Abstract
This study provides a timely comparative genomic and transcriptomic analysis of the terpene synthase (TPS) gene family in Medicago truncatula (bears glandular and non‐glandular trichomes) and Arabidopsis thaliana (bears non‐glandular trichomes). The authors’ efforts aimed to gain insight into TPS function, phylogenetic relationships and the role of trichomes in terpene biosynthesis and function. In silico analysis identified 33 and 23 putative full‐length TPS genes in Arabidopsis and Medicago, respectively. All AtTPS and MtTPS fall into the five established angiosperm TPS subfamilies, with lineage‐specific expansion of Subfamily A in Arabidopsis and Subfamily G in Medicago. Large amounts of tandem duplication have occurred in both species, but only one syntenic duplication seems to have occurred in Arabidopsis, with no such duplication apparent in Medicago. Expression analysis indicates that there is much more trichome‐localised TPS expression in Medicago than in Arabidopsis. However, TPS genes were expressed in non‐glandular trichomes in both species. One trichome‐specific gene has been identified in each Medicago and Arabidopsis along with flower‐, seed‐, stem‐ and root‐specific genes. Of these, MtTPS11 is a promising candidate for trichome‐specific genetic engineering, a technology that may be possible for both plants according to the findings of this manuscript. These results suggest that non‐glandular trichomes may play a role in plant chemical defense and/or ecological communication instead of only in physical defence. Finally, the general lack of correlation between expression patterns and phylogenetic relationships in both species suggests that phylogenetic analysis alone is insufficient to predict gene function even for phylogenetically close paralogs.
Inspec keywords: genomics, genetic engineering, botany, enzymes, evolution (biological), genetics
Other keywords: genomic analysis, transcriptomic analysis, terpene synthase, Medicago truncatula, Arabidopsis thaliana, nonglandular trichomes, phylogenetic relationships, terpene biosynthesis, terpene function, tandem duplication, syntenic duplication, trichome‐localised TPS expression, flower‐speciflc genes, seed‐speciflc genes, root‐speciflc genes, MtTPS11, trichome‐speciflc genetic engineering, plant chemical defense, ecological communication, stem‐speciflc genes, expression analysis
1 Introduction
Terpene synthases (TPSs) synthesise the largest, most diverse class of plant secondary metabolites, terpenes, which function botanically in ecological communication or defence, and are used extensively in the manufacture of medicines, nutritional supplements, pesticides, perfumes and essential oils [1, 2]. Nearly, all monoterpenes and sesquiterpenes, as well as some diterpenes, are released as volatiles at ambient temperature and pressure; TPS products have also been shown to accumulate in glandular trichomes to act in induced plant defence [3, 4].
Past investigations of the TPS families in plants have elucidated that the TPS share a common ancestor based on similar reaction mechanisms, structure, sequential homology and conserved functional motifs, but that the large divergence of these enzymes evolutionarily has been integral to the present diversity of terpenes today [2, 5–12]. TPS comprise a usually mid‐sized family of genes in flowering plants that have diverged into five different subfamilies: TPS‐A, which are primarily sesquiterpene producing; TPS‐B and TPS‐G, which are monoterpene producing; and TPS‐C and TPS‐E/F, which are diterpene producing. These subfamilies can be distinguished from one another based on reaction mechanism as Class I (prenyl diphosphate ionisation), which includes Subfamilies A, B, G and E/F, or Class II (geranylgeranyl disphosphate protonation), which includes Subfamily C [2, 13].
Establishment of these subfamilies by phylogenetic analysis had allowed prediction of function and properties based on sequential and structural homology, and placement of newly discovered TPS into subfamilies, although true discernment of function can only be achieved through expression of active recombinant enzymes [14, 15]. Establishment and in‐depth characterisation of TPS families in model plant species with predictive value for other plants are an important endeavour to facilitate identification and characterisation of new TPS. To these ends, scientists have since identified genome‐wide TPS families in commercially and agriculturally important plant species like citrus, grape and tomato [9–11] and large TPS family constructions have also been undertaken in various gymnosperm species [7, 12, 16].
The TPS family (AtTPS) of the non‐glandular trichome‐bearing Arabidopsis thaliana, a plant not known for its fragrance and lacking obvious secretory cell structures like glandular trichomes, was the first TPS family to be characterised [6]. Owing to these qualities, Arabidopsis is a useful model for studying the biosynthesis, molecular regulation and function of TPS in plants without significant storage of TPS products [13]. The first evaluation of the Arabidopsis TPS family identified 32 AtTPS members and determined their localisation, architecture, major conserved domains and phylogenetic relationships [6]. However, a recent study has hinted that the family may not have been fully described previously through the addition of a new member of Subfamily A [2]. The tissue/organ expression of the original 32 TPS has previously been reported, but the results often contradict each other [2, 3, 13, 17], and so far, functional characterisation of 18 of these genes has been executed [1, 3, 4, 15, 18–22; Huh, Tholl et al., unpublished; Vaughan, Tholl et al., unpublished].
Unlike Arabidopsis, Medicago truncatula bears both glandular trichomes and non‐glandular trichomes, and can therefore serve as a model for studying TPS in plants with significant storage of TPS products. Four MtTPS (sesquiterpene‐producing MtTPS1, MtTPS3 and MtTPS5, and the monoterpene‐producing MtTPS4) have been functionally characterised [23, 25]. A comprehensive genome‐wide extension of these studies can now be conducted thanks to the publication of the Medicago MT3.5 genome assembly in late 2011, which made available the most comprehensive genomic information to date by capturing about 94% of all [26].
Of imperative interest in comparing these two model plants is the presence and characteristics of TPS in the two types of trichomes. Non‐glandular trichomes are generally regarded as non‐secreting cells which function in physical defence by protecting plants from insect damage, reducing or maintaining leaf temperature and preventing water loss, although it has recently been suggested that they may synthesize and harbor artemisinin (a sesquiterpene) in Artemisia annua [27, 28]. Glandular trichomes are known to be a major site for biosynthesis, accumulation and secretion of secondary metabolites like terpenes [27, 28], and have become targets of genetic manipulations aimed at improving resistance against disease, herbivory and other stressors in transgenic crops via altered terpene production [29]. A genomic and transcriptomic analysis of the TPS family in Arabidopsis and Medicago will not only afford insight into the function of the two TPS families, and the role of the two types of trichomes in TPS biosynthesis and function, but will also lead to identification of TPS genes with promise for use in genetic manipulation of crops through trichome‐specific genetic engineering. Plants that are enhanced in their expression of trichome‐specific TPS could result in increased disease resistance, enhanced environmental protection and the improved food safety of transgenic crops as well as the ability to harvest terpenes important for pharmaceuticals, eco‐friendly herbicides and pesticides and other commercial goods.
2 Materials and method
2.1 Gene identification
The databases used for identification of genes are as follows: for Arabidopsis, the TAIR10 genome assembly, and for Medicago both the MT3.5 genome assembly by the International Medicago Genome Annotation Group (IMGAG) (http://www.tofu.cfans.umn.edu/downloads_genome/Mt3.5/) and the Dana Farber Cancer Institute Medicago Gene Index Release 11.0 (http://www.compbio.dfci.harvard.edu/tgi/cgi‐bin/tgi/gimain.pl?gudb=medicago), plus the National Centre for Biotechnology Information and trichome expressed sequence tag (http://www.planttrichome.org/trichomedb/) databases. All of these data sets are publicly available at the indicated web addresses. TPS protein sequences were identified using keyword searches, basic local alignment seacrch tool searches and InterproScan TPS functional domain analysis after the removal of redundant sequences.
2.2 Chromosomal location and duplications
Paralogons in Arabidopsis [30] (http://www.wolfe.gen.tcd.ie/athal/dup), PLAZA 2.0 [31] (http://www.bioinformatics.psb.ugent.be/plaza/) and the Plant Genome Duplication Database [32] (http://www.chibba.agtec.uga.edu/duplication/) were utilised to identify duplication events for the AtTPS family. Duplication was paired with chromosomal localisation information determined using The Arabidopsis Information Resource (TAIR) chromosome map tool [33] (http://www.arabidopsis.org/jsp/ChromosomeMap/tool.jsp).
PLAZA 2.0 and the Plant Genome Duplication Database were utilised to identify duplication events for the MtTPS family. The positioning of each gene as part of the IMGAG annotations was paired with chromosome size data from the Medicago HapMap website (http://www.medicagohapmap.org/) to manually visualise the MtTPS genes at their respective positions along chromosomes. All of these resources are publicly available at the specified web addresses.
2.3 Expression analysis
TPS expression patterns in different organs/tissues in both species were analysed and compared using expression data for 32 out of 33 AtTPS and 14 out of 23 MtTPS. Data were obtained from the Samuel Roberts Noble Foundation via ArrayExpress where the expression data were normalised using the quantile method with robust multichip average, and presence (P)/absence (A) calls for each probe set were obtained using dCHIP [methods in [34]; ArrayExpress database; http://www.mtgea.noble.org/v3/]. If P was a result for at least two‐thirds of the trials, then the numerical means were used in expression heat maps. The log2 transformed means of three to six biological replicates of different tissues were loaded into the freely available multi experiment view program and analysed with the hierarchical cluster module [35] (http://www.tm4.org/).
2.4 Phylogenetics
Dataset was divided into subgroups by USEARCH, and Muscle was employed to conduct protein sequence alignments for each subgroup. Aligned subgroups were combined by T‐coffee to generate the final alignment that was used for phylogenetic analysis. Phylogenetic trees were generated by PhyML and visualised by Figtree.
3 Results and discussion
3.1 Identification of Arabidopsis TPS (AtTPS) and Medicago TPS (MtTPS) genes
The complete set of 33 AtTPS gene models is listed in Table 1. This study confirms the prediction of an additional member within the AtTPS Subfamily A (AT1G48820.1/AtTPS31; described in Data S1 of [2]), which was not included in the 32 putative full‐length AtTPS genes reported by Aubourg et al. [6].
Table 1.
TPS family of A. thaliana
| IDs | Other names | Sizes (aa) | Mass (kDa) | Sub‐families | Exons | TPS conserved motifs | Products | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 200 aa | RxR/DDxxD | RRx(8)W | DXDD | NSE/DTE | |||||||
| AT4G15870.1 | AtTPS01 | 598 | 69.6 | A | 7 | N | Y | Y* | N | Y* | N/A |
| AT2G23230.1 | AtTPS05 | 611 | 70.2 | A | 7 | N | Y | Y* | N | Y* | N/A |
| AT1G70080.1 | AtTPS06 | 611 | 71.1 | A | 7 | N | Y* | Y* | N | Y* | N/A |
| AT4G20200.1 | AtTPS07 | 604 | 70.0 | A | 7 | N | Y | Y* | N | Y* | N/A |
| AT4G20210.1 | AtTPS08 | 600 | 69.1 | A | 7 | N | Y* | Y* | N | Y* | Diterpene |
| AT4G20230.1 | AtTPS09 | 609 | 70.7 | A | 7 | N | Y | Y* | N | Y* | N/A |
| AT5G44630.1 | AtTPS11 | 557 | 64.9 | A | 7 | N | Y | Y* | N | Y* | Sesquiterpene blend |
| AT4G13280.1 | AtTPS12 | 524 | 60.6 | A | 7 | N | Y | Y* | N | Y* | (Z)‐y ‐bisabolene [S] |
| AT4G13300.1 | AtTPS13 | 554 | 64.1 | A | 7 | N | Y | Y* | N | Y* | (Z)‐y ‐bisabolene [S] |
| AT3G29190.1 | AtTPS15 | 601 | 69.7 | A | 7 | N | Y | Y* | N | Y* | N/A |
| AT3G29110.1 | AtTPS16 | 569 | 65.6 | A | 7 | N | Y* | Y* | N | Y* | N/A |
| AT3G14490.1 | AtTPS17 | 601 | 70.2 | A | 7 | N | Y* | Y* | N | Y* | N/A |
| AT3G14520.1 | AtTPS18 | 605 | 69.4 | A | 7 | N | Y | Y* | N | Y* | N/A |
| AT3G14540.1 | AtTPS19 | 602 | 69.2 | A | 7 | N | Y | Y* | N | Y* | N/A |
| AT5G48110.1 | AtTPS20 | 575 | 66.9 | A | 7 | N | Y | Y* | N | Y* | Diterpene |
| AT5G23960.1 | AtTPS21 | 547 | 63.2 | A | 7 | N | Y | Y* | N | Y* | (E)‐β ‐caryophyllene [S] |
| AT1G33750.1 | AtTPS22 | 603 | 69.9 | A | 6 | N | Y* | Y* | N | Y* | Sesquiterpene blend |
| AT3G29410.1 | AtTPS25 | 603 | 70.2 | A | 7 | N | Y | Y* | N | Y* | Sesquiterpene blend |
| AT1G66020.1 | AtTPS26 | 598 | 69.3 | A | 7 | N | Y* | Y* | N | Y* | N/A |
| AT1G48800.1 | AtTPS28 | 603 | 69.7 | A | 7 | N | Y | Y* | N | Y* | N/A |
| AT1G31950.1 | AtTPS29 | 606 | 70.1 | A | 7 | N | Y | Y* | N | Y* | N/A |
| AT3G32030.1 | AtTPS30 | 604 | 69.8 | A | 7 | N | Y* | Y* | N | Y* | N/A |
| AT1G48820.1 | AtTPS31 | 561 | 64.9 | A | 7 | N | Y | Y* | N | Y* | N/A |
| AT4G16730.1 | Ocisb_Arath, AtTPS02 | 589 | 69.1 | B | 7 | N | Y | Y | N | Y* | (E)‐β ‐ocimene/myrcene [M] and (E, E)‐a ‐farnesene [S] |
| AT4G16740.1 | AtTPS03 | 565 | 65.8 | B | 7 | N | Y | Y | N | Y* | (E)‐β ‐ocimene/myrcene [M] |
| AT2G24210.1 | AtTPS10 | 591 | 69.3 | B | 7 | N | Y | Y | N | Y* | Monoterpene blend |
| AT3G25830.1 | AtTPS‐CIN, AtTPS23 | 600 | 70.5 | B | 7 | N | Y | Y | N | Y* | 1,8‐cineole [M] |
| AT3G25810.1 | AtTPS24 | 598 | 69.8 | B | 7 | N | Y | Y | N | Y* | 1,8‐cineole [M] |
| AT3G25820.1 | AtTPS‐CIN, AtTPS27 | 600 | 70.5 | B | 7 | N | Y | Y | N | Y* | (E)‐β ‐ocimene/myrcene [M] and (E, E)‐α ‐farnesene [S] |
| AT4G02780.1 | AtTPSGA1 | 802 | 93.0 | C | 15 | Y* | N | N | Y | N | ent‐copalyl diphosphate [D] |
| AT1G61120.1 | AtTPS04 | 877 | 101.9 | E/F | 12 | Y* | Y* | N | N | Y* | (E, E)‐geranyllinalool [D] |
| AT1G79460.1 | AtTPSGA2 | 785 | 89.6 | E/F | 14 | Y* | Y* | N | N | Y* | ent‐kaurene [D] |
| AT1G61680.1 | AtTPS14 | 569 | 65.4 | G | 7 | N | Y* | N | N | Y* | (+) ‐3S‐linalool [M] |
The complete set of 24 MtTPS gene models, 20 from MT3.5 and 4 from MtGI 10, is listed in Table 2, which includes 24 putative full‐length genes containing all four InterProScan TPS functional domains. Twenty‐three of the set contain conserved motifs essential for TPS function of the identified five subfamilies. One candidate of the set (IMGA|Medtr2g082050.1) contains all of the four InterProScan TPS functional domains and has highest homology with a member of Subfamily A (Medtr2g082060.1) (Fig. 2 b), but is missing DDXXD and RRX8 W motifs (Table 1), and therefore may be a pseudogene.
Table 2.
TPS family of M. truncatula
| IDs | Other names | Sizes (aa) | Mass (kDa) | Subfamilies | Exons | TPS conserved motifs | Products | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 200 aa | RxR/DDxxD | RRx(8)W | DXDD | NSE/DTE | |||||||
| TC172570 | MtTPS1 | 562 | 64.9 | A | ? | N | Y | Y | N | Y | N/A |
| TC172668 | MtTPS2 | 553 | 64.6 | A | ? | N | Y | Y* | N | Y* | N/A |
| Medtr5g062230.1 | MtTPS5 | 553 | 64.4 | A | 7 | N | Y | Y* | N | Y* | N/A |
| Medtr2g081980.1 | MtTPS7 | 561 | 65.1 | A | 7 | N | Y | Y* | N | Y* | N/A |
| Medtr2g082010.1 | MtTPS8 | 572 | 66.7 | A | 7 | N | Y | Y* | N | Y* | N/A |
| Medtr2g082060.1 | MtTPS9 | 557 | 65.3 | A | 8 | N | Y | Y* | N | Y* | N/A |
| Medtr5g073200.1 | MtTPS10 | 548 | 64.1 | A | 7 | N | Y | Y* | N | Y* | N/A |
| Medtr5g073260.1 | MtTPS11 | 539 | 63.1 | A | 7 | N | Y | Y* | N | Y | N/A |
| Medtr5g094620.1 | MtTPS12 | 545 | 64.1 | A | 8 | N | Y | Y* | N | Y* | (E)‐β ‐caryophyllene |
| Medtr6g008560.1 | MtTPS13 | 557 | 64.9 | A | 7 | N | Y | Y* | N | Y* | sesquiterpene blend |
| TC172588 | MtTPS4 | 580 | 67.1 | B | ? | N | Y | Y* | N | Y* | N/A |
| Medtr2g065450.1 | MtTPS14 | 589 | 69.0 | B | 7 | N | Y | Y | N | Y* | N/A |
| Medtr7g010710.1 | MtTPS15 | 553 | 65.0 | B | 7 | N | Y | Y | N | Y* | (E)‐β ‐ocimene |
| Medtr7g011670.1 | MtTPS16 | 850 | 98.4 | C | 17 | Y* | N | N | Y | N | N/A |
| Medtr2g012870.1 | MtTPS17 | 806 | 93.4 | E/F | 12 | Y* | Y | N | N | Y* | N/A |
| Medtr2g012900.1 | MtTPS18 | 835 | 96.3 | E/F | 13 | Y* | Y* | N | N | Y* | N/A |
| Medtr3g058160.1 | MtTPS19 | 835 | 95.8 | E/F | 14 | Y* | Y* | N | N | Y* | N/A |
| TC172576 | MtTPS3 | 573 | 65.8 | G | ? | N | Y* | N | N | Y* | N/A |
| Medtr2g089120.1 | MtTPS6 | 567 | 64.9 | G | 7 | N | Y | N | N | Y* | N/A |
| Medtr2g010960.1 | MtTPS20 | 627 | 72.7 | G | 7 | N | Y* | N | N | Y* | N/A |
| Medtr2g089130.1 | MtTPS21 | 565 | 64.4 | G | 7 | N | Y* | N | N | Y* | N/A |
| Medtr3g052120.1 | MtTPS22 | 524 | 60.5 | G | 7 | N | Y | N | N | Y* | N/A |
| Medtr6g064980.1 | MtTPS23 | 519 | 60.0 | G | 7 | N | Y* | N | N | Y* | (3S)‐(E)‐nerolidol |
| Medtr2g082050.1 | — | 590 | 68.49 | pseudogene | 7 | N | N | N | N | Y | N/A |
Y = motif is present; Y* = motif is present, but in a modified form; and N = motif is absent.
Fig. 2.
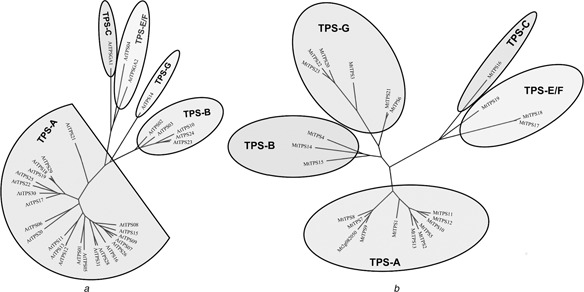
Phylogenetic trees of AtTPS (A) and MtTPS (B) gene families
a AtTPS phylogenetic tree includes 33 genes divided into Subfamilies A, B, C, E/F and G
b MtTPS phylogenetic tree includes 23 full genes and one prospective MT3.5 gene (Medtr2g082050) and is divided into the same subfamilies as above
The AtTPS and MtTPS families conform to the reported size of TPSs with lengths of 500–900 amino acids and masses of about 60–100 kDa. In Subfamilies A, B and G (monoterpene and sesquiterpene synthases), exon numbers range between 7 and 8 in MtTPSs against 6 and 7 in AtTPSs; in Subfamily C, 17 in MtTPSs against 15 in AtTPSs; and in Subfamily E/F (diterpene synthases), 12–14 for both species (Tables 1 and 2).
3.2 Chromosomal location and duplications
In both species, the members of divergent TPS subfamilies are located within the same chromosomal region, whereas the members of the same subfamily are mostly distributed in different chromosomal regions, suggesting that TPS genes were distributed widely in the genome of the common ancestor of the two species (Fig. 1).
Fig. 1.
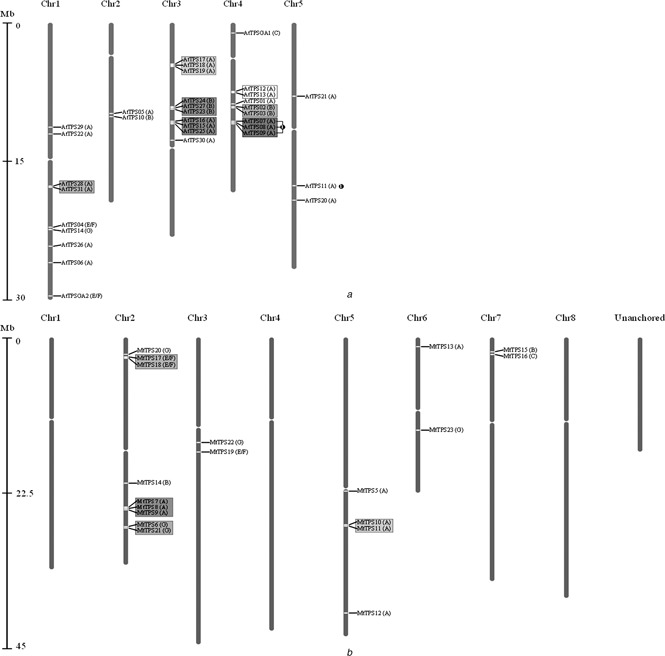
Chromosomal distribution and duplication events of
a AtTPS gene family
b MtTPS gene family
Relative distance from the start of each chromosome is represented by the Mb (Megabase) bar on the left, with the unanchored ‘chromosome’ of Medicago having an arbitrarily defined order. Each gene's placement is designated by a white line across the chromosome. Genes that have experienced tandem duplication events are clustered together on chromosomes and are shown here surrounded by a coloured box grouping them together. Block duplication genes on different chromosomes are marked partners by black circles with matching numbers.
For the 33 putative full‐length AtTPS, seven separate gene clusters (coloured boxes in Fig. 1 a) were detected, among which the clusters of AtTPS28/AtTPS31 (on chromosome 1) and AtTPS16/AtTPS15/AtTPS25 (on chromosome 3) are new, with the other five being previously reported [6]. One new syntenic large‐scale duplication event (AtTPS11 with AtTPS07, AtTPS08 and AtTPS09) was identified in Arabidopsis (Fig. 1 a).
For the 23 putative full‐length MtTPS, four gene clusters (Fig. 1 b, coloured boxes) that exhibit high sequential homology to their partners were identified to be likely the result of local tandem duplication events. It appears that, of the putative full‐length TPS genes currently available in the MT3.5 genome assembly, there is a much less even distribution of TPS throughout the genome of Medicago than in Arabidopsis. No syntenic large‐scale duplications were identified in Medicago.
In summary, the TPS family appears to have experienced large amounts of duplication in both Arabidopsis and Medicago (58 and 39% of genes), the majority of which appear to be a result of localised tandem duplication events. Large‐scale polyploidisation appears to have no significant impact on the two gene families.
3.3 Phylogenetic and comparative analysis of TPS family in Arabidopsis and Medicago
All AtTPS and MtTPS genes can be divided into five subfamilies (Fig. 2) reported for other TPS families in flowering plants [9–11]. Such classification is supported by the conserved motifs, the number of exons and gene length characteristic of each subfamily (Tables 1 and 2). The structural similarity between the MtTPS, AtTPS and other TPS families of flowering plants suggests their shared ancestry. Lineage‐specific expansion has occurred in Subfamily A of Arabidopsis, and in Subfamily G of Medicago (Figs. 2 a and b).
The mixed phylogenetic tree for Subfamily A (Fig. 3), which appears to be the largest and most important subfamily in both species, suggests that most of the Subfamily A genes are more conserved among the same species, except for one member of Arabidopsis (AtTPS21), which shows closer homology with Medicago. There appears to be no putative Arabidopsis /Medicago orthologs according to the phylogenetic tree.
Fig. 3.
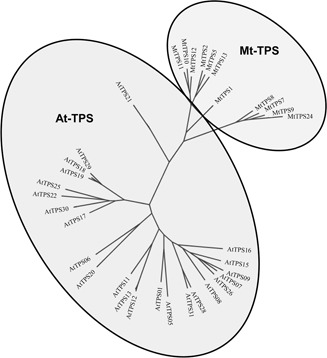
Mixed phylogenetic tree of Subfamily A members from both the AtTPS and MtTPS gene families
Among the seven AtTPS gene clusters (07/08/09, 17/18/19, 23/24/27, 02/03, 12/13, 28/31 and 15/16/25), six phylogenetically close paralogous pairs are found (07 and 09, 18 and 19, 02 and 03, 24 and 23/27, 12 and 13 and 28 and 31) (Fig. 1 a). Among the four MtTPS gene clusters, three phylogenetically close paralogous pairs (17/18, 06/21 and 10/11) are found, and one cluster (07/08/09) also forms a phylogenetically close clade (Fig. 1 b). These results further support a paralogous pattern of TPS gene divergence by localised tandem duplication in both species. The only syntenic large‐scale AtTPS duplication (11 against 07/08/09) does not show phylogenetic closeness (Fig. 2 a), suggesting the rapid divergence of the duplicated genes.
3.4 Expression analysis of TPS genes in tissues of Arabidopsis and Medicago
For Arabidopsis, normalised transcript levels of roots, stems, leaves, flowers, seeds, petioles and non‐glandular trichomes were available for all AtTPS genes (Fig. 4 a) except AtTPS13, a member of Subfamily A. Twelve AtTPS genes were found to have no expression in the analysed tissues. Eleven genes exhibit tissue/organ‐specific expression: AtTPS18, AtTPS19, AtTPS21 and AtTPS24 in the flowers, AtTPS07, AtTPS09, AtTPS12, AtTPS20 and AtTPS26 in the stems, AtTPSGA1 in the roots and AtTPS04 in the non‐glandular trichomes. The remaining nine AtTPS genes exhibited expression in multiple tissue types, with AtTPSGA2 (ent ‐kaurene synthase) being constitutively expressed in all tissues, probably because of its role in gibberellin biosynthesis. Owing to low signal values, the expression of the following genes may be doubtful: AtTPS02, AtTPS07, AtTPS11, AtTPS17, AtTPS18/19 (cross‐hybridising genes), AtTPS20 and AtTPS29.
Fig. 4.
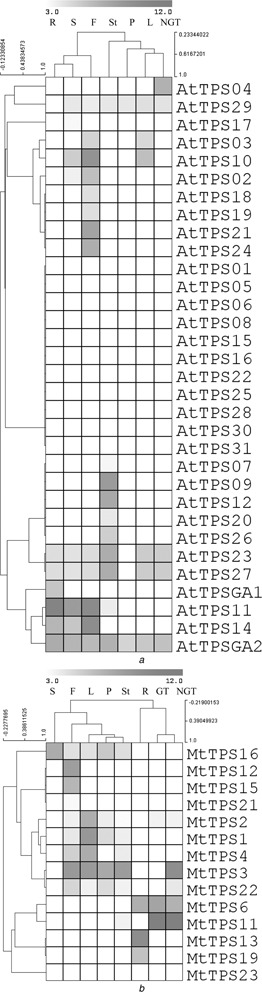
Hierarchical clustering based on Pearson correlation of variable relationship for expression values of
a 32 of the 33 AtTPS genes
b 14 of 23 MtTPS genes
Tissues analysed are abbreviated as follows: root (R), flower (F), seed (S), stem (St), leaf (L), petiole (P), glandular trichome (G) and non‐glandular trichome. Analysis of this log2 transformed data from the ArrayExpress database was performed in the MultiExperiment Viewer. Density of colour indicates the strength of the expression signal intensity, therefore, red indicates expression and brighter red indicates greater expression and closeness of genes/tissues indicates their expression pattern similarity
Normalised transcript levels of roots, stems, leaves, flowers, seeds, petioles, glandular trichomes and non‐glandular trichomes were compared for 14 of 23 MtTPS genes in Medicago (Fig. 4 b); no expression data were available for MtTPS5, MtTPS7, MtTPS8, MtTPS9, MtTPS10, MtTPS14, MtTPS17, MtTPS18 and MtTPS20, limiting the breadth of this analysis. One gene (MtTPS23) did not show expression in any of the tissues analysed. Five genes exhibit tissue/organ‐specific expression: MtTPS13 and MtTPS19 in the roots, and MtTPS12, MtTPS15 and MtTPS21 in the flowers. MtTPS11 has very high expression in both types of trichomes, with the signal level in glandular trichomes (>7000) about double that in non‐glandular trichomes as well as a rather low signal (12.2) in the stem, allowing this gene to be considered to have trichome‐specific expression. The remaining eight TPS genes were expressed in multiple tissue types. Some genes (MtTPS1, MtTPS2, MtTPS4, MtTPS11, MtTPS16, MtTPS21 and MtTPS22) might have doubtful expression because of low signal values.
A comparison of both plants’ expression in the analysed tissues by the number of expressed genes and their expression signal intensity can be found in Table 3. Subfamily A, which primarily syntheses sesquiterpenes, appears to be the most expressed among all the subfamilies in both species. Subfamily G (monoterpene producing) and C (diterpene producing) had much more expression in Medicago than in Arabidopsis, whereas Subfamily E/F (diterpene producing) was more expressed in Arabidopsis than in Medicago (Table 3). The very different distribution of TPS expression in the two species could be related to the difference in the types of trichomes each bears and the presence/absence of root nodules. Although tissue/organ‐specific expression of TPS genes seems to be similarly important in both Arabidopsis and Medicago, trichome‐localised TPS expression appears to be much more important in Medicago. Both species also have high levels of TPS expression in roots and flowers. Our results further infer high levels of monoterpene and sesquiterpene‐producing TPS floral expression in the flowers of both Arabidopsis and Medicago.
Table 3.
Rankings of TPS gene expression based on number of genes and signal intensity in Arabidopsis and Medicago
| Ranks | Number of expressed genes | Gene expression intensities |
|---|---|---|
| A. thaliana | ||
| 1 | Flowers (13/32 genes) | Flowers (A, B, G) |
| 2 | Stems (11/32 genes) | Roots (A, B, C, E/F, G) |
| 3 | Seeds (9/32 genes) | Stems (A, B, C) |
| 4 | Roots (6/32 genes) | Seeds (A, B, E/F, G) |
| 5 | Leaves (6/32 genes) | Leaves (B, E/F) |
| 6 | Non‐glandular trichomes (4/32 genes) | Non‐glandular trichomes (A, B, E/F) |
| 7 | Petioles (2/32 genes) | Petioles (A, E/F) |
| M. truncatula | ||
| 1 | Flowers (9/14 genes) | Glandular trichomes (A, G) |
| 2 | Stems (7/14 genes) | Non‐glandular trichomes (A, G) |
| 3 | Leaves (6/14 genes) | Leaves (A, B, G) |
| 4 | Non‐glandular trichomes (5/14 genes) | Roots (A, E/F, G) |
| 5 | Petioles (5/14 genes) | Flowers (A, B, G) |
| 6 | Glandular trichomes (3/14 genes) | Stems (G) |
| 7 | Roots (3/14 genes) | Petioles (C, G) |
| 8 | Seeds (1/14 genes) | Seeds (C) |
Bolded letters indicate the subfamilies with highest intensity.
In hierarchical clustering, non‐glandular trichome and glandular trichome expression patterns in Medicago are very strongly correlated, suggesting similar functions and possible synergistic effects. MtTPS6 (Subfamily G) and MtTPS11 (Subfamily A) are expressed at similarly high levels in both trichome types and may be working in tandem in volatile plant defence and/or ecological interactions. TPS in these subfamilies produce monoterpenes and sesquiterpenes that function in volatile plant defence [4], making their coordination quite interesting. The expression of more TPS genes (three at high levels) in non‐glandular trichomes than in glandular trichomes in Medicago and the expression of several AtTPS in non‐glandular trichomes of Arabidopsis points to a possible role of non‐glandular trichomes in plant chemical defence and/or ecological communication, contrary to the textbook physical defence view of non‐glandular trichome function. Along this same line of thinking, a recent study in A. annua describes three contigs likely encoding enzymes necessary for sesquiterpene biosynthesis that were found to be strongly expressed in non‐glandular trichomes, thus bringing forth the speculation that glandular trichomes may not be the sole site for the biosynthesis of artemisinin and other sesquiterpenes [28].
The results of this study provide a new perspective for trichome‐specific genetic engineering, which could be done in not only glandular trichomes, but also non‐glandular trichomes. MtTPS11, which was highly and almost exclusively expressed in both types of trichomes, is a promising candidate for further functional study and possible industrial pursuit on trichome‐specific genetic engineering. The other tissue‐specific genes could possibly be used for genetic engineering in specific organs as well.
There appears to be some correlation between paralog duplication events and the expression patterns of these similarly derived TPS in Arabidopsis, although this does not always hold true. Among the four phylogenetically close AtTPS paralogs with expression data available for comparison, three paralogs (AtTPS07 and AtTPS09, AtTPS18 and AtTPS19 and AtTPS02 and AtTPS03) showed clustered expression, whereas the paralogs AtTPS24 and AtTPS27 did not. The phylogenetically distant block duplications (AtTPS07, AtTPS08/AtTPS09 and AtTPS11) did not show clustered expression. It should be noted that some phylogenetically distant non‐paralogous AtTPS also had clustered expression with one another, including AtTPS11 (A) and AtTPS14 (G), AtTPS12 (A) and AtTPS15 (A), AtTPS20 (A) and AtTPS26 (A) and AtTPS21 (A) and AtTPS24 (B). In Medicago, the only phylogenetically close paralog with expression data available for comparison, MtTPS6 and MtTPS21, did not exhibit clustered expression. On the other hand, the genes with clustered expression (MtTPS1 and MtTPS4, MtTPS3 and MtTPS22, MtTPS6 and MtTPS11, MtTPS13 and MtTPS19 and MtTPS12, MtTPS15 and MtTPS21) are all phylogenetically distant. These results suggest that in both species even phylogenetically close paralogs might not have predictive value for one another in regard to function. Our results obtained in plants appear quite different from those obtained in mammals, which showed that paralogs are often a much better predictor of function than are orthologs, even at lower sequence identities [36].
4 Conclusions
In conclusion, the results of this study point to new roles of non‐glandular trichome in plant chemical defence and/or ecological communication instead of only in physical defence in both Arabidopsis and Medicago, and new perspectives for trichome‐specific genetic engineering in non‐glandular trichomes. This study identified one possible trichome‐specific gene in Arabidopsis (AtTPS04), and one highly expressed trichome‐specific gene in Medicago (MtTPS11) that shows promise in trichome‐specific genetic engineering. The general lack of correlation between expression patterns and phylogenetic relationships in both species suggests that phylogenetic analysis alone is insufficient to predict gene function even for phylogenetically close paralogs, which calls for the reconsideration of the use of paralogs for functional prediction in plants.
5 Acknowledgments
We thank Drs. Thomas Sharkey, Carol Hepfer and Judith Thomas for their helpful comments in improving this work. We also thank Jonathan Kettering and Ahmad Ismaeil Hussein for validating data and making corrections. We are grateful for Dr. Yuhong Tang's help with the gene expression analysis. This work was supported by grants and funding provided by Faculty Professional Development Council Grant of Pennsylvania State System [grant no 2011‐MU‐05], Millersville University of Pennsylvania and The Samuel Roberts Noble Foundation.
6 References
- 1. Huang M. Abel C., and Sohrabi R. et al.: ‘Variation of herbivore‐induced volatile terpenes among Arabidopsis ecotypes depends on allelic differences and subcellular targeting of two terpene synthases, TPS02 and TPS031’, Plant Physiol., 2010, 153, (3), pp. 1293–1310 (doi: 10.1104/pp.110.154864) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen F. Tholl D. Bohlmann J., and Pichersky E.: ‘The family of terpene synthases in plants: a mid‐size family of genes for specialized metabolism that is highly diversified throughout the kingdom’, Plant J., 2011, 66, (1), pp. 212–229 (doi: 10.1111/j.1365-313X.2011.04520.x) [DOI] [PubMed] [Google Scholar]
- 3. Chen F. Tholl D. D'Auria J.C. Farooq A. Pichersky E., and Gershenzon J.: ‘Biosynthesis and emission of terpenoid volatiles from arabidopsis flowers’, Plant Cell, 2003, 15, (2), pp. 481–494 (doi: 10.1105/tpc.007989) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen F. Ro D.K., and Petri J. et al.: ‘Characterization of a root‐specific Arabidopsis terpene synthase responsible for the formation of the volatile monoterpene 1,8‐cineole1’, Plant Physiol., 2004, 135, (4), pp. 1956–1966 (doi: 10.1104/pp.104.044388) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Trapp S.C., and Croteau R.B.: ‘Genomic organization of plant terpene synthases and molecular evolutionary implications’, Genetics, 2001, 158, pp. 811–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Aubourg S. Lecharny A., and Bohlmann J.: ‘Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana’, Mol. Genetics Genomics, 2002, 267, (6), pp. 730–745 (doi: 10.1007/s00438-002-0709-y) [DOI] [PubMed] [Google Scholar]
- 7. Martin D.M. Fäldt J., and Bohlmann J.: ‘Functional characterization of nine Norway spruce TPS genes and evolution of gymnosperm terpene synthases of the TPS‐d subfamily’, Plant Physiol., 2004, 5, (4), pp. 1908–1927 (doi: 10.1104/pp.104.042028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tholl D.: ‘Terpene synthases and the regulation, diversity and biological roles of terpene metabolism’, Curr. Opin. Plant Biol., 2006, 9, (3), pp. 297–304 (doi: 10.1016/j.pbi.2006.03.014) [DOI] [PubMed] [Google Scholar]
- 9. Dornelas M.C., and Mazzafera P.: ‘A genomic approach to characterization of the citrus terpene synthase gene family’, Genetics Mol. Biol., 2007, 30, (3), pp. 832–840 (doi: 10.1590/S1415-47572007000500011) [DOI] [Google Scholar]
- 10. Martin D.M. Aubourg S., and Schouwey M.B. et al.: ‘Functional annotation, genome organization and phylogeny of the grapevine (Vitis vinifera) terpene synthase gene family based on genome assembly, FLcDNA cloning, enzyme assays’, BMC Plant Biol., 2010, 10, p. 226 (doi: 10.1186/1471-2229-10-226) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Falara V. Akhtar T.A., and Nguyen T.T.H. et al.: ‘The tomato terpene synthase gene family’, Plant Physiol., 2011, 157, (2), pp. 770–789 (doi: 10.1104/pp.111.179648) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Keeling C.I. Weisshaar S. Ralph S.G. Jancsik S. Hamberger B. Dullat H.K., and Bohlmann J.: ‘Transcriptome mining, functional characterization, and phylogeny of a large terpene synthase gene family in spruce (Picea spp.)’, BMC Plant Biol., 2011, 11, p. 43 (doi: 10.1186/1471-2229-11-43) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tholl D., and Lee S.: ‘Terpene specialized metabolism in arabidopsis thaliana’, The Arabidopsis Book, 2011, 9, p. e0143 (doi: 10.1199/tab.0143) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bohlmann J. Meyer‐Gauen G., and Croteau R.: ‘Plant terpenoid synthases: molecular biology and phylogenetic analysis’, Proc. Natl. Acad. Sci., 1998, 95, (8), pp. 4126–4133 (doi: 10.1073/pnas.95.8.4126) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bohlmann J. Martin D. Oldham N.J., and Gershenzon J.: ‘Terpenoid secondary metabolism in Arabidopsis thaliana: cDNA cloning, characterization, functional expression of a myrcene/(E)‐beta‐ocimene synthase’, Arch. Biochem. Biophys., 2000, 375, (2), pp. 261–269 (doi: 10.1006/abbi.1999.1669) [DOI] [PubMed] [Google Scholar]
- 16. Hall D.E. Yuen M.M., and Jancsik S. et al.: ‘Transcriptome resources and functional characterization of monoterpene synthases for two host species of the mountain pine beetle, lodgepole pine (Pinus contorta) and jack pine (Pinus banksiana)’, BMC Plant Biol., 2013, 13, p. 80 (doi: 10.1186/1471-2229-13-80) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ro D.K. Ehlting J. Keeling C.I. Lin R. Mattheus N., and Bohlmann J.: ‘Microarray expression profiling and functional characterization of AtTPS genes: duplicated Arabidopsis thaliana sesquiterpene synthase genes At4g13280 and At4g13300 encode root‐specific and wound‐inducible (Z)‐gamma‐bisabolene synthases’, Arch. Biochem. Biophys., 2006, 448, (1–2), pp. 104–116 (doi: 10.1016/j.abb.2005.09.019) [DOI] [PubMed] [Google Scholar]
- 18. Fäldt J. Arimura G. Gershenzon J. Takabayashi J., and Bohlmann J.: ‘Functional identification of AtTPS03 as (E)‐beta‐ocimene synthase: a monoterpene synthase catalyzing jasmonate‐ and wound‐induced volatile formation in Arabidopsis thaliana’, Planta, 2003, 216, (5), pp. 745–751 [DOI] [PubMed] [Google Scholar]
- 19. Herde M. Gärtner K., and Köllner T.G. et al.: ‘Identification and regulation of TPS04/GES, an Arabidopsis geranyllinalool synthase catalyzing the first step in the formation of the insect‐induced volatile C16‐homoterpene TMTT’, Plant Cell, 2008, 20, (4), pp. 1152–1168 (doi: 10.1105/tpc.106.049478) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sun T.P., and Kamiya Y.: ‘The Arabidopsis GA1 locus encodes the cyclase ent‐kaurene synthetase A of gibberellin biosynthesis’, Plant Cell, 1994, 6, (10), pp. 1509–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tholl D. Chen F. Petri J. Gershenzon J., and Pichersky E.: ‘Two sesquiterpene synthases are responsible for the complex mixture of sesquiterpenes emitted from Arabidopsis flowers’, Plant J., 2005, 42, pp. 757–771 (doi: 10.1111/j.1365-313X.2005.02417.x) [DOI] [PubMed] [Google Scholar]
- 22. Yamaguchi S. Sun T. Kawaide H., and Kamiya Y.: ‘The GA2 locus of Arabidopsis thaliana encodes ent‐kaurene synthase of gibberellin biosynthesis’, Plant Physiol., 1998, 116, (4), pp. 1271–1278 (doi: 10.1104/pp.116.4.1271) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Arimura G. Garms S., and Maffei M. et al.: ‘Herbivore‐induced terpenoid emission in Medicago truncatula: concerted action of jasmonate, ethylene and calcium signaling’, Planta, 2008, 227, (2), pp. 453–464 (doi: 10.1007/s00425-007-0631-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Navia‐Giné W.G. Yuan J.S. Mauromoustakos A. Murphy J.B. Chen F., and Korth K.L.: ‘Medicago truncatula (E)‐beta‐ocimene synthase is induced by insect herbivory with corresponding increases in emission of volatile ocimene’, Plant Physiol. Biochem., 2009, 47, (5), pp. 416–425 (doi: 10.1016/j.plaphy.2009.01.008) [DOI] [PubMed] [Google Scholar]
- 25. Gomez S.K. Cox M.M., and Bede J.C. et al.: ‘Lepidopteran herbivory and oral factors induce transcripts encoding novel terpene synthases in Medicago truncatula’, Arch. Insect Biochem. Physiol., 2005, 58, pp. 114–127 (doi: 10.1002/arch.20037) [DOI] [PubMed] [Google Scholar]
- 26. Young N.D. Debellé F., and Oldroyd G.E. et al.: ‘The Medicago genome provides insight into the evolution of rhizobial symbioses’, Nature, 2011, 480, pp. 520–524 (doi: 10.1038/480162a) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dai X. Wang G., and Yang D.S. et al.: ‘TrichOME: a comparative omics database for plant trichomes’, Plant Physiol., 2010, 152, (1), pp. 44–54 (doi: 10.1104/pp.109.145813) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang W. Wang Y. Zhang Q. Qi Y., and Guo D.: ‘Global characterization of Artemisia annua glandular trichome transcriptome using 454 pyrosequencing’, BMC Genomics, 2009, 10, p. 465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Aziz N. Paiva N.L. May G.D., and Dixon R.A.: ‘Transcriptome analysis of alfalfa glandular trichomes’, Planta, 2005, 221, (1), pp. 28–38 (doi: 10.1007/s00425-004-1424-1) [DOI] [PubMed] [Google Scholar]
- 30. Blanc G. Hokamp K., and Wolfe K.H.: ‘A recent polyploidy superimposed on older large‐scale duplications in the Arabidopsis genome’, Genome Res., 2003, 13, (2), pp. 137–144 (doi: 10.1101/gr.751803) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Van Bel M. Proost S., and Wischnitzki E. et al.: ‘Dissecting plant genomes with the PLAZA comparative genomics platform’, Plant Physiol., 2012, 158, (2), pp. 590–600 (doi: 10.1104/pp.111.189514) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tang H. Bowers J.E. Wang X. Ming R. Alam M., and Paterson A.H.: ‘Synteny and collinearity in plant genomes’, Science, 2008, 320, pp. 486–488 (doi: 10.1126/science.1153917) [DOI] [PubMed] [Google Scholar]
- 33. Lamesch P. Berardini T.Z., and Li D. et al.: ‘The Arabidopsis information resource (TAIR): improved gene annotation and new tools’, Nucleic Acids Res., 2012, 40, (1), pp. 1202–1210 (doi: 10.1093/nar/gkr1090) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Benedito V.A. Torres‐Jerez I., and Murray J.D. et al.: ‘A gene expression atlas of the model legume Medicago truncatula’, Plant J., 2008, 55, (3), pp. 504–513 (doi: 10.1111/j.1365-313X.2008.03519.x) [DOI] [PubMed] [Google Scholar]
- 35. Saeed A.I. Bhagabati N.K., and Braisted J.C. et al.: ‘TM4 microarray software suite’, Methods Enzymol., 2006, 411, pp. 134–193 (doi: 10.1016/S0076-6879(06)11009-5) [DOI] [PubMed] [Google Scholar]
- 36. Nehrt N.L. Clark W.T. Radivojac P., and Hahn M.W.: ‘Testing the ortholog conjecture with comparative functional genomic data from mammals’, PLoS Comput. Biol., 2011, 7, (6), p. e1002073 (doi: 10.1371/journal.pcbi.1002073) [DOI] [PMC free article] [PubMed] [Google Scholar]


