Highlights
-
•
EWS/FLI is the driver oncogene of Ewing sarcoma.
-
•
EWS-FLI driven transcription yields numerous therapeutic vulnerabilities.
-
•
The intrinsic cellular stress of dysregulated transcription leads to additional therapeutic vulnerabilities.
-
•
A number of compounds targeting various steps in EWS/FLI driven transcription are in various stages of clinical development including those that are now in the clinic.
-
•
The future of EWS/FLI targeting will likely focus on molecularly targeted combination therapies.
Keywords: Ewing sarcoma, EWS/FLI, Transcription factor targeting, Molecular pharmacology
Abstract
EWS/FLI is the defining mutation of Ewing sarcoma. This oncogene drives malignant transformation and progression and occurs in a genetic background characterized by few other recurrent cooperating mutations. In addition, the tumor is absolutely dependent on the continued expression of EWS/FLI to maintain the malignant phenotype. However, EWS/FLI is a transcription factor and therefore a challenging drug target. The difficulty of directly targeting EWS/FLI stems from unique features of this fusion protein as well as the network of interacting proteins required to execute the transcriptional program. This network includes interacting proteins as well as upstream and downstream effectors that together reprogram the epigenome and transcriptome. While the vast number of proteins involved in this process challenge the development of a highly specific inhibitors, they also yield numerous therapeutic opportunities. In this report, we will review how this vast EWS-FLI transcriptional network has been exploited over the last two decades to identify compounds that directly target EWS/FLI and/or associated vulnerabilities.
1. Introduction
1.1. Ewing sarcoma
Ewing sarcoma is a primary bone and soft tissue tumor with a peak incidence in patients under the age of twenty [1]. Current treatment for this disease consists of surgery and/or radiation in combination with a regimen of five different chemotherapeutic agents administered on an interval compressed schedule [2]. While this approach has substantially improved survival for patients with localized tumors, the regimen is associated with significant short and long-term side effects [3]. Further, patients with relapsed and metastatic disease continue to show poor overall survival of 15–25% and 20–30%, respectively [4]. Therefore, there is a clear need for more effective and less toxic therapies for patients with Ewing sarcoma.
1.2. The rationale for EWS/FLI targeted therapies
One promising therapeutic strategy for Ewing sarcoma is to develop a molecularly targeted therapy focused on the defining lesion of the tumor, the EWS/FLI transcription factor. EWS/FLI (or another FET/ETS fusion protein) is found in every case of Ewing sarcoma. In addition, it is the only recurrent somatic mutation occurring in more than 20% of patients [5], [6], [7], [8]. EWS/FLI is generated by either a reciprocal translocation [9] or complex chromosomal rearrangement [10] leading to the fusion of the N-terminus of the FET RNA-binding protein EWSR1 to the C-terminus of the ETS transcription factor FLI1. Most commonly, the protein joins Exon 7 of EWSR1 to Exon 6 of FLI1 (Type 1, 60%) or Exon 7 of EWSR1 to Exon 5 of FLI1 (Type II; 25%) [11], [12], [13], [14]. Alternative fusions are found with the EWSR1 family member FUS and other ETS transcription factors including ETV1, ETV4, ERG and FEV with the highest percentage being ERG occurring in 5–10% of Ewing sarcoma patients [15], [16], [17], [18], [19]. Since over 80% of the tumors (and the majority of preclinical models) are driven by EWS/FLI, the majority of therapeutic efforts have focused on EWS/FLI. Nevertheless, the vast majority of physicians/scientists in the community believe that consideration of fusion type, subtype and partner is therapeutically important and should be collected prospectively in future clinical studies [20].
EWS/FLI is a powerful oncogenic driver that, through a series of complex mechanisms involving several binding partners, binds both high affinity GGAA sequences and GGAA repeat regions on DNA to establish de novo enhancers that drive tumor formation and progression [21], [22], [23], [24], [25], [26]. Indeed, genes induced by EWS/FLI drive proliferation, DNA damage response genes and alter the cell cycle, while repressed genes maintain a block in proliferation and inhibit cell communication [27], [28], [29], [30], [31], [32]. Furthermore, more than 25 years ago Kovar et al. used antisense DNA technology to demonstrate a dependence of this tumor on the continued expression of EWS/FLI [33]. This dependence on EWS/FLI was subsequently confirmed in multiple independent studies using a variety of molecular techniques including siRNA, shRNA and CRISPR/Cas9 both in vitro and in vivo [34], [35], [36]. Therefore, a therapy targeting EWS/FLI should cause a loss of proliferation, reversal of the oncogenic phenotype and lead to clinical responses. Since EWS/FLI is not found in normal cells, highly specific targeting of EWS/FLI could have a wide therapeutic window. Unfortunately, EWS/FLI is a transcription factor and therefore a challenging drug target.
1.3. The challenge of targeting EWS/FLI
Transcription is a highly regulated process involving several distinct steps and protein interactions. EWS/FLI influences many of these well-orchestrated steps. For many genes, RNA polymerase II is paused at the proximal promoter only producing abortive mRNA transcripts until its C-terminal tail is hyperphosphorylated [37]. This pause allows the polymerase to recruit splicing factors involved in RNA processing. Wild-type EWSR1 is known to interact with hyperphosphorylated RNA polymerase II, splicing factors, hnRNPs, RNA helicases, snRNPs, and mRNA to regulate splicing in the presence of DNA damage [38], [39], [40], [41]. EWSR1 also inhibits CDK9 mediated phosphorylation of RNA polymerase II [32]. EWS/FLI maintains the ability to bind both transcriptionally processive RNA polymerase II and splicing factors, first shown with the splicing factor U1C [42]. In addition, it serves to modulate the interactions of wild-type EWSR1 [32]. Therefore, the presence of the fusion protein directly impacts both transcription and RNA processing by numerous mechanisms leading to alterations in expression of individual genes and alternate exon usage as seen with TERT and CCND1 [43], [44].
Unfortunately, drugging this complex process of transcription is a challenge. Transcription factors like EWS/FLI typically have a transactivation domain and a DNA-binding domain. Many transactivation domains are highly dynamic molecular structures known as intrinsically disordered proteins that can collapse or expand their three-dimensional structure in response to solvent interactions, binding of other biological molecules, or post-translational modification [45]. The plasticity of intrinsically disordered proteins enables them to coordinate complex interactions among multiple binding partners to regulate intricate biological processes like transcription and chromatin remodeling. The N-terminus of EWS/FLI contains an intrinsically disordered region that can form ternary complexes with many proteins including but not limited to BARD1, AP-1, and RNAPII [46], [47], [48], [49], [50]. The EWS/FLI N-terminus also interacts with other copies of itself to change conformation in a manner reminiscent to the PrPC → PrPSc transition that drives spongiform encephalopathies [51]. This phase transition within the “prion-like” domain of EWS/FLI directly binds and recruits the SWI/SNF chromatin remodeling complex to DNA binding sites to generate de novo enhancer regions but challenges the development of highly specific small molecule inhibitors [21], [24].
The C-terminus of EWS/FLI is a DNA binding domain that recognizes a GGAA core DNA sequence with a wing-turn-helix structure common among ETS transcriptions factors [21], [52]. The FLI1 DNA binding domain binds DNA as both a monomer and dimer in vitro although the biological relevance of dimerization is unclear because full-length wild-type FLI1 has not been observed to dimerize [53], [54]. In either stoichiometry, the DNA binding domain of FLI1 lacks a concavity amenable to small molecule binding making drug development difficult. In short, direct therapeutic targeting of EWS/FLI is challenging because the N-terminus of EWS/FLI is intrinsically disordered and the active site of the C-terminus lacks an obvious druggable pocket.
In summary, the complex process of transcription coupled with structural features of EWS/FLI challenges the development of highly specific small molecule inhibitors of EWS/FLI. Nevertheless, because of the absolute dependence of this tumor on EWS/FLI, several approaches have been proposed to target the transcriptome, interactome. spliceosome or upstream/downstream signaling.
2. Targeting the EWS/FLI interactome and splicing
RNA processing is tightly linked to transcription. Several studies link EWS/FLI to altered exon usage either directly or indirectly as a dominant negative inhibitor of wild-type EWSR1 as has been shown for TASR-1, TASR-2, FAS, and YBX1 [41], [45], [46], [47]. Therefore, some groups have attempted to directly target the EWS/FLI interactome to modulate splicing or to target the splicing of EWS/FLI itself.
A series of studies of the interactome converged on a surface plasmon resonance screen that identified the compound YK-4-279 as an EWS/FLI binding compound that inhibits the interaction between EWS/FLI and RNA helicase A (DHX9) [48], [55]. Interruption of this interaction showed activity both in vitro and in vivo and reversed EWS/FLI mediated alternative splicing of several important targets such as ARID1A [26]. An analog of YK-4-279, TK-216, is currently undergoing clinical testing (NCT02657005).
An alternative approach to target the EWS/FLI interactome led to the identification of an EWS/FLI targeted peptide. Jully et al. used computer modeling to determine that the junction region of the most common fusion form of EWS/FLI forms a putative ligand binding site [56]. They next generated a peptide that was further modified by incorporating an HIV TAT cell penetrating protein and a nuclear localization signal [57]. This CIEWSPEP protein was readily taken up by EWS502 cells, decreased Ewing sarcoma cell proliferation, led to apoptosis, decreased Matrigel invasion, and reversed EWS/FLI driven gene expression [56]. However, although this system is both elegant and useful in the in vitro setting, it’s translation to patients is challenged by bioavailability and intracellular proteolysis [58].
Finally, the splicing of EWS/FLI itself has been identified as a possible therapeutic vulnerability [59]. A subset of tumors has a cryptic exon near the breakpoint of EWSR1 and FLI1 that requires the protein HNRNPH1 for proper processing into the mature EWS/FLI transcript [60]. HNRNPH1 is an hnRNP that can bind the G-quadraplex formed by exon 8 of the type 1 fusion form of EWS/FLI. This has been targeted with the G-quadruplex stabilizer pyridostatin to limit colony formation of Ewing sarcoma cell lines [61]. Alternatively, proper mRNA processing requires U2 snRNP complex formation which can be disrupted with Pladienolide B to induce mis-splicing, loss of expression of EWS/FLI and a loss of Ewing sarcoma cell viability in tumors with this cryptic exon [60].
3. Oligonucleotide based approaches targeting EWS/FLI
The mRNA of EWS/FLI has also been directly targeted by numerous investigators using oligonucleotide-based approaches. Indeed, many of the pivotal studies that have characterized the biology of EWS/FLI have used nucleic acid approaches to target EWS/FLI mRNA. Antisense oligodeoxynucleotides (oligos) bind complementary mRNA targets to block translation into protein and induce degradation by ribonuclease H [62]. Early studies with these molecules in Ewing sarcoma showed that oligos to either the junction region of EWS/FLI or the start of the coding region effectively led to loss of EWS/FLI mRNA expression and cell proliferation [63], [64], [65]. It was later demonstrated that direct injection of Ewing sarcoma tumors with EWS/FLI oligos in xenografts also leads to a decrease in tumor volume which could be further augmented by utilizing nanoparticle delivery systems or combining them with rapamycin [66], [67], [68], [69], [70], [71].
EWS/FLI silencing has also been achieved using small interfering RNA (siRNA). Gene knockdown of EWS/FLI using siRNA results in decreased Ewing sarcoma cell viability, limits invasive potential, and prevents tumor engraftment in a metastatic xenograft model [72], [73], [74]. Furthermore, the precise control of gene knockdown provided by siRNA has allowed researchers to delineate important relationships between EWS/FLI and downstream targets such as MYC, IGFBP3, NR0B1, STEAP1, CALCB, and several neural crest specific genes [72], [75], [76], [77], [78], [79]. The creation of nanoparticles containing these targeted oligonucleotides remains an ongoing area of investigation [80], [81]. In addition, one such construct is currently being evaluated in the clinic (NCT02736565). Unfortunately, both oligos and siRNA are limited by their half-lives and require repeated administration to achieve and sustain EWS/FLI suppression [62].
Finally, these oligonucleotide-based approaches have been used to evaluate the role EWS/FLI plays in metastasis. Some preclinical studies have demonstrated that silencing EWS/FLI leads to a loss of proliferation but increases cellular migration, particularly in A673 cells [82]. Consistent with this idea, transient suppression of EWS/FLI in A673 cells followed by tail vein injection leads to an increase in the number of mice with lung nodules (although it is notable that the control mice also formed lung tumors in these experiments) [83]. It has been suggested that these effects are driven by EWS/FLI mediated antagonism of WNT, STAG2 or YAP/TAZ signaling [82], [83], [84], [85], [86], [87]. This leads to a model where transient suppression of EWS/FLI allows the cell to become more migratory at the expense of proliferation, second-site colonization and anoikis. With recovery and restoration of EWS/FLI activity the cells regain proliferative potential and second site colonization. However, there are a number of studies that disagree with this model and suggest that suppression of EWS/FLI inhibits metastasis to bone and lung. These studies show that EWS/FLI repression leads to a loss of specific downstream targets that are well-established metastatic effectors such VEGF and CAV1/MMP9 [88], [89]. Importantly this suppression of metastasis with EWS/FLI suppression has been seen in multiple studies and has been linked to suppression of a number of EWS/FLI target genes including CAV1 [90], STEAP1 [91], CHM1 [92], TNC [93], NPY1R [94], PPP1R1A [95] and EZH2 [96]. Therefore, more work needs to be done to understand the role of EWS/FLI in the very complicated metastatic cascade.
4. Therapeutic approaches that target EWS/FLI expression
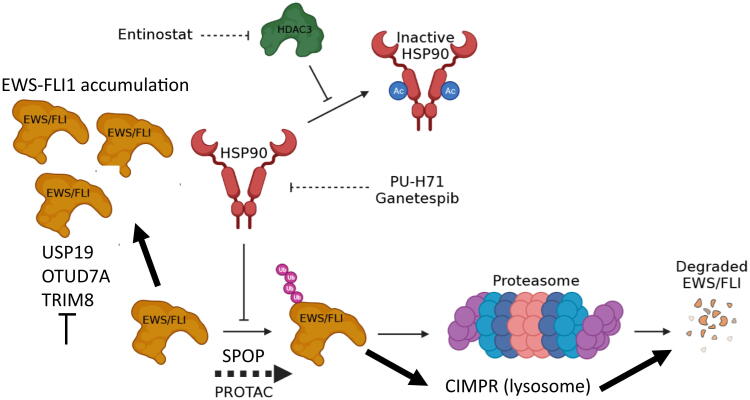
Another approach to target EWS/FLI is to modulate the expression of the protein (Fig. 1). The half-life of the EWS/FLI protein is reported to be less than 4 hours [97]. This means that Ewing sarcoma cells must maintain continued production of EWS/FLI to keep pace with its degradation. Mass spectrometry identified a single lysine at position 380 of EWS/FLI as the substrate for the ubiquitination that controls proteasomal turnover [97]. Facilitating this EWS/FLI ubiquitination or blocking de-ubiquitination may prove a viable therapeutic strategy to deplete the cell of EWS/FLI expression and activity. Towards this end, USP19 was identified as a de-ubiquitinating enzyme that binds EWS/FLI and wild-type EWSR1 to regulate stability [98]. More recently, another report established SPOP as a E3-ligase and OTUD7A as an associated de-ubiquitinase associated with EWS/FL1 stability [99]. Alternatively, Ewing sarcoma cells also do not tolerate an increase in expression of EWS/FLI. This fact is highlighted in a recent publication that identified TRIM8 as an E3 ubiquitin ligase that causes the ubiquitination of EWS/FLI but not of EWSR1 or FLI1. Depletion of TRIM8 impairs Ewing sarcoma tumor growth [100].
Fig. 1.
Therapeutic approaches targeting EWS/FLI expression. Ewing cells do not tolerate either an increase or decrease in EWS/FLI expression. Approaches modulating molecular chaperones or activating E3 ubiquitin ligases may deplete EWS/FLI expression leading to degradation either in the lysosome or proteosome. Inhibition of EWS/FLI associated de-ubiquitinases also could be effective because the cell does not tolerate increased EWS/FLI.
An alternative approach that has been used to modulate EWS/FLI expression is targeting the molecular chaperone, HSP90. PU-H71 and ganetespib are HSP90 inhibitors that increase EWS/FLI turnover, decrease Ewing sarcoma cell viability, and limit progression of Ewing sarcoma tumors in xenografts [101], [102]. These effects have been recapitulated with the histone deacetylase inhibitor entinostat because HSP90 relies on HDAC3 to prevent its deactivation by acetylation [103]. It is not clear how many proteins in addition to EWS/FLI are modulated by these effects of entinostat. Nevertheless, the impressive preclinical results of proteasomal degradation modulators (e.g., entinostat reduced tumor volume more than 97% in mice with CHLA-258 xenografts) support further investigation of these compounds for use in Ewing sarcoma [103].
EWS/FLI may also be sensitive to proteosomal degradation by Proteolysis Targeting Chimeric Molecules (PROTACs). PROTACs are bi-functional compounds where one portion of the compound binds a protein target and the other portion chaperones the protein to an E3 ubiquitin ligase for degradation [104], [105]. Gierisch et al. demonstrated that the degradation of EWS/FLI involves polyubiquitination at lysine-380 which tags the fusion protein for proteasomal destruction [97]. Lysine-380 is located within the DNA-binding domain and is present in wildtype FLI1 and conserved among several other ETS family members such as ETS1 which may limit specificity [106]. However, given the short half-life of EWS/FLI, a PROTAC designed to target a lysine-380 containing motif may be able to establish a therapeutic window [97]. To our knowledge, there is not an EWS/FLI-specific PROTAC developed yet. However, PROTACs targeting BET and CK proteins have been successfully used in Ewing sarcoma cells [99], [107]. And while PROTAC technology has yet to be used in clinics, it is advancing rapidly in preclinical models [108]. With improvements to stability and delivery PROTACS targeting EWS/FLI may be a viable therapy for Ewing sarcoma.
Finally, although the above studies focus on the regulation of EWS/FLI expression by the proteosome, it is likely that steady-state expression of EWS/FLI is more complicated and at least partially dependent on autophagy and the lysosomal system. One report used Bio-ID to biotinylate all proteins with 20–30 nm of EWS/FLI to identify CIMPR as an interacting protein that is known to chaperone clients to the endosome-lysosome system [109]. Activation of this system led to the degradation of EWS/FLI while inhibition of lysosomal activity with either chloroquine or pepstatin A stabilized EWS/FLI expression [109]. The therapeutic implications of these observations is an ongoing area of investigation.
5. Manipulating EWS/FLI induced epigenetic changes
The first studies of epigenetics in Ewing sarcoma clearly demonstrated the importance of epigenetic modulation by EWS/FLI to malignant transformation via the polycomb factors EZH2 and BMI1 [110], [111]. Subsequent studies demonstrated clear dysregulation of developmental programs such as the posterior HOX gene cluster; an effect linked to the trithorax proteins MLL1 and Menin [112], [113]. Further, silencing of EWS/FLI has been linked to widespread epigenetic reprogramming in multiple studies [23], [114], [115]. Therefore, targeting EWS/FLI associated epigenetic processes is a promising therapeutic strategy.
EWS/FLI likely uses multiple mechanisms to alter the epigenetic landscape. However, the effects of EWS/FLI at GGAA microsatellite response element has been best described [21]. The Davis group used chromatin immunoprecipitation sequencing (ChIPseq) to demonstrate increased occupancy of EWS/FLI and nucleosome depletion at GGAA microsatellites that had features of enhancers [116]. Work by Riggi et al. in 2014 validated these findings and combined ChIPseq with assay for transposase-accessible chromatin sequencing (ATACseq) to demonstrate that EWS/FLI exerts its effects on global transcription by binding to heterochromatic GGAA microsatellite regions and recruiting acetyltransferases to establish de novo enhancers that can regulate target gene promoters [23]. Boulay et al. subsequently elaborated on the mechanism of EWS/FLI enhancer remodeling by demonstrating that the intrinsically disordered region of EWS/FLI recruits the chromatin remodeling complex SWI/SNF to GGAA microsatellites [24]. Importantly, the finding of widespread epigenetic remodeling was later shown to occur in a large sample of primary Ewing sarcoma tumors thus establishing the importance of these epigenetic mechanisms to tumorigenesis [118]. Tomazou et al. comprehensively mapped the enhancer landscape using DNA methylation, seven histone post-translational modifications, accessibility assays, and gene expression to describe 4 distinct gene expression clusters with associated histone post-translational modifications [117]. H3K27ac was the mark most associated with EWS/FLI activity and widespread enhancer reprogramming by the fusion protein was evident [117]. Importantly, EWS/FLI repressed genes grouped in the same 4 clusters. However, these genes had distinct transcription factor binding signatures (AP-1 vs. E2F) and near uniform gain of H3K27ac and H3K4me3 at the promoters of these genes [117]. This suggests a common mechanism of repression which has been suggested to involve displacement of wild-type ETS transcription factors by EWS/FLI at the GGAA motif [23].
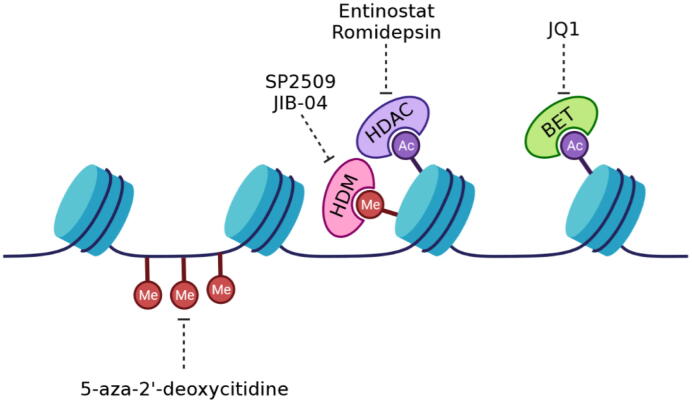
Since widespread epigenetic reprogramming is a prerequisite to Ewing sarcoma tumorigenesis under the influence of EWS/FLI, several efforts have focused on modulating the epigenome in Ewing tumors as a therapeutic approach (Fig. 2). Ewing sarcoma cell viability has been linked to differential methylation at specific targets in tumor relative to mesenchymal stem cells [118], [119], [120], [121], [122]. The DNA demethylating agent 5-aza-2′-deoxycytidine causes a loss of clonogenicity in a dose-dependent manner in TC32 and TC71 Ewing sarcoma cells [123]. A similar effect is observed with histone deacetylase (HDAC) inhibitors like entinostat which also produced a synergistic loss of clonogenicity when combined with 5-aza-2′-deoxycytidine [123]. HDAC inhibitors were shown to be active in Ewing sarcoma both in vitro and in xenografts and to restore the expression of the EWS/FLI repressed target, TGFBR2 even in the continued presence and activity of the fusion protein; an effect that was subsequently demonstrated genome-wide [117], [124]. Entinostat in particular shows increased activity in Ewing sarcoma relative to other HDAC inhibitors likely due to its secondary function in limiting HSP90 activity (see above) [103]. HDAC inhibitors have also been shown to activate TP53 to sensitize Ewing sarcoma cells to TRAIL mediated apoptosis and limit metastasis of Ewing sarcoma tumors [125], [126].
Fig. 2.
EWS/FLI associated epigenetic targets. EWS/FLI requires alterations in chromatin structure to generate its oncogenic transcriptome. Therefore, a variety of compounds and targets have been evaluated to modulated the associated epigenome with great promise.
LSD1 is a histone demethylase overexpressed in Ewing sarcoma [127] and a drug target in this tumor. The LSD1 inhibitor SP2509 reverses the EWS/FLI and EWS/ERG gene signatures as measured by gene-set enrichment analysis [128]. This induces Ewing sarcoma cell apoptosis and reduces tumor volume in xenografts of A673 and SK-N-MC cells [128]. The effect has been shown to be secondary to reversal of LSD1 activity at GGAA microsatellite and non-microsatellite sites co-occupied with EWS/FLI with some preference for the non-microsatellite sites [129]. Importantly, combining SP2509 with an HDAC inhibitor leads to marked synergy in various Ewing sarcoma cell lines and patient-derived xenografts [130], [131].
Inhibitors of a second class of histone demethylases, Jumonji-domain histone demethylases, have also shown efficacy in preclinical Ewing sarcoma models. The pan-Jumonji inhibitor JIB-04 poisons KDM3A to reverse the EWS/FLI transcriptional signature, increase DNA damage, and decrease Ewing sarcoma cell growth in vitro and in vivo [132]. More recently, the same group showed repression of metastatic properties of Ewing sarcoma cells with repression of KDM5A add PHF2 [133].
BET proteins are epigenetic readers of acetylated lysine residues on histones. They play important roles in regulating transcription by interacting with several proteins such as RNA polymerase II [134]. One study demonstrated that the BET inhibitor JQ1 reverses the EWS/FLI transcriptional signature of Ewing sarcoma cells and inhibits tumor growth of Ewing sarcoma xenografts [135]. A second study of JQ1 in Ewing sarcoma xenografts showed a survival benefit as well as decreased vascularization [136]. The effect on angiogenesis was reaffirmed by another study that showed suppression of expression of tumor-derived angiogenic factors as well as direct effects on endothelial cells and tube formation [137]. A third report confirmed the reversal of the EWS/FLI transcriptional signature and a decrease in Ewing sarcoma xenograft tumor growth after JQ1 treatment [138]. Like HDAC inhibitors, other authors used BET inhibitors to modulate the expression of important pathways such as IGF1 [138]. Finally, one paper placed EWS/FLI or EWS/ERG in a functional complex with BRD4, MED1 and RNAPII [107]. Impairment of this complex either by silencing expression of BRD4 or with BET inhibitors reversed EWS-ETS activity [107]. This same paper showed that silencing of EWS/FLI or BRD4 limited invasiveness of Ewing sarcoma cells in a CAM assay [107].
6. Inhibiting downstream targets of EWS/FLI
EWS/FLI downstream targets play a critical role in Ewing sarcoma tumor formation and progression. Therefore, a large body of work by multiple investigators has focused on these effectors as therapeutic targets and as a means to understand the biology and origins of the disease. Perhaps the most well-established Ewing sarcoma cancer susceptibility gene is EGR2 which is a downstream target of the fusion protein [139], [140]. Further, mesenchymal stem cells (MSCs) are among the top contenders for the cell of origin (along with neural crest cells) for Ewing sarcoma [111], [114]. Consistent with this hypothesis, EWS/FLI alters the expression of several pathways important to MSC differentiation and survival of derivative cells [141], [142], [143]. Some of these pathways can be manipulated pharmacologically.
Numerous studies have used comprehensive genome-wide approaches to map the downstream targets of EWS/FLI. Interestingly, while these reports do identify common targets, the exact identity of EWS/FLI targets is quite discordant among the reports. This was highlighted in a meta-analysis which illuminated these differences and highlighted what is perhaps the most agreed upon list of target genes [144]. In addition, the functional characterization of EWS/FLI targets has been captured genome-wide in numerous reports. One study clearly captures EWS/FLI-driven transcription across multiple models describes induced targets as driving proliferation, dysregulating the cell cycle and driving genes associated with the DNA damage response while repressed genes are associated with the repression of cell differentiation and cell-to-cell communication [27]. Finally, the most-well established target which plays a role in the biology of EWS/FLI is NR0B1 [76], [145], [146]. The NR0B1 promoter contains a GGAA microsatellite that if deleted leads to a loss of EWS/FLI responsiveness [147]. Additionally, while both FLI1 and EWS/FLI can bind this region, only EWS/FLI can transactivate and drive expression [21], [148].
One important downstream target of EWS/FLI is the transcription factor GLI1. Indian hedgehog (Ihh) binds to patched molecules (PTCH1/2) on the surface of MSCs to release the inhibition of smoothened [149]. This release leads to expression of glioma-associated oncogene (GLI) transcription factors which are critical regulators of endochondral ossification and highly expressed in membranous bone and teeth [150], [151]. EWS/FLI drives high levels of GLI1 expression in Ewing sarcoma which contributes to tumorigenicity [152], [153]. The promyelocytic leukemia drug arsenic trioxide (ASO) directly inhibits GLI1 and GLI2 [154], [155]. The sensitivity of cells to ASO positively correlates to the expression levels of GLI1 [156], however, ASO has a highly variable IC50 in Ewing sarcoma cell lines and does not produce a response in xenografts [157].
There are also numerous pathways upregulated by EWS/FLI that are not specific to MSCs but promote cell growth, nonetheless. Cholecystokinin (CCK) is a peptide hormone with critical roles in satiety regulation, bile release, bile production, and digestion [158]. It is also highly expressed in several cancers, Ewing sarcoma among them [79], [159], [160]. The classic functions of CCK are obviously absent in Ewing sarcoma tumors, however, the presence of CCK receptors suggest that it may play an autocrine role to promote cell growth or survival. This is further supported by a decrease in Ewing sarcoma cell proliferation in vitro and a modest reduction in tumor volume in xenografts of A673 cells after inducible expression of an shRNA against CCK [161]. Devazepide is an inhibitor of the CCK receptor CCKA that inhibits Ewing sarcoma tumor growth in vivo but requires a millimolar concentration [162].
The mitogen-activated protein kinase (MAPK) pathway is one of the best studied cascades involved in regulating cell growth and proliferation with several pro-cancer mutations in members of this pathway described [163]. EWS/FLI drives constitutive activation of MAPK3/ERK1 and MAPK1/ERK2 [164], [165]. This activation is mediated in part by EWS/FLI driven upregulation of CAV1 and downregulation of sprouty 1 (SPRY1) [166], [167]. Inhibition of the MAPK pathway with the inhibitor U0126 or the microRNA miR-30d limits the invasive potential of Ewing sarcoma cells in vitro [89], [168].
The fact that cancer cells favor anaerobic glycolysis over oxidative phosphorylation (Warburg effect) has been appreciated for almost a century [169]. The lynchpin of the Warburg effect is the lactate dehydrogenase (LDH) enzyme which converts pyruvate to lactate [169]. Ewing sarcoma expresses high levels of LDH, and its serum concentration is of prognostic value in this tumor [170]. LDH inhibitors produced marked necrosis in Ewing sarcoma xenografts but only modest changes to total tumor volume with increased dosing limited by hemolysis [171]. Mature erythrocytes rely exclusively on LDH mediated anaerobic glycolysis for ATP production as they lack mitochondria for oxidative phosphorylation [172]. Therefore, hemolysis may be a universal dose-limiting toxicity of LDH inhibitors [173].
It is also clear that a major function of EWS/FLI is to dysregulate metabolism and a number of studies have linked the activity of the fusion protein to the dysregulation of serine biosynthesis [174], [175], [176] perhaps via glutamine import [177], energy and redox balance via NAMPT [178], [179],and tryptophan metabolism [180]. The peptide hormone insulin-like growth factor 1 (IGF-1) is a key anabolic signaling molecule. The Helman lab recognized the dysregulation of this pathway in patient samples years ago and characterized the loss of imprinting of the IGF2 locus in Ewing patient samples [181], [182]. Numerous studies have explored therapeutic targeting and dysregulation of this pathway in this tumor. EWS/FLI upregulates expression of IGF-1, its receptor IGF1-R, and downregulates the carrier protein IGFBP-3 which appear to be important to tumorigenesis [75], [183], [184], [185]. The largest single-agent study of an anti-IGF-1 antibody in the clinic was of teprotumumab (R1507) through the Sarcoma Alliance for Research through Collaboration (SARC). This study impressively accrued 115 patients across 31 countries in two years and found a response rate of 10–15% [186]. Subsets of patients were exceptional responders and other studies point to specific molecular features that may confer sensitivity [187], [188]. IGF1R targeting was also evaluated in the metastatic setting in combination with chemotherapy in the Children’s Oncology Group. Although the results are yet to be reported, the study highlights the need to find the right combination and/or identify the exceptional responders [189].
Finally, numerous other studies describe important downstream targets that are beyond the scope of this review, these are both induced and repressed targets and include but are not limited to, NPY1R [190], NKX2.2 [191], telomerase [192], TGFBR2[193], HOX genes (see above), and CAV1 [166].
7. Exploiting EWS/FLI induced DNA damage
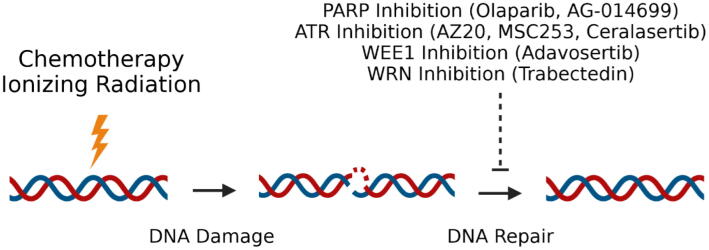
One of the most important observations made by James Ewing in the 1920s about the sarcoma that would one day bear his name is that Ewing sarcoma is exquisitely sensitive to ionizing radiation [194]. Ionizing radiation leads to the accumulation of DNA double strand breaks which must be repaired to avoid cell death. Standard chemotherapy for Ewing sarcoma employs vincristine in combination with four different DNA damaging agents: doxorubicin, cyclophosphamide, etoposide, and ifosfamide [2], [195], [196], [197]. It is known that Ewing sarcoma cells contain high basal levels of DNA damage which are likely driven by high replication and transcriptional stress mediated by EWS/FLI driven formation of R-loops [32]. This suggests that the cells are dependent on specific DNA damage response genes to cope with the increase in basal DNA damage. A number of DNA damage response targeted agents and pathways have been extensively evaluated as therapeutic targets in Ewing sarcoma (Fig. 3).
Fig. 3.
Targeting the DNA damage response. EWS/FLI is known to bind and regulate the expression/activity of multiple proteins involved in the DNA damage response. These pathways have been explored as therapeutic targets both alone, sequentially and in combination with EWS/FLI targeted agents.
One candidate gene that mitigates this DNA damage and is expressed at high levels in Ewing sarcoma cells is poly(ADP-ribose) polymerase (PARP)[198], [199]. PARP aids in the repair of single and double strand DNA breaks [200], [201]. The mechanism of PARP induction in Ewing sarcoma is controversial. One study suggests that ETS1 is the main driver of PARP expression, not EWS/FLI [202]. A second study from the same group claims that EWS/FLI inhibits the expression of PARP mRNA [203]. These reports are challenged by Brenner et al. who report that EWS/FLI can bind PARP-1 directly and induce PARP expression to regulate EWS/FLI production in a positive feedback loop [30]. An additional study demonstrated the interaction between EWS/FLI, PARP and BRCA1 [32]. This study links the sensitivity of the cells to PARP inhibition to an EWS/FLI induced increase in R-loop formation that releases BRCA1 from sites of DNA damage [32]. Independent of the mechanism of PARP hypersensitivity, the observation that Ewing sarcoma cells were particularly sensitive to PARP inhibitors came from a large unbiased screen of 639 cell lines with 130 different agents [204]. This study linked the sensitivity of Ewing sarcoma cells to PARP to EWS/FLI and inspired multiple independent studies of PARP inhibitors both preclinically and in patients. Single agent activity has not been found in the clinic [205] and the results in combination with temozolomide have been unclear with one study showing no activity [206] and another demonstrating a clinical response [207]. This has led to the search for biomarkers of activity such as SLFN11 or PALB2 [208], [209] and the identification of new combinations such as PARP-irinotecan-temozolomide [210]. A number of excellent studies identifying PARP based combinations have been published suggesting the combination of PARP with other chemotherapy [211], ionizing radiation [212], trabectedin [213], NAMPT blockade [179], [214], or CDK12 inhibitors [215].
Like PARP, the ATR pathway is an important regulator of DNA single and double strand break repair but with specific activity during cell cycle checkpoints [216]. Preclinical evidence suggests that inhibition of the ATR pathway is another vulnerability in Ewing sarcoma. Two ATR inhibitors, AZ20 and MSC253, produced significant reduction in tumor volume of subcutaneous A4573 xenografts and increased DNA damage as measured by γH2AX staining [217]. Moreover, the ATR inhibitor ceralasertib is impressively synergistic with adavosertib, which blocks the downstream effector of the ATR pathway WEE1, in several Ewing sarcoma cell lines [218].
Finally, the therapeutic targeting of EWS/FLI in combination with specific DNA damaging agents has been proposed. Grohar et al. proposed using trabectedin to suppress EWS/FLI and the WRN helicase to sensitize to the DNA damaging effects of irinotecan [219]. A similar approach by Lin et al. used mithramycin as a radiation sensitizer in an EWS/FLI restricted manner [220]. Other studies have focused on increasing the activity of DNA damaging agents by exploiting EWS/FLI relationships such as with DNA-PK [30], PARP [32], anti-apoptotic pathways [221], pro-apoptotic pathways [41], or histone demethylases [132]. These methods hold great promise for the treatment of Ewing sarcoma patients.
8. Blocking EWS/FLI mediated transcription
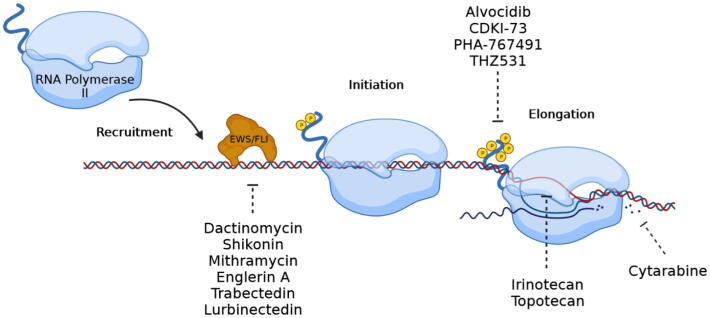
EWS/FLI driven transcription occurs in distinct steps: EWS/FLI expression, chromatin decompaction, DNA binding, productive elongation, termination, RNA processing and protein expression of downstream targets [37], [222], [223]. Interestingly, there are many compounds that have been identified as inhibitors of transcription that work at distinct steps in this process. Recognition and targeting of these steps in series or in parallel may be a clinically important strategy in the anti-Ewing sarcoma armamentarium (Fig. 4).
Fig. 4.
Blocking EWS/FLI driven transcription. Transcription is a multi-step process. Compounds have been identified that interfere with the DNA binding of EWS/FLI, promoter escape or productive elongation with varying degrees of specificity. Sequential targeting of this process has been proposed to increase activity and perhaps specificity.
As discussed above, chromatin remodeling both upstream and downstream of the DNA binding of EWS/FLI is an important area of drug development as LSD1 inhibitors and bromodomain inhibitors have all been shown to perturb the Ewing sarcoma transcriptome [107], [128]. In addition, RNA processing is influenced by EWS/FLI and is the basis for the development of YK-4-279 and its derivative TK-216 [55], [224].
The next most likely step in transcription that is targetable is DNA binding. Several compounds have been identified that work at this step in the process. Chen et al. used a fluorometric proximity assay to measure changes to EWS/FLI DNA binding [225]. Their model used two synthetic biotin conjugated oligos that were bound to streptavidin conjugated donor beads [225]. One oligo contained two GGAA motifs for EWS/FLI binding, the second lacked any GGAA motif but was recognized by p53 and served as a control [225]. With this system, 5200 small molecules were tested and ranked based on the ability to prevent EWS/FLI DNA binding while sparing p53 binding [225]. This led to the characterization of low dose dactinomycin and shikonin as EWS/FLI inhibitors [225], [226].
The expression of NR0B1 in Ewing sarcoma completely depends on EWS/FLI binding its promoter [147]. Grohar et al. leveraged this fact by developing a screen that measured EWS/FLI driven transcription by placing the luciferase gene under the control of the NR0B1 promoter [227]. Over 50,000 compounds were screened using this system. Multiplex RT-qPCR of 11 EWS/FLI induced targets further evaluated the top 200 hits [227]. The top hit from this screen was mithramycin which was shown to interfere with EWS/FLI activity, mRNA, and protein expression and genome-wide expression of downstream targets [227]. The results with mithramycin prompted the opening of a phase I/II clinical trial for patients with relapsed/refractory Ewing sarcoma. Unfortunately, dose-limiting liver toxicity was observed at a plasma concentration approximately 3–5 times lower than what was necessary to inhibit EWS/FLI [228]. This mechanism of acute liver injury may be due to disrupted bile deposition and/or an increase in free radical production [229], [230]. This led to the identification of a less toxic (EC8042) and more potent (EC8105) analog of mithramycin [231]. Finally, an additional screen of natural product extracts using this same system led to the identification of englerin A as a compound that inhibits the DNA binding of EWS/FLI [232].
The ascidian derived compound trabectedin binds CGG DNA repeats within the minor groove [233]. Grohar et al. reported that trabectedin inhibits NR0B1 promoter activity, downstream target expression and reverses the gene-signature of EWS/FLI [234]. In addition, transfer of EWS/FLI to HT1080 cells with the NR0B1 promoter luciferase construct described above leads to the marked induction of NR0B1 luciferase, an effect that can be rescued by co-incubation with trabectedin [234]. Furthermore, trabectedin leads to a redistribution of EWS/FLI away from chromatin and toward the nucleolus [235]. Lurbinectedin, a newer analog of trabectedin with an improved toxicity profile, also redistributes EWS/FLI to the nucleolus [236]. This nucleolar sequestration is observed with a concomitant loss of SWI/SNF binding at EWS/FLI induced targets [235]. These effects trigger an epigenetic switch leading to an increase in H3K9me3 and H3K27me3 most notably at GGAA microsatellites [235]. These results establish this class of compounds as EWS/FLI inhibitors.
Transcriptional elongation is also an intriguing target. Indeed, the nucleoside analog cytarabine (a known elongation terminator) was the first identified EWS/FLI inhibitor that reversed the expression of the EWS/FLI transcriptome [237]. It is not known if the compound interferes with transcription elongation as part of the mechanism of this compound in Ewing sarcoma. Nevertheless, the data in the original report was compelling and cytarabine remains an intriguing compound. Additional compounds that interfere with transcription elongation have also been extensively evaluated in Ewing sarcoma. Camptothecins block topoisomerase I which is necessary to relieve the supercoiling stress caused by the transcription bubble [238]. Two, clinically relevant camptothecins are irinotecan and topotecan. Both compounds are widely used in the setting of relapsed Ewing sarcoma [239], [240]. Importantly, our group demonstrated that irinotecan improves the magnitude and sustains the duration of EWS/FLI suppression by trabectedin [235].
The observation that irinotecan works downstream of trabectedin to improve the magnitude and duration of EWS/FLI suppression is consistent with targeting EWS/FLI at successive steps in transcription. This idea has been proposed by us and other groups. Mateo-Lozano et al. proposed combining highly specific EWS/FLI-targeted antisense oligos with rapamycin, an agent that inhibits translation [68]. More recently, inhibitors of cyclin dependent kinases have provided a new therapeutic opportunity. Cyclin dependent kinase 9 (CDK9) is a CDK critical to proper maintenance of transcription [241]. Other transcriptional CDKs include CDK7, CDK8, CDK12, and CDK13 [242]. Transcriptional CDKs regulate RNA polymerase II processivity by phosphorylating key serine residues on the C-terminal domain of Rpb1. Specific phosphorylation patterns promote RNA polymerase II proximal promoter pausing, pause release, elongation, and termination [37], [222], [223].
The CDK9 inhibitor alvocidib has been granted orphan drug status for acute myeloid leukemia [243]. Preclinical evidence also supports the efficacy of alvocidib in Ewing sarcoma. Alvocidib drives Ewing sarcoma cell apoptosis in a dose dependent manner in vitro and reduces tumor volume in xenografts [244]. However, even though alvocidib is regarded as a CDK9 inhibitor, it is not completely selective. In a cell-free assay, the IC50 values of alvocidib were 20 nM for CDK9, 30 nM for CDK1, 40 nM for CDK2, and 875 nM for CDK7 [245]. This means the preclinical efficacy of alvocidib may not be due entirely to CDK9 inhibition. Li et al. support this notion by connecting the apoptosis seen in Ewing sarcoma cells after alvocidib treatment to inhibition of CDK2 [246]. Therefore, single agent CDK9 inhibitors may not be effective in the setting of Ewing sarcoma. However, CDK9 inhibitors may be perfect candidates for combination therapies. Two preclinical studies support this idea. In one study, CDK9 was shown to bind EWS/FLI through the BET protein BRD4 [247]. Combining the CDK9 inhibitor CDKI-73 with the BET inhibitor JQ1 was more effective at reducing Ewing sarcoma cell proliferation and reducing tumor volume in xenografts than either compound alone [247]. A second study demonstrated synergy between the EWS/FLI inhibitor mithramycin and the CDK9 inhibitor PHA-767491 [230]. Importantly, the synergy occurred at clinically achievable concentrations of mithramycin [230]. Further, the combination may limit toxicity as it attenuated mithramycin induced free radical production by blocking the induction of the BTG2 protein [230]. Synergy was also seen with a sequential targeting approach featuring the histone demethylase inhibitor GSK-J4 and the CDK inhibitor THZ1 [248]. Finally, inhibition of CDK12 (another CDK necessary for maintenance of transcriptional elongation) in combination with the PARP inhibitor olaparib led to reductions in tumor volume and increased survival in both cell line-derived and patient-derived xenografts [215].
9. Conclusion
EWS/FLI is widely regarded as the oncogenic driver in Ewing sarcoma based on multiple independent studies. This transcription factor is a challenging drug target because of the wide range of interacting proteins and vast number of downstream effectors. Indeed, it is not clear if a single molecule that binds to EWS/FLI would be able to completely reverse all of the molecular interactions and associated oncogenic properties of the fusion protein. However, this same complexity of transcription renders the mutation vulnerable to a number of complementary strategies to both directly target the fusion and exploit downstream alterations of the transcriptome. In this review, we summarize these therapeutic efforts focused on or linked to EWS/FLI. The direct therapeutic targeting of EWS/FLI with a high degree of specificity remains challenging due to the unstructured 5′ EWSR1 transactivating domain and the lack of a known high-affinity binding site in the 3′ FLI1 portion of the molecule. It is possible that efforts focused on the EWS/FLI mRNA, steady-state expression levels of EWS/FLI or nuclear/genomic distribution of EWS/FL1 will lead to effective therapies with some degree of specificity. However, it is not clear if specificity is necessary. Indeed, it may be therapeutically inferior in patients. Consistent with this idea, some of the most impressive tumor regressions seen in vivo in Ewing sarcoma xenograft models has been seen by combining EWS/FLI targeting with less-specific agents that inhibit other steps in the transcription process and/or in combination with DNA damaging agents [107], [219], [230], [247], [249]. Nevertheless, many of the approaches summarized in this review that link therapeutic effects to EWS/FLI show promise for Ewing sarcoma patients. Therefore, while EWS/FLI remains the greatest strength of the tumor, it may ultimately prove to be its greatest therapeutic vulnerability.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: PJG receives research support from Janssen Pharmaceutical to support the clinical evaluation of trabectedin in combination with low dose irinotecan.
References
- 1.Ye C., Dai M., Zhang B. Risk factors for metastasis at initial diagnosis with ewing sarcoma. Front. Oncol. 2019;9(1043) doi: 10.3389/fonc.2019.01043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Womer R.B., West D.C., Krailo M.D., Dickman P.S., Pawel B.R., Grier H.E., Marcus K., Sailer S., Healey J.H., Dormans J.P., Weiss A.R. Randomized controlled trial of interval-compressed chemotherapy for the treatment of localized Ewing sarcoma: a report from the Children's Oncology Group. J. Clin. Oncol. 2012;30(33):4148–4154. doi: 10.1200/JCO.2011.41.5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marina N.M., Liu Q., Donaldson S.S., Sklar C.A., Armstrong G.T., Oeffinger K.C., Leisenring W.M., Ginsberg J.P., Henderson T.O., Neglia J.P., Stovall M.A., Yasui Y., Randall R.L., Geller D.S., Robison L.L., Ness K.K. Longitudinal follow-up of adult survivors of Ewing sarcoma: a report from the Childhood Cancer Survivor Study. Cancer. 2017;123(13):2551–2560. doi: 10.1002/cncr.30627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bosma S.E., Ayu O., Fiocco M., Gelderblom H., Dijkstra P.D.S. Prognostic factors for survival in Ewing sarcoma: a systematic review. Surg. Oncol. 2018;27(4):603–610. doi: 10.1016/j.suronc.2018.07.016. [DOI] [PubMed] [Google Scholar]
- 5.Delattre O., Zucman J., Plougastel B., Desmaze C., Melot T., Peter M., Kovar H., Joubert I., de Jong P., Rouleau G., et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359(6391):162–165. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- 6.Tirode F., Surdez D., Ma X., Parker M., Le Deley M.C., Bahrami A., Zhang Z., Lapouble E., Grossetete-Lalami S., Rusch M., Reynaud S., Rio-Frio T., Hedlund E., Wu G., Chen X., Pierron G., Oberlin O., Zaidi S., Lemmon G., Gupta P., Vadodaria B., Easton J., Gut M., Ding L., Mardis E.R., Wilson R.K., Shurtleff S., Laurence V., Michon J., Marec-Berard P., Gut I., Downing J., Dyer M., Zhang J., Delattre O. Genomic landscape of Ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer Discovery. 2014;4(11):1342–1353. doi: 10.1158/2159-8290.CD-14-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brohl A.S., Solomon D.A., Chang W., Wang J., Song Y., Sindiri S., Patidar R., Hurd L., Chen L., Shern J.F., Liao H., Wen X., Gerard J., Kim J.S., Lopez Guerrero J.A., Machado I., Wai D.H., Picci P., Triche T., Horvai A.E., Miettinen M., Wei J.S., Catchpool D., Llombart-Bosch A., Waldman T., Khan J. The genomic landscape of the Ewing Sarcoma family of tumors reveals recurrent STAG2 mutation. PLoS Genet. 2014;10(7) doi: 10.1371/journal.pgen.1004475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crompton B.D., Stewart C., Taylor-Weiner A., Alexe G., Kurek K.C., Calicchio M.L., Kiezun A., Carter S.L., Shukla S.A., Mehta S.S., Thorner A.R., de Torres C., Lavarino C., Suñol M., McKenna A., Sivachenko A., Cibulskis K., Lawrence M.S., Stojanov P., Rosenberg M., Ambrogio L., Auclair D., Seepo S., Blumenstiel B., DeFelice M., Imaz-Rosshandler I., Schwarz-Cruz Y.C.A., Rivera M.N., Rodriguez-Galindo C., Fleming M.D., Golub T.R., Getz G., Mora J., Stegmaier K. The genomic landscape of pediatric Ewing sarcoma. Cancer Discovery. 2014;4(11):1326–1341. doi: 10.1158/2159-8290.CD-13-1037. [DOI] [PubMed] [Google Scholar]
- 9.Grunewald T.G.P., Cidre-Aranaz F., Surdez D., Tomazou E.M., de Alava E., Kovar H., Sorensen P.H., Delattre O., Dirksen U. Ewing sarcoma. Nat. Rev. Dis. Primers. 2018;4(1):5. doi: 10.1038/s41572-018-0003-x. [DOI] [PubMed] [Google Scholar]
- 10.Anderson N.D., de Borja R., Young M.D., Fuligni F., Rosic A., Roberts N.D., Hajjar S., Layeghifard M., Novokmet A., Kowalski P.E., Anaka M., Davidson S., Zarrei M., Id Said B., Schreiner L.C., Marchand R., Sitter J., Gokgoz N., Brunga L., Graham G.T., Fullam A., Pillay N., Toretsky J.A., Yoshida A., Shibata T., Metzler M., Somers G.R., Scherer S.W., Flanagan A.M., Campbell P.J., Schiffman J.D., Shago M., Alexandrov L.B., Wunder J.S., Andrulis I.L., Malkin D., Behjati S., Shlien A. Rearrangement bursts generate canonical gene fusions in bone and soft tissue tumors. Science (New York, N.Y.) 2018;361(6405) doi: 10.1126/science.aam8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Alava E., Kawai A., Healey J.H., Fligman I., Meyers P.A., Huvos A.G., Gerald W.L., Jhanwar S.C., Argani P., Antonescu C.R., Pardo-Mindan F.J., Ginsberg J., Womer R., Lawlor E.R., Wunder J., Andrulis I., Sorensen P.H., Barr F.G., Ladanyi M. EWS-FLI1 fusion transcript structure is an independent determinant of prognosis in Ewing's sarcoma. J. Clin. Oncol. 1998;16(4):1248–1255. doi: 10.1200/JCO.1998.16.4.1248. [DOI] [PubMed] [Google Scholar]
- 12.van Doorninck J.A., Ji L., Schaub B., Shimada H., Wing M.R., Krailo M.D., Lessnick S.L., Marina N., Triche T.J., Sposto R., Womer R.B., Lawlor E.R. Current treatment protocols have eliminated the prognostic advantage of type 1 fusions in Ewing sarcoma: a report from the Children's Oncology Group. J. Clin. Oncol. 2010;28(12):1989–1994. doi: 10.1200/JCO.2009.24.5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zoubek A., Dockhorn-Dworniczak B., Delattre O., Christiansen H., Niggli F., Gatterer-Menz I., Smith T.L., Jurgens H., Gadner H., Kovar H. Does expression of different EWS chimeric transcripts define clinically distinct risk groups of Ewing tumor patients? J. Clin. Oncol. 1996;14(4):1245–1251. doi: 10.1200/JCO.1996.14.4.1245. [DOI] [PubMed] [Google Scholar]
- 14.Asch R.H., Weckstein L.N., Balmaceda J.P., Rojas F., Spitz I.M., Tadir Y. Non-surgical expulsion of non-viable early pregnancy: a new application of RU 486. Hum. Reprod. 1990;5(4):481–483. doi: 10.1093/oxfordjournals.humrep.a137128. [DOI] [PubMed] [Google Scholar]
- 15.Shing D.C., McMullan D.J., Roberts P., Smith K., Chin S.F., Nicholson J., Tillman R.M., Ramani P., Cullinane C., Coleman N. FUS/ERG gene fusions in Ewing's tumors. Cancer Res. 2003;63(15):4568–4576. [PubMed] [Google Scholar]
- 16.Sorensen P.H., Lessnick S.L., Lopez-Terrada D., Liu X.F., Triche T.J., Denny C.T. A second Ewing's sarcoma translocation, t(21;22), fuses the EWS gene to another ETS-family transcription factor, ERG. Nat. Genet. 1994;6(2):146–151. doi: 10.1038/ng0294-146. [DOI] [PubMed] [Google Scholar]
- 17.Jeon I.S., Davis J.N., Braun B.S., Sublett J.E., Roussel M.F., Denny C.T., Shapiro D.N. A variant Ewing's sarcoma translocation (7;22) fuses the EWS gene to the ETS gene ETV1. Oncogene. 1995;10(6):1229–1234. [PubMed] [Google Scholar]
- 18.Peter M., Couturier J., Pacquement H., Michon J., Thomas G., Magdelenat H., Delattre O. A new member of the ETS family fused to EWS in Ewing tumors. Oncogene. 1997;14(10):1159–1164. doi: 10.1038/sj.onc.1200933. [DOI] [PubMed] [Google Scholar]
- 19.Kaneko Y., Yoshida K., Handa M., Toyoda Y., Nishihira H., Tanaka Y., Sasaki Y., Ishida S., Higashino F., Fujinaga K. Fusion of an ETS-family gene, EIAF, to EWS by t(17;22)(q12;q12) chromosome translocation in an undifferentiated sarcoma of infancy. Genes Chromosom. Cancer. 1996;15(2):115–121. doi: 10.1002/(SICI)1098-2264(199602)15:2<115::AID-GCC6>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 20.Kinnaman M.D., Zhu C., Weiser D.A., Mohiuddin S., Hingorani P., Roth M., Gill J., Janeway K.A., Gorlick R., Lessnick S.L., Grohar P.J. Survey of paediatric oncologists and pathologists regarding their views and experiences with variant translocations in ewing and ewing-like sarcoma: a report of the Children's Oncology Group. Sarcoma. 2020;2020:3498549. doi: 10.1155/2020/3498549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gangwal K., Sankar S., Hollenhorst P.C., Kinsey M., Haroldsen S.C., Shah A.A., Boucher K.M., Watkins W.S., Jorde L.B., Graves B.J., Lessnick S.L. Microsatellites as EWS/FLI response elements in Ewing's sarcoma. Proc. Natl. Acad. Sci. USA. 2008;105(29):10149–10154. doi: 10.1073/pnas.0801073105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boone M.A., Taslim C., Crow J.C., Selich-Anderson J., Byrum A.K., Showpnil I.A., Sunkel B.D., Wang M., Stanton B.Z., Theisen E.R., Lessnick S.L. The FLI portion of EWS/FLI contributes a transcriptional regulatory function that is distinct and separable from its DNA-binding function in Ewing sarcoma. Oncogene. 2021 doi: 10.1038/s41388-021-01876-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riggi N., Knoechel B., Gillespie S.M., Rheinbay E., Boulay G., Suva M.L., Rossetti N.E., Boonseng W.E., Oksuz O., Cook E.B., Formey A., Patel A., Gymrek M., Thapar V., Deshpande V., Ting D.T., Hornicek F.J., Nielsen G.P., Stamenkovic I., Aryee M.J., Bernstein B.E., Rivera M.N. EWS-FLI1 utilizes divergent chromatin remodeling mechanisms to directly activate or repress enhancer elements in Ewing sarcoma. Cancer Cell. 2014;26(5):668–681. doi: 10.1016/j.ccell.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boulay G., Sandoval G.J., Riggi N., Iyer S., Buisson R., Naigles B., Awad M.E., Rengarajan S., Volorio A., McBride M.J., Broye L.C., Zou L., Stamenkovic I., Kadoch C., Rivera M.N. Cancer-specific retargeting of BAF complexes by a prion-like domain. Cell. 2017;171(1):163–178.e19. doi: 10.1016/j.cell.2017.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson K.M., Taslim C., Saund R.S., Lessnick S.L. Identification of two types of GGAA-microsatellites and their roles in EWS/FLI binding and gene regulation in Ewing sarcoma. PLoS ONE. 2017;12(11) doi: 10.1371/journal.pone.0186275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Selvanathan S.P., Graham G.T., Grego A.R., Baker T.M., Hogg J.R., Simpson M., Batish M., Crompton B., Stegmaier K., Tomazou E.M., Kovar H., Uren A., Toretsky J.A. EWS-FLI1 modulated alternative splicing of ARID1A reveals novel oncogenic function through the BAF complex. Nucl. Acids Res. 2019;47(18):9619–9636. doi: 10.1093/nar/gkz699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kauer M., Ban J., Kofler R., Walker B., Davis S., Meltzer P., Kovar H. A molecular function map of Ewing's sarcoma. PLoS ONE. 2009;4(4) doi: 10.1371/journal.pone.0005415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwentner R., Papamarkou T., Kauer M.O., Stathopoulos V., Yang F., Bilke S., Meltzer P.S., Girolami M., Kovar H. EWS-FLI1 employs an E2F switch to drive target gene expression. Nucl. Acids Res. 2015;43(5):2780–2789. doi: 10.1093/nar/gkv123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kowalewski A.A., Randall R.L., Lessnick S.L. Cell Cycle deregulation in Ewing's sarcoma pathogenesis. Sarcoma. 2011;2011 doi: 10.1155/2011/598704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brenner J.C., Feng F.Y., Han S., Patel S., Goyal S.V., Bou-Maroun L.M., Liu M., Lonigro R., Prensner J.R., Tomlins S.A., Chinnaiyan A.M. PARP-1 inhibition as a targeted strategy to treat Ewing's sarcoma. Cancer Res. 2012;72(7):1608–1613. doi: 10.1158/0008-5472.CAN-11-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller H.E., Gorthi A., Bassani N., Lawrence L.A., Iskra B.S., Bishop A.J.R. Reconstruction of Ewing sarcoma developmental context from mass-scale transcriptomics reveals characteristics of EWSR1-FLI1 permissibility. Cancers (Basel) 2020;12(4) doi: 10.3390/cancers12040948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gorthi A., Romero J.C., Loranc E., Cao L., Lawrence L.A., Goodale E., Iniguez A.B., Bernard X., Masamsetti V.P., Roston S., Lawlor E.R., Toretsky J.A., Stegmaier K., Lessnick S.L., Chen Y., Bishop A.J.R. EWS-FLI1 increases transcription to cause R-loops and block BRCA1 repair in Ewing sarcoma. Nature. 2018;555(7696):387–391. doi: 10.1038/nature25748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kovar H., Aryee D.N., Jug G., Henöckl C., Schemper M., Delattre O., Thomas G., Gadner H. EWS/FLI-1 antagonists induce growth inhibition of Ewing tumor cells in vitro. Cell Growth Differ. 1996;7(4):429–437. [PubMed] [Google Scholar]
- 34.Toub N., Bertrand J.R., Malvy C., Fattal E., Couvreur P. Antisense oligonucleotide nanocapsules efficiently inhibit EWS-Fli1 expression in a Ewing's sarcoma model. Oligonucleotides. 2006;16(2):158–168. doi: 10.1089/oli.2006.16.158. [DOI] [PubMed] [Google Scholar]
- 35.Tanner J.M., Bensard C., Wei P., Krah N.M., Schell J.C., Gardiner J., Schiffman J., Lessnick S.L., Rutter J. EWS/FLI is a master regulator of metabolic reprogramming in ewing sarcoma. Mol. Cancer Res. 2017;15(11):1517–1530. doi: 10.1158/1541-7786.MCR-17-0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez-Lage M., Torres-Ruiz R., Puig-Serra P., Moreno-Gaona P., Martin M.C., Moya F.J., Quintana-Bustamante O., Garcia-Silva S., Carcaboso A.M., Petazzi P., Bueno C., Mora J., Peinado H., Segovia J.C., Menendez P., Rodriguez-Perales S. In vivo CRISPR/Cas9 targeting of fusion oncogenes for selective elimination of cancer cells. Nat. Commun. 2020;11(1):5060. doi: 10.1038/s41467-020-18875-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haberle V., Stark A. Eukaryotic core promoters and the functional basis of transcription initiation. Nat. Rev. Mol. Cell Biol. 2018;19(10):621–637. doi: 10.1038/s41580-018-0028-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang L., Chansky H.A., Hickstein D.D., EWS Fli-1 fusion protein interacts with hyperphosphorylated RNA polymerase II and interferes with serine-arginine protein-mediated RNA splicing. J. Biol. Chem. 2000;275(48):37612–37618. doi: 10.1074/jbc.M005739200. [DOI] [PubMed] [Google Scholar]
- 39.Pahlich S., Quero L., Roschitzki B., Leemann-Zakaryan R.P., Gehring H. Analysis of Ewing sarcoma (EWS)-binding proteins: interaction with hnRNP M, U, and RNA-helicases p68/72 within protein-RNA complexes. J. Proteome Res. 2009;8(10):4455–4465. doi: 10.1021/pr900235t. [DOI] [PubMed] [Google Scholar]
- 40.Paronetto M.P., Minana B., Valcarcel J. The Ewing sarcoma protein regulates DNA damage-induced alternative splicing. Mol. Cell. 2011;43(3):353–368. doi: 10.1016/j.molcel.2011.05.035. [DOI] [PubMed] [Google Scholar]
- 41.Paronetto M.P., Bernardis I., Volpe E., Bechara E., Sebestyen E., Eyras E., Valcarcel J. Regulation of FAS exon definition and apoptosis by the Ewing sarcoma protein. Cell reports. 2014;7(4):1211–1226. doi: 10.1016/j.celrep.2014.03.077. [DOI] [PubMed] [Google Scholar]
- 42.Knoop L.L., Baker S.J. The splicing factor U1C represses EWS/FLI-mediated transactivation. J. Biol. Chem. 2000;275(32):24865–24871. doi: 10.1074/jbc.M001661200. [DOI] [PubMed] [Google Scholar]
- 43.Sanchez G., Bittencourt D., Laud K., Barbier J., Delattre O., Auboeuf D., Dutertre M. Alteration of cyclin D1 transcript elongation by a mutated transcription factor up-regulates the oncogenic D1b splice isoform in cancer. Proc. Natl. Acad. Sci. USA. 2008;105(16):6004–6009. doi: 10.1073/pnas.0710748105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Selvanathan S.P., Graham G.T., Erkizan H.V., Dirksen U., Natarajan T.G., Dakic A., Yu S., Liu X., Paulsen M.T., Ljungman M.E., Wu C.H., Lawlor E.R., Üren A., Toretsky J.A. Oncogenic fusion protein EWS-FLI1 is a network hub that regulates alternative splicing. Proc. Natl. Acad. Sci. USA. 2015;112(11):E1307–E1316. doi: 10.1073/pnas.1500536112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soranno A. Physical basis of the disorder-order transition. Arch. Biochem. Biophys. 2020;108305 doi: 10.1016/j.abb.2020.108305. [DOI] [PubMed] [Google Scholar]
- 46.Spahn L., Petermann R., Siligan C., Schmid J.A., Aryee D.N., Kovar H. Interaction of the EWS NH2 terminus with BARD1 links the Ewing's sarcoma gene to a common tumor suppressor pathway. Cancer Res. 2002;62(16):4583–4587. [PubMed] [Google Scholar]
- 47.Kim S., Denny C.T., Wisdom R. Cooperative DNA binding with AP-1 proteins is required for transformation by EWS-Ets fusion proteins. Mol. Cell. Biol. 2006;26(7):2467–2478. doi: 10.1128/MCB.26.7.2467-2478.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Erkizan H.V., Uversky V.N., Toretsky J.A. Oncogenic partnerships: EWS-FLI1 protein interactions initiate key pathways of Ewing's sarcoma. Clin. Cancer Res. 2010;16(16):4077–4083. doi: 10.1158/1078-0432.CCR-09-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Selvanathan S.P., Graham G.T., Erkizan H.V., Dirksen U., Natarajan T.G., Dakic A., Yu S., Liu X., Paulsen M.T., Ljungman M.E., Wu C.H., Lawlor E.R., Uren A., Toretsky J.A. Oncogenic fusion protein EWS-FLI1 is a network hub that regulates alternative splicing. Proc. Natl. Acad. Sci. USA. 2015;112(11):E1307–E1316. doi: 10.1073/pnas.1500536112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fidaleo M., Svetoni F., Volpe E., Miñana B., Caporossi D., Paronetto M.P. Genotoxic stress inhibits Ewing sarcoma cell growth by modulating alternative pre-mRNA processing of the RNA helicase DHX9. Oncotarget. 2015;6(31):31740–31757. doi: 10.18632/oncotarget.5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prusiner S.B. Novel proteinaceous infectious particles cause scrapie. Science (New York, N.Y.) 1982;216(4542):136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 52.Mao X., Miesfeldt S., Yang H., Leiden J.M., Thompson C.B. The FLI-1 and chimeric EWS-FLI-1 oncoproteins display similar DNA binding specificities. J. Biol. Chem. 1994;269(27):18216–18222. [PubMed] [Google Scholar]
- 53.Hou C., Tsodikov O.V. Structural basis for dimerization and DNA binding of transcription factor FLI1. Biochemistry. 2015;54(50):7365–7374. doi: 10.1021/acs.biochem.5b01121. [DOI] [PubMed] [Google Scholar]
- 54.Spahn L., Siligan C., Bachmaier R., Schmid J.A., Aryee D.N., Kovar H. Homotypic and heterotypic interactions of EWS, FLI1 and their oncogenic fusion protein. Oncogene. 2003;22(44):6819–6829. doi: 10.1038/sj.onc.1206810. [DOI] [PubMed] [Google Scholar]
- 55.Erkizan H.V., Kong Y., Merchant M., Schlottmann S., Barber-Rotenberg J.S., Yuan L., Abaan O.D., Chou T.H., Dakshanamurthy S., Brown M.L., Uren A., Toretsky J.A. A small molecule blocking oncogenic protein EWS-FLI1 interaction with RNA helicase A inhibits growth of Ewing's sarcoma. Nat. Med. 2009;15(7):750–756. doi: 10.1038/nm.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jully B., Vijayalakshmi R., Gopal G., Sabitha K., Rajkumar T. Junction region of EWS-FLI1 fusion protein has a dominant negative effect in Ewing's sarcoma in vitro. BMC cancer. 2012;12:513. doi: 10.1186/1471-2407-12-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thangaretnam K.P., Gopisetty G., Ramanathan P., Rajkumar T. A polypeptide from the junction region sequence of EWS-FLI1 inhibits Ewing's sarcoma cells, interacts with the EWS-FLI1 and partner proteins. Sci. Rep. 2017;7(1):7172. doi: 10.1038/s41598-017-07482-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu D., Gao Y., Qi Y., Chen L., Ma Y., Li Y. Peptide-based cancer therapy: opportunity and challenge. Cancer Lett. 2014;351(1):13–22. doi: 10.1016/j.canlet.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 59.Patocs B., Nemeth K., Garami M., Arato G., Kovalszky I., Szendroi M., Fekete G. Multiple splice variants of EWSR1-ETS fusion transcripts co-existing in the Ewing sarcoma family of tumors. Cell. Oncol. (Dordrecht) 2013;36(3):191–200. doi: 10.1007/s13402-013-0126-8. [DOI] [PubMed] [Google Scholar]
- 60.Grohar P.J., Kim S., Rangel Rivera G.O., Sen N., Haddock S., Harlow M.L., Maloney N.K., Zhu J., O'Neill M., Jones T.L., Huppi K., Grandin M., Gehlhaus K., Klumpp-Thomas C.A., Buehler E., Helman L.J., Martin S.E., Caplen N.J. Functional genomic screening reveals splicing of the EWS-FLI1 fusion transcript as a vulnerability in ewing sarcoma. Cell Reports. 2016;14(3):598–610. doi: 10.1016/j.celrep.2015.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.C. Neckles, R.E. Boer, N. Aboreden, A.M. Cross, R.L. Walker, B.H. Kim, S. Kim, J.S. Schneekloth, Jr., N.J. Caplen, HNRNPH1-dependent splicing of a fusion oncogene reveals a targetable RNA G-quadruplex interaction, RNA (New York, N.Y.) 25(12) (2019) 1731-1750. [DOI] [PMC free article] [PubMed]
- 62.Bajan S., Hutvagner G. RNA-based therapeutics: from antisense oligonucleotides to miRNAs. Cells. 2020;9(1) doi: 10.3390/cells9010137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Toretsky J.A., Connell Y., Neckers L., Bhat N.K. Inhibition of EWS-FLI-1 fusion protein with antisense oligodeoxynucleotides. J. Neurooncol. 1997;31(1–2):9–16. doi: 10.1023/a:1005716926800. [DOI] [PubMed] [Google Scholar]
- 64.Matsumoto Y., Tanaka K., Nakatani F., Matsunobu T., Matsuda S., Iwamoto Y. Downregulation and forced expression of EWS-Fli1 fusion gene results in changes in the expression of G(1)regulatory genes. Br. J. Cancer. 2001;84(6):768–775. doi: 10.1054/bjoc.2000.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tanaka K., Iwakuma T., Harimaya K., Sato H., Iwamoto Y. EWS-Fli1 antisense oligodeoxynucleotide inhibits proliferation of human Ewing's sarcoma and primitive neuroectodermal tumor cells. J. Clin. Investig. 1997;99(2):239–247. doi: 10.1172/JCI119152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lambert G., Bertrand J.R., Fattal E., Subra F., Pinto-Alphandary H., Malvy C., Auclair C., Couvreur P. EWS fli-1 antisense nanocapsules inhibits ewing sarcoma-related tumor in mice. Biochem. Biophys. Res. Commun. 2000;279(2):401–406. doi: 10.1006/bbrc.2000.3963. [DOI] [PubMed] [Google Scholar]
- 67.Maksimenko A., Malvy C., Lambert G., Bertrand J.R., Fattal E., Maccario J., Couvreur P. Oligonucleotides targeted against a junction oncogene are made efficient by nanotechnologies. Pharm. Res. 2003;20(10):1565–1567. doi: 10.1023/a:1026122914852. [DOI] [PubMed] [Google Scholar]
- 68.Mateo-Lozano S., Gokhale P.C., Soldatenkov V.A., Dritschilo A., Tirado O.M., Notario V. Combined transcriptional and translational targeting of EWS/FLI-1 in Ewing's sarcoma. Clin. Cancer Res. 2006;12(22):6781–6790. doi: 10.1158/1078-0432.CCR-06-0609. [DOI] [PubMed] [Google Scholar]
- 69.Asami S., Chin M., Shichino H., Yoshida Y., Nemoto N., Mugishima H., Suzuki T. Treatment of Ewing's sarcoma using an antisense oligodeoxynucleotide to regulate the cell cycle. Biol. Pharm. Bull. 2008;31(3):391–394. doi: 10.1248/bpb.31.391. [DOI] [PubMed] [Google Scholar]
- 70.Maksimenko A., Lambert G., Bertrand J.R., Fattal E., Couvreur P., Malvy C. Therapeutic potentialities of EWS-Fli-1 mRNA-targeted vectorized antisense oligonucleotides. Ann. N. Y. Acad. Sci. 2003;1002:72–77. doi: 10.1196/annals.1281.017. [DOI] [PubMed] [Google Scholar]
- 71.Maksimenko A., Polard V., Villemeur M., Elhamess H., Couvreur P., Bertrand J.R., Aboubakar M., Gottikh M., Malvy C. In vivo potentialities of EWS-Fli-1 targeted antisense oligonucleotides-nanospheres complexes. Ann. N. Y. Acad. Sci. 2005;1058:52–61. doi: 10.1196/annals.1359.010. [DOI] [PubMed] [Google Scholar]
- 72.Dohjima T., Lee N.S., Li H., Ohno T., Rossi J.J. Small interfering RNAs expressed from a Pol III promoter suppress the EWS/Fli-1 transcript in an Ewing sarcoma cell line. Mol. Therapy. 2003;7(6):811–816. doi: 10.1016/s1525-0016(03)00101-1. [DOI] [PubMed] [Google Scholar]
- 73.Chansky H.A., Barahmand-Pour F., Mei Q., Kahn-Farooqi W., Zielinska-Kwiatkowska A., Blackburn M., Chansky K., Conrad E.U., 3rd, Bruckner J.D., Greenlee T.K., Yang L. Targeting of EWS/FLI-1 by RNA interference attenuates the tumor phenotype of Ewing's sarcoma cells in vitro. J. Orthopaedic Res. 2004;22(4):910–917. doi: 10.1016/j.orthres.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 74.Hu-Lieskovan S., Heidel J.D., Bartlett D.W., Davis M.E., Triche T.J. Sequence-specific knockdown of EWS-FLI1 by targeted, nonviral delivery of small interfering RNA inhibits tumor growth in a murine model of metastatic Ewing's sarcoma. Cancer Res. 2005;65(19):8984–8992. doi: 10.1158/0008-5472.CAN-05-0565. [DOI] [PubMed] [Google Scholar]
- 75.Prieur A., Tirode F., Cohen P., Delattre O. EWS/FLI-1 silencing and gene profiling of Ewing cells reveal downstream oncogenic pathways and a crucial role for repression of insulin-like growth factor binding protein 3. Mol. Cell. Biol. 2004;24(16):7275–7283. doi: 10.1128/MCB.24.16.7275-7283.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mendiola M., Carrillo J., Garcia E., Lalli E., Hernandez T., de Alava E., Tirode F., Delattre O., Garcia-Miguel P., Lopez-Barea F., Pestana A., Alonso J. The orphan nuclear receptor DAX1 is up-regulated by the EWS/FLI oncoprotein and is highly expressed in Ewing tumors. Int. J. Cancer. 2006;118(6):1381–1389. doi: 10.1002/ijc.21578. [DOI] [PubMed] [Google Scholar]
- 77.Grunewald T.G., Diebold I., Esposito I., Plehm S., Hauer K., Thiel U., da Silva-Buttkus P., Neff F., Unland R., Muller-Tidow C., Zobywalski C., Lohrig K., Lewandrowski U., Sickmann A., Prazeres da Costa O., Gorlach A., Cossarizza A., Butt E., Richter G.H., Burdach S. STEAP1 is associated with the invasive and oxidative stress phenotype of Ewing tumors. Mol. Cancer Res. 2012;10(1):52–65. doi: 10.1158/1541-7786.MCR-11-0524. [DOI] [PubMed] [Google Scholar]
- 78.Dallmayer M., Li J., Ohmura S., Alba Rubio R., Baldauf M.C., Holting T.L.B., Musa J., Knott M.M.L., Stein S., Cidre-Aranaz F., Wehweck F.S., Romero-Perez L., Gerke J.S., Orth M.F., Marchetto A., Kirchner T., Bach H., Sannino G., Grunewald T.G.P. Targeting the CALCB/RAMP1 axis inhibits growth of Ewing sarcoma. Cell Death Dis. 2019;10(2):116. doi: 10.1038/s41419-019-1372-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hu-Lieskovan S., Zhang J., Wu L., Shimada H., Schofield D.E., Triche T.J. EWS-FLI1 fusion protein up-regulates critical genes in neural crest development and is responsible for the observed phenotype of Ewing's family of tumors. Cancer Res. 2005;65(11):4633–4644. doi: 10.1158/0008-5472.CAN-04-2857. [DOI] [PubMed] [Google Scholar]
- 80.Elhamess H., Bertrand J.R., Maccario J., Maksimenko A., Malvy C. Antitumor vectorized oligonucleotides in a model of ewing sarcoma: unexpected role of nanoparticles. Oligonucleotides. 2009;19(3):255–264. doi: 10.1089/oli.2009.0197. [DOI] [PubMed] [Google Scholar]
- 81.Volpi S., Cancelli U., Neri M., Corradini R. Multifunctional delivery systems for peptide nucleic acids. Pharmaceuticals (Basel) 2020;14(1) doi: 10.3390/ph14010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chaturvedi A., Hoffman L.M., Welm A.L., Lessnick S.L., Beckerle M.C. The EWS/FLI oncogene drives changes in cellular morphology, adhesion, and migration in ewing sarcoma. Genes Cancer. 2012;3(2):102–116. doi: 10.1177/1947601912457024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Franzetti G.A., Laud-Duval K., van der Ent W., Brisac A., Irondelle M., Aubert S., Dirksen U., Bouvier C., de Pinieux G., Snaar-Jagalska E., Chavrier P., Delattre O. Cell-to-cell heterogeneity of EWSR1-FLI1 activity determines proliferation/migration choices in Ewing sarcoma cells. Oncogene. 2017;36(25):3505–3514. doi: 10.1038/onc.2016.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Surdez D., Zaidi S., Grossetête S., Laud-Duval K., Ferre A.S., Mous L., Vourc'h T., Tirode F., Pierron G., Raynal V., Baulande S., Brunet E., Hill V., Delattre O. STAG2 mutations alter CTCF-anchored loop extrusion, reduce cis-regulatory interactions and EWSR1-FLI1 activity in Ewing sarcoma. Cancer Cell. 2021;39(6):810–826.e9. doi: 10.1016/j.ccell.2021.04.001. [DOI] [PubMed] [Google Scholar]
- 85.Hawkins A.G., Pedersen E.A., Treichel S., Temprine K., Sperring C., Read J.A., Magnuson B., Chugh R., Lawlor E.R. Wnt/β-catenin-activated Ewing sarcoma cells promote the angiogenic switch. JCI Insight. 2020;5(13) doi: 10.1172/jci.insight.135188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Katschnig A.M., Kauer M.O., Schwentner R., Tomazou E.M., Mutz C.N., Linder M., Sibilia M., Alonso J., Aryee D.N.T., Kovar H. EWS-FLI1 perturbs MRTFB/YAP-1/TEAD target gene regulation inhibiting cytoskeletal autoregulatory feedback in Ewing sarcoma. Oncogene. 2017;36(43):5995–6005. doi: 10.1038/onc.2017.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Adane B., Alexe G., Seong B.K.A., Lu D., Hwang E.E., Hnisz D., Lareau C.A., Ross L., Lin S., Dela Cruz F.S., Richardson M., Weintraub A.S., Wang S., Iniguez A.B., Dharia N.V., Conway A.S., Robichaud A.L., Tanenbaum B., Krill-Burger J.M., Vazquez F., Schenone M., Berman J.N., Kung A.L., Carr S.A., Aryee M.J., Young R.A., Crompton B.D., Stegmaier K. STAG2 loss rewires oncogenic and developmental programs to promote metastasis in Ewing sarcoma. Cancer Cell. 2021;39(6):827–844.e10. doi: 10.1016/j.ccell.2021.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fuchs B., Inwards C.Y., Janknecht R. Vascular endothelial growth factor expression is up-regulated by EWS-ETS oncoproteins and Sp1 and may represent an independent predictor of survival in Ewing's sarcoma. Clin. Cancer Res. 2004;10(4):1344–1353. doi: 10.1158/1078-0432.ccr-03-0038. [DOI] [PubMed] [Google Scholar]
- 89.Lagares-Tena L., Garcia-Monclus S., Lopez-Alemany R., Almacellas-Rabaiget O., Huertas-Martinez J., Sainz-Jaspeado M., Mateo-Lozano S., Rodriguez-Galindo C., Rello-Varona S., Herrero-Martin D., Tirado O.M. Caveolin-1 promotes Ewing sarcoma metastasis regulating MMP-9 expression through MAPK/ERK pathway. Oncotarget. 2016;7(35):56889–56903. doi: 10.18632/oncotarget.10872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lagares-Tena L., García-Monclús S., López-Alemany R., Almacellas-Rabaiget O., Huertas-Martínez J., Sáinz-Jaspeado M., Mateo-Lozano S., Rodríguez-Galindo C., Rello-Varona S., Herrero-Martín D., Tirado O.M. Caveolin-1 promotes Ewing sarcoma metastasis regulating MMP-9 expression through MAPK/ERK pathway. Oncotarget. 2016;7(35):56889–56903. doi: 10.18632/oncotarget.10872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Grunewald T.G., Diebold I., Esposito I., Plehm S., Hauer K., Thiel U., da Silva-Buttkus P., Neff F., Unland R., Müller-Tidow C., Zobywalski C., Lohrig K., Lewandrowski U., Sickmann A., Prazeres da Costa O., Görlach A., Cossarizza A., Butt E., Richter G.H., Burdach S. STEAP1 is associated with the invasive and oxidative stress phenotype of Ewing tumors. Mol. Cancer Res. 2012;10(1):52–65. doi: 10.1158/1541-7786.MCR-11-0524. [DOI] [PubMed] [Google Scholar]
- 92.von Heyking K., Calzada-Wack J., Göllner S., Neff F., Schmidt O., Hensel T., Schirmer D., Fasan A., Esposito I., Müller-Tidow C., Sorensen P.H., Burdach S., Richter G.H.S. The endochondral bone protein CHM1 sustains an undifferentiated, invasive phenotype, promoting lung metastasis in Ewing sarcoma. Mol. Oncol. 2017;11(9):1288–1301. doi: 10.1002/1878-0261.12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.He S., Huang Q., Hu J., Li L., Xiao Y., Yu H., Han Z., Wang T., Zhou W., Wei H., Xiao J. EWS-FLI1-mediated tenascin-C expression promotes tumour progression by targeting MALAT1 through integrin α5β1-mediated YAP activation in Ewing sarcoma. Br. J. Cancer. 2019;121(11):922–933. doi: 10.1038/s41416-019-0608-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hong S.H., Tilan J.U., Galli S., Izycka-Swieszewska E., Polk T., Horton M., Mahajan A., Christian D., Jenkins S., Acree R., Connors K., Ledo P., Lu C., Lee Y.C., Rodriguez O., Toretsky J.A., Albanese C., Kitlinska J. High neuropeptide Y release associates with Ewing sarcoma bone dissemination - in vivo model of site-specific metastases. Oncotarget. 2015;6(9):7151–7165. doi: 10.18632/oncotarget.3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Luo W., Xu C., Ayello J., Dela Cruz F., Rosenblum J.M., Lessnick S.L., Cairo M.S. Protein phosphatase 1 regulatory subunit 1A in ewing sarcoma tumorigenesis and metastasis. Oncogene. 2018;37(6):798–809. doi: 10.1038/onc.2017.378. [DOI] [PubMed] [Google Scholar]
- 96.Richter G.H., Plehm S., Fasan A., Rössler S., Unland R., Bennani-Baiti I.M., Hotfilder M., Löwel D., von Luettichau I., Mossbrugger I., Quintanilla-Martinez L., Kovar H., Staege M.S., Müller-Tidow C., Burdach S. EZH2 is a mediator of EWS/FLI driven tumor growth and metastasis blocking endothelial and neuro-ectodermal differentiation. Proc. Natl. Acad. Sci. USA. 2009;106(13):5324–5329. doi: 10.1073/pnas.0810759106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gierisch M.E., Pfistner F., Lopez-Garcia L.A., Harder L., Schafer B.W., Niggli F.K. Proteasomal degradation of the EWS-FLI1 fusion protein is regulated by a single lysine residue. J. Biol. Chem. 2016;291(52):26922–26933. doi: 10.1074/jbc.M116.752063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gierisch M.E., Pedot G., Walser F., Lopez-Garcia L.A., Jaaks P., Niggli F.K., Schafer B.W. USP19 deubiquitinates EWS-FLI1 to regulate Ewing sarcoma growth. Sci. Rep. 2019;9(1):951. doi: 10.1038/s41598-018-37264-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Su S., Chen J., Jiang Y., Wang Y., Vital T., Zhang J., Laggner C., Nguyen K.T., Zhu Z., Prevatte A.W., Barker N.K., Herring L.E., Davis I.J., Liu P. SPOP and OTUD7A control EWS-FLI1 protein stability to govern ewing sarcoma growth. Adv. Sci. (Weinh.) 2021;8(14) doi: 10.1002/advs.202004846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.B.K.A. Seong, N.V. Dharia, S. Lin, K.A. Donovan, S. Chong, A. Robichaud, A. Conway, A. Hamze, L. Ross, G. Alexe, B. Adane, B. Nabet, F.M. Ferguson, B. Stolte, E.J. Wang, J. Sun, X. Darzacq, F. Piccioni, N.S. Gray, E.S. Fischer, K. Stegmaier, TRIM8 modulates the EWS/FLI oncoprotein to promote survival in Ewing sarcoma, Cancer cell 39(9) (2021) 1262-1278 e7. [DOI] [PMC free article] [PubMed]
- 101.Ambati S.R., Lopes E.C., Kosugi K., Mony U., Zehir A., Shah S.K., Taldone T., Moreira A.L., Meyers P.A., Chiosis G., Moore M.A. Pre-clinical efficacy of PU-H71, a novel HSP90 inhibitor, alone and in combination with bortezomib in Ewing sarcoma. Mol. Oncol. 2014;8(2):323–336. doi: 10.1016/j.molonc.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pessetto Z.Y., Chen B., Alturkmani H., Hyter S., Flynn C.A., Baltezor M., Ma Y., Rosenthal H.G., Neville K.A., Weir S.J., Butte A.J., Godwin A.K. In silico and in vitro drug screening identifies new therapeutic approaches for Ewing sarcoma. Oncotarget. 2017;8(3):4079–4095. doi: 10.18632/oncotarget.13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ma Y., Baltezor M., Rajewski L., Crow J., Samuel G., Staggs V.S., Chastain K.M., Toretsky J.A., Weir S.J., Godwin A.K. Targeted inhibition of histone deacetylase leads to suppression of Ewing sarcoma tumor growth through an unappreciated EWS-FLI1/HDAC3/HSP90 signaling axis. J. Mol. Med. (Berlin, Germany) 2019;97(7):957–972. doi: 10.1007/s00109-019-01782-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Paiva S.L., Crews C.M. Targeted protein degradation: elements of PROTAC design. Curr. Opin. Chem. Biol. 2019;50:111–119. doi: 10.1016/j.cbpa.2019.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xia L., Liu W., Song Y., Zhu H., Duan Y. The present and future of novel protein degradation technology. Curr. Top. Med. Chem. 2019;19(20):1784–1788. doi: 10.2174/1568026619666191011162955. [DOI] [PubMed] [Google Scholar]
- 106.Ducker C., Shaw P.E. Ubiquitin-mediated control of ETS transcription factors: roles in cancer and development. Int. J. Mol. Sci. 2021;22(10) doi: 10.3390/ijms22105119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gollavilli P.N., Pawar A., Wilder-Romans K., Natesan R., Engelke C.G., Dommeti V.L., Krishnamurthy P.M., Nallasivam A., Apel I.J., Xu T., Qin Z.S., Feng F.Y., Asangani I.A. EWS/ETS-driven ewing sarcoma requires BET bromodomain proteins. Cancer Res. 2018;78(16):4760–4773. doi: 10.1158/0008-5472.CAN-18-0484. [DOI] [PubMed] [Google Scholar]
- 108.Sun X., Wang J., Yao X., Zheng W., Mao Y., Lan T., Wang L., Sun Y., Zhang X., Zhao Q., Zhao J., Xiao R.P., Zhang X., Ji G., Rao Y. A chemical approach for global protein knockdown from mice to non-human primates. Cell Discov. 2019;5:10. doi: 10.1038/s41421-018-0079-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Elzi D.J., Song M., Hakala K., Weintraub S.T., Shiio Y. Proteomic analysis of the EWS-Fli-1 interactome reveals the role of the lysosome in EWS-Fli-1 turnover. J. Proteome Res. 2014;13(8):3783–3791. doi: 10.1021/pr500387m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Burdach S., Plehm S., Unland R., Dirksen U., Borkhardt A., Staege M.S., Müller-Tidow C., Richter G.H. Epigenetic maintenance of stemness and malignancy in peripheral neuroectodermal tumors by EZH2. Cell Cycle. 2009;8(13):1991–1996. doi: 10.4161/cc.8.13.8929. [DOI] [PubMed] [Google Scholar]
- 111.von Levetzow C., Jiang X., Gwye Y., von Levetzow G., Hung L., Cooper A., Hsu J.H., Lawlor E.R. Modeling initiation of Ewing sarcoma in human neural crest cells. PLoS ONE. 2011;6(4) doi: 10.1371/journal.pone.0019305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Svoboda L.K., Harris A., Bailey N.J., Schwentner R., Tomazou E., von Levetzow C., Magnuson B., Ljungman M., Kovar H., Lawlor E.R. Overexpression of HOX genes is prevalent in Ewing sarcoma and is associated with altered epigenetic regulation of developmental transcription programs. Epigenetics. 2014;9(12):1613–1625. doi: 10.4161/15592294.2014.988048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Svoboda L.K., Bailey N., Van Noord R.A., Krook M.A., Harris A., Cramer C., Jasman B., Patel R.M., Thomas D., Borkin D., Cierpicki T., Grembecka J., Lawlor E.R. Tumorigenicity of Ewing sarcoma is critically dependent on the trithorax proteins MLL1 and menin. Oncotarget. 2017;8(1):458–471. doi: 10.18632/oncotarget.13444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tirode F., Laud-Duval K., Prieur A., Delorme B., Charbord P., Delattre O. Mesenchymal stem cell features of Ewing tumors. Cancer Cell. 2007;11(5):421–429. doi: 10.1016/j.ccr.2007.02.027. [DOI] [PubMed] [Google Scholar]
- 115.Guillon N., Tirode F., Boeva V., Zynovyev A., Barillot E., Delattre O. The oncogenic EWS-FLI1 protein binds in vivo GGAA microsatellite sequences with potential transcriptional activation function. PLoS ONE. 2009;4(3) doi: 10.1371/journal.pone.0004932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Patel M., Simon J.M., Iglesia M.D., Wu S.B., McFadden A.W., Lieb J.D., Davis I.J. Tumor-specific retargeting of an oncogenic transcription factor chimera results in dysregulation of chromatin and transcription. Genome Res. 2012;22(2):259–270. doi: 10.1101/gr.125666.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tomazou E.M., Sheffield N.C., Schmidl C., Schuster M., Schonegger A., Datlinger P., Kubicek S., Bock C., Kovar H. Epigenome mapping reveals distinct modes of gene regulation and widespread enhancer reprogramming by the oncogenic fusion protein EWS-FLI1. Cell Reports. 2015;10(7):1082–1095. doi: 10.1016/j.celrep.2015.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sheffield N.C., Pierron G., Klughammer J., Datlinger P., Schonegger A., Schuster M., Hadler J., Surdez D., Guillemot D., Lapouble E., Freneaux P., Champigneulle J., Bouvier R., Walder D., Ambros I.M., Hutter C., Sorz E., Amaral A.T., de Alava E., Schallmoser K., Strunk D., Rinner B., Liegl-Atzwanger B., Huppertz B., Leithner A., de Pinieux G., Terrier P., Laurence V., Michon J., Ladenstein R., Holter W., Windhager R., Dirksen U., Ambros P.F., Delattre O., Kovar H., Bock C., Tomazou E.M. DNA methylation heterogeneity defines a disease spectrum in Ewing sarcoma. Nat. Med. 2017;23(3):386–395. doi: 10.1038/nm.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Patel N., Black J., Chen X., Marcondes A.M., Grady W.M., Lawlor E.R., Borinstein S.C. DNA methylation and gene expression profiling of ewing sarcoma primary tumors reveal genes that are potential targets of epigenetic inactivation. Sarcoma. 2012;2012 doi: 10.1155/2012/498472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Avigad S., Shukla S., Naumov I., Cohen I.J., Ash S., Meller I., Kollender Y., Issakov J., Yaniv I. Aberrant methylation and reduced expression of RASSF1A in Ewing sarcoma. Pediatr. Blood Cancer. 2009;53(6):1023–1028. doi: 10.1002/pbc.22115. [DOI] [PubMed] [Google Scholar]
- 121.Nestheide S., Bridge J.A., Barnes M., Frayer R., Sumegi J. Pharmacologic inhibition of epigenetic modification reveals targets of aberrant promoter methylation in Ewing sarcoma. Pediatr. Blood Cancer. 2013;60(9):1437–1446. doi: 10.1002/pbc.24526. [DOI] [PubMed] [Google Scholar]
- 122.Huertas-Martínez J., Court F., Rello-Varona S., Herrero-Martín D., Almacellas-Rabaiget O., Sáinz-Jaspeado M., Garcia-Monclús S., Lagares-Tena L., Buj R., Hontecillas-Prieto L., Sastre A., Azorin D., Sanjuan X., López-Alemany R., Moran S., Roma J., Gallego S., Mora J., García Del Muro X., Giangrande P.H., Peinado M.A., Alonso J., de Alava E., Monk D., Esteller M., Tirado O.M. DNA methylation profiling identifies PTRF/Cavin-1 as a novel tumor suppressor in Ewing sarcoma when co-expressed with caveolin-1. Cancer Lett. 2017;386:196–207. doi: 10.1016/j.canlet.2016.11.020. [DOI] [PubMed] [Google Scholar]
- 123.Hurtubise A., Bernstein M.L., Momparler R.L. Preclinical evaluation of the antineoplastic action of 5-aza-2'-deoxycytidine and different histone deacetylase inhibitors on human Ewing's sarcoma cells. Cancer Cell Int. 2008;8:16. doi: 10.1186/1475-2867-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jaboin J., Wild J., Hamidi H., Khanna C., Kim C.J., Robey R., Bates S.E., Thiele C.J. MS-27-275, an inhibitor of histone deacetylase, has marked in vitro and in vivo antitumor activity against pediatric solid tumors. Cancer Res. 2002;62(21):6108–6115. [PubMed] [Google Scholar]
- 125.Sonnemann J., Trommer N., Becker S., Wittig S., Grauel D., Palani C.D., Beck J.F. Histone deacetylase inhibitor-mediated sensitization to TRAIL-induced apoptosis in childhood malignancies is not associated with upregulation of TRAIL receptor expression, but with potentiated caspase-8 activation. Cancer Biol. Ther. 2012;13(6):417–424. doi: 10.4161/cbt.19293. [DOI] [PubMed] [Google Scholar]
- 126.El-Naggar A.M., Somasekharan S.P., Wang Y., Cheng H., Negri G.L., Pan M., Wang X.Q., Delaidelli A., Rafn B., Cran J., Zhang F., Zhang H., Colborne S., Gleave M., Mandinova A., Kedersha N., Hughes C.S., Surdez D., Delattre O., Wang Y., Huntsman D.G., Morin G.B., Sorensen P.H. Class I HDAC inhibitors enhance YB-1 acetylation and oxidative stress to block sarcoma metastasis. EMBO Rep. 2019;20(12) doi: 10.15252/embr.201948375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bennani-Baiti I.M., Machado I., Llombart-Bosch A., Kovar H. Lysine-specific demethylase 1 (LSD1/KDM1A/AOF2/BHC110) is expressed and is an epigenetic drug target in chondrosarcoma, Ewing's sarcoma, osteosarcoma, and rhabdomyosarcoma. Human Pathol. 2012;43(8):1300–1307. doi: 10.1016/j.humpath.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 128.Sankar S., Theisen E.R., Bearss J., Mulvihill T., Hoffman L.M., Sorna V., Beckerle M.C., Sharma S., Lessnick S.L. Reversible LSD1 inhibition interferes with global EWS/ETS transcriptional activity and impedes Ewing sarcoma tumor growth. Clin. Cancer Res. 2014;20(17):4584–4597. doi: 10.1158/1078-0432.CCR-14-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Theisen E.R., Selich-Anderson J., Miller K.R., Tanner J.M., Taslim C., Pishas K.I., Sharma S., Lessnick S.L. Chromatin profiling reveals relocalization of lysine-specific demethylase 1 by an oncogenic fusion protein. Epigenetics. 2021;16(4):405–424. doi: 10.1080/15592294.2020.1805678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Garcia-Dominguez D.J., Hontecillas-Prieto L., Rodriguez-Nunez P., Pascual-Pasto G., Vila-Ubach M., Garcia-Mejias R., Robles M.J., Tirado O.M., Mora J., Carcaboso A.M., de Alava E. The combination of epigenetic drugs SAHA and HCI-2509 synergistically inhibits EWS-FLI1 and tumor growth in Ewing sarcoma. Oncotarget. 2018;9(59):31397–31410. doi: 10.18632/oncotarget.25829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Welch D., Kahen E., Fridley B., Brohl A.S., Cubitt C.L., Reed D.R. Small molecule inhibition of lysine-specific demethylase 1 (LSD1) and histone deacetylase (HDAC) alone and in combination in Ewing sarcoma cell lines. PLoS ONE. 2019;14(9) doi: 10.1371/journal.pone.0222228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Parrish J.K., McCann T.S., Sechler M., Sobral L.M., Ren W., Jones K.L., Tan A.C., Jedlicka P. The Jumonji-domain histone demethylase inhibitor JIB-04 deregulates oncogenic programs and increases DNA damage in Ewing Sarcoma, resulting in impaired cell proliferation and survival, and reduced tumor growth. Oncotarget. 2018;9(69):33110–33123. doi: 10.18632/oncotarget.26011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.McCann T.S., Parrish J.K., Hsieh J., Sechler M., Sobral L.M., Self C., Jones K.L., Goodspeed A., Costello J.C., Jedlicka P. KDM5A and PHF2 positively control expression of pro-metastatic genes repressed by EWS/FLI, and promote growth and metastatic properties in Ewing sarcoma. Oncotarget. 2020;11(43):3818–3831. doi: 10.18632/oncotarget.27737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bechter O., Schoffski P. Make your best BET: The emerging role of BET inhibitor treatment in malignant tumors. Pharmacol. Ther. 2020 doi: 10.1016/j.pharmthera.2020.107479. [DOI] [PubMed] [Google Scholar]
- 135.Hensel T., Giorgi C., Schmidt O., Calzada-Wack J., Neff F., Buch T., Niggli F.K., Schafer B.W., Burdach S., Richter G.H. Targeting the EWS-ETS transcriptional program by BET bromodomain inhibition in Ewing sarcoma. Oncotarget. 2016;7(2):1451–1463. doi: 10.18632/oncotarget.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Jacques C., Lamoureux F., Baud'huin M., Rodriguez Calleja L., Quillard T., Amiaud J., Tirode F., Redini F., Bradner J.E., Heymann D., Ory B. Targeting the epigenetic readers in Ewing sarcoma inhibits the oncogenic transcription factor EWS/FLI. Oncotarget. 2016;7(17):24125–24140. doi: 10.18632/oncotarget.8214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bid H.K., Phelps D.A., Xaio L., Guttridge D.C., Lin J., London C., Baker L.H., Mo X., Houghton P.J. The Bromodomain BET Inhibitor JQ1 Suppresses Tumor Angiogenesis in Models of Childhood Sarcoma. Mol. Cancer Ther. 2016;15(5):1018–1028. doi: 10.1158/1535-7163.MCT-15-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Loganathan S.N., Tang N., Fleming J.T., Ma Y., Guo Y., Borinstein S.C., Chiang C., Wang J. BET bromodomain inhibitors suppress EWS-FLI1-dependent transcription and the IGF1 autocrine mechanism in Ewing sarcoma. Oncotarget. 2016;7(28):43504–43517. doi: 10.18632/oncotarget.9762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Postel-Vinay S., Véron A.S., Tirode F., Pierron G., Reynaud S., Kovar H., Oberlin O., Lapouble E., Ballet S., Lucchesi C., Kontny U., González-Neira A., Picci P., Alonso J., Patino-Garcia A., de Paillerets B.B., Laud K., Dina C., Froguel P., Clavel-Chapelon F., Doz F., Michon J., Chanock S.J., Thomas G., Cox D.G., Delattre O. Common variants near TARDBP and EGR2 are associated with susceptibility to Ewing sarcoma. Nat. Genet. 2012;44(3):323–327. doi: 10.1038/ng.1085. [DOI] [PubMed] [Google Scholar]
- 140.Grünewald T.G., Bernard V., Gilardi-Hebenstreit P., Raynal V., Surdez D., Aynaud M.M., Mirabeau O., Cidre-Aranaz F., Tirode F., Zaidi S., Perot G., Jonker A.H., Lucchesi C., Le Deley M.C., Oberlin O., Marec-Bérard P., Véron A.S., Reynaud S., Lapouble E., Boeva V., Rio Frio T., Alonso J., Bhatia S., Pierron G., Cancel-Tassin G., Cussenot O., Cox D.G., Morton L.M., Machiela M.J., Chanock S.J., Charnay P., Delattre O. Chimeric EWSR1-FLI1 regulates the Ewing sarcoma susceptibility gene EGR2 via a GGAA microsatellite. Nat. Genet. 2015;47(9):1073–1078. doi: 10.1038/ng.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Riggi N., Cironi L., Provero P., Suva M.L., Kaloulis K., Garcia-Echeverria C., Hoffmann F., Trumpp A., Stamenkovic I. Development of Ewing's sarcoma from primary bone marrow-derived mesenchymal progenitor cells. Cancer Res. 2005;65(24):11459–11468. doi: 10.1158/0008-5472.CAN-05-1696. [DOI] [PubMed] [Google Scholar]
- 142.Riggi N., Suva M.L., Suva D., Cironi L., Provero P., Tercier S., Joseph J.M., Stehle J.C., Baumer K., Kindler V., Stamenkovic I. EWS-FLI-1 expression triggers a Ewing's sarcoma initiation program in primary human mesenchymal stem cells. Cancer Res. 2008;68(7):2176–2185. doi: 10.1158/0008-5472.CAN-07-1761. [DOI] [PubMed] [Google Scholar]
- 143.Torchia E.C., Jaishankar S., Baker S.J. Ewing tumor fusion proteins block the differentiation of pluripotent marrow stromal cells. Cancer Res. 2003;63(13):3464–3468. [PubMed] [Google Scholar]
- 144.Hancock J.D., Lessnick S.L. A transcriptional profiling meta-analysis reveals a core EWS-FLI gene expression signature. Cell Cycle. 2008;7(2):250–256. doi: 10.4161/cc.7.2.5229. [DOI] [PubMed] [Google Scholar]
- 145.Kinsey M., Smith R., Lessnick S.L. NR0B1 is required for the oncogenic phenotype mediated by EWS/FLI in Ewing's sarcoma. Mol. Cancer Res. 2006;4(11):851–859. doi: 10.1158/1541-7786.MCR-06-0090. [DOI] [PubMed] [Google Scholar]
- 146.Kinsey M., Smith R., Iyer A.K., McCabe E.R., Lessnick S.L. EWS/FLI and its downstream target NR0B1 interact directly to modulate transcription and oncogenesis in Ewing's sarcoma. Cancer Res. 2009;69(23):9047–9055. doi: 10.1158/0008-5472.CAN-09-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Johnson K.M., Mahler N.R., Saund R.S., Theisen E.R., Taslim C., Callender N.W., Crow J.C., Miller K.R., Lessnick S.L. Role for the EWS domain of EWS/FLI in binding GGAA-microsatellites required for Ewing sarcoma anchorage independent growth. Proc. Natl. Acad. Sci. USA. 2017;114(37):9870–9875. doi: 10.1073/pnas.1701872114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Gangwal K., Close D., Enriquez C.A., Hill C.P., Lessnick S.L. Emergent Properties of EWS/FLI Regulation via GGAA Microsatellites in Ewing's Sarcoma. Genes & cancer. 2010;1(2):177–187. doi: 10.1177/1947601910361495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Komori T. Molecular mechanism of Runx2-dependent bone development. Mol. Cells. 2020;43(2):168–175. doi: 10.14348/molcells.2019.0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zhao H., Feng J., Ho T.V., Grimes W., Urata M., Chai Y. The suture provides a niche for mesenchymal stem cells of craniofacial bones. Nat. Cell Biol. 2015;17(4):386–396. doi: 10.1038/ncb3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zhao H., Feng J., Seidel K., Shi S., Klein O., Sharpe P., Chai Y. Secretion of shh by a neurovascular bundle niche supports mesenchymal stem cell homeostasis in the adult mouse incisor. Cell Stem Cell. 2014;14(2):160–173. doi: 10.1016/j.stem.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Beauchamp E., Bulut G., Abaan O., Chen K., Merchant A., Matsui W., Endo Y., Rubin J.S., Toretsky J., Uren A. GLI1 is a direct transcriptional target of EWS-FLI1 oncoprotein. J. Biol. Chem. 2009;284(14):9074–9082. doi: 10.1074/jbc.M806233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zwerner J.P., Joo J., Warner K.L., Christensen L., Hu-Lieskovan S., Triche T.J., May W.A. The EWS/FLI oncogenic transcription factor deregulates GLI1. Oncogene. 2008;27(23):3282–3291. doi: 10.1038/sj.onc.1210991. [DOI] [PubMed] [Google Scholar]
- 154.Beauchamp E.M., Ringer L., Bulut G., Sajwan K.P., Hall M.D., Lee Y.C., Peaceman D., Ozdemirli M., Rodriguez O., Macdonald T.J., Albanese C., Toretsky J.A., Uren A. Arsenic trioxide inhibits human cancer cell growth and tumor development in mice by blocking Hedgehog/GLI pathway. J. Clin. Investig. 2011;121(1):148–160. doi: 10.1172/JCI42874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kim J., Lee J.J., Kim J., Gardner D., Beachy P.A. Arsenic antagonizes the Hedgehog pathway by preventing ciliary accumulation and reducing stability of the Gli2 transcriptional effector. Proc. Natl. Acad. Sci. USA. 2010;107(30):13432–13437. doi: 10.1073/pnas.1006822107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Li B., Giambelli C., Tang B., Winterbottom E., Long J., Jin K., Wang Z., Fei D.L., Nguyen D.M., Athar M., Wang B., Subbarayan P.R., Wang L., Rai P., Ardalan B., Capobianco A.J., Robbins D.J. Arsenic attenuates GLI signaling, increasing or decreasing its transcriptional program in a context-dependent manner. Mol. Pharmacol. 2016;89(2):226–232. doi: 10.1124/mol.115.100867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Smith M.A., Kang M.H., Reynolds C.P., Kurmasheva R.T., Alexander D., Billups C.A., Toretsky J.A., Houghton P.J. Evaluation of arsenic trioxide by the pediatric preclinical testing program with a focus on Ewing sarcoma. Pediatr. Blood Cancer. 2012;59(4):753–755. doi: 10.1002/pbc.23391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Schalla M.A., Tache Y., Stengel A. Neuroendocrine Peptides of the Gut and Their Role in the Regulation of Food Intake. Compr Physiol. 2021;11(2):1679–1730. doi: 10.1002/cphy.c200007. [DOI] [PubMed] [Google Scholar]
- 159.Rehfeld J.F. Cholecystokinin expression in tumors: biogenetic and diagnostic implications. Future Oncol. (London, England) 2016;12(18):2135–2147. doi: 10.2217/fon-2015-0053. [DOI] [PubMed] [Google Scholar]
- 160.Reubi J.C., Koefoed P., Hansen T., Stauffer E., Rauch D., Nielsen F.C., Rehfeld J.F. Procholecystokinin as marker of human Ewing sarcomas. Clin. Cancer Res. 2004;10(16):5523–5530. doi: 10.1158/1078-0432.CCR-1015-03. [DOI] [PubMed] [Google Scholar]
- 161.Carrillo J., Garcia-Aragoncillo E., Azorin D., Agra N., Sastre A., Gonzalez-Mediero I., Garcia-Miguel P., Pestana A., Gallego S., Segura D., Alonso J. Cholecystokinin down-regulation by RNA interference impairs Ewing tumor growth. Clin. Cancer Res. 2007;13(8):2429–2440. doi: 10.1158/1078-0432.CCR-06-1762. [DOI] [PubMed] [Google Scholar]
- 162.Carrillo J., Agra N., Fernandez N., Pestana A., Alonso J. Devazepide, a nonpeptide antagonist of CCK receptors, induces apoptosis and inhibits Ewing tumor growth. Anticancer Drugs. 2009;20(7):527–533. doi: 10.1097/CAD.0b013e32832c3a4f. [DOI] [PubMed] [Google Scholar]
- 163.Guo Y.J., Pan W.W., Liu S.B., Shen Z.F., Xu Y., Hu L.L. ERK/MAPK signalling pathway and tumorigenesis. Exp. Therapeutic Med. 2020;19(3):1997–2007. doi: 10.3892/etm.2020.8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Silvany R.E., Eliazer S., Wolff N.C., Ilaria R.L., Jr. Interference with the constitutive activation of ERK1 and ERK2 impairs EWS/FLI-1-dependent transformation. Oncogene. 2000;19(39):4523–4530. doi: 10.1038/sj.onc.1203811. [DOI] [PubMed] [Google Scholar]
- 165.Zenali M.J., Zhang P.L., Bendel A.E., Brown R.E. Morphoproteomic confirmation of constitutively activated mTOR, ERK, and NF-kappaB pathways in Ewing family of tumors. Ann. Clin. Lab. Sci. 2009;39(2):160–166. [PubMed] [Google Scholar]
- 166.Tirado O.M., Mateo-Lozano S., Villar J., Dettin L.E., Llort A., Gallego S., Ban J., Kovar H., Notario V. Caveolin-1 (CAV1) is a target of EWS/FLI-1 and a key determinant of the oncogenic phenotype and tumorigenicity of Ewing's sarcoma cells. Cancer Res. 2006;66(20):9937–9947. doi: 10.1158/0008-5472.CAN-06-0927. [DOI] [PubMed] [Google Scholar]
- 167.Cidre-Aranaz F., Grunewald T.G., Surdez D., Garcia-Garcia L., Carlos Lazaro J., Kirchner T., Gonzalez-Gonzalez L., Sastre A., Garcia-Miguel P., Lopez-Perez S.E., Monzon S., Delattre O., Alonso J. EWS-FLI1-mediated suppression of the RAS-antagonist Sprouty 1 (SPRY1) confers aggressiveness to Ewing sarcoma. Oncogene. 2017;36(6):766–776. doi: 10.1038/onc.2016.244. [DOI] [PubMed] [Google Scholar]
- 168.Ye C., Yu X., Liu X., Dai M., Zhang B. miR-30d inhibits cell biological progression of Ewing's sarcoma by suppressing the MEK/ERK and PI3K/Akt pathways in vitro. Oncol. Lett. 2018;15(4):4390–4396. doi: 10.3892/ol.2018.7900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Abbaszadeh Z., Cesmeli S., Biray Avci C. Crucial players in glycolysis: cancer progress. Gene. 2020;726 doi: 10.1016/j.gene.2019.144158. [DOI] [PubMed] [Google Scholar]
- 170.Li S., Yang Q., Wang H., Wang Z., Zuo D., Cai Z., Hua Y. Prognostic significance of serum lactate dehydrogenase levels in Ewing's sarcoma: a meta-analysis. Mol. Clin. Oncol. 2016;5(6):832–838. doi: 10.3892/mco.2016.1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Yeung C., Gibson A.E., Issaq S.H., Oshima N., Baumgart J.T., Edessa L.D., Rai G., Urban D.J., Johnson M.S., Benavides G.A., Squadrito G.L., Yohe M.E., Lei H., Eldridge S., Hamre J., 3rd, Dowdy T., Ruiz-Rodado V., Lita A., Mendoza A., Shern J.F., Larion M., Helman L.J., Stott G.M., Krishna M.C., Hall M.D., Darley-Usmar V., Neckers L.M., Heske C.M. Targeting glycolysis through inhibition of lactate dehydrogenase impairs tumor growth in preclinical models of Ewing sarcoma. Cancer Res. 2019;79(19):5060–5073. doi: 10.1158/0008-5472.CAN-19-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.van Wijk R., van Solinge W.W. The energy-less red blood cell is lost: erythrocyte enzyme abnormalities of glycolysis. Blood. 2005;106(13):4034–4042. doi: 10.1182/blood-2005-04-1622. [DOI] [PubMed] [Google Scholar]
- 173.Melkonian E.A., Schury M.P. StatPearls Publishing StatPearls Publishing LLC.; Treasure Island (FL): 2020. Biochemistry, Anaerobic Glycolysis, StatPearls. [PubMed] [Google Scholar]
- 174.Jiménez J.A., Apfelbaum A.A., Hawkins A.G., Svoboda L.K., Kumar A., Ruiz R.O., Garcia A.X., Haarer E., Nwosu Z.C., Bradin J., Purohit T., Chen D., Cierpicki T., Grembecka J., Lyssiotis C.A., Lawlor E.R. EWS-FLI1 and menin converge to regulate ATF4 activity in Ewing sarcoma. Mol. Cancer Res. 2021;19(7):1182–1195. doi: 10.1158/1541-7786.MCR-20-0679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Issaq S.H., Mendoza A., Kidner R., Rosales T.I., Duveau D.Y., Heske C.M., Rohde J.M., Boxer M.B., Thomas C.J., DeBerardinis R.J., Helman L.J. EWS-FLI1-regulated serine synthesis and exogenous serine are necessary for ewing sarcoma cellular proliferation and tumor growth. Mol. Cancer Ther. 2020;19(7):1520–1529. doi: 10.1158/1535-7163.MCT-19-0748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Svoboda L.K., Teh S.S.K., Sud S., Kerk S., Zebolsky A., Treichel S., Thomas D., Halbrook C.J., Lee H.J., Kremer D., Zhang L., Klossowski S., Bankhead A.R., Magnuson B., Ljungman M., Cierpicki T., Grembecka J., Lyssiotis C.A., Lawlor E.R. Menin regulates the serine biosynthetic pathway in Ewing sarcoma. J. Pathol. 2018;245(3):324–336. doi: 10.1002/path.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Sen N., Cross A.M., Lorenzi P.L., Khan J., Gryder B.E., Kim S., Caplen N.J. EWS-FLI1 reprograms the metabolism of Ewing sarcoma cells via positive regulation of glutamine import and serine-glycine biosynthesis. Mol. Carcinog. 2018;57(10):1342–1357. doi: 10.1002/mc.22849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Mutz C.N., Schwentner R., Aryee D.N.T., Bouchard E.D.J., Mejia E.M., Hatch G.M., Kauer M.O., Katschnig A.M., Ban J., Garten A., Alonso J., Banerji V., Kovar H. EWS-FLI1 confers exquisite sensitivity to NAMPT inhibition in Ewing sarcoma cells. Oncotarget. 2017;8(15):24679–24693. doi: 10.18632/oncotarget.14976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Gibson A.E., Yeung C., Issaq S.H., Collins V.J., Gouzoulis M., Zhang Y., Ji J., Mendoza A., Heske C.M. Inhibition of nicotinamide phosphoribosyltransferase (NAMPT) with OT-82 induces DNA damage, cell death, and suppression of tumor growth in preclinical models of Ewing sarcoma. Oncogenesis. 2020;9(9):80. doi: 10.1038/s41389-020-00264-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Mutz C.N., Schwentner R., Kauer M.O., Katschnig A.M., Kromp F., Aryee D.N., Erhardt S., Goiny M., Alonso J., Fuchs D., Kovar H. EWS-FLI1 impairs aryl hydrocarbon receptor activation by blocking tryptophan breakdown via the kynurenine pathway. FEBS Lett. 2016;590(14):2063–2075. doi: 10.1002/1873-3468.12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Hirschfeld S., Helman L. Diverse roles of insulin-like growth factors in pediatric solid tumors. In Vivo. 1994;8(1):81–90. [PubMed] [Google Scholar]
- 182.Zhan S., Shapiro D.N., Helman L.J. Loss of imprinting of IGF2 in Ewing's sarcoma. Oncogene. 1995;11(12):2503–2507. [PubMed] [Google Scholar]
- 183.Yee D., Favoni R.E., Lebovic G.S., Lombana F., Powell D.R., Reynolds C.P., Rosen N. Insulin-like growth factor I expression by tumors of neuroectodermal origin with the t(11;22) chromosomal translocation. A potential autocrine growth factor. J. Clin. Invest. 1990;86(6):1806–1814. doi: 10.1172/JCI114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Scotlandi K., Benini S., Sarti M., Serra M., Lollini P.L., Maurici D., Picci P., Manara M.C., Baldini N. Insulin-like growth factor I receptor-mediated circuit in Ewing's sarcoma/peripheral neuroectodermal tumor: a possible therapeutic target. Cancer Res. 1996;56(20):4570–4574. [PubMed] [Google Scholar]
- 185.Toretsky J.A., Steinberg S.M., Thakar M., Counts D., Pironis B., Parente C., Eskenazi A., Helman L., Wexler L.H. Insulin-like growth factor type 1 (IGF-1) and IGF binding protein-3 in patients with Ewing sarcoma family of tumors. Cancer. 2001;92(11):2941–2947. doi: 10.1002/1097-0142(20011201)92:11<2941::aid-cncr10072>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 186.Pappo A.S., Patel S.R., Crowley J., Reinke D.K., Kuenkele K.P., Chawla S.P., Toner G.C., Maki R.G., Meyers P.A., Chugh R., Ganjoo K.N., Schuetze S.M., Juergens H., Leahy M.G., Geoerger B., Benjamin R.S., Helman L.J., Baker L.H. R1507, a monoclonal antibody to the insulin-like growth factor 1 receptor, in patients with recurrent or refractory Ewing sarcoma family of tumors: results of a phase II Sarcoma Alliance for Research through Collaboration study. J. Clin. Oncol. 2011;29(34):4541–4547. doi: 10.1200/JCO.2010.34.0000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Amin H.M., Morani A.C., Daw N.C., Lamhamedi-Cherradi S.E., Subbiah V., Menegaz B.A., Vishwamitra D., Eskandari G., George B., Benjamin R.S., Patel S., Song J., Lazar A.J., Wang W.L., Kurzrock R., Pappo A., Anderson P.M., Schwartz G.K., Araujo D., Cuglievan B., Ratan R., McCall D., Mohiuddin S., Livingston J.A., Molina E.R., Naing A., Ludwig J.A. IGF-1R/mTOR targeted therapy for ewing sarcoma: a meta-analysis of five IGF-1R-related trials matched to proteomic and radiologic predictive biomarkers. Cancers (Basel) 2020;12(7) doi: 10.3390/cancers12071768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Lamhamedi-Cherradi S.E., Menegaz B.A., Ramamoorthy V., Vishwamitra D., Wang Y., Maywald R.L., Buford A.S., Fokt I., Skora S., Wang J., Naing A., Lazar A.J., Rohren E.M., Daw N.C., Subbiah V., Benjamin R.S., Ratan R., Priebe W., Mikos A.G., Amin H.M., Ludwig J.A. IGF-1R and mTOR blockade: novel resistance mechanisms and synergistic drug combinations for Ewing sarcoma. J. Natl Cancer Inst. 2016;108(12) doi: 10.1093/jnci/djw182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.S.G. DuBois, Randomized Phase 3 Trial of Ganitumab Added to Interval Compressed Chemotherapy for Patients with Newly Diagnosed Metastatic Ewing Sarcoma: A Report from the Children’s Oncology Group (COG), The Connective Tissue Oncology Society Annual Meeting, Tokyo, Japan, 2019. [DOI] [PMC free article] [PubMed]
- 190.Tilan J.U., Lu C., Galli S., Izycka-Swieszewska E., Earnest J.P., Shabbir A., Everhart L.M., Wang S., Martin S., Horton M., Mahajan A., Christian D., O'Neill A., Wang H., Zhuang T., Czarnecka M., Johnson M.D., Toretsky J.A., Kitlinska J. Hypoxia shifts activity of neuropeptide Y in Ewing sarcoma from growth-inhibitory to growth-promoting effects. Oncotarget. 2013;4(12):2487–2501. doi: 10.18632/oncotarget.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Davis S., Meltzer P.S. Ewing's sarcoma: general insights from a rare model. Cancer Cell. 2006;9(5):331–332. doi: 10.1016/j.ccr.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 192.Takahashi A., Higashino F., Aoyagi M., Yoshida K., Itoh M., Kyo S., Ohno T., Taira T., Ariga H., Nakajima K., Hatta M., Kobayashi M., Sano H., Kohgo T., Shindoh M. EWS/ETS fusions activate telomerase in Ewing's tumors. Cancer Res. 2003;63(23):8338–8344. [PubMed] [Google Scholar]
- 193.Hahm K.B., Cho K., Lee C., Im Y.H., Chang J., Choi S.G., Sorensen P.H., Thiele C.J., Kim S.J. Repression of the gene encoding the TGF-beta type II receptor is a major target of the EWS-FLI1 oncoprotein. Nat. Genet. 1999;23(2):222–227. doi: 10.1038/13854. [DOI] [PubMed] [Google Scholar]
- 194.J. Ewing, Classics in oncology. Diffuse endothelioma of bone. James Ewing. Proceedings of the New York Pathological Society, 1921, CA: a cancer journal for clinicians 22(2) (1972) 95-8. [DOI] [PubMed]
- 195.Taymaz-Nikerel H., Karabekmez M.E., Eraslan S., Kirdar B. Doxorubicin induces an extensive transcriptional and metabolic rewiring in yeast cells. Sci. Rep. 2018;8(1):13672. doi: 10.1038/s41598-018-31939-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Montecucco A., Zanetta F., Biamonti G. Molecular mechanisms of etoposide. EXCLI J. 2015;14:95–108. doi: 10.17179/excli2015-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Johnstone E.C., Lind M.J., Griffin M.J., Boddy A.V. Ifosfamide metabolism and DNA damage in tumour and peripheral blood lymphocytes of breast cancer patients. Cancer Chemother. Pharmacol. 2000;46(6):433–441. doi: 10.1007/s002800000185. [DOI] [PubMed] [Google Scholar]
- 198.Jorgensen T.J., Prasad S.C., Brennan T.P., Dritschilo A. Constraints to DNA unwinding near radiation-induced strand breaks in Ewing's sarcoma cells. Radiat. Res. 1990;123(3):320–324. [PubMed] [Google Scholar]
- 199.Newman R.E., Soldatenkov V.A., Dritschilo A., Notario V. Poly(ADP-ribose) polymerase turnover alterations do not contribute to PARP overexpression in Ewing's sarcoma cells. Oncol. Rep. 2002;9(3):529–532. [PubMed] [Google Scholar]
- 200.Ray Chaudhuri A., Nussenzweig A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol. 2017;18(10):610–621. doi: 10.1038/nrm.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Caron M.C., Sharma A.K., O'Sullivan J., Myler L.R., Ferreira M.T., Rodrigue A., Coulombe Y., Ethier C., Gagne J.P., Langelier M.F., Pascal J.M., Finkelstein I.J., Hendzel M.J., Poirier G.G., Masson J.Y. Poly(ADP-ribose) polymerase-1 antagonizes DNA resection at double-strand breaks. Nat. Commun. 2019;10(1):2954. doi: 10.1038/s41467-019-10741-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Soldatenkov V.A., Albor A., Patel B.K., Dreszer R., Dritschilo A., Notario V. Regulation of the human poly(ADP-ribose) polymerase promoter by the ETS transcription factor. Oncogene. 1999;18(27):3954–3962. doi: 10.1038/sj.onc.1202778. [DOI] [PubMed] [Google Scholar]
- 203.Soldatenkov V.A., Trofimova I.N., Rouzaut A., McDermott F., Dritschilo A., Notario V. Differential regulation of the response to DNA damage in Ewing's sarcoma cells by ETS1 and EWS/FLI-1. Oncogene. 2002;21(18):2890–2895. doi: 10.1038/sj.onc.1205393. [DOI] [PubMed] [Google Scholar]
- 204.Garnett M.J., Edelman E.J., Heidorn S.J., Greenman C.D., Dastur A., Lau K.W., Greninger P., Thompson I.R., Luo X., Soares J., Liu Q., Iorio F., Surdez D., Chen L., Milano R.J., Bignell G.R., Tam A.T., Davies H., Stevenson J.A., Barthorpe S., Lutz S.R., Kogera F., Lawrence K., McLaren-Douglas A., Mitropoulos X., Mironenko T., Thi H., Richardson L., Zhou W., Jewitt F., Zhang T., O'Brien P., Boisvert J.L., Price S., Hur W., Yang W., Deng X., Butler A., Choi H.G., Chang J.W., Baselga J., Stamenkovic I., Engelman J.A., Sharma S.V., Delattre O., Saez-Rodriguez J., Gray N.S., Settleman J., Futreal P.A., Haber D.A., Stratton M.R., Ramaswamy S., McDermott U., Benes C.H. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483(7391):570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Choy E., Butrynski J.E., Harmon D.C., Morgan J.A., George S., Wagner A.J., D'Adamo D., Cote G.M., Flamand Y., Benes C.H., Haber D.A., Baselga J.M., Demetri G.D. Phase II study of olaparib in patients with refractory Ewing sarcoma following failure of standard chemotherapy. BMC Cancer. 2014;14:813. doi: 10.1186/1471-2407-14-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Schafer E.S., Rau R.E., Berg S.L., Liu X., Minard C.G., Bishop A.J.R., Romero J.C., Hicks M.J., Nelson M.D., Jr., Voss S., Reid J.M., Fox E., Weigel B.J., Blaney S.M. Phase 1/2 trial of talazoparib in combination with temozolomide in children and adolescents with refractory/recurrent solid tumors including Ewing sarcoma: A Children's Oncology Group Phase 1 Consortium study (ADVL1411) Pediatr. Blood Cancer. 2020;67(2) doi: 10.1002/pbc.28073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Federico S.M., Pappo A.S., Sahr N., Sykes A., Campagne O., Stewart C.F., Clay M.R., Bahrami A., McCarville M.B., Kaste S.C., Santana V.M., Helmig S., Gartrell J., Shelat A., Brennan R.C., Hawkins D., Godwin K., Bishop M.W., Furman W.L., Stewart E. A phase I trial of talazoparib and irinotecan with and without temozolomide in children and young adults with recurrent or refractory solid malignancies. Eur. J. Cancer. 2020;137:204–213. doi: 10.1016/j.ejca.2020.06.014. [DOI] [PubMed] [Google Scholar]
- 208.Tang S.W., Bilke S., Cao L., Murai J., Sousa F.G., Yamade M., Rajapakse V., Varma S., Helman L.J., Khan J., Meltzer P.S., Pommier Y. SLFN11 is a transcriptional target of EWS-FLI1 and a determinant of drug response in Ewing sarcoma. Clin. Cancer Res. 2015;21(18):4184–4193. doi: 10.1158/1078-0432.CCR-14-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Smith M.A., Hampton O.A., Reynolds C.P., Kang M.H., Maris J.M., Gorlick R., Kolb E.A., Lock R., Carol H., Keir S.T., Wu J., Kurmasheva R.T., Wheeler D.A., Houghton P.J. Initial testing (stage 1) of the PARP inhibitor BMN 673 by the pediatric preclinical testing program: PALB2 mutation predicts exceptional in vivo response to BMN 673. Pediatr. Blood Cancer. 2015;62(1):91–98. doi: 10.1002/pbc.25201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Stewart E., Goshorn R., Bradley C., Griffiths L.M., Benavente C., Twarog N.R., Miller G.M., Caufield W., Freeman B.B., 3rd, Bahrami A., Pappo A., Wu J., Loh A., Karlström Å., Calabrese C., Gordon B., Tsurkan L., Hatfield M.J., Potter P.M., Snyder S.E., Thiagarajan S., Shirinifard A., Sablauer A., Shelat A.A., Dyer M.A. Targeting the DNA repair pathway in Ewing sarcoma. Cell Reports. 2014;9(3):829–841. doi: 10.1016/j.celrep.2014.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Norris R.E., Adamson P.C., Nguyen V.T., Fox E. Preclinical evaluation of the PARP inhibitor, olaparib, in combination with cytotoxic chemotherapy in pediatric solid tumors. Pediatr. Blood Cancer. 2014;61(1):145–150. doi: 10.1002/pbc.24697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Vormoor B., Schlosser Y.T., Blair H., Sharma A., Wilkinson S., Newell D.R., Curtin N. Sensitizing Ewing sarcoma to chemo- and radiotherapy by inhibition of the DNA-repair enzymes DNA protein kinase (DNA-PK) and poly-ADP-ribose polymerase (PARP) 1/2. Oncotarget. 2017;8(69):113418–113430. doi: 10.18632/oncotarget.21300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Ordonez J.L., Amaral A.T., Carcaboso A.M., Herrero-Martin D., del Carmen Garcia-Macias M., Sevillano V., Alonso D., Pascual-Pasto G., San-Segundo L., Vila-Ubach M., Rodrigues T., Fraile S., Teodosio C., Mayo-Iscar A., Aracil M., Galmarini C.M., Tirado O.M., Mora J., de Alava E. The PARP inhibitor olaparib enhances the sensitivity of Ewing sarcoma to trabectedin. Oncotarget. 2015;6(22):18875–18890. doi: 10.18632/oncotarget.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Heske C.M., Davis M.I., Baumgart J.T., Wilson K., Gormally M.V., Chen L., Zhang X., Ceribelli M., Duveau D.Y., Guha R., Ferrer M., Arnaldez F.I., Ji J., Tran H.L., Zhang Y., Mendoza A., Helman L.J., Thomas C.J. Matrix screen identifies synergistic combination of PARP inhibitors and nicotinamide phosphoribosyltransferase (NAMPT) inhibitors in Ewing sarcoma. Clin. Cancer Res. 2017;23(23):7301–7311. doi: 10.1158/1078-0432.CCR-17-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Iniguez A.B., Stolte B., Wang E.J., Conway A.S., Alexe G., Dharia N.V., Kwiatkowski N., Zhang T., Abraham B.J., Mora J., Kalev P., Leggett A., Chowdhury D., Benes C.H., Young R.A., Gray N.S., Stegmaier K. EWS/FLI confers tumor cell synthetic lethality to CDK12 inhibition in Ewing sarcoma. Cancer Cell. 2018;33(2):202–216.e6. doi: 10.1016/j.ccell.2017.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Bradbury A., Hall S., Curtin N., Drew Y. Targeting ATR as Cancer Therapy: a new era for synthetic lethality and synergistic combinations? Pharmacol. Ther. 2020;207 doi: 10.1016/j.pharmthera.2019.107450. [DOI] [PubMed] [Google Scholar]
- 217.Nieto-Soler M., Morgado-Palacin I., Lafarga V., Lecona E., Murga M., Callen E., Azorin D., Alonso J., Lopez-Contreras A.J., Nussenzweig A., Fernandez-Capetillo O. Efficacy of ATR inhibitors as single agents in Ewing sarcoma. Oncotarget. 2016;7(37):58759–58767. doi: 10.18632/oncotarget.11643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Koppenhafer S.L., Goss K.L., Terry W.W., Gordon D.J. Inhibition of the ATR-CHK1 Pathway in Ewing Sarcoma Cells Causes DNA Damage and Apoptosis via the CDK2-Mediated Degradation of RRM2. Mol. Cancer Res. 2020;18(1):91–104. doi: 10.1158/1541-7786.MCR-19-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Grohar P.J., Segars L.E., Yeung C., Pommier Y., D'Incalci M., Mendoza A., Helman L.J. Dual targeting of EWS-FLI1 activity and the associated DNA damage response with trabectedin and SN38 synergistically inhibits Ewing sarcoma cell growth. Clin. Cancer Res. 2014;20(5):1190–1203. doi: 10.1158/1078-0432.CCR-13-0901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Lin M.Y., Damron T.A., Oest M.E., Horton J.A. Mithramycin A radiosensitizes EWS:Fli1(+) Ewing sarcoma cells by inhibiting double strand break repair. Int. J. Radiat. Oncol. Biol. Phys. 2021;109(5):1454–1471. doi: 10.1016/j.ijrobp.2020.12.010. [DOI] [PubMed] [Google Scholar]
- 221.Heisey D.A.R., Lochmann T.L., Floros K.V., Coon C.M., Powell K.M., Jacob S., Calbert M.L., Ghotra M.S., Stein G.T., Maves Y.K., Smith S.C., Benes C.H., Leverson J.D., Souers A.J., Boikos S.A., Faber A.C. The Ewing family of tumors relies on BCL-2 and BCL-X(L) to escape PARP inhibitor toxicity. Clin. Cancer Res. 2019;25(5):1664–1675. doi: 10.1158/1078-0432.CCR-18-0277. [DOI] [PubMed] [Google Scholar]
- 222.Jonkers I., Lis J.T. Getting up to speed with transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2015;16(3):167–177. doi: 10.1038/nrm3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Porrua O., Libri D. Transcription termination and the control of the transcriptome: why, where and how to stop. Nat. Rev. Mol. Cell Biol. 2015;16(3):190–202. doi: 10.1038/nrm3943. [DOI] [PubMed] [Google Scholar]
- 224.Spriano F., Chung E.Y.L., Gaudio E., Tarantelli C., Cascione L., Napoli S., Jessen K., Carrassa L., Priebe V., Sartori G., Graham G., Selvanathan S.P., Cavalli A., Rinaldi A., Kwee I., Testoni M., Genini D., Ye B.H., Zucca E., Stathis A., Lannutti B., Toretsky J.A., Bertoni F. The ETS Inhibitors YK-4-279 and TK-216 Are Novel Antilymphoma Agents. Clin. Cancer Res. 2019;25(16):5167–5176. doi: 10.1158/1078-0432.CCR-18-2718. [DOI] [PubMed] [Google Scholar]
- 225.Chen C., Wonsey D.R., Lemieux M.E., Kung A.L. Differential disruption of EWS-FLI1 binding by DNA-binding agents. PLoS ONE. 2013;8(7) doi: 10.1371/journal.pone.0069714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Chen C., Shanmugasundaram K., Rigby A.C., Kung A.L. Shikonin, a natural product from the root of Lithospermum erythrorhizon, is a cytotoxic DNA-binding agent. Eur. J. Pharm. Sci. 2013;49(1):18–26. doi: 10.1016/j.ejps.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 227.Grohar P.J., Woldemichael G.M., Griffin L.B., Mendoza A., Chen Q.R., Yeung C., Currier D.G., Davis S., Khanna C., Khan J., McMahon J.B., Helman L.J. Identification of an inhibitor of the EWS-FLI1 oncogenic transcription factor by high-throughput screening. J. Natl. Cancer Inst. 2011;103(12):962–978. doi: 10.1093/jnci/djr156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Grohar P.J., Glod J., Peer C.J., Sissung T.M., Arnaldez F.I., Long L., Figg W.D., Whitcomb P., Helman L.J., Widemann B.C. A phase I/II trial and pharmacokinetic study of mithramycin in children and adults with refractory Ewing sarcoma and EWS-FLI1 fusion transcript. Cancer Chemother. Pharmacol. 2017;80(3):645–652. doi: 10.1007/s00280-017-3382-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Sissung T.M., Huang P.A., Hauke R.J., McCrea E.M., Peer C.J., Barbier R.H., Strope J.D., Ley A.M., Zhang M., Hong J.A., Venzon D., Jackson J.P., Brouwer K.R., Grohar P., Glod J., Widemann B.C., Heller T., Schrump D.S., Figg W.D. Severe hepatotoxicity of mithramycin therapy caused by altered expression of hepatocellular bile transporters. Mol. Pharmacol. 2019;96(2):158–167. doi: 10.1124/mol.118.114827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Flores G., Everett J.H., Boguslawski E.A., Oswald B.M., Madaj Z.B., Beddows I., Dikalov S., Adams M., Klumpp-Thomas C.A., Kitchen-Goosen S.M., Martin S.E., Caplen N.J., Helman L.J., Grohar P.J. CDK9 blockade exploits context-dependent transcriptional changes to improve activity and limit toxicity of mithramycin for ewing sarcoma. Mol. Cancer Ther. 2020;19(5):1183–1196. doi: 10.1158/1535-7163.MCT-19-0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Osgood C.L., Maloney N., Kidd C.G., Kitchen-Goosen S., Segars L., Gebregiorgis M., Woldemichael G.M., He M., Sankar S., Lessnick S.L., Kang M., Smith M., Turner L., Madaj Z.B., Winn M.E., Nunez L.E., Gonzalez-Sabin J., Helman L.J., Moris F., Grohar P.J. Identification of mithramycin analogues with improved targeting of the EWS-FLI1 transcription factor. Clin. Cancer Res. 2016;22(16):4105–4118. doi: 10.1158/1078-0432.CCR-15-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Caropreso V., Darvishi E., Turbyville T.J., Ratnayake R., Grohar P.J., McMahon J.B., Woldemichael G.M. Englerin A inhibits EWS-FLI1 DNA binding in ewing sarcoma cells. J. Biol. Chem. 2016;291(19):10058–10066. doi: 10.1074/jbc.M115.701375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Larsen A.K., Galmarini C.M., D'Incalci M. Unique features of trabectedin mechanism of action. Cancer Chemother. Pharmacol. 2016;77(4):663–671. doi: 10.1007/s00280-015-2918-1. [DOI] [PubMed] [Google Scholar]
- 234.P.J. Grohar, L.B. Griffin, C. Yeung, Q.R. Chen, Y. Pommier, C. Khanna, J. Khan, L.J. Helman, Ecteinascidin 743 interferes with the activity of EWS-FLI1 in Ewing sarcoma cells, Neoplasia (New York, N.Y.) 13(2) (2011) 145-53. [DOI] [PMC free article] [PubMed]
- 235.Harlow M.L., Chasse M.H., Boguslawski E.A., Sorensen K.M., Gedminas J.M., Kitchen-Goosen S.M., Rothbart S.B., Taslim C., Lessnick S.L., Peck A.S., Madaj Z.B., Bowman M.J., Grohar P.J. Trabectedin inhibits EWS-FLI1 and evicts SWI/SNF from chromatin in a schedule-dependent manner. Clin. Cancer Res. 2019;25(11):3417–3429. doi: 10.1158/1078-0432.CCR-18-3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Harlow M.L., Maloney N., Roland J., Guillen Navarro M.J., Easton M.K., Kitchen-Goosen S.M., Boguslawski E.A., Madaj Z.B., Johnson B.K., Bowman M.J., D'Incalci M., Winn M.E., Turner L., Hostetter G., Galmarini C.M., Aviles P.M., Grohar P.J. Lurbinectedin inactivates the Ewing sarcoma oncoprotein EWS-FLI1 by redistributing it within the nucleus. Cancer Res. 2016;76(22):6657–6668. doi: 10.1158/0008-5472.CAN-16-0568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Stegmaier K., Wong J.S., Ross K.N., Chow K.T., Peck D., Wright R.D., Lessnick S.L., Kung A.L., Golub T.R. Signature-based small molecule screening identifies cytosine arabinoside as an EWS/FLI modulator in Ewing sarcoma. PLoS Med. 2007;4(4) doi: 10.1371/journal.pmed.0040122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Matsuno Y., Hyodo M., Fujimori H., Shimizu A., Yoshioka K.I. Sensitization of cancer cells to radiation and topoisomerase i inhibitor camptothecin using inhibitors of PARP and other signaling molecules. Cancers (Basel) 2018;10(10) doi: 10.3390/cancers10100364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Casey D.A., Wexler L.H., Merchant M.S., Chou A.J., Merola P.R., Price A.P., Meyers P.A. Irinotecan and temozolomide for Ewing sarcoma: the Memorial Sloan-Kettering experience. Pediatr. Blood Cancer. 2009;53(6):1029–1034. doi: 10.1002/pbc.22206. [DOI] [PubMed] [Google Scholar]
- 240.Hunold A., Weddeling N., Paulussen M., Ranft A., Liebscher C., Jurgens H. Topotecan and cyclophosphamide in patients with refractory or relapsed Ewing tumors. Pediatr. Blood Cancer. 2006;47(6):795–800. doi: 10.1002/pbc.20719. [DOI] [PubMed] [Google Scholar]
- 241.Chou J., Quigley D.A., Robinson T.M., Feng F.Y., Ashworth A. Transcription-associated cyclin-dependent kinases as targets and biomarkers for cancer therapy. Cancer discovery. 2020;10(3):351–370. doi: 10.1158/2159-8290.CD-19-0528. [DOI] [PubMed] [Google Scholar]
- 242.Espinosa J.M. Transcriptional CDKs in the spotlight. Transcription. 2019;10(2):45–46. doi: 10.1080/21541264.2019.1597479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Lee D.J., Zeidner J.F. Cyclin-dependent kinase (CDK) 9 and 4/6 inhibitors in acute myeloid leukemia (AML): a promising therapeutic approach. Expert Opin. Invest. Drugs. 2019;28(11):989–1001. doi: 10.1080/13543784.2019.1678583. [DOI] [PubMed] [Google Scholar]
- 244.Li Y., Tanaka K., Li X., Okada T., Nakamura T., Takasaki M., Yamamoto S., Oda Y., Tsuneyoshi M., Iwamoto Y. Cyclin-dependent kinase inhibitor, flavopiridol, induces apoptosis and inhibits tumor growth in drug-resistant osteosarcoma and Ewing's family tumor cells. Int. J. Cancer. 2007;121(6):1212–1218. doi: 10.1002/ijc.22820. [DOI] [PubMed] [Google Scholar]
- 245.Montagnoli A., Valsasina B., Croci V., Menichincheri M., Rainoldi S., Marchesi V., Tibolla M., Tenca P., Brotherton D., Albanese C., Patton V., Alzani R., Ciavolella A., Sola F., Molinari A., Volpi D., Avanzi N., Fiorentini F., Cattoni M., Healy S., Ballinari D., Pesenti E., Isacchi A., Moll J., Bensimon A., Vanotti E., Santocanale C. A Cdc7 kinase inhibitor restricts initiation of DNA replication and has antitumor activity. Nat. Chem. Biol. 2008;4(6):357–365. doi: 10.1038/nchembio.90. [DOI] [PubMed] [Google Scholar]
- 246.Li X., Tanaka K., Nakatani F., Matsunobu T., Sakimura R., Hanada M., Okada T., Nakamura T., Iwamoto Y. Transactivation of cyclin E gene by EWS-Fli1 and antitumor effects of cyclin dependent kinase inhibitor on Ewing's family tumor cells. Int. J. Cancer. 2005;116(3):385–394. doi: 10.1002/ijc.21010. [DOI] [PubMed] [Google Scholar]
- 247.Richter G.H.S., Hensel T., Schmidt O., Saratov V., von Heyking K., Becker-Dettling F., Prexler C., Yen H.Y., Steiger K., Fulda S., Dirksen U., Weichert W., Wang S., Burdach S., Schafer B.W. Combined inhibition of epigenetic readers and transcription initiation targets the EWS-ETS transcriptional program in Ewing sarcoma. Cancers (Basel) 2020;12(2) doi: 10.3390/cancers12020304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.D.A.R. Heisey, S. Jacob, T.L. Lochmann, R. Kurupi, M.S. Ghotra, M.L. Calbert, M. Shende, Y. Kato Maves, J.E. Koblinski, M.G. Dozmorov, S.A. Boikos, C.H. Benes, A.C. Faber, Pharmaceutical interference of the EWS-FLI1-driven transcriptome by co-targeting H3K27ac and RNA polymerase activity in Ewing Sarcoma, Mol Cancer Ther (2021). [DOI] [PubMed]
- 249.Stewart E., Goshorn R., Bradley C., Griffiths L.M., Benavente C., Twarog N.R., Miller G.M., Caufield W., Freeman B.B., 3rd, Bahrami A., Pappo A., Wu J., Loh A., Karlstrom A., Calabrese C., Gordon B., Tsurkan L., Hatfield M.J., Potter P.M., Snyder S.E., Thiagarajan S., Shirinifard A., Sablauer A., Shelat A.A., Dyer M.A. Targeting the DNA repair pathway in Ewing sarcoma. Cell Reports. 2014;9(3):829–841. doi: 10.1016/j.celrep.2014.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]