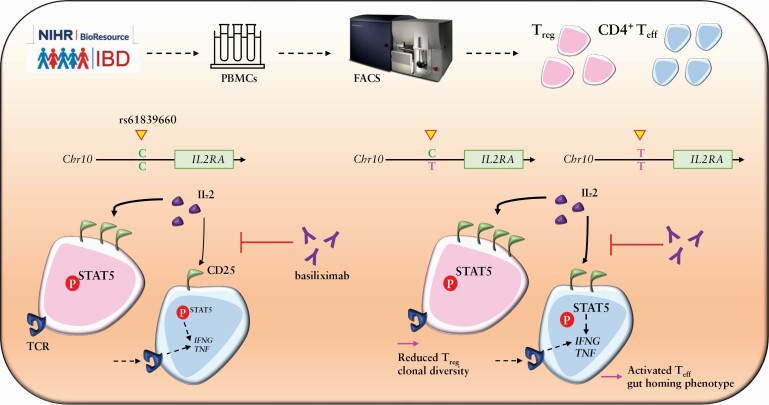
Abstract
Background and Aims
Differential responsiveness to interleukin [IL]-2 between effector CD4+ T cells [Teff] and regulatory T cells [Treg] is a fundamental mechanism of immunoregulation. The single nucleotide polymorphism [SNP] rs61839660, located within IL2RA [CD25], has been associated with the development of Crohn’s disease [CD]. We sought to identify the T cell immune phenotype of IBD patients who carry this SNP.
Methods
Teff and Treg were isolated from individuals homozygous [TT], heterozygous [CT], or wild-type [CC] for the minor allele at rs61839660, and used for phenotyping [flow cytometry, Cytometry Time Of Flight] functional assays or T cell receptor [TCR] sequencing. Phosphorylation of signal transducer and activator of transcription 5 [STAT5] was assessed in response to IL-2, IL-7, and in the presence of basiliximab, a monoclonal antibody directed against CD25. Teff pro-inflammatory cytokine expression levels were assessed by reverse transcription quantitative polymerase chain reaction after IL-2 and/or TCR stimulation.
Results
Presence of the minor T allele enhances CD25 expression, leading to increased STAT5 phosphorylation and pro-inflammatory cytokine transcript expression by Teff in response to IL-2 stimulation in vitro. Teff from TT individuals demonstrate a more activated gut homing phenotype. TCR sequencing analysis suggests that TT patients may have a reduced clonal capacity to mount an optimal regulatory T cell response.
Conclusions
rs61839660 regulates the responsiveness of T cells to IL-2 signalling by modulating CD25 expression. As low-dose IL-2 is being trialled as a selective Treg modulator in CD, these findings highlight the potential for adverse effects in patients with this genotype.
Keywords: Crohn’s disease, T cells, TREGs, CD25, IL-2, basiliximab
Graphical Abstract
Graphical Abstract.
1. Introduction
Crohn’s disease [CD] is a complex immune-mediated disorder with polygenic inheritance, in which inappropriate activation of the intestinal immune system in a genetically susceptible individual triggers chronic inflammation of the gastrointestinal tract. Despite the identification of multiple associated genetic polymorphisms,1 few have been mapped to a mechanistic pathway and none have yet yielded a tractable therapeutic approach for CD patients. Recent years have seen an increasing array of treatment options with the advent of biologic therapy, including anti-tumour necrosis factor [TNF]-α and anti-integrin agents. However, rates of surgery and hospitalisation for CD patients have not significantly changed in the biologic era.2 Therefore, there is a pressing need to explore novel therapeutic pathways. The value of a personalised approach to treatment is increasingly recognised, with the aim of targeting a treatment at the dominant aberrant pathway in a given patient to maximise efficacy and minimise side effects.3 The IL-2 pathway has been identified as potentially therapeutically tractable in numerous autoimmune conditions, including inflammatory bowel disease [IBD], with recruitment to a trial of low-dose IL-2 in CD under way [ClinicalTrials.gov NCT01988506].
Regulatory [Treg] and effector [Teff] CD4+ T cells are important mediators of the immune response in the gut of IBD patients, with defects in Treg number and suppressive function noted in the lamina propria and peripheral blood of patients with active IBD.4,5 Additionally, many identified IBD genetic risk loci map to immune cell enhancer regions, with particular enrichment in CD4+ T cell enhancers.6 Treg are characterised by high constitutive expression of the IL-2 receptor alpha chain [IL2RA, CD25], a component of the high-affinity IL-2 receptor heterotrimer [IL2Rα/β/γ]. The lineage-defining transcription factor Forkhead box P-3 [FOXP3] is essential for their suppressive phenotype and stability.7,8 An additional defining feature of Treg is their low expression, or absence, of the IL-7 receptor alpha chain [CD127].9 In homeostatic conditions, IL-2 is mainly produced by activated CD4+ T cells in secondary lymphoid organs and is consumed at the same site by cells that express CD25, of which the majority are Treg.10 By contrast, CD4+ Teff express CD25 only after activation, and at relatively lower levels compared with Treg, suggesting that Treg can preferentially respond to low concentrations of environmental IL-2. This differential responsiveness to IL-2 signalling is an important mechanism of immunoregulation.
Over 200 risk loci for IBD have been identified through genome-wide association studies [GWAS], with many in key regulatory pathways.1,11 The single nucleotide polymorphism [SNP] rs61839660 denotes a cytosine [C] to thymidine [T] base change on chromosome 10, located within a putative intronic enhancer region [intron 7] of IL2RA. rs61839660 was initially identified through GWAS, and is one of a minority of SNPs to be confidently resolved through fine-mapping as a variant highly associated with CD; 9.4% of CD patients carry the risk allele,12 which equates to almost 18 000 CD patients in the UK.13 rs61839660 is also associated with the development of other autoimmune conditions including ankylosing spondylitis, psoriasis, and primary sclerosing cholangitis,6,14 demonstrating its importance for core immunoregulatory processes.
Previous experimental work has generated conflicting results on the effect of rs61839660 on CD25 expression in CD4+ T cells. A study in healthy human volunteers found that carriage of the rs61839660 minor allele was associated with increased levels of IL2RA messenger mRNA [mRNA] and increased CD25 surface expression on CD4+ memory T cells.14 However, contemporaneous studies in which rs61839660 was delivered into murine CD4+ T cells via CRISPR-based methods, in addition to studies of healthy human carriers, reported that the minor variant of rs61839660 was associated with reduced CD25 transcript levels in CD4+ T cells stimulated with anti-CD3/CD28.15 This group hypothesised, therefore, that rs61839660 impairs the function of the intronic enhancer that regulates CD25 expression in response to T cell receptor [TCR] stimulation of Teff. We sought to define the immune phenotype associated with rs61839660 in CD patients, with the aim of understanding potential therapeutic targets in this patient cohort.
2. Materials and Methods
Due to the Covid-19 pandemic and the closure of many patient-facing services in the UK, we were unable to source TT homozygote patient samples for certain experiments included in our study. Specifically, at certain times during which our work was carried out, the UK-wide IBD BioResource, upon which this study is based, was unable to continue recruitment based on UK Government legislation. Therefore, we were obliged to focus certain experiments on patients who are heterozygous for the minor risk allele at rs61839660, which represents nearly 10% of all patients with Crohn’s disease worldwide. The data from these experiments are presented in Figures 3 and 4 of this study.
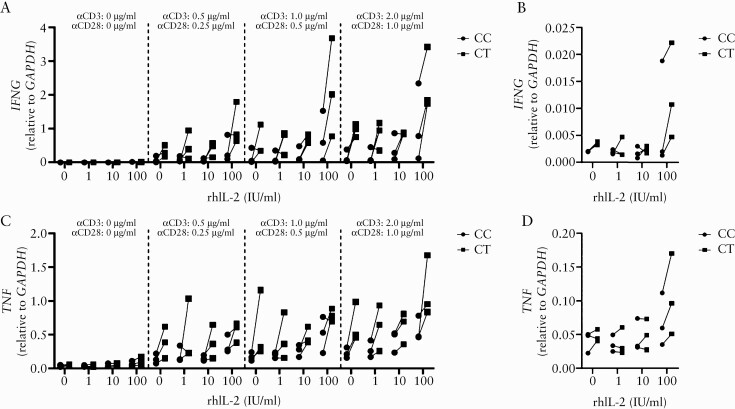
Figure 3.
The rs61839660 minor allele enhances transcription of pro-inflammatory cytokines following exogenous IL-2 and acute TCR stimulation of CD4+ effector T cells. CD4+ effector T cells [Teff] from heterozygous [CT] and major allele homozygous [CC] patients were treated with different doses of recombinant human IL-2 for 24 h before being stimulated for 6 h [still in the presence of IL-2] with different concentrations of plate-bound αCD3/CD28. [a] Relative expression of IFNG in Teff from the two genotypes investigated after the specified stimulations. [b] Relative expression of IFNG after 24 h of incubation with different concentrations of IL-2 in absence of TCR stimulation. [c] Relative expression of TNF in Teff from the two genotypes investigated after the specified stimulations. [d] Relative expression of TNF after 24 h of stimulation with different concentrations of IL-2 in absence of TCR stimulation; n = 3 individual Crohn’s disease patients per genotype. Results shown are the summary from three independent experiments using n = 1 of each genotype per experimental run. Connected points are comparative samples from the same experimental conditions, run in the same experiment. Relative expression values are normalised to GAPDH. TCR, T cell receptor.
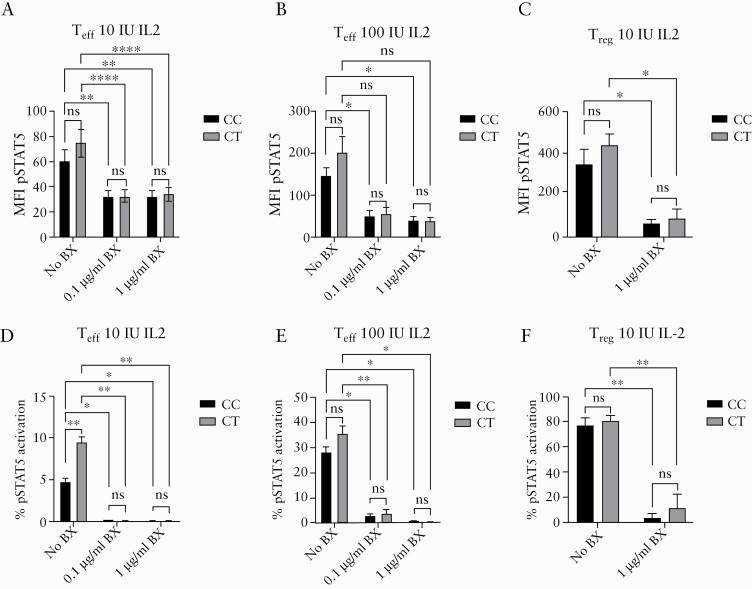
Figure 4.
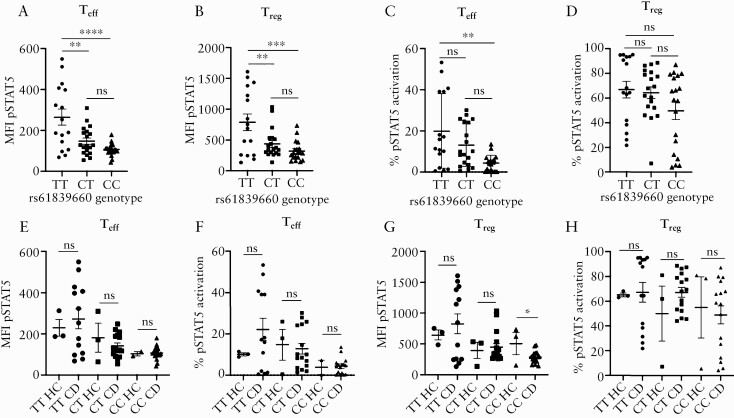
Basiliximab is effective at blocking phosphorylation of STAT5 in response to IL-2 signalling in rs61839660 heterozygote CD4+ T cells. Basiliximab was added to in vitro cultures of CD4+ effector T cells [Teff] and regulatory T cells [Treg] from heterozygous [CT] and major allele homozygous [CC] Crohn’s disease [CD] patients [n = 3 of each genotype], at the specified concentrations. After 6 h, cells were stimulated with 10 or 100 IU/ml of recombinant human IL-2 for 15 min. [a] Basiliximab was effective in reducing signal transducer and activator of transcription 5 [STAT5] phosphorylation [pSTAT5], quantified as a decrease in the mean fluorescence intensity [MFI] of pSTAT5 flow cytometry staining in Teff stimulated with [a] 10 IU/ml IL-2 or [b] with 100 IU/ml of IL-2. There were no significant differences in basiliximab response between the genotype groups. [c] Basiliximab reduced the MFI for pSTAT5 staining in Treg stimulated with 10 IU of IL-2 in CT and CC subjects. [d] Proportion [%] of pSTAT5 stained Teff activated with 10 IU/ml and [e] 100 IU/ml IL-2 following basiliximab treatment. [f] Proportion [%] of pSTAT5 stained Treg after stimulation with 10 IU/ml IL-2 following basiliximab treatment. Mean ± SEM plotted. Statistical analyses performed using two-way ANOVA, Tukey’s multiple comparisons test for comparisons involving more than two groups, and paired t tests for comparisons involving two treatments within the same genotype. *p <0.05, **p <0.01, ****p <0.0001. BX, basiliximab; Ns, not significant; SEM, standard error of the mean; ANOVA, analysis of variance.
Ethics approval for human blood and tissue collection was obtained from NRES Committee – London Riverside [REC reference: 15/LO/0151] and Guy’s and St Thomas’ NHS Trust R&D [R&D REF: RJ115/N122].
Permission was obtained from the NIHR IBD Bioresource to conduct a study of Treg/Teff phenotype and function in patients and healthy controls [HC] carrying the CD-associated single nucleotide polymorphism [SNP] rs61839660. Individuals homozygous [TT], heterozygous [CT], or wild-type [CC] voluntarily provided a whole blood sample. Baseline clinical information collected included age, gender, disease duration and location, and treatment history.
2.1. Peripheral blood mononuclear cell isolation
Peripheral blood was layered over Lymphocyte Separation Medium [LSM 1077; PAA] followed by centrifugation at 2000 rpm for 30 min at 20°C with slow acceleration and no brake. The interface was aspirated and washed with sterile PBS at 1800 rpm for 10 min at 4°C with normal acceleration and deceleration. Cell count and viability were confirmed with trypan blue staining [Sigma-Aldrich].
2.2. Cell sorting
A proportion of peripheral blood mononuclear cells [PBMCs] were stained with Human Regulatory T Cell Sorting Kit [BD Biosciences, San Diego, CA, USA], according to the manufacturer’s instructions. Enriched CD4+ cells were labelled with mouse anti-human CD4-PerCpCy5.5 [clone L200], anti-CD25-phyroerythrin [PE] [clone 2A3], anti-CD127-AlexaFluor 647 [clone 40131.111], and CD45RA-FITC [clone HI100]. Cells were sorted into CD4+CD25hiCD127lo Treg and CD25loCD127hi Teff on a FACSAria [BD Biosciences]. A FOXP3 stain was performed to ensure that the flow sort was selecting a true Treg population [gating strategy, Supplementary Figure 1, available as Supplementary data at ECCO-JCC online].
2.3. Functional pSTAT5 assays
To ascertain the functional relevance of rs61839660, pSTAT5 assays were performed on sorted Treg and Teff of subjects of each genotype. PBMCs were defrosted the day before the experiment and rested overnight in XVIVO-15 [Lonza], at 37°C, 5% CO2. On the day of the experiment, cells were counted and 4 x 106 were kept aside for cytometry time of flight [CyTOF] staining. The remainder were washed and stained for flow sorting as described above. Treg and Teff from each sample were sorted into XVIVO-15 and taken forward into the pSTAT5 assay.
In the first experimental phase, the assay was optimised on Treg and Teff sorted from a healthy control [HC] [blood bank cone]. Three concentrations of recombinant human [rh]IL-2 were used [1, 10, or 100 IU/ml per 1 x 105 cells] and each concentration was assessed at three time points [15, 20, and 30 min]. An unstimulated sample was run in parallel. 10 IU/ml IL-2 for 15 min yielded a robust pSTAT5 response, but not the maximal response. This was selected as the most appropriate initial concentration to assess pSTAT5 response without saturating the pathway, to permit discrimination of differences in Treg and Teff responses.
As a control, 1 x 105 sorted Treg and Teff from each subject were stimulated with recombinant human IL-7 across a logarithmically increasing dose range of concentrations [0, 1, 10, 100 IU/ml] under the same conditions. We hypothesised that, as IL-7 signals through CD127 and not through CD25, there should be no difference in the pSTAT5 response seen across genotypes.
For STAT5 phosphorylation assays, the BD Phosphoflow Protocol for human PBMCs was followed. Cells were washed, re-suspended in X-VIVO15, and rested for 1 h at 37°C, 5% CO2, before stimulation with IL-2 for 15 min in a 37°C water bath. Cells were fixed with warmed Cytofix buffer and incubated at 37°C for 10 min, then centrifuged at 600 x g for 5 min, supernatant discarded, and re-suspended in 100 μl Perm Buffer III. Cells were transferred to a 96-well v bottom plate and incubated on ice for 30 min, before washing and staining with 1 µl pSTAT5 AF488 (BD, clone 47/Stat5[pY694]) in 10 μl Stain Buffer, and incubated at room temperature for 60 min. Finally, cells were washed and re-suspended in 100 μl of PBS for acquisition on a Fortessa [BD]. Standardised acquisition settings were put in place and used in all subsequent experiments. Gates for the pSTAT5-positive population were set against an fluorescence minus one [FMO] control and the unstimulated sample.
2.4. Assessment of pro-inflammatory cytokine transcript expression by CD4+ Teffin response to IL-2 and acute T cell receptor stimulation.
Teff were isolated by fluorescence-activated cell sorting [FACS] as described. Cells were cultured at 37oC/5% CO2 at a density of 4 x 105–1 x 106 cells / ml in U bottom 96-well cell culture plates [Corning Costar] for 24 h with 0, 1, 10, or 100 IU/ml rhIL-2 in 200 µl RPMI [Gibco] + 10% fetal calf serum [FCS] + 2 mM L-glutamine + 10 mM HEPES + 1 x non-essential amino acids + 1 mM sodium pyruvate + 50 U/ml penicillin + 50 µg/ml streptomycin + 50µM β-mercaptoethanol. Following this, cultures were transferred to wells of a 96-well flat-bottom plate, pre-coated with the indicated concentrations of αCD3 [clone OKT3] and αCD28 [clone CD28.2], and briefly centrifuged to quickly settle the cell layer. Following 6 h of culture, samples were removed from the activation plates, washed in ice-cold 1 x PBS [without Ca2+/Mg2+], re-suspended in 700 µl Qiazol [Qiagen], incubated for 30 min at room temperature, vortexed thoroughly for 15 s and frozen at -70oC for a minimum of 24 h before further use.
2.5. Reverse-transcription quantitative polymerase chain reaction
Teff frozen in Qiazol were defrosted and RNA was isolated and purified via chloroform extraction using the miRNeasy Micro Kit, following the manufacturer’s protocol for isolation of total RNA from small samples [Qiagen]. RNA concentration was quantified using a Nanodrop spectrophotometer [Thermofisher Scientific]; 100 ng of total RNA per sample was reverse-transcribed using the RevertAid First Strand cDNA Synthesis Kit and random hexamer primers according to the manufacturer’s protocol. Quantitative polymerase chain reaction [qPCR] was performed using a ViiA 7 real-time PCR thermocycler [Applied Biosystems] in a final volume of 10 µl, using the 2 x Maxima probe/ROX qPCR Master Mix [Thermofisher Scientific] and the cDNA equivalent of 5 ng total RNA per sample. Detection of transcripts for pro-inflammatory cytokines was carried out using FAM-labelled 20 x TaqMan probe assays [Thermofisher Scientific] for IFNG [assay ID: HS00989291_m1] and TNF [assay ID: HS00174128_m1] and relative expression of each gene was calculated using the comparative cycle threshold [Ct] method [2-ΔCt] using GAPDH [assay ID: HS99999905_m1] as the endogenous reference control. Reactions were run in duplicates and the average Ct value for each reaction was used for analysis.
2.6. Basiliximab suppression assays
Optimisation work was performed to identify basiliximab conditions that would permit discrimination of CD25 blockade and subsequent pSTAT5 suppression between samples [HC blood bank cone, two replicates performed]. Basiliximab doses of between 0.001 and 10 µg/ml have been reported to cause marked CD25 blockade and thus reduction of lymphocyte proliferation when applied for up to 48 h to cell culture.16,17 Trials were performed using 0.01, 0.1, and 1.0 µg/ml basiliximab to sorted Treg or Teff in culture. Following the defined treatment period, cells were stimulated with 0–100 IU/ml IL-2 and the functional pSTAT5 assay was performed as described above. An untreated sample was used as a control. A 6-h application of 0.1 and 1.0 µg/ml basiliximab caused marked, but not complete, pSTAT5 suppression, and were taken forward into assays on patient samples.
2.7. Assessment of Tregphenotype using CyTOF
A Cytometry Time of Flight [CyTOF] mass cytometry technique was employed to allow in-depth multi-parameter assessment of the Treg phenotype. Two panels [Supplementary Figure 3, available as Supplementary data at ECCO-JCC online] were designed to assess Treg trafficking capacity [Panel 1] and Treg suppressive ability [Panel 2]. The panels were extensively optimised and dose titration of each antibody was performed on PBMCs isolated from a blood bank cone, with all experiments performed on the same cone to minimise donor variability in marker expression. Antibodies were sourced from Fluidigm and have been validated for human use as per the manufacturer.
Cells for CyTOF staining were defrosted the day before the experiment and rested overnight in complete RPMI supplemented with 10% FCS at 37°C, 5% CO2. Cells were washed and dispersed into FACS staining tubes at 2 x 106 cells per panel. A cisplatin viability stain was performed (cells re-suspended in 1 ml PBS, with 1 μl of cisplatin for 5 min, then reaction quenched with 3 ml MACS buffer, which was used as the cell staining medium [CSM] for all CyTOF experiments). Cells were then centrifuged at 400 x g for 5 min at room temperature, supernatant discarded, pellet dispersed, and 4 μl of FC block [MACS, Miltenyi Biotec] applied per sample. Conjugated antibodies were purchased from Fluidigm. Unconjugated antibodies were purchased from Biolegend and conjugated to the requisite metal isotopes using MaxPar labelling kits [Fluidigm]. A master mix of the extracellular CyTOF antibodies was made up for each panel and the samples were stained in a total volume of 100 μl for 30 min at room temperature. Following staining, cells were washed with CSM and re-suspended in fix/perm buffer [eBioscience FOXP3/Transcription factor staining buffer set] followed by a 1-h incubation at room temperature. Cells were washed with 1x permeabilisation buffer and stained with intracellular/transcription factor antibody master mix for 30 min at room temperature. Cells were then washed again with 1x permeabilisation buffer and re-suspended in 500 μl of 2% paraformaldehyde [PFA], left at room temperature for 1 h, and refrigerated overnight. On the day of acquisition, the cells were pelleted, washed, and resuspended in 1x Ir [Iridium] intercalator solution for 30 min; this acted as a DNA/final viability stain. After two further PBS washes, cells were re-suspended in water at a maximal concentration of 5 x 105 with an internal bead standard [DVS EQ beads, Fluidigm].
2.8. Analysis of CyTOF data
Raw FCS files were normalised using Fluidigm CyTOF Software [https://www.fluidigm.com/software]. Data were uploaded to Cytobank [https://mrc.cytobank.org] and standard pre-processing performed to remove debris, doublets, beads, and dead cells. CD4+ T Cells were manually gated and used as a starting population for the automated unsupervised analysis.
Cytobank’s implementation of the t-Distributed Stochastic Neighbour Embedding [t-SNE] algorithm [known as viSNE]18 was used to transform the data to two dimensions, while still conserving the high dimensional structure of the data. The resulting t-SNE1 and t-SNE2 dimensions were then inputted into a clustering algorithm-spanning tree-progression analysis of density normalised events [SPADE; available on Cytobank],19 which extracts a cellular hierarchy from high-dimensional cytometry data and presents the populations in a branched tree diagram.
Cluster frequencies were quantified based on SPADE population clusters, and different cell populations were identified based on the median expression of known markers using our in-house developed pipeline [CytoClustR; https://github.com/kordastilab/cytoClustR]. For example, Treg and Teff nodes were identified in the SPADE tree based on the expression of CD127, CD25, and FOXP3. Further populations were identified within the Treg and Teff nodes, based on differential expression of one or more of the rest of the markers within those populations. Both marker expressions and cell abundances were analysed with one-way analysis of variance [ANOVA] for differences between the genotype groups for each identified population.
To quantify similarity/difference between the populations as well as expression of different markers, marker enrichment modelling [MEM] scores were calculated for each identified population and the output was visualised using heatmaps. The MEM algorithm objectively describes characteristics of cell populations using large numbers of markers, rather than a few most differentially expressed, and provides each marker with a score which denotes its expression level.20
2.9. TCR sequencing
2.9.1. RNA extraction
1 x 105 Treg and Teff were FACS-purified [BD Aria II] into TRIzol LS reagent [Invitrogen]. Total RNA was extracted as per the manufacturer’s instructions. To aid the precipitation of RNA, GlycoBlue™ Coprecipitant [Invitrogen] was used as per the manufacturer’s instructions. Processed samples were assessed for RNA integrity with an Agilent RNA 6000 Pico Kit in conjunction with an Agilent 2100 Bioanalyser. RNA concentration was quantified using a Qubit RNA HS Assay Kit [Invitrogen] and a Qubit analyser. Paired samples with RIN >9 were shipped to iRepetoire [iRepertoire Inc., Huntsville, AL] for TCR beta chain amplification and sequencing.
2.9.2. Library construction procedure
For the construction of human TCR beta chain libraries, iRepertoire used amplicon rescued multiplex PCR [arm-PCR] to amplify each of the 18 submitted RNA samples. The cDNA synthesis protocol involved initial first-round reverse transcriptase [RT]-PCR using high concentrations of gene-specific primers [iRepertoire’s HTBI-M primers], followed by Beckman Coulter’s SPRIselect bead-cleanup procedure, then PCR2 using universal primers to amplify the cDNA exponentially.
2.10. Statistical analysis
Flow cytometric data were analysed with FlowJo 10.6.0 for MacOsX. Statistical analysis was performed with GraphPad Prism 8.4.1 for MacOsX. Normality was assessed using D’Agostino Pearson testing. Continuous data are presented as mean ± standard error of the mean for normally distributed variablesand as medians and interquartile ranges for skewed variables. Comparison of central tendency was performed using paired parametric and non-parametric tests as appropriate [t test or Wilcoxon signed rank test, respectively]. Similarly, multiple means [or ranks] were compared by one- or two-way ANOVA or Kruskal‐Wallis tests, as appropriate. For comparison of matched values, the Wilcoxon matched pairs signed rank test was used. The Mann‐Whitney test with a two-tailed p-value was used to determine significance level in all unmatched values. The TT, CT, and CC groups were matched by age and gender. For all significance testing, alpha was set to 0.05.
3. Results
3.1. rs61839660 modulates the expression of CD25 on CD4+ T cell subsets
We sought to investigate the effect of rs61839660 on CD25 cell surface expression, downstream signalling, and effector cell function. We obtained peripheral blood samples from genotyped CD patients and healthy controls, either homozygous minor allele [TT], homozygous major allele [CC], or heterozygous [CT], for rs61839660 from the National Institute of Health Research [NIHR] Inflammatory Bowel Disease BioResource,21 a UK-based biobank which holds demographic and genetic information pertaining to almost 50 000 IBD patients. Patient demographics are summarised in Table 1. There were no significant differences between genotype groups in terms of age, sex, or disease distribution. There was no difference in rates of medication use or need for surgery, suggesting that minor allele carriers do not have a more severe disease phenotype.
Table 1.
NIHR IBD Bioresource Crohn’s disease patient demographics.
| TT [n = 14] | CT [n = 21] | CC [n = 22] | p-value | |
|---|---|---|---|---|
| Age [mean] | 50.2 ± 13.58 | 47.2 ± 13.40 | 52.1 ± 14.41 | ns |
| Sex [% female] | 57.1 | 57.1 | 36.4 | ns |
| Disease distribution [% colonic disease] | 71.4 | 33.3 | 36.8 | ns |
| Disease distribution [% perianal disease] | 35.7 | 19.0 | 18.2 | ns |
| Biologic medication [% exposed] | 7.7 | 30.8 | 15.4 | ns |
| Surgery [% yes] | 60.0 | 52.4 | 54.5 | ns |
| Extraintestinal manifestations [% yes] | 28.6 | 38.1 | 18.2 | ns |
Statistical analyses performed using one-way ANOVA for age and chi square analysis for categorical variables.
TT, homozygous minor allele; CT, heterozygous; CC, homozygous major allele, ns, not significant;ANOVA, analysis of variance.
Results considered significant if p </= 0.05.
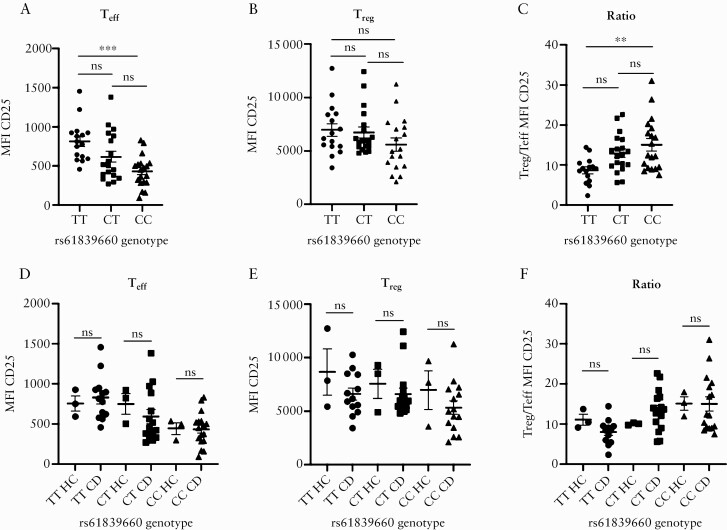
CD4+ Treg and Teff were sorted by flow cytometry [Supplementary Figure 1]. We demonstrated increased CD25 cell surface expression in CD patients and healthy controls, specifically on Teff of TT patients as measured by mean fluorescence intensity [MFI] of flow cytometry staining [Figure 1a], compared with CC subjects. There was a non-significant trend for increased CD25 expression on Treg in TT patients [Figure 1b] as well as a non-significant trend towards enhanced CD25 expression on Teff and Treg from CT heterozygote carriers of the minor allele at rs61839660 [Figure 1a, b, and d]. The ratio of Treg:Teff CD25 expression was significantly reduced in TT patients [Figure 1c] compared with CC patients. No difference was seen in Treg or Teff CD25 expression between CD patients and healthy controls with the same genotype [Figure 1d–f], suggesting that differential CD25 expression between genotypes is conferred by the risk allele, rather than by the presence of inflammatory disease.
Figure 1.
rs61839660 modulates CD25 expression on effector T cells. [a] Mean fluorescence intensity [MFI] of CD25 flow cytometry staining by genotype in CD4+ effector T cells [Teff]. Teff from minor allele homozygotes [TT] demonstrated increased expression of CD25 compared with major allele homozygotes [CC] [p = 0.0002]. [b] MFI of CD25 staining by genotype in Treg. There was no difference in the MFI of CD25 by genotype in regulatory T cells [Treg]. [c] Ratio of CD25 expression in Treg/Teff. TT subjects demonstrated a reduced ratio of CD25 expression compared with CC subjects [p = 0.002]. [d] There was no difference in CD25 expression in Teff or [e] Treg or [f] in the MFI CD25 ratio Treg/Teff between CD patients and healthy controls within each genotype group. Mean ± SEM plotted. Statistical analyses performed using one-way ANOVA, Tukey post hoc test, for comparisons involving more than two groups, and unpaired t- ests for comparisons involving two groups. TT: n = 16, CT: n = 19, CC: n = 19. **p <0.01, ***p <0.001. HC, healthy control; CD, Crohn’s disease; ns, not significant; SEM, standard error of the mean; ANOVA, analysis of variance.
3.2. rs61839660 regulates responsiveness of CD4+ T cells to IL-2 signalling through altered CD25 expression
To determine whether rs61839660 affected the CD4+ T cell response to IL-2 signalling, the extent of STAT5 phosphorylation [pSTAT5] was measured in Teff and Treg following short-term exposure to 10 IU IL-2 [Figure 2]. In optimisation work, this concentration of IL-2 was found to be sufficient to induce pSTAT5 expression in approximately 50% of Treg, with no pSTAT5 expression in Teff in CC patients. Teff and Treg from TT patients had an enhanced pSTAT5 response as measured by mean fluorescence intensity [MFI] of pSTAT5, compared with CT and CC patients [Figure 2a and b]. Teff from TT subjects also exhibited a higher proportion of pSTAT5+ cells compared with Teff from CC patients [Figure 2c]. There was a non-significant increase in the proportion of pSTAT5+ Treg in TT patients [Figure 2d]. There was no difference in Teff or Treg pSTAT5 response to IL-2 signalling between healthy controls and CD patients of the same genotype [Figure 2e, f, g, and h]. As noted for CD25 expression, a non-significant trend for enhanced pSTAT5 staining was observed in CT heterozygote carriers of the minor allele, and this was more apparent in Teff than Treg [Figure 2a–h].
Figure 2.
STAT5 phosphorylation in response to IL-2 is enhanced in carriers of the rs61839660 minor allele. Signal transducer and activator of transcription 5 [STAT5] phosphorylation [pSTAT5] was assessed after 15 min of incubation with 10 IU/ml of recombinant human IL-2 in CD4+ effector T cells [Teff] and regulatory T cells [Treg]. [a] Minor allele homozygotes [TT] demonstrated increased mean fluorescence intensity [MFI] for pSTAT5 flow cytometry staining compared with heterozygous [CT] and major allele homozygous [CC] subjects in Teff [TT vs CC p <0.0001; TT vs CT p = 0.0026] and [b] Treg [TT vs CC p = 0.0004; TT vs CT [p = 0.0085]. [c] The proportion of cells with pSTAT5 staining was also increased in Teff from TT subjects compared with CC subjects [p = 0.0012]. [d] No differences were detected in Treg when comparing the percentage of pSTAT5 stained cells between the genotypes. [e] There was no difference in pSTAT5 staining MFI or [f] proportion of pSTAT5-positive cells in Teff or [g‐h] Treg between healthy controls [HC] and Crohn’s disease [CD] patients in carriers of the minor allele. Mean ± SEM plotted. Statistical analyses performed using one-way ANOVA, Tukey post hoc test, for comparisons involving more than two groups and unpaired t tests for comparisons involving two groups. TT: n = 16, CT: n = 19, CC: n = 19. *p <0.05, ** p <0.01, ***p <0.001, ****p <0.0001. SEM, standard error of the mean; ANOVA, analysis of variance; ns = not significant.
It was noted that among TT subjects, there was a subgroup that exhibited a more pronounced pSTAT5 response to IL-2 in both Teff and Tregs [Figure 2a–d]. We performed a subanalysis on this ‘hyper-responder’ cohort, defining subjects with a Teff MFI pSTAT5 greater than 395 and a Treg MFI pSTAT5 greater than 1100 in response to 10 IU/ml IL-2, as ‘hyper-responders’ [Table 2]. The same five subjects with hyper-responsive Teff also exhibited an exaggerated Treg pSTAT5 response. There was no significant difference in age, sex, disease distribution, disease severity, or proportion of patients exposed to biologic medication between the TT ‘hyper-responders’ and the remaining TT subjects. There was no significant difference in Teff or Treg CD25 expression between the TT ‘hyper-responder’ subgroup and the remaining TT subjects.
Table 2.
Characteristics of the TT ‘hyper-responder’ cohort.
| TT Hyper-responder [n = 5] | TT Other [n = 11] | p value | |
|---|---|---|---|
| Mean Teff CD25 MFI | 795.4 | 825.2 | ns |
| Mean Treg CD25 MFI | 5924 | 7631 | ns |
| Mean Treg/Teff CD25 MFI | 7.46 | 10.94 | ns [0.076] |
| Age [mean] | 33.6 | 39.9 | ns |
| Sex [% female] | 100.0 | 72.7 | ns |
| Disease distribution [% colonic disease] | 20.0 | 50.0 | ns |
| Disease distribution [% perianal disease] | 66.6 | 60.0 | ns |
| Biologic medication [% exposed] | 40.0 | 12.5 | ns |
| Surgery [% yes] | 100.0 | 60.0 | ns |
Statistical analyses performed using unpaired t test to compare means and chi square analysis for categorical variables.
TT, homozygous minor allele; ns, not significant.
Results considered significant if p </= 0.05.
As a control, the pSTAT5 response to IL-7 was examined across a range of doses [0‐1000 IU]. rs61839660 did not affect CD127 surface expression across the genotypes in Teff or Treg [Supplementary Figure 2a and b, available as Supplementary data at ECCO-JCC online]. Importantly, STAT5 phosphorylation downstream of IL-7 signalling was independent of genotype in Teff or Treg [Supplementary Figure 2c–h], indicating that the differences in pSTAT5 response to IL-2 stimulation are likely caused by enhanced CD25 expression in the presence of the minor allele.
3.3. The rs61839660 minor allele enhances transcription of pro-inflammatory cytokines following exogenous IL-2 and acute TCR stimulation of Teffs
Together, our findings suggested that the rs61839660 minor allele may confer a functional ‘hyper-responsive’ state to IL-2 signalling in Teff. To further investigate this proposal, we sought to identify whether presence of the minor allele influenced the functional response to IL-2 by Teff, by measuring cytokine expression [IFNG and TNF] responses by the cells after TCR stimulation.
Teff from cryopreserved PBMCs of CD patients, genotyped as CC and CT at rs61839660, were isolated by FACS and used to conduct the assays. Teff were stimulated for 24 h with a range of IL-2 doses only, or with IL-2 followed by a brief stimulation [6 h] with different concentrations of plate-bound αCD3/αCD28 [Figure 3].
We observed that for both expression of IFNG [Figure 3a and b] and TNF [Figure 3c and d], compared with CC samples, the presence of the minor T allele at rs61839660 enhanced the relative transcript abundance in response to IL-2, together with TCR stimulation. This was most clearly observed at the highest concentration of IL-2 [100 IU/ml] for each of the TCR cross-linking conditions [Figure 3a and c]. Importantly, greater pro-inflammatory cytokine relative transcript abundance was also observed in CT samples treated with 100 IU/ml of rhIL-2 in the absence of TCR crosslinking, despite similar baseline [0 IU IL-2, no TCR cross-linking] transcript abundance levels between the genotypes [Figure 3b and d]. These data support the notion that within the context of CD, presence of the rs61839660 minor allele confers a functional capacity for hyper-responsiveness of CD4+ Teff in response to IL-2 signalling.
3.4. CD25 blockade is effective in suppressing the enhanced pSTAT5 response conferred by the rs61839660 minor allele
Basiliximab, a mouse-human chimeric monoclonal antibody which antagonises CD25, is currently licensed for the prevention of renal allograft rejection. We examined whether basiliximab added to in vitro culture of sorted Treg and Teff could block pSTAT5 activation in a cohort of CD patients with CC and CT rs61839660 genotypes, recruited from our local patient population. Basiliximab was effective in blocking Teff and Treg pSTAT5 activation in both genotypes [Figure 4a–f], confirming that the enhanced pSTAT5 response is dependent on signalling through CD25. Of note, CT Teff also demonstrated elevated baseline pSTAT5 activation in the absence of basiliximab, when compared with CC Teff [Figure 4d], as previously seen in the other sample cohorts used in our study. There was no significant difference in the response to basiliximab between the genotype groups.
3.5. Teffin subjects homozygous for rs61839660 demonstrate an activated, gut-homing phenotype
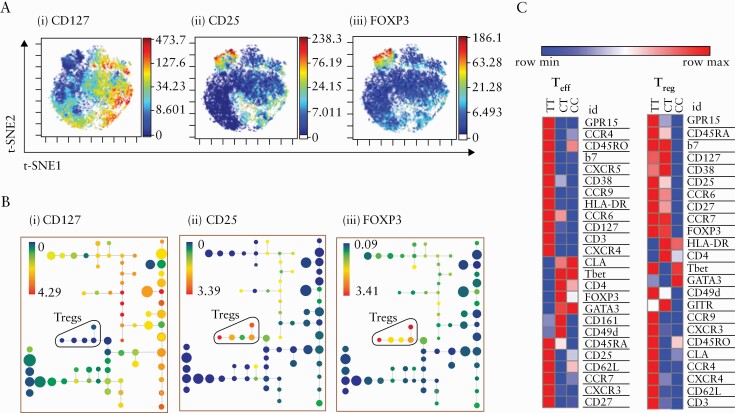
To further elucidate the phenotype of Treg and Teff from patients harbouring the rs61839660 minor variant, we performed an in-depth phenotypic analysis using Cytometry Time of Flight [CyToF] [Panels 1 and 2 and antibody details in Supplementary Figure 3], with unsupervised clustering analysis and Marker Enrichment Modeling [MEM].20 Treg and Teff subsets were identified by CD25, CD127, and FOXP3 viSNE plots [Figure 5a] and SPADE clustering analysis [Figure 5b] and analysed with specific focus on the expression of trafficking and cell activation markers. The number of cells per each SPADE cluster and the Teff and Treg groups per subject were recorded, and tables for the Teff and Treg groups for Panel 1 and Panel 2 may be viewed in Supplementary Data Files 1 and 2, respectively, available as Supplementary data at ECCO-JCC online. The dimensionality reduction was performed in Cytobank with the following setup parameters: equal sampling of 22 500 [Panel 1] and 29 000 [Panel 2] CD4+ cells per subject, perplexity = 30, theta = 0.5, and iterations = 5000. Each group was composed of 10 subjects. The resulting t-SNE1 and t-SNE2 dimensions of the viSNE transformation were used as input for the SPADE clustering. The following markers were used for the viSNE analysis for Panel 1: CCR6, CLA, CD31, CD49d, CD62L, CD161, GITR, FOXP3, b7, CCR7, CD127, T-bet, GATA3, CCR9, CD25, CXCR5, CD38, CXCR4, HLA-DR, and for Panel 2: OX40, PD1, LAG3, ICOS, CD62L, CD137, CD161, CD39, Ki67, FOXP3, CD30, PDL1, CD127, T-bet, GATA3, CD37, CD25, CTLA-4, Granzyme-B, CD120b, HLA-DR, and Perforin.
Figure 5.
CD4+ T cell effectors of minor allele homozygotes at rs61839660 demonstrate a more activated gut-homing phenotype. [a] Identifying regulatory T cells [Treg] and non-Tregs [CD4+ T effectors, Teff] using viSNE map. Cell population islands coloured by expression of [i] CD127, [ii] CD25, [iii] FOXP3. Red-blue colouring denotes highest to lowest level of expression. [b] Identifying Treg and Teff using SPADE [Spanning-tree Progression Analysis of Density-normalised Events] trees. Each node denotes a cluster. Proximity to other nodes denotes expression of similar markers. Treg populations are coloured by the expression of [i] CD127, [ii] CD25, [iii] FOXP3. Red-blue colouring denotes highest to lowest level of expression. [c] Marker Enrichment Modelling [MEM] analyses of Teff for rs61839660 minor allele homozygote [TT], heterozygote [CT], and major allele homozygote [CC] Crohn’s disease patient groups. Scaling is done per row [red = row maximum, blue = row minimum]. Higher MEM score denotes higher expression of a marker in a group, compared with another.
Confirming observations made by flow cytometry, TT Teff demonstrated increased CD25 expression [Figure 5a]. However, this analysis also revealed a comparable increase in CD25 expression on TT Treg, providing an explanation as to why homozygous minor allele Tregs also exhibited hyper-responsiveness during IL-2/STAT5 phosphorylation assays [Figure 2b]. Furthermore, this analysis demonstrated that both Treg and Teff from minor allele homozygotes have a more activated, gut-homing phenotype compared with cells from CT and CC subjects [Figure 5c]. Key markers that were differentially upregulated in TT patients included the chemokine receptor CXCR3, which is typically associated with inflammatory Th1 cell infiltration, and CXCR4, associated with IL-2-mediated immune activation. GPR15, which specifically traffics cells to the colon, was also preferentially expressed on both Treg and Teff of TT patients, as were other markers associated with Treg tissue trafficking such as CCR6 and integrin alpha 7 [Supplementary Figures 4a–c and 5a–d, available as Supplementary data at ECCO-JCC online]. In addition, Teff from the TT group were the only cells to express Ki67 [Supplementary Figure 5c], indicating an enhanced proliferative effector phenotype and cellular activation.
3.6. TT patients may have a reduced capacity to mount a regulatory T cell response
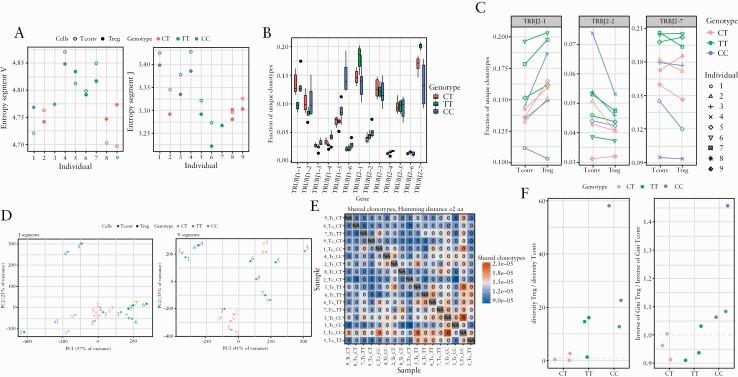
To assess whether the difference in phenotypes observed between the three patient groups was driven by their ability to respond to antigens, we performed T cell receptor [TCR] sequencing of Treg and Teff from three CD patients from each SNP genotype. We assessed the variability displayed in both the V and J segment of the CDR3 region. We found that the J segments in TT patient cells consistently displayed the least variability, those from CC patient cells displayed the greatest variability, and the CT patients had an intermediate phenotype [Figure 6a]. J segments in Teff showed higher variability than Treg across the three genotypically defined groups. Further analysis of individual J segments revealed that TT cells preferentially use two TRBJ segments, segments TRBJ2-1 and TRBJ2-7 [Figure 6b and c]. Principal component analysis of J segment usage revealed distinct clustering by genotype [Figure 6d]. Such differential clustering was not observed on analysis of the V segment usage. We then examined whether any of the individuals cluster together based on shared clonotypes on either an amino acid or a nucleotide level, and did not observe clustering based on genotypes [Figure 6e]. Finally, to assess whether TCR diversity was different between Treg and Teff subsets, we examined their diversity ratio across the three genotypes. We found that this ratio was lower in TT and CT patients compared with CC patients. Whereas Tregs were more diverse than Teff [measured by Gini index] in CC patients, in TT and CT patients, Treg diversity was comparable to or lower than Teff [Figure 6f]. The reduced Treg diversity when compared with Teff in TT patients, coupled with Teff hyper-responsiveness to IL-2, may suggest that TT patients are less efficient at mounting a regulatory response to counter Teff-driven inflammation.
Figure 6.
T cell receptor profiling reveals that rs61839660 minor allele homozygote Crohn’s disease patients may have a reduced capacity to mount a regulatory T cell response. [a] CDR3 region J and V segment entropy for CD4+ effector T cells [Teff, empty circles] and regulatory T cells [Treg, filled circles] among the genotypes. [b] Fractions of unique J segment clonotypes used by each genotype. [c] TT patient CD4+ T cells (Treg and Teff /conventional [Tconv] cells) preferentially use two TRBJ segments [TRBJ2-1 and TRBJ2-7]. Treg and Teff/Tconv from the same patient are denoted by conjoined lines and matching individual symbols. [d] Principal component analysis of J segment usage revealed distinct clustering by genotype.[ e] Heatmap for clustering of shared clonotypes matched by Hamming distance. [f]: TCR diversity ratio for Treg/Teff, measured by Gini index. TCR, T cell receptor.
In summary, we show that the rs61839660 minor allele enhances sensitivity to IL-2 signalling in CD4+ T cells by increasing CD25 expression. We propose that the reduced ratio of Treg:Teff CD25 expression in individuals homozygous [TT] for this SNP may increase the ability of Teff to respond to low environmental IL-2, permitting activation of Teff at levels of IL-2 which would usually only activate Treg. This Teff hyper-responsiveness to IL-2 could therefore be a major driver of inflammation in these subjects. Mass cytometric analysis of immune cells from these patients confirmed that CD25 is elevated in patients homozygous for the risk allele at rs61839660, and that effector T cells from these patients are primed to home to the intestine by virtue of upregulation of gut trafficking molecules such as alpha 4 beta 7 integrin and GPR15.
4. Discussion
Here we show that rs61839660 enhances IL-2 signalling in CD4+ T cells from CD patients by regulating the expression of CD25. Effector T cells from TT patients display the ability to respond to doses of IL-2 that normally only activate regulatory T cells. Additionally, Teff from carriers of the minor allele express elevated transcript levels of pro-inflammatory cytokines in response to IL-2 and/or TCR signalling—supportive of our conclusion that rs61839660 regulates the functional responsiveness of T cells in CD patients via the regulation of sensitivity to IL-2 signalling. Our findings are in contrast to previous work involving CRISPR inactivation of immune enhancers in mice, which proposed that rs61839660 impaired induction of CD25 and therefore disrupted IL-2 signalling in T cells, and found that carriage of the risk allele was associated with reduced CD25 transcript levels in stimulated undifferentiated CD4+ T cells from healthy human subjects.15 However, our findings are supported by a previous study of 323 healthy human subjects, which found that the presence of the rs61839660 minor allele was associated with increased IL2RA transcript levels and surface expression of CD25 on CD4+ memory T cells.14
Basiliximab was found to effectively block Teff hyper-responsiveness in minor allele heterozygote [CT] patients. Previous clinical trials of basiliximab in small cohorts of unselected patients with ulcerative colitis, in whom rs61839660 had not been identified as a risk locus, showed safety but did not demonstrate efficacy compared with conventional treatment.22,23 However, these data suggest that basiliximab could have a role in blocking the hyper-responsive Teff population seen in rs61839660 carriers. Further in vitro experimental work is required to assess the potential therapeutic value of CD25 blockade in this cohort.
Mass cytometric analysis of immune cells from these patients was also performed, which confirmed that CD25 expression is elevated in patients homozygous for the TT allele at rs61839660 and that effector T cells from these patients are primed to home to the intestine by virtue of upregulation of gut-trafficking molecules such as alpha 4 beta 7 integrin and GPR15. Analysis of the TCR repertoire further reveals that diversity in the Treg subset does not mirror diversity in the Teff subset in TT patients. Lack of ability in the Treg compartment to respond to aberrant Teff responses in TT patients, may be another underlying pathogenic mechanism. Unlike in type 1 diabetes mellitus, where rs61839660 is protective,12,14 in CD rs61839660 is linked to pathology. Based on our observations, the mechanism of this likely involves the capacity of rs61839660 to render Teff hyper-responsive to doses of IL-2 which typically activate only Tregs, putatively resulting in an enhanced activation profile of the cells. This, alongside an activated gut-homing profile of TT Teff, potentially explains the association between rs61839660 and CD. However, it will be necessary to follow up our observational analyses with further mechanistic studies to fully confirm or refute these hypotheses.
This study provides a unique perspective on the link between genetic risk and the development of disease, by studying genotyped CD and HC patients recruited across the UK. Recruiting genotyped human subjects across the country presents a logistically complex process; thus a limitation of our study is that insufficient patient numbers could be recruited to adequately study the link between disease severity and the risks allele. Further studies are also needed to define the interplay between the rs61839660 risk allele and other genotypic and environmental factors in patients with CD, to allow for a precise definition of the mechanism of pathogenicity of the rs61839660 risk allele.
This is one of the first works in IBD to link a pathogenic mechanism to a CD-associated genetic variant, advancing the goal of a personalised therapeutic approach. Accurate phenotypic and genetic profiling of CD patients would allow targeted therapeutic intervention, permitting selection of a treatment targeted to correct the aberrant pathway in a given patient. We highlight the vital role of open-access resources, such as the NIHR IBD BioResource, for sourcing genetically selected material for functional analysis of rare variants. We provide a novel mechanistic link between genotype and function relevant to multiple human autoimmune diseases. Over 40 phase 2/3 trials of low-dose IL-2 in the treatment of autoimmune conditions are currently registered, including in CD [ClinicalTrials.gov NCT01988506]. These trials typically use doses of 0.5‐3 x 106 IU/m2 body area, which are delivered subcutaneously under trial-specific regimens. The premise of this treatment is that low doses of IL-2 will preferentially expand the tolerising Treg population, correcting the dysregulated autoimmune response. However, given the exaggerated Teff response to low doses of IL-2 in carriers of rs61839660, these subjects could be at increased risk of deleterious clinical outcomes due to theoretical Teff hyperactivation. With 9.4% CD patients carrying the risk allele, this represents a significant subset of potential trial participants. Furthermore, our findings suggest there is merit in further assessing the role of basiliximab in genetically stratified patients, as an adjunct to existing treatments.
Supplementary Material
Acknowledgements
We acknowledge the help of the IBD Bioresource, supported by the NIHR, Medical Research Council, Crohn’s and Colitis UK, Open Targets, and the Wellcome Trust Sanger Institute.
Funding
This work was supported by an Medical Research Council grant to RGJ and GML [MR/R001413/1], and a Guts UK/forCrohns Development grant to JNC, PMI, and GML [DGC2019_02]. This research was supported by the National Institute for Health [NIHR] Biomedical Research Centres at Guy’s & St Thomas’ NHS Foundation Trust and King’s College London, and Cambridge University Hospital NHS Foundation Trust and the University of Cambridge. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR, or the Department of Health.
Conflict of Interest
GML holds a consultancy position with F. Hoffmann-La Roche AG [Roche]. GML and RG hold a patent for the manufacture of regulatory T cells for the treatment of inflammatory bowel disease [PCT/GB2019/053618].
Author Contributions
Obtaining funding: RG, JNC, PI, RGJ, GML. Study content and design: RG, JNC, LBR, JS, MP, NP, GML. Acquisition of data: RG, JNC, LBR, JS, AH, IJ, ST, AA, OO, MP, NP, RGJ. Analysis and interpretation of data: RG, JNC, LBR, JS, SK, NP, RGJ, PMI, GML. Statistical analysis: RG, JNC, JS, SK, NP. Initial manuscript preparation: RG, JNC. Revised manuscript preparation: LBR, JS, JNC. Design of graphical abstract: LBR. Critical revision of manuscript: LBR, JS, JNC, AH, OO, MP, NP, RGJ, PMI, GML. Overall study supervision: GML.
References
- 1. Jostins L, Ripke S, Weersma RK, et al. ; International IBD Genetics Consortium [IIBDGC]. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012;491:119–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Murthy SK, Begum J, Benchimol EI, et al. Introduction of anti-TNF therapy has not yielded expected declines in hospitalisation and intestinal resection rates in inflammatory bowel diseases: a population-based interrupted time series study. Gut 2020;69:274–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Noor NM, Verstockt B, Parkes M, Lee JC. Personalised medicine in Crohn’s disease. Lancet Gastroenterol Hepatol 2020;5:80–92. [DOI] [PubMed] [Google Scholar]
- 4. Maul J, Loddenkemper C, Mundt P, et al. Peripheral and intestinal regulatory CD4+ CD25[high] T cells in inflammatory bowel disease. Gastroenterology 2005;128:1868–78. [DOI] [PubMed] [Google Scholar]
- 5. Fantini MC, Pallone F, Monteleone G. Common immunologic mechanisms in inflammatory bowel disease and spondylarthropathies. World J Gastroenterol 2009;15:2472–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Farh KKH, Marson A, Zhu J, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 2015;518:337–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fontenot JD, Gavin MA, Rudensky AY. Pillars article: Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. J Immunol 2017;198:986–92. [PubMed] [Google Scholar]
- 8. Hori S, Nomura T, Sakaguchi S. Pillars article: control of regulatory T cell development by the transcription factor Foxp3. J Immunol 2017;198:981–5. [PubMed] [Google Scholar]
- 9. Liu W, Putnam AL, Xu-Yu Z, et al. CD127 expression inversely correlates with FoxP3 and suppressive function of human CD4+ T reg cells. J Exp Med 2006;203:1701–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boyman O, Sprent J. The role of interleukin-2 during homeostasis and activation of the immune system. Nat Rev Immunol 2012;12:180–90. [DOI] [PubMed] [Google Scholar]
- 11. Liu JZ, van Sommeren S, Huang H, et al. ; International Multiple Sclerosis Genetics Consortium; International IBD Genetics Consortium. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet 2015;47:979–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huang H, Fang M, Jostins L, et al. Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature 2017;547:173–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jones GR, Lyons M, Plevris N, et al. IBD prevalence in Lothian, Scotland, derived by capture-recapture methodology. Gut 2019;68:1953–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rainbow DB, Pekalski M, Cutler AJ, et al. A rare IL2RA haplotype identifies SNP rs61839660 as causal for autoimmunity. bioRxiv February 2017:108126. doi: 10.1101/108126. [DOI] [Google Scholar]
- 15. Simeonov DR, Gowen BG, Boontanrart M, et al. Discovery of stimulation-responsive immune enhancers with CRISPR activation. Nature 2017;549:111–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kircher B, Lätzer K, Gastl G, Nachbaur D. Comparative in vitro study of the immunomodulatory activity of humanized and chimeric anti-CD25 monoclonal antibodies. Clin Exp Immunol 2003;134:426–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang Z, Shi BY, Qian YY, Cai M, Wang Q. Short-term anti-CD25 monoclonal antibody administration down-regulated CD25 expression without eliminating the neogenetic functional regulatory T cells in kidney transplantation. Clin Exp Immunol 2009;155:496–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Amir el-AD, Davis KL, Tadmor MD, et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol 2013;31:545–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Qiu P, Simonds EF, Bendall SC, et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol 2011;29:886–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Diggins KE, Greenplate AR, Leelatian N, Wogsland CE, Irish JM. Characterizing cell subsets using marker enrichment modeling. Nat Methods 2017;14:275–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Parkes M; IBD BioResource Investigators . IBD BioResource: an open-access platform of 25 000 patients to accelerate research in Crohn’s and Colitis. Gut 2019;68:1537–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Creed TJ, Probert CS, Norman MN, et al. ; BASBUC Investigators. Basiliximab for the treatment of steroid-resistant ulcerative colitis: further experience in moderate and severe disease. Aliment Pharmacol Ther 2006;23:1435–42. [DOI] [PubMed] [Google Scholar]
- 23. Sands BE, Sandborn WJ, Creed TJ, et al. Basiliximab does not increase efficacy of corticosteroids in patients with steroid-refractory ulcerative colitis. Gastroenterology 2012;143:356–64.e1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.