MIS-C is a novel immunodysregulation syndrome in children with a history of SARS-CoV-2 infection. This study employs a multi-omics approach to explore its immunopathogenesis. The authors show that IFNγ-mediated interactions between T cells, monocytes, and NK cells reside at the heart of the disease.
Abstract
In rare instances, pediatric SARS-CoV-2 infection results in a novel immunodysregulation syndrome termed multisystem inflammatory syndrome in children (MIS-C). We compared MIS-C immunopathology with severe COVID-19 in adults. MIS-C does not result in pneumocyte damage but is associated with vascular endotheliitis and gastrointestinal epithelial injury. In MIS-C, the cytokine release syndrome is characterized by IFNγ and not type I interferon. Persistence of patrolling monocytes differentiates MIS-C from severe COVID-19, which is dominated by HLA-DRlo classical monocytes. IFNγ levels correlate with granzyme B production in CD16+ NK cells and TIM3 expression on CD38+/HLA-DR+ T cells. Single-cell TCR profiling reveals a skewed TCRβ repertoire enriched for TRBV11-2 and a superantigenic signature in TIM3+/CD38+/HLA-DR+ T cells. Using NicheNet, we confirm IFNγ as a central cytokine in the communication between TIM3+/CD38+/HLA-DR+ T cells, CD16+ NK cells, and patrolling monocytes. Normalization of IFNγ, loss of TIM3, quiescence of CD16+ NK cells, and contraction of patrolling monocytes upon clinical resolution highlight their potential role in MIS-C immunopathogenesis.
Graphical Abstract
Introduction
Individuals infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) present a clinical heterogeneous disease. Coronavirus disease 2019 (COVID-19) is primarily characterized by infection of the respiratory tract with no or mild clinical manifestations in 80% of individuals infected with SARS-CoV-2. In adults, the incidence of severe presentation, including an evolution to acute respiratory distress syndrome, is particularly high in the elderly (Zhang et al., 2020b). Pre-existing comorbidities, such as cardiovascular disease, obesity, diabetes mellitus, and chronic lung disease, are known contributing risk factors (Zhang et al., 2020b). In addition, a deficient type I IFN response accounts for at least 10% of severe COVID-19 (Bastard et al., 2020; Zhang et al., 2020c). In children, SARS-CoV-2 causes asymptomatic or mild disease in general, resulting in only 1.5% of all COVID-19 hospitalizations and <0.1% of total deaths (Bialek et al., 2020; Castagnoli et al., 2020; Götzinger et al., 2020; Ludvigsson, 2020).
Multiple months into the pandemic, clinicians started to report a multisystem inflammatory syndrome in previously healthy children (Riphagen et al., 2020; Verdoni et al., 2020), temporally associated with SARS-CoV-2 infection and arising weeks after the peak incidence of COVID-19. This novel syndrome was initially termed “pediatric inflammatory multisystem syndrome temporally associated with SARS-CoV-2 infection” by the Royal College of Pediatrics and Child Health, and later “multisystem inflammatory syndrome in children” (MIS-C) by the Centers for Disease Control and Prevention and the World Health Organization (WHO). MIS-C is characterized by prolonged high-grade fever, conjunctivitis, rash, gastrointestinal symptoms, and cardiac shock (Feldstein et al., 2020), resembling other inflammatory disorders such as Kawasaki disease (KD), toxic shock syndrome (TSS), bacterial sepsis, or hyperinflammatory syndromes such as macrophage activation syndrome (MAS) and hemophagocytic lymphohistiocytosis (HLH; Radia et al., 2021; Hoste et al., 2021; Yasuhara et al., 2021). Patients unequivocally present cardinal features of inflammation (increased C-reactive protein [CRP], ferritin, and erythrocyte sedimentation rate) and coagulopathy (increased D-dimers) and have marked neutrophilia and lymphopenia (Whittaker et al., 2020). In addition, biochemical evidence of myocarditis and cardiac shock (increased troponins and brain natriuretic peptide), hepatitis, and acute kidney injury are often reported (Whittaker et al., 2020). 80% of MIS-C patients test positive for anti–SARS-CoV-2 antibodies and/or have a detectable viral load using PCR (Ahmed et al., 2020; Dufort et al., 2020; Weisberg et al., 2021).
In contrast with these clinical observations, the cascade of immunological events and the identity of critical players of the innate and adaptive immune response in MIS-C remain obscure. Increased concentrations of an array of proinflammatory (e.g., IL-1β, IL-2, IL-6, IL-8, IL-17A, IL-18, IFNγ, TNFα) and regulatory (e.g., IL-1RA, IL-10, sTNFR1, and sTNFR2) cytokines have been documented in MIS-C, but the relative contributions of these are debated (Consiglio et al., 2020; Diorio et al., 2020; Gruber et al., 2020; Syrimi et al., 2021; Vella et al., 2021). The immune cells implicated in MIS-C pathogenesis are also controversial as critical roles have been ascribed to granulocytes and monocytes (Gruber et al., 2020; de Cevins et al., 2021 Preprint; Syrimi et al., 2021), natural killer (NK) cells (Beckmann et al., 2020 Preprint; Gruber et al., 2020; Ramaswamy et al., 2021), γδ T cells (Carter et al., 2020), classical cluster of differentiation (CD) 4 T cells (Carter et al., 2020; Vella et al., 2021), and cytotoxic T cells (Beckmann et al., 2020 Preprint; Ramaswamy et al., 2021; Vella et al., 2021). In addition, several groups suggested a role for autoantibodies (e.g., anti-Jo and anti-La antibodies) inducing tissue damage in MIS-C (Consiglio et al., 2020; Gruber et al., 2020; Ramaswamy et al., 2021).
These immunological findings and analogies with known pediatric syndromes such as KD have fueled several pathophysiological hypotheses for MIS-C (Sancho-Shimizu et al., 2021). Antibody-dependent enhancement, tissue damage by immune complexes formation, and overzealous cytotoxic responses caused by viral superantigens (SAgs) or secondary to viral escape have all been proposed (Consiglio et al., 2020; Iwasaki and Yang, 2020; Moreews et al., 2021; Porritt et al., 2021; Ramaswamy et al., 2021). These hypotheses are not per se exclusive, as they can describe parts of a mechanism or reveal multiple MIS-C subtypes with differing drivers (Brodsky et al., 2020; Sancho-Shimizu et al., 2021). The presence of the same viral etiology in adult COVID-19 and the resembling clinical features of KD and TSS have spurred clinicians to adopt analogous therapeutic approaches (e.g., intravenous Igs [IVIGs], aspirin, corticosteroids, and biologicals targeting IL-1, IL-6, or TNFα; Diorio et al., 2020; Gruber et al., 2020; Whittaker et al., 2020), although evidence-based data for these treatments remain scarce to date. In this context of ill-defined pathophysiological events and many unknowns in clinical management and therapeutic decision-making, we performed a comprehensive characterization of the immune system in a cohort of MIS-C patients and adult COVID-19 patients with signs of cytokine release syndrome (CRS).
Results
Cohort characteristics
We prospectively enrolled 14 patients (P1–14) with MIS-C (Table 1 and Fig. S1 A) according to WHO MIS-C case definition. Demographic characteristics are similar as reported (Hoste et al., 2021; male:female, 1:1; median age, 10 yr; interquartile range [IQR], 8.25–13.5). All patients presented a combination of prolonged, high-grade fever with gastrointestinal manifestations (abdominal pain, 12/14; vomiting, 9/14; and diarrhea, 6/14). The majority (11/14) displayed cardiovascular compromise, including decreased ejection fraction and need for intravenous fluid bolus and/or inotropics. 11 patients fulfilled at least 2/5 KD criteria in addition to prolonged fever. Complete KD was present in only two cases. In all patients, a previous SARS-CoV-2 infection of the patient or in the patient’s household was confirmed. Detailed clinical characterization and routine laboratory markers are presented in Table 1. Patients were stratified into subgroups of severity based on their need for inotropics and/or oxygen therapy (Fig. S1 A). In this study, we compared MIS-C patients with a cohort of adult patients with severe COVID-19 (n = 94) hospitalized with hypoxemia and biochemical evidence of CRS, and two cohorts of matched SARS-CoV-2–naive healthy controls (HCs; based on age, sex, and risk factors; Table 2 and Table S1). In the majority of patients, sampling was performed before treatment initiation or within 12 h after administration. More delayed sampling was performed in P1 (two doses of hydrocortisone) and P9 (two doses of methylprednisolone; Fig. S1 A). Resolution samples (at the second time point of sampling [T2]) were available in 13 cases at day 5 to day 10 after first sampling.
Table 1. MIS-C patient characterization.
| Moderate MIS-C (n = 6) | Severe MIS-C (n = 8) | All (n = 14) | P valuea | |
|---|---|---|---|---|
| Demographics | ||||
| Females, n (%) | 3 (50) | 4 (50) | 7 (50) | 1.000 |
| Age, yr, median (IQR) | 10 (9–11.75) | 10 (7.75–14.25) | 10 (8.25–13.5) | 0.755 |
| Race/ethnicity | ||||
| White/Caucasian, n (%) | 3 (50) | 2 (25) | 5 (35) | 0.514 |
| African/Black, n (%) | 2 (33) | 2 (25) | 4 (28) | |
| Asian/Middle Eastern, n (%) | 1 (16) | 2 (25) | 3 (21) | |
| Other, n (%) | 0 (0) | 2 (25) | 2 (14) | |
| BMI, median (IQR) | 16.4 (16.1–18.1) | 19.1 (16.5–20.6) | 17.8 (16.1–20.3) | 0.491 |
| Admission data | ||||
| ICU stay, median days (IQR) | 2 (1.7–2) | 5.5 (4–7) | 4 (2–6.2) | 0.004 |
| Hospital stay, median days (IQR) | 7.5 (7–8) | 12.5 (9.7–13) | 9 (7.2–12.7) | 0.008 |
| PIM2 score, median (IQR) | 1.3 (1.1–1.6) | 7.9 (7.5–11.8) | 7.3 (1.6–7.9) | 0.002 |
| PRISM score, median (IQR) | 0 (0–2.25) | 3 (1.5–6) | 2 (0–4) | 0.135 |
| pSOFA score, median (IQR) | 0.5 (0–1) | 4.5 (4–6) | 3.5 (1–4.8) | <0.001 |
| Presenting symptoms | ||||
| Fever, median days (IQR) | 6.5 (6–7.7) | 6 (6–6.2) | 6 (6–7) | 0.573 |
| Upper respiratory tract infection, n (%) | 0 (0) | 2 (25) | 2 (14) | 0.186 |
| (Secondary) respiratory distress, n (%) | 0 (0) | 5 (62) | 5 (35) | 0.016 |
| Gastrointestinal, n (%) | 6 (100) | 8 (100) | 14 (100) | 1.000 |
| Abdominal pain, n (%) | 4 (66) | 8 (100) | 12 (85) | 0.078 |
| Vomiting, n (%) | 4 (66) | 5 (62) | 9 (64) | 0.872 |
| Diarrhea, n (%) | 3 (50) | 3 (37) | 6 (42) | 0.640 |
| Myalgia, n (%) | 1 (16) | 2 (25) | 3 (21) | 0.707 |
| Headache, n (%) | 3 (50) | 3 (37) | 6 (42) | 0.640 |
| Encephalopathy, n (%) | 0 (0) | 2 (25) | 2 (14) | 0.186 |
| Kawasaki criteria | ||||
| Incomplete KD, n (%) | 2 (33) | 7 (87) | 9 (64) | 0.036 |
| Complete KD, n (%) | 2 (33) | 0 (0) | 2 (14) | 0.078 |
| Fever ≥5 d, n (%) | 5 (83) | 7 (87) | 12 (85) | 0.825 |
| Extremity changes, n (%) | 2 (33) | 1 (12) | 3 (21) | 0.347 |
| Exanthema, n (%) | 3 (50) | 6 (75) | 9 (64) | 0.334 |
| Non-exudative conjunctivitis, n (%) | 4 (66) | 7 (87) | 11 (78) | 0.347 |
| Lip or mouth changes, n (%) | 4 (66) | 3 (37) | 7 (50) | 0.280 |
| Cervical lymphadenopathy, n (%) | 3 (50) | 2 (25) | 5 (35) | 0.334 |
| Cardiac ultrasound | ||||
| Hyperechogenicity, n (%) | 2 (33) | 1 (12) | 3 (21) | 0.347 |
| Prominent coronaries without dilatation, n (%) | 3 (50) | 1 (12) | 4 (28) | 0.124 |
| Coronary dilatation, n (%) | 0 (0) | 2 (25) | 2 (14) | 0.186 |
| Decreased LV function (FS <25%), n (%) | 0 (0) | 4 (50) | 4 (28) | 0.091 |
| Association with SARS-CoV-2 | ||||
| Previous SARS-CoV-2 contact (suspect or confirmed), n (%) | 5 (83) | 5 (62) | 10 (71) | 0.393 |
| Interval between first contact and onset symptoms, range | 4–6 wk | 11 d–6 wk | 11 d–6 wk | |
| First positive nasopharyngeal SARS-CoV-2 RT-PCR | ||||
| Prior to MIS-C presentation, n (%) | 1 (16) | 1 (12) | 2 (14) | 0.825 |
| At admission, n (%) | 0 (0) | 1 weak positive (Ct 34) (12) | 1 (7) | 0.369 |
| IgG positive (tested during admission) | 5 (83) | 8 (100) | 13 (92) | 0.231 |
| Laboratory markers during admission | ||||
| ESR (max), median (IQR), mm/U | 87 (48–109.5) | 20 (18–29.5) | 42 (20–108) | 0.095 |
| Hb (min), median (IQR), g/dl | 9.3 (8.4–10.2) | 9.2 (8.5–10) | 9.2 (8.3–10.1) | 0.755 |
| WBC (max), median (IQR), × 109/liter | 7.9 (7.86–15.04) | 15.51 (14.83–20.4) | 15.03 (8.8–17.53) | 0.081 |
| Lymphocytes (min), median (IQR), × 109/liter | 0.63 (0.4–0.76) | 0.28 (0.24–0.4) | 0.37 (0.26–0.57) | 0.029 |
| Neutrophils (max), median (IQR), × 109/liter | 6.75 (6.42–10.01) | 13.49 (12.63–17.09) | 12.27 (7.47–14.7) | 0.020 |
| Eosinophils (min), median (IQR), × 109/liter | 0.07 (0.02–0.1) | 0 (0–0.01) | 0.01 (0–0.05) | 0.029 |
| Platelets (min), median (IQR), × 109/liter | 162 (148–189) | 138 (77–192) | 153 (88–193) | 0.662 |
| Creatinine (max), median (IQR), mg/dl | 0.62 (0.54–0.72) | 0.78 (0.53–1.19) | 0.71 (0.53–1.09) | 0.512 |
| Urea (max), median (IQR), mg/dl | 29 (21–48) | 60 (43–76) | 47 (36–68) | 0.056 |
| AST (max), median (IQR), U/liter | 44 (34–60) | 54 (43–94) | 49 (37–70) | 0.445 |
| ALT (max), median (IQR), U/liter | 46 (35–53) | 68 (61–86) | 55 (43–68) | 0.073 |
| GGT (max), median (IQR), U/liter | 45 (27–52) | 75 (55–102) | 55 (44–77) | 0.022 |
| Ferritin (max), median (IQR), µg/liter | 531 (240.5–608.5) | 703 (557.3–885.3) | 592.5 (510.8–858,8) | 0.282 |
| Fibrinogen (max), median (IQR), mg/dl | 606.5 (453.3–757.5) | 660.5 (460–741.8) | 660.5 (452–759.3) | 1.000 |
| D-dimer (max), median (IQR), ng/ml | 2,906 (2,286–3,700) | 3,920 (2,430–4,945) | 3,190 (2,174–4,420) | 0.534 |
| CRP (max), median (IQR), mg/liter | 189.3 (178.3–193.8) | 324.3 (289–390.6) | 286.4 (187.3–376.1) | 0.142 |
| hsTrop (max), median (IQR), pg/ml | 35 (7.5–114.63) | 193 (77–1,980) | 77 (47.2–278.5) | 0.352 |
| NT-proBNP (max), median (IQR), pg/ml | 507 (506–507) | 5,333 (3,142–8,752) | 3,142 (846–7,043) | 0.095 |
| Treatment | ||||
| Low-flow nasal cannula oxygen, n (%) | 0 (0) | 8 (100) | 8 (57) | <0.001 |
| High-flow nasal cannula oxygen, n (%) | 0 (0) | 5 (62) | 5 (35) | 0.016 |
| Mechanical ventilation, n (%) | 0 (0) | 0 (0) | 0 (0) | 1.000 |
| Inotropics, n (%) | 0 (0) | 8 (100) | 8 (57) | <0.001 |
| IVIG, n (%) | 6 (100) | 5 (62) | 11 (78) | 0.091 |
| Corticosteroids, n (%) | 2 (33) | 7 (87) | 9 (64) | 0.036 |
| Combination IVIG and corticosteroids | 2 (33) | 4 (50) | 6 (42) | 0.533 |
| Acetylsalicylic acid, n (%) | 4 (66) | 5 (62) | 9 (64) | 0.872 |
| Antibiotics, n (%) | 6 (100) | 8 (100) | 14 (100) | 1.000 |
| Biologicals, n (%) | 0 (0) | 0 (0) | 0 (0) | 1.000 |
| Outcome | ||||
| Alive at follow-up, n (%) | 6 (100) | 8 (100) | 14 (100) | 1.000 |
| No residual morbidity, n (%) | 0 (0) | 0 (0) | 0 (0) | 1.000 |
Bold and italic text indicates that P values are <0.05 and are considered significant. BMI, body mass index; ESR, erythrocyte sedimentation rate; FS, fractional shortening; Hb, hemoglobin; ICU, intensive care unit; LV, left ventricular; min, minimum; max, maximum; NT-proBNP, N-terminal pro-brain natriuretic peptide; PRISM, pediatric risk of mortality; pSOFA, pediatric sequential organ failure assessment.
Chi-square test (two-sided) for categorical variables, Mann–Whitney U for continuous variables, tested between moderate and severe MIS-C.
Figure S1.
Clinical data from patients with MIS-C and adults with severe COVID-19. (A) Timeline of symptoms, medical interventions, and time points of sampling for each of the 14 MIS-C patients included in the study. Treatment and dosages are denoted according to the legend. If tapering of corticosteroids was performed, partial circles are displayed according to the relative dose reductions that were applied. (B) Principal component (PC) analysis of routine laboratory data (top). Routine variables with missing data for individual patients (e.g., troponin, N-terminal pro-brain natriuretic peptide) were excluded from the analysis. Included variables are denoted in the vectors in the loadings plot (bottom). (C) Violin plots showing endotoxin levels in the plasma of MIS-C patients as compared with HCs. Dotted lines show median and quartiles. (D) Correlation plot of clinical variables of adult patients with severe COVID-19 and their routine laboratory parameters and biomarkers of endothelial and epithelial injury. Significance determined by Spearman’s rank correlation coefficients, indicated by *, P < 0.05; **, P < 0.01; and ***, P < 0.001. BMI, body mass index; PICU, pediatric intensive care unit; NIV, noninvasive ventilation; EU, endotoxin units; neutro, neutrophil; lympho, lymphocyte; eosino, eosinophil.
Table 2. Comparison of MIS-C patients and adults with severe COVID-19.
| MIS-C (n = 14) | Severe adult COVID-19 (n = 94) | P valuea | |
|---|---|---|---|
| Demographics | |||
| Females, n (%) | 7 (50) | 25 (27) | 0.0736 |
| Age, yr, median (IQR) | 10 (8.25–13.5) | 67 (58–74) | <0.0001 |
| Race/ethnicity | |||
| White/Caucasian, n (%) | 5 (35) | 90 (96) | <0.0001 |
| African/Black, n (%) | 4 (28) | 2 (2) | <0.0001 |
| Asian/Middle Eastern, n (%) | 3 (21) | 1 (1) | 0.0002 |
| Other, n (%) | 2 (14) | 1 (1) | 0.005 |
| BMI, median (IQR) | 17.8 (16.1–20.3) | 28.1 (25.8–31.6) | <0.0001 |
| Co-morbidities | |||
| Cardiovascular disease, n (%) | 0 (-) | 32 (36) | 0.0093 |
| Arterial hypertension, n (%) | 0 (-) | 34 (38) | 0.0066 |
| Diabetes mellitus, n (%) | 0 (-) | 28 (31) | 0.0177 |
| Chronic kidney disease, n (%) | 0 (-) | 13 (14) | 0.1379 |
| Chronic lung disease, n (%) | 0 (-) | 13 (14) | 0.1379 |
| Laboratory markers at inclusion | |||
| Hb, median (IQR), g/dl | 10.5 (9.9–11.2) | 13.3 (12.3–14.3) | <0.0001 |
| WBC, median (IQR), × 109/liter | 7.1 (5.7–10.7) | 7 (5.4–9.3) | 0.7004 |
| Lymphocytes, median (IQR), × 109/liter | 0.8 (0.6–1.1) | 0.6 (0.5–0.8) | 0.0766 |
| Eosinophils, median (IQR), × 109/liter | 0.1 (0–0.1) | 0 (0–0) | <0.0001 |
| Platelets, median (IQR), × 109/liter | 194.5 (172.3–229.8) | 218 (155.5–291.5) | 0.4366 |
| Creatinine, median (IQR), mg/dl | 0.6 (0.5–0.7) | 0.9 (0.8–1.1) | <0.0001 |
| Urea, median (IQR), mg/dl | 27 (17.7–37.8) | 28 (22–36) | 0.5339 |
| AST, median (IQR), U/liter | 33 (24–44) | 54 (38–76) | 0.0021 |
| ALT, median (IQR), U/liter | 38 (20–42) | 44 (30–63.5) | 0.1155 |
| GGT, median (IQR), U/liter | 47 (36–59) | 54.5 (28.8–101.5) | 0.2207 |
| Ferritin, median (IQR), µg/liter | 507 (285–736) | 1,704 (1,169–2,744.5) | <0.0001 |
| Fibrinogen, median (IQR), mg/dl | 523 (425–663.5) | 634 (571–728) | 0.0567 |
| D-dimer, median (IQR), ng/ml | 2,555 (2,002.5–4,855) | 1,100 (722.5–1,486.8) | 0.0006 |
| CRP, median (IQR), mg/liter | 286.4 (187.275–376.125) | 130.9 (86–190.7) | <0.0001 |
Bold and italic text indicates that P values are <0.05 and are considered significant. BMI, body mass index; Hb, hemoglobin.
Chi-square test (two-sided) for categorical variables, Mann–Whitney U for continuous variables, tested between adult COVID-19 and MIS-C.
Evidence of enterocyte injury and endotoxemia but not pneumocyte damage in MIS-C
The high frequencies of gastrointestinal symptoms, signs of vasculitis, and relatively few respiratory complaints (Table 1) are in line with previously reported cohorts (Abrams et al., 2020; Miller et al., 2020). Angiopoietin-2 (Ang-2), a biomarker of endothelial cell injury and suggested as a therapeutic target in severe COVID-19 (Hultström et al., 2021 Preprint), was significantly increased in the serum of both MIS-C and severe COVID-19 patients (Fig. 1 A). To allow comparison of both cohorts side by side, we normalized biomarkers relative to the median of their respective age-matched HCs (Fig. 1 D) and found that Ang-2 was more pronounced in MIS-C compared with severe COVID-19. Soluble receptor for advanced glycation end products (sRAGE), released by damaged type I alveolar epithelial cells (Uchida et al., 2006; Jabaudon et al., 2011, 2018), was not increased in MIS-C patients but was significantly elevated during severe COVID-19 (Fig. 1, B and D). Finally, we measured serum fatty acid binding protein 2 (FABP2), originating from damaged enterocytes and used as a biomarker in inflammatory bowel disease and mesenteric ischemia (Pelsers et al., 2003; Derikx et al., 2010). FABP2 was uniformly increased in MIS-C but not in severe COVID-19 (Fig. 1, C and D). In line with a previous report (Yonker et al., 2021), enterocyte damage was paralleled by measurable endotoxins in the systemic circulation in 4 out of 14 MIS-C patients (Fig. S1 B).
Figure 1.
Signature of vascular damage and gastrointestinal barrier disruption but no signs of lung involvement in MIS-C. (A–D) Violin plots depicting serum concentrations of Ang-2, sRAGE, and FABP2 revealing distinct patterns for patients with MIS-C (red), severe adult COVID-19 (orange), and age-matched HCs (blue), which is confirmed in D after normalizing for age (Log2 fold change of individual values versus the median of age-matched HCs). Dashed lines show median and quartiles. Significance determined by Mann–Whitney U tests (and Dunn’s test for multiple testing), indicated by *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001. (E) Correlation plot of clinical variables, routine laboratory parameters, and biomarkers of endothelial and epithelial injury, measured in patients with MIS-C at first sampling time point. Significance determined by Spearman’s rank correlation coefficients, indicated by *, P < 0.05; **, P < 0.01; and ***, P < 0.001. (F) Dot plots displaying concentrations of Ang-2, FABP2, and ferritin in relation to each other in both patients with MIS-C and severe COVID-19. Dashed lines represent simple linear regression with 95% CIs. R2 displays goodness-of-fit, and statistical analysis to test if slope differs from 0 (P < 0.05) is shown. Distinct symbols depict patients with severe MIS-C with (inverted triangle) or without treatment (triangle) and moderate disease (circle). BMI, body mass index; FS%, fractional shortening; ICU, intensive care unit.
Next, we investigated the relationships between these biomarkers, patient characteristics, and markers of inflammation, coagulation, organ damage, and disease severity in MIS-C and COVID-19 (Fig. 1 E and Fig. S1 C). MIS-C severity correlated strongly with clinically validated score systems for critically ill pediatric patients (pSOFA and PIM2). Analysis suggests a positive correlation between disease severity and white blood cell (WBC) and neutrophil count and degree of lymphopenia and eosinopenia. As reported by Syrimi et al. (2021), higher CRP was found in MIS-C patients with compromised heart function, quantified as reduced left ventricular fractional shortening. Although only minimally increased liver enzymes (aspartate transaminase [AST]/alanine transaminase [ALT]/γ-glutamyltransferase [GGT]) were noted, GGT levels correlated with disease severity. Principal component analysis using these markers of inflammation and organ damage clearly segregated MIS-C patients with severe disease (Fig. S1 D). In individual MIS-C patients, levels of Ang-2 and FABP2 increased in parallel, which was not observed in severe COVID-19 (Fig. 1 F). In MIS-C, but not in severe COVID-19, Ang-2 and FABP2 correlated with serum ferritin, an acute phase reactant produced by hepatocytes and activated macrophages and a reported biomarker of MIS-C severity (Abrams et al., 2021; Fig. 1 F).
Together, these data suggest that several markers of inflammation correlate with MIS-C severity. In both MIS-C and severe COVID-19, vascular damage can be observed. MIS-C patients uniquely present with enterocyte damage, which associates with microbial translocation and correlates with ferritin, a hallmark of macrophage and hepatocyte activation.
All MIS-C patients have a strong humoral response against SARS-CoV-2
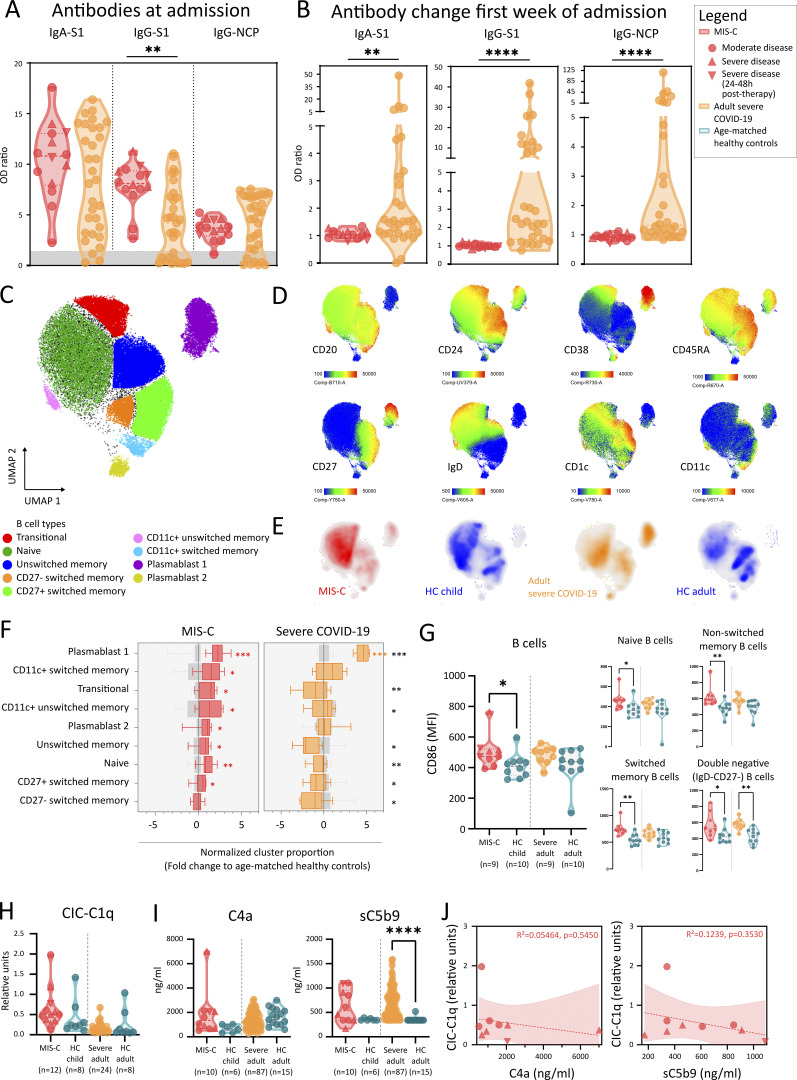
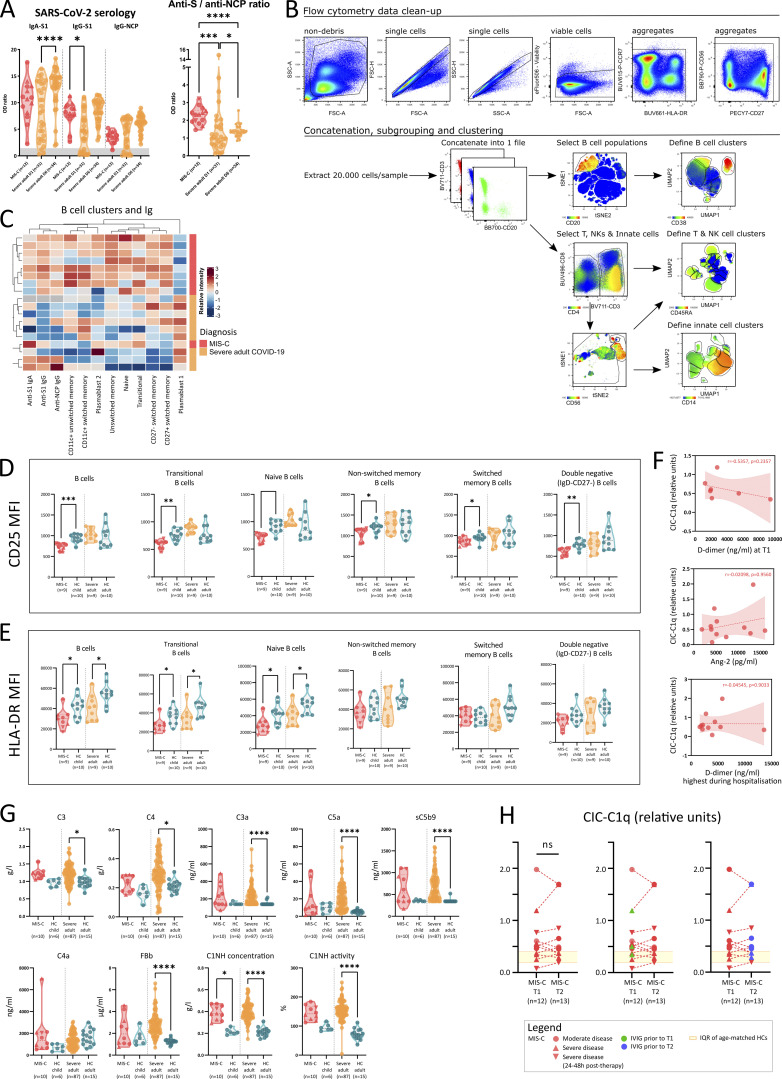
Recent SARS-CoV-2 infection was proven by serology in all but one of the tested cases (Table 1). We investigated the humoral response in more detail by quantifying IgG and IgA directed against the receptor binding domain of the spike (S) protein and IgG specific for the nucleocapsid protein (NCP). We found marked increase of all Igs in MIS-C (Fig. 2 A). A substantial proportion of adults with severe COVID-19 had Ig levels comparable or slightly above values obtained from naive HCs (Fig. 2 A). Sampling at a later time point revealed significant Ig increase in severe COVID-19, whereas Igs in MIS-C did not display obvious changes (Fig. 2 B and Fig. S2 A). This suggests a later disease onset of MIS-C relative to SARS-CoV-2 infection compared with the adult cohort. Comparable with previous studies (Weisberg et al., 2021), children with MIS-C predominantly generated anti-S antibodies (significantly higher as compared with adult COVID-19), resulting in a significantly higher anti-S/anti-NCP ratio in MIS-C (Fig. S2 A).
Figure 2.
Widespread B cell activation and convalescent antibody repertoire but absence of immune complex mediated complement activation. (A) Violin plots of anti–SARS-CoV-2–specific serological assays, quantified as relative OD versus the calibrator for anti-spike 1 (S1) IgA and IgG, and anti-nucleocapsid (NCP) IgG for MIS-C (red) and adults with severe COVID-19 (orange). Area in gray depicts insufficient serological response. Dashed lines show median and quartiles. (B) Fold increase of serology during hospitalization, calculated as ratio of OD values of second and first time point of sampling of patients with MIS-C and severe COVID-19. Significance determined by Mann–Whitney U tests (and Dunn’s test for multiple testing), indicated by *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001. (C) UMAP representation of B lymphocytes (within PBMCs) of 9 MIS-C patients, 9 adults with severe COVID-19, and 20 age-matched HCs. (D) Signature marker expression characterizing B lymphocyte clusters. Legend depicting fluorescence intensity. (E) Density plot of UMAP panels stratified by condition or HC group. (F) Boxplots representing median and ranges of normalized B lymphocyte cluster proportions versus the median of corresponding HCs (Log2 fold change). Significance determined by multiple Mann–Whitney U tests and corrected for multiple testing (FDR of 5%) both within disease and age-matched HCs (in red and orange for MIS-C and severe COVID-19, respectively) or between MIS-C and severe COVID-19 (in black, right), indicated as *, q < 0.05; **, q < 0.01; ***, q < 0.001; and ****, q < 0.0001. (G) MFI of CD86 on B lymphocytes (CD3−CD20+; left) and specific B cell subsets (right): naive (IgD+CD27−), unswitched memory (IgD+CD27+), switched memory (IgD−CD27+), and DN (IgD−CD27−) B cells. (H and I) Violin plots representing values of CICs bound by C1q (H) and (I) concentrations of cleaved complement factor 4 (C4a) and soluble membrane attack complex (sC5b9) in plasma. Dashed lines show median and quartiles. Significance determined by Mann–Whitney U tests (and Dunn’s test for multiple testing), indicated by ****, P < 0.0001. (J) Dot plots displaying concentrations of C4a and sC5b9 in relation to CIC-C1q in patients with MIS-C. Dashed lines represent simple linear regression with 95% CIs. R2 displays goodness-of-fit, and statistical analysis to test if slope differs from 0 (P < 0.05) is shown. Values for individual patients are displayed with MIS-C (red), severe adult COVID-19 (orange), and age-matched HCs (blue). Distinct symbols depict patients with severe MIS-C with (inverted triangle) or without treatment (triangle) and moderate disease (circle).
Figure S2.
Humoral response, complement pathway activation and CICs in MIS-C and severe COVID-19. (A) Violin plots of anti–SARS-CoV-2–specific serological assays, quantified as relative OD versus the calibrator for anti-spike 1 (S1) IgA and IgG, and anti-nucleocapsid (NCP) IgG for MIS-C at inclusion (red) and adults with severe COVID-19 measured on day 1 and day 6 (orange). The zone below the calibrator is denoted in gray. Dotted lines depict median and quartiles. (B) Gating strategy and cluster analysis of FCM data obtained from PBMCs of 9 MIS-C patients, 9 adults with severe COVID-19, and 20 age-matched HCs for further analysis by UMAP. (C) Heatmap of normalized B cell clusters identified by UMAP and anti–SARS-CoV-2 serology of individual MIS-C and severe COVID-19 patients. Rows and columns are ordered by hierarchical clustering methods. (D and E) Violin plots representing MFI of CD25 (D) and (E) HLA-DR as activation markers on multiple stages of B cell development. Dotted lines depict median and quartiles. (F) Dot plots displaying absence of correlation between CICs as measured by C1q quantification at T1, and D-dimer (on T1 or highest during admission) or Ang-2 levels (measured on T1). Dashed line represents simple linear regression with r and P value of Spearman testing. (G) Violin plots of plasma complement factors associated with classical (C4a), alternative (FBb), and terminal complement pathway activation (C3a, C5a, and sC5b9). Dotted lines depict median and quartiles. (H) Paired dot plot graph displaying the evolution of CIC from T1 to T2 for patients with MIS-C (left), having received IVIG therapy before T1 (in green, middle) or between T1 and T2 (blue, right). Significances are determined by Mann–Whitney U tests, corrected for multiple testing by Dunn’s test, between MIS-C or severe COVID-19 with their respective age-matched HCs, indicated by *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001. Values for individual patients are displayed with MIS-C (red), severe adult COVID-19 (orange), and age-matched HCs (blue). Distinct symbols depict patients with severe MIS-C with (inverted triangle) or without treatment (triangle) and moderate disease (circle).
MIS-C patients have distinct B cell expansion and activation
To examine the B cell compartment in more detail, we developed a 26-color/33-antibody flow cytometry (FCM) panel (Table S2). 960,000 cells derived from peripheral blood mononuclear cells (PBMCs) of MIS-C patients (n = 9), severe COVID-19 (n = 9), and age-matched HCs (n = 20) were visualized and clustered using the dimensionality reduction algorithm t-distributed stochastic neighbor embedding. We separated PBMCs into three distinct subgroups (granulocytes/mononuclear phagocytes, T cells/NK cells, and B cells/plasmablasts; Fig. S2 B). From this dataset, we extracted B cell populations and used uniform manifold approximation and projection (UMAP) to reduce dimensionality (Fig. 2 C). By manual gating on the clusters, we identified nine B cell and plasmablast clusters using signature surface markers (Fig. 2 D). Comparing MIS-C patients with age-matched HCs revealed relative increases of clusters containing transitional B cells, naive B cells, CD11c+ B cells, and plasmablasts (Fig. 2, E and F). Expansion of CD11c+ B cells has been described in both viral infections and autoimmune diseases such as systemic lupus erythematosus. These cells typically have a memory phenotype, express TBX1, and have reduced levels of CXCR5, CD21, CD24, and CD38 (Jenks et al., 2018; Rincon-Arevalo et al., 2021). In severe COVID-19, plasmablasts are similarly increased, but transitional, naive, and memory B cells remained unchanged (Fig. 2, E and F). In all but one MIS-C case, hierarchical clustering of B cell populations allowed differentiation of MIS-C from COVID-19 (Fig. S2 C). We examined B cell compartment activation in more detail and found higher CD86 median fluorescence intensity (MFI) in multiple B cell populations in both MIS-C and severe COVID-19 (Fig. 2 G). Compared with moderate cases, the expression of CD86 was more pronounced in severe MIS-C. We observed reduced expression of both CD25 and HLA-DR in MIS-C, while only HLA-DR was down-regulated on B cell populations in severe COVID-19 (Fig. S2, D and E).
Together, we observe that MIS-C is associated with a strong but distinct humoral immune response, separating MIS-C patients from severe COVID-19 cases.
Vascular damage is not associated with circulating immunocomplexes or complement activation
Previous studies have proposed an important role for the humoral response in MIS-C pathogenesis. Both antibody-dependent enhancement and increased levels of circulating autoantibodies inducing organ damage have been suggested (Consiglio et al., 2020; Gruber et al., 2020; Weisberg et al., 2021). One of the consequences of autoantibodies is the formation of immunocomplexes of autoantigen, Igs, and the complement factor C1q. We quantified circulating immunocomplexes (CICs) in the serum and found increased levels of CIC in only a few patients (Fig. 2 H). Next, we examined classical complement activation, which can initiate downstream of CIC. Activation of the complement cascade (increased sC5b-9 formation) through the classical pathway (C4 cleavage) was present in some but not all MIS-C patients (Fig. 2 I). In MIS-C, there was no relationship between CIC and complement activation (C4a or sC5b-9) (Fig. 2 J). Furthermore, CIC did not correlate with coagulopathy (D-dimers) or endotheliitis (Ang-2; Fig. S2 F). In severe COVID-19, on the other hand, significantly increased terminal complement activation (increased sC5b9 complexes) was observed. In accordance with previous studies (Ma et al., 2021), adults displayed clear activation of the alternative complement pathway (increased complement factor Bb [FBb]), which was unaffected in MIS-C (Fig. S2 G). Finally, MIS-C disease severity or IVIG administration before sampling was not associated with the level of CIC (Fig. S2 H).
In conclusion, CIC or complement activation was not uniformly present in MIS-C patients, nor could we identify an association with vascular damage.
MIS-C and severe COVID-19 present with distinct cytokine signatures
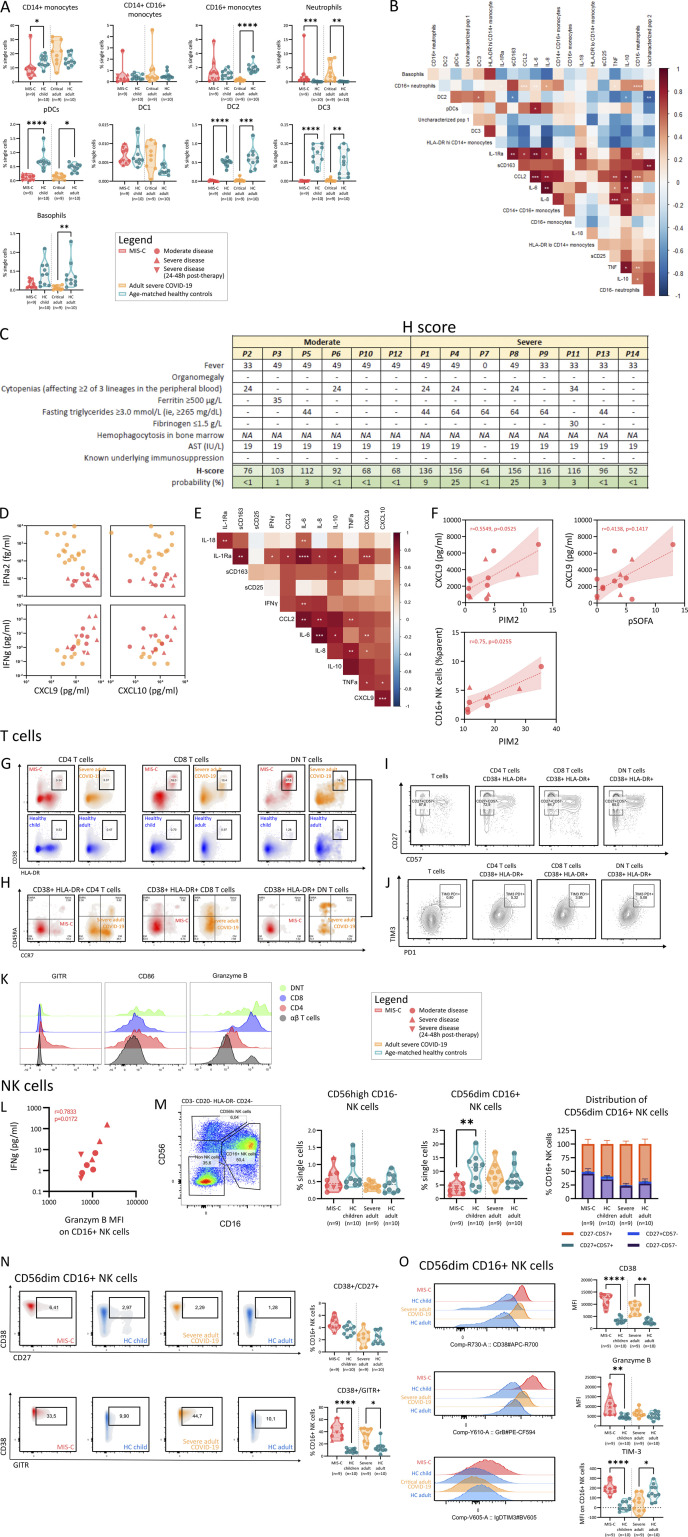
Inappropriate and uncontrolled cytokine release is at the heart of the immunopathology of MIS-C and severe COVID-19 pneumonia with CRS (Fajgenbaum and June, 2020; Leisman et al., 2020). We found significantly increased serum levels of IL-1RA, IL-8, IL-10, TNFα, and CCL2 in MIS-C patients and adults with severe COVID-19. In contrast, serum IL-6 or IL-18 was not significantly enhanced in MIS-C (Fig. 3 A). Levels of IL-1β and GM-CSF were below levels of detection for most samples (data not shown). Comparing the relative increases of MIS-C and severe COVID-19 to age-matched HCs, MIS-C patients had stronger increases in IL-10 and TNFα, whereas IL-6 up-regulation was more pronounced in severe COVID-19 (Fig. 3 B). We quantified sCD163 and sCD25 to measure systemic activation of monocytes/macrophages and T cells, respectively. We found elevated sCD25 as a unique feature of MIS-C as compared with adult COVID-19 patients and HCs (Fig. 3 C). While sCD163 was significantly elevated in adult COVID-19 as well, the relative increase was stronger in MIS-C (Fig. 3 D).
Figure 3.
CRS with marked increase of TNFα, IL-10, sCD25, and sCD163 and profound changes of innate leukocytes. (A–D) Violin plots depicting serum concentrations of IL-1RA, IL-18, IL-6, IL-8, IL-10, TNF, sCD25, and sCD163 comparing MIS-C (red), severe COVID-19 (orange), and respective HCs (blue). (B and D) Normalized values for age (Log2 fold change of individual values versus the median of age-matched HCs). Dashed lines show median and quartiles. Significance determined by Mann–Whitney U tests (and Dunn’s test for multiple testing), indicated by *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001. (E) UMAP representation of innate leukocytes (within PBMCs) of 9 MIS-C patients, 9 adults with severe COVID-19, and 20 age-matched HCs. (F) Signature marker expression characterizing innate leukocyte clusters. Legend depicting fluorescence intensity. (G) Density plot of UMAP panels stratified by condition or HC group. (H) Boxplots representing median and ranges of normalized B lymphocyte cluster proportions versus the median of corresponding HCs (Log2 fold change). Significance determined by multiple Mann–Whitney U tests and corrected for multiple testing (FDR of 5%) both within disease and age-matched HCs (in red and orange for MIS-C and severe COVID-19, respectively) or between MIS-C and severe COVID-19 (in black, right), indicated as *, q < 0.05; **, q < 0.01; ***, q < 0.001; and ****, q < 0.0001. (I) Violin plots depicting percentage of CD16+ monocytes among viable PBMCs. Dashed lines show median and quartiles. Significance determined by Mann–Whitney U tests (and Dunn’s test for multiple testing), indicated by ****, P < 0.0001. Values for individual patients are displayed with MIS-C (red), severe adult COVID-19 (orange), and age-matched HCs (blue). Distinct symbols depict patients with severe MIS-C with (inverted triangle) or without treatment (triangle) and moderate disease (circle).
FCM reveals distinct patterns of innate leukocyte activation
We analyzed the innate leukocyte compartment in PBMCs by FCM and distinguished 12 clusters of innate leukocytes using UMAP (Fig. 3, E and F; and Fig. S2 B). This analysis clustered CD14+ classical monocytes, CD14+/CD16+ inflammatory monocytes, and CD16+ patrolling monocytes. The classical monocyte population could be further subdivided based on CD86 and HLA-DR expression (Fig. 3, E and F). A distinct population of CD24+ but FceR1− cells could be visualized, identifying these cells as low-density neutrophils. Expression of the complement factor C5a receptor (CD88) and the IgG receptor IIIA (CD16) allowed further subdivision of neutrophil populations (Fig. 3, E and F). Additional clusters identified basophils (FceR1+), plasmacytoid dendritic cells (pDCs; CD123+), and type 2 and type 3 conventional DCs (CD1c+ and CD14+, respectively; Fig. 3, E and F). Two additional clusters could not be identified. Among others, these populations could contain innate lymphoid cells or platelets as the panel did not contain the appropriate lineage markers (Table S2). Type I DCs were too rare to be accurately detected.
Both neutrophil populations (CD16+ and CD16−) were significantly increased in MIS-C and severe COVID-19 (Fig. 3, G and H). One of the uncharacterized populations increased similarly in both groups (Fig. 3, G and H). Among the contracted innate cell populations, pDCs and conventional DC clusters were affected in both MIS-C and severe COVID-19 (Fig. 3, G and H). Analyzing monocyte clusters, adults with severe COVID-19 had univocally increased circulating HLA-DRlo classical monocytes. In parallel, patrolling monocytes were lost. In MIS-C patients, patrolling monocytes were preserved, and increases of HLA-DRlo classical monocytes were more moderate (Fig. 3, G and H). We could replicate these findings using conventional gating strategies (Fig. 3 I and Fig. S3 A). We found that relative proportions of neutrophil clusters from MIS-C patients correlated strongly with cytokines such as IL-6, IL-8, IL-10, TNFα, and CCL2 (Fig. S3 B), suggesting an intricate relationship.
Figure S3.
H-score, type II IFN signature, innate leukocyte subsets, and T and NK cell activation in MIS-C. (A) Violin plots of monocytes, neutrophils, DCs, and basophils. Dotted lines show median and quartiles. Significances are determined by Mann–Whitney U tests, corrected for multiple testing by Dunn’s test, indicated by *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001. (B) Correlation plot of innate leukocytes subsets as identified by UMAP analysis and serum cytokines, measured in the MIS-C patients. Significance determined by Spearman’s rank correlation coefficients, indicated by *, P < 0.05; **, P < 0.01; and ***, P < 0.001. (C) H-score (as described previously; Fardet et al., 2014), calculated for the MIS-C patients, including the corresponding probability of having hemophagocytic syndrome. (D) Correlation plot of IFNα2 and IFNγ with their downstream chemokines CXCL9 and CXCL10. (E) Correlation matrix of IFNγ, CXCL9, CXCL10, and innate cytokines, measured in the MIS-C patients. Significance determined by Spearman’s rank correlation coefficients, indicated by *, P < 0.05; **, P < 0.01; and ***, P < 0.001. (F) Correlation plots of clinical severity scores and CXCL9 or CD16+ NK cells. Dashed line represents simple linear regression with r and P value of Spearman testing. (G) Density plots of HLA-DR and CD38 among CD4+, CD8+, and DN T cell populations. One representative plot for each group (MIS-C, severe COVID-19, or matched HCs) is displayed. Percentage of positive cells (from parent) is denoted. (H) Density plots of CCR7 and CD45RA on CD38+/HLA-DR+ T cells. One representative plot for each patient group (MIS-C and severe COVID-19) is displayed. Percentage of parent is denoted per quadrant. (I and J) Contour plots showing expression of CD57 and CD27 (I) or (J) PD-1 and TIM3 among all T cells and on CD38+/HLA-DR+ T cell populations (CD4+, CD8+, DN). One graph representative for MIS-C is displayed. (K) Histograms showing GITR, CD86 and granzyme B (GrB) fluorescence intensities of CD4, CD8, DN, and all αβ T cells of MIS-C patients. One representative plot is displayed. (L) Correlation plots of GrB MFI on CD16+ NK cells and serum IFNγ measured in MIS-C patients. r and P value of two-sided Spearman’s rank testing are shown. (M) Violin plots showing percentage of CD56+ and CD16+ NK cell populations in MIS-C, severe COVID-19, and age-matched HCs. Dotted lines show median and quartiles. Significances are determined by Mann–Whitney U tests, corrected for multiple testing by Dunn’s test, indicated by **, P < 0.01. (N) Density plots showing CD38 and CD27 or CD38 and CD27 fluorescence in CD16+ NK cells in MIS-C, severe COVID-19, and age-matched HCs. One representative plot for each group is displayed. Percentage of positive cells (from parent) is denoted. Violin plots showing percentage of CD38+/CD27+ or CD38+/GITR+ populations among CD16+ NK cells. Dotted lines show median and quartiles. Significances are determined by Mann–Whitney U tests, corrected for multiple testing by Dunn’s test, indicated by *, P < 0.05; and ****, P < 0.0001. (O) Histograms showing CD38, GrB, or TIM3 fluorescence in MIS-C, severe COVID-19, and age-matched HCs. One representative plot for each group is displayed. Violin plots showing MFI of CD38, GrB, and TIM3 in CD16+ NK cells of each group. Dotted lines show median and quartiles. Significances for violin plots are determined by Mann–Whitney U tests, corrected for multiple testing by Dunn’s test, between MIS-C or severe COVID-19 with age-matched HCs, indicated by *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001. Values for individual patients are displayed with MIS-C (red), severe COVID-19 (orange), and age-matched HCs (blue). Distinct symbols depict patients with severe MIS-C with (inverted triangle) or without treatment (triangle) and moderate disease (circle).
In conclusion, cytokine and innate leukocyte profiling in both MIS-C and severe COVID-19 suggested a distinct pattern of innate leukocyte activation in MIS-C with increased TNFα, IL-10, sCD25, and sCD163 levels and preserved patrolling monocytes as key features. While high levels of sCD163, sCD25, and IL-10 have been documented in hyperferritinemic syndromes, such as MAS or HLH (Schaer et al., 2005; Xu et al., 2012; Humblet-Baron et al., 2019), none of the MIS-C patients showed substantially elevated HLH scores (H-scores; Fig. S3 C).
IFNγ and not type I IFN correlates with inflammation in MIS-C
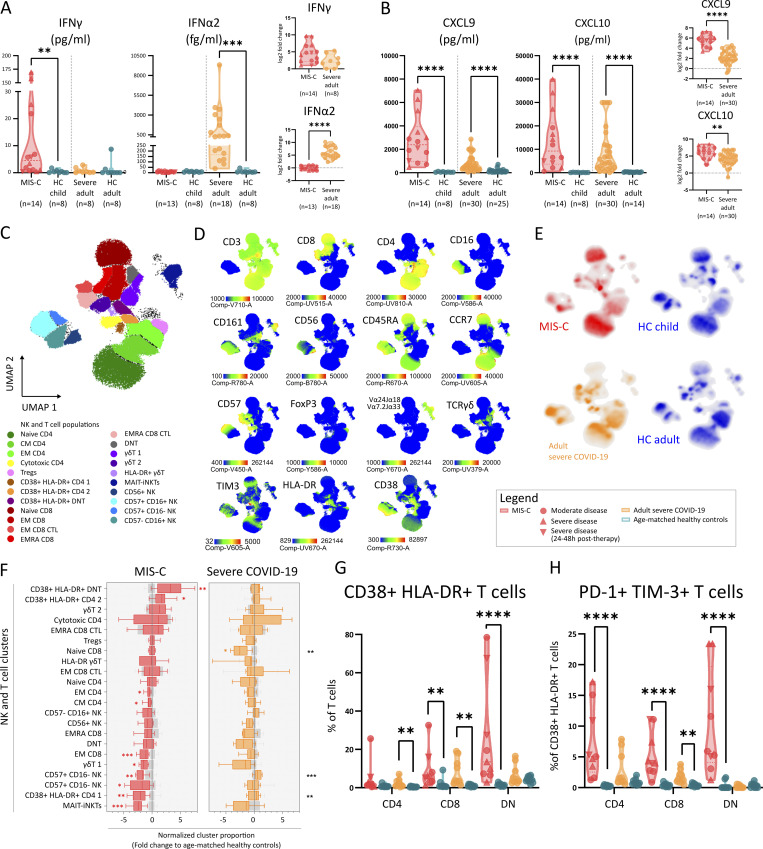
The clinical and immunological correlates with hyperferritinemic disease suggest a potential role for IFNγ in disease pathogenesis. We assessed serum levels of IFN-responsive chemokines and found significantly elevated levels of CXCL9 and CXCL10 in both MIS-C and COVID-19 patients (Fig. 4 B), which is in line with previous observations in MIS-C (Caldarale et al., 2021; Esteve-Sole et al., 2021) and severe COVID-19 (Yang et al., 2020). However, MIS-C patients showed relatively stronger increases compared with severe COVID-19 (Fig. 4 B). We quantified IFNγ and the prototypical type I IFN IFNα2 and found that IFNα2 concentrations were significantly elevated in severe COVID-19. In MIS-C, primarily IFNγ was increased compared with HCs (Fig. 4 A). Analyzing their relationships, we found that IFNγ and not IFNα2 concentrations correlated positively with CXCL9 and CXCL10 (Fig. S3 D). Levels of CXCL9, CXCL10, and IFNγ in MIS-C patients correlated positively with innate cytokines IL-1RA, IL-6, IL-8, IL-10, CCL2, and TNFα (Fig. S3 E). Furthermore, we examined the relationship between IFNγ and its signature cytokines CXCL9 and CXCL10 with clinical scores of disease severity and found a positive correlation between PIM2 and CXCL9 (Fig. S3 F).
Figure 4.
Type II IFN signature in absence of type I IFN and TIM3+ PD1+ activated T cells characterize MIS-C. (A and B) Violin plots depicting serum concentrations of IFNγ and IFNα2, C-X-C motif ligand 9 (CXCL9), and CXCL10 in MIS-C, adults with severe COVID-19, and their respective HCs. Normalized values for age (Log2 fold change of individual values versus the median of age-matched HCs). Dashed lines show median and quartiles. Significance determined by Mann–Whitney U tests (and Dunn’s test for multiple testing), indicated by *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001. (C) UMAP representation of T lymphocytes, NK cells, and iNKT/MAIT cells (within PBMCs) of 9 MIS-C patients, 9 adults with severe COVID-19, and 20 age-matched HCs. (D) Signature marker expression characterizing T lymphocytes and NK cell clusters. Legend depicting fluorescence intensity. (E) Density plot of UMAP panels stratified by condition or HC group. (F) Boxplots representing median and ranges of normalized T/NK lymphocyte cluster proportions versus the median of HCs (Log2 fold change). Significance determined by multiple Mann–Whitney U tests and corrected for multiple testing (FDR of 5%) both within disease and age-matched HCs (in red and orange for MIS-C and severe COVID-19, respectively) or between MIS-C and severe COVID-19 (in black, right), indicated as *, q < 0.05; **, q < 0.01; ***, q < 0.001; and ****, q < 0.0001. (G) Violin plots showing percentage of CD38+/HLA-DR+ double positive CD4+, CD8+, or DN cells (percentage of T cells). (H) Violin plots showing the percentage of PD-1+/TIM3+ double positive CD4+, CD8+, or DN cells (percentage of CD38++/HLA-DR+ T cells). Dashed lines show median and quartiles. Significance determined by Mann–Whitney U tests (and Dunn’s test for multiple testing), indicated by *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001. Values for individual patients are displayed with MIS-C (red), severe adult COVID-19 (orange), and age-matched HCs (blue). Distinct symbols depict patients with severe MIS-C with (inverted triangle) or without treatment (triangle) and moderate disease (circle).
Expansion of activated CD4 and double negative (DN) T cells among IFNγ-producing lymphocyte subsets
Using FCM and UMAP visualization, we analyzed the contribution of known cellular sources of IFNγ, such as NK cells, mucosal-associated invariant T (MAIT) cells, γδ T cells, invariant natural killer T (iNKT) cells, and conventional T cells. We identified 22 distinct clusters encompassing four NK cell populations, three γδ T cell populations, one combined MAIT/iNKT cell cluster, and five CD8, seven CD4, and two DN T cell populations (Fig. 4, C–E). MAIT/iNKT cells and cluster 1 of γδ T cells (containing naive cells) were similarly decreased in MIS-C and severe COVID-19 (Fig. 4 F). We found lower amounts of CD56dimCD16+ NK cells in MIS-C patients, while in accordance with previous reports, naive CD8 T cells contracted in severe COVID-19 but not in MIS-C (Fig. 4 F). Most importantly, activated CD38+HLA-DR+ CD4 T cells and DN T cells were significantly increased in MIS-C patients (Fig. 4 F). We confirmed the activation of these CD4, CD8 and, particularly, DN T cells in MIS-C by manual gating and found increased percentages of CD38+/HLA-DR+ expression among all populations (Fig. 4 G and Fig. S3 G). In MIS-C, most of the cells had an effector memory (EM) phenotype, while in severe COVID-19, the small population of CD38+HLA-DR+ T cells were mostly EM cells reexpressing CD45RA (TEMRA; Fig. S3 H).
Activated T cells in MIS-C express high levels of TIM3, a marker for IFNγ-secreting effector cells
Our data suggests a role for conventional T cells in MIS-C pathogenesis. Analyzing the functional status of these T cells using surface markers such as glucocorticoid-induced TNFR-related protein (GITR), TIM3, PD-1, CD27, and CD57, we found that almost all cells expressed CD27 but not CD57, indicating a functional rather than a senescent phenotype (Fig. S3 I). A subset of the CD38+/HLA-DR+ T cells coexpressed the coinhibitory molecules TIM3 and PD-1, markers of both activation and exhaustion (Fig. S3 J). This PD-1/TIM3 double positive T cell subset was uniquely enriched in MIS-C (Fig. 4 H). TIM3 expression was especially relevant given that it was originally identified as a marker of IFNγ producing T helper type 1 cells (Wolf et al., 2020). In MIS-C, CD38+/HLA-DR+ CD4 T cells also expressed GITR, further supporting an activated status (Fig. S3 K).
We explored a potential role for other IFNγ-producing lymphocytes and found a strong correlation between the activation of NK cells (measured as surface expression of GrB) and serum IFNγ levels (Fig. S3 L). Confirming the unsupervised cluster analysis, we observed decreased numbers of CD56dimCD16+ NK cells (Fig. S3 M). The remaining NK cells were activated (defined as CD38+/CD27+ and CD38+/GITR+) and expressed higher levels of GrB and TIM3, which could not be observed in severe COVID-19 (Fig. S3, N and O).
MIS-C and severe COVID-19 have similar numbers of circulating IFNγ+ SARS-CoV-2–specific T cells
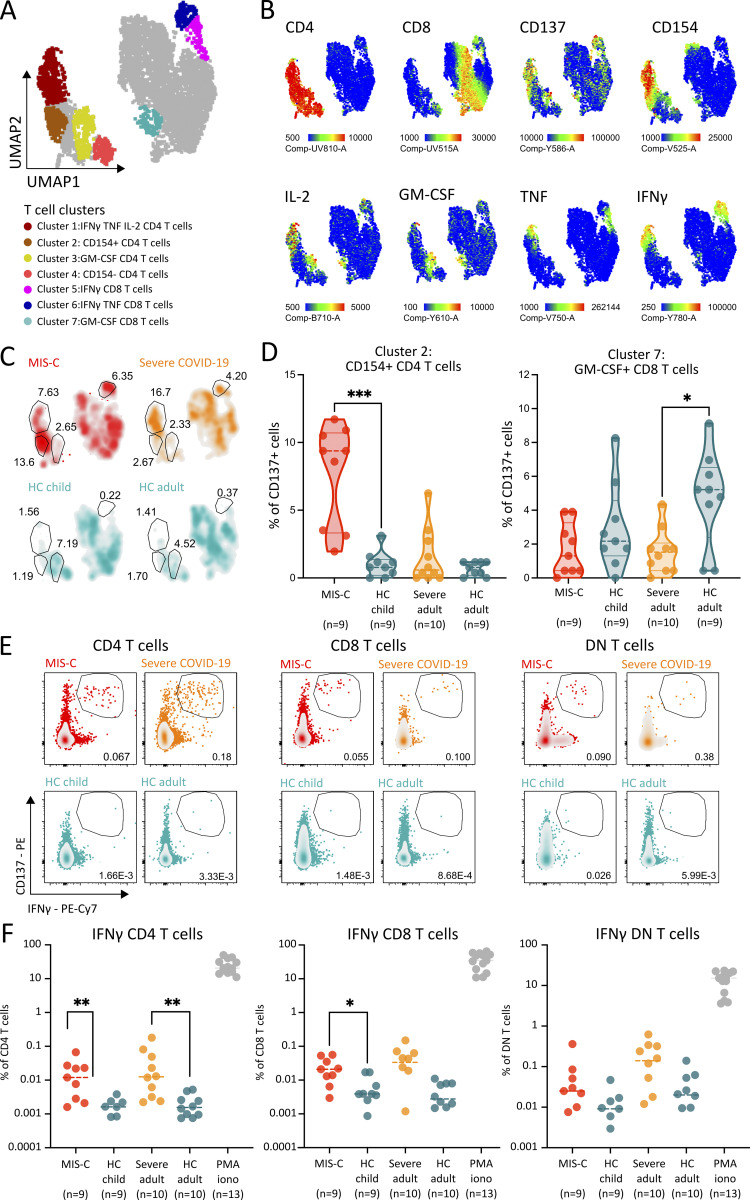
To explore the antigen specificity of the activated T cell populations in MIS-C, we restimulated PBMCs with peptide pools spanning the full SARS-CoV-2 spike protein, and selected epitopes from nucleocapsid, membrane, and open reading frame proteins, as described previously (Weiskopf et al., 2020; da Silva Antunes et al., 2021; Ogbe et al., 2021). SARS-CoV-2–specific T cells were quantified by FCM using activation markers (CD137 and CD154) and intracellular cytokine staining (Table S3). Using UMAP analysis of CD137-expressing cells, we identified seven distinct clusters of activated T cells (Fig. 5, A and B). As compared with HCs, IFNγ/IL-2–positive CD4 and CD8 T cells were increased in MIS-C and severe COVID-19 (Fig. 5 C). Intriguingly, cluster 2 (activated, but cytokine-negative CD4 T cells) and cluster 3 (IL-17A/IL-2–positive CD4 T cells) were enriched in MIS-C and adult HCs, respectively (Fig. 5, C and D). Given our previous findings on IFNγ, we performed manual gating to study the IFNγ+ SARS-CoV-2–specific T cells in more detail. We could identify IFNγ+/CD137+ CD4, CD8, and DN T cells (Fig. 5 E) but found similar increases in both MIS-C and severe COVID-19 (Fig. 5, E and F). Notably, IFNγ+ SARS-CoV-2–specific CD4 and CD8 but not DN T cell populations were significantly increased in MIS-C patients compared with HCs (Fig. 5 F). Moreover, in MIS-C patients, the numbers of CD38+/HLA-DR+ CD4 and CD8 T cells exceeded the T cells responding to the peptides derived from SARS-CoV-2 (Fig. 4, H and L; and Fig. 5 B). These findings suggest that peptide restimulation might not reliably identify the activated TIM3+ T cell population in MIS-C.
Figure 5.
SARS-CoV-2 peptide pool restimulation reveals distinct clusters of antigen specific T cells in MIS-C. (A) UMAP analysis of CD137+ T cells of 9 MIS-C patients, 9 adults with severe COVID-19, and 19 age-matched HCs after overnight restimulation with peptide pools. (B) Signature marker expression characterizing antigen responsive T lymphocytes (CD137+). Legend depicting fluorescence intensity. (C) Density plot of UMAP panels stratified by condition or HC group (percentage of cells are denoted for clusters of interest). (D) Violin plots depicting proportions of activated, cytokine negative T cells (cluster 2) and GM-CSF expressing CD4 cells (cluster 7) in patients with MIS-C, adults with severe COVID-19, and their respective HCs. Dashed lines show median and quartiles. Significance determined by Mann–Whitney U tests (and Dunn’s test for multiple testing), indicated by *, P < 0.05; and ***, P < 0.001. (E) Representative FCM panels for both conditions and HCs, showing CD137+/IFNγ+ double positive T cells after peptide pool restimulation of PBMCs (percentage of cells within the gate is shown). (F) Dot plots showing percentage of CD137+/IFNγ+ double positive T cells after peptide pool restimulation. PBMCs stimulated with PMA and ionomycin (iono) as control are shown. Dotted line depicts median. Significance determined by Mann–Whitney U tests (and Dunn’s test for multiple testing), indicated by *, P < 0.05; **, P < 0.01; and ***, P < 0.001. Values for individual patients are displayed with MIS-C (red), severe adult COVID-19 (orange), and age-matched HCs (blue).
Single-cell TCR profiling identifies an activated and proliferative population with skewed TCR repertoire
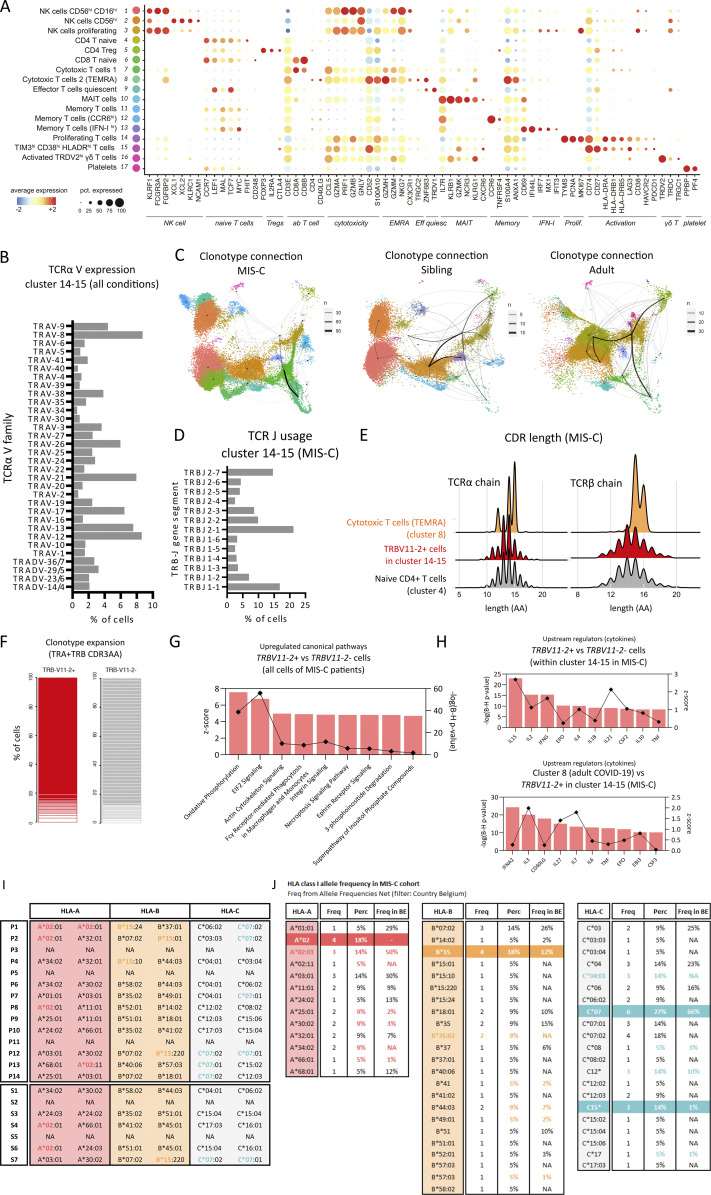
Given the discrepancy between SARS-CoV-2 peptide responsiveness among T cells and the expansion of activated TIM3+ T cells in MIS-C, we set out to better understand their characteristics by performing high-resolution receptor profiling and link this information to the cellular molecular profile of these T cells. We performed single-cell RNA sequencing (scRNA-seq) with 5′ sequencing on purity-sorted viable PBMCs of seven patients with MIS-C (P1, P2, P3, P4, P6, P7, and P10), five healthy siblings (SARS-CoV-2–seronegative sister [12 yr old] and brother [18 yr old] from P1, seronegative brother [13 yr old] from P6, and seropositive brother [7 yr old] and sister [13 yr old] from P10, both of whom had experienced a natural infection at the same time as P10) and four adults with severe COVID-19. By annotating the clusters within the UMAP object enriched for CD3E transcripts, we could identify CD4- and CD8-expressing naive T cells (clusters 4 and 6), memory T cells (clusters 11–13), regulatory T cells (cluster 5), MAIT (cluster 10), NK (clusters 1–3), γδ T (cluster 16), and effector T cell populations in various states of activation (clusters 7–9, 14, and 15; Fig. 6 A and Fig. S4 A). CD38 and genes encoding HLA-DR (HLA-DRA, DRB1, and DRB5) were mostly expressed in the clusters containing effector T cells and NK cells. PDCD1 (PD-1) and HAVCR2 (TIM3) expression was more restricted and was particularly enriched in the proliferating effector cells (cluster 15) and a neighboring cluster that we defined as activated T cells based on CD38 and HLA-DRA expression (cluster 14; Fig. 6 B). We analyzed the relative contribution of each sample and confirmed that both clusters were dominated by cells from MIS-C cases (Fig. 6 A). In contrast, clusters containing TEMRA CD8 T cells (cluster 8) and memory T cells expressing a type I IFN signature (cluster 13) contained predominantly cells of patients with severe COVID-19 (Fig. 6 A). In severe COVID-19, limited data have been published on expansion of T cells expressing specific TCRβ variable segments (TRBV5-6, TRBV14, and TRBV24-1; Cheng et al., 2020). On the other hand, strong evidence for expansion of TRBV11-2-expressing T cells has recently been documented in MIS-C (Moreews et al., 2021; Porritt et al., 2021; Ramaswamy et al., 2021). Plotting TCRβ-V and TCRα-V usage onto the UMAP dimensional reduction plot revealed that cells mapping to clusters 14 and 15 displayed clear skewing of TCRβ-V but not of TCRα-V (Fig. 6 C and Fig. S4 B). The majority used TCRβ-V11 variable gene segments, and T cells expressing TCRβ-V11 were markedly expanded in MIS-C (Fig. 6 D). A clonotype connection plot showed that identical TCR clones were frequently found between clusters 14 and 15, indicating homologous T cells in an activated or proliferative stage (Fig. S4 C). The use of TCRβ-V11 variable gene segments could largely be attributed to TRBV11-2 (Fig. 6, D and E). When analyzing the dataset in individual patient samples, we found that approximately half of the MIS-C patients contributed to most TRBV11-2+ cells (Fig. 6 F), reminiscent of proportions described in previous studies (Moreews et al., 2021; Porritt et al., 2021; Ramaswamy et al., 2021). Nonetheless, in all MIS-C samples, significantly high proportions of the activated or proliferative cells (clusters 14 and 15) consisted of TRBV11-2–expressing cells, supporting a similar disease course in all patients (Fig. 6 G).
Figure 6.
MIS-C is characterized by an activated and proliferative TRBV11-2 expressing T cell population with a skewed TCR repertoire. (A) UMAP plot depicting cluster annotation and numbering on a Seurat object of cells enriched for CD3E transcripts of MIS-C patients (n = 7), adults with severe COVID-19 (n = 4), and HCs (n = 5). The relative proportion of patient or control samples (M = MIS-C, S = healthy sibling, A = adult COVID-19) that contributes to each cluster is visualized in the bar graphs. (B) UMAP plot visualizing expression of signature genes CD38, HLA-DRA, PDCD1, HAVCR2, CCR6 and KI-67. (C) TRB-V expansion plot, highlighting TRB-V counts from all patient conditions on the UMAP shown in A. (D) Bar graphs showing TRBV11 usage within clusters 14 and 15 grouped per condition (M = MIS-C, S = healthy sibling, A = adult COVID-19) and presented as a percentage of all T/NK cells of each group. (E) Doughnut plots showing TRBV11-2 allele usage within clusters 14 and 15 of MIS-C patients. (F) Stacked bar plot presenting the relative frequency of individual patients or HCs that contribute to clusters 14 and 15. (G) Bar plots depicting TRBV11-2 usage for each patient or control within clusters 14 and 15. (H) Circos plots depicting TRA-V–TRB-V pairing within clusters 14 and 15 of MIS-C patients. (I) Stacked bar graphs showing clonotype frequencies within cluster 8 (TEMRA), 14 (proliferating T cells), and 15 (TIM3+ HLAD-DR+ CD38+ T cells) from all patient conditions. Clonotype frequencies of naive T cells (cluster 4) are shown as a reference of unexpanded clones. Clonotypes were defined as the combination of TRA + TRB CDR3AA. (J) Violin plots showing expression of DEGs in TRBV11-2+ versus TRBV11-2− cells of MIS-C patients. (K) Volcano plots showing differentially expressed (DE) genes of TRBV11-2+ positive cells versus TRBV11-2− cells within clusters 14 and 15 and only for MIS-C patients. Genes were selected on a Log2 fold change of ±0.25 and a Benjamini–Hochberg adj P value of 0.05. (L) Analogous volcano plot as in K for DE genes between cells in cluster 8 of adult COVID-19 patients compared with TRBV11-2+ cells in clusters 14 and 15 of MIS-C patients. Treg, regulatory T cell.
Figure S4.
Single-cell TCR profiling reveals an activated and proliferating population with skewed TCR repertoire, suggestive of superantigenic stimulation of TRBV11-2+ cells. (A) Dot plot depicting signature genes defining the UMAP clusters shown in Fig. 5, A and B. (B) TRA-V gene family usage within clusters 14 and 15 among all conditions. (C) Clonotype connection plots linking UMAP clusters with shared clonotypes. Line widths depict number of overlapping clones. UMAPs are split based on patient condition. Clonotypes are defined by TRB + TRA CDR3AA sequences. (D) Barplot depicting TRBJ gene segment usage within clusters 14 and 15 of the MIS-C condition. (E) Ridgeplot depicting CDR3 length (number of amino acids) within TRBV11-2+ cells of clusters 14 and 15 of the MIS-C condition (red) versus cytotoxic T cells (TEMRA) of cluster 8 (orange). The CDR3 length of cluster 4 (naive CD4 T cells) is presented as a reference (gray). (F) Stacked bar graphs showing clonotype expansion within TRBV11-2+ and TRBV11-2− cells of clusters 14 and 15 of the MIS-C condition. Clonotypes are defined by TRB + TRA CDR3AA sequences. (G) Up-regulated canonical pathways as identified by IPA on DEGs of TRBV11-2+ cells as compared with TRBV11-2− cells for MIS-C patients within cluster 14 and 15. Pathways were ordered on their Benjamini–Hochberg-corrected P value (bars) and filtered on pathways with a positive z-score (squares). (H) Upstream regulators with a filter on cytokines as identified by IPA on DEGs of TRBV11-2+ cells as compared with TRBV11-2− cells for MIS-C patients within clusters 14 and 15 (top) and on cells of adult patients with severe COVID-19 within cluster 8 as compared with TRBV11-2+ cells of MIS-C within clusters 14 and 15 (bottom). Cytokines were ordered on their Benjamini–Hochberg-corrected P value (bars) and filtered on cytokines with a positive z-score (squares). (I) HLA clonotypes for MIS-C patients and siblings (J) compared with local reference frequencies. Eff quiesc, quiescent effector; Freq, frequency; NA, nonavailable; pct., percent; Prolif, proliferating; Perc, percent; Tregs, regulatory T cells.
Single-cell TCR profiling suggests a superantigenic stimulation of TRBV11-2 cells
Next, we examined TCRα and β pairing and found that the TCRβ encompassing TRBV11-2 did not pair with a specific TCRα (Fig. 6 H), suggestive of superantigenic stimulation. This was affirmed by the unspecific TCRβ joining segment (TRBJ) usage, an unbiased CDR3 length distribution, and dissimilar CDR3 amino acid sequences in T cells expressing TRBV11-2 (Fig. S4, D–F). We determined cellular expansion in clusters 14 and 15 but failed to observe significant expansion of specific clonotypes (defined TRA/TRB CDR3 region combinations; Fig. 6 I). While no clonotype specific expansion was observed in MIS-C, adults with severe COVID-19 showed clusters of oligoclonal T cells with specific TCRαβ pairs (Fig. 5 I), suggesting a peptide-specific response, rather than superantigenic stimulation. We examined the differentially expressed gene (DEG) of the TRBV11-2+–expressing cells versus TRBV11-2− in MIS-C patients and found 1,333 up-regulated genes and 245 down-regulated genes (Table S4; adjusted [adj] P: <0.05; Log2: ±0.25). The top expressed genes are enriched for granzymes (GZMA, GZMB, GZMK, and GZMH) and cytotoxic function (NKG7, GNLY, and PRF1). Ingenuity pathway analysis (IPA) revealed expressed gene sets associated with canonical pathways, such as oxidative phosphorylation, proliferation, anti-apoptotic pathways, and enhanced protein translation (Fig. S4 G). These enriched gene sets are indicative of the activation of TRBV11-2+ cells and have been previously described in the context of T cell biology (Patil et al., 2018; Zhang et al., 2020a). To study the TRBV11-2+ cells in more detail, we compared TRBV11-2+ and TRBV11-2− cells of MIS-C patients within clusters 14 and 15. This resulted in 122 up-regulated and 73 down-regulated DEGs (Table S5; adj P: <0.05; Log2: ±0.25). Using IPA, analysis of upstream regulators suggested IL-15, IL-2, and IFNγ as major upstream cytokines (Fig. S4 H) and PLZF and XBP1 as predicted regulatory transcription factors. ZBTB16 (encoding PLZF) and XBP1 are both up-regulated in TRBV11-2+ cells (Fig. 6 J). XBP1 orchestrates adaptive metabolic responses such as the unfolded protein response and antioxidant response and promotes T cell function. PLZF controls the effector functions and tissue homing of ILC1 and NKT cells in mice. Disturbed MAIT, γδ T, and effector CD161+ CD8 T cells in a PLZF-deficient patient suggest a broader function in humans (Savage et al., 2008; Eidson et al., 2011; Constantinides et al., 2014). Indeed, compared with activated TRBV11-2− cells, several PLZF targets such as IL7R, KLRB1, and ID2 and chemokines such as CXCR6 and CXCR3 were differentially expressed (Fig. 6, J and K). The gene expression profile further suggested a pronounced cytotoxic profile as GZMA, GZMB, and GNLY were significantly up-regulated in TRBV11-2+ cells (Fig. 6, J and K). Comparing TRBV11-2+ cells in clusters 14 and 15 with the oligoclonal cytotoxic T cells of severe adult COVID-19 in cluster 8, we found that genes associated with T cell activation (CD27 and CD74), cytotoxicity (GZMK), and metabolic activity (GADPH, MT-CO3, and MTRNR2L8) were selectively enriched in MIS-C. In severe COVID-19, genes associated with V(D)J recombination were differentially expressed (Fig. 6 L). IPA confirmed previous observations and indicated that type I IFN signaling was a distinctive feature of severe COVID-19 but not of MIS-C (Fig. S4 H). Finally, previous research suggested that clonal expansion of TRBV11-2 in MIS-C was associated with specific HLA class I haplotypes (Porritt et al., 2021). Genotyping of our cases using whole exome sequencing revealed no HLA bias (Fig. S4 I). Alleles with higher frequencies in the MIS-C cohort were also prevalent in the Belgian population according to the Allele Frequency Net database (Fig. S4 J) and similarly present in healthy siblings.
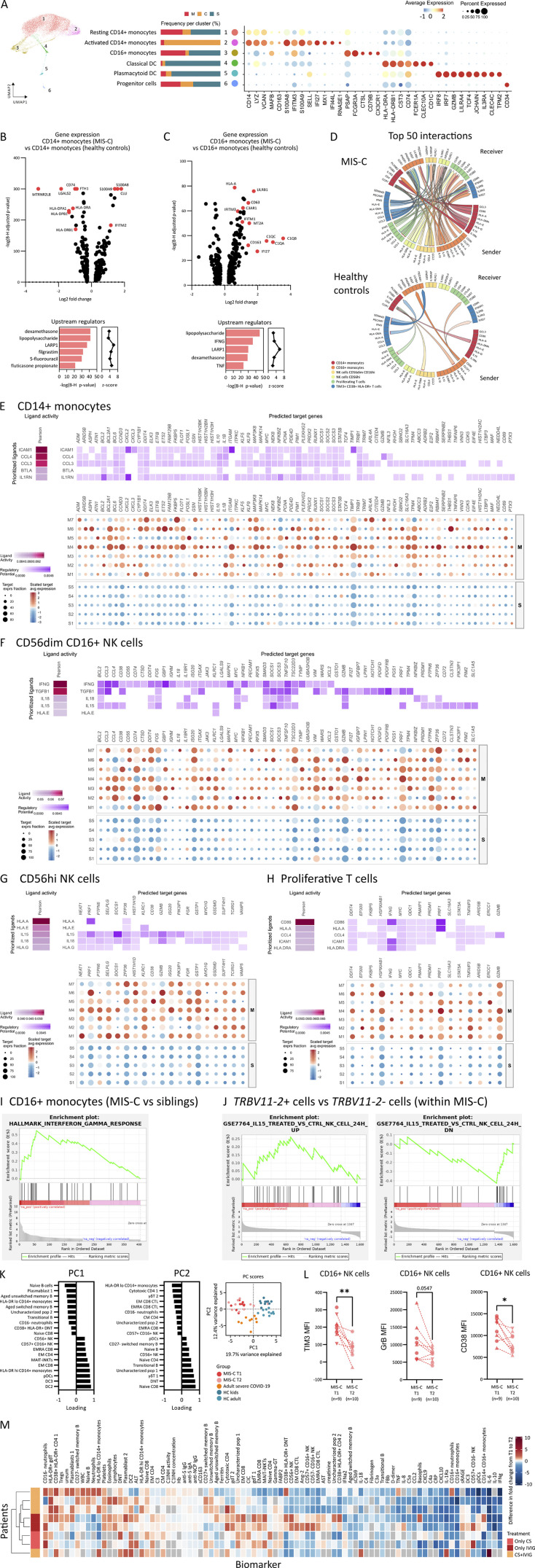
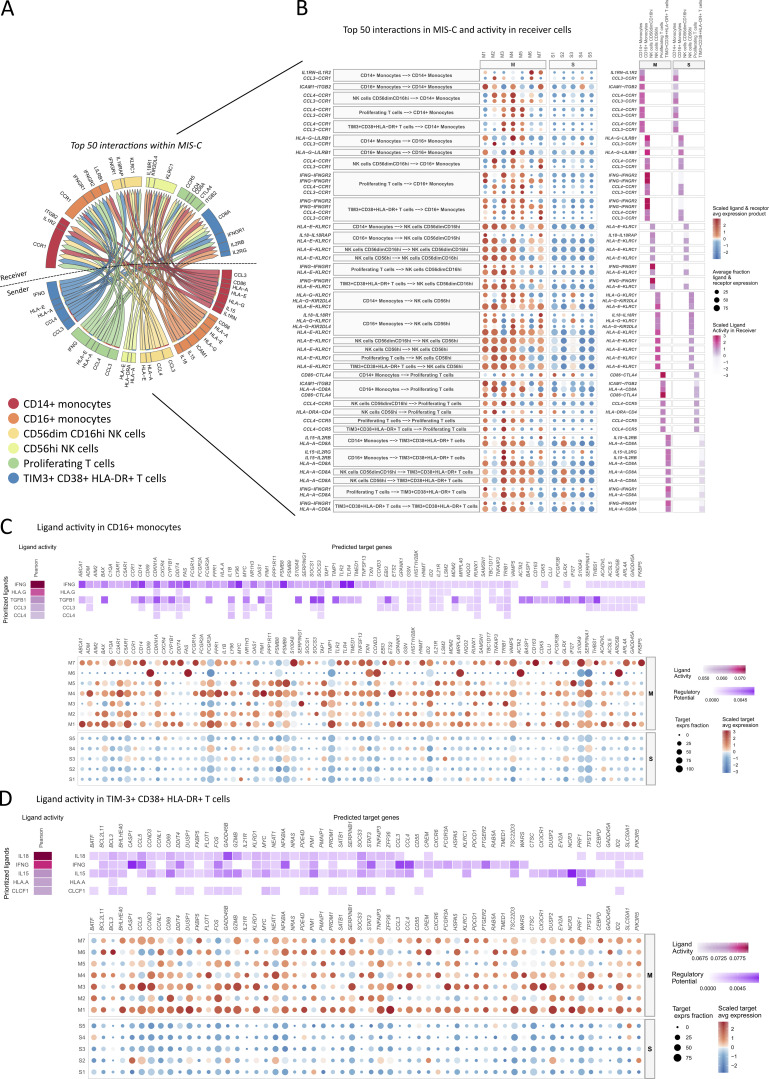
NicheNet reveals T cell–monocyte–NK cell communication through IFNγ and IL-15 signaling
UMAP analysis of the scRNA-seq data of innate leukocytes readily identified progenitor cells, conventional DCs, and pDCs, and both classical and patrolling monocyte populations. The proportion of these clusters obtained by scRNA-seq were in line with the marked differences between MIS-C and severe COVID-19 found by FCM (Fig. S5 A). Comparing MIS-C and HCs, analysis of DEG (adj P: <0.05; Log2: ±0.25) revealed 190 up-regulated and 179 down-regulated and 320 up-regulated and 113 down-regulated in classical and patrolling monocytes, respectively (Table S6 and Table S7). The top up-regulated DEG in patrolling monocytes confirmed activation with increased expression of components of the complement cascade (C1QA,B,C and C3RA1), alarmins (S100A8,9 and 12), and IFN-inducible genes (RNASE2, IFI27, and IFITM2) and identified IFNγ and LPS as upstream regulators (Fig. S5, B and C). Having defined all populations of interest, we used NicheNet (Browaeys et al., 2020), a computational modeling tool using gene expression data as inputs to assess the ligand-to-target signaling pathways between monocyte, NK cell, and activated and proliferative T cell clusters. Assessing the number of interactions in this network, NicheNet confirmed increased ligand-to-target signaling in MIS-C compared with HCs (Fig. S5 D). Among the 50 top interactions, patrolling monocytes and CD16+ NK cells were enriched in “initiating” ligands, whereas “receiving” targets were increased in CD14+ monocytes (Fig. 7 A). Analyzing pairwise interactions, patrolling monocytes and CD16+ NK cells interacted with all other interrogated leukocytes. There was a particular strong interaction between patrolling monocytes and the activated/proliferating T cells in MIS-C as ∼50% of the pairwise interactions originating from these specific leukocytes implicated the respective counterpart (Fig. 7 B). Confirming our findings, IFNG stood out as a ligand originating from activated T cells to interact with the IFNγ receptor (IFNGR1/2) on patrolling monocytes and CD16+ NK cells. Conversely, activated and proliferating T cell clusters received IL15 signaling from patrolling monocytes (Fig. 7 A). The strong ligand activity (based on ligand dependent gene expression) for these ligand-to-target interactions further confirmed the specific activation in MIS-C (Fig. 7 B). To prioritize ligand-to-target interactions in patrolling monocytes and activated T cells, we performed an in-depth analysis on the level of single patients. These analyses identified IFNG as upstream ligand in patrolling monocytes, while prioritizing IL18, IL15, and IFNG in the activated T cells (Fig. 7, C and D). Similar interactions could, to a lesser extent, be described in clusters of classical monocytes, proliferative T cells, and NK cells (Fig. S5, E–H). Finally, gene set enrichment analysis (GSEA) of DEG independently confirmed an IFNG responsive gene set in CD16+ monocytes and IL15-induced response genes in TRBV11.2+ T cells (Fig. S5, I and J).
Figure S5.
scRNA-seq of innate leukocytes, NicheNet analysis, and immune profile of MIS-C patients upon clinical recovery. (A) UMAP plot depicting cluster annotation and numbering on a Seurat object of innate leukocyte cell types. The relative proportion of patient or control samples (M = MIS-C, S = healthy sibling, A = adult COVID-19) that contributes to each cluster is visualized in the bar graphs in the middle panel. Right: Dotplot analysis depicts the signature genes of each the clusters. (B) Volcano plot (top) showing differentially expressed (DE) genes of CD14+ monocytes of MIS-C patients as compared with CD14+ monocytes of HCs. Genes were selected on a Log2 fold change of ±0.25 and a Benjamini–Hochberg adj P value of 0.05. Upstream regulators (bottom) as identified by IPA on DEGs of CD14+ monocytes of MIS-C as compared with HCs. Regulators were sorted on their Benjamini–Hochberg-corrected P value (bars) and filtered on regulators with a positive z-score (squares). (C) Analogous volcano plot (top) and upstream regulators (bottom) as in B for DE genes between CD16+ monocytes of MIS-C as compared with CD16+ monocytes of HCs. (D) Circos plot of the top 50 interactions between clusters of T cells, NK cells, and monocytes identified by NicheNet, connecting sending and receiving cell types of interest by their differential ligand–receptor communication, split for MIS-C and HCs. (E–H) Heatmap and dim plots visualizing NicheNet analysis of (E) CD14+ monocytes, (F) CD56dim CD16+ NK cells, (G) CD56+ NK cells, and (H) proliferative T cells as receiving cells, displaying their respective differential ligand activity and downstream regulation of target genes for all analyzed samples (top) and for MIS-C patients (M) and healthy siblings (S) separately. (I and J) GSEA of IFNγ response in the DEGs between CD16+ monocytes of MIS-C compared with siblings (I) and (J) IL15 up- and down-regulation in TRBV11-2+ cells of MIS-C versus TRBV11-2− cells of MIS-C patients. (K) Loadings of the principal component analysis of UMAP clusters of patients and HC groups. (L) Pairwise analysis of the MFIs of TIM3, granzyme B (GrB), and CD38 on CD16+ NK cells of MIS-C patients on T1 and T2. Significances between T2 and T1 were calculated by Wilcoxon matched-pairs signed-rank test, indicated by *, P < 0.05; **, P < 0.01. Patients with severe MIS-C (T1 sampling after treatment initiation, inverted triangle; T1 before treatment initiation, triangle) and moderate disease (circle) are displayed separately. (M) Heatmap displaying absolute differences between normalized biomarkers on the two time points of sampling in MIS-C (T2 minus T1). Gray boxes denote if a variable was not available for an individual patient. Patients were hierarchically clustered on their variables (corticosteroids [CS], IVIG, or a combination of both).
Figure 7.
NicheNet reveals T cell-monocyte-NK cell communication through IFNγ, IL-15 and IL-18 signaling in MIS-C. (A) Circos plot of the top 50 interactions between clusters of T cells, NK cells, and monocytes in MIS-C identified by NicheNet, connecting sending and receiving cell types of interest by their differential ligand–receptor communication. (B) Visualization of differential interactions found by NicheNet for individual MIS-C patients (M1–M7) and healthy siblings (S1–S5) with a prediction of ligand activity in receiving cells (C and D) Heatmap and dim plots visualizing NicheNet analysis of (C) CD16+ monocytes and (D) TIM3+ CD38+ HLA-DR+ T cells as receiving cells, displaying their respective differential ligand activity and downstream regulation of target genes for all analyzed samples (top) and for MIS-C patients (M) and healthy siblings (S) separately (bottom). avg, average; exprs, expressing.
Together, NicheNet analysis provided evidence for intricate interactions between distinct sets of leukocytes that were specifically activated in MIS-C. Whereas mostly classical monocytes received inflammatory signals from other leukocytes, intense bilateral interaction could be identified between activated/proliferating T cells and patrolling monocytes. In line with our previous observations, NicheNet prioritized IFNγ as one of the top regulators in this communication.
Treatment results in rapid normalization of IFNγ signature and activated leukocyte subsets
MIS-C patients display rapid clinical improvement upon treatment with IVIG and/or systemic corticosteroids. To study immunological correlates, we performed an immunophenotypic analysis at T2. Sorting differences of normalized variables between T2 and T1 samples revealed that recuperation was evident for proinflammatory and immunoregulatory cytokines (Fig. 8 A). Cytokines associated with activation of both innate and adaptive leukocytes resolved rapidly (Fig. 8, A and B), including significant reductions of IFNγ and chemokines CXCL9 and CXCL10 (Fig. 8 C). Lymphocytes, eosinophils, and platelets recovered similarly (Fig. 8 A).
Figure 8.
Clinical resolution is associated with recovery of type II IFN signature and contraction of CD16+ monocytes and TIM3+ T cells. (A) Bar chart depicting ratios of routine laboratory values, serum proteins, and UMAP cell populations measured at sampling time points T2 and T1. Ratios were calculated from normalized values (fold changes to the median of HCs). Bars are ordered on the median and represent the top 25 most negative (blue) and top 10 most positive (red) differences, with whiskers denoting the range. Significant differences between T2 and T1 were calculated by Wilcoxon matched-pairs signed-rank test, indicated by *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001. (B) Heatmap separated by condition (T1 and T2) displaying hierarchical clustered normalized biomarkers (cytokines and tissue marker fold change relative to median of HCs) for individual MIS-C patients. CS, corticosteroids. (C) Dot plot showing type I and type II IFN and downstream chemokines (CXCL9 and CXCL10) concentrations from patients collected at sampling time points T2 and T1. (D) Principal component (PC) analysis of all UMAP clusters (B, T/NK, innate cells), identified by flow cytometry. (E) UMAP representation of innate leukocytes (within PBMCs) with density plots stratified per time point of sampling in MIS-C (n = 9 on T1 and n = 10 on T2). Numbers show percentage of clusters of interest. (F) Evolution of the proportions of innate UMAP clusters by pairing samples from identical patients on both time points. (G) UMAP representation of T lymphocytes, NK cells, and iNKT/MAIT cells (within PBMCs) and density plots for both time points of sampling in MIS-C, similar to E. (H and I) Evolution of the proportions of T lymphocytes as determined by manual gating of FCM data, pairing samples from identical patients on both time points. Significances between T2 and T1 were calculated by Wilcoxon matched-pairs signed-rank test, indicated by *, P < 0.05; **, P < 0.01; and ***, P < 0.001. Values for individual patients are displayed with MIS-C (red), severe adult COVID-19 (orange), and age-matched HCs (blue). Distinct symbols depict patients with severe MIS-C with (inverted triangle) or without treatment (triangle) and moderate disease (circle).
Principal component analysis on innate, B, T, and NK cell clusters generated by FCM did not separate T1 and T2 in MIS-C patients (Fig. 8 D and Fig. S5 K). Nonetheless, analyzing temporal changes within clusters revealed compelling differences (Fig. 8 E). Both patrolling and inflammatory monocytes decreased significantly after treatment (Fig. 8, E and F). Neutrophils, abundantly present at T1 in two distinct clusters, evolved asymmetrically with CD16− neutrophils significantly increasing and CD16+ neutrophils contracting at T2 (Fig. 8, E and F). Basophil and pDC numbers did not recover during admission (Fig. 8, E and F). In contrast, DC2 numbers increased at T2 (Fig. 8 F).
Subtle differences were also observed in T cell clusters (Fig. 8 G). Analyzing UMAP, clusters of activated (CD38+/HLA-DR+) CD4 and DN T cells contracted at T2, although not reaching statistical significance (Fig. 8 H). Analyzing activation status, the percentage of TIM3+/PD-1+ cells dropped significantly by T2 among all T cell populations (Fig. 8 I). In parallel, markers of CD16+ NK cell activation similarly decreased (Fig. S5 L). Finally, hierarchical clustering did not reveal clear immunological differences between patients treated with IVIG, corticosteroids, or both (Fig. S5 M).
Together, immunophenotyping of early resolution samples of MIS-C patients confirms that the type II IFN signature, TIM3+ T cells, patrolling monocytes, and activation of CD16+ NK cells not only distinguished MIS-C from HCs or severe COVID-19 but also resolved completely within days, mirroring clinical resolution.
Discussion
In 14 children with characteristic MIS-C presentation, we performed in-depth immune profiling using FCM, peptide restimulation, scRNA-seq, TCR repertoire analysis, and serum proteomics, and compared findings with HCs and adult patients with severe COVID-19 and signs of CRS. Based on this comprehensive study, we propose that MIS-C is characterized by gut epithelium damage and IFNγ-mediated interactions between SAg–stimulated T cells, NK cells, and patrolling monocytes, activating additional innate leukocytes, like classical monocytes, and inducing pleiotropic release of cytokines.
The T cells implicated in disease pathogenesis were activated (CD38+/HLA-DR+), functional (CD27+CD57−), and coexpressed markers of activation (GITR) and exhaustion (TIM3+/PD-1+). Using HLA-DR, CD38, PDCD1 (PD-1), and HAVCR2 (TIM3) as signature genes in the scRNA-seq data, we identified these cells as a proliferative population with cytotoxic capacity and a skewed TCR repertoire. Whereas recent publications only report on the expansion of T cells harboring TRBV11-2 (Vβ 21.3) in a subset of MIS-C patients (Moreews et al., 2021; Porritt et al., 2021; Ramaswamy et al., 2021), our scRNA-seq analysis revealed activation and/or proliferation of TRBV11-2 T cells in all patients, suggesting a homogeneous disease, with a central role for TRBV11-2 T cells. In line with previous observations, promiscuous usage of Vα and unbiased V(D)J recombination indicate interactions outside the classical CDRs, akin to immunopathology driven by SAgs. To date, the identity of the SAg remains to be discovered, and both viral protein and SAg from a bystander source are plausible. The delayed onset after documented SARS-CoV-2 infection, seropositivity, and the inability to detect viral RNA in nasal swabs of most patients are at first sight compelling arguments against a SAg of viral origin, although viral persistence outside the respiratory tract remains possible. Indeed, it has been documented that during SARS-CoV-2 infection, the gastrointestinal tract harbors viral antigen for a prolonged period, particularly in children (Wu et al., 2020; Xu et al., 2020). A second argument in favor of a role of viral SAg is the finding of widespread presence of SARS-CoV-2 in organs of deceased MIS-C patients (Duarte-Neto et al., 2021). Although one should be careful extrapolating findings from patients with adverse outcome to all MIS-C patients, virus presence in tissue could serve as a persistent source of SAg and explain a late-onset and systemic presentation. Also, previous in silico predictions have hinted toward an interaction of a SAg-like motif on the SARS-CoV-2 spike protein with Vβ 21.3 (Porritt et al., 2021). In contrast with classical SAg, presentation of S protein through interaction with a restricted number of HLA-I proteins was suggested. Although this might explain the relatively rare occurrence of MIS-C and overrepresentation of specific ethnic groups, we could not confirm the presence of HLA bias in our MIS-C cohort.
Aside from SAg identity and source, the striking age specificity of MIS-C (8–12 yr) compared with TSS (Nelson et al., 2016) is enigmatic. It remains to be proven whether host factors (rare genetic variants or polymorphisms) or features specific to the SAg (geographical or seasonal occurrence as in KD or microbiome association as in TSS) determine the age of onset. Similar to TSS, a complex interplay of both host and SAg factors is to be expected. In TSS, the absence of prior developed protective humoral immunity predisposes patients to TSS upon exposure to the SAg produced by the pathogen (typically during tampon usage). Finally, although the significance of gut damage in MIS-C is yet to be determined, changes in the composition and function of the gut microbiome are age dependent and might influence host immune response (Bosco and Noti, 2021).
Analysis of cytokine expression and scRNA-seq data demonstrated that MIS-C was characterized by IFNγ whereas serum concentrations of type I IFN were enhanced in severe COVID-19. In contrast with type I IFN in COVID-19 (Bastard et al., 2020; Zhang et al., 2020c), the role of IFNγ in MIS-C is unknown. Previous reports have similarly detected increased IFNγ and/or CXCL9 in MIS-C patients, although variable differences compared with pediatric COVID-19 have been described (Caldarale et al., 2021; Esteve-Sole et al., 2021; Yonker et al., 2021). While IFNγ is the quintessential cytokine in protective T helper type 1 cell responses, excessive IFNγ is toxic and drives hyperinflammatory disorders such as HLH (Jordan et al., 2004; Humblet-Baron et al., 2019) or MAS (Bracaglia et al., 2017), and rare monogenic syndromes (Reinhardt et al., 2015; Tavernier et al., 2019). Although chronic fever, cytopenia, and hyperferritinemia in MIS-C are reminiscent of HLH, none of our patients reached HLH diagnostic criteria. Additionally, MIS-C shares many characteristics (prolonged high fever, skin rash, hemodynamic shock, and myocardial dysfunction) with diseases that are triggered by SAg, such as TSS. TSS is provoked by the exotoxin TSST-1, which is produced by Staphylococcus aureus and Streptococcus pyogenes, and drives SAg-mediated stimulation of Vβ 2.1+ T cells (Schlievert, 1983). IFNγ is instrumental in TSS pathogenesis as lethality in a murine model is abolished in the absence of IFNγ (Dinges and Schlievert, 2001b). The clinical correlation between TSS and MIS-C might be explained by excessive IFNγ. IFNγ-producing transgenic mice spontaneously develop myocarditis, and high IFNγ concentrations correlate with heart failure (Reifenberg et al., 2007; Levick and Goldspink, 2014). IFNγ induces gastrointestinal epithelial disruption (Madara and Stafford, 1989; Li et al., 2008; Nava et al., 2010) and gut permeability in TSS (Tilahun et al., 2011). All our MIS-C patients presented gastrointestinal symptoms and had increased FABP2, a marker of gut epithelium damage. Furthermore, circulating LPS was found in a proportion of patients, and transcriptome analysis on patrolling monocytes suggested endotoxemia as an upstream driver. This might be highly relevant as the presence of endotoxins in the circulation, such as LPS, was found to contribute to immune perturbation and cytokine production in TSS (Dinges and Schlievert, 2001a). Although a recent case series confirmed gut barrier disruption in MIS-C, some patients displayed normal serum IFNγ levels (Yonker et al., 2021). Furthermore, in animal models of colitis, both protective and deleterious roles of IFNγ have been described (Brasseit et al., 2018; Langer et al., 2019). Future studies should further delineate the role of IFNγ in gut barrier disruption in MIS-C.
The current mainstay of MIS-C therapy is the use of IVIG and systemic corticosteroids. The underlying mechanisms of IVIG in MIS-C are debated. A substantial role for neutralization of viral-derived SAg or clearance of SARS-CoV-2 viral particles seems unlikely given the absence of protective anti–SARS-CoV-2 antibodies in IVIG during the early wave of MIS-C. Other (in)direct anti-inflammatory activities of IVIG have also been proposed (Schwab and Nimmerjahn, 2013). While our data support a potential role for such mechanisms, IVIG resistance in MIS-C is substantial (38–80%; Pouletty et al., 2020; Verdoni et al., 2020; Ouldali et al., 2021). Recent publications provide evidence for a more favorable outcome using systemic steroids in addition to IVIG (Ouldali et al., 2021) and/or less escalation of treatment when first-line steroids are given (McArdle et al., 2021; Son et al., 2021). Our novel insights on IFNγ suggest that putting a brake on excess type 2 IFN might abrogate the systemic hyperinflammation in MIS-C refractory to IVIG and/or corticosteroids. Emapalumab, a monoclonal antibody blocking IFNγ, has shown promising results in pediatric patients with primary HLH (Locatelli et al., 2020). Interfering JAK/STAT signaling using small molecules such as ruxolitinib might represent an attractive alternative (Kale et al., 2020). To our knowledge, none of these compounds have yet been tested in MIS-C. Given the rare and heterogeneous presentation of MIS-C and the rapid recovery of most patients with primary treatment (IVIG and/or corticosteroids; Feldstein et al., 2021), proving the superiority of targeted treatment of IFNγ or JAK/STAT signaling will be challenging, and targeted therapy will likely be limited to treatment-resistant cases.
Our study has some limitations. In general, most of the research on MIS-C has been limited to the analysis of blood-derived immune effectors. To fully understand the pathogenesis, a detailed study on the immunological processes in tissue such as the gastrointestinal epithelial lining is urgently needed. This exploratory research was based on a small cohort, and the use of adults with severe COVID-19 as a control group might influence the disease specificity of our findings. As such, larger studies using pediatric COVID-19 patients as controls are needed to confirm the proposed elements that underlie the pathophysiology of MIS-C. Finally, our cohort was underpowered to assess the treatment-specific effects on the immunological response.
Materials and methods
Extended description of MIS-C cohort
Most MIS-C patients were school-aged children (median age, 10 yr; IQR, 8.25–13.5), and an equal sex distribution (male:female ratio, 1:1) was found. A substantial proportion of individuals were Black (4/14) or of Middle Eastern descent (3/14). Patients had no relevant prior medical history (one patient was born premature, one with β thalassemia minor, and one with idiopathic short stature) or recurring infectious or inflammatory diseases. All patients presented fever at admission with a total median fever duration of 6 d (IQR, 6–7). All patients (14/14) presented with gastrointestinal manifestations, including abdominal pain (12/14), vomiting (9/14), and diarrhea (6/14). Abnormal findings on gastrointestinal imaging (ultrasound and/or computed tomography) were found in half of cases (7/14), and 1 case underwent explorative laparoscopy for presumed appendicitis. Symptoms furthermore included neurological complaints in 6/14, respiratory signs in 5/14, and myalgia in 3/14. Cardiovascular compromise (decreased ejection fraction, need for intravenous fluid bolus and/or inotropics) occurred in 11/14. 11 patients fulfilled at least 2/5 KD criteria in addition to prolonged fever. Non-exudative bilateral conjunctivitis (11/14) and exanthema (9/14) occurred in a high proportion of cases. Complete KD was present in only 2/14. No thrombogenic complications were observed. Echocardiographic abnormalities included hyperechogenicity of the coronaries (3/14), prominently visible coronary arteries without dilatation (4/14), and coronary dilatation with z-scores >2.0 (2/14). At least moderate decreased left ventricular function was present in 4/14 (fractional shortening ≤25%). Substantially elevated inflammatory markers were described in routine laboratory assays, including median peak ferritin 592.5 µg/liter (IQR, 510.8–858.8), median peak CRP 286.4 mg/liter (IQR, 187.3–376.1), and median peak erythrocyte sedimentation rate 42 mm/h (IQR, 20–108). Signs of thrombolysis (median peak D-dimers 3,190 ng/ml and peak fibrinogen 660.5 mg/dl) were found. Four patients presented acute kidney injury (serum creatinine >1.00 mg/dl). Liver function tests (ALT, AST, GGT) were only minimally and transiently elevated in most cases. Hyperleukocytosis (>12 × 109/liter) and lymphopenia (<1 × 109/liter) were present in 9/14 (median highest WBC count, 15.0 × 109/liter; IQR, 8.8–17.5) and 13/14 (median low lymphocytes of 0.4 × 109/liter; IQR, 0.3–0.6), respectively. All patients were hospitalized (median length of stay of 9 d; IQR, 7.25–12.75). 12/14 patients required admission at an intensive care unit (median stay of 4 d; IQR, 2–6.25). Pharmacological treatment included IVIGs at immunomodulatory dosage (2 g/kg) in 11/14 patients and systemic corticosteroids in 9/14. Six patients received a combination of IVIG and corticosteroids. Signs of hemodynamic shock were present in 11/14 at presentation, of which 8 required inotropics. All patients were empirically treated with broad-spectrum intravenous antibiotics. Respiratory support was needed in 8/14, of which 5 required noninvasive ventilation. No patients needed mechanical ventilation, extracorporeal membrane oxygenation, or treatment with biologicals. Sampling for immunological studies was performed before the initiation of specific treatment in all patients or within 12 h after administration, except in P1 (two doses of low-dose hydrocortisone) and P9 (two doses of methylprednisolone; Fig. S1 A). Resolution samples (T2) were available in 13 cases at time points day 5 to day 10 after the first sample. Rapid clinical improvement was seen in all patients (Fig. S1 A), without substantial residual morbidity at time of discharge. No deaths were registered, and all patients are doing clinically well to date.
Study participants
From May 2020 to September 2020, we prospectively included pediatric patients (0–18 yr of age) who fulfilled the MIS-C case definition by the WHO (World Health Organization, 2020) and were either hospitalized in our university hospital (tertiary Children’s Hospital Ghent University Hospital) or in district or other university hospitals within our network.
In parallel, we collaborated on an interventional, randomized, open-label, multi-center study involving adult SARS-CoV-2 PCR-positive patients with hyperinflammation (COV-AID trial; Maes et al., 2020). Inclusion criteria (Table S1) required subjects to present with respiratory failure and have signs of systemic CRS (serum ferritin >2,000 µg/liter or >1,000 µg/liter and rising last 24 h, and/or lymphopenia <800/µl with at least two of the other criteria, including elevated D-dimers, lactate dehydrogenase, CRP, and ferritin) for the randomized initiation of individual or simultaneous blocking of IL-6 (siltuximab), IL-6 receptor (tocilizumab), and IL-1 (anakinra). The full study protocol can be found at ClinicalTrials.gov Identifier NCT04330638. Simultaneously with these inclusions, age-matched healthy individuals without prior medical history and in the absence of recent infection or vaccination (<6 wk) were included for their blood samples to be used as controls.
The patients and family members provided written informed consent for study participation, in accordance with the 1975 Helsinki Declaration. The research protocol was approved by the main ethics committee of Ghent University Hospital, as well as in local committees of participating centers. Demographic and clinical data on the participating subjects were registered in an in-house stored, protected, and anonymized data file.
Sample collection and processing
After parental informed consent, arterial or venous blood samples in children presenting with MIS-C were collected at admission (T1), preferably before specific therapy was initiated (including IVIGs, systemic corticosteroids, or biopharmaceuticals). T1 thus reflects a clinical state of maximal inflammation. A control sample (T2), reflecting resolution, was collected at day 7 after treatment or before discharge from the hospital if sooner than day 7. Sample collection in adult cases participating in the COV-AID trial was performed according to the trial protocol (i.e., before randomization and administration of study medication, at day 6, day 15, and 10–20 wk follow-up).
Peripheral venous blood specimens were collected from healthy individuals and patients using simultaneously obtained EDTA and serum tubes. EDTA blood was diluted 1:2 in HBSS (Thermo Fisher Scientific; 24020117) and PBMCs isolated after gradient centrifugation over Ficoll-Paque (GE Healthcare; 17–1440-02). Cell-free plasma was subsequently transferred from the supernatant, aliquoted, and stored at −80°C. After two washings in cold HBSS, the yielded layer of PBMCs was counted in a Neubauer plate with trypan blue exclusion of dead cells. PBMCs were aliquoted in 90% FCS (Sigma-Aldrich; F7524) and 10% DMSO (Sigma-Aldrich; D2650). Vials were placed in a −80°C freezer using controlled rate freezing in preparation for final storage at −150°C until further use. Serum tubes were spun at 4°C, and cell-free serum was subsequently aliquoted and stored at −80°C until analysis. Differences in the number of patients analyzed per assay occurred as a consequence of sample availability.
SARS-CoV-2 confirmation
During hospital admission, patients were tested on acute or recent COVID-19 by nasopharyngeal swab for SARS-CoV-2 nucleic acid using RT-PCR and/or by performing antibody quantification for anti–SARS-CoV-2 IgM, IgG, or both. The precise modalities (e.g., manufacturer of the assays) of these investigations in routine clinical care depended on the center where the patient was included and varied over time according to the assay’s availability. SARS-CoV-2 seroconversion was confirmed on stored serum samples of included patients with antigen-coated ELISA kits (EUROIMMUN AG) for anti–spike 1 (S1) IgA (EI 2606–9601 A) and IgG (EI 2606–9601 G) and anti-NCP IgG (EI 2606–9601-2 G), according to the manufacturer’s protocol.
Cytokines and tissue biomarkers
Serum cytokines IL-1β, IL-1Ra, IL-6, IL-8 (CXCL8), IL-10, IL-18, CXCL9, CXCL10, TNFα, MCP-1 (CCL2), sCD25, and sCD163 were quantified in duplicate by magnetic bead-based multiplex assay using xMAP technology (Luminex Corporation) and Bio-Plex assays, kits, and standards (Bio-Rad, Human Cytokine Screening Panel #12007919; and Bio-Rad, Human Inflammation Panel #171DL0001) according to manufacturer’s protocol. Multiplex assay samples were diluted at 1:2. Acquisition and analysis was performed on a Bio-Plex 200 reader and using the Bio-Plex Manager software (Bio-Rad). Commercially available ELISA kits were used for the detection of serum Ang-2 (Thermo Fisher Scientific; KHC1641), FABP2 (Thermo Fisher Scientific; EHFABP2), and receptor for advanced glycation end products (RAGE; R&D Systems; DRG00). For IFNγ detection, the Ready-SET-Go! ELISA reagent set (Thermo Fisher Scientific; 88–7316-88) was used. Samples were measured in duplicate and according to the manufacturer’s instructions. Samples were diluted at 1:30 (Ang-2), 1:2 (FABP2), 1:5 (RAGE), and 1:2 (IFNγ). IFNα2 was quantified by Meso Scale Discovery using the S-plex Human IFNα2a kit (Meso Scale Discovery; K151P3S-1) according to the manufacturer’s protocol, with data acquired on a MESO QuickPlex SQ 120. CICs were quantified in serum using a commercially available C1q-coated microplate ELISA (EUROIMMUN AG; 1818–9601 G), according to the manufacturer’s protocol. The lower limits of quantification were used for statistical analysis if samples were not measurable above this concentration.
Complement pathway
Complement components were measured in cell-free plasma. FBb, C3a, C4a, C5a, and SC5b9 were measured using customizable enzyme immunoassay multiplex kits (MicroVue Complement Multiplex; Quidel; A905s), according to the manufacturer's instructions. Data were acquired on a Q-View Imager LS, using the Q-View Software 3.11. C3, C4, and C1-INH protein concentrations were measured by an automated turbidimetric assay on the Optilite analyzer (The Binding Site Group Limited), and C1-INH functional activity was determined using the Berichrom C1-Inhibitor assay (Siemens).
Endotoxin quantification
Lyophilized amebocyte lysate endotoxin concentrations were quantified in plasma of MIS-C patients and HCs using the ToxinSensor Chromogenic LAL Endotoxin Assay Kit (GenScript; L00350) according to the manufacturer’s protocol. For individual MIS-C patients, the highest concentration of T1 and T2 samples was used if multiple samples were available.
Extended immunophenotyping on PBMCs
Cryopreserved PBMCs were thawed in 37°C preheated complete medium (RPMI-1640 medium supplemented with GlutaMAX, 10% FCS, 1% penicillin-streptomycin [10,000 U/ml; Gibco; 15140122], 1 mM sodium pyruvate [Gibco; 11360070], 1% nonessential amino acids [Gibco; 11140035], and 50 µM 2-mercaptoethanol [Gibco; 31350010]). Cells were left to recuperate for 30 min at 37°C and 5% CO2 after removal of DMSO. All patients and controls were analyzed in one single experiment. Acquisition and analysis of labeled cell suspensions was performed with a FACSymphony flowcytometer (BD Biosciences) and subsequent analysis with FlowJo v10.7 (BD Biosciences). A compensation matrix was created based on single stain controls. Antibodies used and gating strategy used to define PBMC populations can be found in Table S2.
Peptide pool restimulation
PBMCs were thawed, left to recuperate, and diluted in complete medium as described above. Cells (2 × 106) were cultured in tubes at 37°C and 5% CO2 for 20 h in the presence of SARS-CoV-2–specific peptide pools (a balanced mixture of commercially available S1 scanning pool 3629–1, S2-N proteins 3620–1, and SNMO proteins 3622, all from MABTech) at a final concentration of 0.5 µg/ml, and brefeldin A (400 ng/ml). As a positive control, cells were stimulated with 200 ng/ml PMA and 1 µg/ml ionomycin. Cells were labeled using antibodies as listed in Table S3.
Single-cell RNA and TCR sequencing
All single-cell experiments have been conducted at a containment laboratory with inward directional airflow (BSL-2/3). After thawing, up to 2 million PBMCs were counted, isolated, and spun down. The cell pellet was resuspended and incubated for 30 min on ice with staining mix in PBS containing 0.04% BSA, L/D-DAPI, CD3-FITC (HIT3A), CD14-PerCP-Cy5-5 (HIB19), CCR5-PE (2D7), Human TruStain FcX (BioLegend; 422302), and TotalSeq-C hashing antibodies (BioLegend) diluted 1:500. Two patient samples were multiplexed per lane using TotalSeq-C Cell Hashing Antibodies. Live cells were sorted with T cells/B cells/innate cells in a 70:10:20 ratio. Sorted single-cell suspensions were resuspended at an estimated final concentration of 2,000 cells/µl and loaded on a Chromium GemCode Single Cell Instrument (10x Genomics) to generate single-cell gel beads in emulsion. The scRNA/Feature Barcoding/TCR libraries were prepared using the GemCode Single Cell 5′ Gel Bead and Library kit, version 1.1 (10x Genomics; 1000165) according to the manufacturer’s instructions. The cDNA content of prefragmentation and post-sample index PCR samples was analyzed using the 2100 BioAnalyzer (Agilent). Sequencing libraries were loaded on an Illumina NovaSeq flow cell at Vlaams Instituut voor Biotechnologie Nucleomics core with sequencing settings according to the recommendations of 10x Genomics, pooled in a 75:20:5 ratio for the gene expression, TCR, and antibody-derived libraries, respectively.
Single-cell data processing and analysis
The Cell Ranger pipeline (10x Genomics; version 5.0.0) was used to perform sample demultiplexing and to generate FASTQ files for read 1, read 2, and the i7 sample index for the gene expression and cell surface protein libraries. Read 2 of the gene expression libraries was mapped to the human reference genome (GRCh38.99). Subsequent barcode processing, unique molecular identifiers filtering, and gene counting were performed using the Cell Ranger suite version 5.00. Outlier cells were identified based on three metrics (library size, number of expressed genes, and mitochondrial proportion), and cells were tagged as outliers when they were three median absolute deviations away from the median value of each metric across all cells. Low-quality cells (low unique molecular identifier counts, high percentage of mitochondrial genes) were removed from the analysis. The number of cells was normalized to match the condition with the lowest number of cells when comparing the different conditions (Hao et al., 2021). Differential gene expression analysis between cell clusters and conditions was performed using the Wilcoxon rank-sum test. P value adjustment was performed using Bonferroni correction. Single-cell TCR sequencing analysis was performed using RStudio (R version 4.0.2). Cells that did not have data for both TRB and TRA CDR3 were excluded from the TCR analysis. T cell clonotypes were defined by combining TRA and TRB CDR3 amino acid information. The following CRAN packages were used for single cell sequence data and TCR analysis: circlize, dplyr, ggraph, ggplot2, ggridges, gridExtra, gtools, magrittr, methods, RColorBrewer, reldist, reshape2, rlang, Seurat, stringr, tibble, tidyr, tidygraph, vegan, and writexl. To predict downstream canonical pathways and upstream regulators, IPA (Qiagen; v62089861) was applied on the single cell data. Dataset files of DEGs (adj P value ≤0.05 and logFC ≥0.25 with a minimum expression in ≥10% of cells) were imported with the Ingenuity Knowledge Base as a reference database. Activity z-scores and P values of overlap (Benjamini–Hochberg-corrected) of canonical pathways and upstream regulators were subsequently used, as calculated by the software. To predict key intercellular signaling events between different immune cells, we used NicheNet (Browaeys et al., 2020; https://github.com/saeyslab/nichenetr). NicheNet is a computational method that uses prior knowledge on ligand–receptor, signal transduction, and gene regulatory networks to prioritize ligand–receptor pairs that could regulate the expression of a gene set of interest. The gene set was determined as the up-regulated genes in MIS-C versus HCs for each cell type of interest (compared with the genomic background). The gene set was determined as the up-regulated genes in MIS-C versus HCs for each cell type of interest. To determine DEGs, muscat (Crowell et al., 2020) was used, a tool for differential state analysis in multi-group/multi-sample datasets using thresholds: P value ≤0.05 and logFC ≥0.50. The analysis included only ligands expressed in ≥5% of cells in at least one cell type of interest and with at least one cognate receptor expressed in ≥5% of cells of the receiver cell type of interest. For each receiver cell type, the five ligands with the highest ligand activity were considered for downstream analyses and visualization. For ligand–target inference via the “get_weighted_ligand_target_links” function, the parameter n was set to 500 to extend the search space for possible target genes compared with the default of 250. Since intercellular signaling occurs in each patient individually, ligand–receptor and target gene expression were visualized per patient separately. To define ligand–receptor pairs more strongly expressed in one group versus the other, the average logFC of ligand and receptor should be larger than 0, and one of the following criteria should be met: logFC ligand >0.5 or logFC receptor >0.5 or average logFC of ligand and receptor >0.25. GSEA using the GSEAPreranked module on GenePattern (http://www.genepattern.org; v3.9.11) by importing dataset files of DEGs (adj P value ≤0.05 and logFC ≥0.25 with a minimum expression in ≥10% of cells) as a ranked list compared with gene sets of interest from MSigDB (v7.4).
HLA typing
HLA alleles were determined from whole exome sequencing data using the HLA-HD algorithm (Kawaguchi et al., 2017). Whole exome sequencing data were generated on the NovaSeq6000 system (Illumina) using the SureSelectXT Low Input Human All Exon V7 kit (Agilent Technologies). HLA allele frequencies were compared with the local population (Belgium), as obtained from the Allele Frequency Net Database.
Statistics
Sets of biomarker values were analyzed using the nonparametric unpaired Mann–Whitney U test or paired Wilcoxon matched-pairs signed-rank test to compare two populations (when indicated) or Kruskal–Wallis comparing multiple populations. All tests were performed as two-sided and Dunn’s corrections applied for multiple testing. To correct for multiple testing to compare all PBMC clusters and multiple correlation testing, the two-stage step-up (Benjamini, Krieger, and Yekutieli) method was applied to calculate q values with a false discovery rate (FDR) of 5%. Results with a P or q value of 0.05 or less were considered significant. Significance levels were denoted as *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001. To compare pediatric with adult samples, the Log2 fold change over the median of age-matched HCs was calculated for each patient. Statistical analyses were performed with Prism v9 (GraphPad software) and R v4.0.3 (including the packages corrplot and ComplexHeatmap).
Online supplemental material
Fig. S1 provides additional clinical data from patients with MIS-C and adults with severe COVID-19. Fig. S2 contains data supporting the humoral response, complement pathway activation, and CICs in MIS-C and severe COVID-19. Fig. S3 entails the H-score, IFN type II signature, innate leukocyte subsets, and T and NK cell activation in MIS-C patients. Fig. S4 provides additional data concerning scRNA-seq analysis of T cells and whole exome sequencing analysis of HLA clonotypes. Fig. S5 contains scRNA-seq data concerning innate immune cells, NicheNet analysis of additional leukocyte populations, GSEA analysis, and clustering of clinical and immunological data of MIS-C patients upon disease resolution. Table S1 describes the clinical and biochemical characteristic of the cohort of adults with severe COVID-19. Table S2 and Table S3 contain detailed information of the antibodies used for FCM. Table S4, Table S5, Table S6, and Table S7 depict the DEG between the leukocytes mentioned in the manuscript.
Supplementary Material
shows inclusion criteria for the adult COVID-19 cohort.
shows the FCM panel, immunophenotyping.
shows the FCM panel, T cell restimulation.
shows DEGs of TRBV11-2+ versus TRBV11-2− cells of MIS-C patients.
shows DEGs of TRBV11-2+ versus TRBV11-2− of cells that are activated and proliferative of MIS-C patients.
shows DEGs of classical monocytes of MIS-C patients versus age-matched HCs.
shows DEGs of patrolling monocytes of MIS-C patients versus age-matched HCs.
Acknowledgments
We thank the patients and their families for participating in this study. We thank the members of the laboratory for helpful discussions. We acknowledge the professional support and committed efforts from all the involved study nurses, caregivers, and clinicians. We thank Helena Flipts for administrative support. The clinical trial team of the Department of Respiratory Medicine at UZ Gent (Stefanie Vermeersch, Benedicte Demeyere, and Anja Delporte) was involved in the recruitment of adult patients with severe COVID-19. We further acknowledge all the clinicians involved in patient recruitment at the different participating sites.
This work was co-funded by the Vlaams Instituut voor Biotechnologie Grand Challenge UGent (S. Bekaert) and the Ghent University Bijzonder Onderzoeksfonds. L. Roels, L. Naesens, V. Bosteels, and K.F.A. Van Damme are beneficiaries of Fonds Wetenschappelijk Onderzoek PhD fellowships. L. Seys, S. Vanhee, and S.J. Tavernier are beneficiaries of a postdoctoral Fonds Wetenschappelijk Onderzoek grant.
Author contributions: L. Hoste, S.J. Tavernier, and F. Haerynck designed the experiments. L. Hoste, L. Naesens, L. Roels, L. Seys, H. Aegerter, U. Smole, and S.J. Tavernier collected and processed patient material. K.F.A. Van Damme., J. Declercq., E. De Leeuw, B. Maes, and C. Bosteels recruited severe COVID-19 adult patients. MIS-C Clinicians recruited MIS-C patients. P. Bastard, L. Hoste, L. Naesens, L. Roels, V. Bosteels, N. Vandamme, and S.J. Tavernier performed the experiments and interpreted immunological and molecular data. K. Verstaen, R. Browaeys, S. Dupont, S. Vanhee, and J. Roels interpreted single-cell sequencing data. K. Claes aided in patient-oriented care. V. Debacker, S. De Prijck, G. Van Isterdael, and M. Vanheerswynghels performed experiments. L. Backers and K.B.M. Claes performed genetic analyses. E. Jouanguy, S.Y. Zhang, J.L. Casanova, B.N. Lambrecht, P. Stordeur, J. Dehoorne, R. Beyaert, K. Vanderkerckhove, J. Willems, G. Mets, S. Janssens, Y. Saeys, P. Schelstraete, and F. Haerynck provided scientific guidance. L. Hoste, S.J. Tavernier, and F. Haerynck drafted the first manuscript. All authors revised the manuscript and gave their final approval of the version to be published.
Data availability
All scRNA-seq datasets described in this article can be accessed via our interactive webserver at http://www.single-cell.be/misc, where all gene–cell counts, cell annotation matrices, and code can also be downloaded. Through this tool, it is possible to evaluate the expression of genes in our datasets and to download DEG lists. Additionally, all scRNA-seq raw data and gene–cell count matrices have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (accession no. GSE184330).
References
- Abrams, J.Y., Godfred-Cato S.E., Oster M.E., Chow E.J., Koumans E.H., Bryant B., Leung J.W., and Belay E.D.. 2020. Multisystem Inflammatory Syndrome in Children (MIS-C) Associated with SARS-CoV-2: A Systematic Review. J. Pediatr. 10.1016/j.jpeds.2020.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrams, J.Y., Oster M.E., Godfred-Cato S.E., Bryant B., Datta S.D., Campbell A.P., Leung J.W., Tsang C.A., Pierce T.J., Kennedy J.L., et al. 2021. Factors linked to severe outcomes in multisystem inflammatory syndrome in children (MIS-C) in the USA: a retrospective surveillance study. Lancet Child Adolesc. Health. 5:323–331. 10.1016/S2352-4642(21)00050-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed, M., Advani S., Moreira A., Zoretic S., Martinez J., Chorath K., Acosta S., Naqvi R., Burmeister-Morton F., Burmeister F., et al. 2020. Multisystem inflammatory syndrome in children: A systematic review. EClinicalMedicine. 26:100527. 10.1016/j.eclinm.2020.100527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastard, P., Rosen L.B., Zhang Q., Michailidis E., Hoffmann H.H., Zhang Y., Dorgham K., Philippot Q., Rosain J., Béziat V., et al. COVID Human Genetic Effort . 2020. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 370:eabd4585. 10.1126/science.abd4585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann, N.D., Comella P.H., Cheng E., Lepow L., Beckmann A.G., Mouskas K., Simons N.W., Hoffman G.E., Francoeur N.J., Del Valle D.M., et al. 2020. Cytotoxic lymphocytes are dysregulated in multisystem inflammatory syndrome in children. medRxiv. 10.1101/2020.08.29.20182899 (Preprint posted September 2, 2020) [DOI]
- Bialek, S., Gierke R., Hughes M., McNamara L.A., Pilishvili T., and Skoff T.. CDC COVID-19 Response Team . 2020. Coronavirus Disease 2019 in Children - United States, February 12-April 2, 2020. MMWR Morb. Mortal. Wkly. Rep. 69:422–426. 10.15585/mmwr.mm6914e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco, N., and Noti M.. 2021. The aging gut microbiome and its impact on host immunity. Genes Immun. 22:289–303. 10.1038/s41435-021-00126-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracaglia, C., de Graaf K., Pires Marafon D., Guilhot F., Ferlin W., Prencipe G., Caiello I., Davì S., Schulert G., Ravelli A., et al. 2017. Elevated circulating levels of interferon-γ and interferon-γ-induced chemokines characterise patients with macrophage activation syndrome complicating systemic juvenile idiopathic arthritis. Ann. Rheum. Dis. 76:166–172. 10.1136/annrheumdis-2015-209020 [DOI] [PubMed] [Google Scholar]
- Brasseit, J., Kwong Chung C.K.C., Noti M., Zysset D., Hoheisel-Dickgreber N., Genitsch V., Corazza N., and Mueller C.. 2018. Divergent roles of interferon-γ and innate lymphoid cells in innate and adaptive immune cell-mediated intestinal inflammation. Front. Immunol. 9:23. 10.3389/fimmu.2018.00023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky, N.N., Ramaswamy A., and Lucas C.L.. 2020. The Mystery of MIS-C Post-SARS-CoV-2 Infection. Trends Microbiol. 28:956–958. 10.1016/j.tim.2020.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browaeys, R., Saelens W., and Saeys Y.. 2020. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat. Methods. 17:159–162. 10.1038/s41592-019-0667-5 [DOI] [PubMed] [Google Scholar]
- Caldarale, F., Giacomelli M., Garrafa E., Tamassia N., Morreale A., Poli P., Timpano S., Baresi G., Zunica F., Cattalini M., et al. 2021. Plasmacytoid Dendritic Cells Depletion and Elevation of IFN-γ Dependent Chemokines CXCL9 and CXCL10 in Children With Multisystem Inflammatory Syndrome. Front. Immunol. 12:654587. 10.3389/fimmu.2021.654587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter, M.J., Fish M., Jennings A., Doores K.J., Wellman P., Seow J., Acors S., Graham C., Timms E., Kenny J., et al. 2020. Peripheral immunophenotypes in children with multisystem inflammatory syndrome associated with SARS-CoV-2 infection. Nat. Med. 26:1701–1707. 10.1038/s41591-020-1054-6 [DOI] [PubMed] [Google Scholar]
- Castagnoli, R., Votto M., Licari A., Brambilla I., Bruno R., Perlini S., Rovida F., Baldanti F., and Marseglia G.L.. 2020. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Infection in Children and Adolescents: A Systematic Review. JAMA Pediatr. 174:882–889. 10.1001/jamapediatrics.2020.1467 [DOI] [PubMed] [Google Scholar]
- Cheng, M.H., Zhang S., Porritt R.A., Noval Rivas M., Paschold L., Willscher E., Binder M., Arditi M., and Bahar I.. 2020. Superantigenic character of an insert unique to SARS-CoV-2 spike supported by skewed TCR repertoire in patients with hyperinflammation. Proc. Natl. Acad. Sci. USA. 117:25254–25262. 10.1073/pnas.2010722117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consiglio, C.R., Cotugno N., Sardh F., Pou C., Amodio D., Rodriguez L., Tan Z., Zicari S., Ruggiero A., Pascucci G.R., et al. CACTUS Study Team . 2020. The Immunology of Multisystem Inflammatory Syndrome in Children with COVID-19. Cell. 183:968–981.e7. 10.1016/j.cell.2020.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinides, M.G., McDonald B.D., Verhoef P.A., and Bendelac A.. 2014. A committed precursor to innate lymphoid cells. Nature. 508:397–401. 10.1038/nature13047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell, H.L., Soneson C., Germain P.-L., Calini D., Collin L., Raposo C., Malhotra D., and Robinson M.D.. 2020. muscat detects subpopulation-specific state transitions from multi-sample multi-condition single-cell transcriptomics data. Nat. Commun. 11:6077. 10.1038/s41467-020-19894-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Antunes, R., Pallikkuth S., Williams E., Dawen Yu E., Mateus J., Quiambao L., Wang E., Rawlings S.A., Stadlbauer D., Jiang K., et al. 2021. Differential T cell reactivity to endemic coronaviruses and SARS-CoV-2 in community and health care workers. J. Infect. Dis. 224:70–80. 10.1093/infdis/jiab176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Cevins, C., Luka M., Smith N., Meynier S., Magérus A., Carbone F., García-Paredes V., Barnabei L., Batignes M., Boullé A., et al. 2021. A monocyte/dendritic cell molecular signature of SARS-CoV2-related multisystem inflammatory syndrome in children (MIS-C) with severe myocarditis. bioRxiv. 10.1101/2021.02.23.432486 (Preprint posted February 23, 2021) [DOI] [PMC free article] [PubMed]
- Derikx, J.P., Luyer M.D., Heineman E., and Buurman W.A.. 2010. Non-invasive markers of gut wall integrity in health and disease. World J. Gastroenterol. 16:5272–5279. 10.3748/wjg.v16.i42.5272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinges, M.M., and Schlievert P.M.. 2001a. Role of T cells and gamma interferon during induction of hypersensitivity to lipopolysaccharide by toxic shock syndrome toxin 1 in mice. Infect. Immun. 69:1256–1264. 10.1128/IAI.69.3.1256-1264.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinges, M.M., and Schlievert P.M.. 2001b. Comparative analysis of lipopolysaccharide-induced tumor necrosis factor alpha activity in serum and lethality in mice and rabbits pretreated with the staphylococcal superantigen toxic shock syndrome toxin 1. Infect. Immun. 69:7169–7172. 10.1128/IAI.69.11.7169-7172.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diorio, C., Henrickson S.E., Vella L.A., McNerney K.O., Chase J., Burudpakdee C., Lee J.H., Jasen C., Balamuth F., Barrett D.M., et al. 2020. Multisystem inflammatory syndrome in children and COVID-19 are distinct presentations of SARS-CoV-2. J. Clin. Invest. 130:5967–5975. 10.1172/JCI140970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte-Neto, A.N., Caldini E.G., Gomes-Gouvêa M.S., Kanamura C.T., de Almeida Monteiro R.A., Ferranti J.F., Ventura A.M.C., Regalio F.A., Fiorenzano D.M., Gibelli M.A.B.C., et al. 2021. An autopsy study of the spectrum of severe COVID-19 in children: From SARS to different phenotypes of MIS-C. EClinicalMedicine. 35:100850. 10.1016/j.eclinm.2021.100850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufort, E.M., Koumans E.H., Chow E.J., Rosenthal E.M., Muse A., Rowlands J., Barranco M.A., Maxted A.M., Rosenberg E.S., Easton D., et al. New York State and Centers for Disease Control and Prevention Multisystem Inflammatory Syndrome in Children Investigation Team . 2020. Multisystem inflammatory syndrome in children in New York state. N. Engl. J. Med. 383:347–358. 10.1056/NEJMoa2021756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eidson, M., Wahlstrom J., Beaulieu A.M., Zaidi B., Carsons S.E., Crow P.K., Yuan J., Wolchok J.D., Horsthemke B., Wieczorek D., and Sant’Angelo D.B.. 2011. Altered development of NKT cells, γδ T cells, CD8 T cells and NK cells in a PLZF deficient patient. PLoS One. 6:e24441. 10.1371/journal.pone.0024441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteve-Sole, A., Anton J., Pino-Ramirez R.M., Sanchez-Manubens J., Fumadó V., Fortuny C., Rios-Barnes M., Sanchez-de-Toledo J., Girona-Alarcón M., Mosquera J.M., et al. 2021. Similarities and differences between the immunopathogenesis of COVID-19-related pediatric multisystem inflammatory syndrome and Kawasaki disease. J. Clin. Invest. 131:e144554. 10.1172/JCI144554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajgenbaum, D.C., and June C.H.. 2020. Cytokine Storm. N. Engl. J. Med. 383:2255–2273. 10.1056/NEJMra2026131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fardet, L., Galicier L., Lambotte O., Marzac C., Aumont C., Chahwan D., Coppo P., and Hejblum G.. 2014. Development and validation of the HScore, a score for the diagnosis of reactive hemophagocytic syndrome. Arthritis Rheumatol. 66:2613–2620. 10.1002/art.38690 [DOI] [PubMed] [Google Scholar]
- Feldstein, L.R., Rose E.B., Horwitz S.M., Collins J.P., Newhams M.M., Son M.B.F., Newburger J.W., Kleinman L.C., Heidemann S.M., Martin A.A., et al. CDC COVID-19 Response Team . 2020. Multisystem Inflammatory Syndrome in U.S. Children and Adolescents. N. Engl. J. Med. 383:334–346. 10.1056/NEJMoa2021680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldstein, L.R., Tenforde M.W., Friedman K.G., Newhams M., Rose E.B., Dapul H., Soma V.L., Maddux A.B., Mourani P.M., Bowens C., et al. Overcoming COVID-19 Investigators . 2021. Characteristics and Outcomes of US Children and Adolescents With Multisystem Inflammatory Syndrome in Children (MIS-C) Compared With Severe Acute COVID-19. JAMA. 325:1074–1087. 10.1001/jama.2021.2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götzinger, F., Santiago-García B., Noguera-Julián A., Lanaspa M., Lancella L., Calò Carducci F.I., Gabrovska N., Velizarova S., Prunk P., Osterman V., et al. ptbnet COVID-19 Study Group . 2020. COVID-19 in children and adolescents in Europe: a multinational, multicentre cohort study. Lancet Child Adolesc. Health. 4:653–661. 10.1016/S2352-4642(20)30177-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber, C.N., Patel R.S., Trachtman R., Lepow L., Amanat F., Krammer F., Wilson K.M., Onel K., Geanon D., Tuballes K., et al. 2020. Mapping Systemic Inflammation and Antibody Responses in Multisystem Inflammatory Syndrome in Children (MIS-C). Cell. 183:982–995.e14. 10.1016/j.cell.2020.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao, Y., Hao S., Andersen-Nissen E., Mauck W.M., Zheng S., Butler A., Lee M.J., Wilk A.J., Darby C., Zagar M., et al. 2021. Integrated analysis of multimodal single-cell data. Cell. 184:3573–3587.e29. 10.1016/j.cell.2021.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoste, L., Van Paemel R., and Haerynck F.. 2021. Multisystem inflammatory syndrome in children related to COVID-19: a systematic review. Eur. J. Pediatr. 180:2019–2034. 10.1007/s00431-021-03993-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultström, M., Fromell K., Larsson A., Quaggin S.E., Betsholtz C., Frithiof R., Lipcsey M., and Jeansson M.. 2021. Elevated Angiopoietin-2 inhibits thrombomodulin-mediated anticoagulation in critically ill COVID-19 patients. medRxiv. 10.1101/2021.01.13.21249429 (Preprint posted May 27, 2021) [DOI] [PMC free article] [PubMed]
- Humblet-Baron, S., Franckaert D., Dooley J., Ailal F., Bousfiha A., Deswarte C., Oleaga-Quintas C., Casanova J.L., Bustamante J., and Liston A.. 2019. IFN-γ and CD25 drive distinct pathologic features during hemophagocytic lymphohistiocytosis. J. Allergy Clin. Immunol. 143:2215–2226.e7. 10.1016/j.jaci.2018.10.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki, A., and Yang Y.. 2020. The potential danger of suboptimal antibody responses in COVID-19. Nat. Rev. Immunol. 20:339–341. 10.1038/s41577-020-0321-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabaudon, M., Futier E., Roszyk L., Chalus E., Guerin R., Petit A., Mrozek S., Perbet S., Cayot-Constantin S., Chartier C., et al. 2011. Soluble form of the receptor for advanced glycation end products is a marker of acute lung injury but not of severe sepsis in critically ill patients. Crit. Care Med. 39:480–488. 10.1097/CCM.0b013e318206b3ca [DOI] [PubMed] [Google Scholar]
- Jabaudon, M., Berthelin P., Pranal T., Roszyk L., Godet T., Faure J.S., Chabanne R., Eisenmann N., Lautrette A., Belville C., et al. 2018. Receptor for advanced glycation end-products and ARDS prediction: a multicentre observational study. Sci. Rep. 8:2603. 10.1038/s41598-018-20994-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenks, S.A., Cashman K.S., Zumaquero E., Marigorta U.M., Patel A.V., Wang X., Tomar D., Woodruff M.C., Simon Z., Bugrovsky R., et al. 2018. Distinct Effector B Cells Induced by Unregulated Toll-like Receptor 7 Contribute to Pathogenic Responses in Systemic Lupus Erythematosus. Immunity. 49:725–739.e6. 10.1016/j.immuni.2018.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan, M.B., Hildeman D., Kappler J., and Marrack P.. 2004. An animal model of hemophagocytic lymphohistiocytosis (HLH): CD8+ T cells and interferon gamma are essential for the disorder. Blood. 104:735–743. 10.1182/blood-2003-10-3413 [DOI] [PubMed] [Google Scholar]
- Kale, S.D., Mehrkens B.N., Stegman M.M., Kastelberg B., Carnes H., McNeill R.J., Rizzo A., Karyala S.V., Coutermarsh-Ott S., Fretz J.A., et al. 2020. “Small” Intestinal Immunopathology Plays a “Big” Role in Lethal Cytokine Release Syndrome, and Its Modulation by Interferon-γ, IL-17A, and a Janus Kinase Inhibitor. Front. Immunol. 11:1311. 10.3389/fimmu.2020.01311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi, S., Higasa K., Shimizu M., Yamada R., and Matsuda F.. 2017. HLA-HD: An accurate HLA typing algorithm for next-generation sequencing data. Hum. Mutat. 38:788–797. 10.1002/humu.23230 [DOI] [PubMed] [Google Scholar]
- Langer, V., Vivi E., Regensburger D., Winkler T.H., Waldner M.J., Rath T., Schmid B., Skottke L., Lee S., Jeon N.L., et al. 2019. IFN-γ drives inflammatory bowel disease pathogenesis through VE-cadherin-directed vascular barrier disruption. J. Clin. Invest. 129:4691–4707. 10.1172/JCI124884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leisman, D.E., Ronner L., Pinotti R., Taylor M.D., Sinha P., Calfee C.S., Hirayama A.V., Mastroiani F., Turtle C.J., Harhay M.O., et al. 2020. Cytokine elevation in severe and critical COVID-19: a rapid systematic review, meta-analysis, and comparison with other inflammatory syndromes. Lancet Respir. Med. 8:1233–1244. 10.1016/S2213-2600(20)30404-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levick, S.P., and Goldspink P.H.. 2014. Could interferon-gamma be a therapeutic target for treating heart failure? Heart Fail. Rev. 19:227–236. 10.1007/s10741-013-9393-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q., Zhang Q., Wang M., Zhao S., Ma J., Luo N., Li N., Li Y., Xu G., and Li J.. 2008. Interferon-γ and tumor necrosis factor-α disrupt epithelial barrier function by altering lipid composition in membrane microdomains of tight junction. Clin. Immunol. 126:67–80. 10.1016/j.clim.2007.08.017 [DOI] [PubMed] [Google Scholar]
- Locatelli, F., Jordan M.B., Allen C., Cesaro S., Rizzari C., Rao A., Degar B., Garrington T.P., Sevilla J., Putti M.-C., et al. 2020. Emapalumab in Children with Primary Hemophagocytic Lymphohistiocytosis. N. Engl. J. Med. 382:1811–1822. 10.1056/NEJMoa1911326 [DOI] [PubMed] [Google Scholar]
- Ludvigsson, J.F. 2020. Systematic review of COVID-19 in children shows milder cases and a better prognosis than adults. Acta Paediatr. 109:1088–1095. 10.1111/apa.15270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L., Sahu S.K., Cano M., Kuppuswamy V., Bajwa J., McPhatter J., Pine A., Meizlish M.L., Goshua G., Chang C.-H., et al. 2021. Increased complement activation is a distinctive feature of severe SARS-CoV-2 infection. Sci. Immunol. 6:eabh2259. 10.1126/sciimmunol.abh2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madara, J.L., and Stafford J.. 1989. Interferon-γ directly affects barrier function of cultured intestinal epithelial monolayers. J. Clin. Invest. 83:724–727. 10.1172/JCI113938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes, B., Bosteels C., De Leeuw E., Declercq J., Van Damme K., Delporte A., Demeyere B., Vermeersch S., Vuylsteke M., Willaert J., et al. 2020. Treatment of severely ill COVID-19 patients with anti-interleukin drugs (COV-AID): A structured summary of a study protocol for a randomised controlled trial. Trials. 21:468. 10.1186/s13063-020-04453-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArdle, A.J., Vito O., Patel H., Seaby E.G., Shah P., Wilson C., Broderick C., Nijman R., Tremoulet A.H., Munblit D., et al. BATS Consortium . 2021. Treatment of Multisystem Inflammatory Syndrome in Children. N. Engl. J. Med. 385:11–22. 10.1056/NEJMoa2102968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, J., Cantor A., Zachariah P., Ahn D., Martinez M., and Margolis K.. 2020. Gastrointestinal symptoms as a major presentation component of a novel multisystem inflammatory syndrome in children (MIS-C) that is related to COVID-19: a single center experience of 44 cases. Gastroenterology. 159:1571–1574.e2. 10.1053/j.gastro.2020.05.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreews, M., Le Gouge K., Khaldi-Plassart S., Pescarmona R., Mathieu A.-L., Malcus C., Djebali S., Bellomo A., Dauwalder O., Perret M., et al. 2021. Polyclonal expansion of TCR Vbeta 21.3+ CD4+ and CD8+ T cells is a hallmark of Multisystem Inflammatory Syndrome in Children. Sci. Immunol. 6:eabh1516. 10.1126/sciimmunol.abh1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nava, P., Koch S., Laukoetter M.G., Lee W.Y., Kolegraff K., Capaldo C.T., Beeman N., Addis C., Gerner-Smidt K., Neumaier I., et al. 2010. Interferon-γ regulates intestinal epithelial homeostasis through converging β-catenin signaling pathways. Immunity. 32:392–402. 10.1016/j.immuni.2010.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, G.E., Pondo T., Toews K.-A., Farley M.M., Lindegren M.L., Lynfield R., Aragon D., Zansky S.M., Watt J.P., Cieslak P.R., et al. 2016. Epidemiology of Invasive Group A Streptococcal Infections in the United States, 2005-2012. Clin. Infect. Dis. 63:478–486. 10.1093/cid/ciw248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogbe, A., Kronsteiner B., Skelly D.T., Pace M., Brown A., Adland E., Adair K., Akhter H.D., Ali M., Ali S.-E., et al. Oxford Protective T Cell Immunology for COVID-19 (OPTIC) Clinical Team . 2021. T cell assays differentiate clinical and subclinical SARS-CoV-2 infections from cross-reactive antiviral responses. Nat. Commun. 12:2055. 10.1038/s41467-021-21856-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouldali, N., Toubiana J., Antona D., Javouhey E., Madhi F., Lorrot M., Léger P.L., Galeotti C., Claude C., Wiedemann A., et al. French Covid-19 Paediatric Inflammation Consortium . 2021. Association of Intravenous Immunoglobulins Plus Methylprednisolone vs Immunoglobulins Alone With Course of Fever in Multisystem Inflammatory Syndrome in Children. JAMA. 325:855–864. 10.1001/jama.2021.0694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil, V.S., Madrigal A., Schmiedel B.J., Clarke J., O’Rourke P., de Silva A.D., Harris E., Peters B., Seumois G., Weiskopf D., et al. 2018. Precursors of human CD4+ cytotoxic T lymphocytes identified by single-cell transcriptome analysis. Sci. Immunol. 3:eaan8664. 10.1126/sciimmunol.aan8664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelsers, M.M.A.L., Namiot Z., Kisielewski W., Namiot A., Januszkiewicz M., Hermens W.T., and Glatz J.F.C.. 2003. Intestinal-type and liver-type fatty acid-binding protein in the intestine. Tissue distribution and clinical utility. Clin. Biochem. 36:529–535. 10.1016/S0009-9120(03)00096-1 [DOI] [PubMed] [Google Scholar]
- Porritt, R.A., Paschold L., Rivas M.N., Cheng M.H., Yonker L.M., Chandnani H., Lopez M., Simnica D., Schultheiß C., Santiskulvong C., et al. 2021. HLA class I-associated expansion of TRBV11-2 T cells in multisystem inflammatory syndrome in children. J. Clin. Invest. 131:e146614. 10.1172/JCI146614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouletty, M., Borocco C., Ouldali N., Caseris M., Basmaci R., Lachaume N., Bensaid P., Pichard S., Kouider H., Morelle G., et al. 2020. Paediatric multisystem inflammatory syndrome temporally associated with SARS-CoV-2 mimicking Kawasaki disease (Kawa-COVID-19): a multicentre cohort. Ann. Rheum. Dis. 79:999–1006. 10.1136/annrheumdis-2020-217960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radia, T., Williams N., Agrawal P., Harman K., Weale J., Cook J., and Gupta A.. 2021. Multi-system inflammatory syndrome in children & adolescents (MIS-C): A systematic review of clinical features and presentation. Paediatr. Respir. Rev. 38:51–57. 10.1016/j.prrv.2020.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy, A., Brodsky N.N., Sumida T.S., Comi M., Asashima H., Hoehn K.B., Li N., Liu Y., Shah A., Ravindra N.G., et al. 2021. Immune dysregulation and autoreactivity correlate with disease severity in SARS-CoV-2-associated multisystem inflammatory syndrome in children. Immunity. 54:1083–1095.e7. 10.1016/j.immuni.2021.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reifenberg, K., Lehr H.A., Torzewski M., Steige G., Wiese E., Küpper I., Becker C., Ott S., Nusser P., Yamamura K., et al. 2007. Interferon-γ induces chronic active myocarditis and cardiomyopathy in transgenic mice. Am. J. Pathol. 171:463–472. 10.2353/ajpath.2007.060906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt, R.L., Liang H.-E., Bao K., Price A.E., Mohrs M., Kelly B.L., and Locksley R.M.. 2015. A novel model for IFN-γ-mediated autoinflammatory syndromes. J. Immunol. 194:2358–2368. 10.4049/jimmunol.1401992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rincon-Arevalo, H., Wiedemann A., Stefanski A.L., Lettau M., Szelinski F., Fuchs S., Frei A.P., Steinberg M., Kam-Thong T., Hatje K., et al. 2021. Deep Phenotyping of CD11c+ B Cells in Systemic Autoimmunity and Controls. Front. Immunol. 12:635615. 10.3389/fimmu.2021.635615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riphagen, S., Gomez X., Gonzalez-Martinez C., Wilkinson N., and Theocharis P.. 2020. Hyperinflammatory shock in children during COVID-19 pandemic. Lancet. 395:1607–1608. 10.1016/S0140-6736(20)31094-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho-Shimizu, V., Brodin P., Cobat A., Biggs C.M., Toubiana J., Lucas C.L., Henrickson S.E., Belot A., Tangye S.G., Milner J.D., et al. MIS-C@CHGE . 2021. SARS-CoV-2-related MIS-C: A key to the viral and genetic causes of Kawasaki disease? J. Exp. Med. 218:e20210446. 10.1084/jem.20210446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage, A.K., Constantinides M.G., Han J., Picard D., Martin E., Li B., Lantz O., and Bendelac A.. 2008. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 29:391–403. 10.1016/j.immuni.2008.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaer, D.J., Schleiffenbaum B., Kurrer M., Imhof A., Bächli E., Fehr J., Moller H.J., Moestrup S.K., and Schaffner A.. 2005. Soluble hemoglobin-haptoglobin scavenger receptor CD163 as a lineage-specific marker in the reactive hemophagocytic syndrome. Eur. J. Haematol. 74:6–10. 10.1111/j.1600-0609.2004.00318.x [DOI] [PubMed] [Google Scholar]
- Schlievert, P.M. 1983. Alteration of immune function by staphylococcal pyrogenic exotoxin type C: possible role in toxic-shock syndrome. J. Infect. Dis. 147:391–398. 10.1093/infdis/147.3.391 [DOI] [PubMed] [Google Scholar]
- Schwab, I., and Nimmerjahn F.. 2013. Intravenous immunoglobulin therapy: how does IgG modulate the immune system? Nat. Rev. Immunol. 13:176–189. 10.1038/nri3401 [DOI] [PubMed] [Google Scholar]
- Son, M.B.F., Murray N., Friedman K., Young C.C., Newhams M.M., Feldstein L.R., Loftis L.L., Tarquinio K.M., Singh A.R., Heidemann S.M., et al. Overcoming COVID-19 Investigators . 2021. Multisystem Inflammatory Syndrome in Children - Initial Therapy and Outcomes. N. Engl. J. Med. 385:23–34. 10.1056/NEJMoa2102605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syrimi, E., Fennell E., Richter A., Vrljicak P., Stark R., Ott S., Murray P.G., Al-Abadi E., Chikermane A., Dawson P., et al. 2021. The immune landscape of SARS-CoV-2-associated Multisystem Inflammatory Syndrome in Children (MIS-C) from acute disease to recovery. iScience. 24:103215. 10.1016/j.isci.2021.103215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavernier, S.J., Athanasopoulos V., Verloo P., Behrens G., Staal J., Bogaert D.J., Naesens L., De Bruyne M., Van Gassen S., Parthoens E., et al. 2019. A human immune dysregulation syndrome characterized by severe hyperinflammation with a homozygous nonsense Roquin-1 mutation. Nat. Commun. 10:4779. 10.1038/s41467-019-12704-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilahun, A.Y., Holz M., Wu T.T., David C.S., and Rajagopalan G.. 2011. Interferon gamma-dependent intestinal pathology contributes to the lethality in bacterial superantigen-induced toxic shock syndrome. PLoS One. 6:e16764. 10.1371/journal.pone.0016764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida, T., Shirasawa M., Ware L.B., Kojima K., Hata Y., Makita K., Mednick G., Matthay Z.A., and Matthay M.A.. 2006. Receptor for advanced glycation end-products is a marker of type I cell injury in acute lung injury. Am. J. Respir. Crit. Care Med. 173:1008–1015. 10.1164/rccm.200509-1477OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vella, L.A., Giles J.R., Baxter A.E., Oldridge D.A., Diorio C., Kuri-Cervantes L., Alanio C., Pampena M.B., Wu J.E., Chen Z., et al. UPenn COVID Processing Unit . 2021. Deep immune profiling of MIS-C demonstrates marked but transient immune activation compared to adult and pediatric COVID-19. Sci. Immunol. 6:eabf7570. 10.1126/sciimmunol.abf7570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdoni, L., Mazza A., Gervasoni A., Martelli L., Ruggeri M., Ciuffreda M., Bonanomi E., and D’Antiga L.. 2020. An outbreak of severe Kawasaki-like disease at the Italian epicentre of the SARS-CoV-2 epidemic: an observational cohort study. Lancet. 395:1771–1778. 10.1016/S0140-6736(20)31103-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisberg, S.P., Connors T.J., Zhu Y., Baldwin M.R., Lin W.-H., Wontakal S., Szabo P.A., Wells S.B., Dogra P., Gray J., et al. 2021. Distinct antibody responses to SARS-CoV-2 in children and adults across the COVID-19 clinical spectrum. Nat. Immunol. 22:25–31. 10.1038/s41590-020-00826-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf, D., Schmitz K.S., Raadsen M.P., Grifoni A., Okba N.M.A., Endeman H., van den Akker J.P.C., Molenkamp R., Koopmans M.P.G., van Gorp E.C.M., et al. 2020. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 5:eabd2071. 10.1126/sciimmunol.abd2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker, E., Bamford A., Kenny J., Kaforou M., Jones C.E., Shah P., Ramnarayan P., Fraisse A., Miller O., Davies P., et al. PIMS-TS Study Group and EUCLIDS and PERFORM Consortia . 2020. Clinical Characteristics of 58 Children With a Pediatric Inflammatory Multisystem Syndrome Temporally Associated With SARS-CoV-2. JAMA. 324:259–269. 10.1001/jama.2020.10369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf, Y., Anderson A.C., and Kuchroo V.K.. 2020. TIM3 comes of age as an inhibitory receptor. Nat. Rev. Immunol. 20:173–185. 10.1038/s41577-019-0224-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . 2020. Multisystem inflammatory syndrome in children and adolescents with COVID-19. https://www.who.int/news-room/commentaries/detail/multisystem-inflammatory-syndrome-in-children-and-adolescents-with-covid-19 (accessed November 19, 2021)
- Wu, Y., Guo C., Tang L., Hong Z., Zhou J., Dong X., Yin H., Xiao Q., Tang Y., Qu X., et al. 2020. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 5:434–435. 10.1016/S2468-1253(20)30083-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X.J., Tang Y.M., Song H., Yang S.L., Xu W.Q., Zhao N., Shi S.W., Shen H.P., Mao J.Q., Zhang L.Y., and Pan B.H.. 2012. Diagnostic accuracy of a specific cytokine pattern in hemophagocytic lymphohistiocytosis in children. J. Pediatr. 160:984–90.e1. 10.1016/j.jpeds.2011.11.046 [DOI] [PubMed] [Google Scholar]
- Xu, Y., Li X., Zhu B., Liang H., Fang C., Gong Y., Guo Q., Sun X., Zhao D., Shen J., et al. 2020. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat. Med. 26:502–505. 10.1038/s41591-020-0817-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y., Shen C., Li J., Yuan J., Wei J., Huang F., Wang F., Li G., Li Y., Xing L., et al. 2020. Plasma IP-10 and MCP-3 levels are highly associated with disease severity and predict the progression of COVID-19. J. Allergy Clin. Immunol. 146:119–127.e4. 10.1016/j.jaci.2020.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuhara, J., Watanabe K., Takagi H., Sumitomo N., and Kuno T.. 2021. COVID-19 and multisystem inflammatory syndrome in children: A systematic review and meta-analysis. Pediatr. Pulmonol. 56:837–848. 10.1002/ppul.25245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonker, L.M., Gilboa T., Ogata A.F., Senussi Y., Lazarovits R., Boribong B.P., Bartsch Y.C., Loiselle M., Rivas M.N., Porritt R.A., et al. 2021. Multisystem inflammatory syndrome in children is driven by zonulin-dependent loss of gut mucosal barrier. J. Clin. Invest. 131:e149633. 10.1172/JCI149633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J.Y., Wang X.M., Xing X., Xu Z., Zhang C., Song J.W., Fan X., Xia P., Fu J.L., Wang S.Y., et al. 2020a. Single-cell landscape of immunological responses in patients with COVID-19. Nat. Immunol. 21:1107–1118. 10.1038/s41590-020-0762-x [DOI] [PubMed] [Google Scholar]
- Zhang, Q., Bastard P., Bolze A., Jouanguy E., Zhang S.-Y., Cobat A., Notarangelo L.D., Su H.C., Abel L., and Casanova J.-L.. COVID Human Genetic Effort . 2020b. Life-Threatening COVID-19: Defective Interferons Unleash Excessive Inflammation. Med (N Y). 1:14–20. 10.1016/j.medj.2020.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q., Bastard P., Liu Z., Le Pen J., Moncada-Velez M., Chen J., Ogishi M., Sabli I.K.D., Hodeib S., Korol C., et al. NIAID-USUHS/TAGC COVID Immunity Group . 2020c. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 370:eabd4570. 10.1126/science.abd4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows inclusion criteria for the adult COVID-19 cohort.
shows the FCM panel, immunophenotyping.
shows the FCM panel, T cell restimulation.
shows DEGs of TRBV11-2+ versus TRBV11-2− cells of MIS-C patients.
shows DEGs of TRBV11-2+ versus TRBV11-2− of cells that are activated and proliferative of MIS-C patients.
shows DEGs of classical monocytes of MIS-C patients versus age-matched HCs.
shows DEGs of patrolling monocytes of MIS-C patients versus age-matched HCs.
Data Availability Statement
All scRNA-seq datasets described in this article can be accessed via our interactive webserver at http://www.single-cell.be/misc, where all gene–cell counts, cell annotation matrices, and code can also be downloaded. Through this tool, it is possible to evaluate the expression of genes in our datasets and to download DEG lists. Additionally, all scRNA-seq raw data and gene–cell count matrices have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (accession no. GSE184330).