Abstract
A smut fungus that hinders wiregrass restoration efforts in longleaf pine-grassland ecosystems was collected from Aristida stricta and A. beyrichiana (Poaceae) in three states in the southeastern USA. Morphological and phylogenetic characteristics of this fungus were examined. These data show that the specimens from both plant species were infected by the same fungus and represent a new species of Langdonia. The new species differs morphologically from other species of Langdonia by teliospores being solitary and not compacted into spore balls. Spore wall ornamentation and teliospore size also differ from other Langdonia species. Phylogenetic analyses of DNA sequences of the ITS, LSU, and EF-1α supported separation of the species from A. stricta and A. beyrichiana from other Langdonia species. Based on these results, a new species, Langdonia walkerae, is proposed.
Keywords: new taxa, plant pathogens, smut fungi, spore balls, Ustilaginaceae
INTRODUCTION
Aristida, a member of the Poaceae and the Aristidoideae (Barkworth 2003, Cerros-Tlatilpa et al. 2011), is distributed worldwide with approximately 300 species (Barkworth 2003) primarily located in Central, North and South America, Australia, and Africa (Cerros-Tlatilpa et al. 2011). This genus is distinct from other grass genera by its three-awned lemma (Clewell 1989, Barkworth 2003). In the southeastern United States, two Aristida species, A. stricta and A. beyrichiana, are common grasses found in the ground layer of longleaf pine, Pinus palustris, forests (Peet 1993), which extend from the border of Texas and Louisiana eastward to the Atlantic Ocean and from the middle of Florida northward into Virginia (Brockway et al. 1998, Frost 2006). Aristida stricta, called Carolina wiregrass or the pineland threeawn, occurs in North Carolina and the northern part of South Carolina, and is distinguished morphologically by having hairs along the length of the blades, wider culms, and shorter ligules (Peet 1993, Van Eerden 1997). Aristida beyrichiana, called southern wiregrass or Beyrich threeawn, grows in the southern part of South Carolina into Florida and west to Mississippi, and is distinguished morphologically by its soft, short hairs at the base of the blades and the glumes are more unequal in length (Peet 1993, Van Eerden 1997).
These perennial bunchgrasses are adapted to fire, enabling them to persist and reproduce despite frequently recurring fires (Platt 1999, Means 2007) as well as to survive for decades without burning (Shearman et al. 2019). Flowers and seed production are most abundant within a year of burning; without fire, few, if any, are produced each subsequent year (Clewell 1989, Van Eerden 1997, Fill et al. 2012). These native wiregrasses are considered keystone species in the longleaf pine-grass ecosystem as they produce the fine fuels needed for frequent burning, a requirement for sustained ecosystem structure and function (Clewell 1989, Duever 1989, Noss 1989). Thus, the seed viability of wiregrasses is important for ecosystem restoration efforts (Van Eerden 1997) in projects that focus on wiregrass as a primary understory species.
The production of A. stricta and A. beyrichiana seeds has been observed to be affected by a smut fungus that replaces the developing ovaries with teliospores (Van Eerden 1997). Smut fungi are pathogens that primarily infect grasses and occur world-wide. They produce sori, sporocarps in which teliospores are produced, in different organs of their host (Vánky 2013). Previously, many grass-infecting smut fungi, including Sporisorium, were united into the genus Ustilago that has proved to be polyphyletic after the introduction of molecular phylogenetic studies (Begerow et al. 1997, 2006). In the taxonomy of smut fungi, the host plant is one of the criteria usually used to classify groups (Bauer et al. 2001, Begerow et al. 2004). Consequently, McTaggart et al. (2012b) emended the description of the genus Sporisorium and separated all species that occur on Aristida into a new genus, Langdonia, based on the host plant and morphological and phylogenetic differences. Langdonia is a monophyletic group within the Ustilaginaceae (McTaggart et al. 2012b). Currently, eight species have been described on other Aristida species from various countries - including Argentina, Australia, Bolivia, Madagascar, Mexico, Thailand, and the USA (Durán 1987, McTaggart et al. 2012b, Vánky 2012, Denchev et al. 2015). The defining morphological characteristics of the genus Langdonia are sori that can be found in some or all ovaries of the panicle, lack of columella and sterile cells, teliospores compacted into spore balls, and Ustilago-type germination (McTaggart et al. 2012b, Vánky 2013). Based on these criteria, the tentative identification of the fungus causing smut on A. stricta and A. beyrichiana was a species of Langdonia. Therefore, the objectives of this study were to identify the fungus that infects A. stricta and A. beyrichiana and to determine if the same fungus species infects both A. stricta and A. beyrichiana.
MATERIALS AND METHODS
Sample collection and documentation
Naturally infected plants were collected from seven locations in North Carolina, South Carolina, and Florida during the seed production stage (Table 1). In 2017, samples were collected from two locations - one in South Carolina and one in Florida, and in 2018 samples were collected from six locations in all three states, with only one location being common to both years. All sampled sites had been burned by prescription within the year. The locations and the wiregrass species from which each sample was obtained are included in the “specimens examined” section and in Table 1. For collection, samples of infected wiregrass culms, erect stems bearing inflorescences, were cut at ground level, placed in paper bags, and returned to the laboratory where the specimens were kept at room temperature for subsequent use. Plant voucher specimens were deposited in the Clemson University Herbarium (CLEMS), and fungus specimens were deposited in the Washington State University Mycological Herbarium (WSP) and the U.S. National Fungus Collections - Herbarium (BPI) and kept as voucher specimens.
Table 1 .
Smut samples on Aristida beyrichiana and A. stricta collected and examined for this study.
| Site | Voucher | Isolate | State | City/County | Aristida spp. | Date |
|---|---|---|---|---|---|---|
| Silver Bluff Audubon Center | CLEMS0080381 | JK 5017 | SC | Jackson/Aiken | A. beyrichiana | 10 Nov. 2017 |
| Holotype: WSP74240 | ||||||
| Isotype: BPI911222 | ||||||
| CLEMS0080382 | JK 6017 | |||||
| CLEMS0080383 CLEMS0080384 |
SB 1218 | 6 Dec. 2018 | ||||
| Aiken Gopher Tortoise Heritage Preserve | CLEMS0080385 | AGT 1218 | SC | Aiken/Aiken | A. beyrichiana | 6 Dec. 2018 |
| Savannah River Site | CLEMS0080388 WSP74241 BPI 911220 |
SRS 1218 | SC | Barnwell/Barnwell | A. beyrichiana | 4 Dec. 2018 |
| Apalachicola Bluffs and Ravines Preserve | CLEMS0080387 | FLW 1017 | FL | Bristol/Liberty | A. beyrichiana | 31 Oct. 2017 |
| Austin Cary Forest | CLEMS0080386 | FL 1218 | FL | Gainesville/Alachua | A. beyrichiana | 3 Dec. 2018 |
| Carolina Sandhills National Wildlife Refuge | CLEMS0080389 WSP74242 BPI 911221 |
CSN 1118 | SC | Hoffman/Richmond | A. stricta | 27 Nov. 2018 |
| North Carolina Sandhills Game Land | CLEMS0080390 WSP74243 BPI 911219 |
NC 1018 NC 2018 NC 3018 NC 5018 |
NC | Chesterfield/Chesterfield | A. stricta | 29 Nov. 2018 |
Morphological examination
The characteristics of the sori and teliospores were examined on infected plants. Pictures of sori were taken using a Nikon D5100 camera. Teliospore characteristics were studied using a compound microscope (LM; BX60F; Olympus optical Co. Ltd., Japan) equipped with ProgRes C5 camera (JENOPTIK, Germany) and CapturePro software; and a scanning electron microscope (SEM; Hitachi SU6600) at 5.0 kV at the Clemson University Electron Microscopy Facility. For LM, teliospores were mounted in lactic acid (85–90 %, VWR, International, LLC) (Savchenko et al. 2014) and examined at 1 000 × magnification. The diameters of 30 teliospores, oriented in plane view so that they appeared globose, were measured from each sample collection. The colors of the sori and the teliospores were described according to Rayner (1970). For SEM examination, teliospores were dusted on double-sided adhesive carbon tape, mounted on aluminum stubs, and sputter-coated with platinum using a Cressington sputter-coater (ca. 30 nm in 6 min).
Growth on artificial medium
Sori were carefully removed from infected plants and surface-sterilized by immersing in 70 % ethanol for 30 s, rinsing with sterile distilled water, immersing in 3 % sodium hypochlorite for 10 s, and rinsing twice with sterile distilled water. Each sorus was handled individually using aseptic techniques; one sorus was placed in a 2.0 mL centrifuge tube and gently ground with a tissue grinder pestle to release the teliospores. To make a spore suspension, 500 μL sterile distilled water was added to a tube, the contents were thoroughly mixed, 100 μL was pipetted onto a petri dish containing Difco™ malt extract agar (MEA; Becton, Dickinson and Co., Sparks, MD) and the suspension was spread with a L-shaped cell spreader over the agar surface. The MEA was made according to the manufacturer’s instruction and amended with streptomycin (MEA+S; 200 ppm = 200 mg/L). Plates were placed at 28 °C for 2–3 d; and then individual colonies were aseptically subcultured onto MEA+S and incubated for 10 d at 28 °C. The morphological characteristics of shape, color, and diameter of the colony were recorded. The color of the colony was defined according to Rayner (1970).
DNA Extraction, Polymerase chain reaction (PCR), and sequencing
DNA was extracted from 10-d-old cultures using a DNeasy Plant Mini Kit (QIAGEN GmbH, Hilden, Germany) according to the manufacturer’s instructions. DNA concentration was determined using a Thermo Scientific Nanodrop 2000 /2000c spectrophotometer. All DNA samples were stored at -20 °C until used for amplification. Genomic DNA was amplified using an Eppendorf Mastercycler Gradient thermocycler with Thermo Scientific Phusion High-Fidelity DNA Polymerase following the manufacturer’s cycling and reaction conditions. Two nuclear ribosomal DNA regions and one gene locus were amplified and sequenced: the internal transcribed spacer (ITS) region, the large subunit region (LSU), and translation elongation factor-1α locus (EF-1α). The ITS region was amplified with primers M-ITS1 (Stoll et al. 2003) and ITS4 (White et al. 1990) and annealed at 58 °C (McTaggart et al. 2012a); the LSU region was amplified with primers LROR and LR5 (Vilgalys & Hester 1990) at 60 °C; and the EF-1α locus was amplified with primers EF-1αF and EF-1αR (McTaggart et al. 2012a) at 64 °C. The PCR products were analyzed using gel electrophoresis on a 1.0 % agarose gel and sent to Arizona State University Core Laboratories for purification and sequencing using the same forward and reverse PCR primers. AB1 sequence trace files were assembled using Geneious Prime® v. 2020.0.5 software, and a BLAST search was conducted for all resulting sequences to confirm accurate species identification. The sequences were deposited in GenBank and their accession numbers are shown in Table 2.
Table 2 .
Species of fungi used in phylogenetic analyses for this study, including host plant and country of origin.
| Species | Voucher/Isolate | Host | Country |
GenBank accession no.
|
||
|---|---|---|---|---|---|---|
| ITS | LSU | EF-1α | ||||
| Langdonia aristidae | H.U.V.19145 | Aristida urugayensis | Germany | AY740048 | AY740101 | n/a |
| HUV19145 | Aristida urugayensis | Argentina | JN367292 | JN367317 | JN367369 | |
| Langdonia aristidicola | BRIP 26930 | Aristida jerichoensis | Australia | HQ013091 | n/a | HQ013032 |
| Langdonia confusa | BRIP 42670 | Aristida queenslandica | Australia | HQ013095 | HQ013132 | n/a |
| BRIP 52755 | Aristida sp. | Australia | HQ013096 | n/a | n/a | |
| Langdonia fraseriana | BRIP 49668 | Aristida nitidula | Australia | HQ013100 | n/a | n/a |
| Langdonia inopinata | M-0215944 | Aristida adscensionis | Zambia | KY929612 | MF668632 | n/a |
| Langdonia walkerae | JK 5017 | Aristida beyrichiana | USA, SC | MT429291 | MT429279 | MT577017 |
| JK 6017 | Aristida beyrichiana | USA, SC | MT429292 | MT429280 | MT577018 | |
| SB 1218 | Aristida beyrichiana | USA, SC | MT429293 | MT429281 | MT577019 | |
| AGT 1218 | Aristida beyrichiana | USA, SC | MT429294 | MT429282 | MT577020 | |
| SRS 1218 | Aristida beyrichiana | USA, SC | MT429295 | MT429283 | MT577021 | |
| FLW 1017 | Aristida beyrichiana | USA, FL | MT429296 | MT429284 | MT577022 | |
| FL 1218 | Aristida beyrichiana | USA, FL | MT429297 | MT429285 | MT577023 | |
| CSN 1118 | Aristida stricta | USA, SC | MT429298 | MT429286 | MT577024 | |
| NC 1018 | Aristida stricta | USA, NC | MT429299 | MT429287 | MT577025 | |
| NC 2018 | Aristida stricta | USA, NC | MT429300 | MT429288 | MT577026 | |
| NC 3018 | Aristida stricta | USA, NC | MT429301 | MT429289 | MT577027 | |
| NC 5018 | Aristida stricta | USA, NC | MT429302 | MT429290 | MT577028 | |
| Macalpinomyces eragrostiellae | Ust. Exs. 960 | Eragrostiella bifaria | India | AY740036 | AY740089 | n/a |
| Melanotaenium euphorbiae | HUV17733 | Euphorbia heterophylla | Papua New Guinea | JN367289 | JN367314 | JN367365 |
| Sporisorium andropogonis | 56588 (M) | Bothriochloa saccharoides | Ecuador | AY740042 | AY740095 | n/a |
| Sporisorium exsertum | KVU965 | Themeda triandra | Australia | JN367293 | JN367318 | JN367370 |
| Sporisorium manilense | Ust.Exs.854 (M) | Sacciolepis indica | India | AY740059 | AY740112 | n/a |
| Sporisorium moniliferum | BRIP 52504 | Heteropogon contortus | Australia | HQ013104 | n/a | HQ01302 |
| Sporisorium ophiuri | HB 20 | Rottboellia cochinchinensis | Unknown | AY740019 | AJ236136 | n/a |
| Sporisorium sorghi | MP 2036a | Sorghum bicolor | Nicaragua | AY740021 | AF009872 | n/a |
| Sporisorium vermiculum | BRIP 49748 | Sorghum plumosum | Australia | HQ013114 | HQ013134 | HQ01307 |
| Sporisorium wynaadense | BRIP 27640 | Sarga leiocladum | Australia | HQ013116 | HQ013124 | HQ01309 |
| Ustilago bromivora | H.U.V.19322 | Bromus catharticus | Argentina | AY740064 | AY740118 | n/a |
| Ustilago hordei | Ust.exs.784 | Hordeum vulgare | Iran | AY345003 | AF453934 | n/a |
| Ustilago nuda | H.U.V.17782 | Hordeum leporinum | Greece | AY740069 | AJ236139 | n/a |
| Ustilago nunavutica | DAOM 91211 | Puccinellia angustata | Canada | KF381025 | KF381049 | n/a |
| Ustilago sparsa | Ust.exs.892 | Dactyloctenium aegyptium | India | AY345008 | DQ864974 | n/a |
| Ustilago striiformis | HAI 4610 | Milium effusum | Ukraine | KF381021 | KF381046 | n/a |
| Ustilago xerochloae | Ust. Exs. 1000 | Xerochloa imberbis | Australia | AY345012 | AY740150 | n/a |
Note: Species and accession numbers in bold are new sequences from this study.
Phylogenetic analysis
The phylogenetic analysis was conducted using Geneious Prime® v. 2020.0.5 software. The relationships among the isolates of Langdonia species sequenced in this study and other taxa in the Ustilaginaceae were inferred from a phylogenetic tree based on the ITS, LSU, and EF-1α data sets. The sequences for all taxa were first concatenated for each taxon and then aligned using the MUSCLE algorithm. The final data set contained sequences from 12 isolates collected in this study and 43 reference sequences obtained from GenBank (Table 2). Alignments were uploaded to NGphylogeny.fr (https://ngphylogeny.fr; Lemoine et al. 2019) and curated using BMGE (Criscuolo & Gribaldo 2010) to remove poorly aligned positions. The final super matrix contained 1 465 characters, including gaps.
The phylogenetic analysis was conducted using Bayesian inference and maximum likelihood; a GTR model with GAMMA distribution (Nylander 2004) was selected for both analyses. For Bayesian inference, MrBayes v. 3.2.6 was used to conduct a Markov Chain Monte Carlo (MCMC) (Huelsenbeck & Ronquist 2001). Four runs, each consisting of four chains, were conducted for 1 000 000 generations, and the cold chain was heated at a temperature of 0.25. Trees were sampled every 1 000 generations. The standard deviation of split frequencies was 0.01. RAxML v. 8 (Stamatakis 2014) was used for the maximum likelihood using a bootstrapping analysis. RAxML analyses were run with a rapid bootstrap analysis to search for the best-scoring likelihood tree using a random starting tree and 1 000 maximum likelihood bootstrap replicates. Trees were rooted using Melanotaenium euphorbiae (HUV17733). The final alignment and trees were deposited in TreeBASE 23819.
RESULTS
Growth on artificial medium
When teliospores were cultured on MEA+S, germ-tube formation occurred after 6 h incubation at 28 °C, and the germination rate increased steadily over a period of 2–3 d. Spore germination was the Ustilago-type, making a phragmobasisium on which ovoid sporidia (i.e., basidiospores) were produced. Colonies grew slowly; most colonies were visible after 3–5 d, and when subcultured, a colony reached 3–4 cm diam after 14 d. However, more than 50 % of the teliospores remained dormant throughout the incubation period. Young colonies first appeared pale luteous but later became ochreous with age. Colony shape was convex and irregular with undulate edges. At a later stage of growth, the colony surface in some isolates became wrinkled and formed cerebriform sectors (Ulloa & Hanlin 2000).
Phylogenetic analysis
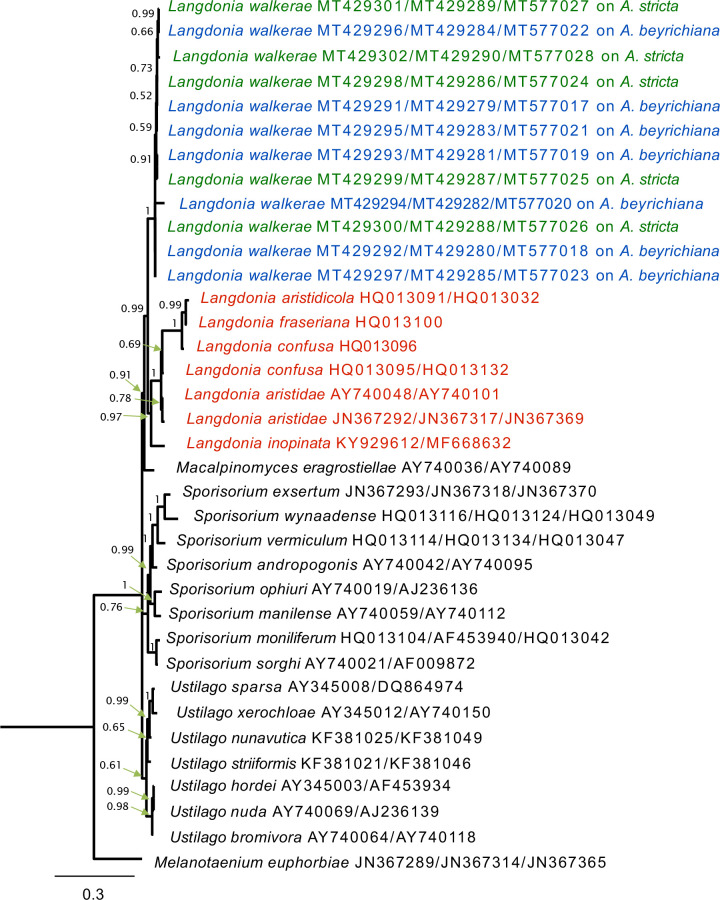
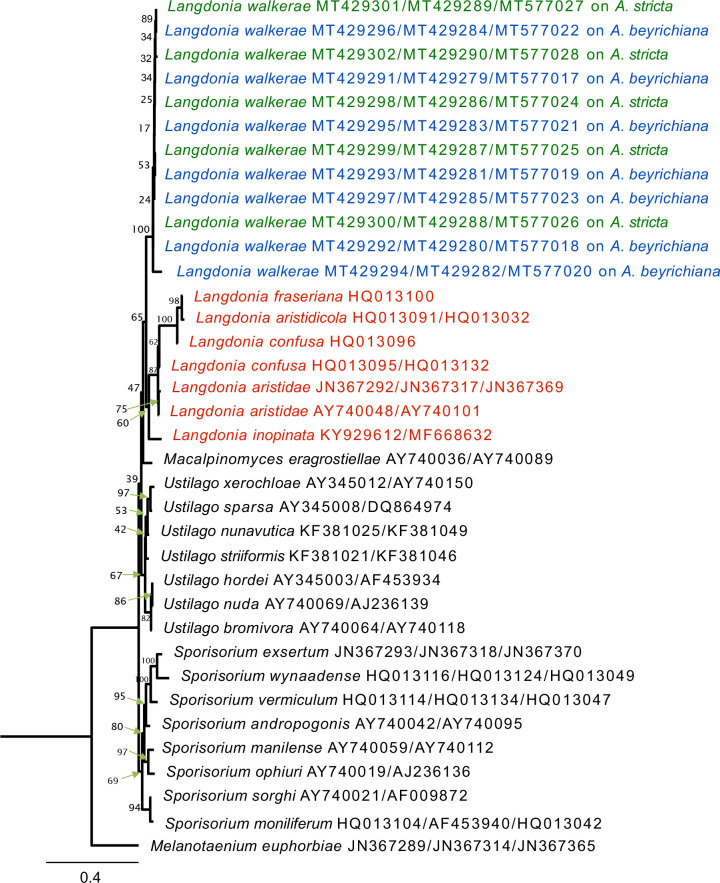
A total of 12 isolates from the culture were sequenced for two nuclear ribosomal DNA regions, ITS and LSU and one gene locus, EF-1α, and included in the final data set with 43 reference sequences obtained from GenBank. The list of taxa, their host plants and geographic sources, and the GenBank accession numbers for these sequences are shown in Table 2. The phylogenetic relationships between the isolates from A. stricta and A. beyrichiana exhibited full support to recognize them as one species based on the MrBayes and RAxML analyses, with values of 1.0 and 100, respectively. The topology of the MrBayes and RAxML trees (Figs 1, 2) indicate that the new species described below forms a highly supported sister group to other species of Langdonia as found from ITS+LSU+EF-1α analyses. Estimates for posteriori probabilities are indicated on branches of the tree (Fig. 1). In both analyses, isolates collected in this study clustered with values of 1.0 and 100 for MrBayes and RAxML, respectively (Figs 1, 2). The ITS+LSU+EF-1α dataset showed that the monophyly of these isolates within the Ustilaginaceae was fully supported. All taxa of Langdonia on Aristida spp. formed a well-supported monophyletic clade in the Ustilaginaceae and were well separated from the representatives of species in other genera of this family. Within this clade of Langdonia species, all isolates from both Aristida species clustered with strong support. In MrBayes and RAxML analyses of the alignment, Macalpinomyces eragrostiellae was sister to the clade of Langdonia species (Fig. 2).
Fig. 1.
Bayesian inference of phylogenetic relationships resulting from the analysis of ITS, LSU and EF-1α sequences. Numbers on the branches are estimates for Posterior Probability from Bayesian inference. The tree was rooted using Melanotaenium euphorbiae.
Fig. 2.
Maximum likelihood of phylogenetic relationships resulting from the analysis of ITS, LSU and EF-1α sequences. Numbers on the branches are estimates of bootstrap support values. The tree was rooted using Melanotaenium euphorbiae.
Taxonomy
Langdonia walkerae Alqurashi, J. Kerrigan & K. G. Savchenko, sp. nov. MycoBank MB 839451. Fig. 3A–F.
Fig. 3.

Sori and teliospores of Langdonia walkerae sp. nov. on Aristida beyrichiana (A–C) and on Aristida stricta (D–F). A. Sori (WSU 74240). B. Teliospores viewed with transmitted light. C. Scanning electron micrograph (SEM) of teliospores. D. Sori. E. Teliospores viewed with transmitted light. F. SEM of teliospores. Scale bars: A, D = 2 mm; B, E = 10 μm; C, F = 5 μm.
Etymology: Named after Dr. Joan L. Walker (USDA-Forest Service), a plant ecologist and conservationist who observed and contemplated the ecological significance of the sori in A. stricta and A. beyrichiana inflorescences as early as 1979 and brought that to our attention for study.
Typus: USA, South Carolina, Silver Bluff Audubon Center, on Aristida beyrichiana, 10 Nov. 2017, J. Kerrigan & A.S. Alqurashi, CLEMS 0080381, CLEMS 0080382 (holotype WSP 74240; isotype BPI 911222).
Sori in some ovaries, covered at first by a dark herbage green and later by a dark brick peridium. Sori are conspicuous, ovoid or symmetrically fusiform with an acute tip and sized between 1–1.5 × 2.5–7 mm. Teliospores are solitary, not compacted into spore balls, dark sienna in color. In the plane view, teliospores are globose, subglobose, or ellipsoidal and sized 8–13 μm diam. In the side view, teliospores are slightly flattened tangentially. Spore walls are uniformly thick, aculeate, and have dense conical spines (Ulloa & Hanlin 2000). Columella, spore balls, and sterile cells are lacking.
Host: Aristida beyrichiana (Poaceae).
Distribution: USA, southern part of South Carolina to southern Florida.
Additional specimens examined: On Aristida beyrichiana. USA, South Carolina, Silver Bluff Audubon Center, 6 Dec. 2018, A.S. Alqurashi & A.H. Alqurashi, CLEMS 0080383 & CLEMS 0080384; South Carolina, Aiken Gopher Tortoise Heritage Preserve, 6 Dec. 2018, A.S. Alqurashi & A.H. Alqurashi, CLEMS 0080385; South Carolina, Savannah River Site, 4 Dec. 2018, L. Lee, CLEMS 0080388, WSP 74241, BPI 911220; Florida, Apalachicola Bluffs and Ravines Preserve, 31 Oct. 2017, J. Walker, CLEMS 0080387; Florida, Austin Cary Forest, 3 Dec. 2018, J. Hong, CLEMS 0080386. On Aristida stricta. USA, North Carolina, North Carolina Sandhills Game Land, 27 Nov. 2018, A.S. Alqurashi, A.H. Alqurashi & B. Beck, CLEMS 0080389, WSP 74242, BPI 911221; South Carolina, Carolina Sandhills National Wildlife Refuge, 29 Nov. 2018, A.S. Alqurashi & A.H. Alqurashi, CLEMS 0080390, WSP 74243, BPI 911219.
Notes: Morphologically, Langdonia walkerae is distinguished from other species of Langdonia by teliospores that are not compacted into spore balls (McTaggart et al. 2012a, b). Phylogenetically, there is a strong support to recognize L. walkerae as a separate species.
DISCUSSION
Smut specimens from two species of wiregrass, A. stricta and A. beyrichiana, collected from longleaf pine ecosystems in three southeastern states — South Carolina, North Carolina, and Florida — were examined and compared in terms of morphology and DNA sequence data. Our findings determined both Aristida species were infected by the same pathogen, which was identified as a new species. Langdonia walkerae is designated as a new species based on its morphological and phylogenetic differences from the other described species of Langdonia. Only eight other species of Langdonia have been erected, including L. aristidae, L. aristidaria, L. aristidicola, L. clandestina, L. confusa, L. fraseriana, L. goniospora, and L. inopinata. All of these species have been reported from two or more Aristida species. For example, L. confusa, originally named Sporisorium confusum, was reported from several species of Aristida including A. dichotoma, A. fendleriana, A. spiciformis, and A. wrightii from several US states (Farr & Rossman 2020). In contrast, one host has been reported to be infected by different Langdonia species. For example, Aristida adscensionis is infected by Sporisorium aristidicola ≡ Langdonia aristidicola, S. consanguineum ≡ L. aristidae, and S. inopinatum ≡ L. inopinata in Zambia, India, and Zimbabwe, respectively (Farr & Rossman 2020). In the present study, sori were found in a proportion of the ovaries of A. stricta and A. beyrichiana. This is similar to four species of Langdonia, namely L. aristidaria (Durán 1987), L. aristidicola, L. clandestina and L. inopinata (Vánky 2012). In the other four species of Langdonia, namely L. confusa, L. aristidae, L. fraseriana and L. goniospora, the sori are produced in all ovaries of an inflorescence on a culm of the host plants (Vánky 2012).
Langdonia walkerae teliospores were solitary in all specimens examined on A. stricta and A. beyrichiana, and no spore balls were observed. This finding conflicts with that from McTaggart et al. (2012b), who considered this an apomorphic characteristic among the species of Langdonia. Based on our data, the genus Langdonia seems to be sharing the recent common ancestor with Macalpinomyces eragrostiellae, another species with well-defined spore balls. It is possible that the ancestors of L. walkerae have had spore balls, but they were lost in the process of species evolution.
The absence of spore balls leads us to infer that this fungus is a new species of Langdonia, and the genus description, which states that teliospores are compacted into spore balls (McTaggart et al. 2012b), needs to be emended. The size of teliospores is distinct from L. confusa and L. aristidae, both of which have larger teliospore diameters, and from L. aristidaria, L. aristidicola, L. clandestina, L. fraseriana, L. goniospora, and L. inopinata, all of which have smaller teliospore diameters (Durán 1987, Vánky 2012, Denchev et al. 2015). Furthermore, the teliospore wall ornamentation of L. walkerae is distinguishable from other described species by having dense conical spines whereas the teliospore wall ornamentation in other species of Langdonia varies from smooth to verruculose (Vánky 2012). For example, the wall ornamentation in Sporisorium confusum ≡ L. confusa on A. dichotoma was densely verrucose-echinulate and in S. consanguineum ≡ L. aristidae on A. arizonica, the teliospores wall were almost smooth to finely verruculose (Vánky 2012). Teliospore germination of L. walkerae is Ustilago-type, and columella and sterile cells are absent, fitting the description of the genus Langdonia. No data are available from previous studies to compare the culture growth of L. walkerae with other species of Langdonia.
Phylogenetic analysis demonstrated that all 12 isolates included in this study, five on A. stricta and seven on A. beyrichiana, form a highly supported monophylum, clustering in one separate phylogenetic clade within the Ustilaginaceae. These isolates were sister to other species of Langdonia and together formed a monophyletic group supporting the results of McTaggart et al. (2012b).
Although many species of Langdonia were reported on different species of Aristida in different regions around the world, only limited molecular data are available in GenBank for some of these species. Thus, additional sequences are needed for a more complete understanding of the phylogenetic relationships within this genus. The current study shows that L. walkerae on A. stricta and A. beyrichiana is clearly distinguishable morphologically and phylogenetically from other species of Langdonia on Aristida spp. More material from additional locations in longleaf pine ecosystems should be collected and analyzed to determine if L. walkerae occurs throughout the natural range of longleaf pine.
ACKNOWLEDGEMENTS
Research was supported by the USDA–FS. Technical Contribution No. 6910 of the Clemson University Experiment Station. This material is based upon work supported by NIFA/USDA, under project number SC-1700560. We thank the curators of Aiken Gopher Tortoise Heritage Preserve, Carolina Sandhills National Wildlife Refuge, North Carolina Sandhills Game Land, and Silver Bluff Audubon Center for permissions to sample. Special thanks to Brady Beck, the southern piedmont management biologist, for his extreme field assistance in collecting the samples. Also, thanks to Dr. Joan Walker, Jessica Hong and Linda Lee for providing samples from Apalachicola Bluffs and Ravines Preserve, Austin Cary Forest, and Savannah River Site, respectively. We thank Dayton Cash and George Wetzel at Clemson University electron microscopy facility. We also thank Karen Bryson, Andrew Gitto and Linus Schmitz for technical assistance and Xinyuan Ma for his assistance in molecular data analysis. Also, thanks to the curators of CLEMSON, BPI and WSP herbaria for their help in depositing the samples. We thank Drs Steven Jeffers, Guido Schnabel, and Joan Walker for their valuable reviews and suggestions for the manuscript.
Footnotes
Citation: Alqurashi AS, Kerrigan J, Savchenko KG (2021). Morphological and molecular characterization of Langdonia walkerae sp. nov. infecting Aristida stricta and A. beyrichiana in longleaf pine-grassland ecosystems in the southeastern USA. Fungal Systematics and Evolution 8: 39–47. doi: 10.3114/fuse.2021.08.04
Corresponding editor: P.W. Crous
Conflict of interest: The authors declare that there is no conflict of interest.
REFERENCES
- Barkworth ME, Capels KM, Long S, Michael B. (2003). Flora of North America, North of Mexico: 25, Part 2. Magnoliophyta: Commelinidae (in Part): Poaceae. Oxford University Press, NY, USA. [Google Scholar]
- Bauer R, Begerow D, Oberwinkler E. et al. (2001). Ustilaginomycetes. In: Systematic and Evolution (McLaughlin DJ, McLaughlin EG, Lemke PA, eds). The Mycota 7: Part B. Springer, Berlin, Heidelberg: 57–83. [Google Scholar]
- Begerow D, Bauer R, Oberwinkler F. (1997). Phylogenetic studies on nuclear large subunit ribosomal DNA sequences of smut fungi and related taxa. Canadian Journal of Botany 75: 2045–2056. [Google Scholar]
- Begerow D, John B, Oberwinkler F. (2004). Evolutionary relationships among β-tubulin gene sequences of basidiomycetous fungi. Part 222 of the series ‘Studies in Heterobasidiomycetes’. Mycological research 108: 1257–1263. [DOI] [PubMed] [Google Scholar]
- Begerow D, Stoll M, Bauer R. (2006). A phylogenetic hypothesis of Ustilaginomycotina based on multiple gene analyses and morphological data. Mycologia 98: 906–916. [DOI] [PubMed] [Google Scholar]
- Brockway DG, Outcalt KW, Wilkins RN. (1998). Restoring longleaf pine wiregrass ecosystems: plant cover, diversity and biomass following low-rate hexazinone application on Florida sandhills. Forest Ecology and Management 103: 159–175. [Google Scholar]
- Cerros-Tlatilpa R, Columbus JT, Barker NP. (2011). Phylogenetic relationships of Aristida and relatives (Poaceae, Aristidoideae) based on noncoding chloroplast (trnL-F, rpl16) and nuclear (ITS) DNA sequences. American Journal of Botany 98: 1868–1886. [DOI] [PubMed] [Google Scholar]
- Clewell AF. (1989). Natural history of wiregrass (Aristida strida Michx., Gramineae). Natural Areas Journal 1: 223–233. [Google Scholar]
- Criscuolo A, Gribaldo S. (2010). BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evolutionary Biology 10: 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denchev TT, Vorontsova MS, Denchev CM. (2015). First record of Langdonia aristidae (Ustilaginales) from Madagascar. Mycobiota 5: 15–20. [Google Scholar]
- Duever LC. (1989). Research priorities for the preservation, management, and restoration of wiregrass ecosystems. Natural Areas Journal 9: 214–218. [Google Scholar]
- Durán R. (1987). Ustilaginales of Mexico: Taxonomy, symptomatology, spore germination, and basidial cytology. Washington State University Press, Pullman, WA, USA. [Google Scholar]
- Farr DF, Rossman AY. (2020). Fungal Databases, systemic mycology and microbiology laboratory, ARS, USDA. Retrieved June 30, 2020, from https://nt.arsgrin.gov/fungaldatabases/
- Fill JM, Welch SM, Waldron JL. et al. (2012). The reproductive response of an endemic bunchgrass indicates historical timing of a keystone process. Ecosphere 3: 1–12. [Google Scholar]
- Frost C. (2007). History and future of the longleaf pine ecosystem. In: The longleaf pine ecosystem (Jose S, Jokela EJ, Miller DL, eds). Springer, NY, USA: 9–48. [Google Scholar]
- Huelsenbeck JP, Ronquist F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- Lemoine F, Correia D, Lefort V. et al. (2019). NGPhylogeny.fr: new generation phylogenetic services for non-specialists. Nucleic Acids Research 47: W260–W265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McTaggart AR, Shivas RG, Geering A. et al. (2012a). Soral synapomorphies are significant for the systematics of the Ustilago-Sporisorium-Macalpinomyces complex (Ustilaginaceae). Persoonia 29: 63–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McTaggart AR, Shivas RG, Geering ADW. et al. (2012b). Taxonomic revision of Ustilago, Sporisorium and Macalpinomyces. Persoonia 29: 116–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Means DB. (2007). Vertebrate faunal diversity of longleaf pine ecosystems. In: The longleaf pine ecosystem (Jose S, Jokela EJ, Miller DL, eds). Springer, NY, USA: 157–213. [Google Scholar]
- Noss RF. (1989). Longleaf pine and wiregrass: keystone components of an endangered ecosystem. Natural Areas Journal 9: 211–213. [Google Scholar]
- Nylander JAA. (2004). MrModeltest v2. (Programme distributed by the author). Evolutionary Biology Centre, Uppsala University, Sweden. [Google Scholar]
- Peet RK. (1993). A taxonomic study of Aristida stricta and A. beyrichiana. Rhodora 1: 25–37. [Google Scholar]
- Platt WJ. (1999). Southeastern pine savannas. In: Savannas, barrens, and rock outcrop plant communities of North America (Anderson RC, Fralish JS, Basking JM, eds). Cambridge University Press, Cambridge, UK: 23–51. [Google Scholar]
- Rayner RW. (1970). A mycological colour chart. British Mycological Society Press, UK. [Google Scholar]
- Savchenko KG, Carris LM, Castlebury LA. et al. (2014). Stripe smuts of grasses: one lineage or high levels of polyphyly? Persoonia 33: 169–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearman TM, Varner JM, Kreye JK. (2019). Pyrogenic flowering of Aristida beyrichiana following 50 years of fire exclusion. Ecosphere 10: e02541. [Google Scholar]
- Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoll M, Piepenbring M, Begerow D, Oberwinkler F. (2003). Molecular phylogeny of Ustilago and Sporisorium species (Basidiomycota, Ustilaginales) based on internal transcribed spacer (ITS) sequences. Canadian Journal of Botany 81: 976–984. [Google Scholar]
- Ulloa M, Hanlin RT. (2000). Illustrated dictionary of mycology. American Phytopathological Society. APS Press, St. Paul, MN, USA. [Google Scholar]
- Van Eerden BP. (1997). Studies on the reproductive biology of wiregrass (Aristida stricta Michx.) in the Carolina sandhills. M.S. thesis. University of Georgia, Athens, USA. [Google Scholar]
- Vánky K. (2012). Smut fungi of the world. The American Phytopathological Society. APS Press, St. Paul, MN, USA. [Google Scholar]
- Vánky K. (2013). Illustrated genera of smut fungi. The American Phytopathological Society. APS Press, St. Paul, MN, USA. [Google Scholar]
- Vilgalys R, Hester M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S. et al. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR protocols: A guide to methods and applications (Innis MA, Gelfand DH, Sninsky JJ, et al., eds). Academic Press, NY, USA: 315–322. [Google Scholar]