Significance
Innate responses against viral infection and other intracellular pathogens rely on immune cells that are capable of lysing infected cells and producing interferon-gamma (IFNγ). These cells encompass two major cell lineages: natural killer (NK) cells and type 1 innate lymphoid cells (ILC1s). While NK cells have been extensively characterized, identification of ILC1s and their distinction from NK cells are less clear. The transcription factor Hobit encoded by Zfp683 has been put forth as a prototypic feature of ILC1s. By analyzing Zfp683 reporter, fate-map, and -deficient mice, we demonstrate that the impact of Hobit on ILC1 identity and transcriptional and functional programs is tissue- and context-dependent. Thus, ILC1s adapt to local stimuli and tailor their responses to the tissue niche.
Keywords: innate lymphoid cells, natural killer cells, Hobit, Eomes, liver
Abstract
Identification of type 1 innate lymphoid cells (ILC1s) has been problematic. The transcription factor Hobit encoded by Zfp683 has been proposed as a major driver of ILC1 programs. Using Zfp683 reporter mice, we showed that correlation of Hobit expression with ILC1s is tissue- and context-dependent. In liver and intestinal mucosa, Zfp683 expression correlated well with ILC1s; in salivary glands, Zfp683 was coexpressed with the natural killer (NK) master transcription factors Eomes and TCF1 in a unique cell population, which we call ILC1-like NK cells; during viral infection, Zfp683 was induced in conventional NK cells of spleen and liver. The impact of Zfp683 deletion on ILC1s and NK cells was also multifaceted, including a marked decrease in granzyme- and interferon-gamma (IFNγ)–producing ILC1s in the liver, slightly fewer ILC1s and more Eomes+ TCF1+ ILC1-like NK cells in salivary glands, and only reduced production of granzyme B by ILC1 in the intestinal mucosa. NK cell–mediated control of viral infection was unaffected. We conclude that Hobit has two major impacts on ILC1s: It sustains liver ILC1 numbers, while promoting ILC1 functional maturation in other tissues by controlling TCF1, Eomes, and granzyme expression.
Innate lymphoid cells (ILCs) comprise a diverse group of lymphocytes devoid of rearranged antigen receptors. ILCs are classified into three groups based on their functional programs. The type 1 ILC (ILC1) program driven by Tbet culminates with production of interferon-gamma (IFNγ); the ILC2 program dictated by GATA3 promotes secretion of interleukin-5 (IL-5) and IL-13; the ILC3 program driven by Rorγt leads to production of IL-17 and IL-22 (1–3). In mice, ILC1s and natural killer (NK) cells both require Tbet, produce IFNγ, and express the cell-surface markers NKp46 and NK1.1. Given these similarities, distinguishing ILC1s from NK cells has been problematic (4–6). One important distinction between ILC1s and NK cells is that ILC1s are mainly tissue-resident, whereas NK cells circulate in the blood (7). Therefore, ILC1s provide a local source of IFNγ for prompt control of intracellular pathogens, whereas NK cells recruited from the blood provide a second wave of IFNγ (8). Moreover, NK cells are thought to be more cytotoxic than ILC1s due to superior production of perforin and granzymes, though recent studies have challenged this view (9, 10). Phenotypically, various specific cell-surface markers have been identified for each cell type. CD49a, CD200R1, CD69, CD103, and CD127 are mainly expressed in ILC1s (11), while NK cells express CD49b, CD62L, as well as NK cell receptors of the immunoglobulin and C-type lectin families with a semiclonal distribution (12, 13). However, the expression of these markers can vary among tissues and can be acquired or modulated in pathological conditions, such as viral infections and tumors.
ILC1s and NK cells follow different developmental trajectories. NK cells originate from an early common innate lymphoid progenitor, whereas all ILCs derive from a downstream Id2+ common helper innate lymphoid progenitor (5, 14, 15); however, recent reports have identified a shared progenitor that gives rise to both ILC1s and NK cells (16). Regardless of their progenitor, NK cells and ILC1s express unique transcription factors that shape their identities. NK cells depend on Eomes for development, whereas ILC1s are Eomes-independent (17). The transcriptional regulator Hobit, encoded by the gene Zfp683, is highly expressed in ILC1s and is required for liver ILC1s but not NK cells (18). In a recent single-cell RNA-sequencing (scRNA-seq) analysis of the ILC1–NK spectrum in multiple tissues, we confirmed that ILC1s and NK cells are generally associated with expression of Hobit and Eomes, respectively, but also noticed a considerable tissue heterogeneity of ILC1s and NK cells (19). For example, salivary glands and uterus contain a unique cell population that expresses both Eomes and Hobit, as well as the transcription factor TCF1. We refer to these cells as “ILC1-like NK cells.” These cells are also unique for their independence from NFIL3 (20–22), a transcriptional repressor required by both NK cells and ILC1s for development (23–27). Thus, Hobit may not be a univocal driver of ILC1s.
To evaluate the association of Hobit with ILC1s and its impact on their development and functions, we generated a Hobit reporter mouse that identifies cells expressing Hobit within the ILC1–NK spectrum, as well as a Hobit fate-map mouse that identifies ILCs and NK cells that have expressed Hobit at any time of their development. Using these tools, we showed that Hobit association with ILC1s and their cell-surface markers varies in different tissues. We also showed that Hobit fate-mapped+ cells can be found in ILC1s and a subset of ILC3s but not in NK cells, demonstrating that ILC1s represent a lineage distinct from NK cells, which can in part convert into ILC3s. scRNA-seq and flow cytometric analyses of mice with a conditional deletion of Zfp683 in the ILC1–NK spectrum showed that the impact of Hobit on ILC1s also varies in different tissues. Lack of Hobit has the following effects: 1) markedly reduced numbers of two ILC1 subsets specialized in either granzyme or IFNγ production in the liver; 2) slightly fewer ILC1s that fail to effectively produce granzyme B along with more Eomes- and TCF1-driven ILC1-like NK cells in the salivary gland; and 3) only defective expression of granzyme B by ILC1s in the small intestine. Finally, we found that murine cytomegalovirus (MCMV) infection induced Hobit expression in spleen and liver NK cells, although it was not necessary to sustain NK cell–mediated control of viral infection in liver. We conclude that the expression and impact of Hobit are tissue- and context-dependent in ILC1s and NK cells.
Results
Hobit Association with ILC1 Surface Markers Varies in Different Organs.
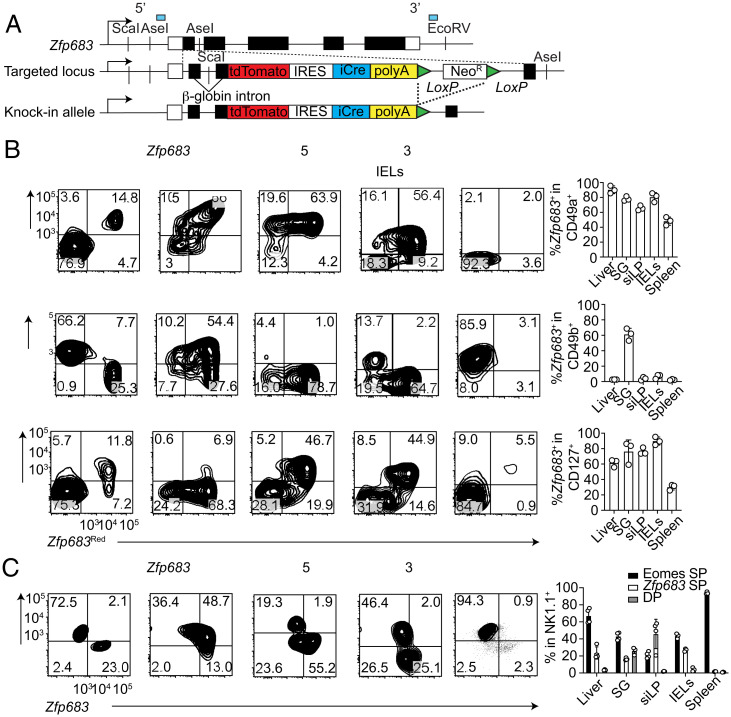
To trace cells expressing Hobit and define their features compared with other innate lymphocytes, we generated knockin/knockout Hobit reporter mice. We inserted a tandem dimer red fluorescent protein (tdTomato) and a Cre recombinase linked by an internal ribosomal entry site (IRES) at the translation initiation site of the Zfp683 gene encoding Hobit (Fig. 1A). We called these mice Zfp683Red. The size of the Zfp683+ population among Lin–NK1.1+ cells differed in various tissues; Zfp683+ cells comprised a large fraction of NK1.1+ cells in liver, salivary glands, intestinal lamina propria, and intestinal epithelium, but were scarce in the spleen (Fig. 1B). We next investigated the association of Hobit with ILC1 markers in these tissues. CD49a and CD49b are often used to identify ILC1s and NK cells, respectively (1). In liver, Zfp683+ cells coincided with CD49a+ cells, whereas CD49b+ cells encompassed Zfp683– cells. In the lamina propria and the epithelial layer of intestinal mucosa, NK1.1+ cells were mainly CD49a+ and ∼60 to 80% of these cells expressed Zfp683 (Fig. 1B). In salivary glands, the majority of NK1.1+ cells expressed both CD49a and CD49b as well as Zfp683, with only relatively small fractions of CD49a+ or CD49b+ cells lacking Zfp683 (Fig. 1B). The spleen contained mainly CD49b+ cells, which were largely Zfp683– (Fig. 1B). The receptor for IL-7 (CD127) is another marker reported to be expressed by most ILCs (11). A large proportion of Zfp683+ cells expressed CD127 in the intestinal lamina propria and epithelium; around 50 to 70% of Zfp683+ cells expressed CD127 in the liver. Only a very small fraction of salivary glands and splenic NK1.1+ cells (2 to 7%) expressed CD127, most of which expressed Zfp683. These results indicate that the commonly used cell markers CD49a and CD127 identify Zfp683+ ILC1s only in part, as the association of these markers with Zfp683 expression varies in different tissues.
Fig. 1.
Zfp683 and Eomes define ILC1s and NK cells across tissues. (A) Schematic of targeting construct. (B) Representative flow cytometric dot plots and quantification of Zfp683+ cells among live, CD45+Lin−NK1.1+CD49a+, CD49b+, or CD127+ cells in the indicated organs. Single-cell suspensions were obtained from heterozygous Zfp683Red mice. IELs, intraepithelial lymphocytes; SG, salivary gland; siLP, small intestinal lamina propria. Data are representative of two independent experiments (n = 3 or 4). Data represent mean ± SEM. (C) Expression of Eomes and Zfp683 in single-cell suspensions from different organs of Zfp683Red heterozygous mice, and their quantification among NK1.1+ cells gated as in B. DP, double-positive; SP, single positive. Data represent mean ± SEM. Data are representative of two independent experiments (n = 3 or 4).
Hobit and Eomes Are Mutually Exclusively Expressed in Most Tissues.
If Hobit and Eomes identify ILC1s and NK cells, respectively, their expression should be mutually exclusive. We examined Lin–NK1.1+ cells for intracellular content of Eomes versus Zfp683 expression in various tissues of Zfp683Red mice. Cells in liver, intestinal lamina propria, intestinal epithelium, and spleen exclusively expressed either Eomes or Zfp683 (Fig. 1C). This pattern corroborated the existence of Zfp683+ ILC1s distinct from Eomes+ NK cells. However, about 30 to 50% of NK1.1+ cells in salivary glands expressed both Eomes and Zfp683 (Fig. 1C), as well as CD49a (SI Appendix, Fig. S1), and therefore could not be rigorously classified as NK cells or ILC1s; we refer to these cells as ILC1-like NK cells.
scRNA-Seq Reveals ILC1 Subsets Differentially Represented in Distinct Tissues.
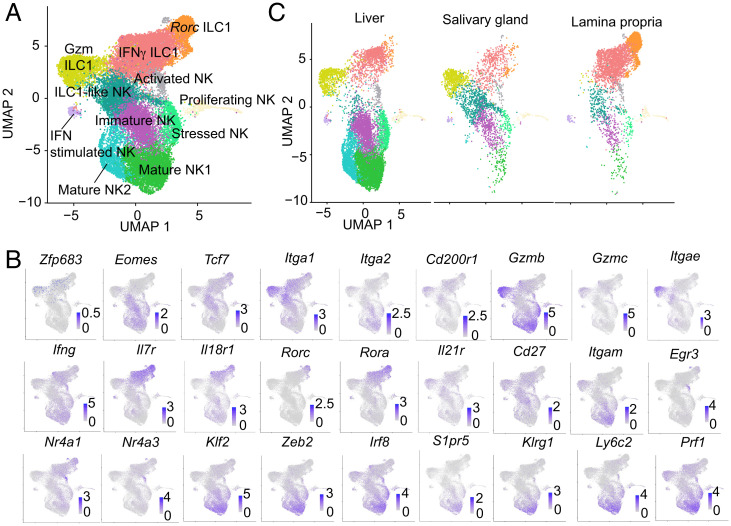
To further investigate the expression of Hobit across tissue ILC1s and its association with different markers, we isolated Lin–NK1.1+ cells from liver, salivary glands, and intestinal lamina propria, which contain the largest fraction of Zfp683+ cells, and performed scRNA-seq using the 10X Chromium platform. We profiled a total of quality-controlled 17,660 cells (SI Appendix, Table S1) that were visualized using the uniform manifold approximation and projection (UMAP) method, regardless of the tissue origin. ILC1 and NK cell clusters were defined based on the top 20 differentially expressed genes (Fig. 2 A and B and SI Appendix, Fig. S2). We promptly identified ILC1 and NK cell clusters based on expression of Zfp683 and Eomes, respectively, as well as ILC1-like NK cells expressing both Zfp683 and Eomes. ILC1-like NK cells also expressed Tcf7, which encodes TCF1. This distinction was consistent with the distribution of Itga1 (for CD49a) and Itga2 (for CD49b). Cd200r1, a marker of ILC1s (8), overlapped with Zfp683 and Itga1. We further distinguished three groups of ILC1s: granzyme-producing ILC1s (Gzm ILC1), IFNγ-producing ILC1s (IFNγ ILC1), and Rorc ILC1s (Rorc ILC1). Gzm+ ILC1s expressed messenger RNA (mRNA) for granzymes (Gzma, Gzmb, Gzmc), as well as Itgae (for CD103), Tyrobp (for DAP12), and the inhibitory receptor Lag3 (Fig. 2 A and B and SI Appendix, Fig. S2). Gzmc was uniquely expressed by ILC1s, whereas Gzmb was expressed in both ILC1s and NK cells. Gzm ILC1s expressed less perforin mRNA (Prf1) than NK cells, suggesting a lower cytotoxic capacity. However, Gzm ILC1s abundantly expressed the IL-21 receptor transcript (Il21r), which has been shown to enhance cytotoxicity (28). Thus, ILC1 lytic ability may depend on the levels of IL-21 in the tissue. IFNγ ILC1s were discriminated by high expression of Ifng, Xcl1, Ltb, as well as Il7r, Il18r1, and Cd226 (Fig. 2 A and B and SI Appendix, Fig. S2). The Rorc ILC1 cluster expressed some ILC3 signature genes, including Rorc, Rora, Maf, Tmem176a, Tmem176b, Lta, and Ltb. Despite this signature, Rorc ILC1s expressed no Il17 or Il22, while Ifng levels were similar to the other ILC1 clusters (Fig. 2 A and B and SI Appendix, Fig. S2). Thus, Rorc ILC1s may encompass ILC1s that originate from conversion of Rorγt+ ILC3s, commonly known as “exILC3s” (29), or from a Rorγt+ progenitor.
Fig. 2.
scRNA-seq delineates previously unappreciated ILC1 and NK cell subpopulations. (A) UMAP plot of 17,660 CD3−NK1.1+ cells from multiple tissues. Each cluster was annotated based on differentially expressed genes. (B) UMAP plots of representative selected genes that discriminate among the identified clusters. (C) UMAP plots of CD3−NK1.1+ cells from the indicated tissues.
NK cells included three major clusters: immature NK (iNK) cells expressing Cd27, mature NK (mNK) cells expressing Itgam (for CD11b), and activated NK cells expressing various cytokines and chemokine genes and the transcription factors Egr3, Nr4a1, and Nr4a3 (Fig. 2B and SI Appendix, Fig. S2). iNK cells expressed Tcf7, indicating a program for proliferation and lymph node homing (30). Among mNK cell signature genes, Klf2, Zeb2, Irf8, S1pr5, Klrg1, Ly6c2, and Prf1 indicated programs for effector function and recirculation (31–34). The distinction between mNK1 and mNK2 clusters was based on slightly different expression levels of mNK signature genes. We also identified a “stressed” NK cell cluster based on high expression of the stress response genes Hspe1 and Hsp90ab1, as well as a cluster of proliferating NK cells expressing Mki67, Tuba1b, Stmn1, and Tubb5 (SI Appendix, Fig. S2). We also detected a small NK cell cluster enriched for the IFN-stimulated genes (Ifit1, Ifit3, Ifi209, Irf7, Iigp1, Ifi208, Isg20), which was termed “IFN-stimulated” NK cells (SI Appendix, Fig. S2). The representation of the ILC1 clusters was skewed in a tissue-specific manner. The liver included all major ILC1 subsets, Gzm ILC1s, IFNγ ILC1s, and Rorc ILC1s; salivary glands contained a relatively large population of ILC1-like NK cells; and IFNγ ILC1s and Rorc ILC1s predominated in the intestinal lamina propria (Fig. 2C). Together, these data show that although all ILC1s express Hobit, they include disparate subsets with specialized effector functions, which are differentially represented in various tissues.
Liver Gzm and IFNγ ILC1s Require Hobit.
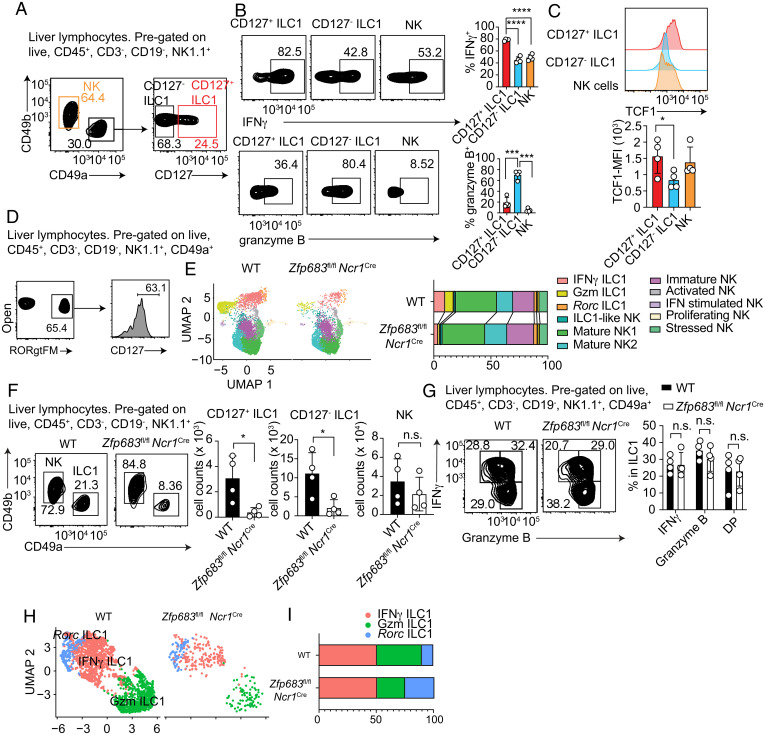
We sought to validate liver Gzm and IFNγ ILC1s by flow cytometry. We examined liver Lin–NK1.1+ for surface expression of CD49a, CD49b, and CD127 and intracellular content of IFNγ and granzyme B. CD49a+CD127+ ILC1s produced more IFNγ but less granzyme B than did CD49a+CD127– ILC1s (Fig. 3 A and B). Since TCF1 limits granzyme B expression in NK cells (35), we also analyzed intracellular levels of TCF1 in ILC1s. In line with elevated granzyme B production, CD49a+CD127– ILC1s expressed less TCF1 than did CD49a+CD127+ ILC1s (Fig. 3C). We conclude that liver ILC1s include a CD127+TCF1hi subset specialized in IFNγ production and a CD127–TCF1lo subset specialized in granzyme production. To validate liver Rorc ILC1s, we examined liver ILC1s in Rorc-Cre mice crossed with Rosa26RYFP mice (RorcFM), in which cells that express or have expressed Rorγt+ can be fate-mapped as YFP+. A considerable percentage of liver CD49a+ ILC1s were RorcFM+ (Fig. 3D), suggesting that they derive, at least in part, from the conversion of Rorγt+ ILC3s or from a Rorγt+ progenitor.
Fig. 3.
CD127 expression identifies at least two liver ILC1 subsets. (A) Representative flow cytometric gating strategy for liver NK cell and ILC1 subsets and expression of CD127 on CD49a+CD49b− ILC1s. (B) Representative flow cytometric dot plots and quantification of IFNγ and granzyme B expression in the indicated populations. It should be noted that liver NK cells include both iNK and mNK cells, which produce distinct amounts of IFNγ; therefore, the values of IFNγ depicted represent an average of the entire NK population. (C) Representative flow cytometric histogram plots and quantification of TCF1 protein expression in the indicated populations. (D) Representative flow cytometric dot plots showing expression of CD127 on RORγt fate-map (RORγtFM) liver CD49a+ ILC1s. (E) UMAP plots and cluster representation among CD3−NK1.1+ cells from wild-type or Zfp683fl/fl Ncr1Cre mice. (F) Representative flow cytometric dot plots and total cell counts of the indicated populations from wild-type or Zfp683fl/fl Ncr1Cremice. (G) Representative flow cytometric dot plots and quantification showing the expression of IFNγ and granzyme B in the indicated populations from wild-type or Zfp683fl/fl Ncr1Cre mice. (H and I) UMAP plots (H) and cluster proportion (I) of liver ILC1s from wild-type or Zfp683fl/fl Ncr1Cre mice. Data from A–D, F, and G are representative of two independent experiments (n = 4). Data represent mean ± SEM. *P < 0.05, ***P < 0.001, ****P < 0.0001; n.s., not significant. MFI, mean fluorescence intensity; WT, wild type.
Since it has been shown that Hobit is required for liver ILC1s (18), we asked whether IFNγ ILC1, Gzm ILC1, and Rorc ILC1 subsets depend on it. We generated Zfp683fl/fl mice and crossed them with Ncr1Cre mice, which express Cre recombinase in NK cells and ILC1s, thereby abrogating Hobit expression in both populations. scRNA-seq analysis of NK1.1+ cells from Zfp683fl/fl Ncr1Cre versus Zfp683fl/fl mice revealed a reduction of both Gzm and IFNγ ILC1s, whereas Rorc ILC1s seemed unaffected (Fig. 3E). Flow cytometric analysis corroborated a marked reduction of both CD127+ and CD127– ILC1s in Zfp683fl/fl Ncr1Cre mice, while NK cells were not significantly affected (Fig. 3F). The few remaining ILC1s present in Zfp683fl/fl Ncr1Cre mice maintained production of IFNγ and granzyme B, perhaps representing cells escaped from Cre recombination (Fig. 3G).
To further examine the impact of Zfp683 deletion in liver ILC1s, we reclustered ILC1s from our scRNA-seq data (Fig. 3 H and I and SI Appendix, Fig. S3). This analysis corroborated the presence of Gzm, IFNγ, and Rorc ILC1s and the strong reduction of Gzm ILC1 and IFNγ ILC1 clusters in the absence of Zfp683. However, we noticed a small relative increase of Rorc ILC1s in Zfp683fl/fl Ncr1Cre samples, suggesting that this subset may be less affected or unaffected by Zfp683 deletion. All ILC1 clusters expressed Zfp683 and Ifng. Rorc ILC1s shared expression of Kit, Il7r, and Tcf7 with IFNγ ILC1s, whereas Gzmb was preferentially expressed in Gzm ILC1s. Together, these data demonstrate that liver ILC1s include two functionally polarized ILC1 subsets that are Hobit-dependent and a third Hobit-independent subset that may derive from Rorγt+ ILC3s or from a Rorγt+ progenitor.
ILC1 Requirement for Hobit Varies in Different Tissues.
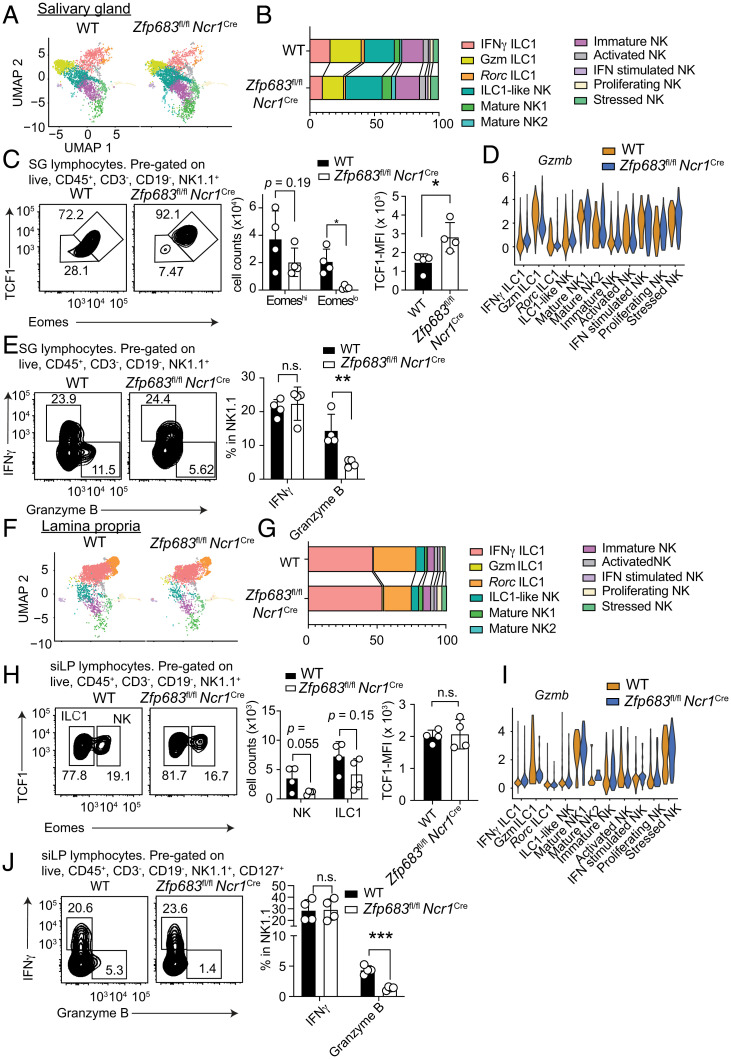
We further examined the impact of Hobit deficiency on ILC1s in salivary glands and intestinal lamina propria by scRNA-seq of Lin–NK1.1+ cells isolated from Zfp683fl/fl Ncr1Cre mice and control Zfp683fl/fl mice. Salivary glands harbored both Gzm and IFNγ ILC1s, as well as ILC1-like NK cells. Fewer Gzm and IFNγ ILC1s and more ILC1-like NK cells were present in Zfp683fl/fl Ncr1Cre mice than in Zfp683fl/fl mice, whereas NK cells were similarly represented in the two (Fig. 4 A and B). We confirmed this observation in Zfp683Red mice. Since the reporter cassette disrupts the Zfp683 allele, homozygous Zfp683Red mice lack functional Hobit-encoding genes; again, fewer ILC1s and more ILC1-like NK cells were found in the salivary glands of homozygous than in heterozygous Zfp683Red mice (SI Appendix, Fig. S4).
Fig. 4.
Hobit differentially regulates expression of granzyme B, TCF1, and Eomes in salivary gland and lamina propria ILC1s. (A and B) UMAP plots (A) and cluster representation (B) of salivary gland CD3−NK1.1+ cells from wild-type or Zfp683fl/fl Ncr1Cre mice. (C) Representative flow cytometric dot plots and quantification of Eomeshi/lo and TCF1 expression cells among NK1.1+ cells in salivary glands from wild-type or Zfp683fl/fl Ncr1Cre mice. (D) Violin plots of granzyme B expression levels in each cluster of salivary gland CD3−NK1.1+ cells. (E) Representative flow cytometric dot plots and quantification of IFNγ and granzyme B in the indicated NK1.1+ populations of salivary glands from wild-type or Zfp683fl/fl Ncr1Cre mice. (F and G) UMAP plots (F) and cluster representation (G) in lamina propria CD3−NK1.1+ cells from wild-type or Zfp683fl/fl Ncr1Cre mice. (H) Representative flow cytometric dot plots showing expression of Eomes and TCF1 and quantification of absolute numbers of ILC1s and NK cells and of TCF1 expression in the indicated populations of lamina propria NK1.1+ cells from wild-type or Zfp683fl/fl Ncr1Cre mice. (I) Violin plots of granzyme B expression levels in each cluster of intestinal lamina propria CD3−NK1.1+ cells. (J) Representative flow cytometric dot plots and quantification of IFNγ and granzyme B in the indicated lamina propria NK1.1+ populations from wild-type or Zfp683fl/fl Ncr1Cre mice. Data represent mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001; n.s., not significant. (C, E, H, and J) Data are representative of two independent experiments (n = 4). (E and J) NK cells and ILC1s were stimulated with IL-12 and IL-18. All flow cytometric plots were pregated with the indicated markers. Data represent mean ± SEM.
Since ILC1s are TCF1lo Eomes–, whereas ILC1-like NK cells are TCF1hi Eomes+ (Fig. 2), we further investigated the impact of Hobit deficiency on TCF1 and Eomes by flow cytometry. A reduction of Eomes– TCF1lo cells paralleled by an increase in Eomes+ TCF1hi ILC1-like NK cells was noted in the salivary glands of Zfp683fl/fl Ncr1Cre mice (Fig. 4C). These results suggest that Hobit negatively regulates expression of Eomes and TCF1 in salivary gland ILC1s and ILC1-like NK cells. We also examined the impact of Zfp683 deletion on effector functions: violin plots indicating the expression of Gzmb in each ILC1–NK subset showed a clear reduction of Gzmb in ILC1s from Zfp683fl/fl Ncr1Cre mice (Fig. 4D). Flow cytometric analysis validated that Zfp683 deletion led to reduced intracellular content of granzyme B, whereas IFNγ was unchanged (Fig. 4E).
scRNA-seq exposed no marked impact of Zfp683 deletion on the abundance of ILC1s in the Lin–NK1.1+ cells of the intestinal lamina propria (Fig. 4 F and G). Moreover, flow cytometric analysis corroborated that Zfp683 deletion had no obvious impact on Eomes or TCF1 expression (Fig. 4H). However, granzyme B mRNA and protein expression were significantly impaired in Zfp683-deficient lamina propria ILC1s, as observed in salivary glands (Fig. 4 I and J). Overall, these results demonstrate that the impact of Zfp683 deletion on the representation of ILC1 subsets and their expression of TCF1, Eomes, and granzyme B varies considerably depending on the tissue.
ILC1s Show Limited Conversion into ILC3s in Steady State.
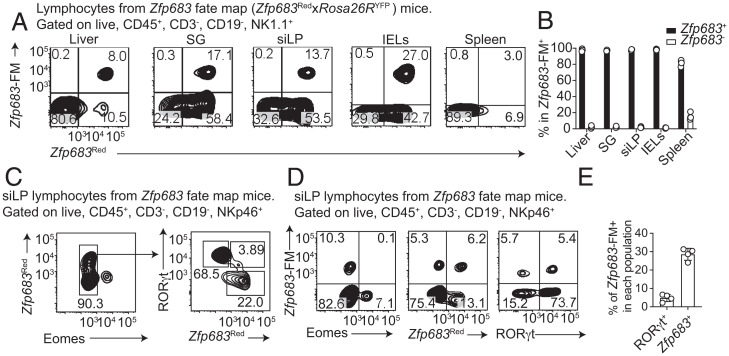
Several studies of ILC1–NK cell lineage relationships have proposed that ILC1s and NK cells are distinct lineages (14, 15, 17), although other reports disagree (16). Prior studies utilized either adoptive cell transfer or ex vivo culture systems but did not analyze lineage relationships in unperturbed physiological conditions. To overcome these limitations, we crossed Zfp683Red mice with Rosa26RYFP mice to generate Zfp683 fate-map (Zfp683FM) mice, in which cells that currently express Zfp683 are Tomato+, while cells that have expressed Zfp683 at any time point of their development are YFP+. We focused on Lin–NK1.1+ cells, which include ILC1s and NK cells. Analysis of Zfp683FM+ (YFP+) versus Zfp683+ (tdTomato+) Lin–NK1.1+ cells in various tissues revealed that Zfp683FM+ cells are almost exclusively detected within the Zfp683+ population, while rarely observed in Zfp683–NK1.1+ cells (Fig. 5 A and B). These data corroborate that ILC1s are a distinct lineage from NK cells. Of note, ∼30 to 50% of Zfp683+ ILC1s were Zfp683FM+, suggesting a partial efficiency in the recombination induced by Zfp683-driven Cre recombinase.
Fig. 5.
NK cells and ILC1s are independent lineages, but a small portion of ILC1s convert to ILC3s. (A and B) Representative flow cytometric dot plots (A) and quantification (B) of Hobit-expressing (Zfp683Red) versus Hobit fate-map (Zfp683FM) among NK1.1+ cells in different organs. Single-cell suspensions were obtained from heterozygous Zfp683Red mice. (C) Representative flow cytometric dot plots showing expression of Hobit (Zfp683Red), Eomes, and RORγt among NKp46+ cells of the small intestinal lamina propria, which include ILC1s, ILC3s, and NK cells. (D and E) Representative flow cytometric dot plots (D) and quantification (E) showing expression of Hobit fate-map (Zfp683FM) cells coexpressing RORγt or Zfp683. Data are representative of at least two independent experiments (n = 3 or 4). Data represent mean ± SEM.
Since it was shown that human ILC1s can convert into ILC3s when cultured with IL-12, IL-23, and IL-1β (36), we next investigated whether this conversion occurs in vivo in our reporter mice. We examined the small intestine lamina propria, in which ILC3s are particularly abundant. First, we assessed the intracellular content of Rorγt and Eomes in Lin–NKp46+ cells of Zfp683FM mice, which include ILC1s, NK cells, and NKp46+NK1.1– ILC3s. Zfp683+ (Tomato+) ILC1s, Eomes+ NK cells, and Rorγt+ ILC3s appeared as distinct populations (Fig. 5C). However, ∼5 to 10% of Rorγt+ ILC3s were Zfp683FM+ (YFP+) (Fig. 5 D and E), suggesting that a small percentage of ILC3s derive from ILC1s. None of the Eomes+ cells were Zfp683FM+, corroborating that ILC1s do not convert into NK cells. We conclude that in the small intestine only a limited percentage of ILC1s generate ILC3s, whereas ILC1s and NK cells remain distinct lineages.
NK Cells Express Hobit during MCMV Infection.
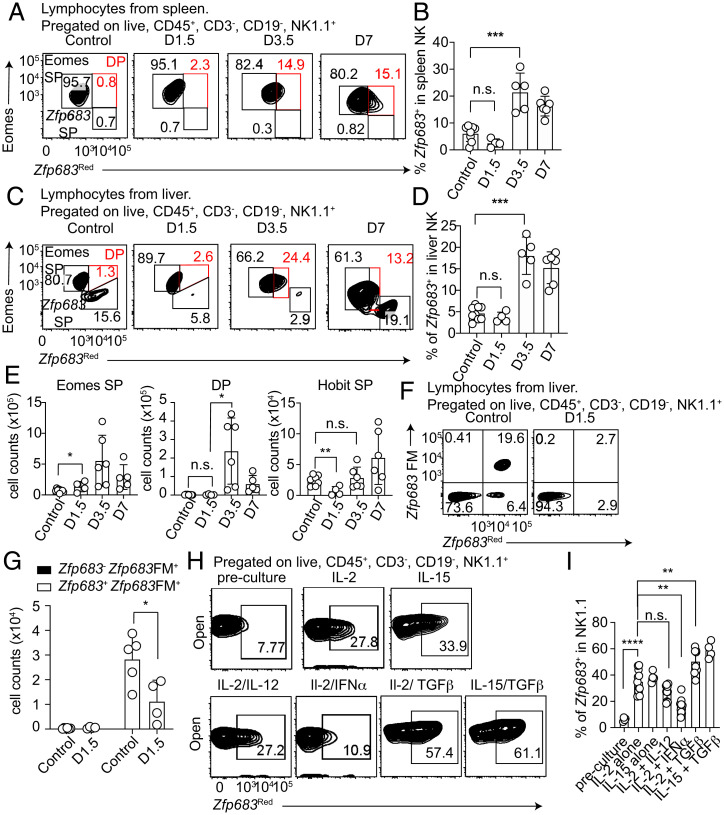
Previous studies indicated that NK cells acquire phenotypical features of ILC1s in the tumor microenvironment (37, 38) and during Toxoplasma gondii infection (39). In these studies, the ILC1-like phenotype was defined based on expression of CD49a, whereas Hobit expression was not assessed. We took advantage of our Zfp683Red mice to study the expression pattern of Hobit during MCMV infection in spleen and liver NK1.1+NKp46+ cells. Corroborating previous reports, CD49a was induced in spleen and liver NK cells as early as 1.5 d postinfection and continued to be expressed until 7 d postinfection (SI Appendix, Fig. S5). On day 1.5 postinfection, spleen and liver Eomes+ NK cells did not express Zfp683; however, Zfp683 appeared in a significant fraction (15 to 30%) of Eomes+ NK cells on day 3.5 postinfection and persisted until day 7 postinfection (Fig. 6 A–D). Thus, NK cells can express Zfp683 and CD49a during MCMV infection.
Fig. 6.
MCMV infection induces Hobit expression in NK cells. (A and B) Representative flow cytometric dot plots (A) and quantification (B) of Hobit-expressing (Zfp683Red) splenic NK cells at different time points after MCMV i.p. infection. (C and D) Representative dot plots of Hobit-expressing (Zfp683Red) and Eomes-expressing liver ILC1s and NK cells (C) and their quantification (D) at the indicated time points after MCMV i.p. infection in heterozygous Zfp683Red mice. (E) Absolute cell numbers of the indicated populations in the liver at the indicated time points post–MCMV i.p. infection. (F and G) Representative flow cytometric dot plots (F) and absolute cell counts (G) of Hobit-expressing (Zfp683Red) or Hobit fate map (Zfp683FM) among liver NK1.1+ cells at 1.5 d post–MCMV infection. (H and I) Representative flow cytometric dot plots (H) and quantification (I) of Hobit-expressing (Zfp683Red) in splenic NK1.1+ cells after 72 h in culture with the indicated cytokines. Data are representative of at least two independent experiments (n = 5 to 8). Data represent mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; n.s., not significant.
Consistent with previous studies (40), we observed that Zfp683+ ILC1s in the liver went through a transient contraction phase at day 1.5 post–MCMV infection and returned to baseline levels in absolute numbers by day 3.5, whereas the numbers of Eomes+ NK cells expanded throughout (Fig. 6E). To test whether the initial ILC1 contraction was due to conversion of ILC1s into NK cells, we examined MCMV infection in Zfp683FM mice. Zfp683FM+ cells were exclusively present in the Zfp683+ population at steady state and entirely disappeared on day 1.5 postinfection (Fig. 6 F and G), indicating that ILC1s remain distinct from NK cells also during MCMV infection.
To identify the cytokines that may control Hobit expression in NK cells, we isolated spleen NK cells from Zfp683Red mice and cultured them with IL-2, IL-15, IL-12, transforming growth factor-beta (TGFβ), and IFNα, either alone or in different combinations. After 72 h, Zfp683 was induced by IL-2 or IL-15 alone, and expression was further enhanced by TGFβ (Fig. 6 H and I). On the contrary, IFNα inhibited IL-2–mediated up-regulation of Zfp683 expression. Although IL-12 is a proinflammatory cytokine that activates NK cells, it had no impact on Zfp683 expression. Thus, while Zfp683 is a faithful ILC1 marker in homeostasis, it is induced in NK cells in pathological conditions that generate an environment rich in cytokines such as IL-15, IL-2, and TGFβ.
It was previously shown that Hobit+ ILC1s provide a very early source of IFNγ for the control of MCMV in the liver 1 d after mice have been infected by hydrodynamic or intraperitoneal (i.p.) injection (8). Given that Hobit is also expressed in spleen and liver NK cells 3.5 d after infection, we wanted to evaluate whether Hobit deficiency impacts control of viral infection at a later time point when NK cells expand and become involved (41). Zfp683fl/fl Ncr1Cre and control mice were infected i.p. with MCMV and viral titers were assessed in the liver and spleen at day 7. No major differences in viral titers were detected at this time point (SI Appendix, Fig. S5). We conclude that NK cell control of MCMV infection is Hobit-independent.
Discussion
Hobit is a transcriptional repressor encoded by Zfp683 that has been proposed as a master transcription factor for tissue-resident memory T cells and ILC1s (18, 42). Using Zfp683Red mice, we showed that the association of Hobit with ILC1s is tissue- and context-dependent. In liver and intestinal mucosa, Zfp683 expression correlated well with ILC1s; however, Zfp683 was coexpressed with Eomes in an ILC1-like NK cell subset uniquely present in salivary glands. Using Zfp683FM mice, we corroborated that ILC1s and NK cells are distinct lineages in the steady state. However, Zfp683 was induced in spleen and liver Eomes+ NK cells during MCMV infection and was up-regulated in vitro by cytokines, such as IL-15 and TGFβ that have been shown to induce markers of tissue residency (43). The impact of Hobit on ILC1s and NK cells was also tissue- and context-dependent. Comparing Zfp683fl/fl mice and Zfp683fl/fl Ncr1Cre mice exposed the variable impacts of Zfp683 deletion: a drastic reduction of Gzm ILC1s and IFNγ ILC1s in the liver; fewer ILC1s and more Eomes+ ILC1-like NK cells, along with decreased granzyme B production by ILC1s, in the salivary glands; and, solely, the reduction of granzyme B+ ILC1s in the small intestine lamina propria. Zfp683 deletion did not affect the ability of NK cells to control MCMV infection in the liver at late time points, when NK cells are the major anti-MCMV effectors (41). We conclude that Hobit predominantly defines ILC1 programs and numbers, whereas it is less impactful when coexpressed with Eomes in ILC1-like NK cells and conventional NK cells.
A recently published study by Friedrich et al. showed that liver ILC1s encompassed a CD127+TCF1hi IFNγ+ subset and a CD127–TCF1lo cytotoxic subset and noted that germline Zfp683 deletion increased the representation of the former versus the latter, indicating that CD127+TCF1hi IFNγ+ ILC1s are the progenitors of CD127–TCF1lo cytotoxic ILC1s and that Hobit is required for this transition (44). While our study identifies similar subsets of liver ILC1s, conditional deletion of Zfp683 in the ILC1–NK lineage markedly reduced both subsets. Thus, our results are more consistent with the possibility that both IFNγ and Gzm ILC1 subsets develop from another progenitor through a Hobit-dependent process. The reason for the discrepancy between the two studies remains unclear. It should be noted that Friedrich et al. divided liver CD127±TCF1hi ILC1s into cKit+ and cKit– subsets and indicated that the cKit– subset was predominantly diminished by Hobit deficiency, while the cKit+ subset was unchanged. This latter subset may overlap with the Rorc ILC1 subset we detected in liver, which was unaffected by lack of Hobit and, in fact, relatively expanded under these conditions. Whether this Rorc ILC1 subset is a precursor developmentally related to other ILC1s, corresponds to a liver ILC1 progenitor recently identified (45), or encompasses exILC3s (29) remains to be investigated.
Hobit is a transcriptional repressor that inhibits the expression of molecules, such as the sphingosine-1-phosphate receptor, CCR7, and L-selectin, that mediate lymphocyte recirculation, facilitating the retention of a pool of memory T cells in tissues (46). Our study shows that Hobit is required for the presence of ILC1s in liver and, to a lesser extent, salivary glands. It remains unknown whether Hobit is necessary for the development of ILC1s or their retention in the tissue after they have developed. In the former case, Hobit may inhibit Eomes expression, directing a Tbet+ progenitor toward an ILC1 fate. In the latter case, one would expect ILC1s to be released from liver and salivary glands into the bloodstream in Hobit-deficient mice. In vivo imaging of tissue ILC1s will be essential to detect their movement in and out of tissues.
Beyond its function in tissue homing and retention, our study demonstrates that Hobit promotes expression of granzyme B while restraining expression of Eomes and TCF1. These effects are likely to be linked. It has been shown that TCF1 promotes Eomes expression in CD8 T cells (47). Moreover, TCF1 curbs the activity of a granzyme B–associated regulatory element, limiting granzyme B expression in TCF1-expressing NK cells (35). Thus, Hobit may directly inhibit expression of TCF1, causing reduced expression of Eomes and increased expression of granzyme B. This model is reminiscent of a similar role of Blimp, a Hobit homolog, in repressing TCF1 in NK cells (30). Thus, Hobit and Blimp may have parallel functions in controlling functional maturation of ILC1s and NK cells, respectively. Future epigenetic studies will be necessary to validate the presence of regulatory elements in the TCF1 region that are bound and repressed by Hobit. We do not exclude that Eomes and Hobit may also cross-regulate each other, as recently reported in CD8 T cells (48).
Although ILC1s have long been considered noncytotoxic, our study shows that ILC1s express granzymes A, B, and C. Granzyme C seems to be uniquely expressed in ILC1s, whereas granzymes A and B are also present in cytotoxic NK cells. Since granzyme C is induced by TGFβ (37), its expression may reflect ILC1 location in TGFβ-rich tissue niches. ILC1s also expressed perforin mRNA, but to lower levels than NK cells, perhaps reflecting a lower cytotoxic capacity. However, Gzm ILC1s highly expressed the receptor for IL-21, a cytokine known to enhance NK cell cytotoxicity and inhibit NK cell survival and expansion (28). Thus, IL-21 may increase the lytic capacity of ILC1s while restraining their lifespan. ILC1s have previously been shown to provide an immediate source of IFNγ against MCMV infection in the liver upon hydrodynamic injection, which allows preferential delivery of virus to the liver (8). Although we found that Hobit expression was also induced in spleen and liver NK cells a few days after MCMV infection, Hobit deficiency had no effect on NK cell numbers or their ability to control MCMV infection in our model. It will be important to see whether Hobit impacts ILC1 or NK cell functions in other contexts, such as tumor growth and bacterial infections, or affects long-term memory of NK cells and ILC1s (49, 50).
ILCs have exhibited a certain degree of plasticity (1, 51, 52). Conversion of ILC3s into ILC1s has been shown both in vitro and in vivo using Rorγt fate-map mice (29, 53, 54). The reverse conversion from ILC1s to ILC3s has also been reported (36), although not formally demonstrated in an unperturbed system. Our analysis of Zfp683FM mice demonstrates that some ILC3s, although Zfp683–, have expressed Zfp683 at a certain point, suggesting that they derive from ILC1s or from a Zfp683-expressing progenitor. In contrast, we saw no evidence for ILC1s converting into NK cells. These results corroborate the plasticity of ILCs and reaffirm ILC1s as a lineage distinct from NK cells.
Materials and Methods
Experimental details on animals, cell extraction from tissues, antibody staining for flow cytometry and sorting, cell culture and stimulations, scRNA-seq analyses, MCMV infection, and statistical analyses for this study are described in detail in SI Appendix, Materials and Methods.
All animal studies were approved by the Washington University Institutional Animal Care and Use Committee.
Supplementary Material
Acknowledgments
We thank Richard M. Locksley for targeting vectors, and the Genome Technology Access Center at the McDonnel Genome Institute at Washington University for scRNA-seq on the 10X platform. The Genome Technology Access Center is supported in part by NCI Cancer Center Support Grant #P30 CA91842 to the Siteman Cancer Center at Washington University School of Medicine in St. Louis, MO and Grant# UL1TR002345 from the National Center for Research Resources (NCRR) to the Institute of Clinical and Translational Sciences (ICTS) at Washington University in St. Louis, MO. We thank E. Lantelme and D. Brinja and the Pathology and Immunology Flow Cytometry Core for cell sorting. This work was supported by Grants U01 AI095542, R01 DE025884, R01 AI134035, R01 DK124699, and U19 AI142733 (to M. Colonna) and NIH R01 AI130152 (to T.E.). K.Y. was supported by the Rheumatology Research Foundation Tobé and Stephen E. Malawista, MD Endowment in Academic Rheumatology and by Child Health Research Center K12.
Footnotes
Reviewers: B.P., Sveuciliste u Rijeci Zavod za Histologiju i Embriologiju; and C.R., Deutsches Rheuma-Forschungszentrum.
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2117965118/-/DCSupplemental.
Data Availability
All scRNA-seq data reported in this paper have been deposited in the Gene Expression Omnibus (accession no. GSE185346).
All study data are included in the article and/or SI Appendix.
Change History
January 22, 2022: The byline has been updated.
References
- 1.Colonna M., Innate lymphoid cells: Diversity, plasticity, and unique functions in immunity. Immunity 48, 1104–1117 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vivier E., et al. , Innate lymphoid cells: 10 years on. Cell 174, 1054–1066 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Sonnenberg G. F., Hepworth M. R., Functional interactions between innate lymphoid cells and adaptive immunity. Nat. Rev. Immunol. 19, 599–613 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riggan L., Freud A. G., O’Sullivan T. E., True detective: Unraveling group 1 innate lymphocyte heterogeneity. Trends Immunol. 40, 909–921 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stokic-Trtica V., Diefenbach A., Klose C. S. N., NK cell development in times of innate lymphoid cell diversity. Front. Immunol. 11, 813 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seillet C., Brossay L., Vivier E., Natural killers or ILC1s? That is the question. Curr. Opin. Immunol. 68, 48–53 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gasteiger G., Fan X., Dikiy S., Lee S. Y., Rudensky A. Y., Tissue residency of innate lymphoid cells in lymphoid and nonlymphoid organs. Science 350, 981–985 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weizman O. E., et al. , ILC1 confer early host protection at initial sites of viral infection. Cell 171, 795–808.e12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dadi S., et al. , Cancer immunosurveillance by tissue-resident innate lymphoid cells and innate-like T cells. Cell 164, 365–377 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Censo C., et al. , Granzyme A and CD160 expression delineates ILC1 with graded functions in the mouse liver. Eur. J. Immunol. 51, 2568–2575 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krabbendam L., Bernink J. H., Spits H., Innate lymphoid cells: From helper to killer. Curr. Opin. Immunol. 68, 28–33 (2021). [DOI] [PubMed] [Google Scholar]
- 12.Rahim M. M., Makrigiannis A. P., Ly49 receptors: Evolution, genetic diversity, and impact on immunity. Immunol. Rev. 267, 137–147 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Kirkham C. L., Carlyle J. R., Complexity and diversity of the NKR-P1:Clr (Klrb1:Clec2) recognition systems. Front. Immunol. 5, 214 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Constantinides M. G., McDonald B. D., Verhoef P. A., Bendelac A., A committed precursor to innate lymphoid cells. Nature 508, 397–401 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klose C. S. N., et al. , Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell 157, 340–356 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Xu W., et al. , An Id2RFP-reporter mouse redefines innate lymphoid cell precursor potentials. Immunity 50, 1054–1068.e3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J., et al. , T-bet and Eomes govern differentiation and function of mouse and human NK cells and ILC1. Eur. J. Immunol. 48, 738–750 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Mackay L. K., et al. , Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science 352, 459–463 (2016). [DOI] [PubMed] [Google Scholar]
- 19.McFarland A. P., et al. , Multi-tissue single-cell analysis deconstructs the complex programs of mouse natural killer and type 1 innate lymphoid cells in tissues and circulation. Immunity 54, 1320–1337.e4 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cortez V. S., Fuchs A., Cella M., Gilfillan S., Colonna M., Cutting edge: Salivary gland NK cells develop independently of Nfil3 in steady-state. J. Immunol. 192, 4487–4491 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Sojka D. K., et al. , Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. eLife 3, e01659 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Redhead M. L., et al. , The transcription factor NFIL3 is essential for normal placental and embryonic development but not for uterine natural killer (UNK) cell differentiation in mice. Biol. Reprod. 94, 101 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Gascoyne D. M., et al. , The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat. Immunol. 10, 1118–1124 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Kamizono S., et al. , Nfil3/E4bp4 is required for the development and maturation of NK cells in vivo. J. Exp. Med. 206, 2977–2986 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seillet C., et al. , Nfil3 is required for the development of all innate lymphoid cell subsets. J. Exp. Med. 211, 1733–1740 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geiger T. L., et al. , Nfil3 is crucial for development of innate lymphoid cells and host protection against intestinal pathogens. J. Exp. Med. 211, 1723–1731 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu X., et al. , The basic leucine zipper transcription factor NFIL3 directs the development of a common innate lymphoid cell precursor. eLife 3, e04406 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kasaian M. T., et al. , IL-21 limits NK cell responses and promotes antigen-specific T cell activation: A mediator of the transition from innate to adaptive immunity. Immunity 16, 559–569 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Klose C. S., et al. , A T-bet gradient controls the fate and function of CCR6-RORγt+ innate lymphoid cells. Nature 494, 261–265 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Collins P. L., et al. , Gene regulatory programs conferring phenotypic identities to human NK cells. Cell 176, 348–360.e12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skon C. N., et al. , Transcriptional downregulation of S1pr1 is required for the establishment of resident memory CD8+ T cells. Nat. Immunol. 14, 1285–1293 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Helden M. J., et al. , Terminal NK cell maturation is controlled by concerted actions of T-bet and Zeb2 and is essential for melanoma rejection. J. Exp. Med. 212, 2015–2025 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adams N. M., et al. , Transcription factor IRF8 orchestrates the adaptive natural killer cell response. Immunity 48, 1172–1182.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mace E. M., et al. , Biallelic mutations in IRF8 impair human NK cell maturation and function. J. Clin. Invest. 127, 306–320 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeevan-Raj B., et al. , The transcription factor Tcf1 contributes to normal NK cell development and function by limiting the expression of granzymes. Cell Rep. 20, 613–626 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Bernink J. H., et al. , Interleukin-12 and -23 control plasticity of CD127(+) group 1 and group 3 innate lymphoid cells in the intestinal lamina propria. Immunity 43, 146–160 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Cortez V. S., et al. , SMAD4 impedes the conversion of NK cells into ILC1-like cells by curtailing non-canonical TGF-β signaling. Nat. Immunol. 18, 995–1003 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao Y., et al. , Tumor immunoevasion by the conversion of effector NK cells into type 1 innate lymphoid cells. Nat. Immunol. 18, 1004–1015 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Park E., et al. , Toxoplasma gondii infection drives conversion of NK cells into ILC1-like cells. eLife 8, e47605 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dodard G., et al. , Inflammation-induced lactate leads to rapid loss of hepatic tissue-resident NK cells. Cell Rep. 32, 107855 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lam V. C., Lanier L. L., NK cells in host responses to viral infections. Curr. Opin. Immunol. 44, 43–51 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zundler S., et al. , Hobit- and Blimp-1-driven CD4+ tissue-resident memory T cells control chronic intestinal inflammation. Nat. Immunol. 20, 288–300 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Mackay L. K., et al. , T-box transcription factors combine with the cytokines TGF-β and IL-15 to control tissue-resident memory T cell fate. Immunity 43, 1101–1111 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Friedrich C., et al. , Effector differentiation downstream of lineage commitment in ILC1s is driven by Hobit across tissues. Nat. Immunol. 22, 1256–1267 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bai L., et al. , Liver type 1 innate lymphoid cells develop locally via an interferon-γ-dependent loop. Science 371, eaba4177 (2021). [DOI] [PubMed] [Google Scholar]
- 46.Masopust D., Soerens A. G., Tissue-resident T cells and other resident leukocytes. Annu. Rev. Immunol. 37, 521–546 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen Z., et al. , TCF-1-centered transcriptional network drives an effector versus exhausted CD8 T cell-fate decision. Immunity 51, 840–855.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parga-Vidal L., et al. , Hobit identifies tissue-resident memory T cell precursors that are regulated by Eomes. Sci. Immunol. 6, eabg3533 (2021). [DOI] [PubMed] [Google Scholar]
- 49.Mujal A. M., Delconte R. B., Sun J. C., Natural killer cells: From innate to adaptive features. Annu. Rev. Immunol. 39, 417–447 (2021). [DOI] [PubMed] [Google Scholar]
- 50.Weizman O. E., et al. , Mouse cytomegalovirus-experienced ILC1s acquire a memory response dependent on the viral glycoprotein m12. Nat. Immunol. 20, 1004–1011 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bal S. M., Golebski K., Spits H., Plasticity of innate lymphoid cell subsets. Nat. Rev. Immunol. 20, 552–565 (2020). [DOI] [PubMed] [Google Scholar]
- 52.Bielecki P., et al. , Skin-resident innate lymphoid cells converge on a pathogenic effector state. Nature 592, 128–132 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cella M., Otero K., Colonna M., Expansion of human NK-22 cells with IL-7, IL-2, and IL-1beta reveals intrinsic functional plasticity. Proc. Natl. Acad. Sci. U.S.A. 107, 10961–10966 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cella M., et al. , Subsets of ILC3-ILC1-like cells generate a diversity spectrum of innate lymphoid cells in human mucosal tissues. Nat. Immunol. 20, 980–991 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All scRNA-seq data reported in this paper have been deposited in the Gene Expression Omnibus (accession no. GSE185346).
All study data are included in the article and/or SI Appendix.