Abstract
The ribosome, the site for protein synthesis, is composed of ribosomal RNAs (rRNAs) and ribosomal proteins (RPs). The latter have been shown to have many ribosomal and extraribosomal functions. RPs are implicated in a variety of pathological processes, especially tumorigenesis and cell transformation. In this review, we will focus on the recent advances that shed light on the effects of RPs deregulation in different types of cancer and their roles in regulating the tumor cell fate.
Keywords: apoptosis, Cancer, cell survival, metastasis, ribosomal proteins, Ribosomes
Introduction
Protein synthesis is a highly regulated and coordinated process involving the action of ribosomes and a set of translation factors. Ribosome biogenesis occurs in the nucleolus and requires the action of 80 ribosomal proteins (RPs), 4 ribosomal RNAs (rRNAs), other associated proteins and small nucleolar RNAs (snoRNAs) [1,2]. Ribosome biogenesis is a highly ordered process that requires a substantial amount of energy [3]. Deregulation of RPs can affect ribosomal biogenesis which can impair cell survival, growth and proliferation. Studies have shown that RPs have extraribosomal functions including roles in DNA repair, replication, proliferation, apoptosis and chemoresistance [4]. Indeed, mutations in RPs in animal models induce a wide variety of phenotypes ranging from decrease in body size, defective organs to embryonic lethality suggesting a role of RPs beyond the ribosome structure [4,5]. In mammalian cells, RPs deficiency leads to ribosomal stress; the deficiency in one RP leads to depletion of other RPs with deleterious effects on cell survival. Mutations in RPs lead to poor assembly into the ribosomal subunit and increased cancer risk [4,6]. In fact, several RPs are found overexpressed in various human tumors, including prostate cancer (PCa), colon adenocarcinomas, liver, pancreatic, gastric, lung and breast cancer among others [6,7]. RPs’ patterns of expression in various human tumors are highly specific to tumor types and tissues implicating them as biomarkers for various cancers, while others are involved in chemoresistance [7,8].
In addition, RPs have been linked to p53 [9], a tumor suppressor protein that is involved in the surveillance mechanism that keeps the integrity of the ribosomes and the cell. p53 is involved in the cell stress response, including ribosomal stress, usually inducing apoptosis, cell cycle arrest or senescence. Upon ribosomal stress, several RPs interact with MDM-2, the suppressor of p53, thus stabilizing p53 while deficiency in individual RPs can prompt p53 activation [10].
In normal conditions, p53 expression is low due to its interaction with MDM-2 that causes its ubiquitination and its degradation by the proteasome. However, upon ribosomal stress, induced by radiations, genotoxic agents or oxidizing chemicals, the release of many RPs from the ribosomes into the nucleoplasm may happen leading to the impairment of ribosome biogenesis and to the association of those RPs with MDM-2, suppressing the degradation of p53. In fact, genome-wide analyses across human cancers have tightly linked p53 mutations with RPs deletions which increase the vulnerability to human cancers, especially mutations in RPL5 and RPL11 [11,12].
In addition to the activation of the p53 pathway, some RPs are also shown to be involved in other mechanisms by modulating oncoproteins, or interacting with other signaling pathways such as mTOR, NFκB, wnt/β-catenin and let-7a, among others [13]. It is well known that the down-regulation of the mTOR pathway inhibits the translation of several RPs, while genome-wide analysis in pancreatic cancer cells have shown that RPs are among the top differentially translated genes upon treatment with mTOR inhibitor [14,15].
In the present review, we elucidate the effect of several deregulated RPs on different human cancer types as well as the mechanisms implicated in this process, in order to shed light on the importance of RPs as biomarkers or targets to treat tumor cells.
RPs in colon cancer
Accumulating data have shown that many RPs are overexpressed in colorectal cancer (CRC) [16]. Analyzing the gene-expression profiles for almost 50 genes of 20 CRC tumors have shown that RPS3, S3A, S4X, S27a, L3, L6, L9, S2 are all overexpressed in colon cancer [17]. Kitahara et al. have demonstrated that RPL8, L18, L18a, L29, L6, L3, S19, L7 are all up-regulated in eight CRC tissues compared with normal tissues [18]. RPL35 and RPS23 are found to be overexpressed in both early- and late-stage of CRC [19]. Luo et al. have reported that the expression of RPS2 and RPS12 are increased in colon cancer tissues [20]. RPS15a, RPL18, RPL19, RPL31, RPS19, RPS27, RPL7, RPS11 and RPL15 are confirmed to be overexpressed in human colon cancer tissues compared with normal tissues in several studies [21–28].
Among these RPs, RPS3 has been shown to have high levels of expression in adenocarcinoma tissues compared with normal ones [29]. In one of these tumors, a high expression of RPS6, S8, S12, L5 and P0 is also observed which suggests that RPs could be regulated by mutual coordination. Moreover, Alam et al. have shown that RPS3 modulates the levels of p53 and lactate dehydrogenase (LDH) which affects the growth of colon cancer. The knockdown (KD) of RPS3 by small interfering RNA (siRNA) significantly impedes cell proliferation, invasion and migration while increasing apoptosis rates in colon cancer Caco-2 cells with no effect on normal colon mucosa NCM-460 cells. This has correlated with an increase in p53 expression and decrease in LDH levels exclusively in Caco-2 cells. These results demonstrate that RPS3 is involved in colon cancer growth and progression with minimal effect on normal cells which might suggest it as a potential therapeutic target and prognostic marker for colon cancer [30].
Similarly, the knockdown of RPS9, RPS15a and RPS27 inhibit human colon cancer cell growth and proliferation [21,25,31]. Targeting either RPS9 or RPS15a induces cell cycle arrest at the G2-M phase by down-regulating cyclin-dependent kinase 1 (CDK1) which suggest that these RPs could interact with the p53 pathway [21,31]. On the other hand, RPS27 KD is linked to inhibition of colon tumor progression by impeding the JNK/c-Jun signaling pathway and promoting leptin circulation, especially in obese CRC patients in vivo [25]. In fact, RPS27 up-regulation is associated with bad prognosis in obese leptin-induced CRC patients [25]. Interestingly, RPS27A, a family member of RPS27 similar to c-jun and c-fos, is shown to be also overexpressed in human colonic cancer [32]. In addition, RPS27-like (RPS27L) down-regulation is associated with poor prognosis in feces and tumor tissues of intermediate-stage CRC patients, while its overexpression and high p53 level promote better prognosis by enhancing the DNA repair capacity in LoVo cells [33]. This shows that intermediate-stage CRC patients, with consistent expression of p53 and RPS27L, have a better prognosis than patients whose level of p53 is not able to activate RPS27L.
In addition, Huang et al. have shown that RPL19 has been detected in the feces of late-stage CRC patients and correlate with poor prognosis when co-expressed with carcinoembryonic antigen (CEA). RT-PCR has revealed that RPL19 mRNA is up-regulated in late-stage colon cancer cell lines LOVO, Caco-2, HCT 116 and HT29, which indicates that RPL19 could be a prognostic marker in CRC [22].
On the other hand, the knockdown of RPL9 by siRNA has been linked to apoptosis in colon cancer cells in vitro and in vivo. RPL9 KD inactivates Id-1/nuclear factor-κB (NFкB) signaling pathway in HT29 and HCT116 colon cancer cells when compared with the control by phosphorylating IкB, which is known to prevent the translocation of NFкB into the nucleus and the activation of genes that promote cell survival. These results suggest that RPL9 could be a critical target to treat CRC [34].
RPL29 is up-regulated in colon cancer cells which correlates with the inhibition of apoptosis and promotion of the survival of the cancerous cells. A repression of RPL29 has been associated with cell differentiation in colon cancer cells and has shown that the expression of RPL29 is controlled by β-catenin/Tcf-4 pathway, which is an important pathway that controls the switch between cellular proliferation and differentiation in normal and malignant intestinal epithelial cells [35,36].
Similarly, the mRNA of RPS19 has been shown to be overexpressed in primary colon carcinoma tissues and present in both well and poorly differentiated cell lines [24]. A high level of RPS19 and laminin-binding protein (LBP) mRNAs, combined with low level of HLA-I, are linked to high malignancy of colon carcinoma. However, Chien et al. have demonstrated that RPS19 has a low expression in the feces of CRC patients with bad prognosis [37]. In fact, the mechanism through which RPS19 affects colon cancer is possibly through Bax/p53 pathway as the latter is known to be a negative prognostic factor in colon cancer when it is down-regulated [38].
On the other hand, some RPs are down-regulated in colon cancer. RPS5, in addition to four other genes (BRI3, CHD2, MGC23401 and ZNF148), are shown to be down-regulated in colon cancer and associated with tumor progression [39]. RPS7 is also down-regulated in colon cancer and its overexpression is correlated with good prognosis as it suppresses hypoxia-inducible transcription factor-1α (HIF-1α), metabolic promoting proteins glucose transporter 4 (GLUT4) and lactate dehydrogenase B (LDHB) both in vitro and in vivo [40].
Russo et al. have demonstrated that RPL3 is down-regulated in colon cancer and inversely related to Bcl-2/Bax ratio. The activation and overexpression of RPL3 by the chemotherapeutic drug 5-fluorouracil (5-FU) induces cell apoptosis and regresses cell proliferation in colon cancer, which suggest that the combination of RPL3 and 5-FU could be a novel therapy in colon cancer [41]. This is supported by the fact that the absence of uL3, the cross-domain name of RPL3, can induce chemoresistance by increasing autophagic flux in colon cancer cells [42].
Kasai et al. have shown that ten RPs (Sa, S8, S12, S18, S24, L13a, L18, L28, L32, and L35a) are highly down-regulated in colon cancer cells compared with normal mucosa as confirmed by immunohistochemistry (IHC) analysis using an antibody panel. These RPs are highly expressed in the normal tissues, especially in the mature region of the mucosal epithelia compared with immature epithelial cells. Some contradicting data suggest that some of these RPs, such as RPS24 and RPL28, are overexpressed in CRC and their inhibition could lead to a better prognosis [43,44]. In fact, RPS24 KD by small hairpin shRNA in the human colon cancer HCT116 and HT-29 cell lines leads to an inhibition in proliferation and colony formation as well as induction of cell cycle arrest in S phase and decrease in G2-M phase [43]. Similarly, RPL28 KD decreases HCT116 and HT-29 cell proliferation in vitro [44] while its expression is higher in colorectal tissues compared with normal counterparts and also correlates with decreased survival rates in metastatic cases. These controversial findings need more analysis to pinpoint the exact role of these RPs in CRC by determining in which pathways they could be exerting their effects.
Genomic and functional studies have shown that mutations in the RPS20 gene predispose patients to familial colorectal cancer type X (FCCX) which is a type of hereditary non-polyposis colorectal carcinoma. These mutations are linked to a deregulation in the maturation of pre-rRNA in these patients [45].
All these data indicate that many RPs could work as efficient prognostic markers in colon cancer and could be targeted for potential therapy. Several pathways seem to be involved with p53 being the major protein regulated by these RPs in different settings.
RPs in prostate cancer
Many RPs are also shown to be differentially expressed in PCa. RPS19, RPS21 and RPS24 are found to be up-regulated in human cancerous prostate tissues, gathered from 82 patients, when compared with normal ones suggesting that these RPs could serve as biomarkers for PCa [46]. In addition, RPL21L and RPS21 are overexpressed in PCa with a higher expression in high Gleason grade compared with low Gleason grade cases. In vitro studies have revealed that these RPs promote cell proliferation and metastasis while inhibiting apoptosis in PCa cell lines [47]. This is also confirmed by Fan et al. who reported that many key genes and signaling pathways are implicated in PCa including RPS21 and RPL22L that are significantly up-regulated in PCa, suggesting that they could be used as potential prognostic markers [48].
Moreover, RPL31 is shown to be overexpressed in PCa tissues by affecting the levels of the tumor suppressor p53 and its targets, the cell-cycle negative regulator p21 and the E3 ubiquitin ligase targeting p53, MDM2. Upon the suppression of RPL31 using a lentiviral shRNA library in PCa cells LNCaP and BicR, an enhancement of the levels of p53, p21 and MDM2 has been demonstrated by Western Blot, which is associated with a decrease in cell growth. Transfection with p53 siRNA recovers this suppression of cell growth upon RPL31 knockdown, which further proves that this RP is implicated in PCa growth and development in a p53-dependent manner [49].
Another example of a highly expressed RP in PCa is RPS2. This protein promotes tumorigenesis in all PCa tissues when compared with their normal counterparts. It is proposed that RPS2 blocks the expression of let-7a which is known to inhibit Ras and Myc activation by binding to pre-let-7a. Therefore, the inhibition of let-7a promotes the activation of Ras, and possibly Myc or p53, which may be linked to the tumor growth in PCa [50].
RPS6KB1 is well demonstrated to be up-regulated in PCa and associated with tumor growth and stage. RPS6KB1 expression decreases in the presence of Nexrutine (Nx), a natural compound that inhibits PCa tumor growth in combination with radiotherapy. The KD of RPS6KB1 can increase the sensitivity of PCa cells to radiotherapy and inhibit their survival in vitro and in vivo [51]. Similarly, palmatine, a subfraction of Nx, is able to inhibit PCa growth and invasion by concurrently targeting RPS6 and NFκB/FLIP pathway [52]. The expression of phosphorylated mammalian target of rapamycin complex 1 (p-mTORC1), the upstream regulator of RPS6 is also increased in PCa tissues and correlates with a high expression of p-RPS6 and p70 RPS6 kinase 1 (p70S6K1) as shown by IHC analysis [53]. These data suggest that RPS6 could play an important role in PCa growth and could be targeted for efficient therapy.
RPS7 is reported to be overexpressed in PCa and closely associated with tumor growth and invasion via epithelial–mesenchymal transition (EMT). In fact, knockdown of RPS7 could participate in the up-regulation of the epithelial protein marker E-cadherin and the down-regulation of the mesenchymal protein markers, such as N-cadherin and Snail and subsequently attenuating prostate tumor growth [54]. Moreover, RPS7 overexpression in PCa is associated with poor prognosis. Interestingly, an inhibition of RPS7 and PIM1, a member of PIM family and an important player in the control of cell growth, apoptosis and cell cycle progression, has been linked to the stabilization of the oncogene c-Myc and a reduction in tumor growth in vitro and in vivo [55].
According to Bee et al., the overexpression of RPL19 in PCa could lead to tumor growth and is inversely correlated with the patients’ survival rate. Interestingly, knockdown of RPL19 by siRNA avoids prostatic malignancy in vitro and in vivo in PC-3M PCa cell lines suggesting that RPL19 could be used as a prognostic marker [56,57].
Slot-blot hybridization studies have demonstrated that RPL4, L5, L7a, L23a, L30, L37, S14 and S18 mRNA levels are higher in at least one of the three types of PCa cell lines LNCaP, DU-145 and PC-3 and the overexpression of RPL7a and RPL37 is confirmed in PCa tissues [58]. Using cDNA microarray analysis, it has been reported that among the differentially expressed genes in PCa cell lines LNCaP, RPL10, RPL32 and RPS16 are shown to be overexpressed in these cell lines [59].
All these data indicate that a wide variety of RPs are up-regulated in PCa and could be used as potential biomarkers or targeted for effective treatment.
RPs in breast cancer
RPs are known to have many extraribosomal functions involved in the regulation of breast cancer growth and development. Some RPs have been shown to be overexpressed in breast cancer in different settings. RPS3, which is known to interact with NFκB, is overexpressed in breast cancer by up-regulating the X-linked inhibitor of apoptosis (XIAP) [60]. The knockdown of RPS3 in human breast cancer cells induces a decrease in proliferation and increase in apoptosis which correlates with a reduction in the levels of the XIAP protein, with no effect on NFκB activity. These results suggest that RPS3 is involved in human breast cancer in an NFκB-independent pathway [61].
In breast tumor tissues, a higher expression of RPS15A is detected and correlates with larger tumor size and higher TNM stage [62]. In MDA-MB-231 cell lines, the knockdown of RPS15A suppresses breast cancer proliferation and induces apoptosis by increasing the caspase 3/7 activity, and suppressing the phosphorylated levels of ERK1/2, Bad and Chk1. Indeed, the expression of RPS15A leads to an inhibition of apoptosis by up-regulating phosphorylated ERK1/2, Bad and Chk1 [62]. The knockdown of RPS15A by shRNA in breast cancer cell line ZR-75-30 and BT474 mediates apoptosis via the activation of caspase-3 and PARP cleavage, the up-regulation of Bad and BAX and the down-regulation of Bcl-2 confirming the importance of RPS15 in the modulation of breast cancer development [63].
RPS19 and RPL39 are also demonstrated to be overexpressed in breast cancer. RPS19 induces the production of immunosuppressive cytokines and promotes tumor growth which can be impaired by the blockage of RPS19 [64] while the knockdown of RPL39 in triple-negative breast cancer (TNBC) xenografts reduces significantly primary tumor growth, as well as metastasis [65].
In addition, knockdown of RPS6 is shown to be linked to a reduction in breast cancer cell proliferation and viability [66]. RPS6 expression decreases in cells treated with a combination of epithelial growth factor receptor (EGFR) and mesenchymal–epithelial transition (MET) inhibitors in vitro which correlate with a reduction in cell proliferation and survival as well as induction of cell cycle arrest [66].
RPL24 level is shown to be higher in breast cancer tissues compared with their normal counterparts. The knockdown of RPL24 decreases cell viability by reducing the level of the cell cycle regulator cyclin D1, the anti-apoptotic protein survivin and NBS1 which is involved in the regulation of DNA damage response [67].
Hong, Kim, and Kim have shown that RPL19 is also up-regulated in breast cancer [68]. Overexpression of RPL19 induces pre-activation of the unfolded protein response (UPR), including phosphorylation of pERK-like ER kinase (PERK), phosphorylation of eukaryotic translation initiation factor 2 α (eIF2a), and activation of p38 MAPK-associated stress signaling which enhanced cell death in MCF7 breast cancer cells [68].
An in vivo genome-wide CRISPR activation screen has identified the overexpression of RPL15 in breast cancer patient-derived circulating tumor cells in mice. The high levels of RL15 are linked to an increase in metastatic growth and enhancement of translation of other RPs and cell cycle regulators suggesting the role of RPL15 in breast cancer metastasis [69].
RPL32 is also found to be up-regulated in human breast cancer tissues and cells. The knockdown of RPL32 by a lentivirus-delivered siRNA has a negative effect on the migration and invasion of breast cancer cells in vitro and in vivo. RPL32 KD also decreases the expression levels of matrix metalloproteinase (MMP)-2 and MMP-9, which are important in tumor cell invasion and metastasis. These results indicate that RPL32 could be a novel target to treat patients with advanced breast cancer [70].
On the other hand, the low expression levels of RPS9, RPS14, RPS27, RPL11 and RPL14 are related to a poor overall survival in breast cancer patients, especially in TNBC [71]. These five genes could play a role in the detection and treatment of this type of cancer. RPL5 down-regulation in breast cancer is also associated with a poor prognosis. Knockdown of RPL5 in breast cancer cell lines promotes G2/M cell cycle progression and enhances tumor growth in xenograft mouse model which confirms the potential suppressive role of RPL5 in breast cancer [72].
Other RPs are found to be down-regulated in breast cancer including RPS27L. The knockdown of RPS27L induces autophagy in breast cancer MB231 and SK-BR3 cells by shortening the protein half-life of β-TrCP which is involved in the degradation of DEPTOR, the natural inhibitor of mTOR. The accumulation of DEPTOR inactivates mTORC1 and induces autophagy. The blockage of autophagy induced by RPS27L leads to the regression of breast cancer cell growth by triggering apoptosis [73]. These results point to the involvement of the β-TrCP-DEPTOR-mTOR axis in the progression of breast cancer in patients with low level of RPS27L.
RPs in liver cancer
RPs expression is also modulated in liver cancer. The most common type of this cancer is hepatocellular carcinoma (HCC) where many RPs are known to be differently regulated. Among these, RPL12, L23a, L27, L3, SA and S2 are shown to be up-regulated in HCC. The latter is known to induce tumor proliferation in vitro and in vivo [74–76]. In the case of RPSA, the overexpression of the microRNA-587 (miR-587) inhibits RPSA expression and reduces cell proliferation and invasion in HCC cell line SMMC-7721 [76]. Similarly, RPL39L overexpression is correlated with increased tumor invasion and poor prognosis in HCC cells [77].
RPL36 is found to be up-regulated in 75% of 60 HCC patients especially in early stages of carcinogenesis suggesting its potential role as a biomarker in liver cancer [78]. On the other hand, RPS8 is found to be up-regulated in alcohol-associated HCC and not in non-alcoholic-associated HCC which makes RPS8 a biomarker and a novel therapeutic target for HCC linked to alcohol [79].
Moreover, RPS6K, which phosphorylates RPS6, is up-regulated in HCC patients when compared with normal ones, and this up-regulation is linked to the activation of Akt-mTOR-p70S6K signaling pathway that promotes neoangiogenesis in HCC [80,81]. In fact, the AKT/mTORC1/RPS6 pathway is involved in the regulation of lipogenesis, an oncogenic event in liver cancer in vitro and in vivo [82]. This axis is also dampened upon the knockdown of RPS4 X-linked (RPS4X) in HCC cell lines which correlate with a decrease in cell proliferation, migration and invasion and an increase in apoptosis [83]. These results indicate that the AKT/mTORC1/RPS6 axis is a potential marker in HCC but the precise function of the phosphorylation of RPS6 in this pathway needs further elucidation.
In HCC cells HepG2, the knockdown of RPS15A by shRNA causes cell cycle arrest at G0/G1 phase suggesting that RPS15A could be a potential therapeutic target in liver cancer [84]. Indeed, the overexpression of RPS15A in HCC is linked to an increase in fibroblast growth factor 18 (FGF18) expression following the stimulation of Wnt/β-catenin pathway which induces tumor angiogenesis [85].
It is also reported that RPS3 is up-regulated in HCC and can promote hepatocarcinogenesis in vitro and in vivo by stabilizing its target, the silent information regulator 1 (SIRT1) mRNA at the post-transcriptional level. The inhibition of RPS3/SIRT1 pathway by 5-formylfuran-2-yl methyl 4-hydroxy-2-methylenebutanoate (FMHM) represses HCC progression suggesting that RPS3/SIRT1 could serve as a potential therapeutic target in liver cancer [86].
RPL4 is reported to be up-regulated in HCC cells when compared with normal ones and associates with poor prognosis [87]. Yang et al. have shown that the level of RPL4 mRNA is significantly reduced in HCC cells upon the knockdown of the lncRNA small nucleolar RNA host gene 7 (SNHG7), which plays a pivotal role in HCC metastasis. The restoration of RPL4 level reverses the effect of SNHG7 on liver cancer cells. These results indicate that RPL4 targeting could play a potential role in HCC treatment [88].
In addition, a chronic infection with hepatitis B virus (HBV) could lead to HCC development especially through the factor hepatitis B virus X protein (HBx) which is detected in many cases of HBV-related HCC patients and can activate diverse signaling pathways related to tumorigenesis [89]. Several RPs such as the acidic ribosomal phosphoprotein P0, RPS20, L8, L27a, L21, L31, L35a, L37a, S24, S27a, S8, L37a, S3a, S15a and S5 are found to be up-regulated in HBV-associated HCC [90–92]. Among these RPs, RPS3a has been found to interact with HBx via its chaperoning activity and subsequently stimulate the activation of the oncogenic signaling pathway NFκB in HCC [93]. Similarly, RPS27A is found to be overexpressed in HBx-expressing HCC cell lines and in the liver of transgenic model of HCC. RPS27A inhibition induces significant reduction in cell proliferation and cell cycle arrest either at G0/G1 phase in HCC cell lines [94]. However, RPS27A is down-regulated in virus-induced HCC tissues compared with normal liver tissues and correlates with the overexpression of a cold shock domain family protein, Y box-binding protein (YB-1), that is known to be associated with several RPs and induce carcinogenesis [95]. The differential expression of RPS27A in different settings and the possibility for targeting it for HCC therapy needs to be further explored.
RPL5 and RPL11 are known to interact with MDM-2 and stabilize p53 by inhibiting its ubiquitination and degradation. This pathway seems to be highly involved in HCC progression. A germline copy number variation (CNV)-based genome-wide association study (GWAS) have identified the overexpression of small nucleolar RNA H/ACA box 18-like 5 (SNORA18L5) to be associated with a high risk of HBV-associated HCC [96]. SNORA18L5 expression is found to increase ribosome biogenesis causing RPL5 and RPL11 to stay in the nucleolus, which alter their localization and their interaction with MDM2 inducing p53 ubiquitination and degradation. This correlates with a decrease in p53-dependent cell cycle arrest and apoptosis in HCC cell lines [96]. Moreover, c-Myc inhibition leads to the up-regulation of RPL5 and L11 in HCC [97]. In fact, c-Myc is inhibited by a depletion of the midline 1 interacting protein 1 (MID1IP1), one of the glucose-responsive genes that is found to be overexpressed in HCC cells. However, an attenuation of RPL5 and RPL11 activates c-Myc in MID1IP1-depleted HCC cells. These data clearly suggest that RPL5 and L11 could serve as tumor suppressors in liver cancer.
RPs in pancreatic cancer
RPs may also play a role in the development and progression of pancreatic cancer which is considered one of the most malignant types of tumors. It is reported that RPSA is highly expressed in the invasive human pancreatic cancer cell line PC-1.0 and is linked to Integrin α 6 (ITGA6), a protein involved in the regulation of cell–cell attachment [98]. ITGA6 promotes the activation of PI3K after regulating the phosphorylation level of AKT, while RPSA activates MAPK signaling pathway which consequently stimulates the invasion and the metastasis of pancreatic cancer cells. Knockdown of RPSA and ITGA-6 inhibits these pathways and leads to a reduction in pancreatic cancer cell line invasion and metastasis [98]. RPSA could also form a complex with the transient receptor potential melastatin-related 7 (TRPM7) that regulates pancreatic ductal cancer cell migration [99] suggesting that RPSA could work through different pathways and be a promising target in the treatment of this cancer.
Knockdown of RPL34 is also shown to have a negative effect on pancreatic cancer cells by inhibiting tumor proliferation, metastasis and promoting apoptosis. In vivo assays confirm that RPL34 affects pancreatic cancer cells through MAPK and p53 pathways which suggest that this RP could serve as a potential biomarker to detect and treat pancreatic cancer [100].
In addition, RPS15A is highly expressed in pancreatic cancer cell lines and is inversely correlated with the tumor suppressor miR-519d-3p expression. Inhibition of RPS15A by miR-519d-3p leads to the down-regulation of Wnt/β-catenin pathway in pancreatic cancer which proves the potential role of RPS15A as a target to treat pancreatic cancer [101].
RPs effect has been linked to KRAS, an oncogene mutated in 90% of pancreatic cancer. RPL26 and L29 are up-regulated upon knockdown of KRAS leading to cell proliferation of PANC-1 cell lines [102]. Inhibition of RPL26 and RPL29 in these cells is associated with reduced proliferation, increased apoptosis and blockage of cell cycle, which shed light on the importance of targeting these RPs as a potential therapy of pancreatic cancer. Muro et al. have discovered that the use of serum anti-RPL29 antibody retards pancreatic cancer cells proliferation in vitro suggesting that serum anti-RPL29 could be a biomarker in this cancer [103].
In addition, pancreatic acinar cells that express mutant KRAS or are treated with 7,12-dimethylbenz(a)anthracene (DMBA) show increase in RPS6 phosphorylation and expression. Phosphorylated P-RPS6 induces DNA damage triggered by the expression of the mutant KRAS and reduces p53-mediated tumor suppression which reveals that p-RPS6 is an important target in pancreatic cancer at initial stages [104].
RPL39 knockdown by siRNA in pancreatic cancer is also linked to cancer cell regression and apoptosis enhancement in vivo and in vitro [105]. RPL39 is overexpressed in the most aggressive pancreatic cancer cell lines PANC-1 and MIA PaCa-2. The effect of RPL39 on pancreatic cancer cell apoptosis seems to be dependent on caspase 8 activation suggesting that targeting RPL39 could be a potential treatment in pancreatic cancer [105].
RPL10 is another example of the deregulated RPs in pancreatic cancer. When pancreatic cell lines are treated with dimethylaminoparthenolide (DMAPT), a water-soluble agent that has anti-tumor activities, RPL10 expression is reduced. DMAPT can directly bind to RPL10 which consequently down-regulate the expression of p65 and IKKγ that are part of the NFκB signaling pathway inhibiting cell proliferation [106].
RPS8, RPL15 and RPL21 could also serve as biomarkers in pancreatic cancer. RPS8 overexpression is associated with short survival times in pancreatic cancer patients compared with the ones who express a lower level of RPS8 [107]. Yan et al. have reported that a low expression of RPL15 is related to a poor prognosis, as well as cell invasion and metastasis in pancreatic cancer patients. A high expression of this RP is inversely correlated with cell differentiation and metastatic stage classifications suggesting that RPL15 is an important indicator in this cancer [108]. In its turn, RPL21 KD by siRNA inhibits cell proliferation and DNA replication as well as induces G1 cell cycle arrest in pancreatic cancer cells PANC-1 and Bxpc-3, in vitro and in vivo. RPL21 KD promotes apoptosis in these two types of cells but not in normal ones by increasing caspase 8 activity, suggesting that targeting RPL21 could be an effective anti-cancer therapy [109].
RPs in gastric cancer
Many RPs are found affected in gastric cancer (GC) including RPS13, RPL23, RPS27, RPL6, RPL13 and RPL15 [110–114]. Indeed, RPS13 and RPL6 are overexpressed in GC and can enhance colony formation in vitro as well as tumor formation in vivo and stimulate the G1/S transition in GC cells by inhibiting p27kip1 due to the up-regulation of cyclin E expression [115,116]. Similarly, RPS13 and RPL23 are overexpressed in GC and can promote multidrug resistance in GC cells by inhibiting drug-induced apoptosis. RPL23 may induce multidrug resistance by regulating glutathione S-transferase-mediated drug-detoxifying system [110].
RPS15A is also found to be up-regulated in GC. The knockdown of RPS15A by shRNA is linked to a reduction in cell proliferation, metastasis and to an arrest in the G0/G1 phase of the cell cycle [117]. RPS15A can activate the NFκB pathway through Akt/IKK-β signaling axis, thus promoting GC metastasis, suggesting that RPS15A could be a biomarker in GC [118].
In its turn, RPL15 is overexpressed in GC and associated with an increase in cell proliferation [114]. RPL15 can act as an interacting partner of p56, a gene encoding antiviral proteins activated by Type I interferons. Overexpression of p56 inhibits the growth of GC cells that overexpress RPL15 [119].
Some RPs are involved in the modulation of GC resistance to therapy. High levels of RPL11 in GC patients treated with 5-FU is associated with good prognosis. In vitro, it was demonstrated that RPL11 increases the sensitivity of GC cell lines to 5-FU by activating the p53 pathway suggesting the potential role of increased level of RPL11 in combination with 5-FU in the efficient treatment of GC [120]. In HER2-Amplified Gastric Cancer, RPS6 seems to be involved in resistance to therapy. Targeting RPS6 by siRNA or drug inhibitors induces tumor regression in resistant cells and models both in vitro and in vivo [121]. The PI3/AKT/mTOR/RPS6 pathway confers resistance in HER2-Amplified Gastric Cancer through the stimulation of the nuclear factor erythroid 2-related factor 2 (NRF2), a factor involved in chemo- and radioresistance in different tumors [121].
RPs in lung cancer
RPs also play a critical role in lung cancer. For example, RPL9 is shown to be overexpressed in different types of lung cancer tissues with the highest expression in small cell lung carcinoma while RPS3A is highly expressed in squamous cell carcinoma indicating that these RPs could be considered as potential biomarkers in specific types of lung cancer [122,123]. Similarly, RPL34 is up-regulated in non-small cell lung carcinoma (NSCLC) tissues when compared with normal ones. Knockdown of RPL34 by a lentivirus-mediated shRNA in NSCLC cell line H1299 is correlated to a significant decrease in cancer cell proliferation and increase in apoptosis and S-phase arrest, suggesting that RPL34 may play an important role in the development of NSCLC [124].
Hyper-phosphorylated RPS6 (p-RPS6) is found to be associated with an unfavorable survival of NSCLC patients. The knockdown of RPS6, along with p-RPS6 alterations, induces cell cycle arrest at G0/G1 phase especially through the abnormal regulation of AKT2/mTOR/p70S6K signaling pathway suggesting that targeting RPS6 could be an effective therapy for NSCLC [125].
On the contrary, it has been reported that RPL22 is down-regulated in NSCLC suggesting that the low expression of RPL22 could play a potential role in the carcinogenesis of NSCLC [126].
In vitro, RPL19 is overexpressed in lung cancer H1224L cell line and positively correlates with interferon γ (IFN-γ) production. Inhibition of RPL19 by siRNA is linked to a regression in lung cancer progression and suppression in cyclinD1 and D3 syntheses [127]. Similarly, down-regulation of RPS15A by siRNA is found to be linked with a decrease in lung cancer A549 cells proliferation through G0/G1 cell cycle arrest suggesting that these RPs could function as potential biomarkers in lung cancer [128].
Some RPs have been correlated with apoptosis modulation in lung cancer. Yang et al. have demonstrated that phosphorylation of RPS3 and anti-apoptotic TRAF2 protein leads to radioresistance in NSCLC. Indeed, ionizing radiation (IR), in radioresistant NSCLC cells, leads to the phosphorylation of RPS3 and TRAF2 by casein kinase 2α (CK2α) and PKC, respectively, which promote the dissociation of RPS3–TRAF2 complex and the activation of NFκB. This activation is linked to a significant up-regulation of prosurvival genes, including cIAP1, cIAP2 and survivin. However, this phenomenon is not detected in radiosensible NSCLC cells [129]. In addition, IR can induce a dissociation of the macrophage migration inhibitory factor (MIF) from RPS3, which can lead to an increase in pro-inflammatory cytokines secretion and a modulation in the expression of EMT marker proteins in vitro and in vivo [130].
RPS29 has been shown to induce apoptosis and improve the effect of anticancer drugs by down-regulating the expression of anti-apoptotic proteins Bcl-2, Bcl-X(L) and survivin, while up-regulating the expression of pro-apoptotic proteins p53 and Bax as confirmed by Western blot analysis in H520 cells. These modulations correlate with the release of cytochrome c from the mitochondria and the activation of initiator caspase-8 and 9 and effector caspase-3 suggesting the potential role of RPS29 as a biomarker in NSCLC [131].
5-FU chemotherapy treatment induces the production of RPL3 in Calu-6 cells that is found to have a role in negatively regulating the activation of NFκB through IκB-α up-regulation. Furthermore, RPL3 significantly enhances the apoptosis of Calu-6 cells which lead to the overexpression of the pro-apoptotic protein Bax and the inhibition of the anti-apoptotic protein Bcl-2 [132].
Other RPs can modulate the progression of lung cancer by affecting the DNA repair mechanisms. RPS27L has been shown to directly bind to two proteins involved in DNA interstrand cross-link repair, FANCD2 and FANCI, which protect them from degradation in lung cancer cells. The KD of RPS27L abrogates the level of FANCD2 and FANCI which sensitizes the cells to mitomycin C treatment reducing cell viability and colony formation [133].
These results indicate that the effects of RPs on lung cancer are dependent on the types of cancer and the different pathways through which these proteins are involved.
RPs in other types of cancer
In addition to the major types of cancers mentioned above in which RPs can play a critical extraribsosomal role, these proteins have been also detected in other types of tumors. In here, we mention some examples including RPL34 that is up-regulated in esophageal cancer and its KD inhibits cancer cell proliferation, migration and invasion in vitro by down-regulating the protein expression level of p-PI3K and p-Akt [134]. RPS15A is overexpressed in renal cell carcinoma and related to tumor growth as its inhibition promotes apoptosis [135]. Moreover, mutations affecting RPL5 and RPL10 are present in 9.8% of pediatric T-cell acute lymphoblastic leukemia (T-ALLs) [136]. RPL10 mutation in T-ALL cells affects proliferation of these cells by increasing the reactive oxygen species (ROS) levels and BCL-2 expression highlighting the involvement of mutated RPs in tumorigenesis [137].
In nasopharyngeal cancer, it has been discovered that four RP genes uS8 (S8), uS4 (S9), eS31 (S27a) and uL14 (L23) are down-regulated in cancer cell lines suggesting that these RPs could serve as biomarkers in the carcinogenesis of nasopharyngeal cancer [138]. Expression of RPS7 is found to be linked with ovarian tumorigenesis and metastasis suppression. Silencing of RPS7 enhances ovarian cancer cell migration and invasion through PI3K/AKT and MAPK signal pathways [139]. RPS6 is found to be highly expressed in glioblastoma (GBM) tissues especially in the stem cell niche. RPS6 KD significantly dampens the stem cell-like features of GBM including a decrease in tumorsphere formation and the stem cell marker STAT3 expression indicating the importance of RPS6 in the maintenance of stem cell characteristics of GBM which are known to be the major reason for cancer relapse and resistance to therapy [140].
In cervical cancer, RPLP0 KD decreases cell viability and proliferation while increasing apoptosis level. These effects correlate with a modulation of cell cycle and apoptosis-associated proteins. In cells overexpressing the tumor suppressor gene phospholipase A and acyltransferase 4 (PLAAT4), RPLPO levels are decreased suggesting that the ratio of RPLP0 to PLAAT4 is an important regulator of cervical cancer growth and development [141].
Conclusion and perspectives
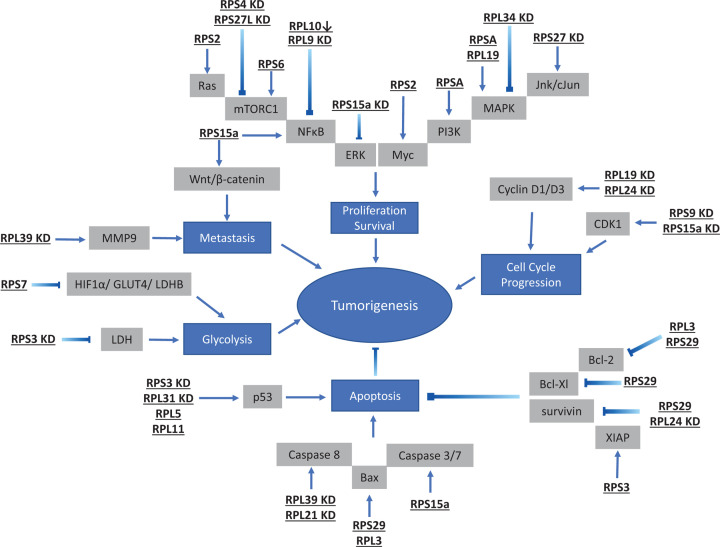
The link between RPs and cancer is clearly established in this review. Table 1 summarizes the level of expression of the major RPs in several types of cancer with an emphasis on the target pathways involved. In conclusion, RPs seem to play crucial roles in different settings ranging from cancer onset and progression to metastasis. These proteins are involved in many pathways including those involved in cell survival, cell cycle progression, DNA repair and apoptosis (Figure 1). In the future, more work is needed to efficiently use RPs as biomarkers for either good or bad prognosis depending on the tumor type and stage. More importantly, it is crucial to start identifying drugs that can target these different proteins for efficient treatment of specific types of cancer.
Table 1. Expression of RPs in common human cancers with an emphasis on the targets involved in this deregulation.
| Type of cancer | Ribosomal proteins | Level of expression | Targets | Reference |
|---|---|---|---|---|
| Colon cancer | RPS3 | Increased | p53 and LDH | [29,30] |
| RPS9, RPS15a | Increased | CDK1 | [21,31] | |
| RPS27 | Increased | JNK/c-Jun | [25] | |
| RPL9 | Increased | NFκB | [34] | |
| RPS2, S3A, S4X, S5, S6, S8, S11, S12, S19, S23, S24, S27a, L3, L5, L6, L7, L8, L15, L18, L18a, L19, L28, L29, L31, L35, and P0 | Increased | Not available | ||
| RPS5 | Decreased | HIF-1α, GLUT4, LDHB | [40] | |
| RPSa, S7, S8, S12, S18, S24, L3, L13a, L18, L28, L32, and L35a | Decreased | Not available | ||
| Prostate cancer | RPL31 | Increased | p53 and its targets: p21 and MDM2 | [48] |
| RPS2 | Increased | let-7a | [50] | |
| RPS7 | Increased | E-cadherin, N-cadherin and Snail; c-Myc | [54,55] | |
| RPS6, S6K, S14S16, S18, S19, S21, S24, L4, L5, L7a, L10, L19, L21L, L22L, L23a, L30, L32, and L37 | Increased | Not available | ||
| Breast cancer | RPS3 | Increased | XIAP | [60] |
| RPS15A | Increased | Caspase 3/7; ERK1/2, Bad and Chk1 | [62] | |
| RPL24 | Increased | Cyclin D1, survivin and NBS1 | [67] | |
| RPL19 | Increased | UPR, including PERK, eIF2a, and p38 MAPK-associated stress signaling pathway | [68] | |
| RPL32 | Increased | MMP-2 and MMP-9 | [70] | |
| RPS6, S19, L15, and L39 | Increased | Not available | ||
| RPL5 | Decreased | G2/M cell cycle | [72] | |
| RPS27L | Decreased | β-TrCP | [73] | |
| RPS9, S14, S27, S27L, L11 and L14 | Decreased | Not available | ||
| Liver cancer | RPS6K | Increased | Akt-mTOR-p70S6K signaling pathway | [80,81] |
| RPS15A | Increased | Wnt/β-catenin pathway | [85] | |
| RPS3 | Increased | SIRT1 | [86] | |
| RPS3a | Increased | NFκB | [93] | |
| RPL5, L11 | Increased | MDM2 | [96] | |
| RPSA, S2, S4X, S5, S6, S20, S24, S27A, S8, L4, L5, L8, L11, L12, L21, L23a, L27, L27a, L30, L31, L35a, L36, L37a, L39L and P0 | Increased | Not available | ||
| RPS27A | Decreased | YB-1 | [95] | |
| Pancreatic cancer | RPSA | Increased | MAPK; TRPM7 | [98] |
| RPL34 | Increased | MAPK and p53 | [100] | |
| RPS15A | Increased | Wnt/β-catenin pathway | [101] | |
| P-RPS6 | Increased | p53 | [104] | |
| RPL10 | Increased | p65 and IKKγ | [106] | |
| RPL21 | Increased | Caspase 8 | [109] | |
| RPS6KA2, S8, L26, L29, L34 and L39 | Increased | Not available | ||
| RPL15 | Decreased | Not available | ||
| Gastric cancer | RPS13, L6 | Increased | p27kip1 | [115,116] |
| RPL23 | Increased | Glutathione S-transferase-mediated drug-detoxifying system | [110] | |
| RPS15A | Increased | NFκB pathway through Akt/IKK-β signaling axis | [117] | |
| RPL15 | Increased | p56 | [119] | |
| RPS6 | Increased | PI3/AKT/mTOR/RPS6 pathway | [121] | |
| RPS27, L13 and L23 | Increased | Not available | ||
| RPL11 | Decreased | p53 pathway | [120] | |
| Lung cancer | RPS6 | Increased | AKT2/mTOR/p70S6K | [125] |
| RPL19 | Increased | cyclinD1 and D3 | [127] | |
| RPS27L | Increased | FANCD2 and FANCI | [133] | |
| RPS3, S3A, S15A, S27L, L9, L19 and L34 | Increased | Not available | ||
| RPL3 | Decreased | IκB-α | [132] | |
| RPS29 | Decreased | Down-regulating Bcl-2, Bcl-X(L) and survivin; up-regulating p53 and Bax | [131] | |
| RPL22 | Decreased | Not available |
Figure 1. A representation of the most recent findings involving various RPs in the major known oncogenic pathways.
Abbreviations
- CRC
colorectal cancer
- DMAPT
dimethylaminoparthenolide
- EMT
epithelial–mesenchymal transition
- GBM
glioblastoma
- GC
gastric cancer
- HBV
hepatitis B virus
- HBx
hepatitis B virus X protein
- HCC
hepatocellular carcinoma
- IHC
immunohistochemistry
- IR
ionizing radiation
- ITGA6
Integrin α 6
- KD
knockdown
- LDH
lactate dehydrogenase
- MID1IP1
midline 1 interacting protein 1
- NFκB
nuclear factor-κB
- NSCLC
non-small cell lung carcinoma/cancer
- Nx
Nexrutine
- PCa
prostate cancer
- RP
ribosomal protein
- RPS27L
RPS27-like
- RPSA
ribosomal protein A
- rRNA
ribosomal RNA
- siRNA
small interfering RNA
- SNHG7
small nucleolar RNA host gene 7
- SNORA18L5
small nucleolar RNA H/ACA box 18-like 5
- TNBC
triple-negative breast cancer
- TNM
tumor, nodes and metastasis
- T-ALL
T-cell acute lymphoblastic leukemia
- XIAP
X-linked inhibitor of apoptosis
- 5-FU
5-fluorouracil
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Author Contribution
W.E.K. performed the literature review, gathered the data and wrote the manuscript. Z.N. designed, reviewed and corrected the manuscript. The final version of the manuscript was reviewed and approved by both authors.
References
- 1.Sollner-Webb B. and Mougey E.B. (1991) News from the nucleolus: rRNA gene expression. Trends Biochem. Sci. 16, 58–62 10.1016/0968-0004(91)90025-Q [DOI] [PubMed] [Google Scholar]
- 2.Uechi T., Tanaka T. and Kenmochi N. (2001) A complete map of the human ribosomal protein genes: assignment of 80 genes to the cytogenetic map and implications for human disorders. Genomics 72, 223–230 10.1006/geno.2000.6470 [DOI] [PubMed] [Google Scholar]
- 3.Lafontaine D.L. and Tollervey D. (2001) The function and synthesis of ribosomes. Nat. Rev. Mol. Cell Biol. 2, 514–520 10.1038/35080045 [DOI] [PubMed] [Google Scholar]
- 4.Zhou X., Liao W.-J., Liao J.-M., Liao P. and Lu H. (2015) Ribosomal proteins: functions beyond the ribosome. J. Mol. Cell Biol. 7, 92–104 10.1093/jmcb/mjv014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Las Heras-Rubio A., Perucho L., Paciucci R., Vilardell J. and LLeonart M.E. (2014) Ribosomal proteins as novel players in tumorigenesis. Cancer Metastasis Rev. 33, 115–141 [DOI] [PubMed] [Google Scholar]
- 6.Penzo M., Montanaro L., Treré D. and Derenzini M. (2019) The ribosome biogenesis-cancer connection. Cells 8, 55 10.3390/cells8010055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goudarzi K.M. and Lindström M.S. (2016) Role of ribosomal protein mutations in tumor development (Review). Int. J. Oncol. 48, 1313–1324 10.3892/ijo.2016.3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dolezal J.M., Dash A.P. and Prochownik E.V. (2018) Diagnostic and prognostic implications of ribosomal protein transcript expression patterns in human cancers. BMC Cancer 18, 275 10.1186/s12885-018-4178-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chakraborty A., Uechi T. and Kenmochi N. (2011) Guarding the ‘translation apparatus’: defective ribosome biogenesis and the p53 signaling pathway. Wiley Interdiscip. Rev. RNA 2, 507–522 10.1002/wrna.73 [DOI] [PubMed] [Google Scholar]
- 10.Deisenroth C., Franklin D.A. and Zhang Y. (2016) The evolution of the ribosomal protein-MDM2-p53 pathway. Cold Spring Harb. Perspect. Med. 6, a026138 10.1101/cshperspect.a026138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ajore R. and Raiser D. (2017) Deletion of ribosomal protein genes is a common vulnerability in human cancer, especially in concert with TP53 mutations. EMBO Mol. Med. 9, 498–507 10.15252/emmm.201606660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang J., Brajanovski N., Chan K.T., Xuan J., Pearson R.B. and Sanij E. (2021) Ribosomal proteins and human diseases: molecular mechanisms and targeted therapy. Signal Transduct. Target. Ther. 6, 323 10.1038/s41392-021-00728-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oršolić I., Bursać S., Jurada D., Drmić Hofman I., Dembić Z., Bartek J.et al. (2020) Cancer-associated mutations in the ribosomal protein L5 gene dysregulate the HDM2/p53-mediated ribosome biogenesis checkpoint. Oncogene 10.1038/s41388-020-1231-6 [DOI] [PubMed] [Google Scholar]
- 14.Fonseca B.D.et al. (2014) The ever-evolving role of mTOR in translation. Sem. Cell Dev. Biol. 36C, 102–112 10.1016/j.semcdb.2014.09.014 [DOI] [PubMed] [Google Scholar]
- 15.Xiao Z., Zou Q., Liu Y.et al. (2016) Genome-wide assessment of differential translations with ribosome profiling data. Nat. Commun. 7, 11194 10.1038/ncomms11194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Molavi G., Samadi N., Hosseingholi E.Z.et al. (2019) The roles of moonlight ribosomal proteins in the development of human cancers: MOLAVI et al. J. Cell. Physiol. 234, 8327–8341 10.1002/jcp.27722 [DOI] [PubMed] [Google Scholar]
- 17.Lin Y.-M., Furukawa Y., Tsunoda T., Yue C.-T., Yang K.-C. and Nakamura Y. (2002) Molecular diagnosis of colorectal tumors by expression profiles of 50 genes expressed differentially in adenomas and carcinomas. Oncogene 21, 4120–4128 10.1038/sj.onc.1205518 [DOI] [PubMed] [Google Scholar]
- 18.Kitahara O., Furukawa Y., Tanaka T., Kihara C., Ono K., Yanagawa R.et al. (2001) Alterations of gene expression during colorectal carcinogenesis revealed by cDNA microarrays after laser-capture microdissection of tumor tissues and normal epithelia. Cancer Res. 61, 3544–3549 [PubMed] [Google Scholar]
- 19.Lau T.P., Roslani A.C., Lian L.H., Chai H.C., Lee P.C., Hilmi I.et al. (2014) Pair-wise comparison analysis of differential expression of mRNAs in early and advanced stage primary colorectal adenocarcinomas. BMJ Open 4, e004930 10.1136/bmjopen-2014-004930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo M.J. and Lai M.D. (2001) Identification of differentially expressed genes in normal mucosa, adenoma and adenocarcinoma of colon by SSH. World J. Gastroenterol. 7, 726–731 10.3748/wjg.v7.i5.726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J., Wei Y., Feng Q., Ren L., He G., Chang W.et al. (2016) Ribosomal protein S15A promotes malignant transformation and predicts poor outcome in colorectal cancer through misregulation of p53 signaling pathway. Int. J. Oncol. 48, 1628–1638 10.3892/ijo.2016.3366 [DOI] [PubMed] [Google Scholar]
- 22.Huang C.-J., Chien C.-C., Yang S.-H., Chang C.-C., Sun H.-L., Cheng Y.-C.et al. (2008) Faecal ribosomal protein L19 is a genetic prognostic factor for survival in colorectal cancer. J. Cell. Mol. Med. 12, 1936–1943 10.1111/j.1582-4934.2008.00253.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chester K.A., Robson L., Begent R.H.J., Talbot I.C., Pringle J.H., Primrose L.et al. (1989) Identification of a human ribosomal protein mRNA with increased expression in colorectal tumours. Biochim. Biophys. Acta Gene Struct. Expr. 1009, 297–300 10.1016/0167-4781(89)90119-X [DOI] [PubMed] [Google Scholar]
- 24.Kondoh N., Schweinfest C.W., Henderson K.W. and Papas T.S. (1992) Differential expression of S19 ribosomal protein, laminin-binding protein, and human lymphocyte antigen class I messenger RNAs associated with colon carcinoma progression and differentiation. Cancer Res. 52, 791–796 [PubMed] [Google Scholar]
- 25.Cao D., Luo Y., Qin S., Yu M., Mu Y., Ye G.et al. (2019) Metallopanstimulin-1 (MPS-1) mediates the promotion effect of leptin on colorectal cancer through activation of JNK/c-Jun signaling pathway. Cell Death Dis. 10, 655 10.1038/s41419-019-1911-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kasai H., Nadano D., Hidaka E., Higuchi K., Kawakubo M., Sato T.-A.et al. (2003) Differential expression of ribosomal proteins in human normal and neoplastic colorectum. J. Histochem. Cytochem. 51, 567–574 10.1177/002215540305100502 [DOI] [PubMed] [Google Scholar]
- 27.Dong Z., Jiang H., Liang S., Wang Y., Jiang W. and Zhu C. (2019) Ribosomal protein L15 is involved in colon carcinogenesis. Int. J. Med. Sci. 16, 1132–1141 10.7150/ijms.34386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barnard G.F., Staniunas R.J., Mori M., Puder M., Jessup M.J., Steele G.D. Jret al. (1993) Gastric and hepatocellular carcinomas do not overexpress the same ribosomal protein messenger RNAs as colonic carcinoma. Cancer Res. 53, 4048–4052 [PubMed] [Google Scholar]
- 29.Pogue-Geile K., Geiser J.R., Shu M., Miller C., Wool I.G., Meisler A.I.et al. (1991) Ribosomal protein genes are overexpressed in colorectal cancer: isolation of a cDNA clone encoding the human S3 ribosomal protein. Mol. Cell. Biol. 11, 3842–3849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alam E., Maaliki L. and Nasr Z. (2020) Ribosomal protein S3 selectively affects colon cancer growth by modulating the levels of p53 and lactate dehydrogenase. Mol. Biol. Rep. 47, 6083–6090 10.1007/s11033-020-05683-1 [DOI] [PubMed] [Google Scholar]
- 31.Iizumi Y., Oishi M., Taniguchi T., Goi W., Sowa Y. and Sakai T. (2013) The flavonoid apigenin downregulates CDK1 by directly targeting ribosomal protein S9. PLoS ONE 8, e73219 10.1371/journal.pone.0073219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong J.M., Mafune K., Yow H., Rivers E.N., Ravikumar T.S., Steele G.D. Jret al. (1993) Ubiquitin-ribosomal protein S27a gene overexpressed in human colorectal carcinoma is an early growth response gene. Cancer Res. 53, 1916–1920 [PubMed] [Google Scholar]
- 33.Huang C.-J., Yang S.-H., Lee C.-L., Cheng Y.-C., Tai S.-Y. and Chien C.-C. (2013) Ribosomal protein S27-like in colorectal cancer: A candidate for predicting prognoses. PLoS ONE 8, e67043 10.1371/journal.pone.0067043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baik I.H., Jo G.-H., Seo D., Ko M.J., Cho C.H., Lee M.G.et al. (2016) Knockdown of RPL9 expression inhibits colorectal carcinoma growth via the inactivation of Id-1/NF-κB signaling axis. Int. J. Oncol. 49, 1953–1962 10.3892/ijo.2016.3688 [DOI] [PubMed] [Google Scholar]
- 35.Liu J.-J., Zhang J., Ramanan S., Julian J., Carson D.D. and Hooi S.C. (2004) Heparin/heparan sulfate interacting protein plays a role in apoptosis induced by anticancer drugs. Carcinogenesis 25, 873–879 10.1093/carcin/bgh081 [DOI] [PubMed] [Google Scholar]
- 36.Liu J.-J., Huang B.H., Zhang J., Carson D.D. and Hooi S.C. (2006) Repression of HIP/RPL29 expression induces differentiation in colon cancer cells. J. Cell. Physiol. 207, 287–292 10.1002/jcp.20589 [DOI] [PubMed] [Google Scholar]
- 37.Chien C.-C., Tu T.-C., Huang C.-J., Yang S.-H. and Lee C.-L. (2012) Lowly expressed ribosomal protein s19 in the feces of patients with colorectal cancer. ISRN Gastroenterol. 2012, 394545 10.5402/2012/394545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sturm I., Köhne C.H., Wolff G., Petrowsky H., Hillebrand T., Hauptmann S.et al. (1999) Analysis of the p53/BAX pathway in colorectal cancer: low BAX is a negative prognostic factor in patients with resected liver metastases. J. Clin. Oncol. 17, 1364–1374 10.1200/JCO.1999.17.5.1364 [DOI] [PubMed] [Google Scholar]
- 39.Bandrés E., Malumbres R., Cubedo E., Honorato B., Zarate R., Labarga A.et al. (2007) A gene signature of 8 genes could identify the risk of recurrence and progression in Dukes' B colon cancer patients. Oncol. Rep. 17, 1089–1094 10.3892/or.17.5.1089 [DOI] [PubMed] [Google Scholar]
- 40.Zhang W., Tong D., Liu F., Li D., Li J., Cheng X.et al. (2016) RPS7 inhibits colorectal cancer growth via decreasing HIF-1α-mediated glycolysis. Oncotarget 7, 5800–5814 10.18632/oncotarget.6807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Russo A., Maiolino S., Pagliara V., Ungaro F., Tatangelo F., Leone A.et al. (2016) Enhancement of 5-FU sensitivity by the proapoptotic rpL3 gene in p53 null colon cancer cells through combined polymer nanoparticles. Oncotarget 7, 79670–79687 10.18632/oncotarget.13216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pecoraro A., Carotenuto P., Franco B., De Cegli R., Russo G. and Russo A. (2020) Role of uL3 in the crosstalk between nucleolar stress and autophagy in colon cancer cells. Int. J. Mol. Sci. 21, 2143 10.3390/ijms21062143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y., Sui J., Li X., Cao F., He J., Yang B.et al. (2015) RPS24 knockdown inhibits colorectal cancer cell migration and proliferation in vitro. Gene 571, 286–291 10.1016/j.gene.2015.06.084 [DOI] [PubMed] [Google Scholar]
- 44.Labriet A., Lévesque É., Cecchin E., De Mattia E., Villeneuve L., Rouleau M.et al. (2019) Germline variability and tumor expression level of ribosomal protein gene RPL28 are associated with survival of metastatic colorectal cancer patients. Sci. Rep. 9, 13008 10.1038/s41598-019-49477-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nieminen T.T., O'Donohue M.F., Wu Y., Lohi H., Scherer S.W., Paterson A.D.et al. (2014) Germline mutation of RPS20, encoding a ribosomal protein, causes predisposition to hereditary nonpolyposis colorectal carcinoma without DNA mismatch repair deficiency. Gastroenterology 147, 595.e5–598.e5 10.1053/j.gastro.2014.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arthurs C., Murtaza B.N., Thomson C., Dickens K., Henrique R., Patel H.R.H.et al. (2017) Expression of ribosomal proteins in normal and cancerous human prostate tissue. PLoS ONE 12, e0186047 10.1371/journal.pone.0186047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liang Z., Mou Q., Pan Z., Zhang Q., Gao G., Cao Y.et al. (2019) Identification of candidate diagnostic and prognostic biomarkers for human prostate cancer: RPL22L1 and RPS21. Med. Oncol. 36, 56 10.1007/s12032-019-1283-z [DOI] [PubMed] [Google Scholar]
- 48.Fan S., Liang Z., Gao Z., Pan Z., Han S., Liu X.et al. (2018) Identification of the key genes and pathways in prostate cancer. Oncol. Lett. 16, 6663–6669 10.3892/ol.2018.9491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maruyama Y., Miyazaki T., Ikeda K., Okumura T., Sato W., Horie-Inoue K.et al. (2014) Short hairpin RNA library-based functional screening identified ribosomal protein L31 that modulates prostate cancer cell growth via p53 pathway. PloS ONE 9, e108743 10.1371/journal.pone.0108743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang M., Hu Y., Amantagelo M.D. and Stearns M.E. (2012) Retraction: role of ribosomal protein RPS2 in controlling let-7a expression in human prostate cancer. Mol. Cancer Res. 10, 570 10.1158/1541-7786.MCR-12-0085 [DOI] [PubMed] [Google Scholar]
- 51.Hussain S.S., Huang S.-B., Bedolla R.G., Rivas P., Basler J.W., Swanson G.P.et al. (2018) Suppression of ribosomal protein RPS6KB1 by Nexrutine increases sensitivity of prostate tumors to radiation. Cancer Lett. 433, 232–241 10.1016/j.canlet.2018.07.009 [DOI] [PubMed] [Google Scholar]
- 52.Hambright H.G., Batth I.S., Xie J., Ghosh R. and Kumar A.P. (2015) Palmatine inhibits growth and invasion in prostate cancer cell: potential role for rpS6/NFκB/FLIP. Mol. Carcinog. 54, 1227–1234 10.1002/mc.22192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Evren S., Dermen A., Lockwood G., Fleshner N. and Sweet J. (2010) Immunohistochemical examination of the mTORC1 pathway in high grade prostatic intraepithelial neoplasia (HGPIN) and prostatic adenocarcinomas (PCa): a tissue microarray study (TMA). Prostate 70, 1429–1436 10.1002/pros.21178 [DOI] [PubMed] [Google Scholar]
- 54.Wen Y., An Z., Qiao B., Zhang C. and Zhang Z. (2019) RPS7 promotes cell migration through targeting epithelial-mesenchymal transition in prostate cancer. Urol. Oncol. 37, 297.e1–297.e7 10.1016/j.urolonc.2019.01.011 [DOI] [PubMed] [Google Scholar]
- 55.Zhang C., Qie Y., Yang T., Wang L., Du E., Liu Y.et al. (2019) Kinase PIM1 promotes prostate cancer cell growth via c-Myc-RPS7-driven ribosomal stress. Carcinogenesis 40, 52–60 10.1093/carcin/bgy126 [DOI] [PubMed] [Google Scholar]
- 56.Bee A., Ke Y., Forootan S., Lin K., Beesley C., Forrest S.E.et al. (2006) Ribosomal protein l19 is a prognostic marker for human prostate cancer. Clin. Cancer Res. 12, 2061–2065 10.1158/1078-0432.CCR-05-2445 [DOI] [PubMed] [Google Scholar]
- 57.Bee A., Brewer D., Beesley C., Dodson A., Forootan S., Dickinson T.et al. (2011) siRNA knockdown of ribosomal protein gene RPL19 abrogates the aggressive phenotype of human prostate cancer. PLoS ONE 6, e22672 10.1371/journal.pone.0022672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vaarala M.H., Porvari K.S., Kyllönen A.P., Mustonen M.V., Lukkarinen O. and Vihko P.T. (1998) Several genes encoding ribosomal proteins are over-expressed in prostate-cancer cell lines: confirmation of L7a and L37 over-expression in prostate-cancer tissue samples. Int. J. Cancer 78, 27–32 [DOI] [PubMed] [Google Scholar]
- 59.Karan D., Kelly D.L., Rizzino A., Lin M.-F. and Batra S.K. (2002) Expression profile of differentially-regulated genes during progression of androgen-independent growth in human prostate cancer cells. Carcinogenesis 23, 967–975 10.1093/carcin/23.6.967 [DOI] [PubMed] [Google Scholar]
- 60.Stehlik C., de Martin R., Kumabashiri I., Schmid J.A., Binder B.R. and Lipp J. (1998) Nuclear factor (NF)-kappaB-regulated X-chromosome-linked iap gene expression protects endothelial cells from tumor necrosis factor alpha-induced apoptosis. J. Exp. Med. 188, 211–216 10.1084/jem.188.1.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ono H., Iizumi Y., Goi W., Sowa Y., Taguchi T. and Sakai T. (2017) Ribosomal protein S3 regulates XIAP expression independently of the NF-κB pathway in breast cancer cells. Oncol. Rep. 38, 3205–3210 10.3892/or.2017.6008 [DOI] [PubMed] [Google Scholar]
- 62.Kong L., Wei Q., Hu X., Chen L. and Li J. (2020) Ribosomal protein small subunit 15A (RPS15A) inhibits the apoptosis of breast cancer MDA-MB-231 cells via upregulating phosphorylated ERK1/2, Bad, and Chk1. J. Cell. Biochem. 121, 587–595 10.1002/jcb.29304 [DOI] [PubMed] [Google Scholar]
- 63.Feng W., Liang C., Wang C., Yu X., Li Q. and Yang H. (2018) Knockdown of ribosomal protein S15A inhibits proliferation of breast cancer cells through induction of apoptosis in vitro. Cytotechnology 70, 1315–1323 10.1007/s10616-018-0221-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Markiewski M.M., Vadrevu S.K., Sharma S.K., Chintala N.K., Ghouse S., Cho J.-H.et al. (2017) The ribosomal protein S19 suppresses antitumor immune responses via the complement C5a receptor 1. J. Immunol. 198, 2989–2999 10.4049/jimmunol.1602057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dave B., Granados-Principal S., Zhu R., Benz S., Rabizadeh S., Soon-Shiong P.et al. (2014) Targeting RPL39 and MLF2 reduces tumor initiation and metastasis in breast cancer by inhibiting nitric oxide synthase signaling. Proc. Natl. Acad. Sci. U.S.A. 111, 8838–8843 10.1073/pnas.1320769111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yi Y.W., You K., Bae E.J., Kwak S.-J., Seong Y.-S. and Bae I. (2015) Dual inhibition of EGFR and MET induces synthetic lethality in triple-negative breast cancer cells through downregulation of ribosomal protein S6. Int. J. Oncol. 47, 122–132 10.3892/ijo.2015.2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wilson-Edell K.A., Kehasse A., Scott G.K., Yau C., Rothschild D.E., Schilling B.et al. (2014) RPL24: a potential therapeutic target whose depletion or acetylation inhibits polysome assembly and cancer cell growth. Oncotarget 5, 5165–5176 10.18632/oncotarget.2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hong M., Kim H. and Kim I. (2014) Ribosomal protein L19 overexpression activates the unfolded protein response and sensitizes MCF7 breast cancer cells to endoplasmic reticulum stress-induced cell death. Biochem. Biophys. Res. Commun. 450, 673–678 10.1016/j.bbrc.2014.06.036 [DOI] [PubMed] [Google Scholar]
- 69.Ebright R.Y., Lee S., Wittner B.S., Niederhoffer K.L., Nicholson B.T., Bardia A.et al. (2020) Deregulation of ribosomal protein expression and translation promotes breast cancer metastasis. Science 367, 1468–1473 10.1126/science.aay0939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu L., Wang L., Jiang C., Zhu Q., Chen R., Wang J.et al. (2020) Biological effect of ribosomal protein L32 on human breast cancer cell behavior. Mol. Med. Rep. 22, 2478–2486 10.3892/mmr.2020.11302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lin Z., Peng R., Sun Y., Zhang L. and Zhang Z. (2021) Identification of ribosomal protein family in triple-negative breast cancer by bioinformatics analysis. Biosci. Rep. 41, 10.1042/BSR20200869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fancello L., Kampen K.R., Hofman I.J.F., Verbeeck J. and De Keersmaecker K. (2017) The ribosomal protein gene RPL5 is a haploinsufficient tumor suppressor in multiple cancer types. Oncotarget 8, 14462–14478 10.18632/oncotarget.14895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xiong X., Liu X., Li H., He H., Sun Y. and Zhao Y. (2018) Ribosomal protein S27-like regulates autophagy via the β-TrCP-DEPTOR-mTORC1 axis. Cell Death Dis. 9, 1131 10.1038/s41419-018-1168-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kondoh N., Shuda M., Tanaka K., Wakatsuki T., Hada A. and Yamamoto M. (2001) Enhanced expression of S8, L12, L23a, L27 and L30 ribosomal protein mRNAs in human hepatocellular carcinoma. Anticancer Res. 21, 2429–2433 [PubMed] [Google Scholar]
- 75.Kowalczyk P., Woszczyński M. and Ostrowski J. (2002) Increased expression of ribosomal protein S2 in liver tumors, posthepactomized livers, and proliferating hepatocytes in vitro. Acta Biochim. Pol. 49, 615–624 10.18388/abp.2002_3770 [DOI] [PubMed] [Google Scholar]
- 76.Chen M., Wang D., Liu J., Zhou Z., Ding Z., Liu L.et al. (2020) MicroRNA-587 functions as a tumor suppressor in hepatocellular carcinoma by targeting ribosomal protein SA. Biomed Res. Int. 2020, 3280530 10.1155/2020/3280530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wong Q.W.-L., Li J., Ng S.R., Lim S.G., Yang H. and Vardy L.A. (2014) RPL39L is an example of a recently evolved ribosomal protein paralog that shows highly specific tissue expression patterns and is upregulated in ESCs and HCC tumors. RNA Biol. 11, 33–41 10.4161/rna.27427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Song M.J., Jung C.K., Park C.-H., Hur W., Choi J.E., Bae S.H.et al. (2011) RPL36 as a prognostic marker in hepatocellular carcinoma: RPL36 expression in HCC. Pathol. Int. 61, 638–644 10.1111/j.1440-1827.2011.02716.x [DOI] [PubMed] [Google Scholar]
- 79.Bi N., Sun Y., Lei S., Zeng Z., Zhang Y., Sun C.et al. (2020) Identification of 40S ribosomal protein S8 as a novel biomarker for alcohol-associated hepatocellular carcinoma using weighted gene co-expression network analysis. Oncol. Rep. 44, 611–627 10.3892/or.2020.7634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li W., Tan D., Zhang Z., Liang J.J. and Brown R.E. (2008) Activation of Akt-mTOR-p70S6K pathway in angiogenesis in hepatocellular carcinoma. Oncol. Rep. 20, 713–719 [PubMed] [Google Scholar]
- 81.Xie X., Guo P., Yu H., Wang Y. and Chen G. (2018) Ribosomal proteins: insight into molecular roles and functions in hepatocellular carcinoma. Oncogene 37, 277–285 10.1038/onc.2017.343 [DOI] [PubMed] [Google Scholar]
- 82.Calvisi D.F., Wang C., Ho C., Ladu S., Lee S.A., Mattu S.et al. (2011) Increased lipogenesis, induced by AKT-mTORC1-RPS6 signaling, promotes development of human hepatocellular carcinoma. Gastroenterology 140, 1071–1083 10.1053/j.gastro.2010.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhou C., Liu C., Liu W., Chen W., Yin Y., Li C.-W.et al. (2020) SLFN11 inhibits hepatocellular carcinoma tumorigenesis and metastasis by targeting RPS4X via mTOR pathway. Theranostics 10, 4627–4643 10.7150/thno.42869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xu M., Wang Y., Chen L., Pan B., Chen F., Fang Y.et al. (2014) Down-regulation of ribosomal protein S15A mRNA with a short hairpin RNA inhibits human hepatic cancer cell growth in vitro. Gene 536, 84–89 10.1016/j.gene.2013.11.075 [DOI] [PubMed] [Google Scholar]
- 85.Guo P., Wang Y., Dai C., Tao C., Wu F., Xie X.et al. (2018) Ribosomal protein S15a promotes tumor angiogenesis via enhancing Wnt/β-catenin-induced FGF18 expression in hepatocellular carcinoma. Oncogene 37, 1220–1236 10.1038/s41388-017-0017-y [DOI] [PubMed] [Google Scholar]
- 86.Zhao L., Cao J., Hu K., Wang P., Li G., He X.et al. (2019) RNA-binding protein RPS3 contributes to hepatocarcinogenesis by post-transcriptionally up-regulating SIRT1. Nucleic Acids Res. 47, 2011–2028 10.1093/nar/gky1209 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 87.Yoon S.Y., Kim J.-M., Oh J.-H., Jeon Y.-J., Lee D.-S., Kim J.H.et al. (2006) Gene expression profiling of human HBV- and/or HCV-associated hepatocellular carcinoma cells using expressed sequence tags. Int. J. Oncol. 29, 315–327 10.3892/ijo.29.2.315 [DOI] [PubMed] [Google Scholar]
- 88.Yang X., Sun L., Wang L., Yao B., Mo H. and Yang W. (2019) LncRNA SNHG7 accelerates the proliferation, migration and invasion of hepatocellular carcinoma cells via regulating miR-122-5p and RPL4. Biomed. Pharmacother. 118, 109386 10.1186/s12885-018-4178-z [DOI] [PubMed] [Google Scholar]
- 89.Tang H., Oishi N., Kaneko S. and Murakami S. (2006) Molecular functions and biological roles of hepatitis B virus x protein. Cancer Sci. 97, 977–983 10.1111/j.1349-7006.2006.00299.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kim M.Y., Park E., Park J.H., Park D.H., Moon W.S., Cho B.H.et al. (2001) Expression profile of nine novel genes differentially expressed in hepatitis B virus-associated hepatocellular carcinomas. Oncogene 20, 4568–4575 10.1038/sj.onc.1204626 [DOI] [PubMed] [Google Scholar]
- 91.Lian Z., Liu J., Li L., Li X., Tufan N.L.S., Wu M.-C.et al. (2004) Human S15a expression is upregulated by hepatitis B virus X protein: S15a is upregulated by HBx. Mol. Carcinog. 40, 34–46 10.1002/mc.20012 [DOI] [PubMed] [Google Scholar]
- 92.Lee C.-F., Ling Z.-Q., Zhao T. and Lee K.-R. (2008) Distinct expression patterns in hepatitis B virus- and hepatitis C virus-infected hepatocellular carcinoma. World J. Gastroenterol. 14, 6072–6077 10.3748/wjg.14.6072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lim K.-H., Kim K.-H., Choi S.I., Park E.-S., Park S.H., Ryu K.et al. (2011) RPS3a over-expressed in HBV-associated hepatocellular carcinoma enhances the HBx-induced NF-κB signaling via its novel chaperoning function. PLoS ONE 6, e22258 10.1371/journal.pone.0022258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fatima G., Mathan G. and Kumar V. (2012) The HBx protein of hepatitis B virus regulates the expression, intracellular distribution and functions of ribosomal protein S27a. J. Gen. Virol. 93, 706–715 10.1099/vir.0.035691-0 [DOI] [PubMed] [Google Scholar]
- 95.Gunasekaran V.P. and Ganeshan M. (2014) Inverse correlation of ribosomal protein S27A and multifunctional protein YB-1 in hepatocellular carcinoma. Clin. Biochem. 47, 1262–1264 10.1016/j.clinbiochem.2014.05.004 [DOI] [PubMed] [Google Scholar]
- 96.Cao P., Yang A., Wang R., Xia X., Zhai Y., Li Y.et al. (2018) Germline duplication of SNORA18L5 increases risk for HBV-related hepatocellular carcinoma by altering localization of ribosomal proteins and decreasing levels of p53. Gastroenterology 155, 542–556 10.1053/j.gastro.2018.04.020 [DOI] [PubMed] [Google Scholar]
- 97.Jung J.H., Lee H.-J., Kim J.-H., Sim D.Y., Im E., Kim S.et al. (2020) Colocalization of MID1IP1 and c-Myc is critically involved in liver cancer growth via regulation of ribosomal protein L5 and L11 and CNOT2. Cells 9, 985 10.3390/cells9040985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wu Y., Tan X., Liu P., Yang Y., Huang Y., Liu X.et al. (2019) ITGA6 and RPSA synergistically promote pancreatic cancer invasion and metastasis via PI3K and MAPK signaling pathways. Exp. Cell. Res. 379, 30–47 10.1016/j.yexcr.2019.03.022 [DOI] [PubMed] [Google Scholar]
- 99.Lefebvre T., Rybarczyk P., Bretaudeau C., Vanlaeys A., Cousin R., Brassart-Pasco S.et al. (2020) TRPM7/RPSA complex regulates pancreatic cancer cell migration. Front. Cell Dev. Biol. 8, 549 10.3389/fcell.2020.00549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wei F., Ding L., Wei Z., Zhang Y., Li Y., Qinghua L.et al. (2016) Ribosomal protein L34 promotes the proliferation, invasion and metastasis of pancreatic cancer cells. Oncotarget 7, 85259–85272 10.18632/oncotarget.13269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liang J., Liu Y., Zhang L., Tan J., Li E. and Li F. (2019) Overexpression of microRNA-519d-3p suppressed the growth of pancreatic cancer cells by inhibiting ribosomal protein S15A-mediated Wnt/β-catenin signaling. Chem. Biol. Interact. 304, 1–9 10.1016/j.cbi.2019.02.026 [DOI] [PubMed] [Google Scholar]
- 102.Li C., Ge M., Yin Y., Luo M. and Chen D. (2012) Silencing expression of ribosomal protein L26 and L29 by RNA interfering inhibits proliferation of human pancreatic cancer PANC-1 cells. Mol. Cell. Biochem. 370, 127–139 10.1007/s11010-012-1404-x [DOI] [PubMed] [Google Scholar]
- 103.Muro S., Miyake Y., Kato H., Tsutsumi K. and Yamamoto K. (2015) Serum anti-60S ribosomal protein L29 antibody as a novel prognostic marker for unresectable pancreatic cancer. Digestion 91, 164–173 10.1159/000371545 [DOI] [PubMed] [Google Scholar]
- 104.Khalaileh A., Dreazen A., Khatib A., Apel R., Swisa A., Kidess-Bassir N.et al. (2013) Phosphorylation of ribosomal protein S6 attenuates DNA damage and tumor suppression during development of pancreatic cancer. Cancer Res. 73, 1811–1820 10.1158/0008-5472.CAN-12-2014 [DOI] [PubMed] [Google Scholar]
- 105.Li C., Chen D., Luo M., Ge M. and Zhu J. (2014) Knockdown of ribosomal protein L39 by RNA interference inhibits the growth of human pancreatic cancer cells in vitro and in vivo. Biotechnol. J. 9, 652–663 10.1002/biot.201300321 [DOI] [PubMed] [Google Scholar]
- 106.Shi C., Wang Y., Guo Y., Chen Y. and Liu N. (2017) Cooperative down-regulation of ribosomal protein L10 and NF-κB signaling pathway is responsible for the anti-proliferative effects by DMAPT in pancreatic cancer cells. Oncotarget 8, 35009–35018 10.18632/oncotarget.16557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chen R., Dawson D.W., Pan S., Ottenhof N.A., de Wilde R.F., Wolfgang C.L.et al. (2015) Proteins associated with pancreatic cancer survival in patients with resectable pancreatic ductal adenocarcinoma. Lab. Invest. 95, 43–55 10.1038/labinvest.2014.128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yan T.-T., Fu X.-L., Li J., Bian Y.-N., Liu D.J., Hua R.et al. (2015) Downregulation of RPL15 may predict poor survival and associate with tumor progression in pancreatic ductal adenocarcinoma. Oncotarget 6, 37028–37042 10.18632/oncotarget.5939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li C., Ge M., Chen D., Sun T., Jiang H., Xie Y.et al. (2020) RPL21 siRNA blocks proliferation in pancreatic cancer cells by inhibiting DNA replication and inducing G1 arrest and apoptosis. Front. Oncol. 10, 1730 10.3389/fonc.2020.01730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shi Y., Zhai H., Wang X., Han Z., Liu C., Lan M.et al. (2004) Ribosomal proteins S13 and L23 promote multidrug resistance in gastric cancer cells by suppressing drug-induced apoptosis. Exp. Cell. Res. 296, 337–346 10.1016/j.yexcr.2004.02.009 [DOI] [PubMed] [Google Scholar]
- 111.Yang Z.-Y., Jiang H., Qu Y., Wei M., Yan M., Zhu Z.-G.et al. (2013) Metallopanstimulin-1 regulates invasion and migration of gastric cancer cells partially through integrin β4. Carcinogenesis 34, 2851–2860 10.1093/carcin/bgt226 [DOI] [PubMed] [Google Scholar]
- 112.Wu Q., Gou Y., Wang Q., Jin H., Cui L., Zhang Y.et al. (2011) Downregulation of RPL6 by siRNA inhibits proliferation and cell cycle progression of human gastric cancer cell lines. PLoS ONE 6, e26401 10.1371/journal.pone.0026401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kobayashi T., Sasaki Y., Oshima Y., Yamamoto H., Mita H., Suzuki H.et al. (2006) Activation of the ribosomal protein L13 gene in human gastrointestinal cancer. Int. J. Mol. Med. 18, 161–170 10.3892/ijmm.18.1.161 [DOI] [PubMed] [Google Scholar]
- 114.Wang H., Zhao L.-N., Li K.-Z., Ling R., Li X.-J. and Wang L. (2006) Overexpression of ribosomal protein L15 is associated with cell proliferation in gastric cancer. BMC Cancer 6, 91 10.1186/1471-2407-6-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Guo X., Shi Y., Gou Y., Li J., Han S., Zhang Y.et al. (2011) Human ribosomal protein S13 promotes gastric cancer growth through down-regulating p27(Kip1). J. Cell. Mol. Med. 15, 296–306 10.1111/j.1582-4934.2009.00969.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gou Y., Shi Y., Zhang Y., Nie Y., Wang J., Song J.et al. (2010) Ribosomal protein L6 promotes growth and cell cycle progression through upregulating cyclin E in gastric cancer cells. Biochem. Biophys. Res. Commun. 393, 788–793 10.1016/j.bbrc.2010.02.083 [DOI] [PubMed] [Google Scholar]
- 117.Shi D. and Liu J. (2018) RPS15a silencing suppresses cell proliferation and migration of gastric cancer. Yonsei Med. J. 59, 1166–1173 10.3349/ymj.2018.59.10.1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Liu C., He X., Liu X., Yu J., Zhang M., Yu F.et al. (2019) RPS15A promotes gastric cancer progression via activation of the Akt/IKK-β/NF-κB signalling pathway. J. Cell. Mol. Med. 23, 2207–2218 10.1111/jcmm.14141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hsu Y.-A., Lin H.-J., Sheu J.J.C., Shieh F.-K., Chen S.-Y., Lai C.-H.et al. (2011) A novel interaction between interferon-inducible protein p56 and ribosomal protein L15 in gastric cancer cells. DNA Cell Biol. 30, 671–679 10.1089/dna.2010.1149 [DOI] [PubMed] [Google Scholar]
- 120.Kawahata T., Kawahara K., Shimokawa M., Sakiyama A., Shiraishi T., Minami K.et al. (2020) Involvement of ribosomal protein L11 expression in sensitivity of gastric cancer against 5-FU. Oncol. Lett. 19, 2258–2264 10.3892/ol.2020.11352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gambardella V., Gimeno-Valiente F., Tarazona N., Martinez-Ciarpaglini C., Roda D., Fleitas T.et al. (2019) NRF2 through RPS6 activation is related to anti-HER2 drug resistance in HER2-amplified gastric cancer. Clin. Cancer Res. 25, 1639–1649 10.1158/1078-0432.CCR-18-2421 [DOI] [PubMed] [Google Scholar]
- 122.Dlamini Z. and Mphahlele L. (2006) Molecular evaluation of ribosomal protein L9 gene (RPL9) in lung cancer. Cancer Res. 66, 77016424008 [Google Scholar]
- 123.Slizhikova D.K., Vinogradova T.V. and Sverdlov E.D. (2005) The NOLA2 and RPS3A genes as highly informative markers for human squamous cell lung cancer. Bioorg. Khim 31, 195–199 [DOI] [PubMed] [Google Scholar]
- 124.Yang S., Cui J., Yang Y., Liu Z., Yan H., Tang C.et al. (2016) Over-expressed RPL34 promotes malignant proliferation of non-small cell lung cancer cells. Gene 576, 421–428 10.1016/j.gene.2015.10.053 [DOI] [PubMed] [Google Scholar]
- 125.Chen B., Tan Z., Gao J., Wu W., Liu L., Jin W.et al. (2015) Hyperphosphorylation of ribosomal protein S6 predicts unfavorable clinical survival in non-small cell lung cancer. J. Exp. Clin. Cancer Res. 34, 126 10.1186/s13046-015-0239-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yang M., Sun H., Wang H., Zhang S., Yu X. and Zhang L. (2013) Down-regulation of ribosomal protein L22 in non-small cell lung cancer. Med. Oncol. 30, 646 10.1007/s12032-013-0646-0 [DOI] [PubMed] [Google Scholar]
- 127.Kuroda K., Takenoyama M., Baba T., Shigematsu Y., Shiota H., Ichiki Y.et al. (2010) Identification of ribosomal protein L19 as a novel tumor antigen recognized by autologous cytotoxic T lymphocytes in lung adenocarcinoma. Cancer Sci. 101, 46–53 10.1111/j.1349-7006.2009.01351.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhao X., Shen L., Feng Y., Yu H., Wu X., Chang J.et al. (2015) Decreased expression of RPS15A suppresses proliferation of lung cancer cells. Tumour Biol. 36, 6733–6740 10.1007/s13277-015-3371-9 [DOI] [PubMed] [Google Scholar]
- 129.Yang H.J., Youn H., Seong K.M., Jin Y.-W., Kim J. and Youn B. (2013) Phosphorylation of ribosomal protein S3 and antiapoptotic TRAF2 protein mediates radioresistance in non-small cell lung cancer cells. J. Biol. Chem. 288, 2965–2975 10.1074/jbc.M112.385989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Youn H., Son B., Kim W., Jun S.Y., Lee J.S., Lee J.-M.et al. (2015) Dissociation of MIF-rpS3 complex and sequential NF-κB activation is involved in IR-induced metastatic conversion of NSCLC: METASTATIC CONVERSION BY MIF-rpS3DISSOCIATION. J. Cell. Biochem. 116, 2504–2516 10.1002/jcb.25195 [DOI] [PubMed] [Google Scholar]
- 131.Khanna N., Sen S., Sharma H. and Singh N. (2003) S29 ribosomal protein induces apoptosis in H520 cells and sensitizes them to chemotherapy. Biochem. Biophys. Res. Commun. 304, 26–35 10.1016/S0006-291X(03)00532-1 [DOI] [PubMed] [Google Scholar]
- 132.Russo A., Saide A., Cagliani R., Cantile M., Botti G. and Russo G. (2016) rpL3 promotes the apoptosis of p53 mutated lung cancer cells by down-regulating CBS and NFκB upon 5-FU treatment. Sci. Rep. 6, 38369 10.1038/srep38369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sun S., He H., Ma Y., Xu J., Chen G., Sun Y.et al. (2020) Inactivation of ribosomal protein S27-like impairs DNA interstrand cross-link repair by destabilization of FANCD2 and FANCI. Cell Death Dis. 11, 852 10.1038/s41419-020-03082-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Fan H., Li J., Jia Y., Wu J., Yuan L., Li M.et al. (2017) Silencing of ribosomal protein L34 (RPL34) inhibits the proliferation and invasion of esophageal cancer cells. Oncol. Res. 25, 1061–1068 10.3727/096504016X14830466773541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Liang J., Liu Z., Zou Z., Wang X., Tang Y., Zhou C.et al. (2019) Knockdown of ribosomal protein S15A inhibits human kidney cancer cell growth in vitro and in vivo. Mol. Med. Rep. 19, 1117–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.De Keersmaecker K., Atak Z.K., Li N., Vicente C., Patchett S., Girardi T.et al. (2013) Exome sequencing identifies mutation in CNOT3 and ribosomal genes RPL5 and RPL10 in T-cell acute lymphoblastic leukemia. Nat. Genet. 45, 186–190 10.1038/ng.2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kampen K.R., Sulima S.O., Verbelen B., Girardi T., Vereecke S., Rinaldi G.et al. (2019) The ribosomal RPL10 R98S mutation drives IRES-dependent BCL-2 translation in T-ALL. Leukemia 33, 319–332 10.1038/s41375-018-0176-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sim E.U.-H., Ng K.-L., Lee C.-W. and Narayanan K. (2017) The uS8, uS4, eS31, and uL14 ribosomal protein genes are dysregulated in nasopharyngeal carcinoma cell lines. Biomed Res. Int. 2017, 1–8 10.1155/2017/4876954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang Z., Hou J., Lu L., Qi Z., Sun J., Gao W.et al. (2013) Small ribosomal protein subunit S7 suppresses ovarian tumorigenesis through regulation of the PI3K/AKT and MAPK pathways. PLoS ONE 8, e79117 10.1371/journal.pone.0079117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Shirakawa Y., Hide T., Yamaoka M., Ito Y., Ito N., Ohta K.et al. (2020) Ribosomal protein S6 promotes stem-like characters in glioma cells. Cancer Sci. 111, 2041–2051 10.1111/cas.14399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wang C.-H., Wang L.-K., Wu C.-C., Chen M.-L., Lee M.-C., Lin Y.-Y.et al. (2019) The ribosomal protein RPLP0 mediates PLAAT4-induced cell cycle arrest and cell apoptosis. Cell Biochem. Biophys. 77, 253–260 10.1007/s12013-019-00876-3 [DOI] [PubMed] [Google Scholar]