Abstract
Objectives
To investigate potential genetic biomarkers of peri-implantitis and target genes for the therapy of peri-implantitis by bioinformatics analysis of publicly available data.
Methods
The GSE33774 microarray dataset was downloaded from the Gene Expression Omnibus (GEO). The differentially expressed genes (DEGs) between peri-implantitis and healthy gingival tissues were identified using the GEO2R tool. GO enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were performed using the DAVID database and the Metascape tool, and the results were expressed as a bubble diagram. The protein-protein interaction network of DEGs was constructed using the Search Tool for the Retrieval of Interacting Genes (STRING) and visualized using Cytoscape. The hub genes were screened by the cytoHubba plugin of Cytoscape. The potential target genes associated with peri-implantitis were obtained from the DisGeNET database and the Open Targets Platform. The intersecting genes were identified using the Venn diagram web tool.
Results
Between the peri-implantitis group and the healthy group, 205 DEGs were investigated including 140 upregulated genes and 65 downregulated genes. These DEGs were mainly enriched in functions such as the immune response, inflammatory response, cell adhesion, receptor activity, and protease binding. The results of KEGG pathway enrichment analysis revealed that DEGs were mainly involved in the cytokine-cytokine receptor interaction, pathways in cancer, and the PI3K-Akt signaling pathway. The intersecting genes, including IL6, TLR4, FN1, IL1β, CXCL8, MMP9, and SPP1, were revealed as potential genetic biomarkers and target genes of peri-implantitis.
Conclusions
This study provides supportive evidence that IL6, TLR4, FN1, IL1β, CXCL8, MMP9, and SPP1 might be used as potential target biomarkers for peri-implantitis which may provide further therapeutic potentials for peri-implantitis.
1. Introduction
In recent decades, dental implants have been widely used for the restoration of missing teeth with high success rates [1]. Peri-implantitis is a common complication of dental implants that can result in implant failure. Peri-implantitis can be defined as an inflammation of the peri-implant connective tissue and progressive loss of the supporting bone around the implants [2], and it is considered to be the leading cause of implant failure. According to previous literature, the approximate prevalence of peri-implantitis is 22% (range: 1%–47%) and that of peri-implant mucositis is 43% (range:19%–65%) [3, 4]. Studies have shown that bacterial infection could be the cause of the peri-implantitis and subsequent implant failure, and the various gene polymorphisms may be associated with the occurrence of peri-implantitis [2].
Although the underlying pathogenic mechanisms of peri-implantitis remain unclear, the excessive inflammatory response due to the microbial biofilms on implants and their toxins is believed to play an important role in the occurrence of peri-implantitis [5, 6]. Therefore, the immune-inflammatory response elicited by the bacterial biofilm may be responsible for the gingival recession and alveolar bone loss associated with peri-implantitis. Lipopolysaccharide can induce the cells of gingival and osseous tissues to overexpress proinflammatory cytokines including interleukin- (IL-) 1β and IL-6 [7]. Different methods have been used for treating peri-implantitis, such as mechanical debridement, implant surface modifications, adjunctive antibiotic therapy, and surgery. Gene therapy might be considered as a therapeutic option in regenerative medicine for peri-implant tissues [8].
In recent years, host modulation therapies are considered as a potential alternative for the treatment of peri-implantitis [9]. This treatment method based on the effects of inflammatory regulation not only promotes the efficacy of traditional management approaches for peri-implantitis but also reduces the risk of systemic disease and inflammation. Proinflammatory cytokines, such as IL-1β, IL-6, and TNF-α, have been used as biomarkers to identify periodontitis and peri-implantitis [10, 11]. There are also many other well-accepted biomarkers of tissue destruction and systemic inflammation, including matrix metalloproteinase- (MMP-) 8, MMP-9, high-sensitivity C-reactive protein, TNF-α, and IL-6, which are easily detected in both oral fluids and blood samples [12–15]. Certain cytokine inhibitors such as TNF-α antagonists and IL-1 receptor antagonists have exhibited anti-inflammatory effects in periodontal diseases and may be used for treating peri-implantitis [16, 17]. Further investigation of the underlying mechanism of peri-implantitis is needed to develop rational treatment strategies.
In recent years, bioinformatics methods have been widely used to analyze microarray data to identify differentially expressed genes (DEGs). Numerous bioinformatics tools and approaches have been developed, which could help us to better understand the underlying mechanisms [18–20]. Scientific literature on peri-implantitis has increased rapidly in recent years and particularly in the last 5 years [21]. However, few studies have focused on the application of bioinformatics analysis to gain insights on peri-implantitis. The study of Becker et al. showed that peri-implantitis and periodontitis show different mRNA signatures, although they share similar clinical characteristics [22]. In the present study, to better understand the potential molecular biomarkers and the potential therapeutic agents for peri-implantitis, we used the GSE33774 microarray dataset and bioinformatics tools. We downloaded mRNA expression profiles from Gene Expression Omnibus (GEO, http://www. http://ncbi.nlm.nih.gov/geo/). We identified the DEGs between samples of peri-implantitis and healthy samples. Gene Ontology (GO) is a major bioinformatics tool to annotate genes and analyze the biological process of these genes [23]. Kyoto Encyclopedia of Genes and Genomes (KEGG) is a widely used database that stores extensive data on genomes, biological pathways, diseases, chemical substances, and drugs. GO analysis is a common useful method for large-scale functional enrichment research, wherein gene functions can be classified into biological process (BP), molecular function (MF), and cellular component (CC). In the present study, KEGG pathway and GO functional enrichment analyses were performed. The DisGeNET platform and the Open Targets Platform (OTP) were used to further investigate the coexpressed genes associated with peri-implantitis. The findings of this study may help to predict the molecular mechanism and the potential therapeutic targets of peri-implantitis.
2. Materials and Methods
2.1. Data Source and Identification of DEGs
The gene expression dataset of GSE33774 analyzed in this study was obtained from the GEO database (http://www.ncbi.nlm.nih.gov/geo/) [24, 25]. GSE33774 was based on the Agilent GPL platform GPL6244 (Affymetrix Human Gene 1.0 ST Array (transcript (gene) version)). The GSE33774 dataset contains seven gingival tissue samples of peri-implantitis, seven gingival tissue samples of periodontitis, and eight healthy gingival samples [22]. All of the data are freely available online, and this study did not involve any experiments on humans or animals.
The DEGs between peri-implantitis samples and healthy samples were analyzed using the GEO2R online analysis tool (http://www.ncbi.nlm.nih.gov/geo/geo2r). The DEGs with the threshold criterion of ∣log(fold change) | ≥1.0 and P value < 0.05 were considered to be significantly differentially expressed.
2.2. GO and KEGG Pathway Enrichment Analyses of DEGs
To analyze the function of DEGs, biological analyses were performed using the online database DAVID [26, 27]. P < 0.05 was considered to be statistically significant. The Metascape tool (https://metascape.org) [28] was also used to perform functional and pathway enrichment analysis of DEGs, including BP, CC, MF, and KEGG pathway enrichment analysis. A P value of <0.05 was considered to be the cut-off criterion.
2.3. Protein-Protein Interaction (PPI) Network Construction and Hub Gene Identification
The PPI network of DEGs was constructed using the Search Tool for the Retrieval of Interacting Genes (STRING, http://string-db.org) (version 11.0) online database [29], and an interaction with a combined score of >0.40 (medium confidence interaction score) was considered to be statistically significant. Subsequently, the PPI network was visualized by Cytoscape software (version 3.8.0) [30]. Hub genes were identified and visualized using the CytoHubba plugin of Cytoscape [31]. The top ten genes with high degree of connectivity in the PPI network were identified as hub genes.
2.4. Prediction of the Gene-Disease Associations
To accurately predict the gene-disease associations of peri-implantitis, the DisGeNET platform (http://www.disgenet.org/) and the Open Targets Platform (OTP) (https://www.targetvalidation.org/) were used. DisGeNET is an online database that includes a collection of genes associated with human diseases based on expert-curated databases and scientific literature, and this database is publicly accessible [32]. The OTP provides evidence for human target-disease associations and tools that provide evidence-based systematic prioritization of targets for disease treatment [33]. The genes associated with peri-implantitis were exported from the DisGeNET database and the OTP. The following search terms were used in the DisGeNET database and the OTP (UMLS:C2936258) and peri-implantitis (EFO: 1001390), respectively. The intersecting genes of the top 10 hub genes and the disease-associated genes obtained from the DisGeNET database and the OTP were identified using the Venn diagram web tool (bioinformatics.psb.ugent.be/webtools/Venn/). A gene-disease network around peri-implantitis was generated by the DisGeNET Cytoscape plugin.
3. Results
3.1. Identification of DEGs
The microarray expression profile of GSE33774 was selected in this study. We identified 205 DEGs including 140 upregulated genes and 65 downregulated genes based on the criteria of P < 0.05 and ∣logFC | ≥1.0. All DEGs were identified by comparing samples of peri-implantitis with healthy gingival samples. Subsequently, the volcano plots were generated for the identified DEGs (Figure 1).
Figure 1.
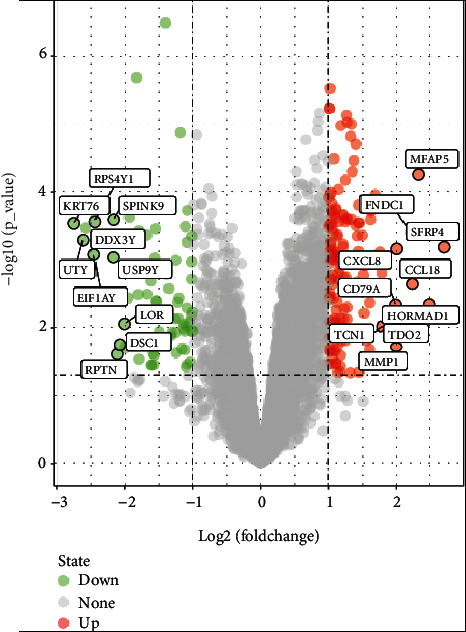
Volcano maps of differentially expressed genes. Red and green spots represent differentially expressed genes: red spots represent upregulated genes and green spots represent downregulated genes.
3.2. Functional Enrichment Analysis of DEGs
GO enrichment analysis and the KEGG pathway enrichment analysis of DEGs were performed using the online database DAVID (Table 1). The enriched GO terms were divided into BP, CC, and MF ontologies. The results indicated that for BP analysis, the DEGs were mainly enriched in immune response, inflammatory response, signal transduction, and cell adhesion. For the CC terms, the DEGs were enriched in plasma membrane, integral component of membrane, extracellular exosome, extracellular space, and extracellular region. The MF analysis showed that the DEGs were enriched in receptor activity, protease binding, RNA polymerase II regulatory region sequence-specific DNA binding, and transmembrane signaling receptor activity. The results of the KEGG pathway enrichment analysis showed that DEGs were mainly enriched in cytokine-cytokine receptor interaction, pathways in cancer, amoebiasis, phagosome, and the PI3K-Akt signaling pathway. The results obtained by enrichment analysis were illustrated by a bubble diagram (Figure 2). The results of the enrichment analysis of DEGs performed by Metascape are shown in Figure 3. DEGs are mainly concentrated in response to the bacterium and immune effector process. DEGs associated with innate immune responses and defense responses may play an important role in inflammation associated with peri-implantitis.
Table 1.
The results of GO function analysis and KEGG pathway enrichment analysis (top 5 terms are listed).
| Category | Term | Count | P value |
|---|---|---|---|
| BP term | GO:0006955~immune response | 23 | 4.62E − 10 |
| BP term | GO:0006954~inflammatory response | 20 | 1.51E − 08 |
| BP term | GO:0007165~signal transduction | 20 | 0.032080 |
| BP term | GO:0007155~cell adhesion | 19 | 1.38E − 06 |
| BP term | GO:0007186~G-protein-coupled receptor signaling pathway | 17 | 0.024583 |
| CC term | GO:0016021~integral component of membrane | 68 | 0.008933 |
| CC term | GO:0070062~extracellular exosome | 43 | 0.004559 |
| CC term | GO:0005615~extracellular space | 40 | 1.38E − 09 |
| CC term | GO:0005576~extracellular region | 38 | 1.42E − 06 |
| MF term | GO:0004872~receptor activity | 11 | 7.84E − 05 |
| MF term | GO:0004252~serine-type endopeptidase activity | 10 | 0.001235 |
| MF term | GO:0002020~protease binding | 7 | 5.84E − 04 |
| MF term | GO:0000977~RNA polymerase II regulatory region sequence-specific DNA binding | 7 | 0.019798 |
| MF term | GO:0004888~transmembrane signaling receptor activity | 7 | 0.022448 |
| KEGG pathway | hsa04060:cytokine-cytokine receptor interaction | 14 | 8.47E − 06 |
| KEGG pathway | hsa05200:pathways in cancer | 12 | 0.009391 |
| KEGG pathway | hsa05146:amoebiasis | 11 | 6.57E − 07 |
| KEGG pathway | hsa04145:phagosome | 11 | 1.54E − 05 |
| KEGG pathway | hsa04151:PI3K-Akt signaling pathway | 11 | 0.010471 |
Figure 2.
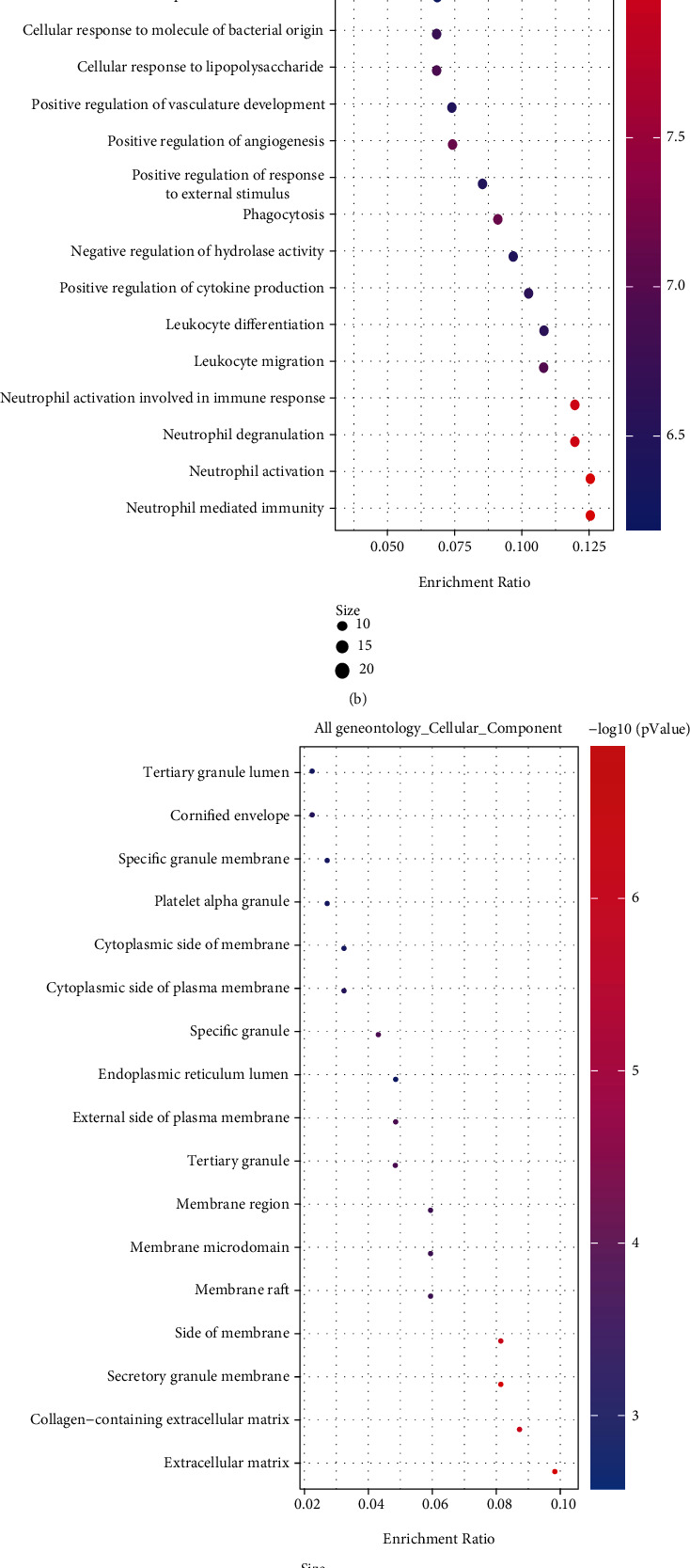
Results of GO and KEGG analysis results of differentially expressed genes: (a) KEGG pathway enrichment results; (b) GO biological process enrichment results; (c) GO cell component enrichment results; (d) GO molecular function enrichment results. The x-axis represents gene ratio, and the y-axis represents GO terms. The size of each circle indicates gene count. The color of circles represents different -log10(P.values).
Figure 3.
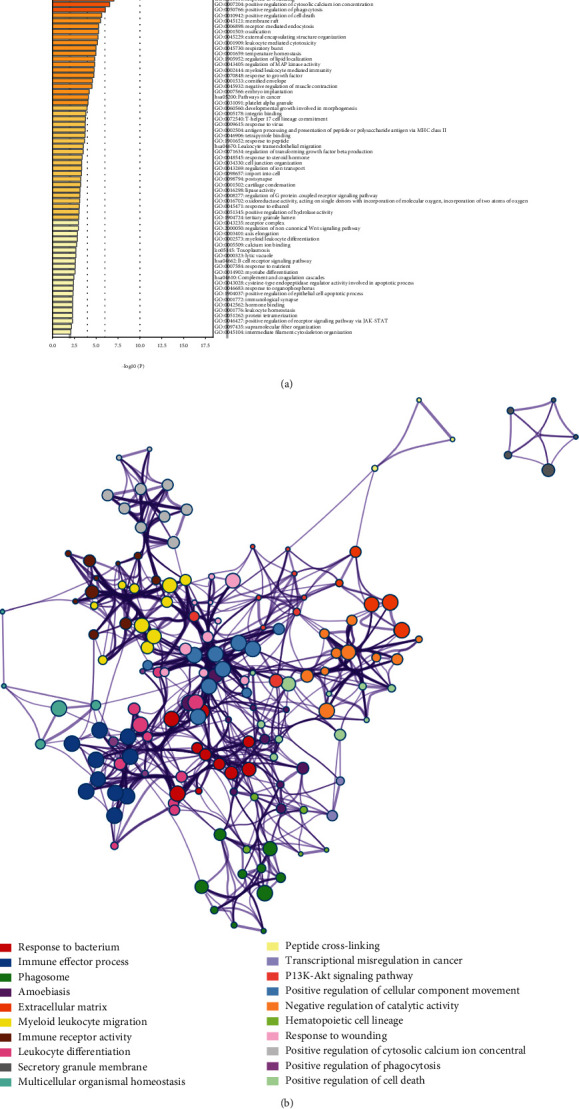
DEG enrichment results obtained using Metascape: (a) significantly enriched GO terms and KEGG pathways of DEGs; (b) network contact of GO terms and KEGG pathways.
3.3. Analysis of the PPI Network and Identification of Hub Genes
Protein interactions among the DEGs were predicted with STRING tools. A total of 143 nodes and 601 edges were involved in the PPI network, as shown in Figure 4(a). The top 10 genes according to their degree of connectivity in the PPI network were identified as hub genes (Figures 4(b) and 4(c)). The results showed that IL6, TLR4, FN1, IL1β, MMP9, CXCL8, CXCR4, CXCL1, PECAM1, and SPP1 were identified as hub genes (Table 2). Among these genes, IL-6 and TLR4 showed the highest node degrees, suggesting that they may play important roles in peri-implantitis. All the 10 hub genes were upregulated in peri-implantitis. As shown in Figure 4(c), all hub genes interact with each other directly. The hub genes were closely related to the results of GO and KEGG pathway enrichment analyses (Table 3). As shown in Table 3, CXCL8, CXCL1, IL-6, IL-1β, and TLR4 are involved in the immune response (GO:0006955), while CXCL8, CXCR4, CXCL1, IL-6, IL-1β, SPP1, and TLR4 are involved in the inflammatory response (GO:0006954). CXCL8, CXCL1, IL-6, and IL-1β are directly involved in the cytokine-cytokine receptor interaction pathway (hsa04060).
Figure 4.
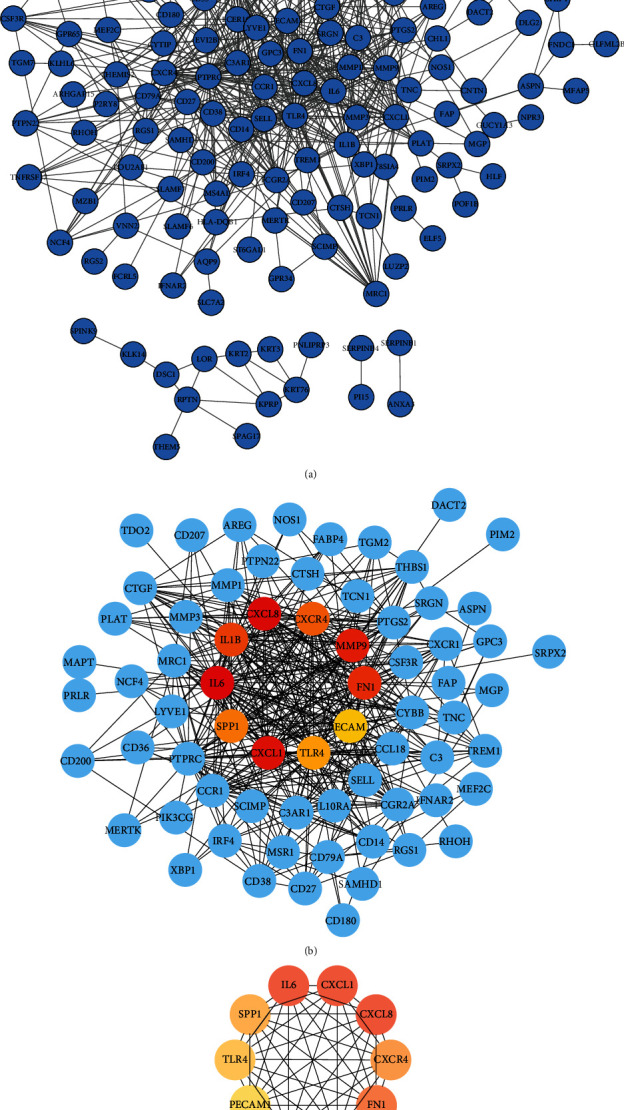
(a) Protein-protein interaction network of DEGs. (b) The top 10 hub genes in the PPI network with neighbors and expanded network. (c) Subnetwork of the top 10 hub genes from the PPI network.
Table 2.
Top 10 hub genes with higher degree of connectivity.
| Gene symbol | Gene description | Degree |
|---|---|---|
| IL-6 | Interleukin-6 | 46 |
| TLR4 | Toll-like receptor 4 | 43 |
| FN1 | Fibronectin 1 | 33 |
| IL-1β | Interleukin-1 beta | 32 |
| CXCL8 | C-X-C motif chemokine ligand 8 | 32 |
| MMP-9 | Matrix metallopeptidase-9 | 31 |
| CXCR4 | C-X-C motif chemokine receptor 4 | 30 |
| CXCL1 | C-X-C motif chemokine ligand 1 | 27 |
| PECAM1 | Platelet and endothelial cell adhesion molecule 1 | 26 |
| SPP1 | Secreted phosphoprotein 1 | 22 |
Table 3.
Top 10 hub genes that were closely associated with the results of GO and KEGG analyses.
| Term | Genes |
|---|---|
| GO:0006955~immune response | CCR1, XBP1, CXCL8, GPR65, AQP9, NCF4, CXCL1, SERPINB9, SAMHD1, THBS1, LAX1, C3, IL-6, IGKC, RGS1, IL-1β, PXDN, ENPP2, CD27, CD36, CCL18, TLR4, HLA-DQB1 |
| GO:0006954~inflammatory response | CCR1, CXCL8, CD180, CYBB, CXCR4, CXCL1, PTGS2, THBS1, PIK3CG, C3, IL-6, THEMIS2, CXCR1, IL-1β, SPP1, C3AR1, CD27, CD14, CCL18, TLR4 |
| GO:0007165~signal transduction | CD53, GABRP, SH3GL3, CSF3R, CXCL8, CXCL1, ABCC9, HTR3A, ARHGAP15, LYVE1, C3, EVI2A, CHL1, RGS1, IL-1β, MRC1, PECAM1, TNFRSF17, CD38, CCL18 |
| GO:0007155~cell adhesion | NLGN4Y, CCR1, CSF3R, LAMB4, TNC, FN1, LYVE1, THBS1, CTGF, THEMIS2, SELL, FAP, CHL1, BOC, SPP1, PECAM1, CNTN1, SLAMF7, CD36 |
| GO:0007186~G-protein-coupled receptor signaling pathway | CXCL8, GPR34, GPR65, CXCR4, CXCL1, AREG, PIK3CG, C3, SFRP4, SFRP2, CXCR1, RGS1, C3AR1, ENPP2, CCL18, ADGRL4, TGM2 |
| GO:0005886~plasma membrane | CSF3R, GPR65, AQP9, SLC2A3, TREM1, PIK3CG, CTGF, RGS5, RGS2, CHL1, RGS1, BOC, MRC1, ENPP2, C3AR1, CD38, CD36, LRRFIP1, TGM2, GUCY1A3, CD53, IFNAR2, FCER1G, FCRL5, ANXA3, DCC, CYBB, RHOH, HTR3A, ABCC9, PRLR, LAX1, CDHR1, PECAM1, SLC27A6, PLIN2, MAPT, DSC1, TLR4, HLA-DQB1, SLC47A2, NPR3, KCNA3, CXCR4, SAMHD1, SLC7A2, RASGRP3, P2RY8, C3, CD79A, IGKC, CXCR1, GPC3, SLAMF7, TNFRSF17, SLAMF6, CD14, CCR1, MSR1, GABRP, IL-10RA, MERTK, LYVE1, PTPRC, FCGR2A, DLG2, SELL, FAP, VNN2, CD207, CNTN1, CD27, ADGRL4, CDK14, F2RL2, CD200 |
| GO:0016021~integral component of membrane | CSF3R, DENND5B, GPR65, AQP9, LRMP, SLC2A3, TREM1, AREG, MS4A4A, AADACL2, CHL1, ENPP2, C3AR1, CD38, CD36, UTY, GUCY1A3, CD53, IFNAR2, ST6GAL1, FCER1G, GPR34, FCRL5, DCC, CYBB, HTR3A, ABCC9, PRLR, LAX1, SFRP4, SFRP2, CDHR1, PECAM1, SLC27A6, CFAP54, DSC1, HLA-DQB1, SLC47A2, NPR3, CXCR4, SEL1L3, P2RY8, CD79A, CXCR1, SLITRK3, SLAMF7, TNFRSF17, SLAMF6, CCR1, MSR1, GABRP, XBP1, IL-10RA, MERTK, LYVE1, SCIMP, TMEM156, PTPRC, FCGR2A, EVI2A, SELL, FAP, CD207, ESYT3, EVI2B, ADGRL4, F2RL2, CD200 |
| GO:0070062~extracellular exosome | KPRP, SH3GL3, NPR3, CXCR4, PLAT, SLC2A3, THBS1, C3, IGKC, CHL1, GPC3, SPP1, CTSH, CD38, SLAMF6, CD14, TGM2, CD53, SERPINB4, SERPINB1, ST6GAL1, KRT3, ANXA3, KRT2, FN1, KLK14, SERPINB9, KRT76, LYVE1, MMP-9, ALDH1A3, PTPRC, FCGR2A, FABP4, IL-1β, PXDN, MGP, PI15, PECAM1, CNTN1, CD27, DSC1, MS4A1 |
| GO:0005615~extracellular space | CXCL8, TNC, PLAT, CXCL1, THBS1, AREG, CTGF, C3, SRPX2, IGKC, GPC3, SPP1, ENPP2, CTSH, CD14, CD36, CCL18, NLGN4Y, SRGN, IFNAR2, SERPINB4, SERPINB1, KRT2, MMP-3, FN1, KLK14, SERPINB9, IGFL1, MERTK, MMP-9, SFRP4, IL-6, SFRP2, FAP, TCN1, IL1-β, PXDN, PECAM1, MS4A1, BPIFC |
| GO:0005576~extracellular region | CSF3R, OLFML2B, CXCL8, LUZP2, TNC, PLAT, CXCL1, TREM1, THBS1, CTGF, C3, AADACL2, IGKC, SPP1, LIPM, LIPK, CD14, SRGN, IFNAR2, MMP-1, MMP-3, FN1, MMP-9, PRLR, MFAP5, GZMK, SFRP4, IL-6, SFRP2, FNDC1, TCN1, IL-1β, MZB1, CD27, PLIN2, SPINK9, F2RL2, PNLIPRP3 |
| GO:0004872~receptor activity | GUCY1A3, NLGN4Y, CSF3R, IL-10RA, CD180, MRC1, TNFRSF17, TREM1, TLR4, LYVE1, CD200 |
| GO:0004252~serine-type endopeptidase activity | GZMK, C3, IGKC, FAP, MMP-1, MMP-3, KLK14, CTSH, PLAT, MMP-9 |
| GO:0002020~protease binding | SERPINB4, XBP1, SELL, FAP, CHL1, FN1, SERPINB9 |
| GO:0000977~RNA polymerase II regulatory region sequence-specific DNA binding | MEF2C, XBP1, HLF, ELF5, EAF2, ZFY, BARX2 |
| GO:0004888~transmembrane signaling receptor activity | CD79A, EVI2A, DCC, MRC1, CD27, TLR4, LYVE1 |
| hsa04060:cytokine-cytokine receptor interaction | CCR1, IFNAR2, CSF3R, CXCL8, IL-10RA, CXCR4, CXCL1, PRLR, IL-6, CXCR1, IL-1β, TNFRSF17, CD27, CCL18 |
| hsa05200:pathways in cancer | IL-6, CSF3R, CXCL8, MMP-1, DCC, LAMB4, FN1, CXCR4, PTGS2, MMP-9, PIK3CG, RASGRP3 |
| hsa05146:amoebiasis | SERPINB4, IL-6, SERPINB1, CXCL8, IL-1β, LAMB4, FN1, SERPINB9, CD14, TLR4, PIK3CG |
| hsa04145:phagosome | C3, MSR1, FCGR2A, NCF4, MRC1, CD14, CD36, NOS1, THBS1, TLR4, HLA-DQB1 |
| hsa04151:PI3K-Akt signaling pathway | IFNAR2, IL-6, CSF3R, LAMB4, SPP1, TNC, FN1, THBS1, PRLR, TLR4, PIK3CG |
3.4. The Potential Target Genes Associated with Peri-Implantitis
Sixty-two target genes associated with peri-implantitis were downloaded from the DisGeNET database, and 217 potential target genes were obtained from the OTP. The intersecting genes, including IL-6, TLR4, FN1, IL-1β, CXCL8, MMP-9, and SPP1, were identified using the Venn diagram web tool (Figure 5(a)). The gene-disease network around the peri-implantitis was generated by using the DisGeNET Cytoscape plugin (Figure 5(b)).
Figure 5.
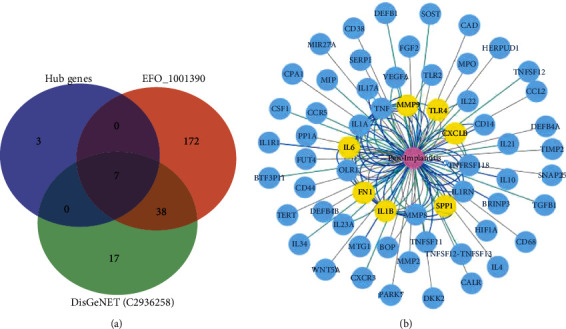
(a) Venn diagram of the intersecting genes of the top 10 hub genes and the associated genes obtained from the DisGeNET database (UMLS: C2936258) and the OTP (EFO: 1001390). (b) The gene-disease networks generated using the DisGeNET Cytoscape plugin, yellow nodes represent the intersecting genes.
4. Discussion
In the present study, bioinformatics methods were used to analyze the critical genes and pathways that were associated with peri-implantitis using the GSE33774 microarray and bioinformatics tools. By using the GSE33774 microarray dataset, Becker et al. found that peri-implantitis and periodontitis show different mRNA signatures [22]. In the present study, we investigated the DEGs between seven gingival tissue samples of peri-implantitis and eight healthy gingival samples by using the GSE33774 dataset. We examined a total of 13,057 DEGs, of which 205 DEGs were considered for further studies and 10 hub genes were identified. Potential disease-related genes were collected from the DisGeNET database and the OTP. The DisGeNET database has been used to study a variety of biomedical issues, and it contains one of the largest publicly available collections of genes and variants related to human diseases for investigating the molecular basis of specific diseases [32]. The OTP provides disease-centric or target-centric workflows that facilitate target selection and validation [33]. The validity was verified by intersecting the hub genes with the potential target genes obtained from the DisGeNET and OTP. Finally, the potential target genes, namely, IL-6, TLR4, FN1, IL-1β, CXCL8, MMP-9, and SPP1, were found to be associated with peri-implantitis, and our results were consistent with those of previous studies [16, 34].
A previous study showed that osteoclastogenesis-related cytokines may be associated with the occurrence and the severity of peri-implantitis [35]. Moreover, the proinflammatory cytokines, such as IL-1β, IL-6, and TNF-α, have been used as biomarkers to diagnose periodontitis and peri-implantitis [11]. Analysis of cytokine levels may help to confirm the early diagnosis of peri-implantitis in high-risk patients. In vitro experiments have shown that the levels of proinflammatory cytokines increase in peri-implantitis, and these levels significantly decrease after clinical treatment [34]. In the present study, our results showed that the proinflammatory cytokines IL-6, IL-1β, and CXCL8 were upregulated in peri-implantitis. The monitoring of TNF-α, CXCL8, and IL-1β levels could be considered as one of the diagnostic elements [36].
For the BP terms, the DEGs were enriched in the immune response and inflammatory response, and this result was consistent with that of previous studies [37]. For the CC terms of GO, the DEGs were enriched in the integral component of the membrane, which included 68 DEGs. In the MF analysis, the DEGs were the most significantly enriched in the immune response, inflammatory response, signal transduction, and cytokine-cytokine receptor interaction pathway. The hub genes play an important role in understanding the biological mechanism of peri-implantitis. The hub genes were closely related to the results of GO and KEGG pathway enrichment analyses as shown in Table 3. Our findings showed that the CXCL8, IL-6, IL-1β, and TLR4 genes are related to immune response (GO:0006955) and inflammatory response (GO:0006954). The CXCL8, IL-6, and IL-1β genes are directly involved in the cytokine-cytokine receptor interaction pathway (hsa04060). The hub gene IL-6 is a cytokine that stimulates immune response and is upregulated in peri-implantitis [37]. SPP1 is also known as OPN, and it is a type of osteoimmunoinflammatory marker related to the inflammation and regulation of cytokine production [17].
Deng et al. [38] found that TLR4 signaling may mediate inflammation and bone resorption in peri-implantitis through the regulation of B cell infiltration, the RANKL/OPG ratio, and differential inflammatory cytokine production. Previous studies have shown that the anti-inflammatory microRNA miR-146a enhances the inhibition of peri-implant bone resorption through the regulation of TLR2/4 signaling [39] and Wnt5a involved in TLR4 signaling induces the production of inflammatory cytokines and causes breakdown of extracellular matrix in peri-implantitis [40]. The microRNAs miR-146a and miR-146b are the most common members of the miR-146 family in periodontitis lesions, and miR-146a may protect gingival tissue from immune-mediated periodontal inflammation [41]. Correspondingly, the KEGG pathway analysis showed that these DEGs were mapped to cytokine-cytokine receptor interaction, pathways in cancer, amoebiasis, phagosome, and the PI3K-Akt signaling pathway, all of which were consistent with the results of previous studies.
Regarding the other target genes, the expression level of FN1 can reflect the progress of periodontitis or peri-implantitis. The mRNA expression of FN1 in the peri-implantitis group was significantly higher than that in the control group [42]. Cellular fibronectin occurs abundantly in the periodontium and may be associated with the state of implants [40]. MMP-9 is involved in the progression of peri-implantitis and is correlated with LOX-1 and the ERK1/2-mediated signaling pathway [43]. However, the regulatory mechanisms of MMP-9 in peri-implantitis need to be well elucidated. The LOX-1/MMP-9 signaling pathway and OPN may be potential drug targets to decrease the levels of proinflammatory cytokines and increase apoptosis in peri-implantitis [37, 43].
Host modulation therapy with anti-inflammatory drugs has been used as a potential method for treating periodontitis [44]. Based on the present literature, many immunoinflammatory molecules can be considered as potential biomarkers for diagnosis of peri-implantitis [45].
5. Conclusion
Our bioinformatics analysis identified 205 DEGs between gingival tissues of peri-implantitis and healthy tissues based on the gene expression datasets obtained from the GEO database, and the potential therapeutic target genes were validated by the analysis of the DisGeNET database and the OTP. We found that IL-6, TLR4, FN1, IL-1β, CXCL8, MMP-9, and SPP1 might be used as potential biomarkers for the diagnosis of peri-implantitis. Further studies are needed to reveal the potential association of these genes with peri-implantitis and to determine potential therapeutic drug targets for peri-implantitis.
Abbreviations
- GEO:
Gene Expression Omnibus
- DEGs:
Differentially expressed genes
- GO:
Gene Ontology
- KEGG:
Kyoto Encyclopedia of Genes and Genomes
- DAVID:
The Database for Annotation, Visualization, and Integrated Discovery
- BP:
Biological process
- MF:
Molecular function
- CC:
Cellular component
- PPI:
Protein-protein interaction
- STRING:
Search Tool for the Retrieval of Interacting Genes Database
- OTP:
The Open Targets Platform
- IL-6:
Interleukin-6
- TNF-α:
Tumor necrosis factor α
- TLR4:
Toll-like receptor 4
- FN1:
Fibronectin 1
- IL-1β:
Interleukin 1 beta
- CXCL8:
C-X-C motif chemokine ligand 8
- MMP-9:
Matrix metallopeptidase-9
- SPP1:
Secreted phosphoprotein 1
- CXCL1:
C-X-C motif chemokine ligand 1
- PECAM1:
Platelet and endothelial cell adhesion molecule 1
- MMP-8:
Matrix metalloproteinase-8.
Data Availability
The datasets analyzed during the current study are available in the Gene Expression Omnibus with the accession GSE33774.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Authors' Contributions
Xiaogen Zhang and Zhifa Wang contributed equally to this work.
References
- 1.Alghamdi H. S., Jansen J. A. The development and future of dental implants. Dental Materials Journal . 2020;39(2):167–172. doi: 10.4012/dmj.2019-140. [DOI] [PubMed] [Google Scholar]
- 2.Schwarz F., Derks J., Monje A., Wang H. L. Peri-implantitis. Journal of Periodontology . 2018;89:S267–S290. doi: 10.1002/JPER.16-0350. [DOI] [PubMed] [Google Scholar]
- 3.Derks J., Tomasi C. Peri-implant health and disease. A systematic review of current epidemiology. Journal of Clinical Periodontology . 2015;42(Suppl 16):S158–S171. doi: 10.1111/jcpe.12334. [DOI] [PubMed] [Google Scholar]
- 4.Salvi G. E., Cosgarea R., Sculean A. Prevalence of periimplant diseases. Implant Dentistry . 2019;28(2):100–102. doi: 10.1097/ID.0000000000000872. [DOI] [PubMed] [Google Scholar]
- 5.Li H., Chen Z., Zhong X., Li J., Li W. Mangiferin alleviates experimental peri-implantitis via suppressing interleukin-6 production and toll-like receptor 2 signaling pathway. Journal of Orthopaedic Surgery and Research . 2019;14(1):p. 325. doi: 10.1186/s13018-019-1387-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lafaurie G. I., Sabogal M. A., Castillo D. M., et al. Microbiome and microbial biofilm profiles of peri-implantitis: a systematic review. Journal of Periodontology . 2017;88(10):1066–1089. doi: 10.1902/jop.2017.170123. [DOI] [PubMed] [Google Scholar]
- 7.Ghassib I., Chen Z., Zhu J., Wang H. L. Use of IL-1 β, IL-6, TNF-α, and MMP-8 biomarkers to distinguish peri-implant diseases: a systematic review and meta-analysis. Clinical Implant Dentistry and Related Research . 2019;21(1):190–207. doi: 10.1111/cid.12694. [DOI] [PubMed] [Google Scholar]
- 8.Goker F., Larsson L., del Fabbro M., Asa’ad F. Gene delivery therapeutics in the treatment of periodontitis and peri-implantitis: a state of the art review. International Journal of Molecular Sciences . 2019;20(14):p. 3551. doi: 10.3390/ijms20143551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sorsa T., Bacigalupo J., Könönen M., Pärnänen P., Räisänen I. T. Host-modulation therapy and chair-side diagnostics in the treatment of peri-implantitis. Biosensors . 2020;10(5):p. 44. doi: 10.3390/bios10050044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duarte P. M., de Mendonça A. C., Máximo M. B. B., Santos V. R., Bastos M. F., Nociti Júnior F. H. Differential cytokine expressions affect the severity of peri-implant disease. Clinical Oral Implants Research . 2009;20(5):514–520. doi: 10.1111/j.1600-0501.2008.01680.x. [DOI] [PubMed] [Google Scholar]
- 11.Alassy H., Parachuru P., Wolff L. Peri-implantitis diagnosis and prognosis using biomarkers in peri-implant crevicular fluid: a narrative review. Diagnostics . 2019;9(4):p. 214. doi: 10.3390/diagnostics9040214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown D. L., Desai K. K., Vakili B. A., Nouneh C., Lee H. M., Golub L. M. Clinical and biochemical results of the metalloproteinase inhibition with subantimicrobial doses of doxycycline to prevent acute coronary syndromes (MIDAS) pilot trial. Arteriosclerosis, Thrombosis, and Vascular Biology . 2004;24(4):733–738. doi: 10.1161/01.ATV.0000121571.78696.dc. [DOI] [PubMed] [Google Scholar]
- 13.Alassiri S., Parnanen P., Rathnayake N., et al. The ability of quantitative, specific, and sensitive point-of-care/chair-side oral fluid immunotests for aMMP-8 to detect periodontal and peri-implant diseases. Disease Markers . 2018;2018:5. doi: 10.1155/2018/1306396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leppilahti J. M., Harjunmaa U., Järnstedt J., et al. Diagnosis of newly delivered mothers for periodontitis with a novel oral-rinse aMMP-8 point-of-care test in a rural Malawian population. Diagnostics . 2018;8(3):p. 67. doi: 10.3390/diagnostics8030067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeh H., Lu J., Chang S., Ge M. C. Identification of microbiota in peri-implantitis pockets by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Scientific Reports . 2019;9(1):p. 774. doi: 10.1038/s41598-018-37450-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He K., Jian F., He T., Tang H., Huang B., Wei N. Analysis of the association of TNF-α, IL-1A, and IL-1B polymorphisms with peri-implantitis in a Chinese non-smoking population. Clinical Oral Investigations . 2020;24(2):693–699. doi: 10.1007/s00784-019-02968-z. [DOI] [PubMed] [Google Scholar]
- 17.Che C., Liu J., Yang J., Ma L., Bai N., Zhang Q. Osteopontin is essential for IL-1β production and apoptosis in peri- implantitis. Clinical Implant Dentistry and Related Research . 2018;20(3):384–392. doi: 10.1111/cid.12592. [DOI] [PubMed] [Google Scholar]
- 18.Udhaya Kumar S., Thirumal Kumar D., Bithia R., et al. Analysis of differentially expressed genes and molecular pathways in familial hypercholesterolemia involved in atherosclerosis: a systematic and bioinformatics approach. Frontiers in Genetics . 2020;11:p. 734. doi: 10.3389/fgene.2020.00734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wan J., Jiang S., Jiang Y., et al. Data mining and expression analysis of differential lncRNA ADAMTS9-AS1 in prostate cancer. Frontiers in Genetics . 2020;10:p. 1377. doi: 10.3389/fgene.2019.01377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu D., Zhang B., Yang L., Huang S., Xin W. Development of an immune-related risk signature for predicting prognosis in lung squamous cell carcinoma. Frontiers in Genetics . 2020;11:p. 978. doi: 10.3389/fgene.2020.00978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tarazona-Álvarez B., López-Roldan A., Vidal-Infer A., López-Padilla D., Alonso-Arroyo A. Bibliometric analysis of the scientific production of literature on peri- implant diseases in the Web of Science. Clinical Implant Dentistry and Related Research . 2021;23(4):625–634. doi: 10.1111/cid.13024. [DOI] [PubMed] [Google Scholar]
- 22.Becker S. T., Beck-Broichsitter B. E., Graetz C., Dörfer C. E., Wiltfang J., Häsler R. Peri-implantitis versus periodontitis: functional differences indicated by transcriptome profiling. Clinical Implant Dentistry and Related Research . 2014;16(3):401–411. doi: 10.1111/cid.12001. [DOI] [PubMed] [Google Scholar]
- 23.Ashburner M., Ball C. A., Blake J. A., et al. Gene Ontology: tool for the unification of biology. Nature Genetics . 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edgar R., Domrachev M., Lash A. E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Research . 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barrett T., Troup D. B., Wilhite S. E., et al. NCBI GEO: mining tens of millions of expression profiles--database and tools update. Nucleic Acids Research . 2007;35(Database):D760–D765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang D. W., Sherman B. T., Tan Q., et al. DAVID bioinformatics resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Research . 2007;35(suppl_2):W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang D. W., Sherman B. T., Tan Q., et al. The DAVID gene functional classification tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biology . 2007;8(9):p. R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Y., Zhou B., Pache L., et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Communications . 2019;10(1) doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szklarczyk D., Gable A. L., Lyon D., et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Research . 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smoot M. E., Ono K., Ruscheinski J., Wang P. L., Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics . 2011;27(3):431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maere S., Heymans K., Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics . 2005;21(16):3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 32.Piñero J., Bravo À., Queralt-Rosinach N., et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Research . 2017;45(D1):D833–D839. doi: 10.1093/nar/gkw943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ochoa D., Hercules A., Carmona M., et al. Open Targets Platform: supporting systematic drug-target identification and prioritisation. Nucleic Acids Research . 2021;49(D1):D1302–D1310. doi: 10.1093/nar/gkaa1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Renvert S., Widen C., Persson R. G. Cytokine and microbial profiles in relation to the clinical outcome following treatment of peri-implantitis. Clinical Oral Implants Research . 2017;28(9):1127–1132. doi: 10.1111/clr.12927. [DOI] [PubMed] [Google Scholar]
- 35.al-Sowygh Z. H., Aldamkh M. K., Binmahfooz A. M., et al. Assessment of matrix metalloproteinase-8 and -9 levels in the peri-implant sulcular fluid among waterpipe (narghile) smokers and never-smokers with peri-implantitis. Inhalation Toxicology . 2018;30(2):72–77. doi: 10.1080/08958378.2018.1449273. [DOI] [PubMed] [Google Scholar]
- 36.Aleksandrowicz P., Brzezińska-Błaszczyk E., Kozłowska E., Żelechowska P., Borgonovo A. E., Agier J. Analysis of IL-1β, CXCL8, and TNF-α levels in the crevicular fluid of patients with periodontitis or healthy implants. BMC Oral Health . 2021;21(1):p. 120. doi: 10.1186/s12903-021-01478-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baseri M., Radmand F., Hamedi R., Yousefi M., Kafil H. S. Immunological aspects of dental implant rejection. BioMed Research International . 2020;2020 doi: 10.1155/2020/7279509.7279509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deng S., Hu Y., Zhou J., et al. TLR4 mediates alveolar bone resorption in experimental peri-implantitis through regulation of CD45+cell infiltration, RANKL/OPG ratio, and inflammatory cytokine production. Journal of Periodontology . 2020;91(5):671–682. doi: 10.1002/JPER.18-0748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan K., Hu Y., Wang Y., et al. RANKL blockade alleviates peri-implant bone loss and is enhanced by anti-inflammatory microRNA-146a through TLR2/4 signaling. International journal of implant dentistry . 2020;6(1):p. 15. doi: 10.1186/s40729-020-00210-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Q., Liu J., Ma L., Bai N., Xu H. Wnt5a is involved in LOX-1 and TLR4 induced host inflammatory response in peri-implantitis. Journal of Periodontal Research . 2020;55(2):199–208. doi: 10.1111/jre.12702. [DOI] [PubMed] [Google Scholar]
- 41.Yuan Y., Zhang H., Huang H. MicroRNAs in inflammatory alveolar bone defect: a review. Journal of Periodontal Research . 2021;56(2):219–225. doi: 10.1111/jre.12819. [DOI] [PubMed] [Google Scholar]
- 42.Wu Y. Y., Cao H. H., Kang N., Gong P., Ou G. M. Expression of cellular fibronectin mRNA in adult periodontitis and peri- implantitis: a real-time polymerase chain reaction study. International Journal of Oral Science . 2013;5(4):212–216. doi: 10.1038/ijos.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Q., Xu H., Bai N., Tan F., Xu H., Liu J. Matrix metalloproteinase 9 is regulated by LOX-1 and erk1/2 pathway in dental peri-implantitis. Current Pharmaceutical Biotechnology . 2020;21(9):862–871. doi: 10.2174/1389201021666200221121139. [DOI] [PubMed] [Google Scholar]
- 44.Preshaw P. M. Host modulation therapy with anti-inflammatory agents. Periodontology 2000 . 2018;76(1):131–149. doi: 10.1111/prd.12148. [DOI] [PubMed] [Google Scholar]
- 45.Corrêa M. G., Pimentel S. P., Ribeiro F. V., Cirano F. R., Casati M. Z. Host response and peri-implantitis. Brazilian Oral Research . 2019;33, article e66(suppl 1):p. e066. doi: 10.1590/1807-3107bor-2019.vol33.0066. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analyzed during the current study are available in the Gene Expression Omnibus with the accession GSE33774.


