Abstract
Ndn is located on chromosome 7C, an imprinted region of the mouse genome. Imprinting of Ndn and adjacent paternally expressed genes is regulated by a regional imprinting control element known as the imprinting center (IC). An IC also controls imprint resetting of target genes in the region of conserved synteny on human chromosome 15q11-q13, which is deleted or rearranged in the neurodevelopmental disorder Prader-Willi syndrome. Epigenetic modifications such as DNA methylation, which occur in gametes and can be stably propagated, are presumed to establish and maintain the imprint in target genes of the IC. While most DNA becomes substantially demethylated by the blastocyst stage, some imprinted genes have regions that escape global demethylation and may maintain the imprint. We have now analyzed the methylation of 39 CpG dinucleotide sequences in the 5′ end of Ndn by sodium bisulfite sequencing in gametes and in preimplantation and adult tissues. While sperm DNA is completely unmethylated across this region, oocyte DNA is partially methylated. A distinctive but unstable maternal methylation pattern persists until the morula stage and is lost in the blastocyst stage, where low levels of methylation are present on most DNA strands of either parental origin. The methylation pattern is then substantially remodeled, and fewer than half of maternally derived DNA strands in adult brain resemble the oocyte pattern. We postulate that for Ndn, DNA methylation may initially preserve a gametic imprint during preimplantation development, but other epigenetic events may maintain the imprint later in embryonic development.
DNA methylation is an important form of gene regulation during mammalian development and has been implicated in such diverse processes as genomic imprinting (18), X-inactivation (4), and differential gene expression (9). Imprinting is a form of gene regulation whereby certain genes are restricted in expression to only one parental allele (32). Molecular control of imprinting requires an epigenetic modification of DNA in the haploid genome, leading to hemizygous expression in the diploid embryo. The initial mark which differentiates the two parental alleles likely originates in the gametes when the two alleles are still separate. After zygote formation, the parental alleles maintain their identity so that one allele eventually becomes preferentially expressed. Methylation of CpG dinucleotides is proposed to be one mechanism for differentially marking the parental chromosomes, since methylation can be stably inherited in somatic cells yet can be removed and reset in the next generation according to the parent of origin (15, 25).
The developmental stages prior to blastocyst formation are of particular importance in genomic imprinting (28). Genome-wide, oocyte DNA tends to be hypomethylated while sperm DNA tends to be hypermethylated (21). During preimplantation development, the overall level of methylation decreases. Most methylation moieties present on the original parental chromosomes are removed from the DNA by the morula stage, giving rise to a predominantly unmethylated genome which remains this way at least through blastulation. A wave of de novo methylation follows, leading to an overall increase in genome methylation levels as the newly implanted embryo develops and differentiates. In imprinted genes, gamete-derived methylated CpG sequences are predicted to be preserved during preimplantation development and must therefore be specifically recognized and protected from the global, generalized demethylation that takes place at these stages (24, 34). Allelic differences present in germ cells could be rapidly expanded during preimplantation development and therefore serve as a primary imprinting signal. Such methylated sites may or may not be retained in all somatic cells as a marker of parental identity.
All imprinted genes analyzed to date have displayed allele-specific DNA methylation patterns (22, 25). For some of these genes, differentially methylated regions (DMRs) in the gametes precede allele-specific methylation differences in adult tissues. However, studies of methylation in early embryogenesis have been limited to a small number of imprinted genes (3, 7, 10, 20, 27, 29, 34, 38). The interpretation of these studies is complicated because the DMRs have been implicated in imprinting control of other genes. For example, the DMR upstream of H19 is implicated in reciprocal imprinting of Igf2 (3, 5, 13, 31), and the upstream region of Snrpn contains an essential regional imprinting element (39). Although DNA methylation is essential for normal development, its exact role in imprinting at all developmental stages remains controversial, and other epigenetic events have been invoked to explain the development of the final imprinted expression pattern (15, 32). For genes that are not involved in the imprinting of other genes but are the target of an imprinting center (IC), it is not known whether the establishment of allele-specific methylation in the gametes is required for allele-specific expression of genes. For these genes, it is also not known whether maintenance of gamete-specific methylation patterns throughout development is necessary for proper imprinted gene expression.
To better understand the role of DNA methylation in the maintenance of imprinted gene expression, we have analyzed allele-specific DNA methylation patterns in the imprinted Ndn gene. Ndn is an ideal gene for imprinting study because it has a simple, intronless gene structure and is a target of the IC rather than being intimately involved in imprinting other genes. The Ndn gene product, necdin, is a protein expressed in terminally differentiated neurons and is proposed to have a role in neuronal development (40).
Ndn is located on mouse chromosome 7 in a region of conserved synteny with human 15q11-q13, the region commonly deleted in Prader-Willi syndrome (PWS) (23). PWS results from the loss of expression of genes, including the human Ndn orthologue NDN, which are normally active on the paternal copy of chromosome 15q11-q13 (16, 19). Imprinting in the PWS region is controlled by the IC, located in the 5′ end of the SNRPN gene, about 1 Mb from the NDN locus. Ndn is also paternally expressed and under the control of an IC (39).
Ndn contains a CpG-rich region that extends from immediately upstream of the transcription start site into the first half of the open reading frame. In this study, we have analyzed the developmental and adult methylation profiles of 39 individual CpG sites in the 5′ end of the Ndn gene by sodium bisulfite sequencing (SBS) (11). This technique enables methylation analysis of individual DNA strands even in limited tissue samples. Our results show that while the completely unmethylated pattern seen in sperm becomes partially methylated during development, a distinctive oocyte pattern persists in the majority of clones at least until the morula stage and is lost in many of the blastocyst clones. In adult tissues, the gametic methylation patterns are rarely discernible. This suggests that gametic methylation patterns are lost around the time of implantation, although some DNA strands apparently escape global demethylation and persist in adult tissues. The heterogeneous nature of the maternal methylation pattern after the morula stage suggests that for Ndn, methylation may be important for early stages of imprint establishment. However, a second process, perhaps involving more dispersed methylation or other epigenetic modification, is likely to be important in imprint maintenance in somatic cells.
MATERIALS AND METHODS
Collection of oocytes, early embryos, and sperm.
Early embryos and gametes were collected from C57BL/6 mice essentially as described previously (14). Some of the blastocysts were derived from crosses between C57BL/6 females and Mus spretus (SPRET) males. Five-week-old C57BL/6 females were superovulated with pregnant mare's serum and human chorionic gonadotropin and were mated with 2- to 7-month-old males. Oocytes; 2-cell, 4-cell, and 8-cell embryos; and morulae were collected from the oviducts by flushing out through the infundibulum. Oocytes were washed with careful inspection to remove maternal cells, and 2-cell, 4-cell, and 8-cell embryos were collected around 32, 38, and 56 h postcoitum (p.c.), respectively. Morulae were collected at about 62 h p.c. Blastocysts either were collected around 3.5 days p.c. by flushing out the uterus or were collected at the morula stage (2.5 to 3 days) and were cultured for 24 h. Sperm was collected from the epididymus of 5-month-old males.
DNA extraction.
Liver, heart, brain, and testes were dissected from 6-week-old F1 progeny of a cross between C57BL/6 females and SPRET males or between C57BL/6 females and Mus musculus castaneus (CAST) males. Tissues were crushed under liquid nitrogen, and DNA was extracted by proteinase K-sodium dodecyl sulfate (SDS) digestion, phenol-chloroform extraction, and ethanol precipitation (1). To extract DNA from the oocytes and embryos, 30 to 200 cells were suspended in ∼3 μl of Dulbecco modified Eagle medium (DMEM). Approximately 40 oocytes and 3 to 37 early embryos were combined for each DNA preparation, with the number of embryos pooled being inversely proportional to the age of the embryo because total cell number increases with age. The cell suspension was made up to 18 μl with a 1 mM SDS–280-μg/ml proteinase K solution containing 1 or 2 μg of salmon sperm carrier DNA in phosphate-buffered saline. This mixture was covered in mineral oil and was incubated at 37 or 50°C for 30 to 90 min and then at 98°C for 15 min. Sperm was first treated with 10 mM EDTA, 100 mM NaCl, 2% SDS, 20 μg of proteinase K/ml, and 10 mM Tris-Cl (pH 8) overnight at 37°C to digest the nonsperm cells. The sperm sample was centrifuged at 600 × g for 10 min at room temperature, and the supernatant was removed. Sperm was then processed as for tissues with the addition of 39 mM dithiothreitol to the proteinase K-SDS extraction buffer.
SBS.
Early embryo and oocyte DNA samples were denatured by the addition of 2 μl of fresh 3 M NaOH, were incubated for 20 to 30 min at 42°C and then for 3 min at 95°C, and were placed on ice. DNA from adult tissues or sperm (200 ng) was added to 2 μg of salmon sperm carrier, was denatured in 0.3 M NaOH at 42°C for 30 min and then at 95°C for 3 min, and was placed on ice. Salmon sperm carrier DNA (1 or 2 μg) was added to early embryo samples and oocyte samples. To all samples, 255 μl of 40.5% (wt/vol) sodium bisulfite at pH 5, 15 μl of 20 mM hydroquinone, and up to 300 μl of H2O were added. The samples were covered in mineral oil and were incubated at 55°C for ∼16 h. The treated DNA was mixed with 100 μl of a purification buffer containing 50 mM KCl, 10 mM Tris-HCl (pH 8.8), 1.5 mM MgCl2, and 0.1% Triton X-100, followed by 1 ml of a resin containing 4 M guanidine thiocyanate, 20 mM EDTA, 20 g of diatomaceous earth (Sigma D-5384)/liter, and 50 mM Tris-Cl (pH 7.5). Samples were centrifuged through a Wizard Miniprep column (Promega, Inc.) and were eluted in 50 μl of H2O. To complete the reaction, fresh 3 M NaOH was added until reaching a concentration of 0.3 M, and the samples were incubated at 37°C for 20 min and placed on ice. The DNA was precipitated in 3 M ammonium acetate and 3 volumes of 95% ethanol at 4°C for 15 min to 1 h or at −20°C overnight and was then centrifuged at 4°C at 20,000 × g in a microcentrifuge for 10 min. DNA pellets were washed twice with 70% ethanol and resuspended in 30 μl of H2O. For early oocytes and early embryos, an additional 1 or 2 μg of salmon sperm carrier DNA was added at the ethanol precipitation step.
PCR.
Primers to amplify the region of study from C57BL/6, SPRET, and CAST mice for detection of polymorphisms were as follows: NEC11F, 5′TCATTCTCCAGGACCTTCAC; and NEC12R, 5′CTTCGGATCAGAGCAGGAC. PCR was performed in 1.5 mM MgCl2 as follows: 5 min at 94°C; 30 s at 94°C, 30 s at 50°C, and 30 s at 72°C cycled 30 times; and 10 min at 72°C, yielding a 401-bp PCR product. Nested PCR was used to amplify a 560-bp Ndn product from sodium bisulfite-treated DNA. For the majority of clones, the first-round PCR forward primer was NEC43F, 5′TTTTGTGTTATATAGGAGATTAGGAAATTT/GTTTATA. T/G indicates that the primer was degenerate at this position, where T represents the C57BL/6 sequence and G represents the SPRET sequence. The reverse primer was NEC45R, 5′TCTAACCTACTCCAAAACCTCCCTATATC. For some samples the first-round PCR was done with the forward primer NEC78F, 5′TATTTAGTTTTGTGTTATATAGGAGATTAGG, and the reverse primer NEC79R, 5′ATTTCTTATAACTACCCATAACCTCTTTCA. Second-round nested primers were NEC41F, 5′TTTTTTAGATTTTAGTGGTTGGGTTTTG, and NEC48R, 5′CACCTTCTACACCAACTAAACAAAAGT. First-round PCR was performed in 1.5 mM MgCl2 as follows: 5 min at 94°C; 2 min at 94°C, 2 min at 58°C, and 2 min at 72°C cycled 2 times; 30 s at 94°C, 30 s at 58°C, and 1 min at 72°C cycled 35 times; and 10 min at 72°C. Second-round PCR was performed in 1.5 mM MgCl2 as follows: 5 min at 94°C; 30 s at 94°C, 30 s at 58°C, and 1 min at 72°C cycled 35 times; and then 10 min at 72°C. For some of the samples, a seminested PCR was used under the reaction conditions described above. The first- and second-round PCR forward primer was NEC53F, 5′ATATTTAATTTGATTTTGTTTAAATTTAGTGTG. The reverse primers were NEC47R, 5′CATTCCAAACCACACCCTCTC, for the first round and NEC48R for the second round. A 709-bp product resulted. In some seminested PCRs the first- and second-round forward PCR primer was NEC62F, 5′TTATTTAGTTTTGTGTTATATAGGAGATTAGGG, resulting in a 615-bp product. Control primers amplified a 544-bp product from H19 region B, a CpG-rich region located 2.6 kb upstream of the transcription start site (38). The first-round PCR was performed in 2 mM MgCl2 as follows: 5 min at 94°C; 30 s at 94°C, 30 s at 55°C, and 1 min at 72°C cycled 35 times; and then 10 min at 72°C. The second-round PCR was performed in 2 mM MgCl2 as follows: 5 min at 94°C; 30 s at 94°C, 30 s at 58°C, and 1 min at 72°C cycled 35 times; and then 10 min at 72°C.
Cloning and sequencing.
PCR products were electrophoresed on 2% agarose gels, gel purified using the QIAquick Gel Extraction Kit (Qiagen Inc.), and cloned into the pGEM–T vector (Promega Corp.). Plasmid DNA was prepared by alkaline lysis (26). Recombinant clones were sequenced using an Amersham kit with fluorescently labeled M13 forward and reverse primers and were analyzed on a LiCor automated sequencer.
Genomic structure analysis.
The mouse sequence (GenBank accession number AC026388) corresponds to the working draft sequence of M. musculus chromosome 7 clone RP23–426B15. The human sequence (GenBank accession number AC006596) corresponds to the complete sequence of human chromosome 15 PAC clone pDJ181P7. Repeatmasker (http://ftp.genome.washington.edu/cgi-bin/RepeatMasker) was used to annotate the sequences for murine and human repeats. The PipMaker gene analysis program (http://nog.cse.psu.edu/pipmaker/) was used to compare the sequences of the genomic regions surrounding Ndn and NDN and to detect regions of high CpG content.
RESULTS
Identification of repetitive elements in CpG-rich regions.
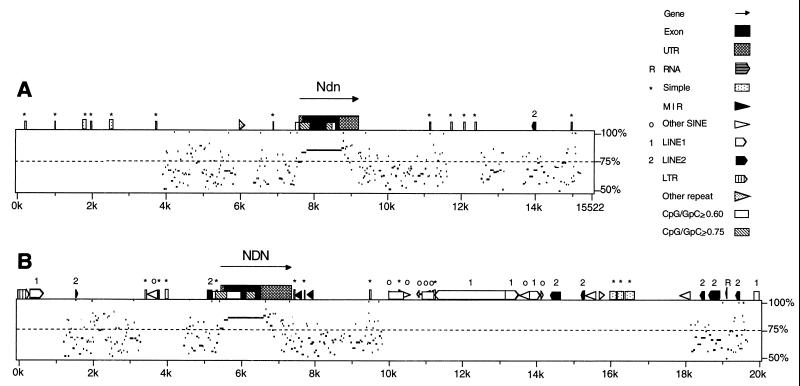
The mouse and human Ndn and NDN genes are both composed of single exons containing small open reading frames encoding 325 and 321 amino acids, respectively. The genomic sequence of a 15,522-bp region surrounding Ndn and of a 104-kb region surrounding NDN was retrieved from GenBank. We first analyzed the genomic sequence surrounding Ndn (15,522 bp) and NDN (20,000 bp) for the presence of repetitive elements using RepeatMasker Web-based software (Fig. 1). With the repeat-masked sequence as input, we used PipMaker Web-based software to compute the percentage of identity between mouse and human Ndn and NDN (Fig. 1). As expected, significant nucleotide identity extends through the open reading frame. Short regions immediately flanking the open reading frame and isolated regions several kilobases on either side of the gene which may correspond to gene regulatory elements also show some sequence similarity.
FIG. 1.
PipMaker sequence alignments. (A) Sequence (15,522 bp) surrounding mouse Ndn was compared to sequence (20 kb) surrounding human NDN. (B) Twenty kilobases of sequence surrounding human NDN was compared to Ndn. The arrow and full-height box indicate the position of Ndn and NDN, with the open reading frame shaded black and the untranslated regions shaded gray. Half-height boxes outside Ndn and NDN indicate repetitive elements. Low boxes within Ndn and NDN indicate CpG-rich regions, within which white shading indicates a CpG/GpC ratio of ≥0.60 and gray shading indicates a CpG/GpC ratio of ≥0.75. On the plot below each sequence schematic, regions of homology are shown by black dashes and percent nucleotide identity is indicated on the right. See http://nog.cse.psu.edu/pipmaker/ for details of mouse and human repeat types. UTR, untranslated region; LTR, long terminal repeat.
PipMaker analysis indicated the locations of CpG islands, defined as having a CpG/GpC ratio greater than or equal to 0.6. PipMaker analysis also distinguished denser CpG islands with a ratio of at least 0.75 from islands meeting the 0.60 criterion. Two CpG islands, located in equivalent positions in human and mouse Ndn and NDN, were found (Fig. 1). The 5′ CpG island begins in the putative promoter and extends into the open reading frame, while a 3′ CpG island is located in the 3′ portion of the open reading frame. We predicted that if a gamete-specific methylation imprint exists in Ndn, then the 5′ CpG island represents the most probable location for the imprint. This prediction is based on two recent gene-targeting experiments that created null mutations of Ndn in the mouse (12, 35). In both cases, the lacZ gene replaced the Ndn gene starting at the BamHI site located between CpG positions 23 and 24 (Fig. 2) and terminating at the stop codon. Mice inheriting the deleted allele maintain proper parent-of-origin gene expression of the reporter gene and, in one case, show an imprinted phenotype due to loss of necdin gene expression on germ line transmission (12). This implies that the region downstream of the BamHI site in the open reading frame is not necessary for proper imprinting of Ndn.
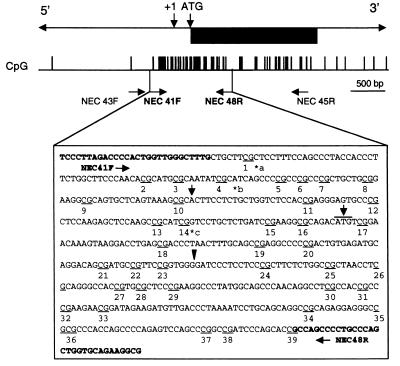
FIG. 2.
Map of the region of mouse necdin analyzed for CpG methylation status. The gene structure is shown at the top, where the black box represents the open reading frame and the +1 and ATG are the transcriptional and translational start sites, respectively. CpG dinucleotides are shown as vertical bars below the map. The arrowhead points to the BamHI restriction site used to insert the lacZ gene into the Ndn knockout mouse. The relative positions of the primers used for nested PCR are shown, and the second set of primers (NEC41F-NEC48R) is in boldface. The 560-bp region analyzed covers CpG sites 1 to 39 and is underlined in the C57BL/6 type sequence. For some of the samples, other primers were used to amplify the same region (see Materials and Methods). Single-nucleotide polymorphisms between C57BL/6 and SPRET and between C57BL/6 and CAST are indicated by an asterisk and are as follows: a, CAST, CGGTC; and b, SPRET and CAST, CGCGT. A 5-bp insertion was found in SPRET at c, SPRET CGCATCGCATCG. The changes from the C57BL/6 sequence are in boldface. CpG dinucleotides are underlined.
Strategy for SBS.
To analyze the developmental pattern of CpG methylation within the 5′ CpG island, we used SBS, a sensitive technique for single CpG dinucleotide methylation analysis. Sodium bisulfite treatment of DNA converts all unmethylated cytosines to uracil. Upon DNA sequencing of cloned PCR products, bases appear as cytosine only if they were methylated in the original DNA sample and were thus protected from bisulfite-moderated conversion. Brain, liver, heart, and testis samples were prepared from 6-week-old interspecific F1 mice. Gametes and early embryos were isolated from C57BL/6 mice and F1 interspecific crosses. DNA isolated from all samples was then processed for sodium bisulfite treatment. Bisulfite-modified DNA was amplified by nested PCR, and cloned products were sequenced from both ends. Because of previous reports of difficulties in obtaining complete conversion of sodium bisulfite-treated DNA, particularly with samples derived from small numbers of cells, we performed control experiments using primers specific to the H19 gene (38). We amplified sodium bisulfite-treated DNA from sperm and adult brain with H19 region B primers and sequenced the cloned PCR products. These experiments demonstrated that we had obtained a high conversion efficiency (>99%), based on the complete conversion of C residues not present in CpG dinucleotides. In addition, we replicated the observation that sperm DNA is almost completely methylated at each CpG in H19 region B. We also sequenced seven adult brain clones that were completely unmethylated throughout region B and were presumed to be maternally derived, consistent with the previous observation that maternally derived clones are mostly unmethylated in this region. These experiments validated the SBS in our hands.
Ndn methylation analysis in adult tissues.
PCR primers were designed to amplify the 5′ CpG-rich region in Ndn (Fig. 2). Single-site polymorphisms among C57BL/6, SPRET, and CAST mice were determined by direct sequencing of PCR products generated from genomic DNA. All experiments on adult tissues were done on samples from F1 interspecific mice, and the single-site polymorphisms were used to identify the parent of origin of cloned PCR products in these experiments.
We analyzed the methylation patterns in Ndn by PCR amplification of sodium bisulfite-treated DNA and sequencing of individual PCR clones. In the 5′ CpG island, we assayed 39 CpG dinucleotides. The CpG sites are approximately evenly distributed over this interval. The predicted transcriptional start site is between CpG sites 10 and 11, and the translational start site is between CpG sites 16 and 17. DNA samples were derived from adult brain, in which Ndn is transcribed, and from liver and heart, in which Ndn is not active. In order to obtain a representative sample, at least seven clones from each parental allele were sequenced. We noted a parental bias in the distribution of clones obtained, with maternally derived alleles appearing about five times more frequently than paternally derived clones. A similar bias in parental alleles had also been noted in studies of xist (20), but in our case the presence of an interspecies polymorphism under the forward primer for the first round of PCR may be the cause of the bias. To enrich for paternally derived clones, some PCRs were performed with a first-round forward primer that contained only the SPRET (paternal) allele of this polymorphism.
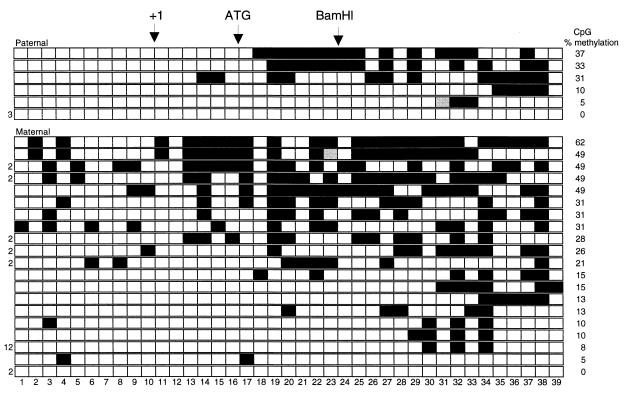
SBS analysis of adult brain revealed a mosaic pattern of DNA methylation on both parental alleles (Fig. 3). Additional clones derived from PCR products that contained only CpG dinucleotides 1 to 20 were also included in subsequent analysis (data not shown). Most sites showed variable methylation between clones, and no sites were consistently methylated, although a few sites (numbered 1, 12, and 39) were unmethylated in all or almost all clones on both parental alleles. The average level of methylation was 22% in the maternal clones and 17% in the paternal clones. The average level of methylation in liver and heart was much lower. For example, the average levels of methylation in maternal and paternal heart clones were 10% and 0.4%, respectively.
FIG. 3.
Heterogeneity in methylation analysis of adult brain. The CpG dinucleotides are numbered across the bottom. Each row represents a single clone derived from a (C57BL/6 × CAST)F1 mouse or a (C57BL/6 × SPRET)F1 mouse. Black boxes represent methylated CpG sites, white boxes are unmethylated CpG sites, and gray boxes were ambiguous in the sequence analysis. In cases where clones with the same profile appeared more than once, the number of times that it was found is indicated at the left. Paternally derived clones are at the top, with maternally derived clones below, as determined by polymorphism analysis. The clones are ordered by the percent methylation for each clone, shown to the right.
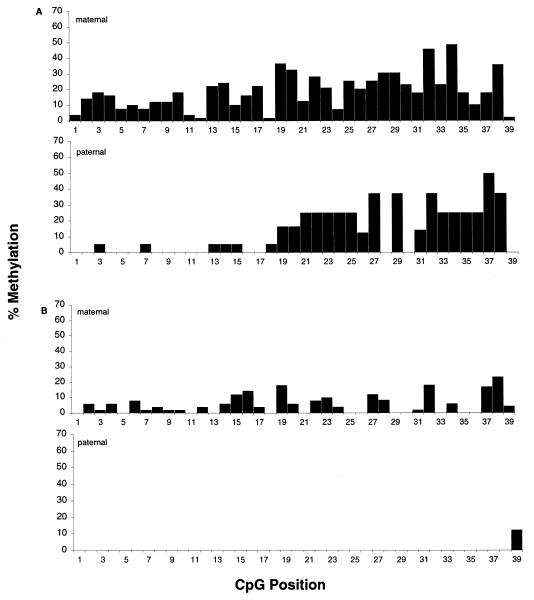
The proportion of clones methylated at each individual CpG dinucleotide in heart and brain was calculated for each parental allele (Fig. 4). In the adult brain, paternally inherited CpG sites 1 to 17 were hypomethylated compared to the maternal allele sites, while in sites 18 through 39, methylation levels were more equivalent (Fig. 4A). In the adult heart (Fig. 4B) and liver (data not shown), the maternal allele was more highly methylated than the paternal alleles throughout the region studied.
FIG. 4.
Methylation profile of 39 CpG dinucleotides in adult brain and heart. The percentage of clones methylated at each CpG dinucleotide position was calculated and plotted against the CpG position (1 to 39). Profiles are of maternal and paternal clones from adult brain (A) and of maternal and paternal clones from adult heart (B).
Developmental analysis of Ndn methylation.
We next performed SBS analysis on gametes and early embryos. The parental origin of the cloned PCR products from blastocysts was inferred from interspecific polymorphisms, although analysis of earlier stages was performed on inbred C57BL/6 crosses where the parent of origin could not be determined. Because of previous reports that individual PCRs may not contain clones that adequately represent the initial DNA strands when amplifying from limited starting material (38), multiple PCRs were carried out with each sample. We sequenced between 6 and 25 clones from at least two PCRs from each of the gamete samples and from each developmental stage. The only exception was the morula data, which came from a single PCR.
The sperm and oocyte methylation patterns define the initial methylation patterns, which are subsequently remodeled in the early embryo. In sperm, all 39 CpG sites within the region were unmethylated in all clones (Fig. 5). In testis, which contains a substantial proportion of germ cells, we detected clones that were completely unmethylated, with both maternally and paternally derived alleles represented (data not shown). The lack of partially methylated clones is either because the somatic cell DNA is unmethylated or because, in the sample of clones sequenced, there were no somatic cell DNA clones represented. In contrast, oocyte DNA did not display a single pattern of methylated CpG sites. Furthermore, there were no CpG sites consistently methylated in all oocyte clones. Two of the 45 oocyte clones were completely unmethylated and were distinct from the other clones. This may represent a true diversity in the oocyte population; alternatively, these clones may represent contamination from adhering maternal cells, despite precautions taken to eliminate those cells. If these two clones are excluded, the only methylated site conserved among the remaining 43 oocyte clones is at position 19. Other CpG sites that are methylated in a consistent pattern in most clones are at positions 13 and 14, 15 to 17, 22, 23, 32, and 35 to 39.
FIG. 5.
Methylation in gametes and embryos. The presentation is the same as in Fig. 3, with the CpG dinucleotides numbered across the top. Paternal (p) and maternal (m) designations are given in the clone sets that came from interspecific crosses. (A) The clones are grouped according to the tissue source. CpG sites 1 to 39 are numbered along the top. (B) Clones from blastocysts derived from (C57BL/6 × M. spretus) crosses.
Analysis of 2-cell, 4-cell, and 8-cell, morula, and blastocyst DNA derived from C57BL/6J inbred crosses was then performed. The sites noted as consistently methylated in most oocyte clones were in general highly methylated at all stages up to the morula in the presumed maternally derived clones. Overall, methylation at CpG positions 13 to 17 was found at all stages except for the blastocyst stage (Fig. 5). Methylation at positions 15 to 17 was found in blastocysts, and methylation at positions 35 to 38 was found in all early developmental stages. Completely or almost completely unmethylated clones were present in the 2-cell, 4-cell, morula, and blastocyst stages. These likely represent paternal clones since they are unmethylated as in the male gametes. It is also possible that some unmethylated maternal DNA strands exist at these embryonic stages. Many clones with low levels of methylation could not be assigned a putative parent of origin because they could be derived from either de novo methylation of the paternal allele or demethylation of the maternal allele.
The overall methylation levels were low in the blastocyst samples, and the putative maternal alleles retained less of the oocyte-specific methylation than in the earlier developmental stages. To gain more information on the parent of origin of the partially methylated clones, we next analyzed blastocysts from a C57BL/6J × SPRET cross (Fig. 5B). Maternal and paternal clones were both sparsely methylated, indicating that de novo methylation occurs in the paternal allele and that demethylation occurs in the maternal allele, leading to equivalent levels of methylation in both parental alleles. No single CpG dinucleotide was consistently methylated in all preimplantation stages.
DISCUSSION
In this study, we have analyzed the developmental patterns of CpG methylation in the imprinted murine Ndn gene. Previous analysis of nonimprinted genes by restriction enzyme analysis (17) or SBS (37) suggested that partial removal of gametic methylation was found by the morula stage and that little methylation is detectable by the blastocyst stage. During this time frame, allele-specific methylation of a core DMR 4 kb upstream of the imprinted H19 gene remains distinct until the blastocyst stage and is only somewhat remodeled in midgestation embryos (33). The promoter-proximal region of H19 showed more variability in methylation levels and a lack of differential methylation at the blastocyst stage. A similar study investigated preimplantation embryos only to the 8-cell stage but did not assess remodeling during the critical 8-cell-to-blastocyst interval (38).
Our studies found that the 5′ CpG-rich region of Ndn is unmethylated in sperm but that the paternal allele becomes gradually methylated, resulting in partial mosaic methylation of the majority of DNA strands in adult tissues. The maternal allele is partially methylated in the majority of oocytes and remains at about the same level of methylation, at least to the morula stage. A reduced level of methylation of the maternal allele is seen in blastocysts, and de novo methylation occurs, subsequently resulting in a mosaic pattern of methylation in adult tissues. Analysis of the maternally derived adult brain clones reveals that about 35% have methylation patterns the same as or similar to the major pattern in oocytes, with overall higher levels of methylation at positions 19, 32, and 38 (Fig. 3). Many paternal clones are also highly methylated in positions 19 to 38. The distinction between overall parental methylation levels is confined to sites 1 to 17 in the more 5′ region. However, no well-defined pattern emerges to suggest that CpG methylation of either allele is the signal that carries the imprint through from the blastocyst stage to adult tissues. Thus, the methylation patterns seen in Ndn resemble the H19 promoter-proximal region in that no core region of differential methylation is maintained to the blastocyst stage. Differential methylation at a distant but linked site may play an important role in the imprinting of necdin.
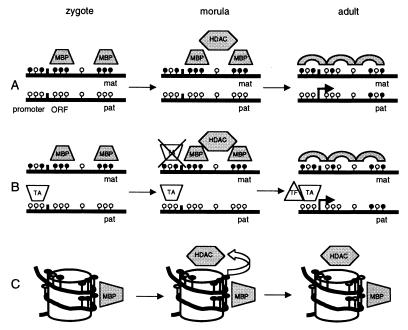
Other DNA modifications, such as histone acetylation and CpG binding proteins, may be involved in imprinting, although when these other modifications come into play is not yet known. One interpretation of our data is that the DNA methylation patterns originating in the oocyte are important early in imprinting but that these methylation patterns are required only to initiate a series of additional epigenetic events leading to the silencing of the maternal allele. Interestingly, the most prominent oocyte pattern, the methylation of CpG sites 13 through 22, covers 115 bp of DNA, in two segments of 36 bp (CpG sites 13 to 17) and 39 bp (CpG sites 19 to 22), with an intervening, unmethylated space of 40 bp. Hypermethylation on the maternal allele in preimplantation embryos may block binding of a transiently expressed transcriptional activator or chromatin remodeling factor, which is free to bind to the unmethylated paternal allele and maintain an active chromatin state (Fig. 6B). Alternatively, a methylated maternal allele in the early embryo may bind methyl-CpG binding proteins and promote subsequent epigenetic events (Fig. 6A). Methyl-CpG binding proteins have been shown to preferentially bind methylated CpG sites and recruit histone deacetylases (36). If the two prominently methylated oocyte DNA segments were located on the outer part of the nucleosome as it winds around the core particle twice, the methylated DNA could potentially be a recognizable structural target for subsequent epigenetic events (Fig. 6C). Alternatively, the positions that consistently lack methylation may reflect specific DNA-protein contact points. Studies of chromatin structure in early embryos are limited by difficulties in obtaining sufficient experimental material but would help to prove or disprove this hypothesis.
FIG. 6.
Models for imprinting of target genes. The gametic methylation represents a simplified version of the average methylation seen at these stages. Black and white lollipops represent methylated and unmethylated CpG dinucleotides, respectively. (A) The remodeling of the maternal allele to a closed chromatin conformation may involve methyl binding proteins (MBPs) that recognize a target of densely methylated CpG sites, which then recruit histone deacetylases (HDAC). Since the closed chromatin conformation is maintained by a methylation-independent mechanism, the methylation state seen in the adult tissues can be variable. ORF, open reading frame; mat, maternal; pat, paternal. (B) The hypermethylation on the maternal allele blocks the binding of a transiently expressed transcriptional activator (TA) which binds only to the paternal allele. The TA keeps the paternal allele in a transcriptionally permissive state and allows transcription factors (TFs) to bind. In the absence of this TA, the maternal allele is remodeled to a closed chromatin state. (C) The positions of methylated CpG sites 13 to 22 found in the early embryo samples may become closely situated on the surface of a nucleosome and act as a target for MBPs.
Most imprinted genes studied so far have the same allele-specific methylation pattern in all tissues irrespective of gene expression. In Ndn, we noted tissue-specific differences. Mainly, relative hypermethylation of the maternal allele in the first 17 CpG dinucleotides is apparent in brain, where Ndn is expressed. In liver and heart, which do not express Ndn, the maternal allele is relatively hypermethylated compared to the paternal allele throughout the region analyzed and overall methylation levels are much lower than in brain. The paternal allele has become methylated to about the same level as the maternal allele in brain DNA, in the 3′ end of the region analyzed. This suggests that there may be tissue-specific hypermethylation of Ndn in the brain, with the 5′ end (CpG dinucleotides 1 to 17) being protected from this hypermethylation on the expressed paternal allele. The paternal allele-specific hypomethylation of the 5′ region may be a consequence of transcription. Alternatively, hypermethylation on the maternal allele may be a remnant of the initial gametic pattern or a reflection of other allele-specific factors, such as chromatin structure. Finally, the methylation differences between tissues may be due to the binding of tissue-specific repressors and/or differences in chromatin structure between tissues which may change accessibility to methyltransferases.
Our study contrasts with previous studies of the Snrpn and H19 upstream DMRs in which maintenance of differential DNA methylation is proposed to be important throughout development. For Ndn, no other CpG-rich clusters are found upstream or downstream of the region analyzed over a distance of about 15 kb, although it is possible that isolated CpG sites outside the region studied may play a role in the imprinting process. As for Snrpn and H19, their DMRs are presumed to have a direct role in the imprinting process, rather than being a target of a separate IC (2). In addition, the Snrpn IC and its human equivalent have recently been shown to be important in imprint maintenance as well as in establishment, which could explain why they show a stable differential methylation throughout development (6). Another imprinted gene, Mash2, was found to be unaffected by loss of methylation in mice deficient for the DNA methyltransferase gene (Dnmt) (8, 30) and, like Ndn, is not known to contain an IC for other genes. Mash2 and Ndn may fall into a category of genes whose imprinting is maintained not by methylation but by some other imprinting control mechanism, such as allele-specific chromatin structure. In support of this hypothesis, Ndn gene expression does not respond to demethylation during treatment of androgenetic and parthenogenetic mouse fibroblasts with aza-cytidine (C. Stewart, personal communication). Further developmental characterization of imprinted genes that are targets of ICs will be an important step towards understanding the role of DNA methylation in imprinting and how genes under the control of an IC are regulated.
ACKNOWLEDGMENTS
This work was supported by a grant from the Canadian Institutes of Health Research (CIHR). R.W. is a Scholar of the CIHR and of the Alberta Heritage Foundation for Medical Research (AHFMR). M.L.H. is supported by a Studentship from the AHFMR.
We also thank Peter Dickie for advice and assistance with the mouse embryo collection, Mark Kelly and Jocelyn Carroll for technical help, David Rodenhiser for advice on primer design and SBS, and Colin Stewart for helpful discussions.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. Vol. 1. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1993. [Google Scholar]
- 2.Barlow D P. Competition—a common motif for the imprinting mechanism? EMBO J. 1997;16:6899–6905. doi: 10.1093/emboj/16.23.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartolomei S M, Webber A L, Brunkow M E, Tilghman S M. Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes Dev. 1993;7:1663–1673. doi: 10.1101/gad.7.9.1663. [DOI] [PubMed] [Google Scholar]
- 4.Beard C, Li E, Jaenisch R. Loss of methylation activates Xist in somatic but not in embryonic cells. Genes Dev. 1995;9:2325–2334. doi: 10.1101/gad.9.19.2325. [DOI] [PubMed] [Google Scholar]
- 5.Bell A C, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 6.Bielinska B, Blaydes S M, Buiting K, Yang T, Krajewska-Walasek M, Horsthemke B, Brannan C I. De novo deletions of SNRPN exon 1 in early human and mouse embryos result in a paternal to maternal imprint switch. Nat Genet. 2000;25:74–78. doi: 10.1038/75629. [DOI] [PubMed] [Google Scholar]
- 7.Birger Y, Shemer R, Perk J, Razin A. The imprinting box of the mouse Igf2r gene. Nature. 1999;397:84–88. doi: 10.1038/16291. [DOI] [PubMed] [Google Scholar]
- 8.Caspary T, Cleary M A, Baker C C, Guan X J, Tilghman S M. Multiple mechanisms regulate imprinting of the mouse distal chromosome 7 gene cluster. Mol Cell Biol. 1998;18:3466–3474. doi: 10.1128/mcb.18.6.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eden S, Cedar H. Role of DNA methylation in the regulation of transcription. Curr Opin Genet Dev. 1994;4:255–259. doi: 10.1016/s0959-437x(05)80052-8. [DOI] [PubMed] [Google Scholar]
- 10.Feil R, Walter J, Allen N D, Reik W. Developmental control of allelic methylation in the imprinted mouse Igf2 and H19 genes. Development. 1994;120:2933–2943. doi: 10.1242/dev.120.10.2933. [DOI] [PubMed] [Google Scholar]
- 11.Frommer M, McDonald L E, Millar D S, Collis C M, Watt F, Grigg G W, Molloy P L, Paul C L. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerard M, Hernandez L, Wevrick R, Stewart C L. Disruption of the mouse necdin gene results in early post-natal lethality. Nat Genet. 1999;23:199–202. doi: 10.1038/13828. [DOI] [PubMed] [Google Scholar]
- 13.Hark A T, Schoenherr C J, Katz D J, Ingram R S, Levorse J M, Tilghman S M. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 14.Hogan B, Beddington R, Costanini F, Lacy E. Manipulating the mouse embryo. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 15.Jaenisch R. DNA methylation and imprinting: why bother? Trends Genet. 1997;13:323–329. doi: 10.1016/s0168-9525(97)01180-3. [DOI] [PubMed] [Google Scholar]
- 16.Jay P, Rougeulle C, Massacrier A, Moncla A, Mattei M G, Malzac P, Roeckel N, Taviaux S, Lefranc J L, Cau P, Berta P, Lalande M, Muscatelli F. The human necdin gene, NDN, is maternally imprinted and located in the Prader-Willi syndrome chromosomal region. Nat Genet. 1997;17:357–361. doi: 10.1038/ng1197-357. [DOI] [PubMed] [Google Scholar]
- 17.Kafri T, Ariel M, Brandeis M, Shemer R, Urven L, McCarrey J, Cedar H, Razin A. Developmental pattern of gene-specific DNA methylation in the mouse embryo and germ line. Genes Dev. 1992;6:705–714. doi: 10.1101/gad.6.5.705. [DOI] [PubMed] [Google Scholar]
- 18.Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 19.MacDonald H R, Wevrick R. The necdin gene is deleted in Prader-Willi syndrome and is imprinted in human and mouse. Hum Mol Genet. 1997;6:1873–1878. doi: 10.1093/hmg/6.11.1873. [DOI] [PubMed] [Google Scholar]
- 20.McDonald L E, Paterson C A, Kay G F. Bisulfite genomic sequencing-derived methylation profile of the Xist gene throughout early mouse development. Genomics. 1998;54:379–386. doi: 10.1006/geno.1998.5570. [DOI] [PubMed] [Google Scholar]
- 21.Monk M, Boubelik M, Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development. 1987;99:371–382. doi: 10.1242/dev.99.3.371. [DOI] [PubMed] [Google Scholar]
- 22.Neumann B, Kubicka P, Barlow D P. Characteristics of imprinted genes. Nat Genet. 1995;9:12–13. doi: 10.1038/ng0195-12. [DOI] [PubMed] [Google Scholar]
- 23.Nicholls R D. The impact of genomic imprinting for neurobehavioral and developmental disorders. J Clin Investig. 2000;105:413–418. doi: 10.1172/JCI9460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olek A, Walter J. The pre-implantation ontogeny of the H19 methylation imprint. Nat Genet. 1997;17:275–276. doi: 10.1038/ng1197-275. [DOI] [PubMed] [Google Scholar]
- 25.Razin A, Cedar H. DNA methylation and genomic imprinting. Cell. 1994;77:473–476. doi: 10.1016/0092-8674(94)90208-9. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Shemer R, Birger Y, Riggs A D, Razin A. Structure of the imprinted mouse Snrpn gene and establishment of its parental-specific methylation pattern. Proc Natl Acad Sci USA. 1997;94:10267–10272. doi: 10.1073/pnas.94.19.10267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Solter D. Imprinting. Int J Dev Biol. 1998;42:951–954. [PubMed] [Google Scholar]
- 29.Stoger R, Kubicka P, Liu C G, Kafri T, Razin A, Cedar H, Barlow D P. Maternal-specific methylation of the imprinted mouse Igf2r locus identifies the expressed locus as carrying the imprinting signal. Cell. 1993;73:61–71. doi: 10.1016/0092-8674(93)90160-r. [DOI] [PubMed] [Google Scholar]
- 30.Tanaka M, Puchyr M, Gertsenstein M, Harpal K, Jaenisch R, Rossant J, Nagy A. Parental origin-specific expression of Mash2 is established at the time of implantation with its imprinting mechanism highly resistant to genome-wide demethylation. Mech Dev. 1999;87:129–142. doi: 10.1016/s0925-4773(99)00158-6. [DOI] [PubMed] [Google Scholar]
- 31.Thorvaldsen J L, Duran K L, Bartolomei M S. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 1998;12:3693–3702. doi: 10.1101/gad.12.23.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tilghman S M. The sins of the fathers and mothers: genomic imprinting in mammalian development. Cell. 1999;96:185–193. doi: 10.1016/s0092-8674(00)80559-0. [DOI] [PubMed] [Google Scholar]
- 33.Tremblay K D, Duran K L, Bartolomei M S. A 5′ 2-kilobase-pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Mol Cell Biol. 1997;17:4322–4329. doi: 10.1128/mcb.17.8.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tremblay K D, Saam J R, Ingram R S, Tilghman S M, Bartolomei M S. A paternal-specific methylation imprint marks the alleles of the mouse H19 gene. Nat Genet. 1995;9:407–413. doi: 10.1038/ng0495-407. [DOI] [PubMed] [Google Scholar]
- 35.Tsai T F, Armstrong D, Beaudet A L. Necdin-deficient mice do not show lethality or the obesity and infertility of Prader-Willi syndrome. Nat Genet. 1999;22:15–16. doi: 10.1038/8722. [DOI] [PubMed] [Google Scholar]
- 36.Wade P A, Gegonne A, Jones P L, Ballestar E, Aubry F, Wolffe A P. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat Genet. 1999;23:62–66. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- 37.Warnecke P M, Clark S J. DNA methylation profile of the mouse skeletal α-actin promoter during development and differentiation. Mol Cell Biol. 1999;19:164–172. doi: 10.1128/mcb.19.1.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Warnecke P M, Mann J R, Frommer M, Clark S J. Bisulfite sequencing in preimplantation embryos: DNA methylation profile of the upstream region of the mouse imprinted H19 gene. Genomics. 1998;51:182–190. doi: 10.1006/geno.1998.5371. [DOI] [PubMed] [Google Scholar]
- 39.Yang T, Adamson T E, Resnick J L, Leff S, Wevrick R, Francke U, Jenkins N A, Copeland N G, Brannan C I. A mouse model for Prader-Willi syndrome imprinting-centre mutations. Nat Genet. 1998;19:25–31. doi: 10.1038/ng0598-25. [DOI] [PubMed] [Google Scholar]
- 40.Yoshikawa K. Cell cycle regulators in neural stem cells and postmitotic neurons. Neurosci Res. 2000;37:1–14. doi: 10.1016/s0168-0102(00)00101-2. [DOI] [PubMed] [Google Scholar]