Abstract
During the early stages of budding, cell wall remodeling and polarized secretion are concentrated at the bud tip (apical growth). The CBK1 gene, encoding a putative serine/threonine protein kinase, was identified in a screen designed to isolate mutations that affect apical growth. Analysis of cbk1Δ cells reveals that Cbk1p is required for efficient apical growth, proper mating projection morphology, bipolar bud site selection in diploid cells, and cell separation. Epitope-tagged Cbk1p localizes to both sides of the bud neck in late anaphase, just prior to cell separation. CBK1 and another gene, HYM1, were previously identified in a screen for genes involved in transcriptional repression and proposed to function in the same pathway. Deletion of HYM1 causes phenotypes similar to those observed in cbk1Δ cells and disrupts the bud neck localization of Cbk1p. Whole-genome transcriptional analysis of cbk1Δ suggests that the kinase regulates the expression of a number of genes with cell wall-related functions, including two genes required for efficient cell separation: the chitinase-encoding gene CTS1 and the glucanase-encoding gene SCW11. The Ace2p transcription factor is required for expression of CTS1 and has been shown to physically interact with Cbk1p. Analysis of ace2Δ cells reveals that Ace2p is required for cell separation but not for polarized growth. Our results suggest that Cbk1p and Hym1p function to regulate two distinct cell morphogenesis pathways: an ACE2-independent pathway that is required for efficient apical growth and mating projection formation and an ACE2-dependent pathway that is required for efficient cell separation following cytokinesis. Cbk1p is most closely related to the Neurospora crassa Cot-1; Schizosaccharomyces pombe Orb6; Caenorhabditis elegans, Drosophila, and human Ndr; and Drosophila and mammalian WARTS/LATS kinases. Many Cbk1-related kinases have been shown to regulate cellular morphology.
The proper control of polarized growth and cellular morphology is crucial for both eukaryotic developmental processes and the specialized function of diverse cell types. For example, the formation of polarized cell structures is important for such diverse processes as the interaction of helper T cells with antigen-presenting B cells (25), nutrient absorption by the microvilli of epithelial cells (37), and flower pollen tube growth (3). The mechanisms by which cells mediate and regulate polarized cell growth are poorly understood.
In the budding yeast Saccharomyces cerevisiae, polarized growth is required for budding during vegetative growth and for projection formation during the mating response. Bud growth occurs in two phases, apical growth and isotropic growth (28). Apical bud growth occurs in G1, when cell wall deposition is restricted to the tip of the bud. Upon entry into mitosis, buds switch to isotropic growth, in which growth still occurs in the bud but is no longer restricted to the tip and instead occurs throughout the entire bud surface (28). The balance between apical and isotropic bud growth determines the shape of the resulting cell. The identification of components important for these different growth phases and how they are regulated is therefore important for understanding how cell shape is controlled in yeast.
Yeast polarized cell growth involves the coordinated function of polarity-regulating proteins, organization of cytoskeletal components, and regulation of signal transduction cascades (11, 30). Numerous components important for polarized cell growth in yeast are known; however, the number that have been shown to function specifically during apical growth is limited. Thus far, only a few proteins, the polarity components Spa2p, Pea2p, Bud6p, and Bni1p and the Pak1 homolog Ste20p, have been shown to be specifically required for apical growth (44). Many yeast proteins important for polarized growth have functional homologs in other organisms, suggesting that the molecular mechanisms underlying these processes are highly conserved among eukaryotes.
One important aspect of polarized growth is cell wall synthesis and remodeling (7). Yeast cells maintain a rigid cell wall that protects them from osmotic and mechanical stress. This wall must be partially degraded in a localized fashion for polarized cell growth to occur. Localized degradation of cell wall components must also occur following cytokinesis to allow separation of mother and daughter cells. It is likely that cell wall remodeling is tightly controlled both spatially and temporally by the cell polarization and cell cycle regulatory machinery; failure to do so would presumably result in cell lysis. How cells accomplish this control is not well understood.
To identify genes involved in apical growth, we have employed a transposon-based mutagenesis system (42) to screen for mutations that alter the elongated bud morphology of cells arrested during the apical growth phase. Here, the characterization of one gene identified in this screen, CBK1, is reported. This gene is predicted to encode a highly conserved protein kinase; we demonstrate that CBK1 is important for apical growth, mating projection formation, cell separation, and the transcriptional regulation of many cell wall-related genes.
MATERIALS AND METHODS
Yeast strains and media.
Yeast strains used in this study (Table 1) were congenic with S288c and W303. Standard genetic methods and growth media were used as described previously (18). Deletions of the entire protein coding regions of CBK1, ACE2, and HYM1 were produced by the PCR method described by Baudin et al. (2). Deletions were confirmed by PCR. Strains containing 3xHA-tagged CBK1 alleles were generated using plasmids from an ordered collection of transposon-mutagenized genomic clones. The details of this method have been described previously (43) and can be accessed online at http://ygac.med.yale.edu/. Correct tagging was confirmed by PCR and immunoblot analysis.
TABLE 1.
Yeast strains used in this study
| Straina | Genotype | Source |
|---|---|---|
| YSB124 | MATaleu2Δ98 ura3-52 lys2-801 ade2-101 his3-Δ200 | This laboratory |
| YSB171 | MATabar1Δ::LEU2 leu2Δ98 ura3-52 lys2-801 ade2-101 his3-Δ200 | This study |
| YSB127 | MATa/MATα leu2Δ98/leu2Δ98 ura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 his3-Δ200/his3-Δ200 | This study |
| YSB126 | MATacdc34-2 leu2Δ98 ura3-52 lys2-801 ade2-101 his3-Δ200 | This study |
| YSB89 | MATacdc34-2 cbk1-TnURA3-4D8 leu2Δ98 ura3-52 lys2-801 ade2-101 his3-Δ200 | This study |
| YSB131 | MATacbk1Δ::URA3 leu2Δ98 ura3-52 lys2-801 ade2-101 his3-Δ200 | This study |
| YSB144 | MATa/MATα cbk1Δ::URA3/cbk1Δ::URA3 leu2Δ98/leu2Δ98 ura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 his3-Δ200/his3-Δ200 | This study |
| YSB169 | MATacbk1Δ::URA3 bar1Δ::Leu2 leu2Δ98 ura3-52 lys2-801 ade2-101 his3-Δ200 | This study |
| YSB190 | MATaace2Δ::HIS3 leu2Δ98 ura3-52 lys2-801 ade2-101 his3-Δ200 | This study |
| YSB192 | MATa/MATα ace2Δ::HIS3/ace2Δ::HIS3 leu2Δ98/leu2Δ98 ura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 his3-Δ200/his3-Δ200 | This study |
| YSB169 | MATacbk1-mTn110F9 leu2Δ98 ura3-52 lys2-801 ade2-101 his3-Δ200b | This study |
| YSB151 | MATacbk1-mTn4D8 leu2Δ98 ura3-52 lys2-801 ade2-101 his3-Δ200 | This study |
| YSB153 | MATacbk1-mTn3F1 leu2Δ98 ura3-52 lys2-801 ade2-101 his3-Δ200 | This study |
| YSB209 | MATa/MATα cbk1-mTn3F1/cbk1-mTn3F1 leu2Δ98/leu2Δ98 ura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 his3-Δ200/his3-Δ200 | This study |
| YSB148 | MATa/MATα cbk1-mTn4D8/cbk1-mTn4D8 leu2Δ98/leu2Δ98 ura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 his3-Δ200/his3-Δ200 | This study |
| YSB183 | MATahym1Δ::LEU2 leu2Δ98 ura3-52 lys2-801 ade2-101 his3-Δ200 | This study |
| YSB189 | MATa/MATα hym1Δ::LEU2/hym1Δ::LEU2 leu2Δ98/leu2Δ98 ura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 his3-Δ200/his3-Δ200 | This study |
| YSB194 | MATacbk1-mTn4D8 hym1Δ::URA3 leu2Δ98 ura3-52 lys2-801 ade2-101 his3-Δ200 | This study |
| YSB200 | MATa/MATα cbk1Δ::URA3/cbk1Δ::URA3 hym1Δ::LEU2/hym1Δ::LEU2 leu2Δ98/leu2Δ98 ura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 his3-Δ200/his3-Δ200 | This study |
| YSB201 | MATacbk1Δ::URA3 hym1Δ::LEU2 leu2Δ98 ura3-52 lys2-801 ade2-101 his3-Δ200 | This study |
| DDY759 | MATaade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 | A. Sachs |
| DDY2080 | MATacbk1Δ::KanMX3 ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 | This study |
| DDY2081 | MATaade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 pFUS1-lacZ-URA3 | This study |
| DDY2082 | MATacbk1Δ::KanMX3 ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 pFUS1-lacZ-URA3 | This study |
Strains with the prefix YSB are from the S288C genetic background; strains with the prefix DDY are from the W303 genetic backgrround.
mTn indicates excision of the sequence between lox sites (43).
Morphological analysis and fluorescence microscopy.
Exponentially growing cells were treated as indicated and fixed for 1 h with 3.7% formaldehyde. Fixed cells were washed and resuspended in phosphate-buffered saline (PBS) plus 1.2 M sorbitol. For cell shape analysis, the length (long axis relative to the birth pole) and width (maximum distance perpendicular to the length) of budded cells were measured from images recorded using a Sensys charge-coupled device camera and Imagepoint Lab Spectrum software (Signal Analytics Corporation). To quantify the severity of cell separation defects, exponentially growing cells were fixed and the number of cells in random clusters was recorded and averaged. For quantitative analysis of mating projection formation defects, early log phase cells were treated with α-factor (Sigma, St. Louis, Mo.) at 5 μg/ml for 2 h, fixed, and analyzed. Mating projections that had a length (distance from base to tip of projection) greater than half the diameter of the cell were scored as normal. We stained F-actin with rhodamine-phalloidin (Molecular Probes, Eugene, Oreg.) using previously described methods (31). To visualize localization of cell wall chitin, cells were treated with calcofluor at a final concentration of 2 μg/ml. Stained cells were analyzed by fluorescence microscopy using a Leitz Aristoplan microscope. Images were captured using a Sensys charge-coupled device camera and Imagepoint Lab Spectrum software (Signal Analytics Corporation); subsequent image processing was done using Adobe PhotoShop software (Adobe Systems, Sunnyvale, Calif.).
Electron microscopy.
Exponentially growing cells were harvested by filtration, frozen at high pressure, cryofixed, and embedded in Epon-Araldite resin as described elsewhere (33); freeze substitution was done in 2% osmium tetroxide plus 0.1% uranyl acetate. Serial sections were examined and micrographs were taken using a Philips CM10 electron microscope. Negatives were digitally scanned directly at a 1,600-dot-per-in. resolution using an Epson ES-1200C transparency scanner (Epson, Long Beach, Calif.) to create 8-bit tagged image files, and these images were processed using Adobe Photoshop software (Adobe Systems, Sunnyvale, Calif.).
FITC-ConA analysis of apical growth.
To label cell wall mannoproteins, exponentially growing cells were collected, washed once with PBS, and incubated in the dark with fluorescein isothiocyanate (FITC)-concanavalin A (ConA) (Polysciences Inc., Warrington, Pa.) at a final concentration of 100 μg/ml for 10 min at 25°C. The cells were then washed with PBS and returned to growth at 30°C in fresh YPAD medium. After 25 min, cells were fixed, washed with PBS, and observed by fluorescence microscopy. Buds with completely faded staining at the tip were categorized as undergoing apical growth, while buds with uniformly decreased staining throughout the surface were categorized as undergoing isotropic growth. Buds with a gradient of staining that became increasingly faded toward the bud tip were categorized as gradient.
Halo and FUS1-lacZ induction assays.
For analysis of pheromone-induced growth arrest, sterile filter disks were saturated with α-factor at 5 μg/ml and placed in the middle of freshly plated lawns of exponentially growing MATa bar1Δ or MATa cbk1Δ bar1Δ cells. Plates were photographed after 2 days of growth at 30°C. Analysis of FUS1-lacZ reporter construct induction was carried out using yeast strains (DDY2081 and DDY2082) carrying a centromeric plasmid-borne FUS1-lacZ reporter construct as described elsewhere (51). Results show the averages and standard deviations of three independent trials for each condition.
Analysis of bud site selection.
Cells maintained in exponential growth for at least 4 consecutive generations were fixed in 3.7% formaldehyde, washed with PBS, stained with calcofluor at a final concentration of 2 μg/ml, and visualized by fluorescence microscopy. In wild-type diploid cells, bud site selection for the first three budding events was determined by analyzing the position of newly emerging buds in cells with zero, one, or two bud scars (13). Buds emerging from the one-third of the cell opposite the birth scar (a chitin-poor region of daughter cell walls where separation from the mother occurred) were scored as distal. Buds emerging from the one-third of the cell closest to the birth scar were scored as proximal. Buds emerging within the middle third of the cell were scored as medial. The vast majority of cbk1Δ cells fail to separate promptly following cytokinesis and therefore do not have birth scars. To quantify bud site selection in cbk1Δ cells, we analyzed small clusters of cells in which a progenitor cell with a birth scar could be clearly identified. The budding history in these clusters can be accurately determined by analyzing the pattern of the chains of cells that emerge from the progenitor cell. The sites chosen for the first three budding events were categorized by the criteria described above. At least 100 cells of each type (first, second, and third buds) were analyzed for each strain.
Gene expression analysis.
Northern analysis of CTS1 mRNA levels was conducted by blotting total RNA as described elsewhere (1). 32P-labelled CTS1, ACT1, and EGT2 probe was produced by random hexamer priming in the presence of [α-32P]dCTP of a gel-purified PCR fragment produced from yeast genomic DNA using primers specific for the appropriate genes. Blots were scanned and quantified using a PhosphorImager and ImageQuant software (Molecular Dynamics, San Jose, Calif.). Whole-genome transcriptional analyses were carried out using genomic DNA microarrays generously provided by Joe DeRisi (University of California, San Francisco), using methods described elsewhere (8); detailed protocols are available at http://zenith.berkeley.edu/∼seidel/Protocols/. Briefly, poly(A) mRNA was prepared from asynchronously growing cbk1Δ mutant (DDY2080) and wild-type (DDY759) cells, as well as from asynchronous cbk1Δ using a Qiagen Oligotex kit as specified by the manufacturer. Cy3- and Cy5-labeled cDNA probes were prepared and hybridized to DNA arrays. Arrays were scanned using a GSI Luminomics Scannarray 3000 slide scanner, and the resulting images were processed using Scanalyze software (M. Eisen; freely available at http://www.microarrays.org). Spots with a correlation coefficient of less than 0.35 were not included in the expression analysis. The complete set of data is available at http://zenith.berkeley.edu/amad/cgi-bin/index.pl.
Indirect immunolocalization of epitope-tagged Cbk1p.
Indirect immunofluorescence was performed as described previously (14). Exponentially growing cells were fixed with 3.7% formaldehyde for 20 min, washed twice with PBS, and spheroplasted at 37°C for various amounts of time with Zymolyase 100T at 5 μg/ml in PBS–1.2 M sorbitol. Cells were then washed twice with PBS-sorbitol and transferred to polylysine-coated slides. Cells were washed once with PBS–0.1% bovine serum albumin (BSA), twice with PBS–0.1% BSA–0.1% NP-40, and once again with PBS–0.1% BSA. Cells were incubated overnight at 4°C with monoclonal anti-hemagglutinin (HA) (16B12; Covance) and rabbit anti-yeast β-tubulin (Tub2p) antibodies (gift of F. Solomon, Massachusetts Institute of Technology). After four washes (as previously), cells were incubated with Cy3-conjugated goat anti-mouse antibodies (Jackson ImmunoResearch Laboratories, Inc., West Grove, Pa.) and FITC-conjugated goat anti-rabbit antibodies at room temperature for 1.5 h. Cells were then washed, mounted with a solution containing 4′,6′-diamidino-2-phenylindole (DAPI) and analyzed by fluorescence microscopy as described above.
RESULTS
Cbk1p belongs to a family of closely related eukaryotic protein kinases.
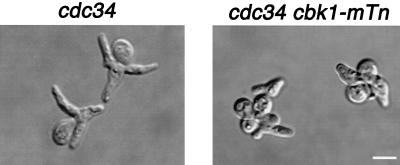
CBK1 was identified in a screen for genes involved in the apical growth phase of bud formation. The details and full results of this screen will be presented elsewhere. When grown at the restrictive temperature, cdc34-2 cells arrest in the apical growth phase and form multiple highly elongated buds (17) (Fig. 1). We introduced an ordered collection of transposon disruption alleles (43) into cdc34-2 cells and screened for mutants with altered bud morphology. We found that cdc34-2 cbk1-mTn cells form multiple buds that are much less elongated than those of cdc34-2 cells (Fig. 1). The average bud length of cdc34-2 cells grown at the restrictive temperature for 7 h is 3.8 ± 1.5 μm (n = 103), while the average bud length of cdc34-2 cbk1-mTn cells is 2.5 ± 1.0 μm (n = 95). This phenotype suggests that the CBK1 gene is required for efficient apical growth.
FIG. 1.
Morphology of cdc34-2 and cdc34-2 cbk1Δ cells grown at the restrictive temperature. Cells were grown to mid-logarithmic phase at 25°C in YPAD, shifted to 37°C for 7 h, and then fixed with formaldehyde and photographed. Bar, 5.0 μm.
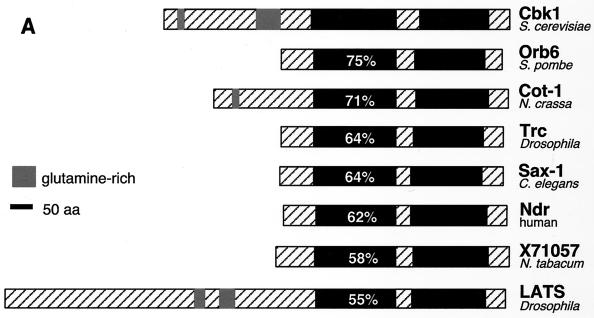
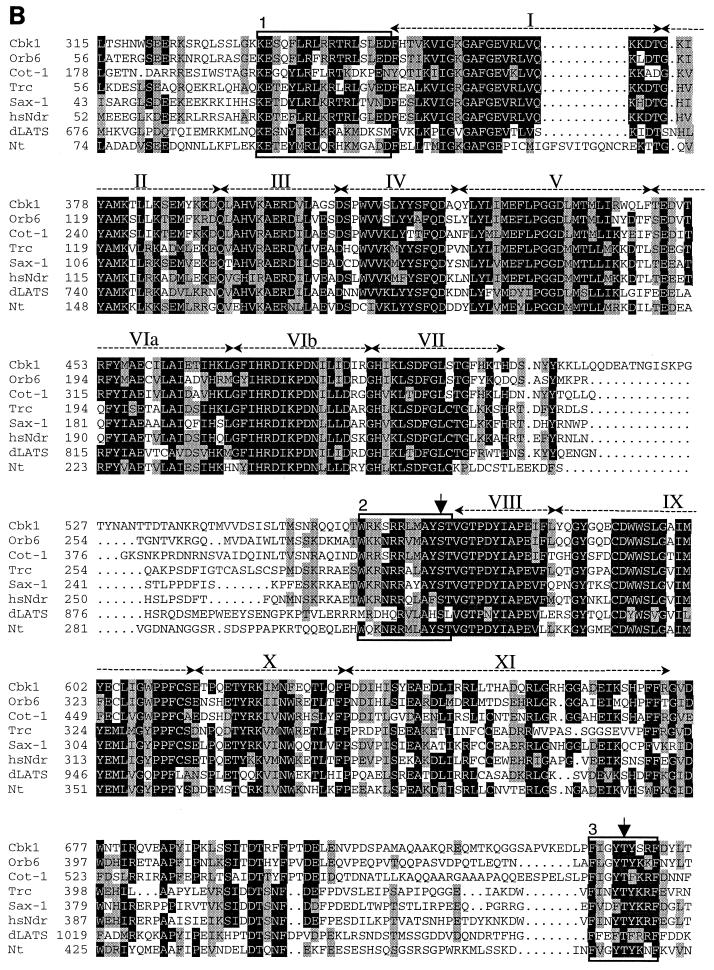
CBK1 encodes a serine/threonine kinase belonging to the AGC (protein kinase A, protein kinase G, protein kinase C) group of kinases. Among this group of kinases, those most closely related to Cbk1p include the fungal kinases Orb6 (53), Cot-1 (55), and Ukc1 (12); the Drosophila, Caenorhabditis elegans, and human Ndr kinases (15, 34, 56); the Drosophila and mammalian WARTS/LATS kinases (21, 38, 47, 54); and a kinase from the plant Nicotiana tabacum (GenBank accession no. X71057) (Fig. 2A and B). Sequence analysis reveals several notable similarities between the Cbk1-related kinases. The protein kinase catalytic domain is comprised of 12 highly conserved subdomains (19). All of the Cbk1p-related kinases have an insert between subdomains VII and VIII of their kinase domains. The size of this insert varies, and its sequence is not conserved (Fig. 2B). A feature common to many AGC kinases is that they require phosphorylation at a conserved site (consensus sequence T[F/L]CGT [bold indicates the phosphorylation site]) within the “activation loop” between subdomains VII and VIII for full kinase activity (40). The Cbk1-related kinases possess a conserved motif at this location (Fig. 2B). Although this motif does not fit the AGC family activation loop phosphorylation site consensus sequence, it has been shown to be a site of activating phosphorylation in human Ndr (36). The Cbk1p-related kinases also have two regions of similarity outside of their kinase domains that are likely to be functionally important: one immediately preceding kinase subdomain I and one near the carboxy terminus (Fig. 2B). The region immediately preceding the catalytic domain overlaps a region responsible for Ca2+-dependent binding and activation of human Ndr by the Ca2+ receptor S100B (35). The carboxy-terminal motif matches the consensus sequence of a conserved regulatory site termed the “hydrophobic motif” (FXX[F/Y][S/T][F/Y], where X is any residue and bold indicates the phosphorylated residue). This motif was first recognized in p70 S6 kinase (39) and regulates the stability and/or activity of many AGC kinases (4). This motif has been shown to be a site of activating phosphorylation in human Ndr kinase (36).
FIG. 2.
Sequence comparison of Cbk1p-related protein kinases. (A) The predicted amino acid (aa) sequences of Cbk1p from S. cerevisiae, Orb6 from S. pombe (53), Cot-1 from N. crassa (55), human Ndr (34), Drosophila Trc (Ndr) (15), C. elegans SAX-1 (Ndr) (56), N. tabacum kinase X71057 (GenBank accession no. X71057), and Drosophila melanogaster LATS (21, 54) were compared using the program CLUSTALW (48). Kinase domains (19) are shown in black, and the percent amino acid identity of the kinase domains with the Cbk1p kinase domain is shown in white. Regions outside the putative catalytic domain are crosshatched. The insert between subdomains VII and VIII of the catalytic domain (crosshatched region in the middle of the kinase domain) is characteristic of this family of protein kinases. Several members of this kinase family also have glutamine-rich regions in the sequence N terminal of their catalytic domains. (B) Multiple-sequence alignment of the catalytic domain and surrounding sequences of the Cbk1p-related kinases was performed using CLUSTALW. Shading was done using MacBoxShade (M. Baron). Shaded residues are conserved in at least five of the eight sequences. Black shading indicates identity with Cbk1p, while grey shading indicates similarity. Kinase subdomains are labeled above the sequence in roman numerals. Numbered boxed sequences indicate functionally significant regions discussed in the text. Boxed region 1, directly preceding the kinase domain, overlaps the Ca2+/S100B-binding domain of Ndr. Boxed region 2, directly preceding kinase subdomain VIII, indicates a conserved motif that overlaps the activation loop phosphorylation site in human Ndr. Boxed region 3, near the carboxy terminus, corresponds to the hydrophobic motif that is found in many AGC family kinases. Residues corresponding to experimentally determined sites of regulatory phosphorylation in human Ndr are marked with arrows.
Disruption of CBK1 alters cellular morphology and causes cell separation defects.
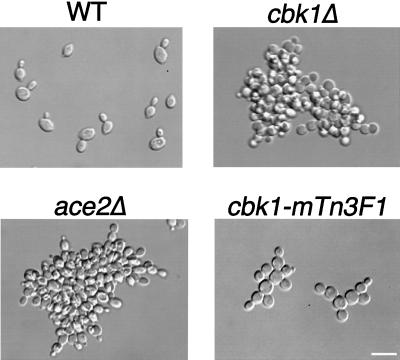
In order to further analyze its cellular function, we constructed strains lacking the entire CBK1 open reading frame in both the S288C and W303 genetic backgrounds. The two strains produced similar results. For brevity, the data presented here are for S288C background strains only, except where otherwise noted. cbk1Δ cells have no obvious defects in growth rate when incubated on solid medium or in liquid culture, regardless of temperature or genetic background. In addition, cytological analysis indicates that cell cycle progression is not affected in cbk1Δ cells (data not shown). However, cbk1Δ cells have altered cellular morphologies (Fig. 3). Wild-type diploid cells are oval shaped (Fig. 3), with an average length/width ratio of 1.4 ± 0.2 (n = 102). In contrast, cbk1Δ diploid cells are rounder (Fig. 3), with an average length/width ratio of 1.1 ± 0.1 (n = 107). In addition to the changes in cell shape, cbk1Δ mutants form clumps of cells that are resistant to sonication. The cell separation defect is most severe in diploid cbk1Δ mutants, with the average clump containing 90 ± 64 (n = 40) cells (Fig. 3). Similar phenotypes are observed in haploid cbk1Δ cells (data not shown).
FIG. 3.
Morphology of wild-type (WT) and cbk1Δ, ace2Δ, and cbk1-mTn3F1 mutant homozygous diploid cells. Cells were grown to early logarithmic phase in YPAD and then fixed with formaldehyde and photographed. Bar, 10 μm.
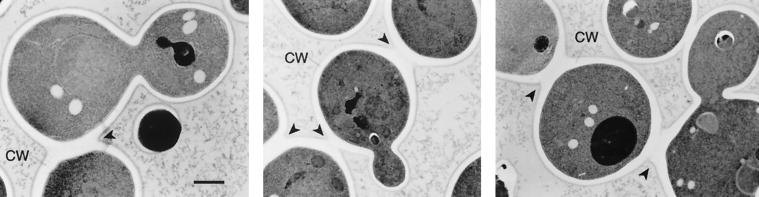
Staining of cbk1Δ cells with the chitin-binding dye calcofluor reveals that the aggregated cells are associated with one another by chitin-rich junctions (data not shown). Since chitin is enriched at the bud neck between mother and daughter cells, this suggests that cbk1Δ cells adhere to one another either because they fail to complete cytokinesis or because they fail to complete mother-daughter separation following cytokinesis. Electron microscopy of cbk1Δ cells using a fixation technique that preserves the cell wall favors the latter explanation. Cells connected by shared cell wall material were often observed (Fig. 4). However, no cells were found to have more than one cytoplasmic connection to a second cell body (n = 200 single sections of distinct cells; five cells were sectioned completely). Furthermore, treatment with Zymolyase, which removes the yeast cell wall, disrupts the cell clumps (data not shown). Therefore, we concluded that cbk1Δ cells remain associated with one another due to a failure to degrade the septum connecting mother and daughter cells.
FIG. 4.
Electron microscopy of high-pressure-frozen–freeze-substituted cbk1Δ cells. Black arrowheads indicate regions of shared cell wall (CW) material. No cytoplasmic connections were observed in adjacent sections at any of these sites. Bar, 0.5 μm.
Efficiency of apical growth is diminished in cbk1Δ cells.
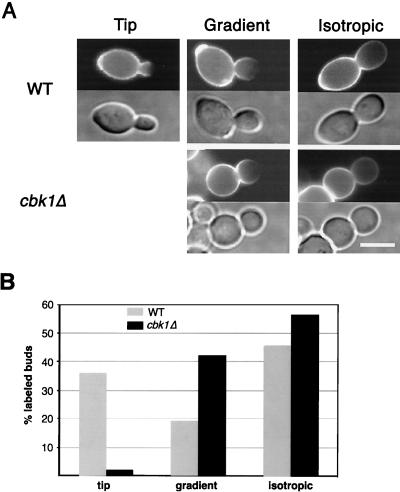
The decreased hyperpolarized growth observed in cdc34-2 cbk1-mTn cells (Fig. 1) and the round shape of cbk1Δ cells (Fig. 3) suggest that Cbk1p plays a role in maintaining apical growth. To test this directly, we visually monitored cell surface growth with FITC-ConA, which binds cell wall mannose (50). Exponentially growing wild-type or cbk1Δ diploid cells were labeled with FITC-ConA for 10 min and then introduced to fresh medium for 25 min. The cells were then fixed and scored for staining pattern. In these experiments, regions where new cell wall growth has occurred have reduced staining intensity. Of the wild-type cells with labeled buds, 36% (n = 132) had a completely unlabeled region at the tip of the bud (Fig. 5), indicating that these cells were in the apical growth phase during the time following FITC-ConA labeling. Nineteen percent exhibited a gradient of staining that decreases toward the bud tip, leaving the tip still partially labeled (Fig. 5), indicating that some apical growth occurred after the time of labeling. Forty-five percent had uniform staining throughout the bud that was of reduced intensity (Fig. 5), indicative of isotropic growth. Thus, in wild-type cells, 55% of buds experienced some apical growth in the 25-min period after they were labeled. In comparison, only 2% (n = 127) of labeled cbk1Δ buds were completely unlabeled at the tip. However, 42% had a detectable gradient of staining (Fig. 5), indicating that some apical growth had occurred. Fifty-six percent of labeled cbk1Δ buds had uniformly decreased staining and were thus growing isotropically in the time following labeling (Fig. 5). Therefore, in cbk1Δ cells, 44% of buds underwent a period of at least partial apical growth after they were labeled. Thus, there was a striking difference in the fractions of wild-type and cbk1Δ cells with completely unlabeled bud tips (36% in the wild type versus 2% in the cbk1Δ mutant). This suggests that cbk1Δ cells are less efficient in restricting growth to a small region at the bud tip. In summary, these results suggest that the duration of the apical growth phase in cbk1Δ cells is similar to that in the wild type but the ability to concentrate apical growth at the bud tip is decreased during this phase.
FIG. 5.
Analysis of apical growth in wild-type (WT) and cbk1Δ mutant cells by FITC-ConA pulse-labeling. (A) Exponentially growing wild-type and cbk1Δ mutant diploid cells were pulse-labeled with FITC-ConA for 10 min, washed, and returned to growth in fresh medium. After 25 min, cells were fixed and observed by fluorescence microscopy. FITC-ConA fluorescence images are shown above differential interference contrast images. Staining that is completely faded at the bud tip indicates apical growth (Tip), while uniformly decreased staining throughout the bud indicates isotropic growth (Isotropic). Cells with a gradient of staining that was not completely faded at the bud tip were also observed (Gradient). (B) The pattern of fading in labeled buds was quantitated for wild-type and cbk1Δ diploid cells. Labeled buds were divided into three categories as described above: labeled buds with a completely faded tip (tip), labeled buds with staining that faded toward the tip but with staining at the tip still visible (gradient), and labeled buds with uniformly faded staining (isotropic). Bar, 5.0 μm.
CBK1 is required for the formation of normal mating projections in response to pheromone.
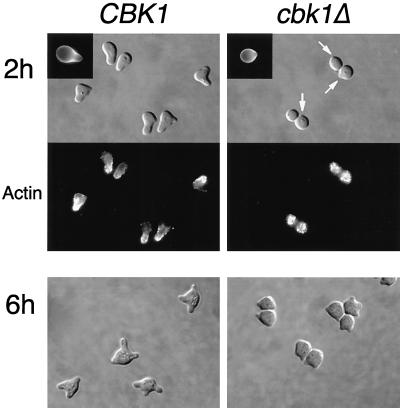
Since polarized growth is critical for mating projection formation, we tested the ability of cbk1Δ cells to form mating projections in response to pheromone. After a 2-h treatment with α-factor mating pheromone, 93% (n = 148) of wild-type cells form normal-sized (at least half a cell diameter in length) mating projections (Fig. 6). In contrast, only 8% (n = 203) of cbk1Δ cells formed normal mating projections (Fig. 6). Interestingly, a majority (74%) of the cbk1Δ cells developed one or more small surface bumps (Fig. 6, arrows). While much smaller than normal mating projections, these small protrusions were flanked by regions of increased chitin deposition, as seen in normal mating projections (Fig. 6, insets). In order to rule out the possibility that the mating projection defect observed in cbk1Δ mutants is due simply to a delayed response to pheromone, we tested the effect of longer treatments with pheromone. After a 6-h incubation with α-factor, nearly all wild-type cells formed at least one normal-sized projection (98%, n = 160) and 66% formed two or more normal-sized projections (Fig. 6). In contrast, only 9% (n = 175) of the cbk1Δ cells formed at least one normal-sized mating projection (Fig. 6). As with the 2-h induction, most (76%) of the cbk1Δ cells formed small surface bumps and 58% had two or more of these apparently defective mating projections (Fig. 6). These results indicate that Cbk1p is not required for the establishment of a mating projection but is required for maintenance of persistent polarized growth of the structure.
FIG. 6.
Pheromone-induced morphology and actin reorganization in wild-type and cbk1Δ mutant cells. Exponentially growing cells were incubated with α-factor (5 μg/ml) for 2 or 6 h, fixed with formaldehyde, and stained with rhodamine-phalloidin to visualize the actin cytoskeleton. White arrows point out small protrusions on the cell surface. Upper left insets show calcofluor staining of chitin. Strains used for the 6-h time point were bar1Δ mutants. Bar, 5.0 μm.
After the site of a new mating projection has been selected through pheromone signaling, the actin cytoskeleton and polarized growth are directed to the tip of the growing projection. We therefore examined the ability of cbk1Δ mutants to polarize their actin cytoskeleton in response to pheromone by fluorescence microscopy of cells stained with rhodamine-phalloidin. In wild-type cells treated with α-factor, actin patches concentrate at the tips of growing projections (Fig. 6). In contrast, actin patches are distributed evenly throughout the cell surface of pheromone-treated cbk1Δ cells (Fig. 6), indicating that these cells are unable to maintain actin cytoskeleton polarization. Interestingly, cells that have apparently initiated projection formation (as evidenced by a small surface bump) do not contain a polarized actin cytoskeleton at this site. Again, this suggests that polarized growth was initiated at these sites but then aborted, suggestive of a role for Cbk1p in maintaining polarized growth processes.
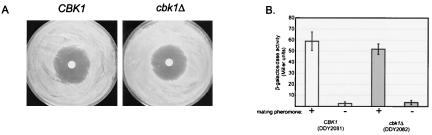
cbk1Δ mutants were also analyzed for other defects in the mating pheromone response. The ability to arrest growth in response to pheromone was analyzed by α-factor halo assays. Within the sensitivity limits of the assay, pheromone-induced growth arrest in cbk1Δ cells is indistinguishable from that in the wild type (Fig. 7A). A previous study reported that cbk1Δ cells display increased resistance to α-factor-induced growth arrest (16). Rather than halo assays, this investigation used α-factor-containing solid medium to determine α-factor sensitivity. We repeated our experiments using this methodology and found no increased resistance to α-factor-induced growth arrest in cbk1Δ cells (data not shown). Furthermore, we found that transcriptional induction of a FUS1-lacZ reporter construct, which is activated in wild-type cells upon exposure to mating pheromone, was not significantly different in wild-type versus cbk1Δ cells (Fig. 7B). Thus, in contrast to previous reports (16), we did not find evidence of increased resistance to mating pheromone using our strain background. In summary, Cbk1p is required for persistent polarized growth of the mating projections formed in response to pheromone but is apparently not necessary for other aspects of the mating response.
FIG. 7.
Pheromone-induced growth arrest and transcriptional induction in wild-type and cbk1Δ mutant cells. (A) Sterile filter disks were saturated with α-factor at 5 μg/ml and placed in the middle of freshly plated lawns of wild-type or cbk1Δ mutant cells. Pictures were taken after 2 days of incubation at 30°C. The strains used were bar1Δ mutants. (B) Expression of FUS1-lacZ reporter fusion upon exposure to α-factor at 5 μg/ml in YPAD medium. Open bars represent wild-type cells; gray bars represent cbk1Δ mutant cells. β-Galactosidase expression levels were measured in Miller units as previously described (55).
Deletion of CBK1 alters the pattern of bud site selection in diploids.
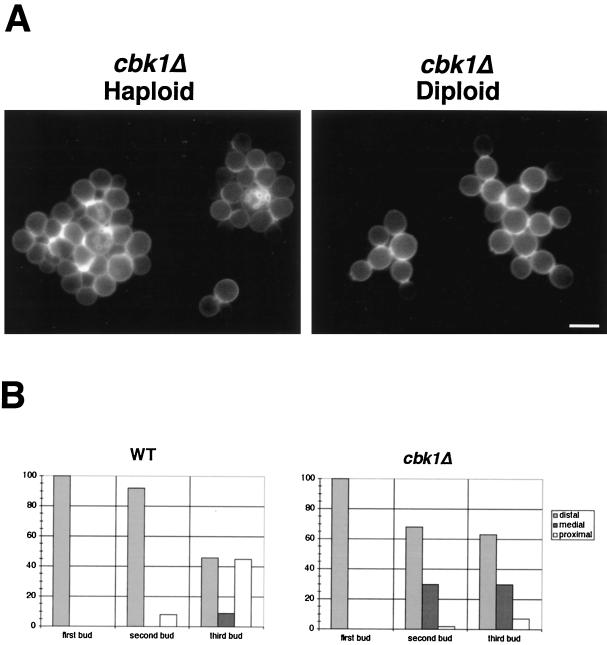
Mutants defective in polarized growth often have bud site selection defects (30, 44). We therefore examined the budding pattern of haploid and diploid cbk1Δ cells by staining with calcofluor, which stains the chitin-rich bud scars (20). Although the separation defect of these cells made analysis difficult, chains of adjacent bud scars can be clearly seen in haploid cbk1Δ cells. Clusters of heavy calcofluor staining were found in the middle of globular clumps, indicative of a normal haploid axial budding pattern (Fig. 8A).
FIG. 8.
Bud site selection in wild-type (WT) and cbk1Δ mutant cells. (A) Bud scars in haploid and diploid cbk1Δ cells were stained with calcofluor and photographed. Note the chitin-rich junctions between attached cells. Bar, 5.0 μm. (B) The graphs show the placement of the first three buds in wild-type and cbk1Δ mutant diploid cells. Bud scars were scored as distal (opposite the birth site), proximal (near the birth site), or medial (see Materials and Methods). At least 100 cells of each type (first, second, and third buds) were analyzed for both strains.
However, cbk1Δ diploid cells have a defect in the diploid bipolar budding pattern. In the bipolar pattern, daughter cells generally bud at the distal pole (opposite the site of septation) and mother cells bud at either the proximal pole (adjacent to the site of septation) or the distal pole (45). To compare the budding pattern of cbk1Δ diploids to that of an isogenic wild-type strain, we stained cells with calcofluor and recorded the position of emerging buds in cells with zero, one, or two bud scars (see Materials and Methods for a description of the classification scheme used). In wild-type diploids, the first two buds form predominantly at sites distal to the birth scar (>90%) while 46% of third buds emerge at proximal sites (Fig. 8B). As in the wild type, the first bud in cbk1Δ diploids forms exclusively at distal sites. However, 30% of the second and third buds form at medial sites. Medial sites are never selected in wild-type cells during the first two budding events, and only 7% of cells choose these nonbipolar sites for the third bud. Most notably, only 7% of third buds in cbk1Δ cells emerge at a proximal site (Fig. 8B). Thus, disruption of CBK1 in diploids results in the increased selection of medial bud sites and severely impairs the ability to select proximal bud sites.
Deletion of CBK1 affects expression of a range of cell wall-modifying enzymes.
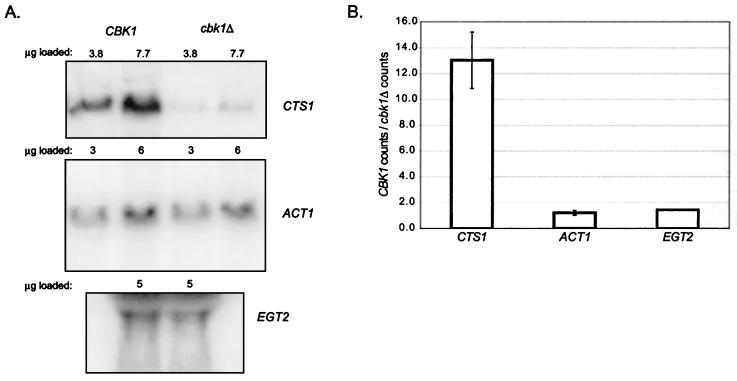
Cell separation following cytokinesis requires degradation of the chitin-rich septum between mother and daughter cells. This event is dependent upon expression of a chitinase encoded by the CTS1 gene (26). Strikingly, RNA blot analysis of mRNA abundance indicates that expression of CTS1 is reduced approximately 13-fold in cbk1Δ cells (Fig. 9). In contrast, expression of the actin and EGT2 (early G1 transcript) (23) genes was not affected in cbk1Δ cells. The normal transcription of EGT2, which is temporally coregulated in late M/early G1 with CTS1 (23), indicates that the effect of CBK1 deletion is not general for late M/early G1 genes.
FIG. 9.
Northern analysis of CTS1 mRNA levels in wild-type and cbk1Δ mutant cells. (A) Northern blots showing CTS1 (top), ACT1 (middle), and EGT2 (bottom) message levels in wild-type and cbk1Δ mutant cells. The amount of total RNA loaded in each lane is indicated above the corresponding lane. (B) Relative expression of CTS1, ACT1, and EGT2 mRNAs in wild-type and cbk1Δ cells, expressed as the ratio of PhosphorImager counts (arbitrary units) of mRNA from wild-type cells over mRNA from cbk1Δ mutant cells.
To examine the effects of CBK1 deletion on gene expression more broadly, we examined transcriptional differences between wild-type (DDY759) and cbk1Δ (DDY2080) cells using a whole-genome DNA array (a generous gift from J. DeRisi). We compared hybridization of a probe derived from poly(A) RNA isolated from asynchronous wild-type and cbk1Δ cells (Tables 2 and 3). Similar to the results of our RNA blot analysis, we found that CTS1 expression was markedly reduced in cbk1Δ cells. Additionally, YER124c, a gene of unknown function, was repressed, as were several other genes involved in cell wall physiology, notably, SCW11 (24) (Table 2). The SCW11 gene, like CTS1, is required for efficient cell separation (6, 27), and thus, both of these genes may participate in cell wall degradation. Conversely, the expression of a chitin synthase encoded by CHS1 (5) was significantly elevated in cbk1Δ cells, as was the expression of the ECM3 (extracellular mutant), ECM5, and ECM8 genes (Table 3). ECM3, ECM5, and ECM8 are likely to function in cell wall biosynthesis, since mutations in these genes result in hypersensitivity to calcofluor, which interferes with cell wall assembly (29). These results suggest that Cbk1p may, among other things, regulate the balance of cell wall synthetic and degradative activities in a manner that is important for local remodeling of the cell wall during polarized growth and cell separation.
TABLE 2.
Genes with >2.2-fold higher expression in wild-type cells
| Gene or ORFa | Cy5 + Cy3b | Cy5/Cy3 ratio | Function |
|---|---|---|---|
| YER124C | 9,125 | 12.7 | Unknown |
| CTS1 | 11,941 | 11.9 | Endochitinase; cell wall biogenesis |
| SCW11 | 1,705 | 4.8 | Glucanase (putative); cell wall biogenesis |
| PHO11 | 16,442 | 4.7 | Secreted acid phosphatase; phosphate metabolism |
| PHO12 | 11,676 | 4.4 | Secreted acid phosphatase; phosphate metabolism |
| MEP2 | 6,128 | 4.1 | Transport |
| BTN2 | 7,784 | 3.3 | Unknown |
| IMP3 | 5,317 | 3.2 | rRNA processing |
| GIT1 | 1,112 | 3.2 | Unknown |
| MET17 | 16,309 | 2.9 | Methionine biosynthesis |
| YHR049W | 1,182 | 2.9 | Unknown |
| YHR146W | 7,286 | 2.6 | Unknown |
| CBK1 | 1,311 | 2.5 | Deleted in Cy3-labeled strain |
| YLR302C | 2,518 | 2.4 | Unknown |
| YNR067C | 3,827 | 2.4 | Similar to glucanases |
| ALD6 | 7,400 | 2.3 | Ethanol utilization |
| GSY1 | 1,978 | 2.3 | Glycogen metabolism |
| TIP1 | 9,937 | 2.3 | Cell wall mannoprotein |
| HHT1 | 33,333 | 2.3 | Histone H3; chromatin structure |
ORF; open reading frame.
Arbitrary units.
TABLE 3.
Genes with >2.2-fold higher expression in cbk1Δ mutant cells
| Gene or ORFa | Cy5 + Cy3b | Cy3/Cy5 ratio | Function |
|---|---|---|---|
| CHA1 | 2,454 | 4.3 | Hydroxy amino acid metabolism |
| YLR416C | 291 | 4.0 | Unknown |
| YGR138C | 803 | 3.2 | Unknown |
| YPK2 | 1,123 | 3.1 | Protein kinase; sphingolipid metabolism |
| PLB3 | 3,350 | 3.1 | Plasma membrane phospholipase |
| YLR414C | 4,486 | 3.0 | Unknown |
| YOR137C | 1,853 | 3.0 | Unknown |
| YBL098W | 2,767 | 2.9 | Unknown |
| ECM5 | 248 | 2.8 | Cell wall biogenesis |
| GPD2 | 7,721 | 2.7 | Glycerol metabolism |
| YMR085W | 717 | 2.7 | Unknown |
| ECM3 | 1,108 | 2.7 | Cell wall biogenesis |
| NCA3 | 3,593 | 2.7 | ATP synthesis |
| SPI1 | 6,282 | 2.6 | Cell wall localized |
| YMR316C-A | 5,929 | 2.6 | Unknown |
| YJL163C | 2,354 | 2.5 | Unknown; hydrophobic |
| YGR263C | 1,712 | 2.5 | Unknown |
| YER166W | 1,803 | 2.5 | Unknown |
| APS1 | 1,286 | 2.5 | Secretion |
| ECM8 | 3,796 | 2.4 | Cell wall biogenesis |
| YCR068W | 746 | 2.4 | Unknown |
| YDL039C | 1,106 | 2.4 | Unknown |
| YGR146C | 4,270 | 2.3 | Unknown |
| CHS1 | 1,676 | 2.3 | Chitin synthase; cell wall biogenesis |
| ISF1 | 743 | 2.3 | RNA splicing; mitochondrial |
| YHR100C | 7,854 | 2.3 | Unknown |
| YLR312C | 745 | 2.3 | Unknown |
| YAK1 | 1,638 | 2.3 | Serine-threonine protein kinase |
| YNL288W | 1,622 | 2.3 | Unknown |
| BOP1 | 3,211 | 2.3 | Unknown |
| HSP30 | 21,055 | 2.3 | Plasma membrane heat shock protein |
| YGR190C | 2,361 | 2.3 | Unknown |
| YPL067C | 1,490 | 2.3 | Unknown |
| EFT1 | 9,683 | 2.3 | Protein synthesis |
| YDL038C | 3,739 | 2.3 | Related to cell wall mannoproteins |
| YLR297W | 1,346 | 2.3 | Unknown |
| HXT3 | 1,413 | 2.3 | Transport |
| FOL1 | 1,155 | 2.3 | Folate biosynthesis |
| YPS1 | 3,294 | 2.3 | Cell surface-localized protease |
ORF, open reading frame.
Arbitrary units.
Genetic evidence suggests that Cbk1p functions in different pathways to regulate morphogenesis and cell separation.
The expression of CTS1 is regulated by the transcription factor Ace2p (9). Our results indicate that CTS1 expression is greatly reduced in cbk1Δ mutants; other recent data suggest that Cbk1p may act as an upstream activator of the transcription factor Ace2p to promote the expression of CTS1 (41). To analyze the relationship between CBK1 and ACE2, we constructed ace2Δ strains and compared their phenotypes to cbk1Δ mutant phenotypes. The cell separation defect of ace2Δ cells is comparable in severity to that observed in cbk1Δ cells, with ace2Δ diploids forming clumps containing an average of 115 ± 54 cells (n = 37) (Fig. 3). However, unlike cbk1Δ cells, ace2Δ cells are not rounder than wild-type cells (Fig. 3); ace2Δ diploid cells have an average length/width ratio of 1.4 ± 0.2 (n = 101). Additionally, 82% (n = 210) of ace2Δ cells form normal-sized mating projections after a 2-h treatment with α-factor (Fig. 7; also data not shown). These results suggest that Cbk1p acts through downstream components other than Ace2p to promote polarized growth.
Since cbk1Δ strains have phenotypes that are distinct from that of ace2Δ cells, we hypothesized that it might be possible to create an allele of CBK1 that is specifically defective for either polarized growth or cell separation and colony morphology. We examined three yeast strains in which the chromosomal CBK1 locus was tagged in frame with a DNA fragment encoding three copies of the HA epitope at different positions (Fig. 10A). Strains containing CBK1 tagged at amino acid 21 or 259 in the amino-terminal nonkinase domain have wild-type morphologies, form normal mating projections in response to pheromone, and undergo efficient cell separation, indicating that the tagged Cbk1p in these strains is functional (data not shown). In contrast, strains with CBK1 tagged at amino acid 567 (subsequently referred to as cbk1-mTn3F1) within the kinase domain have partial defects. Interestingly, this insertion lies within the previously described putative activation loop phosphorylation motif. Diploid cells homozygous for the cbk1-nTn3F1 allele are significantly rounder than wild-type cells (Fig. 3). The average length/width ratio of cbk1-nTn3F1 diploid cells is 1.1 ± 0.1 (n = 110), which is similar to the 1.1 ratio of cbk1Δ diploid cells but distinct from the 1.4 ratio of oval-shaped wild-type diploid cells. Haploid cells containing the cbk1-mTn3F1 allele are also defective for mating projection formation. After a 2-h treatment with α-factor, only 10% (n = 258) of cbk1-mTn3F1 cells form normal-sized (at least half a cell diameter in length) mating projections. As with cbk1Δ cells, most of the pheromone-treated cbk1-mTn3F1 cells form small surface bumps (data not shown). Thus, the cbk1-mTn3F1 allele produces polarized growth defects that are comparable in severity to those observed in cbk1Δ strains. However, cbk1-mTn3F1 cells have much milder cell separation defects than cbk1Δ cells (Fig. 3). cbk1-mTn3F1 diploids form smaller clusters that contain an average of 7.7 ± 4.8 cells (n = 77), compared with an average of 90 cells/clump for cbk1Δ mutants. These data suggest that Cbk1p may function in two distinct pathways: one that is required for efficient polarized growth and another that promotes cell separation.
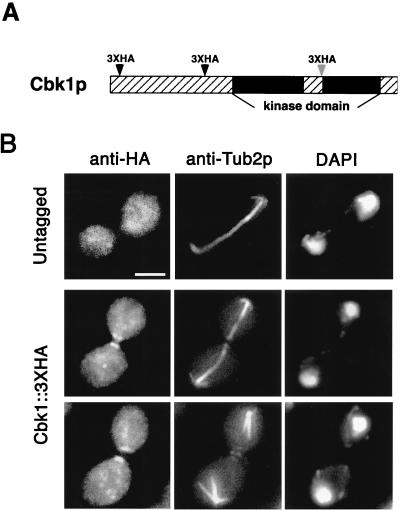
FIG. 10.
Localization of Cbk1p by indirect immunofluorescence staining. (A) The chromosomal CBK1 gene was tagged with a triple HA epitope (3xHA) at the indicated positions (see Materials and Methods). Dark arrowheads indicate that the tagged protein retained its function, and the light arrowhead represents the cbk1-mTn3F1 allele, which is partially functional (details are discussed in Results). (B) CBK1::3xHA and untagged haploid strains were stained with anti-HA antibodies, anti-Tub2p antibodies (microtubule staining), and DAPI (DNA staining). Cbk1p::3xHA localizes to both sides of the bud neck late in anaphase (note long spindle length) and remains as a patch on the surface of each recently separated cell. No polarized localization was detected in the untagged strain. For these pictures, the strain with CBK1 tagged at amino acid 259 was used. Bar, 5.0 μm.
Cbk1p localizes to both sides of the bud neck during late anaphase.
To determine the intracellular location of Cbk1p, we examined the localization of Cbk1p::3xHA in both functionally tagged strains (amino acids 21 and 259) by indirect immunofluorescence assay using antibodies directed against the HA epitope. The cells were also stained with anti-Tub2p antibodies to help determine the cell cycle stage. In both tagged strains, Cbk1p::3xHA localizes to both sides of the bud neck during late anaphase (Fig. 10B). Some unbudded cells have a single patch of staining. It is likely that most of the patches seen in unbudded cells are sites of recent cell separation, but it is possible that some represent incipient bud sites. Staining is occasionally observed at the tips of very small buds (data not shown) but is never detected at the tips of larger buds. To determine more definitively if Cbk1p localizes to apical growth sites, we examined Cbk1p::3xHA localization in cdc34-2 cells grown at the restrictive temperature. As a control, we simultaneously visualized the polarized-growth component Spa2p using anti-Spa2p serum (14). While Spa2p localizes to the tips of actively growing buds in cdc34-2 cells grown at the restrictive temperature, no staining above the background is observed for Cbk1p::3xHA (data not shown). Immunoblot analysis indicates that Cbk1p is present at wild-type levels in cdc34-2 cells grown at the restrictive temperature (data not shown). Localization of Cbk1p::3xHA is not detected in α-factor-treated cells.
hym1Δ mutants are phenotypically similar to cbk1Δ mutants, and Hym1p is required for Cbk1p localization.
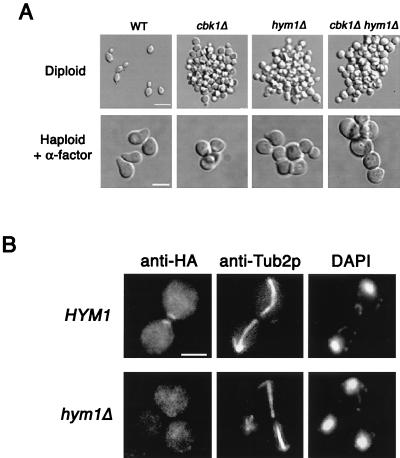
Recently, CBK1 was identified in a screen for genes involved in Sin3p-mediated transcriptional repression (10). The same screen identified another gene, HYM1, which, based on genetic analysis, was proposed to function in a pathway with CBK1 (10). Hym1p is an evolutionarily conserved protein that has homologs in worms, flies, plants, fish, mice, and humans. Interestingly, the Hym1p homolog HymA is important for the morphological development of specialized structures called conidiophores in Aspergillus nidulans (22). To further characterize the function of HYM1 and its relationship to CBK1, we deleted HYM1 and analyzed the resulting phenotypes. As described previously (10), we found that hym1Δ mutants have cell separation defects (average number of cells per clump, 95 ± 58) that are similar to phenotypes observed in cbk1Δ mutants (Fig. 11A). In addition, hym1Δ cells, like cbk1Δ cells, are rounder than wild-type cells (Fig. 11A). The average length/width ratio of hym1Δ diploid cells is 1.1 ± 0.1 (n = 103). We tested the ability of haploid hym1Δ cells to form mating projections in response to pheromone. After a 2-h treatment with α-factor, only 6% (n = 199) of hym1Δ cells form mating projections that are at least half a cell diameter in length (Fig. 11A). As observed with cbk1Δ cells, α-factor-treated hym1Δ cells form small surface bumps (Fig. 11A). Thus, based on the phenotypes we tested, hym1Δ mutants are similar to cbk1Δ mutants.
FIG. 11.
hym1Δ and cbk1Δ mutants have similar phenotypes, and Hym1p is required for normal Cbk1p localization. (A) Morphology of vegetatively growing and pheromone-treated wild-type (WT) and cbk1Δ, hym1Δ, and cbk1Δ hym1Δ mutant cells. Exponentially growing diploid cells were fixed with formaldehyde and photographed. Bar, 10 μm. Exponentially growing haploid cells were treated with α-factor at 5 μg/ml for 2 h, fixed, and then photographed. Bar, 5.0 μm. (B) Cbk1p::3xHA was visualized by indirect immunofluorescence assay in wild-type and hym1Δ mutant cells. No polarized localization of Cbk1p::3xHA is observed in hym1Δ mutant cells. Bar, 5.0 μm.
We examined the phenotypes of cells with both genes deleted. Homozygous cbk1Δ hym1Δ diploid cells form clumps containing an average of 105 ± 65 (n = 44) cells. This is comparable to results obtained with the respective single mutants (Fig. 11A). The average length/width ratio of cbk1Δ hym1Δ diploid cells (1.1 ± 0.1, n = 111) is also not significantly different from that of the single mutants. Similar to the results obtained with the single mutants, 8% (n = 257) of cbk1Δ hym1Δ cells form normal-sized mating projections after a 2-h treatment with α-factor (Fig. 11A). Hence, we concluded that the phenotypic effects caused by deletion of CBK1 and HYM1 are not additive.
To determine the epistasis relationship of CBK1 and HYM1, cbk1Δ cells were transformed with a high-copy plasmid expressing Hym1p and hym1Δ cells were transformed with a high-copy plasmid expressing Cbk1p. In each case, overexpression did not suppress any of the mutant phenotypes (data not shown).
To further investigate the relationship between Cbk1p and Hym1p, we examined the localization of Cbk1p::3xHA in hym1Δ cells. In contrast to wild-type cells, in which Cbk1p::3xHA localizes to both sides of the bud neck during late anaphase and persists as a patch at sites of recent cell separation (Fig. 10B and 11B), in hym1Δ cells, polarized localization of Cbk1p::3xHA is never observed (Fig. 11B). Hym1p is not required for the expression of Cbk1p, since Western blot analysis indicates that hym1Δ cells possess wild-type levels of Cbk1p (data not shown). Thus, Hym1p is required for the proper localization of Cbk1p.
DISCUSSION
Cbk1p and Hym1p are important for apical bud growth, mating projection formation, and cell separation.
CBK1 was identified in a screen for genes involved in the apical growth phase of bud formation. Analysis of cbk1Δ mutants indicates that Cbk1p is important for apical bud growth, mating projection formation, and cell separation following cytokinesis. cbk1Δ cells are rounder than wild-type cells and form abnormally small mating projections in response to pheromone treatment. In addition, direct visualization of the incorporation of new cell wall material using FITC-ConA suggests that cbk1Δ cells have a decreased ability to concentrate growth at the bud tip during the apical growth phase. These results suggest that Cbk1p is required to maintain polarized growth processes. cbk1Δ mutants also have a severe defect in postcytokinesis cell separation.
Recently, CBK1 and another gene, HYM1, were identified in a screen for genes involved in Sin3p-mediated transcriptional repression (10). Based on analysis of cbk1Δ hym1Δ double mutants, it was proposed that CBK1 and HYM1 function in the same genetic pathway (10). Our results support this conclusion. cbk1Δ and hym1Δ mutants have similar phenotypes, and simultaneous deletion of both genes does not increase the severity of the cell shape, cell separation, or mating projection morphology defects. In addition, we found that Hym1p is essential for normal Cbk1p localization. Since overexpression of Cbk1p does not suppress hym1Δ phenotypes and Hym1p overexpression does not suppress cbk1Δ phenotypes, it is possible that Hym1p and Cbk1p function together directly as a complex. Regardless, these results strongly suggest that Cbk1p and Hym1p act in a common pathway.
Cbk1-related kinases in N. crassa (55), Schizosaccharomyces pombe (53), C. elegans (56), and Drosophila (15) have been shown to play a role in the regulation of cellular morphology. Hym1p has homologs in many organisms, and the A. nidulans Hym1p homolog HymA is important for morphological development (22). Thus, it is possible that Cbk1p and Hym1p act in an evolutionarily conserved pathway that regulates cellular morphogenesis.
Cbk1p regulates the expression of a number of genes with cell wall-related functions.
Efficient cell separation is dependent on the expression of a chitinase encoded by the gene CTS1 (26); transcription of this gene is dependent on the transcription factor Ace2p (9). We found that CTS1 expression is greatly reduced in cbk1Δ cells. To determine if deletion of CBK1 affects the expression of other genes, we conducted a genomewide comparison of mRNA transcript levels between wild-type and cbk1Δ cells. Our results suggest that Cbk1p may exert transcriptional control over a broad range of cell wall modification activities. Expression of the glucanase-encoding gene SCW11, which is also required for efficient mother-daughter separation, as well as the putative glucanase-encoding gene YNR067c, is reduced in cbk1Δ mutants, while expression of the CHS1 chitin synthase gene and the ECM3, ECM5, and ECM8 cell wall integrity genes is elevated. Strikingly, expression of YER124c, a gene of unknown function, was reduced 12.7-fold in cbk1Δ cells.
Intriguingly, some of the genes whose transcription is reduced in cbk1Δ cells (YER124c, CTS1, SCW11, YNR067c, and TIP1) are expressed in the G1 phase of the cell cycle (46). Expression of the G1-transcribed EGT2 gene is not affected in cbk1Δ cells (Fig. 9 and data not shown). Since EGT2 expression can be mediated by either Ace2p or Swi5p (32), this result suggests that Cbk1p is not generally required for the activity of this class of transcription factors. Thus, Cbk1p appears to be important for the expression of a subset of G1-expressed genes with cell wall-related functions. During polarized growth, cells must maintain a localized dynamic balance between cell wall degradation and synthesis. Thus, it is possible that the polarized growth defects observed in cbk1Δ cells are caused by misregulation of the expression of genes important for these processes.
Cbk1p and Hym1p appear to function in Ace2p-dependent and -independent pathways that regulate cell morphogenesis and cell separation.
Recently, it has been reported that Cbk1p may promote cell separation by acting through Ace2p to promote the expression of CTS1. The cell separation defects of cbk1Δ cells can be suppressed by dominant gain-of-function ACE2 alleles and partially suppressed by overexpression of CTS1 (41). Additionally, Ace2p interacts with Cbk1p in the two-hybrid system (41). Our results are consistent with this model and suggest that the expression of at least one other gene important for cell separation, SCW11, is also regulated by Cbk1p. Low expression of SCW11 in cbk1Δ mutants provides a possible explanation for the incomplete suppression of cbk1Δ cell separation defects by CTS1 overexpression.
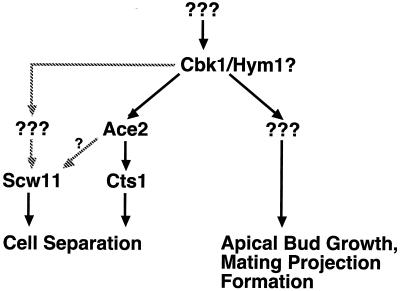
Importantly, our results indicate that factors other than Ace2p act downstream of Cbk1p to promote both apical growth and mating projection formation. We found that ace2Δ cells are normally shaped and form normal mating projections in response to pheromone treatment. In addition, we identified a cbk1 allele that causes severe apical growth and mating projection formation defects but retains nearly wild-type cell separation activity. Taken together, these results suggest that Cbk1p and Hym1p function in two pathways: an Ace2p-independent pathway required for efficient apical growth and mating projection formation and an Ace2p-dependent pathway that promotes cell separation (Fig. 12).
FIG. 12.
Model for the regulation of morphogenesis by Cbk1p and Hym1p. Cbk1p functions in two distinct cell morphogenesis pathways: an ACE2-independent pathway that is required for apical growth and mating projection formation and an ACE2-dependent pathway that is required for cell separation. Based on genetic analysis, Hym1p is proposed to act at the same level as Cbk1p. Expression of SCW11 may be Ace2p dependent or require another, unknown, transcription factor. Downstream targets of Cbk1p that promote apical growth and mating projection formation await identification.
Cbk1p and the coordination of gene expression with morphogenesis.
Our results and the recent results of Racki et al. (41) strongly suggest that Cbk1p promotes cell separation by stimulating the transcription of genes such as CTS1 and SCW11, which encode enzymes required to digest the chitinous septum between mother and daughter cells. Additionally, our results suggest that Cbk1p regulates the transcription of a number of genes with cell wall-related functions and may thereby control the dynamic balance between cell wall degradation and synthesis during polarized growth.
Cbk1p appears at both sides of the bud neck very late in anaphase. As noted above, the transcript levels of several genes whose expression is dependent on Cbk1p peak during G1 and Cbk1 participates in morphogenic events (cell separation, bud site selection, and apical growth) that occur at the end of G1. Thus, it is intriguing to speculate that Cbk1p is activated in late anaphase, perhaps upon its localization to the bud neck. Activated Cbk1p may then positively regulate the function of Ace2p and other, unidentified, transcription factors, thereby coordinating the timing of gene expression with morphogenic events.
Direct regulation of cortical activities by Cbk1p.
Although our results and the results of others (10) indicate that Cbk1p is involved in the regulation of transcription (a nuclear event), the polarized localization of Cbk1p suggests that it may also function to directly mediate cortical activities. Proper organization of the actin cytoskeleton is essential for polarized growth in yeast (30), and other Cbk1p-related kinases are known to affect actin organization. When N. crassa Cot-1 mutants are shifted to the restrictive temperature, actin polarity is lost and actin patches become uniformly located throughout the hyphae (49). Orb6 has also been shown to be involved in actin organization in fission yeast (53). Recently, it has been suggested that the Drosophila Ndr kinase (Trc) may regulate the actin cytoskeleton (15). Consistent with these results, we have shown that pheromone-treated cbk1Δ cells are unable to maintain a polarized actin cytoskeleton in growing mating projections. However, vegetatively growing cbk1Δ cells appear to have normal actin organization (data not shown). Thus, our results suggest that Cbk1p functions to promote pheromone-induced polarization of the actin cytoskeleton. The mechanisms by which Cbk1p and related kinases control organization of the actin cytoskeleton are unknown and await further characterization.
Several lines of evidence suggest that Cbk1p may function in a pathway with the PAK homolog Ste20p to regulate polarized growth and morphogenesis. In fission yeast, a temperature-sensitive orb6 allele was found to be synthetically lethal in combination with a temperature-sensitive allele of the Ste20p homolog Pak1 (Shk1) (52, 53). In addition, Cbk1p has recently been shown to interact with Ste20p, Ste5p, and Ste50p in a two-hybrid assay (16). Further genetic and biochemical analysis is required to determine the functional relationship between Cbk1p and Ste20p.
ACKNOWLEDGMENTS
The first two authors contributed equally to this work.
We thank K. McDonald for assistance with the preparation of samples for electron microscopy, J. DeRisi for providing yeast genomic DNA arrays, J. Thorner for providing a FUS1-lacZ reporter construct, W. Racki and C. Herbert for communication of results prior to publication, and B. Manning, C. Horak, J. Hanrahan, R. Stewart, and G. Michaud for critical reading of the manuscript.
This research was supported by National Institutes of Health grants GM36494 to M.S. and GM50399 to D.G.D. S.B. was supported by a National Institutes of Health training grant. E.L.W. was supported by an American Cancer Society postdoctoral fellowship.
REFERENCES
- 1.Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 2000. [Google Scholar]
- 2.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bedinger P A, Hardeman K J, Loukides C A. Travelling in style: the cell biology of pollen. Trends Cell Biol. 1994;4:132–138. doi: 10.1016/0962-8924(94)90068-x. [DOI] [PubMed] [Google Scholar]
- 4.Belham C, Wu S, Avruch J. Intracellular signalling: PDK1—a kinase at the hub of things. Curr Biol. 1999;9:R93–R96. doi: 10.1016/s0960-9822(99)80058-x. [DOI] [PubMed] [Google Scholar]
- 5.Bulawa C E, Slater M, Cabib E, Au-Young J, Sburlati A, Adair W L, Jr, Robbins P W. The S. cerevisiae structural gene for chitin synthase is not required for chitin synthesis in vivo. Cell. 1986;46:213–225. doi: 10.1016/0092-8674(86)90738-5. [DOI] [PubMed] [Google Scholar]
- 6.Cappellaro C, Mrsa V, Tanner W. New potential cell wall glucanases of Saccharomyces cerevisiae and their involvement in mating. J Bacteriol. 1998;180:5030–5037. doi: 10.1128/jb.180.19.5030-5037.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cid V J, Durán A, del Rey F, Snyder M P, Nombela C, Sánchez M. Molecular basis of cell integrity and morphogenesis in Saccharomyces cerevisiae. Microbiol Rev. 1995;59:345–386. doi: 10.1128/mr.59.3.345-386.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeRisi J, Iyer V, Brown P. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 9.Dohrmann P R, Butler G, Tamai K, Dorland S, Greene J R, Thiele D J, Stillman D J. Parallel pathways of gene regulation: homologous regulators SW15 and ACE2 differentially control transcription of HO and chitinase. Genes Dev. 1992;6:93–104. doi: 10.1101/gad.6.1.93. [DOI] [PubMed] [Google Scholar]
- 10.Dorland S, Deegenaars M L, Stillman D J. Roles for the Saccharomyces cerevisiae SDS3, CBK1 and HYM1 genes in transcriptional repression by SIN3. Genetics. 2000;154:573–586. doi: 10.1093/genetics/154.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drubin D G, Nelson W J. Origins of cell polarity. Cell. 1996;84:335–344. doi: 10.1016/s0092-8674(00)81278-7. [DOI] [PubMed] [Google Scholar]
- 12.Durrenberger F, Kronstad J. The ukc1 gene encodes a protein kinase involved in morphogenesis, pathogenicity and pigment formation in Ustilago maydis. Mol Gen Genet. 1999;261:281–289. doi: 10.1007/s004380050968. [DOI] [PubMed] [Google Scholar]
- 13.Flescher E G, Madden K, Snyder M. Components required for cytokinesis are important for bud site selection in yeast. J Cell Biol. 1993;122:373–386. doi: 10.1083/jcb.122.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gehrung S, Snyder M. The SPA2 gene of Saccharomyces cerevisiae is important for pheromone-induced morphogenesis and efficient mating. J Cell Biol. 1990;111:1451–1464. doi: 10.1083/jcb.111.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geng W, He B, Wang M, Adler P N. The tricornered gene, which is required for the integrity of epidermal cell extensions, encodes the drosophila nuclear DBF2-related kinase. Genetics. 2000;156:1817–1828. doi: 10.1093/genetics/156.4.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geyer C R, Colman-Lerner A, Brent R. “Mutagenesis” by peptide aptamers identifies genetic network members and pathway connections. Proc Natl Acad Sci USA. 1999;96:8567–8572. doi: 10.1073/pnas.96.15.8567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goebl M G, Yochem J, Jentsch S, McGrath J P, Varshavsky A, Byers B. The yeast cell cycle gene CDC34 encodes a ubiquitin-conjugating enzyme. Science. 1988;241:1331–1335. doi: 10.1126/science.2842867. [DOI] [PubMed] [Google Scholar]
- 18.Guthrie C, Fink G R. Guide to yeast genetics and molecular biology. Methods Enzymol. 1991;194:1–933. [PubMed] [Google Scholar]
- 19.Hanks S K, Hunter T. Protein kinases. 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- 20.Hayashibe M, Katohda S. Initiation of budding and chitin-ring. J Gen Appl Microbiol. 1973;19:23–29. [Google Scholar]
- 21.Justice R W, Zilian O, Woods D F, Noll M, Bryant P J. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 1995;9:534–546. doi: 10.1101/gad.9.5.534. [DOI] [PubMed] [Google Scholar]
- 22.Karos M, Fischer R. Molecular characterization of HymA, an evolutionarily highly conserved and highly expressed protein of Aspergillus nidulans. Mol Gen Genet. 1999;260:510–521. doi: 10.1007/s004380050924. [DOI] [PubMed] [Google Scholar]
- 23.Kovacech B, Nasmyth K, Schuster T. EGT2 gene transcription is induced predominantly by Swi5 in early G1. Mol Cell Biol. 1996;16:3264–3274. doi: 10.1128/mcb.16.7.3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kowalski L R, Kondo K, Inouye M. Cold-shock induction of a family of TIP1-related proteins associated with the membrane in Saccharomyces cerevisiae. Mol Microbiol. 1995;15:341–353. doi: 10.1111/j.1365-2958.1995.tb02248.x. [DOI] [PubMed] [Google Scholar]
- 25.Kupfer A, Swain S L, Janeway C A, Jr, Singer S J. The specific direct interaction of helper T cells and antigen-presenting B cells. Proc Natl Acad Sci USA. 1986;83:6080–6083. doi: 10.1073/pnas.83.16.6080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuranda M J, Robbins P W. Chitinase is required for cell separation during growth of Saccharomyces cerevisiae. J Biol Chem. 1991;266:19758–19767. [PubMed] [Google Scholar]
- 27.Kuranda M J, Robbins P W. Cloning and heterologous expression of glycosidase genes from Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1987;84:2585–2589. doi: 10.1073/pnas.84.9.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lew D J, Reed S I. Cell cycle control of morphogenesis in budding yeast. Curr Opin Genet Dev. 1995;5:17–23. doi: 10.1016/s0959-437x(95)90048-9. [DOI] [PubMed] [Google Scholar]
- 29.Lussier M, White A M, Sheraton J, di Paolo T, Treadwell J, Southard S B, Horenstein C I, Chen-Weiner J, Ram A F, Kapteyn J C, Roemer T W, Vo D H, Bondoc D C, Hall J, Zhong W W, Sdicu A M, Davies J, Klis F M, Robbins P W, Bussey H. Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics. 1997;147:435–450. doi: 10.1093/genetics/147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Madden K, Snyder M. Cell polarity and morphogenesis in budding yeast. Annu Rev Microbiol. 1998;52:687–744. doi: 10.1146/annurev.micro.52.1.687. [DOI] [PubMed] [Google Scholar]
- 31.Manning B D, Padmanabha R, Snyder M. The Rho-GEF Rom2p localizes to sites of polarized cell growth and participates in cytoskeletal functions in Saccharomyces cerevisiae. Mol Biol Cell. 1997;8:1829–1844. doi: 10.1091/mbc.8.10.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McBride H J, Yu Y, Stillman D J. Distinct regions of the Swi5 and Ace2 transcription factors are required for specific gene activation. J Biol Chem. 1999;274:21029–21036. doi: 10.1074/jbc.274.30.21029. [DOI] [PubMed] [Google Scholar]
- 33.McDonald K. High pressure freezing for preservation of high resolution fine structure and antigenicity for immunolabeling. Methods Mol Biol. 1999;117:77–97. doi: 10.1385/1-59259-201-5:77. [DOI] [PubMed] [Google Scholar]
- 34.Millward T, Cron P, Hemmings B A. Molecular cloning and characterization of a conserved nuclear serine(threonine) protein kinase. Proc Natl Acad Sci USA. 1995;92:5022–5026. doi: 10.1073/pnas.92.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Millward T A, Heizmann C W, Schafer B W, Hemmings B A. Calcium regulation of Ndr protein kinase mediated by S100 calcium-binding proteins. EMBO J. 1998;17:5913–5922. doi: 10.1093/emboj/17.20.5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Millward T A, Hess D, Hemmings B A. Ndr protein kinase is regulated by phosphorylation on two conserved sequence motifs. J Biol Chem. 1999;274:33847–33850. doi: 10.1074/jbc.274.48.33847. [DOI] [PubMed] [Google Scholar]
- 37.Mooseker M S. Organization, chemistry, and assembly of the cytoskeletal apparatus of the intestinal brush border. Annu Rev Cell Biol. 1985;1:209–241. doi: 10.1146/annurev.cb.01.110185.001233. [DOI] [PubMed] [Google Scholar]
- 38.Nishiyama Y, Hirota T, Morisaki T, Hara T, Marumoto T, Iida S, Makino K, Yamamoto H, Hiraoka T, Kitamura N, Saya H. A human homolog of Drosophila warts tumor suppressor, h-warts, localized to mitotic apparatus and specifically phosphorylated during mitosis. FEBS Lett. 1999;459:159–165. doi: 10.1016/s0014-5793(99)01224-7. [DOI] [PubMed] [Google Scholar]
- 39.Pearson R B, Dennis P B, Han J W, Williamson N A, Kozma S C, Wettenhall R E, Thomas G. The principal target of rapamycin-induced p70s6k inactivation is a novel phosphorylation site within a conserved hydrophobic domain. EMBO J. 1995;14:5279–5287. doi: 10.1002/j.1460-2075.1995.tb00212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peterson R T, Schreiber S L. Kinase phosphorylation: keeping it all in the family. Curr Biol. 1999;9:R521–R524. doi: 10.1016/s0960-9822(99)80326-1. [DOI] [PubMed] [Google Scholar]
- 41.Racki W, Becam A, Nasr F, Herbert C J. Cbk1p, a protein similar to the human myotonic dystrophy kinase, is essential for normal morphogenesis in Saccharomyces cerevisiae. EMBO J. 2000;19:4524–4532. doi: 10.1093/emboj/19.17.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ross-Macdonald P, Coelho P S, Roemer T, Agarwal S, Kumar A, Jansen R, Cheung K H, Sheehan A, Symoniatis D, Umansky L, Heidtman M, Nelson F K, Iwasaki H, Hager K, Gerstein M, Miller P, Roeder G S, Snyder M. Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature. 1999;402:413–418. doi: 10.1038/46558. [DOI] [PubMed] [Google Scholar]
- 43.Ross-Macdonald P, Sheehan A, Roeder G S, Snyder M. A multipurpose transposon system for analyzing protein production, localization, and function in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1997;94:190–195. doi: 10.1073/pnas.94.1.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sheu Y-J, Barral Y, Snyder M. Polarized growth controls cell shape and bipolar bud site selection in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:5235–5247. doi: 10.1128/mcb.20.14.5235-5247.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Snyder M, Gehrung S, Page B D. Studies concerning the temporal and genetic control of cell polarity in Saccharomyces cerevisiae. J Cell Biol. 1991;114:515–532. doi: 10.1083/jcb.114.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spellman P T, Sherlock G, Zhang M Q, Iyer V R, Anders K, Eisen M B, Brown P O, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tao W, Zhang S, Turenchalk G S, Stewart R A, St. John M A, Chen W, Xu T. Human homologue of the Drosophila melanogaster lats tumour suppressor modulates CDC2 activity. Nat Genet. 1999;21:177–181. doi: 10.1038/5960. [DOI] [PubMed] [Google Scholar]
- 48.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tinsley J H, Lee I H, Minke P F, Plamann M. Analysis of actin and actin-related protein 3 (ARP3) gene expression following induction of hyphal tip formation and apolar growth in Neurospora. Mol Gen Genet. 1998;259:601–609. doi: 10.1007/s004380050853. [DOI] [PubMed] [Google Scholar]
- 50.Tkacz J S, Lampen J O. Wall replication in saccharomyces species: use of fluorescein-conjugated concanavalin A to reveal the site of mannan insertion. J Gen Microbiol. 1972;72:243–247. doi: 10.1099/00221287-72-2-243. [DOI] [PubMed] [Google Scholar]
- 51.Trueheart J, Boeke J D, Fink G R. Two genes required for cell fusion during yeast conjugation: evidence for a pheromone-induced surface protein. Mol Cell Biol. 1987;7:2316–2328. doi: 10.1128/mcb.7.7.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Verde F, Mata J, Nurse P. Fission yeast cell morphogenesis: identification of new genes and analysis of their role during the cell cycle. J Cell Biol. 1995;131:1529–1538. doi: 10.1083/jcb.131.6.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Verde F, Wiley D J, Nurse P. Fission yeast orb6, a ser/thr protein kinase related to mammalian rho kinase and myotonic dystrophy kinase, is required for maintenance of cell polarity and coordinates cell morphogenesis with the cell cycle. Proc Natl Acad Sci USA. 1998;95:7526–7531. doi: 10.1073/pnas.95.13.7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu T, Wang W, Zhang S, Stewart R A, Yu W. Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development. 1995;121:1053–1063. doi: 10.1242/dev.121.4.1053. [DOI] [PubMed] [Google Scholar]
- 55.Yarden O, Plamann M, Ebbole D J, Yanofsky C. cot-1, a gene required for hyphal elongation in Neurospora crassa, encodes a protein kinase. EMBO J. 1992;11:2159–2166. doi: 10.1002/j.1460-2075.1992.tb05275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zallen J A, Peckol E L, Tobin D M, Bargmann C I. Neuronal cell shape and neurite initiation are regulated by the ndr kinase SAX-1, a member of the Orb6/COT-1/Warts serine/threonine kinase family. Mol Biol Cell. 2000;11:3177–3190. doi: 10.1091/mbc.11.9.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]