Abstract
We describe the first species of Lentinula from Africa, Lentinula madagasikarensis sp. nov. The new taxon, which was collected from central Madagascar, is strikingly similar to L. edodes, the shiitake mushroom. A BLAST search using ITS sequences from L. madagasikarensis as the query retrieves a mix of Lentinula, Gymnopus, Marasmiellus, and other members of Omphalotaceae as the top hits. A 28S phylogeny of the Omphalotaceae confirms placement of L. madagasikarensis within Lentinula. An ITS phylogeny places L. madagasikarensis as the sister group of L. aciculospora, which is a neotropical species. Lentinula madagasikarensis is characterized by robust basidiomata with vinaceous pilei, prominent floccose scales near the pileus margin, florets of sphaeropedunculate cheilocystidia, and subcylindrical basidiospores. This report constitutes a 4 000-mile, trans-oceanic range extension for Lentinula.
Keywords: Africa, biogeography, edible mushrooms, Omphalotaceae, systematics, 1 new taxon
INTRODUCTION
Lentinula is a group of lignicolous agarics that includes the shiitake mushroom, L. edodes. Shiitake is a traditional food in East Asia and it has reportedly been cultivated in China for ca. 1 000 years (Chang & Miles 1987). However, the genus Lentinula has a broad distribution that spans much of South Asia, Australasia, and tropical and subtropical regions of the Americas (Pegler 1983). Lentinula has been resolved as a monophyletic group within the Omphalotaceae (Agaricales), which also contains Gymnopus, Rhodocollybia, and other collybioid mushrooms (Wilson & Desjardin 2005, Matheny et al. 2006, He et al. 2019, Oliviera et al. 2019).
Seven species have been formally described in Lentinula. Pegler (1983) included five species in his monograph of Lentinula: L. edodes in eastern Asia; L. lateritia in southeast Asia and Australasia; L. novae-zelandiae in New Zealand; L. boryana from the Gulf Coast of North America to South America; and L. guarapiensis from Paraguay. The latter is known only from the type collection (Spegazzini 1883). Using a combination of morphology, mating compatibility, and ITS sequences, Mata & Petersen (2000) described a new species from Costa Rica, L. aciculospora. Mata et al. (2001) then segregated a previous synonym, Armillaria raphanica, as L. raphanica from Pegler’s broad concept of L. boryana. Lentinula raphanica is reported by Mata et al. (2001) from Florida, Louisiana, Puerto Rico, Costa Rica, Venezuela, and Brazil (i.e., overlapping with L. boryana s. str.). Phylogenetic analyses of ITS sequences suggest that L. lateritia and L. edodes may each contain multiple species-level lineages, but these have not been formally described (Hibbett et al. 1998).
Most species of Lentinula grow on wood of Fagales, specifically Fagaceae or Nothofagaceae (Pegler 1983). The absence of Lentinula from Europe and most of North America (where oaks and their relatives abound, and outdoor log cultivation of shiitake is successful) is puzzling. Historical biogeographic analyses have suggested that the present distribution of Lentinula could reflect an ancient trans-Beringian distribution, which would require local extinction in North America (Hibbett 2001), but alternative dispersal routes via the southern hemisphere cannot be rejected.
During a study of the wild edible mushrooms of Madagascar, Buyck (2008) made two collections of a fungus that closely resembled L. edodes. Here, we present morphological and molecular analyses of the Malagasy material, which we describe as L. madagasikarensis.
MATERIALS AND METHODS
Sampling and phylogenetic inference
Two collections were made by Buyck (2008) in native forests of central Madagascar with endemic Uapaca densifolia (Phyllanthaceae) and Sarcolaenaceae, and introduced Eucalyptus robusta in 2006 and 2008. Both collections have been deposited at the fungarium (PC) of the Muséum National d’Histoire Naturelle, Paris. Genomic DNA was extracted from both collections in 2020 using the Extraction Solution-based method of Looney et al. (2020).
Two nuclear loci were amplified and sequenced with standard primer sets: ITS1F-ITS4 (White et al. 1990) for ITS, and LROR-LR5 (Vilgalys & Hester 1990) for 28S. Both strands were sequenced using the Sanger method, by Psomagen, New York, NY, and sequences have been deposited into GenBank (accession Nos. MW810299–MW810302).
Two datasets were used to infer relationships at different scales, Dataset 1 was used to assess the placement within the Omphalotaceae and Dataset 2 was used to infer placement within Lentinula. Dataset 1 used the 28S Omphalotaceae matrix from Oliveira et al. (2019) as a starting alignment. Dataset 2 included 60 ITS sequences, representing all known lineages of Lentinula, which were downloaded using emerencia (Nilsson et al. 2005), and six sequences of Gymnopus spp. as the outgroup.
Sequences of the Madagascar material were manually aligned with the two datasets in AliView v. 1.27 (Larsson 2014). Phylogenetic inference under Maximum Likelihood was performed in RAxML-VI-HPC using raxmlGUI v. 2.0 with autoMRE bootstopping criteria (Stamatakis 2006, Edler et al. 2020). Bayesian analysis was performed in MrBayes v. 3.2.7a using Metropolis coupled MCMC under a GTR +G model through the CIPRES Science gateway (Ronquist et al. 2012). The analysis consisted of 3 runs of 50 M generations and four chains sampled every 5 000 generations. Finally, a distance matrix of nine representative sequences of different taxa was generated from the final ITS alignment in R using the dist.dna function of the ‘ape’ package. Alignments and resulting trees have been deposited in TreeBASE (https://treebase.org/) under study number 28094.
Morphological analysis
Morphological field notes were taken on fresh basidiomata using color standards given in parentheses (Kornerup & Wanscher 1978). Microscopic observations were made on a Nikon Eclipse e600 compound microscope with a mounted SPOT RT Slider digital camera. Dried specimens were rehydrated in 3 % KOH and observed in phase contrast or stained with phloxine and by bright field microscopy. Melzer’s reagent was used for testing amyloidity. At least 20 observations were made for each feature. Microscopic feature measurements are given as lower limit–[arithmetic mean]–upper limit with n = total number of observations and Q-value for basidiospores calculated as arithmetic mean of the lengths divided by arithmetic mean of the widths for the lower limit–[arithmetic mean]–upper limit.
RESULTS
Molecular systematics
BLAST searches using ITS sequences from L. madagasikarensis as the queries retrieved sequences of Lentinula, Gymnopus, Marasmiellus, and other members of Omphalotaceae as the top hits (results not shown). Phylogenetic analyses of Dataset 1 recovered all of the major clades of Omphalotaceae identified by Oliveira et al. (2019), but often with weak bootstrap support (BS) (Fig. 1). Lentinula was strongly supported as monophyletic (BS 91 %). The sister group of Lentinula was not resolved with confidence, but a clade containing Lentinula and Clades A–J of Oliveira et al. (2019) was strongly supported (BS 99 %).
Fig. 1.
Maximum-likelihood phylogeny of Omphalotaceae based on the 28S sequence dataset of Oliveira et al. (2019) with 788 bootstrap replicates according to autoMRE bootstopping criteria. Major clades from the original study are labeled and collapsed with number of samples included in each clade. BS values ≥ 50 % are included with the branches.
The Lentinula-focused ITS analyses of Dataset 2 placed L. madagasikarensis as sister to L. aciculospora with moderate BS (84 %) (Fig. 2). Two major groups were resolved in Lentinula, one weakly-supported group containing all Asian-Australasian species (BS 56 %) and one strongly-supported clade containing all American species, plus L. madagasikarensis (BS 98 %). There are three major groups in the Americas, L. aciculospora (BS 100 %), L. boryana (BS 86 %), and L. raphanica (BS 100 %).
Fig. 2.
Maximum-likelihood phylogeny of Lentinula based on Dataset 2 of ITS. BS values ≥ 50 % / Bayesian PP values ≥ 0.80 are included with the branches. Sequence labels include GenBank accession numbers and collection numbers. G1–G5 in the Asian-Australasian clade correspond to ITS-based groups from Hibbett et al. (1998). Country codes: AUS = Australia, BRA = Brazil, COL = Colombia, CRI = Costa Rica, IND = India, KOR = South Korea, MEX = Mexico, NPL = Nepal, NZL = New Zealand, PHL = Philippines, PNG = Papua New Guinea, POL = Poland (possible cultivar), PRI = Puerto Rico, RUS = Russia, USA = continental USA, VEN = Venezuela.
The Asian-Australasian clade contains five lineages that have previously been termed Groups 1–5 based on ITS data (Hibbett et al. 1998). Each of the ITS-based groups are moderately to strongly supported as monophyletic, but relationships among them are not well resolved (Fig. 2). Groups 1 and 5 correspond to L. edodes sensu lato, including cultivated shiitake mushrooms. Based on sampling in the present study and that of Hibbett et al. (1998), Group 1 includes isolates from China, Japan, South Korea, Philippines, India, and the Russian Far East, whereas Group 5 includes isolates from China, Nepal and India. Group 2 and Group 4 both include collections from Papua New Guinea; Group 2 also includes collections from Australia. Group 3 is equivalent to L. novae-zelandieae, from New Zealand.
Pairwise distances of ITS sequences of L. madagasikarensis to other species of Lentinula ranged from 11 % vs. L. edodes (Group 5), L. lateritia (Group 2), L. novae-zelandiae, and L. aciculospora, to 20 % vs. L. raphanica (Fig. 3). Pairwise sequence distances among three isolates from the Americas are 11–20 %, whereas those among five Asian-Australasian isolates are 1–4 % (Fig. 3).
Fig. 3.
Distance-matrix heat map of representative sequences of species of Lentinula based on pairwise distances from the ITS multi-sequence alignment given as proportion of divergence.
Taxonomy
Lentinula madagasikarensis Buyck, Randrianjohany & Looney, sp. nov. MycoBank MB 839129. Fig. 4A–G.
Fig. 4.
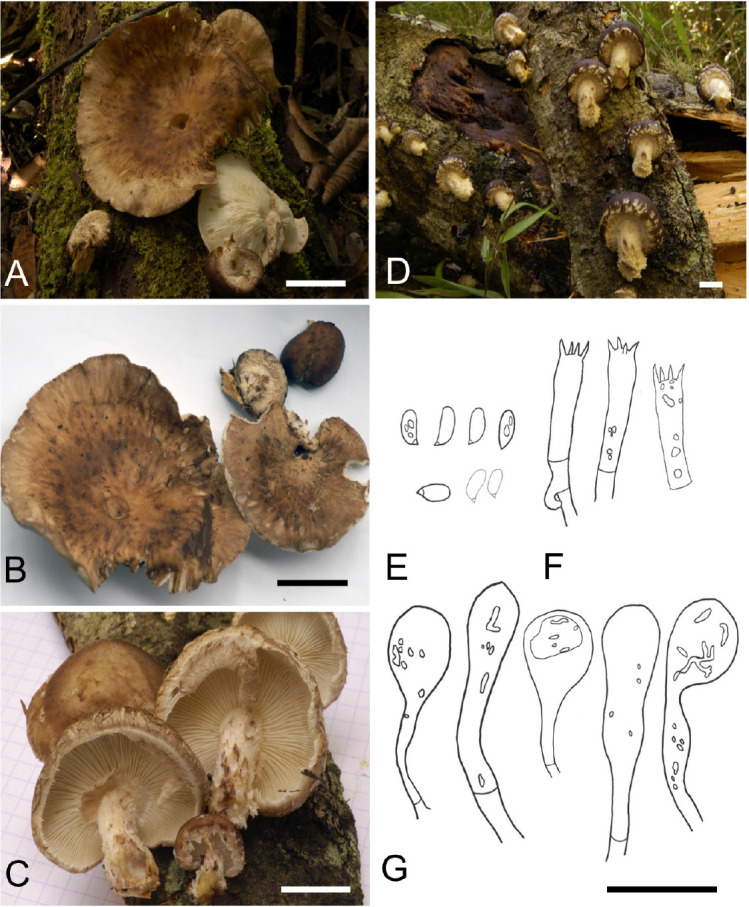
A, B. Field photograph of Lentinula madagasikarensis (type specimen, PC coll. BB06.007). C, D. Field photographs of Lentinula madagasikarensis (paratype specimen, PC coll. BB08.120). E. Basidiospores. F. Basidia. G. Cheilocystidia. Scale bars: A–D = 2 cm, E–G = 20 μm.
Etymology: The specific epithet is derived from Madagasikara, which is the Malagasy name for Madagascar. To our knowledge, there is no Malagasy name for this species.
Diagnosis: Robust basidiomata with vinaceous pileus color and large, tufted scales near pileus margin merging into an appendiculate margin. Fibrous, ivory-colored stipe with squamules. Small and narrow basidiospores. Sphaeropedunculate cheilocystidia forming scattered florets.
Typus: Madagascar, Moramanga district, Alaotra-Mangoro region, Andasibe, 18°56’S 48°25’E, on E. robusta log, 20 Jan. 2006, B. Buyck & V. Hofstetter 06.007 (holotype PC0142531).
Description: Pileus convex to hemisphaerical when young, soon umbonate to applanate or broadly depressed, firm and fleshy, up to 6–7 cm diam.; surface smooth, sometimes scurfy or with minute scales or granules and almost greasy at the touch in the central portion, toward the margin rapidly becoming covered with a paler, hairy-fibrillose scurf, building more or less concentrically arranged, thick, floccose deposits with fibrillose extremities pointing downward; dark reddish brown (9EF6-8) to vinaceous or purplish brown (14EF5-8) at center when young, at maturity becoming paler, pale reddish brown to ochraceous brown (3A5-6, 4B5-8, 5C5-7) but retaining locally some darker shades. Lamellae adnato- or adnexo-sinuate with a decurrent tooth, later frequently subfree, unequal with up to 4–5 series of lamellulae of different lengths, whitish to ivory, discolouring brownish after injury, crowded; edge concolorous, entire or irregularly serrulate. Stipe up to 4 × 2.5 cm; stout and firm, always shorter than the pileus diameter, central to mostly eccentric, cylindrical or a little widening to almost bulbous near the base, strongly fibrillose-squamose over the entire surface from thick fibrils or squamae pointing out- or upward, solid. Partial veil a thick cottony tissue, strongly fibrillose-squamose on the lower surface from fibrils pointing downward, fugacious, soon breaking up and limited to remnants on the pileal margin or sometimes leaving a (partial) ring on upper part of the stipe. Context firm-fleshy, pale coloured, not discolouring but often colored yellowish in the stipe near the attachment to the woody host surface. Spore print white. Basidiospores oblong to subcylindrical, 5.0–[6.0]–7.0 × 2.0–[3.0]–3.5 μm; Q = 1.7–[2.1]–2.4 (n = 30), hyaline, inamyloid, thin-walled, smooth; hilar appendage conspicuous; contents heterogenous or homogeneous. Hymenium composed mostly of basidia and basidioles. Basidia 11.0–[19.0]–26 × 4.0–[5.0]–5.5 μm (n = 40), oblong to cylindrical, clavate with a median constriction, with a clamp connection at the base, four-sterigmate; sterigmata slender, up to 3 μm long. Pleurocystidia absent. Cheilocystidia 15.0–[28]–46 × 7.0–[11.0]–16.0 μm (n = 20), clavate to sphaeropedunculate, inflated apically without lobes, thin-walled, smooth, forming dense clusters or florets, infrequent. Lamellar trama regular to subregular; hyphae 4.0–15.0 μm diam., thin- to thick-walled. Subhymenium rudimentary. Epicutis 30–50 μm thick; hyphae 3.0–7.0 μm diam, brown in mass, hyaline singly, repent, interwoven and subregular cutis; hyphae in scales erumpent, ending in eroded hyphal fragments. Subpellis composed of frequently branching, irregular hyphae 5.0–11.0 μm diam, hyaline, thick-walled. Stipitipellis composed of interwoven hyphae 3.0–7.0 μm diam, branching, with floccons formed of obtuse terminal end cells. Caulocystidia absent.
Habit and habitat: Gregarious to scattered on corticate logs of E. robusta and unidentified hardwood in humid, mixed mountain forests at higher elevations of the central plateau and eastern escarpment of Madagascar.
Additional specimen examined: Madagascar, Ankazobe district, Analamanga region, Ambohitantely Special Reserve, 18.161°S 47.302°E, in primary forest dominated by U. densifolia and Sarcolaenaceae, 22 Jan. 2008, B. Buyck & V. Hofstetter no. 08.120 (paratype PC0142532).
Notes: Lentinula madagasikarensis can readily be distinguished from its apparent sister species, L. aciculospora, by gross morphological characters including a darker, vinaceous pileus color in L. madagasikarensis and larger tufted scales concentrated near the pileus margin forming a distinctive appendiculate margin as well as microscopic differences in basidiospore size, which is smaller in L. aciculospora (5.6–8.8 × 1.6–2.8 μm), and shape of cheilocystidia that are gnarled and bluntly lobed in L. aciculospora (Mata & Petersen 2000). Lentinula madagasikarensis closely resembles L. boryana (Mata et al. 2001) but differs microscopically because the first never has appendages on cheilocystidia and caulocystidia are absent. Macroscopically, L. madagasikarensis differs from L. boryana in size, color, and robustness of the pilei, which are smaller (1.8–2.5 cm) and paler (light brown to golden brown or grayish orange) in L. boryana (Mata et al. 2001). Morphologically, L. madagasikarensis closely resembles L. edodes in pileus color and the robust aspect of the basidiomata, which also has a dark vinaceous brown and > 5 cm diam pileus (Pegler 1983). Microscopic characters are similar between the two species with similar size of the basidiospores, 5–6.5(–7) × 3–3.7 μm in L. edodes and general shape of cheilocystidia, also clavate without any apical appendages in L. edodes (Pegler 1983). However, basidiospores of L. edodes are slightly more ovoid (Q = 1.78) as reported by Pegler (1983) and cheilocystidia are not reported as sphaeropedunculate or forming florets as is typically found in L. madagasikarensis. The geographic distribution also separates L. edodes from L. madagasikarensis with the first broadly distributed in Asia and the latter known only from Madagascar.
DISCUSSION
Buyck (2008: 516) described the Malagasy Lentinula as a “shiitake look-alike.” Our analyses confirm that his collections, from two different localities in Madagascar, represent a new species of Lentinula. Lentinula madagasikarensis was discovered 4 000 miles from the nearest well-documented wild populations of Lentinula, which are those of L. edodes Group 5 in India and Nepal. Its apparent sister group, L. aciculospora, occurs 9 000 miles away, in Costa Rica (Mata & Petersen 2000), Ecuador (Andrade et al. 2012), Panama (Piepenbring 2008), and Nicaragua (Gómez 2018). However, several collections identified as L. edodes or “L. edodes-like” have been reported from the Democratic Republic of the Congo (DRC), which is less than 2 000 miles from Madagascar (Global Biodiversity Information Facility [www.gbif.org] occurrence records 1836956264, 1840736410, 1840736444). As of this writing, we have not been able to study the material from the DRC.
At its narrowest point, the Mozambique Channel separates Madagascar and mainland Africa by less than 300 miles. Based on their geographic proximity, one would expect a Lentinula species from the DRC and L. madagasikarensis to be closely related. Even if they are, the presence of Lentinula in Madagascar is probably due to long-distance dispersal; Madagascar began to separate from continental Africa during the Late Jurassic Period, about 150-M-yr ago (de Wit 2003), whereas Lentinula is about 50-M-yr-old, according to a molecular clock analysis by Varga et al. (2019).
Both collections of L. madagasikarensis were found in similar habitats in the highlands of the Central Plateau at 1 500–2 000 m alt. One collection was found growing on wood of E. robusta and the other was on a fallen log of an “unidentified native tree” (Buyck 2008: 516), but both were in dense forests largely dominated by U. densifolia and various species of Sarcolaenaceae. All other species or species complexes of Lentinula are reported to occur on Fagales, particularly Fagaceae or Nothofagaceae, but some are also capable of growing on other hardwood substrates (Pegler 1983, Mata & Petersen 2000, McKenzie et al. 2000, Mata et al. 2001, Piepenbring 2008). There are no native Fagaceae or Nothofagaceae in Madagascar or Sub-Saharan Africa (Manos & Stanford 2001, Knapp et al. 2005). A recent discovery of 52-M-yr-old Castanopsis fossils from Patagonia shows that Fagaceae was present in the southern hemisphere in the early Eocene (Wilf et al. 2019), but that was late in the breakup of Gondwana, long after the opening of the South Atlantic Ocean. Nothofagus has an 80-M-yr fossil record, but there is no evidence of this group having existed in Africa or Madagascar (Knapp et al. 2005).
The phylogenetic placement of L. madagasikarensis is important for understanding host shifts and historical biogeography of Lentinula. The distribution of host ranges in Lentinula suggests that the ancestor of the genus decayed Fagales. If so, then L. madagasikarensis would represent an expansion onto hosts that are indigenous to Madagascar, as well as introduced eucalypts. Based on the ITS phylogeny, L. madagasikarensis is closely related to L. aciculospora within the American-African clade (Fig. 2), suggesting that it (and possibly the entire genus) is derived from an ancestor in the neotropics. However, many branches in the ITS phylogeny of Lentinula are not strongly resolved (Fig. 2). We defer formal analyses of historical biogeography and evolution of substrate ranges until we have a genome sequence for L. madagasikarensis, which we are currently pursuing.
ACKNOWLEDGEMENTS
BB expresses sincere thanks to the Committee for Research and Exploration of the National Geographic Society for funding part of the fieldwork through grant #7921-05. Colleagues of the CNRE at Antananarivo are acknowledged for excellent field assistance. Jadson J. S. Oliveira kindly provided the starting alignment for Dataset 1. Research at Clark University was supported by National Science Foundation award DEB-1456588 (DSH, PI).
Footnotes
Citation: Looney BP, Buyck B, Menolli Jr N, Randrianjohany E, Hibbett D (2021). Lentinula madagasikarensis sp. nov., a relative of shiitake mushrooms from Madagascar. Fungal Systematics and Evolution 8: 1–8. doi: 10.3114/fuse.2021.08.01
Corresponding editor: P.W. Crous
Conflict of interest: The authors declare that there is no conflict of interest.
REFERENCES
- Andrade JM, Malagoìn O, Piepenbring M, et al. (2012). Etnomicología y valor nutricional de macrohongos silvestres de la comunidad indígena Saraguro en el Sur del Ecuador. Boletín de la Sociedad Micologíca de Madrid 36: 193–201. [Google Scholar]
- Bazzicalupo AL, Buyck B, Saar I, et al. (2017). Troubles with mycorrhizal mushroom identification where morphological differentiation lags behind barcode sequence divergence. Taxon 66: 791–810. [Google Scholar]
- Berkeley MJ. (1877). Contribution to the botany of H.M.S. ‘Challenger.’ XXXVIII. Enumeration of the fungi during the expedition of H.M.S. ‘Challenger,’ 1874–75. Journal of the Linnean Society, Botany 16: 38–54. [Google Scholar]
- Berkeley MJ, Curtis MA. (1869). Fungi Cubenses (Hymenomycetes). Journal of the Linnean Society, Botany 10: 280–342. [Google Scholar]
- Buyck B. (2008). The edible mushrooms of Madagascar: an evolving enigma. Economic Botany 62: 509–520. [Google Scholar]
- Chang ST, Miles PG. (1987). Historical record of the early cultivation of Lentinus in China. The Mushroom Journal for the Tropics 7: 31–37. [Google Scholar]
- Edler D, Klein J, Antonelli A, et al. (2020). raxmlGUI 2.0: a graphical interface and toolkit for phylogenetic analyses using RAxML. Methods in Ecology and Evolution 12: 373–377. [Google Scholar]
- Gómez ILS. (2018). Hongos del Caribe nicaragüense. Resumen de especies de dos estudios hechos en la Reserva Bioloìgica Indio Maíz. Wani 73: 44–54. [Google Scholar]
- Harrower E, Ammirati JF, Cappuccino AA, et al. (2011) Cortinarius species diversity in British Columbia and molecular phylogenetic comparison with European specimen sequences. Botany 89: 799–810. [Google Scholar]
- He MQ, Zhao RL, Hyde KD. et al. (2019). Notes, outline and divergence times of Basidiomycota. Fungal Diversity 99: 105–367. [Google Scholar]
- Hibbett DS. (2001). Shiitake mushrooms and molecular clocks: historical biogeography of Lentinula. Journal of Biogeography 28: 231–241. [Google Scholar]
- Hibbett DS, Hansen K, Donoghue MJ. (1998). Phylogeny and biogeography of Lentinula inferred from an expanded rDNA dataset. Mycological Research 102: 1041–1049. [Google Scholar]
- Hughes KW, Petersen RH, Lickey EB. (2009). Using heterozygosity to estimate a percentage DNA sequence similarity for environmental species’ delimitation across basidiomycete fungi. The New Phytologist 182: 795–798. [DOI] [PubMed] [Google Scholar]
- Knapp M, Stöckler K, Havell D, et al. (2005). Relaxed molecular clock provides evidence for long-distance dispersal of Nothofagus (southern beech). PLoS Biology 3: e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornerup A, Wanscher JH. (1978). Methuen Handbook of Colour. 3rd ed., Methuen Co., London. [Google Scholar]
- Larsson A. (2014). AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30: 3276–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looney BP, Adamčík S, Matheny PB. (2020). Coalescent-based delimitation and species-tree estimations reveal Appalachian origin and Neogene diversification in Russula subsection Roseinae. Molecular Phylogenetics and Evolution 147: 106787. [DOI] [PubMed] [Google Scholar]
- Manos PS, Standofr AM. (2001). The historical biogeography of Fagaceae: tracking the tertiary history of temperate and subtropical forests of the northern hemisphere. International Journal of Plant Sciences 162: S77–S93. [Google Scholar]
- Mata JL, Petersen RH. (2000). A new species of Lentinula (Agaricales) from Central America. Mycoscience 41: 351–355. [Google Scholar]
- Mata JL, Petersen RH, Hughes KW. (2001). The genus Lentinula in the Americas. Mycologia 93: 1102–1112. [Google Scholar]
- McKenzie EHC, Buchanan PK, Johnston PR. (2000). Checklist of fungi on Nothofagus species in New Zealand. New Zealand Journal of Botany 38: 635–720. [Google Scholar]
- McLoughlin S. (2001). The breakup history of Gondwana and its impact on pre-Cenozoic floristic provincialism. Australian Journal of Botany 49: 271–300. [Google Scholar]
- Nilsson RH, Kristiansson E, Ryberg M, et al. (2005). Approaching the taxonomic affiliation of unidentified sequences in public databases–an example from the mycorrhizal fungi. BMC Bioinformatics 6: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira JJ, Vargas-Isla R, Cabral TS, et al. (2019). Progress on the phylogeny of the Omphalotaceae: Gymnopus s. str., Marasmiellus s. str., Paragymnopus gen. nov. and Pusillomyces gen. nov. Mycological Progress 18: 713–739. [Google Scholar]
- Pegler DN. (1984). The genus Lentinula (Tricholomataceae tribe Collybieae). Sydowia 36: 227–239. [Google Scholar]
- Piepenbring M. (2008). Reportes nuevos de Agaricales para Panamá. Acta Biologica Panamensis 1: 22–38. [Google Scholar]
- Rehner SA, Buckley E. (2005). A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97: 84–98. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, Van Der Mark P, et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spegazzini C. (1883). Fungi guarantici. Anales de la Sociedad Científica Argentina 16: 242–248. [Google Scholar]
- Stamatakis A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- Varga T, Krizsán K, Földi C, et al. (2019). Megaphylogeny resolves global patterns of mushroom evolution. Nature Ecology & Evolution 3: 668–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilgalys R, Hester M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee SB, et al. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR protocols: a guide to methods and applications (Innis MA, Gelfand DH, Sninsky JJ, et al., eds). Academic Press, Inc., New York: 315–322. [Google Scholar]
- Wilf P, Nixon KC, Gandolfo MA, et al. (2019). Eocene Fagaceae from Patagonia and Gondwanan legacy in Asian rainforests. Science 364: eaaw5139. [DOI] [PubMed] [Google Scholar]
- Wilson AW, Desjardin DE. (2005) Phylogenetic relationships in the gymnopoid and marasmioid fungi (Basidiomycetes, euagarics clade). Mycologia 97: 667–679. [DOI] [PubMed] [Google Scholar]
- Wilson DS, Pollard D, DeConto RM, et al. (2013). Initiation of the West Antarctic Ice Sheet and estimates of total Antarctic ice volume in the earliest Oligocene. Geophysical Research Letters 40: 4305–4309. [Google Scholar]
- Wit MJD. (2003). Madagascar: heads it’s a continent, tails it’s an island. Annual Review of Earth and Planetary Sciences 31: 213–248. [Google Scholar]





