Significance
Phase separation of low-complexity (LC) domains appended to most RNA-binding proteins (RBPs) emerges as a principle underlying spatiotemporal protein recruitment. Yet, how LC domains regulate the function of RBPs in cells remains unclear. An alternative-splicing regulator, hnRNPH1, contains two LC domains (LC1 and LC2). Here, we show that phase separation of the LC1 can exert control over hnRNPH1 function in RNA-splicing possibly by facilitating interactions between hnRNPH1 and a variety of RBPs. In contrast, the LC2 lacking in vitro phase properties, is required for aberrant transcriptional activation in the context of fusion oncoproteins. These results have broad implications for understanding how phase separation contributes to distinct roles of LC domains in control of physiological as well as oncogenic functions.
Keywords: hnRNPH1, low-complexity domain, phase separation, splicing, transcription
Abstract
Heterogeneous nuclear ribonucleoproteins (hnRNPs) represent a large family of RNA-binding proteins that control key events in RNA biogenesis under both normal and diseased cellular conditions. The low-complexity (LC) domain of hnRNPs can become liquid-like droplets or reversible amyloid-like polymers by phase separation. Yet, whether phase separation of the LC domains contributes to physiological functions of hnRNPs remains unclear. hnRNPH1 contains two LC domains, LC1 and LC2. Here, we show that reversible phase separation of the LC1 domain is critical for both interaction with different kinds of RNA-binding proteins and control of the alternative-splicing activity of hnRNPH1. Interestingly, although not required for phase separation, the LC2 domain contributes to the robust transcriptional activation of hnRNPH1 when fused to the DNA-binding domain, as found recently in acute lymphoblastic leukemia. Our data suggest that the ability of the LC1 domain to phase-separate into reversible polymers or liquid-like droplets is essential for function of hnRNPH1 as an alternative RNA-splicing regulator, whereas the LC2 domain may contribute to the aberrant transcriptional activity responsible for cancer transformation.
Many RNA-binding proteins (RBPs) contain low-complexity (LC) domains as well as RNA-recognition motifs (RRMs). In contrast to the structurally well-defined RRMs, LC domains are not amenable to crystallography. These regions have therefore been classified by their amino acid profiles as they contain only a small subset of the 20 available amino acids. The LC domains in RBPs can undergo phase separation into liquid-like droplets (LLDs) or amyloid-like reversible polymers (1–5). Considerable interest in the phase properties of LC domains stems from the discoveries that abnormally formed FUS/EWS/TAF15 (FET) fusion oncoproteins promote aberrant gene transcription through phase separation of their LC domains and that amyotrophic lateral sclerosis (ALS)– or frontotemporal dementia–causing mutations in RBPs alter phase behavior, possibly promoting fibrillization of aggregation-prone proteins that underlies the pathologies of relevant diseases (3, 6). In addition to playing a role in a pathological context, a growing body of evidence has revealed that phase separation under normal conditions allows LC domains to function as scaffolds for the formation of reversible, membraneless, intracellular organelles such as nucleoli, stress granules, and neuronal transport granules (7, 8). Despite many studies focusing on the role of phase separation in the formation of pathological aggregates and membraneless organelles, its role in the physiological functions of RBPs has still not been carefully explored and many questions remain. The molecular code that drives phase separation often remains enigmatic (9). This has been a major barrier to understanding how phase separation regulates the transcription and RNA-processing functions of RBPs in the context of living cells.
The heterogeneous nuclear ribonucleoproteins (hnRNPs) represent a large family of RBPs contributing to multiple aspects of nucleic acid metabolism including alternative-splicing, messenger RNA (mRNA) stabilization, and transcriptional and translational regulation (10). hnRNPH1 is a prototypical hnRNP consisting of RRMs that occupy the N-terminal half of the protein and LC domains that occupy the C-terminal half. The LC domains appended to most RBPs, including hnRNPs, contain repeats of amino acid triplets in which tyrosine (Y) residues are flanked by either glycine (G) or serine (S) residues. Some hnRNP LC domains also contain repeats of phenylalanine and glycine residues (FG repeats) in addition to [G/S]Y[G/S] triplets. hnRNPH1 and its homolog hnRNPF are known to be involved in the control of alternative RNA-splicing of a variety of transcripts at distinct splice sites in cooperation with DEAD-box RNA helicases 5 and 17 (11, 12). The impact of hnRNPH1 binding on alternative-splicing is associated with several diseases, including ALS. hnRNPH1 colocalizes with RNA foci in brain tissues of familial ALS patients (10, 13). Expanding evidence demonstrates a potential role of hnRNPH1 in extensive alternative-splicing defects in patients with familial and sporadic ALS (14). hnRNPH1 splicing activity also shows clinical significance in congenital myasthenic syndrome (15) and cystic fibrosis (16). While the function of hnRNPH1 in regulating alternative-splicing is established, recent genomic analysis identified a novel fusion gene involving hnRNPH1 in acute lymphoblastic leukemia (17). The C-terminal half of hnRNPH1 containing the LC domains is rearranged with the N-terminal DNA-binding domain (DBD) of a transcriptional activator, MEF2D (myocyte-specific enhancer factor 2D), implying its role in tumorigenesis. Thus, understanding the regulatory mechanisms of hnRNPH1 is important for both basic and translational research.
To function as a regulator of alternative RNA-splicing, hnRNPH1 needs to form RNP complexes with different kinds of RNAs and other RBPs. Although RNA-binding by hnRNPH1 is known to be mediated by RRMs (18), control of protein-binding remains largely unclear. Remarkably, assembly and disassembly of hnRNP complexes occur rapidly; hnRNP complexes are successively remodeled through loss or acquisition of diverse RBPs as a consequence of specific processes (19). This dynamic nature is not readily explained by traditional lock-and-key models of protein–protein interaction. Given that dynamic low-affinity interactions are characteristic of phase-separated condensates (20), we hypothesized that phase separation of LC domains mediates protein–protein interactions of hnRNPH1 to regulate normal splicing activity. We thus set out to determine the phase separation properties of hnRNPH1 in regulating RNA-splicing and cancer. Our results show that the ability of the LC1 domain to phase-separate is essential for the physiological function of hnRNPH1 as a splicing regulator, whereas the LC2, which does not undergo phase separation, may contribute to aberrant transcriptional activity in leukemia-associated fusion proteins. Our observations reveal that distinct LC domains in hnRNPH1 may help organize and specialize its different functions.
Results
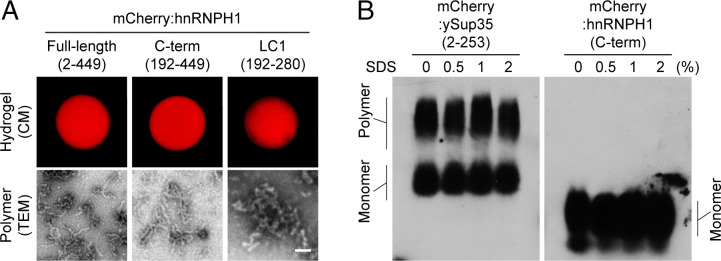
To investigate the phase separation of hnRNPH1, we first cloned different domains of hnRNPH1 (as shown in SI Appendix, Fig. S1A) into bacterial expression constructs with an N-terminal mCherry tag. Purified, recombinant proteins comprising mCherry-linked hnRNPH1 domains were then concentrated up to 100 mg/mL and prepared as small droplets on glass-bottomed chambers. Upon incubation at room temperature for 1 wk, pure solutions of full-length (amino acids 2 through 449) and C-terminal half (amino acids 192 through 449) hnRNPH1 became hydrogel-like state that could be visualized by confocal microscopy (CM) (Fig. 1A, Top Left and Middle). The LC1 domain (amino acids 192 through 280) of hnRNPH1 also became a hydrogel-like state; however, gelation took longer than for the full-length or C-terminal half and hydrogel droplets were not as solid, easily detaching upon addition of buffer (Fig. 1A, Top Right). The other domains of hnRNPH1 (N-terminal half, RRM3, and LC2 domain) did not become hydrogel. We next used a transmission electron microscope (TEM) to identify the nature of hydrogel droplets composed of the full-length, C-terminal half, and LC1 domain of hnRNPH1. TEM images revealed that the hnRNPH1 hydrogel droplets were composed of polymeric fibers (Fig. 1A, Bottom), which were shorter and more tangled in the case of the LC1 domain (Fig. 1A, Bottom Right).
Fig. 1.
In vitro phase separation of hnRNPH1. (A) Hydrogel droplets composed of mCherry-linked full-length (amino acids 2 through 449, Top Left), C-terminal half (C-term, amino acids 192 through 449, Top Middle), or LC1 domain (amino acids 192 through 280, Top Right) of hnRNPH1 were imaged by confocal microscopy (CM) (Materials and Methods). Polymers of each hydrogel droplet sample shown in Bottom panels were visualized by TEM (Materials and Methods). (Scale bar, 0.1 µm) (B) The polymer samples of mCherry-linked ySup35 or hnRNPH1 C terminus were incubated with indicated levels of SDS. The samples were then migrated through the agarose gel containing SDS, and the polymers or monomers were visualized by Western blotting using anti-mCherry antibodies.
In contrast to the extreme stability of pathogenic amyloid fibers (21, 22), phase separation–driven polymeric fibers are known to be labile to disassembly when exposed to sodium dodecyl sulfate (SDS) (1, 4). To examine the biochemical properties of polymeric fibers formed from the C-terminal half of hnRNPH1 in comparison with polymers composed of the typical prion, yeast Sup35 (ySup35) protein (SI Appendix, Fig. S2A) (23), we subjected polymer samples to semidenaturing detergent agarose gel electrophoresis (SDDAGE). When incubated in buffer containing no SDS or 0.5%, 1%, or 2% SDS, the ySup35 polymers did not depolymerize, even in the buffer with the highest SDS concentration (2%), instead migrating as large polymers by electrophoresis (Fig. 1B, Left). However, polymers of hnRNPH1 C-terminal half almost fully disassembled into monomers in gelation buffer containing no SDS (Fig. 1B, Right). In the previous studies, it was reported that the reversible amyloid-like polymers and LLDs formed by phase separation of LC domains are melted specifically by an aliphatic alcohol, 1,6-HD (hexanediol), but not by 2,5-HD (24, 25). As shown in SI Appendix, Fig. S2B, the hydrogel droplets composed of the C-terminal half of hnRNPH1 was melted by incubation with 15% level of 1,6-HD but not by 2,5-HD, indicating that the hydrogel droplet of hnRNPH1 C-terminal half is composed of labile polymers.
LC1 Domain of hnRNPH1 Drives Phase Separation.
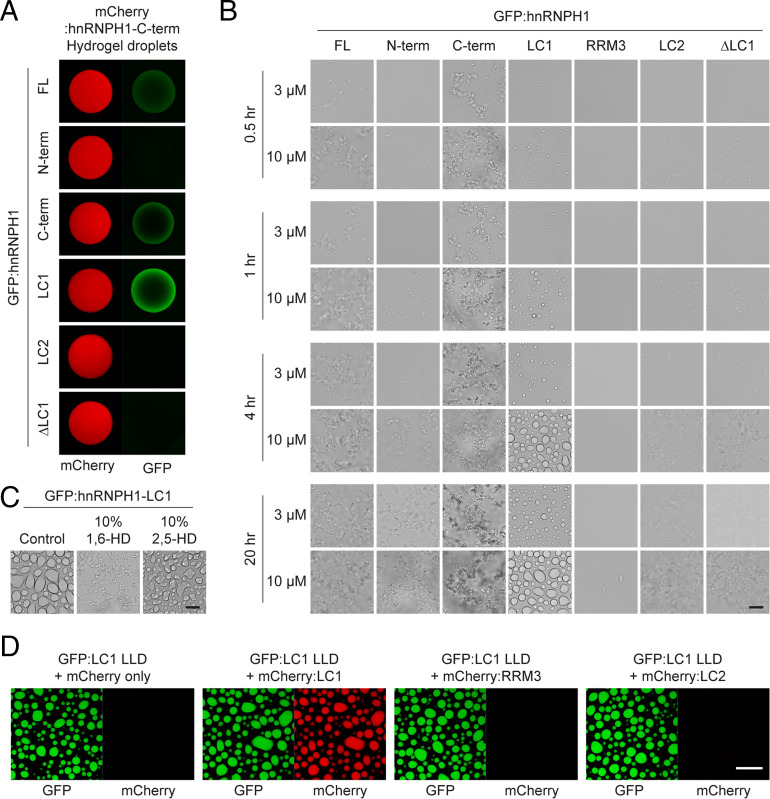
To further assess the subregions of hnRNPH1 responsible for phase separation, we used a hydrogel-binding assay. The hydrogel-binding assay is a reliable technique for identifying protein regions responsible for phase separation or phase separation–dependent association between proteins (2, 24). We therefore incubated mCherry hydrogel droplets composed of the hnRNPH1 C-terminal half with soluble GFP-linked proteins representing different regions of hnRNPH1 (Fig. 2A). Consistent with our polymerization and hydrogel formation assays (Fig. 1), the full-length, C-terminal half, and LC1 domain of hnRNPH1 were retained on mCherry hydrogel droplets composed of the hnRNPH1 C-terminal half but the N-terminal half, and LC2 domain of hnRNPH1 were not. Furthermore, ΔLC1 hnRNPH1 did not bind mCherry hydrogel droplets composed of the hnRNPH1 C-terminal half. These data suggest that the LC1 domain is required for phase separation–dependent self-association of hnRHNPH1.
Fig. 2.
Hydrogel binding and phase separation of the LC1 domain of hnRNPH1. (A) Hydrogel droplets composed of mCherry-linked C-terminal half of hnRNPH1 were challenged with soluble proteins of GFP-linked full-length (FL), N-terminal half (N-term), C-terminal half (C-term), LC1 domain, LC2 domain, or ΔLC1 hnRNPH1. Upon overnight incubation, trapping of the GFP-linked proteins by the mCherry hydrogel droplets was detected using confocal microscopy. (B) Phase separation of GFP-linked different regions of hnRNPH1 proteins into LLDs. Different concentrations (3 or 10 µM) of GFP-linked recombinant proteins were incubated in the presence of 10% PEG for indicated time periods. The LLDs were visualized using light microscopy. (Scale bar, 10 µm.) (C) Melting of LLDs formed from the hnRNPH1 LC1 domain by 1,6- or 2,5-HDs. The droplets were visualized using light microscopy. (Scale bar, 10 µm.) (D) Incorporation of 1 µM of mCherry alone, mCherry-linked LC1, RRM3, or LC2 domains of hnRNPH1 into LLDs composed of 10 µM GFP-linked LC1 domain of hnRNPH1. Upon overnight incubation, the mCherry signals incorporated to the GFP LLDs were assessed using confocal microscopy. (Scale bar, 20 µm.)
Recent studies have shown that the LC domains of a variety of RNA regulatory proteins can undergo phase separation into LLDs (4, 26). To examine whether hnRNPH1 can also become LLDs, we subjected purified, recombinant proteins of different GFP-linked regions of hnRNPH1 to an in vitro LLD formation assay. When dialyzed in buffer containing a physiological concentration of salt (150 mM NaCl), all the hnRNPH1 proteins tested remained clear. However, upon addition of the crowding agent, 10% polyethylene glycol (PEG), we observed formation of spherical-shaped LLDs in a concentration- and time-dependent manner for the LC1 domain of hnRNPH1 (Fig. 2B). We could observe the LC1 domain LLDs from 30 min after addition of 10% PEG (Fig. 2B). We validated the formation of labile LLDs by monitoring the effects of the aliphatic alcohols and as shown in Fig. 2C, the LC1 LLDs were melted substantially by 10% level of 1,6-HD but not by 2,5-HD following overnight incubation. Interestingly, robust formation of the aggregation-like precipitates was observed for full-length and C-terminal half of hnRNPH1, and it was also concentration- and time-dependent (Fig. 2B). The aggregation-like precipitates were melted upon addition of 0.5% SDS or 10% 1,6-HD but not by 10% 2,5-HD, implying the reversible and labile nature of the precipitates produced from each protein sample (SI Appendix, Fig. S3A). The N-terminal half, RRM3, LC2, and ΔLC1 of hnRNPH1 formed only small precipitates at high concentrations and after longer incubation time, suggesting that the formation of the labile aggregation-like precipitates is LC1 domain dependent (Fig. 2B and SI Appendix, Fig. S3A).
Given that the LC1 domain of hnRNPH1 is sufficient for phase separation into LLDs, while full-length or C-terminal half exhibited stronger hydrogel formation and were prone to precipitate in LLD formation assays, we asked whether RRM3 or LC2 domains modulate phase behavior of the LC1 domain. To test this, mCherry-linked recombinant proteins of the C terminus (LC1/RRM3/LC2), ΔLC1 (RRM3/LC2), ΔRRM3 (LC1/LC2), and ΔLC2 (LC1/RRM3) and LC1 alone of hnRNPH1 were subjected to hydrogel formation. Of note, mCherry-linked ΔRRM3 (LC1/LC2) became hydrogel droplets more effectively than the C terminus (LC1/RRM3/LC2), indicating that the LC2 domain enhances phase separation (SI Appendix, Fig. S3B). The other mCherry-linked proteins did not become hydrogel droplets until 10 d of incubation (SI Appendix, Fig. S3B). We confirmed that the hydrogel droplets composed of mCherry-linked ΔRRM3 (LC1/LC2) were melted upon incubation with 15% level of 1,6-HD but not by 2,5-HD (SI Appendix, Fig. S3B). To further understand the contribution of the LC1, RRM3, and LC2 to phase separation, we next applied mCherry-linked recombinant proteins of the LC1, RRM3, or LC2 domains to the LLDs composed of the GFP-linked LC1 domain and examined the incorporation of the mCherry-linked proteins into the LLDs. As shown in Fig. 2D, mCherry-linked LC1 domain exhibited strong incorporation into LLDs composed of GFP-linked LC1 domain, while no such incorporation was observed for the mCherry alone, mCherry-linked RRM3, or LC2 domains. Collectively, these data suggest that the LC1 domain is critical for driving phase separation of hnRNPH1, while the LC2 domain is capable of modulating phase behavior but dispensable for phase separation.
The LC1 Domain Is Required for Interactions between hnRNPH1 and Other RBPs.
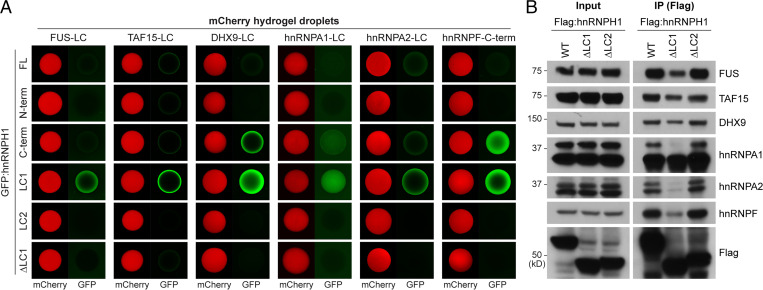
For hnRNPH1 to function as a regulator of RNA-processing in cells, it needs to form a complex through interactions with a variety of RNAs and RNA regulatory proteins (27). Considering that the hnRNPH1 LC1 domain underwent phase separation into reversible polymeric fibers or LLDs and was sufficient to drive homotypic association in hydrogel-binding assays, we hypothesized that the LC1 domain contributes to hnRNPH1 interactions with other RBPs harboring LC domains by forming dynamic LC–LC interactions. To test this possibility, we first challenged hydrogel droplets composed of LC domains appended to different types of RBPs (FUS, TAF15, DHX9, hnRNPA1, or hnRNPA2) with soluble GFP-linked proteins of different regions of hnRNPH1. As shown in Fig. 3A, the LC1 domain of hnRNPH1 was avidly trapped by all hydrogel droplets tested. Weak trapping was observed when the full-length or C-terminal half of hnRNPH1 were exposed to hydrogel droplets, and no trapping was observed when the N-terminal half, LC2 domain, or ΔLC1 hnRNPH1 were exposed. Given that neither the N-terminal half nor the LC2 domain are, on their own, capable of phase separation in vitro, these data tentatively indicate that hnRNPH1 makes use of the LC1 domain to adhere to existing hydrogel droplets of different LC domains. We also tested whether the LC1 domain is important for hnRNPH1 interaction with hnRNPF, its coregulator for RNA-splicing, which contains LC domain in the C-terminal domain (SI Appendix, Fig. S1A). As shown in SI Appendix, Fig. S4, the C-terminal region of hnRNPF underwent phase separation into hydrogel droplet, which was melted upon incubation with 15% level of 1,6-HD but not by 2,5-HD. In hydrogel-binding assays, hydrogel droplets composed of the hnRNPF C-terminal region trapped the GFP-linked C-terminal half and LC1 domain of hnRNPH1 avidly, the full-length hnRNPH1 weakly, and the N-terminal half, LC2, and ΔLC1 of hnRNPH1 poorly (Fig. 3A, Right), implying a role for the hnRNPH1 LC1 domain in the interactions between hnRNPH1 and hnRNPF.
Fig. 3.
The LC1 domain of hnRNPH1 is required for interactions with different kinds of RNA regulatory proteins. (A) Different regions of hnRNPH1 in the form of GFP-linked protein at equal molar concentrations were incubated with mCherry hydrogel droplets composed of LC domains of FUS, TAF15, DHX9, hnRNPA1, hnRNPA2, and hnRNPF. The intensity of the hydrogel binding was analyzed using confocal microscopy. (B) HEK-293T cells were transfected with Flag-tagged WT, ΔLC1, or ΔLC2 hnRNPH1 constructs. Immunoprecipitation was performed using anti-Flag antibodies, and coprecipitated proteins were analyzed by Western blotting using antibodies for indicated proteins. Representative images are shown from more than three independent immunoprecipitation experiments.
To further validate the internal domain of hnRNPH1 responsible for interaction with other RBPs in living cells, we subjected Flag-tagged full-length, ΔLC1 domain, or ΔLC2 domain hnRNPH1 to pull-down assays in human embryonic kidney (HEK)-293T cells. As shown in Fig. 3B, full-length hnRNPH1 coprecipitated with a variety of RBPs including FUS, TAF15, DHX9, hnRNPA1, hnRNPA2, and hnRNPF. Consistent with the in vitro hydrogel-binding assay, interactions between hnRNPH1 and these RBPs were reduced by deletion of the hnRNPH1 LC1 domain (ΔLC1) but not by deletion of the LC2 domain (ΔLC2). These data suggest that the LC1 domain but not the LC2 domain is critical for interactions between hnRNPH1 and other RBPs in cells.
Tyrosine Residues in LC1 Are Important Determinants for Phase Separation of hnRNPH1.
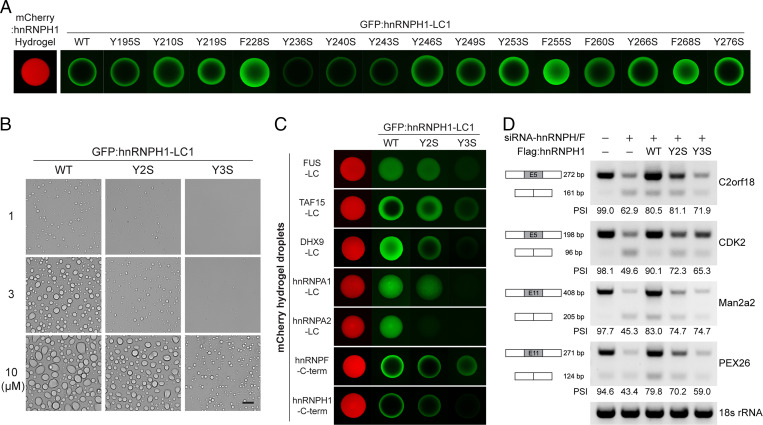
Many LC domains that undergo phase separation contain repeats of aromatic amino acids—tyrosine and/or phenylalanine residues. Previous studies have revealed that repetitive tyrosine or phenylalanine residues are critical for phase separation and function of LC domains (1, 3, 5, 28). Tyrosine-to-serine (Y-to-S) substitutions within the [G/S]Y[G/S] triplet repeats of FUS and TAF15 LC domains effectively disrupt the formation of amyloid-like polymers as well as the transcriptional activity of the LC domains (3). In the case of hnRNPA2, a single phenylalanine-to-serine (F-to-S) substitution is sufficient to interrupt incorporation of the LC domain into preexisting LLDs or hydrogel droplets (5). The LC1 domain of hnRNPH1 contains 11 repeats of the triplet sequence [G/S]Y[G/S] and four phenylalanine residues (SI Appendix, Fig. S1B). To identify the sequence determinants for phase separation of the hnRNPH1 LC1 domain, we generated 11 mutant LC1 domains with a single Y-to-S (Y195S, Y210S, Y219S, Y236S, Y240S, Y243S, Y246S, Y249S, Y253S, Y266S, and Y276S) substitution and four with a single F-to-S (F228S, F255S, F260S, and F268S) substitution. Wild-type (WT) or mutant LC1 domains were produced recombinantly in the form of fusion proteins with an N-terminal GFP tag and applied to chamber slides containing hydrogel droplets composed of the mCherry-linked hnRNPH1 C-terminal half. Following overnight incubation, hydrogel-trapping of WT or mutant GFP-tagged LC1 domains of hnRNPH1 was analyzed using confocal microscopy. As shown in Fig. 4A, the intensity of hydrogel binding was decreased among LC1 domains with Y236S, Y240S, or Y243S substitutions compared with that of the WT LC1, suggesting that residues Y236, Y240, and Y243 are critical to trapping by hydrogel droplets of the hnRNPH1 C-terminal half. We then tested the effect of the Y/F-to-S substitutions on LLD formation of the hnRNPH1 LC1 domain. GFP-linked recombinant proteins of WT or mutant hnRNPH1 LC1 domains were dialyzed in buffer containing 150 mM NaCl and 10% PEG. As shown in Fig. 4B and SI Appendix, Fig. S5A, WT LC1 formed stable droplets at all protein concentrations (1 to 10 µM), and single Y/F-to-S substitutions did not lead to observable changes in the formation of LLDs except for Y210S and Y219S, which exhibited slight decreases in the size of LLDs. We therefore generated a double Y-to-S mutant, Y210/219S (Y2S), and a triple Y-to-S mutant, Y236/240/243S (Y3S), and examined their effects on LLD formation. As shown in Fig. 4B, the Y2S LC1 mutant formed droplets at all protein concentrations, but the LLDs were smaller than those of the WT LC1 domain. Further, the Y3S LC1 mutant failed to form LLDs at 1 µM and formed much smaller droplets at 3 and 10 µM compared with those of the WT LC1 or Y2S LC1 mutant (Fig. 4B). These results suggest that the three tyrosine residues, Y236, Y240, and Y243, in the LC1 domain are critical for phase separation into LLDs and polymer formation.
Fig. 4.
Effects of Y-to-S mutations on phase separation and function of hnRNPH1. (A) The recombinant proteins of the WT or Y/F-to-S mutant hnRNPH1 LC1 domains with N-terminal GFP tag were incubated with hydrogel droplets composed of the mCherry-linked hnRNPH1 C-terminal half. Upon overnight incubation, hydrogel-trapping of the GFP-linked LC1 domains was analyzed using confocal microscopy. (B) For LLD formation assay, 1, 3, or 10 µM of the purified recombinant proteins of GFP-linked WT, Y210/219S (Y2S), and Y236/240/243S (Y3S) were incubated in buffer containing 150 mM NaCl and 10% level of the crowding agent, PEG. The LLDs were visualized by light microscopy. (Scale bar, 10 µm.) (C) GFP-linked proteins of WT, Y2S, or Y3S LC1 domain were incubated with different kinds of mCherry hydrogel droplets. Hydrogel binding of the GFP-linked proteins was analyzed by confocal microscopy. (D) RT-PCR was performed to analyze the splicing of the exon 5 of C2orf18, exon 5 of CDK2, exon 11 of Man2a2, and exon 11 of PEX26 transcripts. The mean value of PSI levels obtained from three independent experiments is shown.
To test whether the phase separation–deficient Y-to-S mutations impact hnRNPH1–RBP interactions, we examined trapping of GFP-linked WT or mutant hnRNPH1 LC domains by the six mCherry-linked hydrogel droplets presented in Fig. 3A, and hydrogel droplets composed of the hnRNPH1 C-terminal half using confocal microscopy. As shown in Fig. 4C, sequential diminishment in hydrogel-trapping was observed for Y2S or Y3S mutants. Consistent with the in vitro hydrogel-binding assays, Y2S and Y3S substitutions induced a reduction in interaction between hnRNPH1 and different kinds of RBPs as analyzed by immunoprecipitation assays (SI Appendix, Fig. S5B) without altering the subcellular localization of hnRNPH1 (SI Appendix, Fig. S5C). It is possible that the reduction in RBP interactions of hnRNPH1 mutants reflects the changes in their RNA-binding ability. This proved not to be the case. When we conducted RT-PCR following immunoprecipitation of Flag-tagged WT, ΔLC1, Y2S, and Y3S hnRNPH1 expressed in HEK-293T cells for human telomerase RNA component (hTERC), which has been reported to bind hnRNPH1 (29) ΔLC1 but neither Y2S nor Y3S reduced RNA-binding ability (SI Appendix, Fig. S5D). We also examined the effect of the phenylalanine residues (F228, F255, and F268) on hydrogel-trapping, RBP interaction, and alternative RNA-splicing (SI Appendix, Fig. S5 E–G). Although substitution of all three phenylalanine residues to serine (F3S) somewhat decreased hydrogel-trapping of the LC1 domain of hnRNPH1, they did not affect RBP-binding and -splicing activity of hnRNPH1. These results indicate that the effects of Y-to-S mutations on hnRNPH1 phase behavior and protein interactions are correlated, supporting the conclusion that phase separation is essential for protein–protein interactions of hnRNPH1.
Phase Separation of the LC1 Domain Is Required for Splicing Regulation by hnRNPH1.
Knowing that the Y-to-S substitutions in the LC1 domain interfere with phase separation and interaction with other proteins, we hypothesized that diminished phase separation ability caused by these LC1 mutations might affect the function of hnRNPH1 as an alternative RNA-splicing regulator. To test this possibility, we analyzed changes in splicing pattern of transcripts produced from C2orf18, CDK2, Man2a2, and PEX26 genes upon knockdown (KD) of endogenous hnRNPH1 expression using RT-PCR. A small interfering RNA (siRNA) targeting hnRNPH1, its paralog hnRNPH2, and the homolog hnRNPF was used for KD because these proteins have functional redundancy in control of alternative RNA-splicing in cells (12). KD of the endogenously expressed hnRNPH1/2 and hnRNPF was validated using immunoblotting (SI Appendix, Fig. S6A). As shown in Fig. 4D, KD of hnRNPH/F resulted in a significant decrease in the percent spliced in (PSI) levels of C2orf18 exon 5, CDK2 exon 5, Man2a2 exon 11, and PEX26 exon 11. Upon transient transfection of the WT hnRNPH1 gene, the PSI levels of those exons were increased (Fig. 4D, third lane); whereas the Y2S and Y3S hnRNPH1 mutants failed to restore PSI levels as much as the WT hnRNPH1 (Fig. 4D, fourth and fifth lanes). Expression of the transiently expressed hnRNPH1 proteins was analyzed by Western blotting (SI Appendix, Fig. S6B). Thus, these data indicate that phase separation mediated by the LC1 domain might be required for the biological function of hnRNPH1 in regulating RNA-splicing.
The hnRNPH1 LC2 Domain Can Function as a Transcriptional Activation Domain.
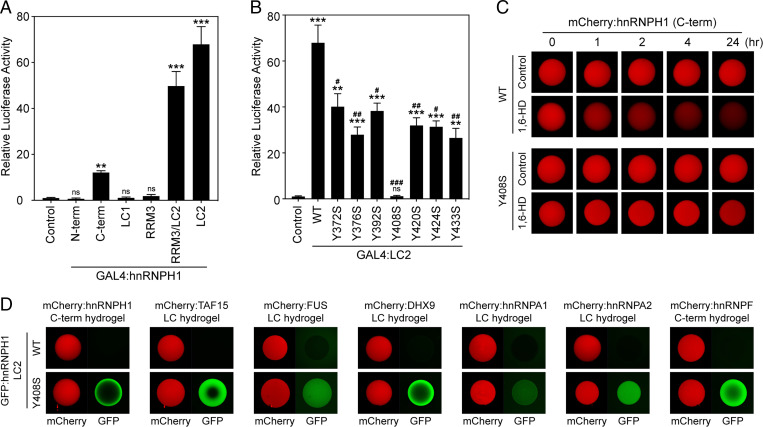
B-cell precursor acute lymphoblastic leukemia (BCP-ALL) is the most-common pediatric cancer, and its prognosis in adults is generally unfavorable (30). Chimeric fusion proteins involving transcription factors are hallmarks of BCP-ALL. A recent human genetic study reported the presence of genes encoding fusions between the N-terminal DBD of the MEF2D transcription factor and the C-terminal regions of two different hnRNPs, hnRNPH1 and hnRNPUL1, in BCP-ALL (17). Remarkably, both MEF2D-hnRNPH1 and MEF2D-hnRNPUL1 retain the LC domains of the hnRNP protein. Although MEF2D-BCL9, the most-common MEF2D fusion protein found in ALL, results in enhanced transcriptional activity (31), little is known about the mechanisms by which MEF2D–hnRNP fusion genes contribute to tumorigenesis (17). We hypothesized that the LC domains in the C terminus of hnRNPH1 might activate transcription, because LC domains from the FET (FUS, EWS, and TAF15) protein family have been shown to function as potent transcriptional activation domains in oncogenic fusion proteins (3, 32). To test this possibility, we linked hnRNPH1 genes of different lengths and containing different subregions to the GAL4-DBD and assayed for transcriptional activation of the firefly luciferase gene as a reporter. As shown in Fig. 5A, the C-terminal half of hnRNPH1 linked to GAL4 DBD activated transcription of luciferase downstream of the upstream activation sequence (Materials and Methods) by more than 10-fold compared with the GAL4 control, while the N-terminal half of hnRNPH1 did not, suggesting that the C terminus of hnRNPH1 indeed gained transcriptional activity through the formation of a fusion gene with the DBD. Further dissecting the C-terminal half of hnRNPH1 for transcriptional activity revealed the highest transcriptional activity in cells transfected with the LC2 domain, while LC1 or RRM3 alone did not transactivate the luciferase reporter gene (Fig. 5A).
Fig. 5.
The LC2 domain of hnRNPH1 is required for transcriptional activity as a fusion protein with DBD. (A) The indicated regions of hnRNPH1 were linked to the DBD of GAL4 and assayed for activation of GAL4-dependent firefly luciferase reporter gene in HEK-293T cells. Relative luciferase activity compared to GAL4-DBD-only control. (B) Transcriptional activity of the WT or Y-to-S mutant LC2 domains of hnRNPH1 linked to GAL4-DBD were analyzed in HEK-293T cells. (A and B) One-way ANOVA was used to evaluate statistical significance. Mean ± SEM from three independent assays; ns, not significant versus control; *P < 0.01, **P < 0.001, and ***P < 0.0001 versus control; and #P < 0.01, ##P < 0.001, and ###P < 0.0001 versus WT. (C) Melting of mCherry hydrogel droplets composed of WT or Y408S C-terminal half of hnRNPH1 15% levels of an aliphatic alcohol, 1,6-HD. The hydrogel droplets were imaged by confocal microscopy at indicated time points after addition of 1,6-HD. (D) Different kinds of mCherry hydrogel droplets were challenged with GFP-linked proteins of WT or Y408S LC2 domains of hnRNPH1. Trapping of the GFP-linked proteins by the hydrogel droplets was analyzed by confocal microscopy.
The Y-to-S mutations inhibiting in vitro phase separation of FUS and TAF15 LC domains also abolished the transcriptional activity of these LC domains (3), while Y-to-F mutations, which enhanced phase separation of AKAP95 to form less-dynamic droplets, impaired AKAP95 functions in gene expression and tumorigenesis (33). Thus, mutations perturbing phase separation in opposite directions may actually be deleterious to protein function. Similarly, although in vitro phase separation into a hydrogel-like state or LLDs was not observed for the hnRNPH1 LC2 domain, it is still possible that mutations altering the phase property of the LC2 domain can affect transcriptional activity. To explore this possibility, we first generated GAL4-linked mutant LC2 domains where tyrosine residues were mutated to serine (SI Appendix, Fig. S1B). In GAL4-dependent transcriptional reporter assays, all seven mutant LC2 domains showed decreased transcriptional activity compared with the WT one: in particular, the Y408S mutation almost completely abolished LC2-mediated transcription of the luciferase reporter gene (Fig. 5B). Since the C-terminal half without LC1 (RRM3/LC2) was considerably more active than WT C-terminal half of hnRNPH1 as shown in Fig. 5A, it was hypothesized that the phase separation capability of the LC1 domain might interfere with the transcriptional activity of the LC2 domain. To test this possibility, we conducted luciferase reporter assays using WT, Y408S, Y2S, and Y3S C-terminal half of hnRNPH1 linked to GAL4-DBD. While Y408S mutation significantly decreased the transcriptional activity of the hnRNPH1 C-terminal half, C-terminal variants carrying either Y2S or Y3S exhibited transcriptional activation capacity indistinguishable from WT, suggesting that phase separation property of LC1 is separate from the LC2-mediated transcriptional activation (SI Appendix, Fig. S7A).
We then examined the effects of the Y408S mutation on the phase behavior of the LC2 domain. To do this, we generated hydrogel droplets using mCherry-linked hnRNPH1 C-terminal halfs having WT or Y408S mutant LC2 domains (Fig. 5C). The C-terminal half of hnRNPH1 harboring Y408S formed hydrogel droplets that were fully analogous to those formed from WT protein. However, unlike the WT hydrogel droplets, which were melted substantially by 1,6-HD, hydrogel droplets composed of the C terminus with Y408S mutation were resistant to 1,6-HD even after overnight exposure at 37 °C (Fig. 5C). Moreover, in hydrogel-binding assays using mCherry hydrogel droplets composed of different types of LC domain, the GFP-linked Y408S LC2 domain exhibited avid hydrogel-trapping, which was not observed for the GFP-linked WT LC2 domain (Fig. 5D). However, Y408S LC2 domain did not form hydrogel droplets on its own, like the WT LC2 domain (SI Appendix, Fig. S7B). These results suggest that the Y408S mutation not only promotes phase separation of the C-terminal half of hnRNPH1 into less-labile droplets but also induces the LC2 domain to favor hydrogel-dependent interactions with other LC domains of a variety of RBPs. Taken together, these data indicate that the transcriptional activation capacity of the hnRNPH1 C-terminal requires its LC2 domain and is impaired by perturbation of the phase separation property. We propose a model in which LC1 drives dynamic protein–protein interactions, regulating splicing function of hnRNPH1 under physiological conditions, while LC2, which shows little tendency to phase-separate, is important for the transcriptional activation capacity of hnRNPH1 when divorced from its normal setting.
Discussion
The proper function of hnRNPH1 in splicing regulation is highly dependent on its multivalent interactions with RBPs including hnRNPF, a coregulator of alternative-splicing (29). Here, we showed that the C-terminal half of hnRNPH1, which contains two LC domains (LC1 and LC2), became hydrogel droplets composed of reversible polymers by phase separation (Fig. 1). Of the two LC domains of hnRNPH1, the LC1 domain was not only required for phase separation–dependent interactions with a variety of other LC domains but also sufficient for phase separation into reversible polymers as well as LLDs in vitro (Figs. 1 and 2). Aliphatic alcohols showed similar abilities to melt both hydrogels and LLDs composed of the LC1 domain, implying that the LC1 domains are disposed similarly in hydrogel droplets and LLDs (Figs. 2 and 5) (2). Despite the sequence similarity of the tyrosine triplets, in vitro phase separation was not observed for the LC2 domain. The LC1 domain was also required for interaction with other RBPs including FUS, TAF15, DHX9, hnRNPA1, hnRNPA2, and hnRNPF as revealed by in vitro hydrogel-binding assays and pull-down assays in living cells (Fig. 3). By investigating the aromatic amino acid residues of [G/S]Y[G/S] triplets and FG dipeptide repeats within the LC1 domain, we demonstrated that the tyrosine residues might be the key drivers of phase separation (Fig. 4 A and B). Notably, the Y210/219S (Y2S) and Y236/240/243S (Y3S) mutations that abrogate phase separation of the LC1 domain prevented both trapping of the LC1 domain by the hydrogel droplets composed of the LC domains appended to different kinds of RBPs in vitro and RBP interaction of hnRNPH1 in vivo without altering the subcellular localization and the RNA-binding ability of hnRNPH1 (Fig. 4C and SI Appendix, Fig. S5). In addition, the hnRNPH1 genes containing Y2S or Y3S mutations failed to restore the splicing of endogenous target gene transcripts in HEK-293T cells (Fig. 4D). Thus, our data link the phase properties of hnRNPH1 to its physiological function in splicing via its ability to compartmentalize multiple proteins.
A causal role of hnRNPH1 phase separation in its splicing activities is consistent with previous studies showing a role for phase separation of Rbfox proteins in their splicing activity (34) and the importance of LC domain-mediated interactions between the hnRNPA and D families in splicing regulation (35). The present study extends earlier work in several ways. First, we established a role for phase separation in the protein–protein interactions of hnRNPH1. Multivalent interactions between hnRNPs and other RBPs together with their distinct RNA-binding specificities underlie the mechanisms of hnRNP-dependent control of alternative RNA-splicing (36). Using in vitro hydrogel-binding assays and pull-down assays in cells, we demonstrated that hnRNPH1 recruits other related RBPs containing LC domain possibly via the phase separation process of the LC1 domain (Fig. 3). Second, we uncovered the sequence determinants for hnRNPH1 phase properties and provided evidence that both hnRNPH1 interactions with RBPs in vitro and the splicing activity of hnRNPH1 in HEK-293T cells are disrupted in phase separation–deficient mutants (Fig. 4). Interestingly, the LC domains enriched in [G/S]Y[G/S] motifs have been implicated in the formation of membraneless organelles and fibrillar-like structures in vivo (1, 28, 37, 38). Moreover, aberrant assembly of these structures can lead to formation of protein aggregates implicated in multisystem degenerative diseases (6). However, how protein assemblies formed by the LC domains containing [G/S]Y[G/S] and other types of repeat motifs regulate the physiological functions of RBPs are poorly understood. Our results demonstrate that phase separation control of multivalent interactions of hnRNPH1 might represent an important mechanism underlying splicing regulation.
The diverse functions played by hnRNPH1 are not limited to mRNA biogenesis. Genomic approaches have recently identified MEF2D–hnRNPH1 fusion as a novel rearrangement in BCP-ALL (17). However, the roles of the MEF2D–hnRNPH1 fusion molecule in the pathogenesis of leukemia remains unclear. The resulting gene product is a fusion protein in which a DBD of MEF2D is connected to the C-terminal region of hnRNPH1, retaining LC1 domain, LC2 domain, and RRM3. By employing a simple, functional read-out (transcriptional activation), we demonstrated that the LC2 domain has strong transcriptional activation capacity, whereas the LC1 domains and RRM3 show no detectable transcriptional activity (Fig. 5A). Furthermore, evaluation of single–amino acid substitutions of all seven tyrosine residues within the hnRNPH1 LC2 domain with respect to their abilities to activate transcription revealed that the transcriptional activity is strongly dependent upon tyrosine residues within the LC2 domain (Fig. 5B and SI Appendix, Fig. S1B). Evidence that the LC2 domain of hnRNPH1 is able to function as a transcriptional activation domain is consistent with recent studies of the cancer-causing fusion proteins linking FET (FUS, EWS, and TAF15) LC domains to a variety of different DBDs (3, 32, 39, 40). In the form of fusion genes with the GAL4-DBD, the LC domains of FUS and TAF15 exhibited strong transcriptional activity, which was in correlation with the phase separation capabilities of the LC domains (3). However, transcriptional regulation of hnRNPH1 and FET proteins shows different dependencies on phase separation propensity, suggesting a context-dependent regulation of protein function by phase properties. Collectively, our findings reveal that the hnRNPH1 C-terminal region gains an activity affecting gene expression when fused to DBDs, giving rise to the intriguing possibility that MEF2D–hnRNPH1 may contribute to the process of oncogenesis through an aberrant transcription-dependent mechanism.
How, then, might one conceptualize the manner in which intact hnRNPH1 proteins regulate RNA alternative-splicing in the context of their normal cellular function, while isolated parts of hnRNPH1 proteins that are translocated onto DBDs in cancer can achieve transcriptional activation? One possibility is that intact hnRNPH1 proteins are able to bind to RNAs via RRMs in their N-terminal regions. Perhaps the presence of RNAs might contribute to phase separation of hnRNPH1 into labile structures that are required for recruitment of RBPs implicated in splicing regulation to an hnRNPH1 complex. Further studies are required to understand the dynamic behavior of the LC domains under physiological and pathological conditions. Importantly, mutations of key residues in the LC1 and LC2 domains perturb the phase separation property of hnRNPH1 in opposite directions. Blocking of splicing function by Y-to-S mutations in the LC1 domain probably results from a failure in phase separation for recruiting key proteins into local proximity. By contrast, the Y408S mutation in the LC2 domain, which induces phase separation into less-labile hydrogel droplets, may slow down biochemical reaction kinetics inside by restricting movement and interactions of other macromolecules such as transcription factors and coactivators. Enhanced homotypic and heterotypic bindings to hydrogel droplets composed of different LC domains and resistance to 1,6-HD-mediated melting (Fig. 5) suggest that the Y408S mutation of the LC2 domain may restrict hnRNPH1 dynamics. Taken together, our findings suggest that the two LC domains in hnRNPH1 take on distinct roles: the LC1 domain regulates normal splicing activity, whereas the LC2 domain is required for aberrant transcriptional activation, providing a potential therapeutic approach directed against the oncogenic function of hnRNPH1.
Materials and Methods
Cloning.
For construction of bacterial expression plasmids, DNA fragments encoding different lengths of hnRNPH1 (SI Appendix, Fig. S1), FUS LC domain (residues 2 through 214), TAF15 LC domain (residues 2 through 208), hnRNPA1 LC domain (residues 190 through 319), hnRNPA2 LC domain (residues 181 through 341), hnRNPF LC domain (residues 192 through 415), and DHX9 LC domain (residues 1,151 through 1,270) were amplified by PCR using a complementary DNA (cDNA) library made from HEK-293T cells as a template. The amplified DNA fragments were inserted into pHis-GFP or pHis-mCherry parallel vectors. For generation of the mammalian expression plasmids of hnRNPH1, DNA fragments encoding WT, ΔLC1, ΔLC2, Y210/219S (Y2S), and Y236/240/243S (Y3S) were subcloned into pCMV10-3xFlag vector.
Purification of Recombinant Proteins.
The bacterial expression plasmids were transformed into Escherichia coli BL21 (DE3). Transformed bacterial cells were grown onto the Luria broth (LB) agar plate containing 100 µg ampicillin (Amp). A single colony was inoculated in 30 mL of LBAmp media and grown at 37 °C for overnight without shaking. The precultured bacterial cells were then transferred into 1 L of LBAmp media and grown at 37 °C with vigorous shaking until an optical density at 600 nm (OD600) of 0.7 was reached. Expression of the recombinant proteins was induced by addition of 0.5 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG). The bacterial cells were cultured for overnight at 16 °C with shaking. The bacterial cells were harvested and kept frozen until purification. For purification, the frozen cells were resuspended in a lysis buffer (50 mM Tris⋅HCl pH7.5, 500 mM NaCl, 20 mM β-mercaptoethanol (BME), 1% Tritone X-100, protease inhibitor mixture [PIC, Sigma-Aldrich, Germany], and 0.4 mg/mL lysozyme) and kept on ice for 30 min. The cell lysates were then further lysed by sonication at 60% power for total 3 min (Model 705 Sonic Dismembrator, Fisher). The cell lysates were then subjected to centrifugation at 20,000 rpm for 1 h at 4 °C, and the supernatant was mixed with the nickel nitrilotriacetic acid (Ni-NTA) agarose beads (Qiagen, Germany). Upon 30 min of incubation at 4 °C with gentle rotation, the Ni-NTA agarose beads were packed into a glass column (Bio-Rad). The Ni-NTA resin was washed with 500 mL of a wash buffer (20 mM Tris⋅HCl pH7.5, 500 mM NaCl, 20 mM imidazole, 20 mM BME, and 0.1 mM phenylmethylsulfonyl fluoride (PMSF)). Bound proteins were eluted using a wash buffer containing 200 mM imidazole. The purified proteins were kept frozen until use.
Hydrogel-Binding Assay.
For hydrogel droplet formation, the recombinant proteins of mCherry-linked LC domains of hnRNPH1, FUS, TAF15, hnRNPA2, hnRNPA1, hnRNPF, and DHX9 were dialyzed in gelation buffer (20 mM Tris⋅HCl pH7.5, 200 mM NaCl, 20 mM BME, and 0.1 mM PMSF). Dialyzed protein solution was subjected to brief sonication for 1 s at 3% power for three times. Upon removal of the precipitates by centrifugation for 3 min at maximum speed, the protein solution was concentrated to 80 to 100 mg/mL using Amicon Ultra (Merck Millipore). Small droplets (0.5 µl) of the protein solution were deposited on to a glass-bottomed confocal dishes (SPL, Korea). The confocal dishes were sealed with parafilm (Bemis) and incubated for 1 wk at room temperature until protein solution adopted a gel-like state. For hydrogel-binding assays, solutions of GFP-linked proteins at low concentration (1 or 3 µM) were applied to the confocal dishes containing different kinds of mCherry hydrogel droplets. Upon overnight incubation at 4 °C, trapping of the GFP-linked proteins by mCherry hydrogel droplets was analyzed by confocal microscopy (LSM 510, Zeiss, Germany).
TEM for Polymer Detection.
The recombinant proteins of mCherry-linked hnRNPH1 or ySup35 were dialyzed in gelation buffer and then concentrated and sonicated as described for hydrogel droplet formation. The protein samples were transferred to the surface of a TEM grid (Electron Microscopy Science). The polymer samples were stained with 10 µl of 2% uranyl acetate for 10 s and were washed with 20 µl of distilled water four times. The samples were dried for 1 min in the air at room temperature. TEM images were obtained at 120 kV using Zeiss LIBRA 120 electron microscope.
SDDAGE.
The polymer samples of mCherry-linked hnRNPH1 or ySup35 were diluted to 10 mg/mL in a gelation buffer. Upon brief sonication for 1 s at 1% power, the polymer samples were further diluted to 0.1 mg/mL in a gelation buffer. The samples were then exposed to indicated amounts of SDS and then incubated for 10 min at 37 °C. The samples were mixed with loading dye and subjected to electrophoresis using 1.5% agarose gel containing 0.1% SDS. Following electrophoresis, separated samples were transferred to nitrocellulose membranes by downward capillary transfer and were visualized by Western blotting using anti-mCherry antibodies (Abcam).
Melting of Hydrogel Droplets by Aliphatic Alcohols.
Hydrogel droplets composed of mCherry:hnRNPH1 C-terminal domain (residues 192 through 449) were exposed to 2 mL of 15% 1,6- or 2,5-hexandiol (Sigma-Aldrich, Germany). Upon incubation at 37 °C for indicated time periods, images of mCherry hydrogel droplets were obtained by confocal microscopy (LSM 510, Zeiss, Germany).
LLD Formation.
For LLD formation, the N-terminal His tags of all of the recombinant proteins used in the assays were precleaved using tobacco etch virus protease, and cut proteins were further purified by gel filtration chromatography using HiLoad 16/600 Superdex 75 pg column coupled to a fast protein liquid chromatography (FPLC) Äkta Pure (GE Healthcare). The purified GFP-linked hnRNPH1 proteins were dialyzed in LLD buffer (20 mM Tris⋅HCl pH7.5, 150 mM NaCl, 20 mM BME and 0.1 mM PMSF) for overnight at room temperature. The dialyzed proteins were mixed with buffer containing the crowding reagent, PEG (Sigma-Aldrich, Germany) to reach indicated concentration of proteins and 10% PEG. The samples were then transferred to glass-bottomed 96 well plates (PerkinElmer), which were precoated with 3% bovine serum albumin and washed three times with pure water. The samples were incubated at room temperature for overnight, and the LLD formation was analyzed by light microscopy (DMi8, Leica, Germany).
For LLD incorporation assays, mCherry-linked recombinant proteins (1 µM) were mixed with 10 µM of GFP:hnRNPH1-LC1. Upon addition of 10% PEG, the LLD samples were transferred to glass-bottomed 96-well plates (PerkinElmer). The intensity of GFP and mCherry signals were analyzed by confocal microscopy (LSM710, Zeiss, Germany).
Luciferase Reporter Assay.
The GAL4/UAS system was used to analyze the transcriptional activity of different domains of hnRNPH1. For this, at first, fusion genes linking GAL4 DBD, and different kinds of DNA fragments of hnRNPH1 as indicated in Fig. 5 A and B were cloned into pCMV10-3×Flag vector. The minimal promoter region containing six repeats of UAS were subcloned into pGL3-basic vector (Promega). For reporter assays, HEK-293T cells were reverse transfected with 10 ng of pGL3-basic-UAS and 5 ng of pRL-TK (Promega) in 96 well plates (50,000 cells per well) using lipofectamine 2000 (Invitrogen). Upon overnight incubation, the cells were forward transfected with indicated GAL4-hnRNPH1 plasmids using lipofectamine 2000. Transfection was performed according to the manufacturer’s protocol. Luciferase assay was performed at 36 h after forward transfection using Dual-Glo Luciferase Assay System (Promega) according to the manufacturer’s protocol. Luciferase activity was measured using the VICTOR ×3 Plate Reader (PerkinElmer, USA).
siRNA-Mediated KD of hnRNPH1.
The control siRNA and siRNA targeting hnRNPH/F (5′-GGAAGAAAUUGUUCAGUUC-3′) were synthesized from GenePharma, China. To validate the KD of hnRNPH/F, HEK-293T cells were reverse transfected with the indicated amounts of siRNA using lipofectamine RNAiMAX (Thermo Fisher Scientific) according to the manufacture’s protocol. The transfected cells were lysed at 72 h upon siRNA transfection, and the lysates were analyzed by Western blotting. For reintroduction of WT or mutant hnRNPH1 after KD, gene constructs were generated to escape from siRNA-mediated KD by modifying the nucleotide sequence of the siRNA target region. For overexpression of the sequence-modified hnRNPH1, at first, HEK-293T cells were reverse transfected with 10 µM of siRNAs. Upon 24 h of siRNA transfection, cells were forward transfected with indicated hnRNP constructs using lipofectamine 2000. Western blotting and RNA preparation was performed at 48 h after hnRNP transfection.
Immunoprecipitation.
HEK-293T cells were transfected with indicated Flag-tagged hnRNPH1 constructs and incubated for 48 h at 37 °C. The transfected cells were resuspended in IP buffer (50 mM Tris⋅HCl pH8.0, 150 mM NaCl, 1% Triton X-100, 1 mM ethylenediaminetetraacetic acid (EDTA), and PIC). The collected cells were lysed by sonication twice for 10 s (2 s on/10 s off ice) at 3% power. The cell lysates were subjected to centrifugation at 15,000 rpm for 15 min at 4 °C and 2 mg of the cleared cell lysates was incubated with 4 µg of anti-Flag antibodies at 4 °C with rotation. Upon overnight incubation, the immune complexes were supplemented with protein G magnetic beads (Bio-Rad) for 2 h at 4 °C with rotation and washed three times with ice-cold phosphate-buffered saline (PBS). The bound proteins were eluted using 2×SDS sample buffer by boiling at 95 °C for 10 min. For Western blotting, the eluted samples were separated using NuPAGE 4 to 12% Bis-Tris Gel (Thermo Fisher Scientific) and separated proteins were transferred onto the nitrocellulose membrane (Bio-Rad) and detected using indicated antibodies.
For assessment of the RNA coprecipitated with hnRNPH1 protein, HEK-293T cells were transfected with Flag-tagged WT, ΔLC1, Y2S, or Y3S hnRNPH1 plasmids. At 48 h after transfection, culture medium was replaced to ice-cold PBS. The cells were then exposed to ultraviolet (UV) irradiation at 400 mJ/cm2 power (XL-1500, Spectronics) to induce crosslinking between proteins and RNAs. The cells were then lysed in IP buffer containing RNase inhibitor. Upon immunoprecipitation, RNAs were prepared from the input and the IP samples. Immunoprecipitated proteins were analyzed by Western blotting using Flag antibody. The purified RNAs were subjected to RT-PCR (RNA Preparation and RT-PCR).
Antibodies.
The following primary antibodies were used for Western blotting and immunoprecipitation: anti-hnRNPH1 (A300-511A, Bethyl Laboratories), anti-Flag M2 (F3165, Sigma-Aldrich, Germany), anti-TAF15 (NB100-566, Novus Biologicals), anti-FUS (A300-294, Bethyl Laboratories), anti-hnRNPA2 (SC-53531, Santa Cruz), anti-hnRNPA1 (8443, Cell Signaling Technology), anti-hnRNPH2 (ab179439, Abcam), anti-hnRNPF (ab50982, Abcam), and anti-DHX9 (PA5-19542, Thermo Fisher Scientific).
RNA Preparation and RT-PCR.
Total RNA was extracted using TRIzol (Thermo Fisher Scientific) according to the manufacturer’s instruction. Upon incubation with TRIzol for 5 min at room temperature, the cells lysates were mixed with chloroform. The samples were then incubated for 3 min and centrifuged for 15 min at 12,000 × g at 4 °C. The upper aqueous phase containing RNA was transferred to a new tube and mixed with isopropanol by gentle inverting. After 10 min of incubation at room temperature, the sample was centrifuged for 10 min at 12,000 × g at 4 °C. The RNA pellet was washed using ice-cold 75% ethanol and then resuspended in RNase-free water. For reverse transcription, at first, 2 µg of total RNA was treated with Turbo DNase (AM2239, Ambion) for 30 min at 37 °C to remove DNA. Reverse transcription was performed using SuperScript IV Reverse Transcriptase (Invitrogen) according to the manufacturer’s protocol. For PCR, the RT samples were diluted to contain 20 ng/μL of RNA in RNase-free water, and 2.5 µl of RT sample was used as templated for PCR. Primers used for PCR were as follows:
C2orf18-F, 5′-CTGACCTCCTGAGCAAGCAC-3′
C2orf18-R, 5′-GTCCAATGCATCCTCCAGTG-3′
CDK2-F, 5′-GTCCCTGTTCGTACTTACAC-3′
CDK2-R, 5′-CAGAGTCCGAAAGATCCGGA-3′
Man2a2-F, 5′-CCTCTTGGAGATGACTTCCG-3′
Man2a2-R, 5′-CGTCAGGAGGGTGAAATCAG-3′
PEX26-F, 5′-GCAGAAACAGGAACACTCAG-3′
PEX26-R, 5′-AAATGCAGCCTTCCGGATCC-3′
hTERC-F, 5′-CCCTAACTGAGAAGGGCGTA-3′
hTERC-R, 5′-AGAATGAACGGTGGAAGGCG-3′.
Supplementary Material
Acknowledgments
We thank Hana Cho for stimulating discussion and helpful comments on the composition of our manuscript. This work was supported by the National Research Foundation of Korea Grant No. NRF-2019R1A2C2003767 awarded to I.K.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2109668118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Kato M., et al. , Cell-free formation of RNA granules: Low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kato M., McKnight S. L., A solid-state conceptualization of information transfer from gene to message to protein. Annu. Rev. Biochem. 87, 351–390 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Kwon I., et al. , Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell 155, 1049–1060 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin Y., Protter D. S., Rosen M. K., Parker R., Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell 60, 208–219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiang S., et al. , The LC domain of hnRNPA2 adopts similar conformations in hydrogel polymers, liquid-like droplets, and nuclei. Cell 163, 829–839 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor J. P., Brown R. H. Jr., Cleveland D. W., Decoding ALS: From genes to mechanism. Nature 539, 197–206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boeynaems S., et al. , Protein phase separation: A new phase in cell biology. Trends Cell Biol. 28, 420–435 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitrea D. M., Kriwacki R. W., Phase separation in biology; functional organization of a higher order. Cell Commun. Signal. 14, 1 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harrison A. F., Shorter J., RNA-binding proteins with prion-like domains in health and disease. Biochem. J. 474, 1417–1438 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geuens T., Bouhy D., Timmerman V., The hnRNP family: Insights into their role in health and disease. Hum. Genet. 135, 851–867 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nazim M., et al. , Competitive regulation of alternative splicing and alternative polyadenylation by hnRNP H and CstF64 determines acetylcholinesterase isoforms. Nucleic Acids Res. 45, 1455–1468 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dardenne E., et al. , RNA helicases DDX5 and DDX17 dynamically orchestrate transcription, miRNA, and splicing programs in cell differentiation. Cell Rep. 7, 1900–1913 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Cooper-Knock J., et al. , Sequestration of multiple RNA recognition motif-containing proteins by C9orf72 repeat expansions. Brain 137, 2040–2051 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prudencio M., et al. , Distinct brain transcriptome profiles in C9orf72-associated and sporadic ALS. Nat. Neurosci. 18, 1175–1182 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masuda A., et al. , hnRNP H enhances skipping of a nonfunctional exon P3A in CHRNA1 and a mutation disrupting its binding causes congenital myasthenic syndrome. Hum. Mol. Genet. 17, 4022–4035 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pagani F., Buratti E., Stuani C., Baralle F. E., Missense, nonsense, and neutral mutations define juxtaposed regulatory elements of splicing in cystic fibrosis transmembrane regulator exon 9. J. Biol. Chem. 278, 26580–26588 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Ohki K., et al. , Tokyo Children’s Cancer Study Group (TCCSG), Clinical and molecular characteristics of MEF2D fusion-positive B-cell precursor acute lymphoblastic leukemia in childhood, including a novel translocation resulting in MEF2D-HNRNPH1 gene fusion. Haematologica 104, 128–137 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neckles C., et al. , HNRNPH1-dependent splicing of a fusion oncogene reveals a targetable RNA G-quadruplex interaction. RNA 25, 1731–1750 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chaudhury A., Chander P., Howe P. H., Heterogeneous nuclear ribonucleoproteins (hnRNPs) in cellular processes: Focus on hnRNP E1's multifunctional regulatory roles. RNA 16, 1449–1462 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boija A., et al. , Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175, 1842–1855.e1816 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dobson C. M., Protein folding and misfolding. Nature 426, 884–890 (2003). [DOI] [PubMed] [Google Scholar]
- 22.McKinley M. P., Bolton D. C., Prusiner S. B., A protease-resistant protein is a structural component of the scrapie prion. Cell 35, 57–62 (1983). [DOI] [PubMed] [Google Scholar]
- 23.Wickner R. B., [URE3] as an altered URE2 protein: Evidence for a prion analog in Saccharomyces cerevisiae. Science 264, 566–569 (1994). [DOI] [PubMed] [Google Scholar]
- 24.Kato M., Lin Y., McKnight S. L., Cross-β polymerization and hydrogel formation by low-complexity sequence proteins. Methods 126, 3–11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin Y., et al. , Toxic PR Poly-dipeptides encoded by the C9orf72 repeat expansion target LC domain polymers. Cell 167, 789–802.e12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hondele M., et al. , DEAD-box ATPases are global regulators of phase-separated organelles. Nature 573, 144–148 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gautrey H., et al. , SRSF3 and hnRNP H1 regulate a splicing hotspot of HER2 in breast cancer cells. RNA Biol. 12, 1139–1151 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim E., Kwon I., Phase transition of fibrillarin LC domain regulates localization and protein interaction of fibrillarin. Biochem. J. 478, 799–810 (2021). [DOI] [PubMed] [Google Scholar]
- 29.Xu C., et al. , HnRNP F/H associate with hTERC and telomerase holoenzyme to modulate telomerase function and promote cell proliferation. Cell Death Differ. 27, 1998–2013 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moorman A. V., The clinical relevance of chromosomal and genomic abnormalities in B-cell precursor acute lymphoblastic leukaemia. Blood Rev. 26, 123–135 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Gu Z., et al. , Genomic analyses identify recurrent MEF2D fusions in acute lymphoblastic leukaemia. Nat. Commun. 7, 13331 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bertolotti A., Bell B., Tora L., The N-terminal domain of human TAFII68 displays transactivation and oncogenic properties. Oncogene 18, 8000–8010 (1999). [DOI] [PubMed] [Google Scholar]
- 33.Li W., et al. , Biophysical properties of AKAP95 protein condensates regulate splicing and tumorigenesis. Nat. Cell Biol. 22, 960–972 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun S., Zhang Z., Fregoso O., Krainer A. R., Mechanisms of activation and repression by the alternative splicing factors RBFOX1/2. RNA 18, 274–283 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gueroussov S., et al. , Regulatory expansion in mammals of multivalent hnRNP assemblies that globally control alternative splicing. Cell 170, 324–339.e23 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Martinez-Contreras R., et al. , hnRNP proteins and splicing control. Adv. Exp. Med. Biol. 623, 123–147 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Wu H., Fuxreiter M., The structure and dynamics of higher-order assemblies: Amyloids, signalosomes, and granules. Cell 165, 1055–1066 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weber S. C., Brangwynne C. P., Getting RNA and protein in phase. Cell 149, 1188–1191 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Arvand A., Denny C. T., Biology of EWS/ETS fusions in Ewing’s family tumors. Oncogene 20, 5747–5754 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Gangwal K., et al. , Microsatellites as EWS/FLI response elements in Ewing’s sarcoma. Proc. Natl. Acad. Sci. U.S.A. 105, 10149–10154 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.