Abstract
Vitiligo is an autoimmune skin disease characterized by the targeted destruction of melanocytes by T cells. Cytokine signaling between keratinocytes and T cells results in CD8+ T cell infiltration of vitiligo lesions, but the full scope of signals required to coordinate autoimmune responses is not completely understood. We performed single-cell RNA sequencing (scRNA-seq) on affected and unaffected skin from vitiligo patients, as well as healthy controls, to define the role of each cell type in coordinating autoimmunity during disease progression. We confirmed that type 1 cytokine signaling occupied a central role in disease, but we also discovered that this pathway was used by regulatory T cells (Tregs) to restrain disease progression in non-lesional skin. We determined that CCL5-CCR5 signaling served as a chemokine circuit between effector CD8+ T cells and Tregs, and mechanistic studies in a mouse model of vitiligo revealed that CCR5 expression on Tregs was required to suppress disease in vivo but not in vitro. CCR5 was not required for Treg recruitment to skin but appeared to facilitate Treg function by properly positioning these cells within the skin. Our data provides critical insights into the pathogenesis of vitiligo and uncovers potential opportunities for therapeutic interventions.
One Sentence Summary:
scRNA-seq of human vitiligo samples reveals complex autoimmune networks, including a role for CCR5 in Treg function.
Vitiligo Allies and Villains
Vitiligo is an autoimmune skin disease defined by T cell-mediated destruction of melanocytes. Gellatly et al. (p. 0000) used single cell RNA sequencing (scRNA-seq) to characterize immune cell subsets that are associated with skin lesions in vitiligo. Type 1 cytokine signaling is a key driver of disease progression this signaling pathway was also used by regulatory T cells (Tregs) to limit disease in non-lesional skin. CCR5-CCL5 signaling was critical to effector CD8+ T cell and Treg function, and mouse studies showed that disease suppression required CCR5 expression on Tregs. These studies reveal the complex interactions between immune cell subsets that are involved in vitiligo progression and containment.
INTRODUCTION
Vitiligo is an autoimmune disease of the skin in which CD8+ T cells lead to the focal elimination of melanocytes, resulting in the formation of depigmented white spots. It affects ~1% of the world population, and has a significant impact on patients’ quality of life (1). Current treatments for vitiligo are moderately effective at returning pigment to the skin but are cumbersome, and more effective targeted therapies are needed (2). Vitiligo appears in a heterogeneous pattern, with patches of activity scattered within otherwise normal-appearing skin (1). How and why immune cells initiate autoimmune responses at one site in the skin while sparing others is largely unknown. Previous studies suggested that regulatory T cells (Tregs) may be defective in vitiligo patients; however, these studies disagree on whether the defect is due to decreased systemic Treg numbers, decreased ability to home to the skin, or decreased function (3-10). Thus, it is not clear what role Tregs play in the onset, progression, and distribution of vitiligo lesions.
Recent mechanistic studies in mice identified several signaling pathways (e.g. IFN-γ and IL-15) as key drivers of inflammation and disease (11-13). Initial drug repurposing studies have shown that blocking these interactions is effective in vitiligo treatment (2). With the goal of expanding the treatment options and to ask fundamental questions about the initiation and progression of vitiligo, we sought to build a comprehensive view of the signaling pathways among cells of the epidermis using single cell RNA-sequencing (scRNA-seq), flow cytometry, and ELISA. We used suction blistering (14) to sample active human vitiligo lesions, unaffected non-lesional skin, and healthy control skin.
RESULTS
scRNA-seq of suction blisters identifies key cell types in the skin of vitiligo subjects
We performed suction blistering followed by scRNA-seq on 10 subjects with active vitiligo (treatment-naïve for at least 6 months) and 7 healthy individuals (table S1, table S2). For each vitiligo subject, we collected 2 sets of blisters, one lesional sample from affected skin and one non-lesional sample from unaffected skin, located at least 10cm away from any visible lesion (Fig. 1A). After stringent filtering of the scRNA-seq data, we detected 32,405 cells, each with at least 500 detectable transcripts across 14,759 genes (table S3). Unbiased spectral clustering (15) grouped cells into 10 distinct clusters that were well represented across all assayed samples (fig. S1A).
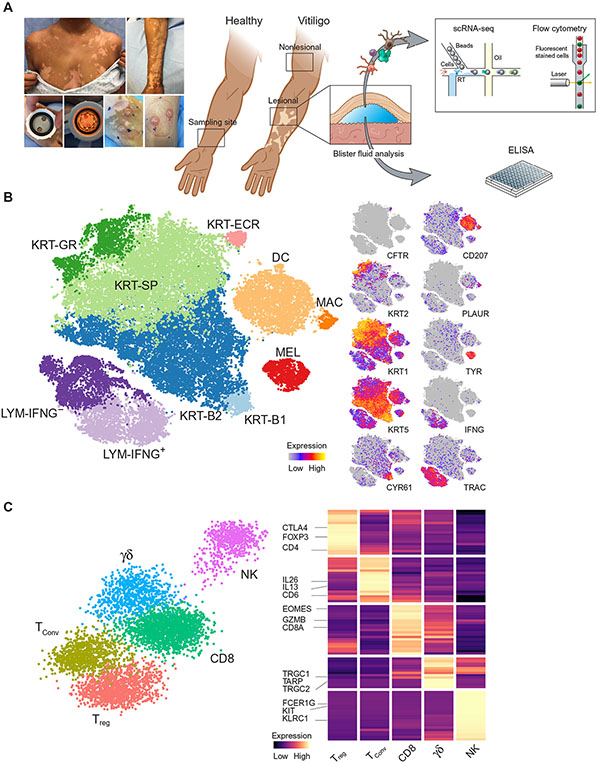
Fig. 1: scRNA-seq of vitiligo skin reveals epidermal cell types and immune cells.
Suction blister biopsies were obtained from healthy skin as well as non-lesional and active lesions from vitiligo subjects. The supernatant was analyzed by ELISA while the skin cells were analyzed by flow cytometry or single cell RNA-sequencing (A). Unbiased clustering of the scRNA-seq data resulted in 10 distinct clusters, consisting of 5 main cell types. Melanocytes (MEL), dendritic DCs (DC), and Macrophages were each identified as a single cluster. Keratinocytes (KRT) consisted of 5 clusters, Basal-1 (B1), Basal-2 (B2), Spinous (SP), Granular (GR), and Eccrine (ECR). Lymphocytes consisted of 2 clusters, IFNG− and IFNG+ (B). Refined clustering of the original TRAC+ cluster revealed 5 distinct cell types, Tregs (FOXP3+/CTLA4+), TConv (FOXP3−/IL13+/IL26+), CD8+ T cells (TRAC+/CD8+), γδ T cells (TRGC1+/TRDC+), and NK cells (FCER1G+/KIT+) (C).
The 10 clusters represented the five broad cell types typically found in the epidermis as was evident by cluster-specific expression of genes that included known cell type specific markers: melanocytes (TYR+/DCT+/KIT+), keratinocytes (KRT1+/KRT2+/KRT5+), lymphocytes (TRAC+/CD3+), macrophages (CD83+/CD87+/CSF1R+), and Langerin-positive dendritic cells (CD207+/CD1A+) (Fig. 1B, fig. S1B). The large number of keratinocytes (66% of all cells) enabled subclassification corresponding to their differentiation states, including two groups of basal keratinocytes (KRT-B1, KRT-B2) that were both positive for KRT5 and KRT14, whereas the KRT-B1 group also expressed CYR61 and ADRB2. We found spinous keratinocytes (KRT-SP) expressing KRT1 and KRT10, granular keratinocytes (KRT-GR) expressing KRT2 and KLK11 (16), and a population of eccrine keratinocytes (KRT-ECR) expressing CFTR and MUCL1 (17) (Fig. 1B, fig. S1B). We detected all epidermal cells that were previously reported in scRNA-seq datasets from skin punch biopsies, but no cell types from deeper within the dermis such as fibroblasts or fat cells (18, 19).
Initially, our TRAC+ lymphocyte cluster had split based on IFNG expression, with LYM-IFNG+ and LYM-IFNG− clusters (Fig. 1B). Because this cluster did not segregate into established functional subtypes (e.g., CD4+ and CD8+ T cells), we re-analyzed these cells independently using a supervised clustering approach that relied on known lymphocytes markers.
This resulted in five clear lymphocyte subsets based on established cellular markers, representing 2 groups of CD4+ T cells (one with high FOXP3 expression and the other with low FOXP3 expression), CD8+ T cells, γδ T cells, and NK cells (Fig. 1C, fig. S1C). To determine whether the CD4+/FOXP3+ cell cluster represented true Tregs, we tested for the enrichment of an expression signature that distinguishes Tregs from conventional T (TConv) cells (20, 21) using gene set enrichment analysis (GSEA). This method is advantageous due to the fact that FOXP3 alone is often insufficient to fully classify a T cell as a Treg, because effector T cells can express FOXP3 in certain conditions (22). The CD4+/FOXP3+ cluster had significantly (p = 1.2E−3) higher expression of the gene set reported to be increased within Tregs and significant (p = 2.2E−2) downregulation of the gene set that was decreased within Tregs (fig. S1D). Based on these findings, we categorized the CD4+/FOXP3+ cluster as true Tregs, and the other CD4+ cluster as TConv cells due to their lack of expression of genes tied to Treg identity. The TConv cells expressed multiple cytokines that represented diverse inflammatory pathways, and these were expressed at increased levels compared with Tregs and included IL13 (type 2) (23) and IL26 (type 17) (24) (fig. S1C). The last 3 groups of cells that originally clustered with lymphocytes were CD8+ T cells marked by CD8A/EOMES, γδ T cells marked by TRGC1/TRGC2/TRDC, and NK cells marked by KIT/FCER1G/KLRC1 (Fig. 1C, fig. S1E). NK and γδ T cells had not been previously identified via scRNA-seq analysis of mouse or human skin samples in healthy skin (16, 18), thus we confirmed their identity using flow cytometry on blister fluid for canonical markers (table S4, fig. S1F, G), which revealed their presence at low abundance.
Lesional skin is characterized by T cell infiltration, and CD8+ T cells are the primary source of IFN-γ
The cell type proportions inferred from our single cell data recapitulated previously reported changes in the cell type composition within vitiligo lesions, with a large influx of lymphocytes and a concomitant loss of melanocytes specific to lesional skin (Fig. 2A) (25). Specifically, we found the greatest increase in the proportion of both CD8+ T cells and Tregs, with no dramatic changes in the ratio of Tregs to CD8+ T cells (Fig. 2B).
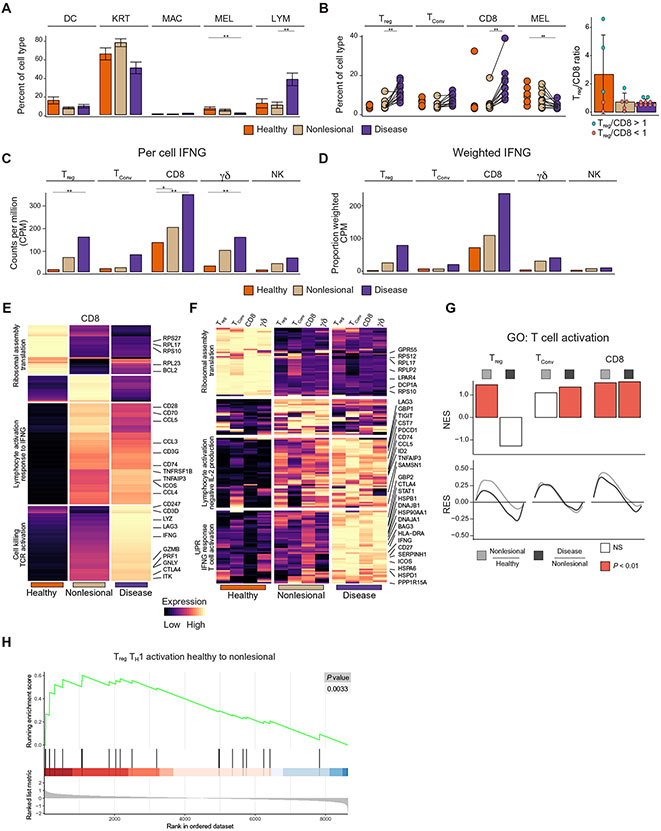
Fig. 2: Activation of immune cells reveals subclinical inflammation in non-lesional vitiligo skin.
Barplots reflecting the fraction of each cell type in each skin condition across all donors (healthy, non-lesional and lesional); error bars indicate standard error of the mean (A). Dotplot showing the paired proportions of lymphocytes and melanocytes within each patient, with an inset showing the ratio of Treg to CD8 T cells for patients where over 30 cells of these types were detected (B). Barplot of CPM (counts per million) values of IFNG in the T cell subsets and NK cells (C). Barplot of the weighted (CPM X Cell proportion) CPM values of IFNG in the T cell subsets and NK cells (D). K-means clustering of all differentially expressed genes within CD8+ T cells (E). K-means clustering of all genes found differentially expressed in more than two T Cell subsets (F). GSEA of the T cell activation pathway within Treg, TConv, and CD8+ T cells between all skin transitions. NES = normalized enrichment score, RES = running enrichment score (G). GSEA of the Th1 Activation pathways within Tregs in the transition from healthy to non-lesional skin (H). *FDR < 0.1, **FDR < 0.01
IFN-γ signaling is central to vitiligo pathogenesis (26), yet the source of this cytokine in inflamed skin is not well understood. Consistent with previous studies, we found that T cells had higher IFNG expression in both lesional and non-lesional skin of vitiligo patients (27), with CD8+ T cells having a ~2 fold significant (FDR = 1.1E-8) increase in IFNG expression on a per cell basis between affected and healthy skin (Fig. 2C). Although only a modest per-cell increase was found, when coupled with the large influx of CD8+ T cells within lesional tissue (Fig. 2B), it became clear that CD8+ T cells were the predominant source of IFN-γ that accounted for the previously reported amplification of interferon stimulated genes, including CXCL9 and CXCL10 (Fig. 2D) (14, 28). Interestingly, Tregs represented the second major producer of IFN-γ within vitiligo lesions (Fig. 2D).
T cells in non-lesional skin are in a subclinical state of activation
Given the role of CD8+ T cells in autoimmunity and in vitiligo in particular (2, 29), we focused on differentially expressed genes within this group. Clustering of the aggregated gene expression profile within CD8+ T cells revealed a series of transcriptional transitions between healthy, non-lesional, and lesional skin. We used Gene Ontology (GO) to determine whether there were significantly enriched gene sets within each bin of the resulting heatmap. Despite normal melanocyte numbers within the non-lesional skin of vitiligo patients, there is a significant response to IFNγ (FDR = 1.9E−3) and T cell activation (FDR = 3.3E−6) within CD8+ T cells in non-lesional skin (Fig. 2E), including transcripts for costimulatory molecules CD28 and ICOS, as well as chemokines CCL3/4/5. This contrasts expression of transcripts for cytotoxic molecules such as GZMB, GNLY, and PRF1, which are not upregulated until the final transition between non-lesional and lesional skin (Fig. 2E). GZMB, GNLY, PRF1 and transcripts for other cytotoxic molecules were also found to be highly expressed within NK cells in lesional skin, raising the possibility that they participate in melanocyte clearance (fig. S2A).
We found several pathways that were dysregulated within all T cell subsets, which suggested global activation patterns within the lymphocytes of vitiligo patients. A strong and conserved downregulation of ribosomal proteins, which has been suggested to influence T cell differentiation and activation (30), and upregulation of immune checkpoint inhibitors (LAG3, TIGIT, PDCD1), even in non-lesional skin (Fig. 2F, fig. S2B). In the final transition to lesional skin, there was strongly induced expression of genes associated with the unfolded protein response (UPR), IFN-γ, and T cell activation responses, HSPs, STAT1, and IFNG (Fig. 2F, fig. S2C, D). HSPs have been previously implicated in vitiligo (31) and have been proposed to potentiate cytotoxic T cell responses (32), whereas T cell activation coincides with IFN-γ sensing in vitiligo skin. CD8+ T cells upregulate CTLA4 (fig. S2D) and downregulate BCL2 (fig. S2E), a trend consistent with that of melanocyte-specific CD8+ T cells rendered anergic by Treg suppression in vitro (6). This observation suggests that despite the presence of depigmentation, activated Tregs may attempt to suppress CD8+ T cells in lesional skin.
GSEA analysis showed that Tregs are fully activated in non-lesional skin, and CD8+ T cells become significantly more activated with increases from healthy to non-lesional skin (p = 7.2E−3) and then again from non-lesional to lesional skin (p = 4.5E−2) (Fig. 2G). In addition to the activation state of Tregs in non-lesional skin and clear response to IFN-γ, we noted that Tregs unexpectedly expressed a type 1 inflammatory profile, including IFNG and TBX21, as well as chemokine receptors CXCR3 and CCR5 (fig. S2F), despite maintaining their Treg identity (fig. S1D). To confirm this upregulation of type 1-specific markers in Tregs, we conducted GSEA using a published list of expressed genes characteristic of naive CD4+ T cells polarized into Th1 cells in vitro (33). The Th1 cell gene set was significantly (p = 3.3E−3) upregulated in the transition from healthy to non-lesional skin within Tregs (Fig. 2H). It is unclear whether this transition to a type 1 inflammatory program in Tregs indicates that they are losing their suppressive function or developing enhanced suppression by adapting to a type 1 inflammatory environment. It should be noted that expression of IL10, an immunosuppressive cytokine used by Tregs, was increased within lesional skin, although this did not reach significance (fig. S2G) (34, 35).
Cell type specific responses to IFNγ lead to a subclinical inflammatory state within non-lesional skin
One of the key events that mediates lesion formation in vitiligo is increased IFNG expression. It was surprising to find elevated IFNG expression in non-lesional skin, whereas depigmentation was only observed in lesional skin. We conducted GSEA comparing healthy to non-lesional, as well as non-lesional to lesional skin to determine the skin state in which each cell type responds to the IFN-γ signal. CD8+ T cells exhibited a strong upregulation of IFN-γ response genes even when comparing non-lesional to healthy skin (Fig. 3A). This indicated that CD8+ T cells respond to IFN-γ even within non-lesional skin, suggesting that non-lesional skin is subclinically active rather than simply uninvolved.
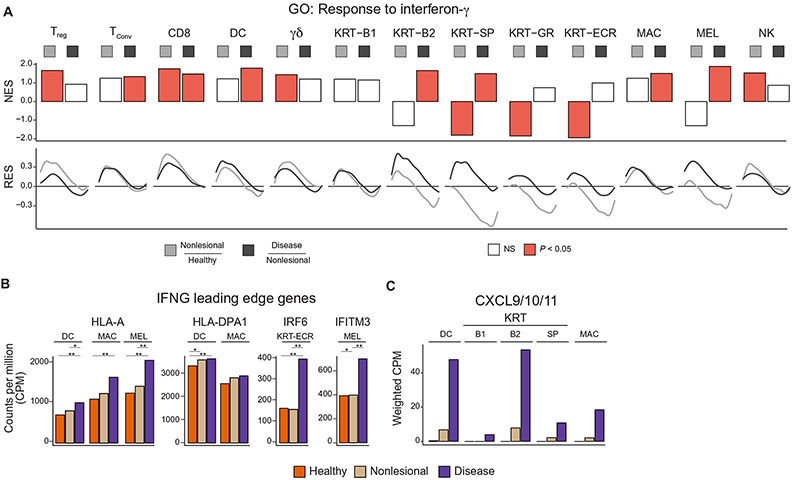
Fig. 3: Response to IFNG in healthy and vitiligo skin.
GSEA enrichment plot showing the IFNG response within all cell types detected within the epidermis and p value for the IFNG response between healthy to non-lesional, and non-lesional to lesional skin. NES = normalized enrichment score, RES = running enrichment score (A). Barplots showing the cell type specific expression of IFNG responsive genes, HLA-A, HLA-DPA1, IRF6, and IFITM3 (B). Barplot showing the weighted CPM expression levels of CXCL9/10/11 within APCs and keratinocytes (C). *p < 0.1, **p < 0.01
CD8+ T cells were not the only lymphocyte with dramatic upregulation of IFN-γ response pathways in non-lesional skin. Four of the major producers of IFN-γ, CD8+, Treg, γδ T cells, and NK cells, showed increased upregulation of IFN-γ response pathways in non-lesional skin, but only CD8+ T cells further increased this response in the transition from non-lesional to lesional skin. In contrast, Tregs reached maximal IFN-γ response within non-lesional skin and did not further increase this response in lesional skin. CD8+ T cells sensed IFN-γ even within non-lesional skin, but melanocytes themselves did not respond to the IFN-γ signal until the transition to lesional skin, coinciding with melanocyte loss and depigmentation. It is still unclear for what reason CD8+ T cells respond to IFN-γ within non-lesional skin, whereas melanocytes only responded within lesional skin.
Given the critical importance of IFN-γ-induced changes in the skin and the differences in the stage at which each cell types responded to IFN-γ, we sought to define the shared as well as cell type-specific responses to IFNγ. We used the combination of all leading-edge genes found by GSEA within the IFN-γ response GO term, across all cell types and skin type comparisons (fig. S3A). Clustering of the resulting aggregate bulk heatmap revealed both conserved as well as cell type specific IFN-γ responses. Cluster 1 represented the conserved, core IFN-γ-responsive genes that were induced across most cell types, including MHC class I molecules and IRF1/2 (Fig. 3B, fig. S3A). Other clusters revealed changes in expression specific to individual cell types. Cluster 2 was specific to antigen presenting cells and consisted of MHC class II genes (Fig. 3B, fig. S3A), Cluster 4 had KRT-ECR specific genes such as IRF6 (Fig. 3B, fig. S3A), and Cluster 6 contained melanocyte-specific genes such as IFITM3 (Fig. 3B, fig. S3A). Both shared and cell type specific IFN-γ responses are generated within each cell type of the skin during vitiligo progression.
We found that KRT-B2 keratinocytes had the most significant (p = 4.4E−3) response to IFNγ within lesions, whereas other keratinocytes subtypes had variable responses. This same KRT-B2 group is also responsible for most of the production of the CXCR3 ligands CXCL9/10/11 when factoring in the large number of these cells within the skin (Fig. 3C, S3B, C). This is in contrast to antigen presenting cells (APCs), including dendritic cells and macrophages, that upregulated CXCL9/10/11 to the greatest extent on a per-cell basis (fig. S3B), supporting the hypothesis that APCs may also have a role in T cell recruitment (12).
Receptor and ligand mapping reveal cell type-specific signaling programs
To better characterize the events that lead to T cell recruitment and ultimately to melanocyte elimination from the skin, we systematically analyzed all cellular communications among all cell types within the epidermis utilizing a published receptor and ligand database (36) to identify all possible cell-to-cell signaling events. We independently clustered normalized expression values of all expressed ligands and receptors, across each cell type and in each condition. This revealed that most ligands (Fig. 4A) and receptors (Fig. 4B) have strong cell type-specific expression, reflecting unique signaling programs and the potential for distinct “private conversations" employed between cell types. Gene ontology (GO) analysis revealed that melanocytes expressed many molecules necessary for the maintenance of the extracellular matrix (ECM) and cell-cell adhesion, including fibronectins, collagens, and laminins. These genes are necessary for melanocyte adherence to the basement membrane of the epidermis. Most keratinocyte-specific ligands were enriched in general processes associated with skin development and function (Fig. 4A), such as the WNT signaling pathway (WNT3) and bone morphogenic proteins (BMP4). Within the immune cells, there was enrichment in biological processes related to their function. Macrophages and dendritic cells expressed immune response genes such as CCL22 and C3 (DCs), as well as the CXCR3 ligands CXCL9-11 and IL1B (MACs). Interestingly, even within T cells there was specificity in ligand expression with IFNG, GZMB, and CCL5 enriched in CD8+ T cells, reflecting their role in cytotoxicity and T cell recruitment. IL13 (a type 2 cytokine) and IL26 (a type 17 cytokine) were specifically expressed by TConv cells, and NCAM1 and CCL1 were expressed by NK cells.
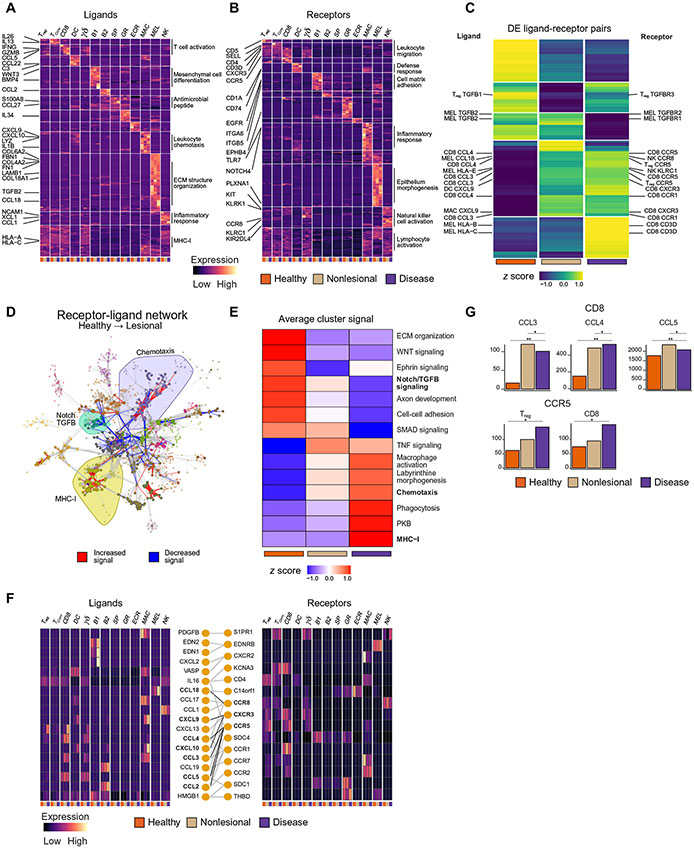
Fig. 4: Ligand receptor mapping establishes communications between cells and their change in vitiligo.
K-means clustering of all expressed ligands (A) and receptors (B) across each cell type and each skin condition, as annotated by a published database with representative GO terms for each k group. Each box represents the aggregate bulk CPM value for a given gene expressed in that cell type in each condition. K-means clustering of all ligand-receptor pairs in which at least one of the genes is significantly (p < 1.0E−2) DE (C). Plot of the full receptor and ligand network showing the 18 identified clusters within the network, with the Chemotaxis, TGFB / Notch Signaling, and MHC-I clusters highlighted. Edge color denotes whether the given interaction is increased or decreased in signal between healthy and lesional skin (D). K-means clustering of the meta-edges of each cluster within the network across each of the skin conditions (E). Chemokine network heatmaps showing the expression of ligands and receptors, as well as their annotated interactions (F). Barplots showing CCL3/4/5 expression within CD8+ T cells and CCR5 expression on Tregs and CD8+ T cells (G)
For receptors, clustering revealed expression characteristic of specific cell types, such as Toll-like receptors (e.g., TLR2/TLR7) in macrophages, CD1A in DCs, classic T cell markers (e.g., CD3D/G, CD4, CD5) and cytotoxicity-specific markers (KLRK1) in T cells, as well as NK-specific markers (e.g., KLRC1, KIR2DL4) and the chemokine receptor CCR8 in NK cells (Fig. 4B). Melanocytes primarily expressed integral membrane proteins involved in epithelial morphogenesis and axonogenesis, reflecting their neural crest origin. Keratinocytes were enriched in functions such as wound healing and hemidesmosome assembly. The hemidesmosome is a keratinocyte-specific structure responsible for adherence to the basement membrane, and the basal keratinocytes (KRT-B1 cluster) expressed ITGA6 and COL17A1, specifically reflecting their role in basement membrane production and adherence (37). We found melanocytes expressed the cognate ligands for these receptors including FN1, LAMBA4, and LAMB1, reflecting their adhesion to the basal keratinocyte layer.
Signaling changes within vitiligo skin identify complex chemokine circuits that may drive lymphocyte localization
We computed connection strengths for each individual ligand-receptor pair to determine how a given communication may change in association with disease. This connection strength was defined as the product of the ligand and receptor CPM. Each ligand-receptor pair had three values: one for healthy, one for non-lesional skin, and one for lesional skin. To understand which cell signals contributed to vitiligo pathogenesis, we clustered the connection strength values for any ligand-receptor pair in which either the ligand or the receptor was significantly (p < 1.0E−2) differentially expressed between any skin state. In total, 2907 connections were found to be dysregulated (Fig. 4C), revealing previously known signals such as CXCL9 to CXCR3, as well as many communications that had previously not been implicated in vitiligo pathogenesis
Given the natural graph representation of the ligand and receptor data, we defined a network in which nodes are defined as a cell type expressing a ligand or receptor, and edges are drawn between nodes to reflect an annotated interaction between a given ligand-receptor pair. Thus, we integrated two sources of information: a database of ligands and receptors that determined the interactions or edges of the network, and the single cell data that determined the sources of each ligand or receptor. The resulting graph consists of 2,202 nodes and 15,986 edges. Louvain clustering on the resulting ligand and receptor graph produced 18 distinct clusters of defined cell-cell communications. Nodes within each cluster tended to belong to specific biological processes and were annotated by the most highly enriched GO category within that cluster (fig. S4A, B). Coloring edges of the graph by the net direction of change in the connection strength between healthy and lesional skin showed certain clusters of the network had consistent upward (MHC-1, chemotaxis) or downward (Notch/TGF-β signaling) changes in signal (Fig. 4D). By averaging over all connection strength values within each cluster of the graph, we captured the general trends in the change of signal for the 14 clusters that had dramatic changes (Fig. 4E, fig. S4C).
There was a melanocyte-specific expression and loss of TGFB2 signaling in lesional skin, within the TGF-β signaling cluster, whereas TGFB1 was lost within Tregs (fig. S4D). TGF-β1 has been implicated in immune regulation, and its loss may lead to a lack of tolerance in lesional tissue (38), whereas TGF-β2 has been shown to be essential for melanocyte development, and its loss may contribute to aberrant differentiation and growth of melanocytes (39). IFN-γ has been shown to directly affect TGFB expression (40), and this may explain its loss in lesional skin. MHC-I signaling was increased within lesional skin, with HLA-A, HLA-B, and HLA-E all being upregulated in melanocytes (fig. S4E). CD8+ T cells expressed CD3D, the cognate receptor for HLA-A/B, and NK cells highly expressed KLRC1, the cognate receptor for HLA-E. This suggested an antigen recognition paradigm between melanocytes and NK cells in vitiligo that has not been previously reported and supports our previous data that NK cells develop a cytotoxic profile within lesional skin (fig. S2A) and thus may participate in melanocyte killing.
We aimed to expand on the previously known intercellular chemotactic signals in vitiligo given the enrichment of chemotaxis genes during disease progression and their amenability to therapeutic targeting. Chemokine expression may inform pathways that govern migration of key immune cells in the skin from the bloodstream, within the skin to the epidermis, or promote their aggregation into discrete groups around specific chemokine sources within the epidermis. To address this, we extracted the chemotaxis cluster from the full network and made a heatmap of all expressed ligands and receptors within this cluster (Fig. 4F). This showed both the cell type specificity of CXCR3 ligand expression (highest expression in APCs) as well as the expression changes within the CXCL9/10/11-CXCR3 chemokine circuit. It also revealed chemokine circuits that had not been previously described in vitiligo. One example is CCL18, a melanocyte-specific chemokine that was highly induced only in lesional skin and can bind to CCR8, which is primarily expressed on NK cells (Fig. 4F, fig. S4F). To confirm this finding, we stimulated cultured melanocytes with IFN-γ and analyzed the mRNA and protein levels of CCL18 in the samples. We found that CCL18 is induced in IFN-γ-treated melanocytes, which was confirmed via qPCR (fig. S4G) and ELISA (fig. S4H). CCL18 was also confirmed to be increased in the lesional blister fluid as measured by ELISA in 11 out of 14 patients (table S4, fig. S4I). These data provide an additional link between melanocytes and NK cells in vitiligo lesional skin.
We also found the chemokine-receptor CCL3/4/5-CCR5 interaction to be among the most dysregulated communications involving Tregs in both non-lesional and lesional skin when compared with healthy skin (Fig. 4D, F, G). CCR5 is the receptor for CCL3/4/5 and is upregulated in activated CD8+ T cells to facilitate their migration to sites of inflammation (41), but the role of CCR5 in Treg function is less well-defined.
Tregs upregulate CCR5 protein, cluster with CD8+ T cells in vitiligo lesions, and require CCR5 for optimal suppression in a mouse model of vitiligo
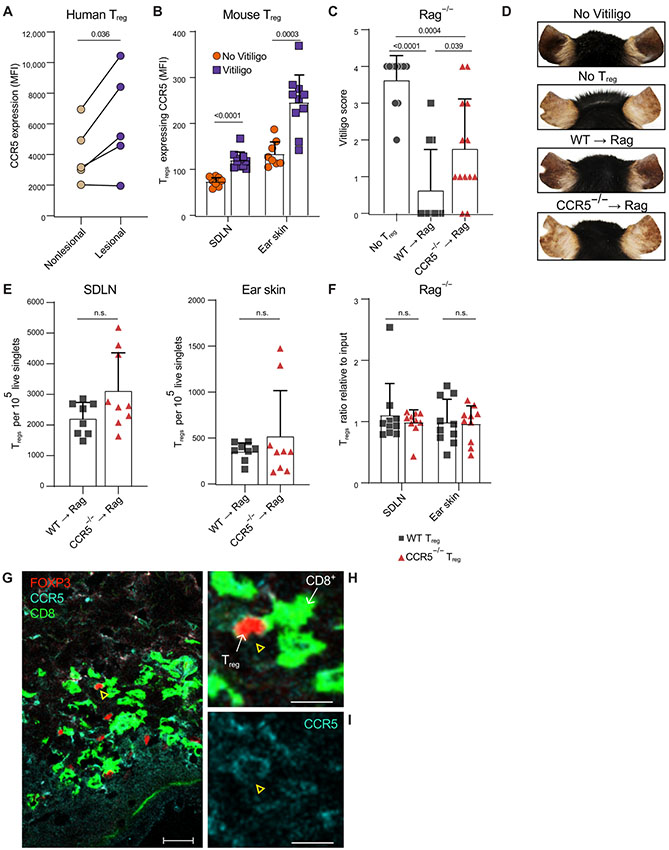
We sought to investigate the function of CCR5 in Tregs during the initiation and progression of vitiligo. Skin cells were isolated from vitiligo patients using suction blistering, and flow cytometry was used to measure CCR5 protein expression on CD4+FOXP3+ Tregs in lesional and non-lesional skin (table S4). We confirmed that nearly all Tregs in patient skin express CCR5, but those in lesional skin express significantly (p = 3.6E−2) higher levels of CCR5 protein compared to those in non-lesional skin (Fig. 5A, fig. S5A-B). CCR5 expression was not unique to Tregs, as CD4+ and CD8+ T cells also expressed CCR5 in patient skin, likely a reflection of the type 1 signature of vitiligo (fig. S5C-D). Previous studies performed in mouse models of vitiligo revealed that Tregs can suppress depigmentation (13, 42). To investigate the functional significance of CCR5 expression in Tregs during disease progression, we used an adoptive transfer mouse model of vitiligo. In this model, CD8+ T cells are adoptively transferred into sub-lethally irradiated Krt14-Kitl mice that retain melanocytes in the epidermis (43), and mice develop epidermal depigmentation comparable to human vitiligo 5 to 7 weeks after melanocyte targeting. After mice developed vitiligo, we used flow cytometry to compare CCR5 expression on Tregs in the skin draining lymph nodes (SDLNs) and ear skin from mice with vitiligo, as well as non-vitiligo controls. Tregs expressed significantly higher levels of CCR5 during vitiligo progression in both SDLNs (p < 0.0001) and ear skin (p = 3.0E−4), which is similar to human vitiligo (Fig. 5B, fig. S5E).
Fig. 5: Treg CCR5 is required for optimal suppression of effector CD8+ T cells in the skin.
Graph summarizing flow cytometry data comparing the average CCR5 expression (MFI) by CD4+FOXP3+ Tregs in blister fluid from the non-lesional and lesional skin of the same patient (A) (N=5, paired T-test). Quantification of CCR5 expression by Tregs in the skin draining lymph nodes (SDLN) and ear skin of uninduced FOXP3-GFP mice (No Vitiligo) and FOXP3-GFP with vitiligo (Vitiligo) (B) (3 experiments, N≥3, student T test). Ear vitiligo scores (C) and representative images (D) of ears from Rag−/− mice that received no Tregs, WT (WT→Rag) or CCR5−/− (CCR5−/−→Rag) Tregs 5 weeks post vitiligo induction (4 experiments, N≥2, student T test). Graphs depicts quantification of the average number of Tregs in the SDLNs and ear skin of Rag−/− mice that received WT or CCR5−/− Tregs 5 weeks post vitiligo induction (E) (3 experiments, N≥2, student T test). Quantification of WT and CCR5−/− Tregs in the SDLNs and ear skin of Rag−/− mice in a competitive Treg-adoptive transfer model of vitiligo (F). Graph depicts the proportion of WT or CCR5−/− Tregs in the indicated tissue relative to their proportion at input; (3 experiments, N≥3, student T test). Representative images of lesional vitiligo skin stained with CD8 (cell surface, AF488, green fluorescence), FOXP3 (nucleus, AF555, red fluorescence) and CCR5 (cell surface, AF647 cyan fluorescence); N=3 patients, 40x image, bar = 25μm (G). CCR5, FOXP3, and CD8 expression in the area defined by the white box is magnified in (H) and CCR5 expression in (I); yellow arrow highlights a FOXP3+ cell expressing CCR5, bar = 10μm. Bars depict the mean ± SD in all graphs.
CCR5-expressing Tregs in human skin also expressed type 1 proinflammatory genes like TBX21 and IFNG, thus it is possible that Tregs may lose their suppressive status and use CCR5 to promote depigmentation. To investigate the role of CCR5 in Treg function during vitiligo, we modified our mouse model to incorporate Rag−/− hosts, similar to a previously reported mouse model of melanoma immunotherapy (44). This permitted us to selectively transfer WT or CCR5 KO Tregs along with CD8+ T effector cells into the same host and measure disease severity and progression. We generated Krt14-Kitl×Rag−/− (Rag−/−) mice that lack all T and B cells, and we induced vitiligo in these hosts by adoptively transferring melanocyte-targeting CD8+ T cells with Treg-depleted CD4+ T cells to support their engraftment. Together with these effector cells, we transferred either no Tregs, WT Tregs, or CCR5−/− Tregs to test their ability to suppress vitiligo. After 5 weeks, we found that hosts without Tregs exhibited extensive depigmentation compared with hosts reconstituted with WT Tregs, supporting previous findings that describe a role for Tregs in suppressing effector T cell (Teff)-mediated depigmentation in vitiligo. Hosts reconstituted with CCR5−/− Tregs exhibited significantly (p = 3.9E−2) higher vitiligo scores compared with hosts that received WT Tregs (Fig. 5C, D). CCR5−/− Tregs have been reported to suppress Teff responses normally in vitro (45). We confirmed this, as CCR5−/− Tregs and WT Tregs exhibited a comparable ability to suppress Teff cytokine expression in response to αCD3/αCD28 in vitro (fig. S5F-G), demonstrating that CCR5-dependent Treg function was limited to the in vivo setting.
As a chemokine receptor, CCR5 may be able to promote Treg migration to the skin. We used flow cytometry to quantify Tregs in Rag−/− hosts and found that comparable numbers of WT and CCR5−/− Tregs migrate to the skin and SDLNs (Fig. 5E, fig. S5H,). To directly compare the ability of CCR5−/− Tregs and WT Tregs to migrate to the skin during vitiligo, we induced vitiligo in Rag−/− hosts and reconstituted hosts with equal numbers of Thy1.1+Thy1.2+ WT or Thy1.2+ CCR5−/− Tregs. We used flow cytometry to quantify Treg numbers in the SDLNs and ear skin and observed that the proportion of WT Tregs to CCR5−/− Tregs in both SDLNs and ear skin was comparable to the proportion at input (Fig. 5F, fig. S5I). Together, these observations revealed CCR5 was dispensable for Treg migration to the skin during vitiligo but was required specifically for Treg function within the skin microenvironment.
CCL5 is a ligand for CCR5 and is highly expressed by CD8+ T cells in vitiligo lesional skin (Fig. 2E, 3C). It is possible that CD8+ T cells in the skin secrete CCL5 that attracts CCR5-expressing Tregs, which may require CCR5 to properly colocalize with Teffs and efficiently suppress their activity. We examined skin biopsies from the lesional skin of active vitiligo patients and found that Tregs and CD8+ T cells infiltrated vitiligo lesional skin, and clustered together within the skin (Fig. 5G). CCR5 was expressed by Tregs in contact with CD8+ T cells within these clusters (Fig. 5G-I, fig. S5J-N), which suggested that CCR5 may be dispensable for Treg migration to the skin, but it may facilitate Treg function by properly positioning Tregs near CD8+ T cells within the skin to suppress them.
Cell type specific expression of genome wide associated variants of vitiligo
Vitiligo, as all autoimmune diseases, results from a combination of genetic and environmental factors (26). To date, over 50 distinct loci have been associated with vitiligo (46). However, the cells in which they act to influence the risk of developing vitiligo is unknown. Our scRNA-seq data allow us to compile a first-in-kind expression map of GWAS-associated gene expression in the epidermis. Such a map may also indicate cell types that tend to be affected by common human variations identified through GWAS. We utilized aggregate bulk expression values to confirm that expression of suspected melanocyte specific GWAS hits, MC1R, TYR, PMEL, and OCA2, are specific to the melanocyte cluster (fig. S6). For other genes, we observed a trend in cell type-specific expression, with a large group having T cell-specific (CD44, IL2RA, GZMB, CTLA4, FASLG, IRF3, IKZF4), melanocyte-specific (listed above), and macrophage-specific expression (CD80, IFIH1, TICAM1, ZMIZ1), with very few vitiligo associated genes expressed in dendritic cells or keratinocytes. A similar approach identified the expression of specific familial kidney disease risk loci to only podocytes (47), revealing expression of key genes in key cell types can drive whole-organ disease. These results support more prominent roles for T cells, melanocytes, and macrophages during the initiation of vitiligo, with less impact from other cell types.
DISCUSSION
An enhanced understanding of vitiligo pathogenesis through translational research has accelerated our development of new treatments. Cellular communications that promote T cell recruitment and maintenance in the skin are key targets for emerging treatments, and this knowledge may improve our approach to related autoimmune diseases that are more difficult to study in humans (48). Our current study provides a comprehensive view of intercellular signaling pathways among cells within the epidermis during vitiligo by combining suction blister biopsies with single cell RNA sequencing (scRNA-seq). This approach revealed the cellular sources of key pathogenic signals identified in previous studies (11, 27), and also identified many signals that had not been previously implicated in vitiligo.
We chose to collect samples by suction blistering (14), as opposed to conventional punch biopsies, before performing scRNA-seq. In doing so, we avoided disassociation protocols. We identified epidermal cell types and immune cells identified in previous studies performed on skin biopsies (16, 18, 19) but were unable to detect fibroblasts and other cells found deeper in the dermis of the skin. More recently, suction blistering and conventional biopsies were performed and directly compared in the same study (49), in which suction blister biopsy samples revealed many of the same pathways as in traditional biopsies but underrepresented some subtypes. Rojahn et al. 2020 also reported that scRNA-seq after suction blistering allowed for better resolution of epidermal cell types compared to traditional biopsy (49). Although suction blistering itself can induce inflammation in the skin at late time points (50), our rapid collection method limited this inflammation, and comparison of lesional skin to control skin highlighted pathways specific to vitiligo pathogenesis.
Our receptor and ligand analysis of the single cell data enabled us to build a comprehensive interactome that detailed individual cell to cell communications found within the epidermis. Some of the signals we discovered have not been previously described in the context of vitiligo or other autoimmune diseases. We found evidence of NK cells directly communicating with melanocytes, which may be relevant for the progression of vitiligo (Fig. 4F, S4E). CCL18 reportedly binds to CCR8, suggesting that CCL18 expressed by melanocytes in lesional skin could bind to and recruit CCR8-expressing NK cells to melanocyte targets. These observations strongly implicate NK cells as potential partners in elimination of melanocytes in vitiligo, although the possibility remains that melanocyte produced CCL18 may also contribute to recruitment of Treg, TConv, or γδT cells that also express CCR8 at low levels. Future studies that incorporate spatial information would be able to confirm the interactions inferred in our study.
Our previous studies identified two key mechanistic keratinocyte-T cell communications that are currently under research as targets for emerging treatments (48). The IFN-γ-CXCL10-CXCR3 axis promotes disease progression (11, 12) whereas IL15-IL15Rb promotes disease maintenance (51), and both pathways are being investigated in clinical trials as targets for emerging treatments (48). The current data reveals many additional disrupted clusters of cellular communication, including “private conversations” between skin cells. These “private conversations” include induced chemokine signaling that represents distinct T cell-T cell communications (CCL3/4/5-CCR5), melanocyte-T cell communications (HLAs-CD3D), melanocyte-NK cell communications (HLA-E-KLRC1 and CCL18-CCR8), and others (fig. S4D, F).
Previous studies identifying Tregs in patient skin drew conflicting results concerning their deficiency in number or function in lesional skin, due to use of low-resolution techniques that relied on few gene markers on a bulk population, such as histology (3, 4, 52-54). However, with the extensive expression data we have gathered, we confidently defined individual Tregs based on the expression of a large panel of Treg identity genes (fig. S1D). Our data reveal increased Treg number in lesional skin, indicating that lesions do not form because of a deficiency in Tregs in the skin. We observed that CD8+ T cells are activated and respond to IFN-γ in non-lesional skin (Fig. 3A) but do not acquire an effector phenotype as they do in lesional skin. CD8+ T cells become further activated in lesional skin whereas the activation status of Tregs is unchanged (Fig. 2G). Thus, while Tregs are sufficiently activated to suppress CD8+ T cell mediated depigmentation in non-lesional skin, they may be unable to sufficiently increase this suppression to match increased CD8+ T cell activity in lesional skin. Our data suggests Treg activation/function prevents widespread depigmentation of the skin by keeping autoreactive CD8+ T cells within non-lesional skin in check. Identification of the precise event that allows Tregs to become overwhelmed by CD8+ T cells in lesional skin requires additional studies.
We observed that Tregs within vitiligo lesions adopt a type 1 immune profile, including expression of IFN-γ as well as TBX21, CXCR3, and CCR5, each of which may be a cause or effect of IFN-γ production (Fig. 2C, D, S4B, 4D). IFN-γ production by Tregs has also been reported in the peripheral blood of diabetes patients (55) and a mouse model of GVHD (35). Additionally, other studies reported that Tregs required TBX21 and IFN-γ to facilitate their suppressive functions in the context of type 1 inflammation (34, 35). Effector Tregs adopt the expression profile of their environment (56) and Tregs that we observed in vitiligo skin may be adopting a type 1-like expression profile to specialize their function. In contrast, Feng et al. report that IFN-γ-producing Tregs represented a transition state to terminally differentiated effectors (57), and another recent study reported that a type 1 expression profile in Tregs supported the formation of resident memory T cells (58). The IFN-γ-producing Tregs that we observed may be transitioning to effectors without suppressive capabilities.(57)
We discovered that CCR5 was one of the most upregulated genes in non-lesional and lesional Tregs (fig. S4B, Fig. 5A, B) and confirmed increased expression of CCR5 protein on Tregs in both vitiligo patients and a mouse model of vitiligo. A majority of Tregs, CD4+ T cells, and CD8+ T cells in patient skin expressed CCR5 (fig. S5B-D), suggesting that CCR5 may play a role in the function of multiple T cell subsets, consistent with the type 1 signature that characterizes vitiligo skin. We determined that Tregs required the chemokine receptor CCR5 for optimal function in vivo, but not for migration to the skin. This observation does not rule out the possibility that CCR5 contributes to CD4+ or CD8+ T cell migration to the skin, and this should be explored in future studies. Notably, hosts with CCR5−/− Tregs exhibited less depigmentation than those without any Tregs (Fig. 5C, D). This suggests that Treg suppression is coordinated by numerous signals and CCR5 is likely one of many receptors that promote Treg function. Other studies using animal models of inflammation suggest that CCR5 was solely required for Treg recruitment to inflamed tissues (45, 59, 60), but our observations in patient skin suggest that CCR5 helps Tregs colocalize with and subsequently suppress CCL5-secreting CD8+ T cells within the skin. Similarly, Collins et al reported that CD8+ T cells utilize CCL5 to induce the clustering of CD4 memory cells in response to immune challenge in the skin (61) and our data illustrates how Tregs adapting a Th1-like expression profile can facilitate their function in Th1-dominated microenvironments like vitiligo lesions.
Our observations do not rule out the possibility that CCR5 signaling enhances Treg function without inducing migration, as some groups report migration-independent functions of chemokine receptor signaling (62-64) (65). These observations challenge conventional thinking that chemokine receptor signaling merely directs cell migration and understanding how CCR5 contributes to Treg function during vitiligo could improve the strategies that we use to treat patients. Since our results suggest a role for CCR5 in Treg suppression, treatments that spare CCR5 signaling while blocking effector pathways would potentially yield better treatment results. These observations may apply to autoimmune diseases in other organs characterized by type 1 inflammation as well, such as type 1 diabetes, however focused studies in these patients will be important to confirm this.
In summary, scRNA-seq analysis of suction blister infiltrates of both vitiligo and healthy skin provides unprecedented insight into the intercellular signaling pathways that drive vitiligo. These new observations promote a comprehensive model for understanding the initiation and progression of vitiligo directly within human skin, as well as new therapeutic opportunities.
MATERIALS AND METHODS
Study Design
Subjects with rapidly progressing, active vitiligo and healthy controls were recruited under an institutional review board (IRB) approved protocol (H-14848) at the University of Massachusetts Medical School. Subjects with a diagnosis of vitiligo by clinical exam performed by a dermatologist and treatment naive for more than 6 months were included for sampling. Objective clinical signs of disease activity including confetti (66) depigmentation and trichrome vitiligo (67) were used to identify patients. Healthy individuals were included based on the absence of any other autoimmune or inflammatory skin disease. Skin types sampled were healthy skin, non-lesional skin (unaffected tissue from vitiligo patients), and lesional skin (affected depigmenting tissue from vitiligo patients). Patient samples were used for scRNA-seq, flow cytometry, ELISA, or immunohistochemistry where applicable.
Blistering
Suction Blister Biopsies
Lesional suction blister biopsies were taken from sites with clinical signs of disease activity described above. Blisters were induced specifically over confetti and trichrome lesions, and incorporated lesional and perilesional skin to ensure capture of immune infiltrates. Spreading active non-confetti, non-trichrome lesions were sampled at the perilesional border with overlap of depigmented and pigmented skin. Non-lesional blister biopsies were taken from uninvolved skin on the same subjects as determined by Wood’s lamp examination and were greater 10 cm from the nearest depigmented macule.
Suction blisters (up to 1 cm in diameter) were induced using the Negative Pressure Instrument Model NP-4 (Electronic Diversities, Finksburg, MD). A negative pressure of 10-15 mm Hg was applied at a constant temperature of 40 degrees C for 30-60 minutes until blisters formed. The blister fluid was then collected via aspiration through a 0.5mL or 1.0mL insulin syringe. The blister fluid was immediately stored on ice and prepared for analysis as previously described (14).
Blister Processing
The blister fluid was centrifuged at 350 x G for 10 minutes at 4 degrees C. Supernatant representing cell-free blister fluid was flash frozen and stored for protein analysis. For single-cell RNA-sequencing, the cell pellet was resuspended at a density of ~80,000 cells/mL in 15% optiprep/PBS. For flow cytometry, the pellet was resuspended in a 1% solution of FBS.
scRNA-seq
scRNA-seq was performed using the inDrop protocol according to Zillionis et al, (68) with only slight modifications. The raw fastq files were processed through publicly available pipelines (https://github.com/garber-lab/inDrop_Processing), resulting in the final UMI table where rows are genes, and columns are cells. The resulting DGE matrices were loaded into R (V 4.0.0, (69)) for filtering, dimensionality reduction, and downstream analysis (see supplementary Materials and Methods).
Flow Cytometry
Blister fluid was stained using Human Trustain (clone 93), anti-human TCR γδ (B1), CD8 (SK1), CD56 (5.1H11), CD314 (1D11), and CD45 (H130), CD4 (OKT4), FOXP3 (FJK-16s), and CCR5 (HEK/1/85a) (1:20 dilution, Biolegend), and anti-human CD3 (OKT3) (1:200 dilution). Mouse skin samples were stained with anti-mouse CD3 (17A2), CD8b (YTS156.7.7), CD4 (GK1.5), CD45 (30-F11), CD45.1 (A20), CD45.2 (104), CD90.1 (OX-7), CD90.2 (53-2.1), CCR5 (HEK/1/85a) (Biolegend, San Diego, CA). All samples were stained with eBioscience viability fixable dye eFluor 455UV (1:1000 dilution, thermofisher scientific).
For extracellular staining cells were washed in FACS buffer (1% FBS in PBS) and then incubated with antibodies at 4° C in the dark for 20 minutes. CCR5 staining was done at 37° C in the dark for 30 minutes. For intracellular FOXP3 staining, surface staining was performed, and cells were fixed and permeabilized using the FOXP3/Transcription factor staining buffer set from eBioscience (Santa Clara, CA) according to the manufacturer’s recommendations. Human samples were washed in 1% FBS and fixed using 2% paraformaldehyde prior to analysis. Samples were analyzed with a BD LSR II flow cytometer (BD Biosciences) and FlowJo (Tree Star Inc.).
Immunofluorescence
Biopsies from lesional skin were put directly into optimal cutting temperature (OCT) medium and frozen at −80°C. Sections cut from blocks were put on slides and fixed with cold acetone for 10 minutes at −20°C before they were left to dry overnight. Slides were then incubated with the following antibodies at a 1:100 dilution for an hour at room temperature: anti-human CD8 rat monoclonal antibody, clone YTC182.20 (BioRad); anti-human FOXP3 mouse monoclonal antibody, clone 236A/E7 (Abcam); anti-human CCR5 rabbit monoclonal antibody, clone 2H14L10 (Invitrogen). The following primary antibodies were used for isotype controls: purified mouse IgG (Tonbo); purified rat IgG (Biolegend); Purified rabbit IgG (R&D). After primary antibody staining, slides were washed and incubated with the following secondary antibodies at a 1:500 dilution for an hour at room temperature: Alexa Fluor 488 donkey anti-rat IgG (Invitrogen); Alexa Fluor 555 donkey anti-mouse IgG (Invitrogen); Alexa Fluor 647 goat anti-rabbit IgG (Invitrogen). Slides were washed and cover-slipped with Prolong Gold antifade reagent (Invitrogen).
Mouse Studies
Vitiligo was induced through adoptive transfer of CD8+ T cells isolated from PMEL mice (Teffs) as described by Harris et al (43). The scoring system used to quantify depigmentation was described by Harris et al (43). To induce vitiligo in Rag−/− mice, 106 Teffs, 106 Treg-depleted CD4s and no Tregs or 105 WT Tregs or 105 CCR5−/− Tregs were adoptively transferred into sublethally irradiated Rag−/− hosts, infected with 106 PFU of rVV-hPMEL (N. Restifo, National Cancer Institute, NIH) the day of adoptive transfer. In the competitive Treg-adoptive transfer model, vitiligo was induced by adoptive transfer of 106 Teffs, 106 Treg-depleted CD4+ T cells, 50,000 WT Tregs and 50,000 CCR5−/− Tregs. Treg number was quantified by flow cytometry and the proportion of WT or CCR5−/− Tregs recovered from total Tregs in mouse tissue was divided by their proportion at input and reported as “Treg ratio relative to input”. Ear epidermis and skin-draining lymph nodes harvested from mice were mechanically disrupted using 70-μm cell strainers. Whole ear skin was incubated with dispase (Roche) to separate epidermal and dermal tissue. Dermal tissue was incubated with collagenase IV and DNase I (Sigma-Aldrich) and dissociated by gentle agitation. All tissues were washed with and resuspended as single cell suspensions in PBS with 1% FBS for staining. See supplementary Materials and Methods for more details.
Statistical Analysis
For all scRNA-seq data, EdgeR (70) was used to detect gene expression changes using the glmFit() function, and resulting p values were corrected using false discovery rate correction. Gene ontology and gene set enrichment analysis were performed using clusterProfiler (71). For flow cytometry data, all statistical analyses were performed with GraphPad Prism software. Dual comparisons for human data were made with a paired Student’s T test (parametric data) and dual comparisons for mouse data were made with an unpaired Student’s t test (parametric data).
Supplementary Material
Figure S1: Identification and validation of clustered cells
Figure S2: Gene expression of T cell subsets in healthy and vitiligo skin
Figure S3: Antigen presenting cells are significant producers of IFNG dependent chemokines
Figure S4: Melanocyte response to IFNG leads to increased immune surveillance
Figure S5: CCR5 is expressed by Tregs but not required for Treg-mediated suppression in vitro
Figure S6: Cell Type specific expression of GWAS risk loci
Table S1: Patient Data Summary for scRNA-seq Samples
Table S2: Patient Data for scRNA-seq Samples
Table S3: Summary Statistics for scRNA-seq Data
Table S4: Additional Patient Data
Acknowledgments:
We thank the patients of J.E.H. for generous donation of their time and tissues. Equipment used for flow cytometry were maintained by the University of Massachusetts Medical School Flow Cytometry Core.
Funding:
Dermatology Foundation and American Skin Association (JEH)
National Institutes of Health T32 AI095213 (JPS & KIE)
National Institutes of Health AI132152 (JPS)
National Institutes of Health GM10700 (JPS)
Hartford Foundation Vitiligo Grant and others (JEH)
National Institutes of Health R61 AR07302 (JEH & MG)
National Institutes of Health R01 AR069114 (JEH)
National Institutes of Health UL1-TR001453 (JEH)
This work was supported in part by Rheos Medicines
Footnotes
Competing interests: JEH is scientific founder of Villaris Therapeutics, Inc, which develops therapeutic treatments for vitiligo. JEH and JPS are inventors on patent #62489191 “Diagnosis and Treatment of Vitiligo” which covers targeting IL-15 and Trm for treatment of vitiligo. AGP is a consultant at Abbvie, Arcutis, Avita, Chromaderm, Incyte, Pfizer, Viela Bio, Villaris, Immune Tolerance Network, and TWI. AGP holds stock of Zerigo Health, Tara Medical. AGP is on the Board of Directors of the Global Vitiligo Foundation.
Data and materials availability: All data associated with this study are in the paper or supplementary materials. scRNA-seq data has been desposited to dbGAP, accession number phs002455.v1.p1. Functions used for analysis of the scRNA-seq data are part of an available on github, https://github.com/garber-lab/SignallingSingleCell. Data, code, and materials will be made available upon request.
References and Notes
- 1.Rodrigues M, Ezzedine K, Hamzavi I, Pandya AG, Harris JE, Vitiligo Working Group, Current and emerging treatments for vitiligo. J. Am. Acad. Dermatol. 77, 17–29 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Frisoli ML, Harris JE, Vitiligo: Mechanistic insights lead to novel treatments. J. Allergy Clin. Immunol. 140, 654–662 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Klarquist J, Denman CJ, Hernandez C, Wainwright DA, Strickland FM, Overbeck A, Mehrotra S, Nishimura MI, Le Poole IC, Reduced skin homing by functional Treg in vitiligo. Pigment Cell Melanoma Res. 23, 276–286 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lili Y, Yi W, Ji Y, Yue S, Weimin S, Ming L, Global activation of CD8+ cytotoxic T lymphocytes correlates with an impairment in regulatory T cells in patients with generalized vitiligo. PLoS One. 7, e37513 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dwivedi M, Laddha NC, Arora P, Marfatia YS, Begum R, Decreased regulatory T-cells and CD4(+) /CD8(+) ratio correlate with disease onset and progression in patients with generalized vitiligo. Pigment Cell Melanoma Res. 26, 586–591 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Maeda Y, Nishikawa H, Sugiyama D, Ha D, Hamaguchi M, Saito T, Nishioka M, Wing JB, Adeegbe D, Katayama I, Sakaguchi S, Detection of self-reactive CD8+ T cells with an anergic phenotype in healthy individuals. Science. 346, 1536–1540 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Hegab DS, Attia MAS, Decreased Circulating T Regulatory Cells in Egyptian Patients with Nonsegmental Vitiligo: Correlation with Disease Activity. Dermatology Research and Practice. 2015 (2015), pp. 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tembhre MK, Parihar AS, Sharma VK, Sharma A, Chattopadhyay P, Gupta S, Alteration in regulatory T cells and programmed cell death 1-expressing regulatory T cells in active generalized vitiligo and their clinical correlation. Br. J. Dermatol 172, 940–950 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Giri PS, Dwivedi M, Laddha NC, Begum R, Bharti AH, Altered expression of nuclear factor of activated T cells, forkhead box P3, and immune-suppressive genes in regulatory T cells of generalized vitiligo patients. Pigment Cell Melanoma Res. (2020), doi: 10.1111/pcmr.12862. [DOI] [PubMed] [Google Scholar]
- 10.Giri PS, Dwivedi M, Begum R, Decreased suppression of CD8 and CD4 T cells by peripheral regulatory T cells in generalized vitiligo due to reduced NFATC1 and FOXP3 proteins. Exp. Dermatol. 29, 759–775 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Rashighi M, Agarwal P, Richmond JM, Harris TH, Dresser K, Su M-W, Zhou Y, Deng A, Hunter CA, Luster AD, Harris JE, CXCL10 is critical for the progression and maintenance of depigmentation in a mouse model of vitiligo. Sci. Transl. Med 6, 223ra23 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richmond JM, Bangari DS, Essien KI, Currimbhoy SD, Groom JR, Pandya AG, Youd ME, Luster AD, Harris JE, Keratinocyte-Derived Chemokines Orchestrate T-Cell Positioning in the Epidermis during Vitiligo and May Serve as Biomarkers of Disease. J. Invest. Dermatol 137, 350–358 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gregg RK, Nichols L, Chen Y, Lu B, Engelhard VH, Mechanisms of spatial and temporal development of autoimmune vitiligo in tyrosinase-specific TCR transgenic mice. J. Immunol 184, 1909–1917 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strassner JP, Rashighi M, Ahmed Refat M, Richmond JM, Harris JE, Suction blistering the lesional skin of vitiligo patients reveals useful biomarkers of disease activity. J. Am. Acad. Dermatol 76, 847–855.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jianbo Shi J Malik, Normalized cuts and image segmentation. IEEE Trans. Pattern Anal. Mach. Intell 22, 888–905 (2000). [Google Scholar]
- 16.Cheng JB, Sedgewick AJ, Finnegan AI, Harirchian P, Lee J, Kwon S, Fassett MS, Golovato J, Gray M, Ghadially R, Liao W, Perez White BE, Mauro TM, Mully T, Kim EA, Sbitany H, Neuhaus IM, Grekin RC, Yu SS, Gray JW, Purdom E, Paus R, Vaske CJ, Benz SC, Song JS, Cho RJ, Transcriptional Programming of Normal and Inflamed Human Epidermis at Single-Cell Resolution. Cell Rep. 25, 871–883 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Na CH, Sharma N, Madugundu AK, Chen R, Aksit MA, Rosson GD, Cutting GR, Pandey A, Integrated Transcriptomic and Proteomic Analysis of Human Eccrine Sweat Glands Identifies Missing and Novel Proteins. Mol. Cell. Proteomics 18, 1382–1395 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joost S, Zeisel A, Jacob T, Sun X, La Manno G, Lönnerberg P, Linnarsson S, Kasper M, Single-Cell Transcriptomics Reveals that Differentiation and Spatial Signatures Shape Epidermal and Hair Follicle Heterogeneity. Cell Syst. 3, 221–237.e9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Philippeos C, Telerman SB, Oulès B, Pisco AO, Shaw TJ, Elgueta R, Lombardi G, Driskell RR, Soldin M, Lynch MD, Watt FM, Spatial and Single-Cell Transcriptional Profiling Identifies Functionally Distinct Human Dermal Fibroblast Subpopulations. J. Invest. Dermatol 138, 811–825 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feuerer M, Hill JA, Mathis D, Benoist C, Foxp3+ regulatory T cells: differentiation, specification, subphenotypes. Nat. Immunol 10, 689–695 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Pfoertner S, Jeron A, Probst-Kepper M, Guzman CA, Hansen W, Westendorf AM, Toepfer T, Schrader AJ, Franzke A, Buer J, Geffers R, Signatures of human regulatory T cells: an encounter with old friends and new players. Genome Biol. 7, R54 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang J, Ioan-Facsinay A, van der Voort EIH, Huizinga TWJ, Toes REM, Transient expression of FOXP3 in human activated nonregulatory CD4 T cells. European Journal of Immunology. 37 (2007), pp. 129–138. [DOI] [PubMed] [Google Scholar]
- 23.Zhu J, Yamane H, Paul WE, Differentiation of Effector CD4 T Cell Populations. Annual Review of Immunology. 28 (2010), pp. 445–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Donnelly RP, Sheikh F, Dickensheets H, Savan R, Young HA, Walter MR, Interleukin-26: an IL-10-related cytokine produced by Th17 cells. Cytokine Growth Factor Rev. 21, 393–401 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van den Boorn JG, Konijnenberg D, Dellemijn TAM, van der Veen JPW, Bos JD, Melief CJM, Vyth-Dreese FA, Luiten RM, Autoimmune destruction of skin melanocytes by perilesional T cells from vitiligo patients. J. Invest. Dermatol 129, 2220–2232 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Harris JE, Cellular stress and innate inflammation in organ-specific autoimmunity: lessons learned from vitiligo. Immunol. Rev 269, 11–25 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wańkowicz-Kalińska A, van den Wijngaard RMJGJ, Tigges BJ, Westerhof W, Ogg GS, Cerundolo V, Storkus WJ, Das PK, Immunopolarization of CD4+ and CD8+ T cells to Type-1-like is associated with melanocyte loss in human vitiligo. Lab. Invest 83, 683–695 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Wang XX, Wang QQ, Wu JQ, Jiang M, Chen L, Zhang CF, Xiang LH, Increased expression of CXCR3 and its ligands in patients with vitiligo and CXCL10 as a potential clinical marker for vitiligo. Br. J. Dermatol 174, 1318–1326 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Ogg GS, Rod Dunbar P, Romero P, Chen JL, Cerundolo V, High frequency of skin-homing melanocyte-specific cytotoxic T lymphocytes in autoimmune vitiligo. J. Exp. Med 188, 1203–1208 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Araki K, Morita M, Bederman AG, Konieczny BT, Kissick HT, Sonenberg N, Ahmed R, Translation is actively regulated during the differentiation of CD8+ effector T cells. Nat. Immunol 18, 1046–1057 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mosenson JA, Zloza A, Nieland JD, Garrett-Mayer E, Eby JM, Huelsmann EJ, Kumar P, Denman CJ, Lacek AT, Kohlhapp FJ, Alamiri A, Hughes T, Bines SD, Kaufman HL, Overbeck A, Mehrotra S, Hernandez C, Nishimura MI, Guevara-Patino JA, Le Poole IC, Mutant HSP70 reverses autoimmune depigmentation in vitiligo. Sci. Transl. Med 5, 174ra28 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Figueiredo C, Wittmann M, Wang D, Dressel R, Seltsam A, Blasczyk R, Eiz-Vesper B, Heat shock protein 70 (HSP70) induces cytotoxicity of T-helper cells. Blood. 113, 3008–3016 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Äijö T, Edelman SM, Lönnberg T, Larjo A, Kallionpää H, Tuomela S, Engström E, Lahesmaa R, Lähdesmäki H, An integrative computational systems biology approach identifies differentially regulated dynamic transcriptome signatures which drive the initiation of human T helper cell differentiation. BMC Genomics. 13 (2012), p. 572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koch MA, Tucker-Heard G, Perdue NR, Killebrew JR, Urdahl KB, Campbell DJ, The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat. Immunol 10, 595–602 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koenecke C, Lee C-W, Thamm K, Föhse L, Schafferus M, Mittrücker H-W, Floess S, Huehn J, Ganser A, Förster R, Prinz I, IFN-γ production by allogeneic Foxp3+ regulatory T cells is essential for preventing experimental graft-versus-host disease. J. Immunol 189, 2890–2896 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Ramilowski JA, Goldberg T, Harshbarger J, Kloppmann E, Lizio M, Satagopam VP, Itoh M, Kawaji H, Carninci P, Rost B, Forrest ARR, A draft network of ligand-receptor-mediated multicellular signalling in human. Nat. Commun 6, 7866 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borradori L, Sonnenberg A, Structure and function of hemidesmosomes: more than simple adhesion complexes. J. Invest. Dermatol 112, 411–418 (1999). [DOI] [PubMed] [Google Scholar]
- 38.Gorelik L, Flavell RA, Abrogation of TGFbeta signaling in T cells leads to spontaneous T cell differentiation and autoimmune disease. Immunity. 12, 171–181 (2000). [DOI] [PubMed] [Google Scholar]
- 39.Nishimura EK, Suzuki M, Igras V, Du J, Lonning S, Miyachi Y, Roes J, Beermann F, Fisher DE, Key roles for transforming growth factor beta in melanocyte stem cell maintenance. Cell Stem Cell. 6, 130–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nagineni CN, Cherukuri KS, Kutty V, Detrick B, Hooks JJ, Interferon-gamma differentially regulates TGF-beta1 and TGF-beta2 expression in human retinal pigment epithelial cells through JAK-STAT pathway. J. Cell. Physiol 210, 192–200 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Ferguson AR, Engelhard VH, CD8 T cells activated in distinct lymphoid organs differentially express adhesion proteins and coexpress multiple chemokine receptors. J. Immunol 184, 4079–4086 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chatterjee S, Eby JM, Al-Khami AA, Soloshchenko M, Kang H-K, Kaur N, Naga OS, Murali A, Nishimura MI, Caroline Le Poole I, Mehrotra S, A quantitative increase in regulatory T cells controls development of vitiligo. J. Invest. Dermatol 134, 1285–1294 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harris JE, Harris TH, Weninger W, Wherry EJ, Hunter CA, Turka LA, A mouse model of vitiligo with focused epidermal depigmentation requires IFN-γ for autoreactive CD8+ T-cell accumulation in the skin. J. Invest. Dermatol 132, 1869–1876 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Antony PA, Piccirillo CA, Akpinarli A, Finkelstein SE, Speiss PJ, Surman DR, Palmer DC, Chan C-C, Klebanoff CA, Overwijk WW, Rosenberg SA, Restifo NP, CD8+ T cell immunity against a tumor/self-antigen is augmented by CD4+ T helper cells and hindered by naturally occurring T regulatory cells. J. Immunol 174, 2591–2601 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wysocki CA, Jiang Q, Panoskaltsis-Mortari A, Taylor PA, McKinnon KP, Su L, Blazar BR, Serody JS, Critical role for CCR5 in the function of donor CD4+CD25+ regulatory T cells during acute graft-versus-host disease. Blood. 106, 3300–3307 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spritz RA, Andersen GHL, Genetics of Vitiligo. Dermatol. Clin 35, 245–255 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park J, Shrestha R, Qiu C, Kondo A, Huang S, Werth M, Li M, Barasch J, Suszták K, Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science. 360, 758–763 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frisoli ML, Essien K, Harris JE, Vitiligo: Mechanisms of Pathogenesis and Treatment. Annu. Rev. Immunol 38, 621–648 (2020). [DOI] [PubMed] [Google Scholar]
- 49.Rojahn TB, Vorstandlechner V, Krausgruber T, Bauer WM, Alkon N, Bangert C, Thaler FM, Sadeghyar F, Fortelny N, Gernedl V, Rindler K, Elbe-Bürger A, Bock C, Mildner M, Brunner PM, Single-cell transcriptomics combined with interstitial fluid proteomics defines cell type-specific immune regulation in atopic dermatitis. J. Allergy Clin. Immunol (2020), doi: 10.1016/j.jaci.2020.03.041. [DOI] [PubMed] [Google Scholar]
- 50.Kuhns DB, DeCarlo E, Hawk DM, Gallin JI, Dynamics of the cellular and humoral components of the inflammatory response elicited in skin blisters in humans. J. Clin. Invest 89, 1734–1740 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Richmond JM, Strassner JP, Zapata L Jr, Garg M, Riding RL, Refat MA, Fan X, Azzolino V, Tovar-Garza A, Tsurushita N, Pandya AG, Tso JY, Harris JE, Antibody blockade of IL-15 signaling has the potential to durably reverse vitiligo. Sci. Transl. Med 10 (2018), doi: 10.1126/scitranslmed.aam7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ben Ahmed M, Zaraa I, Rekik R, Elbeldi-Ferchiou A, Kourda N, Belhadj Hmida N, Abdeladhim M, Karoui O, Ben Osman A, Mokni M, Louzir H, Functional defects of peripheral regulatory T lymphocytes in patients with progressive vitiligo. Pigment Cell Melanoma Res. 25, 99–109 (2012). [DOI] [PubMed] [Google Scholar]
- 53.Terras S, Gambichler T, Moritz RKC, Altmeyer P, Lambert J, Immunohistochemical analysis of FOXP3+ regulatory T cells in healthy human skin and autoimmune dermatoses. Int. J. Dermatol 53, 294–299 (2014). [DOI] [PubMed] [Google Scholar]
- 54.Abdallah M, Lotfi R, Othman W, Galal R, Assessment of tissue FOXP3+, CD4+ and CD8+ T-cells in active and stable nonsegmental vitiligo. Int. J. Dermatol 53, 940–946 (2014). [DOI] [PubMed] [Google Scholar]
- 55.McClymont SA, Putnam AL, Lee MR, Esensten JH, Liu W, Hulme MA, Hoffmüller U, Baron U, Olek S, Bluestone JA, Brusko TM, Plasticity of human regulatory T cells in healthy subjects and patients with type 1 diabetes. J. Immunol 186, 3918–3926 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cretney E, Kallies A, Nutt SL, Differentiation and function of Foxp3 effector regulatory T cells. Trends in Immunology. 34 (2013), pp. 74–80. [DOI] [PubMed] [Google Scholar]
- 57.Feng T, Cao AT, Weaver CT, Elson CO, Cong Y, Interleukin-12 converts Foxp3+ regulatory T cells to interferon-γ-producing Foxp3+ T cells that inhibit colitis. Gastroenterology. 140, 2031–2043 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ferreira C, Barros L, Baptista M, Blankenhaus B, Barros A, Figueiredo-Campos P, Konjar Š, Lainé A, Kamenjarin N, Stojanovic A, Cerwenka A, Probst HC, Marie JC, Veldhoen M, Type 1 Treg cells promote the generation of CD8+ tissue-resident memory T cells. Nat. Immunol. (2020), doi: 10.1038/s41590-020-0674-9. [DOI] [PubMed] [Google Scholar]
- 59.Dobaczewski M, Xia Y, Bujak M, Gonzalez-Quesada C, Frangogiannis NG, CCR5 signaling suppresses inflammation and reduces adverse remodeling of the infarcted heart, mediating recruitment of regulatory T cells. Am. J. Pathol 176, 2177–2187 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yurchenko E, Tritt M, Hay V, Shevach EM, Belkaid Y, Piccirillo CA, CCR5-dependent homing of naturally occurring CD4+ regulatory T cells to sites of Leishmania major infection favors pathogen persistence. J. Exp. Med 203, 2451–2460 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Collins N, Jiang X, Zaid A, Macleod BL, Li J, Park CO, Haque A, Bedoui S, Heath WR, Mueller SN, Kupper TS, Gebhardt T, Carbone FR, Skin CD4(+) memory T cells exhibit combined cluster-mediated retention and equilibration with the circulation. Nat. Commun 7, 11514 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chang L-Y, Lin Y-C, Mahalingam J, Huang C-T, Chen T-W, Kang C-W, Peng H-M, Chu Y-Y, Chiang J-M, Dutta A, Day Y-J, Chen T-C, Yeh C-T, Lin C-Y, Tumor-derived chemokine CCL5 enhances TGF-β-mediated killing of CD8(+) T cells in colon cancer by T-regulatory cells. Cancer Res. 72, 1092–1102 (2012). [DOI] [PubMed] [Google Scholar]
- 63.Mueller A, Strange PG, CCL3, acting via the chemokine receptor CCR5, leads to independent activation of Janus kinase 2 (JAK2) and Gi proteins. FEBS Lett. 570, 126–132 (2004). [DOI] [PubMed] [Google Scholar]
- 64.Wong M, Uddin S, Majchrzak B, Huynh T, Proudfoot AEI, Platanias LC, Fish EN, RANTES Activates Jak2 and Jak3 to Regulate Engagement of Multiple Signaling Pathways in T Cells. Journal of Biological Chemistry. 276 (2001), pp. 11427–11431. [DOI] [PubMed] [Google Scholar]
- 65.Barsheshet Y, Wildbaum G, Levy E, Vitenshtein A, Akinseye C, Griggs J, Lira SA, Karin N, CCR8 FOXp3 Treg cells as master drivers of immune regulation. Proceedings of the National Academy of Sciences. 114 (2017), pp. 6086–6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sosa JJ, Currimbhoy SD, Ukoha U, Sirignano S, O’Leary R, Vandergriff T, Hynan LS, Pandya AG, Confetti-like depigmentation: A potential sign of rapidly progressing vitiligo. J. Am. Acad. Dermatol 73, 272–275 (2015). [DOI] [PubMed] [Google Scholar]
- 67.Hann SK, Kim YS, Yoo JH, Chun YS, Clinical and histopathologic characteristics of trichrome vitiligo. J. Am. Acad. Dermatol 42, 589–596 (2000). [DOI] [PubMed] [Google Scholar]
- 68.Zilionis R, Nainys J, Veres A, Savova V, Zemmour D, Klein AM, Mazutis L, Single-cell barcoding and sequencing using droplet microfluidics. Nat. Protoc. 12, 44–73 (2017). [DOI] [PubMed] [Google Scholar]
- 69.R Core Team, R: A Language and Environment for Statistical Computing R Foundation for Statistical Computing (2020; http://www.R-project.org/). [Google Scholar]
- 70.Robinson MD, Oshlack A, A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu G, Wang L-G, Han Y, He Q-Y, clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Identification and validation of clustered cells
Figure S2: Gene expression of T cell subsets in healthy and vitiligo skin
Figure S3: Antigen presenting cells are significant producers of IFNG dependent chemokines
Figure S4: Melanocyte response to IFNG leads to increased immune surveillance
Figure S5: CCR5 is expressed by Tregs but not required for Treg-mediated suppression in vitro
Figure S6: Cell Type specific expression of GWAS risk loci
Table S1: Patient Data Summary for scRNA-seq Samples
Table S2: Patient Data for scRNA-seq Samples
Table S3: Summary Statistics for scRNA-seq Data
Table S4: Additional Patient Data