Abstract
Accumulating evidence indicates that increased ribosome biogenesis is a hallmark of cancer. It is well established that inhibition of any steps of ribosome biogenesis induces nucleolar stress characterized by p53 activation and subsequent cell cycle arrest and/or cell death. However, cells derived from solid tumors have demonstrated different degrees of sensitivity to ribosome biogenesis inhibition, where cytostatic effects rather than apoptosis are observed. The reason for this is not clear, and the p53-specific transcriptional program induced after nucleolar stress has not been previously investigated. Here we demonstrate that blocking rRNA synthesis by depletion of essential rRNA processing factors such as LAS1L, PELP1, and NOP2 or by inhibition of RNA Pol I with the specific small molecule inhibitor CX-5461, mainly induce cell cycle arrest accompanied by autophagy in solid tumor–derived cell lines. Using gene expression analysis, we find that p53 orchestrates a transcriptional program involved in promoting metabolic remodeling and autophagy to help cells survive under nucleolar stress. Importantly, our study demonstrates that blocking autophagy significantly sensitizes cancer cells to RNA Pol I inhibition by CX-5461, suggesting that interfering with autophagy should be considered a strategy to heighten the responsiveness of ribosome biogenesis–targeted therapies in p53-positive tumors.
INTRODUCTION
The synthesis of ribosomes is aberrantly increased in almost all cancers, and several studies have demonstrated that deregulation of ribosome biogenesis in cancer is not solely a consequence of increased proliferation, but is a key event in oncogenesis (Barna et al., 2008; Chan et al., 2011; Delloye-Bourgeois et al., 2012; Pelletier et al., 2018; Penzo et al., 2019). Biogenesis of ribosomes is initiated in the nucleolus by the transcription of a long 47S precursor ribosomal RNA (rRNA) that undergoes concomitant site-specific modifications, processing into mature 18S, 5.8S, and 28S rRNA species, and assembly with ribosomal proteins to form the 40S and 60S ribosomal subunits (Hadjiolova et al., 1993; Aubert et al., 2018; Bohnsack and Bohnsack, 2019). The fourth rRNA (5S), which is part of the 60S subunit, is transcribed independently by RNA Pol III (Ciganda and Williams, 2011). These steps involve a large assortment of processing and maturation factors in addition to the RNA and protein components of the ribosome itself. It is estimated that more than 200 additional accessory proteins and non-coding small RNAs engage in intricate reactions of processing, assembly and nuclear export to generate functional cytoplasmic 40S and 60S subunits (Bohnsack and Bohnsack, 2019). The integrity of the nucleolus relies on the precise coordination of these reactions, and defects in any of these steps ultimately trigger a nucleolar stress characterized by a p53-regulated G1 cell cycle arrest and/or induction of apoptosis (Pestov et al., 2001; Lohrum et al., 2003; Rubbi and Milner, 2003; Zhang et al., 2003; Bhat et al., 2004; Dai and Lu, 2004; Dai et al., 2004; Jin et al., 2004; Yuan et al., 2005; Chen et al., 2007; Fumagalli et al., 2009; Zhu et al., 2009; Holzel et al., 2010; Golomb et al., 2014). Tumors appears to be more sensitive than normal cells to nucleolar stress, indicating that pathways involved in regulating ribosome production constitute a therapeutic vulnerability for cancers (Bywater et al., 2012).
In general, activation of p53 represents a primordial response mechanism elicited by diverse type of stresses or loss of cellular homeostasis (Kastenhuber and Lowe, 2017; Zhang and Lozano, 2017). It is now well established that ribosomal proteins (RPs) play a critical role in mediating p53-signaling response triggered by nucleolar stress (Deisenroth et al., 2016). One of the main mechanisms responsible for the activation of p53 after disruption of rRNA synthesis is the binding of unassembled RPs to MDM2, the E3 ubiquitin–protein ligase that normally targets p53 for proteasomal degradation in the absence of cellular stresses. Binding of free RPs to MDM2 inhibits its E3 ligase activity and consequently leads to the stabilization of p53. At least 16 different RPs from both the large and the small ribosomal subunits have been shown to bind to MDM2 (Chakraborty et al., 2011). Among these, RPL11/uL5 and RPL5/uL18, as part of the 5S ribonucleoprotein (5S RNP), are absolutely essential to stabilize p53 after the induction of a nucleolar stress (Sun et al., 2010; Bursać et al., 2012; Fumagalli et al., 2012; Sloan et al., 2013).
p53 is well characterized by its ability to inhibit cell proliferation and induce apoptosis by transcriptional activation of the cyclin-dependent kinase inhibitor p21CIP1 and proapoptotic proteins, respectively. However, numerous studies have demonstrated that upon activation, p53 engages in an intricate anti-proliferative transcriptional program that spans a wide variety of biological processes including metabolism, reactive oxygen species (ROS) control, and autophagy (Nikulenkov et al., 2012; Menendez et al., 2013; Schlereth et al., 2013; Allen et al., 2014; Wang et al., 2014). The p53-driven transcriptional program and its subsequent cellular outcome (survival or cell death) appear to be highly tissue-specific and dependent on the type of stress encountered. Studies examining the consequences of nucleolar stress after inhibition of rRNA synthesis in cells derived from solid tumors have demonstrated different degree of sensitivity where cytostatic effects rather than apoptosis are observed (Pestov et al., 2001; Li et al., 2016; Cornelison et al., 2017; Ismael et al., 2019). These effects have mainly been attributed to the transcriptional activation of p21CIP1, which leads to a G1-cell cycle arrest with minimal cell death observed. Besides these observations, the extent to which the p53-specific transcriptional program is activated after nucleolar stress is currently unknown.
Given the growing interest in targeting ribosome synthesis as a novel therapeutic approach for cancer, it is becoming crucial to understand the p53 specific transcriptional response to nucleolar stress. Here, we utilized gene expression analysis to assess transcriptomic changes after inhibition of ribosome biogenesis. To identify p53-induced transcripts, the data were compared with published global run-on (GRO) sequencing/ChIP-seq studies of p53 targets (Nikulenkov et al., 2012; Menendez et al., 2013; Schlereth et al., 2013; Allen et al., 2014; Wang et al., 2014) and verified in wild type (WT) vs. isogenic p53 −/− cells. We uncovered that nucleolar stress leads to the activation of a p53-transcriptional survival program that includes genes involved in autophagy, metabolism, and ROS control. Importantly, inhibition of autophagy dramatically reduced the viability of cancer cells treated with the selective RNA pol I inhibitor CX-5461 (Drygin et al., 2011; Bywater et al., 2012). These results demonstrate that targeting p53-regulated pathways could enhance the efficacy of ribosome biogenesis drugs used for the treatment of cancers with active p53.
RESULTS
Inhibition of rRNA processing and ribosome biogenesis induces autophagy
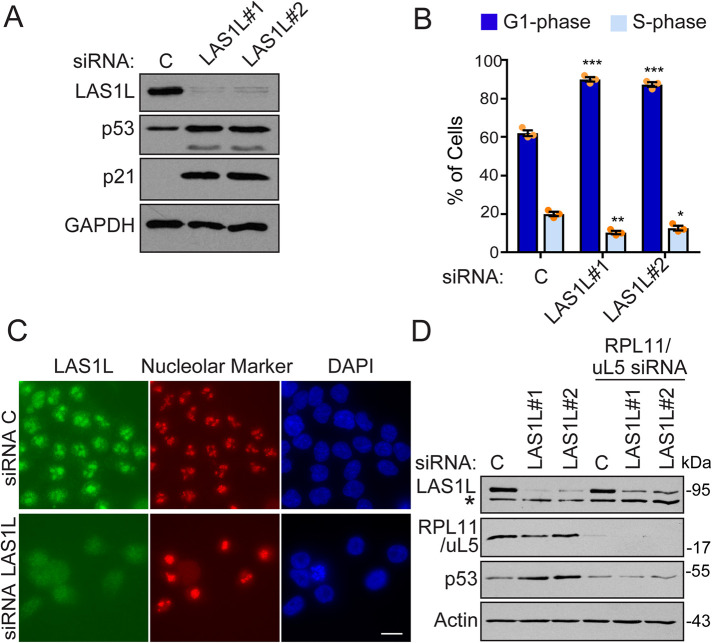
Apart from the induction of the cyclin-CDK inhibitor p21CIP1, very little is known about the cellular response and the p53-dependent transcriptional program initiated upon nucleolar stress. To further investigate how cells respond and adapt to nucleolar stress, we inhibited rRNA processing in HCT116 colon carcinoma cells (p53 WT) by RNAi depletion of LAS1L, an endoribonuclease essential for processing of the 28S rRNA and the synthesis of the 60S ribosomal sub-unit (Castle et al., 2010, 2013; Schillewaert et al., 2012; Gasse et al., 2015). As expected, knockdowns of LAS1L lead to protein stabilization of p53, induction of p21CIP1, and a G1-cell cycle arrest (Figure 1, A and B). Disruption of normal nucleolar morphology typical of nucleolar stress was also observed in LAS1L depleted cells (Figure 1C). In response to nucleolar stress, the ribosomal proteins RPL11/uL5 and RPL5/uL18 have been shown to be essential for p53 activation, as they bind to and inactivate MDM2, the E3 ubiquitin ligase that targets p53 for proteasome-mediated degradation (Sun et al., 2010; Bursać et al., 2012; Fumagalli et al., 2012; Sloan et al., 2013). Knockdown of both LAS1L and RPL11/uL5 abolished the stabilization p53 observed, indicating that inhibition of rRNA processing activates p53 through the sequestration of MDM2 by ribosomal proteins (Figure 1D).
FIGURE 1:
Inhibition of rRNA processing through depletion of LAS1L causes nucleolar stress. (A) HCT116 cells were transfected with control (represented by the letter C) or two different-LAS1L specific siRNAs. After 72 h, a portion of the cells were analyzed by Western blotting with the indicated antibodies. (B) RNAi-treated cells from C were labeled with BrdU and PI and analyzed by flow cytometry for DNA content. The results are graphed as percentages of cells in G1- and S-phase. The data are presented as means of three independent experiments ± SEM. Differences between groups (RNAi vs. nontargeting control) were evaluated using two-tailed Student t tests among replicate experiments. **P = 0.003, *P = 0.012: comparing to S-phase of the control; ***P < 0.001: comparing to G1-phase of the control. (C) Immunofluorescence analysis of HCT116 cells transfected with control C or LAS1L #1 siRNA. Seventy-two h after transfection, cells were fixed and immunostained with anti-LAS1L (green) and anti-Fibrillarin (red nucleolar marker). DNA was visualized by staining with Hoechst 33342 (blue). Scale bar is representative of all panels: 10 µm. (D) HCT116 cells were transfected with C or two different LAS1L specific siRNAs with or without cotransfection of RPL11/uL5 siRNA. Seventy-two h later, cells were analyzed by Western blotting with the indicated antibodies. Nonspecific bands were annotated with *.
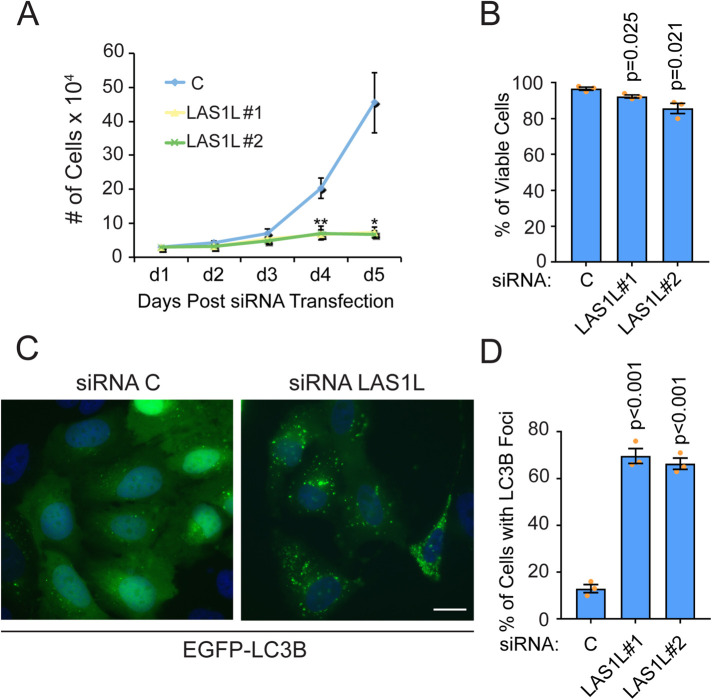
Although depletion of LAS1L dramatically affects cell proliferation (Figure 2A), only a small but significant drop in cell viability is observed at day 5 after LAS1L knockdowns (Figure 2B), suggesting that p53 is not likely to activate the transcription of apoptotic genes in response to nucleolar stress. Instead, we observed a dramatic increase in cells undergoing autophagy as monitored by the relocalization of EGFP-LC3B into punctate autophagosomes by fluorescence microscopy (Figure 2, C and D). Taken together, these observations indicate that inhibition of rRNA processing triggers a classical nucleolar stress response whereby p53 protein levels accumulate as a result of MDM2 inhibition by unassembled ribosomal proteins. Activated p53 likely promotes a prosurvival stress response that involves the transcriptional programing of autophagy-promoting pathways.
FIGURE 2:
Inhibition of rRNA processing induces autophagy. (A) HCT116 cells were transfected with nontargeting control (C) or LAS1L#1 and LAS1L#2 siRNA. Twenty-four h after transfection, cells were replated and counted at days 2, 3, 4, and 5 posttransfection. Cell count is represented as number of cells/ml × 104. The data are presented as means of three independent experiments ± SEM. *P = 0.026 (LAS1L#1 vs. C), P = 0.024 (LAS1L#2 vs. C), ** P = 0.008 (LAS1L#1 vs. C), P = 0.002 (LAS1L#2 vs. C). (B) Cells from A were harvested at day 5 and viability was determined by trypan blue exclusion. The data are presented as means of three independent experiments ± SEM. (C) HCT116 cells stably expressing EGFP-LC3B were transfected with C or LAS1L#1 and LAS1L#2 siRNA. Seventy-two h after, cells were fixed, stained with Hoechst 33342 (blue), and imaged by fluorescence microscopy (only the data from LAS1L siRNA #1 are represented). Scale bar is representative of all panels: 10 µm. The number of cells with more than 10 EGFP-LC3B foci was counted (200 cells per condition). (D) Quantification of the data from panel C. The data are presented as means of three independent experiments ± SEM. For all panels, statistical significance comparing knockdowns with C was calculated using two-tailed Student’s t tests among biological replicates.
p53 activates a survival transcriptional program in response to nucleolar stress
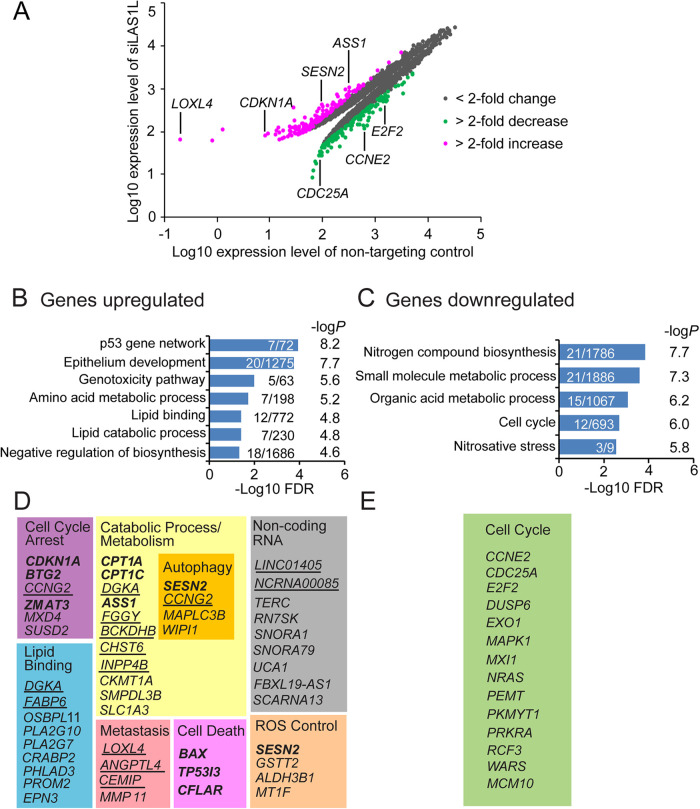
To investigate changes in gene expression induced by nucleolar stress, we performed gene expression analysis comparing RNA from HCT116 (p53 WT) cells treated with control (duplicate experiment) vs. LAS1L siRNAs (two different siRNAs). By setting an arbitrary fold change (FC), cutoffs of >2, and significance p-values of <0.01, we identified 157 up-regulated and 128 down-regulated genes in LAS1L-depleted cells (Figure 3A and Supplemental Table S1). Of these 157 up-regulated genes, 11 are known validated direct p53 targets and 51 are p53 targets predicted by one or more published microarray, ChIP-seq, or GRO-seq studies (Supplemental Table S2). A gene set enrichment analysis (GSEA) from the Molecular Signatures Database (Subramanian et al., 2005; Liberzon et al., 2011) revealed that up-regulated genes were enriched for p53 gene network, epithelium development, genotoxicity pathway, and metabolic processes including lipid binding and catabolic process/metabolism categories that comprise genes involved in promoting autophagy, such as SESN2, MAPLC3B, WIPI1, and CCNG2 (Maiuri et al., 2009; Mourgues et al., 2015; Proikas-Cezanne et al., 2015; Bento et al., 2016; Figure 3, B and D). A closer inspection of the up-regulated gene list showed enrichment for cell cycle arrest, ROS control, cell migration, and noncoding RNAs (Figure 3D). Genes that were down-regulated showed enrichment for nitrogen compound biosynthesis, small molecule/organic acid metabolic processes, cell cycle, and nitrosative stress categories (Figure 3C). Genes enriched for cell cycle regulation included CCNE2 (CYCLIN E2), CDC25A, and E2F2 (Figure 3E), which is in accordance with the G1-cell cycle arrest observed upon LAS1L depletion (Figure 1B). Only a few apoptotic genes were up-regulated (BAX, TP53I3, CFLAR; Figure 3D) suggesting that instead of cell death, nucleolar stress leads to p53-mediated metabolic pathway remodeling to support cell survival by promoting the maintenance of cellular energy levels.
FIGURE 3:
Nucleolar stress promotes the induction of a survival transcriptional program. (A–C) HCT116 cells were transfected with nontargeting control (in duplicate) or LAS1L#1 and LAS1L#2 siRNA. Seventy-two h later, total RNAs from two independent biological replicates were extracted and subjected to microarray analysis to identify transcriptional changes. The data were analyzed with an arbitrary fold change (FC) cut-offs of >2 and significance p-values of <0.01. A, scatterplot of all genes with significant change (p < 0.01). Genes found up-regulated are listed in B and C. Gene set enrichment analysis (GSEA) was determined using the Molecular Signatures Database for up-regulated and down-regulated genes separately using the ontology and canonical pathways gene sets. Overlaps were computed with an FDR q-value less than 0.05. The number of genes overlapping the particular term over the total term size is indicated in the bar plot. (D, E) Partial list of genes found up-regulated and down-regulated according to their cellular functions. Genes in bold represent known validated direct p53 targets and genes underlined are p53 targets predicted by one or more published microarray, ChIP-seq, or GRO-seq studies.
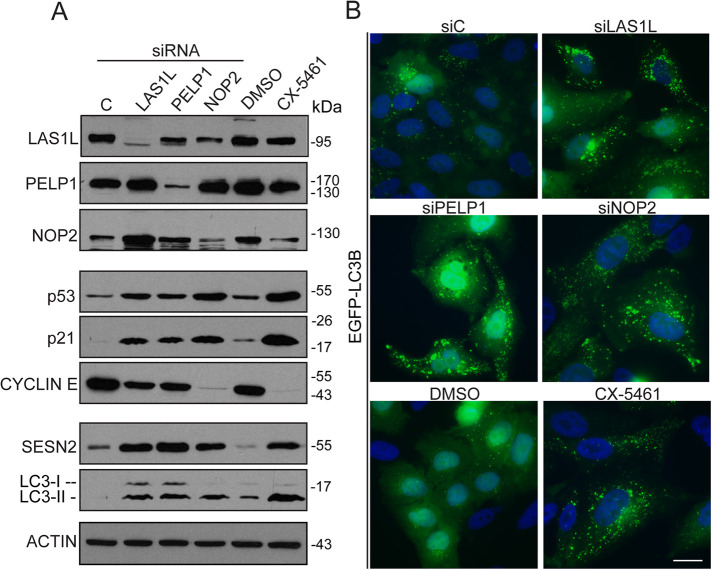
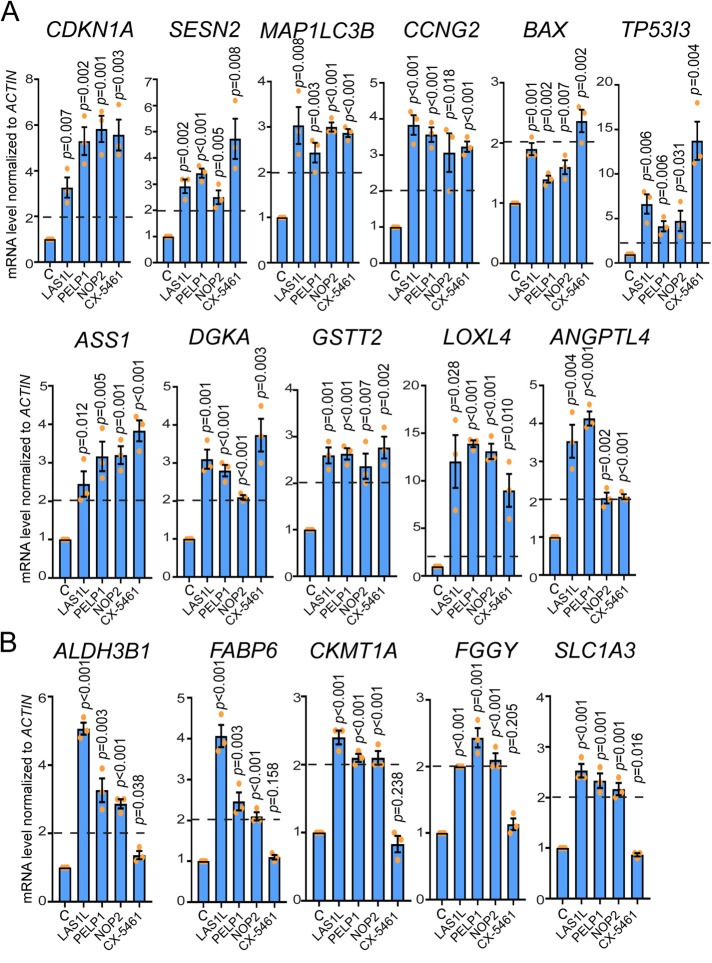
To determine whether changes in metabolic gene expression observed after LAS1L depletion were associated with nucleolar stress in general, we tested for autophagy induction after RNAi knockdown of two other essential 60S ribosome biogenesis factors (PELP1 and NOP2; Hong et al., 1997; Finkbeiner et al., 2011; Castle et al., 2012; Bourgeois et al., 2015) and after inhibition of RNA Pol I with CX-5461. Similarly to depletion of LAS1L, knockdowns of PELP1 and NOP2, as well as inhibition of rRNA transcription by CX-5461, leads to p53 stabilization, induction of p21CIP1, and down-regulation of cyclin E protein (Figure 4A). The well-characterized p53 target and positive regulator of autophagy SESTRIN 2 (Budanov et al., 2004; Budanov and Karin, 2008), whose gene expression (SESN2) was found increased in our gene expression analysis (Figure 3 and Supplementary Table S1), was also dramatically up-regulated at the protein level after inhibition of ribosome synthesis (Figure 4A). Autophagy is associated with conversion of the microtubule-associated protein 1 light chain 3 (LC3) from the cytosolic LC3-I to the autophagosome-associated LC3-II form (Bento et al., 2016). Increased levels of LC3-II were clearly detected in cells depleted of LAS1L, PELP1, or NOP2 and after inhibition of RNA Pol I by CX-5461 (Figure 4A). In addition, fluorescence microscopy showed the redistribution of EGFP-LC3 into punctate autophagosomes after nucleolar stress (Figure 4B and Supplemental Figure S1), suggesting that the increase in LC3B gene (MAPLC3B) expression observed is associated with induction of autophagy.
FIGURE 4:
Nucleolar stress increases SESTRIN 2 protein level and promotes autophagy. (A) HCT116 cells were transfected with nontargeting control (siC), LAS1L siRNA #1, PELP1 siRNA, or NOP2 siRNA or treated with DMSO or 500 nM CX-5461. Seventy-two h later (for siRNA treatment) or 24 h later (CX-5461), cells were analyzed by Western blotting with the indicated antibodies. (B) HCT116 cells stably expressing EGFP-LC3B were transfected with nontargeting control (C) or LAS1L siRNA #1, PELP1 siRNA, or NOP2 siRNA or treated with DMSO or 500 nM CX-5461. Seventy-two h after (for siRNA treatment) or 24 h after (CX-5461), cells were fixed, stained with Hoechst 33342 (blue), and imaged by fluorescence microscopy to visualize the relocalization of EGFP-LC3B to punctate autophagosomes. Scale bar is representative of all panels: 10 µm.
We next used qRT-PCR to confirm the gene expression results in selected subsets of genes from different biological processes categories found to be up-regulated by LAS1L depletion (Figure 3B). In agreement with the induction of autophagy observed in Figure 4, depletion of LAS1L, PELP1, and NOP2 or inhibition of rRNA transcription by CX-5461 leads to increased transcripts of SESN2, MAP1BLC3, and CCNG2, which are known to regulate autophagy positively (Maiuri et al., 2009; Mourgues et al., 2015; Bento et al., 2016; Figure 5A). As expected, p21CIP1 (CDKN1A) transcript was up-regulated above twofold, whereas transcription of the proapoptotic gene BAX only showed a modest increase after nucleolar stress (Figure 5A). However, transcription of TP53I3 (PIG3), another proapoptotic gene, was substantially increased, particularly in the CX-5461 treated cells (Figure 5A). Because PIG3 expression alone is insufficient to induce apoptosis (Polyak et al., 1997), this increase is unlikely to by itself override the overall transcriptional output of survival genes up-regulated by nucleolar stress. Genes involved in central metabolism regulation (ASS1, DGKA; Merida et al., 2008; Haines et al., 2011), ROS control (GSTT2; Tew and Townsend, 2012), and, surprisingly, cell migration (LOXL4, ANGPTL4; Minn et al., 2005; Payne et al., 2005; Padua et al., 2008) also showed higher transcript levels after inhibition of ribosome synthesis (Figure 5A). Interestingly, a subset of genes (ALDH3B1, FABP6, CKMT1A, FGGY, and SLC1A3) were only found to be up-regulated by depletion of LAS1L, PELP1, or NOP2 and not by inhibition of Pol I transcription by CX-5461 (Figure 5B). In addition to inhibition of Pol I activity, CX-5461 has also been shown to cause DNA damage by blocking replication forks and inducing single-strand DNA breaks, which could explain the differences in gene transcription observed (Xu et al., 2017; Bruno et al., 2020).
FIGURE 5:
Q-RT-PCR validates the induction of mRNA transcripts after nucleolar stress. (A, B) Total RNAs from HCT116 cells treated with nontargeting control (C), LAS1L siRNA #1, PELP1 siRNA, NOP2 siRNA, DMSO, or 500 nM CX-5461 were analyzed 72 h after (for siRNA treatment) or 24 h after (CX-5461) by qRT-PCR to assess changes in gene expression. mRNA expression was normalized to beta-ACTIN values. Changes in gene expression are presented as fold change of siRNA-treated samples over control siRNA or CX-5461-treated samples over DMSO. A, genes that were confirmed to be up-regulated for all conditions. B, genes that were found to be up-regulated only in cells treated with LAS1L, PELP1, and NOP2 siRNA and not by CX-5461 treatment. The data are presented as means of three independent experiments ± SEM. Statistical significance comparing knockdowns with nontargeting control or CX-5461 to DMSO was calculated using two-tailed Student’s t tests among biological replicates.
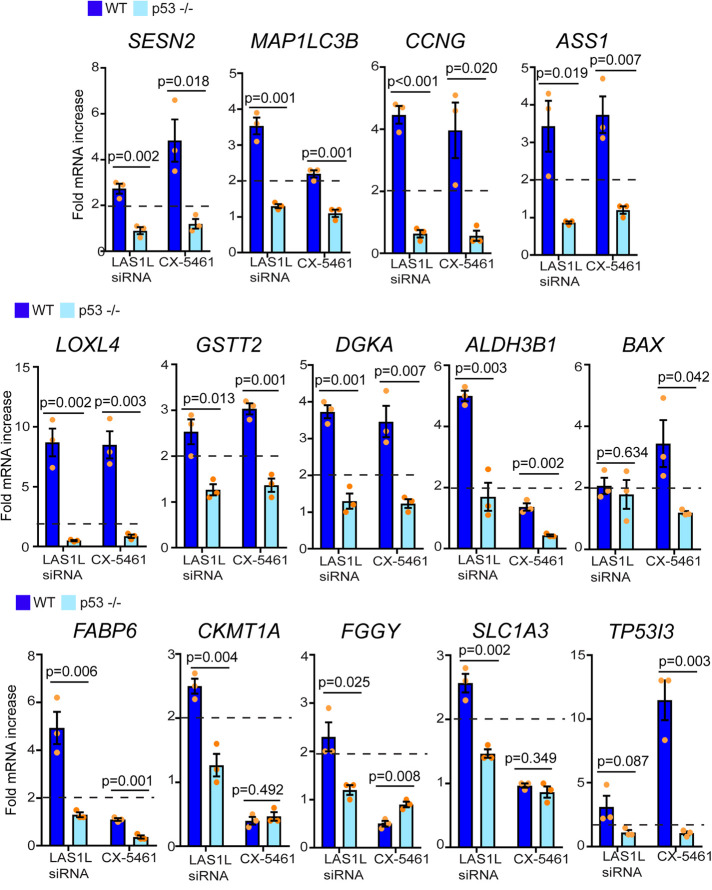
To assess whether the sets of genes found to be up-regulated after nucleolar stress are p53-regulated, we performed qRT-PCR analysis comparing their expression level in isogenic HCT116 p53 +/+ and p53 −/− cell lines. Expectedly, inhibition of ribosome synthesis by LAS1L knockdown or treatment with CX-5461 induced the expression and stabilization of p21CIP1 protein only in p53 WT cells (Supplemental Figure S2A). Additionally, the up-regulation of SESTRIN 2 and LC3B proteins was not observed in p53 null cells after ribosomal stress (Supplemental Figure S2B). Nucleolar stress increased the expression of SESN2 (SESTRIN 2) as well as the expression of the predicted p53 target genes CCNG2, ASS1, LOXL4, DGKA, BAX, FABP6, FGGY, and TP53I3 in a p53-dependent manner (Figure 6). Although MAP1LC3B (LC3B), GSTT2, ALDH3B1, CKMT1A,and SLC1A3 have not been predicted by any microarray, ChIP-seq, or GRO-seq studies to be p53 targets, their up-regulation following inhibition of ribosome biogenesis was observed only in p53 WT cells, suggesting that their transcriptional regulation is p53-dependent (Figure 6). Whether they are direct transcription targets of p53 remains to be determined. Altogether, these results suggest that p53 activates the transcription of genes mainly involved in cell cycle arrest, metabolism, ROS control, and autophagy to promote the survival of cancer cells after nucleolar stress.
FIGURE 6:
p53-dependent gene expression after nucleolar stress. HCT116 or HCT116 p53 –/– cells were treated with nontargeting control, LAS1L siRNA #1, DMSO, or 500 nM CX-5461. Seventy-two h after (for siRNA treatment) of 24 h after (CX-5461), total RNA was extracted and analyzed by qRT-PCR to assess changes in gene expression. mRNA expression was normalized to beta-ACTIN values. Changes in gene expression are presented as fold change of LAS1L siRNA over control siRNA or CX-5461 over DMSO. The data are presented as means of three independent experiments ± SEM. Statistical significance comparing knockdowns with nontargeting control or CX-5461 to DMSO was calculated using two-tailed Student’s t tests among biological replicates.
Inhibition of autophagy sensitizes p53 positive cancer cells to nucleolar stress
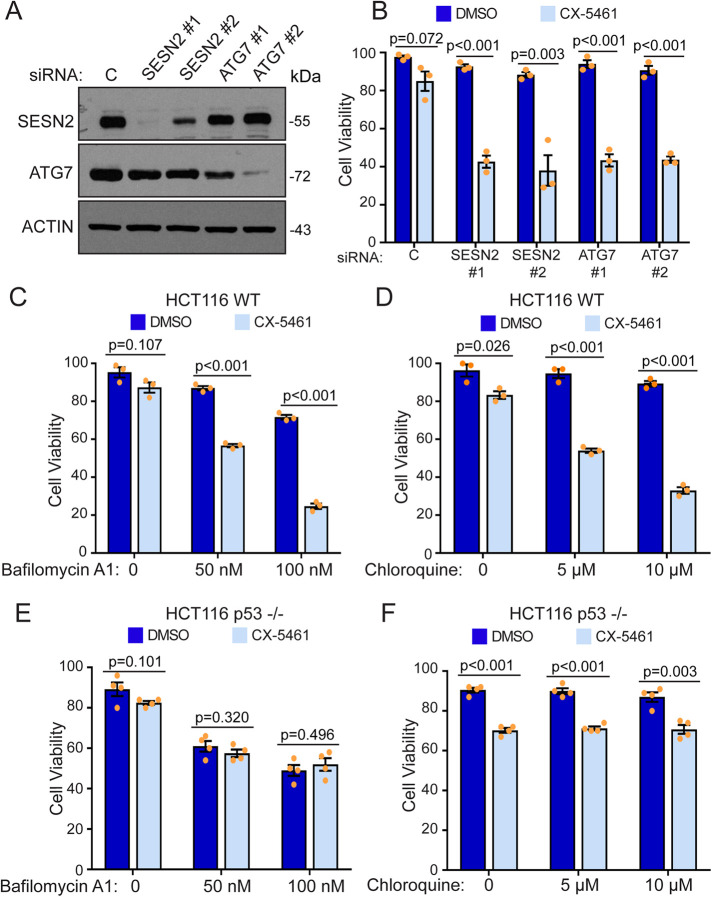
Increased ribosome biogenesis is a hallmark of cancer and inhibition of rRNA synthesis is now emerging as a prime therapeutic approach for cancer. The rDNA transcription inhibitor CX-5461 has been shown to have p53-dependent anti-tumorigenic activity in B-lymphoma models, and results of a phase I dose escalation study in patients with advanced hematologic cancer have shown antitumor activity in both WT and p53 mutant malignancies (Bywater et al., 2012; Khot et al., 2019). However, in some solid tumors, CX-5461 appears to have variable cytostatic effects characterized by the inhibition of cell growth and induction of autophagy (Drygin et al., 2011; Li et al., 2016; Cornelison et al., 2017; Ismael et al., 2019). In agreement with these observations, our data demonstrating that inhibition of ribosome synthesis induces the expression of autophagy promoting genes suggest that interfering with this pathway could reduce the survival of these cells after nucleolar stress. To test this, we used RNAi to knock down ATG7, an E1-like activating enzyme essential for autophagy, and SESTRIN 2, which was shown to promote autophagy by activating an AMPK-dependent inhibition of mTOR signaling (Tanida et al., 1999; Komatsu et al., 2001; Budanov and Karin, 2008; Maiuri et al., 2009). As shown in Figure 7, A and B, depletion of SESTRIN 2 or ATG7 with two different siRNAs considerably increased the sensitivity of HCT116 cells to CX-5461 treatment, suggesting that autophagy is required for cell survival after inhibition of rRNA synthesis. To further confirm this, we next tested whether pharmacological inhibition of autophagy could also sensitize cells to nucleolar stress. Cells were treated concomitantly with CX-5461 and increasing doses of bafilomycin A1 (0, 50 nM, 100 nM), a macrolide antibiotic that reversibly inhibits late-phase autophagy, or chloroquine (0, 5 µM, 10 µM), which inhibits lysosomal hydrolases and prevents autophagosomal fusion and degradation of vesicle content. Although bafilomycin treatment by itself showed a moderate reduction in cell viability, cotreatment with bafilomycin A1 and CX-5461 dramatically impaired cell survival after only 24 h (Figure 7C). Similarly, chloroquine significantly sensitized cells to CX-5461 treatment (Figure 7D). We also observed a similar trend in cell sensitization in two other p53 positive cell lines (MCF7 and U2OS), where CX-5461 also up-regulated the expression of CNKN1A (p21CIP1), SESN2 (SESTRIN 2), MAP1LC3B (LC3B), and CCNG2 (cyclin G2) (Supplemental Figure S3).
FIGURE 7:
Inhibition of autophagy sensitizes cancer cells to nucleolar stress. (A) HCT116 cells were treated with nontargeting control (C), SESN2 #1, SESN2 #2, ATG7 #1, or ATG7 #2 siRNAs. Seventy-two h later, cells were collected and knockdowns were monitored by Western blotting. (B) Cells from panel A were treated with DMSO or 500 nM CX-5461 for 24 h. HCT116 (C, D) or HCT116 p53 –/– cells were treated with vehicle (DMSO) or 500 nM CX-5461 for 24 h in the presence of bafilomycin (0, 50 nM, 100 nM; C, E) or chloroquine (0, 5 µM, 10 µM; D, F). Cell viability, B–F, was determined by trypan blue exclusion. The data are presented as means of four, E, F, or three, B–D, independent experiments ± SEM. Statistical significance comparing CX-5461 to DMSO was calculated using two-tailed Student’s t tests among biological replicates.
As our data indicate that the up-regulation of autophagy genes in response to nucleolar stress is p53-dependent (Figure 6), we next tested the effect of inhibiting autophagy on the sensitization of HCT 116 p53 –/– cells to CX-5461 treatment. Compared with p53 WT cells, HCT 116 p53 –/– cells showed a greater reduction in cell viability after treatment with bafilomycin A1 alone (Figure 7E). However, cotreatment of bafilomycin A1 with CX-5461 did not further decrease cell survival, contrary to what was observed for p53 WT cells (Figure 7E). Similarly, chloroquine did not show any dose-dependent sensitization to CX-5461 treatment (Figure 7F). Both bafilomycin A1 and chloroquine have been shown to also reduce mTORC1 activity by acting as lysosomal storage disorders mimetics, which could explain their effects on p53 –/– cell viability (Fedele and Proud, 2020). Together, these observations suggest that blocking autophagy could be used as a strategy to impair the survival of p53-positive cancer cells after inhibition of ribosome biogenesis.
DISCUSSION
The data presented in this study demonstrate that inhibition of ribosome biogenesis leads to the establishment of a survival transcriptional program characterized by cell cycle arrest and activation of autophagy-promoting pathways in p53-positive cancer cells. Our data also uncovered several genes involved in the regulation of central metabolism and ROS control that are up-regulated in a p53-dependent manner in response to rRNA synthesis inhibition. These findings suggest that inhibition of ribosome biogenesis elicits a transient metabolic stress whereby p53 regulates an adaptive transcriptional program by promoting catabolism, metabolic rewiring, and protection against ROS, while inhibiting cellular proliferation.
Although the roles of p53 have mostly been studied in response to genotoxic stress, it is now well established that p53 also plays a major function in the transcriptional regulation of metabolic homeostasis in response to energetic stress (Napoli and Flores, 2017; Labuschagne et al., 2018; Lacroix et al., 2020). For example, p53 has been shown to activate fatty acid oxidation (FAO) (Goldstein and Rotter, 2012) and rearrange arginine metabolism through direct transcriptional activation of carnitine palmitoyltransferases (CPT1A, CPT1C) and argininosuccinate synthase 1 (ASS1), respectively, which we found up-regulated in a p53-dependent manner after inhibition of ribosome synthesis (Zaugg et al., 2011). Our findings suggest that inhibition of ribosome synthesis leads to a p53-dependent transcriptional program similar to what is observed during energetic stress.
Induction of autophagy by p53 has been shown to occur by different processes, and one of the best-understood mechanisms is inhibition of the mTORC1 pathway, a major negative regulator of autophagy. Nuclear p53 can also promote autophagy by transcriptionally activating target genes directly involved in promoting this process, such as LKB1, ULK1/2, ATG4, ATG7, and ATG10 (Kenzelmann Broz et al., 2013). Our findings suggest that activation of autophagy in response to nucleolar stress is mediated by the p53-dependent transcriptional up-regulation of SESTRIN 2, which inhibits mTORC1 activity through AMPK stimulation or by impairing mTORC1 localization to lysosomes (Parmigiani et al., 2014; Kim et al., 2015a, 2015b). Although not characterized as a p53 direct target, LC3B (MAP1LC3B, microtubule-associated protein 1 light-chain 3 transcript), required for elongation and maturation of the autophagosome, was also up-regulated in a p53-dependent manner after inhibition of ribosome biogenesis. This gene may represent a putative novel direct p53 target; however, further testing for p53 binding to the MAP1LC3B promoter region after nucleolar stress will be required to confirm this.
Several drugs used for cancer treatment have been shown to induce autophagy in tumors as a response mechanism against cellular stress (Thorburn et al., 2014). Survival through induction of autophagy is often considered an underlying cause of therapeutic resistance in multiple cancer types. Thus, there has been a growing interest in developing clinical trials aiming at blocking autophagy to potentiate the effect of a cancer drug (Levy et al., 2017). Targeting rRNA production has emerged as a promising therapeutic for cancer and much of the efforts have focused on developing selective inhibitors that block or disassemble the Pol I transcription machinery. Although it also exerts a cytotoxic effect as a G-quadruplex stabilizer and topoisomerase II poison (Xu et al., 2017; Bruno et al., 2020), the selective Pol I inhibitor CX-5461 is currently in clinical trial for the treatment of breast and advanced hematologic cancers. Studies have suggested that CX-5461 induces autophagy via inhibition of mTOR signaling in osteosarcoma and leukemia cells (Li et al., 2016; Okamoto et al., 2020). Our data corroborate these findings and suggest that inhibiting autophagy may be a viable strategy to heighten the responsiveness of ribosome biogenesis-targeted therapies, at least in p53 WT tumors. The majority of p53 mutations found in cancers are missense substitutions leading to the expression of mutant proteins with unique protumorigenic gain-of-function activities (Muller and Vousden, 2013). Interestingly, mutant p53 proteins have been shown to have the ability to hypertransactivate p53-target genes, such as CDKN1A (p21CIP1) and SESN2 (SESTRIN 2), after metabolic stress, thus promoting cell cycle arrest and cell survival (Tran et al., 2017). It will be important to evaluate the transcriptional program activated by these mutant p53 proteins after nucleolar stress to determine whether inhibition of rRNA synthesis would beneficially synergize with concomitant inhibition of autophagy or metabolic processes in the treatment of tumors harboring gain-of-function p53 mutations.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Cell culture, RNA interference, and drug treatments
Cells (HCT116 WT, HCT116 p53 –/–, MCF7, and U2OS) were cultured at 37°C with 5% CO2 in DMEM high glucose (Hyclone) supplemented with 5% fetal bovine serum (FBS, Hyclone) and penicillin–streptomycin. All cells used were obtained from ATCC, which performs short-tandem repeat profiling to confirm the identity of their cell lines. Cell lines are tested for mycoplasma contamination every 6 months by PCR assay with appropriate positive controls (Young et al., 2010). Synthetic short interfering RNA (siRNA) oligonucleotides (Sigma) were delivered into cells with Lipofectamine RNAi Max (Invitrogen). siRNA oligonucleotides (40 nM) were transfected using the reverse transfection protocol according to the manufacturer’s instructions. The following siRNA oligonucleotide targeting sequences were used:
nontargeting scramble: 5′-GAUCAUACGUGCGAUCAGAdTdT-3′;
human LAS1L #1: 5′-CUGAUACGCUGUAAGCUCUdTdT-3′;
human LAS1L #2: 5′-CAUUUAUACCCAGAGUGGAdTdT-3′;
human PELP1 5′-GGAAUGAAGGCUUGUAUGAdTdT-3′;
human NOP2 5′-CACCUGUUCUAUCACAGUAdTdT-3′;
human SESTRIN 2 #1 5′-CCUACAAUACCAUCGCCAUdTdT-3′;
human SESTRIN 2 #2 5′-CGAAGAAUGUACAACCUCUdTdT-3′;
human ATG7 #1 5′-CAGCUAUUGGAACACUGUAdTdT-3′;
human ATG7 #2 5′-GAGAUAUGGGAAUCCAUAAdTdT-3′;
human RPL11/uL5 5′-GGUGCGGGAGUAUGAGUUAdTdT-3′
CX-5461 (Cellagen Technologies) was reconstituted in DMSO and used at a final concentration of 500 nM. Bafilomycin A1 (Sigma) was reconstituted in DMSO and used at final concentrations of 50 and 100 nM. Chloroquine (Sigma) was reconstituted in sterile dH2O and used at final concentrations of 5 and 10 µM.
RNA, microarray, genes set enrichment analysis, and quantitative RT-PCR analysis
HCT116 cells were treated with control siRNA (in duplicate), LAS1L siRNA #1, or LAS1L siRNA #2 for 72 h. Total RNAs, extracted with Trizol reagent (Invitrogen), were amplified, labeled, and hybridized to an Illumina HumanHT-12 v4 expression bead chip kit using standard protocols at the Microarray Core Lab, UT McGovern Medical School. All analyses were performed using GenomeStudio software (Illumina). Genes set enrichment analysis (GSEA) was performed separately on up-regulated and down-regulated genes using the ontology and canonical pathways gene sets. The p value from the hypergeometric distribution for k-1, K, N-K, and n, where k is the number of genes in the intersection of the query set with a set from MSigDB (Subramanian et al., 2005; Liberzon et al., 2011, 2015), K is the number of genes in the set from MSigDB, N is the total number of genes in the universe (all known human gene symbols), and n is the number of genes in the query set, is represented. A cut off for FDR q-value less than 0.05 was selected. Only the top 10 gene sets were considered.
For quantitative RT-PCR analysis, complementary DNA (cDNA) was synthesized using 1 μg of total RNA with the QuantiTect reverse transcription kit (Qiagen). Real-time PCR analysis was performed on a Light Cycler 480 real-time PCR instrument. Values were normalized to beta-ACTIN mRNA. A qPCR standard curve was performed to test the efficiency of each primer pairs. The primers used for the quantitative PCR are listed in Supplemental Table S3.
Flow cytometry and cell survival assays
Cell cycle progression was assayed by DNA content using propidium iodide (PI) and BrdU labeling (anti-BrdU: BD Biosciences, Cat# 347580, goat anti-mouse Alexa-Fluo488, Invitrogen, Cat# A11001) followed by flow cytometry analysis as described previously (Castle et al., 2010). Analyses were performed on a BD LSRFortessa. For the survival assay, cells were transfected with nontargeting control, SESTRIN 2, or ATG7 siRNAs for 3 d. On day 2 posttransfections, cells were treated with vehicle (dimethyl sulfoxide, DMSO) or CX-5461 (500 nM) for 24 h. Another set of cells were grown in a monolayer and treated for 24 h with DMSO or CX-5461 (500 nM) in conjunction with either bafilomycin A1 (0, 50 nM, or 100 nM) or chloroquine (0, 5 µM, or 10 µM). Cell viability for both sets of experiments was determined by trypan blue exclusion.
Immunoblotting and antibodies
For immunoblotting, cells were lysed in RIPA buffer (25 mM Tris-HCl pH 7.6, 150 mM NaCl, 1% NP-40, 1% Triton X-100, 1% sodium deoxycholate, and 0.1% SDS) plus protease inhibitors (aprotinin, leupeptin, AEBSF) and phosphatase inhibitor cocktail (ThermoFisher) for 15 min on ice. Lysates were cleared by centrifugation at 22,000 × g for 10 min at 4°C. Protein concentrations were evaluated with a BCA kit (Pierce). Proteins were separated by SDS-polyacrylamide gel electrophoresis (SDS–PAGE) and transferred to a nitrocellulose membrane (Bio-Rad). The following antibodies were used: LAS1L (Cat#: A304-438A, Bethyl Laboratories), p53 (Cat#: SC-126, Santa Cruz Biotechnology), p21CIP1 (Cat#: 556430, BD Biosciences), β-ACTIN (Cat#: SC-69879, Santa Cruz Biotechnology), GAPDH (Cat#: 47724, Santa Cruz Biotechnology); RPL11/uL5 (Cat#: SAB1402896, Sigma), LC3B (Cat#: 3868, Cell Signaling), PELP1 (Cat#: A300-180A, Bethyl Laboratories), NOP2 (Cat#: A302-018A, Bethyl Laboratories), Cyclin E (Cat#: SC-198, Santa Cruz Biotechnology), SESTRIN 2 (Cat#: 10795-1-AP, Proteintech), and ATG7 (Cat#: SC-376212, Santa Cruz Biotechnology).
Fluorescence microscopy
For the detection of LAS1L by immunofluorescence, cells grown on cover glass were fixed with 4% paraformaldehyde at room temperature for 15 min. Cells were then permeabilized at room temperature with 0.1% Triton for 10 min. All samples were blocked with 1% BSA for 30 min, washed with PBS, and incubated with an anti-LAS1L antibody (Cat#: AV34629, Sigma) for 1 h. Cells were washed with PBS containing 0.05% Tween-20 and incubated with secondary antibody for 1 h (Alexa-Fluor 488 goat anti-rabbit antibody, Cat#: A11001, Invitrogen). Cells were washed again with PBS containing 0.05% Tween-20, stained with DAPI (Molecular Probes), and mounted on slides with Vectashield (Vector Labs). To visualize LC3 localization to autophagosomes, HCT116 cells were transfected with a plasmid expressing pEGFP-LC3 (a gift from Toren Finkel (Addgene plasmid #24920; Lee et al., 2008) and pools of stable clones were established after selection with G418. Fluorescence microscopy was performed on a Zeiss Axioskop 40 fluorescence microscope with a Plan-APOCHROMAT 63×/1.4 NA oil DIC objective. Images were acquired with an Axiocam MRm camera using the Axiovision Release 4.6 software with a capturing resolution of 1388 × 1040 pixels. All microscopy was performed at room temperature, and all images were prepared in Adobe Photoshop and Adobe Illustrator.
Statistical analyses
Results are represented as mean ± SEM (standard error of the mean) except as otherwise specified. Differences between groups were considered significant at p < 0.05 by a two-tailed Student’s t test. Data shown are average values of three independent experiments except as otherwise specified (each qPCR sample was performed with technical triplicates).
Supplementary Material
Acknowledgments
We thank the members of the Denicourt and Breton laboratories for their suggestions on the manuscript. This work was supported by NIH Grant R01-CA230746.
Abbreviations used:
- ATG
autophagy-related genes
- BrdU
Bromodeoxyuridine
- CDK
cyclin-dependent kinase
- ChIP-Seq
chromatin immunoprecipitation sequencing
- DMSO
dimethyl sulfoxide
- EGFP
enhanced green fluorescent protein
- FDR
false discovery rate
- GRO-Seq
global run-on sequencing
- GSEA
gene set enrichment analysis
- LC3
microtubule-associated protein 1 light chain 3
- MDM2
mouse double minute 2 homolog
- mTORC1
mammalian target of rapamycin complex 1
- PI
propidium iodide
- qRT-PCR
quantitative reverse transcription polymerase chain reaction
- RNP
ribonucleoprotein
- ROS
reactive oxygen species
- RPs
ribosomal proteins
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E21-05-0251).
REFERENCES
- Allen MA, Andrysik Z, Dengler VL, Mellert HS, Guarnieri A, Freeman JA, Sullivan KD, Galbraith MD, Luo X, Kraus WL, et al. (2014). Global analysis of p53-regulated transcription identifies its direct targets and unexpected regulatory mechanisms. eLife 3, e02200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert M, O’Donohue MF, Lebaron S, Gleizes PE (2018). Pre-ribosomal RNA processing in human cells: from mechanisms to congenital diseases. Biomolecules 8, 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barna M, Pusic A, Zollo O, Costa M, Kondrashov N, Rego E, Rao PH, Ruggero D (2008). Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature 456, 971–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bento CF, Renna M, Ghislat G, Puri C, Ashkenazi A, Vicinanza M, Menzies FM, Rubinsztein DC (2016). Mammalian autophagy: how does it work? Annu Rev Biochem 85, 685–713. [DOI] [PubMed] [Google Scholar]
- Bhat KP, Itahana K, Jin A, Zhang Y (2004). Essential role of ribosomal protein L11 in mediating growth inhibition-induced p53 activation. EMBO J 23, 2402–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack KE, Bohnsack MT (2019). Uncovering the assembly pathway of human ribosomes and its emerging links to disease. EMBO J 38, e100278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeois G, Ney M, Gaspar I, Aigueperse C, Schaefer M, Kellner S, Helm M, Motorin Y (2015). Eukaryotic rRNA modification by yeast 5-methylcytosine-methyltransferases and human proliferation-associated antigen p120. PLoS One 10, e0133321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno PM, Lu M, Dennis KA, Inam H, Moore CJ, Sheehe J, Elledge SJ, Hemann MT, Pritchard JR (2020). The primary mechanism of cytotoxicity of the chemotherapeutic agent CX-5461 is topoisomerase II poisoning. Proc Natl Acad Sci USA 117, 4053–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budanov AV, Karin M (2008). p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell 134, 451–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budanov AV, Sablina AA, Feinstein E, Koonin EV, Chumakov PM (2004). Regeneration of peroxiredoxins by p53-regulated sestrins, homologs of bacterial AhpD. Science 304, 596–600. [DOI] [PubMed] [Google Scholar]
- Bursać S, Brdovcˇak MC, Pfannkuchen M, Orsolic´ I, Golomb L, Zhu Y, Katz C, Daftuar L, Grabušic´ K, Vukelic´ I, et al. (2012). Mutual protection of ribosomal proteins L5 and L11 from degradation is essential for p53 activation upon ribosomal biogenesis stress. Proc Natl Acad Sci USA 109, 20467–20472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bywater MJ, Poortinga G, Sanij E, Hein N, Peck A, Cullinane C, Wall M, Cluse L, Drygin D, Anderes K, et al. (2012). Inhibition of RNA polymerase I as a therapeutic strategy to promote cancer-specific activation of p53. Cancer Cell 22, 51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle CD, Cassimere EK, Denicourt C (2012). LAS1L interacts with the mammalian Rix1 complex to regulate ribosome biogenesis. Mol Biol Cell 23, 716–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle CD, Cassimere EK, Lee J, Denicourt C (2010). Las1L is a nucleolar protein required for cell proliferation and ribosome biogenesis. Mol Cell Biol 30, 4404–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle CD, Sardana R, Dandekar V, Borgianini V, Johnson AW, Denicourt C (2013). Las1 interacts with Grc3 polynucleotide kinase and is required for ribosome synthesis in Saccharomyces cerevisiae. Nucleic Acids Res 41, 1135–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty A, Uechi T, Kenmochi N (2011). Guarding the ‘translation apparatus’: defective ribosome biogenesis and the p53 signaling pathway. Wiley interdisciplinary reviews. RNA 2, 507–522. [DOI] [PubMed] [Google Scholar]
- Chan JC, Hannan KM, Riddell K, Ng PY, Peck A, Lee RS, Hung S, Astle MV, Bywater M, Wall M, et al. (2011). AKT promotes rRNA synthesis and cooperates with c-MYC to stimulate ribosome biogenesis in cancer. Science Signaling 4, ra56. [DOI] [PubMed] [Google Scholar]
- Chen D, Zhang Z, Li M, Wang W, Li Y, Rayburn ER, Hill DL, Wang H, Zhang R (2007). Ribosomal protein S7 as a novel modulator of p53-MDM2 interaction: binding to MDM2, stabilization of p53 protein, and activation of p53 function. Oncogene 26, 5029–5037. [DOI] [PubMed] [Google Scholar]
- Ciganda M, Williams N (2011). Eukaryotic 5S rRNA biogenesis. RNA 2, 523–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelison R, Dobbin ZC, Katre AA, Jeong DH, Zhang Y, Chen D, Petrova Y, Llaneza DC, Steg AD, Parsons L, et al. (2017). Targeting RNA-polymerase I in both chemosensitive and chemoresistant populations in epithelial ovarian cancer. Clin Cancer Res 23, 6529–6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai MS, Lu H (2004). Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J Biol Chem 279, 44475–44482. [DOI] [PubMed] [Google Scholar]
- Dai MS, Zeng SX, Jin Y, Sun XX, David L, Lu H (2004). Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol Cell Biol 24, 7654–7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisenroth C, Franklin DA, Zhang Y (2016). The evolution of the ribosomal protein-MDM2-p53 pathway. Cold Spring Harb Perspect Med 6, a026138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delloye-Bourgeois C, Goldschneider D, Paradisi A, Therizols G, Belin S, Hacot S, Rosa-Calatrava M, Scoazec JY, Diaz JJ, Bernet A, Mehlen P (2012). Nucleolar localization of a netrin-1 isoform enhances tumor cell proliferation. Science signaling 5, ra57. [DOI] [PubMed] [Google Scholar]
- Drygin D, Lin A, Bliesath J, Ho CB, O’Brien SE, Proffitt C, Omori M, Haddach M, Schwaebe MK, Siddiqui-Jain A, et al. (2011). Targeting RNA polymerase I with an oral small molecule CX-5461 inhibits ribosomal RNA synthesis and solid tumor growth. Cancer Res 71, 1418–1430. [DOI] [PubMed] [Google Scholar]
- Fedele AO, Proud CG (2020). Chloroquine and bafilomycin A mimic lysosomal storage disorders and impair mTORC1 signalling. Biosci Rep 40, BSR20200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkbeiner E, Haindl M, Muller S (2011). The SUMO system controls nucleolar partitioning of a novel mammalian ribosome biogenesis complex. EMBO J 30, 1067–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli S, Di Cara A, Neb-Gulati A, Natt F, Schwemberger S, Hall J, Babcock GF, Bernardi R, Pandolfi PP, Thomas G (2009). Absence of nucleolar disruption after impairment of 40S ribosome biogenesis reveals an rpL11-translation-dependent mechanism of p53 induction. Nat Cell Biol 11, 501–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli S, Ivanenkov VV, Teng T, Thomas G (2012). Suprainduction of p53 by disruption of 40S and 60S ribosome biogenesis leads to the activation of a novel G2/M checkpoint. Genes Dev 26, 1028–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasse L, Flemming D, Hurt E (2015). Coordinated ribosomal ITS2 RNA processing by the Las1 complex integrating endonuclease, polynucleotide kinase, and exonuclease activities. Mol Cell 60, 808–815. [DOI] [PubMed] [Google Scholar]
- Goldstein I, Rotter V (2012). Regulation of lipid metabolism by p53—fighting two villains with one sword. Trends Endocrinol Metab 23, 567–575. [DOI] [PubMed] [Google Scholar]
- Golomb L, Volarevic S, Oren M (2014). p53 and ribosome biogenesis stress: the essentials. FEBS Letters 588, 2571–2579. [DOI] [PubMed] [Google Scholar]
- Hadjiolova KV, Nicoloso M, Mazan S, Hadjiolov AA, Bachellerie JP (1993). Alternative pre-rRNA processing pathways in human cells and their alteration by cycloheximide inhibition of protein synthesis. Eur J Biochem 212, 211–215. [DOI] [PubMed] [Google Scholar]
- Haines RJ, Pendleton LC, Eichler DC (2011). Argininosuccinate synthase: at the center of arginine metabolism. Int J Biochem Mol Biol 2, 8–23. [PMC free article] [PubMed] [Google Scholar]
- Holzel M, Orban M, Hochstatter J, Rohrmoser M, Harasim T, Malamoussi A, Kremmer E, Langst G, Eick D (2010). Defects in 18 S or 28 S rRNA processing activate the p53 pathway. J Biol Chem 285, 6364–6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong B, Brockenbrough JS, Wu P, Aris JP (1997). Nop2p is required for pre-rRNA processing and 60S ribosome subunit synthesis in yeast. Mol Cell Biol 17, 378–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismael M, Webb R, Ajaz M, Kirkby KJ, Coley HM (2019). The targeting of RNA polymerase I transcription using CX-5461 in combination with radiation enhances tumour cell killing effects in human solid cancers. Cancers (Basel) 11, 1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin A, Itahana K, O’Keefe K, Zhang Y (2004). Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Mol Cell Biol 24, 7669–7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastenhuber ER, Lowe SW (2017). Putting p53 in context. Cell 170, 1062–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenzelmann Broz D, Spano Mello S, Bieging KT, Jiang D, Dusek RL, Brady CA, Sidow A, Attardi LD (2013). Global genomic profiling reveals an extensive p53-regulated autophagy program contributing to key p53 responses. Genes Dev 27, 1016–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khot A, Brajanovski N, Cameron DP, Hein N, Maclachlan KH, Sanij E, Lim J, Soong J, Link E, Blombery P, et al. (2019). First-in-human RNA polymerase I transcription inhibitor CX-5461 in patients with advanced hematologic cancers: results of a phase I dose-escalation study. Cancer Discov 9, 1036–1049. [DOI] [PubMed] [Google Scholar]
- Kim H, An S, Ro SH, Teixeira F, Park GJ, Kim C, Cho CS, Kim JS, Jakob U, Lee JH, Cho US (2015a). Janus-faced Sestrin2 controls ROS and mTOR signalling through two separate functional domains. Nature communications 6, 10025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Ro SH, Kim M, Park HW, Semple IA, Park H, Cho US, Wang W, Guan KL, Karin M, Lee JH (2015b). Sestrin2 inhibits mTORC1 through modulation of GATOR complexes. Sci Rep 5, 9502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Tanida I, Ueno T, Ohsumi M, Ohsumi Y, Kominami E (2001). The C-terminal region of an Apg7p/Cvt2p is required for homodimerization and is essential for its E1 activity and E1-E2 complex formation. J Biol Chem 276, 9846–9854. [DOI] [PubMed] [Google Scholar]
- Labuschagne CF, Zani F, Vousden KH (2018). Control of metabolism by p53—cancer and beyond. Biochim Biophys Acta 1870, 32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix M, Riscal R, Arena G, Linares LK, Le Cam L (2020). Metabolic functions of the tumor suppressor p53: implications in normal physiology, metabolic disorders, and cancer. Mol Metab 33, 2–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, Tsokos M, Alt FW, Finkel T (2008). A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci USA 105, 3374–3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy JMM, Towers CG, Thorburn A (2017). Targeting autophagy in cancer. Nat Rev Cancer 17, 528–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Li Y, Zhao J, Fan S, Wang L, Li X (2016). CX-5461 induces autophagy and inhibits tumor growth via mammalian target of rapamycin-related signaling pathways in osteosarcoma. Onco Targets Ther 9, 5985–5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P (2015). The molecular signatures database (MSigDB) hallmark gene set collection. Cell systems 1, 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP (2011). Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH (2003). Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell 3, 577–587. [DOI] [PubMed] [Google Scholar]
- Maiuri MC, Malik SA, Morselli E, Kepp O, Criollo A, Mouchel PL, Carnuccio R, Kroemer G (2009). Stimulation of autophagy by the p53 target gene Sestrin2. Cell cycle (Georgetown, Tex.) 8, 1571–1576. [DOI] [PubMed] [Google Scholar]
- Menendez D, Nguyen TA, Freudenberg JM, Mathew VJ, Anderson CW, Jothi R, Resnick MA (2013). Diverse stresses dramatically alter genome-wide p53 binding and transactivation landscape in human cancer cells. Nucleic Acids Res 41, 7286–7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merida I, Avila-Flores A, Merino E (2008). Diacylglycerol kinases: at the hub of cell signalling. Biochem J 409, 1–18. [DOI] [PubMed] [Google Scholar]
- Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massague J (2005). Genes that mediate breast cancer metastasis to lung. Nature 436, 518–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourgues L, Imbert V, Nebout M, Colosetti P, Neffati Z, Lagadec P, Verhoeyen E, Peng C, Duprez E, Legros L, et al. (2015). The BMI1 polycomb protein represses cyclin G2-induced autophagy to support proliferation in chronic myeloid leukemia cells. Leukemia 29, 1993–2002. [DOI] [PubMed] [Google Scholar]
- Muller PA, Vousden KH (2013). p53 mutations in cancer. Nat Cell Biol 15, 2–8. [DOI] [PubMed] [Google Scholar]
- Napoli M, Flores ER (2017). The p53 family orchestrates the regulation of metabolism: physiological regulation and implications for cancer therapy. Br J Cancer 116, 149–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikulenkov F, Spinnler C, Li H, Tonelli C, Shi Y, Turunen M, Kivioja T, Ignatiev I, Kel A, Taipale J, Selivanova G (2012). Insights into p53 transcriptional function via genome-wide chromatin occupancy and gene expression analysis. Cell Death Differ 19, 1992–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto S, Miyano K, Kajikawa M, Yamauchi A, Kuribayashi F (2020). The rRNA synthesis inhibitor CX-5461 may induce autophagy that inhibits anticancer drug-induced cell damage to leukemia cells. Biosci Biotechnol Biochem 84, 2319–2326. [DOI] [PubMed] [Google Scholar]
- Padua D, Zhang XH, Wang Q, Nadal C, Gerald WL, Gomis RR, Massague J (2008). TGFbeta primes breast tumors for lung metastasis seeding through angiopoietin-like 4. Cell 133, 66–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmigiani A, Nourbakhsh A, Ding B, Wang W, Kim YC, Akopiants K, Guan KL, Karin M, Budanov AV (2014). Sestrins inhibit mTORC1 kinase activation through the GATOR complex. Cell Rep 9, 1281–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne SL, Fogelgren B, Hess AR, Seftor EA, Wiley EL, Fong SF, Csiszar K, Hendrix MJ, Kirschmann DA (2005). Lysyl oxidase regulates breast cancer cell migration and adhesion through a hydrogen peroxide-mediated mechanism. Cancer Res 65, 11429–11436. [DOI] [PubMed] [Google Scholar]
- Pelletier J, Thomas G, Volarevic S (2018). Ribosome biogenesis in cancer: new players and therapeutic avenues. Nat Rev Cancer 18, 51–63. [DOI] [PubMed] [Google Scholar]
- Penzo M, Montanaro L, Trere D, Derenzini M (2019). The ribosome biogenesis–cancer connection. Cells 8, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestov DG, Strezoska Z, Lau LF (2001). Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: effects of nucleolar protein Bop1 on G(1)/S transition. Mol Cell Biol 21, 4246–4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak K, Xia Y, Zweier JL, Kinzler KW, Vogelstein B (1997). A model for p53-induced apoptosis. Nature 389, 300–305. [DOI] [PubMed] [Google Scholar]
- Proikas-Cezanne T, Takacs Z, Donnes P, Kohlbacher O (2015). WIPI proteins: essential PtdIns3P effectors at the nascent autophagosome. J Cell Sci 128, 207–217. [DOI] [PubMed] [Google Scholar]
- Rubbi CP, Milner J (2003). Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J 22, 6068–6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schillewaert S, Wacheul L, Lhomme F, Lafontaine DL (2012). The evolutionarily conserved protein Las1 is required for pre-rRNA processing at both ends of ITS2. Mol Cell Biol 32, 430–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlereth K, Heyl C, Krampitz AM, Mernberger M, Finkernagel F, Scharfe M, Jarek M, Leich E, Rosenwald A, Stiewe T (2013). Characterization of the p53 cistrome–DNA binding cooperativity dissects p53’s tumor suppressor functions. PLoS Genet 9, e1003726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan KE, Bohnsack MT, Watkins NJ (2013). The 5S RNP couples p53 homeostasis to ribosome biogenesis and nucleolar stress. Cell Rep 5, 237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XX, Wang YG, Xirodimas DP, Dai MS (2010). Perturbation of 60 S ribosomal biogenesis results in ribosomal protein L5- and L11-dependent p53 activation. J Biol Chem 285, 25812–25821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I, Mizushima N, Kiyooka M, Ohsumi M, Ueno T, Ohsumi Y, Kominami E (1999). Apg7p/Cvt2p: a novel protein-activating enzyme essential for autophagy. Mol Biol Cell 10, 1367–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tew KD, Townsend DM (2012). Glutathione-s-transferases as determinants of cell survival and death. Antioxid Redox Signal 17, 1728–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorburn A, Thamm DH, Gustafson DL (2014). Autophagy and cancer therapy. Mol Pharmacol 85, 830–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran TQ, Lowman XH, Reid MA, Mendez-Dorantes C, Pan M, Yang Y, Kong M (2017). Tumor-associated mutant p53 promotes cancer cell survival upon glutamine deprivation through p21 induction. Oncogene 36, 1991–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Niu D, Lam TH, Xiao Z, Ren EC (2014). Mapping the p53 transcriptome universe using p53 natural polymorphs. Cell Death Differ. 21, 521–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Di Antonio M, McKinney S, Mathew V, Ho B, O’Neil NJ, Santos ND, Silvester J, Wei V, Garcia J, et al. (2017). CX-5461 is a DNA G-quadruplex stabilizer with selective lethality in BRCA1/2 deficient tumours. Nature communications 8, 14432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young L, Sung J, Stacey G, Masters JR (2010). Detection of mycoplasma in cell cultures. Nat Protoc 5, 929–934. [DOI] [PubMed] [Google Scholar]
- Yuan X, Zhou Y, Casanova E, Chai M, Kiss E, Grone HJ, Schutz G, Grummt I (2005). Genetic inactivation of the transcription factor TIF-IA leads to nucleolar disruption, cell cycle arrest, and p53-mediated apoptosis. Mol Cell 19, 77–87. [DOI] [PubMed] [Google Scholar]
- Zaugg K, Yao Y, Reilly PT, Kannan K, Kiarash R, Mason J, Huang P, Sawyer SK, Fuerth B, Faubert B, et al. (2011). Carnitine palmitoyltransferase 1C promotes cell survival and tumor growth under conditions of metabolic stress. Genes Dev 25, 1041–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lozano G (2017). p53: multiple facets of a Rubik’s cube. Annu Rev Cancer Biol 1, 185–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wolf GW, Bhat K, Jin A, Allio T, Burkhart WA, Xiong Y (2003). Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol Cell Biol 23, 8902–8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Poyurovsky MV, Li Y, Biderman L, Stahl J, Jacq X, Prives C (2009). Ribosomal protein S7 is both a regulator and a substrate of MDM2. Mol Cell 35, 316–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.