Significance
Trophoblast cell–guided uterine spiral artery remodeling is a key event in successful hemochorial placentation. Connections between coagulopathies and diseases of placentation are compelling. Tissue factor pathway inhibitor (TFPI) is a prominent regulator of blood coagulation and an intriguing constituent of trophoblast cells situated at the uterine–placental interface. The actions of TFPI extend beyond controlling hemostasis and directly affect trophoblast cell development. TFPI facilitates the differentiation of rat and human trophoblast stem cells into the invasive/extravillous trophoblast cell lineage and promotes intrauterine trophoblast invasion and trophoblast-guided uterine spiral artery remodeling at the maternal–fetal interface. Thus, TFPI is a conserved regulator of a fundamental event determining the efficacy of the hemochorial placenta.
Keywords: placenta, hemostasis, trophoblast cell, uterine spiral artery
Abstract
Hemochorial placentation is characterized by the development of trophoblast cells specialized to interact with the uterine vascular bed. We utilized trophoblast stem (TS) cell and mutant rat models to investigate regulatory mechanisms controlling trophoblast cell development. TS cell differentiation was characterized by acquisition of transcript signatures indicative of an endothelial cell-like phenotype, which was highlighted by the expression of anticoagulation factors including tissue factor pathway inhibitor (TFPI). TFPI localized to invasive endovascular trophoblast cells of the rat placentation site. Disruption of TFPI in rat TS cells interfered with development of the endothelial cell-like endovascular trophoblast cell phenotype. Similarly, TFPI was expressed in human invasive/extravillous trophoblast (EVT) cells situated within first-trimester human placental tissues and following differentiation of human TS cells. TFPI was required for human TS cell differentiation to EVT cells. We next investigated the physiological relevance of TFPI at the placentation site. Genome-edited global TFPI loss-of-function rat models revealed critical roles for TFPI in embryonic development, resulting in homogeneous midgestation lethality prohibiting analysis of the role of TFPI as a regulator of the late-gestation wave of intrauterine trophoblast cell invasion. In vivo trophoblast-specific TFPI knockdown was compatible with pregnancy but had profound effects at the uterine–placental interface, including restriction of the depth of intrauterine trophoblast cell invasion while leading to the accumulation of natural killer cells and increased fibrin deposition. Collectively, the experimentation implicates TFPI as a conserved regulator of invasive/EVT cell development, uterine spiral artery remodeling, and hemostasis at the maternal–fetal interface.
The placenta develops in concert with the embryo to manage the environment in which the embryo develops (1, 2). Hemochorial placentation, as observed in humans, rats, and mice, is a process whereby extraembryonic cells, referred to as invasive trophoblast cells, breach the uterine parenchyma, permitting maternal blood direct access to the trophoblast–embryonic barrier (3–7). This critical event requires the acquisition of specialized trophoblast cell properties that facilitate intravasation into the uterine vasculature and orchestration of maternal adaptive responses ensuring the effective redirection of nutrients to the developing embryo (8, 9). The extent of trophoblast penetration into the uterus varies among species. Rats and humans possess deep placentation with extensive intrauterine trophoblast cell invasion (10–12), whereas mice are among species with shallow placentation and limited intrauterine trophoblast cell invasion (11, 13, 14). In humans, failures in trophoblast cell restructuring of the maternal environment are at the core of pregnancy-related diseases such as miscarriage, preeclampsia, intrauterine growth restriction, and preterm birth (15). Elucidating mechanisms underlying the differentiation of trophoblast stem (TS) and progenitor cells into trophoblast cells with specialized invasive properties is needed to better understand and develop treatment for these diseases.
The goal of this report was to identify conserved regulators of invasive trophoblast cell development and deep hemochorial placentation. The research approach took advantage of similarities in deep intrauterine trophoblast cell invasion observed in rats and humans (10, 12). A key component of the investigation was the utilization of rat and human TS cells (16, 17) to identify candidate regulators and to test their in vivo efficacy in genetically manipulated rats. We identified tissue factor pathway inhibitor (TFPI) expression as a prominent feature of differentiating TS cells and demonstrated its involvement in the development of the invasive trophoblast cell lineage and trophoblast cell–guided uterine spiral artery remodeling. TFPI is a Kunitz domain–containing protease inhibitor and critical regulator of blood coagulation (18–20). Until now, the contributions of TFPI to the regulation of pregnancy and placentation were largely unknown.
Results
Differentiating Rat TS Cells Exhibit an Endothelial Cell–Like Phenotype.
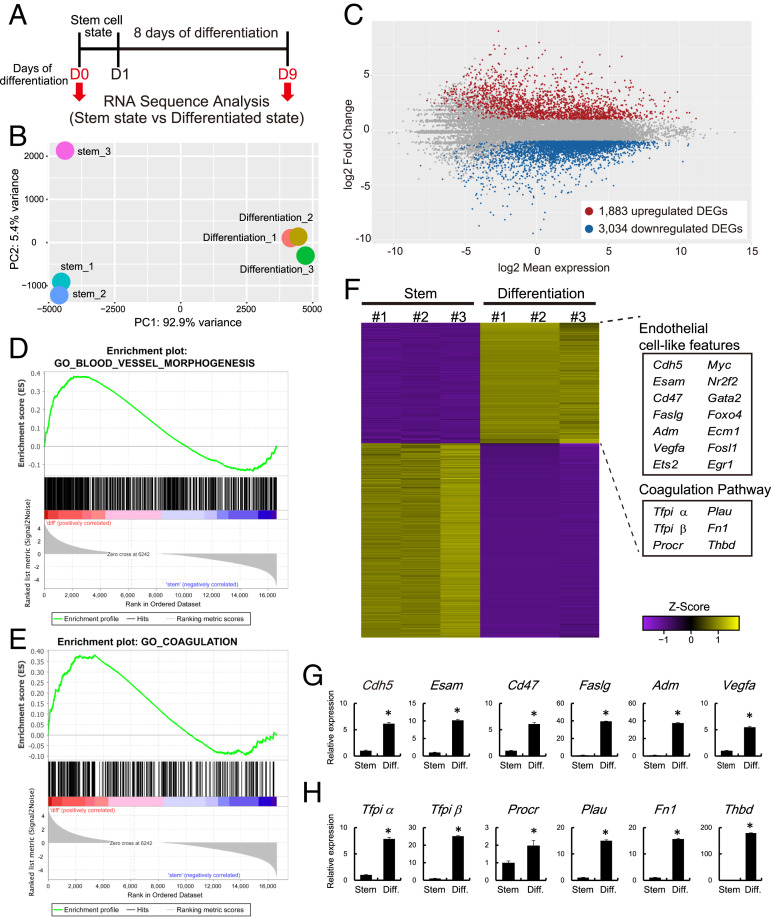
As a first step in the identification of regulatory events controlling trophoblast cell differentiation, we examined the transcriptomes of rat TS cells maintained in the stem state or induced to differentiate via mitogen withdrawal (Fig. 1A). RNA sequencing (RNA-seq) identified 4,917 differentially expressed genes (DEGs), including 1,883 transcripts up-regulated and 3,034 transcripts down-regulated in differentiated TS cells (Fig. 1 B and C and Dataset S1). TS cell stem and differentiation states exhibited expected trophoblast signatures (e.g., stem state: Cdh1, Cldn4, Phlda2, Id2, Bmp4, Esrrb, Bambi, Pcsk6, Esrp1, and Glut3; differentiated state: Prl2a1, Prl4a1, Hsd17b2, Pgf, Tgfb3, and Ets2), which were validated by RT-qPCR (SI Appendix, Fig. S1). Interestingly, the TS cell differentiation state was also characterized by the up-regulation of transcripts characteristic of an endothelial cell–like phenotype (Fig. 1 D–F), including the expression of endothelial cell–adhesion molecules (e.g., Cdh5, Esam, Ecm1, and Cd47), cytokines/growth factors (e.g., Faslg, Adm, and Vegfa), endothelial cell–associated transcription factors (e.g., Gata2, Foxo4, Nr2f2, Egr1, Myc), and components of the coagulation pathway (e.g., Tfpi, Procr, Plau, Fn1, and Thbd; Fig. 1F), which were validated by RT-qPCR (Fig. 1 G and H and SI Appendix, Fig. S1). Tfpi alpha and beta transcripts encode secreted and membrane-linked TFPI proteins, respectively (19). Acquisition of endothelial cell–like features is consistent with previous reports for differentiating mouse TS cells (21) and human trophoblast cells (22, 23).
Fig. 1.
Transcriptome analysis of rat TS cells maintained in stem and differentiation states. (A) Schematic representation of RNA-seq analysis for stem and differentiated rat TS cells. Differentiation was induced by mitogen withdrawal. (B) PCA plot of stem and differentiated rat TS cells. (C) Bland-Altman plot showing the global transcriptomic changes in differentiated rat TS cells. Colored dots indicate DEGs (≥2-fold with a false discovery rate of P < 0.05; red: up-regulated, blue: down-regulated). (D and E) GSEA of DEGs associated with rat TS cells in the stem and differentiated states. Results for “Blood vessel morphogenesis” (D) and the “Coagulation” (E) gene sets are shown. (F) Heatmap showing the expression patterns of stem and differentiated rat TS cells. These DEGs are the same genes as those shown in C. Z score-transformed RPKM are shown. The endothelial cell–associated genes and the coagulation regulatory factors were up-regulated in differentiated rat TS cells. (G) RT-qPCR transcript validation for endothelial cell–associated genes. (H) RT-qPCR transcripts validation for coagulation regulatory factors. Asterisks in G and H denote P < 0.05.
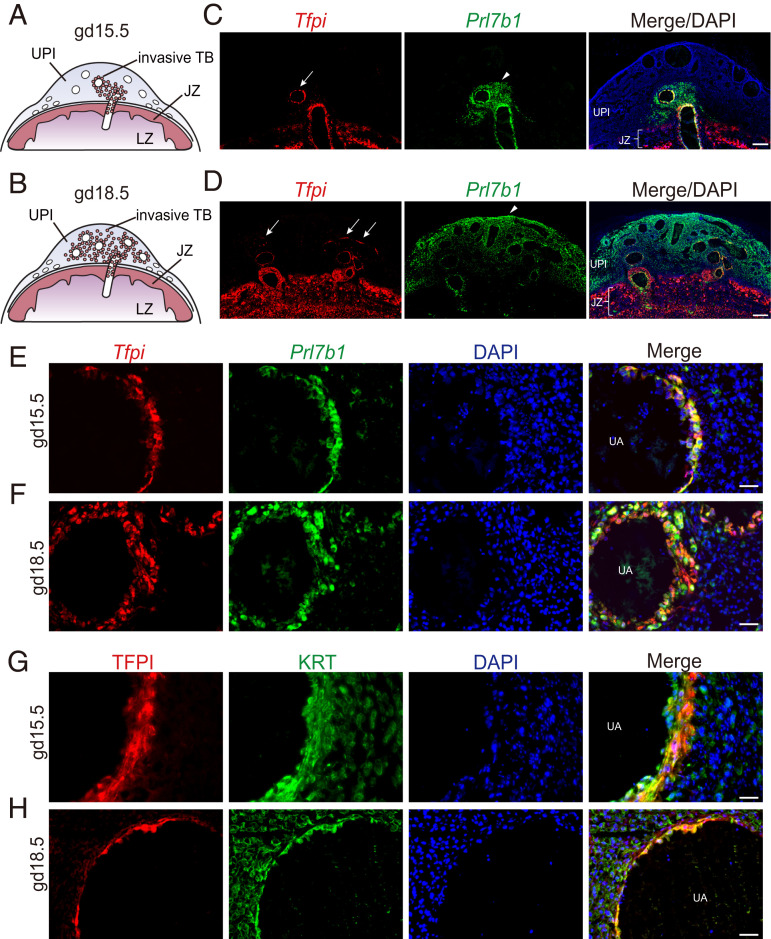
The established ties between coagulopathies and pregnancy-related diseases (24–27) drew our attention to transcripts encoding components of the coagulation regulatory cascade. Tfpi transcripts were among the most highly up-regulated transcripts accompanying TS cell differentiation. In the developing rat placenta, Tfpi transcripts were colocalized with prolactin family 7, subfamily b, member 1 (Prl7b1) transcripts, a known marker of invasive trophoblast cells in rats (28), to endovascular invasive trophoblast cells and, to a lesser extent, to interstitial invasive trophoblast cells (Fig. 2 A–F). Tfpi transcripts were also expressed in trophoblast cells within the junctional zone, including trophoblast giant cells and trophoblast cells lining channels in which maternal blood transits the junctional zone, and the labyrinth zone of the rat placenta (Fig. 2D and SI Appendix, Fig. S2 B and C). Thus, TFPI is strategically positioned throughout the placenta, including the labyrinth zone, to facilitate blood flow. TFPI protein was identified in endovascular invasive trophoblast cells (Fig. 2 G and H) and trophoblast giant cells (SI Appendix, Fig. S2C). The presence of TFPI in endovascular invasive trophoblast cells placed it in a compelling location for regulating uterine spiral artery adaptations to pregnancy.
Fig. 2.
Localization of TFPI within the rat uterine-placental interface (UPI). (A and B) Schematic representations of gd 15.5 (A) and 18.5 (B) placentation sites, consisting of the junctional zone (JZ), the labyrinth zone (LZ), the UPI, and invasive trophoblast cells (TB). (C and D) Detection of Tfpi (red) and Prl7b1 (green) transcripts by in situ hybridization within gd 15.5 (C) and 18.5 (D) placentation sites. (Scale bar: 500 μm.) DAPI marks cell nuclei (blue). Arrows in the Tfpi images show localization associated with uterine spiral arterioles, and arrowheads in the Prl7b1 images demarcate the depth of intrauterine trophoblast cell invasion. (E and F) Higher-magnification images of Tfpi (red) and Prl7b1 (green) transcripts detected by in situ hybridization at the gd 15.5 (E) and 18.5 (F) uterine–placental interface. (Scale bar: 50 μm.) (G and H) Immunohistochemical detection of TFPI (red) and cytokeratin (KRT, green) proteins within endovascular trophoblast cells of gd 15.5 (G) and 18.5 (H) placentation sites. (Scale bar: 50 μm.)
TFPI Is an Intrinsic Regulator of the Trophoblast Endothelial Cell–Like Phenotype.
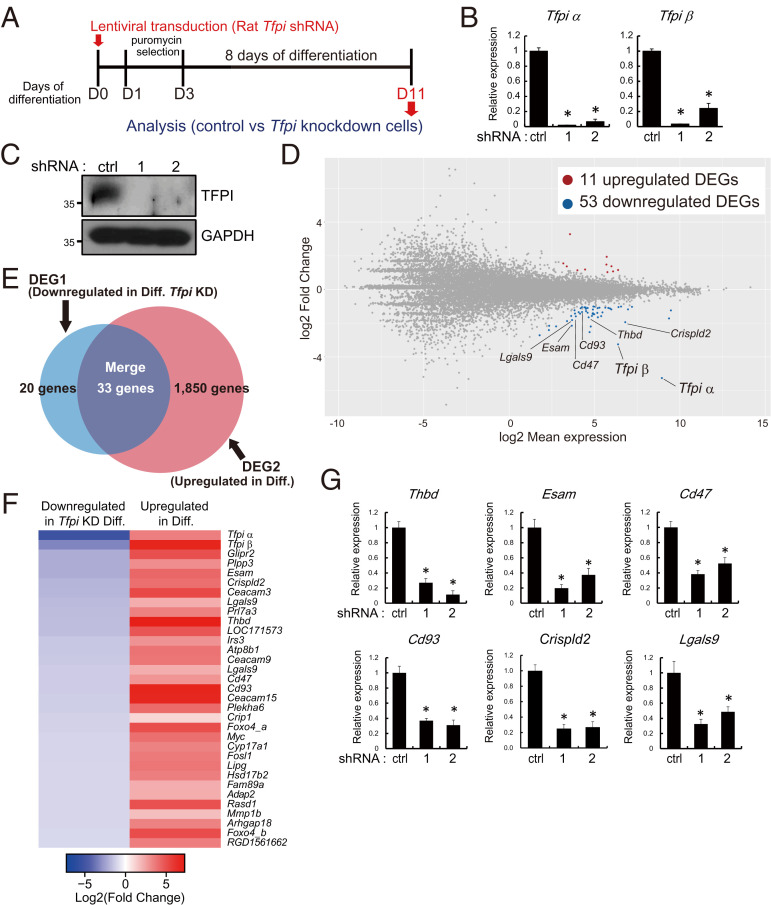
A cell-autonomous role for TFPI in regulating the migration of endothelial cells and angiogenesis has been described (29–32). Accordingly, we next used a loss-of-function approach to investigate a role for TFPI in the regulation of rat TS cell differentiation. Control and Tfpi short hairpin RNAs (shRNAs) were delivered to rat TS cells via lentiviral-mediated transduction (Fig. 3A). Inhibition of Tfpi messenger RNA (mRNA) and protein expression in Tfpi shRNA transduced cells were confirmed (Fig. 3 B and C). Rat TS cells stably expressing control or Tfpi shRNAs were induced to differentiate, and their transcriptomes were interrogated by RNA-seq. Knockdown of TFPI was associated with 11 up-regulated transcripts and 53 down-regulated transcripts (Fig. 3D and Dataset S2). Down-regulated transcripts overlapped with a subset of differentiation-induced transcripts shown in Fig. 1C (Fig. 3 E and F). Among these down-regulated transcripts were transcripts characteristic of endothelial cells (e.g., Thbd, Esam, Cd47, Cd93, Crispld2, and Lgals9; Fig. 3F), which were validated by RT-qPCR (Fig. 3G). These findings indicated that TFPI potentially has an intrinsic role in the regulation of rat TS cell differentiation, including contributing to the acquisition of the endothelial cell–like phenotype.
Fig. 3.
TFPI is an intrinsic regulator of the trophoblast endothelial cell–like phenotype. (A) Schematic representation of lentiviral vector-mediated TFPI knockdown on differentiated rat TS cells. (B and C) Efficiency of Tfpi shRNA treatment (shRNA-1 and 2) efficiency was determined by RT-qPCR (B) and Western blotting (C). (D) MA plot showing the global transcriptomic changes in differentiated rat TS cells exposed to control or Tfpi-specific shRNAs. Colored dots indicate DEGs (≥2-fold with an FDR of P < 0.05; red: up-regulated, blue: down-regulated). Transcripts characteristic of an endothelial cell–like phenotype were down-regulated. (E) Venn diagram showing the number of down-regulated transcripts associated with the control shRNA (Ctrl) versus TFPI knockdown (KD, DEG1, red) and up-regulated transcripts associated with stem state versus differentiated (Diff) state (DEG2, blue). DEG1 and DEG2 datasets overlapped (33 of 53 transcripts). (F) Heatmap showing the 33 DEGs shared in the DEG1 and DEG2 datasets. Left column shows the Log2 (fold change) of transcripts down-regulated by TFPI knockdown, whereas the right column shows the Log2 (fold change) of transcripts up-regulated in differentiated trophoblast cells (ascending order). (G) RT-qPCR validation of selected down-regulated transcripts in Tfpi shRNA–treated cells. Comparisons were performed on control versus Tfpi-specific shRNA-exposed cells. Asterisks denote P < 0.05.
TFPI Expression in the Human Placentation Site.
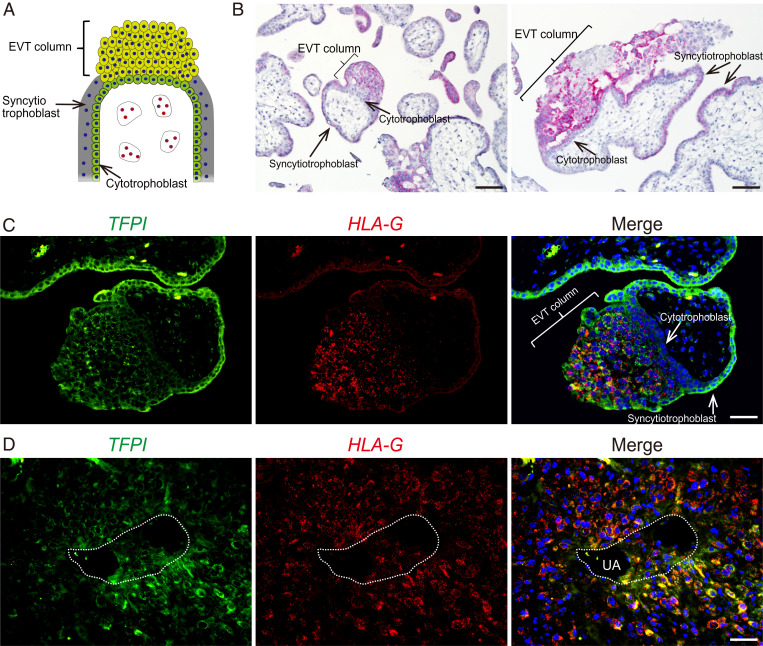
TFPI expression has previously been demonstrated within the human placenta (33–35), suggesting that some aspects of its contributions to trophoblast cell biology could be conserved. However, limited evidence exists for the expression of TFPI in invasive trophoblast cells of the human placentation site (referred to as extravillous trophoblast [EVT], Fig. 4A). TFPI transcripts were localized to the EVT column (Fig. 4 B–D). Duplex in situ hybridization was performed to simultaneously localize TFPI and major histocompatibility complex, class I, G (HLA-G) transcripts in first-trimester placental tissues. HLA-G is a known marker of EVT cells (36). TFPI transcripts were localized to trophoblast cells within EVT columns, especially distal regions, which also express HLA-G transcripts (Fig. 4C). TFPI transcripts were also present in syncytiotrophoblast of villous structures (Fig. 4C), consistent with earlier reports (33, 35). We also investigated TFPI transcript expression in first-trimester uterine decidual tissue infiltrated with EVT cells. TFPI transcripts were detected in EVT cells, including endovascular trophoblast cells present within arterioles located in the uterine decidua (Fig. 4D). The reliability of the localization experiments is strengthened by single-cell RNA-seq analysis of first-trimester human placentation sites showing that TFPI is expressed in cell clusters possessing features of EVT cells (37, 38). Thus, TFPI exhibits parallel expression patterns in rat and human placentation sites.
Fig. 4.
TFPI is expressed in human placentation sites. (A) Schematic diagram of the structure of a human villous. (B) Two images showing localization of TFPI transcripts (red) to the EVT column and syncytiotrophoblast in the first-trimester human placenta. (C and D) Duplex in situ hybridization of TFPI (green) and HLA-G (red) in the first-trimester human placenta (C) and in first-trimester EVT cells associated with a uterine spiral arteriole (UA) (D). Note that TFPI is expressed in HLA-G–positive EVT cells, HLA-G–positive endovascular trophoblast cells, and syncytiotrophoblast cells. (Scale bar: 50 μm.)
TFPI Is a Regulator of EVT Cell Differentiation.
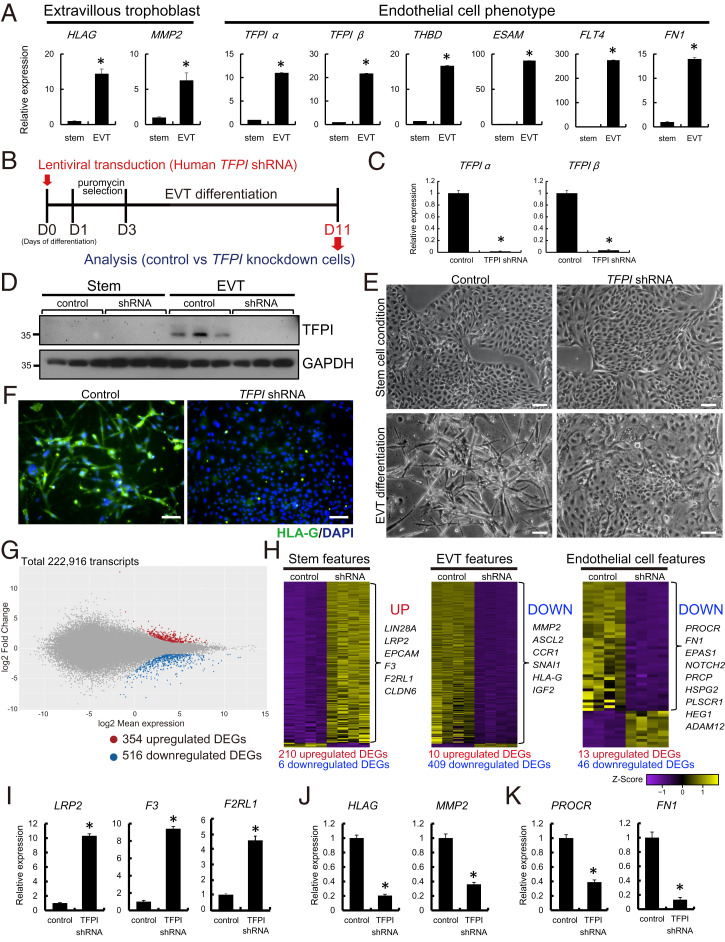
Since TFPI transcripts were readily detected in human EVT cells, we next explored a role for TFPI in EVT cell differentiation using a human TS cell culture system (17). Human TS cells were maintained in the stem state or induced to differentiate into EVT cells (17). EVT cell differentiation was characterized by up-regulation of HLA-G and MMP2 transcripts and a prominent up-regulation of endothelial cell–like transcripts, which included increased expression of TFPI, THBD, ESAM, FLT4, and FN1 (Fig. 5A). A loss-of-function approach using lentiviral vector-delivered control or TFPI shRNAs into human TS cells was implemented to investigate a role for TFPI in EVT cell differentiation (Fig. 5B). Inhibition of TFPI mRNA and TFPI protein expression was confirmed in TFPI shRNA–expressing human TS cells (Fig. 5 C and D). Loss of TFPI interfered with EVT cell differentiation. The elongated/spindle-shaped cells characteristic of in vitro EVT cell differentiation were not observed following TFPI knockdown (Fig. 5E and SI Appendix, Fig. S3). HLA-G protein expression was notably decreased in TFPI shRNA–expressing EVT-differentiated cells (Fig. 5F). Instead, TFPI knockdown cells more closely resembled human TS cells in the stem state (Fig. 5 E and F). To further investigate the impact of TFPI knockdown on EVT cell differentiation, we performed RNA-seq analysis. We identified 354 up-regulated and 516 down-regulated transcripts in TFPI knockdown versus control-transduced human TS cells induced to differentiate into EVT cells (Fig. 5G and Dataset S3). Inhibition of TFPI expression led to a striking down-regulation of transcripts characteristic of EVT cell differentiation (e.g., MMP2, ASCL2, CCR1, SNAI1, HLA-G, and IGF2), including transcripts characteristic of an endothelial cell–like phenotype (e.g., PROCR, FN1, EPAS1, NOTCH2, PRCP, HSPG2, PLSCR1, HEG1, and ADAM12). Surprisingly, TFPI disruption also led to an up-regulation of transcripts associated with the human TS cell stem state (e.g., LIN28A, LRP2, F3, EPCAM, F2RL1, and CLDN6; Fig. 5H), which was consistent with morphological observations (Fig. 5E). These differential patterns of gene expression were validated by RT-qPCR (Fig. 5 I–K).
Fig. 5.
TFPI is a regulator of human TS cell differentiation into EVT cells. (A) RT-qPCR analysis of stem cell and differentiated EVT cell states of human TS cells. Endothelial cell–associated transcripts, including TFPI alpha and beta, were significantly up-regulated in EVT cells. (B) Schematic representation of lentiviral vector-mediated TFPI knockdown on human TS cells and analysis on EVT cell differentiated state. (C and D) Efficiency of TFPI shRNA (#1) treatment was determined by RT-qPCR for TFPI alpha and beta (C) and Western blotting for TFPI (D). (E) Representative phase contrast images of stem state human TS cells and differentiated EVT cells transduced with lentivirus containing control or TFPI shRNA. (Scale bar: 200 μm.) (F) Immunocytochemistry of HLA-G expression (green) of differentiated EVT cells treated with control and TFPI shRNAs. (Scale bar: 100 μm.) DAPI marks cell nuclei (blue). (G) MA plot showing the global transcriptomic changes in differentiated human EVT cells exposed to control or TFPI-specific shRNAs. Colored dots indicate DEGs (≥2-fold with a false discovery rate of P < 0.05; red: up-regulated, blue: down-regulated). (H) Heatmap representation of Z-score–transformed RPKM values of transcripts predominantly expressed in the stem cell state (Stem features, Left), differentiated EVT cell state (EVT features, Middle). Classification of stem and EVT cell features were based on RNA-seq analysis of human TS cells in the stem and differentiated EVT cell states (17). Endothelial cell–associated transcripts (Endothelial cell features, Right) were determined from “Blood vessel morphogenesis” and “Coagulation” gene sets extracted from GO:0050817 and GO:0048514. (I–K) RT-qPCR validation of selected up- or down-regulated transcripts, including stem cell (I), EVT cell (J), and endothelial cell (K) characteristic transcripts in EVT cells transduced with control or TFPI shRNAs. Asterisks denote P < 0.05.
The results obtained with rat and human TS cells reveal a potential conserved action of TFPI in the regulation of invasive trophoblast cell development. However, it is important to acknowledge that comparisons between rat and human TS cells are challenging. Rat and human TS cells are fundamentally different in their derivation, maintenance, and developmental potential.
In Vivo Evaluation of a Role for TFPI in the Regulation of Intrauterine Trophoblast Cell Invasion.
Compelling results from in vitro experiments demonstrating a role for TFPI in trophoblast cell differentiation led to the evaluation of the in vivo actions of TFPI in hemochorial placentation. A Tfpi mutant mouse generated using standard homologous recombination in mouse embryonic stem cells was first reported in 1997 (39). A heterogenous phenotype was observed in embryos possessing deficits in TFPI. About 60% of Tfpi mutant embryos died at midgestation, and the remaining survived for various durations during the last half of the pregnancy. Vascular abnormalities were noted in the yolk sac and placenta associated with the dying embryos. Subsequent reports verified these observations in the TFPI-deficient mouse (40–42). The mouse is an excellent animal model for elucidating many aspects of hemochorial placentation (43), but it has limitations as a tool for investigating the trophoblast cell–uterine interface (11, 13, 14). In contrast, the rat exhibits extensive intrauterine trophoblast cell invasion and thus represents a more informative model for studying deep placentation, a characteristic of human placentation (11–13). In the rat, invasion is restricted to the movement of trophoblast cells within decidual arterioles until gestation day (gd) 13.5, when extensive intrauterine interstitial and endovascular trophoblast cell invasion is initiated and proceeds through the remainder of pregnancy to fill the mesometrial compartment (11, 12). We proceeded with two strategies to investigate the involvement of TFPI in the regulation of trophoblast invasion and uterine spiral artery remodeling within the rat placentation site: 1) CRISPR-Cas9 genome editing to disrupt the Tfpi gene and 2) trophoblast-specific Tfpi silencing.
CRISPR-Cas9 genome editing of the Tfpi gene.
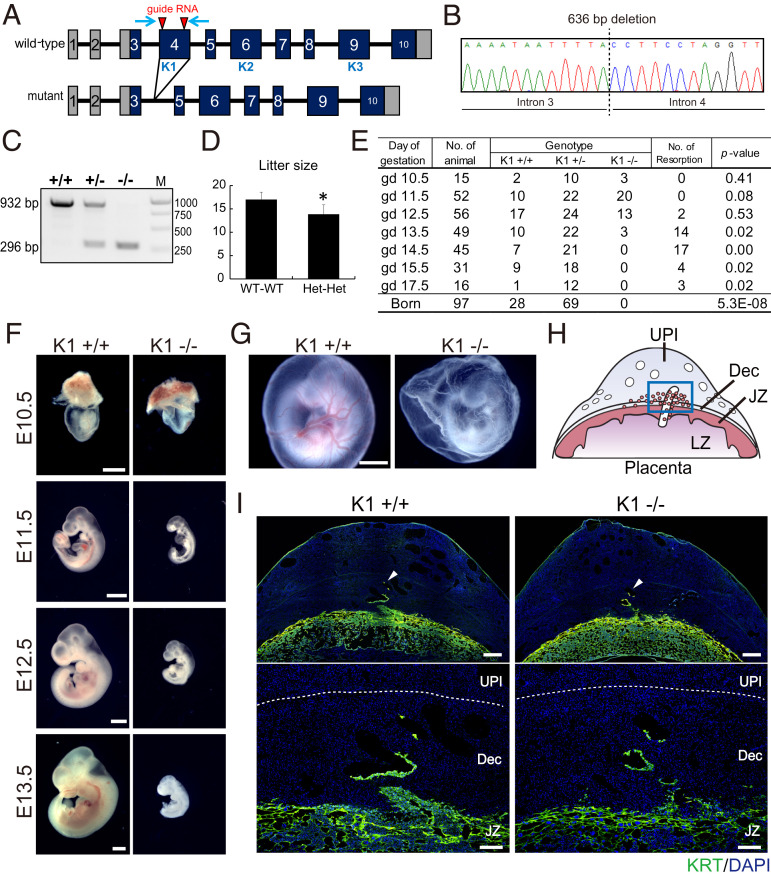
Two mutant rat models were generated. One mutant Tfpi rat model possessed a complete deletion of Exon 4 (encoding the first Kunitz domain, TfpiK1, Fig. 6 A–C), which is the same region targeted in previously reported Tfpi mutant mice (39–41). A second mutant Tfpi rat model possessed a one-base-pair insertion within Exon 4, resulting in a frameshift and a premature stop codon (Tfpi1bp; SI Appendix, Fig. S4 A–C). At homozygosity, both Tfpi mutations were associated with intrauterine lethality and showed indistinguishable homogenous phenotypes (Fig. 6 D and E and SI Appendix, Fig. S4 D and E), differing from the heterogenous phenotypes described for the mouse (34–36). The etiology of the differences in rat versus mouse Tfpi mutant phenotypes may reflect genetic and/or environment-dependent factors. Tfpi rat mutants died between gd 13.5 and gd 14.5 (Fig. 6E and SI Appendix, Fig. S4E) and showed embryonic growth restriction (Fig. 6F and SI Appendix, Fig. S4F) and avascular yolk sacs at gd 11.5 (Fig. 6G and SI Appendix, Fig. S4G). Both wild-type and Tfpi mutant placentation sites similarly exhibited evidence of trophoblast cell invasion into decidual arterioles at gd 12.5 (Fig. 6 H and I and SI Appendix, Fig. S4H); however, unlike the mouse, embryonic death uniformly occurred at midgestation and thus precluded assessment of a role for TFPI in the major wave of intrauterine trophoblast invasion, which occurs during the last third of gestation (13).
Fig. 6.
Phenotypic analysis of Kunitz domain 1 (K1) Tfpi mutant rat. (A) Schematic representation of the rat Tfpi gene and the disruption of the K1 domain using CRISPR-Cas9 system. The red arrowheads indicate target sites for the guide RNAs used in genome editing. The blue arrows indicate positioning of the primer set used to amplify the Tfpi K1 domain. K1, Kunitz domain 1; K2, Kunitz domain 2; K3, Kunitz domain 3. (B) DNA sequence analysis showing a 636-bp deletion within the Tfpi locus, resulting in the deletion of the entire Exon 4, leading to a deletion of the K1 domain and an in-frame mutation. (C) Genotyping of wild type (+/+), heterozygous (+/−), and homozygous mutant (−/−) Tfpi alleles. The wild-type allele (932 bp) and mutant allele (296 bp) were detected by PCR. M denotes molecular size markers. (D) Litter sizes from Tfpi K1 heterozygous intercrosses. *P < 0.05. (E) Table showing the Mendelian ratios for gd 10.5 through 17.5 and postdelivery. (F and G) Macroscopic analysis of wild-type and K1 homozygous mutant embryos at E10.5, E11.5, E12.5, and E13.5 (F) and yolk sac at E11.5 (G). (Scale bar: 1 mm.) (H) Schematic representation of a midgestation placentation site, consisting of the uterine–placental interface (UPI), junctional zone (JZ), and labyrinth zone (LZ). The blue box in H corresponds to the uterine–placental interface for I. (I) Cytokeratin (KRT) immunohistochemical analysis for wild-type and Tfpi K1 mutant placentas (gd 12.5). The lower panels are high-magnification images of the upper panels. Arrowheads in the upper panels demarcate the depth of intrauterine trophoblast cell invasion. The demarcation of the decidua (Dec) and UPI is shown as a dashed white line. (Scale bars of upper panels: 500 µm. Scale bars of lower panels: 200 µm.)
Trophoblast-specific Tfpi silencing.
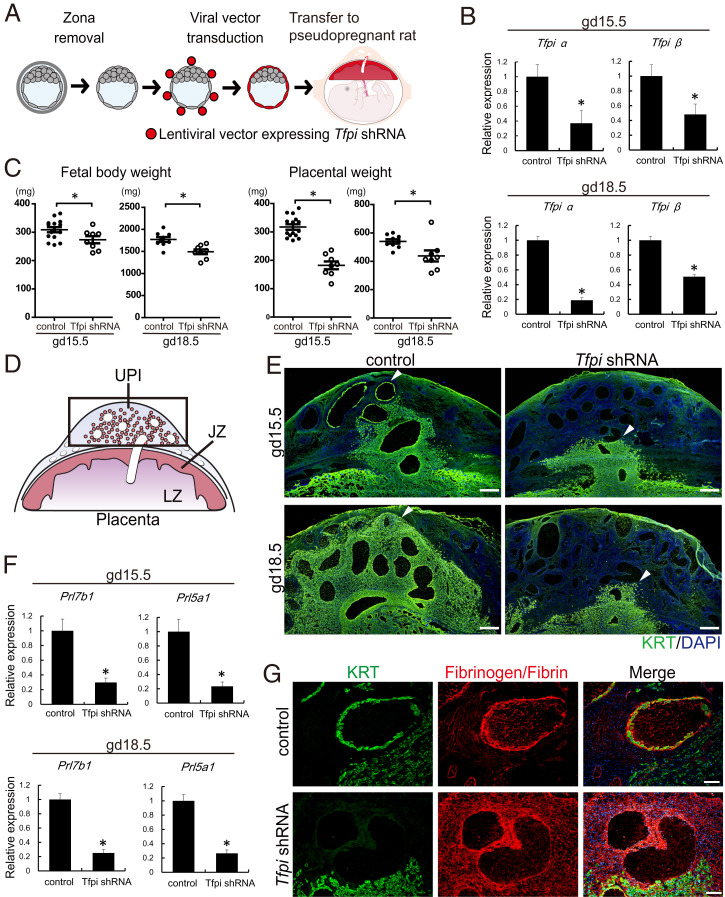
Since Tfpi rat mutants exhibited embryonic death before expansion of the invasive trophoblast cell lineage, an alternative loss-of-function strategy was necessary. Lentiviral trophoblast-specific delivery of control and Tfpi shRNAs was performed (Fig. 7A). Lentiviral delivery of Tfpi shRNAs was effective (Fig. 7B) and led to significant decreases in placental and fetal weights (Fig. 7C). Furthermore, in vivo knockdown of TFPI arrested intrauterine trophoblast cell invasion on gd 15.5 and gd 18.5, as assessed by cytokeratin immunostaining (Fig. 7 D and E) and RT-qPCR assessment of invasive trophoblast-specific transcripts (Prl5a1 and Prl7b1; Fig. 7F). Furthermore, disruption of TFPI was associated with abnormal retention of natural killer (NK) cells at the uterine–placental interface (SI Appendix, Fig. S5) and a prominent fibrin/fibrinogen deposition within placentation sites (Fig. 7G). NK cells and excess fibrin accumulation each possess restraining effects on intrauterine trophoblast cell invasion (44–48).
Fig. 7.
Effects of trophoblast-specific Tfpi knockdown on the uterine–placental interface (UPI). (A) Schematic diagram of in vivo lentiviral vector-mediated trophoblast-specific Tfpi knockdown. Rat blastocysts were transduced with lentivirus-expressing control or Tfpi shRNAs and subsequently transferred to pseudopregnant animals. (B) Efficiency of the Tfpi knockdown in the junctional zone of rat placentation sites were determined by RT-qPCR (gd 15.5 and 18.5). (C) Fetal and placental weights of control and Tfpi shRNA-transduced embryos (gd 15.5 and gd 18.5). (D) Schematic representation of a midgestation placentation site. The black box corresponds to the location of the UPI. (E) Immunohistochemisty of KRT (green) within the metrial gland at gd 15.5 and 18.5 placentation sites. Depth of intrauterine trophoblast cell invasion was decreased in Tfpi shRNA-transduced placentation sites compared to control. (Scale bar: 500 μm.) Arrowheads demarcate the depth of intrauterine trophoblast cell invasion. (F) Quantification of invasive trophoblast cell-specific transcripts (Prl5a1 and Prl7b1) within the UPI (gd 15. 5 and gd 18.5) of control and Tfpi shRNA-exposed placentation sites as measured by RT-qPCR. Asterisks denote P < 0.05. (G) Immunohistochemisty of KRT (green) and fibrinogen (red) within the gd 15.5 UPI of control and Tfpi shRNA-exposed placentation sites. Fibrinogen deposition was observed in placentation sites from blastocysts transduced with Tfpi shRNA. (Scale bar: 100 μm.)
Collectively, the experimentation indicates that TFPI exhibits a conserved role in the regulation of invasive trophoblast cell lineage development, a fundamental property of hemochorial placentation.
Discussion
Optimal hemochorial placentation involves uterine vascular remodeling and achieving an appropriate hemostatic balance (4, 7, 49, 50). TFPI counteracts prothrombotic challenges (18), including those triggered by placentation. Endothelial cells, platelets, and other cell types, including trophoblast cells, are sources of TFPI (19, 21, 33–36, 51). In this report, TFPI was identified as a major transcript up-regulated during TS cell differentiation, a property conserved in rats and humans. At the placentation site, TFPI was prominently positioned in trophoblast cells lining uterine spiral arterioles and other spaces within the placenta serving as conduits for blood delivery. Thus, TFPI is positioned to prevent activation of blood coagulation cascades that could impair blood flow through the placenta. The “Great Obstetric Syndromes,” including preeclampsia, intrauterine growth restriction, placenta abruption, and preterm birth, are characterized by vascular pathologies and a failed balance of pro- and anticoagulation activities, including dysregulated TFPI (15, 27, 35, 50, 52, 53). This critical function of TFPI in maintaining blood flow at the maternal–fetal interface is now coupled to the role of TFPI in regulating development of the invasive trophoblast cell lineage.
Interconnectivity between blood coagulation and placentation is also observed in antiphospholipid syndrome (54, 55). Pathologies characteristic of antiphospholipid syndrome negatively affect pregnancy and placentation (54, 55). Most interestingly, antiphospholipid syndrome can be associated with auto-antibodies to TFPI (56, 57) and impairments in trophoblast cell invasion (58–60). Thus far, a direct linkage between TFPI neutralization and abnormalities in deep intrauterine trophoblast cell invasion or trophoblast cell–guided uterine spiral artery remodeling has not been reported.
Invasive trophoblast cells are defined based on their destination. Some invasive trophoblast cells are directed to the uterine vasculature, where they replace endothelial cells lining vessels and are termed endovascular invasive trophoblast cells, while others termed interstitial invasive trophoblast cells migrate to positions between the uterine vasculature (61). A third destination for invasive trophoblast cells is within the walls of blood vessels. These cells are termed intramural invasive trophoblast cells (4). Evidence supports invasive trophoblast cells entering arterial, venous, and lymphatic vessels and uterine glands (61–64). TFPI specifically marks endovascular invasive trophoblast cells and possesses intrinsic actions on the development of invasive trophoblast cells. Trophoblast cell deficits in TFPI lead to a failure in invasive trophoblast cell differentiation. TFPI-depleted human trophoblast cells retain aspects of stem state cell behavior, including the up-regulation of trophoblast cell stem state–associated transcripts (e.g., EPCAM, LIN28A, LRP2, CLDN6, F3, and F2RL1).
The reciprocal relationship of TFPI with F3 and F2RL1, which encode tissue factor and protease activated receptor 2 (PAR2), respectively, during the transition of TS cells from the stem state to differentiated EVT cells, is intriguing. Tissue factor–factor VIIa complexes activate proteases that signal through PAR2 and influence cell proliferation, survival, and metabolism (65–67). Contributions of tissue factor and PAR2 to the regulation of TS cells are yet to be determined.
Among the invasive trophoblast cell lineages, TFPI is prominently expressed in endovascular invasive trophoblast cells. However, TFPI regulates development of both endovascular and interstitial invasive trophoblast cells. The relationship of endovascular and interstitial invasive trophoblast cells is not well understood (4, 61, 62, 68). In addition to endovascular invasive trophoblast cells, TFPI is also expressed in the EVT column and junctional zone, which are sources of invasive trophoblast progenitor cells in humans and rats, respectively. Our current findings are consistent with TFPI acting on progenitor cells seeding both endovascular and interstitial invasive trophoblast cell development. It is also important to appreciate that in vivo TFPI deficits may further thwart trophoblast cell invasion indirectly through the retention and restraining actions of NK cells (44, 45) and excessive fibrin accumulation (46–48) within the uterine parenchyma.
We have an understanding of how TFPI regulates blood coagulation (18–20) but only rudimentary knowledge of how TFPI acts to modulate other cellular functions (29–32), especially trophoblast cell lineage development. TFPI is a Kunitz domain protease inhibitor and physically associates with factor VIIa and factor Xa to regulate the tissue factor–mediated protease cascade (18–20, 69, 70). Kunitz domains are a common feature of expanded protein families specifically expressed by ruminant trophoblast cells during early pregnancy (71–73). Select members of the ruminant trophoblast Kunitz domain protein family act as serine protease inhibitors; however, their target proteases and precise roles in the biology of ruminant pregnancy are not well defined (72). The first Kunitz domain of TFPI is essential for successful prenatal development (39); however, the contribution of any of the three Kunitz domains within TFPI to regulating the trophoblast cell lineage is not known. TFPI-α has been shown to physically interact with other proteins present in the extracellular matrix, including thrombospondin 1 (THBS1, refs. 74 and 75), syndecan 4 (SDC4, refs. 75 and 76), glypicans (75, 77, 78), and laminin (75). TFPI likely interacts with these proteins through its carboxyl terminus (74, 75, 79). THBS1, SDC4, glypican 3 (GPC3), and laminins have been connected to trophoblast cell biology. THBS1 promotes trophoblast cell outgrowth from mouse blastocysts (80) and is dysregulated in pregnancy disorders (81, 82), while SDC4 facilitates anticoagulant processes in the developing placenta (83) and has recently been shown to regulate EVT cell invasive properties (84). The actions of THBS1 and SDC4 may be linked (85). Placental GPC3 expression is diminished in preeclampsia and intrauterine growth restriction (86, 87), whereas laminin controls TS cell dynamics (88). Whether TFPI actions on invasive trophoblast cell development involve its interaction with SDC4, THBS1, GPC3, or laminin remains to be determined. High-throughput analysis of the TFPI protein interactome has implicated TFPI in an assortment of additional cellular functions (89); however, each requires validation in biologically relevant cell types, including trophoblast cells.
Trophoblast-guided uterine spiral arteriole remodeling and hemostatic control at the maternal–fetal interface are fundamental to establishing a fully functional hemochorial placenta and a successful pregnancy. Remarkably, these developmental processes are intertwined and each exquisitely regulated through the conserved actions of TFPI.
Materials and Methods
Animals and Tissue Collection.
Holtzman Sprague Dawley rat breeding stocks were obtained from Envigo. Animals were housed in an environmentally controlled facility with lights on from 6:00 AM to 8:00 PM and allowed free access to food and water. Virgin female rats at 8 to 10 wk of age were cohabited with adult males (>3 mo of age). The presence of sperm in the vaginal lavage was considered gd 0.5. Rat embryos were collected by flushing uteri with Roswell Park Memorial Institute (RPMI) medium 1640 (11875-093, Thermo Fisher) at gd 4.5. Pseudopregnant female rats were generated by mating with vasectomized males. Detection of seminal plugs was considered day 0.5 of pseudopregnancy. Rat placental tissues were collected from gd 10.5 through gd 18.5. Dissections into placental compartments (junctional zone, labyrinth zone, and metrial gland) were performed as previously described (90, 91). Conceptuses for histological analysis were frozen in dry ice−cooled heptane and stored at −80 °C. Tissue samples for protein or RNA extraction were frozen in liquid nitrogen and stored at −80 °C. The University of Kansas Medical Center (KUMC) Animal Care and Use Committee approved protocols for the care and use of animals.
Human Placentation Site Specimens.
Sections of paraffin-embedded first-trimester placenta and placental bed were produced from deidentified specimens obtained at the Lunenfeld-Tanenbaum Research Institute (Mount Sinai Hospital, Toronto, Canada) or St. Mary’s Hospital, Manchester, United Kingdom with written informed consent. Prior approval was granted by the respective local human research ethics review committees at the Mount Sinai Hospital, Central Manchester Health Trust, and KUMC.
Rat and Human Trophoblast Stem Cells.
Blastocyst-derived rat TS cells (16) were cultured in rat TS cell medium (RPMI 1640, 20% (vol/vol) fetal bovine serum [FBS, Thermo Fisher], 100 μm 2-mercaptoethanol [M7522, Sigma-Aldrich], 1 mM sodium pyruvate [11360-070, Thermo Fisher], 50 μM penicillin [15140122, Thermo Fisher], and 50 U/mL streptomycin [15140122, Thermo Fisher]) supplemented with 70% rat embryonic fibroblast (REF)-conditioned medium prepared as previously described (16), fibroblast growth factor 4 (FGF4, 25 ng/mL; 100-31, Peprotech), and heparin (1 μg/mL; H3149, Sigma-Aldrich). For induction of differentiation, rat TS cells were cultured for 8 d in rat TS cell medium without FGF4, heparin, or REF-conditioned medium.
Human TS cells (17) were routinely maintained in six-well plates precoated with 5 μg/mL collagen IV (CB40233, Fisher) containing Basal Human TS Cell Medium (DMEM/F12 [11320033, Thermo Fisher], with 100 μm 2-mercaptoethanol, 0.2% [vol/vol] FBS, 50 μM penicillin, 50 U/mL streptomycin, 0.3% bovine serum albumin [BP9704100, Fisher], 1% Insulin-Transferrin-Selenium-Ethanolamine solution [vol/vol, Thermo Fisher]) supplemented with 1.5 μg/mL L-ascorbic acid (A8960, Sigma-Aldrich), 50 ng/mL epidermal growth factor (E9644, Sigma-Aldrich), 2 μM CHIR99021 (04-0004, Reprocell), 0.5 μM A83-01 (04-0014, Reprocell), 1 μM SB431542 (04-0010, Reprocell), 0.8 mM valproic acid (P4543, Sigma-Aldrich), and 5 μM Y27632 (04-0012-02, Reprocell). For induction of EVT cell differentiation, human TS cells were plated onto a 6-well plate precoated with 1 μg/mL collagen IV at a density of 1 × 105 cells per well and cultured in EVT Cell Differentiation Medium, which consists of Basal Human TS Cell Medium supplemented with 100 ng/mL of neuregulin 1 (NRG1, 5218SC, Cell Signaling), 7.5 μM A83-01, 2.5 μM Y27632, 4% KnockOut Serum Replacement (KSR, 10828028, Thermo Fisher), and 2% Matrigel (CB-40234, Fisher). On day 3 of EVT cell differentiation, the medium was replaced with the EVT Cell Differentiation Medium without NRG1, and the Matrigel concentration was decreased to 0.5%. On day 6 of EVT cell differentiation, the medium was replaced with EVT Cell Differentiation Medium without NRG1 or KSR and with Matrigel at a concentration of 0.5%. Cells were cultured for two additional days before analysis.
RNA-seq Analysis.
RNA-seq was performed as previously described (91). Rat TS cells in the stem or differentiated states (n = 3/group), rat differentiated TS cells expressing control shRNA versus Tfpi shRNA (n = 3/group), and human EVT differentiated TS cells expressing control shRNA versus TFPI shRNA (n = 4/group) were analyzed. Complementary DNA (cDNA) libraries from total RNA samples (500 ng input) were prepared from Illumina TruSeq RNA sample preparation kits according to the manufacturer’s instructions. RNA integrity was assessed using an Agilent 2100 Bioanalyzer. Libraries were clustered onto a TruSeq paired-end flow cell and were sequenced (100-bp paired-end reads) using a TruSeq 200-cycle SBS kit (Illumina). Samples were run on Illumina HiSeq2500 sequencers located at the KUMC Genome Sequencing Facility or at the Genomic Medicine Center at Children’s Mercy. Reads from fastq files were mapped to RGSC 6.0/rn6 genome (rat) or GRCh37/hg19 genome (human) using CLC Bio Genomics Workbench 7.0 (Qiagen), and transcript abundance was expressed as reads per kilobase of transcript per million mapped reads (RPKM). DEGs were identified using false discovery rate (FDR) and fold change (FC) versus control groups (FDR < 0.05; FC ≥ 2). Principal component analysis (PCA) was performed by R (https://www.r-project.org/) using the RNA-seq dataset of rat TS cells (stem versus differentiated). Gene set enrichment analysis (GSEA) was run by GSEA version 4.0.3 Mac App from the Broad Institute (https://www.gsea-msigdb.org/gsea/index.jsp). Heatmaps were generated using Z-score by R. The gene sets, which exhibit stem and EVT features, were identified using previously reported human TS cell RNA-seq data (17). The gene list identified as “endothelial cell feature” was extracted from GO_COAGULATION (GO:0050817) and GO_BLOOD_VESSEL_MORPHOGENESIS (GO:0048514) in AmiGO2 (http://amigo.geneontology.org/amigo). Statistical significance was calculated by empirical analysis of digital gene expression, followed by Bonferroni’s correction. Transcripts with an adjusted P value of <0.05 were considered differentially regulated.
RT-qPCR.
RT-qPCR was performed as previously described (91). Total RNA was extracted from cells and tissues using TRIzol reagent (15596018, Thermo-Fisher). cDNA was synthesized from total RNA for each sample using High Capacity cDNA Reverse Transcription kit (4368814, Thermo-Fisher), diluted five times with water, and subjected to qPCR. PCR primer sequences are presented in SI Appendix, Table S1. Real-time PCR amplification of cDNAs for qPCR was carried out in a reaction mixture containing SYBR GREEN PCR Master Mix (4309155, Thermo-Fisher) and primers (250 nM each). Amplification and fluorescence detection were carried out using QuantStudio 7 Flex Real-time PCR System (Thermo-Fisher). Cycling conditions included an initial step (95 °C for 10 min) and 40 cycles of a two-step PCR (92 °C for 15 s and then 60 °C for 1 min), followed by a dissociation step (95 °C for 15 s, 60 °C for 15 s, and then 95 °C for 15 s). The comparative cycle threshold method was used for relative quantification of the amount of mRNA for each sample normalized to a housekeeping gene (Gapdh for rat samples and POLR2A for human samples).
Immunohistochemistry.
Frozen tissue sections (10-μm) were prepared and incubated with 10% normal goat serum (50062Z, Thermo Fisher) for 1 h to block nonspecific reactivity. Sections were then incubated overnight with the following primary antibodies: anti-mouse TFPI antibody (1:500, ref. 92), anti-human cytokeratin (1:500, F3418, Sigma-Aldrich), or anti-rat fibrinogen (1:500, 11352-05011, Assaypro) and followed by a 2-h incubation with Alexa 488–tagged goat anti-mouse immunoglobulin G (IgG) (1:200, A11001, Thermo Fisher) or Alexa 568–tagged goat anti-rabbit IgG (1:200, A11011, Thermo Fisher). Nuclei were visualized with DAPI (Molecular Probes). Immunostained sections were mounted in Fluoromount-G (0100-01, SouthernBiotech), examined, and captured on a Nikon 80i upright microscope (Nikon) with a Photometrics CoolSNAP-ES monochrome camera (Roper).
In Situ Hybridization.
Detection of rat transcripts for Tfpi, Prl7b1, and Krt8 was performed on cryosections of rat placentation sites, and detection of human transcripts for TFPI and HLA-G was performed on paraffin-embedded human placenta tissue sections. The RNAscope 2.5 HD Detection kit (RED) and the RNAscope Multiplex Fluorescent Reagent Kit version 2 (Advanced Cell Diagnostics) were used for the in situ hybridization analysis, according to the manufacturer’s instructions. Probes were prepared to detect rat Tfpi (NM_017200.1; 878371, target region: 2 to 1,138), rat Prl7b1 (NM_153738.1, 860181-C2, target region: 28 to 900), rat Krt8 (NM_199370.1, 873041-C2, target region: 134 to 1,472), human TFPI (NM_006287.5, 562981, target region: 309 to 936), and human HLA-G (NM_002127.5, 426691-C2, target region: 14 to 1,329). Fluorescence images were captured on a Nikon 80i upright microscope (Nikon) with a Photometrics CoolSNAP-ES monochrome camera (Roper).
Western Blot Analysis.
Cell lysates were prepared in radioimmunopreciptation assay buffer (sc-24948A, Santa Cruz Biotechnology). Protein concentrations were determined using the detergent compatible (DC) protein assay (5000112JA, Bio-Rad). Proteins were separated by sodium dodecyl sulphate–polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membranes. Immunoreactive proteins were detected with rabbit antibodies to mouse TFPI (1:500, ref. 91), human TFPI (1:1,000, ref. 93), and glyceraldehyde 3-phosphate dehydrogenase (1:300, ab9485, Abcam). Immunoreactive proteins were visualized by enhanced chemiluminescence according to the manufacturer’s instructions (Amersham).
Generation of Rat Models with Tfpi Mutations.
Mutations at the rat Tfpi locus were generated using CRISPR-Cas9 genome editing system. Two guide RNAs targeting Exon 4 of the Tfpi gene (target sequence: 5′-ACTGCCGGGAGATAGTTACA-3′, 5′-AGGCTTGTGGGAAACCTACC-3′; NM_017200.1) were assembled with crispr RNA, transactivating crRNA, and Cas9 nuclease V3 (Integrated DNA Technologies). The genome editing constructs were electroporated into one-cell rat embryos using NEPA21 electroporator (Bulldog Bio). The electroporated embryos were transferred to oviducts of rats on day 0.5 of pseudopregnancy. Offspring were screened for mutations at specific target sites within the Tfpi gene by PCR, and precise boundaries of deletions determined by DNA sequencing. Founders were backcrossed with wild-type rats to evaluate germ-line transmission. Two mutant rat strains were generated: 1) 636-bp deletion including all of Exon 4 (TfpiK1), which encodes Kunitz domain 1; and 2) 1-bp insertion within Exon 4 (Tfpi1bp). Phenotypic analyses were performed on offspring from intercrosses of heterozygous rats. Genotyping for the TfpiK1 strain was performed by PCR (Primers: forward: 5′-AAAATGTGGCTAGAAGATTC-3′, reverse: 5′-ACAGTGGGCTAGAAACCTAG-3′), whereas genotyping for Tfpi1bp was performed by sequencing of a PCR template generated using the same primer set.
shRNA Constructs and Production of Lentivirus.
shRNA constructs were subcloned into pLKO.1. shRNA sequences used in the analyses are shown in SI Appendix, Table S2. Lentiviral packaging vectors were obtained from Addgene and included pMDLg/pRRE (plasmid 12251), pRSV-Rev (plasmid 12253), and pMD2.G (plasmid 12259). Lentiviral particles were produced following transient transfection of the shRNA-pLKO.1 vector and packaging plasmids into human embryonic kidney (HEK)293FT (Thermo Fisher) cells using Attractene (301005, Qiagen) in Opti-MEM I (51985-034, Thermo Fisher). Thereafter, cells were maintained in Dulbecco's Modified Eagle Medium (11995-065, Thermo Fisher) with 10% FBS. Culture supernatants containing lentiviral particles were collected every 24 h for 2 d. For in vivo lentiviral transduction, the collected supernatants were concentrated by ultracentrifugation (30,000 × g). The titration of lentiviral particles was determined by measurement of p24 mRNA levels by RT-qPCR (631235, Clontech).
In Vitro Lentiviral Transduction.
Rat and human TS cells were incubated with lentiviral particles and selected with puromycin dihydrochloride (5 μg/mL, A11138-03, Thermo Fisher) for 2 d. Puromycin was removed during cell differentiation.
Ex Vivo Lentiviral-Mediated Trophoblast-Specific Transduction.
Lentiviral-mediated trophoblast transduction was performed as previously described (94). Rat blastocysts were collected on gd 4.5 and incubated in mR1ECM medium (R0114, CytoSpring). Zona pellucidae were removed with acidic Tyrode’s solution (MR-004-D, EMD Millipore) and incubated with concentrated lentiviral particles in mR1ECM medium at a concentration of 6 × 108 IFU/mL for 5 h. The transduced blastocysts were washed with fresh mR1ECM medium and transferred to uteri of gd 3.5 pseudopregnant rats for subsequent evaluation of control and Tfpi knockdown and placentation site phenotypes on gd 15.5 and gd 18.5.
Statistical Analyses.
All values are presented as the mean ± SEM of at least three independent experiments. Comparisons of two means were analyzed by Student’s t test, whereas the statistical analyses of Mendelian ratios of gene-modified rats were performed by χ2 test.
Supplementary Material
Acknowledgments
This study was supported by the Lalor Foundation (M.M., K.M.V., and A.M.-I.), American Heart Association (M.M.), an NIH National Research Service Award, HD096809 (K.M.V.), postdoctoral fellowships, NIH grants (HD020676, HD079363, and HD099638), and the Sosland Foundation. We thank Brandi Miller and Stacy Oxley for their assistance. We also thank Dr. Robert D. Simari for providing TFPI antibodies for detection of rat TFPI.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2111267118/-/DCSupplemental.
Data Availability
RNA-seq datasets are available at the Gene Expression Omnibus website (https://www.ncbi.nlm.nih.gov/geo/); rat TS cells: stem versus differentiated, GSE171753; rat TS cells: control versus TFPI knockdown, GSE171754; human TS cells: control versus TFPI knockdown, GSE171223). All data generated and analyzed during this study are included in the published article and the online supporting files. Resources generated and analyzed during the current study are available from the corresponding author upon reasonable request.
References
- 1.Burton G. J., Fowden A. L., Thornburg K. L., Placental origins of chronic disease. Physiol. Rev. 96, 1509–1565 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roberts R. M., Green J. A., Schulz L. C., The evolution of the placenta. Reproduction 152, R179–R189 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaufmann P., Black S., Huppertz B., Endovascular trophoblast invasion: Implications for the pathogenesis of intrauterine growth retardation and preeclampsia. Biol. Reprod. 69, 1–7 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Pijnenborg R., Vercruysse L., Hanssens M., The uterine spiral arteries in human pregnancy: Facts and controversies. Placenta 27, 939–958 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Harris L. K., Aplin J. D., Vascular remodeling and extracellular matrix breakdown in the uterine spiral arteries during pregnancy. Reprod. Sci. 14, 28–34 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Maltepe E., Fisher S. J., Placenta: The forgotten organ. Annu. Rev. Cell Dev. Biol. 31, 523–552 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Knöfler M., et al. , Human placenta and trophoblast development: Key molecular mechanisms and model systems. Cell. Mol. Life Sci. 76, 3479–3496 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Red-Horse K., et al. , Trophoblast differentiation during embryo implantation and formation of the maternal-fetal interface. J. Clin. Invest. 114, 744–754 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Velicky P., Knöfler M., Pollheimer J., Function and control of human invasive trophoblast subtypes: Intrinsic vs. maternal control. Cell Adhes. Migr. 10, 154–162 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pijnenborg R., Robertson W. B., Brosens I., Dixon G., Review article: Trophoblast invasion and the establishment of haemochorial placentation in man and laboratory animals. Placenta 2, 71–91 (1981). [DOI] [PubMed] [Google Scholar]
- 11.Pijnenborg R., Vercruysse L., “Animal models of deep trophoblast invasion” in Placental Bed Disorders: Basic Science and its Translation to Obstetrics, Pijnenborg R., Brosens I., Romero R., Eds. (Cambridge University Press, 2010), pp. 127–139. [Google Scholar]
- 12.Soares M. J., Chakraborty D., Karim Rumi M. A., Konno T., Renaud S. J., Rat placentation: An experimental model for investigating the hemochorial maternal-fetal interface. Placenta 33, 233–243 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ain R., Canham L. N., Soares M. J., Gestation stage-dependent intrauterine trophoblast cell invasion in the rat and mouse: Novel endocrine phenotype and regulation. Dev. Biol. 260, 176–190 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Soncin F., Natale D., Parast M. M., Signaling pathways in mouse and human trophoblast differentiation: A comparative review. Cell. Mol. Life Sci. 72, 1291–1302 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brosens I., Pijnenborg R., Vercruysse L., Romero R., The “Great Obstetrical Syndromes” are associated with disorders of deep placentation. Am. J. Obstet. Gynecol. 204, 193–201 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Asanoma K., et al. , FGF4-dependent stem cells derived from rat blastocysts differentiate along the trophoblast lineage. Dev. Biol. 351, 110–119 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okae H., et al. , Derivation of human trophoblast stem cells. Cell Stem Cell 22, 50–63.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Wood J. P., Ellery P. E. R., Maroney S. A., Mast A. E., Biology of tissue factor pathway inhibitor. Blood 123, 2934–2943 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maroney S. A., Mast A. E., New insights into the biology of tissue factor pathway inhibitor. J. Thromb. Haemost. 13, S200–S207 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mast A. E., Tissue factor pathway inhibitor: Multiple anticoagulant activities for a single protein. Arterioscler. Thromb. Vasc. Biol. 36, 9–14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sood R., Kalloway S., Mast A. E., Hillard C. J., Weiler H., Fetomaternal cross talk in the placental vascular bed: Control of coagulation by trophoblast cells. Blood 107, 3173–3180 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou Y., et al. , Human cytotrophoblasts adopt a vascular phenotype as they differentiate. A strategy for successful endovascular invasion? J. Clin. Invest. 99, 2139–2151 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harris L. K., Review: Trophoblast-vascular cell interactions in early pregnancy: How to remodel a vessel. Placenta 31, S93–S98 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Sheppard B. L., Bonnar J., Uteroplacental hemostasis in intrauterine fetal growth retardation. Semin. Thromb. Hemost. 25, 443–446 (1999). [DOI] [PubMed] [Google Scholar]
- 25.Bick R. L., Recurrent miscarriage syndrome and infertility caused by blood coagulation protein or platelet defects. Hematol. Oncol. Clin. North Am. 14, 1117–1131 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Brenner B., Haemostatic changes in pregnancy. Thromb. Res. 114, 409–414 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Greer I. A., Aharon A., Brenner B., Gris J. C., Coagulation and placenta-mediated complications. Rambam Maimonides Med. J. 5, e0034 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiemers D. O., Ain R., Ohboshi S., Soares M. J., Migratory trophoblast cells express a newly identified member of the prolactin gene family. J. Endocrinol. 179, 335–346 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Singh R., et al. , Tissue factor pathway inhibitor deficiency enhances neointimal proliferation and formation in a murine model of vascular remodelling. Thromb. Haemost. 89, 747–751 (2003). [PubMed] [Google Scholar]
- 30.Holroyd E. W., Simari R. D., Interdependent biological systems, multi-functional molecules: The evolving role of tissue factor pathway inhibitor beyond anti-coagulation. Thromb. Res. 125, S57–S59 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holroyd E. W., et al. , Tissue factor pathway inhibitor blocks angiogenesis via its carboxyl terminus. Arterioscler. Thromb. Vasc. Biol. 32, 704–711 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maroney S. A., et al. , Comparison of the inhibitory activities of human tissue factor pathway inhibitor (TFPI)α and TFPIβ. J. Thromb. Haemost. 11, 911–918 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edstrom C. S., Calhoun D. A., Christensen R. D., Expression of tissue factor pathway inhibitor in human fetal and placental tissues. Early Hum. Dev. 59, 77–84 (2000). [DOI] [PubMed] [Google Scholar]
- 34.Mast A. E., Acharya N., Malecha M. J., Hall C. L., Dietzen D. J., Characterization of the association of tissue factor pathway inhibitor with human placenta. Arterioscler. Thromb. Vasc. Biol. 22, 2099–2104 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Aharon A., Lanir N., Drugan A., Brenner B., Placental TFPI is decreased in gestational vascular complications and can be restored by maternal enoxaparin treatment. J. Thromb. Haemost. 3, 2355–2357 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Ferreira L. M. R., Meissner T. B., Tilburgs T., Strominger J. L., HLA-G: At the interface of maternal-fetal tolerance. Trends Immunol. 38, 272–286 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Suryawanshi H., et al. , A single-cell survey of the human first-trimester placenta and decidua. Sci. Adv. 4, eaau4788 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vento-Tormo R., et al. , Single-cell reconstruction of the early maternal-fetal interface in humans. Nature 563, 347–353 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang Z. F., Higuchi D., Lasky N., Broze G. J. Jr., Tissue factor pathway inhibitor gene disruption produces intrauterine lethality in mice. Blood 90, 944–951 (1997). [PubMed] [Google Scholar]
- 40.Pedersen B., Holscher T., Sato Y., Pawlinski R., Mackman N., A balance between tissue factor and tissue factor pathway inhibitor is required for embryonic development and hemostasis in adult mice. Blood 105, 2777–2782 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Castillo M. M., et al. , Maintaining extraembryonic expression allows generation of mice with severe tissue factor pathway inhibitor deficiency. Blood Adv. 3, 489–498 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maroney S. A., et al. , Tissue factor pathway inhibitor is required for cerebrovascular development in mice. Blood 137, 258–268 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rossant J., Cross J. C., Placental development: Lessons from mouse mutants. Nat. Rev. Genet. 2, 538–548 (2001). [DOI] [PubMed] [Google Scholar]
- 44.Chakraborty D., Rumi M. A. K., Konno T., Soares M. J., Natural killer cells direct hemochorial placentation by regulating hypoxia-inducible factor dependent trophoblast lineage decisions. Proc. Natl. Acad. Sci. U.S.A. 108, 16295–16300 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Renaud S. J., Scott R. L., Chakraborty D., Rumi M. A., Soares M. J., Natural killer-cell deficiency alters placental development in rats. Biol. Reprod. 96, 145–158 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pijnenborg R., et al. , Placental bed spiral arteries in the hypertensive disorders of pregnancy. Br. J. Obstet. Gynaecol. 98, 648–655 (1991). [DOI] [PubMed] [Google Scholar]
- 47.Jindal P., et al. , Placental pathology of recurrent spontaneous abortion: The role of histopathological examination of products of conception in routine clinical practice: A mini review. Hum. Reprod. 22, 313–316 (2007). [DOI] [PubMed] [Google Scholar]
- 48.von Steinburg S. P., Krüger A., Fischer T., Mario Schneider K. T., Schmitt M., Placental expression of proteases and their inhibitors in patients with HELLP syndrome. Biol. Chem. 390, 1199–1204 (2009). [DOI] [PubMed] [Google Scholar]
- 49.Kashif M., Isermann B., Role of the coagulation system in development. Thromb. Res. 131, S14–S17 (2013). [DOI] [PubMed] [Google Scholar]
- 50.Mastrolia S. A., Mazor M., Loverro G., Klaitman V., Erez O., Placental vascular pathology and increased thrombin generation as mechanisms of disease in obstetrical syndromes. PeerJ 2, e653 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Girard T. J., Tuley E., Broze G. J. Jr., TFPIβ is the GPI-anchored TFPI isoform on human endothelial cells and placental microsomes. Blood 119, 1256–1262 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brosens I., Puttemans P., Benagiano G., Placental bed research: I. The placental bed: From spiral arteries remodeling to the great obstetrical syndromes. Am. J. Obstet. Gynecol. 221, 437–456 (2019). [DOI] [PubMed] [Google Scholar]
- 53.Guerra-Shinohara E. M., et al. , Polymorphisms in antithrombin and in tissue factor pathway inhibitor genes are associated with recurrent pregnancy loss. Thromb. Haemost. 108, 693–700 (2012). [DOI] [PubMed] [Google Scholar]
- 54.Schreiber K., Hunt B. J., Pregnancy and antiphospholipid syndrome. Semin. Thromb. Hemost. 42, 780–788 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Tong M., Viall C. A., Chamley L. W., Antiphospholipid antibodies and the placenta: A systematic review of their in vitro effects and modulation by treatment. Hum. Reprod. Update 21, 97–118 (2015). [DOI] [PubMed] [Google Scholar]
- 56.Forastiero R. R., Martinuzzo M. E., Broze G. J. Jr., High titers of autoantibodies to tissue factor pathway inhibitor are associated with the antiphospholipid syndrome. J. Thromb. Haemost. 1, 718–724 (2003). [DOI] [PubMed] [Google Scholar]
- 57.Adams M. J., Donohoe S., Mackie I. J., Machin S. J., Anti-tissue factor pathway inhibitor activity in patients with primary antiphospholipid syndrome. Br. J. Haematol. 114, 375–379 (2001). [DOI] [PubMed] [Google Scholar]
- 58.Sebire N. J., et al. , Defective endovascular trophoblast invasion in primary antiphospholipid antibody syndrome-associated early pregnancy failure. Hum. Reprod. 17, 1067–1071 (2002). [DOI] [PubMed] [Google Scholar]
- 59.Poulton K., et al. , Purified IgG from patients with obstetric but not IgG from non-obstetric antiphospholipid syndrome inhibit trophoblast invasion. Am. J. Reprod. Immunol. 73, 390–401 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Blank M., et al. , The efficacy of specific IVIG anti-idiotypic antibodies in antiphospholipid syndrome (APS): Trophoblast invasiveness and APS animal model. Int. Immunol. 19, 857–865 (2007). [DOI] [PubMed] [Google Scholar]
- 61.Soares M. J., Chakraborty D., Kubota K., Renaud S. J., Rumi M. A. K., Adaptive mechanisms controlling uterine spiral artery remodeling during the establishment of pregnancy. Int. J. Dev. Biol. 58, 247–259 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pollheimer J., Vondra S., Baltayeva J., Beristain A. G., Knöfler M., Regulation of placental extravillous trophoblasts by the maternal uterine environment. Front. Immunol. 9, 2597 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Red-Horse K., et al. , Cytotrophoblast induction of arterial apoptosis and lymphangiogenesis in an in vivo model of human placentation. J. Clin. Invest. 116, 2643–2652 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moser G., et al. , Extravillous trophoblasts invade more than uterine arteries: Evidence for the invasion of uterine veins. Histochem. Cell Biol. 147, 353–366 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mackman N., The many faces of tissue factor. J. Thromb. Haemost. 7, 136–139 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Åberg M., Eriksson O., Siegbahn A., Tissue factor noncoagulant signaling: Mechanisms and implications for cell migration and apoptosis. Semin. Thromb. Hemost. 41, 691–699 (2015). [DOI] [PubMed] [Google Scholar]
- 67.Zelaya H., Rothmeier A. S., Ruf W., Tissue factor at the crossroad of coagulation and cell signaling. J. Thromb. Haemost. 16, 1941–1952 (2018). [DOI] [PubMed] [Google Scholar]
- 68.Sato Y., Endovascular trophoblast and spiral artery remodeling. Mol. Cell. Endocrinol. 503, 110699 (2020). [DOI] [PubMed] [Google Scholar]
- 69.Broze G. J. Jr., et al. , The lipoprotein-associated coagulation inhibitor that inhibits the factor VII-tissue factor complex also inhibits factor Xa: Insight into its possible mechanism of action. Blood 71, 335–343 (1988). [PubMed] [Google Scholar]
- 70.Girard T. J., et al. , Functional significance of the Kunitz-type inhibitory domains of lipoprotein-associated coagulation inhibitor. Nature 338, 518–520 (1989). [DOI] [PubMed] [Google Scholar]
- 71.MacLean J. A. II, et al. , Family of Kunitz proteins from trophoblast: Expression of the trophoblast Kunitz domain proteins (TKDP) in cattle and sheep. Mol. Reprod. Dev. 65, 30–40 (2003). [DOI] [PubMed] [Google Scholar]
- 72.MacLean J. A. II, Roberts R. M., Green J. A., Atypical Kunitz-type serine proteinase inhibitors produced by the ruminant placenta. Biol. Reprod. 71, 455–463 (2004). [DOI] [PubMed] [Google Scholar]
- 73.Chakrabarty A., Green J. A., Roberts R. M., Origin and evolution of the TKDP gene family. Gene 373, 35–43 (2006). [DOI] [PubMed] [Google Scholar]
- 74.Mast A. E., et al. , Tissue factor pathway inhibitor binds to platelet thrombospondin-1. J. Biol. Chem. 275, 31715–31721 (2000). [DOI] [PubMed] [Google Scholar]
- 75.Peterson J. A., Maroney S. A., Martinez N. D., Mast A. E., Major reservoir for heparin-releasable TFPIα (tissue factor pathway inhibitor α) is extracellular matrix. Arterioscler. Thromb. Vasc. Biol. 41, 1942–1955 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kojima T., et al. , Human ryudocan from endothelium-like cells binds basic fibroblast growth factor, midkine, and tissue factor pathway inhibitor. J. Biol. Chem. 271, 5914–5920 (1996). [DOI] [PubMed] [Google Scholar]
- 77.Mast A. E., et al. , Glypican-3 is a binding protein on the HepG2 cell surface for tissue factor pathway inhibitor. Biochem. J. 327, 577–583 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Khurana S., et al. , Glypican-3-mediated inhibition of CD26 by TFPI: A novel mechanism in hematopoietic stem cell homing and maintenance. Blood 121, 2587–2595 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mine S., Yamazaki T., Miyata T., Hara S., Kato H., Structural mechanism for heparin-binding of the third Kunitz domain of human tissue factor pathway inhibitor. Biochemistry 41, 78–85 (2002). [DOI] [PubMed] [Google Scholar]
- 80.O’Shea K. S., Liu L. H., Kinnunen L. H., Dixit V. M., Role of the extracellular matrix protein thrombospondin in the early development of the mouse embryo. J. Cell Biol. 111, 2713–2723 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Andraweera P. H., et al. , Scope Consortium, A functional variant in the thrombospondin-1 gene and the risk of small for gestational age infants. J. Thromb. Haemost. 9, 2221–2228 (2011). [DOI] [PubMed] [Google Scholar]
- 82.Stenczer B., et al. , Circulating levels of thrombospondin-1 are decreased in HELLP syndrome. Thromb. Res. 129, 470–473 (2012). [DOI] [PubMed] [Google Scholar]
- 83.Ishiguro K., et al. , Syndecan-4 deficiency impairs the fetal vessels in the placental labyrinth. Dev. Dyn. 219, 539–544 (2000). [DOI] [PubMed] [Google Scholar]
- 84.Jeyarajah M. J., Jaju Bhattad G., Kops B. F., Renaud S. J., Syndecan-4 regulates extravillous trophoblast migration by coordinating protein kinase C activation. Sci. Rep. 9, 10175 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ferrari do Outeiro-Bernstein M. A., et al. , A recombinant NH(2)-terminal heparin-binding domain of the adhesive glycoprotein, thrombospondin-1, promotes endothelial tube formation and cell survival: A possible role for syndecan-4 proteoglycan. Matrix Biol. 21, 311–324 (2002). [DOI] [PubMed] [Google Scholar]
- 86.Chui A., et al. , The expression of placental proteoglycans in pre-eclampsia. Gynecol. Obstet. Invest. 73, 277–284 (2012). [DOI] [PubMed] [Google Scholar]
- 87.Gunatillake T., et al. , Decreased placental glypican expression is associated with human fetal growth restriction. Placenta 76, 6–9 (2019). [DOI] [PubMed] [Google Scholar]
- 88.Kiyozumi D., Nakano I., Sato-Nishiuchi R., Tanaka S., Sekiguchi K., Laminin is the ECM niche for trophoblast stem cells. Life Sci. Alliance 3, e201900515 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Havugimana P. C., et al. , A census of human soluble protein complexes. Cell 150, 1068–1081 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ain R., Konno T., Canham L. N., Soares M. J., Phenotypic analysis of the rat placenta. Methods Mol. Med. 121, 295–313 (2006). [DOI] [PubMed] [Google Scholar]
- 91.Chakraborty D., et al. , HIF-KDM3A-MMP12 regulatory circuit ensures trophoblast plasticity and placental adaptations to hypoxia. Proc. Natl. Acad. Sci. U.S.A. 113, E7212–E7221 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pan S., Kleppe L. S., Witt T. A., Mueske C. S., Simari R. D., The effect of vascular smooth muscle cell-targeted expression of tissue factor pathway inhibitor in a murine model of arterial thrombosis. Thromb. Haemost. 92, 495–502 (2004). [DOI] [PubMed] [Google Scholar]
- 93.Wood J. P., et al. , TFPIα interacts with FVa and FXa to inhibit prothrombinase during the initiation of coagulation. Blood Adv. 1, 2692–2702 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chakraborty D., Muto M., Soares M. J., Ex vivo trophoblast-specific genetic manipulation using lentiviral delivery. Bio Protoc. 7, e2652 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq datasets are available at the Gene Expression Omnibus website (https://www.ncbi.nlm.nih.gov/geo/); rat TS cells: stem versus differentiated, GSE171753; rat TS cells: control versus TFPI knockdown, GSE171754; human TS cells: control versus TFPI knockdown, GSE171223). All data generated and analyzed during this study are included in the published article and the online supporting files. Resources generated and analyzed during the current study are available from the corresponding author upon reasonable request.