosis. Briefly the LFA strips were tested using, 10-fold serial dilutions of the SARS-CoV-2 genomic RNA with concentrations ranging from 67250 copies/μL to 0.001 copies/μL as shown in Fig. 2D. It was observed that the test provided a visible detectable signal even when SARS-CoV-2 genomic RNA quantities are as low as 0.02 copies/μL.
It is also evident from the results represented in Fig. 2D that a clear test line with no change in band intensity was observed till 8 copies/μL. This might be due to the efficient RNA-ASO hybridization which is further chemically augmented by the cysteamine coated AuNPs. While visual distinction of the bands was not obvious at a higher concentration, the change in band intensity was found at lower viral RNA concentrations with limit of detection of the assay as 0.02 copies/μL. This observation corroborates with the findings in Fig. 4 A where we did not observe any significant difference in test line while evaluating the patients’ samples. It is worth mentioning here that changing the RNA concentration from 67250 copies/μL to 8 copies/μL (Fig. 2D) should also have an impact on the LFA band intensity, however, these variations are not captured by human eye and also not captured efficiently using the digital camera as reflected in the photographs. Furthermore, to test specificity of the developed LFA approach towards the selective detection of SARS-CoV-2, SARS-CoV-2 genomic RNA and 6 other viral RNA types of common pathogenic microorganisms (Zika, Influenza A, Influenza B, FIPV, SARS-CoV, and MERS-CoV), obtained from BEI resources, NIAID, NIH, were used. No positive test lines (red T line) were seen in any case other than for SARS-CoV-2 genomic RNA (Fig. 2E). Therefore, the results demonstrated that the developed methodology for SARS-CoV-2 had little to no cross-reactivity with other infectious pathogens.
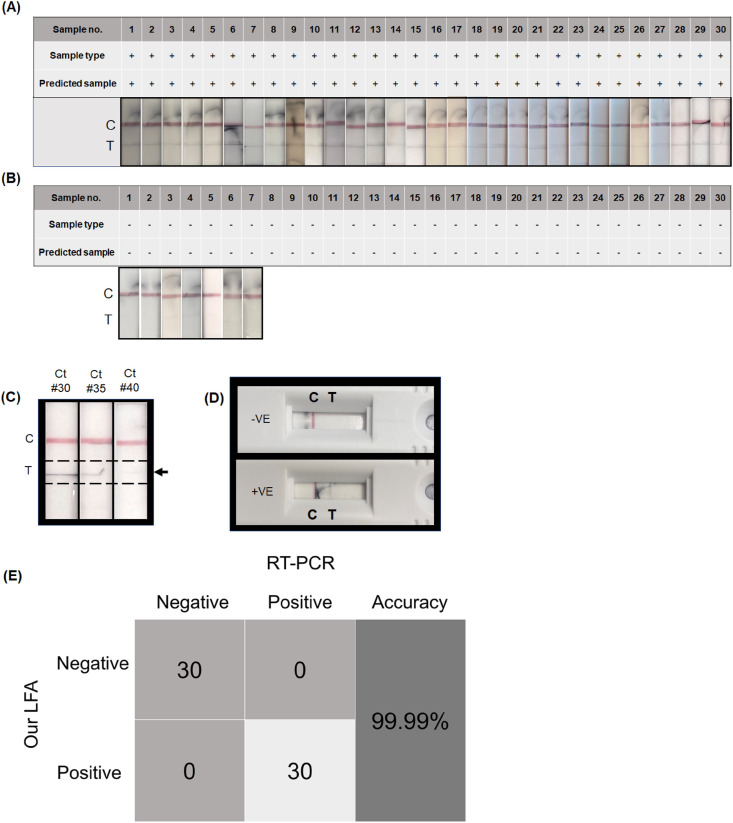
Fig. 4.
Diagnostic results of de-identified clinical nasal swab samples. (A) Results from 30 COVID-19 positive samples without RNA extraction and amplification; (B) Diagnostic results of 30 negative de-identified clinical nasal swab samples without RNA extraction and amplification; (C) Analytical performance of the LFA with samples having equivalent Ct values of 30, 35 and 40 respectively. Here the genomic SARS-CoV-2 RNA was diluted to prepare the samples. (D) Schematic representation of diagnostic results using the developed lateral flow assay (clinical -ve nasal swab sample #6 and clinical + ve nasal swab sample #2); (E) Confusion matrix analysis of clinical nasal swab samples without extraction of RNA and amplification. Clinical reliability of the developed LFA was validated using the optimized conditions.
2.8. Clinical application of the LFA
Finally, the performance of the LFA system was evaluated to detect SARS-CoV-2 in de-identified clinical nasal swab samples. Briefly, nasal swab samples from symptomatic as well as asymptomatic subjects were obtained and stored in viral transport media (VTM) until further use. The results were evaluated using a gold standard RT-PCR kit for SARS-CoV-2 detection in order to prove repeatability and demonstrate effective detection of SARS-CoV-2 in clinical samples. The quantity of SARS-CoV-2 RNA contained in each clinical sample is shown in Table S5 as copies/μL. The conversion of Ct number to copy number has been performed with the following equation: Copy number/μL = Fig. 4A–B demonstrates diagnostics results of 30 positive and 30 negative de-identified clinical nasal swab samples without RNA extraction and amplification. It was observed that the positive samples show both a red C and T lines (Fig. 4A) whereas only the red C line (Fig. 4B) is visible in case of negative samples. To evaluate the performance of the developed lateral flow assay in samples having higher Ct values, we used genomic SARS-CoV-2 RNA and diluted it to a pre-determined copy numbers equivalent to Ct values of 30, 35 and 40 respectively. The samples with Ct values of 30, 35 and 40 will have ∼235, ∼32 and ∼4 copies/μL of SARS-CoV-2 RNA. It was seen that a band on test line was observed for the samples with Ct values of 30 and 35, whereas the band seems to be quite faint for Ct value 40 (Fig. 4C). Fig. 4D shows the complete LFA strip integrated in a cassette. The confusion matrix shown in Fig. 4E demonstrates the developed LFA system's performance in diagnosing COVID-19 in clinical samples. It can be seen that the developed LFA was able to assign the COVID-19 clinical samples to their respective positive and negative groups with almost 99.9% specificity, accuracy and sensitivity.
3. Conclusion
Overall, we describe herein an easy-to-use, rapid nucleic acid based LFA system for the POC detection of SARS-CoV-2. The developed approach utilizes four steps which are schematically represented in Fig. 1A. These include: (a) sample nasal swab collection from the patient and RNA isolation through NAP-10 column; (b) incubation of the isolated RNA with biotin and FAM labeled ASOs; (c) sample loading onto the lateral flow strip and signal augmentation with cationic cysteamine capped AuNPs; and (d) visual read out as per the kit instruction. The complete working principle of the developed LFA platform has been represented in Fig. 2B. The diagnosis can be completed by naked eye within 30 min of operation. The assay has been found to be quite selective towards SARS-CoV-2 with minimal crosstalk with other pathogenic viruses. Moreover, the assay achieved an accuracy and specificity of almost 99.99% when tested with 60 clinical samples with a LOD of 0.02 copies/μL. We envisage that the newly developed inexpensive RNA extraction and amplification free lateral flow assay could harbinger a unique platform for rapid and POC diagnosis of COVID-19. A long-term vision for this project is to develop a multiplexed Flu/COVID-19 home use test (HUT) where samples will be collected by the consumers immediately prior to conducting the assay. The final fully integrated version of the test will therefore not require sample storing in VTM or other stabilizing media. Our future clinical studies will include useability and direct testing of samples, without storing them into VTM. The useability and patient testing are out of the scope of this rapid communication reporting the feasibility of the concept. Moreover, it can also be proposed that developed LFA method could be expanded beyond COVID-19 detection, simply by altering its targeting antisense oligonucleotides, to become a global health technology that contributes to providing low-cost diagnostics (Table S6) to a variety of emerging infectious diseases around the world in a portable (point-of-care) and inexpensive way.
CRediT authorship contribution statement
Ketan Dighe: Conceptualization, Methodology, Investigation, Writing – original draft. Parikshit Moitra: Conceptualization, Methodology, Investigation, Writing – original draft. Maha Alafeef: Conceptualization, Writing – original draft. Nivetha Gunaseelan: Investigation. Dipanjan Pan: Supervision, Conceptualization, Writing – review & editing.
Declaration of competing interest
The authors declare the following competing financial interest(s): Prof. Pan founded or co-founded three university-affiliated start-up companies. However, this work was not supported by any of these entities.
Acknowledgements
The authors gratefully acknowledge the receipt of funding from the National Institute of Biomedical Imaging and Bioengineering (NIBIB) R03EB028026, R03 EB028026-02S2, and R03 EB028026-02S1, the University of Maryland Baltimore (UMB), and University of Maryland Baltimore County (UMBC).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113900.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Alafeef M., Dighe K., Pan D. Label-free pathogen detection based on yttrium-doped carbon nanoparticles up to single-cell resolution. ACS Appl. Mater. Interfaces. 2019;11:42943–42955. doi: 10.1021/acsami.9b14110. [DOI] [PubMed] [Google Scholar]
- Alafeef M., Dighe K., Moitra P., Pan D. Rapid, ultrasensitive, and quantitative detection of SARS-CoV-2 using antisense oligonucleotides directed electrochemical biosensor chip. ACS Nano. 2020;14:17028–17045. doi: 10.1021/acsnano.0c06392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alafeef M., Moitra P., Pan D. Nano-enabled sensing approaches for pathogenic bacterial detection. Biosens. Bioelectron. 2020;165 doi: 10.1016/j.bios.2020.112276. [DOI] [PubMed] [Google Scholar]
- Alafeef M., Moitra P., Dighe K., Pan D. RNA-extraction-free nano-amplified colorimetric test for point-of-care clinical diagnosis of COVID-19. Nat. Protoc. 2021;16:3141–3162. doi: 10.1038/s41596-021-00546-w. [DOI] [PubMed] [Google Scholar]
- Binnicker M.J. Challenges and controversies to testing for COVID-19. J. Clin. Microbiol. 2020;58:1–7. doi: 10.1128/JCM.01695-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W., Yang J., Fan G., Xu L., Zhang B., Liu R. Chest CT findings of coronavirus disease 2019 (COVID-19) J. College Phys. Surg. Pak. 2020;30 doi: 10.29271/jcpsp.2020.Supp1.S53. S53–S55. [DOI] [PubMed] [Google Scholar]
- Carter L.J., Garner L.v., Smoot J.W., Li Y., Zhou Q., Saveson C.J., Sasso J.M., Gregg A.C., Soares D.J., Beskid T.R., Jervey S.R., Liu C. Assay techniques and test development for COVID-19 diagnosis. ACS Cent. Sci. 2020;6:591–605. doi: 10.1021/acscentsci.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherkaoui D., Huang D., Miller B.S., Turbé V., McKendry R.A. Harnessing recombinase polymerase amplification for rapid multi-gene detection of SARS-CoV-2 in resource-limited settings. Biosens. Bioelectron. 2021;189 doi: 10.1016/j.bios.2021.113328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H., Liu Y., Lv M., Zhong J. A case report of COVID-19 with false negative RT-PCR test: necessity of chest CT. Jpn. J. Radiol. 2020;38:409–410. doi: 10.1007/s11604-020-00967-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W., Newbigging A.M., Le C., Pang B., Peng H., Cao Y., Wu J., Abbas G., Song J., Wang D.B., Cui M., Tao J., Tyrrell D.L., Zhang X.E., Zhang H., Le X.C. Molecular diagnosis of COVID-19: challenges and research needs. Anal. Chem. 2020;92:10196–10209. doi: 10.1021/acs.analchem.0c02060. [DOI] [PubMed] [Google Scholar]
- Gearheart L.A., Ploehn H.J., Murphy C.J. Oligonucleotide adsorption to gold nanoparticles: a surface-enhanced Raman spectroscopy study of intrinsically bent DNA. J. Phys. Chem. B. 2001;105:12609–12615. doi: 10.1021/jp0106606. [DOI] [Google Scholar]
- Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., Marrama D., de Silva A.M., Frazier A., Carlin A.F., Greenbaum J.A., Peters B., Krammer F., Smith D.M., Crotty S., Sette A. Targets of T Cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501. doi: 10.1016/j.cell.2020.05.015. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Luo L., Luo Z., Lyu J., Ng M., Shen X., Wen Z. Diagnostic performance between CT and initial real-time RT-PCR for clinically suspected 2019 coronavirus disease (COVID-19) patients outside Wuhan, China. Respir. Med. 2020;168 doi: 10.1016/j.rmed.2020.105980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Guo H., Zhou P., Shi Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2020;19:141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J.A., Mousavi M.Z., Zhao Y., Hubarevich A., Omeis F., Giovannini G., Schütte M., Garoli D., de Angelis F. SERS discrimination of single DNA bases in single oligonucleotides by electro-plasmonic trapping. Nat. Commun. 2019;10:5321. doi: 10.1038/s41467-019-13242-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Fan C., Li M., Nie H.-L., Wang F.-B., Wang H., Wang R., Xia J., Zheng X., Zuo X., Huang J. COVID-19: a call for physical scientists and engineers. ACS Nano. 2020;14:3747–3754. doi: 10.1021/acsnano.0c02618. [DOI] [PubMed] [Google Scholar]
- Internet Resource . 2021. Detection of Variant SARS-CoV-2 Strains on the ePlex® RP2 Panel.https://genmarkdx.com/detection-of-variant-sars-cov-2-strains-on-eplex-rp2-panel/ [Google Scholar]
- Kumar K., Moitra P., Bashir M., Kondaiah P., Bhattacharya S. Natural tripeptide capped pH-sensitive gold nanoparticles for efficacious doxorubicin delivery both: in vitro and in vivo. Nanoscale. 2020;12:1067–1074. doi: 10.1039/c9nr08475d. [DOI] [PubMed] [Google Scholar]
- Liu G., Rusling J.F. COVID-19 antibody tests and their limitations. ACS Sens. 2021;6:593–612. doi: 10.1021/acssensors.0c02621. [DOI] [PubMed] [Google Scholar]
- Lukas H., Xu C., Yu Y., Gao W. Emerging telemedicine tools for remote covid-19 diagnosis, monitoring, and management. ACS Nano. 2020;14:16180–16193. doi: 10.1021/acsnano.0c08494. [DOI] [PubMed] [Google Scholar]
- Moitra P., Alafeef M., Dighe K., Frieman M.B., Pan D. Selective naked-eye detection of SARS-CoV-2 mediated by N gene targeted antisense oligonucleotide capped plasmonic nanoparticles. ACS Nano. 2020;14:7617–7627. doi: 10.1021/acsnano.0c03822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moitra P., Alafeef M., Dighe K., Ray P., Chang J., Thole A., Punshon-Smith B., Tolosa M., Ramamurthy S.S., Ge X., Frey D.D., Pan D., Rao G. Rapid and low-cost sampling for detection of airborne SARS-CoV-2 in dehumidifier condensate. Biotechnol. Bioeng. 2021:3029–3036. doi: 10.1002/bit.27812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostadhossein F., Sar D., Tripathi I., Soares J., Remsen E.E., Pan D. Oligodots: structurally defined fluorescent nanoprobes for multiscale dual-color imaging in vitro and in vivo. ACS Appl. Mater. Interfaces. 2020;12:10183–10192. doi: 10.1021/acsami.0c00705. [DOI] [PubMed] [Google Scholar]
- Pokhrel P., Hu C., Mao H. Detecting the coronavirus (CoVID-19) ACS Sens. 2020;5:2283–2297. doi: 10.1021/ACSSENSORS.0C01153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi N., Cortade D.L., Ng E., Wang S.X. Diagnostics for SARS-CoV-2 detection: a comprehensive review of the FDA-EUA COVID-19 testing landscape. Biosens. Bioelectron. 2020;165 doi: 10.1016/j.bios.2020.112454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong Y., Wang F., Tian J., Liang X., Wang J., Li X., Zhang D., Liu J., Zeng H., Zhou Y., Shi Y. Clinical and CT features of mild-to-moderate COVID-19 cases after two sequential negative nucleic acid testing results: a retrospective analysis. BMC Infect. Dis. 2021;21:1–10. doi: 10.1186/s12879-021-06013-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SARS-CoV-2 Viral Mutations . FDA press Release; 2021. Impact on COVID-19 Tests. [Google Scholar]
- Schwartz-Duval A.S., Konopka C.J., Moitra P., Daza E.A., Srivastava I., Johnson E.v., Kampert T.L., Fayn S., Haran A., Dobrucki L.W., Pan D. Intratumoral generation of photothermal gold nanoparticles through a vectorized biomineralization of ionic gold. Nat. Commun. 2020;11:4530. doi: 10.1038/s41467-020-17595-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava I., Misra S.K., Bangru S., Boateng K.A., Soares J.A.N.T., Schwartz-Duval A.S., Kalsotra A., Pan D. Complementary oligonucleotide conjugated multicolor carbon dots for intracellular recognition of biological events. ACS Appl. Mater. Interfaces. 2020;12:16137–16149. doi: 10.1021/acsami.0c02463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torreggiani A., Fini G. The binding of biotin analogues by streptavidin: a Raman spectroscopic study. Biospectroscopy. 1998;4:197–208. doi: 10.1002/(SICI)1520-6343(1998)4:3%3C197::AID-BSPY5%3E3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Udugama B., Kadhiresan P., Kozlowski H.N., Malekjahani A., Osborne M., Li V.Y.C., Chen H., Mubareka S., Gubbay J.B., Chan W.C.W. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020;14:3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- Valera E., Jankelow A., Lim J., Kindratenko V., Ganguli A., White K., Kumar J., Bashir R. COVID-19 point-of-care diagnostics: present and future. ACS Nano. 2021;15:7899–7906. doi: 10.1021/acsnano.1c02981. [DOI] [PubMed] [Google Scholar]
- Vandenberg O., Martiny D., Rochas O., van Belkum A., Kozlakidis Z. Considerations for diagnostic COVID-19 tests. Nat. Rev. Microbiol. 2021;19:171–183. doi: 10.1038/s41579-020-00461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Hozumi Y., Yin C., Wei G.W. Decoding SARS-CoV-2 transmission and evolution and ramifications for COVID-19 diagnosis, vaccine, and medicine. J. Chem. Inf. Model. 2020;60:5853–5865. doi: 10.1021/acs.jcim.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yüce M., Filiztekin E., Özkaya K.G. COVID-19 diagnosis —a review of current methods. Biosens. Bioelectron. 2021;172 doi: 10.1016/j.bios.2020.112752. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.