Abstract
CTCF is a widely expressed and highly conserved multi-Zn-finger (ZF) nuclear factor. Binding to various CTCF target sites (CTSs) is mediated by combinatorial contributions of different ZFs. Different CTSs mediate distinct CTCF functions in transcriptional regulation, including promoter repression or activation and hormone-responsive gene silencing. In addition, the necessary and sufficient core sequences of diverse enhancer-blocking (insulator) elements, including CpG methylation-sensitive ones, have recently been pinpointed to CTSs. To determine whether a posttranslational modification may modulate CTCF functions, we studied CTCF phosphorylation. We demonstrated that most of the modifications that occur at the carboxy terminus in vivo can be reproduced in vitro with casein kinase II (CKII). Major modification sites map to four serines within the S604KKEDS609S610DS612E motif that is highly conserved in vertebrates. Specific mutations of these serines abrogate phosphorylation of CTCF in vivo and CKII-induced phosphorylation in vitro. In addition, we showed that completely preventing phosphorylation by substituting all serines within this site resulted in markedly enhanced repression of the CTS-bearing vertebrate c-myc promoters, but did not alter CTCF nuclear localization or in vitro DNA-binding characteristics assayed with c-myc CTSs. Moreover, these substitutions manifested a profound effect on negative cell growth regulation by wild-type CTCF. CKII may thus be responsible for attenuation of CTCF activity, either acting on its own or by providing the signal for phosphorylation by other kinases and for CTCF-interacting protein partners.
A search for conserved transcription factors that could similarly regulate highly divergent promoters of avian and mammalian c-myc oncogenes resulted in the identification and cloning of a single-copy nonredundant gene encoding CTCF, a protein with a DNA-binding domain containing 11 Zn fingers (ZFs) (20, 26, 30–33). The protein binds to a number of remarkably different, ∼50- to ∼60-bp-long CTCF target sites (CTSs) in the promoters of chicken and human c-myc genes (20, 26).
The CTCF protein displays 100% identity of all 11 ZFs between avian and human amino acid sequences (20). Among numerous ZF factors, CTCF stands out as a unique protein with no known family members. An extensive search for sequence homologies outside of the ZF domain, in the N- and C-terminal regions, which together account for approximately two-thirds of the entire protein, showed no significant similarities to any previously described protein modules, including those often found associated with a variety of ZF factors.
CTCF appears to act differently upon binding to functionally distinct regulatory elements, which include (i) enhancer-blocking (insulator) elements in the two boundaries flanking the β-globin locus, in the T-cell receptor α/γ locus site, and in a matrix-attachment 5′ boundary of the chicken lysozyme gene (7, 8, 49); (ii) promoter-proximal sites in chicken, mouse, and human c-myc proto-oncogenes (20, 26); (iii) negative thyroid response elements which require CTCF for efficient regulation, as exemplified by the S-2.4 lysozyme gene transcriptional silencer (12), the 144 genomic element (5), and human c-myc (42); and (iv) the crucial regulatory site of the amyloid protein precursor (APP) gene promoter (45, 62, 65).
Moreover, CTCF was recently found to be a parent of an origin-specific and methylation-sensitive structural and functional component of the chromatin insulator upstream of the H19 gene (6, 22, 25, 57), thus suggesting a new important role for a CpG-containing subset of CTSs in control of genomic imprinting (25; reviewed in references 2 and 64).
Neither CpG methylation-dependent nor methylation-independent CTS-containing insulators act as general transcriptional repressors or activators, but rather they block functional interactions between an enhancer and a promoter. However, binding of CTCF to the CTS in the APP promoter activates transcription (62, 65), while binding to chicken or human c-myc major promoters represses it (20, 32). The diverse variety of functions that CTCF can perform suggests a very complex mode of multilevel regulation of CTCF itself.
Posttranslational modification by phosphorylation is commonly found to attenuate transcription factor function (reviewed in reference 23). Here we show that most, if not all, strong phosphorylation sites in CTCF map to the C-terminal region (CTCF-C). This region, fused in frame to the GAL4 DNA-binding domain, behaved as a trans repressor when targeted to a model promoter in cotransfection experiments (20).
We demonstrate here that CTCF-C is serine phosphorylated in vitro by casein kinase II (CKII) within the acidic amino acid motif that is strictly conserved in frog, chicken, mouse, rat, and human proteins. The same motif is found to be similarly phosphorylated in vivo. Selectively replacing serines in this motif by nonphosphorylatable amino acids completely eliminates phosphorylation of CTCF in vivo without affecting cellular localization, DNA-binding activity, or sequence specificity of CTCF. However, in a transcriptional activity reporter system utilizing the very first CTS-containing promoter identified by us in vertebrate c-myc oncogene 5′ noncoding regions (20, 26, 30–33), the same Ser mutations result in stronger inhibition of the chicken and human c-myc promoter reporters than that seen with the wild-type (wt) CTCF. Phosphorylation at the CKII sites appears to be of great physiological significance, because the inability to phosphorylate the mutated C terminus noticeably abrogates profound inhibition of cell proliferation by wt CTCF in clonogenicity assays.
Thus, in signal transduction pathways that may require a change in the trans-repressing activity of CTCF binding to promoters like c-myc, reversible phosphorylation of the region downstream of the 11-ZF DNA-binding domain appears to play an essential regulatory role(s). In this region, CKII seems to serve a priming role in vivo. We suggest that regulated dephosphorylation at CKII sites and/or CKII-induced activity of another kinase, leading to regulated phosphorylation-dependent interaction with CTCF protein partners, may occur to selectively change the activity of a subset of CTS-driven genes, some of which are involved in cell growth control.
MATERIALS AND METHODS
Expression vectors.
To obtain glutathione S-transferase (GST) fusion bacterial expression constructs pGST-N, pGST-Zns and pGST-C (Fig. 1B) producing, respectively, the amino-terminal region (CTCF-N), the 11-ZF domain (CTCF-ZF), and the carboxy-terminal domain (CTCF-C) of the chicken CTCF protein individually fused in frame with the GST open reading frame, three DNA fragments from the chicken CTCF cDNA clone p900 (26), the SmaI-HindIII fragment (positions 117 to 830), the PvuII-EcoRI fragment (positions 541 to 2037) and the MscI-EcoRI fragment (positions 1908 to 3786), were ligated in frame into the pGEX-2TK or pGEX-3X vectors provided with the GST purification kit (Pharmacia), and GST-fused CTCF proteins were expressed and purified as described in the manufacturer's manual.
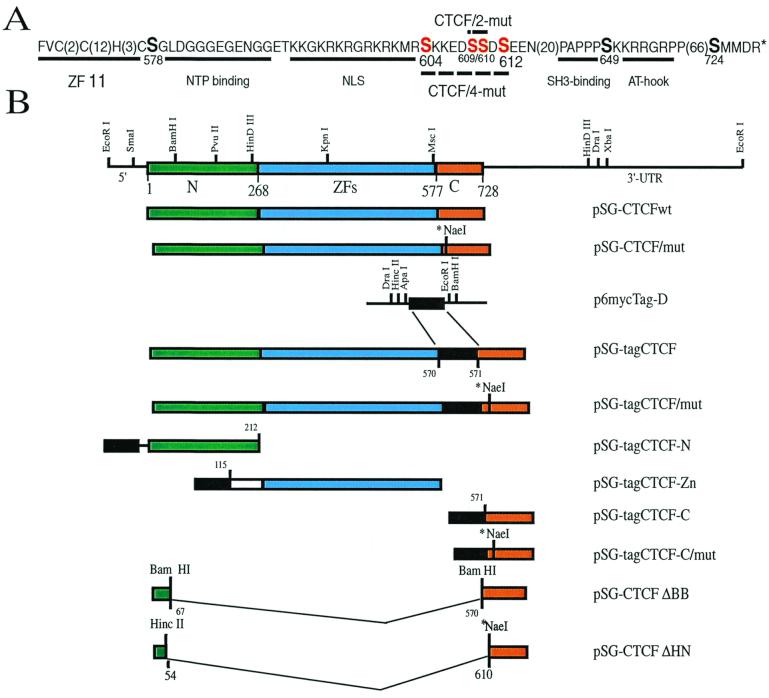
FIG. 1.
(A) Structural features of the C-terminal CTCF-C amino acid sequence with several sites matching the CKII target consensus motif (S/T)XX(E/D) and other serines shown in a bigger font size. Amino acid numbering is according to Klenova et al. (26). The numbers of residues not shown are indicated in brackets. The underlined 11th ZF shows the C-terminal CTCF ZF of the C2HC type. Also underlined are putative motifs for (i) the β-sheet–(GXGXXG)–α-helix pocket matching the Rossman fold associated with many nucleotide triphosphate-binding sites (58), (ii) the NLS (18), (iii) the PXXP type of SH3-binding motif (55), and (iv) the AT-hook consensus likely involved in AT-rich DNA binding (3) and/or protein-protein interactions in chromatin (13, 16). The serines mutated as described in Materials and Methods are highlighted in red. (B) Schematic representation of the full-length chicken CTCF cDNA, of the three structurally distinct domains (CTCF-N, CTCF-ZFs, and CTCF-C), and of a number of different forms of CTCF expression vectors constructed for the study. Restriction sites used to engineer a variety of the myc-tagged and other expression constructs are also shown. The asterisk-marked NaeI site indicates the presence of the double mutation replacing Ser-609 and -610, while the presence of “mut” in the construct names indicates the presence of either 2 or 4 Ser mutations as depicted in panel A.
Vectors for eukaryotic cell expression, based on the simian virus 40 promoter-enhancer containing plasmid pSG5 (Stratagene), are schematically shown in Fig. 1B. To obtain vectors pSG-tagCTCF-N, pSG-tagCTCF-ZFs, and pSG-tagCTCF-C producing three CTCF domains tagged with six myc-epitope tags for the 9E10 monoclonal antibody (19), each of the three DNA fragments described above was initially fused in frame at the N-terminal open reading frame ends, with the DNA fragment encoding six repeats of the myc-tag epitope (see Fig. 1B for details) isolated from the plasmid p6MYC-Tags-D, which was kindly provided by M. Roth (Fred Hutchinson Cancer Research Center, Seattle, Wash.).
The CTCF cDNA expression vector p-HHc, referred to here as pCTCFwt, contains the chicken CTCF cDNA clone described previously (27). To obtain the pCTCF/2-mut vector, a two-step PCR-mediated procedure was used to specifically mutate two codons, for Ser-609 and Ser-610, within the CTCF-C region (Fig. 1A). We employed the external rightward primer 1, 5′-TATGCATGCAGACAGGAGCG-3′, starting at nucleotide position 1703 of the CTCF cDNA sequence (numbering is according to reference 26), and the external leftward primer 4, 5′-GCTGTACAGCAGTATATTCT-3′, starting at nucleotide position 3753. Two internal self-complementary primers containing Ala and Gly codons within a new NaeI site (GCC-GGC in place of the TCC-TCC of the Ser-609 and Ser-610 codons) (the leftward primer from position 2036, 5′-CTCACTATCGCCGGCATCTTC-3′, with the mutated nucleotides underlined and the new codons indicated in italics, together with the complementary rightward primer, starting at cDNA position 2018) were also utilized for the site-directed mutagenesis, as described previously (39).
The same method was also employed to produce vectors to express the CTCF/4-mut protein in which Ser-604, -609, -610, and -612 are replaced by nonphosphorylatable amino acids (Fig. 1A). To generate pCTCF/4-mut (Fig. 1A), two internal self-complementary primers carrying Ala codons to replace the Ser codons 604, 609, and 612 and a Gly codon to replace Ser 610 were prepared. The leftward primer starting at position 2039 had the sequence 5′-TTCCTCAGCA TCGCCGGCATCTTCTTTCTTAGCGCGCAT-3′, and the rightward primer, starting at cDNA position 2003, and the corresponding complementary primers were used. Fragment swapping using unique restriction sites generated the myc-tagged versions of the mutated expression vectors (Fig. 1B). Two CTCF mutants with large internal deletions were also prepared (Fig. 1B). In the first, the BamHI-BamHI DNA fragment encoding amino acids 67 to 570 was removed in frame from the full-length expression vector, resulting in the construct pSG-CTCF-ΔBB, leaving in place the C terminus with the potential CKII sites as indicated in Fig. 1. In the pSG-tagCTCF-ΔHN construct, amino acids 54 to 610, inclusive, were removed (Fig. 1B) by cutting and religating the plasmid in frame at HincII-NaeI sites.
DNA-protein interaction assays.
Different forms of CTCF proteins were synthesized with the TnT reticulocyte lysate-coupled in vitro transcription-translation system from Promega (5, 20). In this system, CKII is known to be fully active. Because CTCF binding to DNA requires flanking segments of any sequence in addition to ∼50 bp of a given CTS (32), 120- to 200-bp-long DNA fragments bearing the c-myc CTSs V and A (characterized previously by DNase I footprinting and by methylation interference techniques) (20, 26) were 32P labeled, gel purified, and employed as DNA probes for electrophoretic mobility shift assays (EMSAs) with equal amounts of the in vitro-translated CTCF proteins as previously described (5, 20). Affinity and kinetic measurements by surface plasmon resonance were carried out essentially as described earlier (25) using the BIAcore CM-5 chip (BIAcore, Uppsala, Sweden). Binding parameters for the interaction of each DNA probe with immobilized wt, CTCF/2-mut, and CTCF/4-mut proteins were analyzed using the BIAcore software supplied by the manufacturer.
Methylation interference assays with 32P-labeled gel-purified DNA probes were performed as described in detail earlier (20, 32). For DNase I footprinting, 1 μl (1 Kunitz unit) of ultrapure DNase I (New England Biolabs) dissolved in phosphate-buffered saline supplemented with 1 mM CaCl2 and 1 mM MgCl2 was added to each of 10 32P-DNA-CTCF binding mixtures, with 20-μl final volumes prepared as described for EMSA reactions, mixed at room temperature for 30 s, and immediately loaded on EMSA gels already running at 300 cV. Gel-separated nuclease-treated CTCF-bound and free DNA probes from 10 reactions were isolated and analyzed on a sequencing gel as described earlier (33, 44).
Reporter constructs.
pPst2myc-CAT, the reporter construct containing the wt 3.3-kb PstI-PstI fragment of the chicken c-myc 5′ noncoding region, including the first exon and intron sequences from the pCc-mycPst2 plasmid (33), and pP2myc-CAT, containing the ApaI-PvuII fragment (from −121 to +352 relative to the major P2 promoter) of the human c-myc 5′ noncoding region, were previously described (20, 26).
Cells and transfections.
The HD3 cell line derived from chicken erythroblasts was maintained as described earlier (44) in Dulbecco's minimal essential medium (DMEM) supplemented with 8% fetal bovine serum and 2% chicken serum at a density of ∼106 cells/ml. COS-6, Swiss 3T3, and PC3 cells were grown in DMEM supplemented with 10% fetal calf serum and replated at low (40 to 50% confluence) density.
For chloramphenicol acetyltransferase (CAT) assays, cell lines were transfected with 1 μg of the appropriate plasmid by the calcium phosphate precipitation method with 2× DNA precipitation buffer (5′-3′, Inc., Boulder, Colo.) according to the method in the manufacturer's manual. Cell extracts were prepared at 48 h posttransfection, and CAT activity was assayed as previously described (51). HD3 cells were transfected with Lipofectamine (Gibco-BRL) according to the manufacturer's instructions.
Western and Northern blotting and indirect immunofluorescence microscopy.
Total cell lysates, prepared for loading in sodium dodecyl sulfate (SDS)-containing buffer with brief treatment with DNase I to reduce viscosity as described previously (26), were run through SDS–10% polyacrylamide gel electrophoresis (PAGE) gels and transferred onto Immobilon P membranes (Millipore, Bedford, Mass.) by semidry blotting. Membranes were probed with either the monoclonal 9E10 anti-myc-tag antibody (19) or the polyclonal affinity-purified anti-CTCF antibodies Ab1 and Ab2 (26). The primary antibody-bound proteins were visualized by the enhanced chemiluminescence (ECL) procedure using the detection system and protocol from Amersham Life Science, Inc. For indirect immunofluorescence analyses with monoclonal 9E10 anti-myc-tag antibody, the protocol described elsewhere (56) was adapted.
For Northern blot analysis, 10 μg of total cellular RNA isolated by the guanidinium thiocyanate extraction method was separated on a 1% agarose gel containing 2.2 M formaldehyde and transferred to a nylon membrane. RNA blots were hybridized with chicken CTCF cDNA probes that were 32P labeled as described by the supplier of a random priming kit (Stratagene). After overnight hybridization, blots were washed in 0.1× SSC–1% SDS (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at 65°C and autoradiographed.
Cell labeling and immunoprecipitation.
COS-6 cells transfected with the vectors expressing different CTCF forms shown in Fig. 1 were incubated for 6 h in phosphate-free growth medium supplemented with dialyzed fetal calf serum and then metabolically labeled with [32P]orthophosphate (Amersham International, PLC), added to achieve a final concentration of 1 mCi/ml. Cells were incubated for an additional 12 h. For immunoprecipitation, the radiolabeled cells were washed twice with ice-cold phosphate-buffered saline and lysed on dishes in 500 μl of high-salt radioimmunoprecipitation assay (RIPA) buffer (0.4 M NaCl, 50 mM Tris-HCl [pH 7.5], 1% NP-40, 0.1% SDS, 0.1% sodium deoxycholate, 1 mM EDTA) supplemented with protease inhibitors (leupeptin, 20 μg/ml; aprotinin, 25 μg/ml; phenylmethylsulfonyl fluoride, 1 mM; and benzamidine, 10 μg/ml) and phosphatase inhibitors (NaF, 50 mM; sodium vanadate, 100 μM; and sodium pyrophosphate, 400 mM), as described by Towatari et. al. (59). After incubation for 1 h at 4°C, 500 μl of the RIPA buffer without NaCl was added to cell lysates, which were then clarified by centrifugation at 17,000 × g for 20 min. CTCF and the myc-epitope-tagged proteins were immunoprecipitated either by the anti-CTCF polyclonal affinity-purified antibody Ab1 or Ab2 or by the 9E10 monoclonal antibody, respectively, and incubated with protein G-Sepharose-CL4B or protein A-Sepharose-4B-Fast (Sigma). The Sepharose beads were washed five times with 1 ml of the RIPA buffer containing 0.2 M NaCl, immobilized proteins were resolved by SDS–10% PAGE and transferred to an Immobilon P membrane, and 32P-labeled proteins were visualized by autoradiography. In some experiments, these membranes were also analyzed with specific antibodies by the ECL-mediated Western blot procedure as described above.
Kinase assays.
To analyze in vitro protein phosphorylation by CKII, a modified procedure described by Marais et al. (35) was employed. Briefly, purified GST-CTCF-N, GST-CTCF-Zn and GST-CTCF-C proteins were incubated with CKII (0.5 μl of the 117-U/ml stock of the human CKII α- and β-subunits coexpressed in insect cells [D. Neri, H. Petrul, G. Winter, Y. Light, R. Marais, K. E. Brotton, and A. M. Greighton, submitted for publication]) in 20 μl of kinase buffer (20 mM HEPES [pH 7.5], 20 mM Tris-HCl [pH 7.5], 10 mM MgCl2, 2 mM EDTA, 100 mM NaCl, 0.1 mM ATP, 0.1% NP-40, 1 mM dithiothreitol, 5 μg of leupeptin per ml, 5 μg of aprotinin per ml, 5 μg of pepstatin per ml, 1 mM benzamidine, 50 μg of phenylmethylsulfonyl fluoride per ml) in the presence of 0.5 μCi of [γ-32P]ATP (Amersham) for 60 min at 30°C.
Specific protein immunocomplexes attached to the Sepharose beads were initially washed twice in buffer containing 40 mM Tris-HCl (pH 7.5), 100 mM NaCl, 10 mM MgCl2, and 2 mM EDTA and then once in RIPA buffer. After adding 50 μl of the CKII buffer supplemented with 1 μl of CKII enzyme stock solution and 1 μCi of [γ-32P]ATP, the bead suspension was incubated for 60 min at 30°C. Phosphorylation reactions were stopped by mixing with an equal volume of the SDS-containing gel-loading buffer. The samples were boiled for 2 min and separated by SDS–10% PAGE. Finally, gels (stained, when needed, with Coomassie blue) were dried and subjected to autoradiography.
Two-dimensional tryptic phosphopeptide and phosphoamino acid analyses.
These experiments were carried out as described in detail by Boyle et al. (11). In brief, 32P-labeled protein bands cut from SDS-PAGE gels were ground in 1 ml of 50 mM ammonium bicarbonate (pH 7.3) solution, 50 μl of 2-mercaptoethanol and 10 μl of 10% SDS was added, and the samples were boiled for 5 min and then incubated overnight. The suspension was intensively stirred and clarified in a Spin-X centrifuge filter unit (Costar), and then 20 μg of a carrier protein (RNase A) was added to the chilled eluate and peptides were precipitated with 250 μl of 100% ice-cold trichloroacetic acid. The precipitate was collected by centrifugation, oxidized in 100 μl of ice-cold performic acid, and freeze-dried. The oxidized protein pellet was resuspended in 50 μl of ammonium bicarbonate (pH 8.0) and digested overnight with 10 μg of tosylsulfonyl phenylalanyl chloromethyl ketone-treated trypsin (Sigma). Peptide products were lyophilized twice from water and dissolved in buffer (pH 1.9) containing formic acid, glacial acetic acid, and deionized water at 50:156:1,794 (vol/vol). First-dimension resolution by electrophoresis was performed on cellulose thin-layer plates (Kodak) at pH 1.9 for 30 min at 1.0 kV. For second-dimension fractionation, the plates were subjected to chromatography in the buffer containing isobutyric acid, n-butanol, pyridine, glacial acetic acid, and deionized water (1,250:38:96:58:558 [vol/vol]). For phosphoamino acid analysis, labeled protein was blotted onto polyvinylidene difluoride (Immobilon) membranes. The radioactive band was excised; the protein was hydrolyzed in 5.7 M HCl for 1 h at 110°C and then electrophoresed on a cellulose thin-layer plate at pH 1.9 with phosphoserine and phosphothreonine standards. The plate was dried at 65°C for 20 min, the phosphoamino acids were detected by the ninhydrin spray reagent (Sigma), and then the plate was autoradiographed.
Cell clonogenicity assay.
Chicken HD3 cells were cotransfected with Lipofectamine (Gibco BRL) according to the manufacturer's instructions with 1 mg of the pSV40neo plasmid and 10 mg of either pSG5 (the empty vector control), pCTCFwt, or pCTCF/4-mut. Approximately 107 cells from each transfection were plated into methyl cellulose-containing medium in the presence of 3.2 mg of G418 sulfate (Gibco BRL) per ml under established selection conditions (53). In 20 days, the resistant colonies were counted, and several colonies from each series were randomly picked and expanded in medium containing 1 mg of G418 per ml. Expression of exogenous CTCF was analyzed by Northern hybridization and Western blotting.
The adherent prostate cancer cell line, PC3, and the no-myc TGR15 cell line, expressing no MYC family genes (36), kindly provided by John Sedivy, were transfected by standard calcium phosphate-mediated procedures and selected for G418-resistant colonies on 9-cm-diameter tissue culture plates.
RESULTS
CTCF is phosphorylated in vivo.
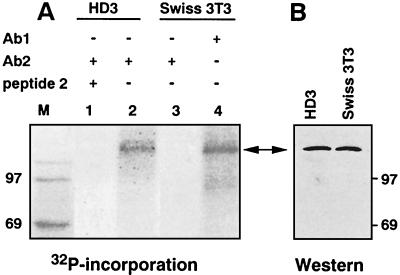
Employing affinity-purified species-specific anti-CTCF polyclonal antibodies (26) for immunoprecipitation from chicken and mouse cell extracts prepared from 32P-labeled exponentially growing cell lines, we found that CTCF is phosphorylated in vivo (Fig. 2). The 32P-CTCF band comigrates with in vitro-translated CTCF (reference 20 and data not shown), and a band of the same size is detected by immunoblotting (Fig. 2B; also see Fig. 6E and F). No protein was precipitated from the mouse fibroblast extract by the chicken-specific antibody Ab2 (Fig. 2, lane 3) or from the chicken HD3 cell extracts blocked with an excess of the chicken CTCF peptide 2 (26) (Fig. 2, lane 1).
FIG. 2.
CTCF is phosphorylated in vivo. (A) Exponentially growing chicken HD3 and mouse Swiss 3T3 cells were metabolically labeled with [32P]orthophosphate, and CTCF-containing material was immunoprecipitated from cell extracts with 20 μl of protein A-Sepharose 4B-Fast and affinity-purified rabbit polyclonal antibodies (Ab2) specific for the chicken CTCF peptide 2 (lanes 1 to 3) or with pan-specific Ab1 antibodies that recognize CTCF in all vertebrates (26) (lane 4). The Ab2 antibody-blocking peptide 2 (26) was included as the specificity control with chicken cell extracts (lane 1). Immobilized proteins resolved by SDS-PAGE were transferred to membranes and exposed to an X-ray film. (B) The same membranes as in panel A were probed with Ab1 antibodies by ECL immunoblotting assays. The two-headed arrow indicates the position of CTCF. It migrates aberrantly in SDS gels as the 130- to 160-kDa CTCF band (see reference 27 and Results for more details). Lane M, 14C-labeled molecular mass markers.
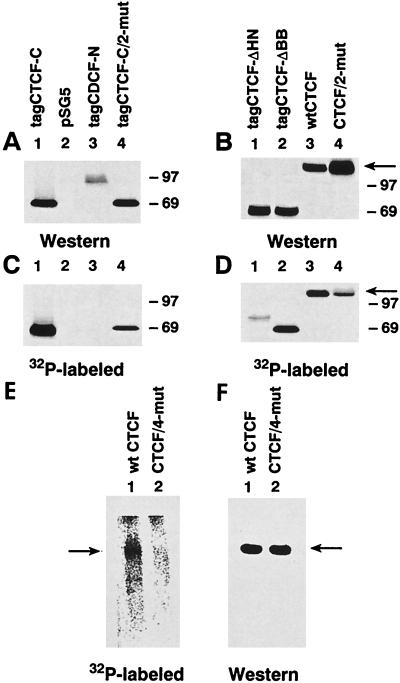
FIG. 6.
In vivo phosphorylation of the C-terminal CTCF domain containing specific mutations of major CKII sites and deletions depicted in Fig. 1A. COS-6 cells were transfected with the expression vectors listed at the top of panels A and B and schematically shown in Fig. 1B. The empty pSG5 vector was also used as a negative control (panels A and C, lanes 2). Transfected cells were metabolically labeled with [32P]orthophosphate. The six-myc-tag-containing CTCF forms and the N-terminal peptide 2 containing CTCF proteins were immunoprecipitated with either the 9E10 antibody (A and C) or anti-CTCF antibody Ab2 (B and D), respectively, and then washed on beads, separated by SDS-PAGE, blotted, and autoradiographed (C and D). The same membranes were also probed by an ECL Western blot procedure with either 9E10 antibody (A) or Ab2 (B). The 130- to 160-kDa full-length CTCF band detected by Western blotting (panel B, lanes 3 and 4) and by phosphate incorporation (panel D, lane 3) is shown by an arrow. Note that 32P incorporation into this band is specifically reduced by replacing the CKII target serines in the CTCF/2-mut protein (panel D, lane 4). (E and F) The CTCF/4-mut protein with simultaneous mutations of Ser-604, -609, -610, and -612 shows complete loss of phosphorylation in vivo. COS-6 cells were transfected with two expression vectors, pSG-CTCF, producing the wt CTCF protein, and pSG-CTCF/4-mut, in which serines 604, 609, 610, and 612 were substituted with nonphosphorylatable amino acids. Transfected cells were metabolically labeled with 32P. CTCF protein was immunoprecipitated with anti-CTCF chicken-specific Ab2 antibodies and then washed on protein A-Sepharose-4B-Fast, separated by SDS-PAGE, blotted, and autoradiographed (E). The same membrane was then probed as a Western blot with the Ab2 antibodies developed by the ECL (F). The arrows indicate the position of the 130- to 160-kDa full-length wt (panels E and F, lanes 1) or four-site-mutated CTCF band (panels E and F, lanes 2).
The carboxy-terminal domain of CTCF is predominantly phosphorylated in vivo.
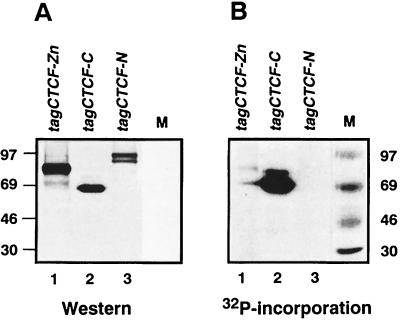
To determine the region of CTCF phosphorylation, the N-terminal region upstream of the 11 ZFs (CTCF-N), the 11-ZF domain (CTCF-ZF), and the C-terminal domain (CTCF-C) were transiently expressed as myc-epitope-tagged proteins in COS-6 cells undergoing 32P metabolic labeling. Immunoblotting with the 9E10 anti-myc-tag antibody revealed that each of the three CTCF domains was equally well expressed (Fig. 3A). Only the CTCF-C protein incorporated 32P at high levels in vivo (Fig. 3B, lane 2); phosphorylation of the CTCF-ZF domain was very low (Fig. 3B, lane 1) and was undetectable for CTCF-N (Fig. 3B, lane 3). Apparent molecular masses of CTCF-derived polypeptides as seen on SDS gels differ from the predicted molecular masses because of aberrant electrophoretic mobility, previously analyzed in detail using matrix-assisted laser desorption and ionization mass spectrometry of the gel-resolved CTCF subdomains (27). For example, although the predicted molecular mass for the CTCF-C domain based on amino acid number and composition should be about 17 kDa (or 28 kDa when fused with the six-myc-tag polypeptide), tag-CTCF-C migrates as a band of 69 kDa (Fig. 3 and 4).
FIG. 3.
Expression versus phosphorylation levels for three CTCF domains expressed in COS-6 cells. COS-6 cells were transfected with the pSG5-based expression plasmids, producing three epitope-tagged CTCF domains (tagCTCF-Zn, tagCTCF-C, and tagCTCF-N, schematically shown in Fig. 1B), and metabolically labeled with 32P. The tagged proteins were immunoprecipitated with the 9E10 anti-tag antibody and analyzed by Western blot probed with the 9E10 antibody (A) or autoradiographed (B) to reveal [32P]phosphate incorporation. Positions of the 14C-labeled protein markers are shown on a side of each panel. See text for a detailed explanation of an unusual aberrant electrophoretic migration in SDS-containing gels characteristic for the full-length CTCF protein (Fig. 2) and for its separate domains (27).
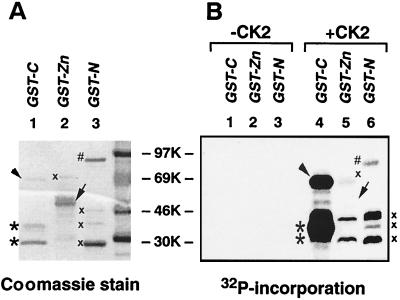
FIG. 4.
Phosphorylation of three bacterially expressed CTCF domains by purified CKII in vitro. (A) Proteins were resolved by SDS-PAGE, and gels were stained with Coomassie blue and autoradiographed. (B) Three GST-fused recombinant CTCF proteins expressed in bacteria (1 to 5 μg) were incubated in the CKII reaction buffer in the absence (lanes 1 to 3) or presence (lanes 4 to 6) of the kinase. An arrowhead and two asterisks in lanes 1 and 4 indicate the positions of bacterially expressed cognate CTCF-C and degraded CTCF-C proteins, respectively. Arrows at the right side of lanes 2 and 5 show CTCF-Zn protein that did not incorporate phosphate. Several 32P-labeled proteins that do not correspond to the GST-ZF and GST-N bands visualized by Coomassie blue gel staining are marked by an “x”. Therefore, these bands do not present phosphorylation of the CTCF-ZF or CTCF-N protein domains as discussed in Results.
The phosphorylation pattern of the three CTCF domains shown in Fig. 3 is most likely to mimic the distribution of protein kinase target sequences within the full-length CTCF protein. Therefore, these results suggest that the major in vivo phosphorylation serine/threonine sites map within the carboxy-terminal domain of CTCF.
CKII efficiently phosphorylates CTCF-C in vitro.
CTCF-C possesses a number of potential phosphorylation sites matching the CKII phosphorylation consensus motif, (S/T)XX(E/D) (1). To determine if CKII could account for the preferential phosphorylation of this region in vivo, we tested if CTCF-C would serve as a substrate for recombinant purified CKII in vitro. The three GST fusion proteins produced in bacteria (Fig. 4A) were treated with CKII in vitro in the presence of [γ-32P]ATP. Without CKII, no 32P was incorporated into any of the proteins (Fig. 4B, lanes 1 to 3). However, in the presence of the enzyme, the 69-kDa carboxy-terminal domain, GST-CTCF-C, and apparent degradation products of ∼45 kDa labeled intensely (Fig. 4B, lane 4). Using the ZF and N-terminal domains as CKII substrates, several labeled nonspecific protein bands (see lanes 2 and 3 of panel A and lanes 5 and 6 of panel B) were observed. In GST-CTCF-Zn protein preparations, a 32P-labeled band corresponds to a slower migrating contaminating protein of bacterial origin, while the ZF domain-GST fusion protein itself cannot serve as a substrate for CKII, as shown by the lack of 32P labeling at the same position of the same gel autoradiograph (panel B, lane 5). However, some weak 32P incorporation was detected with the GST-CTCF-N protein of the expected size and mobility (27) (panel B, lane 6).
Thus, among three bacterially expressed distinct CTCF domains, the C-terminal region was the primary target for phosphorylation by CKII. Identical results were obtained with tagged CTCF domains purified from other transfected cell lines by immunoprecipitation and then incubated with CKII in vitro (data not shown). Thus, the C-terminal CTCF region, which is preferentially phosphorylated in vivo (Fig. 3), also contains major CKII target sites, detected in vitro (Fig. 4). These results suggest that CKII, an unknown kinase(s) with similar sequence specificity, and/or CKII plus CKII phosphorylation-initiated secondary modifications, result in preferential phosphorylation of the carboxy-terminal region of CTCF in vivo.
Mapping of in vitro and in vivo phosphorylation sites within the carboxy-terminal CTCF domain.
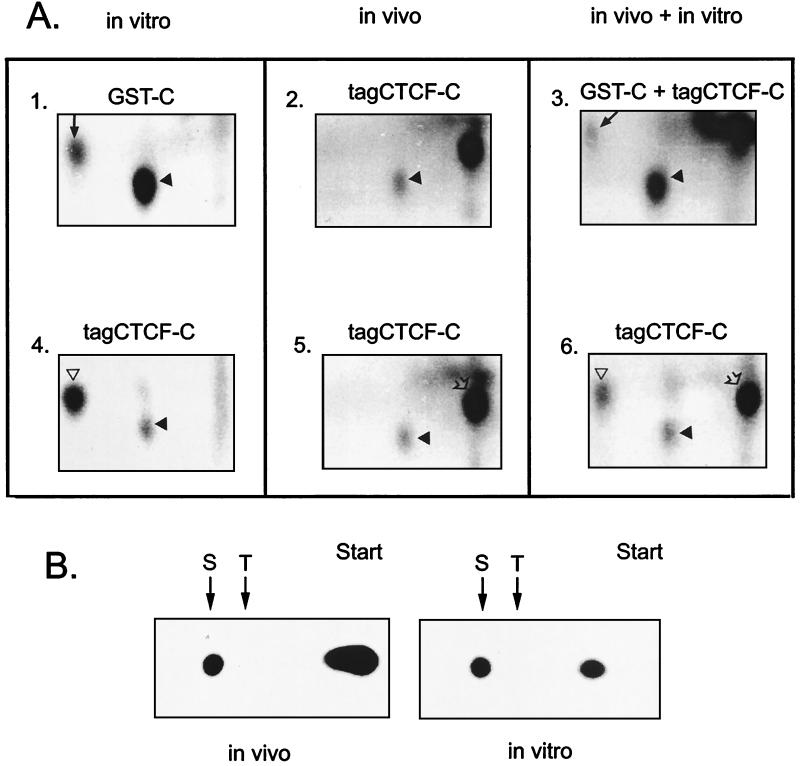
The in vivo and in vitro CKII-driven phosphorylation sites in CTCF-C were determined by tryptic peptide mapping utilizing in vivo-phosphorylated tagCTCF-C protein immunoprecipitated with the 9E10 antibody (Fig. 5A, panel 2). This was compared with the protein precipitate phosphorylated by recombinant CKII in vitro (Fig. 5A, panel 4). Specifically labeled peptides of the same size (Fig. 5A) were detected in the tagCTCF-C proteins phosphorylated by CKII in vivo or in vitro.
FIG. 5.
(A) Phosphopeptide analyses of the CTCF-C domain expressed in bacteria or in COS-6 cells and phosphorylated in vivo and in vitro. Bacterially expressed GST-C protein (panel 1) and a COS-6 cell-derived tagCTCF-C protein immunocomplex on beads (panel 4) were 32P labeled in vitro with recombinant CKII and then electrophoresed though an SDS gel, labeled bands were identified, and 32P-proteins were eluted and precipitated. The metabolically 32P-labeled tagCTCF-C protein expressed in COS-6 cells (panels 2 and 5) was immunoprecipitated and also purified from an SDS gel. After trypsin digestion and lyophilization, peptides were resolved in the first dimension by electrophoresis at pH 1.9 (horizontal direction, with the anode to the right of the gels shown) and then in the second dimension by ascending chromatography. An arrow in panels 1 and 3 and an open triangle in panels 4 and 6 indicate the position of free phosphate. An under-digested material which was unresolved by the acid electrophoresis is indicated by the open arrowheads at the top right corners of panels 5 and 6. (B) Phosphoamino acid analyses of the in vivo- and in vitro-phosphorylated C-terminal CTCF domain. The 32P-tagCTCF-C protein immunoprecipitated from transfected metabolically labeled COS-6 cells (in vivo panels) and “cold” tagCTCF-C protein labeled with [γ-32P]ATP and CKII (in vitro panels) were hydrolyzed and electrophoresed at pH 1.9 with phosphoserine (S) and phosphothreonine (T) standards.
Second, bacterially expressed purified GST-CTCF-C protein was treated with CKII and 32P in vitro (Fig. 5A, panel 1). The labeling was compared with that of tagCTCF-C protein phosphorylated by CKII in vivo (Fig. 5A, panel 5) or in vitro (Fig. 5A, panel 4). The phosphorylation site(s) utilized in vivo localized to the same single peptide as it did in vitro (Fig. 5A). By mixing equal amounts of tryptic digests labeled in vitro and in vivo (Fig. 5A, panels 3 and 6) and analyzing the mixture in the same experiment, we showed exact comigration of the major phosphopeptide labeled by CKII from both GST-CTCF-C and tagCTCF-C, as well as the peptide derived from the in vivo-phosphorylated CTCF-C.
Since our reciprocal phosphoamino acid analyses of phosphorylated CTCF-C demonstrated that only serines were extensively phosphorylated (Fig. 5B), we focused further analyses only on potential CKII target sites with the SXX(D/E) consensus sequence (37); serines at positions 578, 604, 610, and 724 (Fig. 1) fit this consensus. In addition, Ser-609 and Ser-612 could also serve as de novo targets for so-called cascade reactions when initial modification of one serine creates a suitable context for phosphorylation of another (43, 47).
To determine which of the C-terminal serines are phosphorylated in vivo, we analyzed two deletion mutants and several site-specific mutants of CTCF. In the pSG-CTCF-ΔBB expression construct, the DNA fragment encoding amino acids 67 to 570 upstream of the three potential CTCF-C CKII sites was removed as shown in Fig. 1B. In the pSG-tagCTCF-ΔHN vector, two more CKII sites were removed by deleting the DNA fragment encoding amino acids 54 to 610 (Fig. 1B). In the pSG-tagCTCF-C/2mut and the pSG-tagCTCF/2mut expression vectors, serines at positions 609 and 610 were simultaneously replaced by nonphosphorylatable residues by creating a new NaeI restriction site as described in Materials and Methods. The N-terminal CTCF epitope recognized by chicken CTCF-specific Ab2 antibodies was retained at the N termini of the deleted and of the mutated tagCTCF constructs. These constructs, together with the empty vector and the pSG-tagCTCF-N construct (Fig. 1B) expressing the N-terminal CTCF region, were transfected into COS-6 cells. Transfected cells were metabolically labeled with 32P. Proteins were immunoprecipitated with the anti-CTCF Ab2 antibodies, resolved by SDS-PAGE, blotted onto a membrane, and visualized by autoradiography. The same membranes were also probed with the Ab2 or 9E10 antibodies by the ECL Western blot procedure to compare levels of protein synthesis with efficiency of phosphorylation. Western blot analyses (Fig. 6A and B) demonstrated that all constructs produced comparable levels of homogeneous proteins of expected sizes (27). Only proteins which harbor the expected C-terminal CKII sites, namely the tagCTCF-C (Fig. 6, panel C, lane 1), tagCTCF-ΔBB (panel D, lane 2), and tagCTCF (panel D, lane 3) proteins were efficiently labeled in vivo. The tagCTCF-N protein (Fig. 6C, lane 3) was not phosphorylated, as one would predict based on the results shown in Fig. 3. Deletion of amino acids 54 to 610 (Fig. 1B) containing the three potential CKII Ser residues results in a complete loss of 32P incorporation into the tagCTCF-ΔHN protein (Fig. 6D, lane 1). A labeled band seen in Fig. 6D, lane 1, does not correspond to the tagCTCF-ΔHN protein detected by Western blot analysis (Fig. 6B). It may represent a phosphoprotein that specifically coimmunoprecipitates with the tagCTCF-ΔHN protein or may be entirely nonspecific. The tagCTCF-ΔHN protein is clearly not phosphorylated, as seen by comparing lanes 1 of Fig. 6B and D. Thus, CTCF Ser-724 is not a CKII target, or structural changes resulting from deleting amino acids 54 to 610 prevented phosphorylation.
The site-specific substitution of serines 609 and 610 in tagCTCF/2mut (Fig. 6D, compare lanes 3 and 4) and in tagCTCF-C/2mut (Fig. 6C, compare lanes 1 and 4) resulted in a dramatic inhibition of phosphate incorporation, indicating that the EDSS610DSEE sequence of the CTCF-C domain is the major CKII target site in vivo. However, the same lanes in Fig. 6 show some residual incorporation of 32P in tagCTCF-c/2mut in vivo, thus indicating the presence of other phosphorylation sites. Similar to wt CTCF-C (Fig. 5B), phosphoamino acid analysis of tagCTCF-C/2mut revealed only these phosphoamino acids (data not shown). Therefore, additional sites have to be other serines on the same peptide.
To evaluate if sites in addition to Ser-609 and -610 in CTCF-C may contribute to the residual Ser-specific phosphorylation (Fig. 6D, lane 4), we generated multimutant CTCF-C and full-length CTCF proteins with additional Ser-to-Ala mutations at Ser-578, Ser-604, and Ser-612 (Fig. 1A). Expression constructs with all possible combinations of these additional mutations were assayed for phosphorylation in vivo and in vitro (data not shown). The multimutant CTCF protein, termed CTCF/4-mut, with alterations of the Ser-604, -609, -610, and -612 residues, showed complete absence of CTCF phosphorylation in COS-6 cells (Fig. 6E, lane 2) when compared to wt CTCF (Fig. 6E, lane 1). Similar results were obtained in Swiss 3T3 fibroblasts (data not shown). Thus, the major posttranslational modification of CTCF is likely due to CKII phosphorylation of the strictly conserved S604KKEDS609S610DS612E sequence of the C terminus.
Mutating phosphorylation sites does not affect CTCF cellular localization or binding characteristics to c-myc CTSs.
Indirect immunofluorescence microscopy detected endogenous CTCF only in the nucleus (26). Nuclear transport of proteins as large as CTCF usually requires specific nuclear localization signals (NLS) (18). A putative NLS sequence is located within CTCF-C downstream of the 11th ZF and close to the CKII phosphorylation sites (26) (schematically shown in Fig. 1A). Close proximity of phosphorylation sites to the NLS consensus motif suggests that nuclear transport may be regulated by phosphorylation (reviewed in reference 23). The arrangement of the last ZF-NLS-spacer-phosphorylation sites motif in CTCF (Fig. 1A) is remarkably similar to that in the yeast SW15 transcription factor (24) and in several other transcription factors containing NLS regulated by phosphorylation (24). Therefore, we tested whether nuclear translocation of CTCF was regulated by phosphorylation.
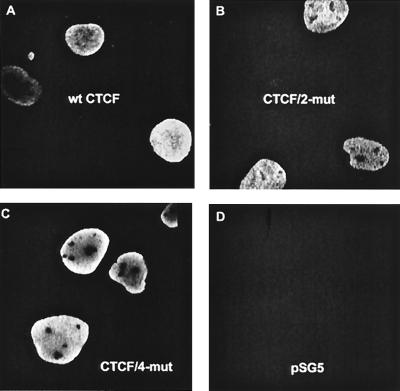
Cellular localization of the wt tagCTCF, full-length tagCTCF/4-mut, and tagCTCF-C proteins by indirect immunofluorescence microscopy demonstrated that the mutations had no obvious effect on CTCF cellular localization (Fig. 7).
FIG. 7.
Disabling phosphorylation at the C-terminal CKII sites does not affect cellular localization of CTCF. Swiss 3T3 cells transiently transfected with vectors expressing wt tagCTCF (A), full-length tagCTCF/2-mut (B), tagCTCF/4-mut (C), or no CTCF (D) were analyzed by indirect immunofluorescence microscopy, and several pictures of random groups of positive cells were taken.
In addition to nuclear localization, the DNA-binding characteristics of many transcription factors have been shown to depend on phosphorylation mediated by CKII (see references 1 and 10 for reviews). There is a possibility that the substitutions introduced at the serine-rich motif which is outside but close to the 11-ZF DNA-binding domain (Fig. 1A) may alter the entire DNA-binding conformation, resulting in altered affinity and different patterns of DNase I footprints and/or CTCF-contacting nucleotides. Indeed, in certain transcription factors, an intramolecular folding of the full-length protein may occur in a way that brings together regions outside and inside of a DNA-binding domain to regulate interactions with DNA (15). Phosphorylation at CKII target sites in the C-terminal regions of ZF factors SP1 (4) and MAZ (also called Pur-1 and ZF87) (60) regulates binding to the respective target sites either negatively (for SP1) or positively (for MAZ). Therefore, one could suggest that the C-terminal phosphorylation in CTCF might cause alterations in the overall structure of the protein, resulting in a change in DNA binding.
To directly address these issues, we first used EMSA to analyze the interactions of the c-myc site V DNA (CTS V) with equal amounts of recombinant full-length wt CTCF, CTCF/2-mut, CTCF/4-mut, and CTCF-ZF proteins. The results shown in Fig. 8A rule out any significant influence of the Ser phosphorylation on CTCF interaction with this CTS. To assess possible effects on DNA-binding kinetics and affinity not readily revealed by EMSA, we evaluated the variant CTCF proteins for these characteristics by surface plasmon resonance as described for other CTCF-binding sites (25). When assayed identically, binding of CTCF to CTS A was of lower affinity than for CTSs in the Igf2/H19 imprinting control region (25). Little or no formation of nondissociating complexes was detected. Furthermore, pairwise comparisons of wt CTCF, CTCF/2-mut, and CTCF/4-mut proteins showed no statistically significant deviations from the average value of the apparent binding affinity constant, in the range of ∼1011 M−1.
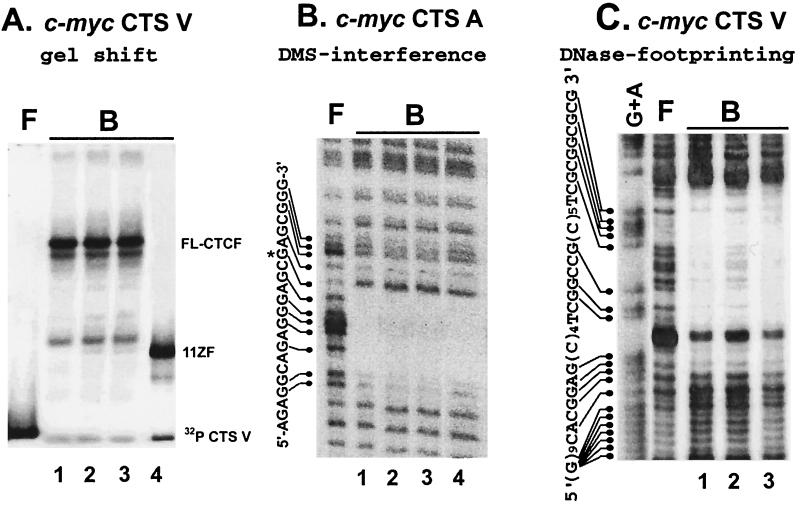
FIG. 8.
Mutating of the major C-terminal CKII sites does not affect interaction of CTCF with c-myc CTSs assayed by EMSAs (A), methylation interference (B), and DNase I footprinting (C). Lanes: 1, full-length (FL) wtCTCF; 2, CTCF/2-mut; 3, CTCF/4-mut; 4, 11ZF protein. See the text for more details. F, free probe; B, protein-bound probe.
Next, a DMS interference assay with the human c-myc CTS A and DNase I footprinting analysis with the chicken c-myc CTS V revealed no differences among the DNA recognition patterns produced by the full-length wt CTCF, CTCF/2-mut, CTCF/4-mut, or the 11-ZF domain (Fig. 8B and C). In sum, these results provide strong evidence that the C-terminal phosphorylation does not affect CTCF-DNA interactions in vitro, at least with the target sequences tested. Phosphorylation at the C-terminal side of the ZFs would, therefore, be unlikely to play a direct role in modulating sequence-specific interactions of CTCF with the c-myc promoter-proximal regions in vivo.
Mutating CTCF at the major phosphorylation sites results in enhanced repression of the chicken and human c-myc promoters.
In some cells, a substantial proportion of total cellular CTCF is not phosphorylated (reference 17 and data not shown). Although mutating different combinations of CKII target serines, resulting in only a partial loss of the C-terminal phosphorylation, would possibly have somewhat different functional significance, the identification of all possible phosphorylation sites clustered within one short acidic motif (Fig. 1A) permitted the generation of the CTCF/4-mut protein. The latter could not be phosphorylated (Fig. 6E and F), thus allowing us to study the function of nonphosphorylated CTCF.
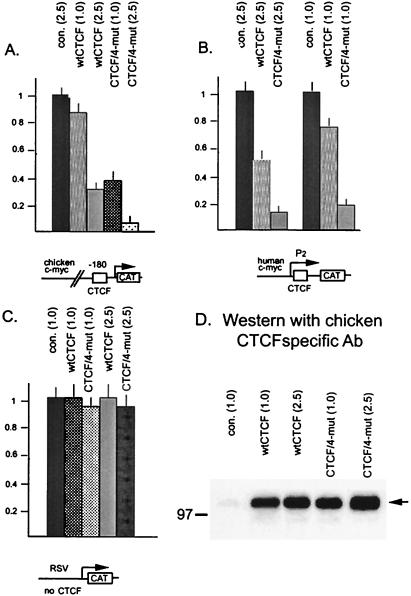
We asked whether C-terminal phosphorylation could modulate CTS-dependent transcriptional repression of c-myc gene promoters. These are the first targets for repression by CTCF characterized by us (20, 32). Increasing amounts of expression vectors for the wt CTCF and for the CTCF/4-mut proteins were cotransfected with reporter constructs containing promoters with either of two well-characterized CTSs that differ greatly in sequence: the chicken c-myc promoter (32) and the major P2 promoter of human c-myc (20). The Rous sarcoma virus (RSV) 5′ long terminal repeat (LTR) promoter that lacks a CTS served as a negative control. Very similar results were obtained with both c-myc promoters. With the chicken c-myc construct containing CTS V upstream of the promoter, transfection of 1 μg of pSG-CTCFwt resulted in only marginal repression, while the same amount of pSG-CTCF/4-mut resulted in about threefold repression (Fig. 9A). At higher inputs of these expression vectors (2.5 μg), wt CTCF produced a 2.5-fold repression, while the CTCF/4-mut protein resulted in stronger (10-fold) repression. With the human c-myc construct containing CTS A immediately downstream of the major P2 promoter, repression by CTCF was more pronounced at the lower input of the expression vectors (1.0 μg versus 2.5 μg) (Fig. 9B), an effect noticed earlier (20). Again, mutations disabling C-terminal phosphorylation of CTCF led to enhanced transcriptional repression (Fig. 9B).
FIG. 9.
Mutating the carboxy-terminal CKII target serines converts CTCF to a stronger repressor of the chicken and human c-myc gene promoters bearing different target sites, CTS V (32) or CTS A (20), respectively. Specific CTCF-binding promoter-reporter constructs, the pPst2myc-CAT plasmid containing 3.3 kb of the 5′ noncoding region of the chicken c-myc (A) or the P2 promoter-containing pP2myc-CAT plasmid (B), were utilized. The pRSV-CAT construct containing the RSV LTR promoter without CTSs (data not shown) was utilized as a control (C). The reporters were cotransfected in COS-6 cells along with the expression vectors for the wt and mutant CTCF forms, as indicated at each bar of each panel. The reporter activity was measured as described in Materials and Methods. The CAT values were normalized to the total cell extract protein concentration and were expressed in relative units by taking the activity of pPst2myc-CAT cotransfected with the empty expression vector pSG5 at an input of 1.0 μg per plate as 1.0 U. The results shown represent the mean values with the standard deviations obtained in the cotransfection experiments repeated two times in triplicate plates for each combination of the reporter and effector plasmids. (D) Accumulation of the wt and mutant chicken CTCF proteins produced in COS-6 cells from transiently transfected expression vectors for CTCF without myc tags as described in Materials and Methods. con., control. Numbers in parentheses indicate the amount of effector vectors (in micrograms) used for each transfection.
Both effects (repression of the c-myc promoter by wt CTCF and greatly enhanced repression by the CTCF/4-mut protein) appeared to be specific for the c-myc targets, because no repression of the RSV LTR promoter lacking a CTS was observed (Fig. 9C). Other promoters that lack CTSs, from the human immunodeficiency virus and murine sarcoma virus LTRs and the chicken β-actin gene, also showed no repression by either wt or mutant CTCF proteins (data not shown). Thus, enhancement of transcriptional repression by nonphosphorylatable CTCF seems to require CTS DNA-protein interaction. Moreover, the same effect of the mutations on repression of c-myc promoters was observed in both chicken and human cell lines (data not shown). Also, if expression vectors pSG-tagCTCF and pSG-tagCTCF/4-mut were employed in place of untagged CTCF vectors in identical cotransfection experiments, we obtained the same results as those shown in Fig. 9, indicating that the inclusion of the tag epitope does not alter the transcriptional properties of CTCF. Using chicken CTCF-specific antibodies for the experiments in COS-6 cells, we demonstrated that at equal DNA input in each transfection, the expression vectors produced similar levels of wt and mutated CTCF proteins (Fig. 9D). We therefore conclude that the markedly enhanced repression of c-myc promoters by mutated CTCF is specifically due to lack of phosphorylation.
Mutations of the phosphorylation sites result in marked but partial relief of the profound cell growth inhibition produced by wt CTCF.
In addition to vertebrate c-myc oncogenes, different CTSs have been identified in the Igf2/H19 intergenic region where CTCF discriminates imprinted alleles in vivo (25). This interaction was shown to control enhancer-blocking (insulator) activity that regulates Igf2 expression (reviewed in reference 2). Also, two novel functional CTSs were recently found in the promoters of the p19ARF gene (C.-F. Qi, S. Vatolin, D. I. Loukinov, E. M. Pugacheva, Z.-S. Li, T. McCarty, W. Quitschke, A. Vostrov, V. V. Lobanenkov, and H. C. Morse III, submitted for publication) and of the Polo-like kinase (PLK) (unpublished data) genes. The myc, p19ARF, PLK, and Igf2 genes are among the most studied examples of cellular oncogenes and/or mitogenic growth factor genes that regulate cell proliferation (see references 40, 46, 52, and 54 for more details and references). Regulation by CTCF of several important proliferation-controlling genes suggested that CTCF might, in turn, regulate cell growth.
To investigate whether targeted substitutions at the phosphorylation sites might have any effect on growth characteristics of cells ectopically expressing CTCF, we stably transfected HD3 erythroblasts or PC3 prostate cancer cells with the pSG-CTCF/4-mut expression plasmid and the pSG5 empty control vector together with a neoR-expressing plasmid. Since the cells were cotransfected at a 10:1 ratio of the CTCF expression construct relative to the neoR construct, all neomycin-resistant cells were expected to incorporate the CTCF-expressing vector.
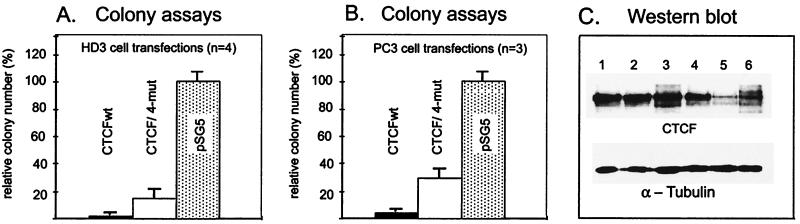
Compared to neomycin-resistant cells expressing control vector pSG5, there was practically no colony formation by cells stably transfected with wt CTCF (Fig. 10A and B). In contrast, cells stably transfected with the vector producing the CTCF/4-mut protein developed a significant number of G418-resistant colonies (Fig. 10A and B). The few colonies obtained with the wt CTCF vector expressed levels of exogenous CTCF mRNA too low to be detected by Northern blot hybridization. CTCF/4-mut was expressed at higher levels in cell lines derived from G418-resistant cell colonies. Due to the partial truncation of the CTCF cDNA untranslated regions (UTRs) in our pSG-based constructs (Fig. 1), we could distinguish the CTCF/4-mut message of 3.3 kb from the abundantly expressed endogenous CTCF mRNA of 3.8 kb by Northern blot hybridization. When compared to one another, similarly low levels of exogenous CTCF/4-mut mRNA were detected in each of these cell lines (data not shown). These findings suggest that while induced expression of the wt CTCF is almost completely nonpermissive for colony formation, mutations at the CKII sites alleviate this effect by about 20 to 30%.
FIG. 10.
Mutations at the major CKII sites result in significant, though incomplete, reversal of the profound block of the cell clonogenic capacity caused by wt CTCF. (A, B) Following stable transfection of HD3 or PC3 cells with pCTCFwt, pCTCF/4-mut, and pSG5 control vector, total numbers of cell colonies formed after 20 days in methyl cellulose-containing selection medium were counted. In each experiment, a 100% value was assigned to the number of colonies formed with the pSG5 control vector. The error bars represent the standard deviation of three or four independent transfections with each expression vector. (C) HD3 cell colonies which survive stable expression of exogenous CTCF/4-mut detected by Northern blotting (data not shown) have increased levels of CTCF protein (lanes 1 to 4) in comparison with the colonies from cells that stably express empty control vectors (lanes 5 and 6). The total amount of protein loaded on the gel and probed for CTCF by immunoblotting was nearly equal in all lanes as judged by reprobing the same membranes with an anti-α-tubulin antibody (panel C, lower part).
Four randomly selected HD3 cell lines with low CTCF/4-mut mRNA levels showed increased levels of CTCF protein in comparison to clones with the control vector (Fig. 10C, compare lanes 1 to 4 with lanes 5 and 6). Although the standard Western blot analysis could not distinguish the wt CTCF from the comigrating CTCF/4-mut protein (Fig. 10C), we surmise that this finding may indicate increased stability of the CTCF/4-mut protein.
The noticeable increase in the clonogenic capacity of CTCF/4-mut relative to the wt protein (Fig. 10) indicates a significant contribution of phosphorylation to CTCF-induced cell growth inhibition. However, in all cell lines tested, the relief from growth suppression obtained by mutating these sites was only partial. Therefore, a mechanism unrelated to the C-terminal phosphorylation also appears to contribute to the suppression of cell proliferation induced by wt CTCF.
DISCUSSION
To determine whether there is a posttranslational modification that could modulate any of the multiple CTS functions in gene regulation, we asked initially if CTCF is phosphorylated in vivo. This CTCF modification has recently been suggested to have possible functional relevance (17), but neither the sites of phosphorylation nor the kinase(s) involved was defined. Our main experiments were designed to (i) identify the kinase(s), (ii) map the phosphorylation sites, and (iii) determine if CTCF phosphorylation plays a role in regulating gene transcription from CTS-containing promoters and in regulating cell proliferation.
We showed that the CTCF C terminus, and to a much lesser extent, the ZF domain, were phosphorylated in vivo. Phosphorylation within the C2H2-type ZF arrays may regulate DNA binding, as has been shown for WT1 (50), but this possibility remains to be tested for CTCF. Since there are no tyrosines in the CTCF C terminus and endogenous CTCF could not be immunoprecipitated from several cell lines by antiphosphotyrosine antibodies (Brad Nelson and V. V. Lobanenkov, unpublished data), it is likely that only serine/threonine phosphorylation events are involved in CTCF regulation.
By studying how deletions and site-specific mutations affect CTCF phosphorylation, we demonstrated that most of the modifications occurring at the carboxy terminus in vivo can be reproduced in vitro with CKII kinase. Phosphopeptide and phosphoamino acid analyses were combined with amino acid sequence inspection to identify targets for phosphorylation. Comparative analyses of deletions and point mutations of C-terminal phosphorylation sites in vitro and in vivo suggested that CKII or a kinase with similar specificity phosphorylates CTCF within a relatively short C-terminal motif and/or it phosphorylates and initiates a further phosphorylation cascade in CTCF in vivo.
Replacing Ser-609 and Ser-610 with nonphosphorylatable amino acids in the S604KKEDS609S610DS612E sequence significantly reduced in vivo phosphorylation of full-length CTCF, while further mutating all four serines within this site completely abolished phosphorylation (Fig. 6). Since the sequence containing these four Ser residues has been maintained unchanged for the estimated ∼300 million years of evolution from birds to humans, the C-terminal phosphorylation of CTCF, which is likely to involve priming by CKII and perhaps further phosphorylation by additional kinases, would be expected to have important functions.
Phosphorylation is commonly found to attenuate transcription factor function by a number of different mechanisms (reviewed in references 10 and 23). It can regulate (i) DNA-binding activity, and even DNA sequence specificity; (ii) subcellular localization; (iii) trans activation or trans repressing activity; (iv) protein stability; (v) intermolecular folding; (vi) homo- and heterodimerization; and (vii) affinity for chromatin. Often, alternative functions of the same protein can be modulated when different parts of the protein are phosphorylated.
The major modification target sites map outside of the CTCF 11-ZF domain, which by itself is sufficient for specific binding to a variety of CTSs (5, 12, 20, 25, 26). Nevertheless, we wondered if phosphorylation of the C terminus might modulate DNA binding through intermolecular association between the ZF and C-terminal CTCF domains. A mechanistically similar example of such unusual regulation is provided by Ets-1-mediated transcriptional repression-derepression (15).
However, using the CTS-containing DNA fragments from the chicken and human c-myc promoters as the probes in gel-shift assays, in DMS interference, in DNase I footprinting experiments (Fig. 8), and in affinity measurements by the BIAcore plasmon resonance technique, we demonstrated that phosphorylation of the CTCF C terminus has very little, if any, effect on its interaction with these two CTSs.
While these experiments provide evidence for efficient binding of the Ser-mutated forms of CTCF to the c-myc CTSs in vitro, they do not exclude two feasible and intriguing possibilities. First, phosphorylation may affect CTCF binding to other CTSs not tested here, and second, certain forms of CTCF differentially phosphorylated at individual CKII sites may have different DNA-binding characteristics in vivo. We believe that these possibilities merit further investigation to elucidate how the same protein with multiple DNA sequence specificities can perform mechanistically rather different roles in gene regulation.
Besides binding to the c-myc CTSs, we examined whether phosphorylation would influence CTCF subcellular localization but found no effect (Fig. 7). This observation does not preclude the possibility that differentially phosphorylated forms of CTCF may differ in fine-level subnuclear compartmentalization.
CTCF is a candidate tumor suppressor gene at a cancer-associated hot spot on chromosome 16q22.1 (21), with established function as a c-myc promoter repressor (20) (Fig. 8) and as a parent of origin-specific and methylation-sensitive functional components of the chromatin insulator(s) upstream of the H19 gene that controls the Igf2 gene (reference 25 and references therein). As mentioned above, CTCF also regulates promoters of PLK and p19ARF genes. These four CTCF target genes are frequently noted to be overexpressed and/or deregulated in cancers (reviewed in references 40, 46, 52, 54, and 61), thus supporting the notion that CTCF, in turn, may be a tumor suppressor gene (21).
In line with the suggested function of CTCF as a tumor suppressor, we found that enforced constitutive or inducible expression of wt CTCF resulted in nearly complete inhibition of cell proliferation. The effect was shown to involve greatly reduced DNA replication and cell division delay unaccompanied by immediate cell death, which together manifest as a near complete loss of cell clonogenicity (G. N. Filippova, J. E. J. Rasko, J. León, Y.-J. Hu, P. E. Neiman, S. Collins, V. V. Lobanenkov, Abstr. 14th Annu. Meet. Oncogenes: Oncogenes, Tumor Suppressors, Signal Transduction Cancer Dev., p. 15, 1998; J. E. J. Rasko, E. M. Klenova, J. León, G. N. Filippova, D. I. Loukinov, S. Vatolin, Y.-J. Hu, J. Ulmer, M. Ward, A. Robinson, P. E. Neiman, H. C. Morse III, S. Collins, and V. V. Lobanenkov, submitted for publication).
Therefore, we attempted to determine whether the phosphorylation sites we mapped would contribute to the two physiologically important, and likely tightly intertwined, functions of CTCF: c-myc promoter repression and cell growth inhibition. We showed that when cotransfected with chicken or human c-myc promoter-reporter constructs, CTCF proteins mutated at the conserved C-terminal modification target motif induced significantly greater repression of the promoters then vectors producing wt CTCF (Fig. 8).
The part played by CTCF in inducing growth arrest should not be attributed solely to its role in repressing expression of c-myc. Support for this idea comes from our experiments (data not shown) demonstrating that the wt CTCF efficiently inhibits colony formation in a no-myc TGR15 cell line expressing no MYC family genes (36). Thus, recognition of target sites found in the promoters of other genes, such as PLK and/or p19ARF, with prominent roles in growth regulation (52, 54) are likely to be of importance. The effects of phosphorylation or other posttranscriptional modifications of CTCF on the regulation of these genes are not known, but if studies of the myc promoters provide a paradigm, they will likely be important.
CTCF is a truly multifunctional factor, because depending on the context, distinct 50- to 60-bp-long CTSs bound through combinatorial contributions of ZFs, mediate a variety of distinct functions. In this study, the c-myc promoter was employed as a model of the CTCF-binding regulatory target to demonstrate that the presence of posttranslational modifications at the strictly conserved C-terminal serines, which we mapped for the first time, can affect transcriptional activity of the protein. Attempts to investigate possible effects of phosphorylation on CTCF-driven regulation of diverse chromatin insulators (reviewed in references 2 and 64) and of negative nuclear receptor-binding hormone-responsive elements (5, 12) are currently underway. It is clear, however, that for better understanding of why under-phosphorylated CTCF partially loses its ability to inhibit cell growth, direct comparison of the whole expression spectrum of target genes, for example by a microarray technology, will be required.
CKII is usually viewed as an absolutely essential kinase that constitutively and efficiently phosphorylates nuclear substrates under different cell growth conditions (1). Functional regulation of proteins through CKII target sites is believed to occur through specific dephosphorylation in a cascade of signal transduction aimed at the function of a substrate. This model has been suggested to explain the function of CKII in phosphorylating JUN (29), MAX (10), MYB (41), MAZ (60), and the high-mobility group (HMG) 1 and 2 proteins (63). Thus, at least for those CTS-containing genes which have promoters negatively regulated by CTCF, constitutive phosphorylation by CKII may prevent efficient CTS-specific transcriptional repression.
Also, CKII can act as a priming kinase that produces specific substrates for other kinases. Inactivation of the Drosophila transcription repressor Even-skipped (Eve) is regulated by a glycogen synthase kinase 3b (GSK-3b) homologue and CKII together, but not by either kinase alone, through phosphorylation-induced conformational changes in Eve that interfere with repressive binding of this factor to the TATA box binding protein (28).
As a first step towards a model for the function of CKII sites in CTCF, it is tempting to speculate that specific dephosphorylation and/or shielding of the conserved S604KKEDS609S610DS612E motif by specifically binding proteins occurs in response to stimuli which operate through repression-derepression of gene promoters bearing different CTSs. For repression through some CTSs, CTCF may recruit histone deacetylases through direct binding to SIN3A (34). However, unless other regions in CTCF can interact with the C terminus in a phosphorylation-dependent manner to mask association with other proteins, it is unlikely that dephosphorylation of the CKII sites is involved in histone deacetylase-mediated repression of negatively controlled CTS-containing genes. This view is supported by the observation that deacetylases appear to interact with CTCF through ZF subsets rather than through the C terminus (34). Experiments aimed at identifying a protein(s) that would directly interact with the CTCF C terminus in a phosphorylation-dependent manner are currently in progress. An affinity chromatography method based on matrix-immobilized purified recombinant CTCF has recently been developed to identify CTCF-binding nuclear proteins (14). Preliminary results of these experiments demonstrate that the large subunit of RNA polymerase II (Pol II) is one such protein. Compared with the unphosphorylated form, CTCF modified at the CKII sites appears to bind to Pol II less efficiently, thus explaining how repression through the negative CTSs may be modulated. Although at present it remains unclear if the C-terminal phosphorylation may affect CTCF functions associated with promoters that are activated by (62, 65) rather than repressed by CTCF, phosphorylation-regulated CTCF-Pol II interactions are likely to be involved as well.
The participation of CTCF CKII sites in repression-derepression of the dominant oncogene c-myc, which positively regulates cell growth, and results related to CTCF-induced inhibition of S phase and cell division encouraged us to evaluate the possibility that the contribution of CTCF to cell proliferation control might be regulated via the same CKII sites which affect c-myc promoter activity in vitro.
Indeed, in the present study we demonstrated by colony assays that forms of CTCF mutated at CKII sites markedly relieve the inhibition of cell proliferation caused by the wt CTCF (at least 20-fold compared to the effect of the wt CTCF). However, the inability of the dephosphorylated CTCF/4-mut protein to completely reverse cell growth inhibition indicates that mechanisms in addition to C-terminal phosphorylation are involved in the regulation of cell growth. It remains to be seen whether these mechanisms would cooperate with phosphorylation of CKII sites.
A better understanding of CTCF regulation by CKII is complicated by the fact that the function of CKII itself remains enigmatic. CKII does not have an obvious modulator that would point to the pathways it may control. Of the more than 100 known protein substrates phosphorylated by CKII, many are nuclear proteins. Like CTCF, most of them are involved in DNA replication and transcription. Examples include DNA topoisomerases and ligases, nuclear onco-proteins MYC, N-MYC, MYB, JUN, ERBA, E1A, and the SV40 large T antigen, tumor suppressor proteins p53 and Rb, transcription factors SP1, SRF, ATFI, MAX, UBF, ERF, CREM, PU.1, MAZ, and others (for a review, see references 10 and 23). Phosphorylation at CKII sites was shown to be critically important for the biological functions of some of these proteins. For instance, targeting of the DNA ligase I to the replication machinery requires cell cycle-regulated phosphorylation at the SXXE CKII sites within the PCNA-interacting region (48). Mutation of the CKII phosphorylation site abolished the antiproliferative activity of the wt p53 (38), while DNA damage triggers phosphorylation at the p53 CKII site (9). This later finding implies the existence of a common regulatory pathway that involves phosphorylation at specific CKII sites in suppression of cell growth by a known tumor suppressor gene, p53, and a candidate tumor suppressor gene, CTCF.
ACKNOWLEDGMENTS
We are grateful to R. Marais for the kind gift of purified recombinant CKII and for sharing unpublished data; to M. Roth for the p6MycTags-D plasmid; to C. Heath for helpful advice and encouragement; to T. Zarkowska and M. Towatari for helping with the phosphoamino acid analyses; to H. Paterson for assistance with confocal microscopy; to J. Frampton for helping with a cell clonogenicity assay in HD3 cells; and to R. Nicolas, Y.-J. Hu, J. Moore, S. J. Thomas, G. Loring, L. Mueller, and S. Fagerlie for excellent technical assistance. We also thank D. Levens, J. Breen, and R. Ohlsson for critically reading the manuscript.
Initial work presented here was funded in part by the NCI/NIH RO1 grants CA20068 to P.E.N. and CA68360 and CA71732 to V.V.L., by The Cancer Research Campaign grants to G.H.G., by the Human Frontier Science Program long-term fellowship and The Royal Society research grants to E.M.K., by the UICC fellowship to I.C., and, more lately, by the NCI/NIH RO1 grant CA68360 to G.N.F., by the NIAID/NIH intramural research funding to V.V.L. and H.C.M. III, and by a research grant from The Research and Equipment Committee of the University of Oxford and a Royal Society research grant to E.M.K.
REFERENCES
- 1.Allende J E, Allende C C. Protein kinases. 4. Protein kinase CK2: an enzyme with multiple substrates and a puzzling regulation. FASEB J. 1995;9:313–323. doi: 10.1096/fasebj.9.5.7896000. [DOI] [PubMed] [Google Scholar]
- 2.Allshire R, Bickmore W. Pausing for thought on the boundaries of imprinting. Cell. 2000;102:705–708. doi: 10.1016/s0092-8674(00)00058-1. [DOI] [PubMed] [Google Scholar]
- 3.Aravind L, Landsman D. AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res. 1998;26:4413–4421. doi: 10.1093/nar/26.19.4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armstrong S A, Barry D A, Leggett R W, Mueller C R. Casein kinase II-mediated phosphorylation of the C terminus of Sp1 decreases its DNA binding activity. J Biol Chem. 1997;272:13489–13495. doi: 10.1074/jbc.272.21.13489. [DOI] [PubMed] [Google Scholar]
- 5.Awad T A, Bigler J, Ulmer J E, Hu Y J, Moore J M, Lutz M, Neiman P E, Collins S J, Renkawitz R, Lobanenkov V V, Filippova G N. Negative transcriptional regulation mediated by thyroid hormone response element 144 requires binding of the multivalent factor CTCF to a novel target DNA sequence. J Biol Chem. 1999;274:27092–27098. doi: 10.1074/jbc.274.38.27092. [DOI] [PubMed] [Google Scholar]
- 6.Bell A C, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 7.Bell A C, Felsenfeld G. Stopped at the border: boundaries and insulators. Curr Opin Genet Dev. 1999;9:191–198. doi: 10.1016/S0959-437X(99)80029-X. [DOI] [PubMed] [Google Scholar]
- 8.Bell A C, West A G, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 9.Blaydes J P, Hupp T R. DNA damage triggers DRB-resistant phosphorylation of human p53 at the CK2 site. Oncogene. 1998;17:1045–1052. doi: 10.1038/sj.onc.1202014. [DOI] [PubMed] [Google Scholar]
- 10.Bousset K, Oelgeschlager M H, Henriksson M, Schreek S, Burkhardt H, Litchfield D W, Luscher-Firzlaff J M, Luscher B. Regulation of transcription factors c-Myc, Max, and c-Myb by casein kinase II. Cell Mol Biol Res. 1994;40:501–511. [PubMed] [Google Scholar]
- 11.Boyle W J, van der Geer P, Hunter T. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 1991;201:110–149. doi: 10.1016/0076-6879(91)01013-r. [DOI] [PubMed] [Google Scholar]
- 12.Burcin M, Arnold R, Lutz M, Kaiser B, Runge D, Lottspeich F, Filippova G N, Lobanenkov V V, Renkawitz R. Negative protein 1, which is required for function of the chicken lysozyme gene silencer in conjunction with hormone receptors, is identical to the multivalent zinc finger repressor CTCF. Mol Cell Biol. 1997;17:1281–1288. doi: 10.1128/mcb.17.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cairns B R, Schlichter A, Erdjument-Bromage H, Tempst P, Kornberg R D, Winston F. Two functionally distinct forms of the RSC nucleosome-remodeling complex, containing essential AT hook, BAH, and bromodomains. Mol Cell. 1999;4:715–723. doi: 10.1016/s1097-2765(00)80382-2. [DOI] [PubMed] [Google Scholar]
- 14.Chernukhin I V, Shamsuddin S, Robinson A F, Carne A F, Paul A, El-Kady A I, Lobanenkov V V, Klenova E M. Physical and functional interaction between two pluripotent proteins, the Y-box DNA/RNA-binding factor, YB-1, and the multivalent zinc finger factor, CTCF. J Biol Chem. 2000;275:29915–29921. doi: 10.1074/jbc.M001538200. [DOI] [PubMed] [Google Scholar]
- 15.Cowley D O, Graves B J. Phosphorylation represses Ets-1 DNA binding by reinforcing autoinhibition. Genes Dev. 2000;14:366–376. [PMC free article] [PubMed] [Google Scholar]
- 16.Currie R A. Functional interaction between the DNA binding subunit trimerization domain of NF-Y and the high mobility group protein HMG-I(Y) J Biol Chem. 1997;272:30880–30888. doi: 10.1074/jbc.272.49.30880. [DOI] [PubMed] [Google Scholar]
- 17.Delgado M D, Chernukhin I V, Bigas A, Klenova E M, Leon J. Differential expression and phosphorylation of CTCF, a c-myc transcriptional regulator, during differentiation of human myeloid cells. FEBS Lett. 1999;444:5–10. doi: 10.1016/s0014-5793(99)00013-7. [DOI] [PubMed] [Google Scholar]
- 18.Dingwall C, Laskey R A. Nuclear targeting sequences—a consensus? Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 19.Evan G I, Lewis G K, Ramsay G, Bishop J M. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Filippova G, Fagerlie S, Klenova E, Myers C, Dehner Y, Goodwin G, Neiman P, Collins S, Lobanenkov V. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol Cell Biol. 1996;16:2802–2813. doi: 10.1128/mcb.16.6.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Filippova G N, Lindblom A, Meincke L J, Klenova E M, Neiman P E, Collins S J, Doggett N A, Lobanenkov V V. A widely expressed transcription factor with multiple DNA sequence specificity, CTCF, is localized at chromosome segment 16q22.1 within one of the smallest regions of overlap for common deletions in breast and prostate cancers. Genes Chromosomes Cancer. 1998;22:26–36. [PubMed] [Google Scholar]
- 22.Hark A T, Schoenherr C J, Katz D J, Ingram R S, Levorse J M, Tilghman S M. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 23.Hunter T, Karin M. The regulation of transcription by phosphorylation. Cell. 1992;70:375–387. doi: 10.1016/0092-8674(92)90162-6. [DOI] [PubMed] [Google Scholar]
- 24.Jans D A, Moll T, Nasmyth K, Jans P. Cyclin-dependent kinase site-regulated signal-dependent nuclear localization of the SW15 yeast transcription factor in mammalian cells. J Biol Chem. 1995;270:17064–17067. doi: 10.1074/jbc.270.29.17064. [DOI] [PubMed] [Google Scholar]
- 25.Kanduri C, Pant V, Loukinov D, Pugacheva E, Qi C-F, Wolffe A, Ohlsson R, Lobanenkov V. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr Biol. 2000;10:853–856. doi: 10.1016/s0960-9822(00)00597-2. [DOI] [PubMed] [Google Scholar]
- 26.Klenova E M, Nicolas R H, Paterson H F, Carne A F, Heath C M, Goodwin G H, Neiman P E, Lobanenkov V V. CTCF, a conserved nuclear factor required for optimal transcriptional activity of the chicken c-myc gene, is an 11-Zn-finger protein differentially expressed in multiple forms. Mol Cell Biol. 1993;13:7612–7624. doi: 10.1128/mcb.13.12.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klenova E M, Nicolas R H, U S, Carne A F, Lee R E, Lobanenkov V V, Goodwin G H. Molecular weight abnormalities of the CTCF transcription factor: CTCF migrates aberrantly in SDS-PAGE and the size of the expressed protein is affected by the UTRs and sequences within the coding region of the CTCF gene. Nucleic Acids Res. 1997;25:466–474. doi: 10.1093/nar/25.3.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li C, Manley J L. Allosteric regulation of even-skipped repression activity by phosphorylation. Mol Cell. 1999;3:77–86. doi: 10.1016/s1097-2765(00)80176-8. [DOI] [PubMed] [Google Scholar]
- 29.Lin A, Frost J, Deng T, Smeal T, al-Alawi N, Kikkawa U, Hunter T, Brenner D, Karin M. Casein kinase II is a negative regulator of c-Jun DNA binding and AP-1 activity. Cell. 1992;70:777–789. doi: 10.1016/0092-8674(92)90311-y. [DOI] [PubMed] [Google Scholar]
- 30.Lobanenkov V V, Adler V V, Klenova E M, Nicolas R H, Goodwin G H. CCCTC-binding factor (CTCF): a novel sequence-specific DNA-binding protein which interacts with the 5′-flanking sequence of the chicken c-myc gene. In: Papas T S, editor. Gene regulation and AIDS: transcriptional activation, retroviruses and pathogens. Woodlands, Tex: Portfolio Publishing Corp.; 1989. pp. 45–68. [Google Scholar]
- 31.Lobanenkov V V, Goodwin G H. CCCTC-binding protein: a new nuclear protein factor with interaction with 5′-flanking sequence of chicken c-myc oncogene correlates with repression of the gene. Proc Acad Sci USSR (Moscow) 1989;309:741–745. [PubMed] [Google Scholar]
- 32.Lobanenkov V V, Nicolas R H, Adler V V, Paterson H, Klenova E M, Polotskaja A V, Goodwin G H. A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5′-flanking sequence of the chicken c-myc gene. Oncogene. 1990;5:1743–1753. [PubMed] [Google Scholar]
- 33.Lobanenkov V V, Nicolas R H, Plumb M A, Wright C A, Goodwin G H. Sequence-specific DNA-binding proteins which interact with (G + C)-rich sequences flanking the chicken c-myc gene. Eur J Biochem. 1986;159:181–188. doi: 10.1111/j.1432-1033.1986.tb09850.x. [DOI] [PubMed] [Google Scholar]
- 34.Lutz M, Burke L J, Barreto G, Goeman F, Greb H, Arnold R, Schultheib H, Brehm A, Kouzarides T, Lobanenkov V, Renkawitz R. Transcriptional repression by the insulator protein CTCF involves histone deacetylases. Nucleic Acids Res. 2000;28:1703–1713. doi: 10.1093/nar/28.8.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marais R M, Hsuan J J, McGuigan C, Wynne J, Treisman R. Casein kinase II phosphorylation increases the rate of serum response factor-binding site exchange. EMBO J. 1992;11:97–105. doi: 10.1002/j.1460-2075.1992.tb05032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mateyak M K, Obaya A J, Adachi S, Sedivy J M. Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 1997;8:1039–1048. [PubMed] [Google Scholar]
- 37.Meggio F, Marin O, Pinna L A. Substrate specificity of protein kinase CK2. Cell Mol Biol Res. 1994;40:401–409. [PubMed] [Google Scholar]
- 38.Milne D M, Palmer R H, Meek D W. Mutation of the casein kinase II phosphorylation site abolishes the anti-proliferative activity of p53. Nucleic Acids Res. 1992;20:5565–5570. doi: 10.1093/nar/20.21.5565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nelson R M, Long G L. A general method of site-specific mutagenesis. Anal Biochem. 1989;180:147–151. doi: 10.1016/0003-2697(89)90103-6. [DOI] [PubMed] [Google Scholar]
- 40.Nesbit C E, Tersak J M, Prochownik E V. Myc oncogenes and human neoplastic disease. Oncogene. 1999;18:3004–3076. doi: 10.1038/sj.onc.1202746. [DOI] [PubMed] [Google Scholar]
- 41.Oelgeschläger M, Krieg J, Lüscher-Firzlaff J M, Lüscher B. Casein kinase II phosphorylation site mutations in c-Myb affect DNA binding and transcriptional cooperativity with NF-M. Mol Cell Biol. 1995;15:5966–5974. doi: 10.1128/mcb.15.11.5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perez-Juste G, Garcia-Silva S, Aranda A. An element in the region responsible for premature termination of transcription mediates repression of c-myc gene expression by thyroid hormone in neuroblastoma cells. J Biol Chem. 2000;275:1307–1314. doi: 10.1074/jbc.275.2.1307. [DOI] [PubMed] [Google Scholar]
- 43.Pinna L A. Casein kinase 2: an “eminence grise” in cellular regulation? Biochim Biophys Acta. 1990;1054:267–284. doi: 10.1016/0167-4889(90)90098-x. [DOI] [PubMed] [Google Scholar]
- 44.Plumb M A, Lobanenkov V V, Nicolas R H, Wright C A, Zavou S, Goodwin G H. Characterisation of chicken erythroid nuclear proteins which bind to the nuclease hypersensitive regions upstream of the beta A- and beta H-globin genes. Nucleic Acids Res. 1986;14:7675–7693. doi: 10.1093/nar/14.19.7675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quitschke W W. Two nuclear factor binding domains activate expression from the human amyloid beta-protein precursor promoter. J Biol Chem. 1994;269:21229–21233. [PubMed] [Google Scholar]
- 46.Reik W, Constancia M, Dean W, Davies K, Bowden L, Murrell A, Feil R, Walter J, Kelsey G. Igf2 imprinting in development and disease. Int J Dev Biol. 2000;44:145–150. [PubMed] [Google Scholar]
- 47.Roach P J. Multisite and hierarchal protein phosphorylation. J Biol Chem. 1991;266:14139–14142. [PubMed] [Google Scholar]
- 48.Rossi R, Villa A, Negri C, Scovassi I, Ciarrocchi G, Biamonti G, Montecucco A. The replication factory targeting sequence/PCNA-binding site is required in G(1) to control the phosphorylation status of DNA ligase I. EMBO J. 1999;18:5745–5754. doi: 10.1093/emboj/18.20.5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saitoh N, Bell A C, Recillas-Targa F, West A G, Simpson M, Pikaart M, Felsenfeld G. Structural and functional conservation at the boundaries of the chicken beta-globin domain. EMBO J. 2000;19:2315–2322. doi: 10.1093/emboj/19.10.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sakamoto Y, Yoshida M, Semba K, Hunter T. Inhibition of the DNA-binding and transcriptional repression activity of the Wilms' tumor gene product, WT1, by cAMP-dependent protein kinase-mediated phosphorylation of Ser-365 and Ser-393 in the zinc finger domain. Oncogene. 1997;15:2001–2012. doi: 10.1038/sj.onc.1201391. [DOI] [PubMed] [Google Scholar]
- 51.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 52.Sherr C J. The Pezcoller lecture: cancer cell cycles revisited. Cancer Res. 2000;60:3689–3695. [PubMed] [Google Scholar]
- 53.Sieweke M H, Tekotte H, Frampton J, Graf T. MafB is an interaction partner and repressor of Ets-1 that inhibits erythroid differentiation. Cell. 1996;85:49–60. doi: 10.1016/s0092-8674(00)81081-8. [DOI] [PubMed] [Google Scholar]
- 54.Smith M R, Wilson M L, Hamanaka R, Chase D, Kung H, Longo D L, Ferris D K. Malignant transformation of mammalian cells initiated by constitutive expression of the polo-like kinase. Biochem Biophys Res Commun. 1997;234:397–405. doi: 10.1006/bbrc.1997.6633. [DOI] [PubMed] [Google Scholar]
- 55.Sparks A B, Rider J E, Hoffman N G, Fowlkes D M, Quillam L A, Kay B K. Distinct ligand preferences of Src homology 3 domains from Src, Yes, Abl, Cortactin, p53bp2, PLCgamma, Crk, and Grb2. Proc Natl Acad Sci USA. 1996;93:1540–1544. doi: 10.1073/pnas.93.4.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spector D L, Goldman R D, Leinwand L A. Cells. A laboratory manual. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- 57.Szabo P E, Tang S-H E, Rentsendorj A, Pfeifer G P, Mann J R. Maternal-specific footprints at putative CTCF sites in the H19 imprinting control region give evidence for insulator function. Curr Biol. 2000;10:607–610. doi: 10.1016/s0960-9822(00)00489-9. [DOI] [PubMed] [Google Scholar]
- 58.Taylor S S, Buechler J A, Slice L W, Knighton D K, Durgerian S, Ringheim G E, Neitzel J J, Yonemoto W M, Sowadski J M, Dospmann W. cAMP-dependent protein kinase: a framework for a diverse family of enzymes. Cold Spring Harbor Symp Quant Biol. 1988;53:121–130. doi: 10.1101/sqb.1988.053.01.018. [DOI] [PubMed] [Google Scholar]
- 59.Towatari M, May G E, Marais R, Perkins G R, Marshall C J, Cowley S, Enver T. Regulation of GATA-2 phosphorylation by mitogen-activated protein kinase and interleukin-3. J Biol Chem. 1995;270:4101–4107. doi: 10.1074/jbc.270.8.4101. [DOI] [PubMed] [Google Scholar]
- 60.Tsutsui H, Geltinger C, Murata T, Itakura K, Wada T, Handa H, Yokoyama K K. The DNA-binding and transcriptional activities of MAZ, a myc-associated zinc finger protein, are regulated by casein kinase II. Biochem Biophys Res Commun. 1999;262:198–205. doi: 10.1006/bbrc.1999.1130. [DOI] [PubMed] [Google Scholar]
- 61.Tycko B. Genomic imprinting and cancer. In: Ohlsson R, editor. Genomic imprinting: an interdisciplinary approach. Vol. 25. New York, N.Y: Springer-Verlag; 1999. pp. 133–170. [Google Scholar]
- 62.Vostrov A, Quitschke W. The zinc finger protein CTCF binds to the APBbeta domain of the amyloid beta-protein precursor promoter. Evidence for a role in transcriptional activation. J Biol Chem. 1997;272:33353–33359. doi: 10.1074/jbc.272.52.33353. [DOI] [PubMed] [Google Scholar]
- 63.Wisniewski J R, Szewczuk Z, Petry I, Schwanbeck R, Renner U. Constitutive phosphorylation of the acidic tails of the high mobility group 1 proteins by casein kinase II alters their conformation, stability, and DNA binding specificity. J Biol Chem. 1999;274:20116–20122. [PubMed] [Google Scholar]
- 64.Wolffe A P. Imprinting insulation. Curr Biol. 2000;10:R463–R465. doi: 10.1016/s0960-9822(00)00534-0. [DOI] [PubMed] [Google Scholar]
- 65.Yang Y, Quitschke W, Vostrov A, Brewer G. CTCF is essential for up-regulating expression from the amyloid precursor protein promoter during differentiation of primary hippocampal neurons. J Neurochem. 1999;73:2286–2298. doi: 10.1046/j.1471-4159.1999.0732286.x. [DOI] [PubMed] [Google Scholar]