Abstract
Members of the type II nuclear hormone receptor subfamily (e.g., thyroid hormone receptors [TRs], retinoic acid receptors, retinoid X receptors [RXRs], vitamin D receptor, and the peroxisome proliferator-activated receptors) bind to their response sequences with or without ligand. In the absence of ligand, these DNA-bound receptors mediate different degrees of repression or silencing of gene expression which is thought to result from the association of their ligand binding domains (LBDs) with corepressors. Two related corepressors, N-CoR and SMRT, interact to various degrees with the LBDs of these type II receptors in the absence of their cognate ligands. N-CoR and SMRT have been proposed to act by recruiting class I histone deacetylases (HDAC I) through an association with Sin3, although they have also been shown to recruit class II HDACs through a Sin3-independent mechanism. In this study, we used a biochemical approach to identify novel nuclear factors that interact with unliganded full-length TR and RXR. We found that the DNA binding domains (DBDs) of TR and RXR associate with two proteins which we identified as PSF (polypyrimidine tract-binding protein-associated splicing factor) and NonO/p54nrb. Our studies indicate that PSF is a novel repressor which interacts with Sin3A and mediates silencing through the recruitment of HDACs to the receptor DBD. In vivo studies with TR showed that although N-CoR fully dissociates in the presence of ligand, the levels of TR-bound PSF and Sin3A appear to remain unchanged, indicating that Sin3A can be recruited to the receptor independent of N-CoR or SMRT. RXR was not detected to bind N-CoR although it bound PSF and Sin3A as effectively as TR, and this association with RXR did not change with ligand. Our studies point to a novel PSF/Sin3-mediated pathway for nuclear hormone receptors, and possibly other transcription factors, which may fine-tune the transcriptional response as well as play an important role in mediating the repressive effects of those type II receptors which only weakly interact with N-CoR and SMRT.
Nuclear hormone receptors comprise a superfamily of ligand-dependent transcription factors which play important roles in cell growth, differentiation, development, and homeostasis (52). The nuclear hormone receptor superfamily consists of type I and type II receptor subfamilies. Type I receptors mediate the effects of glucocorticoids, estrogens, mineralocorticoids, progestins, and androgens, while the type II receptors mediate the actions of thyroid hormone (TRs), all-trans retinoic acid (RA) (RARs), 9-cis RA (RARs and RXRs), and vitamin D (VDR). The peroxisome proliferator-activated receptors (PPARs) are members of the type II subfamily which mediate the effects of a wide variety of physiologically important lipid-derived ligands. Type I and type II receptors have similar modular structures consisting of a variable-sized N-terminal A/B domain, a 68- to 70-amino-acid DNA binding C domain (DBD), and a ∼300-amino-acid ligand binding domain (LBD) consisting of the D (hinge), E, and F regions (7, 52). The DBD is highly conserved among members of type I and type II receptor subfamilies.
Type I receptors are thought to interact with their cognate DNA sequences in regulated genes only in the presence of ligand, while type II receptors appear to bind their cognate regulatory sequences in the presence or absence of ligand. Transcriptional activation is thought to be mediated by a ligand-dependent conformational change in the LBD which recruits coactivators to the DNA-bound receptor (52). Coactivators, identified by yeast two-hybrid screens, generally fall into two main groups: the p160/SRC family (SRC-1/NCoA-1 [39, 57, 77], TIF-2/GRIP-1/NCoA-2 [32, 33, 77, 79], AIB1/p/CIP/ACTR/RAC3/TRAM-1 [2, 11, 46, 74, 77]) and the CBP (CREB binding protein)/p300 family (9, 30, 39). Coactivators which fall outside of these groups include PGC-1 (60), ARA70 (88), p/CAF (5, 84), hNRC/ASC-2/RAP250/TRBP (6, 42, 44, 51), and NRIF3 (45), which exhibits specificity for only the TRs and the RXRs. Using a biochemical approach, the DRIPs (VDR-interacting proteins) (63, 64) and TRAPs (TR-associated proteins) were identified as factors from HeLa cells which associate with TR and VDR in the presence of ligand (23, 37). The DRIPs and TRAPs are similar, if not identical, multiprotein complexes which are human homologues of yeast mediator/RNA polymerase II holoenzyme factors (37). Members of the p160/SRC family (11, 69), CBP/p300 (3, 56), and p/CAF (84) are thought to act through an intrinsic histone acetyltransferase activity which leads to an increase in the level of histone acetylation.
In the absence of ligand, the binding of a number of type II receptors (e.g., TRs and RARs) to their target DNA sequences leads to repression or silencing of basal gene expression. Stimulation of gene transcription by ligand is considered to result from both the reversal of repression (8) and the recruitment of coactivators to the DNA-bound receptor (57, 68). Repression was first noted for unliganded TR and for v-ErbA (14), the retroviral counterpart of the chicken TRα isoform (cTRα) of the avian erythroblastosis virus which does not bind ligand. Evidence that repression results from a ligand-dependent reversible interaction of a cellular repressor(s) with the LBD of certain type II receptors first came from studies using Gal4-TR LBD-VP16 chimeras (8). Insertion of the TR LBD between the Gal4 DBD and the VP16 activation domain was found to completely repress the activity of VP16. This repression could be relieved by coexpression of the LBD of TR or RAR which competed for a cellular repressor(s), and this apparent derepression was reversed by ligand. Interestingly, the repression of Gal4-TR LBD-VP16 was not reversed by expression of the LBD of RXR, suggesting that the RXR LBD only weakly interacted with the cellular repressor(s) (8).
Two related candidate corepressors, N-CoR and SMRT, which interact with the LBDs of TR and RAR in the absence of ligand and dissociate in the presence of ligand, have been cloned (12, 35). The receptor interaction domains of SMRT and N-CoR are found in their C-terminal regions, while the domains mediating repression are found in the N-terminal half of each protein (12, 35, 67, 90). The repressor domains were found to interact with Sin3 in vitro, suggesting that repression is mediated through the recruitment of class I histone deacetylases (HDAC1 to -3) to the promoter-bound unliganded receptor (31, 43, 54, 82). These studies, along with an analysis of the mechanism of repression by many other factors, have suggested that gene silencing is an important component of gene regulation and development, which is mediated through the recruitment of class I HDACs through distinct multiprotein complexes coordinated by either Sin3 (31, 43, 54) or Mi-2/NuRD (41, 80, 92). Factors thought to mediate repression through Sin3 include N-CoR/SMRT, Mad, Mnt, MeCP2, UME6, and Ski/Sno (41). Recent studies, however, have also raised the possibility for a direct in vivo interaction between N-CoR/SMRT with class II HDACs (HDAC4 to -7) independent of Sin3 (36, 40).
Almost all studies involved in the identification of nuclear receptor corepressors and coactivators have utilized the receptor LBD rather than full-length receptor. We have used full-length TRs and RXRs, in both yeast two-hybrid (45, 51) and biochemical (59) approaches to identify novel corepressors and coactivators that might require regions of these receptors other than the LBD. We previously reported that three proteins (p55, p65, and p100) from HeLa cell nuclear extracts bound in vitro to glutathione S-transferase (GST)–TR and to GST-RXR independent of ligand binding (59). One of these proteins (p65) was identified as TLS (translocated in liposarcoma). The receptor DBD and not the LBD was found to be necessary for the association of TLS with TR or RXR, although the affinity of interaction was enhanced by hinge region residues just C terminal to the DBD (59).
In this paper we report the identification of p100 as PSF-A and p55 as NonO/p54nrb. PSF is expressed as multiple cDNAs designated A, B, C, and F (58). The A, B, and C PSF cDNAs express the same protein but differ in their 3′ untranslated regions. The F cDNA encodes a shorter form of PSF. Thus, PSF protein is expressed as two alternatively spliced forms, indicated as PSF-A and PSF-F (58). NonO (mouse) (85) and p54nrb (human) (21) are highly conserved and differ by only one amino acid. Thus, throughout the paper we generally refer to p55 as NonO/p54nrb. Interestingly, TLS, PSF, and NonO/p54nrb each contain RNA recognition motifs (RRMs), although the region of interaction of these proteins with TR and RXR does not involve the RRM of these proteins. The association of PSF-A and NonO/p54nrb with TR and RXR was found to require their DBDs with a segment of the hinge region just C terminal to the DBD enhancing the interaction. PSF-A, NonO/p54nrb, or TLS does not interact with the LBD of TR or RXR (domains D, E, and F). The association of PSF-A, NonO/p54nrb, or TLS with the DBD also occurs in the presence of cognate DNA. The finding that TLS, PSF-A, and NonO/p54nrb interact with the DBD of nuclear hormone receptors provides further evidence for a multifunctional role of the DBD in nuclear receptor function.
Prior to the cloning of PSF and NonO/p54nrb, p100, and p55 were identified as part of a DNA-binding heterodimer, and UV cross-linking suggested that p100 was the DNA-binding component of the complex (91). The subsequent cloning of PSF (58) and NonO/p54nrb (21, 85) indicated that the components of the DNA-binding heterodimer were the same factors. PSF (polypyrimidine tract-binding protein [PTB]-associated splicing factor) was cloned as a putative splicing factor in association with PTB (58), although most of the PSF in the nucleus does not appear to be associated with PTB (53). NonO/p54nrb and PSF exhibit about 70% identity within their RRMs (21, 85).
Since PSF and NonO/p54nrb contain highly conserved RRMs, and PSF has been reported to play a possible role in RNA splicing (27, 48, 50, 58, 76), we anticipated that PSF-A and/or NonO/p54nrb might enhance the regulation of gene expression by nuclear hormone receptors by somehow coupling transcriptional activation with RNA splicing. Surprisingly, we found that PSF functions as a transcriptional repressor and recruits Sin3A to the receptor DBD. These studies provide evidence for multifunctional activities of PSF and define a new pathway for silencing of gene expression by nuclear hormone receptors and possibly other transcriptional regulators.
MATERIALS AND METHODS
Plasmids and antibodies.
pGEX2T-mRXRα expressing the full-length murine receptor fused to GST was provided by Paul T. van der Saag (22). Deletion mutants of mRXRα were constructed by PCR amplification of mRXRα using a 5′ primer containing a BgIII site linked to the first codon of mRXRα and a 3′ primer containing a stop codon and an EcoRI site. The BgIII-EcoRI digest of the PCR products were cloned into BamHI-EcoRI-digested pGEX2T. Different deletion mutants of cTRα fused to GST were described previously (18).
pEBG, a mammalian GST expression vector, was used to express GST fusions of receptors in 293T cells (59, 75). cTRα (amino acids 1 to 408) was excised from pGEX2T-cTRα with BamHI and cloned into the BamHI site of pEBG. mRXRα was removed from pGEX2T-mRXRα by complete digestion with AvrII and partial digestion with BamHI. The AvrII site is present just after the mRXRα stop codon. The gel-purified 1.5-kb fragment was then cloned into BamHI-SpeI site of pEBG.
Gal4–PSF-A (amino acids 1 to 707) was constructed by excising full-length PSF-A from a pET-PSF-A vector with NdeI and XhoI, blunt ending the NdeI-XhoI fragment with Klenow enzyme, and ligating it to SmaI-cut and dephosphorylated pSG424 (66). The various Gal4–PSF-A deletion mutants were constructed by PCR using primers containing a BamHI site at the 5′ end and a SacI site at the 3′ end. The PCR products were purified, digested with BamHI and SacI, and cloned into the BamHI-SacI sites of pSG424. Gal4-RXR(1-450)-VP16 and Gal4-RXR LBD(206-450)-VP16 were constructed by PCR amplification of mRXRα and cloning the sequences encoding amino acids 1 to 450 or 206 to 450 between Gal4 and VP16. Full-length PSF-A was cloned into pcDNA3.1(+) after excising PSF-A from pGEX4T1-PSF-A by partial digestion with EcoRI. The 2.2-kb fragment was gel purified and cloned into EcoRI site of pcDNA3.1(+). Similarly, NonO was cloned into pSG424 as well as pcDNA3.1(+).
In vitro 35S labeling of PSF-A and NonO cloned in pcDNA3.1(+) was carried out using coupled in vitro transcription-translation with T7 polymerase and rabbit reticulocyte lysates. Similarly, 35S-cTRα was expressed from a pEX vector using T7 polymerase (25). The GST-SMRT and GST-Sin3A clones (83) used in our study were obtained from Martin Privalsky, University of California, Davis. Antibodies against Sin3A (K-20) and GST were purchased from Santa Cruz Biotechnology, Inc. PSF and NonO antibodies were developed in the P. W. Tucker laboratory, University of Texas, Austin, and TLS antibodies were from David Ron, NYU School of Medicine. Other PSF antibody was from James G. Patton, Vanderbilt University. Antibodies directed against N-CoR was a gift from Mitchell Lazar, University of Pennsylvania School of Medicine.
Preparation of GST fusion proteins.
For the preparation of GST fusion proteins in Escherichia coli, 5 ml of overnight cultures were diluted 100 times and grown further for about 4 h. This culture was induced with 1 mM isopropyl-β-d-thiogalactopyranoside for about 1 h at 37°C and then chilled at 4°C for 10 min before pelleting of the bacteria. The cell pellet from each 500-ml culture was resuspended in 20 ml of phosphate-buffered saline (PBS) containing EDTA (50 mM, pH 8.0), dithiothreitol (DTT; 1 mM), leupeptin (20 μg/ml) and pepstatin (20 μg/ml). The bacteria were then incubated with lysozyme (1 mg/ml) for 10 min on ice, and the cells were disrupted using a sonicator. The bacterial suspension was centrifuged, and the supernatant was incubated with 500 μl of a 50% slurry of glutathione-agarose beads for 30 min. The beads were then washed in PBS three times (the last wash contained 50 μM ZnCl2 and 2 mM DTT), suspended in 250 μl of PBS containing 50 μM ZnCl2, 2 mM DTT, and 50% glycerol, and stored at −20°C. The addition of 50 μM ZnCl2 and 2 mM DTT was only for the GST-receptor preparations.
Preparation of nuclear extracts.
Nuclear extracts were prepared from HeLa cells by the method described by Dignam et al. (20). The final extract was dialyzed overnight at 4°C in a mixture of 20 mM HEPES (pH 7.8), 20% glycerol, 0.1 M KCl, 0.2 mM EDTA, 0.5 mM DTT, phenylmethylsulfonyl fluoride. The dialyzed nuclear fraction was centrifuged at 10,000 rpm to remove any precipitate and then stored at −80°C in aliquots. The protein concentration of dialyzed nuclear extract ranged between 3 and 5 mg/ml. The samples were used for GST-receptor binding studies or for immunoprecipitation.
Purification and sequencing of p100 and p55.
To isolate p100 and p55 for sequencing, the nuclear extracts were first partially purified on DEAE-Sephadex columns as previously described (59). Nuclear extract (∼4 mg of protein) was diluted with DEAE buffer (20 mM Tris-C1 [pH 7.8], 10% glycerol, 2 mM DTT, 100 mM KCl) in a 1:1 ratio and passed through a 1-ml packed column of DEAE-Sephadex equilibrated in the same buffer. The flowthrough was collected and combined with the eluted fraction (2 bed volumes eluted with buffer containing 100 mM KCl) of the column. The samples were then incubated in the elution buffer for 30 min to 1 h at 4°C with 1 μg of GST-mRXRα(140–240), and the bound proteins were then analyzed by electrophoresis in sodium dodecyl sulfate (SDS)-gels. The proteins in the gel were transferred to nitrocellulose and visualized by staining with Ponceau S. The protein bands of interest (p100 and p55) were cut out, eluted, and cleaved with endoproteinase Glu-C. Peptide fragments were isolated by high-pressure liquid chromatography followed by automated sequence analysis.
In vitro protein binding assays.
The nuclear extract was diluted with an equal volume of protein binding buffer (20 mM HEPES [pH 7.8], 1 mM MgCl2; 10 μM ZnCl2, 2 mM DTT, 10% glycerol, 0.05% Triton X-100, 40 μg of leupeptin/ml, 100 mM KCl) and centrifuged for 10 min. The supernatant was incubated with equal amounts of GST fusion proteins for 1 h at 4°C, and the beads were then washed three times with the same buffer. The beads were suspended in an equal volume of 2× SDS sample buffer and boiled, and the eluted proteins were analyzed by electrophoresis in SDS gels. About 300-ng aliquots of GST fusion proteins were used for Western blotting or for studies with 35S-labeled proteins. In one experiment, nuclear extract was incubated with GST-RXR(140–240) in the absence or presence of a 5- and 10-fold molar excess of a duplex oligonucleotide containing a high-affinity hormone response element recognition sequence (HRE) for RXR or TR (AGGTCA TGACCT) or an oligonucleotide containing a G→C change (HREm) which has no affinity for receptors (ACGTCA TGACGT [changed nucleotides are underlined]) (24). The exact sequences of the oligonucleotides were (AGCTT AGGTCA TGACCT AAGCT) for the HRE and (AGCTT ACGTCA TGACGT AAGCT) for the HREm. For Western blotting, the bound proteins were transferred to nitrocellulose in Tris-glycine buffer containing 20% methanol and 0.1% SDS at a constant voltage of 40 V over a period of 8 to 10 h at 4°C. The membranes were blocked in 10% milk and then probed with the antibody of interest. The immunoreactive bands were detected by using the Super Signal chemiluminescence system from Pierce and an appropriate second antibody linked to peroxidase. For protein binding assays using 35S-protein labeled in vitro, the reticulocyte lysates were incubated in protein binding buffer containing RNase A (15 μg/ml) at room temperature for 15 min. GST fusion proteins bound to glutathione-agarose beads were added, and the samples were incubated for an additional h at 4°C. The beads were washed three times with protein binding buffer, and the bound proteins were analyzed by electrophoresis in SDS-gels followed by autoradiography.
Immunoprecepitation.
HeLa nuclear extract (∼200 μg of protein) was diluted with an equal volume of buffer (20 mM HEPES [pH 7.8], 1 mM MgCl2, 10% glycerol, 0.05% Triton X-100, 100 mM KCl, 40 μg each of leupeptin, pepstatin, and antipain per ml). The buffered extract was precleared by incubation with protein A-Sepharose followed by centrifugation before incubating the extract with antibody for immunoprecipitation. The antibody-associated proteins were then bound to protein A-Sepharose beads and washed three times in the same buffer. The samples were then fractionated in SDS–8% PAGE gels followed by Western blotting with the antibody of interest. The blots were developed using the Pierce Super Signal chemiluminescence system.
Cell culture and transfection.
For chloramphenicol acetyltransferase (CAT) assay studies in 293T cells, the cells were transfected with 1 μg of the ΔMTV-HRE-CAT reporter gene (24) along with vectors expressing cTRα and/or PSF-A or NonO. After incubation for 48 h with or without T3, the cells were harvested for assay of CAT activity. Similar transfection studies were also carried out in HeLa cells with the Gal4 reporters, G5pBLCat2 (61) or pMC110 (65), and Gal4 fusions of PSF-A and NonO or the Ga14-VP16 chimeras of RXR. These CAT reporter experiments were performed several times with duplicate or triplicate samples. The experiments shown are representative studies which were repeated at least three times with similar results. For in vivo protein-protein interaction assays, 293T cells were transfected with pEBG, pEBG-mRXRα, or pEBG-cTRα. Forty-eight hours after transfection, ligand was added to the medium for 2 h before harvesting the cells, and the nuclear extracts were prepared by the method of Dignam et al. (20). Nuclear extracts expressing equivalent amounts of GST protein (determined by Western blotting with antibody against GST) were incubated with glutathione-agarose beads for 1 h at 4°C. The bound proteins were then analyzed by electrophoresis in SDS-gels followed by Western blotting using antibody against the protein of interest.
RESULTS
The association of PSF and NonO/p54nrb with TR and RXR requires the receptor DBD.
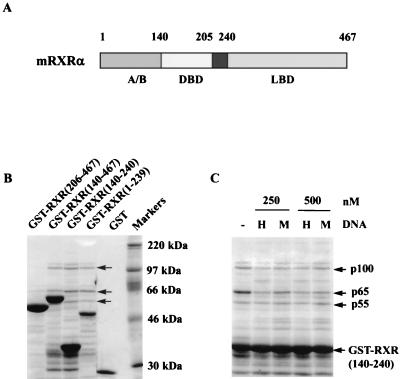
We previously reported that the DBDs of TR and RXR are required for the in vitro binding of a ∼65-kDa nuclear protein, which we identified as TLS, and two other nuclear proteins (∼100 and ∼55 kDa) (59). Figure 1 illustrates the domain structure of mRXRα and the association of HeLa cell nuclear extract proteins with different domains of mRXRα (amino acids 1 to 239, 140 to 240, 140 to 467, and 206 to 467) expressed as GST fusion proteins. TLS (p65) and the p100 and p55 proteins bind to only those fusion proteins containing the mRXRα DBD (amino acids 140 to 205) (Fig. 1B). A less abundant protein which migrates just above the 100-kDa protein was also identified in the experiment in Fig. 1 but is not consistently found in all experiments. Most of the other proteins in the gel, other than the predominant GST fusion protein, are not derived from the nuclear extract since, as previously described, they are also found after electrophoresis of the GST fusion protein preparation (59). In a previous study we found that a high-affinity HRE (AGGTCA TGACCT) binding sequence for TR and RXR did not interfere with the association of TLS with TR (59). Figure 1C illustrates that a 10-fold molar excess of the HRE over GST-RXR (140–240) does not inhibit the binding of TLS (p65), p55, or p100, suggesting that these proteins bind to the DBD independent of DNA binding. Incubation with RNase did not affect the binding of TLS, p55, or p100 to RXR(140–240) (59), suggesting that RNA does not play a role in the binding of these proteins to RXR(140–240).
FIG. 1.
The binding of p100 and p55 to mRXRα requires the receptor DBD region. (A) mRXRα structural domains. (B) Different regions of mRXRα, expressed in E. coli as GST fusion proteins and bound to glutathione-agarose beads, were incubated with partially purified HeLa cell nuclear extracts. The proteins binding to different regions of mRXRα were analyzed by SDS-gel electrophoresis and staining the gel with Coomassie brilliant blue. Three prominent proteins p100, p65, and p55 (arrows) were identified to associate with the GST-RXR fusion proteins containing the mRXRα DBD (amino acids 140 to 205) but not with GST-RXR expressing only the LBD (amino acids 206 to 467). p65 was previously identified as TLS (59). (C) GST-RXR(140–240), bound to glutathione-agarose, was initially incubated at 4°C without (−) or with a high-affinity RXR and TR HRE (H) or a mutant control sequence, HREm (M), which does not bind receptors. One milliliter of partially purified HeLa cell nuclear extracts was then added, and the samples were further incubated for 1 h. The concentration of GST-RXR(140–240) was 50 nM, while those of the oligonucleotides were 250 and 500 nM as indicated. The beads were washed and electrophoresed, and the gel was stained with Coomassie brilliant blue. The HRE does not affect the association of p55, p65, or p100 with GST-RXR.
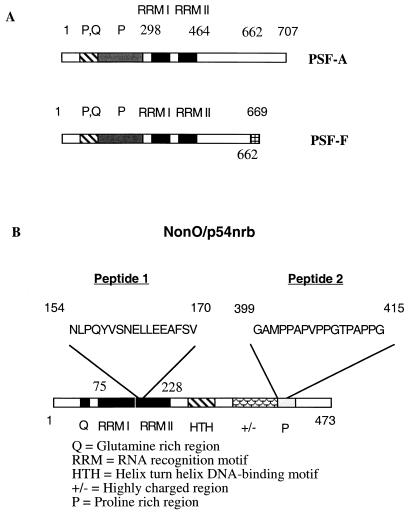
For large-scale purification of p100 and p55, the proteins which bound to GST-mRXRα(140–240) were transferred to nitrocellulose (hereafter, mRXRα is referred to as RXR). The proteins of interest were detected by Ponceau S staining, eluted, and sequenced. The sequence of the derived peptides indicated that p100 is PSF-A (58) and p55 is NonO/p54nrb (21, 85) (Fig. 2). For p100, several overlapping peptides matched exactly with the sequence of PSF-A from amino acid residues 300 to 700. No peptides specific for PSF-F were detected. PSF isoforms of 76 and 72 kDa have been reported (58) and are designated as PSF-A and PSF-F, respectively (Fig. 2A). These two isoforms are identical through amino acid 662 but thereafter diverge, with PSF-F containing 669 amino acids and PSF-A containing 707 amino acids (58). PSF contains two RRMs (RRMs I and II, amino acids 298 to 464) and an unusual N-terminal region rich in proline and glutamine residues and appears to migrate anomalously as a ∼100-kDa protein in SDS-gels.
FIG. 2.
p100 is PSF-A, and p55 is NonO/p54nrb. (A) Domain structures of PSF-A and PSF-F. p100 was identified as PSF-A, a 707-amino-acid protein with two predicted RNA binding domains (RRM I and RRM II) and other regions enriched for proline (P) or proline and glutamine (P, Q). The sequence of several overlapping peptides, obtained after digestion of purified p100, spanned amino acids 300 to 700 and matched the known sequence of PSF-A. PSF-F (669 amino acids), a shorter spliced version of PSF-A, is identical up to amino acid 662 of PSF-A and then diverges with only seven additional amino acids at the C-terminal end (VRMIDVG). (B) Structure of NonO/p54nrb. NonO/p54nrb is a 473-amino-acid protein with a number of structural features indicated in the diagram. Two peptide sequences obtained after digestion of purified p55 exactly matched a sequence in the RRM II domain and the proline-rich region at the C terminus of NonO/p54nrb.
Two peptide sequences obtained from purified p55 confirmed that the protein is NonO/p54nrb. One peptide sequence was within one of the NonO/p54nrb RRMs (amino acids 154 to 170), and the other was within the C-terminal proline rich region (amino acids 399 to 415) (Fig. 2B). NonO/p54nrb is a nuclear DNA and RNA-binding protein of 473 amino acids (21, 85). PSF and NonO/p54nrb are closely related proteins with over 70% identity in an internal 320-amino-acid region (amino acids 54 to 374 of NonO/p54nrb and amino acids 277 to 597 of PSF) containing two consensus RRMs. The function of NonO/p54nrb is unknown. However, NonO/p54nrb has been reported to bind to and enhance the in vitro transcription of an intracisternal A particle proximal enhancer element-driven reporter gene (4) as well as the DNA binding of Oct2 and E47 to their cognate sites (86).
PSF and NonO/p54nrb exist as monomers and as higher-molecular-weight protein complexes.
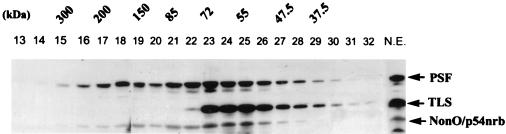
To investigate possible interactions of TLS, PSF, and NonO/p54nrb, nuclear protein extracts of HeLa cells were partially purified on DEAE-Sephadex and then subjected to size exclusion fast protein liquid chromatography (FPLC) using a Superose 6 column. As shown in Fig. 3, NonO/p54nrb (∼55 kDa) and PSF (∼72 kDa) elute as free proteins and as part of a larger complex(es) (∼100 to 300 kDa), which is consistent with the notion that these proteins may exist as a heterotetrameric complex in the cell (91). Whether the ∼100- to 300-kDa complex identified by FPLC consists of just PSF and NonO/p54nrb or includes other proteins was not determined. However, we show later (see Fig. 9 and 10) that PSF can interact with Sin3A. In contrast with PSF and NonO/p54nrb, TLS elutes only as a monomer and is not a component of a higher-molecular-weight complex with PSF or NonO/p54nrb.
FIG. 3.
PSF and NonO/p54nrb exist as monomers and as higher-molecular-weight protein complexes. Partially purified HeLa nuclear extract (N.E.) was fractionated with a Superose 6 FPLC column to study the elution profile of TLS, PSF, and NonO/p54nrb. Each fraction (indicated by numbers above the lanes) was concentrated and analyzed by Western blotting with antibodies against TLS, PSF, and NonO. TLS elutes in a pattern consistent with its size, while PSF and NonO/p54nrb elute both as part of a larger complex in addition to apparent free uncomplexed forms. NonO/p54nrb elutes as ∼55 kDa while PSF elutes at ∼80 kDa, sizes which are predicted by their amino acid sequences (PSF migrates anomalously as ∼100 kDa in SDS-gels).
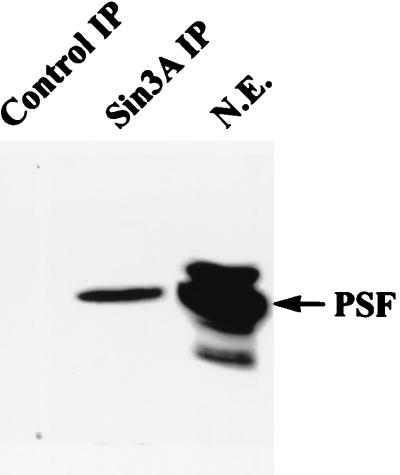
FIG. 9.
PSF immunoprecipitates with Sin3A. Nuclear extracts prepared from HeLa cells were immunoprecipitated with anti-Sin3A antibody or preimmune serum, and the samples were then analyzed by Western blotting with anti-PSF antibody. N.E., nuclear extract; Sin3A IP, immunoprecipitation with Sin3A antibody; Control IP, immunoprecipitation with preimmune serum.
FIG. 10.
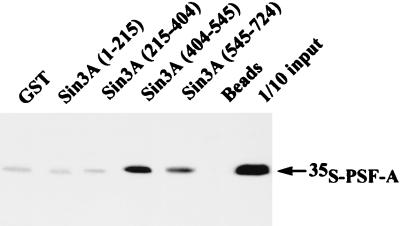
PSF-A interacts with Sin3A in vitro. Different domains of Sin3A were expressed as GST-fusion proteins in E. coli. Equivalent amounts of these fusion proteins were incubated with in vitro-synthesized 35S-PSF-A. The glutathione-agarose beads were washed and the bound 35S-PSF-A analyzed by SDS-gel electrophoresis followed by autoradiography. PSF-A binds to the region of Sin3A (amino acids 404 to 545) between the PAH2 and PAH3 domains.
Physical and functional interaction of PSF-A with TR and RXR.
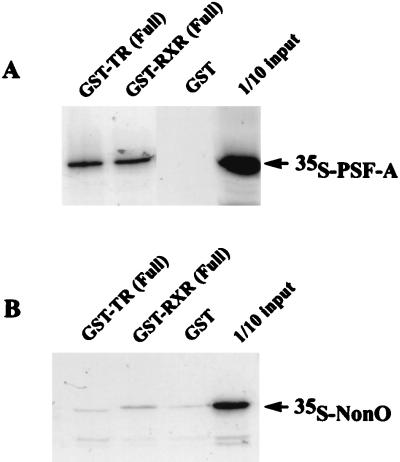
To study the physical interaction of receptors and PSF, we first carried out binding studies using GST fusions of full-length cTRα (hereafter, cTRα is referred to as TR) and full-length RXR expressed in bacteria and 35S-labeled PSF-A synthesized with reticulocyte lysates. As shown in Fig. 4A, PSF-A binds to GST-RXR and GST-TR but not to GST alone, suggesting a direct interaction of PSF-A and these receptors. In contrast, 35S-labeled NonO does not bind directly to GST-TR or GST-RXR (Fig. 4B), suggesting that the apparent association of NonO/p54nrb with the receptors likely results from its interaction with receptor-bound PSF-A.
FIG. 4.
In vitro binding of 35S-labeled PSF-A to full-length GST-TR and GST-RXR. PSF-A and NonO were synthesized in vitro using rabbit reticulocyte lysates and radiolabeled with [35S]methionine. (A) Equal amounts of 35S-PSF-A were incubated with different GST proteins (∼300 ng) bound to glutathione-agarose beads. PSF-A binds to GST-TR and GST-RXR but not to GST alone. (B) In contrast with the findings for 35S-PSF-A, 35S-NonO does not bind to full length GST-TR or GST-RXR.
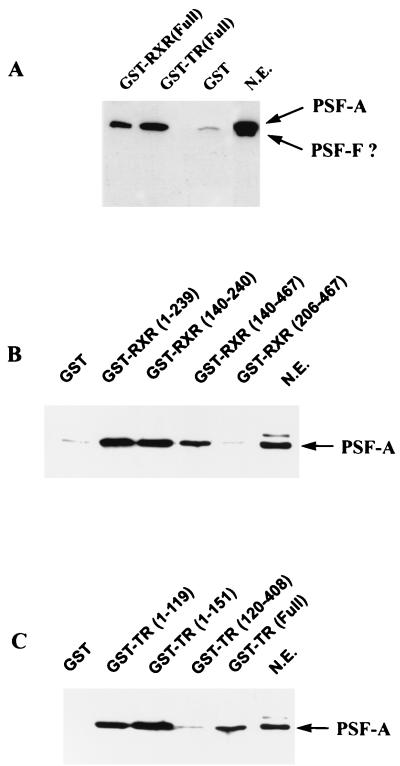
Several approaches were also undertaken to provide support for in vivo interactions of PSF with TR and RXR. Full-length TR and RXR were cloned into a GST mammalian expression vector (pEBG) (59, 75) and transfected into 293T cells. These GST-receptor chimeras are biologically active and lead to ligand-dependent transcriptional activation in transfection experiments (not shown). For analysis of receptor-PSF interactions, nuclear extracts were prepared from cells transfected with pEBG alone, pEBG-TR, or pEBG-RXR. Nuclear extracts expressing equivalent amounts of GST or GST fusion proteins (determined by Western blotting with antibody against GST) were incubated with glutathione-agarose beads for 1 h, washed, and analyzed by Western blotting for PSF (Fig. 5A). PSF appears to electrophorese as a broad, partially resolved doublet, although a very short autoradiographic exposure shows two distinct bands. The size of these bands are consistent with the proteins being PSF-A and PSF-F, although we cannot exclude the possibility that the lower-molecular-weight species is a degradation product of PSF-A. Assuming that the proteins detected by Western blot are PSF-A and PSF-F, only PSF-A appears to bind to GST-TR and GST-RXR (Fig. 5A), which is consistent with our peptide sequencing results. The C-terminal amino acids present in PSF-A and not in PSF-F are TERFGQGGAGPVGGQGPRGMGPGTPAGYGRGREEYEGPNKKPRF. This region, which is outside the RRM and is glycine rich (30%), appears to be important for the interaction of PSF-A with the receptors. Thus, the RNA binding and receptor binding functions of PSF appear to reside in two separate domains of PSF-A.
FIG. 5.
RXR and TR selectively bind PSF-A. (A) 293T cells were transfected with the mammalian GST expression vector pEBG alone or pEBG expressing full-length TR or RXR. Nuclear extracts expressing equivalent amounts of GST, GST-RXR, or GST-TR were incubated with glutathione-agarose beads. The bound proteins were then analyzed by Western blotting with anti-PSF antibody. In this experiment, the gel was electrophoresed for a longer time than normal. Under these conditions, PSF electrophoreses as a broad, partially resolved doublet, although a very short autoradiographic exposure shows two distinct bands. The size of these bands are consistent with the proteins being PSF-A and PSF-F. Since we cannot precisely determine whether the lower-molecular-weight species is PSF-F or a degradation product of PSF-A, we have indicated the band with a ?. Although both forms are present in the nuclear extract (N.E.), only PSF-A binds to the receptors. (B and C) PSF-A binding to RXR and TR involves the DBDs of the receptors. An equal amount of HeLa nuclear extract (500 μg) was incubated with equimolar amounts of bacterial expressed GST fusion proteins containing different regions of RXR or TR as indicated. The glutathione-agarose beads were washed, and the bound proteins were analyzed by Western blotting with anti-PSF antibody. PSF-A binds only to the GST-receptor fusion proteins which contain the DBD of RXR or TR. The LBDs of RXR(206–467) and TR(120–408) do not bind PSF-A.
To further analyze the regions of TR and RXR involved in the association of endogenous HeLa cell PSF-A, in vitro binding studies were performed with various deletion mutants of TR and RXR expressed as GST fusion proteins in bacteria. Equimolar amounts of GST fusion proteins were incubated with HeLa nuclear extracts (500 μg of protein), and the association of PSF was determined by Western blotting with anti-PSF antibody. As shown in Fig. 5B, PSF-A binds to GST-RXRs (amino acids 1 to 239, 140 to 240, and 140 to 467) containing the RXR DBD (amino acids 140 to 205) but not to the RXR LBD alone (amino acids 206 to 467). Similar results were also found for TR (Fig. 5C). PSF-A binds to GST-TRs containing amino acids 1 to 119, 1 to 151 and 1 to 408 (full length), which each include the TR DBD (amino acids 51 to 119), but not to amino acids 120 to 408, which contain only the TR LBD. These results confirmed our previous observations (Fig. 1) indicating that the binding of PSF-A to TR and RXR involves their DBDs whereas the LBDs of the receptors do not interact with PSF-A.
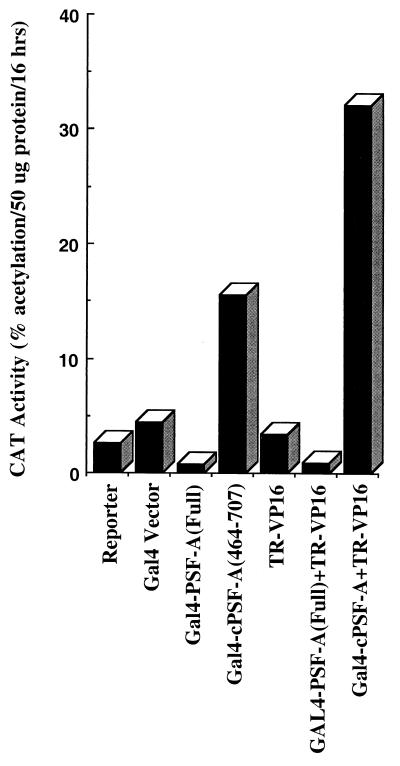
To provide evidence for a functional in vivo interaction of TR with PSF-A, HeLa cells were transfected with a Gal4-responsive thymidine kinase-CAT reporter (G5pBLCat2) along with vectors expressing Gal4 fused to full-length PSF-A (Gal4–PSF-A) or to the C-terminal region of PSF-A (Gal4–cPSF-A, amino acids 464 to 707) and/or a cTRα construct (TR-VP16, amino acids 1 to 221). This TR-VP16 does not bind ligand but contains the DBD (amino acids 51 to 119) along with the 50-amino-acid N-terminal A/B domain of TRα and about 100 amino acids of the LBD fused to the transactivation domain of VP16. Since this TR-VP16 has very little of the LBD, it would not be expected to bind to corepressors such as N-CoR or SMRT. The expectation of this study was that if TR-VP16 interacted with Gal4–PSF-A or GAL4–cPSF-A, this would lead to increased activation of G5pBLCat2 when the Gal4–PSF-A fusions were expressed with TR-VP16. Surprisingly, we found that the expression of Gal4–PSF-A resulted in a four- to five-fold repression of the CAT reporter gene (Fig. 6), while expression of Gal4–cPSF-A resulted in a threefold increase. This weak activation function of Gal4–cPSF-A appears to be repressed in the context of full-length PSF-A in Gal4–PSF-A, suggesting that the N-terminal half of PSF harbors a repression domain.
FIG. 6.
In vivo interaction of TR with the C-terminal domain of PSF-A (cPSF-A). HeLa cells were transfected with a CAT reporter (G5pBLCat2) with the Gal4 DBD or Gal4 DBD fusion of full-length PSF-A or the C-terminal region of PSF-A (amino acids 464 to 707) (cPSF-A) with or without TR-VP16 (amino acids 1 to 221 of TR fused to the VP16 activation domain). Gal4–cPSF-A exhibits a threefold increase in activity compared to Gal4 alone, and its interaction with TR is evident by further activation after expression of TR-VP16. In contrast, expression of Gal4–PSF-A (full length) leads to repression compared to Gal4 alone or when expressed with TR-VP16.
Expression of TR-VP16 with Gal4–cPSF-A further enhanced the extent of activation, providing additional support for an in vivo interaction between the truncated TR-VP16 protein and the C-terminal region of PSF-A. However, the repressor activity of full-length PSF-A in Gal4–PSF-A completely masks the activation function of TR-VP16. We did not observe the same extent of repression by Gal4–PSF-A on a Gal4-CAT reporter gene regulated by the simian virus 40 early promoter (G5pSV-Cat) as opposed to G5pBLCat2, indicating that promoter context may play a role in repression, possibly resulting from differences in the transcription factors that bind specifically to these promoters. In similar experiments with G5pBLCat2, Gal4-NonO (full length), which is 70% identical in RRM II, also resulted in repression. However, the extent of repression was less (∼1.5- to 2-fold inhibition) than with Gal4–PSF-A, although Western blotting using Gal4 DBD antibody indicated that Gal4-NonO was expressed at similar levels as Gal4-PSF-A (not shown).
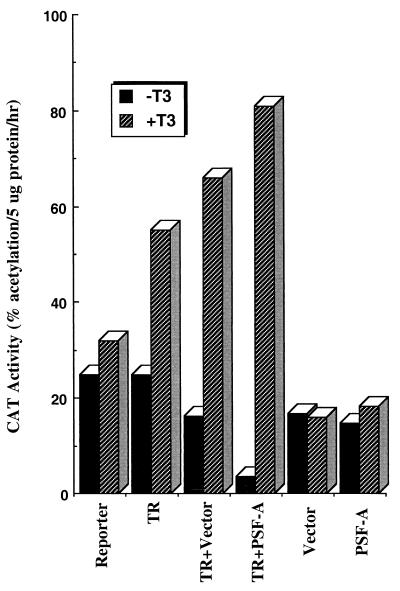
PSF-A enhances TR-mediated repression in the absence of ligand.
Since Gal4–PSF-A exhibits repressor activity, we sought to study the effect of PSF-A on wild-type TR activity in mammalian cells. To study the effect of PSF-A on the repressive function of TR, transfection studies were performed with 293T cells (Fig. 7), where we found that expression of unliganded TR does not lead to as much repression as in other cell types. The reason for this is unclear. Nevertheless, this gave us the opportunity to examine whether expression of PSF-A can lead to repression in this system. Transfection studies were performed with pcDNA vectors expressing full-length PSF-A along with full-length cTRα in the presence or absence of T3. Since PSF-A is a moderately abundant protein, cells were transfected with 30 μg of PSF-A expression vector to express this protein at a level higher than the endogenous amount. In this study, expression of PSF-A with TR resulted in a fourfold enhancement in the level of repression compared to unliganded receptor alone. However, expression of PSF-A alone had little or no effect on the basal activity of the reporter. These studies suggest that PSF-A enhances promoter repression through interaction with unliganded receptor.
FIG. 7.
PSF-A enhances the extent of transcriptional repression by unliganded TR. 293T cells were cotransfected with the ΔMTV-HRE-CAT reporter and with vectors expressing full-length PSF-A and/or cTRα with or without T3. PSF-A represses the basal activity of the CAT reporter only when the receptor is expressed in the absence of hormone, suggesting that it enhances the gene silencing effect of unliganded TR. PSF-A alone had no effect on the basal activity of ΔMTV-HRE-CAT.
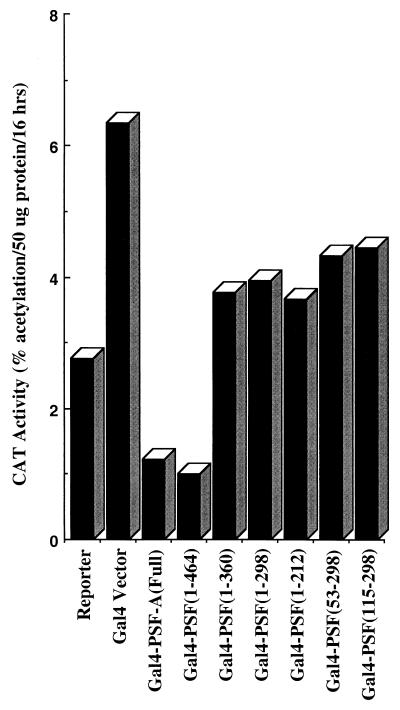
The repressor activity of PSF-A resides in the N-terminal half of the protein.
The Gal4–PSF-A and Gal4–cPSF-A results in Fig. 6 suggest that the repressor activity of PSF-A resides in the N-terminal region of the protein (amino acids 1 to 464). To further specify the domain(s) of PSF-A necessary for repression, a series of Gal4–PSF-A chimeras spanning different regions in the N terminus of PSF-A were created by PCR. These Gal4–PSF-A chimeras were found to express correctly and at similar levels as determined by Western blotting using antibody against the Gal4 DBD (data not shown). As shown in Fig. 8, the repressor activity of PSF-A requires residues containing RRM II of PSF-A (amino acids 361 to 464).
FIG. 8.
The repressor activity of PSF-A requires amino acids 361 to 464 of the protein. PSF-A deletion mutants, created by PCR, spanning various regions of the N-terminal domain of PSF-A (amino acids 1 to 464) were cloned as Gal4 fusion constructs. The region mediating the repressor activity of PSF-A was functionally mapped by transfection studies using the G5pBLCat2 reporter in HeLa cells.
PSF-A directly associates with Sin3A.
Since a number of factors are thought to mediate repression through interaction with Sin3A (31, 41, 43, 54), we examined whether Sin3A associates with PSF. HeLa nuclear extracts were immunoprecipitated with anti-Sin3A antibody followed by Western blotting for PSF. As shown in Fig. 9, PSF is immunoprecipitated by antibodies against Sin3A, suggesting that PSF and Sin3A associate directly or indirectly in vivo. To provide evidence for a direct interaction of Sin3A and PSF, we first constructed GST fusions of PSF-A. However, we found that GST–PSF-A proteins are poorly expressed in E. coli, and thus we examined for an association of Sin3A with PSF-A using GST-mSin3A proteins (83) and 35S-PSF-A. As shown in Fig. 10, 35S-PSF-A binds to amino acids 404 to 545 of mSin3A, a region encompassing the paired ampipathic helix PAH2 and PAH3 domains (81, 83). GST-Sin3A binding studies were also performed using 35S-NonO. Although NonO/p54nrb and PSF share 70% structural identity over a 320-amino-acid region residing in their RRMs, NonO/p54nrb does not bind to Sin3A in our in vitro binding assays (data not presented). This suggests that the lower level of repression seen with Gal4-NonO may be mediated through its ability to form a complex with PSF (91). However, the role of NonO/p54nrb in mediating repression by PSF-A is unclear.
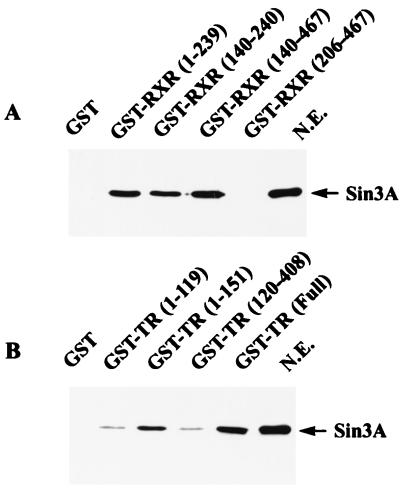
Association of Sin3A with TR and RXR involves the receptor DBD regions.
Since PSF-A interacts with Sin3A, we examined whether Sin3A associates with the same regions of RXR and TR that interacts with PSF-A. HeLa cell nuclear extracts were incubated in vitro with GST fusions of RXR and TR as described earlier for PSF-A (Fig. 5), followed by Western blotting with antibodies against Sin3A (Fig. 11). Sin3A associates with GST-RXRs (amino acids 1 to 239, 140 to 240, and 140 to 467) which contain the DBD (amino acids 140 to 205) but does not associate with only the LBD (amino acids 206 to 467) (Fig. 11A). Thus, the pattern of binding of Sin3A with the GST-RXRs is identical to that found for the binding of PSF-A (Fig. 5B), suggesting that Sin3A binds to RXR through an association with PSF-A. The pattern of binding of Sin3A with the GST-TRs (Fig. 11B) was generally similar but showed some differences from that for PSF-A (Fig. 5C). Sin3A associated with GST-TRs expressing amino acids 1 to 151 which contains the A/B domain (amino acids 1 to 50), the DBD (amino acids 51 to 119), and part of the hinge region (amino acids 120 to 151). GST-TR(1–119) bound less Sin3A than GST-TR(1–151) and somewhat less but detectable binding was found with GST-TR(120–408) containing the LBD.
FIG. 11.
Efficient binding of Sin3A to RXR and TR involves the DBD and the proximal hinge (D) region of the receptors. HeLa nuclear extracts were incubated with GST proteins expressing different domains of RXR or TR. The bound proteins were analyzed by Western blotting using anti-Sin3A antibody. (A) Sin3A binds to GST-fusion proteins expressing the RXR DBD but not the RXR LBD (amino acids 206 to 467). (B) Sin3A binds efficiently to GST-TR (full length) and to GST-TR(1–151) (which include the DBD and the proximal hinge region) but much less efficiently to GST-TR(1–119) (lacking the hinge region) and to GST expressing only the LBD [GST-TR(120–408)]. N.E, nuclear extract.
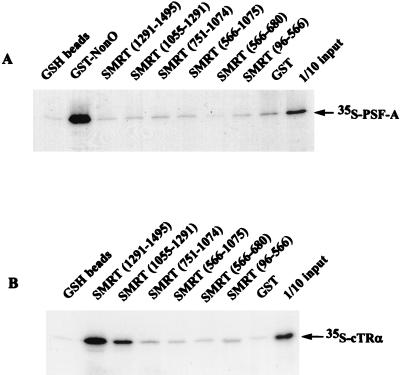
These findings imply that the association of Sin3A requires the receptor DBD as well as the N-terminal part of the hinge region. Thus, both the DBD and the hinge region (the proximal part of the hinge region is thought to participate in DNA binding and thus may be considered as part of the DBD as well as the LBD) appear to participate in high-affinity binding of Sin3A to receptors, possibly through its association with PSF-A. Since SMRT and N-CoR have been reported to associate with Sin3A in vitro, we considered the possibility that such corepressors might also interact with PSF-A and thus recruit Sin3A to the receptor DBD region. To explore this, in vitro binding studies were carried out with 35S-PSF-A and GST-SMRT fusions encompassing different domains of SMRT (Fig. 12A). 35S-PSF-A did not bind to any of the GST-SMRT proteins, although GST-SMRT (amino acids 1055 to 1291) and GST-SMRT (amino acids 1291 to 1495) bound 35S-TR, which served as a positive control (Fig. 12B).
FIG. 12.
PSF-A does not interact directly with SMRT. Different domains of SMRT, expressed as GST fusion proteins in E. coli, were incubated with in vitro-synthesized 35S-PSF-A. The samples were then analyzed by electrophoresis followed by autoradiography. In the glutathione control, samples were incubated with glutathione-agarose (GSH) beads without bound GST protein. (A) PSF-A does not bind to any of the GST-SMRT fusion proteins, while 35S-PSF-A binds to GST-NonO (positive control). (B) The GST-SMRT proteins which do not bind PSF-A bind 35S-cTRα (positive control). As previously described (83), the C-terminal domain of SMRT (amino acids 1291 to 1495 and amino acids 1055 to 1291) strongly interacts with TR.
Transcriptional repression mediated by PSF-A involves HDAC activity.
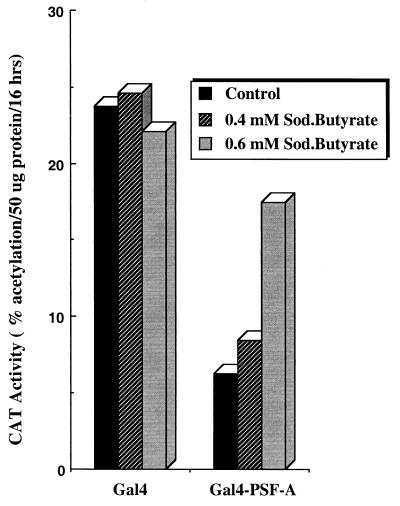
Since PSF-A associates with Sin3A, and Sin3A interacts with class I HDACs, we examined if sodium butyrate, an inhibitor of HDACs (70), can reverse the repression mediated by PSF-A. Sodium butyrate was used instead of the HDAC inhibitor trichostatin A (89) because we found that trichostatin A, at concentrations which inhibit HDACs, is toxic in HeLa cells. HeLa cells were transfected with G5pBLCat2 along with a vector expressing Gal4–PSF-A (full length) or the Gal4 DBD alone. Sodium butyrate was initially tested over a concentration range of 0.5 to 2.5 mM. Concentrations of sodium butyrate greater than 1 mM inhibited cell growth. Thus, we used two concentrations of sodium butyrate (0.4 and 0.6 mM). As shown in Fig. 13, expression of Gal4–PSF-A leads to a fivefold inhibition of transcriptional activity. However, addition of 0.4 and 0.6 mM sodium butyrate reversed this repression 1.5- and 3-fold, respectively, while having no effect on cells expressing only the Gal4 DBD. These findings suggest that histone deacetylation plays a role in the repressor activity of PSF and provide support for a model where PSF-A mediates repression through the recruitment of a Sin3A-HDAC complex.
FIG. 13.
Effect of sodium butyrate on PSF-A-mediated transcriptional repression. HeLa cells were transfected with equimolar quantities of vector expressing the Gal4 DBD or Gal4-PSF-A (full length) with the G5pBLCat2 reporter. The cells were then incubated without or with either 0.4 or 0.6 mM sodium butyrate for 48 h before harvesting of cells for assay of CAT activity.
PSF-A and Sin3A do not dissociate from ligand-bound TR or RXR in vivo.
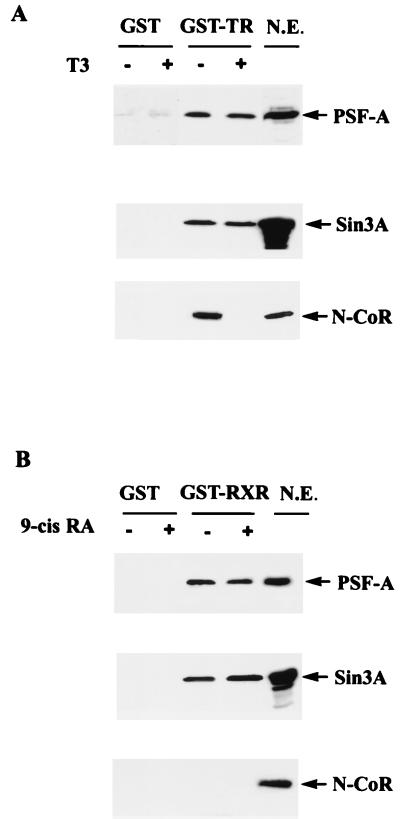
To test for possible in vivo interactions of PSF and Sin3A, and the role of ligand on their binding to receptors, pEBG vectors expressing GST, GST-TR (full length), or GST-RXR (full length) were transiently expressed in 293T cells. Forty-eight hours after transfection, the cells were incubated without or with T3 or 9-cis RA for 2 h prior to harvesting. Nuclear extracts containing equivalent amounts of GST, GST-TR, or GST-RXR (determined by Western blotting with antibody against GST) were incubated with glutathione-agarose beads followed by Western blotting with antibodies against PSF, Sin3A, and N-CoR.
PSF-A and Sin3A bound to both TR and RXR, and this association was not affected by incubation with T3 or 9-cis RA (Fig. 14). N-CoR bound to unliganded TR, and this association was completely reversed by T3 incubation (Fig. 14A). N-CoR was not detected to associate with RXR either in the presence or absence of ligand (Fig. 14B), which is consistent with the notion that the LBD of RXR exhibits very low levels of repressor activity (61). The finding that ligand did not alter the binding of Sin3A to TR in vivo was surprising since Sin3A has been reported to bind to N-CoR/SMRT in vitro. However, this finding is consistent with our in vitro binding studies indicating that the association of Sin3A with TR and RXR does not require the LBD of these receptors. This finding is also consistent with recent studies indicating that N-CoR/SMRT may mediate repression via a direct interaction with class II HDACs (36, 40).
FIG. 14.
PSF-A and Sin3A remain bound to TR and RXR in the presence of ligand. GST-TR and GST-RXR expressing full-length receptors or GST alone were transiently expressed in 293T cells. Forty-eight hours after transfection, the cells were incubated with T3 or 9-cis RA for 2 h before harvesting of cells for preparation of nuclear extracts. Nuclear extracts expressing equivalent amounts of GST-TR, GST-RXR, or GST were incubated with glutathione-agarose beads, washed, and then analyzed for the association of PSF, Sin3A, or N-CoR by Western blotting. PSF-A and Sin3A remain associated with TR or RXR in the presence or absence of ligand (A and B). N-CoR associates with TR only in the absence of ligand (A), while N-CoR does not interact with RXR in the presence or absence of ligand (B). N.E., aliquot of nuclear extract which was not incubated with glutathione-agarose.
The DBD and PSF mediate transcriptional repression in vivo.
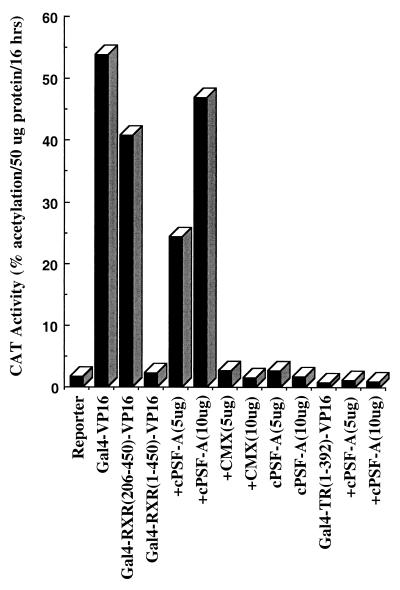
The finding that unliganded RXR does not bind N-CoR, while it associates with PSF-A and Sin3A through the DBD, allowed us to design experiments to (i) functionally assess the importance of this interaction in vivo and (ii) examine the possibility that the association of Sin3A with RXR occurs via an interaction with PSF-A. Our previous studies showed that Gal4-RXR LBD-VP16 is transcriptionally active while the activity of Gal4-TR LBD-VP16 is repressed (8, 61). This difference in transcriptional activity is consistent with our present finding that N-CoR binds to unliganded TR but not to RXR. The finding that Sin3A binds to the receptor DBD predicts that the transcriptional activity of a Gal4-VP16 chimera expressing both the DBD and LBD of RXR would be repressed compared with a chimera expressing only the LBD. This prediction was confirmed by comparing the transcriptional activities of Gal4-RXR(1–450)-VP16 and Gal4-RXR LBD(206–450)-VP16 (Fig. 15). Gal4-RXR LBD(206–450)-VP16 was as active as Gal4-VP16, while the activity of Gal4-RXR(1–450)-VP16 was markedly repressed.
FIG. 15.
Repression of Gal4-RXR(1–450)-VP16 is reversed by the C-terminal region of PSF-A (cPSF-A). Gal4-RXR(1–450)-VP16 or Gal4-RXR LBD(206–450)-VP16 was transfected along with a CAT reporter plasmid (pMC110) in HeLa cells. Gal4-RXR LBD(206–450)-VP16 was as active as Gal4-VP16, while the activity of Gal4-RXR(1–450)-VP16 was markedly repressed. cPSF-A (5 and 10 μg) reversed the repression of Gal4-RXR(1–450)-VP16, resulting in an activity similar to that of Gal4-RXR LBD(206–450)-VP16. The cPSF-A expression vector (CMX) had no effect on Gal4-RXR(1–450)-VP16. cPSF-A did not reverse the repression of Gal4-TR(1–392)-VP16, which, unlike RXR, also binds N-CoR/SMRT corepressors in the LBD.
If this repression is mediated through PSF-A, expression of cPSF-A, which interacts with receptors but lacks the PSF-A repressor domain, would be expected to block the binding of endogenous PSF-A and thus relieve the repression of Gal4-RXR(1–450)-VP16. Figure 15 shows that the expression of cPSF-A results in a marked increase in the transcriptional activity of Gal4-RXR(1–450)-VP16 which is comparable to the activities of Gal4-VP16 or Gal4-RXR LBD(206–450)-VP16. Cotransfection of Gal4-RXR(1–450)-VP16 with the control vector (CMX) used to express cPSF-A had no effect on the activity of Gal4-RXR(1–450)-VP16. A comparison of the effect of the cPSF-A and control CMX vectors indicated that cPSF-A did not further enhance the activities of Gal4-VP16 or Gal4-RXR LBD(206–450)-VP16 (not illustrated). We also studied the effect of cPSF-A on the activity of Gal4-TR(1–392)-VP16. In contrast with Gal4-RXR(1–450)-VP16, expression of cPSF-A did not lead to reversal of repression of Gal4-TR(1–392)-VP16. This is consistent with our finding that N-CoR/SMRT interacts with the LBD of TR but not RXR and thus maintains Gal4-TR(1–392)-VP16 in a repressed state.
DISCUSSION
Regulation of gene expression by certain members of the type II receptor subfamily is thought to result from both the ligand-mediated release of a corepressor(s) and the recruitment of a coactivator(s) to the LBD. In this study we used a biochemical approach to identify novel factors that might play a role in receptor function. We purified and sequenced two proteins which associate with the DBDs of TR and RXR and identified them as PSF-A (p100) and NonO/p54nrb (p55). In vitro binding studies indicate that PSF-A interacts directly with the receptor DBDs whereas NonO/p54nrb appears to associate with the DBDs as a result of its interaction with PSF-A. Although both PSF-A and NonO/p54nrb contain RRMs, these domains are not involved in their interactions with TR or RXR. RNA does not appear to play a role in the interaction of PSF-A and NonO/p54nrb with receptors since RNase incubation does not interfere with the association. In addition, the association of these proteins with the DBD can occur even in the presence of a high-affinity cognate DNA recognition sequence. Although the binding of PSF-A and NonO/p54nrb requires the DBD, a segment of the D hinge region just C terminal to the DBD appears to enhance the interaction. PSF-A and NonO/p54nrb do not interact with the LBDs of TR or RXR (domains D, E, and F).
The finding that PSF-A and NonO/p54nrb (and TLS) form a complex with the DBDs of nuclear hormone receptors suggests a multifunctional role for the DBD in nuclear receptor actions. Indeed, the DBD of TR has been shown to functionally interact in vivo with p53 (62, 87), which blocks TR binding to DNA, and with human immunodeficiency virus type 1 tat (18, 19), which does not block TR binding to DNA. In addition, the transcriptional activator p/CAF (5) has been reported to associate with the DBD of RAR or RXR. Thus, in addition to its role in DNA binding, the DBD of nuclear receptors appears to play an important role in receptor protein-protein interactions.
PSF was initially cloned as a component thought to be involved in splicing (58) and NonO as a DNA-binding protein (85), each with tandem RRMs which exhibit about 70% identity (21). Although tandem RRMs are thought to be involved in RNA binding, they may also specifically recognize both single-stranded and double-stranded DNA sequences (1, 16, 17, 55). NonO/p54nrb and PSF exist both as free forms and as part of a higher-molecular-weight complex(es), while TLS does not appear to interact with PSF or NonO/p54nrb (Fig. 3) (91). Although PSF is thought to play a role in RNA splicing (27, 48, 50, 58, 76), we found that PSF-A acts as a transcriptional repressor, possibly through the recruitment of HDACs. This conclusion is supported by the finding that Gal4–PSF-A exhibited repressor activity and that repression could be reversed by sodium butyrate, an inhibitor of HDACs.
The level of expression of PSF in various tissues, and its possible regulation by physiologic events, has not been well defined. However, evidence that the level of PSF may be dynamically regulated comes from studies in certain tissues indicating that the expression of PSF is cell type specific and developmentally regulated. For example, PSF mRNA and protein are highly expressed in differentiating neurons of the embryonic and neonatal cortex and cerebellum, while their expression virtually disappears in adult brain tissue (10). This implies that PSF is not absolutely essential for splicing and also suggests that PSF may influence the neural differentiation process. Although PSF was first identified as a putative splicing factor in association with PTB (58), antibodies against PSF do not immunoprecipitate PTB, suggesting that not all PSF is associated with PTB in the cell (53). This finding is also supported by confocal microscopy studies indicating that the majority of PSF and PTB are not associated in the nucleus (53). Although a body of evidence from different laboratories support a role for PSF in splicing (27, 48, 50, 58, 76), the above results support our findings that PSF may also mediate effects on gene expression independent of its role as a PTB-associated splicing factor. This conclusion is also supported by studies indicating that (i) PSF can influence the activity of topoisomerase I (71, 72) and (ii) PSF may inhibit expression of the gene encoding the porcine p450 cholesterol side chain cleavage enzyme by binding to a DNA sequence (CTGAGTC) which is 5′ to the Sp1 binding site of the gene promoter (78).
Several lines of evidence indicate that the repressor activity of PSF-A is mediated through Sin3A and involves the receptor DBD. First, PSF-A was found to bind to Sin3A in vitro. Second, immunoprecipitation of nuclear extracts with antibody to Sin3A also immunoprecipitates PSF. Third, in vitro binding studies of nuclear extracts with various regions of GST-RXR indicated that Sin3A and PSF-A interact similarly with various RXR constructs which contain the DBD but not with constructs which contain only the RXR LBD (amino acids 206 to 467). In general, similar results were found for the various domains of TR [i.e., as with PSF-A, Sin3A binds less efficiently to GST-TR(120–408) containing the LBD than GST-TR(1–408) (full length) or GST-TR(1–151) containing the DBD (amino acids 51 to 119)]. Fourth, expression of cPSF-A, which would be expected to block the binding of endogenous PSF-A to the DBD of RXR, increases the activity of the repressed Gal4-RXR(1–450)-VP16 chimera to a level similar to that found for Gal4-VP16.
The markedly reduced binding of Sin3A to the LBD compared with full-length receptor or the receptor DBD raised the possibility that the binding of Sin3A with receptors may not be affected by ligand in vivo. Indeed, we found that PSF-A and Sin3A bind to full-length TR or RXR in vivo but that ligand does not dissociate PSF-A or Sin3A from the receptors. Interestingly, N-CoR bound to TR but not to RXR, which is consistent with the finding that Gal4-RXR LBD-VP16 is transcriptionally active while Gal4-TR LBD-VP16 is silent (8, 61). Surprisingly, although T3 mediated a complete dissociation of N-CoR, very little if any reduction of Sin3A binding occurred, suggesting that Sin3A does not associate with LBD bound N-CoR/SMRT in vivo. This finding is consistent with a recent report indicating that N-CoR/SMRT may mediate repression independent of Sin3 through a direct interaction with class II HDACs (36, 40).
The finding that the corepressors N-CoR/SMRT do not bind or only weakly associate with the LBD of RXR compared with TR suggests that PSF-A may play a more important role in mediating repression by unliganded RXRs. Similarly, N-CoR/SMRT has been reported to only weakly associate with VDR (38, 73) and possibly the PPARs (29). Thus, PSF-A may also play a role in mediating effects of those unliganded receptors and possibly a number of orphan receptors as well. The activation of gene expression by ligand is thought to reflect a shift in the equilibrium of receptor from an inactive or repressed to an active state (68). Thus, the extent of transcriptional activation is dependent, in part, on the absolute and/or relative levels of repressor(s) and activator(s) that interact with the receptor in the absence or presence of ligand. Although activation by ligand through the recruitment of coactivators appears sufficient to overcome the extent of PSF-A-mediated repression through Sin3A, the DBD-bound PSF-A–Sin3A may act to fine-tune the transcriptional response. The extent of this effect would be dependent, in part, on the level and/or distribution of PSF-A in the cell and whether this level or its interaction with the receptor DBD is modulated by developmental or physiologic events.
The observation that PSF acts as a repressor may also explain the mechanism underlying the development of some papillary renal cell carcinomas involving the translocation of PSF t(X;1) or inversion of NonO/p54nrb inv(X) with the TFE3 gene (13). In each case, the rearrangement results in the fusion of almost the entire PSF or NonO/p54nrb protein with the DBD of TFE3, a member of basic helix-loop-helix family of transcription factors. These chimeras are reminiscent of PML-human RARα and PLZF-human RARα (15, 28, 34, 47) and AML1-ETO (26, 49), which mediate a differentiation block leading to leukemia. This block is thought to result from the recruitment of a repressor (e.g., N-CoR/SMRT) to a gene recognized by the DBD of the fusion protein. Whether PSF-TFE3 leads to the development of renal cell carcinoma through repression of a TFE3-regulated gene is unknown but is a possibility raised by our findings. The finding that PSF and NonO/p54nrb can interact to form a complex (91) suggests that p54nrb-TFE3 might also act as a repressor through the recruitment of PSF to the DNA-bound chimera.
Although our study has focused on the role of PSF-A on influencing the activity of nuclear hormone receptors via its association with the receptor DBD, other effects of PSF may be mediated by either PSF-A and/or PSF-F. Furthermore, PSF-F would be expected to also function as a repressor, and its unique C terminus may target it to other transcription factors. Thus, in addition to its possible role as a splicing factor, PSF may mediate repression (i) through its association with nuclear hormone receptors, (ii) through its association with other transcription factors, or (iii) by direct binding to specific genes. The finding that PSF can interact with multiple factors (e.g., nuclear hormone receptors, Sin3A, NonO/p54nrb, and topoisomerase I) and bind to specific DNA sequences suggests that PSF is a multifunctional protein that mediates diverse activities in the cell. Our studies establish that PSF can mediate repression through the Sin3 pathway and define a new mechanism for silencing of gene expression by nuclear hormone receptors and possibly other transcription factors.
ACKNOWLEDGMENTS
Plasmid containing NonO cDNA and antibodies to NonO and PSF as well as pET-PSF1 were from the P.W.T. laboratory and have not been previously described. We also thank James Patton for antibodies to PSF, Mitchell Lazar for antibodies to N-CoR, and Martin Privalsky for the GST-Sin3 and GST-SMRT clones. We also thank Dansheng Li, Muktar Mahajan, and Bruce Raaka of the H.H.S. laboratory for critically reading the manuscript.
This research was supported by NIH grants DK16636 (H.H.S.), AI18016 (P.W.T.), and AR02083 (M.M.). H.H.S is a member of the NYUMC Cancer Center (CA16087). Sequence analysis and database searches were through the NYUMC Research Computing Resource, which received support from the National Science Foundation (DIR-8908095).
REFERENCES
- 1.Akhmedov A T, Lopez B S. Human 100-kDa homologous DNA-pairing protein is the splicing factor PSF and promotes DNA strand invasion. Nucleic Acids Res. 2000;28:3022–3030. doi: 10.1093/nar/28.16.3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anzick S L, Kononen J, Walker R L, Azorsa D O, Tanner M M, Guan X Y, Sauter G, Kallioniemi O P, Trent J M, Meltzer P S. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- 3.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 4.Basu A, Dong B, Krainer A R, Howe C C. The intracisternal A-particle proximal enhancer-binding protein activates transcription and is identical to the RNA- and DNA-binding protein p54nrb/NonO. Mol Cell Biol. 1997;17:677–686. doi: 10.1128/mcb.17.2.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanco J C G, Minucci S, Lu J, Yang X J, Walker K K, Chen H, Evans R M, Nakatani V, Ozato K. The histone acetylase PCAF is a nuclear receptor coactivator. Genes Dev. 1998;12:1638–1651. doi: 10.1101/gad.12.11.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caira F, Antonson P, Pelto-Huikko M, Treuter E, Gustafsson J A. Cloning and characterization of RAP250, a novel nuclear receptor coactivator. J Biol Chem. 2000;275:5308–5317. doi: 10.1074/jbc.275.8.5308. [DOI] [PubMed] [Google Scholar]
- 7.Carson-Jurica M A, Schrader W T, O'Malley B W. Steroid receptor family: structure and functions. Endocrine Rev. 1990;11:201–218. doi: 10.1210/edrv-11-2-201. [DOI] [PubMed] [Google Scholar]
- 8.Casanova J, Helmer E, Selmi-Ruby S, Qi J-S, Au-Fliegner M, Desai-Yajnik V, Koudinova N, Yarm F, Raaka B M, Samuels H H. Functional evidence for ligand-dependent dissociation of thyroid hormone and retinoic acid receptors from an inhibitory cellular factor. Mol Cell Biol. 1994;14:5756–5765. doi: 10.1128/mcb.14.9.5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chakravarti D, LaMorte V J, Nelson M C, Nakajima T, Schulman I G, Juguilon H, Montminy M, Evans R M. Role of CBP/P300 in nuclear receptor signalling. Nature. 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 10.Chanas-Sacre G, Mazy-Servais C, Wattiez R, Pirard S, Rogister B, Patton J G, Belachew S, Malgrange B, Moonen G, Leprince P. Identification of PSF, the polypyrimidine tract-binding protein-associated splicing factor, as a developmentally regulated neuronal protein. J Neurosci Res. 1999;57:62–73. doi: 10.1002/(SICI)1097-4547(19990701)57:1<62::AID-JNR7>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 11.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 12.Chen J D, Evans R M. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 13.Clark J, Lu Y J, Sidhar S K, Parker C, Gill S, Smedley D, Hamoudi R, Linehan W M, Shipley J, Cooper C S. Fusion of splicing factor genes PSF and NonO (p54nrb) to the TFE3 gene in papillary renal cell carcinoma. Oncogene. 1997;15:2233–2239. doi: 10.1038/sj.onc.1201394. [DOI] [PubMed] [Google Scholar]
- 14.Damm K, Thompson C C, Evans R M. Protein encoded by v-erbA functions as a thyroid hormone receptor antagonist. Nature. 1989;339:593–597. doi: 10.1038/339593a0. [DOI] [PubMed] [Google Scholar]
- 15.David G, Alland L, Hong S H, Wong C W, DePinho R A, Dejean A. Histone deacetylase associated with mSin3A mediates repression by the acute promyelocytic leukemia-associated PLZF protein. Oncogene. 1998;16:2549–2556. doi: 10.1038/sj.onc.1202043. [DOI] [PubMed] [Google Scholar]
- 16.DeAngelo D J, DeFalco J, Rybacki L, Childs G. The embryonic enhancer-binding protein SSAP contains a novel DNA-binding domain which has homology to several RNA-binding proteins. Mol Cell Biol. 1995;15:1254–1264. doi: 10.1128/mcb.15.3.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeFalco J, Childs G. The embryonic transcription factor stage specific activator protein contains a potent bipartite activation domain that interacts with several RNA polymerase II basal transcription factors. Proc Natl Acad Sci USA. 1996;93:5802–5807. doi: 10.1073/pnas.93.12.5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desai-Yajnik V, Hadzic E, Modlinger P, Malhotra S, Gechlik G, Samuels H H. Interactions of thyroid hormone receptor with the human immunodeficiency virus type 1 (HIV-1) long terminal repeat and the HIV-1 Tat transactivator. J Virol. 1995;69:5103–5112. doi: 10.1128/jvi.69.8.5103-5112.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Desai-Yajnik V, Samuels H H. The NF-κB and Sp1 DNA motifs of the human immunodeficiency virus type 1 long terminal repeat function as novel thyroid hormone response elements. Mol Cell Biol. 1993;13:5057–5069. doi: 10.1128/mcb.13.8.5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong B, Horowitz D S, Kobayashi R, Krainer A R. Purification and cDNA cloning of HeLa cell p54nrb, a nuclear protein with two RNA recognition motifs and extensive homology to human splicing factor PSF and Drosophila NONA/BJ6. Nucleic Acids Res. 1993;21:4085–4092. doi: 10.1093/nar/21.17.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Folkers G E, van der Leede B M, van der Saag P T. The retinoic acid receptor-β2 contains two separate cell-specific transactivation domains, at the N-terminus and in the ligand binding domain. Mol Endocrinol. 1993;7:616–627. doi: 10.1210/mend.7.4.8389001. [DOI] [PubMed] [Google Scholar]
- 23.Fondell J D, Ge H, Roeder R G. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc Natl Acad Sci USA. 1996;93:8329–8333. doi: 10.1073/pnas.93.16.8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forman B M, Casanova J, Raaka B M, Ghysdael J, Samuels H H. Half-site spacing and orientation determines whether thyroid hormone and retinoic acid receptors and related factors bind to DNA response elements as monomers, homodimers, or heterodimers. Mol Endocrinol. 1992;6:429–442. doi: 10.1210/mend.6.3.1316541. [DOI] [PubMed] [Google Scholar]
- 25.Forman B M, Samuels H H. pEXPRESS: A family of expression vectors containing a single transcription unit active in prokaryotes, eukaryotes and in vitro. Gene. 1991;105:9–15. doi: 10.1016/0378-1119(91)90507-8. [DOI] [PubMed] [Google Scholar]
- 26.Gelmetti V, Zhang J, Fanelli M, Minucci S, Pelicci P G, Lazar M A. Aberrant recruitment of the nuclear receptor corepressor-histone deacetylase complex by the acute myeloid leukemia fusion partner ETO. Mol Cell Biol. 1998;18:7185–7191. doi: 10.1128/mcb.18.12.7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gozani O, Patton J G, Reed R. A novel set of spliceosome-associated proteins and the essential splicing factor PSF bind stably to pre-mRNA prior to catalytic step II of the splicing reaction. EMBO J. 1994;13:3356–3367. doi: 10.1002/j.1460-2075.1994.tb06638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grignani F, De Matteis S, Nervi C, Tomassoni L, Gelmetti V, Cioce M, Fanelli M, Ruthardt M, Ferrara F F, Zamir I, Seiser C, Lazar M A, Minucci S, Pelicci P G. Fusion proteins of the retinoic acid receptor-alpha recruit histone deacetylase in promyelocytic leukaemia. Nature. 1998;391:815–818. doi: 10.1038/35901. [DOI] [PubMed] [Google Scholar]
- 29.Gurnell M, Wentworth J M, Agostini M, Adams M, Collingwood T N, Provenzano C, Browne P O, Rajanayagam O, Burris T P, Schwabe J W, Lazar M A, Chatterjee V K. A dominant-negative peroxisome proliferator-activated receptor gamma (PPARgamma) mutant is a constitutive repressor and inhibits PPARgamma- mediated adipogenesis. J Biol Chem. 2000;275:5754–5759. doi: 10.1074/jbc.275.8.5754. [DOI] [PubMed] [Google Scholar]
- 30.Hanstein B, Eckner R, DiRenzo J, Halachmi S, Liu H, Searcy B, Kurokawa R, Brown M. p300 is a component of an estrogen receptor coactivator complex. Proc Natl Acad Sci USA. 1996;93:11540–11545. doi: 10.1073/pnas.93.21.11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heinzel T, Lavinsky R M, Mullen T M, Soderstrom M, Laherty C D, Torchia J, Yang W M, Brard G, Ngo S D, Davie J R, Seto E, Eisenman R N, Rose D W, Glass C K, Rosenfeld M G. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 32.Hong H, Kohli K, Garabedian M, Stallcup M R. GRIP1, a transcriptional coactivator for the AF-2 transactivation domain of steroid, thyroid, retinoid, and vitamin D receptors. Mol Cell Biol. 1997;17:2735–2744. doi: 10.1128/mcb.17.5.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hong H, Kohli K, Trivedi A, Johnson D L, Stallcup M R. GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc Natl Acad Sci USA. 1996;93:4948–4952. doi: 10.1073/pnas.93.10.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hong S H, David G, Wong C W, Dejean A, Privalsky M L. SMRT corepressor interacts with PLZF and with the PML-retinoic acid receptor alpha (RARalpha) and PLZF-RARalpha oncoproteins associated with acute promyelocytic leukemia. Proc Natl Acad Sci USA. 1997;94:9028–9033. doi: 10.1073/pnas.94.17.9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horlein A J, Naar A M, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamil Y, Soderstrom M, Glass C K, Rosenfeld M G. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 36.Huang E Y, Zhang J, Miska E A, Guenther M G, Kouzarides T, Lazar M A. Nuclear receptor corepressors partner with class II histone deacetylases in a Sin3-independent repression pathway. Genes Dev. 2000;14:45–54. [PMC free article] [PubMed] [Google Scholar]
- 37.Ito M, Yuan C X, Malik S, Gu W, Fondell J D, Yamamura S, Fu Z Y, Zhang X, Qin J, Roeder R G. Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol Cell. 1999;3:361–370. doi: 10.1016/s1097-2765(00)80463-3. [DOI] [PubMed] [Google Scholar]
- 38.Jimenez-Lara A M, Aranda A. The vitamin D receptor binds in a transcriptionally inactive form and without a defined polarity on a retinoic acid response element. FASEB J. 1999;13:1073–1081. doi: 10.1096/fasebj.13.9.1073. [DOI] [PubMed] [Google Scholar]
- 39.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S-C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 40.Kao H Y, Downes M, Ordentlich P, Evans R M. Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes Dev. 2000;14:55–66. [PMC free article] [PubMed] [Google Scholar]
- 41.Knoepfler P S, Eisenman R N. Sin meets NuRD and other tails of repression. Cell. 1999;99:447–450. doi: 10.1016/s0092-8674(00)81531-7. [DOI] [PubMed] [Google Scholar]
- 42.Ko L, Cardona G R, Chin W W. Thyroid hormone receptor-binding protein, an LXXLL motif-containing protein, functions as a general coactivator. Proc Natl Acad Sci USA. 2000;97:6212–6217. doi: 10.1073/pnas.97.11.6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laherty C D, Yang W M, Sun J M, Davie J R, Seto E, Eisenman R N. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 44.Lee S K, Anzick S L, Choi J E, Bubendorf L, Guan X Y, Jung Y K, Kallioniemi O P, Kononen L, Trent J M, Azorsa D, Jhun B H, Cheong J H, Lee Y C, Meltzer P S, Lee J W. A nuclear factor, ASC-2, is a cancer-amplified transcriptional coactivator essential for ligand-dependent transactivation by nuclear receptors in vivo. J Biol Chem. 1999;274:34283–34293. doi: 10.1074/jbc.274.48.34283. [DOI] [PubMed] [Google Scholar]
- 45.Li D, Desai-Yajnik V, Lo E, Schapira M, Abagyan R, Samuels H H. NRIF3 is a novel coactivator mediating functional specificity of nuclear hormone receptors. Mol Cell Biol. 1999;19:7191–7202. doi: 10.1128/mcb.19.10.7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H, Gomes P J, Chen J D. RAC3, a steroid/nuclear receptor-associated coactivator that is related to SRC-1 and TIF2. Proc Natl Acad Sci USA. 1997;94:8479–8484. doi: 10.1073/pnas.94.16.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin R J, Nagy L, Inoue S, Shao W, Miller W H, Jr, Evans R M. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature. 1998;391:811–814. doi: 10.1038/35895. [DOI] [PubMed] [Google Scholar]
- 48.Lindsey L A, Crow A J, Garcia-Blanco M A. A mammalian activity required for the second step of pre-messenger RNA splicing. J Biol Chem. 1995;270:13415–13421. doi: 10.1074/jbc.270.22.13415. [DOI] [PubMed] [Google Scholar]
- 49.Lutterbach B, Westendorf J J, Linggi B, Pattern A, Moniwa M, Davie J R, Huynh K D, Bardwell V J, Lavinsky R M, Rosenfeld M G, Glass C, Seto E, Hiebert S W. ETO, a target of t(8;21) in acute leukemia, interacts with the N-CoR and mSin3 corepressors. Mol Cell Biol. 1998;18:7176–7184. doi: 10.1128/mcb.18.12.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lutz C S, Cooke C, O'Connor J P, Kobayashi R, Alwine J C. The snRNP-free U1A (SF-A) complex(es): identification of the largest subunit as PSF, the polypyrimidine-tract binding protein-associated splicing factor. RNA. 1998;4:1493–1499. doi: 10.1017/s1355838298981183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mahajan M A, Samuels H H. A new family of nuclear receptor coregulators that integrate nuclear receptor signaling through CREB-binding protein. Mol Cell Biol. 2000;20:5048–5063. doi: 10.1128/mcb.20.14.5048-5063.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McKenna N J, Lanz R B, O'Malley B W. Nuclear receptor coregulators: cellular and molecular biology. Endocrine Rev. 1999;20:321–344. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- 53.Meissner M, Dechat T, Gerner C, Grimm R, Foisner R, Sauermann G. Differential nuclear localization and nuclear matrix association of the splicing factors PSF and PTB. J Cell Biochem. 2000;76:559–566. [PubMed] [Google Scholar]
- 54.Nagy L, Kao H V, Chakravarti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 55.Newberry E P, Latifi T, Towler D A. The RRM domain of MINT, a novel Msx2 binding protein, recognizes and regulates the rat osteocalcin promoter. Biochemistry. 1999;38:10678–10690. doi: 10.1021/bi990967j. [DOI] [PubMed] [Google Scholar]
- 56.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 57.Onate S A, Tsai S Y, Tsai M-J, O'Malley B W. Sequence and characterization of a coactivator of the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 58.Patton J G, Porro E B, Galceran J, Tempst P, Nadal-Ginard B. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Dev. 1993;7:393–406. doi: 10.1101/gad.7.3.393. [DOI] [PubMed] [Google Scholar]
- 59.Powers C A, Mathur M, Raaka B M, Ron D, Samuels H H. TLS (translocated-in-liposarcoma) is a high-affinity interactor for steroid, thyroid hormone, and retinoid receptors. Mol Endocrinol. 1998;12:4–18. doi: 10.1210/mend.12.1.0043. [DOI] [PubMed] [Google Scholar]
- 60.Puigserver P, Wu Z, Park C W, Graves R, Wright M, Spiegelman B M. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 61.Qi J-S, Desai-Yajnik V, Greene M E, Raaka B M, Samuels H H. The ligand binding domains of the thyroid hormone/retinoid receptor gene subfamily function in vivo to mediate heterodimerization, gene silencing, and transactivation. Mol Cell Biol. 1995;15:1817–1825. doi: 10.1128/mcb.15.3.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qi J-S, Desai-Yajnik V, Yuan Y, Samuels H H. Constitutive activation of gene expression by thyroid hormone receptor results from reversal of p53-mediated repression. Mol Cell Biol. 1997;17:7195–7207. doi: 10.1128/mcb.17.12.7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rachez C, Lemon B D, Suldan Z, Bromleigh V, Gamble M, Naar A M, Erdjument-Bromage H, Tempst P, Freedman L P. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature. 1999;398:824–828. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- 64.Rachez C, Suldan Z, Ward J, Chang C P, Burakov D, Erdjument-Bromage H, Tempst P, Freedman L P. A novel protein complex that interacts with the vitamin D3 receptor in a ligand-dependent manner and enhances VDR transactivation in a cell-free system. Genes Dev. 1998;12:1787–1800. doi: 10.1101/gad.12.12.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 66.Sadowski I, Ptashne M. A vector for expressing GAL4(1-147) fusions in mammalian cells. Nucleic Acids Res. 1989;17:7539. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sande S, Privalsky M L. Identification of TRACs (T3 receptor-associating cofactors), a family of cofactors that associate with, and modulate the activity of, nuclear hormone receptors. Mol Endocrinol. 1996;10:813–825. doi: 10.1210/mend.10.7.8813722. [DOI] [PubMed] [Google Scholar]
- 68.Schulman I G, Juguilon H, Evans R M. Activation and repression by nuclear hormone receptors: hormone modulates an equilibrium between active and repressive states. Mol Cell Biol. 1996;16:3807–3813. doi: 10.1128/mcb.16.7.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M J, O'Malley B W. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 70.Stanley F, Samuels H H. n-Butyrate effects thyroid hormone stimulation of prolactin production and mRNA levels in GH1 cells. J Biol Chem. 1984;259:9768–9775. [PubMed] [Google Scholar]
- 71.Straub T, Grue P, Uhse A, Lisby M, Knudsen B R, Tange T O, Westergaard O, Boege F. The RNA-splicing factor PSF/p54 controls DNA-topoisomerase I activity by a direct interaction. J Biol Chem. 1998;273:26261–26264. doi: 10.1074/jbc.273.41.26261. [DOI] [PubMed] [Google Scholar]
- 72.Straub T, Knudsen B R, Boege F. PSF/p54nrb stimulates “jumping” of DNA topoisomerase I between separate DNA helices. Biochemistry. 2000;39:7552–7558. doi: 10.1021/bi992898e. [DOI] [PubMed] [Google Scholar]
- 73.Tagami T, Lutz W H, Kumar R, Jameson J L. The interaction of the vitamin D receptor with nuclear receptor corepressors and coactivators. Biochem Biophys Res Commun. 1998;253:358–363. doi: 10.1006/bbrc.1998.9799. [DOI] [PubMed] [Google Scholar]
- 74.Takeshita A, Cardona G R, Koibuchi N, Suen C S, Chin W W. TRAM-1, a novel 160-kDa thyroid hormone receptor activator molecule, exhibits distinct properties from steroid receptor coactivator-1. J Biol Chem. 1997;272:27629–27634. doi: 10.1074/jbc.272.44.27629. [DOI] [PubMed] [Google Scholar]
- 75.Tanaka M, Gupta R, Mayer B J. Differential inhibition of signaling pathways by dominant-negative SH2/SH3 adapter proteins. Mol Cell Biol. 1995;15:6829–6837. doi: 10.1128/mcb.15.12.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tiscornia G, Mahadevan M S. Myotonic dystrophy: the role of the CUG triplet repeats in splicing of a novel DMPK exon and altered cytoplasmic DMPK mRNA isoform ratios. Mol Cell. 2000;5:959–967. doi: 10.1016/s1097-2765(00)80261-0. [DOI] [PubMed] [Google Scholar]
- 77.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 78.Urban R J, Bodenburg Y, Kurosky A, Wood T G, Gasic S. Polypyrimidine tract-binding protein-associated splicing factor is a negative regulator of transcriptional activity of the porcine p450scc insulin-like growth factor response element. Mol Endocrinol. 2000;14:774–782. doi: 10.1210/mend.14.6.0485. [DOI] [PubMed] [Google Scholar]
- 79.Voegel J J, Heine M J S, Zechel C, Chambon P, Gronemeyer H. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 1996;15:3667–3675. [PMC free article] [PubMed] [Google Scholar]
- 80.von Zelewsky T, Palladino F, Brunschwig K, Tobler H, Hajnal A, Muller F. The C. elegans Mi-2 chromatin-remodelling proteins function in vulval cell fate determination. Development. 2000;127:5277–5284. doi: 10.1242/dev.127.24.5277. [DOI] [PubMed] [Google Scholar]
- 81.Wang H, Stillman D J. Transcriptional repression in Saccharomyces cerevisiae by a SIN3-LexA fusion protein. Mol Cell Biol. 1993;13:1805–1814. doi: 10.1128/mcb.13.3.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wen Y D, Perissi V, Staszewski L M, Yang W M, Krones A, Glass C K, Rosenfeld M G, Seto E. The histone deacetylase-3 complex contains nuclear receptor corepressors. Proc Natl Acad Sci USA. 2000;97:7202–7207. doi: 10.1073/pnas.97.13.7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wong C W, Privalsky M L. Transcriptional repression by the SMRT-mSin3 corepressor: multiple interactions, multiple mechanisms, and a potential role for TFIIB. Mol Cell Biol. 1998;18:5500–5510. doi: 10.1128/mcb.18.9.5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 85.Yang Y S, Hanke J H, Carayannopoulos L, Craft C M, Capra J D, Tucker P W. NonO, a non-POU-domain-containing, octamer-binding protein, is the mammalian homolog of Drosophila nonAdiss. Mol Cell Biol. 1993;13:5593–5603. doi: 10.1128/mcb.13.9.5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yang Y S, Yang M C, Tucker P W, Capra J D. NonO enhances the association of many DNA-binding proteins to their targets. Nucleic Acids Res. 1997;25:2284–2292. doi: 10.1093/nar/25.12.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yap N, Yu C L, Cheng S Y. Modulation of the transcriptional activity of thyroid hormone receptors by the tumor suppressor p53. Proc Natl Acad Sci USA. 1996;93:4273–4277. doi: 10.1073/pnas.93.9.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yeh S, Chang C. Cloning and characterization of a specific coactivator, ARA70, for the androgen receptor in human prostate cells. Proc Natl Acad Sci USA. 1996;93:5517–5521. doi: 10.1073/pnas.93.11.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yoshida M, Kijima M, Akita M, Beppu T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J Biol Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]
- 90.Zamir I, Dawson J, Lavinsky R M, Glass C K, Rosenfeld M G, Lazar M A. Cloning and characterization of a corepressor and potential component of the nuclear hormone receptor repression complex. Proc Natl Acad Sci USA. 1997;94:14400–14405. doi: 10.1073/pnas.94.26.14400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang W W, Zhang L X, Busch R K, Farres J, Busch H. Purification and characterization of a DNA-binding heterodimer of 52 and 100 kDa from HeLa cells. Biochem J. 1993;290:267–272. doi: 10.1042/bj2900267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang Y, Ng H H, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]