Abstract
Background
Accumulating studies indicated that dysregulated long non-coding RNA human histocompatibility leukocyte antigen (HLA) Complex P5 (HCP5) may functions as an potential prognostic predictor in multiple cancers. This meta-analysis was performed to systematically collect studies and conduct an evidence-based evaluation of the prognostic role of HCP5 in malignancies.
Methods
Four databases (PubMed, Web of Science, Embase and Cochrane library) were comprehensively retrieved from their initiation date to November 9, 2021. Hazard ratio (HR) or odds ratio (OR) with 95% confidence interval (CI) were used to assess the associations between the expression level of HCP5 and prognosis or clinical characteristics. Moreover, results were validated by Gene Expression Profiling Interactive Analysis 2 (GEPIA2) and the National Genomics Data Center (NGDC). Subsequently, the molecular mechanism of HCP5 was predicted based on MEM and StarBase databases. The study protocol was registered at PROSPERO (ID: CRD42021274208).
Results
9 studies, containing 641 patients, were included in this meta-analysis. Our results revealed that HCP5 overexpression was associated with poor overall survival (OS), tumor type, histological differentiation, and lymph node metastasis in most cancers, but was not associated with age, gender and tumor size; down-regulation of HCP5 was associated with worse OS, advanced tumor stage, positive distal metastasis and lymph node metastasis in skin cutaneous melanoma (SKCM). HCP5 was significantly up-regulated in four cancers and down-regulated in SKCM, which was validated by the GEPIA2 cohort. HCP5 expression in various types of cancer was also verified in NGDC. Further functional prediction revealed that HCP5 may participate in some cancer-related pathways.
Conclusion
There is a significantly association between dysregulation of HCP5 and both prognosis and clinicopathological features in various cancers. HCP5 may be functions as a novel potential prognostic biomarker and therapeutic target in multiple human cancers.
Keywords: lncRNA HCP5, Meta-analysis, Cancer, Prognosis, Bioinformatics
Introduction
Cancer is the leading cause of death in every country of the world and an important barrier to extending life expectancy [1]. According to the latest global cancer statistics reported in CA cancer journals, an estimated 19.3 million new cancer cases and almost 10.0 million cancer deaths occurred worldwide in 2020 [2]. Despite increasing number of treatment methods for cancer in recent years, the overall prognosis of most cancers remains poor; one of the major causes for this is lack of sensitive and specific biomarker for tumor early diagnosis, most patients are already at an advanced stage when they are initially diagnosed [3]. Early diagnosis and treatment are important to improve the prognosis in cancer patients, therefore, it is of great clinical significance to search novel biomarkers and therapeutic targets of cancer.
Mutations in the non-coding genome were considered to be a major determinant of cancer. Long non-coding RNAs (lncRNAs) are a type of non-coding RNAs with more than 200 nucleotides in length and transcribed by RNA polymerase II [4]. Increasing studies had confirmed that lncRNAs are dysregulated in different cancers, which regulated the progression of malignancies by function as oncogenes or tumor suppressor [5]. Particularly, some lncRNAs have been reported to be markedly associated with the clinicopathological characteristics of cancer patients and may serve as potential therapeutic targets or prognostic biomarkers to predict the clinical outcomes [6, 7].
LncRNA human histocompatibility leukocyte antigen (HLA), complex P5 (HCP5), is primarily found expressed in immune system cells, participating in adaptive and innate immune responses [8, 9]. Recently, several studies proposed that HCP5 could functions as a competitive endogenous RNA (ceRNA) affect the distribution of microRNAs on their targets, thereby participating in the occurrence and progression of tumors. For example, HCP5 expression level was increased in anaplastic thyroid cancer cell lines, HCP5 knockdown inhibited the cell viability and induced apoptosis via sponging miR-128-3p [10]. Furthermore, HCP5 functions as a ceRNA to sponge miR-29b-3p, miR-140-5p, miR-139-5p, and miR-186-5p, which consequently promotes cell growth, metastasis, invasion and epithelial–mesenchymal transition in hepatocellular carcinoma, clear cell renal cell carcinoma, colorectal cancer, or gastric cancer [9, 11–13]. Changes in ceRNA regulation may affect the expression of oncogenes or tumor suppressors, which might provide a potential therapeutic target for cancers [14].
Previous studies have described HCP5 as a cancer promoter in most malignancies, while the expression level of HCP5 has also been found to be downregulated in some tumor tissues. For example, one study found that HCP5 level was decreased in skin cutaneous melanoma, suggesting HCP5 could function as a tumor suppressor and suppresses melanoma development by regulating RARRES3 gene expression via sponging miR-1286 [15]. Clinically, most evidences have showed that HCP5 overexpression is associated with the clinicopathological characteristics and prognosis of various cancers, including gastric cancer [16, 17], non-small cell lung cancer [18], renal cell carcinoma [19] and osteosarcoma [20]. Meanwhile, only one study reported that downregulation of HCP5 was associated with a poor prognosis, advanced tumor stage, positive distal metastasis and lymph node metastasis [15].
Collectively, most studies suggested that HCP5 is more frequently acting as an tumor oncogene in various malignancies, and associated with the prognosis and clinicopathological characteristics. However, results of these previous studies were controversial due to the limitations of the small sample size of single study, the prognostic value of lncRNA HCP5 remains unclear. Therefore, we systematically selected relevant literatures and performed this meta-analysis to conduct an evidence-based evaluation of the prognostic role of lncRNA HCP5 in various cancers.
Materials and methods
The Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines was used to guide this meta-analysis. This meta-analysis has been registered with PROSPERO (ID: CRD42021274208).
Literature search and selection
PubMed, Web of Science, Embase and Cochrane library from their initiation date to November 9, 2021 were searched by two investigators (Shaopu Hu and Mengxue Ge). All covered literatures that evaluated the clinicopathological and prognostic value of HCP5 in cancer patients, without language limitation, were collected. The terms of (((((Tumour) OR (tumor)) OR (neoplasms)) OR (cancer)) OR (carcinoma)) AND ((((long non coding RNA HCP5) OR (lncRNA HCP5)) OR (Human histocompatibility leukocyte antigen (HLA) complex P5)) OR (HCP5)) were used as the search strategy.
Inclusion and exclusion criteria
Inclusion criteria were as the following: (1) studies evaluated the expression level of HCP5 in cancers; (2) patients were divided into low HCP5 expression group and high HCP5 expression group; (3) hazard ratios (HR) and 95% confidence interval (CI) for prognostic indicators were described or could be indirectly calculated according to the survival curves; (4) the relationship between HCP5 and prognosis or clinicopathological features was reported.
Exclusion criteria were as the following: (1) studies evaluated a group of lncRNAs rather than a single HCP5; (2) non-clinical study, editorials, letters, expert opinions, reviews and case reports; (3) necessary data cannot be extracted; (4) prognosis or clinicopathological data from bioinformatics analysis.
Data extraction and quality assessment
The data of included studies were extracted independently by two reviewers (SH and MG). If there were any divergences, a discussion was performed with another investigator (Kaiwen Hu). The following information were extracted from each study: first author, publication year, region, sample size, tumor type, detection method, outcome, follow-up months, HRs and 95% CIs for OS. We followed the methods of Hu et al. [21]. The quality of included studies was assessed independently by two reviewers (SH and MG). Any divergences were resolved through discussion with a third investigator. The Newcastle–Ottawa Scale (NOS) was applied to assess the quality of all studies. The total scores for different studies ranged from 0 to 9. The study was considered to be high quality if the score was > 6 [22].
Validation of bioinformatics database
Gene Expression Profiling Interactive Analysis 2 (GEPIA2, http://gepia2.cancer-pku.cn/), which is based on RNA sequencing expression data from the The Cancer Genome Atlas (TCGA) and the GTEx projects [23], was utilized to further verify the expression level and survival analysis of HCP5 in various cancers. One-way ANOVA was used for differential expression analysis. Kaplan–Meier method and log-rank test were used for survival analysis [24]. National Genomics Data Center (NGDC, https://bigd.big.ac.cn) was utilized to further verify the expression level of HCP5 in various cancer types [25].
Prediction the functions and pathways of HCP5
MEM (http://biit.cs.ut.ee/mem/index.cgi) was used to predict the target genes of HCP5 based on Affymetrix Gene Chip Human Genome U133 Plus 2.0 Array platform. Then, Database for Annotation, visualization and Integrated Discovery (DAVID, https://david.ncifcrf.gov/) was applied to perform GO and KEGG analyses based on the target genes. Moreover, StarBase (http://starbase.sysu.edu.cn/) was utilized to predict the miRNA-HCP5 interactions, with was supported by ago CLIP-seq data. Finally, Cytoscape software (version 3.6.0, https://cytoscape.org/) was used to construct the network of HCP5-target genes and miRNA-HCP5 interactions.
Statistical analysis
Statistical analysis in this meta-analysis were performed by REVIEW MANAGER 5.3 software, which followed the methods of Hu et al. 2018 [21]. Based on the reported Kaplan–Meier curve in each included study, the HRs and 95% CIs were estimated using the software of Engauge Digitizer 10.0 [21]. The survival results were calculated by log HR and standard error (SE) values. Moreover, the association between HCP5 expression levels and the tumor clinicopathological parameters (age, gender, differentiation, LNM, tumor size and TNM stage) were evaluated by ORs and 95% CIs. The heterogeneity of the eligible studies was evaluated by the Q and I2 test. The application of effects model depends on the heterogeneity among the studies. If no significant heterogeneity (I2 < 50%, P > 0.1) in the included studies, fixed-effects model was used to analyze the results; while random-effects model was applied for meta-analysis if significant heterogeneity (I2 ≥ 50%, P ≤ 0.1) existed in the eligible studies [21]. Funnel plot was used to evaluate the potential publication bias.
Results
Literature screening
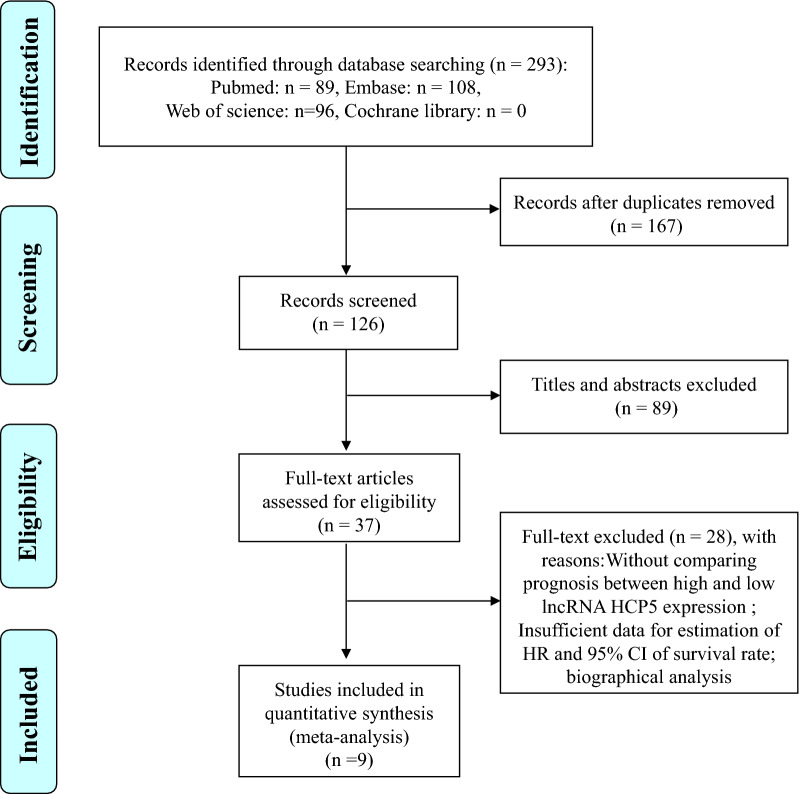
The detailed process of the literature identification and selection was presented in Fig. 1. Initially, a total of 293 publications were retrieved by the search strategy, while 167 of the duplicated articles were excluded. After reviewing the titles and abstracts, 89 literatures were removed because of not cancer related articles or not original research works (reviews, case reports, letters and editorials). After checking the full text of remaining articles, 28 studies were further excluded due to the reason of these studies without comparing the prognosis between high and low HCP5 expression or providing insufficient data for estimation of HR and 95% CI for survival rate or data from biographical analysis. Finally, nine literatures [13, 15–20, 26, 27] coincided with the inclusion criteria were included in the present meta-analysis.
Fig. 1.
Flow diagram of process for the literature identification and selection
Study characteristics and quality assessment
The primary characteristics of nine included studies were shown in Table 1. All studies were from China and enrolled a total of 641 patients. The types of cancer in these studies included: non-small cell lung cancer, colorectal cancer, oral squamous cell carcinoma, clear cell renal cell carcinoma, gastric cancer, skin cutaneous melanoma and osteosarcoma. All included studies [13, 15–20, 26, 27] evaluated the association between HCP5 expression levels and overall survival, the follow-up months ranged from 36 to 105. Five studies [13, 15, 16, 19, 26] reported the clinicopathological parameters. The method of all studies to measure the expression of HCP5 was real-time quantitative polymerase chain reaction. The NOS outcome showed all scores were ≥ 6, which suggested that these studies displayed a medium-high quality.
Table 1.
Characteristics of the included studies
| Study | Year | Region | Tumor type | Sample size (low/high) | HR (95% CI) | Outcome | Method | Follow-up months | NOS |
|---|---|---|---|---|---|---|---|---|---|
| Li [18] | 2020 | China | NSCLC | 63 (31/32) | 0.44 (0.23, 0.84) | OS | qRT-PCR | 60 | 7 |
| Yang [13] | 2019 | China | CRC | 135 (43/92) | 0.36 (0.16, 0.81) | CP, OS | qRT-PCR | 60 | 7 |
| Zhao [26] | 2019 | China | OSCC | 73 (37/36) | 0.63 (0.35, 1.13) | CP, OS | qRT-PCR | 60 | 7 |
| Hao [19] | 2020 | China | ccRCC | 66 (33/33) | 0.55 (0.28, 1.08) | CP, OS | qRT-PCR | 60 | 7 |
| Liang [17] | 2021 | China | GC | 36 (18/18) | 0.47 (0.24, 0.92) | OS | qRT-PCR | 60 | 6 |
| Qin [16] | 2021 | China | GC | 98 (49/49) | 0.36 (0.13, 1.00) | CP, OS | qRT-PCR | 36 | 6 |
| Zhang [27] | 2020 | China | ccRCC | 76 (38/38) | 0.33 (0.09, 1.19) | OS | qRT-PCR | 60 | 7 |
| Tu [20] | 2021 | China | OSC | 40 (21/19) | 0.33 (0.09, 1.21) | OS | qRT-PCR | 60 | 6 |
| Wei [15] | 2019 | China | SKCM | 54 (27/27) | 2.97 (1.64, 5.36) | CP, OS | qRT-PCR | 105 | 7 |
NSCLC non-small cell lung cancer, CRC colorectal cancer, OSCC Oral squamous cell carcinoma, ccRCC clear cell renal cell carcinoma, GC gastric cancer, SKCM skin cutaneous melanoma, OSC osteosarcoma, CP clinicopathological parameters, OS overall survival, HR hazard ratios, CI confidence intervals, NA not available, NOS Newcastle–Ottawa Scale
Association between HCP5 expression levels and OS
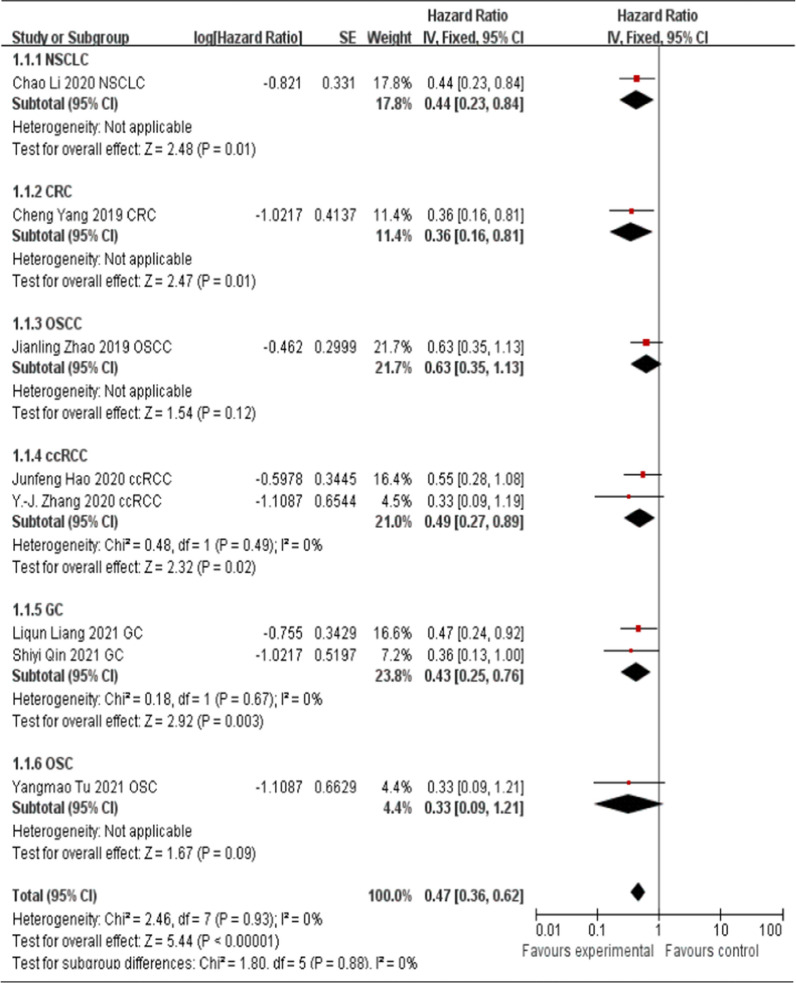
There were nine studies evaluated the association between HCP5 expression levels and OS in this meta-analysis. Eight [13, 16–20, 26, 27] of them reported that HCP5 was up-regulated in cancer tissues and function as the oncogenes. Therefore, these eight studies were pooled for analysis. As shown in Fig. 2, fixed-effects model was used (I2 = 0%, P = 0.93). The pooled HR = 0.47 (95% CI 0.36–0.62, P < 0.00001), which indicated that higher HCP5 expression in the tumor tissues of patients with non-small cell lung cancer, colorectal cancer, oral squamous cell carcinoma, clear cell renal cell carcinoma, gastric cancer and osteosarcoma were associated with a poor OS. Only one study [15] reported that HCP5 function as the tumor suppressor in skin cutaneous melanoma, down-regulation of HCP5 was associated with a poor OS (HR = 2.97, 95% CI 1.64–5.36), advanced tumor stage, positive distal metastasis and lymph node metastasis. Therefore, the data of this study could not be pooled for follow-up analysis.
Fig. 2.
Forest plot for the association between HCP5 expression levels and OS
Association between HCP5 expression levels and tumor types
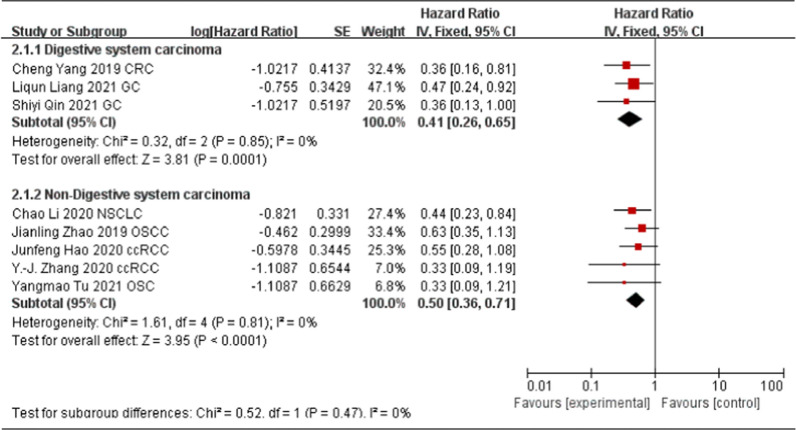
To further explore the potential prognostic value of HCP5, we evaluated the association between up-regulated HCP5 and tumor types. As shown in Fig. 3, three studies [13, 16, 17] explored the digestive system carcinoma and five studies [18–20, 26, 27] explored the non-digestive system carcinoma. The results of the forest plot suggested that regardless of the digestive system carcinoma or non-digestive system carcinoma, up-regulated HCP5 expression in tumor tissues was correlated with poor OS (digestive system carcinoma, HR = 0.41; 95% CI 0.26–0.65, P = 0.0001; non-digestive system carcinoma, HR = 0.50; 95% CI 0.36–0.71, P < 0.0001, Fig. 3).
Fig. 3.
Forest plot for the association between HCP5 expression levels and tumor type
Associations between HCP5 expression levels and clinicopathological parameters
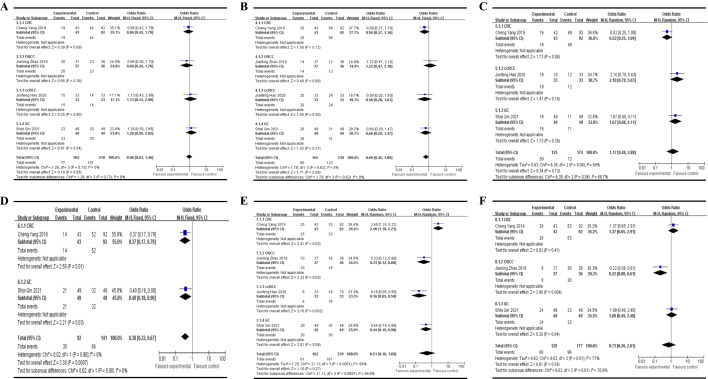
There were five studies evaluated the association between HCP5 expression levels and clinicopathological parameters in this meta-analysis. One study [15] reported HCP5 function as the tumor suppressor in skin cutaneous melanoma has been described in the above part, we conducted further meta-analysis for other five studies. As shown in Table 2, there were no statistically significant relationships between HCP5 overexpression and age (total OR = 0.96, 95% CI 0.63–1.46, P = 0.85, Fixed model, Fig. 4A), gender (total OR = 0.69, 95% CI 0.45–1.06, P = 0.09, Fixed model, Fig. 4B) and tumor size (total OR = 1.17, 95% CI 0.48–2.88, P = 0.73, Random model, Fig. 4C); However, HCP5 overexpression was statistically correlated with poor histological differentiation in CRC and GC (total OR = 0.38, 95% CI 0.22–0.67, P = 0.0007, fixed model, Fig. 4D), positive lymph node metastasis in CRC (OR = 2.48, 95% CI 1.18–5.21, P = 0.02), OSCC (OR = 0.33, 95% CI 0.12–0.88, P = 0.03), ccRCC (OR = 0.16, 95% CI 0.05–0.50, P = 0.002), and GC (OR = 0.44, 95% CI 0.19–0.98, P = 0.04) (Fig. 4E), and advanced TNM stage in OSCC (OR = 0.22, 95% CI 0.08–0.61, P = 0.004, Fig. 4F).
Table 2.
Meta-analysis results for the association between HCP5 expression and clinicopathological characteristics
| Characteristics | Cancer type | OR (95% CI) | P value | I2% | Model |
|---|---|---|---|---|---|
| Age (old vs young) | CRC | 0.86 (0.42, 1.79) | 0.69 | ||
| OSCC | 0.66 (0.26, 1.70) | 0.39 | |||
| ccRCC | 1.13 (0.43, 2.99) | 0.80 | |||
| GC | 1.28 (0.58, 2.85) | 0.54 | |||
| Total | 0.96 (0.63, 1.46) | 0.85 | 0 | Fixed | |
| Gender (male vs female) | CRC | 0.56 (0.27, 1.16) | 0.12 | ||
| OSCC | 1.22 (0.47, 3.18) | 0.69 | |||
| ccRCC | 0.58 (0.20, 1.63) | 0.30 | |||
| GC | 0.66 (0.29, 1.47) | 0.31 | |||
| Total | 0.69 (0.45, 1.06) | 0.09 | 0 | Fixed | |
| Tumor size (large vs small) | CRC | 0.52 (0.25, 1.09) | 0.08 | ||
| ccRCC | 2.10 (0.78, 5.63) | 0.14 | |||
| GC | 1.67 (0.68, 4.11) | 0.26 | |||
| Total | 1.17 (0.48, 2.88) | 0.73 | 69 | Random | |
| Differentiation (poor vs well) | CRC | 0.37 (0.17, 0.79) | 0.01* | ||
| GC | 0.40 (0.18, 0.90) | 0.03* | |||
| Total | 0.38 (0.22, 0.67) | 0.0007* | 0 | Fixed | |
| Lymph node metastasis (yes vs no) | CRC | 2.48 (1.18, 5.21) | 0.02* | ||
| OSCC | 0.33 (0.12, 0.88) | 0.03* | |||
| ccRCC | 0.16 (0.05, 0.50) | 0.002* | |||
| GC | 0.44 (0.19, 0.98) | 0.04* | |||
| Total | 0.51 (0.16, 1.69) | 0.27 | 86 | Random | |
| TNM stage (III–IV vs I–II) | CRC | 1.37 (0.65, 2.91) | 0.41 | ||
| OSCC | 0.22 (0.08, 0.61) | 0.004* | |||
| GC | 1.09 (0.49, 2.40) | 0.84 | |||
| Total | 0.73 (0.26, 2.01) | 0.54 | 77 | Random |
OR odds ratio, CI confidence interval, CRC colorectal cancer, OSCC Oral squamous cell carcinoma, ccRCC clear cell renal cell carcinoma, GC gastric cancer
*P < 0.05
Fig. 4.
Forest plot for the association between HCP5 expression levels and age (A), gender (B), tumor size (C), tumor differentiation (D), Lymph node metastasis (E) and TNM stage (F)
Publication bias
A funnel plot has been made to analyze whether our results were influenced by the potential publication bias. Studies reported the association between up-regulated HCP5 expression levels and OS were chosen for analysis, as most of the studies measured it. A total of eight studies [13, 16–20, 26, 27] were analyzed, the results showed that there was no obvious publication bias in the research (Fig. 5).
Fig. 5.
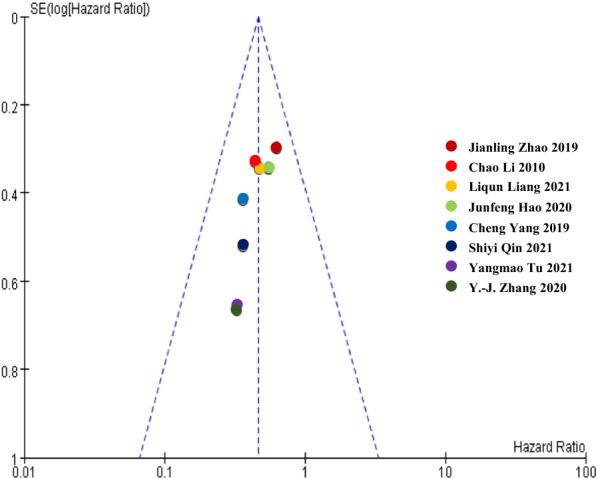
Funnel plots for the association between HCP5 expression levels and OS
Validation results of HCP5 in public databases
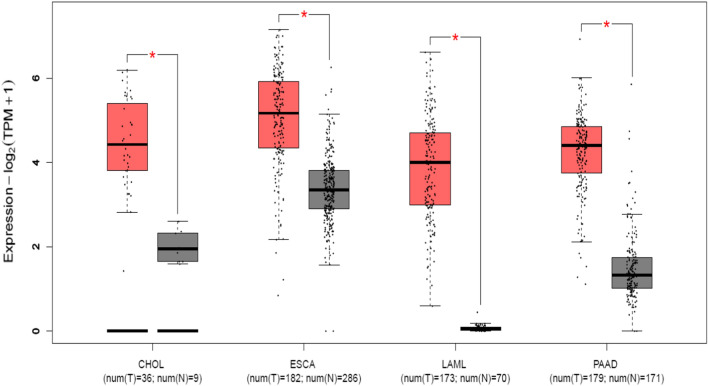
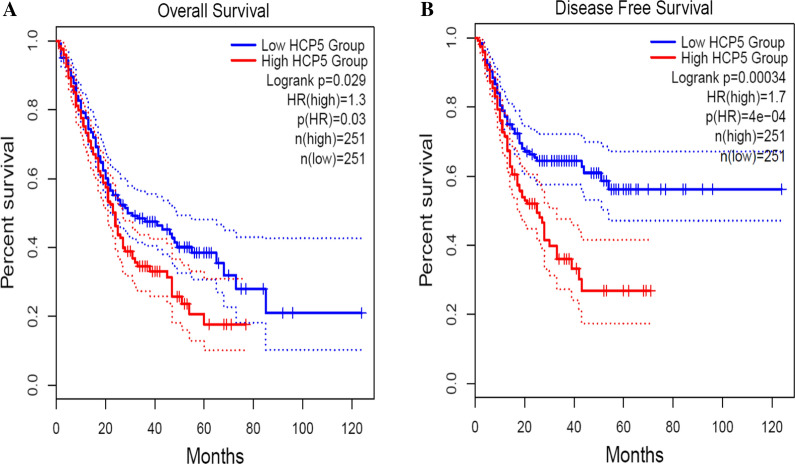
To further verify our results, we used GEPIA 2 to evaluate the expression level and survival analysis of HCP5 in various cancers. The results indicated that HCP5 was up-regulated in most cancers, including cholangio carcinoma (CHOL), esophageal carcinoma (ESCA), acute myeloid leukemia (LAML) and pancreatic adenocarcinoma (PAAD) (|log2FC| Cutoff: 1, P-value Cutoff: 0.01, Fig. 6). Moreover, we merged the expression and prognosis data for four type of cancer, including CHOL, ESCA, LAML and PAAD. As shown in Fig. 7, 502 patients were grouped into high (n = 251) and low (n = 251) HCP5 level groups with the median expression level of HCP5 in four cancer types as the cutoff value. The HCP5 high expression group had shorter OS and DFS than the HCP5 low expression group, confirming that up-regulated HCP5 is correlated with poor OS in most human cancers.
Fig. 6.
The expression levels of HCP5 in four types of cancer tissues and normal tissues in GEPIA2 cohort. Red box plots, HCP5 expression level in cancer tissues; grey box plots, HCP5 expression level in normal tissues; *log2FC| > 1 and P < 0.01
Fig. 7.
Overall survival plots (A) and Disease free survival plots (B) of HCP5 in GEPIA2 cohort, including CHOL, ESCA, LAML and PAAD (n = 502)
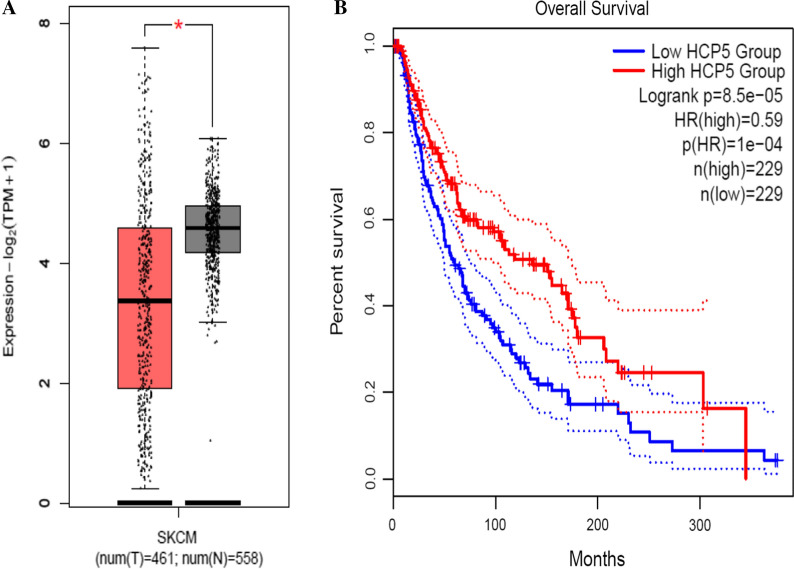
Meanwhile, we evaluated the expression level and survival analysis of HCP5 in skin cutaneous melanoma (SKCM). As shown in Fig. 8, HCP5 was down-regulated in SKCM, HCP5 high expression group had longer OS than the HCP5 low expression group. This result is consistent with the reported literature, confirming that down-regulated HCP5 is correlated with well OS in SKCM.
Fig. 8.
The expression levels (A) and Overall survival plots (B) of HCP5 in SKCM in GEPIA2 cohort
NGDC was used to further validate the expression of HCP5 in various cancers. Ten types of cancer (breast cancer, cervical cancer, colon cancer, esophageal tumor, gastric cancer, liver tumor, serous ovarian cancer, bladder cancer, ovarian cancer and prostate cancer) with 27 datasets were collected. As shown in Table 3, HCP5 was more frequently over-expressed in multiple cancer tissues compared with these normal tissues, and the differences were statistically significant (P < 0.05). Regrettably, the expression of HCP5 in SKCM could not be further validated via NGDC since lack SKCM related datasets.
Table 3.
Differential expression of HCP5 in various cancers
| Tumor Type | GSE ID | Sample size | Expression | P-value | |
|---|---|---|---|---|---|
| Tumor | Normal | ||||
| Breast cancer | GSE42568 | 104 | 17 | High | 0.0004 |
| Triple-negative breast cancer | GSE76250 | 165 | 33 | High | 0.0001 |
| GSE65194 | 55 | 11 | High | 0.0037 | |
| GSE65212 | 55 | 11 | High | 0.0037 | |
| Cervical cancer | GSE29570 | 45 | 17 | High | 0.0000 |
| GSE63678 | 5 | 5 | High | 0.0007 | |
| GSE67522 | 20 | 22 | High | 0.0270 | |
| GSE39001 | 43 | 12 | High | 0.0376 | |
| Colon cancer | GSE28000 | 81 | 34 | High | 0.0152 |
| GSE84984 | 7 | 6 | High | 0.0152 | |
| Esophageal tumor | GSE29001 | 21 | 24 | High | 0.0000 |
| GSE5364 | 16 | 13 | High | 0.0053 | |
| Gastric cancer | GSE63089 | 45 | 45 | High | 0.0002 |
| GSE49515 | 3 | 10 | High | 0.0004 | |
| GSE36076 | 3 | 9 | High | 0.0006 | |
| GSE58828 | 3 | 3 | High | 0.0040 | |
| GSE56807 | 5 | 5 | High | 0.0051 | |
| Liver tumor | GSE14520 | 225 | 220 | High | 0.0000 |
| GSE11819 | 8 | 8 | High | 0.0000 | |
| Serous ovarian cancer | GSE36668 | 4 | 4 | High | 0.0016 |
| GSE16708 | 17 | 9 | High | 0.0026 | |
| Bladder cancer | GSE89006 | 4 | 4 | Low | 0.0000 |
| Ovarian cancer | GSE52037 | 10 | 10 | Low | 0.0001 |
| GSE18520 | 53 | 10 | Low | 0.0003 | |
| Prostate cancer | GSE29079 | 47 | 48 | Low | 0.0000 |
| GSE38241 | 18 | 21 | Low | 0.0000 | |
| GSE17951 | 109 | 45 | Low | 0.0006 | |
Prediction results of the functions and pathways of HCP5
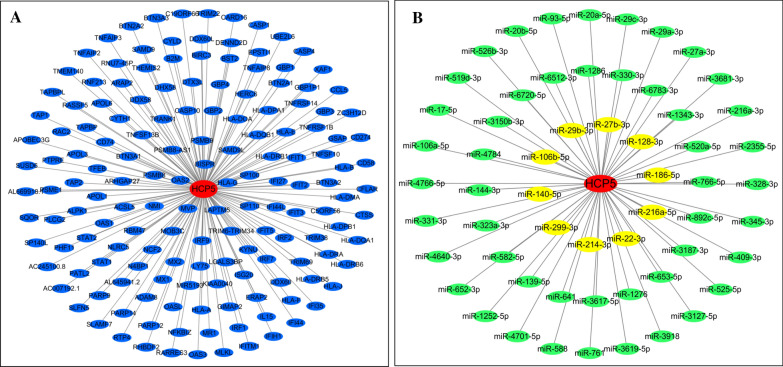
Based on MEM database, a total of 157 target genes of HCP5 were selected and collected to perform the GO and KEGG pathway analyses (Fig. 9A). The most strongly enriched GO terms were identified as follows: immune response, defense response to virus, peptide antigen binding and integral component of lumenal side of endoplasmic reticulum membrane (Table 4). Besides, the KEGG pathway analysis result confirmed that these co-expressed genes were significantly involved in Cell adhesion molecules (CAMs), TNF signaling pathway, NF-kappa B signaling pathway, Apoptosis and Viral carcinogenesis (Table 5). Altogether, these results revealed that HCP5 main participate in the biological mechanism of cancer and autoimmune disease. Moreover, a total of 58 miRNAs were predicted via StarBase, which may exist interactions with HCP5 (Fig. 9B). Ten miRNAs (miR-27b-3p, miR-29b-3p, miR-128-3p, miR-106b-5p, miR-186-5p, miR-140-5p, miR-216a-5p, miR-299-3p, miR-22-3p and miR-213-3p) have been reported, additional studies are needed to explore the potential mechanisms of other miRNAs-HCP5 interactions.
Fig. 9.
Network analysis between HCP5 and target genes (A), miRNA-HCP5 interactions (B). Red circle, lncRNA HCP5; Blue circle, target genes of HCP5; Green circle, miRNAs that interacts with HCP5; Yellow circle, miRNAs that interact with HCP5 have been validated
Table 4.
Top five enrichment GO terms (BP, CC and MF) of the potential genes of HCP5
| GO ID | Term | Ontology | Count | P-value |
|---|---|---|---|---|
| GO:0060337 | Type I interferon signaling pathway | BP | 29 | 3.90E−43 |
| GO:0060333 | Interferon-gamma-mediated signaling pathway | BP | 28 | 1.57E−39 |
| GO:0051607 | Defense response to virus | BP | 26 | 1.89E−25 |
| GO:0006955 | Immune response | BP | 31 | 1.04E−20 |
| GO:0009615 | Response to virus | BP | 20 | 1.20E−20 |
| GO:0042605 | Peptide antigen binding | MF | 13 | 1.12E−18 |
| GO:0032395 | MHC class II receptor activity | MF | 8 | 1.09E−11 |
| GO:0003725 | Double-stranded RNA binding | MF | 8 | 5.45E−07 |
| GO:0046978 | TAP1 binding | MF | 4 | 1.99E−06 |
| GO:0001730 | 2ʹ-5ʹ-oligoadenylate synthetase activity | MF | 4 | 1.99E−06 |
| GO:0071556 | Integral component of lumenal side of endoplasmic reticulum membrane | CC | 14 | 9.17E−21 |
| GO:0012507 | ER to Golgi transport vesicle membrane | CC | 14 | 7.32E−17 |
| GO:0042613 | MHC class II protein complex | CC | 11 | 2.64E−16 |
| GO:0042612 | MHC class I protein complex | CC | 8 | 3.94E−13 |
| GO:0030658 | Transport vesicle membrane | CC | 8 | 1.27E−08 |
GO Gene Ontology, BP biological process, CC cellular component, MF molecular function
Table 5.
KEGG pathway enrichment analysis of the potential genes of HCP5
| KEGG ID | Term | Count | P value | Genes |
|---|---|---|---|---|
| hsa04145 | Phagosome | 18 | 9.74E−15 | HLA-DRB5, NCF2, HLA-B, TAP2, TAP1, HLA-A, HLA-F, HLA-G, CTSS, HLA-DMA, HLA-DPB1, HLA-DRA, HLA-DOA, HLA-DQA1, HLA-DRB1, HLA-DPA1, HLA-DQB1, HLA-E |
| hsa04514 | Cell adhesion molecules (CAMs) | 16 | 1.31E−12 | CD274, HLA-DRB5, HLA-B, HLA-A, HLA-F, HLA-G, HLA-E, HLA-DMA, HLA-DPB1, HLA-DRA, CD58, HLA-DOA, HLA-DQA1, HLA-DRB1, HLA-DPA1, HLA-DQB1 |
| hsa04668 | TNF signaling pathway | 8 | 4.89E−05 | MLKL, IL15, CASP10, CCL5, TNFAIP3,CFLAR, TNFRSF1B, BIRC3 |
| hsa04064 | NF-kappa B signaling pathway | 6 | 0.001138 | DDX58, PLCG2, TNFAIP3, CFLAR,TNFSF13B, BIRC3 |
| hsa05203 | Viral carcinogenesis | 8 | 0.002532 | SP100, IRF7, HLA-B, HLA-A, HLA-F,HLA-G, IRF9, HLA-E |
| hsa04210 | Apoptosis | 4 | 0.018872 | CASP10, TNFSF10, CFLAR, BIRC3 |
| hsa04144 | Endocytosis | 7 | 0.022316 | HLA-B, ARAP2, HLA-A, HLA-F, HLA-G,CYTH1, HLA-E |
KEGG Kyoto Encyclopedia of Genes and Genomes
Functional mechanisms of HCP5 in various cancers
The known functional mechanisms of HCP5 in various cancers were summarized in Table 6. Briefly elaborated as follows:
In thyroid carcinoma, HCP5 promoted the proliferation, migration, invasiveness and angiogenic ability of follicular thyroid carcinoma cells via sponging miR-22-3p, miR-186-5p, miR-216a-5p and activating ST6GAL2. In addition, HCP5 knockdown reduced cell viability, while elevated apoptotic rate via sponging miR-128-3p in anaplastic thyroid cancer [10, 14].
In bladder cancer, HCP5 upregulation could promote cell invasion and migration via sponging miR-29b-3p [28].
In triple negative breast cancer, HCP5 as a ceRNA to regulate BIRC3 by sponging miR-219a-5p, thereby promoting cancer progression. In addition, overexpression of HCP5 promoted cisplatin resistance by inhibiting PTEN expression [29, 30].
In cervical cancer, HCP5 promoted the development of cervical cancer through increasing MACC1 expression by microRNA-15a adsorption [31].
In clear cell renal cell carcinoma, HCP5 promoted proliferation and metastasis of clear cell renal cell carcinoma via targeting miR-140-5p/IGF1R pathway [12].
In colon cancer, HCP5 enhanced cell proliferation and migration of colon cancer cells by inhibiting the AP1G1 expression and activating the PI3K/AKT pathway [32].
In colorectal cancer, HCP5 contributes to epithelial–mesenchymal transition in colorectal cancer through HCP5/miR-139-5p/ZEB1 axis. In addition, HCP5 knockdown inhibited viabilities, migration and invasion, while inducing apoptosis by miR-299-3p/PFN1/AKT axis [13, 33].
In cutaneous squamous cell carcinoma, HCP5 promoted autophagy and reduced apoptosis by competitively bind to miR-138-5p to regulate EZH2 [34].
In esophageal squamous cell carcinoma, HCP5 promoted cellular activities via modulating the miR-139-5p/PDE4A pathway and stimulating the PI3K/AKT/mTOR signaling pathway [35].
In gastric cancer, HCP5 was highly expressed in gastric cancer cell lines, HCP5 silencing inhibited cell proliferation, migration, invasion, and promoted cell apoptosis via regulation of miR-299-3p/SMAD5 axis [36]. Meanwhile, HCP5 induced EMT processes via the miR-186-5p/WNT5A axis under hypoxia [9], and contributes to cisplatin resistance through miR-519d/HMGA1 and miR-128/HMGA2 axis [17, 37].
In pancreatic cancer, HCP5 silencing inhibited proliferation, migration, and invasion by downregulating CDK8 via sponging miR-140-5p [38].
Table 6.
Summary of functional characterization of HCP5 in various cancers
| Cancers | Expression | Related genes | Role | References |
|---|---|---|---|---|
| Thyroid carcinoma | Upregulated | miR-22-3p, miR-186-5p, miR-216a-5p, ST6GAL2 | Oncogene | [14] |
| Anaplastic thyroid cancer | Upregulated | miR-128-3p | Oncogene | [10] |
| Bladder cancer | Upregulated | miR-29b-3p, HMGB1 | Oncogene | [28] |
| Breast cancer | Upregulated | miR-219a-5p, BIRC3 | Oncogene | [29] |
| TNBC | Upregulated | PTEN | Oncogene | [30] |
| Cervical cancer | Upregulated | miR-15a, MACC1 | Oncogene | [31] |
| ccRCC | Upregulated | miR-140-5p, IGF1R | Oncogene | [12] |
| Colon cancer | Upregulated | PI3K/AKT/AP1G1 | Oncogene | [32] |
| Colorectal cancer | Upregulated | miR-299-3p, PFN1/AKT | Oncogene | [33] |
| Colorectal cancer | Upregulated | miR-139-5p, ZEB1 | Oncogene | [13] |
| CSCC | Upregulated | miR-138-5p, EZH2 | Oncogene | [34] |
| ESCC | Upregulated |
miR-139-5p/PDE4A PI3K/AKT/mTOR |
Oncogene | [35] |
| Gastric cancer | Upregulated | miR-299-3p, SMAD5 | Oncogene | [36] |
| Gastric cancer | Upregulated | miR-186-5p, WNT5A | Oncogene | [9] |
| Gastric cancer | Upregulated | miR-519d, HMGA1 | Oncogene | [37] |
| Gastric cancer | Upregulated | miR-128, HMGA2 | Oncogene | [17] |
| Glioma | Upregulated | miR-139, RUNX1 | Oncogene | [42] |
| Glioma | Upregulated | miR-128 | Oncogene | [43] |
| Hepatocellular carcinoma | Upregulated | miR-29b-3p, DNMT3A | Oncogene | [11] |
| Large B-cell lymphoma | Upregulated | miR-27b-3p, MET | Oncogene | [44] |
| Lung adenocarcinoma | Upregulated | miR-203, SNAI | Oncogene | [45] |
| NSCLC | Upregulated | miR-320, Survivin | Oncogene | [18] |
| Multiple myeloma | Upregulated | miR-128-3p, PLAGL2 | Oncogene | [46] |
| Neuroblastoma | Upregulated | miR-186-5p, MAP3K2 | Oncogene | [47] |
| OSCC | Upregulated | miR-140-5p, SOX4 | Oncogene | [48] |
| Osteosarcoma | Upregulated | miR-101, EPHA7 | Oncogene | [20] |
| Ovarian cancer | Upregulated | miR-525-5p, PRC1 | Oncogene | [49] |
| Pancreatic cancer | Upregulated | miR-214-3p, HDGF | Oncogene | [8] |
| Pancreatic cancer | Upregulated | miR-140-5p, CDK8 | Oncogene | [38] |
| Prostate cancer | Upregulated | miR-4656, CEMIP | Oncogene | [50] |
| Renal cell carcinoma | Upregulated | miR-214-3p, MAPK1 | Oncogene | [19] |
| Retinoblastoma | Upregulated | miR-3619-5p, HDAC9 | Oncogene | [51] |
| Skin cutaneous melanoma | Downregulated | miR-12, RARRES3 | Suppressor | [15] |
TNBC triple-negative breast cancer, ccRCC clear cell renal cell carcinoma, CSCC cutaneous squamous cell carcinoma, ESCC esophageal squamous cell carcinoma, NSCLC non-small cell lung cancer cells, OSCC oral squamous cell carcinoma
Discussion
With the rapid development of high-throughput sequencing technology, the field of lncRNAs has attracted extensive attention. Countless studies have revealed that lncRNAs could participate in various chemical and biological processes, such as chromosome remodeling, transcription, cancer metastasis and posttranscriptional processing [39]. Accumulating studies have shown that lncRNAs were abnormally expressed in various cancers, and function as oncogene or tumor suppressor based on these expression level, participate in the initiation and progression of cancer. Previous meta-analyses have indicated that DLX6-AS1 [40], PVT1 [41], SNHG15 [22] and GHET1 [24] are related to the clinicopathological features and prognosis of various cancers. These evidences indicated that lncRNA may be a specific prognostic biomarker and therapeutic target for cancer.
HCP5, a promising novel cancer-related lncRNA, numerous studies have revealed that HCP5 was dysregulated in various cancers, and that HCP5 has the potential to become a diagnostic biomarker and therapeutic target. Li et al. found that HCP5 was significantly overexpression in NSCLC tissues, and the overall survival rate of NSCLC patients in high HCP5 group was significantly lower [18]. Yang et al. reported that HCP5 expression was higher in colorectal cancer tissues than in adjacent normal tissue, and that HCP5 overexpression was correlated with low TNM stage, poor differentiation and low tumor depth invasion [13]. Zhao et al. found that HCP5 expression was increased in oral squamous cell carcinoma tissues compared with adjacent normal tissues, and that high expression of HCP5 was closely associated with lymph node metastasis, advanced TNM stage and the unfavorable overall survival in oral squamous cell carcinoma patients [26]. Hao et al. [19] showed that HCP5 expression was enhanced in neoplasm tissues of renal cell carcinoma patients, and that high HCP5 was related to Fuhrman neoplasm grade and lymphatic metastasis. Tu et al. [20] demonstrated HCP5 was up-regulated both in osteosarcoma tissues and cell lines and high expression of HCP5 was associated to low survival in osteosarcoma patients.
This is the first meta-analysis to systematically collect studies and to evaluate the potential functions of HCP5 as therapeutic target and prognostic biomarker for human cancers. We found that HCP5 overexpression increased the risk of shorter overall survival in most cancers (non-small cell lung cancer, colorectal cancer, oral squamous cell carcinoma, clear cell renal cell carcinoma, gastric cancer and osteosarcoma). Moreover, the relationship between HCP5 expression levels and clinical attributes has been analyzed according to the specific cancer types. The results suggest that there were no statistically significant relationships between HCP5 overexpression and age, gender and tumor size. However, HCP5 overexpression was statistically correlated with poor histological differentiation in CRC and GC, positive lymph node metastasis in CRC, OSCC, ccRCC and GC, and advanced TNM stage in OSCC. In addition, only one study reported that down-regulation of HCP5 was associated with worse OS, advanced tumor stage, positive distal metastasis and lymph node metastasis in skin cutaneous melanoma, the number of study restricted the prognostic meta-analysis. Furthermore, we used two public databases (GEPIA 2 and NGDC) to validate HCP5 expression level in various cancers. The results revealed that HCP5 was significantly up-regulated in four cancers (CHOL, ESCA, LAML and PAAD), and that strongly associated with poor prognosis; HCP5 was down-regulated in SKCM, and that HCP5 high expression group had longer OS than the HCP5 low expression group. These results are consistent with the findings presented in this study.
In sum up, dysregulation of HCP5 expression was closely related to tumor prognosis, thus may be a potential prognostic biomarker in cancers. Then, we used bioinformatics databases to predict the function mechanism of HCP5. According to GO and KEGG analyses, we found that the most strongly enriched functional terms were immune response, defense response to virus, peptide antigen binding and integral component of lumenal side of endoplasmic reticulum membrane. Also, the HCP5 target genes were significantly related to TNF signaling pathway, NF-kappa B signaling pathway and Apoptosis. Up to now, no studies were found on HCP5 and TNF signaling pathway or NF-kappa B signaling pathway. Moreover, a total of 58 miRNAs-HCP5 interactions were predicted, which could provide reference for future molecular mechanism research of HCP5.
Nonetheless, several limitations in the present study should be emphasized. First, our meta analysis included only nine studies, limited sample sizes, and all from China, there was the possibility of selection bias for positive results in our research of the literatures; Second, only one study reported the association between down-regulated of HCP5 with both prognosis and clinicopathological features in SKCM, the reliability of the conclusion may be limited. Third, no standard cut-off value to measure the expression level of HCP5, and the data of HRs and 95% CIs in some studies were extracted by the software of Engauge Digitizer, which might generate statistical errors. Finally, the prediction results of the functions and pathways of HCP5 needs to be validated further in experiments.
Conclusion
In summary, based on current reported literature, there is significantly association between dysregulation of HCP5 and both prognosis and advanced clinicopathological features in most cancers. HCP5 may be functions as a novel potential prognostic biomarker and therapeutic target in multiple human cancers. However, there were still several limitations in our study, high-quality and multicenter studies are still needed to confirm these conclusions.
Acknowledgements
Not applicable.
Abbreviations
- lncRNA
Long noncoding RNA
- HCP5
Human histocompatibility leukocyte antigen (HLA) complex P5
- HR
Hazard ratio
- OR
Odds ratio
- NSCLC
Non-small cell lung cancer
- CRC
Colorectal cancer
- OSCC
Oral squamous cell carcinoma
- ccRCC
Clear cell renal cell carcinoma
- GC
Gastric cancer
- SKCM
Skin cutaneous melanoma
- OSC
Osteosarcoma
- CP
Clinicopathological parameters
- OS
Overall survival
- CI
Confidence intervals
- NOS
Newcastle–Ottawa Scale
- GO
Gene Ontology
- BP
Biological process
- CC
Cellular component
- MF
Molecular function
Authors’ contributions
KH conceptualized the study. SH and MG collected and analysed the data. SH wrote the manuscript. SH and MG arranged the tables and figures. LG and MJ revised the paper. All authors have reviewed the final manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by Grants from the Fundamental Research Funds for the Central Universities (2021-JYB-XJSJJ-066, 2020-JYB-ZDGG-123), National Natural Science Foundation of China (No. 82174458) and New Innovation Project-Yiqilin Leading Talent Project.
Availability of data and materials
All data are included in this article.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Shao-pu Hu and Meng-xue Ge contributed equally to this article
Contributor Information
Min Jiang, Email: dongfangjm@126.com.
Kai-wen Hu, Email: kaiwenh@163.com.
References
- 1.Bray F, Laversanne M, Weiderpass E, Soerjomataram I. The ever-increasing importance of cancer as a leading cause of premature death worldwide. Cancer. 2021;127(16):3029–3030. doi: 10.1002/cncr.33587. [DOI] [PubMed] [Google Scholar]
- 2.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 3.Shi SB, Cheng QH, Gong SY, Lu TT, Guo SF, Song SM, Yang YP, Cui Q, Yang KH, Qian YW. PCAT6 may be a new prognostic biomarker in various cancers: a meta-analysis and bioinformatics analysis. Cancer Cell Int. 2021;21(1):370. doi: 10.1186/s12935-021-02079-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morlando M, Fatica A. Alteration of epigenetic regulation by long noncoding RNAs in cancer. Int J Mol Sci. 2018;19(2):570. doi: 10.3390/ijms19020570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li J, Li Z, Leng K, Xu Y, Ji D, Huang L, Cui Y, Jiang X. ZEB1-AS1: a crucial cancer-related long non-coding RNA. Cell Prolif. 2018;51(1):e12423. doi: 10.1111/cpr.12423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu L, Zhang L, Zheng S. Role of the long non-coding RNA HOTAIR in hepatocellular carcinoma. Oncol Lett. 2017;14(2):1233–1239. doi: 10.3892/ol.2017.6312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chandra Gupta S, Nandan Tripathi Y. Potential of long non-coding RNAs in cancer patients: from biomarkers to therapeutic targets. Int J Cancer. 2017;140(9):1955–1967. doi: 10.1002/ijc.30546. [DOI] [PubMed] [Google Scholar]
- 8.Liu Y, Wang J, Dong L, Xia L, Zhu H, Li Z, Yu X. Long noncoding RNA HCP5 regulates pancreatic cancer gemcitabine (GEM) resistance by sponging Hsa-miR-214-3p to target HDGF. Onco Targets Ther. 2019;12:8207–8216. doi: 10.2147/OTT.S222703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao M, Liu L, Yang Y, Li M, Ma Q, Chang Z. LncRNA HCP5 induces gastric cancer cell proliferation, invasion, and EMT processes through the miR-186–5p/WNT5A axis under hypoxia. Front Cell Dev Biol. 2021;9:663654. doi: 10.3389/fcell.2021.663654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J, Zhao D, Meng Q. Knockdown of HCP5 exerts tumor-suppressive functions by up-regulating tumor suppressor miR-128–3p in anaplastic thyroid cancer. Biomed Pharmacother. 2019;116:108966. doi: 10.1016/j.biopha.2019.108966. [DOI] [PubMed] [Google Scholar]
- 11.Zhou Y, Li K, Dai T, Wang H, Hua Z, Bian W, Wang H, Chen F, Ai X. Long non-coding RNA HCP5 functions as a sponge of miR-29b-3p and promotes cell growth and metastasis in hepatocellular carcinoma through upregulating DNMT3A. Aging. 2021;13(12):16267–16286. doi: 10.18632/aging.203155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang YJ, Lu C. Long non-coding RNA HCP5 promotes proliferation and metastasis of clear cell renal cell carcinoma via targeting miR-140-5p/IGF1R pathway. Eur Rev Med Pharmacol Sci. 2020;24(6):2965–2975. doi: 10.26355/eurrev_202003_20661. [DOI] [PubMed] [Google Scholar]
- 13.Yang C, Sun J, Liu W, Yang Y, Chu Z, Yang T, Gui Y, Wang D. Long noncoding RNA HCP5 contributes to epithelial-mesenchymal transition in colorectal cancer through ZEB1 activation and interacting with miR-139-5p. Am J Transl Res. 2019;11(2):953–963. [PMC free article] [PubMed] [Google Scholar]
- 14.Liang L, Xu J, Wang M, Xu G, Zhang N, Wang G, Zhao Y. LncRNA HCP5 promotes follicular thyroid carcinoma progression via miRNAs sponge. Cell Death Dis. 2018;9(3):372. doi: 10.1038/s41419-018-0382-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei X, Gu X, Ma M, Lou C. Long noncoding RNA HCP5 suppresses skin cutaneous melanoma development by regulating RARRES3 gene expression via sponging miR-12. Onco Targets Ther. 2019;12:6323–6335. doi: 10.2147/OTT.S195796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qin SY, Yang L, Kong S, Xu YH, Liang B, Ju SQ. LncRNA HCP5: a potential biomarker for diagnosing gastric cancer. Front Oncol. 2021;11:2207. doi: 10.3389/fonc.2021.684531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang L, Kang H, Jia J. HCP5 contributes to cisplatin resistance in gastric cancer through miR-128/HMGA2 axis. Cell Cycle (Georgetown, Tex) 2021;20(11):1080–1090. doi: 10.1080/15384101.2021.1924948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li C, Lei Z, Peng B, Zhu J, Chen L. LncRNA HCP5 stimulates the proliferation of non-small cell lung cancer cells by up-regulating survivin through the down-regulation of miR-320. Cancer Manage Res. 2020;12:1129–1134. doi: 10.2147/CMAR.S222221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hao JF, Chen P, Li HY, Li YJ, Zhang YL. Effects of LncRNA HCP5/miR-214-3p/MAPK1 molecular network on renal cell carcinoma cells. Cancer Manag Res. 2020;12:13347–13356. doi: 10.2147/CMAR.S274426. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Tu Y, Cai Q, Zhu X, Xu M. Down-regulation of HCP5 inhibits cell proliferation, migration, and invasion through regulating EPHA7 by competitively binding miR-101 in osteosarcoma. Braz J Med Biol Res. 2021;54(2):e9161. doi: 10.1590/1414-431X20209161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu S, Chang J, Li Y, Wang W, Guo M, Zou EC, Wang Y, Yang Y. Long non-coding RNA XIST as a potential prognostic biomarker in human cancers: a meta-analysis. Oncotarget. 2018;9(17):13911–13919. doi: 10.18632/oncotarget.23744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen C, Feng Y, Wang J, Liang Y, Zou W. Long non-coding RNA SNHG15 in various cancers: a meta and bioinformatic analysis. BMC Cancer. 2020;20(1):1156. doi: 10.1186/s12885-020-07649-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47(W1):W556–W560. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang D, Zhang H, Fang X, Zhang X, Liu H. Prognostic value of long non-coding RNA GHET1 in cancers: a systematic review and meta-analysis. Cancer Cell Int. 2020;20:109. doi: 10.1186/s12935-020-01189-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Partners C-NMa Database Resources of the National Genomics Data Center, China National Center for Bioinformation in 2021. Nucleic Acids Res. 2021;49(D1):D18–D28. doi: 10.1093/nar/gkaa1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao J, Bai X, Feng C, Shang X, Xi Y. Long non-coding RNA HCP5 facilitates cell invasion and epithelial-mesenchymal transition in oral squamous cell carcinoma by miR-140-5p/ SOX4 axis. Cancer Manage Res. 2019;11:10455–10462. doi: 10.2147/CMAR.S230324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang YJ, Lu C. Long non-coding RNA HCP5 promotes proliferation and metastasis of clear cell renal cell carcinoma via targeting miR-1 40–5p/IGF1R pathway. Eur Rev Med Pharmacol Sci. 2020;24(6):2965–2976. doi: 10.26355/eurrev_202003_20661. [DOI] [PubMed] [Google Scholar]
- 28.Zhao C, Li Y, Hu X, Wang R, He W, Wang L, Qi L, Tong S. LncRNA HCP5 promotes cell invasion and migration by sponging miR-29b-3p in human bladder cancer. Onco Targets Ther. 2020;13:11827–11838. doi: 10.2147/OTT.S249770. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Wang L, Luan T, Zhou S, Lin J, Yang Y, Liu W, Tong X, Jiang W. LncRNA HCP5 promotes triple negative breast cancer progression as a ceRNA to regulate BIRC3 by sponging miR-219a-5p. Cancer Med. 2019;8(9):4389–4403. doi: 10.1002/cam4.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu J, Chen H, Ye M, Wang B, Zhang Y, Sheng J, Meng T, Chen H. Long noncoding RNA HCP5 contributes to cisplatin resistance in human triple-negative breast cancer via regulation of PTEN expression. Biomed Pharmacother. 2019;115:108869. doi: 10.1016/j.biopha.2019.108869. [DOI] [PubMed] [Google Scholar]
- 31.Yu Y, Shen HM, Fang DM, Meng QJ, Xin YH. LncRNA HCP5 promotes the development of cervical cancer by regulating MACC1 via suppression of microRNA-15a. Eur Rev Med Pharmacol Sci. 2018;22(15):4812–4819. doi: 10.26355/eurrev_201808_15616. [DOI] [PubMed] [Google Scholar]
- 32.Yun WK, Hu YM, Zhao CB, Yu DY, Tang JB. HCP5 promotes colon cancer development by activating AP1G1 via PI3K/AKT pathway. Eur Rev Med Pharmacol Sci. 2019;23(7):2786–2793. doi: 10.26355/eurrev_201904_17553. [DOI] [PubMed] [Google Scholar]
- 33.Bai N, Ma Y, Zhao J, Li B. Knockdown of lncRNA HCP5 suppresses the progression of colorectal cancer by miR-299-3p/PFN1/AKT Axis. Cancer Manag Res. 2020;12:4747–4758. doi: 10.2147/CMAR.S255866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zou S, Gao Y, Zhang S. lncRNA HCP5 acts as a ceRNA to regulate EZH2 by sponging miR-138-5p in cutaneous squamous cell carcinoma. Int J Oncol. 2021;59(2):1–13. doi: 10.3892/ijo.2021.5236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu J, Ma J, Guan B, Li J, Wang Y, Hu S. LncRNA HCP5 promotes malignant cell behaviors in esophageal squamous cell carcinoma via the PI3K/AKT/mTOR signaling. Cell Cycle (Georgetown, Tex) 2021;20(14):1374–1388. doi: 10.1080/15384101.2021.1944512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yin D, Lu X. Silencing of long non-coding RNA HCP5 inhibits proliferation, invasion, migration, and promotes apoptosis via regulation of miR-299-3p/SMAD5 axis in gastric cancer cells. Bioengineered. 2021;12(1):225–239. doi: 10.1080/21655979.2020.1863619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Z, Wang H. HCP5 promotes proliferation and contributes to cisplatin resistance in gastric cancer through miR-519d/HMGA1 axis. Cancer Manage Res. 2021;13:787–794. doi: 10.2147/CMAR.S289997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuan B, Guan Q, Yan T, Zhang X, Xu W, Li J. LncRNA HCP5 regulates pancreatic cancer progression by miR-140-5p/CDK8 axis. Cancer Biother Radiopharm. 2020;35(9):711–719. doi: 10.1089/cbr.2019.3294. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y, Huang JC, Cai KT, Yu XB, Chen YR, Pan WY, He ZL, Lv J, Feng ZB, Chen G. Long noncoding RNA HOTTIP promotes hepatocellular carcinoma tumorigenesis and development: a comprehensive investigation based on bioinformatics, qRTPCR and metaanalysis of 393 cases. Int J Oncol. 2017;51(6):1705–1721. doi: 10.3892/ijo.2017.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tian S, Liu J, Kong S, Peng L. LncRNA DLX6-AS1 as a potential molecular biomarker in the clinicopathology and prognosis of various cancers: a meta-analysis. Biosci Rep. 2020;40(8):BSR20193532. doi: 10.1042/BSR20193532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen X, Yang Y, Cao Y, Wu C, Wu S, Su Z, Jin H, Wang D, Zhang G, Fan W, et al. lncRNA PVT1 identified as an independent biomarker for prognosis surveillance of solid tumors based on transcriptome data and meta-analysis. Cancer Manag Res. 2018;10:2711–2727. doi: 10.2147/CMAR.S166260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Teng H, Wang P, Xue Y, Liu X, Ma J, Cai H, Xi Z, Li Z, Liu Y. Role of HCP5-miR-139-RUNX1 feedback loop in regulating malignant behavior of glioma cells. Mol Ther J Am Soc Gene Ther. 2016;24(10):1806–1822. doi: 10.1038/mt.2016.103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Wang C, Yu G, Xu Y, Liu C, Sun Q, Li W, Sun J, Jiang Y, Ye L. Knockdown of long non-coding RNA HCP5 increases radiosensitivity through cellular senescence by regulating microRNA-128 in gliomas. Cancer Manag Res. 2021;13:3723–3737. doi: 10.2147/CMAR.S301333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu L, Zhao J, Liu Y, Liu X, Lu Q, Zeng Z, Zhu L, Tong X, Xu Q. Geniposide inhibits proliferation and induces apoptosis of diffuse large B-cell lymphoma cells by inactivating the HCP5/miR-27b-3p/MET axis. Int J Med Sci. 2020;17(17):2735–2743. doi: 10.7150/ijms.51329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang L, Wang R, Fang L, Ge X, Chen L, Zhou M, Zhou Y, Xiong W, Hu Y, Tang X, et al. HCP5 is a SMAD3-responsive long non-coding RNA that promotes lung adenocarcinoma metastasis via miR-203/SNAI axis. Theranostics. 2019;9(9):2460–2474. doi: 10.7150/thno.31097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu Q, Ran R, Song M, Li X, Wu Z, Dai G, Xia R. LncRNA HCP5 acts as a miR-128–3p sponge to promote the progression of multiple myeloma through activating Wnt/β-catenin/cyclin D1 signaling via PLAGL2. Cell Biol Toxicol. 2021. [DOI] [PubMed]
- 47.Zhu K, Wang L, Zhang X, Sun H, Chen T, Sun C, Zhang F, Zhu Y, Yu X, He X, et al. LncRNA HCP5 promotes neuroblastoma proliferation by regulating miR-186-5p/MAP3K2 signal axis. J Pediatr Surg. 2021;56(4):778–787. doi: 10.1016/j.jpedsurg.2020.10.011. [DOI] [PubMed] [Google Scholar]
- 48.Zhao J, Bai X, Feng C, Shang X, Xi Y. Long non-coding RNA HCP5 facilitates cell invasion and epithelial-mesenchymal transition in oral squamous cell carcinoma By miR-140-5p/SOX4 Axis. Cancer Manag Res. 2019;11:10455–10462. doi: 10.2147/CMAR.S230324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang L, He M, Fu L, Jin Y. Role of lncRNAHCP5/microRNA-525–5p/PRC1 crosstalk in the malignant behaviors of ovarian cancer cells. Exp Cell Res. 2020;394(1):112129. doi: 10.1016/j.yexcr.2020.112129. [DOI] [PubMed] [Google Scholar]
- 50.Hu R, Lu Z. Long non-coding RNA HCP5 promotes prostate cancer cell proliferation by acting as the sponge of miR-4656 to modulate CEMIP expression. Oncol Rep. 2020;43(1):328–336. doi: 10.3892/or.2019.7404. [DOI] [PubMed] [Google Scholar]
- 51.Zhu Y, Hao F. Knockdown of long non-coding RNA HCP5 suppresses the malignant behavior of retinoblastoma by sponging miR-3619-5p to target HDAC9. Int J Mol Med. 2021;47(5):74. doi: 10.3892/ijmm.2021.4907. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are included in this article.