Abstract
An H1N2 influenza virus was isolated from a pig during an outbreak of respiratory disease and abortion on an Indiana farm in November 1999. Results of phylogenetic analyses indicate that this virus is a reassortant between a recent classical H1 swine virus and the reassortant H3N2 viruses that have emerged among American pigs since 1998.
H3N2 influenza viruses have caused widespread outbreaks of respiratory disease and, in some cases, abortion among pigs throughout the major swine production regions of the United States since 1998. These are reassortant viruses containing hemagglutinin (HA), neuraminidase (NA), and PB1 polymerase genes of human influenza virus origin; matrix (M), nonstructural (NS), and nucleoprotein (NP) genes of classical swine virus origin; and PA and PB2 polymerase genes of swine or avian virus origin (16a, 43). The emergence of these viruses has been remarkable because, in contrast to Europe and Asia, where human H3N2 (4, 17, 23, 24, 29, 36, 37) and avian (2, 3, 5, 19, 31, 33, 39), influenza viruses have been regularly isolated from pigs, influenza among American pigs has historically been due almost exclusively to infection with classical H1N1 swine influenza viruses (7, 8, 13). We now report the isolation of an H1N2 influenza virus, A/Swine/Indiana/9K035/99 (Sw/IN/99), that appears, based upon results of phylogenetic analyses, to be a second-generation reassortant between one of the recent reassortant H3N2 viruses and a classical H1 swine influenza virus.
Influenza-like illness spread throughout the farm of origin of Sw/IN/99 over a 6-week period, beginning in November 1999. Clinical signs in the pigs included temperatures up to 40.6°C, lethargy, lack of appetite, and dyspnea. In addition, 20 of approximately 600 sows on the farm aborted. This herd had been assembled for 4 years, but there had been no prior influenza-like illness and the pigs had not been vaccinated against influenza virus. The Sw/IN/99 virus was isolated in Madin-Darby canine kidney (MDCK) cells from lung tissue from a sow that died during the outbreak. The subtype of Sw/IN/99 was determined to be H1N2 by hemagglutination-inhibition and neuraminidase-inhibition assays (18) using subtype-monospecific antisera and by reverse transcription (RT)-PCR (see below). There was no detectable H3 hemagglutination-inhibition or N1 neuraminidase-inhibition activity in the MDCK cell culture supernatant nor could H3 HA or N1 NA gene sequences be amplified by RT-PCR.
To genotype Sw/In/99 and understand its evolutionary origin, the full-length protein coding region sequences of all eight of its viral RNA segments were determined by cycle sequencing (ABI Big Dye; PE Applied Biosystems, Foster City, Calif.) following amplification by RT-PCR. Amplifications were conducted with avian myeloblastosis virus reverse transcriptase (Promega Corporation, Madison, Wis.) and Pfu polymerase (Stratagene, La Jolla, Calif.) as suggested by the enzyme suppliers, except that the RT reactions were performed using 1 μg of primer per reaction and reaction conditions of 49.5°C for 60 min for the polymerase genes and 48.5°C for 45 min for the remaining genes. Amplifications of the NA, NP, M, and NS genes were accomplished by using the SZANA+/−, SZANP+/−, SZAM+/−, and SZANS+/− primers described by Zou (44). Amplifications of the HA and polymerase gene segments were conducted with the following primers: H1HA-1.1F (5′-AGCAAAAGCAGGGGAAAATAA-3′) and H1HA-1770R (5′-CAAGGGTGTTTTTTCTCATGTCTC-3′); HPA-1F (5′-AGCAAAAGCAGGTACTGAT-3′) and HPA-2202R (5′-GGATAGCAAATAGTAGCATTGCC-3′); HPB1-5F (5′- AAAGCAGGCAAACCATTTGAATGGA-3′) and HPB1-2340R (5′-GTAGAAACAAGGCATTTTTTCATG-3′); HPB2-8F (5′-GCAGGTCAATTATATTGAATATGGAAA-3′) and HPB2-2337R (5′-GAAACAAGGTCGTTTTTAAACTATTCG-3′) (F denotes a forward primer, and R denotes a reverse primer.) Sequence comparisons to reference viruses were conducted by using DNASTAR software (version 4.0 for Win32). The phylogenetic relationships of Sw/IN/99 to selected reference strains were estimated from the nucleotide sequences by the method of maximum parsimony (PAUP software, v.4.0b2; David Swofford, Smithsonian Institution) using the tree-bisection-reconnection branch swapping algorithm and with the MULTREES option in effect. The sequences of the Sw/IN/99 genes were analyzed in relation to the available reference virus sequences in GenBank, with the “gaps treated as missing” PAUP rule in effect.
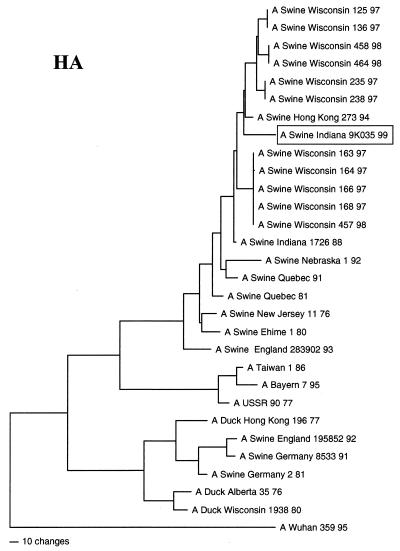
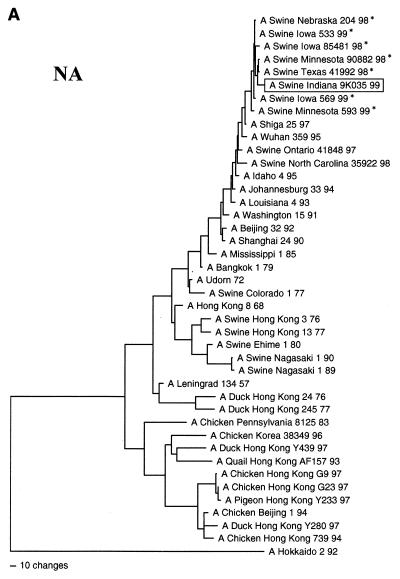
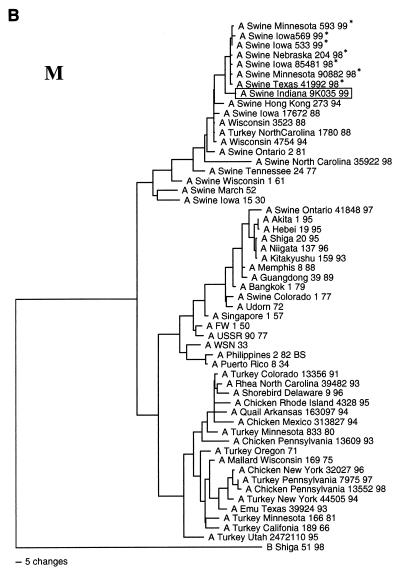
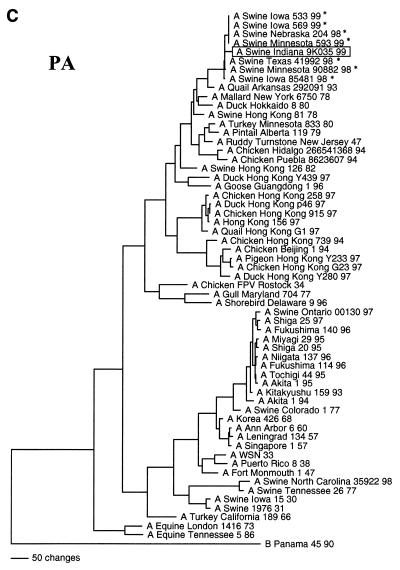
The genotype of Sw/IN/99 was initially inferred by pairwise comparisons of each gene segment to the sequences of reference influenza viruses available in GenBank. Table 1 lists the reference viruses with the highest level of sequence identity to Sw/IN/99 for each gene segment. (It is important to note that to clarify the overall genotype of Sw/IN/99, these analyses excluded viruses known to have been isolated following interspecies transmission, as well as the H3N2 reassortant viruses isolated recently from pigs in the United States [16a, 43].) These results demonstrate that Sw/IN/99 is a reassortant virus with HA, M, NS, and NP genes most closely related to genes from the swine influenza virus lineage, NA and PB1 genes most closely related to genes from the human influenza virus lineage, and PA and PB2 genes most closely related to genes from the avian influenza virus lineage. The results of phylogenetic analyses confirmed this genotype and further indicated that Sw/IN/99 most likely arose from the reassortment of a classical H1 swine influenza virus with one of the recent reassortant H3N2 swine isolates (16a, 43). Specifically, the HA gene of Sw/IN/99 is within the clade of contemporary classical swine H1N1 viruses (Fig. 1). The NA (Fig. 2A), M (Fig. 2B), PA (Fig. 2C), and NP, NS, PB1, and PB2 (data not shown) genes are all phylogenetically most closely related to the homologous genes from the H3N2 reassortant viruses isolated from pigs in the midwestern United States in 1998–1999 and only more distantly related to older human (NA and PB1), swine (M, NP, and NS), and avian (PA and PB2) virus isolates. Pairwise amino acid sequence comparison of the HA proteins of Sw/IN/99 and a representative virus from the recent classical H1N1 clade, A/Swine/Wisconsin/235/97 (27), revealed 24 amino acid differences in HA1 and 4 amino acid differences in HA2. Only 8 of the HA1 differences are in previously defined antigenic sites (6, 21, 22, 28, 32, 41). The remainder are spread throughout HA1, although none affected potential glycosylation (N∼P∼S/T∼P) (28) or previously defined receptor binding (14, 40) sites.
TABLE 1.
Sequence homology of each gene from Sw/IN/99 and reference virus sequences available in GenBank
| Gene | Nucleotide positions of Sw/IN/99 compared | Virus with the highest degree of homologya | Nucleotide sequence identity (%) | Influenza virus lineage |
|---|---|---|---|---|
| HA | 33–1,065 | A/Swine/Hong Kong/273/94 (H1N1) [U45452]b (11)c | 95.2 | Swine |
| NA | 21–1,411 | A/Shiga/25/97 (H3N2) [AF038264] (20) | 98.1 | Human |
| M | 1–975 | A/Swine/Iowa/17672/88 (H1N1) [M63522] (15) | 97.3 | Swine |
| NP | 41–1,543 | A/Swine/Iowa/17672/88 (H1N1) [M63768] (9) | 97.5 | Swine |
| NS | 22–867 | A/Swine/Hong Kong/273/94 (H1N1) [U49490] (11) | 97.3 | Swine |
| PA | 43–1,436 | A/Quail/Arkansas/292091/93 (H9N2) [AF156456] (12) | 93.7 | Avian |
| PB1 | 13–2,292 | A/Shiga/20/95 (H3N2) [U71130] (20) | 98.6 | Human |
| PB2 | 46–2,309 | A/Shorebird/Delaware/9/96 (H9N2) [AF156441] (12) | 96.0 | Avian |
To clarify the overall genotype of Sw/IN/99, these analyses excluded viruses known to have been isolated following interspecies transmission, as well as the H3N2 reassortant viruses isolated recently from pigs in the United States (16a, 43).
The numbers in brackets are the GenBank accession numbers for the reference virus sequences.
The numbers in parentheses are the references for each sequence.
FIG. 1.
Nucleotide phylogenetic tree for the HA gene of Sw/IN/99. The evolutionary relationship of this gene to HA genes from reference viruses was estimated by the method of maximum parsimony (PAUP software, v.4.0b2; David Swofford, Smithsonian Institution) using the tree-bisection-reconnection branch swapping algorithm and with the MULTREES option in effect. The tree shown represents the best (score = 1,385) of 9,698 rearrangements that were generated. Horizontal line distances are proportional to the minimum number of nucleotide changes needed to join nodes and gene sequences. The vertical lines are simply for spacing the branches and labels. The tree is rooted to an H3 HA sequence. The accession numbers for the reference virus sequences used in this analysis are available upon request.
FIG. 2.
Nucleotide phylogenetic trees for the NA (A), M (B), and PA (C) genes of Sw/IN/99. The evolutionary relationships were estimated by the method of maximum parsimony as described in the legend to Fig. 1. The score and number of rearrangements for each tree are as follows: NA, score = 2,011 of 475,664 rearrangements; M, score = 1,020 of 10,892,448 rearrangements; and PA, score = 3,850 of 2,345,030 rearrangements. The reassortant H3N2 viruses isolated from pigs in the Midwestern United States since 1998 (16a, 43) are marked by an asterisk. The NA tree is rooted to an influenza A virus of a different subtype (N1), whereas the M and PA trees are rooted to influenza B viruses.
To our knowledge, Sw/IN/99 is the first H1N2 virus isolated from a pig in the United States. H1N2 viruses have been isolated previously from pigs in France in 1987 and 1988 (10), in Japan in 1978–1980 (25, 26, 38, 42) and 1989–1992 (16, 30), and in the United Kingdom since 1994 (1, 2; J. H. Brown, Programme Abstr. Eur. Soc. Vet. Virol. Symp. Anim. Influenza Viruses, Gent, Belgium, 1999, abstr. 31. European Society for Veterinary Virology, Universiteit Gent, Gent, Belgium). Antigenic and/or genetic characterization demonstrated that the viruses from Japan and France were the products of reassortment between classical swine H1N1 viruses and human H3N2 viruses circulating in pigs (10, 16, 25, 26, 30, 38, 42). In contrast, the H1N2 viruses in the United Kingdom have been suggested to be the result of multiple reassortment events. Reassortment between human H1N1 and H3N2 viruses is thought to have created an H1N2 subtype virus, which subsequently acquired all six of its internal viral protein genes through a second reassortment with a wholly-avian H1N1 virus (16). The H1N2 viruses in Japan and the United Kingdom caused large-scale outbreaks of disease and spread throughout the swine populations of each country (1, 2, 16, 30; Brown, Programme Abstr. Eur. Soc. Vet. Virol. Symp. Anim. Influenza Viruses, 1999, abstr. 31), whereas the viruses in France apparently did not (10). Whether the Sw/IN/99 virus will spread within the regional or national swine population of the United States remains to be determined. In addition, documentation of its relative virulence and abortigenic potential compared to classical swine H1N1 and the contemporary reassortant H3N2 viruses awaits experimental infection studies. Nonetheless, the isolation of Sw/IN/99 emphasizes the potential for pigs to serve as “mixing vessel” (34, 36, 39) hosts for the generation of new reassortant genotypes of viruses. If both H1N1 and H3N2 viruses continue to cocirculate within the swine population of the United States, we should expect to isolate additional reassortant viruses with unique combinations of surface and/or internal protein genes.
Nucleotide sequence accession numbers.
The GenBank numbers assigned to the gene sequences of Sw/IN/99 determined in this study are as follows: the HA gene, AF250124; the NA gene, AF250126; the M gene, AF250125; the NP gene, AF250127; the NS gene, AF250128; the PA gene, AF250129; the PB1 gene, AF250130; the PB2 gene, AF250131. The GenBank accession numbers for all of the reference virus sequences used in the phylogenetic analyses are available upon request.
Acknowledgments
This work was supported by USDA Agricultural Experiment Station grant WIS04080 and in part by a USDA NRICGP grant.
We thank Melissa Schutten for technical assistance and Yoshihiro Kawaoka and Diane Larsen from the University of Wisconsin—Madison for reviewing the manuscript and for many helpful discussions.
REFERENCES
- 1.Brown I H, Chakraverty P, Harris P A, Alexander D J. Disease outbreaks in pigs in Great Britain due to an influenza A virus of H1N2 subtype. Vet Rec. 1995;136:328–329. doi: 10.1136/vr.136.13.328. [DOI] [PubMed] [Google Scholar]
- 2.Brown I H, Harris P A, McCauley J W, Alexander D J. Multiple genetic reassortment of avian and human influenza A viruses in European pigs, resulting in emergence of an H1N2 virus of novel genotype. J Gen Virol. 1998;79:2947–2955. doi: 10.1099/0022-1317-79-12-2947. [DOI] [PubMed] [Google Scholar]
- 3.Campitelli L, Donatelli I, Foni E, Castrucci M R, Fabiani C, Kawaoka Y, Krauss S, Webster R G. Continued evolution of H1N1 and H3N2 influenza viruses in pigs in Italy. Virology. 1997;232:310–318. doi: 10.1006/viro.1997.8514. [DOI] [PubMed] [Google Scholar]
- 4.Castrucci M R, Campitelli L, Ruggieri A, Barigazzi G, Sidoli L, Daniels R, Oxford J S, Donatelli I. Antigenic and sequence analysis of H3 influenza virus haemagglutinins from pigs in Italy. J Gen Virol. 1994;75:371–379. doi: 10.1099/0022-1317-75-2-371. [DOI] [PubMed] [Google Scholar]
- 5.Castrucci M R, Donatelli I, Sidoli L, Barigazzi G, Kawaoka Y, Webster R G. Genetic reassortment between avian and human influenza viruses in Italian pigs. Virology. 1993;193:503–506. doi: 10.1006/viro.1993.1155. [DOI] [PubMed] [Google Scholar]
- 6.Caton A J, Brownlee G G, Yewdell J W, Gerhard W. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype) Cell. 1982;31:417–427. doi: 10.1016/0092-8674(82)90135-0. [DOI] [PubMed] [Google Scholar]
- 7.Chambers T M, Hinshaw V S, Kawaoka Y, Easterday B C, Webster R G. Influenza viral infection of swine in the United States 1988–1989. Arch Virol. 1991;116:261–265. doi: 10.1007/BF01319247. [DOI] [PubMed] [Google Scholar]
- 8.Easterday B C, Hinshaw V S. Swine influenza. In: Leman A D, Straw B E, Mengeling W L, D'Allaire S D, Taylor D J, editors. Diseases of swine. Ames: Iowa State Press; 1992. pp. 349–357. [Google Scholar]
- 9.Gorman O T, Bean W J, Kawaoka Y, Donatelli I, Guo Y, Webster R G. Evolution of influenza A virus nucleoprotein genes: implications for the origins of H1N1 human and classical swine viruses. J Virol. 1991;65:3704–3714. doi: 10.1128/jvi.65.7.3704-3714.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gourreau J M, Kaiser C, Valette M, Douglas A R, Labie J, Aymard M. Isolation of two H1N2 influenza viruses from swine in France. Arch Virol. 1994;135:365–382. doi: 10.1007/BF01310021. [DOI] [PubMed] [Google Scholar]
- 11.Guan Y, Shortridge K F, Krauss S, Li P H, Kawaoka Y, Webster R G. Emergence of avian H1N1 influenza viruses in pigs in China. J Virol. 1996;70:8041–8046. doi: 10.1128/jvi.70.11.8041-8046.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guan Y, Shortridge K F, Krauss S, Webster R G. Molecular characterization of H9N2 influenza viruses: were they the donors of the “internal” genes of H5N1 viruses in Hong Kong. Proc Natl Acad Sci USA. 1999;96:9363–9367. doi: 10.1073/pnas.96.16.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hinshaw V S, Bean W J, Jr, Webster R G, Easterday B C. The prevalence of influenza viruses in swine and the antigenic and genetic relatedness of influenza viruses from man and swine. Virology. 1978;84:51–62. doi: 10.1016/0042-6822(78)90217-9. [DOI] [PubMed] [Google Scholar]
- 14.Inkster M D, Hinshaw V S, Schulze I T. The hemagglutinins of duck and human H1 influenza viruses differ in sequence conservation and in glycosylation. J Virol. 1993;67:7436–7443. doi: 10.1128/jvi.67.12.7436-7443.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ito T, Gorman O T, Kawaoka Y, Bean W J, Webster R G. Evolutionary analysis of the influenza A virus M gene with comparison of the M1 and M2 proteins. J Virol. 1991;65:5491–5498. doi: 10.1128/jvi.65.10.5491-5498.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito T, Kawaoka Y, Vines A, Ishikawa H, Asai T, Kida H. Continued circulation of reassortant H1N2 influenza viruses in pigs in Japan. Arch Virol. 1998;143:1773–1782. doi: 10.1007/s007050050415. [DOI] [PubMed] [Google Scholar]
- 16a.Karasin, A. I., M. M. Schutten, L. A. Cooper, C. B. Smith, K. Subbarao, G. A. Anderson, S. Carman, and C. W. Olsen. Genetic characterization of H3N2 influenza viruses isolated from pigs in North America, 1977–1999: evidence for wholly human and reassortant genotypes. Virus Res., in press. [DOI] [PubMed]
- 17.Katsuda K, Shirahata T, Kida H, Goto H. Antigenic and genetic analyses of the hemagglutinin of influenza viruses isolated from pigs in 1993. J Vet Med Sci. 1995;57:1023–1027. doi: 10.1292/jvms.57.1023. [DOI] [PubMed] [Google Scholar]
- 18.Kendal A P, Pereira M S, Skehel J J. Concepts and procedures for laboratory-based influenza surveillance. Atlanta, Ga: Centers for Disease Control; 1982. [Google Scholar]
- 19.Kida H, Ito T, Yasuda J, Shimizu Y, Itakura C, Shortridge K F, Kawaoka Y, Webster R G. Potential for transmission of avian influenza viruses to pigs. J Gen Virol. 1994;75:2183–2188. doi: 10.1099/0022-1317-75-9-2183. [DOI] [PubMed] [Google Scholar]
- 20.Lindstrom S E, Hiromoto Y, Nerome R, Omoe K, Sugita S, Yamazaki Y, Takahashi T, Nerome K. Phylogenetic analysis of the entire genome of influenza A (H3N2) viruses from Japan: evidence for genetic reassortment of the six internal genes. J Virol. 1998;72:8021–8031. doi: 10.1128/jvi.72.10.8021-8031.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lubeck M D, Gerhard W. Topological mapping of antigenic sites on the influenza A/PR/8/34 virus hemagglutinin using monoclonal antibodies. Virology. 1981;113:64–72. doi: 10.1016/0042-6822(81)90136-7. [DOI] [PubMed] [Google Scholar]
- 22.Luoh S M, McGregor M W, Hinshaw V S. Hemagglutinin mutations related to antigenic variation in H1 swine influenza viruses. J Virol. 1992;66:1066–1073. doi: 10.1128/jvi.66.2.1066-1073.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mancini G, Donatelli I, Rozera C, Ruiz G A, Butto S. Antigenic and biochemical analysis of influenza A H3N2 viruses isolated from pigs. Arch Virol. 1985;83:157–167. doi: 10.1007/BF01309913. [DOI] [PubMed] [Google Scholar]
- 24.Nakajima K, Nakajima S, Shortridge K F, Kendal A P. Further genetic evidence for maintenance of early Hong Kong-like influenza A (H3N2) strains in swine until 1976. Virology. 1982;116:562–572. doi: 10.1016/0042-6822(82)90148-9. [DOI] [PubMed] [Google Scholar]
- 25.Nerome K, Ishida M, Oya A, Oda K. The possible origin of H1N1 (Hsw1N1) virus in the swine population of Japan and antigenic analysis of the isolates. J Gen Virol. 1982;62:171–175. doi: 10.1099/0022-1317-62-1-171. [DOI] [PubMed] [Google Scholar]
- 26.Nerome K, Yoshioka Y, Sakamoto S, Yasuhara M, Oya A. Characterization of a 1980 - swine recombinant influenza virus possessing H1 haemagglutinin and N2 neuraminidase similar to that of the earliest Hong Kong (H3N2) virus. Arch Virol. 1985;86:197–211. doi: 10.1007/BF01309825. [DOI] [PubMed] [Google Scholar]
- 27.Olsen, C. W., S. Carey, L. Hinshaw, and A. I. Karasin. Virologic and serologic surveillance for human, swine and avian influenza virus infections among pigs in the north-central United States. Arch. Virol., in press. [DOI] [PMC free article] [PubMed]
- 28.Olsen C W, McGregor M W, Cooley A J, Schantz B, Hotze B, Hinshaw V S. Antigenic and genetic analysis of a recently isolated H1N1 swine influenza virus. Am J Vet Res. 1993;54:1630–1636. [PubMed] [Google Scholar]
- 29.Ottis K, Sidoli L, Bachman P A, Webster R G, Kaplan M M. Human influenza A viruses in pigs: isolation of a H3N2 strain antigenically related to A/England/42/72 and evidence for continuous circulation of human viruses in the pig population. Arch Virol. 1982;73:103–108. doi: 10.1007/BF01314719. [DOI] [PubMed] [Google Scholar]
- 30.Ouchi A, Nerome K, Kanegae Y, Ishida M, Nerome R, Hayashi K, Hashimoto T, Kaji M, Kaji Y, Inaba Y. Large outbreak of swine influenza in southern Japan caused by reassortant (H1N2) influenza viruses: its epizootic background and characterization of the causative viruses. J Gen Virol. 1996;77:1751–1759. doi: 10.1099/0022-1317-77-8-1751. [DOI] [PubMed] [Google Scholar]
- 31.Pensaert M, Ottis K, Vandeputte J, Kaplan M M, Bachmann P A. Evidence for the natural transmission of influenza A virus from wild ducks to swine and its potential importance for man. Bull WHO. 1981;59:75–78. [PMC free article] [PubMed] [Google Scholar]
- 32.Raymond F L, Caton A J, Cox N J, Kendal A P, Brownlee G G. The antigenicity and evolution of influenza H1 haemagglutinin, from 1950–1957 and 1977–1983: two pathways from one gene. Virology. 1986;148:275–287. doi: 10.1016/0042-6822(86)90325-9. [DOI] [PubMed] [Google Scholar]
- 33.Scholtissek C, Burger H, Bachmann P A, Hannoun C. Genetic relatedness of hemagglutinins of the H1 subtype of influenza A viruses isolated from swine and birds. Virology. 1983;129:521–523. doi: 10.1016/0042-6822(83)90194-0. [DOI] [PubMed] [Google Scholar]
- 34.Scholtissek C, Burger H, Kistner O, Shortridge K. The nucleoprotein as a possible major factor in determining host specificity of influenza H3N2 viruses. Virology. 1985;147:287–294. doi: 10.1016/0042-6822(85)90131-x. [DOI] [PubMed] [Google Scholar]
- 35.Scholtissek C, Naylor E. Fish farming and influenza pandemics. Nature. 1988;331:215. doi: 10.1038/331215a0. [DOI] [PubMed] [Google Scholar]
- 36.Shortridge K, Cherry A, Kendal A. Further studies of the antigenic properties of H3N2 strains of influenza A isolated from swine in Southeast Asia. J Gen Virol. 1979;44:251–254. doi: 10.1099/0022-1317-44-1-251. [DOI] [PubMed] [Google Scholar]
- 37.Shortridge K F, Webster R G, Butterfield W K, Campbell C H. Persistence of Hong Kong influenza virus variants in pigs. Science. 1977;196:1454–1455. doi: 10.1126/science.867041. [DOI] [PubMed] [Google Scholar]
- 38.Sugimura T, Yonemochi H, Ogawa T, Tanaka K, Kumagai T. Isolation of a recombinant influenza virus (Hsw1N2) from swine in Japan. Arch Virol. 1980;66:217–274. doi: 10.1007/BF01314741. [DOI] [PubMed] [Google Scholar]
- 39.Webster R G, Bean W J, Gorman O T, Chambers T M, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weis W, Brown J H, Cusack S, Paulson J C, Skehel J J, Wiley D C. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature. 1988;333:426–431. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- 41.Winter G, Fields S, Brownlee G G. Nucleotide sequence of the haemagglutinin gene of a human influenza virus H1 subtype. Nature. 1981;292:72–75. doi: 10.1038/292072a0. [DOI] [PubMed] [Google Scholar]
- 42.Yasuhara M, Hirahara T, Nakai M, Sasaki N, Kata J, Watanabe T, Morikawa M. Further isolation of a recombinant virus (H1N2), formerly (Hsw1N2), from a pig in Japan in 1980. Microbiol Immunol. 1983;27:43–50. doi: 10.1111/j.1348-0421.1983.tb03566.x. [DOI] [PubMed] [Google Scholar]
- 43.Zhou N N, Senne D A, Landgraf J S, Swenson S L, Erickson G, Rossow K, Liu L, Yoon K-J, Krauss S, Webster R G. Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. J Virol. 1999;73:8851–8856. doi: 10.1128/jvi.73.10.8851-8856.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zou S. A practical approach to genetic screening for influenza virus variants. J Clin Microbiol. 1997;35:2623–2627. doi: 10.1128/jcm.35.10.2623-2627.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]