Abstract
Melanoma is the most lethal skin cancer that originates from the malignant transformation of melanocytes. Although melanoma has long been regarded as a cancerous malignancy with few therapeutic options, increased biological understanding and unprecedented innovations in therapies targeting mutated driver genes and immune checkpoints have substantially improved the prognosis of patients. However, the low response rate and inevitable occurrence of resistance to currently available targeted therapies have posed the obstacle in the path of melanoma management to obtain further amelioration. Therefore, it is necessary to understand the mechanisms underlying melanoma pathogenesis more comprehensively, which might lead to more substantial progress in therapeutic approaches and expand clinical options for melanoma therapy. In this review, we firstly make a brief introduction to melanoma epidemiology, clinical subtypes, risk factors, and current therapies. Then, the signal pathways orchestrating melanoma pathogenesis, including genetic mutations, key transcriptional regulators, epigenetic dysregulations, metabolic reprogramming, crucial metastasis-related signals, tumor-promoting inflammatory pathways, and pro-angiogenic factors, have been systemically reviewed and discussed. Subsequently, we outline current progresses in therapies targeting mutated driver genes and immune checkpoints, as well as the mechanisms underlying the treatment resistance. Finally, the prospects and challenges in the development of melanoma therapy, especially immunotherapy and related ongoing clinical trials, are summarized and discussed.
Subject terms: Skin cancer, Drug development
Introduction
Melanoma is the most lethal type of skin cancer that originates from the malignant transformation of melanocytes. Actually, melanocytes are of neuroectodermal origin and then migrate extensively to reside throughout the body, including skin, uveal, mucosa, inner ear, and rectum, displaying as highly dendritic cells to manufacture melanin to defend against photodamage.1 Due to the relatively wide distribution of melanocytes, melanoma can occur ubiquitously regardless of the anatomical location or the types of organs and tissues. Worldwide, the incidence of melanoma occupies around 1.7% of all newly-diagnosed primary malignant cancers, and patients dying from melanoma account for nearly 0.7% of all cancer mortality.2 Of note, the incidence and mortality of melanoma vary among different countries, being relatively high in Australia, New Zealand, Europe, and Northern America, and lowest in Africa. The discrepancy is associated with ethnicity, lifestyle, and genetic background.2 The prevalent subtypes and pathogenesis of melanoma in different populations are distinct. White populations with fair skin mainly suffer from cutaneous melanoma, of which the etiology is largely attributed to ultraviolet (UV) exposure. However, pigmented populations from Asia and Africa mainly develop acral and mucosal melanomas at relative lower incidence rates. Trauma and chronic inflammation have been documented as risk factors of acral melanoma, which is supported by the reports that the occurrence of acral melanoma is frequently at the lesions with trauma, infection, and chronic ulcer.3 In 2021, there are estimated 106,110 cases of melanoma emerging and 7180 deaths arising from this disease in the United States. Although the mortality rate of melanoma patients is prominently reduced in the past few decades due to early diagnosis, proper screening approaches, improved surgery principle and revolutionary advances of targeted therapy and immunotherapy, the prognosis of patients, in particular those with distant metastasis, remains unoptimistic with the 5-year survival rate around 27%.4
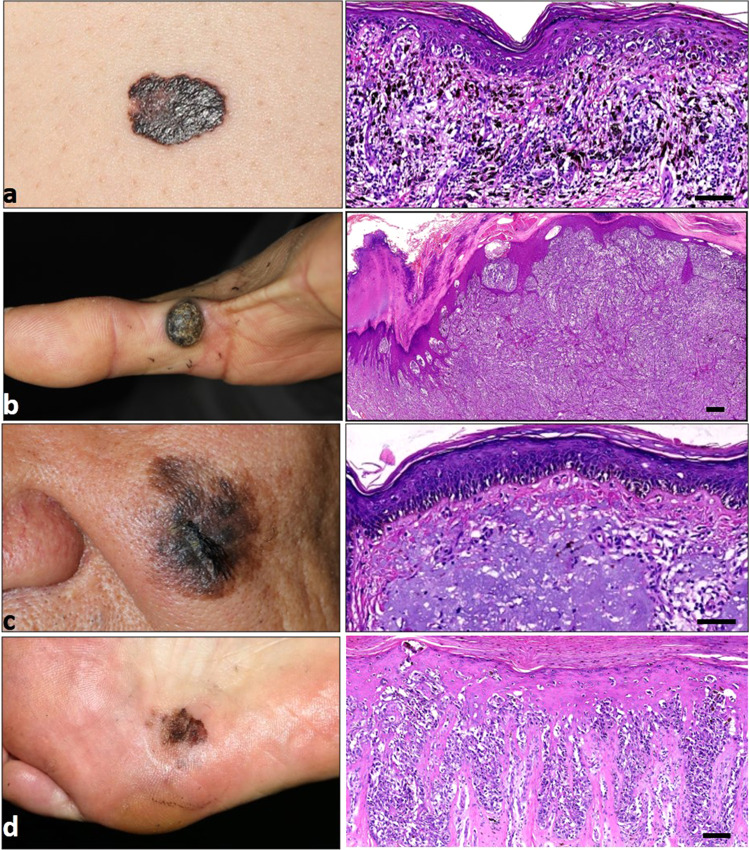
Clinically, melanoma cases are stratified as the following major subtypes according to their histopathological characteristics, namely, superficial spreading melanoma, nodular melanoma, lentigo maligna melanoma, and acral lentiginous melanoma. To be specific, superficial spreading melanoma usually refers to melanoma in a radial or horizontal growth phase with tumor cells distributing as a nest or solitary units displaying in a pagetoid pattern, whereas nodular melanoma generally occurs in the vertical growth phase. In addition, lentigo maligna melanoma has a significant sign of chronic UV radiation (UVR), and has cells individually distributing alongside the dermal–epidermal junction and skin appendages. Moreover, acral lentiginous melanoma histologically presents as tumor cells in single units along the dermal–epidermal junction and as confluent foci, and commonly occurs at acral sites5 (Fig. 1). There are also some other subtype variants defined by clinical or histological characteristics including ocular melanoma, mucosal melanoma, acral melanoma, spitzoid melanoma, and desmoplastic melanoma.
Fig. 1.
Clinical and corresponding histopathological images of melanomas. Generally, melanomas are classified into four main types according to the histopathological characteristics, namely, superficial spreading melanoma (a), nodular melanoma (b), lentigo maligna melanoma (c), and acral lentiginous melanoma (d). The corresponding histopathological image of the same patient is displayed on the right. Scale bar = 100 μm
The risk factors of melanoma supported by strong epidemiologic evidence include UVR, multiple moles, family history (a family history/personal history of melanoma), and fair skin, eye, and hair.1 Epidemiologic studies by systemic review and meta-analysis have unveiled that intense intermittent UVR exposure through either sunburns or indoor tanning for those before 35 years old, even during childhood, confers the highest risk.6 Intense UVR can induce genetic alterations like DNA damage and genetic mutation, reactive oxygen species (ROS) accumulation, and oxidative stress, as well as inflammatory responses involving macrophages and neutrophils infiltration, which are related to malignant switch of melanocytes.7–10 Moreover, the phenotypic characteristics of fair skin, eye, and hair indicate the insufficient ability of melanocytes to generate eumelanin that have stronger photoprotective capacity than pheomelanin, rendering individuals more vulnerable to sun exposure and other environmental stress on the skin.11 Of note, UV exposure accounts for two distinct etiological mechanisms for melanoma pathogenesis.12 On one hand, early sun exposure and proneness to nevi tend to induce melanoma carcinogenesis driven by BRAF mutation. Patients with these risk factors are usually diagnosed at a young age and generally display superficially spreading melanoma on the trunk. On the other, chronic sun exposure often leads to melanoma harboring NRAS mutation, without any involvement of nevi proneness. Apart from environmental UVR and some phenotypic characteristics in individuals that are associated with the carcinogenesis of cutaneous melanoma, some other factors are documented to be associated with non-cutaneous melanoma, especially trauma and chronic inflammation for acral melanoma. A previous study conducted by our group that involves 685 Chinese patients with melanoma has revealed a prominent correlation between acral melanoma and the history of trauma on the lesion.13 In addition, some reports also indicated the generation of acral melanoma after trauma or from lesions of infection and chronic ulcer.14–16 The pro-tumorigenic effect of trauma and chronic inflammation might result from the increased cytokines and ROS that can induce genetic instability or activate oncogenic pathways in melanocytes.17
The therapeutic approaches of melanoma undergo a dramatic evolution in the past few decades due to the progress in the understanding of melanoma pathogenesis and thereby revolutionary advances of targeted therapies that specifically intervene mutant driver genes and immune checkpoints. Historically, there were only dacarbazine chemotherapy and high-dose interleukin-2 (IL-2) approved by Food and Drug Administration (FDA) as treatment agents for metastatic melanoma before 2010. Interferon-α2b (IFN-α2b) was also employed as adjuvant agent, whereas the usage was largely limited due to the frequent occurrence of severe adverse effects.1,18 Since ten years ago, a series of therapeutic agents and combinatorial approaches have been approved by FDA, including immunotherapy (single-agent ipilimumab, nivolumab, pembrolizumab, and combination of ipilimumab and nivolumab), targeted therapy (single-agent vemurafenib and dabrafenib, combinations of dabrafenib plus trametinib, vemurafenib plus cobimetinib, and encorafenib plus binimetinib) as well as one intralesional modified oncolytic herpes virus talimogene laherparepvec (T-VEC) (Fig. 2 and Table 1).1,18–21 The above-mentioned therapeutic approaches have gained evident and encouraging responses in treating patients with advanced melanoma and some of them have also been approved in the adjuvant setting. Compared to 10 years ago, the 5-year survival has gained considerable improvement from <5% to around 30% in patients with advanced melanoma who accept the combination of BRAF inhibitor and MEK inhibitor or single anti-PD-1 antibody.21–23 Although current therapies have revolutionized the standard of management for patients with advanced melanomas, low response rate and inevitable occurrence of treatment resistance retard forward improvement of therapeutic outcome.24 Therefore, it is necessary to understand the molecular mechanisms underlying melanoma pathogenesis more comprehensively, which might lead to the innovations of more applicable therapeutic approaches and provide additional clinical options for melanoma therapy.
Fig. 2.
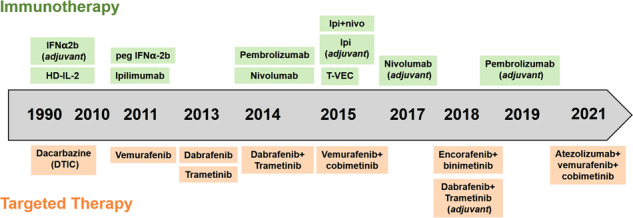
Timeline for FDA-approved therapies for metastatic melanoma. HD high-dose, Ipi Ipilimumab, T-VEC talimogene laherparepvec
Table 1.
FDA-approved therapies for melanoma
| Agent | Mechanism | Type | Indications | Time |
|---|---|---|---|---|
| Dacarbazine | Alkylating agent | Chemotherapy | Metastatic melanoma | 1975 |
| IL-2 | Cytokine | Immunotherapy | Metastatic melanoma | 1998 |
| IFN-alfa-2b | Cytokine | Immunotherapy | Adjuvant therapy for high risk of recurrent melanoma | 1995 |
| Pegylated IFN-alfa-2b | Cytokine | Immunotherapy | Resected stage III melanoma | 2011 |
| T-VEC | Oncolytic virus | Immunotherapy | For the local treatment of unresectable cutaneous, subcutaneous, and nodal lesions in patients with melanoma recurrent after initial surgery | 2015 |
| Vemurafenib | BRAF inhibitor | Targeted therapy | Unresectable/metastatic melanoma with BRAF V600E/K mutation | 2011 |
| Cobimetinib | MEK inhibitor | Targeted therapy | Unresectable/metastatic melanoma with BRAF V600E/K mutation | 2015 |
| Dabrafenib | BRAF inhibitor | Targeted therapy | Unresectable/metastatic melanoma with BRAF V600E/K mutation | 2013 |
| Trametinib | MEK inhibitor | Targeted therapy | Unresectable/metastatic melanoma with BRAF V600E/K mutation | 2013 |
| Dabrafenib + tremetinib | BRAF inhibitor + MEK inhibitor | Targeted therapy |
Unresectable/metastatic melanoma with BRAF V600E/K mutation Adjuvant treatment of patients with melanoma with BRAF V600E/K mutation |
2015 |
| Vemurafenib + cobimetinib | BRAF inhibitor + MEK inhibitor | Targeted therapy | Unresectable/metastatic melanoma with BRAF V600E/K mutation | 2015 |
| Encorafenib + binimetinib | BRAF inhibitor + MEK inhibitor | Targeted therapy | Unresectable/metastatic melanoma with BRAF V600E/K mutation | 2018 |
| Ipilimumab | Anti-CTLA-4 antibody | Immunotherapy |
Unresectable/metastatic melanoma Adjuvant treatment of resected stage III melanoma |
2011 |
| Nivolumab | Anti-PD-1 antibody | Immunotherapy |
Unresectable/metastatic melanoma Adjuvant treatment of resected stage III melanoma |
2014 |
| Pembrolizumab | Anti-PD-1 antibody | Immunotherapy |
Unresectable/metastatic melanoma Adjuvant treatment of resected stage III melanoma |
2014 |
| Nivolumab + ipilimuab | Anti-CTLA-4 antibody + anti-PD-1 antibody | Immunotherapy | Unresectable/metastatic melanoma | 2015 |
| Atezolumab + vemurafenib + cobimetinib | Anti-PD-L1 antibody + BRAF inhibior + MEK inhibitor | Combination therapy | Unresectable or metastatic melanoma with BRAF V600E/K mutation | 2020 |
In this review, we systemically summarized the signal pathways driving melanoma pathogenesis, including the main mutated driver genes and signals, key transcriptional factors and downstream molecular biology, epigenetic modification, metabolic reprogramming, crucial metastasis-related signals, and tumor-promoting inflammatory pathways and pro-angiogenic factors. Then, current progress in therapies targeting mutant driver genes and immune checkpoints, and novel combined therapeutic approaches were introduced, with some discussions about the mechanism underlying treatment resistance. Finally, the ongoing clinical trials and future perspectives of clinical advances were concluded.
Signal pathways driving melanoma pathogenesis
Mutated driver genes and downstream signal pathways
In the past few decades, the methods to dig into the genomic alterations in driving melanoma carcinogenesis have evolved and helped to gain more and more encouraging insights. Multiple mutated driver genes have been identified and are enriched in various signal pathways that are pivotal contributors to melanoma carcinogenesis and development, including mitogen-activated protein kinase (MAPK) pathway, protein kinase B (AKT) pathway, cell-cycle regulation pathway, pigmentation-related pathway, p53 pathway, epigenetic factors, and some other pathways.
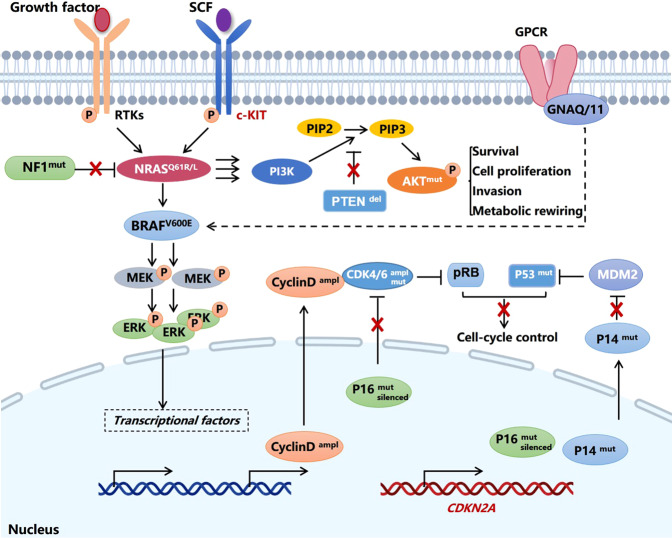
MAPK pathway is frequently activated in cancer to enable tumor cell rapid proliferation. In response to extracellular binding of growth factors to receptor tyrosine kinases (RTKs), intracellular sequential activation of Ras, Raf, MEK, and ERK occurs to regulate a plenty of oncogenic biological activities. For melanoma, mutations in the key signal components, including BRAF, NRAS, NF1, and KIT, are responsible for the hyper-activation of the MAPK pathway. BRAF is a serine/threonine kinase belonging to Raf family and implicated in the signal transduction in the MAPK pathway. In 2002, a genome-wide screening discovered a point mutation of BRAF occurring more frequently in melanoma than other types of solid tumors. The substitution from valine to glutamic acid at codon 600 (V600E), which leads to constitutive activation of the kinase activity of BRAF protein and downstream MAPK pathway, can be detectable in ~50% of melanomas.25 Other variants including V600K, V600D, and V600R occupy around 12%, 5%, and 1% of BRAF mutations, respectively.26,27 The presence of BRAF mutations has great potential in predicting an unfavorable prognosis in melanoma patients.28,29 In other subtypes of melanoma like acral melanoma and mucosal melanoma, the incidence of BRAF mutation is around 20% and 6%, which is much lower than that in cutaneous melanoma.30,31 NRAS is a small GTP-binding protein belonging to Ras family and transduces upstream RTK activation to promote the activity of downstream Raf. The mutations of NRAS usually occur at G12, G13, and Q61 sites, and are found in around 25% of cases of melanomas.32 Compared to BRAFV600E mutation which can be effectively targeted by some agents like vemurafenib and dabrafenib, therapeutic options for melanoma harboring NRAS mutations lag behind. A recent study has identified STK19 as a novel NRAS activator by enhancing its phosphorylation and binding to downstream effectors. Therefore, the blockade of STK19 kinase activity might be a promising strategy to treat melanomas harboring NRAS mutations.33 Moreover, through the technology of whole-exome sequencing, NF1, that encodes a negative regulator of RAS, has been identified as the third most frequently mutated gene in melanoma after BRAF and NRAS.34,35 The loss of NF1 or inactivating NF1 mutation are present in 46% of melanomas expressing wild-type BRAF and RAS, which leads to constitute activation of Ras by lessening its intrinsic GTPase activity and induces hyper-activation of MAPK pathway.36 What’s more, KIT mutation and amplification, that cause receptor dimerization, auto-phosphorylation of tyrosine residues, and the activation of downstream oncogenic pathways, are mainly found in mucosal and acral melanomas (10–20% of these types).37
Uncontrolled cell-cycle progression is a hallmark characteristic of melanoma development, and multiple components in this process are found to be mutated, including cyclin-dependent kinase inhibitor 2A (CDKN2A), retinoblastoma-associated protein (RB), Cyclin D1, and cyclin-dependent kinase 4/6 (CDK4/6). To be specific, the risk loci in CDKN2A is identified in about 40% of familial melanomas. Physiologically, CDKN2A encodes two proteins p16Ink4a and p14Arf through distinct translational programs. Wild-type p16Ink4a restrains cell-cycle progression by abrogating cyclin-dependent kinase 4 (CDK4) or CDK6-mediated phosphorylation and inactivation of RB. In addition, wild-type p14Arf can prevent E3 ubiquitin ligase MDM2-mediated degradation of p53 to control cell cycle. Therefore, germline mutation of CDKN2A induces the functional deficiency of both p16Ink4a and p14Arf, which leads to uncontrolled cell-cycle progression by impairing the function of downstream RB1 and p53, respectively (Fig. 3). Besides, the aberrations of CCND1 and CDK4/6 pathways have been documented to occur more frequently in acral melanoma than cutaneous melanoma, with the incidence being around 40% and 80%, respectively.38,39 The amplification rates of CDK4 and CCND1 are also frequently found in mucosal melanoma, with the incidence being 47.0% and 27.7%, respectively.40 Therefore, targeting the CDK4/6 pathway by specific inhibitor is hopeful to bring encouraging outcome for the treatment of acral melanoma and mucosal melanoma.
Fig. 3.
Mutated driver genes and downstream signal pathways in melanoma. Ampl amplification, CDK cyclin-dependent kinase, Del deletion, GPCR G protein-coupled receptor, Mut mutation, P (in a pink circle) phosphate, p14ARF and p16INK4A splice variant encoded by CDKN2A gene, PIP2 phosphatidylinositol-(4,5)-bisphosphate, PIP3 phosphatidylinositol-(3,4,5)-trisphosphate, PTEN phosphatidylinositol‑3,4,5‑trisphosphate 3‑phosphatase, and dual-specificity protein phosphatase, RB retinoblastoma-associated protein, RTK receptor tyrosine kinase, SCF stem cell factor
The dysregulated activation of the AKT pathway occurs in around 70% of total melanomas, which is the result of AKT3 amplification and PTEN loss by epigenetic silencing or deletion as previously described.41,42 Generally, the activation of the AKT pathway is initiated by activated phosphoinositide 3-kinase (PI3K) after the stimulation by exogenous growth factors, followed by increased generation of the second messenger phosphatidylinositol-3,4,5-trisphosphate (PIP3) that can promote the translocation of AKT to the plasma membrane for its subsequent phosphorylation and activation. Since that intracellular level of PIP3 is negatively regulated by the phosphatase PTEN, the functional deficiency of PTEN can induce the upregulation of PIP3 level and promote AKT activation.43 It has been documented that the predominant AKT isoform in melanoma is AKT3. SiRNA transfection targeting AKT3 or overexpression of PTEN can effectively suppress AKT3 activity to reduce the tumorigenic potential of melanoma cells.42 Therefore, the hyper-activation of AKT pathway is a pivotal oncogenic event for melanoma carcinogenesis and development.
As for sporadic cases of melanomas that account for around 90% of total melanomas, they are mostly driven by the mutations in genes implicated in pigmentation process like MC1R (encoding melanocortin-1 receptor), TYR (encoding tyrosinase), TYRP1 (encoding tyrosinase-related protein-1), PAX3 (encoding paired box 3), EDNRB (encoding endothelin receptor type B), ASIP (encoding agouti signaling protein), OCA2 (encoding oculocutaneous albinism II), SLC45A2 (encoding solute carrier family 45 member 2) and SOX10 (encoding SRY-box transcription factor 10), which dictates the causal relationship between UV radiation and increased risk of melanoma.44–46 In response to the stimulation of α‑melanocyte-stimulating hormone (α-MSH), MC1R can be activated to transduce downstream signals to induce the expression of microphthalmia-associated transcription factor (MITF), a master regulator of the generation of melanin.47 TYR and TYRP1 are the main downstream targets of MITF implicated in melanin production, whereas SOX10 and PAX3 are a canonical melanocytic lineage-specific transcriptional factor of MITF.48 The functional deficiency of the above-mentioned genes causes unbalanced production of UV‑protective eumelanin and less-protective pheomelanin in melanocytes,49,50 thus contributing to melanoma carcinogenesis in highly-risky individuals encountering environmental insults.
Apart from MAPK pathway, cell-cycle regulation pathway, AKT pathway, and pigmentation-related pathway, genetic mutations in some other pathways are also implicated in the carcinogenesis of melanoma. The activating mutations in NOTCH2, CTNNB1 can lead to aberrant activation of Notch and Wnt pathway respectively to facilitate the pathogenesis of melanomas of different subtypes, including cutaneous, acral, and mucosal melanomas.51–53 In addition, GNAQ/GNA11 mutations are the major genetic drivers in uveal melanoma with the incidence of 80–90%,54,55 which contribute to the hyper-activation of downstream MAPK pathway. What’s more, mutations in TP53, ARID1B, ARID2, and TERT and the amplification of MDM2 have also been identified in melanoma, which are involved in the regulation of p53 pathway, SWI/SNF chromatin remodeling complex, and telomerase activity respectively.31,56–59
Taken together, the above-mentioned high-frequency mutations depict the framework of the melanoma mutational landscape, and more importantly, provide multiple druggable targets for precisely intervening specific signaling pathways. Although the mutational landscape in cutaneous melanoma has been extensively investigated, the genetic aberrations of acral melanoma and mucosal melanoma are far from understood. Further investigations are needed to clarify the mutational landscape of non-cutaneous melanoma in the future.
Key transcriptional signal pathways implicated in melanoma development
Melanoma originates from epidermal melanocytes and shares many molecular similarities with melanocyte precursors, indicating that the developmental program of melanocytes, especially the transcriptional regulation program, is utilized by melanoma cells to facilitate tumor progression.60 In fact, multiple key transcriptional factors and signal pathways responsible for the formation of melanocytic lineage, including SOX10, MITF, Notch, and Wnt-β-catenin, are greatly implicated in the malignant characteristics of melanoma cells.
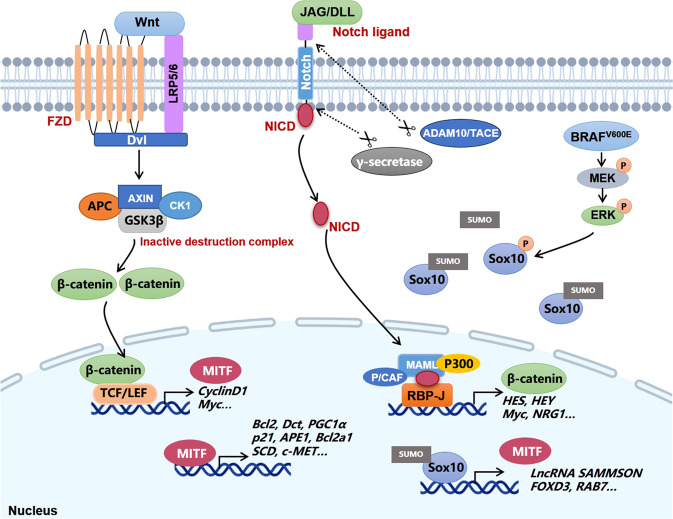
SOX10 is a neural crest transcription factor essential for the initiation and development of Schwann cells and melanocytes. It has been identified as a specific marker with relatively high sensitivity and specificity for the diagnosis of melanocytic and Schwannian tumors, including metastatic melanoma occurring in sentinel lymph nodes.61–63 Preliminary studies have demonstrated that haplo-insufficiency of SOX10 inhibits mutant NRAS-driven formation of congenital nevus and melanoma in transgenic mice model. Genetic knockdown of SOX10 expression robustly abolishes the proliferative and migratory capacity of melanoma cells in vitro and the growth of melanoma in vivo, implicating the fundamental role played by SOX10 in maintaining melanoma cell survival.64–66 As a transcriptional factor, SOX10 activates various targets like MITF, long non-coding RNA (lncRNA) SAMMSON, forkhead box D3 (FOXD3), and RAB7 to regulate cell proliferation, mitochondrial function, and endolysosomal pathway, therefore affecting various biological activities in melanoma67–70 (Fig. 4). Meanwhile, SOX10 expression is under the control of multiple species-conserved regulatory sequences in the upstream region of its encoding gene that can be bound by several other transcriptional factors.71 Besides, multiple post-translational modification paradigms like sumoylation, ubiquitination, and phosphorylation also participate in the regulation of SOX10 transcriptional activity and protein stability.67,72–74 In a previous study on the mechanism underlying vemurafenib resistance, using a chromatin-regulator-focused shRNA library, the authors identified the loss of SOX10 expression as a crucial cause of the resistance to targeted therapy via the activation of transforming growth factor-β (TGF-β) signaling and subsequent upregulation of epithelial growth factor receptor (EGFR) and platelet-derived growth factor receptor β (PDGFRβ), linking SOX10 to a slow-growth resistance phenotype.75,76 However, two recent reports have demonstrated that the depletion of SOX10 expression or its downstream SAMMSON sensitizes BRAF-mutant melanoma to MAPK-inhibition agents,67,77 supporting that SOX10 upregulation contrarily triggers the resistance to targeted therapy. The above-mentioned paradoxical role of SOX10 in the resistance to MAPK inhibition-targeted therapy can be associated with the different characteristics between adaptive resistance and acquired resistance occurring at different phases after treatment. To be specific, during the early phase of inhibitor treatment, adaptive response would be readjusted to enable tumor cell survival, and increased transcriptional activity of SOX10 is responsible for the upregulation of two cyto-protective factors FOXD3 and SAMMSON, rendering adaptive resistance. Nevertheless, mutational acquired resistance will prevail after a long-term inhibitor treatment, with the upregulation of multiple receptor tyrosine kinases (RTKs) greatly involved in. The downregulation of SOX10 predominantly tends to induce the senescent phenotype and compensatory reactivation of receptor tyrosine kinases in this phase, rather than to suppress tumor cell survival.76–78 Therefore, the biological effect of SOX10 in melanoma targeted therapy is context-dependent, raising the notion that the intervention of SOX10 expression for overcoming resistance to targeted therapy should take the phases and paradigms of drug resistance into consideration.
Fig. 4.
Key transcriptional factors and signal pathways in melanoma. ADAM10 ADAM metallopeptidase domain 10, APC adenomatosis polyposis coli, DLL delta-like canonical Notch ligand, DVL Disheveled segment polarity protein, FZD frizzled class receptor, GSK3β Glycogen synthase kinase 3β, JAG jagged canonical Notch ligand, LEF-1 lymphoid enhancer binding factor 1, LRP5/6 low density lipoprotein receptor-related protein 5/6, MAML mastermind-like transcriptional coactivator, NICD Notch intracellular domain, P/CAF P300/CBP-associated factor, RBPJ recombination signal binding protein for immunoglobulin κ J region, TACE TNFα-converting enzyme
MITF is an extensively studied melanocytic lineage-specific transcriptional factor, which coordinates many signal pathways of melanoma biology including cell proliferation, survival, metastasis, metabolism, phenotypic plasticity, antitumor immunity, and therapeutic resistance.79 Initial investigations regarding the role of MITF in melanoma cell proliferation had obtained confusing conclusions,80,81 which were then largely reconciled by the “rheostat model” proposed by Carreira et al. To be specific, low MITF expression is associated with slow-cycling state characterized by the upregulation of cell-cycle inhibitor and the potentiation of invasive and metastatic capacity. In contrast, moderate MITF expression induces phenotype switch from invasive to proliferative state, endowing melanoma cells with a stronger growth advantage. However, when MITF expression is further upregulated, a differentiation-associated G1 arrest would be re-established.82 This model suggests that the biological function of MITF is highly correlated with its activity level. Since that MITF is an integrated transcriptional factor governing the expressions of a plenty of target genes implicated in cell-cycle progression (CDK2, CCND1, p21, p16, and p27), cell differentiation (TYR, TYPR1, DCT, RAB27, MYO5a), and invasive capacity (GMPR, DIAPH1),79 the cellular phenotype regulated by MITF is possibly attributed to the mixture of dysregulated molecules involved in distinct biological aspects. The transcription of genes responsible for cell-cycle arrest and invasion predominates when MITF is expressed at a low level, whereas moderate to high MITF expression mainly potentiates downstream targets that facilitate cell proliferation and de-differentiation. Different from the paradoxical role of MITF in cell proliferation, many reports have provided compelling evidence to support the essential role of MITF in melanoma cell survival. Through the regulation of a series of transcriptional targets like Bcl2, Bcl2a1, ML-IAP, HIF1α, c-MET, APE1, p21, and BRAC1, MITF plays its pro-survival role under normal or stressful conditions by regulating oxidative stress, cell senescence, DNA damage repair, and oncogenic pathways (Fig. 4).83–88 While the correlation between low MITF expression and high invasive capacity of melanoma cell has long been observed, the underlying mechanism is only recently elucidated by Bianchi-Smiraglia et al. Their study has disclosed that MITF suppresses the invasive capacity by reducing intracellular GTP pools and subsequent amounts of active (GTP-bound) RAC1, RHO-A, and RHO-C, mainly via the transcriptional regulation of guanosine monophosphate reductase (GMPR).89 In addition, downregulation of MITF mediates the facilitative role of signal transducer and activator of transcription 3 (STAT3) in melanoma metastasis under the transcriptional control of cAMP-response element-binding protein (CREB),90 further confirming the enhanced invasive phenotype in MITFlow melanomas. The regulation of melanoma cell phenotypic plasticity by MITF is also attributed to the alteration of cell metabolism. MITF is recently discovered to be the lineage-restricted transcriptional activator of the key lipogenic enzyme stearoyl-CoA desaturase (SCD). By promoting the conversion from saturated fatty acids to mono-unsaturated fatty acids, SCD upregulation is required for highly-expressed MITF-driven melanoma cell proliferation. In contrast, the suppression of SCD in MITFlow melanoma cells accentuates the invasive and metastatic capacity by activating inflammation-related signaling and induces de-differentiation state.91 In addition to the above-mentioned biological effects, MITF expression can also dictate the sensitivity of BRAF-mutant melanoma to MAPK- inhibition-targeted therapy, though the role is paradoxical in terms of the phase of drug resistance establishment. While low MITF expression is highly associated with intrinsic/acquired resistance to targeted therapy and especially renders increased resistance in aged microenvironment,92,93 the upregulation of MITF is considered as a protective factor to defend against BRAF inhibitor-induced cell apoptosis and mediates adaptive resistance within a relative short duration after treatment.78,94 Based on this, a drug-repositioning screening has identified the HIV1-protease inhibitor nelfinavir as a promising drug to overcome the adaptive resistance to MAPK pathway inhibitors via the downregulation of MITF expression.95 Therefore, the intervention of MITF expression to modulate the outcome of targeted therapy should also take the phase of drug resistance establishment into account.
Notch signal pathway has been documented as a cardinal signal pathway implicated in stem cell self-renewal, cell differentiation, and cell fate decisions in many organs.96 The oral administration of a γ-secretase inhibitor (GSI) that can block Notch signal in mice has been reported to impair hair pigmentation, even 20 weeks after discontinuing the treatment, indicating that Notch is essential for maintaining the homeostasis of melanocyte.96 Compared with normal melanocytes, the expression and activity of Notch are significantly higher in melanoma. Melanocytes transfected with truncated Notch transgene construct (N(IC)) containing enhanced Notch activity display augmented cell proliferation and malignant characteristics similar to melanoma.97 The upregulation of Notch in melanoma is the result of both intracellular AKT pathway activation and the presence of tumor microenvironment factors like hypoxia.98 As a cardinal driver of cancer pathogenesis, Notch can exert its oncogenic role in melanoma via the activation of MAPK pathway, the upregulation of N-cadherin expression, and the potentiation of β-catenin signaling,99,100 indicating that it acts as a nexus node coordinating multiple carcinogenic signals. Supplementary to intrinsic effect on tumor cell behavior, Notch activation is also highly associated with angiogenesis, and participates in the crosstalk between tumor cells and endothelial cells to facilitate tumor migration as well.101,102 In particular, sustained Notch1 activation leads to the senescence of endothelial cells and increase of vascular cell-adhesion molecule 1 (VCAM1) expression, thereby contributing to neutrophil infiltration, tumor cell adhesion to endothelium, intravasation, lung colonization, and postsurgical metastasis.103 Moreover, a recent study demonstrates that the activation of Notch renders the resistance of BRAF-mutant melanoma cells to MEK inhibitors like Cobimetinib.104 Therefore, targeting Notch can be exploited to not only restrain the progression of melanoma, but also increase the efficacy of targeted therapy.
Wnt signal is an evolutionarily conserved pathway implicated in embryonic development, tissue regeneration, and cell homeostasis. Previously, the notification of the importance of Wnt in melanocyte development comes from the observation of the absence of melanoblasts in Wnt-deficient mice.105 Moreover, the activation of β-catenin downstream Wnt greatly contributes to the cell fate decision from the loss of glial derivatives to the expansion of melanocytes, further supporting the fundamental role of Wnt-β-catenin in melanocyte development. Wnt signaling has two main types of pathways, namely, β-catenin-dependent pathway (canonical) and β-catenin-independent pathway (non-canonical), with their function in melanoma emphasizing cell proliferation and cell polarity/migration respectively.106 Takeda et al. has firstly reported that MITF is activated by canonical Wnt-β-catenin signaling via LEF-1107 (Fig. 4), which is then proved to be required for the pro-proliferative effect of Wnt in melanoma.108 Additional evidence supports the conclusion that the oncogenic role of the Wnt-β-catenin pathway is mediated by the suppression of p16 and the overcome of oncogene-induced senescence. Therefore, co-operation of NRAS mutation and Wnt-β-catenin activation can lead to melanoma formation with high penetrance and short latency in mice.109 Later on, the role of Wnt in melanoma metastasis receives more attention. Damsky et al. has provided evidence that β-catenin is the central mediator of tumor metastasis to lymph node and lung in established melanoma transgenic mice model induced by both BRAF mutation and PTEN deficiency.110 Moreover, Wnt3a derived from tumor-infiltrating fibroblast leads to β-catenin activation in melanoma cell, which diminishes tumor cell adhesion and enhances migration to form liver metastasis.111 Alternative Wnt ligands like Wnt5a derived from myeloid-derived suppressor cells (MDSC) and Wnt11 also participate in melanoma metastasis.112,113 However, the exact role of Wnt-β-catenin signal in melanoma development remains controversial even with extensive investigations, namely, β-catenin cannot be fully defined as an oncogene according to available results.114–116 In particular, nuclear β-catenin expression has been previously unveiled to be downregulated during melanoma progression.117,118 Moreover, a recent study has proved that temporal activation of Wnt/β-catenin signal is sufficient to suppress SOX10 expression via the proteasome degradation and therefore blocks the growth of melanoma.119 These reports indicate the tumor-suppressive role of Wnt/β-catenin in melanoma. Therefore, more investigations are needed to forwardly clarify the exact role of Wnt in different contexts, distinct biological activities and heterogeneous genetic backgrounds in melanoma.
In aggregate, the above-mentioned key transcriptional programs driven by SOX10, MITF, Notch, and Wnt-β-catenin signals that share molecular similarities with melanocyte precursors contribute greatly to the malignant switch from melanocyte to melanoma. Of note, these factors that dictate the plasticity and differentiation state of melanoma cells are also decisive for the efficacy of targeted therapy. To overcome the de-differentiation characteristic of tumor cell might be a useful strategy to improve the outcome of patients that receive MAPK inhibition-targeted therapy.120
Epigenetic alterations and the downstream signal pathways
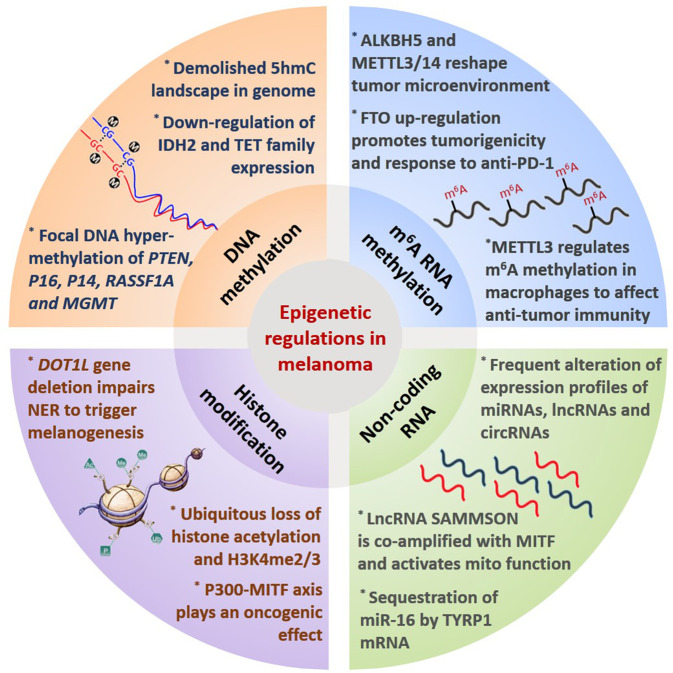
Aside from genetic and transcriptional modulation, epigenetics is emerging as another crucial regulatory paradigm of melanoma biology and signal pathways. Epigenetic modification refers to heritable changes in gene expression without an alteration in the genome sequence, of which the core mechanism is the covalent modifications of either histone tails or nucleosome complexes that can re-shape chromatin structure and modulate gene expression.121 DNA methylation, histone modification, non-coding RNA, and newly-discovered N6-methyladenosine (m6A) RNA methylation are the main types of epigenetic modification, and their dysregulations are highly correlated with melanoma development.122
As the most intensively investigated epigenetic modification in cancer,122 DNA methylation involves the addition of a methyl group to the 5 position of cytosine by DNA methyltransferase to form 5-methylcytosine (5-mC). This process is dynamically regulated by DNA methyltransferases (DNMTs) that transfer the methyl group to the cytosine residue and ten-eleven translocation (TET) family that indirectly promotes DNA de-methylation via the oxidative catalysis of 5-mC to form 5-hmC.123–125 Lian et al. has discovered that loss of 5-hmC triggered by the downregulation of isocitrate dehydrogenase 2 (IDH2) and TET family members is an epigenetic hallmark of melanoma progression, with potent diagnostic and prognostic implications. Re-establishment of 5-hmC landscape is capable of suppressing melanoma growth and improving the survival of the preclinical mice model.126 In addition to the general downregulation of 5-hmC content, focal DNA hyper-methylation of the promoters of some specific tumor suppressors has also been well illustrated in melanoma, in particular PTEN, P16INK4A, P14ARF, RASSF1A, and MGMT (occurring in ~60%, 30%, 80%, 55%, and 30% of melanomas, respectively)127–134 (Fig. 5), which is associated with functional deficiency of these genes during melanoma progression. Besides, a series of alternative genes have been identified to be differentially-methylated at the promoter region between melanoma and benign nevi by genome-wide promoter methylation analysis.135–137 These genes are generally enriched in signal pathways orchestrating cell differentiation, immune-related function, epithelial-to-mesenchymal transition, PI3K/mTOR signaling, metastasis, and cellular metabolism that are all hallmark characteristics of cancer biology.138,139
Fig. 5.
Epigenetic regulation in melanoma. Main paradigms of epigenetic modification and representative effects on cancer biology in melanoma
Chromatin is dynamically switched between two states, namely, dense heterochromatin state of deficient transcriptional activity and relaxed euchromatin state with active transcriptional capacity.140 Histone modifications refer to a class of post-translational modifications (PTMs) typically occurring on the N-terminal “tails” of histones, which can shape the structure of chromatin and thereby modulate the accessibility of DNA for gene transcription or DNA damage repair.141,142 The histone PTMs typically include acetylation, methylation, phosphorylation, and ubiquitination.143 To be specific, histone acetylation is defined by the addition of acetyl-CoA to lysine residues that can be reversely regulated by histone acetyltransferases (HATs) and histone deacetylases (HDACs).144 Besides, histone methylation frequently occurs at lysine or arginine residues at specific sites on histones H3 and H4 that can also be dynamically regulated by lysine-specific demethylase (LSD) and multiple methyltransferases.145,146 Recently, systematic epigenomic profiling of 35 epigenetic modifications and transcriptomic analysis has revealed the ubiquitous loss of histone acetylations and H3K4me2/3 on regulatory regions proximal to specific cancer-regulatory genes in tumor-driving pathways in melanoma (Fig. 5). In parallel, the expressions of HATs and HDACs in tumorigenic transition are also ubiquitously dysregulated.147 On this basis, the restoration of histone acetylation with HDAC inhibitors has prominent tumor suppressor potential,147,148 and can also synergistically increase the efficacy of radiotherapy, MAPK pathway-targeted therapy, and immunotherapy via the regulation of DNA damage repair, intracellular ROS generation, and programmed death ligand-1(PD-L1) expression, respectively.149–152 Some novel selective inhibitors of HDAC families have been developed and exploited in first-in-human study to testify the therapeutic effect on patients with refractory solid tumors including melanoma.153 Aside from HDAC, investigations regarding histone acetyltransferases demonstrate the pivotal role of P300 in melanoma development. Selective pharmacological inhibition of P300 displays a prominent tumor-suppressive effect.154 The expression of MITF can be a promising predictor of the therapeutic vulnerability to P300 inhibition155,156 (Fig. 5). Of note, the activation of P300 also contributes to the resistance to BRAF-targeted therapy by activating ERK signal and mitochondrial oxidative phosphorylation,157,158 highlighting targeting P300 as a valuable synergistic therapeutic approach to sensitize melanoma cells toward MAPK pathway inhibition.
Supplementary to histone acetylation, histone methylation also impacts chromatin condensation and gene transcription to regulate melanoma biology,145 with some enzyme families like SET domain bifurcated 1 (SETDB1), disruptor of telomeric silencing 1-like proteins (Dot1L), enhancer of Zeste homolog 2 (EZH2) and LSD1 implicated in. SETDB1 is responsible for the methylation of histone H3 on lysine 9 (H3K9) and is recurrently amplified in melanoma to play an oncogenic role.159,160 Metabolic reprogramming endows melanoma cells with increased histone H3 trimethylation and paralleled higher metastatic capacity that can be reversed by the pharmacological inhibition of SETDB1.161 Different from SETDB1, DOT1L gene is located in a frequently deleted region and undergoes somatic mutation that compromises its methyltransferase enzyme activity which leads to reduced H3K79 methylation. The loss of function of Dot1L accelerates UVR-triggered melanoma development by impairing the recruitment of nucleotide excision repair (NER) machinery XPC and hindering DNA damage repair162 (Fig. 5), indicating that Dot1L is a protector against melanomagenesis. Moreover, the expression of another histone methyltransferases EZH2 is upregulated in melanoma, which promotes H3K27 trimethylation to silence multiple tumor suppressors.163,164 Deficiency of cilium construction and peroxisome proliferator-activated receptor γ coactivator-1 (PGC1α) resulting from EZH2 upregulation significantly activates both Wnt/β-catenin and Yes-associated protein (YAP) signals to drive melanoma metastasis.165,166 Notably, the dysregulation of EZH2 is also greatly implicated in the resistance to immunotherapy by downregulating antigen presentation, IFN-γ gene signature, and lymphocytes infiltration,167–169 implying that EZH2 might be versatile player in melanoma pathogenesis. What’s more, it has been reported that two different types of H3K9 demethylases, LSD1 and JMJD2C, abolish oncogenic RAS- or BRAF-induced senescence by promoting the expression of E2F target genes, cooperatively driving melanomagenesis. Specific inhibition of highly-expressed H3K9-active demethylases restores oncogene-induced senescence and suppresses melanoma development.170 In addition, the ablation of LSD1 enhances tumor immunogenicity via the upregulation of endogenous retroviral element (ERV) transcripts and the downregulation of RNA-induced silencing complex. This mechanism is also responsible for the diminished resistance to anti-PD-1 immunotherapy in melanoma, suggesting LSD1 as a promising immunotherapy target.171 It seems that H3K9 methylation regulated by SETDB1, LSD1, and JMJD2C plays contrary roles in melanoma carcinogenesis and development respectively. While the restoration of H3K9 methylation by targeting LSD1 and JMJD2C abrogates the transition from melanocyte to melanoma triggered by driver mutations, increased H3K9 methylation induced by SETDB1 displays an oncogenic effect in an established tumor. What’s more, the loss of H3K27 trimethylation due to DOT1L gene deletion also contributes to UVR-triggered melanomagenesis. Therefore, these reports highly indicate the bi-modal role of histone methylation during the whole process of melanoma pathogenesis, namely, being suppressive for carcinogenesis whereas facilitative for progression. Taken together, histone modification coordinates various hallmark characteristics of cancer including cell metabolism, genome instability, and immune evasion in melanoma, and histone methylation-targeted therapy should be based on the progression stage.
In addition to DNA methylation and histone modification, non-coding RNA and m6A RNA methylation are additional pivotal paradigms of epigenetic modification. The expression profiles of microRNAs, lncRNAs, and circRNAs in melanoma have been extensively investigated and are found to be related to multiple cancer characteristics like metastasis, cellular metabolism, migration and invasion, and antitumor immunity.172–180 Investigations of miRNAs in melanoma initially focused on their roles in hallmark characteristics of cancer biology.181–184 Then, more and more attentions were paid to their roles in the tumor microenvironment including angiogenesis, metastatic niche formation, and T cell dysfunction.182,185,186 It should be noted that the biological functions of some non-coding RNAs are of high specificity in melanocytic lineage. For example, Gilot et al. has demonstrated that the sequestration of miR-16 by the mRNA of melanocyte specifically-expressed TYRP1 can promote tumor growth by relieving the suppression of downstream tumor-promoting factors like RAB17, highlighting miRNA displacement as a promising therapeutic approach.187 The crosstalk between melanocytic lineage-specific factor and non-coding RNA is further extended by the investigation conducted by Leucci et al., which demonstrates that recently annotated lncRNA SAMMSON is co-amplified with MITF and plays an oncogenic role. As a transcriptional target of SOX10, the expression of SAMMSON is detectable in more than 90% of melanomas. The knockdown of SAMMSON disrupts mitochondrial functions by targeting downstream p32 and can thereby increase the efficacy of MAPK inhibition-targeted therapy69 (Fig. 5). These reports emphasize that the function of non-coding RNAs can be exerted in a cancer-type-specific manner. The current progress of lncRNAs in melanoma pathogenesis is summarized in Table 2. Apart from non-coding RNA, m6A RNA methylation is another crucial chemical modification discovered in mRNA and non-coding RNA in eukaryotic cells,188,189 the process of which is dynamically regulated by a series of “writers”, “readers”, and “erasers”.190 In melanoma, the expression of m6A demethylase FTO is significantly upregulated to contribute to not only tumorigenicity but also increased response to anti-PD-1 blockade by orchestrating the expressions of PD-1, CXCR4, and SOX10.191 Other reports also provide evidence that m6A demethylase alkylation repair homolog 5 (ALKBH5) and methyltransferases METTL3/14 regulate the response to anti-PD-1 blockade by re-shaping tumor microenvironment through the regulation of metabolism and chemokine secretion.192,193 The role of m6A methylation in tumor-infiltrating macrophages and related impact on tumor progression have also been elucidated in melanoma. Loss of METTL3 in myeloid cells can prominently impair YTH N6-methyladenosine RNA binding protein-1 (YTHDF1)-mediated translation of sprouty related EVH1 domain-containing 2 (SPRED2), which promotes the activation of nuclear factor kappa B (NF-kB) and STAT3 through ERK pathway, leading to increased tumor growth and metastasis. This regulatory mechanism is also implicated in the regulation of the response to anti-PD-1 immunotherapy194 (Fig. 5). Given the pivotal role of non-coding RNA and m6A methylation in melanoma pathogenesis elucidated by previous reports, further investigations in this area can bring more insights leading to innovative advances for melanoma therapy.
Table 2.
Current progress of lncRNAs in melanoma pathogenesis
| LncRNA | Expression status | Role in melanoma pathogenesis | Binding partner/target | Effect on downstream signal pathways | Reference (PMID number) |
|---|---|---|---|---|---|
| BASP1-AS1 | Upregulated | Promoter | YBX1 | Notch activation | 34533860 |
| SAMMSON | Upregulated | Promoter | P32 | Promote mitochondrial function | 27008969 |
| NCK1-AS1 | Upregulated | Promoter | miR-526b-5p | ADAM15 upregulation | 34247598 |
| FUT8-AS1 | Downregulated | Suppressor | NF90 | NRAS/MAPK | 34094894 |
| LINC00470 | Upregulated | Promoter | N.A. | Promote APE1 expression | 33875645 |
| LINC01291 | Upregulated | Promoter | miR-625-5p | Promote IGF-1R expression | 33674778 |
| TINCR | Downregulated | Suppressor | ATF4 mRNA | Prevent ATF4 translation and expression | 33586907 |
| MIR205HG | Upregulated | Promoter | miR-299-3p | Promote VEGF-A expression | 33535182 |
| LINC00518 | Upregulated | Promoter | N.A. | Affect multiple events like EMT and hypoxia-like response | 33371395 |
| NEAT1 | Upregulated | Promoter | miR-200b-3p | SMAD2 activation | 33202380 |
| LHFPL3-AS1 | Upregulated | Promoter | miR-181a-5p | Promote BCL2 expression and stem cells survival | 33149126 |
| TTN-AS1 | Upregulated | Promoter | TTN | Promote TTN expression | 32820147 |
| LHFPL3-AS1 | Upregulated | Promoter | miR-580-3p | STAT3 activation | 32753471 |
| LINC00520 | Upregulated | Promoter | miR-125b-5p | Promote EIF5A2 expression | 32466797 |
| SRA | Upregulated | Promoter | N.A. | P38 activation and EMT | 31945347 |
| MEG3 | Downregulated | Suppressor | miR-21 | E-cadherin upregulation | 31938020 |
| LINC-PINT | Downregulated | Suppressor | EZH2 | Increased H3K27 trimethylation and epigenetic rewiring | 31921860 |
| DIRC3 | Downregulated | Suppressor | N.A. | IGFBP5 upregulation | 31881017 |
| LINC00518 | Upregulated | Promoter | miR-204-5p | AP1S2 upregulation | 31712557 |
| FOXD3-AS1 | Upregulated | Promoter | N.A. | MAP3K2 activation | 31541886 |
| ZNNT1 | N.A. | Suppressor | N.A. | Autophagy activation | 31462126 |
| Linc00961 | Downregulated | Suppressor | miR‑367 | PTEN upregulation | 31364744 |
| LNMAT1 | Upregulated | Promoter | EZH2 | Downregulation of CADM1 | 31334110 |
| SLNCR1 | Upregulated | Promoter | AR and EGR1 | Downregulation of P21 | 31116991 |
| CPS1-IT1 | Downregulated | Suppressor | BRG1 | Downregulation of CYR61 | 31111478 |
| OIP5-AS1 | Upregulated | Promoter | miR-217 | Promote GLS expression and glutamine catabolism | 30779126 |
| CASC15 | Upregulated | Promoter | EZH2 | PDCD4 downregulation | 30013768 |
| LncRNA-ATB | Upregulated | Promoter | miR-590-5p | Promote YAP1 expression | 29956757 |
| KCNQ1OT1 | Upregulated | Promoter | miR-153 | Suppress MET expression | 29667930 |
| CASC2 | Downregulated | Suppressor | miR-18a-5p | RUNX1 downregulation | 29422114 |
| HOXD-AS1 | Upregulated | Promoter | EZH2 | RUNX3 upregulation | 29312805 |
| FALEC | Upregulated | Promoter | EZH2 | p21 downregulation | 29196104 |
| BANCR | Upregulated | Promoter | miR‑204 | Notch2 upregulation | 29075789 |
| CCAT1 | Upregulated | Promoter | miR-33a | N.A. | 28409554 |
| PVT1 | Upregulated | Promoter | miR-26b | N.A. | 28409552 |
| RHPN1-AS1 | Upregulated | Promoter | N.A. | N.A. | 28124977 |
| NKILA | Downregulated | Suppressor | N.A. | Suppression of NF-ĸB | 28123845 |
| ANRIL | Upregulated | Promoter | PRC1 | Repress the expression of CDKN2A | 20541999 |
| SLNCR1 | Upregulated | Promoter | N.A. | MMP9 upregulation | 27210747 |
| SAMMSON | Upregulated | Promoter | p32 | Increased mitochondrial function | 27008969 |
| CDR1as | Downregulated | Suppressor | IGF2BP3 | N.A. | 31935372 |
| GAS5 | Downregulated | Suppressor | E2F4 | Repress E2F4 expression | 32308561 |
| HEIH | Upregulated | Promoter | EZH2 | Inhibition of miR-200 cluster | 28487474 |
| TSLNC8 | Upregulated | Promoter | PP1α | MAPK reactivation | 33389075 |
| KCNQ1OT1 | Upregulated | Promoter | miR-153 | Repress MET expression | 29667930 |
| LINC00459 | Downregulated | Suppressor | miR-218 | DKK3 activation | 31844121 |
| LINC01158 | Upregulated | Promoter | miR-650 | MGMT upregulation | 33816296 |
| MALAT1 | Upregulated | Promoter | miR-34a | c-Myc, MET | 31101802 |
| ZEB1-AS1 | Upregulated | Promoter | miR-1224-5p | N.A. | 30651872 |
In general, compared to genetic variations, epigenetic modifications are more accessible and easily reversible, providing more options to develop promising drugs like HDAC inhibitors and EZH2 inhibitors. However, the non-specific characteristic of epigenetic modification determines that intervening epigenetics might not be as precise as that of targeted therapy for cancer treatment, and can lead to more side effects.195 Additional investigations are needed to improve the specificity of epigenetic modulation-based cancer therapy, as well as lowering the toxicity.
Signal pathways implicated in metabolic reprogramming
Metabolic reprogramming is a hallmark characteristic of cancer. The metabolism of melanoma cells is of rather high plasticity and builds a bridge that connects oncogenic factors to energetic supplement. Multiple paradigms of cellular metabolism like glycolysis, lipid metabolism, amino acid metabolism, nucleotide metabolism, oxidative phosphorylation and autophagy participate in not only the malignant behavior of melanoma cells, but also the re-establishment of the tumor microenvironment and the regulation of tumor-infiltrating immune cells.196 Some key metabolic enzymes have been considered as promising intervening targets to restrain melanoma progression, as well as to synergize with targeted therapy and immunotherapy.197,198
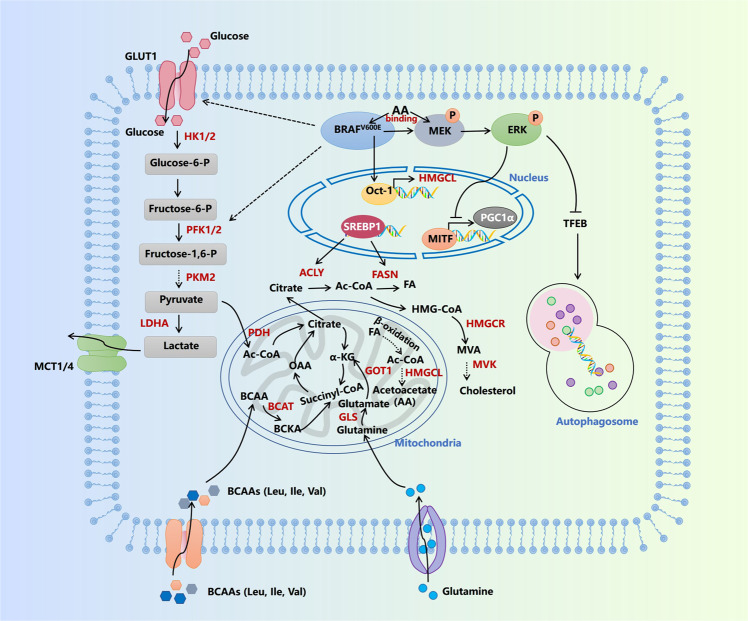
Aerobic glycolysis, also termed as the Warburg effect, is the most common metabolic characteristic in many cancers, with no exception in melanoma. Metabolite profiling reveals that the most frequently-occurred BRAF mutation endows melanoma cells with enhanced glycolytic capacity, which can be attributed to the upregulation of a network of transcriptional factors, glucose transporters, and kinases controlling glycolysis including HIF1α, MYC, Glut 1, Glut 3, and Hexokinase 2 (HK2).161,199 Moreover, BRAF negatively regulates MITF–PGC1α axis to suppress oxidative phosphorylation, which indicates that BRAF-driven glycolysis also partially results from the compensatory activation after the inhibition of mitochondrial function.78,200 Supplementary to transcriptional regulation, the downstream kinase of hyper-activated MAPK pathway ribosomal protein S6 kinase (RSK) can directly phosphorylate and activate 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 2 (PFKFB2), an enzyme that catalyzes the synthesis of fructose-2,6-bisphosphate during glycolysis, to facilitate BRAF-driven glycolytic metabolism201 (Fig. 6). Enhanced glycolysis provides enriched metabolic intermediate as building bricks to promote the synthesis of macromolecules like proteins, nucleic acid, and lipid acid.202 Moreover, the accumulation of intracellular lactate, the end product of glycolysis, leads to extracellular acidification via monocarboxylate transporter 4 (MCT-4), which impedes the function of tumor-infiltrating CD8+T lymphocytes.203 Therefore, the inhibition of glycolysis and lactate production can increase the treatment efficacy of anti-PD-1 immunotherapy via the reactivation of antitumor immunity.204,205 In addition to enhanced glycolysis, there are some data supporting a specific role of BRAF mutation on oncogenic metabolism. By using a shRNA library covering the known metabolism-related enzymes and protein factors in the human genome, 3-hydroxy-3-methylglutaryl-CoA (HMGCL), the rate-limiting enzyme implicated in ketogenesis, is identified as a “synthetic lethal” partner of BRAF mutation in melanoma cells.206 To be specific, mutant BRAF can transcriptionally upregulate HMGCL via Oct-1, and the end product of ketogenesis acetoacetate (AA) catalyzed by HMGCL further promotes the interaction between BRAFV600E and MEK1 to amplify MAPK activation (Fig. 6). Therefore, BRAF-mutant melanomas are addicted to ketogenesis.
Fig. 6.
Signal pathways of metabolic reprogramming in melanoma. AA acetoacetate, ACLY ATP-citrate lyase, BCAT branched-chain amino acid transaminase, BCKA branched-chain keto acids, FA fatty acid, FASN fatty acid synthase, GLS glutaminase, GLUT1 glucose transporter type 1, GOT1 glutamicoxaloacetic transaminase 1, HK1/2 hexokinase 1/2, HMGCL 3-hydroxy-3-methylglutaryl-CoA lyase, HMGCR 3-hydroxy-3-methylglutaryl-CoA reductase, LDHA lactate dehydrogenase A, MCT1/4 monocarboxylate transporter, MVA mevalonate, MVK mevalonate kinase, OAA oxaloacetate, PDH pyruvate dehydrogenase, PFK1/2 phosphofructokinase 1/2, PKM2 pyruvate kinase M, SREBP-1 sterol regulatory element-binding transcription factor 1, TFEB transcription factor EB
Mitochondrial oxidative phosphorylation is generally considered to be suppressed during tumor progression as a result of the Warburg effect. However, previous studies have provided compelling evidence that mitochondrial function is essential for maintaining tumor cell survival in melanoma and contributes greatly to melanoma growth. Vazquez et al. firstly found that a subset of melanomas harboring higher PGC1α expression has stronger capacity to defend against oxidative stress and rely on the mitochondrial function to survive and develop.207 This metabolic alteration is induced by MITF-mediated transcriptional upregulation, which is negatively regulated by BRAF.200 Upon the inhibition of the MAPK pathway by BRAF-targeted therapeutic agent, mitochondrial oxidative phosphorylation would be activated as a result of MITF–PGC1α axis activation, rendering the resistance to treatment.158,208 Moreover, lncRNA SAMMSON that co-expresses with MITF promotes melanoma growth by directly regulating mitochondrial master regulator p32, the inhibition of which significantly impedes the survival capacity of tumor cells.209 In contrast to the essential effect on tumor growth, PGC1α orchestrates a transcriptional axis that suppresses melanoma metastasis.210 Moreover, mitochondrial gatekeeper pyruvate dehydrogenase (PDH) is proved as a crucial mediator of BRAF-induced senescence. The suppression of PDH and relevant mitochondrial function is capable of abrogating BRAF-induced senescence, thereby licensing BRAF-driven melanoma development.211 Therefore, mitochondrial function switches from tumor suppressor to oncogenic factor during melanoma carcinogenesis and development. The function of mitochondria in melanoma pathogenesis seems paradoxical and is rather similar to that of histone methylation. Increased mitochondrial oxidative phosphorylation is essential for melanoma cell proliferation, whereas acts as an obstacle in melanomagenesis. Moreover, mitochondria master regulator PGC1α is also related to the phenotype switch between proliferative and invasive state, which is due to the discrepancy of downstream predominant activated signal pathways. Therefore, the intervention of PGC1α expression for melanoma therapy should take the clinical stage into consideration. Of note, a recent study using proteomics analysis demonstrates that the sensitivity to immunotherapy is highly related to higher oxidative phosphorylation and lipid metabolism in tumor cells,212 expanding the pathologic implication of mitochondrial function in melanoma.
The disorder of lipid metabolism is another hallmark metabolic characteristic of melanoma. Potentiation of lipogenesis and enhanced lipid uptake endow tumor cells with proliferative advantage by not only constituting the structure of multiple biological membranes but also providing an energy source.213 De novo fatty acid synthesis is regulated by a series of enzymes including ATP-citrate lyase (ACLY), acetyl-CoA carboxylase (ACC), fatty acid synthase (FASN), and acyl-CoA synthetase (ACS), whereas cholesterol biogenesis is orchestrated by acetyl-CoA acetyltransferase 2 (ACAT2), 3-hydroxy-3-methylglutaryl-CoA synthase (HMGCS), 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR) and mevalonate kinase (MVK). These two pathways are governed by sterol regulatory element-binding protein-1 (SREBP-1) and SREBP-2 respectively in a transcription-dependent manner214 (Fig. 6). The expressions of these lipid metabolism regulatory factors are generally increased in melanoma, and the inhibition of them can lead to prominent tumor regression,158,215–222 indicating the essential role of lipid biosynthesis for melanoma cell proliferation. In particular, integrative analysis with the use of positron emission tomography (PET), desorption electrospray ionization-mass spectrometry (DESI-MS), nonimaging MS and transcriptomic analyses in the zebrafish melanoma model provides direct evidence of increased lipid uptake and dysregulated glycerophospholipid metabolism.215 Targeted inhibition of fatty acid receptor CD36 can restrain the potential of metastasis-initiating cells and thereby impair tumor metastasis.223 What should be noted is that the lipogenic enzymes ACLY and SREBP-2 also have some lipogenesis-independent tumorigenic functions, with the engagement of mitochondrial oxidative phosphorylation and iron metabolism respectively.158,216 In addition to this, some metabolic intermediates in lipid metabolism like acetyl-CoA and palmitic acid are highly related to histone modification and non-histone modification,158,224 highlighting the close relationship between metabolism and protein modification. To be specific, palmitic acid can induce palmitoylation of cysteine residues of MC1R to trigger its activation and regulate downstream pigmentation and cell-cycle arrest under UVB radiation, which plays a protective role in preventing melanomagenesis.224,225 Moreover, increased acetyl-CoA produced by ACLY promotes histone acetylation and activates the transcription of the MITF–PGC1α axis to facilitate melanoma growth.158 More importantly, the upregulation of lipogenic enzymes like ACLY and SREBP-1 can also mediate the resistance to targeted therapy in melanoma,158,226 indicating that targeting lipogenesis is also a promising therapeutic approach to reinforce the treatment efficacy of MAPK inhibition.
Autophagy is a crucial metabolic process characterized by delivering intracellular proteins and organelles into autolysosomes for digesting and recycling, which physiologically provides intermediates as building bricks for macromolecules biogenesis, and sufficient ATP for cell survival and homeostasis.227 Liu et al. initially discovered that low expression of autophagy regulator autophagy-related 5 (ATG5) helps to overcome BRAF-induced senescence to promote the malignant transformation of early-stage melanoma.228,229 Besides, the deficiency of autophagy also facilitates tumor metastasis by stabilizing Twist family BHLH transcription factor 1 (TWIST1) protein through p62 accumulation.230 These reports indicate autophagy as a potential tumor suppressor in melanoma. In contrary, by employing a well-established melanoma mice model induced by both BRAF mutation and loss of PTEN, Xie et al. reported that ATG7 deficiency could restrain tumor progression by increasing oxidative stress and inducing cell senescence.231 This finding has been further supported by Rosenfeldt et al, while the tumorigenic role of ATG7 in vivo relies on the status of PTEN.232 Moreover, autophagy-driven ATP secretion also contributes to the invasive and migratory capacity of melanoma cells through the purinergic receptor P2RX7.233 The above-mentioned paradoxical roles of autophagy in melanoma have been clarified by our group based on the results of the fluctuation of autophagy level during melanoma progression. In particular, autophagy level is downregulated in the early stage but upregulated in metastatic stage, which is under the control of histone deacetylase sirtuin 6 (SIRT6) via epigenetic modulation of insulin-like growth factor (IGF)-AKT pathway. More importantly, autophagy indeed plays a bi-modal role in melanoma growth at different clinical stages, namely, tumor suppressor at early stage but tumor promoter at advanced stage.234 Other regulators including BRAF, miR-23a, transcription factor EB (TFEB), unfolded protein response (UPR) machineries, and receptor-interacting serine/threonine kinase 1 (RIPK1) are also responsible for increased autophagy level under either normal or stressful conditions in melanoma.181,235–237 Intriguingly, the activation of autophagy has also been observed in melanomas resistant to BRAF-targeted agents, which significantly hinders the treatment effect and renders therapeutic resistance.238 Therefore, targeting autophagy should be emphasized as a potential therapeutic approach for not only restraining tumor progression, but also increasing the efficacy of targeted therapy in melanoma.239,240
The dysregulation of amino acid metabolism, especially that of serine, glutamine, and branched-chain amino acid, is also greatly involved in melanoma pathogenesis. The rate-limiting enzyme of de novo serine synthesis is 3-phosphoglycerate dehydrogenase (PHGDH), of which the copy number is reported to be significantly increased in melanoma to ensure tumor cell survival and proliferation under low physiological serine concentrations.241,242 Supplement of dietary serine or genetic overexpression of PHGDH can efficiently foster melanoma progression via the enhancement of intracellular serine level.242 More intriguingly, PHGDH is also decisive in mediating melanoma metastasis to the brain. With the use of proteomics, metabolomics, and multiple brain metastasis models, it has been revealed that the metastatic lesions of melanoma is sensitive to the limitation of serine synthesis in nutrient-limited environment, making PHGDH an attractive target to control melanoma distant metastasis.243 Recent study regarding targeted therapy forwardly shows that PHGDH upregulation also renders the resistance to MEK inhibitor in melanoma harboring NRAS mutation. Targeting PHGDH can re-sensitize resistant tumors to MAPK inhibition via the reduction of glutathione and the increment of oxidative stress.244,245 Therefore, PHGDH inhibition is a lethal partner with MAPK inhibitor for melanoma therapy. Apart from serine metabolism, melanoma cells are also addicted to glutamine for fueling tumor progression. Glutamine metabolism is orchestrated by a series of machineries including glutamine importer, glutaminases 2 (GLS2) and glutamicoxaloacetic transaminase 1 (GOT1) to promote glutamine uptake and subsequent formation of glutamate and α-ketoglutarate (α-KG) (Fig. 6). Targeting either glutamine transport or glutaminase can obtain prominent regression of tumor.246,247 Moreover, glutamine acts as a compensated source supporting melanoma cell survival in case of the inhibition of lactate dehydrogenase A (LDHA)-mediated Warburg effect through activating transcription factor 4 (ATF4)-mediated upregulation of glutamine transporter,248 or after the suppression of mitochondrial oxidative phosphorylation by PGC1α knockdown.249 In the tumor microenvironment, decreased α-KG level caused by regional loss of glutamine leads to histone hyper-methylation and thereafter cell de-differentiation to render melanoma cells more resistance to targeted therapy.250 Nevertheless, melanoma cells with the acquired resistance to BRAF inhibitor are highly dependent on glutamine for cell proliferation,251 indicating that glutamine metabolism might play contrary roles in different phases of treatment resistance. Although the blockade of intra-tumoral glutamine metabolism results in impeded tumor cell proliferation, dietary glutamine supplementation could suppress melanoma growth in vivo via α-KG-mediated epigenetic rewiring,252 indicating a proposed model that excessive glutamine uptake might otherwise abrogate the essential effect of glutamine on supporting tumor cell survival. In addition to serine and glutamine, branched-chain amino acids (BCAAs) comprising of leucine (Leu), isoleucine (Ile), and valine (Val), as well as their metabolism, are implicated in cancer pathogenesis. After the intake by tumor cells, branched-chain amino acid transaminase 1 and 2 (BCAT1/2) transfers nitrogen of BCAAs to α-ketoglutarate (αKG) to produce glutamine and branched-chain keto acid (BCKA), which is then metabolized by branched-chain alpha-keto acid dehydrogenase complex (BCKDH) and some other enzymes to produce tricarboxylic acid (TCA) cycle intermediates acetyl-CoA and/or succinyl-CoA, thus fueling tumor cell proliferation and providing some building bricks to biogenesis253 (Fig. 6). As is similar to other types of cancers, the expression of BCAT1 is significantly upregulated in melanoma. Genetic knockdown of BCAT1 expression impairs the proliferative capacity of melanoma cells via the suppression of mitochondrial oxidative phosphorylation.254 Moreover, genome-wide CRISPR/Cas9 knockout screening assay has identified that dihydrolipoamide branched-chain transacylase (DBT), a subunit of BCKDH, is implicated in the regulation of cell apoptosis induced by BRAFV600E overexpression in melanocytes. The inhibition of DBT contributes to BCAAs accumulation and attenuates oncogene-induced apoptosis of melanocytes, suggesting that DBT is a gatekeeper mediating mutant BRAF-driven malignant transformation from melanocytes to melanoma.255 What’s more important, melanoma cells harboring BRAF mutation and hyper-activation of the MAPK pathway are highly dependent on leucine for survival. The inhibition of autophagy mimicking activated RAS-MEK signaling renders tumor cells to leucine deprivation, so that dietary leucine deprivation and autophagy inhibition could synergistically suppress melanoma growth.256 Therefore, BCAAs metabolism is a promising therapeutic target for not only preventing the malignant transformation of melanocytes but also suppressing melanoma progression.
In aggregate, melanoma cells are of high metabolic flexibility and have a complicated metabolic network with the involvement of multiple paradigms including glycolysis, mitochondrial oxidative phosphorylation, lipid metabolism, autophagy, and amino acid metabolism. These metabolic characteristics endow tumor cell with growth advantages by supplying not only sufficient energy, but also abundant metabolic intermediates for the synthesis of various macromolecules, which are essential for rapid cell proliferation. Of note, there are some metabolic alterations specific to oncogenic mutations or melanocytic lineage, including mutated BRAF-driven ketogenesis and SOX10-mediated SAMMSON upregulation and mitochondrial oxidative phosphorylation, which might provide an accessible target for a specific subgroup of patients with melanoma. However, the frequent occurrence of compensated activation of alternative pathways prominently hinders the efficacy of targeting one single metabolic paradigm.249,257 Combinatorial suppression of multiple metabolic pathways might be more efficient in controlling melanoma growth, which needs more investigations in the future.
Key signal pathways in tumor metastasis
Tumor metastasis is the most important cause of the unoptimistic prognosis of melanoma patients.258 Generally, the occurrence of metastasis mainly includes the following key steps, namely, invasion, intravasation, circulation, extravasation and colonization at secondary tumor sites,259 which are orchestrated by a series of distinct biological principles.260 Herein, we emphasize epithelial–mesenchymal transition (EMT), melanoma cell adhesion, and exosomes to delineate the signal pathways landscape and hope to generate mechanistic insights of melanoma metastasis.
EMT refers to a cellular program generally characterized by the downregulation of multiple epithelial markers like E-cadherin, laminin, cytokeratin, and the upregulation of mesenchymal markers like N-cadherin, vimentin, and α-SMA. EMT is physiologically fundamental in embryogenesis, fibrosis, and wound healing.260 In cancer, the occurrence of EMT is accompanied by morphologic switch from an epithelioid toward a mesenchymal/spindle cell shape, endowing tumor cells with enhanced invasive and migratory capacity. Previously, a high-throughput gene-expression profile has identified EMT as a major determinant of melanoma metastasis.261 Subsequent mounting evidence has revealed the pivotal role of EMT in facilitating melanoma metastasis and the upstream regulatory network.262 To be specific, under the control of some canonical oncogenic pathways like BRAF/MEK, AKT/mTOR, Wnt/β-catenin and TGF-β, the switch of the expressions of EMT-inducing transcriptional factors (EMT-TF) including MITF, SOX2, Snail, Slug, TWIST1, zinc finger E-box binding homeobox 1/2 (ZEB1/2) and NF-κB governs the irreversible process of EMT.262 In response to the activation of MEK/ERK in melanomas harboring BRAF or NRAS mutation, molecular network of EMT-TF is profoundly re-organized to be in favor of TWIST1 and ZEB1, whereas the expressions of Snail2 and ZEB2 are deficient. This alteration is highly attributed to ERK activation and the resultant induction of AP-1 family member FRA1, cooperating with BRAF mutation to drive the gain of invasive ability and tumor metastasis.263 In addition to BRAF/MEK cascade, AKT/mTOR signal is greatly engaged in melanoma metastasis as well. The activation of AKT and mTOR downstream effector 4E-BP1 is in strongly positive correlation with the aggressiveness of melanoma, and the poor prognosis of patients in two independent cohorts.264–267 Loss of PTEN is the main reason for hyper-activation of AKT pathway, and several integrated molecular and clinical analysis all point out the close relationship between AKT activation and the occurrence of metastasis to specific regions like brain, lymph nodes, and lungs, accompanied with the upregulation of β-catenin and the downregulation of E-cadherin.110,268–270 Moreover, overexpression of AKT dramatically induces invasive phenotype of melanoma, not only promoting the conversion from radial to vertical growth, but also contributing to metastasis to distant organs in established melanoma transgenic mice model,42,271–273 with EMT responsible for this phenotypic switch.274–276 Aside from MAPK and AKT pathway, Wnt/β-catenin has also been closely related to EMT and tumor metastasis in melanoma. β-catenin is a central mediator of metastasis in established transgenic mice model harboring both BRAF gain-of-function mutation and PTEN loss-of-function mutation via simultaneous activation of MAPK and AKT pathways.110 Clinically, the activation of non-canonical Wnt5a signaling is positively related to the clinical stage and survival of patients, and strong cytoplasmic Wnt5a staining is an independent risk factor for reduced metastasis-free and overall survival in multivariate analysis.277,278 Dissanayake et al. has demonstrated that Wnt5a suppresses PKC signaling to initiate EMT by increasing the expressions of Snail and vimentin, as well as the downregulation of E-cadherin.279 Moreover, Sinnberg et al. has proved that β-catenin is frequently expressed at the invasive front of melanoma, and Wnt/β-catenin promotes neural crest migration of melanoma cells and induces an invasive phenotype.111 Therefore, targeting Wnt/β-catenin could be exploited as a promising approach to impede melanoma metastasis by attenuating EMT. Of note, TGF-β is another vital player in melanoma metastasis. In tumor microenvironment, mesenchymal stem cell-derived TGF-β promotes melanoma cell EMT in paracrine/autocrine-dependent manner.280 What should be noted is that TGF-β-mediated transcriptional profile also facilitates ameboid phenotype independent of the EMT process to promote melanoma cell dissemination.281 Downstream the above-mentioned oncogenic pathways, EMT-TFs are indeed master regulator of melanoma phenotype switching in a transcriptional regulation-dependent manner. Wels et al. has discovered the specifically regulatory effect of Slug, rather than Snail or Twist, on the upregulation of ZEB1 and downregulation of E-cadherin, thus resulting in decreased adhesion to human keratinocytes, and enhanced migration of melanoma cells.282 Another investigation with the employment of in vivo fate mapping technology demonstrates that melanoma cells undergo a conversion in state where ZEB2 expression is replaced by ZEB1 expression associated with gain of an invasive phenotype, suggesting that reversible switching of the ZEB2/ZEB1 ratio could enhance melanoma metastatic dissemination.283 Meanwhile, loss of ZEB2 expression results in prominent downregulation of MITF and relevant differentiated phenotype, concomitant with an upregulation of ZEB1 and mesenchymal characteristic of melanoma cells. The crosstalk among these three crucial EMT-TFs orchestrates the transcriptional program of phenotypic plasticity.284 As the melanocytic lineage-specific transcriptional factor, MITF exerts an integrated effect on suppressing focal adhesion and N-cadherin expression, thus ameliorating the local detachment and dissemination of tumor cell.285 What’s more, the stabilization and accumulation of TWIST1 due to autophagy deficiency and increased SQSTM1-TWIST1 interaction is also an important trigger of EMT in melanoma.230 Taken together, the regulatory network of EMT-TFs is a pivotal contributor and potent therapeutic target of melanoma metastasis.
The adhesiveness and intercellular communication within the tumor microenvironment are also decisive for tumor cell dissemination, with cell-adhesion molecules (CAM) governing this process. CAM includes a large family of proteins located on cell surface like integrin, cadherin, IgSF, connexin and mucin that regulate attractive or repulsive forces to the extracellular matrix (ECM), stroma and other cancer cells, thus affecting the invasive and migratory capacity.286 Integrins are heterodimeric proteins on the cell membrane that adhere to the ECM and can also sense and transduce extrinsic signalings.287 Besides, at least 18 α- and 8 β-subunits constitute the presently-known 24 heterodimers of the superfamily integrins.288 Initial reports have demonstrated the dysregulation of multiple integrins in regulating melanoma cell survival, tumor growth, tumor metastasis, and the association with clinical characteristics.289–294 To be specific, previous reports from Herlyn’s group have reported that increased expression of integrin ɑvβ3 is associated with growth and the conversion from radial growth phase (RGP) to vertical growth phase (VGP) in melanoma.295,296 In addition, integrins like ɑ2β1, ɑ5β1, and ɑvβ3 can directly stimulate the expression and function of matrix metallopeptidases (MMPs), which facilitates the degradation of collagen and fibronectin, so as to contribute to tumor cell invasion and progression.297–299 Of note, targeted blockade of either integrins β1 or ɑvβ3 leads to prominent suppression of tumor cell adhesion and migration.300 What’s more, integrins also play important role in promoting angiogenesis, which is implicated in supplying sufficient nutrient and orienting the spread of tumor cells to distant organs. In particular, fibroblast growth factor 2 (FGF-2) and vascular endothelial growth factor A (VEGF-A) promotes angiogenesis via the regulation of integrin ɑvβ3 and ɑvβ5 respectively, which is supported by the evidence that pharmacological blockade of ɑvβ3 and ɑvβ5 ameliorates increased angiogenesis induced by FGF-2 and VEGF-A.301 In addition to these, integrin-based signaling transduction and associated intermediaries have recently attracted more attention due to their impact on melanoma pathogenesis. For example, increased expression of integrin-linked kinase (ILK) is highly associated with progression of melanoma and the poor prognosis of patients.302 Genetic knockdown of ILK expression results in significant impairment of melanoma cell migration and the formation of anchorage-independent colonies in soft agar as well, indicating the indispensable role of ILK in melanoma development.303 In addition, the upregulation of ILK also contributes to melanoma angiogenesis via the enhancement of NF-κB and IL-6 signaling.304 Recently, Gil et al. has demonstrated that the deficiency of ILK regulates the endosomal recycling of N-cadherin and reduces membrane N-cadherin expression to ameliorate melanoma metastasis.305 Huang et al. further pointed out that distinct integrins on melanoma cell specifically direct circulating melanoma cells to different organs and the establishment of metastases at specific organ sites.306 Besides, exosomes derived from tumor cell that contained different integrins contribute to the formation of pre-metastatic niche in targeted organs and promoted organ-specific metastases.306 Therefore, the pharmacological intervention of integrins has been broadly investigated in preclinical and clinical trials.307–310 What should also be mentioned regarding CAM is cadherin that refers to a family of calcium-dependent cell-adhesion proteins. The “cadherin switching” during tumor progression is generally characterized by the loss of E-cadherin and the increase of N-cadherin,311 which highly contributes to increased interactions between melanoma cells and dermal fibroblasts/vascular endothelial cells, and the impaired junction to keratinocytes. The E-cadherin/N-cadherin switch is highly associated with low or absent PTEN expression and disease progression in melanoma.312 Of note, N-cadherin-regulated cell-adhesion results in the potentiation of AKT-β-catenin signaling to antagonize the expressions of pro-apoptotic factors, exerting pivotal effect on tumor cell survival in addition to migratory ability.313 Aside from these, two reports have demonstrated that P-cadherin counteracts the invasion and migration of melanomas via the increase of cell-cell interaction, suggesting that more members of cadherins might play a role in melanoma pathogenesis.314,315
Exosomes are extracellular vesicles transporting proteins, nuclear acids, and metabolites that can mediate intercellular communication in the tumor microenvironment.316 Due to the resistance to proteolytic and nuclease activity, exosomes are relatively stable and cargos in them are protected from various stress and degradation.317 The capacity of exosomes to carry nuclear acids and proteins endows them with the function to regulate the metastatic ability of tumor cells in the primary region and remote organs or tissues that provide the soil the formation of metastases.318 Initially, Hood et al. discovered that exosomes released by melanoma cells define microanatomic responses in sentinel lymph nodes that licenses metastasis of melanoma cells. To be specific, homing of melanoma cells-derived exosomes to sentinel lymph nodes exerts integrated effects on melanoma cell recruitment, extracellular matrix deposition, and vascular proliferation in the lymph nodes, thus helping microanatomic niche preparation to facilitate tumor cell lymphatic metastasis.319 Later, it was unveiled that exosomes from highly metastatic melanoma could educate bone marrow progenitor cells toward a pro-vasculogenic phenotype and trigger vascular leakiness at pre-metastatic sites through the receptor tyrosine kinase MET. The formation and trafficking of exosomes are under the control of RAB27A, the knockdown of which significantly diminishes tumor metastasis.320,321 Given the great implication of exosomes in regulating tumor biology, the profiles of mRNA, miRNA and protein in melanoma cells-derived exosomes have been systemically analyzed, displaying specific signature related to metastatic potential.322,323 Of note, exosomes from melanoma cells can induce the phenotype switching of melanocytes, endowing them with increased invasive and metastatic capacity. For example, let-7i transferred by melanoma cell exosomes can induce epithelial–mesenchymal transition in primary melanocytes via the activation of MAPK signaling.324 In addition, exosomal miR-106b-5p derived from melanoma cell contributes to the EMT process of melanocytes by targeting EphA4 to activate the ERK pathway.325 Apart from these, exosomes also play a role in regulating angiogenesis and immune cell function in tumor microenvironment and preparation of a hospitable metastatic niche in distant organs to facilitate metastasis. For example, exosomes secreted by metastatic melanoma cells can instigate a pro-inflammatory gene signature in both lung fibroblasts and brain astrocytes, which promotes the formation of an inflammatory metastatic niche, suggesting that the reprogramming of stromal cells by tumor cell exosomes is a general mechanism in distant organs.326 Moreover, a series of reports have demonstrated that melanoma cell exosomes regulate the function of tumor-infiltrating immune cells to shape tumor microenvironment toward a pro-tumorigenic state. Gerloff et al. has reported that melanoma-derived exosomal miR-125b-5p induces a phenotype switch of tumor-associated macrophages toward a tumor-promoting state by targeting lysosomal acid lipase A.327 In addition, pre-metastatic tumors are capable of producing exosomes to potentiate immune surveillance by patrolling monocytes at the metastatic niche, indicating that exosomes from poorly metastatic melanoma cells can also potently inhibit metastasis to distant organ.328 In addition, some recent studies also demonstrate the involvement of tumor cell exosomes in regulating angiogenesis, metastatic niche formation, and mesenchymal stem cell oncogenic reprogramming.329–331 Therefore, exosomes exert a facilitative role in tumor metastasis via the regulation of multiple downstream biological activities.
The formation of metastasis is an integrative process with rather a complexity and mainly accounts for the mortality of melanoma patients. Extensive investigations have been conducted to elucidate the underlying mechanisms, which are far more than EMT, cell-adhesion alteration, and exosomes that we mentioned above. Metabolic rewiring, pre-metastatic niche formation, and the existence of dormancy have also been documented as critical characteristics of melanoma metastasis,332 which should also be taken into consideration for melanoma therapy.
Signal pathways regulating oncogenic inflammation and angiogenesis
The inflammatory signal pathway is highly related to tumor carcinogenesis and progression, with no exception in melanoma.333 Inflammatory factors including tumor necrosis factor (TNFα), IFN-γ, interleukins, and related regulatory signalings such as Janus kinase (JAK)-STAT, NF-κB, and inflammasome have attracted more and more attention in the investigation of melanoma biology and tumor microenvironment.
A previous study using in situ hybridization assay firstly verified the existence of TNFα in melanoma cells in the tumor microenvironment,334 and the expression status of TNFα is related to driver mutation of the oncogene.335 In contrast to the fact that high-dose exogenous TNFα can induce apoptosis of melanoma which has been employed in various clinical trials and cancer therapy,336,337 TNFα derived from tumor cell or tumor microenvironment exerted a prominent regulatory role in tumor cell survival, proliferation, invasion, metastasis, and immune escape. Melanoma stimulated with recombinant TNFα displays downregulation of oncogenic factor c-myc, which thereby delays cell proliferation, indicating that TNFα antagonizes the outgrowth of tumor.338 However, tumor cells-secreted TNFα promotes downstream activation of RIPK1-NF-κB cascade in an autocrine manner to enable tumor cell survival, which is based on the observation that the deficiency of TNFαR1 or neutralizing TNFα in culture supernatant abrogates the activation of NF-κB signaling and restrains the proliferation of melanoma cell,339 highlighting TNFα as an oncogenic inflammatory factor. This conclusion is further supported by the results that in response to targeted inhibition of the MAPK pathway, TNFα enabled tumor cell survival by inducing c-FLIP upregulation and NF-κB activation.340,341 Therefore, the blockade of TNFα could be a promising synergized therapeutic approach with targeted therapy. Besides, Zhu et al. has provided evidence that TNFα stimulates the migratory potential of melanoma cells via the upregulation of fibronectin and integrin expressions, counteracting the suppressive effect of α-MSH.342,343 Consistent with this, TNFα promotes the expression of MMP2 and MMP9 to facilitate tumor cell migration.344,345 More importantly, it has been revealed that TNFα determines the phenotypic plasticity of melanoma cells by antagonizing MITF expression via downstream c-Jun. Dedifferentiated state of melanoma cells characterized by low MITF level has a higher inflammatory responsiveness and pathway activity. Clinically, the expression ratio of MITF and c-Jun could reflect the recruitment and infiltration of myeloid cells in tumor microenvironment, thus dictating the sensitivity to myeloid cells-directed immunotherapy.346 Some recent studies have also emphasized the role of TNFα in anti-tumor immunity and immunotherapy. Bertrand et al. demonstrated that the blockade of TNFα or TNFαR1 could enhance CD8+T cells-dependent antitumor immunity in established melanoma.347 Moreover, genome-wide screening uncovered that the ablation of TRAF2 could lower the TNFα cytotoxicity threshold in tumors by redirecting TNFα signaling to favor RIPK1-dependent apoptosis, thus increasing the susceptibility of tumors to immunotherapy.348 Further investigation also revealed that TNFα blockade could overcome the resistance to anti-PD-1 in melanoma via the prevention of cell death of tumor-infiltrating lymphocytes.347 In aggregate, targeting TNFα might be of high translational potential to synergize with anti-PD-1 antibody in treating melanoma. Of note, previous studies have shown the bifurcated functions of TNFα on melanoma pathogenesis, which is possibly determined by the source and the dosage of TNFα for the stimulation of melanoma cell. While high-dose exogenous TNFα mainly induces tumor cell apoptosis, endogenous low-concentration TNFα derived from tumor cell or tumor microenvironment contrarily plays an oncogenic role.
IFN-γ is enriched in the tumor microenvironment due to the infiltration of cytotoxic CD8+T lymphocytes. Physiologically, IFN-γ could activate JAK-STAT signaling to promote the expression of genes to defend against pathogen and infection. Recent studies mainly concentrate on the role of IFN-γ signaling in the regulation of tumor immune evasion and the implication in immunotherapy for melanoma. To be specific, IFN-γ could induce the expressions of multiple immune checkpoints including cytotoxic T lymphocyte antigen-4 (CTLA-4), PD-L1, and PD-L1 via JAK-STAT-dependent transcriptional cascade.349–351 The facilitation of PD-L1 by IFN-γ in melanoma cells is highly related to p53 expression.350 These data indicate that IFN-γ might modulate tumoral immune checkpoint to terminate the immune surveillance of tumor cell performed by lymphocytes, namely, immune evasion. Th1/IFNγ gene signature in the tumor microenvironment has been regarded as an independent biomarker to predict the prognosis of resectable high-risk melanoma patients.352 Moreover, the status of IFN-γ is associated with the response or resistance to immunotherapy. For instance, upregulated IFN-γ-related mRNA profile could predict better response to immunotherapy and better survival of patients.353 In line with this, Grasso et al. has also demonstrated that conserved IFN-γ transcriptome could drive the amplification of antitumor immune response and better treatment outcome of immune checkpoint blockade.354 Intriguingly, IFN-γ secreted by tumor-infiltrating lymphocytes after immunotherapy or radiotherapy could exert its direct effect on tumor cells via the downregulation of glutamate-cystine antiporter system Xc- to trigger ferroptosis, a novel cell death modality characterized by excessive lipid oxidation, further supporting the facilitative role of IFN-γ in immunotherapy.355,356 However, there are two studies raising the notion that sustained activation of IFN signaling might be the cause of resistance to immune checkpoint blockades.357,358 The discrepancy could be related to the different phases and characteristics of intrinsic resistance and adaptive resistance in immunotherapy. Aside from IFN-γ, IFN-α is another crucial interferon with integrated functions of tumor control and immune regulation.359–362 In particular, IFN-α could stimulate host anti-tumor immunity by promoting the expression of major histocompatibility complexes (MHC) on tumor cell membrane, dendritic cell maturation, the cytotoxicity of natural killer (NK) cell, as well as the capacity of CD8+T cells for eradicating tumor cells.361–363 Although high-dose IFN-α has been approved for the treatment of resected melanoma as adjuvant therapy, the adverse effect is considerable and then is replaced by anti-PD-1 and anti-CTLA-4 immunotherapy.364 What should be noted is that IFN-α1b is reported to exert a better safety profile compared to IFN-α1a and is more tolerable for melanoma treatment. Prolonged usage of IFN-α1b in patients with unresectable metastatic melanoma has gained encouraging outcome.365
Interleukins and chemokines are two alternative pivotal types of inflammation-related factors in melanoma pathogenesis, especially in the regulation of immune cell function in the tumor microenvironment.366 Different interleukins exert distinct, even contrary effects on antitumor immunity. IL-2 is documented as a potent activator of both CD8+T cells and NK cells through the binding to the heterotrimeric receptor consisting of three subunits including α, β, and γ.367,368 In 1998, high-dose IL-2 was approved for melanoma treatment and obtained a considerable objective response in 15–20% patients with advanced melanoma.369 However, the increased risk of severe adverse effects including capillary, leak syndrome, gastrointestinal side effects, fever and chills limited the continuous usage of IL-2 to prolong the survival of patients. Other interleukins like IL-15 and IL-10 also have immune stimulatory function and antitumor capacity.370–372 Novel IL-15 super agonist complex and PEGylated formulation of recombinant IL-10 have been processed to clinical trials for treating melanoma, revealing promising therapeutic effect and satisfying tolerance.373,374
Inflammasomes are a class of cytosolic multiprotein complexes classically consisting of the NOD-like receptor (NLR) sensor protein, the adaptor protein ASC and the downstream effector caspase-1.375 Generally, inflammasomes act as a sensor and responder to pathogen-associated molecular patterns (PAMPs) via the maturation of pro-inflammatory IL-1β and IL-18 to activate immune cells, thus defending against various pathogens.376 For melanoma, Okamoto et al. firstly unveiled that NLR family pyrin domain-containing 3 (NALP3) inflammasome is constitutively assembled and activated, which is responsible for the spontaneous secretion of IL-1β from melanoma cells. The increased IL-1β could promote angiogenesis and modulate immune cells to promote melanoma progression.377 Bioinformatics analysis of pan-cancer data further demonstrates that NLRP3 inflammasome gene signature could be regarded as an independent prognostic factor of melanoma, with better predictive credibility than either tumor mutation burden (TMB) or tumorous glycolytic activity.378 Through a series of functional and mechanistic studies, Tengesdal et al. has proved that tumor-associated NLRP3/IL-1β signaling promotes the expansion of myeloid-derived suppressor cells (MDSCs), thereby resulting in ameliorated natural killer and CD8+ T cell activity and increased presence of Treg cells in tumor microenvironment. The combination of NLRP3 inhibition and anti-PD-1 antibody has obtained a synergized effect via the suppression of the function of MDSCs.379 In parallel with this, the activation of NLRP3 is also responsible for the role of tumorous PD-L1 in the resistance to anti-PD-1 immunotherapy through the regulation of MDSCs.380 In aggregate, inflammasomes play an oncogenic role in melanoma via the simultaneous effect on both tumor cell behavior and antitumor immunity. Targeting inflammasome is a promising strategy as a monotherapy or a combined option with immunotherapy for melanoma.
Angiogenesis is defined as the formation of new blood vessels derived from pre-existed neoplastic vasculature, which is mainly responsible for supplying sufficient nutrient and oxygen to ensure the rapid proliferation of tumor cells in cancer carcinogenesis.381 The first piece of evidence on angiogenesis in melanoma is the increased blood supply after the transplantation of melanoma cells into the cheek pouches of hamsters discovered by Warren et al.382. Then, the status of angiogenesis is found to be highly correlated with melanoma progression, especially the transition from the radial growth phase to the vertical growth phase.383 The development of a rich vascular architecture within the tumor microenvironment is orchestrated by angiogenic switch, which means the potentiation of pro-angiogenic factors and the suppression of anti-angiogenic factors.384 There are several main factors contributing to neovascularization in melanoma, including VEGF-A, basic fibroblast growth factor (bFGF), placental growth factor (PlGF), angiopoietin (Ang), IL-8, and PDGF. These factors are produced mainly by tumor cells, with alternative types of cells like endothelial cells and immune cells also participating in.385 To be specific, VEGF-A secreted from melanoma cells exerts its effect on angiogenesis through the binding to the receptor VEGFR on endothelial cells. Consequently, nitric oxide synthase (NOS) and PI3K/AKT signaling are activated to increase the permeability of vessel and promote endothelial cell proliferation and tube formation.386,387 In addition, the resultant activation of downstream focal adhesion kinase (FAK) contributes to melanoma cell extravasation across the vessel barrier.388 Similar to other cancers, the expression level of VEGF-A in melanoma is also under the transcriptional regulation of HIF1α, which is induced by local tissue hypoxia if the pace of angiogenesis does not meet the demand of nutrient and oxygen required for tumor cell proliferation.389 Apart from VEGF-A, bFGF is another principle pro-angiogenesis factor that is physiologically implicated in the modulation of wound healing via stimulating the proliferation of endothelial cells and the migration of macrophages and fibroblasts.385 Mechanistically, bFGF secreted from melanoma cells could not only interact with the receptor on the surface of endothelial cells to facilitate neovascularization, but also activate tumor cells in an autocrine way to promote their proliferation.381 The suppression of bFGF activity through the employment of targeted antibody or antisense oligodeoxynucleotides leads to prominent regression of angiogenesis and diminishment of tumor growth.390,391 Therefore, bFGF is a multi-effect therapeutic target for melanoma treatment. What’s more, PIGF, as a member of the VEGF family, has also been documented as a crucial promoter of angiogenesis in melanoma. Aside from the canonical receptor NRP-1 and NRP-2, PIGF can also bind to VEGFR via the formation of heterodimers with VEGF-A, so that the similar downstream pathways responsible for angiogenesis would be activated as that of VEGF-A alone.392 PlGF is also capable of directly interacting with VEGFR-1-positive hematopoietic precursors and pericytes and smooth muscle cells as well, so as to increase the recruitment and migration of hematopoietic precursors from bone marrow and promote the maturation of newly formed blood vessels as well.392 The above-mentioned mechanisms indeed mediate PIGF-driven melanoma growth and metastasis in a transgenic mice model in vivo, indicating that PIGF is of high potential as a therapeutic target in inhibiting melanoma progression via the obstruction of angiogenesis.393 Additionally, mounting evidence also reveals that other pro-angiogenic factors Ang, IL-8, and PDGF play a crucial role in facilitating the establishment of neovascularization and melanoma progression through multiple downstream mechanisms.394–400 In aggregate, the pro-angiogenic mechanism in melanoma is of rather complexity and highly interconnected due to the co-existence of multiple stimulatory ligands and receptors that exert their function through both paracrine and autocrine manners, which might be the reason for the limited therapeutic efficacy of treatment with a monoclonal antibody neutralizing VEGF-A in melanoma.401 Therefore, simultaneous suppression of multiple angiogenic pathways may be more effective.
Current progresses in targeted therapy in melanoma
MAPK inhibition-targeted therapy
The identification of BRAF mutation in melanoma in 2002 has opened a new era for understanding oncogenic events of melanomagenesis and provided the molecular basis for developing targeted therapy.25 Over 50% of cutaneous melanomas harbor BRAF mutation, which can induce a robust increase of its kinase activity and constitutive enhancement of downstream MEK-ERK signaling cascade.402 About ten years ago, vemurafenib and dabrafenib had been approved by FDA for the treatment of advanced melanoma harboring BRAF mutations. As a result, the two BRAF-targeted agents achieve a considerable objective response rate and some patients can even gain complete regression of tumor.20,403 Vemurafenib was the first-in-class agent which provided dramatic improvement of treatment outcome. Compared with dacarbazine chemotherapy in the phase III BRIM3 trial, vemurafenib single-agent treatment significantly increased the objective response rate (ORR) from 5 to 48%.404 In addition, the median progression-free survival (PFS) and overall survival (OS) were also substantially extended to 5.3 and 13.3 months respectively.405 The second BRAF-targeted agent dabrafenib was then developed which exhibited similar therapeutic effects as that of vemurafenib, with an objective response rate of 50% and progression-free survival being 5.1 months.19 It should be noted that the application of BRAF-targeted agent can induce the onset of keratoacanthoma and squamous-cell carcinoma, in around 15–20% of patients.19,405
Given the molecular rationale that BRAF mutation robustly triggers the hyper-activation of downstream MEK-ERK pathway, MEK-targeted agent trametinib has then been developed for melanoma targeted therapy. According to the result from the phase III METRIC trial, trametinib single-agent treatment obtained an ORR of 22% and a median PFS of 4.8 months,406 which was not as ideal as that of BRAF-targeted therapy in melanoma harboring BRAF mutation. The application of MEK-targeted agent has been then extended to melanomas lacking BRAF mutation, since that some other populations of melanomas are also dependent on MAPK signal pathway to survive and growth. To be specific, NRASQ61R-mutant melanoma receiving trametinib single-agent treatment obtained an ORR of 20% and a median PFS of 4 months in a phase II study.407 Forwardly, MEK inhibitor monotherapy has been approved in non-BRAF-mutation melanoma settings based on the results of phase III study, whereas the effect was not that satisfactory. Therefore, the combinations with CDK4/6 inhibition, MDM2 inhibition, and PI3K/AKT-pathway inhibition based on some mechanistic insights has been evaluated in recent studies.408–410
For patients with BRAF-mutant melanoma receiving MAPK inhibition-targeted therapy, drug resistance would inevitably occur within 6–12 months, which significantly hinders the treatment efficacy and result in frequent recurrence.406 Based on the distinct mechanisms and characteristics, resistance to MAPK inhibition therapy is classified to three types including intrinsic resistance, adaptive resistance and acquired resistance. To be specific, intrinsic resistance refers to the innate capacity of tumor cells to resist the toxicity of targeted inhibitors, which is decisive for the innate response to treatment. However, in the early phase after the application of targeted therapy, especially the first 24–48 h, multiple protective signaling pathways would be rapidly activated to mitigate the pro-apoptotic effect of targeted agent, which is defined as adaptive resistance. Later on, long-term MAPK inhibition treatment can lead to intracellular mutational alterations and establishment of mutational clones, contributing to the acquired resistance that is irreversible.78
Signal pathways of resistance to MAPK inhibition-targeted therapy
Approximately 20% of melanoma patients are intrinsically insensitive to targeted therapy even their tumors harbor BRAF mutation. Previous investigations have demonstrated multiple genomic and non-genomic alterations rendering intrinsic resistance, including loss of PTEN, loss of NF1, CCND1 amplification, COT upregulation, RAC1 mutation, eIF4F activation, low MITF expression, and high AXL expression.411,412 Mechanistically, deletions or mutations of PTEN can trigger the activation of AKT signaling and thereby the suppression of downstream pro-apoptotic signaling.413 Melanoma cell lines with loss of PTEN are more resistant to BRAF inhibitor.413 More importantly, patients with melanoma carrying wild-type PTEN are reported to have better survival after targeted therapy.414 Besides, loss of NF1 gene that encodes neurofibromin can result in the activation of downstream RAS, PI3K-AKT-mTOR and MAPK pathways through multiple mechanisms, so as to defend against the inhibition of MAPK in response to targeted therapy.415,416 RAC1 mutation is also documented responsible for downstream RAS activation that underlies the intrinsic resistance to BRAF inhibition.417 In a previous clinical study enrolling 45 patients receiving the treatment with BRAF inhibitors, three out of them who had RAC1 mutation revealed no prominent response. The deficiency of RAC1 can amplify the inhibitory effect of BRAF-targeted agent on melanoma cell survival.417 In addition, low MITF/AXL ratio that indicates the phenotypic plasticity of melanoma cells is reported to determine the intrinsic response to targeted therapy. Drug cocktails containing AXL inhibitor can promote melanoma cell elimination by MAPK inhibition.93
During early phase after targeted therapy, intracellular protective signaling pathways are activated to enable tumor cell survival. The establishment of adaptive resistance not only impairs the therapeutic efficacy, but also offers sufficient time to develop acquired resistance.78 Therefore, it is necessary to restrain adaptive resistance so as to delay or block the occurrence of irreversible acquired resistance. Multiple mechanisms are responsible for this process, including resetting of ERK1/2 pathway activation, upregulation of RTKs, MITF upregulation, and metabolic rewiring. To be specific, BRAF-targeted inhibitors would downregulate the expressions of sprouty RTK signaling antagonist 2/4 (SPRY2/4) and dual-specificity phosphatases (DUSPs), which leads to the relief of the feedback suppression on Ras and reactivation of the ERK signaling, thus rendering the treatment resistance.415,418 Besides, adaptive upregulation of multiple RTKs including Erb-B2 receptor tyrosine kinase 3 (ERBB3), PDGFR, EGFR, and FGFR contributes to cell survival and protects tumor cells from apoptosis induced by BRAF inhibition.419–421 A recent study has revealed that it is the downregulation of SOX10 that is responsible for the upregulation of PDGFR and EGFR via the potentiation of TGFβ signal. Meanwhile, the upregulation of ERBB3 after BRAF inhibitor treatment is highly attributed to FOXD3-mediated transcriptional activation.421 What’s more, canonical melanocytic lineage-specific transcriptional factor MITF is also reported to be induced and to promote downstream PGC1α-dependent oxidative phosphorylation.200,422 Therefore, the suppression of MITF and mitochondrial function could be promising in overcoming adaptive resistance of melanoma cells to targeted therapy.
After long-term treatment with MAPK inhibition agents, intracellular mutational alterations would occur to promote the establishment of mutational clones that are irreversibly resistant to targeted therapy. There are a considerable number of alterations of signaling pathways enriched mainly in MAPK signaling, PI3K/AKT signaling and PDGF signaling that mediate this process.423 In particular, the occurrence of RAS, MEK, and NF1 mutations, the amplification of BRAF, the upregulation of COT1 and the alternative splicing of BRAF mutation all contribute to hyper-activation of MAPK cascade,420,424–428 which can impede the suppression of MAPK pathway by BRAF inhibitor. Moreover, IGF-1R upregulation, PTEN loss, PIK3CA missense mutation, and AKT mutation all prominently promote the activation of pro-survival PI3K-AKT pathway, giving compensatory protective action upon the blockade of MAPK pathway.413,428–430 What’s more, the upregulation of EGFR and PDGFRβ has also been documented as a crucial mechanism that activated downstream protective factors to enable tumor cell survival.75,420 In particular, whole-exome sequencing (WES) of melanomas with acquired resistance to BRAF inhibitors has revealed that BRAF gene amplification is identified in around 20% of patients, leading to robust upregulation of BRAF protein expression and the reactivation of ERK in response to BRAF inhibition.426 Besides, p61BRAFV600E splice variant is identified in a subgroup of patients with acquired resistance, resulting in the expression of truncated BRAF proteins that lack the N-terminal RAS-binding domain whereas keep the kinase domain. This alteration helps to form homodimers that is resistant to BRAF inhibitor treatment.431 These reports have pointed out the great implication of BRAF amplification and splicing in acquired resistance. Additionally, the activation of YAP/TAZ pathway also renders acquired resistance to targeted therapy via the transcriptional activation of cell-cycle facilitators.432,433 Downregulation of dual-specificity MAPK phosphatases and ring finger protein 125 (RNF125) can mediate acquired resistance to BRAF inhibitor via the activation of MER-ERK signal pathway and EGFR respectively.434,435 Of note, the signaling pathways contributing to acquired resistance are quite complex, indicating the intricate molecular network in mutational clones induced by prolonged treatment with MAPK inhibition agent.423 Therefore, to overcome the acquired resistance through the usage of single inhibitory agent targeting one specific pathway may be difficult to obtain an extensive benefit with broad coverage.
Combinatorial MAPK inhibition-targeted therapy and alternative targeted agents
The inevitable occurrence of resistance to single-agent targeted therapy has prompted the development of combinatorial regimens. Since that a series of mechanisms facilitate MAPK pathway hyper-activation and make melanoma cells refractory to sole BRAF inhibitor, combined inhibition of MEK is subsequently evaluated, and encouraging outcomes have been obtained in two phase III clinical trials COMBI-v and COMBI-d.436,437 Compared to previous single BRAF-targeted therapy, combined inhibition of both BRAF and MEK lead to the upregulation of clinical response rate from 50% to 60–70%. In addition, the profile of adverse effects is also changed in these two different therapeutic paradigms. The incidence of keratoacanthoma and squamous-cell carcinoma is significantly reduced in combinatorial group compared with BRAF inhibitor single-agent group. After the treatment with vemurafenib or dabrafenib, approximately 15–30% of total patients would develop keratoacanthoma and squamous-cell carcinoma, which is due to the acquisition of RAS mutations and paradoxical activation of CRAF in keratinocytes.438,439 Therefore, based on improved treatment outcome and mitigated adverse effects, the combinatorial therapy dabrafenib plus trametinib has been approved by FDA for treating patients with unresectable or metastatic melanoma harboring BRAFV600E or BRAFV600K mutations.437,440 Later on, the other two BRAFi/MEKi combinations including vemurafenib plus cobimetinib and encorafenib plus binimetinib have been also approved by FDA. Results from clinical trials have revealed that the therapeutic efficacy of these approaches are comparable,21,440–442 and indirect side-by-side analysis also supports this conclusion.443 The development of these combinations provides more options for patients with advanced melanoma harboring BRAF mutation.
It has been reported that various alterations like RAS mutations, feedback reactivation of receptor tyrosine kinases and RAS, BRAF amplification and BRAFV600E splice variants are responsible for acquired resistance to BRAF-targeted therapy by facilitating the dimerization of RAF proteins and subsequent activation of ERK signal,420,426,430,444 causing the so-called paradoxical effect. Therefore, next-generation RAF inhibitors proposed as pan-RAF inhibitors have been developed to obtain the equipotent suppression on both of RAF monomers and dimers,445–448 which is different from vemurafenib or dabrafenib that could only suppress one protomer within the RAF dimer. These pan-RAF inhibitors like TAK-632, LY3009120 and AZ-628 favor catalytic inhibition of both RAF protomers within the dimer via the stabilization of the αC-helix toward the active IN position.447,449,450 Ongoing clinical trials have revealed that these agents are well-tolerant, but can also disturb the aberration of essential MAPK signal in normal tissues and induce additional AEs.451 Of note, due to the lack of selectivity toward the suppression of mutant RAF dimers in cancer cells compared to the inhibition of wild-type RAF dimers in normal cells, LY3009120 showed limited efficacy at its maximum tolerated dose,452 which prompts the generation of selective RAF dimer inhibitors to suppress resistant RAF dimers in tumors more effectively.
Moreover, paradox breakers that are a class of BRAF inhibitors have been recently developed by Plexxikon, aiming to obtain the inhibition of ERK1/2 in BRAFV600E cells without driving paradoxical activation of ERK1/2 in RAS-mutant cells.453 After the investigation of hundreds of vemurafenib derivatives for biochemical activity and cell activity, two chemicals named PLX7904 and its further optimized analog PLX8394 are found to be capable of evading the paradoxical MAPK pathway activation. To be specific, these two drugs can directly suppress the formation of BRAF:CRAF heterodimers normally observed in RAS-mutated cells receiving the treatment of RAF inhibitors, meanwhile maintaining the binding affinity for the dimer partner. Not surprisingly, PLX8394 now has entered the clinical trial stage (ClinicalTrials.gov identifiers: NCT02012231 and NCT02428712).453
Aside from BRAF and MEK inhibitors, alternative molecular targets and related agents have also been investigated in preclinical studies and clinical trials, especially ERK and AKT. In 2013, Morris et al. has identified a selective inhibitor of ERK1/2 called SCH772984 that can eradicate tumor cells harboring BRAF, NRAS, or KRAS mutation within nanomolar concentration. The treatment effect of SCH772984 on melanoma has been well confirmed in preclinical xenograft tumor model.454 Besides, the combination of vemurafenib and SCH772984 has been proved to have synergistic effect on suppressing BRAF-mutant melanoma progression, which can help to delay the onset of the resistance to targeted therapy.455 In this case, SCH772984 could be broadly used in a wide variety of melanomas of distinct genetic backgrounds, only if they manifested hyper-activation of ERK signaling. What’s more, the frequent occurrence of AKT hyper-activation and its crucial tumorigenic role in melanoma have encouraged the investigations of targeting AKT in treating melanoma. GSK2141795 has been discovered as an orally bioavailable and highly potent AKT-specific inhibitor, which exhibits prominent antitumor effects in combination with the MEK inhibitor.456 Of note, the initial combination of AKT inhibitor GSK2141795 and MAPK inhibitors revealed superior growth inhibitory effects compared with the later addition of AKT inhibitors to tumors with acquired resistance to MAPK inhibitors.457 However, dual MEK/AKT inhibition with Trametinib and GSK2141795 did not yield clinical benefits in metastatic NRAS-mutant and wild-type melanoma,458 indicating the limited spectrum of application of AKTi in melanoma with rather high heterogeneity. What should also be noticed is belvarafenib, a potent and selective RAF dimer (type II) inhibitor, exhibits clinical efficacy in patients with BRAFV600E and NRAS-mutant melanomas. The first-in-human phase I study of belvarafenib for melanoma treatment has been conducted (NCT02405065, NCT03118817). In addition, by generating belvarafenib-resistant NRAS-mutant melanoma cells and analyzing circulating tumor DNA from patients treated with belvarafenib, new recurrent mutations in ARAF have been identified to confer the treatment resistance. The combination of RAF plus MEK inhibition may be used to delay ARAF-driven resistance.459
Present advances in therapies targeting tumor immunology and ongoing clinical trials
Immune escape and related signal pathways
Apart from directly targeting driver genes that contribute to tumor intrinsic malignancy, therapies targeting the immune system, which is generally thought to eliminate cancer cells, have shown promising efficacy in melanoma treatment. The importance of the host immune system in eradicating cancer cells has long been appreciated, with instances of spontaneous melanoma regression.460 Thomas and Burnet initially introduced the notion of immunosurveillance, demonstrating that the immune system is responsible for eliminating malignant cells via recognition of tumor-associated antigens,461 and it was further experimentally confirmed by the observation of increased incidence of melanoma in immune-deficient patients.462 However, immunosurveillance is only one part of the complicated interplay between cancer and host immune system. The relationship between cancer and immune cells is composed of three phases: elimination, equilibrium, and escape.462 Elimination indicates a classical view of immunosurveillance. During the early stage of tumorigenesis, the innate immune cells present tumor antigens through binding to MHC and activate adaptive immune cells via co-stimulatory signal, as well as releasing cytokines to eradicate cancer cells. However, the plasticity of melanoma enables transformed tumor cells to evade immunosurveillance,463 indicating the relationship between the two moving to the next phase, equilibrium. In this stage, the host immune system could control the outgrowth of tumor through eliminating immune-sensitive tumors. The last phase, escape, refers to the outgrowth of tumors that have got rid of the restrain of immune system.
Although melanoma represents one of the most immunogenic tumors, which could have elicited adaptive antitumor immune response due to its high mutational burden,464 yet plasticity of melanoma allows it to evade the immunosurveillance of the immune system.465,466 Mechanisms involved in tumor immune escape mainly include: developing lesions in antigen processing, increased resistance to cytotoxicity induced by immune cells and development of immunosuppressive tumor environment.467–469 Tumor cells escape T cell recognition through downregulation of tumor antigens and MHC, as well as impaired antigen processing. On the other hand, immunosuppressive cytokines, such as TGF-β and granulocyte-macrophage colony-stimulating factor (GM-CSF), were released to facilitate the recruitment of MDSCs or the suppression of antigen-presenting cells (APCs). Moreover, melanoma cells recruit immunosuppressive regulatory Treg cells through secreting relevant chemokines, and inhibit the natural cytotoxicity of NK cells against tumor cells through metabolic reprogramming.470,471 Last but not the least, melanoma cells directly induced the exhaustion of cytotoxic T cells by expressing co-inhibitory molecules PD-L1, which binds to the co-inhibitory receptor PD-1 located on the surface of T cells.472–474 Recently, by linking the antigenic specificity of T cell receptors (TCRs) and the cellular phenotype of melanoma-infiltrating lymphocytes at single-cell resolution, the interplay between tumor cell phenotypic characteristics and TCR properties is revealed. Melanoma-reactive lymphocytes predominantly displayed an exhausted state that encompassed diverse levels of differentiation but rarely acquired memory properties, suggesting that tumor specificity shapes the expression state of intra-tumoral CD8+T cells.475 The targets of immunotherapies in the treatment of melanoma mainly focus on the mechanisms associated with the formation of an immunosuppressive environment.
The era of therapies targeting tumor immunology
Cytokines
Since the 1950s, immunotherapy has been an appealing area of cancer treatment research, and attempts to reactivate immune response against tumor cells have been made. However, “Coley’s toxin”476,477 and immune stimulants, such as Bacillus Calmette Guerin,1 which meant to augment non-specific immune responses, have all failed to show significant response in melanoma.423
IFNα-2b, a member of the type I interferon family, demonstrates multiple antitumor activities including the suppression of proliferation and angiogenesis, and the enhancement of antitumor immune response through increasing the cytotoxicity of NK cells and tumor antigen processing.478 In 1995, high-dose IFNα-2b became the first exogenous cytokine approved by FDA for the adjuvant treatment of advanced melanoma.479 Over a decade later, Pegylated IFNα-2b, polyethylene glycol-modified IFNα-2b with a longer half-life also obtained FDA approval, although still with low efficacy and sever toxicity.423,480 Gao et al. found that another type I interferon member, IFNα-1b, which was approved by Chinese Food and Drug Administration for cancer treatment, possessed improved safety and efficacy especially in patients with unresectable metastatic melanoma.365,423
IL-2 is the second cytokine approved by FDA in 1998 for the use of patients with stage IV melanoma.481 As a cytokine, IL-2 could activate antitumor response of cytotoxic T cells and NK cells. Although high-dose IL-2 shows promising results in melanoma with an 18% objective response rate, the severe side effects and low response rates of the regimen restrict the use of it in larger population.481–483 Attempts to improve response rate of high-dose IL-2 have been made by evaluating the efficacy of high-dose IL-2 combined with IFNα, which, however, showed a minimally improved ORR of 25%.484 Since the severe toxicity of high-dose IL-2 and IFN-γ leads to the intolerance of patients to treatment and restrains the therapeutic efficacy in a large population, the combination of chemotherapies, targeted therapies or other immunotherapies with low-dose IL-2 or IFN-γ has been employed in some investigations to avoid the side effects brought by single high-dose cytokines therapy, which can meanwhile achieve better treatment response.481,485–488 The reasons for relatively low response rate of IL-2 probably attributed to the activation of immunosuppressive Tregs.489 Based on this, many modified forms of IL-2, including PEGylation form, antibody-cytokine conjugates, and fusion proteins began to emerge, aiming to extend the half-life of IL-2 and elevate the ORR of patients through inhibiting the binding of IL-2 to CD25 (receptor of IL-2) that lead to the activation of Tregs.490 A preclinical study has revealed that engineered IL-2, a CD25-mimobody, shows lower toxicity and increased potency to IL-2, and it still needs further assessment as a therapeutic agent.491
Oncolytic virus
Oncolytic virus therapy, a new class of antitumor immunotherapy, can lead to tumor regression through selective kill of tumor cells, induction of immunogenic cell death, and stimulation of systemic antitumor immune response.492 Until recent years, clinical benefits have been observed with intra-tumoral administration of oncolytic virus. In 2015, FDA approved the first oncolytic virus therapy, T-VEC for the local treatment of patients with recurrent melanoma that cannot be surgically removed, based on results from a phase III study of patients with metastatic melanoma lesions in skin and nodes. T-VEC is a genetically modified herpes simplex virus type 1 (HSV-1) with reduced virulence and selective proliferation in tumors, encoding GM-CSF, which promotes the priming of T cell responses.493 A randomized open-label phase III clinical trial evaluating the efficacy and safety of T-VEC compared with GM-CSF revealed promising durable clinical benefits with a higher ORR (31.5% vs 6.4%), higher durable response rates (19.0% vs 1.4%), and longer median OS (23.3 months vs 18.9 months) versus GM-CSF alone, as well as favorable safety profile, especially in IIIB-IVM1a melanoma patients.494,495 Subsequent multi-center studies also demonstrated high rates of complete and durable response in advanced melanoma patients administrated with T-VEC,496,497 even for the local lesions of patients who developed acquired resistance to immune checkpoint blockade.498
Therapies targeting immune checkpoints
In the last few decades, immune checkpoint blockades (ICBs) therapy has led to important clinical advances, which holds great promise in cancer treatment. Normally, T cell activation requires two signals upon recognition of tumor antigen presented on the surface of APCs. TCR specifically binds to an antigen in the context of MHC and a co-stimulatory signal transduced by CD28 on T cell surface which could be stimulated by B7 molecules (CD80 and CD86) on the APCs.499,500 The co-receptors on the T cells engaged in secondary signal could be stimulatory or inhibitory. Co-stimulatory molecules such as CD28 and B7 mediate T cell activation, while co-inhibitory molecules including PD-1, CTLA-4, PD-L1, and PD-L2, which are known as “immune checkpoints”, function as T cell brakes.501,502 To date, the most well-studied immune checkpoints are CTLA-4, PD-1, and PD-L1, the blockade of which has shown promising effects against various cancers through reinvigorating antitumor immunity.503–508
Upon T cell activation, CTLA-4 expression is initiated which could bind to B7 molecules with much higher affinity than CD28, resulting in an inhibited immune response.509–511 Preclinical studies have demonstrated that CTLA-4 blockade with an antagonistic antibody leads to improved T cell function and regression of tumor cells in mouse models.512–514 Ipilimumab, a fully human monoclonal antibody against CTLA-4, is the first approved immune checkpoint by the FDA for the use in patients with advanced melanoma in 2011.515,516 Although only a small part of melanoma patients benefits from ipilimumab treatment and noticeable side effects associated with immune-related adverse events (irAE) could occur, patients with unresectable advanced melanoma treated with ipilimumab have a long-term survival effect.517–519
PD-1 is another co-inhibitory receptor expressed on the surface of T cells upon T cell activation, which could bind to its receptor including PD-L1 and PD-L2, on the surface of tumor cells or other immune cells within the tumor environment to control the cytolytic function of effector T cells through activation of the tyrosine phosphatase SHP1/2 signaling.24,520 As such, antibodies against PD-1/PD-L1 are supposed to be effective for the treatment of cancers via the invigoration of infiltrating CD8+T cells.521 Pembrolizumab and nivolumab are the two PD-1 inhibitors approved by FDA in 2014. Both nivolumab and pembrolizumab demonstrated improved survival benefits versus ipilimumab522–526 and chemotherapies, such as dacarbazine,527–529 with a prolonged progression-free survival and overall survival and elevated response rates. Clinical trials have revealed the response rate for ipilimumab-refractory patients treated with nivolumab or pembrolizumab was 30%, and 1-and 2-year survival rates were 68.4% and 31.2%.530,531 Due to the improved efficacy and much more tolerable toxicity of nivolumab and pembrolizumab than that of ipilimumab, PD-1 inhibitors have become the first-line treatment of patients with BRAF-wide-type metastatic melanoma.532 Nivolumab and pembrolizumab have also shown clinical benefits in patients with other types of melanoma including untreated melanoma brain metastases,533–535 uveal melanoma,536 acral lentiginous melanoma and mucosal melanoma,537 resulting in 46%, 19%, 18.8%, and 20.8% ORR, respectively. In addition, pembrolizumab has shown antitumor activity in aged melanoma patients,538 but not in pediatric melanomas.539 Toripalimab, a selective recombinant human PD-1 monoclonal antibody, has been approved by China FDA in 2018 for the treatment of a variety of cancers, including melanoma.540 Treatment with toripalimab resulted in an ORR of 20.7% in patients with advanced melanoma, most of which are acral and mucosal melanoma.540–542 Base on the promising clinical results seen with toripalimab-treated mucosal melanoma, FDA has granted toripalimab a fast track designation for use in the frontline treatment of patients with mucosal melanoma. HX008, another humanized IgG4 monoclonal antibody against PD-1 showed favorable clinical benefits for Asian melanoma patients, who had been treated with chemotherapy, targeted therapy, or immunotherapy, with an ORR of 20.2% according to the results from a phase II study.543
Given that PD-1 and CTLA-4 participate in different processes of T cell recognition and cytotoxic T cell reinvigoration, combinations of antibodies against PD-1 and CTLA-4 are of great potential to induce tumor regression synergistically. Clinical trials of combined nivolumab and ipilimumab versus nivolumab or ipilimumab monotherapy in previously untreated melanoma have revealed that patients treated with nivolumab plus ipilimumab obtained durable and sustained clinical benefits.544–547 The 3-, 4- and 5-years overall survival rates were 58%, 53%, and 52% for the patients treated with nivolumab and ipilimumab, 52%, 46%, 44% for the nivolumab group, as compared with 34%, 30%, 26% for the ipilimumab group, respectively.525,548,549 Based on the results of the trials, FDA-approved combined nivolumab and ipilimumab for the frontline use of patients with advanced melanoma in 2015. However, the combination of nivolumab and ipilimumab also leads to increased treatment-related adverse events compared with nivolumab and ipilimumab (59% vs 23% and 28%).548 A subsequent clinical trial was conducted in melanoma patients with sequential administration of nivolumab and ipilimumab as well as the reverse sequence, and demonstrated that nivolumab followed by ipilimumab have a lower toxicity and similar clinical benefits with co-administration of nivolumab and ipilimumab.550 Studies of modulated dosing regimen for nivolumab plus ipilimumab trying to decrease the toxicity display a lower incidence of treatment-related adverse events without weakening the antitumor activity for the treatment of higher/standard-dose nivolumab and a lower-dose ipilimumab.551 Of note, improved antitumor response and tolerable safety are seen in anti-PD-1/PD-L1-refractory melanoma patients with pembrolizumab plus low-dose ipilimumab.552 The treatment of other types of melanoma excluding cutaneous melanoma is of great challenge for their relatively insensitive response to existing therapies. The combination of nivolumab and ipilimumab has been subsequently evaluated in non-cutaneous melanoma including acral, mucosal, and uveal melanoma, and the combination regimen shows sustained and improved response compared with either single agent, although with elevated toxicity but manageable safety profile.553–555 Clinical trials have also been conducted in another population of patients with brain metastasis of melanoma, which were generally excluded from clinical trials for their poor prognosis. In two phase II studies, combination of nivolumab and ipilimumab displays similar intracranial antitumor effects in a relatively small population.534,556 Another clinical trial of 380 patients with asymptomatic and symptomatic melanoma brain metastasis also shows that the 2- and 3-years overall survival rates are 41% and 30% with the combination regimen, respectively.557 The combination of another standard-dose anti-PD-1, pembrolizumab, and lower-dose ipilimumab provides robust clinical benefits with a 3-year ORR of 62.1%,558,559 while studies of standard-dose pembrolizumab plus alternate-dose ipilimumab shows that pembrolizumab 200 mg plus ipilimumab 50 mg could prominently reduce the toxicity of the combination therapy.560
PD-L1 and PD-L2 that are PD-1 ligands expressed on tumors cells contribute to cancer cell evasion. Most melanomas were reported to highly express PD-L1.561 Actually, PD-L1 is expressed in tumor cells and myeloid cells in the tumor environment mainly mediated by constitutive activation of oncogenic signal pathways in tumor cells and IFNγ signaling.499,562 The expression of PD-L2 is highly upregulated in certain B cell lymphoma.501 Given that PD-1 binds to both PD-L1 and PD-L2, and PD-1 interacts with either CD80 or PD-L1, the antibodies targeting PD-1 or PD-L1 may lead to different antitumor effects and toxicities,563 which has been supported by relevant clinical trials of anti-PD-L1. Although single use of antibodies against PD-L1 such as durvalumab, avelumab, and atezolizumab has been approved for the use in patients other than melanoma, yet investigations of PD-L1 antibodies in combination with targeted therapy or/and chemotherapy are still underway in the treatment of melanoma. Notably, FDA has approved atezolizumab for unresectable or metastatic melanoma harboring BRAF mutation in combinatorial regimens with targeted therapies.
Adjuvant and neoadjuvant therapy
For advanced melanoma patients with high risk of recurrence, adjuvant therapy is offered to lower the risk of recurrence after the surgery. High-dose IFNα-2b of pegylated IFN had been the sole approved adjuvant therapy in the treatment of melanoma for a long time. However, adjuvant IFNα-2b showed marginally significant and slightly diminished positive effects on the recurrence-free survival (RFS) and OS of resected stage III melanoma patients, concomitant with intolerable toxicity.564,565 In 2005, adjuvant ipilimumab at a dose of 10 mg/kg resulted in significantly higher rates of RFS and OS, although with higher rates of immune-related adverse events than with placebo, which led to the approval of adjuvant treatment of ipilimumab for stage III melanoma patients.566 A phase III clinical trial demonstrated that adjuvant ipilimumab (10 mg/kg) was not superior in efficacy to IFNα-2b, but ipilimumab (3 mg/kg) significantly improved the OS with lower toxicity compared with high-dose IFNα-2b.567 In 2017, adjuvant anti-PD-1, nivolumab, showed longer RFS and lower rates of high-grade adverse events than adjuvant with ipilimumab among patients with resected stage III or IV melanoma in a phase III trial, Checkmate 238.568 Comparisons of another anti-PD-1, pembrolizumab with placebo also showed improved efficacy and favorable safety profile.569–571 Based on those trials, nivolumab and pembrolizumab had been approved for adjuvant treatment of patients with unresected stage III melanoma. Clinical trials of combining nivolumab and ipilimumab or nivolumab monotherapy in patients with stage IV melanoma are still ongoing.572 Besides, combined targeted therapies like BRAF inhibitor dabrafenib plus MEK inhibitor trametinib have demonstrated improved RFS and tolerable toxicity in patients with stage III BRAFV600R/K-mutant melanoma compared with the adjuvant use of placebo.573 Both anti-PD-1 treatment and BRAFi plus MEKi therapy are the frontline options for adjuvant therapy. However, how to choose between the two still needs further investigations.
Although no neoadjuvant treatment has been approved by the FDA, neoadjuvant therapy appears promising in the treatment of patients with high-risk melanoma. Ongoing clinical trials of neoadjuvant ipilimumab plus nivolumab have showed high pathological response rates in patients with macroscopic stage III melanoma, and it needs further investigation to preserve efficacy and reduce toxicity.574
Signal pathways of resistance to ICBs
Although therapies targeting immune checkpoints have achieved better outcomes in patients of a variety of cancer types, only a minority of patients obtains a durable benefit from the treatment of ICBs, and some initial responders even have their tumors progressing on after a period of response. Unveiling the mechanism underlying the patients who do not respond or sustainably respond to ICBs is appealing for scientists. Resistance to ICBs could be classified into two categories: primary resistance and acquired resistance. Primary resistance refers to patients who do not respond to the initial ICB therapy. Acquired resistance means the cases in which patients have response to ICB therapy initially, but have tumor progression after a duration of therapy.575 Investigations to find out possible predictors of response to immunotherapy blockade have revealed that PD-L1 expression, tumor mutational burden,576,577 tumor intrinsic oncogenes, such as IFN-γ, p53, and Wnt signaling,578–581 signatures of T cell dysfunction and antigen presentation expression,582–586 gut microbiota and its derived metabolites587–589 are all significantly associated with clinical benefit. PD-L1 expression has been shown to identify melanoma patients who are more likely to respond to PD-1 inhibitors.590 However, it is not recommended to take PD-L1 expression into account for treatment decisions because of the imperfect correlation between PD-L1 expression and clinical benefits from PD-1 inhibitors.525,591 Higher tumor mutational burden is associated with better response to ICBs, which is thought to enhance antitumor immune response through augmenting neoantigen formation. IFN-γ signaling plays a pivotal role in stimulating antitumor response mainly through activating cytolytic T cells and promoting tumor antigen presentation,592 and IFN-γ signaling profile is related to response to ICBs.353 Activation of Wnt or Braf signaling as well as loss function of PTEN partially mediates ICBs resistance. The relationship of host gut microbiome and resistance to ICBs is complex and not elucidated. High level of microbiome-derived metabolites, especially short-chain fatty acids, is reported to mediate the resistance to CTLA-4 antibody via restraining the function of DCs and T cells.589 However, mechanisms of acquired resistance are far from fully understood. Similar to primary resistance, defects in antigen presentation and IFN-γ signaling, neoantigen depletion, and anergic T cells in tumor all contribute to acquired resistance to ICBs therapies.593 To overcome the resistance to ICBs, substantial efforts have been made on combinatorial approaches to broaden the responders and lower the toxicities.
Combinatorial therapies targeting tumor immunology and mutated driver genes
With the disclosure of mechanisms underlying cancer growth and interactions between cancer cells and tumor environment, therapies targeting different intra-and extra-tumor processes undoubtedly increased. The pluralistic targets in cancer treatment provide a great potential for combinatorial regimens. While monotherapy may not display an optimistic clinical benefit or safety profile, harnessing combination approaches to maximize the antitumor effects while minimizing toxicities seems to be a promising strategy for cancer treatment. There are many clinical trials of combinatorial regimens under development, and most of them are combining ICBs, anti-PD-1/PD-L1/CTLA-4, along with targeted therapy or other immunomodulatory approaches.
Recently, engineered forms of cytokines with lower adverse effects and enhanced efficacy in combination with ICBs have also raised great interest. NKTR-214 (Bempegaldesleukin), a pegylated IL-2, preferentially binds to CD122 other than CD25, which leads to enhanced activation of T cells and NK cells and reduced toxicity resulting from Tregs activation.594 NKRT-214 plus nivolumab is well tolerated and has promising clinical efficacy with an overall ORR of 59.5%.595 Combination of pembrolizumab and pegylated IFNa-2b shows improved response rates in advanced melanoma.596,597 The combination of ICBs and T-VEC is appealing for the reason that T-VEC could activate antitumor immune response through promoting the expression of IFN-γ and immune checkpoints such as PD-L1, as well as stimulating tumor antigen presentation.598 Patients with advanced melanoma treated with combination of T-VEC and pembrolizumab have an elevated ORR of 62%.598 Consistently, combination of T-VEC and ipilimumab shows greater antitumor activity in the treatment of advanced unresectable melanoma without additional safety concerns versus ipilimumab.599,600 Adoptive cell therapy (ACT) is also a promising therapy for the patients who are refractory or non-tolerant to current first-line therapies. With the development of cellular therapy, ACT could be divided into three types according to a different mechanism of action: isolated tumor-infiltrating T cells from resected tumor, T cells with engineered chimeric antigen receptors (CAR-Ts) and T cells with engineered T cell receptor (TCR-Ts).601 The initial study of ACT using high-dose IL-2 combined with autologous TILs expanded in vitro after the treatment of chemotherapy achieved 60% objective regression of melanomas.602 Systematic analysis of the efficacy of ACT with TIL plus high-dose IL-2 showed durable clinical benefits in the treatment of advanced melanoma.487 Notably, adoptive transfer of TILs has achieved an ORR of 24% in patients who are refractory to PD-1 antibody603 and showed antitumor effects in acral and mucosal melanoma patients.604 Recently, long-term follow-up of lifileucel (LN-144) cryopreserved autologous TIL therapy reveals a promising effectiveness, with an ORR of 36.4% in pretreated melanoma patients who failed on first-class targeted therapy or ICBs.605 A trial of adoptive transfer of TIL engineered with IL-12 has demonstrated antitumor activity but with high toxicities.606 Combining ACT with TIL and IFN-α provides better median OS and disease-free survival in Chinese resected stage III melanoma patients.607 Although MART1-specific TCR-Ts shows clinical potency for melanoma patients, the specific loss of MART1 of tumor cells renders low response rates of the therapy.601 CAR-Ts have been approved for the treatment of hematologic tumor, but it did not show the same antitumor effects, not to mention severer toxicities, in melanoma treatment.608
The improvement of targeted therapies and immune checkpoint inhibitors has shed a light on the treatment of melanoma patients. BRAF and MEK inhibitors displayed high ORR, yet only lower than half of patients with BRAFV600-mutated melanoma could obtain long-term benefits form BRAF inhibitors.609 The ORR of ICBs is relatively lower than targeted therapies, although ICBs provide durable responses.610,611 Given that complementary clinical profiles led by targeted therapies and ICBs, the proposal that combinatorial regimen of these two therapies might provide durable response and elevated ORR, as well as lower toxicity are of great interest. Ample evidence has proved that BRAF and MEK inhibitors could promote the priming and function of tumor infiltration T cells, facilitate antigen presentation and modulate the tumor environment to be harmful for tumor cells in mouse model and in vitro.612–617 However, the combination of CTLA-4 inhibitor, ipilimumab, with BRAF inhibitor, vemurafenib, or dabrafenib and trametinib fails for severe liver or gastric toxicity.618,619 Subsequent studies focused on the combination of BRAF/MEK inhibitors and PD-1/PD-L1 blockades, which have better tolerable safety profiles than CTLA-4 blockades. Clinical trials of the triple combination of PD-L1 antibody, atezolizumab, BRAF inhibitor, vemurafenib and MEK inhibitor, cobimetinib result in significantly prolonged progression-free survival compared with the combination of vemurafenib and cobimetinib (15.1 months vs 10.6 months), and manageable toxicity.620,621 Based on the favorable results of the triplet combination, the FDA-approved atezolizumab in combination with cobimetinib and vemurafenib for patients with BRAFV600 mutated unresectable or metastatic melanoma in 2020. A phase II study of pembrolizumab, dabrafenib, and trametinib in BRAF-mutant melanoma has revealed improved PFS and OS. A nearly 3 years follow-up suggested a 16.9-months median PFS of the treatment of triplet combination and 10.7-months of the treatment with dabrafenib and trametinib.622 Combination of an investigational PD-1 antibody, spartalizumab, dabrafenib and trametinib leads to an ORR of 78% in advanced BRAF-mutant melanoma, including 44% complete response, which suggests the triplet combinatorial regimen is promising, although in a relatively small population.623 With the successful trials of combined targeted therapies and ICBs, numerous studies of combinatorial therapies begin to emerge. Clinical trials of combining PD-1/PD-L1/CTLA-4 blockades with MAPK inhibitors, EGFR inhibitors or BRAF and MEK inhibitor are underway in BRAF-mutant and wide-type melanoma.624–628 Recent studies of combined VEGF inhibitor, apatinib, and an investigational humanized IgG4 monoclonal antibody against PD-1, camrelizumab demonstrates an ORR of 22.2% in advanced untreated acral melanoma patients,629 and longer follow-up time is needed to confirm the efficacy of the combinatorial regimen. A retrospective study evaluating the efficacy and safety of VEGF inhibitor, axitinib plus anti-PD-1 provides improved clinical benefits, with an ORR of 24.5%.630 The latest results of trials evaluating atezolizumab in combination with VEGF inhibitor, bevacizumab, for the use in patients with advanced mucosal melanoma shows promising benefit with an unconfirmed ORR of 42.9% in a relatively small population, and phase III trials with large population was needed to confirm the benefits.631
Novel targeted therapies for re-activating antitumor immunity and ongoing clinical trials
As we learn more about the mechanisms underlying cancer evasion, other new inhibitors and stimulatory immune checkpoints regulating function of T cells have sprung out in recent years. Blocking antibodies specific for those inhibitory receptors are under investigations, although most of them are far from clinical application. The majority of ongoing investigations about newly-discovered immune checkpoints are combination therapies with PD-1 or PD-1 plus CTLA-4 antibodies to broaden the responders or reduce toxicities (Table 3).
Table 3.
Ongoing mono- or combination clinical trials for melanoma treatment
| Target | Agents | Combinations | Trial identifier | Phase | Type of tumor | Status and results |
|---|---|---|---|---|---|---|
| CTLA-4 | Ipilimumab | Plus TLR9 agonist IMO-2125 | NCT03445533 | III | Anti-PD-1 refractory melanoma | Active, not recruiting |
| Plus VEGFR antagonist bevacizumab | NCT00790010 | I | Unresectable stage III/IV melanoma | Active, not recruiting | ||
| Plus VEGFR inhibitor axitinib | NCT04996823 | II | Advanced melanoma | Recruiting | ||
| Plus multi-kinase inhibitor cabozantinib | NCT04091750 | II | Advanced melanoma | Recruiting | ||
| Plus CCR4 antagonist FLX475 | NCT04894994 | II | Advanced melanoma refractory to anti-PD-1/PD-L1 antibody | Recruiting | ||
| PD-1 | Nivolumab | Plus multi-kinase inhibitor cabozantinib | NCT04091750 | II | Unresectable advanced melanoma | Recruiting |
| Plus HDAC inhibitor tinostamustine | NCT03903458 | I | Refractory, locally advanced, or metastatic melanoma | Recruiting | ||
| Plus encorafenib and binimetinib | NCT04511013 | II | BRAF V600 mutant melanoma with brain metastasis | Recruiting | ||
| Pembrolizumab | Plus dabrafenib and trametinib | NCT02130466 | I/II | Advanced melanoma | Completed | |
| Plus encorafenib and binimetinib | NCT02902042 | I/II | Unresectable/metastatic BRAF V600 mutant melanoma | Completed | ||
| NCT04657991 | III | Unresectable/metastatic locally advanced BRAF V600 mutant melanoma | Recruiting | |||
| Plus HDAC inhibitor 4SC-202 | NCT03278665 | I/II | Anti-PD-1 therapy refractory/non-responding melanoma | Recruiting | ||
| HX008 | Monotherapy | NCT04749485 | II | Locally advanced or metastatic melanoma refractory to the standard treatments | Active, not recruiting | |
| Plus anti-PD-L1 antibody LP002 | NCT04756934 | I | Advanced or metastatic melanoma who have failed previous anti-PD-1/PD-L1 | Recruiting | ||
| Plus oncolytic virus HG52 | NCT04616443 | Ib/II | Melanoma | Recruiting | ||
| Camrelizumab/SHR-1210 | Plus VEGF inhibitor apatinib | NCT03955354 | II | Advanced acral melanoma | Recruiting | |
| Plus VEGFR antagonist bevacizumab | NCT04091217 | II | Unresectable locally advanced or metastatic mucosal melanoma | Recruiting | ||
| Spartalizumab | Plus dabrafenib and trametinib | NCT02967692 | III | Previously untreated unresectable/metastatic BRAF V600 mutant melanoma | Active, not recruiting | |
| PD-1 and CTLA-4 | Nivolumab plus ipilimumab | Plus BRAF/MEK inhibitor, vemurafenib, and cobimetinib | NCT02968303 | II | Unresectable/metastatic melanoma | Recruiting |
| Plus dabrafenib and trametinib | NCT01940809 | I | Unresectable/metastatic BRAF V600 mutant melanoma | Active, not recruiting | ||
| Plus dabrafenib and trametinib | NCT02224781 | III | Unresectable/metastatic BRAF V600 mutant melanoma | Recruiting | ||
| PD-L1 | Atezolizumab | Plus cobimetinib | NCT03273153 | III | Previously untreated advanced BRAF V600 wild-type melanoma | Completed |
| NCT01988896 | I | Locally advanced/metastatic solid tumors | Completed | |||
| Avelumab | Plus other cancer immunotherapies | NCT02554812 | II | Advanced tumors | Active, not recruiting | |
| LAG-3 | Relatlimab/BMS-986016 | Plus nivolumab | NCT03470922 | III | Advanced melanoma | Active, not recruiting |
| Plus nivolumab | NCT04552223 | II | Uveal melanoma | Recruiting | ||
| IMP-321/Eftilagimod alfa | Plus pembrolizumab | NCT02676869 | I | Unresectable/metastatic melanoma | Completed | |
| TIM-3 | TSR-022 | Plus PD-1 inhibitor dostarlimab (TSR-042) | NCT04139902 | II | Resectable regionally advanced or oligometastatic melanoma | Recruiting |
| INCAGN02390 | Plus anti-LAG-3, INCAGN02385 and anti-PD-1 INCMGA00012 | NCT04370704 | I/II | Advanced tumors | Recruiting | |
| LY3321367 | Plus anti-PD-L1 antibody LY3300054 | NCT02791334 | I | Advanced refractory solid tumors | Active, not recruiting | |
| TIGIT | Vibostolimab | Plus pembrolizumab | NCT04303169 | I/II | Neoadjuvant therapy to advanced melanoma | Recruiting |
| Plus pembrolizumab | NCT04305054 | I/II | Advanced melanoma | Recruiting | ||
| Plus pembrolizumab and anti-CTLA-4, quavonlimab | NCT04305041 | I/II | PD-1 refractory melanoma | Recruiting | ||
| 4-1BB | PF-05082566 | Plus OX40 agonist PF-04518600 | NCT02315066 | I | Locally advanced/metastatic melanoma | Completed |
| Plus CD20 antibody rituximab | NCT01307267 | I | Advanced tumors | Completed | ||
| OX40 | MEDI0562 | Plus tremelimumab or durvalumab | NCT02705482 | I | Advanced solid tumors | Completed |
| INCAGN01949 | Plus nivolumab or/and ipilimumab | NCT03241173 | I/II | Advanced/metastatic tumors | Completed | |
| INBRX-106 | Plus pembrolizumab | NCT04198766 | I | Locally advanced/metastatic solid tumors | Recruiting | |
| GSK3174998 | Plus pembrolizumab | NCT02528357 | I | Advanced solid tumors | Completed | |
| CD40 | CP-870893 | Plus tremelimumab or durvalumab | NCT01103635 | I | Metastatic melanoma | Completed |
| APX005M | Plus nivolumab and cabiralizumab | NCT03502330 | I | Advanced melanoma | Recruiting | |
| Plus nivolumab | NCT03123783 | I/II | Metastatic melanoma | Completed | ||
| Plus pembrolizumab | NCT02706353 | I/II | Metastatic melanoma | Recruiting | ||
| JNJ-64457107/ADC-1013 | monotherapy | NCT02829099 | I | Advanced tumors | Active, not recruiting | |
| CDX-1140 | Monotherapy | NCT03329950 | I | Advanced tumors | Recruiting | |
| GITR | TRX518 | Plus gemcitabine, pembrolizumab, or nivolumab | NCT02628574 | I | Advanced solid tumors | Completed |
| GWN323 | Plus spartalizumab | NCT02740270 | I | Advanced solid tumors and lymphomas | Completed | |
| MK-4166 | Plus pembrolizumab | NCT02132754 | I | Advanced solid tumors | Completed | |
| IDO1 | Epacadostat | Plus pembrolizumab | NCT02752074 | III | Unresectable/metastatic melanoma | Completed |
| IL-2 | NKTR-214 | Plus nivolumab | NCT03635983 | III | Previously untreated inoperable or metastatic melanoma | Recruiting |
| IL-6 | Tocilizumab | Plus nivolumab and ipilimumab | NCT03999749 | II | Unresectable/metastatic melanoma | Recruiting |
| TNF-a | Inflimab/certolizumab | Plus nivolumab and ipilimumab | NCT03293784 | I | Advanced melanoma | Recruiting |
| Oncolytic virus | T-VEC | Plus nivolumab | NCT04330430 | II | Resectable early metastatic melanoma with injectable disease (NIVEC) | Recruiting |
| Plus ipilimumab | NCT01740297 | I/II | Advanced/unresectable melanoma | Completed | ||
| ACT | Lifileucel | Monotherapy | NCT02360579 | II | Metastatic melanoma | Active, not recruiting |
LAG-3
Lymphocyte activation gene-3 (LAG-3) is a T cells-associated inhibitor receptor that co-expressed with PD-1 on anergic or exhaustion T cells.632 Preclinical studies have shown that LAG-3 and PD-1 blockades synergistically stimulate T cell responses and decrease tumor burden in murine model.633–635 There are two LAG-3 inhibitors in clinical trials, IMP-321 and relatlimab (BMS-986016). The latest results of a phase III clinical trials combining relatlimab and nivolumab have demonstrated an improved antitumor activity in patients who showed tumor progression on PD-1 treatment, with a prominently prolonged median PFS versus nivolumab monotherapy (10.1 months vs 4.6 months) companied with higher incidence of treatment-related adverse events in the treatment of advanced melanoma.636,637 Clinical trials aiming to evaluate the safety and efficacy of relatlimab plus nivolumab are recruiting melanoma patients whose disease progressed on PD-1 monotherapy or naive to prior immunotherapy, as well as relatlimab in uveal melanoma.638
TIM-3
T cell immunoglobulin and mucin domain-containing protein 3 (TIM-3), with multiple ligands including galectin-9, high mobility group box 1 (HMGB1), and carcinoembryonic antigen-related cell-adhesion molecule 1 (CEACAM-1), functions as a co-inhibitory receptor on dysfunctional T cells.639 Co-inhibition of PD-1 and TIM-3 has demonstrated antitumor activity in preclinical studies,640 which leads to the development of TIM-3 blockades for clinical application. Phase I/II clinical trials have been initiated with the single use of TIM-3 antibody or combination with PD-1 blockade in melanoma. TIM-3 antibodies in clinical study mainly contain sabatolimab (MBG453), TSR-022, INCAGN02390, and LY3321367. While no response was seen with sabatolimab in advanced solid tumor including melanoma, patients undergoing the treatment of sabatolimab plus PD-1 antibody exhibit better response signs like elevated expression of immune markers.641,642 LY3321367 has demonstrated promising antitumor activity in single use or in combination therapies with PD-L1 antibody for the treatment of advanced cancers, with 68.2% and 88.2% response rate, respectively (NCT03099109). Studies on a bispecific antibody targeting both TIM-3 and PD-1 are also recruiting patients with advanced tumors including melanoma.
TIGIT
T cell immunoglobulin and ITIM domain (TIGIT), a promising new target for cancer immunotherapy, is upregulated mainly on activated T cells and NK cells as a co-inhibitory receptor.643 TIGIT impedes T cell and NK cell antitumor activities through the competing with CD226 binding to CD155 and CD122, two ligands on the surface of melanoma cells and APCs.644–647 Despite numerous clinical studies on TIGIT blockades, promising clinical results were merely seen with dual therapy of PD-1 blockades and only one TIGIT inhibitor, vibostolimab, for the treatment of advanced melanoma or PD-1-refractory melanoma.
T cells and NK cells express several cell surface co-stimulatory receptors which belong to TNFR family that induce the effector function of T cells and NK cells in tumor environment.648 Members of TNFR family including CD137, OX40, GITR, CD40, and CD27 have long been considered as viable immunotherapy targets.
CD137 (4-1BB), induced upon TCR stimulation, demonstrates co-stimulatory activities through boosting T cell proliferation, facilitating memory differentiation, and enhancing effector functions of both T cells and NK cells once binding to its ligand CD137L expressed on APCs.649 Although agonistic CD137 antibodies alone could not reinvigorate antitumor immunity against melanoma, CD137 agonists synergistically suppress melanoma with chemotherapy, radiotherapy, and other immunotherapy modalities including adoptive T cell therapy, ICBs, virotherapy and vaccines in mouse model and in vitro.650–660 However, clinical trials of the first agonistic CD137 antibody, urelumab (BMS-663513), have been hampered for severe liver toxicity in the treatment of advanced melanoma.661 Clinical trials of combinatorial regimen with urelumab and nivolumab are underway against melanoma. A phase Ib clinical study of utoliumab (CD137 agonist) plus pembrolizumab revealed improved safety but no synergic effects in advanced solid tumors.662 Combinatorial approaches of either optimized (LVGN6051) or lower dose (urelumab) of CD137 agonist and other immune modulators are also ongoing.663 Bispecific antibody (INBRX-105) targeting PD-L1 and 4-1BB also simultaneously enter the clinical use with tolerable safety and improved efficacy profiles.
OX40
OX40 (CD134) that belongs to TNFR superfamily 4 is mainly expressed on CD4+ T cells, CD8+ T cells, neutrophils, and NK cells driven by TCR-engaged activation.664 OX40 agonists promoted effector T cell expansion and survival, as well as depleted tumor-infiltrating Tregs.665 Preclinical studies have revealed that MEDI6383, a human OX40 ligand fusion protein, has the potential to boost antitumor immunity in human cancers,666,667 and clinical studies to evaluate the efficacy and safety of MEDI6383 is ongoing.668 Another CD134 agonist, MEDI0562, in combination with durvalumab or tremelimumab has demonstrated tolerable safety and clinical benefits, with median overall survival of 17.4 and 11.9 months for MEDI0562 plus durvalumab and MEDI0562 plus tremelimumab, respectively, in the treatment of advanced solid tumors.669,670 Combinatorial regimens of PF-8600 (OX40 agonist) and utomilumab (4-1BB agonist) has demonstrated a tolerable safety profile and clinical benefits with 70% melanoma patients achieving stable disease.671 A phase I dose-escalation trial of INBRX-106, a novel hexavalent OX40 agonist, has revealed the safety profile and clinical benefits in patients with a range of cancer types. Clinical trials of INBRX-106 with or without pembrolizumab are recruiting for the treatment of advanced tumors including melanoma. MOXR0916, an agonist monoclonal antibody targeting OX40, is under clinical investigations in combination with atezolizumab.672 GSK3174998 and BMS-986178, both of which are humanized IgG1 agonistic OX40 monoclonal antibodies, have very modest combination therapeutic activities with ICBs against advanced solid tumors, although with a tolerable safety profile.673,674 Other forms of immunotherapy modalities targeting OX40L are being developed to overcome the low efficacy of OX40 agonists. SL-279252, a first-in-class agonist redirected checkpoint fusion protein including PD-1 and OX40L, the clinical trial of which is also ongoing. DNX-2440, a tumor-selective conditionally replicative oncolytic adenovirus expressing OX40L is being developed for the treatment of cancers with liver metastasis.
GITR
Similar to CD137 and CD134, glucocorticoid-induced TNFR related protein (GITR) modulates T cell activation by providing a co-stimulatory signaling. However, clinical trials of GITR agonists, TRX518, AMG228, MK-1248, MK-4166, BMS986156, MEDI18730, and GWN323 showed limited antitumor activities in monotherapy, although with tolerable safety profiles.675–681 These studies have implicated that GITR agonists alone could not reactivate the cytolytic function of T cells in tumor environment, and combinatorial approaches of GITR agonism and other immunomodulatory therapies are of great interest. Responses were observed in ICBs-naive melanoma patients (ORR, 62%) with the combination of MK-4166 and Pembrolizumab.677
CD40
CD40, also known as TNFRSF5, is mainly expressed on DCs, B cells, and macrophages whereas its ligand, CD154, is transiently expressed by activated T cells.682 CD40 agonists could lead to T cell activation through increased antigen presentation and elevated level of critical T cell stimulatory cytokines.683 CP-870893, a fully human IgG2 monoclonal antibody against CD40, exhibited well tolerable safety profile and clinical benefits. 27% of melanoma patients had objective partial responses with the treatment of CP-870893, which, however, was not reproducible.684,685 Combined regimens containing CP- 870893 with tremelimumab or the immune stimulant, oncovir poly IC:LC, along with a melanoma vaccine, NY-ESO-1/gp100, are under evaluation in a phase I trial in patients with melanoma. Another CD40 agonist, APX005M and the inhibitor of another macrophage polarizing regulator CSF1R, cabiraluzumab with or without nivolumab demonstrated tolerable safety profile, warranting further investigations into the optimization of the dosing and selection of patients.686 The investigations on the combinations of APX005M and nivolumab or pembrolizumab are still ongoing. Other CD40 agonists entering the clinic including SEA-CD40, ADC-1013, and CDX-1140 are being tested for single use or in combination with either chemotherapy, vaccines, or ICBs in early clinical trials in patients with advanced melanoma.687,688
IDO1
Indoleamine 2,3-dioxygenase 1 (IDO1) is an intracellular IFNγ-inducible enzyme that converts tryptophan to kynurenine, which leads to a suppressed tumor environment through impairing T cell proliferation and activity due to amino acid deficiency and promoting the differentiation of Tregs.682 IDO1 is overexpressed in a variety types of cancers including melanoma.689 Low level of IDO1 in melanoma metastasis is associated with improved overall survival and could predict the outcome of metastatic melanoma patients with immunotherapies.690–692 Preclinical studies have implicated that pharmacological inhibition of IDO1 enhanced T cell response and impeded tumor growth.693–698 Epacadostat, a selective reversible IDO1 inhibitor, has no effect as monotherapy or in combination with pembrolizumab for the treatment of multiple types of solid tumors. Although epacadostat achieved a favorable response rate of 55% in combination with pembrolizumab for the use in patients with solid tumors from a phase I/II trial, which led to a phase III study to evaluate the efficacy of epacadostat in 706 unresectable melanoma patients, the results from the larger trial revealed that epacadostat plus pembrolizumab did not provide an improved overall survival or progression-free survival.699,700 Due to the negative results of the trial, multiple clinical trials of epacadostat were halted. Another IDO1 inhibitor, BMS-986205, in combination with ipilimumab showed modest clinical benefit with an ORR of 26%.701 Clinical trials of other two IDO1 inhibitors, indoximod and navoximod demonstrated tolerable safety and antitumor efficacy worthy of further evaluations in advanced melanoma.702,703
Agents targeting other immune inhibitor receptors, such as B7-H3, BTLA, and VISTA, stimulatory receptor including CD27 and CD70 and checkpoints on NK cells containing NKG2A and KIR family are also under evaluation for further clinical application in melanoma patients. Besides, other immunomodulatory molecules, TLR, TNFα and IL-10 are also appealing for potential combination with ICBs to treat melanoma.704,705
Conclusion and future perspectives
Melanoma is the most lethal skin cancer that results from the malignant transformation of melanocytes. Intense UVR, multiple moles, family history, and fair skin are the main risk factors associated with increased incidence of melanoma. In the past few decades, the therapeutic approaches have gained revolutionary advances due to a deeper understanding of the molecular mechanisms underlying melanoma pathogenesis. In particular, the wide application of targeted therapy and immunotherapy has substantially improved the 5-year-survival of patients with advanced melanoma from <10% to around 30%. Nevertheless, the prognosis of patients remains suboptimal because of the low response rate and frequent occurrence of treatment resistance to currently available therapies. Therefore, it is of necessity to obtain a more comprehensive understanding of the mechanisms driving distinct aspects of melanoma biology, including mutated driver genes, transcriptional regulation of tumor biology, dysregulated epigenetic modifications, metabolic reprogramming, metastasis-associated modifiers, and tumor-promoting inflammatory signals and angiogenesis (Table 4), which might bring about more promising and innovative therapeutic strategies.
Table 4.
Summary of signaling pathways and potential therapeutic targets in the present review
| Mutated driver genes | |
| MAPK pathway | BRAF, NRAS, MEK, ERK, KIT, STK19 |
| Cell-cycle regulation pathway | CDK4/6, MDM2, Cyclin D1, Rb |
| AKT pathway | PI3K, Akt |
| Pigmentation-related pathway | MC1R, Tyrp1, Pax3, Ednrb, MITF, SOX10 |
| Other pathways | GNAQ/GNA11, Notch2, β-catenin, ARID1B, ARID2, TERT |
| Key transcriptional pathways | |
| SOX10 pathway | Sox10, MITF, lncRNA SAMMSON, FOXD3, Rab7 |
| MITF pathway | MITF, Bcl2, Bcl2a1, ML-IAP, HIF1α, c-Met, APE1, p21, BRAC1, SCD |
| Notch and Wnt pathways | Notch, β-catenin, LEF-1 |
| Epigenetic regulation-related pathways | |
| DNA methylation | DNMTs, TET family, IDH2 |
| Histone acetylation and methylation | LSD family, HDAC family, EP300, SETDB1, Dot1L, EZH2, JMJD2C |
| Non-coding RNA and m6A RNA methylation | SAMMSON, FTO, ALKBH5, METTL3/14, YTHDF1 |
| Metabolic reprogramming | |
| Aerobic glycolysis | HIF1α, MYC, Glut 1, Glut 3, HK2, PFKFB2, MCT-4, HMGCL, Oct-1 |
| Oxidative phosphorylation | MITF, PGC1α, lncRNA SAMMSON, p32 |
| Lipid metabolism | ACLY, ACC, FASN, ACS, ACAT2, HMGCS, HMGCR, MVK, SREBP-1, SREBP-2, CD36 |
| Autophagy | Atg5, Twist1, p62, Atg7, SIRT6, Akt, IGF, miR-23a, TFEB, UPR pathway, RIPK1 |
| Amino acid metabolism | PHGDH, GLS2, GOT1, BCAT1/2, BCKDH, DBT |
| Key signal pathways in tumor metastasis | |
| EMT process | N-cadherin, vimentin, α-SMA, SOX2, Snail, Slug, Twist, ZEB1/2, NF-κB, FRA1 |
| CAM | integrin, cadherin, IgSF, connexin, mucin, ILK |
| Exosomes | Rab27A, let-7i, miR-106b-5p, miR-125b-5p, |
| Signal pathways in oncogenic inflammation and angiogenesis | |
| Inflammatory factors | TNFα, IFN-γ, interleukins, JAK-STAT, NF-κB, NALP3 inflammasome, c-Jun |
| Angiogenesis | VEGF-A, bFGF, PlGF, Ang, IL-8, PDGF, VEGFR, NOS, FAK |
The low response rate to immunotherapy and inevitable establishment of resistance to targeted therapy and immunotherapy significantly hinder the treatment efficacy. Recently, more and more investigations have emphasized elucidating the underlying mechanisms, so as to develop novel combined therapy to improve the patients’ outcome. Of note, immunotherapy has been the leading edge of melanoma treatment, with more and more targets revealing encouraging translational potential not only as a single agent but also in combined therapy. Severe adverse effects should be noted and overcome during the process of immunotherapy innovations. More importantly, the heterogeneity of melanomas needs to be taken into consideration and this characteristic determines the fact that a single molecular or pathway might not yield more therapeutic choices. More attention needs to be paid on the study of minority subgroup of melanoma, for example, acral melanoma, in which current therapies display poor effects. The path to clinical translation of novel therapeutic approaches is still in demand of great efforts in the future.
Acknowledgements
This work has received funding from the National Natural Science Foundation of China (Nos. 81625020, 81902791), Support Program of Young Talents in Shaanxi Province (No. 20200303). and Young Eagle Project of Fourth Military Medical University (No. 2019cyjhgwn). We thank Drs. Weigang Zhang, Jianru Chen, Pan Kang, Sen Guo, and Yuqi Yang for their kind help.
Author contributions
W.G and C.L designed the review. W.G and H.W. drafted the manuscript and prepared the figures. C.L revised the draft. All authors read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Weinan Guo, Huina Wang
References
- 1.Lo JA, Fisher DE. The melanoma revolution: from UV carcinogenesis to a new era in therapeutics. Science. 2014;346:945–949. doi: 10.1126/science.1253735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schadendorf D, et al. Melanoma. Lancet. 2018;392:971–984. doi: 10.1016/S0140-6736(18)31559-9. [DOI] [PubMed] [Google Scholar]
- 3.Liu L, Zhang W, Gao T, Li C. Is UV an etiological factor of acral melanoma? J. Expo. Sci. Environ. Epidemiol. 2016;26:539–545. doi: 10.1038/jes.2015.60. [DOI] [PubMed] [Google Scholar]
- 4.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J. Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 5.Schadendorf D, et al. Melanoma. Nat. Rev. Dis. Prim. 2015;1:15003. doi: 10.1038/nrdp.2015.3. [DOI] [PubMed] [Google Scholar]
- 6.Boniol M, Autier P, Boyle P, Gandini S. Cutaneous melanoma attributable to sunbed use: systematic review and meta-analysis. BMJ. 2012;345:e4757. doi: 10.1136/bmj.e4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bald T, et al. Ultraviolet-radiation-induced inflammation promotes angiotropism and metastasis in melanoma. Nature. 2014;507:109–113. doi: 10.1038/nature13111. [DOI] [PubMed] [Google Scholar]
- 8.Li C, et al. Polymorphisms in the DNA repair genes XPC, XPD, and XPG and risk of cutaneous melanoma: a case-control analysis. Cancer Epidemiol. Biomark. Prev. 2006;15:2526–2532. doi: 10.1158/1055-9965.EPI-06-0672. [DOI] [PubMed] [Google Scholar]
- 9.Li C, et al. Polymorphisms of nucleotide excision repair genes predict melanoma survival. J. Invest. Dermatol. 2013;133:1813–1821. doi: 10.1038/jid.2012.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zaidi MR, et al. Interferon-gamma links ultraviolet radiation to melanomagenesis in mice. Nature. 2011;469:548–553. doi: 10.1038/nature09666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rees JL. Genetics of hair and skin color. Annu. Rev. Genet. 2003;37:67–90. doi: 10.1146/annurev.genet.37.110801.143233. [DOI] [PubMed] [Google Scholar]
- 12.Armstrong BK, Cust AE. Sun exposure and skin cancer, and the puzzle of cutaneous melanoma: A perspective on Fears et al. Mathematical models of age and ultraviolet effects on the incidence of skin cancer among whites in the United States. American Journal of Epidemiology 1977; 105: 420-427. Cancer Epidemiol. 2017;48:147–156. doi: 10.1016/j.canep.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 13.Zhang N, et al. The association between trauma and melanoma in the Chinese population: a retrospective study. J. Eur. Acad. Dermatol. Venereol. 2014;28:597–603. doi: 10.1111/jdv.12141. [DOI] [PubMed] [Google Scholar]
- 14.Lesage C, Journet-Tollhupp J, Bernard P, Grange F. Post-traumatic acral melanoma: an underestimated reality? Ann. Dermatol. Venereol. 2012;139:727–731. doi: 10.1016/j.annder.2012.06.034. [DOI] [PubMed] [Google Scholar]
- 15.Juten PG, Hinnen JW. A 71-year-old woman with a pigmented nail bed, which persisted after trauma. Acta Chir. Belg. 2010;110:475–478. doi: 10.1080/00015458.2010.11680659. [DOI] [PubMed] [Google Scholar]
- 16.Ghariani N, et al. Post traumatic amelanotic subungual melanoma. Dermatol. Online J. 2008;14:13. [PubMed] [Google Scholar]
- 17.Ohnishi S, et al. DNA damage in inflammation-related carcinogenesis and cancer stem cells. Oxid. Med. Cell. Longev. 2013;2013:387014. doi: 10.1155/2013/387014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jenkins RW, Fisher DE. Treatment of advanced melanoma in 2020 and beyond. J. Invest. Dermatol. 2021;141:23–31. doi: 10.1016/j.jid.2020.03.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hauschild A, et al. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet. 2012;380:358–365. doi: 10.1016/S0140-6736(12)60868-X. [DOI] [PubMed] [Google Scholar]
- 20.Chapman PB, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robert C, et al. Five-year outcomes with dabrafenib plus trametinib in metastatic melanoma. N. Engl. J. Med. 2019;381:626–636. doi: 10.1056/NEJMoa1904059. [DOI] [PubMed] [Google Scholar]
- 22.Hamid O, et al. Five-year survival outcomes for patients with advanced melanoma treated with pembrolizumab in KEYNOTE-001. Ann. Oncol. 2019;30:582–588. doi: 10.1093/annonc/mdz011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dickson PV, Gershenwald JE. Staging and prognosis of cutaneous melanoma. Surg. Oncol. Clin. N. Am. 2011;20:1–17. doi: 10.1016/j.soc.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luke JJ, Flaherty KT, Ribas A, Long GV. Targeted agents and immunotherapies: optimizing outcomes in melanoma. Nat. Rev. Clin. Oncol. 2017;14:463–482. doi: 10.1038/nrclinonc.2017.43. [DOI] [PubMed] [Google Scholar]
- 25.Davies H, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 26.Lovly CM, et al. Routine multiplex mutational profiling of melanomas enables enrollment in genotype-driven therapeutic trials. PLoS ONE. 2012;7:e35309. doi: 10.1371/journal.pone.0035309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rubinstein JC, et al. Incidence of the V600K mutation among melanoma patients with BRAF mutations, and potential therapeutic response to the specific BRAF inhibitor PLX4032. J. Transl. Med. 2010;8:67. doi: 10.1186/1479-5876-8-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mann GJ, et al. BRAF mutation, NRAS mutation, and the absence of an immune-related expressed gene profile predict poor outcome in patients with stage III melanoma. J. Invest. Dermatol. 2013;133:509–517. doi: 10.1038/jid.2012.283. [DOI] [PubMed] [Google Scholar]
- 29.Long GV, et al. Prognostic and clinicopathologic associations of oncogenic BRAF in metastatic melanoma. J. Clin. Oncol. 2011;29:1239–1246. doi: 10.1200/JCO.2010.32.4327. [DOI] [PubMed] [Google Scholar]
- 30.Nassar KW, Tan AC. The mutational landscape of mucosal melanoma. Semin. Cancer Biol. 2020;61:139–148. doi: 10.1016/j.semcancer.2019.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yeh I, et al. Targeted genomic profiling of acral melanoma. J. Natl Cancer Inst. 2019;111:1068–1077. doi: 10.1093/jnci/djz005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Randic T, et al. NRAS mutant melanoma: towards better therapies. Cancer Treat. Rev. 2021;99:102238. doi: 10.1016/j.ctrv.2021.102238. [DOI] [PubMed] [Google Scholar]
- 33.Yin C, et al. Pharmacological targeting of STK19 inhibits oncogenic NRAS-driven melanomagenesis. Cell. 2019;176:1113–1127.e1116. doi: 10.1016/j.cell.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 34.Nissan MH, et al. Loss of NF1 in cutaneous melanoma is associated with RAS activation and MEK dependence. Cancer Res. 2014;74:2340–2350. doi: 10.1158/0008-5472.CAN-13-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krauthammer M, et al. Exome sequencing identifies recurrent mutations in NF1 and RASopathy genes in sun-exposed melanomas. Nat. Genet. 2015;47:996–1002. doi: 10.1038/ng.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andersen LB, et al. Mutations in the neurofibromatosis 1 gene in sporadic malignant melanoma cell lines. Nat. Genet. 1993;3:118–121. doi: 10.1038/ng0293-118. [DOI] [PubMed] [Google Scholar]
- 37.Handolias D, et al. Mutations in KIT occur at low frequency in melanomas arising from anatomical sites associated with chronic and intermittent sun exposure. Pigment Cell Melanoma Res. 2010;23:210–215. doi: 10.1111/j.1755-148X.2010.00671.x. [DOI] [PubMed] [Google Scholar]
- 38.Kong Y, et al. Frequent genetic aberrations in the CDK4 pathway in acral melanoma indicate the potential for CDK4/6 inhibitors in targeted therapy. Clin. Cancer Res. 2017;23:6946–6957. doi: 10.1158/1078-0432.CCR-17-0070. [DOI] [PubMed] [Google Scholar]
- 39.Sauter ER, et al. Cyclin D1 is a candidate oncogene in cutaneous melanoma. Cancer Res. 2002;62:3200–3206. [PubMed] [Google Scholar]
- 40.Xu L, et al. Frequent genetic aberrations in the cell cycle related genes in mucosal melanoma indicate the potential for targeted therapy. J. Transl. Med. 2019;17:245. doi: 10.1186/s12967-019-1987-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stark M, Hayward N. Genome-wide loss of heterozygosity and copy number analysis in melanoma using high-density single-nucleotide polymorphism arrays. Cancer Res. 2007;67:2632–2642. doi: 10.1158/0008-5472.CAN-06-4152. [DOI] [PubMed] [Google Scholar]
- 42.Stahl JM, et al. Deregulated Akt3 activity promotes development of malignant melanoma. Cancer Res. 2004;64:7002–7010. doi: 10.1158/0008-5472.CAN-04-1399. [DOI] [PubMed] [Google Scholar]
- 43.Robertson GP. Functional and therapeutic significance of Akt deregulation in malignant melanoma. Cancer Metastasis Rev. 2005;24:273–285. doi: 10.1007/s10555-005-1577-9. [DOI] [PubMed] [Google Scholar]
- 44.Hawryluk EB, Tsao H. Melanoma: clinical features and genomic insights. Cold Spring Harb. Perspect. Med. 2014;4:a015388. doi: 10.1101/cshperspect.a015388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ward KA, Lazovich D, Hordinsky MK. Germline melanoma susceptibility and prognostic genes: a review of the literature. J. Am. Acad. Dermatol. 2012;67:1055–1067. doi: 10.1016/j.jaad.2012.02.042. [DOI] [PubMed] [Google Scholar]
- 46.Bishop DT, et al. Genome-wide association study identifies three loci associated with melanoma risk. Nat. Genet. 2009;41:920–925. doi: 10.1038/ng.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garcia-Borron JC, Sanchez-Laorden BL, Jimenez-Cervantes C. Melanocortin-1 receptor structure and functional regulation. Pigment Cell Res. 2005;18:393–410. doi: 10.1111/j.1600-0749.2005.00278.x. [DOI] [PubMed] [Google Scholar]
- 48.Zhou S, et al. Epigenetic regulation of melanogenesis. Ageing Res. Rev. 2021;69:101349. doi: 10.1016/j.arr.2021.101349. [DOI] [PubMed] [Google Scholar]
- 49.Maresca V, Flori E, Picardo M. Skin phototype: a new perspective. Pigment Cell Melanoma Res. 2015;28:378–389. doi: 10.1111/pcmr.12365. [DOI] [PubMed] [Google Scholar]
- 50.Scherer D, Kumar R. Genetics of pigmentation in skin cancer-a review. Mutat. Res. 2010;705:141–153. doi: 10.1016/j.mrrev.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 51.Newell F, et al. Whole-genome sequencing of acral melanoma reveals genomic complexity and diversity. Nat. Commun. 2020;11:5259. doi: 10.1038/s41467-020-18988-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jonsson G, et al. Genomic profiling of malignant melanoma using tiling-resolution arrayCGH. Oncogene. 2007;26:4738–4748. doi: 10.1038/sj.onc.1210252. [DOI] [PubMed] [Google Scholar]
- 53.Worm J, et al. Genetic and epigenetic alterations of the APC gene in malignant melanoma. Oncogene. 2004;23:5215–5226. doi: 10.1038/sj.onc.1207647. [DOI] [PubMed] [Google Scholar]
- 54.Van Raamsdonk CD, et al. Mutations in GNA11 in uveal melanoma. N. Engl. J. Med. 2010;363:2191–2199. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van Raamsdonk CD, et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457:599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Horn S, et al. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339:959–961. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- 57.Newell F, et al. Whole-genome landscape of mucosal melanoma reveals diverse drivers and therapeutic targets. Nat. Commun. 2019;10:3163. doi: 10.1038/s41467-019-11107-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Forschner A, et al. MDM2, MDM4 and EGFR amplifications and hyperprogression in metastatic acral and mucosal melanoma. Cancers. 2020;12:540. doi: 10.3390/cancers12030540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abou Alaiwi S, et al. Mammalian SWI/SNF complex genomic alterations and immune checkpoint blockade in solid tumors. Cancer Immunol. Res. 2020;8:1075–1084. doi: 10.1158/2326-6066.CIR-19-0866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wessely A, Steeb T, Berking C, Heppt MV. How neural crest transcription factors contribute to melanoma heterogeneity, cellular plasticity, and treatment resistance. Int. J. Mol. Sci. 2021;22:5761. doi: 10.3390/ijms22115761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nonaka D, Chiriboga L, Rubin BP. Sox10: a pan-schwannian and melanocytic marker. Am. J. Surg. Pathol. 2008;32:1291–1298. doi: 10.1097/PAS.0b013e3181658c14. [DOI] [PubMed] [Google Scholar]
- 62.Szumera-Cieckiewicz A, et al. SOX10 is as specific as S100 protein in detecting metastases of melanoma in lymph nodes and is recommended for sentinel lymph node assessment. Eur. J. Cancer. 2020;137:175–182. doi: 10.1016/j.ejca.2020.06.037. [DOI] [PubMed] [Google Scholar]
- 63.Blochin E, Nonaka D. Diagnostic value of Sox10 immunohistochemical staining for the detection of metastatic melanoma in sentinel lymph nodes. Histopathology. 2009;55:626–628. doi: 10.1111/j.1365-2559.2009.03415.x. [DOI] [PubMed] [Google Scholar]
- 64.Graf SA, et al. SOX10 promotes melanoma cell invasion by regulating melanoma inhibitory activity. J. Invest. Dermatol. 2014;134:2212–2220. doi: 10.1038/jid.2014.128. [DOI] [PubMed] [Google Scholar]
- 65.Cronin JC, et al. SOX10 ablation arrests cell cycle, induces senescence, and suppresses melanomagenesis. Cancer Res. 2013;73:5709–5718. doi: 10.1158/0008-5472.CAN-12-4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shakhova O, et al. Sox10 promotes the formation and maintenance of giant congenital naevi and melanoma. Nat. Cell Biol. 2012;14:882–890. doi: 10.1038/ncb2535. [DOI] [PubMed] [Google Scholar]
- 67.Han S, et al. ERK-mediated phosphorylation regulates SOX10 sumoylation and targets expression in mutant BRAF melanoma. Nat. Commun. 2018;9:28. doi: 10.1038/s41467-017-02354-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alonso-Curbelo D, et al. RAB7 controls melanoma progression by exploiting a lineage-specific wiring of the endolysosomal pathway. Cancer Cell. 2014;26:61–76. doi: 10.1016/j.ccr.2014.04.030. [DOI] [PubMed] [Google Scholar]
- 69.Leucci E, et al. Melanoma addiction to the long non-coding RNA SAMMSON. Nature. 2016;531:518–522. doi: 10.1038/nature17161. [DOI] [PubMed] [Google Scholar]
- 70.Harris ML, Baxter LL, Loftus SK, Pavan WJ. Sox proteins in melanocyte development and melanoma. Pigment Cell Melanoma Res. 2010;23:496–513. doi: 10.1111/j.1755-148X.2010.00711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Antonellis A, et al. Identification of neural crest and glial enhancers at the mouse Sox10 locus through transgenesis in zebrafish. PLoS Genet. 2008;4:e1000174. doi: 10.1371/journal.pgen.1000174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lv XB, et al. Regulation of SOX10 stability via ubiquitination-mediated degradation by Fbxw7alpha modulates melanoma cell migration. Oncotarget. 2015;6:36370–36382. doi: 10.18632/oncotarget.5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Taylor KM, Labonne C. SoxE factors function equivalently during neural crest and inner ear development and their activity is regulated by SUMOylation. Dev. Cell. 2005;9:593–603. doi: 10.1016/j.devcel.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 74.Girard M, Goossens M. Sumoylation of the SOX10 transcription factor regulates its transcriptional activity. FEBS Lett. 2006;580:1635–1641. doi: 10.1016/j.febslet.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 75.Sun C, et al. Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature. 2014;508:118–122. doi: 10.1038/nature13121. [DOI] [PubMed] [Google Scholar]
- 76.Zhang G, Herlyn M. Linking SOX10 to a slow-growth resistance phenotype. Cell Res. 2014;24:906–907. doi: 10.1038/cr.2014.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Han S, et al. LncRNA SAMMSON mediates adaptive resistance to RAF inhibition in BRAF-mutant melanoma cells. Cancer Res. 2021;81:2918–2929. doi: 10.1158/0008-5472.CAN-20-3145. [DOI] [PubMed] [Google Scholar]
- 78.Smith MP, et al. Inhibiting drivers of non-mutational drug tolerance is a salvage strategy for targeted melanoma therapy. Cancer Cell. 2016;29:270–284. doi: 10.1016/j.ccell.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Goding CR, Arnheiter H. MITF-the first 25 years. Genes Dev. 2019;33:983–1007. doi: 10.1101/gad.324657.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Loercher AE, Tank EM, Delston RB, Harbour JW. MITF links differentiation with cell cycle arrest in melanocytes by transcriptional activation of INK4A. J. Cell Biol. 2005;168:35–40. doi: 10.1083/jcb.200410115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Garraway LA, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 82.Carreira S, et al. Mitf regulation of Dia1 controls melanoma proliferation and invasiveness. Genes Dev. 2006;20:3426–3439. doi: 10.1101/gad.406406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hartman ML, Czyz M. Pro-survival role of MITF in melanoma. J. Invest. Dermatol. 2015;135:352–358. doi: 10.1038/jid.2014.319. [DOI] [PubMed] [Google Scholar]
- 84.Beuret L, et al. BRCA1 is a new MITF target gene. Pigment Cell Melanoma Res. 2011;24:725–727. doi: 10.1111/j.1755-148X.2011.00862.x. [DOI] [PubMed] [Google Scholar]
- 85.Carreira S, et al. Mitf cooperates with Rb1 and activates p21Cip1 expression to regulate cell cycle progression. Nature. 2005;433:764–769. doi: 10.1038/nature03269. [DOI] [PubMed] [Google Scholar]
- 86.Lazar I, et al. The clinical effect of the inhibitor of apopotosis protein livin in melanoma. Oncology. 2012;82:197–204. doi: 10.1159/000334234. [DOI] [PubMed] [Google Scholar]
- 87.Haq R, et al. BCL2A1 is a lineage-specific antiapoptotic melanoma oncogene that confers resistance to BRAF inhibition. Proc. Natl Acad. Sci. USA. 2013;110:4321–4326. doi: 10.1073/pnas.1205575110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.McGill GG, et al. Bcl2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability. Cell. 2002;109:707–718. doi: 10.1016/s0092-8674(02)00762-6. [DOI] [PubMed] [Google Scholar]
- 89.Bianchi-Smiraglia A, et al. Microphthalmia-associated transcription factor suppresses invasion by reducing intracellular GTP pools. Oncogene. 2017;36:84–96. doi: 10.1038/onc.2016.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Swoboda A, et al. STAT3 promotes melanoma metastasis by CEBP-induced repression of the MITF pathway. Oncogene. 2021;40:1091–1105. doi: 10.1038/s41388-020-01584-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vivas-Garcia Y, et al. Lineage-restricted regulation of SCD and fatty acid saturation by MITF controls melanoma phenotypic plasticity. Mol. Cell. 2020;77:120–137.e129. doi: 10.1016/j.molcel.2019.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kaur A, et al. sFRP2 in the aged microenvironment drives melanoma metastasis and therapy resistance. Nature. 2016;532:250–254. doi: 10.1038/nature17392. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 93.Muller J, et al. Low MITF/AXL ratio predicts early resistance to multiple targeted drugs in melanoma. Nat. Commun. 2014;5:5712. doi: 10.1038/ncomms6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.MITF drives a reversible drug-tolerant state in melanoma. Cancer Discov. 6, OF11 (2016). [DOI] [PubMed]
- 95.Kim H, Ronai ZA. HIV drug to aid melanoma therapies? Cancer Cell. 2016;29:245–246. doi: 10.1016/j.ccell.2016.02.014. [DOI] [PubMed] [Google Scholar]
- 96.Asnaghi L, et al. Notch signaling promotes growth and invasion in uveal melanoma. Clin. Cancer Res. 2012;18:654–665. doi: 10.1158/1078-0432.CCR-11-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pinnix CC, et al. Active Notch1 confers a transformed phenotype to primary human melanocytes. Cancer Res. 2009;69:5312–5320. doi: 10.1158/0008-5472.CAN-08-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bedogni B, et al. Notch1 is an effector of Akt and hypoxia in melanoma development. J. Clin. Invest. 2008;118:3660–3670. doi: 10.1172/JCI36157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Balint K, et al. Activation of Notch1 signaling is required for beta-catenin-mediated human primary melanoma progression. J. Clin. Invest. 2005;115:3166–3176. doi: 10.1172/JCI25001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liu ZJ, et al. Notch1 signaling promotes primary melanoma progression by activating mitogen-activated protein kinase/phosphatidylinositol 3-kinase-Akt pathways and up-regulating N-cadherin expression. Cancer Res. 2006;66:4182–4190. doi: 10.1158/0008-5472.CAN-05-3589. [DOI] [PubMed] [Google Scholar]
- 101.Murtas D, et al. Activated Notch1 expression is associated with angiogenesis in cutaneous melanoma. Clin. Exp. Med. 2015;15:351–360. doi: 10.1007/s10238-014-0300-y. [DOI] [PubMed] [Google Scholar]
- 102.Howard JD, et al. Notch signaling mediates melanoma-endothelial cell communication and melanoma cell migration. Pigment Cell Melanoma Res. 2013;26:697–707. doi: 10.1111/pcmr.12131. [DOI] [PubMed] [Google Scholar]
- 103.Wieland E, et al. Endothelial Notch1 activity facilitates metastasis. Cancer Cell. 2017;31:355–367. doi: 10.1016/j.ccell.2017.01.007. [DOI] [PubMed] [Google Scholar]
- 104.Porcelli L, et al. Active notch protects MAPK activated melanoma cell lines from MEK inhibitor cobimetinib. Biomed. Pharmacother. 2021;133:111006. doi: 10.1016/j.biopha.2020.111006. [DOI] [PubMed] [Google Scholar]
- 105.Ikeya M, et al. Wnt signalling required for expansion of neural crest and CNS progenitors. Nature. 1997;389:966–970. doi: 10.1038/40146. [DOI] [PubMed] [Google Scholar]
- 106.Grumolato L, et al. Canonical and noncanonical Wnts use a common mechanism to activate completely unrelated coreceptors. Genes Dev. 2010;24:2517–2530. doi: 10.1101/gad.1957710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Takeda K, et al. Induction of melanocyte-specific microphthalmia-associated transcription factor by Wnt-3a. J. Biol. Chem. 2000;275:14013–14016. doi: 10.1074/jbc.c000113200. [DOI] [PubMed] [Google Scholar]
- 108.Widlund HR, et al. Beta-catenin-induced melanoma growth requires the downstream target Microphthalmia-associated transcription factor. J. Cell Biol. 2002;158:1079–1087. doi: 10.1083/jcb.200202049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Delmas V, et al. Beta-catenin induces immortalization of melanocytes by suppressing p16INK4a expression and cooperates with N-Ras in melanoma development. Genes Dev. 2007;21:2923–2935. doi: 10.1101/gad.450107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Damsky WE, et al. beta-catenin signaling controls metastasis in Braf-activated Pten-deficient melanomas. Cancer Cell. 2011;20:741–754. doi: 10.1016/j.ccr.2011.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sinnberg T, et al. Wnt-signaling enhances neural crest migration of melanoma cells and induces an invasive phenotype. Mol. Cancer. 2018;17:59. doi: 10.1186/s12943-018-0773-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Douglass SM, et al. Myeloid-derived suppressor cells are a major source of Wnt5A in the melanoma microenvironment and depend on Wnt5A for full suppressive activity. Cancer Res. 2021;81:658–670. doi: 10.1158/0008-5472.CAN-20-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rodriguez-Hernandez I, et al. WNT11-FZD7-DAAM1 signalling supports tumour initiating abilities and melanoma amoeboid invasion. Nat. Commun. 2020;11:5315. doi: 10.1038/s41467-020-18951-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Webster MR, Weeraratna AT. A Wnt-er migration: the confusing role of beta-catenin in melanoma metastasis. Sci. Signal. 2013;6:pe11. doi: 10.1126/scisignal.2004114. [DOI] [PubMed] [Google Scholar]
- 115.Gallagher SJ, et al. Beta-catenin inhibits melanocyte migration but induces melanoma metastasis. Oncogene. 2013;32:2230–2238. doi: 10.1038/onc.2012.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chien AJ, et al. Activated Wnt/beta-catenin signaling in melanoma is associated with decreased proliferation in patient tumors and a murine melanoma model. Proc. Natl Acad. Sci. USA. 2009;106:1193–1198. doi: 10.1073/pnas.0811902106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Maelandsmo GM, et al. Reduced beta-catenin expression in the cytoplasm of advanced-stage superficial spreading malignant melanoma. Clin. Cancer Res. 2003;9:3383–3388. [PubMed] [Google Scholar]
- 118.Kageshita T, et al. Loss of beta-catenin expression associated with disease progression in malignant melanoma. Br. J. Dermatol. 2001;145:210–216. doi: 10.1046/j.1365-2133.2001.04336.x. [DOI] [PubMed] [Google Scholar]
- 119.Uka R, et al. Temporal activation of WNT/beta-catenin signaling is sufficient to inhibit SOX10 expression and block melanoma growth. Oncogene. 2020;39:4132–4154. doi: 10.1038/s41388-020-1267-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tsoi J, et al. Multi-stage differentiation defines melanoma subtypes with differential vulnerability to drug-induced iron-dependent oxidative stress. Cancer Cell. 2018;33:890–904.e895. doi: 10.1016/j.ccell.2018.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 122.Moran B, Silva R, Perry AS, Gallagher WM. Epigenetics of malignant melanoma. Semin. Cancer Biol. 2018;51:80–88. doi: 10.1016/j.semcancer.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 123.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lei H, et al. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122:3195–3205. doi: 10.1242/dev.122.10.3195. [DOI] [PubMed] [Google Scholar]
- 125.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 126.Lian CG, et al. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell. 2012;150:1135–1146. doi: 10.1016/j.cell.2012.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tuominen R, et al. MGMT promoter methylation is associated with temozolomide response and prolonged progression-free survival in disseminated cutaneous melanoma. Int. J. Cancer. 2015;136:2844–2853. doi: 10.1002/ijc.29332. [DOI] [PubMed] [Google Scholar]
- 128.Kohonen-Corish MR, et al. Promoter hypermethylation of the O(6)-methylguanine DNA methyltransferase gene and microsatellite instability in metastatic melanoma. J. Invest. Dermatol. 2006;126:167–171. doi: 10.1038/sj.jid.5700005. [DOI] [PubMed] [Google Scholar]
- 129.Maat W, et al. Epigenetic inactivation of RASSF1a in uveal melanoma. Invest. Ophthalmol. Vis. Sci. 2007;48:486–490. doi: 10.1167/iovs.06-0781. [DOI] [PubMed] [Google Scholar]
- 130.Spugnardi M, et al. Epigenetic inactivation of RAS association domain family protein 1 (RASSF1A) in malignant cutaneous melanoma. Cancer Res. 2003;63:1639–1643. [PubMed] [Google Scholar]
- 131.Jonsson A, et al. High frequency of p16(INK4A) promoter methylation in NRAS-mutated cutaneous melanoma. J. Invest. Dermatol. 2010;130:2809–2817. doi: 10.1038/jid.2010.216. [DOI] [PubMed] [Google Scholar]
- 132.Venza M, et al. Epigenetic regulation of p14ARF and p16INK4A expression in cutaneous and uveal melanoma. Biochim. Biophys. Acta. 2015;1849:247–256. doi: 10.1016/j.bbagrm.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 133.Mirmohammadsadegh A, et al. Epigenetic silencing of the PTEN gene in melanoma. Cancer Res. 2006;66:6546–6552. doi: 10.1158/0008-5472.CAN-06-0384. [DOI] [PubMed] [Google Scholar]
- 134.Lahtz C, et al. Methylation of PTEN as a prognostic factor in malignant melanoma of the skin. J. Invest. Dermatol. 2010;130:620–622. doi: 10.1038/jid.2009.226. [DOI] [PubMed] [Google Scholar]
- 135.Gao L, et al. Genome-wide promoter methylation analysis identifies epigenetic silencing of MAPK13 in primary cutaneous melanoma. Pigment Cell Melanoma Res. 2013;26:542–554. doi: 10.1111/pcmr.12096. [DOI] [PubMed] [Google Scholar]
- 136.Koga Y, et al. Genome-wide screen of promoter methylation identifies novel markers in melanoma. Genome Res. 2009;19:1462–1470. doi: 10.1101/gr.091447.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Conway K, et al. DNA-methylation profiling distinguishes malignant melanomas from benign nevi. Pigment Cell Melanoma Res. 2011;24:352–360. doi: 10.1111/j.1755-148X.2011.00828.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sarkar D, et al. Epigenetic regulation in human melanoma: past and future. Epigenetics. 2015;10:103–121. doi: 10.1080/15592294.2014.1003746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Micevic G, Theodosakis N, Bosenberg M. Aberrant DNA methylation in melanoma: biomarker and therapeutic opportunities. Clin. Epigenet. 2017;9:34. doi: 10.1186/s13148-017-0332-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Altemose N, Miga KH, Maggioni M, Willard HF. Genomic characterization of large heterochromatic gaps in the human genome assembly. PLoS Comput. Biol. 2014;10:e1003628. doi: 10.1371/journal.pcbi.1003628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Carrozza MJ, Utley RT, Workman JL, Cote J. The diverse functions of histone acetyltransferase complexes. Trends Genet. 2003;19:321–329. doi: 10.1016/S0168-9525(03)00115-X. [DOI] [PubMed] [Google Scholar]
- 142.Nurse NP, Jimenez-Useche I, Smith IT, Yuan C. Clipping of flexible tails of histones H3 and H4 affects the structure and dynamics of the nucleosome. Biophys. J. 2013;104:1081–1088. doi: 10.1016/j.bpj.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhao S, Allis CD, Wang GG. The language of chromatin modification in human cancers. Nat. Rev. Cancer. 2021;21:413–430. doi: 10.1038/s41568-021-00357-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bennett RL, Licht JD. Targeting epigenetics in cancer. Annu. Rev. Pharmacol. Toxicol. 2018;58:187–207. doi: 10.1146/annurev-pharmtox-010716-105106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hashimoto H, Vertino PM, Cheng X. Molecular coupling of DNA methylation and histone methylation. Epigenomics. 2010;2:657–669. doi: 10.2217/epi.10.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Shi Y, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 147.Fiziev P, et al. Systematic epigenomic analysis reveals chromatin states associated with melanoma progression. Cell Rep. 2017;19:875–889. doi: 10.1016/j.celrep.2017.03.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Landreville S, et al. Histone deacetylase inhibitors induce growth arrest and differentiation in uveal melanoma. Clin. Cancer Res. 2012;18:408–416. doi: 10.1158/1078-0432.CCR-11-0946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Woods DM, et al. HDAC inhibition upregulates PD-1 ligands in melanoma and augments immunotherapy with PD-1 blockade. Cancer Immunol. Res. 2015;3:1375–1385. doi: 10.1158/2326-6066.CIR-15-0077-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lombard DB, Cierpicki T, Grembecka J. Combined MAPK pathway and HDAC inhibition breaks melanoma. Cancer Discov. 2019;9:469–471. doi: 10.1158/2159-8290.CD-19-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wang L, et al. An acquired vulnerability of drug-resistant melanoma with therapeutic potential. Cell. 2018;173:1413–1425.e1414. doi: 10.1016/j.cell.2018.04.012. [DOI] [PubMed] [Google Scholar]
- 152.Munshi A, et al. Histone deacetylase inhibitors radiosensitize human melanoma cells by suppressing DNA repair activity. Clin. Cancer Res. 2005;11:4912–4922. doi: 10.1158/1078-0432.CCR-04-2088. [DOI] [PubMed] [Google Scholar]
- 153.Tsimberidou AM, et al. Preclinical development and first-in-human study of KA2507, a selective and potent inhibitor of histone deacetylase 6, for patients with refractory solid tumors. Clin. Cancer Res. 2021;27:3584–3594. doi: 10.1158/1078-0432.CCR-21-0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yan G, et al. Selective inhibition of p300 HAT blocks cell cycle progression, induces cellular senescence, and inhibits the DNA damage response in melanoma cells. J. Invest. Dermatol. 2013;133:2444–2452. doi: 10.1038/jid.2013.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wang R, et al. Targeting lineage-specific MITF pathway in human melanoma cell lines by A-485, the selective small-molecule inhibitor of p300/CBP. Mol. Cancer Ther. 2018;17:2543–2550. doi: 10.1158/1535-7163.MCT-18-0511. [DOI] [PubMed] [Google Scholar]
- 156.Kim E, et al. MITF expression predicts therapeutic vulnerability to p300 inhibition in human melanoma. Cancer Res. 2019;79:2649–2661. doi: 10.1158/0008-5472.CAN-18-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zhang F, et al. Targeting the p300/NONO axis sensitizes melanoma cells to BRAF inhibitors. Oncogene. 2021;40:4137–4150. doi: 10.1038/s41388-021-01834-1. [DOI] [PubMed] [Google Scholar]
- 158.Guo W, et al. ATP-citrate lyase epigenetically potentiates oxidative phosphorylation to promote melanoma growth and adaptive resistance to MAPK inhibition. Clin. Cancer Res. 2020;26:2725–2739. doi: 10.1158/1078-0432.CCR-19-1359. [DOI] [PubMed] [Google Scholar]
- 159.Orouji E, et al. Histone methyltransferase SETDB1 contributes to melanoma tumorigenesis and serves as a new potential therapeutic target. Int. J. Cancer. 2019;145:3462–3477. doi: 10.1002/ijc.32432. [DOI] [PubMed] [Google Scholar]
- 160.Ceol CJ, et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature. 2011;471:513–517. doi: 10.1038/nature09806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Shi X, et al. The abundance of metabolites related to protein methylation correlates with the metastatic capacity of human melanoma xenografts. Sci. Adv. 2017;3:eaao5268. doi: 10.1126/sciadv.aao5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zhu B, et al. The protective role of DOT1L in UV-induced melanomagenesis. Nat. Commun. 2018;9:259. doi: 10.1038/s41467-017-02687-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zingg D, et al. The epigenetic modifier EZH2 controls melanoma growth and metastasis through silencing of distinct tumour suppressors. Nat. Commun. 2015;6:6051. doi: 10.1038/ncomms7051. [DOI] [PubMed] [Google Scholar]
- 164.Bachmann IM, et al. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J. Clin. Oncol. 2006;24:268–273. doi: 10.1200/JCO.2005.01.5180. [DOI] [PubMed] [Google Scholar]
- 165.Luo C, et al. H3K27me3-mediated PGC1alpha gene silencing promotes melanoma invasion through WNT5A and YAP. J. Clin. Invest. 2020;130:853–862. doi: 10.1172/JCI130038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zingg D, et al. EZH2-mediated primary cilium deconstruction drives metastatic melanoma formation. Cancer Cell. 2018;34:69–84.e14. doi: 10.1016/j.ccell.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 167.Emran AA, et al. Targeting DNA methylation and EZH2 activity to overcome melanoma resistance to immunotherapy. Trends Immunol. 2019;40:328–344. doi: 10.1016/j.it.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 168.Hoffmann F, et al. H3K27me3 and EZH2 expression in melanoma: relevance for melanoma progression and response to immune checkpoint blockade. Clin. Epigenet. 2020;12:24. doi: 10.1186/s13148-020-0818-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Tiffen J, et al. EZH2 cooperates with DNA methylation to downregulate key tumor suppressors and IFN gene signatures in melanoma. J. Invest. Dermatol. 2020;140:2442–2454.e2445. doi: 10.1016/j.jid.2020.02.042. [DOI] [PubMed] [Google Scholar]
- 170.Yu Y, et al. Targeting the senescence-overriding cooperative activity of structurally unrelated H3K9 demethylases in melanoma. Cancer Cell. 2018;33:322–336.e328. doi: 10.1016/j.ccell.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Sheng W, et al. LSD1 ablation stimulates anti-tumor immunity and enables checkpoint blockade. Cell. 2018;174:549–563.e519. doi: 10.1016/j.cell.2018.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ju H, et al. Altered expression pattern of circular RNAs in metastatic oral mucosal melanoma. Am. J. Cancer Res. 2018;8:1788–1800. [PMC free article] [PubMed] [Google Scholar]
- 173.Yu X, et al. Long non-coding RNAs in melanoma. Cell Prolif. 2018;51:e12457. doi: 10.1111/cpr.12457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Zhang J, et al. Identification of lncRNA-mRNA regulatory module to explore the pathogenesis and prognosis of melanoma. Front. Cell Dev. Biol. 2020;8:615671. doi: 10.3389/fcell.2020.615671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Montico, B. et al. The pleiotropic role of circular and long noncoding RNAs in cutaneous melanoma. Mol. Oncol. 10.1002/1878-0261.13034 (2021). [DOI] [PMC free article] [PubMed]
- 176.Xiao B, et al. Identification of epithelial-mesenchymal transition-related prognostic lncRNAs biomarkers associated with melanoma microenvironment. Front. Cell Dev. Biol. 2021;9:679133. doi: 10.3389/fcell.2021.679133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Vignard V, et al. MicroRNAs in tumor exosomes drive immune escape in melanoma. Cancer Immunol. Res. 2020;8:255–267. doi: 10.1158/2326-6066.CIR-19-0522. [DOI] [PubMed] [Google Scholar]
- 178.Thyagarajan A, Tsai KY, Sahu RP. MicroRNA heterogeneity in melanoma progression. Semin. Cancer Biol. 2019;59:208–220. doi: 10.1016/j.semcancer.2019.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Hanniford D, et al. Identification of metastasis-suppressive microRNAs in primary melanoma. J. Natl Cancer Inst. 2015;107:dju494. doi: 10.1093/jnci/dju494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Molnar V, et al. Changes in miRNA expression in solid tumors: an miRNA profiling in melanomas. Semin. Cancer Biol. 2008;18:111–122. doi: 10.1016/j.semcancer.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 181.Guo W, et al. Down-regulated miR-23a contributes to the metastasis of cutaneous melanoma by promoting autophagy. Theranostics. 2017;7:2231–2249. doi: 10.7150/thno.18835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Pencheva N, et al. Convergent multi-miRNA targeting of ApoE drives LRP1/LRP8-dependent melanoma metastasis and angiogenesis. Cell. 2012;151:1068–1082. doi: 10.1016/j.cell.2012.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Gaziel-Sovran A, et al. miR-30b/30d regulation of GalNAc transferases enhances invasion and immunosuppression during metastasis. Cancer Cell. 2011;20:104–118. doi: 10.1016/j.ccr.2011.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Schultz J, et al. MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res. 2008;18:549–557. doi: 10.1038/cr.2008.45. [DOI] [PubMed] [Google Scholar]
- 185.Zhao E, et al. Cancer mediates effector T cell dysfunction by targeting microRNAs and EZH2 via glycolysis restriction. Nat. Immunol. 2016;17:95–103. doi: 10.1038/ni.3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Dror S, et al. Melanoma miRNA trafficking controls tumour primary niche formation. Nat. Cell Biol. 2016;18:1006–1017. doi: 10.1038/ncb3399. [DOI] [PubMed] [Google Scholar]
- 187.Gilot D, et al. A non-coding function of TYRP1 mRNA promotes melanoma growth. Nat. Cell Biol. 2017;19:1348–1357. doi: 10.1038/ncb3623. [DOI] [PubMed] [Google Scholar]
- 188.Meyer KD, Jaffrey SR. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 2014;15:313–326. doi: 10.1038/nrm3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017;18:31–42. doi: 10.1038/nrm.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Sun T, Wu R, Ming L. The role of m6A RNA methylation in cancer. Biomed. Pharmacother. 2019;112:108613. doi: 10.1016/j.biopha.2019.108613. [DOI] [PubMed] [Google Scholar]
- 191.Yang S, et al. m(6)A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat. Commun. 2019;10:2782. doi: 10.1038/s41467-019-10669-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Wang L, et al. m(6) A RNA methyltransferases METTL3/14 regulate immune responses to anti-PD-1 therapy. EMBO J. 2020;39:e104514. doi: 10.15252/embj.2020104514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Li N, et al. ALKBH5 regulates anti-PD-1 therapy response by modulating lactate and suppressive immune cell accumulation in tumor microenvironment. Proc. Natl Acad. Sci. USA. 2020;117:20159–20170. doi: 10.1073/pnas.1918986117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Yin H, et al. RNA m6A methylation orchestrates cancer growth and metastasis via macrophage reprogramming. Nat. Commun. 2021;12:1394. doi: 10.1038/s41467-021-21514-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Xiong K, et al. Identification of HDAC9 as a viable therapeutic target for the treatment of gastric cancer. Exp. Mol. Med. 2019;51:1–15. doi: 10.1038/s12276-019-0301-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Ruocco MR, et al. Metabolic flexibility in melanoma: a potential therapeutic target. Semin. Cancer Biol. 2019;59:187–207. doi: 10.1016/j.semcancer.2019.07.016. [DOI] [PubMed] [Google Scholar]
- 197.Avagliano A, et al. Metabolic plasticity of melanoma cells and their crosstalk with tumor microenvironment. Front. Oncol. 2020;10:722. doi: 10.3389/fonc.2020.00722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Yang R, et al. The identification of the metabolism subtypes of skin cutaneous melanoma associated with the tumor microenvironment and the immunotherapy. Front. Cell Dev. Biol. 2021;9:707677. doi: 10.3389/fcell.2021.707677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Parmenter TJ, et al. Response of BRAF-mutant melanoma to BRAF inhibition is mediated by a network of transcriptional regulators of glycolysis. Cancer Discov. 2014;4:423–433. doi: 10.1158/2159-8290.CD-13-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Haq R, et al. Oncogenic BRAF regulates oxidative metabolism via PGC1alpha and MITF. Cancer Cell. 2013;23:302–315. doi: 10.1016/j.ccr.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Houles T, et al. RSK regulates PFK-2 activity to promote metabolic rewiring in melanoma. Cancer Res. 2018;78:2191–2204. doi: 10.1158/0008-5472.CAN-17-2215. [DOI] [PubMed] [Google Scholar]
- 202.Ratnikov BI, et al. Metabolic rewiring in melanoma. Oncogene. 2017;36:147–157. doi: 10.1038/onc.2016.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Brand A, et al. LDHA-associated lactic acid production blunts tumor immunosurveillance by T and NK cells. Cell Metab. 2016;24:657–671. doi: 10.1016/j.cmet.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 204.Lacroix R, et al. Targeting tumor-associated acidity in cancer immunotherapy. Cancer Immunol. Immunother. 2018;67:1331–1348. doi: 10.1007/s00262-018-2195-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Daneshmandi S, Wegiel B, Seth P. Blockade of lactate dehydrogenase-A (LDH-A) improves efficacy of anti-programmed cell death-1 (PD-1) therapy in melanoma. Cancers. 2019;11:450. doi: 10.3390/cancers11040450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Kang HB, et al. Metabolic rewiring by oncogenic BRAF V600E links ketogenesis pathway to BRAF-MEK1 signaling. Mol. Cell. 2015;59:345–358. doi: 10.1016/j.molcel.2015.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Vazquez F, et al. PGC1alpha expression defines a subset of human melanoma tumors with increased mitochondrial capacity and resistance to oxidative stress. Cancer Cell. 2013;23:287–301. doi: 10.1016/j.ccr.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Haq R, Fisher DE, Widlund HR. Molecular pathways: BRAF induces bioenergetic adaptation by attenuating oxidative phosphorylation. Clin. Cancer Res. 2014;20:2257–2263. doi: 10.1158/1078-0432.CCR-13-0898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Goding CR. Targeting the lncRNA SAMMSON reveals metabolic vulnerability in melanoma. Cancer Cell. 2016;29:619–621. doi: 10.1016/j.ccell.2016.04.010. [DOI] [PubMed] [Google Scholar]
- 210.Luo C, et al. A PGC1alpha-mediated transcriptional axis suppresses melanoma metastasis. Nature. 2016;537:422–426. doi: 10.1038/nature19347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Kaplon J, et al. A key role for mitochondrial gatekeeper pyruvate dehydrogenase in oncogene-induced senescence. Nature. 2013;498:109–112. doi: 10.1038/nature12154. [DOI] [PubMed] [Google Scholar]
- 212.Harel M, et al. Proteomics of melanoma response to immunotherapy reveals mitochondrial dependence. Cell. 2019;179:236–250.e218. doi: 10.1016/j.cell.2019.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Snaebjornsson MT, Janaki-Raman S, Schulze A. Greasing the wheels of the cancer machine: the role of lipid metabolism in cancer. Cell Metab. 2020;31:62–76. doi: 10.1016/j.cmet.2019.11.010. [DOI] [PubMed] [Google Scholar]
- 214.Currie E, et al. Cellular fatty acid metabolism and cancer. Cell Metab. 2013;18:153–161. doi: 10.1016/j.cmet.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Henderson F, et al. Enhanced fatty acid scavenging and glycerophospholipid metabolism accompany melanocyte neoplasia progression in zebrafish. Cancer Res. 2019;79:2136–2151. doi: 10.1158/0008-5472.CAN-18-2409. [DOI] [PubMed] [Google Scholar]
- 216.Hong X, et al. The lipogenic regulator SREBP2 induces transferrin in circulating melanoma cells and suppresses ferroptosis. Cancer Discov. 2021;11:678–695. doi: 10.1158/2159-8290.CD-19-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Wu S, Naar AM. SREBP1-dependent de novo fatty acid synthesis gene expression is elevated in malignant melanoma and represents a cellular survival trait. Sci. Rep. 2019;9:10369. doi: 10.1038/s41598-019-46594-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Katuru R, et al. Mevalonate depletion mediates the suppressive impact of geranylgeraniol on murine B16 melanoma cells. Exp. Biol. Med. 2011;236:604–613. doi: 10.1258/ebm.2011.010379. [DOI] [PubMed] [Google Scholar]
- 219.Favero GM, et al. Simvastatin impairs murine melanoma growth. Lipids Health Dis. 2010;9:142. doi: 10.1186/1476-511X-9-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Zanfardino M, et al. Simvastatin reduces melanoma progression in a murine model. Int. J. Oncol. 2013;43:1763–1770. doi: 10.3892/ijo.2013.2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Wu J, et al. Iciartin, a novel FASN inhibitor, exerts anti-melanoma activities through IGF-1R/STAT3 signaling. Oncotarget. 2016;7:51251–51269. doi: 10.18632/oncotarget.9984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Saab J, et al. Fatty acid synthase and acetyl-CoA carboxylase are expressed in nodal metastatic melanoma but not in benign intracapsular nodal nevi. Am. J. Dermatopathol. 2018;40:259–264. doi: 10.1097/DAD.0000000000000939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Pascual G, et al. Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature. 2017;541:41–45. doi: 10.1038/nature20791. [DOI] [PubMed] [Google Scholar]
- 224.Chen S, et al. Palmitoylation-dependent activation of MC1R prevents melanomagenesis. Nature. 2017;549:399–403. doi: 10.1038/nature23887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Chen S, et al. Targeting MC1R depalmitoylation to prevent melanomagenesis in redheads. Nat. Commun. 2019;10:877. doi: 10.1038/s41467-019-08691-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Talebi A, et al. Sustained SREBP-1-dependent lipogenesis as a key mediator of resistance to BRAF-targeted therapy. Nat. Commun. 2018;9:2500. doi: 10.1038/s41467-018-04664-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Klionsky DJ, et al. Autophagy in major human diseases. EMBO J. 2021;40:e108863. doi: 10.15252/embj.2021108863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Liu H, He Z, Simon HU. Autophagy suppresses melanoma tumorigenesis by inducing senescence. Autophagy. 2014;10:372–373. doi: 10.4161/auto.27163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Liu H, et al. Down-regulation of autophagy-related protein 5 (ATG5) contributes to the pathogenesis of early-stage cutaneous melanoma. Sci. Transl. Med. 2013;5:202ra123. doi: 10.1126/scitranslmed.3005864. [DOI] [PubMed] [Google Scholar]
- 230.Qiang L, He YY. Autophagy deficiency stabilizes TWIST1 to promote epithelial-mesenchymal transition. Autophagy. 2014;10:1864–1865. doi: 10.4161/auto.32171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Xie X, et al. Atg7 overcomes senescence and promotes growth of BrafV600E-driven melanoma. Cancer Discov. 2015;5:410–423. doi: 10.1158/2159-8290.CD-14-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Rosenfeldt MT, et al. Loss of autophagy affects melanoma development in a manner dependent on PTEN status. Cell Death Differ. 2021;28:1437–1439. doi: 10.1038/s41418-021-00746-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Martin S, et al. An autophagy-driven pathway of ATP secretion supports the aggressive phenotype of BRAF(V600E) inhibitor-resistant metastatic melanoma cells. Autophagy. 2017;13:1512–1527. doi: 10.1080/15548627.2017.1332550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Wang L, et al. Aberrant SIRT6 expression contributes to melanoma growth: Role of the autophagy paradox and IGF-AKT signaling. Autophagy. 2018;14:518–533. doi: 10.1080/15548627.2017.1384886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Li S, et al. Transcriptional regulation of autophagy-lysosomal function in BRAF-driven melanoma progression and chemoresistance. Nat. Commun. 2019;10:1693. doi: 10.1038/s41467-019-09634-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Corazzari M, et al. Oncogenic BRAF induces chronic ER stress condition resulting in increased basal autophagy and apoptotic resistance of cutaneous melanoma. Cell Death Differ. 2015;22:946–958. doi: 10.1038/cdd.2014.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Luan Q, et al. RIPK1 regulates survival of human melanoma cells upon endoplasmic reticulum stress through autophagy. Autophagy. 2015;11:975–994. doi: 10.1080/15548627.2015.1049800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Ma XH, et al. Targeting ER stress-induced autophagy overcomes BRAF inhibitor resistance in melanoma. J. Clin. Invest. 2014;124:1406–1417. doi: 10.1172/JCI70454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Truong A, et al. Chloroquine sensitizes GNAQ/11-mutated melanoma to MEK1/2 inhibition. Clin. Cancer Res. 2020;26:6374–6386. doi: 10.1158/1078-0432.CCR-20-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Goodall ML, et al. Development of potent autophagy inhibitors that sensitize oncogenic BRAF V600E mutant melanoma tumor cells to vemurafenib. Autophagy. 2014;10:1120–1136. doi: 10.4161/auto.28594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Locasale JW, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat. Genet. 2011;43:869–874. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Sullivan MR, et al. Increased serine synthesis provides an advantage for tumors arising in tissues where serine levels are limiting. Cell Metab. 2019;29:1410–1421.e1414. doi: 10.1016/j.cmet.2019.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Ngo B, et al. Limited environmental serine and glycine confer brain metastasis sensitivity to PHGDH inhibition. Cancer Discov. 2020;10:1352–1373. doi: 10.1158/2159-8290.CD-19-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Doepner M, Lee IY, Ridky TW. Drug resistant melanoma may be vulnerable to inhibitors of serine synthesis. J. Invest. Dermatol. 2020;140:2114–2116. doi: 10.1016/j.jid.2020.05.103. [DOI] [PubMed] [Google Scholar]
- 245.Nguyen MQ, et al. Targeting PHGDH upregulation reduces glutathione levels and resensitizes resistant NRAS-mutant melanoma to MAPK kinase inhibition. J. Invest. Dermatol. 2020;140:2242–2252.e2247. doi: 10.1016/j.jid.2020.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Luan W, et al. miR-137 inhibits glutamine catabolism and growth of malignant melanoma by targeting glutaminase. Biochem. Biophys. Res. Commun. 2018;495:46–52. doi: 10.1016/j.bbrc.2017.10.152. [DOI] [PubMed] [Google Scholar]
- 247.Wang Q, et al. Targeting glutamine transport to suppress melanoma cell growth. Int. J. Cancer. 2014;135:1060–1071. doi: 10.1002/ijc.28749. [DOI] [PubMed] [Google Scholar]
- 248.Pathria G, et al. Targeting the Warburg effect via LDHA inhibition engages ATF4 signaling for cancer cell survival. EMBO J. 2018;37:e99735. doi: 10.15252/embj.201899735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Lim JH, Luo C, Vazquez F, Puigserver P. Targeting mitochondrial oxidative metabolism in melanoma causes metabolic compensation through glucose and glutamine utilization. Cancer Res. 2014;74:3535–3545. doi: 10.1158/0008-5472.CAN-13-2893-T. [DOI] [PubMed] [Google Scholar]
- 250.Pan M, et al. Regional glutamine deficiency in tumours promotes dedifferentiation through inhibition of histone demethylation. Nat. Cell Biol. 2016;18:1090–1101. doi: 10.1038/ncb3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Baenke F, et al. Resistance to BRAF inhibitors induces glutamine dependency in melanoma cells. Mol. Oncol. 2016;10:73–84. doi: 10.1016/j.molonc.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Ishak Gabra MB, et al. Dietary glutamine supplementation suppresses epigenetically-activated oncogenic pathways to inhibit melanoma tumour growth. Nat. Commun. 2020;11:3326. doi: 10.1038/s41467-020-17181-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Sivanand S, Vander Heiden MG. Emerging roles for branched-chain amino acid metabolism in cancer. Cancer Cell. 2020;37:147–156. doi: 10.1016/j.ccell.2019.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Zhang B, et al. BCAT1 knockdown-mediated suppression of melanoma cell proliferation and migration is associated with reduced oxidative phosphorylation. Am. J. Cancer Res. 2021;11:2670–2683. [PMC free article] [PubMed] [Google Scholar]
- 255.Ko T, Sharma R, Li S. Genome-wide screening identifies novel genes implicated in cellular sensitivity to BRAF(V600E) expression. Oncogene. 2020;39:723–738. doi: 10.1038/s41388-019-1022-0. [DOI] [PubMed] [Google Scholar]
- 256.Sheen JH, Zoncu R, Kim D, Sabatini DM. Defective regulation of autophagy upon leucine deprivation reveals a targetable liability of human melanoma cells in vitro and in vivo. Cancer Cell. 2011;19:613–628. doi: 10.1016/j.ccr.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Fernandez MR, Cleveland JL. ATF4-amino acid circuits: a recipe for resistance in melanoma. EMBO J. 2018;37:e100600. doi: 10.15252/embj.2018100600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Carr S, Smith C, Wernberg J. Epidemiology and risk factors of melanoma. Surg. Clin. North Am. 2020;100:1–12. doi: 10.1016/j.suc.2019.09.005. [DOI] [PubMed] [Google Scholar]
- 259.Lambert AW, Pattabiraman DR, Weinberg RA. Emerging biological principles of metastasis. Cell. 2017;168:670–691. doi: 10.1016/j.cell.2016.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Fares J, et al. Molecular principles of metastasis: a hallmark of cancer revisited. Signal Transduct. Target Ther. 2020;5:28. doi: 10.1038/s41392-020-0134-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Alonso SR, et al. A high-throughput study in melanoma identifies epithelial-mesenchymal transition as a major determinant of metastasis. Cancer Res. 2007;67:3450–3460. doi: 10.1158/0008-5472.CAN-06-3481. [DOI] [PubMed] [Google Scholar]
- 262.Pearlman RL, Montes de Oca MK, Pal HC, Afaq F. Potential therapeutic targets of epithelial-mesenchymal transition in melanoma. Cancer Lett. 2017;391:125–140. doi: 10.1016/j.canlet.2017.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Caramel J, et al. A switch in the expression of embryonic EMT-inducers drives the development of malignant melanoma. Cancer Cell. 2013;24:466–480. doi: 10.1016/j.ccr.2013.08.018. [DOI] [PubMed] [Google Scholar]
- 264.Tehranian, C. et al. The PI3K/Akt/mTOR pathway as a preventive target in melanoma brain metastasis. Neuro Oncol. 10.1093/neuonc/noab159 (2021). [DOI] [PMC free article] [PubMed]
- 265.Bucheit AD, et al. Complete loss of PTEN protein expression correlates with shorter time to brain metastasis and survival in stage IIIB/C melanoma patients with BRAFV600 mutations. Clin. Cancer Res. 2014;20:5527–5536. doi: 10.1158/1078-0432.CCR-14-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Dai DL, Martinka M, Li G. Prognostic significance of activated Akt expression in melanoma: a clinicopathologic study of 292 cases. J. Clin. Oncol. 2005;23:1473–1482. doi: 10.1200/JCO.2005.07.168. [DOI] [PubMed] [Google Scholar]
- 267.O’Reilly KE, et al. Phosphorylated 4E-BP1 is associated with poor survival in melanoma. Clin. Cancer Res. 2009;15:2872–2878. doi: 10.1158/1078-0432.CCR-08-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Chen G, et al. Molecular profiling of patient-matched brain and extracranial melanoma metastases implicates the PI3K pathway as a therapeutic target. Clin. Cancer Res. 2014;20:5537–5546. doi: 10.1158/1078-0432.CCR-13-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Davies MA, et al. Integrated molecular and clinical analysis of AKT activation in metastatic melanoma. Clin. Cancer Res. 2009;15:7538–7546. doi: 10.1158/1078-0432.CCR-09-1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Nogueira C, et al. Cooperative interactions of PTEN deficiency and RAS activation in melanoma metastasis. Oncogene. 2010;29:6222–6232. doi: 10.1038/onc.2010.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Yoo JH, et al. The small GTPase ARF6 activates PI3K in melanoma to induce a prometastatic state. Cancer Res. 2019;79:2892–2908. doi: 10.1158/0008-5472.CAN-18-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Cho JH, et al. AKT1 activation promotes development of melanoma metastases. Cell Rep. 2015;13:898–905. doi: 10.1016/j.celrep.2015.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Govindarajan B, et al. Overexpression of Akt converts radial growth melanoma to vertical growth melanoma. J. Clin. Invest. 2007;117:719–729. doi: 10.1172/JCI30102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Fenouille N, et al. The epithelial-mesenchymal transition (EMT) regulatory factor SLUG (SNAI2) is a downstream target of SPARC and AKT in promoting melanoma cell invasion. PLoS ONE. 2012;7:e40378. doi: 10.1371/journal.pone.0040378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Lionarons DA, et al. RAC1(P29S) induces a mesenchymal phenotypic switch via serum response factor to promote melanoma development and therapy resistance. Cancer Cell. 2019;36:68–83.e69. doi: 10.1016/j.ccell.2019.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Wei CY, et al. TRIM44 activates the AKT/mTOR signal pathway to induce melanoma progression by stabilizing TLR4. J. Exp. Clin. Cancer Res. 2019;38:137. doi: 10.1186/s13046-019-1138-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Sadeghi RS, et al. Wnt5a signaling induced phosphorylation increases APT1 activity and promotes melanoma metastatic behavior. Elife. 2018;7:e34362. doi: 10.7554/eLife.34362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Da Forno PD, et al. WNT5A expression increases during melanoma progression and correlates with outcome. Clin. Cancer Res. 2008;14:5825–5832. doi: 10.1158/1078-0432.CCR-07-5104. [DOI] [PubMed] [Google Scholar]
- 279.Dissanayake SK, et al. The Wnt5A/protein kinase C pathway mediates motility in melanoma cells via the inhibition of metastasis suppressors and initiation of an epithelial to mesenchymal transition. J. Biol. Chem. 2007;282:17259–17271. doi: 10.1074/jbc.M700075200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Lv C, et al. Mesenchymal stem cells induce epithelial mesenchymal transition in melanoma by paracrine secretion of transforming growth factor-beta. Melanoma Res. 2017;27:74–84. doi: 10.1097/CMR.0000000000000325. [DOI] [PubMed] [Google Scholar]
- 281.Cantelli G, et al. TGF-beta-induced transcription sustains amoeboid melanoma migration and dissemination. Curr. Biol. 2015;25:2899–2914. doi: 10.1016/j.cub.2015.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Wels C, et al. Transcriptional activation of ZEB1 by Slug leads to cooperative regulation of the epithelial-mesenchymal transition-like phenotype in melanoma. J. Invest. Dermatol. 2011;131:1877–1885. doi: 10.1038/jid.2011.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Vandamme N, et al. The EMT transcription factor ZEB2 promotes proliferation of primary and metastatic melanoma while suppressing an invasive, mesenchymal-like phenotype. Cancer Res. 2020;80:2983–2995. doi: 10.1158/0008-5472.CAN-19-2373. [DOI] [PubMed] [Google Scholar]
- 284.Denecker G, et al. Identification of a ZEB2-MITF-ZEB1 transcriptional network that controls melanogenesis and melanoma progression. Cell Death Differ. 2014;21:1250–1261. doi: 10.1038/cdd.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Dilshat R, et al. MITF reprograms the extracellular matrix and focal adhesion in melanoma. Elife. 2021;10:e63093. doi: 10.7554/eLife.63093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Smart JA, Oleksak JE, Hartsough EJ. Cell adhesion molecules in plasticity and metastasis. Mol. Cancer Res. 2021;19:25–37. doi: 10.1158/1541-7786.MCR-20-0595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Giancotti FG, Ruoslahti E. Integrin signaling. Science. 1999;285:1028–1032. doi: 10.1126/science.285.5430.1028. [DOI] [PubMed] [Google Scholar]
- 288.Takada Y, Ye X, Simon S. The integrins. Genome Biol. 2007;8:215. doi: 10.1186/gb-2007-8-5-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Ramos DM, Berston ED, Kramer RH. Analysis of integrin receptors for laminin and type IV collagen on metastatic B16 melanoma cells. Cancer Res. 1990;50:728–734. [PubMed] [Google Scholar]
- 290.Iida J, et al. Spreading and focal contact formation of human melanoma cells in response to the stimulation of both melanoma-associated proteoglycan (NG2) and alpha 4 beta 1 integrin. Cancer Res. 1995;55:2177–2185. [PubMed] [Google Scholar]
- 291.Felding-Habermann B, Mueller BM, Romerdahl CA, Cheresh DA. Involvement of integrin alpha V gene expression in human melanoma tumorigenicity. J. Clin. Invest. 1992;89:2018–2022. doi: 10.1172/JCI115811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Natali PG, et al. Clinical significance of alpha(v)beta3 integrin and intercellular adhesion molecule-1 expression in cutaneous malignant melanoma lesions. Cancer Res. 1997;57:1554–1560. [PubMed] [Google Scholar]
- 293.Petitclerc E, et al. Integrin alpha(v)beta3 promotes M21 melanoma growth in human skin by regulating tumor cell survival. Cancer Res. 1999;59:2724–2730. [PubMed] [Google Scholar]
- 294.Montgomery AM, Reisfeld RA, Cheresh DA. Integrin alpha v beta 3 rescues melanoma cells from apoptosis in three-dimensional dermal collagen. Proc. Natl Acad. Sci. USA. 1994;91:8856–8860. doi: 10.1073/pnas.91.19.8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Van Belle PA, et al. Progression-related expression of beta3 integrin in melanomas and nevi. Hum. Pathol. 1999;30:562–567. doi: 10.1016/s0046-8177(99)90202-2. [DOI] [PubMed] [Google Scholar]
- 296.Hsu MY, et al. Adenoviral gene transfer of beta3 integrin subunit induces conversion from radial to vertical growth phase in primary human melanoma. Am. J. Pathol. 1998;153:1435–1442. doi: 10.1016/s0002-9440(10)65730-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Jiao Y, et al. Matrix metalloproteinase-2 promotes alphavbeta3 integrin-mediated adhesion and migration of human melanoma cells by cleaving fibronectin. PLoS ONE. 2012;7:e41591. doi: 10.1371/journal.pone.0041591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Vuoriluoto K, et al. Syndecan-1 and -4 differentially regulate oncogenic K-ras dependent cell invasion into collagen through alpha2beta1 integrin and MT1-MMP. Matrix Biol. 2011;30:207–217. doi: 10.1016/j.matbio.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 299.Mitra A, Chakrabarti J, Chatterjee A. Binding of alpha5 monoclonal antibody to cell surface alpha5beta1 integrin modulates MMP-2 and MMP-7 activity in B16F10 melanoma cells. J. Environ. Pathol. Toxicol. Oncol. 2003;22:167–178. doi: 10.1615/jenvpathtoxoncol.v22.i3.20. [DOI] [PubMed] [Google Scholar]
- 300.Zeng DF, et al. Autoantibody against integrin alphav beta3 contributes to thrombocytopenia by blocking the migration and adhesion of megakaryocytes. J. Thromb. Haemost. 2018;16:1843–1856. doi: 10.1111/jth.14214. [DOI] [PubMed] [Google Scholar]
- 301.Friedlander M, et al. Definition of two angiogenic pathways by distinct alpha v integrins. Science. 1995;270:1500–1502. doi: 10.1126/science.270.5241.1500. [DOI] [PubMed] [Google Scholar]
- 302.Dai DL, et al. Increased expression of integrin-linked kinase is correlated with melanoma progression and poor patient survival. Clin. Cancer Res. 2003;9:4409–4414. [PubMed] [Google Scholar]
- 303.Wong RP, Ng P, Dedhar S, Li G. The role of integrin-linked kinase in melanoma cell migration, invasion, and tumor growth. Mol. Cancer Ther. 2007;6:1692–1700. doi: 10.1158/1535-7163.MCT-07-0134. [DOI] [PubMed] [Google Scholar]
- 304.Wani AA, Jafarnejad SM, Zhou J, Li G. Integrin-linked kinase regulates melanoma angiogenesis by activating NF-kappaB/interleukin-6 signaling pathway. Oncogene. 2011;30:2778–2788. doi: 10.1038/onc.2010.644. [DOI] [PubMed] [Google Scholar]
- 305.Gil D, Zarzycka M, Ciolczyk-Wierzbicka D, Laidler P. Integrin linked kinase regulates endosomal recycling of N-cadherin in melanoma cells. Cell Signal. 2020;72:109642. doi: 10.1016/j.cellsig.2020.109642. [DOI] [PubMed] [Google Scholar]
- 306.Huang R, Rofstad EK. Integrins as therapeutic targets in the organ-specific metastasis of human malignant melanoma. J. Exp. Clin. Cancer Res. 2018;37:92. doi: 10.1186/s13046-018-0763-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Jiang Y, et al. Abituzumab targeting of alphaV-class integrins inhibits prostate cancer progression. Mol. Cancer Res. 2017;15:875–883. doi: 10.1158/1541-7786.MCR-16-0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Uhl W, et al. Safety, tolerability, and pharmacokinetics of the novel alphav-integrin antibody EMD 525797 (DI17E6) in healthy subjects after ascending single intravenous doses. Invest. N. Drugs. 2014;32:347–354. doi: 10.1007/s10637-013-0038-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Kim KB, et al. A randomized phase II study of cilengitide (EMD 121974) in patients with metastatic melanoma. Melanoma Res. 2012;22:294–301. doi: 10.1097/CMR.0b013e32835312e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Weiler SME, et al. TAZ target gene ITGAV regulates invasion and feeds back positively on YAP and TAZ in liver cancer cells. Cancer Lett. 2020;473:164–175. doi: 10.1016/j.canlet.2019.12.044. [DOI] [PubMed] [Google Scholar]
- 311.Hazan RB, et al. Cadherin switch in tumor progression. Ann. N. Y. Acad. Sci. 2004;1014:155–163. doi: 10.1196/annals.1294.016. [DOI] [PubMed] [Google Scholar]
- 312.Lade-Keller J, et al. E- to N-cadherin switch in melanoma is associated with decreased expression of phosphatase and tensin homolog and cancer progression. Br. J. Dermatol. 2013;169:618–628. doi: 10.1111/bjd.12426. [DOI] [PubMed] [Google Scholar]
- 313.Li G, Satyamoorthy K, Herlyn M. N-cadherin-mediated intercellular interactions promote survival and migration of melanoma cells. Cancer Res. 2001;61:3819–3825. [PubMed] [Google Scholar]
- 314.Van Marck V, et al. P-cadherin promotes cell-cell adhesion and counteracts invasion in human melanoma. Cancer Res. 2005;65:8774–8783. doi: 10.1158/0008-5472.CAN-04-4414. [DOI] [PubMed] [Google Scholar]
- 315.Jacobs K, et al. P-cadherin expression reduces melanoma growth, invasion, and responsiveness to growth factors in nude mice. Eur. J. Cancer Prev. 2011;20:207–216. doi: 10.1097/CEJ.0b013e3283429e8b. [DOI] [PubMed] [Google Scholar]
- 316.Gowda R, et al. The role of exosomes in metastasis and progression of melanoma. Cancer Treat. Rev. 2020;85:101975. doi: 10.1016/j.ctrv.2020.101975. [DOI] [PubMed] [Google Scholar]
- 317.Whitehead B, et al. Tumour exosomes display differential mechanical and complement activation properties dependent on malignant state: implications in endothelial leakiness. J. Extracell. Vesicles. 2015;4:29685. doi: 10.3402/jev.v4.29685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Azmi AS, Bao B, Sarkar FH. Exosomes in cancer development, metastasis, and drug resistance: a comprehensive review. Cancer Metastasis Rev. 2013;32:623–642. doi: 10.1007/s10555-013-9441-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Hood JL, San RS, Wickline SA. Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res. 2011;71:3792–3801. doi: 10.1158/0008-5472.CAN-10-4455. [DOI] [PubMed] [Google Scholar]
- 320.Guo D, et al. RAB27A promotes melanoma cell invasion and metastasis via regulation of pro-invasive exosomes. Int. J. Cancer. 2019;144:3070–3085. doi: 10.1002/ijc.32064. [DOI] [PubMed] [Google Scholar]
- 321.Peinado H, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Lazar I, et al. Proteome characterization of melanoma exosomes reveals a specific signature for metastatic cell lines. Pigment Cell Melanoma Res. 2015;28:464–475. doi: 10.1111/pcmr.12380. [DOI] [PubMed] [Google Scholar]
- 323.Xiao D, et al. Identifying mRNA, microRNA and protein profiles of melanoma exosomes. PLoS ONE. 2012;7:e46874. doi: 10.1371/journal.pone.0046874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Xiao D, et al. Melanoma cell-derived exosomes promote epithelial-mesenchymal transition in primary melanocytes through paracrine/autocrine signaling in the tumor microenvironment. Cancer Lett. 2016;376:318–327. doi: 10.1016/j.canlet.2016.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Luan W, et al. Exosomal miR-106b-5p derived from melanoma cell promotes primary melanocytes epithelial-mesenchymal transition through targeting EphA4. J. Exp. Clin. Cancer Res. 2021;40:107. doi: 10.1186/s13046-021-01906-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Gener Lahav T, et al. Melanoma-derived extracellular vesicles instigate proinflammatory signaling in the metastatic microenvironment. Int. J. Cancer. 2019;145:2521–2534. doi: 10.1002/ijc.32521. [DOI] [PubMed] [Google Scholar]
- 327.Gerloff D, et al. Melanoma-derived exosomal miR-125b-5p educates tumor associated macrophages (TAMs) by targeting lysosomal acid lipase A (LIPA) Cancers. 2020;12:464. doi: 10.3390/cancers12020464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Plebanek MP, et al. Pre-metastatic cancer exosomes induce immune surveillance by patrolling monocytes at the metastatic niche. Nat. Commun. 2017;8:1319. doi: 10.1038/s41467-017-01433-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Gyukity-Sebestyen E, et al. Melanoma-derived exosomes induce PD-1 overexpression and tumor progression via mesenchymal stem cell oncogenic reprogramming. Front. Immunol. 2019;10:2459. doi: 10.3389/fimmu.2019.02459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Tsering T, et al. Uveal melanoma-derived extracellular vesicles display transforming potential and carry protein cargo involved in metastatic niche preparation. Cancers. 2020;12:2923. doi: 10.3390/cancers12102923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Biagioni A, et al. uPAR-expressing melanoma exosomes promote angiogenesis by VE-cadherin, EGFR and uPAR overexpression and rise of ERK1,2 signaling in endothelial cells. Cell. Mol. Life Sci. 2021;78:3057–3072. doi: 10.1007/s00018-020-03707-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Vandyck, H H, et al. Rethinking the biology of metastatic melanoma: a holistic approach. Cancer Metastasis Rev. 2021;40:603–624. doi: 10.1007/s10555-021-09960-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Shivakumar K, et al. Synthetic hydrogel microspheres as substrata for cell adhesion and growth. Vitr. Cell. Dev. Biol. 1989;25:353–357. doi: 10.1007/BF02624598. [DOI] [PubMed] [Google Scholar]
- 334.Bergenwald C, Westermark G, Sander B. Variable expression of tumor necrosis factor alpha in human malignant melanoma localized by in situ hybridization for mRNA. Cancer Immunol. Immunother. 1997;44:335–340. doi: 10.1007/s002620050391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Castelli C, et al. Expression of interleukin 1 alpha, interleukin 6, and tumor necrosis factor alpha genes in human melanoma clones is associated with that of mutated N-RAS oncogene. Cancer Res. 1994;54:4785–4790. [PubMed] [Google Scholar]
- 336.Lienard D, et al. High-dose recombinant tumor necrosis factor alpha in combination with interferon gamma and melphalan in isolation perfusion of the limbs for melanoma and sarcoma. J. Clin. Oncol. 1992;10:52–60. doi: 10.1200/JCO.1992.10.1.52. [DOI] [PubMed] [Google Scholar]
- 337.Eggermont AM, ten Hagen TL. Tumor necrosis factor-based isolated limb perfusion for soft tissue sarcoma and melanoma: ten years of successful antivascular therapy. Curr. Oncol. Rep. 2003;5:79–80. doi: 10.1007/s11912-003-0091-x. [DOI] [PubMed] [Google Scholar]
- 338.Osanto S, Jansen R, Vloemans M. Downmodulation of c-myc expression by interferon gamma and tumour necrosis factor alpha precedes growth arrest in human melanoma cells. Eur. J. Cancer. 1992;28A:1622–1627. doi: 10.1016/0959-8049(92)90055-7. [DOI] [PubMed] [Google Scholar]
- 339.Liu XY, et al. RIP1 kinase is an oncogenic driver in melanoma. Cancer Res. 2015;75:1736–1748. doi: 10.1158/0008-5472.CAN-14-2199. [DOI] [PubMed] [Google Scholar]
- 340.Gray-Schopfer VC, Karasarides M, Hayward R, Marais R. Tumor necrosis factor-alpha blocks apoptosis in melanoma cells when BRAF signaling is inhibited. Cancer Res. 2007;67:122–129. doi: 10.1158/0008-5472.CAN-06-1880. [DOI] [PubMed] [Google Scholar]
- 341.Shao Y, Le K, Cheng H, Aplin AE. NF-kappaB regulation of c-FLIP promotes TNFalpha-mediated RAF inhibitor resistance in melanoma. J. Invest. Dermatol. 2015;135:1839–1848. doi: 10.1038/jid.2015.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Zhu N, et al. Melanoma cell migration is upregulated by tumour necrosis factor-alpha and suppressed by alpha-melanocyte-stimulating hormone. Br. J. Cancer. 2004;90:1457–1463. doi: 10.1038/sj.bjc.6601698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Zhu N, et al. Melanoma cell attachment, invasion, and integrin expression is upregulated by tumor necrosis factor alpha and suppressed by alpha melanocyte stimulating hormone. J. Invest. Dermatol. 2002;119:1165–1171. doi: 10.1046/j.1523-1747.2002.19516.x. [DOI] [PubMed] [Google Scholar]
- 344.Rossi S, et al. TNF-alpha and metalloproteases as key players in melanoma cells aggressiveness. J. Exp. Clin. Cancer Res. 2018;37:326. doi: 10.1186/s13046-018-0982-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Katerinaki E, Evans GS, Lorigan PC, MacNeil S. TNF-alpha increases human melanoma cell invasion and migration in vitro: the role of proteolytic enzymes. Br. J. Cancer. 2003;89:1123–1129. doi: 10.1038/sj.bjc.6601257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Riesenberg S, et al. MITF and c-Jun antagonism interconnects melanoma dedifferentiation with pro-inflammatory cytokine responsiveness and myeloid cell recruitment. Nat. Commun. 2015;6:8755. doi: 10.1038/ncomms9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Bertrand F, et al. TNFalpha blockade overcomes resistance to anti-PD-1 in experimental melanoma. Nat. Commun. 2017;8:2256. doi: 10.1038/s41467-017-02358-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Vredevoogd DW, et al. Augmenting immunotherapy impact by lowering tumor TNF cytotoxicity threshold. Cell. 2019;178:585–599.e515. doi: 10.1016/j.cell.2019.06.014. [DOI] [PubMed] [Google Scholar]
- 349.Garcia-Diaz A, et al. Interferon receptor signaling pathways regulating PD-L1 and PD-L2 expression. Cell Rep. 2017;19:1189–1201. doi: 10.1016/j.celrep.2017.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Thiem A, et al. IFN-gamma-induced PD-L1 expression in melanoma depends on p53 expression. J. Exp. Clin. Cancer Res. 2019;38:397. doi: 10.1186/s13046-019-1403-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Mo X, et al. Interferon-gamma signaling in melanocytes and melanoma cells regulates expression of CTLA-4. Cancer Res. 2018;78:436–450. doi: 10.1158/0008-5472.CAN-17-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Dizier B, et al. A Th1/IFNgamma gene signature is prognostic in the adjuvant setting of resectable high-risk melanoma but not in non-small cell lung cancer. Clin. Cancer Res. 2020;26:1725–1735. doi: 10.1158/1078-0432.CCR-18-3717. [DOI] [PubMed] [Google Scholar]
- 353.Ayers M, et al. IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Invest. 2017;127:2930–2940. doi: 10.1172/JCI91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Grasso CS, et al. Conserved interferon-gamma signaling drives clinical response to immune checkpoint blockade therapy in melanoma. Cancer Cell. 2021;39:122. doi: 10.1016/j.ccell.2020.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Lang X, et al. Radiotherapy and immunotherapy promote tumoral lipid oxidation and ferroptosis via synergistic repression of SLC7A11. Cancer Discov. 2019;9:1673–1685. doi: 10.1158/2159-8290.CD-19-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Wang W, et al. CD8(+) T cells regulate tumour ferroptosis during cancer immunotherapy. Nature. 2019;569:270–274. doi: 10.1038/s41586-019-1170-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Benci JL, et al. Tumor interferon signaling regulates a multigenic resistance program to immune checkpoint blockade. Cell. 2016;167:1540–1554.e1512. doi: 10.1016/j.cell.2016.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Jacquelot N, et al. Sustained type I interferon signaling as a mechanism of resistance to PD-1 blockade. Cell Res. 2019;29:846–861. doi: 10.1038/s41422-019-0224-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Chin YE, et al. Activation of the STAT signaling pathway can cause expression of caspase 1 and apoptosis. Mol. Cell. Biol. 1997;17:5328–5337. doi: 10.1128/mcb.17.9.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Gresser I, Bourali C. Exogenous interferon and inducers of interferon in the treatment Balb-c mice inoculated with RC19 tumour cells. Nature. 1969;223:844–845. doi: 10.1038/223844a0. [DOI] [PubMed] [Google Scholar]
- 361.von Stamm U, et al. Effects of systemic interferon-alpha (IFN-alpha) on the antigenic phenotype of melanoma metastases. EORTC melanoma group cooperative study No. 18852. Melanoma Res. 1993;3:173–180. doi: 10.1097/00008390-199306000-00005. [DOI] [PubMed] [Google Scholar]
- 362.Biron CA. Interferons alpha and beta as immune regulators-a new look. Immunity. 2001;14:661–664. doi: 10.1016/s1074-7613(01)00154-6. [DOI] [PubMed] [Google Scholar]
- 363.Martin-Henao GA, et al. L-selectin expression is low on CD34+ cells from patients with chronic myeloid leukemia and interferon-a up-regulates this expression. Haematologica. 2000;85:139–146. [PubMed] [Google Scholar]
- 364.Christofyllakis K, et al. Adjuvant therapy of high-risk (Stages IIC-IV) malignant melanoma in the post interferon-alpha era: a systematic review and meta-analysis. Front. Oncol. 2020;10:637161. doi: 10.3389/fonc.2020.637161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Shi Q, et al. Interferon-alpha1b for the treatment of metastatic melanoma: results of a retrospective study. Anticancer Drugs. 2021;32:1105–1110. doi: 10.1097/CAD.0000000000001120. [DOI] [PubMed] [Google Scholar]
- 366.Bentebibel SE, Diab A. Cytokines in the treatment of melanoma. Curr. Oncol. Rep. 2021;23:83. doi: 10.1007/s11912-021-01064-4. [DOI] [PubMed] [Google Scholar]
- 367.Rosenberg SA. IL-2: the first effective immunotherapy for human cancer. J. Immunol. 2014;192:5451–5458. doi: 10.4049/jimmunol.1490019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Boyman O, Sprent J. The role of interleukin-2 during homeostasis and activation of the immune system. Nat. Rev. Immunol. 2012;12:180–190. doi: 10.1038/nri3156. [DOI] [PubMed] [Google Scholar]
- 369.Atkins MB, Regan M, McDermott D. Update on the role of interleukin 2 and other cytokines in the treatment of patients with stage IV renal carcinoma. Clin. Cancer Res. 2004;10:6342S–6346S. doi: 10.1158/1078-0432.CCR-040029. [DOI] [PubMed] [Google Scholar]
- 370.Emmerich J, et al. IL-10 directly activates and expands tumor-resident CD8(+) T cells without de novo infiltration from secondary lymphoid organs. Cancer Res. 2012;72:3570–3581. doi: 10.1158/0008-5472.CAN-12-0721. [DOI] [PubMed] [Google Scholar]
- 371.Kobayashi H, et al. Role of trans-cellular IL-15 presentation in the activation of NK cell-mediated killing, which leads to enhanced tumor immunosurveillance. Blood. 2005;105:721–727. doi: 10.1182/blood-2003-12-4187. [DOI] [PubMed] [Google Scholar]
- 372.Dubois S, Mariner J, Waldmann TA, Tagaya Y. IL-15Ralpha recycles and presents IL-15 In trans to neighboring cells. Immunity. 2002;17:537–547. doi: 10.1016/s1074-7613(02)00429-6. [DOI] [PubMed] [Google Scholar]
- 373.Chan IH, et al. The potentiation of IFN-gamma and induction of cytotoxic proteins by pegylated IL-10 in human CD8 T cells. J. Interferon Cytokine Res. 2015;35:948–955. doi: 10.1089/jir.2014.0221. [DOI] [PubMed] [Google Scholar]
- 374.Wrangle JM, et al. ALT-803, an IL-15 superagonist, in combination with nivolumab in patients with metastatic non-small cell lung cancer: a non-randomised, open-label, phase 1b trial. Lancet Oncol. 2018;19:694–704. doi: 10.1016/S1470-2045(18)30148-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Strowig T, Henao-Mejia J, Elinav E, Flavell R. Inflammasomes in health and disease. Nature. 2012;481:278–286. doi: 10.1038/nature10759. [DOI] [PubMed] [Google Scholar]
- 376.Pandey A, Shen C, Feng S, Man SM. Cell biology of inflammasome activation. Trends Cell Biol. 2021;31:924–939. doi: 10.1016/j.tcb.2021.06.010. [DOI] [PubMed] [Google Scholar]
- 377.Okamoto M, et al. Constitutively active inflammasome in human melanoma cells mediating autoinflammation via caspase-1 processing and secretion of interleukin-1beta. J. Biol. Chem. 2010;285:6477–6488. doi: 10.1074/jbc.M109.064907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Ju M, et al. Pan-cancer analysis of NLRP3 inflammasome with potential implications in prognosis and immunotherapy in human cancer. Brief. Bioinform. 2021;22:bbaa345. doi: 10.1093/bib/bbaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Tengesdal IW, et al. Targeting tumor-derived NLRP3 reduces melanoma progression by limiting MDSCs expansion. Proc. Natl Acad. Sci. USA. 2021;118:e2000915118. doi: 10.1073/pnas.2000915118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Theivanthiran B, et al. A tumor-intrinsic PD-L1/NLRP3 inflammasome signaling pathway drives resistance to anti-PD-1 immunotherapy. J. Clin. Invest. 2020;130:2570–2586. doi: 10.1172/JCI133055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Emmett MS, Dewing D, Pritchard-Jones RO. Angiogenesis and melanoma - from basic science to clinical trials. Am. J. Cancer Res. 2011;1:852–868. [PMC free article] [PubMed] [Google Scholar]
- 382.Warren BA, Shubik P. The growth of the blood supply to melanoma transplants in the hamster cheek pouch. Lab. Invest. 1966;15:464–478. [PubMed] [Google Scholar]
- 383.Marcoval J, et al. Angiogenesis and malignant melanoma. Angiogenesis is related to the development of vertical (tumorigenic) growth phase. J. Cutan. Pathol. 1997;24:212–218. doi: 10.1111/j.1600-0560.1997.tb01583.x. [DOI] [PubMed] [Google Scholar]
- 384.Mabeta P. Paradigms of vascularization in melanoma: clinical significance and potential for therapeutic targeting. Biomed. Pharmacother. 2020;127:110135. doi: 10.1016/j.biopha.2020.110135. [DOI] [PubMed] [Google Scholar]
- 385.Cho WC, Jour G, Aung PP. Role of angiogenesis in melanoma progression: update on key angiogenic mechanisms and other associated components. Semin. Cancer Biol. 2019;59:175–186. doi: 10.1016/j.semcancer.2019.06.015. [DOI] [PubMed] [Google Scholar]
- 386.Lacal PM, et al. Human melanoma cells secrete and respond to placenta growth factor and vascular endothelial growth factor. J. Invest. Dermatol. 2000;115:1000–1007. doi: 10.1046/j.1523-1747.2000.00199.x. [DOI] [PubMed] [Google Scholar]
- 387.Claffey KP, et al. Expression of vascular permeability factor/vascular endothelial growth factor by melanoma cells increases tumor growth, angiogenesis, and experimental metastasis. Cancer Res. 1996;56:172–181. [PubMed] [Google Scholar]
- 388.Jouve N, et al. CD146 mediates VEGF-induced melanoma cell extravasation through FAK activation. Int. J. Cancer. 2015;137:50–60. doi: 10.1002/ijc.29370. [DOI] [PubMed] [Google Scholar]
- 389.Giatromanolaki A, et al. Hypoxia-inducible factors 1alpha and 2alpha are related to vascular endothelial growth factor expression and a poorer prognosis in nodular malignant melanomas of the skin. Melanoma Res. 2003;13:493–501. doi: 10.1097/00008390-200310000-00008. [DOI] [PubMed] [Google Scholar]
- 390.Becker D, Meier CB, Herlyn M. Proliferation of human malignant melanomas is inhibited by antisense oligodeoxynucleotides targeted against basic fibroblast growth factor. EMBO J. 1989;8:3685–3691. doi: 10.1002/j.1460-2075.1989.tb08543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Rodeck U, Becker D, Herlyn M. Basic fibroblast growth factor in human melanoma. Cancer Cells. 1991;3:308–311. [PubMed] [Google Scholar]
- 392.Mahabeleshwar GH, Byzova TV. Angiogenesis in melanoma. Semin. Oncol. 2007;34:555–565. doi: 10.1053/j.seminoncol.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Marcellini M, et al. Increased melanoma growth and metastasis spreading in mice overexpressing placenta growth factor. Am. J. Pathol. 2006;169:643–654. doi: 10.2353/ajpath.2006.051041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Luca M, et al. Expression of interleukin-8 by human melanoma cells up-regulates MMP-2 activity and increases tumor growth and metastasis. Am. J. Pathol. 1997;151:1105–1113. [PMC free article] [PubMed] [Google Scholar]
- 395.Li A, et al. Autocrine role of interleukin-8 in induction of endothelial cell proliferation, survival, migration and MMP-2 production and angiogenesis. Angiogenesis. 2005;8:63–71. doi: 10.1007/s10456-005-5208-4. [DOI] [PubMed] [Google Scholar]
- 396.Huang S, et al. Fully humanized neutralizing antibodies to interleukin-8 (ABX-IL8) inhibit angiogenesis, tumor growth, and metastasis of human melanoma. Am. J. Pathol. 2002;161:125–134. doi: 10.1016/S0002-9440(10)64164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Singh S, et al. CXCL8 and its cognate receptors in melanoma progression and metastasis. Future Oncol. 2010;6:111–116. doi: 10.2217/fon.09.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Jendreyko N, Popkov M, Rader C, Barbas CF. 3rd. Phenotypic knockout of VEGF-R2 and Tie-2 with an intradiabody reduces tumor growth and angiogenesis in vivo. Proc. Natl Acad. Sci. USA. 2005;102:8293–8298. doi: 10.1073/pnas.0503168102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Fagiani E, Christofori G. Angiopoietins in angiogenesis. Cancer Lett. 2013;328:18–26. doi: 10.1016/j.canlet.2012.08.018. [DOI] [PubMed] [Google Scholar]
- 400.Hansen TM, Singh H, Tahir TA, Brindle NP. Effects of angiopoietins-1 and -2 on the receptor tyrosine kinase Tie2 are differentially regulated at the endothelial cell surface. Cell Signal. 2010;22:527–532. doi: 10.1016/j.cellsig.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Varker KA, et al. A randomized phase 2 trial of bevacizumab with or without daily low-dose interferon alfa-2b in metastatic malignant melanoma. Ann. Surg. Oncol. 2007;14:2367–2376. doi: 10.1245/s10434-007-9389-5. [DOI] [PubMed] [Google Scholar]
- 402.Curtin JA, et al. Distinct sets of genetic alterations in melanoma. N. Engl. J. Med. 2005;353:2135–2147. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 403.Ballantyne AD, Garnock-Jones KP. Dabrafenib: first global approval. Drugs. 2013;73:1367–1376. doi: 10.1007/s40265-013-0095-2. [DOI] [PubMed] [Google Scholar]
- 404.Eisenhauer EA, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur. J. Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 405.McArthur GA, et al. Safety and efficacy of vemurafenib in BRAF(V600E) and BRAF(V600K) mutation-positive melanoma (BRIM-3): extended follow-up of a phase 3, randomised, open-label study. Lancet Oncol. 2014;15:323–332. doi: 10.1016/S1470-2045(14)70012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Flaherty KT, et al. Improved survival with MEK inhibition in BRAF-mutated melanoma. N. Engl. J. Med. 2012;367:107–114. doi: 10.1056/NEJMoa1203421. [DOI] [PubMed] [Google Scholar]
- 407.Ascierto PA, et al. MEK162 for patients with advanced melanoma harbouring NRAS or Val600 BRAF mutations: a non-randomised, open-label phase 2 study. Lancet Oncol. 2013;14:249–256. doi: 10.1016/S1470-2045(13)70024-X. [DOI] [PubMed] [Google Scholar]
- 408.Atefi M, et al. Reversing melanoma cross-resistance to BRAF and MEK inhibitors by co-targeting the AKT/mTOR pathway. PLoS ONE. 2011;6:e28973. doi: 10.1371/journal.pone.0028973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Ji Z, et al. p53 rescue through HDM2 antagonism suppresses melanoma growth and potentiates MEK inhibition. J. Invest. Dermatol. 2012;132:356–364. doi: 10.1038/jid.2011.313. [DOI] [PubMed] [Google Scholar]
- 410.Kwong LN, et al. Oncogenic NRAS signaling differentially regulates survival and proliferation in melanoma. Nat. Med. 2012;18:1503–1510. doi: 10.1038/nm.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Boussemart L, et al. eIF4F is a nexus of resistance to anti-BRAF and anti-MEK cancer therapies. Nature. 2014;513:105–109. doi: 10.1038/nature13572. [DOI] [PubMed] [Google Scholar]
- 412.Tian Y, Guo W. A review of the molecular pathways involved in resistance to BRAF inhibitors in patients with advanced-stage melanoma. Med. Sci. Monit. 2020;26:e920957. doi: 10.12659/MSM.920957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Paraiso KH, et al. PTEN loss confers BRAF inhibitor resistance to melanoma cells through the suppression of BIM expression. Cancer Res. 2011;71:2750–2760. doi: 10.1158/0008-5472.CAN-10-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Nathanson KL, et al. Tumor genetic analyses of patients with metastatic melanoma treated with the BRAF inhibitor dabrafenib (GSK2118436) Clin. Cancer Res. 2013;19:4868–4878. doi: 10.1158/1078-0432.CCR-13-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Pratilas CA, et al. (V600E)BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc. Natl Acad. Sci. USA. 2009;106:4519–4524. doi: 10.1073/pnas.0900780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Kiuru M, Busam KJ. The NF1 gene in tumor syndromes and melanoma. Lab. Invest. 2017;97:146–157. doi: 10.1038/labinvest.2016.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Watson IR, et al. The RAC1 P29S hotspot mutation in melanoma confers resistance to pharmacological inhibition of RAF. Cancer Res. 2014;74:4845–4852. doi: 10.1158/0008-5472.CAN-14-1232-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Lito P, et al. Relief of profound feedback inhibition of mitogenic signaling by RAF inhibitors attenuates their activity in BRAFV600E melanomas. Cancer Cell. 2012;22:668–682. doi: 10.1016/j.ccr.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Grimm J, et al. BRAF inhibition causes resilience of melanoma cell lines by inducing the secretion of FGF1. Oncogenesis. 2018;7:71. doi: 10.1038/s41389-018-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Nazarian R, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Abel EV, et al. Melanoma adapts to RAF/MEK inhibitors through FOXD3-mediated upregulation of ERBB3. J. Clin. Invest. 2013;123:2155–2168. doi: 10.1172/JCI65780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Zhang G, et al. Targeting mitochondrial biogenesis to overcome drug resistance to MAPK inhibitors. J. Clin. Invest. 2016;126:1834–1856. doi: 10.1172/JCI82661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Lin WM, Fisher DE. Signaling and immune regulation in melanoma development and responses to therapy. Annu. Rev. Pathol. 2017;12:75–102. doi: 10.1146/annurev-pathol-052016-100208. [DOI] [PubMed] [Google Scholar]
- 424.Johannessen CM, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Whittaker SR, et al. A genome-scale RNA interference screen implicates NF1 loss in resistance to RAF inhibition. Cancer Discov. 2013;3:350–362. doi: 10.1158/2159-8290.CD-12-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Shi H, et al. Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nat. Commun. 2012;3:724. doi: 10.1038/ncomms1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Emery CM, et al. MEK1 mutations confer resistance to MEK and B-RAF inhibition. Proc. Natl Acad. Sci. USA. 2009;106:20411–20416. doi: 10.1073/pnas.0905833106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Hugo W, et al. Non-genomic and immune evolution of melanoma acquiring MAPKi resistance. Cell. 2015;162:1271–1285. doi: 10.1016/j.cell.2015.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Van Allen EM, et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Disco. 2014;4:94–109. doi: 10.1158/2159-8290.CD-13-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Villanueva J, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–695. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Wang J, et al. A secondary mutation in BRAF confers resistance to RAF inhibition in a BRAF(V600E)-mutant brain tumor. Cancer Discov. 2018;8:1130–1141. doi: 10.1158/2159-8290.CD-17-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Fisher ML, et al. Inhibition of YAP function overcomes BRAF inhibitor resistance in melanoma cancer stem cells. Oncotarget. 2017;8:110257–110272. doi: 10.18632/oncotarget.22628. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 433.Kim MH, et al. Actin remodeling confers BRAF inhibitor resistance to melanoma cells through YAP/TAZ activation. EMBO J. 2016;35:462–478. doi: 10.15252/embj.201592081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Kim H, et al. Downregulation of the ubiquitin ligase RNF125 underlies resistance of melanoma cells to BRAF inhibitors via JAK1 deregulation. Cell Rep. 2015;11:1458–1473. doi: 10.1016/j.celrep.2015.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Kidger AM, Keyse SM. The regulation of oncogenic Ras/ERK signalling by dual-specificity mitogen activated protein kinase phosphatases (MKPs) Semin. Cell Dev. Biol. 2016;50:125–132. doi: 10.1016/j.semcdb.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Long GV, et al. Dabrafenib and trametinib versus dabrafenib and placebo for Val600 BRAF-mutant melanoma: a multicentre, double-blind, phase 3 randomised controlled trial. Lancet. 2015;386:444–451. doi: 10.1016/S0140-6736(15)60898-4. [DOI] [PubMed] [Google Scholar]
- 437.Robert C, et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. N. Engl. J. Med. 2015;372:30–39. doi: 10.1056/NEJMoa1412690. [DOI] [PubMed] [Google Scholar]
- 438.Su F, et al. RAS mutations in cutaneous squamous-cell carcinomas in patients treated with BRAF inhibitors. N. Engl. J. Med. 2012;366:207–215. doi: 10.1056/NEJMoa1105358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Flaherty KT, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N. Engl. J. Med. 2012;367:1694–1703. doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Long GV, et al. Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N. Engl. J. Med. 2014;371:1877–1888. doi: 10.1056/NEJMoa1406037. [DOI] [PubMed] [Google Scholar]
- 441.Larkin J, et al. Combined vemurafenib and cobimetinib in BRAF-mutated melanoma. N. Engl. J. Med. 2014;371:1867–1876. doi: 10.1056/NEJMoa1408868. [DOI] [PubMed] [Google Scholar]
- 442.Dummer R, et al. Encorafenib plus binimetinib versus vemurafenib or encorafenib in patients with BRAF-mutant melanoma (COLUMBUS): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2018;19:603–615. doi: 10.1016/S1470-2045(18)30142-6. [DOI] [PubMed] [Google Scholar]
- 443.Hamid O, et al. Efficacy, safety, and tolerability of approved combination BRAF and MEK inhibitor regimens for BRAF-mutant melanoma. Cancers. 2019;11:1642. doi: 10.3390/cancers11111642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Poulikakos PI, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480:387–390. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Agianian B, Gavathiotis E. Current insights of BRAF inhibitors in cancer. J. Med. Chem. 2018;61:5775–5793. doi: 10.1021/acs.jmedchem.7b01306. [DOI] [PubMed] [Google Scholar]
- 446.Karoulia Z, Gavathiotis E, Poulikakos PI. New perspectives for targeting RAF kinase in human cancer. Nat. Rev. Cancer. 2017;17:676–691. doi: 10.1038/nrc.2017.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Karoulia Z, et al. An integrated model of RAF inhibitor action predicts inhibitor activity against oncogenic BRAF signaling. Cancer Cell. 2016;30:485–498. doi: 10.1016/j.ccell.2016.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Yao Z, et al. BRAF mutants evade ERK-dependent feedback by different mechanisms that determine their sensitivity to pharmacologic inhibition. Cancer Cell. 2015;28:370–383. doi: 10.1016/j.ccell.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Peng SB, et al. Inhibition of RAF isoforms and active dimers by LY3009120 leads to anti-tumor activities in RAS or BRAF mutant cancers. Cancer Cell. 2015;28:384–398. doi: 10.1016/j.ccell.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 450.Okaniwa M, et al. Discovery of a selective kinase inhibitor (TAK-632) targeting pan-RAF inhibition: design, synthesis, and biological evaluation of C-7-substituted 1,3-benzothiazole derivatives. J. Med. Chem. 2013;56:6478–6494. doi: 10.1021/jm400778d. [DOI] [PubMed] [Google Scholar]
- 451.Anforth R, Fernandez-Penas P, Long GV. Cutaneous toxicities of RAF inhibitors. Lancet Oncol. 2013;14:e11–18. doi: 10.1016/S1470-2045(12)70413-8. [DOI] [PubMed] [Google Scholar]
- 452.Sullivan RJ, et al. A phase I study of LY3009120, a Pan-RAF Inhibitor, in patients with advanced or metastatic cancer. Mol. Cancer Ther. 2020;19:460–467. doi: 10.1158/1535-7163.MCT-19-0681. [DOI] [PubMed] [Google Scholar]
- 453.Zhang C, et al. RAF inhibitors that evade paradoxical MAPK pathway activation. Nature. 2015;526:583–586. doi: 10.1038/nature14982. [DOI] [PubMed] [Google Scholar]
- 454.Morris EJ, et al. Discovery of a novel ERK inhibitor with activity in models of acquired resistance to BRAF and MEK inhibitors. Cancer Discov. 2013;3:742–750. doi: 10.1158/2159-8290.CD-13-0070. [DOI] [PubMed] [Google Scholar]
- 455.Wong DJ, et al. Antitumor activity of the ERK inhibitor SCH772984 [corrected] against BRAF mutant, NRAS mutant and wild-type melanoma. Mol. Cancer. 2014;13:194. doi: 10.1186/1476-4598-13-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Dumble M, et al. Discovery of novel AKT inhibitors with enhanced anti-tumor effects in combination with the MEK inhibitor. PLoS ONE. 2014;9:e100880. doi: 10.1371/journal.pone.0100880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Lassen A, et al. Effects of AKT inhibitor therapy in response and resistance to BRAF inhibition in melanoma. Mol. Cancer. 2014;13:83. doi: 10.1186/1476-4598-13-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Algazi AP, et al. Dual MEK/AKT inhibition with trametinib and GSK2141795 does not yield clinical benefit in metastatic NRAS-mutant and wild-type melanoma. Pigment Cell Melanoma Res. 2018;31:110–114. doi: 10.1111/pcmr.12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Yen I, et al. ARAF mutations confer resistance to the RAF inhibitor belvarafenib in melanoma. Nature. 2021;594:418–423. doi: 10.1038/s41586-021-03515-1. [DOI] [PubMed] [Google Scholar]
- 460.Papac RJ. Spontaneous regression of cancer. Cancer Treat. Rev. 1996;22:395–423. doi: 10.1016/s0305-7372(96)90023-7. [DOI] [PubMed] [Google Scholar]
- 461.Parish CR. Cancer immunotherapy: the past, the present and the future. Immunol. Cell Biol. 2003;81:106–113. doi: 10.1046/j.0818-9641.2003.01151.x. [DOI] [PubMed] [Google Scholar]
- 462.Dunn GP, Old LJ, Schreiber RD. The immunobiology of cancer immunosurveillance and immunoediting. Immunity. 2004;21:137–148. doi: 10.1016/j.immuni.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 463.Arozarena I, Wellbrock C. Phenotype plasticity as enabler of melanoma progression and therapy resistance. Nat. Rev. Cancer. 2019;19:377–391. doi: 10.1038/s41568-019-0154-4. [DOI] [PubMed] [Google Scholar]
- 464.Margolis N, Markovits E, Markel G. Reprogramming lymphocytes for the treatment of melanoma: From biology to therapy. Adv. Drug Deliv. Rev. 2019;141:104–124. doi: 10.1016/j.addr.2019.06.005. [DOI] [PubMed] [Google Scholar]
- 465.Marzagalli M, Ebelt ND, Manuel ER. Unraveling the crosstalk between melanoma and immune cells in the tumor microenvironment. Semin. Cancer Biol. 2019;59:236–250. doi: 10.1016/j.semcancer.2019.08.002. [DOI] [PubMed] [Google Scholar]
- 466.Muenst S, et al. The immune system and cancer evasion strategies: therapeutic concepts. J. Intern. Med. 2016;279:541–562. doi: 10.1111/joim.12470. [DOI] [PubMed] [Google Scholar]
- 467.Garner H, de Visser KE. Immune crosstalk in cancer progression and metastatic spread: a complex conversation. Nat. Rev. Immunol. 2020;20:483–497. doi: 10.1038/s41577-019-0271-z. [DOI] [PubMed] [Google Scholar]
- 468.Rothlin CV, Ghosh S. Lifting the innate immune barriers to antitumor immunity. J. Immunother. Cancer. 2020;8:e000695. doi: 10.1136/jitc-2020-000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469.Cao J, Yan Q. Cancer epigenetics, tumor immunity, and immunotherapy. Trends Cancer. 2020;6:580–592. doi: 10.1016/j.trecan.2020.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Allen BM, et al. Systemic dysfunction and plasticity of the immune macroenvironment in cancer models. Nat. Med. 2020;26:1125–1134. doi: 10.1038/s41591-020-0892-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Zhai L, et al. IDO1 in cancer: a Gemini of immune checkpoints. Cell. Mol. Immunol. 2018;15:447–457. doi: 10.1038/cmi.2017.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Qin W, et al. The diverse function of PD-1/PD-L pathway beyond cancer. Front. Immunol. 2019;10:2298. doi: 10.3389/fimmu.2019.02298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 473.Balar AV, Weber JS. PD-1 and PD-L1 antibodies in cancer: current status and future directions. Cancer Immunol. Immunother. 2017;66:551–564. doi: 10.1007/s00262-017-1954-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474.Han Y, Liu D, Li L. PD-1/PD-L1 pathway: current researches in cancer. Am. J. Cancer Res. 2020;10:727–742. [PMC free article] [PubMed] [Google Scholar]
- 475.Oliveira G, et al. Phenotype, specificity and avidity of antitumour CD8(+) T cells in melanoma. Nature. 2021;596:119–125. doi: 10.1038/s41586-021-03704-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476.Parish CR, et al. Unexpected new roles for heparanase in Type 1 diabetes and immune gene regulation. Matrix Biol. 2013;32:228–233. doi: 10.1016/j.matbio.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 477.Park JM, Fisher DE. Testimony from the bedside: from Coley’s toxins to targeted immunotherapy. Cancer Cell. 2010;18:9–10. doi: 10.1016/j.ccr.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 478.Sabel MS, Sondak VK. Is there a role for adjuvant high-dose interferon-alpha-2b in the management of melanoma? Drugs. 2003;63:1053–1058. doi: 10.2165/00003495-200363110-00001. [DOI] [PubMed] [Google Scholar]
- 479.Kirkwood JM, et al. Next generation of immunotherapy for melanoma. J. Clin. Oncol. 2008;26:3445–3455. doi: 10.1200/JCO.2007.14.6423. [DOI] [PubMed] [Google Scholar]
- 480.Achkar T, Tarhini AA. The use of immunotherapy in the treatment of melanoma. J. Hematol. Oncol. 2017;10:88. doi: 10.1186/s13045-017-0458-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481.Jiang T, Zhou C, Ren S. Role of IL-2 in cancer immunotherapy. Oncoimmunology. 2016;5:e1163462. doi: 10.1080/2162402X.2016.1163462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Atkins MB, et al. High-dose recombinant interleukin 2 therapy for patients with metastatic melanoma: analysis of 270 patients treated between 1985 and 1993. J. Clin. Oncol. 1999;17:2105–2116. doi: 10.1200/JCO.1999.17.7.2105. [DOI] [PubMed] [Google Scholar]
- 483.Davar D, et al. High-dose interleukin-2 (HD IL-2) for advanced melanoma: a single center experience from the University of Pittsburgh Cancer Institute. J. Immunother. Cancer. 2017;5:74. doi: 10.1186/s40425-017-0279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484.Bastholt L, et al. High-dose interleukin-2 and interferon as first-line immunotherapy for metastatic melanoma: long-term follow-up in a large unselected Danish patient cohort. Eur. J. Cancer. 2019;115:61–67. doi: 10.1016/j.ejca.2019.03.023. [DOI] [PubMed] [Google Scholar]
- 485.Clark JI, et al. A multi-center phase II study of high dose interleukin-2 sequenced with vemurafenib in patients with BRAF-V600 mutation positive metastatic melanoma. J. Immunother. Cancer. 2018;6:76. doi: 10.1186/s40425-018-0387-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 486.Shi VY, et al. 100% Complete response rate in patients with cutaneous metastatic melanoma treated with intralesional interleukin (IL)-2, imiquimod, and topical retinoid combination therapy: results of a case series. J. Am. Acad. Dermatol. 2015;73:645–654. doi: 10.1016/j.jaad.2015.06.060. [DOI] [PubMed] [Google Scholar]
- 487.Dafni U, et al. Efficacy of adoptive therapy with tumor-infiltrating lymphocytes and recombinant interleukin-2 in advanced cutaneous melanoma: a systematic review and meta-analysis. Ann. Oncol. 2019;30:1902–1913. doi: 10.1093/annonc/mdz398. [DOI] [PubMed] [Google Scholar]
- 488.Buchbinder EI, et al. Therapy with high-dose Interleukin-2 (HD IL-2) in metastatic melanoma and renal cell carcinoma following PD1 or PDL1 inhibition. J. Immunother. Cancer. 2019;7:49. doi: 10.1186/s40425-019-0522-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Sivori S, et al. NK cells and ILCs in tumor immunotherapy. Mol. Asp. Med. 2021;80:100870. doi: 10.1016/j.mam.2020.100870. [DOI] [PubMed] [Google Scholar]
- 490.Demaria O, et al. Harnessing innate immunity in cancer therapy. Nature. 2019;574:45–56. doi: 10.1038/s41586-019-1593-5. [DOI] [PubMed] [Google Scholar]
- 491.Arenas-Ramirez N, et al. Improved cancer immunotherapy by a CD25-mimobody conferring selectivity to human interleukin-2. Sci. Transl. Med. 2016;8:367ra166. doi: 10.1126/scitranslmed.aag3187. [DOI] [PubMed] [Google Scholar]
- 492.Kaufman HL, Kohlhapp FJ, Zloza A. Oncolytic viruses: a new class of immunotherapy drugs. Nat. Rev. Drug Discov. 2015;14:642–662. doi: 10.1038/nrd4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 493.Bommareddy PK, Patel A, Hossain S, Kaufman HL. Talimogene Laherparepvec (T-VEC) and other oncolytic viruses for the treatment of melanoma. Am. J. Clin. Dermatol. 2017;18:1–15. doi: 10.1007/s40257-016-0238-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 494.Andtbacka RHI, et al. Final analyses of OPTiM: a randomized phase III trial of talimogene laherparepvec versus granulocyte-macrophage colony-stimulating factor in unresectable stage III-IV melanoma. J. Immunother. Cancer. 2019;7:145. doi: 10.1186/s40425-019-0623-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 495.Andtbacka RH, et al. Talimogene Laherparepvec improves durable response rate in patients with advanced melanoma. J. Clin. Oncol. 2015;33:2780–2788. doi: 10.1200/JCO.2014.58.3377. [DOI] [PubMed] [Google Scholar]
- 496.Stahlie EHA, et al. T-VEC for stage IIIB-IVM1a melanoma achieves high rates of complete and durable responses and is associated with tumor load: a clinical prediction model. Cancer Immunol. Immunother. 2021;70:2291–2300. doi: 10.1007/s00262-020-02839-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 497.Ressler JM, et al. Real-life use of talimogene laherparepvec (T-VEC) in melanoma patients in centers in Austria, Switzerland and Germany. J. Immunother. Cancer. 2021;9:e001701. doi: 10.1136/jitc-2020-001701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498.Frohlich A, et al. Talimogene laherparepvec treatment to overcome loco-regional acquired resistance to immune checkpoint blockade in tumor stage IIIB-IV M1c melanoma patients. Cancer Immunol. Immunother. 2020;69:759–769. doi: 10.1007/s00262-020-02487-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 499.Sun C, Mezzadra R, Schumacher TN. Regulation and function of the PD-L1 checkpoint. Immunity. 2018;48:434–452. doi: 10.1016/j.immuni.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 500.Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348:56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 501.Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 2015;27:450–461. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502.Sharpe AH. Mechanisms of costimulation. Immunol. Rev. 2009;229:5–11. doi: 10.1111/j.1600-065X.2009.00784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 503.Brahmer JR, et al. Phase I study of single-agent anti-programmed death-1 (MDX-1106) in refractory solid tumors: safety, clinical activity, pharmacodynamics, and immunologic correlates. J. Clin. Oncol. 2010;28:3167–3175. doi: 10.1200/JCO.2009.26.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504.Brahmer JR, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N. Engl. J. Med. 2012;366:2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 505.Hamanishi J, et al. Safety and antitumor activity of anti-PD-1 antibody, nivolumab, in patients with platinum-resistant ovarian cancer. J. Clin. Oncol. 2015;33:4015–4022. doi: 10.1200/JCO.2015.62.3397. [DOI] [PubMed] [Google Scholar]
- 506.Yang X, Yin R, Xu L. Neoadjuvant PD-1 blockade in resectable. Lung Cancer N. Engl. J. Med. 2018;379:e14. doi: 10.1056/NEJMc1808251. [DOI] [PubMed] [Google Scholar]
- 507.Constantinidou A, Alifieris C, Trafalis DT. Targeting programmed cell death -1 (PD-1) and ligand (PD-L1): A new era in cancer active immunotherapy. Pharmacol. Ther. 2019;194:84–106. doi: 10.1016/j.pharmthera.2018.09.008. [DOI] [PubMed] [Google Scholar]
- 508.Topalian SL, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 509.Liu Y, Zheng P. How does an anti-CTLA-4 antibody promote cancer immunity? Trends Immunol. 2018;39:953–956. doi: 10.1016/j.it.2018.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510.Walunas TL, et al. CTLA-4 can function as a negative regulator of T cell activation. Immunity. 1994;1:405–413. [PubMed] [Google Scholar]
- 511.Brunet JF, et al. A new member of the immunoglobulin superfamily-CTLA-4. Nature. 1987;328:267–270. doi: 10.1038/328267a0. [DOI] [PubMed] [Google Scholar]
- 512.Kroemer G, Zitvogel L. Immune checkpoint inhibitors. J. Exp. Med. 2021;218:e20192179. doi: 10.1084/jem.20201979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 513.van Elsas A, et al. Elucidating the autoimmune and antitumor effector mechanisms of a treatment based on cytotoxic T lymphocyte antigen-4 blockade in combination with a B16 melanoma vaccine: comparison of prophylaxis and therapy. J. Exp. Med. 2001;194:481–489. doi: 10.1084/jem.194.4.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514.van Elsas A, Hurwitz AA, Allison JP. Combination immunotherapy of B16 melanoma using anti-cytotoxic T lymphocyte-associated antigen 4 (CTLA-4) and granulocyte/macrophage colony-stimulating factor (GM-CSF)-producing vaccines induces rejection of subcutaneous and metastatic tumors accompanied by autoimmune depigmentation. J. Exp. Med. 1999;190:355–366. doi: 10.1084/jem.190.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 515.Lipson EJ, Drake CG. Ipilimumab: an anti-CTLA-4 antibody for metastatic melanoma. Clin. Cancer Res. 2011;17:6958–6962. doi: 10.1158/1078-0432.CCR-11-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 516.Ledford H. Melanoma drug wins US approval. Nature. 2011;471:561. doi: 10.1038/471561a. [DOI] [PubMed] [Google Scholar]
- 517.Schadendorf D, et al. Pooled analysis of long-term survival data from phase II and phase III trials of ipilimumab in unresectable or metastatic melanoma. J. Clin. Oncol. 2015;33:1889–1894. doi: 10.1200/JCO.2014.56.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 518.Robert C, et al. Ipilimumab plus dacarbazine for previously untreated metastatic melanoma. N. Engl. J. Med. 2011;364:2517–2526. doi: 10.1056/NEJMoa1104621. [DOI] [PubMed] [Google Scholar]
- 519.Hodi FS, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 2010;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 520.Postow MA, Callahan MK, Wolchok JD. Immune checkpoint blockade in cancer therapy. J. Clin. Oncol. 2015;33:1974–1982. doi: 10.1200/JCO.2014.59.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 521.Sharpe AH, Pauken KE. The diverse functions of the PD1 inhibitory pathway. Nat. Rev. Immunol. 2018;18:153–167. doi: 10.1038/nri.2017.108. [DOI] [PubMed] [Google Scholar]
- 522.Robert C, et al. Pembrolizumab versus ipilimumab in advanced melanoma. N. Engl. J. Med. 2015;372:2521–2532. doi: 10.1056/NEJMoa1503093. [DOI] [PubMed] [Google Scholar]
- 523.Schachter J, et al. Pembrolizumab versus ipilimumab for advanced melanoma: final overall survival results of a multicentre, randomised, open-label phase 3 study (KEYNOTE-006) Lancet. 2017;390:1853–1862. doi: 10.1016/S0140-6736(17)31601-X. [DOI] [PubMed] [Google Scholar]
- 524.Postow MA, et al. Nivolumab and ipilimumab versus ipilimumab in untreated melanoma. N. Engl. J. Med. 2015;372:2006–2017. doi: 10.1056/NEJMoa1414428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 525.Hodi FS, et al. Nivolumab plus ipilimumab or nivolumab alone versus ipilimumab alone in advanced melanoma (CheckMate 067): 4-year outcomes of a multicentre, randomised, phase 3 trial. Lancet Oncol. 2018;19:1480–1492. doi: 10.1016/S1470-2045(18)30700-9. [DOI] [PubMed] [Google Scholar]
- 526.Ascierto PA, et al. Adjuvant nivolumab versus ipilimumab in resected stage IIIB-C and stage IV melanoma (CheckMate 238): 4-year results from a multicentre, double-blind, randomised, controlled, phase 3 trial. Lancet Oncol. 2020;21:1465–1477. doi: 10.1016/S1470-2045(20)30494-0. [DOI] [PubMed] [Google Scholar]
- 527.Larkin J, et al. Overall survival in patients with advanced melanoma who received nivolumab versus investigator’s choice chemotherapy in CheckMate 037: a randomized, controlled, open-label phase III trial. J. Clin. Oncol. 2018;36:383–390. doi: 10.1200/JCO.2016.71.8023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 528.Weber JS, et al. Nivolumab versus chemotherapy in patients with advanced melanoma who progressed after anti-CTLA-4 treatment (CheckMate 037): a randomised, controlled, open-label, phase 3 trial. Lancet Oncol. 2015;16:375–384. doi: 10.1016/S1470-2045(15)70076-8. [DOI] [PubMed] [Google Scholar]
- 529.Robert C, et al. Five-year outcomes with nivolumab in patients with wild-type BRAF advanced melanoma. J. Clin. Oncol. 2020;38:3937–3946. doi: 10.1200/JCO.20.00995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 530.Topalian SL, et al. Survival, durable tumor remission, and long-term safety in patients with advanced melanoma receiving nivolumab. J. Clin. Oncol. 2014;32:1020–1030. doi: 10.1200/JCO.2013.53.0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 531.Weber J, et al. Phase I/II study of metastatic melanoma patients treated with nivolumab who had progressed after ipilimumab. Cancer Immunol. Res. 2016;4:345–353. doi: 10.1158/2326-6066.CIR-15-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 532.Beaver JA, et al. FDA approval of nivolumab for the first-line treatment of patients with BRAF(V600) wild-type unresectable or metastatic melanoma. Clin. Cancer Res. 2017;23:3479–3483. doi: 10.1158/1078-0432.CCR-16-0714. [DOI] [PubMed] [Google Scholar]
- 533.Kluger HM, et al. Long-term survival of patients with melanoma with active brain metastases treated with pembrolizumab on a phase II trial. J. Clin. Oncol. 2019;37:52–60. doi: 10.1200/JCO.18.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 534.Long GV, et al. Combination nivolumab and ipilimumab or nivolumab alone in melanoma brain metastases: a multicentre randomised phase 2 study. Lancet Oncol. 2018;19:672–681. doi: 10.1016/S1470-2045(18)30139-6. [DOI] [PubMed] [Google Scholar]
- 535.Goldberg SB, et al. Pembrolizumab for patients with melanoma or non-small-cell lung cancer and untreated brain metastases: early analysis of a non-randomised, open-label, phase 2 trial. Lancet Oncol. 2016;17:976–983. doi: 10.1016/S1470-2045(16)30053-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 536.Johnson DB, et al. Response to anti-PD-1 in uveal melanoma without high-volume liver metastasis. J. Natl Compr. Canc. Netw. 2019;17:114–117. doi: 10.6004/jnccn.2018.7070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 537.Maeda T, et al. Efficacy of nivolumab monotherapy against acral lentiginous melanoma and mucosal melanoma in Asian patients. Br. J. Dermatol. 2019;180:1230–1231. doi: 10.1111/bjd.17434. [DOI] [PubMed] [Google Scholar]
- 538.Ibrahim T, Mateus C, Baz M, Robert C. Older melanoma patients aged 75 and above retain responsiveness to anti-PD1 therapy: results of a retrospective single-institution cohort study. Cancer Immunol. Immunother. 2018;67:1571–1578. doi: 10.1007/s00262-018-2219-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 539.Geoerger B, et al. Pembrolizumab in paediatric patients with advanced melanoma or a PD-L1-positive, advanced, relapsed, or refractory solid tumour or lymphoma (KEYNOTE-051): interim analysis of an open-label, single-arm, phase 1-2 trial. Lancet Oncol. 2020;21:121–133. doi: 10.1016/S1470-2045(19)30671-0. [DOI] [PubMed] [Google Scholar]
- 540.Keam SJ. Toripalimab: first global approval. Drugs. 2019;79:573–578. doi: 10.1007/s40265-019-01076-2. [DOI] [PubMed] [Google Scholar]
- 541.Tang B, et al. Safety, efficacy, and biomarker analysis of toripalimab in previously treated advanced melanoma: results of the POLARIS-01 multicenter phase II trial. Clin. Cancer Res. 2020;26:4250–4259. doi: 10.1158/1078-0432.CCR-19-3922. [DOI] [PubMed] [Google Scholar]
- 542.Tang B, et al. Safety and clinical activity with an anti-PD-1 antibody JS001 in advanced melanoma or urologic cancer patients. J. Hematol. Oncol. 2019;12:7. doi: 10.1186/s13045-018-0693-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 543.Lian B, et al. Safety and efficacy of HX008: a humanized immunoglobulin G4 monoclonal antibody in patients with locally advanced or metastatic melanoma—a single-arm, multicenter, phase II study. J. Clin. Oncol. 2021;39:9554–9554. doi: 10.1186/s12885-022-10473-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 544.Wolchok JD, et al. Nivolumab plus ipilimumab in advanced melanoma. N. Engl. J. Med. 2013;369:122–133. doi: 10.1056/NEJMoa1302369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 545.Callahan MK, et al. Nivolumab plus ipilimumab in patients with advanced melanoma: updated survival, response, and safety data in a phase I dose-escalation study. J. Clin. Oncol. 2018;36:391–398. doi: 10.1200/JCO.2017.72.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 546.Hodi FS, et al. Combined nivolumab and ipilimumab versus ipilimumab alone in patients with advanced melanoma: 2-year overall survival outcomes in a multicentre, randomised, controlled, phase 2 trial. Lancet Oncol. 2016;17:1558–1568. doi: 10.1016/S1470-2045(16)30366-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 547.Larkin J, et al. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N. Engl. J. Med. 2015;373:23–34. doi: 10.1056/NEJMoa1504030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 548.Larkin J, et al. Five-year survival with combined nivolumab and ipilimumab in advanced melanoma. N. Engl. J. Med. 2019;381:1535–1546. doi: 10.1056/NEJMoa1910836. [DOI] [PubMed] [Google Scholar]
- 549.Wolchok JD, et al. Overall survival with combined nivolumab and ipilimumab in advanced melanoma. N. Engl. J. Med. 2017;377:1345–1356. doi: 10.1056/NEJMoa1709684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 550.Weber JS, et al. Sequential administration of nivolumab and ipilimumab with a planned switch in patients with advanced melanoma (CheckMate 064): an open-label, randomised, phase 2 trial. Lancet Oncol. 2016;17:943–955. doi: 10.1016/S1470-2045(16)30126-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 551.Lebbe C, et al. Evaluation of two dosing regimens for nivolumab in combination with ipilimumab in patients with advanced melanoma: results from the phase IIIb/IV CheckMate 511 trial. J. Clin. Oncol. 2019;37:867–875. doi: 10.1200/JCO.18.01998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 552.Olson DJ, et al. Pembrolizumab plus ipilimumab following Anti-PD-1/L1 failure in melanoma. J. Clin. Oncol. 2021;39:2647–2655. doi: 10.1200/JCO.21.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 553.Pelster MS, et al. Nivolumab and ipilimumab in metastatic uveal melanoma: results from a single-arm phase II study. J. Clin. Oncol. 2021;39:599–607. doi: 10.1200/JCO.20.00605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 554.Namikawa K, et al. Efficacy and safety of nivolumab in combination with ipilimumab in Japanese patients with advanced melanoma: an open-label, single-arm, multicentre phase II study. Eur. J. Cancer. 2018;105:114–126. doi: 10.1016/j.ejca.2018.09.025. [DOI] [PubMed] [Google Scholar]
- 555.D’Angelo SP, et al. Efficacy and safety of nivolumab alone or in combination with ipilimumab in patients with mucosal melanoma: a pooled analysis. J. Clin. Oncol. 2017;35:226–235. doi: 10.1200/JCO.2016.67.9258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 556.Tawbi HA, et al. Combined nivolumab and ipilimumab in melanoma metastatic to the brain. N. Engl. J. Med. 2018;379:722–730. doi: 10.1056/NEJMoa1805453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 557.Amaral T, et al. Combined immunotherapy with nivolumab and ipilimumab with and without local therapy in patients with melanoma brain metastasis: a DeCOG* study in 380 patients. J. Immunother. Cancer. 2020;8:e000333. doi: 10.1136/jitc-2019-000333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 558.Carlino MS, et al. Long-term follow-up of standard-dose pembrolizumab plus reduced-dose ipilimumab in patients with advanced melanoma: KEYNOTE-029 part 1B. Clin. Cancer Res. 2020;26:5086–5091. doi: 10.1158/1078-0432.CCR-20-0177. [DOI] [PubMed] [Google Scholar]
- 559.Long GV, et al. Standard-dose pembrolizumab in combination with reduced-dose ipilimumab for patients with advanced melanoma (KEYNOTE-029): an open-label, phase 1b trial. Lancet Oncol. 2017;18:1202–1210. doi: 10.1016/S1470-2045(17)30428-X. [DOI] [PubMed] [Google Scholar]
- 560.Long, G. V. et al. Standard-dose pembrolizumab plus alternate-dose ipilimumab in advanced melanoma: KEYNOTE-029 Cohort 1C, a phase 2 randomized study of two dosing schedules. Clin. Cancer Res. 10.1158/1078-0432.CCR-21-0793 (2021). [DOI] [PMC free article] [PubMed]
- 561.Dong H, et al. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat. Med. 2002;8:793–800. doi: 10.1038/nm730. [DOI] [PubMed] [Google Scholar]
- 562.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 563.Smyth MJ, Ngiow SF, Ribas A, Teng MW. Combination cancer immunotherapies tailored to the tumour microenvironment. Nat. Rev. Clin. Oncol. 2016;13:143–158. doi: 10.1038/nrclinonc.2015.209. [DOI] [PubMed] [Google Scholar]
- 564.Eggermont AM, et al. Long-term results of the randomized phase III trial EORTC 18991 of adjuvant therapy with pegylated interferon alfa-2b versus observation in resected stage III melanoma. J. Clin. Oncol. 2012;30:3810–3818. doi: 10.1200/JCO.2011.41.3799. [DOI] [PubMed] [Google Scholar]
- 565.Hancock BW, et al. Adjuvant interferon in high-risk melanoma: the AIM HIGH Study-United Kingdom Coordinating Committee on Cancer Research randomized study of adjuvant low-dose extended-duration interferon Alfa-2a in high-risk resected malignant melanoma. J. Clin. Oncol. 2004;22:53–61. doi: 10.1200/JCO.2004.03.185. [DOI] [PubMed] [Google Scholar]
- 566.Eggermont AM, et al. Prolonged survival in stage III melanoma with ipilimumab adjuvant therapy. N. Engl. J. Med. 2016;375:1845–1855. doi: 10.1056/NEJMoa1611299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 567.Tarhini AA, et al. Phase III study of adjuvant ipilimumab (3 or 10 mg/kg) versus high-dose interferon alfa-2b for resected high-risk melanoma: North American Intergroup E1609. J. Clin. Oncol. 2020;38:567–575. doi: 10.1200/JCO.19.01381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 568.Weber J, et al. Adjuvant nivolumab versus ipilimumab in resected stage III or IV melanoma. N. Engl. J. Med. 2017;377:1824–1835. doi: 10.1056/NEJMoa1709030. [DOI] [PubMed] [Google Scholar]
- 569.Eggermont AMM, et al. Adjuvant pembrolizumab versus placebo in resected stage III melanoma (EORTC 1325-MG/KEYNOTE-054): distant metastasis-free survival results from a double-blind, randomised, controlled, phase 3 trial. Lancet Oncol. 2021;22:643–654. doi: 10.1016/S1470-2045(21)00065-6. [DOI] [PubMed] [Google Scholar]
- 570.Eggermont AMM, et al. Longer follow-up confirms recurrence-free survival benefit of adjuvant pembrolizumab in high-risk stage III melanoma: updated results from the EORTC 1325-MG/KEYNOTE-054 Trial. J. Clin. Oncol. 2020;38:3925–3936. doi: 10.1200/JCO.20.02110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 571.Eggermont AMM, et al. Adjuvant pembrolizumab versus placebo in resected stage III melanoma. N. Engl. J. Med. 2018;378:1789–1801. doi: 10.1056/NEJMoa1802357. [DOI] [PubMed] [Google Scholar]
- 572.Zimmer L, et al. Adjuvant nivolumab plus ipilimumab or nivolumab monotherapy versus placebo in patients with resected stage IV melanoma with no evidence of disease (IMMUNED): a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet. 2020;395:1558–1568. doi: 10.1016/S0140-6736(20)30417-7. [DOI] [PubMed] [Google Scholar]
- 573.Long GV, et al. Adjuvant dabrafenib plus trametinib in stage III BRAF-mutated melanoma. N. Engl. J. Med. 2017;377:1813–1823. doi: 10.1056/NEJMoa1708539. [DOI] [PubMed] [Google Scholar]
- 574.Rozeman EA, et al. Identification of the optimal combination dosing schedule of neoadjuvant ipilimumab plus nivolumab in macroscopic stage III melanoma (OpACIN-neo): a multicentre, phase 2, randomised, controlled trial. Lancet Oncol. 2019;20:948–960. doi: 10.1016/S1470-2045(19)30151-2. [DOI] [PubMed] [Google Scholar]
- 575.Bagchi S, Yuan R, Engleman EG. Immune checkpoint inhibitors for the treatment of cancer: clinical impact and mechanisms of response and resistance. Annu. Rev. Pathol. 2021;16:223–249. doi: 10.1146/annurev-pathol-042020-042741. [DOI] [PubMed] [Google Scholar]
- 576.Cristescu R, et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science. 2018;362:eaar3593. doi: 10.1126/science.aar3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 577.Snyder A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med. 2014;371:2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 578.Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature. 2015;523:231–235. doi: 10.1038/nature14404. [DOI] [PubMed] [Google Scholar]
- 579.Jensen C, et al. Non-invasive biomarkers derived from the extracellular matrix associate with response to immune checkpoint blockade (anti-CTLA-4) in metastatic melanoma patients. J. Immunother. Cancer. 2018;6:152. doi: 10.1186/s40425-018-0474-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 580.Reilley MJ, et al. TLR9 activation cooperates with T cell checkpoint blockade to regress poorly immunogenic melanoma. J. Immunother. Cancer. 2019;7:323. doi: 10.1186/s40425-019-0811-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 581.Xiao W, et al. TP53 mutation as potential negative predictor for response of anti-CTLA-4 therapy in metastatic melanoma. EBioMedicine. 2018;32:119–124. doi: 10.1016/j.ebiom.2018.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 582.Sade-Feldman M, et al. Resistance to checkpoint blockade therapy through inactivation of antigen presentation. Nat. Commun. 2017;8:1136. doi: 10.1038/s41467-017-01062-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 583.Rodig SJ, et al. MHC proteins confer differential sensitivity to CTLA-4 and PD-1 blockade in untreated metastatic melanoma. Sci. Transl. Med. 2018;10:eaar3342. doi: 10.1126/scitranslmed.aar3342. [DOI] [PubMed] [Google Scholar]
- 584.Grasso CS, et al. Conserved interferon-gamma signaling drives clinical response to immune checkpoint blockade therapy in melanoma. Cancer Cell. 2020;38:500–515.e503. doi: 10.1016/j.ccell.2020.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 585.Jiang P, et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 2018;24:1550–1558. doi: 10.1038/s41591-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 586.Jacquelot N, et al. Predictors of responses to immune checkpoint blockade in advanced melanoma. Nat. Commun. 2017;8:592. doi: 10.1038/s41467-017-00608-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 587.Vetizou M, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350:1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 588.Mager LF, et al. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science. 2020;369:1481–1489. doi: 10.1126/science.abc3421. [DOI] [PubMed] [Google Scholar]
- 589.Coutzac C, et al. Systemic short chain fatty acids limit antitumor effect of CTLA-4 blockade in hosts with cancer. Nat. Commun. 2020;11:2168. doi: 10.1038/s41467-020-16079-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 590.Coit DG, et al. Cutaneous melanoma, version 2.2019, NCCN clinical practice guidelines in oncology. J. Natl Compr. Canc. Netw. 2019;17:367–402. doi: 10.6004/jnccn.2019.0018. [DOI] [PubMed] [Google Scholar]
- 591.Barrios, D M, et al. Immune checkpoint inhibitors to treat cutaneous malignancies. J. Am. Acad. Dermatol. 2020;83:1239–1253. doi: 10.1016/j.jaad.2020.03.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 592.Perez-Ruiz E, et al. Cancer immunotherapy resistance based on immune checkpoints inhibitors: targets, biomarkers, and remedies. Drug Resist. Updat. 2020;53:100718. doi: 10.1016/j.drup.2020.100718. [DOI] [PubMed] [Google Scholar]
- 593.Schoenfeld AJ, Hellmann MD. Acquired resistance to immune checkpoint inhibitors. Cancer Cell. 2020;37:443–455. doi: 10.1016/j.ccell.2020.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 594.Sullivan RJ. Back to the future: rethinking and retooling IL2 in the immune checkpoint inhibitor era. Cancer Discov. 2019;9:694–695. doi: 10.1158/2159-8290.CD-19-0412. [DOI] [PubMed] [Google Scholar]
- 595.Diab A, et al. Bempegaldesleukin (NKTR-214) plus nivolumab in patients with advanced solid tumors: phase I dose-escalation study of safety, efficacy, and immune activation (PIVOT-02) Cancer Discov. 2020;10:1158–1173. doi: 10.1158/2159-8290.CD-19-1510. [DOI] [PubMed] [Google Scholar]
- 596.Davar D, et al. Phase Ib/II study of pembrolizumab and pegylated-interferon Alfa-2b in advanced melanoma. J. Clin. Oncol. 2018;36:JCO1800632. doi: 10.1200/JCO.18.00632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 597.Atkins MB, et al. Pembrolizumab plus pegylated interferon alfa-2b or ipilimumab for advanced melanoma or renal cell carcinoma: dose-finding results from the phase Ib KEYNOTE-029 study. Clin. Cancer Res. 2018;24:1805–1815. doi: 10.1158/1078-0432.CCR-17-3436. [DOI] [PubMed] [Google Scholar]
- 598.Ribas A, et al. Oncolytic virotherapy promotes intratumoral T cell infiltration and improves anti-PD-1 immunotherapy. Cell. 2017;170:1109–1119.e1110. doi: 10.1016/j.cell.2017.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 599.Puzanov I, et al. Talimogene laherparepvec in combination with ipilimumab in previously untreated, unresectable stage IIIB-IV melanoma. J. Clin. Oncol. 2016;34:2619–2626. doi: 10.1200/JCO.2016.67.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 600.Chesney J, et al. Randomized, open-label phase II study evaluating the efficacy and safety of talimogene laherparepvec in combination with ipilimumab versus ipilimumab alone in patients with advanced, unresectable melanoma. J. Clin. Oncol. 2018;36:1658–1667. doi: 10.1200/JCO.2017.73.7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 601.Rohaan MW, Wilgenhof S, Haanen J. Adoptive cellular therapies: the current landscape. Virchows Arch. 2019;474:449–461. doi: 10.1007/s00428-018-2484-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 602.Rosenberg SA, et al. Use of tumor-infiltrating lymphocytes and interleukin-2 in the immunotherapy of patients with metastatic melanoma. A preliminary report. N. Engl. J. Med. 1988;319:1676–1680. doi: 10.1056/NEJM198812223192527. [DOI] [PubMed] [Google Scholar]
- 603.Seitter, S. J. et al. Impact of prior treatment on the efficacy of adoptive transfer of tumor-infiltrating lymphocytes in patients with metastatic melanoma. Clin. Cancer Res. 10.1158/1078-0432.CCR-21-1171 (2021). [DOI] [PMC free article] [PubMed]
- 604.Hirai I, et al. Adoptive cell therapy using tumor-infiltrating lymphocytes for melanoma refractory to immune-checkpoint inhibitors. Cancer Sci. 2021;112:3163–3172. doi: 10.1111/cas.15009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 605.Sarnaik A, et al. Long-term follow up of lifileucel (LN-144) cryopreserved autologous tumor infiltrating lymphocyte therapy in patients with advanced melanoma progressed on multiple prior therapies. J. Clin. Oncol. 2020;38:10006–10006. [Google Scholar]
- 606.Zhang L, et al. Tumor-infiltrating lymphocytes genetically engineered with an inducible gene encoding interleukin-12 for the immunotherapy of metastatic melanoma. Clin. Cancer Res. 2015;21:2278–2288. doi: 10.1158/1078-0432.CCR-14-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 607.Li W, et al. Efficacy of tumor-infiltrating lymphocytes combined with IFN-alpha in Chinese resected stage III malignant melanoma. J. Immunol. Res. 2017;2017:1092507. doi: 10.1155/2017/1092507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 608.Siddiqi HF, Staser KW, Nambudiri VE. Research techniques made simple: CAR T-cell therapy. J. Invest. Dermatol. 2018;138:2501–2504 e2501. doi: 10.1016/j.jid.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 609.Ribas A. Triple therapy for BRAF(V600)-mutated melanoma. Lancet. 2020;395:1814–1815. doi: 10.1016/S0140-6736(20)31285-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 610.Yu C, et al. Combination of immunotherapy with targeted therapy: theory and practice in metastatic melanoma. Front. Immunol. 2019;10:990. doi: 10.3389/fimmu.2019.00990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 611.Khair DO, et al. Combining immune checkpoint inhibitors: established and emerging targets and strategies to improve outcomes in melanoma. Front. Immunol. 2019;10:453. doi: 10.3389/fimmu.2019.00453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 612.Vella LJ, et al. MEK inhibition, alone or in combination with BRAF inhibition, affects multiple functions of isolated normal human lymphocytes and dendritic cells. Cancer Immunol. Res. 2014;2:351–360. doi: 10.1158/2326-6066.CIR-13-0181. [DOI] [PubMed] [Google Scholar]
- 613.Hu-Lieskovan S, et al. Improved antitumor activity of immunotherapy with BRAF and MEK inhibitors in BRAF(V600E) melanoma. Sci. Transl. Med. 2015;7:279ra241. doi: 10.1126/scitranslmed.aaa4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 614.Wang T, et al. BRAF inhibition stimulates melanoma-associated macrophages to drive tumor growth. Clin. Cancer Res. 2015;21:1652–1664. doi: 10.1158/1078-0432.CCR-14-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 615.Steinberg SM, et al. BRAF inhibition alleviates immune suppression in murine autochthonous melanoma. Cancer Immunol. Res. 2014;2:1044–1050. doi: 10.1158/2326-6066.CIR-14-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 616.Cooper ZA, et al. Response to BRAF inhibition in melanoma is enhanced when combined with immune checkpoint blockade. Cancer Immunol. Res. 2014;2:643–654. doi: 10.1158/2326-6066.CIR-13-0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 617.Frederick DT, et al. BRAF inhibition is associated with enhanced melanoma antigen expression and a more favorable tumor microenvironment in patients with metastatic melanoma. Clin. Cancer Res. 2013;19:1225–1231. doi: 10.1158/1078-0432.CCR-12-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 618.Minor DR, et al. Severe gastrointestinal toxicity with administration of trametinib in combination with dabrafenib and ipilimumab. Pigment Cell Melanoma Res. 2015;28:611–612. doi: 10.1111/pcmr.12383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 619.Ribas A, et al. Hepatotoxicity with combination of vemurafenib and ipilimumab. N. Engl. J. Med. 2013;368:1365–1366. doi: 10.1056/NEJMc1302338. [DOI] [PubMed] [Google Scholar]
- 620.Sullivan RJ, et al. Atezolizumab plus cobimetinib and vemurafenib in BRAF-mutated melanoma patients. Nat. Med. 2019;25:929–935. doi: 10.1038/s41591-019-0474-7. [DOI] [PubMed] [Google Scholar]
- 621.Gutzmer R, et al. Atezolizumab, vemurafenib, and cobimetinib as first-line treatment for unresectable advanced BRAF(V600) mutation-positive melanoma (IMspire150): primary analysis of the randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2020;395:1835–1844. doi: 10.1016/S0140-6736(20)30934-X. [DOI] [PubMed] [Google Scholar]
- 622.Ferrucci PF, et al. KEYNOTE-022 part 3: a randomized, double-blind, phase 2 study of pembrolizumab, dabrafenib, and trametinib in BRAF-mutant melanoma. J. Immunother. Cancer. 2020;8:e001806. doi: 10.1136/jitc-2020-001806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 623.Dummer R, et al. Combined PD-1, BRAF and MEK inhibition in advanced BRAF-mutant melanoma: safety run-in and biomarker cohorts of COMBI-i. Nat. Med. 2020;26:1557–1563. doi: 10.1038/s41591-020-1082-2. [DOI] [PubMed] [Google Scholar]
- 624.Zimmer L, et al. Triplet therapy with pembrolizumab (PEM), encorafenib (ENC) and binimetinib (BIN) in advanced, BRAF V600 mutant melanoma: final results from the dose-finding phase I part of the IMMU-Target trial. J. Clin. Oncol. 2021;39:9532–9532. [Google Scholar]
- 625.Gogas H, et al. Cobimetinib plus atezolizumab in BRAF(V600) wild-type melanoma: primary results from the randomized phase III IMspire170 study. Ann. Oncol. 2021;32:384–394. doi: 10.1016/j.annonc.2020.12.004. [DOI] [PubMed] [Google Scholar]
- 626.Ribas A, et al. Combined BRAF and MEK inhibition with PD-1 blockade immunotherapy in BRAF-mutant melanoma. Nat. Med. 2019;25:936–940. doi: 10.1038/s41591-019-0476-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 627.Ribas A, et al. PD-L1 blockade in combination with inhibition of MAPK oncogenic signaling in patients with advanced melanoma. Nat. Commun. 2020;11:6262. doi: 10.1038/s41467-020-19810-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 628.Hodi FS, et al. Bevacizumab plus ipilimumab in patients with metastatic melanoma. Cancer Immunol. Res. 2014;2:632–642. doi: 10.1158/2326-6066.CIR-14-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 629.Wang X, et al. Apatinib in combination with camrelizumab, a humanized immunoglobulin G4 monoclonal antibody against programmed cell death-1, in patients with metastatic acral melanoma. J. Clin. Oncol. 2021;39:9539–9539. doi: 10.1200/JCO.19.00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 630.Tang B, et al. Real-world efficacy and safety of axitinib in combination with anti-programmed cell death-1 antibody for advanced mucosal melanoma. Eur. J. Cancer. 2021;156:83–92. doi: 10.1016/j.ejca.2021.07.018. [DOI] [PubMed] [Google Scholar]
- 631.Si L, et al. Atezolizumab in combination with bevacizumab in patients with unresectable locally advanced or metastatic mucosal melanoma: interim analysis of an open-label phase II trial. J. Clin. Oncol. 2021;39:9511–9511. [Google Scholar]
- 632.Grosso JF, et al. Functionally distinct LAG-3 and PD-1 subsets on activated and chronically stimulated CD8 T cells. J. Immunol. 2009;182:6659–6669. doi: 10.4049/jimmunol.0804211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 633.Weiss SA, Wolchok JD, Sznol M. Immunotherapy of melanoma: facts and hopes. Clin. Cancer Res. 2019;25:5191–5201. doi: 10.1158/1078-0432.CCR-18-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 634.Lichtenegger FS, et al. Targeting LAG-3 and PD-1 to enhance T cell activation by antigen-presenting cells. Front. Immunol. 2018;9:385. doi: 10.3389/fimmu.2018.00385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 635.Goding SR, et al. Restoring immune function of tumor-specific CD4 + T cells during recurrence of melanoma. J. Immunol. 2013;190:4899–4909. doi: 10.4049/jimmunol.1300271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 636.Lipson EJ, et al. Relatlimab (RELA) plus nivolumab (NIVO) versus NIVO in first-line advanced melanoma: primary phase III results from RELATIVITY-047 (CA224-047) J. Clin. Oncol. 2021;39:9503–9503. [Google Scholar]
- 637.Lee CH, et al. Lenvatinib plus pembrolizumab in patients with either treatment-naive or previously treated metastatic renal cell carcinoma (Study 111/KEYNOTE-146): a phase 1b/2 study. Lancet Oncol. 2021;22:946–958. doi: 10.1016/S1470-2045(21)00241-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 638.Lutzky J, et al. NCT04552223: a phase II study of nivolumab plus BMS-986016 (relatlimab) in patients with metastatic uveal melanoma (UM) (CA224-094) J. Clin. Oncol. 2021;39:TPS9590–TPS9590. [Google Scholar]
- 639.Wolf Y, Anderson AC, Kuchroo VK. TIM3 comes of age as an inhibitory receptor. Nat. Rev. Immunol. 2020;20:173–185. doi: 10.1038/s41577-019-0224-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 640.Acharya N, Sabatos-Peyton C, Anderson AC. Tim-3 finds its place in the cancer immunotherapy landscape. J. Immunother. Cancer. 2020;8:e000911. doi: 10.1136/jitc-2020-000911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 641.Das M, Zhu C, Kuchroo VK. Tim-3 and its role in regulating anti-tumor immunity. Immunol. Rev. 2017;276:97–111. doi: 10.1111/imr.12520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 642.Curigliano G, et al. Phase I/Ib clinical trial of sabatolimab, an Anti-TIM-3 antibody, alone and in combination with spartalizumab, an anti-PD-1 antibody, in advanced solid tumors. Clin. Cancer Res. 2021;27:3620–3629. doi: 10.1158/1078-0432.CCR-20-4746. [DOI] [PubMed] [Google Scholar]
- 643.Yeo J, et al. TIGIT/CD226 axis regulates anti-tumor immunity. Pharmaceuticals. 2021;14:200. doi: 10.3390/ph14030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 644.Inozume T, et al. Melanoma cells control antimelanoma CTL responses via interaction between TIGIT and CD155 in the effector phase. J. Invest. Dermatol. 2016;136:255–263. doi: 10.1038/JID.2015.404. [DOI] [PubMed] [Google Scholar]
- 645.Chauvin JM, et al. TIGIT and PD-1 impair tumor antigen-specific CD8(+) T cells in melanoma patients. J. Clin. Invest. 2015;125:2046–2058. doi: 10.1172/JCI80445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 646.Johnston RJ, et al. The immunoreceptor TIGIT regulates antitumor and antiviral CD8(+) T cell effector function. Cancer Cell. 2014;26:923–937. doi: 10.1016/j.ccell.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 647.Zhang Q, et al. Blockade of the checkpoint receptor TIGIT prevents NK cell exhaustion and elicits potent anti-tumor immunity. Nat. Immunol. 2018;19:723–732. doi: 10.1038/s41590-018-0132-0. [DOI] [PubMed] [Google Scholar]
- 648.Melero I, et al. Agonist antibodies to TNFR molecules that costimulate T and NK cells. Clin. Cancer Res. 2013;19:1044–1053. doi: 10.1158/1078-0432.CCR-12-2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 649.Etxeberria I, Glez-Vaz J, Teijeira A, Melero I. New emerging targets in cancer immunotherapy: CD137/4-1BB costimulatory axis. ESMO Open. 2020;4:e000733. doi: 10.1136/esmoopen-2020-000733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 650.Sanmamed MF, Etxeberria I, Otano I, Melero I. Twists and turns to translating 4-1BB cancer immunotherapy. Sci. Transl. Med. 2019;11:eaax4738. doi: 10.1126/scitranslmed.aax4738. [DOI] [PubMed] [Google Scholar]
- 651.Huang JH, et al. Therapeutic and tumor-specific immunity induced by combination of dendritic cells and oncolytic adenovirus expressing IL-12 and 4-1BBL. Mol. Ther. 2010;18:264–274. doi: 10.1038/mt.2009.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 652.Xiao W, et al. Chimeric antigen receptor-modified T-cell therapy for platelet-derived growth factor receptor alpha-positive rhabdomyosarcoma. Cancer. 2020;126:2093–2100. doi: 10.1002/cncr.32764. [DOI] [PubMed] [Google Scholar]
- 653.Wang R, et al. Transgenic 4-1BBL-engineered vaccine stimulates potent Gag-specific therapeutic and long-term immunity via increased priming of CD44(+)CD62L(high) IL-7R(+) CTLs with up- and downregulation of anti- and pro-apoptosis genes. Cell. Mol. Immunol. 2015;12:456–465. doi: 10.1038/cmi.2014.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 654.Chacon JA, et al. Manipulating the tumor microenvironment ex vivo for enhanced expansion of tumor-infiltrating lymphocytes for adoptive cell therapy. Clin. Cancer Res. 2015;21:611–621. doi: 10.1158/1078-0432.CCR-14-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 655.Yu J, et al. Anti-GD2/4-1BB chimeric antigen receptor T cell therapy for the treatment of Chinese melanoma patients. J. Hematol. Oncol. 2018;11:1. doi: 10.1186/s13045-017-0548-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 656.Weigelin B, et al. Focusing and sustaining the antitumor CTL effector killer response by agonist anti-CD137 mAb. Proc. Natl Acad. Sci. USA. 2015;112:7551–7556. doi: 10.1073/pnas.1506357112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 657.Horton BL, et al. Intratumoral CD8(+) T-cell apoptosis is a major component of T-cell dysfunction and impedes antitumor immunity. Cancer Immunol. Res. 2018;6:14–24. doi: 10.1158/2326-6066.CIR-17-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 658.Wilcox RA, et al. Provision of antigen and CD137 signaling breaks immunological ignorance, promoting regression of poorly immunogenic tumors. J. Clin. Invest. 2002;109:651–659. doi: 10.1172/JCI14184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 659.Menk AV, et al. 4-1BB costimulation induces T cell mitochondrial function and biogenesis enabling cancer immunotherapeutic responses. J. Exp. Med. 2018;215:1091–1100. doi: 10.1084/jem.20171068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 660.Kim JA, et al. Divergent effects of 4-1BB antibodies on antitumor immunity and on tumor-reactive T-cell generation. Cancer Res. 2001;61:2031–2037. [PubMed] [Google Scholar]
- 661.Segal NH, et al. Results from an integrated safety analysis of urelumab, an agonist anti-CD137 monoclonal antibody. Clin. Cancer Res. 2017;23:1929–1936. doi: 10.1158/1078-0432.CCR-16-1272. [DOI] [PubMed] [Google Scholar]
- 662.Tolcher AW, et al. Phase Ib study of utomilumab (PF-05082566), a 4-1BB/CD137 agonist, in combination with pembrolizumab (MK-3475) in patients with advanced solid tumors. Clin. Cancer Res. 2017;23:5349–5357. doi: 10.1158/1078-0432.CCR-17-1243. [DOI] [PubMed] [Google Scholar]
- 663.Qi X, et al. Optimization of 4-1BB antibody for cancer immunotherapy by balancing agonistic strength with FcgammaR affinity. Nat. Commun. 2019;10:2141. doi: 10.1038/s41467-019-10088-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 664.Choi Y, et al. T-cell agonists in cancer immunotherapy. J. Immunother. Cancer. 2020;8:e000966. doi: 10.1136/jitc-2020-000966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 665.Fu Y, Lin Q, Zhang Z, Zhang L. Therapeutic strategies for the costimulatory molecule OX40 in T-cell-mediated immunity. Acta Pharm. Sin. B. 2020;10:414–433. doi: 10.1016/j.apsb.2019.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 666.Hammond SA, et al. Human OX40 ligand fusion protein (MEDI6383) as a potent OX40 agonist and immuno-modulator in vitro and in vivo. J. Clin. Oncol. 2015;33:3056–3056. [Google Scholar]
- 667.Oberst MD, et al. Potent immune modulation by MEDI6383, an engineered human OX40 ligand IgG4P Fc fusion protein. Mol. Cancer Ther. 2018;17:1024–1038. doi: 10.1158/1535-7163.MCT-17-0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 668.Bauer TM, et al. A phase I study of MEDI6383, an OX40 agonist, in adult patients with select advanced solid tumors. J. Clin. Oncol. 2015;33:TPS3093–TPS3093. [Google Scholar]
- 669.Glisson BS, et al. Safety and clinical activity of MEDI0562, a humanized OX40 agonist monoclonal antibody, in adult patients with advanced solid tumors. Clin. Cancer Res. 2020;26:5358–5367. doi: 10.1158/1078-0432.CCR-19-3070. [DOI] [PubMed] [Google Scholar]
- 670.Goldman JW, et al. Safety and tolerability of MEDI0562 in combination with durvalumab or tremelimumab in patients with advanced solid tumors. J. Clin. Oncol. 2020;38:3003–3003. doi: 10.1158/1078-0432.CCR-21-3016. [DOI] [PubMed] [Google Scholar]
- 671.Chiappori A, et al. P860 Results from a combination of OX40 (PF-04518600) and 4–1BB (utomilumab) agonistic antibodies in melanoma and non-small cell lung cancer in a phase 1 dose expansion cohort. J. Immunother. Cancer. 2020;8:A9–A10. [Google Scholar]
- 672.Infante JR, et al. A phase Ib dose escalation study of the OX40 agonist MOXR0916 and the PD-L1 inhibitor atezolizumab in patients with advanced solid tumors. J. Clin. Oncol. 2016;34:101–101. [Google Scholar]
- 673.Postel-Vinay, S. et al. A first-in-human phase I study of the OX40 agonist GSK3174998 (GSK998) +/- pembrolizumab in patients (Pts) with selected advanced solid tumors (ENGAGE-1). Cancer Res. 80, Abstract nr CT150 (2020). [DOI] [PMC free article] [PubMed]
- 674.Gutierrez M, et al. OX40 agonist BMS-986178 alone or in combination with nivolumab and/or ipilimumab in patients with advanced solid tumors. Clin. Cancer Res. 2021;27:460–472. doi: 10.1158/1078-0432.CCR-20-1830. [DOI] [PubMed] [Google Scholar]
- 675.Heinhuis KM, et al. Safety, tolerability, and potential clinical activity of a glucocorticoid-induced TNF receptor-related protein agonist alone or in combination with nivolumab for patients with advanced solid tumors: a phase 1/2a DOSE-ESCALATION AND COHORT-EXPANSION CLINICAL TRIal. JAMA Oncol. 2020;6:100–107. doi: 10.1001/jamaoncol.2019.3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 676.Hashimoto K. CD137 as an attractive T cell co-stimulatory target in the TNFRSF for immuno-oncology drug development. Cancers. 2021;13:2288. doi: 10.3390/cancers13102288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 677.Papadopoulos KP, et al. Phase I study of MK-4166, an anti-human glucocorticoid-induced TNF receptor antibody, alone or with pembrolizumab in advanced solid tumors. Clin. Cancer Res. 2021;27:1904–1911. doi: 10.1158/1078-0432.CCR-20-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 678.Piha-Paul SA, et al. First-in-human phase I/Ib open-label dose-escalation study of GWN323 (anti-GITR) as a single agent and in combination with spartalizumab (anti-PD-1) in patients with advanced solid tumors and lymphomas. J. Immunother. Cancer. 2021;9:e002863. doi: 10.1136/jitc-2021-002863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 679.Geva R, et al. First-in-human phase 1 study of MK-1248, an anti-glucocorticoid-induced tumor necrosis factor receptor agonist monoclonal antibody, as monotherapy or with pembrolizumab in patients with advanced solid tumors. Cancer. 2020;126:4926–4935. doi: 10.1002/cncr.33133. [DOI] [PubMed] [Google Scholar]
- 680.Zappasodi R, et al. Rational design of anti-GITR-based combination immunotherapy. Nat. Med. 2019;25:759–766. doi: 10.1038/s41591-019-0420-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 681.Tran B, et al. Dose escalation results from a first-in-human, phase 1 study of glucocorticoid-induced TNF receptor-related protein agonist AMG 228 in patients with advanced solid tumors. J. Immunother. Cancer. 2018;6:93. doi: 10.1186/s40425-018-0407-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 682.Marin-Acevedo JA, et al. Next generation of immune checkpoint therapy in cancer: new developments and challenges. J. Hematol. Oncol. 2018;11:39. doi: 10.1186/s13045-018-0582-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 683.Vonderheide RH. CD40 agonist antibodies in cancer immunotherapy. Annu. Rev. Med. 2020;71:47–58. doi: 10.1146/annurev-med-062518-045435. [DOI] [PubMed] [Google Scholar]
- 684.He S, Xu J, Wu J. The emerging role of co-stimulatory molecules and their agonistic mAb-based combination therapies in melanoma. Int. Immunopharmacol. 2020;89:107097. doi: 10.1016/j.intimp.2020.107097. [DOI] [PubMed] [Google Scholar]
- 685.Vonderheide RH, et al. Clinical activity and immune modulation in cancer patients treated with CP-870,893, a novel CD40 agonist monoclonal antibody. J. Clin. Oncol. 2007;25:876–883. doi: 10.1200/JCO.2006.08.3311. [DOI] [PubMed] [Google Scholar]
- 686.Weiss SA, et al. A phase I study of APX005M and cabiralizumab with or without nivolumab in patients with melanoma, kidney cancer, or non-small cell lung cancer resistant to anti-PD-1/PD-L1. Clin. Cancer Res. 2021;27:4757–4767. doi: 10.1158/1078-0432.CCR-21-0903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 687.Vitale LA, et al. Development of CDX-1140, an agonist CD40 antibody for cancer immunotherapy. Cancer Immunol. Immunother. 2019;68:233–245. doi: 10.1007/s00262-018-2267-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 688.Irenaeus SMM, et al. First-in-human study with intratumoral administration of a CD40 agonistic antibody, ADC-1013, in advanced solid malignancies. Int. J. Cancer. 2019;145:1189–1199. doi: 10.1002/ijc.32141. [DOI] [PubMed] [Google Scholar]
- 689.Spranger S, et al. Up-regulation of PD-L1, IDO, and T(regs) in the melanoma tumor microenvironment is driven by CD8(+) T cells. Sci. Transl. Med. 2013;5:200ra116. doi: 10.1126/scitranslmed.3006504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 690.Hennequart M, et al. Constitutive IDO1 expression in human tumors is driven by cyclooxygenase-2 and mediates intrinsic immune resistance. Cancer Immunol. Res. 2017;5:695–709. doi: 10.1158/2326-6066.CIR-16-0400. [DOI] [PubMed] [Google Scholar]
- 691.Johnson DB, et al. Quantitative spatial profiling of PD-1/PD-L1 interaction and HLA-DR/IDO-1 predicts improved outcomes of anti-PD-1 therapies in metastatic melanoma. Clin. Cancer Res. 2018;24:5250–5260. doi: 10.1158/1078-0432.CCR-18-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 692.Lynch KT, et al. IDO1 expression in melanoma metastases is low and associated with improved overall survival. Am. J. Surg. Pathol. 2021;45:787–795. doi: 10.1097/PAS.0000000000001622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 693.Sorensen RB, et al. Spontaneous cytotoxic T-Cell reactivity against indoleamine 2,3-dioxygenase-2. Cancer Res. 2011;71:2038–2044. doi: 10.1158/0008-5472.CAN-10-3403. [DOI] [PubMed] [Google Scholar]
- 694.Jallad MN, Jurjus AR, Rahal EA, Abdelnoor AM. Triple immunotherapy overcomes immune evasion by tumor in a melanoma mouse model. Front. Oncol. 2020;10:839. doi: 10.3389/fonc.2020.00839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 695.Hou DY, et al. Inhibition of indoleamine 2,3-dioxygenase in dendritic cells by stereoisomers of 1-methyl-tryptophan correlates with antitumor responses. Cancer Res. 2007;67:792–801. doi: 10.1158/0008-5472.CAN-06-2925. [DOI] [PubMed] [Google Scholar]
- 696.Shi J, et al. Carboxyamidotriazole combined with IDO1-Kyn-AhR pathway inhibitors profoundly enhances cancer immunotherapy. J. Immunother. Cancer. 2019;7:246. doi: 10.1186/s40425-019-0725-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 697.Grobben Y, et al. Targeting indoleamine 2,3-dioxygenase in cancer models using the novel small molecule inhibitor NTRC 3883-0. Front. Immunol. 2020;11:609490. doi: 10.3389/fimmu.2020.609490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 698.Yue EW, et al. Discovery of potent competitive inhibitors of indoleamine 2,3-dioxygenase with in vivo pharmacodynamic activity and efficacy in a mouse melanoma model. J. Med. Chem. 2009;52:7364–7367. doi: 10.1021/jm900518f. [DOI] [PubMed] [Google Scholar]
- 699.Muller AJ, Manfredi MG, Zakharia Y, Prendergast GC. Inhibiting IDO pathways to treat cancer: lessons from the ECHO-301 trial and beyond. Semin. Immunopathol. 2019;41:41–48. doi: 10.1007/s00281-018-0702-0. [DOI] [PubMed] [Google Scholar]
- 700.Mitchell TC, et al. Epacadostat plus pembrolizumab in patients with advanced solid tumors: phase I results from a multicenter, open-label phase I/II trial (ECHO-202/KEYNOTE-037) J. Clin. Oncol. 2018;36:3223–3230. doi: 10.1200/JCO.2018.78.9602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 701.Gibney GT, et al. Phase 1/2 study of epacadostat in combination with ipilimumab in patients with unresectable or metastatic melanoma. J. Immunother. Cancer. 2019;7:80. doi: 10.1186/s40425-019-0562-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 702.Zakharia Y, et al. Phase II trial of the IDO pathway inhibitor indoximod plus pembrolizumab for the treatment of patients with advanced melanoma. J. Immunother. Cancer. 2021;9:e002057. doi: 10.1136/jitc-2020-002057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 703.Jung KH, et al. Phase I study of the indoleamine 2,3-dioxygenase 1 (IDO1) inhibitor navoximod (GDC-0919) administered with PD-L1 inhibitor (atezolizumab) in advanced solid tumors. Clin. Cancer Res. 2019;25:3220–3228. doi: 10.1158/1078-0432.CCR-18-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 704.Montfort A, et al. Combining nivolumab and ipilimumab with infliximab or certolizumab in patients with advanced melanoma: first results of a phase Ib clinical trial. Clin. Cancer Res. 2021;27:1037–1047. doi: 10.1158/1078-0432.CCR-20-3449. [DOI] [PubMed] [Google Scholar]
- 705.Ribas A, et al. SD-101 in combination with pembrolizumab in advanced melanoma: results of a phase Ib, multicenter study. Cancer Discov. 2018;8:1250–1257. doi: 10.1158/2159-8290.CD-18-0280. [DOI] [PMC free article] [PubMed] [Google Scholar]