Abstract
Accurate bone segmentation and anatomical landmark localization are essential tasks in computer-aided surgical simulation for patients with craniomaxillofacial (CMF) deformities. To leverage the complementarity between the two tasks, we propose an efficient end-to-end deep network, i.e., multi-task dynamic transformer network (DTNet), to concurrently segment CMF bones and localize large-scale landmarks in one-pass from large volumes of cone-beam computed tomography (CBCT) data. Our DTNet was evaluated quantitatively using CBCTs of patients with CMF deformities. The results demonstrated that our method outperforms the other state-of-the-art methods in both tasks of the bony segmentation and the landmark digitization. Our DTNet features three main technical contributions. First, a collaborative two-branch architecture is designed to efficiently capture both fine-grained image details and complete global context for high-resolution volume-to-volume prediction. Second, leveraging anatomical dependencies between landmarks, regionalized dynamic learners (RDLs) are designed in the concept of “learns to learn” to jointly regress large-scale 3D heatmaps of all landmarks under limited computational costs. Third, adaptive transformer modules (ATMs) are designed for the flexible learning of task-specific feature embedding from common feature bases.
Keywords: Craniomaxillofacial (CMF), Multi-task learning, Segmentation, Landmark localization
1. Introduction
Cone-beam computed tomography (CBCT) scans are routinely used in quantifying deformity and planning orthognathic surgery for patients with jaw deformity. The planning pipeline starts from accurately segmenting the bones from the background and digitizing (localizing) anatomical landmarks onto three-dimensionally (3D) reconstructed models [18]. In current clinical practice, surgeons have to manually segment the bones and soft tissues, and digitize the landmarks. This task is very time-consuming and highly depends on surgeons’ experience. Thus there is an urgent need, from surgeons, to develop reliable and fully automatic method for segmentation and landmark digitization.
Automated CMF bone segmentation and landmark localization are practically challenging. This is mainly because that CBCT scans typically have severe image artifacts (e.g., caused by amalgam dental fillings, and orthodontic braces), significant appearance variations, and a large volume (typically around 600 × 600 × 500 voxels, 0.4 mm3 isotropically per voxel). Most of the traditional methods, including atlas- [15], model- [3], and learning-based methods [17], formulate the segmentation and the localization as two independent tasks, despite of the fact that the bony landmarks are the anatomically meaningful points located on the skeletal surface—both tasks are naturally associated [20]. Recently, deep learning methods leveraging fully convolutional neural networks (FCNs) [7,8,10,14] have been proposed to perform CMF bone segmentation and landmark localization in a unified framework. Torosdagli et al. [16] applied a dense variant of the U-Net [14] to segment mandible, based on which they designed another U-Net to regress the geodesic maps revealing landmark locations. Zhang et al. [21] designed a different cascade of two 3D U-Nets, where the first U-Net provides contextual guidance (i.e., voxel-wise displacements to landmarks) to assist the training of the second U-Net with two parallel outputs for the concurrent segmentation of bony structures and regression of landmark heatmaps.
To a degree, these methods did achieve the performance in both segmentation and localization. However, they still have a number of technical limitations that hampered them to be used in real clinical settings. First, they are typically implemented using small image patches (e.g., 96 × 96 × 96) sampled from a large CBCT volume to compensate the bottleneck of the memory size of graphic processing unit (GPU). It can significantly sacrifice the global view of the whole bony structure, which is important for both semantic segmentation [2] and discrimination between different landmarks [22]. Second, while the efficacy of heatmap regression has been verified in other landmark detection tasks [12,19], these methods can only jointly localize a limited number of CMF landmarks (e.g., 15 landmarks in [21]), due to the heavy memory consumption for the respective 3D heatmaps. That problem greatly limits its capability of efficiently addressing the realistic demand of large-scale landmarks (e.g., more than 60 on the mandible) for orthognathic surgical planning. Third, both [16] and [21] consist of multiple steps (or networks) implemented in an isolated fashion, while potential heterogeneity between different steps may lead to suboptimal results.
In this paper, we propose an end-to-end deep learning method, called multi-task dynamic transformer network (DTNet), for concurrent bone segmentation and landmark localization from CBCT images. Once a large clinical CBCT volume is directly input into DTNet, it can efficiently output the segmentation result and large-scale heatmaps for the landmarks in one-pass (Fig. 1).
Fig.1.
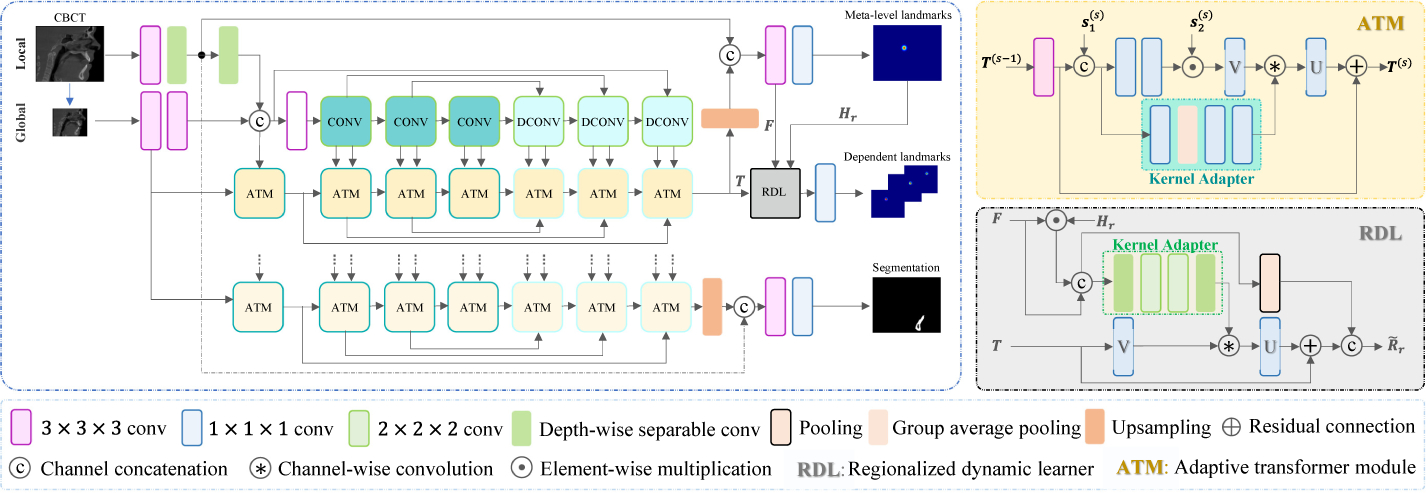
End-to-end DTNet for concurrent jaw segmentation and large-scale landmark localization. For illustration simplicity, here we only show the landmarks from one anatomical region and the common part of all ATMs (i.e., pooling and up-sampling for the encoding and decoding path are ignored).
The main technical contributions of our work are: First, our DTNet contains two collaborative branches, i.e., a light-weight branch to capture the local image details from the high-resolution input, and another deep branch to learn the global context from the down-sampled input. Second, we design a regionalized dynamic learner (RDL) in the “learning-to-learn” framework to leverage the stable dependency between spatially neighboring landmarks. The RDL first localizes a meta-level landmark in each predefined anatomical region by regressing the high-resolution heatmap. Based on that, it further predicts region-specific dynamic convolutional kernels to efficiently localize other neighboring landmarks in the down-sampled image space. Third, as an end-to-end network, our DTNet extends the state-of-the-art multi-task architecture (i.e., MTAN [9]) by introducing adaptive transformer modules (ATMs) to jointly learn task-oriented feature embedding from common feature pools.
2. Multi-task Dynamic Transformer Network
Our DTNet contains three important components, i.e., collaborative local and global branches, adaptive transformer modules, and regionalized dynamic learners besides the fundamental convolutional (Conv), deconvolutional (DConv), pooling, and upsampling operations. The schematic diagram is shown in Fig. 1.
Collaborative Local and Global Branches:
Inspired by recent advances in real-time high-resolution segmentation [2,13], our DTNet designs a two-branch architecture to efficiently learn the local details and global context for high-resolution volume-to-volume predictions from a large 3D input (size: L × W × H). A light-weight local branch works on the original input to capture local details, while a deep global branch works on a significantly down-sampled input (size: ) to learn both shared and task-specific contextual representations.
Specifically, the local branch consists of only two general 3 × 3 × 3 Conv layers (both with unit stride and zero-padding) and a 5 × 5 × 5 depth-wise separable Conv layer [5] (5 strides without padding) to control the number of learnable parameters. The second Conv layer outputs feature maps describing fine-grained image details, which are transferred by a long-range skip connection to the top of DTNet for high-resolution inference. The third Conv layer transfer these local features into the global branch to assist the learning of deep contextual feature representations from the down-sampled input.
The global-branch network adopts FCNs as the backbone to develop a multi-task architecture for concurrent bone segmentation and landmark localization. Inspired by [9], it consists of a single 3D U-Net to learn common feature representations shared between two tasks, based on which two light-weight subnetworks are further designed to learn adaptive feature embedding for each specific task. To begin with, the backbone applies two Conv layers to learn from the down-sampled input the local features, which are further merged with the corresponding features from the local branch, forming multi-scale representations of image details to assist the construction of the subsequent Conv and DeConv blocks with residual connections [4]. Each Conv block in the encoding path contains two convolutional layers, where the first two of them are followed by pooling. Symmetrically, each DeConv block in the decoding path starts with bilinear up-sampling, followed by two Conv layers. Built upon the shared U-Net, task-specific subnetworks apply a series of adaptive transformer modules (ATMs) to learn from each Conv/DConv block the respective feature embedding.
Adaptive Transformer Modules:
We assume that the feature maps produced by each block of U-Net form a common feature pool shared across complementary tasks, upon which ATM learns task-oriented feature embedding in a “learning-to-learn” fashion [1]. According to the diagram shown in Fig. 1, ATM in the encoding path combines task-specific features T(s−1) from the preceding scale and initial common features at current scale (e.g., from the 1st Conv layer of a Conv block) to predict a task adapter, which further applies on the following common features (e.g., from the 2nd Conv layer of a Conv block), yielding task-specific feature representations T(s) at current scale. The task adapter performs joint feature selection and transformation from , using online predicted spatial attentions and dynamic Conv [1]. Specifically, T(s−1) is first processed by a general 3 × 3 × 3 Conv to obtain , which is merged with via channel-wise concatenation, such as [; ]. The attention map A(s) in terms of the merged features is then defined as
| (1) |
where and are the kernel weights of two 1 × 1 × 1 Convs followed by sigmoid and ReLU activations, respectively, and * denotes the Conv operator. Since and have the same size, the task-relevant features are selected via element-wise multiplication, i.e., . Regarding (with mi channels) as a set of bases, a dynamic 1 × 1 × 1 Conv is further learned for task-oriented transformation of the selected common features. Considering that predicting all learnable parameters W(s) is computationally infeasible even for 1 × 1 × 1 kernels, the dynamic Conv is factorized in an analog of SVD [11]. Therefore, the output T(s) (with mo channels) is defined as
| (2) |
where * and *c denote general and channel-wise Convs, respectively; ⊕ denotes the residual connection; U(s) and V(s) are the parameter matrices of two 1 × 1 × 1 Convs with mo and mi output channels, respectively; and is a diagonal matrix with only mi learnable parameters, which are one-shot determined by a light-weight kernel predictor in terms of [; ]. The kernel predictor has an architecture similar to squeeze-and-excitation [6], but learns the output coefficients from distinct input sources. It starts with a 1 × 1 × 1 Conv, followed by global average pooling (GAP) and another two 1 × 1 × 1 Convs to predict .
Notably, the encoding and decoding ATMs have similar structures, except that the former contains pooling while the latter contains bilinear up-sampling. In each of them, we set mo < mi, which effectively controls the number of learnable parameters and realizes task-oriented low-dimensional feature embedding.
Regionalized Dynamic Learners:
Since jointly regressing large-scale 3D heatmaps in high-resolution image space is memory infeasible, our DTNet integrates an efficient localization module by explicitly modeling the dependencies between spatially neighboring landmarks. To this end, we separate the 3D model of the jaw as several predefined anatomical regions, with each of them grouping a set of landmarks that have stable intra-region displacements. For each region, a meta-level landmark is first localized by regressing its high-resolution heatmap. Leveraging the strong guidance provided by this meta-level landmark, an RDL is further constructed to learn dynamically region-aware representations for the localization of other dependent landmarks under very limited memory costs.
Specifically, let F be a high-resolution representation, produced by fusing the localization-related global-branch representation (i.e., T from the last ATM) and the local-branch image details. As shown in Fig. 1, a 1 × 1 × 1 Conv layer with sigmoid activations works on F to predict meta-level heatmaps , corresponding to R different anatomical regions. To detect other dependent landmarks located within each (e.g., the r-th) region, the respective RDL combines Hr and F to learn a region-aware dynamic Conv layer, which applies on T to regressing the heatmaps of these dependent landmarks in significantly down-sampled image space. In more detail, a set of 3 × 3 × 3 channel-wise Conv kernels is first predicted as , where Hr provides high-resolution regional attention, and fr(·) is a light-weight kernel adapter consisting of depth-wise separable Convs and general 2 × 2 × 2 Convs with double strides. The region-aware representation for down-sampled heatmaps regression is finally defined as
| (3) |
where denotes residual SVD-factorized Conv [11] of T, and and are meta-level feature representations after average pooling.
Implementation:
Following [16], we attempt to segment the mandible and localize the associated landmarks, as it is the most frequently deformed bony structure that needs orthognathic surgical correction. The respective DTNet was implemented with PyTorch on a general GPU (i.e., NVIDIA TITAN Xp, 12 GBytes). It takes as input a big CBCT volume (size: 160 × 160 × 160), concurrently output in one-pass the segmentation map of the mandible and 64 heatmaps of all landmarks located on the mandible, costing less than 40 seconds (including loading data and saving results). Using the Adam optimizer, the network was trained by minimizing the combination of three Dice losses for jaw segmentation, high-resolution heatmap regression, and down-sampled heatmap regression, respectively. Dropout and group normalization were used in the network to accelerate training and improve generalization capacity. Training samples were mirror flipped for data augmentation.
3. Experiments
Dataset and Experimental Setup:
Our method was evaluated quantitatively using 77 sets of CBCT images of patients with non-syndromic CMF deformities, and 63 sets of CT images of normal subjects taken for the reason other than CMF deformities. All personal informations were de-identified. The study was approved by Institutional Review Board. The ground-truth of the segmented bones and digitalized landmarks were manually established by two experienced CMF surgeons. The 64 landmarks on the mandible were divided to 6 groups based on anatomical regions, including Ramus Upper Right (RUR), Ramus Low Right (RLR), Ramus Upper Left (RUL), Ramus Low Left (RLL), Distal Mandible (DMand) and Lower Teeth (LT). For each anatomical region, one landmark was preselected as the meta-level landmark based on a surgeon’s experience, including SIG-R, Go-R, SIG-L, Go-L, Pg, and L0 (Fig. 2). All images were spatially normalized to the same resolution of 0.8 × 0.8 × 0.8 mm3, and the gray-scale values were also normalized using histogram matching to reach similar gray-scale distributions. The 3D heatmap of each landmark was generated using a Gaussian filter with the standard deviation of 3mm. We randomly split 20 CBCT images for performance evaluation, and used the remaining 57 patients’ CBCT and 63 normal CT images for network training.
Fig.2.
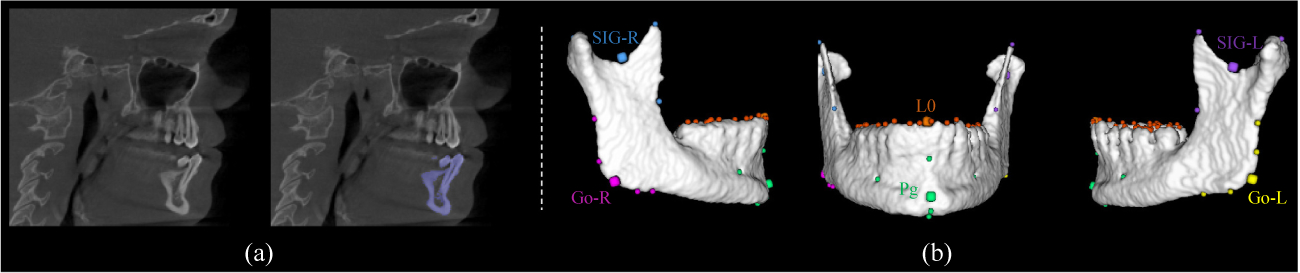
An representative subject in our dataset, where (a) shows the slice-view of the ground-truth mandible segmentation, and (b) shows the 6 anatomical regions with different colors. The dilated points denote the meta-level landmarks for each region.
Using the same settings in loss function and optimizer, our DTNet was compared to the other state-of-the-art methods, including MTAN [9], multi-task U-Net (MTU-Net) [21], and the original mono-task U-Net [14]. All competing networks were adjusted to a comparable number of learnable parameters. Specifically, we first replaced the global-branch subnetwork of our DTNet with MTAN, MTU-Net and U-Net to evaluate the efficacy of the proposed ATMs and RDLs for adaptive multi-task learning and large-scale landmark localization, respectively. In addition, we compared DTNet to the U-Net implemented with smaller patches (size: 96 × 96 × 96), denoted as SU-Net, to evaluate the efficacy of the proposed collaborative two-branch architecture in capturing local details and global context for high-resolution inference. Finally, the segmented results of the mandible were quantitatively compared to the ground truth in dice similarity coefficient (DSC), sensitivity (SEN) and positive prediction value (PPV). The landmark localization results were also quantitatively evaluated by calculating the root mean squared error (RMSE, in mm) between algorithm-detected and ground-truth landmark coordinates.
Results:
Table 1 presents the quantitative results obtained by different automated methods for both mandible segmentation and landmark localization. From Table 1, we can have the following observations. First, compared with the mono-task U-Net, the three multi-task networks (i.e., MTU-Net, MTAN, and our DTNet) led to better segmentation and localization results in most cases. It suggests that the two tasks are correlated and can provide each other auxiliary information for performance enhancement. Second, compared with the state-of-the-art multi-task architectures (i.e., MTU-Net and MTAN), our DTNet has superior performance with respect to all metrics, which implies that the proposed ATM modules are effective, and more powerful than simple task-specific attention, in extracting task-specific representations from shared feature bases for adaptive multi-task learning. Third, by performing intra-method comparisons between the localization results of the meta-level landmarks and all landmarks, we can observe that there is no big differences in terms of RMSE. It indicates that the proposed RDL can generally work with different architectures for the efficient localization of large-scale landmarks. In addition to the quantitative comparisons, the representative examples of mandible segmentation and landmark localization are shown in Fig. 3 and Fig. 4 (a), respectively. These qualitative comparisons between our DTNet and other multi-task methods further justify the efficacy of our DTNet, especially in segmenting low-contract structures and localizing landmarks in challenging anatomical regions (e.g., on the lower teeth).
Table 1.
The segmentation and localization results (mean ± standard deviation) obtained by our DTNet and other three competing methods. The localization results were quantified in terms of RMSE for the 6 meta-level and all 64 landmarks, respectively. Notably, all competing methods leveraged our two-branch architecture and RDLs to deal with large input CBCT volume and large-scale landmarks.
| Method | Mandible segmentation (%) | Landmark localization (mm) | |||
|---|---|---|---|---|---|
| DSC | SEN | PPV | Meta-level | All | |
| U-Net | 92.49 ± 2.13 | 92.47 ± 5.13 | 93.32 ± 4.43 | 2.18 ± 0.54 | 2.99 ± 0.33 |
| MTU-Net | 92.34 ± 2.06 | 93.59 ± 4.42 | 91.53 ± 5.10 | 2.01 ± 0.55 | 2.85 ± 0.40 |
| MTAN | 92.66 ± 2.23 | 94.17 ± 4.16 | 91.43 ± 4.84 | 2.02 ± 0.51 | 2.91 ± 0.45 |
| DTNet | 93.95 ± 1.30 | 94.24 ± 1.37 | 93.68 ± 1.78 | 1.95 ± 0.43 | 2.52 ± 0.31 |
Fig.3.
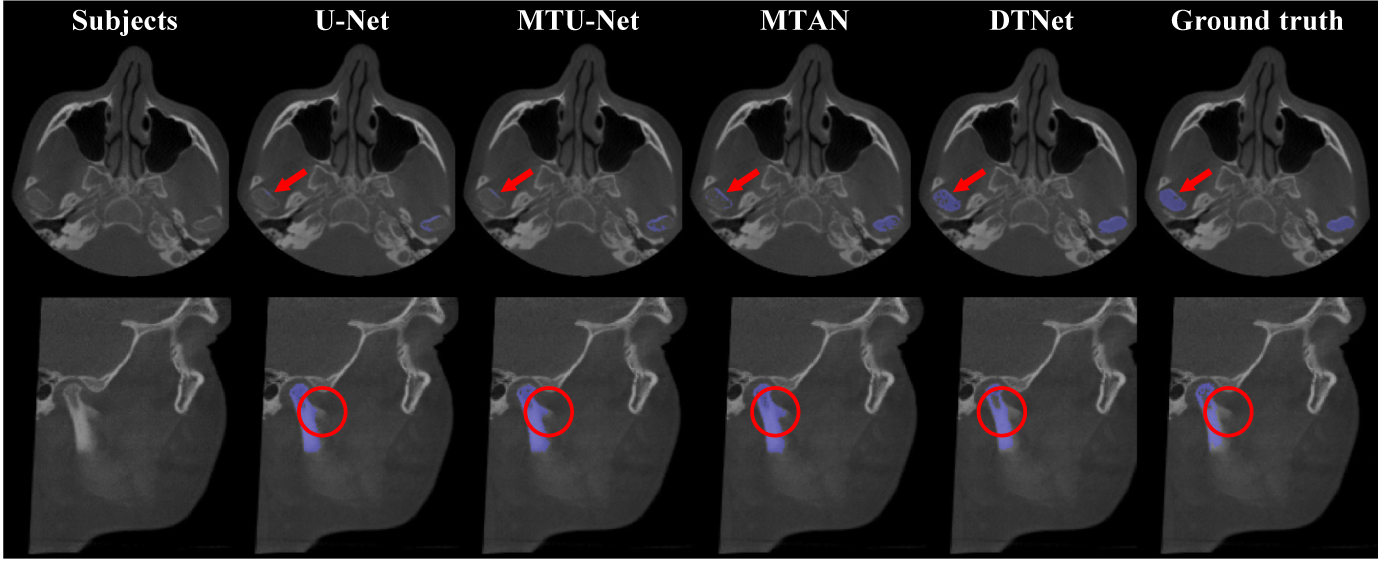
Representative results obtained by our DTNet and other competing methods for the segmentation of mandible. The marks in red indicate the challenging bony structures with low-contrast boundary.
Fig.4.
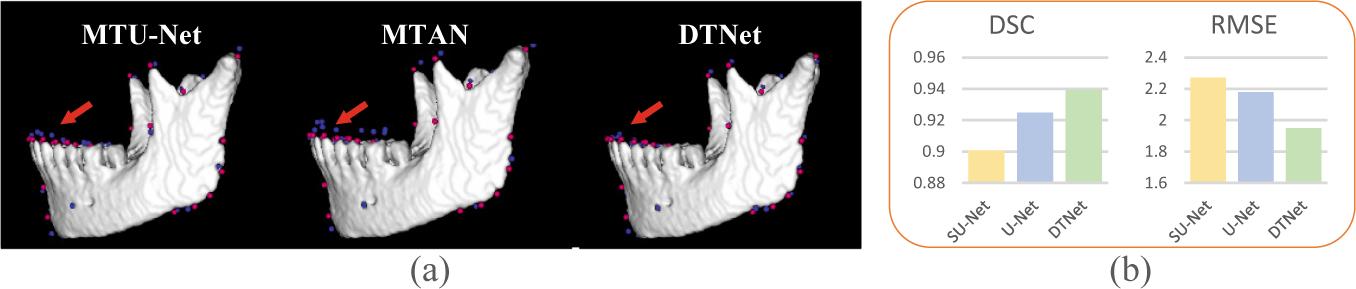
(a) Representative localization results obtained by different multi-task networks, where blue and red points denote the detected and ground-truth landmarks, respectively. (b) A comparison between small-patch U-Net (i.e., SU-Net) and two-branch networks (i.e., U-Net and our DTNet).
To evaluate the efficacy of the collaborative two-branch design in capturing both local details and global context from large CBCT images, our DTNet and the above two-branch U-Net were further compared with SU-Net, which was implemented with smaller image patches. The corresponding results of the mandible segmentation and meta-level landmark localization are summarized in Fig. 4 (b), from which we can see that both U-Net and our DTNet largely outperformed SU-Net. It implies that the integration of local image details and global context is important for high-resolution volume-to-volume inference, which also verifies the effectiveness of our collaborative two-branch design to this end.
4. Conclusion
In this paper, we have proposed a multi-task deep neural network, DTNet, for concurrent mandible segmentation and large-scale landmark localization in one-pass for large-volume CBCT images. Our DTNet uses a collaborative two-branch architecture to efficiently capture both local image details and global context for high-resolution volume-to-volume inference. Adaptive transformer modules are designed in the “learning-to-learn” framework to learn dynamically task-specific feature embeddings from a common feature pool. Regional dynamic learners are also proposed to leverage the local dependencies among the neighboring landmarks for efficient large-scale localization. The experimental results confirm the performance of our DTNet on real-patient CBCT data.
Acknowledgements.
This work was supported in part by NIH grants (R01 DE022676, R01 DE027251 and R01 DE021863).
References
- 1.Bertinetto L, et al. : Learning feed-forward one-shot learners. In: NeurIPS, pp.523–531 (2016) [Google Scholar]
- 2.Chen W, et al. : Collaborative global-local networks for memory-efficient segmentation of ultra-high resolution images. In: CVPR, pp. 8924–8933 (2019) [Google Scholar]
- 3.Gupta A, et al. : A knowledge-based algorithm for automatic detection of cephalometric landmarks on CBCT images. Int. J. Comput. Assist. Radiol. Surg 10(11), 1737–1752 (2015) [DOI] [PubMed] [Google Scholar]
- 4.He K, et al. : Deep residual learning for image recognition. In: CVPR, pp. 770–778 (2016) [Google Scholar]
- 5.Howard AG, et al. : MobileNets: efficient convolutional neural networks for mobile vision applications. arXiv preprint arXiv:1704.04861 (2017) [Google Scholar]
- 6.Hu J, et al. : Squeeze-and-excitation networks. In: CVPR, pp. 7132–7141 (2018) [Google Scholar]
- 7.Lian C, et al. : Multi-channel multi-scale fully convolutional network for 3D perivascular spaces segmentation in 7T MR images. Med. Image Anal 46, 106–117 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lian C, et al. : Hierarchical fully convolutional network for joint atrophy localization and Alzheimer’s disease diagnosis using structural MRI. IEEE Trans. Pattern Anal. Mach. Intell 42(4), 880–893 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu S, et al. : End-to-end multi-task learning with attention. In: CVPR, pp. 1871–1880 (2019) [Google Scholar]
- 10.Long J, et al. : Fully convolutional networks for semantic segmentation. In: CVPR,pp. 3431–3440 (2015) [DOI] [PubMed] [Google Scholar]
- 11.Nie X, et al. : Human pose estimation with parsing induced learner. In: CVPR, pp. 2100–2108 (2018) [Google Scholar]
- 12.Payer C, Štern D, Bischof H, Urschler M: Regressing heatmaps for multiple landmark localization using CNNs. In: Ourselin S, Joskowicz L, Sabuncu MR, Unal G, Wells W (eds.) MICCAI 2016. LNCS, vol. 9901, pp. 230–238. Springer, Cham; (2016). 10.1007/978-3-319-46723-8_27 [DOI] [Google Scholar]
- 13.Poudel RP, et al. : ContextNet: exploring context and detail for semantic segmentation in real-time. In: BMVC (2018) [Google Scholar]
- 14.Ronneberger O, Fischer P, Brox T: U-Net: convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells WM, Frangi AF (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham; (2015). 10.1007/978-3-319-24574-4_28 [DOI] [Google Scholar]
- 15.Shahidi S, et al. : The accuracy of a designed software for automated localization of craniofacial landmarks on CBCT images. BMC Med. Imaging 14(1), 32 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torosdagli N, et al. : Deep geodesic learning for segmentation and anatomical landmarking. IEEE Trans. Med. Imaging 38(4), 919–931 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang L, et al. : Automated segmentation of dental CBCT image with prior-guided sequential random forests. Med. Phys 43(1), 336–346 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xia JJ, et al. : New clinical protocol to evaluate craniomaxillofacial deformity and plan surgical correction. J. Oral Maxillofac. Surg 67(10), 2093–2106 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang D, et al. : Automatic vertebra labeling in large-scale 3D CT using deep image-to-image network with message passing and sparsity regularization. In: Niethammer M, et al. (eds.) IPMI 2017. LNCS, vol. 10265, pp. 633–644. Springer, Cham; (2017). 10.1007/978-3-319-59050-9_50 [DOI] [Google Scholar]
- 20.Zhang J, et al. : Automatic craniomaxillofacial landmark digitization via segmentation-guided partially-joint regression forest model and multiscale statistical features. IEEE Trans. Biomed. Eng 63(9), 1820–1829 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J, et al. : Context-guided fully convolutional networks for joint craniomaxillofacial bone segmentation and landmark digitization. Med. Image Anal 60, 101621 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhong Z, Li J, Zhang Z, Jiao Z, Gao X: An attention-guided deep regression model for landmark detection in cephalograms. In: Shen D, et al. (eds.) MICCAI 2019. LNCS, vol. 11769, pp. 540–548. Springer, Cham; (2019). 10.1007/978-3-030-32226-7_60 [DOI] [Google Scholar]


