Abstract
MicroRNAs (miRNAs) are a class of small endogenous non‐coding genes that play important roles in post‐transcriptional regulation as well as other important biological processes. Accumulating evidence indicated that miRNAs were extensively involved in the pathology of cancer. However, determining which miRNAs are related to a specific cancer is problematic because one miRNA may target multiple genes and one gene may be targeted by multiple miRNAs. The authors proposed a new approach, named miR_SubPath, to identify cancer‐associated miRNAs by three steps. The targeted genes were determined based on differentially expressed genes in significant dysfunctional subpathways. Then the candidate miRNAs were determined according to miRNA–genes associations. Finally, these candidate miRNAs were ranked based on their relations with some seed miRNAs in a functional similarity network. Results on real‐world datasets showed that the proposed miR_SubPath method was more robust and could identify more cancer‐related miRNAs than a prior approach, miR_Path, miR_Clust and Zhang's method.
Inspec keywords: bioinformatics, cancer, genetics, molecular biophysics, genomics, RNA, cellular biophysics, macromolecules, medical computing, biology computing
Other keywords: identifying cancer, MicroRNAs, noncoding genes, important biological processes, specific cancer, multiple genes, gene, multiple miRNAs, targeted genes, differentially expressed genes, candidate miRNAs, miRNA–genes associations, seed miRNAs, cancer‐related miRNAs
1 Introduction
MicroRNAs (miRNAs) are a class of small (∼22 nt) non‐coding RNAs that may degrade their target genes or suppress their expression at the post‐transcriptional stage [1, 2]. A single miRNA can usually regulate many genes, and different miRNAs could also target the same gene [3]. MiRNAs are involving in many critical processes, such as cell development, proliferation, differentiation, apoptosis, signal transduction and viral infection [4–6]. Therefore, they have critical influence in the initiation and progression of human cancers [7]. More and more miRNAs have been identified to associate to cancers. For example, miR‐21 played a pivotal role in gastric cancer pathogenesis and progression [8], and altered expression of miR‐21, miR‐31, miR‐143 and miR‐145 were found to be related to colorectal cancer [9]. MiR‐205 could suppress cell growth and invasion in breast cancer [10]. The up‐regulation of miR‐155 and down‐regulation of let‐7a were used to predict survival in patients with lung cancer [11].
The human miRNA disease database (HMDD) and miR2Disease database have collected hundreds of miRNAs related to various diseases [12, 13]. However, there are still many cancer‐related miRNAs not being identified, so many approaches have been proposed to tackle this problem. For example, Kuo et al. developed a statistical approach, imputed miRNA regulation based on weighted ranked expression and putative miRNA targets, to detect differentially expressed miRNAs between normal and cancerous samples [14]. Li et al. [15] proposed to identify cancer related miRNAs through the functional consistency between the target genes and cancer. Zhang et al. [16] proposed to predict cancer‐related miRNAs by investigating matched gene and miRNA expression data as well as miRNA‐gene regulations. Other approaches, such as RWRMDA and RLSMDA, identify cancer miRNA through the Random Walk strategy in the miRNA–miRNA functional similarity network [17, 18]. Recently, Zhao et al. proposed two novel approaches, miR_Clust and miR_Path, to predict cancer‐associated miRNAs based solely on differentially expressed genes (DEGs) [19]. For each miRNA, they first clustered all its possible target genes into different groups according to the Pearson's correlation coefficients of their expressions. MiR_Clust ranked each miRNA according to the discriminant power of its clusters for separating the cancers form the controls. MiR_Path ranked each miRNA based on the enrichment score of each cluster on the dysfunctional pathways.
In recent years, more and more works showed that cancers were caused by the dysfunction in some small networks of the pathways, named subpathway, instead of the entire pathway [20–22]. Then a lot of methods had been proposed to detect cancer‐related genes based on subpathways, such as DEgraph [23], the clipper approach [24], PATHWAYS [25], and sub‐SPIA [26]. In this paper, we proposed a new approach, named miR_SubPath, to identify cancer‐associated miRNAs based on genes significantly differentially expressed in some subpathways. Results on real‐world datasets showed that the proposed miR_SubPath method was more robust and could identify more cancer‐related miRNAs than prior approaches, miR_Path, miR_Clust and Zhang's method.
2 Materials and methods
2.1 Gene expression datasets
We used eight cancer datasets downloaded from NCBI's Gene Expression Omnibus (GEO) [27]: GSE7670 and GSE10072 for lung cancer, GSE9348 and GSE20916 for colon cancer, GSE13911 and GSE19826 for gastric cancer, and GSE37290 and GSE69428 for ovarian cancer. The number of samples and the platform of these datasets are presented in Table 1.
Table 1.
Eight gene expression datasets for four different types of cancers
| Cancer | GEO accession number | Number of samples (case/control) | platform |
|---|---|---|---|
| colon | GSE9348 | 82 (70/12) | GPL570 |
| GSE20916 | 145 (101/44) | GPL570 | |
| lung | GSE7670 | 66 (36/30) | GPL96 |
| GSE10072 | 107 (58/59) | GPL96 | |
| gastric | GSE13911 | 69 (38/31) | GPL570 |
| GSE19826 | 27 (12/15) | GPL570 | |
| ovarian | GSE37290 | 20 (10/10) | GPL570 |
| GSE69428 | 20 (10/10) | GPL570 |
2.2 miRNA networks
In this paper, we used the relations in three miRNA networks to identify cancer related miRNAs: miRNA–gene associations, miRNA–miRNA functional similarity network and disease‐miRNA networks. The relationship between miRNA and their targeting genes was the same as the one used by Zhao et al. [16]. First, they downloaded all candidate miRNAs from miRBase (version 16) [28]. Only those miRNA–gene relations affirmed by at least two tools, including PicTar [29], miRanda (version 3.0) [30], microT (version 5.0) [31], or TargetScan (release 6.2) [32], were retained. Second, they added these experimentally determined relations in TarBase (version 6.0) [33] to the candidate miRNAs. As miRNAs with similar functions are more likely associated with similar diseases, we downloaded the miRNA–miRNA functional similarity network including 271 miRNAs (http://cmbi.bjmu.edu.cn/misim). In this network, miRNAs in the same family or cluster were assigned a higher score than those outside. Concerning the disease–miRNA association, we combined HMDD (10,368 curated and experimentally supported miRNA–disease associations between 572 miRNAs and 378 diseases) and miR2Diseas (containing 1939 miRNA–disease associations between 299 miRNAs and 94 diseases). These miRNAs related with a specific disease were used as seed to identify other potential cancer‐related miRNAs in the functional similarity network.
2.3 Methods
2.3.1 Identifying cancer‐related candidate genes
The Kyoto Encyclopaedia of Genes and Genomes (KEGG; http://www.kegg.jp/kegg/xml/) is an important pathway database that includes both metabolic and signalling pathways. We downloaded 137 signalling pathways from the KEGG database, and the gene network for signalling pathway was reconstructed by graphite package [34]. The DEGs were mapped onto each pathway network using the Limma package for R [35]. We applied sub‐SPIA method to detect cancer‐related subpathways with p ‐values = 0.01 corrected by false discovery rate.
2.3.2 Rank cancer‐associated candidate miRNA
The miRNAs targeting genes in the identified significant subpathways were considered as potential candidate cancer‐related miRNAs. We ranked them based on their relations with seed miRNAs in the functional similarity network. Given a seed set S with N verified cancer‐related miRNAs, miRNA r was scored as
The item was adopted to emphasise that the more cancer‐related miRNAs associated with r the more likely it is associated with the cancer.
Fig. 1 shows the three steps to identify cancer‐related miRNAs in this paper:
Step 1 : Identify significant cancer‐related subpathways by sub_SPIA method based on DEGs.
Step 2 : Identify potential cancer‐related miRNAs based on genes in the subpathways and the miRNA‐gene relations.
Step 3 : Rank candidate miRNAs based on their similarity score with seed miRNAs in the functional similarity network.
Fig. 1.
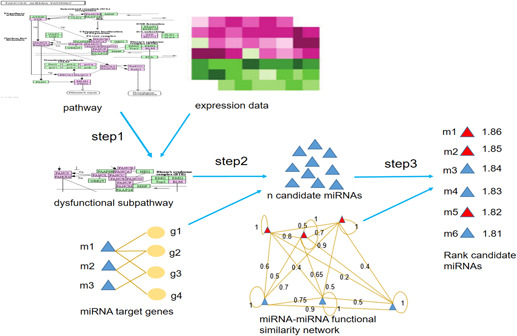
Main workflow to identify cancer‐related miRNAs based on subpathways and functional similarity network
As the potential targeted genes are determined based on significant dysfunctional subpathways, we call the proposed method as miR_SubPath.
3 Result
We applied the proposed miR_SubPath to the eight cancer datasets. For each dataset, N seeds were randomly generated to calculate the scores of all candidate miRNAs. The final score for each candidate miRNA was the average value of 1000 random tests. For fair comparison, we compared our results with miR_Path, miR‐Clust and Zhang's method based on the results of the top 100 miRNAs. The results for the miR_Path method were obtained by the code and data files provided at http://comp‐sysbio.org/miR_Path/ [19]. Zhang's pipeline identified cancer miRNAs that regulated the top 30% genes differentially expressed between normal and control samples were regarded as cancer miRNAs.
3.1 Performance and evaluation
We implemented miR_SubPath by setting the seed number to N = 10, 20 and 30, respectively. As the results of miR_SubPath on the eight datasets for N > 10 have no obvious difference, we only present the precision, recall, F1, and mean of the two approaches for the eight cancer datasets for N = 10 in Table 2. The precision scores of miR_SubPath are all higher than the other three methods on the eight datasets, while the recall scores of miR_SubPath are all higher than them apart from datasets GSE10072 and GSE9348. Concerning the average F1 score, miR_SubPath performs about 14, 16 and 19% higher than that of miR_Path, miR_Clust and Zhang's method on the eight datasets. Furthermore, the F1 scores of miR_SubPath in two independent datasets of a cancer are very close while that of other three methods differ a lot. These observations demonstrate that miR_SubPath performs better than other three methods when using only a few known cancer‐related miRNAs as seeds, resulting in the identification of an increased number of potential miRNAs.
Table 2.
Performance of different approaches over eight cancer dataset
| Index | Method | Colon cancer | Lung cancer | Gastric cancer | Ovarian cancer | Mean | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| GSE9348 | GSE20916 | GSE7670 | GSE10072 | GSE13911 | GSE19826 | GSE37290 | GSE69428 | |||
| precision | miR_SubPath miR_Path | 0.66 | 0.69 | 0.71 | 0.72 | 0.57 | 0.56 | 0.63 | 0.62 | 0.64 |
| 0.51 | 0.49 | 0.39 | 0.48 | 0.39 | 0.40 | 0.38 | 0.40 | 0.43 | ||
| miR_Clust | 0.47 | 0.46 | 0.38 | 0.46 | 0.37 | 0.36 | 0.37 | 0.39 | 0.41 | |
| zhang | 0.43 | 0.44 | 0.33 | 0.42 | 0.36 | 0.37 | 0.35 | 0.35 | 0.38 | |
| recall | miR_SubPath miR_Path | 0.61 | 0.75 | 0.66 | 0.67 | 0.61 | 0.62 | 0.60 | 0.61 | 0.64 |
| 0.94 | 0.48 | 0.66 | 0.88 | 0.57 | 0.57 | 0.31 | 0.42 | 0.60 | ||
| miR_Clust | 0.92 | 0.41 | 0.69 | 0.90 | 0.61 | 0.62 | 0.34 | 0.45 | 0.61 | |
| zhang | 0.84 | 0.47 | 0.38 | 0.79 | 0.53 | 0.55 | 0.43 | 0.43 | 0.55 | |
| F1 | miR_SubPath miR_Path | 0.63 | 0.71 | 0.68 | 0.69 | 0.58 | 0.58 | 0.61 | 0.61 | 0.63 |
| 0.66 | 0.48 | 0.49 | 0.62 | 0.46 | 0.47 | 0.34 | 0.40 | 0.49 | ||
| miR_Clust | 0.62 | 0.43 | 0.49 | 0.60 | 0.46 | 0.45 | 0.35 | 0.41 | 0.47 | |
| zhang | 0.57 | 0.45 | 0.35 | 0.55 | 0.42 | 0.44 | 0.39 | 0.39 | 0.44 | |
3.2 Identification of novel cancer‐related miRNAs
Although the precision, recall, F1, and mean of miR_SubPath had no obvious difference for N > 10, the ranks of the miRNAs might be different given different seeds. In this section, we used all of the cancer‐related miRNAs in the HMDD and miR2Disease databases as seeds to identify potential cancer‐related miRNAs. Among the top 100 miRNAs detected from each of the eight datasets, we found a number of novel miRNAs not recorded in HMDD or miR2Disease that were reported to be associated with a specific cancer in previous publications. Table 3 presents the rank of these potential miRNAs, and the supporting literatures are listed in Table 4. Interestingly, the ranks of most miRNAs from the two independent datasets for a specific cancer are very close. The overlap of the miRNAs in the two independent of a cancer is over 88. All these results indicate the proposed miR_SubPath method is very robust to rank candidate miRNAs.
Table 3.
Rank of the predicted cancer‐related miRNAs supported by at least one publication
| No. | Rank (colon cancer) | Rank (lung cancer) | ||||
|---|---|---|---|---|---|---|
| miRNA | GSE9348 | GSE20919 | miRNA | GSE7670 | GSE10072 | |
| 1 | Mir‐16 | 61 | 55 | Mir‐92a | 49 | 50 |
| 2 | Mir‐101 | 40 | 38 | Mir‐194 | 64 | 66 |
| 3 | Mir‐9 | 43 | 44 | Mir‐30c | 25 | 24 |
| 4 | Mir‐29c | 93 | 85 | Mir‐153 | 97 | 95 |
| 5 | Mir‐7 | 72 | 68 | Mir‐302b | 59 | 60 |
| 6 | Mir‐218 | 29 | 37 | Mir‐373 | 74 | 74 |
| 7 | Mir‐30b | 46 | 52 | Mir‐367 | 58 | 58 |
| 8 | Mir‐204 | 66 | 67 | Mir‐24 | 47 | 48 |
| 9 | Mir‐125b | 64 | 62 | Mir‐135a | 81 | 81 |
| 10 | Mir‐92a | 58 | 51 | Mir‐133a | 71 | 71 |
| 11 | Mir‐214 | 33 | 31 | Mir‐181a | 84 | 83 |
| 12 | Mir‐302a | 62 | 64 | Mir‐23b | 85 | 85 |
| No. | Rank (gastric cancer) | Rank (ovarian cancer) | ||||
|---|---|---|---|---|---|---|
| miRNA | GSE13911 | GSE19826 | miRNA | GSE37290 | GSE69428 | |
| 1 | Mir‐19 | 10 | 10 | Mir‐132 | 25 | 28 |
| 2 | Let‐7c | 7 | 7 | Mir‐194 | 34 | 37 |
| 3 | Let‐7b | 30 | 27 | Mir‐29b | 42 | 41 |
| 4 | Let‐7f | 6 | 6 | Mir‐205 | 46 | 45 |
| 5 | Let‐7i | 19 | 20 | Mir‐153 | 53 | 53 |
| 6 | Mir‐218 | 29 | 28 | Mir‐7 | 60 | 60 |
| 7 | Mir‐92a | 63 | 60 | Mir‐196a | 62 | 62 |
| 8 | Mir‐7 | 59 | 55 | Mir‐1 | 64 | 64 |
| 9 | Mir‐23b | 85 | 83 | Mir‐92a | 66 | 66 |
| 10 | Mir‐30a | 79 | 74 | Mir‐98 | 69 | 69 |
| 11 | Mir‐24 | 62 | 61 | Mir‐106a | 71 | 70 |
| 12 | Mir‐101 | 48 | 46 | Mir‐373 | 72 | 71 |
| 13 | Mir‐194 | 50 | 49 | Mir‐372 | 75 | 75 |
| 14 | Mir‐196a | 49 | 48 | Mir‐181b | 78 | 77 |
| 15 | Mir‐125b | 48 | 65 | Mir‐203 | 80 | 82 |
| 16 | Mir‐1 | 73 | 69 | Mir‐181a | 82 | 83 |
| 17 | Mir‐133a | 67 | 66 | Mir‐135a | 89 | 89 |
| 18 | Mir‐367 | 47 | 45 | Mir‐15a | 91 | 91 |
| 19 | Mir‐215 | 56 | 54 | Mir‐130b | 98 | 95 |
| 20 | Mir‐15a | 97 | 95 | — | — | — |
| 21 | Mir‐181a | 86 | 84 | — | — | — |
| 22 | Mir‐203 | 80 | 80 | — | — | — |
Table 4.
PubMed IDs for the identified novel miRNA in PubMed database for cancer
| Cancer | MiRNA/PubMed ID | MiRNA/PubMed ID | MiRNA/PubMed ID |
|---|---|---|---|
| colon cancer | Mir‐204/27095441 | Mir ‐302a/26191138 | Mir‐9/26983891/25940709 |
| Mir‐29c/26187445/25193986 | Mir‐106/25623762/25435873 | Mir‐125b/26693202/26038573 | |
| Mir‐30b/24593661/24293274 | Mir‐214/27811858/27537384 | Mir‐7/27919977/26648422 | |
| Mir‐92a/27565378/27131314 | Mir‐101/27435782/26071354 | Mir‐21827779719/27462788 | |
| lung cancer | Mir‐302b/27160836 | Mir‐194/27035759/26909612 | Mir‐135a/27525941/26235874 |
| Mir‐367/22835608 | Mir‐153/26339455/25475731 | Mir‐133a/27282282/25903369 | |
| Mir‐92a/26432332/23820254 | Mir‐373/25591738/25063738 | Mir‐181a/27802900/26323677 | |
| Mir‐23b/27268921/24966325 | Mir‐24/25725584/23794259 | Mir‐30c/25119247/25249344 | |
| gastric cancer | Mir‐19/26762410 | Mir‐367/25489984 | Let‐7c/26701848/25549793 |
| Let‐7i/25549793/23107361 | Let‐7f/25549793/21533124 | Mir‐92a/26790436/26499948 | |
| Mir‐15a/26894855/25743273 | Mir‐7/24573489/22614005 | Mir‐30a/27876712/27212164 | |
| Let‐7b/27497248/26564501 | Mir‐23b/26835790/26041881 | Mir‐24/26758252/26045155 | |
| Mir‐194/27874950/25412959 | Mir‐1/27349337/2587449 | Mir‐125b/26504803/25240408 | |
| Mir‐181a/26793992/26589846 | Mir‐203/27542403/27142767 | Mir‐218/27696291/27642088 | |
| Mir‐215/26716895/24981590 | Mir‐133a/26629938/25815687 | Mir‐196a/27420607/25374225 | |
| ovarian cancer | Mir‐194/27486333 | Mir‐153/25954928 | Mir‐1/27354590 |
| Mir‐92a/25448599 | Mir‐98/21109987 | Mir‐372/28456593 | |
| Mir‐98/21109987 | Mir‐372/28456593 | Mir‐181b/24735543 | |
| Mir‐132/27812929/27186275 | Mir‐29b/26512921/25738313 | Mir‐205/28145479/26275944 | |
| Mir‐196a/27890373/26097603 | Mir‐181a/27249598/24394555 | Mir‐135a/24607788/24016480 | |
| Mir‐130b/27048832/26573160 | Mir‐203/27655286/27347348 | Mir‐106a/27510094/27393101 |
A total of 12 of the 31 miRNAs were found to be related to colon cancer. For example, decreased expression of mir‐218 (ranked 29 in GSE9348 and 37 in GSE20916) was associated with poor prognosis in patients with colorectal cancer [36]. It could inhibit cell‐cycle progression, the invasion and migration of colon cancer cells by targeting the PI3K/Akt/mTOR signalling pathway [37]. Mir‐9 (ranked 43 in GSE9348 and 44 in GSE20916) suppressed cell migration and invasion via down‐regulation of the TM4SF1 transmembrane protein in colorectal cancer [38]. Its up‐regulation promoted cell motility to induce metastasis of colorectal cancer [39].
A total of 12 of the 28 miRNAs were found to be associated with lung cancer. Mir‐373 (ranked 74 in GSE7670 and 74 GSE10072) could affect human lung cancer cell growth and the expression of E‐cadherin [40]. Mir‐133a (ranked 71 in GSE7670 and 71 in GSE10072) could suppress multiple oncogenic membrane receptors and cell invasion in non‐small cell lung carcinomas [41]. A clinical study also observed the down‐regulation of mir‐133a in non‐small cell lung cancer [42]. It could regulate some novel molecular networks in lung squamous cell carcinomas [43].
A total of 22 of the 36 miRNAs were found to be associated with gastric cancer. For example, let‐7b (ranked 30 in GSE13911 and 27 in GSE19826) could inhibit cell proliferation, migration, and invasion of cancer cells by targeting CTHRC1 [44, 45]. Silencing of let‐7b could activate AKT signalling to promote gastric carcinogenesis [46]. Mir‐203 (ranked 80 in GSE13911 and 80 in GSE19826) could suppress growth of gastric cancer by targeting PIBF1/Akt signalling [47], and inhibit tumour invasion and metastasis in gastric cancer by regulating the serine/threonine kinase ATM [48]. It could promote the proliferation and invasion of gastric cancer cells by targeting the calcium/calmodulin‐dependent serine protein kinase CASK [49].
A total of 19 of the 37 miRNAs were found to be associated with ovarian cancer. For example, mir‐194 (ranked 34 in GSE37290 and 37 in GSE69428) could promote the growth, migration and invasion of ovarian carcinoma cells by targeting the protein tyrosine phosphatase PTPN12 [50]. Mir‐181b (ranked 78 in GSE37290 and 77 in GSE69428) could promote cell growth and invasion in ovarian cancer by targeting the serine/threonine‐protein kinase LATS2 [51]. Mir‐130b (ranked 98 in GSE37290 and 95 in GSE69428) was a tumour suppressor by regulating the transcription factor RUNX3 in epithelial ovarian cancer [52].
4 Discussion
As important regulators of gene expression, the aberrant function of miRNAs may drive the initiation and development of numerous cancers. Identifying potential cancer‐miRNAs is critical to our understanding of the pathogenesis of cancer and for designing new targeted therapies. However, only a limited number of miRNAs is known to be directly related to cancer, which hinders the development of miRNA‐based therapeutic strategies. The performance of previous methods based on miRNA–gene interactions was affected by two factors: background noise present in the data and the lack of context. Integrating other information, such as pathway data, known miRNA‐diseases relations, and miRNA–miRNA similarities, may help to improve the identification validity.
In this paper, we developed a new method, named miR_SubPath, to identify cancer‐related miRNAs. Compared with miR_Path and miR_Clust, the proposed miR_SubPath determines the candidate miRNAs based on genes in the significantly differentially expressed subpathways, while the former two check if the genes in a highly correlated cluster targeted by a miRNA could classify the controls and the cases or are significantly different in some pathways. If only a few of the targeted genes of a miRNA are significantly differentially expressed, then both miR_Path and miR_Clust may lose to consider them. However, our proposed miR‐SubPath will consider an miRNA even if only one of its target gene is significantly differentially expressed in a subpathway. This is one of main reasons that the proposed miR‐SubPath could identify more cancer‐related miRNAs. Moreover, the targeted genes of the candidate miRNAs determined by miR‐SubPath generally functions in a co‐ordinately way while those targeted genes of miRNAs from miR_Path and miR_Clust may not.
The scoring method of miR_SubPath is also different from that of miR_Path and miR_Clust. They rank an miRNA based one the classification performance of one of its targeted cluster or a combined score of the cluster considered both the enrichment and significance of the cluster. The proposed miR_SubPath rank a candidate miRNA based its similarity with known seed miRNAs in functional similarity network. This scoring method improves both the accuracy and robustness of the miR_SubPath.
Results from eight datasets, consisting of four cancers types, showed that the F1 score for miR_SubPath was dramatically higher than that of miR_Path, miR_Clust and Zhang's method. This demonstrated that the proposed miR_SubPath method could identify an increased number of cancer‐related miRNAs. Furthermore, both the F ‐score and the ranks of the new novel miRNAs among the top 100 miRNAs from the two independent datasets were very close. This indirectly demonstrated that the proposed miR_SubPath was relatively robust. Concerning the implementation of the proposed MiR‐SubPath, sub‐SPIA implemented by our group using R can be freely downloaded from https://github.com/eshinesimida/subpathway‐analysis. Interested readers for the ranking process could ask us for the R code with email. Finally, it is important to point out that the number of cancer‐related miRNAs identified using this method was relatively small, as there were only 271 miRNAs included in the miRNA–miRNA functional similarity network. Identification of cancer‐related miRNA outside of this dataset is the goal of our future research.
5 Acknowledgments
This work was funded in part by the National Science Foundation of China (grant nos. 61572367 and 61573017) and the Zhejiang Provincial Natural Science Foundation of China (grant nos. LQ17C060001 and LY13F020022).
6 References
- 1. Ambros V.: ‘The functions of animal microRNAs’, Nature, 2004, 431, (7006), p. 350 [DOI] [PubMed] [Google Scholar]
- 2. Bartel D.P.: ‘MicroRNAs: genomics, biogenesis, mechanism, and function’, Cell, 2004, 116, (2), pp. 281–297 [DOI] [PubMed] [Google Scholar]
- 3. Esquela‐Kerscher A., and Slack F.J.: ‘Oncomirs‐microRNAs with a role in cancer’, Nat. Rev. Cancer, 2006, 6, (4), pp. 259–269 [DOI] [PubMed] [Google Scholar]
- 4. Cheng A.M. Byrom M.W., and Shelton J. et al.: ‘Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis’, Nucleic Acids Res., 2005, 33, (4), pp. 1290–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Karp X., and Ambros V.: ‘Developmental biology. Encountering microRNAs in cell fate signaling’, Science, 2005, 310, (5752), p. 1288 [DOI] [PubMed] [Google Scholar]
- 6. Miska E.A.: ‘How microRNAs control cell division, differentiation and death’, Curr. Opin. Genet. Develop., 2005, 15, (5), pp. 563–568 [DOI] [PubMed] [Google Scholar]
- 7. Calin G.A., and Croce C.M.: ‘MicroRNA signatures in human cancers’, Nat. Rev. Cancer, 2006, 6, (11), p. 857 [DOI] [PubMed] [Google Scholar]
- 8. Zhang Z. Li Z., and Gao C. et al.: ‘miR‐21 plays a pivotal role in gastric cancer pathogenesis and progression’, Lab. Invest., 2008, 88, (12), p. 1358 [DOI] [PubMed] [Google Scholar]
- 9. Slaby O. Svoboda M., and Fabian P. et al.: ‘Altered expression of miR‐21, miR‐31, miR‐143 and miR‐145 is related to clinicopathologic features of colorectal cancer’, Oncology, 2007, 72, (5–6), pp. 397–402 [DOI] [PubMed] [Google Scholar]
- 10. Wu H. Zhu S., and Mo Y.‐Y.: ‘Suppression of cell growth and invasion by miR‐205 in breast cancer’, Cell Res., 2009, 19, (4), p. 439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yanaihara N. Caplen N., and Bowman E. et al.: ‘Unique microRNA molecular profiles in lung cancer diagnosis and prognosis’, Cancer Cell, 2006, 9, (3), pp. 189–198 [DOI] [PubMed] [Google Scholar]
- 12. Li Y. Qiu C., and Tu J. et al.: ‘HMDD v2.0: a database for experimentally supported human microRNA and disease associations’, Nucleic Acids Res., 2013, 42, (D1), pp. D1070–D1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jiang Q. Wang Y., and Hao Y. et al.: ‘Mir2disease: a manually curated database for microRNA deregulation in human disease’, Nucleic Acids Res., 2008, 37, (Suppl. 1), pp. D98–D104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kuo T.‐Y. Hsi E., and Yang I.‐P. et al.: ‘Computational analysis of mRNA expression profiles identifies microRNA‐29a/c as predictor of colorectal cancer early recurrence’, PLoS One, 2012, 7, (2), p. e31587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li X. Wang Q., and Zheng Y. et al.: ‘Prioritizing human cancer microRNAs based on genes’ functional consistency between microRNA and cancer’, Nucleic Acids Res., 2011, 39, (22), pp. e153–e153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang W. Zang J., and Jing X. et al.: ‘Identification of candidate miRNA biomarkers from miRNA regulatory network with application to prostate cancer’, J. Translat. Med., 2014, 12, (1), p. 66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen X., and Yan G.‐Y.: ‘Semi‐supervised learning for potential human microRNA‐disease associations inference’, Sci. Rep., 2014, 4, p. 5501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen X. Liu M.‐X., and Yan G.‐Y.: ‘RWRMDA: predicting novel human microRNA–disease associations’, Mol. Biosyst., 2012, 8, (10), pp. 2792–2798 [DOI] [PubMed] [Google Scholar]
- 19. Zhao X.M. Liu K.Q., and Zhu G. et al.: ‘Identifying cancer‐related microRNAs based on gene expression data’, Bioinformatics, 2015, 31, (8), p. 1226 [DOI] [PubMed] [Google Scholar]
- 20. Li C. Li X., and Miao Y. et al.: ‘Subpathwayminer: a software package for flexible identification of pathways’, Nucleic Acids Res., 2009, 37, (19), p. e131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li C. Shang D., and Wang Y. et al.: ‘Characterizing the network of drugs and their affected metabolic subpathways’, PLoS One, 2012, 7, (10), p. e47326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li X. Li C., and Shang D. et al.: ‘The implications of relationships between human diseases and metabolic subpathways’, PLoS One, 2011, 6, (6), p. e21131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jacob L. Neuvial P., and Dudoit S.: ‘More power via graph‐structured tests for differential expression of gene networks’, Ann. Appl. Stat., 2012, 6, (2), pp. 561–600 [Google Scholar]
- 24. Martini P. Sales G., and Massa M.S. et al.: ‘Along signal paths: an empirical gene set approach exploiting pathway topology’, Nucleic Acids Res., 2012, 41, (1), pp. e19–e19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sebastian‐Leon P. Vidal E., and Minguez P. et al.: ‘Understanding disease mechanisms with models of signaling pathway activities’, BMC Syst. Biol., 2014, 8, (1), p. 121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li X. Shen L., and Shang X. et al.: ‘Subpathway analysis based on signaling‐pathway impact analysis of signaling pathway’, PLoS One, 2015, 10, (7), p. e0132813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Edgar R. Domrachev M., and Lash A.E.: ‘Gene expression omnibus: NCBI gene expression and hybridization array data repository’, Nucleic Acids Res., 2002, 30, (1), pp. 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Griffiths‐Jones S. Grocock R.J., and Van Dongen S. et al.: ‘miRBase: microRNA sequences, targets and gene nomenclature’, Nucleic Acids Res., 2006, 34, (Suppl. 1), pp. D140–D144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Krek A. Grün D., and Poy M.N. et al.: ‘Combinatorial microRNA target predictions’, Nat. Genet., 2005, 37, (5), pp. 495–500 [DOI] [PubMed] [Google Scholar]
- 30. John B. Enright A.J., and Aravin A. et al.: ‘Human microRNA targets’, PLoS Biol., 2004, 2, (11), p. e363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Maragkakis M. Alexiou P., and Papadopoulos G.L. et al.: ‘Accurate microRNA target prediction correlates with protein repression levels’, BMC Bioinf., 2009, 10, (1), p. 295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lewis B.P. Burge C.B., and Bartel D.P.: ‘Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets’, Cell, 2005, 120, (1), pp. 15–20 [DOI] [PubMed] [Google Scholar]
- 33. Sethupathy P. Corda B., and Hatzigeorgiou A.G.: ‘Tarbase: a comprehensive database of experimentally supported animal microRNA targets’, RNA, 2006, 12, (2), pp. 192–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sales G. Calura E., and Cavalieri D. et al.: ‘Graphite – a bioconductor package to convert pathway topology to gene network’, BMC Bioinf., 2012, 13, (1), p. 20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Smyth G.K.: ‘Linear models and empirical Bayes methods for assessing differential expression in microarray experiments’, Stat. Appl. Genetics Mol. Biol., 2004, 3, (1), pp. 1–25 [DOI] [PubMed] [Google Scholar]
- 36. Yu H. Gao G., and Jiang L. et al.: ‘Decreased expression of miR‐218 is associated with poor prognosis in patients with colorectal cancer’, Int. J. Clin. Exp. Pathol., 2013, 6, (12), p. 2904 [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang X. Shi H., and Tang H. et al.: ‘miR‐218 inhibits the invasion and migration of colon cancer cells by targeting the PI3K/Akt/mTOR signaling pathway’, Int. J. Mol. Med., 2015, 35, (5), pp. 1301–1308 [DOI] [PubMed] [Google Scholar]
- 38. Park Y.R. Lee S.T., and Kim S.L. et al.: ‘MicroRNA‐9 suppresses cell migration and invasion through downregulation of TM4SF1 in colorectal cancer’, Int. J. Oncol., 2016, 48, (5), pp. 2135–2143 [DOI] [PubMed] [Google Scholar]
- 39. Zhu L. Chen H., and Zhou D. et al.: ‘MicroRNA‐9 up‐regulation is involved in colorectal cancer metastasis via promoting cell motility’, Med. Oncol., 2012, 29, (2), pp. 1037–1043 [DOI] [PubMed] [Google Scholar]
- 40. Wu W. He X., and Kong J. et al.: ‘Mir‐373 affects human lung cancer cells’ growth and its E‐cadherin expression’, Oncol. Res. Featuring Preclin. Clin. Cancer Therapeut., 2012, 20, (4), pp. 163–170 [DOI] [PubMed] [Google Scholar]
- 41. Wang L.‐K. Hsiao T.‐H., and Hong T.‐M. et al.: ‘MicroRNA‐133a suppresses multiple oncogenic membrane receptors and cell invasion in non‐small cell lung carcinoma’, PLoS One, 2014, 9, (5), p. e96765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lan D. Zhang X., and He R. et al.: ‘MiR‐133a is downregulated in non‐small cell lung cancer: a study of clinical significance’, European J. Med. Res., 2015, 20, (1), p. 50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Moriya Y. Nohata N., and Kinoshita T. et al.: ‘Tumor suppressive microRNA‐133a regulates novel molecular networks in lung squamous cell carcinoma’, J. Human Genetics, 2012, 57, (1), p. 38 [DOI] [PubMed] [Google Scholar]
- 44. Yu J. Feng J., and Zhi X. et al.: ‘Let‐7b inhibits cell proliferation, migration, and invasion through targeting Cthrc1 in gastric cancer’, Tumor Biol., 2015, 36, (5), pp. 3221–3229 [DOI] [PubMed] [Google Scholar]
- 45. Han X. Chen Y., and Yao N. et al.: ‘MicroRNA let‐7b suppresses human gastric cancer malignancy by targeting ING1’, Cancer Gene Ther., 2015, 22, (3), p. 122 [DOI] [PubMed] [Google Scholar]
- 46. Kang W. Tong J.H., and Lung R.W. et al.: ‘let‐7b/g silencing activates AKT signaling to promote gastric carcinogenesis’, J. Translat. Med., 2014, 12, (1), p. 281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chu S.‐J. Wang G., and Zhang P.‐F. et al.: ‘Retracted article: MicroRNA‐203 suppresses gastric cancer growth by targeting PIBF1/Akt signaling’, J. Exp. Clin. Cancer Res., 2016, 35, (1), p. 47 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48. Zhou P. Jiang N., and Zhang G.‐X. et al.: ‘MiR‐203 inhibits tumor invasion and metastasis in gastric cancer by ATM’, Acta Biochim. Biophys. Sin, 2016, 48, (8), pp. 696–703 [DOI] [PubMed] [Google Scholar]
- 49. Zhou X. Xu G., and Yin C. et al.: ‘Down‐regulation of miR‐203 induced by Helicobacter pylori infection promotes the proliferation and invasion of gastric cancer by targeting CASK’, OncoTarget, 2014, 5, (22), p. 11631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liang T. Li L., and Cheng Y. et al.: ‘MicroRNA‐194 promotes the growth, migration, and invasion of ovarian carcinoma cells by targeting protein tyrosine phosphatase nonreceptor type 12’, OncoTargets Ther., 2016, 9, p. 4307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xia Y., and Gao Y.: ‘MicroRNA‐181b promotes ovarian cancer cell growth and invasion by targeting LATS2’, Biochem. Biophys. Res. Commun., 2014, 447, (3), pp. 446–451 [DOI] [PubMed] [Google Scholar]
- 52. Paudel D. Zhou W., and Ouyang Y. et al.: ‘MicroRNA‐130b functions as a tumor suppressor by regulating RUNX3 in epithelial ovarian cancer’, Gene, 2016, 586, (1), pp. 48–55 [DOI] [PubMed] [Google Scholar]


