Abstract
Glioblastoma (GBM) is the most common primary brain cancer in adults where tumor cell heterogeneity and sex differences influence clinical outcomes. Here, we functionally characterize three male and three female patient-derived GBM cell lines, identify pro-tumorigenic BTICs, and create novel male and female preclinical models of GBM. Cell lines were evaluated on the following features: proliferation, stemness, migration, tumorigenesis, clinical characteristics, and sensitivity to radiation, TMZ, rhTRAIL, and rhBMP4. All cell lines were classified as GBM according to epigenetic subtyping, were heterogenous and functionally distinct from one another, and re-capitulated features of the original patient tumor. In establishing male and female preclinical models, it was found that two male derived GBM cell lines (QNS108 and QNS120) and one female derived GBM cell line (QNS315) grew at a faster rate in female mice brains. One male derived GBM cell line (QNS108) decreased survival in female mice in comparison to male mice. However, no survival differences were observed for mice injected with a female derived cell line (QNS315). In summary, a panel of 6 GBM patient-derived cell lines were functionally characterized, and it was shown that BTIC lines can be used to construct sex-specific models with differential phenotypes for additional studies.
Keywords: Glioblastoma, Heterogeneity, Brain tumor-initiating cells, Sex Characteristics, Models, Animal
Introduction
Glioblastoma (GBM) is the most common and aggressive malignant brain tumor arising from the central nervous system among adult patients with a relative 1-year survival rate of 40.8% and 5-year survival rate of 6.8% for all ages (1). Although the 1-year survival rate for GBM has improved over the last decade, modern medical interventions remain ineffective for long-term survival where tumor resection coupled with chemo- and radiotherapy are the standard form of treatment for the disease (2–4).
It is well established that a subset of GBM cells, known as brain tumor-initiating cells (BTICs), are the perpetrators of recurrence and invasion throughout the brain parenchyma (5–8). Traditional identification of BTICs required that these cellular populations exhibit stem cell markers, tumorigenic capacity, and self-renewal potential (6, 8–12). It is now understood that BTICs are composed of several different cell types within the tumor bulk and that BTIC and non-BTIC populations work together within an encompassing environment to promote tumor growth (13–15). In addition, GBM cellular populations display inter- and intra-tumoral heterogeneity, which makes therapeutic targeting challenging (5, 7, 14, 16).
Recent findings have shown that sex differences play a major role in GBM treatment outcomes where female patients have a survival advantage over male patients (17–22). Soon after, it was shown that GBM tumor cells in men and women carry different molecular signatures which affect their tumorigenic behavior (17, 20, 23). It is likely that for future treatments to work against GBM, BTICs must be targeted in a population-specific and sex-specific manner where heterogeneity is accounted for.
Several systems used to classify or characterize GBM tumor cells and BTICs have been described, but they often rely solely on genetic and epigenetic classification to model cellular heterogeneity and the ability of these cells to initiate tumor formation (24–29). Although epigenetic and genetic classifications inform which genes can be targeted, the changing genetic or epigenetic landscape and presence of mutations provides alternative cellular mechanisms that can compensate for any lost function derived from targeting a specific pathway (7, 30–32). In terms of functional characteristics, description of BTIC tumorigenesis alone fails to account for the varied functional behaviors of different BTIC populations. Other functional features include metrics such as proliferation, migration, characteristics of the types of tumors formed, and varying degrees of stemness. Lastly, older methods of characterization have failed to account for sex differences as the majority of in vivo GBM preclinical models are comprised of male or female mice alone, instead of parallel male vs. female studies (12, 24, 29).
For these reasons, we chose to characterize our own collection of GBM tumor cells by incorporating three approaches of epigenetic subtyping, measurement of several functional behaviors, and preclinical modeling with both male and female mice. Our approach allows for better observation of the varied functional behaviors and sex differences that may be found among GBM tumor cell populations. Observations gleaned from our preclinical models will be used to improve our understanding of brain cancer and allow us to design novel therapeutic strategies better targeted towards GBM in a population-, functional-, and sex-specific manner.
Methods
All material, software, and equipment used in the following methods are listed in Supplementary Tables S1 and S2.
Clinical Characterization and Diagnosis:
All patient studies were approved by the Mayo Clinic, Jacksonville FL Institutional Review Board (IRB) and were done in accordance with U.S. Common Rule. Informed written consent was obtained from all patients included in the study. Clinical data and tissue were collected from patients undergoing surgical resection for glioblastoma. Prior to surgery, all patients underwent routine diagnostic and stereotactic magnetic resonance imaging (MRI) of the brain on a 3 Tesla (T) MAGNETOM Skyra (Siemens Healthineers AG, Erlangen, Germany) with exception of patient QNS120, who was scanned on a 1.5T Siemens MAGNETOM Espree. Tissue samples were collected intraoperatively and fixed in formalin. Tissue was stained with H&E for pathological diagnosis and with molecular diagnostic markers according to Mayo Clinic, FL protocols. Further details are listed in supplementary methods.
GBM Tumor Cell Line Isolation and Expansion from Tumor Tissue:
GBM tumor cells/BTICs were collected according to previously reported methods (9, 24, 33–36). In brief, GBM tumor cells were isolated from patient clinical samples, grown in non-adherent conditions for 3 passages and then expanded on adherent flasks. All cells were grown with GBM complete media (DMEM F/12, 10% Neuroplex w/o vitamin A, 5% anti-anti, growth factors: +hEGF (20 ng/ml), +hFGF (20 ng/ml)). GBM base media does not contain growth factors (-hEGF/hFGF). Images of neurospheres and adherent cells were taken at 10x magnification using a light microscope. Further details are listed in supplementary materials. Cells used for in vitro and in vivo experiments were maintained between 9–11 passages to help conserve the molecular and phenotypic characteristics of the original patient tumor.
Short Tandem Repeat (STR) Analysis for Cell Authentication:
As conducted previously by our group, patient-derived cell lines were analyzed using STR analysis to confirm authenticity and sex of GBM tumor cell lines (9, 24). DNA samples were sent to IDEXX BioAnalytics for analysis.
Epigenetic Subtyping:
A novel DNA-methylation classifier system has recently been described, with the goal of being used to standardize histopathological assessment of Central Nervous System (CNS) tumors (37). The same DNA-methylation classifier system was used to conduct epigenetic subtyping for GBM tumor cell lines. DNA was isolated using the Qiagen DNeasy® Blood & Tissue Kit protocol. DNA was processed with bisulfite sequencing and analyzed using the illumina® Infinium HumanMethylationEPIC 450K BeadChip array to generate genome-wide methylation profiles. Methylation profiles were compared to a CNS tumor reference cohort composed of 2,801 samples derived from 91 CNS tumor classes to identify the proper classification. In addition, a chromosome copy number variation profile (CNV) was generated for each GBM tumor cell line.
Differentiation Assay and Immunocytochemistry (ICC):
Immunocytochemistry was performed according to previously published methods (9). In brief, glass coverslips (15 mm2) were inserted in a 24 well plate, coated with poly-ornithine for 15 minutes at room temperature, and then coated with laminin for 1 hour at room temperature. Cells grown as spheres were dissociated into single cells with mechanical trituration and seeded at a density of 5×104 cells per coverslip. Cells adhered to the coverslip for 12 hours in stem cell media. The following day, media was removed, and cells were cultured in either stem cell promoting GBM complete media (+hEGF/hFGF) or differentiation promoting GBM base media (-hEGF/hFGF) for 10 days. Media was changed every 3–4 days. At the end of treatment, cells were washed 1x with PBS, fixed with 4% paraformaldehyde for 20 minutes, and washed again with PBS. After fixing, cells were blocked with a 0.1% Triton/PBS and 10% Normal Goat Serum (NGS) solution for 1 hour at room temperature on a gentle shake cycle. Primary antibodies for Glial Fibrillary Acidic Protein (GFAP), Anti-Beta-Tubulin III protein (TUBB3) clone referred to as Tuj1, and Nestin (NES) were diluted (1:500) in 0.1% Triton/PBS with 2% NGS. Cells in all conditions were co-stained with GFAP and Nestin antibodies or with GFAP and Tuj1 antibodies overnight at 4°C. The following day, primary antibodies were removed, and cells were washed 3x with 0.1% Triton/PBS for 10 minutes per wash. Secondary antibodies, Alexa Fluor ® 488 goat anti-mouse IgG and Alexa Fluor ® 594 goat anti-rabbit IgG were diluted together (1:500) in 0.1% Triton/PBS and added to the cells for 1 hour at room temperature in the dark on a gentle shake cycle. After secondary staining, cells were washed 3x with 0.1% Triton/PBS with 10-minute washes. Cover slips were mounted onto glass slides using DAPI-mounting media. Cells were imaged using a Zeiss LSM 800 Confocal Microscope at 10x magnification. All images were analyzed with ZEN 2.3 System software. Expression of each marker was quantified by measuring fluorescence intensity and correcting for background fluorescence in each image with ImageJ (38). Five representative high-powered fields were analyzed for Tuj1 and Nestin expression and ten representative high-powered fields were analyzed for GFAP expression. The Corrected Total Cell Fluorescence (CTCF) was calculated with the following equation: CTCF = Integrated Density – (Area of selected cell x Mean fluorescence of background readings), as described previously (38).
Sphere Forming Unit (SFU) Assay:
GBM tumor cells were suspended as single cells in 6-well plates at a density of 500 cells/well in triplicate. Single cells grew in non-adherent conditions in stem cell media for 14 days. The number and size of spheres were measured under phase contrast microscopy at 10x magnification. Spheres were counted using 50 μm as the cutoff longest diameter for consideration as a distinct sphere as previously reported (39). Sphere sizes were obtained by measuring the longest diameter of the spheres.
Measurement of CD133 Status with Flow Cytometry:
CD133 expression in BTICs was determined using flow cytometry. Cells were harvested and re-suspended in 2% FBS containing PBS. A total of 5×105 cells were labeled with PE-conjugated CD133/1 Miltenyi antibody (PROM1), (1:20) for 30 min at 4°C. Cells were washed and re-suspended in 2% FBS containing PBS. Flow cytometric data was collected using the Attune NxT (Invitrogen) and was analyzed using Attune NxT software and FlowJo 10.6 software. A minimum of 1.0×104 live cell events were gated per sample. Mouse IgG1 PE (Miltenyi biotec) was used as an isotype control for flow cytometry experiments.
Transwell Migration Assay:
The relative migration capacity of primary GBM cell lines was measured using the Boyden chamber assay as previously described (40, 41). Briefly, CoStar© 6.5 mm inserts containing an 8.0 μm pore polycarbonate membrane was used as a scaffold. A total of 3×104 cells were seeded in the top chamber of the insert and were incubated at 37°C in 5% CO2 for a period of 24 hours in three replicates. An FBS gradient of 2% was established for each group. After 24 hours of incubation, the inserts were washed with PBS and cells were fixed with 4% PFA. The upper side of the insert was swabbed to remove cells that did not migrate through the membrane. The migrated cells attached to the opposite side of the membrane were then permeabilized using a PBS/Triton solution. The membranes were cut, mounted, and stained with DAPI. Membranes were visualized at 10x magnification using the Zeiss LSM 800 Confocal microscope. The number of migrating cells were quantified by examining 9 random high-powered fields per membrane which were analyzed using Arivis Vision4D x64 3.0 software. All experiments were performed in triplicate.
Extreme Limiting Dilution Assay:
Cells were isolated as single cells and seeded via FACS cell seeding at different concentrations of 100, 50, 25, 12, 6, and 3 cells per well (10 wells per condition) in 200μL of complete GBM media in ultra-low attachment 96 well plates (Corning Life Sciences, Costar, Corning, NY, USA). Plates were centrifuged at 300xg for 5 minutes at 4°C and incubated at 37°C in 5% CO2 right after. Cells were incubated for 21 days and 25μL of GBM complete media was added every 5 days to each well. For this assay, spheroids were defined as homogeneous and rounded aggregates of cells measuring at least 50μm in diameter with poor cell-to-cell definition and a smooth surface. After the incubation period, spheroids were counted in each well and analysis was carried out utilizing the online extreme limiting dilution assay (ELDA) webtool (http://bioinf.wehi.edu.au/software/elda/) (42). The percentage of wells negative for spheroids for each plating density was calculated and plotted against the number of cells seeded per well. The ELDA experiments were carried out in triplicate.
Growth Kinetic Assay:
GBM tumor cell in vitro growth kinetics were measured using the BioSpa 8 automated incubator and Cytation™ 5 microplate reader (BioTek®). GBM tumor cell lines were seeded at a density of 1000 cells/well in 96-well laminin coated plates with 48 replicates per cell line. Cells adhered for 4 hours before being inserted into the automated incubator. Each cell line was monitored for 5 days, with images taken by the Cytation™ 5 microplate reader (BioTek®) every 12 hours. Cell number was counted at each time point and analyzed with Gen5™ software (v. 3.4) to calculate doubling time. Growth rate was plotted over the course of five days. An exponential growth equation with a least-squares fit model was used to generate the growth curves and a comparison of fits was performed to determine statistical significance.
Therapeutic Sensitivity Assay with Temozolomide (TMZ), Radiation, rhBMP4, and rhTRAIL:
To test the effects of standard and novel therapeutics used to treat GBM, temozolomide (TMZ), radiation, recombinant bone morphogenetic protein 4 (rhBMP4), and recombinant human TNF-related apoptosis-inducing ligand (TNFSF10), referred to as (rhTRAIL), were administered in several doses to a panel of GBM tumor cell lines (2, 41, 43–46). The therapeutics rhTRAIL and rhBMP4 were chosen as they are emerging therapies shown to have anti-tumor effects against GBM (41, 43–49). Cells were seeded on laminin-coated 96-well plates in replicates of 6 for each condition, with a density of 1.0×104 cells per well. Cells were adhered for 12 hours in GBM complete media before applying treatment. Fresh media was added to wells prior to treatments. Cells were treated in a dose-dependent manner. Cells were irradiated in doses of 2 Gy, 5 Gy, and 10 Gy, with a control group receiving no treatment. Radiotherapy was applied by using the X-RAD SmART© irradiator with a 40 mm2 collimator. For the TMZ dosing regimen, the drug was dissolved in DMSO to make a stock solution of 33 mg/ml. Stock TMZ solution was added to GBM complete media to form working solutions that were administered to cells in doses of 250 μM, 500 μM, 750 μM, and 1000 μM with a control group that received GBM complete media with 0.6% v/v DMSO. For rhTRAIL and rhBMP4 treatments, the protein was dissolved in GBM complete media and administered to cells in concentrations of 25, 50, 75, and 100 ng/ml as reported previously (41, 43–47, 49). Cells were grown for 4 days post treatment, before removing media and freezing plates at −80°C. Cell line viability was measured with the CyQUANT® Cell Proliferation Assay Kit according to the manufacturer’s protocol. A cell number standard curve was generated using QNS120 tumor cells to establish a linear detection range from 0 to 50,000 cells. The Synergy HTX multi-mode reader was used to measure fluorescence after adding the CyQUANT® reagent to the wells to detect the nuclei of total cells present (480 nm excitation, 520 nm emission maxima).
Transduction of Target Cells:
GBM tumor cells were transduced with a lentivirus to express Green Fluorescent Protein and luciferin (GFP/Luc) for in vivo experiments. Preparation of lentiviral particle production and titration are in supplementary methods. Patient-derived GBM tumor cells were plated in 6 well plates (0.5×106 cell per well). Lentiviral particles (LPs) were added directly to the culture media plus 4 μg/mL of polybrene (Sigma-Aldrich). Different multiplicity of infections (MOÍS) ranging between 1 and 3, were used for the transduction according to each primary cell culture. Cells were centrifuged at 1200xg, 60 min, 35°C, and incubated with LPs for 48h. Transduction media was then replaced with fresh GBM complete media to remove the virus and allow the cells to express the reporter gene (EGFP) for transduction efficiency calculations using confocal microscopy. After transduction, cells were allowed to expand until enough was available for intracranial injections.
BTIC Tumorigenicity and in vivo Modeling:
All animal procedures were approved by Mayo Clinic, Jacksonville FL, IACUC. Patient-derived GBM tumor cells between passage 9–11 were implanted intracranially in 6-week old male and female (n=5 male mice, n=5 female mice per cell line for a total of 10) athymic nude mice to assess tumor forming ability, as described previously by our group (41, 45, 50). Cells implanted in mice were transduced before injection and expressed GFP/Luc. Luciferin was prepared at a concentration of 15 mg/ml and 0.1 ml luciferin/gram (mouse bodyweight) was injected intraperitoneally (IP) ten minutes before imaging. Tumor growth was tracked with weekly bioluminescent (BLI) measurements of the gbm tumor cell luciferase/luciferin signal with the IVIS® Spectrum XENOGEN for a total of 15–17 weeks or until the first mouse in a cohort reached humane endpoint. At the end of in vivo studies, mouse brains were collected and preserved in 4% PFA and prepared for histology and confocal microscopy. Brains were imaged at 10x under confocal microscopy to detect GFP+ tumor cells. Brains were stained with H&E and with Nestin, GFAP, Human-Nuclei, and Ki-67 (MKI67) antibodies. Further details are included in supplementary methods.
Survival Study:
Following prior protocols, animal survival was tracked over the course of the study to determine the effect of tumor burden and aggression on overall health of mice across injected cell lines and sexes (36, 41). In brief, mice were euthanized through intracardiac perfusion using 30% saline to flush out blood and with 4% PFA to fix tissue. Brains were collected for confocal imaging and H&E staining. A Kaplan-Meier (KM) analysis was performed for each cell line model and survival was compared between male and female mice. Two mice that were identified as outliers based on BLI growth kinetics or death not due to tumor burden were excluded from the KM survival analysis of QNS108 and QNS315. The final sample size for the KM analysis of QNS108 and QNS315 was n=4 per male and female group.
Statistical Analysis:
All statistical analysis was performed using GraphPad PRISM 8.0.0 (© 2018 GraphPad Software). One-way ANOVA with Turkey’s multiple comparisons test was performed to analyze migration capacity and SFU assays. Unpaired t-tests were performed to compare fluorescence intensity of ICC images. For the ELDA assay, an online webtool (http://bioinf.wehi.edu.au/software/elda/) was used to conduct pair-wise comparisons, overall test for differences, and goodness of fit tests for stem cell frequencies of each GBM cell line as described previously (42).The KM survival analyses performed to generate survival curves were compared with the log-rank test. Growth kinetics were modeled and compared using nonlinear regression following an exponential growth equation with a least square fit. p-values <0.05 were considered significant.
Results
Cell Line Authentication:
The patient-derived GBM tumor cell lines (QNS108, QNS120, QNS140, QNS166, QNS315) were authenticated with STR fragment analysis and found to have a unique identity and sex was determined (Supplementary Fig. S1).
Patient Characteristics and Epigenetic Subtyping:
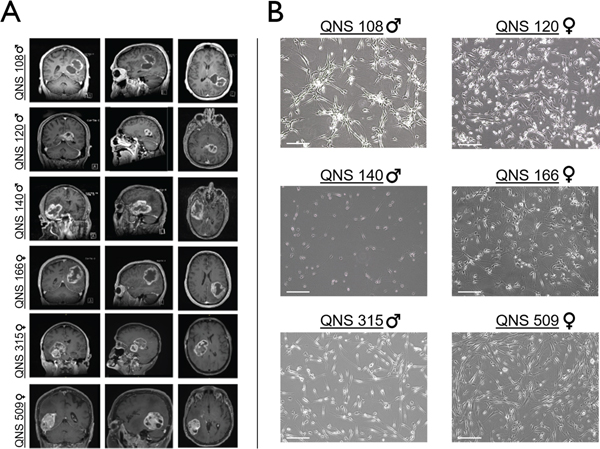
Six GBM tumor cell lines were collected in total. GBM cell lines QNS108, QNS120, and QNS140 were derived from male patients ranging in age from 59–64 years while cell lines QNS166, QNS315, and QNS509 were derived from female patients ranging in age from 59 −75 years. Clinical pathology indicated that isolated tumor tissue retained ATRX expression for all patients. All patients retained intact p53 expression. Tumor tissue from patients QNS108, QNS166, and QNS509 were MGMT methylation-negative while patients QNS120, QNS140, and QNS315 were MGMT methylation-positive (Supplementary Table S3). Pre-operative axial, sagittal, and coronal contrast enhanced T1-weighted magnetic resonance imaging (MRI) was taken for each patient (Fig. 1A). All lesions showed an infiltrative appearance with ring-enhancement and central necrosis (Fig. 1A). There was diffusion restriction present within the solid portions of the lesions (mean 831×10−6 mm2/s; range 586–1079). There was extension to the ependymal surface in all cases, with patient QNS120 showing macroscopic subependymal spread (Fig. 1A). None had imaging findings of leptomeningeal dissemination or multifocal parenchymal lesions. All tumors were located proximal to the lateral ventricles (Fig. 1A). Patient QNS108 presented with two lesions located in the left parietal and temporal lobes, where the primary tumor lies adjacent to a cystic formation. Patient QNS120 displayed a butterfly pattern of invasion spread across the left parietal, temporal, and occipital lobes with extension into the right hemisphere across the splenium of the corpus callosum. Patients QNS509 and QNS166 presented with lesions as large masses containing necrotic cores located in the right temporal and left inferior- parietal lobes respectively (Fig. 1A). Each GBM tumor cell line isolated from patient tumor tissue was successfully grown as oncospheres, one feature of stemness (6, 24), and exhibited unique morphologies when expanded as attached cells on adherent surfaces (Fig. 1B).
Figure 1. Preoperative clinical MRI of patient tumors and patient-derived GBM tumor cell morphologies.
A) Coronal, sagittal, and axial preoperative MRI depicting patient tumor characteristics. B) Light microscopy images demonstrating patient-derived GBM tumor cell morphologies. Scale bar = 50μm.
In addition to using patient pathology to establish a tumor diagnosis, a novel epigenetic subtyping system developed to automate tissue diagnosis of CNS tumors was incorporated into the characterization of patient-derived GBM tumor cells (37). The epigenetic subtyping analysis and CNV profiles of the cellular DNA matched the original patient pathological reports with regards to GBM IDH-WT diagnosis (Supplementary Fig. S2). However, the MGMT methylation status of two female GBM cell lines (QNS166 and QNS509) was discordant with what was reported for original patient pathology. The discordance may be a result of intratumoral heterogeneity or a result of methylation status changing during cell line expansion from original patient tissue (Supplementary Table S4). Each cell line was primarily classified as a Classical/RTK2 subtype GBM based on the reported cutoff score of at least >= 0.5 to indicate a match. However, each cell line exhibited a mixture of another subtype with varied cutoff scores that fell below 0.5 (Supplementary Table S4). For QNS108, QNS140, QNS166, and QNS509 the secondary classification was Proneural/RTK1, with cutoff scores of 0.1, 0.24, 0.11, and 0.14 respectively. QNS120 and QNS315 exhibited a secondary classification with a mesenchymal subtype at a cutoff score of 0.16 and 0.05 respectively (Supplementary Table S4).
Assessment of Stem-like Characteristics:
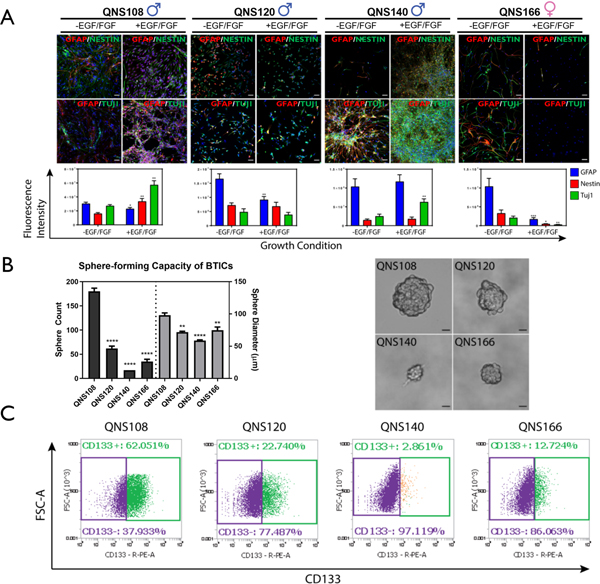
After cell line expansion, four GBM tumor cells were assessed for stem-like characteristics, multipotentiality, and differentiation potential. ICC staining and confocal imaging was used to detect the expression of stem cell markers. GBM tumor cell lines exhibited an aberrant co-expression of astrocytic (GFAP), neuronal (Tuj1), and stem-like (Nestin) markers, as has been observed by others as a distinct characteristic exhibited by BTICs (6, 7, 51). QNS108 expressed all three markers when grown in differentiation promoting GBM base (-EGF/FGF) media with a similar degree of expression (Fig. 2A). When grown in stem cell promoting GBM complete (+EGF/FGF) media, all markers were still co-expressed for QNS108, but expression of Nestin and Tuj1 was elevated (Nestin p=0.0039**, TUJ1 p=0.0012** compared to expression in -EGF/FGF media) while expression of GFAP was decreased (GFAP p=0.0170* compared to expression in -EGF/FGF media), (Fig. 2A). QNS120 displayed greater expression of GFAP in GBM base (-EGF/FGF) media (p = 0.0024** compared to expression in +EGF/FGF media), (Fig. 2A), but otherwise had similar co-expression and morphology between the two culture conditions with no change in the expression of Nestin. QNS140 had unique morphology with small, compact cell bodies containing axon-like projections that clustered into tangled fibers (Fig. 1B; Fig. 2A). Only TUJ1 expression varied for QNS140, with higher expression in GBM complete (+EGF/FGF) media (p=0.0096** compared to expression in -EGF/FGF media), (Fig. 2A). QNS166 expressed the three markers in GBM base (-EGF/FGF) media but had depleted expression of those markers in GBM complete (+EGF/FGF) media (GFAP p<0.001***, TUJ1 p=0.0022**, Nestin p=0.0106* compared to expression in -EGF/FGF media), (Fig. 2A). The sphere forming capacity, a characteristic feature of stem cells, was measured and compared between four of the GBM tumor cell lines by performing a sphere forming unit (SFU) assay. Cells were seeded at a density of 500 cells in six well plates and allowed to grow over two weeks. Spheres that formed in each well were counted. The number of spheres formed by QNS108 (180 SFU) was significantly greater than three other cell lines followed by QNS120 (62 SFU), QNS166 (35 SFU), and QNS140 (17 SFU) (p<0.0001****), (Fig. 2B; Supplementary Table S5). QNS108 also formed the largest spheres with an average diameter of 98.06 μm (p<0.0001**** vs QNS140, p<0.01** vs QNS120 and QNS166). QNS120 (71.86 μm) and QNS166 (74.87 μm) formed similar sized spheres with QNS140 (58.82 μm) growing the smallest. Flow cytometry was performed to measure CD133 expression of GBM tumor cell lines and compared to a mouse IgG1 isotype control (Supplementary Fig. S3). It was observed that CD133 expression, a feature of stemness which was measured with flow cytometry, was proportional to relative sphere counts, but not sphere size, for four cell lines. CD133 expression was highest for QNS108 (62.05%) followed by QNS120 (22.74%), QNS166 (12.72%), and QNS140 (2.86%), (Fig. 2C; Supplementary Table S5).
Figure 2. Differential stem-like properties of GBM tumor cells.
A) Fluorescence intensity and expression of neuronal, astrocytic and stem markers in growth factor depleted (-EGF/FGF) and growth factor enriched media (+EGF/FGF). Scale bar = 50 μm. B) Relative sphere-forming capacity of GBM tumor cells. Sphere count and sphere diameter of each group was compared to QNS108. Scale bar = 20 μm. C) Expression of the stem marker CD133 for each GBM tumor cell line. α = 0.05, *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
Stem Cell Frequency:
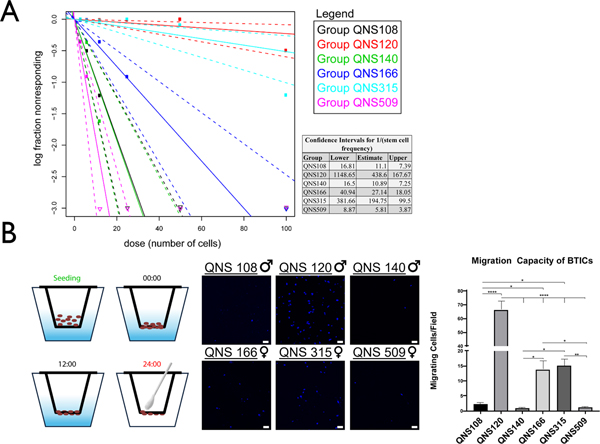
In addition to the SFU assay, the stem cell frequency for all six cell lines was measured with an ELDA assay using an online webtool (http://bioinf.wehi.edu.au/software/elda/) (42). Cells were plated over a concentration gradient ranging from 3–100 cells per well and the stem cell frequency was compared between cell lines. The relative order of greatest to lowest stem cell frequency per cell line was: QNS509, QNS140, QNS108, QNS166, QNS315, and QNS120 (Fig. 3A).
Figure 3. Measure of stem cell frequencies and tumor cell migration.
A) Extreme limiting dilution analysis was conducted with the online webtool (http://bioinf.wehi.edu.au/software/elda/) to measure relative stem cell frequencies for each cell line. The inverse stem cell frequency (1/stem cell frequency) and confidence intervals for each cell line were calculated. B) Relative tumor cell line migration was measured with a transwell assay over a 24-hour time frame and presented as number of migrating cells per field of view. Migration capacity was compared with a 1-way ANOVA. All experiments were done in triplicate. α = 0.05, *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
Migratory Capacity:
The relative migration was measured and compared between each GBM tumor cell line described in the current study. Cells were seeded in a two chamber Transwell and allowed to migrate from a top chamber to a lower chamber over a 24-hour incubation period. The order of relative migratory ability between BTIC lines from greatest to least was QNS120 (mean: 66.36 migrated cells), QNS315 (mean: 15.07 migrated cells), QNS166 (mean: 13.78 migrated cells), QNS108 (mean: 2.22 migrated cells), QNS509 (1.07 migrated cells), and QNS140 (0.8 migrated cells), (Fig. 3B; Supplementary Table S5).
BTIC Therapeutic Sensitivity:
The therapeutic response for each GBM tumor cell line to standard of care (TMZ and radiation) and experimental treatments (rhBMP4, rhTRAIL) was tested at different concentrations as was done previously (41, 44–46, 49). GBM tumor cell lines responded differently to administered treatments. The order of sensitivity to TMZ was: QNS166 (IC50 = 670 μM) > QNS120 (IC50 = 708.8 μM) > QNS108 (IC50 = >1000 μM) = QNS140 (IC50 = >1000 μM) (Supplementary Fig. S4A; Supplementary Table S5). The order of sensitivity to radiation was: QNS120 (IC50 = 6.582 Gy) > QNS140 (IC50 = >10 Gy) = QNS166 (IC50 = >10 Gy) > QNS108 (IC50 = N/A, could not be calculated), (Supplementary Fig. S4B; Supplementary Table S5). GBM tumor cell lines displayed resistance towards rhBMP4 and rhTRAIL. The IC50 values were not detectable for QNS108 and QNS120 against rhBMP4 and QNS140 and QNS166 had large IC50 values of >100 ng/ul (Supplementary Fig. S4C; Supplementary Table S5). The IC50 values were undetectable for all cell lines against rhTRAIL (Supplementary Fig. S4D; Supplementary Table S5).
Growth Kinetics Observed in vitro and across Sex in vivo:
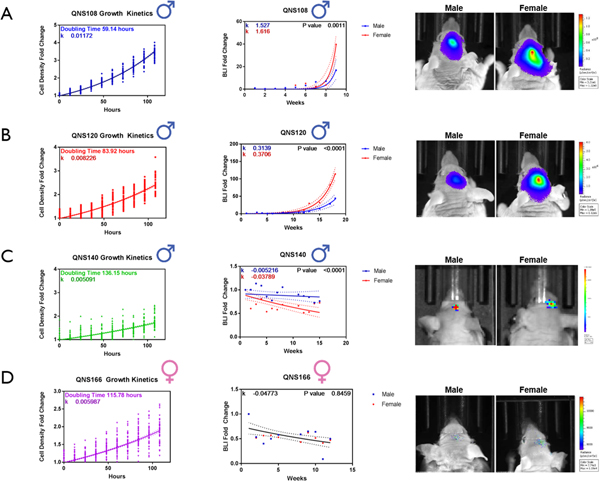
The in vitro growth kinetics were measured for 4 GBM tumor cell lines. Cells were seeded in 96 well-plates at a density of 1000 cells/well, and cell number per well was recorded by an automated plate reader every 12 hours over 5 days. QNS108 exhibited the fastest rate of growth (59.14 hr doubling time), followed by QNS120 (83.92 hr doubling time). QNS166 had the third fastest rate of growth (115.78 hr doubling time) and QNS140 had the slowest rate of growth (136.15 hr doubling time), (Fig. 4). The rate of in vitro growth was significantly different between the four cell lines tested (p<0.0001****). (Supplementary Table S5).
Figure 4. Brain Tumor Initiating Cells (BTICs) demonstrate differential growth kinetics in vitro that mimic tumor growth rates in vivo.
BTICs engineered to express GFP/luciferin were intracranially injected into male and female nude mice to assess tumorigenic capacity. Tumor growth was recorded by measuring the bioluminescence (BLI) signal every week post-injection until first animal death or end of study at week 15–17. BLI signal was plotted as the average fold change from week 1 post-injection for A) QNS108 B) QNS120 C) QNS140 D) QNS166. P values indicate the similarity or difference in growth rate between sex in each BTIC line. α = 0.05, *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
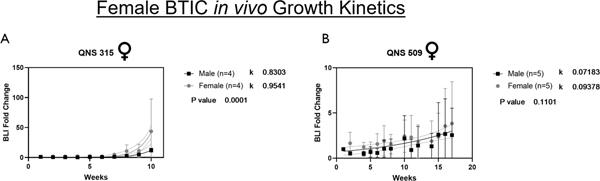
The in vivo growth kinetics were calculated through weekly IVIS imaging to record BLI signal of mice intracranially implanted with (GFP/Luc+) GBM tumor cells. The relative in vitro growth kinetics for the four GBM tumor cell lines measured matched their relative in vivo growth kinetics (Fig. 4A–D). The in vivo growth rate for two additional female derived cell lines (QNS315 and QNS509) was measured and compared to the original four. For in vivo tumor growth kinetics, the rate of growth was significantly greater for female mice vs. male mice, when injected with male derived QNS108 and QNS120 GBM tumor cells (QNS108: p = 0.0011***; QNS120: p < 0.001***), (Fig. 4A–B). Similarly, the in vivo tumor growth rate was significantly greater in female mice vs. male mice when injected with female derived QNS315 GBM tumor cells (p=0.0001****), (Fig. 5A). No growth differences were observed for mice injected with female derived QNS509 GBM tumor cells (Fig. 5B). The tumor growth rate after cell line implantation was greater in female mice for each instance where a significant difference was observed, regardless of the sex of the original patient donor.
Figure 5. Female derived GBM cell lines expressing GFP/luciferin were intracranially injected into male and female nude mice to assess tumorigenic capacity.
Tumor growth was recorded by measuring the bioluminescence (BLI) signal every week post-injection until first animal death or end of study at week 15–17. BLI signal was plotted as the average fold change from week 1 post-injection for A) QNS315 and B) QNS509. P values indicate the similarity or difference in growth rate between sex in each BTIC line. α = 0.05, *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
Overall, QNS108 had the fastest in vivo growth kinetics (male growth curve slope: k =1.527; female growth curve slope: k = 1.616) followed by QNS315 (male growth curve slope: k= 0.8303, female growth curve slope: k=0.9451), QNS120 (male growth curve slope: k = 0.3139; female growth curve slope: k = 0.3706), and QNS509 (male growth curve slope: k=0.07183, female growth curve slope: k=0.09378), while QNS166 and QNS140 displayed minimal BLI tumor signal overall (Fig. 4; Fig. 5; Supplementary Table S5).
BTIC Tumorigenesis:
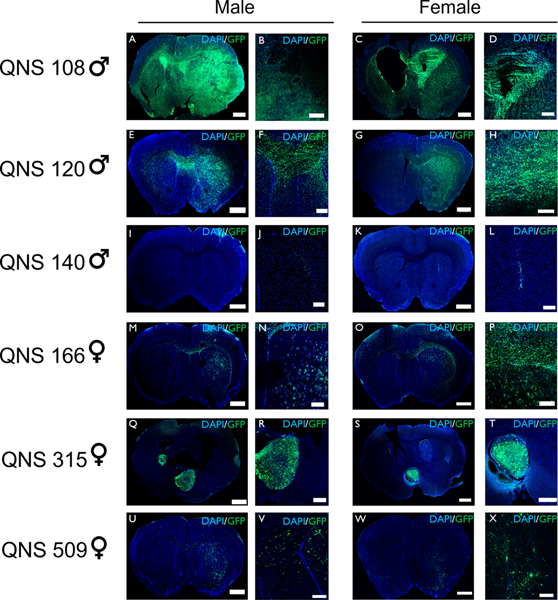
Following intracranial implantation into mice, it was shown that five GBM tumor lines (QNS108, QNS120, QNS166, QNS315, QNS509) exhibited tumor growth according to confocal imaging and were classified as BTICs whereas QNS140 displayed no tumor growth and was classified as a non-BTIC line. QNS108 tumor growth presented with invasion, proliferation, and cystic lesions spread throughout the brain. The cysts observed in murine QNS108 histology may also be indicative of ventricular enlargement (Fig. 6A–D; Supplementary Fig. S5; Supplementary Fig. S6). The appearance of cysts in murine brains matched the cystic formations observed in the original patient. Murine brains implanted with QNS108 tumors exhibited severe mass effect and midline shift (Fig. 6A–D). QNS120 tumorigenesis presented with a diffuse-migratory pattern with invasion into the opposite hemisphere across the corpus callosum (Fig. 6E–H). The QNS120 invasion pattern resembled a butterfly shape that matched the original patient tumor. QNS140 failed to initiate tumor growth, but some cells were able to survive at the site of injection even after 15 weeks indicated by weak fluorescent signal (Fig. 6I–L). QNS166 displayed little to moderate growth in discrete circular patches close to the injection site marked by a diffuse migratory spread rather than a tumor bulk (Fig. 6M–P). QNS315 displayed growth at multiple sites, including the lateral ventricle (Fig. 6Q–T). Growth for QNS509 was diffuse and spread out within the hemisphere of injection with no large tumor bulk forming (Fig. 6U–X). H&E histology staining was performed with tissue collected from in vivo murine studies and with original in situ patient tumor for all cell lines. H&E staining for tumor formed by QNS108 and QNS120 exhibited growth patterns like those observed with confocal imaging and in situ patient histology exhibited common features of GBM which included the presence of necrosis, mitotic figures, and invasion (Supplementary Fig. S5). In addition to H&E, murine tumor tissue for QNS108 and QNS120 was stained for human nuclei, Ki-67, Nestin, and GFAP. QNS108 and QNS120 tumors exhibited positive expression for all markers tested, demonstrating that the aberrant co-expression of stem cell and astrocytic markers by BTICs (Nestin and GFAP) was retained in vivo (Supplementary Fig. S6; Supplementary Fig. S7). However, little tumor formation was observed for H&E stains of QNS166 and QNS140 mouse brains, but in situ patient histology presented with necrosis, tumor cell invasion, and hypervascularity (Supplementary Fig. S8). Multiple lesions and tumor invasion of ventricles was observed in the H&E staining of QNS315 murine brains (Supplementary Fig. S9A–H). The QNS315 in situ patient histology presented with a primitive component, mitotic figures, and vascular proliferation (Supplementary Fig. S9I–K). The lack of clear tumor cells in H&E stains of QNS166 brains can be explained by the diffuse migratory spread of these cells and were only detected by identifying the GFP+ signal in confocal imaging. Similar observations to QNS166 were made for QNS509 murine histology (Supplementary Fig. S9L–S) The QNS509 in situ patient histology presented with necrosis, tumor cell infiltration, and vascular proliferation (Supplementary Fig. S9T–V).
Figure 6. BTICs displayed in vivo tumorigenesis and invasion.
Tumor cells were transduced to express EGFP (green) and nuclei were stained with DAPI (blue). A-D) QNS108 formed tumors with whole brain invasion and cystic lesions in male and female mice (scale bars: A=1000 μm, B=500 μm, C=1000 μm, D=500 μm). E-H) QNS120 formed tumors with butterfly invasion across the corpus callosum in male and female mice (scale bars: E=1000 μm, F=500 μm, G=1000 μm, H=500 μm). I-L) QNS140 (non-BTIC) formed no observable tumor mass in male and female mice (scale bars: I=1000 μm, J=500 μm, K=1000 μm, L=500 μm). M-P) QNS166 formed spotted patterns of tumor cells with migration across the corpus callosum in male and female mice (scale bars: M=1000 μm, N=500 μm, O=1000 μm, P=500 μm). Q-T) QNS315 grew within the lateral ventricles (scale bars: Q=1000 μm, R=500 μm, S=1000 μm, T=500 μm). U-X) QNS509 proliferated diffusely in the hemisphere of injection (scale bars: U=1000 μm, V=500 μm, W=1000 μm, X=500 μm).
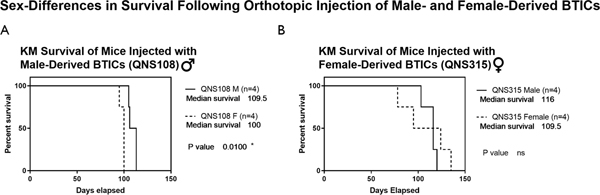
BTIC Tumor Aggression and Effect on Survival Across Sex:
After establishing orthotopic in vivo BTIC models, a KM survival analysis was performed for each cohort of male and female mice as a measure of BTIC tumor aggression. Of the cell lines tested, QNS108 and QNS315 caused animal death within a 15 to 17-week time course due to tumor burden. A sex-specific effect was observed for mice injected with QNS108, a male patient-derived cell line, where female mice died before male mice and a survival difference was significant (p=0.01*), (Fig. 7A). However, no difference in survival between male and female mice was observed when injected with the female derived QNS315 GBM cell line (Fig 7B).
Figure 7. Survival study of male and female mice injected with either male or female patient-derived BTICs.
A) Comparison of overall survival between male and female mice implanted with male patient-derived QNS108 where a survival difference was observed. B) Comparison of overall survival between male and female mice implanted with female patient-derived QNS315 where no survival difference was observed. α = 0.05, *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
Discussion
GBM is a disease marked by intra- and inter-tumoral heterogeneity that exists within an evolving tumor landscape (7, 31). Recently, it has been shown that sex-based differences are implicated as a major determinant affecting GBM tumorigenesis and clinical outcomes (17, 21). As such, patient sex is one variable among several that produce a complex interplay that influence GBM tumor growth. Other variables include cell populations, cell plasticity, and microenvironmental interactions (17, 19–21). The purpose of the present study was to functionally characterize our collected GBM tumor cell lines, classify tumorigenic BTIC populations, and construct novel male and female GBM preclinical models.
Study of Functional Behaviors and Heterogeneity Between Cell Lines:
To better define patient- derived GBM tumor cells, a series of functional behaviors per cell line were characterized and catalogued. It was observed through epigenetic subtyping that each cell-line possessed a CNV profile consistent with GBM tumor cells (Supplementary Fig. S2). It was observed that MGMT methylation was discordant between patient histology and subtyping results for two cell lines (QNS166 and QNS509) (Supplementary Table S4). The discordance illustrates how GBM tumor cell populations can vary within the tumor bulk or how epigenetic alterations can occur when cells are expanded in culture. It is known that DNA methylation patterns can shift between original patient GBM tissue and cultured cancer stem cells (52). Of the four GBM tumor cell lines tested for stem cell markers, all expressed CD133 and Nestin, along with aberrant co-expression of Tuj1 and GFAP in some cases (Fig. 2). All cell lines had varied stem cell frequencies and sphere forming capacity (Fig. 3A). Surprisingly, cell lines that were most migratory also had the lowest stem cell frequency which may suggest that there is an inverse relationship that should be studied further (Fig. 3). In our panel of cell lines, 5 were identified as BTICs because they could form tumors in vivo. QNS140 was not a BTIC but was included as a point of comparison as a non-stem GBM tumor cell. Overall, cell lines exhibited differential migration, sphere forming capacity, stem cell frequencies, expression of stem cell markers, and response to therapies (Figs. 1–3; Supplementary Fig. S4). It should be noted that several cell lines were resistant to conventional (TMZ, radiation) and experimental therapy (rhBMP4, rhTRAIL), as in several cases the IC50 values could not be calculated or had to be estimated from the therapeutic response curve (Supplementary Fig. S4). Cancer stem cells are known to be more resistant to treatment than non-stem cell populations at baseline and certain molecular signatures can increase resistance further (53). Future study of our cell lines should include a broader range of therapeutic concentrations administered over different time courses. For in vivo characterization, each tumorigenic cell displayed a unique growth pattern that recapitulated patient tumor growth. For instance, migration across the corpus callosum was observed for QNS120 in the patient MRI and murine histology (Fig. 1A; Fig. 6; Supplementary Fig. S5; Supplementary Fig. S7). QNS108 tumors in mice recapitulated formation of cysts that were also seen in patient MRIs (Fig. 1A). The findings illustrated the heterogenous nature of different GBM tumor cells and how different functional behaviors influence tumorigenesis.
Sex Differences in GBM:
The role of sex was emphasized in an analysis of the Ohio Brain Tumor Study (OBTS), where it was shown that female patients had a longer overall median survival of 22.5 months compared to men who had an overall median survival of 15 months (21). As such, patient sex is one variable among several that produce a complex interplay that influence GBM tumor growth.
In incorporating sex as a variable of study, we found that two male derived cell lines (QNS108 and QNS120) and one female derived cell line (QNS315) exhibited faster tumor growth in female mice brains (Fig. 4A–B; Fig. 5A). The observation that three GBM cell lines from both male and female patients grew at a faster rate in female mouse brains makes us hypothesize that sex-based microenvironments influence overall tumorigenesis. The immune system is one factor shown to contribute to sex differences in neural microenvironments and tumorigenesis (54–56). As shown by Bayik et al., the immune system as an extrinsic microenvironmental feature, caused sexual dimorphism where male immune systems promoted tumor growth and female immune systems had an anti-tumor effect (55). In the study, female mice had a survival advantage over male mice because of the immunological difference. The increased tumor growth rate in female mice observed in our study is opposite to what was observed by Bayik et al. where they used immunocompetent mouse models. The difference in results is likely due to our use of immunodeficient mice. In the context of immunodeficient female mice, there may be faster tumor growth since the anti-tumor effect of a female-immune system is missing. Likewise, the pro-tumorigenic immune environment of male mice is lacking, which may explain the slower rate of tumor growth. The differences illustrate the importance of using sex-defined microenvironments in preclinical modeling. Sex differences can provide novel therapeutic targets and recent studies have demonstrated the need of designing such sex-specific therapies (17, 55, 57).
In addition to microenvironmental features, a survival difference was observed between male and female mice, where male mice had a survival advantage when implanted with male derived QNS108 cells. (Fig. 7A). No survival difference was observed in mice implanted with female derived QNS315 cells (Fig. 7B). We are unable to make definitive conclusions about the observed trends, but the results suggest that male cell lines may interact with immunodeficient female brains to promote tumor growth while female derived cell lines do not. Possible rationale for the observed trends is that GBM tumor cells have been shown to carry intrinsic molecular properties that differ across sex, and these intrinsic molecular differences influence tumorigenic behavior and survival (57).
Two similar studies explore the role of how the microenvironment and sex-specific cell intrinsic properties interact. In the first study conducted by Sun et al., male and female derived astrocytes were implanted into male and female mice and induced to transform into mesenchymal GBM tumors (20). Male derived cells consistently formed larger tumors, and no difference in tumor growth was observed between male and female mice hosts (20). In a second study conducted by Villa et al., microglia-derived from rats were implanted into animals of the opposite sex (58). The group demonstrated that the cells retained sex-defined molecular signatures as well as functional behaviors irrespective of the environment in which they were placed (58). Although the cells were not tumor stem cells, the study demonstrates the principle that cells derived from male or female neural microenvironments are able to retain sex-intrinsic properties, a feature that may be possessed by BTICs and should be explored further in future studies. One key limitation about the studies mentioned is that only male derived, mouse derived, commercial, or desexualized GBM cell lines were used for in vivo modeling in male and female mice (20, 55, 57). The limitation is that primary female GBM BTICs are not used to initiate tumor formation as they have been found to be unstable and less tumorigenic than male cell lines (20, 57). The reason as to why female GBM cell lines tend to be less tumorigenic is a question for future study.
Limitations and Future Directions:
Our study is limited by the number of tumor models established (n=3) to describe sex differences and more models are required to study the role of sex in GBM tumor biology. Tumor evolution and intra-tumoral heterogeneity are factors that are not fully addressed with our current models, which only functionally describe heterogeneity of BTIC populations from different patient samples. Other limitations include the difficulty of engrafting BTIC lines into murine models. Future directions include characterizing a larger number of cell lines following the procedures presented here and testing multi-modal treatments with our preclinical models.
Conclusion:
GBM is defined by a changing heterogeneous cellular landscape. The presentation of changing variables that promote GBM tumorigenesis have rendered current clinical interventions ineffective. The functional characterization of different BTIC and non-BTIC populations establishes the feasibility of creating a panel of preclinical models that portray different phenotypic features of GBM. As part of the characterization, sex-specific preclinical models of GBM were established from primary derived BTICs. Our use of primary derived female GBM cell lines for in vivo modeling is timely. Current studies primarily use male derived, murine, or commercial cell lines to establish in vivo models of GBM when studying sex differences. Here, we provide a panel of primary male and female derived GBM tumor cell lines that can be used as a novel resource to test next-generation therapeutics informed by BTIC heterogeneity and sex differences.
Supplementary Material
Acknowledgements:
AQH was supported by the Mayo Clinic Professorship, a Clinician Investigator award, the Florida Department of Health Cancer Research Chair Fund, and the NIH (R43CA221490, R01CA200399, R01CA195503, and R01CA216855). CAG was supported by the NIH and NCI Diversity Supplement Program (R01CA195503-04S1). AGB was supported by the Howard Hughes Medical Institute (HHMI) Medical Research Fellows Program. RSE was supported by the Mayo Clinic Convergence Pilot Award. AC was supported by the Eagles 5th District Cancer Telethon Award. HGC, AC, and NZ were supported by the NIH (R03NS109444, K01NS110930). The authors would like to thank Dr. Steven S. Rosenfeld, Rita West, and Amanda Luu for assistance in conducting cell line kinetic growth experiments and Brandy Edenfield for assistance with murine histology staining.
Footnotes
Conflict of Interest/Disclosures: The authors declare no potential conflicts of interest.
References
- 1.Ostrom QT, Cioffi G, Gittleman H, Patil N, Waite K, Kruchko C, et al. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2012–2016. Neuro Oncol. 2019;21:v1–v100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJB, et al. Radiotherapy plus Concomitant and Adjuvant Temozolomide for Glioblastoma. New England Journal of Medicine. 2005;352:987–96. [DOI] [PubMed] [Google Scholar]
- 3.Lara-Velazquez M, Al-Kharboosh R, Jeanneret S, Vazquez-Ramos C, Mahato D, Tavanaiepour D, et al. Advances in Brain Tumor Surgery for Glioblastoma in Adults. Brain Sci. 2017;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cantrell JN, Waddle MR, Rotman M, Peterson JL, Ruiz-Garcia H, Heckman MG, et al. Progress Toward Long-Term Survivors of Glioblastoma. Mayo Clin Proc. 2019;94:1278–86. [DOI] [PubMed] [Google Scholar]
- 5.Gimple RC, Bhargava S, Dixit D, Rich JN. Glioblastoma stem cells: lessons from the tumor hierarchy in a lethal cancer. Genes Dev. 2019;33:591–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, et al. Isolation and Characterization of Tumorigenic, Stem-like Neural Precursors from Human Glioblastoma. Cancer Research. 2004;64:7011–21. [DOI] [PubMed] [Google Scholar]
- 7.Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CL, Rich JN. Cancer stem cells in glioblastoma. Genes Dev. 2015;29:1203–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. [DOI] [PubMed] [Google Scholar]
- 9.Chaichana KL, Guerrero-Cazares H, Capilla-Gonzalez V, Zamora-Berridi G, Achanta P, Gonzalez-Perez O, et al. Intra-operatively obtained human tissue: protocols and techniques for the study of neural stem cells. J Neurosci Methods. 2009;180:116–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanai N, Tramontin AD, Quinones-Hinojosa A, Barbaro NM, Gupta N, Kunwar S, et al. Unique astrocyte ribbon in adult human brain contains neural stem cells but lacks chain migration. Nature. 2004;427:740–4. [DOI] [PubMed] [Google Scholar]
- 11.Reifenberger G, Wirsching HG, Knobbe-Thomsen CB, Weller M. Advances in the molecular genetics of gliomas - implications for classification and therapy. Nat Rev Clin Oncol. 2017;14:434–52. [DOI] [PubMed] [Google Scholar]
- 12.Xie Y, Bergstrom T, Jiang Y, Johansson P, Marinescu VD, Lindberg N, et al. The Human Glioblastoma Cell Culture Resource: Validated Cell Models Representing All Molecular Subtypes. EBioMedicine. 2015;2:1351–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhaduri A, Di Lullo E, Jung D, Muller S, Crouch EE, Espinosa CS, et al. Outer Radial Glia-like Cancer Stem Cells Contribute to Heterogeneity of Glioblastoma. Cell Stem Cell. 2020;26:48–63 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prager BC, Xie Q, Bao S, Rich JN. Cancer Stem Cells: The Architects of the Tumor Ecosystem. Cell Stem Cell. 2019;24:41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X, Prager BC, Wu Q, Kim LJY, Gimple RC, Shi Y, et al. Reciprocal Signaling between Glioblastoma Stem Cells and Differentiated Tumor Cells Promotes Malignant Progression. Cell Stem Cell. 2018;22:514–28 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aldape K, Brindle KM, Chesler L, Chopra R, Gajjar A, Gilbert MR, et al. Challenges to curing primary brain tumours. Nat Rev Clin Oncol. 2019;16:509–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang W, Warrington NM, Taylor SJ, Whitmire P, Carrasco E, Singleton KW, et al. Sex differences in GBM revealed by analysis of patient imaging, transcriptome, and survival data. Sci Transl Med. 2019;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trifiletti DM, Alonso C, Grover S, Fadul CE, Sheehan JP, Showalter TN. Prognostic Implications of Extent of Resection in Glioblastoma: Analysis from a Large Database. World Neurosurg. 2017;103:330–40. [DOI] [PubMed] [Google Scholar]
- 19.Ostrom QT, Coleman W, Huang W, Rubin JB, Lathia JD, Berens ME, et al. Sex-specific gene and pathway modeling of inherited glioma risk. Neuro Oncol. 2019;21:71–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun T, Warrington NM, Luo J, Brooks MD, Dahiya S, Snyder SC, et al. Sexually dimorphic RB inactivation underlies mesenchymal glioblastoma prevalence in males. J Clin Invest. 2014;124:4123–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ostrom QT, Rubin JB, Lathia JD, Berens ME, Barnholtz-Sloan JS. Females have the survival advantage in glioblastoma. Neuro Oncol. 2018;20:576–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gittleman H, Ostrom QT, Stetson LC, Waite K, Hodges TR, Wright CH, et al. Sex is an important prognostic factor for glioblastoma but not for nonglioblastoma. Neurooncol Pract. 2019;6:451–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johansen ML, Stetson LC, Vadmal V, Waite K, Berens ME, Connor JR, et al. Gliomas display distinct sex-based differential methylation patterns based on molecular subtype. Neuro-Oncology Advances. 2020;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Binder ZA, Wilson KM, Salmasi V, Orr BA, Eberhart CG, Siu IM, et al. Establishment and Biological Characterization of a Panel of Glioblastoma Multiforme (GBM) and GBM Variant Oncosphere Cell Lines. PLoS One. 2016;11:e0150271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guerrero-Cazares H, Chen L, Quinones-Hinojosa A. Glioblastoma heterogeneity and more accurate representation in research models. World Neurosurg. 2012;78:594–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14:275–91. [DOI] [PubMed] [Google Scholar]
- 27.Patrizii M, Bartucci M, Pine SR, Sabaawy HE. Utility of Glioblastoma Patient-Derived Orthotopic Xenografts in Drug Discovery and Personalized Therapy. Front Oncol. 2018;8:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klughammer J, Kiesel B, Roetzer T, Fortelny N, Nemc A, Nenning KH, et al. The DNA methylation landscape of glioblastoma disease progression shows extensive heterogeneity in time and space. Nat Med. 2018;24:1611–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vaubel RA, Tian S, Remonde D, Schroeder MA, Mladek AC, Kitange GJ, et al. Genomic and phenotypic characterization of a broad panel of patient derived xenografts reflects the diversity of glioblastoma. Clinical Cancer Research. 2019:clincanres.0909.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dirkse A, Golebiewska A, Buder T, Nazarov PV, Muller A, Poovathingal S, et al. Stem cell-associated heterogeneity in Glioblastoma results from intrinsic tumor plasticity shaped by the microenvironment. Nat Commun. 2019;10:1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saygin C, Matei D, Majeti R, Reizes O, Lathia JD. Targeting Cancer Stemness in the Clinic: From Hype to Hope. Cell Stem Cell. 2019;24:25–40. [DOI] [PubMed] [Google Scholar]
- 32.Huang S Tumor progression: chance and necessity in Darwinian and Lamarckian somatic (mutationless) evolution. Prog Biophys Mol Biol. 2012;110:69–86. [DOI] [PubMed] [Google Scholar]
- 33.Auvergne R, Wu C, Connell A, Au S, Cornwell A, Osipovitch M, et al. PAR1 inhibition suppresses the self-renewal and growth of A2B5-defined glioma progenitor cells and their derived gliomas in vivo. Oncogene. 2016;35:3817–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tilghman J, Wu H, Sang Y, Shi X, Guerrero-Cazares H, Quinones-Hinojosa A, et al. HMMR Maintains the Stemness and Tumorigenicity of Glioblastoma Stem-like Cells. Cancer Research. 2014;74:3168–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y, Li A, Glas M, Lal B, Ying M, Sang Y, et al. c-Met signaling induces a reprogramming network and supports the glioblastoma stem-like phenotype. Proc Natl Acad Sci U S A. 2011;108:9951–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ying M, Wang S, Sang Y, Sun P, Lal B, Goodwin CR, et al. Regulation of glioblastoma stem cells by retinoic acid: role for Notch pathway inhibition. Oncogene. 2011;30:3454–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Capper D, Jones DTW, Sill M, Hovestadt V, Schrimpf D, Sturm D, et al. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555:469–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun P, Xia S, Lal B, Eberhart CG, Quinones-Hinojosa A, Maciaczyk J, et al. DNER, an epigenetically modulated gene, regulates glioblastoma-derived neurosphere cell differentiation and tumor propagation. Stem Cells. 2009;27:1473–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feng Y, Zhu M, Dangelmajer S, Lee YM, Wijesekera O, Castellanos CX, et al. Hypoxia-cultured human adipose-derived mesenchymal stem cells are non-oncogenic and have enhanced viability, motility, and tropism to brain cancer. Cell Death Dis. 2015;6:e1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Q, Wijesekera O, Salas SJ, Wang JY, Zhu M, Aprhys C, et al. Mesenchymal stem cells from human fat engineered to secrete BMP4 are nononcogenic, suppress brain cancer, and prolong survival. Clin Cancer Res. 2014;20:2375–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347:70–8. [DOI] [PubMed] [Google Scholar]
- 43.Stuckey DW, Shah K. TRAIL on trial: preclinical advances in cancer therapy. Trends Mol Med. 2013;19:685–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tzeng SY, Wilson DR, Hansen SK, Quinones-Hinojosa A, Green JJ. Polymeric nanoparticle-based delivery of TRAIL DNA for cancer-specific killing. Bioeng Transl Med. 2016;1:149–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mangraviti A, Tzeng SY, Gullotti D, Kozielski KL, Kim JE, Seng M, et al. Non-virally engineered human adipose mesenchymal stem cells produce BMP4, target brain tumors, and extend survival. Biomaterials. 2016;100:53–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Piccirillo SG, Vescovi AL. Bone morphogenetic proteins regulate tumorigenicity in human glioblastoma stem cells. Ernst Schering Found Symp Proc. 2006:59–81. [DOI] [PubMed] [Google Scholar]
- 47.Jiang X, Fitch S, Wang C, Wilson C, Li J, Grant GA, et al. Nanoparticle engineered TRAIL-overexpressing adipose-derived stem cells target and eradicate glioblastoma via intracranial delivery. Proceedings of the National Academy of Sciences. 2016:201615396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Redjal N, Zhu Y, Shah K. Combination of systemic chemotherapy with local stem cell delivered S-TRAIL in resected brain tumors. Stem Cells. 2015;33:101–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Piccirillo SG, Reynolds BA, Zanetti N, Lamorte G, Binda E, Broggi G, et al. Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature. 2006;444:761–5. [DOI] [PubMed] [Google Scholar]
- 50.Cancer-selective nanoparticles for combinatorial siRNA delivery to primary human GBM in vitro and in vivo. Biomaterials. 2019;209:79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O’Brien CA, Kreso A, Dick JE. Cancer stem cells in solid tumors: an overview. Semin Radiat Oncol. 2009;19:71–7. [DOI] [PubMed] [Google Scholar]
- 52.Lee EJ, Rath P, Liu J, Ryu D, Pei L, Noonepalle SK, et al. Identification of Global DNA Methylation Signatures in Glioblastoma-Derived Cancer Stem Cells. J Genet Genomics. 2015;42:355–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beier D, Schulz JB, Beier CP. Chemoresistance of glioblastoma cancer stem cells--much more complex than expected. Mol Cancer. 2011;10:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Turaga SM, Silver DJ, Bayik D, Paouri E, Peng S, Lauko A, et al. JAM-A functions as a female microglial tumor suppressor in glioblastoma. Neuro Oncol. 2020;22:1591–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bayik D, Zhou Y, Park C, Hong C, Vail D, Silver DJ, et al. Myeloid-Derived Suppressor Cell Subsets Drive Glioblastoma Growth in a Sex-Specific Manner. Cancer Discov. 2020;10:1210–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Toonen JA, Solga AC, Ma Y, Gutmann DH. Estrogen activation of microglia underlies the sexually dimorphic differences in Nf1 optic glioma-induced retinal pathology. J Exp Med. 2017;214:17–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kfoury N, Sun T, Yu K, Rockwell N, Tinkum KL, Qi Z, et al. Cooperative p16 and p21 action protects female astrocytes from transformation. Acta Neuropathol Commun. 2018;6:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Villa A, Gelosa P, Castiglioni L, Cimino M, Rizzi N, Pepe G, et al. Sex-Specific Features of Microglia from Adult Mice. Cell Rep. 2018;23:3501–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.