Abstract
Background
Primary ovarian insufficiency (POI) affects 1% of women and is associated with significant medical consequences. A genetic cause for POI can be found in up to 30% of women, elucidating key roles for these genes in human ovary development.
Objective
We aimed to identify the genetic mechanism underlying early-onset POI in 2 sisters from a consanguineous pedigree.
Methods
Genome sequencing and variant filtering using an autosomal recessive model was performed in the 2 affected sisters and their unaffected family members. Quantitative reverse transcriptase PCR (qRT-PCR) and RNA sequencing were used to study the expression of key genes at critical stages of human fetal gonad development (Carnegie Stage 22/23, 9 weeks post conception (wpc), 11 wpc, 15/16 wpc, 19/20 wpc) and in adult tissue.
Results
Only 1 homozygous variant cosegregating with the POI phenotype was found: a single nucleotide substitution in zinc finger SWIM-type containing 7 (ZSWIM7), NM_001042697.2: c.173C > G; resulting in predicted loss-of-function p.(Ser58*). qRT-PCR demonstrated higher expression of ZSWIM7 in the 15/16 wpc ovary compared with testis, corresponding to peak meiosis in the fetal ovary. RNA sequencing of fetal gonad samples showed that ZSWIM7 has a similar temporal expression profile in the developing ovary to other homologous recombination genes.
Main conclusions
Disruption of ZSWIM7 is associated with POI in humans. ZSWIM7 is likely to be important for human homologous recombination; these findings expand the range of genes associated with POI in women.
Keywords: primary ovarian insufficiency, meiosis, ovary development, primary amenorrhea, delayed puberty, genetics, NGS
Primary ovarian insufficiency (POI) is an important and relatively common condition, affecting 1% of women and causing significant medical, psychosocial, and economic sequelae (1). It arises when a primary defect within the ovary results in ovary dysfunction and disruption of the resting follicle pool (2, 3). POI is diagnosed in women who present before the age of 40 years with amenorrhea of more than 4 months’ duration, estrogen deficiency, and raised FSH concentrations measured twice at least 1 month apart (1, 4). The majority of women with POI experience normal pubertal development and present in adulthood with secondary amenorrhea or oligomenorrhea (5-7). Adolescents presenting with primary amenorrhea and delayed puberty, accounting for 10% of presentations of this condition, represent the most severe end of the POI spectrum.
Establishing an underlying cause for a POI diagnosis can be difficult and in 50% to 80% of women a cause is not found (3, 8). Iatrogenic POI secondary to surgical oophorectomy, chemotherapy, or radiotherapy accounts for a substantial proportion (up to 30%) (8). Environmental toxins and an underlying autoimmune etiology have also been proposed (9, 10). So far, variants in more than 60 different genes have been associated with the pathogenesis of POI, with each gene responsible for only a small subset of cases (11). The advent of next-generation sequencing has proven to be a useful tool in expanding the number of known genetically mediated causes of POI. This has provided a genetic diagnosis for a proportion of affected women (~5%-30%) and allows targeted genetic counselling of patients and family members (5, 12). Notably, identified genes often relate to the known complex biological processes underpinning normal ovary development and function, including sex differentiation, oogenesis, folliculogenesis, and steroidogenesis.
Oogenesis is particularly critical for normal germ cell development and is dependent on meiosis, which encompasses a series of critically regulated processes that result in a diploid germ cell undergoing 1 round of DNA replication followed by 2 cell divisions to produce a haploid ovum capable of being fertilized. In females, meiosis commences in fetal life but is complete only after fertilization of a metaphase II oocyte during adult life. Meiosis begins with homologous chromosomes moving close to one another with the subsequent formation of the synaptonemal complex (SC). Cohesins form protein complexes to stabilize the SC. Homologous recombination follows, initiated by SPO11-mediated double-strand DNA breaks. Resected tails are bound by single-strand DNA-binding proteins to prevent reannealing and to recruit RAD51 and DMC1, which catalyze strand invasion and exchange (13, 14). DNA exchange intermediates are then joined via DNA repair mechanisms, which results in the formation of either crossover or noncrossover intermediates (15). Pathogenic variants in several meiosis genes have already been associated with POI. These include SYCE1 and SYCP3 (SC) (16, 17); STAG3, REC8, and SMC1B (cohesin complex) (18, 19); MEIOB and BRCA2 (strand invasion) (20, 21); MCM8, MCM9, MSH4, MSH5, and HFM1 (DNA repair; stabilization and intermediate processing) (22-26); FANCM, BLM (crossover regulation); and MLH3 (crossover resolution) (27). Because meiosis is a complex process involving the coordinated interaction of many genes, it is likely that other factors involved in meiosis represent candidate genes for POI. Here, we associate an autosomal recessive pathogenic variant in the meiosis-associated gene zinc finger SWIM-type containing 7 (ZSWIM7), also known as SWS1, with primary ovarian insufficiency.
Methods
Participants and Genetic Analysis
A family with 2 sisters affected with POI was recruited as part of an ovarian dysgenesis research project at University College London Hospitals (08/H071/69). In an attempt to find a genetic cause of POI, 2 affected and 2 unaffected members (mother and sibling) of the family underwent whole-genome sequencing.
After obtaining informed consent, genomic DNA was isolated from frozen whole blood using a QIAamp DNA Blood Mini kit (Qiagen). Library preparation and whole-genome sequencing were performed at BGI Genomics (Shenzhen, China) using the BGISEQ-500 platform at ~30× coverage. Sequencing reads were aligned with Burrows-Wheeler Aligner v0.7.17 to human genome build 38 (GRCh38.p1), not including alternate assemblies (GCA_000001405.15_GRCh38_no_alt_analysis_set.fna), and read duplicates were marked with Sambamba (28, 29). Variant calling across the exonic regions with 100-bp padding was performed using Genome Analysis Toolkit v4.0.3.0 according to the best practices workflow for joint (multisample) calling (30-32). Variant filtering was performed through the use of Ingenuity Variant Analysis software (Qiagen). We focused our analysis on coding and splice region (7 bases into an intron) variants with read depth ≥ 5 that are not present or rare (allele frequency ≤ 0.01%) in the Genome Aggregation Database (gnomAD v2.1.1, Cambridge, MA; https://gnomad.broadinstitute.org, accessed March 2021 (33)) using an autosomal recessive mode of inheritance because of consanguinity in the family. Synonymous changes were excluded, unless they were predicted to affect splicing using MaxEntScan. Sanger sequencing was used to verify the presence of the variant and to perform segregation analysis. The target region was amplified using primers (forward 5′-CAAGTTGGGGCAAAAGCCTT-3′ and reverse 5′-CCTTTGGGCAAGTTACTGAGG- 3′). PCR products were purified with ExoSap and sequenced using Big Dye Terminator Cycle Sequencing Kit v3.1 (Life Technologies, Foster City, CA). Result electropherograms were analyzed with Geneious software (Biomatters Ltd., Auckland, New Zealand).
The American College of Medical Genetics variant classification guidelines and in silico software prediction tools Decipher, PROVEAN, MutationTaster, and CADD scoring were used to assess variant pathogenicity (34-38).
Quantitative Reverse Transcriptase PCR Analysis of ZSWIM7
Quantitative reverse transcriptase PCR (qRT-PCR) was used to study the expression of ZSWIM7 during fetal gonadal development. Human fetal tissue samples were obtained with ethical approval (REC references 18/LO/0822; 18/NE/0290) and informed consent from the Human Developmental Biology Resource (http://www.hdbr.org). Four ovary and testis samples were included at each of 5 developmental stages: Carnegie Stage (CS) 22/23, 7.5 to 8 weeks postconception (wpc), 9 wpc, 11 wpc, 15 to 16 wpc, and 19 to 20 wpc. Four adult ovary (catalog number CS500008, Origene) and 4 adult testis samples (catalog number CS502309, Origene) were also included. RNA was quantified using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific) and reverse transcribed using the SuperScript III Reverse Transcriptase kit (Thermo Fisher Scientific). qRT-PCR was performed using Taqman Fast Advanced MasterMix (Applied Biosystems) and TaqMan assays (ZSWIM7: Hs04984973_m1) on the ABI StepOne Plus System (Applied Biosystems). The relative expression of ZSWIM7 was calculated using the comparative Ct method and GAPDH (Hs02786624_g1) was used as a housekeeping gene control. Experiments were conducted in triplicate on 3 occasions. Data are expressed as mean ± SEM. Two-way ANOVA and independent samples t tests were used for statistical analysis (GraphPad Prism v9.0.0). Further analysis of ZSWIM7 expression in adult tissues was performed using GTEx data (v.8; accessed March 2021), the Human Protein Atlas (v20.1; accessed March 2021), and FANTOM5 (accessed March 2021).
RNA Expression in Human Fetal Gonad Development
The expression of ZSWIM7 and associated DNA repair genes in fetal development was studied using bulk RNA sequencing. Five ovary (4 at 15-16 wpc), 5 testes, and 2 46,XX control tissues were included at each of 4 developmental stages: CS22, 9 wpc, 11 wpc, and 15 to 16 wpc. RNA was extracted using the AllPrep DNA/RNA Mini Kit (Qiagen) using the kit manufacturer’s instructions. Libraries were prepared using the KAPA RNA HyperPrep Kit with RiboErase (Illumina) and sequenced with the Illumina HiSeq 4000 platform using 2 × 75-bp paired-end sequencing kit. Quality control of sequencing reads was performed using FastQC (Babraham Bioinformatics) and the reads were aligned to human genome build 38 (GRCh38.p1) using STAR aligner (v2.5.2a) (39, 40). featureCounts (Subread package) and DESeq2 (Bioconductor) were used for gene expression quantification and differential gene expression analysis, respectively (41, 42). The Benjamini-Hochberg approach was used to adjust for multiple testing with cutoff adjusted P values of 0.05 (43).
Results
POI Pedigree
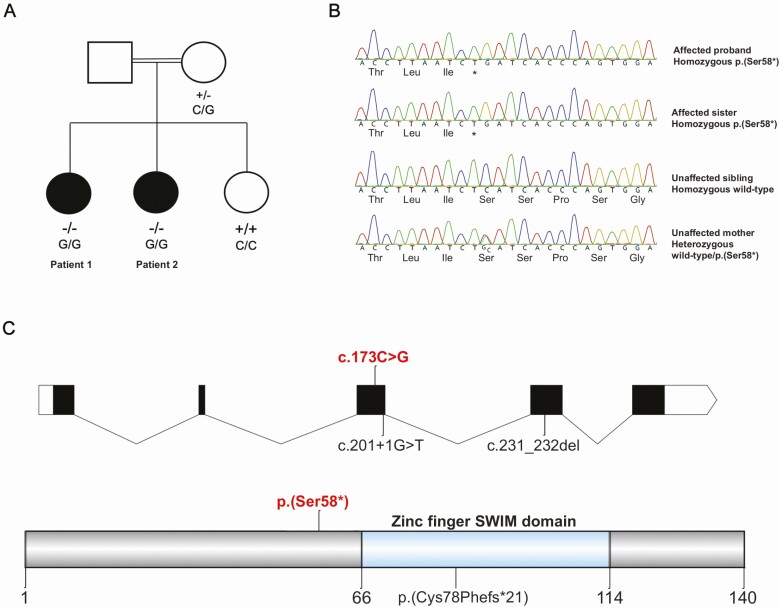
Two sisters from a consanguineous pedigree presented in adolescence with absent puberty and primary amenorrhea (Fig. 1A). Their parents were first cousins originating from Turkey. Both had raised gonadotropin and low estradiol concentrations consistent with a diagnosis of POI (Table 1). Extended characterization for the etiology of ovarian insufficiency in both girls demonstrated 46,XX karyotypes and negative Fragile X screening (FRAXA premutation analysis). Both were normotensive at presentation. Pelvic ultrasound did not identify ovaries. There was no history of previous ovarian surgery nor chemo-/radiotherapy. Both patients were commenced on estrogen replacement and subsequently progressed through puberty as expected, with normal breast development, development of secondary sexual characteristics, and menarche. Their parents had normal fertility and there was no family history of delayed puberty nor infertility in the extended family. A sister was unaffected and developed in puberty normally.
Figure 1.
(A) Affected kindred with the ZSWIM7 variant (NM_001042697.2:c.173C > G, p.(Ser58*)). Solid symbols indicate affected family members. Genotype is indicated underneath tested family members. (B) Sanger sequencing of the affected proband, an affected sister, an unaffected sister, and unaffected mother. NC_000017.11g:g.15987294G > C; NM_001042697.2:c.173C > G; NP_001036162.1:p.(Ser58*). (C) Domains of the human ZSWIM7 protein. The p.(Ser58*) stop-gain variant reported in this study is proximal (N-terminal) to the zinc finger SWIM domain. Previously reported ZSWIM7 variants (associated with male infertility) are also indicated (homozygous splice variant c.201 + 1G > T; homozygous frameshift variant c.231_232del; p.(Cys78Phefs*21)).
Table 1.
Clinical characteristics of 2 women with primary ovarian insufficiency
| Patient 1 | Patient 2 | |
|---|---|---|
| Menarche | Primary amenorrhea | Primary amenorrhea |
| Age at presentation, y | 15 | 12 |
| Pubertal induction required | Yes | Yes |
| FSH at diagnosis, IU/L (follicular range: 3.5-12.5) | 94.8 | 77.8 |
| LH at diagnosis, IU/L (follicular range: 2.4-12.6) | 17.2 | 14.3 |
| E2 at diagnosis (follicular range: 98-571 pmol/L; undetectable < 20 pmol/L) | Undetectable | Undetectable |
| Pubertal stage at diagnosis | B2 P1 A1 | B1 P1 A1 |
| Karyotype | 46,XX | 46,XX |
| Ovaries on imaging at diagnosis (normal range 5-7 cm3) | Small prepubertal uterus; streak ovaries | Small prepubertal uterus; no ovaries seen |
| Fragile X genetic testing (FRAXA analysis) | Negative | Negative |
| Weight, kg | 65.9 | Not available |
| Height, cm | 1.65 | Not available |
| Body mass index, centile | 92nd | - |
Homozygous Stop Gain Variant Identified in ZSWIM7
Analysis of coding and splice region variants using a model consistent with autosomal recessive inheritance revealed only 1 biallelic variant that cosegregated with the primary ovarian insufficiency phenotype.
This single nucleotide substitution occurred in ZSWIM7 (NC_000017.11: g.15987294G > C, NM_001042697.2: c.173C > G) and is predicted to result in a stop-gain change at codon 58, p.(Ser58*). The 2 affected sisters were homozygous for this variant (allele fraction 1.0; read depths 65 and 54), whereas the mother was heterozygous (read depth 43, allele fraction 0.53). DNA from the father of the proband was not available (Fig. 1A). The presence of a larger structural variant, such as a deletion, at the locus was not evident from the mapped sequencing reads. The ZSWIM7 variant was confirmed with Sanger sequencing (Fig. 1B). The variant is novel and has not been previously documented or reported in public databases (gnomAD and dbSNP). There is no ZSWIM7-associated phenotype in OMIM (MIM 614535). ZSWIM7 encodes for a protein known to be important for meiotic homologous recombination during prophase I (Fig. 1C) (44). No homozygous loss-of-function ZSWIM7 variants are reported in the gnomAD database. According to Decipher, the ZSWIM7 transcript carrying a premature termination codon at position 58 is predicted to undergo nonsense mediated decay (36). Using the American College of Medical Genetics classification system, this variant is classed as pathogenic (1a) because: (1) it is a stop-gain mutation subject to nonsense-mediated decay (very strong evidence: PVS1); (2) there are existing animal models supportive of a damaging effect of disruption in this gene (strong evidence: PS3); (3) it cosegregates with disease in family members (supporting evidence: PP1); and (4) the variant is predicted pathogenic using in silico tools (supporting evidence: PP3; Table 2).
Table 2.
Analysis of the c.173C > G variant
| Variant | |
|---|---|
| Gene symbol | ZSWIM7 |
| Full gene name | Zinc finger swim domain-containing protein 7 |
| Position | Exonic |
| Cytoband | 17p12 |
| Region | Chr17:15987294 |
| Transcript identification | NM_001042697.2 |
| Exon involved | Exon 3 |
| cDNA variant | c.173C > G |
| Protein variant | p.S58* |
| Translation impact | Stop gain |
| Genotype | Homozygous |
| CADD score | 39 (pathogenic) |
| PROVEAN score | -7.28 (deleterious) |
| MutationTaster | Disease causing |
| DECIPHER | Predicted nonsense-mediated decay |
| Allele frequency in gnomAD | 0 |
| American College of Medical Genetics classification | Predicted pathogenic (1a) |
Expression of ZSWIM7 in Human Ovary and Testis
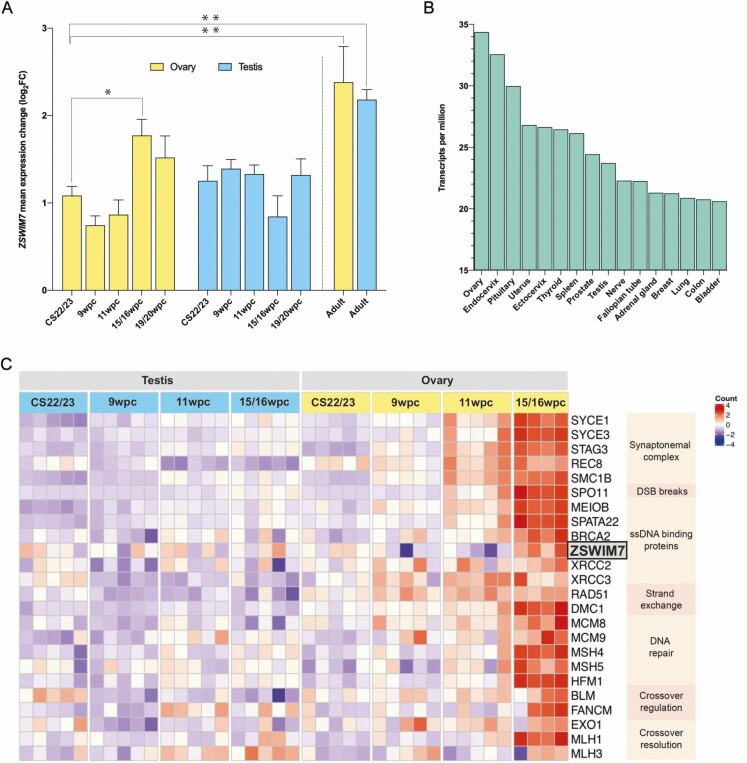
To investigate a role for ZSWIM7 in female meiosis, the expression of this gene was quantified by qRT-PCR of human gonadal tissue at critical stages of fetal gonad development, including sex differentiation, germ cell expansion, and meiotic entry (CS22/23 [7.5-8 wpc], 9 wpc, 11 wpc, 15-16 wpc, and 19-20 wpc) (Fig. 2A; left panel). There was higher expression of ZSWIM7 in the 15 to 16 wpc ovary compared with testis, corresponding with the peak onset of meiosis in the fetal ovary.
Figure 2.
(A) qRT-PCR mean expression (log2) of ZSWIM7 in various tissues compared with reference (GAPDH) and relative to the expression of ZSWIM7 in a Carnegie Stage (CS) 22-ovary sample. Four fetal ovary and fetal testis tissue samples were included at each of the following stages: CS22/CS23, 9 wpc, 11wpc, 15 to 16 wpc, and 19 to 20 wpc. Bar heights indicate mean expression. Error bars indicate mean ± SEM. Independent samples t tests and 1-way ANOVA testing were used to assess differences in ZSWIM7 mean expression change between tissue stages (*P < 0.05; **P < 0.01). There was significantly higher ZSWIM7 expression in the 15 to 16 wpc ovary compared with the 15 to 16 wpc testis (P = 0.02). There was significantly higher ZSWIM7 expression in the adult ovary compared with the CS22/23 ovary (P = 0.004) and in the adult testes compared with the CS22/23 testes (P = 0.003) compared with the CS22/23 testes. (B) ZSWIM7 expression across adult tissues from the GTEx database (v8). Data are expressed in transcripts per million. (C) Heatmap representing differential gene expression of key meiotic genes during prophase I across 4 developmental timepoints (CS22/23, 9 wpc, 11 wpc, 15-16 wpc). The intensity of gene expression is indicated by a color scale: violet for lowest expression and red for highest expression. ZSWIM7 is highlighted in gray. GAPDH, glyceraldehyde 3-phophate dehydrogenase; qRT-PCR, quantitative reverse transcriptase PCR; wpc, weeks postconception.
qRT-PCR analysis of adult ovary and testis showed relatively strong expression in the adult testis, where meiosis is actively occurring, but also in the adult ovary (Fig. 2A; right panel). This observation was supported following analysis of publicly available RNA expression datasets (GTEx v.8; Human Protein Atlas v20.1; FANTOM5), where ZSWIM7 was found to be expressed in adult ovarian tissue (45-47). ZSWIM7 expression across several tissues is displayed in Fig. 2B. These data suggest a potential long-term role for ZSWIM7 beyond the developmental period.
ZSWIM7 Shows Similar Temporal Expression to Other Homologous Recombination Genes in the Human Fetal Ovary
To investigate the temporal expression of ZSWIM7 during human fetal gonad development further, a time-series analysis using RNA sequencing of groups of ovary and testis samples between CS22/23 and 15 to 16 wpc was undertaken (Fig. 2C). ZSWIM7 showed a peak of increased expression in the ovary at 15 to 16 wpc compared with CS22 (FC 1.25, Padj < 0.05), 9 wpc (FC 1.46, Padj < 0.05), and 11 wpc (FC 1.48, Padj < 0.05), consistent with an increase in meiosis across this timeframe. Higher expression of ZSWIM7 was observed in the 15 to 16 wpc ovary compared with the testis at the same stage (FC 1.27, Padj = 0.05).
Finally, the developmental time-series RNAseq dataset was used to study human fetal gonadal expression of genes postulated to interact with ZSWIM7, as well as genes known to be important for prophase I of meiosis (Fig. 2C). Consistent upregulation of these genes was seen coinciding with peak meiosis in the 15 to 16 wpc ovary, when compared with testis and earlier premeiotic ovarian tissue (Fig. 2C).
Discussion
Here, we report a homozygous, stop-gain mutation in ZSWIM7, an emerging key factor in the meiotic pathway, in 2 sisters with early-onset POI from a consanguineous family. The predicted pathogenic variant is proximal to the zinc finger domain of ZSWIM7 and likely results in nonsense-mediated decay or protein truncation with loss of this critical domain.
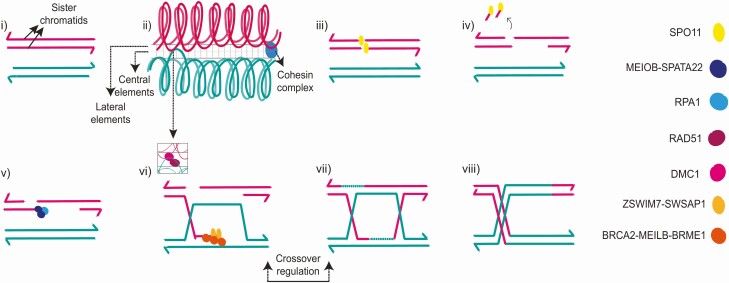
Functional studies have demonstrated ZSWIM7, also known as SWS1, to be a key regulator of meiotic homologous recombination. ZSWIM7/SWS1 interacts with SWSAP1 to form the highly conserved human Shu complex (48-50). This complex is required to regulate the recruitment of the strand-exchange protein RAD51 and its homolog DMC1 to meiotic intermediates during homologous recombination (Fig. 3). Loss of this complex in yeast, Caenorhabditis elegans, and in mice results in preserved viability but impaired fertility in males and females (44, 51-56). For example, Sws1/Swsap1 knock-out mice are grossly morphologically normal but show severe defects in meiotic progression in both sexes with reduced Rad51 and Dmc1 foci formation (44). The weights of both ovary and testis are reduced and histologically show fewer postmeiotic germ cells, evidence of germ cell apoptosis, and higher numbers of early prophase meiocytes (44). Germ cells from Zswim7-depleted mice can form double-stranded DNA breaks normally, but demonstrate synapsis defects and an increase in MEIOB foci (44, 57). Loss of the BRCA2 C-terminus aggravates the phenotype of Sws1/Swsap1-deficient mice, with hybrid knockout models displaying increased synapsis defects. Furthermore, disruption of DMC1 in mice results in synaptic defects similar to those seen in Zswim7 knockout animals (58, 59). These findings suggest that the Sws1/Swsap1 complex is essential for homologous recombination and that abnormal ZSWIM7 results in aberrant progression through prophase I (Fig. 3) (44). They also suggest that ZSWIM7 may have overlapping biological roles with other homologous recombination genes.
Figure 3.
Genes associated with primary ovarian insufficiency (POI) and their relationship with the stages of meiosis I prophase. Genes associated with POI are italicized and in bold in this legend text. (i) Meiosis I begins with homologous chromosomes moving physically close to one another. (ii) The synaptonemal complex (SC) forms between closely apposed homologous chromosomes, composed of central (SYCE1) and lateral (SYCE3) elements and stabilized by cohesin complexes (STAG3, REC8, SMC1B) surrounding the chromatid. (iii) Homologous recombination is initiated by double strand DNA breaks (DSBs) (SPO11). (iv) DSBs are recognized, partially degraded, and act as substrate for homology searching (EXO1). (v) Resected DNA ends are recognized and bound by single-strand DNA (ssDNA) binding proteins (MEIOB) which prevent reannealing. (vi) Downstream binding proteins are then localized to the single-strand tails (BRCA2), which recruit strand-exchange nucleoprotein filaments (RAD51), stabilize and remodel the growing DNA filament (ZSWIM7), and catalyze strand invasion, exchange, and homologous pairing. (vii) DNA exchange intermediates are further processed and joined via DNA repair (MCM8, MCM9, MSH4, MSH5, HFM1). Crossover (pictured) or noncrossover (not pictured) intermediates result. Crossover formation is tightly regulated (BLM, FANCM). (viii) Resolution of crossover and noncrossover intermediates (MLH1, MLH3, EXO1) complete the homologous recombination process. Prophase I concludes with diplotene and diakinesis.
Several homologous recombination genes have been implicated in the pathogenesis of POI and azoospermia, including some which are postulated to interact with ZSWIM7 as described previously. Missense variants and single-nucleotide polymorphisms in RAD51 have been associated with human ovarian dysgenesis and with lower age of menopause, respectively (60, 61). A homozygote mutation in XRCC2, a paralog of RAD51, was identified within a consanguineous pedigree in which males presented with azoospermia and infertility (62). Biallelic BRCA2 variants, within the C-terminus domain, have been reported in both familial and sporadic POI (21, 63). Reduced foci of DMC1 and RAD51 have been noted in fibroblasts of patients with POI and homozygote BRCA2 variants (21, 44). Recently, a homozygous splice variant (c.201 + 1G > T) (64) and a homozygous frameshift variant (c.231_232del; p.(Cys78Phefs*21)) (57, 64) in ZSWIM7 itself have been shown to be associated with human male infertility in 4 unrelated patients (Fig. 1C). Both variants likely result in either nonsense mediated decay or a truncated protein missing the zinc-finger binding domain of ZSWIM7.
Despite a clear emerging role for ZSWIM7 in mammalian meiosis, the expression of ZSWIM7 during human gonadal development has not been explored. Here, we demonstrate ZSWIM7 expression in the developing human ovary with highest expression at 15 to 16 wpc that corresponds with the known peak of meiotic activity (65). We also show increased ZSWIM7 expression in the adult testis and ovary. Significant ZSWIM7 expression in the adult testis is expected given the known postpubertal timing of male meiosis, but the relatively high expression of ZSWIM7 in the adult ovary, and well as degree of ubiquitous expression elsewhere, suggests that ZSWIM7 is involved in recombinational DNA repair pathways outside of meiosis (66). We also demonstrate that ZSWIM7 is expressed during the onset of human meiosis in fetal life and that this expression coincides with the expression of other known meiotic genes within the prophase I pathway.
To our knowledge, the stop-gain variant identified here in ZSWIM7 is the first variant within this gene to be associated with human POI. Our association of ZSWIM7 with POI suggests that disruption of this gene can result in both POI and male factor infertility, which has important clinical implications when counseling patients and their families. Zswim7 (Sws1)/Swsap1 mutant mice reproduce the infertility phenotype, demonstrating marked meiotic abnormalities. This emphasizes the value of drawing on knowledge from existing model systems when clarifying human biology. In the future, knowing the specific genetic cause of a POI diagnosis may help in the development of personalized targeted treatments, especially in males in whom the onset of meiosis occurs only after puberty and in adulthood.
Taken together, these data provide evidence for a role for ZSWIM7 in human female meiosis, implicate it in the pathogenesis of POI, and emphasize the importance of genes associated with homologous recombination and specifically meiosis prophase I in this condition. A broader mechanistic understanding of POI can be gained from considering meiotic genes as functional partners and this approach could extend the list of potential candidate genes for POI.
Acknowledgments
This study is dedicated to the memory of Professor Maria Bitner-Glindzicz who co-initiated this work on the genetics of primary ovarian insufficiency and who made outstanding contributions to the field of human genetics, and to the care of young people and their families, throughout her career. RNA sequencing was conducted in collaboration with UCL Genomics. We are grateful to Dr Paola Niola and Dr Tony Brooks (UCL Genomics, Zayed Centre for Research) for helpful discussions and technical support. The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by National Cancer Institute, National Human Genome Research Institute, National Heart, Lung, and Blood Institute, National Institute on Drug Abuse, National Institute of Mental Health, and National Institute of Neurological Disorders and Stroke. The data used for the analyses described in this manuscript were obtained from the GTEx Portal on March 3, 2021.
This study makes use of data generated by the DECIPHER community. A full list of centers who contributed to the generation of the data is available from https://deciphergenomics.org/about/stats and via email from contact@deciphergenomics.org. Funding for the DECIPHER project was provided by Wellcome.
Glossary
Abbreviations
- CS
Carnegie Stage
- gnomAD
Genome Aggregation Database
- POI
primary ovarian insufficiency
- qRT-PCR
quantitative reverse transcriptase PCR
- SC
synaptonemal complex
- wpc
weeks post conception
- ZSWIM7
zinc finger SWIM-type containing 7
Funding
This research was funded in whole, or in part, by the Wellcome Trust (S.M.B. 216362/Z/19/Z; J.C.A. 209328/Z/17/Z). For the purpose of Open Access, the author has applied a CC BY public copyright license to any Author Accepted Manuscript version arising from this submission. M.T.D. and J.C.A. (V2518) received additional funding from Great Ormond Street Hospital Children’s Charity. M.T.D. and J.C.A. have research support from the National Institute for Health Research, Great Ormond Street Hospital Biomedical Research Centre (IS-BRC-1215-20012). The views expressed are those of the authors and not necessarily those of the National Health Service, National Institute for Health Research, or Department of Health. Human fetal material was provided by the Joint MRC/Wellcome Trust (MR/R006237/1) Human Developmental Biology Resource (http://www.hdbr.org).
Additional Information
Disclosures: The authors have nothing to disclose.
Data Availability
Restrictions apply to the availability of genome sequencing data generated and analyzed during this study to preserve patient confidentiality. The corresponding author will on request detail the restrictions and any conditions under which access to some data may be provided. RNA sequencing data generated and analyzed during this study are included in the data repository BioStudies under accession number S-BSST693 at https://www.ebi.ac.uk/biostudies/.
References
- 1. European Society for Human Reproduction and Embryology (ESHRE) Guideline Group on POI, Webber L, Davies M, et al. ESHRE guideline: management of women with premature ovarian insufficiency. Hum Reprod. 2016;31(5):926-937. [DOI] [PubMed] [Google Scholar]
- 2. De Vos M, Devroey P, Fauser BC. Primary ovarian insufficiency. Lancet. 2010;376(9744):911-921. [DOI] [PubMed] [Google Scholar]
- 3. Nelson LM. Clinical practice. Primary ovarian insufficiency. N Engl J Med. 2009;360(6):606-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Luborsky JL, Meyer P, Sowers MF, Gold EB, Santoro N. Premature menopause in a multi-ethnic population study of the menopause transition. Hum Reprod. 2003;18(1):199-206. [DOI] [PubMed] [Google Scholar]
- 5. Tucker EJ, Grover SR, Bachelot A, Touraine P, Sinclair AH. Premature ovarian insufficiency: new perspectives on genetic cause and phenotypic spectrum. Endocr Rev. 2016;37(6):609-635. [DOI] [PubMed] [Google Scholar]
- 6. Beck-Peccoz P, Persani L. Premature ovarian failure. Orphanet J Rare Dis. 2006;1:9. doi: 10.1186/1750-1172-1-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gordon CM, Kanaoka T, Nelson LM. Update on primary ovarian insufficiency in adolescents. Curr Opin Pediatr. 2015;27(4):511-519. [DOI] [PubMed] [Google Scholar]
- 8. Maclaran K, Panay N. Premature ovarian failure. J Fam Plann Reprod Health Care. 2011;37(1):35-42. [DOI] [PubMed] [Google Scholar]
- 9. Silva CA, Yamakami LY, Aikawa NE, Araujo DB, Carvalho JF, Bonfá E. Autoimmune primary ovarian insufficiency. Autoimmun Rev. 2014;13(4-5):427-430. [DOI] [PubMed] [Google Scholar]
- 10. Craig ZR, Wang W, Flaws JA. Endocrine-disrupting chemicals in ovarian function: effects on steroidogenesis, metabolism and nuclear receptor signaling. Reproduction. 2011;142(5): 633-646. [DOI] [PubMed] [Google Scholar]
- 11. Huhtaniemi I, Hovatta O, La Marca A, et al. Advances in the molecular pathophysiology, genetics, and treatment of primary ovarian insufficiency. Trends Endocrinol Metab. 2018;29(6):400-419. [DOI] [PubMed] [Google Scholar]
- 12. Eskenazi S, Bachelot A, Hugon-Rodin J, et al. Next generation sequencing should be proposed to every woman with “idiopathic” primary ovarian insufficiency. J Endocr Soc. 2021;5(7):bvab032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang J, Gurusaran M, Fujiwara Y, et al. The BRCA2-MEILB2-BRME1 complex governs meiotic recombination and impairs the mitotic BRCA2-RAD51 function in cancer cells. Nat Commun. 2020;11(1):2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Takemoto K, Tani N, Takada-Horisawa Y, et al. Meiosis-specific C19orf57/4930432K21Rik/BRME1 modulates localization of RAD51 and DMC1 to DSBs in mouse meiotic recombination. Cell Rep. 2020;31(8):107686. [DOI] [PubMed] [Google Scholar]
- 15. Brown MS, Bishop DK. DNA strand exchange and RecA homologs in meiosis. Cold Spring Harb Perspect Biol. 2014;7(1):a016659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. de Vries L, Behar DM, Smirin-Yosef P, Lagovsky I, Tzur S, Basel-Vanagaite L. Exome sequencing reveals SYCE1 mutation associated with autosomal recessive primary ovarian insufficiency. J Clin Endocrinol Metab. 2014;99(10):E2129-E2132. [DOI] [PubMed] [Google Scholar]
- 17. Geisinger A, Benavente R. Mutations in genes coding for synaptonemal complex proteins and their impact on human fertility. Cytogenet Genome Res. 2016;150(2):77-85. [DOI] [PubMed] [Google Scholar]
- 18. Caburet S, Arboleda VA, Llano E, et al. Mutant cohesin in premature ovarian failure. N Engl J Med. 2014;370(10):943-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bouilly J, Beau I, Barraud S, et al. Identification of multiple gene mutations accounts for a new genetic architecture of primary ovarian insufficiency. J Clin Endocrinol Metab. 2016;101(12):4541-4550. [DOI] [PubMed] [Google Scholar]
- 20. Caburet S, Todeschini AL, Petrillo C, et al. A truncating MEIOB mutation responsible for familial primary ovarian insufficiency abolishes its interaction with its partner SPATA22 and their recruitment to DNA double-strand breaks. Ebiomedicine. 2019;42:524-531. doi: 10.1016/j.ebiom.2019.03.075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Caburet S, Heddar A, Dardillac E, et al. Homozygous hypomorphic BRCA2 variant in primary ovarian insufficiency without cancer or Fanconi anaemia trait. J Med Genet. 2020. doi: 10.1136/jmedgenet-2019-106672. [DOI] [PubMed] [Google Scholar]
- 22. Guo T, Zhao S, Zhao S, et al. Mutations in MSH5 in primary ovarian insufficiency. Hum Mol Genet. 2017;26(8): 1452-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. AlAsiri S, Basit S, Wood-Trageser MA, et al. Exome sequencing reveals MCM8 mutation underlies ovarian failure and chromosomal instability. J Clin Invest. 2015;125(1):258-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wood-Trageser MA, Gurbuz F, Yatsenko SA, et al. MCM9 mutations are associated with ovarian failure, short stature, and chromosomal instability. Am J Hum Genet. 2014;95(6):754-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Goldberg Y, Halpern N, Hubert A, et al. Mutated MCM9 is associated with predisposition to hereditary mixed polyposis and colorectal cancer in addition to primary ovarian failure. Cancer Genet. 2015;208(12):621-624. [DOI] [PubMed] [Google Scholar]
- 26. Wang J, Zhang W, Jiang H, Wu BL; Primary Ovarian Insufficiency Collaboration. Mutations in HFM1 in recessive primary ovarian insufficiency. N Engl J Med. 2014;370(10):972-974. [DOI] [PubMed] [Google Scholar]
- 27. Patiño LC, Beau I, Carlosama C, et al. New mutations in non-syndromic primary ovarian insufficiency patients identified via whole-exome sequencing. Hum Reprod. 2017;32(7):1512-1520. [DOI] [PubMed] [Google Scholar]
- 28. Tarasov A, Vilella AJ, Cuppen E, Nijman IJ, Prins P. Sambamba: fast processing of NGS alignment formats. Bioinformatics. 2015;31(12):2032-2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Van der Auwera GA, Carneiro MO, Hartl C, et al. From FastQ data to high confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11.10.1-11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McKenna A, Hanna M, Banks E, et al. The genome analysis toolkit: a mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Karczewski KJ, Francioli LC, Tiao G, et al. ; Genome Aggregation Database Consortium. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581(7809):434-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Richards S, Aziz N, Bale S, et al. ; ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. Plos One. 2012;7(10):e46688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Firth HV, Richards SM, Bevan AP, et al. DECIPHER: database of chromosomal imbalance and phenotype in humans using ensembl resources. Am J Hum Genet. 2009;84(4):524-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014;11(4):361-362. [DOI] [PubMed] [Google Scholar]
- 38. Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Andrews S. FastQC: a quality control tool for high throughput sequence data. Accessed April 13, 2021. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 41. Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923-930. [DOI] [PubMed] [Google Scholar]
- 42. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B (Methodological). 1995;57(1):289-300. [Google Scholar]
- 44. Abreu CM, Prakash R, Romanienko PJ, Roig I, Keeney S, Jasin M. Shu complex SWS1-SWSAP1 promotes early steps in mouse meiotic recombination. Nat Commun. 2018;9(1):3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348(6235):648-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lizio M, Harshbarger J, Shimoji H, et al. ; FANTOM consortium. Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol. 2015;16(22). doi: 10.1186/s13059-014-0560-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Uhlén M, Fagerberg L, Hallström BM, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. [DOI] [PubMed] [Google Scholar]
- 48. Martino J, Brunette GJ, Barroso-González J, et al. The human Shu complex functions with PDS5B and SPIDR to promote homologous recombination. Nucleic Acids Res. 2019;47(19):10151-10165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liu T, Wan L, Wu Y, Chen J, Huang J. hSWS1·SWSAP1 is an evolutionarily conserved complex required for efficient homologous recombination repair. J Biol Chem. 2011;286(48):41758-41766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Martín V, Chahwan C, Gao H, et al. Sws1 is a conserved regulator of homologous recombination in eukaryotic cells. Embo J. 2006;25(11):2564-2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sasanuma H, Tawaramoto MS, Lao JP, et al. A new protein complex promoting the assembly of Rad51 filaments. Nat Commun. 2013;4(1):1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lorenz A, Mehats A, Osman F, Whitby MC. Rad51/Dmc1 paralogs and mediators oppose DNA helicases to limit hybrid DNA formation and promote crossovers during meiotic recombination. Nucleic Acids Res. 2014;42(22):13723-13735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. McClendon TB, Sullivan MR, Bernstein KA, Yanowitz JL. Promotion of homologous recombination by SWS-1 in complex with RAD-51 paralogs in Caenorhabditis elegans. Genetics. 2016;203(1):133-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Taylor MRG, Špírek M, Chaurasiya KR, et al. Rad51 paralogs remodel pre-synaptic Rad51 filaments to stimulate homologous recombination. Cell. 2015;162(2):271-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ward JD, Barber LJ, Petalcorin MI, Yanowitz J, Boulton SJ. Replication blocking lesions present a unique substrate for homologous recombination. Embo J. 2007;26(14):3384-3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yanowitz JL. Genome integrity is regulated by the Caenorhabditis elegans Rad51D homolog rfs-1. Genetics. 2008;179(1):249-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Li Y, Wu Y, Zhou J, et al. A recurrent ZSWIM7 mutation causes male infertility resulting from decreased meiotic recombination. Hum Reprod. 2021. doi: 10.1093/humrep/deab046. [DOI] [PubMed] [Google Scholar]
- 58. Yoshida K, Kondoh G, Matsuda Y, Habu T, Nishimune Y, Morita T. The mouse RecA-like gene Dmc1 is required for homologous chromosome synapsis during meiosis. Mol Cell. 1998;1(5):707-718. [DOI] [PubMed] [Google Scholar]
- 59. Pittman DL, Cobb J, Schimenti KJ, et al. Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol Cell. 1998;1(5):697-705. [DOI] [PubMed] [Google Scholar]
- 60. Luo W, Guo T, Li G, et al. Variants in homologous recombination genes EXO1 and RAD51 related with premature ovarian insufficiency. J Clin Endocrinol Metab. 2020. doi: 10.1210/clinem/dgaa505. [DOI] [PubMed] [Google Scholar]
- 61. Day FR, Ruth KS, Thompson DJ, et al. ; PRACTICAL consortium; kConFab Investigators; AOCS Investigators; Generation Scotland; EPIC-InterAct Consortium; LifeLines Cohort Study. Large-scale genomic analyses link reproductive aging to hypothalamic signaling, breast cancer susceptibility and BRCA1-mediated DNA repair. Nat Genet. 2015;47(11):1294-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yang Y, Guo J, Dai L, et al. XRCC2 mutation causes meiotic arrest, azoospermia and infertility. J Med Genet. 2018;55(9):628-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tsui V, Crismani W. The fanconi anemia pathway and fertility. Trends Genet. 2019;35(3):199-214. [DOI] [PubMed] [Google Scholar]
- 64. Alhathal N, Maddirevula S, Coskun S, et al. A genomics approach to male infertility. Genet Med. 2020;22(12):1967-1975. [DOI] [PubMed] [Google Scholar]
- 65. Childs AJ, Cowan G, Kinnell HL, Anderson RA, Saunders PT. Retinoic acid signalling and the control of meiotic entry in the human fetal gonad. Plos One. 2011;6(6):e20249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kowalczykowski SC. An overview of the molecular mechanisms of recombinational DNA repair. Cold Spring Harb Perspect Biol. 2015;7(11). doi: 10.1101/cshperspect.a016410. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Restrictions apply to the availability of genome sequencing data generated and analyzed during this study to preserve patient confidentiality. The corresponding author will on request detail the restrictions and any conditions under which access to some data may be provided. RNA sequencing data generated and analyzed during this study are included in the data repository BioStudies under accession number S-BSST693 at https://www.ebi.ac.uk/biostudies/.