Abstract
Soil fungi play a crucial role in soil quality and fertility in being able to break down organic matter but are frequently also observed to play a role as important plant pathogens. As part of a Citizen Science Project initiated by the Westerdijk Fungal Biodiversity Institute and the Utrecht University Museum, which aimed to describe novel fungal species from Dutch garden soil, the diversity of fusarioid fungi (Fusarium and other fusarioid genera), which are members of Nectriaceae (Hypocreales) was investigated. Preliminary analyses of ITS and LSU sequences from more than 4 750 isolates obtained indicated that 109 strains belong to this generic complex. Based on multi-locus phylogenies of combinations of cmdA, tef1, rpb1, rpb2 and tub2 alignments, and morphological characteristics, 25 species were identified, namely 22 in Fusarium and three in Neocosmospora. Furthermore, two species were described as new namely F. vanleeuwenii from the Fusarium oxysporum species complex (FOSC), and F. wereldwijsianum from the Fusarium incarnatum-equiseti species complex (FIESC). Other species encountered in this study include in the FOSC: F. curvatum, F. nirenbergiae, F. oxysporum and three undescribed Fusarium spp.; in the FIESC: F. clavus, F. croceum, F. equiseti, F. flagelliforme and F. toxicum; Fusarium tricinctum species complex: F. flocciferum and F. torulosum; the Fusarium sambucinum species complex: F. culmorum and F. graminearum; the Fusarium redolens species complex: F. redolens; and the Fusarium fujikuroi species complex: F. verticillioides. Three species of Neocosmospora were encountered, namely N. solani, N. stercicola and N. tonkinensis. Although soil fungal diversity has been well studied in the Netherlands, this study revealed two new species, and eight new records: F. clavus, F. croceum, F. flagelliforme, F. odoratissimum, F. tardicrescens, F. toxicum, F. triseptatum and N. stercicola.
Keywords: biodiversity, Fusarium, multi-gene phylogeny, new taxa, systematics
INTRODUCTION
Fusarium and allied fusarioid genera in Nectriaceae are highly diverse in morphology and ecology, and have a worldwide distribution, commonly occurring on plants and plant products, in air, water and soil. Macroconidia are typically borne in sporodochia, and taxa have in the past been identified as Fusarium if their macroconidia were curved, septate, had a pointed apex, and basal cell with a foot-like notch near the attachment point (Wollenweber & Reinking 1935, Snyder & Hansen 1940, Geiser et al. 2021). However, recent studies have shown that this morphology has evolved several times within Sordariomycetes, and that within Nectriaceae alone up to 20 genera share the fusarioid macromorphology. These genera are distinct phylogenetically and biologically, and have sexual morphs other than Gibberella, which is restricted to Fusarium s. str. (Gräfenhan et al. 2011, Rossman & Seifert 2011, Schroers et al. 2011, Rossman et al. 2013, Lombard et al. 2015, Sandoval-Denis et al. 2019, Crous et al. 2021a).
Species of fusarioid fungi can produce several different spore types, namely macro-, meso- and microconidia, ascospores and chlamydospores (Crous et al. 2021b). Chlamydospores can occur singly or in clusters, forming microsclerotia that have thick, pigmented, smooth to rough walls. They form in hyphae or conidia, either terminally or intercalary, and are the resting spores that make fusarioid taxa highly adapted to survive in soils for extended periods of time. In agricultural soils, chlamydospores commonly occur in plant debris of previous crops, awaiting fresh nutrients and favourable conditions to reactivate (Couteaudier & Alabouvette 1990).
The genus Fusarium s. str. contains 17 species complexes that correlate to different phylogenetic lineages (Crous et al. 2021b). Common soil-borne fusarioid fungi include the Fusarium oxysporum species complex (FOSC; Lombard et al. 2019) and species of Neocosmospora (formerly known as the Fusarium solani species complex; Sandoval-Denis et al. 2018, 2019). The FOSC contains many plant pathogenic taxa, several of which are host specific, which paved the way for “special forms” to be recognised as “formae speciales”, and “races” to help distinguish them (Snyder & Hansen 1940). Such formae speciales, however, are frequently seen to represent distinct phylogenetic species (Lombard et al. 2019, Maryani et al. 2019a, b). Despite this terminology being a dated approach to dealing with the diversity in Fusarium, plant pathologists still use it to help distinguish the diversity they encounter in the field, and more than 144 f. spp. have been named in the FOSC to date (Lombard et al. 2019), with additional subspecific classifications including haplotypes, races and vegetative compatibility groups also being used.
Species of Fusarium produce a range of trichothecenes (mycotoxins) in different ecological niches, that are of concern to animal and human health when such contaminated products are consumed (O’Donnell et al. 2018). These compounds are common throughout Fusarium s. str. and are observed in well-known plant pathogenic species such as F. culmorum, F. graminearum, F. sporotrichioides and F. tricinctum (Bamburg et al. 1968, Tatsuno et al. 1968, Yoshizawa & Morooka 1973, Jiménez et al. 1997), but again absent from species of Neocosmospora (Crous et al. 2021b). Because of the threat and great losses caused by soilborne fusarioid fungi in plant, human and animal health, it is imperative that we gain knowledge of the diversity of fusarioid fungi in soil to better understand their function and impact in different terrestrial ecosystems.
The present Citizen Science Project was initiated by the Westerdijk Fungal Biodiversity Institute (WI) and the Utrecht University Museum, aiming to investigate the diversity of fungi in Dutch garden soil collected by children in their home gardens and schoolgrounds from different regions in the Netherlands (Crous et al. 2017, 2018, 2021a; Groenewald et al. 2018, Giraldo et al. 2019, Hernández-Restrepo et al. 2020, Hou et al. 2020). During this project thousands of isolates were obtained from 404 soil samples. Of these, 109 isolates were found to represent fusarioid fungi, and selected for this study. The aim of the present study was to investigate the diversity of fusarioid fungi from Dutch garden soil, describe and illustrate novel species, and compare them with known taxa.
MATERIALS AND METHODS
Isolates
Soil samples collected from garden soils in the urban environment followed the methods of Groenewald et al. (2018) and Giraldo et al. (2019). Colonies were sub-cultured on 2 % potato-dextrose agar (PDA), oatmeal agar (OA), malt extract agar (MEA) (Crous et al. 2019b), synthetic nutrient-poor agar (SNA; Nirenberg 1976), carnation leaf agar (CLA; Fisher et al. 1982), and incubated at 25 °C under continuous near-ultraviolet light to promote sporulation. Reference strains and specimens of the studied fungi are maintained in the culture collection (CBS) of the Westerdijk Fungal Biodiversity Institute (WI), Utrecht, the Netherlands.
DNA extraction, amplification (PCR) and phylogeny
Protocols for genomic DNA isolation, PCR amplification of partial calmodulin (cmdA) gene, internal transcribed spacer regions with intervening 5.8S nrRNA gene (ITS), partial 28S nrRNA gene (LSU), DNA-directed RNA polymerase II largest (rpb1) and second largest subunit (rpb2) genes, and translation elongation factor 1-alpha (tef1) gene, and sequencing of the novel strains (Table 1) followed Crous et al. (2021b). The two parts of rpb2 listed in Table 1 corresponded to the sequences generated using primer pairs RPB2-5f2 / fRPB2-7cR and fRPB2-7cf / RPB2-11ar (see Crous et al. (2021b) for primer details). Partial beta-tubulin (tub2) gene sequences were not generated during the course of this study.
Table 1 .
Collection details and GenBank accession numbers of isolates treated in this study, and associated ex-type strains where applicable. Species names in bold highlight taxonomic novelties. The ITS and LSU sequences were not used in analyses but are provided for completeness.
| Species complex and Species | Culture or working collection number(s) | Country and Substrate | Collector(s) and Collection date | School or educational institution |
GenBank accession number(s)1
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| tef1 | rpb2 part 1 | rpb2 part 2 | cmdA | rpb1 | ITS | LSU | |||||
| Fusarium fujikuroi species complex | |||||||||||
| Fusarium verticillioides | JW 145017 | Netherlands: Soil | A.E. Jansen; 2017 | – | MZ921825 | MZ921693 | – | MZ921513 | MZ921624 | – | MZ890483 |
| Fusarium incarnatum-equiseti species complex | |||||||||||
| Fusarium clavus | JW 288002 | Netherlands: Soil | Group 8, OBS de Toonladder; 2017 | – | MZ921826 | MZ921694 | – | MZ921514 | MZ921625 | – | MZ890484 |
| NL19-041003 | Netherlands: Soil | L. Oegema, R. van Stee & D. Kwast; 2019 | RSG Simon Vestdijk | MZ921827 | MZ921695 | – | MZ921515 | – | MZ890340 | MZ890485 | |
| NL19-048011 | Netherlands: Soil | S. Goinga & J. de Groot; 10 Oct. 2019 | RSG Simon Vestdijk | MZ921828 | MZ921696 | – | MZ921516 | – | MZ890341 | MZ890486 | |
| NL19-056012 | Netherlands: Soil | S. Verhage, S. Moens & K. Basting; 29 Oct. 2019 | Zwin college | MZ921829 | MZ921697 | – | MZ921517 | – | MZ890342 | MZ890487 | |
| NL19-056013 | Netherlands: Soil | S. Verhage, S. Moens & K. Basting; 29 Oct. 2019 | Zwin college | MZ921830 | MZ921698 | – | MZ921518 | – | MZ890343 | MZ890488 | |
| Fusarium croceum | NL19-059006 | Netherlands: Soil | A. van Strien, I. Beemsterboer & S. Groosman; 23 Oct. 2019 | Zwin college | MZ921831 | MZ921699 | – | MZ921519 | – | MZ890344 | MZ890489 |
| NL19-060011 | Netherlands: Soil | T. Vercruisse; 27 Oct. 2019 | Zwin college | MZ921832 | MZ921700 | – | MZ921520 | – | MZ890345 | MZ890490 | |
| Fusarium equiseti | CBS 148218 = NL19-25004 | Netherlands: Soil | C. Dijkstra & L. Kruit; 6 Jun. 2019 | Het Hogeland College Warffum | MZ921833 | MZ921701 | – | MZ921521 | – | MZ890346 | MZ890491 |
| CBS 148383 = NL19-008004 | Netherlands: Soil | S. de Boer; 17 Dec. 2019 | GSG ’t Schylger Jouw | MZ921834 | MZ921702 | – | MZ921522 | – | MZ890347 | MZ890492 | |
| NL19-045005 | Netherlands: Soil | E.-A. Duinstra, R. Jagersma & M. Postmus; 9 Oct. 2019 | RSG Simon Vestdijk | MZ921835 | MZ921703 | – | MZ921523 | MZ921626 | MZ890348 | MZ890493 | |
| NL19-047003 | Netherlands: Soil | S. Kuiper, N. Zijlstra & E. Schot; 10 Oct. 2019 | RSG Simon Vestdijk | MZ921836 | MZ921704 | – | MZ921524 | MZ921627 | MZ890349 | – | |
| NL19-059004 | Netherlands: Soil | A. van Strien, I. Beemsterboer & S. Groosman; 23 Oct. 2019 | Zwin college | MZ921837 | MZ921705 | – | MZ921525 | – | MZ890350 | MZ890494 | |
| NL19-97009 | Netherlands: Soil | S. Frederikze, J. Mes & S. El Maghnouji; 31 Jul. 2019 | ACB Wereldwijs | MZ921838 | MZ921706 | – | MZ921526 | MZ921628 | MZ890351 | MZ890495 | |
| Fusarium flagelliforme | NL19-041004 | Netherlands: Soil | L. Oegema, R. van Stee & D. Kwast; 2019 | RSG Simon Vestdijk | MZ921839 | MZ921707 | – | MZ921527 | MZ921629 | MZ890352 | MZ890496 |
| NL19-047004 | Netherlands: Soil | S. Kuiper, N. Zijlstra & E. Schot; 10 Oct. 2019 | RSG Simon Vestdijk | MZ921840 | MZ921708 | – | MZ921528 | – | MZ890353 | MZ890497 | |
| NL19-050003 | Netherlands: Soil | T. van der Schoot & J. Koel; 10 Oct. 2019 | RSG Simon Vestdijk | MZ921841 | MZ921709 | – | MZ921529 | – | MZ890354 | MZ890498 | |
| NL19-052002 | Netherlands: Soil | M. Stellemans, L. de Winde & N. Quist; 21 Oct. 2019 | Zwin college | MZ921842 | MZ921710 | – | MZ921530 | – | MZ890355 | MZ890499 | |
| NL19-068002 | Netherlands: Soil | S. Walraven & M. Bekooy; 28 Oct. 2019 | Zwin college | MZ921843 | MZ921711 | – | MZ921531 | – | MZ890356 | MZ890500 | |
| NL19-97010 | Netherlands: Soil | S. Frederikze, J. Mes & S. El Maghnouji; 31 Jul. 2019 | ACB Wereldwijs | MZ921844 | MZ921712 | – | MZ921532 | MZ921630 | MZ890357 | MZ890501 | |
| Fusarium toxicum | NL19-041005 | Netherlands: Soil | L. Oegema, R. van Stee & D. Kwast; 2019 | RSG Simon Vestdijk | MZ921845 | MZ921713 | – | MZ921533 | MZ921631 | MZ890358 | MZ890502 |
| NL19-041006 | Netherlands: Soil | L. Oegema, R. van Stee & D. Kwast; 2019 | RSG Simon Vestdijk | MZ921846 | MZ921714 | – | MZ921534 | MZ921632 | MZ890359 | MZ890503 | |
| NL19-050001 | Netherlands: Soil | T. van der Schoot & J. Koel; 10 Oct. 2019 | RSG Simon Vestdijk | MZ921847 | MZ921715 | – | MZ921535 | MZ921633 | MZ890360 | MZ890504 | |
| Fusarium wereldwijsianum sp. nov. | CBS 148219 = NL19-99002 | Netherlands: Soil | S. Frederikze, J. Mes & S. El Maghnouji; 31 Jul. 2019 | ACB Wereldwijs | MZ921848 | MZ921716 | – | MZ921536 | MZ921634 | MZ890361 | MZ890505 |
| CBS 148220 = NL19-99003 | Netherlands: Soil | S. Frederikze, J. Mes & S. El Maghnouji; 31 Jul. 2019 | ACB Wereldwijs | MZ921849 | MZ921717 | – | MZ921537 | MZ921635 | MZ890362 | MZ890506 | |
| CBS 148244 = NL19-94009, ex-type | Netherlands: Soil | S. Frederikze, J. Mes & S. El Maghnouji; 31 Jul. 2019 | ACB Wereldwijs | MZ921850 | MZ921718 | – | MZ921538 | MZ921636 | MZ890363 | MZ890507 | |
| CBS 148385 = NL19-057012 | Netherlands: Soil | F. Guilliet, T. Bron & I. Geernaert; Oct. 2019 | Zwin college | MZ921851 | MZ921719 | – | MZ921539 | – | MZ890364 | MZ890508 | |
| CBS 148386 = NL19-059003 | Netherlands: Soil | A. van Strien, I. Beemsterboer & S. Groosman; 23 Oct. 2019 | Zwin college | MZ921852 | MZ921720 | – | MZ921540 | – | MZ890365 | MZ890509 | |
| Fusarium oxysporum species complex | |||||||||||
| Fusarium curvatum | JW 39001 | Netherlands: Soil | R. Ramanand; 2017 | – | MZ921853 | MZ921721 | – | MZ921541 | – | MZ890366 | MZ890510 |
| Fusarium nirenbergiae | CBS 148373 = JW 5042 | Netherlands: Soil | F. & R. Niemeijer; 2017 | – | MZ921867 | MZ921735 | – | MZ921555 | MZ921646 | MZ890378 | MZ890522 |
| CBS 148379 = JW 124027 | Netherlands: Soil | S. Vermeulen; 2017 | – | MZ921868 | MZ921736 | – | MZ921556 | MZ921647 | – | MZ890523 | |
| CBS 148381 = JW 288013 | Netherlands: Soil | Group 8, OBS de Toonladder; 2017 | – | MZ921870 | MZ921738 | – | MZ921558 | – | – | MZ890525 | |
| CBS 148382 = JW 289011 | Netherlands: Soil | KMN Spelerij; 2017 | – | MZ921871 | MZ921739 | – | MZ921559 | – | – | MZ890526 | |
| CBS 148384 = NL19-048010 | Netherlands: Soil | S. Goinga & J. de Groot; 10 Oct. 2019 | RSG Simon Vestdijk | MZ921873 | MZ921741 | – | MZ921561 | – | MZ890381 | MZ890528 | |
| CBS 148387 = NL19-100010 | Netherlands: Soil | S. Frederikze, J. Mes & S. El Maghnouji; 31 Jul. 2019 | ACB Wereldwijs | MZ921875 | MZ921744 | – | MZ921564 | – | MZ890384 | MZ890531 | |
| CBS 148388 = BE19-004016 | Belgium: Soil | T. Antheunis; 2019 | Viso Cor Mariae | MZ921866 | MZ921734 | – | MZ921554 | – | MZ890377 | MZ890521 | |
| JW 192006 | Netherlands: Soil | L. Borsboom; 2017 | – | MZ921869 | MZ921737 | – | MZ921557 | – | MZ890379 | MZ890524 | |
| NL19-045004 | Netherlands: Soil | E.-A. Duinstra, R. Jagersma & M. Postmus; 9 Oct. 2019 | RSG Simon Vestdijk | MZ921872 | MZ921740 | – | MZ921560 | MZ921648 | MZ890380 | MZ890527 | |
| NL19-053002 | Netherlands: Soil | L. van Eetveldt, G. Jones & F. Walraven; 25 Oct. 2019 | Zwin college | MZ921874 | MZ921742 | – | MZ921562 | – | MZ890382 | MZ890529 | |
| NL19-053003 | Netherlands: Soil | L. van Eetveldt, G. Jones & F. Walraven; 25 Oct. 2019 | Zwin college | – | MZ921743 | – | MZ921563 | MZ921649 | MZ890383 | MZ890530 | |
| NL19-28011 | Netherlands: Soil | H. Meertens & D. Zaagman; 6 Jun. 2019 | Het Hogeland College Warffum | MZ921876 | MZ921745 | – | MZ921565 | MZ921650 | MZ890385 | MZ890532 | |
| NL19-91009 | Netherlands: Soil | S. Frederikze, J. Mes & S. El Maghnouji; 31 Jul. 2019 | ACB Wereldwijs | MZ921877 | MZ921746 | – | MZ921566 | MZ921651 | MZ890386 | MZ890533 | |
| NL19-91010 | Netherlands: Soil | S. Frederikze, J. Mes & S. El Maghnouji; 31 Jul. 2019 | ACB Wereldwijs | MZ921878 | MZ921747 | – | MZ921567 | MZ921652 | MZ890387 | MZ890534 | |
| NL19-99011 | Netherlands: Soil | S. Frederikze, J. Mes & S. El Maghnouji; 31 Jul. 2019 | ACB Wereldwijs | MZ921879 | MZ921748 | – | MZ921568 | MZ921653 | MZ890388 | MZ890535 | |
| Fusarium odoratissimum | JW 54001 | Netherlands: Soil | I., M. & L. Zoert; 2017 | – | MZ921880 | MZ921749 | – | MZ921569 | MZ921654 | – | – |
| Fusarium oxysporum | JW 11005 | Netherlands: Soil | M. Francisca; 2017 | – | MZ921881 | MZ921750 | – | MZ921570 | MZ921655 | MZ890389 | MZ890536 |
| JW 231014 | Netherlands: Soil | D. Pol, R. Verf, J. Wilks & M. de Ruiter; 2017 | – | MZ921882 | MZ921751 | – | MZ921571 | MZ921656 | MZ890390 | – | |
| JW 257006 | Netherlands: Soil | KSU de Achtbaan; 2017 | – | MZ921883 | MZ921752 | – | MZ921572 | MZ921657 | – | MZ890537 | |
| NL19-94002 | Netherlands: Soil | S. Frederikze, J. Mes & S. El Maghnouji; 31 Jul. 2019 | ACB Wereldwijs | MZ921884 | MZ921753 | – | MZ921573 | MZ921658 | MZ890391 | MZ890538 | |
| NL19-94008 | Netherlands: Soil | S. Frederikze, J. Mes & S. El Maghnouji; 31 Jul. 2019 | ACB Wereldwijs | MZ921885 | MZ921754 | – | MZ921574 | MZ921659 | MZ890392 | MZ890539 | |
| Fusarium sp. 1 | CBS 148204 = JW 191014 | Netherlands: Soil | T. & K. Wesselink; 2017 | – | MZ921858 | MZ921726 | – | MZ921546 | MZ921641 | MZ890371 | MZ890514 |
| CBS 148216 = JW 53002 | Netherlands: Soil | K. Brennand; 2017 | – | MZ921863 | MZ921731 | – | MZ921551 | MZ921645 | – | – | |
| CBS 148217 = NL19-25001 | Netherlands: Soil | C. Dijkstra & L. Kruit; 6 Jun. 2019 | Het Hogeland College Warffum | MZ921864 | MZ921732 | – | MZ921552 | – | MZ890375 | MZ890519 | |
| Fusarium sp. 2 | CBS 130323 = NRRL 26677 | Australia: Subungual debris of 40-year-old female with nail infection | Unknown | – | MH485018 | MH484927 | – | MH484745 | – | – | – |
| CBS 148185 = JW 1072 | Netherlands: Soil | J. van Dijk; 2017 | – | MZ921854 | MZ921722 | – | MZ921542 | MZ921637 | MZ890367 | MZ890511 | |
| CBS 128.81 = BBA 63925 = NRRL 36233 | USA: Chrysanthemum sp. | Unknown | MH484975 | MH484884 | – | MH484702 | – | – | – | ||
| CBS 680.89 = IPO 11179 = NRRL 26221 | Netherlands: Cucumis sativus, in greenhouse on rockwool | N. Hubbeling; – | – | MH484980 | MH484889 | – | MH484707 | – | – | – | |
| Fusarium sp. 3 | CBS 148198 = JW 4030 | Netherlands: Soil | F. Wiegerinck; 2017 | – | MZ921855 | MZ921723 | – | MZ921543 | MZ921638 | MZ890368 | MZ890512 |
| CBS 148199 = JW 9002 | Netherlands: Soil | A.-S. den Boer; 2017 | – | MZ921856 | MZ921724 | – | MZ921544 | MZ921639 | MZ890369 | MZ890513 | |
| CBS 148200 = JW 10005 | Netherlands: Soil | M.J. van Leeuwen; 2017 | – | MZ921857 | MZ921725 | – | MZ921545 | MZ921640 | MZ890370 | – | |
| CBS 148205 = JW 204009 | Netherlands: Soil | I. Kleij; 2017 | – | MZ921859 | MZ921727 | – | MZ921547 | MZ921642 | – | MZ890515 | |
| CBS 148206 = JW 210014 | Netherlands: Soil | N. Keij; 2017 | – | MZ921860 | MZ921728 | – | MZ921548 | MZ921643 | MZ890372 | MZ890516 | |
| CBS 148207 = JW 210019 | Netherlands: Soil | N. Keij; 2017 | – | MZ921861 | MZ921729 | – | MZ921549 | MZ921644 | MZ890373 | MZ890517 | |
| CBS 148208 = JW 231016 | Netherlands: Soil | D. Pol, R. Verf, J. Wilks & M. de Ruiter; 2017 | – | MZ921862 | MZ921730 | – | MZ921550 | – | MZ890374 | MZ890518 | |
| CBS 148222 = BE19-004006 | Belgium: Soil | T. Antheunis; 2019 | Viso Cor Mariae | MZ921865 | MZ921733 | – | MZ921553 | – | MZ890376 | MZ890520 | |
| Fusarium tardicrescens | JW 6021 | Netherlands: Soil | H.W. Vos; 2017 | – | MZ921886 | MZ921755 | – | MZ921575 | MZ921660 | MZ890393 | MZ890540 |
| JW 6043 | Netherlands: Soil | H.W. Vos; 2017 | – | MZ921887 | MZ921756 | – | MZ921576 | MZ921661 | MZ890394 | – | |
| Fusarium triseptatum | CBS 148380 = JW 277008 | Netherlands: Soil | Lukasschool; 2017 | – | MZ921888 | MZ921757 | – | MZ921577 | MZ921662 | – | MZ890541 |
| JW 277009 | Netherlands: Soil | Lukasschool; 2017 | – | MZ921889 | MZ921758 | – | MZ921578 | – | – | MZ890542 | |
| Fusarium vanleeuwenii sp. nov. | CBS 148372 = JW 10008, ex-type | Netherlands: Soil | M.J. van Leeuwen; 2017 | – | MZ921896 | MZ921765 | – | MZ921585 | MZ921669 | MZ890401 | – |
| CBS 148374 = JW 10001 | Netherlands: Soil | M.J. van Leeuwen; 2017 | – | MZ921890 | MZ921759 | – | MZ921579 | MZ921663 | MZ890395 | MZ890543 | |
| CBS 148375 = JW 10003 | Netherlands: Soil | M.J. van Leeuwen; 2017 | – | MZ921892 | MZ921761 | – | MZ921581 | MZ921665 | MZ890397 | – | |
| CBS 148376 = JW 10004 | Netherlands: Soil | M.J. van Leeuwen; 2017 | – | MZ921893 | MZ921762 | – | MZ921582 | MZ921666 | MZ890398 | – | |
| CBS 148377 = JW 10006 | Netherlands: Soil | M.J. van Leeuwen; 2017 | – | MZ921894 | MZ921763 | – | MZ921583 | MZ921667 | MZ890399 | – | |
| CBS 148378 = JW 10007 | Netherlands: Soil | M.J. van Leeuwen; 2017 | – | MZ921895 | MZ921764 | – | MZ921584 | MZ921668 | MZ890400 | – | |
| JW 10002 | Netherlands: Soil | M.J. van Leeuwen; 2017 | – | MZ921891 | MZ921760 | – | MZ921580 | MZ921664 | MZ890396 | MZ890544 | |
| JW 10009 | Netherlands: Soil | M.J. van Leeuwen; 2017 | – | MZ921897 | MZ921766 | – | MZ921586 | MZ921670 | MZ890402 | – | |
| Fusarium redolens species complex | |||||||||||
| Fusarium redolens | NL19-003007 | Netherlands: Soil | B. Wulp; 17 Dec. 2019 | GSG ’t Schylger Jouw | MZ921898 | MZ921767 | – | MZ921671 | MZ890403 | MZ890545 | |
| Fusarium sambucinum species complex | |||||||||||
| Fusarium culmorum | BE19-002002 | Belgium: Soil | S. Vanopbroeke; 2019 | Viso Cor Mariae | MZ921899 | MZ921768 | MZ921802 | MZ921587 | – | MZ890404 | MZ890546 |
| BE19-009002 | Belgium: Soil | N. Caen; 2019 | Viso Cor Mariae | MZ921900 | MZ921769 | MZ921803 | MZ921588 | – | MZ890405 | MZ890547 | |
| NL19-047005 | Netherlands: Soil | S. Kuiper, N. Zijlstra & E. Schot; 10 Oct. 2019 | RSG Simon Vestdijk | MZ921901 | MZ921770 | MZ921804 | MZ921589 | – | MZ890406 | MZ890548 | |
| NL19-060003 | Netherlands: Soil | T. Vercruisse; 27 Oct. 2019 | Zwin college | MZ921902 | MZ921771 | MZ921805 | MZ921590 | – | MZ890407 | MZ890549 | |
| NL19-076001 | Netherlands: Soil | W. Vercouteren, S. Meas & R. Verhije; 6 Nov. 2019 | Zwin college | MZ921903 | MZ921772 | MZ921806 | MZ921591 | – | MZ890408 | MZ890550 | |
| NL19-25005 | Netherlands: Soil | C. Dijkstra & L. Kruit; 6 Jun. 2019 | Het Hogeland College Warffum | MZ921904 | MZ921773 | MZ921807 | – | – | MZ890409 | MZ890551 | |
| NL19-93013 | Netherlands: Soil | S. Frederikze, J. Mes & S. El Maghnouji; 31 Jul. 2019 | ACB Wereldwijs | MZ921905 | MZ921774 | MZ921808 | MZ921592 | MZ921672 | MZ890410 | MZ890552 | |
| Fusarium graminearum | NL19-100008 | Netherlands: Soil | S. Frederikze, J. Mes & S. El Maghnouji; 31 Jul. 2019 | ACB Wereldwijs | MZ921906 | MZ921775 | MZ921809 | MZ921593 | MZ921673 | MZ890411 | MZ890553 |
| Fusarium tricinctum species complex | |||||||||||
| Fusarium acuminatum | JW 288021 | Netherlands: Soil | Group 8, OBS de Toonladder; 2017 | – | MZ921907 | MZ921776 | MZ921810 | MZ921594 | MZ921674 | – | MZ890554 |
| JW 289003 | Netherlands: Soil | KMN Spelerij; 2017 | – | MZ921908 | MZ921777 | MZ921811 | MZ921595 | MZ921675 | – | MZ890555 | |
| NL19-048014 | Netherlands: Soil | S. Goinga & J. de Groot; 10 Oct. 2019 | RSG Simon Vestdijk | MZ921909 | MZ921778 | MZ921812 | MZ921596 | – | MZ890412 | MZ890556 | |
| NL19-077002 | Netherlands: Soil | R. van der Wel & T. Wolfret; 5 Nov. 2019 | Zwin college | MZ921910 | MZ921779 | MZ921813 | MZ921597 | – | MZ890413 | MZ890557 | |
| Fusarium flocciferum | CBS 143231 = JW 14004 | Netherlands: Soil | D. Peters; 2017 | – | MG386159 | MG386149 | MG386149 | MZ921598 | MG386138 | MG386078 | MG386131 |
| CBS 143667 = JW 14005, ex-type of F. petersiae | Netherlands: Soil | D. Peters; 2017 | – | MG386160 | MG386150 | MG386150 | MZ921599 | MG386139 | MG386079 | MG386132 | |
| CBS 147837 = NL19-100011 | Netherlands: Soil | S. Frederikze, J. Mes & S. El Maghnouji; 31 Jul. 2019 | ACB Wereldwijs | – | MZ921780 | MZ921814 | MZ921600 | – | MZ890416 | MZ890558 | |
| CBS 821.68 = NRRL 28450, ex-epitype | Germany: Greenhouse soil | D. Bredemeier; 1966 | – | MW928837 | MW928824 | MW928824 | – | MW928807 | – | – | |
| JW 5026 | Netherlands: Soil | F. & R. Niemeijer; 2017 | – | MZ921911 | MZ921781 | MZ921815 | MZ921601 | MZ921676 | MZ890417 | MZ890559 | |
| JW 18005 | Netherlands: Soil | W. van der Heijden; 2017 | – | MZ921912 | MZ921782 | MZ921816 | MZ921602 | MZ921677 | MZ890418 | – | |
| JW 248008 | Netherlands: Soil | J.-W. Koolen; 2017 | – | MZ921913 | MZ921783 | MZ921817 | MZ921603 | MZ921678 | MZ890419 | MZ890560 | |
| JW 267001 | Netherlands: Soil | Basisschool de Baanbreker; 2017 | – | MZ921914 | MZ921784 | MZ921818 | MZ921604 | – | – | MZ890561 | |
| NL19-048012 | Netherlands: Soil | S. Goinga & J. de Groot; 10 Oct. 2019 | RSG Simon Vestdijk | MZ921915 | MZ921785 | MZ921819 | MZ921605 | – | MZ890420 | MZ890562 | |
| NL19-048013 | Netherlands: Soil | S. Goinga & J. de Groot; 10 Oct. 2019 | RSG Simon Vestdijk | MZ921916 | MZ921786 | MZ921820 | MZ921606 | – | MZ890421 | MZ890563 | |
| NL19-97008 | Netherlands: Soil | S. Frederikze, J. Mes & S. El Maghnouji; 31 Jul. 2019 | ACB Wereldwijs | MZ921917 | MZ921787 | MZ921821 | MZ921607 | MZ921679 | MZ890422 | MZ890564 | |
| Fusarium torulosum | JW 24001 | Netherlands: Soil | J. van der Stel; 2017 | – | MZ921918 | MZ921788 | MZ921822 | MZ921608 | MZ921680 | MZ890423 | MZ890565 |
| Neocosmospora | |||||||||||
| Neocosmospora solani | JW 1075 | Netherlands: Soil | J. van Dijk; 2017 | – | MZ921919 | MZ921789 | – | MZ921609 | MZ921681 | MZ890424 | MZ890566 |
| JW 14011 | Netherlands: Soil | D. Peters; 2017 | – | MZ921920 | MZ921790 | – | MZ921610 | – | MZ890425 | MZ890567 | |
| JW 191039 | Netherlands: Soil | T. & K. Wesselink; 2017 | – | MZ921921 | MZ921791 | – | MZ921611 | MZ921682 | MZ890426 | MZ890568 | |
| JW 232018 | Netherlands: Soil | M. van Meijl; 2017 | – | MZ921922 | MZ921792 | – | MZ921612 | – | MZ890427 | MZ890569 | |
| JW 288011 | Netherlands: Soil | Group 8, OBS de Toonladder; 2017 | – | MZ921923 | MZ921793 | – | MZ921613 | MZ921683 | – | MZ890570 | |
| Neocosmospora stercicola | JW 1093 | Netherlands: Soil | J. van Dijk; 2017 | – | MZ921924 | MZ921794 | – | MZ921614 | MZ921684 | MZ890428 | MZ890571 |
| JW 75001 | Netherlands: Soil | O. Terpstra; 2017 | – | MZ921925 | MZ921795 | – | MZ921615 | MZ921685 | MZ890429 | MZ890572 | |
| JW 235004 | Netherlands: Soil | T. Tuinier; 2017 | – | MZ921926 | MZ921796 | – | MZ921616 | MZ921686 | MZ890430 | MZ890573 | |
| JW 235009 | Netherlands: Soil | T. Tuinier; 2017 | – | MZ921927 | MZ921797 | – | MZ921617 | MZ921687 | MZ890431 | MZ890574 | |
| Neocosmospora tonkinensis | JW 234010 | Netherlands: Soil | T. Vanmeulebrouk; 2017 | – | MZ921928 | MZ921798 | – | MZ921618 | MZ921688 | – | MZ890575 |
| JW 236012 | Netherlands: Soil | A. Vanmeulebrouk; 2017 | – | – | MZ921799 | – | MZ921619 | – | – | MZ890576 | |
1 cmdA: partial calmodulin gene; ITS: internal transcribed spacer regions with intervening 5.8S nrRNA gene; rpb1: partial DNA-directed RNA polymerase II largest subunit gene; rpb2: partial DNA-directed RNA polymerase II second largest subunit gene; tef1: partial translation elongation factor 1-alpha gene; tub2: partial beta-tubulin gene.
Initial identifications to genus level were made using megablast searches (Zhang et al. 2000) of the ITS sequences against NCBI's GenBank nucleotide database, after which tef1 sequences were used to further identify the Fusarium species complexes. Reference sequences (Supplementary Table S1) from Crous et al. (2021b) and based on megablast searches were then used to construct single-gene and multi-gene alignments for Neocosmospora and the different Fusarium species complexes. Phylogenetic analyses using RAxML Blackbox v. 1.0.0 (https://raxml-ng.vital-it.ch/#/; Kozlov et al. 2019), IQ-TREE v. 2.1.3 (Nguyen et al. 2015, Minh et al. 2020) and MrBayes v. 3.2.7 (Ronquist & Huelsenbeck 2003) followed Crous et al. (2021b), with the exception that trees were saved every 10 or 100 generations (Table 2). All resulting trees were printed with Geneious v. 11.1.5 and the layout of the trees was done in Adobe Illustrator v. CC 2018.
Table 2 .
Summary of phylogenetic information for the different analyses in this study1.
| Analysis | Locus2 | Number of strains (incl. outgroup) | Length incl. gaps | BI unique site patterns | Model (AIC) | Model (BIC) | BI sample frequency | Number of sampled trees (BI) | ML -InL (R) | ML -InL (IQ-TREE) |
|---|---|---|---|---|---|---|---|---|---|---|
| Fusarium citricola & F. tricinctum species complexes | cmdA | 23 | 669 | 118 | SYM+G | K2P+G4 | – | – | – | -1717.014 |
| rpb1 | 27 | 1 787 | 370 | GTR+I | TNe+G4 | – | – | – | -4717.229 | |
| rpb2 (part 1) | 42 | 910 | 226 | SYM+G | TNe+G4 | – | – | – | -2795.892 | |
| rpb2 (part 2) | 34 | 628 | 104 | GTR+G | TNe+G4 | – | – | – | -1747.846 | |
| tef1 | 41 | 756 | 256 | GTR+I | TIM2e+G4 | – | – | – | -2469.915 | |
| Combined | 45 | 4 750 | 1 074 | – | – | 10 | 78 002 | -13596.815643 | -13617.725 | |
|
| ||||||||||
| Fusarium incarnatum-equiseti species complex | cmdA | 72 | 661 | 157 | SYM+G | TNe+R3 | – | – | – | -2121.606 |
| rpb1 | 29 | 1 729 | 226 | SYM+I | TNe+R2 | – | – | -4384.505 | ||
| rpb2 (part 1) | 73 | 886 | 174 | GTR+I | TNe+G4 | – | – | -2770.072 | ||
| tef1 | 73 | 743 | 262 | GTR+G | TNe+R3 | – | – | -3166.37 | ||
| Combined | 73 | 4 019 | 819 | – | – | 10 | 1 285 502 | -13545.426546 | -13066.849 | |
|
| ||||||||||
| Fusarium oxysporum species complex | cmdA | 117 | 608 | 53 | K80 | K2P | – | – | – | -1074.196 |
| rpb1 | 73 | 1451 | 216 | SYM+I+G | TNe+R2 | – | – | – | -3381.188 | |
| rpb2 (part 1) | 154 | 882 | 111 | HKY+G | K2P+I | – | – | – | -2070.032 | |
| tef1 | 154 | 584 | 158 | HKY+G | TNe+G4 | – | – | – | -1733.616 | |
| tub2 | 74 | 577 | 141 | SYM+G | TIMe+R2 | – | – | – | -1812.746 | |
| Combined | 155 | 4 102 | 679 | – | – | 100 | 80 178 | -10424.792230 | -10.414.344 | |
|
| ||||||||||
| Fusarium redolens & F. fujikuroi species complexes | cmdA | 17 | 690 | 149 | SYM+I | TIM3e+G4 | – | – | – | -1842.997 |
| rpb1 | 29 | 1 788 | 329 | SYM+I | TNe+G4 | – | – | – | -5168.810 | |
| rpb2 (part 1) | 34 | 904 | 297 | SYM+I+G | TIM2e+I+G4 | – | – | – | -4009.430 | |
| tef1 | 33 | 763 | 323 | GTR+G | TIM2e+G4 | – | – | – | -3494.236 | |
| Combined | 34 | 4 145 | 1 098 | – | – | 10 | 64 502 | -15323.869379 | -15336.259 | |
|
| ||||||||||
| Fusarium sambucinum species complex | cmdA | 14 | 661 | 71 | SYM | TNe+I | – | – | – | -1499.250 |
| rpb1 | 33 | 1 793 | 240 | SYM+I | TNe+G4 | – | – | – | -4257.654 | |
| rpb2 (part 1) | 38 | 905 | 212 | SYM+G | TIM2e+I | – | – | – | -2940.257 | |
| rpb2 (part 2) | 38 | 629 | 133 | GTR+G | TNe+I | – | – | – | -2089.607 | |
| tef1 | 38 | 755 | 197 | GTR+G | TIM2e+R2 | – | – | – | -2285.191 | |
| Combined | 39 | 4 743 | 853 | – | – | 10 | 30 752 | -13596.206653 | -13604.873 | |
|
| ||||||||||
| Neocosmospora | cmdA | 42 | 674 | 221 | SYM+I+G | K2P+G4 | – | – | – | -2585.283 |
| rpb1 | 42 | 1 687 | 524 | GTR+I+G | TIM3e+I+G4 | – | – | – | -6296.579 | |
| rpb2 (part 1) | 69 | 866 | 241 | SYM+G | TNe+G4 | – | – | – | -2990.034 | |
| tef1 | 76 | 752 | 300 | GTR+G | TN+F+G4 | – | – | – | -3000.621 | |
| Combined | 77 | 3 979 | 1 286 | – | – | 10 | 859 502 | -15410.126704 | -15411.266 | |
1 BI: Bayesian inference; Model (AIC): Evolutionary model selected by MrModeltest under the Akaike Information Criterion; Model (BIC): Evolutionary model selected by ModelFinder in IQ-TREE; BI sample frequency: Number of nth generations sampled; ML -InL (R): Log-likelihood of final tree in RAxML; ML -InL(IQ-TREE): Log-likelihood of consensus tree in IQ-TREE.
2 cmdA: partial calmodulin gene, tef1: partial translation elongation factor 1-alpha gene; rpb1: partial DNA-directed RNA polymerase II largest subunit gene; rpb2: partial DNA-directed RNA polymerase II second largest subunit gene; tub2: partial beta-tubulin gene.
Morphology
Slide preparations were mounted in water, from colonies sporulating on CLA, following the protocols described by Crous et al. (2021b). Observations were made with a Nikon SMZ25 dissection-microscope, and with a Zeiss Axio Imager 2 light microscope using differential interference contrast (DIC) illumination and images recorded on a Nikon DS-Ri2 camera with associated software. Colony characters and pigment production were noted after 7 d of growth on MEA, PDA and OA incubated at 25 °C. Colony colours (surface and reverse) were scored using the colour charts of Rayner (1970).
RESULTS
Phylogeny
Six multigene alignments were generated in the present study and subjected to the three phylogenetic analyses described above. Statistical values for the alignments and phylogenetic trees are summarised in Table 2. Sequences derived in this study were deposited in GenBank (Table 1), the alignments in TreeBASE (www.treebase.org; study number 28680), and taxonomic novelties in MycoBank (www.MycoBank.org; Crous et al. 2004).
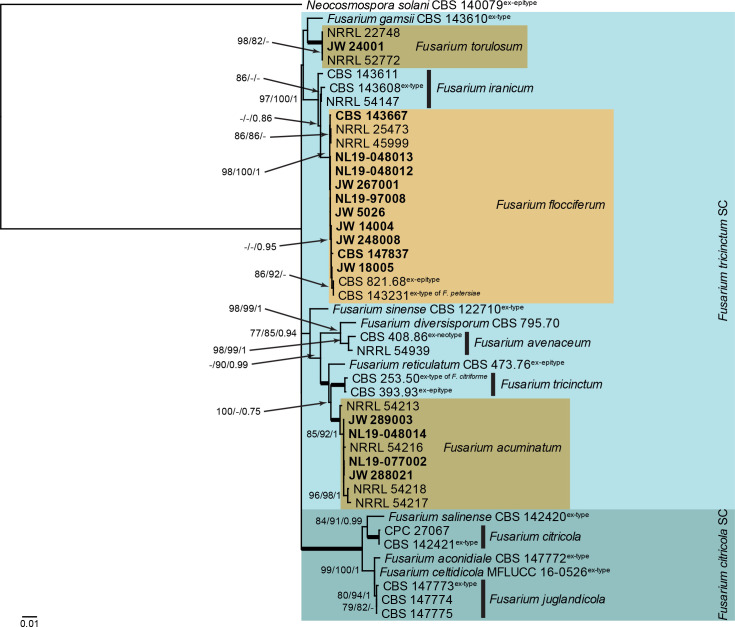
Fusarium citricola and F. tricinctum species complexes (Fig. 1): Novel isolates from Dutch soils clustered with three known species, namely F. acuminatum, F. flocciferum and F. torulosum (all three in the F. tricinctum species complex). The three phylogenetic analyses (RAxML, IQ-TREE, and MrBayes) overall displayed the same species clades and mainly differed with regards to the backbone relationships between species clades/lineages [data not shown, trees available in TreeBASE and support and posterior probability (PP) values are superimposed on the presented figure]. The loci cmdA and rpb1 are not well-represented in the dataset, with roughly half of the strains having a sequence present (Tables 1, 2, Supplementary Table S1).
Fig. 1.
The RAxML consensus tree inferred from the combined F. citricola/tricinctum species complexes tef1, rpb2 (parts 1 and 2), rpb1 and cmdA sequence alignment. Thickened lines indicate branches with full support (RAxML & IQ-TREE bootstrap = 100 %; PP = 1.0) with support values of other branches indicated at the branches (RAxML > 74 % / IQ-TREE > 84 % / PP > 0.74). The tree is rooted to Neocosmospora solani (CBS 140079, ex-epitype culture). The scale bar indicates the number of expected changes per site. Species complexes are indicated on the right and highlighted with coloured blocks. Species clades containing the novel citizen science strains (in bold) are highlighted with coloured blocks.
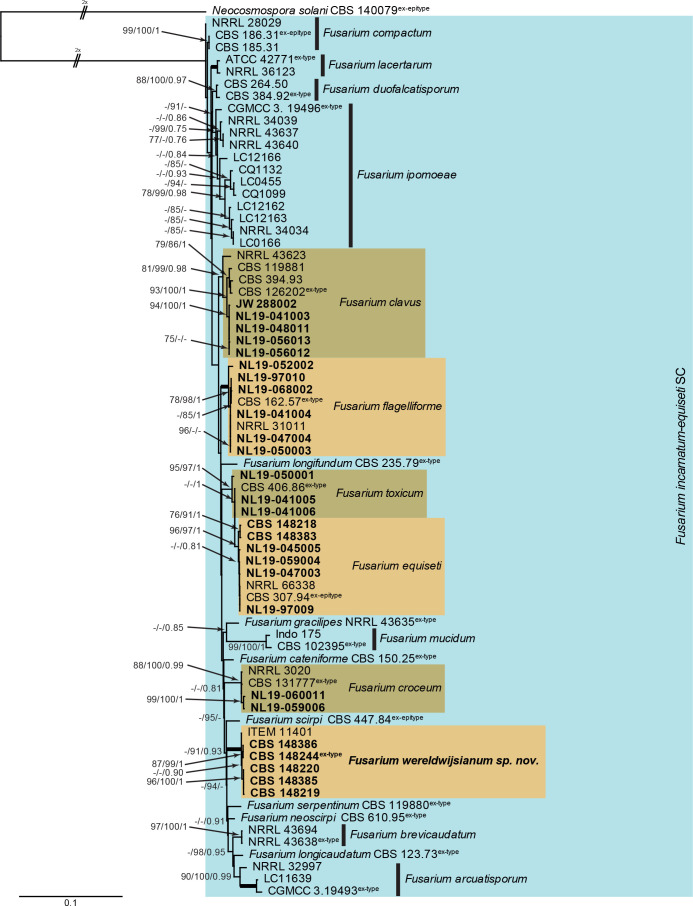
Fusarium incarnatum-equiseti species complex (Fig. 2): Novel isolates from Dutch soils clustered with five known species, namely F. clavus, F. croceum, F. equiseti, F. flagelliforme and F. toxicum, as well as a species clade not associated with any known species. The three phylogenetic analyses (RAxML, IQ-TREE and MrBayes) overall displayed the same species clades and mainly differed with regards to the backbone relationships between species clades/lineages (data not shown, trees available in TreeBASE and support and PP values are superimposed on the presented figure). The locus rpb1 is not well-represented in the dataset, with less than half of the strains having a sequence present (Tables 1, 2, Supplementary Table S1).
Fig. 2.
The RAxML consensus tree inferred from the combined F. incarnatum-equiseti species complex tef1, rpb2 (first part), cmdA and rpb1 sequence alignment. Thickened lines indicate branches with full support (RAxML & IQ-TREE bootstrap = 100 %; PP = 1.0) with support values of other branches indicated at the branches (RAxML > 74 % / IQ-TREE > 84 % / PP > 0.74). The tree is rooted to Neocosmospora solani (CBS 140079, ex-epitype culture) and the two basal branches were halved to facilitate layout. The scale bar indicates the number of expected changes per site. The F. incarnatum-equiseti species complex is indicated on the right and highlighted with a coloured block. Species clades containing the novel citizen science strains (in bold) are highlighted with coloured blocks and the novelty described in the present study is printed in bold font.
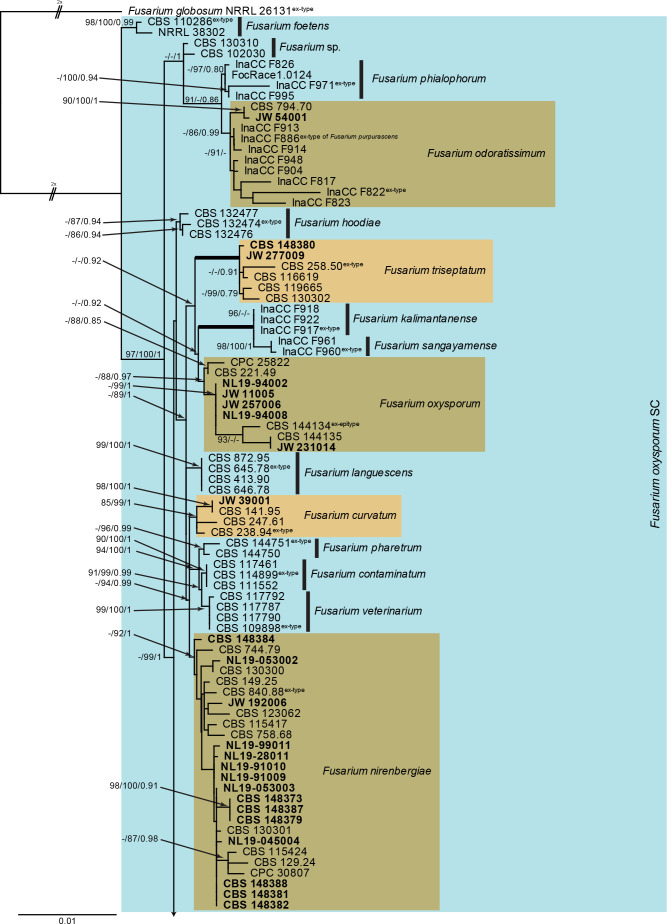
Fusarium oxysporum species complex (Fig. 3): Novel isolates from Dutch soils clustered with six known species, namely F. curvatum, F. nirenbergiae, F. odoratissimum, F. oxysporum and F. triseptatum, as well as four species clades not associated with any known species. The three phylogenetic analyses (RAxML, IQ-TREE and MrBayes) overall displayed the same species clades and mainly differed with regards to the backbone relationships between species clades/lineages (data not shown, trees available in TreeBASE and support and PP values are superimposed on the presented figure). The loci rpb1 and tub2 are not well-represented in the dataset, with roughly half of the strains having a sequence present (Tables 1, 2, Supplementary Table S1).
Fig. 3.
The RAxML consensus tree inferred from the combined F. oxysporum species complex tef1, rpb2 (first part), tub2, cmdA and rpb1 sequence alignment. Thickened lines indicate branches with full support (RAxML & IQ-TREE bootstrap = 100 %; PP = 1.0) with support values of other branches indicated at the branches (RAxML > 74 % / IQ-TREE > 84 % / PP > 0.74). The tree is rooted to Fusarium globosum (NRRL 26131) and the two basal branches were halved to facilitate layout. The scale bar indicates the number of expected changes per site. The F. oxysporum species complex is indicated on the right and highlighted with a coloured block. Species clades containing the novel citizen science strains (in bold) are highlighted with coloured blocks and the novelty described in the present study is printed in bold font.
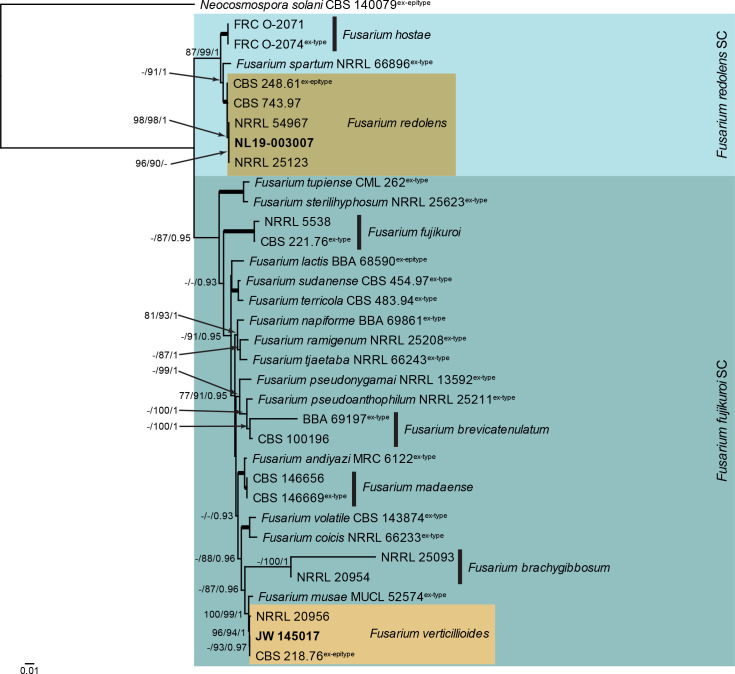
Fusarium fujikuroi and F. redolens species complexes (Fig. 4): Novel isolates from Dutch soils clustered with two known species, namely F. redolens (F. redolens species complex) and F. verticillioides (F. fujikuroi species complex). The three phylogenetic analyses (RAxML, IQ-TREE and MrBayes) had the same overall topology and same species clades/lineages (data not shown, trees available in TreeBASE and support and PP values are superimposed on the presented figure). The locus cmdA is not well-represented in the dataset, with roughly half of the strains having a sequence present (Tables 1, 2, Supplementary Table S1).
Fig. 4.
The RAxML consensus tree inferred from the combined F. redolens/fujikuroi species complexes tef1, rpb2 (first part), rpb1 and cmdA sequence alignment. Thickened lines indicate branches with full support (RAxML & IQ-TREE bootstrap = 100 %; PP = 1.0) with support values of other branches indicated at the branches (RAxML > 74 % / IQ-TREE > 84 % / PP > 0.74). The tree is rooted to Neocosmospora solani (CBS 140079, ex-epitype culture). The scale bar indicates the number of expected changes per site. Species complexes are indicated on the right and highlighted with coloured blocks. Species clades containing the novel citizen science strains (in bold) are highlighted with coloured blocks.
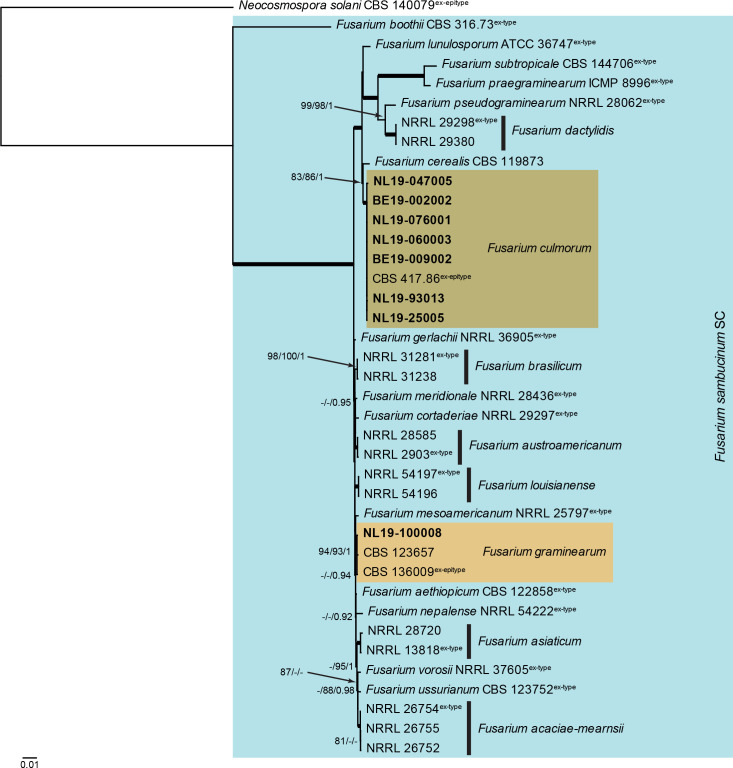
Fusarium sambucinum species complex (Fig. 5): Novel isolates from Dutch soils clustered with two known species, namely F. culmorum and F. graminearum. The three phylogenetic analyses (RAxML, IQ-TREE and MrBayes) overall displayed the same species clades and the Bayesian phylogeny mainly differed with regards to the backbone relationships between species clades/lineages in the lower half of the tree (data not shown, trees available in TreeBASE and support and PP values are superimposed on the presented figure). The locus cmdA is not well-represented in the dataset, with less than half of the strains having a sequence present (Tables 1, 2, Supplementary Table S1).
Fig. 5.
The RAxML consensus tree inferred from the combined F. sambucinum species complex tef1, rpb2 (parts 1 and 2), rpb1 and cmdA sequence alignment. Thickened lines indicate branches with full support (RAxML & IQ-TREE bootstrap = 100 %; PP = 1.0) with support values of other branches indicated at the branches (RAxML > 74 % / IQ-TREE > 84 % / PP > 0.74). The tree is rooted to Neocosmospora solani (CBS 140079, ex-epitype culture). The scale bar indicates the number of expected changes per site. The Fusarium sambucinum species complex is indicated on the right and highlighted with a coloured block. Species clades containing the novel citizen science strains (in bold) are highlighted with coloured blocks.
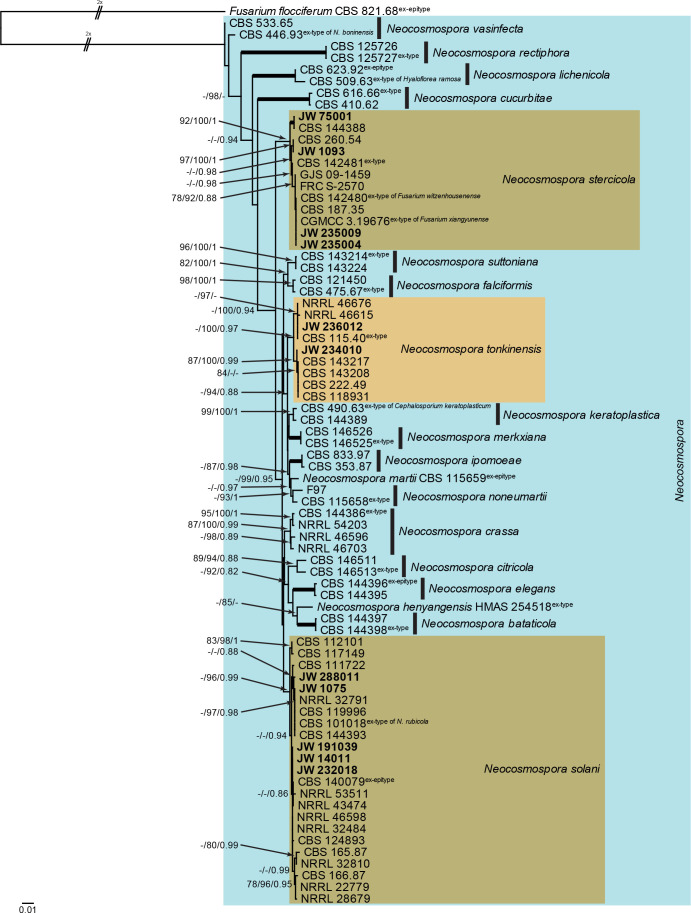
Neocosmospora (Fig. 6): Novel isolates from Dutch soils clustered with three known species, namely N. solani, N. stercicola and N. tonkinensis. The three phylogenetic analyses (RAxML, IQ-TREE and MrBayes) had the same overall topology, except for swapping around between N. rectiphora and N. vasinfecta as being the most basal species, and had the same species clades/lineages (data not shown, trees available in TreeBASE and support and PP values are superimposed on the presented figure). The loci cmdA and rpb1 are not well-represented in the dataset, with roughly half of the strains having a sequence present (Tables 1, 2, Supplementary Table S1).
Fig. 6.
The RAxML consensus tree inferred from the combined Neocosmospora tef1, rpb2 (first part), rpb1 and cmdA sequence alignment. Thickened lines indicate branches with full support (RAxML & IQ-TREE bootstrap = 100 %; PP = 1.0) with support values of other branches indicated at the branches (RAxML > 74 % / IQ-TREE > 84 % / PP > 0.74). The tree is rooted to Fusarium flocciferum (CBS 821.68, ex-epitype culture) and the two basal branches were halved to facilitate layout. The scale bar indicates the number of expected changes per site. The genus Neocosmospora is indicated on the right and highlighted with a coloured block. Species clades containing the novel citizen science strains (in bold) are highlighted with coloured blocks.
Based on these phylogenetic trees, several taxonomic decisions were made, and the individual and combined trees are discussed under the Notes in the Taxonomy section below, where applicable.
Taxonomy
Fusarium flocciferum Corda, in Sturm, Deutschl. Fl., Abt. 3, Pilze Deutschl. 2: 17. 1828.
New synonym: Fusarium petersiae L. Lombard, Persoonia 39: 457. 2017.
Additional synonyms see Crous et al. (2021b)
Material examined: Germany, from greenhouse soil, 1966, D. Bredemeier, ex-epitype culture of F. flocciferum CBS 821.68 = NRRL 28450. Netherlands, Friesland Province, Harlingen, from soil, 10 Oct. 2019, S. Goinga & J. de Groot, cultures NL19-048012, NL19-048013; Gelderland Province, Arnhem, from soil, Mar. 2017, D. Peters (holotype of F. petersiae CBS H-23233, culture ex-type CBS 143231 = JW 14004); ibid., culture JW 14005 = CBS 143667; Nijmegen, from soil, 2017, J.W. Koolen, culture JW 248008; North Brabant Province, Valkenswaard, from soil, 2017, W. van der Heijden, culture JW 18005; Utrecht Province, Utrecht, from soil, 2017, students of Basisschool de Baanbreker, culture JW267001; Bilthoven, Planetenplein, from garden soil, 31 Jul. 2019, S. Frederikze, J. Mes & S. Maghnouji, cultures NL19-97008, NL19-100011 = CBS 147837; Nieuwegein, from soil, 2017, F. & R. Niemeijer, culture JW 5026.
Notes: Fusarium petersiae was described from soil collected in this citizen science project (Crous et al. 2017). In the original publication, it was distinguished from F. flocciferum by the formation of sporodochia, up to 5-septate macroconidia, and the lack of conidiophores in aerial mycelium. Fusarium flocciferum was originally circumscribed as lacking sporodochia in culture and producing abundant 1–3-septate macroconidia on aerial conidiophores (Booth 1971). As we have shown here (Fig. 1), however, F. petersiae (CBS 143231) is phylogenetically identical to F. flocciferum (ex-type CBS 821.68) and is therefore reduced to synonymy.
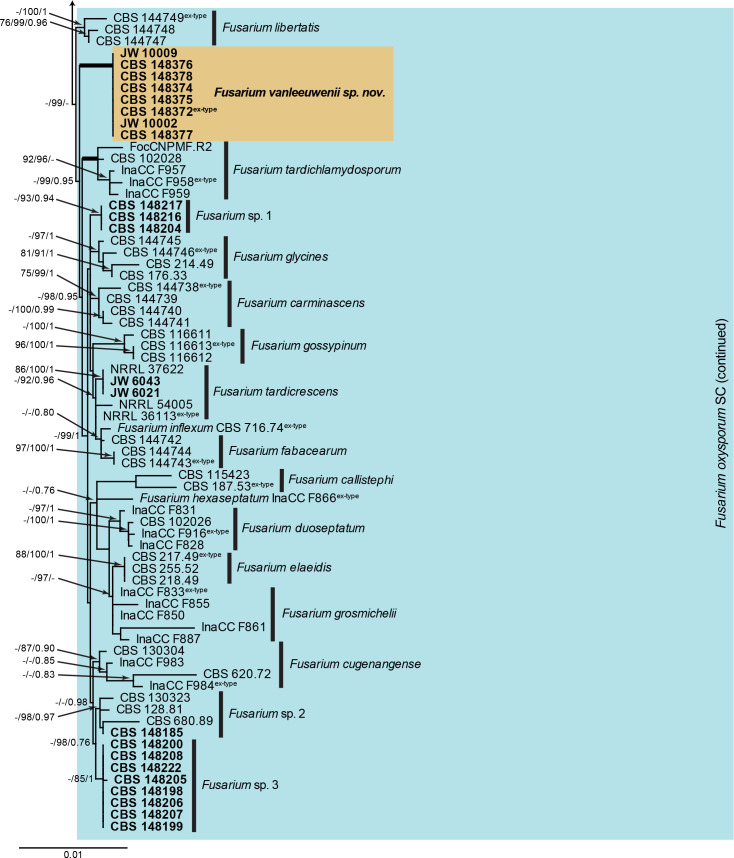
Fusarium sp. 1. Fig. 7.
Fig. 7.
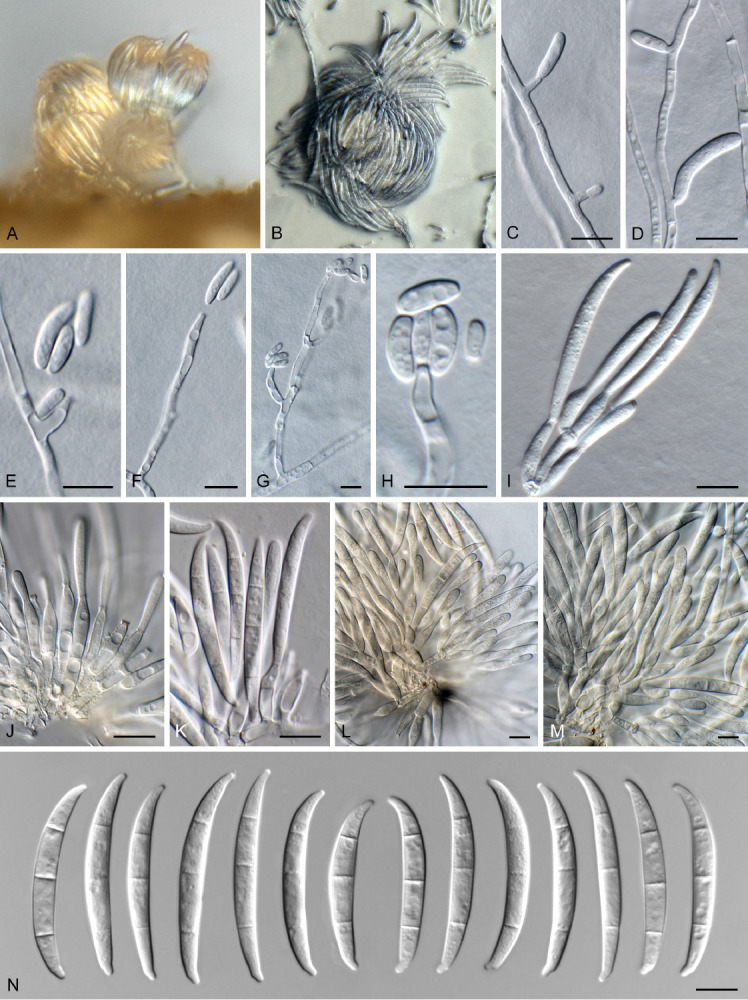
Fusarium sp. 1 (CBS 148217). A. Sporodochium on CLA. B. Sporodochium on SNA. C–H. Aerial conidiophores with microconidia. I–M. Sporodochial conidiophores. N. Macroconidia. Scale bars = 10 μm.
CBS 148217 (= NL19-25001): Aerial conidiophores sparingly branched, with terminal or intercalary conidiogenous cells, giving rise to macro- and microconidia; aerial conidiogenous cells monophialidic, subulate to subcylindrical, smooth and thin-walled, 5–30 × 2–3.5 μm, with flared collarette and minute periclinal thickening at apex. Microconidia aggregating in false heads, ellipsoid to subcylindrical, falcate, 0–1-septate, 5–20 × 3–4 μm. Sporodochia pale luteous to orange, abundant on CLA. Sporodochial conidiophores densely aggregated, verticillately branched, consisting of a short stipe bearing whorls of 2–3 monophialides; sporodochial conidiogenous cells monophialidic, subulate to subcylindrical, 10–15 × 4–5 μm, smooth- and thin-walled, with periclinal thickening at apex and minute, flared collarette. Sporodochial conidia falcate, curved dorsiventrally, sides almost parallel, tapering towards both ends; apical cell papillate and curved; basal cell foot-shaped, notch poorly developed, 3(–5)-septate, hyaline, smooth-walled, guttulate; 3-septate conidia (33–)43–45(–48) × (3.5–)4(–5) μm, 5-septate conidia rare, up to 60 μm long. Chlamydospores not observed.
Culture characteristics: Colonies spreading, with cottony aerial mycelium. On PDA surface and reverse pale vinaceous. On OA surface pale vinaceous, reverse rosy buff.
Isolates examined: Netherlands, Groningen Province, Warffum, from garden soil, 6 Jun. 2019, C. Dijkstra & L. Kruit, culture NL19-25001 = CBS 148217; Limburg Province, Ell, 2017, K. Brennand, culture JW 53002 = CBS 148216; Utrecht Province, Amersfoort, 2017, T. & K. Wesselink, culture JW 191014 = CBS 148204.
Notes: Fusarium sp. 1 (CBS 148217) is related (Fig. 3) to F. tardichlamydosporum [macroconidia (36–)37–43(–45) × (4–)5–6(–7) μm (av. 40 × 5 μm), 3–5-septate; Maryani et al. (2019a)], F. carminascens [3-septate macroconidia: (21–)26–36(–40) × 3–5 μm (av. 31 × 4 μm); 4-septate macroconidia: (31–)33–43(–44) × 4–5 μm (av. 38 × 4 μm); Lombard et al. 2019]; and F. vanleeuwenii [3-septate macroconidia (32–)45–50(–52) × (3.5–)4(–4.5) μm, 4–5-septate conidia 52–60 × 4.5–5 μm, 7–8-septate conidia rare, 65–75 × 5–6 μm] in the FOSC (see elsewhere in this paper). It is morphologically distinct from these species based on the dimensions of its macroconidia. The species is undisguisable from other included species on cmdA (intermingled with numerous species), rpb1 (intermingled with F. keijii and F. joseae), rpb2 (intermingled with numerous species), and tef1 (intermingled with F. cugenangense), and can best be identified using a multi-gene phylogenetic analysis. No tub2 sequences were available for comparison. The species clade is well-supported in two of the analyses (IQ-TREE bootstrap support value = 99 %; Bayesian PP = 0.95). This species is unnamed at present, pending further data.
Fusarium sp. 2. Fig. 8.
Fig. 8.
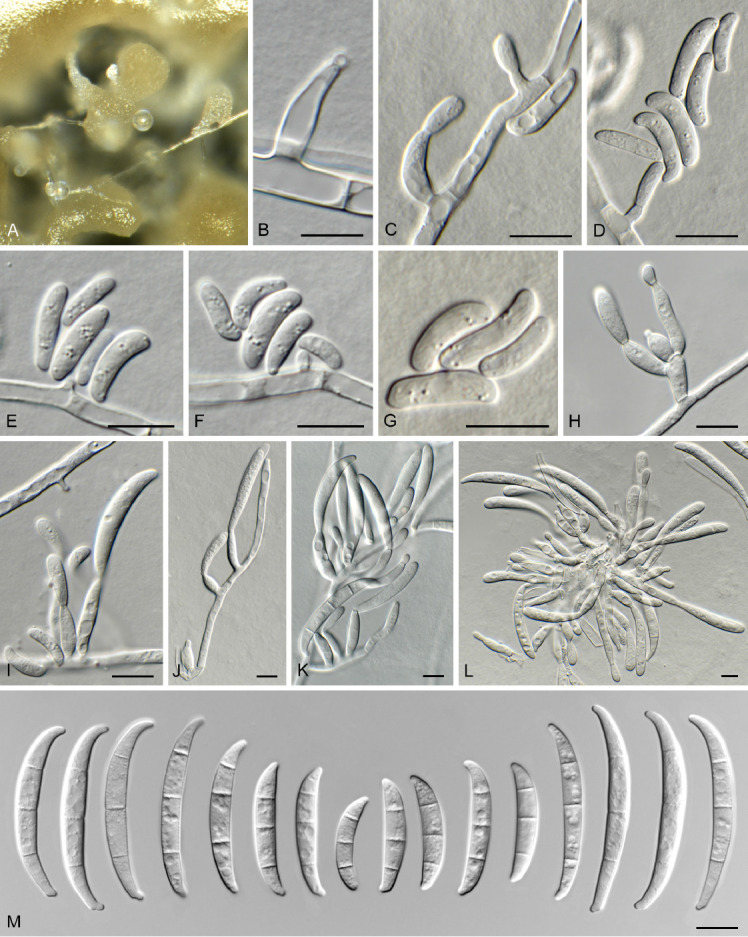
Fusarium sp. 2 (CBS 148185). A. Sporodochia on CLA. B–H. Aerial conidiophores with microconidia. I, J. Aerial conidiophores with macroconidia. K, L. Sporodochial conidiophores. M. Macroconidia. Scale bars = 10 μm.
CBS 148185 (= JW 1072): Aerial conidiophores sparingly branched, 2–20 μm tall, mostly reduced to conidiogenous cells on hyphae; aerial conidiogenous cells monophialidic, subulate to subcylindrical, smooth and thin-walled, 2–20 × 2–6 μm, with flared collarette and minute periclinal thickening at apex. Microconidia aggregating in false heads, falcate, subcylindrical to reniform, (0–)1(–2)-septate, (10–)13–15(–20) × (3–)3.5–4 μm. Sporodochia pale luteous, abundant on CLA. Sporodochial conidiophores densely aggregated, verticillately branched, consisting of a short stipe bearing whorls of 2–3 monophialides; sporodochial conidiogenous cells monophialidic, subulate to subcylindrical, 9–22 × 3–5 μm, smooth- and thin-walled, with periclinal thickening at apex and minute, flared collarette. Sporodochial conidia falcate, moderately curved dorsiventrally, sides almost parallel, tapering towards both ends; apical cell blunt to papillate and curved; basal cell foot-shaped, notch poorly developed, 3(–6)-septate, hyaline, smooth-walled, guttulate; 3-septate conidia (30–)38–43(–47) × 4–5(–6) μm, 4-septate conidia 45–47 × 4.5–5 μm, 5-septate conidia 50–65 × 5 μm. Chlamydospores not observed.
Culture characteristics: Colonies flat, spreading, with cottony aerial mycelium. On PDA surface rosy vinaceous, reverse greyish rose. On OA surface and reverse greyish rose.
Isolates examined: Australia, Subungual debris of 40-year-old female with nail infection, collection date unknown, collector unknown, culture CBS 130323 =NRRL 26677. Netherlands, North Holland Province, Amsterdam, from garden soil, Mar. 2017, J.F.T.M. van Dijk, culture CBS 148185 = JW 1072; Zuid-Holland Province, Nootdorp, Cucumis sativus, in greenhouse on rockwool, No. 1979, collection date unknown, N. Hubbeling, culture CBS 680.89 = IPO 11179 = NRRL 26221. USA, on Chrysantemum sp., collection date unknown, collector unknown, culture CBS 128.81 =NRRL 36233 = BBA63925.
Notes: Fusarium sp. 2. (CBS 148185) is related (Fig. 3) to F. cugenangense (FOSC; associated with banana, but non-pathogenic on Gros Michel (AAA) and Cavendish (AAA); Maryani et al. 2019a) and Fusarium sp. 3 (see below). It is distinguished morphologically from F. cugenangense which has smaller micro- (av. 12 × 5 μm), and larger macroconidia (44–)47–54(–57) × (5–)6–7(–8) μm (av. 53 × 7 μm), 3–6-septate (Maryani et al. 2019a). Fusarium sp. 3 is similar to Fusarium sp. 2, but has larger macroconidia, e.g. 3-septate macroconidia (33–)43–50(–55) × (3.5–)4(–4.5) μm, 5-septate macroconidia 65–75 × 4–5 μm, and produces chlamydospores. This species can readily be distinguished from other included species based on tef1, but is undisguisable from other included species on cmdA, rpb1, rpb2 and tub2. This species clade is supported in two of the analyses (IQ-TREE bootstrap support value = 94 %; Bayesian PP = 0.98), but is left unnamed, pending further data.
Fusarium sp. 3. Fig. 9.
Fig. 9.
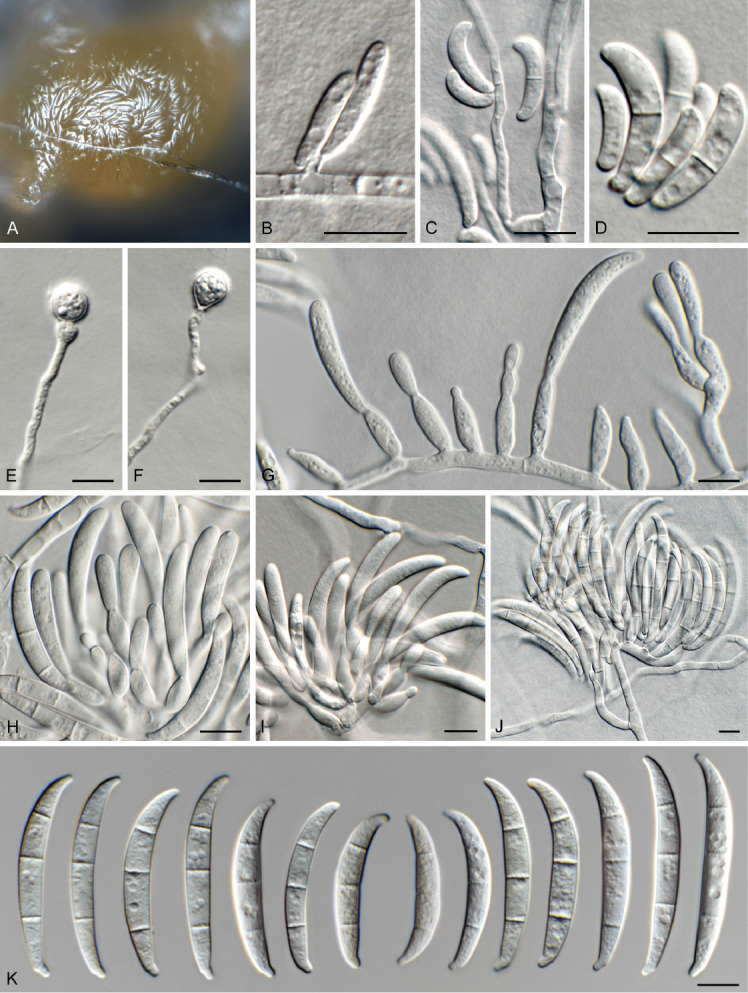
Fusarium sp. 3 (CBS 148207). A. Sporodochium on CLA. B, C, G. Aerial conidiophores with conidia. D. Microconidia. E, F. Chlamydospores. H–J. Sporodochial conidiophores. K. Macroconidia. Scale bars = 10 μm.
CBS 148207 (= JW 210019): Aerial conidiophores sparingly branched, mostly reduced to monophialides; aerial conidiogenous cells monophialidic, subcylindrical, smooth and thin-walled, 2–15 × 3–4 μm, with minute collarette at apex. Microconidia aggregating in false heads, ellipsoid to subcylindrical, falcate, 0–1-septate, (8–)10–17(–28) × (2.5–)3(–3.5) μm. Sporodochia pale white, sparse on CLA. Sporodochial conidiophores densely aggregated, verticillately branched, consisting of a short stipe bearing whorls of 2–3 monophialides; sporodochial conidiogenous cells monophialidic, subulate to subcylindrical, 5–15 × 3–5 μm, smooth- and thin-walled, with periclinal thickening at apex and minute, flared collarette. Sporodochial conidia straight to falcate, curved dorsiventrally, sides almost parallel, tapering towards both ends; apical cell blunt or papillate and curved; basal cell foot-shaped, notch poorly developed, 3(–5)-septate, hyaline, smooth-walled, guttulate; 3-septate conidia (33–)43–50(–55) × (3.5–)4(–4.5) μm, 5-septate conidia rare, 65–75 × 4–5 μm. Chlamydospores sparingly formed on CLA, subglobose to globose, pale brown, thick-walled, terminal or intercalary, 6–8 μm diam.
Culture characteristics: Colonies flat, spreading, with sparse aerial mycelium. On PDA surface and reverse pale vinaceous. On OA surface and reverse livid vinaceous.
Isolates examined: Belgium, East Flanders, Brakel, from garden soil, 2019, T. Antheunis, culture BE 19_004006 = CBS 148222. Netherlands, Friesland Province, Heerenveen, from garden soil, 2017, N. Keij, culture JW 210019 = CBS 148207; Friesland Province, Heerenveen, from garden soil, 2017, N. Keij, culture JW 210014 = CBS 148206; Friesland Province, Leeuwarden, from garden soil, 2017, D. Pol, R. Verf, J. Wilks & M. de Ruiter, culture JW 231016 = CBS 148208; Gelderland Province, Geldermalsen, from garden soil, 2017, A.-S. den Boer, culture JW 9002 = CBS 148199; Gelderland Province, Culemborg, from garden soil, 2017, I. Kleij, culture JW 204009 = CBS 148205; Utrecht Province, Amersfoort, from garden soil, 2017, F. Wiegerinck, culture JW 4030 = CBS 148198; Utrecht Province, Utrecht, from garden soil, 2017, M.J. van Leeuwen, culture JW 10005 = CBS 148200.
Notes: Fusarium sp. 3 (CBS 148207) is closely related (Fig. 3) to Fusarium sp. 2 [3-septate macroconidia (30–)38–43(–47) × 4–5(–6) μm] in the FOSC, and can be distinguished morphologically in having larger 3-septate macroconidia, and in producing chlamydospores, which were not observed in Fusarium sp. 2. This species can readily be distinguished from other included species based on cmdA and tef1, but is undisguisable from other included species on rpb1 and rpb2. No tub2 sequences were available for comparison. The species clade is poorly to fully supported in two of the analyses (IQ-TREE bootstrap support value = 85 %; Bayesian PP = 1), but is left unnamed, pending further data.
Fusarium vanleeuwenii Crous & Sand.-Den., sp. nov. MycoBank MB 840894. Fig. 10.
Fig. 10.
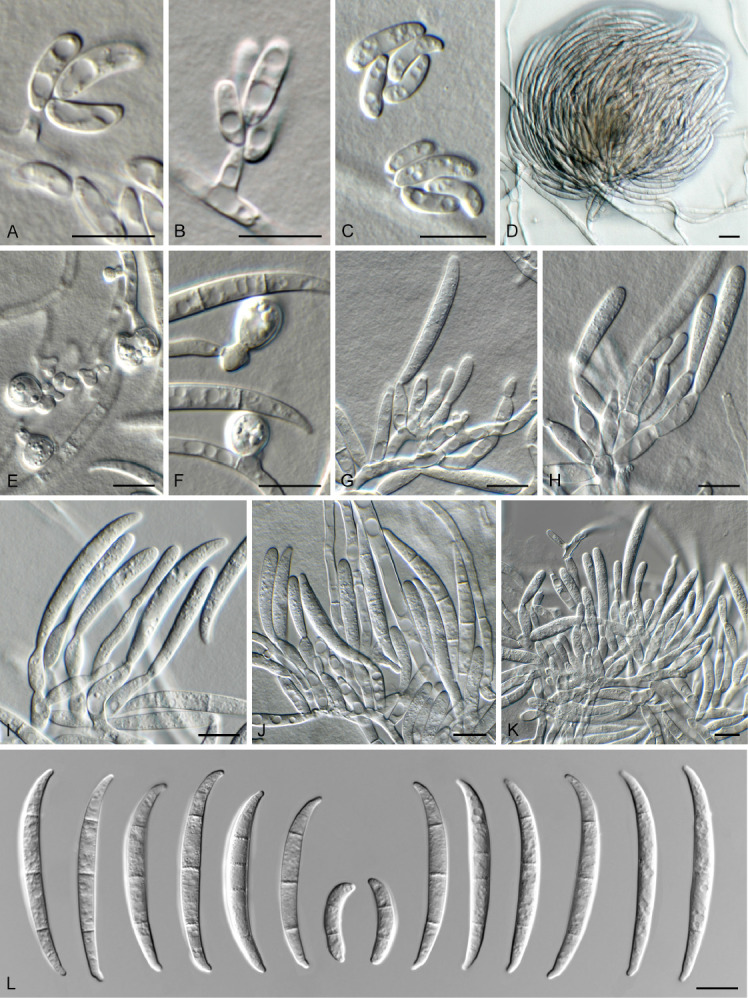
Fusarium vanleeuwenii (CBS 148372). A–C. Aerial conidiophores with microconidia. D. Sporodochium on SNA. E, F. Chlamydospores. G–K. Sporodochial conidiophores. L. Macroconidia. Scale bars = 10 μm.
Etymology: Named after the collector, Maurits Jesse van Leeuwen. This sample was collected during a Citizen Science project of the Westerdijk Fungal Biodiversity Institute.
Typus: Netherlands, Utrecht Province, Utrecht, from garden soil, 2017, M.J. van Leeuwen, (holotype CBS H-24786, culture ex-type CBS 148372 = JW 10008).
Aerial conidiophores irregularly branched, up to 70 μm tall, or reduced to conidiogenous cells on hyphae; conidiogenous cells monophialidic, subulate to subcylindrical, smooth and thin-walled in branched clusters, 10–25 × 4–5 μm; at times reduced to conidiogenous pegs on hyphae, erect, 2–10 × 1.5–2.5 μm, with flared collarette and minute periclinal thickening at apex. Microconidia aggregating in mucoid droplets, 0(–2)-septate, ellipsoid to subcylindrical, reniform to somewhat falcate, apical cell becoming hooked, guttulate, (7–)10–14(–18) × 2.5–4 μm. Sporodochial conidiophores in moderate numbers on CLA, pale yellow, densely aggregated, irregularly branched, typically in whorls of 2–4 phialides; sporodochial conidiogenous cells monophialidic, subulate to subcylindrical, 9–18 × 3–4.5 μm, with periclinal thickening at apex and inconspicuous collarette. Sporodochial conidia falcate, moderately curved, more so on outer than inner plane, widest in middle; apical cell papillate to hooked; basal cell foot-shaped, notch poorly developed, (1–)3(–8)-septate, hyaline, smooth-walled, guttulate; 1-septate conidia 15–20 × 3–4 μm, 2-septate conidia 20–25 × 3–4 μm, 3-septate conidia (32–)45–50(–52) × (3.5–)4(–4.5) μm, 4–5-septate conidia 52–60 × 4.5–5 μm, 7–8-septate conidia rare, 65–75 × 5–6 μm. Chlamydospores sparse after 1 wk, globose to subglobose, 7–8 μm diam, formed terminally or intercalary, single, smooth-walled, subhyaline.
Culture characteristics: Colonies erumpent, spreading, covering dish in 7 d, with moderate aerial mycelium. On PDA surface vinaceous, reverse rosy vinaceous. On OA surface livid red, reverse greyish rose. On MEA surface and reverse dark vinaceous.
Additional isolates examined: Netherlands, Utrecht Province, Utrecht, from garden soil, 2017, M.J. van Leeuwen, cultures CBS 148374 = JW 10001, JW 10002, CBS 148375 = JW 10003, CBS 148376 = JW 10004, CBS 148377 = JW 10006, CBS 148378 = JW 10007, JW 10009.
Notes: Fusarium vanleeuwenii is distantly related (Fig. 3) to F. tardichlamydosporum, a species in the FOSC associated with Panama disease of banana, pathogenic on Gros Michel (AAA) (Foc-Race1) (Maryani et al. 2019a). Morphologically, the two species are very similar, but F. tardichlamydosporum has smaller micro- (3–)5–9(–15) × (2–)5(–9) μm, and macroconidia (36–)37–43(–45) × (4–)5–6(–7) μm (av. 40 × 5 μm), 3–5-septate (Maryani et al. 2019a).
Fusarium vanleeuwenii is characteristic in that it has sparse chlamydospores, the aerial conidiophores are reduced to conidiogenous pegs on hyphae, and the reniform microconidia tend to have hooked apical cells. This species can readily be distinguished from other included species based on cmdA, rpb1, and rpb2, but is intermingled with F. foetens and F. oxysporum on tef1. No tub2 sequences were available for comparison. The species clade is fully supported in all analyses (RAxML bootstrap support value = 100 %; IQ-TREE bootstrap support value = 100 %; Bayesian PP = 1).
Fusarium wereldwijsianum Crous & Sand.-Den., sp. nov. MycoBank MB 840895. Fig. 11.
Fig. 11.
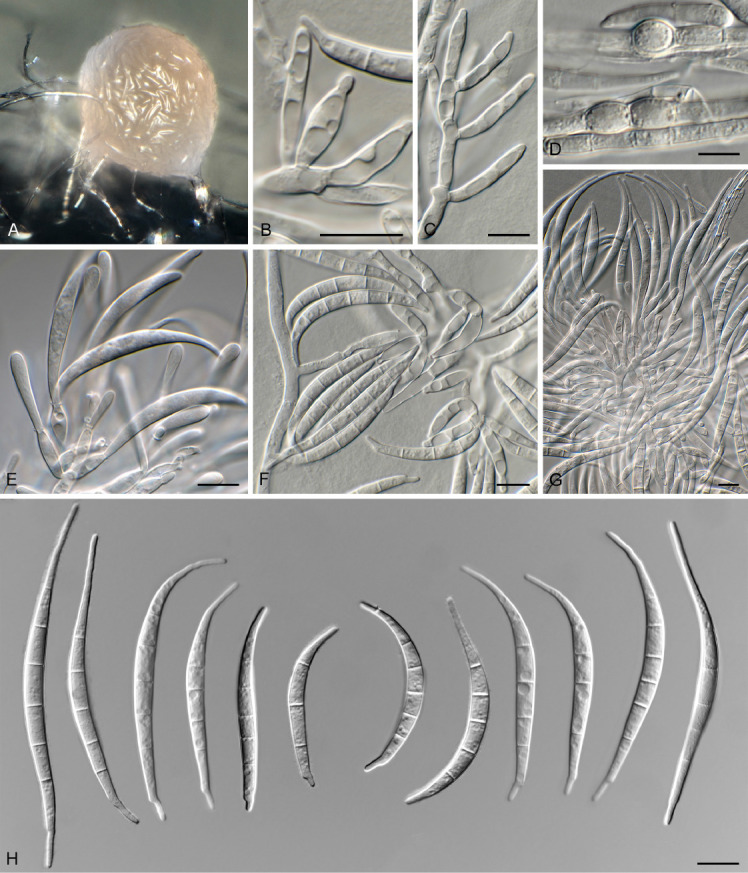
Fusarium wereldwijsianum (CBS 148244). A. Sporodochium on CLA. B, C, E–G. Sporodochial conidiophores. D. Chlamydospores. H. Macroconidia. Scale bars = 10 μm.
Etymology: Named after the school “Wereldwijs” (Bilthoven, the Netherlands) where the sample was collected. This sample was collected during a Citizen Science project of the Westerdijk Fungal Biodiversity Institute.
Typus: Netherlands, Utrecht Province, Bilthoven, Planetenplein, from garden soil, 31 Jul. 2019, S. Frederikze, J. Mes & S. Maghnouji (holotype CBS H-24787, culture ex-type CBS 148244 = NL19-94009).
Aerial conidiophores sparingly branched, 5–20 μm tall, bearing terminal and lateral monophialides, but mostly reduced to conidiogenous cells on hyphae; aerial conidiogenous cells monophialidic, subulate to subcylindrical, smooth and thin-walled, 5–15 × 3.5–4 μm, with flared collarette and minute periclinal thickening at apex. Aerial conidia aggregating in false heads, falcate, 1–3-septate, apex obtuse to acutely rounded, base obtuse to notched, (16–)20–22(–25) × 3–3.5(–4) μm. Sporodochia orange, abundant on CLA. Sporodochial conidiophores densely aggregated, verticillately branched, consisting of a short stipe bearing whorls of 2–4 monophialides; sporodochial conidiogenous cells monophialidic, subulate to subcylindrical, 10–20 × 3.5–4 μm, smooth- and thin-walled, with periclinal thickening at apex and minute, flared collarette. Sporodochial conidia falcate, curved dorsiventrally, tapering towards both ends; apical cell elongated, curved, whip-like; basal cell foot-shaped, notch well developed, 3(–5)-septate, hyaline, smooth-walled, guttulate; 3-septate conidia (40–)45–60(–65) × 4(–5) μm, 5-septate conidia (45–)55–65 × 4–4.5(–5) μm. Chlamydospores on SNA after 1 wk sparse, solitary, intercalary or terminal, subglobose, 6–8 μm diam, becoming brown with age.
Culture characteristics: Colonies spreading, with cottony aerial mycelium. On PDA surface and reverse rosy buff. On OA surface buff to rosy buff, reverse rosy buff to rosy vinaceous.
Additional isolates examined: Netherlands, Utrecht Province, Bilthoven, Planetenplein, 31 Jul. 2019, S. Frederikze, J. Mes & S. Maghnouji, cultures cultures CBS 148219 = NL19-99003, CBS 148220 = NL19-99002; Zeeland Province, Oostburg, 23 Oct. 2019, A. van Strien, I. Beemsterboer & S. Groosman, culture CBS 148386 = NL19-059003; Zeeland Province, Oostburg, Oct. 2019, F. Guilliet, T. Bron & I. Geernaert, culture CBS 148385 = NL19-057012.
Notes: Fusarium wereldwijsianum is a member of the F. incarnatum-equiseti species complex (FIESC; Wang et al. 2019, Xia et al. 2019), clustering among F. scirpi, F. serpentinum and F. neoscirpi (Fig. 2). It can be distinguished morphologically from F. scirpi which commonly has polyphialides, and 6–7-septate macroconidia (Leslie & Summerell 2006). Fusarium wereldwijsianum is further distinguished from F. neoscirpi which has smaller macroconidia [3-septate conidia: (28–)32–42(–46) × 4–5 μm (av. 37 × 4 μm); 5-septate conidia: (47–)50–58(–64) × 4–6 μm (av. 54 × 5 μm); Xia et al. 2019], and lacks chlamydospores. It is also distinct from F. serpentinum which has larger, (3–)5–7(–8)-septate macroconidia [3-septate conidia: (42–)43–51(–54) × 4–6 μm; 5-septate conidia: (57–)67–85(–92) × 4–6 μm; Xia et al. 2019]. Fusarium wereldwijsianum can readily be distinguished from other included species based on cmdA, rpb1, and tef1, but less readily so on rpb2. The species clade is fully supported in all analyses (RAxML bootstrap support value = 100 %; IQ-TREE bootstrap support value = 100 %; Bayesian PP = 1).
DISCUSSION
The present study focused on fusarioid fungi that were isolated from soil in the Netherlands during a Citizen Science project, which already has revealed numerous new species of filamentous fungi and yeasts (Crous et al. 2017, 2018, Groenewald et al. 2018, Giraldo et al. 2019, Hou et al. 2020, Crous et al. 2021a).
Fusarium and allied fusarioid genera are common soil inhabitants, and therefore it should not be seen as surprising that the present study identified 25 taxa, including 22 Fusarium spp., and three species of Neocosmospora. One new species was described from the FOSC, namely F. vanleeuwenii, and one from the FIESC, namely F. wereldwijsianum. Furthermore, F. petersiae (Crous et al. 2017) was also reduced to synonymy under F. flocciferum, which was found to be morphologically more variable than suspected when it was first described (Booth 1971).
Although the various soil samples were collected from garden soils in the urban environment, it was somewhat surprising to also encounter a well-known pathogen of banana, such as F. odoratissimum (syn. F. purpurascens sensu Crous et al. 2021b). Some Dutch isolates clustered with named subclades such as F. callistephi (CBS 187.53) or F. tardicrescens (JW 6021, JW 6043) (Maryani et al. 2019a), or appeared to represent new taxa, which we prefer to leave unnamed for now, pending more data to help resolve species boundaries within this clade. The identification of JW 6021 and JW 6043 as F. tardicrescens is based on the rpb1 and tef1 association with strain NRRL 37622 (see TreeBASE), a strain previously identified as belonging to that species (Maryani et al. 2019a).
Other species isolated that belong to the FOSC include: F. curvatum, described from Beaucarnia sp. and Hedera helix in the Netherlands, but also known from Matthiola incana in Germany (Lombard et al. 2019); F. nirenbergiae, described from Dianthus caryophyllus and Solanum lycopersicum in the Netherlands, but also known from numerous other plant and animal hosts, including humans, in countries such as Brazil, Italy, South Africa and the USA (Lombard et al. 2019); F. oxysporum, originally described from a rotten tuber of Solanum tuberosum, but having a wide host range with a worldwide distribution (Lombard et al. 2019), and F. triseptatum, known from hosts such as Ipomoea batatas, humans (USA), wilted Gossypium hirsutum (Ivory Coast), and sago starch (Papua New Guinea) (Lombard et al. 2019).
Five species from the FIESC isolated include: F. clavus, known from desert soil in Namibia, but also from various plant hosts in Germany, Iran, Russia and the USA (Xia et al. 2019); F. croceum, described from soil in the Czech Republic, but also known from Triticum in Iran (Xia et al. 2019); F. equiseti, a saprobe or secondary invader, common in cool to temperate or hot and arid climates (Leslie & Summerell 2006); F. flagelliforme known from Pinus nigra seedlings in Croatia, and various plant hosts in Germany (Xia et al. 2019), and F. toxicum, known from soil collected in Germany, but also isolated from a dog in the USA (Xia et al. 2019).
The Fusarium tricinctum species complex (FTSC) was represented by three species: F. acuminatum, a soil saprobe associated with roots and crowns of plants in temperate regions (Leslie & Summerell 2006), F. torulosum, occurring in soil in temperate regions, and from a number of plant hosts including cereals, tomatoes, beet root and trees (Leslie & Summerell 2006), and F. flocciferum, a common species in temperate regions, occurring in soil, and roots, fruits, stems and twigs of various plant hosts in Europe, North America and Iran (Gerlach & Nirenberg 1982, Torbati et al. 2018). The Fusarium sambucinum species complex (FSAMSC) was represented by two species: Fusarium culmorum, a species commonly found in temperate climates, associated with cereal crowns and grain, and plant debris in soil, and F. graminearum, a species primarily associated with maize, wheat and barley, but also other plant hosts (Leslie & Summerell 2006). The Fusarium redolens species complex (FRSC) was represented by a single species, F. redolens, which is a common soilborne fungus found in temperate areas. Likewise, the Fusarium fujikuroi species complex (FFSC) was also associated with a single species, F. verticillioides, which is a common pathogen of maize with a worldwide distribution (Leslie & Summerell 2006).
Finally, three species of Neocosmospora were also encountered in this study. These include N. solani, a common soil inhabitant, which is known from several plant species and has a global distribution. Less well-known species include N. stercicola, known from soil, and various other plant hosts in Europe (Sandoval-Denis et al. 2019), and N. tonkinensis, known from Musa sapientum in Vietnam, and various plant hosts in Europe, including Euphorbia fulgens in the Netherlands, and a turtle head lesion and human cornea in the USA (Sandoval-Denis 2019).
These findings underline the fact that fusarioid fungi are common soil inhabitants and are generally widely distributed. The ability of these fungi to produce chlamydospores (resting spores) in hyphae, macroconidia, and plant debris, make them well suited to survive adverse conditions for extended periods of time in the soil environment. Although many are saprobic, they appear to also can switch to an opportunistic or pathogenic lifestyle under more favourable conditions, and once in contact with their ideal host(s). It is therefore probable that several of the species described here as presumed saprobes, will in time be shown to be pathogens under favourable conditions.
In conclusion, this study has revealed a high number of fusarioid taxa in the urban soil environment, underlining the importance of this substrate for the discovery of novel taxa, and for gaining a better understanding of species diversity of fusarioid taxa in soil.
ACKNOWLEDGEMENTS
This study was financially supported by the Utrecht University Museum and the Royal Dutch Academy of Arts and Sciences for promoting the Citizen Science project, and for providing a platform to facilitate interaction with various Dutch primary schools. We are grateful to all the children and parents who participated in this project, collecting samples in their gardens and submitting them to the Westerdijk Institute for analyses. We are thankful to the staff from the Westerdijk Institute: Manon Verweij, Karin Schagen and Mariëtte Oosterwegel, for promoting the project and establishing communication with the collectors and schools. We also thank Marjan Vermaas for assistance with the photographic plates.
Footnotes
Citation: Crous PW, Hernández-Restrepo M, van Iperen AL, Starink-Willemse M, Sandoval-Denis M, Groenewald JZ (2021). Citizen science project reveals novel fusarioid fungi (Nectriaceae, Sordariomycetes) from urban soils. Fungal Systematics and Evolution 8: 101–127. doi: 10.3114/fuse.2021.08.09
Corresponding editor: L. Cai
Conflict of interest: The authors declare that there is no conflict of interest.
Supplementary Material: http://fuse-journal.org/
Collection details and GenBank accession numbers of strains used in the phylogenetic trees.
REFERENCES
- Bamburg JR, Riggs NV, Strong FM. (1968). The structures of toxins from two strains of Fusarium tricinctum. Tetrahedron 24: 3329–3336. [DOI] [PubMed] [Google Scholar]
- Booth C. (1971). The genus Fusarium. Commonwealth Mycological Institute, Kew, Surrey, England. [Google Scholar]
- Couteaudier Y, Alabouvette C. (1990). Survival and inoculum potential of conidia and chlamydospores of Fusarium oxysporum f.sp. lini in soil. Canadian Journal of Microbiology 36: 551–556. [DOI] [PubMed] [Google Scholar]
- Crous PW, Gams W, Stalpers JA, et al. (2004). MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology 50: 19–22. [Google Scholar]
- Crous PW, Hernández-Restrepo M, Schumacher RK, et al. (2021a). New and Interesting Fungi. 4. Fungal Systematics and Evolution 7: 255–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Lombard L, Sandoval-Denis M, et al. (2021b). Fusarium: more than a node or a foot-shaped basal cell. Studies in Mycology 98: 100116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Luangsa-ard JJ, Wingfield MJ, et al. (2018). Fungal Planet description sheets: 785–867. Persoonia 41: 238–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Verkleij GJM, Groenewald JZ, et al. (2019). Fungal Biodiversity. [Westerdijk Laboratory Manual Series No. 1]. Westerdijk Fungal Biodiversity Institute publishing, Utrecht, Netherlands. [Google Scholar]
- Crous PW, Wingfield MJ, Burgess TI. et al. 2017. Fungal Planet description sheets: 625–715. Persoonia 39: 270–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher NL, Burgess LW, Toussoun TA. et al. (1982). Carnation leaves as a substrate and for preserving cultures of Fusarium species. Phytopathology 72: 151–153. [Google Scholar]
- Geiser DM, Al-Hatmi AMS, Aoki T, et al. (2021). Phylogenomic analysis of a 55.1 kb 19-gene dataset resolves a monophyletic Fusarium that includes the Fusarium solani species complex. Phytopathology DOI:10.1094/PHYTO-08-20-0330-LE. [DOI] [PubMed] [Google Scholar]
- Gerlach W, Nirenberg H. (1982). The genus Fusarium – a pictorial atlas. Mitteilungen aus der Biologischen Bundesanstalt für Land- und Forstwirtschaft Berlin-Dahlem 209: 1–406. [Google Scholar]
- Giraldo A, Hernández-Restrepo M, Crous PW. (2019). New plecto-sphaerellaceous species from Dutch garden soil. Mycological Progress 18: 1135–1154. [Google Scholar]
- Gräfenhan T, Schroers H-J, Nirenberg HI, et al. (2011). An overview of the taxonomy, phylogeny, and typification of nectriaceous fungi in Cosmospora, Acremonium, Fusarium, Stilbella and Volutella. Studies in Mycology 68: 79–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenewald M, Lombard L, de Vries M, et al. (2018). Diversity of yeast species from Dutch garden soil and the description of six novel Ascomycetes. Federation of European Microbiological Societies Yeast Research 18: foy076. [DOI] [PubMed] [Google Scholar]
- Hernández-Restrepo M, Giraldo A, van Doorn R, et al. (2020). The Genera of Fungi – G6: Arthrographis, Kramasamuha, Melnikomyces, Thysanorea, and Verruconis. Fungal Systematics and Evolution 6: 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou LW, Hernández-Restrepo M, Groenewald JZ, et al. (2020). Citizen science project reveals high diversity in Didymellaceae (Pleosporales, Dothideomycetes). MycoKeys 65: 49–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez M, Huerta T, Mateo R. (1997). Mycotoxin production by Fusarium species isolated from bananas. Applied and Environmental Microbiology 63: 364–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov AM, Darriba D, Flouri T, et al. (2019). RAxML-NG: A fast, scalable, and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35: 4453–4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie JF, Summerell BA. (2006). The Fusarium Laboratory Manual. Blackwell Publishing Professional, USA. [Google Scholar]
- Lombard L, Sandoval-Denis M, Lamprecht SC, et al. (2019). Epitypification of Fusarium oxysporum – clearing the taxonomic chaos. Persoonia 43: 1–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard L, van der Merwe NA, Groenewald JZ, et al. (2015). Generic concepts in Nectriaceae. Studies in Mycology 80: 189–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maryani N, Lombard L, Poerba YS. et al. (2019a). Phylogeny and genetic diversity of the banana Fusarium wilt pathogen Fusarium oxysporum f. sp. cubense in the Indonesian centre of origin. Studies in Mycology 92: 155–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maryani N, Sandoval-Denis M, Lombard L, et al. (2019b). New endemic Fusarium species hitch-hiking with pathogenic Fusarium strains causing Panama disease in small-holder banana plots in Indonesia. Persoonia 43: 48–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh Q, Schmidt HA, Chernomor O. et al. (2020). IQ-TREE2: new models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution 37: 1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, et al. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32: 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nirenberg HI. (1976). Untersuchungen über die morphologische und biologische Differenzierung in der Fusarium-Sektion Liseola. Mitteilungen aus der Biologischen Bundesanstalt für Land- und Forstwirtschaft Berlin-Dahlem 169: 1–117. [Google Scholar]
- O’Donnell K, McCormick SP, Busman M, et al. (2018). Marasas et al. 1984 “Toxigenic Fusarium species: Identity and mycotoxicology” revisited. Mycologia 110: 1058–1080. [DOI] [PubMed] [Google Scholar]
- Rayner RW. (1970). A mycological colour chart. CMI and British Mycological Society, Kew, Surrey, UK. [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Rossman AY, Seifert KA. eds (2011). Phylogenetic revision of taxonomic concepts in the Hypocreales and other Ascomycota - A tribute to Gary J. Samuels. Studies in Mycology 68: 1–256.21523187 [Google Scholar]
- Rossman AY, Seifert KA, Samuels GJ. et al. (2013). Genera of Bionectriaceae, Hypocreaceae and Nectriaceae (Hypocreales) proposed for acceptance and rejection. IMA Fungus 4: 41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval-Denis M, Guarnaccia V, Polizzi G, et al. (2018). Symptomatic Citrus trees reveal a new pathogenic lineage in Fusarium and two new Neocosmospora species. Persoonia 40: 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval-Denis M, Lombard L, Crous PW. (2019). Back to the roots: a reappraisal of Neocosmospora. Persoonia 43: 90–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroers H-J, Gräfenhan T, Nirenberg HI, et al. (2011). A revision of Cyanonectria and Geejayessis gen. nov., and related species with fusarium-like anamorphs. Studies in Mycology 68: 115–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder WC, Hansen HN. (1940). The species concept in Fusarium. American Journal of Botany 27: 64–67. [Google Scholar]
- Tatsuno T, Saito M, Enomoto M. et al. (1968). Nivalenol, a toxic principle of Fusarium nivale. Chemical and Pharmaceutical Bulletin 16: 2519–2520. [DOI] [PubMed] [Google Scholar]
- Torbati M, Arzanlou M, Sandoval-Denis M, et al. (2019). Multigene phylogeny reveals new fungicolous species in the Fusarium tricinctum species complex and novel hosts in the genus Fusarium from Iran. Mycological Progress 18: 119–133. [Google Scholar]
- Wang MM, Chen Q, Diao YZ. et al. (2019). Fusarium incarnatum-equiseti complex from China. Persoonia 43: 70–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollenweber HW, Reinking OA. (1935). Die Fusarien. Verlagsbuchandlung Paul Parey, Berlin, Germany. [Google Scholar]
- Xia JW, Sandoval-Denis M, Crous PW, et al. (2019). Numbers to names – reappraisal of the Fusarium incarnatum-equiseti species complex. Persoonia 43: 186–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa T, Morooka N. (1973). Deoxynivalenol and its monoacetate: new mycotoxins from Fusarium roseum and moldy barley. Agricultural and Biological Chemistry 37: 2933–2934. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Collection details and GenBank accession numbers of strains used in the phylogenetic trees.