Abstract
As a shortcut for drug development, drug repositioning draws more and more attention in pharmaceutical industry to identify new indications for marketed drugs or drugs failed in late clinical trial phase. At the same time, the abundant high‐throughput data pushes the computationally repositioning drugs a hot topic in the area of systems biology. Here, the authors propose a general framework for repositioning drug by incorporating various functional information. The framework starts with the identification of differentially expressed gene sets under disease state and drug treatment. Then the disease and drug are associated by the overlap of these two gene sets via biological function. The authors provide two strategies to assess the functional overlap. In the first strategy, functional relevance are evaluated by leveraging genes’ lethality information to reveal drug's potential of curing diseases. In the second strategy, biological process perturbation profiles are identified by mapping differentially expressed genes into pathways and gene ontology (GO) terms. Their associations are assessed and used to rank drugs’ potential of curing diseases. The preliminary results on prostate cancer demonstrate that our new framework improves the drug repositioning efficiency and various function information could complement each other. Importantly, the new framework will enhance the biological interpretation and rationale of drug repositioning and provide insights into drug action mechanisms.
Inspec keywords: biological organs, cancer, drug delivery systems, drugs, medical computing, pharmaceutical industry, gene therapy
Other keywords: drug repositioning framework, functional information, drug development, pharmaceutical industry, marketed drugs, clinical trial phase, high‐throughput data, systems biology, gene expression sets, disease state, drug treatment, biological function, gene lethality information leveraging, biological process perturbation profiles, prostate cancer, drug action mechanisms
1 Introduction
The high incidence of complex diseases, such as cancer, diabetes, calls for much more effective drugs. However, the cost and time to develop a new drug increase annually and the number of drugs approved by US Food and Drug Administration (FDA) declined steadily [1]. One way to fill in this gap is drug repositioning, which makes full use of marketed drugs or drugs failed in late clinical trial phases to explore their new indications. Since the safety profiles and pharmacokinetics profiles of these drugs are generally available [2], drug repositioning has the advantage to reduce the costs and risks in early development stage [3], which will shorten routes to approval for therapeutic indications [3]. Given these advantages, drug repositioning has been an important strategy for drug development. Successful examples of drug repositioning include but not limit to the indication of sildenafil for erecile dysfunction and pulmonary hypertension, thalidomide for severe erythema nodosum leprosum [3, 4].
Most of the above‐mentioned successful examples were occasionally discovered in clinical observation. The unclear underlying molecular mechanisms hinder the repositioning drugs in large scale. Fortunately, the rapidly accumulated high‐throughput data for drugs greatly facilitate cellular systems modelling and molecular mechanisms uncovering. Thus, many computational approaches have been proposed to reposition drugs against various diseases from the perspective of systems biology [5].
Based on the strategies utilised, these methods mainly fall into two categories. The first category transfers the known indications of two drugs having a sufficient high similarity measured from various data sources. For example the overlap or sequence similarity among drugs’ targets (primary target plus off‐target) was used for transferring indication among drugs [6–13]. Besides, the similarity between drugs’ side effect profiles was utilised for transferring indications among drugs [14]. Different from the above‐mentioned methods, Jin et al. [15] proposed a network‐based method to facilitate drug repositioning for cancer therapy by leveraging transcriptional data. In their work, the similarity between drugs’ strength of perturbing the active network underlying disease state was measured. Those measures were further exploited to guide indication transferring among drugs. With many types of drug related data available, integrating drug–drug similarities and disease–disease similarities are in pressing need. For example, various similarities between drugs and diseases were mined and integrated to infer novel drug–disease associations indirectly [16].
The aim of drug treatment is to restore the cellular disease state to normal state. To this end, the methods in the second category aim to measure the degree of reverse for predicting new drug–disease associations. Therefore many efforts started to measure the degree of reverse. In Lamb et al.'s work, metric based on gene set enrichment analysis was employed to measure the anti‐correlation of drug and disease at transcriptional level [17]. In this framework, different modified versions of gene set enrichment analysis were used to measure the association between the transcriptional expression profiles under drug administration and disease state [3, 18]. In Hu and Agarwal's work, simple Pearson correlation coefficient was employed to quantify the anti‐correlation between the gene expression signatures [19] under two conditions and the overlap between opposite regulated gene sets was quantified as the anti‐correlation [20]. Besides, the relevance of molecular basis or molecular activities of drugs and diseases were identified and used for measuring the relevance of biological processes perturbed in these two conditions [5, 21–23].
Taken together, similarities in molecular basis or molecular activities are used as the surrogate of molecular mechanisms relevance underlying disease development and drug treatment. However, molecular activities only indicate the main players in a specific biological process related to drug treatment and disease. In other words, this information alone lacks the biological interpretation. Fortunately, gene ontology and pathway provide higher level functional information and will greatly enhance the interpretability to associate drug and disease. It is believed that interplay between disease state and drug perturbation could be inferred more accurately by combining functional information. As a result, the drug–disease association strength could be assessed more accurately. With this in mind, we propose a general framework for repositioning drugs by leveraging cellular function information. In addition to various functional information, the molecular activity profile at transcriptional level is integrated as well. The preliminary results demonstrate that various functional information could complement each other and are effective in repositioning drugs against prostate cancer.
2 Methods and materials
2.1 Materials
In this work, the expression data of prostate cancer was downloaded from gene expression omnibus database [24] with accession number GDS1439. This dataset comprises of six benign samples and 13 disease samples. Subsequently, GDS1439 in soft format were re‐annotated with latest corresponding general public license (GPL) annotation file downloaded from AILUN's website [25]. We averaged the expression values when multiple microarray probes mapped to the same Entrez GeneID.
Expression profiles of 1741 gene from human PC3 cell lines treated with drugs were obtained. More specifically, we fetched the treatment/control expression ratio matrix from Connectivity Map 02 (CMAP) [17]. Then, we annotated each profile with latest corresponding GPL annotation file downloaded from AILUN's website [25]. The geometric mean of ratios for multiple probes was designated as the ratio of that Entrez Gene. This two annotated matrices were merged into a new matrix named PC ratio matrix by removing absent genes. We retained those common genes when the disease expression dataset and drug induced expression profiles were from different microarray platforms.
In this work, the human essential gene (EG) list was obtained from DEG database [26]. Subsequently, symbols of EGs were transformed into Entrez GeneID by R package named biomaRt [27, 28]. The human‐specific filtered GO, gene_association.goa_human were downloaded from GO database [29]. Subsequently, gene2go file was downloaded from NCBI ftp site. We extracted the mapping between genes with Entrez ID and human‐specific filtered biological processes. As a result, 1698 GO terms were used for gene annotation. In addition, we fetched 1452 human canonical pathways from MsigDB (version 3.1) [30]. Finally, we got a list of drugs approved by FDA or under clinical trial for prostate cancer from the supplementary materials of Jin et al.'s work [15]. We took it as the gold standard dataset for prediction.
2.2 Methods
We assume that a cell is a complex networked system. When some key components are perturbed under certain conditions, some biological processes will be further perturbed, and mediate activities of downstream components. Therefore microarray data that snapshots genes’ transient transcriptional level is an important clue for inferring underlying mechanisms. The perturbed biological processes that dictate disease state and drug response can be revealed as well. On the other hand, functional annotations of biological molecules is partially known. Distinct sets of cellular biological molecules are known to coordinate each other to form a biochemical pathways. These knowledge could be leveraged for supervising the inference process for underlying mechanisms that dictate disease state and drug response. Therefore we can measure the relevance of inferred molecule mechanisms to predict drug's new role of treating diseases. Fig. 1 illustrates the strategy of our framework and the details are addressed in the following.
Fig. 1.
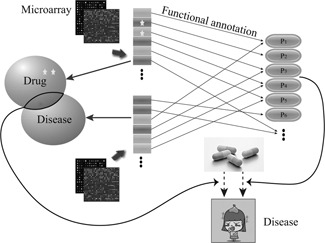
Flowchart of our framework
Star denotes EG and pi denotes pathway or a set of biological process ontologies
2.2.1 Evaluating drug–disease associations based on gene's lethality
We compiled the list of aberrantly regulated genes by comparing disease samples to control samples with R package named limma [31] and further ranked them by absolute value of t ‐statistic. Similarly, ranked list of genes for each drug treatment were generated through ranking the genes by the absolute value of logarithm of treatment/control expression ratio.
Similar to Lamb's work [17] and Shigemiu's work [20], the top k % genes with positive t ‐statistic were defined as up‐regulated genes in prostate cancer (shortened as PCU), and the top k % genes with negative t ‐statistic were defined as down‐regulated genes in prostate cancer (PCD). In the latter category, we defined the top k % genes with treatment/control expression ratio higher than 1 as up‐regulated genes by bioactive compounds (DU), and the top k % genes with treatment/control expression ratio smaller than 1 as down‐regulated genes by bioactive compounds (DD). Here k ranges from 10 to 30 in increments of 5.
The overlap or similarity between DU and PCD was measured by Jaccard index [32] as efficacy score named , that is
| (1) |
Similarly, we also employed Jaccard index to evaluate the overlap between set of genes that fall in DU, whereas outside PCD and set of EG as side effect score named , that is
| (2) |
Further, drug effect in the up‐regulated genes was measured as , which combines with a balancing factor λ varying from 0 to 1
| (3) |
In a similar procedure, the overlap or similarity between DD and PCU, the overlap between set of genes that fall in DD and outside PCU and set of EG were measured by Jaccard index as , respectively
| (4) |
| (5) |
Further, drug effect in down‐regulated genes was measured as scoredown, which combines with a balancing factor α
| (6) |
Finally, the association strength between drug and prostate cancer was evaluated by combing scoreup and scoredown, that is
| (7) |
With fixed k, λ and α, the strength of association between prostate cancer and all compounds in PC ratio matrix were measured and ranked by S.
2.2.2 Evaluating drug–disease associations based on gene's general functional information
With ranked lists of genes under disease state and drug treatment, we further introduced a score to formally measure the differential expression degree of each gene under two conditions. Suppose there are M genes and ri is the rank of gene i under disease state or drug treatment, then differential expression score of gene i was defined as DE i = ((M − ri + 1)/M).
Next, we extracted functional perturbation profile by mapping genes onto pathways or annotating them with GO terms. We employed the score that originally developed by Pham et al. in [33] to evaluate the perturbation degree of pathways and GO terms. For the i th pathway/biological process Pi , it's perturbation degree under disease state was measured as
| (8) |
where is the Jaccard index between top k % differential expression genes Gk and i th pathway/biological process Pi and med{DE x |x ∈ Gk ∩ Pi } is the median of DE score over all genes in Gk ∩ Pi . Collectively, a vector PER o was formulated to measure all the pathway/biological process's perturbation degree, that is
| (9) |
where H is the number of canonical pathways.
On the other hand, we defined functional perturbation profile that dictate d th drug's response, that is
| (10) |
Finally, the association strength between disease and d th drug was measured as the sum of dot product of functional perturbation profiles underlying disease development and drug response, that is
| (11) |
2.2.3 Score collapsing
Each instance in PC matrix corresponds to a score measured by S or AS. It is noteworthy that multiple instances may correspond to the same drug and with the same dose. As a result, we offer three options for such cases. The first option is to simply keep the scores of multiple instances. The second option is to calculate the maximum of individual instances's score that correspond to the same drug with specified dose as the repositioned score of specified drug–dose pair. The last option is to calculate the maximum of individual instances's score that correspond to the same drug as the repositioned score of specified drug.
2.2.4 Prediction assessment
Whether S defined in (7) or AS defined in (11) is used to measure the strength of association between drug and prostate cancer, the F 1 score defined below [34] was adopted as repositioning performance index
| (12) |
where precision is the ratio of true positives in predicted positives and recall is the ratio of true positives that can be predicted correctly. The threshold used to make future prediction is chosen when the highest F 1 score is achieved. Then a drug was repositioned against prostate cancer if its score is above the threshold. Subsequently, the values of parameters that give maximum F 1 score were determined in a similar way.
3 Results and discussion
After evaluating and ranking the strength of association between drug and prostate cancer by S defined in (7), we performed drug repositioning experiments with different choices of collapsing score. The repositioning efficiency was measured by F 1 score defined in (12). With three options of score collapsing, the drug repositioning experiments were performed with 105 combinations of parameters. In this parameter settings, k varies from 10 to 30 by 5 and λ equals to α and ranges from 0 to 1 by 0.05. All the experiments's F 1 scores under each option were summarised in Figs. 2, 3–4, respectively. Besides, the AS defined in (11) was also applied to evaluate the strength of association between drug and prostate cancer. The repositioning efficiency was still measured by F 1 score. All the experiments's F 1 scores under different combinations of parameters and options of score collapsing were summarised in Supplementary Tables 1 –6. Collectively, all the F 1 scores are higher than 0.1. This means that parts of FDA approved or clinical trial drugs for prostate cancer were identified successfully. The moderate F 1 scores show that considerable drugs or compounds were newly repositioned against prostate cancer. As in Sirota et al.'s work [3], these predictions may be followed by experiments on animal models and clinical models. We took a close look at Fig. 4. It summarised repositioning effort with score‐collapsing option 3. The global optimal F 1 score was obtained with k = 10 and λ = α = 0.2. Under this parameter setting, we detected 132 unique bioactive compounds for prostate cancer. Sixteen of these are FDA approved or undergoing clinical trial, which were summarised in Table 1. That means we recovered 16 of the 45 FDA approved or clinical trial compounds for prostate cancer in PC ratio matrix.
Fig. 2.
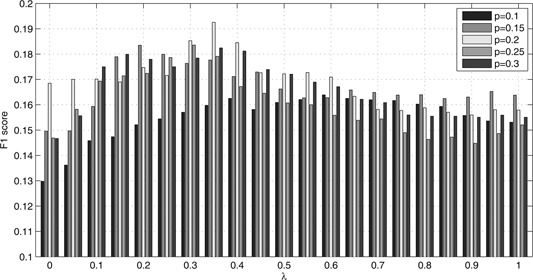
F1 scores for repositioning experiments based on gene's lethality and under score‐collapsing option 1, in which p = (k/100)
Here, X ‐axis represents the value of λ, and Y ‐axis represents the highest F1 score
Fig. 3.

F1 scores for repositioning experiments based on gene's lethality and under score‐collapsing option 2, in which p = (k/100)
Fig. 4.
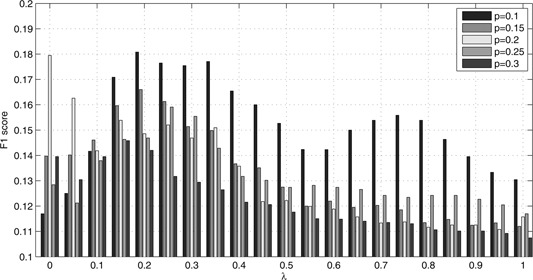
F1 scores for repositioning experiments based on gene's lethality and under score‐collapsing option 3, in which p = (k/100)
Table 1.
Drugs in the gold‐standard set are repositioned under score‐collapsing option 3, utilising gene's lethality information
| Drug name | Status |
|---|---|
| sirolimus | clinical trial |
| paclitaxel | clinical trial |
| phentolamine | clinical trial |
| tanespimycin | clinical trial |
| doxorubicin | clinical trial |
| methylprednisolone | clinical trial |
| estradiol | clinical trial |
| vinblastine | clinical trial |
| valproic acid | clinical trial |
| metformin | clinical trial |
| theophylline | clinical trial |
| diethylstilbestrol | clinical trial |
| tamoxifen | clinical trial |
| azacitidine | clinical trial |
| dexamethasone | clinical trial |
| fulvestrant | clinical trial |
It can be seen from these figures that parameters and score‐collapsing choices indeed affect the repositioning efficiency. Comparatively, the parameter λ has more pronounced effect. Optimal F 1 score was obtained with 0 < λ = α < 1 with fixed k. λ = α = 1 or λ = α = 0 means only efficacy score or side effect was utilised to measure drug–disease association. Therefore this result demonstrated the necessity of measuring drug–disease association with integrated efficacy and side effect information to some extent.
To demonstrate the superiority of repositioning drug by integrating both measures, we took a close look at Fig. 4. It summarised repositioning effort with score‐collapsing option 3. The global optimal F 1 score was obtained with k = 10 and λ = α = 0.2. Under this parameter setting, we detected 16 FDA approved or clinical trial drugs for prostate cancer. On the contrary, 9 of these 45 compounds were among the top 132 compounds sorted by drug–disease score with k = 10 and λ = α = 0. With k = 10 and λ = α = 1, 10 of these 45 compounds were among the top 132 compounds. Besides, five drugs, azacitidine, dexamethasone,estradiol, metformin, and tamoxifen were repositioned against prostate cancer successfully with k = 10 and λ = α = 0.2. Whereas they could not be repositioned under the other two parameter settings. This implies the superiority of repositioning drug by integrating both efficacy and side effect measures.
To demonstrate that the two strategies are complementary, we took a close look at a special case. After mapping genes into pathways, 47 unique bioactive compounds for prostate cancer were identified under score‐collapsing option 3 with k = 10. In the 47 compounds, seven drugs were FDA approved or undergoing clinical trial (Table 2). That also means 7 of 45 FDA approved or clinical trial compounds for prostate cancer were recovered. In the seven compounds, two drugs alprostadic and rofecoxib, could not be repositioned based on gene's lethality.
Table 2.
Drugs in the gold‐standard set are repositioned with k = 10 and under score‐collapsing option 3, utilising pathway information
| Drug name | Status |
|---|---|
| alprostadil | clinical trial |
| fulvestrant | clinical trial |
| rofecoxib | clinical trial |
| valproic acid | clinical trial |
| tamoxifen | clinical trial |
| tanespimycin | clinical trial |
| paclitaxel | clinical trial |
4 Conclusions and future work
A cellular system is a complex networked system. Perturbation will propagate through this networked system. Therefore disease development and drug treatment will induce complicated effects. Microarray data could be used to infer molecular mechanisms or perturbed biological processes that dictate disease state and drug response. However, microarray data or differential expression alone could not reflect the overall functional consequence or phenotype variation. Functional annotations could be leveraged to supervise the inference process. In this paper, we developed a new framework for identifying such repositioned drugs against prostate cancer based on functional information. In our framework, molecular activity profile at the transcription level is utilised by incorporating various functional information. In the first strategy, functional relevance of differentially expressed genes under disease state and drug treatment is evaluated by leveraging genes’ lethality information, and then drugs are ranked by their diseases curing potential. In the second strategy, biological process perturbation profiles under disease state and drug treatment are identified by mapping differentially expressed genes into pathways and GO annotations. Associations of biological process perturbation profiles are assessed and used for ranking drug's potential of curing diseases.
Two drugs may induce the same number of genes to make expression changes. If one drug induces more EG, its side effect may be more severe. Our first strategy could address this situation and prioritise more safe candidates. The repositioning experiments verified this point to some extent. During disease development, some biological processes will be perturbed and only subset of the related genes are abnormally regulated. Then, a drug may perturb the same set of biological processes but target different genes. In this case, the relevance of differentially expressed genes under disease state and drug treatment is very small and some differential expression profile‐based methods may fail. Our second strategy could address this case. To some extent, the repositioning experiments show that the second strategy indeed could predict candidates missed by the first strategy. Collectively, our preliminary results show that various functional information could complement each other and our new framework works well in repositioning drugs against prostate cancer.
However, there are several issues that limit the repositioning performance. First, the number of differentially expression genes that constitute disease signature or drug signature were chosen empirically, which cannot guarantee the resulted signature's biological relevance. Second, the directions in which biological processes perturbed under disease state and drug treatment were not fully taken into account. We will develop parameter optimisation model and design new scheme to measure the degree and direction of functional profile simultaneously. In addition, it is promising to evaluate functional relevance from other aspects and improve the performance in the near future. Finally, the framework is general by only relying on differential expression genes and functional annotations. As long as high quality molecule activity data for diseases is available, it could be employed to reposition drugs against other diseases. In addition, it is also a future topic to integrate information of not only network [35–41] but also dynamics [42, 43] of biomolecules for improving the effectiveness and efficiency of drug repositioning.
5 Acknowledgments
ZW is supported by PhD start funds of University of Shanghai for Science and Technology (Grant no. 1D‐10‐303‐003), Science Foundation for the Youth Scholars by Educational Commission of Shanghai (Grant no. 51‐12‐303‐105). YW is supported by NSFC under Grants nos. 11131009, 61171007. LC is supported by NSFC under Grants nos. 91029301, 61134013 and 61072149; and by the Chief Scientist Program of Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences with Grant no. 2009CSP002; by Shanghai Pujiang Program; and by the FIRST program from JSPS initiated by CSTP. The authors wish to gratefully thank Dr. Xingming Zhao for helpful discussion.
6 References
- 1. Mullard A.: ‘2011 FDA drug approvals’, Nat. Rev. Drug Discovery, 2012, 11, pp. 91–94 (doi: 10.1038/nrd3657) [DOI] [PubMed] [Google Scholar]
- 2. Ashburn T.T., and Thor K.B.: ‘Drug repositioning: identifying and developing new uses for existing drugs’, Nat. Rev. Drug Discovery, 2004, 4, pp. 673–683 (doi: 10.1038/nrd1468) [DOI] [PubMed] [Google Scholar]
- 3. Sirota M. Dudley J.T., and Kim J. et al.: ‘Discovery and preclinical validation of drug indications using compendia of public gene expression data’, Sci. Transl. Med., 2011, 3, (96), pp. 96ra77 (doi: 10.1126/scitranslmed.3001318) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aronson J.: ‘Old drugs‐new uses’, Br. J. Clin. Pharmacol., 2007, 64, (5), pp. 563–565 (doi: 10.1111/j.1365-2125.2007.03058.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wu Z. Wang Y., and Chen L.: ‘Network‐based drug repositioning’, Mol. BioSyst., 2013, 9, (6), pp. 1268–1281 (doi: 10.1039/c3mb25382a) [DOI] [PubMed] [Google Scholar]
- 6. Lum P. Derry J., and Schadt E.: ‘Integrative genomics and drug development’, Pharmacogenomics, 2009, 10, (2), pp. 203–212 (doi: 10.2217/14622416.10.2.203) [DOI] [PubMed] [Google Scholar]
- 7. Luo H. Chen J., and Shi L. et al.: ‘DRAR‐CPI: a server for identifying drug repositioning potential and adverse drug reactions via the chemical‐protein interactome’, Nucleic Acids Res., 2011, 1, (7), pp. W492–W498 (doi: 10.1093/nar/gkr299) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yang L. Wang K., and Chen J. et al.: ‘Exploring off‐targets and off‐systems for adverse drug reactions via chemical‐protein interactome‐clozapine‐induced agranulocytosis as a case study’, PLoS Comput. Biol., 2011, 7, (3), pp. e10002016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schadt E. Friend S., and Shaywitz D.: ‘A network view of disease and compound screening’, Nat. Rev. Drug Discovery, 2009, 8, pp. 286–295 (doi: 10.1038/nrd2826) [DOI] [PubMed] [Google Scholar]
- 10. Qu X.A. Gudivada R.C. Jegga A.G. Neumann E.K., and Aronow B.J.: ‘Inferring novel disease indications for known drugs by semantically linking drug action and disease mechanism relationships’, BMC Bioinf., 2009, 10, (S5), pp. S4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Keiser M.J. Setola V., and Irwin J.J. et al.: ‘Predicting new molecular targets for known drugs’, Nature, 2009, 462, pp. 171–181 (doi: 10.1038/nature08506) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xie L. Li J. Xie L., and Bourne P.: ‘Drug discovery using chemical systems biology: identification of the protein‐ligand binding network to explain the side effects of CETP inhibitors’, PLoS Comput. Biol., 2009, 5, (5), pp. e1000387 (doi: 10.1371/journal.pcbi.1000387) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kinnings S.L. Liu N. Buchmeier N. Tonge P.J. Xie L., and Bourne P.E.: ‘Drug discovery using chemical systems biology: repositioning the safe medicine comtan to treat multi‐drug and extensively drug resistant tuberculosis’, PLoS Comput. Biol., 2009, 5, (7), pp. e1000423 (doi: 10.1371/journal.pcbi.1000423) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang L., and Agarwal P.: ‘Systematic drug repositioning based on clinical side effects’, PLoS One, 2011, 6, (12), pp. e28025 (doi: 10.1371/journal.pone.0028025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jin G. Fu C. Zhao H. Cui K. Chang J., and Wong S.: ‘A novel method of transcriptional response analysis to facilitate drug repositioning for cancer therapy’, Cancer Res., 2012, 72, (1), pp. 33–44 (doi: 10.1158/0008-5472.CAN-11-2333) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gottlieb A. Stein G. Ruppin E., and Sharan R.: ‘PREDICT: a method for inferring novel drug indications with application to personalized medicine’, Mol. Syst. Biol., 2011, 7, pp. 496 (doi: 10.1038/msb.2011.26) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lamb J. Crawford E.D., and Peck D. et al.: ‘The connectivity map: using gene expression signatures to connect small molecular, genes and disease’, Science, 2006, 313, (5795), pp. 1929–1935 (doi: 10.1126/science.1132939) [DOI] [PubMed] [Google Scholar]
- 18. Iorio F. Bosotti R., and Scacheri E. et al.: ‘Discovery of drug mode of action and drug repositioning from transcriptional responses’, Proc. Natl. Acad. Sci. USA., 2010, 107, (33), pp. 14621–14626 (doi: 10.1073/pnas.1000138107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hu G., and Agarwal P.: ‘Human disease‐drug based on genomic expression profiles’, PLoS One, 2009, 4, (8), pp. e6536 (doi: 10.1371/journal.pone.0006536) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shigemiu D. Hu Z.J. Hung J.H. Huang C.L. Wang Y.J., and DeLisi C.: ‘Using functional signatures to identify repositioned drugs for breast, myelogenous leukemia and prostate cancer’, PLoS Comput. Biol., 2012, 8, (2), pp. e1002347 (doi: 10.1371/journal.pcbi.1002347) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hao Y. Yang L.L. Cao Z.W. Tang K.L., and Li Y.X.: ‘A pathway profile‐based method for drug repositioning’, Chin. Sci. Bull., 2012, 57, (17), pp. 2106–2112 (doi: 10.1007/s11434-012-4982-9) [DOI] [Google Scholar]
- 22. Zhao S.W., and Li S.: ‘A co‐module approach for elucidating drug‐disease associations and revealing their molecular basis’, Bioinformatics, 2012, 28, (7), pp. 955–961 (doi: 10.1093/bioinformatics/bts057) [DOI] [PubMed] [Google Scholar]
- 23. Hsiao T.H. Chen H.H. Chen Y. Chen Y.H., and Chuang E.Y.: ‘Using gene sets to identify putative drugs for breast cancer’. Proc. IEEE Int. Conf. on Bioinformatics and Biomedicine, Philadelphia, USA, October 2012, pp. 1–4 [Google Scholar]
- 24. Barrett T. Suzek T.O., and Troup D.B. et al.: ‘NCBI GEO: mining millions of expression profiles‐databases and tools’, Nucleic Acids Res., 2005, 33, (Database issue), pp. D562–D566 (doi: 10.1093/nar/gki022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen R. Li L., and Butte J.: ‘AILUN: reannotating gene expression data automatically’, Nat. Methods, 2007, 4, (11), pp. 879 (doi: 10.1038/nmeth1107-879) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang R. Ou H.Y., and Zhang C.T.: ‘DEG: a database of essential genes’, Nucleic Acids Res., 2004, 32, (s1), pp. D271–D272 (doi: 10.1093/nar/gkh024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Durinck S. Moreau Y., and Kasprzyk A. et al.: ‘BioMart and bioconductor: a powerful link between biological databases and microarray data analysis’, Bioinformatics, 2005, 21, (16), pp. 3439–3440 (doi: 10.1093/bioinformatics/bti525) [DOI] [PubMed] [Google Scholar]
- 28. Durinck S. Spellman P.T. Birney E., and Huber W.: ‘Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt’, Nat. Protoc., 2009, 4, (8), pp. 1184–1191 (doi: 10.1038/nprot.2009.97) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. http://www.geneontology.org/, accessed October 2012
- 30. Subramanian A. Tamayo P., and Mootha V.K. et al.: ‘Gene set enrichment analysis: a knowledge‐based approach for interpreting genome‐wide expression profiles’, Proc. Natl. Acad. Sci. USA., 2005, 102, (43), pp. 15545–15550 (doi: 10.1073/pnas.0506580102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Smyth G.K.: ‘Linear models and empirical Bayes methods for assessing differential expression in microarray experiments’, Stat. Appl. Genet. Mol. Biol., 2004, 3, (1), pp. 3 [DOI] [PubMed] [Google Scholar]
- 32. Lipkus A.H.: ‘A proof of the triangle inequality for the tanimoto distance’, J. Math. Chem., 1999, 26, (1), pp. 263–265 (doi: 10.1023/A:1019154432472) [DOI] [Google Scholar]
- 33. Pham L. Christadore L. Schaus S., and Kolaczykc E.D.: ‘Network‐based prediction for sources of transcriptional dysregulation using latent pathway identification analysis’, Proc. Natl. Acad. Sci. USA, 2011, 108, (32), pp. 13347–13352 (doi: 10.1073/pnas.1100891108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. http://en.wikipedia.org/wiki/F1_score, accessed March 2012
- 35. He D. Liu Z. Honda M. Kaneko S., and Chen L.: ‘Coexpression network analysis in chronic hepatitis B and C hepatic lesion reveals distinct patterns of disease progression to hepatocellular carcinoma’, J. Mol. Cell Biol., 2012, 4, (3), pp. 140–152 (doi: 10.1093/jmcb/mjs011) [DOI] [PubMed] [Google Scholar]
- 36. Zhu H. Rao R. Liu J. Zeng T., and Chen L.: ‘Reconstructing dynamic gene regulatory networks from sample‐based transcriptional data’, Nucleic Acids Res., 2012, 40, (21), pp. 10657–10667 (doi: 10.1093/nar/gks860) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wen Z. Liu Z. Liu Z. Zhang Y., and Chen L.: ‘An integrated approach to identify causal network modules of complex diseases with application to colorectal cancer’, J. Am. Med. Inform. Assoc., 2013, 20, (4), pp. 659–667. doi: 10.1136/amiajnl‐2012‐001168 (doi: 10.1136/amiajnl-2012-001168) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu X. Liu Z. Zhao X., and Chen L.: ‘Identifying disease genes and module biomarkers with differential interactions’, J. Am. Med. Inform. Assoc., 2011, 19, (2), pp. 241–248 (doi: 10.1136/amiajnl-2011-000658) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang X. Zhao X., and He K. et al.: ‘Inferring gene regulatory networks from gene expression data by PC‐algorithm based on conditional mutual information’, Bioinformatics, 2012, 28, (1), pp. 98–104 (doi: 10.1093/bioinformatics/btr626) [DOI] [PubMed] [Google Scholar]
- 40. Zhang X. Liu K., and Liu Z. et al.: ‘NARROMI: a noise and redundancy reduction technique improves accuracy of gene regulatory network inference’, Bioinformatics, 2013, 29, (1), pp. 106–113 (doi: 10.1093/bioinformatics/bts619) [DOI] [PubMed] [Google Scholar]
- 41. Wang J. Sun Y. Zheng S. Zhang X. Zhou H., and Chen L.: ‘APG: an active protein‐gene network model to quantify regulatory signals in complex biological systems’, Sci. Rep., 2013, 3, pp. 1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen L. Liu R. Liu Z. Li M., and Aihara K.: ‘Detecting early‐warning signals for sudden deterioration of complex diseases by dynamical network biomarkers’, Sci. Rep., 2012, 2, pp. 342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu R. Li M. Liu Z. Wu J. Chen L., and Aihara K.: ‘Identifying critical transitions and their leading networks in complex diseases’, Sci. Rep., 2012, 2, pp. 813 [DOI] [PMC free article] [PubMed] [Google Scholar]


