Abstract
Basically, all mammalian tissues are constantly exposed to a variety of environmental mechanical signals. Depending on the signal strength, mechanics intervenes in a multitude of cellular processes and is thus capable of inducing simple cellular adaptations but also complex differentiation processes and even apoptosis. The underlying recognition typically depends on mechanosensitive proteins, which most often sense the mechanical signal for the induction of a cellular signaling cascade by changing their protein conformation. However, the fate of mechanosensors after mechanical stress application is still poorly understood, and it remains unclear whether protein degradation pathways affect the mechanosensitivity of cells. Here, we show that cyclic stretch induces autophagosome formation in a time-dependent manner. Formation depends on the cochaperone BAG family molecular chaperone regulator 3 (BAG3) and thus likely involves BAG3-mediated chaperone-assisted selective autophagy. Furthermore, we demonstrate that strain-induced cell reorientation is clearly delayed upon inhibition of autophagy, suggesting a bidirectional cross-talk between mechanotransduction and autophagic degradation. The strength of the observed delay depends on stable adhesion structures and stress fiber formation in a Ras homologue family member A (RhoA)-dependent manner.
INTRODUCTION
Mammalian tissues are constantly exposed to environmental mechanical signals generated by respiration, heart and muscle contraction, body movement, or pulsating blood flow. The resulting environmental strain indispensably has to be recognized by tissues in order to ensure tissue integrity, development, and survival (Moore et al., 2010; DuFort et al., 2011; Hoffman et al., 2011; Yusko and Asbury, 2014). Mechanosensitivity and thereby induced cellular responses thus represent vital mechanisms at the single cell as well as the multicellular level (Discher et al., 2005; Janmey and Miller, 2011).
Different strain frequencies and wave forms as well as amplitudes of up to 20% and more can be naturally measured to induce reorientation of the cytoskeleton as well as the whole cell body away from the strain direction in the case of uniaxial strain (Hayakawa et al., 2000, 2001; Neidlinger-Wilke et al., 2002; Jungbauer et al., 2008; Tondon et al., 2012; Noethel et al., 2018; Zielinski et al., 2018). This mechanism has been identified for most adherent cell types on single cell and monolayer levels and is primarily caused by actin filament rupture upon straining above a certain threshold (Faust et al., 2011). Reorientation away from the strain direction therefore leads to a reduction of strain in the living system to regain cellular homeostasis (DuFort et al., 2011; Yusko and Asbury, 2014). The processes of strain sensing and subsequent signal transduction depend on multiple elementary processes, including mechanically activated ion channels, tension-dependent conformational changes of proteins, and tension-dependent binding affinities (Kanzaki et al., 1999; Hayakawa et al., 2008, 2011).
Most intensively characterized is strain-induced conformational unfolding of mechanosensory proteins. This unfolding leads to subsequent exposure of previously hidden binding domains for recruitment of binding proteins or phosphorylation to induce signaling pathways and subsequent cellular responses (Bakolitsa et al., 2004; Izard and Vonrhein, 2004; Sawada et al., 2006; Humphries et al., 2007; del Rio et al., 2009; Carisey et al., 2013). Because mechanosensation primarily defines a cellular protective mechanism, most mechanosensitive proteins are positioned at sites that experience mechanical load. Thus, a considerable number of mechanosensitive proteins are found in focal adhesions (FAs) and cell–cell adhesions as well as at the actin cytoskeleton itself (Riveline et al., 2001; Hynes, 2002; Yonemura et al., 2010; Hoffman et al., 2012; Rognoni et al., 2012). Of the proteins forming the integrin adhesome (Zaidel-Bar et al., 2007), strain-induced unfolding and subsequent binding or phosphorylation of previously hidden domains was demonstrated, especially for those with multiple protein binding sites such as talin, vinculin, paxillin, FA kinase (FAK), or p130Cas (Sawada et al., 2006; del Rio et al., 2009; Michael et al., 2009; Pasapera et al., 2010; Carisey et al., 2013). However, very little is known about the fate of unfolded mechanosensory proteins after stress release. For some proteins, such as talin, cycles of unfolding and refolding have been demonstrated (Yao et al., 2016). However, it remains unclear whether this refolding always occurs to a fully functional initial state and whether complete refolding is also possible at high strain amplitudes. It appears likely that at least in some instances a protein quality control system will become engaged, involving the recognition of strain unfolded proteins by molecular chaperones, chaperone-assisted refolding, or targeting onto protein degradation pathways (Arndt et al., 2010; Ulbricht et al., 2013; Balchin et al., 2016; Dikic, 2017; Kathage et al., 2017).
With autophagy, an important lyososome-mediated process for recycling of cytosolic cargoes is known (Yu et al., 2018) and a number of stress conditions are known to stimulate this pathway. Also for mechanical stress several studies indicate a close link between the regulation of FA or actin cytoskeletal stability and various selective autophagy pathways (Kenific et al., 2016b). Specifically, studies on the actin-associated mechanosensory protein filamin (Rognoni et al., 2012) have shown its recognition by the cochaperone BAG family molecular chaperone regulator 3 (BAG3) and subsequent degradation by the chaperone-assisted selective autophagy (CASA) (Arndt et al., 2010; Ulbricht et al., 2013; Ulbricht et al., 2015; Kathage et al., 2017). At least for stressed skeletal muscle cells, recognition of filamin additionally has been shown to depend on direct interaction with filamin-A–interacting protein 1 (FILIP1). This interaction induces the degradation of filamin but is prohibited by filamin phosphorylation (Reimann et al., 2020).
Further evidence for a critical involvement of autophagy in the turnover of mechanosensors has been obtained in a study that revealed degradation of the FA core protein paxilin in cancer cells dependent on the Src-kinase and the autophagic membrane marker microtubule-associated proteins 1A/1B light chain 3B (LC3B) (Sharifi et al., 2016). Furthermore, FAK depletion in cancer cells is associated with autophagy-associated degradation of active Src. At the same time, FAK is known to be a negative regulator of beclin-1–dependent autophagy (Sandilands et al., 2011; Cheng et al., 2017).
On the basis of these findings, we address the question of whether induction of cyclic stretch–induced cell reorientation causes increased autophagic activity. We report a strain-induced reorientation process that is dependent on BAG3-mediated autophagosome (AP) formation. Furthermore, Ras homologue family member A (RhoA) activation and therefore reinforced mechanosensitive cell structures further strengthen this response, revealing a tight interplay between mechanoadaptation and autophagic protein degradation.
RESULTS
AP formation can be mechanically induced by cyclic stretch
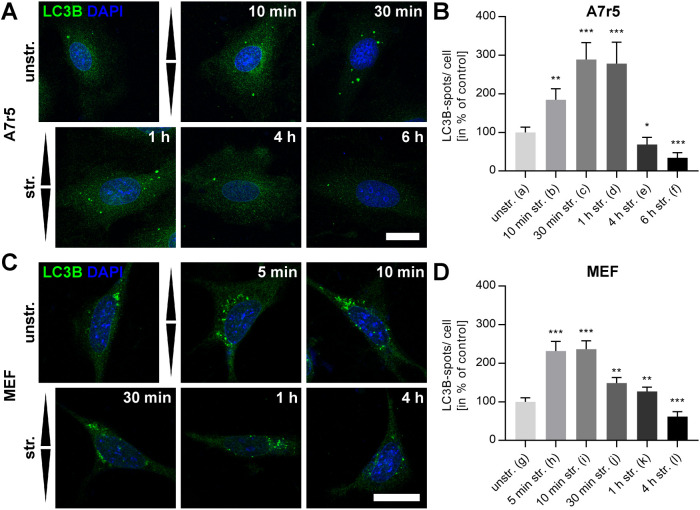
To address the overall capability of mammalian cells to produce APs, rat smooth muscle (A7r5) cells and mouse embryonic fibroblast (MEF) cells were transfected with green fluorescent protein (GFP)-LC3B as autophagy marker protein on glass (Figure 1A). In transfected A7r5 and MEF cells, LC3B-positive vesicular structures were readily detectable. To enable stress application by mechanical straining, cells were cultivated on elastomeric silicone rubber substrates. After 1 h of cyclic stretch, A7r5 and MEF cells exhibited increased numbers of APs compared with unstretched control cells (Figure 1B). Blocking the lysosomal proteolysis of APs by chloroquine (CQ) and subsequent immunostaining against LC3B showed a significant increase (A7r5: p < 0.0001, MEF: p < 0.0001) of LC3B spots in both cell types, with an even stronger accumulation in A7r5 cells. Furthermore, due to a better signal-to-noise ratio, we decided to perform immunostainings of LC3B instead of transfection of GFP-LC3B constructs for all subsequent quantitative analyses.
FIGURE 1:
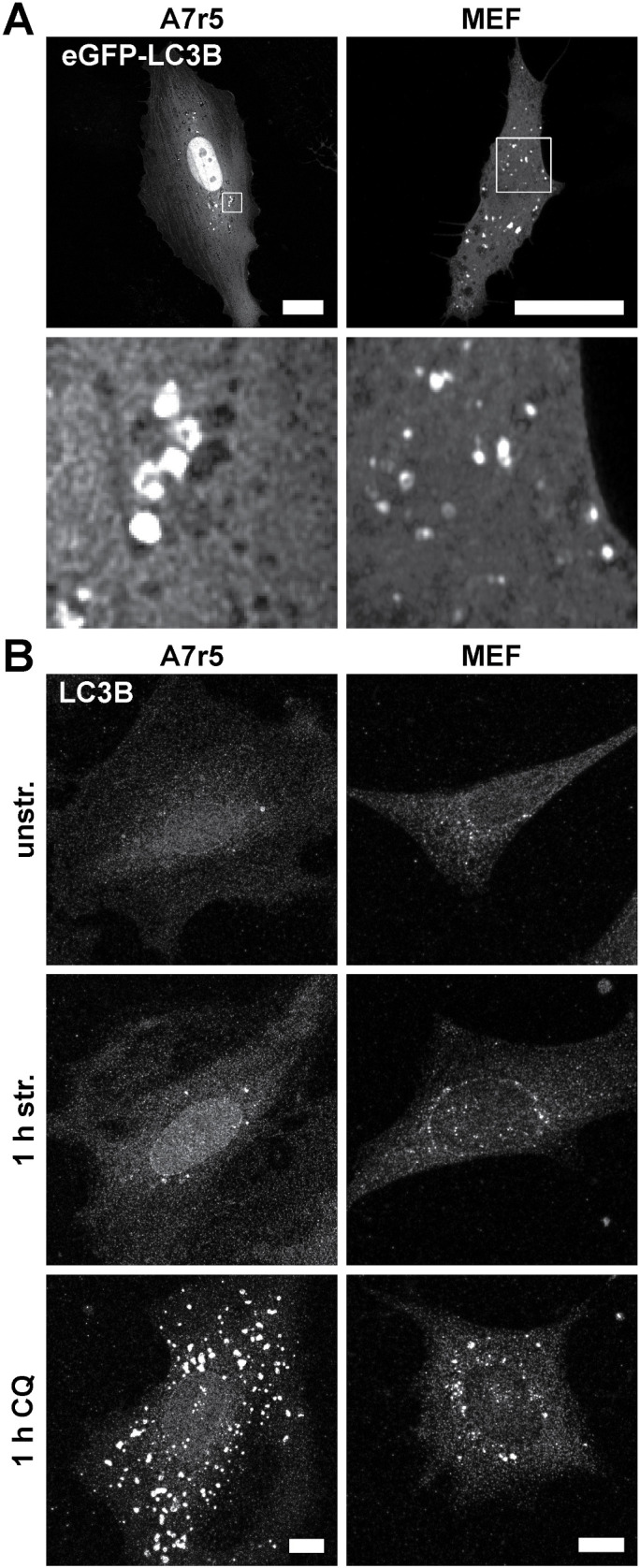
Mammalian cells have different capacities to produce APs. (A) GFP-LC3B–transfected A7r5 and MEF cells cultivated on FN-coated glass showed LC3B spots as markers for APs 24 h after transfection. Scale bar is 20 µm. (B) LC3B spots could be induced in A7r5 and MEF cells by mechanical cyclic stretch (str.) shown by immunostainings of LC3B after fixation. Further, LC3B spots accumulated by blocking lysosomal proteolysis of APs with CQ. Control cells were cultivated on 50 kPa elastomeric substrates coated with FN in the absence of stretch (unstr.). The indicated treatments were performed for 1 h. Scale bar is 10 µm. Note that for all subsequent LC3B quantifications, immunostainings at the indicated time points were performed because of the better signal-to-noise ratio.
Stretch-induced autophagy is a time-dependent process
Because cyclic strain induces cellular reorientation as an adaptation mechanism to regain stable homeostasis, we next quantified stretch-induced AP formation over time for A7r5 and MEF cells (Figure 2). A7r5 cells revealed only low numbers of LC3B spots under unstretched conditions (1.2 spots per cell), which significantly (p < 0.001) increased by almost 100% already after 10 min of cyclic uniaxial stretch with an amplitude of 20% and a frequency of 300 mHz as applied in all subsequent stretch experiments. Highest numbers of APs were identified after 30–60 min, with an almost threefold increased number (Figure 2, A and B). Interestingly, application of long cyclic stretching induced a reverse behavior, with a reduction of AP numbers to numbers significantly (p < 0.0001) below those found for the unstretched control (1/3 after 6 h). An identical significant trend could also be identified for MEF cells. However, increase and reduction showed an accelerated time course with the maximum number of LC3B spots already after 5–10 min and an immediately following reduction, which also ended up at significantly (p < 0.0001) lower numbers of APs after 4 h (60%) compared with control conditions before stretch was applied (n = 16) (Figure 2, C and D). In both cell types, a transient induction of autophagy apparently accompanies the adaptation to cyclic uniaxial strain.
FIGURE 2:
LC3B spots can be mechanically induced by cyclic stretch. (A, C) LC3B (green) immunostainings display a time-dependent increase in the number of LC3B spots in A7r5 and MEF cells after cyclic stretch (str.) for the indicated time on FN-coated elastomeric substrates or for unstretched (unstr.) control. Arrowheads illustrate stretch direction. Scale bar is 20 µm. (B, D) Quantification of LC3B spots per cell shown in A and C in the percent of unstretched control is given for all time points. Data represent mean values ± SEM of analyzed cells (n). Significances are given for all values in comparison to unstretched control (na = 72, nb = 54, nc = 55, nd = 50, ne = 54, nf = 55, ng = 72, nh = 74, ni = 75, nj = 69, nk = 71, nl = 66).
Cyclic strain–induced mechanoresponse is dependent on autophagy
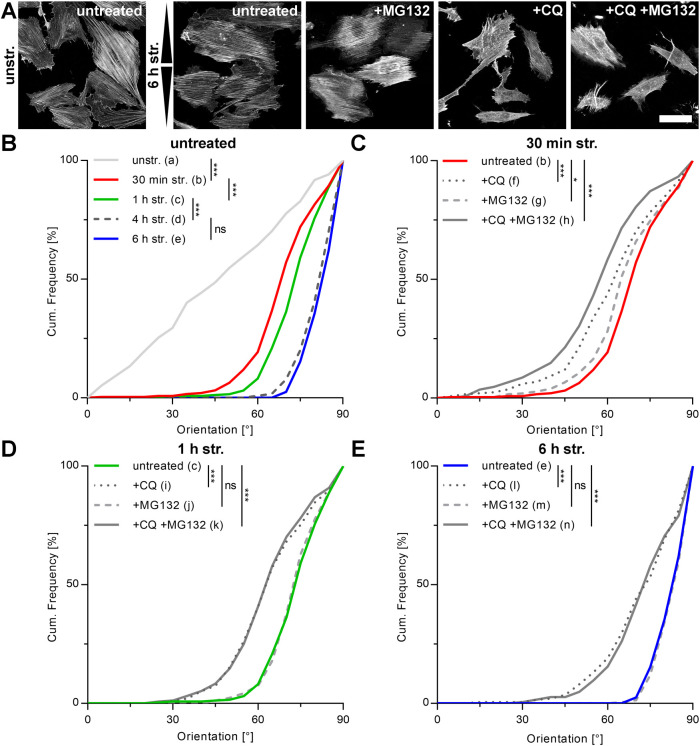
The time course of APs formed under cyclic strain suggested a relationship to cytoskeletal reorientation behavior. To analyze this hypothesis in more detail, we characterized actin orientation based on actin filament gray value gradients in A7r5 and MEF cells over time in the presence of CQ, MG132, or both (Figures 3 and 4). Cumulative frequency plots for actin stress fiber main orientation per cell showed for A7r5 cells an equal distribution of all angles under unstretched conditions (Figure 3, A and B). Cyclic strain induced reorientation in the perpendicular direction relative to strain under control conditions. Here, beginning with a very rapid response within the first 30 min, main actin orientation angles per cell approached their maxima in the range of 68°–82° relative to stretch direction within 4 h. This orientation remained nearly stable even with further prolonged cyclic straining (Figure 3, A and B). Strain-induced cytoskeletal dynamics therefore correlated well with AP formation. Blocking proteasome activity by MG132 showed no effect on the time course of actin reorientation (Figure 3, A and C–E). In contrast, the addition of CQ strongly impaired the cellular mechanoresponse. Here, not only the reorientation behavior was slowed down at every time point analyzed, but also the final average reorientation steady state after 6 h was shifted by more than 10° from 83° for untreated control conditions to 71° upon CQ supplementation (Figure 3, A and C–E). Parallel supplementation with CQ and MG132 resulted in almost identical impairments in mechanoresponse (72°) as found for CQ treatment only, confirming the autophagy-specific influence on mechanosensitivity and response.
FIGURE 3:
Upon stretch, A7r5 actin fibers reorient perpendicular to the direction of stretch, which can be decelerated by blocking lysosomal proteolysis of APs with CQ. (A) Immunofluorescence micrographs of actin cytoskeleton after 6 h of cyclic stretch (str.) after 1 h pretreatment with MG132 and/or CQ to inhibit proteasomal and lysosomal degradation, respectively. Cells were cultivated on FN-coated elastomeric substrates and pretreated with the mentioned inhibitors 1 h before stretch. Control cells were cultivated unstretched (unstr.) in normal growth medium. Arrowheads illustrate stretch direction. Scale bar is 50 µm. (B–E) After fixation and staining, actin fibers were evaluated as angular distribution from 0° to 90° to the direction of stretch and are plotted as cumulative frequencies of all analyzed cells (n). Plots are given for (B) untreated cells, for time points from 30 min to 6 h of cyclic stretch, and for (C) 30 min, (D), 1 h, and (E) 6 h cyclic stretch in the presence and absence of the mentioned inhibitors (na = 292, nb = 254, nc = 259, nd = 244, ne = 203, nf = 249, ng = 256, nh = 257, ni = 255, nj = 256, nk = 252, nl = 199, nm = 219, nn = 187).
FIGURE 4:
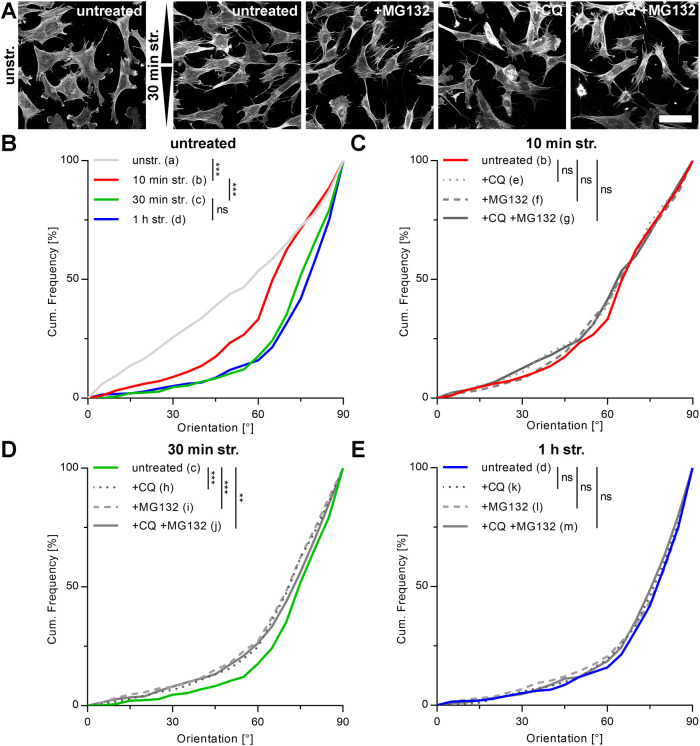
Cyclic stretch in MEF cells leads to time-restricted actin fiber reorientation perpendicular to the direction of stretch, which can be slightly decelerated by blocking autophagy and/or proteasomal degradation. (A) Immunofluorescence micrographs of actin cytoskeleton in the presence and absence of the proteasome inhibitor MG132 and/or the lysosomal proteolysis inhibitor CQ after 30 min of cyclic stretch (str.). Cells were pretreated with the mentioned inhibitors 1 h before stretch. Control cells were cultivated in normal growth medium on FN-coated elastomeric substrates without stretch (unstr.). Arrowheads illustrate stretch direction. Scale bar is 50 µm. (B–E) Subsequent to fixation and staining, actin fibers were evaluated as angular distribution from 0° to 90° to the direction of stretch and are plotted as cumulative frequencies of all analyzed cells (n). Plots are shown for (B) untreated control cells at all time points and for (C) 10 min, (D) 30 min, and (E) 1 h of cyclic stretch in the presence and absence of the indicated inhibitors (na = 336, nb = 280, nc = 437, nd = 396, ne = 304, nf = 287, ng = 270, nh = 451, ni = 428, nj = 438, nk = 455, nl = 428, nm = 429).
Identical experiments on MEF cells showed a much faster reorientation behavior upon cyclic straining for control conditions. Here, already after 30 min main actin angles reached stable distributions that remained largely unchanged with further stretching (Figure 4, A and B). These data correlate well with the much faster LC3B-spot induction upon strain application in MEF cells shown before. Furthermore, with an average actin reorientation angle of 72°, reorientation was less pronounced in MEF compared with A7r5 cells. Interestingly and in contrast to A7r5 cells, neither CQ-, MG132-, nor CQ/MG132-treated MEF cells were impaired in steady state reorientation distribution after 1 h of stretch (Figure 4E). The rate of cellular reorientation toward this reorientation distribution was also similar in the presence of inhibitors and was delayed only after 30 min of stretch (Figure 4, C and D). Ongoing autophagic degradation is apparently a prerequisite for cytoskeleton reorientation in smooth muscle cells but not in embryonic fibroblasts under standard growth conditions.
RhoA activation shifts MEF cells to an autophagy-dependent mechanoresponse
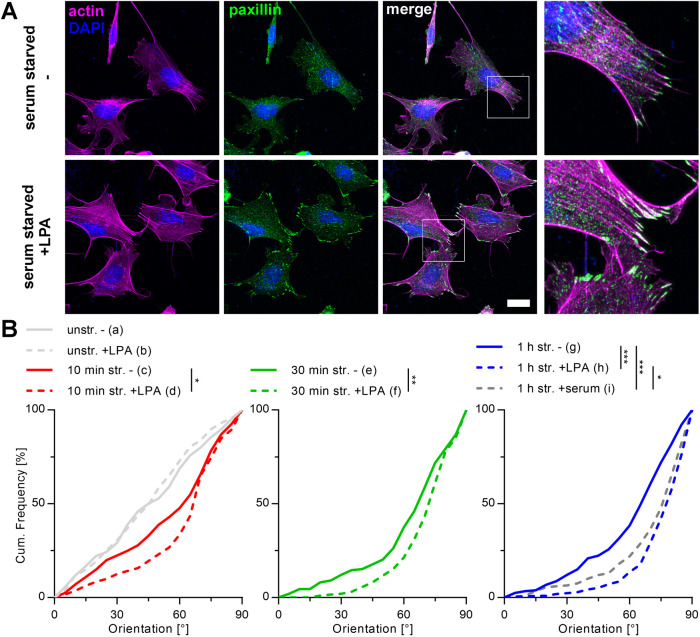
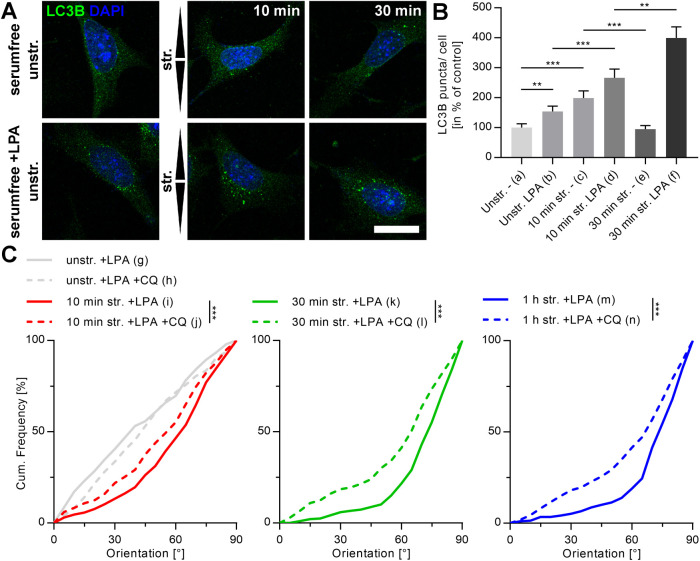
To further analyze the interdependence between autophagy, the spatial organization of the mechanosensitive apparatus, and the mechanoresponse, we next activated RhoA in MEF cells by lysophosphatidic acid (LPA) to increase cell contractility. Efficient activation was confirmed after 30 min of treatment by strongly reinforced FAs and clear stress fiber formation (Figure 5A). When cyclically strained, such cells reoriented significantly faster compared with nontreated MEF cells. This resulted in a stronger overall reorientation behavior in the perpendicular direction relative to stretch with an average reorientation angle for LPA-treated MEF that was similar to that for A7r5 cells (78°) (Figure 5B). Starting from an already massively formed stress fiber network and stable FAs, the same experiments performed on A7r5 smooth muscle cells resulted in neither any further cytoskeletal reinforcement nor changes in strain-induced mechanoresponse (Supplemental Figure S1). In addition, the actin dynamics of A7r5 cells after reorientation due to 4 h of stretch application remains active as shown by a slow loss of reorientation order over time. Furthermore, we could even invert actin reorientation by cyclic stretch in the orthogonal direction after fully completed first reorientation (Supplemental Figure S2). Vice versa, the use of very soft 5 kPa stretching chambers visibly reduced actin stress fiber intensity and order in A7r5 cells to result in a slowed but still detectable reorientation under strain. Interestingly, AP formation to significantly higher numbers in the presence of strain remained unaltered (Supplemental Figure S3).
FIGURE 5:
RhoA activation in MEF cells reinforces actin stress fibers and FAs, thus promoting stronger actin reorientation. (A) Actin (magenta) and paxillin (green) micrographs of MEF cells cultivated on FN-coated elastomeric substrates in the presence and absence of LPA. Cells were fixed and immunostained after 30 min incubation with LPA. Scale bar is 20 µm. (B) Actin reorientation of MEF cells after 10 min, 30 min, and 1 h of cyclic stretch (str.) after 30 min pretreatment with LPA in serum-free medium. Control cells were cultivated in serum-free medium (–). After fixation and staining, the actin fibers were evaluated as angular distribution from 0° to 90° to the direction of stretch and are plotted as cumulative frequencies of all analyzed cells (n) (na = 144, nb = 203, nc = 201, nd = 191, ne = 198, nf = 197, ng = 206, nh = 212, ni = 200).
To further test whether a reinforced cytoskeleton also induces mechanoresponse dependence on autophagy in MEF cells, the formation of APs after RhoA activation was analyzed in a first step. Here, an increased formation of LC3B spots was already clearly visible under unstretched control conditions. When cyclic stretch was applied, this effect was further significantly increased (Figure 6, A and B). In addition, while in untreated cells AP formation peaked at stretch durations of 5–10 min (Figures 2D and 6B), RhoA activation shifted this peak to later time points with fourfold increased numbers of LC3B spots after 30 min. Such time courses resembled those found for A7r5 cells.
FIGURE 6:
RhoA-activated MEF cells indicate higher numbers of stretch-induced LC3B spots, and actin reorientation is decelerated by blocking their lysosomal degradation. (A) LC3B (green) immunostainings after 10 and 30 min of cyclic stretch (str.) and unstretched control (unstr.) in the presence and absence of LPA 30 min before cyclic stretch. Cells were cultivated on FN-coated elastomeric substrates. Arrowheads illustrate direction of stretch. Scale bar is 20 µm. (B) Quantification of LC3B spots per cell shown in A in percent to unstretched control in the absence of LPA. Data represent mean values ± SEM of all analyzed cells (n). (C) Actin reorientation of RhoA-activated MEF cells after 10 min, 30 min, and 1 h cyclic stretch in the presence and absence of CQ to inhibit lysosomal degradation of APs. Angle distributions of actin fibers were evaluated (0° is the direction of stretch) and are displayed as cumulative frequency plots of all analyzed cells (n) (na = 98, nb = 73, nc = 50, nd = 58, ne = 52, nf = 49, ng = 303, nh = 292 ni = 301, nj = 315, nk = 286, nl = 313, nm = 290, nn = 321).
Owing to observed increased numbers of stretch-induced LC3B spots in RhoA-activated MEF cells, we next analyzed whether inhibition with CQ would now also impair mechanoresponsive behavior as we could observe before for A7r5 cells. Indeed, actin reorientation was significantly decelerated for all measured time points in comparison to untreated control (Figure 6C). Furthermore, almost identical reorientation distributions for MEF cells after 1 h compared with the corresponding distributions after 30 min argue that not only the reorientation time course but also the reorientation steady state distribution were impaired upon AP block.
Initially observed differences between A7r5 and MEF cells in the sensitivity of actin reorientation toward autophagy inhibition thus cannot be attributed to distinct mechanoresponses but seem to reflect lower force generation in MEFs under standard growth conditions. Upon elevated force generation, actin reorientation in MEFs depends on ongoing autophagy similar to initial findings for A7r5 cells. Our work points at a force-dependent interplay between mechanoresponse and autophagy conserved between different cell types.
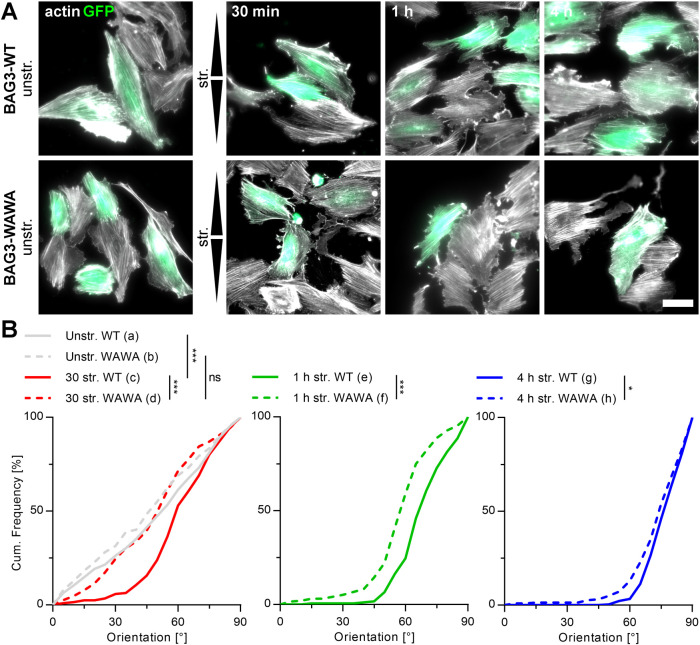
Actin reorientation in A7r5 cells is dependent on BAG3-mediated autophagy
Because earlier work had already identified CASA as a strain-induced autophagy pathway, we determined the contribution of CASA to the observed mechanoresponse to cyclic stretch. For this purpose, we compared actin reorientation of A7r5 cells upon transient expression of either BAG3-wild type (WT) or its inactive, dominant-negative form, carrying tryptophan-to-alanine substitutions at positions 26 and 49 (WAWA). The mutant variant contains alanine substitutions in the WW domain of BAG3, which disrupt the interaction of the cochaperone with several of its binding partners (Ulbricht et al., 2013; Kathage et al., 2017). For identification, cells were cotransfected with GFP-plasmids and only GFP-expressing cells were subsequently analyzed (Figure 7A). The expression of neither BAG3-WT nor BAG3-WAWA caused changes of A7r5 cell morphology, characterized by uniform distributions of all actin orientation angles under unstretched conditions (Figure 7, A and B). However, BAG3-WAWA–expressing A7r5 cells displayed a strongly decelerated actin reorientation after 30 min and 1 h of cyclic stretch compared with BAG3-WT cells. The mechanosensitivity of BAG3-WAWA cells was reduced to such an extent that no response could be detected after 30 min compared with that of control BAG3-WT cells. Also steady state distributions after long cyclic straining of 4 h pointed to a significantly reduced mechanoresponse in the absence of functional BAG3 (Figure 7B).
FIGURE 7:
Stretch-induced actin reorientation in A7r5 cells is BAG3 mediated. (A) Micrographs of GFP- and BAG3-WT– or BAG3-WAWA–cotransfected A7r5 cells (green) on FN-coated elastomeric substrates after 30 min, 1 h, and 4 h of cyclic stretch (str.). Control cells were cultivated unstretched (unstr.). After transfection the cells were cultivated for 24 h, stretched for the indicated times and subsequently fixed and actin fibers (white) immunostained. Arrowheads illustrate direction of stretch. Scale bar is 50 µm. (B) Actin fiber reorientation under conditions mentioned in A were evaluated as angular distribution from 0° to 90° to the direction of stretch; cumulative frequency plots are given for 30 min, 1 h, and 4 h of cyclic stretch of all analyzed BAG3-WT and BAG3-WAWA A7r5 cells (n) (na = 298, nb = 242, nc = 206, nd = 192, ne = 183, nf = 195, ng = 275, nh = 223).
DISCUSSION
Mechanosensing of single cells highly depends on FAs connected to the actin cytoskeleton (Zaidel-Bar et al., 2007). Here, various proteins have been shown to undergo conformational changes under tension to enable transfer of mechanical signals into a chemical signal cascade. In the case of strain these cascades can subsequently trigger disassembly (Ehrlicher et al., 2011), induced dynamics (Sigaut et al., 2018), and reinforcement of structures (Marshall et al., 2003; Yakovenko et al., 2008). This is specifically evident in the cytoskeletal and cellular reorientation in a perpendicular direction to strain as a nearly universal response to cyclic stretch (Hayakawa et al., 2000; Noethel et al., 2018; Zielinski et al., 2018). However, it has remained largely unclear how cellular systems can distinguish different strengths of the mechanical signal, because for only a few proteins, such as p130Cas or fibronectin (FN), a stepwise force-dependent unfolding behavior has been shown (Paci and Karplus, 1999; Lu et al., 2013). For other mechanosensors such as talin, repeated unfolding and refolding has been demonstrated (Yao et al., 2016). Nevertheless, it is still uncertain whether frequently repeated unfolding/refolding cycles or very intense protein unfolding due to high strain amplitudes actually always allow error-free refolding. This is also important because another study additionally showed that stretching of cells caused locally distinctly different deformations, whereby proteins in the contact area between FAs and actin filaments were exposed to significant elongation (Kirchenbuchler et al., 2010). This behavior suggests that even at low and intermediate strain amplitudes, certain proteins become significantly more unfolded, potentially to the point of irreversible structural change.
Moreover, it remained largely unclear whether mechanosensitivity and subsequent cell responses are associated with the degradation of specific proteins. For phosphorylated p130Cas, specific regulation by the ubiquitin–proteasome system has been demonstrated in FAs of motile cells (Teckchandani and Cooper, 2016). Paxillin, on the other hand, is able to interact directly with LC3 to trigger the degradation of FAs via autophagy in tumor cells (Sharifi et al., 2016). Autophagy has also been shown to influence a number of other adhesion and migration processes (Kenific et al., 2016b). Here, NBR1-mediated selective autophagy appears to be of particular importance for motile cells (Kenific et al., 2016a). Consistent with its function as an autophagy cargo receptor, NBR1 biochemically interacts with FA mechanosensors vinculin and zyxin and promotes the targeting of APs to dynamic FAs at the leading edge of migrating cells. Here, binding between NBR1 and mechanosensors is induced by their tension-dependent conformational unfolding. A similar situation is also assumed for binding of NBR1 to the mechanosensor titin in sarcomeres (Lange et al., 2005). However, whether these bindings involve the recognition of irreversibly unfolded mechanosensors or are a simple regulatory mechanism of FA or sarcomer dynamics remains unclear.
In contrast, a clear link between misfolding and autophagy has been demonstrated for the protein filamin in both skeletal muscle (Arndt et al., 2010; Kathage et al., 2017) and smooth muscle (Ulbricht et al., 2013, 2015) cells. Filamin binds to FAs in a tension-dependent manner and induces FA reinforcement (Gehler et al., 2009). In addition, unfolded filamin was shown to be bound by the cochaperone BAG3 to subsequently recruit the CASA machinery (Ulbricht et al., 2013). While different hard substrate elasticities were used in that work to regulate cellular tension, our studies for cyclic stretch demonstrated the strong link between autophagy, mechanosensitivity, and subsequent cell response even on soft substrates under cyclic stretch. Elasticities of 50 kPa that mimic the in vivo elasticities of, for example, basement membranes (Tilleman et al., 2004; Pawlaczyk et al., 2013) but also the use of 10 times softer substrates confirm this interplay. In this context, it is also interesting that Kim et al. (2013) postulate a kinetic context between force-induced protein unfolding and chaperone binding. According to this, the rate of domain refolding of mechanosensors correlates with the rate of chaperone binding to possibly induce the recruitment of degradation factors in the case of slow protein refolding. Slowed domain refolding by several minutes at increased forces even when the applied force was reduced has already been shown for filamin (Rognoni et al., 2014). This kinetic-based hypothesis could thus also explain why static stretch and cyclic straining similarly induce AP formation.
In contrast, we could not see any effect on actin reorientation and therefore mechanoresponse by blocking proteasomal degradation pathways. In turn, proteasomal inhibition is known to up-regulate autophagy by transcriptional up-regulation of BAG3 (Wang et al., 2008), thus amplifying BAG3 necessity also for mechanoresponse. Our analyses with a dominant-negative BAG3 mutant furthermore established the mutual dependence on BAG3. However, it still needs to be shown at this stage whether BAG3 also recruits the CASA machinery after cyclic straining or whether other autophagy factors are involved to induce protein degradation (Gamerdinger et al., 2011). Nevertheless, the BAG3 mutant used here with the inactivated WW domain and thus blocked interaction to SYNPO PPxY motifs (Ji et al., 2019), which is required to induce autophagy (Merabova et al., 2015), argues for a regulatory mechanism similar to that described for filamin.
Interestingly, the induction of autophagy upon application of cyclic stretch occurred in a time-dependent manner with a sharp increase in AP number and subsequent decrease to numbers clearly less than those of unstretched cells. This time-dependent behavior clearly correlates with the time course of cytoskeletal reorientation (Hayakawa et al., 2000, 2001; Faust et al., 2011; Zielinski et al., 2018) and strongly suggests that indeed cellular stress levels and thus mechanical deformation of proteins trigger autophagy. Reorientation, and thus minimization of cellular elongation and stress (Faust et al., 2011), might therefore subsequently also reduce autophagy processes that are below control levels due to homogeneously aligned cells. This hypothesis is further supported by the fact that the strength of the mechanosensitive apparatus made of FAs and stress fibers is associated with the influence of autophagy on mechanosensitivity and response as seen for the different cell types used here. Also, activation of RhoA and thus reinforcement of mechanosensitive structures (Nobes and Hall, 1995) confirmed that both autophagy-dependent mechanoresponse and number of mechanically induced APs depend on the strength of the mechanosensitive apparatus. It therefore seems that with increasing cytoskeletal order from MEF to smooth muscle cells and ultimately skeletal muscles (Ulbricht et al., 2013), the correlation between autophagy and response to mechanical stress becomes increasingly important while AP formation itself is present in all of these cell types. With BAG3 and SYNPO, just first regulators have been identified and further experiments are necessary to identify possibly even cell type–specific regulators. Interestingly, a p62-dependent degradation mechanism via autophagy has also been described for activated RhoA itself (Belaid et al., 2013). Whether this effect needs to be integrated into a deeper understanding of the regulatory mechanism identified here is unclear.
While induction of autophagy under cyclic stretch is readily explicable, we can only speculate how blocking lysosomal degradation of APs using CQ (Mauthe et al., 2018) negatively affects the mechanoresponse of cells. One possibility might be a concentration dilution of important autophagy factors that recognize unfolded mechanosensors and prime them for subsequent degradation. Such a mechanism could result in misfolded proteins that remain in FAs and on the cytoskeleton and thus in a deficient mechanoresponse. Further studies also need to identify which proteins in particular are degraded by the induction of autophagy under cyclic stretch and which proteins regulate cyclic stretch–dependent autophagy. This will help to understand more precisely the BAG3-dependent regulatory mechanism of mechanoresponse revealed here.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Cell culture
MEF and rat smooth muscle (A7r5) cells were purchased from the American Type Culture Collection and cultivated under sterile conditions at 37°C and 5% (vol/vol) CO2 in a humidified atmosphere. MEF cells were cultivated in DMEM/GlutaMAX medium (Life Technologies, Carlsbad, CA) supplemented with 10% (vol/vol) fetal bovine serum (FBS) (Life Technologies) and 1% penicillin/streptomycin (Sigma, St. Louis, MO). A7r5 cells were cultivated in DMEM/low glucose/pyruvate/no glutamine, no phenol red medium (Life Technologies) supplemented with 10% (vol/vol) FBS Premium (P30-3302; Pan Biotech, Aidenbach, Germany), 1% (vol/vol) penicillin/streptomycin (Sigma) and 2 mM GlutaMAX (Thermo Fisher, Waltham, MA). Murine keratinocytes were cultivated as described before (Noethel et al., 2018).
Elastic substrate preparation
Elastic silicone substrates for stretch experiments were prepared and calibrated as described earlier (Faust et al., 2011). Cell straining experiments were performed on 50 kPa elastomeric stretching chambers (Sylgard 184; Dow Corning GmbH; mixing ratio of 1 to 40 cross-linker to base) to mimic natural elasticities except as otherwise stated. For some experiments, elastomeric chambers were overlaid with an additional layer of 5 kPa soft elastomer (mixing ratio of 1 to 60, thickness of approximately 500 µm). Elasticities were determined by indentation as described before (Ulbricht et al., 2013). To promote cell attachment, chambers were coated with human FN (20 µg/ml) (Corning, Tewksbury, MA) in phosphate-buffered saline at 37°C for 1 h before cell seeding. Cells were seeded for single cells 18 h before cell straining at a density of 10,000 cells/cm2 for MEF cells and 3000 cells/cm2 for A7r5 cells. When indicated, cells were cultivated on glass substrates. Therefore, glass coverslips (1.5# high-precision coverslips with a thickness of 170 ± 5 µm; Paul Marienfeld, Lauda-Königshofen, Germany) were fitted underneath Petri dishes with a predrilled hole for confocal imaging. Glass substrates were coated with FN as described above.
Cell culture supplementation
The lysosomal proteolysis inhibitor CQ diphosphate salt (Sigma) and the proteasome inhibitor MG132 (Peptanova, Sandhausen, Germany) were used, respectively. Medium was supplemented with 100 µM CQ and/or 10 µM MG132 1 h before straining experiments. Control cells were incubated in normal growth medium. For Rho-kinase activation LPA (Santa Cruz Biotechnology; Santa Cruz, CA) was utilized to increase cell contractility. Preincubation in 50 µM LPA took place 30 min before straining. LPA concentration was kept constant during stretching. All LPA experiments were performed in serum-free medium.
Cell straining
Uniaxial stretch was applied to six elastic substrates simultaneously as described before (Noethel et al., 2018) adapted from a single chamber stretcher previously used (Faust et al., 2011). To mimic physiological conditions that also induce cytoskeletal reorientation in vivo (Wong et al., 1983), a cyclic stretch amplitude of 20% was applied. As strain frequency, 300 mHz was applied as used before in other studies (Hayakawa et al., 2000). Such a frequency minimized medium flow in the chambers upon stretching and therefore ensured homogeneous cell response.
Plasmid transfection
Transfection with GFP-LC3B–, BAG3-WT–, BAG3-WAWA–,and GFP-plasmid DNA was performed by use of Lipofectamine 3000 (Thermo Fisher) according to the manufacturer’s protocol directly on the elastomeric substrates. For each substrate, 500 ng of DNA was used. All plasmids used here are already published (Ulbricht et al., 2013; Kathage et al., 2017).
Immunocytochemistry
Cells for LC3B immunocytochemistry were fixed in ice-cold methanol (VWR, Radnor, PA) for 10 min at –20°C, subsequently washed twice with cytoskeleton buffer (CB; 150 mM NaCl, 5 mM MgCl2, 5 mM ethylene glycol-bis(β-aminoethylether)-N,N,N’,N’-tetraacetic acid (EGTA), 5 mM glucose, 10 mM 2-(N-morpholino)ethanesulfonic acid, pH 6.1) at room temperature (22°C), as were all the following incubations unless otherwise indicated. Staining of paxillin and actin fibers was performed after fixation in 3.7% (vol/vol) paraformaldehyde in CB for 15 min at 37°C. After fixation, the cells were washed twice with 100 mM glycine in CB. Cell membranes were permeabilized for 10 min with 0.05% (vol/vol) Triton X-100 (Sigma) in CB, rinsed three times with CB, and afterward blocked for 1 h with 5% milk powder (Roth, Karlsruhe, Germany). Subsequently, cells were incubated with 0.2% (vol/vol) anti-LC3B antibody from rabbit (2775; Cell Signaling, Danvers, MA) or 0.2% (vol/vol) anti-paxillin antibody from mouse (Invitrogen; AHO0492) in 1% milk powder in CB at 4°C for 16 h. Unbound primary antibody was removed and cells rinsed three times with CB before incubation with 0.2% (vol/vol) Alexa Fluor 488 anti-rabbit antibody from goat (A11034; Life Technologies) or 0.2% (vol/vol) Alexa Fluor 488 anti-mouse antibody from donkey (Invitrogen; A21202), 0.1% 4′,6-diamidin-2-phenylindol (Invitrogen), and 1% milk powder in CB for 1 h. Actin fibers were stained with 0.2% (vol/vol) Alexa Fluor 546 Phalloidin (Invitrogen; A22283) simultaneously with secondary antibody incubation. Subsequently, cells were washed three times with CB for 5 min each and once with deionized H2O before adding Fluoromount Aqueous Mounting Medium (Sigma) with a glass coverslip on top. Chambers were stuck on a microslide (VWR, Radnor, PA) and dried overnight. Afterward the cells were analyzed upside down.
Microscopy
Actin fiber, LC3B, and paxillin immunostainings were analyzed on a confocal laser scanning microscope (LSM880; Carl Zeiss, Jena, Germany) using Airyscan Mode with a 40× EC-Plan-Neofluar/ph3/1.3 NA oil objective (Carl Zeiss) or a 63× Plan Apochromat/1.4 NA oil objective (Carl Zeiss). Z-stacks of single cells for LC3B and paxillin and images of several cells for actin fiber detection were performed using appropriate settings for excitation and emission. All cells were imaged completely from bottom to top with an optical layer thickness of 159 nm and a layer overlap of 50% based on optimal Z-resolution settings in the Zen Black software (Carl Zeiss). Images were processed with the same software. BAG3-WT–, BAG3-WAWA–, and GFP-plasmid DNA cotransfected cells after actin immunostaining were imaged with a widefield microscope (AxioObserver; Carl Zeiss) using a 40× EC-Plan-Neofluar/ph3/1.3 NA oil objective (Carl Zeiss) and the appropriate filter set.
LC3B-spot analysis
LC3B-spot analysis is described in detail in Supplemental Figure S4. In short, APs were stained immunocytochemically for LC3B and recorded by Z-stack confocal microscopy. Subsequently, intensity projections of the signals were smoothed and a cell mask was calculated based on the mean gray value as threshold. Within the cell mask, the signal intensity of LC3B spots had to be above a manually selected threshold that was kept constant for each single experiment. Furthermore, signals with an area of fewer than 70 pixels for A7r5 and 20 pixels for MEF cells, respectively, were rejected. To separate clusters of spots the watershed algorithm was used.
Analysis of actin fiber orientation
Actin fiber orientation was analyzed as described before (Faust et al., 2011; Noethel et al., 2018). In short, we performed a maximum-intensity projection of recorded z-stacks and marked single cells manually. Cortical actin signals were removed, and the orientation of actin fibers in the inner area was determined from gray value gradients. The main actin fiber orientation of each cell was analyzed as the maximum of the distribution of all angles in the cell area. Cell main actin fiber orientations were plotted as cumulative distributions from parallel (0°) to perpendicular (90°) to the direction of stretch.
Statistical analysis
The actin fiber orientation for all indicated experiments was statistically analyzed using bootstrapping as used by us earlier (Noethel et al., 2018). In short, the bootstrap sample size was set to 5000 and the cumulative frequency as well as the upper and lower 95% confidence intervals were analyzed for classes from 0° to 90° in 5° sections (Supplemental Tables S1–S8). The resulting values were plotted as cumulative frequency distributions (sampled in 5° steps). The Kolmogorov–Smirnov test was used to determine statistical significances between individual sample sets (ns p > 0.05, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001). All statistically analyzed experiments were performed at least three times individually from each other.
Supplementary Material
Acknowledgments
This work received funding from the Deutsche Forschungsgemeinschaft (FOR2734).
Abbreviations used:
- APs
autophagosomes
- BAG3
BAG family molecular chaperone regulator 3
- CASA
chaperone-assisted selective autophagy
- CQ
chloroquine
- FA
focal adhesion
- FAK
FA kinase
- GFP
green fluorescent protein
- LC3B
microtubule-associated proteins 1A/1B light chain 3B
- LPA
lysophosphatidic acid
- MEF
mouse embryonic fibroblast
- RhoA
Ras homologue family member A
- Src
cellular sarcoma kinase
- WAWA
BAG3-mutation carrying tryptophan-to-alanine substitutions at positions 26 and 49
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E21-05-0254).
REFERENCES
- Arndt V, Dick N, Tawo R, Dreiseidler M, Wenzel D, Hesse M, Furst DO, Saftig P, Saint R, Fleischmann BK, et al. (2010). Chaperone-assisted selective autophagy is essential for muscle maintenance. Curr Biol 20, 143–148. [DOI] [PubMed] [Google Scholar]
- Bakolitsa C, Cohen DM, Bankston LA, Bobkov AA, Cadwell GW, Jennings L, Critchley DR, Craig SW, Liddington RC (2004). Structural basis for vinculin activation at sites of cell adhesion. Nature 430, 583–586. [DOI] [PubMed] [Google Scholar]
- Balchin D, Hayer-Hartl M, Hartl FU (2016). In vivo aspects of protein folding and quality control. Science 353, aac4354. [DOI] [PubMed] [Google Scholar]
- Belaid A, Cerezo M, Chargui A, Corcelle-Termeau E, Pedeutour F, Giuliano S, Ilie M, Rubera I, Tauc M, Barale S, et al. (2013). Autophagy plays a critical role in the degradation of active RHOA, the control of cell cytokinesis, and genomic stability. Cancer Res 73, 4311–4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carisey A, Tsang R, Greiner AM, Nijenhuis N, Heath N, Nazgiewicz A, Kemkemer R, Derby B, Spatz J, Ballestrem C (2013). Vinculin regulates the recruitment and release of core focal adhesion proteins in a force-dependent manner. Curr Biol 23, 271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Zhu Q, Dee R, Opheim Z, Mack CP, Cyr DM, Taylor JM (2017). Focal adhesion kinase-mediated phosphorylation of Beclin1 protein suppresses cardiomyocyte autophagy and initiates hypertrophic growth. J Biol Chem 292, 2065–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Rio A, Perez-Jimenez R, Liu R, Roca-Cusachs P, Fernandez JM, Sheetz MP (2009). Stretching single talin rod molecules activates vinculin binding. Science 323, 638–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikic I (2017). Proteasomal and autophagic degradation systems. Annu Rev Biochem 86, 193–224. [DOI] [PubMed] [Google Scholar]
- Discher DE, Janmey P, Wang YL (2005). Tissue cells feel and respond to the stiffness of their substrate. Science 310, 1139–1143. [DOI] [PubMed] [Google Scholar]
- DuFort CC, Paszek MJ, Weaver VM (2011). Balancing forces: architectural control of mechanotransduction. Nat Rev Mol Cell Biol 12, 308–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlicher AJ, Nakamura F, Hartwig JH, Weitz DA, Stossel TP (2011). Mechanical strain in actin networks regulates FilGAP and integrin binding to filamin A. Nature 478, 260–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust U, Hampe N, Rubner W, Kirchgessner N, Safran S, Hoffmann B, Merkel R (2011). Cyclic stress at mHz frequencies aligns fibroblasts in direction of zero strain. PLoS One 6, e28963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamerdinger M, Kaya AM, Wolfrum U, Clement AM, Behl C (2011). BAG3 mediates chaperone-based aggresome-targeting and selective autophagy of misfolded proteins. EMBO Rep 12, 149–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehler S, Baldassarre M, Lad Y, Leight JL, Wozniak MA, Riching KM, Eliceiri KW, Weaver VM, Calderwood DA, Keely PJ (2009). Filamin A-beta1 integrin complex tunes epithelial cell response to matrix tension. Mol Biol Cell 20, 3224–3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa K, Hosokawa A, Yabusaki K, Obinata T (2000). Orientation of smooth muscle-derived A10 cells in culture by cyclic stretching: relationship between stress fiber rearrangement and cell reorientation. Zoolog Sci 17, 617–624. [DOI] [PubMed] [Google Scholar]
- Hayakawa K, Sato N, Obinata T (2001). Dynamic reorientation of cultured cells and stress fibers under mechanical stress from periodic stretching. Exp Cell Res 268, 104–114. [DOI] [PubMed] [Google Scholar]
- Hayakawa K, Tatsumi H, Sokabe M (2008). Actin stress fibers transmit and focus force to activate mechanosensitive channels. J Cell Sci 121, 496–503. [DOI] [PubMed] [Google Scholar]
- Hayakawa K, Tatsumi H, Sokabe M (2011). Actin filaments function as a tension sensor by tension-dependent binding of cofilin to the filament. J Cell Biol 195, 721–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman BD, Grashoff C, Schwartz MA (2011). Dynamic molecular processes mediate cellular mechanotransduction. Nature 475, 316–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman LM, Jensen CC, Chaturvedi A, Yoshigi M, Beckerle MC (2012). Stretch-induced actin remodeling requires targeting of zyxin to stress fibers and recruitment of actin regulators. Mol Biol Cell 23, 1846–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries JD, Wang P, Streuli C, Geiger B, Humphries MJ, Ballestrem C (2007). Vinculin controls focal adhesion formation by direct interactions with talin and actin. J Cell Biol 179, 1043–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes RO (2002). Integrins: bidirectional, allosteric signaling machines. Cell 110, 673–687. [DOI] [PubMed] [Google Scholar]
- Izard T, Vonrhein C (2004). Structural basis for amplifying vinculin activation by talin. J Biol Chem 279, 27667–27678. [DOI] [PubMed] [Google Scholar]
- Janmey PA, Miller RT (2011). Mechanisms of mechanical signaling in development and disease. J Cell Sci 124, 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji C, Tang M, Zeidler C, Hohfeld J, Johnson GV (2019). BAG3 and SYNPO (synaptopodin) facilitate phospho-MAPT/Tau degradation via autophagy in neuronal processes. Autophagy 15, 1199–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungbauer S, Gao H, Spatz JP, Kemkemer R (2008). Two characteristic regimes in frequency-dependent dynamic reorientation of fibroblasts on cyclically stretched substrates. Biophys J 95, 3470–3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzaki M, Nagasawa M, Kojima I, Sato C, Naruse K, Sokabe M, Iida H (1999). Molecular identification of a eukaryotic, stretch-activated nonselective cation channel. Science 285, 882–886. [DOI] [PubMed] [Google Scholar]
- Kathage B, Gehlert S, Ulbricht A, Ludecke L, Tapia VE, Orfanos Z, Wenzel D, Bloch W, Volkmer R, Fleischmann BK, et al. (2017). The cochaperone BAG3 coordinates protein synthesis and autophagy under mechanical strain through spatial regulation of mTORC1. Biochim Biophys Acta 1864, 62–75. [DOI] [PubMed] [Google Scholar]
- Kenific CM, Stehbens SJ, Goldsmith J, Leidal AM, Faure N, Ye J, Wittmann T, Debnath J (2016a). NBR1 enables autophagy-dependent focal adhesion turnover. J Cell Biol 212, 577–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenific CM, Wittmann T, Debnath J (2016b). Autophagy in adhesion and migration. J Cell Sci 129, 3685–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU (2013). Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem 82, 323–355. [DOI] [PubMed] [Google Scholar]
- Kirchenbuchler D, Born S, Kirchgessner N, Houben S, Hoffmann B, Merkel R (2010). Substrate, focal adhesions, and actin filaments: a mechanical unit with a weak spot for mechanosensitive proteins. J Phys Condens Matter 22, 194109. [DOI] [PubMed] [Google Scholar]
- Lange S, Xiang F, Yakovenko A, Vihola A, Hackman P, Rostkova E, Kristensen J, Brandmeier B, Franzen G, Hedberg B, et al. (2005). The kinase domain of titin controls muscle gene expression and protein turnover. Science 308, 1599–1603. [DOI] [PubMed] [Google Scholar]
- Lu C, Wu F, Qiu W, Liu R (2013). P130Cas substrate domain is intrinsically disordered as characterized by single-molecule force measurements. Biophys Chem 180–181, 37–43. [DOI] [PubMed] [Google Scholar]
- Marshall BT, Long M, Piper JW, Yago T, McEver RP, Zhu C (2003). Direct observation of catch bonds involving cell-adhesion molecules. Nature 423, 190–193. [DOI] [PubMed] [Google Scholar]
- Mauthe M, Orhon I, Rocchi C, Zhou X, Luhr M, Hijlkema KJ, Coppes RP, Engedal N, Mari M, Reggiori F (2018). Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy 14, 1435–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merabova N, Sariyer IK, Saribas AS, Knezevic T, Gordon J, Turco MC, Rosati A, Weaver M, Landry J, Khalili K (2015). WW domain of BAG3 is required for the induction of autophagy in glioma cells. J Cell Physiol 230, 831–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael KE, Dumbauld DW, Burns KL, Hanks SK, Garcia AJ (2009). Focal adhesion kinase modulates cell adhesion strengthening via integrin activation. Mol Biol Cell 20, 2508–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore SW, Roca-Cusachs P, Sheetz MP (2010). Stretchy proteins on stretchy substrates: the important elements of integrin-mediated rigidity sensing. Dev Cell 19, 194–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidlinger-Wilke C, Grood E, Claes L, Brand R (2002). Fibroblast orientation to stretch begins within three hours. J Orthop Res 20, 953–956. [DOI] [PubMed] [Google Scholar]
- Nobes CD, Hall A (1995). Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell 81, 53–62. [DOI] [PubMed] [Google Scholar]
- Noethel B, Ramms L, Dreissen G, Hoffmann M, Springer R, Rubsam M, Ziegler WH, Niessen CM, Merkel R, Hoffmann B (2018). Transition of responsive mechanosensitive elements from focal adhesions to adherens junctions on epithelial differentiation. Mol Biol Cell 29, 2317–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paci E, Karplus M (1999). Forced unfolding of fibronectin type 3 modules: an analysis by biased molecular dynamics simulations. J Mol Biol 288, 441–459. [DOI] [PubMed] [Google Scholar]
- Pasapera AM, Schneider IC, Rericha E, Schlaepfer DD, Waterman CM (2010). Myosin II activity regulates vinculin recruitment to focal adhesions through FAK-mediated paxillin phosphorylation. J Cell Biol 188, 877–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlaczyk M, Lelonkiewicz M, Wieczorowski M (2013). Age-dependent biomechanical properties of the skin. Postepy DermatolAlergol 30, 302–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimann L, Schwable AN, Fricke AL, Muhlhauser WWD, Leber Y, Lohanadan K, Puchinger MG, Schauble S, Faessler E, Wiese H, et al. (2020). Phosphoproteomics identifies dual-site phosphorylation in an extended basophilic motif regulating FILIP1-mediated degradation of filamin-C. Commun Biol 3, 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riveline D, Zamir E, Balaban NQ, Schwarz US, Ishizaki T, Narumiya S, Kam Z, Geiger B, Bershadsky AD (2001). Focal contacts as mechanosensors: externally applied local mechanical force induces growth of focal contacts by an mDia1-dependent and ROCK-independent mechanism. J Cell Biol 153, 1175–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rognoni L, Most T, Zoldak G, Rief M (2014). Force-dependent isomerization kinetics of a highly conserved proline switch modulates the mechanosensing region of filamin. Proc Natl Acad Sci USA 111, 5568–5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rognoni L, Stigler J, Pelz B, Ylanne J, Rief M (2012). Dynamic force sensing of filamin revealed in single-molecule experiments. Proc Natl Acad Sci USA 109, 19679–19684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandilands E, Serrels B, McEwan DG, Morton JP, Macagno JP, McLeod K, Stevens C, Brunton VG, Langdon WY, Vidal M, et al. (2011). Autophagic targeting of Src promotes cancer cell survival following reduced FAK signalling. Nat Cell Biol 14, 51–60. [DOI] [PubMed] [Google Scholar]
- Sawada Y, Tamada M, Dubin-Thaler BJ, Cherniavskaya O, Sakai R, Tanaka S, Sheetz MP (2006). Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell 127, 1015–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharifi MN, Mowers EE, Drake LE, Collier C, Chen H, Zamora M, Mui S, Macleod KF (2016). Autophagy promotes focal adhesion disassembly and cell motility of metastatic tumor cells through the direct interaction of paxillin with LC3. Cell Rep 15, 1660–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigaut L, von Bilderling C, Bianchi M, Burdisso JE, Gastaldi L, Pietrasanta LI (2018). Live cell imaging reveals focal adhesions mechanoresponses in mammary epithelial cells under sustained equibiaxial stress. Sci Rep 8, 9788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teckchandani A, Cooper JA (2016). The ubiquitin-proteasome system regulates focal adhesions at the leading edge of migrating cells. eLife 5, e17440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilleman TR, Tilleman MM, Neumann MH (2004). The elastic properties of cancerous skin: Poisson’s ratio and Young’s modulus. Isr Med Asso J 6, 753–755. [PubMed] [Google Scholar]
- Tondon A, Hsu HJ, Kaunas R (2012). Dependence of cyclic stretch-induced stress fiber reorientation on stretch waveform. J Biomech 45, 728–735. [DOI] [PubMed] [Google Scholar]
- Ulbricht A, Eppler FJ, Tapia VE, van der Ven PF, Hampe N, Hersch N, Vakeel P, Stadel D, Haas A, Saftig P, et al. (2013). Cellular mechanotransduction relies on tension-induced and chaperone-assisted autophagy. Curr Biol 23, 430–435. [DOI] [PubMed] [Google Scholar]
- Ulbricht A, Gehlert S, Leciejewski B, Schiffer T, Bloch W, Hohfeld J (2015). Induction and adaptation of chaperone-assisted selective autophagy CASA in response to resistance exercise in human skeletal muscle. Autophagy 11, 538–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HQ, Liu HM, Zhang HY, Guan Y, Du ZX (2008). Transcriptional upregulation of BAG3 upon proteasome inhibition. Biochem Biophys Res Commun 365, 381–385. [DOI] [PubMed] [Google Scholar]
- Wong AJ, Pollard TD, Herman IM (1983). Actin filament stress fibers in vascular endothelial cells in vivo. Science 219, 867–869. [DOI] [PubMed] [Google Scholar]
- Yakovenko O, Sharma S, Forero M, Tchesnokova V, Aprikian P, Kidd B, Mach A, Vogel V, Sokurenko E, Thomas WE (2008). FimH forms catch bonds that are enhanced by mechanical force due to allosteric regulation. J Biol Chem 283, 11596–11605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M, Goult BT, Klapholz B, Hu X, Toseland CP, Guo Y, Cong P, Sheetz MP, Yan J (2016). The mechanical response of talin. Nat Commun 7, 11966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonemura S, Wada Y, Watanabe T, Nagafuchi A, Shibata M (2010). alpha-Catenin as a tension transducer that induces adherens junction development. Nat Cell Biol 12, 533–542. [DOI] [PubMed] [Google Scholar]
- Yu C, Li WB, Liu JB, Lu JW, Feng JF (2018). Autophagy: novel applications of nonsteroidal anti-inflammatory drugs for primary cancer. Cancer Med 7, 471–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusko EC, Asbury CL (2014). Force is a signal that cells cannot ignore. Mol Biol Cell 25, 3717–3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidel-Bar R, Itzkovitz S, Ma’ayan A, Iyengar R, Geiger B (2007). Functional atlas of the integrin adhesome. Nat Cell Biol 9, 858–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski A, Linnartz C, Pleschka C, Dreissen G, Springer R, Merkel R, Hoffmann B (2018). Reorientation dynamics and structural interdependencies of actin, microtubules and intermediate filaments upon cyclic stretch application. Cytoskeleton (Hoboken) 75, 385–394. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.