Abstract
Keratin intermediate filaments form dynamic polymer networks that organize in specific ways dependent on the cell type, the stage of the cell cycle, and the state of the cell. In differentiated cells of the epidermis, they are organized by desmosomes, cell–cell adhesion complexes that provide essential mechanical integrity to this tissue. Despite this, we know little about how keratin organization is controlled and whether desmosomes locally regulate keratin dynamics in addition to binding preassembled filaments. Ndel1 is a desmosome-associated protein in the differentiated epidermis, though its function at these structures has not been examined. Here, we show that Ndel1 binds directly to keratin subunits through a motif conserved in all intermediate filament proteins. Further, Ndel1 was necessary for robust desmosome–keratin association and sufficient to reorganize keratins at distinct cellular sites. Lis1, a Ndel1 binding protein, was required for desmosomal localization of Ndel1, but not for its effects on keratin filaments. Finally, we use mouse genetics to demonstrate that loss of Ndel1 results in desmosome defects in the epidermis. Our data thus identify Ndel1 as a desmosome-associated protein that promotes local assembly/reorganization of keratin filaments and is essential for robust desmosome formation.
INTRODUCTION
Cytoskeletal polymers assemble and are organized to control cell shape, motility, and adhesion. While a great deal of research has identified a large number of proteins that regulate the assembly, dynamics, and organization of both microtubules and F-actin, very few proteins that regulate intermediate filament (IF) organization are known. Yet we understand that intermediate filaments are dynamic structures that respond to changes in a cell’s state (Windoffer et al., 2011). For example, IFs remodel during cell division, in response to externally applied forces on cadherins, at the leading edge of migrating cells, and they quickly form attachments with desmosomes and hemidesmosomes during the assembly of these cell adhesion structures (Liao and Omary, 1996; Coulombe and Wong, 2004; Ridge et al., 2005; Windoffer et al., 2006; Weber et al., 2012). Further emphasizing the need to understand how IFs organize is the spectrum of diseases caused by mutations in these proteins, including blistering disorders in the epidermis caused by mutations in keratins and myopathies/cardiomyopathies associated with desmin mutations (Smith, 2003; Eriksson et al., 2009; Clemen et al., 2013).
The association of desmosome cell–cell adhesion complexes with IFs provides robust mechanical strength to tissues that experience high levels of external forces (Green and Simpson, 2007). Disruption of desmosomes results in a range of disorders of the skin from mild cases presenting with skin thickening to lethal blistering (Kottke et al., 2006). Mutations in genes for desmosomal proteins also cause arrhythmogenic cardiomyopathy—a heart defect characterized by fibro-fatty infiltrates (Delmar and McKenna, 2010; Costa et al., 2020). The major connection between desmosomes and IFs is provided by desmoplakin—a core component of the desmosomal plaque that directly binds to the epithelial keratin filaments and the desmin filaments in cardiac muscle (Stappenbeck et al., 1993; Kouklis et al., 1994; Bornslaeger et al., 1996; Gallicano et al., 1998). Until recently, there were no data that desmosomes actively control IF assembly or organization beyond simply binding preexisting filaments. However, Leube and colleagues recently imaged events that appear to be the de novo nucleation of keratin filaments at desmosomes (Moch et al., 2020). This extended prior work demonstrating that keratin filaments assembled near focal complexes at the leading edge of migrating cells (Windoffer et al., 2006). These findings are intriguing as other cytoskeleton organizing structures—including centrosomes and adherens junctions—also influence the assembly, organization, and dynamics of the polymers they bind (Chhabra and Higgs, 2007; Kollman et al., 2011). Centrosomes recruit γ-tubulin to nucleate microtubules, while both the Arp2/3 complex and formins have been reported to promote local actin assembly at adherens junctions (Moritz et al., 1995; Zheng et al., 1995; Kobielak et al., 2004; Tang and Brieher, 2012; Verma et al., 2012). At present there is only a limited repertoire of proteins known to control keratin organization. These include proteins that bind to preformed filaments, like desmoplakin, as well as other plakin family members, for example, plectin, that can cross-link keratins with other cytoskeletal structures (Sonnenberg and Liem, 2007; Suozzi et al., 2012). In addition, keratin assembly status can be controlled by posttranslational modifications (Omary et al., 1998). This lack of understanding of polymer assembly is largely true for other IF proteins as well. However, the assembly of neurofilaments, an IF polymer quite distinct from keratins, is controlled by Ndel1—a protein whose function has been best studied in neurodevelopment and in its association with Lis1 and dynein (Niethammer et al., 2000; Nguyen et al., 2004; Chansard et al., 2011). Ndel1 promoted the assembly of neurofilaments from purified subunits, and its disruption resulted in neurofilament disorganization in neurons (Nguyen et al., 2004). In addition, Ndel1 has been implicated in controlling the organization of both nuclear lamins and vimentin, two other types of IFs, although these effects have been attributed, at least in part, to dynein-directed transport (Shim et al., 2008; Ma et al., 2009).
In the epidermis, Ndel1, and its binding partner Lis1, localize to centrosomes in epidermal progenitor cells (Sumigray et al., 2011). Upon differentiation, both of these proteins relocalize to desmosomes where Lis1 is necessary to control desmosome-dependent microtubule reorganization (Lechler and Fuchs, 2007; Sumigray et al., 2011). In addition, loss of Lis1 resulted in desmosome defects although the underlying mechanism is unknown. What role Ndel1 provides at desmosomes, and whether it is important for either microtubule or keratin filament organization, has not yet been addressed. Here we demonstrate that Ndel1 directly interacts with keratin subunits and promotes their local assembly/organization within the cell. Further, Ndel1 is required for proper desmosome function in both cultured keratinocytes and intact epidermis.
RESULTS
Ndel1 directly interacts with keratin subunits
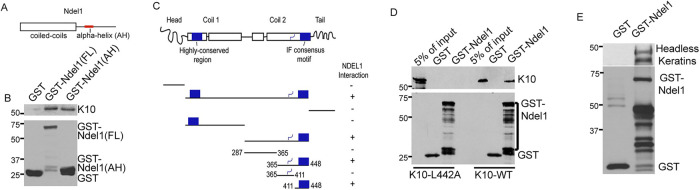
To test the possibility that Ndel1 binds to keratins, we purified GST-tagged Ndel1 and keratin 10 (K10) from bacterial lysates. Ndel1 contains an N-terminal coiled-coil domain that is required for its interaction with Lis1 and an alpha-helix (AH) near in its C-terminus that is important for interaction with a number of proteins, including DISC1 and dynein/dynactin (Figure 1A) (Soares et al., 2012). K10 and keratin 1 (K1) are the most abundant keratins in the differentiated cells of the epidermis (Roop et al., 1988). We saw an interaction of the full-length Ndel1 protein, as well as the isolated AH, with K10 (Figure 1B). Using this binding assay, we performed structure/function analysis of K10 to define the region(s) of the protein required for this interaction. These studies demonstrated that the IF consensus motif of K10 was both necessary and sufficient for interaction with Ndel1 (Figure 1C). This short sequence is found in all intermediate filament proteins, suggesting that Ndel1 is likely to have direct interactions with diverse IF protein families (Supplemental Figure 1) (Geisler and Weber, 1982; Geisler et al., 1983). We performed mutagenesis across this conserved IF motif to identify important amino acids for Ndel1 binding. A conservative change of leucine 442 to alanine was sufficient to disrupt the interaction between Ndel1 and K10 both with the isolated peptide (unpublished data) and with the full-length protein (Figure 1D). This leucine residue is absolutely conserved in the consensus motif across both vertebrate and invertebrate IFs (Supplemental Figure 1). We also found that the highly homologous Ndel1 paralogue, Nde1, could similarly bind to K10, as well as K1 and K5 (Supplemental Figure 2). Despite this biochemical interaction with keratin subunits, we have been unable to detect colocation of Ndel1 with keratin filaments in keratinocytes, in conditions where desmosomes are either formed or not and irrespective of whether we looked at endogenous protein, exogenous protein, or the C-terminal fragment of Ndel1 that can directly bind to keratin subunits (Sumigray et al., 2011; and unpublished data).
FIGURE 1:
Ndel1 directly binds to keratin subunits. (A) Diagram of the domains of Ndel1. (B) GST-pull down assay showing interactions between Ndel1 and keratin 10 (K10). Purified proteins were used to demonstrate a direct interaction between GST-Ndel1, both full length (FL) and the isolated alpha-helical region (AH), and keratin 10. (C) Diagram of keratin domain structure and truncation mutants tested for keratin 10 interaction. Plus marks indicate Ndel1 interaction. Note that the IF consensus motif is sufficient for interaction. (D) Full-length keratin 10 or a point mutant, keratin 10(L442A), was mixed with glutathione-agarose beads bound to GST or GST-Ndel1. Specific interaction of WT but not mutant keratin was noted with GST-Ndel1. Note that the multiple bands in the GST-Ndel1 lane are degradation products. (E) Purified headless K1 and K10 (K1 amino acids 185–637 and K10 amino acids 129–561) were allowed to interact and then mixed with glutathione agarose bound to either GST or GST-Ndel1. A pan-keratin antibody detected association of both headless keratins with GST-Ndel1.
To gain additional information on the Ndel1/keratin interaction, we purified headless keratins, which are unable to polymerize into filaments although they can associate into soluble dimers and tetramers (Hatzfeld and Burba, 1994). Upon centrifugation, the majority of the mutant keratins remained in solution (unpublished data). When the headless keratins were incubated with GST-Ndel1, they were able to effectively interact as shown by GST-Ndel’s ability to pull down the headless keratin subunits (Figure 1E). This demonstrates that Ndel1 is likely to interact with physiologically relevant keratin subunits. Subunit binding proteins can have diverse effects on polymer assembly; they can either sequester monomers, inhibiting polymerization (as is the case for thymosin β-4 for actin and stathmin for tubulin) (Goldschmidt-Clermont et al., 1992; Belmont and Mitchison, 1996), or act to promote polymerization (VCA domains, formins, and profilin in F-actin assembly, TOG domains in tubulin assembly) (Firat-Karalar and Welch, 2011; Gunzelmann et al., 2018). Therefore, we sought to analyze whether Ndel1 might affect keratin organization in cells.
Ndel1 affects keratin organization in keratinocytes
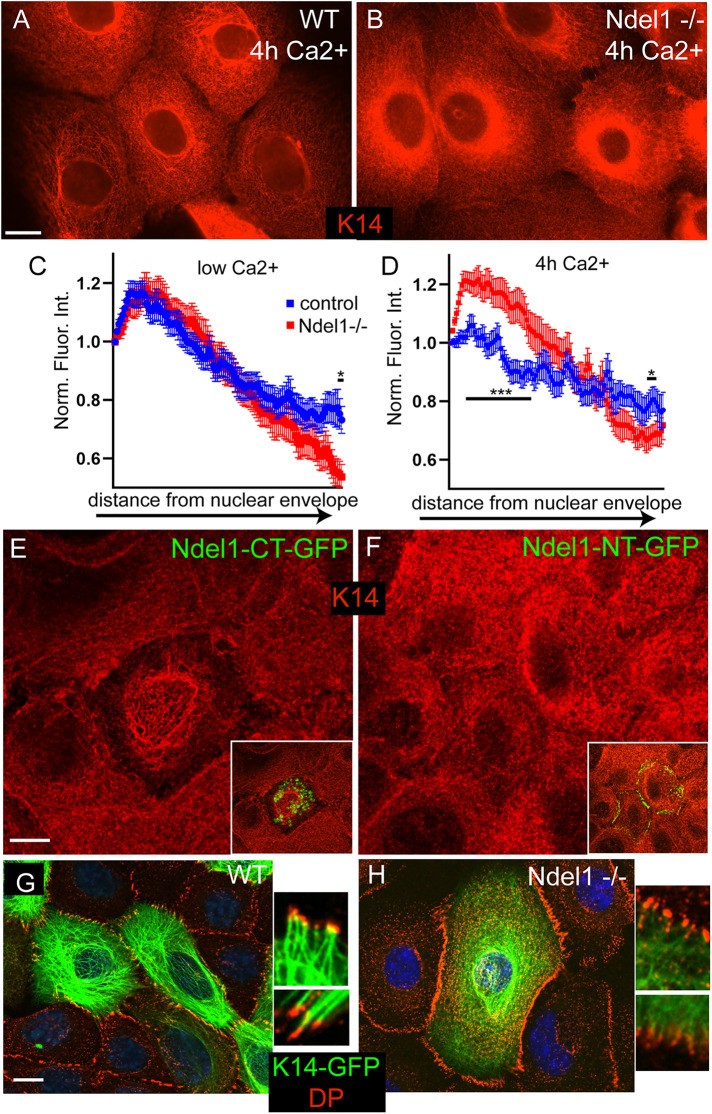
To understand the effects that loss of Ndel1 has in cells, we isolated Ndel1 null keratinocytes (described in more detail below) along with wild-type (WT) controls from the back skin of postnatal day 0 (p0) mice and established cell lines from them. Immunofluorescence analysis of keratin organization after calcium-induced desmosome formation revealed an increase in the perinuclear localization of keratins and a decrease in cortical filaments in Ndel1 null cells when compared with keratin networks in WT cells (Figure 2, A–D). In contrast, there were only minor alterations in keratin organization in low-calcium conditions, though a decrease in peripheral keratin was noted (Figure 2C). In addition, we also examined the effect of expression of a GFP-tagged carboxy-terminal fragment of Ndel1 (Ndel1-CT-GFP) that lacks desmosome targeting activity but retains its keratin interaction domain. Ndel1-CT-GFP did not localize to desmosomes but did cause disruption of the keratin filament network (Figure 2E; keratin disrupted in 67 ± 6% of cells, 60 cells from three experiments). In contrast, an amino-terminal fragment of Ndel1 that lacks its keratin-interaction domain (Ndel1-NT-GFP) localized to desmosomes but did not disrupt keratin organization appreciably (Figure 2F) (15 ± 5% of cells, 60 cells from three experiments, p = 0.0003; Student’s t test).
FIGURE 2:
Ndel1 regulates keratin organization. (A, B) Endogenous keratin organization in WT and Ndel1 KO cells stained with anti-K5/14 antibodies. Cells were switched to high-calcium-containing medium for 4 h before fixation. (C, D) Quantitation of keratin intensity, in low-calcium media and after switch to high calcium for 4 h, in line plots from the nuclear periphery toward the cell edge. (E) WT cells were transfected with Ndel1-CT-GFP and stained for keratin 14 (red). Note the partial collapse of the keratin network. (F) WT cells were transfected with Ndel1-NT-GFP and stained for keratin 14 (red). (G, H) WT and Ndel1 null cells were transfected with keratin 14-GFP (green), grown to confluency, switched to high-calcium-containing media for 4 h, and then fixed and stained for DP (red). Note the higher levels of peripheral keratin filaments and increased desmosome–keratin attachment in WT cells as compared with Ndel1 null cells. Scale bars are 10 μm (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
We were most interested in the connections of keratins to the desmosomes, as Ndel1 localizes to these structures and they are important sites for keratin assembly/organization in keratinocytes (Sumigray et al., 2011; Moch et al., 2020). We therefore transfected cells with GFP-tagged keratin 14 (K14-GFP) for better resolution of keratin filaments (especially at the cell periphery) than we could achieve through indirect immunofluorescence. After 4 h in Ca2+-containing media, clear and strong connections between well-organized keratin filaments and desmosomes were observed at the desmosome in WT cells (Figure 2G). While some keratin attachment was noted in Ndel1 null cells, there were fewer filaments at the cortex and the keratin network was overall poorly organized, possessing the same increase in perinuclear localization we previously observed (Figure 2H). These data are consistent with Ndel1’s keratin binding activity playing an important role at the desmosome to promote keratin organization and attachment. However, our data cannot distinguish whether Ndel1 is directly acting on the desmosome or on keratin filaments to cause this effect.
Ndel1 promotes local keratin filament assembly
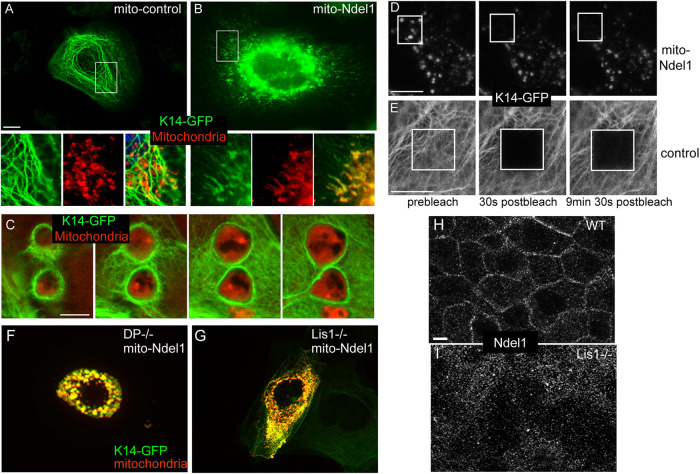
Our data suggest that Ndel1 could be acting at desmosomes locally promote keratin filament assembly/organization, which we hypothesize is required for the efficient attachment of desmosomes to the keratin network. This is supported not only by our biochemical and cell biological data, but also by the published finding that Ndel1 can promote neurofilament assembly kinetics (Nguyen et al., 2004). Unfortunately, the biochemical nature of keratins has prevented us from performing similar kinetic assays for assembly. To test whether Ndel1 is able to affect the local organization of keratins within the cell, we have taken a mistargeting approach. We fused Ndel1 to a mitochondrial membrane targeting sequence (mito-Ndel1) and expressed it in WT keratinocytes. We then coexpressed mito-Ndel1 with K14-GFP to report on keratin organization. When a control construct of mito-PAGFP (mito-photoactivable-GFP) was transfected, keratin organization appeared normal and there was little overlap between mitochondria and the keratin network (Figure 3A). In contrast, expression of mito-Ndel1 induced a dramatic reorganization of the keratin network. Most prominently, we noted small structures in the cell that were enriched in K14-GFP and that were stained by the mitochondrial dye MitoTracker-Red (Figure 3B). Therefore, Ndel1 generated a local change in keratin organization when targeted to mitochondria. Upon extended incubation times after transfection, we noted defects in mitochondrial morphology (Figure 3C). Mitochondria became either larger and round or formed elongated tubules and also showed a high concentration of keratins around them, which is likely responsible for these morphological changes. We thus wanted to determine whether the effects on mitochondrial morphology were dependent on keratin recruitment. We performed the same experiment in WT cells, keratin null cells, and keratin null cells in which the keratin network has been rescued by cotransfection with K5-GFP. We found that keratin null cells were less sensitive to defects in mitochondria morphology due to the expression of mitochondrially targeted Nde1 and that reexpression of K5 restored this sensitivity to WT levels (Supplemental Figure 3A). These data demonstrate that mitochondrial morphology defects are at least partly due to keratin recruitment.
FIGURE 3:
Targeting Ndel1 to mitochondria results in keratin filament reorganization around these organelles. (A, B) WT keratinocytes were cotransfected with keratin 14-GFP (green) and mito-PAGFP (control) or mito-Ndel1 constructs. Mitochondria were labeled by incubating cells with MitoTracker-Red for 30 min before fixation. mito-Ndel1 induced a reorganization of keratins onto the surface of mitochondria. Scale bar is 10 μm. (C) Z-stack through enlarged mitochondria surrounded by keratin filaments. Red, mitochondria; green, keratin 14-GFP. Scale bar is 5 μm. (D, E) FRAP analysis of keratin 14-GFP in control cell and cells transfected with mito-Ndel1. Scale bars in D and E are 10 μm. (F) Desmoplakin null keratinocytes were cotransfected with keratin 14-GFP (green) and mito-Ndel1 constructs. (G) Lis1fl/fl keratinocytes were incubated with adenoviral-Cre recombinase for 24 h and then cotransfected with keratin 14-GFP (green) and mito-Ndel1 constructs. (H, I) Ndel1 localization in WT (H) and Lis1 null (I) keratinocytes was analyzed using an immunofluorescent stain against Ndel1.
It was difficult to discern true filamentous structures around the mitochondria in many cells due to the local concentration of keratins. To determine whether the keratins associated with these mitochondria formed stable structures, a property of filamentous keratin, we performed fluorescence recovery after photobleaching (FRAP) on these cells. In both control and mito-Ndel1 cells, K14-GFP recovery was very slow, consistent with the majority of the mitochondrially associated keratin being in the filamentous form (Figure 3, D and E; representative graphs in Supplemental Figure 3, B and C). This suggests that Ndel1 promotes the local assembly/reorganization of keratin filaments at specific subcellular sites.
Desmoplakin and Lis1 are required for Ndel1 targeting, but not keratin filament assembly
One trivial explanation for Ndel1’s effects on keratin organization could be that it recruits desmoplakin to mitochondria. To test this, we examined the localization of GFP-tagged desmoplakin (DPI-GFP) in mito-Ndel1–expressing cells. We were never saw any localization of DPI-GFP at mitochondria in these experiments (Supplemental Figure 3D). To directly test whether desmoplakin is required for Ndel1’s keratin reorganization activity, we expressed mito-Ndel1 in desmoplakin null keratinocytes. These cells showed no change in keratin accumulation around mitochondria, demonstrating that desmoplakin is not required for this activity (Figure 3F).
Lis1 binds to Ndel1, and its loss results in desmosome defects (Sumigray et al., 2011). To test whether Ndel1’s keratin assembly activity required Lis1, we expressed full-length mito-Ndel1 in Lis1 null cells. To perform this experiment, Lis1 was ablated in Lis1 flox/flox cells by infection with high-titer adenoviral-Cre recombinase. We waited 24 h after infection to ensure complete loss of Lis1 ( Sumigray et al., 2011). K14-GFP localized around mitochondria in these cells, demonstrating that Lis1 is not required for mito-Ndel1’s effects on keratin organization (Figure 3G). Neither loss of desmoplakin (Vasioukhin et al., 2001) nor loss of Lis1 (unpublished data) results in a major defect in keratin organization in low-calcium conditions. In addition, because Lis1 and Ndel1 interact with and regulate dynein activity, we tested the roles of both microtubules and dynein in keratin assembly on mitochondria. Neither nocodazole treatment, which disrupts microtubules, nore treatment with the dynein inhibitor, ciliobrevin D, affected keratin reorganization induced by mito-Ndel1 (Supplemental Figure 4, C and D). These drug treatments similarly did not disrupt keratin organization in WT cells (Supplemental Figure 3, G–I). Therefore, Ndel1 is unlikely to act via localized transport of keratin subunits to specific cellular sites in keratinocytes.
The above data demonstrated that neither desmoplakin nor Lis1 was required for Ndel1’s keratin assembly activity. It did, however, raise the question of why loss of Lis1 resulted in desmosome defects (Sumigray et al., 2011). We asked whether Lis1 might function to target Ndel1 to the desmosome in the same way that desmoplakin is required for Ndel1 localization. Analysis of Ndel1 by immunofluorescence in WT and Lis1 null cells revealed an essential role for Lis1 in recruiting Ndel1 to the cell cortex (Figure 3, H and I).
Ndel1 is essential for desmosome stability
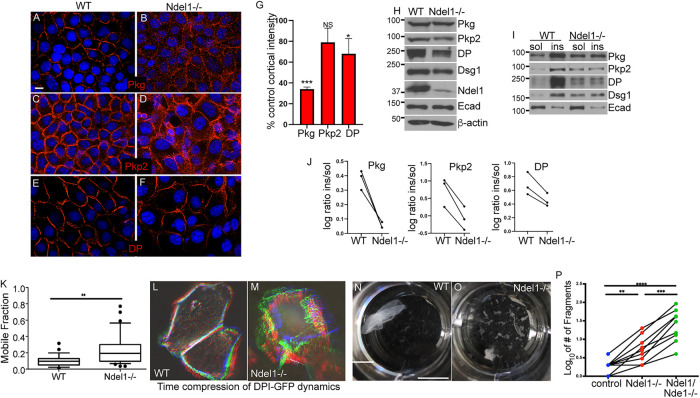
Having demonstrated that Ndel1 is capable of affecting local keratin organization, we next looked at the consequences that loss of Ndel1 has on desmosomes. These adhesions are the major organizers of keratin filaments in differentiated epidermis and require robust keratin attachments. The localizations of the core desmosome components plakoglobin (Pkg) and desmoplakin (DP) were decreased at cell–cell junctions, and cytoplasmic pools were increased in Ndel1 null keratinocytes when compared with control keratinocytes (Figure 4, A–G). Pkp2 also trended toward a decrease in cortical levels, but this did not reach statistical significance (Figure 4, C and D). When we examined the total levels of desmosomal proteins in epidermal lysates, we noted minor decreases in the amounts of several desmosomal proteins, including DP and Pkp2 (Figure 4H). Desmosomes are typically very insoluble in keratinocytes due in part to their interactions with the keratin filament network. When we isolated soluble and insoluble pools of desmosomal proteins from Ndel1 null cells, we observed an increase in the soluble pools of all of the desmosome components tested when compared with control cells (Figure 4, I and J). Therefore, Ndel1 cells have perturbed desmosomes in cultured cells as revealed by both cell biological and biochemical measures.
FIGURE 4:
Ndel1 is required for desmosome stability. (A–F) Immunofluorescence analysis of desmosomal proteins in WT and Ndel1 null keratinocytes, as indicated. Scale bar is 10 μm. (G) Quantitation of cortical intensity of desmosomal proteins compared with control keratinocytes. Six images were analyzed from three separate experiments. P values are 0.003 for Pkg, 0.118 for Pkp2, and 0.044 for DP; one-sample t-test. (H) Western blot analysis of desmosomal and adherens junction protein levels in lysates prepared from WT and Ndel1 null keratinocytes. (I) Fractionation of proteins into soluble and insoluble components in buffer containing 1% Triton X-100. Proteins levels were then analyzed by Western blotting. (J) Quantitation of desmosomal proteins, as indicated, in control and Ndel1 null cells. n = 3 for this paired analysis. P values are 0.024, 0.018, and 0.030 for Pkg, Pkp2, and DP, respectively; paired t test. (K) FRAP was measured for DPI-GFP transfected into WT and Ndel1 null cells. FRAP was performed on cells grown in high-calcium-containing media for 24 h to ensure desmosome assembly. The boxes and whiskers represent the 25–75 and the 10–90 percentiles of the mobile fraction, respectively. n = 15, p < 0.01; Student’s t test. (L, M) These images are time-compressed stacks of DP-GFP dynamics. Movies were aligned for drift, maximum pixel intensities are shown, and the time points were color coded as RGB images. Note the increased dynamics of DPI-GFP in the Ndel1 null cells. Scale bar is 10 μm. (N, O) Dispase-based dissociation assays were done with confluent monolayers of keratinocytes incubated for 72 h after Ca2+ switch and treated with dispase. The monolayers of keratinocytes were then subjected to mechanical stress. Scale bar is 1 cm. (P) Quantitation of dispase assay. Data shown are from nine paired experiments. ANOVA was performed (p < 0.0001). The p value for WT vs. Ndel1 null was p = 0.0041, for WT vs. Ndel1/Nde1 null p < 0.0001, and between Ndel1 null and Ndel1/Nde1 null p = 0.0001.
To characterize the dynamics of membrane adhesion complexes, we transfected WT and Ndel1 null cells with DPI-GFP, a construct encoding a fluorescently tagged version of the core desmosomal protein that provides attachment to keratins. Time-lapse imaging was performed 24 h after calcium induction of desmosomes. WT cells consistently showed very stable junctions, with little movement (Figure 4L). In contrast, Ndel1 null cells showed highly dynamic adhesions that changed considerably over the 2 h of imaging (Figure 4M). Time-lapse compressions were generated and color coded to demonstrate the changes in cell shape and desmosome positioning (Figure 4, L and M). In addition, FRAP analysis revealed that DPI-GFP had a larger mobile fraction in Ndel1 null cells as compared with WT cells, a phenotype similar to that observed in Lis1 null cells (Figure 4K) (Sumigray et al., 2011).
The above data demonstrated that the localization and stability of desmosomes were disrupted in the Ndel1 null keratinocytes. The adhesive activity of keratinocytes is largely due to the presence of robust desmosomes (Vasioukhin et al., 2001). To functionally test the strength of desmosomal adhesions in cultured cells, confluent sheets of keratinocytes were detached from the plate they were grown in by treatment with Dispase. The sheets were then subjected to mechanical forces to examine the epithelial sheets’ resistances to disruption as quantified by the resultant number of sheet fragments. While WT cell sheets were quite mechanically robust, Ndel1 null cell sheets were notably fragile and fragmented into more pieces, consistent with a defect in desmosomal adhesions (Figure 4, N–P).
Ndel1 has a highly homologous paralogue, Nde1, that also localizes to desmosomes (Sumigray et al., 2011; Soares et al., 2012). To test whether Nde1 might have partially redundant functions with Ndel1, we used CRISPR/Cas9 genome editing to knock out Nde1 in Ndel1 null cells. Western blotting confirmed the loss of protein (Supplemental Figure 4, A and B). Staining for desmosomal proteins revealed similar changes in localization as described above for Ndel1 null cells (Supplemental Figure 4C). Further, quantitation of cortical desmoplakin levels in Nde1;Ndel1 dKO cells revealed a decrease over both the control and Ndel1 null cells (Supplemental Figure 4, C and D). The keratin concentration at the cell periphery was also decreased in the dKO cells (Supplemental Figure 4G). These mutant cells showed an enhanced mechanical fragility as compared with both the control and Ndel1 null cells (Figure 4, N–P). Similar results were found for a second clone with independent mutations (unpublished data). Together these data demonstrate that Ndel1 and its paralogue Nde1 are desmosome-associated proteins that are required for desmosome function.
In contrast, we found that Ndel1 and Nde1 were not required either for microtubule reorganization in cultured keratinocytes (Supplemental Figure 4E) or for the recruitment of Lis1 to the cell cortex (Supplemental Figure 4F).
Ndel1 depletion in the epidermis leads to skin defects
To test the functional role of Ndel1 in the epidermis, we have generated mice in which Ndel1 is lost in the epidermis through recombination driven by Cre recombinase under the control of the keratin 14 promoter (referred to as Ndel1 cKO) (Vasioukhin et al., 1999). Cre expression begins during midembryogenesis in epidermal progenitors, and Ndel1 cKO pups had dramatically reduced levels of Ndel1 in epidermal lysates at birth (Figure 5A). Unlike ablation of its binding partner, Lis1, loss of Ndel1 in the epidermis did not result in neonatal lethality (Sumigray et al., 2011). This may be due, in part, to the presence of Nde1, which has overlapping functions with Ndel1 in cultured keratinocytes (Figure 4; Supplemental Figure 4).
FIGURE 5:
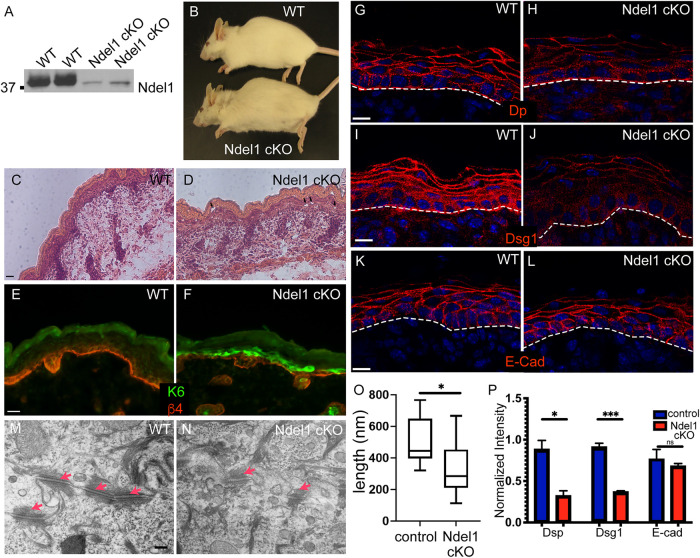
Loss of Ndel1 in the epidermis results in desmosome defects. (A) Western blots of epidermal lysates from two WT and two Ndel1 cKO neonatal mice. (B) Photo of 5-mo-old WT and Ndel1 cKO mice. (C, D) Hematoxylin- and eosin-stained sections from WT and Ndel1 cKO p0 mice. Black arrowheads indicate microblisters in the epidermis. (E, F) Staining for the stress marker keratin 6 (green) and the basement membrane marker β-4 integrin (red) in WT and Ndel1 cKO p0 skin. (G–L) Staining of desmosomal and adherens junction components in WT and Ndel1 cKO p0 skin sections as indicated. All scale bars are 10 μm. (M, N) TEM of spinous cells in control and Ndel1 null epidermis. Red arrows indicate desmosomes. (O) Quantitation of desmosome length in control and Ndel1 null epidermis. n = 25 desmosomes for each, p value is <0.05; Student’s t test. (P) Quantitation of cortical levels of desmoplakin (Dsp), Dsg1, and E-cadherin in control and Ndel1 null epidermis. n = 3, p values are 0.0108 for Dsp, 0.0004 for Dsg1, and 0.534 for E-cad; Student’s t test.
We were unable to reliably identify Ndel1 cKO pups at birth by any gross phenotype. However, adult Ndel1 cKO mice were identifiable by a ragged appearance to their hair coat (Figure 5B). Histologic analysis of p0 pups demonstrated a relatively normal-looking stratified epidermis (Figure 5, C and D). There were some focal regions with microblisters and small cell–cell separations in Ndel1 cKO epidermis compared with the epidermis from WT littermates (Figure 5, C and D). Ablation of some desmosomal genes, such as plakoglobin, similarly results in milder phenotypes than the blistering seen upon complete loss of desmoplakin (Li et al., 2012). Consistent with a mild effect, several further observations demonstrated that the skin was adversely affected by the loss of Ndel1. This included an up-regulation of the stress-induced protein keratin 6 in focal regions of the Ndel1 cKO epidermis (Figure 5, E and F). Because of Ndel1’s localization to desmosomes and our findings in cultured keratinocytes, we examined the status of these cell adhesion structures. Staining for desmoplakin and desmoglein 1 (Dsg1) showed decreased junctional staining (Figure 5, G–J, and P). This effect was specific to desmosomes as normal localization of the adherens junction component E-cadherin was observed (Figure 5, K, L, and P). Total levels of keratins in epidermal lysates were not changed, however (Supplemental Figure 5). Because of the decrease in junctional desmosome components that we saw through immunofluorescence, we next took advantage of the electron dense nature of desmosomes and analyzed them via transmission electron microscopy (TEM). Strikingly, we found that the desmosomes in Ndel1 were smaller and less electron dense than those in the control epidermis (Figure 5, M and N). Quantification of mutant desmosomes revealed that they were roughly half the length of the control desmosomes (Figure 5O). Thus, our data show that Ndel1 is a keratin binding protein that regulates keratin organization and desmosome structure/function in cultured keratinocytes and intact epidermis.
DISCUSSION
Cells have developed many ways to control the local assembly and organization of cytoskeletal filaments. The identification of factors that promote assembly/reorganization of microtubules and F-actin has allowed a much fuller understanding of how these cytoskeletal networks form and the signaling pathways that govern local assembly. This regulation occurs through both localization and activation of these organizers at specific cellular sites. Our data suggest that Ndel1 promotes the local assembly and/or reorganization of keratin filaments. Evidence in this paper as well as previously published data suggest that Ndel1 may be a general factor that promotes local IF assembly and organization across diverse cell types that express different IF types. In addition to Ndel1’s reported localization at desmosomes (Sumigray et al., 2011), Ndel1 has been reported to localize to nascent adhesions at the leading edge of migrating cells, sites where keratin filament nucleation has also been observed (Windoffer et al., 2006; Shan et al., 2009). More broadly, Ndel1 localizes to the mitotic spindle, where it has been further implicated in nuclear lamin assembly/organization (Ma et al., 2009), as well as at centrosomes, another region that has long been appreciated as a focus for IF organization in certain cell types (Kalnins et al., 1985; Niethammer et al., 2000; Sasaki et al., 2000). Thus, in diverse cell types and subcellular locales, Ndel1 promotes IF assembly/organization. The molecular mechanism of Ndel1 and its regulation by signaling pathways are clear and important areas of future investigation.
Two models have been proposed to explain Ndel1’s role in IF assembly. First, Ndel1 could directly control the nucleation and/or elongation of IF filaments, as suggested by the finding that purified Ndel1 promotes neurofilament assembly in vitro (Nguyen et al., 2004). A second, and not mutually exclusive, possibility is that Ndel1 works in a more canonical Lis1/dynein-dependent manner to transport IF subunits or small filaments to distinct cellular locations (Shim et al., 2008; Ma et al., 2009). This has been proposed for both nuclear lamins and vimentin. While our data support the idea that Ndel1 can promote local assembly of keratins, there is currently no kinetic assay for keratin assembly in vitro, precluding direct testing of the first model. We did not find any effect on keratin assembly activity in cells devoid of Lis1 or in cells treated either with nocodazole to disrupt microtubules or with dynein inhibitors. Therefore, our data are more consistent with a more direct role for Ndel1 in keratin filament assembly/organization, such as has been reported for neurofilaments (Nguyen et al., 2004). On the basis of our data, we suggest a model in which desmoplakin recruits Lis1 to desmosomes, where Lis1 is required to localize Ndel1. While desmoplakin binds directly to preexisting keratin filaments and does not require Ndel1 for this activity, the recruitment of Ndel1 may provide a mechanism to increase the number of keratin filaments available for binding by promoting their assembly/reorganization near desmosomes.
Ndel1 ablation resulted in desmosome defects in addition to keratin organization defects. While we cannot rule out that Ndel1 has additional structural roles in desmosome stability, the desmosome defects may be secondary to keratin misorganization. A number of studies have reported roles for keratins in desmosome formation and/or stability. For example, defects in desmosome structure have been reported in samples from human patients with desmoplakin mutations (Wan et al., 2004). In addition, keratin 8 controls the desmosomal localization of desmoplakin in hepatocytes, the ablation of keratins 1 and 10 in the epidermis resulted in smaller desmosomes, and the loss of all keratins led to desmosome defects in embryonic epithelia (Loranger et al., 2006; Vijayaraj et al., 2009). Our data further support an important role for desmosome–keratin attachments in regulating desmosome stability.
The loss of Ndel1 has a much less severe phenotype than loss of Lis1 or desmoplakin in the epidermis (Vasioukhin et al., 2001; Sumigray et al., 2011). Its severity is much more similar to that of loss of plakoglobin, which is not lethal when conditionally ablated in the epidermis (Li et al., 2012). This suggests a partial desmosome loss of function. In fact, our data demonstrate that the desmosomes are present and are functionally able to direct microtubule reorganization, though they are deficient in robust keratin attachment. We demonstrated that Nde1, a homologue of Ndel1, acts in a somewhat functionally redundant manner. Nde1 shows a localization pattern similar to that of Ndel1 in keratinocytes and also binds to keratin subunits (Sumigray et al., 2011). Future genetics approaches are needed to determine the phenotype of combined Ndel1/Nde1 loss in the epidermis in vivo.
While the desmosome has often been portrayed as a rather passive structure, our data highlight its more active role in controlling both keratin and microtubule organization. Strikingly, these are both mediated by the recruitment of a protein complex that often resides at the centrosome and that contains both Lis1 and Ndel1.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Mice
K14-Cre (Vasioukhin et al., 1999) and Ndel1 fl/fl (Sasaki et al., 2005) have been described. All animal work was approved by the Duke University Institution Animal Care and Use Committee.
Keratinocyte cultures
Ndel1 null keratinocytes were isolated from back skins of newborn pups. All keratinocytes were maintained in low Ca2+ (0.05 mM) E media containing 15% fetal bovine serum (Hyclone) in a humidified incubator (37°C and 7.5% CO2). For microtubule organization assays, cells were treated with Taxol after the induction of differentiation for 1 h at 10 µM. Nocodazole (10 µM; Sigma) and ciliobrevin D (50 µM; Millipore) were added to cells for 1 h before fixation. MitoTracker Red (Invitrogen) was added to cells as per the manufacturer’s instructions. All transfections were performed with Mirus TransIT transfection media according to the manufacturer’s protocols. Cell adhesion was induced by the addition of 1.2 mM calcium to the media.
Immunofluorescence staining
Keratinocytes were grown to confluence on glass coverslips in low-Ca2+ media until confluent. After 24 h under terminal differentiating conditions (1.2 mM CaCl2, where appropriate), cells were fixed with either 4% paraformaldehyde at 37°C for 10 min or in methanol for 2 min at –20°C. After fixation, cells were washed with phosphate-buffered saline and then blocking buffer containing 5% bovine serum albumen, 5% normal donkey serum, and 5% normal goat serum. Staining of cryosections of back skin was performed in the same way as explained above. Imaging was performed on an AxioImager Z1 microscope with Apotome attachment and MRm camera (Carl Zeiss). A 63×, 1.4 NA Plan Apotchromat objective and Immersol 518F immersion oil were used (Carl Zeiss). Imaging was performed at room temperature with either 12 mm round coverslips (no. 1) for cells or no. 1.5 coverslips (VWR) for tissue sections. The following primary antibodies were used: rabbit anti-Ndel1 (Sumigray et al., 2011), mouse anti–β-tubulin and β-actin (Sigma-Aldrich), rabbit anti–keratin 6 and rabbit anti–keratin 10 (Covance), anti–keratin 5/14 (generated in lab), mouse anti-DP1/2 (Chemicon), guinea pig anti-plakophillin2 (Progen), mouse anti-desmoglein1 and rat anti-CD104 (BD Biosciences), mouse anti-CD3 (Millipore), guinea pig anti-Dsc1 (gift from I. King, Medical Research Council, London, England, UK), and rat anti–E-cadherin and rabbit anti–keratin 1 (gifts of C. Jamora, University of California, San Diego).
FRAP analysis
Mouse keratinocytes were grown on 35-mm glass-bottom culture dishes (no. 1.5; MatTek Corporation) and transfected with DPI-GFP (provided by K. Green, Northwestern University, Chicago, IL) using TransIT-keratinocyte transfection reagent. Calcium was added to cells to 1.2 mM, and junctions were allowed to form for 24 h before imaging. A Zeiss 710 confocal scanning light microscope with a 63×, 1.4 NA objective and Zen software were used to determine the mobile fraction of desmoplakin at cell cortical regions. Imaging was performed at 37°C. Regions of interest were bleached, and only those cells in which 70% or more bleach was accomplished were analyzed. The fluorescence intensities were calculated by MetaMorph image analysis software and normalized to background intensity. The mobile fraction was determined as mf = Imax – Io/1 – Io (Shen et al., 2008). Three different experiments were performed, and statistical analysis was performed with combined data.
Dispase-based dissociation assay
Mouse keratinocytes in six-well plates were grown to confluence in low-Ca2+ media until confluent. After 24 h under differentiating conditions (1.2 mM CaCl2), the cells were washed twice with phosphate-buffered saline (PBS) and incubated with 2.4 U/ml dispase (Roche) for 1 h at 37°C. Floating monolayer sheets were rotated on an orbital shaker (300 rpm) for 1 h. Pictures of the resulting sheet fragments were taken with a Canon Powershot camera, and the number of fragments was determined by ImageJ software using the cell counter function.
Protein–protein interactions
GST fusion proteins were produced in bacterial cells upon induction with 1 mM isopropryl β-d-1-thiogalactopyranoside. The Escherichia coli cells were harvested and lysed by sonication for 3 × 10 s in ice-cold lysis buffer containing 50 mM Tris-HCl, pH 7.4, 2 mM MgCl2, 50 mM NaCl2, 10% glycerol, 1% Triton-X, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitors. After centrifugation at 12,000 rpm for 10 min at 4°C, the supernatants were obtained and incubated with glutathione-Sepharose beads for 6 h at 4°C and then washed extensively with lysis buffer. Keratins were isolated from bacterial inclusion bodies, solubilized in 8 M urea, and then dialyzed into PBS. GST fusion proteins (0.5 mg/ml) bound to beads were mixed with keratins (0.3 mg/ml). After washing, proteins on beads were eluted through the addition of sample buffer and boiling. Eluted proteins were analyzed by Western blotting.
Preparation of protein lysates
Keratinocytes were washed twice with ice-cold PBS and scraped into lysis buffer containing 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 50 mM NaF, 1 mM sodium pyrophosphate, 1% NP-40, 0.1% sodium deoxycholate. After incubation for 30 min on ice, lysates were separated into soluble and insoluble fractions by centrifugation at 15,000 × g for 10 min.
Electron microscopy analysis
Neonatal back skin was fixed in 4% glutaraldehyde, 1 mM CaCl2, and 0.05 M cacodylic acid (pH 7.4) for 2 h at room temperature and then overnight at 4°C. Samples were washed in 0.1 M sodium cacodylate buffer containing 7.5% sucrose, postfixed in 1% osmium tetroxide in 0.15 M sodium cacodylate buffer for 1 h, and then washed in two changes of 0.11 M veronal acetate buffer for 15 min each. Samples were placed into en bloc stain (0.5% uranyl acetate in veronal acetate buffer) for 1 h, washed in veronal acetate buffer, and then dehydrated in a series of 70, 95, and 100% ethanol. Finally, samples were prepared in 50/50 propylene oxide:Epon resin followed by two 30-min immersions in 100% Epon resin for embedding. Skin samples were sectioned and imaged with a CM12 transmission electron microscope (Phillips) run at 80 kV with an XR60 camera (Advanced Microscopy Techniques, Woburn, MA). Image acquisition was done using 2Vu software (Advanced Microscopy Techniques). TEM images were visualized and analyzed using FIJI software.
Supplementary Material
Acknowledgments
We thank Shinji Hirotsune for Ndel1 floxed mice; Pierre Coulombe for keratin constructs; Henry Foote for assistance with imaging, analysis, and statistics; and Julie Underwood for excellent care of the mice. We are grateful to members of the Lechler lab for valuable comments throughout this work and on the manuscript. This work was supported by National Institutes of Health–National Institute of Arthritis and Musculoskeletal and Skin Diseases grants (R01AR055926 and R01AR067203 to T. L.) and by a Basil O’Connor Award from the March of Dimes.
Abbreviations used:
- AH
alpha-helix
- cKO
conditional knockout
- DP
desmoplakin
- FRAP
fluorescence recovery after photobleaching
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- IF
intermediate filament
- K10
keratin 10
- KO
knockout
- Pkg
plakoglobin
- PKP2
plakophilin 2
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E21-02-0087).
REFERENCES
- Belmont LD, Mitchison TJ (1996). Identification of a protein that interacts with tubulin dimers and increases the catastrophe rate of microtubules. Cell 84, 623–631. [DOI] [PubMed] [Google Scholar]
- Bornslaeger EA, Corcoran CM, Stappenbeck TS, Green KJ (1996). Breaking the connection: displacement of the desmosomal plaque protein desmoplakin from cell-cell interfaces disrupts anchorage of intermediate filament bundles and alters intercellular junction assembly. J Cell Biol 134, 985–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chansard M, Hong JH, Park YU, Park SK, Nguyen MD (2011). Ndel1, Nudel (Noodle): flexible in the cell? Cytoskeleton (Hoboken) 68, 540–554. [DOI] [PubMed] [Google Scholar]
- Chhabra ES, Higgs HN (2007). The many faces of actin: matching assembly factors with cellular structures. Nat Cell Biol 9, 1110–1121. [DOI] [PubMed] [Google Scholar]
- Clemen CS, Herrmann H, Strelkov SV, Schroder R (2013). Desminopathies: pathology and mechanisms. Acta Neuropathol 125, 47–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa S, Cerrone M, Saguner AM, Brunckhorst C, Delmar M, Duru F (2020). Arrhythmogenic cardiomyopathy: an in-depth look at molecular mechanisms and clinical correlates. Trends Cardiovasc Med, doi: 10.1016/j.tcm.2020.07.006. [DOI] [PubMed] [Google Scholar]
- Coulombe PA, Wong P (2004). Cytoplasmic intermediate filaments revealed as dynamic and multipurpose scaffolds. Nat Cell Biol 6, 699–706. [DOI] [PubMed] [Google Scholar]
- Delmar M, McKenna WJ (2010). The cardiac desmosome and arrhythmogenic cardiomyopathies: from gene to disease. Circ Res 107, 700–714. [DOI] [PubMed] [Google Scholar]
- Eriksson JE, Dechat T, Grin B, Helfand B, Mendez M, Pallari HM, Goldman RD (2009). Introducing intermediate filaments: from discovery to disease. J Clin Invest 119, 1763–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firat-Karalar EN, Welch MD (2011). New mechanisms and functions of actin nucleation. Curr Opin Cell Biol 23, 4–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallicano GI, Kouklis P, Bauer C, Yin M, Vasioukhin V, Degenstein L, Fuchs E (1998). Desmoplakin is required early in development for assembly of desmosomes and cytoskeletal linkage. J Cell Biol 143, 2009–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler N, Kaufmann E, Fischer S, Plessmann U, Weber K (1983). Neurofilament architecture combines structural principles of intermediate filaments with carboxy-terminal extensions increasing in size between triplet proteins. EMBO J 2, 1295–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler N, Weber K (1982). The amino acid sequence of chicken muscle desmin provides a common structural model for intermediate filament proteins. EMBO J 1, 1649–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt-Clermont PJ, Furman MI, Wachsstock D, Safer D, Nachmias VT, Pollard TD (1992). The control of actin nucleotide exchange by thymosin beta 4 and profilin. A potential regulatory mechanism for actin polymerization in cells. Mol Biol Cell 3, 1015–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green KJ, Simpson CL (2007). Desmosomes: new perspectives on a classic. J Invest Dermatol 127, 2499–2515. [DOI] [PubMed] [Google Scholar]
- Gunzelmann J, Ruthnick D, Lin TC, Zhang W, Neuner A, Jakle U, Schiebel E (2018). The microtubule polymerase Stu2 promotes oligomerization of the gamma-TuSC for cytoplasmic microtubule nucleation. eLife 7, e39932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzfeld M, Burba M (1994). Function of type I and type II keratin head domains: their role in dimer, tetramer and filament formation. J Cell Sci 107 (Pt 7), 1959–1972. [DOI] [PubMed] [Google Scholar]
- Kalnins VI, Subrahmanyan L, Fedoroff S (1985). Assembly of glial intermediate filament protein is initiated in the centriolar region. Brain Res 345, 322–326. [DOI] [PubMed] [Google Scholar]
- Kobielak A, Pasolli HA, Fuchs E (2004). Mammalian formin-1 participates in adherens junctions and polymerization of linear actin cables. Nat Cell Biol 6, 21–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman JM, Merdes A, Mourey L, Agard DA (2011). Microtubule nucleation by gamma-tubulin complexes. Nat Rev Mol Cell Biol 12, 709–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottke MD, Delva E, Kowalczyk AP (2006). The desmosome: cell science lessons from human diseases. J Cell Sci 119, 797–806. [DOI] [PubMed] [Google Scholar]
- Kouklis PD, Hutton E, Fuchs E (1994). Making a connection: direct binding between keratin intermediate filaments and desmosomal proteins. J Cell Biol 127, 1049–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechler T, Fuchs E (2007). Desmoplakin: an unexpected regulator of microtubule organization in the epidermis. J Cell Biol 176, 147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Zhang W, Liu Y, Haneline LS, Shou W (2012). Lack of plakoglobin in epidermis leads to keratoderma. J Biol Chem 287, 10435–10443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J, Omary MB (1996). 14-3-3 proteins associate with phosphorylated simple epithelial keratins during cell cycle progression and act as a solubility cofactor. J Cell Biol 133, 345–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loranger A, Gilbert S, Brouard JS, Magin TM, Marceau N (2006). Keratin 8 modulation of desmoplakin deposition at desmosomes in hepatocytes. Exp Cell Res 312, 4108–4119. [DOI] [PubMed] [Google Scholar]
- Ma L, Tsai MY, Wang S, Lu B, Chen R, Iii JR, Zhu X, Zheng Y (2009). Requirement for Nudel and dynein for assembly of the lamin B spindle matrix. Nat Cell Biol 11, 247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moch M, Schwarz N, Windoffer R, Leube RE (2020). The keratin-desmosome scaffold: pivotal role of desmosomes for keratin network morphogenesis. Cell Mol Life Sci 77, 543–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz M, Braunfeld MB, Sedat JW, Alberts B, Agard DA (1995). Microtubule nucleation by gamma-tubulin-containing rings in the centrosome. Nature 378, 638–640. [DOI] [PubMed] [Google Scholar]
- Nguyen MD, Shu T, Sanada K, Lariviere RC, Tseng HC, Park SK, Julien JP, Tsai LH (2004). A NUDEL-dependent mechanism of neurofilament assembly regulates the integrity of CNS neurons. Nat Cell Biol 6, 595–608. [DOI] [PubMed] [Google Scholar]
- Niethammer M, Smith DS, Ayala R, Peng J, Ko J, Lee MS, Morabito M, Tsai LH (2000). NUDEL is a novel Cdk5 substrate that associates with LIS1 and cytoplasmic dynein. Neuron 28, 697–711. [DOI] [PubMed] [Google Scholar]
- Omary MB, Ku NO, Liao J, Price D (1998). Keratin modifications and solubility properties in epithelial cells and in vitro. Subcell Biochem 31, 105–140. [PubMed] [Google Scholar]
- Ridge KM, Linz L, Flitney FW, Kuczmarski ER, Chou YH, Omary MB, Sznajder JI, Goldman RD (2005). Keratin 8 phosphorylation by protein kinase C delta regulates shear stress-mediated disassembly of keratin intermediate filaments in alveolar epithelial cells. J Biol Chem 280, 30400–30405. [DOI] [PubMed] [Google Scholar]
- Roop DR, Krieg TM, Mehrel T, Cheng CK, Yuspa SH (1988). Transcriptional control of high molecular weight keratin gene expression in multistage mouse skin carcinogenesis. Cancer Res 48, 3245–3252. [PubMed] [Google Scholar]
- Sasaki S, Mori D, Toyo-oka K, Chen A, Garrett-Beal L, Muramatsu M, Miyagawa S, Hiraiwa N, Yoshiki A, Wynshaw-Boris A, et al. (2005). Complete loss of Ndel1 results in neuronal migration defects and early embryonic lethality. Mol Cell Biol 25, 7812–7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki S, Shionoya A, Ishida M, Gambello MJ, Yingling J, Wynshaw-Boris A, Hirotsune S (2000). A LIS1/NUDEL/cytoplasmic dynein heavy chain complex in the developing and adult nervous system. Neuron 28, 681–696. [DOI] [PubMed] [Google Scholar]
- Shan Y, Yu L, Li Y, Pan Y, Zhang Q, Wang F, Chen J, Zhu X (2009). Nudel and FAK as antagonizing strength modulators of nascent adhesions through paxillin. PLoS Biol 7, e1000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Weber CR, Turner JR (2008). The tight junction molecular complex undergoes rapid and continuous molecular remodeling at steady state. J Cell Biol 181, 683–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim SY, Samuels BA, Wang J, Neumayer G, Belzil C, Ayala R, Shi Y, Shi Y, Tsai LH, Nguyen MD (2008). Ndel1 controls the dynein-mediated transport of vimentin during neurite outgrowth. J Biol Chem 283, 12232–12240. [DOI] [PubMed] [Google Scholar]
- Smith F (2003). The molecular genetics of keratin disorders. Am J Clin Dermatol 4, 347–364. [DOI] [PubMed] [Google Scholar]
- Soares DC, Bradshaw NJ, Zou J, Kennaway CK, Hamilton RS, Chen ZA, Wear MA, Blackburn EA, Bramham J, Bottcher B, et al. (2012). The mitosis and neurodevelopment proteins NDE1 and NDEL1 form dimers, tetramers, and polymers with a folded back structure in solution. J Biol Chem 287, 32381–32393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg A, Liem RK (2007). Plakins in development and disease. Exp Cell Res 313, 2189–2203. [DOI] [PubMed] [Google Scholar]
- Stappenbeck TS, Bornslaeger EA, Corcoran CM, Luu HH, Virata ML, Green KJ (1993). Functional analysis of desmoplakin domains: specification of the interaction with keratin versus vimentin intermediate filament networks. J Cell Biol 123, 691–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumigray KD, Chen H, Lechler T (2011). Lis1 is essential for cortical microtubule organization and desmosome stability in the epidermis. J Cell Biol 194, 631–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suozzi KC, Wu X, Fuchs E (2012). Spectraplakins: master orchestrators of cytoskeletal dynamics. J Cell Biol 197, 465–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang VW, Brieher WM (2012). alpha-Actinin-4/FSGS1 is required for Arp2/3-dependent actin assembly at the adherens junction. J Cell Biol 196, 115–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasioukhin V, Bowers E, Bauer C, Degenstein L, Fuchs E (2001). Desmoplakin is essential in epidermal sheet formation. Nat Cell Biol 3, 1076–1085. [DOI] [PubMed] [Google Scholar]
- Vasioukhin V, Degenstein L, Wise B, Fuchs E (1999). The magical touch: genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc Natl Acad Sci USA 96, 8551–8556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S, Han SP, Michael M, Gomez GA, Yang Z, Teasdale RD, Ratheesh A, Kovacs EM, Ali RG, Yap AS (2012). A WAVE2-Arp2/3 actin nucleator apparatus supports junctional tension at the epithelial zonula adherens. Mol Biol Cell 23, 4601–4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayaraj P, Kroger C, Reuter U, Windoffer R, Leube RE, Magin TM (2009). Keratins regulate protein biosynthesis through localization of GLUT1 and -3 upstream of AMP kinase and Raptor. J Cell Biol 187, 175–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan H, Dopping-Hepenstal PJ, Gratian MJ, Stone MG, Zhu G, Purkis PE, South AP, Keane F, Armstrong DK, Buxton RS, et al. (2004). Striate palmoplantar keratoderma arising from desmoplakin and desmoglein 1 mutations is associated with contrasting perturbations of desmosomes and the keratin filament network. Br J Dermatol 150, 878–891. [DOI] [PubMed] [Google Scholar]
- Weber GF, Bjerke MA, DeSimone DW (2012). A mechanoresponsive cadherin-keratin complex directs polarized protrusive behavior and collective cell migration. Dev Cell 22, 104–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windoffer R, Beil M, Magin TM, Leube RE (2011). Cytoskeleton in motion: the dynamics of keratin intermediate filaments in epithelia. J Cell Biol 194, 669–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windoffer R, Kolsch A, Woll S, Leube RE (2006). Focal adhesions are hotspots for keratin filament precursor formation. J Cell Biol 173, 341–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Wong ML, Alberts B, Mitchison T (1995). Nucleation of microtubule assembly by a gamma-tubulin-containing ring complex. Nature 378, 578–583. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.