Abstract
Objective
To study the N6-methyladenosine (m6A) modification pattern of nucleus pulposus (NP) tissue during intervertebral disc degeneration (IDD).
Methods
A standing mouse model was generated, and staining and imaging methods were used to evaluate the IDD model. Methylated RNA immunoprecipitation with next-generation sequencing (MeRIP-seq) was used to analyze m6A methylation-associated transcripts in the NP, and real-time quantitative polymerase chain reaction (qRT-PCR) was used to detect the expression of methylation-related enzymes and conduct bio-informatics analysis.
Results
The standing mouse model caused IDD. Continuous axial pressure changed the expression of related methylases in degenerated NP tissue. Relative to the control group, the expression levels of KIAA1429, METTL14, METTL3, METTL4, WTAP, DGCR8, EIF3A and YTHDC1 in the experimental group were higher, while those of FTO, ELAVL1, HNRNPC1 and SRSF2 were lower. We identified 985 differentially expressed genes through MeRIP-Seq, among which 363 genes were significantly up-regulated, and 622 genes were significantly down-regulated. In addition, among the 9648 genes counted, 1319 m6A peaks with significant differences in methylation were identified, among which 933 were significantly up-regulated, and 386 were significantly down-regulated. Genes and pathways that were enriched in IDD have been identified.
Conclusion
The results of this study elucidated the m6A methylation pattern of NP tissue in degenerated lumbar intervertebral disc of mice and provided new perspectives and clues for research on and the treatment of lumbar disc degeneration.
The Translational potential of this article
As one of the important causes of low back and leg pain, intervertebral disc degeneration brings a huge economic burden to the society, family and medical system. Therefore, understanding the molecular and cellular mechanisms of intervertebral disc degeneration is of great significance for guiding clinical treatment. In this study, methylated RNA immunoprecipitation with next-generation sequencing on mice lumbar nucleus pulposus tissues found that differentially expressed genes and changes in the expression of related methylases, confirming that RNA methylation is involved in intervertebral disc degeneration. The process provides new vision and clues for future research on intervertebral disc degeneration.
Keywords: Intervertebral disc degeneration, nucleus pulposus, m6A methylation, Methylated RNA immunoprecipitation with next-generation sequencing
1. Introduction
With the rapid development of the social economy and changes in people's lifestyles, the incidence of low back pain(LBP) is increasing. Studies have shown that the prevalence of LBP in the United States is approximately 60%–90%, which was the secondly most common reasons for medical treatment [1]. It can occur at all ages and has become an important factor affecting quality of life and causing disability in modern people, and it imposes a huge economic burden on families, society and the medical system [2]. There are many causes of LBP, including lumbar disc degeneration [3], lumbar facet joint degeneration [4], lumbar laminectomy syndrome [5], tumors [6], etc. IDD is considered to be one of the main causes of LBP. IDD often leads to annulus fibrosus (AF) rupture, and NP herniation and stimulates or compresses nerve roots or the cauda equina, with corresponding clinical symptoms. There are various reasons for IDD, including smoking [7], high mechanical stress [8,9], sex [10], and age [11], but the exact cellular and molecular mechanisms are still unclear. The NP is one of the components of the intervertebral disc (IVD). The normal metabolism of NP cells and the extracellular matrix they secrete play an important role in the maintenance of IVD integrity. An increasing number of studies have shown that NP cells dysfunction plays a vital role in the pathogenesis of IDD.
In recent years, many studies have shown that epigenetic modifications such as DNA methylation and the regulation of gene expression by miRNA and lncRNA, plays a critical role in the pathophysiological response in many human diseases, such as IDD. DNA methylation, which is the most widely studied type of epigenetic mark, can regulate gene expression and the cell phenotype without changing the DNA sequence [12]. Tajerian and colleagues [13] found that DNA methylation could inhibit the expression of the SPARC gene in aged mice, resulting in the reduction of corresponding protein secretion, which in turn promoted the occurrence of IDD and chronic LBP. Ikuno et al. [14] carried out a genome-wide DNA methylation profile analysis of human NP for the first time and proved that the DNA methylation profile showed significant expression differences between the early and later periods of IDD.
N6-methyladenosine (m6A) is one of the most abundant internal RNA modifications in higher eukaryotic cells [15]. It was first discovered in the 1970s [16] and has been shown to regulate RNA translation [17], transportation, splicing [18], and stability [19] and to promote the maturation of miRNA by adding a methyl group to the N atom at the 6th position of the adenine base to produce m6A adenosine. It is mainly located in the coding sequence (CDS) and 3′ UTR of mRNA, and lncRNA and miRNA are also modified by m6A. In mammals, there are three basic mechanisms in m6A modification, mediated by “writers”, “erasers” and “readers”. “Writers” are methylases (mainly METL3, METL14 and WTAP), “erasers” are demethylases (including ALKBH5 and FTO), and “readers” are methylation recognition enzymes. Abnormalities in the above-mentioned enzymes cause a series of corresponding diseases, including sperm development disorders [20,21], tumors [22], circulatory disorders [23], etc. In recent years, an increasing number of studies have confirmed that RNA m6A modification plays a key biological role in the metabolism of mammalian cells and many human diseases. Weng et al. [24] found that m6A methylation plays a major role in the self-renewal of leukemia stem cells/initiating cells (LSCs/LICs) and the development and maintenance of acute myeloid leukemia (AML). Li and colleagues [25] found that the knockout of METTL3 significantly inhibited the migration, stem cell frequency and self-renewal of colorectal cancer cells, and successfully inhibited the occurrence and metastasis of colorectal cancer in vitro cell models and human tumor xenograft models. In addition, it was found that m6A methylation could inhibit virus replication and regulate the interaction with the host immune system during AIDS infection [26]. It could induce protein translation in adult mouse dorsal root ganglia and affect the regeneration of functional axons in the peripheral nervous system [27], which plays an important role in the maintenance of nerve function in rats with traumatic brain injury [28].
Many studies have found that mRNA, miRNA and lncRNA play critical role in IDD. Dong et al. [29] found that miR-640 could mediate IDD by enhancing NF-κB signal activity and inhibiting Wnt signaling. Zhao et al. [30] revealed that miRNA-143 could promote IDD by directly targeting Bcl-2 to mediate NP cell apoptosis. Another study found that the lncRNA HOTAIR could promote NP cell apoptosis, senescence, and extracellular matrix catabolism by stimulating autophagy and thereby mediate the IDD [31]. Recently, a study showed that m6A modification could mediate the progression of osteoarthritis by regulating chondrocyte NF-κB signal transduction and extracellular matrix synthesis [32]. Wang et al. [33] established a rat IDD model through transient static compression of the tail and confirmed that the m6A demethylation modification of mRNA and lncRNA may play an important role in the process of IDD; these authors emphasized that degeneration may be caused by the demethylation of lncRNALOC102555094 through the loc102555094/miR-431/GSK-3β/Wnt signaling pathway. However, the above-mentioned research mainly focused on the NP tissue of humans or rats and failed to fully elucidate the potential function and mechanism of RNA m6A modification in mouse degenerative NP.
Because of ethical issues, healthy IVD tissue is difficult to obtain in clinical practice. Therefore, it is necessary to establish a reliable animal model for simulating IDD. Previous animal models of IDD present some disadvantages, such as invasiveness [34,35], inconsistency with mechanical stress-induced IDD [36], and causing IDD via the compression of rat caudal vertebrae rather than lumbar vertebrae [37]. To explore the pattern of m6A modification and its roles in the NP after IDD in mice, we established a new bipedal mouse model to simulate the IDD based on the water-escape behavior [38]. Methylated RNA immunoprecipitation with next-generation sequencing (MeRIP-seq) was performed on NP tissue to analyze the m6A methylation profile, explore the changes and differential expression of m6A methylation levels in mRNA, and determine the potential role and related mechanism of m6A methylation modification in NP degeneration in mice, which aims to provide a new perspective and clues for the treatment of lumbar disc degeneration in the future.
2. Methods and materials
2.1. Animals and models
Thirty C57BL/6 male mice (each weighing approximately 15–20 g, 7 weeks old) were purchased from Hunan Slac Jingda Laboratory Animal Co., Ltd (Hunan, China) and randomly separated into control group and experimental group with 15 mice, respectively. All experimental mice were approved by the Medical Ethics Committee of the First Affiliated Hospital of the University of South China and were fed in an animal room with constant temperature and humidity in the Animal Experiment Department of University of South China, where they had sufficient food, water, and activity space and the ventilation was good. In the process of experimental modeling, we took advantage of the water-escape behavior of mice [39] and placed them in a 500 ml beaker containing water to induce bipedal standing, to increase vertical stress on the mouse spine. The level of the water added to the beaker was limited by the position of the ankle joint of the mice, which could reduce the influence of the physical environment, including factors such as hair condition and temperature on the experimental mice [38]. The mice stood for 8 h every day and rested for 1 h in the middle of this period, and their standing behavior was observed (Fig. 1). The rest of the time, the mice were placed in a cage and allowed to move freely and forage. The control group received the same treatment as the experimental group except that no water was added to the beaker. At 6 weeks and 10 weeks, 5 mice were randomly selected from each group and sacrificed, and the lumbar vertebrae were collected to observe the degree of lumbar disc degeneration. At 10 weeks, the remaining mice in each group were sacrificed, and the NP was removed from the lumbar intervertebral disc and stored in dry ice.
Figure 1.
Bipedal standing of mice. Experimental group (Left), Control group (Right).
2.2. Imaging assessment
Lumbar specimens of mice were fixed in 4% paraformaldehyde at 4 °C for 48 h, and micro-CT scan was then performed. The intervertebral height index (DHI%) was obtained by averaging the intervertebral disc height (the average of the measured values for the anterior, middle, and posterior IVD (25%, 50%, 75%)) and dividing the obtained value by the mean adjacent vertebral body height to evaluate the degree of IDD (Fig. 2).
Figure 2.
Micro-CT scan and DHI% measurement simulation image of lumbar vertebrae in mice. A-C. The line represents the height of the anterior 25%, 50% and 75% of the IVD; D-H. The line represents the height of the anterior and posterior 25%, 50% and 75% of the adjacent vertebral body. DHI%:Disc Height Index.
2.3. Histology
Lumbar spine specimens were collected for histological analysis after scanning. The specimens were placed in EDTA decalcification solution and decalcified at room temperature for two weeks. After decalcification, the specimens were dehydrated in an alcohol gradient and then embedded in paraffin. Serial sectioning was performed in the coronal plane to obtain sections with a thickness of approximately 4 μm. After the sections were deparaffinized and hydrated, hematoxylin-eosin staining (H&E) and safranin green staining were performed according to the standard process to observe the degree of IDD.
2.4. RNA extraction & quality control
At 10 weeks, 5 mice from each of the experimental and control group were sacrificed. Lumbar NP tissue were collected under a microscope, mixed, and then ground into powder for RNA extraction. First, TRIzol reagent was used to extract total RNA from the NP, and DNA digestion of the extracted RNA was then performed to remove DNA interference. Second, the NanodropTM OneC spectrophotometer was used to detect the A260/A280 ratio to determine RNA quality, the integrity of the RNA was confirmed by 1.5% agarose gel electrophoresis, and the qualified RNA was retained. Finally, quantitative analysis was performed on the qualified RNA using a Qubit3.0 fluorometer and a QubitTM RNA broad-spectrum Analysis Kit to complete the RNA quality control procedure.
2.5. m6A immunoprecipitation
Next, the MeRIP experiment was performed. ZnCl2 (20 mM) was added to 5 μg of RNA, followed by incubation at 94 °C for 5 min until the RNA fragments were mainly distributed in the 100–200 nt range. Then, 10% of the RNA fragments were saved as the “input”, and the rest of the RNA fragments were immunoprecipitated (IP) with specific anti-m6A antibodies. A stranded RNA sequencing library was constructed by using the KC-Digital™ Stranded mRNA Library Prep Kit for Illumina® through five PCR cycles. Then, the SMARTer Stranded Total RNA-Seq Kit version 2 was used to remove ribosomal cDNA, and ten PCR cycles were performed. Next, we used the PE150 model on the NovaSeq 6000 sequencer to enrich, quantify and finally sequence the library products corresponding to the size range of 200–500 bp.
2.6. Data processing and bio-informatics analyses
Trimmomatic was used to filter the raw sequencing data to discard reads with low quality and those contaminated by adaptor sequences. To eliminate the repetitive bias introduced during library preparation and sequencing, we used internal scripts to further process the clean reads. First, the clean reads were clustered according to UMI sequences, and the reads with the same UMI sequence are grouped into the same cluster. Then, the reads in the same cluster were compared in pairs, and the reads with more than 95% sequence identity were extracted into a new sub-cluster. After all the sub-cluster were generated, multiple sequence alignment was performed on each sub-cluster to obtain a consistent sequence. This eliminated any errors and biases introduced by PCR amplification or sequencing.
The obtained deduplicated consensus sequence was used for m6A site analysis, and STAR software with the default parameters was used to map the sequence to the mouse reference genome (https://ftp.ncbi.nih. gov/blast/db) to obtain qualified data. For bio-informatics analysis, we used RSeQC for read distribution analysis, exomePeak for peak calling analysis, and deep Tools for peak distribution analysis. The Fisher test was used to identify the differential m6A peaks with a Python script, and Homer was used to identify sequence motifs that were rich in m6A peak regions. In addition, KOBAS software was used to perform an enrichment analysis of genes annotated by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses, and a corrected p value of 0.05 was used as the basis for judging statistically significant enrichment.
3. Results
3.1. Standing model of IDD in mice
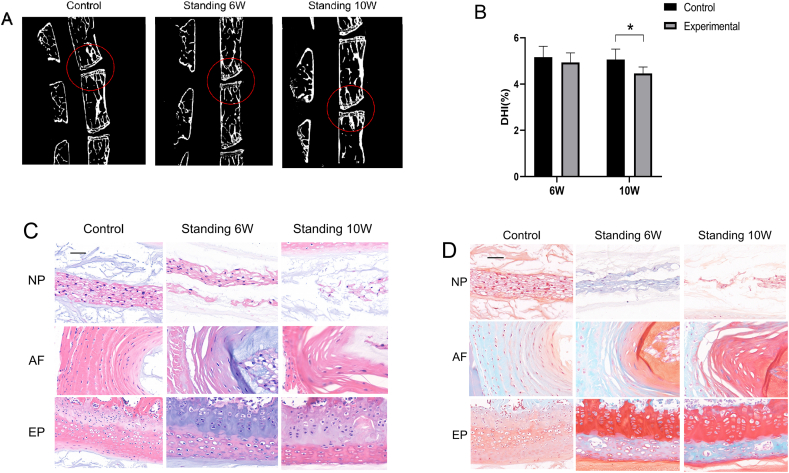
Compared with the control group, the average height of the IVD of the experimental group mice was significantly reduced, as observed by computed tomography (Fig. 3A). At 6 weeks, the DHI% of the control group was (5.2 ± 0.5)%, while that of the experimental group was (4.9% ± 0.4)%, the difference between the two groups was not statistically significant (P > 0.05). At 10 weeks, the DHI% of the control group was (5.1 ± 0.5)%, that of the experimental group was (4.5 ± 0.3)%, and the difference between the two groups was statistically significant (P < 0.05) (Fig. 3B). The results of HE staining and safranin green staining showed that relative to the control group, with prolonged standing time, the number of NP cells and the amount of extracellular matrix in the experimental group gradually decreased, the fibrous ring arrangement appeared disordered, lacerations occurred, and the height of the end-plate gradually decreased (Fig. 3C and D).
Figure 3.
A. Micro-CT results show that the intervertebral height of the experimental group was decreased compared with that of the control group. B. The DHI% of the mice in the experimental and control groups is shown. C-D. HE staining and safranin green staining results show that the number of NP cells and the amount of extracellular matrix decreased, the arrangement of the annulus fibrosus was disordered, and the height of the end-plate was decreased. ∗p < 0.05 compared with the control group; NP = NP; AF = annulus fibrosus; EP = end-plate (scale bars = 20 mm); magnification: 40x.
3.2. Review of methylated RNA immunoprecipitation sequencing results
To determine the m6A methylation levels of the samples from the experimental group and the control group, we performed an MeRIP-seq analysis of the mRNA of lumbar NP tissue to obtain raw sequencing data. We found that the IP samples produced an average of 26.45 Gb of sequencing data, and the input samples generated an average of 24.99 Gb of sequencing data (Table 1). After removing adapters, low-quality reads and contaminating bases from the raw sequencing data, an average of 23.77 Gb and 18.37 Gb of clean sequencing data were generated from the IP samples and input samples, respectively (Table 1). By comparing the obtained clean data with mouse genes, it was found that an average of 93.8% of the genes were mapped to mouse genes, 4.72% of the genes in repeated alignments were removed, and only the clean data (95.28%) for the remaining alignments were retained for further analysis (Table 2).
Table 1.
Raw and clean data obtained by sequencing in the experimental and control groups.
| Sample | Raw_bases(G) | Raw_reads(rRNA) | Raw_reads | Clean_reads | Clean2raw_read_ratio(%) |
|---|---|---|---|---|---|
| DZZ_IP | 31.54 | 210255524 | 79740816 | 71449622 | 89.6 |
| DZZ_input | 20.79 | 138604264 | 31878474 | 24949854 | 78.27 |
| SYZ_IP | 21.36 | 142403552 | 55404536 | 50018662 | 90.28 |
| SYZ_input | 28.31 | 188734778 | 39246710 | 28379178 | 72.31 |
Table 2.
Comparison of the rate of sequencing reads among mouse genes.
| Sample | Total reads | Total mapped(%) | Non-unique(%) | Unique(%) |
|---|---|---|---|---|
| DZZ_IP | 62382600 | 59065845(94.68) | 2587210(4.38) | 56478635(95.62) |
| DZZ_input | 21817616 | 20385748(93.44) | 911145(4.47) | 19474603(95.53) |
| SYZ_IP | 44196052 | 41706066(94.37) | 1989312(4.77) | 39716754(95.23) |
| SYZ_input | 24801870 | 22995563(92.72) | 1211116(5.27) | 21784447(94.73) |
4. Review of the methylation map of m6A in experimental and control mice
4.1. Changes in m6A methylation after IDD
To obtain comprehensive transcript information, we compared the generated sequences to understand their distribution in different regions of the reference genome. We found that the sequencing reads were mainly distributed in the 5′ UTR, CDS region, 3′ UTR, intergenic region and intron region, among which the CDS and intron region were the most frequent sites of the reads (Fig. 4).
Figure 4.
Distribution of reads in different regions of the reference genome.
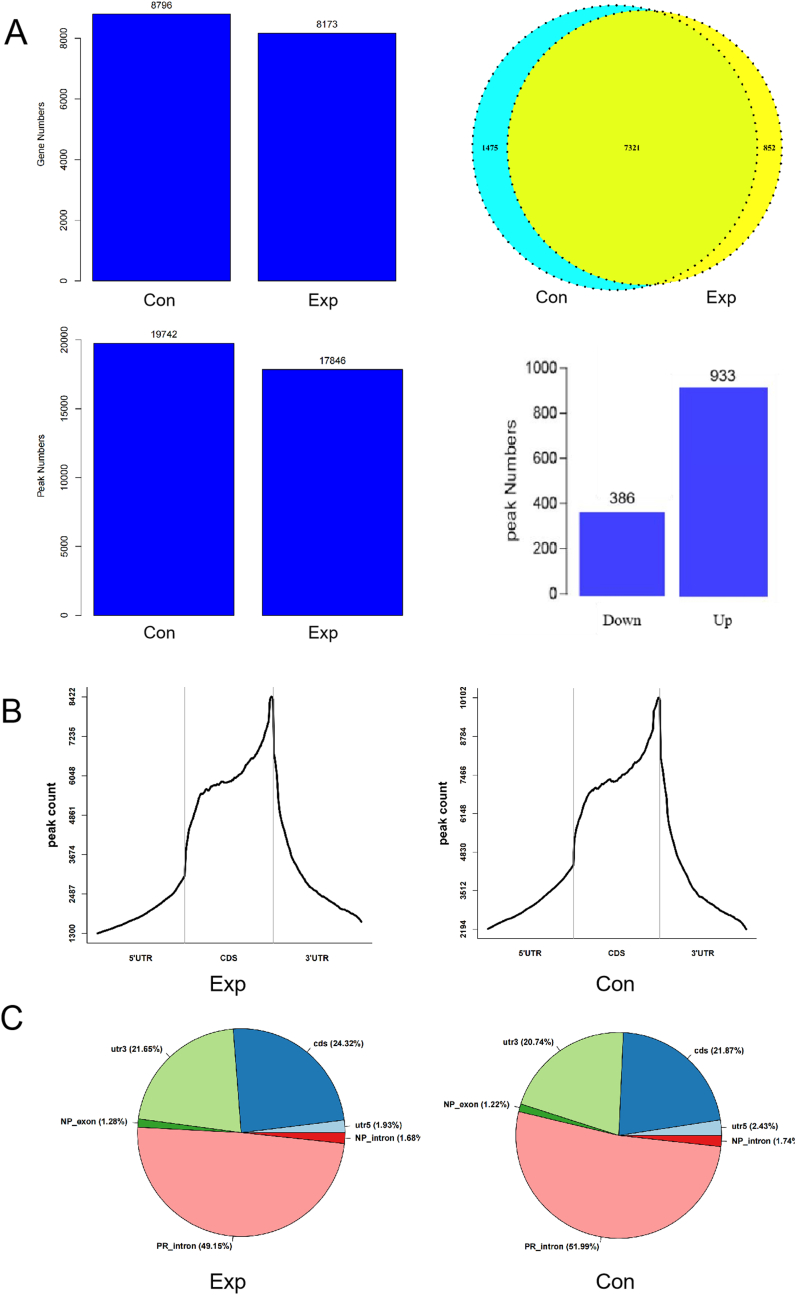
To understand the m6A peaks between the two groups, we annotated the obtained sequencing data and found 8173 and 8796 genes in the experimental and control group exhibited, respectively. There were 7321 identical genes in the two groups, 852 unique genes in the experimental group, and 1475 unique genes in the control group. In addition, 17846 different peaks were detected in 8173 genes in the experimental group, while in the control group, 19742 different peaks were detected in 8796 genes. We also found that 933 m6A peaks were significantly up-regulated and that 386 m6A peaks were significantly down-regulated (Fig. 5A). In addition, we found that the number of m6A peaks in the 5′ UTR, CDS region and 3′ UTR of the two groups were highly similar: they were mainly distributed in CDS region and were significantly enriched at the end of the CDS region and the beginning of the 3′ UTR (Fig. 5B). The distribution statistics of the m6A peaks in each functional region of the genes showed that the peaks with significant changes in the experimental group were mainly distributed in CDS region (24.32%), intron (50.83%), and 3′ UTR (21.65%), while only 1.93% and 1.28% occurred in 5′ UTR and exon region, respectively. The peaks in the control group were similar to those in the experimental group (CDS (21.87%), intron (53. 73%) and 3′ UTR (20. 74%), with only 2.43% and 1.22% occurring in 5′ UTR and exons, respectively) (Fig. 5C).
Figure 5.
A. The numbers of methylated genes and m6A peaks in the experimental and control groups. B. Distribution of m6A peaks in the 5′ UTR, CDS regions and 3′ UTR of mRNA. C. The distribution of m6A peaks in different gene is contexts shown in pie charts.
4.2. Biological information on m6A methylation revealed by GO and KEGG analyses
To elucidate the function of m6A methylation in lumbar disc degeneration, we conducted GO and KEGG analyses to identify gene functions and analyze the significant enrichment of pathways related to genes with significant changes in m6A levels. The GO data-based is a comprehensive database established by the Gene Ontology Federation that classifies and aggregates all gene-related research results available from around the world, with the aim of defining and describing the main biological functions of differentialyl expressed genes. KEGG is a large-scale knowledge base in which gene functions are systematically analyzed and genomics information is connected with functional information. According to the significant enrichment of related gene pathways, the most important biochemical metabolic pathways and signal transduction pathways involving differentially expressed genes can be determined.
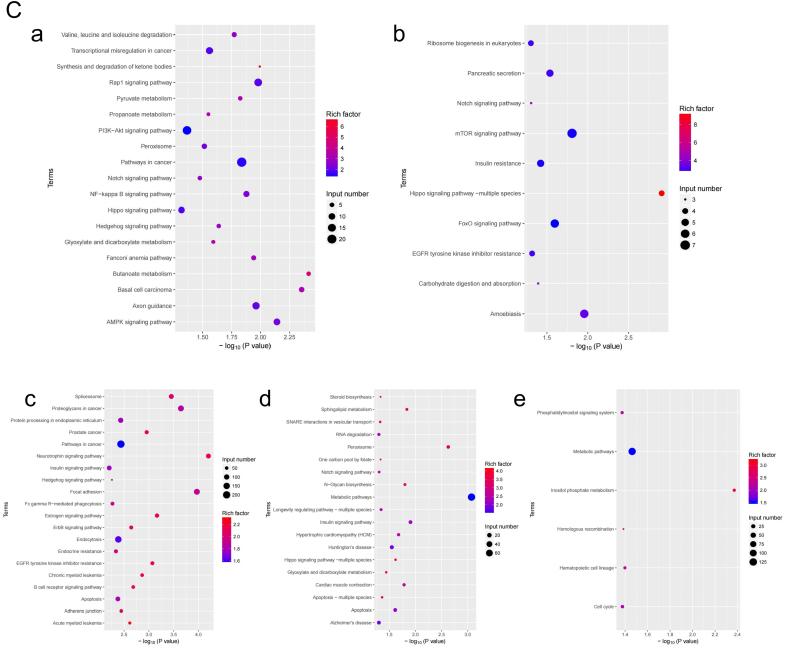
Through GO analysis, it was found that m6A modifications were involved in a variety of biological processes (BP), cellular components (CC) and molecular functions (MF). BP categories such as cellular macro-molecule metabolic process, macro-molecule bio-synthetic process, and nucleic acid metabolic process, CC categories such as nucleoplasm part and postsynaptic specialization, and MF categories such as metal ion binding, transferase activity, and DNA binding, were significantly enriched in genes associated with the hypermethylated m6A peak (Fig. 6A). BP categories such as intracellular signal transduction, regulation of intracellular signal transduction, and DNA metabolic process, CC categories such as nucleoplasm, plasma membrane region, and apical part of cell, and MF categories such as ion binding, cation binding, and metal ion binding were associated with genes related to the hypomethylated m6A peak (Fig. 6B).
Figure 6.
GO and KEGG analyses provided biological information associated with m6A methylation. A. Major GO terms significantly enriched in genes associated with the hypermethylated m6A peak. B. Major GO terms significantly enriched in genes associated with the hypomethylated m6A peak. C. a. Enriched pathways related to hypermethylated m6A-modified genes; b. Enriched pathways related to hypomethylated m6A-modified genes; c-e: Common and unique pathways of the experimental and control groups.
Through KEGG analysis, we annotated the most important biochemical metabolic pathways and signal transduction pathways in which the differentially expressed genes were involved and provided biological information associated with the high- and low-m6A mRNA gene-related pathways. In addition, we found common and unique gene pathways in the experimental and control group (Fig. 6C).
4.3. Changes in gene transcription and correlation analysis of MeRIP-seq and mRNA-seq results after lumbar disc degeneration
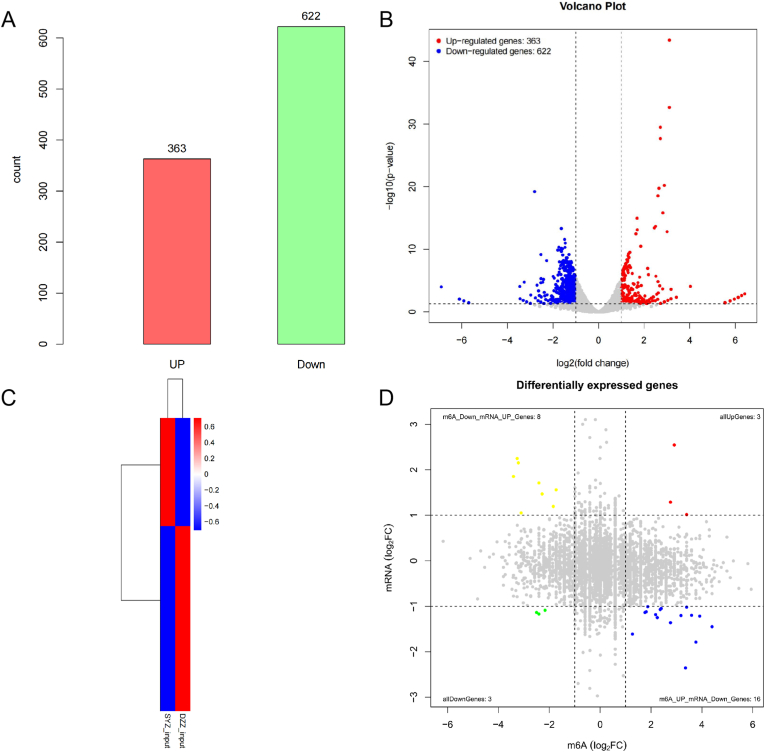
To explore the changes in genomics transcripts after disc degeneration, we sequenced the NP RNA of the experimental and control group to reveal the extent of mRNA changes during lumbar disc degeneration. Compared with the control group, we found that there were 985 genes with significant differentially expression in the experimental group (Log2 fold change≥2.0, p < 0.05), 363 of which were significantly up-regulated, while 622 were significantly down-regulated. The figure (Fig. 7A and B) shows the significantly up-regulated or down-regulated genes, and the table shows the fold changes of the top 10 differentially expressed up-regulated and down-regulated genes (Table 3, Table 4). The heat map shows the relative gene expression levels of the experimental and control group (Fig. 7C).
Figure 7.
Changes in gene transcription and correlation analysis of meRIP-seq and mRNA-seq results associated with lumbar disc degeneration. A-B. The expression levels of genes after IDD. A total of 363 genes showed significantly up-regulated, and 622 genes showed significantly down-regulated in mouse NP after IDD. C. The heat map shows the relative expression levels of the input between the experimental group and control group. D. Four-quadrant diagram showing the relationship between m6A methylation and mRNA expression.
Table 3.
Top 10 significantly upregulated genes among the differentially expressed genes.
| Geneid | SYZ_input | DZZ_input | logFC | logCPM | PValue |
|---|---|---|---|---|---|
| Gm10800 | 4094.582058 | 475.9551513 | 3.104726528 | 10.55917023 | 3.89E-44 |
| Gm21738 | 224.7223516 | 26.11008228 | 3.10383807 | 6.424746802 | 2.11E-33 |
| Gm26870 | 96.61018033 | 14.77272737 | 2.70854167 | 7.23280584 | 3.04E-30 |
| Gm10801 | 359.9109532 | 55.0059753 | 2.708979474 | 6.714950885 | 1.98E-28 |
| Gm10717 | 13.18440142 | 1.784998598 | 2.880731094 | 4.875087764 | 6.17E-21 |
| Gm10718 | 93.18994692 | 14.87993936 | 2.644093401 | 5.220330892 | 1.78E-20 |
| Gm11168 | 29.52102974 | 4.850567014 | 2.602539868 | 5.04417977 | 2.89E-19 |
| Gm10721 | 48.59390984 | 6.858948273 | 2.818603868 | 4.249801919 | 1.52E-16 |
| Myh4 | 652.1364845 | 202.2528952 | 1.688995013 | 11.32781121 | 1.07E-15 |
| Gm10722 | 49.93752025 | 8.810745766 | 2.498096843 | 4.292121084 | 2.07E-14 |
Table 4.
Top 10 significantly downregulated genes among the differentially expressed genes.
| Geneid | SYZ_input | DZZ_input | logFC | logCPM | PValue | |
|---|---|---|---|---|---|---|
| Ppbp | 6.06743667 | 42.54622081 | 2.805607243 | 4.731617832 | 6.01E-20 | |
| Ltf | 47.01472015 | 145.4730741 | 1.629431072 | 8.37639472 | 4.61E-14 | |
| Ngp | 211.6949508 | 593.449872 | 1.487057881 | 8.943160582 | 2.58E-12 | |
| Slc4a1 | 35.53190653 | 97.68795024 | 1.458945625 | 8.349028553 | 9.80E-12 | |
| Gm42743 | 11.10087934 | 37.07452194 | 1.738387495 | 5.196557724 | 4.45E-11 | |
| S100a8 | 105.3976697 | 292.0241702 | 1.469958744 | 7.11045771 | 4.80E-11 | |
| Hist2h2bb | 64.90246234 | 200.2259319 | 1.624389173 | 5.666290083 | 6.56E-11 | |
| Gm20628 | 9.304975584 | 27.89101178 | 1.582996871 | 5.908573294 | 7.93E-11 | |
| Gypa | 10.57369037 | 33.52088276 | 1.663434714 | 5.358248894 | 1.29E-10 | |
| Hist1h3b | 21.19688359 | 73.39351223 | 1.790030109 | 4.887391348 | 1.32E-10 | |
To further clarify the relationships between methylated genes and gene expression, we conducted a joint analysis of the MeRIP-seq and mRNA-seq results to identify overlapping genes between the two groups, and the identified important genes were roughly divided into four categories: 3 genes the up-regulation of both mRNA expression and m6A peaks, 3 genes showed the down-regulation of both mRNA expression and m6A peaks, 16 genes showed up-regulated m6A peaks and down-regulated mRNA expression, and 8 genes showed down-regulated m6A peaks and up-regulated mRNA expression. The relationship between m6A peaks and mRNA expression is shown in a quadrant graph (Fig. 7D).
4.4. Changes in m6A related enzymes after IDD
To explore the changes in m6A-related enzymes in the NP after IDD, we used qRT-PCR to detect the genes related to methylases, demethylases and methylation-binding proteins, including METTL3, METL14, WTAP, VIRMA, FTO, ALKBH5, YTHDF1, YTHDF2 and YTHDF3. As shown in the figure, relative to the control group, the expression levels of KIAA1429, METTL14, METTL3, METTL4, WTAP, DGCR8, EIF3A and YTHDC1 in the experimental group were up-regulated, while those of FTO, ELAVL1, HNRNPC1 and SRSF2 were down-regulated, and the expression levels of ALKBH5, RBM15, RBM15B, YTHDC2, YTHDF1, YTHDF2, YTHDF3 and EIF3B did not differ significantly between the two groups. The dynamic changes in m6A methylation in the NP tissue after IDD may be caused by the aforementioned enzymes with significant expression differences (Fig. 8).
Figure 8.
The relative expression of m6A-related enzymes in the experimental and control groups is shown.
5. Discussion
Lumbar disc herniation (LDH) has become the main cause of LBP in the human population and seriously affects quality of life [40]. As one of the causes of LDH, IDD has been widely studied. As the most abundant and common posttranscriptional modification in higher eukaryotic cells, m6A plays an important role in many biological reactions in the body [15]. Because posttranscriptional mRNA sequences can undergo conversion between a methylated or demethylated status, methylation can be used as a potential therapeutic target for the treatment of neoplastic [24,25] and non-neoplastic diseases [27,28] and presents great research and development prospects. In our study, we elucidated the m6A-related epigenetic mechanism of NP cells in the degenerated IVD of standing mice. First, we verified that standing induced by the water-escape behavior of mice can lead to IDD. Second, we performed qRT-PCR analysis of the NP and found changes in the expression of m6A-related enzymes. Finally, we performed MeRIP-seq and analyzed differentially expressed genes. A total of 985 differential expressed genes were identified, including 363 genes that were significantly up-regulated and 622 genes that were significantly down-regulated. In addition, 21779 m6A peaks were identified in 9648 genes. There were 1319 methylated m6A peaks showing significant differences, including 933 peaks showing significant up-regulated and 386 peaks showing significant down-regulated, and the related enriched genes and pathways were identified. Based on the above findings, we speculate that the changes in m6A modification in NP cells under continuous stress may be the reason for IDD, which further leads to the occurrence of LDH.
LBP caused by IDD has become an important factor affecting the quality of life of modern people, resulting in disability in millions of people every year and imposing a huge economic burden on families, society and the medical system [2]. The etiologies of IDD are diverse, including smoking [7], high mechanical stress [8,9], sex [10], and age [11], but the specific cellular and molecular mechanisms involved are still unclear. Related bio-mechanical changes are considered to be one of the important factors leading to IDD because they can change the cell metabolism of the IVD [41]. Many studies have shown that an increase in disordered mechanical stress will lead to the extensive loss of notochord cells in the IVD, which in turn causes IDD [8,9]. In addition, because the arrangement direction of collagen fibers in the AF was easily changed by mechanical stress [42,43], an increase in disordered mechanical stress will change the force of the AF, impair its cellular function and reshape its structure, eventually leading to IDD. The physiological curvature and structural integrity of the spine play a critical role in weight bearing and stress dispersion in the body. The NP can resist and redistribute pressure, while the AF mainly endures tension. In this experiment, the mice stood up due to hydrophobia, which changed the original walking motion of the limbs and increased stress on the spine and IVD, leading to IDD.
Previous studies have shown that the up-regulation or down-regulation of the posttranscriptional modification methylation levels of genes is closely related to the coordinated activities of methylases, demethylases or methylation recognition enzymes. Studies have shown that the hypermethylation of CpG islands in the promoter region of miR-129–5 P can inhibit the expression of miR-129–5 P and induce autophagy in NP cells, which leads to IDD. In contrast, hypomethylation can promote the expression of miR-129–5p to inhibit beclin-1 and then block autophagy in the NP to delay IDD [44]. Another study used a static compression device to establish a model of caudal IDD in rats [33]. qRT-PCR analysis of the caudal NP revealed that expression was significantly increased in the degenerated IVD, while the levels of methylases (METTL3 and METTL14) and methylation recognition enzymes (YTHDF1 and YTHDF2) were not significantly different between the experimental and control groups. In our study, we found that the expression of METTL14 and METTL3 in the experimental group was significantly up-regulated, while that of FTO was significantly down-regulated. There was no significant difference in the expression of YTHDF1 and YTHDF2 between the two groups. These findings differ from the results found in previous studies, which may be related to different tissue sources. The role of FTO in IDD remains to be studied.
Posttranscriptional epigenetic modification plays a key role in mammals. Studies [45] have confirmed that m6A methylation can reduce the stability of mRNA to adapt to corresponding biological functions. Another study [17] showed that m6A modification could bypass 5′ cap binding protein to increase translation efficiency under stress. In addition, m6A modification not only improves translation efficiency but also inhibits translation through posttranscriptional modification [46]. To further understand the role of m6A modification in the NP under IDD, we performed a correlation analysis between MeRIP-seq and mRNA-seq results to determine the relationship between m6A peaks and mRNA expression levels. We identified 30 genes whose m6A peaks and mRNA levels both changed significantly, including 3 genes whose mRNA expression and m6A peaks were both up-regulated, 3 genes whose mRNA expression and m6A peaks were both down-regulated, 16 genes whose m6A peaks were up-regulated and mRNA expression was down-regulated, and 8 genes whose m6A peaks were down-regulated and mRNA expression was up-regulated. Our results show that the hypermethylation or hypomethylation of m6A not only leads to high mRNA expression but also induces low mRNA expression.
There are a variety of methods for m6A detection, including m6A dot blotting, colorimetry, liquid chromatography-tandem mass spectrometry [47], MeRIP-seq [48] and epitranscriptomics micro-array analysis [49]. The first three methods can only qualitatively measure the overall level of m6A, while the last two combined with high-throughput sequencing can be used to quantitatively analyze the level of m6A, and MeRIP-seq is one of the most common detection methods applied in m6A research. A study in which an epitranscriptomics micro-array was used to analyze the IVD of rats subjected to passive smoking identified differentially expressed genes under the tested conditions. Therefore, the authors speculated that circadian rhythm changes in the peripheral biological clock (IVD clock) may be related to IDD [50]. Tang et al. [51] used gene micro-array analysis to determine the existence of tissue-specific gene expression and age-related differential expression in the NP of rats of different ages. Wang et al. [33] performed epitranscriptomics micro-array analysis on rat NP to investigate the relationship between m6A modification and IDD. They found that the Wnt signaling pathway participates in IDD by regulating the metabolism of NP cells. However, there are no reports illustrating the m6A modification pattern under IDD induced by standing in mice. In our study, we observed changes in the m6A peaks of the NP in mice through MeRIP-seq and found that 933 m6A peaks were significantly up-regulated and that 386 m6A peaks were significantly down-regulated. In addition, GO and KEGG pathway analyses were used to identify the potential mechanisms and functions of differential methylated mRNA. The GO enrichment results show that it is related to cell biosynthesis and metabolism, such as: intracellular macromolecule biosynthesis and metabolism. Due to the mechanical load can directly affect the metabolism of IVD cells and disrupt the balance of matrix metabolism, it is considered to be one of the main predisposing factors for IDD [52]. Studies have reported that static compression increases the glucose consumption of NP cells [53], and found that the oxygen consumption of degenerated IVD cells was significantly increased [54], indicating that the glucose metabolism of IVD cells has changed during the process of IDD. In addition, the study has confirmed that nutritional disorders of IVD cells was also one of the factors leading to IDD [55]. Therefore, bipedal mouse model could increase the stress of the IVD, possibly by changing the bio-synthetic metabolism of the IVD cells to induce degeneration. Our KEGG analysis showed that the Wnt signaling pathway and MAPK signaling pathway were involved in IDD. Previous studies have shown that these pathways can accelerate the IDD, which is consistent with our results.
However, our research still has some limitations. First, we only detected the changes in enzymes and genes related to m6A modification in the NP of degenerative lumbar IVD in mice; we did not verify which enzymes caused the changes, and we do not know whether the same m6A modification occurs in humans. Second, after annotating the related genes and signaling pathways involved, we did not discuss in depth what genes and signaling pathways play an important role in IDD. Finally, we only explored abnormal m6A modification in the NP and its potential mechanism in IDD, and we did not study the AF or end-plate. Therefore, our subsequent research will focus on solving the above problems.
6. Conclusion
We are the first to use MeRIP-seq to study the posttranscriptional m6A modification of the NP in IDD in mice. The results indicate that m6A modification plays an important role in the process of IDD. In addition, we identified differentially expressed m6A peaks and their corresponding gene functions and possible signaling pathways. Finally, considering these results combined with the analysis of MeRIP-seq and mRNA-seq results, we described the relationship between the methylation status of m6A and transcription.
Ethics approval
The study protocol was approved by the Institutional Review Board at The First Affiliated Hospital of University of South China, Hunan, P.R. China.
Author contribution
All authors participated in this article. BZ, HXC, YC and YGY designed the study. SL, JHT, MXZ, CW, JBX, YX, XLL contributed to the conception. BZ, HXC and SL performed experiments. BZ, YC and YGY performed bioinformatics analysis and interpreted the data. All authors were involved in drafting and revision of the manuscript, read and approved the final manuscript.
Funding
This work was supported by the Clinical Research Center for Minimally Invasive Spine Surgery in Hunan province (2017SK4004), Health commission of Hunan Province of China (A2017016), the General Project of the China Postdoctoral Science Foundation (2017M623363) and Hengyang Key Laboratory of Orthopedic Implant Research and Development (2018KJ115).
Declaration of competing interest
The authors declare that they have no competing interests.
Acknowledgments
No honorarium, grant, or other forms of payment were given to anyone to produce the manuscript. Each author listed on the manuscript has seen and approved the submission of this version of the manuscript and takes full responsibility for the manuscript. We also thank American Journal Experts for assistance in preparation of this manuscript.
Contributor Information
Bin Zhu, Email: 627520854@qq.com.
Hao-xiang Chen, Email: 1003096308@qq.com.
Shan Li, Email: Lishan333@hotmail.com.
Jing-hua Tan, Email: 41581015@qq.com.
Yong Xie, Email: 493509012@qq.com.
Ming-xiang Zou, Email: zouwei8887@126.com.
Cheng Wang, Email: hnwangcheng1978@sina.com.
Jing-bo Xue, Email: xuejingbo0218@sina.com.
Xue-lin Li, Email: liduan19@163.com.
Yong Cao, Email: caoyong1912@163.com.
Yi-guo Yan, Email: yanyiguo@live.cn.
References
- 1.Brodke D., Ritter S. Nonsurgical management of low back pain and lumbar disk degeneration. Instr Course Lect. 2005;54:279–286. [PubMed] [Google Scholar]
- 2.Wu A., Lyn M., Zheng X., Huang J., Wang X., Zhao J., et al. Global low back pain prevalence and years lived with disability from 1990 to 2017: estimates from the Global Burden of Disease Study 2017.[J] Ann Transl Med. 2020;8:299. doi: 10.21037/atm.2020.02.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lv B., Yuan J., Ding H., Wan B., Jiang Q., Luo Y., et al. Relationship between endplate defects, modic change, disc degeneration, and facet joint degeneration in patients with low back pain. [J] .Biomed Res Int. 2019;2019:9369853. doi: 10.1155/2019/9369853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogduk N. Evidence-informed management of chronic low back pain with facet injections and radiofrequency neurotomy. Spine J : official journal of the North American Spine Society. 2008;8(1):56–64. doi: 10.1016/j.spinee.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 5.Taylor R., Taylor R. The economic impact of failed back surgery syndrome. British journal of pain. 2012;6(4):174–181. doi: 10.1177/2049463712470887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown Jasmine, Sandesh Lakkol, Sophia Lazenby, et al. Common neoplastic causes of paediatric and adolescent back pain.[J] Br J Hosp Med. 2020;81:1–6. doi: 10.12968/hmed.2020.0026. [DOI] [PubMed] [Google Scholar]
- 7.Jing D., Wu W., Deng X., Peng Y., Yang W., Huang D., et al. FoxO1a mediated cadmium-induced annulus fibrosus cells apoptosis contributes to intervertebral disc degeneration in smoking. [J] .J Cell Physiol. 2021;236:677–687. doi: 10.1002/jcp.29895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu F., Bao R., Yao S., Zhou C., Luo H., Zhang Z., et al. Aberrant spinal mechanical loading stress triggers intervertebral disc degeneration by inducing pyroptosis and nerve ingrowth. [J] .Sci Rep. 2021;11:772. doi: 10.1038/s41598-020-80756-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodrigues-Pinto R., Richardson S., Hoyland J. An understanding of intervertebral disc development, maturation and cell phenotype provides clues to direct cell-based tissue regeneration therapies for disc degeneration. Eur Spine J : official publication of the European Spine Society, the European Spinal Deformity Society, and the European Section of the Cervical Spine Research Society. 2014;23(9):1803–1814. doi: 10.1007/s00586-014-3305-z. [DOI] [PubMed] [Google Scholar]
- 10.Mosley G.E., Wang Mi, Nasser P., Lai A., Charen D.A., Zhang B., et al. Males and females exhibit distinct relationships between intervertebral disc degeneration and pain in a rat model. [J] .Sci Rep. 2020;10:15120. doi: 10.1038/s41598-020-72081-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takashi Ohnishi, Hideki Sudo, Takeru Tsujimoto, et al. Age-related spontaneous lumbar intervertebral disc degeneration in a mouse model. [J] .J Orthop Res. 2018;36:224–232. doi: 10.1002/jor.23634. [DOI] [PubMed] [Google Scholar]
- 12.Dor Y., Cedar H. Principles of DNA methylation and their implications for biology and medicine. Lancet (London, England) 2018;392(10149):777–786. doi: 10.1016/S0140-6736(18)31268-6. [DOI] [PubMed] [Google Scholar]
- 13.Tajerian M., Alvarado S., Millecamps M., Dashwood T., Anderson K.M., Haglund Lt, et al. DNA methylation of SPARC and chronic low back pain. Mol Pain. 2011;7:65. doi: 10.1186/1744-8069-7-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikuno A., Akeda K., Takebayashi S.I., Shimaoka M., Okumura K., Sudo A., et al. Genome-wide analysis of DNA methylation profile identifies differentially methylated loci associated with human intervertebral disc degeneration. PLoS One. 2019;14(9):e0222188. doi: 10.1371/journal.pone.0222188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu Y., Dominissini D., Rechavi G., et al. Gene expression regulation mediated through reversible m⁶A RNA methylation. Nat Rev Genet. 2014;15(5):293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- 16.Wei C., Gershowitz A., Moss B. Methylated nucleotides block 5' terminus of HeLa cell messenger RNA. Cell. 1975;4(4):379–386. doi: 10.1016/0092-8674(75)90158-0. [DOI] [PubMed] [Google Scholar]
- 17.Meyer K., Patil D., Zhou J., Zinoviev A., Skabkin M.A., Elemento O., et al. 5’ UTR m(6)A promotes cap-independent translation. Cell. 2015;163(4):999–1010. doi: 10.1016/j.cell.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Molinie B., Wang J., Lim K.S., Hillebrand R., Lu Z.X., Van W.N., et al. m(6)A-LAIC-seq reveals the census and complexity of the m(6)A epitranscriptome. Nat Methods. 2016;13(8):692–698. doi: 10.1038/nmeth.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X., Lu Z., Gomez A., Hon G.C., Yue Y., Han D., et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505(7481):117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao S., Lu J., Chen Y., Wang Z., Cao J., Dong Y., et al. Exploration of the potential roles of m6A regulators in the uterus in pregnancy and infertility.[J] J Reprod Immunol. 2021;146:103341. doi: 10.1016/j.jri.2021.103341. [DOI] [PubMed] [Google Scholar]
- 21.Yang Y., Huang W., Huang J.T., Shen F., Xiong J., Yuan E.F., et al. Increased N6-methyladenosine in human sperm RNA as a risk factor for asthenozoospermia. Sci Rep. 2016;6:24345. doi: 10.1038/srep24345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan Y., Yan G., He M., Lei H., Li L., Wang Y., et al. ALKBH5 suppresses tumor progression via an mA-dependent epigenetic silencing of pre-miR-181b-1/YAP signaling axis in osteosarcoma. [J] .Cell Death Dis. 2021;12:60. doi: 10.1038/s41419-020-03315-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu Q., Yuan X., Han R., Zhang H., Xiu R., et al. Epitranscriptomic mechanisms of N6-methyladenosine methylation regulating mammalian hypertension development by determined spontaneously hypertensive rats pericytes. Epigenomics. 2019;11(12):1359–1370. doi: 10.2217/epi-2019-0148. [DOI] [PubMed] [Google Scholar]
- 24.Weng H., Huang H., Wu H., Qin X., Zhao B.S., Dong L., et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA mA modification. Cell stem cell. 2018;22(2):191–205. doi: 10.1016/j.stem.2017.11.016. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li T., Hu P., Zuo Z., Lin J.F., Li X., Wu Q.N., et al. METTL3 facilitates tumor progression via an mA-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol Cancer. 2019;18(1):112. doi: 10.1186/s12943-019-1038-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lichinchi G., Gao S., Saletore Y., Gonzalez G.M., Bansal V., Wang Y., et al. Dynamics of the human and viral m(6)A RNA methylomes during HIV-1 infection of T cells. Nature microbiology. 2016;1:16011. doi: 10.1038/nmicrobiol.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weng Y., Wang X., An R., Cassin J., Vissers C., Liu Y., et al. Epitranscriptomic mA regulation of axon regeneration in the adult mammalian nervous system. Neuron. 2018;97(2):313–325. doi: 10.1016/j.neuron.2017.12.036. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu J., Zhang Y., Ma H., Zeng R., Liu R., Wang P., et al. Epitranscriptomic profiling of N6-methyladenosine-related RNA methylation in rat cerebral cortex following traumatic brain injury. Mol Brain. 2020;13(1):11. doi: 10.1186/s13041-020-0554-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dong W., Liu J., Lv Y., Wang F., Liu T., Sun S., et al. miR-640 aggravates intervertebral disc degeneration via NF-κB and WNT signalling pathway. Cell Prolif. 2019;52(5):e12664. doi: 10.1111/cpr.12664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao K., Zhang Y., Kang L., Song Y., Wang K., Li S., et al. Epigenetic silencing of miRNA-143 regulates apoptosis by targeting BCL2 in human intervertebral disc degeneration. Gene. 2017;628:259–266. doi: 10.1016/j.gene.2017.07.043. [DOI] [PubMed] [Google Scholar]
- 31.Zhan S., Wang K., Xiang Q., Song Y., Li S., Liang H., et al. lncRNA HOTAIR upregulates autophagy to promote apoptosis and senescence of nucleus pulposus cells. J Cell Physiol. 2020;235(3):2195–2208. doi: 10.1002/jcp.29129. [DOI] [PubMed] [Google Scholar]
- 32.Liu Q., Li M., Jiang L., Jiang R., Fu B. METTL3 promotes experimental osteoarthritis development by regulating inflammatory response and apoptosis in chondrocyte. Biochem Biophys Res Commun. 2019;516(1):22–27. doi: 10.1016/j.bbrc.2019.05.168. [DOI] [PubMed] [Google Scholar]
- 33.Wang X., Chen N., Du Z., Ling Z., Zhang P., Yang J., et al. Bioinformatics analysis integrating metabolomics of mA RNA microarray in intervertebral disc degeneration. Epigenomics. 2020;12(16):1419–1441. doi: 10.2217/epi-2020-0101. [DOI] [PubMed] [Google Scholar]
- 34.Goff C., Landmesser W. Bipedal rats and mice; laboratory animals for orthopaedic research. The Journal of bone and joint surgery. American volume. 1957;(3):616–622. [PubMed] [Google Scholar]
- 35.Wang Y., Wu Y., Deng M., Kong Q. World Neurosurg; 2021. Establishment of a rabbit intervertebral disc degeneration model by percutaneous posterolateral puncturing of lumbar discs under local anesthesia. [J] [undefined: undefined] [DOI] [PubMed] [Google Scholar]
- 36.Hutton W., Yoon S., Elmer W., Li J., Murakami H., Minamide A., et al. Effect of tail suspension (or simulated weightlessness) on the lumbar intervertebral disc: study of proteoglycans and collagen. Spine. 2002;27(12):1286–1290. doi: 10.1097/00007632-200206150-00008. [DOI] [PubMed] [Google Scholar]
- 37.Yurube T., Hirata H., Ito M., Terashima Y., Kakiuchi Y., Kuroda R., et al. Involvement of autophagy in rat tail static compression-induced intervertebral disc degeneration and notochordal cell disappearance. [J] .Int J Mol Sci. 2021;22 doi: 10.3390/ijms22115648. [undefined] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ao X., Wang L., Shao Y., Chen X., Zhang J., Ch J., et al. Clinical orthopaedics and related research; 2019. Development and characterization of a novel bipedal standing mouse model of intervertebral disc and facet joint degeneration. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pistell P., Ingram D. Development of a water-escape motivated version of the Stone T-maze for mice. Neuroscience. 2010;166(1):61–72. doi: 10.1016/j.neuroscience.2009.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Montano D. Upper body and lower limbs musculoskeletal symptoms and health inequalities in Europe: an analysis of cross-sectional data. BMC Muscoskel Disord. 2014;15:285. doi: 10.1186/1471-2474-15-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wuertz K., Godburn K., MacLean J., Barbir A., Donnelly J.S., Roughley P.J., et al. In vivo remodeling of intervertebral discs in response to short- and long-term dynamic compression. J Orthop Res : official publication of the Orthopaedic Research Society. 2009;27(9):1235–1242. doi: 10.1002/jor.20867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chiquet M. Regulation of extracellular matrix gene expression by mechanical stress. Matrix Biol : journal of the International Society for Matrix Biology. 1999;18(5):417–426. doi: 10.1016/s0945-053x(99)00039-6. [DOI] [PubMed] [Google Scholar]
- 43.Chiquet M., Gelman L., Lutz R., Maier S. From mechanotransduction to extracellular matrix gene expression in fibroblasts. Biochim Biophys Acta. 2009;1793(5):911–920. doi: 10.1016/j.bbamcr.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 44.Zhao K., Zhang Y., Kang L., Song Y., Wang K., Li S., et al. Methylation of microRNA-129-5P modulates nucleus pulposus cell autophagy by targeting Beclin-1 in intervertebral disc degeneration. Oncotarget. 2017;8(49):86264–86276. doi: 10.18632/oncotarget.21137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Geula S., Moshitch-Moshkovitz S., Dominissini D., Mansour A.A., Kol N., Salmon-Divo M., et al. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science (New York, N.Y.) 2015;347(6225):1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- 46.Wang X., Zhao B., Roundtree I., Lu Z., Han D., Ma H., et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161(6):1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Han J., Wang J., Yang X., Yu H., Zhou R., Lu H.C., et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol Cancer. 2019;18(1):110. doi: 10.1186/s12943-019-1036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dominissini D., Moshitch-Moshkovitz S., Salmon-Divon M., Amariglio N., Rechavi G., et al. Transcriptome-wide mapping of N(6)-methyladenosine by m(6)A-seq based on immunocapturing and massively parallel sequencing. Nat Protoc. 2013;8(1):176–189. doi: 10.1038/nprot.2012.148. [DOI] [PubMed] [Google Scholar]
- 49.Chokkalla A., Mehta S., Kim T., Chelluboina B., Kim J., Vemuganti R., et al. Transient focal ischemia significantly alters the mA epitranscriptomic tagging of RNAs in the brain. Stroke. 2019;50(10):2912–2921. doi: 10.1161/STROKEAHA.119.026433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Numaguchi S., Esumi M., Sakamoto M., Endo M., Ebihara T., Soma H., et al. Passive cigarette smoking changes the circadian rhythm of clock genes in rat intervertebral discs. J Orthop Res : official publication of the Orthopaedic Research Society. 2016;34(1):39–47. doi: 10.1002/jor.22941. [DOI] [PubMed] [Google Scholar]
- 51.Tang X., Jing L., Chen J. Changes in the molecular phenotype of nucleus pulposus cells with intervertebral disc aging. PLoS One. 2012;7(12):e52020. doi: 10.1371/journal.pone.0052020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hirata H., Yurube T., Kakutani K., Maeno K., Takada T., Yamamoto J., et al. A rat tail temporary static compression model reproduces different stages of intervertebral disc degeneration with decreased notochordal cell phenotype. J Orthop Res. 2014;32(3):455–463. doi: 10.1002/jor.22533. [DOI] [PubMed] [Google Scholar]
- 53.Fernando H.N., Czamanski J., Yuan T.Y., Gu W., Salahadin A., Huang C.Y. Mechanical loading affects the energy metabolism of intervertebral disc cells. J Orthop Res. 2011;29(11):1634–1641. doi: 10.1002/jor.21430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cisewski S.E., Wu Y., Damon B.J., Sachs B.L., Kern M.J., Yao H. Comparison of oxygen consumption rates of nondegenerate and degenerate human intervertebral disc cells. Spine43. 2018;(2):E60–E67. doi: 10.1097/BRS.0000000000002252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grunhagen T., Shirazi-Adl A., Fairbank J.C., Urban J.P. Intervertebral disk nutrition: a review of factors influencing concentrations of nutrients and metabolites. Orthop Clin N Am. 2011;42:465–477. doi: 10.1016/j.ocl.2011.07.010. [DOI] [PubMed] [Google Scholar]