Abstract
With No Lysine (K) WNK kinases regulate electro-neutral cotransporters that are controlled by osmotic stress and chloride. We showed previously that autophosphorylation of WNK1 is inhibited by chloride, raising the possibility that WNKs are activated by osmotic stress. Here we demonstrate that unphosphorylated WNK isoforms 3 and 1 autophosphorylate in response to osmotic pressure in vitro, applied with the crowding agent polyethylene glycol (PEG)400 or osmolyte ethylene glycol (EG), and that this activation is opposed by chloride. Small angle x-ray scattering of WNK3 in the presence and absence of PEG400, static light scattering in EG, and crystallography of WNK1 were used to understand the mechanism. Osmosensing in WNK3 and WNK1 appears to occur through a conformational equilibrium between an inactive, unphosphorylated, chloride-binding dimer and an autophosphorylation-competent monomer. An improved structure of the inactive kinase domain of WNK1, and a comparison with the structure of a monophosphorylated form of WNK1, suggests that large cavities, greater hydration, and specific bound water may participate in the osmosensing mechanism. Our prior work showed that osmolytes have effects on the structure of phosphorylated WNK1, suggestive of multiple stages of osmotic regulation in WNKs.
INTRODUCTION
With No Lysine (K) (WNK) kinases are soluble intracellular serine/threonine kinases, noted for their unique constellation of catalytic residues (Xu et al., 2000). WNKs regulate the cation chloride cotransporters Na+ Cl– (NCC) and Na+ K+ 2Cl– (NKCCs) through a single intervening kinase, oxidative stress responsive 1/Ste20-related proline/alanine-rich kinase (OSR1/SPAK) (Richardson and Alessi, 2008). The cotransporters have pleiotropic functions in transepithelial transport, blood pressure regulation, cell volume control, and neuronal functions (Russell, 2000; Ben-Ari, 2017). Cotransporter regulation involves an osmotically activated protein kinase named the volume-regulated protein kinase, or V-kinase (Lytle and Forbush, 1992; Parker, 1993). Phosphorylation of the V-kinase is inhibited by chloride (Haas et al., 1995).
WNK1 is associated with the familial form of hypertension pseudohypoaldosteronism II or Gordon’s Syndrome. Gordon’s Syndrome is treated with thiazide diuretics that target NCC (Wilson et al., 2001). This linkage indicates that WNK kinases are the long-anticipated V-kinases and intracellular eukaryotic osmosensors (Kahle et al., 2005; Richardson and Alessi, 2008; de Los Heros et al., 2018). Studies in cells and animals show that WNK kinases are activated by osmotic stresses (Xu et al., 2000; Lenertz et al., 2005; Zagorska et al., 2007). WNKs are required for organism level recovery from osmotic challenge (Choe and Strange, 2007; Bergaya et al., 2011; Roy et al., 2015).
WNKs autophosphorylate in vitro (Thastrup et al., 2012). We demonstrated that chloride inhibits both the in vitro autophosphorylation and the activity of WNK1 (Piala et al., 2014). This direct chloride inhibition, a feature of the hypothesized V-kinase, suggests that WNKs are also regulated by osmotic stress. Thus, we were prompted to inquire whether the autophosphorylation and activity of WNKs is stimulated by osmotic stress in vitro. A further impetus for this study came from our structure of unphosphorylated WNK1 (Min et al., 2004). This inactive form of WNK1 is highly hydrated, suggesting a potential osmosensor mechanism. We focus on WNK isoforms WNK1, a WNK implicated in blood pressure regulation (Wilson et al., 2001), and WNK3, which has important roles in the brain and vasculature (Zeniya et al., 2013; Begum et al., 2015). WNK1 and WNK3 exhibit 94% sequence identity in their kinase domains (Xu et al., 2000; Verissimo and Jordan, 2001).
We demonstrated recently that a phosphorylated form of WNK1 does not adopt the canonical structure of an active kinase, and that an osmolyte induced conformational changes toward a more fully active configuration (Akella et al., 2020). Here we address the effect of osmolytes and crowding agents on the unphosphorylated forms of WNKs in vitro. In studies of uWNK3 (unphosphorylated kinase domain of WNK3) and uWNK1 (unphosphorylated kinase domain of WNK1) we show that two crowding agents/osmolytes, among several tested, significantly enhance autophosphorylation.
Osmosensing bacterial histidine kinases autophosphorylate in response to osmotic stress in cells (Wood, 2006; Meena et al., 2010) and to osmolytes and salts in vitro (Wang et al., 2012). Osmosensing domains of histidine kinases are either juxtamembrane or completely soluble and do not rely on membrane (Meena et al., 2010; Wang et al., 2012). Many soluble serine/threonine protein kinases are activated by osmotic stress in cells (Zhou et al., 2016). Two MAP kinases, HOG1 and p38, exhibit osmotic stress-induced autophosphorylation in cells independent of MAP2Ks (Maayan et al., 2012), suggestive of direct activation. However, few structural insights concerning mechanisms for osmosensing are available for either histidine kinases or Ser/Thr protein kinases (Meena et al., 2010; Parkinson, 2010; Maayan et al., 2012; Wang et al., 2012; Airola et al., 2013).
Osmotic stress puts a demand on solvent that affects protein hydration (Parsegian et al., 1994; LiCata and Allewell, 1997; Reid and Rand, 1997; Zhou et al., 2008). Most studies of osmotic effects have been on model proteins such as hemoglobin (Ross and Minton, 1977). The concept of an equilibrium between two states, a more hydrated conformation and less hydrated conformation, was used to explain the effects of crowding agents on substrate binding and catalysis in hexokinase and aspartate transcarbamylase (LiCata and Allewell, 1997; Reid and Rand, 1997).
Here we show that polyethylene glycol 400 (PEG400) and ethylene glycol (EG) promote autophosphorylation of uWNK3 and uWNK1. The dual effect of osmotic pressure and chloride on these WNKs recapitulates the regulation of NKCCs (Haas et al., 1995).
We further show that uWNK3, in the absence of crowding agents or osmolytes, is dimeric in solution using static light scattering (SLS). SAXS data offer evidence that uWNK3 adopts an asymmetric dimer configuration observed by crystallography in uWNK1 (Min et al., 2004). SLS and SAXS analysis implicate a shift in the conformational equilibrium from dimer to a mixture of monomer and dimer in the activation of uWNKs by crowding agents and osmolytes. Further, we present a new refinement of uWNK1 and highlight the well-formed water-filled cavities in the uWNK1 dimer that suggest a potential mechanism for WNK osmosensing. This is the first report to show that a eukaryotic intracellular protein kinase is directly activated by osmotic pressure in vitro. The present study of WNKs, together with data on HOG1 and EnvZ, challenges the notion that osmosensors must reside at the plasma membrane or on the cell surface.
RESULTS AND DISCUSSION
WNK1 and WNK3 exhibit enhanced autophosphorylation and activity in the presence of crowding agents
The kinase domains of WNK1 and WNK3 are phosphorylated as expressed in bacteria (referred here as pWNK1 and pWNK3), as assessed by mass spectrometry. pWNK1 and pWNK3 are phosphorylated on the primary activating sites (S382 in WNK1 and S308 in WNK3) as well as a second site in the Activation Loop (S378 in WNK1 and S304 in WNK3) and other minor sites (Supplemental Table S1) (Xu et al., 2002; Zagorska et al., 2007; Piala et al., 2014). pWNK1 and pWNK3 were dephosphorylated using phosphatases, and the phosphatases were eliminated by purification (see Materials and Methods and Supplemental Table S1). Rephosphorylation of uWNK3 and uWNK1 was tracked by LCMS over time (Figure 1A). uWNK3 yielded 80% rephosphorylation on the primary activation site (Supplemental Table S1). uWNK3 autophosphorylates faster than uWNK1 as measured in 150 mM Cl–. WNKs are known to transautophosphorylate (Thastrup et al., 2012). Both uWNK3 and uWNK1 show a time lag in autophosphorylation, as expected for transautophosphorylation. Because higher activity was observed for uWNK3, and because it expresses better than WNK1, it was used for most activity assays and biophysical analysis.
FIGURE 1:
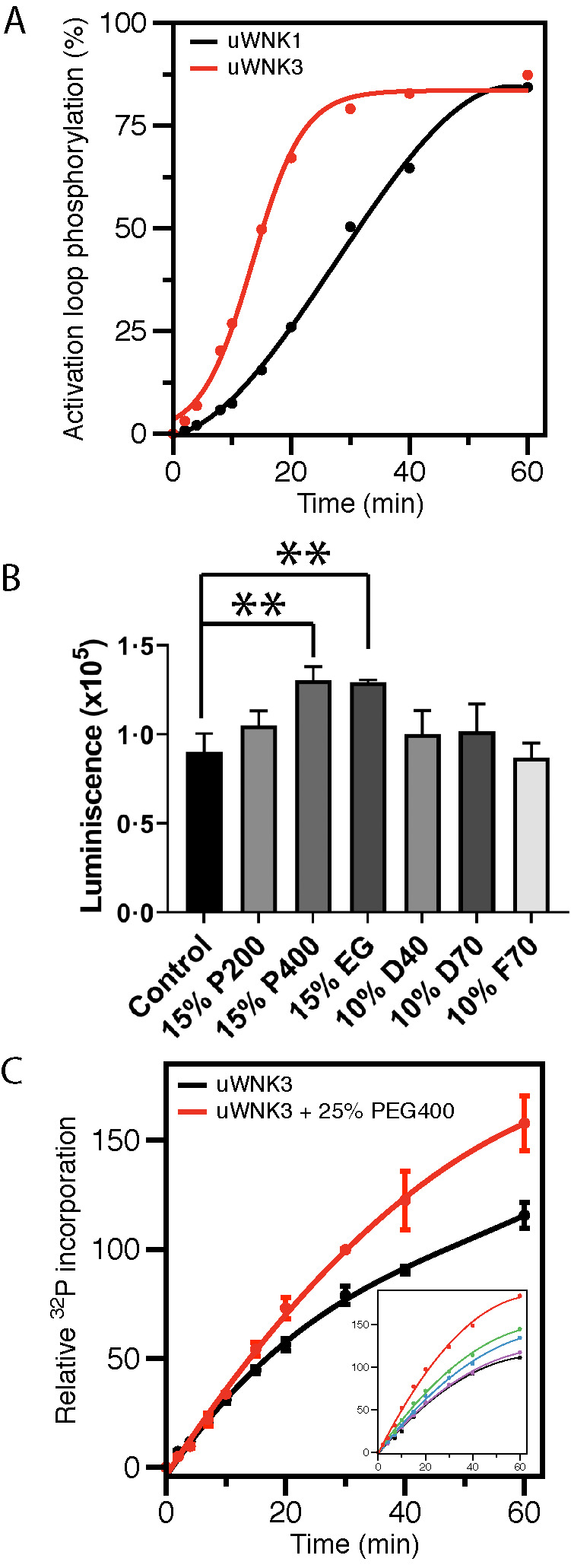
Activity of uWNK3 and uWNK1. (A) In vitro autophosphorylation of the primary activating phosphorylation sites of uWNK3 (Ser 308) (red) (R-value = 0.996) and uWNK1 (Ser382) (black) (R-value = 0.998) tracked by mass spectrometry over time. (B) Total phosphorylation (both gOSR1 peptide and uWNK3 autophosphorylation) in the presence of crowding agents, PEG200/400 (P200/P400), dextran40/70 (D40/D70), and Ficoll70 (F70), and an osmolyte, EG at 15 min, 25°C. Luminescence is proportional to ATP consumption as measured using ADP-Glo. **P < 0.01. (C) Time-course of uWNK3 autophosphorylation with (red) and without (black) 25% vol/vol PEG400 tracked by 32P incorporation. Inset shows progress curves over 60 min with increasing PEG400. Colors for the inset: black (0% PEG400), purple (5%), blue (10%), green (15%), and red (25%). Error bars are SDs from triplicate measurements.
To determine if demands on solvent promote WNK autophosphorylation and activity in vitro, we used gOSR1 (GST-OSR1 peptide, see Materials and Methods) as a substrate with ADP-Glo as the readout. Multiple crowding agents or osmolytes were surveyed (Figure 1B): EG (62 Da), PEG200 (200 Da), PEG400 (400 Da), Dextran40 (40 kDa), Dextran70 (70 kDa), and Ficoll70 (70 kDa). Both PEG400 and EG enhanced activity by about 30% whereas the other tested agents showed minimal effects.
We used γ32P-ATP and autoradiography as an alternative measure of uWNK3 autophosphorylation/gOSR1 substrate phosphorylation in the presence or absence of 25% PEG400. Assays were conducted over 60 min (Figure 1C; autoradiography in Supplemental Figure S1A). A 50% increase in autophosphorylation was observed in the presence of PEG400. The inset in Figure 1C shows 32P incorporation is PEG400 dose dependent (autoradiography in Supplemental Figure S1B).
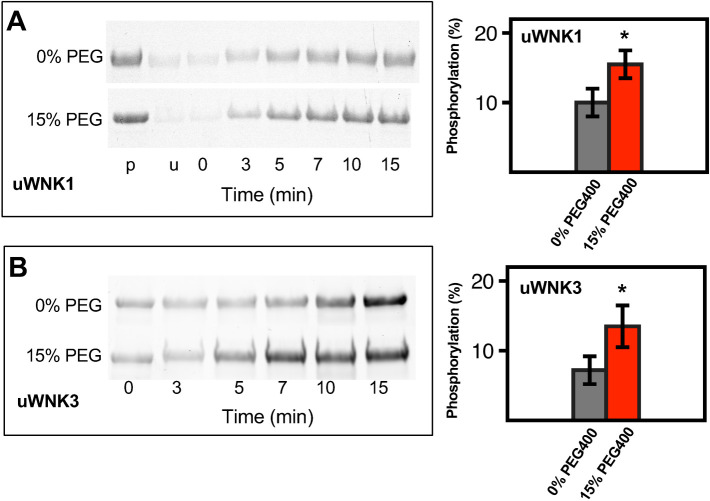
Pro-Q Diamond phospho-protein stain of SDS–PAGE gels provides an additional measure of autophosphorylation. As with 32P, the stain intensity reflects total phosphorylation. Pro-Q Diamond stain was used on both uWNK1 (Figure 2A) and uWNK3 (Figure 2B); 15% (vol/vol) PEG400 again enhanced phosphorylation of both uWNK1 and uWNK3. Coomassie-stained gel loading controls are shown in Supplemental Figure S1, C and D.
FIGURE 2:
Effects of PEG400 on uWNK1 and uWNK3 autophosphorylation and substrate phosphorylation. (A) Time-course of uWNK1 autophosphorylation with and without PEG400 visualized by Pro-Q Diamond phospho-protein stain at indicated times. Densitometry of triplicated gels shown in associated bar graph. *p < 0.1 (right). (B) Time-course of uWNK3 autophosphorylation with and without PEG400.
Data presented in Figures 1 and 2 were collected at 150 mM Cl–. Chloride inhibits WNK1 autophosphorylation (Piala et al., 2014). WNK3 and WNK1 have similar chloride sensitivity (Terker et al., 2016). To test whether chloride opposes PEG400-induced autophosphorylation, we measured uWNK3 autophosphorylation in 15% PEG400 at two different chloride concentrations, 50 and 150 mM. As expected, increasing the Cl– decreases uWNK3 autophosphorylation in the presence of PEG400 as measured over time (Supplemental Figure S1E).
SEC-SAXS and SLS show dimer to monomer transitions in uWNK3 in crowding agents and osmolytes
The structural data on WNKs show a range of dimeric or monomeric configurations dependent on phosphorylation state and crystallization conditions (Min et al., 2004; Piala et al., 2014; Yamada et al., 2016a; Akella et al., 2020). Chloride binds to an inactive dimeric configuration of uWNK1 (PDB file 3FPQ) that buries the primary Activation Loop phosphorylation site. In contrast, fully active pWNK3 is monomeric (PDB file 5O26). This raises the question of whether the activators PEG and EG promote a monomeric configuration. Our model, discussed below, is that a monomer may be autophosphorylation competent.
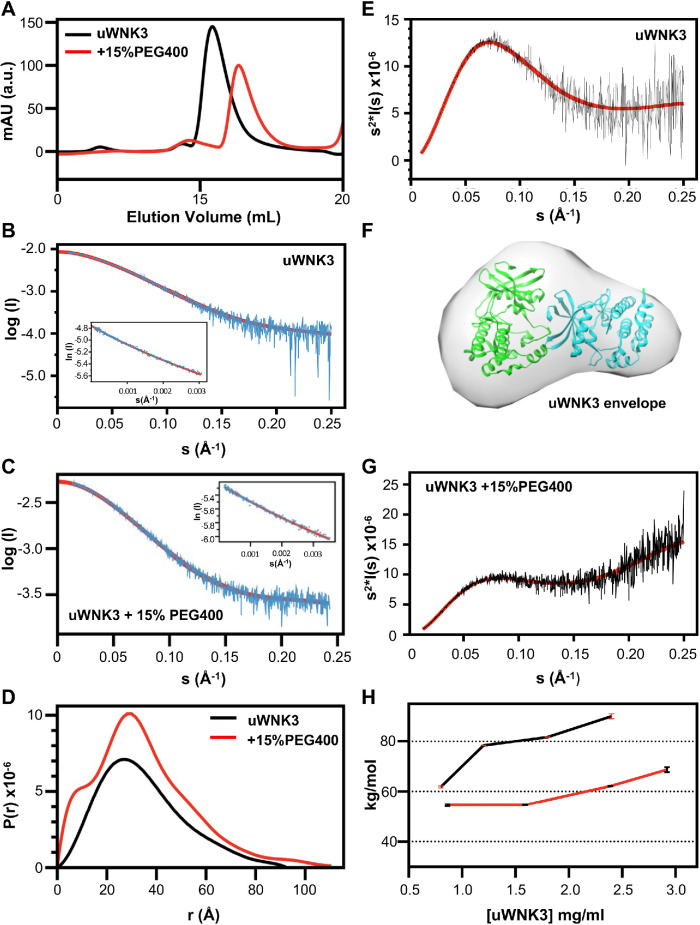
To test whether uWNK3 adopts the dimer seen for unphosphorylated uWNK1, and to test whether PEG400 affects the dimer, size exclusion chromatography in line with small angle x-ray scattering (SEC-SAXS) data were collected in the absence and presence of 15% PEG400.
SEC-profiles for uWNK3 ∓ 15% PEG400 (Figure 3A) gave single peaks. The SEC profile shifted to higher elution volumes in PEG400, suggesting a reduction in molecular weight. The raw SAXS scattering profile and Guinier plots for uWNK3 are shown without PEG400 (Figure 3B) and with PEG400 (Figure 3C). Data collection and derived structural parameters are in Supplemental Table S2. Scattering profiles were truncated at s = 0.25 Å–1. The linearity of the Guinier plots (inserts in Figure 3, B and C) indicate an absence of aggregates ∓ PEG400 (Putnam et al., 2007). The pairwise distribution functions (P(r)) ∓ PEG400 were calculated from the scattering curves. P(r) versus r (Figure 3D) for uWNK3 shows a Dmax of 95Å, consistent with the crystallographically observed uWNK1 dimer. CRYSOL3, which models explicit waters into cavities (Franke et al., 2017), was used to calculate the goodness of fit (χ2) between the observed scattering curves versus dimeric and monomeric models of uWNK1. CRYSOL3 analysis of the uWNK3 profile in the absence of PEG400 gave a good χ2 of 2.6 for the dimer and a poor χ2 of 30 for the monomer (Supplemental Table S3).
FIGURE 3:
Size and shape of uWNK3 ∓ 15% PEG400 from SEC-SAXS and SLS. (A) uWNK3 SEC elution profile (no PEG, black, 15% PEG400, red). uWNK3 scattering profile vs. s and Guinier plot (insert), (B) without PEG400, and (C) with PEG400. Scattering data are cyan, best fit from PRIMUS is red and shown extrapolated to I(0). The scattering curve was truncated at s = 0.25 Å–1. Fit to the Guinier approximation (red). (D) Pairwise distance distribution functions for uWNK3 (black) and uWNK3 with 15% PEG400 (red). (E) Kratky plot (I(s)*s2 vs. s) for uWNK3 (black) with averaged plot (red). (F) Envelope for uWNK3 (generated in DAMMIF) superimposed in CHIMERA on PDB file 6CN9 (dimeric WNK1). (G) Kratky plot uWNK3 in 15% PEG400. (H) SLS of uWNK3 (black) and uWNK3 in 10% EG (red).
The Kratky plot (Figure 3E) indicates that uWNK3 in the absence of PEG400 is well folded. The DAMMIF-generated envelope fit to the uWNK3 scattering curve, from an average of 15 multiple ab initio models (Figure 3F), is asymmetric and resembles the crystallographic uWNK1 dimer. However, the Rg and molecular weight calculated from the Porod volume (calculated from the scattering curve; Supplemental Table S2) corresponds to a monomer, suggesting a mixture of dimer and monomer.
PEG400 induces structural changes in uWNK3. The P(r) versus r plots showed a higher percentage of short distances (Figure 3D), suggesting disorder. Partial unfolding and conformational heterogeneity is indicated from the Kratky plot divergence at high s (Figure 3G) (Konarev et al., 2003). CRYSOL3 gave a poor fit for uWNK3 in PEG400 for both dimer and monomer molecular models. The ATSAS module OLIGOMER (Konarev et al., 2003) gave a dimer/monomer ratio of 70/30 for uWNK3 and 40/60 for uWNK3 in 15% PEG400.
SLS was used as another measure of the uWNK3 oligomeric state in solution (Figure 3H). SLS was measured over uWNK3 concentrations ranging from 0.8 to 2.4 mg/ml. The scattering indicates an 80 kDa dimer between 1.2 to 1.8 mg/ml. The scattering at 0.8 mg/ml scales to 60 kDa, suggesting a 50/50 mixture of monomer and dimer. To assess whether osmolyte changes the oligomeric state, SLS was conducted over approximately the same range of concentrations in 10% EG, from 0.8 to 2.9 mg/ml. The scattering gave a uWNK3 average molecular weight of 55 kDa at 1 mg/ ml, which can be interpreted as 60% monomer and 40% dimer. The proportion of dimer increased at higher concentration and crosses the 60 kDa at 2 mg/ml, indicating the Kd in EG is about 2 mg/ml.
The SAXS and SLS data taken together indicate that 1) that uWNK3 is significantly dimeric in solution; 2) the dimer is asymmetric, resembling the crystallographic dimer of uWNK1; and 3) demands on solvent induced by PEG400 or EG promote conformational changes and a greater proportion of monomer.
Is a conformational equilibrium involved in WNK osmosensing?
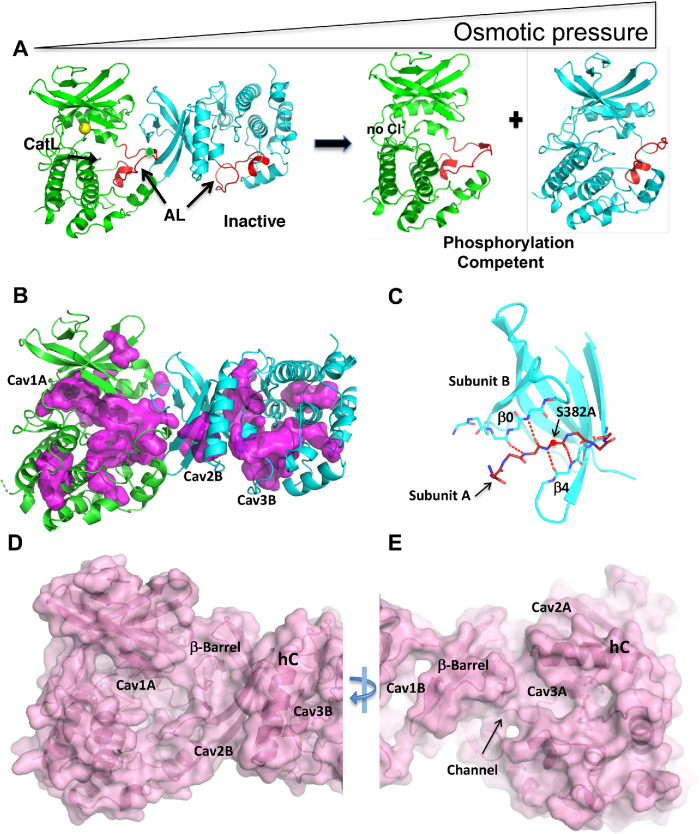
We propose a model in which an osmotic pressure-sensing dimer is in equilibrium with an autophosphorylation-competent monomer (Figure 4A). We showed previously that the chloride ion binds to one Subunit of an unphosphorylated and inactive asymmetric dimer promoting inhibition (Min et al., 2004; Piala et al., 2014). The dimer traps the phosphorylation site (Figure 4A). The inhibited dimer is also favored in low osmotic pressure as demonstrated in the SAXS and SLS data.
FIGURE 4:
Model for osmotic pressure-induced autophosphorylation involving changes in oligomerization and cavities. (A) Inactive dimer to phosphorylation-competent monomer equilibrium model based on the structure of WNK1SA (PDB file 6CN9). Subunit A is green, Subunit B is cyan, Activation Loops are red, and chloride is yellow. (B) WNK1SA cavities, displayed in magenta, for Subunits A and B as calculated in PyMOL. Cavities in WNK1SA (Cav1A, Cav2B, and Cav3B) are labeled. (C) Dimer interface. The Activation Loop encompassing the phosphorylation site at position 382 (alanine in WNK1SA) is inserted as a β-strand in the β-sheet of Subunit B. (D) Surface rendering of WNK1SA (same view as B) calculated in PyMOL highlighting the Cav1A indentation. (E) Surface rendering rotated 180° about the y-axis from D showing a channel and Cav3A.
Unique features of the WNK1 dimer that may contribute to osmosensing
Since osmotic pressure is a demand on solvent, we examined water structures and cavities in WNK1SA (WNK1/S382A). The crystal structure of WNK1SA (PDB file 3FPQ) was re-refined (Supplemental Table S4) (see Materials and Methods) to improve the water shell. The revised structure, now including 938 water molecules, was deposited in the PDB with a new accession number, 6CN9.
The structure of WNK1SA has more cavities than normal. Cavities were visualized in PyMOL (Figure 4B) and volumes calculated in POCASA (Yu et al., 2010). Large cavity volumes, 1900 Å3 in total, are present in the WNK1SA dimer. By comparison, PAK6 (p21-activated kinase-6; Supplemental Figure S2A) (Eswaran et al., 2007), a kinase of similar size (about 300 amino acids), has 450 Å3 of cavities in a monomer (or 900 Å3 in two copies). There are three large cavities in both A and B Subunits (Figure 4B). The largest cavity, Cav1A, has a volume of 650 Å3.
The WNK dimer interface is unique. The inactive dimer forms between the Activation Loop of one Subunit and the N-terminal domain β-sheet of another Subunit (Min et al., 2004; Piala et al., 2014). In the dimer, the Activation Loop (residues F379-S382A) of Subunit A becomes the seventh strand of a 7-stranded β-barrel, between strands β0 (residues V212-M214) and β4 (S289-V291) of Subunit B (Figure 4C). The β−barrel is irregular with fewer hydrogen bonds than normal. The phosphorylation site (S382A, alanine in this structure) is trapped in the interface. Thus, this dimeric configuration is unlikely to support autophosphorylation.
The dimer interaction stabilizes large cavities which can be seen in surface representations (standard view Figure 4D, 180o about y, Figure 4E). Cav 1A contains a cluster of charges described below. Cav2A,B and Cav3A,B arise from displacement of helix C from the body of the kinase (Piala et al., 2014). Cav3A also makes a channel between the two Subunits. In Figure 4E, the Subunit B β-barrel appears structurally isolated, with cavities on either side, separating it both from the C-terminal domain of Subunit B and from Subunit A.
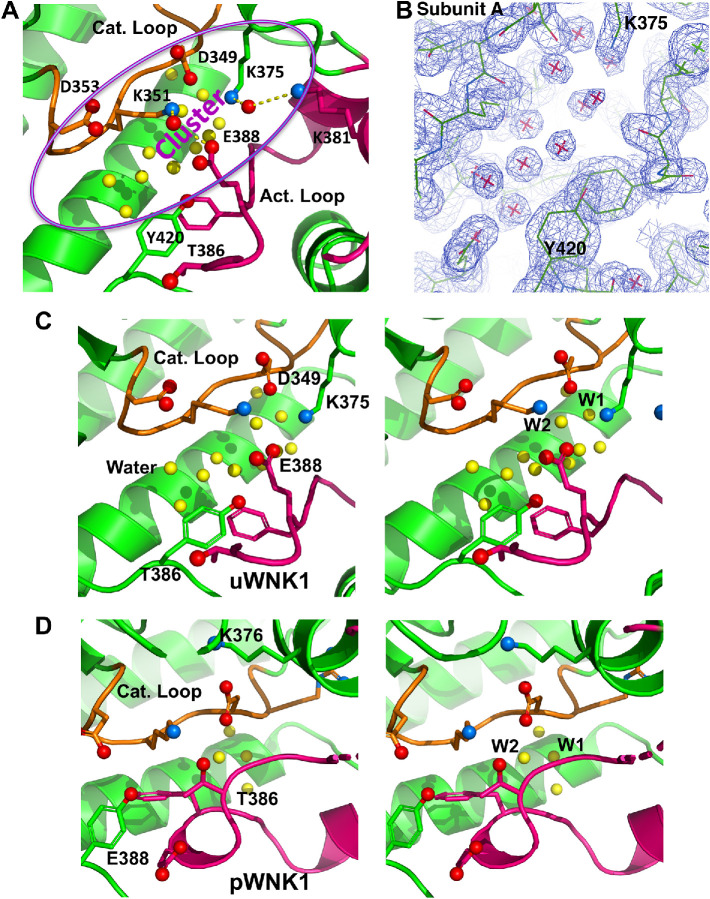
Unusual interactions span the largest cavity (Cav1A) between the Activation Loop and the Catalytic Loop (Figure 5A; alternative view in Supplemental Figure S2B). K375, K381, and E388 in the Activation Loop make charged interactions with conserved catalytic residues, D349 and K351, forming an ionic cluster. Although the Cαs of E388 in the Activation Loop and K351 in the Catalytic Loop are 11Å apart, the side chains are in contact through a water-mediated hydrogen-bonding network. In contrast, canonical active kinases lack a similar cavity because the backbones of the Activation Loop and Catalytic Loop contact each other (Supplemental Figure S2C) (Taylor et al., 1999; Goldsmith et al., 2007). A similar but nonidentical cluster is present in Subunit B (not shown). Charged residues in the Activation Loop that participate in the ionic cluster are conserved among WNKs, including K381 and E388.
FIGURE 5:
Cluster of charged residues in the Catalytic Loop and the Activation Loop in WNK1SA with associated buried water. (A) Cluster of charged residues in the WNK1SA active site with participating residues rendered in ball and stick and highlighted in purple oval. Buried waters underneath ion pairs are yellow. (B) Electron density on water molecules (contoured at 1σ). Same orientation as in A and centered on the water. (C) Stereogram of WNK1SA Catalytic Loop and Activation Loop (similar view as in A). (D) Stereogram of pWNK1 Catalytic Loop and Activation Loop (PDB 5W7T) viewed from the same perspective as C.
Well-ordered water is trapped beneath the ionic cluster, as seen in the structure in Figure 5A and in the corresponding electron density in Figure 5B. The same ion pairs in Subunit A (Figure 5, A and B) are present in Subunit B and in the complex of WNK1SA with an inhibitor (5DRB, PDB file 5DRB) (Yamada et al., 2016b). Both Subunit B and 5DRB have similar buried waters in this cavity (Cav1A). The conservation of the water structure in inactive WNKs suggests its mechanistic importance.
The WNK1SA Cluster is shown in stereo in Figure 5C and is contrasted with pWNK1 (Figure 5D) (PDB file 5W7T; Akella et al., 2020). The pWNK1 stereogram is similar to canonical active kinases (Supplemental Figure S3C). The two stereograms (Figure 5, C and D) show that T386 and E388 exchange places between WNK1SA and pWNK1. E388 binds the catalytic residues in WNK1SA (Figure 5C), and T386 binds the catalytic residues in pWNK1 (Figure 5D). pWNK1 also has less bound water.
Water conservation in the active sites of canonical active protein kinases has been described and reveals two waters in positions similar to the WNK1SA ionic cluster (Knight et al., 2009). Our W2 (shown in Figure 5C) overlaps with Dw defined by Knight et al. (2009), and W1 overlaps with an unnamed water in PKA (PDB file 3FJQ). The water structure is completely distinct from another conserved water cluster involved in cooperative substrate binding in PKA (Setny and Wisniewska, 2018).
The WNK1SA structure may have more bound water overall. Model-building and refinement of the revised structure included waters with 0.8 σ electron density (see Materials and Methods). Over 850 water molecules were identified, about 1.5 waters per residue (Supplemental Table S2). The additional waters over the 350 waters in the original refinement did not raise the R-free (0.22) significantly and raised the overall B-factor only slightly from 24 to 27 Å2. Although many factors contribute to the number of waters observed in crystal structures, on average there are 1.3 waters per amino acid in a recent analysis of structures solved at 1.5 Å resolution or better (Carugo, 2017). Thus, there appears to be more waters in WNK1SA than in proteins of comparable size (Carugo, 2017). The excess bound water may be due to an atypical abundance of charged amino acids in WNKs (Supplemental Figure S2D). WNK1 and WNK3 kinase domains comprise 10% lysine and 10% glutamic acid, which is twice the average found in proteins of comparable size (Carugo, 2008).
These data show that the WNK1SA structure has more water overall than pWNK1, and that it also has specifically trapped water, suggestive that bound water may be part of an osmosensing mechanism. The mechanism appears to be a dimer to monomer equilibrium that involves changes in the amount of bound water. Osmotic pressure, a demand on solvent (Zhou et al., 2008), shifts the equilibrium from a water-trapping dimer to a monomer that may have less bound water. This interpretation parallels an analysis of crowding agents on the enzymes aspartate transcarbamylase (LiCata and Allewell, 1997) and hexokinase (Reid and Rand, 1997). We posit that the induced monomer is competent to autophosphorylate.
WNK kinase domain dedimerization is just one of many factors in WNK conformational regulation (Dbouk et al., 2016; Shekarabi et al., 2017). In addition to regulation by chloride (Piala et al., 2014), we recently discovered that WNK3, WNK4, and Drosophila WNK are inhibited by potassium (Pleinis et al., 2021). Further, WNK oligomerization has been observed in HEK293 and other cells (Lenertz et al., 2005), data indicating formation of high molecular weight complexes. Whether a dimer to monomer equilibrium is involved in potassium regulation and how kinase domain interactions relate to higher order assemblies require further study both in vitro and in vivo.
Osmotically induced autophosphorylation has been observed in other protein kinases. The bacterial histidine kinase EnvZ (Wang et al., 2012) and the HAMP-domain containing hybrid histidine kinases (Airola et al., 2013) function by autophosphorylation. The MAP kinases HOG1 and p38α also have been demonstrated to autoactivate by autophosphorylation (independent of MAP2Ks) in cells subjected to osmotic pressure (Maayan et al., 2012). The osmosensing mechanisms are likely distinct, but again involves autophosphorylation.
Here we have offered evidence that the WNK kinases 1 and 3 are direct osmosensors. The osmosensor feature of WNKs has been anticipated, as noted above. The idea that bound water may play a role in osmosensing aligns with studies of osmotic effects on enzymes (LiCata and Allewell, 1997). WNKs catalyze autophosphorylation and substrate phosphorylation. Thus, the ΔG of the conformational equilibrium serves as an activation energy, increasing signal sensitivity. Our data suggest how a direct demand on solvent may lead to cellular decision making through hydration-driven conformational equilibria.
Direct activation of WNKs by osmotic signals may be sufficient to activate the cascade leading to NKCC phosphorylation. However, other osmotic signals function through WNKs, such as angiotensin II activation of WNK3 (Zeniya et al., 2013). WNKs have the capacity to integrate multiple input and output signals (Shekarabi et al., 2017; Gallolu Kankanamalage et al., 2018) and are proving to be much more complex than originally anticipated for the V-kinase ( Lytle and Forbush, 1992; Parker, 1993). Here we were able to define molecular mechanisms for one form of direct WNK regulation.
MATERIALS AND METHODS
Chemical reagents
EG and PEG200 were obtained from Hampton Research (CA); PEG400, Dextran40 and 70, and Ficoll70 and BioXtra ATP (99% pure) were from Sigma-Aldrich (MO). ADP-Glo is from Promega (WI).
Protein reagents
The clone for WNK3 (118-409) in a pET29b vector was described previously (Akella et al., 2020). Protein expression was carried out in the Escherichia coli strain BL21 DE3, and purification of WNKs involved Ni-NTA affinity, anion exchange on Mono-Q (GE Healthcare), and gel filtration, with details available in Min et al. (2004) and Piala et al. (2014). WNK1SA (194-483) was similar to that of WNK3 as described previously (Min et al., 2004; Piala et al., 2014). We also used wt WNK1 (194-483), which is phosphorylated (pWNK1) as expressed in E. coli (Akella et al., 2020). The pWNK1 and pWNK3 proteins were dephosphorylated using phosphatases to make uWNK1 and uWNK3. pWNK1 was dephosphorylated using λ−phosphatase and shrimp alkaline phosphatase in 10:1 and 5:1 (kinase/phosphatase) ratios, respectively. pWNK3 was dephosphorylated using PP1cγ and λ-phosphatase in a 10:1 ratio in 0.5 mM MnCl2. Phosphatases were removed using Ni-NTA and gel filtration chromatography. The state of phosphorylation was confirmed by mass spectrometry. Residual activity of phosphatase was checked using p-nitrophenyl phosphate (G-Biosciences, St. Louis, MO) following published protocols (Zhou and Zhang, 1999). The kinases were buffer exchanged into 50 mM HEPES, pH 7.4, and 150 mM NaCl. The phosphatase inhibitor orthovanadate prepared as described (Zhou and Zhang, 1999) was added to the protein solutions to 2 mM. The GST-OSR1(314-344) (gOSR) encompassing the phosphorylation site S325 was a gift of Clinton Taylor and Melanie Cobb (Taylor et al., 2018). The gOSR1 was expressed similarly and purified on a glutathione sepharose column (GE Healthcare) and buffer-exchanged into 20 mM HEPES, pH 7.4, 50 mM NaCl, and 5% glycerol.
Generic assay conditions
Autophosphorylation assays were conducted in general in 20 mM HEPES, pH 7.4, 20 mM MgCl2, 5 mM ATP, and usually in 150 mM NaCl at room temperature. Concentrations of uWNK3 or uWNK1 were 4 μM. Substrate phosphorylation assays contained 40 μM gOSR1.
Mass spectrometry
Proteins were digested in an 8:1 M ratio with sequence-grade chymotrypsin (Roche, Basel, Switzerland) in the presence of 100 mM Tris, pH 8.0, and 25 mM CaCl2 (100 μl total reaction volume) at 30°C overnight. Following digestion, the peptide mixture was separated by HPLC (Agilent 1100) on a RP-C18 column (Phenomenex Aeris Widepore 150 × 2.1 mm) using an acetonitrile-water gradient from 4 to 28% acetonitrile containing 0.2% formic acid throughout. Mass spectrometric analysis was performed on an LCQ DECA XP ion-trap mass spectrometer (ThermoFinnigan) with the HPLC coupled inline to an orthogonal electrospray ionization source. MS detector responses were obtained by integration under ion traces corresponding to m/z ranges for Activation Loop peptides. Synthetic isotopically labeled WNK Activation Loop peptide standards for WNKs (21st Century Biochemicals) were used to confirm HPLC elution times and for quantitation. MS/MS spectra were acquired in a data-dependent mode and analyzed using Mass Matrix (Xu et al., 2010) and MASCOT software (Matrix Science) (Koenig et al., 2008).
ADP-Glo
ADP-Glo reagent (Promega) was used as a readout for autophosphorylation and activity measurements in the presence of crowding agents, such as PEGs which are incompatible with mass spectrometry; 50 µl reactions contained 40 mM HEPES (pH 7.4), 10 mM MgCl2, 4 µM pWNK, and 40 µM gOSR1. Final chloride concentration was maintained at 150 mM. The reaction was started by the addition of 5.2 mM ATP. Reactions were stopped after 15 min by the addition of 50 µl of ADP-Glo reagent. Manufacturers protocol was followed for the remaining steps of ATP depletion (40 min), followed by conversion of ADP to ATP (1 h); 100 μl aliquots from each reaction were transferred to a 96-well plate which was centrifuged for 2 min at 800 rpm. Luminescence was read on a CLARIOstar plate reader and data were analyzed using MARS software (both reader and software, BMG Labtech, Ortenberg, Germany). Data were further processed using GraphPad-Prism software to obtain P values.
32P Assays
Reaction conditions were the same as stated above, with exceptions. Reactions were initiated by the addition of uWNK1 or uWNK3 to a final concentration of 4 µM. Assays were stopped using protocols described by Racker (Racker, 1991). Reactions were terminated at time points indicated by spotting aliquots on 3MM filter paper followed by immediate immersion in termination solution (10% trichloroacetic acid and 10 mM pyrophosphate). Extensive washing with fresh termination solution to remove unincorporated radioactivity was followed by drying the filter paper. 32P incorporation was measured by autoradiography using a phosphor screen (Kodak) and Typhoon 9200 imager (General Electric). Additional aliquots of the reaction mixtures were diluted then spotted on unwashed filter paper to establish the ratio of CPM to cold ATP concentration. ImageQuant software (GE Healthcare) was used to quantify the autoradiograms.
Pro-Q Diamond Gel-based phosphorylation assays
Reaction conditions were similar to those used in with 32P, except the PEG400 was set at 15% vol/vol to limit band distortion, the uWNK1 and uWNK3 were 4 μM, and the gOSR1 was 40 μM; 50 μl reactions were stopped in Laemmli buffer. To visualize phospho-proteins, 12% precast Bio-Rad gels were run with 30 μl of the reaction aliquot. Pro-Q Diamond (Life Technologies) stain was applied to the gels using the manufacturer’s protocol which includes fixing, staining, and wash steps. The Pro-Q stain reflects total phosphorylation of the uWNK1 or uWNK3 and gOSR1. Gels were imaged using a ChemiDoc MP imaging system (Bio-Rad); volumes were calculated in ImageLab (Bio-Rad). The gels were then stained with Coomassie (Bio-Rad) to obtain loading controls.
SEC-SAXS
SEC-SAXS (Brosey and Tainer, 2019) data were collected at the BIOCAT (sector18-ID) beamline at Argonne National Laboratory (http://www.bio.aps.anl.gov/pages/about-biocat.html); 300 μl of 5 mg ml–1 sample of uWNK3 were injected on a 24-ml Superadex-200 (GE Healthcare) column equilibrated with 50 mM HEPES, pH 7.4, 150 mM NaCl, 1 mM EDTA, and 1 mM TCEP (Tris[2-carboxyethyl]phosphine) in line with a SAXS flow cell. SAXS data of uWNK3 –/+ PEG400 were collected in 0.5-s exposures every second. Scattering intensity was recorded using a Pilatus3 1M (Dectris) detector which was placed ∼3.5 m from the sample giving us access to a q-range of ∼0.004 Å–1 to 0.4 Å–1. Frames flanking the peak were averaged to obtain the buffer background for subtraction. Data were reduced using BioXTAS RAW 1.6.0 (Hopkins et al., 2017). Data analysis was carried out using the ATSAS package (Version 3.0.1) (Franke et al., 2017). Data processing including buffer subtraction, merging, extrapolation of intensity to zero concentration (I0), curve fitting, and evaluation of radius of gyration (Rg) were performed using the PRIMUS module (Konarev et al., 2003). The GNOM module (Svergun, 1992) was used to obtain I0, Rg, the distance distribution P(r), maximum dimension (Dmax), Porod volume (Vρ), and excluded volume (Ve). Rg and I0 calculated by PRIMUS and GNOM were compared. Protein molecular weights were estimated from the Porod volumes (Vp). Fifteen bead-models comprised of spheres within a radius of Dmax/2 were generated and refined against the scattering curve in DAMMIF (Franke and Svergun, 2009). DAMAVER and DAMFILT were used to generate an average envelope (Konarev et al., 2003). CRYSOL3 was used to calculate the Goodness of Fit between the scattering curve and crystallographic dimer or monomer models as described (Svergun et al., 1998; Franke et al., 2017). CRYSOL3 fills cavities with explicit waters of hydration. The ATSAS module OLIGOMER was used to estimate the fraction of dimer and monomer based on the scattering profiles (Konarev et al., 2003).
SLS
SLS was conducted on a Wyatt DynaPro Nanostar DLS instrument operated at 25°C. Immediately prior to data collection, aggregates and dust were removed by 10-min ultracentrifugation at 16,100 × g; 5 μl uWNK3 in buffer (50 mM HEPES, pH 7.4, and 150 mM NaCl, 1 mM EDTA, and 1 mM TCEP) was loaded into a quartz cuvette and light scattering at 90° was monitored until the SLS detector voltage was stable. Then the scattering was recorded monitored for 5 s and repeated 10 times, with three technical replicates each. Four different uWNK3 concentrations were used (0.8, 1.2, 1.8, and 2.4 mg/ml). Analysis of uWNK3 in 10% EG used the Wyatt reference for this solvent. The uWNK3 concentrations analyzed (in mg/ml) were 0.86, 1.6, 2.4, and 2.9. The data were analyzed using Dynamics version 7.5.0.17 (Wyatt Technologies).
Re-refinement of WNK1SA
The structure of unphosphorylated WNK1SA reported previously was re-refined in CCP4 (Cowtan et al., 2011) using data to 2.0 Å (Supplemental Table S4). The model was built in COOT (Emsley and Cowtan, 2004) based on |2Fo-Fc| maps. Restrained refinement in REFMAC5 (CCP4 suite) of WNK1SA yielded a slightly improved R-factor and R-free of 0.16 and 0.22, respectively (from 0.19 and 0.23 originally reported with 345 waters). Two shells of water were built into |2Fo-Fc| maps contoured at 0.8 σ. Occupancies were set to 1 or 0.5 (938 waters total, 257 at 0.5 occupancy), based on the B-factor refinement. The improved structure was deposited (PDB file 6CN9).
Displaying cavities and calculating volumes
Cavities were visualized in PyMOL (Version 2.0 Schrodinger, LLC) using the Cavitycull option with the radius parameter set to 5 water molecules. Cavity volumes in the re-refined WNK1SA structure were calculated using Pocket Cavity Search Application (POCASA 1.1) (http://altair.sci.hokudai.ac.jp/g6/service/pocasa) (Yu et al., 2010). POCASA defines cavities using a simple grid-scanning algorithm to identify grid points in the solvent surrounded by protein in at least one direction (Yu et al., 2010). POCASA was run with the recommended probe radius of 2 Å and grid size of 1 Å.
Statistics
R-values (RMSD of points to the lines) reported in Figure 1 were calculated in DataGraph (Visual Data Tools) with curves fit to the form L/(1+ exp (-h/x-xo)) where L and h and xo are fitted variables. Figure 2 reports SD for triplicate Pro-Q Diamond stained gels. Standard error is shown for the 32P progress curves. Significance, where reported, was calculated using t test and p values in Excel and GraphPad Prism (*indicates a t test of < 0.1, **< 0.01, and ***< 0.0001). The goodness of fit between the scattering curve and crystallographic dimer or monomer models were calculated as described (Svergun et al., 1998).
Supplementary Material
Acknowledgments
We thank Melanie Cobb and Clinton Taylor for the vector encoding GST-OSR1 and for γ 32P-ATP. Crystallographic results shown in this report are derived from work performed at Argonne National Laboratory, Structural Biology Center (SBC) at the Advanced Photon Source. SBC is operated by the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357. We thank Robert Rambo (Diamond Light Source) who assisted with SAXS interpretation. We also thank Diana Tomchick and Zhe Chen (Structural Biology Lab, UTSW) for assistance with beamline data collection. This research was supported by the American Heart Association 16CSA28530002 (E.J.G.), NIH DK110358 (E.J.G.), Cancer Prevention and Research Institute of Texas RP190421 (E.J.G.), the Welch Foundation I1128 (E.J.G.), NIH P41 GM103622 (S.C.), and NIGMS grant 1S10OD018090-01 (S.C.).
Abbreviations used:
- EG
ethylene glycol
- NCC
Na + Cl –
- NKCC
Na + K + 2Cl –
- OSR1/SPAK
oxidative stress responsive 1/Ste20-related proline/alanine-rich kinase
- PAK6
p21-activated kinase-6
- PEG
polyethylene glycol
- SAXS
small angle x-ray scattering
- SEC
size exclusion chromatography
- SLS
static light scattering
- TCEP
Tris(2-carboxyethyl)phosphine
- V-kinase
volume-regulated protein kinase
- WNK
With No Lysine (K)
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-01-0089) on March 10, 2021.
REFERENCES
- Airola MV, Sukomon N, Samanta D, Borbat PP, Freed JH, Watts KJ, Crane BR (2013). HAMP domain conformers that propagate opposite signals in bacterial chemoreceptors. PLoS Biol 11, e1001479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akella R, Drozdz MA, Humphreys JM, Jiou J, Durbacz MZ, Mohammed ZJ, He H, Liwocha J, Sekulski K, Goldsmith EJ (2020). A phosphorylated intermediate in the activation of WNK kinases. Biochemistry 59, 1747–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begum G, Yuan H, Kahle KT, Li L, Wang S, Shi Y, Shmukler BE, Yang S-S, Lin S-H, Alper SL, Sun D (2015). Inhibition of WNK3 kinase signaling reduces brain damage and accelerates neurological recovery after stroke. Stroke 46, 1956–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ari Y (2017). NKCC1 chloride importer antagonists attenuate many neurological and psychiatric disorders. Trends Neurosci 40, 536–554. [DOI] [PubMed] [Google Scholar]
- Bergaya S, Faure S, Baudrie V, Rio M, Escoubet B, Bonnin P, Henrion D, Loirand G, Achard JM, Jeunemaitre X, Hadchouel J (2011). WNK1 regulates vasoconstriction and blood pressure response to alpha 1-adrenergic stimulation in mice. Hypertension 58, 439–445. [DOI] [PubMed] [Google Scholar]
- Brosey CA, Tainer JA (2019). Evolving SAXS versatility: solution X-ray scattering for macromolecular architecture, functional landscapes, and integrative structural biology. Curr Opin Struct Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carugo O (2008). Amino acid composition and protein dimension. Protein Sci 17, 2187–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carugo O (2017). Protein hydration: Investigation of globular protein crystal structures. Int J Biol Macromol 99, 160–165. [DOI] [PubMed] [Google Scholar]
- Choe KP, Strange K (2007). Evolutionarily conserved WNK and Ste20 kinases are essential for acute volume recovery and survival after hypertonic shrinkage in Caenorhabditis elegans. Am J Physiol Cell Physiol 293, C915–C927. [DOI] [PubMed] [Google Scholar]
- Cowtan K, Emsley P, Wilson KS (2011). From crystal to structure with CCP4. Acta Crystallogr D Biol Crystallogr 67, 233–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dbouk HA, Huang CL, Cobb MH (2016). Hypertension: the missing WNKs. Am J Physiol Renal Physiol 311, F16–F27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Los Heros P, Pacheco-Alvarez D, Gamba G (2018). Role of WNK kinases in the modulation of cell volume. Curr Top Membr 81, 207–235. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Eswaran J, Lee WH, Debreczeni JE, Filippakopoulos P, Turnbull A, Fedorov O, Deacon SW, Peterson JR, Knapp S (2007). Crystal Structures of the p21-activated kinases PAK4, PAK5, and PAK6 reveal catalytic domain plasticity of active group II PAKs. Structure 15, 201–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke D, Petoukhov MV, Konarev PV, Panjkovich A, Tuukkanen A, Mertens HDT, Kikhney AG, Hajizadeh NR, Franklin JM, Jeffries CM, Svergun DI (2017). ATSAS 2.8: a comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J Appl Crystallogr 50, 1212–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke D, Svergun DI (2009). DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J Appl Crystallogr 42, 342–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallolu Kankanamalage S, Karra AS, Cobb MH (2018). WNK pathways in cancer signaling networks. Cell Commun Signal 16, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith EJ, Akella R, Min X, Zhou T, Humphreys JM (2007). Substrate and docking interactions in serine/threonine protein kinases. Chem Rev 107, 5065–5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas M, McBrayer D, Lytle C (1995). [Cl-]i-dependent phosphorylation of the Na-K-Cl cotransport protein of dog tracheal epithelial cells. J Biol Chem 270, 28955–28961. [DOI] [PubMed] [Google Scholar]
- Hopkins JB, Gillilan RE, Skou S (2017). BioXTAS RAW. J Appl Crystallogr 50, 954–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahle KT, Rinehart J, de Los Heros P, Louvi A, Meade P, Vazquez N, Hebert SC, Gamba G, Gimenez I, Lifton RP (2005). WNK3 modulates transport of Cl- in and out of cells: implications for control of cell volume and neuronal excitability. Proc Natl Acad Sci USA 102, 16783–16788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight JD, Hamelberg D, McCammon JA, Kothary R (2009). The role of conserved water molecules in the catalytic domain of protein kinases. Proteins 76, 527–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig T, Menze BH, Kirchner M, Monigatti F, Parker KC, Patterson T, Steen JJ, Hamprecht FA, Steen H (2008). Robust prediction of the MASCOT score for an improved quality assessment in mass spectrometric proteomics. J Proteome Res 7, 3708–3717. [DOI] [PubMed] [Google Scholar]
- Konarev PV, Volkov VV, Sokolova AV, Koch MH, Svergun DI (2003). PRIMUS: a Windows PC-based system for small-angle scattering analysis. J Appl Crystallogr 36, 1277–1282. [Google Scholar]
- Lenertz LY, Lee BH, Min X, Xu BE, Wedin K, Earnest S, Goldsmith EJ, Cobb MH (2005). Properties of WNK1 and implications for other family members. J Biol Chem 280, 26653–26658. [DOI] [PubMed] [Google Scholar]
- LiCata VJ, Allewell NM (1997). Functionally linked hydration changes in Escherichia coli aspartate transcarbamylase and its catalytic Subunit. Biochemistry 36, 10161–10167. [DOI] [PubMed] [Google Scholar]
- Lytle C, Forbush B 3rd (1992). The Na-K-Cl cotransport protein of shark rectal gland. II. Regulation by direct phosphorylation. J Biol Chem 267, 25438–25443. [PubMed] [Google Scholar]
- Maayan I, Beenstock J, Marbach I, Tabachnick S, Livnah O, Engelberg D (2012). Osmostress induces autophosphorylation of Hog1 via a C-terminal regulatory region that is conserved in p38alpha. PLoS One 7, e44749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meena N, Kaur H, Mondal AK (2010). Interactions among HAMP domain repeats act as an osmosensing molecular switch in group III hybrid histidine kinases from fungi. J Biol Chem 285, 12121–12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min XS, Lee BH, Cobb MH, Goldsmith EJ (2004). Crystal structure of the kinase domain of WNK1, a kinase that causes a hereditary form of hypertension. Structure 12, 1303–1311. [DOI] [PubMed] [Google Scholar]
- Parker JC (1993). In defense of cell volume? Am J Physiol 265, C1191–C1200. [DOI] [PubMed] [Google Scholar]
- Parkinson JS (2010). Signaling mechanisms of HAMP domains in chemoreceptors and sensor kinases. Annu Rev Microbiol 64, 101–122. [DOI] [PubMed] [Google Scholar]
- Parsegian VA, Rand RP, Rau DC (1994). Macromolecules and water: probing with osmotic stress. Methods Enzymol 259, 43–94. [DOI] [PubMed] [Google Scholar]
- Piala AT, Moon TM, Akella R, He H, Cobb MH, Goldsmith EJ (2014). Chloride sensing by WNK1 involves inhibition of autophosphorylation. Sci Signal 7, ra41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleinis JM, Norrell L, Akella R, Humphreys JM, He H, Sun Q, Zhang F, Sosa-Pagan J, Morrison DE, Schellinger JN, et al. (2021). WNKs are potassium-sensitive kinases. Am J Physiol Cell Physiol, 10.1152/ajpcell.00456.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam CD, Hammel M, Hura GL, Tainer JA (2007). X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys 40, 191–285. [DOI] [PubMed] [Google Scholar]
- Racker E (1991). Use of synthetic amino acid polymers for assay of protein-tyrosine and protein-serine kinases. Methods Enzymol 200, 107–111. [DOI] [PubMed] [Google Scholar]
- Reid C, Rand RP (1997). Probing protein hydration and conformational states in solution. Biophys J 72, 1022–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson C, Alessi DR (2008). The regulation of salt transport and blood pressure by the WNK-SPAK/OSR1 signalling pathway. J Cell Sci 121, 3293–3304. [DOI] [PubMed] [Google Scholar]
- Ross PD, Minton AP (1977). Analysis of non-ideal behavior in concentrated hemoglobin solutions. J Mol Biol 112, 437–452. [DOI] [PubMed] [Google Scholar]
- Roy A, Goodman JH, Begum G, Donnelly BF, Pittman G, Weinman EJ, Sun D, Subramanya AR (2015). Generation of WNK1 knockout cell lines by CRISPR/Cas-mediated genome editing. Am J Physiol Renal Physiol 308, F366–F376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell JM (2000). Sodium-potassium-chloride cotransport. Physiol Rev 80, 211–276. [DOI] [PubMed] [Google Scholar]
- Setny P, Wisniewska MD (2018). Water-mediated conformational preselection mechanism in substrate binding cooperativity to protein kinase A. Proc Natl Acad Sci USA 115, 3852–3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekarabi M, Zhang J, Khanna AR, Ellison DH, Delpire E, Kahle KT (2017). WNK Kinase Signaling in Ion Homeostasis and Human Disease. Cell Metab 25, 285–299. [DOI] [PubMed] [Google Scholar]
- Svergun DI (1992). Determination of the regularization parameter in indirect-transform methods using perpetual criteria. J Appl Crystallogr 25, 495–503. [Google Scholar]
- Svergun DI, Richard S, Koch MH, Sayers Z, Kuprin S, Zaccai G (1998). Protein hydration in solution: experimental observation by x-ray and neutron scattering. Proc Natl Acad Sci USA 95, 2267–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CAT, An SW, Kankanamalage SG, Stippec S, Earnest S, Trivedi AT, Yang JZ, Mirzaei H, Huang CL, Cobb MH (2018). OSR1 regulates a subset of inward rectifier potassium channels via a binding motif variant. Proc Natl Acad Sci USA 115, 3840–3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SS, Radzio-Andzelm E, Madhusudan, Cheng X, Ten Eyck L, Narayana N (1999). Catalytic Subunit of cyclic AMP-dependent protein kinase: structure and dynamics of the active site cleft. Pharmacol Ther 82, 133–141. [DOI] [PubMed] [Google Scholar]
- Terker AS, Zhang C, Erspamer KJ, Gamba G, Yang CL, Ellison DH (2016). Unique chloride-sensing properties of WNK4 permit the distal nephron to modulate potassium homeostasis. Kidney Int 89, 127–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thastrup JO, Rafiqi FH, Vitari AC, Pozo-Guisado E, Deak M, Mehellou Y, Alessi DR (2012). SPAK/OSR1 regulate NKCC1 and WNK activity: analysis of WNK isoform interactions and activation by T-loop trans-autophosphorylation. Biochem J 441, 325–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verissimo F, Jordan P (2001). WNK kinases, a novel protein kinase subfamily in multi-cellular organisms. Oncogene 20, 5562–5569. [DOI] [PubMed] [Google Scholar]
- Wang LC, Morgan LK, Godakumbura P, Kenney LJ, Anand GS (2012). The inner membrane histidine kinase EnvZ senses osmolality via helix-coil transitions in the cytoplasm. EMBO J 31, 2648–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson FH, Disse-Nicodeme S, Choate KA, Ishikawa K, Nelson-Williams C, Desitter I, Gunel M, Milford DV, Lipkin GW, Achard JM, et al. (2001). Human hypertension caused by mutations in WNK kinases. Science 293, 1107–1112. [DOI] [PubMed] [Google Scholar]
- Wood JM (2006). Osmosensing by bacteria. Sci STKE 2006, pe43. [DOI] [PubMed] [Google Scholar]
- Xu B, English JM, Wilsbacher JL, Stippec S, Goldsmith EJ, Cobb MH (2000). WNK1, a novel mammalian serine/threonine protein kinase lacking the catalytic lysine in subdomain II. J Biol Chem 275, 16795–16801. [DOI] [PubMed] [Google Scholar]
- Xu H, Hsu PH, Zhang L, Tsai MD, Freitas MA (2010). Database search algorithm for identification of intact cross-links in proteins and peptides using tandem mass spectrometry. J Proteome Res 9, 3384–3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu BE, Min XS, Stippec S, Lee BH, Goldsmith EJ, Cobb MH (2002). Regulation of WNK1 by an autoinhibitory domain and autophosphorylation. J Biol Chem 277, 48456–48462. [DOI] [PubMed] [Google Scholar]
- Yamada K, Park HM, Rigel DF, DiPetrillo K, Whalen EJ, Anisowicz A, Beil M, Berstler J, Brocklehurst CE, Burdick DA, et al. (2016a). Small-molecule WNK inhibition regulates cardiovascular and renal function. Nat Chem Biol 12, 896–898. [DOI] [PubMed] [Google Scholar]
- Yamada K, Zhang JH, Xie X, Reinhardt J, Xie AQ, LaSala D, Kohls D, Yowe D, Burdick D, Yoshisue H, et al. (2016b). Discovery and Characterization of Allosteric WNK Kinase Inhibitors. ACS Chem Biol 11, 3338–3346. [DOI] [PubMed] [Google Scholar]
- Yu J, Zhou Y, Tanaka I, Yao M (2010). Roll: a new algorithm for the detection of protein pockets and cavities with a rolling probe sphere. Bioinformatics 26, 46–52. [DOI] [PubMed] [Google Scholar]
- Zagorska A, Pozo-Guisado E, Boudeau J, Vitari AC, Rafiqi FH, Thastrup J, Deak M, Campbell DG, Morrice NA, Prescott AR, Alessi DR (2007). Regulation of activity and localization of the WNK1 protein kinase by hyperosmotic stress. J Cell Biol 176, 89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeniya M, Sohara E, Kita S, Iwamoto T, Susa K, Mori T, Oi K, Chiga M, Takahashi D, Yang SS, et al. (2013). Dietary salt intake regulates WNK3-SPAK-NKCC1 phosphorylation cascade in mouse aorta through angiotensin II. Hypertension 62, 872–878. [DOI] [PubMed] [Google Scholar]
- Zhou X, Naguro I, Ichijo H, Watanabe K (2016). Mitogen-activated protein kinases as key players in osmotic stress signaling. Biochim Biophys Acta 1860, 2037–2052. [DOI] [PubMed] [Google Scholar]
- Zhou HX, Rivas G, Minton AP (2008). Macromolecular crowding and confinement: biochemical, biophysical, and potential physiological consequences. Annu Rev Biophys 37, 375–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Zhang ZY (1999). Mechanism of mitogen-activated protein kinase phosphatase-3 activation by ERK2. J Biol Chem 274, 35526–35534. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.