ABSTRACT
Disturbances in the primary colonization of the infant gut can result in lifelong consequences and have been associated with a range of host conditions. Although early-life factors have been shown to affect infant gut microbiota development, our current understanding of human gut colonization in early life remains limited. To gain more insights into the unique dynamics of this rapidly evolving ecosystem, we investigated the microbiota over the first year of life in eight densely sampled infants (n = 303 total samples). To evaluate the gut microbiota maturation transition toward an adult configuration, we compared the microbiome composition of the infants to that of the Flemish Gut Flora Project (FGFP) population (n = 1,106). We observed the infant gut microbiota to mature through three distinct, conserved stages of ecosystem development. Across these successional gut microbiota maturation stages, the genus predominance was observed to shift from Escherichia over Bifidobacterium to Bacteroides. Both disease and antibiotic treatment were observed to be associated occasionally with gut microbiota maturation stage regression, a transient setback in microbiota maturation dynamics. Although the studied microbiota trajectories evolved to more adult-like constellations, microbiome community typing against the background of the FGFP cohort clustered all infant samples within the (in adults) potentially dysbiotic Bacteroides 2 (Bact2) enterotype. We confirmed the similarities between infant gut microbial colonization and adult dysbiosis. Profound knowledge about the primary gut colonization process in infants might provide crucial insights into how the secondary colonization of a dysbiotic adult gut can be redirected.
KEYWORDS: infant, microbiota, primary succession, enterotypes
INTRODUCTION
The development of a stable adult large intestinal microbiota sets off with the primary colonization of the infant gut. Disturbances in initial colonization or ecosystem maturation can potentially result in lifelong consequences and have been associated with a broad range of host conditions, including inflammatory bowel disease (1), asthma (2), and type I diabetes (3). Although early-life factors such as birth mode and diet have been shown to affect the development of the infant gut microbiota (4, 5), our current understanding of human gut colonization in early life remains limited.
In full-term vaginally born infants, the first pioneering gut species are believed to be facultative anaerobic bacteria such as Escherichia and other members of the Enterobacteriaceae family (6). During the first days of life, the main bacteria colonizing the infant gut are reported to be Bifidobacterium and Clostridium (6). Feeding type is one of the major factors determining early microbial colonization. The differences in gut microbial composition between breastfed and formula-fed infants are well documented. Increased levels of bifidobacteria are reported in breastfed infants, while formula-fed infants have been associated with a more diverse gut microbiota that is dominated by staphylococci, Bacteroides, clostridia, enterococci, enterobacteria, and the genus Atopobium (7–9). During weaning, the alpha diversity of the gut microbiome increases, resulting in the replacement of Proteobacteria and Actinobacteria by the Firmicutes and Bacteroidetes phyla as the dominant members of the infant microbiota (8, 9). Between the 9th and 18th months of life, the relative abundances of bacterial families such as Lachnospiraceae, Ruminococcaceae, Eubacteriaceae, Rikenellaceae, or Sutterellaceae have been reported to increase, while the relative abundances of Bifidobacteriaceae, Actinomycetaceae, Veillonellaceae, Enterobacteriaceae, Lactobacillaceae, Enterococcaceae, Clostridiales incertae sedis XI, and Fusobacteriaceae decreased during the transition from the infant to the toddler stage (10). Two species, namely, Faecalibacterium prausnitzii and Akkermansia muciniphila, which are rarely observed during early infancy, increase in abundance to adult levels at 12 months and 24 months, respectively (11).
Microbiome monitoring efforts combining high sampling frequency with a prolonged longitudinal design would enable gaining more insights into the unique dynamics of this rapidly evolving ecosystem. Here, we investigated microbiome variation over the first year of life in eight densely sampled infants, analyzing on average 38 samples per participant (n = 303 total samples). We observed the infant gut microbiota to mature through three distinct, conserved stages of ecosystem development. Across these successional gut microbiota maturation stages, the genus predominance was observed to shift from Escherichia over Bifidobacterium to Bacteroides. A stable, reproducible order of successive colonization could be established at the genus level across the BaBel cohort infants. Both disease and antibiotic treatment were observed to be associated occasionally with gut microbiota maturation stage regression, a transient setback in microbiota maturation dynamics. Although the studied microbiota trajectories evolved in terms of both richness and composition to more adult-like constellations, microbiome community typing against the background of the Flemish Gut Flora Project (FGFP) population cohort (n = 1,106) clustered all infant samples within the (in adults) potentially dysbiotic Bacteroides 2 (Bact2) enterotype. While these observations reflect incomplete microbiota maturation within the first year of life, the suggested parallel between primary succession as observed in the healthy infant gut and secondary colonization upon ecosystem disruption could inform novel biotherapeutic approaches based on sequential recolonization of a dysbiotic community.
RESULTS AND DISCUSSION
Increasing numbers of diseases are being linked to gut dysbiosis. This state, characterized by a low diversity and high abundance of facultative anaerobic bacteria in adults, also resembles the microbiome composition of healthy infants (12). Profound knowledge about the primary gut colonization process in infants, going from (nearly) sterile at birth toward a diverse and healthy gut microbiota later in life, might provide crucial insights into how the secondary colonization of a dysbiotic adult gut can be redirected.
The colonization process in the healthy infant gut happens through distinct stages of ecosystem development.
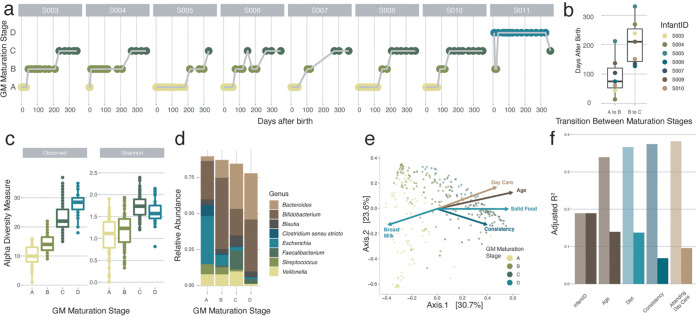
Setting out to map gut microbiota maturation dynamics in eight vaginally delivered, healthy infants from Belgium (BaBel cohort), we analyzed the fecal microbiome profiles of a core data set of 142 samples collected at predefined time points distributed over the first year of life (of the 159 samples at predefined time points, 17 were excluded based on reported disease signs [see Materials and Methods]) (see Table S1a and Fig. S1 in the supplemental material), complemented with 144 post hoc-selected samples associated with clinically relevant events such as disease/drug treatment. Applying Dirichlet multinomial mixture (DMM) modeling to the microbiome profiles, we screened for subcommunities among the infants’ microbiomes. Grouping samples potentially originating from the same community through probabilistic modeling, DMM-based stratification of microbiome data reproducibly identifies community constellations across data sets without making any claims regarding the putative discrete nature of the strata detected (13). In the present data set, community typing revealed the presence of four compositionally distinct clusters or gut microbiota maturation stages, with only one of them restricted to a single individual (Fig. 1a; Table S1b and Fig. S2). Three out of four maturation stages (labeled A, B, and C) comprised almost exclusively samples originating from seven out of eight individuals, reflecting conserved, structured microbiome maturation rather than interindividual variation. Although the time of transition varied between individuals (Fig. 1b), maturation stage A to C succession revealed a strong temporal organization following a conserved pattern across infants (n = 7, Kendall test, Kendall’s corrected w = 1, and P = 5e−4) (Fig. 1a), aligning with an overall increase in microbiome richness (comparison of maturation stage A with stage B and stage B with stage C, n = [182:176], Kruskal-Wallis [KW] test with a post hoc Dunn [phD] test, r = [−0.35:−0.60], and false discovery rate [FDR] < 0.05) (Fig. 1c; Table S1c). However, the highest richness values were noted for the divergent D maturation state (comparison of maturation stages A, B, and C versus stage D, n = [127:119:121], KW test with phD tests, r = [−1:−0.72:−0.18], and FDR < 0.05) (Fig. 1c; Table S1c), observed only in infant S011 and not linked to temporal variation. Focusing on differences in microbiota composition between the gut microbiota maturation stages, we found maturation stage A to be dominated by Escherichia spp. (Fig. 1d). Compared to both stages B and C, maturation stage A was characterized by high proportional abundances of not only Escherichia but also Staphylococcus, Enterococcus, Enterobacter, and Lactobacillus, among others (n = 303, KW with phD tests, r > 0.3, and FDR < 0.05) (Table S1d and Fig. S3). The reported top five (in terms of effect size) maturation stage A-associated genera consist exclusively of facultative anaerobic genera, reflecting the higher oxygen levels present in the infant gut shortly after birth (14). Maturation stage B was dominated by bifidobacteria (Fig. 1d), with Bifidobacterium being the only genus that was proportionally more abundant in stage B than in both stages A and C (n = 303, KW with phD tests, r > 0.4, and FDR < 0.05) (Table S1d and Fig. S3). At the end of their first year of life, all studied infants eventually reached the Bacteroides-dominated C maturation stage (Fig. 1d). With respect to both stages A and B, the higher richness of the C maturation stage was reflected in higher proportions of a broad range of bacteria, including butyrate-producing taxa (15) such as Anaerostipes, Faecalibacterium, and Roseburia (n = 303, KW with phD tests, r > 0.3, and FDR < 0.05) (Table S1d and Fig. S3).
FIG 1.
Detailed overview of the colonization process in the healthy infant gut at the genus level. (a) Overview of the gut microbiota (GM) maturation stage succession of the samples of all the infants over time, colored by the assigned gut microbiota maturation stages determined using the DMM approach (calculated on all samples [n = 303] and shown here for the samples at predefined time points where the infants were not sick [n = 142]). (b) Variation in the timing of the transition between the gut microbiota maturation stages in the different infants. The body of the box plots represents the first and third quartiles of the distribution and the median line. (c) Alpha diversity measures (observed ASV richness and Shannon diversity) of the samples within every gut microbiota maturation stage, increasing from stages A to C (comparisons of gut microbiota maturation stage A with stage B and stage B with stage C, n = [182:176], post hoc Dunn test [phD], r = [−0.35:−0.60], and FDR < 0.05). (d) Mean relative abundances of the most common genera at every gut microbiota maturation stage. (e) Principal-coordinate analysis (PCoA) (Bray-Curtis dissimilarity) representing genus-level microbiome variation in our infant cohort (n = 299). Dots represent one sample and are colored by their assigned gut microbiota maturation stage. The arrows represent the effect size and direction of the post hoc fit of variables significantly associated with microbiota compositional variation (univariate distance-based redundancy analysis [dbRDA]) (infant identification was excluded for clarity). (f) Covariates with nonredundant explanatory power on the genus-level ordination, determined by multivariate dbRDA at the genus level (Bray-Curtis dissimilarity; FDR < 0.05). The light bars represent the cumulative explanatory power (stepwise dbRDA R2), and the darker bars represent the individual univariate explanatory power of the variables (dbRDA R2). Covariates present in fewer than three infants were excluded.
Overview of the collected and selected samples per infant. Gray dots, all samples collected by the parents of the enrolled infants; blue diamonds, samples selected for the study of the longitudinal dynamics at predefined time points with no clinical signs (n = 142); purple diamonds, samples selected for the study of the longitudinal dynamics at predefined time points with clinical signs (n = 17) (see Table S1 in the supplemental material for the signs); green diamonds, additional ad hoc-selected samples at specific external events (n = 144); black filled triangles, three vaccination events in every infant; black open triangles, day care entrance; red triangles, external events around which extra samples were selected. Abbreviations: F, fever; D, diarrhea; V, vomit; AB, antibiotics; PB, probiotics. Download FIG S1, PDF file, 0.4 MB (436.6KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Determination of the optimal number of clusters in the DMM approach. Identification of the optimal number of Dirichlet components in the BaBel data set (n = 303) was based on the Bayesian information criterion (BIC). The optimal number of clusters is 4 (minimum BIC = 37,285.6). Download FIG S2, PDF file, 0.7 MB (767.1KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The most abundant genera that are differentially abundant per gut microbiota maturation stage determined using DMM clustering. (a) Distribution of the relative abundances of the most abundant genera in gut microbiota maturation stage A, which are significantly more abundant in maturation stage A than in stages B and C. (b) Distribution of the relative abundances of the most abundant genera in gut microbiota maturation stage B, which are significantly more abundant in maturation stage B than in stages A and C. (c) Distribution of the relative abundances of the most abundant genera in gut microbiota maturation stage C, which are significantly more abundant in maturation stage C than in stages A and B (n = 303, KW with phD test, r > 0.3, and FDR < 0.05) (see Table S4 in the supplemental material). Download FIG S3, PDF file, 0.6 MB (637.7KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
(a) Description of the infants enrolled in the BaBel cohort (n = 8) and description of the analyzed samples (n = 303). Abbreviations: F, female; M, male; se+, secretor; se−, nonsecretor; Le−, Lewis antigen negative; LeB, Lewis B antigen. Shown are the sample identifier, infant identifier, age at sample collection (days after birth, included as a predefined time point when the infant was healthy (LDA_nosigns, Yes/No), consistency (1, aqueous; 2, soft; 3, solid), food category (only breast milk, no solid food, or solid food), drug category (according to the ATC classification system), disease category (according to the ICD-10 classification), holiday (Yes/No), family stay (Yes/No), attending day care (Yes/No), and initially included as a predefined time point but the infant showed signs (LDA_signs, Yes/No). (b) Gut microbiota maturation stage assignment results from Dirichlet multinomial mixture (DMM) clustering (n = 303). (c) Alpha diversity (genus richness and Shannon index) distribution per gut microbiota maturation stage (n = 303) (Kruskal-Wallis test with post hoc Dunn tests and FDR by the Benjamini-Hochberg method). (d) Genus abundance distribution per gut microbiota maturation stage (n = 303) (Kruskal-Wallis test with post hoc Dunn tests and FDR by the Benjamini-Hochberg method, focusing on the 15 most abundant genera). (e) Covariates with nonredundant explanatory power on the genus-level ordination, determined by multivariate distance-based redundancy analysis (dbRDA) at the genus level (n = 299, top 15 most abundant genera, Bray-Curtis dissimilarity, and FDR < 0.05). (f) Multivariate redundancy analysis carried out on the relative abundances of each genus, after constraining for infant identification and testing the most prevalent covariates, i.e., present in at least 3 infants (n = 299) (multivariate dbRDA; FDR < 0.05). (g) Bifidobacterium abundance distribution per food category (n = 303) (Kruskal-Wallis test with post hoc Dunn tests and FDR by the Benjamini-Hochberg method). (h) Results for event 1 (E1) (antibiotic treatment in infant S004). Multivariate redundancy analysis was carried out on the relative abundances of each genus, after constraining for infant identification and testing all covariates (all genera tested, n = 299, multivariate dbRDA, and FDR < 0.05), and Maaslin2 results (n = 303) are shown. (i) Enterotype assignment results of the infant samples (n = 303). (j) Permutational pairwise MANOVA Adonis test of the infant age bins versus adults from the FGFP data set (n = 1,408 )(Bray-Curtis dissimilarity and FDR by the Benjamini-Hochberg method). (k) Genus richness of the infant age bins versus adults from the FGFP data set (n = 1,408) (Kruskal-Wallis test with post hoc Dunn tests and FDR by the Benjamini-Hochberg method). (l) Contaminating amplicon sequence variants (ASVs) removed in the filtering step using the R package “decontam”. Download Table S1, XLSX file, 0.1 MB (109.8KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Identification of covariates explaining infant gut microbiota variation.
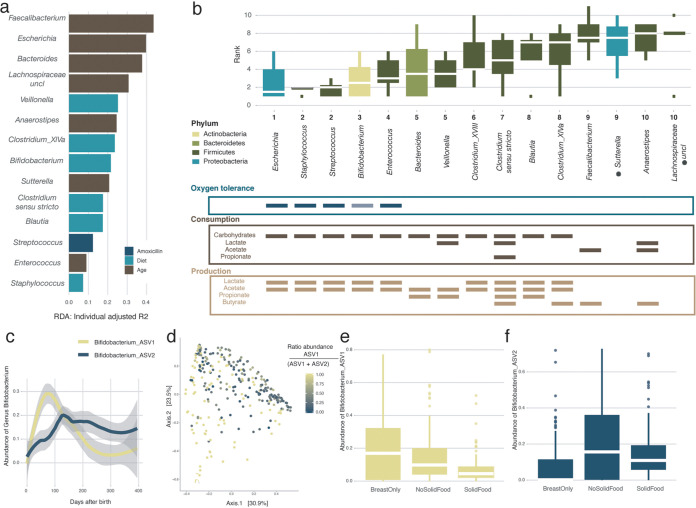
To identify covariates of microbiome diversification within the first year of life, we assessed the nonredundant explanatory power of diet, medication, health status, environment, and infants’ specific characteristics, such as having siblings or their blood group, in the genus-level compositional variation within the BaBel infants. Beyond interindividual variation (n = 299, multivariate stepwise distance-based redundancy analysis [dbRDA] on Bray-Curtis dissimilarity, R2 = 18.9%, and adjusted P [P.adj] = 0.002), microbiome composition was significantly associated with age (R2 = 15.0%), diet (R2 = 2.7%), stool consistency (R2 = 0.8%), and attending day care (R2 = 0.8%) (Fig. 1e and f; Table S1e). Next, we applied a similar approach to assess potential associations between metadata variables and the top 15 most dominant genera (covering on average 92.6% of the samples’ total abundance) as identified based on their average proportional abundance over all samples (n = 299, multivariate stepwise dbRDA with Euclidean distance on composition, constraining for infant identification, and FDR < 0.05). Beyond interindividual variation, we found the effect size of diet to exceed the impact of age in 6 out of 15 genera (Fig. 2a; Table S1f). Among those, we highlight the complex associations between the omnipresent Bifidobacterium spp. and changes in infants’ nutrition (3). While the taxon as a whole was of the lowest abundance in the samples where the infant was weaned (breast milk only:nonsolid food [i.e., breast and formula milk or formula only] versus solid food, n = [236:185]; phD test, r > 0.2; FDR < 0.05) (Table S1g), divergent patterns could be observed when zooming in on the two main amplicon sequence variants (ASVs) detected (Fig. 2c and d): while ASV1 proportions decreased significantly upon weaning (with weaning defined as the first time that solid foods are introduced) (breast milk only:no solid food versus solid food, n = [236:185]; phD test, r > 0.25; FDR < 0.05), ASV2 increased substantially after the addition of formula milk to the diet (breast milk only versus no solid food:solid food, n = [177:236]; phD test, r > 0.3; FDR < 0.05) (Fig. 2e and f; Table S1g).
FIG 2.
Order of appearance of the most common genera in the infant gut. (a) Overview of the covariates with the highest explanatory power for the variation of the top 15 genera in our infant cohort, beyond interinfant variability (note that for Clostridium cluster XVIII, no significance was reached). A multivariate distance-based redundancy analysis (dbRDA) was carried out on the relative abundances of each genus, after constraining for infant identification (FDR < 0.05). The length of the horizontal bars represents the explanatory power of the most significant covariate (stepwise dbRDA R2). (b) Order of appearance (presence defined as an abundance of >0.5%) of the top 15 most abundant genera in the infant gut. The box plots are ordered based on their appearance along the timeline (age) of the infants. The box plots are colored according to the phylum to which the genus belongs. Shown below the box plots are the oxygen tolerance of the different genera (note that Bifidobacterium, while normally assumed to be a strict anaerobe, is found to be oxygen tolerant in the human gut [16]) and the consumption and production of different short-chain fatty acids (SCFAs) by the different genera based on the literature (15, 17, 18). The body of the box plots represents the first and third quartiles of the distribution and the median line. The asterisks indicate the genera for which no information was available. (c) Average relative abundances of the different Bifidobacterium amplicon sequence variants (ASVs) over time averaged over all infants (locally estimated scatterplot smoothing [LOESS]). (d) Genus-level principal-coordinate analysis (PCoA) (Bray-Curtis dissimilarity) (n = 299), colored for the ratio of the two most abundant Bifidobacterium ASVs. (e) Effect of food on the relative abundance of Bifidobacterium ASV1 showing a higher absence during weaning (breast milk only:no solid food versus solid food, n = [236:185], post hoc Dunn [phD] test, r > 0.25, and FDR < 0.05). (f) Effect of food on the relative abundance of Bifidobacterium ASV2 showing an increase in samples where the infants had a formula milk-based diet (with or without solid food) (breast milk only versus no solid food:solid food, n = [177:236], phD, r > 0.3, and FDR < 0.05) (see Table S1g in the supplemental material).
Infant gut microbiota genera appear in a stable, reproducible order.
To assess whether microbiota maturation of the infant gut was determined by a series of successional colonization events conserved across individuals, we zoomed in on the genus rather than the community level, investigating the order of appearance of the top 15 most dominant genera within each 1-year maturation timeline. Defining appearance as the first occurrence of a genus (relative abundance of >0.5%), we established an appearance ranking for the taxa in each infant. We observed the appearance ranking to be significantly conserved across individuals (n = 8, Kendall test, corrected Kendall’s w = 0.523, and P = 2.08e−7) (Fig. 2b). The lowest ranks (i.e., primary colonizers) were mainly attributed to genera that have been described as saccharolytic, oxygen tolerant, and/or lactate and acetate producing (15–19). While such taxa can contribute to colonization resistance of the newborns through acidification of the large intestinal environment (20, 21), they also generate substrates that allow the subsequent recruitment of cross-feeders such as Veillonella and Anaerostipes (22). Ranks correlated negatively with estimated growth rates (GRs), with early colonizers displaying the shortest minimal generation times (GTs) (n = 14, Pearson correlation, r = −0.63, and P = 0.016) (Fig. S4). Only at the end of the first year of life was the appearance of highly oxygen-sensitive butyrate producers, including Faecalibacterium, the hallmark of the healthy adult gut ecosystem (23), observed (data not shown). Microbial production of butyrate is of key importance to create and maintain the anaerobic conditions that characterize a healthy, adult colon environment (24).
Average predicted growth rates for the top genera of the infant gut. (a) Maximum growth rates (MGR) of the top 15 most abundant genera in the infant gut, ordered by their rank of appearance, calculated as reported previously (54). Note that for one genus, unclassified Lachnospiraceae, no growth rate could be obtained. (b) Negative correlation between the ranks of the top genera and their growth rates (Pearson correlation coefficient) (n = 14). Download FIG S4, PDF file, 0.5 MB (509.1KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Effect of external factors on infant gut microbiota maturation.
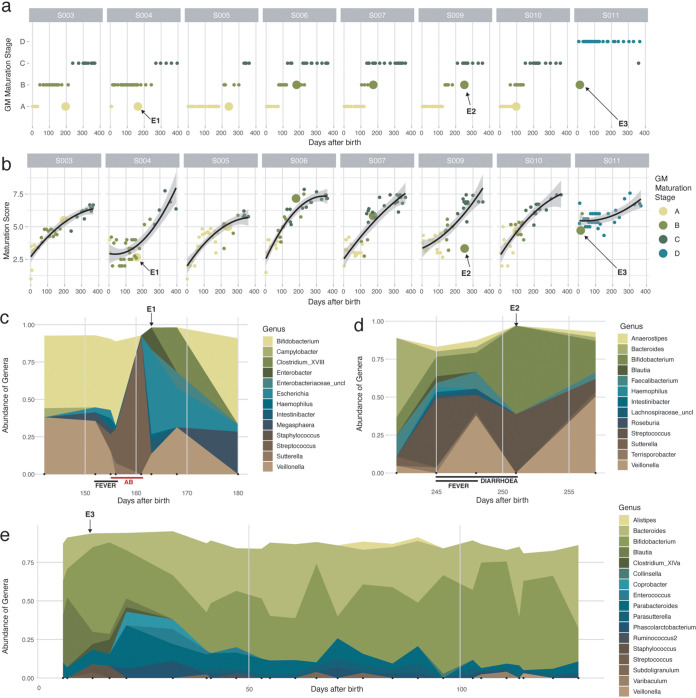
Although the maturation of the infant gut microbiota was identified to be a largely unidirectional process, occasional transient regression toward a preceding gut microbiota maturation stage was observed (Fig. 3a). Hypothesizing maturation stage regression to be associated with disease or medical interventions, we developed an ecosystem maturation index per sample based on the presence/absence of genera belonging to the BaBel average top 15. As discussed above, we ranked each genus according to its order of appearance along the timeline of an infant’s ecosystem maturation process. Next, genera were attributed an overall cohort rank (1 to 10) (Fig. 2b) based on their median order of appearance across individual infants. A sample’s maturation index was calculated by averaging the ranks of the genera present (relative abundance of >0.5%) (Fig. 3b). We identified three time points (events) displaying a lower maturation score than expected (i.e., outside the 95% confidence interval [CI] of the regression of the maturation score) concurring with regression in the maturation stage (Fig. 3a). A first event (E1) (infant S004 at day 163, with regression from maturation stage B to stage A) coincided with the end of a 7-day oral antibiotic treatment (days 155 to 161; amoxicillin with the adjuvant clavulanic acid, a β-lactamase inhibitor) for a urinary tract infection. After treatment initiation, Streptococcus became the predominant genus, falling back below detectable levels 2 days after the last dose of antibiotics (Fig. 3c). Multivariate analysis of the extended BaBel data set (including all eight infants) identified Streptococcus as the genus most significantly increased in abundance during antibiotic treatment (n = 299, dbRDA using all covariates, adjusted R2 = 0.12, and FDR < 0.05; n = 303, MaAsLin2 testing of all covariates on all genera, and FDR = 0.0011) (Fig. 2a; Table S1h). Genera with decreased proportional abundances upon amoxicillin treatment included Bifidobacterium and Veillonella, both decreasing below the detection limits and reappearing after <18 and 6 days after the cessation of treatment, respectively (Fig. 3c). After the disappearance of Streptococcus, Escherichia was the first genus to reestablish, becoming the most dominant member of the gut microbiota <2 days after the last dose of amoxicillin (Fig. 3c). These observations confirm the status of oxygen-tolerant genera as pioneering colonizers in primary succession as well as secondary colonization following antibiotic treatment-associated ecosystem disruption, with gut microbiota maturation stage regression probably being associated with an imbalance in colon oxygen homeostasis (25) (Fig. 3a and c). Of note, two other infants (S003, days 353 to 359, and S010, days 214 to 220) also received amoxicillin (without clavulanic acid), in both cases prescribed to treat an ear infection. However, only less pronounced microbiome alterations were observed upon treatment, possibly due to the absence of an adjuvant (i.e., clavulanic acid) or the fact that the infants’ microbiota had matured to the potentially more stable C maturation stage (Fig. S5). The second event (E2) (infant S009 at day 251, regression from stage C to stage B) coincided with an untreated Cryptosporidium infection (days 248 to 250), accompanied by fever and diarrhea, which was characterized by observed increases in the relative abundances of Bifidobacterium and Streptococcus, while the abundances of the other genera decreased (Fig. 3a, b, and d). E3 (days 13 to 21) cooccurred with the start of a period of severe constipation in infant S011 (Fig. 3e). While the baby’s first samples taken at days 6 and 7 were classified within the infant-specific maturation stage D (Fig. 3a), a transition to the Bifidobacterium-dominated B maturation stage was noted on days 13, 17, and 21. During the period following maturation stage regression, infant S011 suffered from recurrent episodes of severe constipation, including three periods of 6 to 9 days without a bowel movement (defecation on days 32, 40, 41, 47, and 53). However, from day 32, the infant’s fecal microbiome returned to the maturation state D classification. It has been reported that children with functional constipation are more likely to be born by C-section or to have a shorter duration of breastfeeding, suggesting that early gut microbiome events may increase the risk of developing functional constipation later in life (26). For instance, children with a history of necrotizing enterocolitis, who are often premature and treated with prolonged courses of antibiotics, have a higher incidence of functional constipation than healthy children (27). Infant S011 was born vaginally and was solely breastfed during the period in which constipation was reported. The link between the reported constipation and the maturation stage in this study remains unknown.
FIG 3.
Effect of external factors on the infant gut microbiome. (a) Succession of the gut microbiota maturation stages over time, including all 303 time points from the BaBel data set. Time points representing a return to a previous gut microbiota maturation stage (after at least 2 samples from the next gut microbiota maturation stage) are represented with larger dots. (b) Changes in the maturation scores of the samples over time. The maturation score was calculated by averaging the ranks (based on their order of appearance) of the genera present in every sample. The black line represents the quadratic regression with the 95% confidence interval (all P values of the quadratic fits are <0.0002). Three events for which the succession goes back to a previous gut microbiota maturation stage (shown in Fig. 4a) and the maturation score drops (outside the confidence interval) are indicated with arrows. (c) Changes in bacterial abundance during the antibiotic (AB) event in infant S004 (“E1” at day 163) (abundances of >0.02 are shown). The red line indicates the duration of treatment (7 days) with antibiotics (amoxicillin and clavulanic acid). (d) Changes in abundance during Cryptosporidium infection in infant S009 (“E2” at day 251) (abundances of >0.02 are shown). (e) Changes in abundances in the first half-year in infant S011 (“E3” at days 13 to 21) (abundances of >0.05 are shown).
Effect of external factors on the infant gut microbiome. Changes in bacterial abundance during the antibiotic event in infants S003 (a), S004 (b), and S010 (c) are shown. Download FIG S5, PDF file, 0.9 MB (953KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
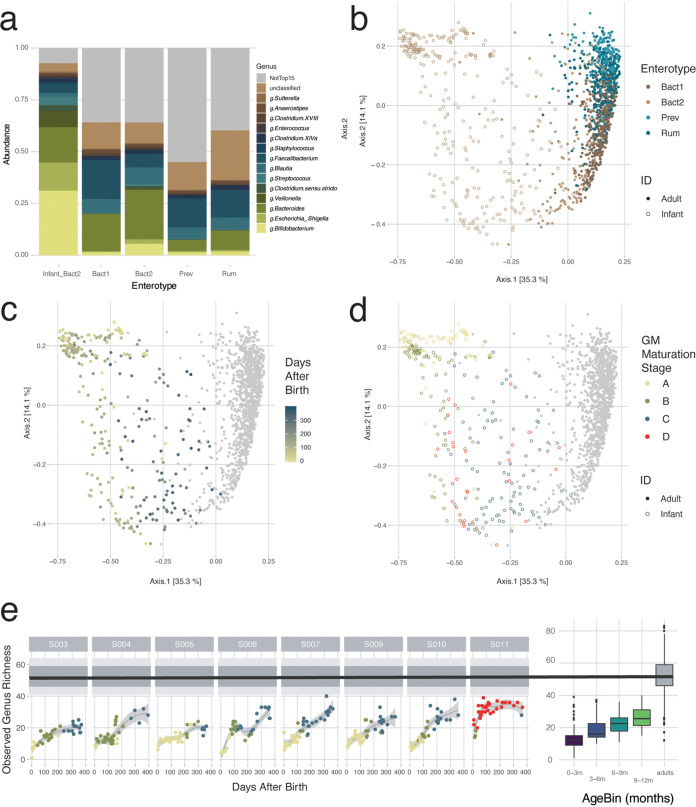
Transition of the infant gut microbiota toward an adult configuration.
To evaluate gut microbiota maturation during the first year of life in terms of ecosystem transition toward an adult configuration, we mapped the microbiome composition of the infant samples onto the background of interindividual variation as observed in the Flemish Gut Flora Project (FGFP) population cohort (n = 1,106) (Fig. 4). Previously, using DMM-based community typing (13), genus-level compositional differentiation of the adult microbiome in the FGFP has been shown to revolve around four enterotypes (28), prevalent, nondiscrete microbiome constellations that can be identified reproducibly across data sets (28–30). Having aligned not only DNA extraction and sequencing methods but also analytical procedures with the FGFP protocols (31), we observed that the fecal microbiomes of Flemish infants differ substantially from those obtained from adults inhabiting the same region (permutational multivariate analysis of variance [MANOVA] Adonis test, n = 1,407, R2 = 0.30, and P = 0.001) (Fig. 4b to d). However, all infant samples were classified as Bacteroides 2 (Bact2) communities (Fig. 4a and b; Table S1i), a recently described low-diversity/low-cell-density constellation characterized by high Bacteroides and low Faecalibacterium proportional abundances. Bact2 communities have previously been linked to loose stools (29), inflammation (29), and reduced well-being (32) and have been hypothesized to reflect ecosystem dysbiosis (28, 29, 33). The similarities of infant microbiota constellations to adult dysbiotic states, as previously noted (12), are likely attributable to convergences between primary (ecosystem development) and secondary (perturbation recovery) succession (12, 34). Like in adult dysbiosis, the infant gut ecosystem has been reported to display low colonization resistance (21, 35), exemplified by the frequency of gastrointestinal infections reported in the present cohort, with BaBel infants experiencing on average 2 (range = [0:3]) episodes of diarrhea during the first year of their life and beyond (36). At the same time, a shift in the infant microbiota composition toward a more adult-like configuration was observed over time. When comparing the microbiota compositions of BaBel age bins (0:3, 3:6, 6:9, and 9:12 months) with the those of the FGFP population cohort, effect sizes in microbiome variation were observed to decrease with increasing infant age (permutational pairwise MANOVA Adonis test, n = [1,206:1,204:1,153:1,159], R2 = [0.228:0.221:0.085:0.067], and FDR < 0.01) (Fig. 4c; Table S1j). Moreover, a detailed analysis of DMM clustering results identified six samples from three infants taken in the last month of their first year having a nonzero probability of not belonging to the Bact2 community type (probability range = [4.34e−6:1.20e−14]) (Table S1i). In all samples, the observed transition toward a more adult microbiome constellation was accompanied by an increase in the observed genus richness over time, although adult richness was not reached (infant age bins versus adults, KW and phD tests, n = [1,207:1,205:1,154:1,160], r = [−0.52:−0.47:−0.32:−0.32], and FDR < 0.05) (Fig. 4e; Table S1k). The fact that microbiome maturation does not reach full adulthood in the first year of life is in agreement with recent reports (37–39). When a composition fully resembling an adult-like composition is reached is still uncertain but probably takes up to the age of 3 years (40).
FIG 4.
Projection of the infant samples to adult samples of the Flemish Gut Flora Project (FGFP) data set. (a) Bar plots showing the average relative abundances of the top 15 most common bacterial genera of the infant samples and the adult samples, per enterotype. (b to d) Projection of the infant samples to the adult FGFP data set, visualized by principal-coordinate analysis (PCoA) (Bray-Curtis dissimilarity), colored for enterotype (b), colored for time after birth (for the infant samples) (c), and colored per gut microbiota maturation stage (d). (e) Observed genus-level richness over time of the BaBel data set (LOESS), compared to the observed genus-level richness of the FGFP data set (the black line is the median, the dark gray area represents the 25 to 75% interquartile range [IQR], and the light gray area represents the 10 to 90% IQR). On the right side, the box plots represent the genus-level richness for the different infant age bins, compared to the adult FGFP data set. The body of the box plots represents the first and third quartiles of the distribution and the median line.
Conclusion.
We show that the maturation of the gut microbiota can be captured in a series of transitions that remain conserved across the BaBel infants, both on the community/gut microbiota maturation stage level and in the order of the appearance of prevalent genera. Throughout the first year of life, successional colonization of the gut microbiota results in a shift from a low-richness, oxygen-tolerant community dominated by pioneering colonizers such as Escherichia to a more diverse community comprising anaerobic butyrogens such as Faecalibacterium, with butyrate being a key metabolite in the maintenance of colonic hypoxia (24). Our analyses confirm previously reported similarities between the infant microbiota and adult dysbiosis (9, 12, 41), likely due to shared features of primary and secondary succession. While temporary regression following ecosystem-disrupting events such as infection or antibiotic treatment can be observed, the microbiota of all studied infants matured to a more adult-like constellation over the first year of their life, as reported previously (42). Given the similarities observed between primary succession and secondary colonization upon disruption, careful dissection of the succession events characterizing gut ecosystem maturation could pave the way for the development of mimicking biotherapeutic strategies in adult microbiome modulation.
MATERIALS AND METHODS
Sample collection.
Between 2013 and 2017, stool samples from eight Belgian healthy infants, i.e., the BaBel infants, were collected starting from birth at a frequency of 2 to 3 samples per week (see Table S1a in the supplemental material). Samples were kept in −20°C freezers at the participants’ homes, and every 3 months, they were transported to our laboratory on dry ice, where they were stored at −80°C until further analysis. Every time a sample was collected, the parents completed a questionnaire containing information about the date, consistency of the stool (aqueous/soft/solid), diet (breast milk/formula milk/vegetables/fruit), clinical signs of disease (diarrhea/vomiting/fever/…), and the location of the infant when the sample was taken (at home/day care/holiday location/…). All infants were vaginally born, the mothers did not take antibiotics during pregnancy or delivery, and no complications during pregnancy were reported. The histo-blood group antigen (HBGA) specificities (ABO group antigens, Lewis antigens, and FUT2 and FUT3 genotypes) were determined as described previously (43), from a saliva sample from each infant collected at the end of the study period. For the investigation of the overall effect of metadata on the microbiome composition, only covariates present in at least three infants were used (infant identification, time after birth, the presence of furry pets, secretor status, Lewis antigens, ABO blood group, diet pattern [breast only/no solid/solid], consistency, diarrhea, fever, respiratory illness and other general sickness signs, painkillers, antibiotics, and day care). In the “breast-only” group, samples are included only when the only component in the infants’ diet was breast milk. In the “no-solid” group, samples are included when no solid foods were introduced yet but the infants also did not consume breast milk only; namely, included are samples where only formula or a combination of formula and breast milk was part of the infants’ diet.
Sample selection.
To study the longitudinal dynamics of the gut microbiome, 21 stool samples from predefined days 0, 3 7, 10, 15, 21, 30, 45, 60, 75, 80, 105, 120, 150, 180, 210, 240, 270, 300, 330, and 360 were selected from each of the eight infants (Fig. S1). When an infant showed clinical signs at any of these time points, we selected the closest available sample without clinical signs present, or this time point was excluded. In total, we included 159 samples at predefined time points, of which 17 fell together with clinical signs (and were not replaceable by a close time point with no signs) and 142 did not fall together with clinical signs (Table S1a and Fig. S1). In addition, we selected 144 additional samples ad hoc from before, during, and after specific external events to study how they influence the gut microbiome (events included vaccination history, type of food consumed, occurrence of diseases, use of antibiotics, and use of pre- or probiotics) (Fig. S1).
To summarize, a total of 303 samples were selected. Of these, 159 were initially selected because they fell together with a predefined time point. Not unexpectedly, of these 159 samples, there were 17 samples that fell together with the presence of clinical signs; the other 142 predefined time points were without any clinical signs reported. Apart from the 159 samples selected at predefined time points, we selected 144 additional samples “ad hoc” based on different external factors (Table S1a and Fig. S1).
16S rRNA gene library preparation and sequencing.
Bacterial profiling was carried out as described previously by Falony and colleagues (31). Briefly, nucleic acids were extracted from frozen fecal aliquots using the RNeasy PowerMicrobiome kit (Qiagen). The manufacturer’s protocol was modified by the addition of a heating step at 90°C for 10 min after vortexing and by the exclusion of DNA removal steps. Microbiome characterization was performed as previously described (44); in short, the extracted DNA was further amplified in triplicate using 16S primers 515F (5′-GTGYCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACNVGGGTWTCTAAT-3′) targeting the V4 region, modified to contain a barcode sequence between each primer and the Illumina adaptor sequences to produce dually barcoded libraries. Deep sequencing was performed on a MiSeq platform (2-by-250 paired-end [PE] reads; Illumina). All samples were randomized, and negative controls were taken along and sequenced.
Sequence read analysis.
After demultiplexing with sdm as part of the LotuS pipeline (45) without allowing for mismatches, fastq sequences were further analyzed per sample using the DADA2 pipeline (v.1.6) (46). Briefly, we removed the primer sequences and the first 10 nucleotides after the primer. After merging paired sequences and removing chimeras, taxonomy was assigned using the formatted RDP training set “rdp_train_set_16.” The decontam (47) R package was used to remove contaminating amplicon sequence variants (ASVs) using the frequency prevalence method (Table S1l). After quality control steps, the ASV table contained on average 46,330 reads per sample (range = 15,427 to 131,451 reads). In total, 197 ASVs were obtained, all belonging to the kingdom Bacteria. No Archaea were detected. All samples were rarefied to 14,668 reads per sample, and ASVs with an overall relative abundance of <0.0001 were removed. From three samples (S009-1, S004-1, and S010-1), the first samples taken from three different infants, we were not able to extract enough DNA to be amplifiable.
Statistical analyses.
All statistical analyses were performed and visualized in R (http://www.R-project.org) using the ggplot2 (48), phyloseq (49), synchrony (50), DirichletMultinomial (51), dunn.test (52), and vegan (53) packages. To test median differences between two or more groups of continuous variables, a Mann-Whitney U test and a Kruskal-Wallis (KW) test were performed, respectively. The KW test was always followed by post hoc Dunn’s (phD) test for all pairs of comparisons between groups. Multiple-testing correction was performed where appropriate using the Benjamini-Hochberg procedure (FDR adjustment set at <0.05). Although it would be interesting to validate our results using data from other studies, no sensible comparison could be made for several reasons, including the fact that for many studies, data sets are not publicly available, or other methodologies were used (e.g., no 16S sequencing or the use of another database, etc.).
DMM clustering to identify the colonization stages.
To determine the stages of the colonization process, a Dirichlet multinomial mixture (DMM)-based approach was followed, as described previously by Holmes et al. (13), using the DirichletMultinomial (51) R package on the genus-level (rarefied) read matrix (n = 303). The optimal number of stages was determined based on the Bayesian information criterion (BIC), and the mean probability for the samples to belong to the assigned Dirichlet component was on average 0.99 (median = 1; standard deviation = 0.05) (Table S1b).
Determination of the order of appearance of the top genera.
Per infant, the 15 most abundant genera (present in more than 3 infants) were ranked based on the first time point at which they were present (with an abundance of >0.5%). Rankings were scored using the Kendal w test using the R function kendall.w of the synchrony package (50) with 10,000 permutations. A final order of appearance was set, based on the order of the medians of the ranks per infant. Finally, a maturation score was calculated for every sample by averaging the ranks of the genera weighted by the presence or absence of that specific genus. Growth rates (GRs) of the different genera were calculated from the predicted generation times (GTs) (GT = 1/GR), as reported previously (54).
Alpha and beta diversity.
Alpha diversity (richness and Shannon diversity) and beta diversity (Bray-Curtis dissimilarity) indices were calculated by using the phyloseq (49) package. Ordinations were visualized by principal-coordinate analysis (PCoA) using Bray-Curtis dissimilarity. The univariate effects of the metadata variables on the first two axes of the ordination were determined using the envfit function of the vegan package (53) (univariate distance-based redundancy analysis [dbRDA]) and plotted as arrows on the PCoA plot (infant identification was excluded for clarity). Community-level differences between groups were tested with the Adonis nonparametric test of the vegan package (53). If more than two groups were compared, a post hoc Adonis test was used in a pairwise way, correcting for multiple testing.
Multivariate analysis of the effect of metadata variables on microbial composition.
To investigate which metadata covariates contribute to the variation in the microbiota community, dbRDA was performed on the genus level (Bray-Curtis distance) using the capscale function in the vegan (53) R package. Covariates found to significantly contribute to the ordination outcome were further implemented with forward model selection on dbRDA using the ordiR2step function in the vegan package (53) to determine the nonredundant cumulative contribution of metadata variables to the variation (stepwise dbRDA). To test the effect of metadata variables on specific genera, the same approach as the one described above was followed by first pruning the community to contain only the genus of interest (for each of the top 15 genera), followed by dbRDA on the Euclidean distances measured on the abundances of that genus and forward model selection as described above, constraining for infant identifier. To confirm the results from the previous step, MaAsLin2 (55) was used, which performs boosted additive general linear models to discover associations between metadata and the relative taxonomic abundances (default settings). Note that for only the dbRDA, four samples were excluded, for which consistency was unknown (n = 299).
Projection to the adult FGFP data set.
While community typing of the infant samples alone revealed clear stages of maturation, we next investigated how the immature infant gut microbiome relates to mature adult microbiomes by classifying the infant samples combined with adult samples. Enterotypes of the infant samples were computed against a background of adult non-disease-associated microbiomes (FGFP data set, genus-level abundance matrix, n = 1,106) by DMM clustering using the DirichletMultinomial package as described previously by Holmes et al. (13) Samples were rarefied to 10,000 reads. To avoid interference by nonindependent samples, enterotyping was performed iteratively on one randomly selected sample from each infant against the FGFP background (n = 42 enterotyping rounds). The optimal number of Dirichlet components based on the BIC was 4 in all iterations, and the clusters were named Prevotella, Bacteroides 1, Bacteroides 2, and Ruminococcaceae, as described previously (28).
Ethics approval.
The study was approved by the IRB at KU Leuven (ML8699, S54745, and B322201215465).
Data availability.
16S sequencing data used in this study are available at the European Nucleotide Archive (ENA) (accession number PRJEB40751). The code to perform analysis and make figures starting from the ASV abundance table is available at https://github.com/Matthijnssenslab/BabyGut16S/.
ACKNOWLEDGMENTS
We thank all participating infants and parents for their contribution. We thank Johan Nordgren for the characterization of the saliva samples.
We declare that we have no competing interests.
This research was supported by the Fonds Wetenschappelijk Onderzoek (Research Foundation Flanders) (1S61618N [L.B.], 1110918N [M.V.-C.], 1S78019N [D.J.], and 1S25720N [L.V.E.]) and by a KU Leuven OT grant (OT-14-113).
The study was conceived by J.M., J.R., and M.V.R. Experiments were designed by J.M., J.R., L.B., M.Z., and R.Y.T. Sampling was set up and carried out by C.S., J.M., C.K.Y., K.F., L.B., and W.D. Experiments were performed by D.J., L.B., L.V.E., and L.R. L.B., M.I.P., and R.Y.T. performed the bioinformatics analyses of the sequenced reads. Statistical analyses were designed and performed by G.F., L.B., M.V.-C., S.V.-S., and W.D. L.B., J.M., G.F., S.V.-S., M.I.P., M.Z., and W.D. drafted the manuscript. All authors revised the article and approved the final version for publication.
Contributor Information
Leen Beller, Email: leen.beller@gmail.com.
Jeroen Raes, Email: jeroen.raes@kuleuven.be.
Jelle Matthijnssens, Email: jelle.matthijnssens@kuleuven.be.
Gary B. Huffnagle, University of Michigan—Ann Arbor
REFERENCES
- 1.Lewis JD, Chen EZ, Baldassano RN, Otley AR, Griffiths AM, Lee D, Bittinger K, Bailey A, Friedman ES, Hoffmann C, Albenberg L, Sinha R, Compher C, Gilroy E, Nessel L, Grant A, Chehoud C, Li H, Wu GD, Bushman FD. 2015. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn’s disease. Cell Host Microbe 18:489–500. doi: 10.1016/j.chom.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arrieta M-C, Arévalo A, Stiemsma L, Dimitriu P, Chico ME, Loor S, Vaca M, Boutin RCT, Morien E, Jin M, Turvey SE, Walter J, Parfrey LW, Cooper PJ, Finlay B. 2018. Associations between infant fungal and bacterial dysbiosis and childhood atopic wheeze in a nonindustrialized setting. J Allergy Clin Immunol 142:424–434.e10. doi: 10.1016/j.jaci.2017.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vatanen T, Franzosa EA, Schwager R, Tripathi S, Arthur TD, Vehik K, Lernmark Å, Hagopian WA, Rewers MJ, She J-X, Toppari J, Ziegler A-G, Akolkar B, Krischer JP, Stewart CJ, Ajami NJ, Petrosino JF, Gevers D, Lähdesmäki H, Vlamakis H, Huttenhower C, Xavier RJ. 2018. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature 562:589–594. doi: 10.1038/s41586-018-0620-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stewart CJ, Ajami NJ, O’Brien JL, Hutchinson DS, Smith DP, Wong MC, Ross MC, Lloyd RE, Doddapaneni H, Metcalf GA, Muzny D, Gibbs RA, Vatanen T, Huttenhower C, Xavier RJ, Rewers M, Hagopian W, Toppari J, Ziegler A-G, She J-X, Akolkar B, Lernmark A, Hyoty H, Vehik K, Krischer JP, Petrosino JF. 2018. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 562:583–588. doi: 10.1038/s41586-018-0617-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, Li Y, Xia Y, Xie H, Zhong H, Khan MT, Zhang J, Li J, Xiao L, Al-Aama J, Zhang D, Lee YS, Kotowska D, Colding C, Tremaroli V, Yin Y, Bergman S, Xu X, Madsen L, Kristiansen K, Dahlgren J, Wang J. 2015. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17:852. doi: 10.1016/j.chom.2015.05.012. [DOI] [PubMed] [Google Scholar]
- 6.Matamoros S, Gras-Leguen C, Le Vacon F, Potel G, de La Cochetiere MF. 2013. Development of intestinal microbiota in infants and its impact on health. Trends Microbiol 21:167–173. doi: 10.1016/j.tim.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 7.Milani C, Duranti S, Bottacini F, Casey E, Turroni F, Mahony J, Belzer C, Delgado Palacio S, Arboleya Montes S, Mancabelli L, Lugli GA, Rodriguez JM, Bode L, de Vos W, Gueimonde M, Margolles A, van Sinderen D, Ventura M. 2017. The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol Mol Biol Rev 81:e00036-17. doi: 10.1128/MMBR.00036-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fallani M, Amarri S, Uusijarvi A, Adam R, Khanna S, Aguilera M, Gil A, Vieites JM, Norin E, Young D, Scott JA, Doré J, Edwards CA, The Infabio Team . 2011. Determinants of the human infant intestinal microbiota after the introduction of first complementary foods in infant samples from five European centres. Microbiology (Reading) 157:1385–1392. doi: 10.1099/mic.0.042143-0. [DOI] [PubMed] [Google Scholar]
- 9.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. 2011. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci USA 108(Suppl):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laursen MF, Andersen LBB, Michaelsen KF, Mølgaard C, Trolle E, Bahl MI, Licht TR. 2016. Infant gut microbiota development is driven by transition to family foods independent of maternal obesity. mSphere 1:e00069-15. doi: 10.1128/mSphere.00069-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yassour M, Vatanen T, Siljander H, Hämäläinen A-M, Härkönen T, Ryhänen SJ, Franzosa EA, Vlamakis H, Huttenhower C, Gevers D, Lander ES, Knip M, DIABIMMUNE Study Group, Xavier RJ. 2016. Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci Transl Med 8:343ra81. doi: 10.1126/scitranslmed.aad0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kriss M, Hazleton KZ, Nusbacher NM, Martin CG, Lozupone CA. 2018. Low diversity gut microbiota dysbiosis: drivers, functional implications and recovery. Curr Opin Microbiol 44:34–40. doi: 10.1016/j.mib.2018.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holmes I, Harris K, Quince C. 2012. Dirichlet multinomial mixtures: generative models for microbial metagenomics. PLoS One 7:e30126. doi: 10.1371/journal.pone.0030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Espey MG. 2013. Role of oxygen gradients in shaping redox relationships between the human intestine and its microbiota. Free Radic Biol Med 55:130–140. doi: 10.1016/j.freeradbiomed.2012.10.554. [DOI] [PubMed] [Google Scholar]
- 15.Rajilić-Stojanović M, de Vos WM. 2014. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol Rev 38:996–1047. doi: 10.1111/1574-6976.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andriantsoanirina V, Allano S, Butel MJ, Aires J. 2013. Tolerance of Bifidobacterium human isolates to bile, acid and oxygen. Anaerobe 21:39–42. doi: 10.1016/j.anaerobe.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 17.Oliphant K, Allen-Vercoe E. 2019. Macronutrient metabolism by the human gut microbiome: major fermentation by-products and their impact on host health. Microbiome 7:91. doi: 10.1186/s40168-019-0704-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramsey M, Hartke A, Huycke M. 15 February 2014. The physiology and metabolism of enterococci. In Gilmore MS, Clewell DB, Ike Y, Shankar N (ed), Enterococci: from commensals to leading causes of drug resistant infection. Massachusetts Eye and Ear Infirmary, Boston, MA. [PubMed] [Google Scholar]
- 19.Vieira-Silva S, Falony G, Darzi Y, Lima-Mendez G, Garcia Yunta R, Okuda S, Vandeputte D, Valles-Colomer M, Hildebrand F, Chaffron S, Raes J. 2016. Species-function relationships shape ecological properties of the human gut microbiome. Nat Microbiol 1:16088. doi: 10.1038/nmicrobiol.2016.88. [DOI] [PubMed] [Google Scholar]
- 20.Roe AJ, O’Byrne C, McLaggan D, Booth IR. 2002. Inhibition of Escherichia coli growth by acetic acid: a problem with methionine biosynthesis and homocysteine toxicity. Microbiology (Reading) 148:2215–2222. doi: 10.1099/00221287-148-7-2215. [DOI] [PubMed] [Google Scholar]
- 21.Ducarmon QR, Zwittink RD, Hornung BVH, van Schaik W, Young VB, Kuijper EJ. 2019. Gut microbiota and colonization resistance against bacterial enteric infection. Microbiol Mol Biol Rev 83:e00007-19. doi: 10.1128/MMBR.00007-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duncan SH, Louis P, Flint HJ. 2004. Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl Environ Microbiol 70:5810–5817. doi: 10.1128/AEM.70.10.5810-5817.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miquel S, Martin R, Bridonneau C, Robert V, Sokol H, Bermúdez-Humarán LG, Thomas M, Langella P. 2014. Ecology and metabolism of the beneficial intestinal commensal bacterium Faecalibacterium prausnitzii. Gut Microbes 5:146–151. doi: 10.4161/gmic.27651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pryde SE, Duncan SH, Hold GL, Stewart CS, Flint HJ. 2002. The microbiology of butyrate formation in the human colon. FEMS Microbiol Lett 217:133–139. doi: 10.1111/j.1574-6968.2002.tb11467.x. [DOI] [PubMed] [Google Scholar]
- 25.Rivera-Chávez F, Lopez CA, Bäumler AJ. 2017. Oxygen as a driver of gut dysbiosis. Free Radic Biol Med 105:93–101. doi: 10.1016/j.freeradbiomed.2016.09.022. [DOI] [PubMed] [Google Scholar]
- 26.de Moraes JG, Motta MEFDA, Beltrão MFDS, Salviano TL, da Silva GAP. 2016. Fecal microbiota and diet of children with chronic constipation. Int J Pediatr 2016:6787269. doi: 10.1155/2016/6787269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen S-M, Huang J-Y, Wu M-C, Chen J-Y. 2020. The risk of developing constipation after neonatal necrotizing enterocolitis. Front Pediatr 8:120. doi: 10.3389/fped.2020.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vandeputte D, Kathagen G, D’Hoe K, Vieira-Silva S, Valles-Colomer M, Sabino J, Wang J, Tito RY, De Commer L, Darzi Y, Vermeire S, Falony G, Raes J. 2017. Quantitative microbiome profiling links gut community variation to microbial load. Nature 551:507–511. doi: 10.1038/nature24460. [DOI] [PubMed] [Google Scholar]
- 29.Vieira-Silva S, Sabino J, Valles-Colomer M, Falony G, Kathagen G, Caenepeel C, Cleynen I, van der Merwe S, Vermeire S, Raes J. 2019. Quantitative microbiome profiling disentangles inflammation- and bile duct obstruction-associated microbiota alterations across PSC/IBD diagnoses. Nat Microbiol 4:1826–1831. doi: 10.1038/s41564-019-0483-9. [DOI] [PubMed] [Google Scholar]
- 30.Ding T, Schloss PD. 2014. Dynamics and associations of microbial community types across the human body. Nature 509:357–360. doi: 10.1038/nature13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Falony G, Joossens M, Vieira-Silva S, Wang J, Darzi Y, Faust K, Kurilshikov A, Bonder MJ, Valles-Colomer M, Vandeputte D, Tito RY, Chaffron S, Rymenans L, Verspecht C, De Sutter L, Lima-Mendez G, D’Hoe K, Jonckheere K, Homola D, Garcia R, Tigchelaar EF, Eeckhaudt L, Fu J, Henckaerts L, Zhernakova A, Wijmenga C, Raes J. 2016. Population-level analysis of gut microbiome variation. Science 352:560–564. doi: 10.1126/science.aad3503. [DOI] [PubMed] [Google Scholar]
- 32.Valles-Colomer M, Falony G, Darzi Y, Tigchelaar EF, Wang J, Tito RY, Schiweck C, Kurilshikov A, Joossens M, Wijmenga C, Claes S, Van Oudenhove L, Zhernakova A, Vieira-Silva S, Raes J. 2019. The neuroactive potential of the human gut microbiota in quality of life and depression. Nat Microbiol 4:623–632. doi: 10.1038/s41564-018-0337-x. [DOI] [PubMed] [Google Scholar]
- 33.Vieira-Silva S, Falony G, Belda E, Nielsen T, Aron-Wisnewsky J, Chakaroun R, Forslund SK, Assmann K, Valles-Colomer M, Nguyen TTD, Proost S, Prifti E, Tremaroli V, Pons N, Le Chatelier E, Andreelli F, Bastard J-P, Coelho LP, Galleron N, Hansen TH, Hulot J-S, Lewinter C, Pedersen HK, Quinquis B, Rouault C, Roume H, Salem J-E, Søndertoft NB, Touch S, MetaCardis Consortium, Dumas M-E, Ehrlich SD, Galan P, Gøtze JP, Hansen T, Holst JJ, Køber L, Letunic I, Nielsen J, Oppert J-M, Stumvoll M, Vestergaard H, Zucker J-D, Bork P, Pedersen O, Bäckhed F, Clément K, Raes J. 2020. Statin therapy is associated with lower prevalence of gut microbiota dysbiosis. Nature 581:310–316. doi: 10.1038/s41586-020-2269-x. [DOI] [PubMed] [Google Scholar]
- 34.Valles-Colomer M, Bacigalupe R, Vieira-Silva S, Suzuki S, Darzi Y, Tito RY, Yamada T, Segata N, Raes J, Falony G. Variation and transmission of the human gut microbiota across generations. Nat Microbiol, in press. [DOI] [PMC free article] [PubMed]
- 35.Houghteling PD, Walker WA. 2015. Why is initial bacterial colonization of the intestine important to infants’ and children’s health? J Pediatr Gastroenterol Nutr 60:294–307. doi: 10.1097/MPG.0000000000000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mughini-Gras L, Pijnacker R, Heusinkveld M, Enserink R, Zuidema R, Duizer E, Kortbeek T, van Pelt W. 2016. Societal burden and correlates of acute gastroenteritis in families with preschool children. Sci Rep 6:22144. doi: 10.1038/srep22144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bergström A, Skov TH, Bahl MI, Roager HM, Christensen LB, Ejlerskov KT, Mølgaard C, Michaelsen KF, Licht TR. 2014. Establishment of intestinal microbiota during early life: a longitudinal, explorative study of a large cohort of Danish infants. Appl Environ Microbiol 80:2889–2900. doi: 10.1128/AEM.00342-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fouhy F, Watkins C, Hill CJ, O’Shea C-A, Nagle B, Dempsey EM, O’Toole PW, Ross RP, Ryan CA, Stanton C. 2019. Perinatal factors affect the gut microbiota up to four years after birth. Nat Commun 10:1517. doi: 10.1038/s41467-019-09252-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Avershina E, Lundgård K, Sekelja M, Dotterud C, Storrø O, Øien T, Johnsen R, Rudi K. 2016. Transition from infant- to adult-like gut microbiota. Environ Microbiol 18:2226–2236. doi: 10.1111/1462-2920.13248. [DOI] [PubMed] [Google Scholar]
- 40.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. 2012. Human gut microbiome viewed across age and geography. Nature 486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lozupone C, Faust K, Raes J, Faith JJ, Frank DN, Zaneveld J, Gordon JI, Knight R. 2012. Identifying genomic and metabolic features that can underlie early successional and opportunistic lifestyles of human gut symbionts. Genome Res 22:1974–1984. doi: 10.1101/gr.138198.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. 2007. Development of the human infant intestinal microbiota. PLoS Biol 5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nordgren J, Sharma S, Bucardo F, Nasir W, Günaydın G, Ouermi D, Nitiema LW, Becker-Dreps S, Simpore J, Hammarström L, Larson G, Svensson L. 2014. Both Lewis and secretor status mediate susceptibility to rotavirus infections in a rotavirus genotype-dependent manner. Clin Infect Dis 59:1567–1573. doi: 10.1093/cid/ciu633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tito RY, Cypers H, Joossens M, Varkas G, Van Praet L, Glorieus E, Van den Bosch F, De Vos M, Raes J, Elewaut D. 2017. Brief report: Dialister as a microbial marker of disease activity in spondyloarthritis. Arthritis Rheumatol 69:114–121. doi: 10.1002/art.39802. [DOI] [PubMed] [Google Scholar]
- 45.Hildebrand F, Tadeo R, Voigt AY, Bork P, Raes J. 2014. LotuS: an efficient and user-friendly OTU processing pipeline. Microbiome 2:30. doi: 10.1186/2049-2618-2-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Callahan B, Davis NM. 2019. decontam: identify contaminants in marker-gene and metagenomics sequencing data.
- 48.Wickham H, Chang W, Henry L, Pedersen TL, Takahashi K, Wilke C, Woo K. 2019. ggplot2: create elegant data visualisations using the grammar of graphics. 2018. R package version 2:2. https://CRAN.R-project.org/package=ggplot2.
- 49.Mcmurdie APJ, Holmes S, Jordan G, Chamberlain S. 2014. Package ‘phyloseq’: handling and analysis of high-throughput microbiome census data.
- 50.Gouhier TC. 2018. synchrony: methods for computing spatial, temporal, and spatiotemporal statistics.
- 51.Morgan M. 2016. DirichletMultinomial: Dirichlet-multinomial mixture model machine learning for microbiome data.
- 52.Dinno A. 2017. dunn.test: Dunn’s test of multiple comparisons using rank sums.
- 53.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Gavin L., Simpson GL, Solymos P, Stevens MHH, Szoecs E, Wagner H. 2019. vegan: community ecology package.
- 54.Vieira-Silva S, Rocha EPC. 2010. The systemic imprint of growth and its uses in ecological (meta)genomics. PLoS Genet 6:e1000808. doi: 10.1371/journal.pgen.1000808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mallick H, Rahnavard A, McIver LJ, Ma S, Zhang Y, Nguyen LH, Tickle TL, Weingart G, Ren B, Schwager EH, Chatterjee S, Thompson KN, Wilkinson JE, Subramanian A, Lu Y, Waldron L, Paulson JN, Franzosa EA, Bravo HC, Huttenhower C. 2021. Multivariable association discovery in population-scale meta-omics studies. bioRxiv doi: 10.1101/2021.01.20.427420. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Overview of the collected and selected samples per infant. Gray dots, all samples collected by the parents of the enrolled infants; blue diamonds, samples selected for the study of the longitudinal dynamics at predefined time points with no clinical signs (n = 142); purple diamonds, samples selected for the study of the longitudinal dynamics at predefined time points with clinical signs (n = 17) (see Table S1 in the supplemental material for the signs); green diamonds, additional ad hoc-selected samples at specific external events (n = 144); black filled triangles, three vaccination events in every infant; black open triangles, day care entrance; red triangles, external events around which extra samples were selected. Abbreviations: F, fever; D, diarrhea; V, vomit; AB, antibiotics; PB, probiotics. Download FIG S1, PDF file, 0.4 MB (436.6KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Determination of the optimal number of clusters in the DMM approach. Identification of the optimal number of Dirichlet components in the BaBel data set (n = 303) was based on the Bayesian information criterion (BIC). The optimal number of clusters is 4 (minimum BIC = 37,285.6). Download FIG S2, PDF file, 0.7 MB (767.1KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The most abundant genera that are differentially abundant per gut microbiota maturation stage determined using DMM clustering. (a) Distribution of the relative abundances of the most abundant genera in gut microbiota maturation stage A, which are significantly more abundant in maturation stage A than in stages B and C. (b) Distribution of the relative abundances of the most abundant genera in gut microbiota maturation stage B, which are significantly more abundant in maturation stage B than in stages A and C. (c) Distribution of the relative abundances of the most abundant genera in gut microbiota maturation stage C, which are significantly more abundant in maturation stage C than in stages A and B (n = 303, KW with phD test, r > 0.3, and FDR < 0.05) (see Table S4 in the supplemental material). Download FIG S3, PDF file, 0.6 MB (637.7KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
(a) Description of the infants enrolled in the BaBel cohort (n = 8) and description of the analyzed samples (n = 303). Abbreviations: F, female; M, male; se+, secretor; se−, nonsecretor; Le−, Lewis antigen negative; LeB, Lewis B antigen. Shown are the sample identifier, infant identifier, age at sample collection (days after birth, included as a predefined time point when the infant was healthy (LDA_nosigns, Yes/No), consistency (1, aqueous; 2, soft; 3, solid), food category (only breast milk, no solid food, or solid food), drug category (according to the ATC classification system), disease category (according to the ICD-10 classification), holiday (Yes/No), family stay (Yes/No), attending day care (Yes/No), and initially included as a predefined time point but the infant showed signs (LDA_signs, Yes/No). (b) Gut microbiota maturation stage assignment results from Dirichlet multinomial mixture (DMM) clustering (n = 303). (c) Alpha diversity (genus richness and Shannon index) distribution per gut microbiota maturation stage (n = 303) (Kruskal-Wallis test with post hoc Dunn tests and FDR by the Benjamini-Hochberg method). (d) Genus abundance distribution per gut microbiota maturation stage (n = 303) (Kruskal-Wallis test with post hoc Dunn tests and FDR by the Benjamini-Hochberg method, focusing on the 15 most abundant genera). (e) Covariates with nonredundant explanatory power on the genus-level ordination, determined by multivariate distance-based redundancy analysis (dbRDA) at the genus level (n = 299, top 15 most abundant genera, Bray-Curtis dissimilarity, and FDR < 0.05). (f) Multivariate redundancy analysis carried out on the relative abundances of each genus, after constraining for infant identification and testing the most prevalent covariates, i.e., present in at least 3 infants (n = 299) (multivariate dbRDA; FDR < 0.05). (g) Bifidobacterium abundance distribution per food category (n = 303) (Kruskal-Wallis test with post hoc Dunn tests and FDR by the Benjamini-Hochberg method). (h) Results for event 1 (E1) (antibiotic treatment in infant S004). Multivariate redundancy analysis was carried out on the relative abundances of each genus, after constraining for infant identification and testing all covariates (all genera tested, n = 299, multivariate dbRDA, and FDR < 0.05), and Maaslin2 results (n = 303) are shown. (i) Enterotype assignment results of the infant samples (n = 303). (j) Permutational pairwise MANOVA Adonis test of the infant age bins versus adults from the FGFP data set (n = 1,408 )(Bray-Curtis dissimilarity and FDR by the Benjamini-Hochberg method). (k) Genus richness of the infant age bins versus adults from the FGFP data set (n = 1,408) (Kruskal-Wallis test with post hoc Dunn tests and FDR by the Benjamini-Hochberg method). (l) Contaminating amplicon sequence variants (ASVs) removed in the filtering step using the R package “decontam”. Download Table S1, XLSX file, 0.1 MB (109.8KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Average predicted growth rates for the top genera of the infant gut. (a) Maximum growth rates (MGR) of the top 15 most abundant genera in the infant gut, ordered by their rank of appearance, calculated as reported previously (54). Note that for one genus, unclassified Lachnospiraceae, no growth rate could be obtained. (b) Negative correlation between the ranks of the top genera and their growth rates (Pearson correlation coefficient) (n = 14). Download FIG S4, PDF file, 0.5 MB (509.1KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Effect of external factors on the infant gut microbiome. Changes in bacterial abundance during the antibiotic event in infants S003 (a), S004 (b), and S010 (c) are shown. Download FIG S5, PDF file, 0.9 MB (953KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Data Availability Statement
16S sequencing data used in this study are available at the European Nucleotide Archive (ENA) (accession number PRJEB40751). The code to perform analysis and make figures starting from the ASV abundance table is available at https://github.com/Matthijnssenslab/BabyGut16S/.