Abstract
Epilepsy is the most common chronic neurologic disorder in the world, affecting 1-2% of the population. Besides, 30% of epilepsy patients are drug-resistant. Genomic mutations seem to play a key role in its etiology and knowledge of strong effect mutations in protein structures might improve prediction and the development of efficacious drugs to treat epilepsy. Several genetic association studies have been undertaken to examine the effect of a range of candidate genes for resistance. Although, few studies have explored the effect of the mutations into protein structure and biophysics in the epilepsy field. Much work remains to be done, but the plans made for exciting developments will hold therapeutic potential for patients with drug-resistance. In summary, we provide a critical review of the perspectives for the development of individualized medicine for epilepsy based on genetic polymorphisms/mutations in light of core elements such as transcriptomics, structural biology, disease model, pharmacogenomics and pharmacokinetics in a manner to improve the success of trial designs of antiepileptic drugs.
Keywords: Genetic epilepsy, brain organoids, transcriptome, single-cell sequencing, pharmacogenomics, drug resistance, drug development, antiepileptic drugs
1. INTRODUCTION
Epilepsy is a disease that affects about 1-2% of the population and is characterized by recurrent, unprovoked seizures triggered by an imbalance between excitation and inhibition in the neuronal circuits [1]. Once regarded as demonic possession, it’s currently known to be caused by brain injuries, infection diseases, genetic conditions and exposure to toxins or drugs. Also, about 30 to 40% of the cases are idiopathic, with the predominance of genetic underlying [2, 3]. One of the first approaches in this direction was framed in a study from 1990 that identified an A to G transition mutation at nucleotide pair 8344 in human mitochondrial DNA as a cause of Myoclonic Epilepsy and Ragged-Red Fiber Disease [4]. The first line of these approaches involved the investigation of membrane proteins and intermediary metabolism in tissues, but the research soon started to tackle mapped loci in families with generalized epilepsy syndromes. In 1995, a study recognized a missense mutation in the CHRNA4 gene, encoding the α4-subunit of a neuronal nicotinic acetylcho-line receptor, the first ion channel missense mutation in an inherited form of epilepsy [5].
Nowadays, most of the epilepsy-associated genes are precisely related to ion channels, receptors, and other components of neuronal signaling. This is conceivable once neuronal hyperexcitability is a main cellular mechanism underlying seizures. The majority of these mutations affect voltage-gated ion channels (such as K+, Na+, and Ca2+) and ligand-gated ion channels (nicotinic acetylcholine and GABAA receptors) [6-8]. A preponderant mutation in this regard was found in the SCN1A gene, which encodes one of the nine α subunits of the voltage-gated sodium channel in mammals that was shown to cause several subtypes of dominant idiopathic generalized epilepsy in humans [8]. Also, the major feature of genetic epilepsy with febrile seizures plus (GEFS+) is a genetic variant in the gene SCN2A, however, mutations in the SCN1B gene (encoding the sodium channel β1 subunit) and in GABAA receptor genes also seems to occur [9, 10].
The first epilepsy predisposition identified gene unrelated directly to ion channel structural proteins was LGI1, a 60-kDa secreted neuronal protein involved in synaptic transmission [11]. The molecular mechanisms connecting the mutations to the epileptic symptoms is not yet totally clear, but a more recent study has evoked the LGI1–ADAM22 ligand-receptor complex, which regulates synaptic transmission and has emerged as a determinant of brain excitability [6].
In the last decades, since the first mutation related to epilepsy was identified, hundreds of genes have been recognized. A 2017’s survey, cross-checking multiple databases (OMIM, HGMD, and EpilepsyGene) and publications, found 977 genes that are associated with epilepsy [12]. Most of them occur under the action of multiple genes (polygenic), such as the most common subtype of epilepsy – the idiopathic generalized epilepsies (IGEs) [13]. All the aspects of neurotransmission are in play, and defects of synaptic inhibition are over-represented. The contributions of somatic mutations and noncoding microRNAs are also being tackled [14]. Even though none of the genes directly explains why, in most cases, the pathogenic molecular mechanism is still unclear. In this way, the application of precise genomic technologies (including genome editing) provides a tremendous impact to prioritize hypothesis-driven discoveries and filling the genetic “proto-map” of this disease. New approaches for discovering genes and pathophysiological pathways may be important to establish novel targets for pharmacotherapy, and the overall development and application of cutting-edge technologies open a new window of possibilities for personalized medicine. Considering that epilepsy treatment is highly refractory and extremely important to compiling state-of-the-art technologies that can improve the drug discovery and treatment in the field of genetic epilepsies, in the next sections, we will focus on how different approaches can improve the development of better drugs to treat genetic epilepsy.
2. GENOMICS AND TRANSCRIPTOMICS TO DRUG DISCOVERY IN EPILEPSY
2.1. Screening the Genome to Better Diagnosis
It is known that pharmaceutical industries can expense a billion of dollars and decades to deliver new drugs, which recently have a rate of around 50% of failure [15]. An alternative to avoiding these high costs and time-consuming is the optimization of the decision making in choosing the molecular target in early-stages of the drug discovery phase, through cutting-edge technologies currently available to measure the biological effect of the target molecules [16, 17].
In the 20th century, the development of the expressed sequencing tags approach generated eminent expectations in the pharmaceutical industry, which had been declining in terms of drug development. Despite the substantial investment made between genomic companies and the pharmaceutical industries, little progress was fulfilled in the drug development field due to the loss in elucidating the biological processes of the drug targets, leading to a large number of false positives [18]. Human genome sequencing has enabled genome-wide association study (GWAS), including whole-genome sequencing and whole-exome sequencing. GWAS is useful in determining individualized disease risks, but the vast majority of GWAS findings alone have little relevance to drug discovery. According to the omnigenic model explanation from Pritchard and colleagues, the sum of these numerous small-effect variants disease-unrelated (called peripheral genes) could play a role in active regulatory variants to a small number of core genes related to the disease, through a highly interconnected network, in which druggable targets would certainly benefit considerably more from the identification of the strong-effect core genes [18, 19].
More than a decade ago, the acknowledgment about the etiology of epileptic disorders was narrow. The genomics era has entered a new phase, the next generation sequencing (NGS) technology has been favoring the availability of sequences generated in public databases together with the improvement of computational tools that allow the analysis of a massive volume of data in record time. The advancement in genomics has been increasing the correlation of genetic variables to clinical symptoms in a robust manner. Thus, the knowledge of genes that are candidates to epilepsy has elevated the clinical diagnosis to a level where the genetic alteration, in some cases, becomes more prevalent than the clinical syndrome itself [20]. Clinical testing with comprehensive gene panels, exomes, and genomes are currently available and have led to plausible diagnostic rates. Although the panel testing provides a diagnosis at a rate of around 15% to 25%, a vast majority of the patients tested will not receive a molecular diagnostic [21]. A recent literature review pointed to 2421 genes included in 107 panels testing for the diagnosis of epilepsy. According to this study, only 153 genes tested across the panels could potentially indicate efficacy in diagnosis, then suggesting a more rational and precise selection of the genes considering a small pool of genes, and more caution in the selection of the epilepsy patients to compose a robust panel testing [22]. Recently, an investigation aiming to analyze if alternative isoforms could be widely relevant to clinical sequencing was performed and as a result, it was found that the resequencing incorporating alternative transcripts might be a useful approach in cases that the result of the gene test is inconclusive [23]. The discovery of new genes and molecular pathways associated with epilepsy play an important role in the diagnosis and genetic counseling of a disease that covers a vast spectrum of heterogeneity and still opens up the opportunity for the development of targeted therapy [20].
Over the past decade, recent advances in NGS have changed the clinical practice of the genetics epilepsy, accomplishing the identification of thousands of mutations associated with this disease. Many of these mutations have been implicated, involving ion channels, and proteins needed for synaptic, regulatory, and developmental functions [24]. The largest whole-exome sequencing study published in 2019, identified ultra-rare deleterious variants associated with epilepsy genes, GABAergic pathways and, cation-channel-encoding genes with at least a 5-fold increase in sample size over previous exome-sequencing studies. This study reinforces that the clinical presentations of genetic generalized epilepsy and non-acquired focal epilepsy with complex inheritance patterns have a combination of both common and rare genetic risk variants [25].
High rates of unrevealing diagnosis in genes testing might be attributed to several causes. As such, the incomplete knowledge of disease architecture, the focus on exonic variation, the challenges in variant pathogenicity interpretation and, technical limitations influencing variant calling. The near future of whole-genome sequencing pinpoints to the analyses of mosaicisms and variants within non-exomic regulatory regions or even intronic variants with deep coverage and accuracy [21]. In this regard, the mechanisms underlying epilepsy will better characterize open paths to new possibilities for novel therapies based on individual specific genotypes [21].
2.2. Using Transcriptomics to Find Drug Targets
RNA-sequencing (RNA-Seq) is the current method of choice to study gene expression and identify novel RNA species, also known as deep sequencing or NGS [26, 27]. Besides, several gene expression analyses have been done in patients with epilepsy and animal model, little is known regarding common transcriptional drivers in the pathophysiology of epilepsy or even drug discovery [28, 29]. Nevertheless, the transcriptomics field using the NGS platform holds the potential to reveal sequence identities that are crucial for the analysis of unknown genes, novel transcript isoforms, and genetic variations [26].
Activity-dependent transcription plays a key role in processes such as synaptic plasticity and its disruption may be intrinsically related to epilepsy. A recent study was performed combining RNA-Seq, chromatin immunoprecipitation, assay for transposase-accessible chromatin with sequencing, and Hi-C to analyze the neuronal responsiveness in status epilepticus [30]. The authors carried out the experiments using mouse tagged nuclei and polysomes of hippocampal excitatory neurons at different time points after synchronous activation during the seizure. Enduring changes were observed in the chromatin occupancy after neuronal activation, guided by the transcription factor AP1. Also, a strong and synchronous activation in all the hippocampal subfields led to robust but transient induction of immediate-early genes in status epilepticus. Bringing together the transcriptional start site and the transcriptional termination site suggest the continuous reloading of the RNAPII complex [30]. This data may underlie the changes in the neuronal circuitry responsiveness in the status epilepticus condition.
A GWAS study employing RNA-Seq platform conjoined with robust bioinformatic tools to analyze differential RNA editing in the hippocampus of an animal model for epilepsy (a mouse model of temporal lobe epilepsy) aiming to identify differential RNA editing events between cases and controls was addressed by the Johnson group [31]. The authors identified 256 RNA sites differentially edited between cases and controls belonging to 87 different genes. These changes were intrinsically associated with pathways and genes previously reported to play a role in this disease. Interestingly, 7 out of 10 differentially RNA-edited sites in exon regions were predicted as deleterious. Also, the degree of differential RNA editing was statistically correlated with the frequency of seizures. This result points to the potential role of RNA differential editing events in epilepsy [32]. Studies using NGS platforms have been providing better discrimination of the disease progression and clarifying epilepsy classifications, by elucidating the molecular dysregulations in the epileptic brain. Additionally, ongoing efforts to fully characterize the transcriptome will help both the increase of the diagnosis rates and ensuring an attained genetic evaluation that can be useful for drug development.
RNA-Seq NGS technology can also be employed in the drug discovery field, which ranges from the prediction of molecular mechanisms and the therapeutic potential of drugs in pharmacodynamics to the identification of genes related to sensitivity or resistance that indicate positive or side-effects to a given drug [16, 17]. The pharmaceutical industries have been obtaining success in drug development using the transcriptomic-based drug discovery to identify more regulatory points and the discrimination between desirable effects and side-effects, providing a high quality of compounds candidates for clinical trials (Fig. 1A). Large-scale RNA-Seq can improve the discovery of new functional genes that can serve as a basis for further studies and the subsequent development of new molecular signatures, all with greater speed, data robustness and lower cost [16, 33]. As an example, RNA-Seq approaches can detect differences in the level of expression between alleles in specific biological processes revealing new molecular mechanisms, which are of paramount importance for the development of new molecular targets [16].
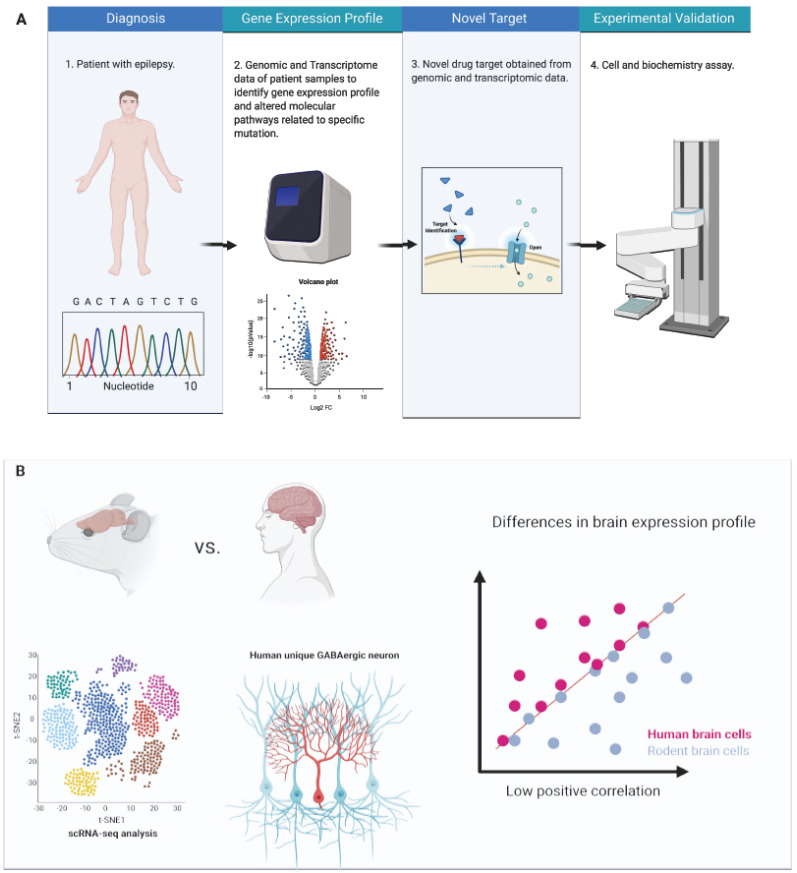
Fig. (1).
Drug discovery strategy based on NGS sequencing. A: Genomic and transcriptomic approaches can be applied to obtain molecular information from patients with epilepsy that may help to discover potential new drug target B: scRNA-Seq comparison between species may help to advance in the choice of the disease model and understand the common and specific features of each model that move towards drug targets. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
Transcriptome analysis also may be a useful tool to investigate the effect of the disease modification status and by the treatment with one or more combined antiepileptic drugs (AEDs) at the RNA levels, providing clues on the disease molecular mechanisms and that triggered by drugs, providing further directions for personalized medicine [34]. In epilepsy, a drug repositioning study used the RNA-Seq approach in brain biopsy to analyze differences between seizing and non-seizing specimens. The data from the gene expression signatures generated by the RNA-Seq platform was crossed with drug screening of anti-epileptic candidate molecules already approved by the Food and Drug Administration (FDA) for human use, with about 50% of them not yet associated with the treatment of epilepsy. In the validation step, using the zebrafish model, three compounds showed potential anti-seizure efficacy [35]. In this way, one of the major focuses of the pharmaceutical industries is to analyze transcripts following molecular targets in several conditions to uncover its mechanisms of action, biological processes and molecular pathways that may predict the effectiveness, toxicity and/or side-effects of the compound, opening new roads for a new formulation or drug repurpose that reduce side-effects and increase the effectiveness of treatment [16, 17, 36].
RNA-Seq analysis was employed by Cui and collaborators to check the gene expression profiles of the cell SCN1A-KO model based on clustered regularly interspaced short palindrome repeats-associated endonuclease Cas9 (CRISPR-Cas9) genome editing technology in order to reveal changes in mRNA levels and its association with Dravet Syndrome (DS). They found some ion channels, such as potassium, sodium, chloride, and calcium, disturbed at gene expression level, suggesting that the poor effectiveness of the AED in patients with DS could be due to the fact that the majority of these drugs act on sodium channels [37].
Another meaningful study evaluated the transcriptional profile of the genome through co-expression genes as candidates for potential regulators of the disease state using the analytical tool Causal Reasoning Analytical Framework for Target discovery in a mouse model of acquired temporal lobe epilepsy. This approach is based on co-expression modules (analysis of regulatory regions between genes) combined with a computational prediction tool for cell surface receptors (based on known interactions between membrane receptors and transcription factors and their target genes with proof of experimental evidence) that influence the co-expression modules. In this study, a potential therapeutic target, Csf1R, was pointed, once again, demonstrating the innovative potential of these cutting-edge technologies combined with advanced computational algorithms for predicting new targets [31]. The search for new therapeutic targets, in this case, occurs through the inverse regulation of these co-expressed genes to keep the expression levels of these targets normalized, bringing the possibility of restoring the individual's health status [31].
More than half of human proteins are regulated by non-coding RNAs. Since the last decade, when the first manuscript related to the study of microRNA and epilepsy was published, successive studies around this topic have been done [38, 39]. Additionally, a curated database-EpimiRBase to provide complete and up-to-date information on publications regarding microRNA and epilepsy was released [40]. Considering that the chemically modified antisense oligonucleotides could inhibit the function of mRNAs in an efficient and specific way, these molecules are potential targets for drug development [41]. There are several challenges to surpass in order to better understand the functionality of the microRNA in the regulation of epileptic processes, such as its pleiotropic functions demanding better experimental designs, analytical tools, and molecular validations [38, 39].
Genetic epilepsy has been urging for biomarkers application for treatment, prediction, prognostic, and pharmacovigilance. Technologies for brain imaging and large-scale clinical screening (such as genomics and transcriptomics) may enhance the possibilities to find potential molecular signatures in this disease. The entire process of epileptogenesis can be covered using these approaches, which can analyze robustly and accurately thousands of different molecules within a unique biological sample. To date, few studies have identified potential signatures in epilepsy. According to more recent reviews, potential biomarkers for epilepsy were suggested, such as in inflammation (IL1-B, IL-6, IL-8, IL-1Ra, IL-17A, IL-6, HMGB1, EPO, HS-CRP, CCL2), complement system (C3, C4, properdin, FH, C1Inh, and Clu) blood-brain barrier dysfunction (GLUT-1, S100B, NSE, GFAP), hypoxia (HIF-1a, COX-2) redox (8OHdG, F2-IsoPs), hormone (ACTH, CRF), growth factor (IGF-1), microRNAs (miR-129, miR-132, miR-134, miR-146a, miR-301a-3p, miR-4521), membrane channels (AQP4), receptor (P2X7R) and NDMA-R antagonists. Despite great advances have been done in the understanding of epileptogenesis and ictiogenesis, no consensus has been achieved in the development of reliable molecular biomarker [ 42- 46].
3. DRUG DEVELOPMENT AT SINGLE-CELL RESOLUTION
Single-cell RNA sequencing (scRNA-Seq) allows the analysis of expression levels in all genes in a large number of individual cells. Briefly, the major steps to perform scRNA-Seq starts with isolation and lysis of single cells or single nuclei. Then, the cDNA is synthesized, amplified, and finally, the preparation of the sequencing library occurs [47]. While traditional transcriptional profiling and proteomics methods are based on estimating the behaviors in ensembles of millions of cells by the signal average, generating bias regarding losses and gains of gene expression, scRNA-Seq is more precise. This technology allows the identification of the cell-to-cell transcriptional variation from the same cell population enabling the analysis of physiology, behavior, phenotype, uncovering new genes and molecular pathways and mechanisms of drug resistance/toxicity at the individual cellular level [48-50].
The closer look at the cellular level of gene expression increases the possibility to select new drug targets with high specificity, empowering the design of drugs targeting specific cell types, thus minimizing undesirable off-target effects [49]. Furthermore, it will be possible to distinguish between molecular markers, which currently correspond to more than one pathology in bulk RNA-Seq studies but may present differences in a single cell expression profiling level never seen before. Also, taking into account the cellular heterogeneity present in tissues, another advantage of scRNA-Seq is the distinction between response mechanisms and drug resistance/toxicity. Such analysis might be affected by several sources of variance such as pre-existent rare resistant cells, emerged resistant cells as an adaptive response to drugs, dynamics of cell status, circadian rhythm, response to stimulus environmental and stress and that could, in turn, be obscured in the analysis of bulk RNA-Seq due to the joint effect of these cells for the maintenance of tissue homeostasis [49, 51].
Regarding the mechanisms of drug resistance and drug discovery, the scRNA-Seq conjoined with parallel CRISPR-based systems, also known as Perturb-seq, CRISP-Seq and CROP-seq, can leverage the field of drug development [52, 53]. This approach provides the screening for genes involved in drug resistance or specific cellular target, combining the resolution of massively parallel scRNA-Seq with the genome editing scale of pooled CRISPR screens resulting in functional information about the influence of specific genetic perturbation on the phenotype measured [52, 54, 55]. To date, the general idea of this technology is to generate barcoded/modified (selectable or fluorescent) RNA Pol II-driven sgRNAs inserted into vectors, allowing the identification of the sequences by amplicon deep sequencing to infer biological interpretation from the genotype quantitative data [56].
Pooled CRISPR-screenings are typically assays based on cellular viability that is designed to a positive or negative selection of essential sgRNAs. The characterization of drug resistance-related molecules can be achieved in the analysis of the variable abundance of genotypes of the positively selected CRISPR-Cas-9 library from differently treated samples. On the other hand, the CRISPR-Cas-9 negative selection library allows the observation of cell death or slow-growing and even if the combination of genes can predict loss of cell viability by synthetic lethality interactions, shedding light on promising candidates or the combination of them for targeted drugs [55]. ScRNA-Seq based on CRISPR-induced perturbations is a cornerstone to obtain high complex phenotype readouts making it possible to expand our knowledge about gene functions and interactions, cell positive or negative feedback mechanisms, transcriptional effects influenced by cellular dynamics and its surrounding environment and finally to uncover complicated survival signals and the drug-resistant mechanisms.
However, the brain is characterized by remarkable cellular diversity and intricate interplays, is an extremely complex organ with high heterogeneity and in epilepsy condition, the use of scRNA-Seq can improve our understanding of pathophysiology at unprecedented resolution and consequently lead to better treatment. For example, a single-cell DNA sequencing study using human brain tissue from the epileptogenic cortical neurodevelopmental malformation patients - focal cortical dysplasia and hemimegalencephaly, found that mutations in excitatory neurons lineage in some cases might be enough to promote these phenotypes, suggesting that abnormal activation of the mTOR pathway in neurons is the main key factor for the development of focal epileptogenic malformations of cortical development. In mouse model analysis, it was observed that the mTOR pathway was not activated by interneuron lineage but in the dorsal telencephalic progenitors. This depth of coverage in next-generation sequencing experiments was responsible for the detection of pathogenic somatic mutations as few as 2% of brain cells; these mutations might be potential targets for mTOR inhibitors drugs in patients with focal cortical dysplasia and hemimegalencephaly [57].
Moreover, it is known that inhibitory interneurons have a fundamental role in epileptic processes and that there are numerous classes of interneurons with characterization, location, and/or function still unknown [58, 59]. As an example, the GABAergic neurons can express neuropeptide Y (NPY) and corticotropin-releasing factor, which consequently will inhibit the epileptiform activity or stimulate seizures, respectively [60]. The study of Harris and collaborators, in addition to the discovery of novel cell classes of inhibitory interneuron from CA1 area of the mouse hippocampus by scRNA-Seq, allowed the observation that cells with targets in proximal or distal dendrites are related to fast and low firing, respectively, and they are driven by a common genetic continuum of gene expression that exists between and within classes. In this vein, this method may be used in further studies to understand how changes in these cell populations and the circuit affect its function in epilepsy. In the sense that instead of examining what is the effect activation of inhibitory interneurons has on seizures, it might start determining the role of a lot more specific and homogenous neuronal populations [58].
Although a lot of AEDs are available in the market, 30-40% of epilepsy patients continue to have seizures. The AEDs appear to work well in the widely used animal models, however, it faces low efficacy in humans. This divergence in efficacy may be related to the brain differences between species (Fig. 1B). The characterization of cell types through scRNA-Seq is useful to show similarities and differences between the human brain and that of other animals, which are often used as models to mimic human epilepsy. Dissimilarities of human and mouse brain at cellular and circuit levels can provide clues related to the current frustrating failure in translating propitious rodent results to deliver efficacious drug treatment against human brain disorder, such as epilepsy [61].
ScRNA-Seq datasets from the human cortex in comparison to similar mouse cortex have been showing an outstanding well-conserved cellular configuration enabling for homologous types the prediction of features of human cell types. However, it was also observed large differences in cell-type homology between human and mouse, such as different morphology, proportions, laminar distributions, and gene expression [62]. For example, interlaminar astrocytes and the inhibitory rosehip neurons present in superficial cortical layers in the human brain have specific features not seen in mice [62]. These observations are essential to establish whether, despite some conserved cellular architecture among humans and mice, there are specialized cell types and system features that cannot be modeled in rodents, in a way to assist in choosing a better model for studying diseases [62]. Another study, combining transcriptomics, morphology, and physiology identified GABAergic interneuron subtypes and described a novel group of human cortical interneurons never observed in the rodent cortex. These cells play a role in local control of distal dendritic computation in cortical pyramidal neurons [61].
The progress achieved in sequencing technologies is enabling the identification of new mutations, functional disruption in molecular pathways, and their role in brain physiology, shedding light on the discovery of better targets for drug delivery in epilepsy. Simultaneously, outstanding advances in characterizing the 3D structures of proteins both experimentally and computationally has been achieved recently. In this way, the integration of genetic variation and protein structure data will build an infrastructure for both large-scale systems that can be extremely helpful to succeed in designing effective drugs for epilepsy.
4. SOLVING THE STRUCTURE OF THE PROBLEM
The three-dimensional structures of biological molecules allow scientists to understand cell biology at the atomic level and answer many important biological questions. X-ray crystallography is a powerful technology to determine the positions of each atom and their interactions in a molecule. The biomolecule of interest is purified, crystallized, and exposed to a beam of x-rays. The diffracted data is used to obtain a 3D image of the atoms within the crystal and their chemical bond interactions. It is a technique widely used for understanding protein structures, and consequently, their biological function [63]. As technological methods have improved, it has become almost usual to deduce crystal structures for macromolecules at subatomic levels. This is demonstrated by the rise in high-resolution structures deposited to the protein data bank (PDB) [64]. X-ray crystallography is the main source of experimental structural biology data. However, there are more structural biology techniques available that can offer complementary information and overcome the main limitation of x-ray crystallography, acquisition of high-quality protein crystals.
Cryogenic Electron Microscopy (Cryo-EM) is a technique used to unravel the 3D structures of macromolecules that work by pointing a beam of electrons at proteins that have been frozen in solution [65]. After that, the lens system converts the scattered signal into a magnified image recorded on the detector, and signal processing is performed to obtain the 3D structure of the sample [66]. Some of the advantages are the need for a small amount of sample and resolution at the atomic level [67].
Specific 3D protein structures and their interactions allow deep comprehension of its role in the body; in this way, it is possible to understand how pathogenic mutations affect protein functions by compromising these structures and interactions. These mutations often change protein structures and affect their binding sites to interact with molecules [68, 69]. Consequently, protein structures are very informative in describing the results of pathogenic mutations and understanding the underlying mechanisms of genetic epilepsies. It is known that epilepsy-related mutations usually occur in genes of ion channels regulating neuronal excitability, neurotransmitter receptors, and some genes beyond the channelopathy hypothesis, such as genes that regulate synaptic vesicle trafficking, mTOR signaling, chromatin remodeling and transcriptional regulation [70-72]. However, it is still unknown how these mutations affect protein activity and neuronal function in the body, thus in this sense, X-ray crystallography and Cryo-EM offer new insights into disease-causing mechanisms and discovery of potential new drugs targets (Fig. 2) [73].
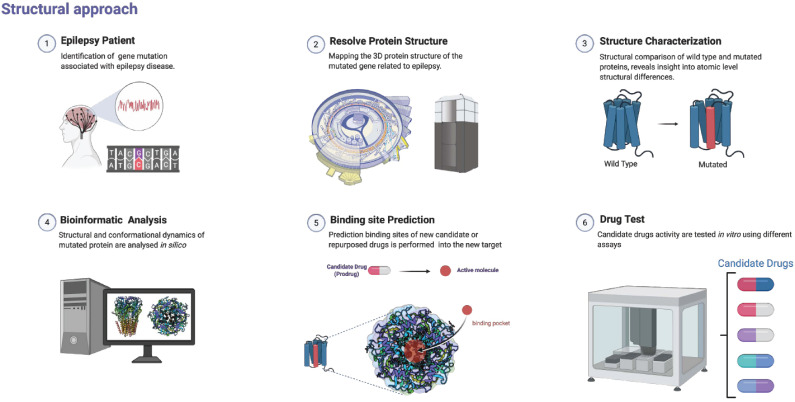
Fig. (2).
Protein structure determination to infer novel drug targets. Genetic epilepsy generates proteins encoded by mutated genes that lead to structural proteins alterations. Analytical tools can be applied to understand differences in protein structures and lead to an improvement in the prediction of binding sites that potentially deliver novel and repurposed drugs. Protein crystal structure was obtained from [73]. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
Much of the work in genetic epilepsies has been done in the channelopathies, for example, mutations in genes encoding voltage-gated sodium (Nav) channels subunits such as SNC1A/SNC2A are related to a wide range of epilepsy syndromes, including DS and GEFS+ [74, 75]. X-ray crystallography has revealed the crystal structure of bacterial sodium channels that provided the first insights on the role of those channels in the electrical signaling of neurons [76, 77]. They have shown that the wild type sodium channels structure differs substantially from mutant channels, the substitution of the Leu 170 equivalent or the Ile 202 equivalent in NaV1.4 radically changes its slow inactivation [77]. A percentage of the Na channels keep activated, causing late sodium currents. Thus, the threshold for depolarization is decreased and more easily reached by a following depolarizing stimulus is more likely to activate sodium channels [78].
Despite the meaningful improvement in the understanding of Nav channel functions and their relevance to diseases using prokaryote models, a structural description of mammalian Nav channels has been challenging. Using the Cryo-EM, the structure of the rabbit voltage-gated calcium (Cav) channel Cav1.1 has been revealed, which was used as a model for studying the structure and function of the mammalian Nav channels [79, 80]. More recently, Pan and collaborators determined the structure of the human Nav1.4-b1 complex, also using Cryo-EM technology. Their structural analysis of functional residues and disease mutations provides strong support for an allosteric blocking mechanism for fast inactivation [81]. The rapid recovery from fast inactivation occurs because more channels are available to be activated upon subsequent depolarization during action potentials, thus contributing to neuronal hyper-excitability [78].
Mutations in other genes encoding voltage ion channels, like potassium (Kv) channels subunits, have also been implicated in epilepsy since mutations in genes such as KCNQ2 and KCNQ3 are related to the benign familial neonatal epilepsy and epileptic encephalopathy (EE) [82, 83]. It is the largest group of ion channels controlling the electrical activity of different cells types in the body, including neurons, Kv channels play an important role in neuronal excitability, controlling the frequency of action potentials, setting the inward-negative resting membrane potential, facilitating membrane repolarization and hyperpolarization. Besides, they also are modulators of neurotransmitter release, regulators of Ca2+ homeostasis, cell proliferation and apoptosis [84, 85]. These channels have been the subject of many diverse studies combining structural biology, genetics, and electrophysiology to understand mechanisms of ion permeation, selectivity, voltage-gating, and more. X-ray crystallography provided the crystal structure of bacterial and mammalian voltage-dependent Kv channels that offered a representation of how membrane voltage influences the open probability of the channel, usually in the body [86-89].
However, mutations lead to diverse structural consequences on the Kv channel affecting membrane hyperexcitability in a way to increase the susceptibility to seizures [90]. For example, a mutation in KCND3 ends in a duplication motif in the S4 segment of the Kv channel and induce voltage domain more positively charged, which leads to a strong depolarizing shift in the voltage dependence in the channel activation and inactivation, causing a severe channel dysfunction in a patient with complex early-onset cerebellar ataxia, intellectual disability, oral apraxia and epilepsy [91]. The crescent number of available Kv channel crystal structures is helping with the follow-up mutagenesis studies to explore the open channel mechanisms and understand drug-channel interaction. Recently, Zhang and collaborators, using computational in-silico algorithms based on structural models to predict the pathogenicity of mutation hotspots in the KCNQ2 gene, have shown defects in voltage and phosphatidylinositol 4,5-bisphosphate (PIP2) dependent activation and axonal expression of Kv7.2 channels. These defects lead to the current decrease and inhibition of neuronal excitability resulting in hyperexcitability and spontaneous seizures of EE pathology [82]. Another study, using Cryo-EM-derived structures of the KNa1.1 channel and computational methods, identified six compounds capable of KNa1.1 channel inhibition better than quinidine, currently used to epileptic encephalopathies of infancy associated with KCNT1 mutations, which demonstrates the potential of Cryo-EM in ion channel drug discovery [92].
Genetic epilepsies can also be caused by mutations in proteins such as TBC1D24 (TBC1 Domain Family Member 24) that are usually related to deafness, onychodystrophy, osteodystrophy, intellectual disability and seizures syndrome. The crystal structure of the N-terminal domain of the TBC1D24 ortholog skywalker of Drosophila melanogaster was used to understand how pathogenic mutations affect the protein function [93]. The structure uncovers that the mutations affect a specific bind site to PIP2, reducing binding and causing defects in presynaptic-vesicle trafficking, which leads to dysfunctional presynaptic protein sorting and disease.
Besides ions channels, epilepsy-linked mutations may also affect neurotransmitter receptors genes such as CHRNA4, encoding a subunit of the nicotinic acetylcholine receptor (nAChRs) that cause autosomal dominant nocturnal frontal lobe epilepsy [94]. X-ray crystallographic and Cryo-EM analyses of nAChRs proteins facilitated the knowledge of how structures impact biological function. Such studies led to the detailed characterization of the structure of the Torpedo marmarota nAChR by Cryo-EM [95]. Also, another study characterized the extracellular domain of the human neuronal α2 nAChR subunit ligated to the agonist epibatidine, which may play as a model for a structure-based drug design approach [96]. Moreover, mutations in several genes encoding for subunits of the γ-aminobutyric acid-A (GABAA) receptor such as GABRA1, GABRB3, and GABRG2 are related to EE [97]. These mutations are associated with a variety of phenotypes; however, they all cause GABA receptor dysfunction through (e.g., reduction of Cl− current), which leads to hyperexcitability in EE [98]. Cryo-EM technology provides a GABAA receptor structure capable of elucidating the underlying principles of receptor assembly and agonist binding, such as benzodiazepine [99]. Benzodiazepines like diazepam increase the action of GABAA receptors at a specific binding site [100]. Some mutations markedly impair the modulation by diazepam, thus understanding the exact location of the binding site to have a better notion of the impact of the mutations on the binding affinity [101].
5. PRECLINICAL MODELS
For epilepsy, the animal models were the main tool to explore pathophysiology and to identify the majority of agents used for the treatment of epilepsy [102, 103]. These models have tremendous value about the current knowledge of the disease. They offered a robust preclinical system to recapitulate epilepsy phenotypes and understand the in vivo pathophysiological outcome of specific disease-associated variants.
More than 20 AEDs have been brought to the clinic on the basis of efficacy in rodent acute and chronic seizure models [104]. Preclinical screening for new AED was first introduced by Putnam and Merritt in 1937, when they tested a number of compounds from Parke Davis against maximal electroshock seizures in cats, leading to the discovery of phenytoin [105]. However, in 1944 Everett and Richards found that phenytoin was ineffective in a pentylenetetrazol mice model, showing that different experimental models of epilepsy may be differentially affected by the same treatment [106].
The use of transgenic animals, especially rodent models, has a great meaning to unravel the specific molecular mechanisms and to identify which genes are implicated with seizure phenotypes. These mutants present disruption of a specific gene (usually a ‘knockout’), which results in the loss of protein function leading to mice that display seizures or non-convulsive phenotypes [107]. Gene alterations can be generated by random transgene integration or by gene targeting, using the CRISPR-Cas9 gene-editing tool, for instance, which makes possible generating either null or point mutation strains. Many transgenic mice examples present loss and gain-of-function mutations in voltage-gated channel subunits genes such as SCN2a, KCNQ2, in transcription factors such as Dlx1 and even in genes related to mTOR pathway such as DEPDC5, which was generated using CRISPR-Cas9 technology [108-114]. However, animal models can be impractical when it comes to high-throughput screening approaches, besides having a genomic background different from humans, which might be a problem to analyze processes thought to be uniquely regulated in humans.
The new technology of induced pluripotent stem cells (iPSCs) used to generate human-derived neuronal cultures and cerebral organoids associate to CRISPR-Cas9 gene editing is a very promising models to investigate the pathophysiology of a diverse number of neurological diseases, such as epilepsy, mostly because it has facilitated the understanding of molecular mechanisms which contribute to unravel gaps in knowledge left by conventional models (Fig. 3) [115-119]. Besides, it represents a new system for preclinical drug screening, and have potential in cell therapy and personalized medicine.
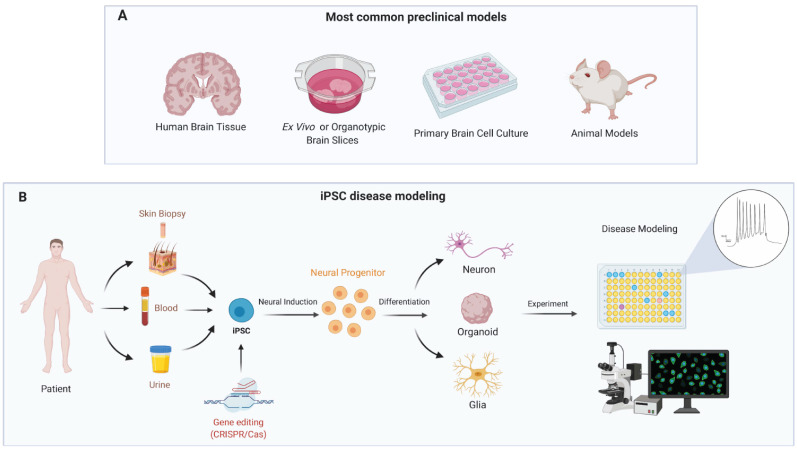
Fig. (3).
Preclinical models for genetic epilepsy. A: General overview of the preclinical models used in epilepsy. B: iPSC-derived brain cells conjoined with gene editing and functional analyses provide clues on molecular pathways of the pathophysiology of epilepsy and can help to identify drug resistance, toxicity, and novel targets compounds. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
The value of these technologies in epilepsy was shown in 2013 when researchers generated neurons from iPSC derived from patients with DS [120, 121]. DS, the mutation in patient-derived neurons revealed a rise of sodium currents compared with cells from healthy patients and hyperexcitability in excitatory and inhibitory forebrain neurons, an opposite result from findings using mouse models [122]. Other publications have shown a decrease in sodium currents in iPCS-derived inhibitory neurons with SCN1A mutations in agreement with the mouse model [123, 124]. The mechanism responsible for a dysfunctional SCN1A gene in DS has not yet been determined, and these results illustrate that the findings from animal models should be carefully interpreted as well as demonstrate the importance of using iPSCs models with a human genetic background to the study of genetic epilepsy. Mutations in SNC1A were also explored combining CRISPR-Cas9 gene-editing technology with iPSC models, using this method is possible to isolate the physiological impacts caused by the mutation by itself [125]. They have endorsed that the mutations cause inactivation of Nav channels, which lead to enhance the thresholds of the action potential in the GABAergic neurons triggering less inhibitory postsynaptic currents. Thus, these models answer in many aspects the molecular basis underlying epileptogenesis in DS. Moreover, scientists used iPSC-derived inhibitory neurons to test cannabidiol drug efficacy on the treatment of DS, demonstrating that iPSC is a good model also for drug discovery [126].
Another epilepsy disease called malignant migrating partial seizures of infancy, which is an early-onset epileptic encephalopathy associated with mutations in the KCNT1 gene that encode a Na+-dependent K+ channel was modeled by Quraishi et al. through the generation of an iPSC line with P924L gain of function variant by gene-editing. They differentiated the iPSC into forebrain neurons which have disclosed an increase in the activity of the Na+-dependent K+ channel along with what was observed in animal models [127, 128]. Mutations in the PCDH19 gene also lead to a childhood EE associated with a range of neurological marks such as epilepsy, autism and intellectual disability [129]. This gene encodes a cell adhesion molecule greatly expressed throughout development and its loss of function causes early neurogenesis of neural stem cells, as shown by researchers who generated iPSC-derived neurons from PDCH19 mutant patients [130].
iPSC can generate cerebral organoids, which represent the most promising way to model genetic epilepsy. The cerebral organoids are three-dimensional (3D) cell cultures that mimic the main features of the microanatomy, architecture and cell-type composition of a part or parts of the brain itself [131]. Mostly, 3D cell cultures mimic in vivo conditions more precisely than 2D. Besides, they can model various aspects of human neurogenesis, neuronal migration, and even early neuronal maturation and connectivity, processes that cannot be perceived in 2D human neurons culture. 3D cell culture presents the potential to study such aspects of the genetic epileptic diseases. In addition, some features of genetic epilepsy disease signs such as the effects of irregular connectivity formation, interneuron migration and spatiotemporal neuronal organization might be more efficiently modeled in a 3D culture system [132].
Brain organoids faithfully recapitulate the disease from progression and development, and this methodology will help to optimize therapy strategies and drug development for each patient. Besides, the cerebral organoids express a great potential to increase our understanding of molecular mechanisms and genetics of epilepsy networks. However, it was not yet possible to observe a seizure in an organoid, which represents a disadvantage in this model. That’s why it might be interesting to pair conventional models with new technologies; some groups have already used combined mouse-human iPSC models by injecting patient-derived neurons into the neonatal mouse brain and into the adult mouse brain [133, 134]. This approach could also be used to expand our knowledge about genetic epilepsy. However, probably the best strategy for drug screening on epilepsy or any other neurological disease will be consequent of a combination of models that offers safeguards against the oversimplification related to a single model.
6. PHARMACOKINETICS & PHARMACOGENOMICS
Inherited genetic variants can significantly influence drug activity and/or metabolism. The individual genetic profile influences key pharmacokinetic aspects, such as drug absorption, distribution, metabolism and excretion. Therefore, the development of new molecules for epilepsy treatment must consider the role of single nucleotide polymorphisms and even frequent variants within the target population. Traditional drug development pipelines do not consider minority genetic profiles due to statistical and cost-effectiveness limitations in clinical testing setups [135]. Usually, rare genetic-derived toxicity and/or performance failure are assessed in the pharmacovigilance phase when approved drugs are tested in the consumer population. The challenge for personalized medicine implementation in the drug development pipeline can be mitigated with currently available methodology and technology.
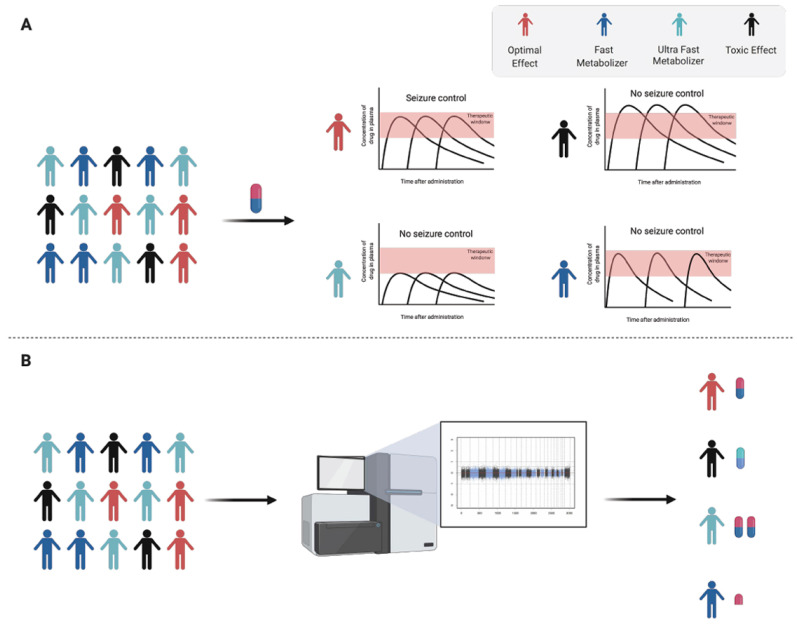
Patients' individual pharmacokinetics profiles can be subdivided according to their drug-specific metabolic ability. Slow metabolizers showed increased and prolonged drug plasma concentration and slow clearance rate, which, if not adjusted, drug concentration can reach toxic levels (Fig. 4A). Fast metabolizers, on the other hand, show reduced plasma concentration and rapid clearance, which can make the therapy ineffective at all or even create gaps in the therapeutic window, increasing the risk of seizures (Fig. 4A). Extreme metabolizers (slow and fast) are a rare subset of individuals. Identification of these individuals is highly recommended due to the high risk of severe toxicity, complete ineffective therapy and improving their therapy outcome (Fig. 4B). In this section, we will discuss how genetic variants can influence epilepsy drug metabolism. For the purpose of this topic, we will briefly point out the pharmacogenetics of classic AED, such as phenytoin, phenobarbital carbamazepine, valproic acid and lamotrigine as an example of how single nucleotide polymorphisms can influence the therapeutic outcome. However, the recent review by Bozina et al., has covered many genetic variants in most used antiseizure treatment [136].
Fig. (4).
Individual pharmacokinetic profile of patients. A: Different types of drug metabolizers and its consequences in drug response. B: Investigating how gene variants can affect the pharmacokinetic profiles is a crucial step in drug development to improve the treatment outcome in personalized medicine. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
Phenytoin is a widely used drug for epilepsy treatment. Genetic polymorphisms of phase I biotransformation enzymes, CYP450 enzymes, specially CYP2C9, CYP2C19 and CYP3A4/3A5, alongside with Phase II genetic polymorphisms of UDP-UGTs can significantly influence clearance rate. Phenytoin is metabolized majorly by CYP2C9, but also by CYP2C19 [137]. Loss of function variants in such critical enzymes results in reduced phenytoin clearance, which leads to potentially neurotoxicity increased plasma concentrations. CYP2C9 gene is highly polymorphic. More than 50 variant alleles have been identified in CYP2C9. Phenobarbital metabolism is influenced mostly through variants in CYP2C19. To this day, more than 25 variant alleles are known in the CYP2C19 gene, of which the most studied are alleles associated with reduced activity, although some show enhanced activity. Phenobarbital clearance can be reduced up to 50%, depending on the genotype [138]. Carbamazepine has a complex multi-stage metabolism, with the participation of several enzymes (e.g., CYP3A4, CYP3A5, CYP2C8, EPHX1 and UGT2B7) [139]. Two variants in the EPHX1 gene are known to influence carbamazepine metabolism (c.337T>C and c.416A>G) [140]. Valproic acid metabolism is influenced by UGT2B7 variants, like 211G>T and 161C>T polymorphisms [141]. Hepatotoxicity of valproic acid is associated with liver biotransformation by CYP2C9 and the generation of toxic metabolites [142]. Lamotrigine is eliminated through glucuronidation in the liver, which is majorly performed by UGT1A4. UGT1A4*3 variants may affect the bioavailability and efficacy of lamotrigine [143].
In the context of the development of new drugs, not only pharmacokinetics enzyme variants must be considered, but also variants that affect the pharmacodynamics as well (i.e., variants in receptors and transporters). Ultimately, such critical and complex structural polymorphisms and biotransformation processes can be modeled using rational drug design strategies associated with recently improved machine learning, artificial intelligence and population-based genomic databases curated and focused on drug metabolism [144-149]. Such data integration can significantly improve the success rate in drug development pipelines. Finally, it is worth noting that new therapeutic molecules can be classified as new molecular entities or structural derivatives from previously approved classes of molecules; the last one can be easier to predict the metabolic route. Although there are available tools for prediction, empirical evaluation is still ultimately necessary.
7. HIGH-THROUGHPUT SCREENING
In the 70s, the National Institute of Neurological Disorders and Stroke sponsored the Anticonvulsant Screening Program (now known as Epilepsy Therapy Screening Program), and it is estimated that this drug development program has screened over 32.000 compounds. Despite the remarkable effort done by the Anticonvulsant Screening Program and pharmaceutical industry during the past decades to release new AED, there is a sense that the old paradigm of drug discovery has not improved the therapeutic efficacy [150]. Over the last two decades, significant technological progress has been achieved in high throughput medicinal chemistry, cell-based assays and High-Throughput Screening (HTS). These advances have opened a new era in drug discovery.
HTS is referred as a massive parallel screening and it was first introduced in the early 1990s, it is a technology where in the best case, it can provide an efficient way to obtain useful data of large number of test samples along with high sensitives miniaturized assays to provide information on test compounds selectivity, specificity and pharmacokinetic characteristics, resulting in the hit identification and lead discovery [151].
The perception of HTS technology has changed dramatically since its concept and is now recognized as a multidisciplinary science, encompassing biological science, engineering, chemistry, biophysics, genomics and information technology. The main goal of HTS assays is the fast and reliable identification of compounds from chemical libraries and its format assays typically fall into two categories, biochemistry and cell-based [152]. Biochemical assays involve the use of cell-free in vitro systems to model the biochemistry of a subset of cellular processes, such as enzyme/substrate reactions, receptor binding, or protein-protein interactions, to more complex models. On the order hand, cell-based assays mimic more closely the in vivo system and can be adapted for targets that are unsuitable for screening in biochemical assays, such as those involving signal transduction pathways, membrane transport, cell division or cytotoxicity [153].
Both assays are fundamental for drug development; the text that follows will focus on the cell-based assay since it has technical advantages that seem to be extremely significant for the development of drugs for genetic epilepsies. The main advantages of cell-based are the capability to measure parameters such as change in membrane potential for ion channels, be able to distinguish between receptor antagonists, agonists, inverse agonists and allosteric modulators, which play an important role in epilepsy treatment since ion channels provide the basis for the regulation of excitability on epilepsy [151].
The most reliable cell assay model to evaluate the efficacy of candidate drugs in epilepsy must be one that mimics the cellular complexity of the nervous system. The evaluation of candidate drugs in brain cells or whole organisms is imperative to obtain more accurate results with increased biological and clinical relevance. The measure of multiple parameters in parallel, such as phenotype, neurotoxic, kinetics and electrophysiologic, will increase considerably the number of significant successes of candidate drugs to treat epilepsy. Over the last years, some groups have made a great effort to improve this technology to impact the discovery of a better treatment for neurological diseases.
Sridharan et al. developed a process resulting in a robust freezer-ready, highly scalable high content screening assay to identify compounds that cause neurotoxicity or regulate neurite outgrowth, which promises a path forward for investigators interested in identifying targets for neuron disease models [154]. The pilot protocol produced acceptable assay statistics and demonstrated that neuronal cells can be produced and screened in high throughput settings. Moreover, Durens and collaborators described a novel method for multiplexing high content screening, Multi-electrode Array (MEA) recording and Ca2+ imaging to examine neuronal activity and cellular composition. The combination of these assays permitted a more efficient interrogation of various properties of samples at both the network and single-cell levels. They developed a staged screening program in organotypic hippocampal culture in vitro models of epilepsy [155]. This model was used in the first stage along with biomarkers of seizures, cell death and electrographic screening. Using this methodology, the authors were able to screen 140 compounds and combinations of compounds, in over 400 separate drug concentration experiments, and identified over 40 hits.
While cell/tissue-based in vitro screening assays can efficiently identify compounds that bind specific targets, this new type of screening is more likely to reliably predict therapeutic outcomes as they maintain the complex neural circuitry involved in the underlying disease process. Dinday and Baraban, developed an alternative whole-organism-based in vivo drug-screening strategy [156]. Using SCN1A mutant zebrafish larvae with a gene homologous to humans, an alternative whole-organism-based model for in vivo drug screening. The authors screened in a two-stage phenotype-based blinded manner, a repurposed library of more than 1000 compounds for drugs that inhibit unprovoked seizure events. During the first stage, they tracked the behavior of the larvae to monitor seizures in freely swimming larval zebrafish. During the second stage, extracellular recording electrodes were used to monitor electroencephalographic activity in agar-immobilized larval zebrafish. Using this two-stage methodology, they were able to identify one compound with potential clinic interest. Ibhazehiebo et al., used a similar screening approach and after conducting a screen of 870 compounds, they identified vorinostat as a potent AED and further showed that selective HDAC1 and HDAC3 inhibition might represent efficacious targets for future drug discovery [157].
Liu and collaborators developed a microfluidic device incorporated with MEA into a miniaturized platform for AED drug discovery, which they named as epilepsy-on-a-chip [158]. They performed a two-stage screening platform, in stage I, lactate and lactate dehydrogenase biomarker were performed to identify potential drug candidates. In stage II, candidate compounds were retested with μflow-MEA to provide electrophysiological confirmation of biomarker results. The authors identified 12 receptors tyrosine kinases inhibitors, and EGFR/ErbB-2 inhibitor and colony-stimulating factor-1 receptor (cFMS) as novel AED compounds, showing the promise and higher-throughput of this new approach.
The use of electrophysiology assays in the drug discovery program on epilepsy is decisive since it is the most reliable data to monitor the effectivity of compounds in epilepsy models. Thus, higher-throughput methodologies that use cell-based assays with membrane potential must become integral components of drug discovery programs in epilepsy. Several groups have developed and introduced in the past years automated platforms for performing electrophysiological studies in mammalian cells (for more information, see [159]). These include both single-channel and multichannel systems for conventional and perforated patch-clamp electrophysiology to allow compatibility with a plate or chip-based formats [159].
The rise of technologies that could be used in HTS that better mimic the in vivo microenvironment could considerably increase the success of drug discovery on epilepsy. The design of organs-on-chip, in particular when used with iPSC technology, has the potential to improve the in vitro drug screening accuracy and in turn, reduce the drug failing rate through clinical trials. The implementation of multiplex screening technologies promises to dramatically impact the delivery of new targeted therapeutics in genetic epilepsy. In addition, the recent advances in computational biology integrated with largely available genomic databases, more accurate cellular and tissue modeling, additional data of cytotoxic assays like the toxicity tests in human liver organoids holds the potential for integration of precision and personalized medicine into drug design pipeline and preclinical evaluation. Turning the new generation of smart-drugs safer and efficient to epilepsy patients [160, 161].
CONCLUSION
The knowledge of gene variants and their influence in the context of molecular changes that trigger genetic epilepsy using non-integrated technologies turns into inaccurate drug therapies. The multifaceted feature of this disease and its subcellular heterogeneity play a crucial role in drug effectiveness, resistance, and toxicity. The employment of organoids as a preclinical model conjoined with analytical tools discussed in this review may help to map the cell subpopulation and subcellular differences in health and disease conditions, providing unprecedented data of new pathways and better understanding those already described. Also, it may support the uncovering of new molecular targets for therapy and even giving clues of molecules related to drug resistance and toxicity. Moreover, these technologies, in addition to reducing costs and time of data acquisition and analysis, may assist in drug discovery and development by mapping the disease phenotype. Their feasibility to measure molecules that can change across time opens new roads to provide an effective picture of the dynamic functional phenotype from a single cell to the entire complex tissue, the brain. Then, being more accurate and robust in order to infer treatment responsiveness.
ACKNOWLEDGEMENTS
Images created with BioRender.com.
LIST OF ABBREVIATIONS
- GEFS+
Genetic Epilepsy with Febrile Seizures
- GWAS
Genome-Wide Association Study
- NGS
Next Generation Sequencing
- RNA-Seq
RNA-sequencing
- AED
Antiepileptic Drug
- CRISPR
Clustered Regularly Interspaced Short Palindrome Repeats
- CRISPR-Cas9
CRISPR-associated endonuclease Cas9
- DS
Dravet Syndrome
- EE
Epileptic Encephalopathy
- HTS
High-Throughput Screening
- MEA
Multi-Electrode Array
CONSENT FOR PUBLICATION
Not applicable.
FUNDING
E.G is a FAPESP fellow (2019/18469-1), V.F.R. is a CNPq (PIBIC) fellow (101229/2020-8).
CONFLICT OF INTEREST
The authors have no conflicts of interest, financial or otherwise.
REFERENCES
- 1.Araujo B.H.S., Torres L.B., Guilhoto L.M.F.F. Cerebal over inhibition could be the basis for the high prevalence of epilepsy in persons with Down syndrome. Epilepsy Behav. 2015;53:120–125. doi: 10.1016/j.yebeh.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 2.Berkovic S.F., Mulley J.C., Scheffer I.E., Petrou S. Human epilepsies: interaction of genetic and acquired factors. Trends Neurosci. 2006;29(7):391–397. doi: 10.1016/j.tins.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 3.Steinlein O.K. Channelopathies can cause epilepsy in man. Eur. J. Pain. 2002;6(Suppl. A):27–34. doi: 10.1053/eujp.2001.0319. [DOI] [PubMed] [Google Scholar]
- 4.Shoffner J.M., Lott M.T., Lezza A.M.S., Seibel P., Ballinger S.W., Wallace D.C. Myoclonic epilepsy and ragged-red fiber disease (MERRF) is associated with a mitochondrial DNA tRNA(Lys) mutation. Cell. 1990;61(6):931–937. doi: 10.1016/0092-8674(90)90059-N. [DOI] [PubMed] [Google Scholar]
- 5.Steinlein O.K., Mulley J.C., Propping P., Wallace R.H., Phillips H.A., Sutherland G.R., Scheffer I.E., Berkovic S.F. A missense mutation in the neuronal nicotinic acetylcholine receptor alpha 4 subunit is associated with autosomal dominant nocturnal frontal lobe epilepsy. Nat. Genet. 1995;11(2):201–203. doi: 10.1038/ng1095-201. [DOI] [PubMed] [Google Scholar]
- 6.Yamagata A., Miyazaki Y., Yokoi N., Shigematsu H., Sato Y., Goto-Ito S., Maeda A., Goto T., Sanbo M., Hirabayashi M., Shirouzu M., Fukata Y., Fukata M., Fukai S. Structural basis of epilepsy-related ligand-receptor complex LGI1-ADAM22. Nat. Commun. 2018;9(1):1546. doi: 10.1038/s41467-018-03947-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meisler M.H., Kearney J., Ottman R., Escayg A. Identification of epilepsy genes in human and mouse. Annu. Rev. Genet. 2001;35:567–588. doi: 10.1146/annurev.genet.35.102401.091142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Escayg A., Goldin A.L. Sodium channel SCN1A and epilepsy: mutations and mechanisms. Epilepsia. 2010;51(9):1650–1658. doi: 10.1111/j.1528-1167.2010.02640.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wallace R.H., Scheffer I.E., Parasivam G., Barnett S., Wallace G.B., Sutherland G.R., Berkovic S.F., Mulley J.C. Generalized epilepsy with febrile seizures plus: mutation of the sodium channel subunit SCN1B. Neurology. 2002;58(9):1426–1429. doi: 10.1212/WNL.58.9.1426. [DOI] [PubMed] [Google Scholar]
- 10.Harkin L.A., Bowser D.N., Dibbens L.M., Singh R., Phillips F., Wallace R.H., Richards M.C., Williams D.A., Mulley J.C., Berkovic S.F., Scheffer I.E., Petrou S. Truncation of the GABA(A)-receptor gamma2 subunit in a family with generalized epilepsy with febrile seizures plus. Am. J. Hum. Genet. 2002;70(2):530–536. doi: 10.1086/338710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu Y.E., Wen L., Silva J., Li Z., Head K., Sossey-Alaoui K., Pao A., Mei L., Cowell J.K. Lgi1 null mutant mice exhibit myoclonic seizures and CA1 neuronal hyperexcitability. Hum. Mol. Genet. 2010;19(9):1702–1711. doi: 10.1093/hmg/ddq047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J., Lin Z.J., Liu L., Xu H.Q., Shi Y.W., Yi Y.H., He N., Liao W.P. Epilepsy-associated genes. Seizure. 2017;44:11–20. doi: 10.1016/j.seizure.2016.11.030. [DOI] [PubMed] [Google Scholar]
- 13.Weber Y.G., Lerche H. Genetic mechanisms in idiopathic epilepsies. Dev. Med. Child Neurol. 2008;50(9):648–654. doi: 10.1111/j.1469-8749.2008.03058.x. [DOI] [PubMed] [Google Scholar]
- 14.Noebels J. Pathway-driven discovery of epilepsy genes. Nat. Neurosci. 2015;18(3):344–350. doi: 10.1038/nn.3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fogel D.B. Factors associated with clinical trials that fail and opportunities for improving the likelihood of success: A review. Contemp. Clin. Trials Commun. 2018;11:156–164. doi: 10.1016/j.conctc.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang X., Kui L., Tang M., Li D., Wei K., Chen W., Miao J., Dong Y. High-Throughput transcriptome profiling in drug and biomarker discovery. Front. Genet. 2020;11:19. doi: 10.3389/fgene.2020.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verbist B., Klambauer G., Vervoort L., Talloen W., Shkedy Z., Thas O., Bender A., Göhlmann H.W.H., Hochreiter S. QSTAR Consortium. Using transcriptomics to guide lead optimization in drug discovery projects: Lessons learned from the QSTAR project. Drug Discov. Today. 2015;20(5):505–513. doi: 10.1016/j.drudis.2014.12.014. [DOI] [PubMed] [Google Scholar]
- 18.Dugger S.A., Platt A., Goldstein D.B. Drug development in the era of precision medicine. Nat. Rev. Drug Discov. 2018;17(3):183–196. doi: 10.1038/nrd.2017.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boyle E.A., Li Y.I., Pritchard J.K. An expanded view of complex traits: from polygenic to omnigenic. Cell. 2017;169(7):1177–1186. doi: 10.1016/j.cell.2017.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maljevic S., Reid C.A., Petrou S. Models for discovery of targeted therapy in genetic epileptic encephalopathies. J. Neurochem. 2017;143(1):30–48. doi: 10.1111/jnc.14134. [DOI] [PubMed] [Google Scholar]
- 21.Myers K.A., Johnstone D.L., Dyment D.A. Epilepsy genetics: Current knowledge, applications, and future directions. Clin. Genet. 2019;95(1):95–111. doi: 10.1111/cge.13414. [DOI] [PubMed] [Google Scholar]
- 22.Pellacani S., Dosi C., Valvo G., Moro F., Mero S., Sicca F., Santorelli F.M. Customized multigene panels in epilepsy: the best things come in small packages. Neurogenetics. 2020;21(1):1–18. doi: 10.1007/s10048-019-00598-x. [DOI] [PubMed] [Google Scholar]
- 23.Bodian D.L., Kothiyal P., Hauser N.S. Pitfalls of clinical exome and gene panel testing: alternative transcripts. Genet. Med. 2019;21(5):1240–1245. doi: 10.1038/s41436-018-0319-7. [DOI] [PubMed] [Google Scholar]
- 24.McTague A., Howell K.B., Cross J.H., Kurian M.A., Scheffer I.E. The genetic landscape of the epileptic encephalopathies of infancy and childhood. Lancet Neurol. 2016;15(3):304–316. doi: 10.1016/S1474-4422(15)00250-1. [DOI] [PubMed] [Google Scholar]
- 25.Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Z., Gerstein M., Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong X., You Y., Wu J.Q. Building an rna sequencing transcriptome of the central nervous system. Neuroscientist. 2016;22(6):579–592. doi: 10.1177/1073858415610541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma A. Genome-wide expression analysis in epilepsy: a synthetic review. Curr. Top. Med. Chem. 2012;12(9):1008–1032. doi: 10.2174/156802612800229189. [DOI] [PubMed] [Google Scholar]
- 29.Dingledine R., Coulter D.A., Fritsch B., Gorter J.A., Lelutiu N., McNamara J., Nadler J.V., Pitkänen A., Rogawski M.A., Skene P., Sloviter R.S., Wang Y., Wadman W.J., Wasterlain C., Roopra A. Transcriptional profile of hippocampal dentate granule cells in four rat epilepsy models. Sci. Data. 2017;4:170061. doi: 10.1038/sdata.2017.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fernandez-Albert J., Lipinski M., Lopez-Cascales M.T., Rowley M.J., Martin-Gonzalez A.M., Del Blanco B., Corces V.G., Barco A. Immediate and deferred epigenomic signatures of in vivo neuronal activation in mouse hippocampus. Nat. Neurosci. 2019;22(10):1718–1730. doi: 10.1038/s41593-019-0476-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Srivastava P.K., van Eyll J., Godard P., Mazzuferi M., Delahaye-Duriez A., Van Steenwinckel J., Gressens P., Danis B., Vandenplas C., Foerch P., Leclercq K., Mairet-Coello G., Cardenas A., Vanclef F., Laaniste L., Niespodziany I., Keaney J., Gasser J., Gillet G., Shkura K., Chong S-A., Behmoaras J., Kadiu I., Petretto E., Kaminski R.M., Johnson M.R. A systems-level framework for drug discovery identifies Csf1R as an anti-epileptic drug target. Nat. Commun. 2018;9(1):3561. doi: 10.1038/s41467-018-06008-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Srivastava P.K., Bagnati M., Delahaye-Duriez A., Ko J-H., Rotival M., Langley S.R., Shkura K., Mazzuferi M., Danis B., van Eyll J., Foerch P., Behmoaras J., Kaminski R.M., Petretto E., Johnson M.R. Genome-wide analysis of differential RNA editing in epilepsy. Genome Res. 2017;27(3):440–450. doi: 10.1101/gr.210740.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ye C., Ho D.J., Neri M., Yang C., Kulkarni T., Randhawa R., Henault M., Mostacci N., Farmer P., Renner S., Ihry R., Mansur L., Keller C.G., McAllister G., Hild M., Jenkins J., Kaykas A. DRUG-seq for miniaturized high-throughput transcriptome profiling in drug discovery. Nat. Commun. 2018;9(1):4307. doi: 10.1038/s41467-018-06500-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schidlitzki A., Bascuñana P., Srivastava P.K., Welzel L., Twele F., Töllner K., Käufer C., Gericke B., Feleke R., Meier M., Polyak A., Ross T.L., Gerhauser I., Bankstahl J.P., Johnson M.R., Bankstahl M., Löscher W. Proof-of-concept that network pharmacology is effective to modify development of acquired temporal lobe epilepsy. Neurobiol. Dis. 2020;134:104664. doi: 10.1016/j.nbd.2019.104664. [DOI] [PubMed] [Google Scholar]
- 35.Brueggeman L., Sturgeon M.L., Martin R.M., Grossbach A.J., Nagahama Y., Zhang A., Howard M.A., III, Kawasaki H., Wu S., Cornell R.A., Michaelson J.J., Bassuk A.G., Michaelson J. Drug repositioning in epilepsy reveals novel antiseizure candidates. Ann. Clin. Transl. Neurol. 2018;6(2):295–309. doi: 10.1002/acn3.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baker N.C., Ekins S., Williams A.J., Tropsha A. A bibliometric review of drug repurposing. Drug Discov. Today. 2018;23(3):661–672. doi: 10.1016/j.drudis.2018.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi X., He W., Guo S., Zhang B., Ren S., Liu K., Sun T., Cui J. RNA-seq Analysis of the SCN1A-KO Model based on CRISPR/Cas9 Genome Editing Technology. Neuroscience. 2019;398:1–11. doi: 10.1016/j.neuroscience.2018.11.052. [DOI] [PubMed] [Google Scholar]
- 38.Bauer S., Schütz V., Strzelczyk A., Rosenow F. Is there a role for microRNAs in epilepsy diagnostics? Expert Rev. Mol. Diagn. 2020;20(7):693–701. doi: 10.1080/14737159.2020.1745065. [DOI] [PubMed] [Google Scholar]
- 39.Brennan G.P., Henshall D.C. microRNAs in the pathophysiology of epilepsy. Neurosci. Lett. 2018;667:47–52. doi: 10.1016/j.neulet.2017.01.017. [DOI] [PubMed] [Google Scholar]
- 40.Mooney C., Becker B.A., Raoof R., Henshall D.C. EpimiRBase: a comprehensive database of microRNA-epilepsy associations. Bioinformatics. 2016;32(9):1436–1438. doi: 10.1093/bioinformatics/btw008. [DOI] [PubMed] [Google Scholar]
- 41.Rupaimoole R., Slack F.J. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017;16(3):203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 42.Kobylarek D., Iwanowski P., Lewandowska Z., Limphaibool N., Szafranek S., Labrzycka A., Kozubski W. Advances in the potential biomarkers of epilepsy. Front. Neurol. 2019;10:685. doi: 10.3389/fneur.2019.00685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Enright N., Simonato M., Henshall D.C. Discovery and validation of blood microRNAs as molecular biomarkers of epilepsy: Ways to close current knowledge gaps. Epilepsia Open. 2018;3(4):427–436. doi: 10.1002/epi4.12275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dixit A.B., Tripathi M., Chandra P.S., Banerjee J. Molecular biomarkers in drug-resistant epilepsy: Facts & possibilities. Int. J. Surg. 2016;36(Pt B):483–491. doi: 10.1016/j.ijsu.2015.08.029. [DOI] [PubMed] [Google Scholar]
- 45.Walker L.E., Janigro D., Heinemann U., Riikonen R., Bernard C., Patel M. WONOEP appraisal: Molecular and cellular biomarkers for epilepsy. Epilepsia. 2016;57(9):1354–1362. doi: 10.1111/epi.13460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Engel J., Jr, Pitkänen A., Loeb J.A., Dudek F.E., Bertram E.H., III, Cole A.J., Moshé S.L., Wiebe S., Jensen F.E., Mody I., Nehlig A., Vezzani A. Epilepsy biomarkers. Epilepsia. 2013;54(Suppl. 4):61–69. doi: 10.1111/epi.12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hedlund E., Deng Q. Single-cell RNA sequencing: Technical advancements and biological applications. Mol. Aspects Med. 2018;59:36–46. doi: 10.1016/j.mam.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 48.Johnson M.R., Kaminski R.M. A systems-level framework for anti-epilepsy drug discovery. Neuropharmacology. 2020;170:107868. doi: 10.1016/j.neuropharm.2019.107868. [DOI] [PubMed] [Google Scholar]
- 49.Wu H., Wang C., Wu S. Single-cell sequencing for drug discovery and drug development. Curr. Top. Med. Chem. 2017;17(15):1769–1777. doi: 10.2174/1568026617666161116145358. [DOI] [PubMed] [Google Scholar]
- 50.Shapiro E., Biezuner T., Linnarsson S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat. Rev. Genet. 2013;14(9):618–630. doi: 10.1038/nrg3542. [DOI] [PubMed] [Google Scholar]
- 51.Mitra A.K., Mukherjee U.K., Harding T., Jang J.S., Stessman H., Li Y., Abyzov A., Jen J., Kumar S., Rajkumar V., Van Ness B. Single-cell analysis of targeted transcriptome predicts drug sensitivity of single cells within human myeloma tumors. Leukemia. 2016;30(5):1094–1102. doi: 10.1038/leu.2015.361. [DOI] [PubMed] [Google Scholar]
- 52.Jaitin D.A., Weiner A., Yofe I., Lara-Astiaso D., Keren-Shaul H., David E., Salame T.M., Tanay A., van Oudenaarden A., Amit I. Dissecting immune circuits by linking CRISPR-pooled screens with single-cell RNA-Seq. Cell. 2016;167(7):1883–1896.e15. doi: 10.1016/j.cell.2016.11.039. [DOI] [PubMed] [Google Scholar]
- 53.Datlinger P., Rendeiro A.F., Schmidl C., Krausgruber T., Traxler P., Klughammer J., Schuster L.C., Kuchler A., Alpar D., Bock C. Pooled CRISPR screening with single-cell transcriptome readout. Nat. Methods. 2017;14(3):297–301. doi: 10.1038/nmeth.4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Duan B., Zhou C., Zhu C., Yu Y., Li G., Zhang S., Zhang C., Ye X., Ma H., Qu S., Zhang Z., Wang P., Sun S., Liu Q. Model-based understanding of single-cell CRISPR screening. Nat. Commun. 2019;10(1):2233. doi: 10.1038/s41467-019-10216-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kurata M., Yamamoto K., Moriarity B.S., Kitagawa M., Largaespada D.A. CRISPR/Cas9 library screening for drug target discovery. J. Hum. Genet. 2018;63(2):179–186. doi: 10.1038/s10038-017-0376-9. [DOI] [PubMed] [Google Scholar]
- 56.Yao J., Dai H.L. Is pooled crispr-screening the dawn of a new era for functional genomics. Adv. Exp. Med. Biol. 2018;1068:171–176. doi: 10.1007/978-981-13-0502-3_14. [DOI] [PubMed] [Google Scholar]
- 57.D’Gama A.M., Woodworth M.B., Hossain A.A., Bizzotto S., Hatem N.E., LaCoursiere C.M., Najm I., Ying Z., Yang E., Barkovich A.J., Kwiatkowski D.J., Vinters H.V., Madsen J.R., Mathern G.W., Blümcke I., Poduri A., Walsh C.A. Somatic mutations activating the mTOR pathway in dorsal telencephalic progenitors cause a continuum of cortical dysplasias. Cell Rep. 2017;21(13):3754–3766. doi: 10.1016/j.celrep.2017.11.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harris K.D., Hochgerner H., Skene N.G., Magno L., Katona L., Bengtsson Gonzales C., Somogyi P., Kessaris N., Linnarsson S., Hjerling-Leffler J. Classes and continua of hippocampal CA1 inhibitory neurons revealed by single-cell transcriptomics. PLoS Biol. 2018;16(6):e2006387. doi: 10.1371/journal.pbio.2006387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu Y-Q., Yu F., Liu W-H., He X-H., Peng B-W. Dysfunction of hippocampal interneurons in epilepsy. Neurosci. Bull. 2014;30(6):985–998. doi: 10.1007/s12264-014-1478-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Clynen E., Swijsen A., Raijmakers M., Hoogland G., Rigo J-M. Neuropeptides as targets for the development of anticonvulsant drugs. Mol. Neurobiol. 2014;50(2):626–646. doi: 10.1007/s12035-014-8669-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boldog E., Bakken T.E., Hodge R.D., Novotny M., Aevermann B.D., Baka J., Bordé S., Close J.L., Diez-Fuertes F., Ding S.L., Faragó N., Kocsis Á.K., Kovács B., Maltzer Z., McCorrison J.M., Miller J.A., Molnár G., Oláh G., Ozsvár A., Rózsa M., Shehata S.I., Smith K.A., Sunkin S.M., Tran D.N., Venepally P., Wall A., Puskás L.G., Barzó P., Steemers F.J., Schork N.J., Scheuermann R.H., Lasken R.S., Lein E.S., Tamás G. Transcriptomic and morphophysiological evidence for a specialized human cortical GABAergic cell type. Nat. Neurosci. 2018;21(9):1185–1195. doi: 10.1038/s41593-018-0205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hodge R.D., Bakken T.E., Miller J.A., Smith K.A., Barkan E.R., Graybuck L.T., Close J.L., Long B., Johansen N., Penn O., Yao Z., Eggermont J., Höllt T., Levi B.P., Shehata S.I., Aevermann B., Beller A., Bertagnolli D., Brouner K., Casper T., Cobbs C., Dalley R., Dee N., Ding S-L., Ellenbogen R.G., Fong O., Garren E., Goldy J., Gwinn R.P., Hirschstein D., Keene C.D., Keshk M., Ko A.L., Lathia K., Mahfouz A., Maltzer Z., McGraw M., Nguyen T.N., Nyhus J., Ojemann J.G., Oldre A., Parry S., Reynolds S., Rimorin C., Shapovalova N.V., Somasundaram S., Szafer A., Thomsen E.R., Tieu M., Quon G., Scheuermann R.H., Yuste R., Sunkin S.M., Lelieveldt B., Feng D., Ng L., Bernard A., Hawrylycz M., Phillips J.W., Tasic B., Zeng H., Jones A.R., Koch C., Lein E.S. Conserved cell types with divergent features in human versus mouse cortex. Nature. 2019;573(7772):61–68. doi: 10.1038/s41586-019-1506-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Parker M.W. Protein structure from x-ray diffraction. J. Biol. Phys. 2003;29(4):341–362. doi: 10.1023/A:1027310719146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The protein data bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shoemaker S.C., Ando N. X-rays in the cryo-electron microscopy era: structural biology’s dynamic future. Biochemistry. 2018;57(3):277–285. doi: 10.1021/acs.biochem.7b01031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lyumkis D. Challenges and opportunities in cryo-EM single-particle analysis. J. Biol. Chem. 2019;294(13):5181–5197. doi: 10.1074/jbc.REV118.005602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yip K.M., Fischer N., Paknia E., Chari A., Stark H. Breaking the next Cryo-EM Resolution Barrier – Atomic Resolution Determination of Proteins! bioRxiv. 2020 doi: 10.1038/s41586-020-2833-4. [DOI] [PubMed] [Google Scholar]
- 68.Jubb H.C., Pandurangan A.P., Turner M.A., Ochoa-Montaño B., Blundell T.L., Ascher D.B. Mutations at protein-protein interfaces: Small changes over big surfaces have large impacts on human health. Prog. Biophys. Mol. Biol. 2017;128:3–13. doi: 10.1016/j.pbiomolbio.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 69.Hijikata A., Tsuji T., Shionyu M., Shirai T. Decoding disease-causing mechanisms of missense mutations from supramolecular structures. Sci. Rep. 2017;7(1):8541. doi: 10.1038/s41598-017-08902-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fukata Y., Fukata M. Epilepsy and synaptic proteins. Curr. Opin. Neurobiol. 2017;45:1–8. doi: 10.1016/j.conb.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 71.Steinlein O.K. Genetic mechanisms that underlie epilepsy. Nat. Rev. Neurosci. 2004;5(5):400–408. doi: 10.1038/nrn1388. [DOI] [PubMed] [Google Scholar]
- 72.Myers C.T., Mefford H.C. Advancing epilepsy genetics in the genomic era. Genome Med. 2015;7:91. doi: 10.1186/s13073-015-0214-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim J.J., Gharpure A., Teng J., Zhuang Y., Howard R.J., Zhu S., Noviello C.M., Walsh R.M., Jr, Lindahl E., Hibbs R.E. Shared structural mechanisms of general anaesthetics and benzodiazepines. Nature. 2020;28:3490–3499. doi: 10.1038/s41586-020-2654-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kaplan D.I., Isom L.L., Petrou S. Role of sodium channels in epilepsy. Cold Spring Harb. Perspect. Med. 2016;6(6):6. doi: 10.1101/cshperspect.a022814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mulley J.C., Scheffer I.E., Petrou S., Dibbens L.M., Berkovic S.F., Harkin L.A. SCN1A mutations and epilepsy. Hum. Mutat. 2005;25(6):535–542. doi: 10.1002/humu.20178. [DOI] [PubMed] [Google Scholar]
- 76.Zhang X., Ren W., DeCaen P., Yan C., Tao X., Tang L., Wang J., Hasegawa K., Kumasaka T., He J., Wang J., Clapham D.E., Yan N. Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel. Nature. 2012;486(7401):130–134. doi: 10.1038/nature11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Payandeh J., Scheuer T., Zheng N., Catterall W.A. The crystal structure of a voltage-gated sodium channel. Nature. 2011;475(7356):353–358. doi: 10.1038/nature10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Abdelsayed M., Sokolov S. Voltage-gated sodium channels: pharmaceutical targets via anticonvulsants to treat epileptic syndromes. Channels (Austin) 2013;7(3):146–152. doi: 10.4161/chan.24380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu J., Yan Z., Li Z., Yan C., Lu S., Dong M., Yan N. Structure of the voltage-gated calcium channel Cav1.1 Complex. Science (80-. ) 2015;350:aad2395. doi: 10.1126/science.aad2395. [DOI] [PubMed] [Google Scholar]
- 80.Wu J., Yan Z., Li Z., Qian X., Lu S., Dong M., Zhou Q., Yan N. Structure of the voltage-gated calcium channel Ca(v)1.1 at 3.6 Å resolution. Nature. 2016;537(7619):191–196. doi: 10.1038/nature19321. [DOI] [PubMed] [Google Scholar]
- 81.Pan X., Li Z., Zhou Q., Shen H., Wu K., Huang X., Chen J., Zhang J., Zhu X., Lei J., Xiong W., Gong H., Xiao B., Yan N. Structure of the human voltage-gated sodium channel nav1.4 in complex with b1. Science. 2018;362(6412):eaau2486. doi: 10.1126/science.aau2486. [DOI] [PubMed] [Google Scholar]
- 82.Zhang J., Kim E.C., Chen C., Procko E., Pant S., Lam K., Patel J., Choi R., Hong M., Joshi D., Bolton E., Tajkhorshid E., Chung H.J. Identifying mutation hotspots reveals pathogenetic mechanisms of KCNQ2 epileptic encephalopathy. Sci. Rep. 2020;10(1):4756. doi: 10.1038/s41598-020-61697-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Miceli F., Striano P., Soldovieri M.V., Fontana A., Nardello R., Robbiano A., Bellini G., Elia M., Zara F., Taglialatela M., Mangano S. A novel KCNQ3 mutation in familial epilepsy with focal seizures and intellectual disability. Epilepsia. 2015;56(2):e15–e20. doi: 10.1111/epi.12887. [DOI] [PubMed] [Google Scholar]
- 84.Shah N.H., Aizenman E. Voltage-gated potassium channels at the crossroads of neuronal function, ischemic tolerance, and neurodegeneration. Transl. Stroke Res. 2014;5(1):38–58. doi: 10.1007/s12975-013-0297-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.D’Adamo M.C., Catacuzzeno L., Di Giovanni G., Franciolini F., Pessia M. K(+) channelepsy: progress in the neurobiology of potassium channels and epilepsy. Front. Cell. Neurosci. 2013;7:134. doi: 10.3389/fncel.2013.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jiang Y., Lee A., Chen J., Ruta V., Cadene M., Chait B.T., MacKinnon R. X-ray structure of a voltage-dependent K+ channel. Nature. 2003;423(6935):33–41. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]
- 87.Doyle D.A., Morais Cabral J., Pfuetzner R.A., Kuo A., Gulbis J.M., Cohen S.L., Chait B.T., MacKinnon R. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280(5360):69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 88.Long S.B., Campbell E.B., Mackinnon R. Voltage sensor of Kv1.2: structural basis of electromechanical coupling. Science. 2005;309(5736):903–908. doi: 10.1126/science.1116270. [DOI] [PubMed] [Google Scholar]
- 89.Long S.B., Campbell E.B., Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309(5736):897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 90.Villa C., Combi R. Potassium channels and human epileptic phenotypes: an updated overview. Front. Cell. Neurosci. 2016;10:81. doi: 10.3389/fncel.2016.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Smets K., Duarri A., Deconinck T., Ceulemans B., van de Warrenburg B.P., Züchner S., Gonzalez M.A., Schüle R., Synofzik M., Van der Aa N., De Jonghe P., Verbeek D.S., Baets J. First de novo KCND3 mutation causes severe Kv4.3 channel dysfunction leading to early onset cerebellar ataxia, intellectual disability, oral apraxia and epilepsy. BMC Med. Genet. 2015;16:51. doi: 10.1186/s12881-015-0200-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cole B.A., Johnson R.M., Dejakaisaya H., Pilati N., Fishwick C.W.G., Muench S.P., Lippiat J.D. Structure-based identification and characterization of inhibitors of the epilepsy-associated KNa1.1 (KCNT1) potassium channel. iScience. 2020;23:101100. doi: 10.1016/j.isci.2020.101100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fischer B., Lüthy K., Paesmans J., De Koninck C., Maes I., Swerts J., Kuenen S., Uytterhoeven V., Verstreken P., Versées W. Skywalker-TBC1D24 has a lipid-binding pocket mutated in epilepsy and required for synaptic function. Nat. Struct. Mol. Biol. 2016;23(11):965–973. doi: 10.1038/nsmb.3297. [DOI] [PubMed] [Google Scholar]
- 94.Bertrand D. Neuronal Nicotinic Acetylcholine Receptors and Epilepsy. Epilepsy Curr. 2002;2(6):191–193. doi: 10.1046/j.1535-7597.2002.00072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4A resolution. J. Mol. Biol. 2005;346(4):967–989. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 96.Kouvatsos N., Giastas P., Chroni-Tzartou D., Poulopoulou C., Tzartos S.J. Crystal structure of a human neuronal nAChR extracellular domain in pentameric assembly: Ligand-bound α2 homopentamer. Proc. Natl. Acad. Sci. USA. 2016;113(34):9635–9640. doi: 10.1073/pnas.1602619113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Absalom N.L., Ahring P.K., Liao V.W., Balle T., Jiang T., Anderson L.L., Arnold J.C., McGregor I.S., Bowen M.T., Chebib M. Functional genomics of epilepsy-associated mutations in the GABAA receptor subunits reveal that one mutation impairs function and two are catastrophic. J. Biol. Chem. 2019;294(15):6157–6171. doi: 10.1074/jbc.RA118.005697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ishii A., Kang J-Q., Schornak C.C., Hernandez C.C., Shen W., Watkins J.C., Macdonald R.L., Hirose S. A de novo missense mutation of GABRB2 causes early myoclonic encephalopathy. J. Med. Genet. 2017;54(3):202–211. doi: 10.1136/jmedgenet-2016-104083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Phulera S., Zhu H., Yu J., Claxton D.P., Yoder N., Yoshioka C., Gouaux E. Cryo-EM structure of the benzodiazepine-sensitive α1β1γ2S tri-heteromeric GABAA receptor in complex with GABA. eLife. 2018;7:7. doi: 10.7554/eLife.39383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Richter L., de Graaf C., Sieghart W., Varagic Z., Mörzinger M., de Esch I.J.P., Ecker G.F., Ernst M. Diazepam-bound GABAA receptor models identify new benzodiazepine binding-site ligands. Nat. Chem. Biol. 2012;8(5):455–464. doi: 10.1038/nchembio.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Amin J., Brooks-Kayal A., Weiss D.S. Two tyrosine residues on the alpha subunit are crucial for benzodiazepine binding and allosteric modulation of gamma-aminobutyric acidA receptors. Mol. Pharmacol. 1997;51(5):833–841. doi: 10.1124/mol.51.5.833. [DOI] [PubMed] [Google Scholar]
- 102.Araujo B., Torres L., Stein M., Cabral F.R., Herai R., Okamoto O., Cavalheiro E. Decreased expression of proteins involved in energy metabolism in the hippocampal granular layer of rats submitted to the pilocarpine epilepsy model. Neurosci. Lett. 2014;561:46–51. doi: 10.1016/j.neulet.2013.12.040. [DOI] [PubMed] [Google Scholar]
- 103.Araujo B.H.S., Torres L.B., Cossa A.C., Naffah-Mazzacoratti M. da G., Cavalheiro E.A. Hippocampal expression and distribution of CB1 receptors in the Amazonian rodent Proechimys: an animal model of resistance to epilepsy. Brain Res. 2010;1335:35–40. doi: 10.1016/j.brainres.2010.03.031. [DOI] [PubMed] [Google Scholar]
- 104.Barker-Haliski M. How do we choose the appropriate animal model for antiseizure therapy development? Expert Opin. Drug Discov. 2019;14(10):947–951. doi: 10.1080/17460441.2019.1636782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Putnam T.J., Merritt H.H. Experimental determination of the anticonvulsant properties of some phenyl derivatives. Science. 1937;85(2213):525–526. doi: 10.1126/science.85.2213.525. [DOI] [PubMed] [Google Scholar]
- 106.Everett G.M., Richards R.K. Comparative Anticonvulsive Action of 3,5,5-Trimethyloxazolidine-2,4-Dione (Tridione), dilantin and phenobarbital. J. Pharmacol. Exp. Ther. 1944;81:402–407. [Google Scholar]
- 107.Baraban S.C. Emerging epilepsy models: insights from mice, flies, worms and fish. Curr. Opin. Neurol. 2007;20(2):164–168. doi: 10.1097/WCO.0b013e328042bae0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ogiwara I., Miyamoto H., Tatsukawa T., Yamagata T., Nakayama T., Atapour N., Miura E., Mazaki E., Ernst S.J., Cao D., Ohtani H., Itohara S., Yanagawa Y., Montal M., Yuzaki M., Inoue Y., Hensch T.K., Noebels J.L., Yamakawa K. Nav1.2 haplodeficiency in excitatory neurons causes absence-like seizures in mice. Commun. Biol. 2018;1:96. doi: 10.1038/s42003-018-0099-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kearney J.A., Plummer N.W., Smith M.R., Kapur J., Cummins T.R., Waxman S.G., Goldin A.L., Meisler M.H. A gain-of-function mutation in the sodium channel gene Scn2a results in seizures and behavioral abnormalities. Neuroscience. 2001;102(2):307–317. doi: 10.1016/S0306-4522(00)00479-6. [DOI] [PubMed] [Google Scholar]
- 110.Planells-Cases R., Caprini M., Zhang J., Rockenstein E.M., Rivera R.R., Murre C., Masliah E., Montal M. Neuronal death and perinatal lethality in voltage-gated sodium channel alpha(II)-deficient mice. Biophys. J. 2000;78(6):2878–2891. doi: 10.1016/S0006-3495(00)76829-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Milh M., Roubertoux P., Biba N., Chavany J., Spiga Ghata A., Fulachier C., Collins S.C., Wagner C., Roux J.C., Yalcin B., Félix M.S., Molinari F., Lenck-Santini P.P., Villard L. A knock-in mouse model for KCNQ2-related epileptic encephalopathy displays spontaneous generalized seizures and cognitive impairment. Epilepsia. 2020;61(5):868–878. doi: 10.1111/epi.16494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Singh N.A., Otto J.F., Dahle E.J., Pappas C., Leslie J.D., Vilaythong A., Noebels J.L., White H.S., Wilcox K.S., Leppert M.F. Mouse models of human KCNQ2 and KCNQ3 mutations for benign familial neonatal convulsions show seizures and neuronal plasticity without synaptic reorganization. J. Physiol. 2008;586(14):3405–3423. doi: 10.1113/jphysiol.2008.154971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cobos I., Calcagnotto M.E., Vilaythong A.J., Thwin M.T., Noebels J.L., Baraban S.C., Rubenstein J.L.R. Mice lacking Dlx1 show subtype-specific loss of interneurons, reduced inhibition and epilepsy. Nat. Neurosci. 2005;8(8):1059–1068. doi: 10.1038/nn1499. [DOI] [PubMed] [Google Scholar]
- 114.Hughes J., Dawson R., Tea M., McAninch D., Piltz S., Jackson D., Stewart L., Ricos M.G., Dibbens L.M., Harvey N.L., Thomas P. Knockout of the epilepsy gene Depdc5 in mice causes severe embryonic dysmorphology with hyperactivity of mTORC1 signalling. Sci. Rep. 2017;7(1):12618. doi: 10.1038/s41598-017-12574-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 116.Hockemeyer D., Jaenisch R. Induced pluripotent stem cells meet genome editing. Cell Stem Cell. 2016;18(5):573–586. doi: 10.1016/j.stem.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Caires-Júnior L.C., Goulart E., Melo U.S., Araujo B.H.S., Alvizi L., Soares-Schanoski A., de Oliveira D.F., Kobayashi G.S., Griesi-Oliveira K., Musso C.M., Amaral M.S., daSilva L.F., Astray R.M., Suárez-Patiño S.F., Ventini D.C., Gomes da Silva S., Yamamoto G.L., Ezquina S., Naslavsky M.S., Telles-Silva K.A., Weinmann K., van der Linden V., van der Linden H., de Oliveira J.R.M., Arrais N.M.R., Melo A., Figueiredo T., Santos S., Meira J.G.C., Passos S.D., de Almeida R.P., Bispo A.J.B., Cavalheiro E.A., Kalil J., Cunha-Neto E., Nakaya H., Andreata-Santos R., de Souza Ferreira L.C., Verjovski-Almeida S., Ho P.L., Passos-Bueno M.R., Zatz M. Discordant congenital Zika syndrome twins show differential in vitro viral susceptibility of neural progenitor cells. Nat. Commun. 2018;9(1):475. doi: 10.1038/s41467-017-02790-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Araujo B.H.S., Kaid C., De Souza J.S., Gomes da Silva S., Goulart E., Caires L.C.J., Musso C.M., Torres L.B., Ferrasa A., Herai R., Zatz M., Okamoto O.K., Cavalheiro E.A. Down syndrome iPSC-Derived astrocytes impair neuronal synaptogenesis and the mTOR pathway in vitro. Mol. Neurobiol. 2018;55(7):5962–5975. doi: 10.1007/s12035-017-0818-6. [DOI] [PubMed] [Google Scholar]
- 119.de Souza J.S., Carromeu C., Torres L.B., Araujo B.H.S., Cugola F.R., Maciel R.M.B., Muotri A.R., Giannocco G. IGF1 neuronal response in the absence of MECP2 is dependent on TRalpha 3. Hum. Mol. Genet. 2017;26(2):270–281. doi: 10.1093/hmg/ddw384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Liu Y., Lopez-Santiago L.F., Yuan Y., Jones J.M., Zhang H., O’Malley H.A., Patino G.A., O’Brien J.E., Rusconi R., Gupta A., Thompson R.C., Natowicz M.R., Meisler M.H., Isom L.L., Parent J.M. Dravet syndrome patient-derived neurons suggest a novel epilepsy mechanism. Ann. Neurol. 2013;74(1):128–139. doi: 10.1002/ana.23897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jiao J., Yang Y., Shi Y., Chen J., Gao R., Fan Y., Yao H., Liao W., Sun X-F., Gao S. Modeling Dravet syndrome using induced pluripotent stem cells (iPSCs) and directly converted neurons. Hum. Mol. Genet. 2013;22(21):4241–4252. doi: 10.1093/hmg/ddt275. [DOI] [PubMed] [Google Scholar]
- 122.Lossin C., Rhodes T.H., Desai R.R., Vanoye C.G., Wang D., Carniciu S., Devinsky O., George A.L., Jr Epilepsy-associated dysfunction in the voltage-gated neuronal sodium channel SCN1A. J. Neurosci. 2003;23(36):11289–11295. doi: 10.1523/JNEUROSCI.23-36-11289.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sun Y., Paşca S.P., Portmann T., Goold C., Worringer K.A., Guan W., Chan K.C., Gai H., Vogt D., Chen Y-J.J., Mao R., Chan K., Rubenstein J.L., Madison D.V., Hallmayer J., Froehlich-Santino W.M., Bernstein J.A., Dolmetsch R.E. A deleterious Nav1.1 mutation selectively impairs telencephalic inhibitory neurons derived from Dravet Syndrome patients. eLife. 2016;5:5. doi: 10.7554/eLife.13073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Higurashi N., Uchida T., Lossin C., Misumi Y., Okada Y., Akamatsu W., Imaizumi Y., Zhang B., Nabeshima K., Mori M.X., Katsurabayashi S., Shirasaka Y., Okano H., Hirose S. A human Dravet syndrome model from patient induced pluripotent stem cells. Mol. Brain. 2013;6:19. doi: 10.1186/1756-6606-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Liu J., Gao C., Chen W., Ma W., Li X., Shi Y., Zhang H., Zhang L., Long Y., Xu H., Guo X., Deng S., Yan X., Yu D., Pan G., Chen Y., Lai L., Liao W., Li Z. CRISPR/Cas9 facilitates investigation of neural circuit disease using human iPSCs: mechanism of epilepsy caused by an SCN1A loss-of-function mutation. Transl. Psychiatry. 2016;6:e703. doi: 10.1038/tp.2015.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sun Y., Dolmetsch R.E. Investigating the therapeutic mechanism of cannabidiol in a human induced pluripotent stem cell (iPSC)-based model of dravet syndrome. Cold Spring Harb. Symp. Quant. Biol. 2018;83:185–191. doi: 10.1101/sqb.2018.83.038174. [DOI] [PubMed] [Google Scholar]
- 127.Quraishi I.H., Stern S., Mangan K.P., Zhang Y., Ali S.R., Mercier M.R., Marchetto M.C., McLachlan M.J., Jones E.M., Gage F.H., Kaczmarek L.K. An Epilepsy-Associated KCNT1 mutation enhances excitability of human ipsc-derived neurons by increasing slack KNa currents. J. Neurosci. 2019;39(37):7438–7449. doi: 10.1523/JNEUROSCI.1628-18.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dilena R., DiFrancesco J.C., Soldovieri M.V., Giacobbe A., Ambrosino P., Mosca I., Galli M.A., Guez S., Fumagalli M., Miceli F., Cattaneo D., Darra F., Gennaro E., Zara F., Striano P., Castellotti B., Gellera C., Varesio C., Veggiotti P., Taglialatela M. Early treatment with quinidine in 2 patients with epilepsy of infancy with migrating focal seizures (EIMFS) Due to Gain-of-Function KCNT1 mutations: functional studies, clinical responses, and critical issues for personalized therapy. Neurotherapeutics. 2018;15(4):1112–1126. doi: 10.1007/s13311-018-0657-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kolc K.L., Sadleir L.G., Scheffer I.E., Ivancevic A., Roberts R., Pham D.H., Gecz J. A systematic review and meta-analysis of 271 PCDH19-variant individuals identifies psychiatric comorbidities, and association of seizure onset and disease severity. Mol. Psychiatry. 2019;24(2):241–251. doi: 10.1038/s41380-018-0066-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Homan C.C., Pederson S., To T-H., Tan C., Piltz S., Corbett M.A., Wolvetang E., Thomas P.Q., Jolly L.A., Gecz J. PCDH19 regulation of neural progenitor cell differentiation suggests asynchrony of neurogenesis as a mechanism contributing to PCDH19 Girls Clustering Epilepsy. Neurobiol. Dis. 2018;116:106–119. doi: 10.1016/j.nbd.2018.05.004. [DOI] [PubMed] [Google Scholar]
- 131.Qian X., Song H., Ming G-L. Brain organoids: advances, applications and challenges. Development. 2019;146(8):146. doi: 10.1242/dev.166074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Simkin D., Kiskinis E. Modeling pediatric epilepsy through iPSC-Based technologies. Epilepsy Curr. 2018;18(4):240–245. doi: 10.5698/1535-7597.18.4.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhang S.C., Wernig M., Duncan I.D., Brüstle O., Thomson J.A. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat. Biotechnol. 2001;19(12):1129–1133. doi: 10.1038/nbt1201-1129. [DOI] [PubMed] [Google Scholar]
- 134.Mansour A.A., Gonçalves J.T., Bloyd C.W., Li H., Fernandes S., Quang D., Johnston S., Parylak S.L., Jin X., Gage F.H. An in vivo model of functional and vascularized human brain organoids. Nat. Biotechnol. 2018;36(5):432–441. doi: 10.1038/nbt.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Krebs K., Milani L. Translating pharmacogenomics into clinical decisions: do not let the perfect be the enemy of the good. Hum. Genomics. 2019;13(1):39. doi: 10.1186/s40246-019-0229-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Božina N., Sporiš I.Š., Božina T., Klarica-Domjanović I., Tvrdeić A., Sporiš D. Pharmacogenetics and the treatment of epilepsy: what do we know? Pharmacogenomics. 2019;20(15):1093–1101. doi: 10.2217/pgs-2019-0085. [DOI] [PubMed] [Google Scholar]
- 137.Löscher W., Klotz U., Zimprich F., Schmidt D. The clinical impact of pharmacogenetics on the treatment of epilepsy. Epilepsia. 2009;50(1):1–23. doi: 10.1111/j.1528-1167.2008.01716.x. [DOI] [PubMed] [Google Scholar]
- 138.Goto S., Seo T., Murata T., Nakada N., Ueda N., Ishitsu T., Nakagawa K. Population estimation of the effects of cytochrome P450 2C9 and 2C19 polymorphisms on phenobarbital clearance in Japanese. Ther. Drug Monit. 2007;29(1):118–121. doi: 10.1097/FTD.0b013e318030def0. [DOI] [PubMed] [Google Scholar]
- 139.Puranik Y.G., Birnbaum A.K., Marino S.E., Ahmed G., Cloyd J.C., Remmel R.P., Leppik I.E., Lamba J.K. Association of carbamazepine major metabolism and transport pathway gene polymorphisms and pharmacokinetics in patients with epilepsy. Pharmacogenomics. 2013;14(1):35–45. doi: 10.2217/pgs.12.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yun W., Zhang F., Hu C., Luo X., Xue P., Wang J., Ge Y., Meng H., Guo Y. Effects of EPHX1, SCN1A and CYP3A4 genetic polymorphisms on plasma carbamazepine concentrations and pharmacoresistance in Chinese patients with epilepsy. Epilepsy Res. 2013;107(3):231–237. doi: 10.1016/j.eplepsyres.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 141.Wang P., Lin X-Q., Cai W-K., Xu G-L., Zhou M-D., Yang M., He G-H. Effect of UGT2B7 genotypes on plasma concentration of valproic acid: a meta-analysis. Eur. J. Clin. Pharmacol. 2018;74(4):433–442. doi: 10.1007/s00228-017-2395-z. [DOI] [PubMed] [Google Scholar]
- 142.Ho P.C., Abbott F.S., Zanger U.M., Chang T.K.H. Influence of CYP2C9 genotypes on the formation of a hepatotoxic metabolite of valproic acid in human liver microsomes. Pharmacogenom. J. 2003;3(6):335–342. doi: 10.1038/sj.tpj.6500210. [DOI] [PubMed] [Google Scholar]
- 143.Gulcebi M.I., Ozkaynakcı A., Goren M.Z., Aker R.G., Ozkara C., Onat F.Y. The relationship between UGT1A4 polymorphism and serum concentration of lamotrigine in patients with epilepsy. Epilepsy Res. 2011;95(1-2):1–8. doi: 10.1016/j.eplepsyres.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 144.Djoumbou-Feunang Y., Fiamoncini J., Gil-de-la-Fuente A., Greiner R., Manach C., Wishart D.S. BioTransformer: a comprehensive computational tool for small molecule metabolism prediction and metabolite identification. J. Cheminform. 2019;11(1):2. doi: 10.1186/s13321-018-0324-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Houy N., Le Grand F. Optimal dynamic regimens with artificial intelligence: The case of temozolomide. PLoS One. 2018;13(6):e0199076. doi: 10.1371/journal.pone.0199076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kumar R., Sharma A., Siddiqui M.H., Tiwari R.K. Prediction of metabolism of drugs using artificial intelligence: how far have we reached? Curr. Drug Metab. 2016;17(2):129–141. doi: 10.2174/1389200216666151103121352. [DOI] [PubMed] [Google Scholar]
- 147.Barbarino J.M., Whirl-Carrillo M., Altman R.B., Klein T.E. PharmGKB: A worldwide resource for pharmacogenomic information. Wiley Interdiscip. Rev. Syst. Biol. Med. 2018;10(4):e1417. doi: 10.1002/wsbm.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kaplun A., Hogan J.D., Schacherer F., Peter A.P., Krishna S., Braun B.R., Nambudiry R., Nitu M.G., Mallelwar R., Albayrak A. PGMD: a comprehensive manually curated pharmacogenomic database. Pharmacogenom. J. 2016;16(2):124–128. doi: 10.1038/tpj.2015.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Relling M.V., Klein T.E. CPIC: Clinical pharmacogenetics implementation consortium of the pharmacogenomics research network. Clin. Pharmacol. Ther. 2011;89(3):464–467. doi: 10.1038/clpt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kehne J.H., Klein B.D., Raeissi S., Sharma S. The national institute of neurological disorders and stroke (NINDS) epilepsy therapy screening program (ETSP). Neurochem. Res. 2017;42(7):1894–1903. doi: 10.1007/s11064-017-2275-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Cronk D. Drug Discovery and Development. Elsevier; 2013. High-throughput screening. pp. 95–117. [DOI] [Google Scholar]
- 152.Hemmilä I.A., Hurskainen P. Novel detection strategies for drug discovery. Drug Discov. Today. 2002;7(18):S150–S156. doi: 10.1016/S1359-6446(02)02390-5. [DOI] [PubMed] [Google Scholar]
- 153.Moore K., Rees S. Cell-based versus isolated target screening: how lucky do you feel? J. Biomol. Screen. 2001;6(2):69–74. doi: 10.1177/108705710100600202. [DOI] [PubMed] [Google Scholar]
- 154.Sridharan B., Hubbs C., Llamosas N., Kilinc M., Singhera F.U., Willems E., Piper D.R., Scampavia L., Rumbaugh G., Spicer T.P. A simple procedure for creating scalable phenotypic screening assays in human neurons. Sci. Rep. 2019;9(1):9000. doi: 10.1038/s41598-019-45265-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Durens M., Nestor J., Williams M., Herold K., Niescier R.F., Lunden J.W., Phillips A.W., Lin Y.C., Dykxhoorn D.M., Nestor M.W. High-throughput screening of human induced pluripotent stem cell-derived brain organoids. J. Neurosci. Methods. 2020;335:108627. doi: 10.1016/j.jneumeth.2020.108627. [DOI] [PubMed] [Google Scholar]
- 156.Dinday M.T., Baraban S.C. Large-scale phenotype-based antiepileptic drug screening in a zebrafish model of dravet syndrome. eNeuro. 2015;2(4):2. doi: 10.1523/ENEURO.0068-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ibhazehiebo K., Gavrilovici C., de la Hoz C.L., Ma S-C., Rehak R., Kaushik G., Meza Santoscoy P.L., Scott L., Nath N., Kim D-Y., Rho J.M., Kurrasch D.M. A novel metabolism-based phenotypic drug discovery platform in zebrafish uncovers HDACs 1 and 3 as a potential combined anti-seizure drug target. Brain. 2018;141(3):744–761. doi: 10.1093/brain/awx364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Liu J., Sternberg A.R., Ghiasvand S., Berdichevsky Y. Epilepsy-on-a-Chip System for antiepileptic drug discovery. IEEE Trans. Biomed. Eng. 2019;66(5):1231–1241. doi: 10.1109/TBME.2018.2871415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Dunlop J., Bowlby M., Peri R., Vasilyev D., Arias R. High-throughput electrophysiology: an emerging paradigm for ion-channel screening and physiology. Nat. Rev. Drug Discov. 2008;7(4):358–368. doi: 10.1038/nrd2552. [DOI] [PubMed] [Google Scholar]
- 160.Goulart E., de Caires-Junior L.C., Telles-Silva K.A., Araujo B.H.S., Rocco S.A., Sforca M., de Sousa I.L., Kobayashi G.S., Musso C.M., Assoni A.F., Oliveira D., Caldini E., Raia S., Lelkes P.I., Zatz M. 3D bioprinting of liver spheroids derived from human induced pluripotent stem cells sustain liver function and viability in vitro. Biofabrication. 2019;12(1):015010. doi: 10.1088/1758-5090/ab4a30. [DOI] [PubMed] [Google Scholar]
- 161.Goulart E., de Caires-Junior L.C., Telles-Silva K.A., Araujo B.H.S., Kobayashi G.S., Musso C.M., Assoni A.F., Oliveira D., Caldini E., Gerstenhaber J.A., Raia S., Lelkes P.I., Zatz M. Adult and iPS-derived non-parenchymal cells regulate liver organoid development through differential modulation of Wnt and TGF-β. Stem Cell Res. Ther. 2019;10(1):258. doi: 10.1186/s13287-019-1367-x. [DOI] [PMC free article] [PubMed] [Google Scholar]