Summary
Palmitoylation is a special kind of lipid modification that targets proteins to membranes. This protocol introduces the acyl-biotin exchange (ABE) assay to determine the palmitoylation of protein cysteines in yeast. Palmitoylation is exchanged by biotinylated compounds so that the palmitoyl proteins can be affinity-purified for downstream assay by western blot. This protocol is easy to perform and can be applied to other biological sources with slight modifications. This protocol is limited to the detection of cysteine-based palmitoylation.
For complete details on the use and execution of this profile, please refer to Lei et al. (2021).
Subject areas: Cell Biology, Model Organisms, Protein Biochemistry, Protein expression and purification
Graphical Abstract

Highlights
-
•
Maintain the status of palmitoylation in proteins under denaturing conditions
-
•
Quantify the specific palmitoylation of each cysteine from yeast extracts
-
•
This protocol is applicable for samples from species other than yeast
Palmitoylation is a special kind of lipid modification that targets proteins to membranes. This protocol introduces the acyl-biotin exchange (ABE) assay to determine the palmitoylation of protein cysteines in yeast. Palmitoylation is exchanged by biotinylated compounds so that the palmitoyl proteins can be affinity-purified for downstream assay by western blot. This protocol is easy to perform and can be applied to other biological sources with slight modifications. This protocol is limited to the detection of cysteine-based palmitoylation.
Before you begin
The protocol below describes the specific steps for detecting S-palmitoylation using yeast cells. However, we also used this protocol in HEK293T cells and MEFs. We describe a protocol for the detection of protein palmitoylation that generally regulates protein localization to specific membranes. There are three types of protein palmitoylation: S-palmitoylation, N-palmitoylation and O-palmitoylation. S-palmitoylation is the most common type of palmitoylation and it can be reversed (depalmitoylation) by acylthioesterase (acyl-protein thioesterase). S-palmitoylation regulates the functioning of proteins by anchoring them to membranes.
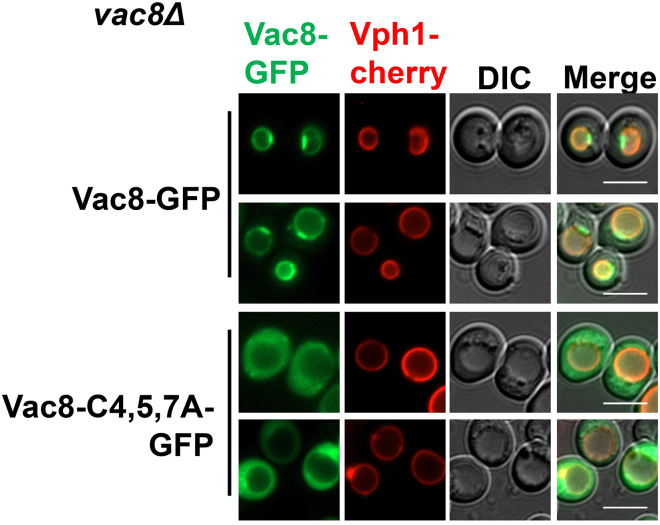
You will need to culture and collect wild-type yeast cells expressing C-terminal GFP- or HA-tagged Vac8 (a protein subject to palmitoylation on N-terminal cysteines) driven by the ADH1 promotor. These plasmids have been used in our recent study on the roles of Vac8/ARMC3 in autophagy initiation and its functions in spermiogenesis (Lei et al., 2021). As a negative control resisting palmitoylation, the C4A/C5A/C7A mutant of Vac8, with three cysteines mutated into alanine, is also used in this protocol. The localization of Vac8-GFP and Vac8C4,5,7A-GFP was observed by fluorescence microscopy (Figure 1).
Figure 1.
Palmitoylation affects the localization of Vac8
C-terminal GFP-tagged Vac8 mutants were expressed in vac8Δ cells expressing Vph1-Cherry, a vacuolar membrane marker, and their cellular localization was observed by fluorescence microscopy.
Scale bars, 5 μm.
The main function of palmitoylation is anchoring proteins to membranes of certain organelles; thus, the localization of a palmitoylated protein is correlated with its palmitoylation status (De and Sadhukhan, 2018; Rowland et al., 2018). The principle of this acyl-biotin exchange (ABE) assay is shown (Figure 2).
Note: The palmitoylation sites in a protein could be predicted by the online tool CSS-Palm 4.0 (http://www.csspalm.biocuckoo.org/) (Ren et al., 2008). For example, Vac8 is predicted to be palmitoylated at cysteines 4, 5 and 7 (Figure 3).
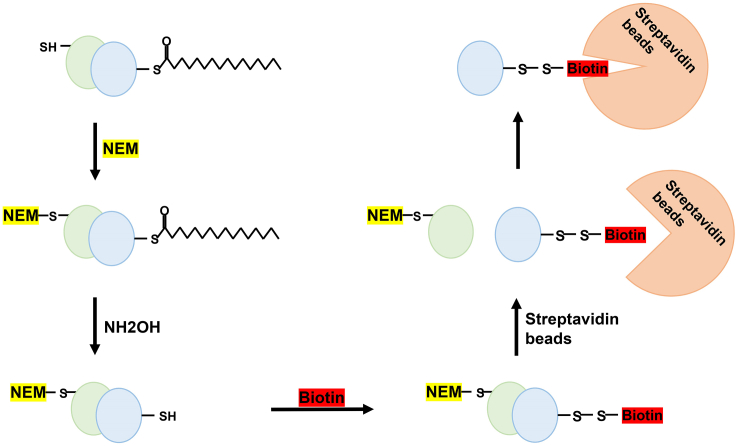
Figure 2.
The schematic of the ABE method
ABE comprises a sequence of four chemical steps: (i) a complete blockade of free thiols with N-ethylmaleimide (NEM); (ii) a treatment by hydroxylamine to release thioester-linked palmitoyl moiety and to restore the modified cysteine to thiols; (iii) biotinylating the exposed thiols using thiol-reactive biotin; and (iv) distinguishing and combining the biotinylated protein with streptavidin beads.
Figure 3.
Palmitoylation site prediction of yeast protein Vac8
The cysteines of Vac8 at N-terminal sites 4,5 and 7 were predicted to be palmitoylated by the online tool CSS-Palm 4.0 (http://www.csspalm.biocuckoo.org/).
Preparation of yeast culture
Timing: 6 days
Preparing competent yeast cells:
-
1.
To prepare competent yeast cells, pick and inoculate a single colony of wild-type yeast S288C (BY4741) cells into 3 mL YPD medium with shaking at 30°C 16 h.
-
2.
The next morning, the yeast cells are inoculated at OD600 (optical density at 600 nm) 0.01 in 50 mL fresh YPD. The yeast cells are grown at 30°C with shaking until the OD600 reaches 0.8 (approximately 6 h).
-
3.
Collect 50 mL of culture into a 50 mL Falcon tube and pellet the cells at 500 g for 5 min at 25°C. Resuspend the pellets and wash twice with sterilized distilled water and once with Sorb buffer.
-
4.
Resuspend the pelleted cells in 360 μL Sorb buffer supplemented with 40 μL salmon-sperm DNA (10 mg/mL) followed by mixing.
-
5.
Store aliquots of the mixture in a −80°C freezer.
Plasmid transformation into the competent yeast cells:
-
6.
Mix 2 μg of plasmids (p415-prADH1-Vac8-HA, p415-prADH1-Vac8C4,5,7A-HA, p415-prADH1-Vac8-GFP and p415-prADH1- Vac8C4,5,7A-GFP), respectively, with 10 μL of competent yeast cells generated as above in 60 μL PEG buffer. Vortex thoroughly.
-
7.
Incubate at 25°C for 30 min.
-
8.
Incubate at 42°C for 15 min followed by cooling down on ice.
-
9.
Collect the yeast cells by centrifuging at 500 g for 5 min and resuspend the yeast cells in sterilized water followed by spreading into Leu-selection plates.
-
10.
Two days later, the colonies are grown on plates, and a single colony is picked and inoculated for culturing in Leu-selection medium.
-
11.
Then, 2.5 mL of 16 h culture medium is transferred into 250 mL of Leu-selection medium in a conical bottle until the next morning. Approximately six and a half hours later (the log-phase culture), the OD value of the yeast culture medium should be measured. When the OD value reaches 0.8, the yeast cells could be collected and frozen in liquid nitrogen before storage in a −80°C freezer.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-HA | Santa Cruz Biotechnology | Cat#sc-7392; RRID: AB_627809 |
| Bacterial and virus strains | ||
| E. coli DH5α | Tsingke | Cat#TSC01 |
| Chemicals, peptides, and recombinant proteins | ||
| Triton X-100 | Sangon Biotech | 9002-93-1 |
| Tris-HCl | Bio Basic Inc. | A600485-0500 |
| NaCl | Bio Basic Inc. | A610476-0001 |
| EDTA | Bio Basic Inc. | A100322-0500 |
| HEPES | Sigma-Aldrich | SLBS9000 |
| Sodium hydroxide solution (NaOH) | Sigma-Aldrich | S8045 |
| Hydroxylamine (HA) HCl | Sigma-Aldrich | 431362 |
| 20%SDS | Solarbio | S1015 |
| N-ethylmaleimide (NEM) | Sigma-Aldrich | 04259 |
| Biotin-HPDP | APE×BIO | A8008 |
| β-Mercaptoethanol (β-ME) | Solarbio | M8210 |
| 5∗Loading buffer | Beyotime | P0015L |
| 1∗ Phosphate buffer saline | Hyclone | SH30256.01 |
| 0.5M DTT | Biotechwell | WB0147 |
| Streptavidin agarose | Merck | 69203 |
| Dimethyl sulfoxide | Sigma-Aldrich | D2650 |
| Methanol | KESHI | 67-56-1 |
| Chloroform | KESHI | 76-03-9 |
| Yeast Extract | Bio Basic Inc. | A610961-0500 |
| Peptone, bacteriological | Bio Basic Inc. | A100636-0500 |
| D-Glucose | Bio Basic Inc. | A600219-0500 |
| Protease Inhibitor Cocktail | Roche | Cat#11836145001 |
| PEG3350 | Solarbio | P8040-1000G |
| Salmon-sperm DNA | Sangon Biotech | B548210-0005 |
| LiAc | Sangon Biotech | A500565-0250 |
| Sorbitol | Sangon Biotech | A610491-0500 |
| Experimental models: Organisms/strains | ||
| Saccharomyces cerevisiae BY4741 | EUROSCARF | BY4741 |
| Recombinant DNA | ||
| p415-prADH1-Vac8-HA | this study | Vac8-HA |
| p415-prADH1-Vac8-C4,5,7A-HA | this study | Vac8-C4,5,7A-HA |
| p415-prADH1-Vac8-GFP | this study | Vac8-GFP |
| p415-prADH1-Vac8-C4,5,7A-GFP | this study | Vac8-C4,5,7A-GFP |
| p416-prADH1-Vph1-Cherry | this study | Vph1-Cherry |
| Software and algorithms | ||
| ZEN2 | ZEISS | N/A |
| DNA STAR sequence assay | DNA STAR | https://www.dnastar.com/ |
| Other | ||
| 0.5 mm Glass beads | BioSpec | 11079105Z |
| Immobilon-P PVDF transfer membrane | Millipore | Cat#IPVH00010 |
| Bicinchoninic Acid solution | SIGMA | Cat#SHBJ4540 |
Materials and equipment
50% glucose
Dissolve 100 g glucose in ddH2O to a 200 mL total volume. After filtering through a 0.2 μm filter, it can be stored at 25°C for at least one year.
YPD
For 1 L YPD, 10 g yeast extract and 20 g peptone are dissolved in 960 mL water. After sterilization in the autoclave, 40 mL of sterile 50% glucose is added. The final concentration is 1% yeast extract, 2% peptone and 2% glucose.
100× proteinase inhibitor buffer
The proteinase inhibitor cocktail (Roche)is dissolved in 0.5 mL ddH2O and the final concentration of the proteinase inhibitor buffer is 100×.
Salmon-sperm DNA
A tube of salmon-sperm DNA (Sangon Biotech) is thawed at −20°C, aliquoted at 500 μL per tube, and stored at −20°C after heat shock at 95°C for 10 min.
Leu-selection plates and medium
Mix 6.7 g YNB (Yeast Nitrogen Base, without amino acid), 2 g prepared corresponding amine acid-deficient powder, 20 g glucose (and 20 g agar when plates are needed) into a conical bottle, add 1 L ddH2O, and then subject to sterilization. The medium could be stored at 25°C, and the plates could be stored at 4°C for at least three months.
PEG buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| EDTA (0.5 M) | 5 mM | 0.4 mL |
| Tris.Hcl (1 M, pH8.0) | 10 mM | 2 mL |
| LiAc (1 M) | 100 mM | 20 mL |
| PEG3350 | 40% | 80 g |
| Total | n/a | 200 mL |
[Storage at 4°C after suction filtration; maximum time for storage: 6 months]
Sorb buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| EDTA (0.5 M) | 5 mM | 0.2 mL |
| Tris.Hcl (1 M, pH8.0) | 10 mM | 11 mL |
| Sorbitol | 18.2% | 200.4 g |
| LiAc | 1.02% | 11.22 g |
| Total | n/a | 1100 mL |
[prepare before using]
Yeast lysis buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| EDTA (0.5 M) | 5 mM | 0.2 mL |
| proteinase inhibitor buffer(100×) | 1× | 0.2 mL |
| PBS | n/a | 19.6 mL |
| Total | n/a | 20 mL |
[prepare before using]
With-HA buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Hydroxylamine (1 M) | 0.7 M | 7 mL |
| HPDP-biotin (4 mM) | 1 mM | 2.5 mL |
| Triton X-100 | 0.2% | 200 μL |
| proteinase inhibitor buffer(100×) | 1× | 0.1 mL |
| PBS | n/a | 0.2 mL |
| Total | n/a | 10 mL |
[prepare before using]
Without-HA buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| HPDP-biotin (4 mM) | 1 mM | 2.5 mL |
| Triton X-100 | 0.2% | 200 μL |
| proteinase inhibitor buffer(100×) | 1× | 0.1 mL |
| Tris-HCl (1M, pH 7.4) | 0.05M | 0.5 mL |
| PBS | n/a | 6.7 mL |
| Total | n/a | 10 mL |
[prepare before using]
Biotin buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| biotin (2 mM) | 0.2 mM | 1 mL |
| NaCl (5 M) | 150 mM | 0.3 mL |
| Tris (1 M, pH 7.4) | 50 mM | 0.5 mL |
| Triton X-100 | 0.2% | 20 μL |
| proteinase inhibitor buffer(100×) | 1× | 0.1 mL |
| EDTA (0.5 M) | 5 mM | 0.1 mL |
| ddH2O | n/a | 7.98 mL |
| Total | n/a | 10 mL |
[prepare before using]
Washing buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| NaCl (5 M) | 0.5 M | 0.1 mL |
| Tris (1 M, pH 7.4) | 50 mM | 2.5 mL |
| Triton X-100 | 0.1% | 50 μL |
| EDTA (0.5 M) | 5 mM | 0.5 mL |
| ddH2O | n/a | 46.85 mL |
| Total | n/a | 50 mL |
[prepare before using]
Elute buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| NaCl (5 M) | 150 mM | 0.3 mL |
| Tris-HCl (1 M, pH 7.4) | 50 mM | 0.5 mL |
| EDTA (0.5 M) | 5 mM | 0.1 mL |
| 5∗ loading buffer | 1∗ loading buffer | 2 mL |
| β-ME | 1% | 0.1 mL |
| ddH2O | n/a | 7 mL |
| Total | n/a | 10 mL |
[prepare before using]
1M NEM
| Reagent | Final concentration | Amount |
|---|---|---|
| N-ethylmaleimide | 1 M | 1.25 g |
| Ethyl alcohol | n/a | Up to 10 mL |
[prepare before using]
4 mM HPDP-biotin
| Reagent | Final concentration | Amount |
|---|---|---|
| HPDP-biotin | 4 mM | 0.022 g |
| DMSO | n/a | Up to 10 mL |
[prepare before using]
1M HA HCl (pH 7.0)
| Reagent | Final concentration | Amount |
|---|---|---|
| Hydroxylamine HCl | 1 M | 1.25 g |
| NaOH (5M) | n/a | pH adjustment |
| ddH2O | n/a | Up to 10 mL |
[prepare before using]
LB buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| NaCl (5 M) | 150 mM | 0.3 mL |
| Tris-HCl (1 M, pH 7.4) | 50 mM | 0.5 mL |
| EDTA (0.5 M) | 5 mM | 0.1 mL |
| ddH2O | n/a | 9.1 mL |
| Total | n/a | 10 mL |
[prepare before using]
SB buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| 20%SDS | 4% | 2 mL |
| Tris-HCl (1 M, pH 7.4) | 50 mM | 0.5 mL |
| EDTA (0.5 M) | 5 mM | 0.1 mL |
| ddH2O | n/a | 7.4 mL |
| Total | n/a | 10 mL |
[prepare before using]
TB buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| 20%SDS | 2% | 1 mL |
| Tris-HCl (1 M, pH 7.4) | 50 mM | 0.5 mL |
| EDTA (0.5 M) | 5 mM | 0.1 mL |
| ddH2O | n/a | 8.4 mL |
| Total | n/a | 10 mL |
[prepare before using]
Step-by-step method details
Yeast protein extract
Timing: 2 h
This step outlines how the yeast protein samples are extracted under denaturing conditions, which will maintain the palmitoylation status of the proteins.
-
1.
First, 600 μL of yeast lysis buffer is added to the yeast pellet, and 100 μL of 0.5 mm glass beads is added.
-
2.
The yeast cells are broken by shaking with a high-throughput tissue grinder (shaking at 60 Hz for 60 s, then a 5 min interval on ice to keep cold, 6 rounds in total).
-
3.Centrifuge at 5000 g for 5 min at 4°C and collect the supernatant in a new 1.5 mL centrifuge tube.
-
a.Quantify the protein concentrations of the supernatant via a bicinchoninic acid (BCA) kit from Sigma. An expected typical concentration is 5 mg/mL.
-
b.One milligram of protein supernatant is placed into each 1.5 mL centrifuge tube. For each sample prepare, three centrifuge tubes are prepared.
-
i.The supernatant is placed in a 1.5 mL centrifuge tube and diluted with lysis buffer to reach a 600 μL total volume.
-
ii.Triton X-100 (6 μL 100% Triton X-100 into 600 μL volume of cell lysates) and final 25 mM N-ethylmaleimide (NEM) are added to the samples.
-
iii.Incubate the protein samples at 4°C for 30 min with gentle full-angle rotation.
-
iv.One of the two tubes of each sample is retained at –80°C for back up and another tube is subject to precipitation with chloroform-methanol (see the next step).
-
i.
-
a.
Pause Point: The collected yeast samples can be stored at –80°C for 2 weeks.
Chloroform–methanol (CM) precipitation and NEM block of samples
Timing: 7 h
In this step, a chloroform-methanol (CM) precipitation assay is used to remove the residual buffer in the protein samples.
-
4.One milligram of protein, diluted with LB buffer (with PI) to a final volume of 1 mL, is transferred into larger polypropylene centrifuge tubes (at least 10 mL).
-
a.A 4-fold volume (4 mL) of methanol is added to the tubes and vortexed thoroughly to mix the solution.
-
b.A 1.5-fold volume (1.5 mL, use a glass pipette) of chloroform is added to the tubes and vortexed thoroughly to mix the solution.
-
c.A 3-fold volume (3 mL) of distilled H2O is added to the tubes and vortexed thoroughly to mix the solution.
-
a.
-
5.
Place the tubes at 4°C for 20 min. Centrifuge (12,000×g, 10 min, and 4°C) to separate the phases. The samples will be separated into two phases, with protein precipitated at the interphase (Figure 4). Discard the top phase (H2O and methanol) carefully using Pasteur pipettes. Add 3 mL methanol followed by gentle up and down mixing to maintain the protein interface. Centrifuge (3,000×g, 10 min, and 4°C) so that protein precipitates can be pelleted to the tube bottom. Discard the supernatant with a Pasteur pipette (remove all of the supernatant).
-
6.
The precipitated proteins are resolubilized in 200 μL SB buffer and incubated at 37°C for enhanced dissolution.
-
7.
Dilute with 800 μL LB buffer with 10 mM NEM and PI, incubate at 25°C for 1–2 h or 16 h at 4°C.
-
8.
Precipitate the protein by repeating the CM assay three times to remove the NEM: add a 4-fold volume (4 mL) of methanol to the tubes, and vortex thoroughly to mix the solution. A 1.5-fold volume (1.5 mL, use a glass pipette) of chloroform is added to the tubes and vortexed thoroughly to mix the solution. A 3-fold volume (3 mL) of distilled H2O is added to the tubes and vortexed thoroughly to mix the solution. Centrifuge (12,000×g, 10 min, and 4°C) to separate the phases. The samples will be separated into two phases, with the protein precipitated at the interphase. Discard the top phase (H2O and methanol) carefully using Pasteur pipettes. Add 3 mL methanol again followed by gentle up and down mixing to maintain the protein interface. Centrifuge (3,000×g, 10 min, and 4°C) so that the protein precipitates can be pelleted to the tube bottom. Discard the supernatant with a Pasteur pipette (remove all of the supernatant).
-
9.
The protein is air dried, dissolved in 450 μL SB buffer, and incubated at 37°C for 20–30 min.
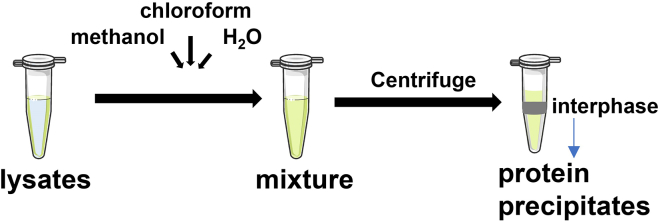
Figure 4.
Pictorial depiction of precipitated proteins in the interphase
Yeast cell lysates were precipitated by sequential adding of methanol, chloroform and H2O. After centrifuge, the proteins were separated in the interphase.
CRITICAL: Any methanol and chloroform remaining in the samples will make it difficult to redissolve the protein precipitates. The protein layer in the interphase should not be disrupted.
Pause Point: The dried protein samples can be stored at –80°C for 1 week.
Note: Any remaining NEM will reduce the acyl-biotin exchange reaction to a large extent. When the cysteine sites released by hydroxylamine are exposed, thiols would be modified by the residual NEM, which results in blocking of the subsequent reaction with the biotin reagent. To ensure that the NEM is removed cleanly, this CM precipitation assay may need to be repeated more than three times.
Acyl-biotin exchange reactions
In this step, the palmitoyl conjugated on cysteine will be removed and then replaced by biotin, which will generate affinity tag for followed recognition and precipitation.
Timing: 2 h
-
10.
The protein samples are divided into two equal aliquots, retaining 50 μL as input for western blotting.
-
11.
One aliquot (200 μL) is mixed with 800 μL of With-HA (with hydroxylamine) buffer, and the other is mixed with 800 μL of Without-HA (without hydroxylamine) buffer. Both aliquots are incubated in the individual buffer for 1–2 h at 25°C by mixing gently.
-
12.
Chloroform and methanol assays are used again to precipitate the proteins as described above in steps 4–5.
-
13.
The protein precipitates obtained by the CM assay are dissolved in 100 μL TB buffer, incubated at 37°C for 10 min and diluted with 900 μL LB buffer.
-
14.
To remove the unreacted biotin, the CM assay is used again to precipitate the protein, which can be precipitated three times continuously if any concern about the unreacted biotin being present is raised.
Note: Any unreacted biotin will interfere with and compete with the streptavidin-agarose in the following steps.
Immunoprecipitation with streptavidin-agarose
In this step, the biotin-cysteine integrated proteins will be recognized and captured by Streptavidin-Agarose.
Timing: 2 h
-
15.
Thirty microliters of streptavidin-agarose beads were added to the samples and incubated at 4°C for 1–2 h.
-
16.
The beads were washed with washing buffer 5 times to remove any nonspecifically bound proteins.
-
17.
Add 60–80 μL Elution buffer and then boil the beads for 5 min as pulldown samples. Load 10 μL of the input samples and Pulldown samples into SDS–polyacrylamide gel electrophoresis (SDS-PAGE) gels followed by western blotting with the anti-HA antibody.
Expected outcomes
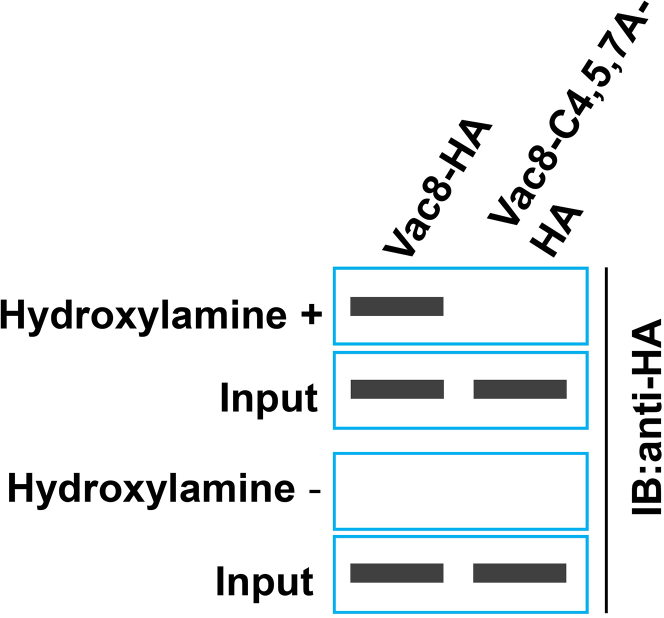
The results of western blotting analysis of the pulldown samples show that wild-type Vac8 is palmitoylated, while the C4A/C5A/C7A mutant of Vac8, with three cysteines mutated to alanine, cannot be palmitoylated (schematic diagrams of blots are shown in Figure 5). This result is consistent with the localization data showing that wild-type Vac8 localizes to the vacuole membranes, while the C4A/C5A/C7A mutant of Vac8 completely loses this distribution.
Figure 5.
Expected detection of palmitoylation of Vac8
The biotin conversion procedure was performed with hydroxylamine (HA) and biotin BMCC as cross-linking agents. The plasmids Vac8-HA and Vac8-C4,5,7A-HA were transferred into yeast, and then the protein was extracted to detect palmitoylation. The cross-linked proteins were analyzed by SDS-PAGE and immunoblotting.
Schematic diagrams (not real blots) are shown.
Limitations
Protein palmitoylation can occur on cysteine residues (S-palmitoylation), on the amino terminus or on the epsilon amino group of lysine (N-palmitoylation), or on serine and threonine residues (O-palmitoylation) (Sobocinska et al., 2018). S-palmitoylation is the most common type of palmitoylation, and this protocol is limited to this kind of modification (Sobocinska et al., 2018). To detect N-, O- or S-palmitoylation, labeling cells or cell extracts with [3H]-palmitate or fluorescent probes can be applied (Gao and Hannoush, 2014). The detection of palmitoylation in plants has been described (Hermsley et al., 2008; Hemsley and Grierson, 2008).
Troubleshooting
Problem 1
Bad morphology of yeast cells and florescence of protein localization (step 2 of before you begin).
Potential solution
Use log-phase (OD600 0.8–1) yeast cells.
Problem 2
Difficult to dissolve the protein precipitates after CM assays (step 9 of step-by-step method details).
Potential solution
The protein precipitates were not air-dried for long enough, as methanol and chloroform remaining in the samples reduces the dissolution of the protein precipitates.
Problem 3
Little palmitoylation is detected although there is enough signal in the Input fraction (step 10 of step-by-step method details).
Potential solution
Palmitoylation is reversible, so the yeast cells need to be frozen in liquid nitrogen immediately after collection. Keep the cell breaking steps (steps 1–3) at 4°C. Residual NEM will block thiols exposed by hydroxylamine to increase the number of chloroform–methanol precipitates. Positive controls that have been shown to be palmitoylated are highly recommended to indicate whether the whole protocol works or not. Similarly, negative controls that contain mutations (cysteine to alanine) are also recommended to indicate the nonspecific detection of palmitoylation.
Problem 4
The nonspecific palmitoylation signal is too strong (step 13 of step-by-step method details).
Potential solution
Keep the protein amounts at 1 mg in step 3, as too much protein input may interfere with the efficiency of NEM blockage of free thiols (step 3) and cause nonspecific binding to the neutravidin-agarose beads (step 13). Increasing the washing times in step 14 is also suggested.
Problem 5
The palmitoylation signal is not stable, e.g., sometimes strong and sometimes weak between repeats (step 17 of step-by-step method details).
Potential solution
Two steps are essential for providing an extended time for stable experimental outcomes due to uneven mixing effects and therefore unstable binding. The first step is NEM blocking. NEM is used to block unmodified free cysteines in protein peptides, and unsaturated NEM blocking might cause false-positive results or increase the background signal in the final WB analysis. The second step is the addition of streptavidin-agarose beads, which are incubated with the protein samples to pull down biotin-labeled proteins. Insufficient incubation time of the beads with the samples might result in inefficient biotin-streptavidin binding and thus a weak signal of protein palmitoylation in the final WB. For both steps, incubations at 4°C for 16 h are recommended.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Kefeng Lu (lukf@scu.edu.cn).
Materials availability
Plasmids and yeast strains used in this study are available from our laboratory upon request.
Acknowledgments
This work was supported by the National Key R&D Program of China under grants 2017YFA0506300 (to K.L.) and the National Natural Science Foundation under grants 31970693 (to K.L.) and 81902997 (to H.L.).
Author contributions
Conceptualization, K.L., E.K., and H.L.; methodology, Y.L. and J.Z.; investigation, Y.L. and J.Z.; writing – original draft, K.L.; writing – review & editing, K.L.; funding acquisition, K.L.; resources, K.L.; supervision, K.L.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Yuqing Lei, Email: 1244837417@qq.com.
Huihui Li, Email: lihuihui@scu.edu.cn.
Eryan Kong, Email: eykong@xxmu.edu.cn.
Kefeng Lu, Email: lukf@scu.edu.cn.
Data and code availability
Source data in this paper is available upon request. This study did not generate new dataset or code.
References
- De I., Sadhukhan S. Emerging roles of DHHC-mediated protein S-palmitoylation in physiological and pathophysiological context. Eur. J. Cell Biol. 2018;97:319–338. doi: 10.1016/j.ejcb.2018.03.005. [DOI] [PubMed] [Google Scholar]
- Gao X., Hannoush R.N. Single-cell in situ imaging of palmitoylation in fatty-acylated proteins. Nat. Protoc. 2014;9:2607–2623. doi: 10.1038/nprot.2014.179. [DOI] [PubMed] [Google Scholar]
- Hemsley P.A., Grierson C.S. Multiple roles for protein palmitoylation in plants. Trends Plant Sci. 2008;13:295–302. doi: 10.1016/j.tplants.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Hermsley P.A., Taylor L., Grierson C.S. Assaying protein palmitoylation in plants. Plant Methods. 2008;4:1–7. doi: 10.1186/1746-4811-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y., Zhang X., Xu Q., Liu S., Li C., Jiang H., Lin H., Kong E., Liu J., Qi S., et al. Autophagic elimination of ribosomes during spermiogenesis provides energy for flagellar motility. Developmental Cell. 2021;56:2313–2328.e7. doi: 10.1016/j.devcel.2021.07.015. [DOI] [PubMed] [Google Scholar]
- Ren J., Wen L., Gao X., Jin C., Xue Y., Yao X. CSS-Palm 2.0: an updated software for palmitoylation sites prediction. Protein Eng. Des. Selection. 2008;21:639–644. doi: 10.1093/protein/gzn039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland E.A., Snowden C.K., Cristea I.M. Protein lipoylation: an evolutionarily conserved metabolic regulator of health and disease. Curr. Opin. Chem. Biol. 2018;42:76–85. doi: 10.1016/j.cbpa.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobocinska J., Roszczenko-Jasinska P., Ciesielska A., Kwiatkowska K. Protein palmitoylation and its role in bacterial and viral infections. Front. Immunol. 2018;9:2003. doi: 10.3389/fimmu.2017.02003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Source data in this paper is available upon request. This study did not generate new dataset or code.