Abstract
The inherited chromosomal instability disorder Nijmegen breakage syndrome (NBS) results from truncating mutations in the NBS1 gene, which encodes the protein nibrin. Nibrin is part of a nuclear multiprotein complex that also contains the DNA repair proteins Mre11 and Rad50. Upon irradiation, this complex redistributes within the nucleus, forming distinct foci that have been implicated as sites of DNA repair. In NBS cells, nibrin is absent and Mre11 and Rad50 are cytoplasmic. In this study, the interacting domains on nibrin and Mre11 were mapped using the yeast two-hybrid system and expression of epitope-tagged constructs in NBS fibroblasts. Deletion of the carboxy-terminal 101 amino acids of nibrin eliminated its ability to interact with Mre11 and to complement the radiation sensitivity of NBS cells. However, this truncated form of nibrin could localize to the nucleus and form radiation-inducible foci. Expression of a carboxy-terminal 354-amino-acid fragment of nibrin was sufficient to direct the nuclear localization of nibrin, as well as that of Mre11 and Rad50. Despite providing some partial complementation of the radiation-sensitive phenotype, the nibrin-Mre11-Rad50 complexes in these cells were unable to form foci. These results indicate that nibrin directs not only the nuclear localization of the nibrin-Mre11-Rad50 complexes but also radiation-induced focus formation. However, direct interaction between nibrin and Mre11 is required for normal cellular survival postirradiation. Distinct domains of nibrin are required for each of these functions, focus formation, nuclear localization, and Mre11 interaction.
The autosomal recessive disorder Nijmegen breakage syndrome (NBS) is characterized by microcephaly, growth retardation, borderline mental retardation, humoral and cellular immunodeficiency, chromosomal instability, radiation sensitivity, and an increased incidence of malignancies, particularly those of lymphoid origin (25). NBS cells cultured in vitro are deficient in the response to treatment with DNA double-strand break (DSB)-inducing agents such as ionizing radiation and radiomimetic compounds. These defective responses include reduction in colony-forming ability postirradiation, a failure to inhibit DNA synthesis in response to acute doses of radiation (radioresistant DNA synthesis), and an increased frequency of chromosomal aberrations (14, 24). Positional cloning studies in NBS families and functional complementation studies identified a single gene, NBS1, that is mutated in most patients with NBS (19, 26).
The NBS1 gene is located on human chromosome 8q21 (6, 20, 22, 26) and encodes a ubiquitously expressed protein of 754 amino acids (aa) termed nibrin or p95. All known NBS1 mutations are clustered between nucleotides 657 and 1142 of the gene, and all are predicted to truncate the nibrin protein. Most (90 to 95%) of reported NBS patients are homozygous for one mutation (657del5); no other mutation has been observed in more than one family. Nibrin is not detectable by Western blotting in NBS cell lines, suggesting that most mutations are null (4). However, the production of a truncated protein product containing the amino-terminal end of nibrin cannot be ruled out. This amino-terminal portion of nibrin contains two adjacent and potentially functional domains, a forkhead-associated (FHA) domain (11) and a breast cancer carboxy-terminal (BRCT) domain (2), which have been observed previously in other proteins involved in DNA damage responses or in cell cycle checkpoint control.
In normal fibroblasts, nibrin is localized in the nucleus in association with two additional proteins, Mre11 and Rad50, which participate in DNA DSB repair (4). Reciprocal coimmunoprecipitation experiments indicate a strong physical association between the three proteins (4, 18). Treatment of cells with DSB-inducing agents, such as ionizing radiation, results in a rapid association between Mre11 and damaged DNA within 30 min of irradiation (21). At later times (8 to 12 h) postirradiation, brightly staining foci containing nibrin, Mre11, and Rad50 are apparent in the nuclei of 60 to 90% of exposed fibroblasts. While such foci are also detectable in unirradiated cells, the average number per cell and the frequency of cells with detectable foci increases in response to irradiation (18). The function of these irradiation-induced foci (IRIF) is unknown, but given the early association of Mre11 with DSB's (21), these foci may represent sites of ongoing repair or of unresolved breaks. In NBS cells, which lack nibrin, Mre11 and Rad50 still interact, but complexes containing these two proteins are confined to the cytoplasm and thus cannot form nuclear foci (4).
In this study, we have mapped the sites of interaction between the nibrin and Mre11 proteins in vitro using yeast two-hybrid analysis and in vivo by expression of epitope- tagged constructs and coimmunoprecipitation. The abilities of in vitro-constructed deletion mutants of nibrin to complement the cellular phenotypes of NBS were assessed by transfection of NBS cell lines.
MATERIALS AND METHODS
Cell lines.
The simian virus 40 (SV40)-transformed fibroblast cell lines GM637 (Coriell Institute, Camden, N.J.) and NBS-ILB1 (16) were grown in Dulbecco modified Eagle medium (DMEM; Life Technologies Inc., Rockville, Md.) supplemented with l-glutamine (Life Technologies), 15% fetal calf serum (FCS; HyClone Laboratories Inc., Logan, Utah), penicillin (100 U/ml), and streptomycin (100 μg/ml) (Life Technologies). NBS-ILB1 cells infected with retroviral expression constructs (5) were maintained in the above medium supplemented with G418 (500 μg/ml; Life Technologies). The SV40-transformed fibroblast cell line MRC5 (12) was maintained in DMEM-F12 medium supplemented with 15% FCS, penicillin (100 U/ml), and streptomycin (100 μg/ml). Phoenix A retroviral packaging cells (P. Achacoso et al., unpublished data) were maintained in DMEM supplemented with 10% heat-inactivated FCS, penicillin (100 U/ml), and streptomycin (100 μg/ml). Cells were grown at 37°C in 5% CO2.
Yeast two-hybrid analysis.
The nibrin 2,265-bp coding region and five different 900- to 1,100-bp overlapping subfragments thereof were cloned into the SalI site of plasmid pAS2-1 (Clontech Laboratories Inc., Palo Alto, Calif.) that expresses the DNA-binding domain of the Gal4 transcription factor. Smaller fragments of nibrin were amplified and cloned in a similar manner. Individual cDNA clones of Mre11 and five overlapping subfragments were generated using a full-length IMAGE consortium cDNA clone of human MRE11 (645656; American Type Culture Collection) as template. The amplified Mre11 fragments were cloned into the XhoI site of plasmid pACT2 (Clontech), which expresses the activation domain of the Gal4 transcription factor. Other Mre11 subfragments were amplified and cloned in a similar fashion.
Saccharomyces cerevisiae strain Y190 was contransformed with nibrin and Mre11 expression plasmids. The transformants were selected for growth on synthetic dropout (SD) medium lacking uridine, tryptophan, and leucine. Individual yeast clones selected for carrying both plasmids were tested for histidine prototrophy by plating on SD medium lacking uridine, tryptophan, leucine, histidine, and lysine and supplemented with 30 mM 3-amino-1,2,4-triazole (Sigma Chemical Co., St. Louis, Mo.). Activation of lacZ reporter gene expression was determined by a β-galactosidase (β-Gal) filter lift assay (1) and quantitated by a liquid assay (Pierce Chemical Co., Rockford, Ill.). For the latter assay, independent cotransformants (three to eight colonies) carrying both plasmids were grown overnight in selective liquid medium until log phase. Optical density at 600 nm (OD600) of the test cultures was determined. Then 70 μl of each individual culture was dispensed in duplicate in individual wells of a 96-well plate, and 70 μl of reagent mix containing equal volumes of 2× β-Gal assay buffer, and Y-PER reagent was added. The reaction mixture was incubated at 30°C until the wells turned yellow; the reaction was stopped by addition of 56 μl of 1 M sodium carbonate, and the total reaction time was recorded. The plates were centrifuged and absorbance of the supernatant was measured at 405 nm. β-Gal activity was calculated as 1,000 × A405/(t × V × OD600), where t is time of incubation (in minutes) and V is volume of cells used in the assay (in milliliters). Relative difference in β-Gal activity was determined by dividing β-Gal activity of the test sample by β-Gal activity of plasmid alone. All data presented are averages of at least three independent colonies.
Retroviral gene expression.
The retroviral construct expressing full-length nibrin (NBS1) was previously described (5). For construction of the Nb652 retrovirus, we took advantage of a spontaneous mutation (1958insA) that arises when nibrin-containing plasmids are passaged in Escherichia coli (5). A cDNA clone containing the 1958insA mutation was isolated from an Epstein-Barr virus-transformed B-cell cDNA library cloned in the pBK-CMV vector (Stratagene). The cDNA was subcloned into the BamHI-XhoI sites of the pBS-SK vector (Stratagene) for further manipulations. For in vivo expression, a BamHI-NcoI (bp −62 to 2287) fragment of the 1958insA cDNA was cloned into the HpaI site of the pLXIN retroviral construct (Clontech) upstream of the internal ribosome entry site-neo gene cassette. For construction of nibrin fragment 5 retroviral constructs, NbFR5 and NbFR5.1 fragments were separately subcloned into the pCMV-Tag3 vector (Stratagene, Cedar Creek, Tex.). SalI-ApaI fragments containing the NbFR5 and NbFR5.1 inserts with the Myc tag were blunted and cloned into the HpaI site of the pLXIN retroviral vector. The 1958insA/pLXIN, NbFR5/pLXIN, and NbFR5.1/pLXIN constructs were introduced into NBS-ILB1 cells by retroviral gene transfer as described elsewhere (5). Bulk-transformed cell lines were selected in G418 (1 mg/ml; Life Technologies).
Immunoprecipitation and immunoblotting.
Cell lysates were prepared from 3 × 106 to 7 × 106 cells in cell lysis buffer (50 mM sodium phosphate [pH 7.2], 0.5% Triton X-100, 2 mM EDTA, 2 mM EGTA, 25 mM sodium fluoride, 25 mM glycerophosphate, 2 mM dithiothreitol, protease inhibitor cocktail tablet [Roche Molecular Biochemicals, Indianapolis, Ind.]). Lysates were sonicated for 3 min at 4°C and cleared by centrifugation. To preclear cell lysates, 5 μg of whole mouse (Pierce) or rabbit (Zymed Laboratories Inc., South San Francisco, Calif.) immunoglobulin G (IgG) and 20 μl of GammaBind Plus-Sepharose beads (Amersham Pharmacia Biotech Inc, Piscataway, N.J.) were added to the supernatant, and tubes were rocked for 1 h at 4°C; then the supernatant was removed, and antibody was added. For immunoprecipitation of Myc-tagged proteins, hybridoma supernatant containing anti-Myc tag antibody 9E10 (9) was used. To immunoprecipitate nibrin, rabbit polyclonal antinibrin antibody (Novus Biologicals, Littleton, Colo.) was added at 1:3,000. After incubation on ice for 1 h, Sepharose beads were added and the tubes were rocked for 1 h at 4°C. The beads were washed four times with lysis buffer and resuspended in lysis buffer with loading dye. After boiling for 5 min, 20 μl of each sample was loaded per lane on a discontinuous sodium dodecyl sulfate-polyacrylamide gel. Electrophoresis was carried out in a Bio-Rad mini-PROTEAN II apparatus (Bio-Rad Laboratories, Hercules, Calif.) using 1× protein gel running buffer containing 25 mM Tris, 192 mM glycine, and 0.1% sodium dodecyl sulfate at 100 V for 2 h. Proteins were transferred to Immobilon P membranes (Millipore Corp., Bedford, Mass.) in transfer buffer with 25 mM Tris, 192 mM glycine, and 15% methanol at 30 V overnight at 4°C.
Nibrin, Mre11, and Rad50 proteins were detected by Western blot analysis. Membranes were blocked with Tris-buffered saline (pH 7.6) containing 0.1% Tween 20 and 10% nonfat milk powder for 2 h at room temperature and washed. Nibrin was detected using the rabbit polyclonal antinibrin antiserum (Novus) at 1:10,000. Mre11 protein was detected using anti-Mre11 monoclonal antibody 292D (kindly provided by Tony DeMaggio, ICOS Corp., Bothell, Wash.) at 0.28 mg/ml. A monoclonal anti-Rad50 antibody (Novus) was used at a dilution of 1:500 to detect Rad50. Biotinylated antibody 9E10 to the Myc epitope tag was used to detect NbFR5 and NbFR5-1 constructs tagged with a Myc epitope. Membranes were probed with primary antibody for 1 h at room temperature. After washing, the blots were probed with horseradish peroxidase-conjugated goat anti-mouse IgG or goat anti-rabbit IgG (Pharmingen, San Diego, Calif.) at a dilution of 1:4,000 for 1 h at room temperature. Bitotinylated anti-Myc antibody was detected with streptavidin-horseradish peroxidase conjugate (Amersham Pharmacia Biotech) at a dilution of 1:1,000. Blots were washed and developed using the Amersham enhanced chemiluminescence system.
Immunofluorescence staining.
Immunofluorescence staining was performed as described elsewhere (30), with a few modifications. Fibroblast cell lines were plated at 1 × 105 to 2 × 105 cells per coverslip in glass vials (Viromed Laboratories Inc., Minneapolis, Minn.) and grown for 24 h. The cells were irradiated with 12 Gy from a Mark 1 model 22 cesium source (J. L. Shepherd and Associates, San Fernando, Calif.). The medium was replaced; cells were incubated for an additional 8 h and then fixed and permeabilized in 4% formaldehyde with 0.1% Triton-X 100 for 10 min. After being washed with phosphate-buffered saline (PBS) twice for 5 min each time, coverslips were blocked in PBS with 10% FCS and incubated at 4°C overnight. Cells were washed and incubated with polyclonal nibrin antibody (Novus) at 1:2,000 dilution and monoclonal Mre11 antibody at 0.7 mg/ml for 1 h at room temperature. After being washed four times with PBS, cells were incubated with goat anti-rabbit IgG conjugated to Alexa 568 (Molecular Probes Inc., Eugene, Oreg.) and goat anti-mouse IgG conjugated to Alexa 488 (Molecular Probes), both at 1:150 dilution, for 1 h at room temperature. Coverslips were washed four times with PBS, and nuclei were counterstained with TOTO-3 iodide (Molecular Probes) at 1 μM for 40 min. After a final wash with PBS, cells were mounted in Vectashield mounting medium (Vector Laboratories Inc., Burlingame, Calif.). Immunofluorescence was analyzed using a Nikon fluorescence microscope and a Bio-Rad confocal imaging system at 488, 568, and 647 nm (TOTO-3 staining). For visualizing foci, individual cells were z planed (18 to 20 sections), and images of individual sections were stacked to produce a final image of the cell. The percentage of cells with nuclear foci was determined by counting a minimum of 100 cells using a 100× oil immersion lens of the Nikon fluorescence microscope.
Colony survival assay.
Cells were exposed to 0, 1, 2, or 3 Gy of ionizing radiation from a J. L. Shepherd Mark I model 22 cesium source and plated at 600 cells per 100-mm-diameter tissue culture dish in quadruplicate for each condition. After 10 days at 37°C, tissue culture dishes were washed once with PBS, stained with 1× Coomasie blue stain (Bio-Rad) for 5 min, and washed a final time with PBS. Colonies per plate were enumerated, and the mean ± standard deviation was determined at each dose of radiation. The survival fraction was calculated as the percentage of the unirradiated control. Results were graphed using GraphPad Prism version 3.00 for Windows (GraphPad Software, San Diego, Calif.). All cell lines were tested in two or more independent experiments.
RESULTS
The carboxy-terminal end of nibrin interacts with the amino-terminal region of Mre11 in vitro.
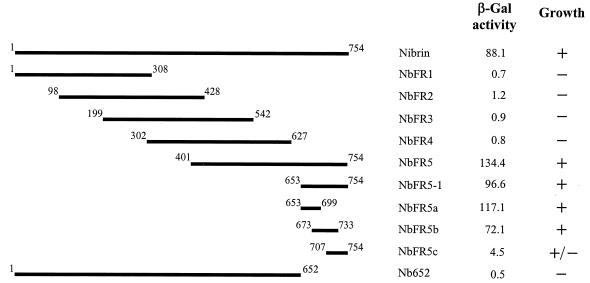
To map regions of nibrin that could potentially interact with Mre11, the full-length NBS1 gene and a series of five subfragments (NbFR1 to -5) overlapping by approximately 300 bp were cloned into plasmid pAS2-1. Yeast strain Y190 was separately cotransformed with a full-length copy of MRE11 cloned into the pACT2 plasmid and each of the individual subclones of NBS1. To assess interaction between nibrin and Mre11, colonies arising from each cotransformation were tested for histidine prototrophy and assayed for β-galactosidase production in a filter assay. β-Gal activity was further quantitated in an o-nitrophenyl-β-d-galactopyranoside assay to provide a relative measure of the strength of interactions between various fragments. There was a strong interaction between full-length nibrin and Mre11 in the yeast two-hybrid analyses as expected (Fig. 1). Of the subfragments of nibrin, only NbFR5, containing the C-terminal 354 amino acids, displayed any significant evidence of interaction with full-length Mre11. Since NbFR4 failed to interact with Mre11, the region of NbFR5 that did not overlap with NbFR4 (aa 653 to 754) and three overlapping subfragments of this region were cloned in pAS2-1 and tested for interaction with Mre11. The C-terminal 101-aa fragment of nibrin, NbFR5-1, interacted strongly with Mre11 (Fig. 1). A reciprocal fragment, Nb652, containing aa 1 to 652 of nibrin displayed no evidence of interaction with Mre11. There was a variable response from the three overlapping subfragments of NbFR5-1, NbFR5a being the strongest, NbFR5b being slightly but not significantly weaker, and NbFR5c showing a significant reduction in β-Gal activity.
FIG. 1.
Interaction of nibrin subfragments with full-length Mre11 in the yeast two-hybrid system. Full-length nibrin and subfragments as indicated in the diagram were expressed as fusion proteins in pAS2-1 and individually tested for interaction with full-length Mre11 expressed in pACT2. Numbers flanking individual fragments indicate amino acid positions. β-Gal activity was measured by liquid assay as described in Materials and Methods. For each combination, three or more independent colonies were tested in duplicate for β-Gal production. The results are normalized to the β-Gal activity of the pAS2-1 vector alone. Growth was assessed by observing the viability of cotransformants on SD medium lacking uracil, lysine, tryptophan, leucine, and histidine and supplemented with 30 mM 3-amino-1,2,4-triazole.
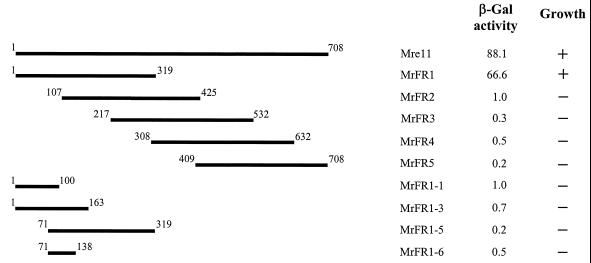
To map the corresponding regions of Mre11 that interact with nibrin, yeast strain Y190 was individually cotransformed with a full-length clone of NBS1 in pAS2-1 and each of five overlapping fragments of Mre11 (MrFR1 to −5) cloned in pACT2. MrFR1 (aa 1 to 319) displayed a significant interaction with full-length nibrin comparable to that observed for full-length Mre11 (Fig. 2). However, it was not possible to further subdivide this fragment and maintain interaction (e.g., fragment MrFR1-1, MrFR1-3, MrFR1-5, or MrFR1-6).
FIG. 2.
Interaction of Mre11 subfragments with full-length nibrin in the yeast two-hybrid system. MRE11 subfragments cloned in pACT2 were tested for interaction with full-length nibrin cloned in pAS2-1. Numbers flanking individual fragments indicate amino acid positions. β-Gal production and growth on selective media were determined as for Fig. 1.
Two-hybrid analysis using full-length interacting partners indicated that aa 653 to 754 of nibrin and 1 to 319 of Mre11 were necessary for interaction. To determine if these regions of nibrin and Mre11 were sufficient to mediate interaction by themselves, subfragments NbFR5-1 (aa 653 to 754) and MrFR1 (aa 1 to 319) were cotransformed into yeast. As expected, the two subfragments interacted with each other (>100-fold excess β-Gal production over vector alone).
Since the stoichiometry of nibrin-Mre11-Rad50 complexes is not known, nibrin and Mre11 were also tested for possible homodimerization in the yeast two-hybrid system. No interaction between full-length copies of nibrin cloned in pAS2-1 and pACT2 was observed. In contrast, interaction between full-length Mre11 molecules in pAS2-1 and pACT2 was observed, indicating that human Mre11 can potentially homodimerize (data not shown).
The carboxy-terminal region of nibrin interacts with Mre11 in vivo.
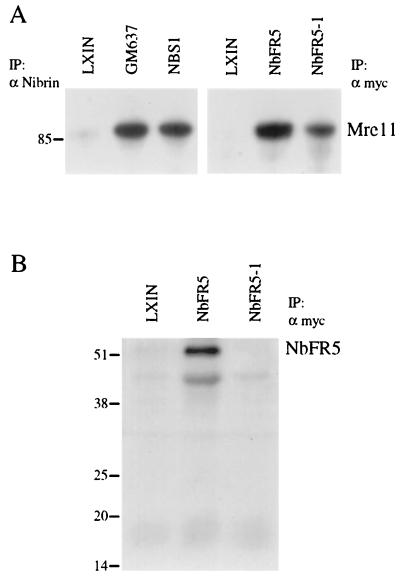
To evaluate whether the C-terminal region of nibrin identified in the yeast two-hybrid screen could interact with Mre11 in vivo, Nb652, NbFR5, and NbFR5-1 were separately cloned in the retroviral expression vector pLXIN. Because NbFR5 and NbFR5-1 were short fragments that might not be detected with antibodies to nibrin, these constructs were tagged with a Myc epitope. Viral supernatants were prepared and used to infect an SV40-transformed fibroblast line from an NBS patient homozygous for the predominant 657del5 mutation that produces no detectable endogenous nibrin protein (NBS-ILB1). For comparison, the full-length NBS1 gene was expressed using the same retroviral vector system. Immunoprecipitation with an antinibrin antibody coprecipitated comparable amounts of the 85-kDa Mre11 protein from a control fibroblast line, GM637, and from NBS1-infected NBS-ILB1 cells but not from cells infected with vector alone (Fig. 3). Cell lysates from NbFR5 and NbFR5-1-infected cells were immunoprecipitated with an antibody to the Myc tag (Fig. 3). NbFR5 was able to interact with and immunoprecipitate the Mre11 protein in the retrovirus-infected NBS-ILB1 cells; however, NbFR5-1 immunoprecipitated significantly less Mre11 (Fig. 3A). Probing with an antibody to the Myc epitope tag revealed the expected 50-kDa band for NbFR5 but not the expected 18-kDa band for NbFR5-1 (Fig. 3B). Similar results were obtained in transient transfections with NbFR5-1, suggesting that this fragment is unstable or rapidly turned over (data not shown).
FIG. 3.
Interaction of NbFR5 and NbFR5-1 with Mre11 by immunoprecipitation and Western blot analysis. (A) Total cell lysates were prepared from normal control cell line GM637 and the NBS-ILB1 cell line separately infected with retrovirus carrying the pLXIN vector alone (LXIN), the NBS1 gene (NBS1), the NbFR5 fragment (NbFR5), or the NbFR5-1 fragment (NbFR5-1). Cell lysates were immunoprecipitated (IP) with an antibody directed against nibrin (α Nibrin) or the Myc epitope tag (α myc) and fractionated on a discontinuous polyacrylamide gel. After electrophoretic transfer to a membrane, the blot was probed with a monoclonal antibody to Mre11. The migration of molecular weight markers is indicated on the left (in kilodaltons). (B) Cell lysates from panel A that were immunoprecipitated with an antibody directed against the Myc epitope tag were fractionated on a protein gel and probed with biotinylated anti-Myc antibody.
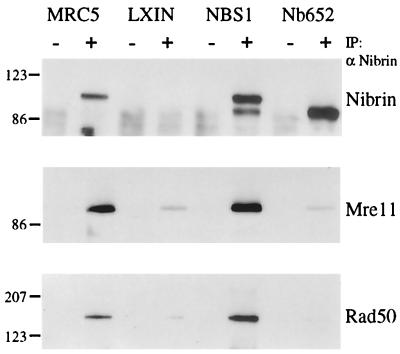
Cell lysates from NBS1 and Nb652 virus-infected cells were immunoprecipitated with an antinibrin antibody, and proteins were detected by Western blot analysis using antibodies against nibrin, Mre11, and Rad50 (Fig. 4). Immunoprecipitation of nibrin from NBS1-infected cells coprecipitated Mre11 and Rad50 in amounts comparable to or greater than that observed in MRC5 cells. In contrast, cells infected with the Nb652 virus produced substantial amounts of the truncated 79-kDa form of nibrin, but antibodies to nibrin did not coprecipitate Mre11 or Rad50 protein above the background level observed in cells infected with pLXIN vector alone.
FIG. 4.
Interaction of the Nb652 mutant with Mre11 and Rad50 by immunoprecipitation and Western blot analysis. Total cell lysates were prepared from a normal control cell line (MRC5) and NBS-ILB1 cells separately infected with the pLXIN vector alone (LXIN), with the NBS1 retrovirus (NBS1), or with the Nb652 retrovirus (Nb652). Lysates were immunoprecipitated (IP) with control rabbit IgG (−) or polyclonal nibrin antiserum (α Nibrin; +). After Western transfer, the blot was probed with the antinibrin antiserum, an Mre11 monoclonal antibody, and a monoclonal anti-Rad50 antibody. The migration of molecular weight markers is indicated on the left (in kilodaltons).
Mre11 binding is not necessary for nuclear focus formation by nibrin.
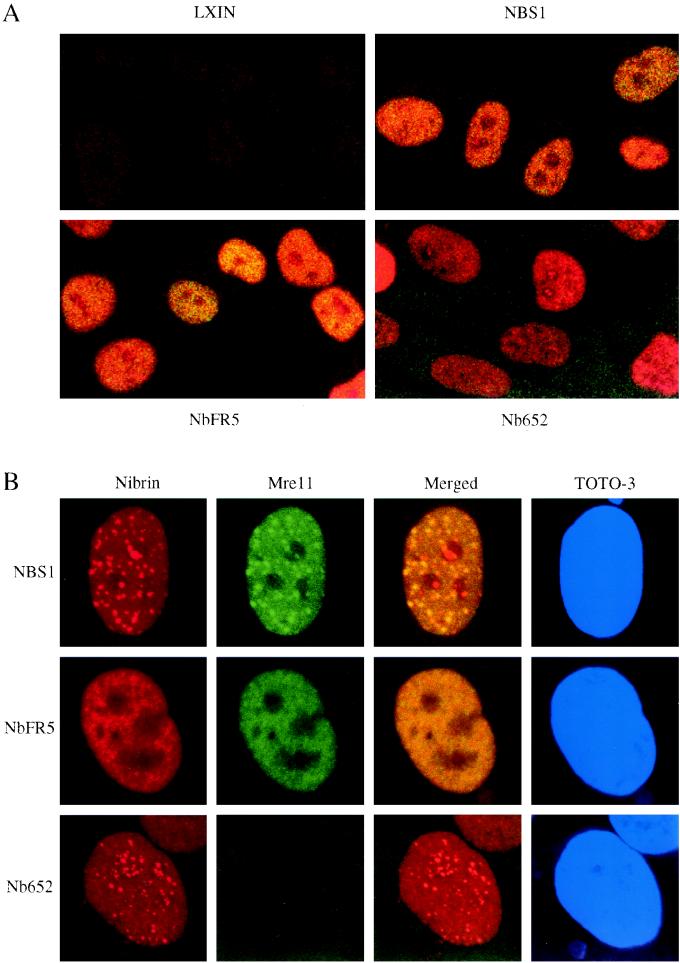
To determine if direct interaction between nibrin and Mre11 is sufficient for nuclear localization of Mre11 and Rad50, as well as for IRIF formation, we assayed the effect of expression of NbFR5 and Nb652 on these phenotypes by staining with antibodies to nibrin and Mre11. In the NBS-ILB1 cell line infected with the NBS1, NbFR5, or Nb652 retrovirus, nibrin expression was exclusively nuclear (Fig. 5A). The NBS1 and NbFR5 constructs, which retain the Mre11 interaction site, were able to relocalize Mre11 to the nucleus, as indicated by the yellow staining in the merged image. In contrast, the Nb652 construct, which lacked the Mre11 binding site, localized to the nucleus, while Mre11 remained in the cytoplasm.
FIG. 5.
Subcellular localization of nibrin and Mre11 detected by immunofluorescence. Cells plated on coverslips were either untreated or exposed to 12 Gy of ionizing radiation. After 8 h, the cells were fixed and stained with antinibrin antiserum (red) and anti-Mre11 monoclonal (green). Merged panels represent overlapped images of nibrin and Mre11 staining results. (A) Localization of nibrin and Mre11 in unirradiated NBS-ILB1 cells expressing vector alone (LXIN), full-length nibrin (NBS1), nibrin fragment 5 (NbFR5), or the truncated 79-kDa form (Nb652). (B) Localization of nibrin and Mre11 in NBS-ILB1 cells expressing full-length nibrin (NBS1), nibrin fragment 5 (NbFR5), or the truncated 79-kDa form (Nb652) after irradiation. Cells were counterstained with the nuclear dye TOTO-3 (blue).
Upon irradiation, the NBS1-infected cell line displayed increased numbers of nuclear foci containing both nibrin and Mre11 (Fig. 5B), as observed in normal cell lines (4, 5, 13, 18). The Nb652-infected NBS-ILB1 cells were able to form nuclear foci containing nibrin, but these foci lacked Mre11, as evidenced by the nonoverlapping staining patterns for nibrin and Mre11 in the merged image (Fig. 5B). These nibrin foci were qualitatively similar to those formed in normal cells by the complex of all three proteins, nibrin, Mre11, and Rad50, and were radiation inducible. Interestingly, the NbFR5 construct, though able to relocalize Mre11 to the nucleus, formed no nuclear foci detectable with antibodies to either nibrin or Mre11 in either irradiated or unirradiated cells (Fig. 5).
The number of cells displaying nuclear foci and the number of foci per cell were quantitated pre- and postirradiation in cells infected with the NBS1, NbFR5, or Nb652 retrovirus (Table 1). NBS-ILB1 cells infected with the NBS1 or Nb652 retrovirus were indistinguishable in terms of number of cells with foci and number of foci per cell whether irradiated or unirradiated. In contrast, no foci were detected in NBS-ILB1 cells infected with the NbFR5 virus.
TABLE 1.
Quantification of nibrin nuclear foci pre- and postirradiationa
| Virus | Irradiation (Gy) | % of cells with focib | No. of foci/celle (mean ± SD) |
|---|---|---|---|
| NBS1 | 0 | 30 | 19.7 ± 7.6 |
| 12 | 64 | 27.6 ± 11.9 | |
| NbFR5 | 0 | <1 | 0 |
| 12 | <1 | 0 | |
| Nb652 | 0 | 31 | 20.9 ± 5.9 |
| 12 | 65 | 27.8 ± 6.9 |
NBS-ILB1 cells infected with the NBS1, NbFR5, or Nb652 retrovirus were either unirradiated or irradiated with 12 Gy of ionizing radiation and analyzed for focus formation 8 h postirradiation.
Cells with >5 foci were considered positive. A minimum of 100 cells were counted for each condition.
Determined from counting of a minimum of 25 cells.
Radiation sensitivity of NBS cells expressing NbFR5 or Nb652.
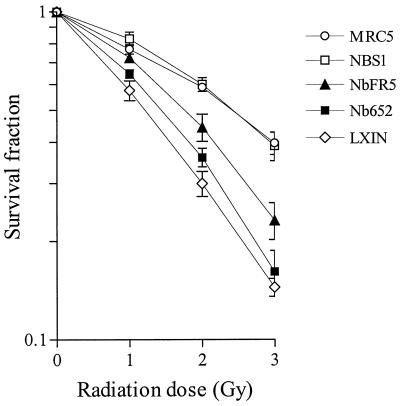
Cells from NBS patients display hypersensitivity to ionizing radiation that can be fully complemented by the introduction of a functional copy of the NBS1 gene (5, 13). To evaluate the role of Mre11-nibrin interaction in radiation sensitivity, we irradiated NBS fibroblasts expressing Nb652 or NbFR5 with increasing doses of X rays and determined their survival relative to normal control cells (MRC5), NBS1-infected cells, and cells infected with empty vector (LXIN) in a standard colony-forming assay. Neither Nb652 nor NbFR5 could fully complement the radiation hypersensitivity of NBS-ILB1 cells to levels observed with NBS1 or normal controls (Fig. 6). However, in multiple assays we consistently observed that NBS cells expressing NbFR5 were less radiation sensitive than cells expressing Nb652 or pLXIN at all doses tested.
FIG. 6.
Radiation sensitivity of NBS-ILB1 cells expressing different nibrin constructs. A normal control cell line (MRC5) and NBS-ILB1 cell line infected with the pLXIN vector (LXIN), NBS1, NbFR5, or Nb652 retrovirus were exposed to 0, 1, 2, or 3 Gy of ionizing radiation. After 10 days, colonies per plate were counted and expressed as the percentage of the unirradiated control. Each data point represents the mean and standard deviation of quadruplicate values.
DISCUSSION
In normal fibroblasts, nibrin is distributed relatively homogeneously in the nucleus in association with the proteins Mre11 and Rad50 (4). Upon irradiation of fibroblasts, this multiprotein complex redistributes, resulting in a high percentage of cells displaying distinct nuclear foci detectable with antibodies to any of the three components of the complex. Other proteins, including BRCA1 and ATM, are likely to be transiently associated with this complex, possibly in a cell type or cell cycle-specific manner (10, 17, 28, 29). In NBS cells, where nibrin is absent, the Mre11 and Rad50 proteins complex with each other but remain cytoplasmic. Based on this phenotype of NBS cells, nibrin was assumed to be necessary for the nuclear localization of Mre11 and Rad50 (4). What the role of nibrin is in the formation of nuclear foci and whether direct or indirect interactions with Mre11 or Rad50 are required for either nuclear localization or focus formation are unclear. In this study, we have mapped the corresponding interaction sites on both nibrin and Mre11 and examined the role of direct nibrin-Mre11 interaction in nuclear localization, focus formation after irradiation, and radiation sensitivity by mutagenesis and expression of nibrin constructs.
Mre11 and nibrin interact strongly in a yeast two-hybrid screen, consistent with the ease with which they can be coimmunoprecipitated from human fibroblasts. Fine mapping of the interaction domains revealed that the C-terminal 101 aa of nibrin were sufficient for this interaction. Two of three subfragments of this 101-aa region displayed some degree of interaction, suggesting that nibrin-Mre11 binding occurs over a broad surface, the primary sequence of which is highly conserved between the mouse and human proteins (27).
Localization of the site of nibrin interaction on Mre11 was more difficult. Yeast two-hybrid screening of fragments of Mre11 localized this site to the amino-terminal 319 aa. However, none of a panel of subfragments from this region revealed any evidence of interaction with nibrin when expressed individually. This N-terminal localization is consistent with a report that a mutation of N to S at position 117 in Mre11, found in two families with an ataxia-telangiectasia-like disorder (ATLD), weakens the interaction between Mre11 and nibrin as assayed by coimmunoprecipitation (23). There are several possible explanations for the difficulty in defining a smaller interaction domain on Mre11 by truncation. The bacterial homologue of Mre11, SbcD, binds Mn2+, and this binding has substantial global effects on the structure of SbcD (8). If the N-terminal region of Mre11 is similarly involved in binding metal ions, then disruption of this binding may have broad effects on the structure of Mre11. Mutational analyses also indicate that Rad50 binding is dependent on several distinct sites in this region of Mre11 (3). Binding of Rad50 may be required for stabilization of Mre11 structure and/or to facilitate nibrin binding.
Yeast two-hybrid analyses also suggested the potential for homodimerization of Mre11 but not nibrin. These results are of interest in terms of defining the stoichiometry of the complex formed by nibrin, Mre11, and Rad50 but should be considered with caution since they could reflect the detection of intramolecular rather than intermolecular interaction sites. Similar results have been described for Mre11 in S. cerevisiae (7, 15).
In this study, a deletion mutant of nibrin lacking the Mre11 interaction domain was constructed and introduced into NBS cells. This mutant form of nibrin, Nb652, failed to interact with Mre11 in vivo and did not restore the nuclear localization of either Mre11 or Rad50. Nb652 did form nuclear foci that, although lacking Mre11 and Rad50, were normal in morphology, frequency, and response to ionizing radiation. However, this ability to form nibrin foci did not result in any significant complementation of the radiation-sensitive phenotype of these cells. These results indicate that nibrin is capable of directing its own nuclear localization and focus formation independent of its interaction with Mre11.
Interestingly, the phenotype of cells expressing Nb652 is distinct from that of cells from ATLD patients with the Mre11 N117S mutation (23). The Mre11 N117S mutation is proposed to weaken the interaction between nibrin and Mre11, based on the reduced amount of nibrin that could be coimmunoprecipitated from patient cell lines with antibodies to Mre11. Neither nibrin nor Mre11 display their characteristic nuclear localization in these cells. A major difference between cells expressing the Nb652 form of nibrin and those expressing the N117S form of Mre11 is that the nibrin truncation completely eliminates interaction between nibrin and Mre11, while the Mre11 N117S mutation retains the interaction but weakens or alters it in some way. The contrasting phenotypes of these mutations with regard to nibrin localization and focus formation suggest that the N117S form of Mre11 may exert some negative effect on nibrin in ATLD cells. For example, the N117S mutated Mre11 might bind to nibrin in an altered conformation that would block access to a nuclear localization signal or to an interaction site for a third protein necessary for transposition to, or retention within, the nucleus.
Both the NbFR5 and Nb652 expression constructs used in this study localized to the nucleus despite deletions that eliminated potential protein-protein interaction sites at the N- and C-terminal ends of the protein. This suggests that residues in the overlapping region between these constructs are sufficient to mediate nuclear localization of nibrin. Within this region, residues 401 to 652, there are two potential nuclear localization signals, both of which are conserved in the mouse homologue of nibrin and one of which is also conserved in the Drosophila homologue.
Are there distinct functions related to DNA repair or cell survival that are attributable to nibrin, or does it simply act as a molecular chaperone, delivering Mre11 and Rad50 to the nucleus? All reported mutations in NBS patients are predicted to prematurely terminate the nibrin protein, and full-length nibrin is not detected in cell lines from these patients by Western blotting (4). These mutations provide no information that might help to partition the cellular phenotypes associated with NBS into those that might be specific to nibrin and others that might be dependent on its interaction with Mre11. In this study, expression of NbFR5 in NBS-ILB1 cells restored the interaction between nibrin and Mre11, allowing nuclear localization of these proteins but not the formation of nuclear foci. These results indicate that sequences in the amino-terminal half of nibrin are required for focus formation. Expression of NbFR5 also failed to fully complement the radiation sensitivity phenotype of NBS cells. The level of radiation sensitivity in NBS cells expressing NbFR5 is comparable to that observed when the same retroviral system was used to express an altered form of nibrin, S343A, in which serine 343, a target of the ATM kinase, had been changed to alanine by site-specific mutagenesis (10). NbFR5 lacks this phosphorylation site, which may explain in part its lack of full complementation. The S343A mutant restores focus formation whereas NbFR5 does not, indicating a requirement for sequences other than just S343 in the amino-terminal half of nibrin for this function.
In summary, direct physical interaction with Mre11 is required for normal cellular survival after radiation exposure but is dispensable for correct nuclear localization and for formation of IRIF by nibrin. Our results define several distinct functional domains within nibrin. Translocation of Mre11 and Rad50 to the nucleus is necessary but not sufficient for focus formation, which is dependent on residues in the amino-terminal half of nibrin. The N-terminal FHA and BRCT domains of nibrin are obvious candidates for this function. Sequences between amino acid residues 401 and 652 of nibrin are sufficient to direct its nuclear localization independent of its interaction with Mre11. Finally, the C-terminal 101 aa of nibrin are necessary for Mre11 binding and for cellular survival after radiation exposure. However, full complementation of radiation sensitivity in NBS cells requires additional amino-terminal sequences of nibrin and/or phosphorylation on multiple serine residues.
ACKNOWLEDGMENTS
This work was supported by grant CA57569 from the National Cancer Institute to P.C. and by an A-T Medical Research Foundation fellowship to A.D.-M.
We thank Tony DeMaggio for the kind gift of the Mre11 monoclonal antibody, Malgorzata Zdzienicka for the NBS-ILB1 fibroblast cell line, Tiong Chia Yeo for isolating the full-length NBS1 cDNA, and Lindsey Johnson for nucleotide sequencing.
REFERENCES
- 1.Bartel P L, Fields S. Analyzing protein-protein interactions using two-hybrid system. Methods Enzymol. 1995;254:241–263. doi: 10.1016/0076-6879(95)54018-0. [DOI] [PubMed] [Google Scholar]
- 2.Bork P, Hofmann K, Bucher P, Neuwald A F, Altschul S F, Koonin E V. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J. 1997;11:68–76. [PubMed] [Google Scholar]
- 3.Bressan D A, Olivares H A, Nelms B E, Petrini J H. Alteration of N-terminal phosphoesterase signature motifs inactivates Saccharomyces cerevisiae Mre11. Genetics. 1998;150:591–600. doi: 10.1093/genetics/150.2.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carney J P, Maser R S, Olivares H, Davis E M, Le Beau M, Yates III J R, Hays L, Morgan W F, Petrini J H. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- 5.Cerosaletti K M, Desai-Mehta A, Yeo T C, Kraakman-van der Zwet M, Zdzienicka M Z, Concannon P. Retroviral expression of the NBS1 gene in cultured Nijmegen breakage syndrome cells restores normal radiation sensitivity and nuclear focus formation. Mutagenesis. 2000;15:281–286. doi: 10.1093/mutage/15.3.281. [DOI] [PubMed] [Google Scholar]
- 6.Cerosaletti K M, Lange E, Stringham H M, Weemaes C M, Smeets D, Solder B, Belohradsky B H, Taylor A M, Karnes P, Elliott A, Komatsu K, Gatti R A, Boehnke M, Concannon P. Fine localization of the Nijmegen breakage syndrome gene to 8q21: evidence for a common founder haplotype. Am J Hum Genet. 1998;63:125–134. doi: 10.1086/301927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chamankhah M, Xiao W. Formation of the yeast Mre11-Rad50-Xrs2 complex is correlated with DNA repair and telomere maintenance. Nucleic Acids Res. 1999;27:2072–2079. doi: 10.1093/nar/27.10.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Connelly J C, de Leau E S, Okely E A, Leach D R. Overexpression, purification, and characterization of the SbcCD protein from Escherichia coli. J Biol Chem. 1997;272:19819–19826. doi: 10.1074/jbc.272.32.19819. [DOI] [PubMed] [Google Scholar]
- 9.Evan G I, Lewis G K, Ramsay G, Bishop J M. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gatei M, Young D, Cerosaletti K M, Desai-Mehta A, Spring K, Kozlov S, Lavin M F, Gatti R A, Concannon P, Khanna K. ATM-dependent phosphorylation of nibrin in response to radiation exposure. Nat Genet. 2000;25:115–119. doi: 10.1038/75508. [DOI] [PubMed] [Google Scholar]
- 11.Hofmann K, Bucher P. The FHA domain: a putative nuclear signalling domain found in protein kinases and transcription factors. Trends Biochem Sci. 1995;20:347–349. doi: 10.1016/s0968-0004(00)89072-6. [DOI] [PubMed] [Google Scholar]
- 12.Huschtscha L I, Holliday R. Limited and unlimited growth of SV40-transformed cells from human diploid MRC-5 fibroblasts. J Cell Sci. 1983;63:77–99. doi: 10.1242/jcs.63.1.77. [DOI] [PubMed] [Google Scholar]
- 13.Ito A, Tauchi H, Kobayashi J, Morishima K, Nakamura A, Hirokawa Y, Matsuura S, Ito K, Komatsu K. Expression of full-length NBS1 protein restores normal radiation responses in cells from Nijmegen breakage syndrome patients. Biochem Biophys Res Commun. 1999;265:716–721. doi: 10.1006/bbrc.1999.1737. [DOI] [PubMed] [Google Scholar]
- 14.Jaspers N G, Taalman R D, Baan C. Patients with an inherited syndrome characterized by immunodeficiency, microcephaly, and chromosomal instability: genetic relationship to ataxia telangiectasia. Am J Hum Genet. 1988;42:66–73. [PMC free article] [PubMed] [Google Scholar]
- 15.Johzuka K, Ogawa H. Interaction of Mre11 and Rad50: two proteins required for DNA repair and meiosis-specific double-strand break formation in Saccharomyces cerevisiae. Genetics. 1995;139:1521–1532. doi: 10.1093/genetics/139.4.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kraakman-van der Zwet M, Overkamp W J, Friedl A A, Klein B, Verhaegh G W, Jaspers N G, Midro A T, Eckardt-Schupp F, Lohman P H, Zdzienicka M Z. Immortalization and characterization of Nijmegen breakage syndrome fibroblasts. Mutat Res. 1999;434:17–27. doi: 10.1016/s0921-8777(99)00009-9. [DOI] [PubMed] [Google Scholar]
- 17.Lim D S, Kim S T, Xu B, Maser R S, Lin J, Petrini J H, Kastan M B. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature. 2000;404:613–617. doi: 10.1038/35007091. [DOI] [PubMed] [Google Scholar]
- 18.Maser R S, Monsen K J, Nelms B E, Petrini J H. hMre11 and hRad50 nuclear foci are induced during the normal cellular response to DNA double-strand breaks. Mol Cell Biol. 1997;17:6087–6096. doi: 10.1128/mcb.17.10.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsuura S, Tauchi H, Nakamura A, Kondo N, Sakamoto S, Endo S, Smeets D, Solder B, Belohradsky B H, Der K, Oshimura V M, Isomura M, Nakamura Y, Komatsu K. Positional cloning of the gene for Nijmegen breakage syndrome. Nat Genet. 1998;19:179–181. doi: 10.1038/549. [DOI] [PubMed] [Google Scholar]
- 20.Matsuura S, Weemaes C, Smeets D, Takami H, Kondo N, Sakamoto S, Yano N, Nakamura A, Tauchi H, Endo S, Oshimura M, Komatsu K. Genetic mapping using microcell-medicated chromosome transfer suggests a locus for Nijmegen breakage syndrome at chromosome 8q21–24. Am J Hum Genet. 1997;60:1487–1494. doi: 10.1086/515461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nelms B E, Maser R S, MacKay J F, Lagally M G, Petrini J H. In situ visualization of DNA double-strand break repair in human fibroblasts. Science. 1998;280:590–592. doi: 10.1126/science.280.5363.590. [DOI] [PubMed] [Google Scholar]
- 22.Saar K, Chrzanowska K H, Stumm M, Jung M, Nurnberg G, Wienker T F, Seemanova E, Wegner R D, Reis A, Sperling K. The gene for the ataxia-telangiectasia variant, Nijmegen breakage syndrome, maps to a 1-cM interval on chromosome 8q21. Am J Hum Genet. 1997;60:605–610. [PMC free article] [PubMed] [Google Scholar]
- 23.Stewart G S, Maser R S, Stankovic T, Bressan D A, Kaplan M I, Jaspers N G, Raams A, Byrd P J, Petrini J H, Taylor A M. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell. 1999;99:577–587. doi: 10.1016/s0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- 24.Taalman R D, Hustinx T W, Weemaes C M, Seemanova E, Schmidt A, Passarge E, Scheres J M. Further delineation of the Nijmegen breakage syndrome. Am J Med Genet. 1989;32:425–431. doi: 10.1002/ajmg.1320320332. [DOI] [PubMed] [Google Scholar]
- 25.van der Burgt I, Chrzanowska K H, Smeets D, Weemaes C. Nijmegen breakage syndrome. J Med Genet. 1996;33:153–156. doi: 10.1136/jmg.33.2.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Varon R, Vissinga C, Platzer M, Cerosaletti K M, Chrzanowska K H, Saar K, Beckmann G, Seemanova E, Cooper P R, Nowak N J, Stumm M, Weemaes C M, Gatti R A, Wilson R K, Digweed M, Rosenthal A, Sperling K, Concannon P, Reis A. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell. 1998;93:467–476. doi: 10.1016/s0092-8674(00)81174-5. [DOI] [PubMed] [Google Scholar]
- 27.Vissinga C S, Yeo T C, Woessner J, Massa H F, Wilson R K, Trask B J, Concannon P. Identification, characterization, and mapping of a mouse homolog of the gene mutated in Nijmegen breakage syndrome. Cytogenet Cell Genet. 1999;87:80–84. doi: 10.1159/000015396. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y, Cortez D, Yazdi P, Neff N, Elledge S J, Qin J. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000;14:927–939. [PMC free article] [PubMed] [Google Scholar]
- 29.Zhong Q, Chen C F, Li S, Chen Y, Wang C C, Xiao J, Chen P L, Sharp Z D, Lee W H. Association of BRCA1 with the hRad50-hMre11–p95 complex and the DNA damage response. Science. 1999;285:747–750. doi: 10.1126/science.285.5428.747. [DOI] [PubMed] [Google Scholar]
- 30.Ziv Y, Bar-Shira A, Pecker I, Russell P, Jorgensen T J, Tsarfati I, Shiloh Y. Recombinant ATM protein complements the cellular A-T phenotype. Oncogene. 1997;15:159–167. doi: 10.1038/sj.onc.1201319. [DOI] [PubMed] [Google Scholar]