Abstract
The mitochondrial gene encoding yeast cytochrome oxidase subunit II (Cox2p) specifies a precursor protein with a 15-amino-acid leader peptide. Deletion of the entire leader peptide coding region is known to block Cox2p accumulation posttranscriptionally. Here, we examined in vivo the role of the pre-Cox2p leader peptide and the mRNA sequence that encodes it in the expression of a mitochondrial reporter gene, ARG8m, fused to the 91st codon of COX2. We found within the coding sequence antagonistic elements that control translation: the positive element includes sequences in the first 14 codons specifying the leader peptide, while the negative element appears to be within codons 15 to 91. Partial deletions, point mutations, and local frameshifts within the leader peptide coding region were placed in both the cox2::ARG8m reporter and in COX2 itself. Surprisingly, the mRNA sequence of the first six codons specifying the leader peptide plays an important role in positively controlling translation, while the amino acid sequence of the leader peptide itself is relatively unconstrained. Two mutations that partially block translation can be suppressed by nearby sequence substitutions that weaken a predicted stem structure and by overproduction of either the COX2 mRNA-specific translational activator Pet111p or the large-subunit mitochondrial ribosomal protein MrpL36p. We propose that regulatory elements embedded in the translated COX2 mRNA sequence could play a role, together with trans-acting factors, in coupling regulated synthesis of nascent pre-Cox2p to its insertion in the mitochondrial inner membrane.
Mitochondrial genetic systems have evolved to supply a limited number of subunits of enzymes that carry out respiration and oxidative phosphorylation in the inner membrane. These subunits, encoded in mitochondrial DNA (mtDNA), are assembled with imported products of nuclear genes by mechanisms that are poorly understood but genetically complex, at least in Saccharomyces cerevisiae (18, 19, 54). It seems likely that the expression of mitochondrial and nuclear genes that encode subunits of energy-transducing enzymes is coordinated with the assembly of those enzymes, although no clear mechanisms for this have been elucidated.
In this study, we have focused closely on expression of the mitochondrial gene encoding cytochrome c oxidase subunit II, Cox2p. Like most, if not all, yeast mitochondrial genes, COX2 is expressed under the control of a nuclearly encoded inner-membrane-bound mRNA-specific translational activator protein, Pet111p (14, 17). Pet111p is tightly associated with the inner mitochondrial membrane and is present at levels that limit COX2 expression (17). This translational activator functionally interacts with the COX2 mRNA 5′ untranslated leader (5′-UTL) and promotes translation by an unknown mechanism (31, 39) that is conserved in other budding yeasts (10). Translation of other coding sequences artificially fused to the COX2 mRNA 5′-UTL is also activated by Pet111p (6, 17, 32), indicating that open reading frame sequences do not play a role in this activation step. Furthermore, its interaction with the COX2 mRNA 5′-UTL appears to be important for correct localization of Cox2p synthesis within the organelle (43).
Cox2p is synthesized as a precursor with a 15-amino-acid leader peptide (41, 46) that is cleaved in the intermembrane space after translocation of the amino terminal domain of pre-Cox2p through the membrane (4, 37, 41, 44). Translocation of the amino-terminal domain is not well understood mechanistically but depends on the highly conserved inner membrane protein Oxalp (5, 21, 22, 27) and is thought to be cotranslational (14, 40). While the leader peptide causes membrane association of a soluble passenger protein fused to it, it does not function as a classical signal sequence since it is incapable of directing translocation of the passenger protein through the inner membrane (21). Nucleic acid sequence comparisons indicate that the pre-Cox2p leader peptides of various budding yeast species, including Neurospora crassa, and plants are not highly conserved (20, 25). Mammalian Cox2p entirely lacks a leader peptide (3, 50).
We previously began a study of the function of the pre-Cox2p leader peptide by deleting the codons that specify it from the S. cerevisiae COX2 mitochondrial gene (53). This deletion dramatically reduced the accumulation of Cox2p and caused a severe respiratory defect, but did not affect COX2 mRNA levels. The defect was bypassed by a chimeric gene whose product had the amino-terminal 251 residues of cytochrome b fused to the remaining COX2 coding sequence (53). However, the mechanism by which the deletion prevented COX2 expression was not established: it could have affected translation, membrane insertion, or both.
Here, we examine in more detail the in vivo function of the pre-Cox2p leader peptide and the mRNA sequence that encodes it by site-directed mutation of the mtDNA sequence. We determine the effects of each mutation on functional expression of COX2, which demands synthesis and assembly of cytochrome oxidase, and on expression of a reporter gene, ARG8m (fused to the 91st codon of the COX2 reading frame), which depends only on translation of the chimeric mRNA. Surprisingly, we find that in the context of the COX2 coding sequence, the mRNA sequence encoding the leader peptide plays an important role in controlling translation, while the amino acid sequence of the leader peptide itself is relatively unconstrained. Our analysis suggests the existence of positive and negative regulatory elements embedded in the translated mRNA sequence specifying the N-terminal portion of pre-Cox2p, which could play a role in coupling regulated synthesis of the nascent Cox2p precursor to its insertion in the inner membrane.
MATERIALS AND METHODS
Strains, media, and genetic techniques.
All strains used in this study are listed in Table 1, with the exception of DFS160p0 (MATα leu2Δ arg8ΔURA3 ura3-52 kar1-1 ade2-101 [rho0]) (6, 49). Strain backgrounds were either D273-10B (ATCC 25627) or DBY947 (36). NB58 comes from the cytoduction of cox2-20 mitochondria (53) into the [rho0] derivative of NB40-3C. cox2-62 and cox2-60 are deletions of sequences −295 to +363 (11) and −63 to +66 (6) relative to AUG, respectively. Fermentable complete medium was YPDA or YPGalA: 1% yeast extract, 2% Bacto-Peptone, 100 mg of adenine/liter, and either 2% glucose or 2% galactose supplemented with 0.1% glucose. Nonfermentable medium was YPEG: 1% yeast extract, 2% bacto-peptone, 100 mg of adenine/liter, 3% ethanol, 3% glycerol. Minimal medium (0.67% yeast nitrogen base without amino acids) was supplemented with 2% glucose and specific amino acids, with uracil and adenine as required. Standard genetic methods were as previously described (15, 42). Yeast nuclear transformation was carried out using the one-step transformation method (8) for both library DNA and the plasmids YEp352 (23), pJM20 (33), pJM57 (31), or pNB107 (see below).
TABLE 1.
Strains used in this study
|
COX2 strains
|
COX2::ARG8m strains
|
||||
|---|---|---|---|---|---|
| COX2 allele | Name | Source or reference | COX2 alleleb | Name | Source or reference |
| Strains isogenic to D273-10Ba | |||||
| COX2 | NB80 | 6 | COX2::ARG8m | NB43 | 6 |
| COX2 | DL2 | 13 | COX2(5′UTL)::ARG8m | HMD22 | 6 |
| cox2-20 | NB58 | This study | cox2-20::ARG8m | NB120 | This study |
| cox2-21::ARG8m | NB121 | This study | |||
| cox2-22 | NB64 | This study | cox2-22::ARG8m | NB122 | This study |
| cox2-22S8a | NB64-S8a | This study | |||
| cox2-23 | NB100 | This study | cox2-23::ARG8m | NB123 | This study |
| cox2-24 | NB65 | This study | cox2-24::ARG8m | NB72 | This study |
| cox2-27 | NB117 | This study | cox2-27::ARG8m | NB127 | This study |
| cox2-28 | NB118 | This study | cox2-28::ARG8m | NB128 | This study |
| cox2-29 | NB119 | This study | cox2-29::ARG8m | NB129 | This study |
| cox2-35 | NB172 | This study | cox2-35::ARG8m | NB135 | This study |
| cox-36 | NB173 | This study | cox-36::ARG8m | NB136 | This study |
| cox2-37 | NB174 | This study | cox2-37::ARG8m | NB137 | This study |
| cox2-38 | NB175 | This study | cox2-38::ARG8m | NB138 | This study |
| cox2-39 | NB176 | This study | cox2-39::ARG8m | NB139 | This study |
| cox2-41 | NB177 | This study | cox2-41::ARG8m | NB179 | This study |
| cox2-43 | NB178 | This study | cox2-43::ARG8m | NB181 | This study |
| cox2-43R1 | NB178R1 | This study | |||
| cox2-60 | NB97 | 6 | cox2-60::ARG8m | NB54 | 6 |
| cox2-62 | NB40-3C | 6 | |||
| cox2-27-S1::ARG8m | NAB3 | This study | |||
| cox2-27-S2::ARG8m | NAB4 | This study | |||
| cox2-27-S3::ARG8m | NAB9 | This study | |||
| cox2-27-S4::ARG8m | NAB5 | This study | |||
| cox2-27-S6::ARG8m | NAB6 | This study | |||
| cox2-27-S7::ARG8m | NAB7 | This study | |||
| cox2-27-S12::ARG8m | NAB8 | This study | |||
| cox2-S1::ARG8m | NAB41 | This study | |||
| cox2-S2::ARG8m | NAB46 | This study | |||
| cox2-S3::ARG8m | NAB45 | This study | |||
| cox2-S4::ARG8m | NAB84 | This study | |||
| Strains isogenic to DBY947c | |||||
| COX2 | HMD21 | This study | |||
| cox2-22 | NB68 | This study | |||
| cox2-24::ARG8m | NB73 | This study | |||
| cox2-27 | NB140 | This study | |||
| cox2-27, S7 | NB140S7 | This study | |||
| cox2-27, S12 | NB140S12 | This study | |||
| cox2-27, S7, S12[rho−] | NB241 | This study | cox2-27, S7, S12::ARG8m[rho−] | NB242 | This study |
| cox2-60 | NB104 | 6 | cox2-60::ARG8m | NB66 | 6 |
All D273-10B-related strains are MATa lys2 leu2-3,112 ura3-52 his3ΔHindIII arg8::hisG and [rho+], with the exception of DL2 (MATa lys2).
All cox2::ARG8m alleles used in this study were based on the fusion of ARG8m after codon 91 of COX2, with the exception of the cox2(UTL)::ARG8m fusion in strain HMD22, which contains no COX2 codons.
All DBY947-related strains are MATα leu2Δ arg8ΔURA3 ura3-52 kar1-1 ade2-101. All are [rho+] except for NB241 and NB242, which are [rho−] as indicated.
Mutagenesis of the leader peptide coding sequence.
The COX2 plasmid pNB69 and the cox2::ARG8m plasmid pNB81, which also contains a marker fragment of COX3, were generated previously (6). They contain two unique sites surrounding the leader peptide coding sequence: PacI at −60 relative to the first COX2 codon and NsiI at +65. These sites were used to replace the wild-type PacI-NsiI fragment by mutant versions of the COX2 leader peptide sequence. Novel mutagenized fragments were created by a two-step PCR (24) using low-error-rate DeepVent polymerase (NE Biolabs, Inc.). We first cloned the cox2-22 PacI-NsiI fragment into the PacI and NsiI sites of both pNB69 (to yield pNB82) and pNB81 (to yield pNB90). We subsequently used pNB82 and pNB90 as backbones in which to insert the other mutagenized PacI-NsiI fragments since cox2-22 introduces a unique SnaBI site between the PacI and NsiI sites (see Fig. 5A), allowing improved cloning efficiency by digestion of the ligation reaction products with SnaBI. Alternatively, some novel mutations were generated by the megaprimer method (45) using Taq polymerase (Boehringer, Inc.). In this case, the template was pNAB1, which contained the COX2 gene in pBKSII+ (Stratagene, Inc.) as a HindIII-BamHI fragment. All final plasmid constructs were confirmed by DNA sequencing using the primer COX2A, which was complementary to COX2 positions −254 to −237 relative to AUG.
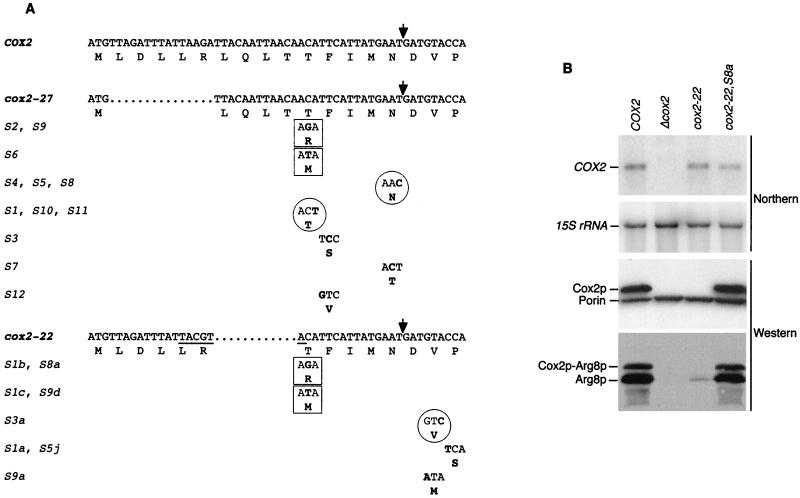
FIG. 5.
Analysis of intragenic suppressors of cox2-22 and cox2-27. (A) Nucleotide sequences of mtDNA from the cox2-22 and cox2-27 mutants, as well as the indicated pseudorevertants, were determined (see Materials and Methods). They are aligned with the sequence of the first 18 codons of the wild-type COX2 open reading frame. The names of the wild-type, mutant, and suppressor alleles are shown with their deduced amino acid sequences. Only the codons modified by the intragenic suppressor mutations are shown for those alleles. Deleted nucleotides are indicated by dots, and nucleotide and/or amino acid substitutions are in boldface. The SnaBI site created by the cox2-22 mutation is underlined. Identical suppressor mutations selected from both the cox2-22 and cox2-27 mutants are boxed. Nucleotide substitutions that do not change the encoded amino acid are circled. Multiple independent isolates of identical suppressor mutations are indicated by names on the same line. The arrow indicates the leader peptide cleavage site. (B) Relative steady-state levels of mRNAs and proteins in cox2-22 and a pseudorevertant strain. For the top two panels, the COX2 mRNA and 15S rRNA were detected on the same blot containing RNA from COX2 (NB80), cox2Δ (NB40-3C), cox2-22 (NB64), and cox2-22S8a (NB64S8A) strains by successive hybridization to COX2 and 15S probes (see Materials and Methods). In the third panel, 100 μg of total proteins from the same strains used in the Northern analysis were probed simultaneously with anti-Cox2p and anti-porin antibodies (both from Molecular Probes, Inc.), as indicated. For the bottom panel, the strains named above were crossed with the cox2-60::ARG8m strain NB66 and diploids were selected on minimal medium supplemented with leucine and arginine. Samples containing 50 μg of total proteins were prepared from mitochondrial recombinants and analyzed by Western blotting using anti-Arg8p antibody.
Mitochondrial transformation and genetic manipulations.
Plasmids carrying modified COX2 sequences were introduced into the [rho0] mitochondria of strain DFS160p0 (Table 1) by high-velocity microprojectile bombardment (7, 56). Mitochondrial synthetic [rho−] transformants were identified, and their altered COX2 sequences were inserted into [rho+] mtDNA by recombination and selection of haploid cytoductants as previously described (6). cox2 mutants were generated by crossing synthetic [rho−] transformants with either [rho+] cox2-60 (NB97) or cox2-62 (NB40-3C) strains, and cox2::ARG8m mutants were generated by crossing synthetic [rho−] transformants with a [rho+] cox2-60::ARG8m strain (NB54) or by recombination with [rho+] cox2 mutant strains. Haploid cytoductants were isolated unless otherwise indicated. In several cases, we also constructed the desired [rho+] strain by directly transforming [rho+] strains (6, 26) carrying the cox2-60 deletion (NB66 or NB104). This approach allows a quicker isolation of desired [rho+] strains, whose phenotypes were identical to those of corresponding mutants constructed by the two-step method. All mitochondrial mutations were verified by DNA sequencing (carried out by the Synthesis and Sequencing Facility in the Cornell Biotechnology Building) following integration into [rho+] mtDNA and PCR amplification from total DNA, as previously described (6).
Selection of suppressors and disruption of MRPL36.
Spontaneous revertants of independent subclones of cox2-22 strain NB64, cox2-27 strain NB140 and cox2-43 strain NB178, were selected on YPEG medium after 1- to 4-week incubations at 28 or 16°C. Revertant phenotypes were confirmed by streak purification. The COX2 upstream region of revertant mtDNA was sequenced as described above. Revertants of cox2-22 were genetically analyzed by two crosses. First, the spontaneous revertants were crossed to a nuclearly wild-type [rho0] strain. Second, [rho0] derivatives of the revertants were generated (15) and crossed with cox2-22 strain NB68. In both cases, the respiratory ability of the diploids was scored. Taken together, the results distinguish nuclear dominant, nuclear recessive, and mitochondrially inherited suppressor mutations.
Dosage-dependent nuclear suppressors of cox2-22 were isolated by transforming strain NB64 with an S. cerevisiae genomic library in the multicopy plasmid YEp13 (35). Roughly 6,000 Leu+ transformants were scored for growth on YPEG medium at 28°C. Eleven such transformants exhibited cosegregation of Leu+ and Pet+ phenotypes after growth under nonselective conditions. Plasmids from these transformants were analyzed by partial sequencing of the inserts by using a YEp13-specific primer. Nine overlapping inserts were from chromosome II (pNB104-type), and two overlapping inserts were from chromosome XIII (pNB105-type). Only the pNB104-type plasmids which were found to also suppress cox2-27 were further analyzed to identify the suppressing gene.
The MRPL36 gene was isolated by cloning the 1.2-kb XhoI-SacI fragment from pNB104/40 into the SalI and SacI sites of YEp352 (23) to yield pNB107. MRPL36 was largely deleted by replacing the 0.65-kb internal BglII-KpnI fragment of pNB107 with the BamHI-KpnI URA3-containing fragment from pNB108 (pNB108 is the 1.56-kb URA3-containing NsiI fragment from YEp352 that was blunt ended and cloned in a YEp351 SmaI site). The resulting plasmid, called pNB109, carried a deletion of the whole MRPL36 coding sequence but the first eight codons. The wild-type strain NB80 was transformed with the 2.1-kb gel-purified PstI-SacI fragment of pNB109, and Ura+ transformants were selected. The disruption was confirmed by PCR analysis and at the genetic level by analysis of the linkage between the deletion marker URA3 and LYS2, a gene tightly linked to MRPL36. Strains carrying mrpL36Δ::URA3 were found to be [rho−] or [rho0] by crossing to a nuclearly wild-type [rho0] strain.
Analysis of cellular RNA and proteins.
Total RNA was prepared and analyzed as previously described (11) except that the samples were supplemented with ethidium bromide before loading 10 μg in each lane. The COX2 probe was the 1.6-kb PacI fragment from pJM2 (32). The cox2::ARG8m probe was the 1.4-kb PacI-BamHI fragment from pNB81, which contained part of the COX2 gene (approximately from the start of the mature mRNA to codon 91) and the whole ARG8m coding sequence. 15S rRNA was probed with either the 2-kb BamHI fragment from plasmid pYJL25 (52) provided by O. Groudinsky or XhoI-linearized plasmid pT82 (47).
Total cellular protein was extracted (58) from cells grown in YPGalA medium to a mid-exponential phase. Fifty to 200 μg of total protein, as indicated, were separated on 10 to 12% polyacrylamide gels and probed with monoclonal anti-Cox2p antibodies (obtained from Molecular Probes Inc.; provided by G. Dujardin) diluted to 1/5,000, CCO6 (38) (obtained from T. Mason) diluted to 1/50, or polyclonal anti-Arg8p antibody (49) diluted to 1/1,000. The monoclonal anti-outer membrane porin antibody from Molecular Probes was provided by G. Dujardin and was diluted to 1/5,000. Secondary anti-mouse or anti-rabbit antibodies were detected using the Pierce Inc. chemiluminescent substrate.
In vivo pulse labeling with [35S]methionine in the presence of cycloheximide was performed as previously described (15), with the following modifications. Cells were grown initially in liquid 1% yeast extract–2% Bacto Peptone–2% raffinose and then transferred to synthetic complete medium lacking Met (0.67% yeast nitrogen base, 0.08% CSM-Met [Bio 101, Inc.], 2% raffinose). Cells were labeled with 5 μCi of [35S]methionine (1,175 Ci/mmol; NEN, Inc.) for 10 min and then chased with unlabeled 2.5 mM methionine for 10 min before chilling and freezing. Crude mitochondria were isolated after the conversion of cells to spheroplasts as previously described (58), except that disruption was by vortexing with glass beads.
RESULTS
Deletion of pre-Cox2p leader peptide coding sequence dramatically decreases translation.
We previously made 13- and 14-codon deletions of mtDNA specifying the pre-Cox2p leader peptide (alleles cox2-20 and cox2-21, respectively) to generate genes that would encode a leaderless Cox2p protein similar to that of animals. We observed that both mutations dramatically decreased Cox2p accumulation and prevented respiratory growth, without altering the level of COX2 mRNA (53). This could have been due to decreased stability of the protein, decreased translation of the mRNA, or both. To distinguish the effects of the leader peptide deletions on Cox2p synthesis from those on Cox2p function, we took advantage of a synthetic mitochondrial reporter gene, ARG8m, which specifies a soluble enzyme necessary for arginine biosynthesis in the mitochondrion (49) by fusing it to the COX2 reading frame (6). This cox2::ARG8m chimeric gene comprises the first 91 codons of COX2, fused to codon 2 of ARG8m. When inserted precisely in place of the native COX2 gene in the mitochondrial genome, cox2::ARG8m complements the Arg− growth phenotype of a nuclear arg8 mutation and directs the synthesis of a chimeric fusion protein. However, since the ARG8m sequence used here specifies the cleavage site for removal of the matrix targeting signal of pre-Arg8p, this chimeric Cox2p-Arg8p fusion protein is largely processed to yield mature Arg8p (6). Thus, translation of the chimeric cox2::ARG8m mRNA can be assayed by scoring for Arg+ growth independently of functional constraints on the structure of Cox2p.
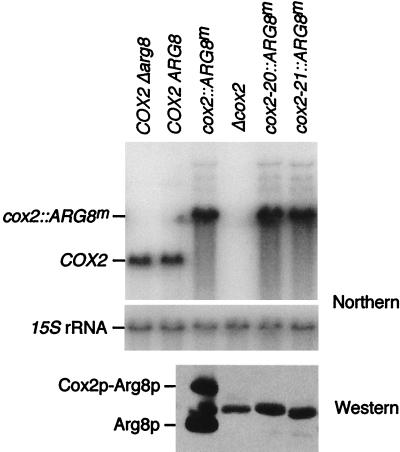
Both of the 13- and 14-codon deletion mutations (cox2-20 and cox2-21, respectively) were inserted into this cox2::ARG8m reporter gene and placed into the mitochondrial genome by transformation and homologous recombination (see Materials and Methods). Neither the cox2-20::ARG8m nor cox2-21::ARG8m construct complemented a nuclear arg8 mutation at the level of growth or directed synthesis of Cox2p-Arg8p, as measured by pulse labeling (Fig. 1; data not shown). Furthermore, neither accumulated detectable steady-state levels of reporter protein (Fig. 2). The lack of Arg8p synthesis was not due to an mRNA synthesis or stability defect, since the chimeric mRNA steady-state level was similar in the mutant and wild-type strains (Fig. 2).
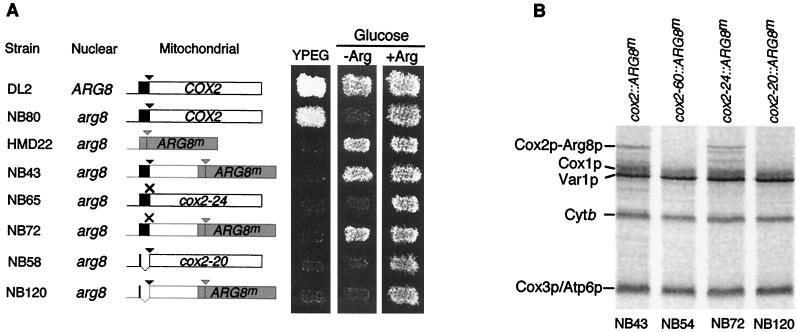
FIG. 1.
The cox2-20 mutation prevents expression of the ARG8m reporter fused to COX2 codon 91. (A) The COX2 leader peptide coding region is in black, other COX2 codons are in white, and the COX2 mRNA 5′-UTL is indicated by the thin line. ARG8mcodons are in grey. Triangles, processing sites of the pre-Cox2p leader peptide (black) and the pre-Arg8p matrix targeting signal (grey); ×, the mutated pre-Cox2p cleavage site of the cox2-24 mutants, in which COX2 codons 15 and 16 were changed from AAT GAT (ND) to AAG CTT (KT) (creating an HindIII site). Strains were patched on complete glucose medium, replica plated to nonfermentable medium (YPEG) and minimal medium lacking arginine (−Arg) or containing arginine (+Arg), and incubated for 2 days at 28°C. In descending order, the strains used were DL2, NB80, HMD22 (this strain contains the only chimeric gene used here entirely lacking COX2 codons), NB43, NB65, NB72, NB58, and NB120 (Table 1). (B) Mitochondrial translation products were radioactively pulse labeled for 10 min in cells treated with cycloheximide (see Materials and Methods). Protein aliquots normalized for radioactivity were analyzed by electrophoresis on sodium dodecyl sulfate–15% polyacrylamide gels and subsequently by phosphorimaging. The positions of the Cox2p-Arg8p fusion protein and products of endogenous mitochondrial genes are indicated. The fusion protein coded by cox2-24::ARG8m migrated slightly slower than the cox2::ARG8m fusion protein on this gel and, more dramatically, on others (not shown) due to the lack of leader peptide processing, and it appears to have yielded novel degradation products. Mature Arg8p comigrates with the upper part of the Cox1p band and is difficult to detect after pulse labeling. Strains were cox2::ARG8m (NB43), cox2-60::ARG8m (NB54; this strain lacks COX2 nucleotides −63 to +66), cox2-24::ARG8m (NB72), and cox2-20::ARG8m (NB120).
FIG. 2.
Deletion of the leader peptide coding region has no effect on cox2::ARG8m mRNA levels but prevents accumulation of reporter protein Arg8p. Northern blot analysis (top two panels) was carried out on total cellular RNAs from the indicated strains: COX2 Δarg8 (NB80), COX2 ARG8 (DL2), cox2::ARG8m (NB43), Δcox2 (NB40-3C), cox2-20::ARG8m (NB120), and cox2-21::ARG8m (NB121) (all strains, except DL2, contain a nuclear arg8::hisG mutation). The COX2 mRNA (0.85 kb) and cox2::ARG8m mRNA (1.7 kb) were detected simultaneously with the COX2 and cox2::ARG8m probes (see Materials and Methods), and 15S rRNA hybridization served as a loading control. Western blot analysis was carried out with 100 μg of total protein from the indicated strains (as above) by using anti-Arg8p antibody (see Materials and Methods). Both the mature Arg8p protein (lower band) and the unprocessed Cox2p-Arg8p fusion protein (upper band) are indicated. The middle band corresponds to a cross-reacting protein present in strains lacking a functional ARG8 gene. For unknown reasons, the strength of this signal is variable (Fig. 5 and 9).
It was unlikely that the leader peptide deletion mutants were lacking the Arg8p reporter protein due to degradation of the chimeric protein, since cleavage of the pre-Arg8p matrix targeting sequence should release mature, wild-type Arg8p from the fusion protein, as observed previously (6). Furthermore, we experimentally verified that a mutation causing instability of Cox2p did not prevent the accumulation of Arg8p encoded by a chimeric gene. A mutation that altered the pre-Cox2p leader peptide processing site from ND to KT (cox2-24) in an otherwise wild-type COX2 gene caused a severe respiratory defect and greatly reduced the level of Cox2p (Fig. 1 and 3C). However, when this mutation was inserted into the cox2::ARG8m reporter gene, the resulting strain was fully Arg+, synthesized pulse-labeled Cox2p-Arg8p (Fig. 1), and contained a normal steady-state level of the processed Arg8p reporter protein (not shown). Thus, the defect in pre-Cox2p processing destabilized Cox2p but had no effect on Arg8p translation, confirming that our cox2::ARG8m reporter system can discriminate between gene expression defects caused by Cox2p instability and those caused by reduced translation.
FIG. 3.
Effect of mutations within the leader peptide coding region on expression of COX2 and the cox2::ARG8m reporter gene. (A) The sequence of the first 18 codons of the wild-type COX2 open reading frame and the amino acids they specify are shown for reference. The arrow indicates the leader peptide cleavage site. Strains carrying the indicated alleles affecting the COX2 leader peptide coding region in either COX2 or cox2::ARG8m were patched on complete glucose medium and replica plated to nonfermentable medium (YPEG) for alleles in COX2 and to minimal medium lacking arginine (−Arg) or containing arginine (+Arg) for alleles in cox2::ARG8m. Incubation was for 2 days at 28°C. Periods indicate deleted residues, and the boxed “R” in bold indicates a mutation of codon 6 from AGA (R) to CGT (R). The arrow marks the leader peptide cleavage site. The indicated alleles of COX2 and cox2::ARG8m, respectively, correspond to the following strains: COX2 (NB80 and NB43), cox2-27 (NB117 and NB127), cox2-23 (NB100 and NB123), cox2-28 (NB118 and NB128), cox2-35 (NB172 and NB135), cox2-22 (NB64 and NB122), cox2-29 (NB119 and NB129), and cox2-20 (NB58 and NB120). (B) Northern blot analysis was used to detect the COX2 mRNA and 15S rRNA in total RNA (see Materials and Methods) prepared from strains with the following alleles: COX2 (NB80), Δcox2 (NB40-3C), cox2-22 (NB64), and cox2-27 (NB117). (C). Western blot analysis probing with the anti-Cox2p monoclonal antibody CCO6 (38) was used to detect Cox2p in 100 μg of total protein (see Materials and Methods) from strains with the following alleles: cox2-22 (NB64), cox2-24 (NB65), COX2 (HMD21), Δcox2 (NB73), and cox2-27 (NB140). A faint band of slightly lower mobility than the wild type, corresponding to unprocessed pre-Cox2p, was detected in the cox2-24 lane after overexposure (not shown).
Taken together, these findings demonstrate that deletion of the pre-Cox2p leader peptide coding sequence from cox2::ARG8m (cox2-20::ARG8m and cox2-21::ARG8m) produces mRNAs that are poorly if at all translated. This result is very surprising since in the complete absence of COX2 codons [the cox2(UTL)::ARG8m allele of strain HMD22], ARG8m is efficiently expressed from the COX2 locus (Fig. 1). Thus, the presence of COX2 codons 15 to 91 in the chimeric mRNA specified by the cox2-20::ARG8m allele apparently blocks translation of the downstream ARG8m sequence. On the other hand, fusion of the COX2 codons 1 to 16 to a variant of ARG8m lacking its matrix targeting signal did not prevent the expression of the reporter (21). Taken together, these results suggest the existence of an element in the sequence of codons 15 to 91 that inhibits translation and a compensating positively acting element within the first 14 codons that specify the leader peptide.
Mutations within the leader peptide coding region that reduce downstream translation.
To define more precisely the leader peptide coding sequences necessary for efficient translation of the COX2 and cox2::ARG8m mRNAs, we constructed a set of mutants harboring small deletions and/or point mutations. In all cases, the initiation codon and the codons for residues 15 and 16 surrounding the leader peptide cleavage site (ND) were left unchanged. First, three small deletions spanning the remaining 13 codons were created: deletion of codons 2 to 6 (residues LDLLR, allele cox2-27), deletion of codons 7 to 10 (residues LQLT, allele cox2-23), and deletion of codons 11 to 14 (residues TFIM, allele cox2-28). None of these deletions had a significant effect on the steady-state levels of either the COX2 or cox2::ARG8m mRNAs (Fig. 3B and data not shown), and neither the codons 7-to-10 nor the codons 11-to-14 deletions detectably affected either respiratory growth or arginine prototrophy (Fig. 3A). In contrast, the codons 2-to-6 deletion, cox2-27, caused decreased respiratory growth at 28°C, prevented arginine prototrophy when inserted into the reporter construct (Fig. 3A), and reduced the steady-state level of Cox2p (Fig. 3C). Both of the growth phenotypes were enhanced when the incubation temperature was lowered to 16°C and were partially suppressed by increasing the temperature to 36°C (not shown). We also deleted codons 7 to 14 in a single mutation (cox2-35) (Fig. 3A). This eight-codon deletion mutation allowed nearly wild-type growth on both nonfermentable medium and minimal medium lacking arginine. These results demonstrate that codons 2 to 6 are most important for allowing the translation of downstream sequences. However, since deletion of these codons does not produce a phenotype as severe as the 13-codon deletion of cox2-20, codons 7 to 14 also contribute to the action of the putative positive element. In addition, these results demonstrate that the leader peptide can be shortened considerably without destroying pre-Cox2p function.
The cox2-22 mutation is a compound allele with both a deletion of codons 7 to 10 (cox2-23) and the translationally silent change of codon 6 from AGA to CGT (cox2-29), both of which specify R (48). Neither cox2-29 alone nor cox2-23 alone affected respiratory growth or arginine prototrophy (Fig. 3A). However, the combination of the two alterations in cox2-22 produced phenotypes similar to that of the codons 2-to-6 deletion, cox2-27, leaky respiratory and Arg+ growth at 28°C (Fig. 3A) and reduced Cox2p levels (Fig. 3C). Both phenotypes were enhanced by growth at a lower temperature and suppressed by growth at higher temperatures (not shown), as in the case of cox2-27. The similarity of the cox2-22 and cox2-27 phenotypes suggests that both mutations could affect COX2 translation by the same mechanism. Interestingly, since the only difference between the phenotypically silent codons 7-to-10 deletion (cox2-23) and cox2-22 was the translationally silent AGA-to-CGT alteration, these results suggest that the mRNA sequence could be important for the function of the putative positive element, independently of the leader peptide amino acid sequence it encodes.
Suppression of mutations within the leader peptide coding region by elevated dosage of MRPL36 and PET111.
To further understand the gene expression defects caused by the cox2-22 and cox2-27 mutations, we screened for nuclear genes that, when present in a high copy number, would suppress them. The cox2-22 strain NB64 was transformed with a 2μm vector genomic library, and plasmids from transformants with increased growth on nonfermentable medium were isolated (see Materials and Methods). We further analyzed those plasmids which suppressed both cox2-22 and cox2-27. These plasmids contained a gene encoding MrpL36p, which had been previously found to copurify with the large subunit of yeast mitochondrial ribosomes (28). A subclone containing only MRPL36 retained suppressor activity for cox2-22 and cox2-27, as well as cox2-22::ARG8m and cox2-27::ARG8m (Fig. 4). Since the phenotype caused by the loss of MrpL36p had not been determined, we deleted the gene (see Materials and Methods). Cells bearing the mrpL36Δ::URA3 allele failed to respire and became [rho−] and/or [rho0]. This phenotype is typical of mutations that block all mitochondrial translation (34) and is consistent with the idea that MrpL36p could play a role in translation elongation as part of the large ribosomal subunit.
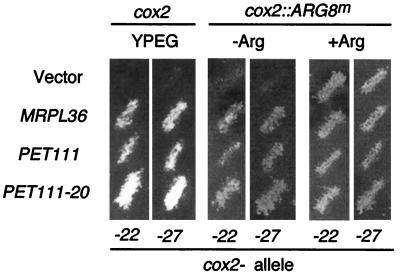
FIG. 4.
Suppression of the cox2-22 and cox2-27 mutations in transformants containing multiple copies of the nuclear genes MRPL36 and PET111. Strains carrying the cox2-22 or cox2-27 alleles (NB64 or NB117) as well as the cox2-22::ARG8m or cox2-27::ARG8m alleles (NB122 or NB127) were transformed with the empty vector (YEp352) and plasmids carrying MRPL36 (pNB107), PET111 (pJM20), or PET111-20 (pJM57), as was indicated (see Materials and Methods). Transformants were grown as patches on minimal medium containing arginine (+Arg) and then were replica plated to nonfermentable medium (YPEG) and minimal medium lacking arginine (−Arg) and incubated at 28°C for 2 or 4 days, respectively.
We also tested the ability of the COX2 mRNA-specific translational activator gene, PET111, to suppress the mutations in the leader peptide coding region. We found that elevated dosage of the wild-type PET111 gene (39), as well as a dominant allele that appears to have increased activity, PET111-20 (31), suppressed cox2-22, cox2-27, cox2-22::ARG8m, and cox2-27::ARG8m (Fig. 4). Taken together, these results are consistent with the idea that the cox2-22 and cox2-27 mutations cause similar translational defects that can be suppressed by improving the translation efficiency either at the level of initiation (PET111) or elongation (MRPL36).
Some intragenic suppressors of cox2-22 and cox2-27 cause translationally silent alterations in mRNA sequence that increase translation.
Both the cox2-22 (NB64) and cox2-27 (NB140) mutant strains yielded spontaneous pseudorevertants exhibiting improved respiratory growth. Genetic analysis of 55 cox2-22 pseudorevertants revealed that the slowest growing 23 contained as-yet-uncharacterized dominant nuclear suppressor mutations, while the most robust 32 contained mitochondrial mutations (see Materials and Methods). Eight of the independently isolated mitochondrial pseudorevertants were further analyzed by DNA sequence analysis of the 500-bp region of mtDNA surrounding their COX2 initiation codons. In parallel, 12 robust independent pseudorevertants of the cox2-27 mutant were also analyzed by sequencing the same region of mtDNA. In each case, a single base substitution was identified in the coding region specifying the leader peptide or the first three amino acids of mature Cox2p (Fig. 5A). These intragenic suppressor mutations fell into three categories: (i) missense substitutions obtained only as suppressors of either cox2-22 or cox2-27, (ii) missense substitutions obtained as suppressors of both original mutations (boxed in Fig. 5A), and (iii) base pair substitutions that do not alter the mutant pre-Cox2p amino acid sequence, obtained only as suppressors of either cox2-22 or cox2-27 (circled in Fig. 5A).
Northern analysis of the pseudorevertant carrying the cox2-22, S8a allele, whose intragenic suppressor is identical to the cox2-27 suppressors S2 and S9, revealed no change in the steady-state level of COX2 mRNA relative to that of the wild type (Fig. 5B). A similar result was obtained for the translationally silent cox2-22 suppressor S3a (not shown). These results support the idea that suppression occurs at the translational level.
Western blot analyses carried out on the original pseudorevertants showed that Cox2p accumulation was restored to wild-type or nearly wild-type levels in cox2-22,S8a and other pseudorevertants of both cox2-22 and cox2-27 (Fig. 5B; data not shown). To monitor the effect of the suppressor mutations on reporter protein levels, cox2-22 was inserted with several of its intragenic suppressors into the cox2::ARG8m gene. Pseudorevertants were crossed with a strain (NB66) that harbors the cox2-60::ARG8m allele (6), a 130-bp deletion in the fusion construct that includes the leader peptide coding region. Reporter protein accumulation in the resulting diploid recombinants closely mirrored the strength of the original haploid suppressors on respiratory medium for cox2-22,S8a (Fig. 5B) and all other cases tested (data not shown). In addition, reporter protein accumulation was similarly increased over that directed by the cox2-27::ARG8m allele in recombinant haploid strains derived from the cox2-27 pseudorevertants by cytoduction (not shown).
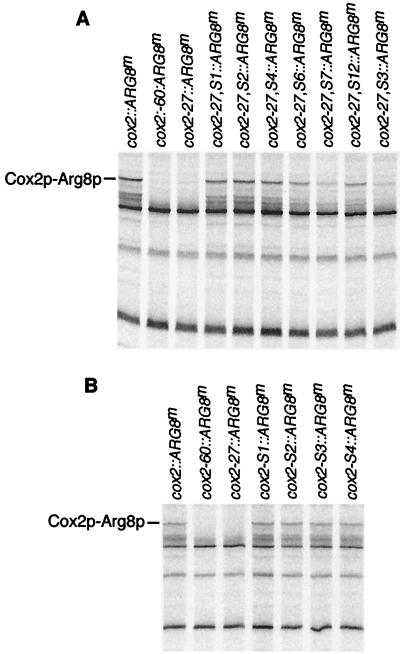
To confirm that the leader peptide mutations and their intragenic suppressors affected translation rates, we monitored relative rates of protein synthesis by pulse labeling cells in the presence of the cytoplasmic translation inhibitor cycloheximide and detecting mitochondrial translation products by autoradiography. The results confirmed that the rate of Arg8p synthesis directed by the mutant cox2-27::ARG8m mRNA was much lower than that of the wild-type reporter mRNA (Fig. 6A). The rate of Arg8p synthesis was restored to various extents in the pseudorevertants (Fig. 6A), largely consistent with the relative steady-state levels of Arg8p observed with the same strains (not shown).
FIG. 6.
Arg8p synthesis is blocked by cox2-27, it was restored in its pseudorevertants, and it was unaffected by intragenic suppressor mutations in an otherwise wild-type context. Mitochondrial translation products were radioactively pulse labeled and analyzed as described for Fig. 1B. The position of mature Arg8p is indicated. (A) Arg8p synthesis in the cox2-27 mutant and pseudorevertants cox2::ARG8m (NB43), cox2-60::ARG8m (NB54), cox2-27::ARG8m (NB127), cox2-27,S1::ARG8m (NAB3), cox2-27,S2::ARG8m (NAB4), cox2-27,S4::ARG8m (NAB5), cox2-27,S6::ARG8m (NAB6), cox2-27,S7::ARG8m (NAB7), cox2-27,S12::ARG8m (NAB8), and cox2-27,S3::ARG8m (NAB9). (B) Arg8p synthesis in strains bearing only intragenic suppressor mutations: cox2::ARG8m (NB43), cox2-60::ARG8m (NB54), cox2-27::ARG8m (NB127), cox2-S1::ARG8m (NAB41), cox2-S2::ARG8m (NAB46), cox2-S3::ARG8m (NAB45), and cox2-S4::ARG8m (NAB84).
To determine whether intragenic suppressor mutations would affect gene expression when placed in a normal context, we constructed haploid strains containing four suppressors of cox2-27 in otherwise wild-type COX2 and cox2::ARG8m genes. In no case did we observe a significant difference from the wild-type rate of synthesis of Arg8p (Fig. 6B) or Cox2p (not shown). Growth phenotypes and steady state levels of Arg8p and Cox2p were also wild type in strains bearing only the suppressor mutations (not shown).
The analysis of translation in the cox2-22 and cox2-27 mutants and their pseudorevertants suggests that both original mutations have similar negative effects on the rate of mRNA translation that are alleviated by intragenic suppressor mutations, some of which work on both mutations. In addition, they argue further that the nucleotide sequence of the mRNA encoding the leader peptide, rather than the peptide sequence itself, strongly influences the rates of COX2 translation.
Evidence that a stem structure in the COX2 mRNA coding region plays a role in regulating translation.
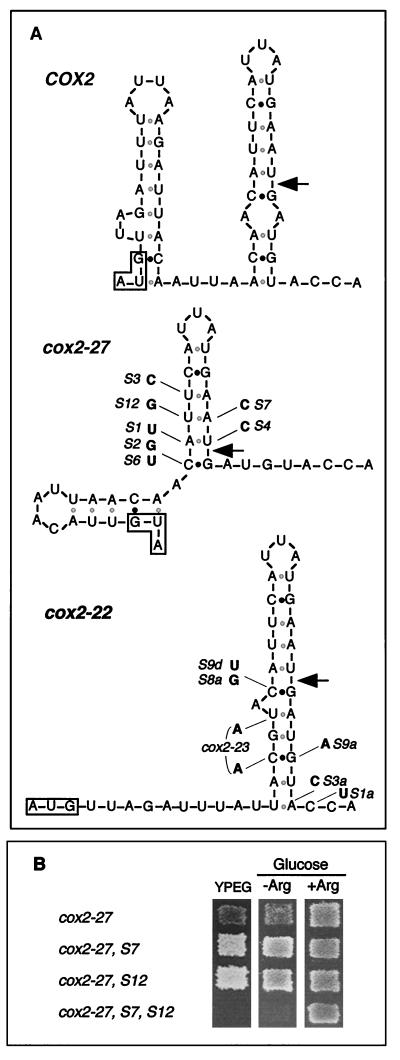
Wild-type COX2 mRNA is predicted to form a relatively stable stem-loop structure extending from codon 10 through the first two bases of codon 17 (Fig. 7A). Similar stems identical to the top part of the wild-type structure are also predicted for the cox2-27 mRNA, which lacks codons 2 to 6, and the compound allele cox2-22 (Fig. 7A). Interestingly, all of the cox2-27 intragenic suppressors and all but one of the cox2-22 intragenic suppressors (Fig. 5) affect base pairs in these predicted stems and would weaken them (Fig. 7A).
FIG. 7.
Evidence for a stem-loop structure in the leader peptide coding region that affects translation. (A) Predicted secondary structure of the 18 first codons of the wild-type COX2 mRNA and the corresponding codons of the cox2-27 and cox2-22 alleles. Structures were generated for 28°C by using the program mfold 3.0 (http://mfold2.wustl.edu/∼mfold/rna/form1-2.3.cgi) with default parameters (29, 59). Similar structures of this region were obtained when folding extended mRNA fragments including the 18 first codons. The initiation codon in each sequence is boxed. Arrows indicate the pre-Cox2p cleavage site position in the encoded polypeptide. Black dots indicate G:C base pairs and grey dots indicate weaker A:U or G:U base pairs. The nucleotide substitutions found in the various cox2-27 and cox2-22 pseudorevertants (Fig. 5) are marked. The cox2-23 allele is also shown on the cox2-22 structure since it can be viewed as an intragenic suppressor of cox2-22 with two substitutions. (B) Phenotypic effects of mutations in the stem structure of the cox2-27 mRNAs. Diploid recombinant cells carrying the indicated alleles were replicated on minimal SD medium lacking arginine (−Arg) and nonfermentable medium (YPEG) and were incubated at 28°C for 2 and 3 days, respectively. The diploid cells were selected for complementing nutritional markers on minimal SD medium containing arginine after mating strains containing the following alleles with the cox2-60::ARG8m strain NB54 (lacking nucleotides −63 to +66): cox2-27, NB140; cox2-27, S7, NB140S7; cox2-27, S12, NB140S12; cox2-27, S7, S12, NB241 for growth on YPEG medium and NB242 for growth on medium lacking arginine.
To test the possible significance of the stem, we constructed triple mutants that restored potential base pairing downstream of the cox2-27 mutation by compensating for several suppressor substitutions. We then examined the effects of the compensating mutations on the expression of the cox2::ARG8m reporter and COX2. For example, a single predicted U:A base pair was disrupted by suppressors S12 (U to G) and S7 (A to C), each of which improved the expression of the cox2-27::ARG8m reporter (Fig. 7). However, when S12 and S7 were placed together in the same mRNA, generating a stronger predicted G:C base pair in the stem, reporter expression and respiratory growth were greatly decreased, to a level below that of cox2-27 alone (Fig. 7B). We observed similar decreases in reporter expression when we tested mutations that restored pairing to the predicted base pair disrupted by both cox2-27 suppressors S1 and S4 (not shown). Disruption of this predicted base pair by combining the S1- and S4-compensating mutations in one mRNA increased reporter expression, as expected (not shown). Similarly, the addition of the S6 suppressor substitution to an mRNA containing S4 and the S4-compensating mutation also increased reporter expression, as expected (not shown). These data strongly support the in vivo existence of the predicted stem and indicate that its stability plays a role in controlling COX2 mRNA translation.
Nucleotide sequence in the mRNA encoding the leader peptide is critical for COX2 translation, while the leader peptide amino acid sequence is not highly constrained.
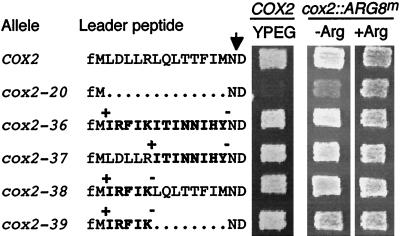
Several results described above suggest that elements in the leader peptide coding region, critical for translation in the context of downstream COX2 codons, are defined at the level of nucleotide sequence rather than amino acid sequence. To investigate this possibility further, we shifted the reading frame in this region to +1 by inserting a single nucleotide at upstream positions and deleting a single nucleotide at downstream positions in both the complete leader peptide coding region and a functional construct with deleted codons 7 to 14 (cox2-35) (Fig. 8). These frameshifts caused wholesale changes in the sequence of the leader peptide, while minimally modifying the mRNA sequence. Surprisingly, all of these frameshift mutations, including cox2-39 (which lacks codons 7 to 14), behaved like the wild type for both respiratory growth and Arg+ prototrophy when placed in the COX2 and cox2::ARG8m genes, respectively (Fig. 8).
FIG. 8.
Shifting the reading frame to +1 in the leader peptide coding region has no effect on respiratory growth or arginine prototrophy. Control strains and strains carrying the indicated frameshift alleles in the COX2 leader peptide coding region were patched on complete glucose medium and replica plated on nonfermentable medium (YPEG) for alleles inserted in the plain COX2 gene and on minimal medium lacking arginine (−Arg) or containing arginine (+Arg) for alleles inserted in the reporter construct. Photographs were taken after a 2-day incubation at 28°C. +, addition of an A nucleotide; for cox2-36, cox2-38, and cox2-39, this is immediately following the initiation codon, and for cox2-37, it is immediately following the sixth codon. −, deletion of the G nucleotide immediately before wild-type codon 15 in cox2-36 and cox2-37 and of the A nucleotide immediately before wild-type codon 7 in cox2-38 and cox2-39. Deleted amino acids are shown by dots, and modified amino acids are in bold letters. The arrow marks the leader peptide cleavage site. Alleles and strains (for COX2 and for cox2::ARG8m, respectively) were as follows: COX2 (NB80 and NB43), cox2-20 (NB58 and NB120), cox2-36 (NB173 and NB136), cox2-37 (NB174 and NB137), cox2-38 (NB175 and NB138), and cox2-39 (NB176 and NB139).
We conclude that the RNA sequence is clearly more important for translation in this context than the leader peptide amino acid sequence. Furthermore, membrane insertion, export of N-tail and C-tail domains and assembly of cytochrome oxidase can all be achieved with a pre-Cox2p leader peptide sequence that is very different from that of the wild type. However, it is important to note that in this context, the charge and hydrophobicity characteristics of the leader peptides generated by these +1 frameshifts are similar to those of the wild type. Thus, one could not exclude functional constraints on the leader peptide amino acid sequence on the basis of these data.
If mRNA sequence in the leader peptide region plays a greater functional role than the amino acid sequence, then multiple synonymous nucleotide changes in the region should affect gene expression. We made six such silent mutations in the first six codons of the functional construct with codons 7 to 14 (cox2-35) deleted to generate the cox2-41 mutation (Fig. 9). In contrast to the corresponding frameshift mutation described above (cox2-39), cox2-41 abolished both respiratory growth and growth in the absence of arginine when placed in the COX2 and cox2::ARG8m genes, respectively (Fig. 9). This mutation had no effect on mRNA levels (Fig. 9B), but prevented accumulation of their protein products (Fig. 9C). Thus, changes in the cox2-41 mRNA sequence abolished translation despite the fact that this allele specifies a functional leader peptide amino acid sequence.
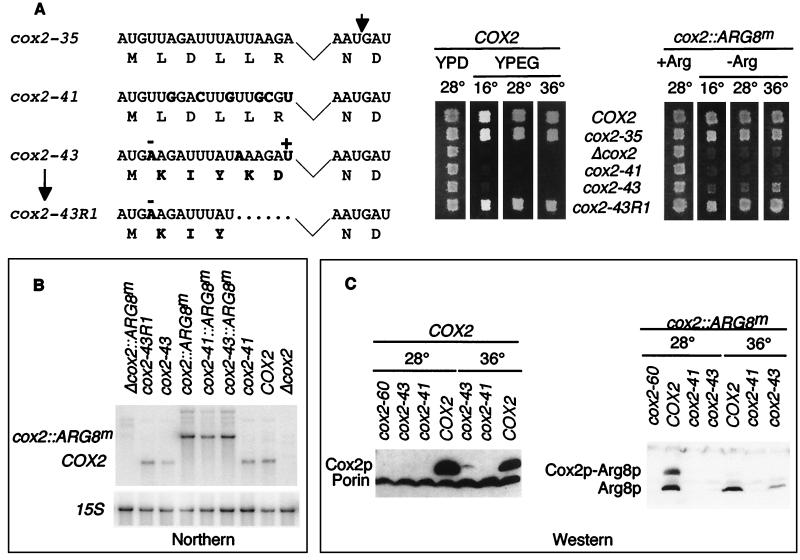
FIG. 9.
Effects of silent substitutions and shifting the reading frame to −1 in the first six codons of a leader peptide coding region lacking codons 7 to 14. (A) Allele names, their nucleotide sequences and predicted amino acid sequences are shown on the left. All are derived from the functional allele cox2-35, which carries a deletion of codons 7 to 14 indicated by the kinked line (Fig. 3). Altered nucleotides and amino acids are shown in bold letters. −, deletion of two T nucleotides and addition of an A nucleotide in the DNA sequence; +, addition of a T nucleotide in cox2-43. The two codons further deleted in cox2-43R1 are indicated by dots. The long arrow indicates the origin of cox43R1 as a spontaneous pseudorevertant of cox2-43. The short arrow indicates the leader peptide cleavage site. On the right, strains carrying the indicated versions of the COX2 leader peptide coding region were patched on complete glucose medium and replica plated on nonfermentable medium (YPEG) or glucose medium (YPD) for alleles inserted in the COX2 gene (left panel) and on minimal medium lacking arginine (−Arg) or containing arginine (+Arg) for alleles inserted in the cox2::ARG8m construct (right panel). Plates were incubated at 16, 28, or 36°C as indicated and were photographed after 1- to 10-day incubations, depending on the medium and temperature. Strains were (for COX2 and for cox2::ARG8m, respectively) COX2, NB80 and NB43; cox2-35, NB172 and NB135; Δcox2, NB97 and NB54; cox2-41, NB177 and NB179; cox2-43, NB178 and NB181; cox2-43R1, NB178R1 and a diploid recombinant from the cross of NB178R1 × NB66. (B) Relative steady-state levels of mutant mRNAs were determined by Northern blot analysis of total RNAs from the indicated strains (as for panel A), which were hybridized simultaneously to the COX2 and cox2::ARG8m probes and were reprobed with the 15S rRNA probe (see Materials and Methods). (C) Relative steady-state levels of Cox2p and Arg8p in cells grown at 28 or 36°C were determined by Western blot analysis. Samples with 200 μg of total proteins from the indicated strains (as in panel A), which were grown at the indicated temperatures, were probed with either anti-Arg8p or anti-Cox2p and anti-porin antibodies (all from Molecular Probes, Inc.) as indicated. The COX2 panel was overexposed to reveal low levels of Cox2p in the cox2-43 mutant grown at 36°C.
The leader peptide amino acid sequence appears to have a role in cytochrome oxidase assembly that is at least partially distinct from the translational role of the mRNA encoding it.
We were unable to simply shift the leader peptide reading frame to −1 due to the occurrence of stop codons in that frame. However, we constructed a compound allele termed cox2-43, which was derived from the functional construct with a deletion of codons 7 to 14 (cox2-35), that was translated in the −1 frame: an upstream nucleotide was deleted, a downstream nucleotide was inserted, and two nucleotide substitutions were made to eliminate the resulting stop codons (Fig. 9A). These changes significantly altered the mRNA sequence and dramatically changed the character of the amino acid sequence.
This compound allele, cox2-43, had little or no effect on the level of COX2 mRNA (Fig. 9B), but prevented detectable Cox2p accumulation in cells grown at 28°C (Fig. 9C). In cox2-43 cells grown at 36°C, there was a low but detectable accumulation of Cox2p, possibly in its precursor form (Fig. 9C), demonstrating that this mRNA is partially translatable. However, this mutation abolished respiratory growth at all three temperatures tested (Fig. 9A), suggesting that the mutant form of pre-Cox2p made in the cox2-43 mutant at 36°C is not functional. Interestingly, a strain bearing the cox2-43::ARG8m construct exhibited weak but significant growth on medium lacking arginine and contained easily detectable levels of Arg8p when grown at 36°C, confirming that this leader peptide mRNA sequence was partially competent in supporting translation at that temperature (Fig. 9A and C). Furthermore, the presence of PET111-20 on a high-copy-number plasmid greatly improved the growth of a cox2-43::ARG8m strain in the absence of arginine at 36°C, but had no effect on the respiratory deficiency of a cox2-43 strain (not shown). In this context, it is important to note that it was previously found that low levels of Cox2p synthesis can be sufficient for detectable respiratory growth (31), while comparably low levels of Arg8p synthesis are insufficient for Arg+ growth (6).
Of all the mutations constructed in this study, cox2-43 is the only one that is competent for translation, albeit only at an elevated temperature, but encodes a nonfunctional form of pre-Cox2p. Thus, while the pre-Cox2p leader peptide amino acid sequence can be altered substantially without destroying function, it nevertheless appears to play a role in assembly of cytochrome oxidase at a step downstream of translation, and some amino acid sequences are incompatible with this role, at least at 36°C.
The cox2-43 mutant strain yielded spontaneous respiring pseudorevertants. We determined the mtDNA sequence in this region for three independent pseudorevertants that exhibited strong respiratory growth at all temperatures. All three contained identical deletions of the two codons immediately preceding the leader peptide cleavage site, as shown for cox2-43R1 (Fig. 9A). The cox2-43R1 allele supported wild-type respiratory growth (Fig. 9A) and wild-type levels of Cox2p (not shown). A strain bearing the cox2-43R1::ARG8m reporter construct grew well on medium lacking arginine at all temperatures (Fig. 9A). Thus, this two-codon deletion generated both an mRNA sequence exhibiting improved translation relative to cox2-43 and a leader peptide amino acid sequence that supports cytochrome oxidase assembly.
DISCUSSION
Leader peptides on precursors of proteins inserted into membranes, or translocated through them, consist of amino acids sequences that generally play a role in protein targeting. However, the nucleotides encoding them are not typically involved in the regulation of precursor synthesis. In this study, we explored genetically the function of the 15-amino-acid pre-Cox2p leader peptide and the mRNA encoding it, specified by S. cerevisiae mtDNA. We examined the effects of mutations on the synthesis and function of pre-Cox2p and on the synthesis of a reporter protein, Arg8p, whose coding sequence was translationally fused to COX2 codon 91. Our data clearly demonstrate that the translation of the COX2 mRNA depends upon the mRNA sequence of codons specifying the pre-Cox2p leader peptide, in addition to its previously known dependence upon mRNA-specific translational activation by Pet111p through the 5′-UTL (17, 31).
First, we found that deletion of 13 leader peptide codons (2 to 14) from the cox2::ARG8m reporter mRNA prevented synthesis of the reporter protein despite normal mRNA levels. However, ARG8m was expressed from the COX2 locus in the complete absence of any COX2 codons. These findings strongly suggest that the first 91 codons of the pre-Cox2p coding sequence contain antagonistic elements that control translation positively and negatively: the positively acting element includes sequences in the first 14 codons specifying the leader peptide, while the negatively acting element appears to be within codons 15 to 91.
Our further analysis of mutations within the leader peptide coding sequence indicates that the positively acting element is complex or multipartite. None of the smaller deletion mutations we made in this region produced phenotypes as strongly negative as the deletion of codons 2 to 14 (cox2-20). Deletion of codons 2 to 6 (cox2-27) caused both leaky respiratory and leaky Arg− phenotypes without affecting mRNA levels, demonstrating the importance of this region for translation. However, residual growth of the cox2-27 and cox2-27::ARG8m strains indicates that the leader peptide codons remaining in this mutation, codons 7 to 14, also have a positive effect on translation in the absence of codons 2 to 6.
The positively acting element embedded in the leader peptide coding sequence functions at the level of nucleotide sequence, not amino acid sequence, since a shift of the reading frame through this region left the mRNA sequence relatively untouched while changing the amino acid sequence and did not detectably affect translation. On the other hand, when we altered the mRNA sequence of codons 2 to 6 (in the absence of codons 7 to 13) without changing the amino acid sequence they encoded (cox2-41), we completely eliminated translation. In this connection, it is noteworthy that the first six codons contain an 11-base sequence (AGAUUUAUUAA) that is also present one base upstream of the initiation codon in the COX2 5′-UTL. It is tempting to speculate that the striking direct repeat of these bases plays a role in the positive regulatory function of the first six codons. The repeats could also play a role in the stringent selection of the COX2 initiation site, which they bracket. The ARG8m coding region lacks this sequence, yet is efficiently expressed in the absence of COX2 codons, indicating that repetition of this sequence in the coding region is not essential for general translation initiation or elongation. However, the sequence repeat could be important for antagonizing the negative element in downstream COX2 codons. A mutation altering the upstream copy of the sequence repeat in the 5′-UTL had a modest negative effect on COX2 expression (11) and greatly reduced cox2::ARG8m expression (N. Bonnefoy, unpublished data), consistent with this possibility.
Two different but overlapping mutations in the leader peptide coding region behave similarly, suggesting that they affect the same mechanism. Translation of the cox2::ARG8m reporter was reduced to a similar degree both by deletion of codons 2 to 6 (cox2-27) and by a compound allele in which codon 6 was changed from AGA to CGU (both encoding R), and codons 7 to 10 (cox2-22) were deleted. Both of these alleles were suppressed by nucleotide substitutions clustered in the downstream part of the leader peptide coding region and, in the case of cox2-22, the first three codons specifying mature Cox2p. Interestingly, we selected two different missense substitutions affecting codon 11 as suppressors of both cox2-27 and cox2-22, supporting the idea that the two original mutations impaired translation by a similar mechanism. In addition, some intragenic suppressors of each of the mutations were silent third-position substitutions in the COX2 coding sequence, consistent with our other data showing the importance of nucleotide sequence.
Similar stem-loop structures are predicted for the COX2 mRNAs of the wild type, cox2-27, and cox2-22 in the sequence corresponding to wild-type codons 10 through 17. This stem appears to exist in vivo and to play a role in reducing the translation of the mutant mRNAs, since all but one of the intragenic suppressors of these mutations that increased translation also weakened the stem. Furthermore, translation was reduced by compensating site-directed mutations that restored pairing with cox2-27 suppressor substitutions. Indeed, generation of a more stable stem, by conversion of a U:A base pair to G:C, reduced expression to a level below that of cox2-27. These results demonstrate that unfolding of the stem is necessary to improve translation of downstream coding sequences in the absence of codons 2 to 6. One possible interpretation of these data is that the residual positively acting sequences in codons 7 to 14 must be unfolded to function in the absence of codons 2 to 6. An alternative interpretation is that in the absence of codons 2 to 6, a stable stem could strengthen the action of the negative element downstream of codon 14 (this stem-loop cannot solely comprise the negative element since cox2-27 mRNAs are translated better than cox2-20 and cox2-41 mRNAs which lack it). These are not mutually exclusive possibilities, considering the potential dynamics of mRNA structure during translation.
We identified two nuclear genes, PET111 and MRPL36, as dosage-dependent suppressors of cox2-27 and cox2-22. Pet111p is the COX2 mRNA-specific translational activator (32, 39, 51), a rate-limiting factor in COX2 expression (17). Elevated Pet111p activity can also suppress mutations in both the mRNA 5′-UTL (31) and the initiation codon (6). Since a fraction of ribosomes are able to pass the leaky translational blocks caused by the cox2-27 and cox2-22 mutations, increased initiation would be expected to improve gene expression.
MrpL36p was previously found to be associated with mitochondrial ribosomal large subunits (28), suggesting that it may function during translation elongation. This protein, which we found to be essential for global mitochondrial gene expression, contains a central 80-residue region exhibiting recognizable similarity to the entire length of the bacterial L31 family of ribosomal proteins in a PSI-BLAST comparison. Little is known about L31, except that it is loosely associated with the large subunit of Escheridria coli ribosomes (12). Loose ribosomal association of MrpL36p could account for the unexpected isolation of a dosage-dependent suppressor encoding a component of the ribosome. Flanking the central L31-like region is a roughly 60-residue N-terminal sequence exhibiting no similarities to known proteins and a 60-residue C-terminal region with weak but nevertheless intriguing similarity in a PSI-BLAST analysis to a short region of E. coli Ffh, a protein subunit of the signal recognition particle. This region of similarity lies within the Ffh M domain, which binds to both the 4.5S RNA and the signal sequence of membrane proteins (16). Thus, one could speculate that MrpL36p might mediate regulatory interactions among the elongating ribosome, positive and negative elements in the COX2 mRNA coding sequence, and the nascent pre-Cox2p polypeptide to coordinate synthesis and translocation of the pre-Cox2p N-tail domain through the inner membrane.
The role of the pre-Cox2p leader peptide amino acid sequence in controlling synthesis and membrane insertion of the protein remains enigmatic. The leader peptide sequences from several fungal and plant species are not strongly conserved (20, 25), and animal forms of Cox2p lack it entirely. While a previous study indicated that the leader peptide causes membrane association of a passenger protein (21), our present findings demonstrate that several amino acid sequences, which were generated by frameshifts and various peptide lengths, can effectively carry out this and/or any other steps necessary for cytochrome oxidase assembly. For example, while the wild-type leader peptide is 15 residues long with an acidic group at the third position, we observed normal Cox2p accumulation and respiratory growth in a strain whose frameshifted leader peptide is 5 residues long, lacks an acidic group, and has a basic group at the second position (cox2-43R1).
Nevertheless, our genetic analysis indicates that the leader peptide amino acid sequence is not completely unconstrained with respect to function in cytochrome oxidase assembly. One allele, cox2-43, allowed synthesis of the reporter protein and Cox2p at reduced rates, but prevented Cox2p from assembling into active cytochrome oxidase. The leader peptide encoded by cox2-43 is strikingly different from the wild type and the other functional sequences we generated, in that it has positively and negatively charged residues (KD) just upstream of the processing site. Spontaneous pseudorevertants of cox2-43 all had identical deletions of the six bases encoding these charged residues, which allowed efficient cytochrome oxidase assembly and greatly improved translation of the reporter protein. It is striking that the pseudorevertant encoding an active form of pre-Cox2p also increased mRNA translation.
We propose that the positive and negative translational regulatory elements specified within the first 91 codons of COX2 could function to ensure that continued translation of the mRNA occurs only if the nascent N terminus has successfully begun the process of membrane insertion leading to cytochrome oxidase assembly. While we cannot yet present a detailed model for how this feedback control mechanism might work, MrpL36p and Pet111p could function to convey information about the state of the nascent pre-Cox2p N terminus to the translating ribosome at the point where it encounters the negative element. We have also isolated dominant nuclear suppressors of both leader peptide mutations cox2-22 and cox2-27, which may identify other components of this system and lead to a better understanding of its mechanism. Passage through this element is likely to involve a dynamic interplay between alternative mRNA secondary structures and bound proteins.
Similar assembly feedback regulation of ATP synthase biogenesis could operate via pre-Atp6p, the only other yeast mitochondrial gene product with a leader peptide (30, 55). Such systems would resemble other translational feedback regulatory loops that couple the synthesis of specific components to the assembly of complexes. For example, translation of the chloroplast mRNA encoding cytochrome f is coupled to assembly of the cytochrome b6/f complex in Chlamydomonas (9, 57). In Caulobacter, translation of the flagellin fljK mRNA is regulated by assembly of the basal body-hook structure (1). In this case, in which the flagellin is transported out of the cell by a type III secretion system, the regulation of fljK translation depends on sequences in both the fljK mRNA 5′-UTL and the first 9 codons of the structural gene. Finally, in the type III secretion system of Yersinia, signals necessary for both the translation and the secretion of Yop proteins have been mapped to the first 15 codons and shown to function at the level of nucleotide sequence rather than amino acid sequence (2). Thus, the regulatory system revealed by this study is likely to have its origins in the bacterial ancestors of mitochondria.
ACKNOWLEDGMENTS
We are grateful to Geneviève Dujardin and Thomas L. Mason for supplying antibodies.
N.B. was a Human Frontier Science Program Organization long-term fellow (LT22/96) during the early stages of this work and is currently supported by the Association Française contre les Myopathies. This work has been supported by an NIH research grant (GM29362) to T.D.F.
REFERENCES
- 1.Anderson D K, Newton A. Posttranscriptional regulation of Caulobacter flagellin genes by a late flagellum assembly checkpoint. J Bacteriol. 1997;179:2281–2288. doi: 10.1128/jb.179.7.2281-2288.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson D M, Schneewind O. A mRNA signal for the type III secretion of Yop proteins by Yersinia enterocolitica. Science. 1997;278:1140–1143. doi: 10.1126/science.278.5340.1140. [DOI] [PubMed] [Google Scholar]
- 3.Anderson S, De Bruijn M H L, Coulson A R, Eperon I C, Sanger F, Young I G. Complete sequence of bovine mitochondrial DNA: conserved features of the mammalian mitochondrial genome. J Mol Biol. 1982;156:683–717. doi: 10.1016/0022-2836(82)90137-1. [DOI] [PubMed] [Google Scholar]
- 4.Behrens M, Michaelis G, Pratje E. Mitochondrial inner membrane protease 1 of Saccharomyces cerevisiae shows sequence similarity to the Escherichia coli leader peptidase. Mol Gen Genet. 1991;228:167–176. doi: 10.1007/BF00282462. [DOI] [PubMed] [Google Scholar]
- 5.Bonnefoy N, Chalvet F, Hamel P, Slonimski P P, Dujardin G. OXA1, a Saccharomyces cerevisiae nuclear gene whose sequence is conserved from prokaryotes to eukaryotes controls cytochrome oxidase biogenesis. J Mol Biol. 1994;239:201–212. doi: 10.1006/jmbi.1994.1363. [DOI] [PubMed] [Google Scholar]
- 6.Bonnefoy N, Fox T D. In vivo analysis of mutated initiation codons in the mitochondrial COX2 gene of Saccharomyces cerevisiae fused to the reporter gene ARG8m reveals lack of downstream reinitiation. Mol Gen Genet. 2000;262:1036–1046. doi: 10.1007/pl00008646. [DOI] [PubMed] [Google Scholar]
- 7.Bonnefoy N, Fox T D. Genetic transformation of Saccharomyces cerevisiae mitochondria. Methods Cell Biol. 2001;65:381–396. doi: 10.1016/s0091-679x(01)65022-2. [DOI] [PubMed] [Google Scholar]
- 8.Chen D C, Yang B C, Kuo T T. One-step transformation of yeast in stationary phase. Curr Genet. 1992;21:83–84. doi: 10.1007/BF00318659. [DOI] [PubMed] [Google Scholar]
- 9.Choquet Y, Stern D B, Wostrikoff K, Kuras R, Girard-Bascou J, Wollman F A. Translation of cytochrome f is autoregulated through the 5′ untranslated region of petA mRNA in Chlamydomonas chloroplasts. Proc Natl Acad Sci USA. 1998;95:4380–4385. doi: 10.1073/pnas.95.8.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costanzo M C, Bonnefoy N, Williams E H, Clark-Walker G D, Fox T D. Highly diverged homologs of Saccharomyces cerevisiae mitochondrial mRNA-specific translational activators have orthologous functions in other budding yeasts. Genetics. 2000;154:999–1012. doi: 10.1093/genetics/154.3.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunstan H M, Green-Willms N S, Fox T D. In vivo analysis of Saccharomyces cerevisiae COX2 mRNA 5′-untranslated leader functions in mitochondrial translation initiation and translational activation. Genetics. 1997;147:87–100. doi: 10.1093/genetics/147.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eistetter A J, Butler P D, Traut R R, Fanning T G. Characterization of Escherichia coli 50S ribosomal protein L31. FEMS Microbiol Lett. 1999;180:345–349. doi: 10.1111/j.1574-6968.1999.tb08816.x. [DOI] [PubMed] [Google Scholar]
- 13.Folley L S, Fox T D. Site-directed mutagenesis of a Saccharomyces cerevisiae mitochondrial translation initiation codon. Genetics. 1991;129:659–668. doi: 10.1093/genetics/129.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fox T D. Genetics of mitochondrial translation. In: Hershey J W B, Matthews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1996. pp. 733–758. [Google Scholar]
- 15.Fox T D, Folley L S, Mulero J J, McMullin T W, Thorsness P E, Hedin L O, Costanzo M C. Analysis and manipulation of yeast mitochondrial genes. Methods Enzymol. 1991;194:149–165. doi: 10.1016/0076-6879(91)94013-3. [DOI] [PubMed] [Google Scholar]
- 16.Freymann D M, Keenan R J, Stroud R M, Walter P. Structure of the conserved GTPase domain of the signal recognition particle. Nature. 1997;385:361–364. doi: 10.1038/385361a0. [DOI] [PubMed] [Google Scholar]
- 17.Green-Willms, N. S., C. A. Butler, H. M. Dunstan, and T. D. Fox. Pet111p, an inner membrane-bound translational activator that limits expression of the Saccharomyces cerevisiae mitochondrial gene COX2. J. Biol. Chem., in press. [DOI] [PubMed]
- 18.Grivell L A. Nucleo-mitochondrial interactions in mitochondrial gene expression. Crit Rev Biochem Mol Biol. 1995;30:121–164. doi: 10.3109/10409239509085141. [DOI] [PubMed] [Google Scholar]
- 19.Grivell L A, Artal-Sanz M, Hakkaart G, de Jong L, Nijtmans L G, van Oosterum K, Siep M, van der Spek H. Mitochondrial assembly in yeast. FEBS Lett. 1999;452:57–60. doi: 10.1016/s0014-5793(99)00532-3. [DOI] [PubMed] [Google Scholar]
- 20.Hardy C M, Clark-Walker G D. Nucleotide sequence of the cytochrome oxidase subunit 2 and Val-tRNA genes and surrounding sequences from Kluyveromyces lactis K8 mitochondrial DNA. Yeast. 1990;6:403–410. doi: 10.1002/yea.320060505. [DOI] [PubMed] [Google Scholar]
- 21.He S, Fox T D. Membrane translocation of mitochondrially coded Cox2p: distinct requirements for export of amino- and carboxy-termini, and dependence on the conserved protein Oxalp. Mol Biol Cell. 1997;8:1449–1460. doi: 10.1091/mbc.8.8.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hell K, Herrmann J, Pratje E, Neupert W, Stuart R A. Oxa1p mediates the export of the N- and C-termini of pCoxII from the mitochondrial matrix to the intermembrane space. FEBS Lett. 1997;418:367–370. doi: 10.1016/s0014-5793(97)01412-9. [DOI] [PubMed] [Google Scholar]
- 23.Hill J E, Myers A M, Koerner T J, Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- 24.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 25.Hoeben P, Weiller G, Clark-Walker G D. Larger rearranged mitochondrial genomes in Dekkera/Brettanomyces yeasts are more closely related than smaller genomes with a conserved gene order. J Mol Evol. 1993;36:263–269. doi: 10.1007/BF00160482. [DOI] [PubMed] [Google Scholar]
- 26.Johnston S A, Anziano P Q, Shark K, Sanford J C, Butow R A. Mitochondrial transformation in yeast by bombardment with microprojectiles. Science. 1988;240:1538–1541. doi: 10.1126/science.2836954. [DOI] [PubMed] [Google Scholar]
- 27.Kermorgant M, Bonnefoy N, Dujardin G. Oxa1p, which is required for cytochrome c oxidase and ATP synthase complex formation, is embedded in the mitochondrial inner membrane. Curr Genet. 1997;31:302–307. doi: 10.1007/s002940050209. [DOI] [PubMed] [Google Scholar]
- 28.Kitakawa M, Graack H-R, Grohmann L, Goldschmidt-Reisin S, Herfurth E, Wittmann-Liebold B, Nishimura T, Isono K. Identification and characterization of genes for mitochondrial ribosomal proteins of Saccharomyces cerevisiae. Eur J Biochem. 1997;245:449–456. doi: 10.1111/j.1432-1033.1997.t01-2-00449.x. [DOI] [PubMed] [Google Scholar]
- 29.Mathews D H, Sabina J, Zuker M, Turner D H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 30.Michon T, Galante M, Velours J. NH2-terminal sequence of the isolated yeast ATP synthase subunit 6 reveals post-translational cleavage. Eur J Biochem. 1988;172:621–625. doi: 10.1111/j.1432-1033.1988.tb13934.x. [DOI] [PubMed] [Google Scholar]
- 31.Mulero J J, Fox T D. Alteration of the Saccharomyces cerevisiae COX2 5′-untranslated leader by mitochondrial gene replacement and functional interaction with the translational activator protein PET111. Mol Biol Cell. 1993;4:1327–1335. doi: 10.1091/mbc.4.12.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mulero J J, Fox T D. PET111 acts in the 5′-leader of the Saccharomyces cerevisiae mitochondrial COX2 mRNA to promote its translation. Genetics. 1993;133:509–516. doi: 10.1093/genetics/133.3.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mulero J J, Fox T D. Reduced but accurate translation from a mutant AUA initiation codon in the mitochondrial COX2 mRNA of Saccharomyces cerevisiae. Mol Gen Genet. 1994;242:383–390. doi: 10.1007/BF00281787. [DOI] [PubMed] [Google Scholar]
- 34.Myers A M, Pape L K, Tzagoloff A. Mitochondrial protein synthesis is required for maintenance of intact mitochondrial genomes in Saccharomyces cerevisiae. EMBO J. 1985;4:2087–2092. doi: 10.1002/j.1460-2075.1985.tb03896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nasmyth K A, Tatchell K. The structure of transposable yeast mating type loci. Cell. 1980;19:753–764. doi: 10.1016/s0092-8674(80)80051-1. [DOI] [PubMed] [Google Scholar]
- 36.Neff N F, Thomas J H, Grisafi P, Botstein D. Isolation of the β-tubulin gene from yeast and demonstration of its essential function in vivo. Cell. 1983;33:211–219. doi: 10.1016/0092-8674(83)90350-1. [DOI] [PubMed] [Google Scholar]
- 37.Nunnari J, Fox T D, Walter P. A mitochondrial protease with two catalytic subunits of nonoverlapping specificities. Science. 1993;262:1997–2004. doi: 10.1126/science.8266095. [DOI] [PubMed] [Google Scholar]
- 38.Pinkham J L, Dudley A M, Mason T L. T7 RNA polymerase-dependent expression of COXII in yeast mitochondria. Mol Cell Biol. 1994;14:4643–4652. doi: 10.1128/mcb.14.7.4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poutre C G, Fox T D. PET111, a Saccharomyces cerevisiae nuclear gene required for translation of the mitochondrial mRNA encoding cytochrome c oxidase subunit II. Genetics. 1987;115:637–647. doi: 10.1093/genetics/115.4.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poyton R O, Duhl D M J, Clarkson G H D. Protein export from the mitochondrial matrix. Trends Cell Biol. 1992;2:369–375. doi: 10.1016/0962-8924(92)90049-s. [DOI] [PubMed] [Google Scholar]
- 41.Pratje E, Mannhaupt G, Michaelis G, Beyreuther K. A nuclear mutation prevents processing of a mitochondrially encoded membrane protein in Saccharomyces cerevisiae. EMBO J. 1983;2:1049–1054. doi: 10.1002/j.1460-2075.1983.tb01544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rose M D, Winston F, Hieter P. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 43.Sanchirico M E, Fox T D, Mason T L. Accumulation of mitochondrially synthesized Saccharomyces cerevisiae Cox2p and Cox3p depends on targeting information in untranslated portions of their mRNAs. EMBO J. 1998;17:5796–5804. doi: 10.1093/emboj/17.19.5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneider A, Behrens M, Scherer P, Pratje E, Michaelis G, Schatz G. Inner membrane protease I, an enzyme mediating intramitochondrial protein sorting in yeast. EMBO J. 1991;10:247–254. doi: 10.1002/j.1460-2075.1991.tb07944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seraphin B, Kandels-Lewis S. An efficient PCR mutagenesis strategy without gel purification step that is amenable to automation. Nucleic Acids Res. 1996;24:3276–3277. doi: 10.1093/nar/24.16.3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sevarino K A, Poyton R O. Mitochondrial biogenesis: identification of a precursor to yeast cytochrome c oxidase subunit II, an integral polypeptide. Proc Natl Acad Sci USA. 1980;77:142–146. doi: 10.1073/pnas.77.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen Z, Fox T D. Substitution of an invariant nucleotide at the base of the highly conserved “530-loop” of 15S rRNA causes suppression of mitochondrial ochre mutations. Nucleic Acids Res. 1989;17:4535–4539. doi: 10.1093/nar/17.12.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sibler A P, Dirheimer G, Martin R P. Codon reading patterns in Saccharomyces cerevisiae mitochondria based on sequences of mitochondrial tRNAs. FEBS Lett. 1986;194:131–138. doi: 10.1016/0014-5793(86)80064-3. [DOI] [PubMed] [Google Scholar]
- 49.Steele D F, Butler C A, Fox T D. Expression of a recoded nuclear gene inserted into yeast mitochondrial DNA is limited by mRNA-specific translational activation. Proc Natl Acad Sci USA. 1996;93:5253–5257. doi: 10.1073/pnas.93.11.5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Steffens G J, Buse G. Studies on cytochrome c oxidase (IV): primary structure and function of subunit II. Hoppe-Seyler's Z Physiol Chem. 1979;360:613–619. [PubMed] [Google Scholar]
- 51.Strick C A, Fox T D. Saccharomyces cerevisiae positive regulatory gene PET111 encodes a mitochondrial protein that is translated from an mRNA with a long 5′ leader. Mol Cell Biol. 1987;7:2728–2734. doi: 10.1128/mcb.7.8.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tian G L, Macadre C, Kruszewska A, Szczesniak B, Ragnini A, Grisanti P, Rinaldi T, Palleschi C, Frontali L, Slonimski P P, Lazowska J. Incipient mitochondrial evolution in yeasts. I. The physical map and gene order of Saccharomyces douglasii mitochondrial DNA discloses a translocation of a segment of 15,000 base-pairs and the presence of new introns in comparison with Saccharomyces cerevisiae. J Mol Biol. 1991;218:735–746. doi: 10.1016/0022-2836(91)90262-5. [DOI] [PubMed] [Google Scholar]
- 53.Torello A T, Overholzer M H, Cameron V L, Bonnefoy N, Fox T D. Deletion of the leader peptide of the mitochondrially encoded precursor of Saccharomyces cerevisiae cytochrome c oxidase subunit II. Genetics. 1997;145:903–910. doi: 10.1093/genetics/145.4.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tzagoloff A, Dieckmann C L. PET genes of Saccharomyces cerevisiae. Microbiol Rev. 1990;54:211–225. doi: 10.1128/mr.54.3.211-225.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Velours J, Spannagel C, Chaignepain S, Vaillier J, Arselin G, Graves P V, Velours G, Camougrand N. Topography of the yeast ATP synthase F0 sector. Biochimie. 1998;80:793–801. doi: 10.1016/s0300-9084(00)88873-2. [DOI] [PubMed] [Google Scholar]
- 56.Wiesenberger G, Costanzo M C, Fox T D. Analysis of the Saccharomyces cerevisiae mitochondrial COX3 mRNA 5′-untranslated leader: translational activation and mRNA processing. Mol Cell Biol. 1995;15:3291–3300. doi: 10.1128/mcb.15.6.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wollman F A, Minai L, Nechushtai R. The biogenesis and assembly of photosynthetic proteins in thylakoid membranes. Biochim Biophys Acta. 1999;1411:21–85. doi: 10.1016/s0005-2728(99)00043-2. [DOI] [PubMed] [Google Scholar]
- 58.Yaffe M P. Analysis of mitochondrial function and assembly. Methods Enzymol. 1991;194:627–643. doi: 10.1016/0076-6879(91)94046-f. [DOI] [PubMed] [Google Scholar]
- 59.Zuker M, Mathews D H, Turner D H. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide. In: Barciszewski J, Clark B F C, editors. RNA biochemistry and biotechnology. Dordrecht, Holland: Kluwer Academic Publishers; 1999. pp. 11–43. [Google Scholar]