Abstract
To decipher the mechanistic roles of Mediator proteins in regulating developmental specific gene expression and compare them to those of TATA-binding protein (TBP)-associated factors (TAFs), we isolated and analyzed a multiprotein complex containing Drosophila Mediator (dMediator) homologs. dMediator interacts with several sequence-specific transcription factors and basal transcription machinery and is critical for activated transcription in response to diverse transcriptional activators. The requirement for dMediator did not depend on a specific core promoter organization. By contrast, TAFs are preferentially utilized by promoters having a specific core element organization. Therefore, Mediator proteins are suggested to act as a pivotal coactivator that integrates promoter-specific activation signals to the basal transcription machinery.
Precise regulation of gene expression is fundamentally required for a broad spectrum of developmental processes in multicellular organisms. Although distinct sequence-specific transcription factors are primarily responsible for this regulation, transcriptional coactivator-corepressor proteins also add a significant secondary layer to the regulation of gene expression. A number of coactivator complexes have been identified in eukaryotes, and their functions in gene activation at specific promoters have been analyzed, primarily in vitro. However, the mechanism by which these transcriptional coactivators regulate gene expression in living organisms is not well understood.
Among eukaryotic transcriptional coactivators, two classes of proteins, Mediator proteins and TATA-binding protein (TBP)-associated factors (TAFs), are central to the process of transcriptional regulation. These proteins were isolated as multiprotein complexes composed of more than 10 polypeptides and associate with the basal transcription machinery (RNA polymerase II [Pol II] and TBP, respectively). Both complexes interact with transcriptional activators and are required for transcriptional activation in reconstituted in vitro systems (3, 8, 40). These facts suggest that Mediator and TAF complexes function as coactivators by relaying transcriptional activation signals from DNA-bound activators to the basal transcription machinery. However, it has not been clearly determined whether Mediator and TAF complexes contribute redundantly or distinctly to the transcriptional activation process, nor have their mechanistic roles in Pol II transcription been clearly deciphered.
Although both Mediator and TAF complexes were initially identified from in vitro assays, recent genetic analyses in yeast suggest that TAFs do not function as general coactivators under physiological conditions. First, the depletion of various TFIID-specific TAFs does not have a significant effect on transcriptional activation of most genes in yeast (12, 28, 41). Second, yeast TAFII145/130 was shown to function as a core promoter selectivity factor rather than a general coactivator (36). These observations are in good agreement with earlier reports that certain TAFs, especially Drosophila TAFII40 (dTAFII40), dTAFII60, dTAFII150, and dTAFII250, directly recognize core promoter elements (7, 39) and thus are likely to act as promoter selectivity factors.
The Mediator complex was first identified in yeast as a multiprotein complex (15, 16) containing functionally distinct modular subassemblies composed of subunits with similar genetic properties (19). Inactivation of individual Mediator proteins causes widely variable effects on yeast gene expression, ranging from deregulation of transcription of a small subset of genes to a genomewide transcriptional defect (12). A number of coactivator complexes related to yeast Mediator have been subsequently isolated in mammals (5, 9, 13, 29, 31, 33, 37). Like yeast Mediator, mammalian complexes play a key role in regulating Pol II transcription in vitro. However, their compositional and functional heterogeneity indicates that there has been a considerable increase in the functional diversity of Mediator complexes during evolution.
In this study, we analyzed the function of the Drosophila Mediator (dMediator) complex in order to gain insight into the mechanism by which these coactivator complexes govern eukaryotic transcriptional activation during Drosophila development. As the first step toward this goal, we isolated a multiprotein complex containing Drosophila homologs of yeast and mammalian Mediator proteins. Our data shows that dMediator interacts physically with several sequence-specific transcription factors and basal transcription machinery and is critically required for transcriptional activation in response to diverse transcriptional activators in vitro. By contrast, the TAF complex is dispensable for transcription of TATA-containing promoters in the presence of dMediator but is required for transcription of TATA-less promoters that depend on an initiator element (Inr) and downstream promoter element (DPE). Our results suggest that dMediator functions as an essential coactivator complex that integrates diverse gene-specific regulatory signals at specific promoters, while TAF complex has its distinct role as a promoter selectivity factor required for the expression of genes with a specific type of core promoter.
MATERIALS AND METHODS
Plasmid constructs.
The transcription templates pG5E4T and pE4T(dl-E)5 were provided by Michael Carey and Albert Courey, respectively. The pG5-Adh40/10-G380, pNP3-Adh40/10-G380, and pAdh-G280 plasmids were provided by James Manley. pG5en and pG5AntpP2 were constructed by cloning PCR-amplified sequences encoding nucleotides -45 to +155 of the engrailed (en) promoter and -45 to +159 of the Antennapedia P2 promoter (AntpP2), respectively, into BamHI/EcoRI-digested pG5E4T.
To construct pGEX-Eve, the BamHI (blunted)-EcoRI fragment from pET-GAL4-EveBCDEF-H6 (24), which encodes amino acids 61 to 246 of Even-skipped, was inserted into EcoRI/SmaI-digested pGEX-4T1 (Amersham Pharmacia Biotech).
Antibodies.
Antisera against dMediator homologs were generated by immunization of rats with glutathione S-transferase (GST) fusion proteins containing either full-length (dMED6, dSOH1, dp34, Trfp, dp28b, and dSRB7) or partial (amino acids 221 to 368 of dRGR1 and amino acids 1 to 168 of dTRAP80) polypeptide. Polyclonal antibodies (Abs) used in immunoblotting and immunoprecipitation were affinity purified from the antisera by antigen affinity chromatography (10).
Buffers.
All buffers used in nuclear protein extraction, column chromatography and dialysis contained 1 mM dithiothreitol (DTT) and protease inhibitors (10 μM phenylmethylsulfonyl fluoride, 20 nM pepstatin A, 6 nM leupeptin, and 20 μM bisbenzamidine) unless otherwise specified. 2× lysis buffer contained 100 mM HEPES-KOH (pH 7.6), 500 mM potassium acetate, 2 mM EDTA, and 20% glycerol. Buffer An contained 50 mM HEPES-KOH (pH 7.6), 1 mM EDTA, 10% glycerol, and n mM potassium acetate. Buffer Sn contained 20 mM HEPES-KOH (pH 7.6), 0.1 mM EDTA, 10% glycerol, and n mM potassium acetate. Buffer Hn contains 100 mM potassium acetate, 10% glycerol, and n mM potassium phosphate (pH 7.6). Buffer IPn is identical to buffer Sn except that it contains nonionic detergent IGEPAL CA-630 (Sigma) up to 0.2% and lacks DTT and protease inhibitors.
Purification of dMediator.
All operations were carried out at 4°C. The amounts of dMediator proteins in nuclear extracts and column fractions were monitored by immunoblotting with anti-dSOH1, anti-dMED6, and anti-dSRB7 Abs. Drosophila embryo nuclei were isolated as described previously (14) and suspended in 2× lysis buffer (1 ml per g of embryos). The resulting suspension was stirred with a magnetic bar for 30 min and centrifuged in an SW41Ti rotor (Beckman) at 30,000 rpm. The clear interface solution was diluted with buffer A0 to adjust the potassium acetate concentration to 100 mM and then applied on a heparin-Sepharose column (1 ml per 20 mg of protein) equilibrated with buffer A100. The resin was washed with buffer A100, and bound proteins were eluted with buffer A300. The eluted proteins were suspended in buffer S60 by dialysis and dilution with S0 and applied on an SP-Sepharose column (1 ml per 20 mg of protein) preequilibrated with buffer S60. The resin was washed with buffer S60, and bound proteins were eluted with buffer S300. The eluted proteins were suspended in buffer H10 by dialysis and applied on a hydroxyapatite HTP column (1 ml per 2 mg of proteins) preequilibrated with buffer H10. The resin was washed with buffer H10, and bound proteins were eluted with a linear gradient from buffer H10 to buffer H500. More than 80% of dMediator proteins were eluted at a potassium phosphate concentration between 350 and 400 mM even though there existed multiple minor peaks of dMediator proteins eluted at lower concentations. The major peak fractions of dMediator proteins (at 350 to 400 mM potassium phosphate concentrations) were pooled, dialyzed in buffer S100, and applied on a Mono S column (1 ml) equilibrated with buffer S100. The resin was washed with buffer S100, and bound proteins were eluted with a linear gradient from buffer S100 to buffer S1000. The peak fractions of dMediator proteins were pooled and incubated with an anti-dSOH1 beads (0.1 ml) that were prepared as described elsewhere (19). The beads were washed with buffer IP300, and bound proteins were eluted twice with 0.1 ml of buffer IP100 containing 5 M urea or with 0.1 ml of 0.1 M glycine-HCl (pH 2.5). The urea-eluted dMediator fractions were dialyzed in buffer A100 and stored at −80°C.
In vitro transcription analyses.
Soluble nuclear fraction was prepared from 0- to 12-h Drosophila embryos as described elsewhere (14) except that 100 mM potassium acetate and 12.5 mM magnesium acetate were used in the nuclear extraction buffer instead of potassium glutamate and magnesium chloride. For depletion of endogenous dMediator and TFIID, 0.4 ml of soluble nuclear fraction was incubated for 2 h at 4°C with 0.1 ml of protein G-agarose beads retaining 0.1 mg of anti-dSOH1 or anti-dTAFII250 Ab. The supernatants were transferred to a fresh tube containing an equal amount of the Ab-bound beads and incubated for another 2 h. The final supernatants were kept frozen at −80°C and used in transcription assays. TFIID was prepared from Drosophila embryo nuclear extract using heparin-Sepharose, Q-Sepharose, hydroxyapatite, and Mono S chromatographies, with dTAFII250 and TBP being monitored by immunoblotting. Transcription reactions were performed in 25 μl containing 50 mM HEPES-KOH (pH 7.6), 50 mM potassium acetate, 10 mM magnesium acetate, 5% glycerol, 0.2 mM EDTA, 1 mM DTT, 1 mM each nucleoside triphosphate, 20 U of RNasin (Promega), 40 μg of soluble nuclear fraction proteins, and 50 ng of plasmid template. After incubation for 30 min at 25°C, the reaction was terminated, and transcribed RNA was isolated and analyzed as described elsewhere (2). For G-less template assay, transcription reactions were carried out as described above except that (i) 50 μM UTP and 0.2 mM 3′-O-methyl-GTP (Pharmacia) were substituted for 1 mM UTP and 1 mM GTP and (ii) 1 U of RNase T1 (Roche) and 10 μCi of [α-32P]UTP were included. Transcribed RNA was isolated as in primer extension assay and analyzed by electrophoresis on a 6% polyacrylamide gel and autoradiography.
Protein-protein interaction assays.
GST fusion proteins used in GST pulldown assays were expressed in Escherichia coli DH5α and purified by glutathione-Sepharose chromatography (Amersham Pharmacia Biotech) as described elsewhere (20). The GST construct containing the Drosophila Pol II C-terminal domain (CTD) was obtained from Arno Greenleaf. FLAG-Dorsal was expressed in Sf9 cells from the baculovirus construct provided by Albert Courey and purified as described elsewhere (43). GST and FLAG pulldown assays were performed by incubating 40 μg of soluble nuclear fraction proteins (Fig. 5B) or 0.2 μg of dMediator proteins in the Mono S fraction 24 (Fig. 6A) and 10 μl beads retaining 1 μg of each GST-fusion protein in 0.3 ml of IP300 buffer for 6 h at 4°C, washing the beads with IP300 buffer, and eluting bound proteins with sodium dodecyl sulfate (SDS) gel sample buffer. One-fifth of the input proteins and the bound proteins were analyzed by immunoblotting.
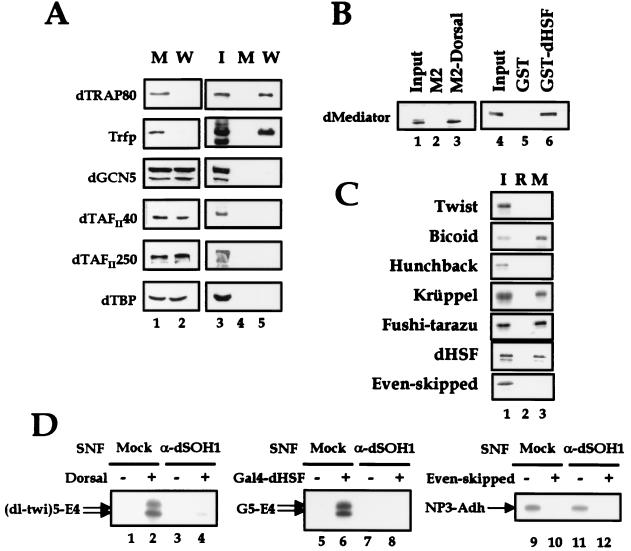
FIG. 5.
Interaction of dMediator with sequence-specific transcription factors. (A) Physical interaction of Drosophila coactivator proteins with the VP16 activation domain. Nuclear extract was incubated with GST beads containing the wild-type (W) and Δ456FP442 (M) VP16 activation domains. Proteins in unbound fractions (lanes 1 and 2), bead-bound proteins (lanes 4 and 5), and the input extract (I; lane 3) were analyzed by SDS-PAGE and then immunoblotted with the Abs specific for the coactivator proteins indicated on the left. (B) Physical interaction of dMediator with Dorsal and dHSF. FLAG-Dorsal and GST-dHSF fusion proteins were immobilized on beads and used in pulldown assays as in panel. A. Binding of dMediator to each bead was monitored by immunoblotting with anti-Trfp (lanes 1 to 3) and anti-dTRAP80 (lanes 4 to 6) Abs. (C) Physical interaction of dMediator with several sequence-specific transcription factors. dMediator was immobilized on anti-dSOH1 beads and incubated with the 35S-labeled transcription factors indicated at the left. The input sample (I) and proteins bound to blank resin (R) and dMediator-immobilized beads (M) were separated by SDS-PAGE and analyzed by autoradiography. (D) Requirement of dMediator for transcriptional regulation by Dorsal, dHSF, and Even-skipped. Plasmid templates containing the respective binding sites for Dorsal, Gal4-dHSF, and Even-skipped were used in transcription reactions. Transcriptional activation by Dorsal and Ga14-dHSF and transcriptional repression by Even-skipped in soluble nuclear fraction (SNF) immunodepleted with anti-β-galactosidase (Mock) and anti-dSOH1 (α-dSOH1) were analyzed as in Fig. 2C. (dl-twi)5, five copies of the Dorsal-Twist binding sites.
FIG. 6.
Interaction of dMediator with the basal transcription machinery. (A) Interaction of dMediator with the Pol II CTD. GST-pulldown assays were carried out as for Fig. 5A and B. dMediator proteins in the input (Mono S fraction 24), GST-bound, and GST-CTD-bound fractions were analyzed by immunoblotting with the antibodies indicated at the left. (B) Phosphorylation of the Pol II CTD by dMediator and TFIIH. GST fusion protein containing Drosophila Pol II CTD was incubated with TFIIH (purified from Drosophila embryo extracts and provided by Gaku Mizuguchi) and/or dMediator and analyzed by immunoblotting with monoclonal Abs that specifically recognize phosphorylated serine 2 (H5) and serine 5 (H14) within the CTD. (C) Physical interaction of dMediator with general transcription factors. 35S-labeled general transcription factors, namely, TBP (T), TFIIB (B), TFIIE-L (EL) and -S (ES), TFIIF-L (FL) and -S (FS), TFIIA-L (AL) and -S (AS), and TFIIS (S), were synthesized in reticulocyte lysates (left) and incubated with bead-bound dMediator as for Fig. 5C. The sizes of molecular weight markers are indicated at the left. Proteins bound to dMediator beads were analyzed by SDS-PAGE and autoradiography (right).
The analyses of transcription factor binding to bead-immobilized dMediator shown in Fig. 5C and 6C were carried out by incubating 35S-labeled proteins synthesized in TnT reticulocyte lysates (Promega) with 10 μl of anti-dSOH1 beads retaining 0.1 μg of dMediator. The beads were washed, and bound proteins along with the input nuclear extracts were analyzed as in the GST pulldown assays.
RESULTS
Drosophila homologs of yeast and mammalian Mediator subunits.
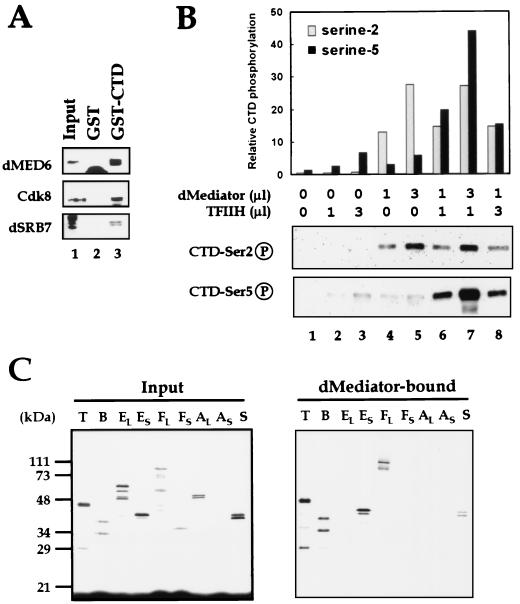
As a first step to identifying the dMediator complex, we cloned the full-length Drosophila MED6 gene by PCR using Drosophila melanogaster embryonic cDNA templates and degenerate primers designed from the conserved regions of the yeast and human MED6 proteins (Fig. 1A) (B. S. Gim and Y. J. Kim, unpublished data). Additionally, we searched the Drosophila expressed sequence tag and genomic databases. We identified the Drosophila homologs of five components with known yeast and mammalian counterparts (Fig. 1A) and of 11 Mediator components known only in mammals (Fig. 1B).
FIG. 1.
Amino acid sequence alignment of dMediator Homologs. (A) Mediator homologs conserved in Drosophila, human, and yeast (denoted by prefixes “d,” “h,” and “y,” respectively). The conserved residues are marked with boxes (black boxes for residues where all three sequences are identical or similar; gray boxes where two of the three are identical or similar). Only the conserved regions are shown. (B) Metazoan-specific Mediator homologs. The conserved amino acid sequences are marked as in panel A. hp28b and hp34 are the human homologs of mouse Mediator proteins p28b and p34 (13).
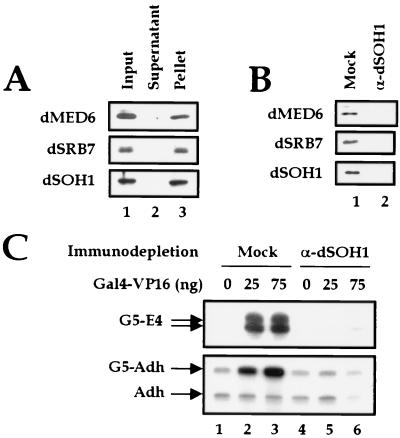
To examine whether the dMediator homologs associate to form a complex as do their yeast and mammalian counterparts, we immunoprecipitated Drosophila embryo nuclear extracts with affinity-purified anti-dMED6 Ab. Immunoblotting revealed that dMED6 protein was precipitated from the extract, together with dSRB7 and dSOH1 (Fig. 2A). When the extract was immunoprecipitated with anti-dSOH1, the same three proteins were coprecipitated (data not shown). This result suggests that these three dMediator homologs associate to form a putative Mediator complex.
FIG. 2.
Identification of the dMediator complex. (A) Immunoprecipitation of nuclear extracts with anti-dMED6 Ab. Equivalent amounts of nuclear extract input, supernatant, and pellet of the immunoprecipitation were resolved by SDS-PAGE and immunoblotted with the antibodies indicated at the left. (B) Immunodepletion of nuclear extracts with anti-β-galactosidase (Mock) and anti-dSOH1 (α-dSOH1). The supernatants obtained after incubation were analyzed as in panel A. (C) Transcriptional activation of the E4 and Adh promoter constructs by Gal4-VP16 in nuclear extracts. Before the in vitro transcription assay, the nuclear extracts were immunodepleted with anti-β-galactosidase (Mock) or anti-dSOH1 (α-dSOH1). The amount (nanograms) of recombinant Gal4-VP16 added to the reactions is indicated at the top. The transcripts from the E4 templates containing five copies of the Gal4 DNA binding sites (G5-E4) and the alcohol dehydrogenase proximal (Adh) templates with or without five copies of the Gal4 binding sites (G5-Adh and Adh, respectively) are indicated by arrows at the left.
To examine whether this putative dMediator complex is the functional analog of yeast and mammalian Mediator complexes, we examined whether dMediator is required for transcriptional activation. To this end, we produced dMediator-deficient nuclear extract by incubating Drosophila embryo soluble nuclear extract with anti-dSOH1 Ab or a control Ab specific for bacterial β-galactosidase. Immunoblot analysis shows that the dSOH1, dMED6, and dSRB7 proteins in the nuclear extract were efficiently depleted by anti-dSOH1 but not by the control Ab (Fig. 2B). The immunodepletion of the putative Mediator complex from the nuclear extract significantly reduced transcriptional activation of Ga14-VP16 without affecting basal transcription, whereas no obvious transcriptional defect was observed in the mock-depleted nuclear extract (Fig. 2C). These results indicate that the dMediator complex is both a structural and functional analog of the yeast and mammalian Mediator complexes.
Purification of dMediator as a multiprotein complex related to human Mediator complexes.
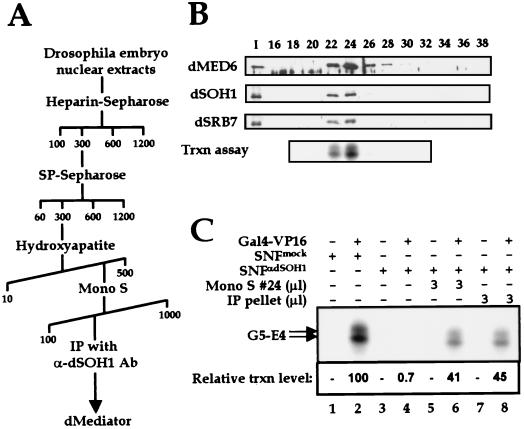
The diversity of Mediator-related coactivator complexes identified in mammals demonstrates that these complexes have great compositional and functional heterogeneity. To test whether the dMediator homologs are part of a multiprotein complex, Drosophila embryo nuclear extract was fractionated using heparin-Sepharose, SP-Sepharose, hydroxyapatite, and Mono S chromatographies (Fig. 3A). dMED6, dSOH1 and dSRB7 were cofractionated during all steps including the last Mono S step (Fig. 3B). The ability of the Mono S fractions to restore transcriptional activation to dMediator-depleted nuclear extract comigrates exactly with the Mediator homologs on the Mono S column (Fig. 3B).
FIG. 3.
Purification of dMediator. (A) Outline of dMediator purification. The numbers indicate the concentrations (millimolar) of potassium acetate (heparin-Sepharose, SP-Sepharose, and Mono S) or potassium phosphate (hydroxyapatite). (B) Immunoblot and transcriptional (Trxn) analyses of Mono S fractions. Equal amounts of the input (I) samples were loaded on the Mono S column, and the eluted fractions (numbers above the lanes) were resolved by SDS-PAGE and immunoblotted with the Abs indicated at the left. Mono S fractions 18 to 32 were added to the transcription reactions containing the Gal4-VP16 protein and soluble nuclear fraction immunodepleted with anti-dSOH1. (C) Transcription assay of dMediator activity. The Gal4-VP16 protein and soluble nuclear fraction (SNF) immunodepleted with anti-β-galactosidase (SNFmock) or anti-dSOH1 (SNFαdSOH1) were used in transcription reactions as in Fig. 2C. The Mono S peak fraction (#24) and immunopurified dMediator (IP pellet) were added to the reactions as described above.
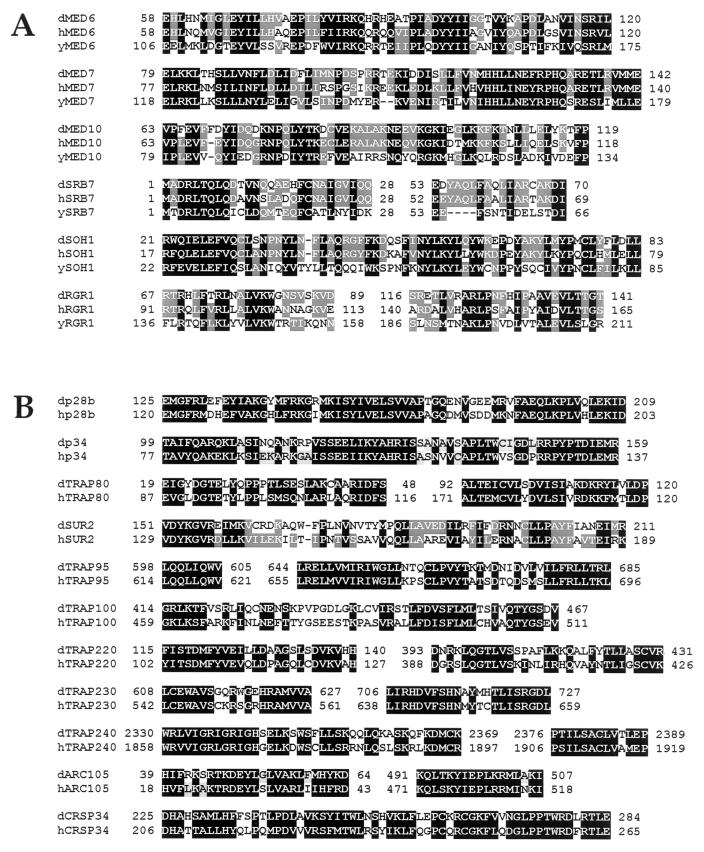
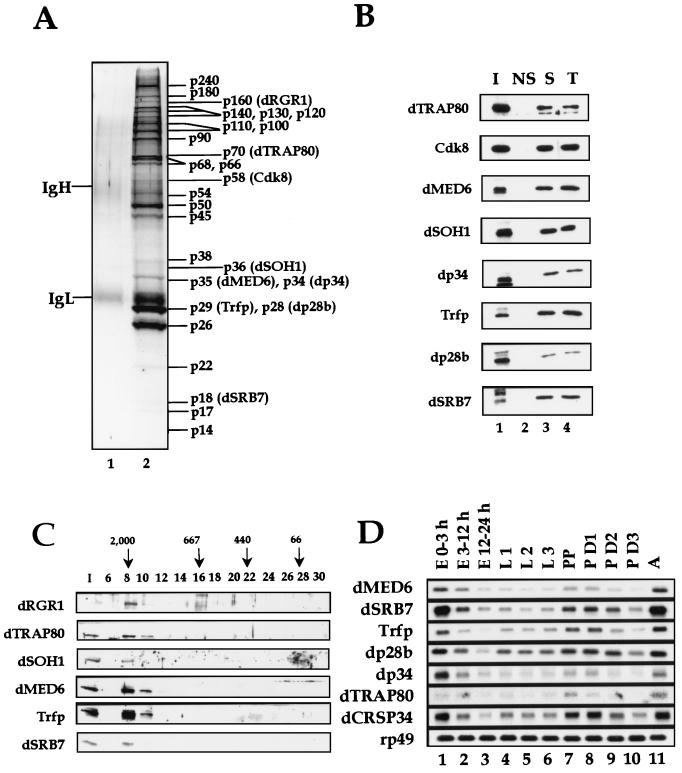
The dMediator complex was immunoprecipitated with anti-dSOH1 Ab from the Mono S fractions. The immunopurified complex as well as the Mono S peak fraction restored transcriptional activation to dMediator-depleted nuclear extract (Fig. 3C, lanes 6 and 8). The restored transcription levels (lanes 6 and 8) were lower than that of the mock-depleted extracts (lane 2). This is presumably due to the partial loss of dMediator activity caused by inactivation of one or more component(s) in a subpopulation of dMediator during purification or, alternatively, by concomitant depletion of other positive factors by the Ab treatment. SDS-polyacrylamide gel electrophoresis (PAGE) and silver staining of the immunoprecipitated dMediator revealed approximately 25 polypeptides of molecular masses ranging from 14 to 240 kDa (Fig. 4A). The coprecipitated polypeptides were identified by immunoblotting with Abs raised against several putative dMediator homologs found from the Drosophila database searches (Fig. 1). These analyses confirmed the presence of dRGR1, dTRAP80, Cdk8, dMED6, dSOH1, dp34, Trfp (13), dp28b, and dSRB7 in the dMediator complex (Fig. 4B, lane 3). When anti-Trfp Ab was used for immunoprecipitation, these polypeptides were also coprecipitated (Fig. 4B, lane 4). Furthermore, mass spectrometric analyses confirmed the presence of dTRAP80, dMED6, and Trfp in the dMediator complex (data not shown). Although we confirmed the identity of only nine dMediator components, the Drosophila genome database contained additional open reading frames homologous to other yeast and mammalian Mediator proteins (Fig. 1). The unidentified components of the dMediator complex may be encoded by some of these homologs. Given the presence of Cdk8 in the dMediator, Drosophila cyclin C (23), the cyclin partner of Cdk8 (18), is also likely a component of the dMediator complex. We are currently raising Abs against these putative dMediator proteins to confirm their presence in the dMediator complex. Female-sterile-homeotic (Fsh), which is a Drosophila homolog of the mouse Mediator component p96a (13), did not cofractionate with, nor was it coprecipitated with, the dMediator proteins (data not shown). This result suggests that Fsh does not stably associate with the Mediator complex in Drosophila. Immunoblot analysis also showed that no detectable amounts of Drosophila CREB binding protein and Pol II were present in the dMediator complex (data not shown).
FIG. 4.
Polypeptide composition and developmental expression pattern of dMediator. (A) Immunoprecipitation of dMediator with anti-dSOH1 beads. Proteins immunoprecipitated from buffer IP100 (lane 1) or the Mono S dMediator peak fraction (lane 2) were electrophoresed on SDS–11.5% polyacrylamide gels and silver stained. Identified dMediator components are indicated at the right. Immunoglobulin heavy (IgH) and light (IgL) chains are indicated at the left. (B) Immunoblot analysis of dMediator proteins immunoprecipitated with nonspecific anti-β-galactosidase (NS), anti-dSOH1 (S), and anti-Trfp (T). The input Mono S fraction (I) is shown for comparison. (C) Superose-6 chromatography of the dMediator complex. The Mono S peak fractions (I) were fractionated on a Superose-6 column, and the fractions (numbers above the lanes) were subjected to SDS-PAGE and analyzed by immunoblotting with the Abs indicated at the left. The elution positions of size markers (in kilodaltons) are indicated by arrows. The size markers used were blue dextran (2,000 kDa; void volume), thyroglobulin (667 kDa), ferritin (440 kDa), and bovine serum albumin (66 kDa). (D) Developmental expression pattern of dMediator homologs. Poly(A) RNA (4 μg per lane) from Drosophila embryos (E), first (L 1)-, second (L 2)-, and third (L 3)-instar larvae, white prepupae (PP), day 1 (P D1), day 2 (P D2), and day 3 (P D3) pupae, and adults (A) were electrophoresed on a formaldehyde agarose gel and probed with radiolabled DNA fragments derived from the dMediator genes indicated at the left. rp49 was used as a positive control.
To estimate the apparent size of the dMediator complex in the Mono S peak fraction, this eluate was further analyzed by Superose-6 gel filtration chromatography. The dMediator complex that we identified migrated at a molecular size of 2 MDa (Fig. 4C). This size is comparable to that observed in the Superose-6 chromatographic analysis of TRAP (9) and human Mediator (5). Taken together, these results suggest that the dMediator complex we purified is a true counterpart of the mammalian Mediator complexes.
Northern blot analysis of developmentally staged Drosophila embryos, larvae, pupae, and adults for the expression of the dMediator genes revealed that many of the components had similar expression patterns (Fig. 4D). Large amounts of the dMediator transcripts were maternally deposited to the embryos (Fig. 4D, lane 1). The expression level gradually decreased during the larval stages (Fig. 4D, lanes 1 to 6) and then increased during mid-pupal metamorphosis (Fig. 4D, lane 7). This result shows that the abundance of dMediator transcripts is correlated with developmental activities.
Interaction of dMediator with gene-specific transcription factors.
Previous studies in yeast and human cells have suggested that transcriptional activator proteins interact with Mediator complexes (5, 9, 11, 20, 29, 31). The requirement of dMediator for the activated transcription in response to Gal4-VP16 (Fig. 2C) indicates that dMediator may also serve as a binding target of transcriptional activators. Because several coactivators, such as TAFs and the GCN5 histone acetyltransferase (HAT) complex, have been suggested to interact directly with transcriptional activators, we examined the relative binding affinities of these coactivator complexes with the VP16 protein. After incubation of nuclear extracts with an excess of GST fusion protein beads containing either wild-type or mutant (Δ456FP442) VP16 activation domain, the supernatants were analyzed by immunoblotting with Abs against the components of the coactivator complexes. Almost all of the dMediator proteins in the nuclear extract (dTRAP80, dMED6, and Trfp) were removed by incubating with GST-VP16 but not with GST-VP16Δ456FP442 (Fig. 5A, lanes 1 and 2). However, the amounts of dGCN5, dTAFII40, dTAFII250, and dTBP in the extract were not reduced at all by the incubation (Fig. 5A, lanes 1 and 2). When the proteins bound to the beads were analyzed, a large amount of dMediator was retained only in the GST beads containing the functional VP16 activation domain (Fig. 5A, lanes 4 and 5). The TFIID and dGCN5 HAT complexes did bind to the wild-type VP16 beads, but the amounts were less than 2% of the total amounts present in the extract (Fig. 5A, lanes 4 and 5, and data not shown). These data indicate that, among known transcriptional coactivator complexes, Mediator is most strongly bound to and most readily recruited to the activation domain.
In addition to the model VP16 activator derived from herpesvirus, dMediator interacts with Drosophila transcriptional activators Dorsal and heat shock factor (dHSF). When dMediator complex was incubated with FLAG-Dorsal or GST-dHSF fusion protein beads, more than 20% of the dMediator input was retained specifically on the beads even after extensive washing (Fig. 5B, lanes 3 and 6). To extend this study to other sequence-specific transcription factors important for Drosophila development, we immobilized dMediator on protein G-agarose beads through anti-dSOH1 Ab and examined the binding of diverse 35S-labeled Drosophila transcription factors. Bicoid, Krüppel, and Fushi-tarazu were retained specifically on the dMediator beads, while Twist and Hunchback were not (Fig. 5C). Therefore, dMediator functions as a binding target for many, but not all, developmental specific transcription factors.
To evaluate the requirement of dMediator for activated transcription in response to the Drosophila activator proteins that interact with dMediator, we tested the ability of dMediator-deficient nuclear extracts to support transcriptional activation by the Dorsal and Gal4-dHSF proteins. The addition of Dorsal or Gal4-dHSF to mock-depleted extract caused 20- and 25-fold increases, respectively, in transcription levels from the adenovirus early region 4 (E4) promoter linked to the appropriate DNA binding site. However, the level of transcriptional activation was reduced significantly (five- and threefold activations, respectively) in nuclear extract that had been depleted by anti-dSOH1 Ab (Fig. 5D, lanes 4 and 8). Therefore, dMediator is absolutely required for transcriptional activation by all the activators tested. Addition of purified dMediator back to depleted extracts partially recovered activation by Dorsal and Gal4-dHSF (data not shown) in much the same way as it did in the case of Gal4-VP16 (Fig. 3C).
One of the functions suggested for Mediator complexes is the negative regulation of Pol II transcription (9, 37). This function may have a mechanistic relationship with transcriptional repression by sequence-specific transcription factors. Therefore, we examined both physical and functional interactions between dMediator and the Drosophila transcriptional repressor Even-skipped. Unlike the strong interaction between dMediator and several transcriptional activators, dMediator did not bind Even-skipped protein (Fig. 5C). As shown previously (24), the addition of Even-skipped protein to nuclear extract caused a 12-fold reduction in transcription from the template containing three copies of the Even-skipped binding site upstream of the alcohol dehydrogenase proximal (Adh) promoter (Fig. 5D, lane 10). Transcriptional repression by Even-skipped still occurred in the dMediator-deficient nuclear extracts (lane 12). Therefore, dMediator is not required for transcriptional repression by the sequence-specific transcription factor Even-skipped.
Physical and functional interaction of dMediator with the basal transcription machinery.
Although the yeast Mediator complex was purified as part of Pol II holoenzyme complexes (15, 16), the dMediator that we isolated does not contain Pol II or other general transcription factors. However, we observed that a small fraction of the dMediator comigrated with Pol II during several chromatographic steps and also coimmunoprecipitated with Pol II from crude fractions at low salt concentrations (lower than 300 mM potassium acetate [data not shown]). Therefore, we tried to confirm the interaction between dMediator and Pol II, in particular, the CTD of Pol II, by GST-pulldown assays. Purified dMediator was incubated with GST-CTD beads. Bound proteins were analyzed by immunoblotting with dMediator Abs. Like yeast Mediator, dMediator bound specifically with GST-CTD (Fig. 6A).
Because the Mediator-CTD interaction in yeast causes a stimulation of CTD phosphorylation by TFIIH (15), we examined the effect of dMediator on the phosphorylation of Drosophila CTD. The phosphorylation states were monitored by immunoblot analyses with Abs that recognize the phosphoserine residues at the second and fifth serines of the CTD. Purified Drosophila TFIIH specifically phosphorylated the fifth serine residue of GST-CTD (Fig. 6B, lanes 2 and 3). On the other hand, dMediator phosphorylated both the second and fifth serine residues (lanes 4 and 5). This may, at least in part, result from the catalytic activity of Cdk8 present in the dMediator complex. When both dMediator and Drosophila TFIIH were present in the reaction, the level of the CTD phosphorylation at serine 5 is increased synergistically, whereas the phosphorylation at serine 2 by dMediator remained unchanged in the presence of TFIIH (lanes 6). Increasing the amount of dMediator further stimulated the synergistic serine 5 phosphorylation while that of TFIIH did not (lanes 7 and 8), indicating that dMediator is the limiting factor for the synergistic serine 5 phosphorylation in our assay condition. All of these data suggest that although we could not observe a stable physical interaction in vitro between dMediator and TFIIH (data not shown), they interact functionally to stimulate the phosphorylation of serine 5 of the CTD.
We next investigated interactions between dMediator and the other Drosophila general transcription factors by incubating individual 35S-labeled TBP, TFIIB, TFIIE, TFIIF, TFIIA, and TFIIS subunits with dMediator-immobilized beads. SDS-PAGE analysis of the dMediator-bound proteins revealed that TBP, TFIIB, the TFIIE small subunit, the TFIIF large subunit, and TFIIS interacted strongly with dMediator, whereas TFIIA, the TFIIE large subunit, and the TFIIF small subunit did not (Fig. 6C). dMediator interacted with heteromeric TFIIE and TFIIF complexes assembled from the large and small subunits (data not shown), presumably through binding to the TFIIE small subunit and TFIIF large subunit, respectively. Therefore, dMediator exhibits a broad spectrum of binding capacities for a subset of general transcription factors and Pol II as well as sequence-specific transcription factors and may influence the function of these binding partners.
Distinct function of Mediator proteins and TAFs.
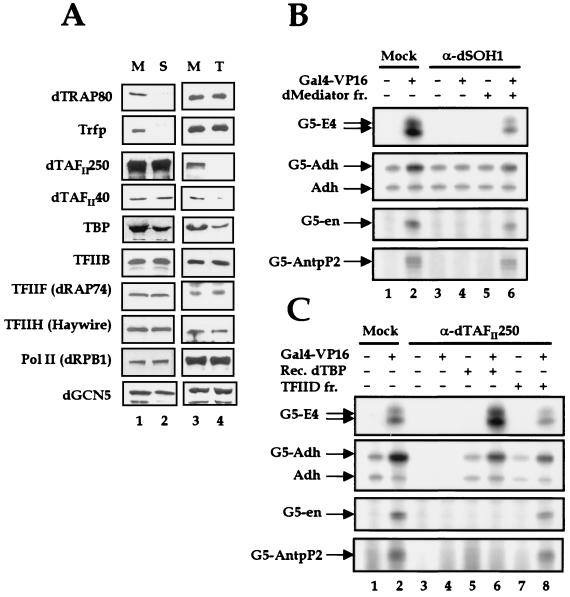
The general requirement of dMediator in transcriptional activation in vitro raises the question about what the functional relationship is between dMediator and TFIID in transcriptional activation. Although the VP16 activator binding assay shown in Fig. 5A clearly indicated that dMediator is the major binding target of the VP16 activator protein in nuclear extracts, whether TAFs provide a redundant, synergistic, or distinct function in transcriptional activation in the presence of dMediator is not known. Therefore, we compared the requirement of dMediator in Pol II transcription to that of TAFs, using immunodepletion studies similar to that carried out in the human system (30). We immunodepleted nuclear extract with an Ab against dMediator (anti-dSOH1), TFIID (anti-dTAFII250), or β-galactosidase (control). Although we observed physical interaction of dMediator with general transcription factors and Pol II (Fig. 6), immunodepletion with anti-dSOH1 did not remove basal transcription factors, TAFs, or Pol II from the nuclear extract (Fig. 7A, lane 2). This shows that the absolute majority of dMediator proteins in nuclear extracts are not engaged in the assembly of the Pol II holoenzyme complex.
FIG. 7.
Differential requirement of mediator proteins and TAFs for transcriptional activation. (A) Immunodepletion of dMediators and TFIID from nuclear extract. Extract was immunodepleted with anti-β-galactosidase (M), anti-dSOH1 (S), and anti-dTAFII250 (T). Each extract was analyzed by immunoblotting to examine the relative amounts of the proteins indicated at the left. (B) Transcriptional activation of the E4, Adh, en, and AntpP2 promoter constructs by Gal4-VP16 in dMediator-deficient extracts. Extracts were immunodepleted with anti-β-galactosidase or anti-dSOH1. Transcription assays were performed and the results analyzed as for Fig. 3C. (C) Different effects of TAF depletion on the activated transcription of the E4, Adh, en, and AntpP2 promoter constructs. The addition of Gal4-VP16, recombinant dTBP, and partially purified TFIID (TFIID fr.) to each reaction is indicated by + on the corresponding lanes.
The activated transcription by Gal4-VP16 from E4, Adh, engrailed (en), and AntpP2 promoters that bear upstream Gal4 binding sites was completely abolished by immunodepleting, and partly restored by adding back, the dMediator proteins (Fig. 7B, lanes 4 and 6). In contrast, the level of basal transcription from the Adh promoter was not affected by the depletion of dMediator (lane 3). Therefore, dMediator is specifically required for transcriptional activation from all four templates, regardless of their differential core promoter architecture (see below).
Nuclear extracts preincubated with the anti-dTAFII250 Ab did not contain a detectable level of the dTAFII250 protein. dTAFII250, the largest component of the TFIID complex, is the core scaffold upon which the other TAFIIs are assembled to form a stable complex with TBP (42). Hence, it is highly likely that the dTAFII250-deficient extracts lack the TFIID complex and the key functions played by TFIID-specific TAFs. On the other hand, they still possessed a residual amount of TBP and dTAFII40 (Fig. 7A, lane 4). The residual TBP probably originated from other TBP-containing complexes such as SL1 and TFIIIB. However, these TBP molecules are not likely to participate in Pol II transcription in an interchangeable fashion as these nuclear extracts did not support either basal or activated transcription from any of the promoters that we tested (Fig. 7C, lanes 3 and 4). The residual dTAFII40 (lane 4) might originate from non-TFIID TAF complexes such as the GCN5 HAT complex, which had not been removed from nuclear extract by the dTAFII250 Ab treatment (lane 4). Addition of recombinant TBP to TFIID-depleted nuclear extracts restored both basal and VP16-activated transcription from the E4 and Adh promoters (Fig. 7C, lanes 5 and 6), demonstrating that TAFs are dispensable for these promoters in the presence of Mediator. The E4 and Adh promoters contain a TATA element at about the −30 position. However, TBP alone was unable to support activated transcription from the en and AntpP2 promoters (Fig. 7C, lanes 5 and 6). The en and AntpP2 core promoters differ from the above two promoters in that they lack a canonical TATA element but instead consist of an Inr and a DPE (6). Therefore, TAFs, or at least dTAFII250, appear to have functions that are redundant in the activated transcription of TATA-containing genes under the condition we used but critical for transcriptional activation from TATA-less, Inr- and DPE-containing promoters. Indeed, addition of partially purified TFIID to TFIID-depleted nuclear extracts restored activated transcription from the en and AntpP2 promoters (lane 8). These observations reveal two distinct roles for Pol II transcription, each played separately by Mediator proteins and TAFs: mediation of activator responses and core promoter selectivity.
DISCUSSION
Mediator proteins and TAFs are differently required for Pol II transcription.
dMediator is generally required for transcriptional activation from both TATA-containing and TATA-less promoters (Fig. 7B) through direct communication with transcriptional activators. The function of dMediator seems to be exclusively related to sequence-specific transcription factors placed at upstream enhancer elements. However, the requirement of TAFs, or at least dTAFII250, in activated transcription appears to be redundant in the in vitro transcription system we used and affected by such factors as the core promoter organization or nucleosomal structure of transcriptional templates. Several TAF components in the TFIID complex indeed have biochemical activities and structural motifs adequate for the recognition of specialized settings of transcription templates. For example, certain TAFs recognize the Inr and DPE sequences located in many Drosophila core promoters and increase the stability of TFIID-promoter interactions (6, 7, 39). In addition, TFIID contains dTAFII250, which has a HAT catalytic activity (27) and also possesses a histone octamer-like module comprising the histone H2B-, H3-, and H4-like TAFs (8). Although not experimentally demonstrated, these TAFs may have some roles in the transcriptional regulation of nucleosomal templates.
Diverse sequence-specific transcription factors interact with and depend on dMediator for transcriptional activation.
The sequence-specific transcription factors which interact physically with dMediator include VP16, Dorsal, dHSF, Bicoid, Krüppel, and Fushi-tarazu (Fig. 5). These factors contain different types of activation domains (acidic and glutamine-rich domains). Most of these transcription factors have been shown to activate transcription either constitutively or inducibly. It is noteworthy that dHSF interacts with and requires dMediator for transcriptional activation (Fig. 5B and D) because previous reports have shown that transcriptional activation by HSF in yeast does not require the function of the Mediator protein Srb4 (21, 26). However, the recent finding that activation by HSF depends on another Mediator protein, Rgr1 (22), suggests that some function of Mediator is required for HSF-mediated transcriptional activation in yeast, as well. As Rgr1, but not Srb4, is conserved between yeast and Drosophila (Fig. 1A), transcriptional activation by HSF might utilize the conserved Rgr1 components of the Mediator complexes.
Although some human Mediator complexes appear to have a negative effect on activated transcription (9, 37), dMediator did not exhibit such an activity in an in vitro transcription system reconstituted with Drosophila transcription factors (data not shown). In addition, Even-skipped, a well-known Drosophila transcriptional repressor, did not interact with, or depend for its transcriptional repression on, dMediator (Fig. 5B and D). Previous reports show that the repression domain of Even-skipped directly targets TBP (1, 38). We also confirmed that the TFIID complex in the nuclear extract specifically interacts with Even-skipped under the same conditions in which Even-skipped failed to interact with dMediator (Fig. 5B and data not shown). Although Krüppel has a well-characterized repressor function in Drosophila development, it can also act as a transcriptional activator under certain conditions (35). Therefore, it is more plausible that the dMediator-Krüppel interaction we observed is a part of the mechanism for transcriptional activation rather than transcriptional repression. Taken together with the fact that dMediator was dispensable for basal transcription, the lack of defect of the dMediator-depleted nuclear extracts on transcriptional repression by Even-skipped protein suggests that dMediator is required mainly for the mediation of transcriptional activation signals to the basal transcription machinery. Very recently, developmental roles of certain dMediator proteins found in the Drosophila genome database have begun to be also identified in genetic studies (4). Genetic interactions between dMediator proteins and a homeotic regulator Sex combs reduced shown in that study (4) implicate dMediator proteins as a transcriptional activator-specific target critical for Drosophila development.
The interactions between basal transcription machineries and dMediator may regulate the transcription initiation process.
Like yeast Mediator, dMediator bound with the CTD repeats of Drosophila Pol II (Fig. 6A). This implies that though dMediator was purified separately from Pol II, these two complexes indeed interact with each other and act together during transcriptional initiation. Besides the physical interaction with Pol II, dMediator also has some binding affinity for TBP, TFIIB, TFIIE, TFIIF, and TFIIS. Such interactions may be involved in the regulation of Pol II preinitiation complex assembly. Related with this idea, it has been reported that in yeast, recruitment of general transcription factors such as TBP, TFIIB, and TFIIH to active promoters requires the function of Mediator (17, 25, 32). Also, TFIIE interacts with the Mediator protein Gal11 (34). Further analyses will be required to clarify whether these interactions, observed both in yeast and Drosophila, participate in the control of the stepwise preinitiation complex assembly in the course of transcription activation or simply reflect the affinities between the components of preassembled Pol II holoenzyme.
dMediator contains the protein kinase component Cdk8, which can phosphorylate serine residues in the CTD. This catalytic kinase subunit seems responsible, at least in part, for the Pol II phosphorylation by dMediator (Fig. 6B). In particular, dMediator and TFIIH synergistically phosphorylate the serine 5 residue of the carboxy-terminal Pol II repeats, suggesting the presence of a functional interaction between these complexes. Given that Pol II phosphorylation at serine 5 by TFIIH has been correlated with transcriptional activation processes (20), the synergy in the serine 5 phosphorylation by TFIIH and dMediator may be intimately linked with the regulatory effects that the Mediator complex exerts on Pol II transcription.
ACKNOWLEDGMENTS
We thank Carl Wu for providing Drosophila embryo nuclear extracts used in the initial stage of dMediator purification and also for helpful comments on the manuscript; Gaku Mizuguchi for purified TFIIH; Hua Xiao, James Kadonaga, Yoshihiro Nakatani, Marc Vigneron, Erich Nigg, James Manley, David Allis, Michael Levine, Arnold Berk, Albert Courey, Pierr Léopold, Peter Becker, Arno Greenleaf, Michael Carey, David Price, Magaret Fuller, and William Zehring for plasmids, baculovirus constructs, and antibodies; our colleagues in the Kim lab for helpful discussion; and Jennifer Macke for substantive editing of the text. J.M.P. thanks Dolph Hatfield for support and encouragement.
J.-G.K. is a National Institutes of Health Fogarty International Center Visiting Fellow and is supported by the Division of Basic Sciences, National Cancer Institute. This work was supported by the Creative Research Initiatives Program from the Korean Ministry of Science and Technology to Y.-J.K.
REFERENCES
- 1.Austin R J, Biggin M D. A domain of the even-skipped protein represses transcription by preventing TFIID binding to a promoter: repression by cooperative blocking. Mol Cell Biol. 1995;15:4683–4693. doi: 10.1128/mcb.15.9.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker P B, Rabindran S K, Wu C. Heat shock-regulated transcription in vitro from a reconstituted chromatin template. Proc Natl Acad Sci USA. 1991;88:4109–4113. doi: 10.1073/pnas.88.10.4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Björklund S, Kim Y-J. Mediator of transcriptional regulation. Trends Biochem Sci. 1996;21:335–337. doi: 10.1016/s0968-0004(96)10051-7. [DOI] [PubMed] [Google Scholar]
- 4.Boube M C, Faucher, Joulia L, Cribbs D L, Bourbon H-M. Drosophila homologs of transcriptional mediator complex subunits are required for adult cell and segment identity specification. Genes Dev. 2000;14:2906–2917. doi: 10.1101/gad.17900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyer T G, Martin M E D, Lees E, Ricciardi R P, Berk A J. Mammalian Srb/Mediator complex is targeted by adenovirus E1A protein. Nature. 1999;399:276–279. doi: 10.1038/20466. [DOI] [PubMed] [Google Scholar]
- 6.Burke T W, Kadonaga J T. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes Dev. 1996;10:711–724. doi: 10.1101/gad.10.6.711. [DOI] [PubMed] [Google Scholar]
- 7.Burke T W, Kadonaga J T. The downstream promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev. 1997;11:3020–3031. doi: 10.1101/gad.11.22.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burley S K, Roeder R G. Biochemistry and structural biology of transcription factor IID (TFIID) Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 9.Gu W, Malik S, Ito M, Yuan C-X, Fondell J D, Zhang X, Martinez E, Qin J, Roeder R G. A novel human SRB/MED-containing cofactor complex, SMCC, involved in transcriptional regulation. Mol Cell. 1999;3:97–108. doi: 10.1016/s1097-2765(00)80178-1. [DOI] [PubMed] [Google Scholar]
- 10.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 11.Hengartner C J, Thompson C M, Zhang J, Chao D M, Liao S-M, Koleske A J, Okamura S, Young R A. Association of an activator with an RNA polymerase II holoenzyme. Genes Dev. 1995;9:897–910. doi: 10.1101/gad.9.8.897. [DOI] [PubMed] [Google Scholar]
- 12.Holstege F C P, Jennings E G, Wyrick J J, Lee T I, Hengartner C J, Green M R, Golub T R, Lander E S, Young R A. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 13.Jiang Y W, Veschambre P, Erdjument-Bromage H, Tempst P, Conaway J W, Conaway R C, Kornberg R D. Mammalian mediator of transcriptional regulation and its possible role as an end-point of signal transduction pathways. Proc Natl Acad Sci USA. 1998;95:8538–8543. doi: 10.1073/pnas.95.15.8538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamakaka R T, Tyree C M, Kadonaga J T. Accurate and efficient RNA polymerase II transcription with a soluble nuclear fraction derived from Drosophila embryos. Proc Natl Acad Sci USA. 1991;88:1024–1028. doi: 10.1073/pnas.88.3.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim Y-J, Björklund S, Li Y, Sayre M H, Kornberg R D. A multiprotein Mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 16.Koleske A J, Young R A. An RNA polymerase II holoenzyme responsive to activators. Nature. 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- 17.Kuras L, Struhl K. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature. 1999;399:609–613. doi: 10.1038/21239. [DOI] [PubMed] [Google Scholar]
- 18.Leclerc V, Tassan J P, O'Farrell P H, Nigg E A, Leopold P. Drosophila Cdk8, a kinase partner of cyclin C that interacts with the large subunit of RNA polymerase II. Mol Biol Cell. 1996;7:505–513. doi: 10.1091/mbc.7.4.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee Y C, Kim Y-J. Requirement of a functional interaction between Mediator components Med6 and Srb4 in RNA polymerase II transcription. Mol Cell Biol. 1998;18:5364–5370. doi: 10.1128/mcb.18.9.5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee Y C, Park J M, Min S, Han S J, Kim Y-J. An activator binding module of yeast RNA polymerase II holoenzyme. Mol Cell Biol. 1999;19:2967–2976. doi: 10.1128/mcb.19.4.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee D-K, Lis J T. Transcriptional activation independent of TFIIH kinase and the RNA polymerase II mediator in vivo. Nature. 1998;393:389–392. doi: 10.1038/30770. [DOI] [PubMed] [Google Scholar]
- 22.Lee D-K, Kim S, Lis J T. Different upstream transcriptional activators have distinct coactivator requirements. Genes Dev. 1999;13:2934–2939. doi: 10.1101/gad.13.22.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leopold P, Farrell P H. An evolutionarily conserved cyclin homolog from Drosophila rescues yeast deficient in G1 cyclins. Cell. 1991;66:1207–1216. doi: 10.1016/0092-8674(91)90043-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li C, Manley J L. Even-skipped represses transcription by binding TATA binding protein and blocking the TFIID-TATA box interaction. Mol Cell Biol. 1998;18:3771–3781. doi: 10.1128/mcb.18.7.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X-Y, Virbasius A, Zhu X, Green M R. Enhancement of TBP binding by activators and general transcription factors. Nature. 1999;399:605–609. doi: 10.1038/21232. [DOI] [PubMed] [Google Scholar]
- 26.McNeil J B, Agah H, Bentley D. Activated transcription independent of the RNA polymerase II holoenzyme in budding yeast. Genes Dev. 1998;12:2510–2521. doi: 10.1101/gad.12.16.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizzen C A, Yang X J, Kokubo T, Brownell J E, Bannister A J, Owen H-T, Workman J, Wang L, Berger S L, Kouzarides T, Nakatani Y, Allis C D. The TAFII250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 28.Moqtaderi Z, Bai Y, Poon D, Weil P A, Struhl K. TBP-associated factors are not generally required for transcriptional activation in yeast. Nature. 1996;383:188–191. doi: 10.1038/383188a0. [DOI] [PubMed] [Google Scholar]
- 29.Näär A M, Beaurang P A, Zhou S, Abraham S, Solomon W, Tjian R. Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature. 1999;398:828–832. doi: 10.1038/19789. [DOI] [PubMed] [Google Scholar]
- 30.Oelgeschläger T, Tao Y, Kang Y K, Roeder R G. Transcription activation via enhanced preinitiation complex assembly in a human cell-free system lacking TAFIIs. Mol Cell. 1998;1:925–931. doi: 10.1016/s1097-2765(00)80092-1. [DOI] [PubMed] [Google Scholar]
- 31.Rachez C, Lemon B D, Suldan Z, Bromleigh V, Gamble M, Näär A M, Erdjument-Bromage H, Tempst P, Freedman L P. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature. 1999;398:824–828. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- 32.Ranish J A, Yudkovsky N, Hahn S. Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev. 1999;13:49–63. doi: 10.1101/gad.13.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ryu S, Zhou S, Ladurner A G, Tjian R. The transcriptional cofactor complex CRSP is required for activity of the enhancer-binding protein Sp1. Nature. 1999;397:446–450. doi: 10.1038/17141. [DOI] [PubMed] [Google Scholar]
- 34.Sakurai H, Kim Y-J, Ohishi T, Kornberg R D, Fukasawa T. The yeast GAL11 protein binds to the transcription factor IIE through GAL11 regions essential for its in vivo function. Proc Natl Acad Sci USA. 1996;93:9488–9492. doi: 10.1073/pnas.93.18.9488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sauer F, Jäckle H. Concentration-dependent transcriptional activation or repression by Krüppel from a single binding site. Nature. 1991;353:563–566. doi: 10.1038/353563a0. [DOI] [PubMed] [Google Scholar]
- 36.Shen W-C, Green M R. Yeast TAFII145 functions as a core promoter selectivity factor, not a general coactivity factor. Cell. 1997;90:615–624. doi: 10.1016/s0092-8674(00)80523-1. [DOI] [PubMed] [Google Scholar]
- 37.Sun X, Zhang Y, Cho H, Rickert P, Lees E, Lane W, Reinberg D. NAT, a human complex containing Srb polypeptides that functions as a negative regulator of activated transcription. Mol Cell. 1998;2:213–222. doi: 10.1016/s1097-2765(00)80131-8. [DOI] [PubMed] [Google Scholar]
- 38.Um M, Li C, Manley J L. The transcriptional repressor Even-skipped interacts directly with TATA-binding protein. Mol Cell Biol. 1995;15:5007–5016. doi: 10.1128/mcb.15.9.5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verrijzer C P, Chen J-L, Yokomori K, Tjian R. Binding of TAFs to core promoter elements directs promoter selectivity by RNA polymerase II. Cell. 1995;81:1115–1125. doi: 10.1016/s0092-8674(05)80016-9. [DOI] [PubMed] [Google Scholar]
- 40.Verrijzer C P, Tjian R. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem Sci. 1996;21:338–342. [PubMed] [Google Scholar]
- 41.Walker S S, Reese J C, Apone L M, Green M R. Transcription activation in cells lacking TAFIIs. Nature. 1996;383:185–188. doi: 10.1038/383185a0. [DOI] [PubMed] [Google Scholar]
- 42.Weinzierl R O J, Dynlacht B D, Tjian R. Largest subunit of Drosophila transcription factor IID directs assembly of a complex containing TBP and a coactivator. Nature. 1993;362:511–517. doi: 10.1038/362511a0. [DOI] [PubMed] [Google Scholar]
- 43.Zhou J, Zwicker J, Szymanski P, Levine M, Tjian R. TAFII mutations disrupt Dorsal activation in the Drosophila embryo. Proc Natl Acad Sci USA. 1998;95:13483–13488. doi: 10.1073/pnas.95.23.13483. [DOI] [PMC free article] [PubMed] [Google Scholar]