Abstract
Acute myeloid leukemia (AML) with an FLT3 internal tandem duplication (FLT3-ITD) mutation is an aggressive hematologic malignancy associated with frequent relapse and poor overall survival. The tyrosine kinase inhibitor gilteritinib is approved for the treatment of relapse/refractory AML with FLT3 mutations, yet its mechanism of action is not completely understood. Here, we sought to identify additional therapeutic targets that can be exploited to enhance gilteritinib's antileukemic effect. Based on unbiased transcriptomic analyses, we identified the glutamine transporter SNAT1 (SLC38A1) as a novel target of gilteritinib that leads to impaired glutamine uptake and utilization within leukemic cells. Using metabolomics and metabolic flux analyses, we found that gilteritinib decreased glutamine metabolism through the TCA cycle and cellular levels of the oncometabolite 2-hydroxyglutarate. In addition, gilteritinib treatment was associated with decreased ATP production and glutathione synthesis and increased reactive oxygen species, resulting in cellular senescence. Finally, we found that the glutaminase inhibitor CB-839 enhanced antileukemic effect of gilteritinib in ex vivo studies using human primary FLT3-ITD–positive AML cells harboring mutations in the enzyme isocitrate dehydrogenase, which catalyzes the oxidative decarboxylation of isocitrate, producing α-ketoglutarate. Collectively, this work has identified a previously unrecognized, gilteritinib-sensitive metabolic pathway downstream of SLC38A1 that causes decreased glutaminolysis and disruption of redox homeostasis. These findings provide a rationale for the development and therapeutic exploration of targeted combinatorial treatment strategies for this subset of relapse/refractory AML.
Introduction
Acute myeloid leukemia (AML) is a heterogeneous hematopoietic malignancy with aberrant proliferation of undifferentiated myeloid cells. Response to therapeutic treatment and overall survival vary depending on disease subtype, cytogenetics, co-occurring mutations, and age (1–3). Internal tandem duplications (ITD) of the FLT3 receptor result in an aggressive subtype of AML (FLT3-ITD+ AML) that is mutated in approximately 30% of adult patients (1, 4). The presence of a FLT3-ITD leads to constitutive activation of the receptor, poor prognosis, and low overall survival (1–3). Gilteritinib, an inhibitor of FLT3, AXL, and ALK (5, 6) approved for the treatment of FLT3-mutated AML (7), induces marrow responses in this disease (8), decreases cell viability (6), inhibits FLT3 downstream signaling (9), and induces G1 cell-cycle arrest and apoptosis (10) in FLT3-ITD+ AML cell lines and patient samples.
Although these cellular responses were seen with gilteritinib treatment, their molecular mechanisms remain poorly understood. We sought to remedy this knowledge deficit using unbiased transcriptomic approaches, and identified a mechanism of gilteritinib activity involving downregulation of the glutamine transporter SNAT1 (SLC38A1) that causes decreased glutaminolysis and disruption of redox homeostasis. These findings provide a rationale for the development and therapeutic exploration of targeted combinatorial treatment strategies for this subset of AML.
Materials and Methods
Cell culture
Human MV4–11 (ATCC), MOLM13 (DSMZ), and MOLM13-RES (11) cells were maintained in RPMI (Thermo Fisher Scientific) with 10% FBS as described previously (11, 12). Cell lines were verified by short tandem repeat (STR) profiling [OSUCCC Genomics Shared Resource (GSR)] and using the Cellosaurus database (Expasy). Cell lines were used between passages 5 and 30 and tested for Mycoplasma (MycoAlert Mycoplasma Detection Kit; Lonza) prior to use. Human primary AML samples (OSUCCC Leukemia Tissue Bank Shared Resource) were cultured in RPMI with 10% FBS and a human cytokine cocktail (10 ng/mL each of SCF, IL3, GMCSF, and FLT3 ligand; Peprotech). Primary murine FLT3ITD/+/IDH2R140Q/+ AML cells (13) were grown in a murine cytokine cocktail (10 ng/mL mSCF and mIL3, and 20 ng/mL mGMCSF; Peprotech) supplemented with 1× GlutaMax (Thermo Fisher Scientific).
In vivo xenograft mouse models
Xenograft mouse models were generated as described previously (13). Female 8- to 12-week-old NSG mice (The Jackson Laboratory stock number: 005557) were intravenously injected with 1 × 106 MOLM13-Luc+ or MOLM13-RES-Luc+ cells. Weekly tumor engraftment was assessed by bioluminescence imaging. Mice were randomized to treatment groups based on leukemia burden. Drug treatments were started on days 7 or 13 after tail vein injection (TVI) of MOLM13 and MOLM13-RES, respectively. Gilteritinib was formulated in 0.5% methylcellulose (Sigma-Aldrich) and administered at 30 mg/kg once daily for 5 days/week for 3 weeks. Mice were observed daily and euthanized when showing signs of progressive disease including hind limb paralysis, weight loss of more than 20%, and/or lethargy. At study endpoint, bone marrow was harvested, and enriched for AML using MACS (Miltenyi Biotec) separation of CD45+ cells. All animal studies were approved by the OSU Institutional Animal Care and Use Committee.
RNA isolation and RT-PCR
RNA was isolated from cells using TRIzol–Chloroform extraction. cDNA was generated from 1 μg of RNA using SuperScript IV First-Strand Synthesis (Thermo Fisher Scientific). Fifty nanograms of cDNA was used in real-time PCR reactions using TaqMan Fast Advanced Master Mix (Thermo Fisher Scientific) in triplicate, and quantified on a QuantStudio 3 system (Life Technologies). Target Ct (threshold cycle) was standardized to the housekeeping gene GAPDH Ct and graphed as 2–ΔCt. TaqMan gene expression arrays used are listed in the RRID table in Supplementary Material.
RNA-seq and bioinformatics analysis
RNA sequencing (RNA-seq) was carried out on 200 ng total RNA using NEBNext Ultra II Directional RNA Library Prep Kit (Illumina). Paired-end 150 bp sequencing was completed on the HiSeq 4000 (OSUCCC GSR). Samples were prepared in triplicate for RNA-seq. Reads were aligned to the human genome (hg19) using Rsubread21, (>90% of ∼25 × 106 unique reads/sample mapped) and translated to expression counts via featurecounts, followed by a standard edgeR pipeline (14) to determine differentially expressed genes (DEG; FDR-corrected P value < 0.1, fold change > 1.3), visualized with volcano plots (ggplot2) and interpreted by gene set enrichment (15) using the enrichment in the Hallmarks, Curated, GO, and Reactome sets. RNA-seq data are available at Gene Expression Omnibus (GEO) under accession numbers GSE180180 and GSE180181.
Western blot analysis
Cells were lysed in RIPA buffer (Cell Signaling Technology) supplemented with 0.1% SDS, phosphatase (Millipore), and protease (Sigma-Aldrich) inhibitors. Equal protein concentrations (determined by BCA) were separated on 4% to 12% Bis-Tris SDS-polyacrylamide gel using MOPS running buffer (Life Technologies) and transferred to nitrocellulose membranes. Membranes were blocked and probed in 4% milk using the antibodies found in the RRID table in Supplementary Materials. Immunoblots were developed with Signal Fire ECL reagent (Cell Signaling Technology) or SuperSignal West Femto Maximum Sensitivity Substrate (Life Technologies) using Odyssey FC Imaging System (Li-Cor Biosciences). Representative images from at least two blots are shown.
In vitro drug treatments
Cell lines were plated at 0.3 × 106 cells/mL in growth media containing DMSO or 100 nmol/L gilteritinib. For time-course studies, assays were performed or samples (RNA, lysates) were collected at 1, 4, 8, or 24 hours. For combination studies, MOLM13 cells were treated with increasing concentrations of gilteritinib and CB-839 for 72 hours and an MTT assay (3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide; Sigma-Aldrich) was performed.
In vitro glutamine deprivation studies
Cell lines were plated at 0.3 × 106 cells/mL in growth media with or without glutamine and an MTT assay was performed. For cell growth and viability measurements, Trypan blue exclusion assay was performed in triplicate using 0.4% Trypan blue and the Nexcelom Cellometer K2 up to 96 hours.
Uptake assays
MOLM13 cells (1.5 × 106 cells/well) were seeded in triplicate in 24-well plates and treated with gilteritinib (100 nmol/L or 10 μmol/L) in warm phenol-red and FBS-free DMEM for up to 8 hours prior to 15-minute uptake of [3H]-glutamine (1 μmol/L) or [Glycine-2–3H]-glutathione (1 μmol/L). Transport was terminated by washing with ice-cold buffer, and lysing cells in 1 mol/L NaOH overnight, followed by neutralization with 2 mol/L HCl for 30 minutes. Intracellular glutamine or glutathione accumulation was quantified by liquid scintillation counting, and results were normalized to total protein concentration determined by BCA.
Cell line derivation and routine tissue culture conditions for maintaining Chinese hamster ovary (CHO; Invitrogen)-human OAT1 stably expressing cells and substrate accumulation and inhibition assay protocols were reported previously (16). For accumulation assay, cells (0.25 × 106 cells/well in duplicate) were equilibrated in transport buffer (10 minutes, room temperature) then exposed to fresh transport buffer containing 5 μmol/L para-aminohippurate (PAH) with tracer 3H-PAH (0.25 μCi/mL) in the presence or absence of gilteritinib (10–600 μmol/L) for 1 minute. The assay was stopped by washing cells with ice-cold transport buffer. Cell lysates were assayed for radioactivity by liquid scintillation counting, results converted to pmol of uptake per mg protein and reported as percent of control uptake. Active hOAT1-mediated uptake was confirmed in every experiment via inhibition by probenecid (500 μmol/L), the prototypical inhibitor of OAT-mediated uptake. To correct for uptake due to diffusion or endogenously expressed transporters in CHO cells, CHO-hOAT1 uptake values were corrected for PAH accumulation in parental CHO cells prior to analysis. The half maximal inhibitory concentration (IC50) value was calculated using nonlinear regression.
Knockdown of SLC38A1
SLC38A1 shRNA plasmids listed in the RRID table in Supplementary Materials were cotransfected into HEK293FT cells (ThermoFisher) with lentiviral packaging plasmids psPAX2 and pMD2.G (Addgene) using jetPRIME transfection reagent (Polyplus-transfection) following the manufacturer's instructions. Lentiviral supernatant was collected 48 hours posttransfection and precipitated in PEG-it virus precipitation solution (System Biosciences). MOLM13 cells (5 × 106 cells/well) were transduced with fresh, concentrated lentivirus using a spinfection protocol at 1500 × g, 32°C for 90 minutes. Transduced cells were resuspended in fresh media and incubated up to 96 hours at 37°C, 5% CO2.
Metabolomics analysis
MOLM13 or MOLM13-RES cells (5 × 106 cells, n = 4) were treated with 100 nmol/L gilteritinib for 24 hours. DMSO concentration was kept at 0.1%. Cells were harvested, washed twice with cold PBS, and snap frozen using liquid nitrogen. Cell pellets were extracted by 2:1:1 chloroform:methanol:water with 10 mmol/L ammonium bicarbonate, containing butylated hydroxytoluene to prevent the metabolites from oxidation. As previously described (17), both polar and apolar metabolites were analyzed on a Q Exactive Hybrid Quadrupole-Orbitrap Mass Spectrometer equipped with an electrospray ionization source, connected to a Vanquish ultra-high performance liquid chromatography (LC) system (Thermo Fisher Scientific). Mobile phases and separation gradients were mentioned previously in detail (17). Features were extracted from the acquired raw files by SIEVE 2.2.0 SP2 (Thermo Fisher Scientific), and then processed using our in-house MATLAB scripts. Data were normalized by probabilistic quotient normalization before statistical analysis (18). Metabolite identification was achieved by matching accurate masses and retention times to an in-house library of metabolite standards that included the IROA300 4, Mass Spectrometry Metabolite Library of Standards (MSMLS; IROA Technologies). Units of measurement for all metabolites are assigned arbitrary units based on their integrated peak area. The analytic method used did not differentiate between L- and D-enantiomers (i.e., L-2-HG vs. D-2-HG). Pooled quality controls (QC) were used to monitor the instrument stability and blanks were used for background subtraction. Features were included in further analysis, if the coefficient of variance was less than 25% in the corresponding QC replicates and signal:noise ratio was higher than 3.
Metabolic flux analysis
Cells (5 × 106 cells, n = 4) were incubated with 13C5, 15N2-glutamine labeled medium containing 100 nmol/L gilteritinib for 24 hours (0.1% DMSO used as control). The sample preparation and data acquisition were the same as mentioned above.
ROS and oxygen consumption assays
Following drug treatment, cell pellets were stained with 150 μL MitoPY1 (10 μmol/L; mitoROS assay) for 90 minutes or 50 μL CM-H2DCFDA (10 μmol/L; cellular ROS assay) for 5 minutes at 37°C. After the cell loading period, cells were washed twice with PBS and lysed in 100 μL 0.2% Triton X-100/PBS and fluorescence was read on a BioTek Synergy HT microplate reader. The relative fluorescence was normalized to μg of protein (determined via BCA).
Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were determined in MOLM13 cells using the XF Real-Time ATP Rate Assay on a Seahorse XFe24 analyzer (OSUCCC Clinical Translational Science Shared Resource). Following a 1- to 24-hour pretreatment in the appropriate experimental conditions, cells were resuspended in Seahorse XF RPMI assay media, seeded (150,000 cells/well) on polylysine coated Seahorse plates, and incubated at 37°C for 30 minutes in a CO2-free incubator. The assay was performed following the manufacturer's instructions with the following changes: 2.5 μmol/L oligomycin and 1 μmol/L rotenone/antimycin A were used. A mitochondrial stress test was performed similar to aforementioned methods with the following: 2.5 μmol/L oligomycin, 2 (DMSO) or 3 (gilteritinib) μmol/L FCCP and 1 μmol/L rotenone/antimycin A. Data were normalized to cell number/well.
Senescence and proliferation assays
Cells were treated under experimental conditions for up to 96 hours before collection. For the senescence assay, CellEvent Senescence Green Flow Cytometry Assay (Thermo Fisher Scientific) was performed according to the manufacturer's instructions, with staining at 10 μmol/L probe for 90 minutes at 37°C. For the proliferation assay, Baseclick EdU Flow Cytometry Kit (Sigma-Aldrich) was performed according to the manufacturer's instructions, with incubation at 10 μmol/L EdU for 90 minutes. Data were acquired on the BD LSRFortessa (BD Biosciences; OSUCCC FLOW Cytometry Shared Resource) and analyzed using FlowJo (FlowJo, LLC).
CRISPR screen
A high-throughput pool-based loss-of-function CRISPR screen was performed in MOLM13 cells similarly as previously described using the human Brunello CRISPR knockout library (gRNA pooled library in lentiCRISPRv2; Addgene; ref. 19) that was a gift from David Root and John Doench (20). MOLM13 cells were cultured in viral-containing media at low multiplicity of infection (MOI), and a 7-day puromycin selection eliminated non-transduced cells as well as essential gene knockout cells, and the remaining cultures were exposed to 2 nmol/L gilteritinib or DMSO vehicle control for 72 hours (n = 4). Quality control was performed on each replicate. Sequencing and downstream analysis were performed identical to Brinton and colleagues (19), which revealed the top positive (resistant) and negative (synergistic) genes by normalizing the post-treatment levels of sgRNA to pre-treatment levels and comparing gilteritinib-treated with DMSO-treated groups. CRISPR screen data referenced in this study are available in the Gene Expression Omnibus (GSE143314).
Ex vivo drug combination treatments, glutamine deprivation, and Seahorse studies in primary samples
Cells/well (100,000) were plated in a 96-well plate with increasing concentrations of drug. Cell viability, via Trypan blue, was measured every 24 hours. If the viability fell below 50% or at 72 hours of treatment, whichever came earlier, activity was measured via CellTiter-Glo (Promega). Synergy was assessed for the combination of gilteritinib and CB-839 using Combenefit software (21).
For glutamine deprivation, cells were plated with or without glutamine in the conditions described above with the same human or murine cytokine cocktail, and their activity was measured at 72 hours using CellTiter-Glo (Promega).
OCR and ECAR values for human primary AML samples were generated using the XF Real-Time ATP Rate Assay on a Seahorse XFe24 analyzer as mentioned above with the following changes: human primary cells were cultured for up to 48 hours in the above human cytokine cocktail and seeded at 200,000 cells/well on polylysine coated 24-well Seahorse plates prior to the assay.
Statistical analysis
Statistics were performed using GraphPad Prism. For two experimental comparisons, two-tailed, unpaired t test was used. All n values are biological replicates as indicated in the figure legends. Unless otherwise noted, data are represented as mean ± SD. Statistical significance is represented by asterisks: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Study approval
All animal studies were reviewed and approved by the OSU Institutional Animal Care and Use Committee. Deidentified, genomically annotated human primary AML samples were obtained through the OSUCCC Leukemia Tissue Bank Shared Resource, under their IRB-approved protocol and honest broker status in accordance with the Declaration of Helsinki; written informed consent was obtained from all patients prior to use of samples.
Results
Gilteritinib downregulates glutamine transporters and metabolic pathway genes
Using an unbiased approach, we wanted to identify gene targets that were dysregulated following gilteritinib treatment. MOLM13 FLT3-ITD+ AML cells and the MOLM13-RES resistant cell line bearing a FLT3 D835Y mutation (13), were xenografted into NSG mice and treated with gilteritinib (Supplementary Fig. S1A and S1B). Gilteritinib, a type I FLT3 TKI, is active against FLT3-ITD as well as FLT3 tyrosine kinase domain (TKD) activating mutations (e.g., D835Y), providing two models for investigation. At leukemia progression, leukemic cells were isolated from the bone marrow and gene expression was analyzed, which indicated that the glutamine transporter SNAT1 (SLC38A1) was in the top 25 significantly downregulated genes in gilteritinib-treated MOLM13 cells compared to vehicle. Because certain AML cell lines are dependent on glutamine (22), this transporter was considered of particular interest and led us to investigate other known glutamine transporters (Supplementary Table S1) and glutamine-dependent pathway genes. Our data showed that additional glutamine transporters (SLC1A5, SLC7A5, and SLC38A5) were also significantly downregulated in MOLM13 cells, and that SLC38A1 was significantly downregulated in MOLM13-RES cells (Fig. 1A). Among these transporters, SLC38A1 was the most significantly downregulated in both datasets, warranting further investigation. Gene set enrichment analysis (GSEA) revealed that oxidative phosphorylation and glycolysis pathways were significantly down and upregulated, respectively (Supplementary Fig. S1C), indicating a dysregulation in metabolic pathways that we wanted to further investigate. We evaluated whether genes involved in amino acid and additional metabolic pathways were also downregulated following gilteritinib treatment, and found genes involved in aspartate/glutamine metabolism were significantly dysregulated following gilteritinib treatment in both MOLM13 and MOLM13-RES xenograft models (Fig. 1B). The influence of gilteritinib treatment on the three glutamine transporters was verified using RT-PCR in these cells (Fig. 1C). In addition, we analyzed two genes involved in aspartate/glutamine metabolism: asparagine synthetase (ASNS) that generates asparagine from aspartate through glutamine-to-glutamate metabolism and glutamic—pyruvic transaminase 2 (GPT2) that catalyzes the reversible transamination of glutamate and pyruvate to generate alanine and α-ketoglutarate (α-KG). Gilteritinib treatment decreased ASNS and GPT2 expression in vivo in MOLM13-RES cells (Supplementary Fig. S1D), indicating that glutamine metabolism may be dysregulated following gilteritinib exposure.
Figure 1.
Gilteritinib downregulates glutamine transporters and glutamine metabolic pathway genes in vivo. Expression of glutamine transporters (A) and genes involved in glycolysis (B), TCA cycle, and aspartate/glutamine metabolism in MOLM13 and MOLM13-RES xenograft models treated with vehicle or gilteritinib, 30 mg/kg once daily for 5 days per week (n = 8–10 mice per treatment cohort). AML cells were isolated from bone marrow at study endpoint and analyzed by RNA-seq. C, Expression of glutamine transporters by RT-PCR in cells obtained from the MOLM13 and MOLM13-RES xenograft models to validate the RNA-seq data. P values are from two-tailed, unpaired t tests. **, P < 0.01; ***, P < 0.001. ns, not significant.
Gilteritinib decreases glutamine uptake and alters glutamine metabolism
To determine the functional significance of transcriptional changes in glutamine transport and metabolism during gilteritinib treatment, we performed a series of in vitro studies in FLT3-ITD+ AML cell lines. Although the in vitro transcriptional data show that all three glutamine transporters were downregulated by gilteritinib (Fig. 2A), SLC38A1 was the most significantly downregulated in vivo with the highest fold change. The importance of SLC38A1 in glutamine uptake is supported by a recent study in which these glutamine transporters were individually silenced in other malignant cells (23). Furthermore, SLC7A5 couples with SLC3A2 as a sodium-independent obligatory amino acid exchanger that transports intracellular glutamine for an extracellular amino acid (i.e., leucine), and therefore may not be responsible for any potential decrease in intracellular glutamine as SLC7A5 was downregulated. Therefore, we focused our initial efforts on a potential role of SLC38A1 in glutamine uptake and utilization in AML.
Figure 2.
Gilteritinib decreases glutamine transporter expression, uptake, and utilization in FLT3-ITD+ AML cells in vitro. A, RT-PCR of glutamine transporters (SLC38A1, SLC1A5, and SLC7A5) in MOLM13 cells treated with 100 nmol/L gilteritinib for 8 hours (n = 9 replicates; error bars, SD). B, Immunoblot of SLC38A1 expression in MOLM13 and MV4–11 cells treated with 100 nmol/L gilteritinib at the indicated time points. Representative of 5 independent experiments. Immunoblot was captured on Li-Cor Odyssey Fc, quantitated by Image Studio Software, and normalized to 0-hour gilteritinib treatment. C, Glutamine uptake assay in MOLM13 cells treated with 100 nmol/L and 10 μmol/L gilteritinib for up to 8 hours followed by a 15-minute, 1 μmol/L 3H-L-glutamine uptake at the indicated time points (n = 6 replicates). Data were normalized to t = 0 and then DMSO treatment at each time point. D, RT-PCR and Western blot of SLC38A1 shRNA knockdown out to 96 hours (left and center), followed by a 15-minute, 1 μmol/L 3H-L-glutamine uptake assay at 96 hours (right). Cells were treated with 10 μmol/L gilteritinib for 8 hours (n = 4). Representative data of two experiments. E, Total pool of key metabolites from metabolomics analysis of MOLM13 and MOLM13-RES cells treated with 100 nmol/L gilteritinib for 24 hours (n = 4). P values are from two-tailed, unpaired t tests. *, P < 0.05; ***, P < 0.001. NT, nontargeting shRNA control; sh38A1, SLC38A1 shRNA.
Treatment of MOLM13 cells with gilteritinib resulted in decreased SLC38A1 transcript in a time-dependent manner in MOLM13, as well as decreased protein expression in both MOLM13 and MV4–11, while cell viability was unaffected (Fig. 2B; Supplementary Fig. S2A and S2B). An uptake assay of extracellular radiolabeled glutamine was impaired in the presence of gilteritinib in a time- and concentration-dependent manner (Fig. 2C), demonstrating that a decrease in SLC38A1 results in reduced cellular glutamine uptake. To determine if SLC38A1 is responsible for glutamine uptake in FLT3-ITD+ AML cells, we utilized RNA interference to silence expression of this transporter (Fig. 2D, left and center). SLC38A1 knockdown resulted in a significant decrease in radiolabeled glutamine uptake compared to a nontargeting control (Fig. 2D, right), regardless of the presence of drug. These results indicate that even partial knockdown of SLC38A1 affects the uptake of glutamine into AML cells.
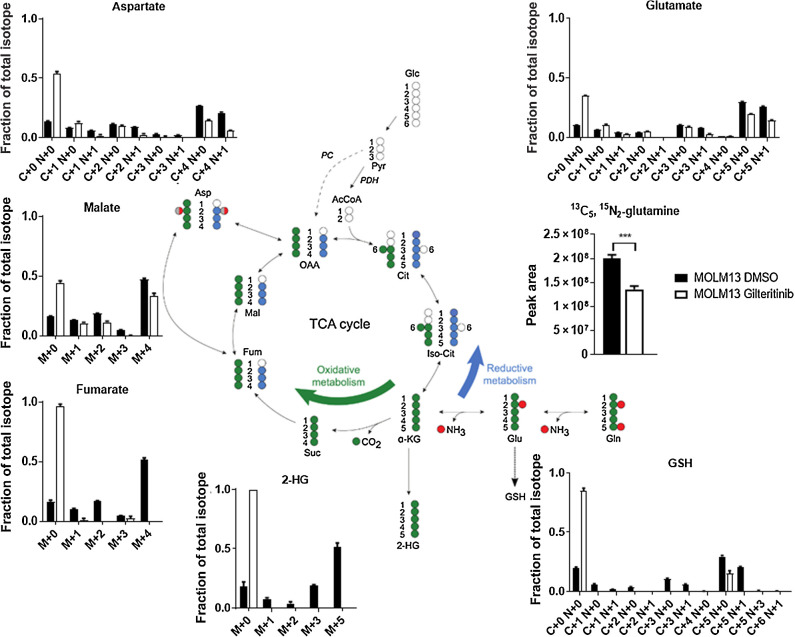
Because glutamine is a metabolic energy precursor, we next analyzed the effects of gilteritinib treatment on key intracellular metabolites in MOLM13 and MOLM13-RES cells in vitro (Fig. 2E; Supplementary Fig. S3). Gilteritinib treatment did not significantly change the total pool of intracellular glutamate, which is produced from the catalysis of glutamine by glutaminase (GLS). This may be attributed to other pathways producing glutamate to compensate for the loss of this amino acid. However, gilteritinib reduced total pools of several key metabolites downstream of glutamate in both cell lines. These included TCA intermediates (fumarate, malate) that are produced from α-KG, which is oxidatively deaminated from glutamate via glutamate dehydrogenase (GLUD), and aspartate, which is produced via glutamic-oxaloacetic transaminase (GOT). Furthermore, gilteritinib treatment in both cell lines significantly decreased the oncometabolite 2-hydroxyglutarate (2-HG), high levels of which are often observed in leukemias, particularly in the context of isocitrate dehydrogenase (IDH) mutations (24–26). Interestingly, gilteritinib treatment increased glycolytic metabolites glucose 6-phosphate (G6P), fructose 1,6-bisphosphate (FBP), glyceraldehyde 3-phosphate (G3P), and the end-product lactate in both cell lines (Supplementary Fig. S3D). In addition, there were changes in NAD/NADH ratios and coenzyme Q with gilteritinib treatment (Supplementary Fig. S3D), indicating a dysregulation of mitochondrial function and oxidative phosphorylation.
Stable isotope-labeling using 13C5, 15N2-glutamine in MOLM13 (Fig. 3; Supplementary Fig. S4A) and MOLM13-RES (Supplementary Fig. S4B and S4C) cells revealed that gilteritinib blocked glutamine anaplerosis. Under normal conditions, glutamine predominately undergoes oxidative metabolism through the TCA cycle. However, gilteritinib treatment reduced the enriched fraction of each TCA intermediate in both cell lines, as well as decreased the enriched fraction of 2-HG. Furthermore, the enriched fraction of aspartate decreased following gilteritinib treatment in both cell lines, indicating that these cells are dependent on the oxidative metabolism of α-KG to generate aspartate.
Figure 3.
Gilteritinib decreases oxidative metabolism in FLT3-ITD+ AML cells. Fractions of total isotopes for key metabolites from metabolic flux analysis of MOLM13 cells treated with 100 nmol/L gilteritinib for 24 hours. P values are from two-tailed, unpaired t tests. ***, P < 0.001.
FLT3-ITD+ AML cells are dependent on glutamine for cell viability and growth
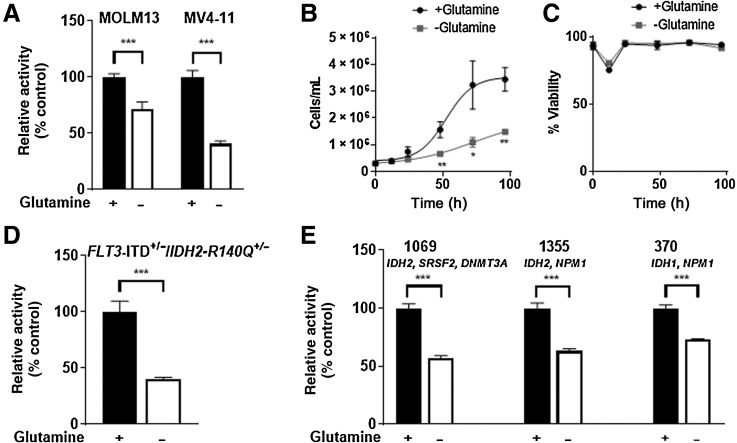
Because glutamine uptake and utilization are decreased in FLT3-ITD+ AML cells following gilteritinib treatment, we investigated the glutamine dependency of these cells, as other AML cell lines are variably affected by glutamine depletion (22). Depleting MOLM13 and MV4–11 cells of glutamine decreased mitochondrial activity (Fig. 4A), and cells grew at a slower rate than cells grown in glutamine-replete media; however, cell viability was unaffected (Fig. 4B and C). When murine primary FLT3-ITD+ cells with a common IDH2 R140Q mutation (FLT3ITD/+/IDH2-R140Q+/−) resulting in neomorphic activity (27) were grown in the presence or absence of glutamine, we found that glutamine depletion reduced ATP production (Fig. 4D). Similarly, glutamine depletion reduced the ATP production of three human primary AML samples with co-occurring FLT3-ITD and IDH mutations (Fig. 4E). Collectively, these findings indicate that FLT3-ITD+ AML cells depend on glutamine for optimal growth.
Figure 4.
FLT3-ITD+ AML cells are dependent on glutamine for growth in vitro and ex vivo. A, MOLM13 and MV4–11 cells were cultured in glutamine-replete or –deplete media for 72 hours (MTT assay, n = 6 replicates; error bars, SD). Data were normalized to +glutamine t = 72. B and C, Cell growth and viability assays of MOLM13 cells grown in glutamine-replete or –deplete media up to 96 hours (Trypan blue exclusion assay, n = 3 replicates; error bars, SD). D and E, Murine (D) and human primary AML cells (E) with the indicated IDH1/2 mutations were cultured in media supplemented with GlutaMax or in glutamine-free media for 72 hours. ATP levels were measured by CellTiter-Glo assay (n = 5–6 replicates; error bars, SD). Data were normalized to +glutamine t = 72. P values are from two-tailed, unpaired t tests. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Gilteritinib decreases glutathione metabolism and reduces metabolic production rate
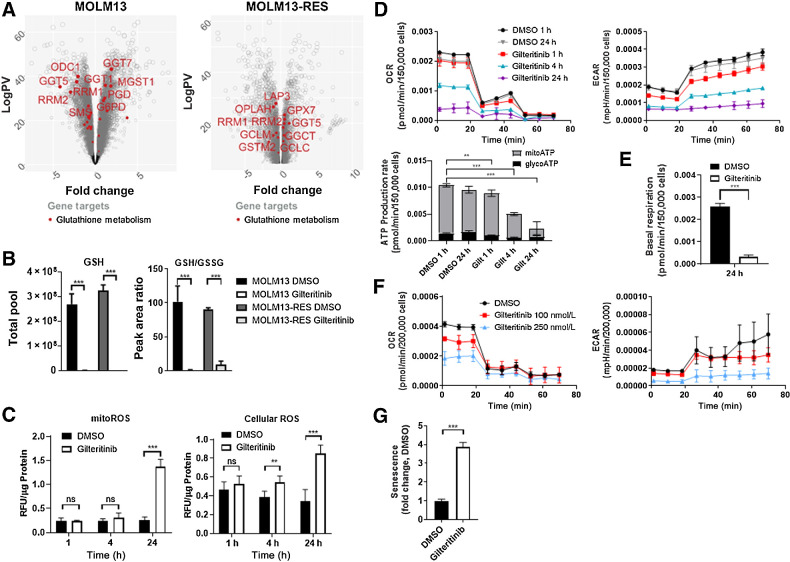
Upon import into the cell, glutamine can be converted to glutamate and then to glutathione (generated from glutamate via two enzymatic reactions: glutamate-cysteine ligase and GSH synthetase). Because ROS and antioxidants, such as glutathione (GSH), are dysregulated in AML (28), we interrogated the GSH and ROS levels in FLT3-ITD+ AML cells following exposure to gilteritinib. RNA-seq data indicated that glutathione metabolism is dysregulated in both MOLM13 and MOLM13-RES xenografts following gilteritinib treatment (Fig. 5A). Metabolomics analysis confirmed a substantial decrease in total abundance of GSH after gilteritinib treatment in vitro (Fig. 5B). Furthermore, metabolic flux analysis of labeled glutamine showed a subsequent decrease in labeled GSH following gilteritinib treatment (Fig. 3; Supplementary Fig. S4). Glutathione is present within cells either in reduced (GSH) or oxidized (GSSG) form, and the ratio of GSH/GSSG can provide insights into the redox state under various conditions. Following gilteritinib treatment, we found that the ratio of GSH/GSSG is significantly decreased, as calculated based on peak areas detected for GSH and GSSG by HPLC-MS (Fig. 5B). The influence of gilteritinib on intracellular glutathione was likely due to glutathione metabolism and not related to direct inhibition of exogenous glutathione uptake, as MOLM13 cells do not take up a substantial amount of exogenous glutathione at baseline, regardless of exposure to gilteritinib. Furthermore, gilteritinib did not inhibit the function of the glutathione transporter OAT1 (IC50 = 79 μmol/L) in engineered cellular models (Supplementary Fig. S5A and S5B).
Figure 5.
Gilteritinib decreases glutathione metabolism and reduces metabolic production rate. A, Expression of genes involved in glutathione metabolism in MOLM13 and MOLM13-RES xenograft models treated with vehicle or gilteritinib, 30 mg/kg once daily for 5 days per week. AML cells were isolated from bone marrow at study endpoint (n = 8–10 mice per treatment cohort) and analyzed by RNA-seq. B, Total pool (GSH) and peak area ratio of glutathione to oxidized glutathione (GSH/GSSG) in MOLM13 and MOLM13-RES cells treated with 100 nmol/L gilteritinib for 24 hours. C, Relative fluorescence normalized to protein of mitochondrial and cellular ROS levels in MOLM13 cells (n = 6–9). D, OCR and ECAR values in MOLM13 cells using the Seahorse XF ATP Production Rate Assay. ATP production rates were calculated using Seahorse Analytics. One representative experiment of two separate experiments (n = 4). E, Basal respiration calculated from OCR values in MOLM13 cells (Seahorse XF substrate oxidation assay) using Seahorse Analytics. One representative experiment of two separate experiments (n = 4). F, OCR and ECAR values in human primary FLT3-ITD+ AML cells (sample 1355 from Fig. 4E) using the Seahorse XF ATP Production Rate Assay. One representative experiment of two separate experiments (n = 4). G, CellEvent Senescence Green FLOW cytometry assay of MOLM13 cells treated with 100 nmol/L gilteritinib for 96 hours. P values are from two-tailed, unpaired t tests. *, P < 0.05; ***, P < 0.001. ns, not significant; RFU, relative fluorescence unit.
Since glutathione metabolism was impaired by gilteritinib in vitro and in vivo, we next evaluated the impact of this process on cellular and mitochondrial ROS levels. Both cellular and mitoROS were significantly increased in gilteritinib-treated cells after 24 hours, with a 5-fold increase in mitoROS (Fig. 5C). Gilteritinib also decreased the oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) in MOLM13 cells in a time-dependent manner, correlating with a reduction in ATP production rate (Fig. 5D). Cells undergoing a mitochondrial stress test confirmed that the cellular respiration was significantly decreased after gilteritinib treatment (Fig. 5E; Supplementary Fig. S5C). We extended these studies to a human primary FLT3-ITD+ AML sample, and observed a concentration-dependent decrease in OCR, ECAR, and ATP production rate (Fig. 5F; Supplementary Fig. S5D). Additionally, a cell proliferation assay revealed that gilteritinib decreased proliferation in MOLM13 cells (Supplementary Fig. S5E). Mitochondrial dysfunction and upregulation of senescence-associated β-galactosidase (SA-β-gal) are hallmarks of senescent cells. To determine whether drug-induced reduction in metabolism was due to the cells undergoing senescence, we used a flow cytometry-based assay and saw a significant increase in senescent-positive cells after a 96-hour exposure to gilteritinib (Fig. 5G; Supplementary Fig. S5F). Taken together, these data indicate that gilteritinib decreased metabolism in FLT3-ITD+ AML cells, and pushed the cells into senescence.
Genome-wide CRISPR knockout screen identifies genes that are synthetically lethal with gilteritinib
A genome-wide CRISPR knockout loss-of-function screen was carried out in MOLM13 cells with selective pressure from 2 nmol/L gilteritinib, as done previously (19) and deposited in the Gene Expression Omnibus (GSE143314), to identify genes that enhance the activity of gilteritinib against AML cells (Fig. 6A). Quality control metrics from the screen are shown in Supplementary Fig. S6A. Because the rationale for this screen was the identification of genes that, once deleted, would enhance the antileukemic activity of gilteritinib, we focused on negatively selected genes in the glutamine transport/metabolism pathway (SLC38A1, SLC1A5, SLC7A5, GLS1, GLS, GCLC, and GSS). Maximum likelihood estimation (MLE) analysis indicated a negative selection of glutaminase (GLS; Fig. 6A), and a KEGG pathway analysis indicated that GLS was positively enriched (Supplementary Fig. S6B). These findings suggest that pharmacologic inhibition of this enzyme might enhance antileukemic activity in combination with gilteritinib.
Figure 6.
Targeting glutaminase enhanced the antileukemic activity of gilteritinib in FLT3-ITD/IDH mutant AML. A, Ranking of enriched sgRNA by maximum likelihood estimation (MLE) analysis by gilteritinib-control beta scores. Beta scores for DMSO and gilteritinib treatment groups were measured with baseline set at day 0, and treatment-control beta score was generated by comparing with each treatment group. GLS indicated by black arrow. B and C, Inhibition of cell viability by gilteritinib in combination with CB-839 was measured by CellTiter-Glo assay (72 hours, n = 3 replicates) in MOLM13 (B) and human primary FLT3-ITD+ AML samples (C). LOEWE analysis was performed for drug synergy using the Combenefit software.
Pharmacologic targeting of GLS enhanced antileukemic activity of gilteritinib
CB-839 is an allosteric GLS inhibitor with intrinsic antileukemic activity in AML (29, 30) that was previously found to reduce 2-HG levels in an IDH1/IDH2-mutant AML cell line (30). Based on the results of our CRISPR screen, we evaluated the efficacy of CB-839 with gilteritinib in FLT3-ITD+ AML cells and found that the drug combination was active in MOLM13 cells (Fig. 6B; Supplementary Fig. S6C and S6D). Because of the downregulation of 2-HG by gilteritinib in MOLM13 and MOLM13-RES cells (Fig. 2E), we anticipated increased activity in AML with concurrent FLT3-ITD and IDH mutations, which was observed in three human primary AML samples (Fig. 6C; Supplementary Fig. S6E and S6F). These results indicate that CB-839 may enhance antileukemic activity in combination with gilteritinib, especially in the context of AML with FLT3-ITD/IDH mutations.
Discussion
Evidence has accumulated in recent years indicating that high expression of glutamine transporters such as SLC38A1 correlates with poor overall survival in various cancers, including AML (31), gastric cancer (32), and hilar cholangiocarcinoma (33). In addition, the glutamine transporters SLC1A5 and SLC7A5 are upregulated in numerous cancers, and often associated with reduced overall survival (34–36). Knockdown or pharmacologically targeting SLC1A5 lead to inhibition of cell proliferation, cell-cycle arrest and increased apoptosis in various cancers (34), but can have varying outcomes in AML cells (37). Collectively, these prior studies warrant further investigation to understand the role of glutamine transporters in AML.
Glutamine is important for anaplerosis processes for rapidly proliferating cells, and many cancer cells become dependent on glutamine as an energy and precursor source for nucleic acid, amino acid and GSH synthesis (38). Inhibition of glutaminolysis demonstrated that this is an important pathway in the growth and survival of AML (29, 30, 39). Interestingly, some AML cell lines had increased apoptotic cells under glutamine deprivation conditions (37), although we observed no change in viability in our studies. There is conflicting evidence whether the FLT3 TKI quizartinib undergoes a metabolic switch to either depend on or impair glutaminolysis in FLT3-ITD+ AML, but cotreatment with the GLS inhibitor CB-839 increased antileukemic response (40, 41). In contrast to our gilteritinib data, quizartinib increased ECAR, indicative of glycolysis utilization, but did not affect GSH/GSSG ratios, suggesting that these FLT3 TKIs may undergo different metabolic processes. It has been postulated that quizartinib impaired glutamine import, but this was not investigated further (41). In our studies presented here, we observed a decrease in TCA intermediates and OXPHOS following gilteritinib treatment. During normal hematopoiesis, ROS is well-regulated in the hypoxic bone marrow microenvironment, controlling the proliferation of hematopoietic stem cells (HSC). However, ROS are elevated in AML, particularly those carrying FLT3 mutations (28). Glutathione is an important antioxidant to regulate oxidative stress, particularly within glutamine-dependent AML cells (22), yet antioxidants are dysregulated in AML (28). Our results indicate that gilteritinib reduced GSH levels and GSH/GSSG ratio, thereby increasing both cellular and mitoROS. The leukemia stem cell (LSC) population may have fewer mitochondria or less mitochondrial activity as well as increased antioxidant pathways, resulting in lower ROS levels (42). It would be interesting to evaluate whether gilteritinib can target this population of AML cells by disrupting GSH and ROS levels. Our results indicate that gilteritinib decreased glutamine transport and metabolism, resulting in decreased basal respiration that pushes the cell towards senescence, which may be a beneficial strategy against highly proliferative AML cells and/or the LSC population. The metabolic mechanism of action is summarized in Supplementary Fig. S7A.
There are several common recurrent mutations in FLT3-ITD+ AML, including those of the IDH1/2 genes (27, 43–45). Under normal conditions, IDH enzymes catalyze the oxidative decarboxylation of isocitrate to α-KG in the TCA cycle. However, mutations in IDH1 or IDH2 can lead to the conversion of α-KG to 2-HG (24–26), which inhibits α-KG–dependent enzymes, resulting in histone and DNA methylation alterations (46) and DNA damage (47). Although MOLM13 FLT3-ITD+ cells do not contain IDH mutations, metabolomics analysis demonstrated that gilteritinib decreased total 2-HG levels, suggesting that inhibition of this oncometabolite may contribute to the mechanism of action in FLT3-ITD/IDH mutant AML cells. Based on the CRISPR screen, we explored the possibility of targeting a second gene involved in glutamine metabolism, GLS, in combination with gilteritinib to further enhance activity in FLT3-ITD+ AML. Using CB-839 to inhibit GLS, we observed an increase in antileukemic activity in combination with gilteritinib in human primary FLT3-ITD+ AML cells with co-occurring IDH mutations, which are inherently more resistant to gilteritinib than the established cell line MOLM13 (13), indicating that this combination may be an effective strategy for further evaluation in this genomic subtype of AML.
One final note is to understand the currently unknown mechanism of gilteritinib's ability to decrease the glutamine transporters. Others have identified potential transcription factors (TF) that regulate the expression of these transporters, such as YAP/TAZ (48, 49), part of the HIPPO pathway. In our in vivo RNA-seq data, the HIPPO pathway was downregulated following gilteritinib treatment (Supplementary Fig. S7B). Future studies will aim to interrogate whether gilteritinib decreases expression or localization of these TFs to the promoter of the glutamine transporter genes, thus decreasing their transcription. In addition, SLC38A1 protein levels decrease rapidly with gilteritinib exposure, indicating a possible role of post-transcriptional/translational regulation that future studies beyond the scope of this study will address.
In conclusion, our data support the premise that gilteritinib inhibits glutamine uptake and utilization by downregulating glutamine transporters, particularly SLC38A1, in FLT3-ITD+ AML. There are drugs that inhibit the glutamine transporters, but these inhibitors often lack efficacy or specificity (50), thus, a clinical grade inhibitor of SLC38A1 has yet to be developed. Consequently, our results, to our knowledge, are the first evidence of a FLT3 TKI decreasing expression of SLC38A1, thereby attenuating glutamine uptake and glutaminolysis to drive the cell towards a senescent state. A drug that decreases expression of glutamine transporters may be therapeutically beneficial to target the cellular metabolism that could be further exploited in combinatorial therapy.
Authors' Disclosures
L.T. Brinton reports personal fees from ZielBio, Inc. outside the submitted work. A. Lodi reports grants from NIH during the conduct of the study. S. Tiziani reports grants from NIH during the conduct of the study. No disclosures were reported by the other authors.
Acknowledgments
We would like to thank the following Ohio State University (OSU) Comprehensive Cancer Center (OSUCCC) Shared Resources: Targeted Validation Shared Resource for acquisition of NSG mice, Genomics Shared Resource for RNA-seq data acquisition, Clinical Translational Science Shared Resource for the use of the Seahorse XF24, and FLOW Cytometry Shared Resource for use of the LSRFortessa. We are grateful to the patients who provided samples for this study and to the OSUCCC Leukemia Tissue Bank (supported by NCIP30 CA016058) for sample procurement. We would also like to acknowledge the OSU University Laboratory Animal Resources (ULAR) for housing and care of animals in this study. We thank Dr. Jack Yalowich for his careful review of this manuscript. This work was supported by the National Institute of Health grants R01 CA138744 (S.D. Baker), P30 CA021765, and R01 CA206210 (S. Tiziani); and the Ohio State University Comprehensive Cancer Center Pelotonia foundation (S.D. Baker, A. Sparreboom).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Therapeutics Online (http://mct.aacrjournals.org/).
Authors' Contributions
M.E. Zavorka Thomas: Conceptualization, data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. X. Lu: Data curation, formal analysis, validation, methodology. Z. Talebi: Investigation. J. Jeon: Formal analysis, investigation. D.R. Buelow: Conceptualization, writing–review and editing. A.A. Gibson: Investigation, methodology. M. Uddin: Investigation. L.T. Brinton: Formal analysis, investigation. J. Nguyen: Formal analysis, investigation. M. Collins: Investigation. A. Lodi: Investigation. S. Sweeney: Resources, formal analysis, investigation. M.J. Campbell: Resources, formal analysis, investigation. D.H. Sweet: Conceptualization, supervision, funding acquisition, investigation, writing–review and editing. A. Sparreboom: Conceptualization, resources, supervision, funding acquisition, writing–review and editing. R. Lapalombella: Resources, formal analysis, funding acquisition, writing–review and editing. S. Tiziani: Conceptualization, resources, formal analysis, supervision, funding acquisition, project administration, writing–review and editing. S.D. Baker: Conceptualization, formal analysis, supervision, funding acquisition, investigation, project administration, writing–review and editing.
References
- 1. Grunwald MR, Levis MJ. FLT3 inhibitors for acute myeloid leukemia: a review of their efficacy and mechanisms of resistance. Int J Hematol 2013;97:683–94. [DOI] [PubMed] [Google Scholar]
- 2. Ferrara F, Schiffer CA. Acute myeloid leukaemia in adults. Lancet 2013;381:484–95. [DOI] [PubMed] [Google Scholar]
- 3. Liu SB, Dong HJ, Bao XB, Qiu QC, Li HZ, Shen HJ, et al. Impact of FLT3-ITD length on prognosis of acute myeloid leukemia. Haematologica 2019;104:e9–e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Daver N, Schlenk RF, Russell NH, Levis MJ. Targeting FLT3 mutations in AML: review of current knowledge and evidence. Leukemia 2019;33:299–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mori M, Kaneko N, Ueno Y, Yamada M, Tanaka R, Saito R, et al. Gilteritinib, a FLT3/AXL inhibitor, shows antileukemic activity in mouse models of FLT3 mutated acute myeloid leukemia. Invest New Drugs 2017;35:556–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lee LY, Hernandez D, Rajkhowa T, Smith SC, Raman JR, Nguyen B, et al. Preclinical studies of gilteritinib, a next-generation FLT3 inhibitor. Blood 2017;129:257–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dhillon S. Gilteritinib: first global approval. Drugs 2019;79:331–9. [DOI] [PubMed] [Google Scholar]
- 8. McMahon CM, Canaani J, Rea B, Sargent RL, Qualtieri JN, Watt CD, et al. Gilteritinib induces differentiation in relapsed and refractory FLT3-mutated acute myeloid leukemia. Blood Adv 2019;3:1581–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kawase T, Nakazawa T, Eguchi T, Tsuzuki H, Ueno Y, Amano Y, et al. Effect of Fms-like tyrosine kinase 3 (FLT3) ligand (FL) on antitumor activity of gilteritinib, a FLT3 inhibitor, in mice xenografted with FL-overexpressing cells. Oncotarget 2019;10:6111–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ueno Y, Mori M, Kamiyama Y, Saito R, Kaneko N, Isshiki E, et al. Evaluation of gilteritinib in combination with chemotherapy in preclinical models of FLT3-ITD(+) acute myeloid leukemia. Oncotarget 2019;10:2530–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zimmerman EI, Turner DC, Buaboonnam J, Hu S, Orwick S, Roberts MS, et al. Crenolanib is active against models of drug-resistant FLT3-ITD-positive acute myeloid leukemia. Blood 2013;122:3607–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jarusiewicz JA, Jeon JY, Connelly MC, Chen Y, Yang L, Baker SD, et al. Discovery of a diaminopyrimidine FLT3 inhibitor active against acute myeloid leukemia. ACS Omega 2017;2:1985–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jeon JY, Buelow DR, Garrison DA, Niu M, Eisenmann ED, Huang KM, et al. TP-0903 is active in models of drug-resistant acute myeloid leukemia. JCI Insight 2020;5:e140169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP. GSEA-P: a desktop application for gene set enrichment analysis. Bioinformatics 2007;23:3251–3. [DOI] [PubMed] [Google Scholar]
- 16. Vanwert AL, Srimaroeng C, Sweet DH. Organic anion transporter 3 (oat3/slc22a8) interacts with carboxyfluoroquinolones, and deletion increases systemic exposure to ciprofloxacin. Mol Pharmacol 2008;74:122–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sweeney SR, Collins M, Pandey R, Chiou J, Lodi A, Tiziani S. Identification of a synergistic combination of dimethylaminoparthenolide and shikonin alters metabolism and inhibits proliferation of pediatric precursor-B cell acute lymphoblastic leukemia. Mol Carcinog 2020;59:399–411. [DOI] [PubMed] [Google Scholar]
- 18. Dieterle F, Ross A, Schlotterbeck G, Senn H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal Chem 2006;78:4281–90. [DOI] [PubMed] [Google Scholar]
- 19. Brinton LT, Sher S, Williams K, Canfield D, Orwick S, Wasmuth R, et al. Cotargeting of XPO1 enhances the antileukemic activity of midostaurin and gilteritinib in acute myeloid leukemia. Cancers (Basel) 2020;12:1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 2016;34:184–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Di Veroli GY, Fornari C, Wang D, Mollard S, Bramhall JL, Richards FM, et al. Combenefit: an interactive platform for the analysis and visualization of drug combinations. Bioinformatics 2016;32:2866–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Goto M, Miwa H, Shikami M, Tsunekawa-Imai N, Suganuma K, Mizuno S, et al. Importance of glutamine metabolism in leukemia cells by energy production through TCA cycle and by redox homeostasis. Cancer Invest 2014;32:241–7. [DOI] [PubMed] [Google Scholar]
- 23. Broer A, Rahimi F, Broer S. Deletion of amino acid transporter ASCT2 (SLC1A5) reveals an essential role for transporters SNAT1 (SLC38A1) and SNAT2 (SLC38A2) to sustain glutaminolysis in cancer cells. J Biol Chem 2016;291:13194–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gross S, Cairns RA, Minden MD, Driggers EM, Bittinger MA, Jang HG, et al. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J Exp Med 2010;207:339–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 2010;17:225–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sellner L, Capper D, Meyer J, Langhans CD, Hartog CM, Pfeifer H, et al. Increased levels of 2-hydroxyglutarate in AML patients with IDH1-R132H and IDH2-R140Q mutations. Eur J Haematol 2010;85:457–9. [DOI] [PubMed] [Google Scholar]
- 27. Medeiros BC, Fathi AT, DiNardo CD, Pollyea DA, Chan SM, Swords R. Isocitrate dehydrogenase mutations in myeloid malignancies. Leukemia 2017;31:272–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sillar JR, Germon ZP, DeIuliis GN, Dun MD. The role of reactive oxygen species in acute myeloid leukaemia. Int J Mol Sci 2019;20:6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jacque N, Ronchetti AM, Larrue C, Meunier G, Birsen R, Willems L, et al. Targeting glutaminolysis has antileukemic activity in acute myeloid leukemia and synergizes with BCL-2 inhibition. Blood 2015;126:1346–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Matre P, Velez J, Jacamo R, Qi Y, Su X, Cai T, et al. Inhibiting glutaminase in acute myeloid leukemia: metabolic dependency of selected AML subtypes. Oncotarget 2016;7:79722–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li Y, Shao H, Da Z, Pan J, Fu B. High expression of SLC38A1 predicts poor prognosis in patients with de novo acute myeloid leukemia. J Cell Physiol 2019;234:20322–8. [DOI] [PubMed] [Google Scholar]
- 32. Xie J, Li P, Gao HF, Qian JX, Yuan LY, Wang JJ. Overexpression of SLC38A1 is associated with poorer prognosis in Chinese patients with gastric cancer. BMC Gastroenterol 2014;14:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yu WL, Cong WM, Zhang Y, Chen Y, Wang F, Yu G. Overexpression of ATA1/SLC38A1 predicts future recurrence and death in Chinese patients with hilar cholangiocarcinoma. J Surg Res 2011;171:663–8. [DOI] [PubMed] [Google Scholar]
- 34. Liu Y, Zhao T, Li Z, Wang L, Yuan S, Sun L. The role of ASCT2 in cancer: a review. Eur J Pharmacol 2018;837:81–7. [DOI] [PubMed] [Google Scholar]
- 35. Scalise M, Pochini L, Galluccio M, Console L, Indiveri C. Glutamine transport and mitochondrial metabolism in cancer cell growth. Front Oncol 2017;7:306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Salisbury TB, Arthur S. The regulation and function of the L-type amino acid transporter 1 (LAT1) in cancer. Int J Mol Sci 2018;19:2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Willems L, Jacque N, Jacquel A, Neveux N, Maciel TT, Lambert M, et al. Inhibiting glutamine uptake represents an attractive new strategy for treating acute myeloid leukemia. Blood 2013;122:3521–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Choi YK, Park KG. Targeting glutamine metabolism for cancer treatment. Biomol Ther 2018;26:19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Emadi A, Jun SA, Tsukamoto T, Fathi AT, Minden MD, Dang CV. Inhibition of glutaminase selectively suppresses the growth of primary acute myeloid leukemia cells with IDH mutations. Exp Hematol 2014;42:247–51. [DOI] [PubMed] [Google Scholar]
- 40. Gallipoli P, Giotopoulos G, Tzelepis K, Costa ASH, Vohra S, Medina-Perez P, et al. Glutaminolysis is a metabolic dependency in FLT3(ITD) acute myeloid leukemia unmasked by FLT3 tyrosine kinase inhibition. Blood 2018;131:1639–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gregory MA, Nemkov T, Reisz JA, Zaberezhnyy V, Hansen KC, D'Alessandro A, et al. Glutaminase inhibition improves FLT3 inhibitor therapy for acute myeloid leukemia. Exp Hematol 2018;58:52–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mattes K, Vellenga E, Schepers H. Differential redox-regulation and mitochondrial dynamics in normal and leukemic hematopoietic stem cells: a potential window for leukemia therapy. Crit Rev Oncol Hematol 2019;144:102814. [DOI] [PubMed] [Google Scholar]
- 43. Patel JP, Gonen M, Figueroa ME, Fernandez H, Sun Z, Racevskis J, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med 2012;366:1079–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med 2010;363:2424–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Panuzzo C, Signorino E, Calabrese C, Ali MS, Petiti J, Bracco E, et al. Landscape of tumor suppressor mutations in acute myeloid leukemia. J Clin Med 2020;9:802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell 2011;19:17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rakheja D, Konoplev S, Medeiros LJ, Chen W. IDH mutations in acute myeloid leukemia. Hum Pathol 2012;43:1541–51. [DOI] [PubMed] [Google Scholar]
- 48. Park YY, Sohn BH, Johnson RL, Kang MH, Kim SB, Shim JJ, et al. Yes-associated protein 1 and transcriptional coactivator with PDZ-binding motif activate the mammalian target of rapamycin complex 1 pathway by regulating amino acid transporters in hepatocellular carcinoma. Hepatology 2016;63:159–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liu P, Calvisi DF, Kiss A, Cigliano A, Schaff Z, Che L, et al. Central role of mTORC1 downstream of YAP/TAZ in hepatoblastoma development. Oncotarget 2017;8:73433–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Broer A, Fairweather S, Broer S. Disruption of amino acid homeostasis by novel ASCT2 inhibitors involves multiple targets. Front Pharmacol 2018;9:785. [DOI] [PMC free article] [PubMed] [Google Scholar]