Abstract
Streptococcus gordonii and Streptococcus sanguinis are primary colonizers of tooth surfaces and are generally associated with oral health, but can also cause infective endocarditis (IE). These species express “Siglec-like” adhesins that bind sialylated glycans on host glycoproteins, which can aid the formation of infected platelet–fibrin thrombi (vegetations) on cardiac valve surfaces. We previously determined that the ability of S. gordonii to bind sialyl T-antigen (sTa) increased pathogenicity, relative to recognition of sialylated core 2 O-glycan structures, in an animal model of IE. However, it is unclear when and where the sTa structure is displayed, and which sTa-modified host factors promote valve colonization. In this study, we identified sialylated glycoproteins in the aortic valve vegetations and plasma of rat and rabbit models of this disease. Glycoproteins that display sTa vs. core 2 O-glycan structures were identified by using recombinant forms of the streptococcal Siglec-like adhesins for lectin blotting and affinity capture, and the O-linked glycans were profiled by mass spectrometry. Proteoglycan 4 (PRG4), also known as lubricin, was a major carrier of sTa in the infected vegetations. Moreover, plasma PRG4 levels were significantly higher in animals with damaged or infected valves, as compared with healthy animals. The combined results demonstrate that, in addition to platelet GPIbα, PRG4 is a highly sialylated mucin-like glycoprotein found in aortic valve vegetations and may contribute to the persistence of oral streptococci in this protected endovascular niche. Moreover, plasma PRG4 could serve as a biomarker for endocardial injury and infection.
Keywords: cardiovascular, ITIH4, O-linked glycan, sialic acid, thrombus
Introduction
Infective endocarditis (IE) is a life-threatening cardiovascular disease in which microbes colonize and persist within platelet–fibrin thrombi (vegetations) on cardiac valve surfaces. Histopathologic studies in humans, along with data from animal models of IE, indicate that IE is preceded by nonbacterial thrombotic endocarditis (NBTE), which is characterized by the formation of “sterile vegetations” on the valve surface (Durack and Beeson 1972; Werdan et al. 2014). In animal models, the intact valve endothelial surface is resistant to bacterial colonization, but microbes readily colonize the sterile vegetations that form on mechanically damaged valves. Subsequent bacterial growth, along with a continued deposition of platelets and other blood components, leads to the formation of macroscopic-infected vegetations. These lesions can embolize and disseminate to other tissues, leading to stroke, metastatic infection and other severe complications.
The precise composition of these cardiac valve vegetations has been only partially characterized. Early studies in rabbit models of IE indicated that infected aortic valve vegetations were composed primarily of platelets and fibrin, with a noted absence of inflammatory cells (Durack and Beeson 1972; Durack 1975). In humans with IE, the protein and cellular composition of vegetations can vary extensively and appears to be somewhat dependent on the infecting bacterial species. In a proteomics analysis, Martin et al. (2020) found that the major components of human IE vegetations included vimentin, hemoglobin B and β-actin, along with fibrinogen α and β subunits. The authors noted extensive proteolysis of the some of the major components and degradation patterns that were highly variable. In some cases, there has been evidence of either neutrophils or neutrophil extracellular traps in human IE vegetations (Sass et al. 2016; Martin et al. 2020). Other studies have implicated additional plasma and clotting factors (Foster et al. 2014). Perhaps, because of heterogeneity of both the host immune and acute-phase responses, as well as the infecting species, it has not been entirely resolved how the various vegetation components facilitate bacterial attachment, proliferation and persistence in IE.
Rabbit or rat IE models have traditionally been used to study bacterial pathogenesis (Liesenborghs et al. 2020). These models involve inserting a trans-aortic catheter prior to infection, which causes mechanical damage of the aortic valve and stimulates the formation of NBTE. Shorter-term infections (1 h or less) have been used to gauge the initial attachment of organisms to sterile vegetations after intravenous challenge. A number of bacterial cell surface adhesins may contribute to the initiation of infection by promoting adherence to fibrinogen, fibronectin and/or membrane proteins of platelets that have deposited on the damaged valve surface (Fitzgerald et al. 2006; Haworth et al. 2017). The latter include platelet glycoprotein Ibα (GPIbα, the receptor for von Willebrand factor [vWF]) and integrin 2bIIIa (the fibrinogen receptor). Longer term infections (18 h or longer) are used to examine bacterial factors that aid the formation of macroscopic vegetations, such as nutrient acquisition, immune evasion and procoagulant activity (Drake et al. 1984; Ge et al. 2008, 2016; Das et al. 2009; Turner et al. 2009; Crump et al. 2014; Baker et al. 2019; Gaytan et al. 2021).
Commensal oral streptococci, including Streptococcus gordonii, Streptococcus sanguinis and Streptococcus oralis, are among the leading causative agents of IE (Kitten et al. 2012; Isaksson et al. 2015; Rasmussen et al. 2016; Chamat-Hedemand et al. 2020). These streptococcal species are primary colonizers of human teeth and gums and express “Siglec-like” cell-surface adhesins that mediate binding to sialylated glycans on MUC7 or other salivary glycoproteins (Plummer and Douglas 2006; Takamatsu et al. 2006; Bensing et al. 2016; Ronis et al. 2019). The Siglec-like adhesins have also been shown to mediate streptococcal binding to platelets via interaction with sialylated glycans on GPIbα (Plummer et al. 2005; Takamatsu et al. 2005; Pyburn et al. 2011). Expression of Siglec-like adhesins by S. gordonii and S. oralis has been linked to enhanced virulence in animal models of IE (Takahashi et al. 2006; Xiong et al. 2008; Pyburn et al. 2011; Bensing et al. 2019a; Gaytan et al. 2021). A few other sialylated ligands have been detected in plasma and on blood cells (Takahashi et al. 2002; Urano-Tashiro et al. 2008; Deng et al. 2014; Haworth et al. 2017; Bensing et al. 2018), but whether these or other sialylated mucin-like proteins are present within the aortic valve vegetations has not been examined.
In a previous study, we noted that binding to a specific sialoglycan structure, a trisaccharide known as sialyl T-antigen (sTa; Figure 1), was correlated with a significant increase in virulence of S. gordonii, as compared with an isogenic strain that recognized 3'sialyllactosamine (3'sLn) and related structures that are found in larger, branched, core 2 sialoglycans. We also determined that the O-linked glycan structures of rat platelet GPIbα are similar to those displayed on human GPIbα. In both cases, most of the O-glycans are di-sialylated core 2 hexasaccharide structures, but the terminal sialic acids of the rat GPIbα include neuraminic acid and N-glycolyl neuraminic acid, in addition to N-acetyl neuraminic acid (Bensing et al. 2019a). sTa is a relatively minor fraction of the O-glycans on GPIbα, and it has not been determined whether any other glycoproteins found in aortic valve vegetations display this structure.
Fig. 1.
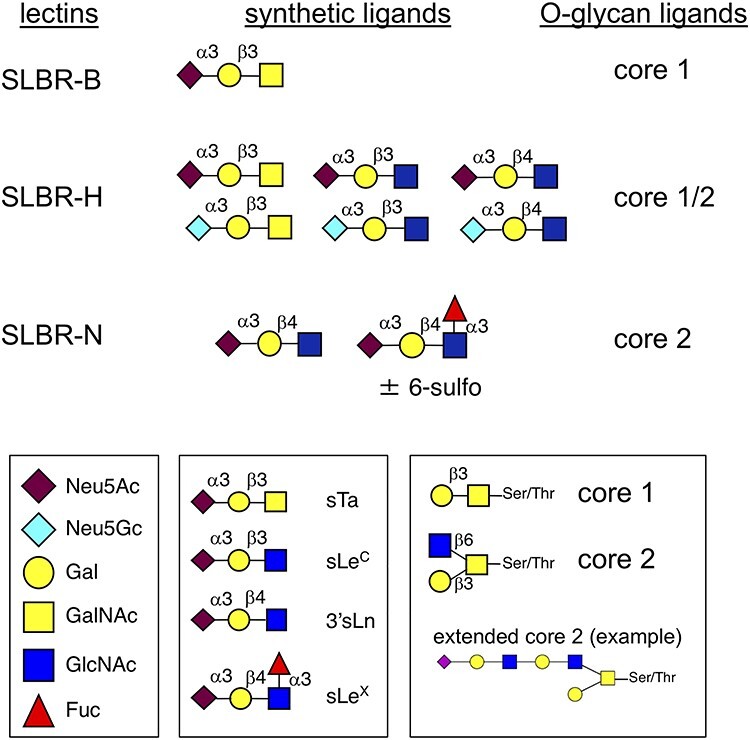
Recombinant streptococcal lectins and sialoglycan structures. The SLBRs are derived from Streptococcus gordonii serine-rich repeat adhesins and recognize different subsets of α2–3 sialylated O-glycans (Bensing et al. 2018). SLBR-B is from GspB; SLBR-H is from Hsa; SLBR-N is from the UB10712 homolog (formerly referred to as the Streptococcus mitis NCTC10712 homolog). The high-affinity synthetic ligands were identified by array and ELISA studies (Bensing et al. 2016, 2019b). The O-glycan subsets were inferred from affinity capture and O-glycan profiling experiments and from cell-based mucin sialoglycan display libraries (Bensing et al. 2018; Narimatsu et al. 2019). The core 2 glycan extensions may include poly-lactosamine, fucose and sulfate.
In this report, we examined aortic valve vegetations and plasma from animals with IE due to S. gordonii, to determine which glycoproteins most prominently display sialylated core 1 and core 2 glycans. We identified several mucin-like proteins modified with sialylated O-glycans, using three well-characterized streptococcal lectins (Siglec-like binding regions, or SLBRs) for detection and affinity capture. We found proteoglycan 4 (PRG4) is the main carrier of sTa in aortic valve vegetations and plasma, in both rat and rabbit models of IE. Moreover, we found that increased plasma PRG4 levels are generally correlated with sustained valve damage.
Results
Aortic valve vegetations have three major sialylated glycoproteins
Previous studies have indicated that sialoglycan binding by streptococci may contribute to both the initial colonization of sterile vegetations and the longer-term persistence within vegetations (Takahashi et al. 2006; Xiong et al. 2008; Pyburn et al. 2011; Gaytan et al. 2021). Accordingly, we looked for sialylated glycoproteins within aortic valve vegetations of rats with IE, using tissues that had been collected 1 or 72 h postinfection with S. gordonii and banked from an earlier study (Bensing et al. 2019a). The homogenized and electrophoretically separated vegetation proteins (Figure 2A) were probed with recombinant streptococcal adhesins that recognize either (a) sTa (SLBR-B), (b) 3'sLn and a number of related structures on branched core 2 O-glycans (SLBR-N) or (c) both sTa and 3'sLn (SLBR-H). The sTa-selective SLBR-B detected a single ~400 kDa glycoprotein (Figure 2B), whereas SLBR-N recognized proteins of 150 and 100–120 kDa (Figure 2C). SLBR-H recognized the 400 and 150 kDa glycoproteins, but not the 100–120 kDa proteins (Figure 2D). Based on the known ligand preferences of these probes (Figure 1), the latter finding suggests that the 100–120 kDa glycoproteins may display fucosylated, sialyl Lewis X structures on the O-linked glycans.
Fig. 2.
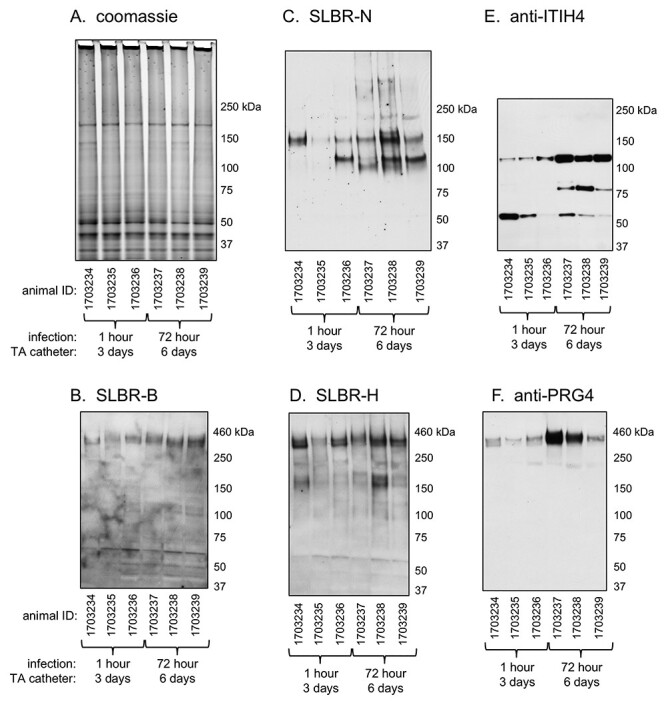
Lectin and western blotting of Streptococcus gordonii-infected rat aortic valve vegetations. All animals were catheterized 3 days prior to infection with S. gordonii, and the transaortic (TA) catheters remained in place for the duration of the experiment. Vegetations were harvested from three animals 1 h after infection (on day 3) or three animals 72 h after infection (on day 6) as indicated. Lanes contain 5 μg homogenized vegetation proteins. Parallel samples were either stained with Coomassie or were transferred to nitrocellulose and probed with antibodies or SLBRs as indicated.
The SLBR-reactive glycoproteins were enriched from the homogenized vegetations by affinity capture using immobilized glutathione S-transferase (GST)-tagged SLBRs, and then identified by mass spectrometry (Table I). The most abundant glycoprotein in the 400 kDa region was an orthologue of human PRG4, also known as lubricin (Supplementary Figure S1). The 150 kDa glycoprotein was identified as platelet GPIbα. The most abundant glycoprotein in the 100–120 kDa region was inter-α-trypsin inhibitor heavy chain 4 (ITIH4). To confirm the presence of PRG4 and ITIH4 in the homogenized vegetations, the homogenized materials were probed with antibodies to these glycoproteins (antibodies that react with rat GPIbα in western blots are not commercially available). ITIH4 and PRG4 were both detected in the vegetation samples (Figure 2E and F, respectively) and were more readily apparent after 72 h compared with the 1 h infections. In addition, the ITIH4 fragmentation pattern differed at 72 vs. 1 h. The apparent proteolysis of ITIH4 in rat vegetations is consistent with the proteolysis of ITIH4 in human IE vegetations (Martin et al. 2020).
Table I.
Identification by mass spectrometry of SLBR-captured rat vegetation glycoproteinsa
| Apparent mass (kDa) | SLBRb | Glycoprotein | UniProt ID | Sequence coverage (%) | Theoretical mass (kDa) |
|---|---|---|---|---|---|
| 400 | H | PRG4 | F1LRA5 | 32c | 116 |
| 150 | N | GPIbα | DZ3QU7 | 28 | 78 |
| 100–120 | N | ITIH4 Fibrinogen α |
Q5EBC0 Q7TQ70 |
37 46 |
104 87 |
aAssignment as an O-glycosylated protein was determined by manual curation of GenBank entries for the human orthologues and published reports.
bSLBR used for affinity capture.
cThe identified peptides are diagrammed in the Supplementary Figure S1.
PRG4 and ITIH4 are readily detected in plasma postinfection
ITIH4, GPIbα and PRG4 are orthologs of sialylated, mucin-like glycoproteins that have been detected in plasma from healthy humans, using the SLBRs (Bensing et al. 2018). We therefore hypothesized that plasma could be a source of the vegetation proteins. We first analyzed via western and lectin blotting plasma that had been collected from the above animals concomitant with vegetation collection. PRG4 and ITIH4 were both readily detected in the plasma of animals sacrificed either 1 or 72 h after infection (Figure 3A and B, respectively). As with the vegetation proteins, the PRG4 in plasma was detected with SLBR-H (Figure 3C), but not SLBR-N (Figure 3D), indicative of modification with core 1 type O-linked sialoglycans. However, in plasma, the SLBR-H reactivity directly paralleled the anti-PRG4 reactivity. This is consistent with less animal-to-animal variation in the O-glycan composition of plasma vs. vegetation-derived PRG4. In addition, there was less apparent variation between animals and between 1 and 72 h postinfection, in the plasma PRG4 levels. The SLBR-H and -N reactive 150 kDa glycoprotein (presumably GPIbα) signal was also fairly consistent between animals. In contrast to the ITIH4 detected in vegetations, the plasma ITIH4 was not recognized by SLBR-N and was not fragmented. These results suggest that the major glycoforms of PRG4 and ITIH4 in plasma are different from vegetations glycoforms and support that the presence of these glycoproteins in vegetations is not simply due to carry over or entrapment of plasma components. However, this does not exclude the possibility that minor glycoforms of PRG4 and ITIH4 may transit from plasma to valve.
Fig. 3.
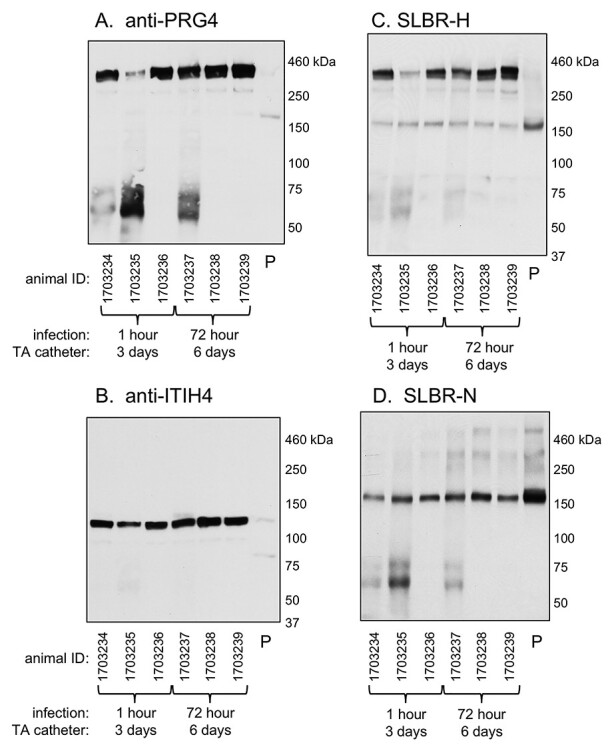
Lectin and western blotting of plasma from IE rats. Plasma from the set of animals from which vegetations were collected and analyzed in Figure 2. Lane 7 (P) contains plasma prepared from commercial Sprague–Dawley rat blood. Lanes contain 0.4 μL of plasma. Samples were probed with antibodies or SLBRs as indicated. Note that the <100 kDa proteins seen in the PRG4 blot were detected with the secondary anti-mouse IgG/IgM alone and are therefore not PRG4 fragments.
PRG4 is increased in plasma of catheterized animals, prior to infection
Since we were unable to detect either PRG4 or ITIH4 in commercial rat blood (Figure 3, lane P), we hypothesized that the presence of these glycoproteins in S. gordonii-infected animals could be due to a host response to either the catheter-induced valve damage or the streptococcal infection. To explore these possibilities, we undertook a longitudinal analysis and analyzed plasma that had been collected from rats at several time points during the course of the experiment: presurgery (i.e. healthy animals, on day 1), postcatherization/preinfection (on day 4) and 1 h postinfection. For each animal, the PRG4 in plasma (detected with SLBR-H or anti-PRG4) appeared to increase sharply postcatheterization, whereas the level of GPIbα remained fairly constant (Figure 4). A further increase in plasma PRG4 was apparent in some animals 1 h after infection with S. gordonii. However, the mean levels pre- and postinfection were not significantly different. Thus, the relatively high levels of PRG4 in plasma of IE rats were primarily correlated with mechanical damage of the aortic valve.
Fig. 4.
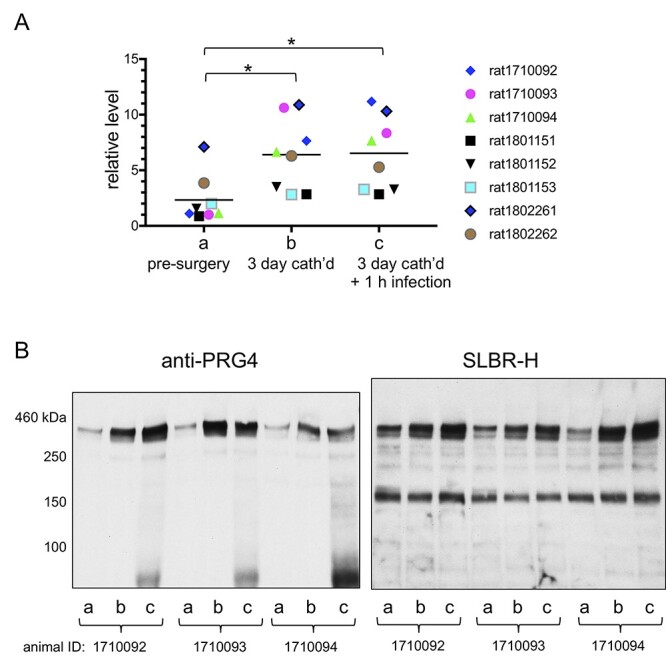
Longitudinal analysis of rat plasma pre- and postinfection. (A) Quantitative analysis of plasma PRG4 from eight animals, collected at three time-points during the experiment: (a) prior to catheterization; (b) 3 days postcatheterization, prior to infection; (c) 1 h after infection with Streptococcus gordonii. Bar indicates mean column value. Statistical significances were determined by two-way ANOVA, followed by Tukey’s post hoc for multiple comparisons. Asterisk indicates P < 0.05. (B) Lectin and western blotting of plasma from three representative animals. Lanes contain 0.4 μL plasma, and blots were probed with anti-PRG4 or SLBR-H. Note that the <100 kDa proteins seen in the PRG4 blot were detected with the secondary anti-mouse IgG/IgM alone and are therefore not PRG4 fragments.
PRG4 is evident in NBTE vegetations
In contrast to the findings for plasma (Figure 3), PRG4 levels in vegetations were distinctly higher in animals infected for 72 vs. 1 h (Figure 2). Increased PRG4 in vegetations could be due either to a host response to the infection or simply to sustained valve trauma (longer time with catheter in place). We therefore sought to compare the valves of catheterized animals that had NBTE vs. IE (i.e. uninfected vs. infected animals, respectively), with undamaged valves from healthy animals. In this case, we collected the aortic valve tissue along with the associated sterile (NBTE) or infected (IE) vegetations 6 days after placement of the trans-aortic catheter (note that IE animals were infected with S. gordonii on day 3). In western blots, PRG4 was more readily detected in valves of NBTE rats compared with healthy valve tissue and was even higher in the valves of two of three animals with IE vs. NBTE (Figure 5). Again, the SLBR-H reactivity did not directly parallel the PRG4 signal, which suggests that the extent of sialylation of the O-glycans on vegetation-associated PRG4 may vary between animals. The results indicate that increased levels of PRG4 in the aortic valve tissue may be due to a host response to both mechanical valve damage and infection.
Fig. 5.
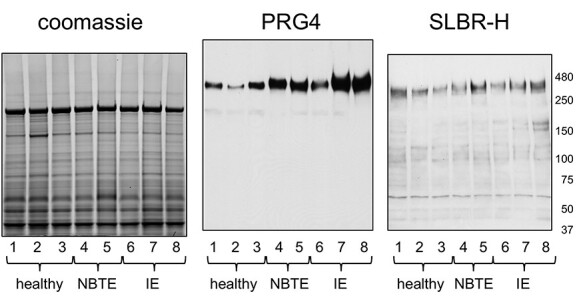
Lectin and western blotting of healthy, NBTE and IE rat aortic valve tissue. Samples were collected from healthy animals (1–3), animals catheterized 6 days (4 and 5) or animals catheterized 6 days and infected with Streptococcus gordonii on day 3 (6–8). Lanes contain 5 μg homogenized tissue. Parallel samples were either stained with Coomassie or transferred to nitrocellulose and probed with anti-PRG4 or SLBR-H as indicated.
O-glycan profiling of rat PRG4
The above results (specifically, the SLBR reactivity) suggested that PRG4 is an sTa-modified glycoprotein present in both plasma and aortic valve vegetations of rats with IE. At 3 vs. 6 days postcatheterization, PRG4 was readily detected in plasma but not in vegetations (Figure 3 vs. Figure 2), which would be consistent with the vegetation PRG4 being deposited from plasma, along with platelets and fibrin. If so, the plasma and vegetation glycoforms should be similar. However, if the plasma and vegetation glycoforms were different, this could indicate that the sources of PRG4 production might be different. It was also unclear whether there might be a change in O-glycosylation associated with the apparent increased levels in plasma concomitant with aortic valve damage. To assess these questions directly, we compared the O-glycans on PRG4 extracted from vegetations, from healthy vs. infected aortic valves and from longitudinally collected plasma. The O-glycans linked to PRG4 were released by beta-elimination and identified by liquid chromatography and tandem mass spectrometry (LC–MS/MS).
The O-glycan structures linked to PRG4 from S. gordonii-infected vegetations were first assessed. PRG4 was captured from homogenized vegetation proteins from two different animals that had been infected with S. gordonii for 72 h. In both vegetation samples, PRG4 displayed a variety of O-linked glycans, with a mix of core 1 and relatively simple core 2 structures (Figure 6 and Supplementary Tables SI and SII). Most of the O-glycans were sialylated. However, one of the two samples had a greater extent of fucosylation and sulfation of the minor structures. In addition, the two samples had very different amounts of sTa (0.7% sTa vs. 40%). These differences are consistent with the different relative levels of SLBR-H vs. PRG4 reactivity seen in Figures 3 and 5.
Fig. 6.
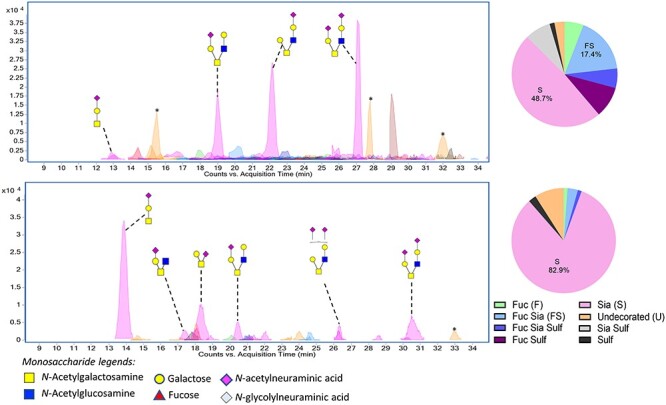
O-glycans on PRG4 from Streptococcus gordonii-infected rat aortic valve vegetations. Upper and lower panels have O-glycans released from PRG4 recovered from the vegetations of two different animals (from 500 μg homogenized vegetation proteins). Asterisks (*) indicate polyhexose O-glycans that were omitted from the pie charts and O-glycan profile tables.
We next compared the O-glycans linked to PRG4 extracted from infected aortic valves with the relatively minimal amount of PRG4 obtained from undamaged aortic valve tissue. In this case, the valve tissue was pooled from the three animals shown in Figure 5 (lanes 6–8 or lanes 1–3, respectively). The O-glycans linked to PRG4 from healthy valve tissue included five major structures (Figure 7, upper panel, and Supplementary Table SIII). Approximately 38% were core 1 structures (18% mono-sialylated and 20% di-sialylated) and 24% were unmodified core 2 or other large structures (not sialylated, fucosylated or sulfated). The O-glycan composition of PRG4 from IE rat valves showed qualitative and quantitative differences compared with that of the healthy valve tissue (Figure 7, lower panel, and Supplementary Table SIV). First was a 10-fold difference in the quantity of O-glycans recovered from damaged vs. healthy valve tissue, consistent with the higher levels detected by western and lectin blotting. Second, the PRG4 recovered from the IE valve tissue had a disproportionate increase in sTa (26 vs. 18%) and decrease in disialylated T-antigen (dsTa; 10 vs. 20%), relative to the other structures, and displayed a disialylated core 2 hexasaccharide, which was absent from the O-glycan profile of PRG4 from the valves of healthy animals. Thus, the pooled infected valve tissue had more sTa and more di-sialylated core 2 hexasaccharide compared with the healthy valve tissue, and those were the two major structures linked to PRG4 in the vegetations of either of two animals (Figure 6).
Fig. 7.
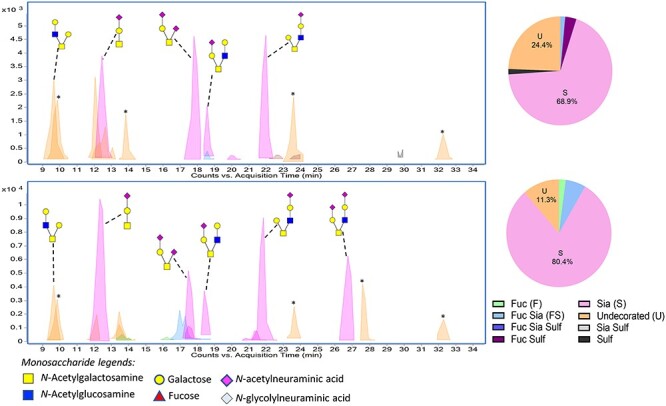
Comparison of PRG4 isolated from intact vs. infected rat valves. PRG4 was enriched from aortic valve tissue pooled from three healthy animals (upper) or three IE animals (lower). The O-glycans were released from the total PRG4 captured from ~1 mg valve tissue. Asterisks (*) indicate polyhexose O-glycans that were omitted from the pie charts and O-glycan profile tables.
We then characterized the O-glycans on rat plasma PRG4 and determined whether the catheter-induced increase in plasma was associated with a change in the O-glycan composition. To do so, PRG4 was recovered from the plasma of a single animal, collected either prior to catheterization, postcatheterization but preinfection, or 72 h postinfection (the animal from which the valve sample shown in Figure 5, lane 7, was harvested). PRG4 recovered from the healthy animal, prior to catheterization displayed O-linked structures similar to those linked to PRG4 in healthy valve tissue, and a similar extent of sialylation, but the sialic acids included minor amounts of Neu5Gc (Figure 8 and Supplementary Table SV). The most abundant structures were sTa (36%) and dsTa (10%). Low levels of sulfated and fucosylated glycans were also detected. Postcatheterization, the O-glycans linked to plasma PRG4 were over 90% sialylated. This included 70% sTa, 8% dsTa and <2% core 2 structures (Figure 8 and Supplementary Table SVI). There was also 20-fold higher recovery of O-glycans, consistent with the increased signal seen in western blots. Postinfection, there was no further change in the O-glycan profile, except for a complete absence of core 2 structures (Figure 8 and Supplementary Table SVII). Thus, the longitudinal analysis indicated a progressive decrease in core 2 structures, but no other change in the O-glycan composition of plasma PRG4, despite the increased levels. The results confirm that PRG4 is an sTa-modified glycoprotein in plasma and vegetations of IE rats. The overall combined results of O-glycan profiling also indicate that, whereas the glycoforms of PRG4 in plasma and valve tissue of healthy animals are fairly similar, the glycoforms in plasma and vegetations of IE animals are quite different.
Fig. 8.
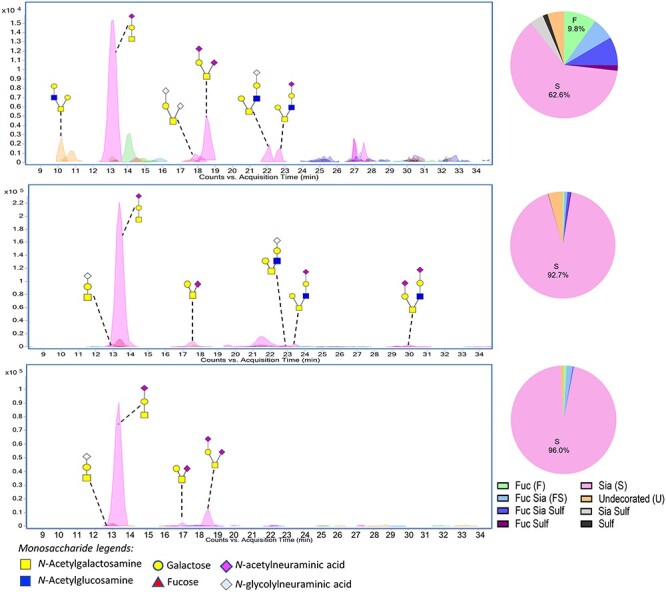
Profile of O-glycans released from rat plasma PRG4. Plasma was collected from the same animal precatheterization (upper panel), 3 days postcatheterization (middle panel) and 72 h postinfection with Streptococcus gordonii (lower panel). The profiles correspond to the total PRG4 recovered from 60 μL plasma.
PRG4 in the rabbit IE model
In addition to rodents, rabbits are another mammal traditionally used to model human endocardial infections. We therefore assessed whether PRG4 or other sialylated glycoproteins were associated with rabbit NBTE and IE. As in the rat model, PRG4 was more readily detected in the valve tissue of NBTE rabbits compared with healthy animals, but was most readily detected in the IE animal tissue (Figure 9A). PRG4 was also higher in the plasma of IE rabbits, compared with the levels presurgery, and compared with NBTE animals (Figure 9B). These results demonstrate that there is a substantial increase of PRG4 in the rabbit valves and plasma concomitant with valve injury. However, there may be a difference in the degree of sialylation (SLBR-H reactivity) in the IE vs. NBTE valve tissue.
Fig. 9.
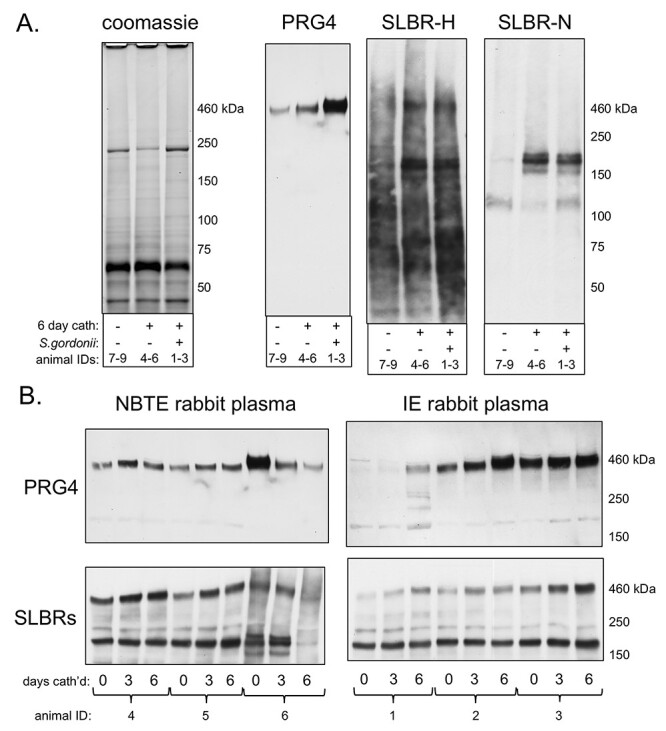
Sialylated glycoproteins in rabbit plasma and aortic valve tissue. (A) Valve tissue was pooled from three untreated (7–9), NBTE (4–6) or IE (1–3) animals. Proteins were either stained with Coomassie or were transferred to nitrocellulose and probed with anti-PRG4, SLBR-H or SLBR-N as indicated. Lanes contain 5 μg homogenized proteins. (B) Longitudinal analysis of plasma collected prior to catheterization (day 0), 3 days postcatheterization and 6 days postcatheterization and uninfected (NBTE) or infected with Streptococcus gordonii on day 3 (IE). Lanes contain 0.4 μL plasma. Blots were probed with anti-PRG4 or a combination of SLBR-H and -N for the detection of sialylated core 1 and core 2 glycans.
Discussion
These studies aimed to identify sialylated glycoproteins in aortic valve vegetations, with a specific interest in sTa-modified proteins that might contribute to the pathogenesis of IE. Three glycoproteins were detected in aortic valve vegetations recovered from IE animals: ITIH4, platelet GPIbα and PRG4. The three sialylated glycoproteins are orthologs of mucin-like proteins previously detected in healthy human plasma (Bensing et al. 2018), but only ITIH4 and GPIbα have been described in endocardial vegetations. The presence of GPIbα in rat IE vegetations was verified by immunohistology (Jung et al. 2012), although there was a distinctly different spatial relationship between GPIbα and streptococci in vegetations produced by S. gordonii vs. Streptococcus mutans. In a proteomics-based analysis of human IE vegetations, ITIH4 was found to be highly abundant and, as seen in the rat tissues here, it was extensively proteolyzed (Martin et al. 2020). In that study, neither GPIbα nor PRG4 was recovered from the vegetations, despite that several other platelet proteins were identified. However, GPIbα and PRG4 are more extensively O-glycosylated than ITIH4 and are thus less amenable to tryptic digestion and ionization, which can render these sialoglycoproteins extremely difficult to detect via shotgun proteomics.
Regarding the means by which the three sialoglycoproteins contribute to IE, GPIbα has a well-documented role in thrombus formation, via interactions with vWF on injured endothelial surfaces. These studies confirm that GPIbα is a sialylated glycoprotein component of infected vegetations, and its presence in NBTE vegetations could facilitate bacterial attachment via Siglec-like adhesins, thereby aiding colonization. In contrast, the functions of ITIH4 and PRG4 in plasma and vegetations are largely unknown. The function of ITIH4 is especially enigmatic, since it lacks a characteristic domain that is essential for protease inhibition by other ITIH family proteins (Zhuo and Kimata 2008). PRG4 was determined to be a major carrier of sTa in rat aortic valve vegetations and plasma and may be more likely to impact later stages of IE, as discussed further below.
The presence of PRG4 in vegetations may be related to a larger role for PRG4 in acute phase responses and innate immunity, as well as in cardiac repair and vascular homeostasis. Although PRG4 has been most extensively characterized as an important lubricating agent in synovial fluid (Jay et al. 2001), it also has reported anti-inflammatory properties and can interact with neutrophils via CD44, toll-like receptors and L-selectin (Alquraini et al. 2015; Al-Sharif et al. 2015; Iqbal et al. 2016; Richendrfer et al. 2020; Bennett et al. 2021). In addition to synovial fluid, PRG4 is expressed in tears and in the liver and heart (Ikegawa et al. 2000; Schmidt et al. 2013; Solka et al. 2016). Increased PRG4 expression in the liver was associated with vasculopathies in a murine model of staphylococcal sepsis (Toledo et al. 2019). In human cardiac tissues, increased PRG4 has been associated with postcardiotomy surgical healing via decreased fibrotic cardiomyocyte adhesions, but has also been correlated with aortic valve stenosis and interstitial cell calcification (Park et al. 2018; Artiach et al. 2020). Thus, PRG4 may be a key component of the innate response to vascular damage from both mechanical and biological causes, and PRG4 in plasma may be a marker of more general cardiovascular pathologies.
The primary function of PRG4 in cardiac valve vegetations is unclear. It is possible that the more general role of PRG4 in cardiac health vs. diseases such as IE is glycoform dependent. Certainly, the O-linked glycans are essential for the rheological properties of synovial PRG4 (Jay et al. 2001). Moreover, aberrant O-glycosylation has been associated with a number of diseases. For example, osteoarthritis and rheumatoid arthritis are correlated with decreased sialylation and increased sulfation in human and equine synovial fluid PRG4 (Estrella et al. 2010; Svala et al. 2017). Other studies have shown that sLeX and sulfated epitopes on the O-linked glycans of synovial PRG4 contribute to inflammation by mediating binding to L-selectin and neutrophils (Jin et al. 2012). Results here agree with an earlier report indicating that PRG4 in plasma and other tissues is more highly sialylated, as compared with the well-characterized synovial fluid form (Solka et al. 2016). More specifically, PRG4 recovered from the intact aortic valves of healthy animals had substantial nonsialylated core 2 glycans, which indicates a similarity to the synovial fluid form of PRG4. In contrast, PRG4 recovered from the IE rat valves and vegetations was modified with mix of core 1 and core 2 O-glycans, nearly all of which were capped with sialic acid, and included both mono- and di-sialylated structures. Our results further suggest that the sTa modification is more prevalent on PRG4 than in plasma and valve tissue of IE vs. healthy animals (Figures 7 and 8). However, the extent of sTa modification in aortic valve vegetations varies from animal to animal, as determined by western and lectin blotting (Figures 2 and 4), as well as the percentage of total O-glycans profiled by mass spectrometry (Figure 6). The ability to separate the differently glycosylated forms of PRG4, or to express recombinants forms thereof, would enable studies of glycoform-specific functions.
A key remaining question is how streptococcal binding to sTa contributes to the severity of IE, as reflected in a higher density of bacteria in aortic valve vegetations, as well as in the kidneys (Bensing et al. 2019a). The answer is likely dependent on whether sTa-modified PRG4 controls adhesion or inflammation in the response to aortic valve damage and IE, and whether streptococcal binding to the sTa epitopes impairs either of these processes. Although synovial and recombinant human PRG4 can directly modulate synovial and vascular endothelial cell migration and proliferation (Al-Sharif et al. 2015; Das et al. 2019; Wang et al. 2020), it is currently unknown whether the more highly sialylated forms of PRG4 directly interact with valve endothelial or interstitial cells, or with other vegetation components. It is possible that sTa-modified PRG4 could promote reendothelialization of the damaged valves. Alternatively, the sTa-modified PRG4 glycoform may stabilize or limit the size of platelet–fibrin thrombi. In the latter case, streptococcal binding to PRG4 could destabilize the vegetations and augment embolization.
Materials and methods
Bacterial strain and media
S. gordonii M99 was isolated from the blood of a patient with IE (Sullam et al. 1987). Todd Hewitt broth or Todd Hewitt agar containing 8% (v/v) sheep blood was used as the culture medium.
Reagents
Dulbecco’s phosphate-buffered saline (DPBS), dithiothreitol (DTT) and horseradish peroxidase-conjugated antibodies were purchased from Sigma St. Louis, MO. Rabbit polyclonal anti-GST, polyacrylamide gradient gels, electrophoretic running and transfer buffers, lithium dodecyl sulfate (LDS) sample buffer, and SimplyBlue SafeStain were from Invitrogen Carlsbad, CA.
Rat and rabbit IE and NBTE tissues
The rat plasma and aortic valve vegetation samples used for the experiments shown in Figures 2–4 were collected in earlier experiments (Bensing et al. 2019a). For the comparison of samples from NBTE vs. IE animals (Figures 5 and 9), NBTE or IE was produced as described previously (Peerschke et al. 2006; Bensing et al. 2019a; Li et al. 2020). In brief, experimental IE was induced by insertion of a sterile vascular catheter via the right carotid artery across the aortic valve and into the left ventricle of anesthetized white New Zealand rabbits (2.2–2.5 kg; female; Irish Farms, Shafter, CA) and Sprague–Dawley rats (250–300 g; female; Envigo, Placentia, CA). Catheters were left in place throughout the study. At 72 h after catheterization, S. gordonii strain M99 was injected intravenously at an inoculum of 5 × 106 colony forming units (CFU) to rabbits and 1 × 105 CFU for rats, the previously established 95% infective dose of this strain in the models. At 72 h postinfection, animals were humanely euthanized by overdose sodium pentobarbital injection. Healthy animals and catheterized but not infected (NBTE) animals were also included for comparison. Aortic valves and associated vegetations were collected with sterile forceps, homogenized in normal saline, quantitatively cultured to confirm the infections (IE animals only), and then stored at −80°C. Blood was collected into vacutainer tubes with ethylenediaminetetraacetic acid (EDTA) anticoagulant, centrifuged to remove cells, and plasma was stored at −80°C. Animals were cared for in accordance with the American Associate for Accreditation of Laboratory Animal Care. The in vivo experiment protocol was reviewed and approved by the Institutional Animal Care and Use Committee of The Lundquist Institute.
Western and far-western (lectin) blotting
Rat or rabbit plasma was diluted 1:10 into 10 mM Tris, 1 mM EDTA, pH 8 (TE), combined with LDS sample buffer and DTT (50 mM final concentration), boiled for 10 min, and loaded (5 μL) to wells of 3–8% polyacrylamide gradient gels. Homogenized valve and vegetation samples were centrifuged briefly to remove insoluble debris, diluted to ~1 mg/mL in DPBS, combined with LDS sample buffer and DTT (100 mM final concentration), boiled for 10 min, and loaded (5 μg) to wells of 3–8% polyacrylamide gradient gels. Following electrophoresis, proteins were transferred to BioTraceNT nitrocellulose (Pall Corporation) and probed with 15 nM GST-SLBRs as described (Bensing et al. 2016), or with anti-PRG4 clone 9G3 (EMD) or anti-ITIH4 (abcam) diluted 1:4000. After incubation with the primary antibodies for 1 h at room temperature, membranes were rinsed three times with DPBS, incubated with peroxidase-conjugated goat anti-mouse IgG/IgM 1:30,000 or anti-rabbit IgG 1:50,000 in DPBS, rinsed an additional three times with DPBS, and then developed with SuperSignal West Pico (Thermo Scientific). Emitted light was detected using CL-X Posure™ film (Thermo Scientific). Where indicated, relative amounts of plasma PRG4 were quantified using ImageJ, with samples normalized to the amount in a healthy animal (the sample shown in Figure 4B, lane 4).
Affinity capture of plasma and vegetation glycoproteins
GST-SLBRs were immobilized on magnetic glutathione resin (Pierce) essentially as described for immobilization on glutathione sepharose, except that the resin was collected using a DynaMag 15 magnet (Invitrogen) rather than by centrifugation. To capture sialylated glycoproteins from homogenized vegetations or valve tissue, samples containing ~2 mg protein were first denatured and reduced by combining with sodium dodecyl sulfate and DTT (0.5% and 40 mM final concentration, respectively) and then heating to 100°C for 10 min. After cooling to room temperature, samples were diluted 1:20 into TEN buffer (TE plus 150 mM NaCl) and combined with ~50 μg GST-SLBR-H or GST-SLBR-N immobilized on 5 μL magnetic glutathione resin. After tumbling 90 min at room temperature, the resin was washed twice with 1 mL TEN. To capture PRG4 from plasma, 200 μL plasma was diluted 1:7 in TEN buffer and combined with ~30 μg GST-SLBR-H immobilized on 8 μL magnetic glutathione resin. After tumbling 2 h at 4°C, the resin was washed twice with 1 mL TEN. The affinity-captured proteins were held on ice prior to further processing.
Identification of captured glycoproteins
The resin-bound GST-SLBRs and affinity-captured plasma proteins were co-eluted into LDS sample buffer supplemented with DTT (50 mM final concentration), separated by electrophoresis in 4–12% polyacrylamide gradient gels, and then stained with SimplyBlue SafeStain. Selected protein bands were excised from the gel and submitted for protein identification by nanoflow LC–MS/MS of tryptic digests (MSBioworks Ann Arbor, MI).
O-glycan profiling
The 480 kDa region corresponding to PRG4 recovered from ~500 μg homogenized tissue or 60 μL plasma was excised from polyacrylamide gels. The excised gel slices were minced, treated by four cycles of rinsing with 100 mM ammonium bicarbonate and dehydration in 100% acetonitrile, and then dried to completion in a vacuum evaporator. The gel pieces were immersed in a mixture of 100 mM NaOH and 1 M NaBH4 and incubated at 45°C for 18 h to release the O-glycans. The supernatant was collected and placed on ice, and the remaining gel pieces were washed with water and sonicated for 30 min to extract the remaining O-glycans. The initial and secondary extracts were combined and acidified to pH 4–6 by drop-wise addition of 10% acetic acid. The O-glycan samples were then enriched using porous graphitized carbon cartridges (Agilent, Santa Clara, CA) and dried prior to analysis by mass spectrometry. Glycan samples were analyzed on an Agilent 6520 Accurate Mass Quadrupole Time-of-Flight mass spectrometer equipped with a porous graphitic carbon microfluidic chip. A binary gradient consisting of (A) 0.1% formic acid in 3% acetonitrile and (B) 1% formic acid in 89% acetonitrile was used to separate the glycans at a flow rate of 0.3 μL/min. Data were processed with Agilent MassHunter B.07 software, using the Find by Molecular Feature algorithm with an in-house library of O-glycan masses and chemical formulae to identify and quantitate the O-glycan signals.
Supplementary Material
Contributor Information
Kemal Solakyildirim, Department of Chemistry, University of California, Davis, CA 95616, USA; Department of Chemistry, Erzincan Binali Yildirim University, Erzincan 24100, Turkey.
Yi Li, Division of Infectious Diseases, Lundquist Institute for Biomedical Innovation at Harbor-UCLA Medical Center, Torrance, CA 90502, USA.
Arnold S Bayer, Division of Infectious Diseases, Lundquist Institute for Biomedical Innovation at Harbor-UCLA Medical Center, Torrance, CA 90502, USA; Department of Medicine, David Geffen School of Medicine at UCLA, Los Angeles, CA 90095, USA.
Paul M Sullam, Department of Medicine, San Francisco Veterans Affairs Medical Center and University of California, San Francisco, CA 94121, USA.
Yan Q Xiong, Division of Infectious Diseases, Lundquist Institute for Biomedical Innovation at Harbor-UCLA Medical Center, Torrance, CA 90502, USA; Department of Medicine, David Geffen School of Medicine at UCLA, Los Angeles, CA 90095, USA.
Carlito B Lebrilla, Department of Chemistry, University of California, Davis, CA 95616, USA.
Barbara A Bensing, Department of Medicine, San Francisco Veterans Affairs Medical Center and University of California, San Francisco, CA 94121, USA.
Funding
The American Heart Association (17SDG33660424 to B.A.B.); the National Institutes of Health (R01AI106987 to P.M.S., R01AI130056 to A.S.B., R01AI139244 to Y.Q.X.).
Conflict of interest statement
None declared.
Abbreviations
3'sLn, 3'sialyllactosamine; DPBS, Dulbecco’s phosphate-buffered saline; dsTa, disialylated T-antigen; DTT, dithiothreitol; GPIbα, platelet glycoprotein Ibα; GST, glutathione S-transferase; IE, infective endocarditis; ITIH4, inter-α-trypsin inhibitor heavy chain 4; NBTE, nonbacterial thrombotic endocarditis; PRG4, proteoglycan 4; SLBR, Siglec-like binding region; sTa, sialyl T-antigen
References
- Al-Sharif A, Jamal M, Zhang LX, Larson K, Schmidt TA, Jay GD, Elsaid KA. 2015. Lubricin/proteoglycan 4 binding to CD44 receptor: A mechanism of the suppression of proinflammatory cytokine-induced synoviocyte proliferation by lubricin. Arthritis Rheumatol. 67:1503–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alquraini A, Garguilo S, D’Souza G, Zhang LX, Schmidt TA, Jay GD, Elsaid KA. 2015. The interaction of lubricin/proteoglycan 4 (PRG4) with toll-like receptors 2 and 4: An anti-inflammatory role of PRG4 in synovial fluid. Arthritis Res Ther. 17:353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artiach G, Carracedo M, Seime T, Plunde O, Laguna-Fernandez A, Matic L, Franco-Cereceda A, Back M. 2020. Proteoglycan 4 is increased in human calcified aortic valves and enhances valvular interstitial cell calcification. Cells. 9:684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SP, Nulton TJ, Kitten T. 2019. Genomic, phenotypic, and virulence analysis of Streptococcus sanguinis oral and infective-endocarditis isolates. Infect Immun. 87:e00703–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M, Chin A, Lee HJ, Morales Cestero E, Strazielle N, Ghersi-Egea JF, Threlkeld SW, Schmidt TA, Richendrfer HA, Szmydynger-Chodobska J, et al. 2021. Proteoglycan 4 reduces neuroinflammation and protects the blood-brain barrier after traumatic brain injury. J Neurotrauma. 38:385–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensing BA, Khedri Z, Deng L, Yu H, Prakobphol A, Fisher SJ, Chen X, Iverson TM, Varki A, Sullam PM. 2016. Novel aspects of sialoglycan recognition by the Siglec-like domains of streptococcal SRR glycoproteins. Glycobiology. 26:1222–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensing BA, Li L, Yakovenko O, Wong M, Barnard KN, Iverson TM, Lebrilla CB, Parrish CR, Thomas WE, Xiong Y, et al. 2019a. Recognition of specific sialoglycan structures by oral streptococci impacts the severity of endocardial infection. PLoS Pathog. 15:e1007896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensing BA, Li Q, Park D, Lebrilla CB, Sullam PM. 2018. Streptococcal Siglec-like adhesins recognize different subsets of human plasma glycoproteins: Implications for infective endocarditis. Glycobiology. 28:601–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensing BA, Loukachevitch LV, Agarwal R, Yamakawa I, Luong K, Hadadianpour A, Yu H, Fialkowski KP, Castro MA, Wawrzak Z, et al. 2019b. Selectivity and engineering of the sialoglycan-binding spectrum in Siglec-like adhesins. bioRxiv. bioRxiv.796912. doi: 10.1101/796912. [DOI] [Google Scholar]
- Chamat-Hedemand S, Dahl A, Ostergaard L, Arpi M, Fosbol E, Boel J, Oestergaard LB, Lauridsen TK, Gislason G, Torp-Pedersen C, et al. 2020. Prevalence of infective endocarditis in streptococcal bloodstream infections is dependent on Streptococcal species. Circulation. 142:720–730. [DOI] [PubMed] [Google Scholar]
- Crump KE, Bainbridge B, Brusko S, Turner LS, Ge X, Stone V, Xu P, Kitten T. 2014. The relationship of the lipoprotein SsaB, manganese and superoxide dismutase in Streptococcus sanguinis virulence for endocarditis. Mol Microbiol. 92:1243–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das N, Schmidt TA, Krawetz RJ, Dufour A. 2019. Proteoglycan 4: From mere lubricant to regulator of tissue homeostasis and inflammation: Does proteoglycan 4 have the ability to buffer the inflammatory response? Bioessays. 41:e1800166. [DOI] [PubMed] [Google Scholar]
- Das S, Kanamoto T, Ge X, Xu P, Unoki T, Munro CL, Kitten T. 2009. Contribution of lipoproteins and lipoprotein processing to endocarditis virulence in Streptococcus sanguinis. J Bacteriol. 191:4166–4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Bensing BA, Thamadilok S, Yu H, Lau K, Chen X, Ruhl S, Sullam PM, Varki A. 2014. Oral streptococci utilize a Siglec-like domain of serine-rich repeat adhesins to preferentially target platelet sialoglycans in human blood. PLoS Pathog. 10:e1004540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake TA, Rodgers GM, Sande MA. 1984. Tissue factor is a major stimulus for vegetation formation in enterococcal endocarditis in rabbits. J Clin Invest. 73:1750–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durack DT. 1975. Experimental bacterial endocarditis. IV. Structure and evolution of very early lesions. J Pathol. 115:81–89. [DOI] [PubMed] [Google Scholar]
- Durack DT, Beeson PB. 1972. Experimental bacterial endocarditis. I. Colonization of a sterile vegetation. Br J Exp Pathol. 53:44–49. [PMC free article] [PubMed] [Google Scholar]
- Estrella RP, Whitelock JM, Packer NH, Karlsson NG. 2010. The glycosylation of human synovial lubricin: Implications for its role in inflammation. Biochem J. 429:359–367. [DOI] [PubMed] [Google Scholar]
- Fitzgerald JR, Foster TJ, Cox D. 2006. The interaction of bacterial pathogens with platelets. Nat Rev Microbiol. 4:445–457. [DOI] [PubMed] [Google Scholar]
- Foster TJ, Geoghegan JA, Ganesh VK, Hook M. 2014. Adhesion, invasion and evasion: The many functions of the surface proteins of Staphylococcus aureus. Nat Rev Microbiol. 12:49–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaytan MO, Singh AK, Woodiga SA, Patel SA, An SS, Vera-Ponce de Leon A, McGrath S, Miller AR, Bush JM, van der Linden M, et al. 2021. A novel sialic acid-binding adhesin present in multiple species contributes to the pathogenesis of infective endocarditis. PLoS Pathog. 17:e1009222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X, Kitten T, Chen Z, Lee SP, Munro CL, Xu P. 2008. Identification of Streptococcus sanguinis genes required for biofilm formation and examination of their role in endocarditis virulence. Infect Immun. 76:2551–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X, Yu Y, Zhang M, Chen L, Chen W, Elrami F, Kong F, Kitten T, Xu P. 2016. Involvement of NADH oxidase in competition and endocarditis virulence in Streptococcus sanguinis. Infect Immun. 84:1470–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haworth JA, Jenkinson HF, Petersen HJ, Back CR, Brittan JL, Kerrigan SW, Nobbs AH. 2017. Concerted functions of Streptococcus gordonii surface proteins PadA and Hsa mediate activation of human platelets and interactions with extracellular matrix. Cell Microbiol. 19:e12667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegawa S, Sano M, Koshizuka Y, Nakamura Y. 2000. Isolation, characterization and mapping of the mouse and human PRG4 (proteoglycan 4) genes. Cytogenet Cell Genet. 90:291–297. [DOI] [PubMed] [Google Scholar]
- Iqbal SM, Leonard C, C. Regmi S, de Rantere D, Tailor P, Ren G, Ishida H, Hsu C, Abubacker S, Pang DS, et al. 2016. Lubricin/proteoglycan 4 binds to and regulates the activity of toll-like receptors in vitro. Sci Rep. 6:18910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaksson J, Rasmussen M, Nilson B, Stadler LS, Kurland S, Olaison L, Ek E, Herrmann B. 2015. Comparison of species identification of endocarditis associated viridans streptococci using rnpB genotyping and 2 MALDI-TOF systems. Diagn Microbiol Infect Dis. 81:240–245. [DOI] [PubMed] [Google Scholar]
- Jay GD, Harris DA, Cha CJ. 2001. Boundary lubrication by lubricin is mediated by O-linked beta(1-3)Gal-GalNAc oligosaccharides. Glycoconj J. 18:807–815. [DOI] [PubMed] [Google Scholar]
- Jin C, Ekwall AK, Bylund J, Bjorkman L, Estrella RP, Whitelock JM, Eisler T, Bokarewa M, Karlsson NG. 2012. Human synovial lubricin expresses sialyl Lewis x determinant and has L-selectin ligand activity. J Biol Chem. 287:35922–35933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CJ, Yeh CY, Shun CT, Hsu RB, Cheng HW, Lin CS, Chia JS. 2012. Platelets enhance biofilm formation and resistance of endocarditis-inducing streptococci on the injured heart valve. J Infect Dis. 205:1066–1075. [DOI] [PubMed] [Google Scholar]
- Kitten T, Munro CL, Zollar NQ, Lee SP, Patel RD. 2012. Oral streptococcal bacteremia in hospitalized patients: Taxonomic identification and clinical characterization. J Clin Microbiol. 50:1039–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Bayer AS, Cheung A, Lu L, Abdelhady W, Donegan NP, Hong JI, Yeaman MR, Xiong YQ. 2020. The stringent response contributes to persistent methicillin-resistant Staphylococcus aureus endovascular infection through the purine biosynthetic pathway. J Infect Dis. 222:1188–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesenborghs L, Meyers S, Vanassche T, Verhamme P. 2020. Coagulation: At the heart of infective endocarditis. J Thromb Haemost. 18:995–1008. [DOI] [PubMed] [Google Scholar]
- Martin DR, Witten JC, Tan CD, Rodriguez ER, Blackstone EH, Pettersson GB, Seifert DE, Willard BB, Apte SS. 2020. Proteomics identifies a convergent innate response to infective endocarditis and extensive proteolysis in vegetation components. JCI Insight. 5:e135317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narimatsu Y, Joshi HJ, Nason R, Van Coillie J, Karlsson R, Sun L, Ye Z, Chen YH, Schjoldager KT, Steentoft C, et al. 2019. An atlas of human glycosylation pathways enables display of the human glycome by gene engineered cells. Mol Cell. 75:394–407.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park DSJ, Regmi SC, Svystonyuk DA, Teng G, Belke D, Turnbull J, Guzzardi DG, Kang S, Cowman MK, Schmidt TA, et al. 2018. Human pericardial proteoglycan 4 (lubricin): Implications for postcardiotomy intrathoracic adhesion formation. J Thorac Cardiovasc Surg. 156:1598–1608.e1. [DOI] [PubMed] [Google Scholar]
- Peerschke EI, Bayer AS, Ghebrehiwet B, Xiong YQ. 2006. gC1qR/p33 blockade reduces Staphylococcus aureus colonization of target tissues in an animal model of infective endocarditis. Infect Immun. 74:4418–4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plummer C, Douglas CW. 2006. Relationship between the ability of oral streptococci to interact with platelet glycoprotein Ibalpha and with the salivary low-molecular-weight mucin, MG2. FEMS Immunol Med Microbiol. 48:390–399. [DOI] [PubMed] [Google Scholar]
- Plummer C, Wu H, Kerrigan SW, Meade G, Cox D, Ian Douglas CW. 2005. A serine-rich glycoprotein of Streptococcus sanguis mediates adhesion to platelets via GPIb. Br J Haematol. 129:101–109. [DOI] [PubMed] [Google Scholar]
- Pyburn TM, Bensing BA, Xiong YQ, Melancon BJ, Tomasiak TM, Ward NJ, Yankovskaya V, Oliver KM, Cecchini G, Sulikowski GA, et al. 2011. A structural model for binding of the serine-rich repeat adhesin GSPB to host carbohydrate receptors. PLoS Pathog. 7:e1002112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen LH, Dargis R, Hojholt K, Christensen JJ, Skovgaard O, Justesen US, Rosenvinge FS, Moser C, Lukjancenko O, Rasmussen S, et al. 2016. Whole genome sequencing as a tool for phylogenetic analysis of clinical strains of Mitis group streptococci. Eur J Clin Microbiol Infect Dis. 35:1615–1625. [DOI] [PubMed] [Google Scholar]
- Richendrfer HA, Levy MM, Elsaid KA, Schmidt TA, Zhang L, Cabezas R, Jay GD. 2020. Recombinant human proteoglycan-4 mediates interleukin-6 response in both human and mouse endothelial cells induced into a sepsis phenotype. Crit Care Explor. 2:e0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronis A, Brockman K, Singh AK, Gaytan MO, Wong A, McGrath S, Owen CD, Magrini V, Wilson RK, van der Linden M, et al. 2019. Streptococcus oralis subsp. dentisani produces monolateral serine-rich repeat protein fibrils, one of which contributes to saliva binding via sialic acid. Infect Immun. 87:e00406-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sass LA, Ziemba KJ, Heiser EA, Mauriello CT, Werner AL, Aguiar MA, Nyalwidhe JO, Cunnion KM. 2016. A 1-year-old with Mycobacterium tuberculosis endocarditis with mass spectrometry analysis of cardiac vegetation composition. J Pediatric Infect Dis Soc. 5:85–88. [DOI] [PubMed] [Google Scholar]
- Schmidt TA, Sullivan DA, Knop E, Richards SM, Knop N, Liu S, Sahin A, Darabad RR, Morrison S, Kam WR, et al. 2013. Transcription, translation, and function of lubricin, a boundary lubricant, at the ocular surface. JAMA Ophthalmol. 131:766–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solka KA, Miller IJ, Schmid TM. 2016. Sialidase unmasks mucin domain epitopes of lubricin. J Histochem Cytochem. 64:647–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullam PM, Valone FH, Mills J. 1987. Mechanisms of platelet aggregation by viridans group streptococci. Infect Immun. 55:1743–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svala E, Jin C, Ruetschi U, Ekman S, Lindahl A, Karlsson NG, Skioldebrand E. 2017. Characterisation of lubricin in synovial fluid from horses with osteoarthritis. Equine Vet J. 49:116–123. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Konishi K, Cisar JO, Yoshikawa M. 2002. Identification and characterization of hsa, the gene encoding the sialic acid-binding adhesin of Streptococcus gordonii DL1. Infect Immun. 70:1209–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Takashima E, Shimazu K, Yagishita H, Aoba T, Konishi K. 2006. Contribution of sialic acid-binding adhesin to pathogenesis of experimental endocarditis caused by Streptococcus gordonii DL1. Infect Immun. 74:740–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamatsu D, Bensing BA, Cheng H, Jarvis GA, Siboo IR, López JA, Griffiss JM, Sullam PM. 2005. Binding of the Streptococcus gordonii surface glycoproteins GspB and Hsa to specific carbohydrate structures on platelet membrane glycoprotein Ibalpha. Mol Microbiol. 58:380–392. [DOI] [PubMed] [Google Scholar]
- Takamatsu D, Bensing BA, Prakobphol A, Fisher SJ, Sullam PM. 2006. Binding of the streptococcal surface glycoproteins GspB and Hsa to human salivary proteins. Infect Immun. 74:1933–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo AG, Golden G, Campos AR, Cuello H, Sorrentino J, Lewis N, Varki N, Nizet V, Smith JW, Esko JD. 2019. Proteomic atlas of organ vasculopathies triggered by Staphylococcus aureus sepsis. Nat Commun. 10:4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner LS, Kanamoto T, Unoki T, Munro CL, Wu H, Kitten T. 2009. Comprehensive evaluation of Streptococcus sanguinis cell wall-anchored proteins in early infective endocarditis. Infect Immun. 77:4966–4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano-Tashiro Y, Yajima A, Takashima E, Takahashi Y, Konishi K. 2008. Binding of the Streptococcus gordonii DL1 surface protein Hsa to the host cell membrane glycoproteins CD11b, CD43, and CD50. Infect Immun. 76:4686–4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Kikuchi S, Schmidt TA, Hoofnagle M, Wight TN, Azuma N, Tang GL, Sobel M, Velamoor GR, Mokadam NA, et al. 2020. Inhibitory effects of PRG4 on migration and proliferation of human venous cells. J Surg Res. 253:53–62. [DOI] [PubMed] [Google Scholar]
- Werdan K, Dietz S, Loffler B, Niemann S, Bushnaq H, Silber RE, Peters G, Muller-Werdan U. 2014. Mechanisms of infective endocarditis: Pathogen-host interaction and risk states. Nat Rev Cardiol. 11:35–50. [DOI] [PubMed] [Google Scholar]
- Xiong YQ, Bensing BA, Bayer AS, Chambers HF, Sullam PM. 2008. Role of the serine-rich surface glycoprotein GspB of Streptococcus gordonii in the pathogenesis of infective endocarditis. Microb Pathog. 45:297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo L, Kimata K. 2008. Structure and function of inter-alpha-trypsin inhibitor heavy chains. Connect Tissue Res. 49:311–320. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


