Abstract
The myogenic basic helix-loop-helix (bHLH) proteins regulate both skeletal muscle specification and differentiation: MyoD and Myf5 establish the muscle lineage, whereas myogenin mediates differentiation. Previously, we demonstrated that MyoD was more efficient than myogenin at initiating the expression of skeletal muscle genes, and in this study we present the molecular basis for this difference. A conserved amphipathic alpha-helix in the carboxy terminus of the myogenic bHLH proteins has distinct activities in MyoD and myogenin: the MyoD helix facilitates the initiation of endogenous gene expression, whereas the myogenin helix functions as a general transcriptional activation domain. Thus, the alternate use of a similar motif for gene initiation and activation provides a molecular basis for the distinction between specification and differentiation within the myogenic bHLH gene family.
Myogenesis is regulated by a family of four transcription factors (Myf5, MyoD, myogenin, and MRF4) that share a common dimerization and DNA binding domain (DBD), the basic helix-loop-helix (bHLH) motif. Genetic studies have demonstrated that MyoD and Myf5 act to establish the skeletal muscle lineage in mice, since disruption of both of these genes resulted in complete absence of skeletal muscle cells (16). In contrast, myogenin is required for the normal differentiation of the myoblasts established by the prior expression of Myf5 or MyoD (6, 12). In this regard, MyoD and Myf5 can be considered determination or specification factors and myogenin can be considered a differentiation factor. MRF4 has been difficult to study because of its proximity to Myf5, but null mutations of MRF4 result in increased expression of myogenin with relatively normal muscle cell differentiation (13, 25).
The difference between the specification of the muscle lineage by MyoD and Myf5 and the terminal differentiation mediated by myogenin might be due to differences in protein sequence or the temporal pattern of gene expression, or both. During embryogenesis, expression of either Myf5 or MyoD is the earliest marker of myoblast specification in the dorsal or ventral dermomyotome, respectively, and precedes expression of myogenin in any given cell (7). Similarly, during the regeneration of adult skeletal muscle, the activated satellite cells initially express Myf5 and MyoD and subsequently express myogenin and MRF4 (19, 24). It is possible that the requirement for MyoD or Myf5 in specifying the muscle lineage reflects gene regulatory sequences that appropriately initiate expression, whereas the proteins encoded by each of the myogenic bHLH genes might have similar functions. Indeed, there is considerable evidence that each of the myogenic bHLH proteins have largely redundant functions. For example, each can initiate myogenesis when artificially expressed in nonmuscle cells, such as fibroblasts (1, 2, 11, 23). Experiments in the developing mouse embryo, however, have shown that myogenin cannot efficiently promote myogenesis when substituted for Myf5 (21). This result suggested that the myogenic specification factors MyoD and Myf5 encode protein functions distinct from the differentiation protein myogenin.
Since one critical aspect of lineage specification is the initiation of tissue-restricted gene expression, we hypothesized that the lineage specification factors may possess a greater intrinsic ability to initiate the expression of silent genes than differentiation factors. Indeed, our previous work demonstrated that MyoD and Myf5 were more efficient than myogenin at initiating expression of endogenous muscle genes (5). In the present study, we extend this initial observation to identify the molecular attributes of MyoD and myogenin that confer the function of specification factor and differentiation factor, respectively. We discovered that a cysteine-rich region amino terminal to the bHLH domain and previously shown necessary for MyoD-mediated chromatin remodeling was functionally conserved in myogenin and was not sufficient to account for their different activities. The major difference between the activities of MyoD and myogenin was encoded in a carboxy-terminal amphipathic alpha-helix conserved during the evolution of the myogenic bHLH proteins. This alpha-helix appears to have evolved distinct functions in MyoD and myogenin, functioning as a specification domain in MyoD, i.e., a domain critical for the efficient initiation of skeletal muscle gene expression, and as a general transcription activation domain in myogenin.
MATERIALS AND METHODS
Plasmids.
Expression vectors for MyoD, Myf5, myogenin, and reporter plasmids were described by Gerber et al. (5). MyoD deletion mutants were generated using a PCR-based approach: primers flanking the desired deletion and primers external to the MyoD coding sequence were used to generate PCR products which encoded the desired deletion following ligation. Each mutant was sequence verified. MyoD helix III mutants were constructed with the use of a shuttle vector in which amino acids 245 to 258 were replaced with an NheI site. Oligonucleotides encoding helix III with the desired mutation(s) were ligated into the NheI-digested shuttle vector. MyoD cysteine-rich motif mutants were created using the Stratagene QuikChange site-directed mutagenesis protocol for single amino acid substitutions. Super myogenin constructs were created using a PCR-based approach as described above for the deletion mutants. Primers were designed which flanked the myogenin histidine-rich motif and the myogenin helix III; the 5′ ends of these primers encoded the amino acid substitutions necessary to create super myogenin. Galactosidase (Gal) fusion proteins were generated by PCR amplifying cDNA encoding MyoD amino acids 170 to 318 and myogenin amino acids 136 to 224 and ligating the PCR products into pSG424 (17). The 4X14DGal-Luc plasmid was a gift of Bob Eisenman (Fred Hutchinson Cancer Research Center, Seattle, Wash.).
Cell culture.
NIH 3T3 cells were obtained from the American Type Culture Collection. Cells were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% bovine calf serum (Hyclone), penicillin, and streptomycin. Differentiation medium was DMEM supplemented with 10 μg of insulin and transferrin per ml.
Transfections.
Transfections were performed using Superfect transfection reagent (Qiagen). Cells were seeded in 6-cm-diameter tissue culture dishes (Corning) at a density of 105 cells/ml on the day prior to transfection. For S1 nuclease protection assays, each plate of cells was transfected with 5 μg of myogenic factor expression vector, 2 μg of 4R-TK-Luc, and 2 μg of p1.7Desmin-CAT. Superfect-DNA complexes were washed from the cells with phosphate-buffered saline after 2 h, and the cells were allowed to grow in DMEM with 10% bovine calf serum for 12 h. Cells were then transferred into differentiation medium for 24 h. RNA was harvested using the RNeasy Mini protocol (Qiagen) and analyzed by S1 nuclease protection assay as described below. For luciferase assays, 2 μg of Gal fusion plasmid, 1 μg of 4X14DGal-Luc, and 1 μg of CS2-βGal were used to transfect 35-mm-diameter tissue culture plates (Corning) using the Superfect protocol. Cell lysates were analyzed for luciferase and beta-galactosidase activity using previously described methods.
S1 nuclease protection assay.
Probe fragments were generated by PCR amplification using a biotinylated T3 primer to prime the sense strand and a gene-specific primer to prime the antisense strand. PCR products were end labeled with 32P using T4 polynucleotide kinase (New England Biolabs). The labeled fragments were immobilized on streptavidin-coated magnetic beads (Dynal), and the radiolabeled antisense DNA probe was eluted by denaturation in weak base. Each probe included 200 to 400 nucleotides of sequence complementary to the gene of interest and 50 to 60 nucleotides of noncomplementary vector sequence. For S1 analysis, 50,000 counts of each probe and total RNA from a transfected 6-cm-diameter plate were ethanol precipitated, dried for 30 min at room temperature, and resuspended in 20 μl of 80% formamide, 400 mM NaCl, 40 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (pH 7.0), and 1 mM EDTA (pH 8.0). The resuspended RNA was denatured at 65°C for 10 min and hybridized for 12 h at 44°C. S1 digestion was performed by adding 200 μl of S1 digestion buffer consisting of 300 mM NaCl, 30 mM Na acetate (pH 5.5), 2 mM ZnSO4, 2 μg of single-stranded DNA, and 400 U of S1 nuclease (Roche) and incubating for 1 h at 37°C. Digested probe was ethanol precipitated, dried, resuspended in 8 μl of formamide loading dye, denatured at 100°C for 3 min, and resolved on a 6% denaturing acrylamide sequencing gel run at 45 W for 5 h.
RESULTS
Conserved motifs in MyoD are necessary to efficiently initiate expression of endogenous skeletal muscle genes.
In our previous studies, we demonstrated that the region between amino acids 63 and 99 was necessary for chromatin remodeling by MyoD, based on nuclease access studies. This region was also necessary for the efficient initiation of endogenous skeletal muscle gene expression, but not for the activation of transfected reporter plasmids driven by skeletal muscle promoters or multimerized MyoD binding sites. A carboxy-terminal region encoded by amino acids 218 to 269 was also shown to be required for efficient endogenous skeletal muscle gene activation (5). Sequence alignment (Fig. 1A) shows that these two regions contain motifs conserved between MyoD and Myf5 and partially conserved with myogenin and MRF4. We therefore sought to determine whether sequence divergence of these conserved motifs could account for the different efficiencies with which MyoD and myogenin initiate the expression of endogenous skeletal muscle genes.
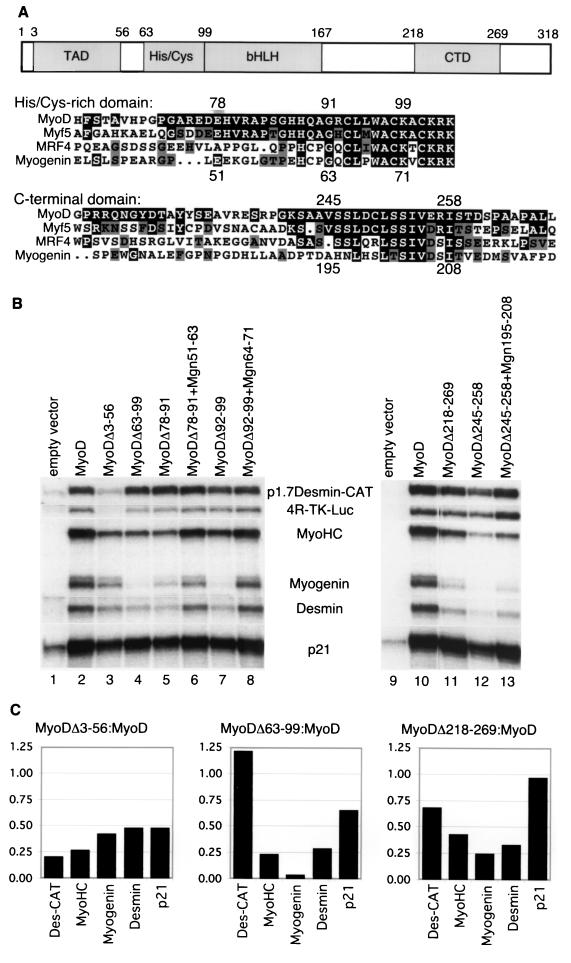
FIG. 1.
The functional differences between MyoD and myogenin map to a C-terminal domain. (A) Schematic diagram of MyoD domains and partial sequence alignment of the murine myogenic bHLH proteins, corresponding to MyoD amino acids 63 to 104 and 218 to 269. MyoD amino acid numbers are indicated along the top, and myogenin amino acid numbers are along the bottom of each alignment. (B) S1 nuclease protection of RNA from NIH 3T3 cells transiently transfected with expression vectors for MyoD or MyoD mutants, as indicated. Protected messages are identified between the two gel images. When normalized to p1.7Des-Cat, MyoDΔ63-99 (lane 4) was 3% as efficient as MyoD at initiating expression of the endogenous myogenin gene, 24% as efficient on the endogenous desmin gene, and 20% as efficient on the endogenous myosin heavy chain gene; it was nearly equal to MyoD at increasing expression from the preinitiated p21 gene (81% relative to MyoD). Each mutant was analyzed multiple times, and the figure represents a typical experiment. (C) Ratio of the signal in select mutants relative to wild-type MyoD. Deletion of the activation domain (MyoDΔ3-56) results in a relatively equal decrease in the expression level of all of the target genes. In comparison, the MyoDΔ63-99 and MyoDΔ218-269 deletions show relatively preserved activity on the p1.7Des-CAT reporter and the endogenous p21 gene compared to their activity on the endogenous MyoHC, myogenin, and desmin genes.
In the current study, we used a quantitative S1 nuclease protection assay to measure the ability of wild-type or mutant myogenic bHLH proteins to activate a set of endogenous target genes. To control for variance in transfection efficiency, protein expression, and possible effects that the mutations might have on general transcriptional activity, we compared the level of activation of the endogenous genes to the level of activation of cotransfected MyoD responsive reporter constructs. Mammalian expression vectors for wild-type and mutant MyoD proteins were transiently transfected into murine NIH 3T3 fibroblasts along with a chloramphenicol acetyltransferase (CAT) reporter gene driven by 1.7 kb of the desmin promoter region (p1.7Des-CAT) and/or a luciferase reporter driven by multimerized MyoD binding sites and a minimal thymidine kinase promoter (p4RTK-LUC). Cells were cultured for 24 h in differentiation medium, and total RNA was used for S1 nuclease assays with probes for myosin heavy chain, myogenin, desmin, p21WAF1/CIP1, luciferase, and CAT.
In the absence of transfected MyoD (Fig. 1B, lane 1), there was a basal amount of p21WAF1/CIP1 mRNA but very low or undetectable amounts of mRNA for the endogenous muscle genes or the transfected reporter genes. Transfection of MyoD (Fig. 1B, lane 2) increased the abundance of p21 mRNA and initiated expression of the endogenous muscle genes (desmin, myogenin, and myosin heavy chain) and the MyoD responsive reporters. Deletion of the MyoD acidic activation domain (MyoDΔ3-56; Fig. 1B, lane 3, and C) (22) reduced the activity on all target genes. In contrast to deletion of the general activation domain and consistent with our prior study, the deletion mutants MyoDΔ63-99 and MyoDΔ218-269 (Fig. 1B, lanes 4 and 11, and C) showed relatively preserved activity on the transfected reporter genes and substantially reduced activity on myosin heavy chain, desmin, and myogenin. Smaller deletions limited to the conserved motifs (a histidine-rich region, MyoDΔ78-91 [lane 5]; a cysteine-rich region, MyoDΔ92-99 [lane 7]; and a carboxy-terminal region, MyoDΔ245-258 [lane 12]) were similar to the larger deletions, indicating that all three conserved motifs were necessary to initiate expression of the skeletal muscle genes. In additional experiments (data not shown), transfection of MyoD with an inactivating mutation of the DNA binding region failed to upregulate p21 or other MyoD target genes, indicating that the regulation of these target genes by MyoD was dependent on DNA binding.
Functional distinction between MyoD and myogenin in a carboxy-terminal domain.
Because the comparable regions of myogenin show partial sequence similarity to the MyoD motifs essential for endogenous gene initiation, we tested the functional activity of the myogenin motifs by substituting them for the MyoD motifs in a chimeric MyoD protein. The region of myogenin comparable to the MyoD histidine-rich region (myogenin amino acids 51 to 63) could partially substitute for the function of this MyoD motif (MyoDΔ78-91+Mgn51-63; Fig. 1B, lane 6), suggesting that the sequence divergence between MyoD and myogenin in this motif was not the primary determinant of the different activities of the two proteins. The myogenin region corresponding to the MyoD cysteine-rich region (myogenin amino acids 64 to 71) differs from MyoD at only two residues and was also functionally similar to the MyoD motif (MyoDΔ92-99+Mgn64-71; Fig. 1B, lane 8). In contrast, substituting the comparable myogenin sequence for the MyoD carboxy-terminal motif (MyoDΔ245-258+Mgn195-208; Fig. 1B, lane 13) had a gene activation profile similar to the deletion of the MyoD sequence. Therefore, this region of myogenin could not functionally substitute for the conserved carboxy-terminal motif in MyoD, and the sequence divergence between MyoD and myogenin in this region could account for their different activities and developmental roles.
Conservation of function of MyoD C-terminal motif.
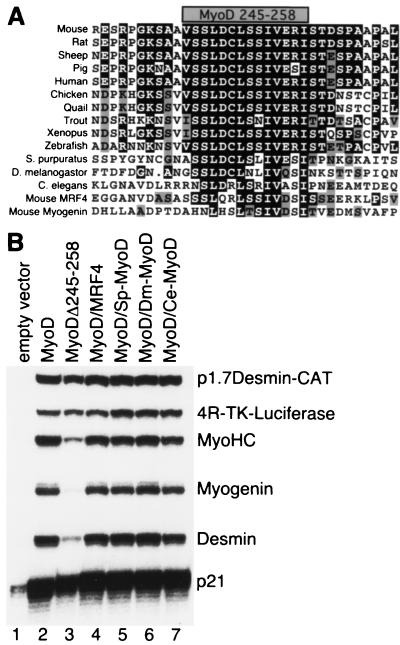
Based on the above results, we hypothesized that the MyoD amino acid 245 to 258 domain encodes an important function that distinguishes MyoD as a specification factor. Therefore, we sought to determine whether the function of this domaïn was conserved in MyoD from diverse species. Sequence alignment demonstrated that the C-terminal motif of MyoD was highly conserved in a variety of vertebrate species, and it was partially conserved in invertebrate MyoD (Fig. 2A). This motif is part of a region that others have named region III (15), domain II (3), or domain III (10). To test for functional conservation, we replaced the murine MyoD C-terminal motif with the comparable regions from three invertebrate MyoD proteins, as well as with the comparable C-terminal motif from murine MRF4. In contrast to the murine myogenin sequence that did not functionally replace the murine MyoD sequence in our assays (Fig. 1A, lane 13; see also Fig. 4, lane 4), the C-terminal motifs from murine MRF4, Strongylocentrotus purpuratus (sea urchin) MyoD, Drosophila melanogaster MyoD and Caenorhabditis elegans MyoD were all capable of functionally substituting for the murine MyoD sequence in this assay (Fig. 2B, lanes 4 through 7) and were more efficient than the murine myogenin sequence. As in previous assays, all of the constructs increased expression of p21WAF1/CIP1 message.
FIG. 2.
Conservation of the function of the MyoD C-terminal motif. (A) Multiple sequence alignment of MyoD proteins from several vertebrate and invertebrate species and from murine MRF4 and myogenin in the region corresponding to the murine MyoD C-terminal domain. The shaded box at the top of the alignment represents the critical motif encoded by amino acids 245 to 258. (B) S1 protection of RNA following transfection of NIH 3T3 cells with MyoD wild-type (MyoD) or chimeric proteins as indicated, showing functional conservation of the MyoD C-terminal domain. For substitution mutants, the sequence aligning with murine MyoD 245 to 258 in panel A was substituted for the murine MyoD sequence to generate chimeric proteins, as follows: MyoD/MRF4, murine MRF4 sequence; MyoD/Sp-MyoD, S. purpuratus MyoD sequence; MyoD/Dm-MyoD, D. melanogaster MyoD sequence; and MyoD/Ce-MyoD, C. elegans MyoD sequence. Chimeric proteins were generated by ligating oligonucleotides encoding the desired C-terminal domain into a murine MyoD shuttle vector, from which the wild-type C-terminal domain was removed and replaced with an NheI site.
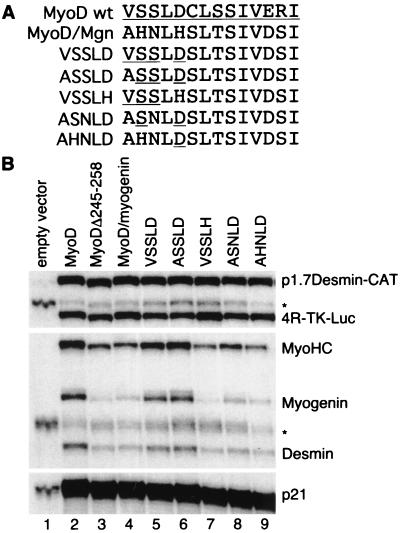
FIG. 4.
Residues on the hydrophilic face of helix III differentiate MyoD and myogenin function. (A) The MyoDΔ245-258–myogenin 195-208 chimera was subjected to back mutagenesis, represented in this schematic. MyoD residues are underlined. Back mutants are identified by the sequence of the first five amino acids of helix III, i.e., VSSLD represents substitution of MyoD amino acids 245 to 249 into the chimeric protein. (B) S1 nuclease protection analysis of back mutations in the chimeric MyoDΔ245-258–myogenin 195-208 protein. ∗, partially digested probe.
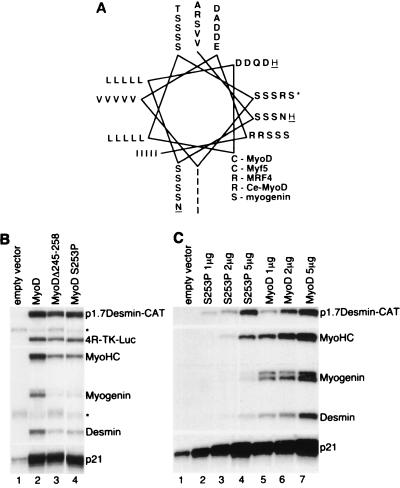
Carboxy-terminal motif of myogenic bHLH proteins can adopt an amphipathic alpha-helical structure.
Since the function of the MyoD carboxy-terminal domain is conserved in invertebrate species but not murine myogenin, we performed experiments to determine whether structural features of this domain could account for its functional characteristics. The C terminus of MyoD was not included in the solved crystal structure (8); however, several secondary structure prediction algorithms suggested the potential for this region to form an amphipathic alpha-helix in all of the myogenic bHLH proteins. A helical wheel model (Fig. 3A) revealed a highly conserved hydrophobic face in all of the myogenic bHLH proteins and a hydrophilic face with greater variability. To determine whether the potential to form an alpha-helix was necessary for function, we introduced a helix-disrupting proline residue at MyoD position 253. This residue was chosen as a central residue on the hydrophilic surface that was demonstrated to functionally tolerate multiple different substitutions, including alanine, glutamate, asparagine, leucine, and arginine (Fig. 2B, lanes 5 to 7; see also Fig. 5, lanes 6 and 7). In contrast to these other substitutions, the MyoD-S253P mutant showed activity identical to the deletion mutant MyoDΔ245-258 (Fig. 3B, lane 4), consistent with the conclusion that an alpha-helical structure is critical for the function of this region. Therefore, all of the MyoD family members (MyoD, MRF4, Myf5, and myogenin) have a carboxy-terminal region capable of forming an amphipathic alpha-helix and, at least for MyoD, a helix-destabilizing mutation is sufficient to disrupt function of this region. Since the myogenic bHLH proteins have two amphipathic alpha-helices forming the HLH region, we will refer to this region as helix III.
FIG. 3.
The function of the carboxy-terminal domain of the myogenic bHLH proteins requires the ability to adopt an amphipathic alpha-helical conformation. (A) Helical wheel representation of the C-terminal motif aligning murine MyoD, Myf5, MRF4, C. elegans MyoD, and murine myogenin, demonstrating the high degree of conservation of the hydrophobic face of the helix and the more variable hydrophilic face. (B) S1 nuclease protection of RNA from NIH 3T3 fibroblasts transfected with wild-type MyoD (MyoD), MyoDΔ245-58, and MyoD-S253P as indicated. Serine-253 (indicated by ∗ in panel A) falls in the middle of the predicted helix on the hydrophilic face. (C) Titration of the MyoD-S253P mutant compared to wild-type MyoD. The indicated amounts of expression plasmid were transfected. Empty expression vector was used to adjust the total quantity of transfected plasmid.
FIG. 5.
Role of putative phosphorylation sites in helix III regulation. Serine residues at MyoD positions 246 and 253 were replaced with alanine or glutamate residues. The ability of the mutants to initiate expression of endogenous skeletal muscle genes was assayed using an S1 nuclease protection assay. ∗, partially digested probe.
These data indicate that the MyoD helix III confers a specific activity necessary to initiate expression of a set of endogenous genes. To determine whether mutation of helix III resulted in a qualitative change in MyoD activity, as opposed to a quantitative decrease in transcription factor potency that revealed different relative sensitivities of the target genes, we transfected cells with increasing amounts of either wild-type MyoD or MyoD-S253P (Fig. 3C, lanes 1 to 7). With increasing concentrations of expression plasmid, MyoD-S253P preferentially activated the transfected reporter and only marginally activated the silent endogenous skeletal muscle genes. In contrast, wild-type MyoD preferentially activated the endogenous skeletal muscle genes. Therefore, the MyoD helix III was required for the efficient initiation of endogenous skeletal muscle gene expression, whereas MyoD helix III was not necessary for the upregulation of a MyoD target gene that was already being transcribed (p21) or for a transiently transfected reporter driven by a skeletal muscle promoter (p1.7Desmin-CAT). Initiation of tissue-specific gene expression is a critical feature of lineage specification, and divergence of the helix III sequence between MyoD and myogenin could therefore be the major distinction between these two proteins, with regard to the ability to specify the skeletal muscle lineage.
Residues on hydrophilic surface of helix III account for different activities of MyoD and myogenin C-terminal motifs.
Since the hydrophobic face of helix III is maintained in all myogenic bHLH proteins, we investigated whether variability on the hydrophilic face might account for the different activities of MyoD and myogenin. To identify the amino acids necessary for the function of the wild-type MyoD helix III motif, we initiated back mutations in the MyoDΔ245-258–myogenin 195-208 chimera (Fig. 4). Back-mutating the first five amino acids of the myogenin C-terminal domain sequence (AHNLH, myogenin amino acids 195 to 199) to the comparable MyoD sequence (VSSLD, amino acids 245 to 249) resulted in nearly full recovery of function (Fig. 4, lane 5), indicating that the other differences between MyoD and myogenin in this region were not functionally significant in the context of our assay. Similar to the VSSLD back mutation, the ASSLD chimera was also comparable to wild-type MyoD (Fig. 4, lane 6), whereas the VSSLH chimera had activity similar to the myogenin sequence or the deletion of MyoD 245-258 (Fig. 4, lane 7), indicating that the positively charged histidine did not functionally substitute for the negatively charged aspartate. Since the aspartate could be replaced by a nonpolar, uncharged alanine residue without loss of function in the MyoD-D249A mutant (data not shown), it appeared that the introduction of the positively charged histidine at this position disrupted the function of the MyoD helix III. In addition to the histidine at position 5 in the myogenin sequence, the histidine and asparagine at positions two and three also decreased the function of the MyoD chimera (Fig. 4, lanes 8 and 9).
Helix III serine mutations do not provide evidence of regulatory phosphorylation site.
Since the charge of the amino acid at the fifth position is a major determinant of the differential functions of the MyoD and myogenin helix III domains, we sought to identify whether potential regulatory modifications such as phosphorylation could modulate helix III activity by altering the charge of other helix III residues. The mouse MyoD helix III sequence contains potential casein kinase II phosphorylation sites at S246 and S253. MyoD-S246A and MyoD-S246E mutants demonstrated equivalent activities in our S1 assays, both of which were slightly less efficient than the MyoD (Fig. 5, lanes 2, 4, and 5). MyoD-S253A and MyoD-S253E also demonstrated equivalent activities, although in this case both had activities slightly greater than wild-type MyoD (Fig. 5, lanes 2, 6, and 7). In addition to S246 and S253, the serine at MyoD position 247 is highly conserved in MyoD from several species (Fig. 2A), but it is not part of a known phosphorylation motif. To assess the requirement for a serine at this site, we tested MyoD-S247A and MyoD-S247E, and both mutants had activities similar to MyoD (data not shown). Although the serine mutations we have studied do not prove that these serines are not phosphorylation sites in some biological contexts, the lack of significant alteration of activity with the different mutations and the lack of evolutionary conservation of S246 and S253 (Fig. 2A) together suggest that phosphorylation of these sites does not constitute a necessary regulatory function.
MyoD helix III increases ability of myogenin to efficiently initiate transcription of endogenous genes.
The above experiments demonstrated that the MyoD helix III was necessary for the efficient initiation of endogenous gene transcription, whereas the myogenin helix III sequence was less efficient in the context of the MyoD protein, despite conservation of the hydrophobic surface of a potential amphipathic alpha-helix. If a major difference in the activities of MyoD and myogenin was due to the residues on the hydrophilic surface of helix III, then substituting these residues into myogenin should increase the ability of myogenin to initiate transcription of endogenous target genes. Therefore, we generated a series of chimeric myogenin proteins that introduced the MyoD helix III region, alone or together with the MyoD histidine-rich region, in the context of the myogenin protein (shown schematically in Fig. 6B).
FIG. 6.
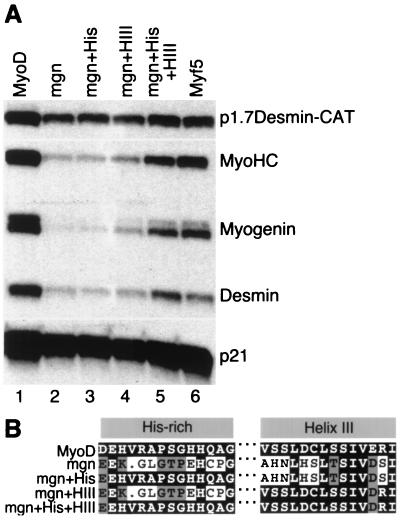
MyoD helix III (HIII) increases the ability of myogenin to efficiently initiate expression of skeletal muscle genes. (A) MyoD helix III increases the ability of myogenin to efficiently initiate expression of skeletal muscle genes. S1 nuclease assay of RNA from NIH 3T3 cells transfected with wild-type MyoD (lane 1), wild-type myogenin (lane 2), myogenin-MyoD chimeras (lanes 3 to 6), and Myf5 (lane 7). (B) Alignment of MyoD, myogenin, and the chimeric proteins: mgn+His was constructed by substituting 10 amino acids from MyoD into the corresponding myogenin region; mgn+HIII required 7 amino acid substitutions.
Wild-type myogenin (Fig. 6A, lane 2) activated the reporter construct and increased expression of the p21 gene, but its activity was substantially reduced compared to that of MyoD on the endogenous skeletal muscle genes. As anticipated, the myogenin chimera that contained only the histidine-rich motif from MyoD (Fig. 6A, lane 3) was essentially the same as wild-type myogenin. Introduction of the MyoD helix III into myogenin resulted in an increase in the ability of the myogenin chimera to initiate expression of the endogenous skeletal muscle genes (Fig. 6A, lane 4), and when both the MyoD histidine-rich motif and the MyoD helix III were introduced into myogenin (Fig. 6A, lane 5), the activity of the chimera was nearly equivalent to that of Myf5 and MyoD (compare lanes 1, 5, and 6). Together, these experiments demonstrated that the MyoD helix III is an important component of the ability of the myogenic bHLH proteins to initiate expression of skeletal muscle genes and that divergent sequence of the helix III in myogenin primarily accounted for its relative inefficiency at activating endogenous genes. Furthermore, the histidine-rich motif can enhance the activity of the MyoD helix III in the context of the myogenin protein.
Distinct functions of myogenin and MyoD helix III motifs.
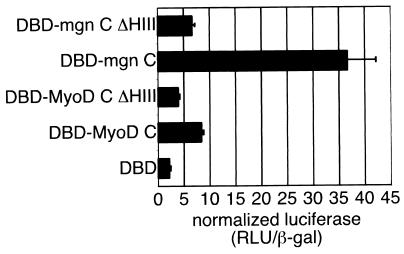
These data indicate that the myogenin helix III is not competent to initiate the expression of endogenous skeletal muscle genes, but two lines of evidence suggest that the myogenin helix III might assume a role in the myogenin protein distinct from that observed for MyoD helix III. First, although the myogenin helix III sequence diverges from that of MyoD, it is conserved in myogenin from several species. Second, the hydrophobic face of helix III is conserved in myogenin, suggesting an evolutionary pressure to maintain this structural motif. Previous studies have identified a carboxy-terminal activation domain in the myogenin protein (18), while a similar activity has not been described for the carboxy terminus of MyoD (22). In order to assess the activation function of the carboxy-terminal domains of myogenin and MyoD, we constructed fusion proteins between the MyoD or myogenin carboxy terminus and the DBD of Gal4 (Gal4 1 to 147) and assayed the ability of the chimeras to promote transcription of a luciferase reporter driven by multimerized Gal4 binding sites (Fig. 7). The MyoD C terminus had very modest activation domain activity, driving expression of the reporter approximately threefold higher than that for the Gal4 DBD alone. In contrast, the myogenin C terminus promoted transcription to a level nearly 15-fold higher than that for Gal4 DBD, and deletion of the myogenin helix III reduced transcription of the reporter to a level similar to that seen with the MyoD C terminus. These results demonstrate that myogenin has a C-terminal activation domain and that myogenin helix III is essential for this activity. Although the myogenin helix III was necessary for the full activation function of the myogenin C terminus, a chimera that introduced the myogenin helix III into the MyoD C terminus did not have increased activation function compared to the wild-type MyoD C terminus (data not shown), indicating that the myogenin helix III is not sufficient in the context of MyoD to function as an activation domain but rather needs additional elements in the myogenin C terminus.
FIG. 7.
Myogenin possess a helix III-dependent C-terminal activation domain. The C termini of MyoD and myogenin were assayed for the ability to function as a general activation domain when fused to the Gal4 DBD (amino acids 1 to 147). Gal fusion proteins were generated by PCR amplifying cDNA encoding MyoD amino acids 170 to 318 (DBD-MyoD C) and myogenin amino acids 136 to 224 (DBD-mgn C) and ligating the PCR products into the pSG424 vector. Similar fusion proteins with deletions of the helix III motif in MyoD (amino acids 245 to 258; DBD-MyoD C ΔHIII) and myogenin (amino acids 195 to 208; DBD-mgn C ΔHIII) were also made. The Gal4-C terminus chimeras were cotransfected into NIH 3T3 cells with a Gal4-responsive luciferase reporter plasmid and a constitutively expressed β-galactosidase reporter to normalize for transfection efficiency. Each transfection was repeated three times. The graph represents the mean normalized luciferase activity (relative light units [RLU]/β-galactosidase activity) for the three experiments. Error bars represent standard deviations.
DISCUSSION
Together, these experiments demonstrate that MyoD and myogenin share a structurally conserved carboxy-terminal alpha-helical motif that performs a distinct function in each protein. In the specification gene, MyoD, helix III is necessary for the efficient initiation of the expression of at least a subset of endogenous skeletal muscle genes, but it does not have significant function as a classical activation domain. In the differentiation gene, myogenin, helix III has activity as a general transcriptional activation domain but cannot facilitate the initiation of skeletal muscle gene expression. This structural motif appears to be a major distinguishing feature between the activities of MyoD and myogenin, since substitution of this motif into myogenin converts it into a protein with activity similar to MyoD and Myf5. The fact that an additional substitution of the MyoD histidine-rich region is necessary for the full activity of the chimeric transcription factor suggests that the helix III activity might be further facilitated or regulated by interactions that are dependent on this region. The mechanism by which the MyoD helix III increases the efficiency of endogenous gene initiation remains unknown. This region might be necessary for the activation of a subset of skeletal muscle promoters, perhaps overcoming promoter-specific negative regulators. Since the transiently transfected desmin promoter does not require these domains for activation, yet these domains are required for the full activation of the endogenous desmin gene, it seems unlikely that simple cis regulatory sequences are sufficient to account for the dependence of the endogenous skeletal muscle genes on these domains. The histidine- and cysteine-rich region is necessary for MyoD-mediated chromatin remodeling (5), and it is possible that helix III also contributes to chromatin remodeling at skeletal muscle gene loci.
A motif similar to helix III exists in the Saccharomyces cerevisiae Pho4 protein. The bHLH transcription factor Pho4 initiates the expression of the Pho5 gene and disrupts a nucleosome array at the Pho5 promoter (20). An amphipathic alpha-helix in the Pho4 protein (amino acids 75 to 99 of Pho4) has been identified as essential for Pho4-mediated chromatin remodeling and Pho5 initiation (9). Inserting the Pho4 amphipathic alpha-helix into MyoD, however, does not functionally substitute for MyoD helix III (D. A. Bergstrom, unpublished data). Currently, it is not known whether factors interact with this domain of Pho4 to mediate chromatin remodeling, but it has been shown that Pho4 chromatin remodeling is independent of SWI/SNF and that RNA polymerase II holoenzyme recruitment is sufficient for chromatin remodeling by a Pho4-Gal11 fusion protein (4). In contrast to the MyoD helix III, which was not necessary as an activation domain, the Pho4 domain was necessary for chromatin remodeling and functioned as an activation domain in fusions to a heterologous DBD (9). It is interesting to speculate that, prior to gene duplication and divergence, the original helix III might have subserved both functions and that evolution has separated the two related functions, gene initiation and gene activation. In this regard, it will be interesting to see if the helix III of invertebrates with a single myogenic bHLH gene, e.g., C. elegans or D. melanogaster, combines the functions of initiation and activation.
The observation that helix III of MRF4 can functionally substitute for helix III of MyoD is consistent with the genetic evidence that MRF4 has a partly overlapping role with MyoD in regulating myogenesis (14). MRF4 is expressed transiently in the murine myotome between embryonic day 9.0 (E9.0) and E11.5, immediately following the onset of Myf5 expression and preceding the expression of MyoD. MRF4 expression initiates again around E16 in the differentiating muscle fibers. The proximity of Myf5 and MRF4 has complicated the interpretation of MRF4 disruptions, since most targeting strategies have also altered the level of Myf5 expression (13, 25). The MRF4 homozygous deletion with the least effect on Myf5 expression results in some alterations of skeletal muscle gene expression, most notably increased levels of myogenin mRNA, but skeletal muscle development is grossly normal. In contrast, the double mutation of MRF4 and MyoD resulted in a severe skeletal muscle deficiency, despite normal expression of myogenin, indicating that either MyoD or MRF4 was necessary for myogenin to mediate normal skeletal muscle cell differentiation (14). Given our current observation that the helix III of both MyoD and MRF4, but not myogenin, is capable of facilitating efficient gene initiation, it is possible that the absence of MyoD-MRF4 helix III function results in the decreased skeletal muscle formation in the double mutant background. Ultimately, this could be tested by introducing the MyoD helix III into myogenin in the MyoD-MRF4 mutant background.
In summary, a major difference in the activities of MyoD and myogenin is encoded by a few residues on the hydrophilic face of a carboxy-terminal amphipathic alpha-helix, helix III. The MyoD helix III is necessary for the efficient initiation of endogenous gene expression at the target loci analyzed. In this regard, it is a domain that imparts the activity of a specification factor to MyoD, although the mechanism of action remains unknown. The myogenin helix III is necessary for the function of its carboxy-terminal activation domain, but it lacks the specification activity of the MyoD helix III. It should be noted that MyoD has an amino-terminal activation domain, but there is evidence that the activity of this domain can be regulated (22). An intuitive argument can be made for the separate regulation of the functions of initiation and full activation of transcription in complex multicellular organisms. One plausible model is that MyoD targets a broad array of genes for initiation, fully activating only a subset of its targets. Myogenin subsequently acts at those genes initiated by MyoD to enhance and maintain transcription, permitting the full level of gene expression needed for terminal differentiation. The separation of these functions might contribute to the spatial and temporal orchestration of skeletal muscle gene expression during commitment and differentiation, and it provides a paradigm for understanding the differential regulation of gene expression during development by individual members of highly related transcription factor families.
ACKNOWLEDGMENTS
We thank M. Groudine and S. Parkhurst for helpful comments on the manuscript.
This work was supported by NIH NIAMSD grant no. AR45113 (S.J.T.) and a Poncin Fellowship (D.A.B.).
REFERENCES
- 1.Braun T, Buschhausen-Denker G, Bober E, Tannich E, Arnold H H. A novel human muscle factor related to but distinct from MyoD1 induces myogenic conversion in 10T1/2 fibroblasts. EMBO J. 1989;8:701–709. doi: 10.1002/j.1460-2075.1989.tb03429.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Edmondson D G, Olson E N. A gene with homology to the myc similarity region of MyoD1 is expressed during myogenesis and is sufficient to activate the muscle differentiation program. Genes Dev. 1989;3:628–640. doi: 10.1101/gad.3.5.628. . (Erratum, 4:1450, 1990.) [DOI] [PubMed] [Google Scholar]
- 3.Fujisawa-Sehara A, Nabeshima Y, Hosoda Y, Obinata T. Myogenin contains two domains conserved among myogenic factors. J Biol Chem. 1990;265:15219–15223. [PubMed] [Google Scholar]
- 4.Gaudreau L, Schmid A, Blaschke D, Ptashne M, Horz W. RNA polymerase II holoenzyme recruitment is sufficient to remodel chromatin at the yeast PHO5 promoter. Cell. 1997;89:55–62. doi: 10.1016/s0092-8674(00)80182-8. [DOI] [PubMed] [Google Scholar]
- 5.Gerber A N, Klesert T R, Bergstrom D A, Tapscott S J. Two domains of MyoD mediate transcriptional activation of genes in repressive chromatin: a mechanism for lineage determination in myogenesis. Genes Dev. 1997;11:436–450. doi: 10.1101/gad.11.4.436. [DOI] [PubMed] [Google Scholar]
- 6.Hasty P, Bradley A, Morris J H, Edmondson D G, Venuti J M, Olson E N, Klein W H. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature. 1993;364:501–506. doi: 10.1038/364501a0. [DOI] [PubMed] [Google Scholar]
- 7.Kablar B, Asakura A, Krastel K, Ying C, May L L, Goldhamer D J, Rudnicki M A. MyoD and Myf-5 define the specification of musculature of distinct embryonic origin. Biochem Cell Biol. 1998;76:1079–1091. [PubMed] [Google Scholar]
- 8.Ma P C, Rould M A, Weintraub H, Pabo C O. Crystal structure of MyoD bHLH domain-DNA complex: perspectives on DNA recognition and implications for transcriptional activation. Cell. 1994;77:451–459. doi: 10.1016/0092-8674(94)90159-7. [DOI] [PubMed] [Google Scholar]
- 9.McAndrew P C, Svaren J, Martin S R, Horz W, Goding C R. Requirements for chromatin modulation and transcription activation by the Pho4 acidic activation domain. Mol Cell Biol. 1998;18:5818–5827. doi: 10.1128/mcb.18.10.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meedel T H, Farmer S C, Lee J J. The single MyoD family gene of Ciona intestinalis encodes two differentially expressed proteins: implications for the evolution of chordate muscle gene regulation. Development. 1997;124:1711–1721. doi: 10.1242/dev.124.9.1711. [DOI] [PubMed] [Google Scholar]
- 11.Miner J H, Wold B. Herculin, a fourth member of the MyoD family of myogenic regulatory genes. Proc Natl Acad Sci USA. 1990;87:1089–1093. doi: 10.1073/pnas.87.3.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nabeshima Y, Hanaoka K, Hayasaka M, Esumi E, Li S, Nonaka I. Myogenin gene disruption results in perinatal lethality because of severe muscle defect. Nature. 1993;364:532–535. doi: 10.1038/364532a0. [DOI] [PubMed] [Google Scholar]
- 13.Olson E N, Arnold H H, Rigby P W, Wold B J. Know your neighbors: three phenotypes in null mutants of the myogenic bHLH gene MRF4. Cell. 1996;85:1–4. doi: 10.1016/s0092-8674(00)81073-9. [DOI] [PubMed] [Google Scholar]
- 14.Rawls A, Valdez M R, Zhang W, Richardson J, Klein W H, Olson E N. Overlapping functions of the myogenic bHLH genes MRF4 and MyoD revealed in double mutant mice. Development. 1998;125:2349–2358. doi: 10.1242/dev.125.13.2349. [DOI] [PubMed] [Google Scholar]
- 15.Rhodes S J, Konieczny S F. Identification of MRF4: a new member of the muscle regulatory factor gene family. Genes Dev. 1989;3:2050–2061. doi: 10.1101/gad.3.12b.2050. [DOI] [PubMed] [Google Scholar]
- 16.Rudnicki M A, Schnegelsberg P N, Stead R H, Braun T, Arnold H H, Jaenisch R. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell. 1993;75:1351–1359. doi: 10.1016/0092-8674(93)90621-v. [DOI] [PubMed] [Google Scholar]
- 17.Sadowski I, Ptashne M. A vector for expressing GAL4(1-147) fusions in mammalian cells. Nucleic Acids Res. 1989;17:7539. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwarz J J, Chakraborty T, Martin J, Zhou J M, Olson E N. The basic region of myogenin cooperates with two transcription activation domains to induce muscle-specific transcription. Mol Cell Biol. 1992;12:266–275. doi: 10.1128/mcb.12.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith C K, II, Janney M J, Allen R E. Temporal expression of myogenic regulatory genes during activation, proliferation, and differentiation of rat skeletal muscle satellite cells. J Cell Physiol. 1994;159:379–385. doi: 10.1002/jcp.1041590222. [DOI] [PubMed] [Google Scholar]
- 20.Svaren J, Horz W. Transcription factors vs nucleosomes: regulation of the PHO5 promoter in yeast. Trends Biochem Sci. 1997;22:93–97. doi: 10.1016/s0968-0004(97)01001-3. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, Jaenisch R. Myogenin can substitute for Myf5 in promoting myogenesis but less efficiently. Development. 1997;124:2507–2513. doi: 10.1242/dev.124.13.2507. [DOI] [PubMed] [Google Scholar]
- 22.Weintraub H, Dwarki V J, Verma I, Davis R, Hollenberg S, Snider L, Lassar A, Tapscott S J. Muscle-specific transcriptional activation by MyoD. Genes Dev. 1991;5:1377–1386. doi: 10.1101/gad.5.8.1377. [DOI] [PubMed] [Google Scholar]
- 23.Weintraub H, Tapscott S J, Davis R L, Thayer M J, Adam M A, Lassar A B, Miller A D. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc Natl Acad Sci USA. 1989;86:5434–5438. doi: 10.1073/pnas.86.14.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yablonka-Reuveni Z, Rivera A J. Temporal expression of regulatory and structural muscle proteins during myogenesis of satellite cells on isolated adult rat fibers. Dev Biol. 1994;164:588–603. doi: 10.1006/dbio.1994.1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoon J K, Olson E N, Arnold H H, Wold B J. Different MRF4 knockout alleles differentially disrupt Myf-5 expression: cis-regulatory interactions at the MRF4/Myf-5 locus. Dev Biol. 1997;188:349–362. doi: 10.1006/dbio.1997.8670. [DOI] [PubMed] [Google Scholar]