Abstract
Background and Aims
Microbial-derived bile acids can modulate host gene expression, and their faecal abundance is decreased in active inflammatory bowel disease [IBD]. We analysed the impact of endoscopic inflammation on microbial genes involved in bile acid biotransformation, and their interaction with host transcriptome in the intestinal mucosa of IBD patients.
Methods
Endoscopic mucosal biopsies were collected from non-inflamed and inflamed terminal ileum, ascending and sigmoid colon of IBD patients. Prediction of imputed metagenome functional content from 16S rRNA profile and real-time quantitative polymerase chain reaction [qPCR] were utsed to assess microbial bile acid biotransformation gene abundance, and RNA-seq was used for host transcriptome analysis. Linear regression and partial Spearman correlation accounting for age, sex, and IBD type were used to assess the association between microbial genes, inflammation, and host transcriptomics in each biopsy location. A Bayesian network [BN] analysis was fitted to infer the direction of interactions between IBD traits and microbial and host genes.
Results
The inferred microbial gene pathway involved in secondary bile acid biosynthesis [ko00121 pathway] was depleted in inflamed terminal ileum of IBD patients compared with non-inflamed tissue. In non-inflamed sigmoid colon, the relative abundance of bile acid-inducible [baiCD] microbial genes was positively correlated with the host Angiopoietin-like 4 [Angptl4] gene expression. The BN analysis suggests that the microbial baiCD gene abundance could affect Angptl4 expression, and this interaction appears to be lost in the presence of inflammation.
Conclusions
Endoscopic inflammation affects the abundance of crucial microbial bile acid-metabolising genes and their interaction with Angptl4 in intestinal mucosa of IBD patients.
Keywords: Enteric microbiota, gene expression, inflammatory bowel disease
1. Introduction
Inflammatory bowel disease [IBD] is characterised by idiopathic chronic inflammation of the gut, including two primary types of disease: ulcerative colitis [UC], defined by continuous inflammation limited to the colonic mucosa, and Crohn’s disease [CD], which can affect any segment of the gastrointestinal tract. IBD is a complex disease where environmental, microbial, immunological, and genomic factors interact to promote intestinal inflammation. However, the mechanism by which these components interact to lead to IBD remains elusive. Characteristic changes in gut microbiome composition and function have been recognised not just as markers of disease but also as actively contributing to pathogenesis.1 One way by which the microbiota exerts its effect on the host is attributed to microbe-derived bile acids.2
Bile acids are amphipathic sterols synthesised in hepatocytes from cholesterol. The human liver produces two primary bile acids, cholic acid [CA] and chenodeoxycholic acid [CDCA]. Before secretion into the intestinal tract, bile acids are conjugated [N-acyl amidation] to either glycine or taurine, producing conjugated bile acids.3 Once in the small intestine, human-derived bile acids undergo chemical diversification via three main microbial pathways: deconjugation, dehydrogenation, and dehydroxylation, which produce an array of secondary bile acids.4 Deconjugation refers to the enzymatic hydrolysis of the amide bond by bile salt hydrolase [BSH] enzymes. The resulting unconjugated bile acids are further exposed to 7α-dehydroxylating enzymes encoded on the microbial bile acid-inducible [bai] operon leading to the formation of the two main secondary bile acids, deoxycholic acid [DCA] and lithocholic acid [LCA]. Finally, oxidation and epimerisation of bile acids by microbial hydroxysteroid dehydrogenases [HSDH] contribute to significantly increase the diversity of secondary bile acids.4
Recent studies have identified significant differences in bile acid composition in stool samples of IBD patients compared with non-IBD subjects.5–9 Furthermore, a depletion of microbial bsh and bai genes has also been observed in stool samples of IBD patients, suggesting a potential role of bile acid-metabolising microbiota in the pathogenesis of IBD.8,10 A significant impact on intestinal gene expression of germ-free mice when monocolonised with bacteria carrying bile acid-transforming enzymes further underlines a potential role of bile acids in modulating the host transcriptomics-gut microbe interaction.11 In IBD patients, studies using intestinal mucosal samples from different locations have identified several host genes that covaried with the relative abundance of mucosa-adherent microorganisms, unravelling distinct interactions between host and gut microbiome.6,12–15 However, a targeted analysis of bacterial genes encoding relevant enzymes involved in bile acid metabolism in intestinal mucosa has not been explored. In this study, we aim to identify the impact of inflammation on the abundance of microbial bile acid-metabolising genes in intestinal mucosa and their interaction with host gene expression in IBD patients.
2. Materials and Methods
2.1. Study design and sample collection
Patients with confirmed UC, IBD-unclassified [IBD-U], and CD were recruited when attending regularly scheduled colonoscopy to assess disease activity or for surveillance. Asymptomatic healthy controls [HC] were recruited during routine, age-related colorectal cancer screening by colonoscopy. Recruitment was carried out at the tertiary IBD referral centre, Mount Sinai Hospital [Toronto, Canada]. All subjects provided written informed consent and the study was approved by the Mount Sinai Hospital Research Ethics Board. Biopsy samples from the terminal ileum, ascending colon, and sigmoid colon were taken using standard forceps. At the time of the procedure, endoscopic scores were calculated by the endoscopist at each biopsy location, using the Mayo endoscopic subscore16 for UC and IBD-U patients [range from 0 to 3 per segment], and the simple endoscopic score [SES-CD]17 for CD patients [range from 0 to 12 per segment]. A Mayo endoscopic subscore >0 and SES-CD >2 were considered endoscopically inflamed mucosa. Clinical and demographic data were recorded, including IBD phenotype, concomitant primary sclerosing cholangitis [PSC], current use of medications, and previous ileocaecal resection. Patients using antibiotics within 3 months before colonoscopy were excluded. UC and IBD-U patients were analysed together, given phenotypic similarities. Two biopsies from each of the terminal ileum, ascending colon, and sigmoid colon were immediately placed into cryovials and flash-frozen in liquid nitrogen for subsequent mucosa-associated microbiome analysis. Two additional samples from each site were immediately placed in RNAlater and sufficient time was provided for RNAlater permeability before storage for further host transcriptomic analysis. Study samples were stored at −80°C.
2.2. Microbial DNA extraction and 16S rRNA gene sequencing
Total microbial DNA was extracted from biopsies using DNeasy blood and tissue kit [Qiagen, Hilden, Germany], as previously described.18 Amplicon sequencing of the V4 hypervariable region of 16s rRNA bacterial DNA was completed using primers 515F/806R19 on an Illumina MiSeq platform [Illumina, San Diego, CA, USA]. Paired-end sequences were subsequently processed using the QIIME 1.9.0 pipeline20 applying a closed reference operational taxonomic unit [OTU]-picking approach, including reference and de novo chimera removal steps. OTUs were then assigned using the Greengenes reference database v13_8.20,21
2.3. Inference of gut microbiota function from 16s rRNA profiles
To predict metagenome functional content from the community structure derived from 16S rRNA gene sequencing data, Phylogenetic Investigation of Communities by Reconstruction of Unobserved States [PICRUSt] V1.0 was applied.22 PICRUSt is a computational approach that uses an extended ancestral-state reconstruction algorithm to predict which microbial gene families are present, and then combines gene families to estimate the functional metagenomic content of the samples. The OTU table was used as the input file for metagenome imputation after a rarefaction step to 9000 sequences per sample. The pre-calculated table of gene counts was used to predict the gene composition of the microorganisms present in the microbiome.23 The clusters of orthologous groups [COG]24 and the Kyoto Encyclopedia of Genes and Genomes [KEGG]25 databases [updated March 2019] were accessed to identify relevant KEGG orthologues and COGs related to microbial secondary bile acid biosynthesis [KEGG pathway, ko00121].24 Relative abundance was calculated dividing KEGG orthologues or COG count in a specific sample by the total KEGG orthologues or COG counts in that sample. To determine which taxa were contributing to these microbial functions, we used the metagenome_contributions.py script in PICRUSt. The generated data were visualised with analyse_contributions package in R v4.0.2, and differential contribution analysis was performed using the linear discriminant analysis [LDA] effect size [LEfSe] pipeline.26 LDA was used to estimate the effect size of each differentially abundant trait. Alpha values <0.05 were used for the Kruskall‐Wallis rank sum test, and a threshold of 2.0 was chosen for logarithmic LDA scores.
2.4. Real-time quantitative polymerase chain reaction of microbial bile acid-metabolising gene abundance
Real-time quantitative polymerase chain reaction [qPCR] was performed using microbial DNA [10 ng/μL] extracted from a subset of mucosal samples to quantify abundance of bsh, baiCD, and 12α-hsdh genes using primers previously described in the literature [Supplementary Table S1, available as Supplementary data at ECCO-JCC online].27–31 For bsh gene, two primer sets corresponding to Bacteroides plebeius [bsh group 1a]27 and Bifidobacterium bifidum [bsh group 2] were amplified.31 For baiCD gene, a primer pair previously designed based on known nucleotide sequences from Clostridium scindens and Clostridium hiranonis was used.29 Finally, two primer sets from Clostridium hylemonae32 and Eggerthella sp.30 were used to amplify 12α-hsdh gene. Each reaction was performed in duplicate using the SSoAdvanced Universal SYBR green supermix [BioRad, Hercules, CA, USA] on the CFX384 real-time PCR detection system, under the following run conditions: initial denaturation at 95°C for 3 min, followed by 40 cycles of 15 s at 95°C and 30 s at 60°C. Fold change was calculated by ΔΔCt method, normalised to the 16S rRNA gene of Eubacteria [housekeeper].33 Mucosal samples from HC were used as the reference group. Outliers were filtered using the interquartile range method.
2.5. Host RNA extraction and RNA-sequencing from mucosal samples
Total RNA was extracted from homogenised tissue with the miRNeasy Mini Kit [Qiagen, Hilden, Germany], following the manufacturer’s instructions. RNA quantity and purity were measured on the Nanodrop ND-1000 instrument [Thermo Fisher Scientific, Waltham, MA, USA]. RNA quality was measured with Bioanalyser 2100 [Agilent, San Francisco, CA, USA] at Princess Margaret Genomics Centre [Toronto, ON, Canada]. Only samples with RNA integrity number [RIN] greater than or equal to 7.5 were considered for further analysis. Complementary DNA [cDNA] libraries were prepared using reagents and protocols provided by Bio-Rad, and sequencing was performed using paired-end runs on a HiSeq 2500 instrument [Illumina, San Diego, CA, USA]. Sequence reads were mapped against Homo sapiens reference genome hg20 [GRC38] using HISAT2-2.1.0 on SciNet, a high-performance computing cluster at the University of Toronto. Trimming was then performed on the mapped reads using Trim Galore.0.3.4 [https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/] and cutadapt 1.2.1 [https://github.com/marcelm/cutadapt]. Quality control was performed and reads with Phred+33 scores less than 20, and those belonging to adaptor sequences were trimmed. Furthermore, paired reads were removed if read became shorter than 20 bases after trimming. Quality-controlled reads were then used to quantify and normalise transcript levels using the StringTie 1.3.3b and Ballgown 2.16.0 pipeline [https://ccb.jhu.edu/software/stringtie/]. A mask was applied to genes coding for ribosomal and haemoglobin subunits.
2.6. Host transcriptome dimensionality reduction and analysis
Principal component analysis [PCA] of the host transcriptomics count data was performed after variant stabilising transformation. Interpretation of the PCA axes was assisted by visual inspection of the top components and loadings. PCAtools package34 implemented in R was used to evaluate correlations between clinical variables and principal components [PCs]. For transcriptomics data dimensionality reduction, the top 500 positive and negative gene loadings explaining the largest variability of PC1 were selected for downstream analysis. Gene count normalisation was performed via the median of ratios method implemented in the Bioconductor package DESeq2 v1.28.1.35 Normalised and log-transformed counts were used for downstream analysis. Finally, a group of functional host genes previously associated with bile acids or bile acid-metabolising microbiota in murine models, including bile acid receptors, were selectively added to the analysis11,36,37 [Supplementary Data, Excel file 1]
2.7. Statistical analysis
Except for PCA, all the analyses were performed separately on each biopsy location. Univariate analyses of imputed microbial genes regarding IBD type and biopsy location were performed by either Kruskal‐Wallis or Mann‐Whitney U test, depending on the number of analysed groups. To identify associations between inferred microbial genes and intestinal inflammation [inflamed and non-inflamed mucosa], a linear regression model was fitted accounting for IBD type [UC/IBDU or CD], age at colonoscopy, and sex. The microbe-microbe gene interactions and the microbial gene-host transcriptomics interactions were assessed by partial Spearman correlation accounting for IBD type, age at colonoscopy, and sex.38 Differential expression of host genes of interest between inflamed and non-inflamed mucosa was analysed by a negative binomial generalised linear model [GLM] implemented in the DESeq2 package in R,35 and p-values were adjusted for false discovery rate [FDR] by the Benjamini‐Hochberg method.39 Although unadjusted p-values <0.05 are reported, only FDR <0.05 were considered statistically significant. To gain further insight into the interaction between microbial genes involved in bile acid metabolism, host genes, and host phenotype, a Bayesian network [BN] probabilistic graphical model was fitted using the bnlearn v4.6.1 package.40 Categorical and continuous variables of interest were discretised [Hartemink’s method] and provided as input to the BN construction. The hill-climbing algorithm with scoring based on the Bayesian Dirichlet equivalence [BDe] score was used for directed acyclic graph [DAG] construction. Consensus networks were obtained by generating 1000 bootstraps of the input data. The empirical frequency for each edge [association], indicating the strength and direction of the association, was determined and edges that met an empirical threshold frequency greater than or equal to 0.6 were used to produce the consensus DAG. The resulting DAGs were visualised using the bnviewer v0.1.6 package implemented in R v4.0.2.
3. Results
3.1. Cohort description
We previously reported the 16S rRNA gene-based mucosa-associated microbiota composition of this cohort consisting of 215 patients with IBD and 48 HC.18 After excluding seven patients with antibiotic exposure at the time of colonoscopy, 114 UC/IBD-U and 94 CD, with a total of 445 biopsy samples available for PICRUSt imputation, were analysed [n = 140 terminal ileum, n = 115 ascending colon, n = 190 sigmoid colon]. Clinical and endoscopic characteristics of this IBD cohort are depicted in Supplementary Table S2, available as Supplementary data at ECCO-JCC online. A total of 165 out of 445 samples [37%] were obtained from endoscopically inflamed mucosa. From this original cohort, 202 biopsy samples [n = 67 terminal ileum, n = 28 ascending colon, n = 111 sigmoid colon], corresponding to 134 participants [n = 52 UC/IBD-U, n = 46 CD, n = 36 HC] were processed for bulk RNA-seq and had microbial DNA available for qPCR analysis. This subset was used for the integrative microbiome-host gene expression analysis. The clinical and endoscopic characteristics of this subcohort are depicted in Table 1. In this subset, 58 out of 151 [38.4%] mucosal samples from IBD patients were collected from inflamed tissue.
Table 1.
Main characteristics of the analysed cohort for microbe-host interaction.
| Characteristics | HC | UC/IBDU | CD | Total IBD cohort |
|---|---|---|---|---|
| [n = 36] | [n = 52] | [n = 46] | [n = 98] | |
| Median age, y [range] | 57 [41–71] | 34 [19–64] | 29 [17–67] | 33 [17–67] |
| Sex, n female [%] | 14 [38.9] | 31 [59.6] | 23 [50.0] | 54 [55.1] |
| Median age at diagnosis of IBD, y [range] | - | 25 [11–56] | 21 [7–57] | 22.5 [7–57] |
| Median duration of IBD, y [range] | - | 8 [1–44] | 8 [0–30] | 8 [0–44] |
| Montreal classification, n | ||||
| CD location, L1/L2/L3 | - | - | 5/15/26 | - |
| CD behaviuor, B1/B2/B3 | - | - | 27/8/11 | - |
| UC extent, E1/E2/E3 | - | 10/16/26 | - | - |
| Primary sclerosing cholangitis, n [%] | - | 3 [5.8] | 1 [2.2] | 4 [4.1] |
| Prior history of Ileocaecal resection, n [%] | - | - | 4 [8.7] | - |
| Medication at colonoscopy | ||||
| 5-aminosalicylates, n [%] | - | 41 [78.8] | 8 [17.4] | 49 [50.0] |
| Immunomodulators, n [%] | - | 3 [5.8] | 10 [21.7] | 13 [13.3] |
| Biologic therapy, n [%] | - | 5 [9.6] | 9 [19.6] | 14 [14.3] |
| Corticosteroids, n [%] | - | 3 [5.8] | 0 | 3 [3.1] |
| Inflamed mucosaa | ||||
| Terminal ileum, n/total samples [%] | 0/23 | 0/23 | 10/21 [46.7] | 10/44 [22.7] |
| Ascending colon, n/total samples [%] | 0/2 | 4/15 [26.7] | 4/11 [36.3] | 8/26 [31.0] |
| Sigmoid colon, n/total samples [%] | 0/30 | 21/41 [51.2] | 19/40 [50.6] | 40/81 [49.3] |
HC, healthy controls; UC, ulcerative colitis; IBDU, inflammatory bowel disease unclassified; CD, Crohn´s disease; y, years.
aPercentages are based on the total number of samples per site as denominator.
3.2. Microbial bile acid-metabolising genes in mucosal samples and univariate analysis against IBD type
A total of 4794 KEGG orthologue gene function categories were imputed in our full set of samples using PICRUSt with a relatively low weighted nearest sequenced taxon index [NSTI, mean 0.057 ± 0.012], indicating a good accuracy of the metagenomic prediction.22 In our samples, we did not detect the KEGG orthologue K15870 corresponding to baiCD gene function, which may imply either low counts of mucosa-associated microbiota carrying out 7α-dehydroxylation or incorrect gene annotation for this function. Conversely, KEGG pathway ko00121 corresponding to overall secondary bile acid biosynthesis and KEGG orthologue K01442 corresponding to microbial bsh gene function [COG3049] were found in all mucosal samples. In the univariate analysis, IBD type was not associated with relative abundance of ko00121 and COG3049 at different biopsy locations [Supplementary Figures S1 to S4, available as Supplementary data at ECCO-JCC online].
3.3. Imputed microbial secondary bile acid-metabolising gene pathway abundance in ileal mucosa is decreased by intestinal inflammation
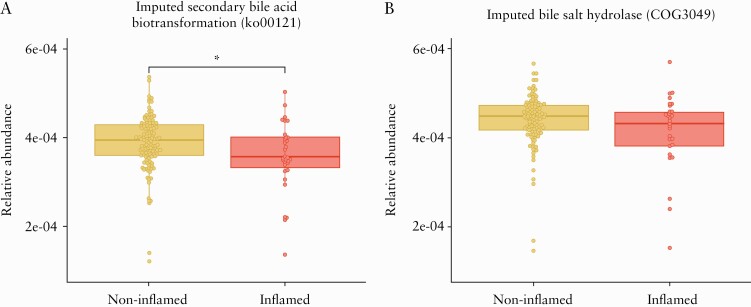
To dissect how inflammation affects the abundance of imputed microbial bile acid metabolism genes in each biopsy location, we applied a multivariable linear regression model to the secondary bile acid metabolism [ko00121] pathway and bsh [COG3049] gene abundance to test their association with non-inflamed and inflamed mucosa while controlling for age, sex, and IBD type. Compared with non-inflamed ileal mucosa, inflamed terminal ileum of IBD patients was associated with a reduced abundance of the ko00121 gene pathway [linear regression, F value = 4.08, p-value = 0.04], but no significant difference was observed for COG3049 gene function in this location [linear regression, F value = 3.19, p-value = 0.07] [Figure 1]. No differences were observed in colonic biopsies for either ko00121 pathway [ascending colon, p-value = 0.35, and sigmoid colon, p-value = 0.23] or COG3049 function [ascending colon, p-value = 0.20, and sigmoid colon, p-value = 0.14] between inflamed and non-inflamed mucosa [Supplementary Figure S5, available as Supplementary data at ECCO-JCC online].
Figure 1.
Inferred bile acid-metabolising gene abundance in terminal ileum samples [n = 140 samples]. Data expressed as relative abundance of imputed gene function from 16S marker gene data using PICRUSt. Differential abundance analysis was performed by linear regression modelling including endoscopic inflammation [non-inflamed and inflamed], IBD type [CD and UC/IBDU], sex, and age as covariates. A] Reduced relative abundance of ko00121 pathway gene abundance is associated with endoscopic inflammation. B] A non-significant numerical reduction of cluster of orthologous groups [COG3049] in inflamed samples was observed; *p-value <0.05 for the linear regression analysis. UC, ulcerative colitis; IBDU, inflammatory bowel disease unclassified; CD, Crohn´s disease.
3.4. Microbial taxa contribution to bile salt hydrolase gene function varies with intestinal inflammation
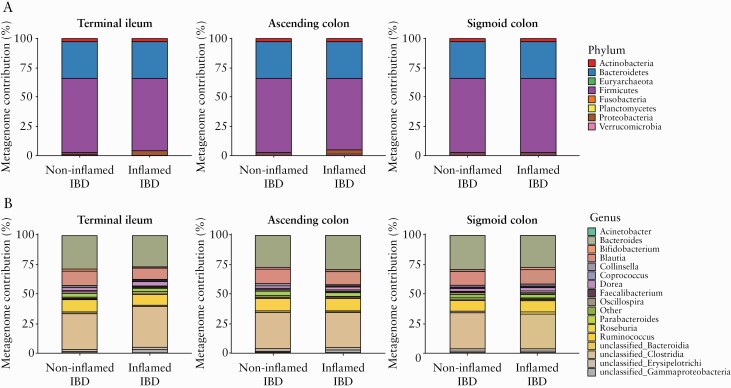
Given the lack of a significant effect of inflammation on imputed bsh relative abundance, we then explored for differences in the taxa contributing to bsh gene [COG3049] function between non-inflamed and inflamed mucosa, using the metagenome_contributions.py script in PICRUSt and the LEfSe pipeline. At the phylum level, an increased relative contribution to bsh gene function of Proteobacteria [LDA = 4.7] and Actinobacteria [LDA = 4.0] was observed in inflamed terminal ileum and sigmoid colon, respectively, compared with non-inflamed mucosa [Figure 2A; Supplementary Figure S6, available as Supplementary data at ECCO-JCC online]. At the genus level, we identified several changes in the taxa contributing to bsh gene function [Figure 2B; Supplementary Figure S6]. Within the Clostridia class, Oscillospira [LDA = 4.0] and Blautia [LDA = 4.2] reduced their contribution, whereas Acinetobacter [LDA = 3.9] and a group of unclassified genera belonging to the class Gammaproteobacteria [LDA = 4.7] increased their contribution in inflamed compared with non-inflamed terminal ileum. Finally, Bifidobacterium [LDA = 3.8] and Faecalibacterium [LDA = 3.8] genera increased their relative contribution to bsh gene abundance in inflamed sigmoid colon compared with non-inflamed tissue.
Figure 2.
Relative metagenome contribution to bile salt hydrolase [bsh] gene abundance [COG3049]. Bar plot with taxa contributing to bsh gene function in intestinal mucosa of healthy controls [HC] and inflammatory bowel disease [IBD] patients with or without endoscopic inflammation at Phylum [A] and Genus [B] level in each biopsy site. Genus contributing to <1% were collapsed as Other. Imputed bsh genes that could not be annotated to the family and/or order level are depicted as unclassified at the Class level.
3.5. Microbial baiCD genes interact with Angptl4 host gene in non-inflamed sigmoid colon mucosa
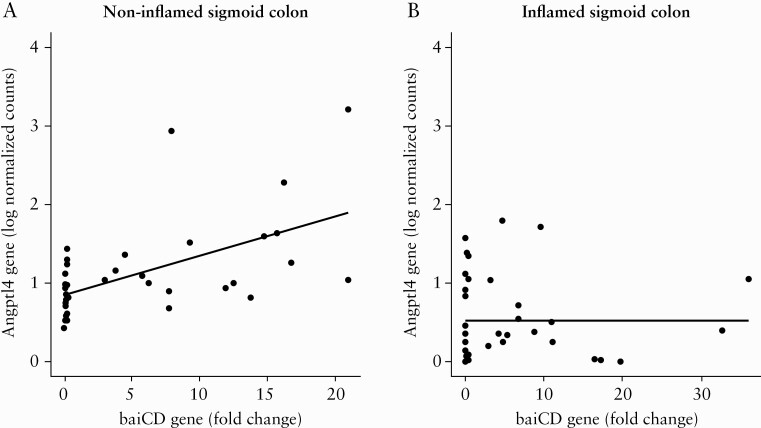
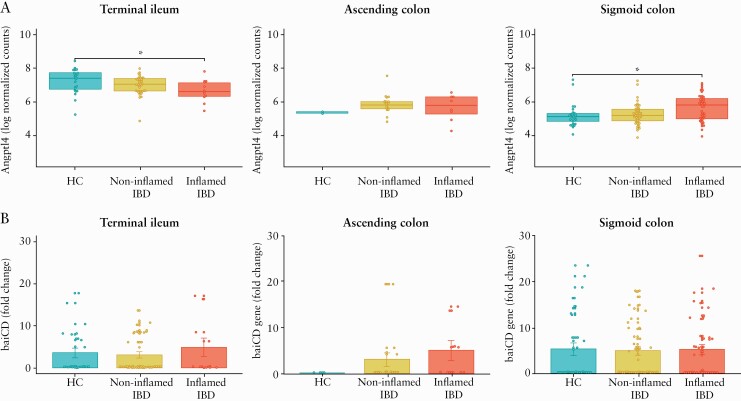
Given the observed reduction of microbial secondary bile acid biosynthesis [ko00121] pathway in our analysis with PICRUSt, using qPCR we then tested the expression of specific mucosa-associated microbial genes which participate in bile acid metabolism. Bsh from B. plebius and B. bifidum, baiCD from C. scindens and C. hiranonis, and 12α-hsdh from C. hylemonae and Eggerthella sp. were analysed. As a measure of microbe-microbe and host-microbe cross-talk, we evaluated partial Spearman correlations among microbial and host genes accounting for age at colonoscopy, sex, and IBD type. Several significant correlations were identified among microbial genes [Supplementary Figure S7, available as Supplementary data at ECCO-JCC online] in non-inflamed and inflamed mucosa. Given these correlations and the double representation of bsh and 12α-hsdh in our microbial gene set, we included only one representative of these gene functions in our microbe-host interaction analysis. For the host genes, the top 500 loadings contributing to the first component of the PCA [Supplementary Figure S8, available as Supplementary data at ECCO-JCC online] were selected for downstream analysis [see Supplementary Data, Excel file 1 for a complete list of the analysed host genes]. The visual inspection of PCA [Supplementary Figure S8] and the correlation analysis between clinical variables and PCA loadings [Supplementary Figure S9, available as Supplementary data at ECCO-JCC online] showed that biopsy location and endoscopic inflammation were the major drivers of host transcriptomics variance; therefore, the three biopsy locations and non-inflamed and inflamed mucosa were independently analysed. Conversely, given the minimal effect of IBD type in differential gene expression [Supplementary Figure S9], this variable along with age and sex, were only included as covariates in our model. A significant positive association between baiCD bacterial gene abundance and expression of Angptl4 host gene in non-inflamed sigmoid colon [n = 39 samples, partial Spearman’s rho = -0.65, p-value = 2.09 × 10-05, FDR = 0.03] was identified [Figure 3A; Supplementary Data, Excel file 2]. The association between baiCD and Angptl4 gene was not observed when inflamed sigmoid colon mucosa was analysed [Figure 3B]. No host gene expression-microbial gene associations were identified in other biopsy locations [see Supplementary Data, Excel file 2]. To understand the dissimilar correlation between baiCD and Angptl4 genes in ileal and colonic mucosa, we analysed the effect of endoscopic inflammation on these genes according to biopsy location. Whereas inflammation significantly downregulated the expression of Angptl4 in the terminal ileum, non-effect or the opposite effect was noticed in the ascending colon and sigmoid colon mucosa, respectively [Figure 4A]. This might indicate a differential regulation of Angptl4 gene according to the spatial location in the intestinal tract. No significant differences were found in the relative abundance of bacterial baiCD genes according to inflammatory status [Figure 4B]. Finally, we did not find correlations between other host genes (including bile acid receptors GPBAR1 [TGR5], NR1H4 [FXR] and VDR) and the studied microbial genes [see Supplementary Data, Excel file 2].
Figure 3.
Partial Spearman correlation between microbial baiCD gene abundance [qPCR fold change] and host Angptla4 gene [log transformation of normalised gene counts] conditioned by age at colonoscopy, sex, and clinical diagnosis. A] Significant association in non-inflamed sigmoid colon mucosa of IBD patients [n = 39 biopsy samples, Spearman’s rho = 0.65, p-value = 2.09 × 10-05, FDR = 0.03]. B] No association between baiCD and Angptl4 in inflamed sigmoid colon mucosa of IBD patients [n = 38 biopsy samples, Spearman’s rho = -0.05, p-value = 0.78, FDR = 0.92]. IBD, inflammatory bowel disease; FDR, false discovery rate; qPCR, quantitative polymerase chain reaction.
Figure 4.
Differential expression of host Angptl4 gene and bacterial baiCD gene according to mucosal inflammation in inflammatory bowel disease patients [IBD]. A] expression of Angptl4 is depicted as log-normalised counts in the terminal ileum, ascending colon, and sigmoid colon. Boxplots depicting the median and interquartile range. Asterisks indicate significant differential expression compared with healthy controls [HC] using a negative binomial generalised linear model [GLM], FDR <0.05. B] Differential expression of baiCD bacterial gene is depicted as fold change calculated by 2^[-ΔΔCt] formula in the terminal ileum, ascending colon, and sigmoid colon. Bar plots include the mean and standard error. FDR, false discovery rate.
3.6. Bayesian network analysis of microbial genes, Angptl4, and host phenotype in sigmoid colon mucosa
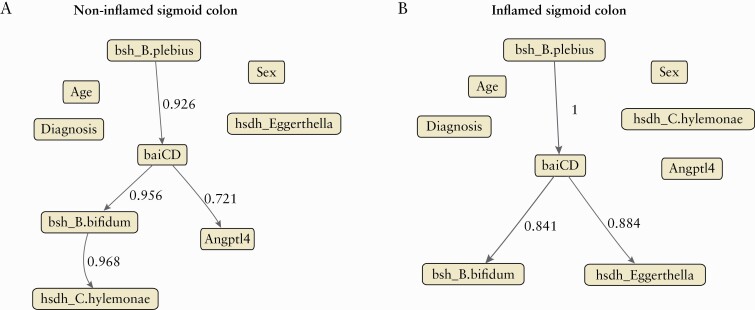
To better understand the interactions and infer the putative direction between the studied microbial genes and Angptl4 host gene in sigmoid colon, taking into account patient characteristics [i.e., age, sex, and IBD type], we used a conditional probabilistic graphical BN model. Similar to previous analysis, each biopsy location and non-inflamed and inflamed mucosa were analysed separately. A total of nine variables were used as the input of the model [Figure 5]. Continuous variables such as age, microbial gene abundance, and Angptl4 gene expression, were discretised before including them in the model. In non-inflamed sigmoid colon mucosa, there were four directed edges that after bootstrap resampling met the empirical threshold frequency of 0.6 [Figure 5A] to be considered significant associations. According to this model, the baiCD microbial gene abundance was the only studied microbial gene putatively affecting Angptl4 host gene expression in non-inflamed sigmoid colon mucosa [empirical bootstrap frequency = 0.721]. When inflamed mucosa was analysed, the influence of baiCD gene abundance in Agptl4 host gene expression was lost [empirical bootstrap frequency = 0.125] [Figure 5B]. No associations between age, sex, or clinical diagnosis and Anptl4 host gene were identified.
Figure 5.
Directed acyclic graph [DAG] depicting a conditional probabilistic Bayesian network. Strength and direction of associations based on the hill-climbing algorithm with 1000 bootstraps. Edges [arrows] connecting the nodes represent probabilistic dependencies, and associations with strength ≥0.6 are shown. Labels of edges indicate the strength of associations. Nodes with no edges indicate conditional independence. A] Non-inflamed sigmoid colon mucosa. B] Inflamed sigmoid colon mucosa. Strength of an interaction is defined as the proportion of bootstrap networks that support the presence of a given edge in a given direction.
4. Discussion
Bile acids are highly abundant endogenous molecules derived from the host-gut microbiota cometabolism, with a recognised effect on immune regulation.41 Previous studies have demonstrated that stool samples from IBD patients show an impairment in the bile acid-metabolising capacity of the gut microbiome compared with non-IBD controls, which has been implicated in the IBD pathogenesis.7,10,42 In the present study, we analysed the effect of inflammation on bile acid-metabolising microbiota in mucosal samples in three different biopsy locations [terminal ileum, ascending colon, and sigmoid colon] using PICRUSt and qPCR. PICRUSt detected imputed microbial secondary bile acid biosynthesis [ko00121 pathway] and bsh [COG3049] functions in the three biopsy sites; however, a significant reduction of ko00121 pathway abundance was only observed in inflamed ileal mucosa of IBD patients compared with non-inflamed tissue. This finding was independent of age, sex, and IBD type [CD or IBD-U]. Moreover, in our univariate analysis, IBD type did not show an effect on ko00121 pathway abundance either. This might imply an effect of inflammation on mucosa-associated microbiota involved in bile acid metabolism, which is common to UC/IBD-U and CD patients. Although no significant reduction of microbial bsh gene abundance was observed with intestinal inflammation in any of the studied biopsy locations, the taxa contributing to this function significantly varied in the presence of inflammation in all the three biopsy sites. These changes in the taxa contributing to bsh gene function in mucosal microbiota may constitute another mechanism affecting the resulting bile acid deconjugation activity.43 For instance, BSH from Bacteroides has a lower enzymatic activity compared with BSH from other taxa; therefore, an increased contribution of Bacteroides to the bsh gene abundance in the microbial community could impair the mucosal microbiota capacity to produce unconjugated bile acids even with no changes in bsh gene abundance.43 We did not observe a significant effect of inflammation in the relative abundance of ko00121 and COG3049 in colonic samples. This could be explained by spatial differences previously reported in the taxonomic profile along the gastrointestinal tract44 producing variable microbial resilience to host inflammation.45
Gut microbes in the small and large bowel deconjugate bile salts to free bile acids by means of BSH enzymes, which makes bile acids accessible for subsequent microbial biotransformation into secondary bile acids.46 After bile acid deconjugation, the minor fraction [~5%] of unconjugated bile acids that overcome ileal reabsorption undergo chemical diversification starting in the terminal ileum and continuing in the colon via two main microbial pathways: dehydroxylation and dehydrogenation reactions.3 Our PICRUSt analysis did not identify the 7α-dehydroxylating function [KEGG orthologue K15870] in mucosal samples, and no KEGG orthologue was identified for hsdh gene function. This emphasises that imputed functions carried out by under-represented species may not be addressed by 16S rRNA-based metagenome-inferring tools.27 However, the abundance of these bacterial genes have been extensively reported in human stool specimens in the context of metabolic diseases, infectious conditions, and IBD using shotgun metagenomics.9,28,43,47
Unfortunately, the low microbial biomass and extremely high fraction of human DNA make the application of shotgun metagenomic sequencing challenging in mucosal samples.48 Consequently, we aim to investigate the presence and abundance of bsh, baiCD, and hsdh microbial genes in human mucosal samples using specific qPCR primers. Unlike stool specimens, mucosal samples provide a more accurate picture of those microbes directly in contact with and more likely to influence the host.6,12–15,49 Leveraging this more suitable niche to study the microbiota-host interaction, we identified that an increased abundance of microbial baiCD genes is associated with the upregulation of the Angptl4 host gene in non-inflamed sigmoid colon biopsies of IBD patients. Microbial baiCD gene is located within a bai operon and encodes a stereo-specific NAD[H]-dependent 3-dehydro-4-bile acid oxidoreductase. This is a critical enzyme in the 7α-dehydroxylation responsible for the biosynthesis of the main secondary bile acids DCA and LCA.50 Although 7α-dehydroxylation is limited to a small subset of gut microbes, this process is extremely efficient, determining that nearly 100% of bile acids in the colon are secondary bile acids.3 Indeed, we used a previously validated primer that amplifies the baiCD gene from C. scindens and C. hiranonis whose 7α-dehydroxylating activity is at least 10 times higher than other species.51 Our analysis of non-inflamed and inflamed mucosa of IBD patients shows that the microbial baiCD-host Angptl4 gene cross-talk in the sigmoid colon is lost in the presence of inflammation. This observation is in line with LePage et al., who identified a significant decrease of transcript/bacterial genus pairs in the colonic mucosa of UC patients in the context of colitis.12 Loss of relevant microbe-host dialogue in active IBD may have a crucial pathogenic role. Although the correlation between Angptl4 host gene and microbial baiCD gene was not statistically significant in ascending colon and terminal ileum, the negative partial Spearman correlation was numerically weaker in inflamed compared with non-inflamed mucosa [ascending colon: -0.13 vs -0.49; terminal ileum: -0.15 vs -0.28].
Transcriptional differences between terminal ileum and colon are pronounced and were noticeable in our PCA. In fact, the differential expression analysis of Agptl4 gene showed an opposite response to inflammation in terminal ileum compared with sigmoid colon. These distinct transcriptomic patterns have been previously described along the proximal-distal axis of the small and large intestine.52 These transcriptional differences might explain divergent relationships between host and bile acid-metabolising microbiota in our three studied biopsy locations.
Although our study design does not allow us to prove biological causality, the BN analysis implies that baiCD microbial genes might influence the expression of the Angptl4 host gene. Interestingly, previous evidence has shown that microbial-derived molecules, including bile acids and short chain fatty acids [SCFA], can modulate Angptl4 expression in entero-endocrine HuTu-80 cell lines.53Angptl4 is a ubiquitously expressed glycoprotein playing a crucial role in lipid homeostasis by inhibiting the enzyme lipoprotein lipase54 and has been implicated as a potential link between the gut microbiota and fat storage.55,56 To the best of our knowledge, a pathogenic role of ANGPTL4 in human IBD has not been demonstrated; however, murine and in vitro models have identified a relevant role of ANGPTL4 in many inflammation-associated diseases, including the capacity of attenuating chemical and dietary saturated fat-induced intestinal inflammation in mice.57,58
Our study has a number of strengths and limitations. This cohort provides us with a high proportion of minimally medicated IBD patients in endoscopic remission [less than 25% of subjects on immunomodulators/biologics at colonoscopy]. This enabled us to evaluate the microbial-host gene interaction in the absence of endoscopic inflammation or potent anti-inflammatory therapy, which may confound or blunt the host-microbe interaction.12 However, our limited sample size impeded some clinically relevant comparisons. For instance, we did not observe differences in bile acid metabolising genes or significant host-microbe interactions when CD and UC/IBD-U patients were separately analysed, which could be determined for the significant reduction in the sample size. However, analysing CD and UC/IBD-U together is common in studies evaluating the effect of inflammation on host transcriptomics and host transcriptomics-microbiome interaction,6,14,59 especially given the limited effect of IBD type on gene expression profile.59 Our unsupervised analysis confirmed this common observation.
Similarly, due to the small group of PSC patients and CD patients with previous ileocaecal resection, we were not able to analyse the influence of liver disease and ileal disease recurrence, where bile acids and bile acid-metabolising microbiota could play a significant pathogenic role. Although our PICRUSt analysis showed an appropriate prediction accuracy [NSTI] of metagenome functions, this approach is only an inference of the secondary bile acid biotransformation and bsh gene abundance, especially considering that many species or strains can carry more than one allele encoding these functions,60 and horizontal gene transmission among bacteria has been identified.47 One of the main limitations in our current approach for studying host gene expression-bile acid-metabolising microbe interactions is the lack of direct bile acid measurement, the end-products of the analysed microbial genes. By using PICRUSt and qPCR, we can only investigate the ex vivo genomic potential of microbes to synthesise bile acids. Though the baiCD gene expression evaluated with the PCR primer used in our study has been significantly correlated with 7α-dehydroxylation activity assays,29 other biologic factors such as hepatic biosynthesis, substrate availability, and entero-hepatic circulation, can also influence the concentration of microbial-derived bile acids. Finally, we emphasise that despite the results of our BN model, no causal inferences can be drawn from our study, especially considering the intricate and bidirectional relationship between gut microbiota, bile acids metabolism, and host. In fact, it has been proposed that ANGPTL4 may influence bile acid absorption through a mechanism dependent on modulation of the gut microbiota composition,61 so an effect of ANGPTL4 on bile acid-metabolising microbiota cannot be ruled out. Moreover, other microbial metabolites synthesised by baiCD-bearing Clostridia, such as SCFA, could also drive the effect on host Angptl4 gene expression.62 Our findings should be further validated in independent IBD cohorts to determine their generalisation.
In conclusion, our study shows for the first time an interaction between microbial baiCD genes and host Angptl4 gene in non-inflamed intestinal mucosa of IBD patients. This study provides evidence that expands the growing realisation of the relevance of bile acid-metabolising microbiota in IBD. Controlled experimental studies determining the impact of specific microbial functions related to bile acid metabolism are needed to support these findings. As far as our knowledge, this is also the first report demonstrating an association between a crucial and specific microbial function involved in secondary bile acid biosynthesis and host Angptl4 gene. Angptl4 has exhibited the capacity to attenuate colonic inflammation in animal models57; therefore, modulation of baiCD abundance in the gut microbiota may be employed to influence Angptl4 expression and modulate the intestinal inflammation in IBD.
Supplementary Data
Supplementary data are available at ECCO-JCC online. Excel files can be accessed through the following Dropbox link: https://www.dropbox.com/sh/k5ot3wuqnucat9x/AACvB2489oZm8Soq-o9MUPq2a?dl=0
Acknowledgements
Some computations were performed on the Niagara supercomputer at the SciNet HPC Consortium. SciNet is funded by: the Canada Foundation for Innovation; the Government of Ontario; Ontario Research Fund‐Research Excellence; and the University of Toronto. We would like to acknowledge the National Institute of Diabetes and Digestive and Kidney Diseases IBD Genetics Consortium [NIDDK IBDGC] for their support.
Funding
This work was supported by the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health [grant number U01DK062423]. CH-R holds a Fellowship Award from the Canadian Institutes of Health Research [CIHR] and the Canadian Association of Gastroenterology [CAG]. WT and JG are recipients of a Fellowship from the Department of Medicine, Mount Sinai Hospital, Toronto.
Conflict of Interest
None declared.
Author Contributions
CH-R: concept and design of the study; acquisition, analysis and interpretation of data; manuscript drafting. KB: analysis and interpretation of data; revising the article critically for important intellectual content. WT: analysis and interpretation of data; revising the article critically for important intellectual content. MF: acquisition of data; revising the article critically for important intellectual content. SN: analysis and interpretation of data; revising the article critically for important intellectual content. JG: analysis and interpretation of data; revising the article critically for important intellectual content. JS: acquisition of data; revising the article critically for important intellectual content. MS: contributed to study concept and design; planning of the study; acquisition, analysis and interpretation of data; critical revision of the manuscript for important intellectual content; and obtained funding, material support, and study supervision. All the authors revised and approved the manuscript. Preliminary results were presented at the Digestive Disease Week in San Diego, May 2019.
References
- 1. Cohen LJ, Cho JH, Gevers D, Chu H. Genetic factors and the intestinal microbiome guide development of microbe-based therapies for inflammatory bowel diseases. Gastroenterology 2019;156:2174–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol 2016;16:341–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res 2006;47:241–59. [DOI] [PubMed] [Google Scholar]
- 4. Winston JA, Theriot CM. Diversification of host bile acids by members of the gut microbiota. Gut Microbes 2020;11:158–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Franzosa EA, Sirota-Madi A, Avila-Pacheco J, et al. Author Correction: gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol 2019;4:898. [DOI] [PubMed] [Google Scholar]
- 6. Lloyd-Price J, Arze C, Ananthakrishnan AN, et al. ; IBDMDB Investigators. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019;569:655–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Heinken A, Ravcheev DA, Baldini F, Heirendt L, Fleming RMT, Thiele I. Systematic assessment of secondary bile acid metabolism in gut microbes reveals distinct metabolic capabilities in inflammatory bowel disease. Microbiome 2019;7:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Labbé A, Ganopolsky JG, Martoni CJ, Prakash S, Jones ML. Bacterial bile metabolising gene abundance in Crohn’s, ulcerative colitis and type 2 diabetes metagenomes. PLoS One 2014;9:e115175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Das P, Marcišauskas S, Ji B, Nielsen J. Metagenomic analysis of bile salt biotransformation in the human gut microbiome. BMC Genomics 2019;20:517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sinha SR, Haileselassie Y, Nguyen LP, et al. Dysbiosis-induced secondary bile acid deficiency promotes intestinal inflammation. Cell Host Microbe 2020;27:659–70.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Joyce SA, MacSharry J, Casey PG, et al. Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proc Natl Acad Sci U S A 2014;111:7421–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lepage P, Häsler R, Spehlmann ME, et al. Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology 2011;141:227–36. [DOI] [PubMed] [Google Scholar]
- 13. Haberman Y, Tickle TL, Dexheimer PJ, et al. Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J Clin Invest 2014;124:3617–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Häsler R, Sheibani-Tezerji R, Sinha A, et al. Uncoupling of mucosal gene regulation, mRNA splicing and adherent microbiota signatures in inflammatory bowel disease. Gut 2017;66:2087–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Quraishi MN, Acharjee A, Beggs AD, et al. A pilot integrative analysis of colonic gene expression, gut microbiota, and immune infiltration in primary sclerosing cholangitis-inflammatory bowel disease: association of disease with bile acid pathways. J Crohns Colitis 2020;14:935–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schroeder KW, Tremaine WJ, Ilstrup DM. Coated oral 5-aminosalicylic acid therapy for mildly to moderately active ulcerative colitis. A randomized study. N Engl J Med 1987;317:1625–9. [DOI] [PubMed] [Google Scholar]
- 17. Daperno M, D’Haens G, Van Assche G, et al. Development and validation of a new, simplified endoscopic activity score for Crohn’s disease: the SES-CD. Gastrointest Endosc 2004;60:505–12. [DOI] [PubMed] [Google Scholar]
- 18. Boland K, Bedrani L, Turpin W, et al. Persistent diarrhea in patients with Crohn’s disease after mucosal healing is associated with lower diversity of the intestinal microbiome and increased dysbiosis. Clin Gastroenterol Hepatol 2021;19:296–304.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Caporaso JG, Lauber CL, Walters WA, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 2012;6:1621–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010;7:335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 2006;72:5069–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Langille MG, Zaneveld J, Caporaso JG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 2013;31:814–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 2004;20:289–90. [DOI] [PubMed] [Google Scholar]
- 24. Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 2000;28:33–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000;28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome Biol 2011;12:R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mullish BH, Pechlivanis A, Barker GF, Thursz MR, Marchesi JR, McDonald JAK. Functional microbiomics: evaluation of gut microbiota-bile acid metabolism interactions in health and disease. Methods 2018;149:49–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mullish BH, McDonald JAK, Pechlivanis A, et al. Microbial bile salt hydrolases mediate the efficacy of faecal microbiota transplant in the treatment of recurrent Clostridioides difficile infection. Gut 2019;68:1791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wells JE, Williams KB, Whitehead TR, Heuman DM, Hylemon PB. Development and application of a polymerase chain reaction assay for the detection and enumeration of bile acid 7α-dehydroxylating bacteria in human feces. Clinica Chimica Acta 2003;331:127–34. [DOI] [PubMed] [Google Scholar]
- 30. Mythen SM, Devendran S, Méndez-García C, Cann I, Ridlon JM. Targeted synthesis and characterization of a gene cluster encoding NAD[P]H-dependent 3α-, 3β-, and 12α-hydroxysteroid dehydrogenases from Eggerthella CAG:298, a gut metagenomic sequence. Appl Environ Microbiol 2018;84. Doi: 10.1128/AEM.02475-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jarocki P, Targoński Z. Genetic diversity of bile salt hydrolases among human intestinal bifidobacteria. Curr Microbiol 2013;67:286–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Doden H, Sallam LA, Devendran S, et al. Metabolism of oxo-bile acids and characterization of recombinant 12α-hydroxysteroid dehydrogenases from bile acid 7α-dehydroxylating human gut bacteria. Appl Environ Microbiol 2018;84. Doi: 10.1128/AEM.00235-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Robertson SJ, Zhou JY, Geddes K, et al. Nod1 and Nod2 signaling does not alter the composition of intestinal bacterial communities at homeostasis. Gut Microbes 2013;4:222–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Blighe K, Lun A. PCAtools: PCAtools: Everything Principal Components Analysis. R package version 2.2.0, 2020. https://github.com/kevinblighe/PCAtools. Accessed 20 April 2021.
- 35. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Song X, Sun X, Oh SF, et al. Microbial bile acid metabolites modulate gut RORγ + regulatory T cell homeostasis. Nature 2019:1–6. Doi: 10.1038/s41586-019-1865-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hang S, Paik D, Yao L, et al. Bile acid metabolites control TH17 and Treg cell differentiation. Nature 2019;576:143–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. R Core Team. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2018. [Google Scholar]
- 39. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 1995;57:289–300. [Google Scholar]
- 40. Scutari M. Learning Bayesian Networks with the bnlearn R Package. J Stat Softw 2010;35:1–22.21603108 [Google Scholar]
- 41. Long SL, Gahan CGM, Joyce SA. Interactions between gut bacteria and bile in health and disease. Mol Aspects Med 2017;56:54–65. [DOI] [PubMed] [Google Scholar]
- 42. Duboc H, Rajca S, Rainteau D, et al. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut 2013;62:531–9. [DOI] [PubMed] [Google Scholar]
- 43. Song Z, Cai Y, Lao X, et al. Taxonomic profiling and populational patterns of bacterial bile salt hydrolase [BSH] genes based on worldwide human gut microbiome. Microbiome 2019;7:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Villmones HC, Haug ES, Ulvestad E, et al. Species level description of the human ileal bacterial microbiota. Sci Rep 2018;8:4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cullen TW, Schofield WB, Barry NA, et al. Gut microbiota. Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science 2015;347:170–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Joyce SA, Shanahan F, Hill C, Gahan CG. Bacterial bile salt hydrolase in host metabolism: potential for influencing gastrointestinal microbe-host crosstalk. Gut Microbes 2014;5:669–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jones BV, Begley M, Hill C, Gahan CG, Marchesi JR. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci U S A 2008;105:13580–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Quince C, Walker AW, Simpson JT, Loman NJ, Segata N. Corrigendum: shotgun metagenomics, from sampling to analysis. Nat Biotechnol 2017;35:1211. [DOI] [PubMed] [Google Scholar]
- 49. Dayama G, Priya S, Niccum DE, Khoruts A, Blekhman R. Interactions between the gut microbiome and host gene regulation in cystic fibrosis. Genome Med 2020;12:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wells JE, Hylemon PB. Identification and characterization of a bile acid 7alpha-dehydroxylation operon in Clostridium sp. strain TO-931, a highly active 7alpha-dehydroxylating strain isolated from human feces. Appl Environ Microbiol 2000;66:1107–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kitahara M, Takamine F, Imamura T, Benno Y. Assignment of Eubacterium sp. VPI 12708 and related strains with high bile acid 7alpha-dehydroxylating activity to Clostridium scindens and proposal of Clostridium hylemonae sp. nov., isolated from human faeces. Int J Syst Evol Microbiol 2000;50[Pt 3]:971–8. [DOI] [PubMed] [Google Scholar]
- 52. LaPointe LC, Dunne R, Brown GS, et al. Map of differential transcript expression in the normal human large intestine. Physiol Genomics 2008;33:50–64. [DOI] [PubMed] [Google Scholar]
- 53. Alex S, Lichtenstein L, Dijk W, Mensink RP, Tan NS, Kersten S. ANGPTL4 is produced by entero-endocrine cells in the human intestinal tract. Histochem Cell Biol 2014;141:383–91. [DOI] [PubMed] [Google Scholar]
- 54. Dijk W, Kersten S. Regulation of lipoprotein lipase by Angptl4. Trends Endocrinol Metab 2014;25:146–55. [DOI] [PubMed] [Google Scholar]
- 55. El Aidy S, Merrifield CA, Derrien M, et al. The gut microbiota elicits a profound metabolic reorientation in the mouse jejunal mucosa during conventionalisation. Gut 2013;62:1306–14. [DOI] [PubMed] [Google Scholar]
- 56. Janssen AWF, Katiraei S, Bartosinska B, Eberhard D, Willems van Dijk K, Kersten S. Loss of angiopoietin-like 4 [ANGPTL4] in mice with diet-induced obesity uncouples visceral obesity from glucose intolerance partly via the gut microbiota. Diabetologia 2018;61:1447–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Phua T, Sng MK, Tan EH, et al. Angiopoietin-like 4 mediates colonic inflammation by regulating chemokine transcript stability via tristetraprolin. Sci Rep 2017;7:44351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lichtenstein L, Mattijssen F, de Wit NJ, et al. Angptl4 protects against severe proinflammatory effects of saturated fat by inhibiting fatty acid uptake into mesenteric lymph node macrophages. Cell Metab 2010;12:580–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hu S, Uniken Venema WT, Westra HJ, et al. Inflammation status modulates the effect of host genetic variation on intestinal gene expression in inflammatory bowel disease. Nat Commun 2021;12:1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lambert JM, Bongers RS, de Vos WM, Kleerebezem M. Functional analysis of four bile salt hydrolase and penicillin acylase family members in Lactobacillus plantarum WCFS1. Appl Environ Microbiol 2008;74:4719–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Janssen AWF, Dijk W, Boekhorst J, et al. ANGPTL4 promotes bile acid absorption during taurocholic acid supplementation via a mechanism dependent on the gut microbiota. Biochim Biophys Acta Mol Cell Biol Lipids 2017;1862:1056–67. [DOI] [PubMed] [Google Scholar]
- 62. Alex S, Lange K, Amolo T, et al. Short-chain fatty acids stimulate angiopoietin-like 4 synthesis in human colon adenocarcinoma cells by activating peroxisome proliferator-activated receptor γ. Mol Cell Biol 2013;33:1303–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.