Abstract
Background
Several members of the bacterial Halomonadacea family are natural producers of polyhydroxyalkanoates (PHA), which are promising materials for use as biodegradable bioplastics. Type-strain species of Cobetia are designated PHA positive, and recent studies have demonstrated relatively high PHA production for a few strains within this genus. Industrially relevant PHA producers may therefore be present among uncharacterized or less explored members. In this study, we characterized PHA production in two marine Cobetia strains. We further analyzed their genomes to elucidate pha genes and metabolic pathways which may facilitate future optimization of PHA production in these strains.
Results
Cobetia sp. MC34 and Cobetia marina DSM 4741T were mesophilic, halotolerant, and produced PHA from four pure substrates. Sodium acetate with- and without co-supplementation of sodium valerate resulted in high PHA production titers, with production of up to 2.5 g poly(3-hydroxybutyrate) (PHB)/L and 2.1 g poly(3-hydroxybutyrate-co-3-hydroxyvalerate) (PHBV)/L in Cobetia sp. MC34, while C. marina DSM 4741T produced 2.4 g PHB/L and 3.7 g PHBV/L. Cobetia marina DSM 4741T also showed production of 2.5 g PHB/L from glycerol. The genome of Cobetia sp. MC34 was sequenced and phylogenetic analyses revealed closest relationship to Cobetia amphilecti. PHA biosynthesis genes were located at separate loci similar to the arrangement in other Halomonadacea. Further genome analyses revealed some differences in acetate- and propanoate metabolism genes between the two strains. Interestingly, only a single PHA polymerase gene (phaC2) was found in Cobetia sp. MC34, in contrast to two copies (phaC1 and phaC2) in C. marina DSM 4741T. In silico analyses based on phaC genes show that the PhaC2 variant is conserved in Cobetia and contains an extended C-terminus with a high isoelectric point and putative DNA-binding domains.
Conclusions
Cobetia sp. MC34 and C. marina DSM 4741T are natural producers of PHB and PHBV from industrially relevant pure substrates including acetate. However, further scale up, optimization of growth conditions, or use of metabolic engineering is required to obtain industrially relevant PHA production titers. The putative role of the Cobetia PhaC2 variant in DNA-binding and the potential implications remains to be addressed by in vitro- or in vivo methods.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12934-021-01713-0.
Keywords: Cobetia spp., Halomonas spp., Halomonadacea, Polyhydroxyalkanoates, PHA, PHB, PHBV, PhaC, Acetate, Genome
Background
The Arctic marine environment constitutes a promising source of cold-adapted microorganisms of potential use for biotechnological production of proteins, green chemicals, polymers, or other valuable compounds [1]. Polyhydroxyalkanoates (PHA) are polyester bioplastics that are gaining renewed commercial interest as green replacements of oil-plastics derived from fossil resources, due to attractive material properties resembling common types of oil-plastics such as polyethylene, but with the advantage of biodegradability in the marine environment as determined by the ASTM standard methods [2, 3]. This biodegradability is furthermore an advantage compared to the bioplastic polylactic acid (PLA) that currently has a high share of the global bioplastic market, but requires high temperature to biodegrade in soil and compost [4]. PHA is produced commercially at low volumes (< 30,000 tons per year) compared to PLA (> 300,000 tons per year) and oil-plastics (> 370 million tons per year) [5]. Industrial production of PHA by bacterial fermentation typically utilizes plant-oils or waste lipids from the agricultural industry or various forms of pure and mixed carbohydrates [6]. A PHA polymer is composed of 3-hydroxyalkanoic acid monomers of varying length, where the excess alkyl chains beyond the 3-hydroxy group take the roles of side chains in the linear polyester. The monomers are classified as either short chain length PHA (scl-PHA) of carbon chain lengths C3 to C5 or as medium chain length PHA (mcl-PHA) of chain lengths C6 to C14, with increased flexibility for the longer chain length polymers [7]. The chain length- and composition determine, together with the molecular weight, the material properties after extraction and processing into a PHA bioplastic product [8, 9]. Establishing substrate utilization and PHA composition phenotypes thus constitute important objectives when characterizing bacterial strains for PHA production.
PHA was discovered as the constituent of intracellular granules found in a Gram-positive Bacillus species, but are now known to be prevalent also in Archaea, Cyanobacteria and in Gram-negative α-, ß-, γ- and δ-proteobacteria [10]. The biological function is generally described as a response to nutrient availability such as when excess carbon is available relative to nitrogen, which was demonstrated already in the 1950s [11]. Recent studies have found several other fitness enhancing effects due to accumulation of PHA in bacteria. These include protection against reactive oxygen species [12] and DNA damage from exposure to UV light [13, 14], and protection against osmotic shock, temperature change, and heavy metals [15]. Ecological studies of the Arctic environment found PHA producing bacteria to be present based on staining with lipophilic dyes and amplification of the PHA polymerase gene phaC, which supports the existence of selective pressure for PHA accumulation in this extreme environment [16, 17]. Taken together, these findings point at cold- and saline environments such as the Arctic marine environment as promising reservoirs for finding novel PHA producing strains of potential use in biotechnology.
Biosynthesis of PHA requires three enzymes utilizing precursors originating from the tricarboxylic acid (TCA) cycle, beta-oxidation-, fatty acid degradation-, and other organism specific pathways [18]. Synthesis of the most common type of PHA, the C4 poly(3-hydroxybutyrate) (PHB), starts with condensation of two acetyl-CoA molecules by the acetyl-CoA acetyltransferase (ß-ketothiolase) PhaA into acetoacetyl-CoA. Subsequent reduction by acetoacetyl-CoA reductase PhaB produces (R)-3-hydroxybutyryl-CoA, which is polymerized into PHB by polyhydroxyalkanoate polymerase PhaC, encoded by the phaC gene. Four different classes of PhaC polymerases are generally described and classified based on their subunit composition and the ability to incorporate scl- or mcl-hydroxyalkanoate precursors, with a fifth class recently proposed primarily based on phylogenetic analyses [19, 20]. A type of proteins known as phasins (PhaP) control size and coagulation of the hydrophobic PHA granules [21], and coats the outer layer together with PhaC, the PHA synthesis repressor PhaR and other PHA regulatory proteins such as the PHA depolymerase enzyme PhaZ. This complex of granules and proteins together form an in vivo subcellular organelle referred to as the carbonosome [22].
The Gram negative γ-proteobacterial family Halomonadaceae consists of 13 genera which are characterized as halotolerant or halophilic, and are typically isolated from extreme environments such as salt lakes [23, 24] or from the marine environment, including the Arctic [25]. Several species of the genus Halomonas are well established as producers of scl-PHA and envisioned as promising candidates for biotech production due to their salt- and alkali tolerance and the applicability of genetic engineering techniques used for strain optimization [26].
Closely related to the Halomonas genus is the Cobetia genus, with five species currently being recognized: Cobetia marina [27], Cobetia pacifica, Cobetia amphilecti and Cobetia crustatorum [28], and Cobetia litoralis [29]. The type strain C. marina DSM 4741T has changed phylogenetic classification several times since its original discovery and classification as Arthrobacter marinus in 1970 [30] and later as Pseudomonas marina [31], Deleya marina [32] and Halomonas marina [33]. Earlier phylogenetic investigations of Cobetia and Halomonas show polyphyletic lineages based on 16S rRNA sequences [34]. Current species classifications of Cobetia are based on DNA hybridization and phenotypic assays [28], however, several strains without species designation are deposited in the National Center for Biotechnology Information (NCBI) database. Strain specific metabolic differences such as temperature tolerance, substrate utilization, and the requirement of sodium ions for growth have been described for strains of C. marina found in three different marine habitats [35]. Despite PHB positive type strain designations for C. marina [27], C. crustatorum [29], C. amphilecti, C. litoralis, and some strains of C. pacifica [28], only a few Cobetia strains have been thoroughly investigated for PHA production. The strain Halomonas sp. SF2003, previously named Cobetia sp. i.4786 [36, 37], is demonstrated to produce PHB from agro-industrial waters rich in sugars and the co-polymer poly(3-hydroxybutyrate-co-3-hydroxyvalerate) (PHBV) when supplied with valeric acid as co-substrate [36, 38]. Similarly, Halomonas marina HMA 103 isolated from a solar saltern in India is capable of producing PHB and PHBV [39]. Most recently, several newly isolated Cobetia strains without formal species designations are demonstrated to produce PHA from polysaccharide substrates originating from brown- and green algae biomass [40, 41]. This demonstrates the potential of using different strains of Cobetia for PHA production.
The aim of this study was to analyze and characterize PHA production for two marine isolates, Cobetia sp. MC34 and the genus type-strain C. marina DSM 4741T. Both strains were found to be putative PHA producers by fluorescent microscopy and Fourier transform infrared spectroscopy (FT-IR) analyses of cells grown on agar plates supplemented with carbon sources. The impact of carbon source, temperature and salinity on growth rate and putative PHA production was analyzed after cultivation in liquid cultures. PHA production was verified and quantified using Gas Chromatography-Flame Ionization Detector (GC-FID) analysis, and the PHA polymer structures were confirmed by Nuclear Magnetic Resonance (NMR) spectroscopy. The genome of Cobetia sp. MC34 was sequenced to enable allocation of Cobetia sp. MC34 to species level, and to allow identification of PHA biosynthesis- and acetate metabolism genes which may form the basis for future targeted studies of PHA metabolism in these strains. Lastly, we performed an in silico study of the Cobetia genus specific PhaC2 enzyme with an intriguing C-terminal end to shed light on its putative function.
Methods
Bacterial strains and media recipes
Cobetia sp. MC34 was isolated in May 2017 from an environmental swab taken from a maturation room for salted fish at a fish-landing facility outside of Tromsø, Norway. The genus type strain C. marina DSM 4741T originally isolated from Woods Hole, USA [30], was obtained from DSMZ (Cat. no. DSM4741). All culture stocks were stored at − 80 °C in 20% glycerol in M4 media and revived on M4 or Marine agar (Difco 2216). The M4 medium contained (per liter 0.2 µm filtered seawater mixed 1:1 with Milli-Q water): 1 g glucose, 1 g malt extract (Merck), 1 g yeast extract (Merck), 1 g peptone (Fluka), and 1 mL glycerol (VWR). The pH of the M4 medium was adjusted to 8.2 using potassium hydroxide before autoclaving. Solid medium was prepared by adding 15 g/L of agar (VWR). MMY agar plates with 1 g/L yeast extract (Merck) was made according to a previously described synthetic seawater medium [42]. Since growth of Cobetia sp. MC34 appeared hampered by development of acidic conditions, two bicarbonate pH buffered PHA production media (MMCY and MMCY_2) were developed. The autoclaved MMCY media contained (per litre): 1 g yeast extract (Merck), 12.9 g sodium chloride, 4 g sodium sulfate, 0.2 g potassium dihydrogen phosphate, 1.4 g magnesium chloride, 0.5 g potassium chloride, 0.25 g ammonium chloride, and 0.112 g calcium chloride. The room tempered media was supplemented to a final concentration of 4 g/L of autoclaved sodium bicarbonate, and sterile filtered vitamin- and trace element solutions as described elsewhere [42]. The MMCY_2 media was made similarly, except that sodium chloride was eliminated and only 2 g/L of sodium bicarbonate was added, to compensate for the addition of sodium from the use of sodium acetate with this media.
Agar plate based screening by fluorescent microscopy and Fourier transform infrared spectroscopy (FT-IR)
A single colony of revived bacteria was re-streaked on MMY agar plates and MMY agar plates supplemented with either 2% (20 g/L) glucose or 2% glycerol (vol/vol). Cells were scraped off after 72 h incubation at 14 °C and re-suspended in 2.5% glutaraldehyde in 1 × PHEM buffer (60 mM PIPES (VWR), 25 mM HEPES (VWR), 10 mM EGTA (Sigma-Aldrich), 4 mM MgSO4·7H2O, pH 6.9), stained with 5 µg/mL Nile-Red (Sigma-Aldrich) for 30 min at room temperature followed by centrifugation at 11,000×g for 1 min. The supernatant was removed and the cells washed twice and re-suspended in 2.5% glutaraldehyde in 1 × PHEM buffer, mounted on agar pads [43] and viewed in a ZEISS Axio Observer 7 microscope with a 100 × NA 1.46 alpha PlanApo oil immersion objective. Images were taken by an Axiocam 506 camera using Colibri LED-fluorescence at wavelengths 559 nm (excitation) and 636 nm (emission). The fluorescent intensity of the images was normalized to 1500 using the ZEN 3.2 software (Carl Zeiss Microscopy GmbH). Cells from the same plates were analyzed by FT-IR as described in the “FT-IR analysis” section.
Cultivation conditions for small-scale liquid cultures
The influence of temperature on the specific growth rate and putative PHA production were tested in 5 mL cultures using MMCY media supplemented with 2% (20 g/L) glucose and incubated at 4 °C, 14 °C, 20 °C, 25 °C, 30 °C, 37 °C, and 42 °C, at 200 rpm. Pre-cultures were prepared in triplicate from single colonies of Cobetia sp. MC34 or C. marina DSM 4741T into 3 mL M4 media with overnight incubation at room temperature (≈ 25 °C, 1270 shakes per minute on a Heidolph Multi Reax shaker), before centrifugation at 11,000×g for 1 min in Eppendorf tubes and re-suspension to a start OD of 0.05 in MMCY media. The specific growth rate, k, was calculated from linear plots corresponding to the equation
| 1 |
where OD600(x) is the optical density at 600 nm corresponding to the time point Tx, sampled during the exponential phase at 2 h intervals. The putative PHA production was measured by FT-IR analysis (as described in “FT-IR analysis” section) when the cultures reached stationary phase, and normalized to production per hour. The results are presented as the average values between duplicate experiments, with error bars depicting the coefficient of variation.
Cultivation conditions for shake-flask production of PHA
Pre-cultures were grown similarly as described for small-scale liquid cultures, except that overnight 3 mL cultures were re-suspended in 50 mL M4 media and grown at 30 °C, 200 rpm, until reaching mid- to late exponential phase (OD600 of 3–4), followed by centrifugation at 11,000×g, 20 °C for 10 min. The pellets were re-suspended in MMCY and MMCY_2 media to a start OD600 of ≈ 0.15 in triplicate 80 mL cultures grown in 250 mL baffled shake-flasks incubated at 30 °C, 200 rpm. Sodium acetate was supplemented to MMCY_2 media (2%, 20 g/L) while glycerol (1.5% vol/vol), glucose and fructose (4%, 40 g/L) were supplemented to MMCY media. Sodium valerate, prepared by adding an equimolar amount of sodium hydroxide to valeric acid, C5, (99%, Alfa Aesar), sodium hexanoate, C6, and sodium octanoate, C8, (TCI) were added to the media at 8 mM concentrations where indicated. At the end of the experiment, 40 mL of bacterial culture was harvested by centrifugation at 13,000×g, 4 °C for 15 min and washed twice in 25 g/L sodium chloride before freezing and lyophilization for determination of cell dry weight (CDW) and subsequent extraction and quantification of PHA (as described in “Quantification of PHA by Gas Chromatography-Flame Ionization Detector (GC-FID)”).
FT-IR analysis
Semi-quantitative analysis of PHA by FT-IR has been demonstrated for mixed cultures of bacteria using a simple model that employs the ratio between the peak areas of the carbonyl-ester and the amide band I when the PHA content is below 44% of the CDW [44]. Bacterial cells for FT-IR analysis were either scraped off from agar plates using an inoculation loop, re-suspended in 96% ethanol and spun down in Eppendorf tubes at 21,000×g for 1 min, or 500 µL were taken from liquid cultures and spun down similarly followed by re-suspension of the pellet in 96% ethanol and a second spin to subsequently remove the supernatant. Spectral information was collected for the full range (650–4000 cm−1) with 64 background scans and 256 sample scans using HappGenzel apodization without zero fill factor on an Agilent Cary 630 ATR FT-IR instrument controlled by Agilent MicroLab PC software. The putative PHA production based on the carbonyl-ester and amide band I-ester regions was calculated as the ratio between the areas of their corresponding spectral regions (C:A 1 ratio) defined as 1763–1705 cm−1 (carbonyl-ester) and 1705–1580 cm−1 (amide band I-ester). The C:A 1 ratio was calculated from raw spectra using the integral function, while spectra obtained from agar plates were baseline corrected to remove negative absorbance values using SpectraGryph, Software for optical spectroscopy v. 1.2.15 (Dr. Friedrich Menges, Oberstdorf, Germany).
Quantification of PHA by Gas Chromatography-Flame Ionization Detector (GC-FID)
The amount of PHA in bacterial pellets was quantified by GC-FID after methanolysis in a 1:1 (vol/vol) mixture of chloroform and 15% concentrated sulfuric acid dissolved in methanol. Approximately 15–30 mg of lyophilized cells were added to 3 mL of the methanolysis mixture containing 0.2% benzoic acid as internal standard, and reacted for 12 h at 100 °C. PHBV (Sigma-Aldrich) was used as external standard. After the reaction, 1.5 mL of Milli-Q water was added to separate the phases followed by careful transfer of the organic phase into GC analysis vials. The samples were analyzed by an Agilent 7820A GC-FID system (Agilent Technologies) using a 30 m long by 0.32 mm i.d. DB-5MS column with 0.25 μm film thickness. The injector temperature was set to 250 °C with a split ratio of 10:1 and the injection volume was 1 μL. The temperature program was set to 50 °C with hold for 5 min, then a linear increase to 250 °C at a rate of 15 °C/min, followed by increase to 310 °C at a rate of 30 °C/min. Hydrogen was used as carrier gas at 0.8 mL/min flow with a column head overpressure of 37 kPa. At least two technical replicates for each PHA extraction were injected.
Nuclear magnetic resonance (NMR)
PHA from approximately 50 mg of lyophilized cells was extracted in screw-cap culture tubes with 2 mL of chloroform at 100 °C for 1 h. The extracts were then allowed to cool to room temperature, filtered through 0.22 μm nylon syringe filters, and dried under a stream of nitrogen gas at room temperature. The dried samples were then dissolved in 0.5 mL CDCl3 (99.8% atom-D, Acros). 1H NMR and 13C NMR spectra were recorded at 298 K with a Bruker DRX-400 spectrometer at 400 MHz and 100 MHz, respectively.
Statistical analysis
The coefficient of variation (standard deviation) for the specific growth rate and the C:A 1 ratio from 5 mL cultures was calculated in Microsoft Excel 2016 using the STDEV.P function from average values of two experiments with 3 biological replicates each. The coefficient of variation (standard deviation) of OD600, the CDW and the PHA content was calculated from a total of 6 biological replicates obtained from two independent experiments using Pandas v. 1.1.3 in Python v. 3.8.8.
DNA extraction and 16S rRNA PCR
DNA from Cobetia sp. MC34 was extracted from an overnight culture using DNeasy Blood & Tissue kit (Qiagen) following the manufacturers’ instructions. The partial 16S rRNA sequence for Cobetia sp. MC34 was amplified by PCR using 10 mM each of the universal bacterial primers fD1/27F (5′AGAGTTTGATCCTGGCTCAG) and rP2/1492R (3′TACGGYTACCTTGTTACGACTT) [45], 10 mM dNTP and 1.25 U Taq polymerase in 1 × Standard Taq buffer (New England Biolabs). Five nanograms of DNA was added as template and the reaction was cycled 30 times (94 °C for 30 s, 55 °C for 30 s and 72 °C for 2 min). The PCR product was treated with 2 µL ExoSap-IT (Affymetrix®, Thermo Fisher) for 15 min at 37 °C followed by 15 min at 80 °C. The PCR product was then sequenced in one direction using primer 515F (5′-GTGCCAGCAGCCGCGGTAA-’3) and BigDye™ Terminator v3.1 Cycle Sequencing Kit following the manufacturers’ instructions (Applied Biosystems™). The full length 16S sequence was later obtained from the annotated whole genome sequence.
Genome sequencing, assembly and annotation
Extracted DNA was eluted in 10 mM Tris–HCl pH 8 and the quality and quantity measured on a Qubit fluorimeter (Invitrogen). Sequencing of the genome was done at The Norwegian Sequencing Centre, Oslo, Norway, using Nextera™ DNA Flex Tagmentation and a Illumina MiSeq. The paired end sequencing reads were quality checked and analyzed for presence of adapter sequences using FastQC [46] and subsequently trimmed with first Trimmomatic v.0.39 (Settings: Phred33, Headcrop 15, average quality threshold 29, minlen 100) [47] and secondly to remove the Nextera specific adapter sequences using BBduk v. 38.22 (Settings: ftm 5, Ktrim R, K 23, Mink 9, Hdist 1, Tbo and Tpe Yes). Kraken2 v. 2.1.1 was used for contamination analysis [48, 49]. The sequencing reads were normalized and error-corrected with BBnorm (Settings: Prefilter true, target 50, min 5 and error correction settings ecc t, keepall passes 1, bits 16 prefilter) and merged with BBmerge [50]. Merged and unmerged reads were assembled using SPAdes v. 3.13.0 run in Python v. 2.7.15 [51]. The assembled contig files were evaluated for genome completeness using BUSCO v. 4.0.4 database oceanospirillales_odb10 [52] and evaluation of genome assembly parameters (number of contigs, N50, etc.) were done with Bandage and Quast v. 5.0.2 [53, 54]. Sequence reads were mapped back onto the assembled genome using Bowtie2 v. 7. 3. 0 [55] and alignment statistic was generated with Samtools v. 1.9 [56]. The assembled genome was first annotated with Prokka v. 1.13 [57] and later, for publishing, with the NCBI Prokaryotic Genome Annotation Pipeline [58]. This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession JADPPY000000000. The version described in this paper is version JADPPY010000000. The sequencing reads are deposited under the NCBI BioProject accession PRJNA678481 and in the NCBI Sequence Read Archive under accession SRR13530190.
Phylogenetic analyses by 16S rRNA and Average Nucleotide Identity (ANI) matrix
The 16S rRNA sequences (Additional file 1: Table S1) were obtained from either NCBI’s 16S RefSeq database (https://www.ncbi.nlm.nih.gov/refseq/targetedloci/16S_process/), the SILVA ribosomal RNA database project (https://www.arb-silva.de/) or from whole genome sequences obtained from Genbank’s Genome Database (https://www.ncbi.nlm.nih.gov/genome/). The C. necator 16S sequence was obtained from NCBI (NR_102851).
The Muscle [59] algorithm (Settings: Gap open -1000, gap extend -1.2, neighbour-joining (NJ) clustering) in MEGA7.0.26 was used to align the 16S rRNA sequences and the Maximum Likelihood (ML) method were used to construct the 16S tree (Settings: Gamma distributed with 2 discrete categories (+ G, parameter = 0.0500), partial deletion with 75% cut-off value and nearest neighbour interchange with default setting for initial tree search) [60, 61]. Evolutionary distances were determined using the Kimura 2-parameter method [62] and the phylogeny tested with 1000 bootstrap replicates [63]. The final tree contains 1520 positions and was visualized using Itol v6 [64].
The ANIb algorithm in PyAni software v. 0.2.10 [65] in Python v. 3. 9. 1 was used to align the whole genome sequences (Additional file 1: Table S1) and generate an ANI matrix used to construct a dendrogram using Pythons Plotly package v. 4. 12. 0 in Python v. 3. 8. 8.
Genome searches
The Basic Local Alignment Search Tool (BLAST) [66] function in CLC MainWorkBench v. 20.0.3 (Settings: Blosum62 matrix, gap cost 10 and 1, word size 2, expect 1.0 and mask low complexity regions) was used to query both DNA- and protein sequences against translated Cobetia genome sequences (Additional file 1: Table S1). PhaZ from Pseudomonas oleovorans (P26495) were obtained from The UniProt Consortium 2020 (https://www.uniprot.org/) while PhaZ from H. bluephagenesis TD01 (EGP18355.1, EGP19590.1, EGP20509.1), and PhaC from Halomonas sp. R5-57 (CEP36398.1 and CEP35484.1), H. bluephagenesis TD01 (EGP20415.1 and EGP19504.1) and Halomonas sp. O-1 (BAO57489.1 and BAO57490.1) were obtained from Genbank (https://www.ncbi.nlm.nih.gov/genbank/). Cobetia spp. phaC sequences were obtained from either the annotated whole genome sequences or from BLAST [66] queries against the translated genomes (Additional file 1: Table S1) using C. marina DSM 4741T PhaC1 and PhaC2. The Kyoto Encyclopedia of Genes and Genomes (KEGG) BlastKOALA v. 2.2 (https://www.kegg.jp/blastkoala/) was used for metabolic pathway analysis based on whole genome sequencing data [67].
PhaC2 in silico analysis
The Cobetia sp. MC34 PhaC2 amino acid sequence (WP_213113863) was used as template to the PSIPRED server v. 4.0 (http://bioinf.cs.ucl.ac.uk/psipred/) which generated the secondary structure prediction and amino acid composition graphic [68, 69]. The Cobetia sp. MC34 PhaC2 was searched against NCBI’s non-redundant protein sequences using protein–protein BLAST algorithm (Protein BLAST) (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome) [66]. Cobetia and Halomonas PhaC2 sequences were searched for domain hits against NCBI’s Conserved Domain Database v. 3.19 (NCBI CDD) using CD-Search (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and Batch CD-search (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi) with the settings “low-complexity filter” and “Composition based statistics adjustment” off [70–74]. Molecular weight (MW) and isoelectric point (pI) values were calculated using Expasy Compute pI/MW tool (https://web.expasy.org/compute_pi/, SIB Swiss Institute of Bioinformatics).
Results
Genome sequencing of Cobetia sp. MC34
Cobetia sp. MC34 was genome sequenced in order to identify and compare PHA biosynthesis genes with the genome sequenced C. marina DSM 4741T [75], and to resolve the evolutionary relationships within the Cobetia genus. The Illumina Paired-end sequencing of Cobetia sp. MC34 generated 1,437,981 raw reads of 301 bases in both directions, which was reduced to 1,010,167 reads after trimming, normalization and merging, plus 38,293 unmerged reads from both directions. The assembled draft genome contained a total of 4,003,645 base pairs (bp) with an N50 of 111,054 bp and a GC content of 62.58%. The genome assembly consisted of 175 contigs exceeding a length of 200 bp, and 109 contigs exceeding a length of 500 bp. The genome contained 3487 genes, of which 3399 were predicted to be protein coding. The quality of the genome assembly was evaluated for contamination and completeness by Kraken2 analysis, which showed contamination to be not present. Busco’s Oceanospirillales database contains 619 essential genes, of which 618 complete single copy orthologs and only one fragmented were found in Cobetia sp. MC34. Hence, the draft genome of Cobetia sp. MC34 was of sufficient quality for identification of PHA biosynthesis genes and phylogenetic studies.
Cobetia sp. MC34 is a strain of Cobetia amphilecti, with close relationship to the PHA producer Halomonas sp. SF2003
BLAST analysis of the complete 16S rRNA gene of Cobetia sp. MC34 revealed high similarity (> 99% identity) to several species annotated as Halomonas and Cobetia and a Maximum Likelihood (ML) tree was constructed to illustrate the phylogenetic relationship of Cobetia sp. MC34 to members of the Halomonadacea family. The 29 sequences (Additional file 1: Table S1) included in the ML tree represent the five recognized species of Cobetia represented by a total of sixteen Cobetia isolates currently available as full genomes, as well as Halomonas sp. SF2003 and nine other verified PHA producing strains of Halomonas. Chromohalobacter salexigens and Zymobacter palmae were included to provide reference to a comprehensive Halomonadacea phylogeny previously published [34].
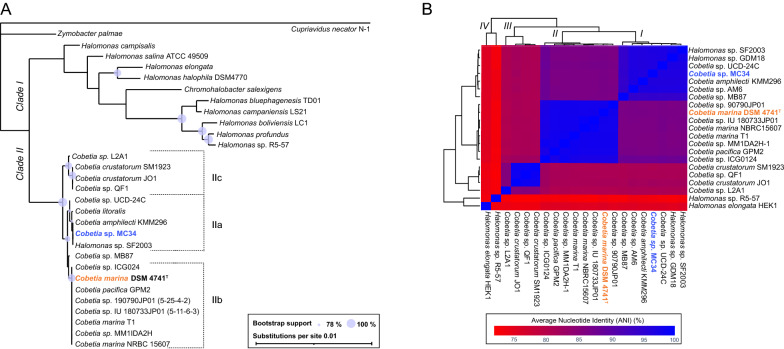
As shown in Fig. 1A, two main clades and two outgroups were resolved in the 16S ML tree. Clade I contains Halomonas sensu stricto sequences and C. salexigens in accordance with earlier studies [34, 76]. Clade II contains three subclades (IIa, IIb and IIc) which include all Cobetia species and Halomonas sp. SF2003. Despite its name, Halomonas sp. SF2003 is shown to belong to the Cobetia genus [76]. Cobetia sp. MC34 allocates to subclade IIa together with Halomonas sp. SF2003 and C. amphilecti KMM296. These three isolates as well as Cobetia sp. AM6 and Halomonas sp. GDM18 (not shown) share identical 16S sequences. Cobetia litoralis and Cobetia sp. UCD-24C contains only two and three nucleotide substitutions compared to Cobetia sp. MC34, respectively, and therefore fell into subclade IIa (bootstrap value of 89%). Subclade IIb (bootstrap value of 96%) is the largest and contains the type strain C. marina DSM 4741T, strains annotated as C. marina, C. pacifica GPM2 and five strains without species designation. Cobetia sp. MB87 has four nucleotide substitutions compared to C. marina DSM 4741T, and does not cluster well with the C. marina group (bootstrap value of 54%). The two strains of C. crustatorum clustered together with Cobetia sp. L2A1 and Cobetia sp. QF1 in subclade IIc (bootstrap value of 94%).
Fig. 1.
Phylogenetic analyses of Cobetia sp. MC34 based on the 16S rRNA gene and the draft genome sequence. A The ML-tree based on 16S rRNA gene sequences include recognized species of Cobetia, genome sequenced strains of Cobetia, known PHA producing Halomonas species and two members of the Halomondacea family. The 16S rRNA gene from C. necator was included as outgroup. Bootstrap support values above 75% (1000 replicates) are depicted as bubbles. The scale bar shows branch length measured as substitutions per site. B The Average Nucleotide Identity (ANI) tree was constructed from whole genome sequences. The ANI similarity values are shown as colors from 75% (red) to 100% (blue). Halomonas elongata HEK1 and the marine isolate Halomonas sp. R5-57 were included as outgroups in the ANI analysis.
Whole genome sequence alignment and calculation of the Average Nucleotide Identity (ANI) can increase phylogenetic resolution compared to use of the 16S rRNA gene tree alone [77]. The draft genome of Cobetia sp. MC34 was therefore compared with sixteen Cobetia genome sequences available in NCBI’s whole genome dendrogram (excluding metagenomes) and the Halomonas isolates SF2003 and GPM18 (Additional file 1: Table S1). Halomonas elongata HEK1 and the Arctic marine isolate Halomonas sp. R5-57 were included as outgroups in the analysis. The genome sequence for C. litoralis is currently not available, and the Cobetia phylogeny is thus incomplete. Figure 1B shows the resultant ANI tree with four clades that further elucidated the phylogenetic relationship between species of Cobetia. Clade I contains C. amphilecti KMM296 and Cobetia sp. MC34, -AM6, Halomonas sp. SF2003 and -GPM18, Cobetia sp. UCD-24C and Cobetia sp. MB87. This is the same clustering as observed in the 16S rRNA gene tree (Fig. 1A). All members of this clade except Cobetia sp. MB87 showed ANI values between 96 and 98%, suggesting they are strains of C. amphilecti, following an ANI species cut-off value of 96% [78]. Closest to the C. amphilecti clade is the C. marina DSM 4741T clade, also in agreement with the 16S rRNA gene tree. The species split of C. amphilecti and C. marina is supported by ANI values below 87%. C. pacifica GPM2 clusters in the C. marina DSM 4741T clade where all members have ANI values above 98%, indicating that they belong to the same species. The third clade contains two strains of C. crustatorum, Cobetia sp. QF1 and Cobetia sp. L2A1, with the latter being most divergent with an ANI value of only 86%. Cobetia sp. QF1 and the two C. crustatorum strains have ANI values of 99%, indicating Cobetia sp. QF1 to be a strain of C. crustatorum. The fourth clade contained the two Halomonas sensu stricto sequences that separate as outgroups relative to the three other clades.
Thus, the 16S rRNA and ANI analysis suggest placement of Cobetia sp. MC34, Cobetia sp. AM6, Halomonas sp. SF2003, and Halomonas sp. GDM18 as strains of the recognized specie C. amphilecti.
Agar plate pre-screening indicate PHA production for Cobetia sp. MC34 and C. marina DSM 4741T
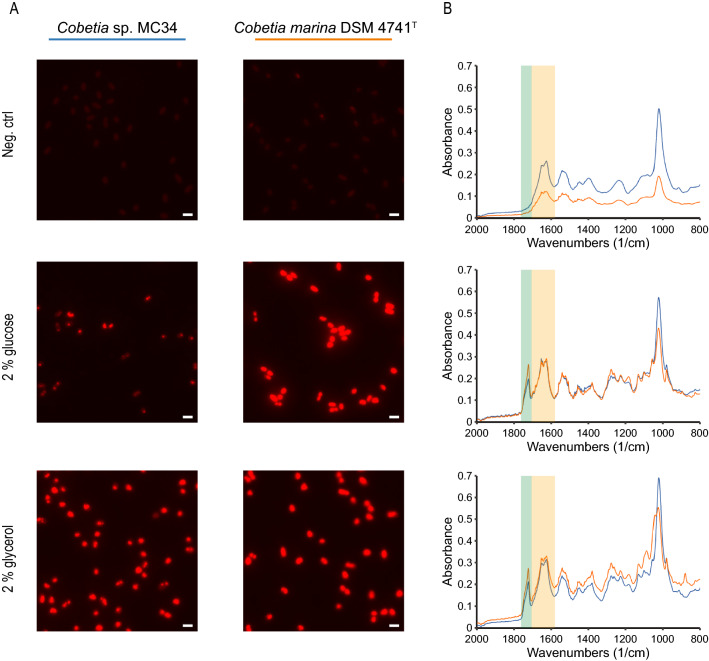
Putative PHA production for Cobetia sp. MC34 and C. marina DSM 4741T was assessed after 72 h incubation at 14 °C on MMY agar plates supplemented with 2% glucose or 2% glycerol by FT-IR and fluorescent microscopy of cells stained with the lipophilic dye Nile-red. As shown in Fig. 2, cells from both strains grown on MMY agar supplemented with 2% glucose and 2% glycerol are short rods with strong intracellular fluorescent staining, typical for cells that produce PHA or other types of storage lipids [79, 80]. Cells grown without an additional carbon substrate showed little fluorescence. FT-IR analysis provides spectral information from vibration of molecules and can identify chemical functional groups even in a complex biological matrix. The carbonyl-ester group is particularly important for identification of PHA and gives a characteristic peak around wavelength 1725 cm−1 for crystalline PHB [81]. As seen in Fig. 2, the spectra of Cobetia sp. MC34 and C. marina DSM 4741T grown on MMY agar supplemented with glucose or glycerol show carbonyl-ester peaks at 1723–1724 cm−1, in contrast to cells grown on the MMY control plates, where this peak is absent. These results suggests that PHA or other types of lipids are produced by the strains when supplemented these carbon sources.
Fig. 2.
Putative PHA production on MMY agar plates supplemented with an excess carbon substrate. A Fluorescent microscopy of Nile-red stained cells grown on MMY agar plates supplemented with 2% glucose and 2% glycerol shows accumulation of intracellular inclusions after 72 h incubation at 14 °C, in contrast to the negative controls. The cells were imaged in a ZEISS Axio Observer 7 microscope at 100X magnification and the fluorescence intensity normalized to 1500 for all images. The scale bar shows 2 μm. B FT-IR spectra of cells from the same agar plates as in A. The cells grown on glucose and glycerol supplemented plates show carbonyl-ester signals centered at wavenumber 1724 cm–1 (Green box, n = 2). The amide band I-ester from proteins is observed in spectra from all plates, including the negative controls (Orange box, n = 2).
Cobetia sp. MC34 and C. marina DSM 4741T exhibit mesophilic growth patterns with high production of PHB from acetate
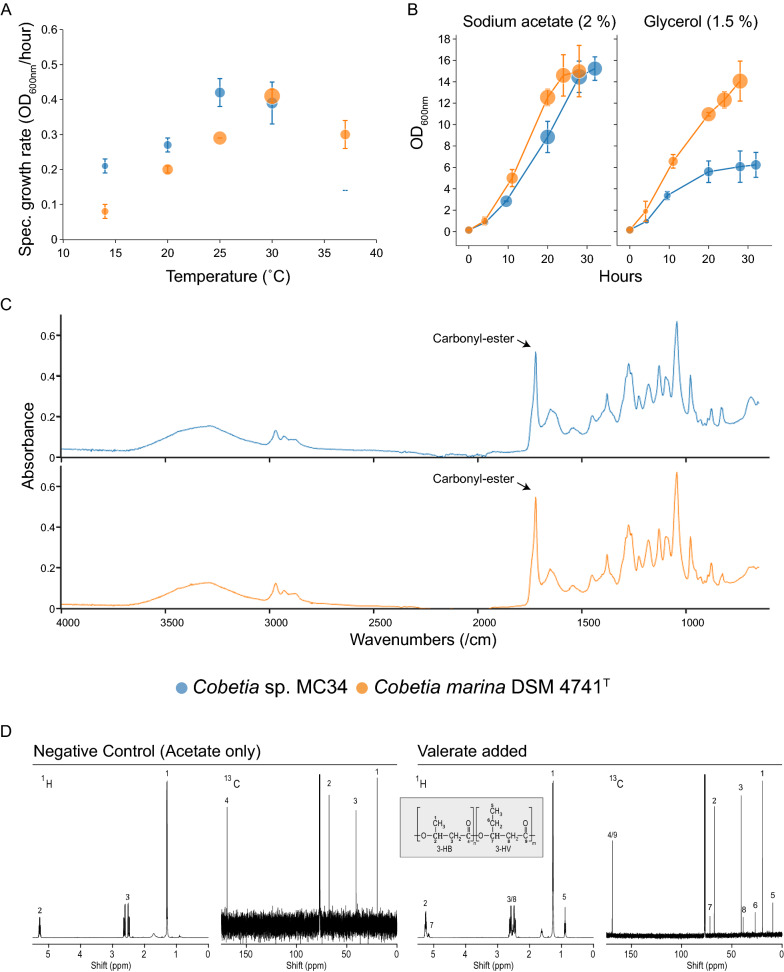
Growth and putative PHA production were further investigated in small scale liquid cultures in MMCY media at different temperatures. Cobetia sp. MC34 and C. marina DSM 4741T both exhibit growth patterns characteristic of mesophilic bacteria with the highest specific growth rate (~ 0.4 OD600/h) observed at 25–30 °C for Cobetia sp. MC34 and at 30 °C for C. marina DSM 4741T (Fig. 3A). A higher specific growth rate for Cobetia sp. MC34 compared to C. marina DSM 4741T was observed at 25 °C and below, and neither of the strains showed any significant growth at 6 °C. Good growth was observed for C. marina DSM 4741T at 37 °C, although not optimal as expected based on the type strain description [27]. This may be due to differences in growth media or other conditions. In contrast, Cobetia sp. MC34 showed poor growth at 37 °C.
Fig. 3.
Cobetia sp. MC34 and Cobetia marina DSM 4741T are mesophilic and produce PHB and PHBV. A Cobetia sp. MC34 (blue) and C. marina DSM 4741T (orange) showed highest growth rate and putative PHA production per hour measured by FT-IR (visualized by sphere area) at 30 °C in 5 mL MMCY media supplemented with 2% glucose. The error bars show coefficient of variation of the specific growth rate between duplicate experiments (n = 6). B Growth curves in 80 mL MMCY and MMCY_2 media supplemented with 2% sodium acetate and 1.5% glycerol, respectively, at 30 °C, 200 rpm. The putative PHA production was measured by FT-IR (visualized by sphere area). The error bars show the coefficient of variation of the OD600 between duplicate experiments (n = 6). C FT-IR spectra of cells grown in 80 mL MMCY_2 media supplemented with 2% sodium acetate at 30 °C, 200 rpm for 28 h (C. marina DSM 4741T) and 32 h (Cobetia sp. MC34). The carbonyl-ester peaks at wavenumber 1724 cm−1 (arrows) indicate PHA production, and high amounts of PHB was confirmed by GC-FID analysis. D NMR confirms PHBV production in Cobetia sp. MC34. When grown on MMCY_2 media supplemented with 20 g/L sodium acetate, 3-HB were produced as indicated by peaks 1, 2 and 3 in the proton 1H plot (negative control) and peaks 1, 2, 3 and 4 in the carbon 13C plot (negative control). Co-supplement with 8 mM sodium valerate resulted in peaks unique to 3-HV as indicated by 5, 7, and 8 in the 1H plot and 5, 6, 7, 8, and 9 in the 13C plot. The spectra were normalized to the non-solvent peak with the highest intensity
The putative PHA production was measured by FT-IR and calculated as the increase in the ratio between the carbonyl-ester region relative to the amide band I-ester region per hour after reaching stationary phase (C:A 1 ratio/h). The highest putative PHA production per hour was observed at 30 °C for both strains (Fig. 3A). Cobetia marina DSM 4741T showed a higher putative PHA production than Cobetia sp. MC34 at all tested temperatures, despite the lower specific growth rate observed at temperatures below 25 °C. The sodium requirement and tolerance to salt in the MMCY media at 30 °C using glucose as carbon source was found to be in the range of 0–10% sodium chloride for Cobetia sp. MC34, with highest specific growth rates obtained within the range of 1–4% sodium chloride (Additional file 1: Fig. S1). Both strains thus classify as halotolerants when grown in MMCY media [82].
PHA production was subsequently determined and characterized after growth in shake-flask cultures at 30 °C, using MMCY and MMCY_2 media supplemented with either 2% sodium acetate, 1.5% glycerol, 4% glucose, or 4% fructose. Samples harvested along the growth curve were analyzed by FT-IR to monitor changes in the C:A 1 ratio in near-real time to determine when to harvest samples for GC-FID analysis. Supplement with acetate resulted in the highest OD600 and highest C:A 1 ratio for Cobetia sp. MC34 while acetate and glycerol were equally good for C. marina DSM 4741T (Fig. 3B and C). Lower maximum OD600 values and lower C:A 1 ratios were found for Cobetia sp. MC34 when glycerol (Fig. 3B), glucose, and fructose were supplemented (Additional file 1: Figs. S2 and S3). An increase in the C:A 1 ratio was, however, still observed as the OD600 increased (Additional file 1: Fig. S3). The FT-IR analyses further showed highest intensity of the carbonyl-ester peak at 1724 ± 2 cm–1 for all conditions, similar to the FT-IR spectra obtained from cells grown on MMY agar and in small scale cultures. Additionally, peaks corresponding to CH3, CH2, CH3/CH2, and C=O-ester groups characteristic of lipids such as from fatty acids and polyesters were detected in all FT-IR spectra [83].
Cells harvested at the end of these growth experiments were lyophilized and subjected to GC-FID analysis for quantification of PHA. As shown in Table 1, the highest production of PHB from the use of a single substrate was found in cells supplemented with acetate, where 72 ± 11% and 61 ± 8.3% PHB/CDW were produced for Cobetia sp. MC34 and C. marina DSM 4741T, respectively. Production of PHB was also detected for both strains when glycerol, glucose, and fructose were used as carbon substrates.
Table 1.
PHA productivity for Cobetia sp. MC34 and C. marina DSM 4741T
| Strain | Carbon substrate | OD600 at harvest | CDW (g/L) | PHB (% of CDW) | PHV (% of CDW) | PHA (g/L) |
|---|---|---|---|---|---|---|
| Cobetia sp. MC34 | Acetate | 15.0 (± 1.3) | 3.4 (± 0.5) | 72 (± 11) | – | 2.5 |
| Acetate + valerate | 14.2 (± 2.1) | 3.4 (± 0.8) | 48 (± 3.4) | 14 (± 2.3) | 2.1 | |
| Glycerol | 6.2 (± 1.4) | 2.5 (± 0.5) | 26 (± 12) | – | 0.7 | |
| Glucose | 6.5 (± 0.3) | 1.8 (± 0.1) | 35 (± 4.8) | – | 0.6 | |
| Fructose | 6.0 (± 0.8) | 3.9 (± 0.8) | 9.9 (± 1.3) | – | 0.4 | |
| Cobetia marina DSM 4741T | Acetate | 15.0 (± 2.9) | 3.9 (± 0.6) | 61 (± 8.3) | – | 2.4 |
| Acetate + valerate | 17.0 (± 1.4) | 4.4 (± 0.3) | 59 (± 11) | 26 (± 2.6) | 3.7 | |
| Glycerol | 14.0 (± 2.1) | 4.0 (± 0.1) | 61 (± 8.1) | – | 2.5 | |
| Glucose | 8.0 (± 0.2) | 2.4 (± 0.3) | 46 (± 2.9) | – | 1.1 | |
| Fructose | 8.1 (± 0.2) | 3.1 (± 0.2) | 28 (± 11) | – | 0.9 |
The cells were harvested from 80 mL cultures in MMCY_2 medium supplemented with 2% sodium acetate with- and without 8 mM sodium valerate, with 1.5% glycerol, 4.0% glucose or 4.0% fructose and were quantified by GC-FID analysis (n = 6). All Cobetia sp. MC34 cultures were grown for 32 h. The C. marina DSM 4741T cultures supplemented with glucose and fructose were grown for 24 h, and those supplemented with acetate, valerate and glycerol were grown for 28 h. Values in parentheses are standard deviations calculated from 6 samples.
Cobetia marina DSM 4741T produced more PHA than Cobetia sp. MC34 when glycerol and fructose were supplemented, with glycerol yielding 61% PHB/CDW, corresponding to a productivity of 2.5 g PHB/L similar to the productivity when acetate was supplemented. On the other hand, Cobetia sp. MC34 produced 26% PHB/CDW corresponding to 0.7 g PHB/L when fed with glycerol. Supplement with fructose resulted in high accumulation of biomass but only 9.9% PHB/CDW for Cobetia sp. MC34 while glucose supplementation resulted in low biomass production but accumulation of 35% PHB/CDW and corresponding productivity values of 0.4 and 0.6 g PHB/L, respectively. This is in contrast to C. marina DSM 4741T that produced approximately 46% and 28% PHB/CDW from glucose and fructose, corresponding to a productivity of 1.1 and 0.9 g PHB/L. Substrates such as sucrose and N-acetylglucosamine were also tested but resulted in limited growth and no PHA production (data not shown).
PHBV produced by Cobetia sp. MC34 and C. marina DSM 4741T from acetate co-supplemented with valerate
The ability to produce co-polymers was tested by co-supplementing acetate (C2) cultures with a second carboxylic acid (C5, C6, and C8) at a C2:C5/6/8 ratio of 31 to 1. Both strains produced the co-polymer PHBV only when valerate (C5) was added as a co-substrate. Highest PHA productivity and a high proportion of the C5 monomer 3-hydroxyvalerate (3-HV) was found when C. marina DSM 4741T was co-fed with acetate and valerate. The contents of 3-hydroxyalkanoate monomers, expressed as percentage of the CDW, was 59 ± 11% of 3-hydroxybutyrate (3-HB) and 26 ± 2.6% of 3-HV in the co-polymer produced by C. marina DSM 4741T, while 48 ± 3.4% of 3-HB and 14 ± 2.3% of 3-HV constituted the CDW of Cobetia sp. MC34 (Table 1). The compositions of PHB and PHBV extracted from Cobetia sp. MC34 were additionally confirmed by proton 1H and carbon 13C NMR analysis. As indicated in Fig. 3D, proton signals at 0.89 ppm (d, 3H) from the methyl group (1) of 3-HB are visible in both the negative control spectrum and the spectrum co-supplemented with valerate, whereas an additional methyl proton signal (5) at 1.27 ppm (t, 3H) is seen for 3-HV. The methylene protons from the main chains of 3-HB and 3-HV (3/8) are visible at 2.55 ppm (m, 4H). The methine protons for 3-HB and 3-HV (2/7) are visible at 5.25 and 5.16 ppm (m, 1H), respectively. A unique signal for the 3-HV methylene side chain (6) is visible at 5.25 ppm (m, 2H). The presence of 3-HV is also confirmed by 13C NMR by four peaks for 3-HV, which are not present in the negative control: The side chain 3-HV methyl carbon (5) at 9.4 ppm, the side chain methylene carbon (6) at 26.9 ppm, the main chain methylene carbon (8) at 38.8 ppm and the methine carbon (7) at 71.9 ppm [84]. Co-supplement with sodium salts of the mcl-precursors hexanoate and octanoate did not result in production of PHA co-polymers (data not shown).
The genetic background for PHA production from acetate
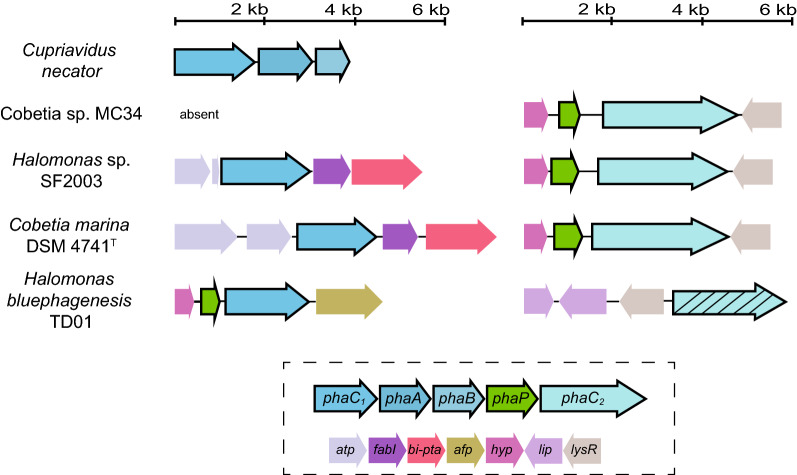
The finding that both Cobetia sp. MC34 and C. marina DSM 4741T are good producers of PHB and PHBV prompted further investigation of their PHA biosynthesis genes. The genes responsible for PHA production are organized in a characteristic phaCAB operon in the model bacterium C. necator [85], whereas the phaA, phaB, and phaC genes are typically located at independent loci in members of Halomonas [86, 87]. Similarly, the annotation of Cobetia sp. MC34 identified these three genes located in separate loci (Table 2). Interestingly, Halomonas sp. SF2003 and C. marina DSM 4741T encode two copies of phaC (phaC1 and phaC2) that differ in length [75, 76] (Table 2). However, only one phaC gene was found in Cobetia sp. MC34 and named phaC2 (IZU87_04860) based on similarity to phaC2 from Halomonas sp. SF2003 (Table 2). A comparative analysis of the phaC2 gene region in Cobetia sp. MC34, C. marina DSM 4741T and Halomonas sp. SF2003 showed conservation and similar gene synteny (Fig. 4). Located upstream of phaC2 is phaP (IZU87_04855) that encodes a phasin and a gene of unknown function hyp (IZU87_04850). Downstream is a LysR family transcriptional regulator gene lysR (IZU87_04865) that is conserved between all three Cobetia strains. This latter gene is also located upstream of the putative phaC2 found in H. bluephagenesis TD01 [86]. The gene synteny downstream of phaC1 is conserved between Halomonas sp. SF2003 and C. marina DSM 4741T and consist of the gene enoyl-[acyl-carrier protein] reductase I fabI (IZU87_11845) and a gene annotated as a bifunctional enoyl-CoA hydratase/phosphate acetyltransferase bi-pta (Fig. 4).
Table 2.
Genes encoding transporters, enzymes, transcriptional regulators and carbonosome proteins involved in PHA synthesis from acetate
| Gene description | Gene(s) | Cobetia sp. MC34 | Cobetia marina DSM 4741T | Halomonas sp. SF2003 |
|---|---|---|---|---|
|
Acetyl-CoA transferase (EC 2.3.1.9) |
phaA (phaA/fadA) |
+ [+] IZU87_07675/(IZU87_02125) |
++ BFX80_16410/(BFX80_14370) |
++ C8233_08335/C8233_10360 |
|
Acetoacetyl-CoA reductase (EC 1.1.1.36) |
phaB |
+ IZU87_12765 |
+ BFX80_01220 |
+ C8233_12530 |
|
Polyhydroxyalkanoate polymerase (EC 2.3.1.-) |
phaC1/phaC2 |
−/+ −/IZU87_04860 |
+/+ BFX80_07630/BFX80_12110 |
+/+ C8233_00780/C8233_06040 |
|
Phosphate acetyltransferase (EC 2.3.1.8) |
pta |
+ IZU87_07815 |
+ BFX80_04540 |
+ C8233_15920 |
| Bifunctional enoyl-CoA hydratase/phosphate acetyltransferase | bi-pta | – |
+ BFX80_07620 |
+ C8233_00770 |
|
Acetate/propionate kinase (EC 2.7.2.1) |
ackA | – |
+ BFX80_07615 |
+ C8233_00765 |
|
Acetate–CoA ligase (EC 6.2.1.1) |
acs |
+ IZU87_06405 |
+ BFX80_02550 |
+ C8233_13850 |
|
Propionyl-CoA synthetase (EC 6.2.1.17) |
prpE |
+ IZU87_15385* |
+ BFX80_09760 |
+ C8233_03035 |
|
Citrate synthase (EC 2.3.3.1) |
gltA |
+ IZU87_01890 |
+ BFX80_14605 |
+ C8233_08570 |
|
Aconitate hydratase (EC 4.2.1.3) |
acnA/acnB |
+/+ IZU87_02425/IZU87_05790 |
+/+ BFX80_01545/BFX80_04820 |
+/+ C8233_12860/C8233_16190 |
|
Isocitrate lyase (EC 4.1.3.1) |
AceA |
+ IZU87_14450 |
+ BFX80_03560 |
+ C8233_14950 |
|
Malate synthase (EC 2.3.3.9) |
AceB (glcB) |
+ IZU87_07285 |
+ BFX80_16805 |
+ C8233_10755 |
| Cation acetate symporter | – |
+ IZU87_06430 |
+ BFX80_02575 |
+ C8233_13875 |
| Phasin | phaP1/P2 |
+/+ IZU87_08295/IZU87_04855 |
+/+ BFX80_04055/BFX80_12115 |
+/+ C8233_15445/C8233_06045 |
| Repressor | phaR |
+ IZU87_03745 |
+ BFX80_15490 |
+ C8233_09450 |
|
3-hydroxybutyrate dehydrogenase (EC 1.1.1.30) |
bdhA |
+ IZU87_16460 |
+ BFX80_07905 |
+ C8233_01125 |
|
Oxoacid transferases (EC 2.8.3) |
scoA/scoB |
+/+ IZU87_09115/IZU87_09110 |
+/+ BFX80_11345/BFX80_11350 |
+/+ C8233_05175/C8233_05180 |
The genes were identified from genome annotations [75, 76] and BLAST search against the different genomes
+ indicates presence and number of gene(s), with locus tags, − indicate absence, [+] indicate ambiguity between the annotation methods Prokka [57] and NCBI’s Prokaryotic Annotation Pipeline [58]
*Annotated as acetate–CoA ligase (EC 6.2.1.1) by NCBI’s Prokaryotic Annotation Pipeline [58], but as propionyl-CoA synthetase (EC 6.2.1.17) by KEGG [67]
Fig. 4.
The phaC2 is the only PhaC polymerase gene in Cobetia sp. MC34. Gene synteny for the up-and downstream region of the PHA polymerase genes phaC1 and phaC2 found in selected Cobetia species. The phaCAB operon in C. necator is included for reference. The gene synteny of the phaC2 region is shared between Cobetia sp. MC34, Halomonas sp. SF2003 and Cobetia marina DSM 4741T. A putative non-homologues phaC2 variant (striped symbol) is also found in H. bluephagenesis TD01 but with different genes located in the upstream region. Gene abbreviations: phaC1/2 Poly(3-hydroxyalkanoate) polymerase subunit PhaC, PHA polymerase (EC 2.3.1.-), phaA Acetyl-CoA acetyltransferase (EC 2.3.1.9), phaB Acetoacetyl-CoA reductase (EC 1.1.1.36), phaP Phasin, atp ATP-metabolism genes, fabI (Enoyl-[acyl-carrier-protein] reductase [NADH], bi-pta bifunctional enoyl-CoA hydratase/phosphate acetyltransferase, afp Aminohydrolase family protein, hyp hypothetical protein of unknown function, lip lipoyl synthase and transferase, lysR LysR family transcriptional regulator
An acetate kinase gene ackA is furthermore found downstream of fabI and bi-pta in both C. marina DSM 4741T and Halomonas sp. SF2003 (not shown). A similar gene synteny for fabI, bi-pta, and ackA is found in H. bluephagenesis TD01 (not shown), but with the difference that phaC1 is located at a separate loci (Fig. 4). The gene fabI (IZU87_11845) is present in the genome of Cobetia sp. MC34, but the ackA and bi-pta genes were not found. This suggests that phaC1 could have been lost during evolution. The possibility that the absence of phaC1 in Cobetia sp. MC34 resulted from a genome assembly artefact was considered and evaluated by investigation of phaC variants found in other members of the Cobetia genus. Two copies of phaC are found in 8 out of 19 whole genome sequences and we consider it unlikely that assembly artefacts are accountable for the presence of only a single copy in the remaining eight genomes.
Two routes to convert propanoate (originating from valerate) to propanoyl-CoA were found in Cobetia sp. MC34: The acetate–CoA ligase Acs (acs, IZU87_06405) and the propionyl-CoA synthetase PrpE (prpE, IZY87_15385), both of which convert propanoate to propanoyl-CoA via a propionyl-adenylate intermediate [67]. The prpE (IZU87_15385) was annotated as one of two acs genes by NCBI’s Prokaryotic Annotation Pipeline [58], but KEGG annotation [67] and its high amino acid sequence similarity to PrpE in C. marina DSM 4741T and Halomonas sp. SF2003 (Additional file 1: Table S2) suggest prpE to be the most correct gene annotation (Table 2). The AckA-Pta found in C. marina DSM 4741T and Halomonas sp. SF2003 constitute a third route for generating propanoyl-CoA from propanoate but via a propanoyl phosphate intermediate [67]. The Acs and Pta-AckA are furthermore responsible for conversion of acetate to acetyl-CoA and therefore likely to be important determinants for acetate turnover into biomass and PHA in Cobetia sp. MC34 and C. marina DSM 4741T [67]. Both strains obtained high PHA production, cell densities (OD600), and high biomass accumulation when supplemented with acetate. These results might therefore be further explained by enzymes belonging to the glyoxylate shunt pathway. In essence, the glyoxylate shunt function as a shortcut in the TCA cycle where it starts with conversion of acetyl-CoA to citrate by the citrate synthase GltA (gltA, IZU87_01890) and via isocitrate by aconitate hydratases AcnA/B (acnA/B, IZU87_02425/IZU87_05790) to glyoxylate by isocitrate lyase AceA (aceA, IZU87_14450), before malate is formed by addition of an acetyl-CoA molecule by AceB (aceB, IZU87_07285) [88]. In contrast to the TCA cycle, the glyoxylate shunt does not lead to loss of CO2 and requires fewer enzymatic steps to generate malate, the precursor for biomass production [89]. In addition, a putative cation acetate symporter (IZU87_06430) was found. Taken together, these genes are likely important for explaining the good growth and high PHA production achieved using acetate as carbon source for both Cobetia sp. MC34 and C. marina DSM 4741T (Table 2).
Of note, genome annotation by Prokka [57] identified the gene fadA (IZU87_02125) as a second phaA copy in Cobetia sp. MC34. The amino acid sequence of this gene share 95.7% similarity with the PhaA annotated in C. marina DSM 4741T and 42.4% to the PhaA annotated in Halomonas sp. SF2003 (Additional file 1: Table S2). We thus consider the function of this gene to be ambiguous. The exact copy number of phaA and phaB remains elusive since no experimental evidence concerning these has been presented in Cobetia. As also shown in Additional file 1: Table S2, the highest percentage of amino acid identity was found between homologs of Cobetia sp. MC34 and Halomonas sp. SF2003, which is in agreement with the phylogenies presented in Fig. 1.
Other insights from genome analysis of Cobetia sp. MC34 related to PHA metabolism
A closer look at the genomes of Cobetia sp. MC34, Halomonas sp. SF2003 and C. marina DSM 4741T gave additional insights in regard to PHA metabolism (Table 2). Copies of the PHA synthesis repressor gene phaR (IZU87_03745) and a second phasin gene, phaP1 (IZU87_08295), were found at independent loci without proximity to any known PHA related genes. The depolymerase PhaZ is important for the biological turnover of PHA and potentially for use in processing of PHA bioplastic waste by enzymatic degradation similar to biorecycling by enzymatic degradation of polyethylene terephthalate (PET) by PET hydrolases [90]. Surprisingly, a phaZ gene was not present in Cobetia sp. MC34, in Halomonas sp. SF2003, nor in C. marina DSM 4741T (Table 2). However, the 3-hydroxybutyrate dehydrogenase BdhA (bdhA, IZU87_16460) which may interconvert the PhaZ hydroxyacyl product (e.g., (R)-3-hydroxybutanoic acid) to an oxoacid (e.g., acetoacetate) was found in all three strains (Table 2). Also, the ScoA/B (scoA/B, IZU87_09115/IZU87_09110) responsible for converting the oxoacid to oxoacyl-CoA (e.g., acetoacetyl-CoA, the substrate for PhaB) were present in all (Table 2). This indicates the existence of one or more yet unidentified PhaZ depolymerases located upstream in the PHA biodegradation pathway. Also, additional BLAST search using PhaZ sequences from P. oleovorans and H. bluephagenesis TD01 against the Cobetia sp. MC34 genome did not find any clear PhaZ matches. Several putative genes involved in fatty acid biosynthesis and fatty acid degradation (ß-oxidation) were found in the genome with a high degree of redundancy, as expected (Additional file 1: Table S3). These genes can have redundant functions in PHA metabolism and are important targets for future studies, e.g., if mcl-PHA production is attempted by genetic engineering techniques.
Cobetia PhaC2 is a class I PhaC polymerase that contains an extended C-terminus with a putative role in DNA-binding
The finding that all whole genome sequences of Cobetia encode one or two PhaCs led us to study them in more detail. PhaC1 and PhaC2 from Halomonas sp. SF2003 are experimentally confirmed to belong to class I PhaC polymerases [87, 91]. The molecular weight of Halomonas sp. SF2003 PhaC1 is calculated to be 71.5 kDa, which is close to the molecular weight for other class I PhaC polymerases such as in C. necator [20]. A homolog of the phaC2 gene in Halomonas sp. SF2003 was found in all Cobetia genomes and is the only phaC gene present in eleven out of nineteen genomes, including in Cobetia sp. MC34. The estimated sizes of the PhaC2 polymerases differ considerably from PhaC1 with predicted molecular weights ranging from 93.6 to 106.4 kDa (Table 3). All PhaC polymerases contain a PhaC box sequence (G/S)XCX(G/A)G with similarity to the lipase box motif found in lipases and PhaZ depolymerases [92]. Most important is the conserved cysteine residue in the center of the box, which constitutes the active center of the enzyme, while the X’s denounce non-conserved amino acids (AA). The PhaC boxes in Halomonas are described to contain either an SYCLG or GYCLG motif, while a GFCVGG motif is found in C. necator PhaC1 [86, 87, 91, 92]. All Cobetia PhaC1 and PhaC2 sequences analyzed in this study contained a GYCLG or SYCIG motif, respectively (Table 3).
Table 3.
Characteristics of PhaC polymerases found in Cobetia and Halomonas
| Strain | PhaC2 MW (kDa)/pIN613/pIC-terminus | PhaC2 box | HC2 domain (E-value) | PhaC1 MW (kDa)/pI | PhaC1 box |
|---|---|---|---|---|---|
| Cobetia sp. MC34 | 105.4/4.63/10.34 | SYCIG | 8.77 × 10–9 | – | – |
| Cobetia amphilecti KMM296 | 103.1/4.67/10.27 | SYCIG | 4.56 × 10–9 | – | – |
| Halomonas sp. SF2003* | 101.8/4.66/10.15 | SYCIG | 4.79 × 10–10 | 71.5/5.27 | GYCLG |
| Halomonas sp. GDM18* | 102.6/4.63/10.24 | SYCIG | 2.99 × 10–8 | 71.5/5.27 | GYCLG |
| Cobetia sp. AM6 | 102.6/4.63/10.34 | SYCIG | 3.55 × 10–9 | 71.5/5.31 | GYCLG |
| Cobetia sp. UCD-24C | 103.9/4.76/10.19 | SYCIG | 2.24 × 10–8 | – | – |
| Cobetia sp. QF1 | 95.1/4.77/10.23 | SYCIG | 3.79 × 10–11 | – | – |
| Cobetia sp. ICG0124 | 106.1/4.74/10.23 | SYCIG | 4.32 × 10–12 | 40–50 kDa* | GYCLG |
| Cobetia sp. L2A1 | 99.9/4.85/10.06 | SYCIG | 1.86 × 10–7 | 65.6/5.43 | GYCLG |
| Cobetia crustatorum SM1923 | 93.6/4.77/10.14 | SYCIG | 2.78 × 10–10 | – | – |
| Cobetia crustatorum JO1 | 95.4/4.77/10.23 | SYCIG | 3.42 × 10–11 | – | – |
| Cobetia marina DSM 4741T | 106.4/4.74/10.35 | SYCIG | 1.77 × 10–10 | 64.0/5.31 | GYCLG |
| Cobetia marina NBRC 15607 | 105.2/4.74/10.36 | SYCIG | 2.85 × 10–10 | 63.9/5.31 | GYCLG |
| Cobetia marina T1 | 105.1/4.74/10.36 | SYCIG | 2.73 × 10–11 | 64.1/5.26 | GYCLG |
| Cobetia sp. MM1IDA2H-1 | 105.0/4.74/10.40 | SYCIG | 3.39 × 10–7 | – | – |
| Cobetia pacifica GPM2 | 105.9/4.74/10.38 | SYCIG | 1.14 × 10–9 | – | – |
|
Cobetia sp. IU 180733JP01 (5-11-6-3) |
100.9/4.74/10.36 | SYCIG | 3.04 × 10–9 | – | – |
|
Cobetia sp. 190790JP01 (5-25-4-2) |
106.2/4.74/10.38 | SYCIG | 3.09 × 10–9 | – | – |
| Cobetia sp. MB87 | 100.4/4.66/10.22 | SYCIG | 2.91 × 10–10 | – | – |
| Halomonas sp. R5-57 | 71.8/4.86/8.54** | SYCVG | – | 66.7/5.29 | GYCLG |
|
Halomonas bluephagenesis TD01 |
92.8/5.39/9.60*** | GNCQAG | – | 69.7/4.80 | SYCVG |
| Halomonas sp. O-1 | 89.0/5.45/10.10*** | GNCQAG | – | 69.4/5.18 | SYCVG |
PhaC1 and PhaC2 sequences found in Cobetia whole genome sequences from genome annotation and by BLAST search. Predicted Molecular weight (MW) is shown for the complete protein sequence while the isoelectric point (pI) is shown for the N-terminal 613 amino acids (pIN613) and for the extended C-terminus (pIC-terminus) for PhaC2 and for the whole protein sequence for PhaC1.Genome accession numbers are listed in Additional file 1: Table S1
*Identified as belonging to Cobetia (this study)
**The C-terminus consist of only 21 amino acids and thus non-homologous to Cobetia PhaC2
***Putative PhaC2, non-homologous to Cobetia PhaC2 (This study)
Further analyses of the Cobetia sp. MC34 PhaC2 (993 AA) reveal high amino acid similarity to putative homologs in Halomonas sp. GDM18 (962 AA, 94.7% identity), C. amphilecti (967 AA, 94.6% identity), Halomonas sp. SF2003 (954 AA, 93.7% identity), and Cobetia sp. AM6 (961 AA, 93.4% identity). Protein BLAST also revealed high sequence similarity for amino acid positions 76–613 to the PHA_synth_I domain (NCBI CDD TIGR01838) in PhaC1 of C. necator, while no domain was identified for the remaining C-terminus similar to previous analysis of Halomonas sp. SF2003 PhaC2 [76]. However, when querying Cobetia sp. MC34 PhaC2 against NCBI’s Conserved Domain Database (NCBI CDD) using less stringent search criteria, many unspecific hits with relatively low e-values were detected. Among these were the “Histone H1-like nucleoprotein HC2 domain” (HC2 Domain, NCBI CDD pfam07382, E-value 8.77 × 10–9), the “transcriptional regulator ICP4 domain” (NCBI CDD PHA03307, E-value 0.25 × 10–13), and the “DNA polymerase III subunit gamma/tau domain” (NCBI CDD PRK07003, E-value 5.20 × 10–11). Investigation of all 19 Cobetia PhaC2 sequences identified similar non-specific hits to the “HC2 domain” in the extended C-terminus in all Cobetia PhaC2 sequences (Table 3). The “HC2 domain” was, however, not the only hit with a low E-value identified in the extended C-terminus of the Cobetia PhaC2 sequences. The domain “Herpes_BLLF1” (NCBI CDD pfam05109) was found in all, and “Endomucin” (NCBI CDD pfam07010) was found in all except C. crustatorum SM1923 PhaC2, in addition to many other non-specific hits found in most but not all PhaC2 C-termini (Additional file 2: S1).
Furthermore, comparison of the isoelectric point (pI) for the N-terminal 613 amino acids versus the extended C-terminus showed a marked difference of more than 5 pH units, with the C-terminus having a pI value of 10.34 for Cobetia sp. MC34 PhaC2, a pattern which was observed in all Cobetia PhaC2 sequences (Table 3). These high pI values for the C-termini indicate a possible role for binding negatively charged molecules such as the DNA backbone. The amino acid AAKP sequence motif is known to be prevalent in DNA-binding domains [93] and five such motifs were indeed identified in the extended C-terminus in Cobetia sp. L2A1 PhaC2, two in PhaC2 of Cobetia sp. QF1, C. crustatorum JO1, and a single AAKP motif in C. crustatorum SM1923 PhaC2.
Halomonas sp. O-1 and H. bluephagenesis TD01 encode putative PhaC2 polymerases that contain “PhaC” domains (NCBI CDD COG3243) and therefore may be homologs to Cobetia PhaC2. However, their PhaC box motifs are different and PHA was not produced when Halomonas sp. O-1 PhaC2 was expressed in E. coli [86, 87]. The domain search did not identify a “HC2 domain” in any of the two. However, hits to “Predicted 5' DNA nuclease, H3TH domain” (NCBI CDD COG3743) and “DNA polymerase III subunits gamma and tau domain” (NCBI CDD PRK14951) were identified for Halomonas sp. O-1 PhaC2, while the “histone H1-like DNA-binding protein” (NCBI CDD NF038053) was identified in H. bluephagenesis TD01 PhaC2 and indicate DNA binding properties as well (Data not shown). Other PhaC variants include the PhaC2 from Halomonas sp. R5-57 that has a slightly different PhaC box and is closer in size to the Cobetia PhaC1 (Table 3).
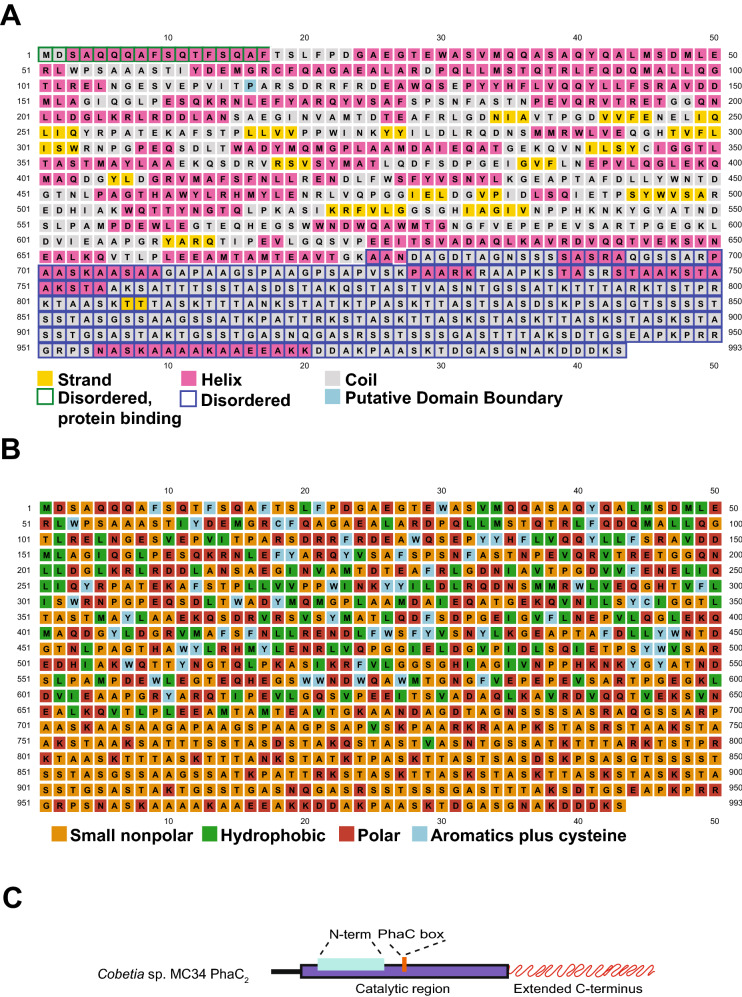
Limited tertiary and quaternary structural information is available for PhaC polymerases with only two crystal structures of the catalytic domain currently published, the PhaC1 from C. necator [94, 95] and the PhaC1 from Chromobacterium sp. USM2, with- and without co-enzyme A [96, 97]. We therefore performed in silico secondary structure predictions by PSIPRED [68, 69] to shed more light on the function of the extended C-terminus of Cobetia sp. MC34 PhaC2. The PSIPRED analysis showed presence of several strand, helix, and coiled structures in the N-terminal domain (amino acids 1 to 674). The extended C-terminus (amino acid position 614 to 993) consisted mostly of unordered coils, although ten helixes and a single strand domain were present (Fig. 5A). Further, the C-terminus domain was found to contain a high proportion of small non-polar and polar amino acids, including positively charged lysine and arginine residues, consistent with a high theoretical pI (Fig. 5B). A generalized structure of PhaC2 from Cobetia sp. MC34 is shown in Fig. 5C.
Fig. 5.
Secondary structure prediction of the PhaC2 polymerase in Cobetia sp. MC34. A The PSIPRED modeling plot predicts a high proportion of strand and helix secondary structures in the N-terminal (amino acids 1 to 674), whereas the extended C-terminus (amino acid 614 to 993) is predicted to consist primarily of unordered coiled domains, short helixes, and one strand domain. B The second PSIPRED plot shows an abundance of small non-polar and polar amino acids in the extended C-terminus including the positively charged lysine and arginine residues. C A generalized structure of the Cobetia sp. MC34 PhaC2 protein. The catalytic region is homologous to class I PhaC polymerases and contains an N-terminal PHA_synth_I domain and a SYCIG l PhaC box. The extended C-terminus (380 amino acids) has an unordered structure with a computed pI of 10.34.
Discussion
In this study, a new strain identified as Cobetia sp. MC34 isolated from a fish-landing facility in the Arctic part of Norway was investigated as a putative PHA producer. The strain showed closest relationship to C. amphilecti and Halomonas sp. SF2003 based on 16S rRNA and whole genome analysis by ANI, which suggest placement of Cobetia sp. MC34 as a strain of C. amphilecti according to current taxonomy [28]. The genus type strain C. marina DSM 4741T included in our study for comparison was the first and only Cobetia specie described after a split from genus Halomonas in 2002 [27]. Since then, the number of species allocated to the Cobetia genus has increased to five, and to 143 species within the Halomonas genus [98, 99]. In addition, the introduction of whole genome sequencing of bacteria and meta-genomes has led to deposition of hundreds of new genomes belonging to the two genera in the NCBI genome database. As a result, allocation of new strains belonging to Cobetia or Halomonas is often confusing and the nomenclature is, at times, used interchangeably. This problem is addressed by the scientific community and new standards for description of the Halomonadacea family appear to be underway [100].
Cobetia sp. MC34 and C. marina DSM 4741T were found to be mesophilic, halotolerant, and PHA producing bacteria. The ability to grow in media with low levels of salt for Cobetia sp. MC34 was somewhat unexpected, given the isolation from a cold marine-associated environment. Cobetia sp. MC34, did however, show a higher specific growth rate than C. marina DSM 4741T in the lower range of the tested temperatures, while C. marina DSM 4741T appeared to have a higher putative PHA production than Cobetia sp. MC34 at all temperatures tested, as well as a lower optimum growth temperature than previously reported [27]. The basic growth- and PHA production phenotypes of Cobetia sp. MC34 and C. marina DSM 4741T were subsequently characterized after growth on the industrially relevant pure substrates acetate, glycerol, glucose and fructose. Acetate proved to be a particularly promising carbon source for PHA production in both strains, and Cobetia sp. MC34 was able to produce 72% PHB/CDW and C. marina DSM 4741T 61% PHB/CDW on this substrate. Co-supplement of acetate with valerate further showed that the same strains were able to produce 14% PHV/CDW and 26% PHV/CDW, respectively, in addition to production of PHB (Table 1). The high natural production of PHB and PHBV when using acetate with- and without valerate in both Cobetia sp. MC34 and C. marina DSM 4741T is interesting from a biotechnological perspective. Acetate is a relatively low-cost carboxylic acid with a global production exceeding 10 million tons per year primarily synthesized from chemical routes, but synthesis of acetate is also possible from green materials such as lignocellulose feedstock or by bacterial fermentation [101]. To our knowledge, our study is the first to show PHA production from acetate in the genus Cobetia. However, others have reported efficient PHA production from acetate both naturally and by heterologous expression of acetate metabolism genes in different bacteria. For example, Halomonas boliviensis is able to produce 88% PHB/CDW when supplemented with a mixture of acetate and butyrate under optimized conditions [100] and H. bluephagenesis TD01 can produce more than 89% PHB/CDW under oxygen limitation when supplemented with a combination of acetic acid and glucose [102]. The latter study also demonstrated increased PHA production by engineering of the NADH/NAD+ pathway [102]. Neither of the studies reported PHA production from use of acetate alone, making a direct comparison with our results difficult. The industrial “workhorse” for PHA production, C. necator, is an efficient producer of PHA from acetate with production of up to 72% PHA/CDW corresponding to 43 g PHA/L achieved in a 3-L bioreactor [103]. Escherichia coli is unable to produce PHA naturally due to lack of pha biosynthesis genes. However, metabolic engineering has enabled E. coli to produce up to 42% PHB/CDW, 35% PHBV/CDW or 58% poly(3-hydroxybutyrate-co-4-hydroxybutyrate)/CDW from acetate with- or without addition of propionate [104]. Strains of Pseudomonas putida can naturally produce 20–30% mcl-PHA/CDW from acetate, and overexpression of acs has been shown to increase the yields [105, 106]. The amounts obtained in Pseudomonas are low compared to our results with Cobetia sp. MC34 and C. marina DSM 4741T, but the production is interesting from an industrial perspective due to the material properties of mcl-PHA. Taken together, these studies imply that there is an industrial potential of using acetate and different bacteria for production of PHA. Glycerol, a by-product form the biodiesel industry, is another substrate of interest for green production of PHA. Our study show that C. marina DSM 4741T produced 61% PHB/CDW from glycerol, significantly more than Cobetia sp. MC34 which produced only 26% PHB/CDW (Table 1). Halomonas sp. SF2003, the close relative to Cobetia sp. MC34, has to our knowledge not been investigated for PHA production on acetate nor glycerol. However, this strain is able to produce PHA in amounts comparable or even higher than Cobetia sp. MC34 from single carbohydrates such as glucose [91]. More important from an industrial perspective is the finding that Halomonas sp. SF2003 is able to produce approximately 78% PHA/CDW from fruit industry process water containing a mixture of fructose and glucose in addition to various ash constituents [36]. This amount is significantly higher than achieved from glucose and fructose in Cobetia sp. MC34.
It should be emphasized that the PHA productivities found in our and the above mentioned studies are still low compared to those achieved in high-density fermentation of a genetically engineered- and artificially selected strain of H. bluephagenesis TD01 where more than 71- and 56 g/L PHA were produced using glucose as substrate [107]. This demonstrates that further optimization and scale up is needed to make Cobetia sp. MC34 and C. marina DSM 4741T into industrially competitive strains for PHA production.
The somewhat lower production of PHB from acetate but higher production of PHV when co-supplemented with valerate in C. marina DSM 4741T compared to Cobetia sp. MC34 could be due to the lack of the acetate kinase ackA in the draft genome of Cobetia sp. MC34 (Table 2). The acetate kinase AckA and the phosphate acetyltransferase Pta in C. marina DSM 4741T offers an additional route to formation of propanoyl-CoA from propanoate via a propanoyl-phosphate intermediate in addition to the Acs and PrpE routes (Table 2) [67]. This may explain why C. marina DSM 4741T was found to produce a higher content of PHV than Cobetia sp. MC34 when co-supplemented with valerate, since propanoyl-CoA is the precursor converted to 3-ketovaleryl-CoA by PhaA, which can be further converted to (R)-3-hydroxyvaleryl-CoA by PhaB, and subsequently to PHBV by PhaC. The apparent lack of ackA in Cobetia sp. MC34 means that only the high-affinity irreversible Acs route is possible for converting acetate to acetyl-CoA, whereas C. marina DSM 4741T can also use the low-affinity but reversible Pta-AckA route [108]. Thus, C. marina DSM 4741T might redirect acetyl-CoA back to acetate whereas Cobetia sp. MC34 must redirect excess acetyl-CoA to other routes, such as PHB production. A closed genome sequence of Cobetia sp. MC34 in combination with transcriptomic data is needed to verify if the apparent lack of ackA can in fact explain the observed differences in PHA production between the two strains using acetate with- and without co-supplement with valerate. Another possible explanation for differences in PHA production, such as when using glycerol, may be due to different phaC copy numbers (Table 2). Having an additional PhaC variant may make C. marina DSM 4741T more robust and versatile in terms of PHA production. Interestingly, the study of Halomonas sp. SF2003 indicates that PhaC2 is capable of producing PHA from a higher number of carbon substrates than PhaC1 [91]. It remains, however, to be shown if the phaC variants in our strains are expressed differently upon feeding with different substrates.
The genome analyses of Cobetia sp. MC34 and C. marina DSM 4741T identified all genes required for PHA production in both strains and the class I PHA polymerase PhaC2 was found to contain a long (> 300 amino acids) extended C-terminus of unknown function. This combined with the finding that phaC2 is the only phaC gene present in Cobetia sp. MC34, as well as in eleven other Cobetia members, made it an interesting candidate for further in silico studies.
The PSIPRED modeling of the secondary structure of Cobetia sp. MC34 PhaC2 predicted that the C-terminal region consist mostly of unordered coils, without apparent structure. Coiled structures are found ubiquitously in protein structures of all domains of life and are attributed to various biological functions. Several functions are therefore possible for the Cobetia PhaC2 extended C-terminus such as oligo- or dimerization domain, activation- or repression domain, chromosome segregating factor, molecular ruler, or as a transcription factor like DNA-binding domain [109]. It is generally established that PhaC polymerases from model organisms function as homo- or hetero-dimers [20]. PhaC2 is expected to localize at the carbonosome surface together with phasins and other proteins, from where PHA is produced and regulated, e.g., via a PhaM-like mediated activation, as described for C. necator [110]. The nucleoid is suggested to serve as scaffold for initiation of PHA synthesis via PhaC-PhaM binding, which allows for the carbonosome to segregate into daughter cells during cell division [110, 111]. The importance of PhaM for subcellular localization of the carbonosome and its distribution to daughter cells in C. necator was most recently demonstrated at the single-cell level using C. necator PhaM expressed in E. coli, where the E. coli gain-of-function mutant exhibited similar “behavior” as observed in C. necator [112]. Thus, although a PhaM homolog is absent in Cobetia sp. MC34, the possibility for the coiled structure in PhaC2 to serve as a nucleoid anchor in replacement of PhaM may be speculated. The PHA synthesis repressor PhaR from Paracoccus denitrificans has also been found to contain a short N-terminal DNA-binding domain, although the high proportion of phasins in the carbonosome suggested that it is more likely to be the free form of PhaR that binds to DNA, rather than a PhaR-carbonosome complex [113]. Similar to Halomonas sp. SF2003 [76], genome analysis of Cobetia sp. MC34 and C. marina DSM 4741T identified phaR. However, interaction of PhaR with the carbonosome and chromosome needs to be addressed by in vivo studies. A DNA binding mechanism for PhaC and the carbonosome is also described in bacteria more closely related to Cobetia, such as in P. oleovorans. Here, PhaF is responsible for repression of phaC and phaI and is found located on PHA granules [114]. PhaF contains AAKP motifs that are also found in alginate transcriptional regulators and in histones [93]. Interestingly, we identified four Cobetia PhaC2 sequences that contain between one and five AAKP motifs in the extended C-terminus. The possibility for the Cobetia PhaC2 extended C-terminus to function in DNA-binding was further substantiated by our domain search, which identified the “Histone H1-like nucleoprotein HC2 domain” (HC2 Domain, NCBI CDD pfam07382) in all Cobetia PhaC2 sequences. Hits to several other domains including “Herpes_BLLF1” (NCBI CDD pfam05109) and “Endomucin” (NCBI CDD pfam07010) which are described in NCBI CDD as types of membrane proteins makes a role in anchoring the Cobetia PhaC2 synthase to the granule surface a possibility as well. Based on these considerations and in silico data, we propose that the extended C-terminus found in Cobetia PhaC2 could be responsible for binding of the PHA carbonosome to the bacterial DNA.
The apparent lack of the PHA depolymerase gene phaZ in the genome of Cobetia sp. MC34 is comparable to recent findings from a consortium of marine bacteria capable of degrading PHA, where the phaZ gene was predicted in only 3 out of 33 strains from 7 genera (3). Annotations of phaZ in Halomonas and Cobetia additionally suffers from limited experimental validation as the three phaZ sequences annotated in H. bluephagenesis TD01 were not supported by non-PHA degrading phenotypes in knock-down experiments [115]. Identification of the Halomonas and Cobetia PhaZ depolymerases may enable research on biological recycling of PHA bioplastic and thus constitute important future biotechnological research targets. Both Cobetia sp. MC34 and C. marina DSM 4741T are natural producers of PHA and potential candidates in the field of blue biotechnology with their salt- and alkali tolerance, mesophilic growth pattern and PHA production from green substrates. This may allow for upscaling experiments using standard fermentation equipment in order to increase biomass- and PHA production to industrially relevant titers. It remains, however, an open question whether megatons industrial scale production of PHA is economically and environmentally competitive with, e.g., chemical recycling of “oil plastics” produced from biological source material. Even if not, production of PHA monomers for use as green platform chemicals convertible into, e.g., crotonic acid [116] might prove just as important in a future green biorefinery that utilizes a spectrum of biological processes for substitution of fossil petroleum.
Conclusion
Cobetia sp. MC34 and Cobetia marina DSM 4741T are producers of scl-PHA from pure substrates obtainable from “green” sources, such as acetate. However, further optimization of growth conditions will be needed to increase biomass- and PHA production to industrially relevant titers. Optimization studies may also include genetic engineering. The observed difference in acetate metabolism genes as well as the presence of one or two copies of phaC in Cobetia warrant further studies to elucidate their impact on PHA biosynthesis. An interesting PhaC2 polymerase variant with an extended C-terminus was present in all Cobetia genomes investigated. Based on in silico analyses, we speculate that the Cobetia PhaC2 extended C-terminus is involved in DNA-binding.
Supplementary Information
Additional file 1: Figure S1. Tolerance- and requirement to sodium chloride in MMCY media supplemented with 2 % glucose at 30 °C, 200 rpm, shows highest specific growth rate in the range of 1-4 % for both strains (n = 6). Figure S2. Representative raw FTIR spectra for Cobetia sp. MC34 (blue) and Cobetia marina DSM4741T (orange) grown in MMCY and MMCY_2 media supplemented with 1.5 % glycerol, 4 % glucose, 4 % fructose or 20 g/l acetate + 8 mM sodium valerate (n = 1). Figure S3. Growth curves measured by optical density (OD600nm) for the strains Cobetia sp. MC34 (blue) and Cobetia marina DSM 4741T (orange) grown in MMCY media supplemented with 40 g/l glucose or fructose, respectively. The carbonyl-ester to amide band I-ester ratio obtained by FTIR is visualized as sphere area and indicate putative PHA production, which was determined at the end of the experiment by chloroform extraction and GC/MS quantification. Error bars represent standard deviation of OD600nm (n = 6). Table S1. List of Halomonadacea sequences used to construct the 16S- and ANI trees, and information about the strains. Table S2. Amino acid sequence similarities of the transporters, enzymes, transcriptional regulators and carbonosome proteins involved in PHA synthesis from acetate. Table S3. Fatty acid biosynthesis (FAS) and fatty acid degradation (ß-oxidation) genes found in Cobetia sp. MC34, derived from the genome annotation.
Acknowledgements
The authors would like to thank the following colleagues at UiT – The Arctic University of Norway: Prof. Arne Smalås, for initiating the MARPLAST project, Susann Skagseth, for help with microbial growth assays, and Seila Pandur for help with strain isolation and preparation of genomic DNA. The authors also acknowledge the Norwegian Sequencing Centre (Oslo) and the Advanced Microscopy Core Facility at UiT – The Arctic University of Norway.
Abbreviations
- AA
Amino acids
- ANI
Average Nucleotide Identity
- BLAST
Basic Local Alignment Search Tool
- bp
Base pairs
- CDW
Cell dry weight
- FT-IR
Fourier transform infrared spectroscopy
- GC-FID
Gas Chromatography-Flame Ionization Detector
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- mcl
Medium chain length
- ML
Maximum likelihood
- MW
Molecular weight
- NCBI
National Center for Biotechnology Information
- (NCBI) CDD
Conserved Domain Database
- NMR
Nuclear magnetic resonance
- PHA
Polyhydroxyalkanoates
- PHB
Poly(3-hydroxybutyrate)
- PHBV
Poly(3-hydroxybutyrate-co-3-hydroxyvalerate)
- pI
Isoelectric point
- PLA
Polylactic acid
- PBS
Phosphate buffered saline
- scl
Short chain length
- TCA
Tricarboxylic acid
- 3-HV
3-Hydroxyvalerate
- 3-HB
3-Hydroxybutyrate
Authors' contributions
HH, BA and KI conceived the study. MC performed the genome assembly and annotation, bioinformatics, phylogenetic analyzes, wet lab experiments, microscopy and FT-IR analyses. PJ performed the GC-FID and NMR. MC and BA identified and performed the in silico analyses of PhaC2. BA and HH did initial isolation of MC34. MC and HH drafted the manuscript. All authors edited and approved the manuscript before submission.
Funding
This work was funded by Grants from the Norwegian Research Council (MarPlast, Project 270308) and the Swedish Research Council for Sustainable Development (Project 2016-02011), both parts of the ERA-NET Marine Biotechnology framework. The work was also funded from a strategic program (MarVal project) at UiT – The Arctic University of Norway. The publication charges for this article have been funded by a grant from the publication fund at UiT – The Arctic University of Norway.
Availability of data and materials
The datasets used and/or analyzed in this study are available from the corresponding author on reasonable request. Genome data for Cobetia sp. MC34 is available from NCBI under accessions listed in the “Methods” section “Genome sequencing and assembly”.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mikkel Christensen, Email: mikkel.christensen@uit.no.
Piotr Jablonski, Email: piotr.jablonski@umu.se.
Bjørn Altermark, Email: bjorn.altermark@uit.no.
Knut Irgum, Email: knut.irgum@umu.se.
Hilde Hansen, Email: hilde.hansen@uit.no.
References
- 1.De Santi C, Altermark B, de Pascale D, Willassen NP. Bioprospecting around Arctic islands: marine bacteria as rich source of biocatalysts. J Basic Microbiol. 2016;56(3):238–253. doi: 10.1002/jobm.201500505. [DOI] [PubMed] [Google Scholar]
- 2.Meereboer KW, Pal AK, Cisneros-Lopez EO, Misra M, Mohanty AK. The effect of natural fillers on the marine biodegradation behaviour of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) (PHBV) Sci Rep. 2021;11(1):911. doi: 10.1038/s41598-020-78122-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vogel FA, Schlundt C, Stote RE, Ratto JA, Amaral-Zettler LA. Comparative genomics of marine bacteria from a historically defined plastic biodegradation consortium with the capacity to biodegrade polyhydroxyalkanoates. Microorganisms. 2021;9(1):186. doi: 10.3390/microorganisms9010186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bugnicourt E, Cinelli P, Lazzeri A, Alvarez V. Polyhydroxyalkanoate (PHA): review of synthesis, characteristics, processing and potential applications in packaging. Express Polym Lett. 2014;8(11):791–808. [Google Scholar]
- 5.European Bioplastics, Nova institute. Bioplastics facts and figures. Report. www.european-bioplastics.org/market/; 2019.
- 6.Tan D, Wang Y, Tong Y, Chen GQ. Grand challenges for industrializing polyhydroxyalkanoates (PHAs) Trends Biotechnol. 2021;39:953–963. doi: 10.1016/j.tibtech.2020.11.010. [DOI] [PubMed] [Google Scholar]
- 7.Rehm BH. Bacterial polymers: biosynthesis, modifications and applications. Nat Rev Microbiol. 2010;8(8):578–592. doi: 10.1038/nrmicro2354. [DOI] [PubMed] [Google Scholar]
- 8.Tsuge T. Fundamental factors determining the molecular weight of polyhydroxyalkanoate during biosynthesis. Polym J. 2016;48(11):1051–1057. [Google Scholar]
- 9.Visakh PM. CHAPTER 1. Polyhydroxyalkanoates (PHAs), their blends, composites and nanocomposites: state of the art, new challenges and opportunities. In: Ipsita R, Visakh PM, editors. Polyhydroxyalkanoate (PHA) based blends, composites and nanocomposites. London: Royal Society of Chemistry; 2014. pp. 1–17. [Google Scholar]
- 10.Kutralam-Muniasamy G, Corona-Hernandez J, Narayanasamy RK, Marsch R, Perez-Guevara F. Phylogenetic diversification and developmental implications of poly-(R)-3-hydroxyalkanoate gene cluster assembly in prokaryotes. FEMS Microbiol Lett. 2017. 364(13). [DOI] [PubMed]
- 11.Macrae RMW, et al. PHB metabolism in washed suspensions of Bacillus cereus and Bacillus megaterium. J General Microbiol. 1958;19:210–22. doi: 10.1099/00221287-19-1-210. [DOI] [PubMed] [Google Scholar]
- 12.Koskimäki JJ, Kajula M, Hokkanen J, Ihantola EL, Kim JH, Hautajarvi H, et al. Methyl-esterified 3-hydroxybutyrate oligomers protect bacteria from hydroxyl radicals. Nat Chem Biol. 2016;12(5):332–338. doi: 10.1038/nchembio.2043. [DOI] [PubMed] [Google Scholar]
- 13.Slaninova E, Sedlacek P, Mravec F, Mullerova L, Samek O, Koller M, et al. Light scattering on PHA granules protects bacterial cells against the harmful effects of UV radiation. Appl Microbiol Biotechnol. 2018;102(4):1923–1931. doi: 10.1007/s00253-018-8760-8. [DOI] [PubMed] [Google Scholar]
- 14.Tribelli PM, Pezzoni M, Brito MG, Montesinos NV, Costa CS, Lopez NI. Response to lethal UVA radiation in the Antarctic bacterium Pseudomonas extremaustralis: polyhydroxybutyrate and cold adaptation as protective factors. Extremophiles. 2020;24(2):265–275. doi: 10.1007/s00792-019-01152-1. [DOI] [PubMed] [Google Scholar]
- 15.Obruca S, Sedlacek P, Koller M, Kucera D, Pernicova I. Involvement of polyhydroxyalkanoates in stress resistance of microbial cells: biotechnological consequences and applications. Biotechnol Adv. 2018;36(3):856–870. doi: 10.1016/j.biotechadv.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 16.Kaartokallio H, Søgaard DH, Norman L, Rysgaard S, Tison JL, Delille B, et al. Short-term variability in bacterial abundance, cell properties, and incorporation of leucine and thymidine in subarctic sea ice. Aquat Microb Ecol. 2013;71(1):57–73. [Google Scholar]
- 17.Pärnänen K, Karkman A, Virta M, Eronen-Rasimus E, Kaartokallio H. Discovery of bacterial polyhydroxyalkanoate synthase (PhaC)-encoding genes from seasonal Baltic Sea ice and cold estuarine waters. Extremophiles. 2015;19(1):197–206. doi: 10.1007/s00792-014-0699-9. [DOI] [PubMed] [Google Scholar]
- 18.Mezzina MP, Manoli MT, Prieto MA, Nikel PI. Engineering native and synthetic pathways in Pseudomonas putida for the production of tailored polyhydroxyalkanoates. Biotechnol J. 2020. 16(3). [DOI] [PubMed]
- 19.Tan IKP, Foong CP, Tan HT, Lim H, Zain NA, Tan YC, et al. Polyhydroxyalkanoate (PHA) synthase genes and PHA-associated gene clusters in Pseudomonas spp. and Janthinobacterium spp. isolated from Antarctica. J Biotechnol. 2020;313:18–28. doi: 10.1016/j.jbiotec.2020.03.006. [DOI] [PubMed] [Google Scholar]
- 20.Mezzolla V, D'Urso OF, Poltronieri P. Role of PhaC type I and type II enzymes during PHA biosynthesis. Polymers (Basel) 2018;10(8):910. doi: 10.3390/polym10080910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maestro B, Sanz JM. Polyhydroxyalkanoate-associated phasins as phylogenetically heterogeneous, multipurpose proteins. Microb Biotechnol. 2017;10(6):1323–1337. doi: 10.1111/1751-7915.12718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jendrossek D. Polyhydroxyalkanoate granules are complex subcellular organelles (carbonosomes) J Bacteriol. 2009;191(10):3195–3202. doi: 10.1128/JB.01723-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yue HL, Yang T, Chen X, Chen Y, Deng H, Wu Q, Chen J, Chen G-Q. A seawater-based open and continuous process for polyhydroxyalkanoates production by recombinant Halomonas campaniensis LS21 grown in mixed substrates. Biotechnol Biofuels. 2014;7:108. [Google Scholar]
- 24.El-Malek FA, Farag A, Omar S, Khairy H. Polyhydroxyalkanoates (PHA) from Halomonas pacifica ASL10 and Halomonas salifodiane ASL11 isolated from Mariout salt lakes. Int J Biol Macromol. 2020;161:1318–1328. doi: 10.1016/j.ijbiomac.2020.07.258. [DOI] [PubMed] [Google Scholar]
- 25.Williamson A, De Santi C, Altermark B, Karlsen C, Hjerde E. Complete genome sequence of Halomonas sp. R5–57. Stand Genomic Sci. 2016;11(1):62. doi: 10.1186/s40793-016-0192-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yin J, Chen JC, Wu Q, Chen GQ. Halophiles, coming stars for industrial biotechnology. Biotechnol Adv. 2015;33(7):1433–1442. doi: 10.1016/j.biotechadv.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 27.Arahal DR, Castillo AM, Ludwig W, Schleifer KH, Ventosa A. Proposal of Cobetia marina gen. nov., comb. Nov., within the family Halomonadaceae, to include the species Halomonas marina. Syst Appl Microbiol. 2002;25(2):207–211. doi: 10.1078/0723-2020-00113. [DOI] [PubMed] [Google Scholar]
- 28.Romanenko LA, Tanaka N, Svetashev VI, Falsen E. Description of Cobetia amphilecti sp. nov., Cobetia litoralis sp. nov. and Cobetia pacifica sp. nov., classification of Halomonas halodurans as a later heterotypic synonym of Cobetia marina and emended descriptions of the genus Cobetia and Cobetia marina. Int J Syst Evol Microbiol. 2013;63(Pt 1):288–297. doi: 10.1099/ijs.0.036863-0. [DOI] [PubMed] [Google Scholar]
- 29.Kim MS, Roh SW, Bae JW. Cobetia crustatorum sp. nov., a novel slightly halophilic bacterium isolated from traditional fermented seafood in Korea. Int J Syst Evol Microbiol. 2010;60:620–626. doi: 10.1099/ijs.0.008847-0. [DOI] [PubMed] [Google Scholar]
- 30.Cobet AB, Wirsen C, Jones GE. The effect of nickel on a marine bacterium Arthrobacter marinus sp. nov. J General Microbiol. 1970;62:159–169. doi: 10.1099/00221287-62-2-159. [DOI] [PubMed] [Google Scholar]
- 31.Baumann L, Baumann P, Mandel M, Allen RD. Taxonomy of aerobic marine eubacteria. J Bacteriol. 1972;110(1):402–429. doi: 10.1128/jb.110.1.402-429.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baumann L, Bowditch RD, Baumann P. Description of Deleya gen. nov. created to accommodate the marine species Alcaligenes aestus, A. pacijicus, A. cupidus, A. venustus, and Pseudomonas marina. Int J Syst Bacteriol. 1983;33(4):793–802. [Google Scholar]
- 33.Dobson SJF. Unification of the Genera Deleya (Baumann et al. 1983), Halomonas (Vreeland et al. 1980), and Halovibrio (Fendrich 1988) and the Species Paracoccus halodenitrificans (Robinson and Gibbons 1952) into a single genus, Halomonas, and placement of the genus Zymobacter in the Family Halomonadaceae. International J Syst Bacteriol. 1996;46(2):550–8.
- 34.de la Haba RR, Arahal DR, Marquez MC, Ventosa A. Phylogenetic relationships within the family Halomonadaceae based on comparative 23S and 16S rRNA gene sequence analysis. Int J Syst Evol Microbiol. 2010;60(Pt 4):737–748. doi: 10.1099/ijs.0.013979-0. [DOI] [PubMed] [Google Scholar]
- 35.Ivanova EPC, Sawabe T, Alexeeva YV, Lysenko AM, Chelomin VP, Mihailov VV. Presence of ecophysiologically diverse populations Cobetia marina strains isolated from marine invertebrate, algae and the environments. Microb Environ. 2005;20(4):200–207. [Google Scholar]
- 36.Elain A, Le Grand A, Corre Y-M, Le Fellic M, Hachet N, Le Tilly V, et al. Valorisation of local agro-industrial processing waters as growth media for polyhydroxyalkanoates (PHA) production. Ind Crops Prod. 2016;80:1–5. [Google Scholar]
- 37.Elain A, Le Fellic M, Corre YM, Le Grand A, Le Tilly V, Audic JL, et al. Rapid and qualitative fluorescence-based method for the assessment of PHA production in marine bacteria during batch culture. World J Microbiol Biotechnol. 2015;31(10):1555–1563. doi: 10.1007/s11274-015-1904-4. [DOI] [PubMed] [Google Scholar]
- 38.Lemechko P, Le Fellic M, Bruzaud S. Production of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) using agro-industrial effluents with tunable proportion of 3-hydroxyvalerate monomer units. Int J Biol Macromol. 2019;128:429–434. doi: 10.1016/j.ijbiomac.2019.01.170. [DOI] [PubMed] [Google Scholar]
- 39.Biswas A, Patra A, Paul AK. Production of poly-3-hydroxyalkanoic acids by a moderately halophilic bacterium, Halomonas marina HMA 103 isolated from solar saltern of Orissa, India. Acta Microbiol Immunol Hung. 2009;56(2):125–143. doi: 10.1556/AMicr.56.2009.2.9. [DOI] [PubMed] [Google Scholar]
- 40.Moriya H, Takita Y, Matsumoto A, Yamahata Y, Nishimukai M, Miyazaki M, et al. Cobetia sp. bacteria, which are capable of utilizing alginate or waste Laminaria sp. for poly(3-hydroxybutyrate) synthesis, isolated from a marine environment. Front Bioeng Biotechnol. 2020;8:974. doi: 10.3389/fbioe.2020.00974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gnaim R, Polikovsky M, Unis R, Sheviryov J, Gozin M, Golberg A. Marine bacteria associated with the green seaweed Ulva sp. for the production of polyhydroxyalkanoates. Bioresour Technol. 2021;328:124815. doi: 10.1016/j.biortech.2021.124815. [DOI] [PubMed] [Google Scholar]
- 42.Xiao N, Jiao N. Formation of polyhydroxyalkanoate in aerobic anoxygenic phototrophic bacteria and its relationship to carbon source and light availability. Appl Environ Microbiol. 2011;77(21):7445–7450. doi: 10.1128/AEM.05955-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Juengert J, Bresan S, Jendrossek D. Determination of polyhydroxybutyrate (PHB) content in Ralstonia eutropha using gas chromatography and nile red staining. Bio-Protoc. 2018;8(5):e2748. doi: 10.21769/BioProtoc.2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Isak I, Patel M, Riddell M, West M, Bowers T, Wijeyekoon S, et al. Quantification of polyhydroxyalkanoates in mixed and pure cultures biomass by Fourier transform infrared spectroscopy: comparison of different approaches. Lett Appl Microbiol. 2016;63(2):139–146. doi: 10.1111/lam.12605. [DOI] [PubMed] [Google Scholar]
- 45.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S Ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173(2):697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010.
- 47.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wood DES. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wood DE, Lu J, Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20(1):257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bushnell B, Rood J, Singer E. BBMerge - accurate paired shotgun read merging via overlap. PLoS ONE. 2017;12(10):e0185056. doi: 10.1371/journal.pone.0185056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seppey M, Manni M, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness. Methods in molecular biology, Springer Protocols. 2019; 1962. [DOI] [PubMed]
- 53.Wick RR, Schultz MB, Zobel J, Holt KE. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 2015;31(20):3350–3352. doi: 10.1093/bioinformatics/btv383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29(8):1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 58.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44(14):6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 61.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 63.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 64.Letunic I, Bork P. Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47(W1):W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pritchard L, Glover RH, Humphris S, Elphinstone JG, Toth IK. Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal Methods. 2016;8(1):12–24. [Google Scholar]
- 66.Altschul SF, Gish PW, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 67.Kanehisa M, Sato Y. KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci. 2020;29(1):28–35. doi: 10.1002/pro.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 69.Buchan DWA, Jones DT. The PSIPRED protein analysis workbench: 20 years on. Nucleic Acids Res. 2019;47(W1):W402–W407. doi: 10.1093/nar/gkz297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32:W327–W331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, et al. CDD: NCBI's conserved domain database. Nucleic Acids Res. 2015;43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, et al. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017;45(D1):D200–D203. doi: 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lu S, Wang J, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, et al. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 2020;48(D1):D265–D268. doi: 10.1093/nar/gkz991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tang X, Xu K, Han X, Mo Z, Mao Y. Complete genome of Cobetia marina JCM 21022T and phylogenomic analysis of the family Halomonadaceae. J Oceanol Limnol. 2017;36(2):528–536. [Google Scholar]
- 76.Thomas T, Elain A, Bazire A, Bruzaud S. Complete genome sequence of the halophilic PHA-producing bacterium Halomonas sp. SF2003: insights into its biotechnological potential. World J Microbiol Biotechnol. 2019;35(3):50. doi: 10.1007/s11274-019-2627-8. [DOI] [PubMed] [Google Scholar]
- 77.Richter MaR-M R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ciufo S, Kannan S, Sharma S, Badretdin A, Clark K, Turner S, et al. Using average nucleotide identity to improve taxonomic assignments in prokaryotic genomes at the NCBI. Int J Syst Evol Microbiol. 2018;68(7):2386–2392. doi: 10.1099/ijsem.0.002809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gorenflo V, Steinbüchel A, Marose S, Rieseberg M, Scheper T. Quantification of bacterial polyhydroxyalkanoic acids by Nile red staining. Appl Microbiol Biotechnol. 1999;51:765–772. doi: 10.1007/s002530051460. [DOI] [PubMed] [Google Scholar]
- 80.Aleman-Nava GS, Cuellar-Bermudez SP, Cuaresma M, Bosma R, Muylaert K, Ritmann BE, et al. How to use Nile Red, a selective fluorescent stain for microalgal neutral lipids. J Microbiol Methods. 2016;128:74–79. doi: 10.1016/j.mimet.2016.07.011. [DOI] [PubMed] [Google Scholar]
- 81.Arcos-Hernandez MV, Gurieff N, Pratt S, Magnusson P, Werker A, Vargas A, et al. Rapid quantification of intracellular PHA using infrared spectroscopy: an application in mixed cultures. J Biotechnol. 2010;150(3):372–379. doi: 10.1016/j.jbiotec.2010.09.939. [DOI] [PubMed] [Google Scholar]
- 82.Oren A. Microbial life at high salt concentrations: phylogenetic and metabolic diversity. Saline Syst. 2008;4:2. doi: 10.1186/1746-1448-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Smirnova M, Miamin U, Kohler A, Valentovich L, Akhremchuk A, Sidarenka A, et al. Isolation and characterization of fast-growing green snow bacteria from coastal East Antarctica. MicrobiologyOpen. 2020;11:e1152. doi: 10.1002/mbo3.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chang CK, Wang HD, Lan JC. Investigation and characterization of plasma-treated poly(3-hydroxybutyrate) and poly(3-hydroxybutyrate-co-3-hydroxyvalerate) biopolymers for an in vitro cellular study of mouse adipose-derived stem cells. Polymers (Basel). 2018;10(4):355. doi: 10.3390/polym10040355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Peoples OP, Sinskey AJ. Poly-β-hydroxybutyrate (PHB) biosynthesis in Alcaligenes eutrophus H16. J Biol Chem. 1989;264(26):15298–15303. [PubMed] [Google Scholar]
- 86.Cai LT, Aibaidula G, Dong X-R, Chen J-C, Tian W-D, Chen G-Q. Comparative genomics study of polyhydroxyalkanoates (PHA) and ectoine relevant genes from Halomonas sp. TD01 revealed extensive horizontal gene transfer events and co-evolutionary relationships. Microb Cell Fact. 2011;10:88. doi: 10.1186/1475-2859-10-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ilham M, Nakanomori S, Kihara T, Hokamura A, Matsusaki H, Tsuge T, et al. Characterization of polyhydroxyalkanoate synthases from Halomonas sp. O-1 and Halomonas elongata DSM2581: site-directed mutagenesis and recombinant expression. Polym Degrad Stability. 2014;109:416–423. [Google Scholar]
- 88.Kornberg HL, Krebs HA. Synthesis of cell constituents from C2-units by a modified tricarboxylic acid cycle. Nature. 1957;179:988–991. doi: 10.1038/179988a0. [DOI] [PubMed] [Google Scholar]
- 89.Dolan SK, Welch M. The glyoxylate shunt, 60 years on. Annu Rev Microbiol. 2018;72:309–330. doi: 10.1146/annurev-micro-090817-062257. [DOI] [PubMed] [Google Scholar]
- 90.Kawai F, Kawabata T, Oda M. Current state and perspectives related to the polyethylene terephthalate hydrolases available for biorecycling. ACS Sustain Chem Eng. 2020;8(24):8894–908. [Google Scholar]
- 91.Thomas T, Sudesh K, Bazire A, Elain A, Tan HT, Lim H, et al. PHA production and PHA synthases of the halophilic bacterium Halomonas sp. SF2003. Bioengineering (Basel). 2020;7(1):1. doi: 10.3390/bioengineering7010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nambu Y, Ishii-Hyakutake M, Harada K, Mizuno S, Tsuge T. Expanded amino acid sequence of the PhaC box in the active center of polyhydroxyalkanoate synthases. FEBS Lett. 2020;594(4):710–716. doi: 10.1002/1873-3468.13651. [DOI] [PubMed] [Google Scholar]
- 93.Medvedkin EA, Klimenko VL, Mitin YV, Matsushima N, Nakayama S, Kretsinger RH. Interactions of (Ala*Ala*Lys*Pro)n and (Lys*Lys*Ser*Pro)n with DNA Proposed coiled-coil structure of AlgR3 and AlgP from Pseudomonas aeruginosa. Protein Eng Design Select. 1995;8(1):63–70. doi: 10.1093/protein/8.1.63. [DOI] [PubMed] [Google Scholar]
- 94.Wittenborn EC, Jost M, Wei Y, Stubbe J, Drennan CL. Structure of the catalytic domain of the class I polyhydroxybutyrate synthase from Cupriavidus necator. J Biol Chem. 2016;291(48):25264–25277. doi: 10.1074/jbc.M116.756833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kim J, Kim YJ, Choi SY, Lee SY, Kim KJ. Crystal structure of Ralstonia eutropha polyhydroxyalkanoate synthase C-terminal domain and reaction mechanisms. Biotechnol J. 2017;12(1):1. doi: 10.1002/biot.201600648. [DOI] [PubMed] [Google Scholar]
- 96.Chek MF, Kim SY, Mori T, Tan HT, Sudesh K, Hakoshima T. Asymmetric open-closed dimer mechanism of polyhydroxyalkanoate synthase PhaC. Iscience. 2020;23(5):101084. doi: 10.1016/j.isci.2020.101084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chek MF, Kim SY, Mori T, Arsad H, Samian MR, Sudesh K, et al. Structure of polyhydroxyalkanoate (PHA) synthase PhaC from Chromobacterium sp USM2, producing biodegradable plastics. Sci Rep. 2017;7(1):5312. doi: 10.1038/s41598-017-05509-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.LPSN List of Prokaryotic names with Standing in Nomenclature [31.05.2021]. https://lpsn.dsmz.de/. [DOI] [PMC free article] [PubMed]
- 99.Parte AC, Sarda Carbasse J, Meier-Kolthoff JP, Reimer LC, Goker M. List of Prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int J Syst Evol Microbiol. 2020;70(11):5607–5612. doi: 10.1099/ijsem.0.004332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Oren A, Ventosa A. International Committee on Systematics of Prokaryotes subcommittee on the taxonomy of Halobacteria and subcommittee on the taxonomy of Halomonadaceae. Minutes of the joint open meeting, 26 June 2019, Cluj-Napoca, Romania. Int J Syst Evol Microbiol. 2019;69(11):3657–3661. doi: 10.1099/ijsem.0.003737. [DOI] [PubMed] [Google Scholar]
- 101.Lim HG, Lee JH, Noh MH, Jung GY. Rediscovering acetate metabolism: its potential sources and utilization for biobased transformation into value-added chemicals. J Agric Food Chem. 2018;66(16):3998–4006. doi: 10.1021/acs.jafc.8b00458. [DOI] [PubMed] [Google Scholar]
- 102.Ling C, Qiao GQ, Shuai BW, Olavarria K, Yin J, Xiang RJ, et al. Engineering NADH/NAD(+) ratio in Halomonas bluephagenesis for enhanced production of polyhydroxyalkanoates (PHA) Metab Eng. 2018;49:275–286. doi: 10.1016/j.ymben.2018.09.007. [DOI] [PubMed] [Google Scholar]
- 103.Garcia-Gonzalez L, De Wever H. Acetic acid as an indirect sink of CO2 for the synthesis of polyhydroxyalkanoates (PHA): comparison with PHA production processes directly using CO2 as feedstock. Appl Sci. 2018;8(9):1416. [Google Scholar]
- 104.Chen J, Li W, Zhang ZZ, Tan TW, Li ZJ. Metabolic engineering of Escherichia coli for the synthesis of polyhydroxyalkanoates using acetate as a main carbon source. Microb Cell Fact. 2018;17(1):102. doi: 10.1186/s12934-018-0949-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yang S, Li S, Jia X. Production of medium chain length polyhydroxyalkanoate from acetate by engineered Pseudomonas putida KT2440. J Ind Microbiol Biotechnol. 2019;46(6):793–800. doi: 10.1007/s10295-019-02159-5. [DOI] [PubMed] [Google Scholar]
- 106.Shahid S, Mosrati R, Ledauphin J, Amiel C, Fontaine P, Gaillard JL, et al. Impact of carbon source and variable nitrogen conditions on bacterial biosynthesis of polyhydroxyalkanoates: evidence of an atypical metabolism in Bacillus megaterium DSM 509. J Biosci Bioeng. 2013;116(3):302–308. doi: 10.1016/j.jbiosc.2013.02.017. [DOI] [PubMed] [Google Scholar]
- 107.Ren Y, Ling C, Hajnal I, Wu Q, Chen GQ. Construction of Halomonas bluephagenesis capable of high cell density growth for efficient PHA production. Appl Microbiol Biotechnol. 2018;102(10):4499–4510. doi: 10.1007/s00253-018-8931-7. [DOI] [PubMed] [Google Scholar]
- 108.Enjalbert B, Millard P, Dinclaux M, Portais JC, Letisse F. Acetate fluxes in Escherichia coli are determined by the thermodynamic control of the Pta-AckA pathway. Sci Rep. 2017;7:42135. doi: 10.1038/srep42135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Truebestein L, Leonard TA. Coiled-coils: the long and short of it. BioEssays. 2016;38(9):903–916. doi: 10.1002/bies.201600062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pfeiffer D, Jendrossek D. PhaM is the physiological activator of poly(3-hydroxybutyrate) (PHB) synthase (PhaC1) in Ralstonia eutropha. Appl Environ Microbiol. 2014;80(2):555–563. doi: 10.1128/AEM.02935-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jendrossek D, Pfeiffer D. New insights in the formation of polyhydroxyalkanoate granules (carbonosomes) and novel functions of poly(3-hydroxybutyrate) Environ Microbiol. 2014;16(8):2357–2373. doi: 10.1111/1462-2920.12356. [DOI] [PubMed] [Google Scholar]
- 112.Choi SY, Oh J, Jung J, Park Y, Lee SY. Three-dimensional label-free visualization and quantification of polyhydroxyalkanoates in individual bacterial cell in its native state. Proc Natl Acad Sci USA. 2021;118(31):1. doi: 10.1073/pnas.2103956118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yamada M, Yamashita K, Wakuda A, Ichimura K, Maehara A, Maeda M, et al. Autoregulator protein PhaR for biosynthesis of polyhydroxybutyrate [P(3HB)] possibly has two separate domains that bind to the target DNA and P(3HB): Functional mapping of amino acid residues responsible for DNA binding. J Bacteriol. 2007;189(3):1118–1127. doi: 10.1128/JB.01550-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Prieto B, Jung K, Witholt B, Kessler B. PhaF, a polyhydroxyalkanoate-granule-associated protein of Pseudomonas oleovorans GPo1 involved in the regulatory expression system for pha genes. J Bacteriol. 1999;181(3):858–868. doi: 10.1128/jb.181.3.858-868.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fu XZ, Tan D, Aibaidula G, Wu Q, Chen JC, Chen GQ. Development of Halomonas TD01 as a host for open production of chemicals. Metab Eng. 2014;23:78–91. doi: 10.1016/j.ymben.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 116.Mamat MRZ, Ariffin H, Hassan MA, Mohd Zahari MAK. Bio-based production of crotonic acid by pyrolysis of poly(3-hydroxybutyrate) inclusions. J Clean Prod. 2014;83:463–472. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Tolerance- and requirement to sodium chloride in MMCY media supplemented with 2 % glucose at 30 °C, 200 rpm, shows highest specific growth rate in the range of 1-4 % for both strains (n = 6). Figure S2. Representative raw FTIR spectra for Cobetia sp. MC34 (blue) and Cobetia marina DSM4741T (orange) grown in MMCY and MMCY_2 media supplemented with 1.5 % glycerol, 4 % glucose, 4 % fructose or 20 g/l acetate + 8 mM sodium valerate (n = 1). Figure S3. Growth curves measured by optical density (OD600nm) for the strains Cobetia sp. MC34 (blue) and Cobetia marina DSM 4741T (orange) grown in MMCY media supplemented with 40 g/l glucose or fructose, respectively. The carbonyl-ester to amide band I-ester ratio obtained by FTIR is visualized as sphere area and indicate putative PHA production, which was determined at the end of the experiment by chloroform extraction and GC/MS quantification. Error bars represent standard deviation of OD600nm (n = 6). Table S1. List of Halomonadacea sequences used to construct the 16S- and ANI trees, and information about the strains. Table S2. Amino acid sequence similarities of the transporters, enzymes, transcriptional regulators and carbonosome proteins involved in PHA synthesis from acetate. Table S3. Fatty acid biosynthesis (FAS) and fatty acid degradation (ß-oxidation) genes found in Cobetia sp. MC34, derived from the genome annotation.
Data Availability Statement
The datasets used and/or analyzed in this study are available from the corresponding author on reasonable request. Genome data for Cobetia sp. MC34 is available from NCBI under accessions listed in the “Methods” section “Genome sequencing and assembly”.