Abstract
The current COVID-19 pandemic urges us to develop ultra-sensitive surface-enhanced Raman scattering (SERS) substrates to identify the infectiousness of SARS-CoV-2 virions in actual environments. Here, a micrometer-sized spherical SnS2 structure with the hierarchical nanostructure of "nano-canyon” morphology was developed as semiconductor-based SERS substrate, and it exhibited an extremely low limit of detection of 10−13 M for methylene blue, which is one of the highest sensitivities among the reported pure semiconductor-based SERS substrates. Such ultra-high SERS sensitivity originated from the synergistic enhancements of the molecular enrichment caused by capillary effect and the charge transfer chemical enhancement boosted by the lattice strain and sulfur vacancies. The novel two-step SERS diagnostic route based on the ultra-sensitive SnS2 substrate was presented to diagnose the infectiousness of SARS-CoV-2 through the identification standard of SERS signals for SARS-CoV-2 S protein and RNA, which could accurately identify non-infectious lysed SARS-CoV-2 virions in actual environments, whereas the current PCR methods cannot.
Keywords: SnS2 hierarchical nanostructures, capillary effect, ultra-low limit of detection, infectiousness of SARS-CoV-2, SERS, lattice strain
Graphical abstract
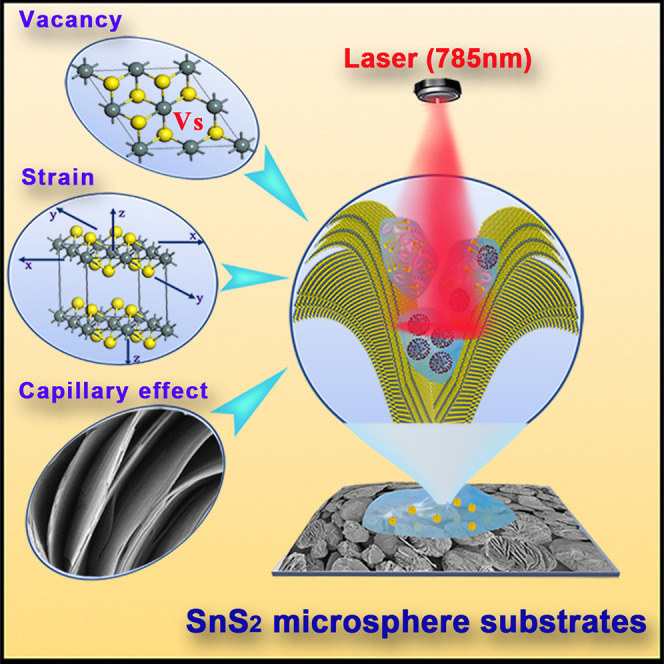
Progress and potential
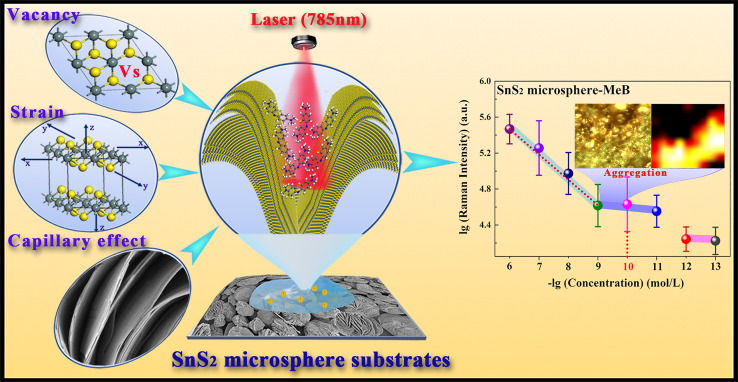
As the COVID-19 epidemic has swept the world, exploiting a rapid and highly sensitive detection method that can identify the infectiousness of SARS-CoV-2 virus and exclude the interference on diagnostic results and false alarms of the non-infectious SARS-CoV-2 virions has become a key scientific challenge. In this work, we designed a micrometer-sized spherical SnS2 structure with the hierarchical nanostructure of "nano-canyon” morphology as an ultra-sensitive SERS substrate to groundbreakingly rapidly detect and identify the infectiousness of SARS-CoV-2 samples on site, which exhibits vital timeliness in patient management that the viral culture method does not have. It is significant to avoid misdiagnosing infectious SARS-CoV-2 in some contaminated patient gathering places.
Motivated by the synergistic contribution of the molecular enrichment caused by capillary effect and the chemical enhancement boosted by lattice strain and sulfur vacancies, the developed ultra-sensitive SnS2 hierarchical nanostructure SERS substrates exhibit an extremely low limit of detection of 10−13 M, which can be applied to complete the identification of infectiousness for SARS-CoV-2 samples, whereas the current PCR methods cannot.
Introduction
Coronavirus disease 2019 (COVID-19), caused by SARS-CoV-2, is a serious threat to human health, where we are facing the dilemma of globalization and time continuity.1 , 2 Since July 2020, identification of SARS-CoV-2 virions on cold chain items has been frequently reported in China and other countries, causing great concern on relevant virus transmission. It can be attributed to the fact that SARS-COV-2 can survive more than 3 weeks below 0°C during cold chain transportation.3 , 4 It is important to note that a portion of SARS-COV-2 virions in the colder environment are lysed, inactive viruses. But the RNA of lysed SARS-CoV-2 might remain and easily be detected by the universally applied polymerase chain reaction (PCR) method,5 resulting in the misdiagnosis of infectiousness for SARS-CoV-2, causing unnecessary social panic. Therefore, it is much more meaningful to determine whether the virus is infectious as identified on contaminated items in various environments.6 However, exploiting a rapid and highly sensitive detection method that can identify lysed, non-infectious SARS-CoV-2 virions to rule out the contamination of infectious SARS-CoV-2 virus is a key scientific challenge.
Surface-enhanced Raman scattering (SERS), a single-molecule spectral detection technology, is currently expanding its promising applications from environmental science7 and food safety8 , 9 toward the biosensing field10 due to its advantages of ultra-high sensitivity, non-destructiveness, excellent repeatability, and accuracy. Motivated by the development of SERS technology, the direct non-labeled detection of various biological samples, such as adenovirus, animal viruses, HIV, and influenza virus, has been successfully developed.10, 11, 12, 13, 14 With respect to SARS-CoV-2 virus, Yeh group15 reported that an Au nanoparticle microfluidic platform was successfully established for rapid and label-free capturing and SERS detection of viruses. In our previous work,16 an ACE2-modified SERS sensor was reported to exhibit a low limit of detection (LOD) of 80 copies/mL for the SARS-CoV-2 in contaminated wastewater in as short as 5 min. In addition, Choo and Ray group17 , 18 applied deoxyribonucleic acid aptamers and viral anti-spike antibodies as receptors to achieve sensitive SERS detection of SARS-CoV-2, and their detection limits reached 10 PFU/mL and 18 copies/mL, respectively. These reports indicated that SERS technology could be developed into a potential method for virus detection. In the SERS biosensor field, to pursue more excellent biocompatibility with biological samples and excellent spectral stability and reproducibility, SERS substrates are now actively being expanded from noble metals to semiconductor materials.19 Recently, Lin20 reported that MCF-7 drug-resistant breast cancer cells could be accurately identified based on highly sensitivity B-TiO2 SERS substrates, which widened the application of semiconductor-based SERS platforms in precise diagnosis of cancer. However, compared with SERS spectra of biological samples mainly originated from electromagnetic enhancement, the SERS enhancement of semiconductor-based substrates is relatively weak21 because it is difficult for the semiconductor-based substrates, in which charge transfer plays a major role, to significantly enhance the chemical bond vibration of the entire biological molecule due to their larger molecular size. Therefore, a top-priority task is to achieve ultra-high SERS sensitivity in semiconductor-based substrates, which can even parallel noble metals with hot spots.
With respect to semiconductor-based substrates, the probability of electronic transition ω lk can be expressed by Fermi's golden rule: , where g(E k) is the electronic density of states, and represents the matrix of electronic transitions on the highest occupied molecular orbital (HOMO)-lowest unoccupied molecular orbital (LUMO).22 Therefore, the SERS sensitivity of semiconductor-based substrates contributed by the charge transfer between probe molecules and semiconductors can be optimized by the following two aspects: (1) increasing the electronic density of states g(E k) near the Fermi level of semiconductor materials to enhance the chemical bond vibrations of molecules and (2) regulating the HOMO and LUMO orbitals of semiconductors to realize the larger probability of charge transfer to significantly enhance the molecular chemical bond vibrations.23, 24, 25 In general, the electronic structures of semiconductors can be modulated by element doping,23 , 26 introducing vacancies,27 , 28 applying external strain fields,29 and constructing heterojunctions30 and amorphous substrates.31 Although motivated by above optimized strategies for SERS performance, the LODs of pure semiconductor-based substrates in most previous publications are still universally lower than 10−10 M level, which is a new bottleneck encountered by semiconductor substrates. SnS2, as a typical two-dimensional layered structure, has a bandgap that varies from 1 to 3 eV with the thickness and morphology of nanosheets. Sulfur vacancies that easily exist on the surface of SnS2 nanosheets are conductive to induce the intermediate energy level near the Fermi level to promote electronic transitions,32 , 33 making them a promising candidate for SERS substrates. However, there are few reports on the SERS applications of SnS2.34 Here, an ultra-high SERS sensitivity of SnS2 nanosheets is realized by simultaneously introducing sulfur vacancies and generating lattice strain through regulating morphology, thereby satisfying sensitivity requirements of a SERS biosensor for the detection of SARS-CoV-2 and other biomolecules.
In this work, we designed a micrometer-sized spherical SnS2 structure with the hierarchical nanostructure of "nano-canyon" morphology. Benefiting from this unique nano-canyon structure, the molecular enrichment caused by capillary effect was generated on the surface of SnS2 microspheres, where the existence of the lattice strain and sulfur vacancies can further boost the chemical enhancement by stimulating a larger probability of charge transfer. Motivated by the synergistic contribution of the above three components to the SERS enhancement (Figure 1 ), the enhancement factor for methylene blue (MeB) can reach up to 3.0 × 108, and its LOD as low as 10−13 M, which is not only much better than most reported pure semiconductor-based SERS substrates, but it also breaks the newly encountered bottleneck of LOD of 10−10 M. Based on this ultra-sensitive SERS detection ability of SnS2 microspheres, the ultra-high SERS sensitivity endowed SnS2 microspheres with the capability to groundbreakingly detect and identify the infectiousness of SARS-CoV-2 based on the identification standard of SERS signals for SARS-CoV-2 S protein and RNA, which could accurately identify non-infectious lysed SARS-CoV-2 virions, whereas the current PCR methods cannot. It is significant to avoid misdiagnosing infectious SARS-CoV-2 in actual environments.
Figure 1.
Schematic diagram of SnS2 microsphere substrates design and the ultra-sensitive SERS performance
Results and discussion
Characterization of SnS2 microsphere
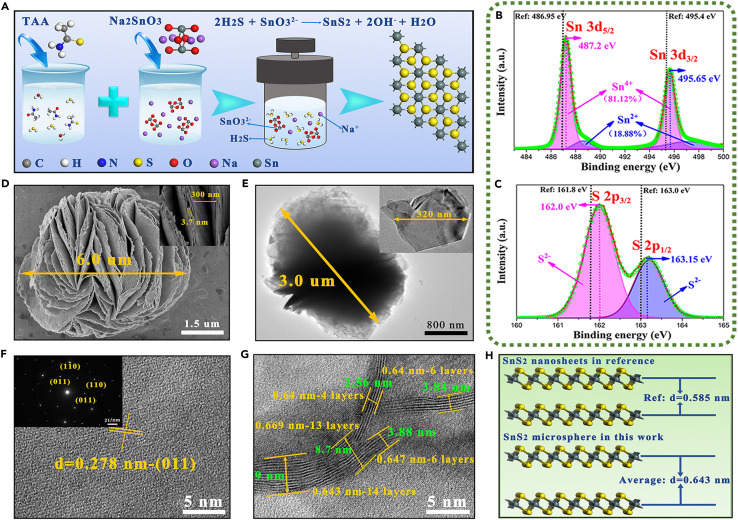
The shape-controlled SnS2 microspheres were first synthesized through a simple one-step hydrothermal reaction, and the schematic illustration of the synthetic process is shown in Figure 2 A. Briefly, the determined concentrations of thioacetamide (TTA) and sodium stannate (Na2SnO3) served as the sulfur source and the tin source, respectively. The X-ray powder diffraction (XRD) pattern in Figure S1A shows that the synthesized brown powder was crystallized with SnS2 phase (PDF#75-0367) with lattice constants corresponding to the hexagonal structure with space group (a = 3.65 Å, b = 3.65 Å, c = 5.90 Å). In a hexagonal SnS2 unit cell, a single Sn atom was covalently bonded to six atoms of S in the octahedral sites of individual layers35 (Figure 2A). The strongest peak indexed to (011) indicated the exposed crystal plane of SnS2 is (011). Additionally, the diffraction peaks corresponding to the (001), (100), and (011) crystal planes all shift to lower angles (Figure S1B and S1C), which indicated the entire crystal lattice of hexagonal SnS2 structure was increased, especially with an increase in interlayer distance. It could be attributed to the fact that the typical layered structure of SnS2 was composed of a three-layered stacked atomic layer (S–Sn–S) connected by van der Waals forces, which was susceptible to generate lattice strain caused by regulating SnS2 morphology. Raman spectra is a perfect method to explore the changes in interlayer distance of SnS2 structure. As shown in Figure S2, the distinct Raman peak at 315 cm−1 was assigned to the vertical plane vibration mode (A1g) of Sn–S bonds. This Raman peak of A1g mode exhibited an obvious red shift in relation to the Raman shift of 312 cm−1 reported in the literature,34 , 36 which also demonstrated the increase in interlayer distance of SnS2 structure. It was mainly attributed to the decrease in van der Waals interactions between the interlayers, resulting in the decrease in restoring forces on atoms.37 , 38 XPS spectra in Figures 2B and 2C shows that Sn and S could be clearly identified in SnS2 samples, among which both Sn3d and S2p peaks presented a significant shift to the higher binding energy in relation to reported literature,33 , 35 indicating the changed electron density difference of SnS2 affected by the lattice strain. In addition to the doublet peaks at 487.2 eV (Sn3d 5/2) and 495.7 eV (Sn3d 3/2) arising from Sn4+, there was also a pair of weaker doublet peaks at 488.7 eV (Sn3d 5/2) and 497.5 eV (Sn3d 3/2), which belonged to Sn2+ (Sn4+:Sn2+ ≈ 4:1), indicating the presence of sulfur vacancy (V S). Additionally, the enlarged characteristic peaks in the S2p region of SnS2 at 162.0 and 163.2 eV were both attributed to S2+. Moreover, there were no S2p peaks between 168 and 170 eV (Figure S3), suggesting that SnS2 was barely oxidized to SnO2.
Figure 2.
Synthesis, crystal phase, morphology, and structure characterization of SnS2 microspheres
(A) Schematics illustrating the synthesis of SnS2 microspheres.
(B and C) Sn3d and S2p XPS spectrum.
(D) SEM images.
(E) TEM images.
(F) HRTEM images.
(G) HRTEM images of cross-section for nanosheets.
(H) Crystal structure diagram.
Then scanning electron microscopy (SEM) and transmission electron microscopy (TEM) were used to study the morphology of SnS2. The low-magnification SEM image (Figure S4) shows that the SnS2 products exhibited a special spherical morphology with the statistical average size of 5.37 μm, which could be called a SnS2 microsphere. The further analysis of high-magnification morphology (Figure 2D) revealed the existence of hierarchical nanostructures on the microsphere surface, and SnS2 microspheres were formed by this hierarchical nanostructure of curling, stacked nanosheets. Furthermore, this hierarchical nanostructure resulted in a special morphology that was similar to a bottomless canyon with wide top and narrow bottom, where the width of these nano-canyons was mostly less than 300 nm. Compared with the reported SnS2 nanostructures such as nanoflowers, nanobelts, and nanosheets,32 , 35 , 36 this unique morphology of SnS2 with the hierarchical nanostructure was reported for the first time to our knowledge, and it was expected to exhibit surprisingly ultra-high sensitivity in the SERS detection field. The TEM image in Figure 2E also demonstrates the spherical morphology with micron-level size, and the shedding nanosheets that formed the hierarchical nanostructure possess a transverse size of about 300 nm. Moreover, there are no other impurity signals in the energy-dispersive X-ray spectroscopy other than the S and Sn signals, which demonstrated the high purity of the synthesized SnS2 microspheres (as shown in Figure S5). Selected area electron diffraction (SAED) and high-resolution TEM (HRTEM) were used to analyze the crystal structure of the SnS2 microspheres (Figure 2F). The clear lattice fringes on SnS2 nanosheets were easily discerned by HRTEM to correspond to the (011) planes of hexagonal structures with an inter-planar spacing of 0.278 nm, which is consistent with the exposed crystal plane obtained from XRD results. The SAED pattern also confirmed the high crystallinity and the hexagonal symmetry structure of SnS2 nanosheets. The cross-section morphology of SnS2 microspheres was adopted to further analyze the detailed information of the thickness and interlayer distance of SnS2 nanosheets. According to the HRTEM and crystal structure diagram of hexagonal SnS2 (Figures 2G, 2H, and S6), the thickness of the single-layer SnS2 nanosheet-formed hierarchical nanostructure was mostly less than 4 nm, which is equivalent to the thickness of 6 atomic layers for hexagonal SnS2. Therefore, the average interlayer distance of SnS2 microspheres was about 0.643 nm, which was far larger than the interlayer distance of 0.59 nm reported in the literature.32 , 35 , 36 Moreover, the larger interlayer distance of 0.669 nm existed at the bend in SnS2 nanosheets, indicating a larger lattice strain. This conclusion confirmed the existence of lattice strain in the SnS2 microspheres once again.
Raman enhancement for SnS2 microspheres
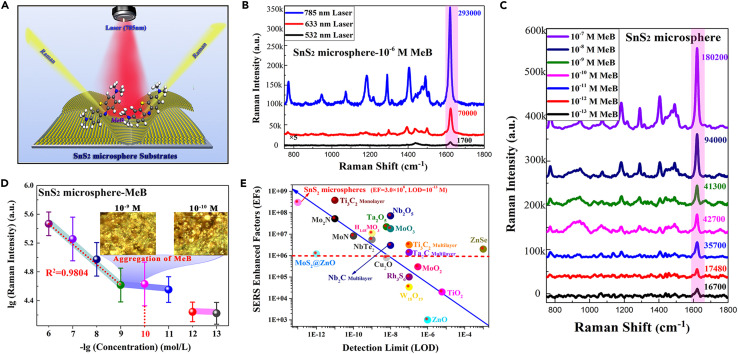
Following extensive research on SnS2 nanostructures with various morphologies, the synthesized SnS2 microspheres with a unique hierarchical nanostructure showed immense attraction for researchers in the photocatalysis and batteries fields, especially in the SERS detection field. The conventional MeB probe molecule was adopted to examine the SERS performance of SnS2 microspheres because MeB exhibited a strong optical absorption peak in the visible light region (Figure S7). The Raman scattering diagram of MeB and SnS2 substrate is presented in Figure 3 A. First, we investigated the optimal excitation wavelength of SnS2 microspheres on MeB molecules. Raman spectra in Figure 3B show that MeB molecules on SnS2 microspheres had an obviously stronger Raman enhancement with the excitation laser of 785 nm than that of the other two excitation lasers of 532 nm and 633 nm. Interestingly, the optimal excitation wavelength of 785 nm is the most popular wavelength of biological Raman detection, which lays a foundation for the bio-detection application of SnS2 microspheres. Under the irradiation of a 785-nm laser, all Raman peaks of MeB molecules were significantly enhanced, especially in the Raman vibration mode of the asymmetric stretching vibration of benzene rings at the main peak of 1,620 cm−1. All these enhanced Raman peaks of MeB and their corresponding Raman vibration modes are listed in Table S1.
Figure 3.
SERS performance of SnS2 microspheres
(A) Raman scattering diagram of MeB and SnS2 substrate.
(B) Raman spectra of 10−6 M MeB on SnS2 microspheres under the excitation laser of 532 nm, 633 nm, and 785 nm.
(C) Raman spectra of MeB with concentration of 10−7–10−13 M on SnS2 microspheres.
(D) Raman intensity of MeB on SnS2 microspheres at 1,620 cm−1 as a function of its concentration of 10−6–10−13 M.
(E) Comparison of SERS performance of different reported semiconductor-based substrates.
Surprisingly, even if the MeB molecules were diluted to an extremely low molar concentration of 10−13 M, an obvious Raman signal of MeB molecules could still be detected by adsorbing molecules to SnS2 substrates (Figure 3C). The relationship trend of Raman intensity at 1,620 cm−1 changing with the MeB concentration of 10−6 to 10−13 M is shown in Figure 3D, and inset graphs show the optical magnification pictures of 10−9 and 10−10 M MeB molecules on SnS2 microspheres. It was found that the linear relationship was satisfactory in the range of 10−6 to 10−9 M with the correlation coefficient of 0.9804. However, when the concentration of MeB was lower than 10−10 M, Raman intensity no longer decreased linearly with the decreased concentration of MeB molecules. The Raman intensity of MeB with lower than 10−10 M decreased little, which could be attributed to a strange phenomenon (inset graphs of Figure 3D) where the low-concentration MeB molecule adsorbed on SnS2 microspheres substrates would automatically enrich during the drying and evaporation process. Beneficial to this enriching phenomenon of low-concentration molecules, the LOD of SnS2 microspheres for MeB could be as low as 10−13 M, and the corresponding SERS enhancement factor at 1,620 cm−1 with the irradiation laser of 785 nm was determined to be 3.0 × 108 (calculation details are shown in SI-3 of the supplemental information). To the best of our knowledge, this SERS performance of SnS2 microspheres is one of the highest sensitivities among the reported pure semiconductor-based SERS substrates (Figure 3E; Table S2), which can even parallel that of the noble metals with hot spots. Encouragingly, the extremely low SERS LOD of 10−13 M for SnS2 microspheres not only breaks the newly encountered bottleneck of detection limits, but it also provides a competitive candidate for a SERS detection application of biomolecules. Additionally, the SERS-enhanced stability of SnS2 microspheres was proved by detecting the Raman spectra of 10−7 M MeB on SnS2 microspheres placed for 5 months (Figure S8). Compared with the Raman intensity of 10−7 M MeB on fresh SnS2 substrates, the average Raman intensity of 10−7 M MeB on SnS2 substrates after 5 months was only discounted 16.7%, and it still maintained a significant Raman enhanced effect. Such excellent SERS-enhanced stability has stimulated SnS2 microspheres to exhibit a promising application prospect in the practical SERS detection of biomolecules. However, with respect to the explanations of enrichment phenomenon for low-concentration probe molecules on SnS2 microspheres, it is worthwhile to further explore that in the following analysis of the SERS-enhanced mechanism.
SERS-enhanced mechanism of SnS2 microspheres
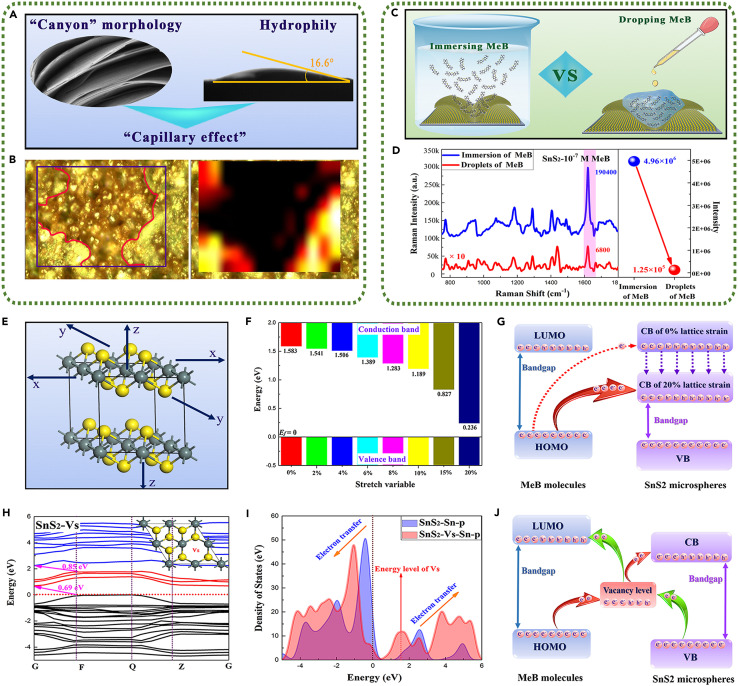
As we expected from the above analysis, SnS2 microspheres with hierarchical nanostructures exhibited ultra-sensitive SERS detection ability. Actually, the extremely low LOD mainly depends on the realization of enrichment phenomenon for low-concentration molecules on SnS2 microspheres. The enrichment phenomenon of molecules is mainly controlled by the superficial microscopic morphology of materials. As seen in SEM images (Figure 2D), the hierarchical nanostructure with a unique nano-canyon morphology existed on the surface of SnS2 microspheres. The excellent hydrophilicity of SnS2 structure would endow this unique nano-canyon morphology with a powerful function (Figure 4 A)—capillary effect—which can break the diffusion limit of molecules in aqueous solution.39 , 40 When a droplet evaporated on the surface of substrates, the large amount of capillary attraction would guide most MeB molecules to enrich on the surface of SnS2 microspheres. Initiated from these views, Raman mapping images with microscope regions of 72 × 48 μm2 for 10−10 M MeB on SnS2 microspheres were measured to provide more intuitive evidence for the enrichment phenomenon of probe molecules (Figures 4B and S9). Comparing the optical magnification picture and Raman mapping image from the same Raman measured area, it was found that when the concentration of MeB molecules decreased to 10−10 M, the enrichment phenomenon of MeB molecules started to emerge on the surface of SnS2 microspheres. Moreover, this molecular enriching area presented an obvious ultra-high Raman intensity at 1,620 cm−1 (refer to the yellow and red areas in Figure 4B). As long as we were able to scan and detect the enriching area of low-concentrations molecules on SnS2 microsphere substrates, the high-intensity Raman signal could be obtained. Therefore, the Raman detection of molecules with ultra-low concentration was achieved due to the capillary effect on the surface of SnS2 microspheres, which endowed SnS2 microspheres with ultra-low LOD that other pure semiconductor-based SERS substrates are unable to surpass. Additionally, benefiting from the unique nano-canyon morphology and capillary effect on the surface of SnS2 microspheres, additional physical enrichment of molecules in the aqueous solution will be generated. As shown in Figure 4C, Raman spectra of immersing SnS2 in the 10−7 M MeB solution and dropping 10−7 M MeB solution on SnS2 were detected to explore the physical enrichment effect of SnS2 microspheres substrates. Raman spectra in Figure 4D showed that the Raman intensity of immersing MeB molecules was far stronger than that of dropping MeB molecules. The calculating results indicated that SnS2 microspheres could achieve about 40-fold physical enrichment for MeB molecules in aqueous solution (SI-2 of the supplemental information), which could significantly improve the SERS sensitivity of substrates.
Figure 4.
The proposed SERS enhancement mechanisms of capillary effect, lattice strain, and sulfur vacancies for SnS2 microspheres
(A) Schematic diagram of combining nano-canyon morphology and hydrophilicity to induce the capillary effect.
(B) Raman mapping image with 72 × 48 μm2 region of 10−10 M MeB on SnS2 microspheres.
(C) Schematic diagram of immersing and dropping MeB molecules to explore physical enrichment.
(D) Raman spectra and SERS intensity of immersing and dropping 10−7 M MeB on SnS2 microspheres.
(E) Schematic diagram of hexagonal SnS2 crystal with the tensile strain in three periodic directions (x, y, and z axis).
(F) Bandgap of hexagonal SnS2 crystal with 0%–20% tensile strain.
(G) Schematic diagram of charge transfers between MeB molecules and the hexagonal SnS2 crystal with 0%–20% tensile strain.
(H) Band structure of SnS2 crystal with sulfur vacancies.
(I) Density of states of Sn p orbitals in the perfect SnS2 crystal and SnS2 crystal with sulfur vacancies.
(J) Schematic diagram of charge transfers between MeB molecules and the hexagonal SnS2 crystal with S vacancies.
Recent research has demonstrated that the chemical mechanism (CM) contributed by charge transfer mainly governs the SERS-enhanced effect of two-dimensional (2D) semiconductor-based substrates. Therefore, in addition to the contribution of the capillary effect to SERS performance, it is necessary to consider the regulation of the electronic structure of hexagonal SnS2 to the CM enhancement. Based on the analyzed results of XRD and HTREM, the hexagonal SnS2 crystal exhibited lattice strain. Therefore, the lattice constants of the hexagonal SnS2 crystal in the three periodic directions applying a tensile strain were changed to explore the influence of lattice strain on the electronic structure (Figure 4E). As shown in Figures 4F and S10, it was found that the bandgap decreased from 1.583 to 0.236 eV while increasing the tensile strain from 0% to 20%, which could be attributed to both electrons of Sn-p orbitals and S-s orbitals in the conduction band shifting to the Fermi level, although the energy of the valence band remained unchanged. The decreased energy level of the conduction band would stimulate the larger probability of charge transfer from the molecular HOMO to the conduction band of semiconductor-based substrates, thus significantly enhancing the chemical bond vibration of molecules41 (Figure 4G). Additionally, the superficial defect states of SnS2 microspheres could effectively break the restriction of the large bandgap on the charge transfer induced by the irradiation of visible light though introducing the intermediate energy level. As mentioned in the XPS results, sulfur vacancies (Vs) existed in the hexagonal SnS2 crystal. Therefore, the SnS2 crystal structure with sulfur vacancies was constructed, and the band structure and density of states are shown in Figures 4H, 4I, and S11. Compared with the electronic structure of the perfect hexagonal SnS2 crystal, the defect energy levels between the conduction band and the Fermi level were introduced due to the existence of sulfur vacancies, thereby simultaneously making the electrons of the valence band shift to the deeper energy level and increasing the excited-state electrons of the conduction band.27 , 28 The introduced defect energy levels served as an intermediate springboard for electronic transitions, which could further promote the charge transfers from the molecular HOMO to the conduction band of the semiconductor or the valence band of the semiconductor to the molecular LUMO (Figure 4J). In conclusion, the ultra-low SERS LOD of SnS2 microspheres mainly contributed by the synergistic enhancements of the molecular enrichment caused by capillary effect and the charge transfer chemical enhancement boosted by the lattice strain and sulfur vacancies.
SERS detection of various SARS-CoV-2 biomarkers and establishing identification standard of SERS signals
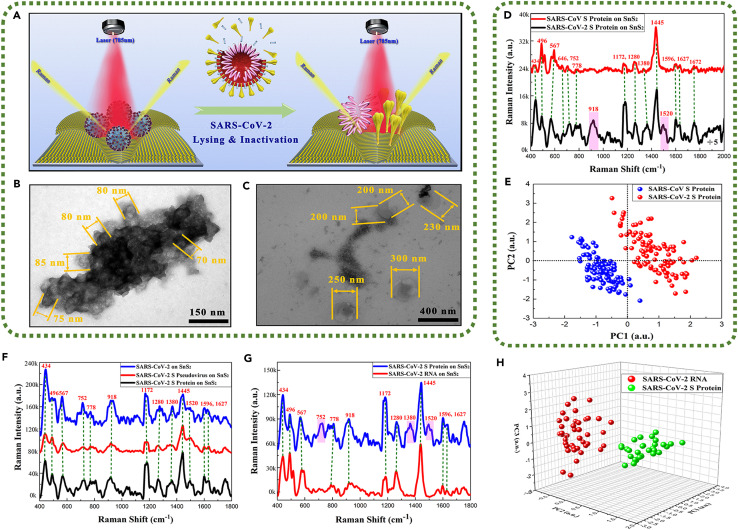
With the COVID-19 pandemic that has swept the world, there has been an urgent solution needed for accurate identification of the infectiousness of SARS-CoV-2. It is well acknowledged that SARS-CoV-2 is an enveloped virus, and the viral surface mainly contains two transmembrane proteins: spike glycoprotein (S) and membrane protein (M), while the larger RNA genome is contained within the envelope42 (Figure S12). Generally, the active or un-lysed coronavirus is covered by spike glycoprotein with a vertical size of about 5 nm. During the semiconductor-based SERS detection process, the SARS-CoV-2 S protein is the main contact with SnS2 microsphere substrates, resulting in detectable characteristic Raman signals that usually contain the surface S protein dominating the SERS-Raman spectra of SARS-CoV-2.16 After the SARS-CoV-2 is lysed and inactivated, its viral structure and spatial configuration are destroyed, and the nucleic acids (RNA) and other proteins originally wrapped in the envelope are exposed or released outside the virus particle (Figure 5 A). Therefore, the infectiousness of SARS-CoV-2 could be identified by analyzing the SERS signal difference in the surface S protein and internal RNA between the coronavirus with complete viral structure and the lysed coronavirus. First, the 120-kV scanning TEM was applied to observe the morphology of the SARS-CoV-2 with complete viral structure and the lysed SARS-CoV-2. The TEM images demonstrated typical intact virions with 70–120 nm size before lysing (Figures 5B and S13A) whereas enlarged and broken virions with about 200–300 nm size after lysing by ultrasound treatment are found (Figures 5C and S13B), suggesting the effective destruction of the integrity of SARS-CoV-2 particles by our lysing method.
Figure 5.
Application of SnS2 microspheres for detecting various physical forms of SARS-CoV-2
(A) Schematic diagram of identifying the lysed SARS-CoV-2.
(B) TEM images of SARS-CoV-2 with complete viral structure.
(C) TEM images of the lysed SARS-CoV-2.
(D) Raman spectra of SARS-CoV-2 S protein and SARS-CoV S protein.
(E) The key features of SERS patterns to classify the SARS-CoV-2 S protein and SARS-CoV S protein.
(F) Raman spectra of physical forms of SARS-CoV-2 including SARS-CoV-2, SARS-CoV-2 S pseudovirus, and SARS-CoV-2 S protein.
(G) Raman spectra of SARS-CoV-2 RNA and SARS-CoV-2 S protein.
(H) The key features of SERS patterns to classify the RNA and S protein of SARS-CoV-2.
We then characterized SERS spectra of five physical forms of SARS-CoV-2: SARS-CoV-2 S pseudovirus, SARS-CoV-2 S protein, SARS-CoV-2 RNA, lysed SARS-CoV-2, and SARS-CoV-2 (SI-5 of the supplemental information). The resultant Raman vibration modes corresponding to their Raman peaks are shown in Table S3. First, the interference of phosphate-buffered saline could be ignored due to its extremely weak Raman peaks in relation to the Raman spectra of SARS-CoV-2 S pseudovirus (Figure S14). In order to confirm the accuracy of obtained Raman spectra for these various coronaviral physical forms on SnS2 microspheres, Au nanoarrays were used as SERS substrates for reference (Figure S15). Analysis results indicated that Raman peaks of SARS-CoV-2 S pseudovirus, SARS-CoV-2 S protein, and SARS-CoV S protein on both substrates matched better with some Raman shifts due to the difference in SERS enhancement mechanisms and the amount of charge transfer between these two SERS substrates. The Raman spectra and the principal component analysis (PCA) results of S protein of SARS-CoV and SARS-CoV-2 (Figures 5D and 5E) both suggested that Raman peaks of these two kinds of S proteins could be completely distinguished in the 2D space, and the SARS-CoV-2 S proteins exhibited the characterized Raman bands at 918 cm−1 and 1,520 cm−1 corresponding to the vibration modes of C-C stretching in skeleton and N-H and C-O in-plane deformation in amide II. Although the S protein of SARS-CoV-2 and SARS-CoV showed up to 74.6% identity in their amino acid sequences (SI-4 of the supplemental information), SnS2 microspheres still showed excellent identification ability for different biomolecules, which provided a promising SERS substrate candidate for practical detection applications of biomolecules. Additionally, three physical forms of SARS-CoV-2 including SARS-CoV-2, SARS-CoV-2 S pseudovirus, and SARS-CoV-2 S protein absorbing on SnS2 microspheres presented almost identical Raman shifts except for the difference in Raman intensity (Figure 5F), which could be attributed to the fact that their main contact with SnS2 microsphere substrates was the SARS-CoV-2 S protein, resulting in the same Raman peaks as SARS-CoV-2 S protein. Therefore, it is reasonable to replace the dangerous active SARS-CoV-2 with the SARS-CoV-2 S protein to obtain a large amount of Raman data for the principal component of machine learning analysis. Finally, the identification standard of SERS signals was established by machine learning and identification techniques to identify the non-infectious SARS-CoV-2. Raman spectra of SARS-CoV-2 RNA on SnS2 microspheres were detected and are shown in Figures 5G and S16. Interestingly, PCA results in Figure 5H showed that SARS-CoV-2 RNA and S protein presented different Raman shifts, and they could be completely distinguished in the three-dimensional space. The SARS-CoV-2 S protein exhibited three characterized Raman bands at 752 cm−1, 1,380 cm−1, and 1,520 cm−1 originated from the vibration modes of the aromatic ring in tryptophan (Trp), C-N stretching in tryptophan (Trp), and N–H and C–O in-plane deformation in amide II, respectively. Therefore, Raman spectra of SARS-CoV-2 RNA and S protein could serve as two principal components of the identification standard to distinguish and classify the unknown SARS-CoV-2 samples. Similarly, the PCA results of SARS-CoV-2 were consistent with the SARS-CoV-2 S protein, showing the completely distinguished Raman peaks from SARS-CoV-2 RNA (Figure S17). Additionally, we further explored the limits of detection for three physical forms of SARS-CoV-2 virus based on SnS2 microsphere SERS substrates. As shown in Figure S18, even if the SARS-CoV-2 S protein, SARS-CoV-2 RNA, and SARS-CoV-2 S pseudovirus were diluted to 10−14 mol/L, 104 copies/mL, and 104 copies/mL, respectively, some obvious, characteristic Raman signal of the above three physical forms of SARS-CoV-2 could still be detected by adsorbing on SnS2 microspheres. To the best of our knowledge, it is the highest detection sensitivity for SARS-CoV-2 particles among the pure semiconductor-based SERS substrates, which also mainly originated from the molecular enrichment phenomenon caused by the capillary effect and the significant chemical enhancement of SnS2 microspheres, but the difference is that virus particles are relatively less affected by capillary attraction due to their larger size. Moreover, the viral loads of SARS-CoV-2 identified from COVID-19 patients’ saliva, stool, urine, or blood and the items in the cold chain environments were in the range of 10–1010 copies/mL, where the currently pandemic SARS-CoV-2 variants generally exhibit a characteristic of high viral load of >105 copies/mL.3 , 43, 44, 45, 46, 47, 48, 49 Therefore, the SERS detection method based on SnS2 microsphere substrates shows great practical application potential.
Diagnosis of the infectiousness of SARS-CoV-2 based on the established identification standard
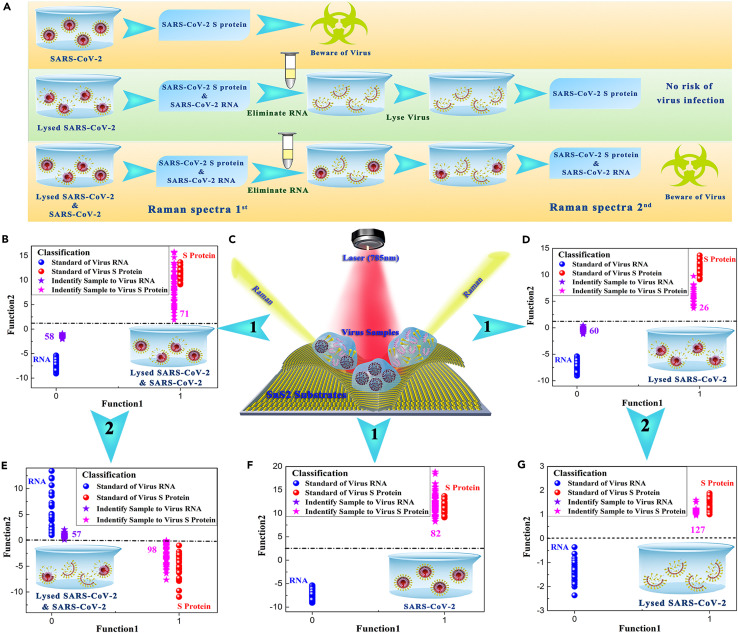
Herein, the SARS-CoV-2 with complete viral structure and the lysed SARS-CoV-2 were adopted to simulate three contamination situations of SARS-CoV-2 in actual environments: the SARS-CoV-2 with complete viral structure, the lysed SARS-CoV-2, and a mixture of the complete-structure viruses and the lysed viruses. The experimental procedure for diagnosing the infectiousness of SARS-CoV-2 is shown in Figure 6 A. Noticeably, the SARS-CoV-2 with complete viral structure showed only the characteristic Raman spectra of SARS-CoV-2 S protein (Figure S19A), whereas the lysed SARS-CoV-2 presented both the characteristic Raman peaks of SARS-CoV-2 S protein and RNA (Figure S19B). Based on the above identification standard of SERS signals for SARS-CoV-2 S protein and RNA, it is possible to complete the diagnosis of the non-infectious virus samples through two-step SERS detections. Multiple testing areas on the surface of SnS2 substrates with these three virus samples were randomly selected to perform Raman detection to obtain at least 80 available SERS spectra, and a more advanced machine learning method of support vector machine (SVM) was adopted to discriminate and classify the obtained SERS spectra (Figure 6C). According to the first SERS detection results (Figures 6B, 6D, and 6F), if all the Raman spectra of virus sample belong to the SARS-CoV-2 S protein, this virus sample is determined to be the SARS-CoV-2 with complete viral structure, suggesting a severe risk of virus infectivity (Figure 6F). SVM analysis results of Figures 6B and 6D showed that 58 and 60 SERS spectra of virus samples could be correctly classified to SARS-CoV-2 RNA, while 71 and 26 SERS spectra could be contributed to SARS-CoV-2 S protein. Such SVM analysis results only indicated that there were some lysed SARS-CoV-2 virions in these two kinds of virus samples, but it could not judge their virus infectivity. Therefore, it is necessary to conduct the second SERS detection after eliminating RNA and re-lysing these two virus samples. Their SVM analysis results are shown in Figures 6E and 6G. If the virus sample still exhibits the characteristic Raman peaks of SARS-CoV-2 RNA at this time (Figures 6E and S20), it is diagnosed as the mixture of the complete-structure virus and the lysed virus, suggesting a moderate risk of virus infectivity. As shown in Figures 6G and S21, we found that 127 SERS spectra of the virus sample were all classified to SARS-CoV-2 S protein, indicating this virus sample belonged to the lysed SARS-CoV-2. Noticeably, this virus sample of all lysed SARS-CoV-2 in actual environments had almost no risk of virus infectivity, but it is frequently misdiagnosed as a source of infectious SARS-CoV-2 by the current commonly used detection method of RT-PCR. In conclusion, the present analysis results demonstrated that it is feasible to diagnose the non-infectious SARS-CoV-2 based on the identification standard of SERS signal for SARS-CoV-2 S protein and RNA, which paves a new path for the identification of SARS-CoV-2-contaminated dangerous items in actual environments.
Figure 6.
Application of SnS2 microspheres for diagnosing the infectiousness of SARS-CoV-2
(A) Experimental procedure for diagnosing the infectiousness of SARS-CoV-2.
(B) SVM analysis results to identify the mixture of the SARS-CoV-2 with complete viral structure and the lysed SARS-CoV-2.
(C) Raman scattering diagram of three contamination situations of the novel coronavirus based on SnS2 substrates.
(D) SVM analysis results to identify the lysed SARS-CoV-2.
(E) SVM analysis results to identify the mixture of the SARS-CoV-2 with complete viral structure and the lysed SARS-CoV-2 after eliminating RNA and re-lysing virus samples.
(F) SVM analysis results to identify the SARS-CoV-2 with complete viral structure.
(G) SVM analysis results to identify the lysed SARS-CoV-2 after eliminating RNA and re-lysing virus samples.
Conclusions
In summary, in response to the challenge of diagnosing the infectiousness of SARS-CoV-2 in the various actual virus-contaminated environments, we developed ultra-sensitive SERS substrate SnS2 microspheres to detect various physical forms of SARS-CoV-2 virus. The first synthesized SnS2 microspheres exhibited a hierarchical nanostructure with a unique nano-canyon morphology, which could generate capillary effect on the surface of microspheres. Based on these SnS2 microsphere substrates, the Raman detection of molecules with ultra-low concentration was achieved through the molecular enrichment caused by capillary effect, which enabled SnS2 microspheres to achieve about 40-fold physical enrichment for molecules. Additionally, benefiting from the contribution of the lattice strain and sulfur vacancies to chemical enhancement, SnS2 microspheres exhibited an ultra-low LOD of 10−13 M and an ultra-high enhancement factor of 3.0 × 108 for MeB. To the best of our knowledge, this remarkable SERS enhancement of SnS2 microspheres is one of the highest sensitivities among the reported pure semiconductor-based SERS substrates, which can even parallel that of the noble metals with hot spots. As a result, various physical forms of SARS-CoV-2 were able to be sensitively detected on SnS2 microspheres, and the identification standard of SARS-CoV-2 RNA and S protein was established by PCA methods. Moreover, based on the advanced machine learning method of SVM, non-infectious lysed SARS-CoV-2 was successfully distinguished, which paved a new path for identifying the infectiousness of SARS-CoV-2 virions and is of significance to avoid misdiagnosing infectious SARS-CoV-2 in actual environments. Furthermore, it is worthwhile to note that recovery of SARS-CoV-2 in viral culture is currently the only approach to confirm the presence of replication-competent virus.50 However, the viral culture method suffers from the defects of long culture time and complicated experimental operation. Meaningfully, the aforementioned two-step SERS detection method can be extended to rapidly diagnose SARS-CoV-2 infectivity on site in some contaminated patient gathering places such as hospitals or at the Centers for Disease Control and Prevention, exhibiting vital timeliness in patient management that the viral culture method does not have.
Experimental procedures
Resource availability
Lead contact
The detailed experimental methods can be found in the supplemental experimental procedures. It is recommended to contact the lead contact directly for further information and requests for resources and materials: Yong Yang (yangyong@mail.sic.ac.cn).
Materials availability
This study did not generate new unique reagents or there are restrictions to availability.
Acknowledgments
This work is supported by the financial support of the National Natural Science Foundation of China (No. 52172167). And the authors also gratefully acknowledge financial support from the Key Research and Development Plan of Anhui Province (No. 202104a07020032).
Author contributions
Conceptualization, Y.S.P. and Y.Y.; methodology, Y.S.P., C.L.L., and X.Y.L.; formal analysis, Y.S.P., C.L.L., and J.J.L.; investigation, Y.S.P, Y.Y.L., and J.W.; resources, Y.G., J.W., and J.H.; writing – original draft, Y.S.P.; writing – review & editing, Y.S.P., Y.Y., J.W., and J.H.; visualization, Y.S.P. and C.L.L.; supervision, Y.Y. and Z.R.H.; funding acquisition, Y.Y., Y.G., and J.H.
Declaration of interests
The authors declare no competing interests.
Published: December 20, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.matt.2021.11.028.
Supplemental information
Data and code availability
All data associated with this study are made publicly available, including the calculation of enrichment multiples, enhancement factor calculations, theoretical results from the DFT calculations, and data analysis results of machine learning methods.
References
- 1.Tang Z.M., Kong N., Zhang X.C., Liu Y., Hu P., Mou S., Liljeström P., Shi J.L., Farokhzad O.C., Tao W., et al. A materials-science perspective on tackling COVID-19. Nat. Rev. Mater. 2020;5:847–860. doi: 10.1038/s41578-020-00247-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tang Z.M., Zhang X.C., Shu Y.Q., Guo M., Zhang H., Tao W. Insights from nanotechnology in COVID-19 treatment. Nano Today. 2021;36:101019. doi: 10.1016/j.nantod.2020.101019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang J., Fang X., Mao Y., Qi H.C., Wu J., Liu X.R., You F.S., Zhao W.C., Chen Y., Zheng L. Real-time, selective, and low-cost detection of trace level SARS-CoV-2 spike-protein for cold-chain food quarantine. NPJ Sci. Food. 2021;5:12. doi: 10.1038/s41538-021-00094-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chi Y.H., Wang Q.X., Chen G.S., Zheng S.L. The long-term presence of SARS-CoV-2 on cold-chain food packaging surfaces indicates a new COVID-19 winter outbreak: a mini review. Front. Public Health. 2021;9:650493. doi: 10.3389/fpubh.2021.650493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zou L.R., Ruan F., Huang M.X., Liang L.J., Huang H.T., Hong Z.S., Yu J.X., Kang M., Song Y.C., Wu J., et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zou Y.Y., Uspal W.E., Tao W. Airborne transmission of COVID-19: aerosol dispersion, lung deposition, and virus-receptor interactions. ACS Nano. 2020;14:16502–16524. doi: 10.1021/acsnano.0c08484. [DOI] [PubMed] [Google Scholar]
- 7.Halvorson R.A., Vikesland P.J. Surface-enhanced Raman spectroscopy (SERS) for environmental analyses. Environ. Sci. Technol. 2010;44:7749–7755. doi: 10.1021/es101228z. [DOI] [PubMed] [Google Scholar]
- 8.Zhang C., Li C.H., Yu J., Jiang S.Z., Xu S.C., Yang C., Liu Y.J., Gao X.G., Liu A.H., Man B.Y. SERS activated platform with three-dimensional hot spots and tunable nanometer gap. Sens. Actuators B. 2018;258:163–171. [Google Scholar]
- 9.Li J.F., Panneerselvam R., Tian Z.Q. Shell-isolated nanoparticle-enhanced Raman spectroscopy. Nature. 2010;464:392–395. doi: 10.1038/nature08907. [DOI] [PubMed] [Google Scholar]
- 10.Zong C., Xu M.X., Xu L.J., Wei T., Ma X., Zheng X.S., Hu R., Ren B. Surface-enhanced Raman spectroscopy for bioanalysis: reliability and challenges. Chem. Rev. 2018;118:4946–4980. doi: 10.1021/acs.chemrev.7b00668. [DOI] [PubMed] [Google Scholar]
- 11.Eom G., Hwang A., Kim H., Yang S., Lee D.K., Song S., Ha K., Jeong J., Jung J., Lim E.K. Diagnosis of Tamiflu-resistant influenza virus in human nasal fluid and saliva using surface-enhanced Raman scattering. ACS Sens. 2019;4:2282–2287. doi: 10.1021/acssensors.9b00697. [DOI] [PubMed] [Google Scholar]
- 12.Zhang X.G., Zhang X.L., Luo C.L., Liu Z.Q., Chen Y.Y., Dong S.L., Jiang C.Z., Yang S.K., Wang F.B., Xiao X.H. Volume-enhanced Raman scattering detection of viruses. Small. 2019;15:1805516. doi: 10.1002/smll.201805516. [DOI] [PubMed] [Google Scholar]
- 13.Shao F., Lu Z.C., Liu C., Han H.Y., Chen K., Li W.T., He Q.G., Peng H., Chen J.N. Hierarchical nanogaps within bioscaffold arrays as a high-performance SERS substrate for animal virus biosensing. ACS Appl. Mater. Interfcaes. 2014;6:6281–6289. doi: 10.1021/am4045212. [DOI] [PubMed] [Google Scholar]
- 14.Li Z.H., Leustean L., Inci F., Zheng M., Demirci U., Wang S.Q. Plasmonic-based platforms for diagnosis of infectious diseases at the point-of-care. Biotechnol. Adv. 2019;37:107440. doi: 10.1016/j.biotechadv.2019.107440. [DOI] [PubMed] [Google Scholar]
- 15.Yeh Y.T., Gulino K., Zhang Y.H., Sabestien A., Chou T.W., Zhou B., Lin Z., Albert I., Lu H.G., Swaminathan V., et al. A rapid and label-free platform for virus capture and identification from clinical samples. PNAS. 2020;117:895–901. doi: 10.1073/pnas.1910113117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Y., Peng Y.S., Lin C.L., Long L., Hu J.Y., He J., Huang Z.R., Li Z.Y., Lombardi J.R., Luo X.Y., et al. Human ACE2-functionalized gold “virus-trap” nanostructures for accurate capture of SARS-CoV-2 and single-virus SERS detection. Nano-micro Lett. 2021;13:109. doi: 10.1007/s40820-021-00620-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen H., Park S.G., Choi N., Kwon H.J., Kang T., Lee M.K., Choo J. Sensitive detection of SARS-CoV-2 using a SERS-based aptasensor. ACS Sens. 2021;6:2378–2385. doi: 10.1021/acssensors.1c00596. [DOI] [PubMed] [Google Scholar]
- 18.Pramanik A., Gao Y., Patibandla S., Mitra D., McCandless M.G., Fassero L.A., Gates K., Tandon R., Ray P.C. The rapid diagnosis and effective inhibition of coronavirus using spike antibody attached gold nanoparticles. Nanoscale Adv. 2021;3:1588–1596. doi: 10.1039/d0na01007c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peng Y.S., Lin C.L., Long L., Masaki T., Tang M., Yang L.L., Liu J.J., Huang Z.R., Li Z.Y., Yang Y., et al. Charge-transfer resonance and electromagnetic enhancement synergistically enabling MXenes with excellent SERS sensitivity for SARS-CoV-2 S protein detection. Nano-micro Lett. 2021;13:52. doi: 10.1007/s40820-020-00565-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin J., Ren W.Z., Li A.R., Yao C.Y., Chen T.X., Ma X.H., Wang X.T., Wu A.G. Crystal–amorphous core–shell structure synergistically enabling TiO2 nanoparticles remarkable SERS sensitivity for cancer cell imaging. ACS Appl. Mater. Interfaces. 2020;12:4204–4211. doi: 10.1021/acsami.9b17150. [DOI] [PubMed] [Google Scholar]
- 21.Yu H.K., Peng Y.S., Yang Y., Li Z.Y. Plasmon-enhanced light-matter interactions and applications. NPJ Comput. Mater. 2019;5:45. [Google Scholar]
- 22.Seo J., Lee J., Kim Y., Koo D., Lee G., Park H. Ultrasensitive plasmon-free surface-enhanced Raman spectroscopy with femtomolar detection limit from 2D van der Waals heterostructure. Nano Lett. 2020;20:1620–1630. doi: 10.1021/acs.nanolett.9b04645. [DOI] [PubMed] [Google Scholar]
- 23.Yang L.L., Peng Y.S., Yang Y., Liu J.J., Huang H.L., Yu B.H., Zhao J.M., Lu Y.L., Huang Z.R., Li Z.Y., Lombardi J.R. A novel ultra-sensitive semiconductor SERS substrate boosted by the coupled resonance effect. Adv. Sci. 2019;6:1900310. doi: 10.1002/advs.201900310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lombardi J.R. Enhanced by organic surfaces. Nat. Mater. 2017;16:878–880. doi: 10.1038/nmat4958. [DOI] [PubMed] [Google Scholar]
- 25.Shan Y.F., Zheng Z.H., Liu J.J., Yang Y., Li Z.Y., Huang Z.R., Jiang D.L. Niobium pentoxide: a promising surface-enhanced Raman scattering active semiconductor substrate. NPJ Comput. Mater. 2017;11:3. [Google Scholar]
- 26.Zheng Z.H., Cong S., Gong W.B., Xuan J.N., Li G.H., Lu W.B., Geng F.X., Zhao Z.G. Semiconductor SERS enhancement enabled by oxygen incorporation. Nat. Commun. 2017;8:1993. doi: 10.1038/s41467-017-02166-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cong S., Yuan Y.Y., Chen Z.G., Hou J.Y., Yang M., Su Y.L., Zhang Y.Y., Li L., Li Q.W., Geng F.X., Zhao Z.G. Noble metal-comparable SERS enhancement from semiconducting metal oxides by making oxygen vacancies. Nat. Commun. 2015;6:7800. doi: 10.1038/ncomms8800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin J., Shang Y., Li X.X., Yu J., Wang X.T., Guo L. Ultrasensitive SERS detection by defect engineering on single Cu2O superstructure particle. Adv. Mater. 2017;29:1604797. doi: 10.1002/adma.201604797. [DOI] [PubMed] [Google Scholar]
- 29.Guo S.Y., Zhang Y.P., Ge Y.Q., Zhang S.L., Zeng H.B., Zhang H. 2D V-V binary materials: status and challenges. Adv. Mater. 2019;39:1902352. doi: 10.1002/adma.201902352. [DOI] [PubMed] [Google Scholar]
- 30.Quan Y.N., Yao J.C., Sun Y.S., Qu X., Su R., Hu M.Y., Chen L., Liu Y., Gao M., Yang J.H. Enhanced semiconductor charge-transfer resonance: unprecedented oxygen bidirectional strategy. Sens. Actuators B. 2021;327:128903. [Google Scholar]
- 31.Wang X.T., Shi W.X., Wang S.X., Zhao H.W., Lin J., Yang Z., Chen M., Guo L. Two-dimensional amorphous TiO2 nanosheets enabling high-efficiency photoinduced charge transfer for excellent SERS activity. J. Am. Chem. Soc. 2019;141:5856–5862. doi: 10.1021/jacs.9b00029. [DOI] [PubMed] [Google Scholar]
- 32.Zhang D.Z., Xu Z.Y., Yang Z.M., Song X.S. High-performance flexible self-powered tin disulfide nanoflowers/reduced graphene oxide nanohybrid-based humidity sensor driven by triboelectric nanogenerator. Nano Energy. 2020;67:104251. [Google Scholar]
- 33.Gong Y.J., Yuan H.T., Wu C.L., Tang P.Z., Yang S.Z., Yang A.K., Li G.D., Liu B.F., Van de Groep J., Cui Y., et al. Spatially controlled doping of two-dimensional SnS2 through intercalation for electronics. Nat. Nanotechnol. 2018;13:294–299. doi: 10.1038/s41565-018-0069-3. [DOI] [PubMed] [Google Scholar]
- 34.Kitadai H., Wang X.Z., Mao N.N., Huang S.X., Ling X. Enhanced Raman scattering on nine 2D van der Waals materials. J. Phys. Chem. Lett. 2019;10:3043–3050. doi: 10.1021/acs.jpclett.9b01146. [DOI] [PubMed] [Google Scholar]
- 35.Zhang M.M., Li X.Y., Fan S.Y., Yin Z.F., Li J.N., Zeng L.B., Tade M.O., Liu S.M. Novel two-dimensional AgInS2/SnS2/RGO dual heterojunctions: high spatial charge and toxicity evaluation. Langmuir. 2020;36:9709–9718. doi: 10.1021/acs.langmuir.0c01072. [DOI] [PubMed] [Google Scholar]
- 36.Mondal C., Ganguly M., Pal J., Roy A., Jana J., Pal T. Morphology controlled synthesis of SnS2 nanomaterial for promoting photocatalytic reduction of aqueous Cr(VI) under visible light. Langmuir. 2014;30:4157–4164. doi: 10.1021/la500509c. [DOI] [PubMed] [Google Scholar]
- 37.Lee C.G., Yan H.G., Brus L.E., Heinz T.F., Hone J., Ryu S. Anomalous lattice vibrations of single- and few-layer MoS2. ACS Nano. 2010;4:2695–2700. doi: 10.1021/nn1003937. [DOI] [PubMed] [Google Scholar]
- 38.Li X.H., Guo S.H., Su J., Ren X.G., Fang Z.Y. Efficient Raman enhancement in molybdenum disulfide by tuning the interlayer spacing. ACS Appl. Mater. Interfaces. 2020;12:28474–28483. doi: 10.1021/acsami.0c04151. [DOI] [PubMed] [Google Scholar]
- 39.Guo H., Qian K., Cai A.J., Tang J., Liu J. Ordered gold nanoparticle arrays on the tip of silver wrinkled structures for single molecule detection. Sens. Actuators B. 2019;300:126846. [Google Scholar]
- 40.Park S.G., Mun C., Xiao X.F., Braun A., Kim S., Giannini V., Maier S.A., Kim D.H. Surface energy-controlled SERS substrates for molecular concentration at plasmonic nanogaps. Adv. Funct. Mater. 2017;27:1703376. [Google Scholar]
- 41.Wang X.T., Shi W.X., Jin Z., Huang W.F., Lin J., Ma G.S., Li S.Z., Guo L. Remarkable SERS activity observed from amorphous ZnO nanocages. Angew. Chem. Int. Ed. 2017;33:9983–9987. doi: 10.1002/anie.201705187. [DOI] [PubMed] [Google Scholar]
- 42.Wrapp D., Wang N.S., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Z.Y., Tang Z.M., Farokhzad N., Chen T.F., Tao W. Sensitive, rapid, low-cost, and multiplexed COVID-19 monitoring by the wireless telemedicine platform. Matter. 2020;3:1818–1820. doi: 10.1016/j.matt.2020.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng S.F., Fan J., Yu F., Feng B.H., Lou B., Zou Q.D., Xie G.L., Lin S., Wang R.N., Yang X.Z., et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ. 2020;369:m1443. doi: 10.1136/bmj.m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peng L., Liu J., Xu W.X., Luo Q.M., Chen D.B., Lei Z.Y., Huang Z.L., Li X.J., Deng K.J., Lin B.L., Gao Z.L. SARS-CoV-2 can be detected in urine, blood, anal swabs, and oropharyngeal swabs specimens. J. Med. Virol. 2020;92:1676–1680. doi: 10.1002/jmv.25936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pan Y., Zhang D., Yang P., Poon L.L., Wang M.Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 2020;20:411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan X., Yang C.M., He Q., Chen J.H., Yu D.M., Li J., Zhai S.Y., Qin Z.F., Du K., Chu Z.H., Qin P.W. Current and perspective diagnostic techniques for COVID-19. ACS Infect. Dis. 2020;6:1998–2016. doi: 10.1021/acsinfecdis.0c00365. [DOI] [PubMed] [Google Scholar]
- 48.Dai M., Li H.N., Yan N., Huang J.Y., Zhao L., Xu S.Q., Wu J.M., Jiang S.B., Pan C.G., Liao M. Long-term survival of SARS-CoV-2 on salmon as a source for international transmission. J. Infect. Dis. 2021;223:537–539. doi: 10.1093/infdis/jiaa712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu H., Fei C.N., Chen Y.L., Luo S.M., Yang T., Yang L., Liu J., Ji X.Y., Wu W.S., Song J. Investigating SARS-CoV-2 persistent contamination in different indoor environments. Environ. Res. 2021;202:111763. doi: 10.1016/j.envres.2021.111763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Binnicker M.J. Can testing predict SARS-CoV-2 infectivity? The potential for certain methods to be surrogates for replication-competent virus. J. Clin. Microbiol. 2021;59 doi: 10.1128/JCM.00469-21. e00469-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data associated with this study are made publicly available, including the calculation of enrichment multiples, enhancement factor calculations, theoretical results from the DFT calculations, and data analysis results of machine learning methods.