Abstract
With the global outbreak of COVID-19, the demand for testing rapidly increased and quickly exceeded the testing capacities of many laboratories. Clinical tests which receive CE (Conformité Européenne) and Food and Drug Administration (FDA) authorisations cannot always be tested thoroughly in a real-world environment. Here we demonstrate the long-term stability of nasopharyngeal swab specimens for SARS-CoV-2 molecular testing across three assays recently approved by the US FDA under Emergency Use Authorization. This study demonstrates that nasopharyngeal swab specimens can be stored under refrigeration or even ambient conditions for 21 days without clinically impacting the results of the real-time reverse transcriptase-PCR testing.
Keywords: virology, polymerase chain reaction, microbiology, pathology department, hospital
Introduction
With the emergence and global spread of COVID-19 caused by SARS-CoV-2, the development of accurate and timely diagnostic tests has been critical to disease containment. To allow for the extensive testing needs required for effective mitigation strategies, the typical regulatory CE (Conformité Européenne) mark or Food and Drug Administration (FDA) approval processes require a more permissive approach.1 2 These streamlined processes may leave individual laboratories and patients at some risk as real-world performance characteristics are assessed within the actual clinical setting. Additionally, the unprecedented rise in necessary testing caused some laboratories to reach maximum testing capacity, creating delayed results and the need for prolonged storage of specimens prior to testing.3 As with every clinical laboratory test, preanalytical sources of error abound. These include errors in sample identification, collection, inadequate sampling, contamination, as well as many others. In the setting of molecular testing for a novel infectious agent, additional potential sources of errors, such as improper storage and interfering substances, need to be considered.4 Given the necessity for possible longer-term storage and the lack of robust preclinical data in the setting of a global pandemic from a novel infectious agent, it is critical to understand the stability of SARS-CoV-2 RNA following collection of patient nasopharyngeal specimens.
There are limited studies investigating the reliability of viral detection by real-time reverse transcriptase PCR (RT-PCR) over an extended period of time. A study by Druce et al 5 in 2012 demonstrated that influenza, enterovirus, herpes simplex virus and adenovirus could be detected by real-time RT-PCR reliably for 7 days when stored under ambient or refrigerated conditions. Another study published in 2016 by Dare et al 6 determined that short delays (up to 4 days) in processing influenza nasal and throat swabs did not significantly affect the ability to detect viral particles by real-time RT-PCR. More recently several studies have demonstrated the stability of SARS-CoV-2 in different types of storage media. One study concluded that SARS-CoV-2 in endotracheal secretions was stable at room temperature in viral transport media and phosphate-buffered saline at room temperature over 18 hours.7 Another study found that SARS-CoV-2 was detectable using the Roche Cobas platform over this 2-week period with a variety of transport media.8
In our current investigation, we expand on these previous findings by assessing the detection of the SARS-CoV-2 in nasopharyngeal swabs on multiple, commonly used emergency authorised platforms. Each of these platforms uses unique DNA probes with distinct fluorophores as well as differing primer regions within the viral genome. These varied assay constructs may be differentially affected by storage conditions and degradation. Additionally, we assessed viral detection daily over a period of 3 weeks enabling extensive precision data to be produced and analysed for each platform, providing essential laboratory performance characteristics in an actual clinical laboratory environment.
Methods
Creating a shared pool
Thirty anonymised remnant nasopharyngeal swab samples collected in viral transport media which were positive for SARS-CoV-2 in our laboratory were combined to create a large pool of positive patient-based material. Given the abundant viable virus present in many of these clinical samples, the decision was made to heat-inactivate the large pooled material at 56°C as previously reported.9 A total of 126 equal aliquots of the pooled, inactivated material were then stored in one of two maintenance conditions, either ambient temperature (18°C–25°C) or within a refrigerator maintained between 2°C and 8°C. Preparation and initial storage of the pooled material occurred on day 0 and testing began the following day.
Instrumentation
This study used three different automated real-time RT-PCR in vitro diagnostic platforms (Luminex ARIES, Panther Fusion and Abbott m2000) currently in use for clinical testing of SARS-CoV-2 at the Department of Pathology, Division of Virology, Montefiore Medical Center, Bronx, New York. All technicians who performed testing were trained in the operation and maintenance of these instruments. All three platforms have been approved for use by the US FDA under the Emergency Use Authorization (EUA).
Each platform’s PCR reaction is designed to amplify two target regions within the SARS-CoV-2 genome (Luminex: open-reading frame 1a (ORF1a)/N; Abbott: RdRp/N; and Hologic: separate ORF1a regions).10–12 However, the Hologic Panther and Abbott m2000 use the same fluorescent channel for reporting of the amplification product, thereby only providing a single cycle threshold (Ct) for the merged fluorescent intensity. The Luminex ARIES platform uses two fluorophores for each amplification probe allowing for individual detection of the amplicons and providing separate Ct values for ORF1a and N. Additionally, whereas the internal amplification control (IC) used by the ARIES amplifies human ribonuclease P within the collected sample, both the Panther Fusion and the m2000 use a spiked exogenous amplification control during sample preparation and would thus not be subject to the effects of long-term storage. All materials were run according to manufacturers’ instructions for use by trained licensed personnel.10–12
Analysis
The Ct for the amplification of each platforms target(s) and the IC were recorded and graphed using GraphPad Prism software. Additional statistical analyses of the recorded data, including linear regression, correlation, coefficient of variation (CV) and paired t-tests, were performed using Microsoft Excel.
Results
Shared pool samples
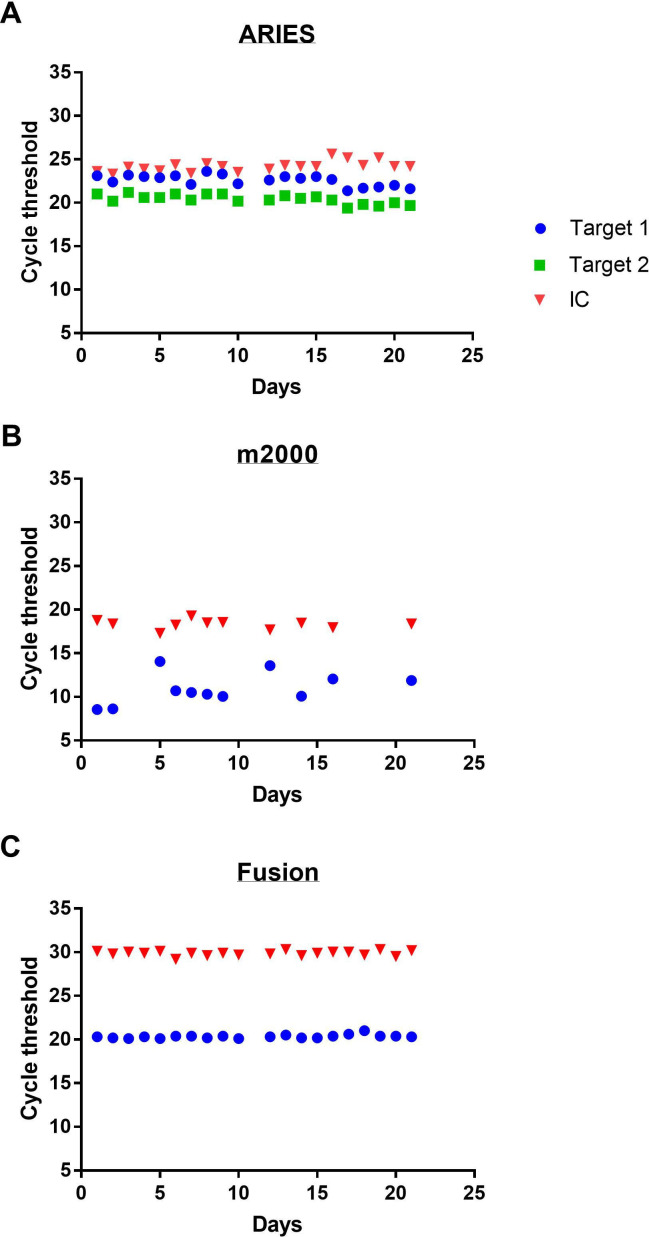
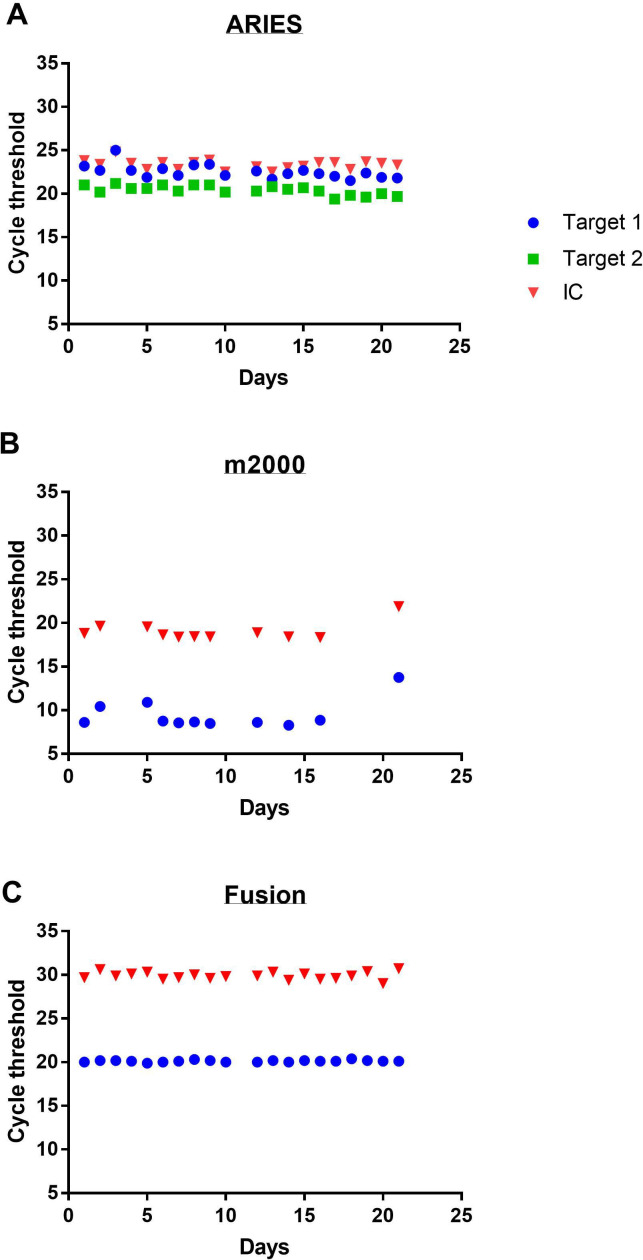
Over the course of 21 days, the pooled materials were tested alongside clinical samples and the Ct recorded for each platform at both ambient (figure 1) and refrigeration (figure 2) temperatures. A total of 102 aliquots were ultimately run for a total of 244 data points. Qualitative detection of the virus was seen across all conditions and platforms demonstrating 100% accuracy with the intended result regardless of sample age or storage condition. While the difference between the means of Ct at the two storage conditions was statistically significant for the Fusion (p<0.0001) and m2000 (p=0.037), there was no statistically significant difference in either target’s Ct on the ARIES (N p=0.26, ORF p=0.72) (table 1). Despite statistical significance, the difference in mean Ct values from the two storage conditions would not be clinically significant. The largest mean Ct deviations between the two temperatures occurred on the m2000 (1.5 cycles); however, a Ct change of this magnitude would only be expected to impact samples with exceedingly low viral burden and only have a minimal impact in the clinical testing environment.
Figure 1.
Cycle threshold of SARS-CoV-2 in viral transport media stored at ambient temperature over time on three automated real-time reverse transcriptase-PCR machines: Luminex ARIES (A), Abbott m2000 (B) and Hologic Panther Fusion (C). Target 1 refers to the N target for the ARIES and both targets for the m2000 and Fusion. Target 2 refers to the ORF target for the ARIES. IC, internal amplification control; ORF, open-reading frame.
Figure 2.
Cycle threshold of SARS-CoV-2 in viral transport media stored under refrigeration over time on three automated real-time reverse transcriptase-PCR machines: Luminex ARIES (A), Abbott m2000 (B) and Hologic Panther Fusion (C). Target 1 refers to the N target for the ARIES and both targets for the m2000 and Fusion. Target 2 refers to the ORF target for the ARIES. IC, internal amplification control; ORF, open-reading frame.
Table 1.
Comparison of the impact of storage temperature on the average cycle threshold
| Mean | SD | ||
| Fusion | |||
| 20°C | 20.34 | 0.2062 | n=20 |
| 4°C | 20.12 | 0.1196 | n=20 |
| Difference | −0.22 | 0.1795 | P<0.0001 |
| m2000 | |||
| 20°C | 10.96 | 1.793 | n=11 |
| 4°C | 9.45 | 1.658 | n=11 |
| Difference | −1.506 | 2.077 | P=0.037 |
| ARIES N target | |||
| 20°C | 20.41 | 0.522 | n=20 |
| 4°C | 20.26 | 0.7687 | n=20 |
| Difference | −0.15 | 0.5835 | P=0.2646 |
| ARIES ORF target | |||
| 20°C | 22.58 | 0.6348 | n=20 |
| 4°C | 22.53 | 0.794 | n=20 |
| Difference | −0.05 | 0.6295 | P=0.7264 |
Significance was calculated using a paired t-test.
ORF, open-reading frame.
Overall consistency of Ct values over the study period for all assays and storage conditions was very good, with the Fusion demonstrating slightly tighter grouping over time with CV of only 0.59% and 1.01% for refrigeration and room temperature, respectively. The slopes of the linear regression through each platform/temperature pair ranged from −0.075 (ARIES 4°C) to 0.129 (m2000 RT), indicating only minimal deviation from the horizontal for all platforms over time. The strengths of these correlations over time as represented by the R2 also demonstrate near 0 values (table 2), indicating very little impact over time.
Table 2.
Statistical analysis of the cycle thresholds
| CV | R2 | ||
| Fusion | |||
| IC | 20°C | 0.921 | 0.0117 |
| 4°C | 1.423 | 0.00834 | |
| Target | 20°C | 1.014 | 0.220 |
| 4°C | 0.599 | 0.0535 | |
| m2000 | |||
| IC | 20°C | 2.924 | 0.022 |
| 4°C | 5.521 | 0.144 | |
| Target | 20°C | 16.362 | 0.192 |
| 4°C | 17.55 | 0.139 | |
| ARIES | |||
| IC | 20°C | 2.482 | 0.360 |
| 4°C | 2.323 | 0.0551 | |
| Target N | 20°C | 2.812 | 0.406 |
| 4°C | 3.525 | 0.343 | |
| Target ORF | 20°C | 2.558 | 0.488 |
| 4°C | 3.794 | 0.390 |
CV, coefficient of variation; IC, internal amplification control; ORF, open-reading frame.
Patient samples
Given the ideal conditions associated with the pooled material and the possibility of RNAse degradation through the heat inactivation step, we next sought to confirm the integrity of our findings using actual stored clinical samples. Seven positive clinical samples which had been held at 4°C were repeated on day 35 after collection and run on Hologic Panther (table 3). These unadulterated samples included some containing abundant mucus which would be expected to contain RNA degrading RNAses. Although we cannot directly compare the Ct values on day 0 and day 35 since the tests were performed on different machines, all samples were positive at both time points and none of the samples was close to the threshold of detection (about Ct value of 40) on day 35, as shown in table 3.
Table 3.
Cycle thresholds (Ct) of true patient samples
| Patient | Ct day 0 | Ct day 35 |
| 1 | 21.28* | 30.6 |
| 2 | 24.6/21.5† | 20 |
| 3 | 28.4/24.5† | 22.9 |
| 4 | 16.8/14.8† | 13.2 |
| 5 | 32.2/28.1† | 29.3 |
| 6 | 16.4* | 27.7 |
| 7 | 25.3/23† | 22.8 |
All samples on day 35 were tested on Hologic Panther.
*Samples tested on the m2000.
†Samples tested on ARIES (N/ORF).
Discussion
Testing delays and laboratory backlogs have contributed to the worsening spread of COVID-19 infections. While the testing situation in the UK and USA is improving, the number of tests performed per 100 000 individuals still lags behind other developed countries such as Denmark and Italy.13 In an effort to broaden testing capabilities, the US FDA allows some manufacturers to market their assays under the less strenuous review process of EUA. It has been demonstrated for some platforms that manufacturers’ claims provided through this more liberal review pathway may significantly overestimate performance characteristics in a true clinical environment.14
For these reasons we sought to evaluate the performance of three separate EUA platforms over a lengthy period of time. Our data support others’ findings that viral RNA may remain stable for molecular testing for some time.8 14 15 All of the tested platforms, each designed to amplify completely different PCR amplicons, performed extremely well, providing precise Ct values throughout the testing period. Most surprisingly, there was only a very minimal, and clinically insignificant, impact of storage condition on the Ct values for all assays. Storage of pooled material at ambient temperature over this extended time course did not alter the test results. This demonstrated stability may allow laboratories with limited cold storage capabilities to maintain specimens at ambient temperatures without an impact on clinical performance. There do exist some recent data from researchers in China who identified an impact on storage conditions. Their data demonstrated that approximately 20% of their throat swabs were impacted after storage at room temperature, under refrigeration for 1–2 days, 1–2 freeze/thaw cycles and 30 or 60 min of 56°C heat inactivation.16 It is important to note that several studies have noted that oropharyngeal collections generally yield less viral RNA than those from the nasopharynx.17 18 The data from these researchers also support these results as more than half of the samples used in that study had significantly elevated Ct values at the upper range of detection, between 35 and 40 cycles. Additionally, most of the samples whose results were listed as not consistent still demonstrated positivity of one out of two SARS-CoV-2 viral target genes and thus would still be reported as presumptive positive in most clinical laboratories. Finally, samples with such late Ct values may have virus present below the limit of detection for the assay, at which point the reproducibility of the original result can be called into question.
Our study uses actual clinical material for pooled experimental testing as no single individual patient could provide sufficient material to perform all 21 days of testing across all conditions and platforms. In fact such pooling is now actually used in many clinical contexts as well, but given this limitation and the fact that some samples with abundant mucus or white blood cell content may be more likely to become degraded, we tested seven unadulterated patient samples which were stored under refrigeration for 35 days prior to retesting. All seven of these samples remained positive at 35 days. These results indicate that our pooled sample’s minimal change to Ct would be unlikely to affect the detectability of the virus in a patient sample stored for 3 weeks. However, testing of additional stored patient samples under multiple conditions would help confirm these results.
While the efforts of manufacturers to produce, and governments to obtain, testing for a global population is admirable, even broader-based test access will be required to further mitigation and containment strategies. Public policy allowing for easier test approval must be balanced against ensuring the safety and efficacy of that testing. With the continued spread of SARS-CoV-2 globally, clinical laboratories will remain a vital resource in evaluating performance characteristics under real-world conditions.
Footnotes
Handling editor: Tahir S Pillay.
Contributors: KAS and DYG wrote the manuscript. JS and EH performed the experiments. MN, DYG, JS and EH conceptualised the project. WS and ASF contributed to writing the manuscript.
Funding: The authors have not declared a specific grant for this research from any funding agency in the public, commercial or not-for-profit sectors.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Ethics statements
Patient consent for publication
Not required.
References
- 1. US Food & Drug Administration . Emergency use authorization, 2020. Available: https://www.fda.gov/emergency-preparedness-and-response/mcm-legal-regulatory-and-policy-framework/emergency-use-authorization
- 2. Care DoHaS . Coronavirus (COVID-19): scaling up our testing programs, 2020. [Google Scholar]
- 3. Richterich P. Severe underestimation of COVID-19 case numbers: effect of epidemic growth rate and test restrictions. medRxiv 2020. [Google Scholar]
- 4. Lippi G, Simundic A-M, Plebani M. Potential preanalytical and analytical vulnerabilities in the laboratory diagnosis of coronavirus disease 2019 (COVID-19). Clin Chem Lab Med 2020;58:1070–6. 10.1515/cclm-2020-0285 [DOI] [PubMed] [Google Scholar]
- 5. Druce J, Garcia K, Tran T, et al. Evaluation of swabs, transport media, and specimen transport conditions for optimal detection of viruses by PCR. J Clin Microbiol 2012;50:1064–5. 10.1128/JCM.06551-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dare R, Zhu Y, Williams JV, et al. Detection of influenza by real time RT-PCR is not affected by delays in respiratory specimen processing. J Med Virol 2016;88:1891–5. 10.1002/jmv.24549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Radbel J, Jagpal S, Roy J, et al. Detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is comparable in clinical samples preserved in saline or viral transport medium. J Mol Diagn 2020;22:871–5. 10.1016/j.jmoldx.2020.04.209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rogers AA, Baumann RE, Borillo GA, et al. Evaluation of transport media and specimen transport conditions for the detection of SARS-CoV-2 by use of real-time reverse transcription-PCR. J Clin Microbiol 2020;58. 10.1128/JCM.00708-20. [Epub ahead of print: 23 Jul 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hu X, An T, Situ B, et al. Heat inactivation of serum interferes with the immunoanalysis of antibodies to SARS-CoV-2. J Clin Lab Anal 2020;34:e23411. 10.1002/jcla.23411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Luminex Corporation . Aries SARS-CoV-2 assay (EUA-IVD), 2020. [Google Scholar]
- 11. Abbott Laboratories . Abbott RealTime SARS-CoV-2: Instructions for use. Abbott molecular technical services 2020.
- 12. Hologic . SARS-CoV-2 assay (Panther fusion system. contract No. AW-21159-001 Rev. 003), 2020. [Google Scholar]
- 13. Roser M, Ritchie H, Ortiz-Ospina E, et al. Coronavirus pandemic (COVID-19) OurWorldInData.org, 2020. Available: https://ourworldindata.org/coronavirus
- 14. Zhen W, Smith E, Manji R, et al. Clinical evaluation of three sample-to-answer platforms for detection of SARS-CoV-2. J Clin Microbiol 2020;58. 10.1128/JCM.00783-20. [Epub ahead of print: 23 Jul 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. van Doremalen N, Bushmaker T, Morris DH, et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N Engl J Med 2020;382:1564–7. 10.1056/NEJMc2004973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li L, Li X, Guo Z, et al. Influence of storage conditions on SARS-CoV-2 nucleic acid detection in throat swabs. J Infect Dis 2020;222:203–5. 10.1093/infdis/jiaa272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yang Y, Yang M, Shen C, et al. Evaluating the accuracy of different respiratory specimens in the laboratory diagnosis and monitoring the viral shedding of 2019-nCoV infections. medRxiv 2020. [Google Scholar]
- 18. Wang W, Xu Y, Gao R, et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 2020;323:1843–4. 10.1001/jama.2020.3786 [DOI] [PMC free article] [PubMed] [Google Scholar]