Abstract
Polyozellus and Pseudotomentella are two genera of closely related, ectomycorrhizal fungi in the order Thelephorales; the former stipitate and the latter corticioid. Both are widespread in the Northern Hemisphere and many species from both genera seem to be restricted to old growth forest. This study aimed to: a) identify genetic regions useful in inferring the phylogenetic relationship between Polyozellus and Pseudotomentella, b) infer this relationship with the regions identified and c) make any taxonomic changes warranted by the result. RPB2, mtSSU and nearly full-length portions of nrLSU and nrSSU were found to be comparatively easy to sequence and provide a strong phylogenetic signal. A STACEY species tree of these three regions revealed that Polyozellus makes Pseudotomentella paraphyletic. As a result, nearly all species currently placed in Pseudotomentella were recombined to Polyozellus. Pseudotomentella larsenii was found to be closer to Tomentellopsis than Polyozellus, but its placement needs further study and it was hence not recombined.
Keywords: Molecular systematics, new taxa, ribosomal tandem repeat, species tree, STACEY, Thelephorales
INTRODUCTION
Thelephorales is a large order of basidiomycetes with a vast number of undescribed species; according to Kirk et al. (2008) 269 described species belong to the order, but following the UNITE ITS sequence database it contains 4 305 Species Hypotheses (SHs), at 1.5 % minimum distance between sister species (Kõljalg et al. 2013, Nilsson et al. 2018).
Nearly all Thelephorales species are ectomycorrhizal, notable exceptions being Lenzitopsis, Odontia and possibly Amaurodon (Miettinen & Kõljalg 2007, He et al. 2019). Thelephorales often constitutes a large proportion of the ectomycorrhizal symbionts found on root tips in any ecosystem where such are present, and are hence important facilitators of forest and shrub growth in for example tundra, boreal and temperate ecosystems in large parts of the world (e.g. Kõljalg et al. 2000, Brundrett 2002, Sønstebø 2002, Taylor & Peterson 2005, Mühlmann & Peintner 2008, Ryberg et al. 2009, Bücking et al. 2012, Botnen et al. 2015). Some genera also form orchid mycorrhiza (e.g. Bidartondo et al. 2004, Jacquemyn et al. 2017).
Although most Thelephorales species are resupinate (Amaurodon, Odontia, Pseudotomentella, Tomentella, Tomentellopsis), some are stipitate hydnoid (Hydnellum, Phellodon, Sarcodon), stipitate poroid (Boletopsis), stipitate smooth (Thelephora) or cantharelloid (Polyozellus, Thelephora; Stalpers 1993, He et al. 2019). A few species are finger-like (Thelephora) and two are lamellate (Lenzitopsis; Stalpers 1993, He et al. 2019). A morphological feature common to all species, except possibly Amaurodon mustialaensis (fine ornamentation sometimes visible in SEM), is the verrucose to echinulate spores (Ginns 1989, Stalpers 1993, He et al. 2019).
Polyozellus was described by Murrill (1910) to accommodate a blackish, cantharelloid fungus, which up until then had been known as Cantharellus multiplex. The genus remained monotypic until Voitk et al. (2018) showed that the morphological concept of this species, Po. multiplex, comprised five molecularly distinct species, four of which were hence previously undescribed.
All presently described Polyozellus species have spathulate to funnel-shaped basidiomata, with a ridged underside, which usually fuse at the base to form irregularly rosette-shaped clumps (Voitk et al. 2018; Fig. 1). They overlap in colour and range from brown to blue, purple or black, depending on species and stage of maturity. The pilei of young specimens are woolly or hirsute on top but become smooth with age, while the hymenium has a consistently matte appearance. Their consistency is soft and brittle. Microscopically all Polyozellus species are similar and share the features of hyaline, inamyloid spores covered in irregular lobes and nodules, thread-like hyphidia and clamped hyphae. The hyphae have a bluish black pigment in the walls, which produces a bluish green solution in KOH. As a genus Polyozellus is easily recognised in the field and by a combination of macroscopical characters and spore size it is possible to distinguish between species. Two species with smaller spores, Po. multiplex and Po. atrolazulinus can be separated from three species with larger spores: Po. mariae, Po. marymargaretae and Po. purpureoniger (Voitk et al. 2018).
Fig. 1.

Basidiomata of species currently placed in Polyozellus and Pseudotomentella. A. Ps. mucidula. B. Ps. media. C. Ps. griseopergamacea. D. Ps. humicola. E. Po. multiplex. F. Po. mariae. Scale bars = 2 cm. Photos A–D. Urmas Kõljalg; E. Michael Burzynski; F. Andrus Voitk.
Based on ITS sequences, the small-spored Polyozellus species form separate phylogenetic clades, while the large-spored species comprise one clade (Voitk et al. 2018). The species have different but overlapping geographical distributions and range from North America to Asia but are not present in Europe (Voitk et al. 2018). The conservation statuses of the new species are not known but prior to their description Po. multiplex was considered a rare but locally abundant species in North America and a good indicator of old-growth forest (United States Forest Service 1994, Baroni 2017). It is also used in wool-dyeing and as a potential medicine against Alzheimer’s disease (Hwang et al. 1997).
Pseudotomentella was described by Svrček (1958), with the pale brown, corticioid species Ps. mucidula as type. Svrček (1958, 1960) described and recombined a further eight species into the genus. Larsen (1967a, b, 1968a, b, 1971a, b, 1974a, b, 1983) subsequently described an additional 13 Pseudotomentella species. Hjortstam (1970 Hjortstam (1974) recombined three of the species described by Svrček into Tomentellopsis and Kõljalg & Larsson (Kõljalg 1996) moved two into Amaurodon. Kõljalg (1996) also synonymised many of the species described by Larsen, and hence reduced the number of names to eight: Ps. atrofusca, Ps. flavovirens, Ps.griseopergamacea, Ps. humicola, Ps. mucidula, Ps. nigra, Ps. tristis and Ps. vepallidospora. Four species have later been described by various authors in separate publications: Ps. armata (Martini & Hentic 2002), Ps. larsenii (Kõljalg & Dunstan 2001), Ps. ochracea (Kõljalg & Larsson 1998) and Ps. rhizopunctata (Martini & Hentic 2003). Svantesson et al. (2019, 2021) described 12 new species from basidiomata previously identified as Ps. tristis, de-synonymised Ps. umbrina from the same and showed in a multi-gene phylogeny that these species form a clade sister to a clade containing Ps. rhizopunctata and Ps. atrofusca.
All Pseudotomentella species have corticioid, resupinate basidiomata, with a quite dense but soft and felt-like texture (Fig. 1). In similarity to Polyozellus, their hymenia have a matt appearance, but vary in colour from white to nearly black, past yellow, green, blue, purple, brown and grey. Microscopically, Pseudotomentella species display considerable variation: some species are simple-septate (e.g. Ps. flavovirens, Ps. griseopergamacea and Ps. mucidula), whereas others have clamped hyphae (e.g. Ps. humicola, the Ps. tristis group and Ps. vepallidospora); some species are monomitic (e.g. the core Ps. tristis group) and others are dimitic (e.g. Ps. humicola, Ps. mucidula and Ps. rhizopunctata); some species have dark-coloured spores (the Ps. tristis group) and others have hyaline spores (e.g. Ps. griseopergamacea, Ps. mucidula); a few species have chlamydospores (Ps. rhizopunctata and Ps. vepallidospora) but the rest do not. All species, however, have spores with bi- or trifurcate verrucae to echinuli. This feature in combination with the texture characteristics of their basidiomata makes Pseudotomentella readily recognisable under the microscope and to the trained eye they can be identified to species group already in the field. For identification to the level of species a combination of macro- and microscopical features is often needed (Kõljalg 1996, Kõljalg & Larsson 1998, Kõljalg & Dunstan 2001, Martini & Hentic 2002, 2003, Svantesson et al. 2019, 2021).
Pseudotomentella basidiomata are formed on the underside of dead wood, stones and turf, often very close to the ground (Kõljalg 1996, Svantesson et al. 2019). The spores of Tomentella sublilacina have been shown to be insect-dispersed (Lilleskov & Bruns 2005) and the similarity in spore-shape and growth habit of basidiomata indicate that this is likely the case for Pseudotomentella as well.
With the exception of Ps. larsenii, Pseudotomentella species are only naturally occurring in the Northern Hemisphere (Kõljalg 1996, Kõljalg & Larsson 1998, Kõljalg & Dunstan 2001, Martini & Hentic 2002, 2003, Svantesson et al. 2019, 2021). Less is known about their habitat than for Polyozellus, but many species are thought to only occur in old-growth forest. Svantesson et al. (2019) showed that all species in the Ps. tristis group, except the very widespread Ps. umbrina, are limited to ground with medium to high pH. Eight Pseudotomentella species are Red Listed in Sweden, three in Denmark and two in Estonia (Moeslund et al. 2019, Saar et al. 2019, SLU Artdatabanken 2020).
Vizzini et al. (2016) published an ITS phylogeny of Thelephorales indicating that, with the exception of Ps. ochracea and Ps. larsenii, Polyozellus and Pseudotomentella form a weakly supported clade together, while the clades of most other Thelephorales genera were strongly supported. Preliminary analyses based on ITS and partial LSU sequences for the current study also displayed a close phylogenetic relationship between the genera Polyozellus and Pseudotomentella. These have, however, in addition shown that the partial LSU region alone has too weak a signal to resolve this part of the Thelephorales tree, while the majority of ITS is too variable to be reliably aligned. The purpose of this article is to explore what genetic markers can resolve the relationship between Polyozellus and Pseudotomentella, establish this relationship with a multi-gene species tree more densely sampled for the taxa of interest and make any nomenclatural changes warranted by the conclusions.
MATERIALS AND METHODS
Taxon sampling
The ingroup consisted of all described species of Polyozellus and Pseudotomentella or representatives from already known clades of described species where such exist, except Ps. nigra, Ps. tenebrosa and the heterotypic synonyms of Ps. flavovirens, Ps. griseopergamacea and Ps. mucidula. The latter are so similar to the species with which they are currently synonymised that it is unclear whether they are separate species. This approach was taken in order to minimise costs and keep sequencing efforts within the time-frame set by a PhD project (pursued by the first author). The identities of Ps. nigra and its heterotypic synonym Ps. tenebrosa are unclear and will be addressed in a separate publication. The sampling hence included one species from each of the Polyozellus clades identified by Voitk et al. (2018), the eight Pseudotomentella species accepted by Kõljalg (1996), except for Ps. nigra, the four Pseudotomentella species subsequently published by various authors, as well as four species from the Ps. tristis group, as delimited by Svantesson et al. (2019). In addition, Tomentella italica was added to the dataset, since Tedersoo et al. (2016) suspected it of belonging to Pseudotomentella. Sequencing was attempted for the types of Ps. mucidula, and Ps. griseopergamacea. In cases (other than for Ps. nigra) where the phylogenetic identity of a species was unclear and several genetically different contenders existed for a name, a representative of each was included in the dataset and were referred to as e.g. Ps. cf. vepallidospora 1 and 2.
In order to further account for as yet undescribed Polyozellus and Pseudotomentella species when inferring the relationship between the two genera, the Compound Cluster function of the UNITE database was used (Kõljalg et al. 2013, Nilsson et al. 2018). Compound Clusters consist of DNA sequences with 80 % or less sequence similarity. The database was queried for sequences belonging to non-singleton Species Hypotheses at the 3 % level. The retrieved sequences were included in the dataset if their taxonomic identity was stated as Polyozellus or Pseudotomentella in UNITE, INSD or both and they were found to belong to other Compound Clusters than those containing sequences of formally described species (or contenders of such). To reveal the phylogenetic placement of the two focal genera two species (type species and one other) were selected from all other genera within Thelephorales. Two specimens of an undescribed species of Auricularia were chosen as outgroup, due to their easy sequenceability and inclusion in an earlier dataset (Table 1; see next subsection).
Table 1 .
DNA sequences included in the STACEY species tree analysis and their vouchers. GenBank (two-letter combination)/ENA (four-letter combination) numbers in boldface and italics indicate sequences generated for this study and as part of Wurzbacher et al. (2019), respectively.
| Species | Voucher | nrLSU 1st part | nrLSU 2nd part | nrSSU | RPB2 | mtSSU |
|---|---|---|---|---|---|---|
| Polyozellus atrolazulinus | TUF117559 | MT737307 | MT732081 | MT732090 | MT724777 | OK586800 |
| Polyozellus marymargaretae | TUF117347 | MT737308 | MT732082 | MT732089 | MT724778 | OK586801 |
| Polyozellus multiplex | TUF115322 | SAMEA4659525 | SAMEA4659525 | SAMEA4659525 | MT724779 | OK586802 |
| Pseudotomentella flavovirens | KHLarsson16727 | OK559566 | OK559602 | OK559682 | OK632648 | OK586803 |
| Pseudotomentella griseopergamacea | SSvantesson401 | SAMEA4659512 | SAMEA4659512 | SAMEA4659512 | OK632653 | OK586810 |
| Pseudotomentella cf. humicola 1 | SSvantesson345 | MK290724 | OK559582 | OK559681 | OK632649 | MK290650 |
| Pseudotomentella cf. humicola 2 | SSvantesson539 | OK559565 | — | OK559680 | OK632650 | OK586804 |
| Pseudotomentella larsenii | TUF100440 | MT737309 | MT732084 | MT732093 | OK632665 | OK586811 |
| Pseudotomentella cf. mucidula 1 | SSvantesson132 | SAMEA4659509 | SAMEA4659509 | SAMEA4659509 | OK632654 | OK586805 |
| Pseudotomentella cf. mucidula 2 | SSvantesson458 | OK559564 | OK559603 | OK559679 | OK632655 | OK586806 |
| Pseudotomentella cf. rhizopunctata 1 | EMartini 10413 | OK559563 | OK559601 | OK559678 | OK632668 | OK586807 |
| Pseudotomentella cf. rhizopunctata 2 | SSvantesson129 | MK290717 | OK559581 | OK559677 | OK632669 | MK290652 |
| Pseudotomentella sciastra | SSvantesson213 | SAMEA4659508 | SAMEA4659508 | SAMEA4659508 | OK632659 | OK586812 |
| Pseudotomentella tristis | SSvantesson193 | MK290679 | OK559580 | OK559676 | OK632658 | MK290662 |
| Pseudotomentella umbrina | SSvantesson351 | SAMEA4659515 | SAMEA4659515 | SAMEA4659515 | OK632667 | MK290659 |
| Pseudotomentella umbrinascens | SSvantesson335 | SAMEA4659496 | SAMEA4659496 | SAMEA4659496 | OK632666 | MK290670 |
| Pseudotomentella cf. vepallidospora 1 | SSvantesson456 | OK559562 | OK559605 | OK559675 | OK632651 | OK586808 |
| Pseudotomentella cf. vepallidospora 2 | SSvantesson493 | OK559561 | OK559604 | OK559674 | OK632652 | OK586809 |
| Amaurodon sumatranus | TUF115407 | SAMEA4659524 | SAMEA4659524 | SAMEA4659524 | OK632661 | OK586789 |
| Amaurodon viridis | TUF115739 | OK559560 | OK559555 | OK559673 | — | OK586790 |
| Boletopsis leucomelaena | MKrikorev140912 | MK602710 | OK559579 | OK559672 | OK632676 | — |
| Hydnellum ferrugineum | ELarsson312-16 | SAMEA4659497 | SAMEA4659497 | SAMEA4659497 | OK632673 | OK586794 |
| Hydnellum suaveolens | ELarsson8-14 | SAMEA4659503 | SAMEA4659503 | SAMEA4659503 | OK632672 | OK586795 |
| Lenzitopsis daii | HSYuan2959 | JN169795 | MT732078 | MT732087 | MT724774 | OK586796 |
| Lenzitopsis oxycedri | TUF115268 | SAMEA4659519 | SAMEA4659519 | SAMEA4659519 | MT724775 | OK586797 |
| Odontia ferruginea | TUF124098 | SAMEA4659527 | SAMEA4659527 | SAMEA4659527 | MT724776 | OK586798 |
| Odontia fibrosa | SSvantesson38 | SAMEA4659510 | SAMEA4659510 | SAMEA4659510 | OK632664 | OK586799 |
| Phellodon melaleucus | RGCarlsson160924 | SAMEA4659494 | SAMEA4659494 | SAMEA4659494 | OK632675 | — |
| Phellodon violascens | RGCarlsson14033 | SAMEA4659505 | SAMEA4659505 | SAMEA4659505 | OK632674 | OK586793 |
| Sarcodon imbricatus | ELarsson384-10 | SAMEA4659502 | SAMEA4659502 | SAMEA4659502 | OK632670 | OK586813 |
| Sarcodon squamosus | ELarsson248-12 | MK602767 | OK559578 | OK559671 | OK632671 | — |
| Thelephora palmata | ATaylor20136 | SAMEA4659526 | SAMEA4659526 | SAMEA4659526 | OK632647 | — |
| Thelephora terrestris | SSvantesson404 | SAMEA4659516 | SAMEA4659516 | SAMEA4659516 | OK632660 | OK586814 |
| Tomentella asperula | SSvantesson392 | OK559559 | OK559556 | OK559670 | OK632662 | OK586815 |
| Tomentella ferruginea | SSvantesson367 | SAMEA4659514 | SAMEA4659514 | SAMEA4659514 | OK632663 | OK586816 |
| Tomentellopsis echinospora | TUF110333 | SAMEA4659521 | SAMEA4659521 | SAMEA4659521 | MT724780 | OK586817 |
| Tomentellopsis zygodesmoides | TUF124075 | MT737311 | — | MT732091 | MT724781 | OK586818 |
| Auricularia sp. | ELarsson17030 | OK559557 | OK559606 | OK559668 | OK632657 | OK586791 |
| ELarsson17032 | OK559558 | OK559607 | OK559669 | OK632656 | OK586792 |
Molecular data
Seven genetic regions were targeted for DNA sequencing: nrLSU, nrSSU, β-tubulin, mtSSU, Tef1α, RPB1 and RPB2. All are unlinked except nrLSU and nrSSU. A majority of the nrLSU and nrSSU sequences were full-length and generated through Nanopore and PacBio sequencing, as part of Wurzbacher et al. (2019; Table 1). These sequences were complemented with sequences derived through Sanger sequencing. Approximately 2 500 bases were amplified from the nrLSU gene with the primers LR0R and LR7, LR7R and LR14 (Hopple & Vilgalys 1999); ca. 1 500 bases from the nrSSU gene with NS1 and NS4, NS3 and NS8 (White et al. 1990); ca. 500 bases from the β-tubulin gene with B36f and B12r (Nagy et al. 2011), ca. 600 bases from the Tef1α gene with EF983F and EF1567R (Rehner & Buckley 2005), ca. 700 bases from the mtSSU gene with MS1 and MS2 (White et al. 1990) and ca. 1 100 bases were amplified from the RPB2 gene with fRPB2-5F and bRPB2-7.1 (Liu et al. 1999, Matheny 2005). RPB1 sequences were obtained from whole genome sequencing by Tedersoo et al. (2016).
The primers used for sequencing were: for nrLSU - Ctb6 (Garbelotto et al. 1997) LR3R, LR5, LR7R, LR8R, LR9, LR14R (Hopple & Vilgalys 1999), for nrSSU - NS1, NS2, NS3, NS4 and NS8 (White et al. 1990), for β-tubulin - B36f and B12r, for Tef1α - EF983F and 1567R, for mtSSU - MS1 and MS2 and for RPB2 - bRPB2-7R, bRPB-6f and fRPB-5f (Matheny 2005).
The DNA sequences were assembled with Sequencher v. 5.4.6 (Gene Codes, Ann Arbor, MI, USA) and lodged in GenBank (Table 1). Alignments were made in AliView v. 1.18 (Larsson 2014), utilising the L-INS-i strategy, as implemented in MAFFT v. 7.017 (Katoh & Standley 2013). Introns and low-quality ends were manually trimmed from the sequences prior to analysis.
Molecular analyses
Gblocks v. 0.91b (Castresana 2000, Talavera & Castresana 2007) was used to trim the alignments of problematic character regions (e.g. missing data, saturated sites and sections with unclear homology). The program has not been evaluated for Bayesian inference (BI), which was employed in this study for generating gene and species trees, but for neighbour joining, parsimony and maximum likelihood (ML) methods. Although they are conceptually very different methods the results of ML and BI tend to be the most similar, and Gblocks was therefore run in the relaxed version of the program, as outlined in Talavera & Castresana (2007), which according to the same study is suitable for ML analysis of alignments created with MAFFT. The resulting alignments were unchanged in length for nrSSU, mtSSU, Tef1α and RPB1, 2 609 bases long for nrLSU (3 405 before) and 1 042 bases long for RPB2 (1 056 before).
RDP4 (Martin et al. 2015) was used to test for recombination. During a first round of testing the methods RDP, GENECONV, Chimaera and MaxChi were used and the significance level set to 0.01. Sequences with significant signs of recombination were submitted to a second round of testing that made use of all recombination methods. Any sequences with a positive result for more than two methods with p-values ≤ 10−5 in the second round were regarded as probable recombinants. Recombined sections of such sequences were removed from the alignments prior to further analysis.
In the phylogenetic analyses the following minimal partitions were assumed: nrLSU, nrSSU, mtSSU and for the protein-encoding genes: first, second and third positions. The automated best-fit tests implemented in PAUP v. 4.0a (Swofford 2002) were used to select optimal substitution models and optimal substitution model partitions. In agreement with the substitution models available in the BI programs used (BEAST v. 2 and STACEY) the tests evaluated models with three substitution schemes and equal or gamma-distributed among-site rate variation, based on BIC score. The partitioning result provided the best fit for keeping all minimal partitions separate, except first and second positions of Tef1α and RPB1. The substitution model GTR+G was output as the optimal model for all partitions except Tef1α and RPB1 first+second positions and RPB2 second positions. For these partitions the optimal models were JC+G, JC+G and K80+G, respectively.
To generate gene trees and assess their concordance prior to the species tree analysis BEAST v. 2.6.2 (Bouckaert et al. 2014, 2019) was used. The xml-files were prepared in the associated software BEAUti v. 2.6.2 (Bouckaert et al. 2014, 2019). The alignments were assigned the optimal partitions and substitution models output by PAUP v.4, but the substitution model was set to HKY+G for RPB2 second partitions, since it is the most similar model to K80+G available in the program. Test runs revealed that all trees retained the same topology if GTR+G was changed to HKY+G, but convergence was considerably faster. This change in substitution models was hence implemented, in order to ensure consistency with the ensuing species tree analysis, which is often slow to converge even under optimal circumstances and where enhanced speed is thus preferable. The trees of the partitions were set as linked inside each genetic linkage group but a separate clock model was assumed for each. The clock models were set as relaxed, lognormal, since all partitions had a coefficient of variation well above 0.1 (i.e. implying a relatively high rate variation among branches) in test runs. The clock rate of each partition was estimated in the run, using a lognormal prior with a mean set to 1 in real space. The growth rate prior was set to lognormal, with a mean of 5 and a standard deviation of 2. These priors were set according to the STACEY package documentation (Jones 2014). The Markov Chain Monte Carlo chains were run until analyses converged well in advance of the 10 % burn-in threshold, had ESS values well above 200 for all parameters, and satisfactory chain mixing, as assessed in Tracer v. 1.6.0 (Rambaut et al. 2014). After discarding the burn-in trees, maximum clade credibility trees were identified by TreeAnnotator v. 2.6.2 (Bouckaert et al. 2014, 2019).
A species tree inferred under the multispecies coalescent model was estimated in STACEY v. 1.2.5 (Jones 2017). Substitution and clock models as well as clock and growth rate priors were set the same as for the gene tree analyses. All individuals were assumed as minimal clusters. The Collapse Height prior was set to 10−5 and a lognormal prior with a mean of -7 and a standard deviation of 2 was set to the PopPriorScale parameter, as per the STACEY package documentation (Jones 2014). The length and result of the analysis was determined and summarized as for the gene trees. The phylograms were visually prepared in FigTree v. 1.4.4 (Rambaut 2012) and Inkscape v. 0.92.3. (https://inkscape.org)
RESULTS
Seven DNA regions were assessed for their functionality in inferring the relationship between the closely related genera Polyozellus and Pseudotomentella. Out of these four could be readily sequenced and had a serviceable phylogenetic signal: nrLSU, nrSSU, RPB2 and mtSSU (Table 1). The BEAST v. 2 gene tree analyses for RPB2, mtSSU and the combined nrDNA regions were run for 30 M, 10 M and 30 M generations, respectively. The gene trees were concordant for all supported nodes, with the exception of the relative placement of Odontia to Tomentella/Thelephora, and could hence be combined into a species tree analysis. The β-tubulin gene proved to be very hard to sequence and RPB1 was found to have a very weak phylogenetic signal. Sequencing of these genes was therefore discontinued, and they were not included in the species tree dataset. Tef1α was found to be paralogous above genus level and therefore unusable in this study. This result will be presented in full in a separate publication. The sequencing attempts of the types of Ps. griseopergamacea and Ps. mucidula were unsuccessful, as was the sequencing of most genetic regions for Ps. armata, Ps. ochracea, T. italica and Boletopsis grisea. They were hence excluded from the analyses. No sequences were found to be recombinants.
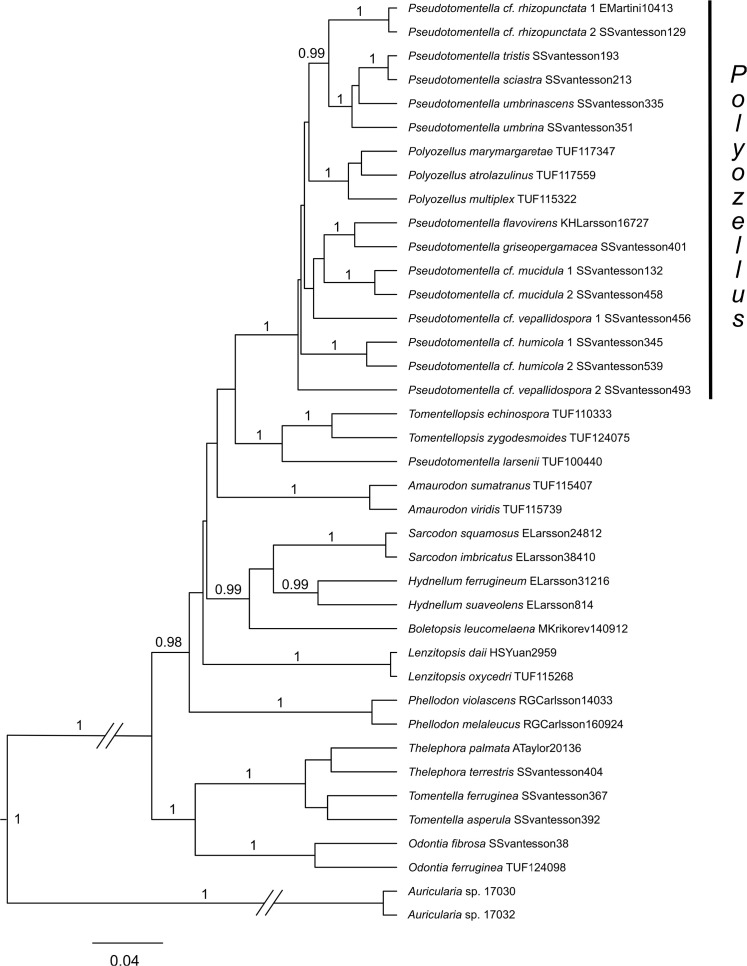
The STACEY species tree analysis was run for 500 M generations. Its resulting phylogram strongly supports Polyozellus as a monophyletic genus but not Pseudotomentella (Fig. 2). The three species of Polyozellus included in the analysis as representatives of three main clades of Polyozellus (see Materials and methods) were retrieved as a fully supported clade and so was the Ps. tristis group. These clades along with all other species of Pseudotomentella, except Ps. larsenii, were found to reside in a fully supported clade with little additional internal structure, i.e. a group corresponding to Polyozellus together with the thus paraphyletic Pseudotomentella. Pseudotomentella larsenii was retrieved as closely related to Tomentellopsis. The sequences of Ps. humicola, Ps. mucidula and Ps. rhizopunctata notably keep together in well-supported clades, while the sequences of Ps. vepallidospora do not.
Fig. 2.
STACEY species tree of Thelephorales with in-depth sampling of Polyozellus and Pseudotomentella, based on nrLSU, nrSSU, RPB2 and mtSSU alignments. Only posterior probability values ≥ 0.95 are shown.
The Polyozellus and Pseudotomentella species included in the analyses were found to belong in eight UNITE Compound Clusters: UCL8_013290, UCL8_012144, UCL8_002955, UCL8_010302, UCL8_012755, UCL8_007778, UCL8_007844, UCL8_008897 and UCL8_018843. Many of these clusters included large numbers of Species Hypotheses with non-singleton DNA-sequences not currently attributed to formally described species. Non-singleton DNA sequences of undescribed species identified as Polyozellus or Pseudotomentella in UNITE or INSD, belonging to other Compound Clusters than the formally described species were not retrieved.
In order to retain monophyly and uphold nomenclatural priority all Pseudotomentella species included in the analysis, except Ps. larsenii, are recombined to Polyozellus, together with close relatives of them within the Ps. tristis group (as shown by Svantesson et al. 2019, 2021). The genus description of Polyozellus is revised accordingly.
Taxonomy
Polyozellus Murrill, N. Amer. Fl. 9: 171. 1910, emend. Svantesson & Kõljalg
Type species: Polyozellus multiplex (Underw.) Murrill, N. Amer. Fl. 9: 171. 1910.
Basionym: Cantharellus multiplex Underw., Bull. Torrey Bot. Club 26: 254. 1899.
Description: Basidiomata annual, of two types:
1. Stipitate, multiple, complex, imbricately foliose; single pilei flabelliform to spathulate, sometimes funnel-shaped; terrestrial. Pileus surface downy to tomentose in active growth, becoming matt with concentric zonation or longitudinal ribbing; black, blue, purple, or brown; eventually glabrous and often shiny, darker in colour, often black. Hymenium composed of irregular, longitudinal, sinuous, anastomosing, decurrent folds, which can vary within basidiomata to be smooth, reticulate or almost poroid; various shades of blue, purple and grey, becoming darker with age. Stipe solid, fibrous, tapering downwards; often multiple, fused, converging to a common subterranean base; matt, scaly or shiny; blue, dark blue, dark purple, dark brown or black. Context soft, brittle, whitish, yellowish, pale grey, various shades of purple and blue or black. Odour faintly pungent, chemical, fruity, mildly sweetish or unremarkable. Taste not recorded for most species, one species (Po. atrolazulinus) mild. Spore deposit white. Basidiomata resistant to decay and often last over a month in the field.
2. Corticioid, resupinate, membranaceous, effused; mature parts continuous, immature parts discontinuous; with a soft, somewhat fibrous and elastic (cottony) texture when fresh and a similar or soft yet compact, fibrous and ± elastic texture when dried; on the underside of wood, stones and debris lying on the ground, common in the roofs of rodent burrows. Hymenium smooth, but sometimes strongly undulating; colour ranging from nearly white, past yellow, green, blue, purple, brown and grey to nearly black. Subiculum well developed, loose, often fibrous, with whitish, yellow, orange, brown or black colours; often forms the outer edge of basidiomata, extending beyond the hymenium. Odour and taste not recorded. Spore deposit white to brown.
Hyphal system monomitic or dimitic, hyphae simple-septate or clamped. Hyphal cords lacking or present. Subicular hyphae, when present, often thick-walled, forming a loose tissue, hyaline or with yellow, orange, brown or black colours. Subhymenial hyphae straight to somewhat sinuous, for some species interwoven and nodulose; thin to thick-walled; often forming a rather dense tissue; hyaline or pale green, yellow, orange or brown in KOH; in some species with a pigment in the walls which has a blue-green reaction in the presence of air and produces a similarly coloured solution; in some species amyloid. Encrustation present or absent, amorphous to granular; hyaline, or with green, orange, brown, purple or black colours in KOH, in some species sometimes dark blue green in the presence of air; when present occurring on the upper parts of subhymenial hyphae and on the lower parts of basidia. Basidia 4-sterigmate, occasionally 2-sterigmate; clavate, narrowly clavate or clavopedunculate, thin-walled, with 1–3 slight constrictions; sterigmata slightly curved; colours and reactions the same as for subhymenial hyphae, but in addition often with granular contents in KOH. Hyphidia present in some species; simple, filiform, not extending beyond basidia. Basidiospores in frontal face with a subcircular, subellipsoid or triangular basic shape; outline angular, nodulose, triangular, subcircular, subellipsoid, heart-shaped or cross-shaped; unlobed or with 3–7 lobes; lateral face with a subcircular, subellipsoid, ellipsoid or ovoid basic shape; outline evenly rounded, angular, lobed or nodulose; apiculus prominent to prolonged (possibly except for in Po. mariae); echinuli in most species long and prominent, bi- or trifurcate, sometimes singularly attached, in some species short and irregularly attached; colours and reactions the same as for subhymenial hyphae but often darker and reactions less frequent. Chlamydospores present or absent. Forming ectomycorrhiza.
Polyozellus abundilobus (Svantesson) Svantesson & Kõljalg, comb. nov. MycoBank MB 836134. UNITE SH: 1152984.08FU.
Basionym: Pseudotomentella abundiloba Svantesson, MycoKeys 50: 24. 2019.
Typus: Norway, Oslo (county), Oslo (municipality), Bygdøy, Hengsåsen, boreonemoral mixed forest on soil with high pH, 22 Sep. 2010, S. Svantesson (holotype O F110312).
Polyozellus alnophilus (Svantesson) Svantesson & Kõljalg, comb. nov. MycoBank MB 836135. UNITE SH: 1564303.08FU.
Basionym: Pseudotomentella alnophila Svantesson, MycoKeys 50: 26. 2019.
Typus: Norway, Buskerud, Ringerike, Juveren N, boreonemoral Alnus incana forest on soil with intermediate pH, 25 Sep. 2010, S. Svantesson & N. Svensson (holotype O F110313).
Polyozellus alobatus (Svantesson) Svantesson & Kõljalg, comb. nov. MycoBank No.: MB 836136. UNITE SH: 1230089.08FU.
Basionym: Pseudotomentella alobata Svantesson, MycoKeys 50: 29. 2019.
Typus: Sweden, Dalsland, Mellerud, Skållerud, Norgekullen SW, coniferous forest on soil with high pH, 20 Sep. 2017, S. Svantesson 425 (holotype GB).
Polyozellus atrofuscus (M.J. Larsen) Svantesson & Kõljalg, comb. nov. MycoBank MB 836137. UNITE SH: 1230079.08FU.
Basionym: Pseudotomentella atrofusca M.J. Larsen, Bull. Torrey Bot. Club 98: 39. 1971.
Typus: USA, Arizona, Fort Valley, Coconino Co., on Pinus ponderosa, 21 Sep. 1967, R.L. Gilbertson 7553 (holotype ARIZ; isotype: SSMF 685–4578).
Polyozellus badjelanndanus (Svantesson) Svantesson & Kõljalg, comb. nov. MycoBank MB 841511. UNITE SH: 1564288.08FU.
Basionym: Pseudotomentella badjelanndana Svantesson, Phytotaxa 497: 69. 2021.
Typus: Sweden, Lule Lappmark, Jokkmokk, Oarjep Slahpetjåhkkå, middle alpine Dryas octopetala heath on ground with high pH, on underside of stones and Dryas twigs, 18 Aug. 2016, S. Svantesson 303 (holotype GB).
Polyozellus flavovirens (Höhn. & Litsch.) Svantesson & Kõljalg, comb. nov. MycoBank MB 836138. UNITE SH: 1184825.08FU.
Basionym: Tomentella flavovirens Höhn. & Litsch., Sitzungsber. Kaiserl. Akad. Wiss., Wien. Math.-Naturwiss. Cl., Abt. 1 116: 831. 1907.
Typus: Germany, Braunlage am Harz, auf nackter Erde [= on bare soil], Lindau, (holotype FH [v. Höhnel herb., sheet 1979]).
Polyozellus griseopergamaceus (M.J. Larsen) Svantesson & Kõljalg, comb. nov. MycoBank MB 836139. UNITE SH: 1184823.08FU.
Basionym: Pseudotomentella griseopergamacea M.J. Larsen, Bull. Torrey Bot. Club 98: 38. 1971.
Typus: USA, New York, Highland Forest, Fabius P.O., Onondaga Co., on Pinus resinosa, 21 Oct. 1961, R. L. Gilbertson 3096 (holotype BPI; isotype: SSMF 695–4961).
Polyozellus humicola (M.J. Larsen) Svantesson & Kõljalg, comb. nov. MycoBank MB 836140. UNITE SH: 1236694.08FU, —.
Basionym: Pseudotomentella humicola M.J. Larsen, Mycologia 60: 547. 1968.
Typus: Canada, Ontario, Algonquin Park, Opeongo Lake, on Thuja occidentalis, 18 Sep. 1939, R. F. Cain (holotype SSMF 8868; isotypes: TRTC 44362 and BPI).
Polyozellus medius (Svantesson & Kõljalg.) Svantesson & Kõljalg, comb. nov. MycoBank MB 836141. UNITE SH: 1185287.08FU.
Basionym: Pseudotomentella media Svantesson & Kõljalg, MycoKeys 50: 33. 2019.
Typus: Estonia, Valga, Otepää, Trommi, 12 Sep. 2012, U. Kõljalg (holotype TUF 115609).
Polyozellus mucidulus (P. Karst.) Svantesson & Kõljalg, comb. nov. MycoBank MB 836142. UNITE SH: 1234281.08FU, 1234278.08FU.
Basionym: Hypochnus mucidulus P. Karst., Bidrag Kännedom Finlands Natur Folk 37: 163. 1882.
Typus: Finland, Haarankorpi, in ligno mucido, 09 Oct. 1878, P. A. Karsten (lectotype H).
Polyozellus pinophilus (Svantesson) Svantesson & Kõljalg, comb. nov. MycoBank MB 836143. UNITE SH: 1185292.08FU.
Basionym: Pseudotomentella pinophila Svantesson, MycoKeys 50: 36. 2019.
Typus: Sweden, Småland, Jönköping, Svarttorp, Ramlaklint, boreonemoral, mixed, old-growth forest, on soil with intermediate pH, 12 Sep. 2016, S. Svantesson 358 (holotype GB).
Polyozellus plurilobus (Svantesson) Svantesson & Kõljalg, comb. nov. MycoBank MB 836144. UNITE SH: 1185290.08FU.
Basionym: Pseudotomentella pluriloba Svantesson, MycoKeys 50: 39. 2019.
Typus: Finland, Uusimaa, Loviisa, Rutosinpyhtää, Marinkylä, rotten trunk on the ground (Picea), 30 Sep. 2010, U. Söderholm 4263 (holotype H 6018127).
Polyozellus rhizopunctatus (E.C. Martini & Hentic) Svantesson & Kõljalg, comb. nov. MycoBank MB 836148. UNITE SH: 1230075.08FU, 1230078.08FU.
Basionym: Pseudotomentella rhizopunctata E.C. Martini & Hentic, Bull. Trimestriel Soc. Mycol. France 119: 20. 2003.
Typus: Switzerland, canton Tessin, Someo, sur écorce d’une branche de Pinus sylvestris, 14 Nov. 1998, E. Zenone em-6886 (holotype PC; isotype: LUG).
Polyozellus rotundisporus (Svantesson) Svantesson & Kõljalg, comb. nov. MycoBank MB 836149. UNITE SH: 1185296.08FU.
Basionym: Pseudotomentella rotundispora Svantesson, MycoKeys 50: 41. 2019.
Typus: Sweden, Västergötland, Götene, Medelplana, Eriksberg, boreonemoral, mixed forest on soil with high pH, 17 Oct. 2016, S. Svantesson 413 (holotype GB).
Polyozellus sciastrus (Svantesson & Kõljalg.) Svantesson & Kõljalg, comb. nov. MycoBank MB 836150. UNITE SH: 1230076.08FU.
Basionym: Pseudotomentella sciastra Svantesson & Kõljalg, MycoKeys 50: 44. 2019.
Typus: Sweden, Småland, Jönköping, Svarttorp, Ramlaklint, boreonemoral, mixed, old-growth forest, on soil with intermediate pH, 12 Sep. 2016, S. Svantesson 359 (holotype GB).
Polyozellus sorjusensis (Svantesson) Svantesson & Kõljalg, comb. nov. MycoBank MB 841513. UNITE SH: 1185284.08FU.
Basionym: Pseudotomentella sorjusensis Svantesson, Phytotaxa 497: 71. 2021.
Typus: Sweden, Lule Lappmark, Jokkmokk, Sårjås N, low alpine heath on ground with intermediate pH, on underside of stone, 17 Aug. 2016, S. Svantesson 298 (holotype GB).
Polyozellus tristis (P. Karst.) Svantesson & Kõljalg, comb. nov. MycoBank MB 836151. UNITE SH: 1230077.08FU.
Basionym: Hypochnus subfuscus subsp. tristis P. Karst., Meddeland. Soc. Fauna Fl. Fenn. 9: 71. 1883.
Typus: Finland, Tavastia australis [= Etelä-Häme], Tammela, Mustiala, ad Betulam, 19 Aug. 1865, P. A. Karsten (lectotype H 6018703 [Herbarium P. A. Karsten 3036]); epitype: Sweden, Västerbotten, Vännäs, Orrböle, boreal, mixed forest on soil with high pH, 28 Aug. 2015, S. Svantesson 193 (GB).
Polyozellus tristoides (Svantesson & K.H. Larss.) Svantesson & Kõljalg, comb. nov. MycoBank MB 836152. UNITE SH: 1230081.08FU.
Basionym: Pseudotomentella tristoides Svantesson & K.H. Larss., MycoKeys 50: 52. 2019.
Typus: Norway, Nord-Tröndelag, Snåsa, Bergsåsen, boreal, deciduous forest on soil with intermediate pH, 28 Aug. 2012, K.-H. Larsson (holotype O F110306).
Polyozellus umbrinus (Fr.) Svantesson & Kõljalg, comb. nov. MycoBank MB 836153. UNITE SH: 1185280.08FU.
Basionym: Thelephora umbrina Fr., Elench. fung. 1: 199. 1828, nom. sanct.
Typus: Sweden, Småland, Femsjö, E. Fries (neotype UPS F003106 [Herb. Fries]); epitype: Sweden, Småland, Hylte, Femsjö, Femsjö Church Nature Reserve, boreonemoral, mixed forest on soil with intermediate pH, 7 Sep. 2016, S. Svantesson 351 (GB).
Polyozellus umbrinascens (Svantesson) Svantesson & Kõljalg, comb. nov. MycoBank MB 836154. UNITE SH: 1185297.08FU.
Basionym: Pseudotomentella umbrinascens Svantesson, MycoKeys 50: 60. 2019.
Typus: Sweden, Bohuslän, Tanum (municipality), Tanum (parish), Greby Kleva, boreonemoral, deciduous forest on soil with high pH, RT90: E1236840, N6518916, 6 Sep. 2016, S. Svantesson 335 (holotype GB).
Polyozellus vepallidosporus (M.J. Larsen) Svantesson & Kõljalg, comb. nov. MycoBank MB 836155. UNITE SH: 1191930.08FU, SH1244119.08FU.
Basionym: Pseudotomentella vepallidospora M.J. Larsen, Canad. J. Bot. 45: 1299. 1967.
Typus: USA, Washington, Quinault, Olympic Peninsula, on rotten conifer log, 15 Oct. 1958, J. L. Lowe 10368 (holotype BPI; isotype: SYRF).
DISCUSSION
Among homobasidiomycetes there is a general evolutionary trend from structurally simple, corticioid basidiomata with flat hymenia to more complex, stipitate forms with gills, tubes, etc., occasionally followed by reversions to simpler forms (Hibbett & Binder 2002, Larsson et al. 2004). Most of these transitions occurred early during fungal evolution and the complex forms have since become well separated from the lineages that have retained a simple basidiome morphology. However, in a few cases species with complex basidiome forms occur nested within the same genus as species with simple forms, for example in Trechispora, as currently circumscribed (Ryvarden 2002, Meiras-Ottoni et al. 2021) and in Thelephorales seemingly also in Tomentella/Thelephora (Vizzini et al. 2016). Here it is documented for Polyozellus/Pseudotomentella.
The authors choose to recombine the type species of Pseudotomentella along with most other species previously placed in the genus to Polyozellus, thus delimiting the latter as a monophyletic genus including species with both stipitate and resupinate basidiomata. Two other alternatives would have been possible: either retaining Pseudotomentella, in the knowledge that is a paraphyletic genus with regards to Polyozellus or keeping both monophyletic and describing many small genera. The first option was not viewed as a fitting solution to the present situation, given the evolutionary reality of basidiome transitions among homobasidiomycetes and the fact that paraphyletic genera are inadvisable in general. A more acceptable way would perhaps have been to create many small genera. However, as already implied genera are a subjective, human construct, to ease our understanding of the relationship between species. In this context it is the view of the authors that it is easier to handle a genus with two distinct but readily identifiable morphologies than to deal with seven genera, where many display small and often overlapping morphology (e.g. the two different contenders for the name Po. vepallidospora). Given additional molecular data from currently undescribed species, this view might change but given that no undescribed species were retrieved from any UNITE Compound Clusters other than those containing the formally described Polyozellus and Pseudotomentella species included in this study, it does not currently seem likely that major changes to the present phylogeny would arise with the inclusion of such.
Four DNA regions, RPB2, mtSSU and the combined nrLSU and nrSSU, were found to be usable in delimiting Polyozellus vs. Pseudotomentella. Additional regions would have been preferable to account for e.g. incomplete lineage sorting, but given the lack of conflict with respect to the ingroup the current dataset was deemed sufficient for the purpose of this study. The ease of sequencing and high information content of the regions used make them good candidates for further systematic studies within Thelephorales. In addition, this result further testifies to the usability of the full tandem repeat as a molecular marker for fungi, especially in cases where the ITS region is too variable to be reliably aligned but the partial LSU region normally used contains too little information to infer a satisfyingly resolved phylogeny (e.g. Krehenwinkel et al. 2019, Wurzbacher et al. 2019, Bradshaw et al. 2020).
The precise phylogenetic identities of Po. humicola, Po. mucidula, Po. rhizopunctata and Po. vepallidospora are currently unknown. Given the unsuccessful sequencing efforts of the types of Po. griseopergamacea and Po. mucidula, epitypification or sequencing with other methods are ways of resolution that should be explored. However, since the several available candidates for these four species were all shown to belong in Polyozellus, as here delimited, lack of knowledge about their definite identity does not pose a problem for their recombination.
The phylogenetic meaning of the heterotypic names currently synonymised with Po. flavovirens, Po. griseopergamacea and Po. mucidula is also unclear. Many of these species, e.g. Pseudotomentella fumosa, Ps. kaniksuensis, and Ps. molybdea, are very similar to the species currently given nomenclatural priority and are therefore unlikely not to belong in Polyozellus. Even so, their possible recombination needs clarification of their status as separate species.
Pseudotomentella larsenii is clearly more closely related to Tomentellopsis than to Polyozellus and is hence not recombined into the latter. This decision means that Polyozellus remains a genus restricted to the Northern Hemisphere. The possible inclusion of Ps. larsenii in Tomentellopsis needs to be addressed by further taxon sampling.
Pseudotomentella armata, Ps. ochracea and T. italica could not be included in the analyses, since sequencing of several gene regions failed. Their phylogenetic placement thus remains to be revealed by further studies.
Concerning conservation, Pseudotomentella is a larger genus than Polyozellus and the recombination of its species into the latter hence warrants more name changes than if the opposite were to be pursued. However, since Polyozellus is both older and has a wider usage – as a food source and indicator of old-growth forest, in dyeing etc. – this was not pursued.
It is clear that the identities of many members of what was formerly Pseudotomentella, and now constitute corticioid Polyozellus species, are deficiently known and will need further study in order to be clarified. Many of these species are rare and probably threatened by extinction due to loss of old growth forest. It is therefore the hope of the authors that their phylogenetically merited incorporation in Polyozellus will bring renewed interest to them and thereby facilitate the amount of attention and study that they deserve.
ACKNOWLEDGEMENTS
Funding for this study was received from The Swedish Taxonomy Initiative (2014-152 4.3), the Norwegian Biodiversity Information Centre (ADB54-09), Stiftelsen Lars Hiertas Minne, Helge Ax:son Johnsons Stiftelse and Kungl. Vetenskaps- och Vitterhets-Samhället i Göteborg. Urmas Kõljalg and Irja Saar were supported by the European Regional Development Fund (Centre of Excellence EcolChange) and the Estonian Research Council (PRG1170). Andrus Voitk and Michael Burzynski are sincerely thanked for the loan of Polyozellus pictures and the curators of herbaria BPI and H for granting and arranging loans.
Footnotes
Citation: Svantesson S,, Kõljalg U, Wurzbacher C, Saar I, Larsson K-H, Larsson E (2021). Polyozellus vs. Pseudotomentella: generic delimitation with a multi-gene dataset. Fungal Systematics and Evolution 8: 143–154. doi: 10.3114/fuse.2021.08.11
Corresponding editor: P.W. Crous
Conflict of interest: The authors declare that there is no conflict of interest.
REFERENCES
- Baroni TJ. (2017). Mushrooms of the Northeastern United States and Eastern Canada. Timber Press, USA. [Google Scholar]
- Bidartondo MI, Burghardt B, Gebauer G, et al. (2004). Changing partners in the dark: isotopic and molecular evidence of ectomycorrhizal liaisons between forest orchids and trees. Proceedings of the Royal Society B: Biological Sciences 271: 1799–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botnen S, Kauserud H, Carlsen T, et al. (2015). Mycorrhizal fungal communities in coastal sand dunes and heaths investigated by pyrosequencing analyses. Mycorrhiza 25: 447–456. [DOI] [PubMed] [Google Scholar]
- Bouckaert R, Heled J, Kühnert D, et al. (2014). BEAST 2: A software platform for Bayesian evolutionary analysis. PLoS Computational Biology 10: e1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert R, Vaughan TG, Barido-Sottani J, et al. (2019). BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Computational Biology 15: e1006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw M, Grewe F, Thomas A, et al. (2020). Characterizing the ribosomal tandem repeat and its utility as a DNA barcode in lichen-forming fungi. BMC Evolutionary Biology 20: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brundrett MC. (2002). Tansley Review No. 134: Coevolution of Roots and Mycorrhizas of Land Plants. New Phytologist 154: 275–304. [DOI] [PubMed] [Google Scholar]
- Bücking H, Liepold E, Ambilwade P. (2012). The Role of the Mycorrhizal Symbiosis in Nutrient Uptake of Plants and the Regulatory Mechanisms Underlying These Transport Processes. In: Plant Science (Dhal NK, Sahu SC, eds.) IntechOpen, Croatia: 107–138. [Google Scholar]
- Castresana J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution 17: 540–552. [DOI] [PubMed] [Google Scholar]
- Garbelotto MM, Lee HK, Slaughter G, et al. (1997). Heterokaryosis is not required for virulence of Heterobasidion annosum. Mycologia 89: 92–102. [Google Scholar]
- Ginns J. (1989). Descriptions and notes for some unusual North American corticioid fungi (Aphyllophorales, Corticiaceae). Memoirs of the New York Botanical Garden 49: 129–137. [Google Scholar]
- He MQ, Zhao RL, Hyde KD, et al. (2019). Notes, outline and divergence times of Basidiomycota. Fungal Diversity 99: 105–367. [Google Scholar]
- Hibbett DS, Binder M. (2002). Evolution of complex fruiting-body morphologies in homobasidiomycetes. Proceedings of the Royal Society B: Biological Sciences 269: 1963–1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjortstam K. (1970). Studies in the Swedish species of the genus Tomentella (Thelephoraceae). II. Svensk Botanisk Tidskrift 64: 421–428. [Google Scholar]
- Hjortstam K. (1970). Studies in the Swedish species of the genus Tomentella (Thelephoraceae). III. The genus Tomentellopsis. Svensk Botanisk Tidskrift 68: 51–56. [Google Scholar]
- Hopple JS, Jr, Vilgalys R. (1999). Phylogenetic relationships in the mushroom genus Coprinus and dark spored allies based on sequence data from the nuclear gene coding for the large ribosomal subunit RNA: divergent domains, outgroups, and monophyly. Molecular Phylogenetics and Evolution 13: 1–19. [DOI] [PubMed] [Google Scholar]
- Hwang J-S, Song K-S, Kim W-O, et al. (1997). Polyozellin, a new inhibitor of prolyl endopeptidase from Polyozellus multiplex. The Journal of Antibiotics 50: 773–777. [DOI] [PubMed] [Google Scholar]
- Jacquemyn H, Duffy KJ, Selosse MA. (2017). Biogeography of Orchid Mycorrhizas. In: Biogeography of Mycorrhizal Symbiosis. Ecological Studies (Analysis and Synthesis), vol. 230 (Tedersoo L, ed.). Springer, Switzerland: 159–177. [Google Scholar]
- Jones G. (2014). STACEY package documentation: species delimitation and species tree estimation with BEAST2. http://www.indriid.com/software.html
- Jones G. (2017). Algorithmic improvements to species delimitation and phylogeny estimation under the multispecies coalescent. Journal of Mathematical Biology 74: 447–467. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. (2013). MAFFT multiple sequence alignment software version 7, improvements in performance and usability. Molecular Biology and Evolution 30: 772–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk PM, Cannon PF, Minter DW, et al. (2008). Ainsworth & Bisby’s Dictionary of the Fungi. 10th edn. CABI Europe, UK. [Google Scholar]
- Kõljalg U. (1996). Tomentella (Basidiomycota) and related genera in Temperate Eurasia. Synopsis Fungorum 9: 1–213. [Google Scholar]
- Kõljalg U, Larsson E. (1998). Pseudotomentella ochracea sp. nov., based on morphological and molecular data. Folia Cryptogamica Estonica 33: 53–56. [Google Scholar]
- Kõljalg U, Dahlberg A, Taylor AF, et al. (2000). Diversity and abundance of resupinate thelephoroid fungi as ectomycorrhizal symbionts in Swedish boreal forests. Molecular Ecology 9: 1985–1996. [DOI] [PubMed] [Google Scholar]
- Kõljalg U, Dunstan W. (2001). Pseudotomentella larsenii sp. nov. (Thelephorales), a common ectomycorrhiza former in dry eucalypt woodland and forests of Western Australia. Harvard Papers in Botany 6: 123–130. [Google Scholar]
- Kõljalg U, Nilsson RH, Abarenkov K, et al. (2013). Towards a unified paradigm for sequence-based identification of Fungi. Molecular Ecology 22: 5271–5277. [DOI] [PubMed] [Google Scholar]
- Krehenwinkel H, Pomerantz A, Henderson JB, et al. (2019). Nanopore sequencing of long ribosomal DNA amplicons enables portable and simple biodiversity assessments with high phylogenetic resolution across broad taxonomic scale. GigaScience 8: giz006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen MJ. (1967a). Tomentella and related genera in North America: III. New species of Tomentella and Pseudotomentella. Canadian Journal of Botany 45: 1297–1307. [Google Scholar]
- Larsen MJ. (1967b). Tomentella and related genera in North America V. New North American records of tomentelloid fungi. Mycopathologia et Mycologia Applicata 32: 37–67. [Google Scholar]
- Larsen MJ. (1968). A new species of Pseudotomentella from North America. Mycologia 60: 547–552. [Google Scholar]
- Larsen MJ. (1971a) [1972]. The genus Pseudotomentella (Basidiomycetes, Thelephoraceae s. str.). Nova Hedwigia 22: 599–619. [Google Scholar]
- Larsen MJ. (1971b). Notes on tomentelloid fungi III. New species of Pseudotomentella. Bulletin of the Torrey Botanical Club 98: 38–41. [Google Scholar]
- Larsen MJ. (1974). Some notes on Pseudotomentella. Mycologia 66: 165–168. [Google Scholar]
- Larsen MJ. (1983). Notes on tomentelloid fungi V. Additional new species of Pseudotomentella. Mycologia 75: 556–562. [Google Scholar]
- Larsson A. (2014). AliView: a fast and lightweight alignment viewer and editor for large data sets. Bioinformatics 30: 3276–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson KH, Larsson E, Kõljalg U. (2004). High phylogenetic diversity among corticioid homobasidiomycetes. Mycological Research 108: 983–1002. [DOI] [PubMed] [Google Scholar]
- Lilleskov EA, Bruns TD. (2005). Spore dispersal of a resupinate ectomycorrhizal fungus, Tomentella sublilacina, via soil food webs. Mycologia 97: 762–769. [DOI] [PubMed] [Google Scholar]
- Liu YL, Whelen S, Hall BD. (1999). Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Molecular Biology and Evolution 16: 1799–1808. [DOI] [PubMed] [Google Scholar]
- Martin DP, Murrell B, Golden M, et al. (2015). RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evolution 1: vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini EC, Hentic R. (2002). Deux nouvelles espèces de champignons tomentelloides. Bulletin trimestriel de la Société Mycologique de France 118: 79–90. [Google Scholar]
- Martini EC, Hentic R. (2003). Pseudotomentella rhizopunctata sp. nov., une nouvelle espèce de champignon tomentelloïde chlamydosporée. Bulletin trimestriel de la Société Mycologique de France 119: 19–29. [Google Scholar]
- Matheny PB. (2005). Improving phylogenetic inference of mushrooms with RPB1 and RPB2 nucleotide sequence (Inocybe, Agaricales). Molecular Phylogenetics and Evolution 35: 1–20. [DOI] [PubMed] [Google Scholar]
- Meiras-Ottoni A, Larsson KH, Gibertoni TB. (2021). Additions to Trechispora and the status of Scytinopogon (Trechisporales, Basidiomycota). Mycological Progress 20: 203–222. [Google Scholar]
- Miettinen O, Kõljalg U. (2007). Amaurodon sumatranus (Thelephorales, Basidiomycota), a new species from Indonesia. Mycotaxon 100: 51–59. [Google Scholar]
- Moeslund JE, Nygaard B, Ejrnæs R, et al. (2019). Rødliste 2019. Aarhus Universitet, DCE – Nationalt Center for Miljø og Energi. www.redlist.au.dk. [Google Scholar]
- Mühlmann O, Peintner U. (2008). Mycobionts of Salix herbacea on a glacier forefront in the Austrian Alps. Mycorrhiza 18: 171–180. [DOI] [PubMed] [Google Scholar]
- Murrill WA. (1910). North American Flora, volume 9, (Agaricales) Polyporaceae–Agaricaceae. The New York Botanical Garden, USA. [Google Scholar]
- Nagy LG, Walther G, Házi J, et al. (2011). Understanding the evolutionary processes of fungal fruiting bodies: Correlated evolution and divergence time in the Psathyrellaceae. Systematic Biology 60: 303–317. [DOI] [PubMed] [Google Scholar]
- Nilsson RH, Larsson K-H, Taylor AFS, et al. (2018). The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Research 47: D259–D264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A. (2012). FigTree 1.4.3. http://tree.bio.ed.ac.uk/software/figtree/
- Rambaut A, Suchard MA, Xie D. et. al (2014). Tracer 1.6. http://tree.bio.ed.ac.uk/software/tracer/
- Rehner SA, Buckley E. (2005). A Beauviera phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97: 84–98. [DOI] [PubMed] [Google Scholar]
- Ryberg M, Larsson E, Molau U. (2009). Ectomycorrhizal diversity on Dryas octopetala and Salix reticulata in an alpine cliff ecosystem. Arctic, Antarctic, and Alpine Research 41: 506–514. [Google Scholar]
- Ryvarden L. (2002). A note on the genus Hydnodon Banker. Synopsis Fungorum 15: 31–33. [Google Scholar]
- Saar I, Oja J, Põldmaa K, et al. (2019). Red List of Estonian Fungi – 2019 update. Folia Cryptogamica Estonica 56: 117–126. [Google Scholar]
- SLU Artdatabanken. (2020). Rödlistade arter i Sverige 2020. SLU Artdatabanken, Sweden. [Google Scholar]
- Sønstebø JH. (2002). Molecular ecology of ectomycorrhizal fungi on Bistorta vivipara (L.) Gray in four alpine tundra communities. Cand. scient. thesis. Department of Biology, University of Oslo, Norway. [Google Scholar]
- Stalpers JA. (1993). The aphyllophoraceous fungi I. Keys to the species of the Thelephorales. Studies in Mycology 35: 1–168. [Google Scholar]
- Svantesson S, Larsson K-H, Kõljalg U, et al. (2019). Solving the taxonomic identity of Pseudotomentella tristis s.l. (Thelephorales, Basidiomycota) – a multi-gene phylogeny and taxonomic review, integrating ecological and geographical data. MycoKeys 50: 1–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svantesson S, Larsson K-H, Larsson E. (2021). Pseudotomentella badjelanndana, Pseudotomentella sorjusensis and Tomentella viridibasidia – three new corticioid Thelephorales species from the Scandes Mountains. Phytotaxa 497: 61–78. [Google Scholar]
- Svrček M. (1958). Contribution to the taxonomy of the resupinate Thelephoraceous Fungi. Ceská Mykologie 12: 66–77. [Google Scholar]
- Svrček M. (1960). Tomentelloideae Čechoslovakiae, Genera resupinata familae Thelephoraceae s.str. Sydowia 14: 170–245. [Google Scholar]
- Swofford DL. (2002). PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods) 4.0a, build 167. Sinauer Associates, Sunderland, MA. https://paup.phylosolutions.com/ [Google Scholar]
- Talavera G, Castresana J. (2007). Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Systematic Biology 56: 564–577. [DOI] [PubMed] [Google Scholar]
- Taylor JH, Peterson CA. (2005). Ectomycorrhizal impacts on nutrient uptake pathways in woody roots. New Forests 30: 203–214. [Google Scholar]
- Tedersoo L, Liiv I, Kivistik PA, et al. (2016). Genomics and metagenomics technologies to recover ribosomal DNA and single-copy genes from old fruit-body and ectomycorrhiza specimens. MycoKeys 13: 1–20. [Google Scholar]
- United States Forest Service (1994). Record of decision for amendments to Forest Service and Bureau of Land Management planning documents within the range of the Northern Spotted Owl: and, standards and guidelines for management of habitat for late-successional and old-growth forest related species within the range of the Northern Spotted Owl. US Department of Agriculture, Forest Service, Ecosystem management, USA. [Google Scholar]
- Vizzini A, Angelini C, Losi C, et al. (2016). Thelephora dominicana (Basidiomycota, Thelephorales), a new species from the Dominican Republic, and preliminary notes on thelephoroid genera. Phytotaxa 265: 27–38. [Google Scholar]
- Voitk A, Saar I, Trudell S, et al. (2017). Polyozellus multiplex (Thelephorales) is a species complex containing four new species. Mycologia 109: 975–992. [DOI] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee L, et al. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR Protocols, A guide to Methods and Applications (Innis MA, Gelfand DH, Sininski JJ, et al., eds.). Academic Press, USA: 315–322. [Google Scholar]
- Wurzbacher C, Larsson E, Bengtsson-Palme J, et al. (2019). Introducing ribosomal tandem repeat barcoding for fungi. Molecular Ecology Resources 19: 118–127. [DOI] [PubMed] [Google Scholar]