Abstract
The curvature of the membrane defines cell shape. Septins are GTP-binding proteins that assemble into heteromeric complexes and polymerize into filaments at areas of micron-scale membrane curvature. An amphipathic helix (AH) domain within the septin complex is necessary and sufficient for septins to preferentially assemble onto micron-scale curvature. Here we report that the nonessential fungal septin, Shs1, also has an AH domain capable of recognizing membrane curvature. In a septin mutant strain lacking a fully functional Cdc12 AH domain (cdc12-6), the C-terminal extension of Shs1, containing an AH domain, becomes essential. Additionally, we find that the Cdc12 AH domain is important for regulating septin filament bundling, suggesting septin AH domains have multiple, distinct functions and that bundling and membrane binding may be coordinately controlled.
INTRODUCTION
Cell shape is critical for function and can be described in terms of membrane curvature. Cell membranes range in curvatures from nanometer to micron scales. How cells use nanometer-sized proteins to sense micron-scale membrane curvature is not well understood. One way cells solve this scaling problem is through a class of cytoskeletal proteins called septins, which are conserved from yeast to humans (Field et al., 1996; Pan et al., 2007). Septins are GTP-binding proteins that form heteromeric complexes (Versele et al., 2004; Low and Macara, 2006; Sirajuddin et al., 2007; Bertin et al., 2008). These complexes associate with the plasma membrane, where they polymerize into filaments and higher-order assemblies such as bundles and rings (Rodal et al., 2005; John et al., 2007; Sirajuddin et al., 2007; Bertin et al., 2008; DeMay et al., 2009; DeMay et al., 2011). Higher-order septin assemblies can act as scaffolds for signaling proteins, organize membrane properties, and influence the organization of other cytoskeletal elements (Lew and Reed, 1995; Longtine et al., 2000; Gladfelter et al., 2001; Spiliotis, 2010; Gilden et al., 2012; Clay et al., 2014; Bridges et al., 2016). In many of these functions, higher-order septin assemblies are built in the context of micron-scale membrane curvature.
Septins can distinguish positive membrane curvatures at the micron scale without any other cellular factors (Bridges et al., 2016). When purified septins are mixed with membranes at a range of different curvatures, septins polymerize into aligned filaments wrapped at an optimal curvature (Tanaka-Takiguchi et al., 2009; Beber et al., 2019b; Cannon et al., 2019). However, even individual septin complexes can distinguish membranes with micron-scale curvature by displaying different kinetics of septin-membrane binding depending on the curvature (Cannon et al., 2019). Septins, like other curvature-sensitive proteins, possess an amphipathic helix (AH) domain (Drin and Antonny, 2010; Kim et al., 2017; Cannon et al., 2019). AH domains detect and bind to lipid packing defects within curved membranes. In the budding yeast Saccharomyces cerevisiae, the AH at the C-terminus of Cdc12 is necessary and sufficient for septins to distinguish between different curvatures of the plasma membrane (Cannon et al., 2019). The mechanisms by which Cdc12-AH membrane binding are coupled to filament polymerization and organization to cause curvature-dependent assembly of septins remains a challenging problem.
Here we report that Shs1, a nonessential mitotic septin, also has an AH domain within its C-terminal extension (CTE). Shs1 occupies the terminal position at either end of a subset of heteromers, adjacent to Cdc12, the only essential septin subunit to possess an AH domain in yeast (Garcia et al., 2011; Finnigan et al., 2015; Cannon et al., 2019). We predicted that the presence of a second AH domain in the septin complex could alter the affinity of the complex binding the membrane or adjust the type of membrane defect detected and thereby influence curvature sensing. The Shs1 AH domain recognizes membrane curvature, although more weakly compared with the Cdc12 AH domain. In the absence of the Cdc12 AH domain, the Shs1 AH domain restores some ability of septins to distinguish between different curvatures. Moreover, Shs1 and its CTE are indispensable in mutants harboring an incomplete Cdc12 AH domain in cells. This suggests septin complexes can contain different numbers of curvature-sensitive AH domains, which may allow for another layer of control of higher-order septin assemblies.
RESULTS AND DISCUSSION
A predicted AH domain of Shs1 differentially binds various membrane curvatures in vitro
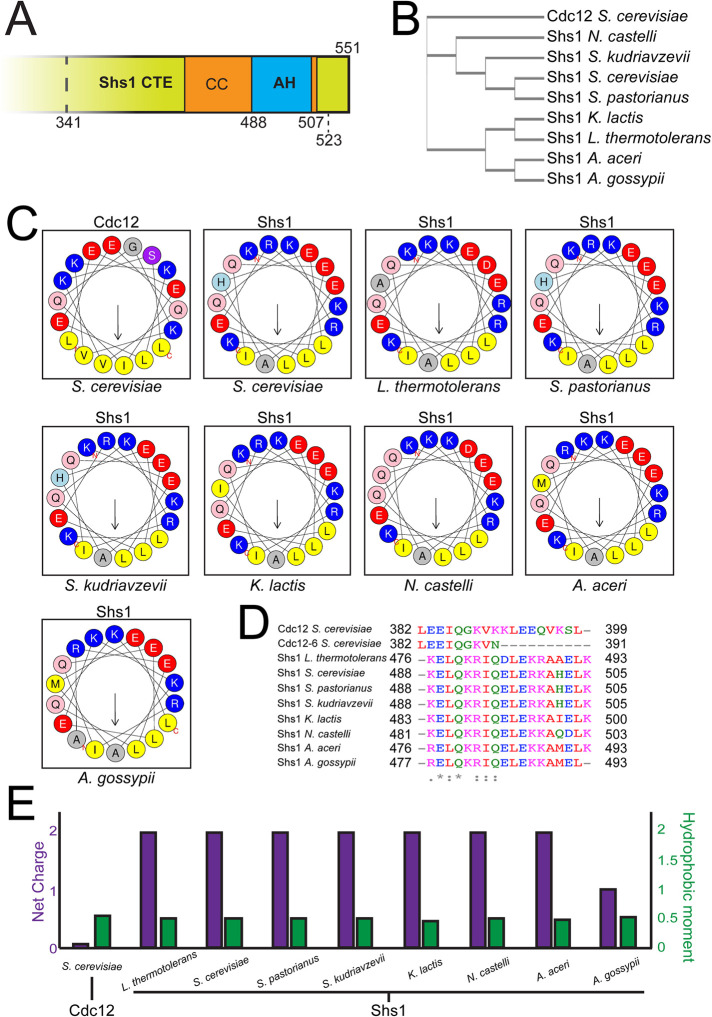
We reported previously that septin complexes harboring a Cdc12 mutant protein, cdc12-6 (K390N point mutation, and truncation of the AH domain including the C-terminal residues 391–407), no longer could distinguish between different membrane curvatures despite still retaining three hydrophobic residues (Adams and Pringle, 1984; Johnson et al., 2015; Cannon et al., 2019). In cdc12-6 mutants, septins quickly disassemble from the bud neck at restrictive temperature. However, at permissive temperature septins remain localized to bud neck, which is a site of positive micron-scale membrane curvature in cells (Gladfelter et al., 2005). We reasoned that another, unidentified AH domain or membrane-binding sequence might enable septins to maintain localization at the bud neck in cdc12-6 mutants at permissive temperature. Screening through protein sequences of all mitotically expressed septins in ascomycetes including S. cerevisiae, Ashbya gossypii, and Kluyveromyces lactis revealed a predicted AH domain at the C-terminus of the coiled coil within the CTE of Shs1 (Figure 1, A–C). Interestingly, the ascomycetes Neurospora crassa, Schizosaccharomyces pombe, and Aspergillus nidulans lack an Shs1 homologue all together, whereas Candida albicans Shs1 does not have an AH. Despite sharing relatively low homology (∼65%) among AH-containing Shs1 polypeptides (Figure 1B), their AH domains are nearly identical (Figure 1, C and D). As the physicochemical properties of AH domains have been shown to affect membrane binding and curvature sensing (Drin et al., 2007; Drin and Antonny, 2010), we compared the net charge and hydrophobic moment of AH domains of Shs1 and Cdc12. Interestingly, both of these properties are different between the AH domain of Shs1 and Cdc12 (Figure 1E). Together, these data suggest that the Shs1 AH domain might impart a distinct curvature preference when combined with Cdc12 in the same septin complex.
FIGURE 1:
Shs1 contains a highly conserved amphipathic helix within its C-terminal domain. (A) cerevisiae Shs1 CTE with a predicted AH domain within the coiled coil. Truncation alleles within Shs1 CTE demarcated (discussed in Figure 3). (B) Cladogram constructed from multiple alignments of Shs1 primary sequences from various ascomycetes compared with Cdc12 from S. cerevisiae. (C) Helical wheels representing amphipathic helices present in septin polypeptides screened ascomycetes. Residues in blue: basic amino acid; red: acidic; yellow: hydrophobic; gray: small side chain; light blue: histidine; pink: asparagine or glutamine; purple: serine or threonine. (D) Sequence alignment of amphipathic helices with indicated residue numbers. Residues in red: hydrophobic, magenta: basic; blue: acidic; green: uncharged. (E) Net charge and hydrophobic moment of septin amphipathic helices.
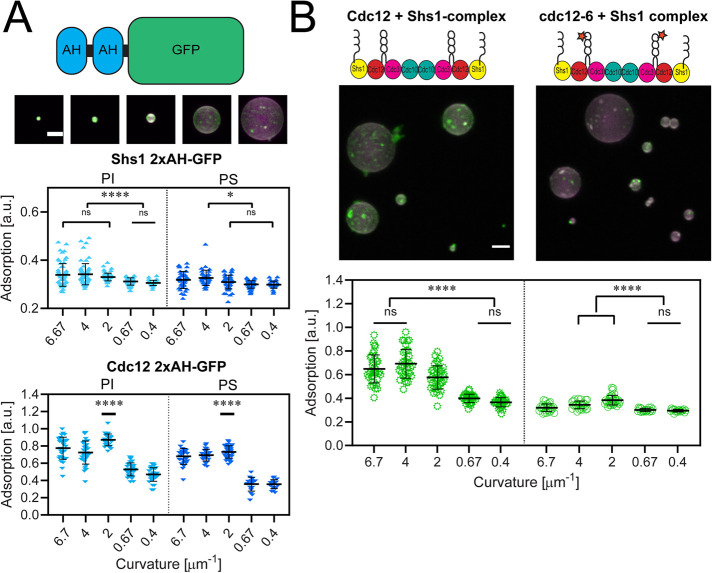
We next assessed whether the S. cerevisiae Shs1 AH domain (residues 488–507) alone is sufficient to distinguish between membrane curvatures (Figure 1D). We purified a polypeptide with a tandem repeat of the AH of Shs1 to mimic the stoichiometry of the septin complex and mixed it with membrane-coated, silica beads ranging in diameter from 300 nm to 5 μm (curvatures of κ = 6.67 μm−1 to κ = 0.4 μm−1, respectively). We chose membrane composition to roughly mimic the anionic charge of the plasma membrane. The Shs1 2xAH polypeptide adsorbed most to higher curvatures (although the preference between curvatures of 6.67 μm−1 and 2 μm−1 were not statistically significant using Tukey’s one-way ANOVA; Figure 2A). In contrast, the Cdc12 2xAH polypeptide adsorbed most to a membrane curvature of 2 μm−1 (Figure 2A; Cannon et al., 2019). The Shs1 2xAH showed less adsorption to all curvatures compared with the Cdc12 2xAH, suggesting potentially different affinities for this composition of bilayers. Uneven adsorption on individual beads likely emerges from the oligomerization of the helices previously documented (Cannon et al., 2019). Curvature sensitivity of the Shs1 2xAH polypeptide was similar whether the anionic lipid used was phosphatidylinositol (PI) or phosphatidylserine (PS; Figure 2A). Thus, the Shs1 AH domain binds differentially between various curvatures, although with a shifted membrane curvature preference and a lower apparent binding affinity compared with the Cdc12 AH domain (Cannon et al., 2019).
FIGURE 2:
Shs1’s amphipathic helix is capable of binding micron-scale membrane curvatures. (A) Top, cartoon depiction of a 2xAH-GFP construct. Middle, maximum projection images of 1.25 µM Shs1 2xAH-GFP (green) adsorbed onto curved supported lipid bilayers (74.5% DOPC, 25% PI, 0.5% rhodamine-PE; magenta). Scale bar, 2 µm. Images contrasted identically. Bottom, plots of 2xAH-GFP peptide adsorption to supported lipid bilayers with different curvatures based on silica bead sizes (as in the image above). Top plot is the Shs1 2xAH-GFP peptide, and the bottom plot is the Cdc12 2xAH-GFP peptide. Experiments on PI membranes (composition is 74.5% DOPC, 25% PI, 0.5% rhodamine-PE) are in light blue, whereas PS membranes (composition is 74.5% DOPC, 25% PS, 0.5% rhodamine-PE) are dark blue. Curvatures (in μm−1) of 6.7, 4, 2, 0.67, and 0.4 represent bead diameters of 0.3, 0.5, 1, 3, and 5 μm, respectively. Each point represents the measured adsorption ratio (GFP fluorescence normalized to lipid fluorescence) on an individual bead (≥30 beads measured across three replicates at each curvature). Black bar represents mean. Error bars represent the SD of the mean. ns, adsorption was not significantly different; *, adsorption was significantly different (p < 0.01); **, p < 0.0053); ****, p < 0.0001. (B) Top, maximum projection images of 50 nM wild-type Cdc12 + Shs1 septin complex (green, panel 1) and mutant cdc12-6 + Shs1 complex (green, panel 2) on curved supported lipid bilayers (magenta). Uneven adsorption of septin complexes on individual beads reflects localized differences in the extent of septin filament polymerization on the supported lipid bilayer. Scale bar, 2 µm. Images are contrasted identically. Bottom, plots quantifying adsorption onto different curvatures (membrane composition is 74.5% DOPC, 25% PI, 0.5% rhodamine-PE). Black bars represent the mean. Error bars are the SD for n ≥ 30 beads at each curvature across three replicates. ns, adsorption was not significantly different; ****, adsorption was significantly different (p < 0.0001).
The presence of an AH on Shs1 has implications for both the valence and spacing of AH domains. Within the septin complex, Cdc12 resides at the penultimate position, with the terminal subunit being either Cdc11 or Shs1 (Versele et al., 2004; Bertin et al., 2008). There is no evidence of “chimeric” complexes with Cdc11/Shs1 on opposing ends (Garcia et al., 2011; Weems and McMurray, 2017; Khan et al., 2018). Thus, Cdc11-capped complexes possess two AH domains, spaced apart by ∼24 nm (the distance between the Cdc12 subunits), whereas Shs1-capped complexes have four AH domains, altering the valency and spacing of AHs within assemblies. These differences could impact affinity as well as the type of membrane defects sensed by complexes composed of Cdc11 versus Shs1. This raises the possibility that septin curvature preference could be different when both types of AHs are combined in the same complex.
To test whether the Shs1 AH domain alters the curvature preference of septin complexes, we purified Shs1-capped complexes (Supplemental Figure 1), which contain four AH sequences, and mixed them with supported lipid bilayers of varying curvature. Shs1-capped complexes preferentially adsorbed onto higher curvatures (κ = 6.67, 4, 2 μm−1, corresponding to bead diameters of 0.3, 0.5, and 1 μm, respectively) on both PI- or PS-containing anionic membranes (Figure 2B and Supplemental Figure 2). In contrast, Cdc11-capped complexes, which possess only the two Cdc12 AH domains, preferentially adsorbed onto curvatures of κ = 2 and 0.67 μm−1 (corresponding to bead diameters of 1 and 3 μm; Cannon et al., 2019). This shift to higher curvatures with Shs1 in the septin complex, and its two additional AH sequences, is consistent with the behavior of tandem Shs1 AH domains (Figure 2A). These data indicate that capping septin complexes with Shs1 leads to more distinct curvature preferences than complexes capped with Cdc11 (Cannon et al., 2019).
Next, we tested whether capping complexes with Shs1 could restore membrane curvature sensitivity of cdc12-6 mutant complexes. Complexes with cdc12-6 but capped with the Cdc11 subunit were previously shown to bind to all membrane curvatures equally, thus lacking curvature sensitivity due to the truncated Cdc12 AH domain (Cannon et al., 2019). Interestingly, septin complexes harboring the cdc12-6 protein but capped with Shs1 (instead of Cdc11) did show modest membrane curvature sensitivity on both PI- or PS-containing anionic membranes (Figure 2B and Supplemental Figure 2). Interestingly, cdc12-6 Shs1-capped septin complexes preferentially adsorbed onto membrane curvatures of κ = 2 μm−1, similar to wild-type Cdc12 complexes capped with Cdc11 (Cannon et al., 2019). Collectively, these data suggest that 1) septin complexes harboring either the Cdc12 or Shs1 AH domain (but not both) have a similar membrane curvature preference (max at κ = 2 μm−1) and 2) complexes harboring both AH domains of Cdc12 and Shs1 shift their curvature preference toward higher curvatures. This points to AH valence (2× versus 4×) being a critical factor in specifying septin curvature preference.
Despite restoration of curvature sensing, we note the adsorption of cdc12-6 Shs1-capped complexes was substantially lower than Cdc12-wt Shs1-capped complexes. This contrasts with cdc12-6 Cdc11-capped complexes, which lack curvature sensitivity but still adsorb at high levels to membranes just independent of curvature (Cannon et al., 2019). The differential adsorption of cdc12-6 complexes with either Cdc11 or Shs1 has several implications. First, it suggests the enhanced, nonspecific binding of cdc12-6 complexes is dependent on Cdc11, possibly reflecting a strong membrane-interaction surface in Cdc11. Second, it had been previously shown that capping septin complexes with Shs1 inhibits filament polymerization (Garcia et al., 2011; Khan et al., 2018). Thus, through the lack of Cdc11 and limited filament formation, cdc12-6 Shs1-capped complexes may be less stably associated with the membrane and yield the lower adsorption levels in this assay. Nonetheless, these data indicate that the Shs1 AH differentially binds various membrane curvatures.
The Shs1 CTE harboring an AH domain is required for normal septin function in the cdc12-6 mutant
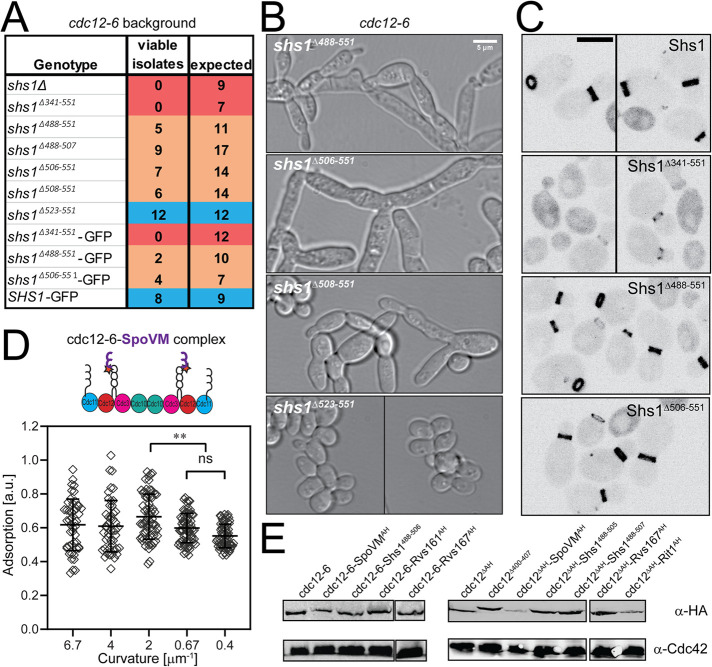
We reasoned that because Shs1 harbors a predicted AH domain, it could become critical for septin localization at the bud neck in cdc12-6 strains at permissive temperature. If the ability of septins to differentiate membrane curvatures is essential, and either the Cdc12 or Shs1 AH domains can sense membrane curvature, then compromising both would produce a synthetic genetic interaction. Consistent with this hypothesis, it had been previously reported that deletion of SHS1 in a cdc12-6 background is inviable (Finnigan et al., 2015), which was corroborated by tetrad dissection in our strain background at 24°C (Figure 3A). To distinguish whether it was the Shs1 AH domain that was essential in cdc12-6 mutants, we generated a panel of C-terminal Shs1 truncations in the cdc12-6 background (Figure 3A). C-terminal truncations of Shs1 that begin immediately after the AH domain (shs1Δ506–551 and shs1Δ508–551) were occasionally viable in the cdc12-6 background at 24°C (Figure 3A). However, viable double mutants had classic septin mutant phenotypes with elongated buds and often failed to separate from the mother (Figure 3B). This may be due to the proximity of the truncation to the AH domain. cdc12-6 mutants expressing Shs1 truncation beginning 15 residues after the AH domain were fully viable without septin defects (Figure 3, A and B). cdc12-6 mutants expressing a more extensive Shs1 truncation (shs1Δ488–551) or an AH deletion (shs1Δ488–507) were sick, displaying fully penetrant septin defects at 24°C (Figure 3, A and B). Truncating the entire Shs1 CTE (shs1Δ341–551) was synthetic lethal with cdc12-6 (Figure 3B). These data indicate that the Shs1 CTE becomes essential in cdc12-6 mutants, and mutants lacking both the full AH domain in Cdc12 and Shs1 have classic septin mutant defects (Hartwell et al., 1970).
FIGURE 3:
Genetic analyses of Shs1 and Cdc12 AH domains. (A) Viability of cdc12-6 cells expressing indicated SHS1 alleles expressed from the endogenous locus (with 3xHA tag, unless indicated with GFP tag, which lacks 3xHA) based on tetrad dissections at permissive temperature (24°C). Cells from genotypes labeled in blue appeared normal, without obvious septin defects. Genotypes in orange were sick, with partially or fully penetrant septin defects. Genotypes labeled in red were inviable. (B) DIC images of cdc12-6 shs1 mutants (see A) at 24°C. Scale bar, 5 μm. (C) Heterozygous diploids expressing the indicated Shs1 protein fused to GFP from the SHS1 locus. Scale bar, 5 μm. (D) Scatter plot quantifying cdc12-6-SpoVM septin complex adsorption onto different membrane curvatures. Black bars represent the mean. Error bars are the SD for more than 30 measured beads at each curvature across three replicates. Adsorption of cdc12-6-SpoVM complexes was significantly greater on a membrane curvature of 2 μm−1 than on curvatures of 0.67 and 0.4 μm−1; ** (p < 0.01 and p < 0.0001, respectively). ns, adsorption was not significantly different. (E) Western blot comparing expression of indicated Cdc12 chimeras fused to 3xHA epitope in heterozygous diploids.
It was possible that truncating Shs1 reduced Shs1 incorporation into septin complexes. To test this hypothesis, we generated GFP-tagged Shs1 truncations (but without an HA tag) expressed from the endogenous SHS1 locus to assess Shs1 expression and localization. In wild-type CDC12 cells, Shs1 truncations were expressed at similar levels to wild-type Shs1-GFP and localized to the bud neck normally, with the exception of Shs1Δ341–551-GFP (Figure 3C). GFP tagging the Shs1 truncations reduced viability in the cdc12-6 background assessed by tetrad dissection (Figure 3A), perhaps indicating perturbations caused by the GFP tag. Shs1Δ341–551-GFP, in which the entire CTE is truncated, was expressed normally in wild-type cells, but localized predominantly in the cytoplasm with reduced signal at the bud neck (Figure 3C). The reduced neck-localized signal of Shs1Δ341–551-GFP suggests a reduced capacity of Shs1 complexes to associate with the membrane, which may limit their ability to incorporate into septin filaments and sense curvature. In cdc12-6 mutants, we hypothesize such a deficiency is lethal. These data suggest that truncating Shs1 through its AH domain in vivo does not interfere with its incorporation into complexes with otherwise wild-type septin subunits (Cdc3, Cdc10, Cdc12). However, we were unable to purify recombinant shs1Δ488–507 cdc12-6 septin complexes in vitro. We therefore cannot rule out the possibility that combining Shs1 CTE truncations with the cdc12-6 subunit interferes with proper septin complex formation.
We reasoned that if the main function of the Cdc12 AH domain is to sense membrane curvature, then the addition of other AH domains might be sufficient to restore cdc12-6 function. We generated chimeric strains expressing different AH domains fused to the C-terminus of either cdc12-6 or Cdc12 lacking its entire AH domain (cdc12ΔAH). Chimeric Cdc12-AH fusion constructs included AH domains from Shs1, Rvs161, Rvs167, mammalian RitC, or Bacillus subtilis SpoVM (Youn et al., 2010; Bendezu et al., 2015; Gill et al., 2015). SpoVM is of particular interest as it has been documented to preferentially adsorb onto membranes with similar curvatures as septins (Gill et al., 2015). Indeed, a recombinant septin complex harboring cdc12-6 fused to SpoVM did restore membrane-curvature sensitivity (Figure 3D). Surprisingly, however, none of the cdc12-6-AH chimeric yeast strains were viable at restrictive temperature (37°C), including the cdc12-6-SpoVM chimeric strain, and none of the cdc12ΔAH-AH chimeras were viable even at permissive temperature as assessed by tetrad dissection. This is despite the fact that Cdc12 AH domain chimeras were expressed normally relative to wild-type Cdc12 (Figure 3E). It remained possible that removing the eight residues C-terminal to the AH domain or the addition of 3xHA tag to the C-terminus interfered with AH domain function. However, strains expressing a truncated Cdc12, but one that kept the endogenous AH domain (Cdc12Δ400–407) fused to the 3xHA tag were mostly viable at 37°C (9 isolates viable of 12 expected), suggesting these modifications do not substantially impact AH domain function. Collectively, these data indicate Cdc12 chimeras with alternate AH domains are nonfunctional in cells even if the chimeric AH domain is predicted to recognize similar membrane curvatures (Gill et al., 2015) even if it can recapitulate curvature sensing in vitro.
There are multiple possibilities as to why cdc12ΔAH mutants are inviable even with wild-type SHS1. One possibility is that cdc12-6, but not Cdc12ΔAH, maintains some limited capacity to recognize membrane curvature in cells (even if not in vitro). Other possibilities revolve around Cdc12ΔAH incorporation into septin complexes. Cdc12ΔAH itself may not fold properly, or it may interfere with complex formation including Shs1-capping. However, we reasoned that the AH domain(s) of Cdc12 and/or Shs1 may have an important function aside from sensing membrane curvature (discussed below).
The Cdc12 AH domain inhibits septin bundling
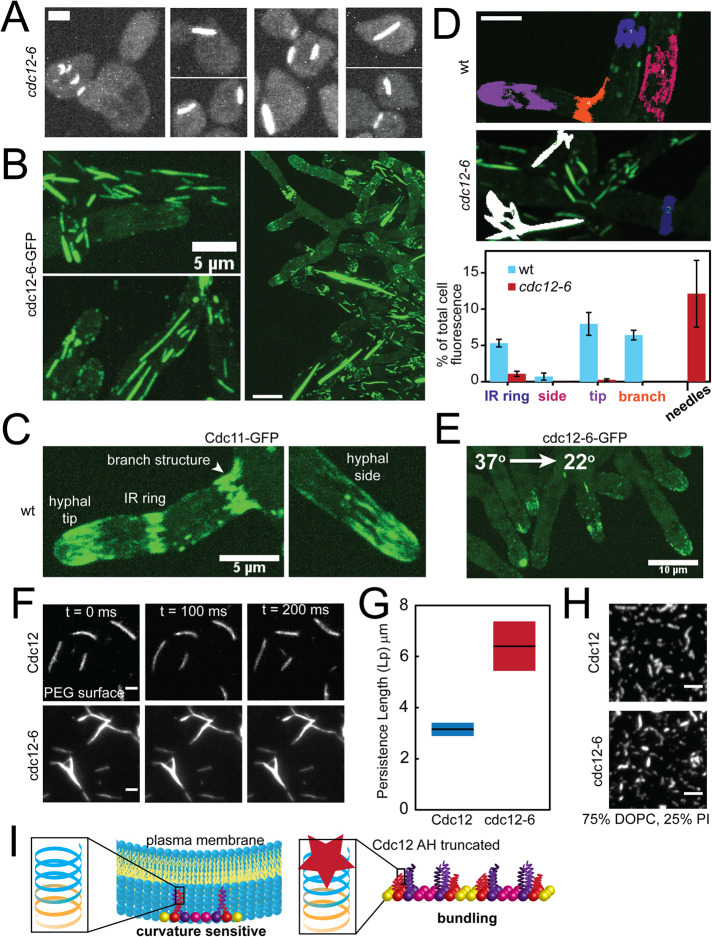
Why are Cdc12-AH domain chimeras nonfunctional? And why are cdc12-6 mutants sensitive to Shs1 CTE truncations, including ones that maintain the Shs1 AH? We reasoned there might be an additional, essential function to the Cdc12 AH domain or a gain of function that occurs when the AH domain is lost. A clue comes from yeast cells exposed to pheromone. In cdc12-6 mutants exposed to pheromone, septins no longer localize filaments to the base of the mating projection (i.e., shmoo), and typically remain cytoplasmic (Giot and Konopka, 1997; Longtine et al., 1998). In a subset of cdc12-6 mutant cells exposed to pheromone, we observed aberrant septin structures that appeared to be highly bundled, and not in association with the cortex (Figure 4A). These bundled structures were not observed in cycling, heterozygous diploids (cdc12-6/CDC12), possibly indicating that ectopic bundled needles are a recessive phenotype of cdc12-6 mutants.
FIGURE 4:
The Cdc12 AH domain inhibits septin bundling. (A) Maximum projections (contrasted identically) of fixed cells with bundled “needles” in cdc12-6 S. cerevisiae mutants expressing Cdc3-mCherry treated with pheromone. Scale bar, 2 µm. (B) Maximum projections of bundled “needles” in A. gossypii expressing cdc12-6-GFP at 22°C. Scale bars, 5 μm (left panels) and 10 μm (right panel). (C) Maximum projection of septin structures in wild-type A. gossypii expressing Cdc11a-GFP at 22°C. Scale bar, 5 μm. (D) Quantification of the relative abundance (based on sum fluorescence) of different septin structures in wild-type and cdc12-6 mutant Ashbya cells. Top, representative, cropped maximum projection images of Ashbya cells with color-coded localized septin structures (blue: IR rings; magenta: hyphal sides; purple: hyphal tips; orange: base of hyphal branches; white: needles). Septin structures were segmented from z-stacks images of an entire cell using the 3D Image Suite plugin in FIJI. Scale bar, 5 μm. Bottom, quantification of the sum fluorescence of different septin structures (as depicted in the above; structures ≥3 μm3) as a percentage of whole cell fluorescence above background in wild-type (blue bars) and cdc12-6 mutant (red bars) cells. N = 5 cells each. (E) Maximum projection of a cdc12-6-GFP A. gossypii mutant imaged at 22°C within 1 min after being shifted down from 37°C for 1 h. Scale bar, 10 μm. (F) TIRF images of wild-type (top) and cdc12-6 (bottom) septin filaments formed in solution at 250 nM on a PEG-coated coverslip. Cdc11-SNAP was fluorescently labeled with AlexaFluor 488 dye. Scale bar, 2 µm. (G) Persistence length as determined by the cosine correlation of tangent angles of filaments (wt, n = 37; cdc12-6, n = 66) polymerized in solution. Lines represent mean; bars represent range. (H) TIRF images of 3 nM wild-type and cdc12-6 septins (Cdc11-SNAP was fluorescently labeled with AlexaFluor 488 dye) polymerized into filaments on planar-supported lipid bilayers. Scale bar, 2 µm. (I) Model for Cdc12 AH domain function. Left, the Cdc12 AH domain enables septins to bind and distinguish micron-scale membrane curvatures. A cartoon depiction of the rod-shaped, nonpolar septin complex is capped with Cdc11 (yellow; CTE not included for clarity), Cdc12 (red shown with CTE), Cdc3 (purple with CTE), and Cdc10 (pink). Left inset, the full-length Cdc12 AH domain (blue) is at the C-terminus of the Cdc12 CTE (orange). Right, the absence of a fully functional Cdc12 AH domain promotes septin bundling in lieu of sensing membrane curvature. We speculate that absence of the AH domain leads to misfolding of the coiled-coil element of Cdc12, promoting coiled-coil interactions between Cdc3 subunits between septin complexes to promote bundling.
We further investigated the bundled septin phenomenon in A. gossypii, an ascomycete fungus closely related to budding yeast (Dietrich et al., 2004). In Ashbya cdc12-6 mutants, bundled needle-like structures were detectable in the cytoplasm of vegetatively growing cells, sometimes extending the entire length of hyphae tens of microns in length (Figure 4B and Supplemental Figure 3). In wild-type Ashbya cells, septins localize to discrete structures such as interregion (IR) rings, hyphal sides, hyphal tips, and hyphal branch points (Figure 4C; DeMay et al., 2009). We compared the relative abundance of different characterized septin structures in wild-type cells and cdc12-6 mutants by quantifying sum fluorescence of the different structures per cell (Figure 4D). Notably, compared with wild-type cells, the majority of septin structures in cdc12-6 mutants were bundled needles. Thus, truncation of the C-terminal AH domain not only blocks membrane-curvature sensing, but also promotes septin bundling. We speculate needles in cdc12-6 mutants may act like a sponge, sequestering septins into inoperative structures and exacerbating the cdc12-6 mutant phenotype in Ashbya.
It was possible the needles are aggregates, and not reflective of septin bundling. However, septin needles were temperature sensitive and disassembled when Ashbya mutants were shifted to 37°C. After shifting down from 37°C to 22°C, septins did not immediately reform into needles, but instead localized to hyphal tips (Figure 4E). This is consistent with a previous observation in cdc12-6 budding yeast mutants in which, after shifting down to permissive temperature, septins did not localize to the bud neck but instead colocalized with polarity factors in the bud (Gladfelter et al., 2005). It also suggests that the septin needles are not irreversible aggregates.
Consistent with observed septin needles in vivo, recombinant cdc12-6 septin complexes (capped with Cdc11) can form needles in solution that were less flexible than wild-type, recombinant filaments (Figure 4, F and G, and Supplemental Movie S1). This indicates the formation of needle-like structures is an intrinsic feature of cdc12-6 septin complexes not dependent on other cellular factors. Interestingly, when incubated on supported planar lipid bilayers, cdc12-6 complexes polymerized into filaments indistinguishable from wild-type complexes (Figure 4H). We note that some cdc12-6 bundles were observed on lipid bilayers (Figure 4H); however, these formed in solution and then bound the membrane. Overall, this suggests that membranes may play an inhibitory role in bundling, and is supported by the observation that needles in cells appeared to be within the cytosol and away from the cortex (Figure 4, A and B).
Movie S1.
Wild-type and cdc12-6 septin filaments in solution. TIRF movies of wild-type (left) and cdc12-6 (right) septin filaments formed in solution as in Figure 4D. Movies acquired and contrasted identically. Scale bar, 10 μm.
The needle-like septin structures in cdc12-6 mutants suggest the AH domain of Cdc12 may also have a role in septin bundling. The coiled-coil element of Cdc12 (harboring the AH domain) can associate with the coiled-coil element of Cdc3 (Versele et al., 2004; Bertin et al., 2008). Homologous coiled-coil elements can dimerize or even oligomerize into larger coiled-coil structures in the absence of the preferred binding partner (Harbury et al., 1993; Lupas and Bassler, 2017). Truncation of the Cdc12 AH domain in cdc12-6 mutants is predicted to misfold the coiled-coil element at the C-terminus of Cdc12 (MultiCoil; Wolf et al., 1997). Although the exact nature of septin needle assembly in cdc12-6 mutants remains speculative, we conjecture that in the absence of properly folded Cdc12 coiled-coil elements (like in cdc12-6 mutants) free Cdc3 coiled-coil elements can oligomerize with one another between adjacent septin complexes, thus promoting bundled septin needle formation (Figure 4I). In wild-type Ashbya, dispersed septin filaments near the tips of hyphae coalesce into discrete bundled structures (IR rings) along the hyphal body (DeMay et al., 2009). This transition is dependent on the kinase Gin4, which is predicted to interact with the coiled-coil element of Cdc3 (Longtine et al., 1998; Mortensen et al., 2002). Thus, in wild-type cells we speculate the Cdc3/Cdc12 coiled-coil may act like toggleable switch, whose regulated disassembly could promote septin bundling through Cdc3 coiled-coil oligomerization.
CONCLUSION
Here we report that an additional AH domain in the nonessential septin, Shs1, can recognize membrane curvature. Shs1 and its CTE harboring the AH domain become essential in mutants lacking a fully functional AH domain in Cdc12 (cdc12-6). Cdc12-6 induces filament bundling into nonphysiological needle-like structures, possibly exacerbating its phenotype. This may indicate a role for the AH domain in septin bundling. Future research should investigate how the Shs1 AH domain might influence septin-membrane–binding affinity and diffusion in the membrane. The role of AH domains in septin biochemistry and biology is only beginning to be understood and is an exciting area of future study.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Yeast strain construction and culturing
Standard molecular genetic techniques were used to generate and culture yeast strains in this study. All yeast strains used in this study are listed in Supplemental Table S1. The temperature-sensitive cdc12-6 mutant was used to assess the function of Shs1 truncations, as the shs1Δ cdc12-6 double mutant is synthetic lethal in our strain background. We received a bar1Δ strain and a diploid strain heterozygous for shs1Δ and cdc12-6 as gifts (D. Lew, Duke University). We generated the untagged Shs1 truncations (Shs1Δ341–551, Shs1Δ488–551, Shs1Δ488–507, Shs1Δ506–551, Shs1Δ508–551, and Shs1Δ523–551) and GFP-tagged Shs1 constructs (Shs1-GFP, Shs1Δ341–551-GFP, Shs1Δ488–551-GFP, Shs1Δ506–551-GFP) using the PCR-based C-terminal modification method (Longtine et al., 1998). We generated cdc12-6, Cdc12ΔAH AH-chimeric (SpoVMAH, Shs1488–505, Shs1488-–507, Rvs161AH, Rvs167AH, S. pombe Rit1AH), and Cdc12Δ400–407 constructs with an HA tag using the PCR-based C-terminal modification method mentioned above. PCR vectors were integrated at their endogenous loci (SHS1 or CDC12, respectively) to generate heterozygous diploids. All transformants were verified for targeted integration by PCR amplification of genomic DNA. Transformed diploids were induced to sporulate, and resulting tetrads were dissected. Tetrad dissections were used to assess genetic interactions.
For imaging, cells were grown to midlog phase at 24°C in complete synthetic medium (CSM; Sunrise Science Products) supplemented with 0.67% yeast nitrogen base, 2% dextrose, and 0.01% adenine, then harvested. Cells were mounted onto CSM + 2% dextrose agarose (2%) pads before imaging. For experiments using yeast pheromone, midlog phase cultures of bar1Δ cells were treated with 50 nM α-factor (GenWay Biotech) for 2 h at 24°C before fixation with 3.7% formaldehyde for 10 min at room temperature before imaging.
A. gossypii culturing
The A. gossypii strain harboring the cdc12-6-GFP allele at the endogenous CDC12 locus (AG884; AgCDC12:cdc12-6-GFP:GEN leu2Δ thr4Δ) was described before (Cannon et al., 2019). A. gossypii was grown from spores in full medium for 16 h at 24°C before harvesting mycelial cells. Cells were mounted on low fluorescence medium agarose (2%) pads and sealed with petroleum jelly for imaging at room temperature (22°C). For experiments at restrictive temperature, cells were grown and harvested, and mounted as above before being placed in humidified chambers at 37°C. Cells were then imaged at 22°C.
Cell microscopy
Confocal images were acquired using a spinning disk (Yokogawa W1) confocal microscope (Nikon Ti-82 stage) using a 100× Plan Apo 1.49 NA oil lens and a Prime 95B CMOS camera (Photometrics). Differential interference contrast (DIC) images were acquired on a custom-built microscope with an inverted Olympus IX-83 body equipped with an Olympus 60× 1.3 NA lens and an EMCCD camera (Andor iXon-888; McQuilken et al., 2017). Live-cell microscopy images were prepared using ImageJ (FIJI). Three-dimensional septin structures in wild-type and cdc12-6 mutant Ashbya were segmented from confocal image z-stacks using the 3D Image Suite plugin in ImageJ/FIJI. The sum fluorescence of segmented structures (that were at least 3 cubic microns) of each category was measured from each cell (five cells from each genotype) and averaged.
Immunoblotting
For Western blot analysis, ∼2 × 107 cells were harvested from midlog phase cultures grown at 24°C and total protein was extracted by TCA precipitation as previously published (Keaton et al., 2008). Electrophoresis and Western blotting were performed as previously described (Bose et al., 2001). A polyclonal rabbit α-HA epitope antibody preparation was used at 1:2000 dilution (Rockland Immunochemicals). A monoclonal mouse α-Cdc42 antibody preparation (gift from P. Brennwald, UNC) was used at 1:500 dilution (Wu et al., 2015). Polyclonal goat antibodies against rabbit (goat anti-rabbit IgG DyLight 680 conjugated; Rockland Immunochemicals) and mouse (goat anti-mouse IgG DyLight 800 conjugated; Rockland Immunochemicals) were used at 1:10,000 dilution as secondary antibodies. Blots were imaged with an ODYSSEY infrared laser scanner (LI-COR Biosciences).
Protein purification
Recombinant S. cerevisiae septin protein complexes were expressed from a duet expression system in BL21 (DE3) Escherichia coli cells and purified as previously described (Supplemental Table S2; Bridges et al., 2016; Cannon et al., 2019). Amphipathic helices conjugated to GFP were purified with a similar protocol as septins; however, induced cultures were grown for 4 h at 37°C instead of 24 h at 22°C.
AH constructs
The 2× AH(Shs1)-GFP construct has the primary sequence: KELQKRIQELERKAHELK-GSGSRSGSGS KELQKRIQELERKAHELK-GSGSSR-GFP tag. The underlined sequence represents the AH domain from Shs1. The 2× AH(Cdc12) construct has been described previously (Cannon et al., 2019). The glycine-serine repeats represent linker sequences between the AH domains and the GFP tag.
Lipid mix preparation
Lipids (from Avanti Polar Lipids) were mixed in chloroform solvent at a ratio of 75 mol% dioleoylphosphatidylcholine (DOPC) and 25 mol% of either PI (liver, bovine) sodium salt or PS in a glass cuvette. Both PI and PS have a charge of −1 at pH 7.4 (Beber et al., 2019a). For lipid mix preparations to be incubated on silica microspheres, trace amounts (>0.1%) of phosphatidylethanolamine-N-(lissamine rhodamine B sulfonyl) (RH-PE) were added. Lipid mixtures were then dried with argon gas to create a lipid film and stored under negative vacuum pressure overnight to evaporate trace chloroform. Lipids were rehydrated in supported lipid bilayer buffer (300 mM KCl, 20 mM HEPES, pH 7.4, 1 mM MgCl2) at 37°C to a final lipid concentration of 5 mM. Lipids were then resuspended over the course of 30 min at 37°C, with vortexing for 10 s every 5 min. Fully resuspended lipids were then bath sonicated at 2-min intervals until the solution clarified to yield small unilamellar vesicles (SUVs).
Preparation of supported lipid bilayers
Lipid bilayers supported on silica microspheres were prepared as previously described (Bridges et al., 2016; Cannon et al., 2019). SUVs were adsorbed onto silica microspheres (Bangs Laboratories) by incubating 50 nM of lipid with different bead sizes (with a summed surface area of 440 mM2) for 1 h at room temperature with gentle rotation. Microspheres were pelleted, then resuspended in in prereaction buffer (33 mM KCl, 50 mM HEPES, pH 7.4) to wash away excess SUVs. This wash step was repeated four additional times. Fluidity of the supported lipid bilayers (including labeled RH-PE) was assessed by fluorescence recovery after photobleaching (FRAP) prepared on large beads (>5 μm), which had an average FRAP t1/2 recovery of 7.4 s.
Planar-supported lipid bilayers were prepared in a similar process as reported previously (Bridges et al., 2014). Number 1.5 coverslips were first cleaned with oxygen plasma for 15 min at the highest power (PE25-JW; Plasma Etch). Chambers for lipid bilayers cut from PCR tubes were glued (Norland optical adhesive; Thor Labs) onto cleaned coverslips. SUVs were incubated at 37°C in chambers on coverslips suspended in supported lipid bilayer buffer with additional 1 mM CaCl2 for 20 min (1 mM final lipid concentration in 50 μl solution). After vesicle fusion, lipid bilayers were washed six times with 150 μl of supported lipid bilayer buffer to rinse away excess SUVs. Immediately before adding septins, bilayers were rinsed six times with 150 μl of buffer containing 50 mM HEPES, pH 7.4, 1 mM BME (Beta-mercaptoethanol), and 0.01% bovine serum albumin (BSA).
Measuring protein adsorption on lipid bilayers supported on silica microspheres
Experiments measuring the adsorption of septins and AH domains on different curvatures were performed as previously published (Cannon et al., 2019). Reactions (with a final buffer composition of 100 mM KCl, 50 mM HEPES, pH 7.4, 1 mM BME, 0.1% methyl cellulose, and 0.01% BSA) were incubated in a plastic chamber (Bridges and Gladfelter, 2016) glued onto a polyethylene glycol (PEG)-coated passivated coverslip for 1 h to let reactions reach equilibria. Confocal images of fluorescent-tagged protein adsorbed onto curved supported bilayers on microspheres were acquired using a spinning disk (Yokogawa W1) confocal microscope (Nikon Ti-82 stage) using a 100× Plan Apo 1.49 NA oil lens and a 95B CMOS camera (Photometric). Raw images were analyzed using Imaris 8.1.2 software (Bitplane AG) as previously described (Cannon et al., 2019). Briefly, fluorescence intensity as a measure of membrane adsorption of constructs (e.g., septins or AH domains) were compared relative to the normalized intensity of the fluorescent lipids. Scatter plots of adsorption were generated using Prism (version 8.3.0; GraphPad). Differences in membrane adsorption between different curvatures were compared by Tukey’s one-way ANOVA.
Total internal reflection fluorescence microscopy
Experiments in which septin filaments were polymerized in solution, reactions were incubated in plastic chambers (glued onto PEG-coated passivated coverslips) for 1 h to reach equilibria. Recombinant protein was diluted to 250 nM final concentration in reaction buffer (75 mM KCl, 50 mM HEPES, pH 7.4, 1 mM BME, 0.1% methyl cellulose, and 0.01% BSA).
For experiments on planar-supported lipid bilayers, recombinant septins were incubated at a final concentration of 3 nM in a buffer with 50 mM KCl, 50 mM HEPES, pH 7.4, 1 mM BME, and 0.01% BSA. On lipid bilayers, septin polymerization was imaged immediately upon incubation.
Images were acquired utilizing a Nikon TiE TIRF (total internal reflection fluorescence) system equipped with a solid-state laser system (15 mW, Nikon LUn-4), a Nikon Ti-82 stage, a 100× Plan Apo 1.49 NA oil lens, and Prime 95B CMOS camera (Photometrics). TIRF microscopy images were processed and analyzed using ImageJ (FIJI).
Persistence length measurements
The cosine correlation of tangent angles along the filament lengths were assembled from single time points and fitted to an exponential to determine the persistence length using a MATLAB GUI previously published (Graham et al., 2014).
AH domain prediction and helical diagram generation
Septin sequences were screened for putative amphipathic helices in silico using Heliquest (Gautier et al., 2008). Amphipathic helices were considered based on their hydrophobic moment, hydrophobicity, and net charge (Eisenberg et al., 1982). Heliquest was used to generate helical wheel diagrams in Figure 1.
Supplementary Material
Acknowledgments
We thank Daniel J. Lew (Duke University) for generously providing cdc12-6 and shs1Δ strains. We thank Wilton Snead for microscopy support. This work was supported by National Institutes of Health (NIH) Grant no. R01GM-130934 to A.S.G. B.L.W. was supported by the NIH Training Grant no. 2T32AI052080-16. K.S.C. and E.J.D.V. were supported in part by a grant from the National Institute of General Medical Sciences under award T32 GM119999.
Abbreviations used:
- AH
amphipathic helix
- CTE
C-terminal extension.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-05-0303).
REFERENCES
- Adams AE, Pringle JR (1984). Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae. J Cell Biol 98, 934–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beber A, Alqabandi M, Prevost C, Viars F, Levy D, Bassereau P, Bertin A, Mangenot S (2019a). Septin-based readout of PI(4,5)P2 incorporation into membranes of giant unilamellar vesicles. Cytoskeleton (Hoboken) 76, 92–103. [DOI] [PubMed] [Google Scholar]
- Beber A, Taveneau C, Nania M, Tsai FC, Di Cicco A, Bassereau P, Levy D, Cabral JT, Isambert H, Mangenot S, Bertin A (2019b). Membrane reshaping by micrometric curvature sensitive septin filaments. Nat Commun 10, 420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendezu FO, Vincenzetti V, Vavylonis D, Wyss R, Vogel H, Martin SG (2015). Spontaneous Cdc42 polarization independent of GDI-mediated extraction and actin-based trafficking. PLoS Biol 13, e1002097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin A, McMurray MA, Grob P, Park SS, Garcia G 3rd, Patanwala I, Ng HL, Alber T, Thorner J, Nogales E (2008). Saccharomyces cerevisiae septins: supramolecular organization of heterooligomers and the mechanism of filament assembly. Proc Natl Acad Sci USA 105, 8274–8279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose I, Irazoqui JE, Moskow JJ, Bardes ES, Zyla TR, Lew DJ (2001). Assembly of scaffold-mediated complexes containing Cdc42p, the exchange factor Cdc24p, and the effector Cla4p required for cell cycle-regulated phosphorylation of Cdc24p. J Biol Chem 276, 7176–7186. [DOI] [PubMed] [Google Scholar]
- Bridges AA, Gladfelter AS (2016). In vitro reconstitution of septin assemblies on supported lipid bilayers. Methods Cell Biol 136, 57–71. [DOI] [PubMed] [Google Scholar]
- Bridges AA, Jentzsch MS, Oakes PW, Occhipinti P, Gladfelter AS (2016). Micron-scale plasma membrane curvature is recognized by the septin cytoskeleton. J Cell Biol 213, 23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges AA, Zhang H, Mehta SB, Occhipinti P, Tani T, Gladfelter AS (2014). Septin assemblies form by diffusion-driven annealing on membranes. Proc Natl Acad Sci USA 111, 2146–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon KS, Woods BL, Crutchley JM, Gladfelter AS (2019). An amphipathic helix enables septins to sense micrometer-scale membrane curvature. J Cell Biol 218, 1128–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay L, Caudron F, Denoth-Lippuner A, Boettcher B, Buvelot Frei S, Snapp EL, Barral Y (2014). A sphingolipid-dependent diffusion barrier confines ER stress to the yeast mother cell. Elife 3, e01883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMay BS, Bai X, Howard L, Occhipinti P, Meseroll RA, Spiliotis ET, Oldenbourg R, Gladfelter AS (2011). Septin filaments exhibit a dynamic, paired organization that is conserved from yeast to mammals. J Cell Biol 193, 1065–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMay BS, Meseroll RA, Occhipinti P, Gladfelter AS (2009). Regulation of distinct septin rings in a single cell by Elm1p and Gin4p kinases. Mol Biol Cell 20, 2311–2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich FS, Voegeli S, Brachat S, Lerch A, Gates K, Steiner S, Mohr C, Pohlmann R, Luedi P, Choi S, et al. (2004). The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science 304, 304–307. [DOI] [PubMed] [Google Scholar]
- Drin G, Antonny B (2010). Amphipathic helices and membrane curvature. FEBS Lett 584, 1840–1847. [DOI] [PubMed] [Google Scholar]
- Drin G, Casella JF, Gautier R, Boehmer T, Schwartz TU, Antonny B (2007). A general amphipathic α-helical motif for sensing membrane curvature. Nat Struct Mol Biol 14, 138–146. [DOI] [PubMed] [Google Scholar]
- Eisenberg D, Weiss RM, Terwilliger TC (1982). The helical hydrophobic moment: a measure of the amphiphilicity of a helix. Nature 299, 371–374. [DOI] [PubMed] [Google Scholar]
- Field CM, Al-Awar O, Rosenblatt J, Wong ML, Alberts B, Mitchison TJ (1996). A purified Drosophila septin complex forms filaments and exhibits GTPase activity. J Cell Biol 133, 605–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnigan GC, Takagi J, Cho C, Thorner J (2015). Comprehensive genetic analysis of paralogous terminal septin subunits Shs1 and Cdc11 in Saccharomyces cerevisiae. Genetics 200, 821–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia G 3rd, Bertin A, Li Z, Song Y, McMurray MA, Thorner J, Nogales E (2011). Subunit-dependent modulation of septin assembly: budding yeast septin Shs1 promotes ring and gauze formation. J Cell Biol 195, 993–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier R, Douguet D, Antonny B, Drin G (2008). HELIQUEST: a web server to screen sequences with specific α-helical properties. Bioinformatics 24, 2101–2102. [DOI] [PubMed] [Google Scholar]
- Gilden JK, Peck S, Chen YC, Krummel MF (2012). The septin cytoskeleton facilitates membrane retraction during motility and blebbing. J Cell Biol 196, 103–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill RL Jr, Castaing JP, Hsin J, Tan IS, Wang X, Huang KC, Tian F, Ramamurthi KS (2015). Structural basis for the geometry-driven localization of a small protein. Proc Natl Acad Sci USA 112, E1908–E1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giot L, Konopka JB (1997). Functional analysis of the interaction between Afr1p and the Cdc12p septin, two proteins involved in pheromone-induced morphogenesis. Mol Biol Cell 8, 987–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladfelter AS, Kozubowski L, Zyla TR, Lew DJ (2005). Interplay between septin organization, cell cycle and cell shape in yeast. J Cell Sci 118, 1617–1628. [DOI] [PubMed] [Google Scholar]
- Gladfelter AS, Pringle JR, Lew DJ (2001). The septin cortex at the yeast mother-bud neck. Curr Opin Microbiol 4, 681–689. [DOI] [PubMed] [Google Scholar]
- Graham JS, McCullough BR, Kang H, Elam WA, Cao W, De EM, Cruz La (2014). Multi-platform compatible software for analysis of polymer bending mechanics. PLoS One 9, e94766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbury PB, Zhang T, Kim PS, Alber T (1993). A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science 262, 1401–1407. [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Culotti J, Reid B (1970). Genetic control of the cell-division cycle in yeast. I. Detection of mutants. Proc Natl Acad Sci USA 66, 352–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John CM, Hite RK, Weirich CS, Fitzgerald DJ, Jawhari H, Faty M, Schlapfer D, Kroschewski R, Winkler FK, Walz T, et al. (2007). The Caenorhabditis elegans septin complex is nonpolar. EMBO J 26, 3296–3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CR, Weems AD, Brewer JM, Thorner J, McMurray MA (2015). Cytosolic chaperones mediate quality control of higher-order septin assembly in budding yeast. Mol Biol Cell 26, 1323–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keaton MA, Szkotnicki L, Marquitz AR, Harrison J, Zyla TR, Lew DJ (2008). Nucleocytoplasmic trafficking of G2/M regulators in yeast. Mol Biol Cell 19, 4006–4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A, Newby J, Gladfelter AS (2018). Control of septin filament flexibility and bundling by subunit composition and nucleotide interactions. Mol Biol Cell 29, 702–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EY, Tyndall ER, Huang KC, Tian F, Ramamurthi KS (2017). Dash-and-recruit mechanism drives membrane curvature recognition by the small bacterial protein SpoVM. Cell Syst 5, 518–526.e513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew DJ, Reed SI (1995). A cell cycle checkpoint monitors cell morphogenesis in budding yeast. J Cell Biol 129, 739–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, Fares H, Pringle JR (1998). Role of the yeast Gin4p protein kinase in septin assembly and the relationship between septin assembly and septin function. J Cell Biol 143, 719–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, Theesfeld CL, McMillan JN, Weaver E, Pringle JR, Lew DJ (2000). Septin-dependent assembly of a cell-cycle-regulatory module in Saccharomyces cerevisiae. Mol Cell Biol 20, 4049–4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low C, Macara IG (2006). Structural analysis of septin 2, 6, and 7 complexes. J Biol Chem 281, 30697–30706. [DOI] [PubMed] [Google Scholar]
- Lupas AN, Bassler J (2017). Coiled coils—a model system for the 21st century. Trends Biochem Sci 42, 130–140. [DOI] [PubMed] [Google Scholar]
- McQuilken M, Jentzsch MS, Verma A, Mehta SB, Oldenbourg R, Gladfelter AS (2017). Analysis of septin reorganization at cytokinesis using polarized fluorescence microscopy. Front Cell Dev Biol 5, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen EM, McDonald H, Yates J 3rd, Kellogg DR (2002). Cell cycle-dependent assembly of a Gin4-septin complex. Mol Biol Cell 13, 2091–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F, Malmberg RL, Momany M (2007). Analysis of septins across kingdoms reveals orthology and new motifs. BMC Evol Biol 7, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodal AA, Kozubowski L, Goode BL, Drubin DG, Hartwig JH (2005). Actin and septin ultrastructures at the budding yeast cell cortex. Mol Biol Cell 16, 372–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirajuddin M, Farkasovsky M, Hauer F, Kuhlmann D, Macara IG, Weyand M, Stark H, Wittinghofer A (2007). Structural insight into filament formation by mammalian septins. Nature 449, 311–315. [DOI] [PubMed] [Google Scholar]
- Spiliotis ET (2010). Regulation of microtubule organization and functions by septin GTPases. Cytoskeleton (Hoboken) 67, 339–345. [DOI] [PubMed] [Google Scholar]
- Tanaka-Takiguchi Y, Kinoshita M, Takiguchi K (2009). Septin-mediated uniform bracing of phospholipid membranes. Curr Biol 19, 140–145. [DOI] [PubMed] [Google Scholar]
- Versele M, Gullbrand B, Shulewitz MJ, Cid VJ, Bahmanyar S, Chen RE, Barth P, Alber T, Thorner J (2004). Protein-protein interactions governing septin heteropentamer assembly and septin filament organization in Saccharomyces cerevisiae. Mol Biol Cell 15, 4568–4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weems A, McMurray M (2017). The step-wise pathway of septin hetero-octamer assembly in budding yeast. Elife 6, e23689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf E, Kim PS, Berger B (1997). MultiCoil: a program for predicting two- and three-stranded coiled coils. Protein Sci 6, 1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CF, Chiou JG, Minakova M, Woods B, Tsygankov D, Zyla TR, Savage NS, Elston TC, Lew DJ (2015). Role of competition between polarity sites in establishing a unique front. Elife 4, e11611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn JY, Friesen H, Kishimoto T, Henne WM, Kurat CF, Ye W, Ceccarelli DF, Sicheri F, Kohlwein SD, McMahon HT, Andrews BJ (2010). Dissecting BAR domain function in the yeast Amphiphysins Rvs161 and Rvs167 during endocytosis. Mol Biol Cell 21, 3054–3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.