Abstract
Background
Streptococcus constellatus is a member of Streptococcus anginosus group (SAG) that tends to cause pyogenic infections in various sites. However, Streptococcus constellatus is easily ignored by routine clinical laboratory tests for its prolonged anaerobic culture environment.
Case presentation
A 71-year-old man was admitted to our hospital due to productive cough, fever, chest pain and shortness of breath for 3 weeks. Chest computed tomography showed patchy opacities and right-sided pleural effusion, so a chest tube was inserted and purulent and hemorrhagic fluid was aspirated. The routine etiological examinations of the pleural effusion were all negative, and next-generation sequencing (NGS) detected Streptococcus constellatus. Intravenous piperacillin-tazobactam 4.5 g every 8 h was used accordingly. The patient recovered and subsequent chest computed tomography confirmed the improvement.
Conclusions
We reported a case of empyema secondary to Streptococcus constellatus infection, which was identified by NGS, instead of bacterial culture. This case highlights the utility of NGS in detecting pathogens negative in traditional bacterial tests.
Keywords: Streptococcus constellatus, Empyema, Next-generation sequencing
Background
Empyema is a common complication of lung bacterial infections, but it is rarely reported to be caused by Streptococcus constellatus (S. constellatus). S. constellatus is one of the Streptococcus anginosus group (SAG), a subgroup of viridans streptococci, widely distributed in the oral cavity, nasopharynx, gastrointestinal tract, and vagina in 15–30% of healthy people [1]. SAG tends to cause pyogenic infections, such as dental, peritonsillar and sinus abscesses [2].
As a member of SAG, S. constellatus induces abscesses mainly in respiratory tract, brain, liver, bone and soft tissues [3–5]. SAG infection has a strong male predominance [6, 7]. The symptoms of pulmonary infection caused by S. constellatus include shortness of breath, chest pain, cough, and fever [6]. Laboratory tests often suggest leucocytosis, neutrophilia, abnormal liver function, and hypo-albuminaemia [6]. The median stay in hospital was 34 days [6].
Here, we reported the use of next-generation sequencing (NGS) in detecting S. constellatus as the pathogen for a case of empyema of unknown causes. This case highlights the use of NGS in etiology diagnosis and guiding the treatment in empyema.
Case presentation
A 71-year-old male was admitted to the hospital due to productive cough along with low grade fever, chest pain and shortness of breath. His past medical history included hypertension and glaucoma, and he took irbesartan regularly (150 mg per day). The patient did not smoke cigarettes, drink alcohol or use recreational drugs. No relevant travel history or contact history were detected. The patient had no food or drug allergies.
Three weeks before admission, the patient began to have productive cough, with chest tightness and a temperature of 38 °C. After 2 weeks of progressive symptoms, the patient visited the local hospital. He reported pleuritic chest pain of visual analogue scale score 2. His vital signs and other physical examination results were reported as normal. Initial blood test showed elevated white blood cell (WBC) count (14.8 × 109/L) and C-reactive protein (CRP) level (86 mg/L) as well as liver enzyme elevation. Other laboratory test results were normal. Chest computed tomography (CT) revealed patchy opacities in both lower lobes and a small amount of right-sided pleural effusion. He was then admitted to the local hospital and received intravenous sulperazon (cefperazone–sulbactam) 2.0 g once every 8 h, but symptomatic improvement was not noted. Repeated chest CT scan revealed increased pleural effusion in the right. Subsequently, the patient was transferred to our hospital for treatment.
On the admission, his temperature was 37.8 °C, pulse rate 109 beats/min, respiratory rate 18 breaths/min, blood pressure 145/87 mmHg, and oxygen saturation 98% on room air. The patient reported no night sweats, weight loss, joint pains, or myalgias. Pulmonary auscultation found decreased breath sounds on both lower fields. No icterus or lymphadenopathy was detected. Thoracocentesis was performed immediately and a chest tube was introduced. Purulent and hemorrhagic fluid was aspirated (Fig. 1A), and laboratory tests of the pleural effusion revealed an exudate (fluid protein 25.70 g/L, serum protein 28.70 g/L [normal: 35.0–55.0 g/L], fluid lactate dehydrogenase 621 U/L, serum lactate dehydrogenase 355 U/L [normal < 243 U/L]). Other pleural fluid analysis showed: WBC 5000/µL with 88% neutrophils, red blood cells 21,000/µL, fluid glucose 7.31 mmol/L, and fluid adenosine deaminase 18 U/L. Pleural fluid smear, gram stain, bacterial culture, acid-fast bacilli culture and smear, and cytology were all negative. Blood culture was also negative. He was diagnosed as empyema and intravenous tazocin (piperacillin-tazobactam) 4.5 g once every 8 h was initiated.
Fig. 1.
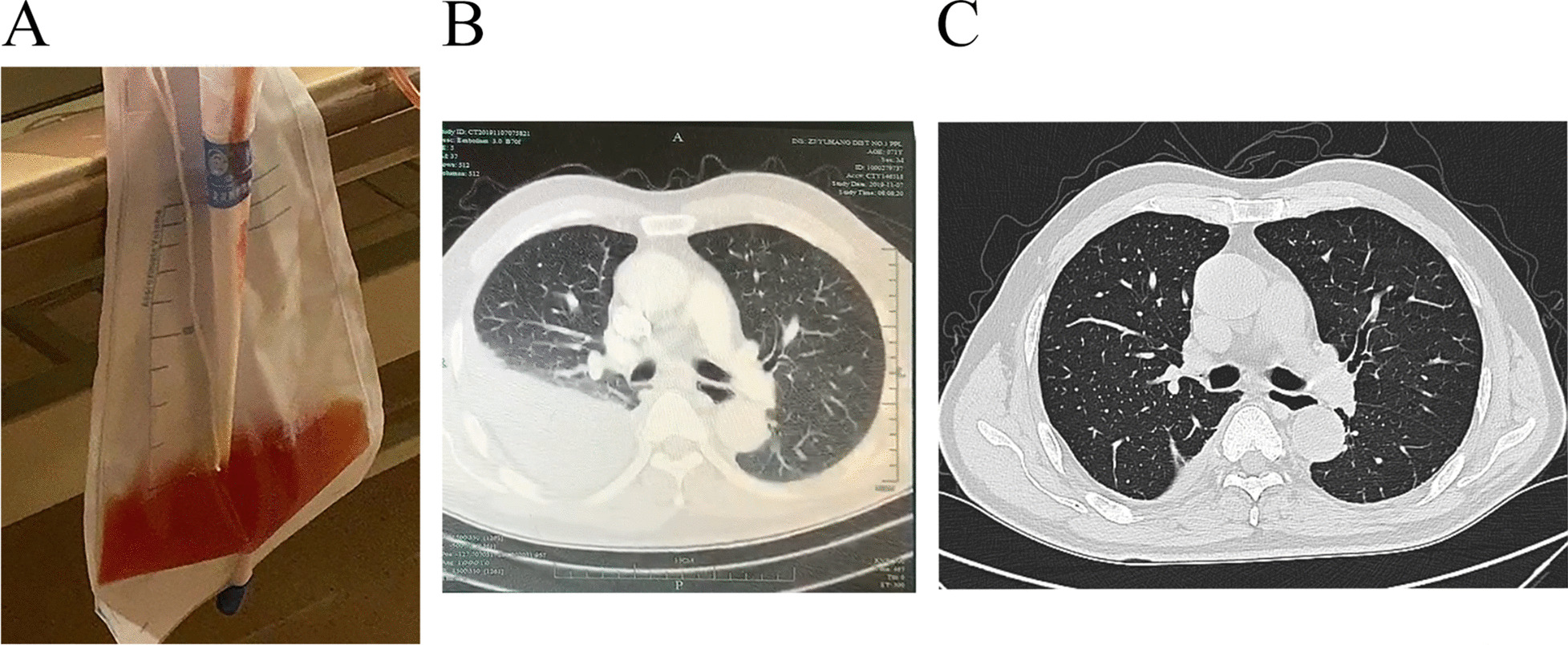
The appearance and CT scan of pleural effusion. A Appearance of the pleural effusion. B Chest CT showed moderate, right-sided pleural effusion on the day of admission. C Chest CT showed significantly reduced, small amount of right-sided pleural effusion 7 days after the admission.
During the treatment, repeated blood and fluid cultures were all negative, so the pleural effusion was sent for NGS. Deoxyribonucleic acid (DNA) was extracted directly from the pleural fluid. The extracted DNA was amplified, purified, and sonicated to a size of 200–300 bp. Fragmented DNA was end repaired to construct DNA libraries, and supplemented with adapter overnight followed by polymerase chain reaction (PCR) amplification. Qualified DNA libraries were sequenced using the BGISEQ-100 platform. We generated high-quality sequencing data by removing low-quality reads, low-complexity reads, and reads shorter than 35 bp. Human data were mapped to a human reference (hg19) and excluded. The remaining sequencing data were aligned to the bacterial, virus, and fungal databases. All pathogen sequence reference databases were from National Center for Biotechnology Information (NCBI) (ftp://ftp.ncbi.nlm.nih.gov/genomes/).
NGS identified 14 out of 19317 reads uniquely corresponding to S. constellatus. Although the number of the reads was small, S. constellatus was the only pathogen detected. Tazocin was used continuously for 12 days, then followed by moxifoxacin 400 mg per day for a total of 14 days. Meanwhile, the pleural effusion was drained continuously through the chest tube. The patient claimed gradually resolved symptoms under antibiotic treatment and effusion drainage, and subsequent CT scanning confirmed the improvement (Fig. 1B, C).
Discussion and conclusions
In this case, we reported a case of empyema caused by S. constellatus. The identification of S. constellatus is sometimes difficult because of the prolonged anaerobic culture environment [8]. During the recent decades, faster and more convenient technical methods have been applied to investigate the pathogens of infectious diseases. For example, real-time PCR assays targeting the cpn60 and 16S ribosomal ribonucleic acid (rRNA) genes were used to detect SAG [1, 8]. Most of the S. constellatus (96%), S. intermedius (94%), and S. anginosus (60%) strains were correctly identified by targeting cpn60 [1]. 16S ribosomal deoxyribonucleic acid (rDNA) sequencing of purulent fluid obtained from a muscle abscess aspirate was successfully used to diagnose pyomyositis caused by SAG [8]. Rapid diagnostic kits [9] and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) [10] were also used in clinical laboratories to identify SAG. In recent years, the application of NGS shed light on detecting pathogens more efficiently. NGS can sequence thousands to billions of DNA fragments simultaneously, making quick identification of the pathogens in culture-negative specimens possible [11]. Compared with 16S rRNA sequencing, which identifies one organism at a time, NGS can provide information on multiple organisms simultaneously [12]. Although NGS has its own drawbacks such as shorter read length and decreased raw accuracy in copy number variants (CNVs) or other structural variants, the development of culture independent NGS technology still offers unbiased, rapid etiology diagnosis for the entire microbial community [13].
In this case, S. constellatus was detected by NGS rather than traditional bacterial culture, and various reasons could contribute to this matter. Frist, S. constellatus is a facultative anaerobic organism sometimes difficult to culture, especially when not specifically requested. Secondly, the patient started with antibiotics before his admission to our hospital. It may reduce culture-dependent bacterial detection. Compared to the difficulty of traditional culture, NGS was able to amplify and detect cell-free DNA and DNA fragments in dead cells, making etiology diagnosis possible. NGS should be considered in the following occasions: pathogens not detected in clinical specimens; patient already receives antibiotics; and suspicious of uncommon pathogens.
According to the literatures, among a collection of 423 clinical SAG, 1.4% of the strains were of intermediate susceptibility to penicillin and none exhibited high-level resistance to gentamicin. All the strains were susceptible to cefotaxime, vancomycin and teicoplanin [14]. Clindamycin, doxycycline, amoxicillin, and metronidazole could be used for S. constellatus infection [15, 16]. Although S. constellatus is not a commonly detected pathogen causing pleural effusion, we still need to pay attention to its pathogenicity in clinical practice.
Collectively, this case reported an empyema secondary to S. constellatus infection, where NGS helped to determine the pathogen. NGS is of great significance to infection of unknown pathogens, and we expect further application of advanced sequencing technologies in infectious diseases in the future.
Acknowledgements
Not applicable.
Abbreviations
- SAG
Streptococcus anginosus group
- NGS
Next-generation sequencing
- WBC
White blood cell
- CRP
C-reactive protein
- CT
Computed tomography
- rRNA
Ribosomal ribonucleic acid
- DNA
Deoxyribonucleic acid
- PCR
Polymerase chain reaction
- NCBI
National Center for Biotechnology Information
- rDNA
Ribosomal deoxyribonucleic acid
- MALDI-TOF MS
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry
- CNV
Copy number variant
Authors’ contributions
JX, HZ and XL collected data and wrote the original draft. LX re-collected the data and revised the manuscript. FX conceived the manuscript and made a revision of it. JX and LX contributed equally to this work. All authors read and approved the final manuscript.
Funding
None.
Availability of data and materials
All data generated or analyzed during this study are included in this published article. All data and materials are available with the corresponding author.
Declarations
Ethics approval and consent to participate
The work has been approved by the Ethnic Committee of Second Affiliated Hospital, Zhejiang University School of Medicine.
Consent for publication
The patient provided written informed consent for their personal or clinical details along with any identifying images to be published in this study. A copy of the written consent is available for review by the Editor of this journal.
Competing interests
The authors declare that they have no conflicts of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Olson AB, Sibley CD, Schmidt L, Wilcox MA, Surette MG, Corbett CR. Development of real-time PCR assays for detection of the Streptococcus milleri group from cystic fibrosis clinical specimens by targeting the cpn60 and 16S rRNA genes. J Clin Microbiol. 2010;48(4):1150–1160. doi: 10.1128/JCM.02082-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gharib SD, Berger DL, Choy G, Huck AE. Case records of the Massachusetts general hospital. Case 21-2015. A 37-year-old American man living in Vietnam, with fever and bacteremia. N Engl J Med. 2015;373(2):174–183. doi: 10.1056/NEJMcpc1411439. [DOI] [PubMed] [Google Scholar]
- 3.Claridge JE, 3rd, Attorri S, Musher DM, Hebert J, Dunbar S. Streptococcus intermedius, Streptococcus constellatus, and Streptococcus anginosus (“Streptococcus milleri group”) are of different clinical importance and are not equally associated with abscess. Clin Infect Dis. 2001;32(10):1511–1515. doi: 10.1086/320163. [DOI] [PubMed] [Google Scholar]
- 4.Inoue M, Okamoto K, Nagao H, Toyoda K. A case of esophageal submucosal abscess originating from a peritonsillar abscess. Nihon Jibiinkoka Gakkai kaiho. 2016;119(7):962–966. doi: 10.3950/jibiinkoka.119.962. [DOI] [PubMed] [Google Scholar]
- 5.Noguchi S, Yatera K, Kawanami T, Yamasaki K, Naito K, Akata K, Shimabukuro I, Ishimoto H, Yoshii C, Mukae H. The clinical features of respiratory infections caused by the Streptococcus anginosus group. BMC Pulm Med. 2015;15:133. doi: 10.1186/s12890-015-0128-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong CA, Donald F, Macfarlane JT. Streptococcus milleri pulmonary disease: a review and clinical description of 25 patients. Thorax. 1995;50(10):1093–1096. doi: 10.1136/thx.50.10.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Porta G, Rodriguez-Carballeira M, Gomez L, Salavert M, Freixas N, Xercavins M, Garau J. Thoracic infection caused by Streptococcus milleri. Eur Respir J. 1998;12(2):357–362. doi: 10.1183/09031936.98.12020357. [DOI] [PubMed] [Google Scholar]
- 8.Walkty A, Embil JM, Nichol K, Karlowsky J. An unusual case of Streptococcus anginosus group pyomyositis diagnosed using direct 16S ribosomal DNA sequencing. Can J Infect Dis Med Microbiol. 2014;25(1):32–34. doi: 10.1155/2014/170517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carroll KC, Reid JL, Thornberg A, Whitfield NN, Trainor D, Lewis S, Wakefield T, Davis TE, Church KG, Samuel L, et al. Clinical performance of the novel GenMark Dx ePlex blood culture ID Gram-positive panel. J Clin Microbiol. 2020;58(4):e01730-19. doi: 10.1128/JCM.01730-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arinto-Garcia R, Pinho MD, Carrico JA, Melo-Cristino J, Ramirez M. Comparing matrix-assisted laser desorption ionization-time of flight mass spectrometry and phenotypic and molecular methods for identification of species within the Streptococcus anginosus group. J Clin Microbiol. 2015;53(11):3580–3588. doi: 10.1128/JCM.01892-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng J, Hu H, Kang Y, Chen W, Fang W, Wang K, et al. Identification of pathogens in culture-negative infective endocarditis cases by metagenomic analysis. Ann Clin Microbiol Antimicrob. 2018;17(1):43. doi: 10.1186/s12941-018-0294-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin JH, Wu ZY, Gong L, Wong CH, Chao WC, Yen CM, et al. Complex microbiome in brain abscess revealed by whole-genome culture-independent and culture-based sequencing. J Clin Med. 2019;8(3):351. doi: 10.3390/jcm8030351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo LY, Feng WY, Guo X, Liu B, Liu G, Dong J. The advantages of next-generation sequencing technology in the detection of different sources of abscess. J Infect. 2019;78(1):75–86. doi: 10.1016/j.jinf.2018.08.002. [DOI] [PubMed] [Google Scholar]
- 14.Jacobs JA, Stobberingh EE. In-vitro antimicrobial susceptibility of the “Streptococcus milleri” group (Streptococcus anginosus, Streptococcus constellatus and Streptococcus intermedius) J Antimicrob Chemother. 1996;37(2):371–375. doi: 10.1093/jac/37.2.371. [DOI] [PubMed] [Google Scholar]
- 15.Rams TE, Feik D, Mortensen JE, Degener JE, van Winkelhoff AJ. Antibiotic susceptibility of periodontal Streptococcus constellatus and Streptococcus intermedius clinical isolates. J Periodontol. 2014;85(12):1792–1798. doi: 10.1902/jop.2014.130291. [DOI] [PubMed] [Google Scholar]
- 16.Rams TE, Degener JE, van Winkelhoff AJ. Antibiotic resistance in human chronic periodontitis microbiota. J Periodontol. 2014;85(1):160–169. doi: 10.1902/jop.2013.130142. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article. All data and materials are available with the corresponding author.


