Abstract
A fragment of the mixed-lineage leukemia (MLL) gene (Mll, HRX, ALL-1) was identified in a yeast genetic screen designed to isolate proteins that interact with the CREB–CREB-binding protein (CBP) complex. When tested for binding to CREB or CBP individually, this MLL fragment interacted directly with CBP, but not with CREB. In vitro binding experiments refined the minimal region of interaction to amino acids 2829 to 2883 of MLL, a potent transcriptional activation domain, and amino acids 581 to 687 of CBP (the CREB-binding or KIX domain). The transactivation activity of MLL was dependent on CBP, as either adenovirus E1A expression, which inhibits CBP activity, or alteration of MLL residues important for CBP interaction proved effective at inhibiting MLL-mediated transactivation. Single amino acid substitutions within the MLL activation domain revealed that five hydrophobic residues, potentially forming a hydrophobic face of an amphipathic helix, were critical for the interaction of MLL with CBP. Using purified components, we found that the MLL activation domain facilitated the binding of CBP to phosphorylated CREB. In contrast with paradigms in which factors compete for limiting quantities of CBP, these results reveal that two distinct transcription factor activation domains can cooperatively target the same motif on CBP.
The interaction of the cyclic AMP response element-binding protein (CREB) with its coactivator, CREB-binding protein (CBP), is one of the best characterized signal-regulated activation domain-coactivator interactions. CREB is a signal-dependent transactivator of the bZIP family which becomes phosphorylated on serine 133 in response to stimuli that result in an increase in intracellular cyclic AMP or Ca2+ (33). CBP, as well as its homologue p300, binds to CREB upon phosphorylation at serine 133 within the kinase-inducible activation domain of CREB (8, 32). CBP recruitment by phosphorylated CREB results in rapid induction of gene expression that is mediated both by direct recruitment of the basal transcription machinery and the intrinsic acetyltransferase activity of CBP (26, 33). A vast number of signal-responsive and developmentally regulated transcription factors interact with different regions of CBP and p300. The CREB-binding (or KIX) domain alone interacts with at least 10 distinct families of transcriptional activators (17). Despite the number of well-established protein interactions involving CBP and p300, the only complex for which structural information is available is the phosphorylated CREB-CBP KIX domain interaction (38, 39).
The solution structure of the phosphorylated CREB-CBP KIX complex revealed that the activation domain of CREB undergoes a dramatic random coil-to-helix transition upon binding to the KIX domain (38). Conversely, the α-3 helix of the KIX domain undergoes a more subtle change upon binding to phosphorylated CREB. Two lines of evidence suggest that this binding event may be regulated at a level beyond serine 133 phosphorylation. First, CREB phosphorylation has been noted in cellular contexts which do not result in the recruitment of CBP or the induction of CREB target genes (4), suggesting that an additional event is required for coactivator recruitment. Second, CREB or CBP KIX domain mutants have been described that exhibit enhanced complex formation compared to the wild type in vitro or in vivo (5, 15). These observations suggest the potential for a third component that regulates CREB-KIX domain interaction by binding to either or both members of the CREB-CBP KIX complex. To isolate proteins that interact with the unique interface formed by the CREB-CBP KIX complex, a genetic screen was devised in which phosphorylated CREB and the CBP KIX domain are coexpressed in yeast to serve as “bait” for such proteins. In contrast to two-component interaction screening, this three-component screen resulted in the isolation of fewer, more specific interacting proteins (63).
In this study, we analyzed a fragment of the mixed-lineage leukemia (MLL) gene (Mll) isolated in the CREB-CBP three-hybrid screen. The MLL gene was originally identified as a gene frequently disrupted by chromosomal translocations in childhood leukemias as well as in therapy-induced acute myeloid leukemias (14, 18, 57). Translocations into the MLL locus generally result in fusion of the N terminus of the MLL protein with one of many diverse fusion partners. Two MLL fusion proteins, MLL-AF9 and MLL-ENL, have been demonstrated to be leukemogenic in murine knock-in or retroviral transduction systems, respectively (10, 52). From these studies, regions within the N-terminal portion of MLL as well as a specific motif within the fusion partner were shown to be important for oncogenesis (52). However, haploinsufficiency at the MLL locus also appears to contribute to leukemia in the knock-in system (13).
The mechanism by which the MLL gene maintains target gene expression patterns is of great interest as a starting point for understanding the altered activity of the leukemogenic fusion proteins. Gene-disruption experiments in the mouse illustrated the functional similarity between MLL and its Drosophila homologue, trithorax (60), most dramatically demonstrated by a posterior shift in Hox gene expression and consequent homeotic transformations of the axial skeleton in Mll heterozygotes (19, 59, 60). Mll, like trithorax, positively regulates Hox genes and counters the repressive effect of chromatin-regulating Polycomb-group proteins such as Bmi-1 (19, 24, 36). Analysis of Mll-null embryos indicates that MLL is not involved in the initial specification of Hox gene expression patterns but is essential for maintaining expression patterns once initiated by other sequence-specific transcription factors (59, 60). The mechanism by which MLL acts on its target genes is incompletely understood, in part due to the limited information available regarding protein complexes in which MLL participates (1, 2, 11, 31, 46).
To gain insight into the mechanism by which MLL regulates its target genes, we evaluated the significance of the isolation of MLL in the CREB-CBP three-hybrid screen. We show that MLL interacts directly with CBP as well as the CREB-CBP complex. The MLL-CBP interaction was found to be essential for transcriptional activation by the MLL minimal activation domain. In addition, we demonstrated that interaction of MLL with the CBP KIX domain facilitates binding of phospho-CREB to CBP. This surprising finding leads us to propose that a unique class of target genes may respond synergistically to both CREB and MLL.
MATERIALS AND METHODS
Yeast three-hybrid screen.
The LexA-CREB-YeACBP bait plasmid was transformed into the L40 yeast strain using standard small-scale transformation protocols (21, 49). This bait strain was then transformed with approximately 50 μg of the library plasmid. The yeast were allowed to grow at 30°C on leu-trp-his-ura-lys deficient plates, and colonies were picked daily for β-galactosidase assays. Plasmids from yeast that were positive for both histidine and β-galactosidase production were isolated and sequenced. Secondary screens were performed using LexA-CREB, LexA-CBP, LexA-CREBM1, and LexA-CREBM1-YeACBP as bait.
Plasmids.
The construction of the yeast three-hybrid bait plasmid and the LexA fusion plasmids has been described previously (63). The parental plasmid pBTM116 and the E9.5 mouse embryo VP16 fusion cDNA library were gifts from Stan Hollenberg (Oregon Health Sciences University), and pAD4 was obtained from Michael Wigler (Cold Spring Harbor Laboratory).
Glutathione S-transferase (GST) fusion proteins encoding portions of MLL were constructed by PCR amplification of the appropriate fragment using a human MLL cDNA clone (graciously provided by Masao Seto, Aichi Cancer Center, Nagoya, Japan), followed by insertion at the BamHI site of pGEX4T-3 (Amersham Pharmacia Biotech). The GST-CBP(KIX) plasmid encoding amino acids 449 to 687 was constructed using pRC/RSV-CBP (29) as a PCR template; the resulting BamHI fragment was inserted into pGEX4T-2. All resulting plasmids were confirmed by sequencing using ABI automated sequencing. Most templates used for in vitro transcription and translation were produced by PCR amplification using specific 5′ primers bearing a T7 polymerase binding site and Kozak sequence and specific 3′ primers bearing a stop codon. All PCR amplification steps were performed using Pfu polymerase (Stratagene). For the production of 35S-labeled MLL C2613-3082 and C2613-2861 (see Fig. 2A), a plasmid template, pCDNA3-C-MLL, was used. For MLL C2613-2861 transcription and translation, the plasmid was first cleaved at a unique XcmI site.
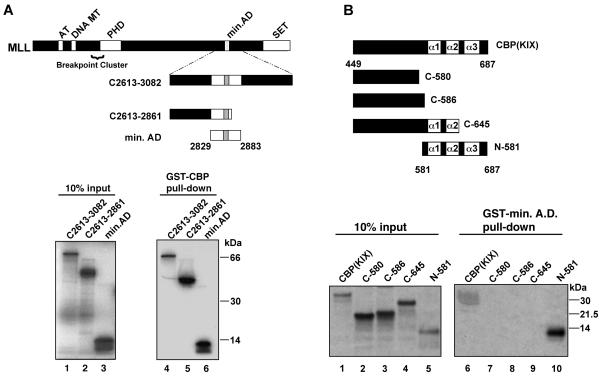
FIG. 2.
In vitro binding assays define minimal interaction domains of MLL and CBP. (A) GST-CBP (encompassing murine CBP amino acids 449 to 687) was tested for interaction with the indicated MLL polypeptides. The diagram shows full-length MLL and the internal polypeptides used in this experiment; numbering is based on GenBank accession number NM005933 (57). 35S-labeled MLL polypeptides were generated by in vitro transcription and translation as described in Materials and Methods. In lanes 1 to 3, 10% of the input 35S-labeled protein was analyzed by SDS-PAGE in parallel with proteins remaining bound to the GST-CBP beads (lanes 4 to 6). Binding to GST was undetectable under these conditions (see Materials and Methods). (B) GST-MLL A.D. (amino acids 2829 to 2883) was tested for binding to the indicated CBP polypeptides. A series of PCR templates encoding portions of the murine CBP KIX domain were transcribed and translated as described in Materials and Methods. Input protein (lanes 1 to 5) was analyzed by SDS-PAGE in parallel with proteins remaining bound to the GST-MLL A.D. beads (lanes 6 to 10). Murine CBP numbering follows as described (8). All binding reactions were repeated in at least three independent experiments.
E1A expression plasmids were kindly provided by Doug Dean (Washington University, St. Louis, Mo.). Site-directed mutagenesis was performed using the Quickchange kit (Stratagene). MLL wild-type and mutant activation domain-GAL4 fusions were produced by transferring a BamHI-to-XbaI fragment from the GST fusion plasmids to a GAL4 plasmid, BXG1 (6), at the BamHI and SpeI sites. Nuclear-localized activation domain polypeptides were produced by inserting the BamHI-to-XbaI fragment described above into a modified version of pCDNA3.1(−) (Invitrogen) containing a Kozak consensus sequence followed by sequences encoding the Flag M2 epitope and a consensus nuclear localization sequence (27). The luciferase-expressing reporter plasmid used was pGL2 basic (Promega), into which a consensus TATA box and initiator element were introduced, as described elsewhere (62). The bacterial expression plasmid encoding mouse CREB has been described previously (41). Histidine-tagged (His6) wild-type and MLL activation domain plasmids were constructed by transferring a BamHI-to-EcoRI fragment from the GAL4 fusion plasmids described above to pET30a+ (Novagen) at the BamHI and EcoRI sites.
Recombinant protein production and purification.
CREB (amino acids 3 to 341) was produced as described previously (41), with the exception that heparin-agarose chromatography was performed immediately following ammonium sulfate precipitation as a final purification step. CREB eluted at 600 mM KCl and was dialyzed before the phosphorylation reaction. Five micrograms of purified protein was phosphorylated using 2 U of recombinant protein kinase A (Calbiochem) following the manufacturer's suggestions. Mock-phosphorylated CREB was produced similarly, with the exception that ATP was omitted from the reaction mixture.
Fusion proteins were induced in DH5α or BL21/DE3 Escherichia coli with 0.1 or 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h. Cells were sonicated four times using 10-s pulses in phosphate buffered saline containing 0.1% Triton X-100, 2 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 2 μg of aprotinin/ml, 2 μg of leupeptin/ml, and 1 μg of pepstatin/ml, all purchased from Sigma (St. Louis, Mo.). Cleared lysates were stored at −80°C for subsequent binding to glutathione-agarose (Sigma). His6-tagged proteins were produced similarly and were purified using Ni-nitrilotriacetic acid resin following the manufacturer's protocol (Qiagen Inc.), with washes of 80 mM imidazole and elution with 250 mM imidazole. Recombinant proteins used in mobility shift assays were dialyzed against HGED.1 (20 mM HEPES [pH 7.9], 20% glycerol, 0.2 mM EDTA, 100 mM KCl) containing 1 mM phenylmethylsulfonyl fluoride and 1 mM dithiothreitol (DTT) before storage at −80°C. Silver staining was performed using a Bio-Rad Silverstain Plus kit.
In vitro binding assays.
35S-labeled MLL and CBP polypeptides were generated by in vitro transcription and translation using the TNT Coupled Wheat Germ Extract System (Promega). In vitro binding assays were performed in ABB buffer (20 mM Tris [pH 8.0], 350 mM NaCl, 0.2% NP-40, 1 mM DTT, and 2 mM EDTA, plus protease inhibitors as described above). Cleared bacterial lysates were bound to glutathione-agarose (Sigma) for 1.5 h at 4°C. The protein-saturated beads were washed three times in ABB buffer, and then 30 μl of beads (50% slurry) were incubated with 10 μl of 35S-labeled polypeptides in 500 μl of ABB buffer for 2 h at 4°C. The binding reaction mixture was washed three times in ABB buffer at ambient temperature. Bound proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), followed by fluorography of fixed and dried gels or staining with Gelcode (Pierce). For the experiment shown below in Fig. 4D, nuclear extracts were prepared as described previously (12), and binding reactions were performed with 30 μL of GST fusion-saturated beads and 800 μg of nuclear extract. Binding reactions were carried out at room temperature for 1 h, and mixtures were washed four times with HGED buffer containing 400 mM KCl. Bound proteins were analyzed on a 4% polyacrylamide SDS gel and transferred to nitrocellulose membranes, and CBP was detected by immunoblotting using the A-21 antibody from Santa Cruz Biotechnology. For the experiment shown below in Fig. 4E, 293 human embryonic kidney (HEK) cell lysates transfected with CBP (kind gift of Andrew Kung) and MLL activation domain plasmids were prepared using radioimmunoprecipitation (RIPA) buffer (50 mM TRIS [pH 8.0], 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% SDS, and 1 mM DTT). Binding reactions were carried out in RIPA buffer at 4°C for 12 to 16 h using 6 μg of anti-V5 antibody (Invitrogen). Bound proteins were precipitated with 30 μl of protein A agarose (Sigma). After three washes in RIPA buffer, bound proteins were eluted with SDS sample buffer and fractionated on 10% NuPage bis-Tris gels (Invitrogen).
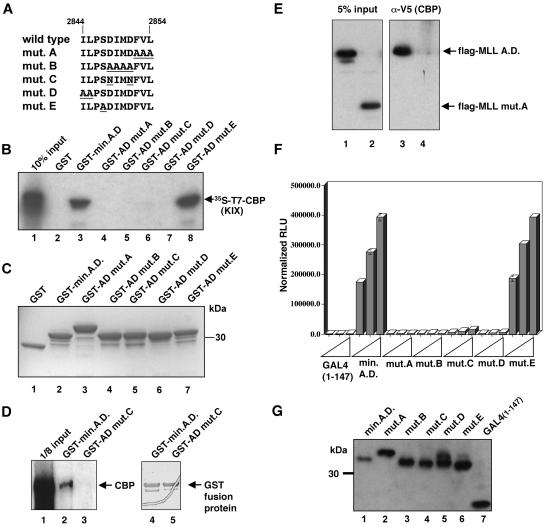
FIG. 4.
CBP binding and transactivation are closely linked in the MLL activation domain. (A) Site-directed mutations introduced into the GST-MLL A.D. (amino acids 2829 to 2883) fusion plasmids are diagrammed with only the mutated region shown (amino acids 2844 to 2854). Altered residues are underlined, and mutant nomenclature is indicated at the left. (B) A subset of site-directed MLL activation domain mutants fail to bind the CBP KIX domain. Individual GST fusion proteins were tested for their ability to bind 35S-labeled CBP KIX polypeptide (arrow) produced by in vitro transcription and translation. Lane 1, input CBP polypeptide; lanes 2 to 8, CBP remaining bound to the indicated GST fusion proteins. (C) GST fusion proteins used in the binding reaction shown in panel B were detected with Gelcode stain. (D) Full-length, native CBP binds the MLL activation domain. Cell extracts were incubated with the GST fusion protein beads as indicated above each lane. After washing, bound proteins were analyzed by immunoblotting with a polyclonal rabbit anti-CBP antibody. Lane 1 represents 100 μg or one-eighth of the total protein used in the binding reaction mixture. The right panel demonstrates similar quantities of GST fusion protein used in the binding reactions. (E) Transfected MLL activation domain peptides interact with full-length CBP. 293 HEK cells were transiently transfected with expression plasmids encoding Flag epitope-tagged, nuclear localization signal-fused MLL activation domain polypeptides and V5 epitope-tagged CBP. Anti-V5 immunoprecipitations were performed, and bound proteins were detected using anti-Flag M2 antibody. Lane 1, lysate from cells transfected with a wild-type MLL activation domain expression plasmid encoding MLL amino acids 2758 to 2864; lane 2, lysate from MLL mutant A expression plasmid encoding MLL amino acids 2829 to 2864, harboring the mutation described in panel A; lanes 3 and 4, proteins coimmunoprecipitated with anti-V5 epitope antibody using the lysates shown in lanes 1 and 2, respectively. (F) MLL activation domain mutants that fail to bind CBP fail to activate transcription. The MLL activation domain mutants shown in panel A were transferred to a GAL4 fusion plasmid for expression in mammalian cells. C33a cells were transiently transfected with 1, 5, or 25 ng of each GAL4 expression plasmid as indicated. The reporter plasmid and internal control were the same as in Fig. 3. Luciferase activity was quantitated 48 h after transfection and normalized to a cotransfected β-galactosidase plasmid. (G) 293 HEK cells were transiently transfected with the GAL4 fusion plasmids indicated above each lane. After 48 h, cell lysates were prepared and analyzed by anti-GAL4 immunoblotting using standard techniques (3).
Transient-transfection assays.
C33a human cervical carcinoma or 293 HEK cells were transiently transfected using Lipofectamine (Life Technologies, Inc.) according to the manufacturer's protocol. For transfection of one well of a six-well plate, 100 ng of reporter plasmid, 100 ng of internal control plasmid, and 2 μg of filler DNA were used. The internal control plasmid expresses β-galactosidase under the control of the murine hsp68 promoter. Cells were washed two times with phosphate-buffered saline and harvested by scraping with a rubber cell scraper in reporter lysis buffer (Promega). Five hundred microliters of lysis buffer was used per well of a six-well dish, of which 5 to 20 μl was used for the luciferase assay and 30 μl was used for the liquid β-galactosidase assay (3). Results are presented as relative luminometer units normalized to β-galactosidase activity. All transfections were performed at least three times, with multiple DNA preparations. GAL4 antibodies were purchased from Santa Cruz Biotechnology.
Electrophoretic mobility shift assays.
The double-stranded CREB oligomer was purchased from Promega and was end labeled with T4 polynucleotide kinase and [γ-32P]ATP (New England Nuclear). Binding reactions were performed as described elsewhere (16). The binding reaction mixtures were electrophoresed on low ionic strength glycerol gels as described previously (25). Dried gels were analyzed by autoradiography using Kodak Biomax MR film. Antibodies for supershift reactions were purchased from Santa Cruz Biotechnology. To transfer mobility shift gels to nitrocellulose, a Novex apparatus and transfer buffer were used according to the manufacturer's suggestions, with the exception that methanol was excluded from the buffer. For immunoblotting, anti-CREB phospho-serine 133 antibody was purchased from New England Biolabs, and anti-His6 antibodies were from Santa Cruz Biotechnology.
RESULTS
A fragment of MLL is isolated in a yeast interaction screen using CREB-CBP complex as bait.
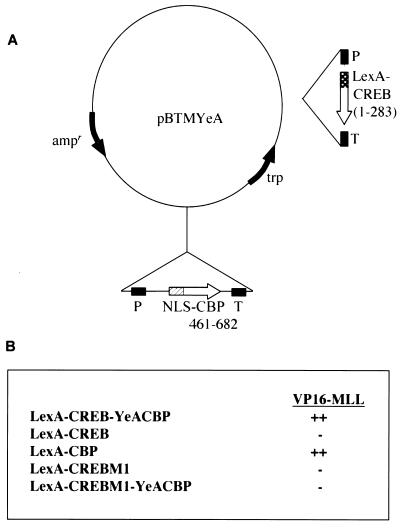
A screening strategy was developed in which CREB and the CREB-binding region of CBP were coexpressed in yeast to identify proteins that specifically recognize the CREB-CBP complex (Fig. 1A; reference 63). In yeast, CREB is constitutively phosphorylated, resulting in a stable nuclear CREB-CBP complex (50). An embryonic day 9.5 murine cDNA library was screened, resulting in the isolation of a clone encoding amino acids 2674 to 2887 of the Mll gene. This fragment was tested for independent interaction with either CREB or CBP by transferring the VP16-MLL fusion to yeast strains expressing LexA-CREB or LexA-CBP fusions. The results summarized in Fig. 1B demonstrate that this fragment of MLL also binds directly to CBP. No interaction was observed between MLL and CREB or a CREB mutant that cannot be phosphorylated (Fig. 1B). Therefore, we conclude that this portion of MLL can interact efficiently and directly with CBP as well as the CBP-CREB complex.
FIG. 1.
The CBP–phospho-CREB complex selects a fragment of MLL in a yeast interaction assay. (A) Bait plasmid, LexA-CREB-YeA CBP, encoding a nuclear-targeted form of the CREB-binding domain of CBP (NLS CBP 461–682) and LexA-fused CREB amino acids 1 to 283. P, promoter; T, terminator; NLS, nuclear localization signal. (B) VP16-MLL interacts with the CREB-CBP complex and CBP alone, but fails to interact with CREB. LexA-CBP, LexA-CREB, and LexA-CREBM1 are LexA fusion constructs encoding amino acids 461 to 682 of CBP, amino acids 1 to 283 of CREB, or amino acids 1 to 283 of CREB bearing a Ser133→Ala mutation, respectively. LexA-CREBM1-YeACBP is a two-component bait plasmid identical to LexA-CREB-YeACBP, with the exception of the described Ser133→Ala mutation. The levels of interaction with VP16-MLL, indicated by + and − signs, were determined by measuring growth of the transformants on a His-minus background. The VP16-MLL clone encodes amino acids 2674 to 2887, based on alignment with GenBank accession number NM005933.
The CBP KIX domain binds to the MLL activation domain.
To confirm whether a direct and specific interaction occurs between MLL and CBP, we employed in vitro binding assays. The CBP KIX domain was expressed as a GST fusion protein and tested for binding to in vitro-transcribed and -translated fragments of the human MLL gene. We first tested a human MLL fragment that is slightly larger than the murine protein isolated in the yeast assay (Fig. 2A, residues 2613 to 3082 of MLL; human and murine MLL are 97% identical in this region). This MLL fragment bound to GST-CBP efficiently, as did several smaller fragments (Fig. 2A, lanes 4 to 6). The MLL fragment did not bind GST, and other N-terminal fragments of MLL did not bind GST-CBP (data not shown). Interestingly, the smallest fragment that bound to GST-CBP coincides with the previously identified activation domain defined by GAL4 fusion experiments (37, 61).
Because the CREB-binding domain of CBP has been shown to interact with several other transcription factors, we sought to further define the minimal region required for binding MLL. The MLL minimal activation domain (Fig. 2A) was expressed as a GST fusion protein, and fragments of CBP (Fig. 2B) were produced by in vitro transcription and translation. In vitro binding experiments demonstrated that a region including all three α-helices of the KIX domain proved necessary and sufficient for interaction with the MLL activation domain (Fig. 2B, lane 10). The three α-helical segments shown are based on the CREB-CBP solution structure (38). In contrast, a region N terminal to the α-1 helix failed to interact with MLL (Fig. 2B, lanes 7 and 8). Deletion of the α-3 helix also abolished MLL interaction (Fig. 2B, lane 9). It has been reported that the structure of the KIX domain is disordered when the α-3 helix is deleted (39), so it is possible that the MLL activation domain recognizes an aspect of the structure formed by the three helices and not necessarily the third helix itself. Therefore, we conclude that the minimal regions mediating the MLL-CBP interaction correspond to CBP amino acids 581 to 687 and MLL amino acids 2829 to 2883.
Physical interaction with CBP correlates with MLL transactivation activity.
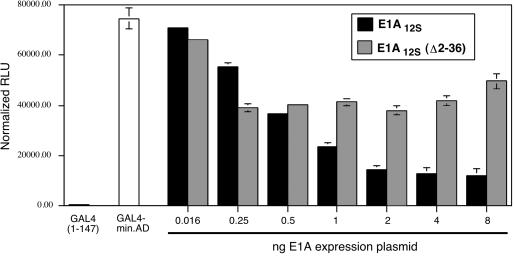
The region of MLL that interacts with CBP corresponds closely to the minimal activation domain (37, 61). Consequently, we used two approaches to test the hypothesis that interaction with CBP is important for transcriptional activation by MLL. As an initial test of CBP and p300 dependence, we determined whether the E1A 12S protein inhibited the transactivation function of the MLL minimal activation domain. This viral protein inhibits CBP- and p300-dependent transactivation by a number of transcription factors that physically interact with CBP or p300 through multiple mechanisms (7, 28, 34, 40, 58) As shown in Fig. 3, we introduced increasing quantities of E1A 12S expression plasmid together with a constant amount of GAL4-MLL minimal activation domain expression plasmid. In parallel, we also transfected a form of E1A 12S which is deficient in CBP binding (E1A Δ2-36) (54). The sensitivity of MLL-mediated transactivation to low levels of E1A, but not E1A Δ2-36 (Fig. 3), suggested that MLL-mediated activation requires the activity of CBP or p300.
FIG. 3.
Adenovirus E1A 12S expression inhibits GAL4-MLL-mediated transactivation. C33a cells were transfected with the GAL4-MLL expression plasmid and a luciferase reporter plasmid containing five GAL4 sites and a synthetic TATA box plus initiator core promoter (62). The first two open bars represent transactivation by GAL4 alone and transactivation by GAL4-MLL (GAL4-min.A.D., encoding MLL amino acids 2829 to 2883) in the absence of E1A expression plasmids. Either adenovirus E1A12S expression plasmid or E1A12SΔ2-36 mutant plasmid was coexpressed with a constant 50 ng of GAL4-MLL expression plasmid. The amount of E1A expression plasmid for each set of transfections is indicated on the x axis. Luciferase activity was measured 48 h after transfection, and all samples were normalized to a cotransfected β-galactosidase plasmid. No inhibition of the internal control was observed under the conditions shown.
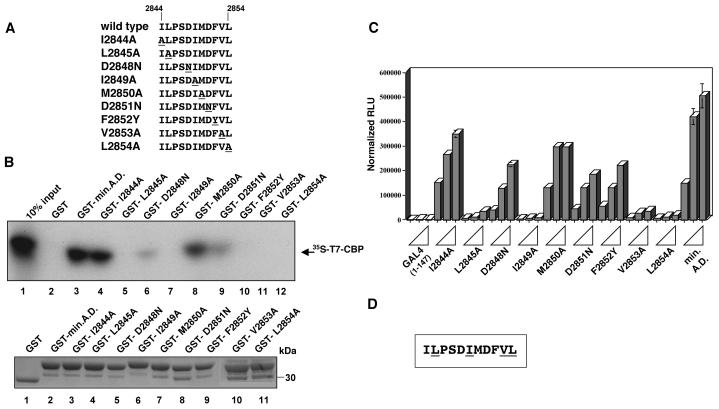
We next asked whether the ability of the MLL minimal activation domain to bind CBP could be separated from transactivation activity by substitution of specific residues. Previous studies demonstrated that block substitution of amino acids within the MLL minimal activation domain strongly reduced transactivation (37). Therefore, we introduced the substitutions shown in Fig. 4A into GST-MLL fusion plasmids and tested these mutants for their ability to bind in vitro translated CBP KIX domain. As shown in Fig. 4B, several of the mutants were severely impaired in their ability to bind the CBP KIX domain (lanes 4 through 7). However, the single amino acid substitution of mutant E had no effect on CBP binding (Fig. 4B, lane 8). Fig. 4C demonstrates that these results were not due to variation in the quantity of GST-fused mutant proteins bound to the affinity resin. We also determined whether the MLL activation domain interacts specifically with native, full-length CBP in cell extracts, by using two approaches. First, we performed in vitro binding experiments using wild-type or mutant GST fusion proteins on beads and C33a cervical carcinoma nuclear extracts as a source of CBP. As shown in Fig. 4D, the wild-type activation domain beads retained CBP (lane 2), whereas mutant C beads failed to interact strongly with CBP (lane 3). Second, we asked whether coexpressed, full-length, epitope-tagged CBP and MLL activation domain polypeptides would interact specifically in mammalian cells. Figure 4E demonstrates that the wild-type MLL activation domain polypeptide, but not a mutant polypeptide, coimmunoprecipitated with CBP. Therefore, this region of MLL is also capable of specifically interacting with native, full-length CBP in cell extracts.
The capacity of each mutant to activate transcription was compared using a transient-transfection assay. Transactivation experiments were performed using subsaturating amounts of GAL4 fusion plasmid, with each expression plasmid tested at three concentrations representing fivefold increments (Fig. 4F). Only mutant E activated transcription as efficiently as the wild-type activation domain, whereas mutants A, B, C, and D were completely inactive (Fig. 4F; reference 37). All GAL4 fusion proteins were stable in the transfected cells, as demonstrated by immunoblotting with anti-GAL4 (Fig. 4G); the aberrant mobility of mutant A was observed for both GAL4 and GST fusion proteins.
Overall, a strict correlation was observed between the ability of MLL to bind CBP and the ability of MLL to activate transcription. However, we could not exclude the possibility that these block substitutions grossly disrupt the structure of the MLL activation domain, resulting in an indirect loss of CBP binding. In fact, the aberrant mobilities of mutants A and C suggested that the structure of this region might be significantly altered by the block substitutions. Secondary structure predictions suggest that this region is likely to adopt an α-helical conformation (41, 44). To more rigorously analyze the connection between CBP binding and transactivation, we generated single amino acid substitutions within the MLL activation domain and subjected the resulting mutants to binding and transactivation assays as described above.
Single amino acid substitution mutants within core of MLL activation domain define residues crucial for both CBP binding and transactivation.
Amino acid substitutions were made within the central core (amino acids 2844 to 2854) of the minimal activation domain of MLL (Fig. 5A). Residues outside this sequence were not important for transactivation (37, 61). When evaluated for binding to the CBP KIX domain, this series of mutants segregated into two categories. The first group (I2844A, D2848N, M2850A, D2851N) demonstrated 20 to 95% binding activity relative to that of the wild type (Fig. 5B). Several of these substitutions had a less dramatic effect on CBP binding than the block substitution from which they were derived. For example, each of the D→N substitutions bound CBP weakly, yet the double mutant (mutant C) (Fig. 4A) demonstrated less than 4% of the wild-type binding activity. All of the remaining single substitutions failed to bind CBP above background levels (L2845A, I2849A, F2852Y, V2853A, L2854A) (lanes 5, 7, 10 to 12). We confirmed that the GST fusion proteins used in these assays were present at similar levels (Fig. 5B, lower panel). Thus, the most important residues for CBP interaction were five hydrophobic residues within a putative α-helical region.
FIG. 5.
Single amino acid substitutions define crucial residues for binding to CBP and for transactivation. (A) Site-directed mutants introduced into the GST-MLL A.D. fusion plasmids are diagrammed with only the mutated region shown (amino acids 2844 to 2854). Altered residues are underlined, and mutant nomenclature is indicated at the left. (B) The top panel shows the results of a representative binding experiment utilizing 35S-labeled CBP KIX polypeptide (arrow) as in Fig. 4B. Lane 1, 10% of 35S-labeled CBP KIX polypeptide used in a binding reaction; lanes 2 to 12, 35S-labeled CBP KIX polypeptide remaining bound to the GST fusion protein as indicated above each lane. The bottom panel shows a Gelcode-stained gel of the GST fusion proteins used in the binding reactions described above. (C) Results of a representative luciferase assay using GAL4-MLL activation domain plasmids as indicated. Each bar reflects the average of triplicate transfections, and each set of three bars reflects transfections using 1, 5, or 25 ng of the indicated GAL4 fusion plasmid. Results are shown in relative luminometer units (RLU) normalized to β-galactosidase activity from the internal control plasmid. All transfections were repeated in at least three independent experiments with multiple DNA preparations. (D) Diagram summarizing the most important residues of the MLL activation domain for binding to CBP and for transactivation. MLL amino acids 2844 to 2854 are shown, with the amino acids underlined that abolish both CBP binding and transactivation when mutated.
We next tested the ability of this new series of mutants to activate transcription. As shown in Fig. 5C, all substitution mutants that failed to bind to CBP were severely impaired in transcriptional activity (L2845A, I2849A, V2853A, and L2854A). Of the remaining mutants, most activated transcription in parallel to the CBP binding results; for example, I2844A, D2848N, M2850A, and D2851N exhibited partial reduction in CBP binding and reduced transactivation (20 to 60% of that in the wild type). One consistent exception to this correlation was mutant F2852Y, which retained 40% of the wild-type transactivation activity (Fig. 5C) but failed to bind CBP (Fig. 5B, lane 10). This one exception may reveal an alternate interaction with a coactivator not assessed here. Figure 5D summarizes the transfection and binding data, with residues important in both assays shown underlined.
Analysis of all substitution mutants revealed a very close correlation between CBP binding and transactivation activity, suggesting that the MLL activation domain must recruit CBP (or a close relative) to activate transcription. This conclusion is consistent with the sensitivity of the MLL activation domain to adenovirus E1A coexpression (Fig. 3). The region spanning L2845 to L2854 can be modeled as an α-helix in which most of the important hydrophobic residues distribute predominantly to one face of the helix, based on secondary structure prediction algorithms (44) and the preference for proline at the second position (42). This arrangement is reminiscent of the predicted structure of other KIX-interacting activation domains of c-Myb, human T-cell lymphotropic virus type 1 Tax, and the known structure of the CREB α-B helix (20, 35). However, the primary sequence of the MLL activation domain differs significantly from that of the putative Myb and Tax helices, leaving open the possibility that these proteins have distinct binding sites on the KIX domain structure.
MLL stabilizes CREB-CBP KIX domain complexes.
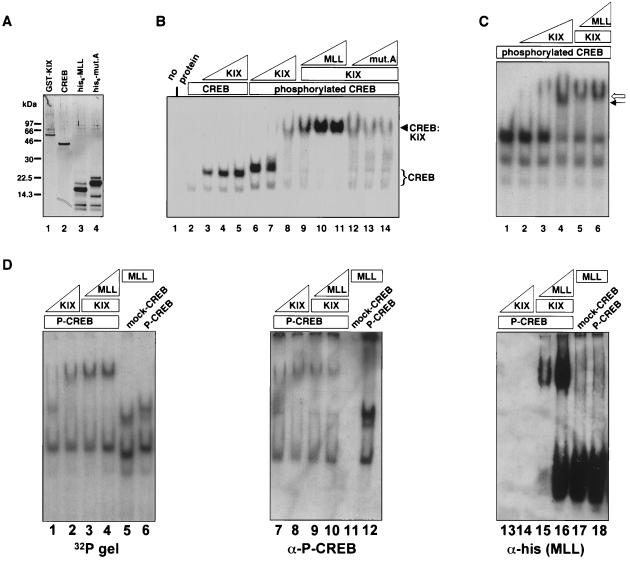
The in vitro binding assays and yeast two-hybrid results (Fig. 1 and 2) demonstrate that MLL interacts strongly and directly with CBP. Isolation of MLL in a three-hybrid screen suggests that CREB and MLL might bind to CBP simultaneously. Consequently, we prepared recombinant CREB, KIX domain, and MLL activation domain polypeptides to determine whether a ternary complex could be detected. Each recombinant protein was purified to near homogeneity (Fig. 6A), and CREB was phosphorylated with recombinant protein kinase A. Both phosphorylated CREB and His6-tagged MLL bound specifically to the GST-KIX polypeptide when tested in an in vitro binding assay (data not shown), demonstrating that the recombinant proteins functioned as expected based on previous interaction assays.
FIG. 6.
The MLL activation domain facilitates phospho-CREB–CBP interaction. (A) Recombinant proteins were produced in E. coli as described in Materials and Methods. One hundred nanograms of each purified protein was electrophoresed on a 10% bis-Tris gel and detected by silver staining. Lane 1, GST-KIX (CBP amino acids 449 to 687); lane 2, nonphosphorylated CREB; lane 3, His6-tagged wild-type MLL activation domain; lane 4, His6-tagged mutant A (see also Fig. 4A) MLL activation domain. The lower bands present in the His6-tagged protein preparations were not immunoreactive with the anti-His6 antibody (data not shown) and are therefore likely to be N-terminal degradation products or contaminating bacterial proteins. (B) Approximately 0.5 pmol of 32P-labeled CREB oligomer was incubated with the proteins indicated above the lanes, and complexes were separated by native gel electrophoresis (see Materials and Methods), such that the unbound probe has been run off the gel. The quantities of proteins used in the binding reaction mixtures were as follows: mock-phosphorylated CREB (mock-CREB) or phosphorylated CREB (phospho-CREB), 0.3 pmol; GST-KIX (KIX), 4, 13, or 40 pmol; His6-MLL or mutant A (mut. A), 1.3, 4, or 12 pmol. GST-KIX was used at 40 pmol in lanes 9 to 14. The CREB-DNA (CREB) and the CREB-KIX-DNA (CREB:KIX) complexes are marked by brackets or arrows, respectively. (C) Mobility shift reactions were performed as described for panel B, with the exception that the gel was run an additional 30 min. Phospho-CREB (0.3 pmol) was present in all binding reaction mixtures. GST-KIX was used at 4, 13, or 40 pmol in lanes 2 to 4 and at 13 pmol in lanes 5 and 6. His6-tagged MLL was present at 4 and 12 pmol in lanes 5 and 6, respectively. (D) Mobility shift reactions were performed in duplicate and electrophoresed on two identical gels as for panel C. One gel (labeled 32P, left panel) was dried and exposed to X-ray film, and the other was transferred to nitrocellulose. Blocked nitrocellulose membranes were probed with anti-CREB phospho-serine 133 antibody (middle panel), then stripped and reprobed with anti-histidine antibody (right panel). The quantities of proteins used in the binding reaction mixtures were as follows: 1.3 pmol of phosphorylated CREB, (lanes 1 to 4, 6, 7 to 10, 12, 13 to 16, and 18); 13 pmol of GST-KIX (lanes 1, 3, 4, 7, 9, 10, 13, 15, 16); 40 pmol of GST-KIX (lanes 2, 8, and 14); 12 pmol of His6-MLL (lanes 3, 9, and 15); 48 pmol of His6-MLL (lanes 4 to 6, 10 to 12, and 16 to 18). For mock-phosphorylated CREB 1.3 pmol was used in lanes 5, 11, and 17.
To detect the CREB-CBP complex, we used a CREB oligonucleotide in an electrophoretic mobility shift assay. The formation of a CREB-CBP complex on DNA was dependent on the phosphorylation of CREB (Fig. 6B, lanes 3 to 5). When an increasing amount of His6-tagged MLL activation domain was introduced into the binding reaction mixture, the complex was stabilized (Fig. 6B, lanes 9 to 11). MLL mutant A, which does not interact with the CBP KIX domain (Fig. 4), does not have this stabilizing effect (Fig. 6B, lanes 12 to 14). The His6-tagged MLL activation domain had no direct effect on CREB, as it did not change the mobility or intensity of the band representing the CREB-DNA complex (data not shown). Interestingly, experiments in which MLL-CBP complex formation was monitored by mobility shift reaction demonstrated that CREB does not facilitate MLL-CBP complex formation (data not shown). To illustrate the difference in mobility of the phospho-CREB–KIX complex and the ternary complex containing MLL, complexes were resolved further by extending the electrophoresis time. Figure 6C demonstrates that the complex formed with the MLL activation domain polypeptide migrates more slowly than the phospho-CREB–KIX complex (lanes 5 and 6 versus lane 4). To confirm that MLL is in the shifted complex, we transferred a duplicate mobility-shift gel to nitrocellulose and blotted with anti-His6 antibody to detect the position of the MLL polypeptide within this native gel. As shown in Fig. 6D, a fraction of MLL migrates with the phospho-CREB–KIX complex (lanes 15 and 16), and this slowly migrating fraction depends on the presence of the KIX polypeptide (lanes 17 and 18).
From these data, we conclude that the MLL minimal activation domain facilitates phospho-CREB–CBP KIX complex formation. Taken together, the mobility shift results and the yeast interaction data demonstrate that the CREB and the MLL activation domain bind to the CBP KIX domain concurrently and cooperatively. The work presented here suggests that MLL may interact with an interface of the KIX domain that is distinct from that shown for CREB (38). MLL interaction with the CBP KIX domain may enhance the percentage of CBP molecules that are appropriately folded for interaction with phosphorylated CREB, or it may alter the thermodynamic or kinetic properties of the KIX domain in a manner that enhances overall binding to CREB.
DISCUSSION
Several other activation domains that interact with the CBP or p300 KIX domain have been studied in detail, including c-Myb, human T-cell lymphotropic virus type 1 Tax, MyoD, SREBP1a, SREBP2, and Drosophila ci (5, 9, 35). The regions of c-Myb or Tax that interact with CBP have been modeled as amphipathic α-helices, after the known structure of the α-B helix of CREB (20, 35). In contrast to the results presented here, the c-Myb activation domain was found to compete with Tax for binding to CBP (9). The Tax protein was also shown to compete with MyoD for binding to the KIX domain of p300 (43). Based on sequence similarity and mutant analysis, this group of activation domains might be predicted to bind to the same structural determinants of the KIX domain (9, 35, 43). Although the MLL activation domain shares primary sequence features with several of the activation domains listed above, it is unlikely to bind to the KIX domain in the manner demonstrated for CREB, based on the data presented here. Determining whether MLL is unique in its ability to bind cooperatively with phospho-CREB or whether a subset of other KIX-interacting activation domains share this property will shed light on the specificity of this regulatory mechanism.
We have demonstrated that a region within MLL has the capacity to bind directly to CBP, and this capacity is essential for its ability to activate transcription. One implication of these results is that the activation domain, as defined in this study and others (37, 61), can recruit CBP in the context of the full-length protein. Due to the large size and low abundance of the MLL protein, immunoprecipitation experiments utilizing extracts containing the full-length MLL and CBP proteins have not been possible, although it is clear that the activation domain itself can interact with full-length CBP (Fig. 4). In addition, transfected full-length MLL and CBP colocalize in a subset of nuclear spots (unpublished data). Further experiments will reveal the importance of this domain within the context of the full-length protein.
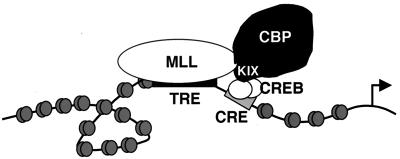
One prediction of these studies is that an overlap in MLL- and CREB-dependent target genes exists such that the cooperative interaction of MLL and CREB with CBP would play a role in regulating these genes (Fig. 7). Murine knockout models for both CREB and MLL may be instructive in identifying potential shared target genes for these two transcriptional activators. One possible function of the MLL protein at such predicted target genes might be to convert the acute, signal-induced activation of CREB into a sustained, developmentally maintained response analogous to the maintenance role of Trithorax-group proteins during Drosophila embryogenesis.
FIG. 7.
Hypothetical target gene upon which MLL (binding the trithorax response element, TRE) and CREB (a dimer binding the CRE) cooperate in recruiting CBP to the promoter region. This simplified diagram does not depict the fact that both MLL and CBP are likely to be incorporated into multiprotein complexes.
Fusion of the MLL gene to either CBP or p300 has been reported in several cases of myeloid leukemia or myeloid dysplasia (22, 45, 53, 55, 56). The data presented here suggest that MLL utilizes CBP to activate transcription. This finding raises intriguing issues regarding the mechanism of MLL-CBP or MLL-p300 fusions in leukemogenesis. One hypothesis for the oncogenic mechanism of the MLL-fusion gene products is that the acquisition of a new C terminus would impart an enhanced or new activity to the MLL molecule (gain-of-function or neomorphic activity) (52, 60). The MLL-CBP physical interaction presented in this study may be regulated during normal hematopoietic development to modulate the maintenance of endogenous MLL target genes. One testable prediction is that fusion of MLL to CBP would result in a form of MLL that could not be uncoupled from CBP, leading to the temporally inappropriate maintenance of MLL target gene expression. Constitutive expression of certain MLL target genes would consequently contribute to leukemogenesis. Attractive candidates for such target genes are HOX genes, which have been shown to play a role in normal or aberrant hematopoiesis through either loss- or gain-of-function experiments (23, 30, 47, 48, 51).
ACKNOWLEDGMENTS
We thank Doug Dean and Antonio Postigo for the E1A expression plasmids and valuable discussions, Adam Shaywitz for insightful suggestions, and Scott Armstrong, Andrew Kung, and Laura Michael for critical evaluation.
P.E. is supported by the Cancer Research Fund of the Damon Runyon-Walter Winchell Foundation (DRG 1467), and J.W. is supported by an H.H.M.I. predoctoral training grant. This work was supported by grants from the NIH.
REFERENCES
- 1.Adler H T, Chinery R, Wu D Y, Kussick S J, Payne J M, Fornace A J, Jr, Tkachuk D C. Leukemic HRX fusion proteins inhibit GADD34-induced apoptosis and associate with the GADD34 and hSNF5/INI1 proteins. Mol Cell Biol. 1999;19:7050–7060. doi: 10.1128/mcb.19.10.7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adler H T, Nallaseth F S, Walter G, Tkachuk D C. HRX leukemic fusion proteins form a heterocomplex with the leukemia-associated protein SET and protein phosphatase 2A. J Biol Chem. 1997;272:28407–28414. doi: 10.1074/jbc.272.45.28407. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1996. [Google Scholar]
- 4.Brindle P, Nakajima T, Montminy M. Multiple protein kinase A-regulated events are required for transcriptional induction by cAMP. Proc Natl Acad Sci USA. 1995;92:10521–10525. doi: 10.1073/pnas.92.23.10521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cardinaux J R, Notis J C, Zhang Q, Vo N, Craig J C, Fass D M, Brennan R G, Goodman R H. Recruitment of CREB binding protein is sufficient for CREB-mediated gene activation. Mol Cell Biol. 2000;20:1546–1552. doi: 10.1128/mcb.20.5.1546-1552.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carey M, Kakidani H, Leatherwood J, Mostashari F, Ptashne M. An amino-terminal fragment of GAL4 binds DNA as a dimer. J Mol Biol. 1989;209:423–432. doi: 10.1016/0022-2836(89)90007-7. [DOI] [PubMed] [Google Scholar]
- 7.Chakravarti D, Ogryzko V, Kao H Y, Nash A, Chen H, Nakatani Y, Evans R M. A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell. 1999;96:393–403. doi: 10.1016/s0092-8674(00)80552-8. [DOI] [PubMed] [Google Scholar]
- 8.Chrivia J C, Kwok R P, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 9.Colgin M A, Nyborg J K. The human T-cell leukemia virus type 1 oncoprotein Tax inhibits the transcriptional activity of c-Myb through competition for the CREB binding protein. J Virol. 1998;72:9396–9399. doi: 10.1128/jvi.72.11.9396-9399.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corral J, Lavenir I, Impey H, Warren A J, Forster A, Larson T A, Bell S, McKenzie A N, King G, Rabbitts T H. An Mll-AF9 fusion gene made by homologous recombination causes acute leukemia in chimeric mice: a method to create fusion oncogenes. Cell. 1996;85:853–861. doi: 10.1016/s0092-8674(00)81269-6. [DOI] [PubMed] [Google Scholar]
- 11.Cui X, De Vivo I, Slany R, Miyamoto A, Firestein R, Cleary M L. Association of SET domain and myotubularin-related proteins modulates growth control. Nat Genet. 1998;18:331–337. doi: 10.1038/ng0498-331. [DOI] [PubMed] [Google Scholar]
- 12.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dobson C L, Warren A J, Pannell R, Forster A, Rabbitts T H. Tumorigenesis in mice with a fusion of the leukaemia oncogene Mll and the bacterial lacZ gene. EMBO J. 2000;19:843–851. doi: 10.1093/emboj/19.5.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Domer P H, Fakharzadeh S S, Chen C S, Jockel J, Johansen L, Silverman G A, Kersey J H, Korsmeyer S J. Acute mixed-lineage leukemia t(4;11)(q21;q23) generates an MLL-AF4 fusion product. Proc Natl Acad Sci USA. 1993;90:7884–7888. doi: 10.1073/pnas.90.16.7884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du K, Asahara H, Jhala U S, Wagner B, Montminy M. Characterization of a CREB gain-of-function mutant with constitutive transcriptional activity in vivo. Mol Cell Biol. 2000;20:4320–4327. doi: 10.1128/mcb.20.12.4320-4327.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ernst P, Hahm K, Trinh L, Davis J N, Roussel M F, Turck C W, Smale S T. A potential role for Elf-1 in terminal transferase gene regulation. Mol Cell Biol. 1996;16:6121–6131. doi: 10.1128/mcb.16.11.6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodman R H, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- 18.Gu Y, Nakamura T, Alder H, Prasad R, Canaani O, Cimino G, Croce C M, Canaani E. The t(4;11) chromosome translocation of human acute leukemias fuses the ALL-1 gene, related to Drosophila trithorax, to the AF-4 gene. Cell. 1992;71:701–708. doi: 10.1016/0092-8674(92)90603-a. [DOI] [PubMed] [Google Scholar]
- 19.Hanson R D, Hess J L, Yu B D, Ernst P, van Lohuizen M, Berns A, van der Lugt N M, Shashikant C S, Ruddle F H, Seto M, Korsmeyer S J. Mammalian trithorax and polycomb-group homologues are antagonistic regulators of homeotic development. Proc Natl Acad Sci USA. 1999;96:14372–14377. doi: 10.1073/pnas.96.25.14372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harrod R, Tang Y, Nicot C, Lu H S, Vassilev A, Nakatani Y, Giam C Z. An exposed KID-like domain in human T-cell lymphotropic virus type 1 Tax is responsible for the recruitment of coactivators CBP/p300. Mol Cell Biol. 1998;18:5052–5061. doi: 10.1128/mcb.18.9.5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hill J, Donald K A, Griffiths D E, Donald G. DMSO-enhanced whole cell yeast transformation. Nucleic Acids Res. 1991;19:5791. doi: 10.1093/nar/19.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ida K, Kitabayashi I, Taki T, Taniwaki M, Noro K, Yamamoto M, Ohki M, Hayashi Y. Adenoviral E1A-associated protein p300 is involved in acute myeloid leukemia with t(11;22)(q23;q13) Blood. 1997;90:4699–4704. [PubMed] [Google Scholar]
- 23.Izon D J, Rozenfeld S, Fong S T, Komuves L, Largman C, Lawrence H J. Loss of function of the homeobox gene Hoxa-9 perturbs early T-cell development and induces apoptosis in primitive thymocytes. Blood. 1998;92:383–393. [PubMed] [Google Scholar]
- 24.Jacobs J J, van Lohuizen M. Cellular memory of transcriptional states by Polycomb-group proteins. Semin Cell Dev Biol. 1999;10:227–235. doi: 10.1006/scdb.1999.0304. [DOI] [PubMed] [Google Scholar]
- 25.Klemsz M J, McKercher S R, Celada A, Van Beveren C, Maki R A. The macrophage and B cell-specific transcription factor PU.1 is related to the ets oncogene. Cell. 1990;61:113–124. doi: 10.1016/0092-8674(90)90219-5. [DOI] [PubMed] [Google Scholar]
- 26.Korzus E, Torchia J, Rose D W, Xu L, Kurokawa R, McInerney E M, Mullen T M, Glass C K, Rosenfeld M G. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 27.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 28.Kurokawa R, Kalafus D, Ogliastro M H, Kioussi C, Xu L, Torchia J, Rosenfeld M G, Glass C K. Differential use of CREB binding protein-coactivator complexes. Science. 1998;279:700–703. doi: 10.1126/science.279.5351.700. [DOI] [PubMed] [Google Scholar]
- 29.Kwok R P, Lundblad J R, Chrivia J C, Richards J P, Bachinger H P, Brennan R G, Roberts S G, Green M R, Goodman R H. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 30.Lawrence H J, Helgason C D, Sauvageau G, Fong S, Izon D J, Humphries R K, Largman C. Mice bearing a targeted interruption of the homeobox gene HOXA9 have defects in myeloid, erythroid, and lymphoid hematopoiesis. Blood. 1997;89:1922–1930. [PubMed] [Google Scholar]
- 31.Leshkowitz D, Rozenblatt O, Nakamura T, Yano T, Dautry F, Croce C M, Canaani E. ALL-1 interacts with unr, a protein containing multiple cold shock domains. Oncogene. 1996;13:2027–2031. [PubMed] [Google Scholar]
- 32.Lundblad J R, Kwok R P, Laurance M E, Harter M L, Goodman R H. Adenoviral E1A-associated protein p300 as a functional homologue of the transcriptional co-activator CBP. Nature. 1995;374:85–88. doi: 10.1038/374085a0. [DOI] [PubMed] [Google Scholar]
- 33.Montminy M. Transcriptional regulation by cyclic AMP. Annu Rev Biochem. 1997;66:807–822. doi: 10.1146/annurev.biochem.66.1.807. [DOI] [PubMed] [Google Scholar]
- 34.Nakajima T, Uchida C, Anderson S F, Parvin J D, Montminy M. Analysis of a cAMP-responsive activator reveals a two-component mechanism for transcriptional induction via signal-dependent factors. Genes Dev. 1997;11:738–747. doi: 10.1101/gad.11.6.738. [DOI] [PubMed] [Google Scholar]
- 35.Parker D, Rivera M, Zor T, Henrion-Caude A, Radhakrishnan I, Kumar A, Shapiro L H, Wright P E, Montminy M, Brindle P K. Role of secondary structure in discrimination between constitutive and inducible activators. Mol Cell Biol. 1999;19:5601–5607. doi: 10.1128/mcb.19.8.5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pirrotta V. Polycombing the genome: PcG, trxG, and chromatin silencing. Cell. 1998;93:333–336. doi: 10.1016/s0092-8674(00)81162-9. [DOI] [PubMed] [Google Scholar]
- 37.Prasad R, Yano T, Sorio C, Nakamura T, Rallapalli R, Gu Y, Leshkowitz D, Croce C M, Canaani E. Domains with transcriptional regulatory activity within the ALL1 and AF4 proteins involved in acute leukemia. Proc Natl Acad Sci USA. 1995;92:12160–12164. doi: 10.1073/pnas.92.26.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Radhakrishnan I, Perez-Alvarado G C, Parker D, Dyson H J, Montminy M R, Wright P E. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interactions. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- 39.Radhakrishnan I, Perez-Alvarado G C, Parker D, Dyson H J, Montminy M R, Wright P E. Structural analyses of CREB-CBP transcriptional activator-coactivator complexes by NMR spectroscopy: implications for mapping the boundaries of structural domains. J Mol Biol. 1999;287:859–865. doi: 10.1006/jmbi.1999.2658. [DOI] [PubMed] [Google Scholar]
- 40.Reid J L, Bannister A J, Zegerman P, Martinez-Balbas M A, Kouzarides T. E1A directly binds and regulates the P/CAF acetyltransferase. EMBO J. 1998;17:4469–4477. doi: 10.1093/emboj/17.15.4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richards J P, Bachinger H P, Goodman R H, Brennan R G. Analysis of the structural properties of cAMP-responsive element-binding protein (CREB) and phosphorylated CREB. J Biol Chem. 1996;271:13716–13723. doi: 10.1074/jbc.271.23.13716. [DOI] [PubMed] [Google Scholar]
- 42.Richardson J S, Richardson D C. Amino acid preferences for specific locations at the ends of alpha helices. Science. 1988;240:1648–1652. doi: 10.1126/science.3381086. . (Erratum, 242:1624.) [DOI] [PubMed] [Google Scholar]
- 43.Riou P, Bex F, Gazzolo L. The human T cell leukemia/lymphotropic virus type 1 tax protein represses MyoD-dependent transcription by inhibiting MyoD-binding to the KIX domain of p300. A potential mechanism for tax-mediated repression of the transcriptional activity of basic helix-loop-helix factors. J Biol Chem. 2000;275:10551–10560. doi: 10.1074/jbc.275.14.10551. [DOI] [PubMed] [Google Scholar]
- 44.Rost B, Sander C. Prediction of protein secondary structure at better than 70% accuracy. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 45.Rowley J D, Reshmi S, Sobulo O, Musvee T, Anastasi J, Raimondi S, Schneider N R, Barredo J C, Cantu E S, Schlegelberger B, Behm F, Doggett N A, Borrow J, Zeleznik-Le N. All patients with the T(11;16)(q23;p13.3) that involves MLL and CBP have treatment-related hematologic disorders. Blood. 1997;90:535–541. [PubMed] [Google Scholar]
- 46.Rozenblatt-Rosen O, Rozovskaia T, Burakov D, Sedkov Y, Tillib S, Blechman J, Nakamura T, Croce C M, Mazo A, Canaani E. The C-terminal SET domains of ALL-1 and Trithorax interact with the INI1 and SNR1 proteins, components of the SWI/SNF complex. Proc Natl Acad Sci USA. 1998;95:4152–4157. doi: 10.1073/pnas.95.8.4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sauvageau G, Thorsteinsdottir U, Eaves C J, Lawrence H J, Largman C, Lansdorp P M, Humphries R K. Overexpression of HOXB4 in hematopoietic cells causes the selective expansion of more primitive populations in vitro and in vivo. Genes Dev. 1995;9:1753–1765. doi: 10.1101/gad.9.14.1753. [DOI] [PubMed] [Google Scholar]
- 48.Sauvageau G, Thorsteinsdottir U, Hough M R, Hugo P, Lawrence H J, Largman C, Humphries R K. Overexpression of HOXB3 in hematopoietic cells causes defective lymphoid development and progressive myeloproliferation. Immunity. 1997;6:13–22. doi: 10.1016/s1074-7613(00)80238-1. [DOI] [PubMed] [Google Scholar]
- 49.Schiestl R H, Gietz R D. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 50.Shih H M, Goldman P S, DeMaggio A J, Hollenberg S M, Goodman R H, Hoekstra M F. A positive genetic selection for disrupting protein-protein interactions: identification of CREB mutations that prevent association with the coactivator CBP. Proc Natl Acad Sci USA. 1996;93:13896–13901. doi: 10.1073/pnas.93.24.13896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shimamoto T, Tang Y, Naot Y, Nardi M, Brulet P, Bieberich C J, Takeshita K. Hematopoietic progenitor cell abnormalities in Hoxc-8 null mutant mice. J Exp Zool. 1999;283:186–193. [PubMed] [Google Scholar]
- 52.Slany R K, Lavau C, Cleary M L. The oncogenic capacity of HRX-ENL requires the transcriptional transactivation activity of ENL and the DNA binding motifs of HRX. Mol Cell Biol. 1998;18:122–129. doi: 10.1128/mcb.18.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sobulo O M, Borrow J, Tomek R, Reshmi S, Harden A, Schlegelberger B, Housman D, Doggett N A, Rowley J D, Zeleznik-Le N J. MLL is fused to CBP, a histone acetyltransferase, in therapy-related acute myeloid leukemia with a t(11;16)(q23;p13.3) Proc Natl Acad Sci USA. 1997;94:8732–8737. doi: 10.1073/pnas.94.16.8732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stein R W, Corrigan M, Yaciuk P, Whelan J, Moran E. Analysis of E1A-mediated growth regulation functions: binding of the 300-kilodalton cellular product correlates with E1A enhancer repression function and DNA synthesis-inducing activity. J Virol. 1990;64:4421–4427. doi: 10.1128/jvi.64.9.4421-4427.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sugita K, Taki T, Hayashi Y, Shimaoka H, Kumazaki H, Inoue H, Konno Y, Taniwaki M, Kurosawa H, Eguchi M. MLL-CBP fusion transcript in a therapy-related acute myeloid leukemia with the t(11;16)(q23;p13) which developed in an acute lymphoblastic leukemia patient with Fanconi anemia. Genes Chromosomes Cancer. 2000;27:264–269. [PubMed] [Google Scholar]
- 56.Taki T, Sako M, Tsuchida M, Hayashi Y. The t(11;16)(q23;p13) translocation in myelodysplastic syndrome fuses the MLL gene to the CBP gene. Blood. 1997;89:3945–3950. [PubMed] [Google Scholar]
- 57.Tkachuk D C, Kohler S, Cleary M L. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell. 1992;71:691–700. doi: 10.1016/0092-8674(92)90602-9. [DOI] [PubMed] [Google Scholar]
- 58.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 59.Yu B D, Hanson R D, Hess J L, Horning S E, Korsmeyer S J. MLL, a mammalian trithorax-group gene, functions as a transcriptional maintenance factor in morphogenesis. Proc Natl Acad Sci USA. 1998;95:10632–10636. doi: 10.1073/pnas.95.18.10632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu B D, Hess J L, Horning S E, Brown G A, Korsmeyer S J. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- 61.Zeleznik-Le N J, Harden A M, Rowley J D. 11q23 translocations split the “AT-hook”; cruciform DNA-binding region and the transcriptional repression domain from the activation domain of the mixed-lineage leukemia (MLL) gene. Proc Natl Acad Sci USA. 1994;91:10610–10614. doi: 10.1073/pnas.91.22.10610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zenzie-Gregory B, O'Shea-Greenfield A, Smale S T. Similar mechanisms for transcription initiation mediated through a TATA box or an initiator element. J Biol Chem. 1992;267:2823–2830. [PubMed] [Google Scholar]
- 63.Zhang Q, Vo N, Goodman R H. Histone binding protein RbAp48 interacts with a complex of CREB binding protein and phosphorylated CREB. Mol Cell Biol. 2000;20:4970–4978. doi: 10.1128/mcb.20.14.4970-4978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]